
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 25 July 2023
Sec. Evolutionary and Genomic Microbiology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1228128
This article is part of the Research Topic Current Progress in Genomic and Genetic Research on Human Viral Diseases View all 9 articles
Over three years’ pandemic of 2019 novel coronavirus disease (COVID-19), multiple variants and novel subvariants have emerged successively, outcompeted earlier variants and become predominant. The sequential emergence of variants reflects the evolutionary process of mutation-selection-adaption of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Amino acid substitution/insertion/deletion in the spike protein causes altered viral antigenicity, transmissibility, and pathogenicity of SARS-CoV-2. Early in the pandemic, D614G mutation conferred virus with advantages over previous variants and increased transmissibility, and it also laid a conservative background for subsequent substantial mutations. The role of genomic recombination in the evolution of SARS-CoV-2 raised increasing concern with the occurrence of novel recombinants such as Deltacron, XBB.1.5, XBB.1.9.1, and XBB.1.16 in the late phase of pandemic. Co-circulation of different variants and co-infection in immunocompromised patients accelerate the emergence of recombinants. Surveillance for SARS-CoV-2 genomic variations, particularly spike protein mutation and recombination, is essential to identify ongoing changes in the viral genome and antigenic epitopes and thus leads to the development of new vaccine strategies and interventions.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a sister clade of SARS-CoV (Coronaviridae Study Group of the International Committee on Taxonomy of Viruses et al., 2020), has posed a global public health threat since its initial outbreak in December 2019 (Hu et al., 2020). On May 5, 2023, the World Health Organization (WHO) declared the end of the 2019 novel coronavirus disease (COVID-19) pandemic as a Public Health Emergency of International Concern. At that time, WHO reported a total of 765,222,932 cases and 6,921,614 deaths worldwide.1 Consistent with other coronaviruses, the genome of SARS-CoV-2 is a single-stranded positive-sense RNA of approximately 30,000 nucleotides, with replication mediated by RNA-dependent RNA polymerase (RdRP) (Vkovski et al., 2020; Li et al., 2020b). The 5’-terminus of the SARS-CoV-2 genome contains two open reading frames (ORFs), while the 3’-terminus contains four major structural proteins coding-gene in the following order: spike protein, envelope protein, membrane protein, and nucleocapsid protein (Bai et al., 2021). Despite the presence of error-correction enzymes, which contribute to a relatively high replication fidelity compared to other RNA viruses, SARS-CoV-2 still undergoes significant mutations (Robson et al., 2020; Domingo et al., 2021; Perales, 2021). The nucleotide mutation rates of SARS-CoV-2 are estimated to be 6.677 × 10–4 and 8.066 × 10–4 substitutions per year for the whole genome and spike protein, respectively (Wang S. et al., 2021).
Amino acid mutations in the spike protein play a crucial role in the evolution of SARS-CoV-2. The spike protein, which forms a trimeric fusion protein on the surface of the coronavirus, exhibits a crown-like appearance and serves as an ideal target for inducing neutralizing antibodies and protective immunity (Kang et al., 2021; Tian et al., 2021). The spike protein is composed of S1 and S2 subunits, and the Receptor Binding Domain (RBD) in the spike interacts with the human receptor angiotensin-converting enzyme 2 (ACE2) receptor when activated to allow the virus to entry into cells (Conceicao et al., 2020; Hoffmann et al., 2020b; Zhang et al., 2021a). Mutations in the spike protein, particularly in the RBD, have led to alterations in spike-ACE2 recognition, resulting in viral immune escape and the failure of neutralizing antibodies (Magazine et al., 2022; Chen et al., 2023). Spike proteins are classified as open and closed forms according to the up and down conformations of the RBD, and mutations in the spike may change the RBD conformation (Walls et al., 2020; Wrapp et al., 2020). The D614G mutation, which represents the substitution of amino acid D (Asp) by G (Gly), is conservative across all major variants (Wassenaar et al., 2022) and predominant in the spike protein during the early stage of pandemic (Chang et al., 2020). The D614G mutation has been shown to enhance furin proteolysis capacity by 50 times (Gobeil et al., 2021). Notably, the Omicron variant harbors more than 60 substitutions, deletions, and insertions, of which 15 rare mutations are found in the spike (He et al., 2021; Ma et al., 2022b). The spike protein of Omicron predominantly adopts closed conformations (Calvaresi et al., 2023), potentially leading to the failure of nearly all anti-spike monoclonal antibodies (Focosi and Casadevall, 2022; Turelli et al., 2022).
In addition to point mutations in the spike protein, viral genomic recombination is common among coronaviruses (Yewdell, 2021), especially during the late pandemic phase when different variants co-circulate. According to the US Centers for Disease Control and Prevention (CDC), the most prevalent circulating strains in the US as of May 13, 2023, were XBB.1.5 (61.5%), XBB.1.9.1 (10.0%), and XBB.1.16 (9.4%) (Ma et al., 2023). The frequent occurrence of recombination makes it challenging to predict the effectiveness of vaccines targeting the spike protein, and recombination may confer altered transmissibility, virulence, and immune escape properties to the virus (Focosi and Maggi, 2022; Carabelli et al., 2023).
The evolution of SARS-CoV-2 within the population follows the mutation-selection-adaptation theory of Darwinian evolution (Goldman, 2021; Figure 1). In this context of hypermutation, both innate and adaptive host immune responses drive mutation selection (Thorne et al., 2021), as we have previously discussed (Shen et al., 2023). The virus evolves to adapt to external selection pressures, and antigenic drift occurs as mutations gradually accumulate, affecting the virus’s immunogenicity (Bano et al., 2021; Shapira et al., 2023). Antigenic drift facilitates viral evasion from host immune response, particularly by affecting antibody neutralization, resulting in viral resistance to previous infection and vaccination (Zhang et al., 2022; Cao et al., 2022c; Planas et al., 2023; Qu et al., 2023). The evolutionary trend tends to lower the pathogenicity but increase the transmissibility of variants, resulting in long-term retention of virus in human hosts (Magiorkinis, 2023). In this review, we provide an overview of SARS-CoV-2, summarize the characteristic amino acid mutations in the spike protein, particularly in novel variants, discuss recent recombination events, and propose future perspectives to guide viral evolution and intervention strategies.
Several nomenclatures have been introduced for SARS-CoV-2 according to genetic relatedness of the sequences, including GISAID,2 Year-Letter (NextStrain) nomenclature,3 and Phylogenetic Assignment of Named Global Outbreak LINeages (Pango lineage) (Rambaut et al., 2020). The GISAID nomenclature system is based on marker mutations within the eight high-level phylogenetic groups, from the early split of S and L, to the further evolution of L into V and G, and later G into GH, GR and GV, and more recently GR into GRY. The Year-Letter nomenclature consists of the year when the clade emerged and a capital letter starting with A for each year, including 19A, 19B, 20A, 20B, 20C, and 20I. The Pango lineage uses an alphabetical prefix and a numerical suffix to identify descendants4 and contains phylogenetic, genetic, and epidemiological information. The first letter represents the lineage label of the variant, with the order from A to Z, then AA to AZ, BA to BZ, etc. The subsequent numbers separated by periods indicate the branches of lineages. When a branch has three more numeric suffixes, a new letter will be used as the lineage label in alphabetical order. For example, C.1 is the branch of B.1.1.1 (O’Toole et al., 2022). The recombinant variants are named in a uniform nomenclature beginning with “X.”
To promote surveillance and research, WHO categorized SARS-CoV-2 variants as three specific classes: variants of concern (VOC), variants of interest (VOI), and variants under monitoring (VUMs).5 VOCs are variants of high mutation and transmission rate. To date, Alpha, Beta, Gamma, Delta, and Omicron are known emerged VOCs and have become dominant in turn globally or regionally. The Alpha variant (B.1.1.7) was discovered in the UK in September 2020 (du Plessis et al., 2021; Galloway et al., 2021). It was proven to be highly transmissible and infectious, and became prevalent a few months later (Davies et al., 2021; Volz et al., 2021). The Beta variant (B.1.351) was first reported in South Africa in October 2020 (Tegally et al., 2021), and the Gamma variant (P.1) was first identified in travelers from Brazil in January 2021 (Fujino et al., 2021). The Delta variant (B.1.617.2) was isolated in India (Mlcochova et al., 2021) and quickly became the most prevalent variant worldwide in June 2021 (Mahase, 2021). The Omicron variant (B.1.1.529/BA sublineages) was first discovered in Botswana, South Africa in November 2021, and outcompeted other VOCs rapidly upon its emergence (He et al., 2021). Five major sublineages of Omicron, BA.1, BA.2, BA.3, BA.4, and BA.5, have been identified so far (Tegally et al., 2022). Most recently, a series of novel Omicron subvariants have emerged, such as BA.2.75 (Saito et al., 2022), BF.7 (Scarpa et al., 2023a), Deltacron (Kreier, 2022), XE (Rahimi and Bezmin Abadi, 2022b), XF (Chakraborty et al., 2022), BQ.1 (Wang et al., 2022b), BQ.1.1 (Wang et al., 2022b), XBB (Imai et al., 2023), XBB.1 (Arora et al., 2023), XBB.1.5 (Tamura et al., 2022), XBB.1.16 (Harris, 2023), and they have raised increasing concern. The timeline of emergence of variants is illustrated in Figure 2A.
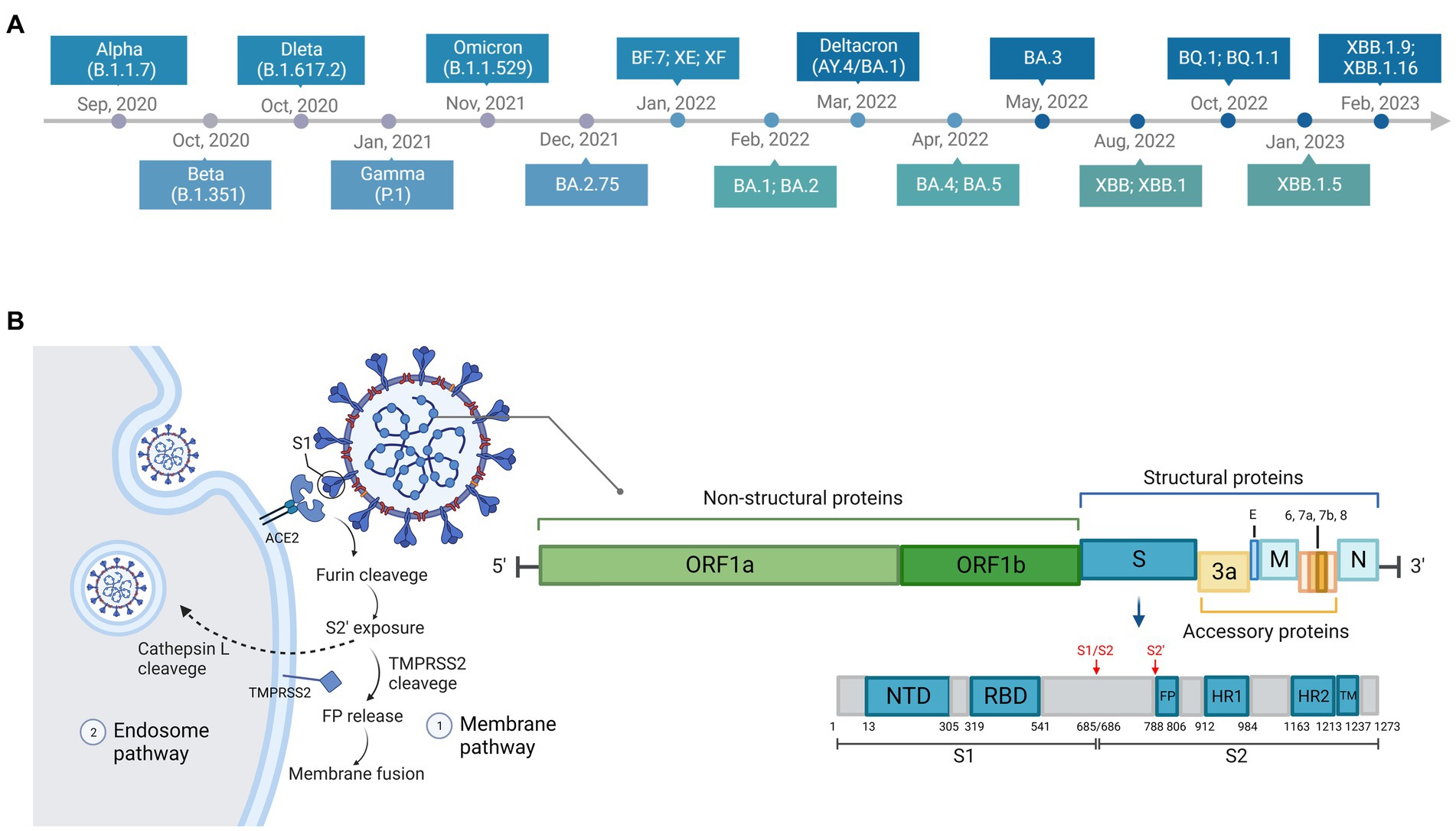
Figure 2. Timeline, structure, and entry pathways of SARS-CoV-2. (A) The chronological order of the emergence of major SARS-CoV-2 variants. (B) There are two pathways for SARS-CoV-2 entering cells: endosome pathway and membrane pathway. ACE2, angiotensin-converting enzyme 2; TMPRSS2, transmembrane protease serine protease 2; S1, subunit 1 of the spike protein; FP, fusion peptide, responsible for membrane fusion; S1/S2, furin cleavage site between S1 and S2 subunit of the spike protein; S2’, another proteolytic site in the subunit 2 of the spike protein.
Two described entry pathways of SARS-COV-2 through the cell membrane or through endosomes (Figure 2B) have been reviewed in detail previously (Shang et al., 2020; Hoffmann et al., 2020b; Rahbar Saadat et al., 2021; Jackson et al., 2021b; Lim, 2023). The two entry pathways differ because S2’ cleavage occurs either at plasma membrane by the transmembrane protease serine protease 2 (TMPRSS2) [such as in the nasal epithelial cells, lungs, and bronchial branches where TMPRSS2 is highly co-expressed with ACE2 (Lukassen et al., 2020; Sungnak et al., 2020)] or within the cell by endolyosomal cathepsins such as Cathepsin L (Bestle et al., 2020; Shang et al., 2020). The proteolytic site between the S1 and S2 subunit of the spike protein, also known as furin cleavage site (FCS), is cleaved by a host protease furin (Lavie et al., 2022). This process of cleavage is essential to the entry pathway and membrane fusion (Bestle et al., 2020; Hossain et al., 2021; Johnson et al., 2021; Peacock et al., 2021; Lavie et al., 2022). Optimization of FCS has been shown to facilitate cell–cell fusion to improve the infectivity (Hoffmann et al., 2020a), increase the transmissibility (Peacock et al., 2021), and promote pathogenesis (Johnson et al., 2021).
Multiple hypotheses have been proposed to explain the origin of VOCs (Mallapaty, 2022), such as (1) circulation in geographically sequencing limited areas; (2) circulation within animal hosts then spillover to humans; and (3) evolution in immunosuppressed chronic infection hosts. In some regions, the limited capacity for genomic sequencing has resulted in a lack of testing for asymptomatic patients. It has been observed that asymptomatic carriers exhibit higher levels of antiviral immunity and lower levels of inflammation compared to symptomatic individuals (Yang et al., 2020b; Le Bert et al., 2021; Ma et al., 2022a). This immunological profile may create an environment conducive to viral evolution under immune pressure. There is evidence supporting the hypothesis of an animal host origin, with white-tailed deer (Hale et al., 2022; Marques et al., 2022) and farmed mink (Koopmans, 2021; Lu et al., 2021) identified as stable animal reservoirs for SARS-CoV-2. These variants have the potential to infect animals and accumulate mutations within animal reservoirs. Subsequently, the virus may undergo further evolution, giving rise to new subvariants that can then spillover to humans. The hypothesis of chronic infection in immunodeficient hosts is widely accepted in many scenarios. Chronic infection in such individuals is associated with ACE2 affinity, immune evasion, and optimization of viral packaging (Choi et al., 2020; Kemp et al., 2021; Harari et al., 2022; Wilkinson et al., 2022). This process drives the mutation profiles of the virus and enhances its fitness (Ghafari et al., 2022; Hill et al., 2022). Extensive immune escape has been observed in SARS-CoV-2 infections in immunocompromised hosts, such as patients with advanced HIV disease (Cele et al., 2022).
