
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 03 August 2023
Sec. Microbial Physiology and Metabolism
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1227206
This article is part of the Research Topic Biology of Myxobacteria View all 16 articles
 Andrew Ahearne1†
Andrew Ahearne1† Kayleigh E. Phillips1†
Kayleigh E. Phillips1† Thomas Knehans1
Thomas Knehans1 Miranda Hoing1
Miranda Hoing1 Scot E. Dowd2
Scot E. Dowd2 David Cole Stevens1*
David Cole Stevens1*Introduction: Natural products discovered from bacteria provide critically needed therapeutic leads for drug discovery, and myxobacteria are an established source for metabolites with unique chemical scaffolds and biological activities. Myxobacterial genomes accommodate an exceptional number and variety of biosynthetic gene clusters (BGCs) which encode for features involved in specialized metabolism.
Methods: In this study, we describe the collection, sequencing, and genome mining of 20 myxobacteria isolated from rhizospheric soil samples collected in North America.
Results: Nine isolates were determined to be novel species of myxobacteria including representatives from the genera Archangium, Myxococcus, Nannocystis, Polyangium, Pyxidicoccus, Sorangium, and Stigmatella. Growth profiles, biochemical assays, and descriptions were provided for all proposed novel species. We assess the BGC content of all isolates and observe differences between Myxococcia and Polyangiia clusters.
Discussion: Continued discovery and sequencing of novel myxobacteria from the environment provide BGCs for the genome mining pipeline. Utilizing complete or near-complete genome sequences, we compare the chromosomal organization of BGCs of related myxobacteria from various genera and suggest that the spatial proximity of hybrid, modular clusters contributes to the metabolic adaptability of myxobacteria.
Myxobacteria are metabolically “gifted” bacteria with large genomes accommodating an exceptional number of biosynthetic gene clusters (BGCs) and the potential to produce highly diverse specialized metabolites (Baltz, 2017, 2021; Herrmann et al., 2017; Bader et al., 2020). Excellent reservoirs of candidate therapeutics, over 100 unique metabolite scaffolds have been discovered from myxobacteria (Herrmann et al., 2017). Extensive metabolomic analysis of ~2,300 myxobacterial extracts revealed a correlation between detected metabolites and taxonomic distance with genus-level hierarchical clustering of metabolite profiles (Hoffmann et al., 2018). Although limited to the metabolic profiles of axenically grown myxobacteria, this observation suggests that the investigation of lesser-studied genera within the phylum Myxococcota might increase the likelihood of metabolite discovery. Ongoing natural product discoveries from novel species of myxobacteria reinforce the need for continued isolation and characterization of environmental myxobacteria (Bader et al., 2022; Haack et al., 2022; Okoth et al., 2022; Zeng et al., 2022). The most recently described Myxococcota belong to the genera Corallococcus, Myxococcus, and Pyxidicoccus (Chambers et al., 2020; Livingstone et al., 2020; Babadi et al., 2022; Inoue et al., 2022; Wang et al., 2022), and comparatively fewer members of lesser-studied myxobacterial taxa have been reported over the last decade (Mohr et al., 2018a; Wang et al., 2021). For example, no new type of strain Stigmatella has been reported since 2007. In this study, we report the isolation and genome sequencing of 20 environmental myxobacteria including representatives from the less well-studied Archangium, Nannocystis, and Polyangium. Complete and near-complete genome data enabled a thorough assessment of BGC content, which revealed (1) significant differences in cluster sizes of Myxococcia and Polyangiia, (2) unique biosynthetic capacity of Nannocystis, and (3) chromosomal organization of myxobacterial BGCs.
Rhizospheric soil samples collected from shrubs and trees were screened for bacterial swarms using standard prey-baiting and filter paper degradation methods (Mohr et al., 2016, 2017; Mohr, 2018) to isolate environmental myxobacteria (Supplementary Figure 1) (Adaikpoh et al., 2020). Morphology screening of visible swarms facilitated the isolation of myxobacteria from multiple genera with a specific focus on lesser-studied myxobacteria. A total of 20 environmental isolates of putative myxobacteria including 8 agarolytic isolates were obtained as monocultures (Figure 1). Lesser-studied myxobacteria include genera with agarolytic phenotypes such as Nannocystis, Polyangium, and Sorangium, hence numerous agarolytic isolates with similar morphologies were advanced for genome sequencing (Mohr, 2018). We have previously discussed 4 of the 20 environmental isolates (SCHIC03, SCPEA02, NCCRE02, and NCSPR01) (Ahearne et al., 2021). Genome sequencing of all isolates provided five complete genomes, seven draft genomes with ≤3 contigs, three draft genomes with 5–8 contigs, and five lower-quality genome assemblies with ≤44 contigs (Table 1). Genome sizes ranged from 9,459,689 to 13,831,693 Mb, and GC content varied from 68.1 to 71.5%. High-quality assemblies enabled subsequent whole-genome comparison approaches for phylogenetic analysis and assessment of biosynthetic gene cluster content and organization.
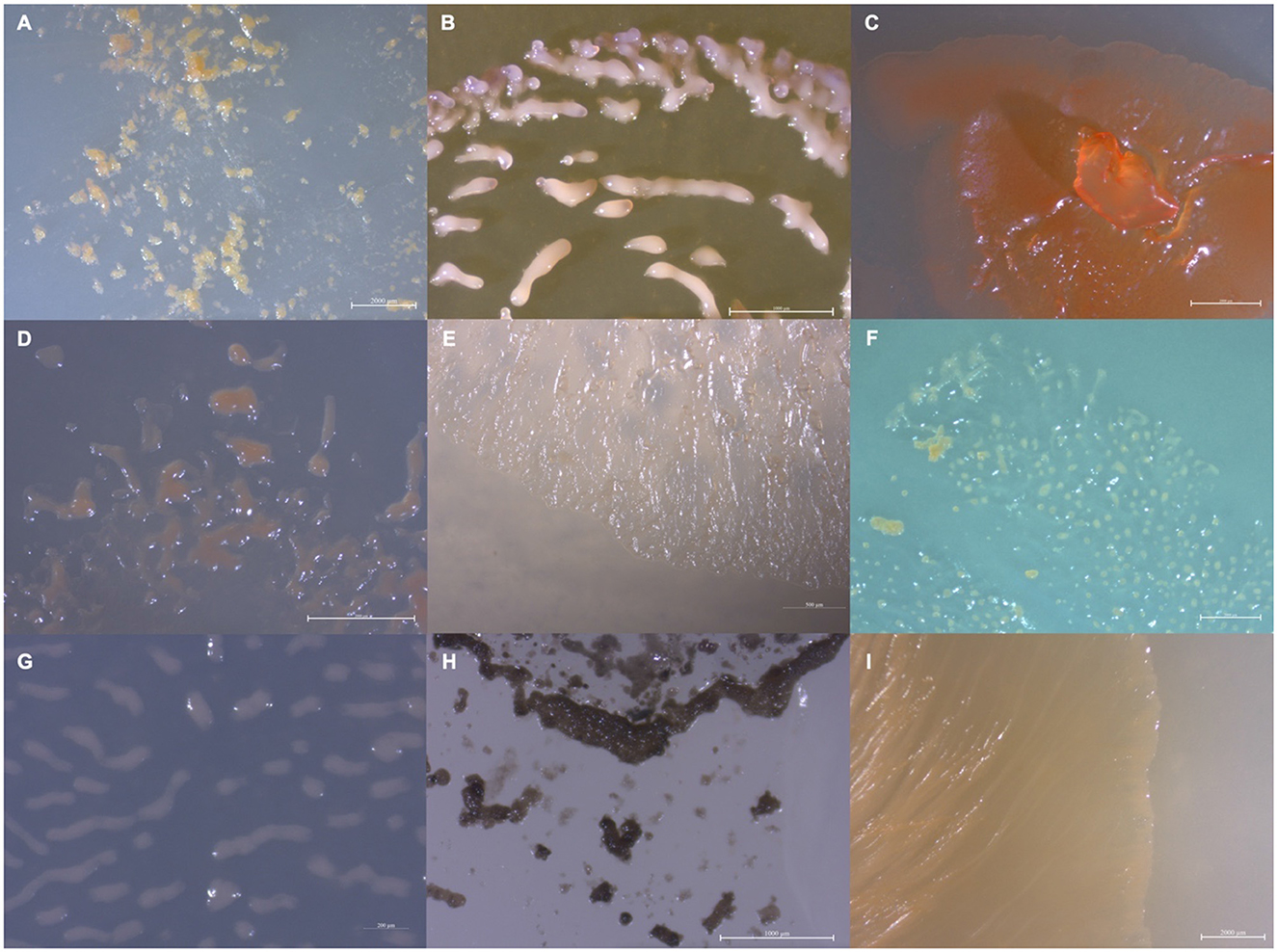
Figure 1. Images depicting the observed variety in isolate morphology (A) MIWBW, (B) SCHIC03, (C) BB15-2, (D) FL3, (E) NCELM, (F) RJM3, (G) SCPEA02, (H) WIWO2, and (I) NCWAL01.
Initial phylogenetic analysis using 16S rRNA sequences of type strain myxobacteria [obtained from the List of Prokaryotic names with Standing in Nomenclature (LPSN)] suggested the environmental isolates included 1 Archangium, 5 Corallococcus, 3 Myxococcus, 6 Nannocystis, 1 Polyangium, 2 Pyxidicoccus, 1 Sorangium, and 1 Stigmatella (Supplementary Figure 2). Utilizing genome data from isolates and the type strain of myxobacteria, sequence similarities were determined using average nucleotide identity (ANI) and digital DNA–DNA hybridization values (dDDH) according to the established methods for the taxonomic assignment of myxobacteria (Chambers et al., 2020; Livingstone et al., 2020). Resulting ANI and dDDH values indicated 10 of the 20 environmental isolates to be novel species with values below the respective cutoffs of 95% and 70% when compared to most similar type strains (Figure 2 and Supplementary Table 1). Isolate MIWBW is most phylogenetically similar to Archangium gephyra DSM2261T and A. gephyra Cbvi76 [previously referred to as Cystobacter violaceus Cbvi76 (Stevens et al., 2014)] (Figure 2A). Of the four published type strain of Archangium, A. gephyra DSM2261T is currently the only representative sufficiently sequenced for comparative genomic analysis. More rigorous analysis comparing sequenced representatives of closely related Cystobacter and Melittangium reinforced MIWBW as a novel species. This analysis also revealed Cystobacter gracilis DSM 14753T to be an outlier within the three genera with ANI values below 77.5 for all included representatives. Isolate SCHIC03 is most phylogenetically similar to Myxococcus stipitatus DSM 14675T when compared to eight Myxococcus-type strains (Figure 2B). As observed by Chambers et al. (2020) the ANI value between the established type strain species Myxococcus xanthus DSM 16526T and Myxococcus virescens DSM 2260T is above the threshold for novel species. Initial 16S rRNA analysis suggested that environmental isolates RBIL2, FL3, BB15-2, and NCELM were all novel Nannocystis species. However, of the three Nannocystis-type strains, there was no genome data for Nannocystis pusilla DSM 14622T (also referred to as N. pusilla Na p29T). Subsequent sequencing of N. pusilla DSM 14622T and comparison including our genome data for N. pusilla DSM 14622T revealed RBIL2 to be a subspecies of N. pusilla that is slightly above the novel species threshold (Figure 2E). Isolates BB15-2, FL3, and NCELM are most phylogenetically similar to Nannocystis exedens DSM 71T and are significantly distinct from each other. Our proposed addition of three Nannocystis doubles the current member total. Isolate RJM3 is most phylogenetically similar to Polyangium fumosum DSM 14688T (Figure 2C). However, only 3 of 10 Polyangium-type strains have sufficient 16S rRNA and genome sequence data suitable for thorough analysis (Lang and Reichenbach, 2013; Wang et al., 2021). Isolate SCPEA02 is most phylogenetically similar to Pyxidicoccus caerfyrddinensis CA032AT (Figure 2G) (Chambers et al., 2020). The proposed addition to Pyxidicoccus will make SCPEA02 only the fourth type of Pyxidicoccus strain. Isolate WIWO2 is most phylogenetically similar to Sorangium cellulosum Soce56, but no type strain of Sorangium has been sufficiently sequenced for comparative genomics (Figure 2D). Alternatively, 16S RNA analysis suggests that WIWO2 is most phylogenetically similar to Sorangium kenyense Soce 375T (Supplementary Figure 2). Isolate NCWAL01 is most phylogenetically similar to Stigmatella aurantiaca DSM17044T and St. aurantiaca DW4_3-1. Interestingly, our analysis indicates ANI values above the threshold for novel species for all three Stigmatella-type strains (Figure 2F).
