- School of Biological Sciences and Biotechnology, Goa University, Taleigao, Goa, India
In Goa, salt production from the local salt pans is an age-old practice. These salt pans harbor a rich diversity of halophilic microbes with immense biotechnological applications, as they tolerate extremely harsh conditions. Detecting the existence of these microbes by a metabarcoding approach could be a primary step to harness their potential. Three salt pans viz. Agarwado, Curca, and Nerul adjoining prominent estuaries of Goa were selected based on their unique geographical locations. The sediments of these salt pans were examined for their bacterial community and function by 16S rRNA amplicon-sequencing. These salt pans were hypersaline (400–450 PSU) and alkaline (pH 7.6–8.25), with 0.036–0.081 mg/L nitrite, 0.0031–0.016 mg/L nitrate, 6.66–15.81 mg/L sulfate, and 20.8–25.6 mg/L sulfide. The relative abundance revealed that the Pseudomonadota was dominant in salt pans of Nerul (13.9%), Curca (19.6%), and Agarwado (32.4%). The predominant genera in Nerul, Curca, and Agarwado salt pan sediments were Rhodopirellula (1.12%), Sulfurivermis (1.28%), and Psychrobacter (25.5%) respectively. The highest alpha diversity (Shannon-diversity Index) was observed in the Nerul salt pan (4.8) followed by Curca (4.3) and Agarwado (2.03). Beta diversity indicated the highest dissimilarity between Agarwado and the other two salt pans (0.73) viz. Nerul and Curca and the lowest dissimilarity was observed between Nerul and Curca salt pans (0.48). Additionally, in the Agarwado salt pan, 125 unique genera were detected, while in Nerul 119, and in Curca 28 distinct genera were noted. The presence of these exclusive microorganisms in a specific salt pan and its absence in the others indicate that the adjacent estuaries play a critical role in determining salt pan bacterial diversity. Further, the functional prediction of bacterial communities indicated the predominance of stress adaptation genes involved in osmotic balance, membrane modification, and DNA repair mechanisms. This is the first study to report the bacterial community structure and its functional genes in these three salt pans using Next-Generation Sequencing. The data generated could be used as a reference by other researchers across the world for bioprospecting these organisms for novel compounds having biotechnological and biomedical potential.
Introduction
Salt production is an aged-old traditional method designed by farmers called Mittkars for over 150 decades. Salt pans are habitats inhabiting a huge community of extremophiles, thriving in extreme salinity, temperature, pH, and minimum nutrient availability (Mani et al., 2012). Halophilic bacteria from these environments have developed strategies and cellular systems that could cope with salt stress. Salt-in and salt-out are the two major strategies observed in halophiles to escape stress. In the salt-in strategy, the intracellular system is adapted to the salt concentration of the surrounding but the cellular energy consumed here is huge. Whereas, in salt-out strategies, organisms avoid adaptation and thereby energy loss by an accumulation of organic compatible solutes, also called a compatible solute strategy (Oren, 1999).
Halophilic bacteria from Goan salt pans are widely studied as a potent source of bioactive compounds (Tonima and Savita, 2011; Fernandes et al., 2019). The potential microorganisms in the salt pans of Goa have attracted the attention of the public as well as the researchers to conserve these habitats, mittkars, and their traditional knowledge of salt making. The culture-dependent approach examines a very minuscule microbial population. Modern methods such as high throughput sequencing and in silico analysis allow us to fill this gap (Janda and Abbott, 2007). The bacterial diversity of Agarwado, Curca, and Nerul salt pans and its comparative analysis has not been elucidated previously by next-generation sequencing. Hence, the present study is the first report accomplished to understand the diversity, distribution, and abundance with a comparative analysis of the bacterial community in these salt pans.
Materials and methods
Sample collection
Sediment samples were collected during the salt harvesting period from the crystallizer pond using a 5.5 cm diameter corer in sterile bags from the salt pans of Curca village (N15.45798, E73.88065) situated at Tiswadi taluka, Agarwado village (N15.640978, E73.76426) situated at Pernem taluka and Nerul village (N15.51979, E73.78909) situated at Bardez taluka (Figures 1A–E). Sediment sample refers to the sediments collected about 2 cm from a surface layer of salt pan. The sediment samples collected in triplicates from each salt pan site were mixed to obtain a single sediment sample. Sediment samples from Agarwado, Curca, and Nerul salt pans were indicated as AC, CC, and NC, respectively. Samples were transported in ice-cold condition to the laboratory and stored at −20°C until further processing. The temperature was recorded on-site using a thermometer of the three salt pan sediment samples. The pore water obtained after centrifugation of sediment samples was used to measure pH, salinity, nitrate, nitrite, sulfate, and sulfide. The pH was recorded by pH meter (Eutech, Germany) and the salinity was measured by refractometer (ATAGO, Japan). The nitrite and nitrate were estimated spectrophotometrically according to APHA (2012). Sulfate was estimated turbidometrically by measuring the precipitation of BaSO4 (Clesceri et al., 1998). For the estimation of sulfide, the sample was fixed in 2% zinc acetate and was further analysed spectrophotometrically (Cline, 1969).

Figure 1. Goan Salt pans in study (A) Location of Agarwado, Curca, and Nerul Salt pans in Goa. Aerial image of (B) Agarwado salt pan (C) Curca salt pan (D) Nerul salt pan [Source: Google earth pro, 2023] (E) Image of crystallizer pond under solar evaporation at Agarwado salt pan.
Environmental DNA extraction, library preparation, and Illumina sequencing
Environmental DNA was extracted from 1 g of each sediment sample using DNeasy PowerSoil Kit (Qiagen). The DNA obtained was checked for its concentration and quality using NanoDrop. The amplicon library was prepared using Nextera XT Index Kit (Illumina inc.) using bacterial specific forward primer 341F (GCCTACG GGNGGCWGCAG) and reverse primer 805R (ACTACHV GGGTATCTAATCC) for V3-V4 hypervariable region of 16S rRNA gene followed by adding Illumina adapters required for cluster generation. Amplicon libraries were purified by AMPureXP beads and quantified using a Qubit Fluorometer. Libraries were then loaded on MiSeq at 10–20 pM concentration for cluster generation and sequencing. The libraries were sequenced at Eurofins Genomics India Pvt. Ltd., Karnataka, India on the Illumina MiSeq platform using 2×300 bp chemistry to obtain paired-end reads.
Sequence analysis by bioinformatics
Raw paired-end reads of AC, CC, and NC were analysed by Quantitative Insights Into Microbial Ecology 2 (QIIME 2), version 2–2023.7 (Bolyen et al., 2019). Reads obtained after sequencing were denoised via the q2-dada2 plugin (Callahan et al., 2016) which involves correction of Illumina sequenced errors, merging of reverse and forward paired-end reads, and clustering them into ASVs (amplicon sequence variants). The rarefaction curve was plotted using QIIME. ASVs obtained were further taxonomically classified into different taxonomic levels using the q2-feature-classifier plugin based on the SILVA (V.138) reference database (Quast et al., 2012). The Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUST2) method (Douglas et al., 2020) was utilized to predict the functional potential of the bacterial communities in the three salt pans mentioned above. The relative abundance of functional protein profiles in the three salt pans was represented in a heatmap constructed using PRIMER7.
Statistical analysis
The alpha (intra-sample) diversity indices [such as Simpson index, Shannon index, Menhinick’s richness index, and Pielou’s evenness (equitability)] were calculated at the genus level for the three sampling sites, similarly the beta diversity was measured as described by Whittaker (1960). Canonical Correspondence analysis (CCA) was done to understand the influence of environmental variables [Temperature (°C), pH, salinity (PSU), nitrate (mg/L), nitrite (mg/L), sulfate (mg/L), sulfide (mg/L)] on bacterial genera. Alpha diversity, beta diversity and CCA analysis were performed by using Paleontological statistics software (PAST), version 4.13 (Hammer et al., 2001).
