
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 22 August 2023
Sec. Infectious Agents and Disease
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1235254
This article is part of the Research TopicNew Insights in the Microbe-Vector InteractionView all 8 articles
 Saowalak Kaewmee1
Saowalak Kaewmee1 Chonlada Mano2
Chonlada Mano2 Thanari Phanitchakun2
Thanari Phanitchakun2 Rinnara Ampol3
Rinnara Ampol3 Thippawan Yasanga4
Thippawan Yasanga4 Urassaya Pattanawong5
Urassaya Pattanawong5 Anuluck Junkum2
Anuluck Junkum2 Padet Siriyasatien3
Padet Siriyasatien3 Paul A. Bates6
Paul A. Bates6 Narissara Jariyapan3*
Narissara Jariyapan3*The prevalence of autochthonous leishmaniasis in Thailand is increasing but the natural vectors that are responsible for transmission remain unknown. Experimental in vivo infections in Culicoides spp. with Leishmania (Mundinia) martiniquensis and Leishmania (Mundinia) orientalis, the major causative pathogens in Thailand, have demonstrated that biting midges can act as competent vectors. Therefore, the isolation and detection of Leishmania and other trypanosomatids were performed in biting midges collected at a field site in an endemic area of leishmaniasis in Tha Ruea and a mixed farm of chickens, goats, and cattle in Khuan Phang, Nakhon Si Thammarat province, southern Thailand. Results showed that Culicoides peregrinus was the abundant species (>84%) found in both locations and only cow blood DNA was detected in engorged females. Microscopic examination revealed various forms of Leishmania promastigotes in the foregut of several C. peregrinus in the absence of bloodmeal remnants, indicating established infections. Molecular identification using ITS1 and 3’UTR HSP70 type I markers showed that the Leishmania parasites found in the midges were L. martiniquensis. The infection rate of L. martiniquensis in the collected flies was 2% in Tha Ruea and 6% in Khuan Phang, but no L. orientalis DNA or parasites were found. Additionally, organisms from two different clades of Crithidia, both possibly new species, were identified using SSU rRNA and gGAPDH genes. Choanomastigotes and promastigotes of both Crithidia spp. were observed in the hindgut of the dissected C. peregrinus. Interestingly, midges infected with both L. martiniquensis and Crithidia were found. Moreover, four strains of Crithidia from one of the clades were successfully isolated into culture. These parasites could grow at 37°C in the culture and infect BALB/c mice macrophages but no multiplication was observed, suggesting they are thermotolerant monoxenous trypanosomatids similar to Cr. thermophila. These findings provide the first evidence of natural infection of L. martiniquensis in C. peregrinus supporting it as a potential vector of L. martiniquensis.
Leishmaniases are vector-borne diseases caused by protozoan parasites of the genus Leishmania (Kinetoplastida, Trypanosomatidae). At least 21 Leishmania species that are members of the subgenera Viannia, Leishmania, and Mundinia have been reported as human pathogens. So far, no vaccine is available for human leishmaniasis (World Health Organization, 2023). In Thailand, human cases of autochthonous leishmaniases are mainly caused by two recently described species, L. (Mundinia) martiniquensis (Pothirat et al., 2014) and L. (Mundinia) orientalis (Jariyapan et al., 2018). The prevalence of autochthonous leishmaniasis in Thailand is increasing and new cases continue to be reported (Anugulruengkitt et al., 2022; Srivarasat et al., 2022), but the natural vectors that are responsible for the transmission of the disease remain unknown. Moreover, more relapse cases caused by L. martiniquensis have been reported (Srivarasat et al., 2022; Mano et al., 2023). Various species of sand flies are known as natural vectors of Leishmania parasites in the subgenera Viannia and Leishmania (Cecílio et al., 2022). For L. martiniquensis and L. orientalis, some species of sand flies have been reported as potential vectors (Chusri et al., 2014; Siripattanapipong et al., 2018; Srisuton et al., 2019; Sriwongpan et al., 2021). However, no natural vectors have been proved.
Recently, various research groups in several countries have reported the detection of Leishmania DNA in biting midges. For example, DNA of L. infantum has been detected in wild-caught Culicoides spp. in Tunisia (Slama et al., 2014), L. (Viannia) braziliensis DNA found in C. ignacioi, C. insignis, and C. foxi and L. (Leishmania) amazonensis DNA detected in C. filariferus and C. flavivenula in Brazil (Rebêlo et al., 2016). Further, in Australia, the natural infection of a day-feeding midge, subgenus Forcipomyia (Lasiohelea) Kieffer, with L. (Mundinia) macropodum parasites has been reported (Dougall et al., 2011). Experimental infections of L. (Mundinia) enriettii and L. orientalis in a laboratory colony of C. sonorensis reveals that both Leishmania species can complete their development to late-stage parasites found in the stomodeal valve, a position suitable for transmission by bite (Seblova et al., 2012; Chanmol et al., 2019). Further, Becvar and colleagues (Becvar et al., 2021) have demonstrated that L. martiniquensis, L. orientalis, and Leishmania (Mundinia) chancei (formerly called L. “Ghana” sp.) (Kwakye-Nuako et al., 2023), all members of subgenus Mundinia, are able to successfully colonize at the stomodeal valve, produce a higher proportion of metacyclic forms than in sand flies, and can be experimentally transmitted to BALB/c mice by C. sonorensis bites. These findings have highlighted Culicoides spp. as potential vectors of the members of the subgenus Mundinia that may participate in leishmaniasis transmission in nature (Becvar et al., 2021).
Biting midges are also probable vectors of avian trypanosomes. DNA of Trypanosoma spp. parasites has been detected in several wild caught Culicoides spp. (Bernotienė et al., 2020). C. alazanicus, C. pictipennis, C. festivipennis, and C. clastrieri are naturally infected with T. bennetti (s. l.) (Svobodová et al., 2017). Experimental infection of C. nubeculosus and C. impunctatus with four closely related haplotypes of T. everetti has shown that these Trypanosoma parasites are able to develop and produce metacyclic trypomastigotes in both biting midge species (Bernotienė et al., 2020).
Besides the dixenous trypanosomatids (Leishmania and Trypanosoma), natural infections of Culicoides biting midges with monoxenous trypanosomatids have been reported. Herpetomonas ztiplika was isolated from a female C. kibunensis caught while attacking buzzard (Buteo buteo) nestlings (Podlipaev S. A. et al., 2004). Several isolates of Sergeia podlipaevi were obtained from females of two species of biting midges C. festivipennis and C. truncorum captured in common buzzard nests (Svobodová et al., 2007). A monoxenous trypanosomatid, Herpetomonas trimorpha, was isolated from the digestive tract of a female biting midge, C. truncorum (Zídková et al., 2010). In addition, Crithidia sp. DNA was detected in one female C. pictipennis and H. ztiplika DNA in two C. obsoletus females (Bernotienė et al., 2020).
Infections with monoxenous trypanosomatids have occasionally been reported in mammals including humans. In mammals a flagellate parasite was isolated from rats (Rattus norvegicus) and stray dogs in Egypt (Morsy et al., 1988) and later found to be a member of the genus Herpetomonas (Podlipaev S. et al., 2004). Recently, mammals including coatis (Nasua nasua), marmosets (Callithrix sp.), bats (Carollia perspicillata, Myotis lavali, M. izecksohni, Artibeus lituratus), crab-eating foxes (Cerdocyon thous), and ocelots (Leopardus pardalis) in Brazil have been found infected in nature by Crithidia mellificae, a monoxenous trypanosomatid classically associated with honeybees (Dario et al., 2021). Also, DNA of Cr. mellificae is detected in a nectar-feeding bat (Anoura caudifer) in Brazil (Rangel et al., 2019).
So far, most infection by monoxenous trypanosomatids in humans has been reported in patients with either HIV or Leishmania co-infection. Leptomonas seymouri-L. donovani co-infection cases have been reported from India (Srivastava et al., 2010; Ghosh et al., 2012; Singh et al., 2013; Thakur et al., 2020). In Brazil, a Crithidia-related species has been isolated from an immunocompetent patient with a fatal visceral leishmaniasis-like illness (Maruyama et al., 2019) and a 9-year-old patient with leishmaniasis caused by L. infantum (Rogerio et al., 2023). Also, a novel thermotolerant monoxenous trypanosomatid closely related to Cr. fasciculata has been isolated from clinical samples of immunocompetent patients suspected of cutaneous leishmaniasis in Iran (Ghobakhloo et al., 2019; Kostygov et al., 2019). However, potential vectors of these monoxenous trypanosomatid parasites remain unknown.
In Thailand, the role of biting midges as potential vectors of Leishmania and Trypanosoma parasites has been investigated and revealed DNA of L. martiniquensis and Trypanosoma spp. in three female C. mahasarakhamense and one female C. huffi, respectively. Blood meal analysis showed C. arakawae, C. mahasarakhamense, C. guttifer, C. huffi, C. fulvus and C. actoni had fed on chickens, whereas C. asiana, C. imicola, C. peregrinus, C. oxystoma and C. shortti had fed on water buffalo and cattle (Jomkumsing et al., 2021; Sunantaraporn et al., 2021). Recently, DNA of L. martiniquensis was detected in C. peregrinus, C. oxystoma, C. mahasarakhamense, C. huffi, C. fordae, and C. fulvus and DNA of L. orientalis in C. peregrinus and C. oxystoma caught near a leishmaniasis patient’s house in southern Thailand. In addition, DNA of Crithidia sp. was detected in Culicoides spp. in those areas (Songumpai et al., 2022; Sunantaraporn et al., 2022).
Observation and isolation of live L. martiniquensis and L. orientalis parasites in natural infections of Culicoides biting midges is still required as it is one of the key criteria used to incriminate a natural vector of leishmaniasis. Therefore, the objectives of this study were (1) to investigate trypanosomatids in Culicoides biting midges collected from an endemic area of leishmaniasis and a mixed farm of chickens, goats, and cattle in southern Thailand, (2) to analyze blood meals of engorged midges, and (3) to characterize any trypanosomatids successfully isolated into culture. Our study provided evidence for C. peregrinus as a potential vector of L. martiniquensis and revealed a novel thermotolerant Crithidia trypanosomatid that could infect mouse cells in vitro.
The use of animals in this study was approved by the animal research ethics committee of Chulalongkorn University Animal Care and Use Protocol (CU-ACUP), Faculty of Medicine, Chulalongkorn University, Bangkok, Thailand (COA No. 023/2564 and COA No. 011/2564).
Collection of biting midges was conducted in two locations, Location 1: Tambon Tha Ruea (TR), an endemic area of leishmaniasis (8°22′42.0”N 99°58′32.0″E) and Location 2: Tambon Khuan Phang (KP), a mixed farm of chickens, goats, and cattle (8°09′35.2”N 99°56′33.8″E), Nakhon Si Thammarat province, southern Thailand. In each location, wild biting midges were collected using five Center for Disease Control and Prevention (CDC) miniature light traps (25 W bulb) with ultraviolet (UV) light for 4 nights in December 2021. Traps were operated from 6.00 pm to 10.00 pm. The midges were kept in plastic boxes covered by two layers of insect nets with moisture papers on the top and transported to the Vector Biology and Vector Borne Disease Research Unit, Department of Parasitology, Faculty of Medicine, Chulalongkorn University.
Live insects were placed on ice to immobilize before investigating under a binocular stereoscopic microscope (SZX10; Olympus, Tokyo, Japan). Males were separated from females by their morphology. Species identification of females was carried out using a taxonomic key according to morphological characters, namely wing spot patterns and head features (palp and antenna) (Dyce et al., 2007; Pramual et al., 2021). In each species, females were grouped by their physiological stage: parous (empty abdomen, with traces of burgundy pigment after blood sucking), engorged (abdomen filled with blood), gravid (abdomen with eggs), and nulliparous (empty abdomen without the presence of blood, they never sucked blood) (Dyce, 1996; Kasičová et al., 2021). Parous (50) and nulliparous (50) females collected from each location were selected to investigate for trypanosomatids. Before dissection, the midges were placed on ice in a small Petri dish containing 0.05% (v/v) Tween 20 in 1× Phosphate Buffered Saline (PBS; 10 mM sodium phosphate, 145 mM sodium chloride, pH 7.2). The whole gut (foregut, midgut, and hindgut) was dissected using sterile fine needles in a drop of 50 μl sterile PBS on a sterile slide under a binocular stereoscopic microscope. Then the gut was transferred into 50 μl sterile PBS on a new sterile slide, covered with a sterile coverslip, and examined for trypanosomatids under a light microscope (Olympus America Inc., USA) at 400 × magnification. Some insects with unidentified trypanosomatids in the whole gut were video recorded and photographed. Each positive sample was divided into three parts in a small volume of PBS for Giemsa’s staining, cultivation, and gDNA extraction. To confirm the species of the infected insects, the female carcass of each positive sample was subjected to gDNA extraction for molecular identification of Culicoides species.
