
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 07 August 2023
Sec. Terrestrial Microbiology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1227132
This article is part of the Research Topic Hazardous Pollutants in Agricultural Soil and Environment View all 15 articles
 Achhada Ujalkaur Avatsingh1
Achhada Ujalkaur Avatsingh1 Shilpa Sharma1
Shilpa Sharma1 Shilippreet Kour1
Shilippreet Kour1 Yukta Arora1
Yukta Arora1 Sheetal Sharma1
Sheetal Sharma1 Divya Joshi2
Divya Joshi2 Prem Prashant Chaudhary3
Prem Prashant Chaudhary3 Kahkashan Perveen4
Kahkashan Perveen4 Mohab Amin Kamal5
Mohab Amin Kamal5 Nasib Singh1*†
Nasib Singh1*†Antibiotic resistance in bacteria has emerged as a serious public health threat worldwide. Aquatic environments including irrigation-purpose wastewaters facilitate the emergence and transmission of antibiotic-resistant bacteria and antibiotic resistance genes leading to detrimental effects on human health and environment sustainability. Considering the paramount threat of ever-increasing antibiotic resistance to human health, there is an urgent need for continuous environmental monitoring of antibiotic-resistant bacteria and antibiotic resistance genes in wastewater being used for irrigation in Indian agro-ecosystems. In this study, the prevalence of antibiotic resistance in Gram-negative bacteria isolated from irrigation-purpose wastewater samples from Sirmaur and Solan districts of Himachal Pradesh was determined. Bacterial isolates of genera Escherichia, Enterobacter, Hafnia, Shigella, Citrobacter, and Klebsiella obtained from 11 different geographical locations were found to exhibit resistance against ampicillin, amoxyclav, cefotaxime, co-trimoxazole, tobramycin, cefpodoxime and ceftazidime. However, all the isolates were sensitive to aminoglycoside antibiotic gentamicin. Enterobacter spp. and Escherichia coli showed predominance among all the isolates. Multidrug-resistance phenotype was observed with isolate AUK-06 (Enterobacter sp.) which exhibited resistant to five antibiotics. Isolate AUK-02 and AUK-09, both E. coli strains showed resistant phenotypes to four antibiotics each. Phenotypic detection revealed that six isolates were positive for extended-spectrum β-lactamases which includes two isolates from Enterobacter spp. and E. coli each and one each from Shigella sp. and Citrobacter sp. Overall, the findings revealed the occurrence of antibiotic resistant and ESBL-positive bacterial isolates in wastewaters utilized for irrigation purpose in the study area and necessitate continuous monitoring and precautionary interventions. The outcomes of the study would be of significant clinical, epidemiological, and agro-environmental importance in designing effective wastewater management and environmental pollution control strategies.
Antibiotics are important anti-infective agents which have been used since the 20th century for the treatment of human infections (Hutchings et al., 2019; Walesch et al., 2023). The β-lactam antibiotics are clinically important antimicrobial medicines and have remained the first-line chemotherapeutic intervention against Gram-positive and Gram-negative bacteria since the 1950s (Hutchings et al., 2019; Soliman et al., 2023). Bacterial resistance to β-lactams has increased substantially in past few decades (Jani et al., 2021; Zaatout et al., 2021; Mutuku et al., 2022; Tan et al., 2023). However, their irrational, injudicious, and excessive use is on steady rise which not only worsen the issue of antibiotic resistance but also resulted in their accumulation in the environment as micro-pollutant (Du and Liu, 2012; Fazaludeen Koya et al., 2022; Gitter et al., 2023). The emergence of antibiotic resistance has threatened the effective treatment of microbial infections (Sanz-García et al., 2023). According to World Health Organization, the majority of pathogenic Gram-negative bacteria, especially those of the family Enterobacteriaceae, are included in the critical-priority group and are represented by multidrug resistant (MDR) bacteria commonly encountered in healthcare settings (Janda and Abbott, 2021; Dantas Palmeira et al., 2022). Further, antibiotic resistance is recognized as one of the top ten threats to public health globally (World Health Organization, 2023). In 2019, an estimated 4·95 million deaths were attributed to antibiotic resistance in bacterial pathogens such as E. coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, Acinetobacter baumannii, and Streptococcus pneumoniae (Antimicrobial Resistance Collaborators, 2022).
Among various mechanisms by which bacteria acquired antibiotic resistance, the production of β-lactamases (EC 3.5.2.6) which cleaves β-lactam antibiotics, is considered the most significant from clinical perspective (Sawa et al., 2020; Castanheira et al., 2021; Noster et al., 2021; Zaatout et al., 2021). These enzymes are categorized into four classes according to their amino acid sequence and catalytic mechanisms (Bush, 2023). Class A β-lactamases are represented by narrow-spectrum β-lactamases (TEM-1, TEM-2, and SHV-1), extended-spectrum β-lactamases also called ESBLs (CTX-M, SHV-2, VEB-1) and serine carbapenemases which includes KPC-1 and SME-1 (Bush and Bradford, 2020; Castanheira et al., 2021; Bush, 2023; Cho et al., 2023). ESBLs are highly diverse, clinically important, and constitutively expressed antibiotic-degradative enzymes capable of hydrolyzing penicillins, cephalosporins (first-, second-, and third-generations), and monobactams (Livermore, 2008; Castanheira et al., 2021; Cho et al., 2023). ESBLs have been reported from several members of Enterobacterales, predominantly from genera Escherichia, Enterobacter, Citrobacter, and Klebsiella (Janda and Abbott, 2021; Cho et al., 2023). Worldwide, the most prevalent ESBL groups are CTX-M-1, CTXM-2, CTX-M-8, CTX-M-9, and CTX-M-25 with blaCTX-M-15 genotype being the predominant (Bush and Bradford, 2020; Castanheira et al., 2021; Bush, 2023; Cho et al., 2023). The ESBL-encoding genes are usually carried by mobile genetic elements viz. plasmids, insertion sequences, integrons, integrative conjugative elements, mobile integrative conjugative elements, transposons and prophages, which have facilitated their wide dissemination to other bacterial species through transformation, conjugation, and transduction (Partridge et al., 2018; Castanheira et al., 2021; Zaatout et al., 2021).
Wastewaters from municipal corporations, hospitals, pharmaceutical companies, animal husbandry, and poultry farms contain a diverse array of antibiotic residues, antibiotic-resistant bacteria (ARB) and antibiotic-resistant genes (ARGs) (Leiva et al., 2021; Uluseker et al., 2021; Larsson and Flach, 2022; Gitter et al., 2023). The combination of all these factors in aquatic environments allows rapid genetic exchange of ARGs from pathogenic to non-pathogenic bacterial strains leading to their genetic evolution and subsequent dissemination to human communities and animals (Liguori et al., 2022; Matthiessen et al., 2022; Muntean et al., 2022; Mutuku et al., 2022). Overuse and misuse of antibiotics have resulted in the widespread occurrence of ARB in surface water of rivers, irrigation-wastewater, soil, meat products, vegetables, etc., which possess the risk of dissemination in vulnerable human populations such as children, elderly, and immunocompromised persons (Uluseker et al., 2021; Zaatout et al., 2021; Gitter et al., 2023). The infections caused by multidrug-resistant bacteria can cause high morbidity and mortality, higher treatment costs and longer hospitalization duration. Over the years, the use of treated wastewater for agricultural purposes have gained widespread acceptance as an alternative irrigation method to relieve pressure on freshwater resources, to increase agricultural production, and to reduce the need for chemical fertilizers (Leiva et al., 2021; Minhas et al., 2022). However, this practice has led to continuous buildup of antibiotic residues in agro-ecosystems. ARB enter the aquatic environments from human and animal waste, fecal matter and hospital discharge where these are able to proliferate due to the availability of the nutrients, antibiotic residues, and inappropriate treatment and disinfection practices. Transmission of ARB from wastewaters into agro-ecosystems represents a serious ecological and public health concern and requires immediate interventions (Bhagat et al., 2020; Helmecke et al., 2020; Tucker et al., 2022). The occurrence of ARGs viz. bla (blaCTX-M, blaTEM), tet (tetO, tetQ, tetW), sul (sul1, sul2), and ermB has been reported from ARB present in the riverine systems, municipal wastewater treatment plants, pharmaceutical industries effluents, and irrigated soils (Karkman et al., 2018; Grenni, 2022; Cho et al., 2023; Shin et al., 2023). Therefore, there is an urgent need for environmental monitoring of ARB and ARGs in aquatic environments for limiting the transmission of antibiotic resistance.
The prevalence and dissemination of ARB carrying ESBLs genes has been reported from diverse agro-ecosystems worldwide. ESBLs genotypes blaCTX-M-1, blaCTX-M-15 and blaCTX-M-14 were found in E. coli, K. pneumoniae, E. hormaechei, and C. freundii from Tunisian farm irrigation water samples (Ben Said et al., 2015). Similarly, agricultural-purpose irrigation water also found to harbor ESBL-positive E. coli strain exhibiting blaCTX-M-55, blaCTX-M-65 and blaCTX-M-15 genotypes (Montero et al., 2021). The predominant prevalence of blaCTX-M, blaCMY and blaSHV in water samples was also reported from Nepal and Canada (Subramanya et al., 2021; Anderson et al., 2023). India being the largest consumer of antibiotics and other antimicrobials, witnessed a significant increase in the prevalence of ARB and ESBL-positive Enterobacterales in recent years (Farooqui et al., 2018; Jani et al., 2021; Fazaludeen Koya et al., 2022). Escherichia coli and other Gram-negative bacteria resistant to β-lactams, flouroquinolones, tetracyclines and other classes of antibiotics have been reported from wastewaters, river waters and wastewater treatment plants from different states of India along with co-prevalence of ESBL genotypes and other Class A β-lactamases including blaCTX-M-15, blaCTX-M-152, blaCTX-M-205, blaSHV and blaTEM (Bajaj et al., 2015; Hanna et al., 2020). Environmental monitoring revealed the presence of bacteria resistant to multiple antibiotics as well as blaCTX-M, blaTEM, blaSHV and other β-lactamase genotypes in water of river Ganga, Gomti, Yamuna and Hindon (Chaturvedi et al., 2020, 2021). The scientific data on the prevalence of antibiotic-resistant Gram-negative bacteria from irrigation-purpose wastewaters are either limited or lacking from Himachal Pradesh. Therefore, this study was aimed to determine the antibacterial susceptibility profiles of Gram-negative bacteria against β-lactams and other classes of antibiotics and the occurrence of MDR and ESBL phenotypes in these isolates from irrigation-purpose wastewaters utilized in different agro-ecosystems of lower Himalayan regions within Himachal Pradesh from Northern India.