Results
Sediment sample analysis
The physical parameters of sediment samples collected from Agarwado (AC), Curca (CC), and Nerul (NC) salt pans possessed temperatures of 35°C (in CC) and 36°C (AC and NC). Salinity observed was 450 PSU in AC and 400 PSU in CC and NC whereas the pH of the sediment samples was found to be 8.25 (AC), 7.6 (CC), and 8 (NC). The environmental DNA extracted from sediment samples AC, CC, and NC were measured by Nanodrop for its concentration and was found to be 28.6 ng/μl, 40.2 ng/μl, and 33.2 ng/μl, respectively.
Relative abundance of bacterial community
A total of 95,299, 104,637, and 240,563 reads were obtained in sediment samples of the Agarwado (AC), Curca (CC), and Nerul (NC) salt pan, respectively. The rarefaction curve of all three samples reached a plateau at this sequencing depth (Figure 2) indicating the information contained in the three samples captured the majority of abundant phylotypes.
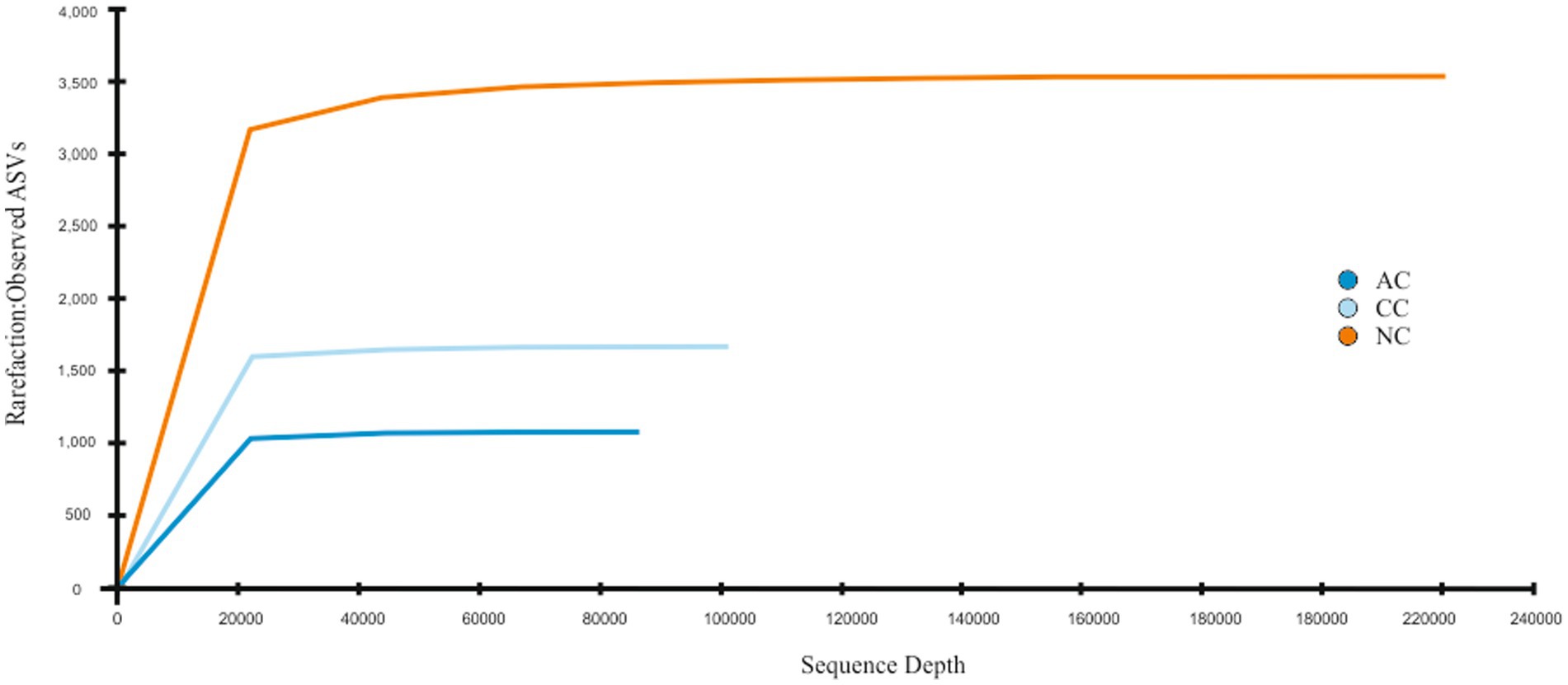
Figure 2. Rarefaction curve: Observed ASVs vs. Sequencing depth for three salt pan samples namely Agarwado salt pan (AC), Curca salt pan (CC), and Nerul salt pan (NC) of Goa, India.
In the bacterial community analysis of the AC sample, 42 phyla and 201 genera were identified. The abundant phyla such as Pseudomonadota (33%), Bacillota (22%), and “Candidatus Patescibacteria” (9.4%), were observed (Figure 3A). Major contribution at the class level was by Gammaproteobacteria (30.7%), and Bacilli (20.7%). At the order level, Pseudomonadales, and Bacillales were found to be abundant with 26.8 and 19.4% relative abundance, respectively. Predominant taxa observed at the family level were Moraxellaceae (26.5%) and Planococcaceae (18.9%) and the major identified genera were Psychrobacter (26%), Halanaerobium (3.4%), and Antarcticibacterium (1.5%) (Figure 3B).
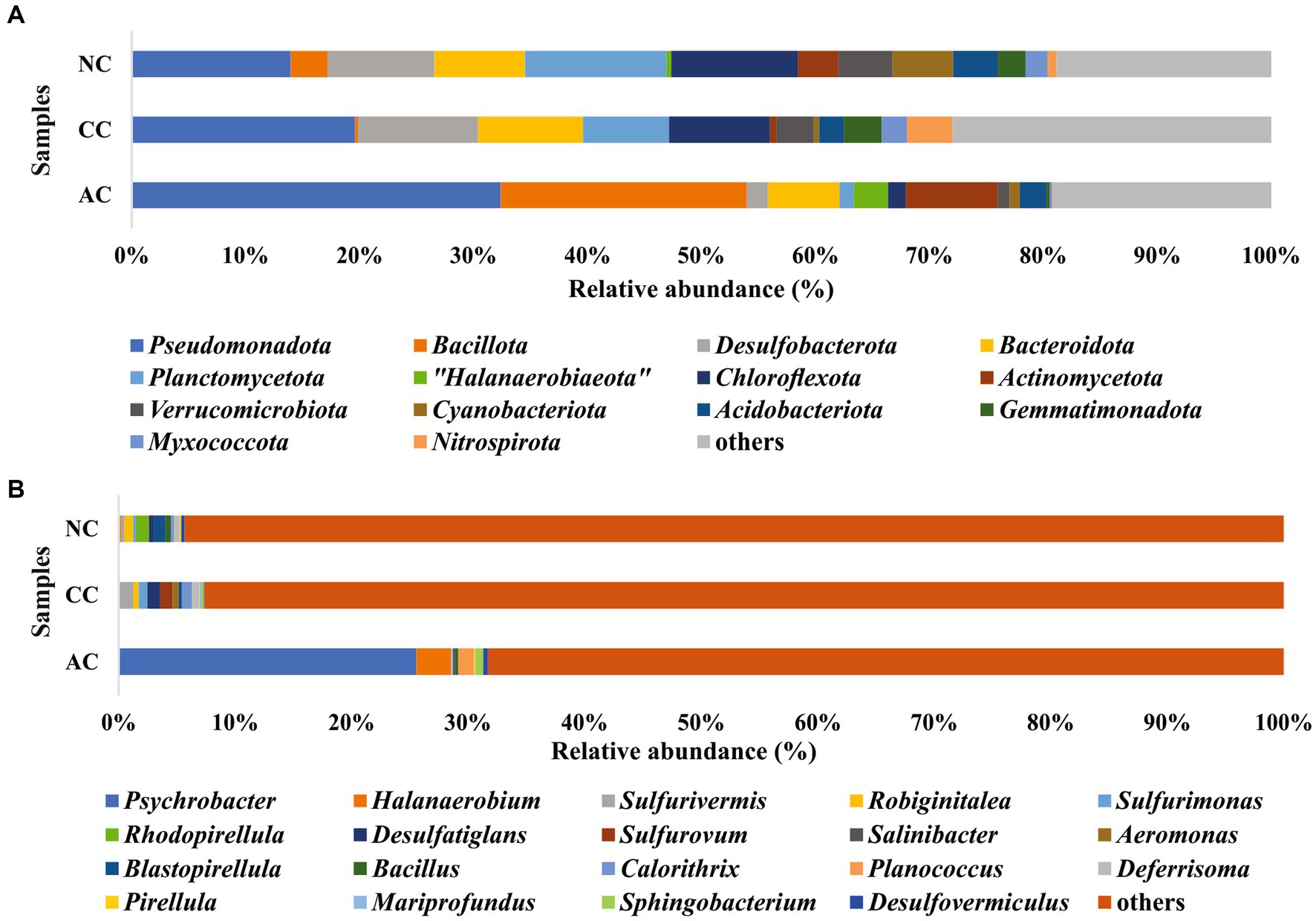
Figure 3. Relative abundance of bacterial communities at (A) phyla level and (B) genus level in sediment samples of Agarwado salt pan (AC), Curca salt pan (CC), and Nerul salt pan (NC) of Goa, India.