Schneider’s Insect medium (SIM) (Sigma-Aldrich, St Louis, MO, USA), pH 6.8 supplemented with 10% (v/v) heat-inactivated fetal bovine serum (hi-FBS) (Life Technologies-Gibco, Grand Island, NY, USA), 100 μg/mL penicillin–streptomycin and 250 μg/mL gentamicin (Sigma-Aldrich, St. Louis, MO, USA) was used to culture trypanosomatids from insects. Each positive sample in PBS was transferred into a 25 ml flask containing 5 mL SIM, pH 6.8 supplemented with 10% (v/v) hi-FBS and 25 μg/ml gentamicin (SIM complete), and incubated at 26°C. An axenic culture was established by serial dilution and subpassage. Promastigotes were mixed with 7.5% (v/v) glycerol in SIM complete and stored at −80°C.
Trypanosomatid samples isolated directly from insects or cultures in SIM with supplements were smeared on microscope slides and air-dried. The slides were then fixed in absolute methanol and stained with 5% (v/v) Giemsa’s stain solution (Sigma-Aldrich, Darmstadt, Germany). The morphological characteristics and morphometry of the trypanosomatids were examined under a light microscope (Olympus America Inc., USA) at 1,000 × magnification. Light microscopy (LM) images of 50 parasites in each stage were used for morphometry, including: cell body length, cell body width, anterior end to kinetoplast distance, anterior end to nucleus distance, and length of flagellum. The measurement results were presented as mean ± standard deviation.
Genomic DNA of biting midges and trypanosomatids was extracted using a genomic DNA purification kit (Thermo Fisher Scientific Inc., Waltham, MA, USA) according to the manufacturer’s instructions. For molecular identification of Culicoides species, LCO1490 primer (5 ́–GGTCAACAAATCATAAAGATATTGG−3 ́) and HCO2198 primer (5 ́–TAAACTTCAGGGTGACCAAAAAA TCA−3 ́) (Folmer et al., 1994) were used to amplify the approximately 658-bp fragment of the mitochondrial cytochrome c oxidase subunit 1 gene (COI) following the polymerase chain reaction (PCR) method described by Harrup et al. (2016). Host blood from engorged females was identified based on the mitochondrial cytochrome b (cyt b) gene sequence using the primers cyt bb1 (5 ́–CCATCMAACATYTCADC ATGAAA−3 ́) and cyt bb2 (5 ́–GCHCCTCAGAATGAYATTTG KCCTCA−3 ́) as described by Radrova et al. (2013). For molecular identification of Leishmania species in gDNA samples extracted from insects and/or cultures, primers LeF (5 ́–TCCGCCCGAAAGTTCACC GATA−3 ́) and LeR (5 ́–CCAAGTCATCCATCGCGACACG−3 ́) (Spanakos et al., 2008), were used to amplify the internal transcript spacer 1 (ITS1) region (approximately 379 bp) and primers, 70-IR-D (5 ́-CCAAGGTCGAGGAGGTCGACTA-3 ́) and 70-IR-M (5 ́-ACG GGTAGGGGGAGGAAAGA −3 ́) (Requena et al., 2012) were used to amplify the 3 ́untranslated region (3 ́UTR) of Leishmania HSP70-type I (HSP70-I) genes as described by Jariyapan et al. (2021).
For molecular identification of trypanosomatids, primers, TRY927F (5 ́–GAAACAAGAAACACGGGAG −3 ́) and TRY927R (5 ́–CTACTGGGCAGCTTGGA−3 ́), were used to amplify approximately 927 bp of the small subunit ribosomal RNA (SSU rRNA) gene (Noyes et al., 1999) of gDNA extracted from insects and cultures. All amplicons were visualized in 1.5% agarose. The PCR products were purified using a GeneJET PCR Purification kit (Thermo Fisher Scientific, CA, USA). PCR conditions and protocols used in this study are provided in Supplementary Data S1.
For trypanosomatids successfully grown in culture, primers M200 (5 ́–ATGGCTCCVVTCAARGTWGGMAT−3 ́) and M201 (5 ́–TAKCCCCACTCRTTRTCRTACCA −3 ́) for the glycosomal glyceraldehyde-3-phosphate dehydrogenase (gGAPDH) gene (Maslov et al., 1996) were used to confirm species identification. The PCR products of M200/M201 primers were cloned into the pGEM-T Easy Vector system (Promega, Madison, WI, USA), following the manufacturer’s instructions. Recombinant plasmid DNA was extracted using the Invisorb Spin Plasmid Mini Kit (STRATEC Molecular, Berlin, Germany), following the manufacturer’s instructions. The purified PCR products were sent for sequencing at the sequencing service of Macrogen Inc., Seoul, Korea.
The nucleotide sequences of the COI gene of Culicoides biting midges, the cyt b gene of vertebrates, ITS1 and 3’UTR-HSP70-I genes of Leishmania parasites, and SSU rRNA and gGAPDH genes of trypanosomatids were analyzed by comparison with the GenBank database using a BLAST search1. The sequences of Leishmania parasites and trypanosomatids were aligned and trimmed using MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms version 10.2.6 (Kumar et al., 2018). The best-fitting nucleotide substitution models for both genes were estimated using the Akaike Information Criterion (AIC) in the jModelTest software (Posada, 2008). Phylogenetic trees were constructed using the Maximum Likelihood (ML) method implemented in MEGA (Kumar et al., 2018) with 1,000 bootstrap-replications. Bootstrap values of ≥70% were taken as an indication support (Hillis and Bull, 1993). For phylogenetic analysis of Leishmania parasites, evolutionary models of GTR + I were chosen for ITS1 (329 bp) and 3’UTR-HSP70-I (817 bp). For phylogenetic analysis of Crithidia sp., evolutionary models of GTR + I + G were chosen for SSU rRNA (720 bp) and gGAPDH (654 bp). Genetic distances were calculated using the Kimura 2-parameter (K2P) model implemented in MEGA (Kumar et al., 2018).
Scanning electron microscopy (SEM) and transmission electron microscopy (TEM) were used to characterize the ultrastructural morphology of the cultured trypanosomatids. For SEM, live trypanosomatids were pelleted at 1,600 × g for 10 min at room temperature and fixed with 2.5% (v/v) glutaraldehyde in 0.1 cacodylate buffer (pH 7.2) overnight at 4°C. Fixed cells were washed twice in PBS and post-fixed with 1% (w/v) osmium tetroxide in PBS for 1 h. The samples were then dehydrated in a graded series of ethanol. Samples were critical point dried in liquid CO2 and coated with gold particles in a sputter-coating apparatus. Samples were observed under a scanning electron microscope, JEOL JSM-6610LV (JEOL, Tokyo, Japan), operated at 15 kV. Samples for TEM were prepared as for SEM, except that 2% (w/v) osmium tetroxide were used to post-fix for 2 h at room temperature. After dehydration in a graded series of ethanol, the samples were incubated overnight in an epoxy resin (PolyBed 812)/acetone solution (1:1), and then embedded in pure resin and polymerized for 48 h at 60°C. ultra-thin sections were stained with 4% (w/v) uranyl acetate and 3% (w/v) lead citrate and observed under a transmission electron microscope, JEM-2200FS (JEOL, Tokyo, Japan), operated at 200 kV.
Trypanosomatids were removed from −80°C, grown in SIM complete at 26°C for 4 days, and subpassaged into new SIM complete for growth analysis. The parasites were counted using a Neubauer chamber (BLAUBRAND, Sigma-Aldrich, Saint Louis, MO, USA) from an initial inoculum of 4 × 104 parasites/mL (day 0). The initial inoculum was transferred into a 25 cm3 flask containing 5 ml of the culture medium for 14 flasks. Seven flasks were incubated at 26°C and the rest were incubated at 37°C with 5% CO2. One flask at each temperature was taken out daily, and a cell scraper (SPL Life Sciences, Gyeonggi, Korea) was applied to remove any adherent cells from the base of the flask. Ten microliters of parasites were collected and mixed at a 1:1 ratio with formaldehyde when all parasites were alive or 0.4% trypan blue solution (Thermo Fisher Scientific, NY, USA) when nonviable forms of parasites were present in the cultures. Live cells were counted using a Neubauer chamber. Also, the cultures were smeared on slides and stained with Giemsa’s staining solution for morphological examination by LM. Experiments were performed on three independent replicates run in duplicate.
To evaluate the ability of trypanosomatids to infect and multiply in mouse macrophages an infection and multiplication assay was performed. Mouse peritoneal exudate macrophages (PEMs) from BALB/c (Mus musculus) (Nomura Siam International Co., Ltd., Bangkok, Thailand) were obtained using the method described previously (Zhang et al., 2008). Round coverslips were placed in 24-well tissue culture plates (ThermoFisher Scientific, Jiangsu, China). PEMs in RPMI 1640 medium (GE Healthcare Life Science-HyClone, UT, United States) supplemented with gentamicin 25 μg/ml and 10% (v/v) hi-FBS were seeded at a density of 2.5 × 105 cells/well and incubated at 37°C with 5% CO2 for 24 h. After the incubation, non-adherent cells were removed by washing three times with pre-warmed serum-free RPMI 1640 medium. Test trypanosomatids (from Day 3 with the greatest number of motile forms) and L. martiniquensis promastigotes (from Day 5) were used to infect PEMs at a ratio of 10:1 cells to macrophages. After incubation for 3 h at 37°C and 5% CO2, the coverslips were washed with pre-warmed serum-free RPMI 1640 medium for three times, replaced with 10% hi-FBS RPMI-1640 medium, and incubated at the same conditions. Coverslips were removed from the culture plates and stained with Giemsa’s staining solution every 24 h until 96 h post-infection. Two hundred macrophages were counted to determine the infection rate and average number of intracellular parasites per macrophage. The infection index, and the intracellular parasite multiplication ratio were determined (Chanmol et al., 2019). Results were expressed as mean ± standard deviation and based on three independent infection experiments each performed in duplicate.
All statistical analyses were performed using GraphPad Prism version 9.1 software. Statistical differences between L. martiniquensis and Crithidia sp. infections at the different time points within one group were determined using two-way ANOVA with Bonferroni’s post hoc multiple comparisons for growth, infection index, and intracellular multiplication ratio. Tests were considered statistically significant if p < 0.05.
A total of 1,094 biting midges of the genus Culicoides were captured in 2 locations, 349 females (parous = 276, engorged = 2, nulliparous = 71) and 6 males from Tha Ruea and 724 females (parous = 527, engorged = 12, nulliparous =185) and 15 males from Khuan Phang (Supplementary Table S1). No gravid females were found in either location. Three species of biting midges were identified in Tha Ruea, C. peregrinus (n = 343, 96.62%), C. mahasarakhamense (n = 9, 2.53%), and C. oxystoma (n = 3, 0.85%). In Khuan Phang seven species were identified, C. peregrinus (n = 623, 84.3%), C. imicola (n = 31, 4.19%), C. shortti (n = 27, 3.65%), C. huffi (n = 25, 3.38%), C. mahasarakhamense (n = 20, 2.7%), C. palpifer (n = 11, 1.5%), and C. oxystoma (n = 2, 0.27%). Thus, in both locations C. peregrinus was the most abundant species. For engorged females (n = 14), all blood host identifications were of Bos indicus cattle (GenBank accession no. OR088254–OR088267).