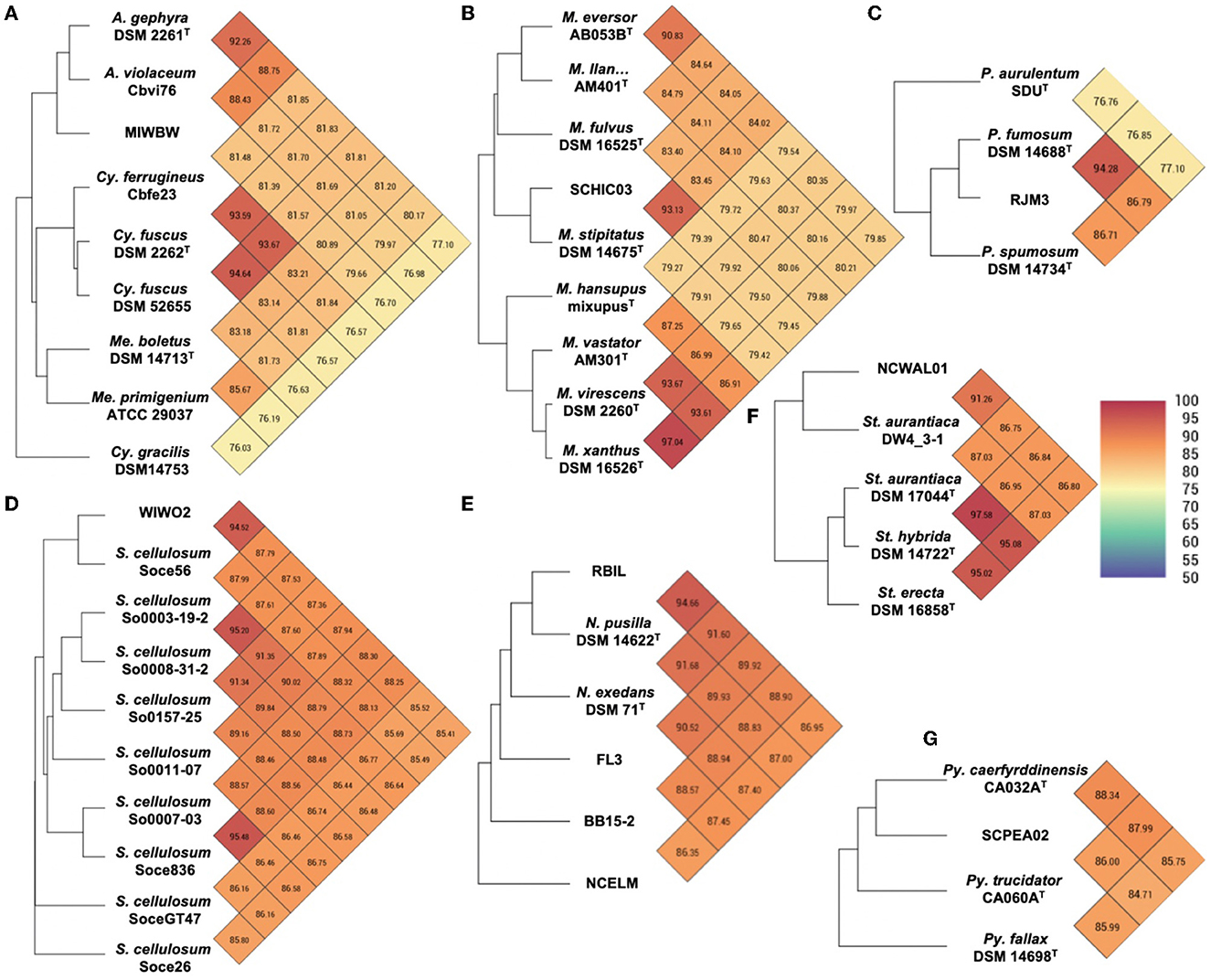
Figure 2. Heatmaps generated from OrthoANI values calculated using OAT for (A) MIWBW and sequenced strains of Archangium, Cystobacter, and Melittangium (B) SCHIC03 and type strain Myxococcus, (C) RJM3 and type strain Polyangium, (D) WIWO2 and sequenced Sorangium, (E) RBIL2, FL3, BB15-2, NCELM, and sequenced Nannocystis, (F) NCWAL01 and sequenced Stigmatella, and (G) SCPEA02 and type strain Pyxidicoccus. The established cutoff for new species designation is <95%. “M. llan…” is an abbreviation of Myxococcus llanfairpwllgwyngyllgogerychwyrndrobwllllantysiliogogogochensis AM401T.
Environmental isolates NCSPR01 and NCRR are highly similar subspecies of Corallococcus coralloides DSM 2259T (Supplementary Table 1 and Supplementary Figure 3A). As previously suggested by Ahearne et al. (2021), isolate NCCRE02 is a subspecies of Corallococcus exiguus DSM 14696T (Supplementary Table 1 and Supplementary Figure 3A). Isolate BB12-1 is likely a subspecies of Corallococcus terminator CA054AT, and isolate BB11-1 is potentially a novel species of Corallococcus (Supplementary Table 1 and Supplementary Figure 3B). However, fragmented genome assemblies for BB11-1 and BB12-1 limited our confidence in precise taxonomic placement. Isolate MISCRS is a subspecies of Myxococcus fulvus DSM 16525T, and isolate NMCA is a subspecies of M. xanthus DSM 16526T (Supplementary Table 1 and Supplementary Figure 3C). Isolate MSG2 is a subspecies of Py. caerfyrddinensis CA032AT (Supplementary Table 1 and Supplementary Figure 3D). Isolate SCPEA04 is a Nannocystis highly similar to NCELM, and isolates RBIL2 and ILAH1 are both subspecies of N. pusilla DSM 14622T (Supplementary Table 1 and Supplementary Figure 3E).
All isolated strains swarmed on VY/2 media, and growth characteristics at various pH values and temperatures were analyzed for all nine novel species (Table 2). All nine strains grew at 25–30°C, and SCPEA02 grew at temperatures up to 40°C. Growth at pH 7 was observed in all strains, and SCHIC03, NCELM, and SCPEA02 all grew at pH 6–9. Agarolytic strains include BB15-2, WIWO2, FL3, NCELM, and RJM3. Metabolic activity was assessed for all strains (Table 3), and none were able to reduce nitrate or metabolize arginine, glucose, or urea. All strains were able to hydrolyze esculin, and all except FL3 and WIWO2 hydrolyzed gelatin. SCPEA02 and SCHIC03 were the only strains that did not exhibit alkaline phosphatase activity. MIWBW was the only strain to demonstrate both trypsin and a-chymotrypsin activity and possessed overlapping characteristics with A. gephyra (Lang et al., 2015). The growth and activity of SCHIC03 were most similar to M. stipitatus and M. fulvus (Chambers et al., 2020). The growth profiles and biochemical activities of BB15-2, FL3, and NCELM were similar to those of other Nannocystis; however, all three demonstrated comparatively limited pH-dependent growth (Mohr et al., 2018a). Unlike Nannocystis konarekensis, N. exedens, and N. pusilla, none of the Nannocystis strains grew at pH 10. Temperature and pH-dependent growth ranges for RJM3 were notably different from those of other Polyangium, which all grow at temperatures above 30°C and pH ranges of 6–8.5 (Wang et al., 2021). The growth profile and biochemical activity of SCPEA02 were closely aligned with the reported activities of Py. caerfyrddinensis (Chambers et al., 2020). The growth profile and biochemical activity of WIWO2 overlapped somewhat with those of the recently described Sorangium species (Mohr et al., 2018b). The characterization and description of all Stigmatella-type strains pre-date the present description methodology; however, St. aurantiaca and NCELM have similar growth and biochemical profiles (Kleinig and Reichenbach, 1969; Reichenbach et al., 1969).
AntiSMASH analysis of BGCs in all sequenced isolates provided notable differences in BGC contents, sizes, and similarities with previously characterized clusters (Medema et al., 2011; Blin et al., 2021b). A total of 735 BGCs were predicted from 20 genome assemblies, and only 36 were identified by antiSMASH to be fragmented clusters (~5%) with the vast majority of fragmented BGCs included in sequenced Corallococcus strains (20 fragmented BGCs). Genome data from sequenced Archangium, Myxococcus, Polyangium, Pyxidicoccus, Sorangium, and Stigmatella provided three or fewer fragmented BGCs total from each genus. Of our isolated strains, MIWBW had the most predicted BGCs with 52 total (zero fragmented), and NMCA1 had the least with 23 total (zero fragmented). The average length of BGCs from all 20 sequenced isolates was ~56 kb. Clusters from Nannocystis strains were significantly shorter (average size ~30 kb) than Corallococcus, Myxococcus, and Pyxidicoccus clusters (Figure 3A). Clusters from Pyxidicoccus strains were significantly longer than those from Archangium, Nannocystis, Polyangium, and Sorangium clusters. Interestingly, Myxococcia clusters were significantly longer than Polyangiia BGCs (Figure 3B).
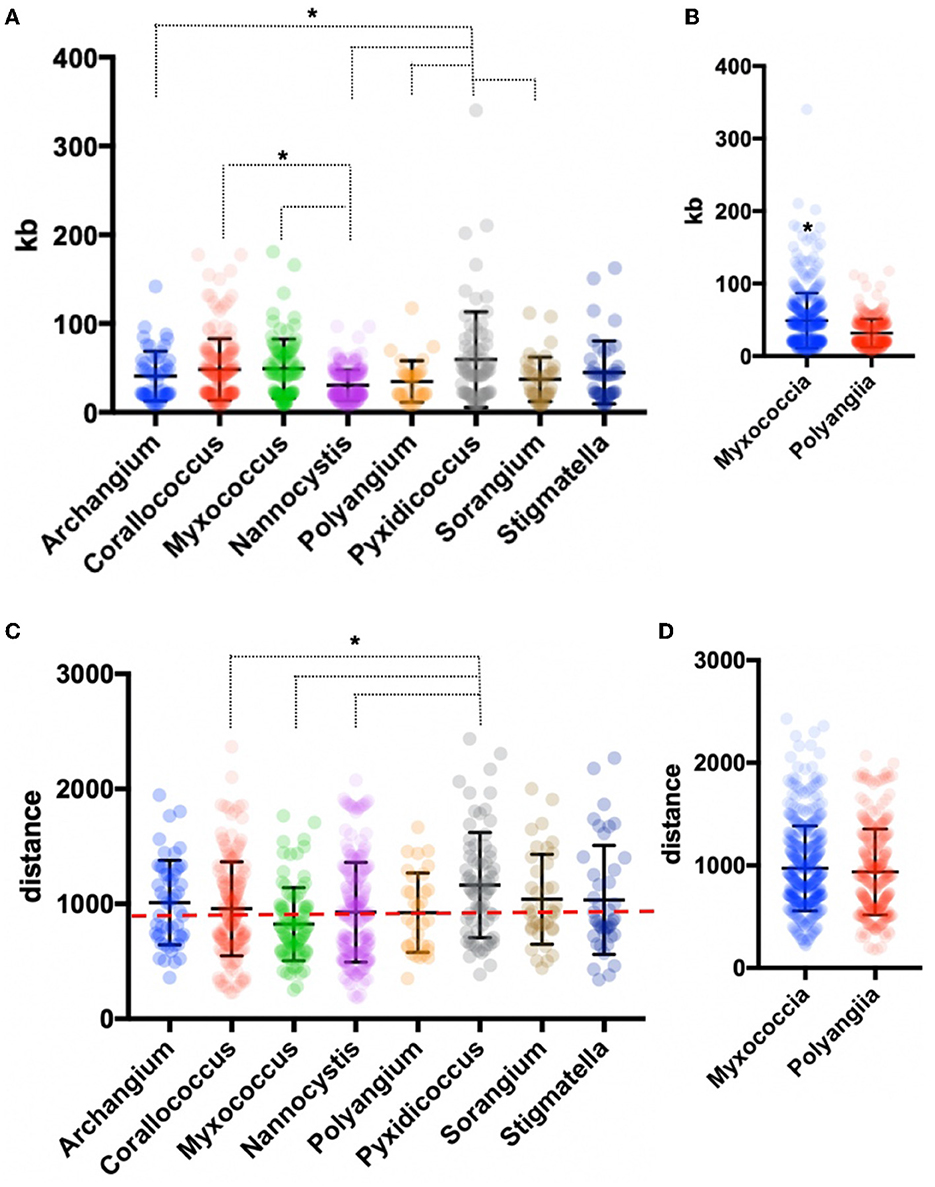
Figure 3. Genus-level analysis of BGCs and BiG-FAM distances. (A) Genus- and (B) class-level comparison of BGC size. (C) Genus- and (D) class-level comparison of BiG-FAM distances of BGCs with the red, dashed line indicating d = 900. Ordinary one-way ANOVA with multiple comparisons was used to determine the significance of genus-level analysis, and Welch's t-test was used to determine the significance of class-level analysis (for genus-level analysis Archangium n = 52, Corallococcus n = 176, Myxococcus n = 91, Nannocystis n = 212, Polyangium n = 32, Pyxidicoccus n = 73, Sorangium n = 39, Stigmatella n = 42; p < 0.05 and class-level analysis Myxococcia n = 434, Polyangiia n = 283; p < 0.0001). Asterisks indicate the associated p values included in the figure descriptions.
All identified BGCs were compared with the 1,225,071 BGCs and 29,955 gene cluster families (GCFs) included in the BiG-FAM database (Kautsar et al., 2021a,b). Utilizing a previously established clustering threshold (T = 900) to determine distance from database GCFs, we evaluated our 735 BGCs for similarity with BiG-FAM BGCs. Clusters below the arbitrary threshold have similarities with BiG-FAM GCFs and are likely less novel than clusters above the threshold (Kautsar et al., 2021a; Waschulin et al., 2022). Clusters from Pyxidicoccus strains (SCPEA02 and MSG2) had the highest average distance (1165), and somewhat predictably Myxococcus strains had the lowest (824) with an average distance below the threshold (Figure 3C). Average distance of Pyxidicoccus BGCs was significantly higher than the average distances of Corallococcus, Myxococcus, and Nannocystis clusters. Although Nannocystis and Polyangium are lesser-studied myxobacteria, the average distances of clusters from members of each genus were just above the threshold for novelty (923 and 924, respectively). No significant difference was observed between the average distances of Myxococcia and Polyangiia clusters (Figure 3D).
