- 1The Second Affiliated Hospital and Yuying Children’s Hospital, Wenzhou Medical University, Wenzhou, China
- 2Key Laboratory of Medical Genetics of Zhejiang Province, Key Laboratory of Laboratory Medicine, Ministry of Education, China, School of Laboratory Medicine and Life Sciences, Wenzhou Medical University, Wenzhou, China
- 3Medical Molecular Biology Laboratory, School of Medicine, Jinhua Polytechnic, Jinhua, China
Background: Aminoglycosides, as important clinical antimicrobials, are used as second-line drugs for treating multidrug-resistant tuberculosis or combined with β-lactam drugs for treating severe infections such as sepsis. Aminoglycoside-modifying enzyme (AME) is the most important mechanism of aminoglycoside resistance and deserves more attention.
Methods: The bacterium Kluyvera intermedia DW18 was isolated from the sewage of an animal farm using the conventional method. The agar dilution method was used to determine the minimum inhibitory concentrations (MICs) of antimicrobials. A novel resistance gene was cloned, and the enzyme was expressed. The kinetic parameters were measured by a SpectraMax M5 multifunctional microplate reader. Bioinformatic analysis was performed to reveal the genetic context of the aph(3′)-Id gene and its phylogenetic relationship with other AMEs.
Results: A novel aminoglycoside 3′-O-phosphotransferase gene designated aph(3′)-Id was identified in K. intermedia DW18 and shared the highest amino acid identity of 77.49% with the functionally characterized aminoglycoside 3′-O-phosphotransferase APH(3′)-Ia. The recombinant plasmid carrying the novel resistance gene (pMD19-aph(3′)-Id/E. coli DH5α) showed 1,024-, 512-, 128- and 16-fold increased MIC levels for kanamycin, ribostamycin, paromomycin and neomycin, respectively, compared with the reference strain DH5α. APH(3′)-Id showed the highest catalytic efficiency for ribostamycin [kcat/Km of (4.96 ± 1.63) × 105 M−1/s−1], followed by paromomycin [kcat/Km of (2.18 ± 0.21) × 105 M−1/s−1], neomycin [kcat/Km of (1.73 ± 0.20) × 105 M−1/s−1], and kanamycin [kcat/Km of (1.10 ± 0.18) × 105 M−1/s−1]. Three conserved functional domains of the aminoglycoside phosphotransferase family and ten amino acid residues responsible for the phosphorylation of kanamycin were found in the amino acid sequence of APH(3′)-Id. No mobile genetic element (MGE) was discovered surrounding the aph(3′)-Id gene.
Conclusion: In this work, a novel aminoglycoside 3’-O-phosphotransferase gene designated aph(3′)-Id encoded in the chromosome of the environmental isolate Kluyvera intermedia DW18 was identified and characterized. These findings will help clinicians select effective antimicrobials to treat infections caused by pathogens with this kind of resistance gene.
Introduction
The genus Kluyvera of the family Enterobacteriaceae was defined in 1981 (Farmer et al., 1981), and at present, it consists of 5 species, including Kluyvera georgiana, Kluyvera ascorbata, Kluyvera cryocrescens, Kluyvera intermedia and Kluyvera sichuanensis. As a rare pathogenic agent, Kluyvera intermedia has the potential to cause a variety of infections in humans, such as abdominal abscesses, septic shock, and soft tissue, urinary and bloodstream infections (Carter and Evans, 2005). In 2005, Enterobacter intermedius was included in Kluyvera cochleae of the genus Kluyvera and was subsequently officially named Kluyvera intermedia after biochemical, DNA hybridization and phylogenetic relationship analyses (Pavan et al., 2005). Besides isolation from human samples, which has certain clinical significance (Prats et al., 1987), strains of K. intermedia have mainly been isolated from surface water and unpolluted soil (Grimont and Grimont, 2006). K. intermedia has been reported to show resistance to some β-lactams (penicillin G, oxacillin, cefoxitin, cefepime and ceftriaxone) and carbapenems (imipenem and meropenem) and was naturally susceptible to most aminoglycosides, such as gentamicin, tobramycin, ribostamycin, spectinomycin and neomycin (Stock, 2002; Ribeiro et al., 2014; Paskova et al., 2018).