Fifty parous and 50 nulliparous females collected from each location were dissected for trypanosomatid infection. Trypanosomatids were observed under light microscopy (LM) in 26 out of the 100 samples of parous females from the two locations. However, no trypanosomatids were found in any nulliparous flies. All infected flies were identified as C. peregrinus by wing spot patterns (Figure 1A) and molecular methods (GenBank accession no. OR077408–OR077433). Light microscopic examination showed trypanosomatids present in the digestive tract of the midges (Figure 1 and Supplementary Videos S1, S2). None of the midges dissected had remains of bloodmeals in their midguts. Although the midges were dissected carefully, due to their small size and delicate nature, the digestive tracts of some midges were torn and trypanosomatids released. However, in most samples with an intact digestive tract trypanosomatids were found in the midgut and hindgut, with choanomastigote and/or promastigote morphologies observed (Figure 1B). Interestingly, trypanosomatids were observed in the foregut, midgut, and hindgut of one C. peregrinus, KP10 (Figure 1C). In this insect, video records revealed typical movement of Leishmania promastigotes in the foregut (Supplementary Video S1) and Crithidia in the hindgut (Supplementary Video S2). Molecular identification (see below) of the KP10 sample revealed that this midge was co-infected with L. martiniquensis and a possible novel Crithidia species (Table 1). Trypanosomatids were examined in slides prepared from infected midges, and various forms of promastigotes and chroanomastigotes were observed (Figure 1D). In the insect KP19 many promastigote forms of Leishmania were observed including procyclic promastigotes (Figures 1E,F), nectomonad promastigotes (Figure 1G), and leptomonad promastigotes (Figure 1H), however, no metacyclic promastigotes were observed. The parasites in KP19 were identified as L. martiniquensis by molecular methods (Table 1). The presence of Leishmania promastigotes in the foregut of midges TR17, KP10, KP17 and KP19 in the absence of bloodmeals are indicative of established infections.
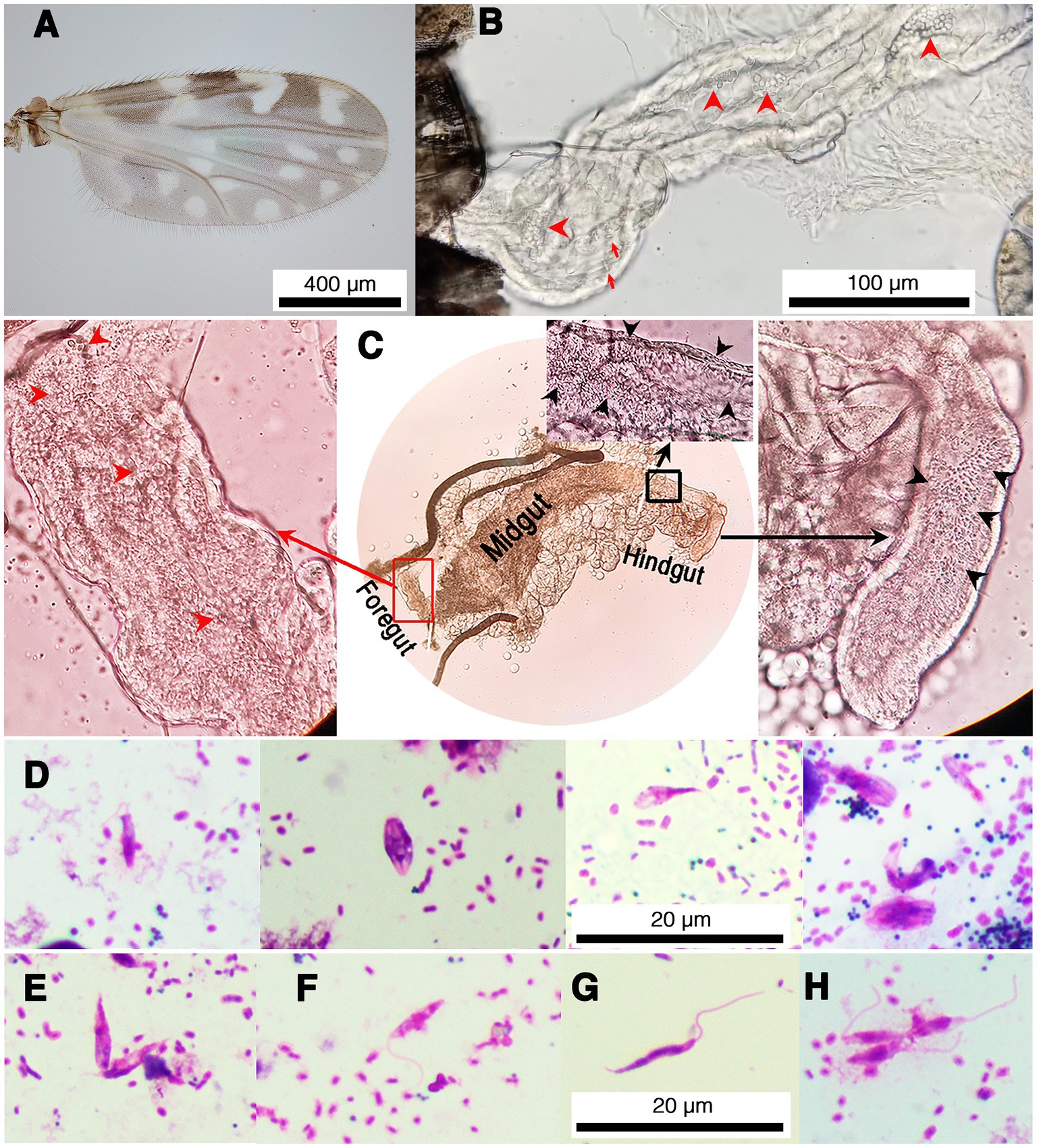
Figure 1. Representative images of trypanosomatids and co-infection of L. martiniquensis and Crithidia sp. in naturally infected C. peregrinus biting midges. (A) Wing pattern of C. peregrinus. (B) Trypanosomatids in the hindgut of the midge KP22. Red arrowheads indicate choanomastigotes and arrows indicate promastigotes. (C) Leishmania parasites and Crithidia sp. co-infected in the midge KP10. Red arrowheads and black arrowheads indicate trypanosomatids in the foregut and the hindgut, respectively. Red rectangle indicates an area in the foregut showing the movement of Leishmania parasites in Supplementary Video S1. Black rectangle indicates an area in the hindgut showing the movement of Crithidia sp. in Supplementary Video S2. (D) Representative of various forms of Crithidia sp. in the midge KP22. (E–H) Representative promastigote forms of Leishmania parasites in the midge KP19. (E) Procyclic promastigotes, aggregated form. (F) Procyclic promastigote. (G) Nectomonad promastigote. (H) Leptomonad promastigotes.
Four isolates of trypanosomatids were successfully isolated into culture, two from C. peregrinus collected in Tha Ruea (TR2 and TR3) and two from C. peregrinus collected in Khuan Phang (KP1 and KP4) (Table 1). Unfortunately, other cultures including all those from midges with L. martiniquensis were too contaminated with bacteria and fungi and had to be discarded.
Molecular methods were used to identify the trypanosomatids. For molecular identification of Leishmania species, 100 gDNA samples extracted from the dissected insects were subjected to the PCR amplification of the ITS1 and 3’UTR-HSP70-I regions. Four samples of the dissected insects, from TR17, KP10, KP17, and KP19, were positive for Leishmania and their ITS1 amplicons were successfully sequenced. Moreover, a second molecular marker, 3’UTR-HSP70-I, was used to confirm the identification of the Leishmania species of the samples. BLAST analysis of the ITS1 and 3’UTR-HSP70-I sequences of the samples revealed that the ITS1 sequences were closest to that of L. martiniquensis, GenBank accession number MK603827.1, with 98.94% identity, whereas the 3’UTR-HSP70-I sequences were very similar (98.64 and 99.77% identity) or identical (100% identity) to that of L. martiniquensis, GenBank accession number MK607435.1, and another one was identical to L. martiniquensis, GenBank accession number MK607437.1 (Table 1). The ITS1 and 3’UTR-HSP70-I sequences were subjected to phylogenetic analyses together with other human Leishmania strains (Figure 2). The results from Maximum Likelihood tree based on amplified section of the ITS1 and 3’UTR-HSP70-I sequences showed that the new sequences fell in one clade with L. martiniquensis with 99% bootstrap values (K2P 0–0.78%) for ITS1 (Figure 2A) and 100% bootstrap values (K2P 0–1.63%) for 3’UTR-HSP70-I (Figure 2B). These results indicate that the trypanosomatid DNAs detected in the C. peregrinus samples, TR17, KP10, KP17, and KP19, were all of L. martiniquensis.
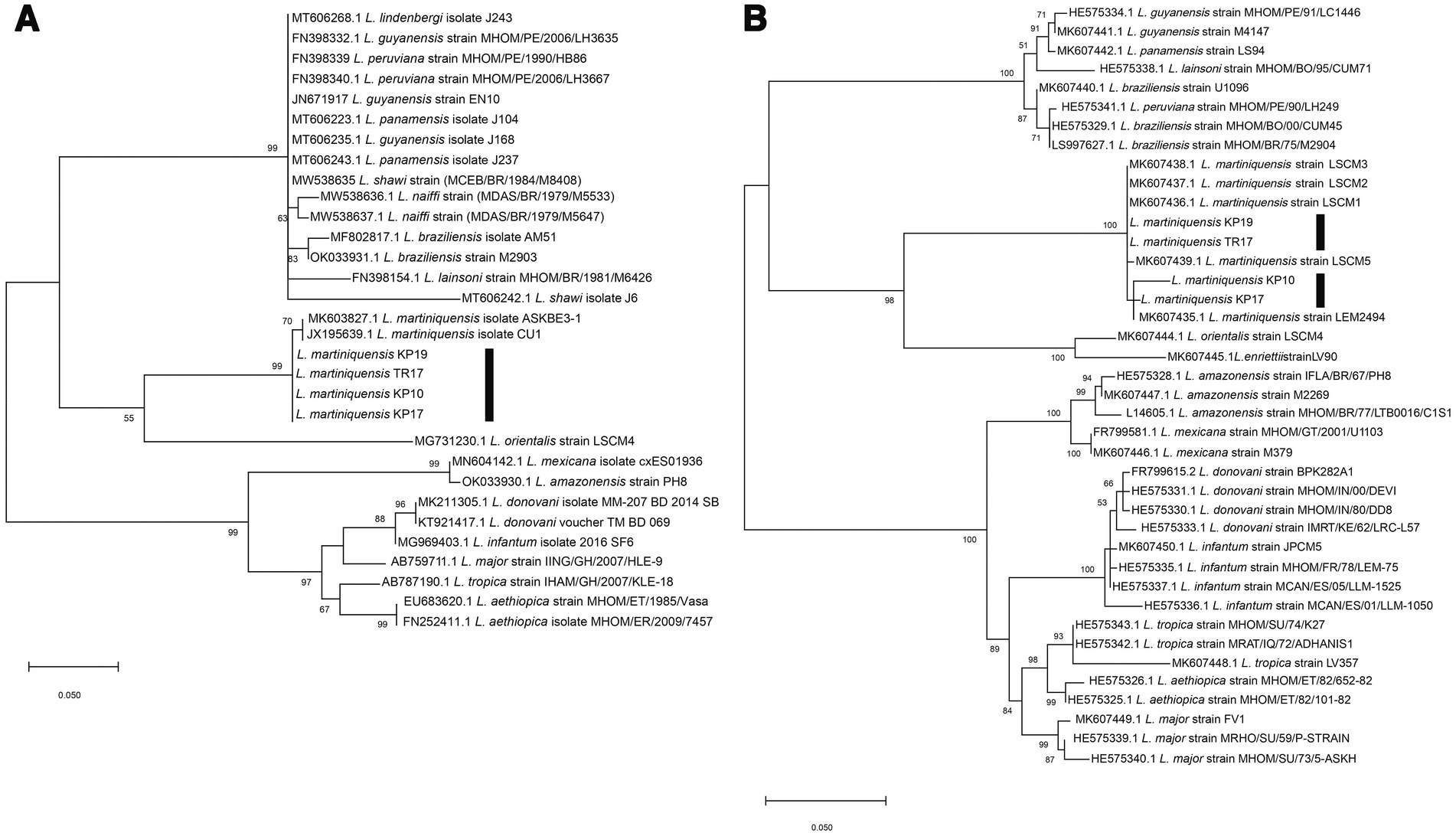
Figure 2. (A) Maximum Likelihood tree based on ITS1 sequences of Leishmania spp. (B) Maximum Likelihood tree based on 3’UTR-HSP70-I sequences of Leishmania spp. Bootstrap values are shown at each node. Bootstrap values of ≥70% correspond to supported clades. Nodes with Bootstrap values of <50% are not shown.
The SSU rRNA of 24 samples were successfully amplified and sequenced allowing the identification of other trypanosomatids. BLAST analysis showed that among these, 19 sequences were similar to each other and to a Crithidia sp. reported from Thailand by Songumpai et al. (2022) (GenBank accession number OP698037–OP698038) with 99.35–100% identity and to Cr. thermophila (GenBank accession number KY264937.1) with 97.00–97.74% identity. The other five sequences were also very similar to another Crithidia sp. reported from Thailand by Songumpai et al. (2022) (GenBank accession number OP217136) and Sunantaraporn et al. (2022) (GenBank accession number MW694347.1) with 99.89–100% and 98.92–99.14% identity, respectively (Table 1). These data showed that two midges, TR17 and KP10, were co-infected with both L. martinquensis and Crithidia, as suspected from dissection for KP10.