The present study was carried out from January, 2022 to July, 2022 in Sirmaur and Solan districts located in the outer Himalayas (Shivalik range) of Himachal Pradesh, India (Figure 1).
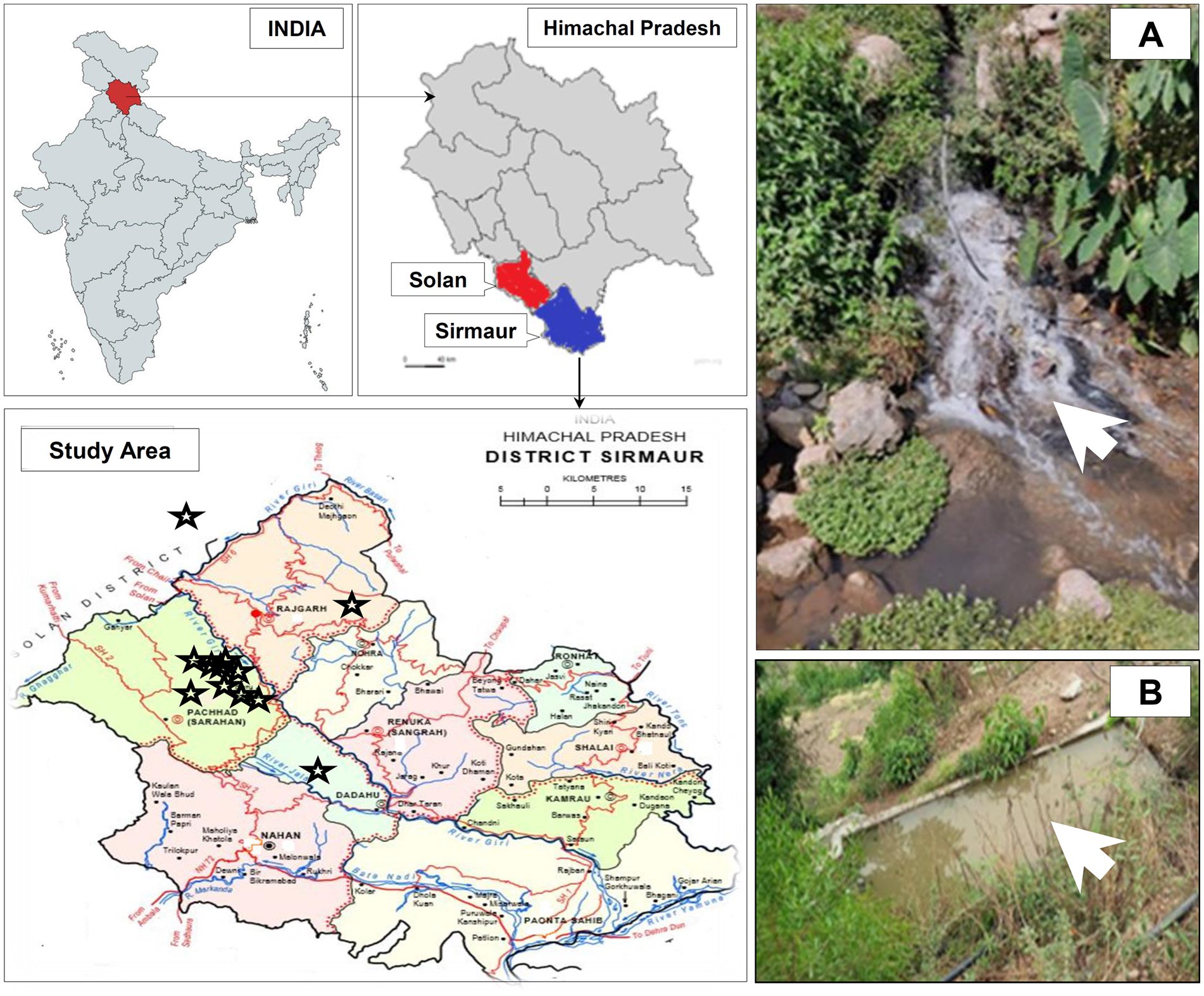
Figure 1. Geographical location of the study area and sampling sites in Sirmaur and Solan districts of Himachal Pradesh, India. The sampling sites are indicated by star symbols. The maps were prepared using https://www.mapchart.net/india.html and https://gadm.org/maps/. Panel (A, B) represents two of the sampling sites from where the wastewater samples were collected during the study.
The irrigation-purpose wastewater samples (50 mL) in triplicate were collected (from January, 2022 to July, 2022) in sterile polystyrene screw-capped test tubes from each sampling site as mentioned in Figure 1 and Table 1, kept in ice and immediately transported to the laboratory for bacteriological analysis under aseptic conditions. All necessary safety guidelines and precautions were followed during the sample collection, transportation, analysis and disposal steps.
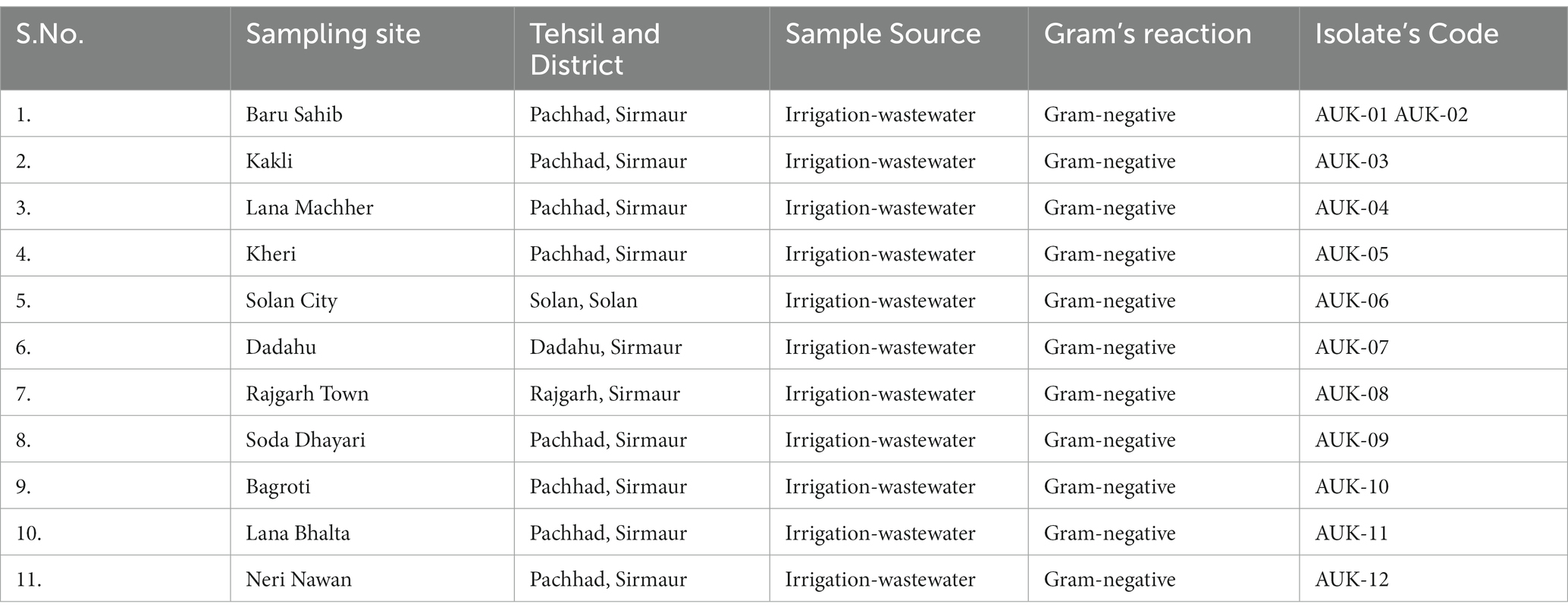
Table 1. List of sampling sites, geographical locations and bacteria isolated from irrigation-purpose wastewaters from lower Himalayan agro-ecosystems located in Himachal Pradesh, India.
Gram-negative bacteria were isolated from wastewater samples on Eosin Methylene Blue agar (HiMedia, India). About 100 μL of wastewater sample was spread on selective agar plates in triplicate and incubated at 35–37°C for 24 h as per the methods described earlier (Chaturvedi et al., 2021; Sivaraman et al., 2021). Morphologically distinct colonies based on appearance, color size, margins, texture, etc. were picked and streaked on selective agar plates for bacterial identification using Gram staining kit (HiMedia, India) and biochemical tests kits (HiMedia, India) according to Bergey’s Manual of Systematic Bacteriology/Determinative Bacteriology, and manufacturer’s instructions.
