
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 07 September 2023
Sec. Microbe and Virus Interactions with Plants
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1233555
This article is part of the Research Topic Plants surviving in extreme environment: Harnessing Soil-Plant–Microbial relationship to enhance crop health and productivity View all 21 articles
 Yanhong Wang1†
Yanhong Wang1† Jiao Xu1†
Jiao Xu1† Qingsong Yuan1
Qingsong Yuan1 Lanping Guo2
Lanping Guo2 Chenghong Xiao1
Chenghong Xiao1 Changgui Yang1
Changgui Yang1 Liangyuan Li1
Liangyuan Li1 Weike Jiang1
Weike Jiang1 Tao Zhou1*
Tao Zhou1*Armillaria members play important roles in the nutrient supply and growth modulation of Gastrodia elata Bl., and they will undergo severe competition with native soil organisms before colonization and become symbiotic with G. elata. Unraveling the response of soil microbial organisms to symbiotic fungi will open up new avenues to illustrate the biological mechanisms driving G. elata’s benefit from Armillaria. For this purpose, Armillaria strains from four main G. elata production areas in China were collected, identified, and co-planted with G. elata in Guizhou Province. The result of the phylogenetic tree indicated that the four Armillaria strains shared the shortest clade with Armillaria gallica. The yields of G. elata were compared to uncover the potential role of these A. gallica strains. Soil microbial DNA was extracted and sequenced using Illumina sequencing of 16S and ITS rRNA gene amplicons to decipher the changes of soil bacterial and fungal communities arising from A. gallica strains. The yield of G. elata symbiosis with the YN strain (A. gallica collected from Yunnan) was four times higher than that of the GZ strain (A. gallica collected from Guizhou) and nearly two times higher than that of the AH and SX strains (A. gallica collected from Shanxi and Anhui). We found that the GZ strain induced changes in the bacterial community, while the YN strain mainly caused changes in the fungal community. Similar patterns were identified in non-metric multidimensional scaling analysis, in which the GZ strain greatly separated from others in bacterial structure, while the YN strain caused significant separation from other strains in fungal structure. This current study revealed the assembly and response of the soil microbial community to A. gallica strains and suggested that exotic strains of A. gallica might be helpful in improving the yield of G. elata by inducing changes in the soil fungal community.
Mycorrhizal symbiosis is found in over 80% of extant land plant species (Wang et al., 2017; Feijen et al., 2018). Orchidaceae showing mycorrhizal association with fungi are essential for seed germination and plant growth (Currah et al., 1997; Brundrett, 2002). It is believed that the ability to recruit saprotrophic lineages of wood and litter-inhibiting fungi is considered instrumental for plant adaptation to various ecological challenges, and many symbiotic fungi have coevolved with plants and showed a high degree of host specificity (Raaijmakers et al., 2009; Crowther et al., 2012). A well-known and highly specific alliance example is Gastrodia elata Bl. and Armillaria (Yuan et al., 2018; Camprubi et al., 2020; Xu et al., 2021). G. elata, also called Tianma in Chinese, is a perennial herb in the Orchidaceae family and one of the most valuable traditional Chinese medicines. Being one of the heterotrophic plants without chlorophyll, the growth and development of G. elata require symbiosis with Armillaria for nutrient supply since the protocorm stage (Yuan et al., 2018). The Armillaria members impose a significant impact on the yield and quality of G. elata; therefore, the isolation and identification of excellent Armillaria strains are of great significance (Yu et al., 2022).
Members of Armillaria generally form linear mycelial organs involved in wood decay and nutrient absorption. Evidence indicates that rhizomorphs of wood-decay species can absorb inorganic nutrients from the external environment (Clipson et al., 1987; Cairney et al., 1988), and the melanized layer of rhizomorphs generally contains calcium, which can provide chemical defense and natural controls (Porter et al., 2022). However, recent studies observed increased diversity of bacteria and fungi during the juvenile tuber to mature tuber periods of G. elata, which implied that Armillaria mellea can affect the structure of the microbial community associated with G. elata and greatly reduce the antifungal and antibacterial activities as a symbiotic association with its host is established (Yuan et al., 2018).
Soil contains large amounts of microbial biomass, and its microbial communities are highly diverse (Torsvik and Vres, 2002; Fierer, 2017). Diversity, structure, and composition of the soil microbial community are central issues in agricultural management, as this information is crucial for understanding and predicting the role played by organisms in maintaining plant health and production (Berendsen et al., 2012). Biotic factors, such as plant cultivars, strongly influence the composition of the soil rhizosphere microbiota and control the assembly of root-associated microbiotas (Tkacz et al., 2015; Zhou and Fong, 2021). Normally, plant cultivars impose an effect on the rhizosphere microbiota by investing a huge amount of root exudates and providing sources of carbonaceous compounds (Bulgarelli et al., 2013). Symbiotic fungi also have the ability to influence soil microbes; however, unlike plants, secretions of fungi through mycelia or rhizomorphs might inhibit the growth of others (Collins et al., 2013). Studies of changes in microbial community composition in response to symbiotic fungi can foster a better understanding of Armillaria utilization in the future.
The geographical areas of G. elata cultivation are mainly spread over the Ta-pieh Mountains, Qinling-Daba Mountains, and Yunnan-Kweichow Plateau in China. These sites are typically among the regions with sufficient rainfall and variable air temperatures in the summer. As global climate change continues and frequent commercial trade occurs among planting regions, it is incredibly significant to choose alternative Armillaria strains that can adapt to the diverse cultivation conditions in the process of G. elata artificial cultivation. However, a knowledge gap exists regarding to what extent G. elata will benefit from and how soil microbial communities vary with different Armillaria strains. For this purpose, four accessions of Armillaria strains were collected around China and tested in Guizhou Province. The genetic relationship of Armillaria strains was identified, and the yields of G. elata and soil microbial community dynamic were compared. Findings from this current study might be helpful in optimizing our strategies to utilize symbiotic fungi and Armillaria strains.
In this study, rhizomorphs of Armillaria were collected from three wild conditions in Lu’an (115°22′E, 31°06′N) Anhui Province, Xiaocaoba (103°51′E, 27°16′N) Yunnan Province, and Bijie (105°00′E, 27°14′N) Guizhou Province, and one commercial Armillaria strain was from Ningqiang (106°30′E, 32°37′N) Shanxi Province (Supplementary Figure S1). All these provinces are the main producing regions of G. elata. First, all the rhizomorphs of Armillaria were rinsed with running water and soaked with 75% ethanol for 45 s, followed by 1% NaOCl for 6 min. Then, the rhizomorphs were rinsed with sterilized water 3–4 times and dried with microbe-free filter paper. Finally, the cortices of rhizomorphs were removed with a sterilized knife, and the mycelia were lightly pulled out. Equal sizes and sections of mycelia for each Armillaria were inoculated in potato dextrose agar (PDA) solid medium and cultured for 20 days. In total, four Armillaria isolates, in accordance with collection sites, were obtained in the laboratory and named as follows: AH strain, SX strain, YN strain, and GZ stain.
The DNA of Armillaria isolates was extracted using the first generation with the Ezup Column Fungi Genomic DNA Purification Kit (Sangon Biotech, Shanghai, China). Then, the intergenic spacer (IGS) regions of ribosomal RNA genes were amplified with primers CNL12 (5’-CTGAACGCCTCTAAGTCAG-3′) and 5SA (5’-CAGAGTCCTATGGCCGTGGAT-3′). The amplicons were subjected to Sanger sequencing, and gene sequence comparisons were done with the basic local alignment search (BLAST) homology comparison.1 The purified Armillaria strains were kept on medium for one generation, then the second generation was used as the primary strain and enlarged by cultivating in sterilized packets with oat bran and cornstalk as nutrient sources. The conditions of Armillaria strain packets used for G. elata artificial cultivation were presented with graphs in Supplementary Figure S2.
The site of G. elata artificial cultivation was located at Gonglongping National Forest Park in Qixingguan district, Bijie, Guizhou Province, China (105°00′38′′E, 27°14′43′′N), 1781 m above sea level. The mean annual temperature and precipitation in this area are 12.8°C and 998 mm, respectively. The soil is classified as brown loam (FAO-UNESCO, 1974).
The trial was performed at a single field site that was newly explored to ensure a uniform growth condition among repetitions. The cultivation of G. elata was carried out according to the WHO guidelines on good agricultural and collection practices (GACP) for medicinal plants.2 In the beginning, holes with a size of 80 cm × 50 cm × 30–50 cm (length × width × depth) were dug in the field. Then, 5–8 pieces of chestnut wood (Ф = 5–10 cm, length = 30 cm) that were fully infected with Armillaria strains were neatly laid out on the bottom of each hole. Subsequently, 0.1 kg of juvenile tubers of G. elata were put evenly on the chestnut wood and crushed with twigs and leaves that were infected with Armillaria strains. Finally, the soil was filled back into the holes and covered with straw or leaves (Yuan et al., 2020). The chestnut wood, twigs, and leaves that were infected with Armillaria strains were prepared ahead of cultivation, and the strains used for the chestnut wood, twig, and leaf infection were in a one-to-one relationship. There are four strains of Armillaria used in this trial in total, namely, AH strain, SX strain, YN strain, and GZ strain; each of them was replicated in three.