Mutation profiles of the variants of concern (VOCs) exhibit certain overlapping patterns, while also assuming distinct roles in the process of viral evolution, thereby suggesting an underlying evolutionary resemblance among these variants. Notably, a common early substitution mutation, namely D614G, is shared by all five VOCs, which has been shown to significantly augment the binding affinity of the viral spike protein to the ACE2 receptor, consequently amplifying viral pathogenicity (Alkhatib et al., 2021; Wang P. et al., 2021; Zhang et al., 2021b; Venkatakrishnan et al., 2022). Moreover, the substitution P681H has been identified in Alpha (Lubinski et al., 2022), Gamma (Fujino et al., 2021), and Omicron (Tian et al., 2022), and has been demonstrated to enhance viral cell entry. Conversely, the substitution P681R, occurring at the same position, has been observed to augment the replication capacity and pathogenicity of the Delta variant (Mlcochova et al., 2021; Saito et al., 2021; Liu et al., 2022). These mutations accumulate in a stepwise manner, progressively modifying the antigenic epitope of the virus, ultimately leading to a transition from “genetic drift” to antigenic drift.
For variant Alpha (B.1.1.7), of eight mutations in the spike protein, D614G, Del H69/V70 (Del H69/V70 represents amino acid deletion mutation in the site 69 and 70 of the spike protein), N501Y, and P681H are most meaningful (Wang P. et al., 2021). The D614G mutation has been found to confer a fitness advantage by promoting efficient replication in primary airway cells, thereby increasing virulence and transmission (Hou et al., 2020; Korber et al., 2020; Ozono et al., 2021; Zhou et al., 2021). It also leads to alterations in spike conformation and enhanced FCS cleavage (Zhang et al., 2020) and leads to alterations in spike conformation and enhanced FCS cleavage (Gobeil et al., 2021; Nguyen et al., 2021). However, it has also been observed that the D614G mutation renders the virus more susceptible to monoclonal antibodies by increasing epitope exposure, suggesting that it does not impede the effectiveness of vaccines (Weissman et al., 2020), indicating it does not impede vaccine effect (Hou et al., 2020; Weissman et al., 2020; Yurkovetskiy et al., 2020; Ozono et al., 2021). Del H69/V70 is associated with diagnostic test failure for probes targeting spike proteins, known as spike gene targeting failure (SGTF) (Bal et al., 2021). SGTF has been utilized as a reliable proxy for monitoring the prevalence of the B.1.1.7 variant (Bal et al., 2021; Borges et al., 2021; Kidd et al., 2021). N501Y has been shown to enhance the binding of the spike protein to human ACE2 receptors, potentially expanding the host range of SARS-CoV-2 (Starr et al., 2020; Chan et al., 2021; Zahradník et al., 2021; Wang et al., 2022c). P681H, which is located adjacent to the FCS, has been found to enhance the efficiency of FCS cleavage during virus entry into cells and contributes to Alpha’s resistance to type I interferons (Lubinski et al., 2022; Lista et al., 2022).
For Beta variant (B.1.351), the combination of E484K and N501Y mutations has a synergistic effect in enhancing the affinity of the spike protein for human ACE2 receptors (Starr et al., 2020; Zahradník et al., 2021). Mutations Del 242–244, K417N, E484K, and N501Y have been shown to confer significant resistance to infection or vaccine-induced neutralizing antibodies (Garcia-Beltran et al., 2021; Hu et al., 2021; Tao et al., 2021; Wibmer et al., 2021; Wang P. et al., 2021).
The Gamma variant (P.1) carries 12 mutations in the spike protein, including K417T, N501Y, and E484K (Faria et al., 2021). These three mutations collectively enhance the affinity of the spike protein for ACE2 receptors, thereby increasing the transmissibility of the Gamma variant. E484K is also associated with reduced neutralization by antibodies (Faria et al., 2021). E484K is also associated with reduced neutralization by antibodies (Cele et al., 2021; Greaney et al., 2021; Wibmer et al., 2021).
The Delta variant (B.1.617.2) harbors several mutations previously reported in other VOCs, including L452R, T478K, E484Q, D614G, and P681R in the spike protein (Liu et al., 2022). These mutations partly explain the rapid global spread of the Delta variant upon its emergence. The L452R mutation has been found to increase infectivity, modestly reduce susceptibility to neutralizing antibodies, and enhance viral fusogenicity, thereby promoting virus replication (Motozono et al., 2021). E484Q exhibits similar reduced sensitivity to vaccine-induced neutralizing antibodies as L452R, but lacks synergistic effects when taken together (Motozono et al., 2021). Similar to P681H in Alpha, P681R in Delta increases FCS cleavage, resulting in enhanced transmissibility (Mlcochova et al., 2021; Saito et al., 2021; Wibmer et al., 2021). Studies have revealed that spike of Delta is more stable and binds with higher affinity to ACE2 than the spike of the wild-type (Gomari et al., 2023).
As discussed above, the evolution of SARS-CoV-2 of pre-Omicron variants has primarily centered around recurrent mutations in key residues of the spike protein, including D614, N501, P681, K417, and E484. However, with the emergence of the Omicron variant and its sublineages, the landscape has undergone a significant shift. The Omicron variant harbors over 30 spike mutations, with 15 of them occurring in the RBD (Kumar et al., 2021). Figure 3 illustrates the mutation profiles of VOCs. In general, Omicron exhibits several distinctive characteristics compared to previous VOCs, including enhanced transmissibility, reduced antibody neutralization capacity (resulting in lower vaccine effectiveness), altered tissue tropism, relatively lower pathogenicity, and an increased likelihood of reinfection.
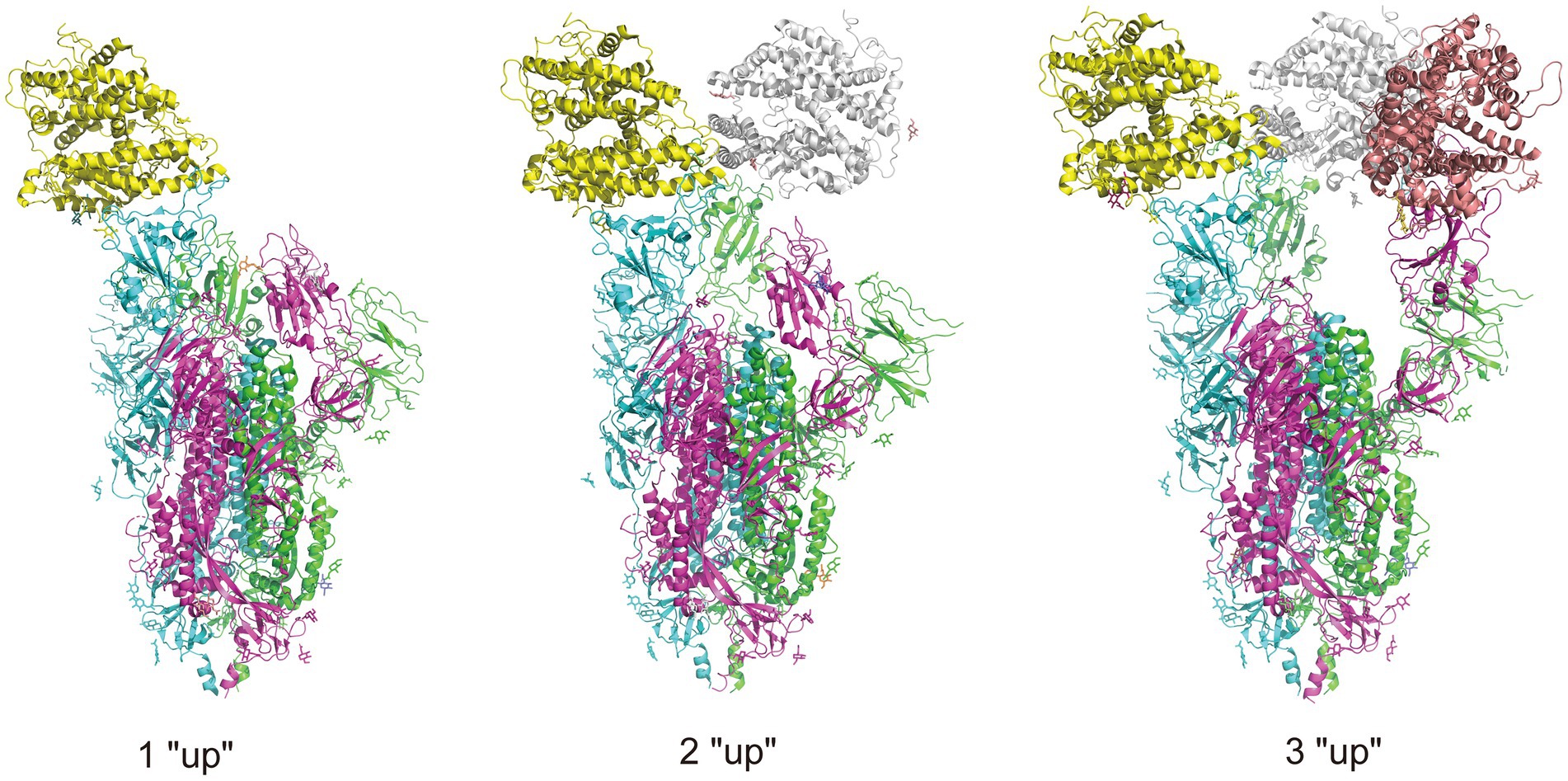
Figure 3. Illustration of RBD conformation of spike protein complexed with ACE2 receptors. There are two RBD conformations: “up” and “down,” and when the RBD is in “up” conformation, the spike protein is open to the ACE2 receptor. The trimeric spike protein is indicated by chain in three colors, purple, green, and blue, and three ACE2 receptors are indicated in yellow, gray, and pink. The complexes are obtained from RCSB.org (7KNE, 7KNH, 7KNI for 1 “up,” 2 “up,” 3 “up,” respectively).
The higher transmissibility may attribute to the altered viral affinity to ACE2 receptor. Multiple experimental observations have demonstrated that the binding affinity between the RBD of the spike protein and ACE2 is significantly higher for Omicron compared to wildtypes (Kumar et al., 2021; Abeywardhana et al., 2022; Cui et al., 2022; Hong et al., 2022). The mutations T478K, Q493R, Q498R, and N501Y collectively contribute to the increased binding affinity through electrostatic effects (Kumar et al., 2021; Abeywardhana et al., 2022). However, another study revealed that Omicron exhibits comparable binding affinity to ACE2 when compared to the wild type SARS-CoV-2 and weaker binding affinity than the Delta variant (Wu et al., 2022). This discrepancy may stem from differences in the surface plasmon resonance methodologies employed in the studies, necessitating further research. The sublineages of Omicron display variations in their ACE2 affinity, with BA.2 exhibiting the highest affinity, followed by BA.3, BA.1, BA.2.75, and BA.5 (Abeywardhana et al., 2022). Furthermore, Omicron variants exhibit reduced sensitivity to neutralizing antibodies induced by triple-dose inactivated vaccines (Ren et al., 2023). Reports indicate that the neutralizing activity against Omicron variants is lost in 90% of immunization serum samples and 43% of convalescent serum samples (Zhang et al., 2022). In contrast to pre-Omicron variants, which primarily exploit TMPRSS2 for cell entry (Hoffmann et al., 2020b), Omicron variants have a propensity for entering nose and throat cells that are deficient in TMPRSS2 via the cathepsin-mediated endosomal pathway (Hui et al., 2022; Meng et al., 2022; Willett et al., 2022; Zhao et al., 2022). This shift in cell entry tropism from the membrane pathway to the endosomal pathway reduces the capacity of Omicron to fuse infected cells and form syncytia, resulting in a lower pathogenicity (Meng et al., 2022; Willett et al., 2022). The Omicron’s propensity to infect upper respiratory tract restricts its clinical manifestation and lowers the disease severity. From a structural standpoint, compared with Delta, Omicron has an inconsistent distribution of electrostatic potential and a geometric reorganization in the FCS of the spike protein. This structural divergence contributes to Omicron’s reduced fusogenicity and consequently lower pathogenicity (Fantini et al., 2022). Moreover, the Omicron variant possesses an enhanced capacity for immune evasion, leading to reinfection of individuals (Chavda et al., 2022; Xia et al., 2022). For pre-Omicron variants, infection-induced protective immunity has limited efficacy against BA.4 and BA.5, but it demonstrates a strong effect in preventing reinfection of BA.1 and BA.2 (Altarawneh et al., 2022).
Notably, the combinatorial mutations in the spike protein appear to have a synergistic effect on the characteristics of Omicron, further complicating its mutation profile. Preliminary findings suggest that certain mutations in Omicron form three distinct clusters, wherein the mutations seem to work in concert to compensate for the detrimental effects of any individual mutation (Martin et al., 2022). Two mutations, N501Y and Q498R, collectively increase the affinity of a variant for the ACE2 receptor by nearly 20-fold (Bate et al., 2022).
BA.2.75, a descendant from BA.2, was first detected in India and Singapore (Saito et al., 2022). Differing from BA.2, BA.2.75 carries 9 additional mutations in the spike protein (147E, W152R, F157L, I210V, G257S, D339H, G446S, N460K, and an R493Q reversion mutation) (Sheward et al., 2022; Kurhade et al., 2023; Qu et al., 2023). BA.2.75 exhibits enhanced resistance to neutralization compared to BA.2 but falls short of the BA.4/5 variant (Qu et al., 2022; Saito et al., 2022; Cao et al., 2022b; Wang L. et al., 2022). The G446S and N460K mutations are primarily responsible for the increased resistance of neutralizing antibodies against BA.2.75 (Qu et al., 2022; Wang et al., 2022a), while the R493Q mutation reduces neutralization resistance (Wang et al., 2022a). Furthermore, the spike protein of BA.2.75 demonstrates significantly higher affinity for ACE2 (Saito et al., 2022), and the N460K mutation, which enhances S processing, leads to increased cell–cell fusion of BA.2.75 compared to BA.2 (Qu et al., 2022).
BA.4.6, a sublineage of BA.4, carries two additional mutations in the spike protein (R346T and N658S) and was initially identified in the US and UK (Hachmann et al., 2022). This subvariant exhibits a notable ability to evade neutralizing antibodies induced by infection or vaccination, with titers lower than those of BA.5 by a factor of 2 to 2.7 (Hachmann et al., 2022; Wang et al., 2022b; Planas et al., 2023).
BF.7 variant (also known as BA.5.2.1.7) is a derivative of BA.5 and has gained attention since the beginning of 2022, particularly in Asia (Kelleni, 2023; Pan et al., 2023; Scarpa et al., 2023a). Compared to BA.5, BF.7 carries an additional R346T mutation in the RBD and shares an identical N-terminal domain (NTD) (Scarpa et al., 2023a). The R346T mutation has been associated with enhancing the virus’s ability to evade neutralizing antibodies generated by vaccines or previous infection (Akif et al., 2023). However, R346T does not greatly increase the affinity of BF.7 to ACE2 (Scarpa et al., 2023a). Although enhanced resistance to neutralization exists (Qu et al., 2023), BF.7 appears to be less virulent, with a low evolutionary rate of 5.62 × 10–4 substitutions/sites/years compared to other Omicron subvariants (Scarpa et al., 2023a).
CH.1.1, a descendant of BA.2.75, has rapidly emerged in the UK. Compared with BA.2.75, CH.1.1 owns additional 4 substitutions (R346T, K444T, L452R, and F486S) in the RBD of the spike protein (Uraki et al., 2023). CH.1.1 does not pose a significant threat to pandemic control. Antiviral drugs (remdesivir, molnupiravir, nirmatrelvir, and ensitrelvir) remain effective against CH.1.1, and an additional dose of bivalent mRNA vaccines may be beneficial in preventing CH.1.1 infection (Uraki et al., 2023).
BQ.1 and BQ.1.1 have evolved from BA.5 (Wang et al., 2022b). Compared with the progenitor BA.5, BQ.1 carries additional K444T and N460K mutations in the spike protein, while BQ.1.1 has an additional R346T mutation (Wang et al., 2022b). Strong resistance to neutralization is observed in the BQ.1 and BQ.1.1 subvariants, largely driven by the N460K mutation (Kurhade et al., 2023; Qu et al., 2023).