It has been recognized that the environment can act as an important source and transmission route of resistance genes (Martínez, 2008; Bengtsson-Palme et al., 2014). Long before antibiotics were used in the clinic, resistance genes were present in bacteria in the natural environment, as evidenced by the discovery of the vancomycin resistance element VanA in 30,000-year-old permafrost (D’Costa et al., 2011). This suggests that resistance genes exist and spread even without modern clinical antibiotic selection pressure. Globally, water and soil can act as very large reservoirs of resistance genes (Suzuki and Hoa, 2012; Schulz et al., 2017), and these resistance genes can be captured by mobile genetic elements (MGEs), such as integrons, transposons, or chromosomal islands, and introduced into clinical pathogens through horizontal transfer (Forsberg et al., 2012; Djordjevic et al., 2013).
Aminoglycosides are highly effective broad-spectrum antibiotics that are active against a variety of gram-positive and gram-negative organisms (Krause et al., 2016). They remain an important part of clinical antimicrobial treatment and can be combined with β-lactam drugs for treating severe sepsis (Dellinger et al., 2013) and second-line drugs for treating multidrug-resistant tuberculosis (Caminero and Scardigli, 2015). The mechanisms of aminoglycoside resistance include aminoglycoside-modifying enzyme (AME) (Ramirez and Tolmasky, 2010), 16S rRNA methylation (Doi and Arakawa, 2007) and efflux pump (Magnet et al., 2001), the most common among which is AME. In susceptible strains, aminoglycosides exert antibacterial effects in two steps: transmembrane entry into cells and interaction with ribosomes leading to protein synthesis errors. The situation differs in strains producing aminoglycoside-modifying enzymes (AMEs); aminoglycosides entering the cell are inactivated by the AMEs so that they cannot interact with ribosomes (Azucena and Mobashery, 2001). AMEs can inactivate aminoglycosides by making different types of modifications to them and are generally divided into three groups, including aminoglycoside acetyltransferases (AACs), aminoglycoside phosphotransferases (APHs) and aminoglycoside nucleotidyltransferases (ANTs) (Azucena and Mobashery, 2001). The designations are composed of the enzyme type and enzyme modification site, with Roman numerals representing unique resistance and lowercase letters representing unique protein names (Shaw et al., 1993).
APHs can catalyze the transfer of a phosphate group from ATP or GTP to the hydroxyl group of aminoglycoside molecules, thus preventing the drug from binding to the aminoacyl-tRNA site (A site) in the prokaryotic 30S ribosomal subunit (Tsai et al., 2013; Costa et al., 2020). It was found that APHs shared similar active sites with eukaryotic protein kinases (ePKs) and common ePK-like folds in the tertiary structure, and the inhibitors of ePKs also showed inhibitory activity against APHs, which suggested that they might have been derived from a common ancestor (Hon et al., 1997; Stogios et al., 2013). Some APHs were also found to have NTPs hydrolase activity and can hydrolyze NTPs in the absence of an antibiotic substrate, but the efficiency of NTP hydrolysis was far less than the catalytic efficiency of the antibiotic substrate, so it had little impact on the modification of antibiotics (Kim C. et al., 2006; Smith et al., 2019). The APH classes and subclasses include APH(4)-I, APH(6)-I, APH(9)-I, APH(3′)-I through IX, APH(3′)-XV, APH(2″)-I through IV, APH(3″)-I and APH(7″)-I (Alcock et al., 2023). The APH(3′)-I subclass, showing resistance to lividomycin, ribostamycin, kanamycin, paromomycin, and neomycin, is composed of three enzymes, and the resistance genes encoding them are generally related to MGEs and are encoded in wide-host-range plasmids (Vakulenko and Mobashery, 2003; Ramirez and Tolmasky, 2010).