Bacterial isolates were evaluated for antibiotic susceptibility profile by Kirby-Bauer disk diffusion method according to standard published methods and Clinical Laboratory Standards Institute (CLSI) guidelines (Drieux et al. 2008; CLSI 2018, 2020). Escherichia coli ATCC 25922 (HiMedia, India) was used as a control. The antibiotics and antibiotics/inhibitor combination were obtained from HiMedia, Mumbai (India). Antimicrobial agents belonging to class penicillins, cephalosporins, aminoglycosides, folate pathway antagonists, and β-lactam/inhibitor combinations were tested. The following antibiotics were evaluated: ampicillin (AMP; 10 μg), amoxyclav (AMC; 30 μg), cefotaxime (CTX; 30 μg), co-trimoxazole (COT; 25 μg), gentamicin (GEN; 10 μg), tobramycin (TOB; 10 μg), cefpodoxime (CPD; 10 μg), ceftazidime (CAZ; 30 μg), cefpodoxime/clavulanic acid (CCL; 10/5 μg), ceftazidime/clavulanic acid (CAC; 30/10 μg) and cefotaxime/clavulanic acid (CEC; 30/10 μg). Bacterial isolates were cultured in Mueller-Hinton broth at 35–37°C and the turbidity was adjusted using 0.5 McFarland standard prior to swabbing on Mueller-Hinton agar plates. The antibiotic hexadiscs were placed on agar surface aseptically, followed by incubation at 35–37°C for 16–18 h. The diameter of the zones of inhibition around the disks was recorded to the nearest mm using Antibiotic Zonescale (HiMedia, India). The zones of inhibition diameters were compared with the Performance Standards for Antimicrobial Testing, 2020, Table 2A of M100 document as published by CLSI in order to classify the bacterial isolates either resistant, intermediate, or susceptible to test antibiotics (Drieux et al., 2008); Tan et al., 2023). Bacterial isolate exhibiting resistance against at least three different antibiotics classes was categorized as multidrug-resistant (Magiorakos et al., 2012). The Multiple Antibiotic Resistance (MAR) index was calculated according the formula (Krumperman, 1983) which is as follows: MAR index = number of antibiotics to which an isolate showed resistance/total number of antibiotics evaluated for susceptibility teesting.
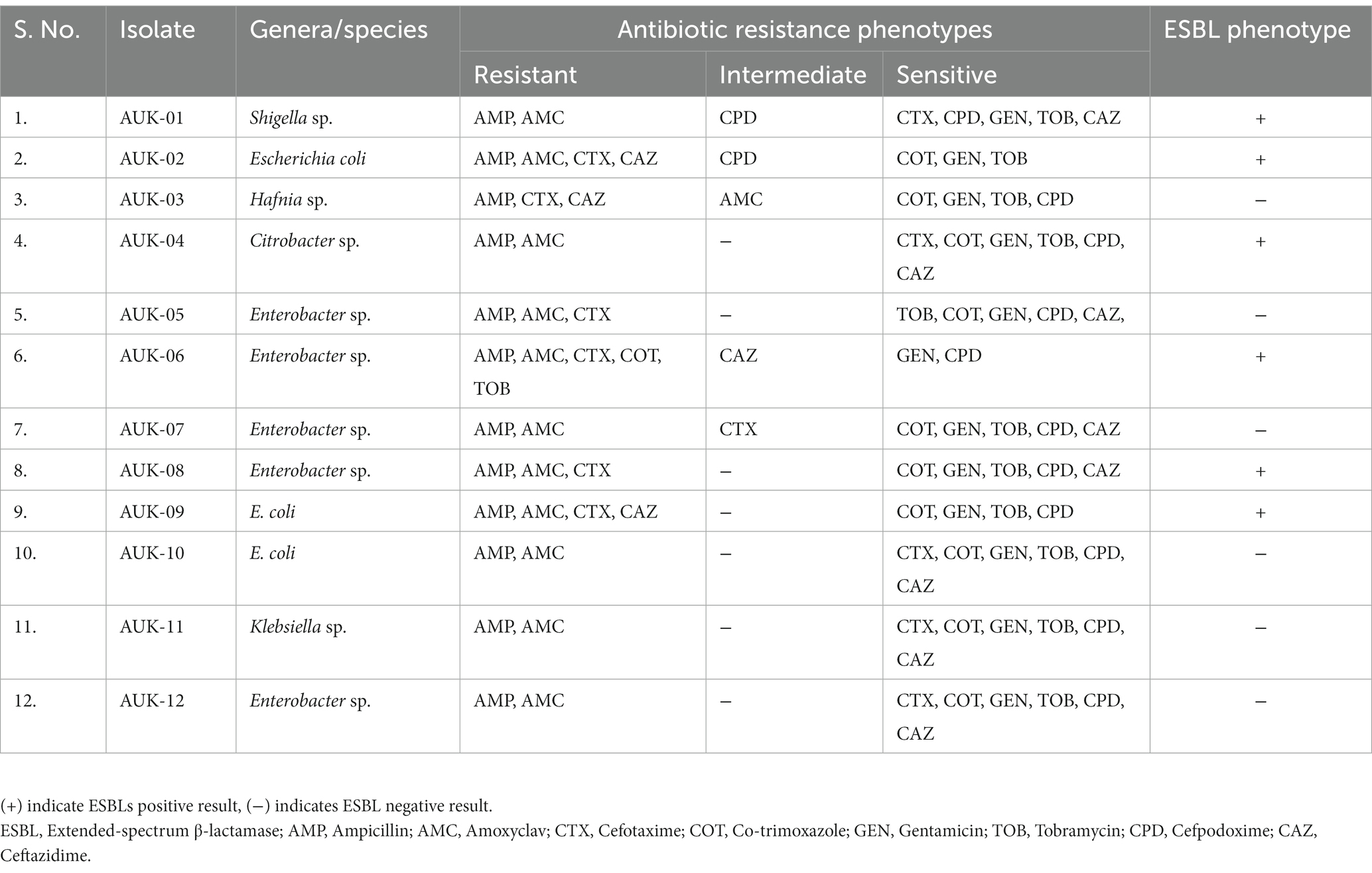
Table 2. Antibiotic susceptibility testing and resistance phenotypes of Gram-negative bacteria isolated from irrigation-purpose wastewaters from lower Himalayan agro-ecosystems located in Himachal Pradesh, India.
Bacterial isolates were tested for ESBLs production by phenotypic confirmatory disk diffusion test (PCDDT) using ESBLs identification kit (HiMedia, India) as per the manufacturer’s instructions. Bacterial isolates were cultured in Mueller-Hinton broth at 35–37°C and the turbidity was adjusted using 0.5 McFarland standard prior to swabbing on Mueller-Hinton agar plates. The disks containing cefotaxime (30 μg) and cefotaxime/clavulanic acid (30/10 μg) were aseptically placed on agar plate containing test bacterial culture at least 24 mm apart and followed by incubation at 35–37°C for 16–18 h. The diameter of the zones of inhibition around the disks was recorded to the nearest mm using Antibiotic Zonescale (HiMedia, India). An increase in the zone of inhibition by ≥5 mm with cefotaxime/clavulanic acid disk was considered ESBL positive result according to the CLSI criteria.
Selected bacterial isolates were subjected to 16S rRNA gene sequencing from Chromus Biotech Pvt. Ltd., India using primer set 27F 5´-AGAGTTTGATCMTGGCTCAG-3′ and 1492R: 5´-GGTTACCTTGTTACGACTT-3′ (Palkova et al., 2021). PCR products were sequenced using BigDye terminator cycle sequencing kit (V3.1) in an ABI Prism 3,730 Genetic Analyzer (Applied Biosystems, United States) The nucleotide sequences were subjected to a similarity search using the BLAST algorithm at National Center for Biotechnology Information (NCBI) GenBank Database1 and the sequences showing the highest scores were retrieved for further analysis. The 16S rRNA gene sequences of isolates AUK-01, AUK-02, AUK-03 and AUK-04 were deposited in NCBI GenBank database under accession numbers ON968448, ON968449, ON968450, and ON968451, respectively.
The 16S rRNA gene sequences were imported to MEGA X v10.2.5 software and phylogenetic trees were constructed on the aligned datasets using the neighbor-joining method (Saitou and Nei, 1987; Tamura et al., 2007). A multiple sequence alignment was performed using the CLUSTAL W program (Thompson et al., 1994) and the data converted to PHYLIP format. All positions containing gaps and missing data were eliminated from the data set (complete deletion option) and branches containing more than 50% gaps were also removed. One sequence from each group was selected as a representative operational taxonomic unit. The phylogenetic tree was constructed by taking the sequences of bacterial strains along with their ten closest type strain matches available in the NCBI database using neighbor joining method with 1,000 bootstrapped replications to estimate evolutionary distance between all pairs of sequences simultaneously.
Experiments were performed in triplicates and data were expressed as mean ± standard deviation. Results were analyzed using Microsoft Excel and Graphpad Prism (Dotmatics).
Wastewaters used for irrigation purpose were collected from 10 different geographical locations in Sirmaur district and one sampling site from Solan district of Himachal Pradesh, India (Table 1). The bacteria growing on selective medium were characterized on the basis of colony features, Gram reaction and biochemical parameters (Supplementary File 1). A total of 12 Gram-negative bacterial isolates were isolated from Baru Sahib (2), Kakli (1), Lana Machher (1), Kheri (1), Solan city (1), Dadahu (1), Rajgarh town (1), Soda Dhayari (1), Bagroti (1), Lana Bhalta (1) and Neri Nawan (1) as shown in Table 1. These isolates were subjected to antibiotic susceptibility testing and detection of ESBL phenotype. The characterization details of the bacterial isolates are provided in Supplementary File 1.
The antibiotic susceptibility testing data are shown in Table 2 and as heatmap in Figure 2. Isolate AUK-06 showed resistant phenotype against five antibiotics viz. ampicillin, amoxicillin/clavulanic acid, cefotaxime, co-trimoxozole and tobramycin (Figure 3). It was susceptible to other antibiotics except ceftazidime where it exhibited intermediate phenotype. AUK-02, AUK-03 and AUK-09 were resistant to four antimicrobials (Table 2 and Figure 2). On the other hand, isolates AUK-04, AUK-10, AUK-11 and AUK-12 were susceptible to all the tested antimicrobials except ampicillin and amoxicillin/clavulanic acid combination. AUK-06 was the only isolate that displayed MDR phenotype as it exhibited resistance to at least three classes of antibiotics viz., β-lactams (ampicillin and amoxyclav), aminoglycosides (tobramycin), and cephalosporins (cefotaxime and ceftazidime). As shown in Figure 2, all the bacterial isolates (n = 12) were resistant to ampicillin. In contrast, gentamicin exhibited antimicrobial action against all the isolates, as none were resistant to this antibiotic. In terms of susceptibility toward each of the tested antibiotic, all of the bacterial isolates were resistant to ampicillin whereas 11 were resistant to amoxyclav and six to cefotaxime. Other tested antibiotics were also able to inhibit the bacteria growth in disk diffusion assays as depicted in Figure 2 and Table 2.