The cultivation was processed in March 2021, and the sampling was conducted in November 2021. When sampling, the twigs and leaves were removed gently without disturbing the soil covered by G. elata. A tensiometer was used to check the tensile strength of mycelia. The conditions of Armillaria mycelia at harvest time can be found in Supplementary Figure S3. At first, Armillaria strains tightly connected with chestnut wood and G. elata were picked out; subsequently, a section of Armillaria rhizomorph was hung on the hook of the tensiometer; then, the other side of the rhizomorph was pulled until it broke; and the real-time data on the tensiometer were recorded. Eight mycelia within one hole were randomly selected for statistical analysis. Soils around mature G. elata tubers were collected. Briefly, the soils loosely attached to G. elata were carefully collected into a 50-ml centrifuge tube with 20 mL of sterile water. After vortex mixing at 200 rpm for 20 min and centrifuging at 11,000 rpm for 10 min, the deposit was maintained at −80°C for subsequent soil microbial DNA extraction. Finally, all fresh tubes including mature and juvenile tubes of G. elata within one hole were collected and weighed, and the data were recorded.
Soil microbial DNA was extracted using the E.Z.N.A.® soil DNA Kit (Omega Bio-tek, Norcross, GA, United States). The 16S rRNA gene was amplified with primers (F: ACTCCTACGGG AGGCAGCA, R: GGACTACHVGGGTWTCT AAT), and the ITS rRNA gene was amplified with primers (F: CTTGGTCATTTA GAGGAAGTAA, R: GCTGCGTTCTTCATCGATGC) with a 12 nt unique barcode at the 5′ end. The PCR reactions were performed in triplicate in 20-μl mixtures, containing 1 μL of 10-fold diluted DNA, 10 μL of SYBR® Premix Ex Taq (Tli RNase H Plus, 2×, Takara Bio, Japan), 1 μL of forward primer and 1 μL of reverse primer at 10 mM, and 7 μL of Milli-Q water. The PCR products were detected by electrophoresis in a 2% agarose gel, and then purified using the agarose gel DNA purification kit (Takara, Dalian, China). Furthermore, equal amounts of purified amplicons were pooled in equimolar amounts and paired-end sequenced by the Illumina NovaSeq platform at Biomarker Technologies Co., Ltd. (Beijing, China).
Raw reads were quality-filtered using Trimmomatic (v0.33) and then clean reads were obtained with Cutadapt (1.9.1) by identifying and removing the primer sequences. Usearch3 was used to merge the paired-end reads into a tag from high-quality clean reads. The operational taxonomic units (OTUs) were defined at the 97% similarity level by Usearch (Edgar, 2013) with the remaining unique sequences. At 70% of the confidence threshold, the taxonomic identity of phylotypes was classified using the Ribosomal Database Project RDP Classifier4, the 16S rRNA gene sequence was analyzed with the Silva (SSU123) 16S rRNA database, and the ITS rRNA gene sequence was analyzed against the UNITE database (Version 6).
The Venn diagrams were constructed to visualize shared and unique OTUs among samples. Based on the calculated Bray–Curtis distance, the NMDS analysis was conducted to investigate the variation of bacterial and fungal community structure among treatments. To assess the significance of the difference in the bacterial and fungal community structure among different Armillaria strains, permutational multivariate ANOVA (PERMANOVA) was conducted using the ADONIS tests in the R vegan package (ADONIS; Oksanen et al., 2014). Based on the pairwise Spearman method and correlation coefficient with a threshold value of r ≥ 0.5, the co-occurrence network analysis of bacteria and fungi was calculated at a p < 0.5 significant level, and the analysis and visualization were performed on the platform BMKCloud.5
The statistical significance of the yield of G. elata, tensile strength of Armillaria mycelia, OTU numbers, alpha diversity of bacterial and fungal communities, and relative abundance at the genus level among different Armillaria strains were tested using a one-way analysis of variance (ANOVA) and Duncan’s multiple range test at a 0.05 significance level (SPSS 20.0 for Windows, IBM, Chicago, IL, United States). The IGS sequence comparisons of Armillaria strains were done with BLAST (see footnote 1), and a phylogenetic tree based on gene sequence comparisons was constructed with Mega (version 11, Tamura et al., 2021).
The gene sequence comparisons of intergenic spacers were done with BLAST homology comparison to classify the origins of Armillaria strains. The results of BLAST indicated that AH, SX, and GZ strains shared more than 98% homology with A. gallica, while the YN strain shared 82% with A. gallica in sequences producing significant alignments. The phylogenetic analysis based on sequences showed that all the Armillaria strains were clustered in three main clades, and the four strains and A. gallica had very close genetic relationships (Figure 1).
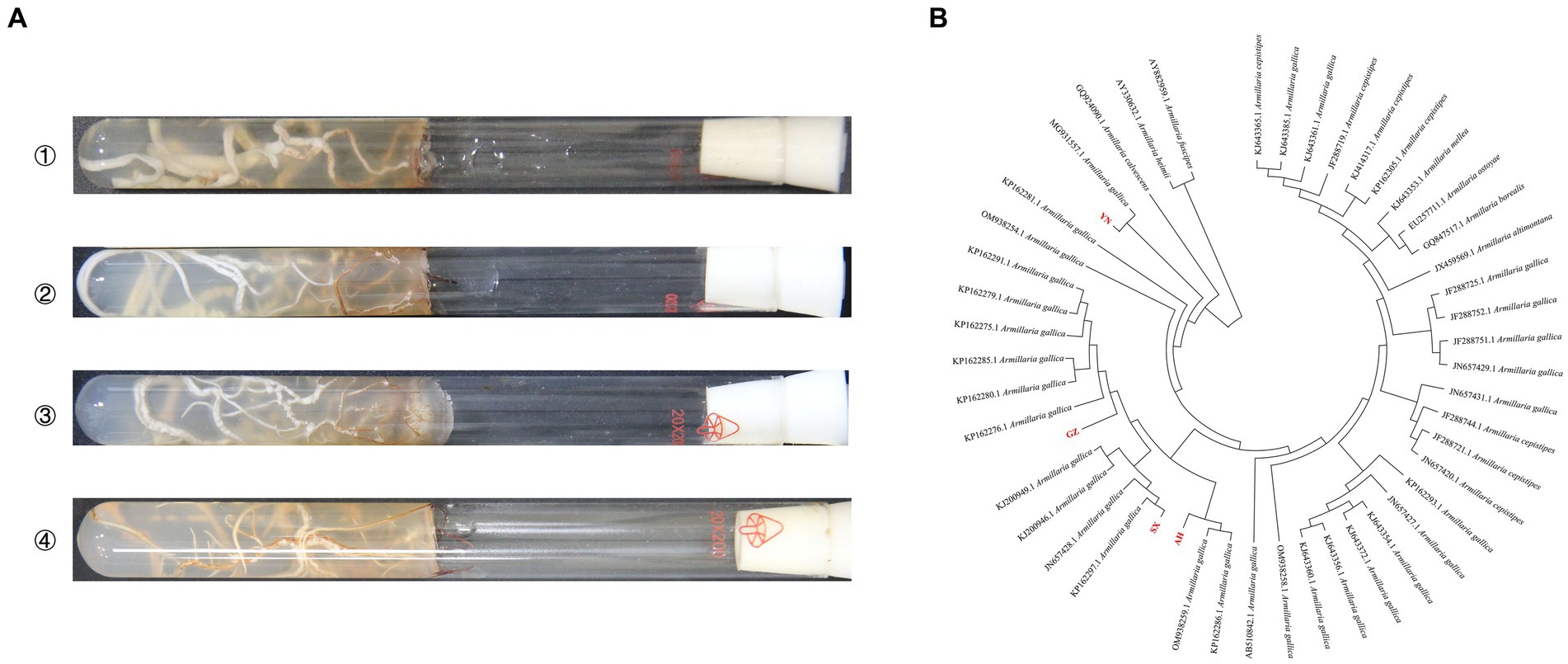
Figure 1. Incubation and identification of Armillaria isolates. (A) Mycelial morphology of four isolates on PDA medium at 23°C for 20 days (①: YN, ②: AH, ③: SX, ④: GZ). (B) Phylogenetic analysis of four Armillaria isolates based on the intergenic spacer.
The yield of G. elata symbiosis with the YN strain was the highest (3.91 kg·m−2) (Figure 2A) and significantly higher than that of other strains (p < 0.05). In addition, the yield of G. elata symbiosis with SX and AH was both 2.38 kg·m−2 and markedly higher compared with the yield of G. elata symbiosis with the GZ strain, which was 0.98 kg·m−2 (p < 0.05).