Of the 735 predicted BGCs, 384 clusters (~50%) had distances above the threshold. The removal of clusters below the threshold revealed differences in the remaining cluster types across genera (Figure 4). Subsequent comparison of these 384 BGCs with cluster similarities identified during antiSMASH analysis revealed that 52 clusters were either highly homologous to characterized clusters deposited in the MiBIG database (Kautsar et al., 2020) or included embedded clusters with high similarity to known clusters. For example, the myxochelin BGC was found to be embedded in 10 clusters that scored above the BiG-FAM threshold (Gaitatzis et al., 2001; Li et al., 2008). Although co-clustering likely impedes the analysis of novelty and similarity to BiG-FAM GCFs, we suggest that such co-clustering does not necessarily preclude the uniqueness of proximal clusters. This analysis also identified clusters that likely produce known metabolites including: 2-methylisoborneol (FL3), alkylpyrone 407/393 (BB11-1 and BB12-1), aurafuron A (NCWAL01), carotenoid (RJM3), chloromyxamide (MSG2), dawenol (BB12-1 and SCHIC03), dkxanthene (MISCRS, NMCA1, and SCHIC03), geosmin (BB12-1, NCRR, and NCSPR01), myxoprincomide (MSG2), nannocystin A (FL3), phenalamide A2 (SCHIC03), pyrronazol B (RBIL2), rhizopodin (NCWAL01 and SCHIC03), ripostatin A/B/C (WIWO2), and VEPE/AEPE/TG-1 (BB11-1, BB12-1, CRE02, MIWBW, MSG2, and NCRR) clusters (Botella et al., 1995; Frank et al., 2007; Jiang et al., 2007; Oliynyk et al., 2007; Meiser et al., 2008; Cortina et al., 2012; Pistorius and Muller, 2012; Bhat et al., 2014; Lorenzen et al., 2014; Osswald et al., 2014; Krastel et al., 2015; Park et al., 2016; Fu et al., 2017; Witte et al., 2017; Gorges et al., 2018; Hug et al., 2019). Modular clusters with high homology but differing organization that likely produce analogs of known metabolites were also identified, including the 2-methylisoborneol (NCELM), fulvuthiacene A/B (MISCRS), lyngbyatoxin A (NCELM and SCPEA04), myxoprincomide (MISCRS, SCPEA02, SCHIC03, and MIWBW), pyrronazol B (BB15-2 and ILAH1), and violacein (SCHIC03) clusters (Cardellina et al., 1979; Edwards and Gerwick, 2004; Jiang et al., 2007; Hoshino, 2011; Cortina et al., 2012; Witte et al., 2017; Panter et al., 2019). Additional cluster similarity was identified across the eight sequenced Nannocystis strains. For example, strain SCPEA04 contained no unique clusters that were not also present in the other seven Nannocystis genomes, and no strain had more than five unique clusters. Five intriguing novel phosphonate clusters from four Nannocystis strains (BB15-2, NCELM, SCPEA04, and RBIL2) and WIWO2 highlight the potential for novel metabolite discovery from the lesser-studied myxobacteria (Supplementary Figure 4). Typically discovered from Streptomyces and Pseudomonas spp. (Rogers and Birnbaum, 1974; Olivares et al., 2017), no phosphonate metabolites have previously been discovered from a myxobacterium. Further differences between Myxococcia and Polyangiia cluster content were revealed with subsequent analysis of BGC relatedness between isolates and sequenced myxobacteria deposited in the antiSMASH database using BiG-SCAPE (Supplementary Figures 5, 6). The resulting gene cluster families included connectivities between clusters of various Myxococcia suggesting an inherited overlap in specialized metabolism. However, all Nannocystis and Sorangium gene cluster families with two or more clusters were exclusively genus-specific, and all BGCs from Polyangium sp. strain RJM3 were present in singleton gene cluster families.
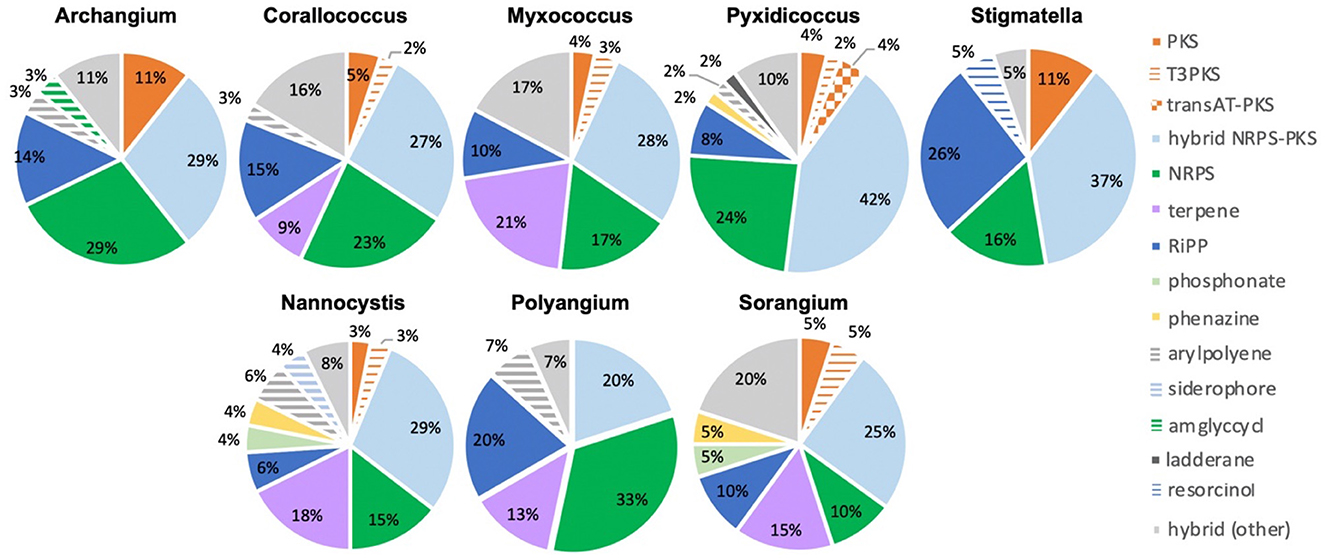
Figure 4. Genus-level distribution of likely novel BGCs by cluster type with cluster novelty assumed for BGCs with distances below the BiG-FAM threshold (d = 900). Cluster type designations provided by antiSMASH analysis.
Additional analysis of myxobacterial BGCs that encode metabolites with reported ecological utility unveiled notable differences and similarities among genera. The myxochelin cluster is present in all sequenced strains, excluding WIWO2 and Nannocystis strains (Figure 5). Myxochelin has been discovered in numerous myxobacteria and functions as a siderophore during iron starvation conditions (Silakowski et al., 2000). One or more alternative siderophore clusters are present in all Nannocystis strains. Carotenoid and VEPE/AEPE/TG-1 clusters were present in all analyzed Myxococcia and notably absent in all Polyangiia. Geosmin serves as a small molecule deterrent or “warning signal” to dissuade predatory nematodes, and the geosmin cluster is present in all strains (Zaroubi et al., 2022). Myxovirescin (Xiao et al., 2011, 2012) and myxoprincomide (Muller et al., 2016) benefit M. xanthus predation on Gram-negative and Gram-positive prey, respectively (Phillips et al., 2022). Myxovirescin has been found to significantly benefit M. xanthus predation on E. coli (Xiao et al., 2011; Ellis et al., 2019). However, none of the investigated strains including the M. xanthus strain NMCA1 possessed a cluster with similarity to the myxovirescin BGC. Aside from the absence of a myxovirescin cluster, NMCA1 and M. xanthus DK1622 share incredible similarity in BGC content including high similarity surrounding the myxovirescin BGC in M. xanthus DK1622 (Supplementary Figure 7). The myxoprincomide cluster or a cluster with high similarity to it is present in all strains excluding members of the class Polyangiia and NCWAL01.
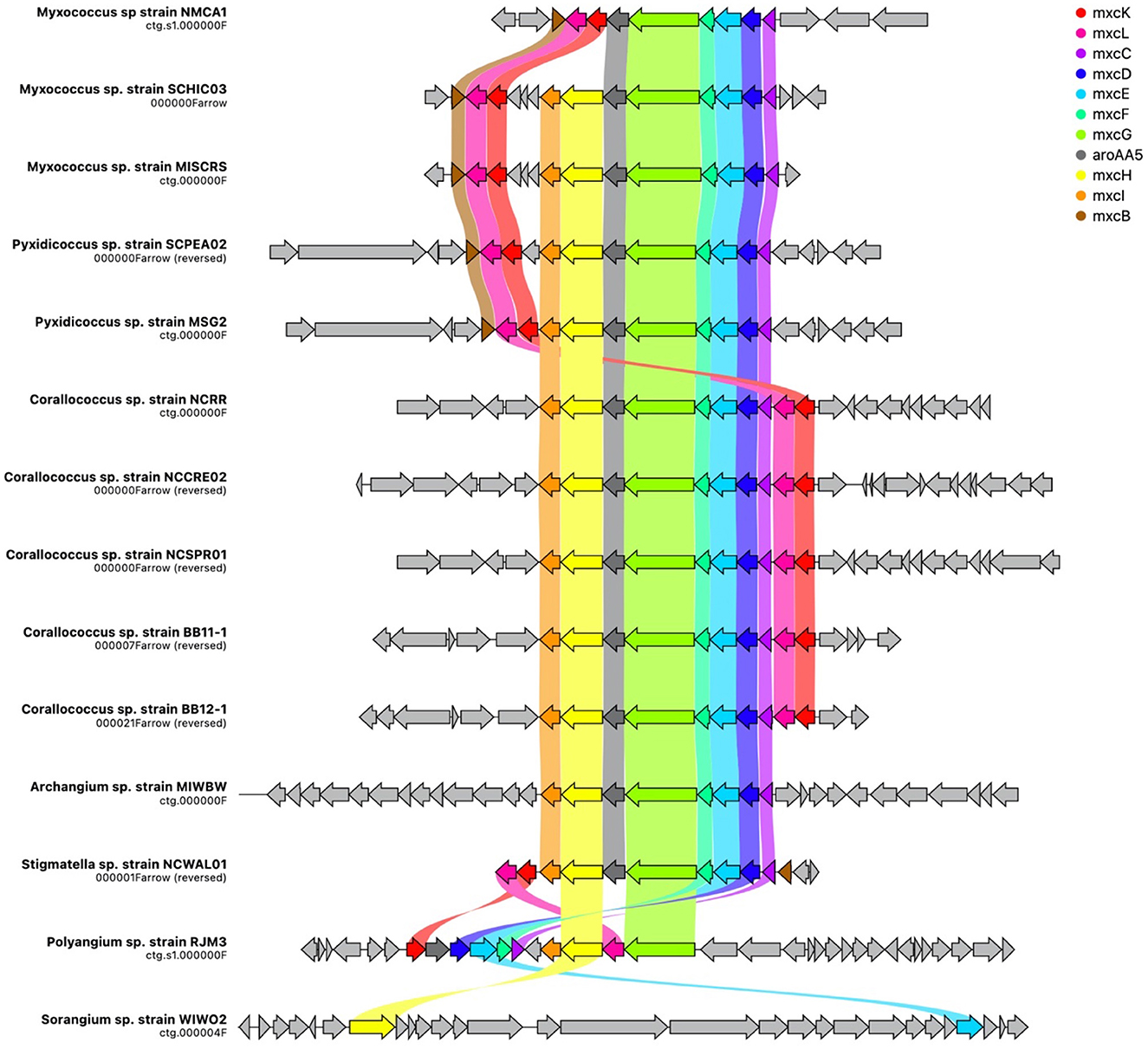
Figure 5. Spatial organization of the myxochelin BGC across all isolates with cluster homology. Gene names assigned by homology to the mxyochelin BGC deposited in the MiBIG database (BGC0001345), and ribbons indicate shared gene identity between clusters. Image generated using CAGECAT (version 1.0) using an identity threshold of 0.49 (van den Belt et al., 2023).