In this study, a novel chromosome-encoded aminoglycoside 3′-O-phosphotransferase gene designated aph(3′)-Id was identified, and its molecular characteristics were characterized through whole-genome sequencing and bioinformatic and kinetic analyses.
Materials and methods
Bacteria and whole-genome sequencing
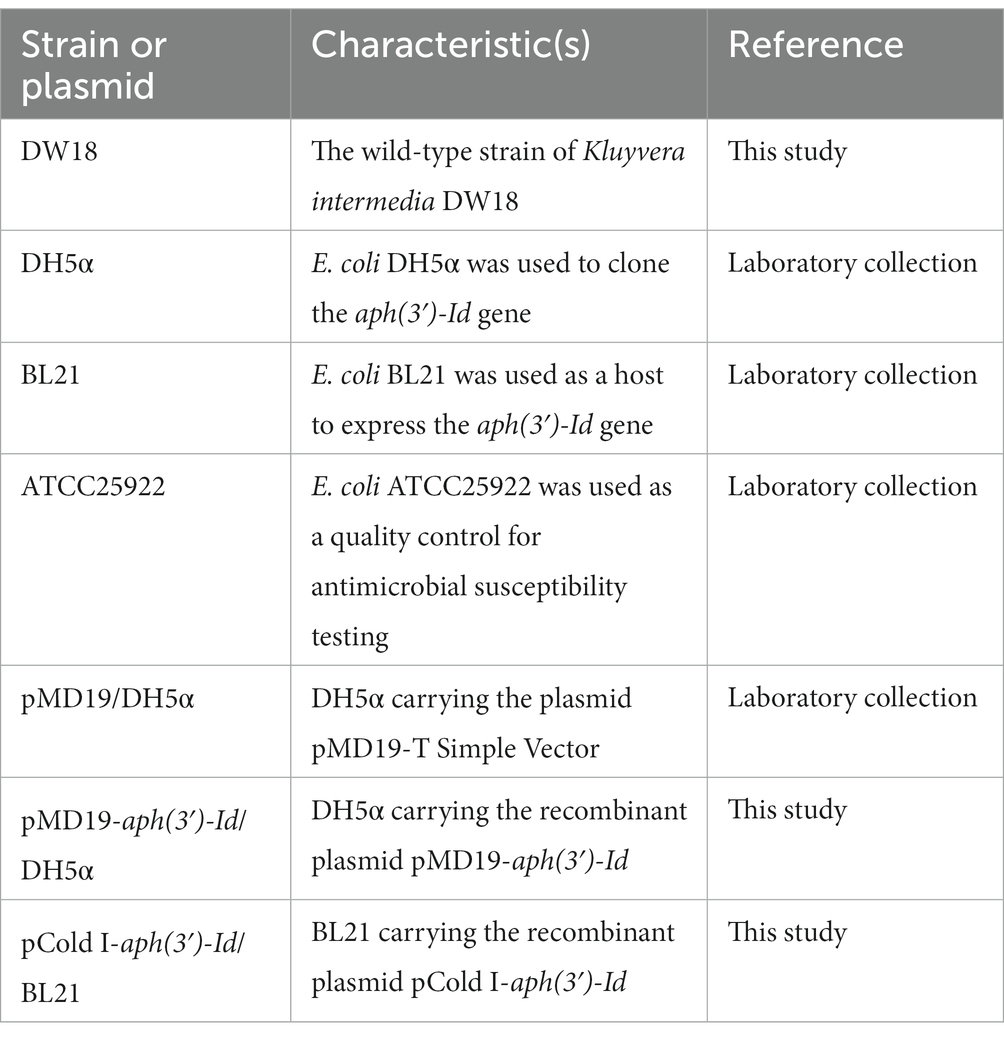
The bacterium DW18 was isolated from the sewage of an animal farm in Wenzhou, China. The collection and isolation methods for DW18 were as follows: a sterile cotton swab was dipped into sewage in the farm sewer line and then immediately placed into a sterile screw-cap specimen collection tube with saline solution. The isolate was streaked on Luria–Bertani agar plates to obtain single colonies, some of which were randomly selected and then preserved in LB broth with 25% glycerol at −80°C. DNA sequencing of DW18 was performed on the Illumina NovaSeq and PacBio RS II platforms (Shanghai Personal Biotechnology Co., Ltd., Shanghai, China). The Illumina short reads were assembled by SKESA v2.4.0 (Souvorov et al., 2018). Unicycler v0.4.8 was used for hybrid assembly of the PacBio long reads and confirming the cyclization of the whole-genome assembly (Wick et al., 2017). Pilon improved the quality of genomic assembly sketches by mapping Illumina short reads onto the assembly to correct possible incorrect assembly (Walker et al., 2014). Species identification was carried out first by 16S rRNA gene homology