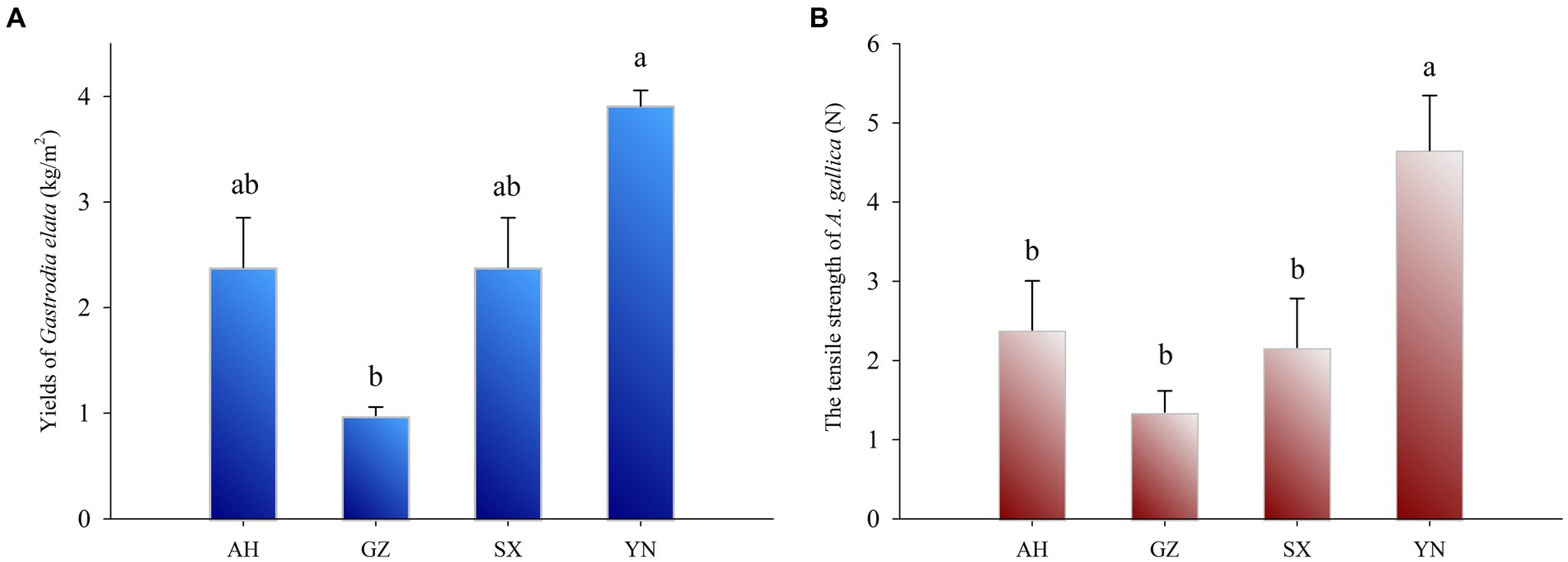
Figure 2. Yields of G. eleta symbiosis with Armillaria strains from different production areas for 8 months (A) and the tensile strength of Armillaria strains at harvest time (B). Lowercase letters a, b, and c within one plot indicate significant differences among treatments, determined using the one-way ANOVA and Duncan’s multiple range test (p < 0.05).
To assess the ability of nutrient supply in different A. gallica strains, we tested the tensile strength of A. gallica mycelia at harvest time using a tensiometer. The tensile strength of the YN strain was significantly higher (p < 0.05) than that of the SX, AH, and GZ strains (Figure 2B), and strong growth of mycelia was observed in the YN strain compared with others (Figure 1A). However, there was no significant difference in the tensile strength among the SX, AH, and GZ strains.
The next-generation sequencing generated 959,747 pairs of reads for 16S rRNA genes and 959,978 pairs of reads for ITS rRNA genes across all samples. After rarefication to the minimum average, 79,764 and 79,666 clean reads were obtained for bacteria and fungi, respectively. Based on 97% similarity, 19,265 and 7,588 OTUs were identified for the bacterial and fungal communities, respectively.
Compared with the SX and YN strains, the GZ strain resulted in increases in soil bacterial OTU numbers (Figure 3A). Compared with others, the YN strain led to significantly higher OTU numbers in soil fungal community (Figure 3C). The Venn diagram analysis showed that there were large numbers of OTUs shared across samples, while less peculiar OTUs were spread in the bacterial community (Figure 3B). In the fungal community, the numbers of peculiar OTUs in soil planted with the YN strain were much higher than those of the AH, SX, and GZ strains, and more numbers of shared OTUs were found across treatments (Figure 3D).
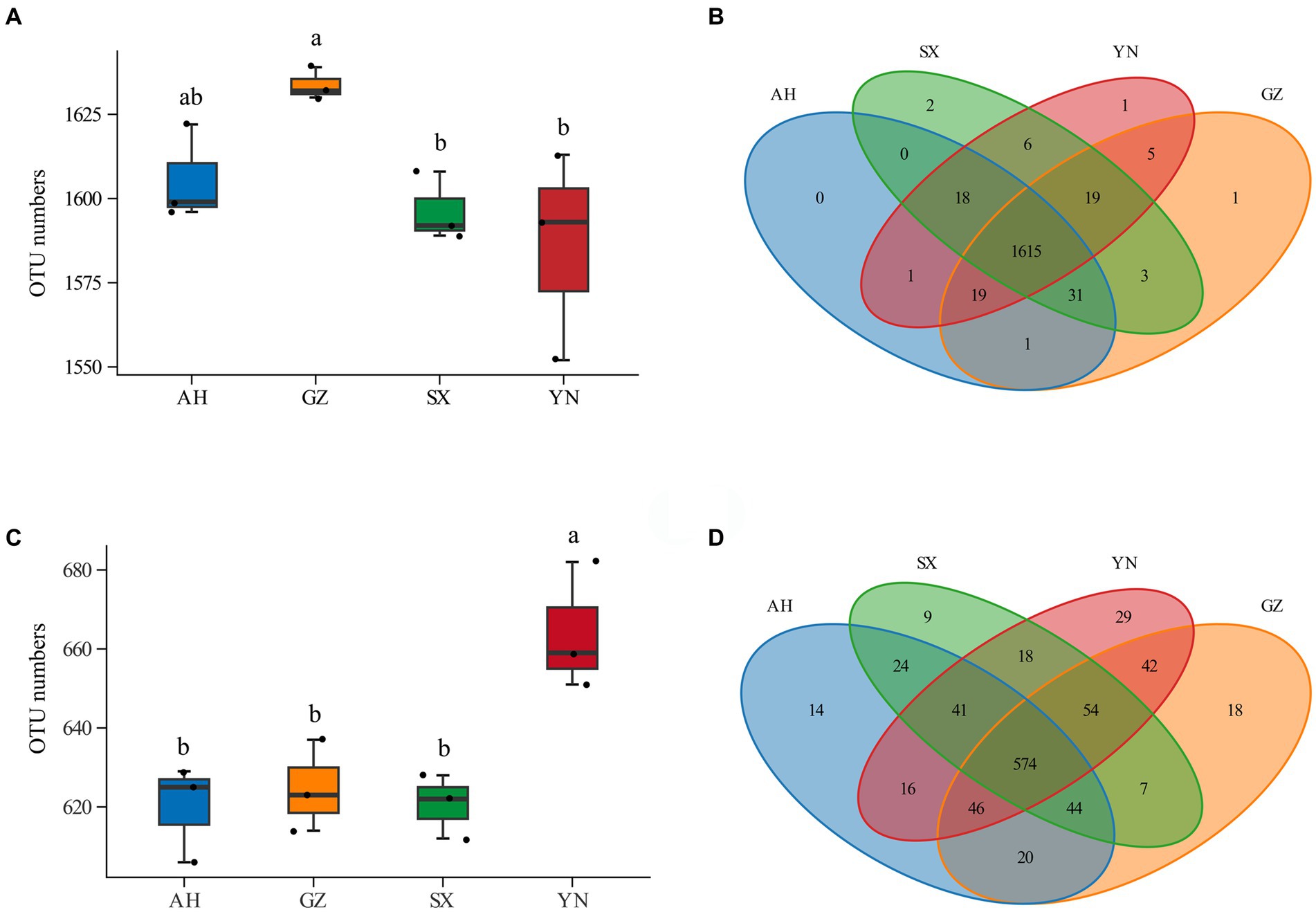
Figure 3. OTU numbers and Venn analysis of bacterial and fungal communities in soils around G. eleta symbiosis with Armillaria strains. (A,B) Represent OTU numbers and the Venn diagram of the bacterial community; (C,D) represent OTU numbers and the Venn diagram of the fungal community. Lowercase letters a and b indicate significant differences among different soils, determined using the one-way ANOVA and Duncan’s multiple range test (p < 0.05).
We analyzed the response of the soil bacterial and fungal communities to different A. gallica strains through Illumina sequencing. The diversity and richness of bacteria and fungi varied among different A. gallica strains and showed different patterns (Figure 4). In particular, the ACE and Simpson indexes of bacterial diversity in soil planted with the GZ strain significantly increased compared with those of other strains (Figures 4A,B), whereas the changes in fungal diversity indexes were different from those of bacteria. Specifically, the ACE and Simpson indexes tended to increase in soil planted with the YN strain, which showed significant differences with others (Figures 4C,D). Overall, the α diversity of the soil bacterial community was significantly influenced by the GZ strain, while that of the soil fungal community was markedly altered by the YN strain.