AntiSMASH analysis of complete or near-complete genome data from FL3, MIWBW, MSG2, NCRR, NCSPR03, NMCA1, SCHIC03, and SCPEA02 provided contiguous sequence data sufficient to observe the genome organization of BGCs. Cluster data from related myxobacteria and complete genome data from the antiSMASH database were used to compare BGC organization between related strains (Figures 6, 7). Similarities between BGC content and genome organization were observed between subspecies (Figures 6A, B) and related strains within the same genus (Figures 6C, 7). Biosynthetic gene clusters were dispersed throughout all genomes, and cluster-dense genomic segments were observed during the comparison of BGC organization. Notably, cluster-dense segments include hybrid-type clusters such as PKS-NRPS clusters or clusters including more than one cluster type. The myxochelin and myxoprincomide BGCs are located within a cluster-dense region in all sequenced environmental Myxococcia (Supplementary Figure 8). Clusters highly similar to carotenoid, geosmin, and VEPE/AEPE/TG-1 BGCs are often located in less-dense segments of analyzed chromosomes. Chromosomal segments with increased adjacency of hybrid and modular clusters were observed for all analyzed myxobacteria albeit less apparent in FL3 and N. exedens DSM 71T (Figure 7C). Other than small differences in cluster content between analyzed subspecies, such as the absence of the myxovirescin BGC from NMCA1 (Figure 6A), numerous inversions of clusters resulting in changes in cluster organization were observed. Apparent BGC inversions were predominantly located within or near cluster-dense regions, and inversions were often relegated to core biosynthetic genes of single clusters with proximal genes unchanged between strains (Supplementary Figure 9). Additional synteny analysis of strains with observed BGC inversions using a set of 10 homologous housekeeping genes revealed highly similar genome organization with no observed inversions (Supplementary Figures 10, 11) (Veltri et al., 2016).
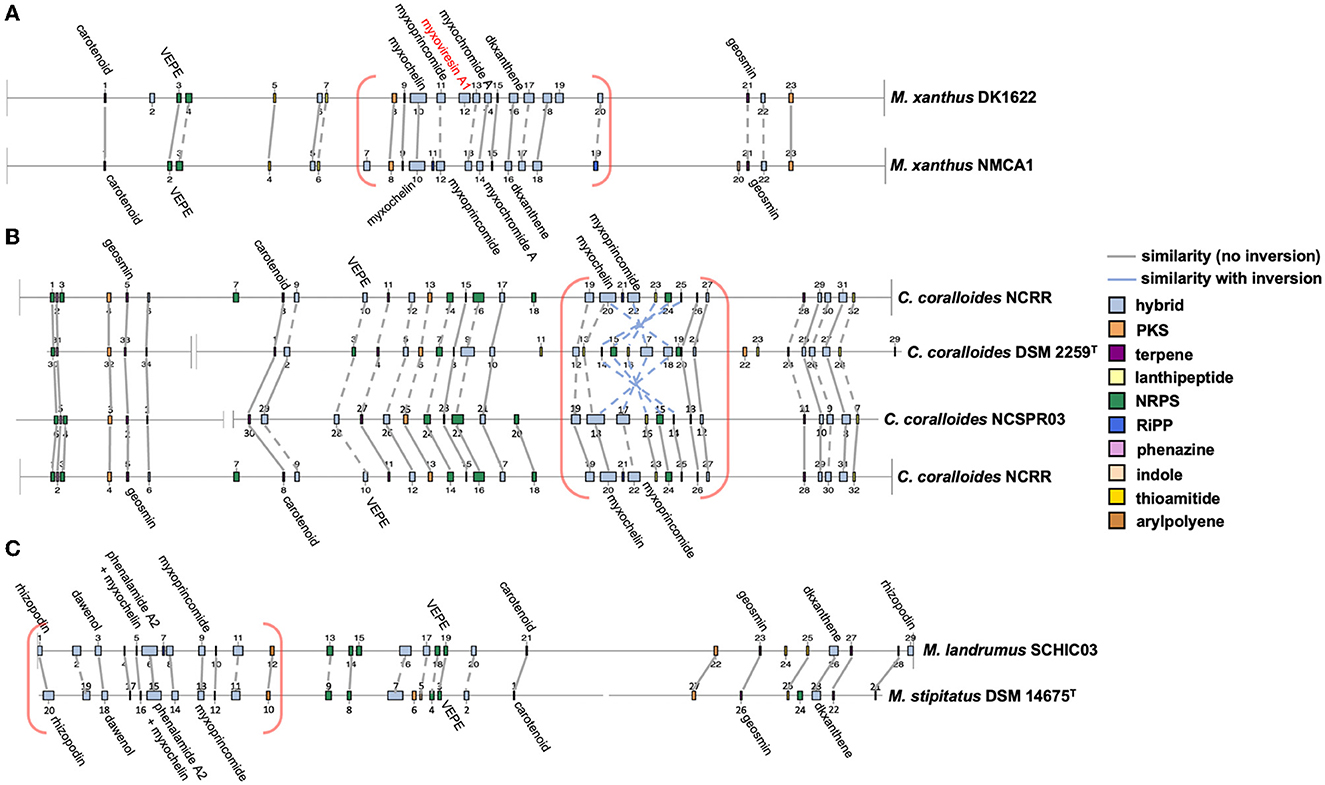
Figure 6. Whole-genome comparisons depicting similar BGC organization between subspecies (A, B) and species (C). Labeled clusters have high similarity (≥66%) with corresponding myxobacterial BGCs deposited in the MIBiG database as determined by antiSMASH analysis. Myxobacterial clusters with no matching cluster between genomes are labeled with red text. Connecting lines between clusters indicate similar gene organization, and dashed lines indicate minor changes in gene organization or domain content of modular clusters. Cluster-dense chromosomal segments are denoted with red brackets. Inversions defined as the inversion of entire clusters or subclusters within multicluster BGCs. BGC data for M. xanthus DK1622, C. coralloides DSM 2259T, and M. stipitatus DSM 14675T obtained from the antiSMASH database (version 3). Numerical labels, BGC coloring (by type), and graphical depictions of genome data taken from antiSMASH (version 6.0) with images altered to depict similarities in cluster organization.
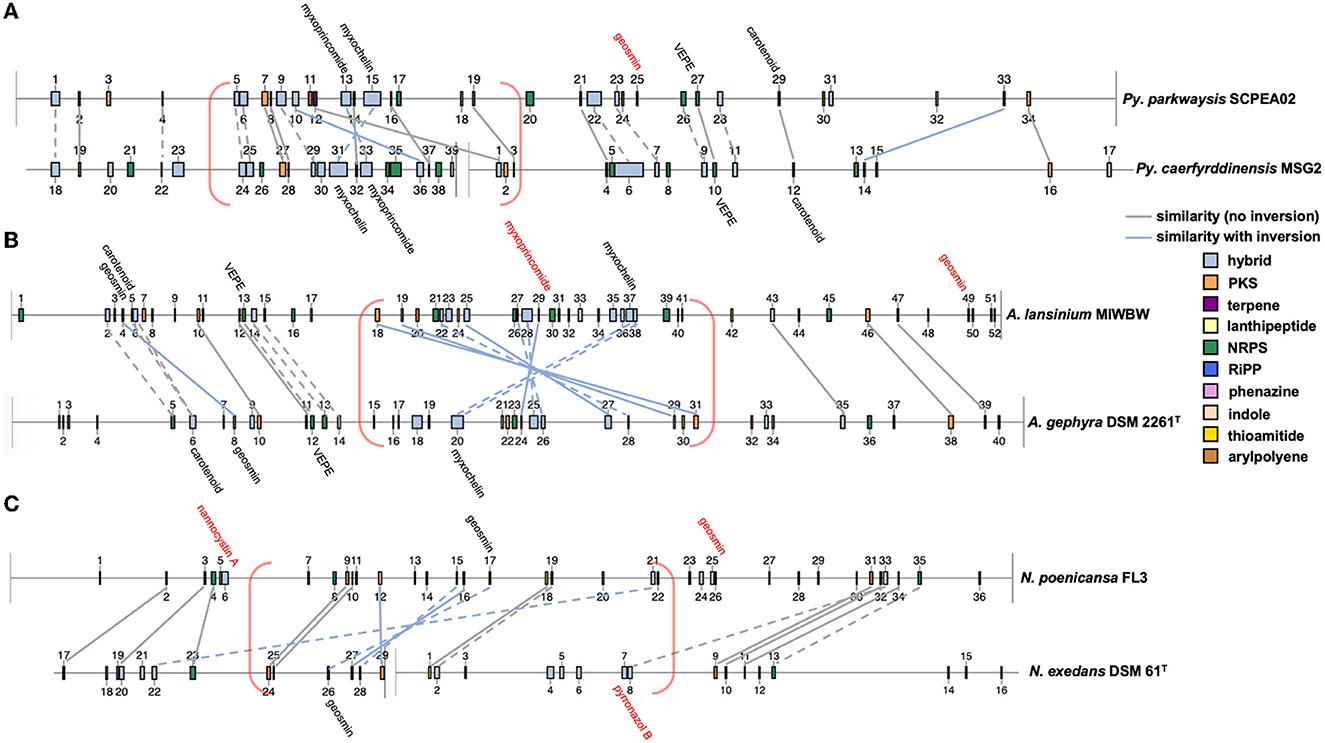
Figure 7. Cluster inversions impacting BGC organization between Pyxidicoccus (A), Archangium (B), and Nannocystis (C). Labeled clusters have high similarity (≥66%) with corresponding myxobacterial BGCs deposited in the MiBIG database as determined by antiSMASH analysis. Myxobacterial clusters with no matching cluster between genomes are labeled with red text. Connecting lines between clusters indicate similar gene organization, and dashed lines indicate slight changes in the gene organization or domain content of modular clusters. Cluster-dense chromosomal segments are denoted with red brackets. Inversions defined as the inversion of entire clusters or subclusters within multicluster BGCs. BGC data for A. gephyra DSM 2261T and N. exedens DSM 61T obtained from the antiSMASH database (version 3). Numerical labels, BGC coloring (by type), and graphical depictions of genome data taken from antiSMASH (version 6.0) with images altered to depict similarities in cluster organization.
We propose nine candidate strains to represent novel species in the genera Archangium, Myxococcus, Nannocystis, Polyangium, Pyxidicoccus, Sorangium, and Stigmatella. Comparative genomics including differences in genome content, phylogeny, and biosynthetic capacities, as well as physiological and biochemical analyses support the following distinctions: Archangium lansinium sp. nov. (MIWBWT), Myxococcus landrumus sp. nov. (SCHIC03T), Nannocystis bainbridge sp. nov. (BB15-2T), Nannocystis poenicansa sp. nov. (FL3T), Nannocystis radixulma sp. nov. (NCELMT), Polyangium mundeleinium sp. nov. (RJM3T), Pyxidicoccus parkwaysis sp. nov. (SCPEA02T), Sorangium aterium sp. nov. (WIWO2T), and Stigmatella ashevillena sp. nov. (NCWAL01T). Corresponding species descriptions for each candidate strain are provided below.
Myxobacteria are excellent resources for the discovery of therapeutics and are suggested to be keystone taxa influencing polymicrobial community structure in soil (Herrmann et al., 2017; Baltz, 2019; Bader et al., 2020; Perez et al., 2020; Petters et al., 2021). Recent discoveries of novel Corallococcus, Myxococcus, and Pyxidicoccus species as well as species from lesser-studied genera indicate an abundance of uncharacterized myxobacteria (Mohr et al., 2012, 2018a,b; Iizuka et al., 2013; Garcia et al., 2014, 2016; Yamamoto et al., 2014; Sood et al., 2015; Awal et al., 2016, 2017; Moradi et al., 2017; Garcia and Muller, 2018; Chambers et al., 2020; Livingstone et al., 2020; Wang et al., 2021, 2022; Zhou et al., 2021; Babadi et al., 2022). Our investigation of rhizospheric soil samples provided 20 environmental myxobacteria including 9 proposed novel species. As an initial attempt to isolate myxobacteria from the soil, we were surprised by the effectiveness of morphology screening to enable the discovery of myxobacteria from a variety of genera. We suspect that improved genome data will clarify the observed discrepancies in type strain differentiation and recommend that high-quality genome data be provided for all newly described type strain myxobacteria. We demonstrate that established comparative genome analysis thresholds for the designation of novel species indicate that M. xanthus DSM 16526T and M. virescens DSM 2260T, St. aurantiaca DSM 17044T, Stigmatella erecta DSM 16858T, and Stigmatella hybrida DSM 14722T are not different species. Data from sequenced Myxococcus and Pyxidicoccus strains align with the previously recommended consideration of Myxococcus/Pyxidicoccus as a single genus (Chambers et al., 2020; Wang et al., 2022). However, we note the significant differences in BGC content between Myxococcus and Pyxidicoccus strains included in this analysis. Overall, our phylogenetic analysis provides further support for comparative genomic approaches to identify and classify myxobacteria. The primary limitation is the absence of quality genome data for established type strains within genera such as Archangium, Polyangium, and Sorangium.
The current type strain Nannocystis includes N. exedens DSM 71T, N. konarekensis DSM 104509T, and N. pusilla DSM 14622T. Our investigation resulted in the discovery of an additional three proposed type strains doubling the current total of Nannocystis (Mohr et al., 2018a). We also report genome data for all three proposed type strain species as well as three Nannocystis subspecies and N. pusilla DSM 14622T. When compared to other myxobacteria, the analysis of all sequenced Nannocystis and our isolates provided notable differences in BGC content, such as the absence of a cluster similar to the myxochelin BGC, the presence of multiple siderophore and phosphonate cluster types, and smaller cluster sizes. The resulting genome data for an additional eight Nannocystis will improve future efforts to characterize and describe the members of this underexplored genus of myxobacteria.