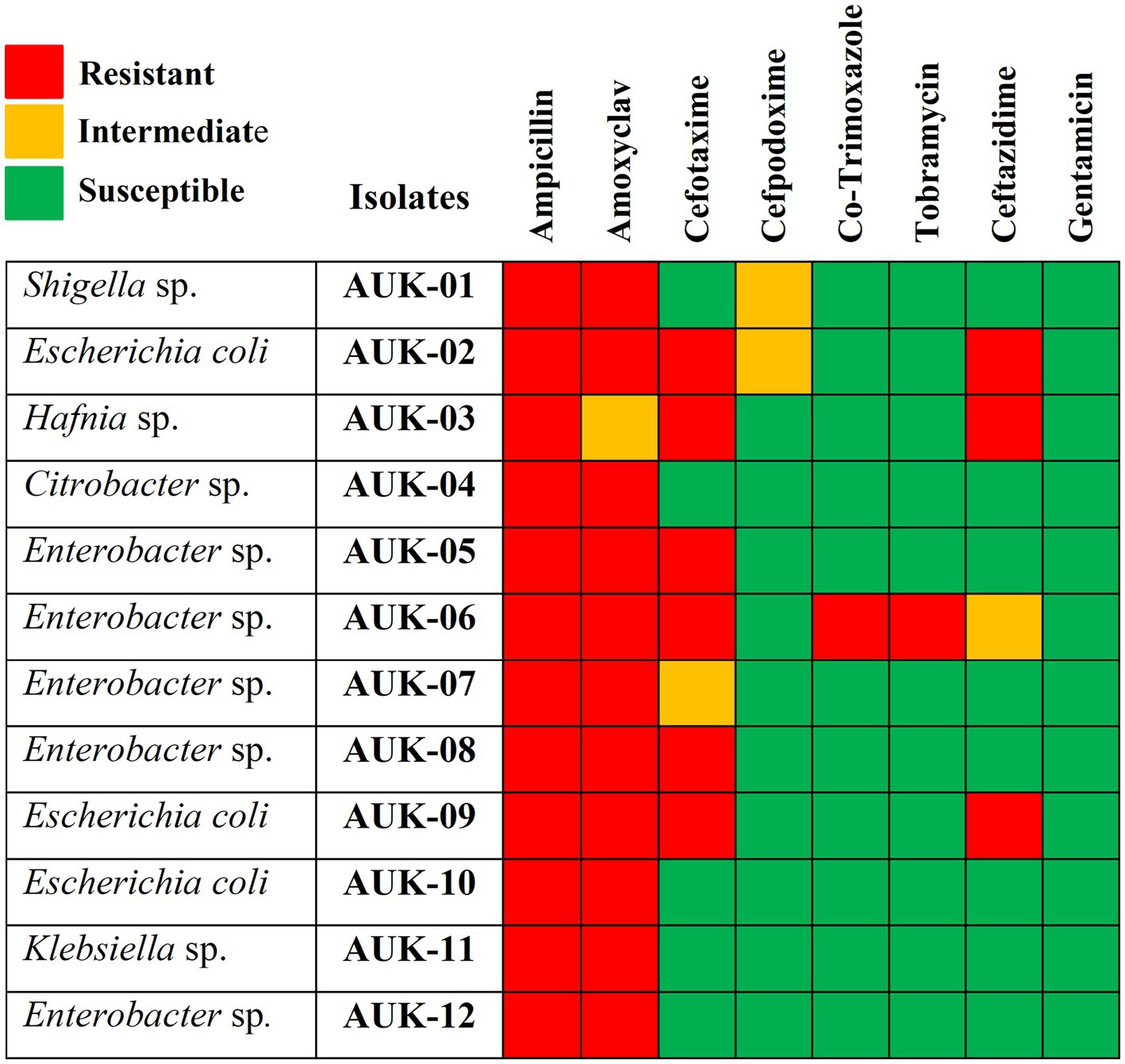
Figure 2. Antibiotic susceptibility heatmap of Gram-negative bacterial isolates as determined by Kirby-Bauer disk diffusion method and interpreted according to CLSI breakpoints. Rows represent bacterial isolates and column represents antimicrobials tested. Red-colored blocks indicate resistance; green blocks indicate susceptible and orange blocks represent intermediate action of the antimicrobial agents.
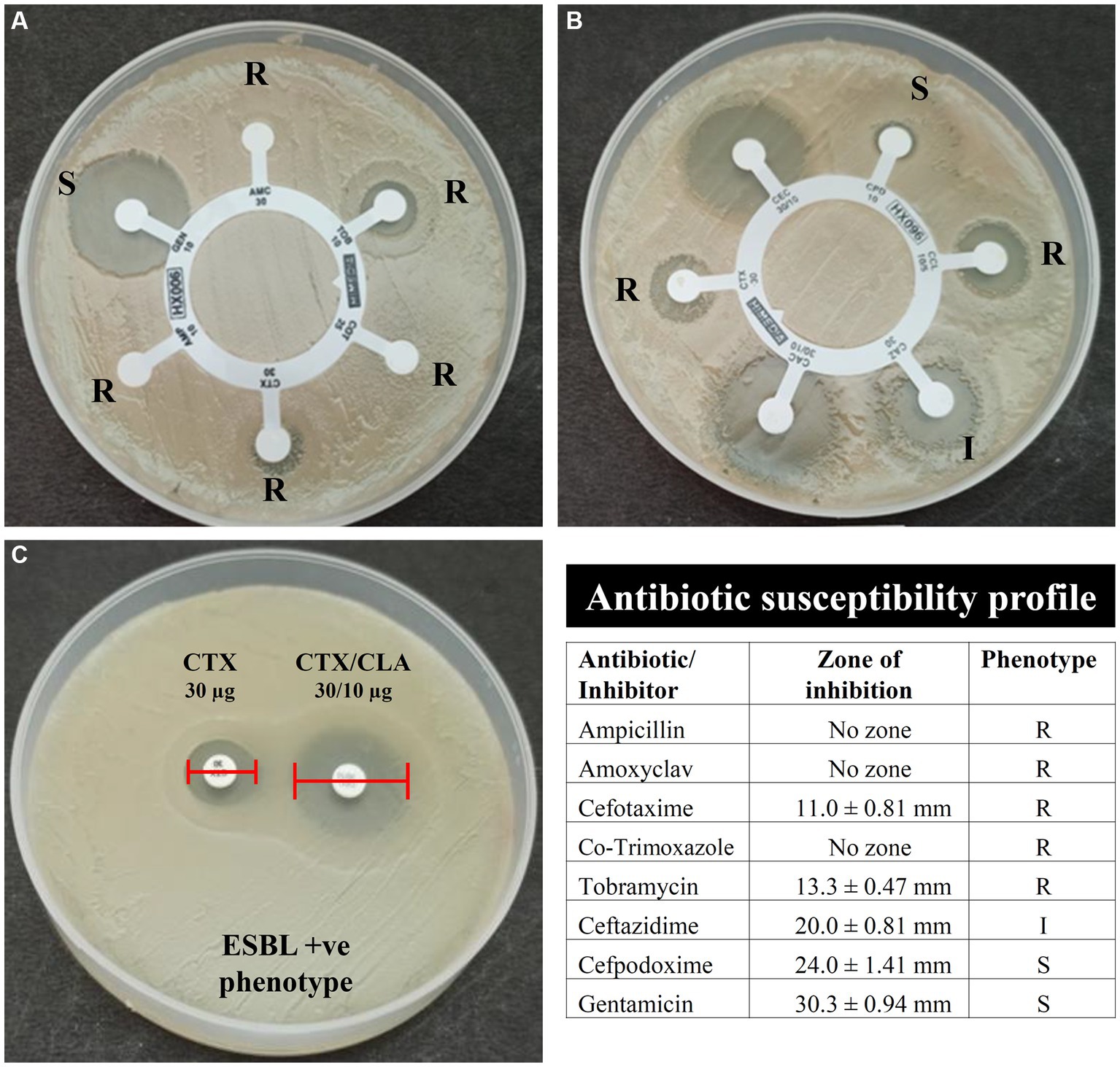
Figure 3. Antibiotic susceptibility and ESBL phenotype of bacterial isolates AUK-06 (Enterobacter sp.) as determined by the Kirby-Bauer disk diffusion method. Here, (A) indicates susceptibility of isolate AUK-06 toward ampicillin (AMP), amoxyclav (AMC), cefotaxime (CTX), co-trimoxazole (COT), gentamicin (GEN), and tobramycin (TOB); panel (B) represent cefpodoxime (CPD), cefpodoxime/CLA (CCL), ceftazidime (CAZ), ceftazidime/CLA (CAC), cefotaxime (CTX), and cefotaxime/CLA (CEC); Phenotypic detection of ESBL production by PCDDT is depicted in (C). The antibiotic susceptibility profile of isolate AUK-06 is shown in tabulated form according to the interpretation criteria mentioned in M100 document, CLSI.