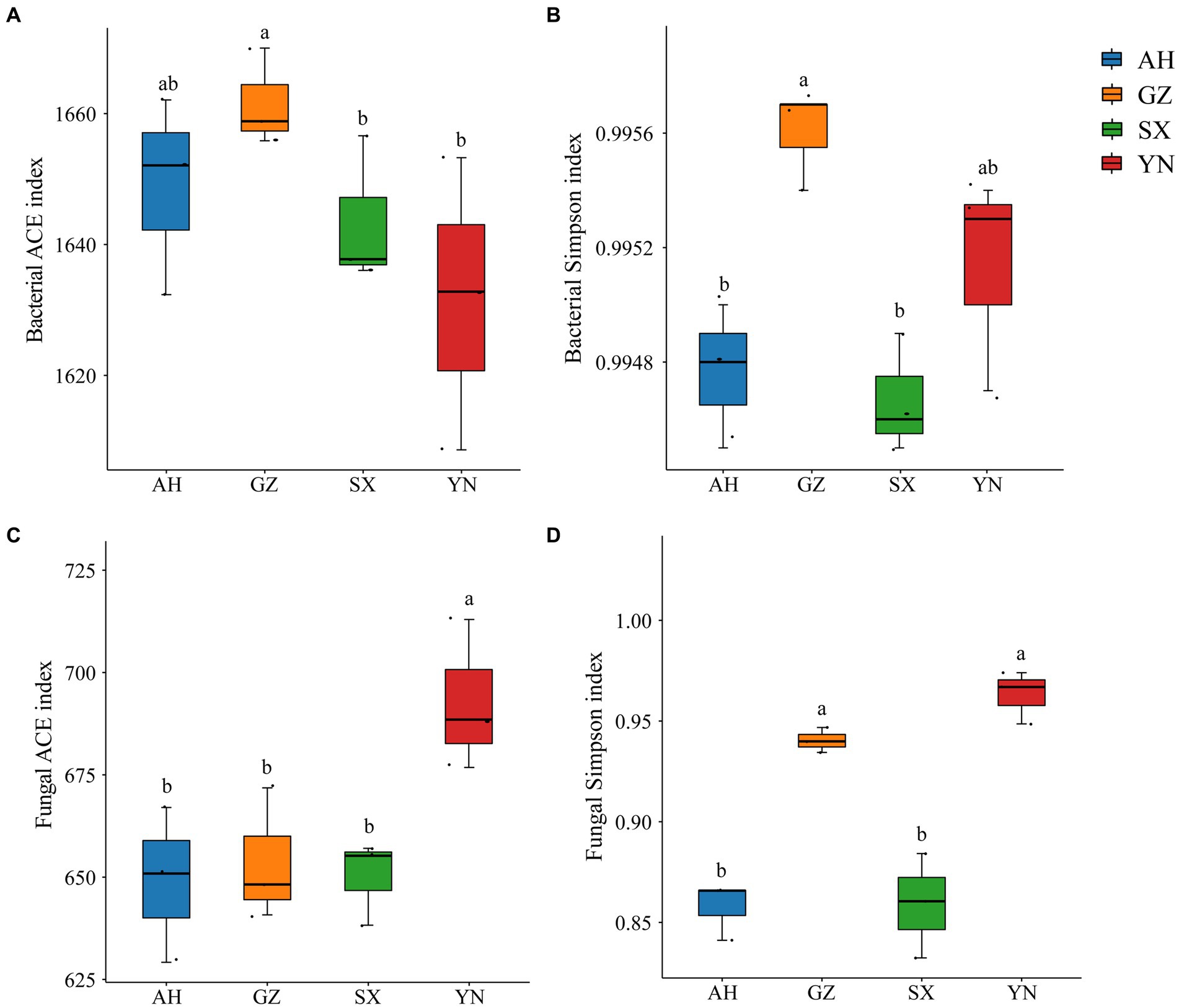
Figure 4. Alfa diversity indices of bacterial and fungal communities in soils around G. eleta symbiosis with Armillaria strains. (A,B) Represent the Ace and Simpson indexes of the bacterial community; (C,D) represent the Ace and Simpson indexes of the fungal community. Lowercase letters a and b indicate significant differences among different soils, determined using the one-way ANOVA and Duncan’s multiple range test (p < 0.05).
Similarly, the community structure of bacteria and fungi significantly changed (Figure 5). The result of NMDS analysis showed that the bacterial community in soil planted with the GZ strain clustered together and was greatly separated from the other strains (R2 = 0.309, p < 0.01) (Figure 5A). Meanwhile, the fungal community structures of YN and SX strains were greatly separated from the others (R2 = 0.401, p < 0.01), while no significant difference was observed between AH and GZ strains (p = 0.1) (Figure 5B).
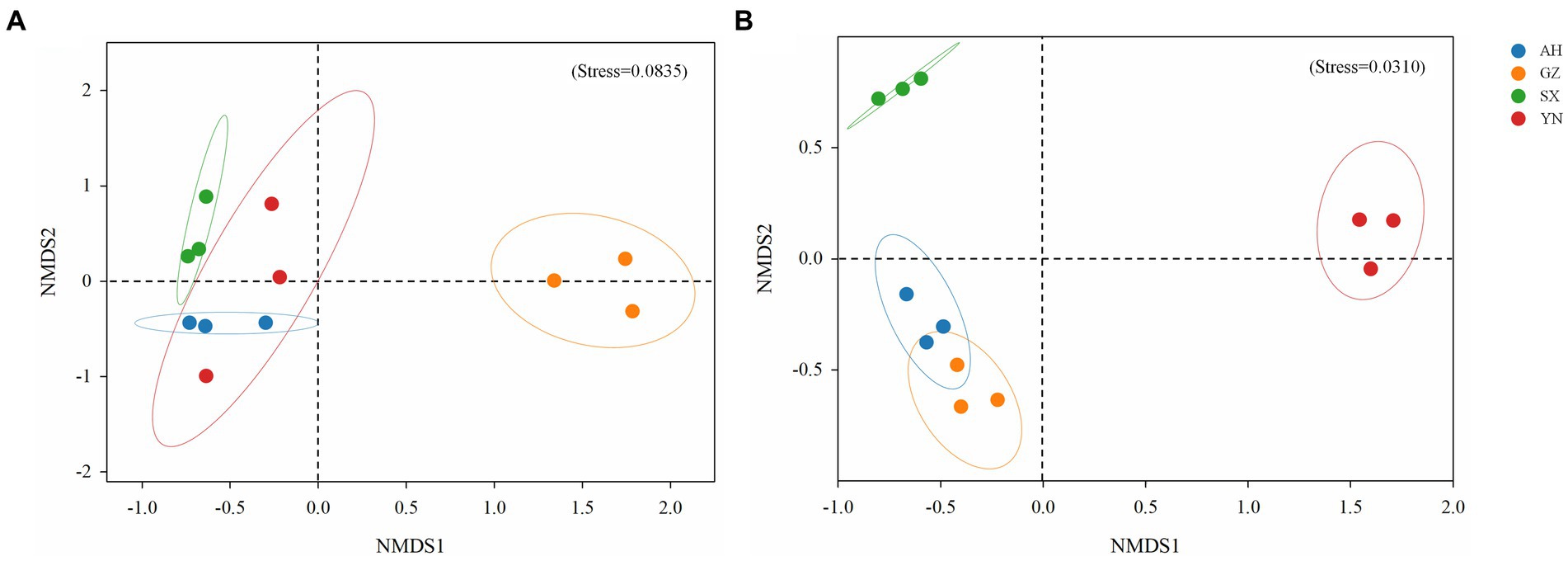
Figure 5. Non-metric multidimensional scaling (NMDS) analysis of bacterial (A) and fungal (B) communities in soils around G. eleta symbiosis with Armillaria strains, calculated based on Bray-Curtis distance.
The bacterial community was predominated by 9 phyla with a minor proportion of others (1.5–2.3%). The phyla Pseudomonadota (38.57–42.34%), Acidobacteriota (27.39–32.69%), and Chloroflexota (6.12–9.77%) were dominant members, followed by Planctomycetota (4.26–5.91%), Actinomycetota (3.83–4.38%), Verrucomicrobiota (3.21–4.16%), Bacteroidota (2.43–4.15%), Gemmatimonadota (1.73–2.81%), and WPS-2 (0.53–1.41%) (Figure 6A). None of the A. gallica strains induced significant changes in the relative abundances of Pseudomonadota (p = 0.344). However, compared with other strains, planting GZ strains significantly increased the abundances of Chloroflexota (p < 0.05) while significantly decreasing the abundances of Acidobacteriota (p < 0.01).