Afforded complete or near-complete genome sequence data, we report the first comparative analysis of spatial organization of myxobacterial BGCs. Dissimilar from Streptomyces localization of BGCs in the extremities of linear genomes (Karoonuthaisiri et al., 2005; Lioy et al., 2021), clusters were distributed throughout the circular genomes of myxobacteria. Observed cluster-rich regions replete with modular BGCs, and noted inversions of biosynthetic genes could, however, contribute to metabolic differentiation similar to how terminal compartments of Streptomyces chromosomes enable spatial reorganization and conditional expression of BGCs during metabolic development and sporulation (Lioy et al., 2021). We suggest that BGC-enriched regions may benefit BGC evolution and contribute to the metabolic adaptability of myxobacteria. Compartmentalization of modular-type clusters with highly homologous domains may benefit module duplication and deletion events associated with the evolution of BGCs (Fischbach et al., 2008; Medema et al., 2014; Chevrette et al., 2020). Vertical inheritance of the myxochelin cluster is apparent in all sequenced Myxococcia. Our data also reveal vertical transfer and the likely concerted evolution of myxoprincomide-type clusters across sequenced Myxococcia (Medema et al., 2014). Alternatively, the presence of the myxovirescin trans-AT PKS cluster within a cluster-rich region of the M. xanthus DK1622 genome and the absence of a homologous cluster in NMCA1 indicate horizontal acquisition. Although the absence of the myxovirescin cluster in NMCA1 provides an alternative explanation, the absence of the myxovirescin cluster in all other sequenced Myxococcus and Myxococcota members currently deposited in the antiSMASH database supports horizontal acquisition by DK1622. Regardless, the presence of clusters in DK1622 and their absence in other myxobacteria demonstrate metabolic adaptability among myxobacterial genomes with BGC-enriched segments. Further investigation of the chromosomal organization of BGCs in myxobacteria is required to determine functional impacts on metabolic adaptability and cluster evolution.
Archangium lansinium (lan.sin'i.um. N.L. neut. adj. lansinium from Lansing, Michigan, USA, referencing the area of isolation).
Vegetative cells glide on solid media. Cells grow as translucent swarms during the early growth phase on VY/4 agar and form non-uniform, clumping fruiting bodies that range from yellow to pale orange over time. Aerobic growth was observed at 20 to 35°C but not at 40°C at a pH range of 6.0 to 8.0. Hydrolyzes gelatin and esculin. Shows an API ZYM positive reaction to alkaline phosphatase, C4 esterase, C8 lipase, C14 lipase, leucine arylamidase, valine arylamidase, cysteine arylamidase, trypsin, α-chymotrypsin, acid phosphatase, napthol-AS-BI-aphosphohydrolase, and N-acetyl-β-glucosaminidase, and a negative reaction to α-galactosidase, β-galactosidase, β-glucuronidase, α-glucosidase, β-glucosidase, α-mannosidase, and α-fucosidase. DNA GC content is 68.1%. The genome assembly of the organism is available at NCBI Assembly (ASM2662663v1). The 16S ribosomal RNA gene sequence is available at GenBank (OP852336.1). Phylogenetically most similar to A. gephyra DSM 2261T.
The type strain (MIWBWT = TSD-326T = NCCB 100916T) was isolated from soil collected in Summer 2021 from the roots of a white basswood tree near the city of Lansing, Michigan, USA (42.73°N, 84.48°W).
Myxococcus landrumus (lan.drum'us. N.L. masc. adj. landrumus from Landrum, South Carolina, USA, referencing the area of isolation).
Vegetative cells glide on solid media. Cells grow as slightly orange swarms and develop rounded, stalking fruiting bodies that range from white to purple on VY/4 media. Aerobic growth was observed at 20 to 35°C but not at 40°C at a pH range of 6.0 to 10.0. Hydrolyzes gelatin and esculin. Shows an API ZYM positive reaction to C4 esterase, C8 lipase, C14 lipase, leucine arylamidase, valine arylamidase, acid phosphatase, and napthol-AS-BI-aphosphohydrolase, and a negative reaction to alkaline phosphatase, cysteine arylamidase, trypsin, α-chymotrypsin, N-acetyl-β-glucosaminidase, α-galactosidase, β-galactosidase, β-glucuronidase, α-glucosidase, β-glucosidase, α-mannosidase, and α-fucosidase. DNA GC content is 68.6%. The genome assembly of the organism is available at NCBI Assembly (ASM1730163v1). The 16S ribosomal RNA gene sequence is available at GenBank (OP852328v1). Phylogenetically most similar to M. stipitatus DSM 14675T.
The type strain (SCHIC03T = TSD-327T = NRRL B-65669T = NCCB 100915T) was isolated from soil collected in Spring 2020 from the roots of a hickory tree near the city of Landrum, South Carolina, USA (35.14°N, -82.16°W).
Nannocystis bainbridge (bain'bridg.ea. N.L. fem. adj. bainbridge from Bainbridge Island, Washington, USA, referencing the area of isolation).
Vegetative cells glide on solid media. Cells are agarolytic and form deep etches in VY/4 agar, and grow as translucent red swarms and form rounded fruiting bodies. Aerobic growth was observed at 20 to 30°C but not at 35 to 40°C at a pH range of 7.0 to 8.0. Hydrolyzes gelatin and esculin. Shows an API ZYM positive reaction to alkaline phosphatase, C4 esterase, C8 lipase, leucine arylamidase, α-chymotrypsin, acid phosphatase, and napthol-AS-BI-aphosphohydrolase, and a negative reaction to C14 lipase, valine arylamidase, cysteine arylamidase, trypsin, N-acetyl-β-glucosaminidase, α-galactosidase, β-galactosidase, β-glucuronidase, α-glucosidase, β-glucosidase, α-mannosidase, and α-fucosidase. DNA GC content is 71.5%. The genome assembly of the organism is available at NCBI Assembly (ASM2696555v1). The 16S ribosomal RNA gene sequence is available at GenBank (OP852329.1). Phylogenetically most similar to N. exedens DSM 71T.
The type strain (BB15-2T = NCCB 100934T) was isolated from soil collected in Summer 2020 from the roots of a blueberry bush near the city of Bainbridge Island, Washington, USA (47.65°N, -122.55°W).
Nannocystis poenicansa (poe.ni.can'sa. N.L. fem. adj. poenicansa the bright red, referencing bright red pigment production).
Vegetative cells glide on solid media. Cells are agarolytic and form deep etches in VY/4 agar. Aerobic growth was observed at 20 to 35°C but not at 40°C at a pH range of 6.0 to 8.0. Hydrolyzes esculin. Shows an API ZYM positive reaction to alkaline phosphatase, C4 esterase, C8 lipase, C14 lipase, leucine arylamidase, cysteine arylamidase, acid phosphatase, napthol-AS-BI-aphosphohydrolase, and β-glucosidase, and a negative reaction to valine arylamidase, trypsin, α-chymotrypsin, N-acetyl-β-glucosaminidase, α-galactosidase, β-galactosidase, β-glucuronidase, α-glucosidase, α-mannosidase, and α-fucosidase. DNA GC content is 71.5%. The complete genome sequence of the organism is available at GenBank (CP114040.1). The 16S ribosomal RNA gene sequence is available at GenBank (OP852330.1). Phylogenetically most similar to N. exedens DSM 71T.
The type strain (FL3T = TSD-332T = NCCB 100918T) was isolated from soil collected in Fall 2020 from the roots of a southern live oak near the city of Palm Coast, Florida, USA (29.59°N, −81.21°W).
Nannocystis radixulma (ra'dix.ul.ma. L. fem. n. poenicansa the root of elm, referencing the isolation from an elm tree rhizosphere).
Vegetative cells glide on solid media. Cells are agarolytic and form deep etches in VY/4 agar. Early growth ranges from translucent swarm perimeters to yellow-pigmented swarm centers, and form textured, clumping fruiting bodies that range from yellow to orange on VY/4 agar. Aerobic growth was observed at 25 to 30°C but not at 20°C or 35 to 40°C at a pH range of 6.0 to 9.0. Hydrolyzes gelatin and esculin. Shows an API ZYM positive reaction to alkaline phosphatase, C4 esterase, C8 lipase, leucine arylamidase, cysteine arylamidase, acid phosphatase, napthol-AS-BI-aphosphohydrolase, and β-glucosidase, and a negative reaction to C14 lipase, valine arylamidase, trypsin, α-chymotrypsin, N-acetyl-β-glucosaminidase, α-galactosidase, β-galactosidase, β-glucuronidase, α-glucosidase, α-mannosidase, and α-fucosidase. DNA GC content is 71.2%. The genome assembly of the organism is available at NCBI Assembly (ASM2836909v1). The 16S ribosomal RNA gene sequence is available at GenBank (OP852331.1). Phylogenetically most similar to N. exedens DSM 71T.
The type strain (NCELMT = NCCB 100919T) was isolated from soil collected in Spring 2020 from the roots of an elm tree near the city of Asheville, North Carolina, USA (35.63°N, −82.55°W).
Polyangium mundelenium (mun.del.en'ni.um. N.L. neut. adj. mundelenium from Mundelein, Illinois, USA, referencing the area of isolation).
Vegetative cells glide on solid media. Cells grow as translucent swarms and develop dispersed rounded, yellow fruiting bodies on VY/4 media. Aerobic growth was observed at 20 to 30°C but not at 20°C or 35 to 40°C at a pH of 7.0. Hydrolyzes gelatin and esculin. Shows an API ZYM positive reaction to alkaline phosphatase, C4 esterase, C8 lipase, leucine arylamidase, valine arylamidase, α-chymotrypsin, acid phosphatase, and napthol-AS-BI-aphosphohydrolase, and a negative reaction to C14 lipase, cysteine arylamidase, trypsin, N-acetyl-β-glucosaminidase, α-galactosidase, β-galactosidase, β-glucuronidase, α-glucosidase, β-glucosidase, α-mannosidase, and α-fucosidase. DNA GC content is 68.6%. The genome assembly of the organism is available at NCBI Assembly (ASM2836910v1). The 16S ribosomal RNA gene sequence is available at GenBank (OP852332.1). Phylogenetically most similar to P. fumosum DSM 14688T.
The type strain (RJM3T = NCCB 100920T) was isolated from soil collected in Fall 2020 from the roots of a red Japanese maple near the village of Mundelein, Illinois, USA (42.26°N, −88.0°W).
Pyxidicoccus parkwaysis (park.way.sis. N.L. masc. adj. parkwaysis from Parkway Farm in Landrum, South Carolina, USA, referencing the area of isolation).
Vegetative cells glide on solid media. Cells grow as mucoid swarms with a slight pink pigmentation and develop, mounded fruiting bodies after 3 weeks of growth on VY/4 media. Aerobic growth was observed at 20 to 40°C at a pH range of 6.0 to 9.0. Hydrolyzes gelatin and esculin. Shows an API ZYM positive reaction to C4 esterase, C8 lipase, leucine arylamidase, acid phosphatase, and napthol-AS-BI-aphosphohydrolase, and a negative reaction to alkaline phosphatase, C14 lipase, valine arylamidase, cysteine arylamidase, trypsin, α-chymotrypsin, N-acetyl-β-glucosaminidase, α-galactosidase, β-galactosidase, β-glucuronidase, α-glucosidase, β-glucosidase, α-mannosidase, and α-fucosidase. DNA GC content is 69.6%. The genome assembly of the organism is available at NCBI Assembly (ASM1730173v1). The 16S ribosomal RNA gene sequence is available at GenBank (OP852333). Phylogenetically most similar to P. caerfyrddinensis CA032AT.
The type strain (SCPEA02T = TSD-328T = NRRL B-65670T = NCCB 100921T) was isolated from soil collected in Spring 2020 from the roots of a peach tree near Parkway Farm in Landrum, South Carolina, USA (35.14°N, −82.12°W).
Sorangium aterium (at'er.i.um. N.L. neut. adj. aterium the flat black, referencing black pigment production).
Vegetative cells are Gram-negative and glide on solid media. Degrades and decomposes cellulose filter paper. Cells are pigmented dark brown to black when grown on VY/4 agar. Dark brown to black fruiting bodies form on ST21 media, and similarly pigmented sporangioles form on VY/4 agar. Aerobic growth was observed at 25 to 30°C but not at 20°C or 35 to 40°C at a pH range of 6.0 to 7.0. Hydrolyzes esculin. Shows an API ZYM positive reaction to alkaline phosphatase, C4 esterase, C8 lipase, leucine arylamidase, acid phosphatase, napthol-AS-BI-aphosphohydrolase, and β-glucosidase, and a negative reaction to C14 lipase, valine arylamidase, cysteine arylamidase, trypsin, α-chymotrypsin, N-acetyl-β-glucosaminidase, α-galactosidase, β-galactosidase, β-glucuronidase, α-glucosidase, α-mannosidase, and α-fucosidase. DNA GC content is 71.1%. The genome assembly of the organism is available at NCBI Assembly (ASM2836893v1). The 16S ribosomal RNA gene sequence is available at GenBank (OP852334.1). Phylogenetically most similar to S. cellulosum Soce56.
The type strain (WIWO2T = NRRL B-65671T = NCCB 100922T) was isolated from soil collected in Fall 2021 from the roots of a white oak near the village of Pleasant Prairie, Wisconsin, USA (42.56°N, −87.94°W).
Stigmatella ashevillena (ash'vill.en.a. N.L. fem. adj. from Asheville, North Carolina, USA, referencing the area of isolation).