XBB variant carries 9 additional changes in the RBD and 5 additional changes in the NTD compared to its progenitor BA.2 (Imai et al., 2023). The R346 position is a critical mutation site (harboring R346T/S/I) that leads to increased immune evasion by neutralizing antibodies (Cao et al., 2021). Similar to BQ.1 and BQ.1.1, the XBB lineage exhibits an exceptionally strong ability to evade antibodies (Arora et al., 2023). BQ and XBB subvariants have rendered all authorized antibodies ineffective, with titers against BQ and XBB significantly lower (Wang et al., 2022a; Chakraborty et al., 2023). A cohort study in Singapore revealed that protection against XBB reinfection was lower and weakened more rapidly compared to protection against BA.4 or BA.5 reinfection in previously vaccinated omicron-infected individuals (Tan et al., 2023), further indicating greater immune evasion in XBB.
XBB.1.5 has a substantial growth advantage over BQ.1.1 and XBB.1, becoming the predominant strain in the US by January 2023 (Tamura et al., 2022). XBB.1.5 is a recombinant of two descendants from BA.2, differing from XBB.1 by an additional F486P mutation in the spike protein (Tamura et al., 2022). Unlike the F486S mutation in XBB.1, which disrupts the local hydrophobic interaction of the spike with ACE2, F486P in XBB.1.5 restores this interaction (Yue et al., 2023). This mechanism enhances the affinity for ACE2 and suggests a higher growth advantage for XBB.1.5 compared to its progenitor XBB.1. F486P makes XBB.1.5 slightly less immune evasive but more infectious than its ancestor XBB.1, likely due to increased binding affinity to human ACE2 (Tamura et al., 2022; Mahase, 2023).
XBB.1.16 is another XBB sublineage harboring the F486P substitution, outcompeting other variants in India by the end of March 2023 (Looi, 2023; Varghese et al., 2023). Compared to XBB.1.5, XBB.1.16 carries two additional substitutions, E180V in the NTD and T478R in the RBD, in the spike protein (Yamasoba et al., 2023). XBB.1.16 exhibits a greater growth advantage compared to XBB.1 and XBB.1.5, but its potential for immune evasion is similar to XBB.1 and XBB.1.5 (Yamasoba et al., 2023). Notably, XBB.1.16 and XBB.1.5 demonstrate similar characteristics in terms of cell line tropism, cell entry efficiency, and neutralization evasion (Nehlmeier et al., 2023).
SARS-CoV-2 infection is partially controlled by the conformation of the spike protein RBD. The RBD located in the S1 subunit of the extracellular domain of the spike is responsible for interacting with ACE2 receptors, and has been shown an important molecular determinant of the COVID-19 pandemic (Shang et al., 2020). The RBD exists in two different conformations: up for receptor binding and down for immune evasion. Accordingly, the spikes are also in open and closed conformations. Compared with the closed-form spike protein, an open-form with an up RBD conformation leads to infection more rapidly (Yin et al., 2022), and binding with antibodies more easily (Berger and Schaffitzel, 2020; Yin et al., 2022). Figure 4 illustrates the different up or down conformations of spike protein complexed with ACE2 receptors. In the early phase of the pandemic, the D614G substitution adjacent to the NTD subdomain leads to a more open and thus receptor-accessible conformations of the spike compared with the wild-type (Benton et al., 2021; Gobeil et al., 2021; Mansbach et al., 2021; Zhang et al., 2021a). The D614G substitution confers the virus an adaptation advantage and higher transmissibility, facilitating the acquisition of further mutations and forming the variants of concern (Korber et al., 2020; Zhang et al., 2020; Plante et al., 2021). It is shown that the conformations of Alpha, Beta and Delta spikes are predominantly open and that the binding of ACE2 increases membrane fusion (Calvaresi et al., 2023). In contrast, substitution of the Omicron spikes results in a predominantly closed conformation that may allow them to evade antibodies (Calvaresi et al., 2023). Other studies show that the mutations in the RBD of Omicron may promote the conformation to change from “down” to “up” and thus increase engagement of ACE2 (Hossen et al., 2022; Ye et al., 2022). This may due to the mutations that reduce the protein–protein interaction affinity of RBD with its neighboring domains (Singh et al., 2022).

Figure 4. Representation of the SARS-CoV-2 spike protein, showing amino acid mutations in VOCs Alpha, Beta, Gamma, Delta and Omicron. Amino acid mutations are colored in orange, Alpha; yellow, Beta; purple, Gamma; green, Delta; red, Omicron; blue, ≥ 2 VOCs. The spike protein structure complexed with ACE2 receptor is obtained from RCSB.org (7KNE). The mutations of VOCs are based on the data from covariants (https://covariants.org, 20I for Alpha, 20H for Beta, 20 J for Gamma, 21A for Delta, and 21 L for Omicron).
Glycosylation is another way to affect the RBD conformation and thus change the spike open state. The SARS-CoV-2 spike gene encodes 22 N-linked glycan sequons per protomer and the trimeric spike protein displays 66 N-linked glycosylation sites. Glycosylated spike has a higher barrier to opening and also energetically favors the down state over the up state (Pang et al., 2022). Inhibition of protein N-glycosylation is shown to block SARS-CoV-2 infection (Casas-Sanchez et al., 2021). The glycosylation sites also have the effect of facilitating immune evasion by shielding specific epitopes from antibody neutralization (Watanabe et al., 2019). It is observed that proximal glycosylation sites (N165, N234, and N343) shield the receptor binding sites on the SARS-CoV-2 spike, especially when the RBD is in the “down” conformation (Watanabe et al., 2020). Sztain et al. (2021) revealed that N-glycan at position N343 facilitates RBD opening, and plays a gating role in the spike protein open state. Although the spike surface is substantially shielded by N-glycans, it presents regions that are vulnerable to neutralizing antibodies such as in the RBM, NTD, and S2 subunit (Chi et al., 2020; Tortorici et al., 2020; Cerutti et al., 2021). Mutations in the spike may affect glycosylation. For example, P681H and P681R were found in Alpha and Delta, respectively, and they decreased O-glycosylation which potentially increases furin cleavage and may influence viral infectivity (Zhang et al., 2021c).
Recombination, a frequently observed evolutionary mechanism in coronaviruses, plays a significant role in the genetic diversity and evolution of these viruses. For example, lineage 5 of Middle East respiratory syndrome coronavirus (MERS-CoV), which caused the MERS-CoV outbreak in South Korea and mass infections in Saudi Arabia in 2015, is putatively a recombinant virus of groups 3 and 5 of clade B, or lineages 3 and 4 (Wang et al., 2015; Sabir et al., 2016). The measurement of recombination versus de novo mutation (R/M) provides insights into the relative impact of these two variations (Patiño-Galindo et al., 2021). In SARS-CoV-2, the R/M ratio is 0.00264 (Turakhia et al., 2022), while in MERS, it is estimated to be 0.25–0.31 (Patiño-Galindo et al., 2021), indicating a low level of recombinant mutations in the early stage of the SARS-CoV-2 pandemic. However, as co-infections and mutation accumulation increase within the population, recombination is expected to play a more prominent role in generating functional genetic diversity (Kim et al., 2020).
Recombination occurs when genetically distinct SARS-CoV-2 variants co-infect the same host during co-circulation (Figure 5A). This process leads to the emergence of recombinant viruses with new properties, such as increased transmissibility or virulence (Li et al., 2020a). Recombination occurs frequently in the later phase of pandemic (Varabyou et al., 2021). Turakhia et al. (2022) developed a method called Recombination Inference using Phylogenetic PLacEmentS (RIPPLES) to detect recombination in pandemic-scale phylogenies. By analyzing a 1.6 million sample tree, they identified 589 recombination events, indicating that approximately 2.7% of sequenced SARS-CoV-2 genomes have detectable recombinant ancestry (Turakhia et al., 2022). The distribution of recombination breakpoints across the SARS-CoV-2 genome is not uniform, with a higher incidence toward the 3’ end compared to the 5’ end, consistent with previous analyses in other human coronaviruses (Patiño-Galindo et al., 2021; Müller et al., 2022). Recombination events often lead to genetic alterations near the breakpoints, and the specific breakpoints vary across the genome (Bolze et al., 2022). For example, a recombinant virus containing genetic material from the Alpha (B.1.1.7) and Epsilon (B.1.429) variants was detected in New York, and recombinant mutations were found in the spike, nucleocapsid, and ORF8 coding regions (Wertheim et al., 2022). In the US, there have been nine reported recombination events between the Delta (AY.119.2) and Omicron (BA.1.1) variants, with the breakpoint located between the NTD and RBD of the spike protein (Lacek et al., 2022a). These recombinants can produce hybridized spike proteins containing characteristic amino acids from both Delta and Omicron (Lacek et al., 2022a). The co-circulation of different variants highlights the importance of ongoing genomic surveillance, with particular attention to recombinants (Jackson et al., 2021a). Figure 5B illustrates different patterns of recombination.
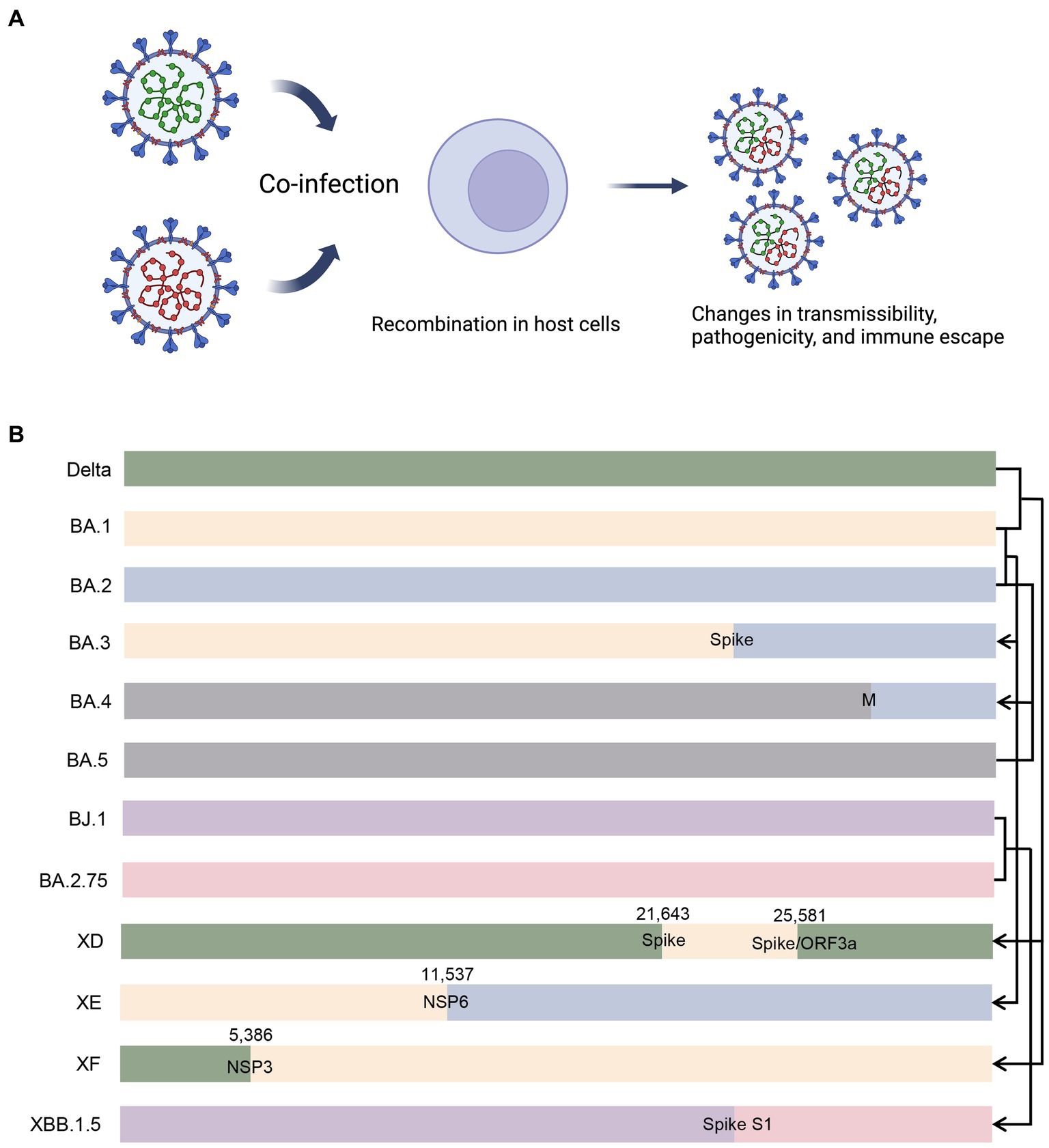
Figure 5. Illustration of recombination in co-infected cells and different recombination patterns. (A) When different variants co-infect an individual, there is possibility that recombinant variants emerge with altered properties. (B) BA.3 is putatively a recombinant of BA.1 and BA.2, and the breakpoint probably lies in the spike protein-coding gene. BA.4 is putatively a recombinant of BA.2 and BA.5, and the breakpoint probably lies in the M protein-coding gene. XD and XF are recombinants of Delta and BA.1, and the breakpoints lie in the spike protein-coding gene/ORF3a and NSP3 protein-coding gene, respectively. XE is a recombinant of BA.1 and BA.2, with breakpoint lying in the NSP6 protein-coding gene. XBB.1.5 is a recombinant od BJ.1 and BA.2.75, and the breakpoint probably lies in the S1 subunit of the spike protein-coding gene. M, membrane protein; ORF3a, open reading frame 3a; NSP, non-structural protein.
Co-infection is common in the later phase of the pandemic. For example, a 17-year-old Portuguese female was reported to be co-infected with two SARS-CoV-2 lineages belonging to distinct clades, differing by six variants (Pedro et al., 2021). Similar co-infection events have been observed, such as B.1.1.28 co-infecting with either B.1.1.248 or B.1.91 lineages (da Silva Francisco et al., 2021), and GH co-infecting with GR clades (Samoilov et al., 2021). In the US, out of 29,719 SARS-CoV-2 positive samples sequenced from November 2021 to February 2022, 20 co-infections were identified (Lacek et al., 2022b). In Brazil, nine co-infection events (0.61%) were identified in the investigated samples from May 2020 to April 2021, although this data is likely an underestimation due to sample limitations. Recombination has been found to occur more frequently in immunodeficient individuals at high risk of severe COVID-19 (Perez-Florido et al., 2023). Immunodeficient individuals are considered incubators for punctuated evolutionary events, possibly due to their vulnerability to chronic and co-infections (Rockett et al., 2022). For instance, a recombinant variant of B.1.160 and Alpha was isolated from a patient with lymphoma who was chronically infected for 14 months. The patient was initially infected with B.1.160, followed by concurrent Alpha infection, and eventually, the recombinant variant emerged (Burel et al., 2022).
Recombination occurs in five major sublineages of Omicron. BA.1, a descendent lineage of B.1.1, shows distinctly different phylogenetic as compared with other VOCs or VOIs. It has caused the fourth epidemic wave in South Africa (Araf et al., 2022; Lino et al., 2022; Saxena et al., 2022; Tian et al., 2022). The spike gene sequencing reveals that the BA.1 subvariant shares nine common amino acid mutations with most VOCs in the spiked proteins (three more than BA.2) (Araf et al., 2022; Ou et al., 2022; Tian et al., 2022), suggesting that Omicron may be derived from the recombinant origin of these VOCs. Three more Alpha-associated mutations (Del 69, Del 70, and Del Y144) were found in BA.1 rather than in BA.2, for BA.1 is phylogenetically closer to Alpha than the other variants (Kumar et al., 2021; Ou et al., 2022). Reverse mutations were also found in some dominant mutations (frequency > 95%) in BA.1 (Ou et al., 2022). Taken together, these support the role of Alpha in Omicron evolution.
Along with BA.1, BA.2 and BA.3 were also isolated in South Africa (Zhou et al., 2022). BA.2 has caused increased global infection, hospitalization, and mortality rate (Chen et al., 2022; Fonager et al., 2022; Rahimi and Bezmin Abadi, 2022a). BA.3 is likely a recombinant derivative of BA.1 and BA.2 due to BA.3 has similar genome in NTD region of the spike protein with BA.1 and BA.2 (Viana et al., 2022). A study revealed that BA.3 shared main mutations with BA.1 and BA.2, and BA.3 seemed to originate later (Wang C. et al., 2022), thus to some extent, corroborating the possibility of recombination.