In the CC sample, 50 phyla and 143 genera were noted. The predominant phyla that contributed to bacterial diversity include Pseudomonadota (20.8%), Desulfobacterota (11.2%), and Bacteroidota (9.8%), (Figure 3A). At the class level, Gammaproteobacteria (17%) and Bacteroidia (8.4%) were found dominant. At the order level, the predominance of Desulfobulbales (5%) and Desulfobacterales (2.8%) were detected. At the family level, Desulfobulbaceae (3.9%) was relatively abundant. Lastly, at the general level, Sulfurivermis (1.3%) and Desulfatiglans (1.1%) presented a major proportion (Figure 3B).
In the NC sediment sample, 53 phyla and 250 genera were recognized. The dominant bacterial phyla include Pseudomonadota (14.7%), Planctomycetota (13%) and Chloroflexota (11.7%) (Figure 3A). The abundant classes observed were Anaerolineae (10.7%) and Gammaproteobacteria (9%). At the order level, Pirellulales with 6.5% relative abundance was found to be the highest followed by Desulfobacterales (3.8%). At the family level, Pirellulaceae (6.5%), was the major family obtained. Further, the most abundant genus identified was Rhodopirellula (1.2%) (Figure 3B).
Correlation of bacterial genera with environmental parameters
The effect of environmental parameters such as temperature, pH, salinity, nitrite, nitrate, sulfate, and sulfide were assessed on the bacterial genera using CCA. The CCA tripod explained 88.67% of the variation in bacterial genera by environmental parameters (Figure 4). The result revealed the positive correlation between genera Blastopirellula, Sva0081 sediment group, Rhodopirellula, Robiginitalea, Pirellula and Bacillus with sulfide and sulfate. However, genera Psychrobacter, Halanaerobium, Planococcus and Sphingobacterium found abundant in AC were positively correlated with nitrite, nitrate, and salinity. Of all the environmental parameters sulfide and sulfate contributed significantly to bacterial genera in NC.
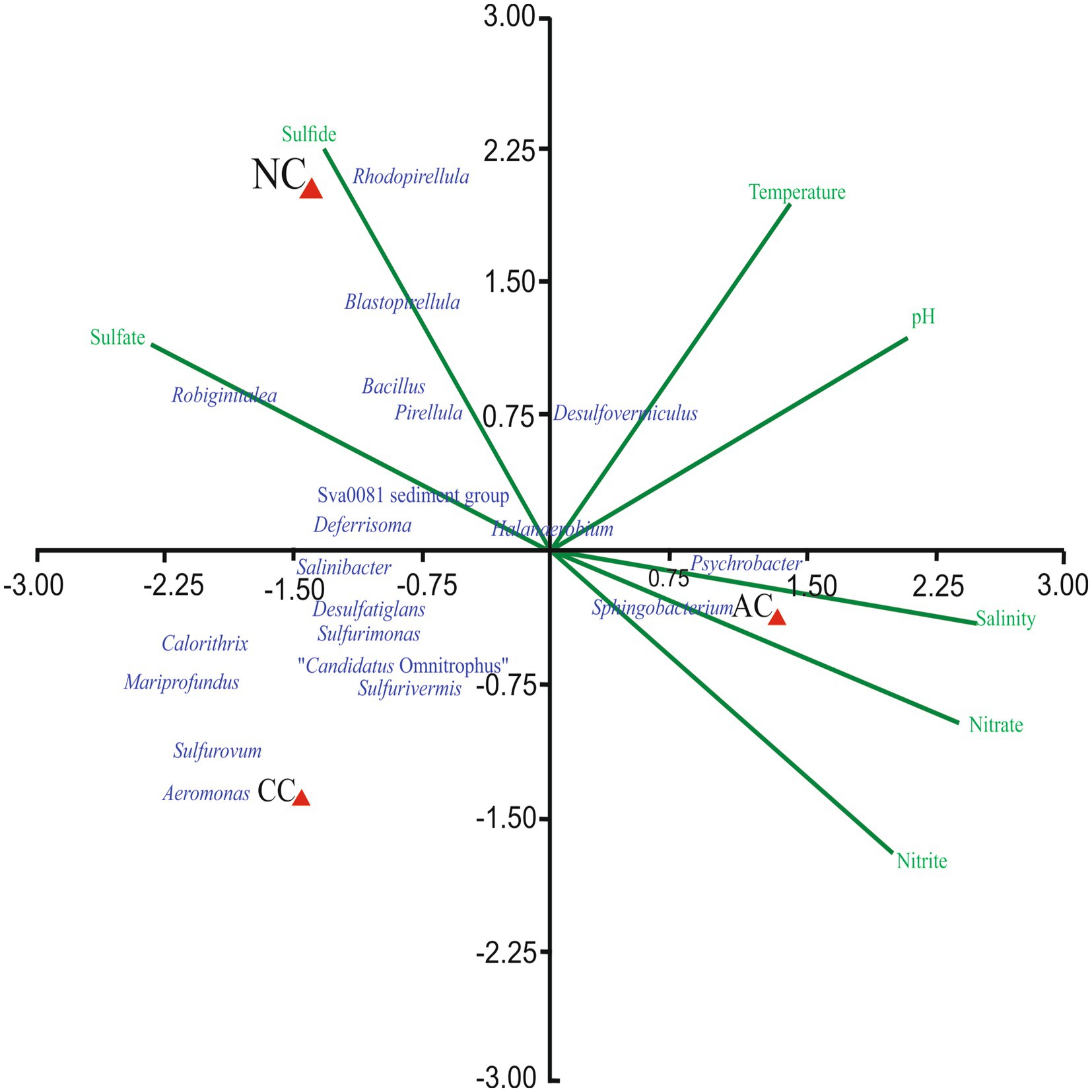
Figure 4. Canonical Correspondence Analysis ordination diagram of bacterial communities at generic-level at Agarwado salt pan (AC), Curca salt pan (CC), and Nerul salt pan (NC) of Goa, India associated with environmental parameters.
Alpha and beta diversity analysis at the genus level
The alpha diversity indices such as Simpson diversity index, Shannon diversity index (Shannon-Wiener index), Dominance, Menhinick’s richness index, and Pielou’s evenness (equitability) were calculated and depicted in Table 1. Similarly, the beta diversity and its pair-wise comparison presented in Table 2 were performed using the mathematical expression S/ā-1, where S is the total number of genera and ā is the average number of genera. The beta diversity was found highest and equal between AC and NC and AC and CC, i.e., 0.73. The lowest beta diversity was observed between CC and NC (0.48).