Phenotypic determination assay for ESBLs production revealed that six bacterial isolates namely AUK-01, AUK-02, AUK-04, AUK-06, AUK-08 and AUK-09 were ESBL-positive which represents half of the total isolates (Table 2). Among these, two isolates were Enterobacter spp., two were E. coli and one each was related to Shigella sp. and Citrobacter sp. Isolate AUK-06 (Enterobacter sp.) which was prevalent in wastewater sample of Solan city had exhibited both MDR and ESBL phenotypes. MAR index values for bacterial isolates ranged from 0.20 ± 0.07 to 0.75 (Figure 4A). The highest MAR index was exhibited by isolate AUK-06 (Enterobacter sp.) followed by isolate AUK-02 (0.58 ± 0.07) and AUK-03 (0.45 ± 0.07). The lowest MAR index was associated with isolates AUK-10, AUK-11 and AUK-12 which correlates with their low antibiotic resistance. In terms of percentage of bacterial isolates having a MAR index value of >0.25 was 66.7% whereas only 33.3% isolates had MAR index ≤0.25 (Figure 4B). These data indicated that the bacterial isolates originated from high antibiotic contamination aquatic environments.
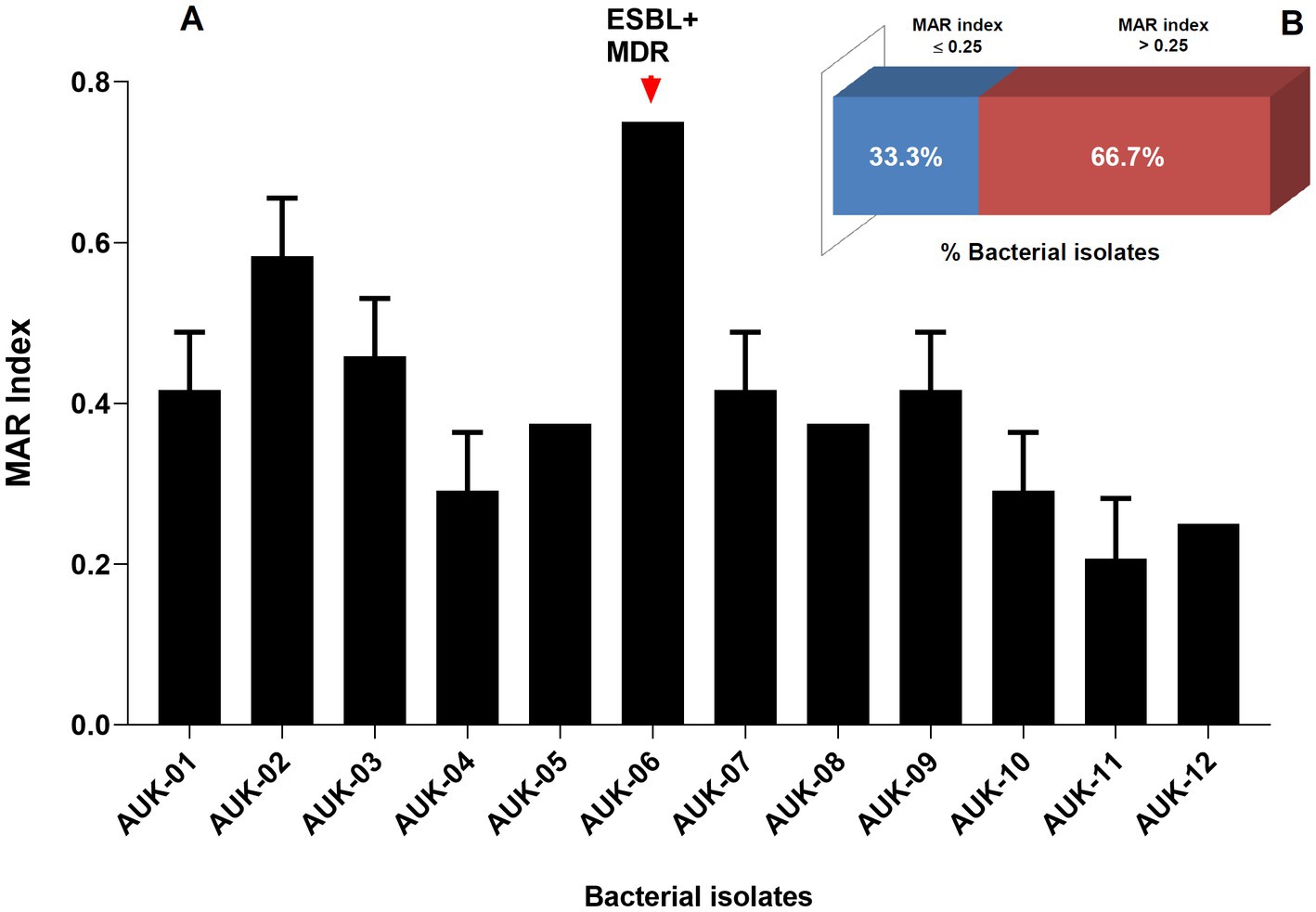
Figure 4. Multiple Antibiotic Resistance (MAR) index of bacteria isolated from irrigation-purpose wastewater samples from lower Himalayan agro-ecosystems located in Himachal Pradesh, India. MAR index was calculated according to the formula described by Krumperman (1983). Here, (A) represent the MAR indices of bacterial isolates and (B) represent the percentage of bacterial isolates having a MAR index ≤0.25 or > 0.25. Data represents mean ± SD of three independent experiments.
Morphological, biochemical, and 16S rRNA gene sequence-based molecular characterization of Gram-negative bacterial isolates showed the dominant prevalence of Enterobacter sp. (41.3%) and E. coli (25%) as depicted in Figure 5. On the other hand, prevalence of Klebsiella sp., Citrobacter koseri, Hafnia paralvei, and Shigella sp. was low, i.e., 8.33% in wastewater samples. Four bacterial isolates, AUK-01, AUK-02, AUK-03, and AUK-04, were characterized by 16S rRNA gene sequencing and were phylogenetically related to Shigella sp., Escherichia coli, Hafnia paralvei, and Citrobacter koseri, respectively (Figure 6). The 16S rRNA gene sequences of these isolates were deposited in the NCBI GenBank database with the following accession numbers: ON968448, ON968449, ON968450, and ON968451. Further, the phylogenetic trees were constructed to assess the evolutionary relatedness among the bacterial isolates and their nearest neighbors available in the database (Figure 6).
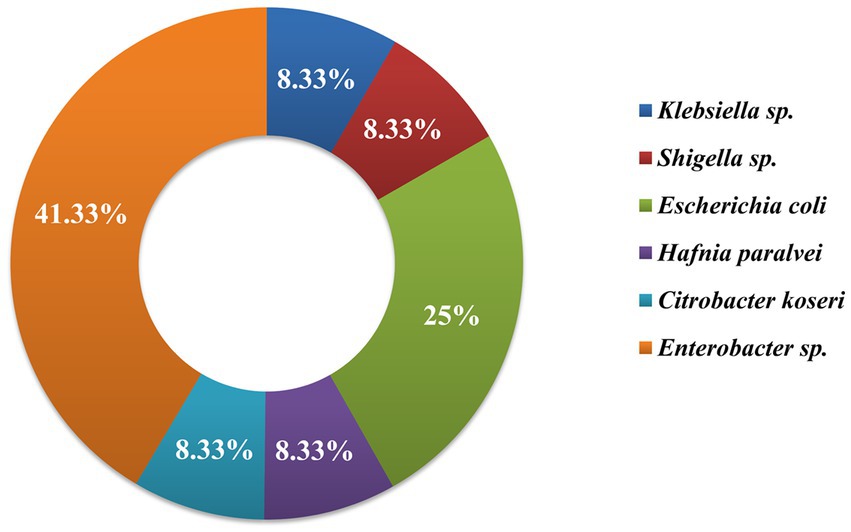
Figure 5. Genera-wise distribution (%) of bacteria (n = 12) isolated from irrigation-purpose wastewaters from lower Himalayan agro-ecosystems located in Himachal Pradesh, India.
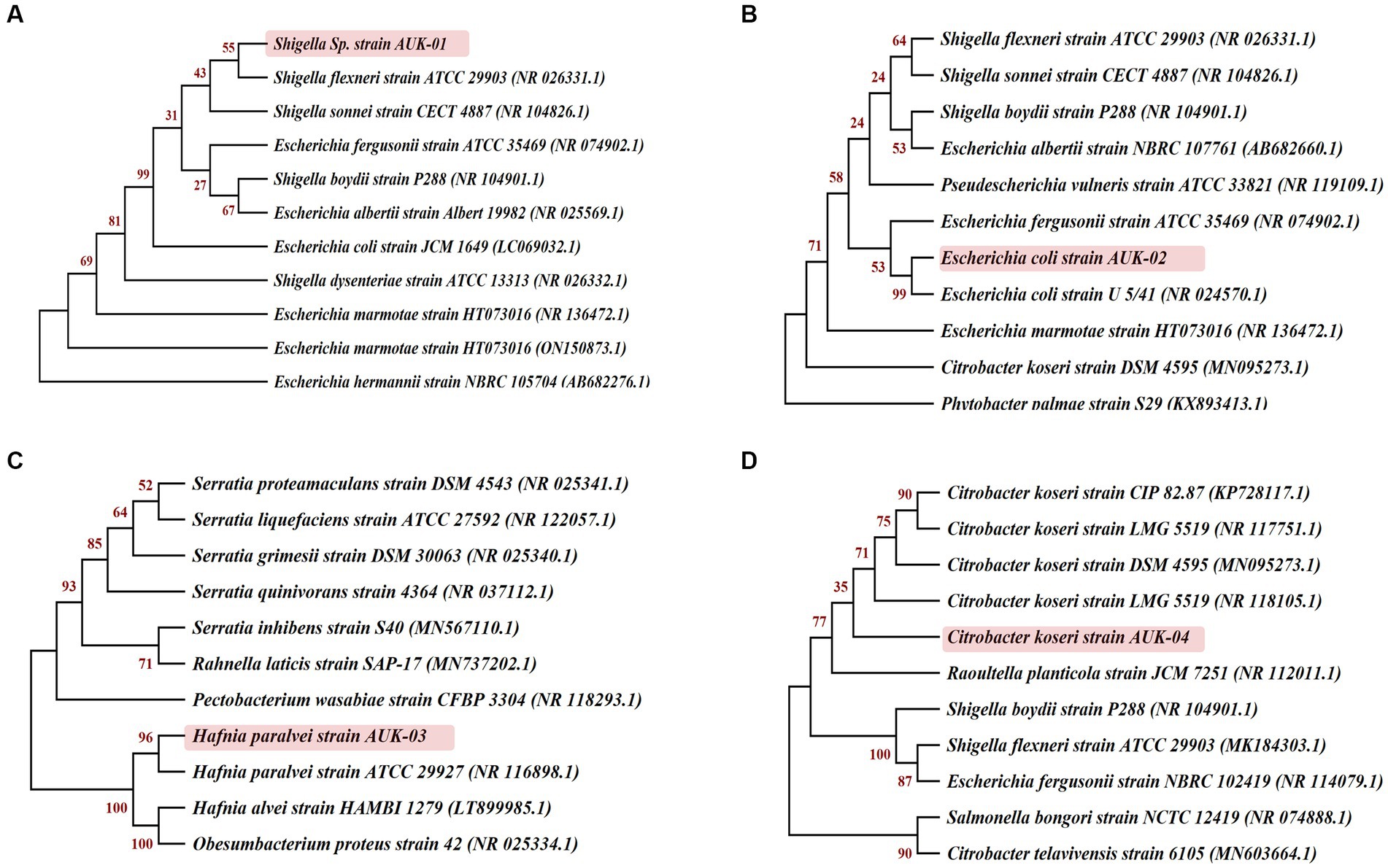
Figure 6. Phylogenetic relationships of selected bacterial isolates obtained from irrigation-purpose wastewater samples were inferred on the basis of 16S rRNA gene sequences by the neighbor-joining method using MEGA X software (version 10.2.5). Bootstrap values expressed as a percentage of 1,000 re-samplings are shown at the nodes. Here (A), (B), (C) and (D) represent the isolates AUK-01, AUK-02, AUK-03, and AUK-04, respectively.