BA.4 and BA.5 were afterwards identified as Omicron lineages in South Africa (Tegally et al., 2022). They were estimated to have originated in mid-December 2021 and early January 2022 (Viana et al., 2022). Their most recent common ancestor was estimated to have originated in mid-November 2021, coinciding with the emergence of BA.2 (Tegally et al., 2022). It deserves to note that BA.4 and BA.5 are close to BA.2 in genomes, and they both have similar spike proteins with BA.2 (Tegally et al., 2022). It is estimated that BA.4 and BA.5 are likely to evolve independently from the common ancestry of BA.2 subvariant (Wang C. et al., 2022). Compared with BA.2, BA.4 and BA.5 own extra mutations Del 69–70, L452R, F486V, and the wild-type amino acid at position Q493 (Ou et al., 2022). BA.4 and BA.5 share mutational profiles from 5’-UTR to envelope protein but differ distinctly from membrane protein to 3’-UTR (Tegally et al., 2022). This mutation pattern suggests that there exists a breakpoint within E and M, which is the possible evidence of recombinant event.
Recombination events raised more concerns when Omicron quickly outcompeted Delta pandemic. Co-circulation of Delta and Omicron provided a grounded basis for recombinant variants. There is growing concern about the possibility that this recombination potential could eventually result in mutations that confer virus on enhanced transmissibility and immune escape properties.
On January 7, 2022, scientists detected a Delta and Omicron recombinant genome, and informally named it as “Deltacron” (Kreier, 2022). Nevertheless, it was later determined as a lab contamination (Kreier, 2022). On March 9, WHO declared the detection of such recombinants in different regions around the world and designated this Deltacron as a VUM (Farheen et al., 2022; Maulud et al., 2022). Generally, Deltacron is referred to as the AY.4/BA.1 recombinant, named XD, and consists of a full-length spike protein of Omicron and backbone of Delta (Mahase, 2022; Wang C. et al., 2022). According to Chinese Center for Disease Control and Prevention, of the 36 amino acid mutations found in the spike protein, 27 are present in BA.1 and 5 in AY.4, while 4 are present in both (Wang and Gao, 2022). Structural analysis of the Deltacron recombinant spike suggests its hybrid content leads to optimization of viral binding to the host cell membrane (Colson et al., 2022a,b). Consequently, this novel recombined virus causes increased disease transmission (Chakraborty et al., 2022; Hosch et al., 2022). The Deltacron recombinant also has the potential to escape neutralization by monoclonal antibody (Evans et al., 2022). Although Delta (AY.45) and BA.1 are sensitive to Sotrovimab neutralization, while an AY.45-BA.1 recombinant, with its breakpoint located adjacent to the Sotrovimab binding site, is resistant to its neutralization (Duerr et al., 2023). Deltacron shows higher transmissibility but lower clinical severity (Moisan et al., 2022). As recombination did not really emerge on a large scale and did not show its power until the appearance of Deltacron, the advent of Deltacron is regarded as a “gray rhino” event, rather than a “black swan” event.
Other than Deltacron (recombinant of AY.4 and BA.1, also known as XD), the UK Health security agency recognized two similar recombinants, XE and XF (Chakraborty et al., 2022). The XE recombinant contains genomic elements from Omicron BA.1 and BA.2 subvariants (Rahimi and Bezmin Abadi, 2022b). The breakpoint lies in the NSP6 protein-coding region of genome, with the 11,537 bp of the BA.1 and 11,537 bp of the BA.2 genomes before and after the break site (Chakraborty et al., 2022). XE appears to be roughly 10% more transmissible than its parent variant BA.2 (Basky and Vogel, 2022). The XF variant contains the genomes of NSP1 to NSP3 from the Delta variant; the breakpoint lies at site 5,386, and the rest genomes from Omicron BA.1 variant (Chakraborty et al., 2022).
XBB, nicknamed Gryphon, is the most recent recombinant. XBB is regarded as the first observed SARS-CoV-2 variant to increase its fitness through recombination rather than substitutions (Tamura et al., 2022). XBB derives from two BA.2 sublineages: BJ.1 (BA.2.10.1) and BM.1.1.1 (BA.2.75) (Arora et al., 2023; Scarpa et al., 2023b). XBB and its first descendant XBB.1 are both evolutionarily close to BA.2 genomes (Scarpa et al., 2023b), suggesting BA.2 acts as their progenitor. The breakpoint lies between position 22,901 and 22,939, a position in the middle of RBD (Scarpa et al., 2023b). The mutation profiles possibly altogether contribute to the greater immune invasion capabilities of XBB than do those of the earlier Omicron variants BA.2 (Imai et al., 2023). The pathogenicity of XBB.1 is comparable to or even lower than that of BA.2.75 (Tamura et al., 2022). Though XBB subvariants exhibit enhanced fusogenicity and substantial immune evasion in elderly population, but the fusion inhibitors EK1 and EK1C4 can potently block either XBB or XBB.1.5 spike protein mediated fusion and viral entry (Xia et al., 2023a).
As a whole, the novel recombinant subvariants demonstrate a higher transmission rate and relatively greater resistance to antibodies compared to earlier variants (Wang et al., 2022a; Brandolini et al., 2023; Faraone et al., 2023). In January 2023, there was a rapid increase in the prevalence of XBB.1.5 in the United States (Callaway, 2023). According to the World Health Organization (WHO), XBB.1.5 accounted for 23-86% of circulating variants throughout the country (XBB.1.5 Updated Risk Assessment, 24 February 2023).6 However, these recombinant variants do not significantly increase the severity of the disease or cause clinical exacerbation (Karyakarte et al., 2023). XBB.1.5 does not carry mutations associated with potential changes in pathogenicity, such as P681R (Mlcochova et al., 2021; Saito et al., 2021). It is important to note that most vaccines are developed based on the spike protein, and the emergence of recombinant variants may pose a risk of vaccine failure (Tamura et al., 2022). Therefore, it is crucial to consider potential new subvariants in the development of novel strategic vaccines.
Various factors drive the viral evolution (Moelling, 2021), including RNA polymerase exchanging accuracy for efficiency (Yewdell, 2021), the selective pressures exerted by host immune system (Milne et al., 2021; Thorne et al., 2021), chronic infection in other species then spillover to human (Lu et al., 2021; Hale et al., 2022; Marques et al., 2022), and prolonged co-infection in immunodeficient hosts (Ou et al., 2022; Rockett et al., 2022). These factors contribute to the mutation-selection-evolution process of SRAS-CoV-2 evolution. Continuous evolution of SARS-CoV-2 has led to rapid and simultaneous emergence of multiple variants that exhibit a growth advantage over previously circulating variants (Wolf et al., 2022). During the evolution of SARS-CoV-2, the spike gene is the only gene that undergo the strong positive selection, while other genes show only weak or temporary positive selection (Lu et al., 2023) Thus, spike mutations contribute highly in its evolution. The mutational process is dynamic, and the mutation spectrum of SARS-CoV-2 may tend to be more similar to that of other animal Sarbecoviruses (Bloom et al., 2022). Here we propose several interventional strategies.
1. Genomic surveillance of SARS-CoV-2, specifically in the spike gene and genomic recombination, is of utmost importance in recognizing its evolutionary trend. Efforts have been made to promote the genomic monitoring. Dadonaite et al. (2023) developed a novel deep mutational scanning (DMS) platform for mapping the effects of spike protein mutations on immune evasion and viral infectivity (Xia et al., 2023b). Saldivar-Espinoza et al. (2023) developed a SARS-CoV-2 Mutation Portal which provides access to a database of SARS-CoV-2 mutations. Sathyaseelan et al. (2023) developed a CoVe-tracker (SARS-CoV-2 evolution tracker)7 for quick surveillance of newly emerging mutations/variants/lineages to facilitate the understanding of viral evolution, transmission, and disease epidemiology. Huang et al. (2023) developed a genomic surveillance framework and a dynamic community-based variant dictionary tree, which enables early detection and continuous investigation of SARS-CoV-2 variants. Outbreak.info is a platform for scalable and dynamic surveillance of SARS-CoV-2 variants and mutations, and it relies on shared virus sequences from the GISAID Initiative (Gangavarapu et al., 2023; Tsueng et al., 2023).
2. As the recently expanding Omicron subvariants are capable of immune evasion from most of the existing neutralizing antibodies, it is imperative to explore broad-spectrum antivirals to combat the emerging variants. Resistance to monoclonal antibody neutralization is dominated by the action of epitope single amino acid substitutions in the spike protein (Cox et al., 2022). Currently, most therapeutic neutralizing antibodies and promising vaccine candidates are designed to target the RBD or use RBD as the sole antigen (Shi et al., 2020; Yang et al., 2020a, 2021; Dai et al., 2022; Han et al., 2022). A novel group of neutralizing antibodies and vaccines targeting S2 subunit of the spike [such as fusion peptide (FP), heptad repeats 1 and 2 (HR1-HR2), and stem helix (SH)] may become the next generation of therapeutic strategies. For example, COV44-62 and COV44-79 were identified as anti-FP antibodies and showed considerable neutralizing capacity (Dacon et al., 2022).
3. Strategies should be implemented to prevent long-term SARS-CoV-2 infection and to limit the spread of emerging, neutralization-resistant variants in immunocompromised patients (Gonzalez-Reiche et al., 2023). It is found that the evolutionary rate of SARS-CoV-2 in chronic infection individual is 2-fold higher than that around the globe (Chaguza et al., 2023). This persistent intrahost evolution may accelerate antigenic alteration and lead to the emergence of genetically distinct subvariants (Smith and Ashby, 2022; Ahmadi et al., 2023; Chaguza et al., 2023). Bendall et al. (2023) observed a tight transmission bottleneck that would limit the development of highly mutated VOCs in the transmission chain of acutely infected individuals, further suggesting that selection for long-term infection in immunocompromised patients may drive SARS-CoV-2 VOC evolution (Braun et al., 2021; Wilkinson et al., 2022). Surveillance by sequencing is recommended for (i) patients carried with SARS-CoV-2, (ii) patients suspected of reinfection, and (iii) patients who are immunocompromised (Landis et al., 2023).
4. Vaccination in large population acts as a valuable measure in decreasing the mortality. However, vaccination alone cannot slow the pace of viral evolution for immune evasion and therefore, vaccine protection against severe and fatal outcomes for COVID-19 patients may not be assured (Van Egeren et al., 2023). Current herd immunity and BA.5 vaccine boosters may not efficiently prevent the infection of Omicron convergent variants (Cao et al., 2022a). However, these may result from the decreased pathogenicity of SARS-CoV-2 via inducing the mutations. The vaccination against SARS-CoV-2 still efficiently decrease the case fatality rate (Wang C. et al., 2022).
In the process of SARS-CoV-2 evolution, external and internal pressures drive the selection of randomly occurring mutations, with the retention of favorable mutations leading to adaptation. SARS-CoV-2 exhibits a trajectory of evolution characterized by increased transmissibility, reduced virulence, and enhanced immune escape, enabling its long-term persistence within the population. The mutation patterns observed in pre-Omicron variants primarily manifest at recurrent amino acid sites within the spike protein, affecting the RBD conformation and glycosylation sites, consequently altering antigenicity. However, the emergence of the Omicron introduced a multitude of novel mutations, resulting in a substantial increase in transmissibility and immune evasion. Remarkably, the severity and clinical manifestation of patients did not escalate further, mainly for Omicron’s tropism for the upper respiratory tract. These changes observed in Omicron are attributed to the ongoing viral evolution. The appearance of the recombinant variant XBB and its subsequent descendants since August 2022 likely stems from the co-circulation of multiple variants and co-infection in the immunocompromised patients during the later stage of the pandemic. Although novel recombinant variants such as XBB.1.5 and XBB.1.16 demonstrate a considerable transmission advantage and outcompete the predecessors, they do not exhibit a significant increase in disease severity and display relatively moderate antibody escape. Although SARS-CoV-2 is no longer regarded as a Public Health Emergency of International Concern, its evolution persists. We strongly recommend for enhanced surveillance of the viral genome, particularly in immunocompromised patients, the development of therapeutics targeting domains beyond the RBD, and the promotion of widespread vaccination.
GC and LF: conceptualization. LF, JX, YZ, JF, and JS: data collection. LF, JX, and GC: writing—original draft preparation. LF, JX, GC, YZ, JF, WL, and JS: writing—review and editing. All authors have read and agreed to the published version of the manuscript.
This work was funded by the National Natural Science Foundation of China (82041022) and Shanghai Commission of Science and Technology (20JC1410200 and 20431900404).