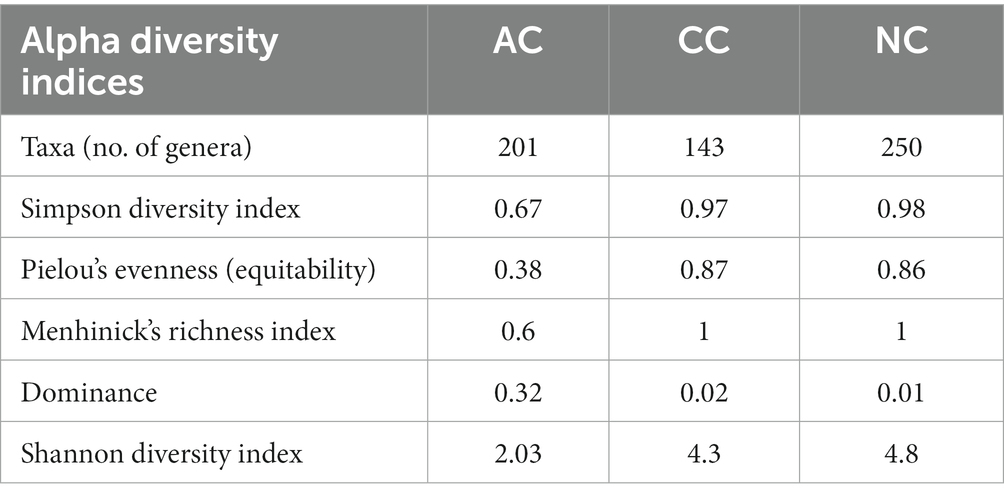
Table 1. Alpha diversity indices in Agarwado salt pan (AC), Curca salt pan (CC) and Nerul salt pan (NC) of Goa, India at genera level.
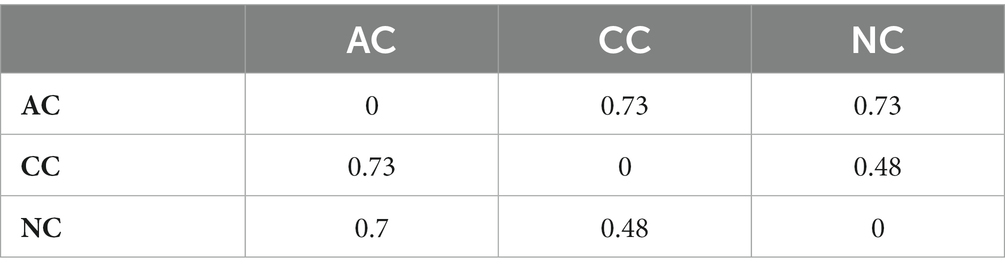
Table 2. Pair-wise comparison of beta diversity between Agarwado salt pan (AC), Curca salt pan (CC), and Nerul salt pan (NC) of Goa, India at the genus level measured by Whittaker using the mathematical expression S/ā-1, where S is the total number of genera and ā is the average number of genera.
Unique and common bacterial communities among the salt pans
The Venn diagram (Figure 5A) represents the number of unique and common phyla in three salt pans. An acceptably small percentage of unique taxon was observed in AC, CC, and NC with 14.2, 6, and 5.6%, respectively. Contrastingly, the common taxa observed in all three salt pans were in a higher proportion ranging from 62 -78%. There were no common phyla found in CC and AC. The phyla found common among all three salt pans are listed in the Supplementary Table S1.
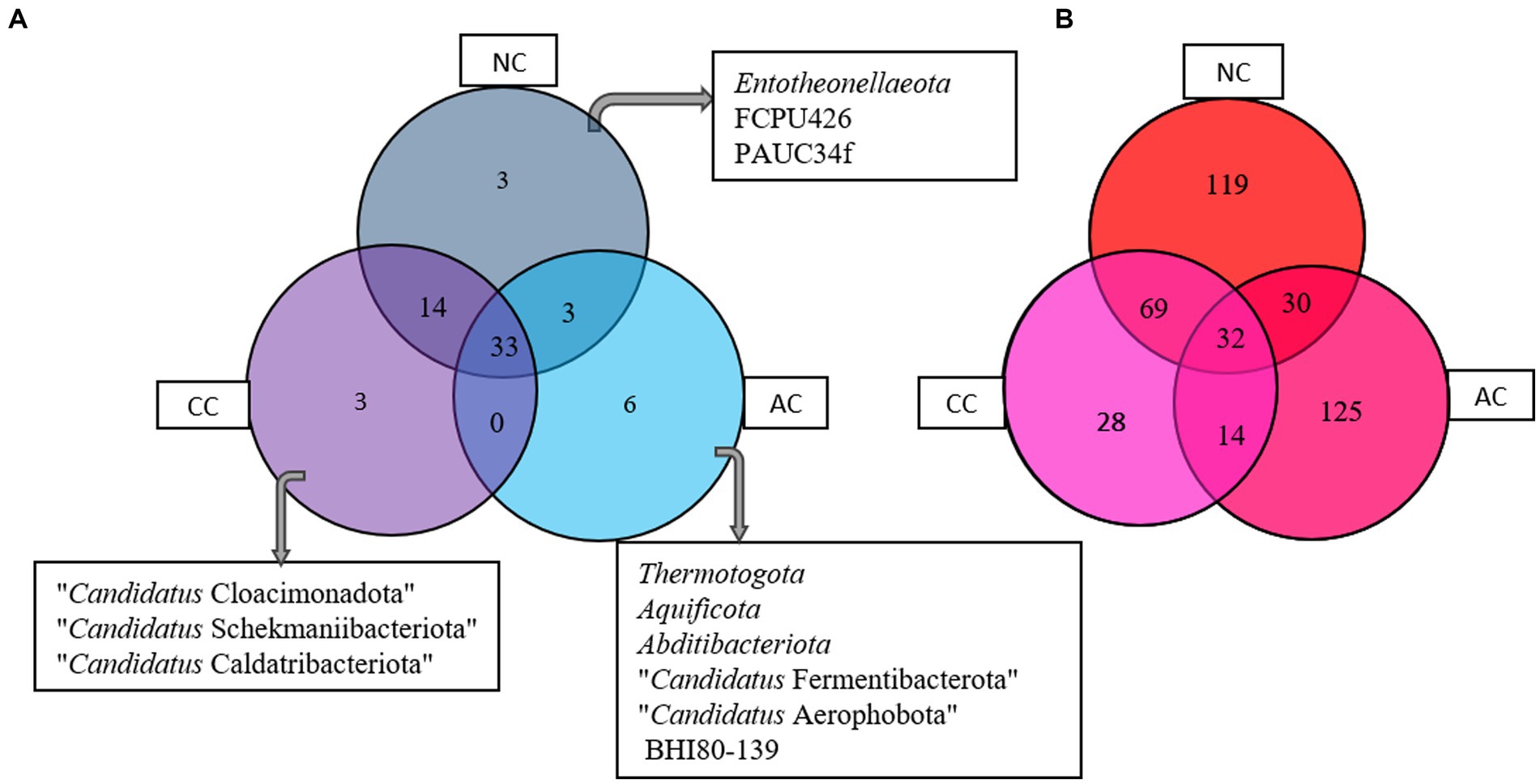
Figure 5. Venn diagram showing common and unique bacterial taxa at (A) phyla and (B) genus level between Agarwado salt pan (AC), Curca salt pan (CC), and Nerul salt pan (NC) of Goa, India.
At the genera level, a total of 201, 143, 250 genera were observed in AC, CC, and NC, respectively. Although the number of genera was found highest in NC, the percentage of exclusivity was observed high in AC (61%) > NC (47%) > CC (19.4%). The common genera found in all three salt pans ranged between 13 - 23%. The Venn diagram (Figure 5B) depicts the number of unique and common genera in the three salt pans and the genera are listed in the Supplementary Table S2.