Wastewater contains high nutrient contents and hence found re-usability in agricultural irrigation systems since it provides organic carbon, nutrients (nitrogen, phosphorus, potassium), and inorganic micronutrients to the crop plants (Alcalde-Sanz and Gawlik, 2017; Minhas et al., 2022). The untreated wastewaters are frequently utilized for irrigation purposes in regions where rain shortfall are common or groundwater availability is limited which also include several states of India, including Himachal Pradesh. Their use in agricultural practices, however represent a serious health risk due to its inherent nature of carrying ARB and ARGs (Wang et al., 2020; Tucker et al., 2022). The antibiotic contents present in wastewaters facilitate mutation and genetic modifications in ARB thus making these multi-drug resistant and difficult to treat under clinical settings (Leiva et al., 2021; Uluseker et al., 2021; Mutuku et al., 2022). In the present study, 12 bacterial isolates belonging to six genera were identified in irrigation-purpose wastewater samples on the basis of morphological, biochemical, and molecular methods from 11 different geographical locations in Sirmaur and Solan districts of Himachal Pradesh. These bacteria belonged to E. coli, Enterobacter sp., Hafnia sp., Shigella sp., Citrobacter sp., and Klebsiella sp. Enterobacter spp. and E. coli were found to be most dominant among 12 bacterial isolates, respectively. The antibiograms of these isolates against penicillins (ampicillin and amoxyclav), aminoglycosides (gentamicin and tobramycin), sulfonamides (co-trimoxazole), and third-generation cephalosporins (cefotaxime, cefpodoxime, and ceftazidime) indicated resistance against all antibiotics except gentamicin. All isolates were resistant to ampicillin. In contrast, gentamicin exhibited antimicrobial action against all the isolates as all were susceptible to this antibiotic. Further, 11 isolates were resistant to amoxyclav whereas six of the isolates displayed resistance to cefotaxime. These findings are supported by previous research reports in which widespread resistance against similar antibiotic classes was reported in bacteria from Enterobacteriaceae. A high frequency of resistance to ciprofloxacin, tetracycline, cefoxitin, amoxicillin/clavulanic acid, cefotaxime, and aztreonam was reported from water samples of the Mondego River in Portugal (Amador et al., 2015). Schmiege et al. (2021); Zagui et al. (2020); Veloo et al. (2022) also observed MDR and ESBL-producing ARB in urban wastewaters. MAR index values for 12 isolates ranged between 0.21–0.75. Ali et al. (2021) found the MAR index value to range between 0.2 and 0.32 in wastewaters from Delhi-NCR region. The MAR index value >0.2 suggest high pollution load and antibiotic exposure in a specific sampling site (Krumperman, 1983; Ali et al., 2021).
Five isolates (AUK-05, AUK-06, AUK-07, AUK-08, and AUK-12) were identified to be Enterobacter spp. which showed their predominant prevalence in irrigation-purpose wastewater samples of the sampling sites. AUK-6 which was obtained from wastewater sample of Solan City was the sole MDR isolate of our study and it also showed ESBL phenotype. Similar to our findings, Ben Said et al. (2015) found the presence of the multidrug-resistant ESBL-producing Enterobacter hormaechei in irrigation water samples from Tunisia. In this study, 50% of bacterial isolates showed ESBL phenotypes, as confirmed by cefotaxime and cefotaxime/clavulanic acid combination-based qualitative methods. Among these isolates, AUK-01, AUK-02, AUK-04, AUK-06, AUK-08, and AUK-09 were ESBL producers. AUK-02 and AUK-09 were identified as E. coli, whereas AUK-06 and AUK-08 were related to Enterobacter spp. Further, Shigella sp. (AUK-01) and Citrobacter sp. (AUK-04) also displayed the ESBL phenotype. Ali et al. (2021) found 106 ESBL-positive isolates (24.3%) out of 436 bacterial isolates isolated from Hauz Khas lake, Ghazipur slaughterhouse, Jasola wastewater treatment plant, and Lodhi garden pond in the Delhi-NCR region. Among these isolates, 42–78% also exhibited MDR phenotypes, depending upon the location. Similarly, Singh et al. (2021) found ESBL-producing strains of E. coli (n = 34) and K. pneumoniae (n = 39) with a prevalence of blaCTX-M-1, blaCTX-M-2, and blaCTX-M-15 genotypes from Manali, Kullu, and Baddi in Himachal Pradesh. In previous studies, antibiotic-resistant strains of E. coli, Enterobacter, Klebsiella, Pseudomonas, Citrobacter, Hafnia, Acinetobacter, Shigella, and Aeromonas have been reported from different regions of India (Lamba and Ahammad, 2017; Chaturvedi et al., 2020, 2021). The prevalence of the ESBL phenotype in 58% of E. coli isolates in irrigation water samples from several provinces of Ecuador was observed (Montero et al., 2021). Similarly, 33% Gram-negative bacterial isolates exhibited MDR phenotypes and ESBL genotypes (blaKPC, blaTEM, blaSHV and blaCTX-M) from hospital sewage and urban wastewater in Brazil (Zagui et al., 2020). Igbinosa et al. (2023) reported that 85.9% of E. coli strains isolated from agricultural farms (irrigation water, soil, manure) were resistant to various antimicrobial classes and exhibited MDR phenotype as well ESBL genotypes which might be due to the use of untreated water for irrigation purposes. Tiwari et al. (2023) showed the presence of β-lactamase (blaGES, blaMOX and blaTEM) producing Enterobacter spp., Aeromonas spp., and Klebsiella sp. and E. coli in untreated municipal wastewater from Helsinki, Finland.
There are two ways that antibiotics can infiltrate agricultural ecosystems: firstly, by fertilizing with animal manures, biosolids, sewage sludge, and sediments that include antibiotics, and secondly, by irrigating with reclaimed water that has been contaminated with antibiotics from sewage treatment plants, wastewater, surface water, or groundwater since these sources are regularly contaminated with antibiotics (Du and Liu, 2012; Leiva et al., 2021; Minhas et al., 2022). In aquatic ecosystems, pathogenic, commensal, and environmental bacteria serve as reservoirs of antibiotic-resistance genes, mobile genetic elements, and bacteriophages, with consequent dissemination through conjugation, transformation, and transduction (von Wintersdorff et al., 2016). Utilization of wastewater for agricultural irrigation may assist in mobilizing clinically relevant antibiotic resistance genes (resistome) from non-pathogenic commensal bacteria to pathogenic strains, thus leading to the spread of resistance. The origin of blaCTX − M genes in Enterobacteriaceae can be traced to the environmental Kluyvera sp. via HGT (Canton and Coque, 2006). Therefore, efficient treatment technologies are essentially required for the removal of microbial pathogens, antibiotic residues and heavy metal hazards from wastewaters (Tucker et al., 2022). Regular monitoring and surveillance of antibiotic residues and bacteria resistant to antibiotics in wastewaters, rivers, ponds and wastewater treatment plants, and effluent treatment sites should be prioritized and included in regulatory action plans.
Wastewaters are being used in agricultural irrigation in several water-scarce countries due to their organic and inorganic nutrients, however this wastewater-based irrigation practices also present serious public health and environmental sustainability concerns. Our findings also confirmed the prevalence of antibiotic resistant Gram-negative bacteria resistant to multiple antibiotics and exhibiting the ESBL phenotype in irrigation-purpose wastewater samples from Himachal Pradesh. Therefore, there is an urgent need for an integrated approach focusing on environmental monitoring, identifying key hotspots of antibiotic resistance, early and rapid detection, continuous surveillance, stringent regulatory guidelines and improved agri-environmental risk assessment models before their use in the agro-ecosystems for ensuring public health and environmental safety.
The datasets presented in this study are deposited in the NCBI repository, accession numbers ON968448, ON968449, ON968450, ON968451: https://www.ncbi.nlm.nih.gov/nuccore/ON968448, https://www.ncbi.nlm.nih.gov/nuccore/ON968449, https://www.ncbi.nlm.nih.gov/nuccore/ON968450, and https://www.ncbi.nlm.nih.gov/nuccore/ON968451.