The authors acknowledge the use of Biorender.com to create Figure 1, 2, and 5.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
2. ^https://www.gisaid.org/references/statements-clarifications/clade-and-lineage-nomenclature-aids-in-genomic-epidemiology-of-active-hcov-19-viruses/
3. ^https://nextstrain.org/ncov
4. ^https://www.pango.network/the-pango-nomenclaturesystem/statement-of-nomenclature-rules/
5. ^https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/
Abeywardhana, S., Premathilaka, M., Bandaranayake, U., Perera, D., and Peiris, L. D. C. (2022). In silico study of SARS-CoV-2 spike protein RBD and human ACE-2 affinity dynamics across variants and omicron subvariants. J. Med. Virol. 95:28406. doi: 10.1002/jmv.28406
Ahmadi, A. S., Zadheidar, S., Sadeghi, K., Nejati, A., Salimi, V., Hajiabdolbaghi, M., et al. (2023). SARS-CoV-2 intrahost evolution in immunocompromised patients in comparison with immunocompetent populations after treatment. J. Med. Virol. 95:e28877. doi: 10.1002/jmv.28877
Akif, A., Bhuiyan, M. A., and Islam, M. R. (2023). SARS-COV-2 omicron subvariant BF.7 is again triggering the Covid fear: what we need to know and what we should do? J. Med. Virol. 95:28551. doi: 10.1002/jmv.28551
Alkhatib, M., Svicher, V., Salpini, R., Ambrosio, F. A., Bellocchi, M. C., Carioti, L., et al. (2021). SARS-CoV-2 variants and their relevant mutational profiles: update summer 2021. Microbiol. Spectr. 9:e0109621. doi: 10.1128/spectrum.01096-21,
Altarawneh, H. N., Chemaitelly, H., Ayoub, H. H., Hasan, M. R., Coyle, P., Yassine, H. M., et al. (2022). Protective Effect of Previous SARS-CoV-2 Infection against Omicron BA.4 and BA.5 Subvariants. N Engl J Med 387, 1620–1622. doi: 10.1056/nejmc2209306
Araf, Y., Akter, F., Tang, Y., Fatemi, R., Parvez, M. D. S. A., Zheng, C., et al. (2022). Omicron variant of SARS-CoV-2: genomics, transmissibility, and responses to current COVID-19 vaccines. J. Med. Virol. 94, 1825–1832. doi: 10.1002/jmv.27588
Arora, P., Cossmann, A., Schulz, S. R., Ramos, G. M., Stankov, M. V., Jäck, H.-M., et al. (2023). Neutralisation sensitivity of the SARS-CoV-2 XBB.1 lineage. Lancet Infect. Dis. 23, 147–148. doi: 10.1016/S1473-3099(22)00831-3
Bai, C., Zhong, Q., and Gao, G. F. (2021). Overview of SARS-CoV-2 genome-encoded proteins. Sci. China Life Sci. 65, 280–294. doi: 10.1007/s11427-021-1964-4
Bal, A., Destras, G., Gaymard, A., Stefic, K., Marlet, J., Eymieux, S., et al. (2021). Two-step strategy for the identification of SARS-CoV-2 variant of concern 202012/01 and other variants with spike deletion H69–V70, France, august to December 2020. Euro Surveill. 26. doi: 10.2807/1560-7917.ES.2021.26.3.2100008
Bano, I., Sharif, M., and Alam, S. (2021). Genetic drift in the genome of SARS COV-2 and its global health concern. J. Med. Virol. 94, 88–98. doi: 10.1002/jmv.27337
Basky, G., and Vogel, L. (2022). XE, XD & XF: what to know about the omicron hybrid variants. CMAJ 194, E654–E655. doi: 10.1503/cmaj.1095998
Bate, N., Savva, C. G., Moody, P. C. E., Brown, E. A., Evans, S. E., Ball, J. K., et al. (2022). In vitro evolution predicts emerging SARS-CoV-2 mutations with high affinity for ACE2 and cross-species binding. PLoS Pathog. 18:e1010733. doi: 10.1371/journal.ppat.1010733
Bendall, E. E., Callear, A. P., Getz, A., Goforth, K., Edwards, D., Monto, A. S., et al. (2023). Rapid transmission and tight bottlenecks constrain the evolution of highly transmissible SARS-CoV-2 variants. Nat. Commun. 14:272. doi: 10.1038/s41467-023-36001-5
Benton, D. J., Wrobel, A. G., Roustan, C., Borg, A., Xu, P., Martin, S. R., et al. (2021). The effect of the D614G substitution on the structure of the spike glycoprotein of SARS-CoV-2. Proc. Natl. Acad. Sci. U. S. A. 118. doi: 10.1073/pnas.2022586118
Berger, I., and Schaffitzel, C. (2020). The SARS-CoV-2 spike protein: balancing stability and infectivity. Cell Res. 30, 1059–1060. doi: 10.1038/s41422-020-00430-4
Bestle, D., Heindl, M. R., Limburg, H., van Lam van, T., Pilgram, O., Moulton, H., et al. (2020). TMPRSS2 and furin are both essential for proteolytic activation of SARS-CoV-2 in human airway cells. Life Sci. Alliance 3:e202000786. doi: 10.26508/lsa.202000786
Bloom, J. D., Beichman, A. C., Neher, R. A., and Harris, K. (2022). Evolution of the SARS-CoV-2 mutational spectrum. bioRxiv. doi: 10.1101/2022.11.19.517207
Bolze, A., Basler, T., White, S., Dei Rossi, A., Wyman, D., Dai, H., et al. (2022). Evidence for SARS-CoV-2 Delta and omicron co-infections and recombination. Med 3, 848–859.e4. doi: 10.1016/j.medj.2022.10.002
Borges, V., Sousa, C., Menezes, L., Gonçalves, A. M., Picão, M., Almeida, J. P., et al. (2021). Tracking SARS-CoV-2 lineage B.1.1.7 dissemination: insights from nationwide spike gene target failure (SGTF) and spike gene late detection (SGTL) data, Portugal, week 49 2020 to week 3 2021. Euro Surveill. 26. doi: 10.2807/1560-7917.ES.2021.26.10.2100130
Brandolini, M., Gatti, G., Grumiro, L., Zannoli, S., Arfilli, V., Cricca, M., et al. (2023). Omicron sub-lineage BA.5 and recombinant XBB evasion from antibody neutralisation in BNT162b2 vaccine recipients. Microorganisms 11:191. doi: 10.3390/microorganisms11010191
Braun, K., Moreno, G., Wagner, C., Accola, M. A., Rehrauer, W. M., Baker, D., et al. (2021). Limited within-host diversity and tight transmission bottlenecks limit SARS-CoV-2 evolution in acutely infected individuals. bioRxiv. doi: 10.1101/2021.04.30.440988
Burel, E., Colson, P., Lagier, J.-C., Levasseur, A., Bedotto, M., Lavrard-Meyer, P., et al. (2022). Sequential appearance and isolation of a SARS-CoV-2 recombinant between two major SARS-CoV-2 variants in a chronically infected immunocompromised patient. Viruses 14:1266. doi: 10.3390/v14061266
Callaway, E. (2023). Coronavirus variant XBB.1.5 rises in the United States — is it a global threat? Nature 613, 222–223. doi: 10.1038/d41586-023-00014-3
Calvaresi, V., Wrobel, A. G., Toporowska, J., Hammerschmid, D., Doores, K. J., Bradshaw, R. T., et al. (2023). Structural dynamics in the evolution of SARS-CoV-2 spike glycoprotein. Nat. Commun. 14:1421. doi: 10.1038/s41467-023-36745-0
Cao, Y., Jian, F., Wang, J., Yu, Y., Song, W., Yisimayi, A., et al. (2022a). Imprinted SARS-CoV-2 humoral immunity induces convergent omicron RBD evolution. Nature 614, 521–529. doi: 10.1038/s41586-022-05644-7
Cao, Y., Song, W., Wang, L., Liu, P., Yue, C., Jian, F., et al. (2022b). Characterization of the enhanced infectivity and antibody evasion of omicron BA.2.75. Cell Host Microbe 30, 1527–1539.e5. doi: 10.1016/j.chom.2022.09.018
Cao, Y., Wang, J., Jian, F., Xiao, T., Song, W., Yisimayi, A., et al. (2021). Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature 602, 657–663. doi: 10.1038/s41586-021-04385-3
Cao, Y., Yisimayi, A., Jian, F., Song, W., Xiao, T., Wang, L., et al. (2022c). BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by omicron infection. Nature 608, 593–602. doi: 10.1038/s41586-022-04980-y
Carabelli, A. M., Peacock, T. P., Thorne, L. G., Harvey, W. T., Hughes, J., COVID-19 Genomics UK Consortium, et al. (2023). SARS-CoV-2 variant biology: immune escape, transmission and fitness. Nat. Rev. Microbiol. 21, 162–177. doi: 10.1038/s41579-022-00841-7
Casas-Sanchez, A., Romero-Ramirez, A., Hargreaves, E., Ellis, C. C., Grajeda, B. I., Estevao, I. L., et al. (2021). Inhibition of protein N-glycosylation blocks SARS-CoV-2 infection. mBio 13:21. doi: 10.1128/mbio.03718-21
Cele, S., Gazy, I., Jackson, L., Hwa, S. H., Tegally, H., Lustig, G., et al. (2021). Escape of SARS-CoV-2 501Y.V2 from neutralization by convalescent plasma. Nature 593, 142–146. doi: 10.1038/s41586-021-03471-w
Cele, S., Karim, F., Lustig, G., San, J. E., Hermanus, T., Tegally, H., et al. (2022). SARS-CoV-2 prolonged infection during advanced HIV disease evolves extensive immune escape. Cell Host Microbe 30, 154–162.e5. doi: 10.1016/j.chom.2022.01.005
Cerutti, G., Guo, Y., Zhou, T., Gorman, J., Lee, M., Rapp, M., et al. (2021). Potent SARS-CoV-2 neutralizing antibodies directed against spike N-terminal domain target a single supersite. Cell Host Microbe 29, 819–833.e7. doi: 10.1016/j.chom.2021.03.005
Chaguza, C., Hahn, A. M., Petrone, M. E., Zhou, S., Ferguson, D., Breban, M. I., et al. (2023). Accelerated SARS-CoV-2 intrahost evolution leading to distinct genotypes during chronic infection. Cell. Rep. Med. 4:100943. doi: 10.1016/j.xcrm.2023.100943
Chakraborty, C., Bhattacharya, M., Chopra, H., Islam, M. A., Saikumar, G., and Dhama, K. (2023). The SARS-CoV-2 omicron recombinant subvariants XBB, XBB.1, and XBB.1.5 are expanding rapidly with unique mutations, antibody evasion, and immune escape properties – an alarming global threat of a surge in COVID-19 cases again? Int. J. Surg. 109, 1041–1043. doi: 10.1097/JS9.0000000000000246
Chakraborty, C., Bhattacharya, M., Sharma, A. R., and Dhama, K. (2022). Recombinant SARS-CoV-2 variants XD, XE, and XF: the emergence of recombinant variants requires an urgent call for research – correspondence. Int. J. Surg. 102:106670. doi: 10.1016/j.ijsu.2022.106670
Chang, T.-J., Yang, D.-M., Wang, M.-L., Liang, K.-H., Tsai, P.-H., Chiou, S.-H., et al. (2020). Genomic analysis and comparative multiple sequences of SARS-CoV2. J. Chin. Med. Assoc. 83, 537–543. doi: 10.1097/jcma.0000000000000335
Chan, K. K., Tan, T. J. C., Narayanan, K. K., and Procko, E. (2021). An engineered decoy receptor for SARS-CoV-2 broadly binds protein S sequence variants. Sci. Adv. 7:eabf1738. doi: 10.1126/sciadv.abf1738
Chavda, V. P., Patel, A. B., and Vaghasiya, D. D. (2022). SARS-CoV-2 variants and vulnerability at the global level. J. Med. Virol. 94, 2986–3005. doi: 10.1002/jmv.27717
Chen, L.-L., Abdullah, S. M. U., Chan, W.-M., Chan, B. P.-C., Ip, J. D., Chu, A. W.-H., et al. (2022). Contribution of low population immunity to the severe omicron BA.2 outbreak in Hong Kong. Nat. Commun. 13:3618. doi: 10.1038/s41467-022-31395-0
Chen, Z., Li, J., Zheng, J., Jin, Y., Zhang, Y., Tang, F., et al. (2023). Emerging omicron subvariants evade neutralizing immunity elicited by vaccine or BA.1/BA.2 infection. J. Med. Virol. 95. doi: 10.1002/jmv.28539
Chi, X., Yan, R., Zhang, J., Zhang, G., Zhang, Y., Hao, M., et al. (2020). A neutralizing human antibody binds to the N-terminal domain of the spike protein of SARS-CoV-2. Science 369, 650–655. doi: 10.1126/science.abc6952
Choi, B., Choudhary, M. C., Regan, J., Sparks, J. A., Padera, R. F., Qiu, X., et al. (2020). Persistence and evolution of SARS-CoV-2 in an immunocompromised host. N. Engl. J. Med. 383, 2291–2293. doi: 10.1056/NEJMc2031364
Colson, P., Delerce, J., Beye, M., Levasseur, A., Boschi, C., Houhamdi, L., et al. (2022a). First cases of infection with the 21L/BA.2 omicron variant in Marseille, France. J. Med. Virol. 94, 3421–3430. doi: 10.1002/jmv.27695
Colson, P., Fournier, P., Delerce, J., Million, M., Bedotto, M., Houhamdi, L., et al. (2022b). Culture and identification of a “Deltamicron” SARS-CoV-2 in a three cases cluster in southern France. J. Med. Virol. 94, 3739–3749. doi: 10.1002/jmv.27789
Conceicao, C., Thakur, N., Human, S., Kelly, J. T., Logan, L., Bialy, D., et al. (2020). The SARS-CoV-2 spike protein has a broad tropism for mammalian ACE2 proteins. PLoS Biol. 18:e3001016. doi: 10.1371/journal.pbio.3001016
Coronaviridae Study Group of the International Committee on Taxonomy of Viruses (2020). The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 5, 536–544. doi: 10.1038/s41564-020-0695-z
Cox, M., Peacock, T. P., Harvey, W. T., Hughes, J., Wright, D. W., Willett, B. J., et al. (2022). SARS-CoV-2 variant evasion of monoclonal antibodies based on in vitro studies. Nat. Rev. Microbiol. 21, 112–124. doi: 10.1038/s41579-022-00809-7
Cui, Z., Liu, P., Wang, N., Wang, L., Fan, K., Zhu, Q., et al. (2022). Structural and functional characterizations of infectivity and immune evasion of SARS-CoV-2 omicron. Cells 185, 860–871.e13. doi: 10.1016/j.cell.2022.01.019
Dacon, C., Tucker, C., Peng, L., Lee, C.-C. D., Lin, T.-H., Yuan, M., et al. (2022). Broadly neutralizing antibodies target the coronavirus fusion peptide. Science 377, 728–735. doi: 10.1126/science.abq3773
Dadonaite, B., Crawford, K. H. D., Radford, C. E., Farrell, A. G., Yu, T. C., Hannon, W. W., et al. (2023). A pseudovirus system enables deep mutational scanning of the full SARS-CoV-2 spike. Cells 186, 1263–1278.e20. doi: 10.1016/j.cell.2023.02.001
Dai, L., Gao, L., Tao, L., Hadinegoro, S. R., Erkin, M., Ying, Z., et al. (2022). Efficacy and safety of the RBD-dimer–based COVID-19 vaccine ZF2001 in adults. N. Engl. J. Med. 386, 2097–2111. doi: 10.1056/NEJMoa2202261
da Silva Francisco, R., Benites, L. F., Lamarca, A. P., de Almeida, L. G. P., Hansen, A. W., Gularte, J. S., et al. (2021). Pervasive transmission of E484K and emergence of VUI-NP13L with evidence of SARS-CoV-2 co-infection events by two different lineages in Rio Grande do Sul, Brazil. medRxiv 15:198345. doi: 10.1101/2021.01.21.21249764
Davies, N. G., Abbott, S., Barnard, R. C., Jarvis, C. I., Kucharski, A. J., Munday, J. D., et al. (2021). Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science 372. doi: 10.1126/science.abg3055
Domingo, E., García-Crespo, C., Lobo-Vega, R., and Perales, C. (2021). Mutation rates, mutation frequencies, and proofreading-repair activities in RNA virus genetics. Viruses 13:1882. doi: 10.3390/v13091882
Duerr, R., Zhou, H., Tada, T., Dimartino, D., Marier, C., Zappile, P., et al. (2023). Delta-omicron recombinant escapes therapeutic antibody neutralization. iScience 26:106075. doi: 10.1016/j.isci.2023.106075
du Plessis, L., McCrone, J. T., Zarebski, A. E., Hill, V., Ruis, C., Gutierrez, B., et al. (2021). Establishment and lineage dynamics of the SARS-CoV-2 epidemic in the UK. Science 371, 708–712. doi: 10.1126/science.abf2946
Evans, J. P., Qu, P., Zeng, C., Zheng, Y.-M., Carlin, C., Bednash, J. S., et al. (2022). Neutralization of the SARS-CoV-2 Deltacron and BA.3 variants. N. Engl. J. Med. 386, 2340–2342. doi: 10.1056/NEJMc2205019