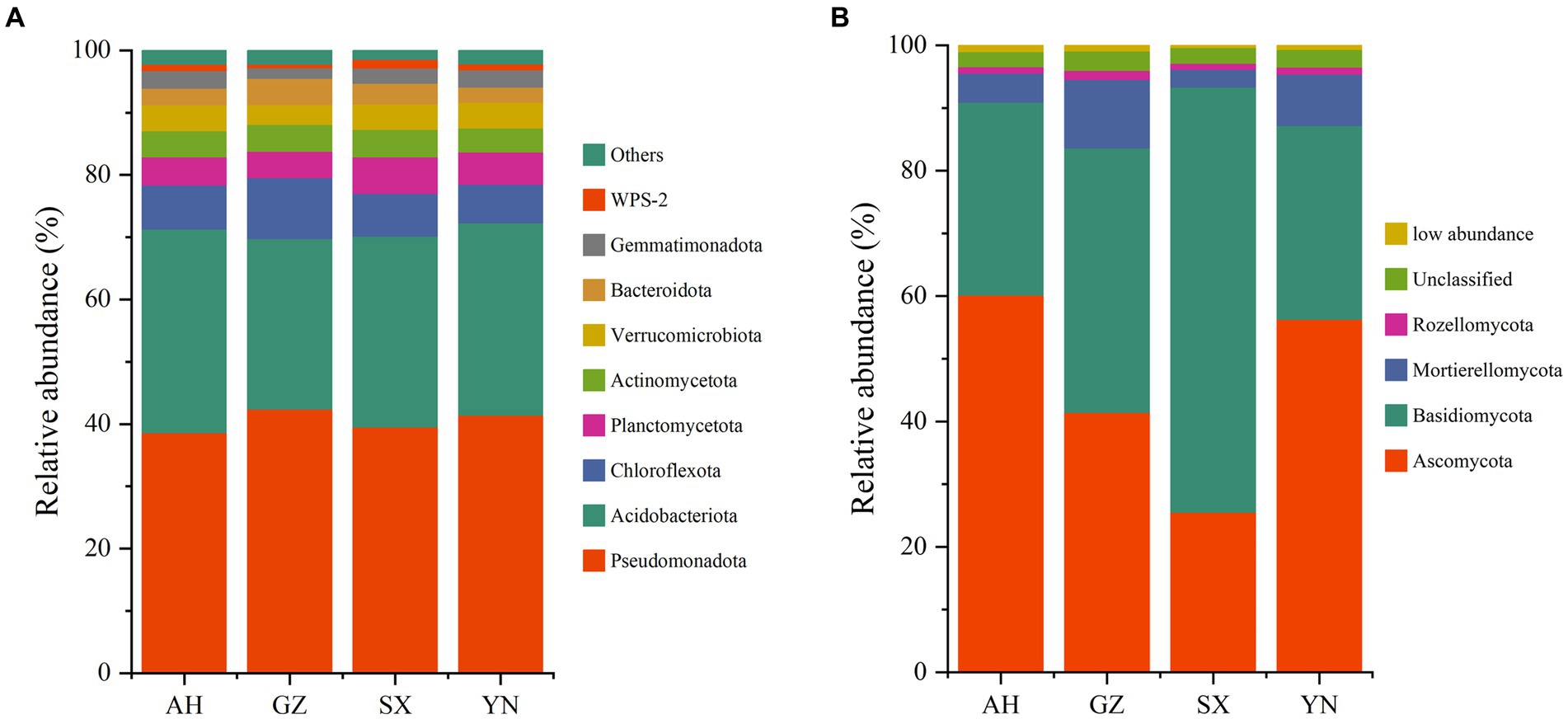
Figure 6. The bacterial (A) and fungal (B) community compositions in soils around G. eleta symbiosis with Armillaria strains at the level of phylum classification.
In the total ITS rRNA amplicons, OTUs in all samples were classified into four phyla, namely, Ascomycota (25.48–60.13%), Basidiomycota (30.73–67.80%), Mortierellomycota (2.85–10.96%), and Rozellomycota (0.95–1.44%), besides the unclassified (2.42–3.09%) and the sum of relative abundances that were lower than 1% (0.40–1.04% in total) (Figure 6B). Notably, the significant role of A. gallica strains in affecting the rhizomicrobiome in three dominant fungal phyla, i.e., Ascomycota, Basidiomycota, and Mortierellomycota, was detectable (p < 0.05) (Figure 6B). Specifically, AH and YN strains significantly increased the relative abundance of Ascomycota and Mortierellomycota (p < 0.01) while decreasing the relative abundance of Basidiomycota compared with other strains.
At the genus level, the number of genera greatly responded to different A. gallica strains. Compared with others, relative abundances of most genera of bacteria were mainly reduced in soil planted with GZ strain, except for the bacterium_c_AD3, bacterium_c_Alphaproteobacteria, and Reyranella, whose relative abundances were significantly increased by GZ strain (p < 0.05) (Figure 7A). However, most fungal groups were enriched by the YN strain. For example, the genera with increased relative abundances in response to the YN strain included Mortierella, Nemania, Agrocybe, Archaeorhizomyces, Armillaria, Pseudoclathrosphaerina, Scopuloides, Leptodontidium, Cladophialophora, Psathyrella, and Saitozyma (p < 0.05) (Figure 7B). Other genera showed negative responses to the YN strain, including Coprinellus, Sebacina, and Russula; however, their relative abundances were significantly increased by the SX strain (p < 0.05).
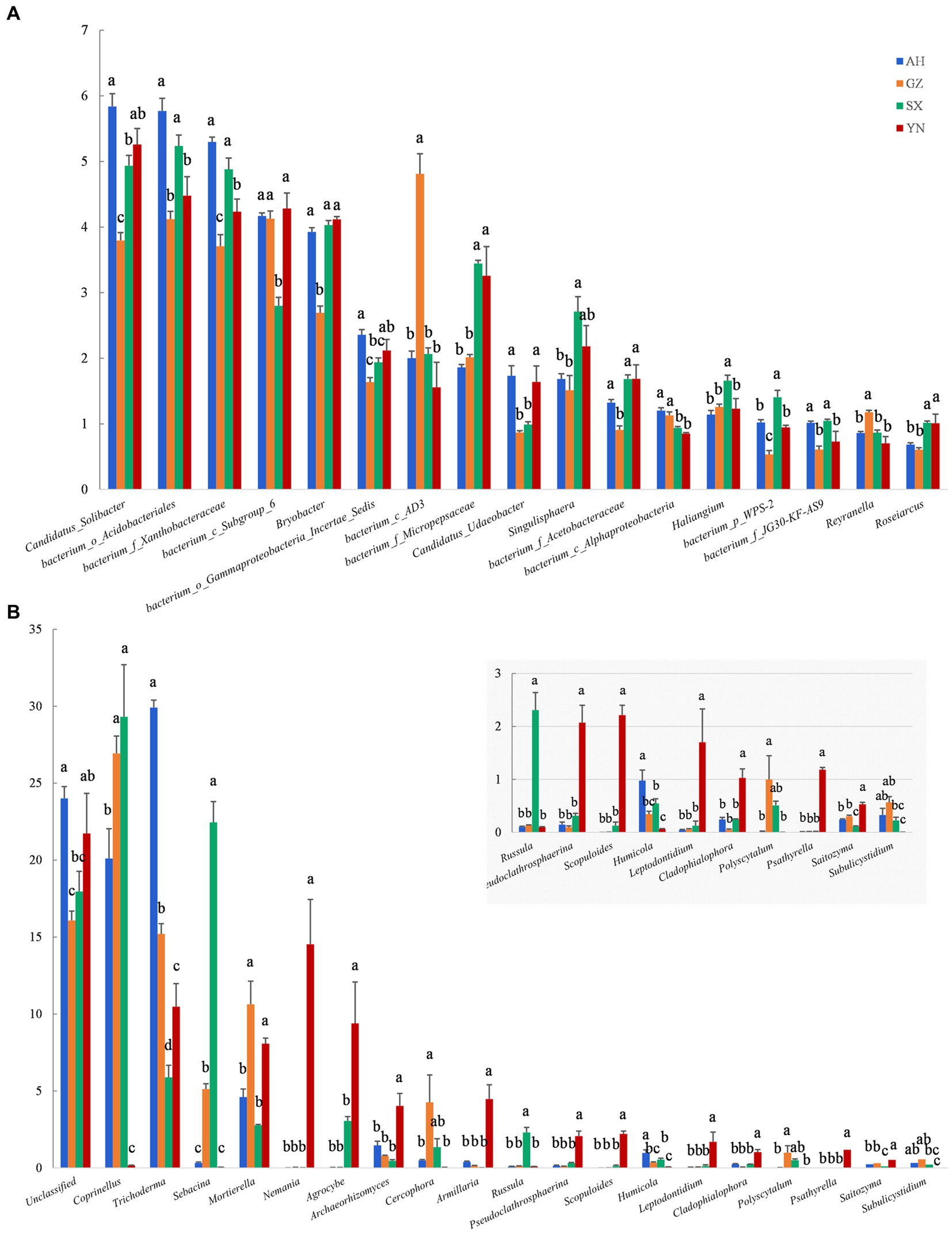
Figure 7. Relative abundance of soil bacterial (A) and fungal (B) communities in soils around G. eleta symbiosis with Armillaria strains at the level of genus classification. Lowercase letters a, b, and c within one genus taxa indicate significant differences among treatments, determined using the one-way ANOVA and Duncan’s multiple range test (p < 0.05).
Networks were constructed, respectively, for bacteria and fungi, and significant correlations were compared. We found that bacterial co-occurrence networks were larger, more connected, and more modular than those in fungal networks, and a larger proportion of bacterial OTUs was included in co-occurrence networks than that of fungal OTUs (Figure 8). In particular, the network for bacteria was composed of 46 nodes and 100 edges, and the modularity was 0.58; all nodes were assigned to 9 phyla. For the fungal community, the network presented 42 nodes and 100 edges; all these nodes were assigned to 4 phyla, i.e., Ascomycota, Basidiomycota, Chytridiomycota, and Mortierellomycota. Moreover, compared with Basidiomycota (14 nodes), Ascomycota (26 nodes) had a more complex correlation with different A. gallica strains.