Phylogenetic analysis of the SSU rRNA sequences demonstrated that representative sequences from the group of 19, namely from insects TR2, TR3, KP1, TR17, KP4, and KP10 clustered together into one distinct clade with 99% bootstrap values (K2P 0.19–0.77%), designated here as Crithidia sp. Clade A (Figure 3A). The corresponding isolates were thus given strain designations Crithidia sp. CLA-TR2, Crithidia sp. CLA-TR3, and so on. Four of these, CLA-TR2, CLA-TR3, CLA-KP1 and CLA-KP4, were the four isolates successfully isolated into culture. Representatives of the group of 5 from insects TR21, TR29, and KP47 also clustered together with 99% bootstrap values (K2P 0–1.35%), and were designated as Crithidia sp. Clade B. The availability of cultures for CLA-TR2, CLA-TR3, CLA-KP1, and CLA-KP4 enabled additional DNA extractions and analysis of their gGAPDH sequences. These gGAPDH sequences also clustered together with 98% bootstrap values (K2P 0.31–0.77%) (Figure 3B). Amongst available sequences they were most similar to those of Cr. fasciculata (GenBank accession number AF047493.1) and Cr. brachyflagelli (GenBank accession number JF717835.1) with 94.39–94.61% identity (Table 1). However, they were almost as close to a clade of Cr. thermophila sequences (K2P 3.77–4.25%), like what was observed in the SSU rRNA analysis (Figure 3A).
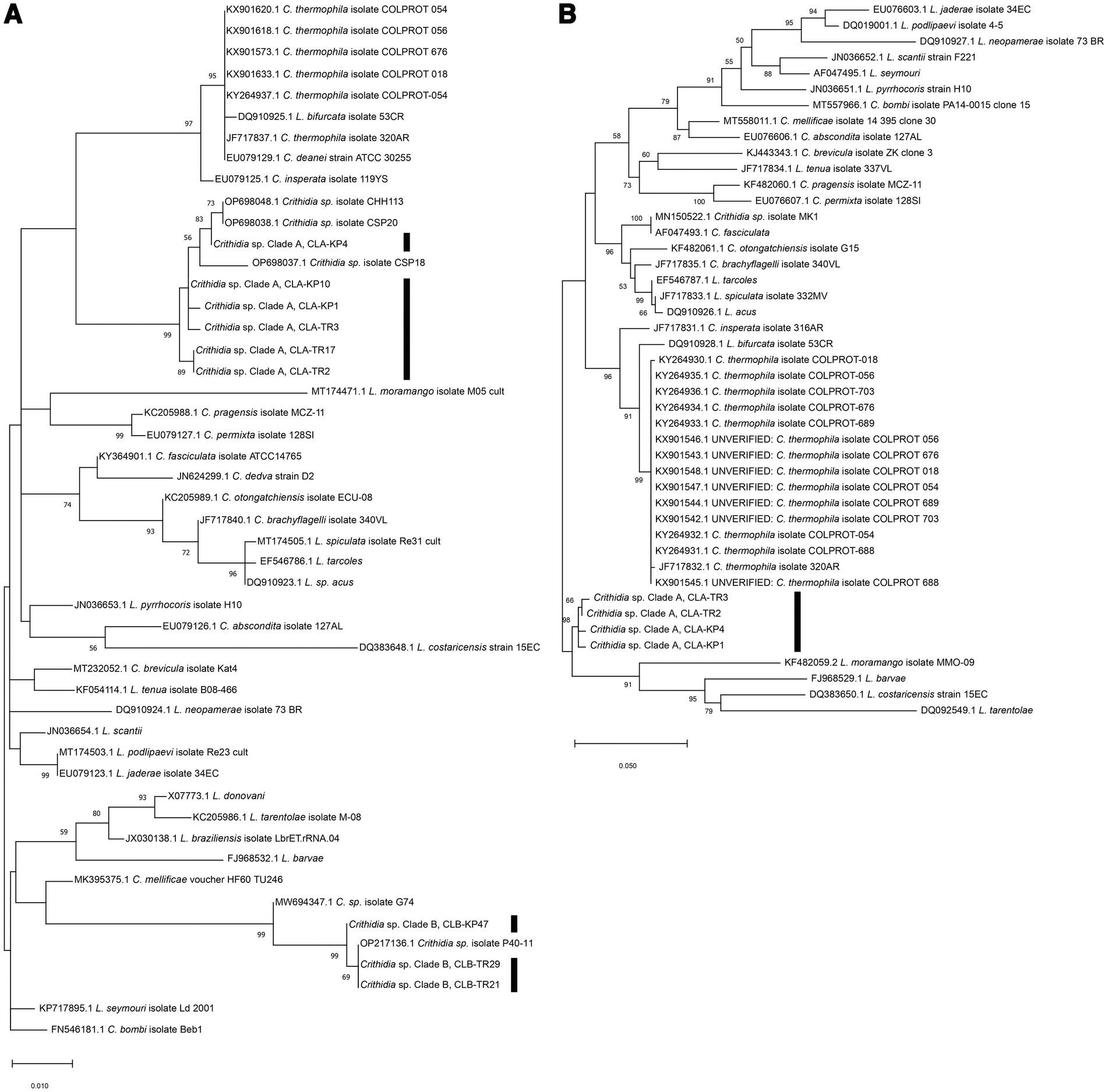
Figure 3. (A) Maximum Likelihood tree based on SSU rRNA sequences of Crithidia spp., Leishmania spp. and Leptomonas spp. (B) Maximum Likelihood tree based on gGAPDH sequences of Crithidia spp., Leishmania spp., and Leptomonas spp. Bootstrap values are shown at each node. Bootstrap values of ≥70% correspond to supported clades. Nodes with Bootstrap values of <50% are not shown.
In summary, from Tha Ruea seven female C. peregrinus were found infected with Crithidia sp. Clade A organisms and four females with Crithidia sp. Clade B organisms. Successful cultures were obtained only from insects TR2 and TR3, both Clade A organisms (Table 1). In this area the infection rate of L. martiniquensis in the 50 parous C. peregrinus was 2%, represented by one insect (TR17) also co-infected with Crithidia sp. Clade A organisms. In Khuan Phang, twelve females were infected with Crithidia sp. Clade A organisms and one female with Crithidia sp. Clade B organisms. In this area the infection rate of the C. peregrinus with L. martiniquensis was 6%, with one insect (TR10) again co-infected with Crithidia sp. Clade A organisms, but also in two other insects (KP17 and KP19) that only contained Leishmania. We also successfully cultured two Crithidia sp. Clade A from two C. peregrinus (KP1 and KP4) (Table 1).
The results of the phylogenetic analysis revealed Crithidia sp. Clade A organisms to be related to Cr. thermophila, an unusual thermotolerant trypanosomatid. To examine whether Clade A organisms might have similar properties further experiments were performed on strain CLA-KP1. Morphological and ultrastructural analyses of cultures in SIM complete at 26°C revealed two distinct morphotypes, choanomastigotes with rounded posterior ends and promastigotes with elongated posterior ends (Figures 4, 5). A wide collar-shaped reservoir (collar-like extension) surrounding the anterior end through which a single flagellum emerges was noted in both morphotypes (Figures 4A,B, 5B). This single flagellum arose from a basal body (kinetosome) located next to a kinetoplast (Figures 5A–C) and was surrounded by the flagellar pocket (Figure 5). The flagellum exhibited nine pairs of peripheral microtubules surrounding two central microtubules, the typical 9 × 2 + 2 axonemal pattern (Figure 5D). The kinetoplast was located just beyond the basal body, anterior or parallel to the nucleus. The nucleus was oval located in the middle of the cell body with visible accumulations of chromatin. Some other structures, such as acidocalcisomes, glycosomes and ribosomes, were observed (Figures 5A–C). Different forms of the choanomastigotes were observed, including those without a free flagellum (haptomonad forms) (Figures 4A–F, 5A) and others with a free flagellum protruding from the flagellar pocket at the anterior end of the cells (Figures 4B–F). A form of choanomastigote comprising attached clusters of cells (Rosette form) could be observed at the bottom of culture flasks (Figures 4A–E). Promastigotes were slender in shape with a free flagellum at the anterior end of the cells. In addition, morphological differences in some details, such as body width, body length, and flagellum length were observed (Figures 4G–J). Comparative morphological measurements of the choanomastigotes and promastigotes determined under LM are shown in Table 2. Promastigotes with the longest body and flagellum length known as a nectomonad form were noted (Figures 4D–J and Table 2). A single layer of subpellicular microtubules was observed around the body except for the membrane inside the area of the flagellar pocket (Figures 5A–C).
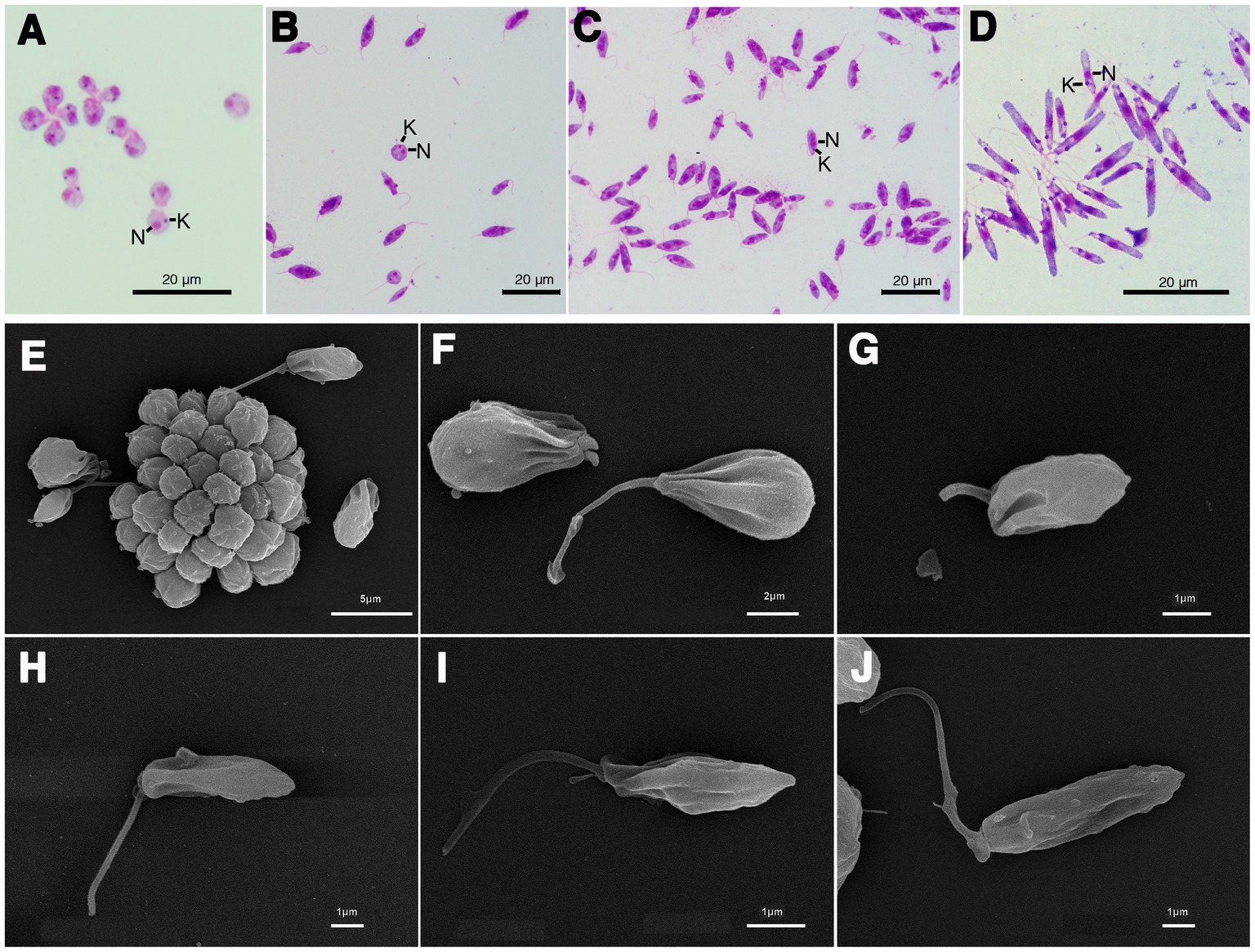
Figure 4. Light microscopy (A–D) and SEM (E–J) of Crithidia sp. CLA-KP1 strain cultured in SIM complete at 26°C. (A) Giemsa-stained haptomonad forms. (B,C) Giemsa-stained choanomastigotes and promastigotes. (D) Giemsa-stained nectomonad forms. (E) A rosette form, adherent, non-motile form. (F) Haptomonad form and choanomastigote with flagellum. (G–I) Promastigotes with different length of flagellum. (J) Nectomonad form. n, nucleus; k, kinetoplast.