Rare taxa in the three salt pans
Rare taxa are defined as those having a relative abundance of ≤0.2% (Hugoni et al., 2013). At the phylum level, rare phyla such as Bdellovibrionota (0.06%), Nitrospirota (0.07%) and “Candidatus Sumerlaeota” (0.07%) were found in AC. In CC, “Candidatus Moduliflexota,” Fusobacteriota, and “Candidatus Acetithermota” are rare taxa with 0.04, 0.07, and 0.06% relative abundance, respectively. Further in NC, “Candidatus Babelota,” Elusimicrobiota, and Deinococcota are a few minor phyla with 0.11, 0.08, and 0.081% relative abundance, respectively. Secondly, “Candidatus Dadaibacteriota” was the only rare (≤ 0.2%) bacterial phyla found common in all three salt pans. Further, at the genus level only 11 taxa were detected common in all three salt pans (AC, CC, NC) with ≤0.2% of relative abundance (Figure 6).
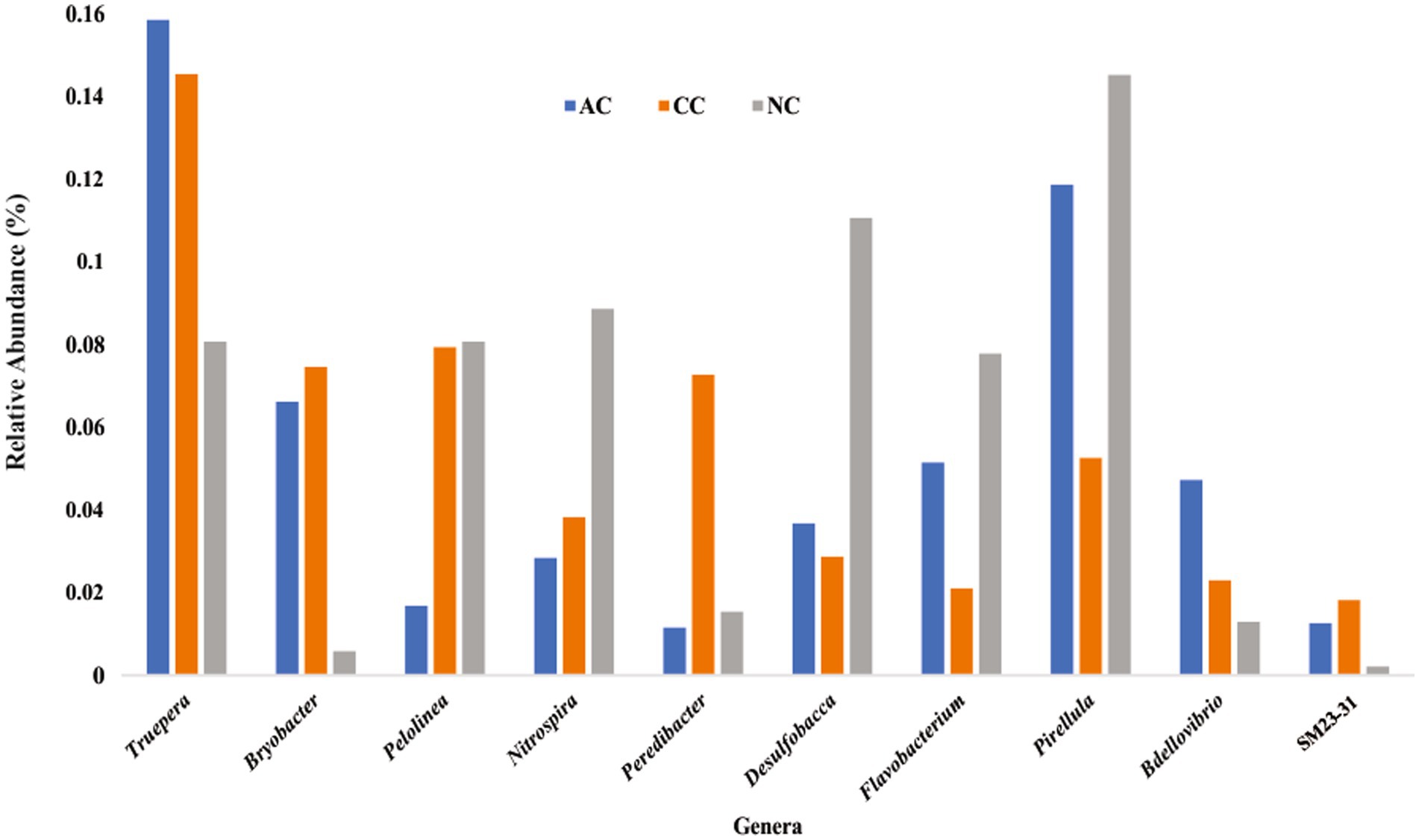
Figure 6. Rare bacterial genera (≤ 0.2% relative abundance) found common in all three sediment samples of Agarwado salt pan (AC), Curca salt pan (CC), and Nerul salt pan (NC) of Goa, India.
Functional gene prediction in three salt pans
A large number of functional genes involved in various biological processes, including cellular metabolism and adaption to extreme environmental conditions, have been found in the indigenous bacterial communities of all the three salt pans. Figure 7 depicts the most abundant genes that were found in all the three salt pans. Further, numerous genes involved in the nitrogen, sulfur, and carbon cycles have also been found to be part of the overall functional gene profile (Supplementary Figures S1–S3).
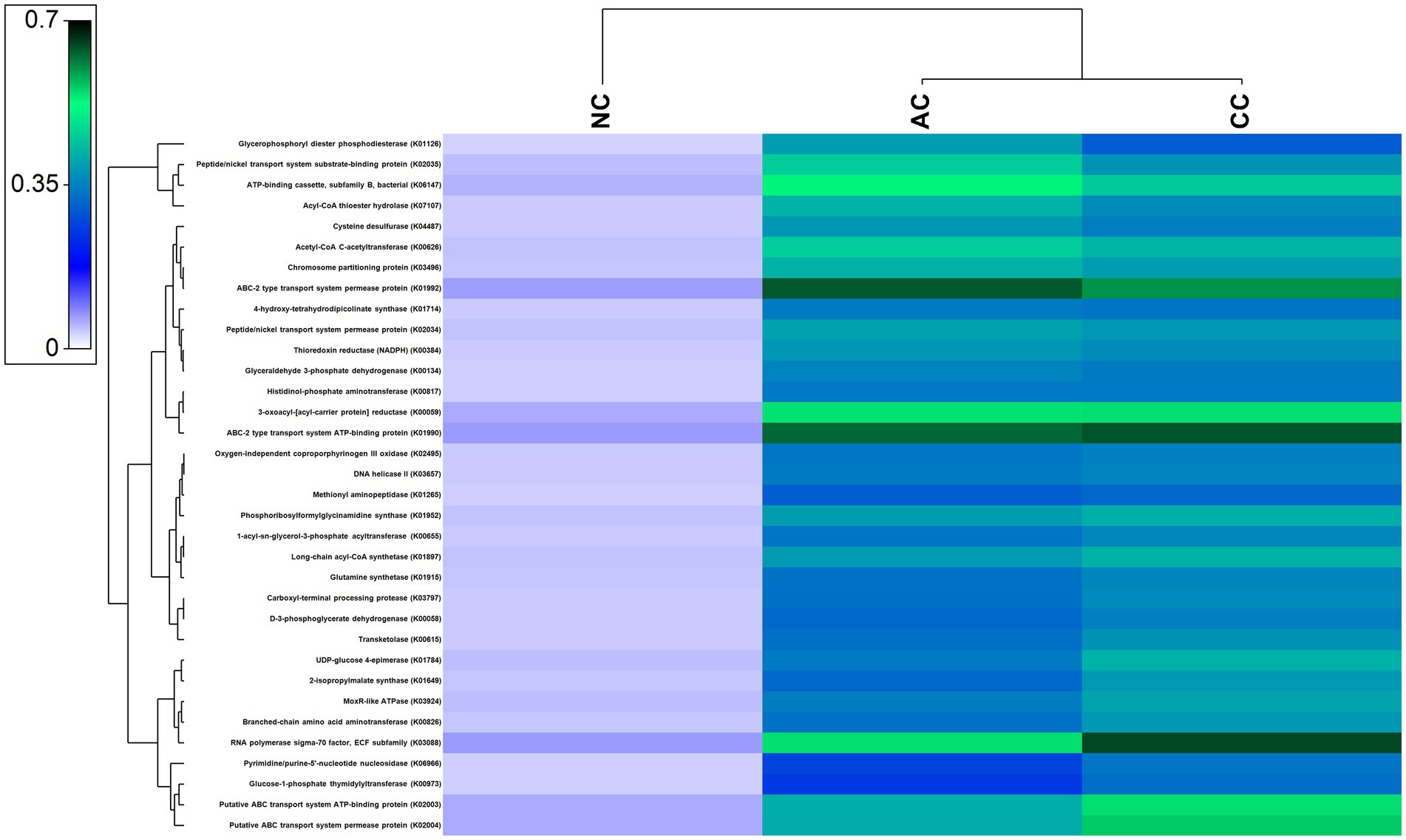
Figure 7. Heatmap showing the abundant functional genes predicted by using PICRUST2 algorithm in the sediment samples of Agarwado salt pan (AC), Curca salt pan (CC), and Nerul salt pan (NC) of Goa, India.