Fantini, J., Yahi, N., Colson, P., Chahinian, H., La Scola, B., and Raoult, D. (2022). The puzzling mutational landscape of the SARS-2-variant omicron. J. Med. Virol. 94, 2019–2025. doi: 10.1002/jmv.27577
Faraone, J. N., Qu, P., Evans, J. P., Zheng, Y.-M., Carlin, C., Anghelina, M., et al. (2023). Neutralization escape of omicron XBB, BR.2, and BA.2.3.20 subvariants. Cell. Rep. Med. 4:101049. doi: 10.1016/j.xcrm.2023.101049
Farheen, S., Araf, Y., Tang, Y., and Zheng, C. (2022). The Deltacron conundrum: its origin and potential health risks. J. Med. Virol. 94, 5096–5102. doi: 10.1002/jmv.27990
Faria, N. R., Mellan, T. A., Whittaker, C., Claro, I. M., Candido, D. D. S., Mishra, S., et al. (2021). Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science 372, 815–821. doi: 10.1126/science.abh2644
Focosi, D., and Casadevall, A. (2022). A critical analysis of the use of Cilgavimab plus Tixagevimab monoclonal antibody cocktail (Evusheld™) for COVID-19 prophylaxis and treatment. Viruses 14:1999. doi: 10.3390/v14091999
Focosi, D., and Maggi, F. (2022). Recombination in coronaviruses, with a focus on SARS-CoV-2. Viruses 14:1239. doi: 10.3390/v14061239
Fonager, J., Bennedbæk, M., Bager, P., Wohlfahrt, J., Ellegaard, K. M., Ingham, A. C., et al. (2022). Molecular epidemiology of the SARS-CoV-2 variant omicron BA.2 sub-lineage in Denmark, 29 November 2021 to 2 January 2022. Euro Surveill. 27:27. doi: 10.2807/1560-7917.es.2022.27.10.2200181
Fujino, T., Nomoto, H., Kutsuna, S., Ujiie, M., Suzuki, T., Sato, R., et al. (2021). Novel SARS-CoV-2 variant in travelers from Brazil to Japan. Emerg. Infect. Dis. 27, 1243–1245. doi: 10.3201/eid2704.210138
Galloway, S. E., Paul, P., MacCannell, D. R., Johansson, M. A., Brooks, J. T., MacNeil, A., et al. (2021). Emergence of SARS-CoV-2 B.1.1.7 lineage — United States, December 29, 2020–January 12, 2021. MMWR Morb. Mortal. Wkly Rep. 70, 95–99. doi: 10.15585/mmwr.mm7003e2
Gangavarapu, K., Latif, A. A., Mullen, J. L., Alkuzweny, M., Hufbauer, E., Tsueng, G., et al. (2023). Outbreak.info genomic reports: scalable and dynamic surveillance of SARS-CoV-2 variants and mutations. Nat. Methods 20, 512–522. doi: 10.1038/s41592-023-01769-3
Garcia-Beltran, W. F., Lam, E. C., St Denis, K., Nitido, A. D., Garcia, Z. H., Hauser, B. M., et al. (2021). Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cells 184:2523. doi: 10.1016/j.cell.2021.04.006
Ghafari, M., Liu, Q., Dhillon, A., Katzourakis, A., and Weissman, D. B. (2022). Investigating the evolutionary origins of the first three SARS-CoV-2 variants of concern. Front. Virol. 2:2555. doi: 10.3389/fviro.2022.942555
Gobeil, S. M.-C., Janowska, K., McDowell, S., Mansouri, K., Parks, R., Manne, K., et al. (2021). D614G mutation alters SARS-CoV-2 spike conformation and enhances protease cleavage at the S1/S2 junction. Cell Rep. 34. doi: 10.1016/j.celrep.2020.108630
Goldman, E. (2021). How the unvaccinated threaten the vaccinated for COVID-19: a Darwinian perspective. Proc. Natl. Acad. Sci. U. S. A. 118:9118. doi: 10.1073/pnas.2114279118
Gomari, M. M., Tarighi, P., Choupani, E., Abkhiz, S., Mohamadzadeh, M., Rostami, N., et al. (2023). Structural evolution of Delta lineage of SARS-CoV-2. Int. J. Biol. Macromol. 226, 1116–1140. doi: 10.1016/j.ijbiomac.2022.11.227
Gonzalez-Reiche, A. S., Alshammary, H., Schaefer, S., Patel, G., Polanco, J., Carreño, J. M., et al. (2023). Sequential intrahost evolution and onward transmission of SARS-CoV-2 variants. Nat. Commun. 14:3235. doi: 10.1038/s41467-023-38867-x
Greaney, A. J., Loes, A. N., Crawford, K. H. D., Starr, T. N., Malone, K. D., Chu, H. Y., et al. (2021). Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe 29, 463–476.e6. doi: 10.1016/j.chom.2021.02.003
Hachmann, N. P., Miller, J., Collier, A. Y., and Barouch, D. H. (2022). Neutralization escape by SARS-CoV-2 omicron subvariant BA.4.6. N. Engl. J. Med. 387, 1904–1906. doi: 10.1056/NEJMc2212117
Hale, V. L., Dennis, P. M., McBride, D. S., Nolting, J. M., Madden, C., Huey, D., et al. (2022). SARS-CoV-2 infection in free-ranging white-tailed deer. Nature 602, 481–486. doi: 10.1038/s41586-021-04353-x
Han, Y., An, Y., Chen, Q., Xu, K., Liu, X., Xu, S., et al. (2022). mRNA vaccines expressing homo-prototype/omicron and hetero-chimeric RBD-dimers against SARS-CoV-2. Cell Res. 32, 1022–1025. doi: 10.1038/s41422-022-00720-z
Harari, S., Tahor, M., Rutsinsky, N., Meijer, S., Miller, D., Henig, O., et al. (2022). Drivers of adaptive evolution during chronic SARS-CoV-2 infections. Nat. Med. 28, 1501–1508. doi: 10.1038/s41591-022-01882-4
Harris, E. (2023). XBB.1.16 deemed COVID-19 variant of interest. JAMA 329:1731. doi: 10.1001/jama.2023.7767
He, X., Hong, W., Pan, X., Lu, G., and Wei, X. (2021, 2020). SARS-CoV-2 omicron variant: characteristics and prevention. MedComm 2, 838–845. doi: 10.1002/mco2.110
Hill, V., du Plessis, L., Peacock, T. P., Aggarwal, D., Colquhoun, R., Carabelli, A. M., et al. (2022). The origins and molecular evolution of SARS-CoV-2 lineage B.1.1.7 in the UK. Virus Evol. 8. doi: 10.1093/ve/veac080
Hoffmann, M., Kleine-Weber, H., and Pöhlmann, S. (2020a). A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol. Cell 78, 779–784.e5. doi: 10.1016/j.molcel.2020.04.022
Hoffmann, M., Kleine-Weber, H., Schroeder, S., Krüger, N., Herrler, T., Erichsen, S., et al. (2020b). SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cells 181, 271–280.e8. doi: 10.1016/j.cell.2020.02.052
Hong, Q., Han, W., Li, J., Xu, S., Wang, Y., Xu, C., et al. (2022). Molecular basis of receptor binding and antibody neutralization of omicron. Nature 604, 546–552. doi: 10.1038/s41586-022-04581-9
Hosch, S., Mpina, M., Nyakurungu, E., Borico, N. S., Obama, T. M. A., Ovona, M. C., et al. (2022). Genomic surveillance enables the identification of co-infections with multiple SARS-CoV-2 lineages in Equatorial Guinea. Front. Public Health 9:8401. doi: 10.3389/fpubh.2021.818401
Hossain, M. G., Tang, Y., Akter, S., and Zheng, C. (2021). Roles of the polybasic furin cleavage site of spike protein in SARS-CoV-2 replication, pathogenesis, and host immune responses and vaccination. J. Med. Virol. 94, 1815–1820. doi: 10.1002/jmv.27539
Hossen, M. L., Baral, P., Sharma, T., Gerstman, B., and Chapagain, P. (2022). Significance of the RBD mutations in the SARS-CoV-2 omicron: from spike opening to antibody escape and cell attachment. Phys. Chem. Chem. Phys. 24, 9123–9129. doi: 10.1039/D2CP00169A
Hou, Y. J., Chiba, S., Halfmann, P., Ehre, C., Kuroda, M., Dinnon, K. H., et al. (2020). SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science 370, 1464–1468. doi: 10.1126/science.abe8499
Huang, Q., Qiu, H., Bible, P. W., Huang, Y., Zheng, F., Gu, J., et al. (2023). Early detection of SARS-CoV-2 variants through dynamic co-mutation network surveillance. Front. Public Health 11:5969. doi: 10.3389/fpubh.2023.1015969
Hu, B., Guo, H., Zhou, P., and Shi, Z.-L. (2020). Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 19, 141–154. doi: 10.1038/s41579-020-00459-7
Hui, K. P. Y., Ho, J. C. W., Cheung, M., Ng, K., Ching, R. H. H., Lai, K., et al. (2022). SARS-CoV-2 omicron variant replication in human bronchus and lung ex vivo. Nature 603, 715–720. doi: 10.1038/s41586-022-04479-6
Hu, J., Peng, P., Wang, K., Fang, L., Luo, F., Jin, A., et al. (2021). Emerging SARS-CoV-2 variants reduce neutralization sensitivity to convalescent sera and monoclonal antibodies. Cell. Mol. Immunol. 18, 1061–1063. doi: 10.1038/s41423-021-00648-1
Imai, M., Ito, M., Kiso, M., Yamayoshi, S., Uraki, R., Fukushi, S., et al. (2023). Efficacy of antiviral agents against omicron subvariants BQ.1.1 and XBB. N. Engl. J. Med. 388, 89–91. doi: 10.1056/NEJMc2214302
Jackson, B., Boni, M. F., Bull, M. J., Colleran, A., Colquhoun, R. M., Darby, A. C., et al. (2021a). Generation and transmission of interlineage recombinants in the SARS-CoV-2 pandemic. Cells 184, 5179–5188.e8. doi: 10.1016/j.cell.2021.08.014
Jackson, C. B., Farzan, M., Chen, B., and Choe, H. (2021b). Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 23, 3–20. doi: 10.1038/s41580-021-00418-x
Johnson, B. A., Xie, X., Bailey, A. L., Kalveram, B., Lokugamage, K. G., Muruato, A., et al. (2021). Loss of furin cleavage site attenuates SARS-CoV-2 pathogenesis. Nature 591, 293–299. doi: 10.1038/s41586-021-03237-4
Kang, Y.-F., Sun, C., Zhuang, Z., Yuan, R.-Y., Zheng, Q., Li, J.-P., et al. (2021). Rapid development of SARS-CoV-2 spike protein receptor-binding domain self-assembled nanoparticle vaccine candidates. ACS Nano 15, 2738–2752. doi: 10.1021/acsnano.0c08379
Karyakarte, R. P., das, R., Dudhate, S., Agarasen, J., Pillai, P., Chandankhede, P. M., et al. (2023). Clinical characteristics and outcomes of laboratory-confirmed SARS-CoV-2 cases infected with omicron subvariants and the XBB recombinant variant. Cureus 15:e35261. doi: 10.7759/cureus.35261
Kelleni, M. T. (2023). Evolution of SARS CoV-2 omicron subvariants BF.7 and XBB.1.5: time to follow Africa and abort all COVID restrictions. J. Infect. 86:405. doi: 10.1016/j.jinf.2023.01.027
Kemp, S. A., Collier, D. A., Datir, R. P., Ferreira, I. A. T. M., Gayed, S., Jahun, A., et al. (2021). SARS-CoV-2 t of chronic infection. Nature 592, 277–282. doi: 10.1038/s41586-021-03291-y
Kidd, M., Richter, A., Best, A., Cumley, N., Mirza, J., Percival, B., et al. (2021). S-variant SARS-CoV-2 lineage B1.1.7 is associated with significantly higher viral load in samples tested by TaqPath polymerase chain reaction. J. Infect. Dis. 223, 1666–1670. doi: 10.1093/infdis/jiab082
Kim, D., Quinn, J., Pinsky, B., Shah, N. H., and Brown, I. (2020). Rates of co-infection between SARS-CoV-2 and other respiratory pathogens. JAMA 323:2085. doi: 10.1001/jama.2020.6266
Koopmans, M. (2021). SARS-CoV-2 and the human-animal interface: outbreaks on mink farms. Lancet Infect. Dis. 21, 18–19. doi: 10.1016/S1473-3099(20)30912-9
Korber, B., Fischer, W. M., Gnanakaran, S., Yoon, H., Theiler, J., Abfalterer, W., et al. (2020). Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cells 182, 812–827.e19. doi: 10.1016/j.cell.2020.06.043
Kreier, F. (2022). Deltacron: the story of the variant that wasn’t. Nature 602:19. doi: 10.1038/d41586-022-00149-9
Kumar, S., Thambiraja, T. S., Karuppanan, K., and Subramaniam, G. (2021). Omicron and Delta variant of SARS-CoV-2: a comparative computational study of spike protein. J. Med. Virol. 94, 1641–1649. doi: 10.1002/jmv.27526
Kurhade, C., Zou, J., Xia, H., Liu, M., Chang, H. C., Ren, P., et al. (2023). Low neutralization of SARS-CoV-2 omicron BA.2.75.2, BQ.1.1 and XBB.1 by parental mRNA vaccine or a BA.5 bivalent booster. Nat. Med. 29, 344–347. doi: 10.1038/s41591-022-02162-x
Lacek, K. A., Rambo-Martin, B. L., Batra, D., Zheng, X., Hassell, N., Sakaguchi, H., et al. (2022a). SARS-CoV-2 Delta–omicron recombinant viruses, United States. Emerg. Infect. Dis. 28, 1442–1445. doi: 10.3201/eid2807.220526
Lacek, K. A., Rambo-Martin, B. L., Batra, D., Zheng, X., Sakaguchi, H., Peacock, T., et al. (2022b). Identification of a novel SARS-CoV-2 Delta-omicron recombinant virus in the United States. bioRxiv. 19.484981. doi: 10.1101/2022.03.19.484981
Landis, J. T., Moorad, R., Pluta, L. J., Caro-Vegas, C., McNamara, R. P., Eason, A. B., et al. (2023). Intra-host evolution provides for the continuous emergence of SARS-CoV-2 variants. MBio 14:e0344822. doi: 10.1128/mbio.03448-22
Lavie, M., Dubuisson, J., and Belouzard, S. (2022). SARS-CoV-2 spike Furin cleavage site and S2′ basic residues modulate the entry process in a host cell-dependent manner. J. Virol. 96:22. doi: 10.1128/jvi.00474-22
Le Bert, N., Clapham, H. E., Tan, A. T., Chia, W. N., Tham, C. Y. L., Lim, J. M., et al. (2021). Highly functional virus-specific cellular immune response in asymptomatic SARS-CoV-2 infection. J. Exp. Med. 218. doi: 10.1084/jem.20202617
Lim, S. P. (2023). Targeting SARS-CoV-2 and host cell receptor interactions. Antivir. Res. 210:105514. doi: 10.1016/j.antiviral.2022.105514
Lino, A., Cardoso, M. A., Martins-Lopes, P., and Gonçalves, H. M. R. (2022). Omicron – the new SARS-CoV-2 challenge? Rev. Med. Virol. 32:e2358. doi: 10.1002/rmv.2358
Lista, M. J., Winstone, H., Wilson, H. D., Dyer, A., Pickering, S., Galao, R. P., et al. (2022). The P681H mutation in the spike glycoprotein of the alpha variant of SARS-CoV-2 escapes IFITM restriction and is necessary for type I interferon resistance. J. Virol. 96:e0125022. doi: 10.1128/jvi.01250-22
Liu, Y., Liu, J., Johnson, B. A., Xia, H., Ku, Z., Schindewolf, C., et al. (2022). Delta spike P681R mutation enhances SARS-CoV-2 fitness over alpha variant. Cell Rep. 39:110829. doi: 10.1016/j.celrep.2022.110829
Li, X., Giorgi, E. E., Marichannegowda, M. H., Foley, B., Xiao, C., Kong, X.-P., et al. (2020a). Emergence of SARS-CoV-2 through recombination and strong purifying selection. Sci. Adv. 6:eabb9153. doi: 10.1126/sciadv.abb9153
Li, Y.-D., Chi, W.-Y., Su, J.-H., Ferrall, L., Hung, C.-F., and Wu, T.-C. (2020b). Coronavirus vaccine development: from SARS and MERS to COVID-19. J. Biomed. Sci. 27:695. doi: 10.1186/s12929-020-00695-2
Looi, M.-K. (2023). What do we know about the Arcturus XBB.1.16 subvariant? Stroke Vasc. Neurol. :p1074. doi: 10.1136/bmj.p1074
Lubinski, B., Fernandes, M. H. V., Frazier, L., Tang, T., Daniel, S., Diel, D. G., et al. (2022). Functional evaluation of the P681H mutation on the proteolytic activation of the SARS-CoV-2 variant B.1.1.7 (alpha) spike. iScience 25:103589. doi: 10.1016/j.isci.2021.103589
Lukassen, S., Chua, R. L., Trefzer, T., Kahn, N. C., Schneider, M. A., Muley, T., et al. (2020). SARS-CoV-2 receptor ACE 2 and TMPRSS 2 are primarily expressed in bronchial transient secretory cells. EMBO J. 39:e105114. doi: 10.15252/embj.20105114
Lu, L., Sikkema, R., Velkers, F., Nieuwenhuijse, D., Fischer, E., Meijer, P., et al. (2021). Adaptation, spread and transmission of SARS-CoV-2 in farmed minks and associated humans in the Netherlands. Nat. Commun. 12:6802. doi: 10.1038/s41467-021-27096-9