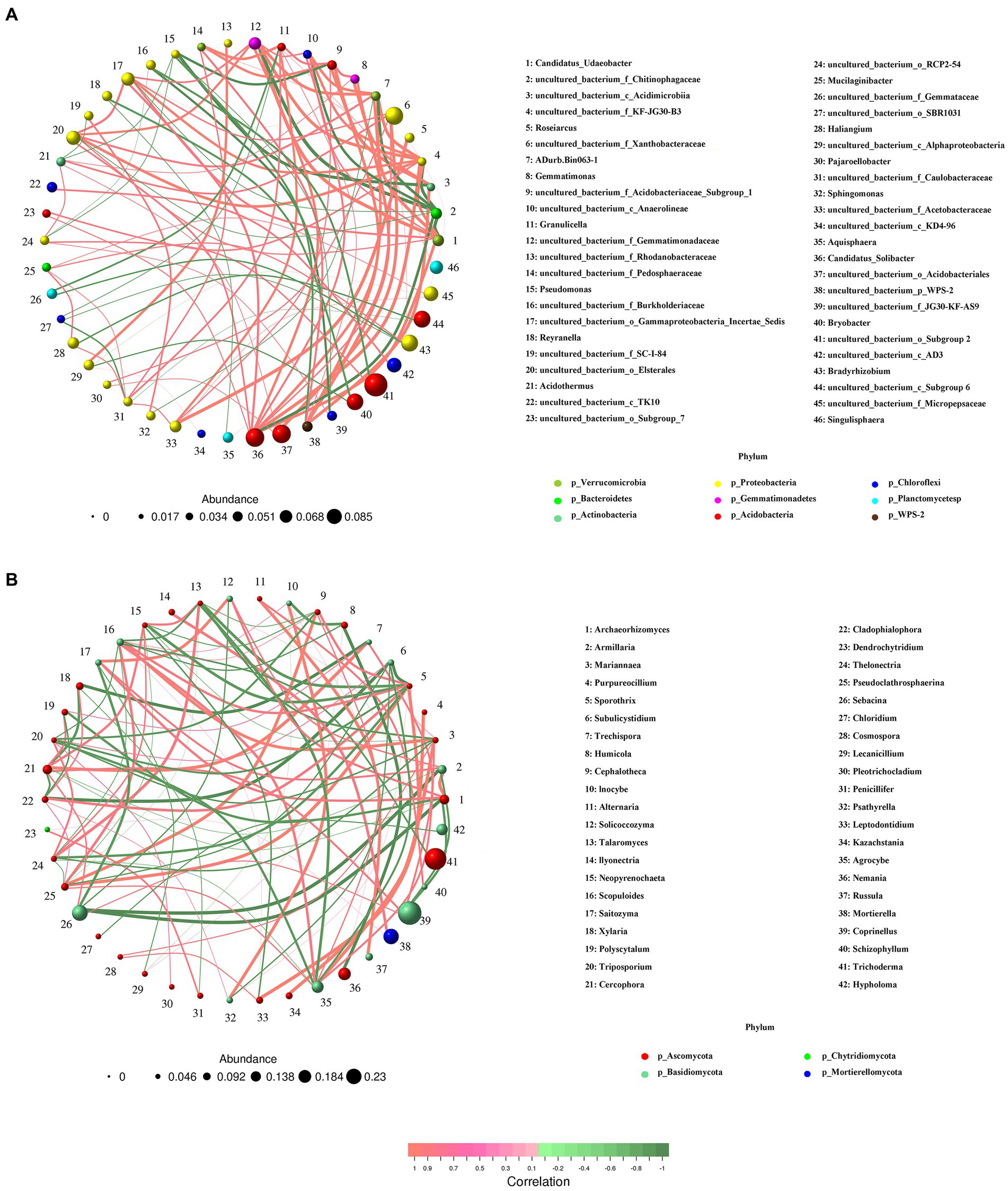
Figure 8. Co-occurrence analysis of soil bacterial (A) and fungal (B) communities in soils around G. eleta symbiosis with Armillaria strains.
The demand for G. elata production is expected to increase further in the future as a result of health preservation and wild resource shortages. Such concerns have led to the development of artificial cultivation of G. elata, in which Armillaria strains are functionally important factors in affecting the growth and yield of G. elata. Soil microbial communities are responsible for nutrient cycling and plant growth; these communities could be affected by abiotic or biotic factors, particularly interactions with other microorganisms (Singh et al., 2009; Kubiak et al., 2017). For these reasons, there has long been interest in selecting the optimum Armillaria strains to co-plant with G. elata in a variety of environments and disentangling the mechanisms behind the high yield of G. elata. This study is one of the few attempts to provide insight into the yield of G. elata co-planted with different Armillaria strains and the microbiome influenced beyond this interaction.
The current study indicated that cultivation of A. gallica strains collected from other regions, especially Yunnan (YN strain), can significantly increase the yield of G. elata compared with that of the original Guizhou local strain (Figure 2A). Our finding is consistent with previous research in which four strains of Armillaria were identified as A. gallica groups, whereas co-planting of these A. gallica strains with G. elata resulted in significant differences in yield and active ingredient content of G. elata (Liu et al., 2019). However, how did that happen if the identification of Armillaria strains indicated that they were in close relation to A. gallica (Figure 1)? Beyond the classification results of Armillaria strains, a possible explanation is that the robust growth condition (Figure 1A) and greater tensile strength of mycelia in YN strain (Figure 2B) contributed to the high yield of G. elata compared with others, which is also evidenced by previous studies that the optimum Armillaria strains that can greatly increase the yield of G. elata are typically the ones with stronger branches of mycelia (Chen et al., 2004; Wang et al., 2020). Therefore, the tensile strength, which can be a measurement of the thickness and strength of Armillaria mycelia, was tested at harvest time, and the results also confirmed that the stronger the Armillaria branches, the higher the yield of G. elata that is harvested (Figure 2). In the meantime, given that Armillaria mycelia are generally considered a special morphological adaptation of this fungus to different environmental conditions (Garett, 1960; Morrison, 2004), the migration of YN, AH, and SX strains from their local habitat partly triggered their strong growth of mycelia. On the contrary, the adaptability of GZ strain to the local condition and its weak growth of mycelia might lead to its poor performance when co-planted with G. elata (Sun et al., 2009). Besides that, the great variation in soil bacterial and fungal communities arising from different A. gallica strains may also play vital roles in controlling the yield of G. elata.
Available studies have shown that the impacts of symbiotic mycorrhizal fungi on the diversity and composition of the soil rhizomicrobiome may induce changes in the ecological environment of soil microorganisms and, potentially, increase stress tolerance and yield of plants (Doubková et al., 2012; Cosme and Wurst, 2013; Selosse et al., 2022). In the current study, we observed that the GZ strain induced great changes in the bacterial community (Figures 3A,B, 4A,B, 5A), while the YN strain had a much stronger impact on the fungal community (Figures 3C,D, 4C,D, 5B). Moreover, a diverse variation and more complex fungal community composition were observed across all soil samples in responding to different A. gallica strains (Figures 6, 7). In a co-culture study, Collins et al. (2013) revealed the defensive and potentially offensive nature of Armillaria members and their ability to influence other microbes. Thus, it is acceptable that greater variation was observed in the fungal community induced by A. gallica strains, considering that soil fungal communities are more sensitive and respond more quickly to a variety of stimuli, including biological disturbance (Xiao et al., 2014).
As also reported, the Armillaria members are known as wood-rotting fungi (Brundrett, 2002), along with an ability to secrete wood-decomposition enzymes such as glycoside hydrolases. When A. gallica exists in soil and attempts symbiosis with G. elata, it will typically secrete enzymes to penetrate plant cell walls. The enzymes can mineralize or decompose most plant cell wall polymers into simple compounds (Floudas et al., 2012; Purahong et al., 2016), which increase their accessibility to other organisms. A previous study conducted with five Armillaria isolates reported a positive effect of an excellent Armillaria strain on the promotion of quality and yield of G. elata Bl. f. glauca in Changbai Mountain (Yu et al., 2022). The researchers found that excellent Armillaria strains increased the yield of G. elata Bl. f. glauca by improving the soil microbial environment, especially by increasing the relative abundances of some beneficial genera such as Mortierella, Agrocybe, and Armillaria. In accordance with this study, the high yield of G. elata co-planted with the YN strain was accompanied by higher relative abundances of Mortierella, Nemania, Agrocybe, Archaeorhizomyces, Armillaria, Pseudoclathrosphaerina, Scopuloides, Leptodontidium, Cladophialophora, Psathyrella, and Saitozyma compared with these in other A. gallica strains (Figure 7B). Species of the genera Mortierella, Nemania, and Leptodontidium were commonly reported as wood-rotting members and are sensitive to readily accessible carbohydrates (Tang et al., 2007; Lindahl et al., 2010; Werner et al., 2016). Their strong co-occurrence with the YN strain implied that they were candidate fungal taxa that might be involved in the high activity of A. gallica. Accordingly, Archaeorhizomyces species are non-pathogenic plant root and rhizosphere-associated fungi that commonly harbor deeper soil horizons with low pH and high nutrient turnover (Rosling et al., 2011; Carrino-Kyker et al., 2016). Previous findings suggest that correlations between Archaeorhizomycetes and other fungal taxa indicate that Archaeorhizomycetes might benefit from carbohydrates or nutrients (Pinto-Figueroa et al., 2019) and may also have potential associations with plant growth (Schadt et al., 2003). Similarly, species of the genus Cladophialophora possessed superior growth promotion activities as well as disease suppression, which significantly increased the growth parameters of strawberry plants (Harsonowati et al., 2020). As a result, the combination of yields of G. elata and responses of these fungal genera due to A. gallica strains in our study may reflect the potential performance of biological and ecological functions in soil fungal communities.