and then whole-genome average nucleotide identity (ANI) analyses (Konstantinidis and Tiedje, 2005; Richter and Rosselló-Móra, 2009). The calculation of ANI was performed by FastANI (Jain et al., 2018). The bacteria and plasmids used in this study are shown in Table 1.
Antimicrobial susceptibility testing
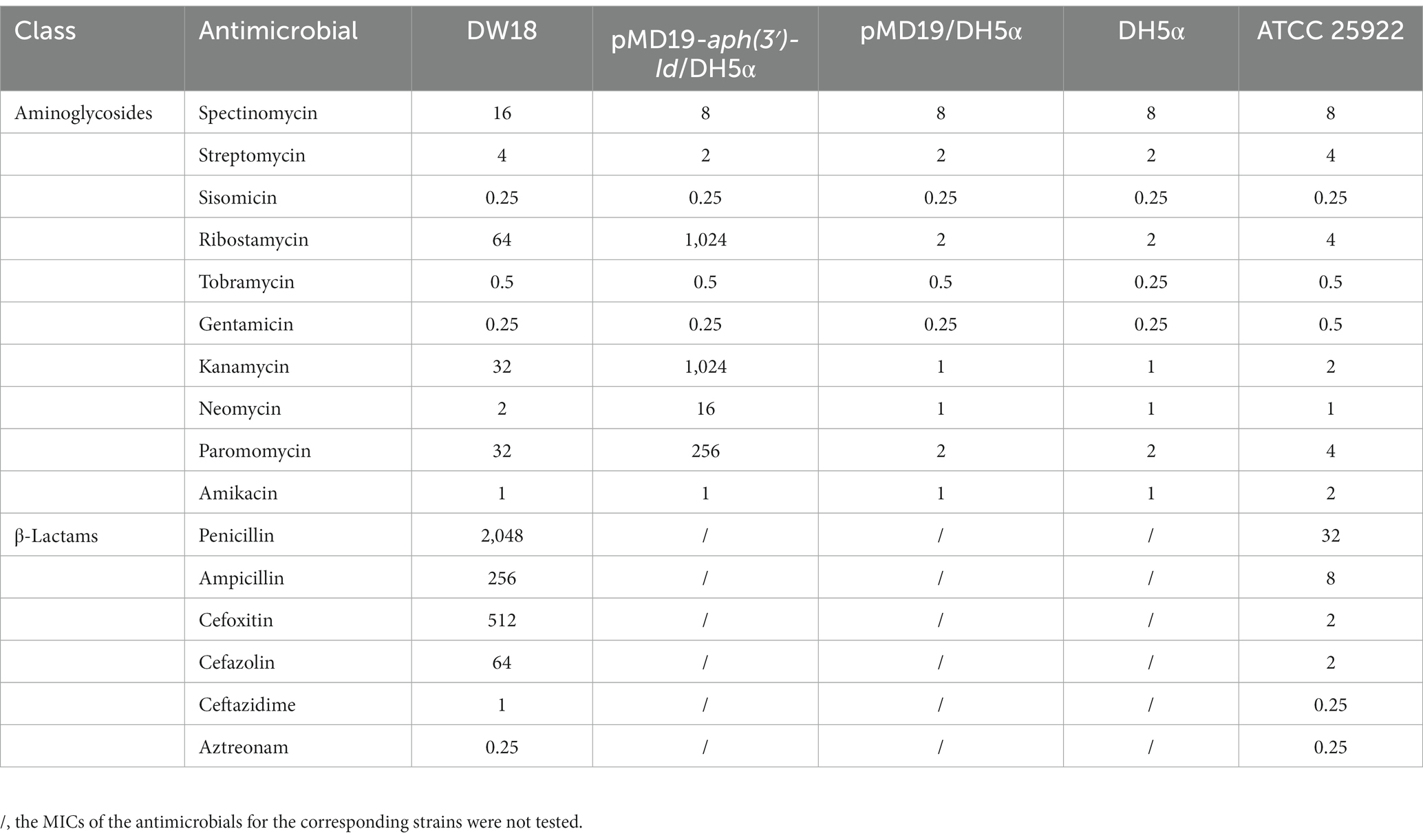
The agar dilution method was used to determine the minimum inhibitory concentrations (MICs) of the antimicrobials. Susceptibility patterns were interpreted on the basis of the Clinical and Laboratory Standards Institute guidelines M100 (32nd Edition, 2022) for Enterobacteriaceae. The reference strain used for quality control was E. coli ATCC 25922. All 16 antimicrobials used in this work are shown in Table 2.
Cloning of the aph(3′)-Id gene
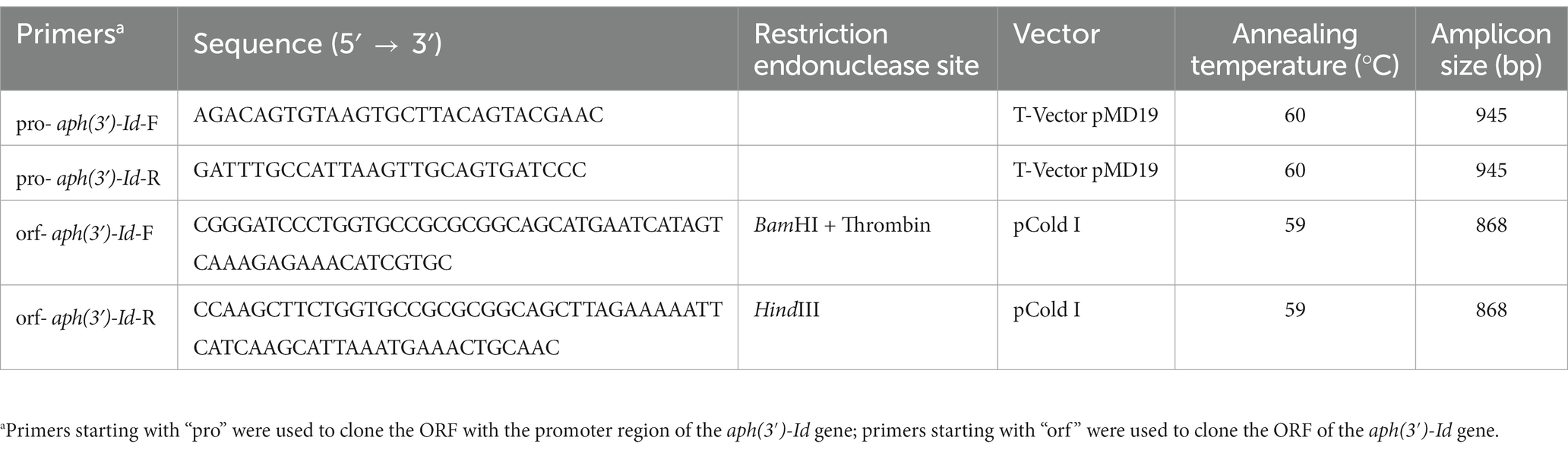
The promoter region and coding sequence of the novel resistance gene aph(3′)-Id were amplified by polymerase chain reaction (PCR). The primers and annealing temperatures used for PCR are listed in Table 3. The PCR products were inserted into T-Vector pMD19 (Simple) (Takara Bio, Inc., Dalian, China) using a T4 DNA ligase cloning kit (Takara Bio, Inc., Dalian, China). The recombinant plasmid was transformed into competent cells of E. coli DH5α by the calcium chloride method, and then LB broth was added, and the cells were resuscitated by shaking at 37°C. The transformants were screened on LB solid culture medium with 100 μg/mL