Discussion
Traditional salt-making is one of the major and unique occupations of Goa due to its lengthy coastline of 107 Km. The three study sites viz. Agarwado, Curca, and Nerul salt pans were selected based on their unique geographical locations. The Agarwado salt pan is in Pernem taluka adjoining the pristine Chapora estuary in the extreme north of Goa. The Nerul and Curca salt pans are connected to the two major lifeline estuaries of Goa, i.e., Mandovi in Bardez taluka and Zuari in Tiswadi taluka, respectively. Seawater from the estuary gushes into the salt pans through a sluice gate during high tide and is evaporated to produce salt. For decades, these three distinct salterns have been conventionally used to produce local salt by traditional methods in Goa. The temperature of 35°C - 36°C, noted from these salt pans during the salt harvesting period favors the evaporation process in the crystallizer pond to achieve the salinity required for salt production. The salinity of these salt pans was found to be in the range of 400–450 PSU and pH 7.6–8.25 indicating a halophilic and alkaline nature.
Comparative analysis of the bacterial community in the salt pan
The bacterial composition of the salt pans of Goa revealed that the three sediment samples of salt pans were dominated by phylum Pseudomonadota. These results were parallel to the work reported by (Gorrasi et al., 2021) in other solar salterns. Within the Pseudomonadota phylum in the AC salt pan, only two classes such as Gammaproteobacteria and Alphaproteobacteria were observed. In addition to these two classes, Zetaproteobacteria was identified in CC and NC. Interestingly the only genera found in our study within the Zetaproteobacteria class was Mariprofundus. This class of bacterium is a model organism for microbial iron oxidation in marine environments (McAllister et al., 2019). The detection of genera Mariprofundus in CC and NC perhaps originated from the mining activities at Mandovi estuary which has deposited several tonnes of iron ore. Secondly, the Cumbarjua Canal interconnects the Mandovi and Zuari estuaries (adjacent to NC and CC respectively) and forms a network that has served as a major route for the transportation of iron ore. Additionally, the discharge of mining rejects from the mining sites has also concentrated iron particles in these estuaries (Gauns, 2011). This demonstrates how the distribution of microorganisms in a particular area is affected by their chemical composition. Similarly, other environmental parameters like salinity, nitrite, nitrate, sulfate, and sulfide also have an influence on bacterial distribution and abundance as depicted by CCA. The dominant phyla observed in our study such as Pseudomonadota, Bacteriodota, Chloroflexota, etc. are found to have ubiquitous genetic machinery and survival weapons (Mehta et al., 2021). Hence, understanding the microbiome composition, evolving genetic machinery, and adaptive mechanisms evolved by these extremophiles make it important in biotechnology industries. The genera Psychrobacter, belonging to the family Moraxellaceae, was the most abundant genera and family, respectively, in the AC with approximately 25.5% relative abundance. Like our studies, the presence of Psychrobacter was also reported in the solar saltern of Bohai Bay, China, revealed by DGGE of the 16S gene (Zhang et al., 2016). The isolation of sulfidogenic bacteria Halanaerobium, reported from the Ribandar salt pan of Goa was observed to be the second abundant genera in AC (Das et al., 2017). The abundant genera observed in the CC and NC salt pans were Sulfurivermis and Rhodopirellula, respectively. The presence of Sulfurivermis shows the occurrence of active sulfur cycles in the salt pan being a sulfur oxidizing bacterium (SOB) that uses nitrate as an electron acceptor in the rTCA pathway (Kojima et al., 2017).
Alpha and Beta diversity analysis of the salt pans
The diversity of any ecosystem depends on two components “richness and evenness” (Soininen et al., 2012). In the Menhinicks richness index, an equal richness value was observed for NC and CC and was higher in comparison to AC, evident for higher community richness in Nerul and Curca salt pans. Similarly, the Pielou’s evenness index for NC and CC was found to be higher as compared to AC, which indicates the distribution of an equal number of taxa in both salt pan (NC and CC) ecosystems than in the Agarwado salt pan. Further, dominance in AC was higher than NC and CC, demonstrating the presence of a more dominant taxon in the Agarwado salt pan community. Lower evenness and higher dominance observed in AC indicate lower alpha diversity. However, in Nerul and Curca salt pans high evenness and lower dominance indicate high alpha diversity. A similar observation was shown by the Shannon diversity index which considers both the richness and evenness of a community. The alpha diversity measured by the Shannon index was NC > CC > AC.
Interestingly equal dissimilarity diversity (beta diversity) was observed between AC and NC and AC and CC, which further indicated that both CC and NC are quite comparable to each other but highly divergent to AC. The similarity between NC and CC was further confirmed by the presence of high number of common genera in both (represented in Figure 5B) in addition to the beta diversity. This is due the adjoining estuaries at Nerul and Curca salt pans that are connected by the Cumbarjua canal and therefore share uniform environmental characteristic (Qasim and Gupta, 1981) influencing bacterial composition.
Bacterial taxa in ecological roles
Pseudomonadota, Planctomycetota, and Chloroflexota observed in the three salt pans are representatives of the commonly occurring phyla that are involved in a variety of ecological roles. The sulfur cycle advances in tandem with the carbon cycle and is most prevalent in hypersaline environments. Using sulfate as an electron acceptor, the sulfate-reducing bacteria break down organic molecules in an anoxic environment thereby cycling carbon anaerobically (Kerkar and Bharathi, 2007). In the current anticipated functional metagenome, genes involved in assimilatory and dissimilatory sulfate reduction and oxidation of sulfur compounds were found. The reductive nature of the salt pans sulfur cycle in the current work is indicated by the presence of the genera Desulfovermiculus and Desulfatiglans (Begmatov et al., 2021) in addition to genes such as adenylylsulfate reductase, subunits A (K00394), adenylylsulfate reductase, subunits B (K00395), dissimilatory sulfite reductase alpha subunit (K11180), dissimilatory sulfite reductase beta subunit (K11181), and sulfate adenylyltransferase (K00958). Likewise, different genes of the Sox system that are involved in sulfur oxidation: SoxA (K17222), SoxX (K17223), SoxB (K17224), SoxY (K17226), SoxZ (K17227), SoxC (K17225) and Sulfane dehydrogenase subunit were observed in the functional gene prediction (Supplementary Figure S3). Thiohalospira is a genus of halophilic, chemolithoautotrophic SOB that utilizes sulfur compounds as electron donors (Sorokin et al., 2020); it has only been found in the Agarwado (AC) salt pan. Secondly, the SOB genus, Sulfuriflexus (class: Gammaproteobacteria) (Kojima and Fukui, 2016; Rana et al., 2020), was exclusive to Curca (CC) sediments. Sphingobacterium and Woeseia functional in carbon-fixing (Lohmann et al., 2020) were also observed in these salt pans along with numerous genes required in carbon metabolism (Supplementary Figure S1). Unique to the Nerul (NC) salt pan, a group at genus level Cm1-21 (family: Nitrosococcaceae) was recently reported as ammonium oxidizers involved in the nitrification process of the nitrogen cycle (Semedo et al., 2021). Besides functional genes (Supplementary Figures S1–S3), bacterial genera like Desulfovibrio (observed in AC and NC sediment samples), Pseudomonas (AC and CC), Nitrospina (CC and NC), and Nitrospira (AC, CC, and NC) noted in our study are implicated in the carbon, nitrogen, and sulfur cycles (Megonigal et al., 2003; Meng et al., 2022; Sun et al., 2022), which illustrates that the movement of nutrients necessary to support the persistent life in these man-made salt pan ecosystems are governed by the presence of its indigenous microbes.