Vegetative cells are Gram-negative and glide on solid media. Cells are yellow during early growth on VY/4 media and form stalked, orange fruiting bodies over time. Aerobic growth was observed at 25 to 30 °C but not at 20°C or 35 to 40°C at a pH range of 6.0 to 9.0. Hydrolyzes gelatin and esculin. Shows an API ZYM positive reaction to alkaline phosphatase, C4 esterase, C8 lipase, C14 lipase, leucine arylamidase, cysteine arylamidase, acid phosphatase, napthol-AS-BI-aphosphohydrolase, α-glucosidase, β-glucosidase, and N-acetyl-β-glucosaminidase, and a negative reaction to valine arylamidase, trypsin, α-chymotrypsin, α-galactosidase, β-galactosidase, β-glucuronidase, α-mannosidase, and α-fucosidase. DNA GC content is 66.9%. The genome assembly of the organism is available at NCBI Assembly (ASM2836897v1). The 16S ribosomal RNA gene sequence is available at GenBank (OP852335.1). Phylogenetically most similar to S. aurantiaca DSM17044T.
The type strain (NCWAL01T = TSD-329T = NCCB 100923T) was isolated from soil collected in Spring 2020 from the roots of a walnut tree near the city of Asheville, North Carolina, USA (35.63°N, −82.55°W).
Bacteriolytic myxobacteria were isolated by the Escherichia coli baiting method (Mohr, 2018). Briefly, an E. coli lawn was grown overnight at 37°C and resuspended in 1 mL of an antifungal solution (250 μg/mL of cycloheximide and nystatin). A 300 μl of the solution was spread across a WAT agar (1.5% agar, 0.1% CaCl2) plate and air-dried. Previously air-dried soil was wetted with the antifungal solution to a mud-like consistency, and a pea-sized amount was placed on the dried E. coli WAT plate. The plate was incubated at 25°C for up to 4 weeks. After 3 days of incubation, the plates were checked daily for the appearance of lytic zones or fruiting bodies in the E. coli lawn. Using a syringe needle, the lytic zones were moved to a plate of VY/4 (Baker's yeast 2.5 g/L, CaCl2 × 2H2O 1.36 g/L, vitamin B12 0.5 mg/L, and agar 15 g/L). The swarm edge was repeatedly used to inoculate a fresh VY/4 plate until pure cultures were obtained. Isolates were cultivated continuously at 25–30°C on VY/4. Isolation of cellulolytic myxobacteria was accomplished using the filter paper method (Mohr, 2018). A small square of autoclaved filter paper was placed in the center of a ST21 agar plate (1 g/L of K2HPO4, 20 mg/L of yeast extract, 14 g/L of agar, 1 g/L of KNO3, 1 g/L of MgSO4 × 7H2O, 1 g/L of CaCl2 × 2H2O, 0.1 g/L of MnSO4 × 7H2O, and 0.2 g/L of FeCl3). A pea-sized amount of soil, wet with the antifungal solution, was placed at the edge of the filter paper. Plates were incubated at 25°C for up to 2 months. After 2 weeks, the plates were checked every 2 days for cellulose degradation and fruiting body formation. Fruiting bodies were moved to a fresh ST21 plate with filter paper repeatedly until pure monocultures were observed. Isolates were cultivated continuously at 25–30°C on VY/4.
All isolates were maintained on VY/4 media. Growth in liquid cultures was achieved using CYH/2 media (0.75 g/L of casitone, 0.75 g/L of yeast extract, 2g/L of starch, 0.5 g/L of soy flour, 0.5 g/L of glucose, 0.5 g/L of MgSO4 × 7H2O, 1 g/L of CaCl2 × 2H2O, 6 g/L of HEPES, 8 mg/L of EDTA-Fe, and 0.5 mg/L of vitamin B12).
Once pure cultures were obtained, DNA isolation was performed using either the Monarch HMW DNA extraction kit for tissue or the Macherey Nagel NucleoBond HMW DNA extraction kit, following the manufacturer's instructions for Gram-negative bacteria. All novel strains were sequenced using PacBio Sequel or Sequel II with a 10 h movie. De novo assembly of the genome was accomplished using the SMRT Analysis Hierarchical Genome Assembly Process (HGAP; SMRT Link 9.0.0 or SMRT Link 10.1.0). Nannocystis pusilla Na p29T was sequenced using an Oxford Nanopore Minion Flongle R9.4.1. Raw data were processed using Guppy (v6.0.1) (Wick et al., 2019). Reads were trimmed using a porechop (v0.2.4) (Wick et al., 2017). The worst 10% of reads were filtered out using filtong (v0.2.1). Flye assembler (v2.9) was used for de novo assembly using the trimmed and filtered reads (Kolmogorov et al., 2019). The correction of the final assembly was achieved by long-read correction using two iterations of Racon (v1.5) (Vaser et al., 2017), followed by two iterations of Medaka (v1.7.2) (Nicholls et al., 2019). Genome assemblies and complete genome data were deposited at NCBI for the following isolates: Corallococcus exiguus strain NCCRE02 (ASM1730297v1), Corallococcus sp. strain NCSPR01 (ASM1730913v1), Corallococcus sp. strain BB11-1 (ASM2662662v1), Corallococcus sp. strain BB12-1 (ASM2662676v1), Corallococcus sp. strain NCRR (ASM2696553v1), Myxococcus sp. strain MISCRS (ASM2662660v1), Myxococcus sp. strain NMCA1 (ASM2681020v1), Nannocystis sp. strain SCPEA4 (ASM2662668v1), Nannocystis sp. strain ILAH1 (ASM2662658v1), Nannocystis sp. strain RBIL2 (ASM2662674v1), and Pyxidicoccus sp. strain MSG2 (ASM2662670v1). Genome assembly for Nannocystis pusilla Na p29T was also deposited at NCBI (ASM2662666v1).
A Zeiss stereo discovery.V12 microscope using Axiocam 105 and a Plano Apo S 1.0X objective was used to observe fruiting bodies and swarming patterns.
OrthoANI calculations and tree generation were achieved using OAT (orthoANI tool v0.93.1) (Lee et al., 2016). dDDH calculations were performed on the type strain genome server (TYGS) website (Meier-Kolthoff and Goker, 2019). Synteny analysis was performed using SimpleSynteny (v1.6) (Veltri et al., 2016).
Enzymatic activity was assessed for myxobacteria utilizing commercial API ZYM (bioMérieux, France) and API NE (bioMérieux, France) kits. Each isolate strain was suspended in 0.85% NaCl to an OD600 of 0.7 and 0.1 for API NE. API ZYM strips were incubated for 4.5 h at 37°C, and API NE strips were incubated for 24 h at 37°C. After incubation, specific reagents were added to the cupule and evaluated according to the manufacturer's instructions.
For most of the myxobacteria tested, strains were grown on VY/4 (pH 7.2) for 5 to 7 days and resuspended in deionized water to an OD600 of 0.5. For the genera Nannocystis and Polyangium, strains were grown for 5 to 7 days in CYH/2 media, centrifuged, washed, and resuspended in sterile distilled water. The optimal growth temperature was tested by inoculating VY/4 plates with 25 μl of the 0.5 OD600 suspension for the given myxobacteria. Plates were incubated at 20, 25, 30, 35, and 40°C for up to 14 days. Optimal pH was assessed by plating the 0.5 OD600 solution on VY/4 plates buffered to pH 5, 6, 7, 8, 9, or 10. The pH conditions were buffered at a pH of 5 to 6 with 25 mM MES buffer, 7 to 8 with 25 mM HEPES buffer, and 9 to 10 with 25 mM TRIS buffer in VY/4 plates and incubated at 25°C for 2 weeks. Comparisons of swarm diameters were used to determine optimal growth conditions.
FASTA files for all sequenced isolates were uploaded for analysis using antiSMASH (version 6.1.1) using relaxed detection strictness with all extra features toggled on (Blin et al., 2021b). Resulting antiSMASH job IDs from analyzed isolates were submitted as queries using the BiG-FAM database v1.0.0 (1,225,071 BGCs and 29,955 GCFs) to assess BGC similarity to database clusters (Kautsar et al., 2021a). All BGCs with >900 distance from model GCFs were subsequently dereplicated manually to remove characterized BGCs not clustered with GCFs within the BiG-FAM database. The antiSMASH database v3.0 (147,571 BGCs) was used to analyze BGCs from myxobacteria (Blin et al., 2021a). BiG-SCAPE v1.1.0 was used to analyze all BGCs.gbk files from sequenced isolates as well as all .gbk files from myxobacteria with sequenced genomes deposited in the antiSMASH database with the “hybrids-off” and “MiBIG” parameters (Kautsar et al., 2020; Navarro-Munoz et al., 2020).
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, ASM2662663v1; https://www.ncbi.nlm.nih.gov/, ASM1730163v1; https://www.ncbi.nlm.nih.gov/, ASM2696555v1; https://www.ncbi.nlm.nih.gov/genbank/, CP114040.1; https://www.ncbi.nlm.nih.gov/, ASM2836909v1; https://www.ncbi.nlm.nih.gov/, ASM2836910v1; https://www.ncbi.nlm.nih.gov/, ASM1730173v1; https://www.ncbi.nlm.nih.gov/, ASM2836893v1; https://www.ncbi.nlm.nih.gov/, ASM2836897v1; https://www.ncbi.nlm.nih.gov/, ASM1730297v1; https://www.ncbi.nlm.nih.gov/, ASM1730913v1; https://www.ncbi.nlm.nih.gov/, ASM2662662v1; https://www.ncbi.nlm.nih.gov/, ASM2662676v1; https://www.ncbi.nlm.nih.gov/, ASM2696553v1; https://www.ncbi.nlm.nih.gov/, ASM2662660v1; https://www.ncbi.nlm.nih.gov/, ASM2681020v1; https://www.ncbi.nlm.nih.gov/, ASM2662668v1; https://www.ncbi.nlm.nih.gov/, ASM2662658v1; https://www.ncbi.nlm.nih.gov/, ASM2662674v1; https://www.ncbi.nlm.nih.gov/, ASM2662670v1; and https://www.ncbi.nlm.nih.gov/, ASM2662666v1.
AA, KP, and TK: isolation of environmental myxobacteria. KP, TK, and MH: growth profiles, biochemical assays, and imaging for isolates. AA, KP, and SD: genome sequencing. AA, KP, and DS: BGC analyses, manuscript preparation, and editing. DS: supervision and administration. All authors have read and approved the final manuscript.
This research was supported by the National Institute of Allergy and Infectious Diseases (1 R15 AI137996) and the National Institute of General Medical Sciences (1 P20 GM130460).
The authors would like to thank Mary Williams and Ashlen Ahearne for coordinating the collection and delivery of soil samples analyzed in this project. The authors also appreciate the Glycoscience Center of Research Excellence Imaging Research Core and Computational Chemistry and Bioinformatics Research Core for assistance in imaging the isolates and assembly of the N. pusilla Na p29T draft genome with specific gratitude for the Imaging Research Core manager, Dr. Ruofan Cao.
SD was employed by Molecular Research LP (MR DNA).