ampicillin. For the expression of the APH(3′)-Id protein, the ORF of aph(3′)-Id was PCR-amplified using forward and reverse primers with BamHI and HindIII restriction sites, respectively, at the 5′-ends. The PCR product was digested with the restriction endonucleases HindIII and BamHI and then cloned into the pCold I vector treated with the same restriction endonucleases. The recombinant plasmid (pCold I-aph(3′)-Id) was transformed into competent E. coli BL21 cells by the calcium chloride method. The transformants were screened on LB agar plates with 100 μg/mL ampicillin. The cloned insert sequences of the recombinant plasmids were confirmed by PCR and Sanger sequencing.
Expression and purification of the APH(3′)-Id enzyme
The recombinant strain (pCold I-aph(3′)-Id/BL21) for the expression of APH(3′)-Id was cultured in 3 mL of LB broth with 100 μg/mL ampicillin at 37°C for 16 h. Then, 1 mL of the bacterial culture was added to 100 mL of LB broth for continuous culture until the OD600 value reached 0.6–0.8 (Qing et al., 2004). After refrigeration for half an hour, a final concentration of 1 mM IPTG was added to induce the expression of APH(3′)-Id. Cultivation was continued for 16 h at 18°C, and the culture was centrifuged to collect cells. The cells were resuspended in 4 mL of nondenatured lysate (50 mM NaH2PO4, 300 mM NaCl) and disrupted by ultrasonication. The recombinant protein in the supernatant collected after centrifugation at 8,000 × g