AA, ShiS, and SK: conducted the experiments, and manuscript preparation. YA, SheS, DJ, PC, KP, MK, and NS: editing, review, and finalization of the manuscript. All authors contributed to the article and approved the submitted version.
The authors acknowledge the necessary facilities provided by Eternal University, Baru Sahib for carrying out the experimental work. The valuable suggestions and expertise from Dr. Ajar Nath Yadav, Dr. Kamal Kishore and Dr. Puneet Negi are also acknowledged. Authors would like to acknowledge the support provided by Researchers Supporting Project Number RSP2023R358, King Saud University, Riyadh, Saudi Arabia.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1227132/full#supplementary-material
Alcalde-Sanz, L., and Gawlik, B. M. (2017). Minimum quality requirements for water reuse in agricultural irrigation and aquifer recharge-towards a legal instrument on water reuse at EU level, EUR 28962 EN, Publications Office of the European Union, Luxembourg. ISBN 978-92-79-77175-0
Ali, A., Sultan, I., Mondal, A. H., Siddiqui, M. T., Gogry, F. A., and Haq, Q. M. R. (2021). Lentic and effluent water of Delhi-NCR: a reservoir of multidrug-resistant bacteria harbouring blaCTX-M, blaTEM and blaSHV type ESBL genes. J. Water Health 19, 592–603. doi: 10.2166/wh.2021.085
Amador, P. P., Fernandes, R. M., Prudêncio, M. C., Barreto, M. P., and Duarte, I. M. (2015). Antibiotic resistance in wastewater: occurrence and fate of Enterobacteriaceae producers of class a and class C β-lactamases. J. Environ. Sci. Health. A Tox. Hazard. Subst. Environ. Eng. 50, 26–39. doi: 10.1080/10934529.2015.964602
Anderson, R. E. V., Chalmers, G., Murray, R., Mataseje, L., Pearl, D. L., Mulvey, M., et al. (2023). Characterization of Escherichia coli and other Enterobacterales resistant to extended-spectrum cephalosporins isolated from dairy manure in Ontario Canada. Appl. Environ. Microbiol. 89:e0186922. doi: 10.1128/aem.01869-22
Antimicrobial Resistance Collaborators (2022). Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399, 629–655. doi: 10.1016/S0140-6736(21)02724-0
Bajaj, P., Singh, N. S., Kanaujia, P. K., and Virdi, J. S. (2015). Distribution and molecular characterization of genes encoding CTX-M and AmpC β-lactamases in Escherichia coli isolated from an Indian urban aquatic environment. Sci. Total Environ. 505, 350–356. doi: 10.1016/j.scitotenv.2014.09.084
Ben Said, L., Jouini, A., Klibi, N., Dziri, R., Alonso, C. A., Boudabous, A., et al. (2015). Detection of extended-spectrum beta-lactamase (ESBL)-producing Enterobacteriaceae in vegetables, soil and water of the farm environment in Tunisia. Int. J. Food Microbiol. 203, 86–92. doi: 10.1016/j.ijfoodmicro.2015.02.023
Bhagat, C., Kumar, M., Tyagi, V. K., and Mohapatra, P. K. (2020). Proclivities for prevalence and treatment of antibiotics in the ambient water: a review. npj Clean Water 3, 1–18. doi: 10.1038/s41545-020-00087-x
Bush, K. (2023). Classification for β-lactamases: historical perspectives. Expert Rev. Anti-Infect. Ther. 21, 513–522. doi: 10.1080/14787210.2023.2194633
Bush, K., and Bradford, P. A. (2020). Epidemiology of β-lactamase-producing pathogens. Clin. Microbiol. Rev. 33, e00047–e00019. doi: 10.1128/CMR.00047-19
Canton, R., and Coque, T. M. (2006). The CTX-M β-lactamase pandemic. Curr. Opin. Microbiol. 9, 466–475. doi: 10.1016/j.mib.2006.08.011
Castanheira, M., Simner, P. J., and Bradford, P. A. (2021). Extended-spectrum β-lactamases: an update on their characteristics, epidemiology and detection. JAC Antimicrob. Resist. 3:dlab092. doi: 10.1093/jacamr/dlab092
Chaturvedi, P., Chaurasia, D., Pandey, A., and Gupta, P. (2020). Co-occurrence of multidrug resistance, β-lactamase and plasmid mediated AmpC genes in bacteria isolated from river ganga, northern India. Environ. Pollut. 267:115502. doi: 10.1016/j.envpol.2020.115502
Chaturvedi, P., Chowdhary, P., Singh, A., Chaurasia, D., Pandey, A., Chandra, R., et al. (2021). Dissemination of antibiotic resistance genes, mobile genetic elements, and efflux genes in anthropogenically impacted riverine environments. Chemosphere 273:129693. doi: 10.1016/j.chemosphere.2021.129693
Cho, S., Jackson, C. R., and Frye, J. G. (2023). Freshwater environment as a reservoir of extended-spectrum β-lactamase-producing Enterobacteriaceae. J. Appl. Microbiol. 134:lxad034. doi: 10.1093/jambio/lxad034
CLSI. (2018). Clinical and laboratory standards institute. M100 performance standards for antimicrobial susceptibility testing. 28th ed. Clinical and Laboratory Standards Institute. Pennsylvania, USA
CLSI. (2020). Clinical and laboratory standards institute. M100 performance standards for antimicrobial susceptibility testing. 30th ed. Clinical and Laboratory Standards Institute. Pennsylvania, USA
Dantas Palmeira, J., do Arte, I., Ragab Mersal, M. M., Carneiro da Mota, C., and Ferreira, H. M. N. (2022). KPC-producing Enterobacterales from Douro River, Portugal-persistent environmental contamination by putative healthcare settings. Antibiotics 12:62. doi: 10.3390/antibiotics12010062
Drieux, L., Brossier, F., Sougakoff, W., and Jarlier, V. (2008). Phenotypic detection of extended-spectrum beta-lactamase production in Enterobacteriaceae: review and bench guide. Clin. Microbiol. Infect. 14, 90–103. doi: 10.1111/j.1469-0691.2007.01846.x
Du, L., and Liu, W. (2012). Occurrence, fate, and ecotoxicity of antibiotics in agro-ecosystems. A review. Agron. Sustain. Dev. 32, 309–327. doi: 10.1007/s13593-011-0062-9
Farooqui, H. H., Selvaraj, S., Mehta, A., and Heymann, D. L. (2018). Community level antibiotic utilization in India and its comparison Vis-à-Vis European countries: evidence from pharmaceutical sales data. PLoS One 13:e0204805. doi: 10.1371/journal.pone.0204805
Fazaludeen Koya, S., Ganesh, S., Selvaraj, S., Wirtz, V. J., Galea, S., and Rockers, P. C. (2022). Antibiotic consumption in India: geographical variations and temporal changes between 2011 and 2019. JAC Antimicrob. Resist. 4:dlac112. doi: 10.1093/jacamr/dlac112
Gitter, A., Oghuan, J., Godbole, A. R., Chavarria, C. A., Monserrat, C., Hu, T., et al. (2023). Not a waste: wastewater surveillance to enhance public health. Front. Chem. Eng. 4:1112876. doi: 10.3389/fceng.2022.1112876
Grenni, P. (2022). Antimicrobial resistance in rivers: a review of the genes detected and new challenges. Environ. Toxicol. Chem. 41, 687–714. doi: 10.1002/etc.5289
Hanna, N., Purohit, M., Diwan, V., Chandran, S. P., Riggi, E., Parashar, V., et al. (2020). Monitoring of water quality, antibiotic residues, and antibiotic-resistant Escherichia coli in the Kshipra River in India over a 3-year period. Int. J. Environ. Res. Public Health 17:7706. doi: 10.3390/ijerph17217706
Helmecke, M., Fries, E., and Schulte, C. (2020). Regulating water reuse for agricultural irrigation: risks related to organic micro-contaminants. Environ. Sci. Eur. 32:4. doi: 10.1186/s12302-019-0283-0
Hutchings, M. I., Truman, A. W., and Wilkinson, B. (2019). Antibiotics: past, present and future. Curr. Opin. Microbiol. 51, 72–80. doi: 10.1016/j.mib.2019.10.008
Igbinosa, E. O., Beshiru, A., Igbinosa, I. H., Cho, G.-S., and Franz, C. M. A. P. (2023). Multidrug-resistant extended spectrum β-lactamase (ESBL)-producing Escherichia coli from farm produce and agricultural environments in Edo state. Nigeria. PLoS ONE 18:e0282835. doi: 10.1371/journal.pone.0282835
Janda, J. M., and Abbott, S. L. (2021). The changing face of the family Enterobacteriaceae (order: "Enterobacterales"): new members, taxonomic issues, geographic expansion, and new diseases and disease syndromes. Clin. Microbiol. Rev. 34, e00174–e00120. doi: 10.1128/CMR.00174-20
Jani, K., Srivastava, V., Sharma, P., Vir, A., and Sharma, A. (2021). Easy access to antibiotics; spread of antimicrobial resistance and implementation of one health approach in India. J. Epidemol. Glob. Health. 11, 444–452. doi: 10.1007/s44197-021-00008-2
Karkman, A., Do, T. T., Walsh, F., and Virta, M. P. J. (2018). Antibiotic-resistance genes in wastewater. Trends Microbiol. 26, 220–228. doi: 10.1016/j.tim.2017.09.005
Krumperman, P. H. (1983). Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl. Environ. Microbiol. 46, 165–170. doi: 10.1128/aem.46.1.165-170.1983
Lamba, M., and Ahammad, S. Z. (2017). Sewage treatment effluents in Delhi: a key contributor of β-lactam resistant bacteria and genes to the environment. Chemosphere 188, 249–256. doi: 10.1016/j.chemosphere.2017.08.133
Larsson, D. G. J., and Flach, C. F. (2022). Antibiotic resistance in the environment. Nat. Rev. Microbiol. 20, 257–269. doi: 10.1038/s41579-021-00649-x