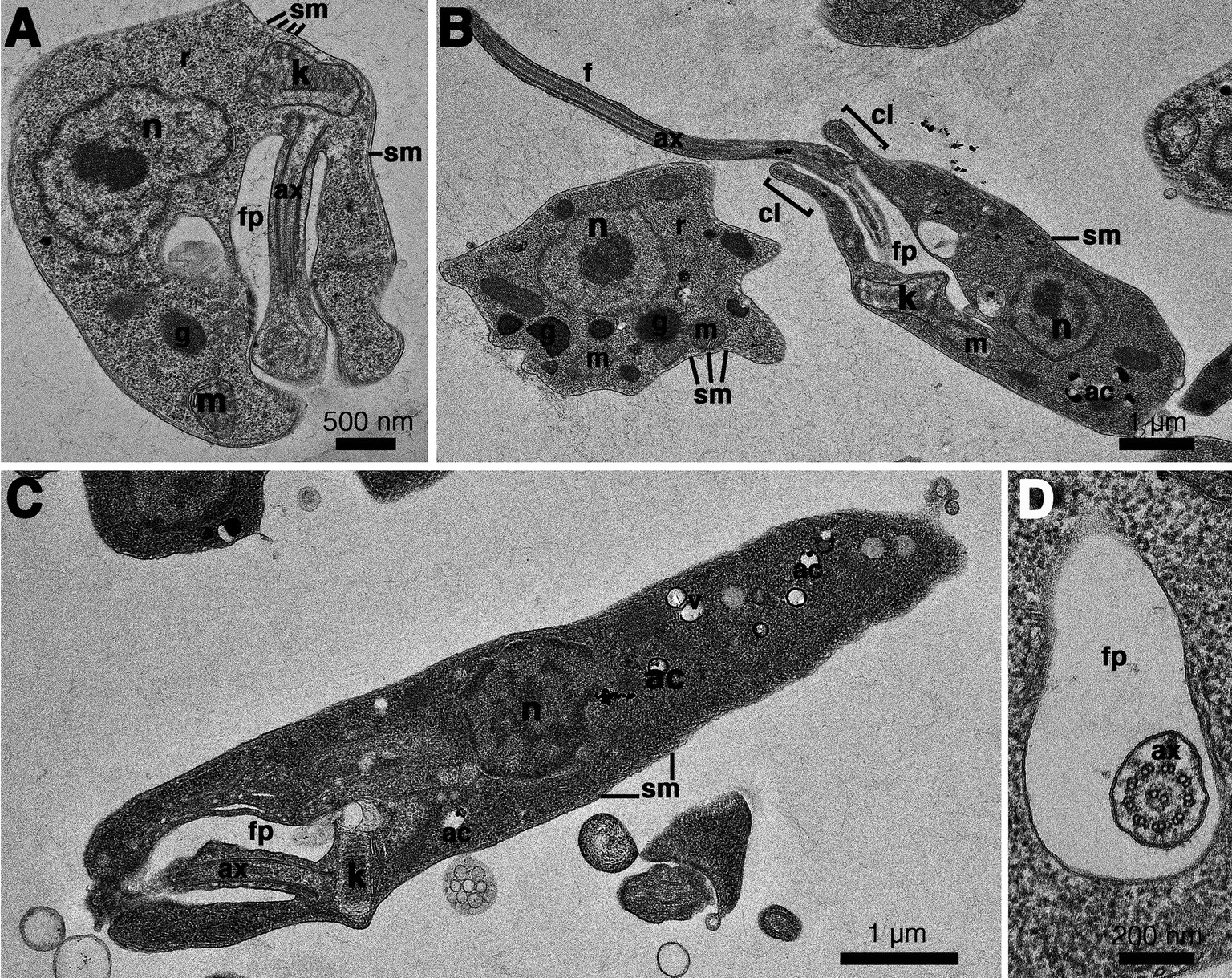
Figure 5. Transmission electron microscopy images of Crithidia sp. CLA-KP1 strain cultured in SIM complete at 26°C. (A) Longitudinal section of a haptomonad form (B,C) Longitudinal section of promastigotes. (D) Cross section of a promastigote showing a flagellum with 9 × 2 + 2 axonemal pattern in the flagellar pocket. ac, acidocalcisome; ax, axoneme; f, flagellum; fp, flagellar pocket; g, glycosome; k, kinetoplast; m, mitochondrion; n, nucleus; r, ribosome; sm, subpellicular microtubules; v, vesicles; cl, collar-like extension.
CLA-KP1 organisms were cultured at 26 and 37°C with an initial cell density of 4 × 104 cells/ml and observed daily for their growth for seven successive days. On the first 4 days, at both temperatures, similar growth patterns were observed. The parasites entered the exponential growth phase after 1 day of the culture and showed rapid multiplication (day 1 to day 4) with a doubling time of approximately 13.5 and 15.6 h for 26°C and 37°C, respectively. After 4 days of culture, at 26°C, the cultures entered the stationary phase and reached their peak on day 6 with a density of 1.9 × 108 cells/ml. The cultures remained in this phase to the last day of the experiments. At 37°C, CLA-KP1 reached maximum growth on day four with a density of 9.8 × 107 cells/mL, remained in stationary phase for 2 days, then the population dropped dramatically after 6 days of culture until the end of the experiment with a density of 1.0 × 106 cells/ml. The average parasite density at 26°C was significantly greater than 37°C after 5 days of culture (Figure 6 and Supplementary Table S2). Both morphotypes were found in the cultures at both temperatures. However, the predominant morphotype at 26°C was promastigotes, whereas at 37°C choanomastigotes were mainly observed.
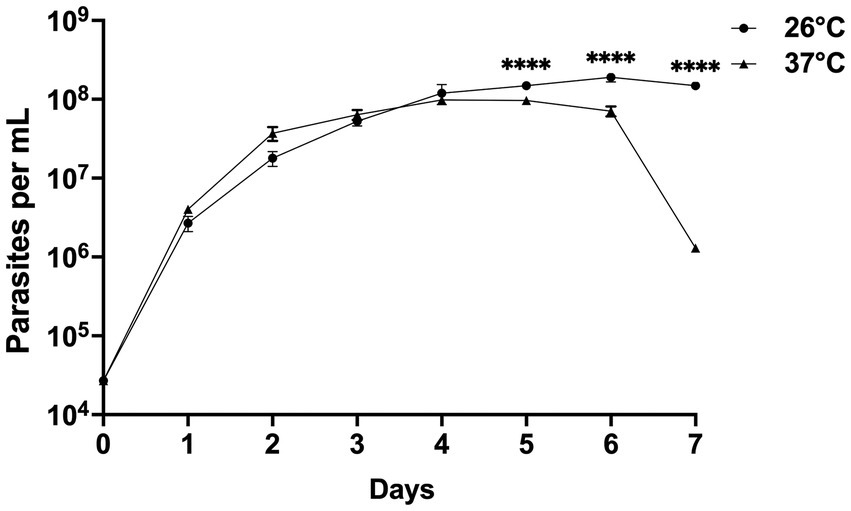
Figure 6. Growth curves of Crithidia sp. CLA-KP1 strain cultured in SIM complete for 7 days at 26 and 37°C. Statistically significant differences between 26 and 37°C are indicated as ****p < 0.0001.
The ability of CLA-KP1 to infect and multiply in PEMs of BALB/c mice was determined and compared with L. martiniquensis (Figure 7). Crithidia sp. CLA-KP1 could infect the PEMs and was found at 24 and 48 h but no parasites were observed at 72 and 96 h. At both time points, the infection rate, the average number of intracellular parasites per cell, and the infection index of CLA-KP1 were statistically significantly lower than that of L. martiniquensis (Figures 7H–J). At 24 h post-infection, the percentage of PEMs infected with CLA-KP1 was 22.27 ± 0.55 and dramatically decreased to 6.68 ± 1.85 at 48 h, which differed from the stable infection rate in Leishmania parasites at the same time points. The average number of intracellular CLA-KP1 parasites within infected PEMs was 2.35 ± 0.09 and 1.56 ± 0.17 parasites/cell at 24 and 48 h, respectively. At both time points, the average number of intracellular parasites/macrophage was approximately 2.6 times lower than that of L. martiniquensis. The infection index of CLA-KP1 was 51.97 ± 0.45 and 10.25 ± 2.10 at 24 h and 48 h, respectively, which was lower than that of the Leishmania parasites approximately 3.5 and 12.5 times, respectively. The intracellular parasite multiplication ratio of CLA-KP1 was only calculated at 48 h post-infection (0.21) because after that the parasites disappeared. A decrease in the intracellular parasite multiplication ratio was also observed in L. martiniquensis after 24 h post-infection, however, the Leishmania parasites were still be found in the PEMs at the end of the experiment (96 h) (Figure 7 and Supplementary Table S3). In summary, these results demonstrated that Crithidia sp. CLA-KP1 could infect and persist in the PEMs up to 48 h but could not multiply within them.
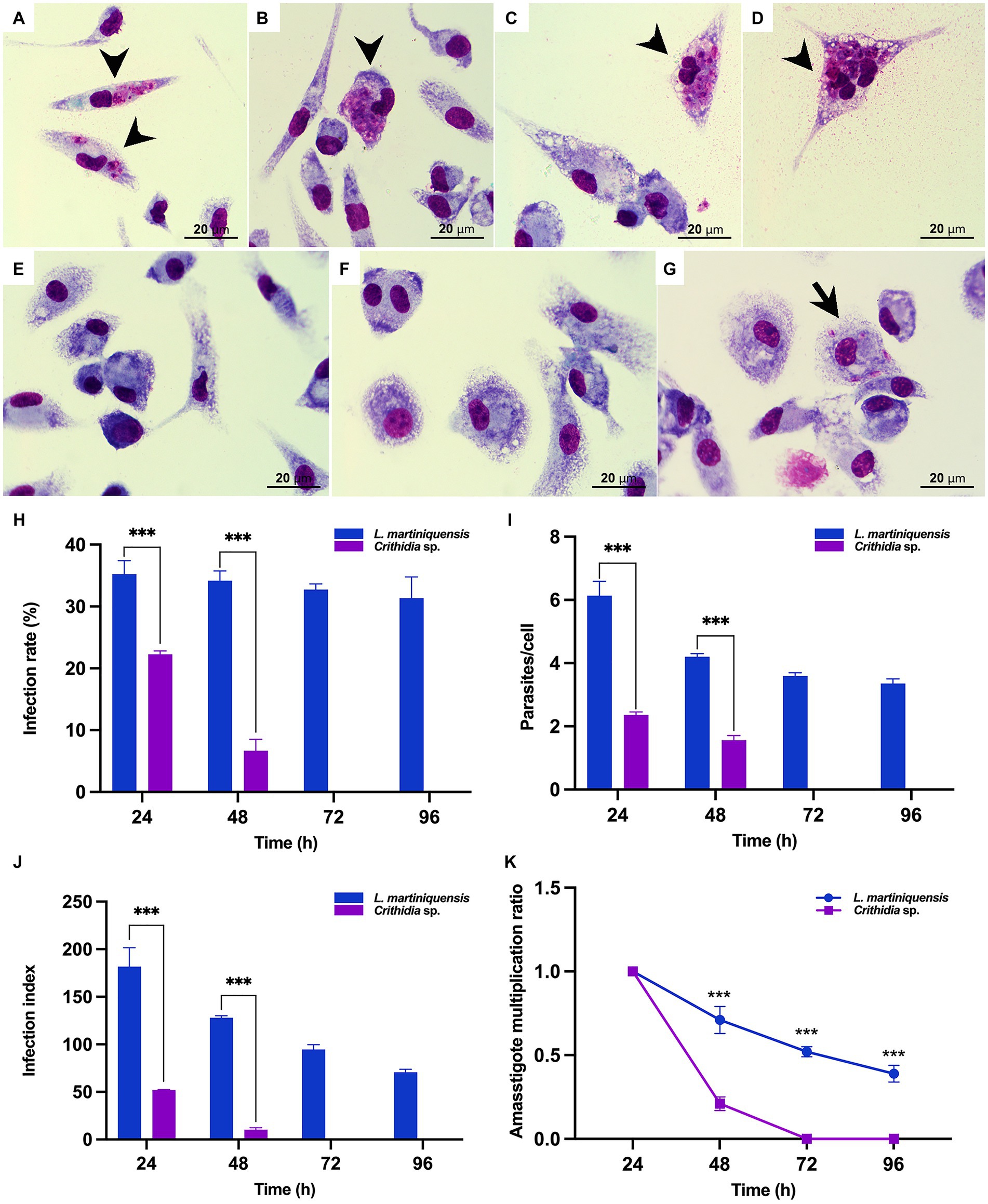
Figure 7. In vitro infection of Crithidia sp. CLA-KP1 in PEMs. (A,B) 24 h after infection. (C,D) 48 h after infection. (E) 72 h after infection. (F) 96 h after infection. (G) L. martiniquensis infection at 96 h (control). (H) Infection rate. (I) Average number of intracellular parasites per macrophage. (J) Infection index. (K) Amastigote multiplication ratio. Results are expressed as the means ± standard deviation from three different experiments run in duplicate. Statistically significant differences between CLA-KP1 and L. martiniquensis are indicated as ***p < 0.001.