It is well known that the interactions of interspecific fungi are complex and dynamic processes (Awan and Asiegbu, 2021). Co-occurrence networks indicated that bacteria such as Candidatus_Solicabcter, bacterium_f_Gemmatimonadaceae were highly central and connected (Figure 8A), suggesting that they might drive the observed A. gallica-induced changes in bacterial networks. Additionally, for fungal networks, the genera Sporothrix (belonging to Ascomycota) and Scopuloides (belonging to Basidiomycota) all interacted with 12 genera, including 7 positive interaction genera and 5 negative interaction genera, implying their important roles in shaping fungal community under such complex environmental conditions. Based on previous studies, soil bacterial communities are less resistant but more resilient to disturbance than fungal communities (de Vries et al., 2018), and changes in functional taxa can alter the response of microbial communities to disturbance and their symbiotic relationships with host plants (Banerjee et al., 2018). In the current study, shifts in the relative abundance of dominant taxa (Figures 6, 7) might later facilitate the stability of networks (Figure 8), and consequently the assemblage of fungal communities in soil substrates and response of soil fungi to different A. gallica strains. Besides that, soil physicochemical properties or nutrient cycling were expected to be altered by symbiotic fungi (Finlay, 2004; Veresoglou et al., 2012), and correlations can be tested between environmental factors and microbial community structure. However, measurements of soil physicochemical properties were not conducted due to the slight amount of soil around G. elata, which is a limitation of this study. Further study is needed to discuss the changes in soil physicochemical properties arising from A. gallica strains and the correlations between them.
The current study emphasizes that soil fungi rather than bacteria were more easily modified by exotic A. gallica strains, and that might have a potentially beneficial impact on the growth and yield of G. elata. Specifically, the dominant advantage of the YN strain was evidenced by its role in triggering changes in the soil fungal community, especially relative abundances of some beneficial genera such as Mortierella, Agrocybe, Neminia, Armillaria, and so on when cultivated in Guizhou Province. Although our study identified these potentially beneficial genera accompanied by a high yield of G. elata, further support is needed to confirm the potential functions and ecosystem services of these soil microbes by isolating and proving their roles in suppressing or fostering A. gallica in a co-culture experiment or mutualistic trial study with G. elata.
The current study revealed that under field conditions in G. elata and A. gallica symbiotic systems, the yield of G. elata significantly differed among A. gallica strains collected from several regions. In addition, the diversity, structure, and composition of soil microbial communities, especially fungal communities, strongly changed in response to different A. gallica strains, which might have a beneficial effect on the yield of G. elata. This comprehensive understanding of the responses that govern the selection and activity of microbial communities by A. gallica strains can be committed to practical management and provide us with new opportunities to increase the yields of G. elata.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, PRJNA951876; https://www.ncbi.nlm.nih.gov/, PRJNA951884.
YW: funding acquisition, methodology, formal analysis, writing—original draft preparation. JX: funding acquisition, methodology, and visualization. QY: funding acquisition, methodology, writing—review and editing. LG: conceptualization and supervision. CX: methodology and software. CY: formal analysis and data curation. LL: resources and data curation. WJ: conceptualization and supervision. TZ: conceptualization, funding acquisition, and supervision. All authors contributed to the article and approved the submitted version.
This study was supported by the Guizhou Provincial Basic Research Program [Natural Science, project No. Z.K. (2021) General 523], the National Natural Science Foundation of China (projects No. 81960694 and No. 32060080), the China Agriculture Research System (CARS-21), the High-level Innovative Talents Program of the Guizhou Province of China [Qian Ke He Platform and Talent (2018) 5638–2], and the Science and Technology Project of the Guizhou Province of China [Qian Ke He Platform and Talent (2019) 5611].
We would like to thank Taosheng Wu who helped us with the layout of Supplementary Figure S1.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1233555/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | Collection sites of Armillaria rhizomorphs from four main producing regions of G. elata.
SUPPLEMENTARY FIGURE S2 | The conditions of Armillaria strains packets used for G. elata artificial cultivation. (A), (B), (C), and (D) represent Armillaria of YN strain, GZ strain, SX strain, and AH strain respectively.
SUPPLEMENTARY FIGURE S3 | The conditions of Armillaria strains mycelia at the harvest time. (A), (B), (C), and (D) represent Armillaria of YN strain, GZ strain, SX strain, and AH strain respectively.
1. ^https://blast.ncbi.nlm.nih.gov/Blast.cgi
2. ^https://www.who.int/medicines/publications/traditional/gacp2004/en/
3. ^version 7.1, http://drive5.com/uparse.
4. ^RDP, V.2.2, http://rdp.cme.msu.edu/.
Awan, H. U. M., and Asiegbu, F. O. (2021). “Interspecific interactions within fungal communities associated with wood decay and forest trees” in Forest microbiology: Tree Microbiome: Phyllosphere, Endosphere and Rhizosphere. eds. F. O. Asiegbu and A. Kovalchuk (Cambridge, USA: Elsevier, Academic Press), 75–108.
Banerjee, S., Schlaeppi, K., and Van Der Heijden, M. G. (2018). Keystone taxa as drivers of microbiome structure and functioning. Nat. Rev. Microbiol. 16, 567–576. doi: 10.1038/s41579-018-0024-1
Berendsen, R. L., Pieterse, C. M. J., and Bakker, P. A. H. M. (2012). The rhizosphere microbiome and plant health. Trends Plant Sci. 17, 478–486. doi: 10.1016/j.tplants.2012.04.001
Brundrett, M. C. (2002). Coevolution of roots and mycorrhizas of land plants. New Phytol. 154, 275–304. doi: 10.1046/j.1469-8137.2002.00397.x
Bulgarelli, D., Schlaeppi, K., Spaepen, S., van Themaat, E. V. L., and Schulze-Lefert, P. (2013). Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 64, 807–838. doi: 10.1146/annurev-arplant-050312-120106
Cairney, J. W. G., Jennings, D. H., Ratcliffe, R. G., and Southon, T. E. (1988). The physiology of basidiomycete linear organs II. Phosphate uptake by rhizomorphs of Armillaria mellea. New Phytol. 109, 327–333. doi: 10.1111/j.1469-8137.1988.tb04202.x
Camprubi, A., Solari, J., Bonini, P., Garcia-Figueres, F., Colosimo, F., Cirino, V., et al. (2020). Plant performance and metabolomic profile of loquat in response to mycorrhizal inoculation, Armillaria mellea and their interaction. Agronomy 10:899. doi: 10.3390/agronomy10060899
Carrino-Kyker, S. R., Kluber, L. A., Petersen, S. M., Coyle, K. P., Hewins, C. R., DeForest, J. L., et al. (2016). Mycorrhizal fungal communities respond to experimental elevation of soil pH and P availability in temperate hardwood forests. FEMS Microbiol. Ecol. 92, 1–19. doi: 10.1093/femsec/fiw024
Chen, M. Y., Li, F. H., and Bian, Y. B. (2004). Effect of different strains of Armillariella mellea on the yield of Gastrodia elata. Acta Edulis Fungi 11, 46–48. doi: 10.16488/j.cnki.1005-9873.2004.01.010
Clipson, N. J. W., Cairney, J. W. G., and Jennings, D. H. (1987). The physiology of basidiomycete linear organs I. phosphate uptake by cords and mycelium in the laboratory and in the field. New Phytol. 105, 449–457. doi: 10.1111/j.1469-8137.1987.tb00882.x
Collins, C., Keane, T. M., Turner, D. J., O’Keeffe, G., Fitzpatrick, D. A., and Doyle, S. (2013). Genomic and proteomic dissection of the ubiquitous plant pathogen, Armillaria mellea: toward a new infection model system. J. Proteome Res. 12, 2552–2570. doi: 10.1021/pr301131t
Cosme, M., and Wurst, S. (2013). Interactions between arbuscular mycorrhizal fungi, rhizobacteria, soil phosphorus and plant cytokinin deficiency change the root morphology, yield and quality of tobacco. Soil Biol. Biochem. 57, 436–443. doi: 10.1016/j.soilbio.2012.09.024
Crowther, T., Boddy, L., and Jones, T. H. (2012). Functional and ecological consequences of saprotrophic fungus–grazer interactions. ISME J. 6, 1992–2001. doi: 10.1038/ismej.2012.53
Currah, R. S., Zelmer, C. D., Hambleton, S., and Richardson, K. A. (1997). “Fungi from orchid mycorrhizas” in Orchid biology: reviews and perspectives VII. eds. J. Arditti and A. M. Pridgeon (Netherlands: Kluwer Academic Press), 117–170.