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer DW is currently organizing a Research Topic with the author DS.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1227206/full#supplementary-material
Adaikpoh, B. I., Akbar, S., Albataineh, H., Misra, S. K., Sharp, J. S., Stevens, D. C., et al. (2020). Myxobacterial response to methyljasmonate exposure indicates contribution to plant recruitment of micropredators. Front. Microbiol. 11, 34. doi: 10.3389/fmicb.2020.00034
Ahearne, A., Albataineh, H., Dowd, S. E., and Stevens, D. C. (2021). Assessment of evolutionary relationships for prioritization of myxobacteria for natural product discovery. Microorganisms 9, 1376. doi: 10.3390/microorganisms9071376
Awal, R. P., Garcia, R., Gemperlein, K., Wink, J., Kunwar, B., Parajuli, N., et al. (2017). Vitiosangium cumulatum gen. nov., sp. nov. and Vitiosangium subalbum sp. nov., soil myxobacteria, and emended descriptions of the genera Archangium and Angiococcus, and of the family Cystobacteraceae. Int. J. Syst. Evol. Microbiol. 67, 1422–1430. doi: 10.1099/ijsem.0.001829
Awal, R. P., Garcia, R., and Muller, R. (2016). Racemicystis crocea gen. nov., sp. nov., a soil myxobacterium in the family Polyangiaceae. Int. J. Syst. Evol. Microbiol. 66, 2389–2395. doi: 10.1099/ijsem.0.001045
Babadi, Z. K., Garcia, R., Ebrahimipour, G. H., Risdian, C., Kampfer, P., Jarek, M., et al. (2022). Corallococcus soli sp. Nov., a soil myxobacterium isolated from subtropical climate, chalus county, iran, and its potential to produce secondary metabolites. Microorganisms 10, 262. doi: 10.3390/microorganisms10071262
Bader, C. D., Panter, F., Garcia, R., Tchesnokov, E. P., Haid, S., Walt, C., et al. (2022). Sandacrabins - structurally unique antiviral RNA polymerase inhibitors from a rare myxobacterium. Chemistry 28, e202104484. doi: 10.1002/chem.202104484
Bader, C. D., Panter, F., and Muller, R. (2020). In depth natural product discovery - Myxobacterial strains that provided multiple secondary metabolites. Biotechnol. Adv. 39, 107480. doi: 10.1016/j.biotechadv.2019.107480
Baltz, R. H. (2017). Gifted microbes for genome mining and natural product discovery. J. Ind. Microbiol. Biotechnol. 44, 573–588. doi: 10.1007/s10295-016-1815-x
Baltz, R. H. (2019). Natural product drug discovery in the genomic era: realities, conjectures, misconceptions, and opportunities. J. Ind. Microbiol. Biotechnol. 46, 281–299. doi: 10.1007/s10295-018-2115-4
Baltz, R. H. (2021). Genome mining for drug discovery: progress at the front end. J. Ind. Microbiol. Biotechnol. 48, kuab044. doi: 10.1093/jimb/kuab044
Bhat, S., Ahrendt, T., Dauth, C., Bode, H. B., and Shimkets, L. J. (2014). Two lipid signals guide fruiting body development of Myxococcus xanthus. mBio 5, e00939–13. doi: 10.1128/mBio.00939-13
Blin, K., Shaw, S., Kautsar, S. A., Medema, M. H., and Weber, T. (2021a). The antiSMASH database version 3: increased taxonomic coverage and new query features for modular enzymes. Nucleic Acids Res. 49, D639–D643. doi: 10.1093/nar/gkaa978
Blin, K., Shaw, S., Kloosterman, A. M., Charlop-Powers, Z., van Wezel, G. P., Medema, M. H., et al. (2021b). antiSMASH 6, 0. improving cluster detection and comparison capabilities. Nucleic Acids Res. 49, W29–W35. doi: 10.1093/nar/gkab335
Botella, J. A., Murillo, F. J., and Ruiz-Vazquez, R. (1995). A cluster of structural and regulatory genes for light-induced carotenogenesis in Myxococcus xanthus. Eur J Biochem 233, 238–48. doi: 10.1111/j.1432-1033.1995.238_1.x
Cardellina, J. H., Marner, F. J., and Moore, R. E. (1979). Seaweed dermatitis: structure of lyngbyatoxin A. Science 204, 193–5. doi: 10.1126/science.107586
Chambers, J., Sparks, N., Sydney, N., Livingstone, P. G., Cookson, A. R., Whitworth, D. E., et al. (2020). Comparative genomics and pan-genomics of the myxococcaceae, including a description of five novel species: Myxococcus eversor sp. nov., Myxococcus llanfairpwllgwyngyllgogerychwyrndrobwllllantysiliogogogochensis sp. nov., Myxococcus vastator sp. nov., Pyxidicoccus caerfyrddinensis sp. nov., and Pyxidicoccus trucidator sp. nov. Genome Biol. Evol. 12, 2289–2302. doi: 10.1093/gbe/evaa212
Chevrette, M. G., Gutierrez-Garcia, K., Selem-Mojica, N., Aguilar-Martinez, C., Yanez-Olvera, A., Ramos-Aboites, H. E., et al. (2020). Evolutionary dynamics of natural product biosynthesis in bacteria. Nat. Prod. Rep. 37, 566–599. doi: 10.1039/C9NP00048H
Cortina, N. S., Krug, D., Plaza, A., Revermann, O., and Muller, R. (2012). Myxoprincomide: a natural product from Myxococcus xanthus discovered by comprehensive analysis of the secondary metabolome. Angew. Chem. Int. Ed. Engl. 51, 811–6. doi: 10.1002/anie.201106305
Edwards, D. J., and Gerwick, W. H. (2004). Lyngbyatoxin biosynthesis: sequence of biosynthetic gene cluster and identification of a novel aromatic prenyltransferase. J. Am. Chem. Soc. 126, 11432–3. doi: 10.1021/ja047876g
Ellis, B. M., Fischer, C. N., Martin, L. B., Bachmann, B. O., and McLean, J. A. (2019). Spatiochemically profiling microbial interactions with membrane scaffolded desorption electrospray ionization-ion mobility-imaging mass spectrometry and unsupervised segmentation. Anal. Chem. 91, 13703–13711. doi: 10.1021/acs.analchem.9b02992
Fischbach, M. A., Walsh, C. T., and Clardy, J. (2008). The evolution of gene collectives: How natural selection drives chemical innovation. Proc. Natl. Acad. Sci. U. S. A. 105, 4601–8. doi: 10.1073/pnas.0709132105
Frank, B., Wenzel, S. C., Bode, H. B., Scharfe, M., Blocker, H., Muller, R., et al. (2007). From genetic diversity to metabolic unity: studies on the biosynthesis of aurafurones and aurafuron-like structures in myxobacteria and streptomycetes. J. Mol. Biol. 374, 24–38. doi: 10.1016/j.jmb.2007.09.015
Fu, C., Auerbach, D., Li, Y., Scheid, U., Luxenburger, E., Garcia, R., et al. (2017). Solving the puzzle of one-carbon loss in ripostatin biosynthesis. Angew. Chem. Int. Ed. Engl. 56, 2192–2197. doi: 10.1002/anie.201609950
Gaitatzis, N., Kunze, B., and Muller, R. (2001). In vitro reconstitution of the myxochelin biosynthetic machinery of Stigmatella aurantiaca Sg a15: biochemical characterization of a reductive release mechanism from nonribosomal peptide synthetases. Proc. Natl. Acad. Sci. U. S. A. 98, 11136–41. doi: 10.1073/pnas.201167098
Garcia, R., Gemperlein, K., and Muller, R. (2014). Minicystis rosea gen. nov., sp. nov., a polyunsaturated fatty acid-rich and steroid-producing soil myxobacterium. Int. J. Syst. Evol. Microbiol. 64, 3733–3742. doi: 10.1099/ijs.0.068270-0
Garcia, R., and Muller, R. (2018). Simulacricoccus ruber gen. nov., sp. nov., a microaerotolerant, non-fruiting, myxospore-forming soil myxobacterium and emended description of the family Myxococcaceae. Int. J. Syst. Evol. Microbiol. 68, 3101–3110. doi: 10.1099/ijsem.0.002936
Garcia, R., Stadler, M., Gemperlein, K., and Muller, R. (2016). Aetherobacter fasciculatus gen. nov., sp. nov. and Aetherobacter rufus sp. nov., novel myxobacteria with promising biotechnological applications. Int. J. Syst. Evol. Microbiol. 66, 928–938. doi: 10.1099/ijsem.0.000813
Gorges, J., Panter, F., Kjaerulff, L., Hoffmann, T., Kazmaier, U., Muller, R., et al. (2018). Structure, total synthesis, and biosynthesis of chloromyxamides: myxobacterial tetrapeptides featuring an uncommon 6-chloromethyl-5-methoxypipecolic acid building block. Angew. Chem. Int. Ed. Engl. 57, 14270–14275. doi: 10.1002/anie.201808028
Haack, P. A., Harmrolfs, K., Bader, C. D., Garcia, R., Gunesch, A. P., Haid, S., et al. (2022). Thiamyxins: structure and biosynthesis of myxobacterial RNA-virus inhibitors. Angew. Chem. Int. Ed. Engl. 61, e202212946. doi: 10.1002/anie.202212946
Herrmann, J., Fayad, A. A., and Muller, R. (2017). Natural products from myxobacteria: novel metabolites and bioactivities. Nat. Prod. Rep. 34, 135–160. doi: 10.1039/C6NP00106H
Hoffmann, T., Krug, D., Bozkurt, N., Duddela, S., Jansen, R., Garcia, R., et al. (2018). Correlating chemical diversity with taxonomic distance for discovery of natural products in myxobacteria. Nat. Commun. 9, 803. doi: 10.1038/s41467-018-03184-1
Hoshino, T. (2011). Violacein and related tryptophan metabolites produced by Chromobacterium violaceum: biosynthetic mechanism and pathway for construction of violacein core. Appl. Microbiol. Biotechnol. 91, 1463–75. doi: 10.1007/s00253-011-3468-z
Hug, J. J., Panter, F., Krug, D., and Muller, R. (2019). Genome mining reveals uncommon alkylpyrones as type III PKS products from myxobacteria. J. Ind. Microbiol. Biotechnol. 46, 319–334. doi: 10.1007/s10295-018-2105-6
Iizuka, T., Jojima, Y., Hayakawa, A., Fujii, T., Yamanaka, S., Fudou, R., et al. (2013). Pseudenhygromyxa salsuginis gen. nov., sp. nov., a myxobacterium isolated from an estuarine marsh. Int. J. Syst. Evol. Microbiol. 63, 1360–1369. doi: 10.1099/ijs.0.040501-0
Inoue, D., Hiroshima, N., Nakamura, S., Ishizawa, H., and Ike, M. (2022). Characterization of Two novel predatory bacteria, bacteriovorax stolpii HI3 and Myxococcus sp. MH1, isolated from a freshwater pond: prey range, and predatory dynamics and efficiency. Microorganisms 10, 816. doi: 10.3390/microorganisms10091816
Jiang, J., He, X., and Cane, D. E. (2007). Biosynthesis of the earthy odorant geosmin by a bifunctional Streptomyces coelicolor enzyme. Nat. Chem. Biol. 3, 711–5. doi: 10.1038/nchembio.2007.29
Karoonuthaisiri, N., Weaver, D., Huang, J., Cohen, S. N., and Kao, C. M. (2005). Regional organization of gene expression in Streptomyces coelicolor. Gene 353, 53–66. doi: 10.1016/j.gene.2005.03.042
Kautsar, S. A., Blin, K., Shaw, S., Navarro-Munoz, J. C., Terlouw, B. R., van der Hooft, J. J. J., et al. (2020). MIBiG 2, 0. a repository for biosynthetic gene clusters of known function. Nucleic Acids Res. 48, D454–D458. doi: 10.1093/nar/gkz882
Kautsar, S. A., Blin, K., Shaw, S., Weber, T., and Medema, M. H. (2021a). BiG-FAM: the biosynthetic gene cluster families database. Nucleic Acids Res. 49, D490–D497. doi: 10.1093/nar/gkaa812
Kautsar, S. A., van der Hooft, J. J. J., de Ridder, D., and Medema, M. H. (2021b). BiG-SLiCE: a highly scalable tool maps the diversity of 1, 2. million biosynthetic gene clusters. Gigascience 10, 154. doi: 10.1093/gigascience/giaa154
Kleinig, H., and Reichenbach, H. (1969). Carotenoid pigments of Stigmatella aurantiaca (Myxobacterales).I. The minor carotenoids. Arch. Mikrobiol. 68, 210–7. doi: 10.1007/BF00409913
Kolmogorov, M., Yuan, J., Lin, Y., and Pevzner, P. A. (2019). Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 37, 540–546. doi: 10.1038/s41587-019-0072-8
Krastel, P., Roggo, S., Schirle, M., Ross, N. T., Perruccio, F., Aspesi, P., et al. (2015). Nannocystin A: an elongation factor 1 inhibitor from myxobacteria with differential anti-cancer properties. Angew. Chem. Int. Ed. Engl. 54, 10149–54. doi: 10.1002/anie.201505069
Lang, E., and Reichenbach, H. (2013). Designation of type strains for seven species of the order Myxococcales and proposal for neotype strains of Cystobacter ferrugineus, Cystobacter minus and Polyangium fumosum. Int. J. Syst. Evol. Microbiol. 63, 4354–60. doi: 10.1099/ijs.0.056440-0
Lang, E., Schumann, P., Tindall, B. J., Mohr, K. I., and Sproer, C. (2015). Reclassification of Angiococcus disciformis, Cystobacter minus and Cystobacter violaceus as Archangium disciforme comb. nov., Archangium minus comb. nov. and Archangium violaceum comb. nov., unification of the families Archangiaceae and Cystobacteraceae, and emended descriptions of the families Myxococcaceae and Archangiaceae. Int. J. Syst. Evol. Microbiol. 65, 4032–4042. doi: 10.1099/ijsem.0.000533
Lee, I., Ouk Kim, Y., Park, S. C., and Chun, J. (2016). OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 66, 1100–1103. doi: 10.1099/ijsem.0.000760
Li, Y., Weissman, K. J., and Muller, R. (2008). Myxochelin biosynthesis: direct evidence for two- and four-electron reduction of a carrier protein-bound thioester. J. Am. Chem. Soc. 130, 7554–5. doi: 10.1021/ja8025278