Significance of halophiles
Salinity is a key factor that governs the microbial community in hypersaline ecosystems such as solar saltern. High salinity imposes stress on microbial cells, challenging their survival (Fei Xi et al., 2014; Song et al., 2022). Numerous genes associated with osmoprotective mechanisms, such as salt-in and salt-out mechanisms, membrane modification, protein adaptation, DNA repair, etc. acquired by residents of hypersaline environment for normal growth and development of cells were observed to be prevalent in our study. These genes include ABC-2 type transport system permease protein (K01992), ABC-2 type transport system ATP-binding protein (K01990), ATP-binding cassette, subfamily B, bacterial (K06147), 1-acyl-sn-glycerol-3-phosphate acyltransferase (K00655), 4-hydroxy-tetrahydrodipicolinate synthase (K01714), and DNA helicase II / ATP-dependent DNA helicase PcrA (K03657) (Sleator and Hill, 2002; Yu et al., 2018; Schmitz et al., 2020).
Salt pans in Goa are influenced by three seasons: pre-monsoon, monsoon, and post-monsoon, where salinity ranges from 30–300 PSU (Mani et al., 2012). Therefore, bacteria with different salt tolerance like slightly, moderately, and extremely halophilic bacteria are expected to inhabit this ecosystem. Bacteria within the moderately halophilic group are very important as they occupy a large proportion of halophiles. As adaptive measures, the production of compatible solutes and hydrolytic enzymes to acclimatize to salinity variations is attracting industries (Baati et al., 2010; Ramos-Cormenzana, 2020). For example, the moderate halophilic genus Micrococcus synthesizes catalase that plays a role in maintaining colour and flavor in cured meats (Onishi and Kamekura, 1972; García-López et al., 1999). Secondly, Rhodopirellula, an abundant genus observed in NC sample is a moderate halophile and was reported to have a role as a food supplement to Daphina magna (indicator used for testing toxicity of water) (da Conceição Marinho et al., 2019). Therefore, profound knowledge and discovery of halophilic bacteria in such an ecosystem will help us to disclose numerous properties, enzymes, and proteins of halophiles that could be substantially important to different biotechnological industries.
Conclusion
Salt pans being polyextremophilic, are dynamic habitats of the resident bacteria potentially capable of tolerating extremely stressful environmental factors, i.e., high salinity (30–450 PSU), high alkalinity (pH 7.6–8.25) and fluctuating temperatures (20°C – 45°C). This is the first study profiling the bacterial communities in the Agarwado, Curca, and Nerul salt pans of Goa. The data reveals that the highest bacterial genera prevail in the Nerul salt pan sediment (250) as compared to Agarwado (201) and Curca (143). However, in the sediments of Agarwado salt pan, 125 distinct genera were found, followed by Nerul (119) and Curca (28) which were exclusively unique to these salt pans. These hypersaline natives are resistant to osmotic stress, demonstrated by the presence of genes in the predicted functional gene profile, that allows them to actively participate in the carbon, nitrogen, and sulfur cycles in these hypersaline environments. A wide microbial community and its function remains unknown, as “unculturable” as it is impractically impossible to mimic these diverse physical, chemical, biological, and mechanical factors which contribute to simulating the salt pan ecosystem in a laboratory. The high throughput sequencing has made it possible to provide an insight of the bacterial community and most of its functions in the salt pans, which could not be explored earlier. Further, a deep understanding of their functional mechanisms in this habitat will help us to assess the existence of these unique bacterial communities, and the adaptive cellular machinery/ biomolecules involved in making this hypersaline environment a conducive environment to thrive in.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
PG: writing original draft preparation, methodology, and software analysis. SK: supervision, experimental design, review and editing. All authors contributed to the article and approved the submitted version.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by a Junior Research Fellowship for a Ph.D. scholar (award letter number: 19/06/2016(i) EU-V), funded by University Grant Commission (UGC), New Delhi, India.
Acknowledgments
The authors are thankful to the School of Biological Sciences and Biotechnology (Discipline of Biotechnology) and Goa Business School (Discipline of Computer Science and Technology), Goa University for the necessary facilities. We are grateful to Venkatachalam S., Project Scientist II at NCPOR, Goa, India for the technical assistance provided.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1230929/full#supplementary-material
References
APHA (2012). Standard methods for the examination of water and waste water. 22 American Public Health Association, American Water Work Association, Water Environment Federation. Washington
Baati, H., Amdouni, R., Gharsallah, N., Sghir, A., and Ammar, E. (2010). Isolation and characterization of moderately halophilic Bacteria from Tunisian solar Saltern. Curr. Microbiol. 60, 157–161. doi: 10.1007/s00284-009-9516-6
Begmatov, S., Savvichev, A. S., Kadnikov, V. V., Beletsky, A. V., Rusanov, I. I., Klyuvitkin, A. A., et al. (2021). Microbial communities involved in methane, sulfur, and nitrogen cycling in the sediments of the Barents Sea. Microorganisms 9:2362. doi: 10.3390/microorganisms9112362
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Clesceri, L. S., Greenberg, A. E., and Eaton, A. D. (1998). Standard methods for examination of water and wastewater. American Public Health Association, Washington.
Cline, J. D. (1969). Spectrophotometric determination of hydrogen sulfide in natural waters1. Limnol. Oceanogr. 14, 454–458. doi: 10.4319/lo.1969.14.3.0454
da Conceição Marinho, M., Lage, O. M., Sousa, C. D., Catita, J., and Antunes, S. C. (2019). Assessment of Rhodopirellula rubra as a supplementary and nutritional food source to the microcrustacean Daphnia magna. Antonie Van Leeuwenhoek 112, 1231–1243. doi: 10.1007/s10482-019-01255-x
Das, K. R., Kerkar, S., Meena, Y., and Mishra, S. (2017). Effects of iron nanoparticles on iron-corroding bacteria 7, 385. doi: 10.1007/s13205-017-1018-9
Douglas, G. M., Maffei, V. J., Zaneveld, J. R., Yurgel, S. N., Brown, J. R., Taylor, C. M., et al. (2020). PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 38, 685–688. doi: 10.1038/s41587-020-0548-6
Fei Xi, X., Wang, L., Jun Hu, J., Shu Tang, Y., Hu, Y., Hua Fu, X., et al. (2014). Salinity influence on soil microbial respiration rate of wetland in the Yangtze River estuary through changing microbial community. J. Environ. Sci. 26, 2562–2570. doi: 10.1016/j.jes.2014.07.016
Fernandes, S., Kerkar, S., Leitao, J., and Mishra, A. (2019). Probiotic role of salt Pan Bacteria in enhancing the growth of Whiteleg shrimp, Litopenaeus vannamei. Probiotics Antimicrob Proteins 11, 1309–1323. doi: 10.1007/s12602-018-9503-y
García-López, M.-L., Santos, J.-Á., and Otero, A. (1999). MICROCOCCUS. Encyclopedia of Food Microbiology (Elsevier). 20, 1344–1350. doi: 10.1006/rwfm.1999.1025
Gauns, R. (2011). Mining havoc: Impact of mining on water resources in Goa - article from dams, Rivers, and People. SANDRP, 10–12.