Lu, X., Chen, Y., and Zhang, G. (2023). Functional evolution of SARS-CoV-2 spike protein: maintaining wide host spectrum and enhancing infectivity via surface charge of spike protein. Comput. Struct. Biotechnol. J. 21, 2068–2074. doi: 10.1016/j.csbj.2023.03.010
Magazine, N., Zhang, T., Wu, Y., McGee, M. C., Veggiani, G., and Huang, W. (2022). Mutations and evolution of the SARS-CoV-2 spike protein. Viruses 14:640. doi: 10.3390/v14030640
Magiorkinis, G. (2023). On the evolution of SARS-CoV-2 and the emergence of variants of concern. Trends Microbiol. 31, 5–8. doi: 10.1016/j.tim.2022.10.008
Mahase, E. (2021). Delta variant: what is happening with transmission, hospital admissions, and restrictions? Stroke Vasc. Neurol. 373:n1513. doi: 10.1136/bmj.n1513
Mahase, E. (2022). COVID-19: what do we know about the delta omicron recombinant variant? Stroke Vasc. Neurol. 376:o792. doi: 10.1136/bmj.o792
Mahase, E. (2023). Covid-19: what do we know about XBB.1.5 and should we be worried? Stroke Vasc. Neurol. 380:p153. doi: 10.1136/bmj.p153
Ma, K. C., Shirk, P., Lambrou, A. S., Hassell, N., Zheng, X., Payne, A. B., et al. (2023). Genomic surveillance for SARS-CoV-2 variants: circulation of omicron lineages — United States, January 2022–May 2023. MMWR Morb. Mortal. Wkly Rep. 72, 651–656. doi: 10.15585/mmwr.mm7224a2
Mallapaty, S. (2022). Where did omicron come from? Three key theories. Nature 602, 26–28. doi: 10.1038/d41586-022-00215-2
Ma, M.-J., Qiu, S.-F., Cui, X.-M., Ni, M., Liu, H.-J., Ye, R.-Z., et al. (2022a). Persistent SARS-CoV-2 infection in asymptomatic young adults. Sig. Transduct. Target Ther. 7:77. doi: 10.1038/s41392-022-00931-1
Mansbach, R. A., Chakraborty, S., Nguyen, K., Montefiori, D. C., Korber, B., and Gnanakaran, S. (2021). The SARS-CoV-2 spike variant D614G favors an open conformational state. Sci. Adv. 7:3671. doi: 10.1126/sciadv.abf3671
Marques, A. D., Sherrill-Mix, S., Everett, J. K., Adhikari, H., Reddy, S., Ellis, J. C., et al. (2022). Multiple introductions of SARS-CoV-2 alpha and Delta variants into white-tailed deer in Pennsylvania. MBio 13:e0210122. doi: 10.1128/mbio.02101-22
Martin, D. P., Lytras, S., Lucaci, A. G., Maier, W., Grüning, B., Shank, S. D., et al. (2022). Selection analysis identifies clusters of unusual mutational changes in omicron lineage BA.1 that likely impact spike function. Mol. Biol. Evol. 39. doi: 10.1093/molbev/msac061
Maulud, S. Q., Hasan, D. A., Ali, R. K., Rashid, R. F., Saied, A. A., Dhawan, M., et al. (2022). Deltacron: apprehending a new phase of the COVID-19 pandemic. Int. J. Surg. 102:106654. doi: 10.1016/j.ijsu.2022.106654
Ma, W., Yang, J., Fu, H., Su, C., Yu, C., Wang, Q., et al. (2022b). Genomic perspectives on the emerging SARS-CoV-2 omicron variant. Genom. Proteom. Bioinformat. 20, 60–69. doi: 10.1016/j.gpb.2022.01.001
Meng, B., Abdullahi, A., Ferreira, I. A. T. M., Goonawardane, N., Saito, A., Kimura, I., et al. (2022). Altered TMPRSS2 usage by SARS-CoV-2 omicron impacts infectivity and fusogenicity. Nature 603, 706–714. doi: 10.1038/s41586-022-04474-x
Milne, G., Hames, T., Scotton, C., Gent, N., Johnsen, A., Anderson, R. M., et al. (2021). Does infection with or vaccination against SARS-CoV-2 lead to lasting immunity? Medicine 9, 1450–1466. doi: 10.1016/S2213-2600(21)00407-0
Mlcochova, P., Kemp, S. A., Dhar, M. S., Papa, G., Meng, B., Ferreira, I. A. T. M., et al. (2021). SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature 599, 114–119. doi: 10.1038/s41586-021-03944-y
Moelling, K. (2021). Within-host and between-host evolution in SARS-CoV-2—new Variant’s source. Viruses 13:751. doi: 10.3390/v13050751
Moisan, A., Mastrovito, B., de Oliveira, F., Martel, M., Hedin, H., Leoz, M., et al. (2022). Evidence of transmission and circulation of Deltacron XD recombinant severe acute respiratory syndrome coronavirus 2 in Northwest France. Clin. Infect. Dis. 75, 1841–1844. doi: 10.1093/cid/ciac360
Motozono, C., Toyoda, M., Zahradnik, J., Saito, A., Nasser, H., Tan, T. S., et al. (2021). SARS-CoV-2 spike L452R variant evades cellular immunity and increases infectivity. Cell Host Microbe 29, 1124–1136.e11. doi: 10.1016/j.chom.2021.06.006
Müller, N. F., Kistler, K. E., and Bedford, T. (2022). A Bayesian approach to infer recombination patterns in coronaviruses. Nat. Commun. 13:31749. doi: 10.1038/s41467-022-31749-8
Nehlmeier, I., Kempf, A., Arora, P., Cossmann, A., Dopfer-Jablonka, A., Stankov, M. V., et al. (2023). Host cell entry and neutralisation sensitivity of the SARS-CoV-2 XBB.1.16 lineage. Cell. Mol. Immunol. 8, 1–3. doi: 10.1038/s41423-023-01030-z
Nguyen, H. T., Zhang, S., Wang, Q., Anang, S., Wang, J., Ding, H., et al. (2021). Spike glycoprotein and host cell determinants of SARS-CoV-2 entry and cytopathic effects. J. Virol. 95. doi: 10.1128/JVI.02304-20
O’Toole, Á., Pybus, O. G., Abram, M. E., Kelly, E. J., and Rambaut, A. (2022). Pango lineage designation and assignment using SARS-CoV-2 spike gene nucleotide sequences. BMC Genomics 23:8358. doi: 10.1186/s12864-022-08358-2
Ou, J., Lan, W., Wu, X., Zhao, T., Duan, B., Yang, P., et al. (2022). Tracking SARS-CoV-2 omicron diverse spike gene mutations identifies multiple inter-variant recombination events. Sig. Transduct. Target Ther. 7:138. doi: 10.1038/s41392-022-00992-2
Ozono, S., Zhang, Y., Ode, H., Sano, K., Tan, T. S., Imai, K., et al. (2021). SARS-CoV-2 D614G spike mutation increases entry efficiency with enhanced ACE2-binding affinity. Nat. Commun. 12:848. doi: 10.1038/s41467-021-21118-2
Pang, Y. T., Acharya, A., Lynch, D. L., Pavlova, A., and Gumbart, J. C. (2022). SARS-CoV-2 spike opening dynamics and energetics reveal the individual roles of glycans and their collective impact. Commun. Biol. 5:4138. doi: 10.1038/s42003-022-04138-6
Pan, Y., Wang, L., Feng, Z., Xu, H., Li, F., Shen, Y., et al. (2023). Characterisation of SARS-CoV-2 variants in Beijing during 2022: an epidemiological and phylogenetic analysis. Lancet 401, 664–672. doi: 10.1016/S0140-6736(23)00129-0
Patiño-Galindo, J. Á., Filip, I., and Rabadan, R. (2021). Global patterns of recombination across human viruses. Mol. Biol. Evol. 38, 2520–2531. doi: 10.1093/molbev/msab046
Peacock, T. P., Goldhill, D. H., Zhou, J., Baillon, L., Frise, R., Swann, O. C., et al. (2021). The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets. Nat. Microbiol. 6, 899–909. doi: 10.1038/s41564-021-00908-w
Pedro, N., Silva, C. N., Magalhães, A. C., Cavadas, B., Rocha, A. M., Moreira, A. C., et al. (2021). Dynamics of a dual SARS-CoV-2 lineage co-infection on a prolonged viral shedding COVID-19 case: insights into clinical severity and disease duration. Microorganisms 9:300. doi: 10.3390/microorganisms9020300
Perales, C. (2021). Mutation rates, mutation frequencies, and proofreading-repair activities in RNA virus genetics. Viruses 13:1882. doi: 10.3390/v13091882
Perez-Florido, J., Casimiro-Soriguer, C. S., Ortuño, F., Fernandez-Rueda, J. L., Aguado, A., Lara, M., et al. (2023). Detection of high level of co-infection and the emergence of novel SARS CoV-2 Delta-omicron and omicron-omicron recombinants in the epidemiological surveillance of Andalusia. IJMS 24:2419. doi: 10.3390/ijms24032419
Planas, D., Bruel, T., Staropoli, I., Guivel-Benhassine, F., Porrot, F., Maes, P., et al. (2023). Resistance of omicron subvariants BA.2.75.2, BA.4.6, and BQ.1.1 to neutralizing antibodies. Nat. Commun. 14:824. doi: 10.1038/s41467-023-36561-6
Plante, J. A., Liu, Y., Liu, J., Xia, H., Johnson, B. A., Lokugamage, K. G., et al. (2021). Spike mutation D614G alters SARS-CoV-2 fitness. Nature 592, 116–121. doi: 10.1038/s41586-020-2895-3
Qu, P., Evans, J. P., Faraone, J. N., Zheng, Y.-M., Carlin, C., Anghelina, M., et al. (2023). Enhanced neutralization resistance of SARS-CoV-2 omicron subvariants BQ.1, BQ.1.1, BA.4.6, BF.7, and BA.2.75.2. Cell Host Microbe 31, 9–17.e3. doi: 10.1016/j.chom.2022.11.012
Qu, P., Evans, J. P., Zheng, Y.-M., Carlin, C., Saif, L. J., Oltz, E. M., et al. (2022). Evasion of neutralizing antibody responses by the SARS-CoV-2 BA.2.75 variant. Cell Host Microbe 30, 1518–1526.e4. doi: 10.1016/j.chom.2022.09.015
Rahbar Saadat, Y., Hosseiniyan Khatibi, S. M., Zununi Vahed, S., and Ardalan, M. (2021). Host serine proteases: a potential targeted therapy for COVID-19 and influenza. Front. Mol. Biosci. 8:5528. doi: 10.3389/fmolb.2021.725528
Rahimi, F., and Bezmin Abadi, A. T. (2022a). The omicron subvariant BA.2: birth of a new challenge during the COVID-19 pandemic. Int. J. Surg. 99:106261. doi: 10.1016/j.ijsu.2022.106261
Rahimi, F., and Bezmin Abadi, A. T. (2022b). Detection of the XE subvariant of SARS-CoV-2: a perspective. Int. J. Surg. 101:106642. doi: 10.1016/j.ijsu.2022.106642
Rambaut, A., Holmes, E. C., O’Toole, Á., Hill, V., McCrone, J. T., Ruis, C., et al. (2020). A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 5, 1403–1407. doi: 10.1038/s41564-020-0770-5
Ren, W., Zhang, Y., Rao, J., Wang, Z., Lan, J., Liu, K., et al. (2023). Evolution of immune evasion and host range expansion by the SARS-CoV-2 B.1.1.529 (omicron) variant. M bio 14. doi: 10.1128/mbio.00416-23
Robson, F., Khan, K. S., le, T. K., Paris, C., Demirbag, S., Barfuss, P., et al. (2020). Coronavirus RNA proofreading: molecular basis and therapeutic targeting. Mol. Cell 79, 710–727. doi: 10.1016/j.molcel.2020.07.027
Rockett, R. J., Draper, J., Gall, M., Sim, E. M., Arnott, A., Agius, J. E., et al. (2022). Co-infection with SARS-CoV-2 omicron and Delta variants revealed by genomic surveillance. Nat. Commun. 13:2745. doi: 10.1038/s41467-022-30518-x
Sabir, J. S. M., Lam, T. T.-Y., Ahmed, M. M. M., Li, L., Shen, Y., Abo-Aba, S. E. M., et al. (2016). Co-circulation of three camel coronavirus species and recombination of MERS-CoVs in Saudi Arabia. Science 351, 81–84. doi: 10.1126/science.aac8608
Saito, A., Irie, T., Suzuki, R., Maemura, T., Nasser, H., Uriu, K., et al. (2021). Enhanced fusogenicity and pathogenicity of SARS-CoV-2 Delta P681R mutation. Nature 602, 300–306. doi: 10.1038/s41586-021-04266-9
Saito, A., Tamura, T., Zahradnik, J., Deguchi, S., Tabata, K., Anraku, Y., et al. (2022). Virological characteristics of the SARS-CoV-2 omicron BA.2.75 variant. Cell Host Microbe 30, 1540–1555.e15. doi: 10.1016/j.chom.2022.10.003
Saldivar-Espinoza, B., Garcia-Segura, P., Novau-Ferré, N., Macip, G., Martínez, R., Puigbò, P., et al. (2023). The mutational landscape of SARS-CoV-2. IJMS 24:9072. doi: 10.3390/ijms24109072
Samoilov, A. E., Kaptelova, V. V., Bukharina, A. Y., Shipulina, O. Y., Korneenko, E. V., Saenko, S. S., et al. (2021). Case report: change of dominant strain during dual SARS-CoV-2 infection. BMC Infect. Dis. 21:959. doi: 10.1186/s12879-021-06664-w
Sathyaseelan, C., Magateshvaren Saras, M. A., Prasad Patro, L. P., Uttamrao, P. P., and Rathinavelan, T. (2023). CoVe-tracker: an interactive SARS-CoV-2 Pan proteome evolution tracker. J. Proteome Res. 22, 1984–1996. doi: 10.1021/acs.jproteome.3c00068
Saxena, S. K., Kumar, S., Ansari, S., Paweska, J. T., Maurya, V. K., Tripathi, A. K., et al. (2022). Characterization of the novel SARS-CoV-2 omicron (B.1.1.529) variant of concern and its global perspective. J. Med. Virol. 94, 1738–1744. doi: 10.1002/jmv.27524
Scarpa, F., Giovanetti, M., Azzena, I., Locci, C., Casu, M., Fiori, P. L., et al. (2023a). Genome-based survey of the SARS-CoV-2 BF.7 variant from Asia. J. Med. Virol. 95:e28714. doi: 10.1002/jmv.28714
Scarpa, F., Sanna, D., Azzena, I., Casu, M., Cossu, P., Fiori, P. L., et al. (2023b). Genome-based comparison between the recombinant SARS-CoV-2 XBB and its parental lineages. J. Med. Virol. 95:e28625. doi: 10.1002/jmv.28625
Shang, J., Wan, Y., Luo, C., Ye, G., Geng, Q., Auerbach, A., et al. (2020). Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. U. S. A. 117, 11727–11734. doi: 10.1073/pnas.2003138117
Shapira, G., Patalon, T., Gazit, S., and Shomron, N. (2023). Immunosuppression as a hub for SARS-CoV-2 mutational drift. Viruses 15:855. doi: 10.3390/v15040855
Shen, J., Fan, J., Zhao, Y., Jiang, D., Niu, Z., Zhang, Z., et al. (2023). Innate and adaptive immunity to SARS-CoV-2 and predisposing factors. Front. Immunol. 14:9326. doi: 10.3389/fimmu.2023.1159326
Sheward, D. J., Kim, C., Fischbach, J., Sato, K., Muschiol, S., Ehling, R. A., et al. (2022). Omicron sublineage BA.2.75.2 exhibits extensive escape from neutralising antibodies. Lancet Infect. Dis. 22, 1538–1540. doi: 10.1016/S1473-3099(22)00663-6
Shi, R., Shan, C., Duan, X., Chen, Z., Liu, P., Song, J., et al. (2020). A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature 584, 120–124. doi: 10.1038/s41586-020-2381-y
Singh, J., Vashishtha, S., Rahman, S. A., Ehtesham, N. Z., Alam, A., Kundu, B., et al. (2022). Energetics of spike protein opening of SARS-CoV-1 and SARS-CoV-2 and its variants of concern: implications in host receptor scanning and transmission. Biochemistry 61, 2188–2197. doi: 10.1021/acs.biochem.2c00301
Smith, C. A., and Ashby, B. (2022). Antigenic evolution of SARS-CoV-2 in immunocompromised hosts. EMPH 11, 90–100. doi: 10.1093/emph/eoac037
Starr, T. N., Greaney, A. J., Hilton, S. K., Ellis, D., Crawford, K. H. D., Dingens, A. S., et al. (2020). Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cells 182, 1295–1310.e20. doi: 10.1016/j.cell.2020.08.012
Sungnak, W., Huang, N., Bécavin, C., Berg, M., Queen, R., Litvinukova, M., et al. (2020). SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 26, 681–687. doi: 10.1038/s41591-020-0868-6
Sztain, T., Ahn, S.-H., Bogetti, A. T., Casalino, L., Goldsmith, J. A., Seitz, E., et al. (2021). A glycan gate controls opening of the SARS-CoV-2 spike protein. Nat. Chem. 13, 963–968. doi: 10.1038/s41557-021-00758-3
Tamura, T., Ito, J., Uriu, K., Zahradnik, J., Kida, I., Nasser, H., et al. (2022). Virological characteristics of the SARS-CoV-2 XBB variant derived from recombination of two omicron subvariants. bioRxiv. 521986. doi: 10.1101/2022.12.27.521986