Evidence that biting midges can act as competent vectors of L. martiniquensis and L. orientalis has been experimentally demonstrated in vivo (Chanmol et al., 2019; Becvar et al., 2021), however, the natural vectors that are responsible for the transmission of leishmaniasis caused by both Leishmania species remain unknown. Here, we show the potential for C. peregrinus to act as a vector for L. martiniquensis in Thailand. Although the number of the biting midges collected was low, C. peregrinus was the prominent midge species found in Tha Ruea and Khuan Phang. Due to the collection of the midges performed only in one month in the dry season, the species composition of Culicoides species found in both areas was less than the previous reports in other provinces in Thailand (Jomkumsing et al., 2021; Sunantaraporn et al., 2021, 2022; Songumpai et al., 2022). The life cycle of Culicoides biting midges varies from 14 days to longer than one year since the development of each Culicoides species depends on latitude, climate temperature, and surrounding environmental conditions (Mellor et al., 2000; Mullen, 2009), therefore, different species of Culicoides are found in different locations and seasons. However, three species, C. peregrinus and C. oxystoma and C. mahasarakhamense, were found in both locations studied corresponding to a previous report that DNA of L. martiniquensis has been detected in these three Culicoides species and some other species, C. huffi, C. fordae, and C. fulvus, collected from Songkhla Province, southern Thailand (Songumpai et al., 2022).
One of the key criteria used to incriminate vectors of leishmaniasis is that the vector species supports the development of parasite life stages after the infecting blood meal has been digested through to the transmissible infective metacyclic stage (Killick-Kendrick, 1999; Lawyer and Perkins, 2000). Thus, the presence of metacyclic forms usually accompanied by other promastigote forms in the anterior midgut or stomodeal valve of the collected insects is important evidence to confirm that the insect is a natural vector of the Leishmania parasites (Killick-Kendrick, 1999; Ready, 2013). In the current study, various promastigote forms, namely procyclic promastigotes, nectomonad promastigotes, and leptomonad promastigotes, of the Leishmania parasites were observed in the foregut of four specimens of C. peregrinus. However, no metacyclic promastigotes were found in the Giemsa-stained slides. However, the presence of parasites in the foregut in the absence of bloodmeal remnants indicate these are established infections. The video record of the midge, insect code KP10, showed the movement of the parasites in the foregut, which was similar to the movement of Leishmania parasites in the foregut of sandflies (Falcão de Oliveira et al., 2017). The results of molecular identification using two molecular targets, ITS1 and 3’UTR HSP70 type I, confirmed that the Leishmania parasites found in the C. peregrinus were L. martiniquensis. Altogether, these findings support C. peregrinus as a natural vector of L. martiniquensis, but as yet do not prove this definitively. The occurrence of L. martiniquensis-Crithidia co-infections in two out of the four Leishmania-positive midges was interesting, but its significance, if any, is not clear. This may simply be co-incidental, but the possibility that the organisms may interact in some way is worth exploring in future studies.
The infection rates of L. martiniquensis in these study areas (2 and 6%) is typical of endemic areas, where only a small proportion of collected vectors are usually found harboring parasites. A recent study focused on the residences of two recently diagnosed visceral leishmaniasis patients in Songkhla Province, Thailand, indicated a local infection rate of 21.2% in Culicoides midges, including C. peregrinus (Songumpai et al., 2022), but no live dissections were performed. Even though no active case was present in our study areas, the infection rate of the parasites in the insects indicated the presence of the parasites circulating in the wild. It implies that vector(s) and animal reservoir host(s) of L. martiniquensis may be present in these locations serving the parasite’s life cycle. No L. orientalis DNA or parasites were found in the insects collected in this study, but further investigation in other seasons and areas that have active cases should be carried out.
To investigate whether the biting midges were anthropophilic or not, engorged females were subjected to blood meal analysis. Cow blood DNA was only detected in all collected engorged C. peregrinus females confirming the predominantly zoophilic behavior of C. peregrinus previously reported (Sunantaraporn et al., 2021, 2022; Kar et al., 2022; Songumpai et al., 2022). In the engorged female samples, no DNAs of Leishmania or other trypanosomatids were found. As the number of samples in our study was low, more samples of engorged females of Culicoides species are required for further investigation of the anthropophilic and/or zoophilic behaviors of the biting midges. Blood meal identification of the midges in the endemic areas would also help investigate reservoir hosts of autochthonous leishmaniasis in Thailand. For definitive vector incrimination transmission by bite needs to be demonstrated, which could be attempted in the field with natural hosts or with wild-caught naturally infected midges in the laboratory. However, neither of these are technically feasible at present. Our preferred approach is to establish a lab colony of C. peregrinus for transmission experiments of L. martiniquensis to mice or hamsters. The experimental results proving that C. peregrinus could transmit the parasites to mice or hamsters would be implied for the transmission of L. martiniquensis from naturally infected C. peregrinus to the reservoir hosts.
In this study, no Leishmania parasites were successfully isolated into cultures. However, this is a challenging procedure, because although dissection and examination for trypanosomatids under a light microscope was performed using sterile techniques, contamination still occurred from bacteria and fungi present inside the digestive tracts of the midges. Also, the amount of each positive sample was limited due to dividing the sample into three parts for Giemsa’s staining, cultivation, and gDNA extraction. These problems made it difficult to culture Leishmania parasites from the dissected insects. A biphasic Novy, McNeil, Nicolle (NNN) culture medium (Nicolle, 1908) containing penicillin–streptomycin and gentamicin was used to establish cultures of L. macropodum from midges of the genus Forcipomyia (Diptera: Ceratopogonidae) (Dougall et al., 2011). Adding antibiotics such as penicillin–streptomycin and gentamicin may help getting rid of some bacteria but not for fungi. Barratt and colleagues (Barratt et al., 2017) have been successfully isolated a novel trypanosomatid, Zelonia australiensis from a black fly, Simulium dycei, by serial dilution to get rid of fungi but only if the parasite cells outnumbered the fungi in the cultures (Barratt et al., 2017). Using other media such as the biphasic NNN with antibiotics and performing serial dilution may allow to obtain pure promastigote cultures easily. Recently, our study regarding Amphotericin B (AmpB)-resistant L. martiniquensis parasites has suggested that the resistant strains could be more efficiently transmitted and maintained in asymptomatic hosts longer than susceptible ones (Mano et al., 2023). Thus, the isolation of parasites from insects is required to investigate not only the vector competence of the insects but also the prevalence of the AmpB-resistant phenotype parasites in natural insect populations. Furthermore, the insect-isolated parasites would be useful sources for genetic analyses to intensively investigate the mechanisms underlying genetic exchange of the parasites in the insects as the sexual cycle of Leishmania parasites has been largely confined to promastigotes developing in the midgut of the vectors (Sadlova et al., 2011; Inbar et al., 2013; Ferreira and Sacks, 2022).
Unlike Leishmania parasites, monoxenous trypanosomatids, insect-restricted parasites, are more easily isolated and cultured in the laboratory (Lukeš and Votýpka, 2020). Here, two clades of novel Crithidia spp. were identified and four isolates of Crithidia sp. Clade A were obtained. Notably, choanomastigotes and promastigotes of both novel Crithidia spp. Clades were found in the hindgut of the dissected C. peregrinus corresponding to the reported developmental habitat of Crithidia spp. in insects (Frolov et al., 2021). The morphology of the forms of Crithidia sp. CLA was similar to those described in Cr. mellificae (Schwarz et al., 2015) and Cr. fasciculata (Filosa et al., 2019). TEM analysis showed typical morphological features of trypanosomatids: an oval nucleus; a kinetoplast; a flagellum with 9 × 2 + 2 axonemal pattern in the flagellar pocket; glycosomes; a reticulated mitochondrion with numerous cristae; and subpellicular microtubules. Cr. thermophila is reclassified and synonymous with Cr. luciliae thermophila and Cr. confusa (Ishemgulova et al., 2017). Only two forms, choanomastigotes and promastigotes with flagellum of Cr. thermophila grown in liquid Brain Heart Infusion (BHI) medium at 23°C have been reported (Ishemgulova et al., 2017). Comparing to Cr. thermophila, the size of choanomastigotes and promastigotes with flagellum of CLA-KP1 was slightly larger than that of Cr. thermophila (Ishemgulova et al., 2017) supporting that they are different species.
The ability to survive at human body temperature and multiply in mammalian macrophage cells is a hallmark of dixenous parasites such as Leishmania but these features are not common in monoxenous trypanosomatids. However, certain monoxenous trypanosomatids have been reported in warm-blooded animals including humans. Cr. mellificae can infect not only insects but also various mammals (Dario et al., 2021). In addition, strains of Crithidia sp. related to Cr. fasciculata have been isolated from lesions in an immunocompetent patient with no underlying diseases (Maruyama et al., 2019) or with leishmaniasis (Ghobakhloo et al., 2019; Kostygov et al., 2019; Rogerio et al., 2023). Moreover, thermotolerance to high temperatures has been demonstrated in vitro in Cr. thermophila (34°C) (Ishemgulova et al., 2017), Cr. mellificae (36–37°C) (Dario et al., 2021), and a Crithidia sp. related to Cr. fasciculata (37°C) (Ghobakhloo et al., 2019). In our study, Crithidia sp. CLA-KP1 was able to grow and survive at 37°C in culture for at least 7 days and the density of the cells at day 7 was still high (1.0 × 106 cells/mL), although the average parasite density at 26°C was significantly greater than 37°C after 5 days of culture. In addition, Crithidia sp. CLA-KP1 could infect and persist appear in the PEMs for up to 48 h. The results suggested that Crithidia sp. CLA-KP1 is a thermotolerant monoxenous trypanosomatid that could live in a wide range of temperature, 26–37°C.
Ghobakhloo and colleagues (Ghobakhloo et al., 2019) have demonstrated that clinical isolates of Crithidia sp. related to Cr. fasciculata are able to infect THP-1 and J774 cells and transform back to promastigotes in culture media but the percentage of infection is much less than that of the clinical isolate of L. major (Ghobakhloo et al., 2019). Another opportunistic trypanosomatid that co-infects in leishmaniasis patients is Le. seymouri. The Le. seymouri parasites isolated from clinical samples can grow well at 35°C but are unable to infect mammalian macrophages in vitro either alone or in co-infection with Leishmania parasites (Kraeva et al., 2015). Although Crithidia sp. CLA-KP1 was isolated from insects, further investigation of in vitro infection using other cell lines such as THP-1, J774, and BMMɸ cells should be performed. Comparative analyses of genomic and transcriptomic profiles of these thermotolerant monoxenous trypanosomatids and dixenous parasites would throw light on the adaptations of trypanosomatids to elevated temperature and tracking the evolution of parasitism of dixenous trypanosomatids.