de Vries, F. T., Griffiths, R. I., Bailey, M., Craig, H., Girlanda, M., Gweon, H. S., et al. (2018). Soil bacterial networks are less stable under drought than fungal networks. Nat. Commun. 9:303. doi: 10.1038/s41467-018-05516-7
Doubková, P., Suda, J., and Sudová, R. (2012). The symbiosis with arbuscular mycorrhizal fungi contributes to plant tolerance to serpentine edaphic stress. Soil Biol. Biochem. 44, 56–64. doi: 10.1016/j.soilbio.2011.09.011
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10:996. doi: 10.1038/nmeth.2604
Feijen, F. A. A., Vos, R. A., Nuytinck, J., and Merckx, V. S. F. T. (2018). Evolutionary dynamics of mycorrhizal symbiosis in land plant diversification. Sci. Rep. 8:10698. doi: 10.1038/s41598-018-28920-x
Fierer, N. (2017). Embracing the unknown: disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 15, 579–590. doi: 10.1038/nrmicro.2017.87
Finlay, R. D. (2004). Mycorrhizal fungi and their multifunctional roles. Mycologist 18, 91–96. doi: 10.1017/S0269915X04002058
Floudas, D., Binder, M., Riley, R., Barry, K., Blanchette, R. A., Henrissat, B., et al. (2012). The Paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science 336, 1715–1719. doi: 10.1126/science.1221748
Garett, S. D. (1960). Rhizomorph behavior in Armillaria mellea (Fr.) Quel., III saprophytic colonization of woody substrates in soil. Ann. Bot. 24, 275–285. doi: 10.1093/oxfordjournals.aob.a083701
Harsonowati, W., Marian, M., and Narisawa, K. (2020). The effectiveness of a dark septate endophytic fungus, Cladophialophora chaetospira SK51, to mitigate strawberry fusarium wilt disease and with growth promotion activities. Front. Microbiol. 11:585 doi: 10.3389/fmicb.2020.00585
Kubiak, K., Zółciak, A., Damszel, M., Lech, P., and Sierota, Z. (2017). Armillaria pathogenesis under climate changes. Forest 8:100. doi: 10.3390/f8040100
Lindahl, B., de Boer, W., and Finlay, R. (2010). Disruption of root carbon transport into forest humus stimulates fungal opportunists at the expense of mycorrhizal fungi. ISME J. 4, 872–881. doi: 10.1038/ismej.2010.19
Liu, T. R., Wang, Z. Q., Chen, X. D., Zhang, W. W., Yang, Y. S., Xu, W. L., et al. (2019). Identification of four Armillaria strains and their effects on quality and yield of Gastrodia elata f. glauca. Zhongguo Zhong Yao Za Zhi 44, 5352–5357. doi: 10.19540/j.cnki.cjcmm.20191009.104
Morrison, D. J. (2004). Rhizomorph growth, habit, saprophytic ability and virulence of 15 Armillaria species. For. Pathol. 34, 15–26. doi: 10.1046/j.1439-0329.2003.00345.x
Oksanen, J., Blanchet, F. G., Kindt, R., Legendre, P., Minchin, P. R., O'Hara, R. B., et al. (2014). Vegan: Community ecology package. Available at: http://CRAN.R-project.org/package=vegan
Pinto-Figueroa, E. A., Seddon, E., Yashiro, E., Buri, A., Niculita-Hirzel, H., van der Meer, J. R., et al. (2019). Archaeorhizomycetes spatial distribution in soils along wide elevational and environmental gradients reveal co-abundance patterns with other fungal saprobes and potential weathering capacities. Front. Microbiol. 10:656. doi: 10.3389/fmicb.2019.00656
Porter, D. L., Bradshaw, A. J., Nielsen, R. H., Newell, P., Dentinger, B. T. M., and Naleway, S. E. (2022). The melanized layer of Armillaria Ostoyae rhizomorphs: its protective role and functions. J. Mech. Behav. Biomed. Mater. 125:104934. doi: 10.1016/j.jmbbm.2021.104934
Purahong, W., Arnstadt, T., Kahl, T., Bauhus, J., Kellner, H., Hofrichter, M., et al. (2016). Are correlations between deadwood fungal community structure, wood physicochemical properties and lignin-modifying enzymes stable across different geographical regions? Fungal Ecol. 22, 98–105. doi: 10.1016/j.funeco.2016.01.002
Raaijmakers, J. M., Paulitz, C. T., Steinberg, C., Alabouvette, C., and Moënne-Loccoz, Y. (2009). The rhizosphere: a playground and battlefield for soilborne pathogens and beneficial microorganisms. Plant Soil 321, 341–361. doi: 10.1007/s11104-008-9568-6
Rosling, A., Cox, F., Cruz-Martinez, K., Ihrmark, K., Grelet, G. A., Lindahl, B. D., et al. (2011). Archaeorhizomycetes: unearthing an ancient class of ubiquitous soil fungi. Science 333, 876–879. doi: 10.1126/science.1206958
Schadt, C. W., Martin, A. P., Lipson, D. A., and Schmidt, S. K. (2003). Seasonal dynamics of previously unknown fungal lineages in tundra soils. Science 301, 1359–1361. doi: 10.1126/science.1086940
Selosse, M. A., Petrolli, R., Mujica, M. I., Laurent, L., Perez-Lamarque, B., Figura, T., et al. (2022). The waiting room hypothesis revisited by orchids: were orchid mycorrhizal fungi recruited among root endophytes? Ann. Bot. 129, 259–270. doi: 10.1093/aob/mcab134
Singh, B. K., Dawson, L. A., Macdonald, C. A., and Buckland, S. M. (2009). Impact of biotic and abiotic interaction on soil microbial communities and functions: a field study. Appl. Soil Ecol. 41, 239–248. doi: 10.1016/j.apsoil.2008.10.003
Sun, S. Q., Ma, Y. H., Meng, Q. J., Zhang, L. Q., Yang, Y., and Shi, J. G. (2009). Influences of wild, degradative, and rejuvenative Armillaria mellea on yield of Gastrodia elata and content of gastrodin. Chin. Tradit. Herb. Drug 40, 1300–1302.
Tamura, K., Stecher, G., and Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Tang, A., Jeewon, R., and Hyde, K. D. (2007). Phylogenetic relationships of Nemania plumbea sp. nov. and related taxa based on ribosomal ITS and RPB2 sequences. Mycol. Res. 111, 392–402. doi: 10.1016/j.mycres.2007.01.009
Tkacz, A., Cheema, J., Chandra, G., Grant, A., and Poole, P. S. (2015). Stability and succession of the rhizosphere microbiota depends upon plant type and soil composition. ISME J. 9, 2349–2359. doi: 10.1038/ismej.2015.41
Torsvik, V., and Vres, L. (2002). Microbial diversity and function in soil: from genes to ecosystems. Curr. Opin. Microbiol. 5, 240–245. doi: 10.1016/S1369-5274(02)00324-7
Veresoglou, S. D., Chen, B. D., and Rillig, M. C. (2012). Arbuscular mycorrhiza and soil nitrogen cycling. Soil Biol. Biochem. 46, 53–62. doi: 10.1016/j.soilbio.2011.11.018
Wang, W. X., Shi, J. C., Xie, Q. J., Jiang, Y. N., Yu, N., and Wang, E. T. (2017). Nutrient exchange and regulation in arbuscular mycorrhizal Symbiosis. Mol. Plant 10, 1147–1158. doi: 10.1016/j.molp.2017.07.012
Wang, Y., Wang, C. Y., Hou, J., Cheng, Z. J., Zhou, M. C., Zou, T., et al. (2020). Effect of different Armillaria mellea strains on agronomic traits, yield and quality of Gastrodia elata Bl. f. elata. J. Microbiol. 40, 59–65. doi: 10.3969/j.issn.1005-7021.2020.06.009
Werner, S., Peršoh, D., and Rambold, G. (2016). New aspects of the biology of Mortierella alliacea. Mycol. Prog. 15, 1293–1301. doi: 10.1007/s11557-016-1243-3
Xiao, H. F., Feng, Y. L., Schaefer, D. A., and Yang, X. D. (2014). Soil fungi rather than bacteria were modified by invasive plants, and that benefited invasive plant growth. Plant Soil 378, 253–264. doi: 10.1007/s11104-014-2040-x
Xu, Y. X., Lei, Y. T., Su, Z. X., Zhao, M., Zhang, J. X., Shen, G. J., et al. (2021). A chromosomescale Gastrodia elata genome and large-scale comparative genomic analysis indicate convergent evolution by gene loss in mycoheterotrophic and parasitic plants. Plant J. 108, 1609–1623. doi: 10.1111/tpj.15528
Yu, E., Gao, Y., Li, Y., Zang, P., Zhao, Y., and He, Z. (2022). An exploration of mechanism of high quality and yield of Gastrodia elata bl. f. Glauca by the isolation, identification and evaluation of Armillaria. BMC Plant Biol. 22, 1–18. doi: 10.1186/s12870-022-04007-8
Yuan, Y., Jin, X. H., Liu, J., Zhao, X., Zhou, J. H., Wang, X., et al. (2018). The Gastrodia elata genome provides insights into plant adaptation to heterotrophy. Nat. Commun. 9:1615. doi: 10.1038/s41467-018-03423-5
Yuan, Q. S., Xu, J., Jiang, W. K., Ou, X. H., Guo, L. P., Xiao, C. H., et al. (2020). Insight to shape of soil microbiome during the ternary cropping system of Gastradia elata. BMC Microbiol. 20:108 doi: 10.1186/s12866-020-01790-y
Keywords: high-throughput sequencing, symbiotic fungi, microbial community, Armillaria , Gastrodia elata Bl.
Citation: Wang Y, Xu J, Yuan Q, Guo L, Xiao C, Yang C, Li L, Jiang W and Zhou T (2023) Effect of symbiotic fungi-Armillaria gallica on the yield of Gastrodia elata Bl. and insight into the response of soil microbial community. Front. Microbiol. 14:1233555. doi: 10.3389/fmicb.2023.1233555
Received: 12 June 2023; Accepted: 21 August 2023;
Published: 07 September 2023.
Edited by:
Dilfuza Egamberdieva, Leibniz Center for Agricultural Landscape Research (ZALF), GermanyReviewed by:
Xiaojing Hu, Chinese Academy of Sciences, ChinaCopyright © 2023 Wang, Xu, Yuan, Guo, Xiao, Yang, Li, Jiang and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tao Zhou, dGFvemhvdTg4QDE2My5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.