Lioy, V. S., Lorenzi, J. N., Najah, S., Poinsignon, T., Leh, H., Saulnier, C., et al. (2021). Dynamics of the compartmentalized Streptomyces chromosome during metabolic differentiation. Nat. Commun. 12, 5221. doi: 10.1038/s41467-021-25462-1
Livingstone, P. G., Ingleby, O., Girdwood, S., Cookson, A. R., Morphew, R. M., Whitworth, D. E., et al. (2020). Predatory organisms with untapped biosynthetic potential: descriptions of novel corallococcus species C. aberystwythensis sp. nov., C. carmarthensis sp. nov., C. exercitus sp. nov., C. interemptor sp. nov., C. llansteffanensis sp. nov., C. praedator sp. nov., C. sicarius sp. nov., and C. terminator sp. nov. Appl. Environ. Microbiol. 86, 19. doi: 10.1128/AEM.01931-19
Lorenzen, W., Ahrendt, T., Bozhuyuk, K. A., and Bode, H. B. (2014). A multifunctional enzyme is involved in bacterial ether lipid biosynthesis. Nat. Chem. Biol. 10, 425–7. doi: 10.1038/nchembio.1526
Medema, M. H., Blin, K., Cimermancic, P., de Jager, V., Zakrzewski, P., Fischbach, M. A., et al. (2011). antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 39, W339–46. doi: 10.1093/nar/gkr466
Medema, M. H., Cimermancic, P., Sali, A., Takano, E., and Fischbach, M. A. (2014). A systematic computational analysis of biosynthetic gene cluster evolution: lessons for engineering biosynthesis. PLoS Comput. Biol. 10, e1004016. doi: 10.1371/journal.pcbi.1004016
Meier-Kolthoff, J. P., and Goker, M. (2019). TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 10, 2182. doi: 10.1038/s41467-019-10210-3
Meiser, P., Weissman, K. J., Bode, H. B., Krug, D., Dickschat, J. S., Sandmann, A., et al. (2008). DKxanthene biosynthesis–understanding the basis for diversity-oriented synthesis in myxobacterial secondary metabolism. Chem. Biol. 15, 771–81. doi: 10.1016/j.chembiol.2008.06.005
Mohr, K. I. (2018). Diversity of myxobacteria-we only see the tip of the iceberg. Microorganisms 6, 84. doi: 10.3390/microorganisms6030084
Mohr, K. I., Garcia, R. O., Gerth, K., Irschik, H., and Muller, R. (2012). Sandaracinus amylolyticus gen. nov., sp. nov., a starch-degrading soil myxobacterium, and description of Sandaracinaceae fam. nov. Int. J. Syst. Evol. Microbiol. 62, 1191–1198. doi: 10.1099/ijs.0.033696-0
Mohr, K. I., Moradi, A., Glaeser, S. P., Kampfer, P., Gemperlein, K., Nubel, U., et al. (2018a). Nannocystis konarekensis sp. nov., a novel myxobacterium from an Iranian desert. Int. J. Syst. Evol. Microbiol. 68, 721–729. doi: 10.1099/ijsem.0.002569
Mohr, K. I., Stechling, M., Wink, J., Wilharm, E., and Stadler, M. (2016). Comparison of myxobacterial diversity and evaluation of isolation success in two niches: Kiritimati Island and German compost. Microbiologyopen 5, 268–78. doi: 10.1002/mbo3.325
Mohr, K. I., Wolf, C., Nubel, U., Szafranska, A. K., Steglich, M., Hennessen, F., et al. (2018b). A polyphasic approach leads to seven new species of the cellulose-decomposing genus Sorangium, Sorangium ambruticinum sp. nov., Sorangium arenae sp. nov., Sorangium bulgaricum sp. nov., Sorangium dawidii sp. nov., Sorangium kenyense sp. nov., Sorangium orientale sp. nov. and Sorangium reichenbachii sp. nov. Int. J. Syst. Evol. Microbiol. 68, 3576–3586. doi: 10.1099/ijsem.0.003034
Mohr, K. I., Zindler, T., Wink, J., Wilharm, E., and Stadler, M. (2017). Myxobacteria in high moor and fen: an astonishing diversity in a neglected extreme habitat. Microbiologyopen 6, 1–14. doi: 10.1002/mbo3.464
Moradi, A., Ebrahimipour, G. H., Mohr, K. I., Kampfer, P., Glaeser, S. P., Hennessen, F., et al. (2017). Racemicystis persica sp. nov., a myxobacterium from soil. Int. J. Syst. Evol. Microbiol. 67, 472–478. doi: 10.1099/ijsem.0.001655
Muller, S., Strack, S. N., Ryan, S. E., Shawgo, M., Walling, A., Harris, S., et al. (2016). Identification of functions affecting predator-prey interactions between Myxococcus xanthus and Bacillus subtilis. J. Bacteriol. 198, 3335–3344. doi: 10.1128/JB.00575-16
Navarro-Munoz, J. C., Selem-Mojica, N., Mullowney, M. W., Kautsar, S. A., Tryon, J. H., and Parkinson, E. I. (2020). A computational framework to explore large-scale biosynthetic diversity. Nat. Chem. Biol. 16, 60–68. doi: 10.1038/s41589-019-0400-9
Nicholls, S. M., Quick, J. C., Tang, S., and Loman, N. J. (2019). Ultra-deep, long-read nanopore sequencing of mock microbial community standards. Gigascience 8, giz043. doi: 10.1093/gigascience/giz043
Okoth, D. A., Hug, J. J., Garcia, R., and Muller, R. (2022). Discovery, Biosynthesis and biological activity of a succinylated myxochelin from the myxobacterial strain MSr12020. Microorganisms 10, 959. doi: 10.3390/microorganisms10101959
Olivares, P., Ulrich, E. C., Chekan, J. R., van der Donk, W. A., and Nair, S. K. (2017). Characterization of two late-stage enzymes involved in fosfomycin biosynthesis in pseudomonads. ACS Chem. Biol. 12, 456–463. doi: 10.1021/acschembio.6b00939
Oliynyk, M., Samborskyy, M., Lester, J. B., Mironenko, T., Scott, N., Dickens, S., et al. (2007). Complete genome sequence of the erythromycin-producing bacterium Saccharopolyspora erythraea NRRL23338. Nat. Biotechnol. 25, 447–53. doi: 10.1038/nbt1297
Osswald, C., Zaburannyi, N., Burgard, C., Hoffmann, T., Wenzel, S. C., Muller, R., et al. (2014). A highly unusual polyketide synthase directs dawenol polyene biosynthesis in Stigmatella aurantiaca. J. Biotechnol. 191, 54–63. doi: 10.1016/j.jbiotec.2014.07.447
Panter, F., Krug, D., and Muller, R. (2019). Novel methoxymethacrylate natural products uncovered by statistics-based mining of the Myxococcus fulvus secondary metabolome. ACS Chem. Biol. 14, 88–98. doi: 10.1021/acschembio.8b00948
Park, S., Hyun, H., Lee, J. S., and Cho, K. (2016). Identification of the phenalamide biosynthetic gene cluster in Myxococcus stipitatus DSM 14675. J. Microbiol. Biotechnol. 26, 1636–42. doi: 10.4014/jmb.1603.03023
Perez, J., Contreras-Moreno, F. J., Marcos-Torres, F. J., Moraleda-Munoz, A., and Munoz-Dorado, J. (2020). The antibiotic crisis: how bacterial predators can help. Comput. Struct. Biotechnol. J. 18, 2547–2555. doi: 10.1016/j.csbj.2020.09.010
Petters, S., Gross, V., Sollinger, A., Pichler, M., Reinhard, A., Bengtsson, M. M., et al. (2021). The soil microbial food web revisited: predatory myxobacteria as keystone taxa? ISME J. doi: 10.1038./s41396-021-00958-2
Phillips, K. E., Akbar, S., and Stevens, D. C. (2022). Concepts and conjectures concerning predatory performance of myxobacteria. Front. Microbiol. 13, 1031346. doi: 10.3389/fmicb.2022.1031346
Pistorius, D., and Muller, R. (2012). Discovery of the rhizopodin biosynthetic gene cluster in Stigmatella aurantiaca Sg a15 by genome mining. Chembiochem 13, 416–26. doi: 10.1002/cbic.201100575
Reichenbach, H., Voelz, H., and Dworkin, M. (1969). Structural changes in Stigmatella aurantiaca during myxospore induction. J. Bacteriol. 97, 905–11. doi: 10.1128/jb.97.2.905-911.1969
Rogers, T. O., and Birnbaum, J. (1974). Biosynthesis of fosfomycin by Streptomyces fradiae. Antimicrob. Agents Chemother. 5, 121–32. doi: 10.1128/AAC.5.2.121
Silakowski, B., Kunze, B., Nordsiek, G., Blocker, H., Hofle, G., Muller, R., et al. (2000). The myxochelin iron transport regulon of the myxobacterium Stigmatella aurantiaca Sg a15. Eur. J. Biochem. 267, 6476–85. doi: 10.1046/j.1432-1327.2000.01740.x
Sood, S., Awal, R. P., Wink, J., Mohr, K. I., Rohde, M., Stadler, M., et al. (2015). Aggregicoccus edonensis gen. nov., sp. nov., an unusually aggregating myxobacterium isolated from a soil sample. Int. J. Syst. Evol. Microbiol. 65, 745–753. doi: 10.1099/ijs.0.061176-0
Stevens, D. C., Young, J., Carmichael, R., Tan, J., and Taylor, R. E. (2014). Draft genome sequence of gephyronic acid producer cystobacter violaceus strain Cb vi76. Genome Announc. 2, 10–128. doi: 10.1128/genomeA.01299-14
van den Belt, M. G., Booth, C., Chooi, T. J., Medema, Y., and Alanjary, M. H. M. (2023). CAGECAT: the comparative gene cluster analysis toolbox for rapid search and visualisation of homologous gene clusters. BMC Inf. 24, 1–8. doi: 10.1101/2023.02.08.527634
Vaser, R., Sovic, I., Nagarajan, N., and Sikic, M. (2017). Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 27, 737–746. doi: 10.1101/gr.214270.116
Veltri, D., Wight, M. M., and Crouch, J. A. (2016). SimpleSynteny: a web-based tool for visualization of microsynteny across multiple species. Nucleic Acids Res. 44, W41–W45. doi: 10.1093/nar/gkw330
Wang, C., Lv, Y., Zhou, L., Zhang, Y., Yao, Q., Zhu, H., et al. (2022). Comparative genomics of Myxococcus and Pyxidicoccus, including the description of four novel species: Myxococcus guangdongensis sp. nov., Myxococcus qinghaiensis sp. nov., Myxococcus dinghuensis sp. nov., and Pyxidicoccus xibeiensis sp. nov. Front. Microbiol. 13, 995049. doi: 10.3389/fmicb.2022.995049
Wang, J., Ran, Q., Du, X., Wu, S., Wang, J., Sheng, D., et al. (2021). Two new Polyangium species, P. aurulentum sp. nov. and P. jinanense sp. nov., isolated from a soil sample. Syst. Appl. Microbiol. 44, 126274. doi: 10.1016/j.syapm.2021.126274
Waschulin, V., Borsetto, C., James, R., Newsham, K. K., Donadio, S., Corre, C., et al. (2022). Biosynthetic potential of uncultured Antarctic soil bacteria revealed through long-read metagenomic sequencing. ISME J. 16, 101–111. doi: 10.1038/s41396-021-01052-3
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Completing bacterial genome assemblies with multiplex MinION sequencing. Microb. Genom. 3, e000132. doi: 10.1099/mgen.0.000132
Wick, R. R., Judd, L. M., and Holt, K. E. (2019). Performance of neural network basecalling tools for Oxford Nanopore sequencing. Genome Biol. 20, 129. doi: 10.1186/s13059-019-1727-y
Witte, S. N. R., Hug, J. J., Geraldy, M. N. E., Muller, R., and Kalesse, M. (2017). Biosynthesis and total synthesis of pyrronazol b: a secondary metabolite from Nannocystis pusilla. Chemistry 23, 15917–15921. doi: 10.1002/chem.201703782
Xiao, Y., Gerth, K., Muller, R., and Wall, D. (2012). Myxobacterium-produced antibiotic TA (myxovirescin) inhibits type II signal peptidase. Antimicrob. Agents Chemother. 56, 2014–21. doi: 10.1128/AAC.06148-11
Xiao, Y., Wei, X., Ebright, R., and Wall, D. (2011). Antibiotic production by myxobacteria plays a role in predation. J. Bacteriol. 193, 4626–33. doi: 10.1128/JB.05052-11
Yamamoto, E., Muramatsu, H., and Nagai, K. (2014). Vulgatibacter incomptus gen. nov., sp. nov. and Labilithrix luteola gen. nov., sp. nov., two myxobacteria isolated from soil in Yakushima Island, and the description of Vulgatibacteraceae fam. nov., Labilitrichaceae fam. nov. and Anaeromyxobacteraceae fam. nov. Int. J. Syst. Evol. Microbiol. 64, 3360–3368. doi: 10.1099/ijs.0.063198-0
Zaroubi, L., Ozugergin, I., Mastronardi, K., Imfeld, A., Law, C., Gelinas, Y., et al. (2022). The ubiquitous soil terpene geosmin acts as a warning chemical. Appl. Environ. Microbiol. 88, e0009322. doi: 10.1128/aem.00093-22
Zeng, H., Birkelbach, J., Hoffmann, J., Popoff, A., Volz, C., Muller, R., et al. (2022). Expanding the ajudazol cytotoxin scaffold: insights from genome mining, biosynthetic investigations, and novel derivatives. J. Nat. Prod. 85, 2610–2619. doi: 10.1021/acs.jnatprod.2c00637
Keywords: myxobacteria, specialized metabolism, genome mining, biosynthetic gene clusters, Nannocystis
Citation: Ahearne A, Phillips KE, Knehans T, Hoing M, Dowd SE and Stevens DC (2023) Chromosomal organization of biosynthetic gene clusters, including those of nine novel species, suggests plasticity of myxobacterial specialized metabolism. Front. Microbiol. 14:1227206. doi: 10.3389/fmicb.2023.1227206
Received: 22 May 2023; Accepted: 11 July 2023;
Published: 03 August 2023.
Edited by:
Veronica Godoy, Northeastern University, United StatesReviewed by:
David Edward Whitworth, Aberystwyth University, United KingdomCopyright © 2023 Ahearne, Phillips, Knehans, Hoing, Dowd and Stevens. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: David Cole Stevens, c3RldmVuc0BvbGVtaXNzLmVkdQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.