In conclusion, here we provide microscopic analyses together with molecular identification of naturally occurring infections of L. martiniquensis in C. peregrinus for the first time. The findings strongly support this biting midge species as a potential vector of L. martiniquensis. However, further isolation and characterization of parasites from more biting midges, including other species, are required to search for more natural infections of both L. martiniquensis and L. orientalis. Also, further investigation of the anthropophilic and/or zoophilic behaviors of the biting midge, C. peregrinus, is needed to investigate reservoir hosts of autochthonous leishmaniasis in Thailand. Finally, for definitive vector incrimination transmission by bite needs to be demonstrated. Four strains of Crithidia sp. CLA were isolated from C. peregrinus and characterized for the first time. The CLA-KP1 strain could infect PEMs but they could not multiply suggesting that it was a thermotolerant monoxenous trypanosomatid capable of surviving in a wide range of temperatures (26–37°C). Analysis of genome sequences of all strains of Crithidia sp. CLA will be performed to compare the evolutionary relationship between this newly isolated trypanosomatid and other related parasites. A description of the detailed molecular characterization and taxonomic assignment will be provided in the future.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
The use of animals in this study was approved by the animal research ethics committee of Chulalongkorn University Animal Care and Use Protocol (CU-ACUP), Faculty of Medicine, Chulalongkorn University, Bangkok, Thailand (COA No. 023/2564 and COA No. 011/2564). The study was conducted in accordance with the local legislation and institutional requirements.
NJ and PS: conceptualization, methodology, and formal analysis. SK, NJ, CM, TP, RA, TY, UP, and AJ: investigation. SK, PS, and NJ: visualization. NJ, SK, TP, and CM: writing – original draft preparation. NJ and PB: writing-review and editing. NJ: funding acquisition. All authors have read and agreed to the published version of the manuscript.
This work was supported by the 90th Anniversary of Chulalongkorn University Fund (Ratchadaphiseksomphot Endowment Fund) (grant number GCUGR1125651016M).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1235254/full#supplementary-material
Anugulruengkitt, S., Songtaweesin, W. N., Thepnarong, N., Tangthanapalakul, A., Sitthisan, M., Chatproedprai, S., et al. (2022). Case report: simple nodular cutaneous leishmaniasis caused by autochthonous Leishmania (Mundinia) orientalis in an 18-month-old girl: the first pediatric case in Thailand and literature review. Am. J. Trop. Med. Hyg. 108, 44–50. doi: 10.4269/ajtmh.22-0385
Barratt, J., Kaufer, A., Peters, B., Craig, D., Lawrence, A., Roberts, T., et al. (2017). Isolation of novel Trypanosomatid, Zelonia australiensis sp. nov. (Kinetoplastida: Trypanosomatidae) provides support for a gondwanan origin of dixenous parasitism in the Leishmaniinae. PLoS Negl. Trop. Dis. 11:e0005215. doi: 10.1371/journal.pntd.0005215
Becvar, T., Vojtkova, B., Siriyasatien, P., Votypka, J., Modry, D., Jahn, P., et al. (2021). Experimental transmission of Leishmania (Mundinia) parasites by biting midges (Diptera: Ceratopogonidae). PLoS Pathog. 17:e1009654. doi: 10.1371/journal.ppat.1009654
Bernotienė, R., Iezhova, T. A., Bukauskaitė, D., Chagas, C. R. F., Kazak, M., and Valkiūnas, G. (2020). Development of Trypanosoma everetti in Culicoides biting midges. Acta Trop. 210:105555. doi: 10.1016/j.actatropica.2020.105555
Cecílio, P., Cordeiro-da-Silva, A., and Oliveira, F. (2022). Sand flies: basic information on the vectors of leishmaniasis and their interactions with Leishmania parasites. Commun. Biol. 5:305. doi: 10.1038/s42003-022-03240-z
Chanmol, W., Jariyapan, N., Somboon, P., Bates, M. D., and Bates, P. A. (2019). Development of Leishmania orientalis in the sand fly Lutzomyia longipalpis (Diptera: Psychodidae) and the biting midge Culicoides soronensis (Diptera: Ceratopogonidae). Acta Trop. 199:105157. doi: 10.1016/j.actatropica.2019.105157
Chusri, S., Thammapalo, S., Chusri, S., Thammapalo, S., Silpapojakul, K., and Siriyasatien, P. (2014). Animal reservoirs and potential vectors of Leishmania siamensis in southern Thailand. Southeast Asian J. Trop. Med. Public Health. 45, 13–19.
Dario, M. A., Lisboa, C. V., Silva, M. V., Herrera, H. M., Rocha, F. L., Furtado, M. C., et al. (2021). Crithidia mellificae infection in different mammalian species in Brazil. Int. J. Parasitol. Parasites Wildl. 15, 58–69. doi: 10.1016/j.ijppaw.2021.04.003
Dougall, A. M., Alexander, B., Holt, D. C., Harris, T., Sultan, A. H., Bates, P. A., et al. (2011). Evidence incriminating midges (Diptera: Ceratopogonidae) as potential vectors of Leishmania in Australia. Int. J. Parasitol. 41, 571–579. doi: 10.1016/j.ijpara.2010.12.008
Dyce, A. L. (1996). Culicoides paragarciai, a new Ornatus group species from Papua New Guinea and the Solomon Islands (Diptera: Ceratopogonidae). Aust. J. Entomol. 35, 313–318. doi: 10.1111/j.1440-6055.1996.tb01410.x
Dyce, A.L., Bellis, G.A., and Muller, M.J. (2007). Pictorial atlas of Australasian Culicoides wings (Diptera: Ceratopogonidae). Canberra, Australia: Australian Biological Resources Study
Falcão de Oliveira, E., Oshiro, E. T., Fernandes, W. S., Murat, P. G., de Medeiros, M. J., Souza, A. I., et al. (2017). Experimental infection and transmission of Leishmania by Lutzomyia cruzi (Diptera: Psychodidae): aspects of the ecology of parasite-vector interactions. PLoS Negl. Trop. Dis. 11:e0005401. doi: 10.1371/journal.pntd.0005401
Ferreira, T. R., and Sacks, D. L. (2022). Experimental hybridization in Leishmania: tools for the study of genetic exchange. Pathogens 11:580. doi: 10.3390/pathogens11050580
Filosa, J. N., Berry, C. T., Ruthel, G., Beverley, S. M., Warren, W. C., Tomlinson, C., et al. (2019). Dramatic changes in gene expression in different forms of Crithidia fasciculata reveal potential mechanisms for insect-specific adhesion in kinetoplastid parasites. PLoS Negl. Trop. Dis. 13:e0007570. doi: 10.1371/journal.pntd.0007570
Folmer, O., Black, M., Hoeh, W., Lutz, R., and Vrijenhoek, R. (1994). DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 3, 294–299.
Frolov, A. O., Kostygov, A. Y., and Yurchenko, V. (2021). Development of monoxenous Trypanosomatids and Phytomonads in insects. Trends Parasitol. 37, 538–551. doi: 10.1016/j.pt.2021.02.004
Ghobakhloo, N., Motazedian, M. H., Naderi, S., and Ebrahimi, S. (2019). Isolation of Crithidia spp. from lesions of immunocompetent patients with suspected cutaneous leishmaniasis in Iran. Tropical Med. Int. Health 24, 116–126. doi: 10.1111/tmi.13042
Ghosh, S., Banerjee, P., Sarkar, A., Datta, S., and Chatterjee, M. (2012). Coinfection of Leptomonas seymouri and Leishmania donovani in Indian leishmaniasis. J. Clin. Microbiol. 50, 2774–2778. doi: 10.1128/JCM.00966-12
Harrup, L. E., Laban, S., Purse, B. V., Reddy, Y. K., Reddy, Y. N., Byregowda, S. M., et al. (2016). DNA barcoding and surveillance sampling strategies for Culicoides biting midges (Diptera: Ceratopogonidae) in southern India. Parasit. Vectors 9:461. doi: 10.1186/s13071-016-1722-z
Hillis, D. M., and Bull, J. J. (1993). An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst. Biol. 42, 182–192. doi: 10.1093/sysbio/42.2.182
Inbar, E., Akopyants, N. S., Charmoy, M., Romano, A., Lawyer, P., Elnaiem, D. E., et al. (2013). The mating competence of geographically diverse Leishmania major strains in their natural and unnatural sand fly vectors. PLoS Genet. 9:e1003672. doi: 10.1371/journal.pgen.1003672
Ishemgulova, A., Butenko, A., Kortišová, L., Boucinha, C., Grybchuk-Ieremenko, A., Morelli, K. A., et al. (2017). Molecular mechanisms of thermal resistance of the insect trypanosomatid Crithidia thermophila. PLoS One 12:e0174165. doi: 10.1371/journal.pone.0174165
Jariyapan, N., Bates, M. D., and Bates, P. A. (2021). Molecular identification of two newly identified human pathogens causing leishmaniasis using PCR-based methods on the 3′ untranslated region of the heat shock protein 70 (type I) gene. PLoS Negl. Trop. Dis. 15:e0009982. doi: 10.1371/journal.pntd.0009982
Jariyapan, N., Daroontum, T., Jaiwong, K., Chanmol, W., Intakhan, N., Sor-Suwan, S., et al. (2018). Leishmania (Mundinia) orientalis n. sp. (Trypanosomatidae), a parasite from Thailand responsible for localised cutaneous leishmaniasis. Parasit. Vectors 11:351. doi: 10.1186/s13071-018-2908-3
Jomkumsing, P., Surapinit, A., Saengpara, T., and Pramual, P. (2021). Genetic variation, DNA barcoding and blood meal identification of Culicoides Latreille biting midges (Diptera: Ceratopogonidae) in Thailand. Acta Trop. 217:105866. doi: 10.1016/j.actatropica
Kar, S., Mondal, B., Ghosh, J., Mazumdar, S. M., and Mazumdar, A. (2022). Host preference of bluetongue virus vectors, Culicoides species associated with livestock in West Bengal, India: potential relevance on bluetongue epidemiology. Acta Trop. 235:106648. doi: 10.1016/j.actatropica.2022.106648
Kasičová, Z., Schreiberová, A., Kimáková, A., and Kočišová, A. (2021). Blood meal analysis: host-feeding patterns of biting midges (Diptera, Ceratopogonidae, Culicoides Latreille) in Slovakia. Parasite 28:58. doi: 10.1051/parasite/2021058
Killick-Kendrick, R. (1999). The biology and control of phlebotomine sand flies. Clin. Dermatol. 17, 279–289. doi: 10.1016/s0738-081x(99)00046-2
Kostygov, A. Y., Butenko, A., and Yurchenko, V. (2019). On monoxenous trypanosomatids from lesions of immunocompetent patients with suspected cutaneous leishmaniasis in Iran. Tropical Med. Int. Health 24, 127–128. doi: 10.1111/tmi.13168
Kraeva, N., Butenko, A., Hlaváčová, J., Kostygov, A., Myškova, J., Grybchuk, D., et al. (2015). Leptomonas seymouri: adaptations to the dixenous life cycle analyzed by genome sequencing, transcriptome profiling and co-infection with Leishmania donovani. PLoS Pathog. 11:e1005127. doi: 10.1371/journal.ppat.1005127
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Kwakye-Nuako, G., Mosore, M. T., Boakye, D., and Bates, P. A. (2023). Description, biology, and medical significance of Lishmania (Mundinia) chancei n. sp. (Kinetoplastea: Trypanosomatidae) from Ghana and Leishmania (Mundinia) procaviensis N. Sp. (Kinetoplastea: Trypanosomatidae) from Namibia. J. Parasitol. 109, 43–50. doi: 10.1645/22-53
Lawyer, P. G., and Perkins, P. V. (2000). “Leishmaniasis and trypanosomiasis” in Medical Entomology. eds. B. F. Eldridge and J. D. Edman (Dordrecht: Springer)
Lukeš, J., and Votýpka, J. (2020). Field isolation and cultivation of Trypanosomatids from insects. Methods Mol. Biol. 2116, 3–21. doi: 10.1007/978-1-0716-0294-2_1
Mano, C., Kongkaew, A., Tippawangkosol, P., Somboon, P., Roytrakul, S., Pescher, P., et al. (2023). Amphotericin B resistance correlates with increased fitness in vitro and in vivo in Leishmania (Mundinia) martiniquensis. Front. Microbiol. 14:1156061. doi: 10.3389/fmicb.2023.1156061
Maruyama, S. R., de Santana, A. K. M., Takamiya, N. T., Takahashi, T. Y., Rogerio, L. A., Oliveira, C. A. B., et al. (2019). Non-leishmania parasite in fatal visceral Leishmaniasis-like disease. Brazil. Emerg. Infect. Dis. 25, 2088–2092. doi: 10.3201/eid2511.181548
Maslov, D. A., Lukes, J., Jirku, M., and Simpson, L. (1996). Phylogeny of trypanosomes as inferred from the small and large subunit rRNAs: implications for the evolution of parasitism in the trypanosomatid protozoa. Mol. Biochem. Parasitol. 75, 197–205. doi: 10.1016/0166-6851(95)02526-x
Mellor, P. S., Boorman, J., and Baylis, M. (2000). Culicoides biting midges: their role as arbovirus vectors. Annu. Rev. Entomol. 45, 307–340. doi: 10.1146/annurev.ento.45.1.307
Morsy, T. A., Schnur, L. F., Feinsod, F. M., Michael, S. A., Saah, A., Salama, M. M., et al. (1988). The discovery and preliminary characterization of a novel trypanosomatid parasite from Rattus norvegicus and stray dogs from Alexandria, Egypt. Ann. Trop. Med. Parasitol. 82, 437–444. doi: 10.1080/00034983.1988.11812273
Mullen, G. R. (2009). Biting midges (Ceratopogonidae). Medical and veterinary entomology. 2nd Edn. ed. G. R. Mullen and L. A. Durden (New York, NY: Academic Press), 169–188.