Tan, C. Y., Chiew, C. J., Pang, D., Lee, V. J., Ong, B., Lye, D. C., et al. (2023). Protective immunity of SARS-CoV-2 infection and vaccines against medically attended symptomatic omicron BA.4, BA.5, and XBB reinfections in Singapore: a national cohort study. Lancet Infect. Dis. 23, 799–805. doi: 10.1016/S1473-3099(23)00060-9
Tao, K., Tzou, P. L., Nouhin, J., Gupta, R. K., de Oliveira, T., Kosakovsky Pond, S. L., et al. (2021). The biological and clinical significance of emerging SARS-CoV-2 variants. Nat. Rev. Genet. 22, 757–773. doi: 10.1038/s41576-021-00408-x
Tegally, H., Moir, M., Everatt, J., Giovanetti, M., Scheepers, C., Wilkinson, E., et al. (2022). Emergence of SARS-CoV-2 omicron lineages BA.4 and BA.5 in South Africa. Nat. Med. 28, 1785–1790. doi: 10.1038/s41591-022-01911-2
Tegally, H., Wilkinson, E., Lessells, R. J., Giandhari, J., Pillay, S., Msomi, N., et al. (2021). Sixteen novel lineages of SARS-CoV-2 in South Africa. Nat. Med. 27, 440–446. doi: 10.1038/s41591-021-01255-3
Thorne, L. G., Bouhaddou, M., Reuschl, A.-K., Zuliani-Alvarez, L., Polacco, B., Pelin, A., et al. (2021). Evolution of enhanced innate immune evasion by SARS-CoV-2. Nature 602, 487–495. doi: 10.1038/s41586-021-04352-y
Tian, D., Sun, Y., Xu, H., and Ye, Q. (2022). The emergence and epidemic characteristics of the highly mutated SARS-CoV-2 omicron variant. J. Med. Virol. 94, 2376–2383. doi: 10.1002/jmv.27643
Tian, J.-H., Patel, N., Haupt, R., Zhou, H., Weston, S., Hammond, H., et al. (2021). SARS-CoV-2 spike glycoprotein vaccine candidate NVX-CoV2373 immunogenicity in baboons and protection in mice. Nat. Commun. 12:372. doi: 10.1038/s41467-020-20653-8
Tortorici, M. A., Beltramello, M., Lempp, F. A., Pinto, D., Dang, H. V., Rosen, L. E., et al. (2020). Ultrapotent human antibodies protect against SARS-CoV-2 challenge via multiple mechanisms. Science 370, 950–957. doi: 10.1126/science.abe3354
Tsueng, G., Mullen, J. L., Alkuzweny, M., Cano, M., Rush, B., Haag, E., et al. (2023). Outbreak.info research library: a standardized, searchable platform to discover and explore COVID-19 resources. Nat. Methods 20, 536–540. doi: 10.1038/s41592-023-01770-w
Turakhia, Y., Thornlow, B., Hinrichs, A., McBroome, J., Ayala, N., Ye, C., et al. (2022). Pandemic-scale phylogenomics reveals the SARS-CoV-2 recombination landscape. Nature 609, 994–997. doi: 10.1038/s41586-022-05189-9
Turelli, P., Fenwick, C., Raclot, C., Genet, V., Pantaleo, G., and Trono, D. (2022). P2G3 human monoclonal antibody neutralizes SARS-CoV-2 omicron subvariants including BA.4 and BA.5 and Bebtelovimab escape mutants. bioRxiv. 501852. doi: 10.1101/2022.07.28.501852
Uraki, R., Ito, M., Kiso, M., Yamayoshi, S., Iwatsuki-Horimoto, K., Sakai-Tagawa, Y., et al. (2023). Efficacy of antivirals and bivalent mRNA vaccines against SARS-CoV-2 isolate CH.1.1. Lancet Infect. Dis. 23, 525–526. doi: 10.1016/S1473-3099(23)00132-9
van Egeren, D., Stoddard, M., White, L. F., Hochberg, N. S., Rogers, M. S., Zetter, B., et al. (2023). Vaccines alone cannot slow the evolution of SARS-CoV-2. Vaccines 11:853. doi: 10.3390/vaccines11040853
Varabyou, A., Pockrandt, C., Salzberg, S. L., and Pertea, M. (2021). Rapid detection of inter-clade recombination in SARS-CoV-2 with Bolotie. Genetics 218:74. doi: 10.1093/genetics/iyab074
Varghese, R., Pai, S., Kumar, D., and Sharma, R. (2023). SARS-CoV-2 XBB.1.16 variant: India in focus? J. Med. Virol. 95:28829. doi: 10.1002/jmv.28829
Venkatakrishnan, A. J., Anand, P., Lenehan, P. J., Suratekar, R., Raghunathan, B., Niesen, M. J. M., et al. (2022). On the origins of Omicron’s unique spike gene insertion. Vaccines 10:1509. doi: 10.3390/vaccines10091509
Viana, R., Moyo, S., Amoako, D. G., Tegally, H., Scheepers, C., Althaus, C. L., et al. (2022). Rapid epidemic expansion of the SARS-CoV-2 omicron variant in southern Africa. Nature 603, 679–686. doi: 10.1038/s41586-022-04411-y
Vkovski, P., Kratzel, A., Steiner, S., Stalder, H., and Thiel, V. (2020). Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 19, 155–170. doi: 10.1038/s41579-020-00468-6
Volz, E., Mishra, S., Chand, M., Barrett, J. C., Johnson, R., Geidelberg, L., et al. (2021). Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England. Nature 593, 266–269. doi: 10.1038/s41586-021-03470-x
Walls, A. C., Park, Y.-J., Tortorici, M. A., Wall, A., McGuire, A. T., and Veesler, D. (2020). Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cells 181, 281–292.e6. doi: 10.1016/j.cell.2020.02.058
Wang, C., Liu, B., Zhang, S., Huang, N., Zhao, T., Lu, Q., et al. (2022). Differences in incidence and fatality of COVID-19 by SARS-CoV-2 omicron variant versus Delta variant in relation to vaccine coverage: a world-wide review. J. Med. Virol. 95:e28118. doi: 10.1002/jmv.28118
Wang, L., and Gao, G. F. (2022). The “wolf” is indeed coming: recombinant “Deltacron” SARS-CoV-2 detected. China. CDC. Wkly. 4, 285–287. doi: 10.46234/ccdcw2022.054
Wang, L., Zhou, H., Li, J., Cheng, Y., Zhang, S., Aliyari, S., et al. (2022). Potential intervariant and intravariant recombination of Delta and omicron variants. J. Med. Virol. 94, 4830–4838. doi: 10.1002/jmv.27939
Wang, P., Nair, M. S., Liu, L., Iketani, S., Luo, Y., Guo, Y., et al. (2021). Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature 593, 130–135. doi: 10.1038/s41586-021-03398-2
Wang, Q., Iketani, S., Li, Z., Guo, Y., Yeh, A. Y., Liu, M., et al. (2022a). Antigenic characterization of the SARS-CoV-2 omicron subvariant BA.2.75. Cell Host Microbe 30, 1512–1517.e4. doi: 10.1016/j.chom.2022.09.002
Wang, Q., Iketani, S., Li, Z., Liu, L., Guo, Y., Huang, Y., et al. (2022b). Alarming antibody evasion properties of rising SARS-CoV-2 BQ and XBB subvariants. Cells 186, 279–286.e8. doi: 10.1016/j.cell.2022.12.018
Wang, Q., Li, Z., Ho, J., Guo, Y., Yeh, A. Y., Mohri, H., et al. (2022c). Resistance of SARS-CoV-2 omicron subvariant BA.4.6 to antibody neutralisation. Lancet Infect. Dis. 22, 1666–1668. doi: 10.1016/s1473-3099(22)00694-6
Wang, S., Xu, X., Wei, C., Li, S., Zhao, J., Zheng, Y., et al. (2021). Molecular evolutionary characteristics of SARS-CoV-2 emerging in the United States. J. Med. Virol. 94, 310–317. doi: 10.1002/jmv.27331
Wang, Y., Liu, D., Shi, W., Lu, R., Wang, W., Zhao, Y., et al. (2015). Origin and possible genetic recombination of the Middle East respiratory syndrome coronavirus from the first imported case in China: Phylogenetics and coalescence analysis. M bio 6, e01280–e01215. doi: 10.1128/mBio.01280-15
Wassenaar, T. M., Wanchai, V., Buzard, G., and Ussery, D. W. (2022). The first three waves of the Covid-19 pandemic hint at a limited genetic repertoire for SARS-CoV-2. FEMS Microbiol. Rev. 46:fuac003. doi: 10.1093/femsre/fuac003
Watanabe, Y., Allen, J. D., Wrapp, D., McLellan, J. S., and Crispin, M. (2020). Site-specific glycan analysis of the SARS-CoV-2 spike. Science :eabb9983. doi: 10.1126/science.abb9983
Watanabe, Y., Bowden, T. A., Wilson, I. A., and Crispin, M. (2019). Exploitation of glycosylation in enveloped virus pathobiology. Biochim. Biophys. Acta Gen. Subj. 1863, 1480–1497. doi: 10.1016/j.bbagen.2019.05.012
Weissman, D., Alameh, M.-G., de Silva, T., Collini, P., Hornsby, H., Brown, R., et al. (2020). D614G spike mutation increases SARS CoV-2 susceptibility to neutralization. Cell Host Microbe 29, 23–31.e4. doi: 10.1016/j.chom.2020.11.012
Wertheim, J. O., Wang, J. C., Leelawong, M., Martin, D. P., Havens, J. L., Chowdhury, M. A., et al. (2022). Capturing intrahost recombination of SARS-CoV-2 during superinfection with alpha and epsilon variants in New York City. medRxiv. 22269300. doi: 10.1101/2022.01.18.22269300
Wibmer, C. K., Ayres, F., Hermanus, T., Madzivhandila, M., Kgagudi, P., Oosthuysen, B., et al. (2021). SARS-CoV-2 501Y.V2 escapes neutralization by south African COVID-19 donor plasma. Nat. Med. 27, 622–625. doi: 10.1038/s41591-021-01285-x
Wilkinson, S. A. J., Richter, A., Casey, A., Osman, H., Mirza, J. D., Stockton, J., et al. (2022). Recurrent SARS-CoV-2 mutations in immunodeficient patients. Virus Evol. 8. doi: 10.1093/ve/veac050
Willett, B. J., Grove, J., MacLean, O. A., Wilkie, C., De Lorenzo, G., Furnon, W., et al. (2022). SARS-CoV-2 Omicron is an immune escape variant with an altered cell entry pathway. Nat. Microbiol. 7, 1161–1179. doi: 10.1038/s41564-022-01143-7
Wolf, J. M., Wolf, L. M., Bello, G. L., Maccari, J. G., and Nasi, L. A. (2022). Molecular evolution of SARS-CoV-2 from December 2019 to august 2022. J. Med. Virol. 95:28366. doi: 10.1002/jmv.28366
Wrapp, D., Wang, N., Corbett, K. S., Goldsmith, J. A., Hsieh, C.-L., Abiona, O., et al. (2020). Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367, 1260–1263. doi: 10.1126/science.abb2507
Wu, L., Zhou, L., Mo, M., Liu, T., Wu, C., Gong, C., et al. (2022). SARS-CoV-2 omicron RBD shows weaker binding affinity than the currently dominant Delta variant to human ACE2. Sig. Transduct. Target Ther. 7:8. doi: 10.1038/s41392-021-00863-2
Xia, S., Jiao, F., Wang, L., Yu, X., Lu, T., Fu, Y., et al. (2023a). SARS-CoV-2 omicron XBB subvariants exhibit enhanced fusogenicity and substantial immune evasion in elderly population, but high sensitivity to pan-coronavirus fusion inhibitors. J. Med. Virol. 95:e28641. doi: 10.1002/jmv.28641
Xia, S., Liu, Z., and Jiang, S. (2023b). High-throughput screening of mutations affecting SARS-CoV-2 spike functions. Trends Immunol. 44, 321–323. doi: 10.1016/j.it.2023.03.010
Xia, S., Wang, L., Zhu, Y., Lu, L., and Jiang, S. (2022). Origin, virological features, immune evasion and intervention of SARS-CoV-2 omicron sublineages. Sig. Transduct. Target Ther. 7:1105. doi: 10.1038/s41392-022-01105-9
Yamasoba, D., Uriu, K., Plianchaisuk, A., Kosugi, Y., Pan, L., Zahradnik, J., et al. (2023). Virological characteristics of the SARS-CoV-2 Omicron XBB.1.16 variant. Lancet 23, 527–528. doi: 10.1101/2023.04.06.535883
Yang, J., Wang, W., Chen, Z., Lu, S., Yang, F., Bi, Z., et al. (2020a). A vaccine targeting the RBD of the S protein of SARS-CoV-2 induces protective immunity. Nature 586, 572–577. doi: 10.1038/s41586-020-2599-8
Yang, P.-H., Ding, Y.-B., Xu, Z., Pu, R., Li, P., Yan, J., et al. (2020b). Increased circulating level of interleukin-6 and CD8+ T cell exhaustion are associated with progression of COVID-19. Infect. Dis. Poverty 9:161. doi: 10.1186/s40249-020-00780-6
Yang, S., Li, Y., Dai, L., Wang, J., He, P., Li, C., et al. (2021). Safety and immunogenicity of a recombinant tandem-repeat dimeric RBD-based protein subunit vaccine (ZF2001) against COVID-19 in adults: two randomised, double-blind, placebo-controlled, phase 1 and 2 trials. Lancet Infect. Dis. 21, 1107–1119. doi: 10.1016/S1473-3099(21)00127-4
Ye, G., Liu, B., and Li, F. (2022). Cryo-EM structure of a SARS-CoV-2 omicron spike protein ectodomain. Nat. Commun. 13:28882. doi: 10.1038/s41467-022-28882-9
Yewdell, J. W. (2021). Antigenic drift: Understanding COVID-19. Immunity 54, 2681–2687. doi: 10.1016/j.immuni.2021.11.016
Yin, W., Xu, Y., Xu, P., Cao, X., Wu, C., Gu, C., et al. (2022). Structures of the omicron spike trimer with ACE2 and an anti-omicron antibody. Science 375, 1048–1053. doi: 10.1126/science.abn8863
Yue, C., Song, W., Wang, L., Jian, F., Chen, X., Gao, F., et al. (2023). ACE2 binding and antibody evasion in enhanced transmissibility of XBB.1.5. Lancet Infect. Dis. 23, 278–280. doi: 10.1016/S1473-3099(23)00010-5
Yurkovetskiy, L., Wang, X., Pascal, K. E., Tomkins-Tinch, C., Nyalile, T. P., Wang, Y., et al. (2020). Structural and functional analysis of the D614G SARS-CoV-2 spike protein variant. Cells 183, 739–751.e8. doi: 10.1016/j.cell.2020.09.032
Zahradník, J., Marciano, S., Shemesh, M., Zoler, E., Harari, D., Chiaravalli, J., et al. (2021). SARS-CoV-2 variant prediction and antiviral drug design are enabled by RBD in vitro evolution. Nat. Microbiol. 6, 1188–1198. doi: 10.1038/s41564-021-00954-4
Zhang, J., Cai, Y., Xiao, T., Lu, J., Peng, H., Sterling, S. M., et al. (2021a). Structural impact on SARS-CoV-2 spike protein by D614G substitution. Science 372, 525–530. doi: 10.1126/science.abf2303
Zhang, L., Jackson, C. B., Mou, H., Ojha, A., Peng, H., Quinlan, B. D., et al. (2020). SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat. Commun. 11:6013. doi: 10.1038/s41467-020-19808-4
Zhang, L., Li, Q., Liang, Z., Li, T., Liu, S., Cui, Q., et al. (2021b). The significant immune escape of pseudotyped SARS-CoV-2 variant omicron. Emerg. Microbes. Infect. 11, 1–5. doi: 10.1080/22221751.2021.2017757
Zhang, L., Mann, M., Syed, Z. A., Reynolds, H. M., Tian, E., Samara, N. L., et al. (2021c). Furin cleavage of the SARS-CoV-2 spike is modulated by O-glycosylation. Proc. Natl. Acad. Sci. U. S. A. 118. doi: 10.1073/pnas.2109905118
Zhang, Y., Ndzouboukou, J. B., Lin, X., Hou, H., Wang, F., Yuan, L., et al. (2022). SARS-CoV-2 evolves to reduce but not abolish neutralizing action. J. Med. Virol. 95:28207. doi: 10.1002/jmv.28207
Zhao, H., Lu, L., Peng, Z., Chen, L.-L., Meng, X., Zhang, C., et al. (2022). SARS-CoV-2 omicron variant shows less efficient replication and fusion activity when compared with Delta variant in TMPRSS2-expressed cells. Emerg. Microbes. Infect. 11, 277–283. doi: 10.1080/22221751.2021.2023329
Zhou, B., Thao, T. T. N., Hoffmann, D., Taddeo, A., Ebert, N., Labroussaa, F., et al. (2021). SARS-CoV-2 spike D614G change enhances replication and transmission. Nature 592, 122–127. doi: 10.1038/s41586-021-03361-1
Keywords: SARS-CoV-2, evolution, Omicron, spike protein, amino acid substitution, recombination
Citation: Fang L, Xu J, Zhao Y, Fan J, Shen J, Liu W and Cao G (2023) The effects of amino acid substitution of spike protein and genomic recombination on the evolution of SARS-CoV-2. Front. Microbiol. 14:1228128. doi: 10.3389/fmicb.2023.1228128
Received: 24 May 2023; Accepted: 03 July 2023;
Published: 25 July 2023.
Edited by:
Hezhao Ji, Public Health Agency of Canada, CanadaReviewed by:
Jasdeep Singh, University of Denver, United StatesCopyright © 2023 Fang, Xu, Zhao, Fan, Shen, Liu and Cao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guangwen Cao, Z2Nhb0BzbW11LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.