Gorrasi, S., Franzetti, A., Ambrosini, R., Pittino, F., Pasqualetti, M., and Fenice, M. (2021). Spatio-temporal variation of the bacterial communities along a salinity gradient within a Thalassohaline environment (saline di Tarquinia Salterns, Italy). Molecules 26:1338. doi: 10.3390/molecules26051338
Hammer, D. A. T., Ryan, P. D., Hammer, Ø., and Harper, D. A. T. (2001). Past: paleontological statistics software package for education and data analysis. Palae. Elec. 4, 1–9.
Hugoni, M., Taib, N., Debroas, D., Domaizon, I., Jouan Dufournel, I., Bronner, G., et al. (2013). Structure of the rare archaeal biosphere and seasonal dynamics of active ecotypes in surface coastal waters. Proc. Natl. Acad. Sci. 110, 6004–6009. doi: 10.1073/pnas.1216863110
Janda, J. M., and Abbott, S. L. (2007). 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J. Clin. Microbiol. 45, 2761–2764. doi: 10.1128/JCM.01228-07
Kerkar, S., and Bharathi, P. A. L. (2007). Stimulation of sulfate-reducing activity at salt-saturation in the salterns of Ribandar, Goa, India. Geomicrobiol J. 24, 101–110. doi: 10.1080/01490450701266597
Kojima, H., and Fukui, M. (2016). Sulfuriflexus mobilis gen. Nov., sp. nov., a sulfur-oxidizing bacterium isolated from a brackish lake sediment. Int. J. Syst. Evol. Microbiol. 66, 3515–3518. doi: 10.1099/ijsem.0.001227
Kojima, H., Watanabe, M., and Fukui, M. (2017). Sulfurivermis fontis gen. Nov., sp. nov., a sulfur-oxidizing autotroph, and proposal of Thioprofundaceae fam. Nov. Int. J. Syst. Evol. Microbiol. 67, 3458–3461. doi: 10.1099/ijsem.0.002137
Lohmann, P., Benk, S., Gleixner, G., Potthast, K., Michalzik, B., Jehmlich, N., et al. (2020). Seasonal patterns of dominant microbes involved in central nutrient cycles in the subsurface. Microorganisms 8:1694. doi: 10.3390/microorganisms8111694
Mani, K., Salgaonkar, B. B., Das, D., and Bragança, J. M. (2012). Community solar salt production in Goa. India. Aquat Biosyst 8:30. doi: 10.1186/2046-9063-8-30
McAllister, S. M., Moore, R. M., Gartman, A., Luther, G. W., Emerson, D., and Chan, C. S. (2019). The Fe(II)-oxidizing Zetaproteobacteria: historical, ecological and genomic perspectives. FEMS Microbiol. Ecol. 95:fiz015. doi: 10.1093/femsec/fiz015
Megonigal, J. P., Hines, M. E., and Visscher, P. T. (2003). Anaerobic metabolism: linkages to trace gases and aerobic processes. Treatise on Geochemistry (Elsevier). 8, 317–424.
Mehta, P., Yadav, M., Ahmed, V., Goyal, K., Pandey, R., and Chauhan, N. S. (2021). Culture-independent exploration of the hypersaline ecosystem indicates the environment-specific microbiome evolution. Front. Microbiol. 12:686549. doi: 10.3389/fmicb.2021.686549
Meng, S., Peng, T., Liu, X., Wang, H., Huang, T., Gu, J.-D., et al. (2022). Ecological role of Bacteria involved in the biogeochemical cycles of mangroves based on functional genes detected through GeoChip 5.0. mSphere 7:21. doi: 10.1128/msphere.00936-21
Onishi, H., and Kamekura, M. (1972). Micrococcus halobius sp. n. Int. J. Syst. Evol. Microbiol. 22, 233–236. doi: 10.1099/00207713-22-4-233
Oren, A. (1999). Bioenergetic aspects of Halophilism. Microbiol. Mol. Biol. Rev. 63, 334–348. doi: 10.1128/MMBR.63.2.334-348.1999
Qasim, S. Z., and Gupta, R. S. (1981). Environmental characteristics of the Mandovi-Zuari estuarine system in Goa. Estuar. Coast. Shelf Sci. 13, 557–578. doi: 10.1016/S0302-3524(81)80058-8
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2012). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Ramos-Cormenzana, A. (2020). Ecology of moderately halophilic Bacteria. The Biology of Halophilic Bacteria Edition (CRC Press), 55–86. doi: 10.1201/9781003069140-3
Rana, K., Rana, N., and Singh, B. (2020). Applications of sulfur oxidizing bacteria. Physiological and Biotechnological Aspects of Extremophiles, (Academic Press), 131–136.
Schmitz, R. A., Dietl, A., Müller, M., Berben, T., Op den Camp, H. J., and Barends, T. R. (2020). Structure of the 4-hydroxy-tetrahydrodipicolinate synthase from the thermoacidophilic methanotroph Methylacidiphilum fumariolicum SolV and the phylogeny of the aminotransferase pathway. Acta Crystallogr, Sect F: Struct Biol Commun 76, 199–208. doi: 10.1107/S2053230X20005294
Semedo, M., Lopes, E., Baptista, M. S., Oller-Ruiz, A., Gilabert, J., Tomasino, M. P., et al. (2021). Depth profile of nitrifying archaeal and bacterial communities in the remote oligotrophic waters of the North Pacific. Front. Microbiol. 12, 1–18. doi: 10.3389/fmicb.2021.624071
Sleator, R. D., and Hill, C. (2002). Bacterial osmoadaptation: the role of osmolytes in bacterial stress and virulence. FEMS Microbiol. Rev. 26, 49–71. doi: 10.1111/j.1574-6976.2002.tb00598.x
Soininen, J., Passy, S., and Hillebrand, H. (2012). The relationship between species richness and evenness: a meta-analysis of studies across aquatic ecosystems. Oecologia 169, 803–809. doi: 10.1007/s00442-011-2236-1
Song, T., Liang, Q., Du, Z., Wang, X., Chen, G., Du, Z., et al. (2022). Salinity gradient controls microbial community structure and assembly in coastal solar Salterns. Genes (Basel) 13:385. doi: 10.3390/genes13020385
Sorokin, D. Y., Merkel, A. Y., and Muyzer, G. (2020). Thiohalospira. Bergey’s Manual of Systematics of Archaea and Bacteria. John Wiley & Sons, Inc.
Sun, X., Kong, T., Li, F., Häggblom, M. M., Kolton, M., Lan, L., et al. (2022). Desulfurivibrio spp. mediate sulfur-oxidation coupled to Sb(V) reduction, a novel biogeochemical process. ISME J. 16, 1547–1556. doi: 10.1038/s41396-022-01201-2
Tonima, K., and Savita, K. (2011). Pharmaceutical potentials of bacteria from saltpans of Goa. Intern J App Pharmaceutics 2, 150–154.
Whittaker, R. H. (1960). Vegetation of the Siskiyou Mountains, Oregon and California. Ecol. Monogr. 30, 279–338. doi: 10.2307/1943563
Yu, J., Loh, K., Song, Z. Y., Yang, H. Q., Zhang, Y., and Lin, S. (2018). Update on glycerol-3-phosphate acyltransferases: the roles in the development of insulin resistance. Nutr. Diabetes 8:34. doi: 10.1038/s41387-018-0045-x
Keywords: salt pan, halophilic, Bacteria, diversity, next generation sequencing
Citation: Gawas P and Kerkar S (2023) Bacterial diversity and community structure of salt pans from Goa, India. Front. Microbiol. 14:1230929. doi: 10.3389/fmicb.2023.1230929
Edited by:
Rafael R de la Haba, University of Sevilla, SpainCopyright © 2023 Gawas and Kerkar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Savita Kerkar, ZHJzYXZpdGFrZXJrYXJAZ21haWwuY29t
 Priti Gawas
Priti Gawas Savita Kerkar
Savita Kerkar