Noyes, H. A., Stevens, J. R., Teixeira, M., Phelan, J., and Holz, P. (1999). A nested PCR for the ssrRNA gene detects Trypanosoma binneyi in the platypus and Trypanosoma sp. in wombats and kangaroos in Australia. Int. J. Parasitol. 29, 331–339. doi: 10.1016/s0020-7519(98)00167-2
Podlipaev, S. A., Sturm, N. R., Fiala, I., Fernandes, O., Westenberger, S. J., Dollet, M., et al. (2004). Diversity of insect trypanosomatids assessed from the spliced leader RNA and 5S rRNA genes and intergenic regions. J. Eukaryot. Microbiol. 51, 283–290. doi: 10.1111/j.1550-7408.2004.tb00568.x
Podlipaev, S., Votýpka, J., Jirků, M., Svobodová, M., and Lukes, J. (2004). Herpetomonas ztiplika n. sp. (Kinetoplastida: Trypanosomatidae): a parasite of the blood-sucking biting midge Culicoides kibunensis Tokunaga, 1937 (Diptera: Ceratopogonidae). J. Parasitol. 90, 342–347. doi: 10.1645/GE-156R.
Posada, D. (2008). jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 25, 1253–1256. doi: 10.1093/molbev/msn083
Pothirat, T., Tantiworawit, A., Chaiwarith, R., Jariyapan, N., Wannasan, A., Siriyasatien, P., et al. (2014). First isolation of Leishmania from northern Thailand: case report, identification as Leishmania martiniquensis and phylogenetic position within the Leishmania enriettii complex. PLoS Negl. Trop. Dis. 8:e3339. doi: 10.1371/journal.pntd.0003339
Pramual, P., Jomkumsing, P., Piraonapicha, K., and Jumpato, W. (2021). Integrative taxonomy uncovers a new Culicoides (Diptera: Ceratopogonidae) biting midge species from Thailand. Acta Trop. 220:105941. doi: 10.1016/j.actatropica.2021.105941
Radrova, J., Seblova, V., and Votypka, J. (2013). Feeding behavior and spatial distribution of Culex mosquitoes (Diptera: Culicidae) in wetland areas of the Czech Republic. J. Med. Entomol. 50, 1097–1104. doi: 10.1603/me13029
Rangel, D. A., Lisboa, C. V., Novaes, R. L. M., Silva, B. A., Souza, R. F., Jansen, A. M., et al. (2019). Isolation and characterization of trypanosomatids, including Crithidia mellificae, in bats from the Atlantic Forest of Rio de Janeiro. Brazil. PLoS Negl. Trop. Dis. 13:e0007527. doi: 10.1371/journal.pntd.0007527
Ready, P. D. (2013). Biology of phlebotomine sand flies as vectors of disease agents. Annu. Rev. Entomol. 58, 227–250. doi: 10.1146/annurev-ento-120811-153557
Rebêlo, J. M., Rodrigues, B. L., Bandeira, M. D., Moraes, J. L., Fonteles, R. S., and Pereira, S. R. (2016). Detection of Leishmania amazonensis and Leishmania braziliensis in Culicoides (Diptera, Ceratopogonidae) in an endemic area of cutaneous leishmaniasis in the Brazilian Amazonia. J. Vector Ecol. 41, 303–308. doi: 10.1111/jvec.12227
Requena, J. M., Chicharro, C., García, L., Parrado, R., Puerta, C. J., and Cañavate, C. (2012). Sequence analysis of the 3′-untranslated region of HSP70 (type I) genes in the genus Leishmania: its usefulness as a molecular marker for species identification. Parasit. Vectors 5:87. doi: 10.1186/1756-3305-5-87
Rogerio, L. A., Takahashi, T. Y., Cardoso, L., Takamiya, N. T., de Melo, E. V., de Jesus, A. R., et al. (2023). Co-infection of Leishmania infantum and a Crithidia-related species in a case of refractory relapsed visceral leishmaniasis with non-ulcerated cutaneous manifestation in Brazil. Int. J. Infect. Dis. 133, 85–88. doi: 10.1016/j.ijid.2023.05.012
Sadlova, J., Yeo, M., Seblova, V., Lewis, M. D., Mauricio, I., Volf, P., et al. (2011). Visualisation of Leishmania donovani fluorescent hybrids during early stage development in the sand fly vector. PLoS One 6:e19851. doi: 10.1371/journal.pone.0019851
Schwarz, R. S., Bauchan, G. R., Murphy, C. A., Ravoet, J., de Graaf, D. C., and Evans, J. D. (2015). Characterization of two species of Trypanosomatidae from the honey bee Apis mellifera: Crithidia mellificae Langridge and McGhee, and Lotmaria passim n. gen., n. sp. J. Eukaryot. Microbiol. 62, 567–583. doi: 10.1111/jeu.12209
Seblova, V., Sadlova, J., Carpenter, S., and Volf, P. (2012). Development of Leishmania parasites in Culicoides nubeculosus (Diptera: Ceratopogonidae) and implications for screening vector competence. J. Med. Entomol. 49, 967–970. doi: 10.1603/me12053
Singh, N., Chikara, S., and Sundar, S. (2013). SOLiD™ sequencing of genomes of clinical isolates of Leishmania donovani from India confirm leptomonas co-infection and raise some key questions. PLoS One 8:e55738. doi: 10.1371/journal.pone.0055738
Siripattanapipong, S., Leelayoova, S., Ninsaeng, U., and Mungthin, M. (2018). Detection of DNA of Leishmania siamensis in Sergentomyia (Neophlebotomus) iyengari (Diptera: Psychodidae) and molecular identification of blood meals of sand flies in an affected area, Southern Thailand. J Med Entomol. 55, 1277–1283. doi: 10.1093/jme/tjy069
Slama, D., Haouas, N., Remadi, L., Mezhoud, H., Babba, H., and Chaker, E. (2014). First detection of Leishmania infantum (Kinetoplastida: Trypanosomatidae) in Culicoides spp. (Diptera: Ceratopogonidae). Parasit. Vectors 7:51. doi: 10.1186/1756-3305-7-51
Songumpai, N., Promrangsee, C., Noopetch, P., Siriyasatien, P., and Preativatanyou, K. (2022). First evidence of co-circulation of emerging Leishmania martiniquensis, Leishmania orientalis, and Crithidia sp. in Culicoides biting midges (Diptera: Ceratopogonidae), the putative vectors for autochthonous transmission in southern Thailand. Trop. Med. Infect. Dis. 7:379. doi: 10.3390/tropicalmed7110379
Spanakos, G., Piperaki, E. T., Menounos, P. G., Tegos, N., Flemetakis, A., and Vakalis, N. C. (2008). Detection and species identification of Old World Leishmania in clinical samples using a PCR-based method. Trans. R. Soc. Trop. Med. Hyg. 102, 46–53. doi: 10.1016/j.trstmh.2007.05.019
Srisuton, P., Phumee, A., Sunantaraporn, S., Boonserm, R., Sor-Suwan, S., Brownell, N., et al. (2019). Detection of Leishmania and Trypanosoma DNA in field-caught sand flies from endemic and non-endemic areas of leishmaniasis in southern Thailand. Insects 10:238. doi: 10.3390/insects10080238
Srivarasat, S., Brownell, N., Siriyasatien, P., Noppakun, N., Asawanonda, P., Rattanakorn, K., et al. (2022). Case report: autochthonous disseminated cutaneous, mucocutaneous, and visceral leishmaniasis caused by Leishmania martiniquensis in a patient with HIV/AIDS from northern Thailand and literature review. Am J Trop Med Hyg. 107, 1196–1202. doi: 10.4269/ajtmh.22-0108
Srivastava, P., Prajapati, V. K., Vanaerschot, M., Van der Auwera, G., Dujardin, J. C., and Sundar, S. (2010). Detection of Leptomonas sp. parasites in clinical isolates of kala-azar patients from India. Infect. Genet. Evol. 10, 1145–1150. doi: 10.1016/j.meegid.2010.07.009
Sriwongpan, P., Nedsuwan, S., Manomat, J., Charoensakulchai, S., Lacharojana, K., Sankwan, J., et al. (2021). Prevalence and associated risk factors of Leishmania infection among immunocompetent hosts, a community-based study in Chiang Rai, Thailand. PLoS Negl. Trop. Dis. 15:e0009545. doi: 10.1371/journal.pntd.0009545
Sunantaraporn, S., Hortiwakul, T., Kraivichian, K., Siriyasatien, P., and Brownell, N. (2022). Molecular identification of host blood meals and detection of blood parasites in Culicoides Latreille (Diptera: Ceratopogonidae) collected from Phatthalung province, Southern Thailand. Insects 13:912. doi: 10.3390/insects13100912
Sunantaraporn, S., Thepparat, A., Phumee, A., Sor-Suwan, S., Boonserm, R., Bellis, G., et al. (2021). Culicoides Latreille (Diptera: Ceratopogonidae) as potential vectors for Leishmania martiniquensis and Trypanosoma sp. in northern Thailand. PLoS Negl. Trop. Dis. 15:e0010014. doi: 10.1371/journal.pntd.0010014
Svobodová, M., Dolnik, O. V., Čepička, I., and Rádrová, J. (2017). Biting midges (Ceratopogonidae) as vectors of avian trypanosomes. Parasit. Vectors 10:224. doi: 10.1186/s13071-017-2158-9
Svobodová, M., Zídková, L., Čepička, I., Oborník, M., Lukeš, J., and Votýpka, J. (2007). Sergeia podlipaevi gen. nov., sp. nov. (Trypanosomatidae, Kinetoplastida), a parasite of biting midges (Ceratopogonidae, Diptera). Int. J. Syst. Evol. Microbiol. 57, 423–432. doi: 10.1099/ijs.0.64557-0
Thakur, L., Kushwaha, H. R., Negi, A., Jain, A., and Jain, M. (2020). Leptomonas seymouri co-infection in cutaneous leishmaniasis cases caused by Leishmania donovani from Himachal Pradesh, India. Front. Cell Infect Microbiol. 10:345. doi: 10.3389/fcimb.2020.00345
World Health Organization. (2023) Fact sheets: Leishmaniasis. Available at: https://www.who.int/news-room/fact-sheets/detail/leishmaniasis (Accessed June 1, 2023)
Zhang, X., Goncalves, R., and Mosser, D. M. (2008). The isolation and characterization of murine macrophages. Curr. Protoc. Immunol. 14, 14.1.1–14.1.14. doi: 10.1002/0471142735.im1401s83
Keywords: Leishmania, leishmaniasis, Crithidia, Trypanosomatids, Leishmania martiniquensis, Culicoides, Culicoides peregrinus, biting midges
Citation: Kaewmee S, Mano C, Phanitchakun T, Ampol R, Yasanga T, Pattanawong U, Junkum A, Siriyasatien P, Bates PA and Jariyapan N (2023) Natural infection with Leishmania (Mundinia) martiniquensis supports Culicoides peregrinus (Diptera: Ceratopogonidae) as a potential vector of leishmaniasis and characterization of a Crithidia sp. isolated from the midges. Front. Microbiol. 14:1235254. doi: 10.3389/fmicb.2023.1235254
Received: 06 June 2023; Accepted: 08 August 2023;
Published: 22 August 2023.
Edited by:
Marcos Rogério André, São Paulo State University, BrazilReviewed by:
Matthew Edward Rogers, University of London, United KingdomCopyright © 2023 Kaewmee, Mano, Phanitchakun, Ampol, Yasanga, Pattanawong, Junkum, Siriyasatien, Bates and Jariyapan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Narissara Jariyapan, bmFyaXNzYXJhLmpAY2h1bGEuYWMudGg=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.