Leiva, A. M., Piña, B., and Vidal, G. (2021). Antibiotic resistance dissemination in wastewater treatment plants: a challenge for the reuse of treated wastewater in agriculture. Rev. Environ. Sci. Biotechnol. 20, 1043–1072. doi: 10.1007/s11157-021-09588-8
Liguori, K., Keenum, I., Davis, B. C., Calarco, J., Milligan, E., Harwood, V. J., et al. (2022). Antimicrobial resistance monitoring of water environments: a framework for standardized methods and quality control. Environ. Sci. Technol. 56, 9149–9160. doi: 10.1021/acs.est.1c08918
Livermore, D. M. (2008). Defining an extended-spectrum beta-lactamase. Clin. Microbiol. Infect. 14, 3–10. doi: 10.1111/j.1469-0691.2007.01857.x
Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Matthiessen, L. E., Hald, T., and Vigre, H. (2022). System mapping of antimicrobial resistance to combat a rising Global Health crisis. Front. Public Health 10:816943. doi: 10.3389/fpubh.2022.816943
Minhas, P. S., Saha, J. K., Dotaniya, M. L., Sarkar, A., and Saha, M. (2022). Wastewater irrigation in India: current status, impacts and response options. Sci. Total Environ. 808:152001. doi: 10.1016/j.scitotenv.2021.152001
Montero, L., Irazabal, J., Cardenas, P., Graham, J. P., and Trueba, G. (2021). Extended-spectrum beta-lactamase producing-Escherichia coli isolated from irrigation waters and produce in Ecuador. Front. Microbiol. 12:709418. doi: 10.3389/fmicb.2021.709418
Muntean, M. M., Muntean, A. A., Preda, M., Manolescu, L. S. C., Dragomirescu, C., Popa, M. I., et al. (2022). Phenotypic and genotypic detection methods for antimicrobial resistance in ESKAPE pathogens (review). Exp. Ther. Med. 24:508. doi: 10.3892/etm.2022.11435
Mutuku, C., Gazdag, Z., and Melegh, S. (2022). Occurrence of antibiotics and bacterial resistance genes in wastewater: resistance mechanisms and antimicrobial resistance control approaches. World J. Microbiol. Biotechnol. 38:152. doi: 10.1007/s11274-022-03334-0
Noster, J., Thelen, P., and Hamprecht, A. (2021). Detection of multidrug-resistant Enterobacterales-from ESBLs to Carbapenemases. Antibiotics (Basel). 10:1140. doi: 10.3390/antibiotics10091140
Palkova, L., Tomova, A., Repiska, G., Babinska, K., Bokor, B., Mikula, I., et al. (2021). Evaluation of 16S rRNA primer sets for characterisation of microbiota in paediatric patients with autism spectrum disorder. Sci. Rep. 11:6781. doi: 10.1038/s41598-021-86378-w
Partridge, S. R., Kwong, S. M., Firth, N., and Jensen, S. O. (2018). Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 31, e00088–e00017. doi: 10.1128/CMR.00088-17
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Sanz-García, F., Gil-Gil, T., Laborda, P., Blanco, P., Ochoa-Sánchez, L. E., Baquero, F., et al. (2023). Translating eco-evolutionary biology into therapy to tackle antibiotic resistance. Nat. Rev. Microbiol. doi: 10.1038/s41579-023-00902-5 [Epub head of print].
Sawa, T., Kooguchi, K., and Moriyama, K. (2020). Molecular diversity of extended-spectrum β-lactamases and carbapenemases, and antimicrobial resistance. J. Intensive Care 8:13. doi: 10.1186/s40560-020-0429-6
Schmiege, D., Zacharias, N., Sib, E., Falkenberg, T., Moebus, S., Evers, M., et al. (2021). Prevalence of multidrug-resistant and extended-spectrum beta-lactamase-producing Escherichia coli in urban community wastewater. Sci. Total Environ. 785:147269. doi: 10.1016/j.scitotenv.2021.147269
Shin, H., Kim, Y., Han, S., and Hur, H. G. (2023). Resistome study in aquatic environments. J. Microbiol. Biotechnol. 33, 277–287. doi: 10.4014/jmb.2210.10044
Singh, A., Keshri, A. K., Rawat, S. S., Swami, D., Uday, K. V., and Prasad, A. (2021). Identification and characterization of colistin-resistant E. coli and K. pneumoniae isolated from lower Himalayan region of India. SN Appl. Sci. 3, 1–9. doi: 10.1007/s42452-021-04596-3
Sivaraman, G. K., Rajan, V., Vijayan, A., Elangovan, R., Prendiville, A., and Bachmann, T. T. (2021). Antibiotic resistance profiles and molecular characteristics of extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli and Klebsiella pneumoniae isolated from shrimp aquaculture farms in Kerala. India. Front. Microbiol. 12:622891. doi: 10.3389/fmicb.2021.622891
Soliman, A. M., Nariya, H., Tanaka, D., Shimamoto, T., and Shimamoto, T. (2023). A novel single-tube eicosaplex/octaplex PCR system for the detection of extended-spectrum β-lactamases, plasmid-mediated AmpC β-lactamases, and integrons in gram-negative bacteria. Antibiotics (Basel). 12:90. doi: 10.3390/antibiotics12010090
Subramanya, S. H., Bairy, I., Metok, Y., Baral, B. P., Gautam, D., and Nayak, N. (2021). Detection and characterization of ESBL-producing Enterobacteriaceae from the gut of subsistence farmers, their livestock, and the surrounding environment in rural Nepal. Sci. Rep. 11:2091. doi: 10.1038/s41598-021-81315-3
Tamura, K., Dudley, J., Nei, M., and Kumar, S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599. doi: 10.1093/molbev/msm092
Tan, H. S., Yan, P., Agustie, H. A., Loh, H. S., Rayamajhi, N., and Fang, C. M. (2023). Characterisation of ESBL/AmpC-producing Enterobacteriaceae isolated from poultry farms in peninsular Malaysia. Lett. Appl. Microbiol. 76:ovac044. doi: 10.1093/lambio/ovac044
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Tiwari, A., Paakkanen, J., Al-Mustapha, A. I., Kirveskari, J., Hendriksen, R. S., and Heikinheimo, A. (2023). Phenotypic and genotypic characteristics of beta-lactamase dominant with CARBA, AmpC, and ESBL-producing bacteria in municipal wastewater influent in Helsinki. Finland. J. Glob. Antimicrob. Resist. 33, 345–352. doi: 10.1016/j.jgar.2023.04.015
Tucker, K., Stone, W., Botes, M., Feil, E. J., and Wolfaardt, G. M. (2022). Wastewater treatment works: a last line of defense for preventing antibiotic resistance entry into the environment. Front. Water 4:883282. doi: 10.3389/frwa.2022.883282
Uluseker, C., Kaster, K. M., Thorsen, K., Basiry, D., Shobana, S., Jain, M., et al. (2021). A review on occurrence and spread of antibiotic resistance in wastewaters and in wastewater treatment plants: mechanisms and perspectives. Front. Microbiol. 12:717809. doi: 10.3389/fmicb.2021.717809
Veloo, Y., Thahir, S. S. A., Rajendiran, S., Hock, L. K., Ahmad, N., Muthu, V., et al. (2022). Multidrug-resistant gram-negative bacteria and extended-spectrum β-lactamase-producing Klebsiella pneumoniae from the poultry farm environment. Microbiol. Spectr. 10:e0269421. doi: 10.1128/spectrum.02694-21
von Wintersdorff, C. J. H., Penders, J., van Niekerk, J. M., Mills, N. D., Majumder, S., van Alphen, L. B., et al. (2016). Dissemination of antimicrobial resistance in microbial ecosystems through horizontal gene transfer. Front. Microbiol. 7:173. doi: 10.3389/fmicb.2016.00173
Walesch, S., Birkelbach, J., Jézéquel, G., Haeckl, F. P. J., Hegemann, J. D., Hesterkamp, T., et al. (2023). Fighting antibiotic resistance-strategies and (pre)clinical developments to find new antibacterials. EMBO Rep. 24:e56033. doi: 10.15252/embr.202256033
Wang, J., Chu, L., Wojnárovits, L., and Takács, E. (2020). Occurrence and fate of antibiotics, antibiotic resistant genes (ARGs) and antibiotic resistant bacteria (ARB) in municipal wastewater treatment plant: an overview. Sci. Total Environ. 744:140997. doi: 10.1016/j.scitotenv.2020.140997
World Health Organization (2023). Antimicrobial resistance. Available at: https://www.who.int/health-topics/antimicrobial-resistance (Accessed 6th April, 2023)
Zaatout, N., Bouras, S., and Slimani, N. (2021). Prevalence of extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae in wastewater: a systematic review and meta-analysis. J. Water Health 19, 705–723. doi: 10.2166/wh.2021.112
Keywords: wastewater, antibiotics, pollution, antibiotic-resistant bacteria, Enterobacterales, extended-spectrum beta-lactamases, agro-ecosystems
Citation: Avatsingh AU, Sharma S, Kour S, Arora Y, Sharma S, Joshi D, Chaudhary PP, Perveen K, Kamal MA and Singh N (2023) Prevalence of antibiotic-resistant Gram-negative bacteria having extended-spectrum β-lactamase phenotypes in polluted irrigation-purpose wastewaters from Indian agro-ecosystems. Front. Microbiol. 14:1227132. doi: 10.3389/fmicb.2023.1227132
Received: 22 May 2023; Accepted: 25 July 2023;
Published: 07 August 2023.
Edited by:
Deep Chandra Suyal, Management and Technology, IndiaReviewed by:
Arun Karnwal, Lovely Professional University, IndiaCopyright © 2023 Avatsingh, Sharma, Kour, Arora, Sharma, Joshi, Chaudhary, Perveen, Kamal and Singh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nasib Singh, ZHJuYXNpYm1pY3JvQGV0ZXJuYWx1bml2ZXJzaXR5LmVkdS5pbg==
†ORCID: Nasib Singh https://orcid.org/0000-0003-3890-2766
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.