for 30 min was purified using His-tag Purification Resin and eluted by the nondenatured eluent (300 mM NaCl, 50 mM imidazole, 50 mM NaH2PO4) according to the instructions of the His-tag Protein Purification Kit (Reductant & Chelator-resistant) (Beyotime, Shanghai, China). The His tag of the recombinant protein was removed by thrombin with incubation at 20°C for 16 h. The purity of APH(3′)-Id was confirmed by SDS–PAGE, and the concentration was spectrophotometrically examined at 562 nm by the standard curve established using a BCA protein assay kit (Beyotime, Shanghai, China). The protein was stored in dissolution buffer (0.5 M NaCl, 20 mM Tris–HCl; pH 8.0) and preserved at −80°C for subsequent enzyme kinetics analysis.
Enzyme kinetics determination
The kinetic parameters of APH(3′)-Id were determined as described in previous reports (Toth et al., 2013; Lu et al., 2021) with slight modifications. The phosphorylation of aminoglycosides was coupled to NADH oxidation by pyruvate kinase (PK) and lactate dehydrogenase (LDH). The reduction of NADH at 340 nm was monitored by a SpectraMax M5 multifunctional microplate reader (Molecular Devices, United States) to reflect the speed of aminoglycoside phosphorylation. The reaction mixture contained 1 mM phosphoenolpyruvate, 1 mM MgCl2, 100 mM HEPES (pH 7.5), 600 μM NADH, 2 mM KCl, a commercial mixture of PK and LDH (Sigma P0294; 18–26 U/mL PK and 25–35 U/mL LDH in final concentration), 1 mM ATP, 30–40 nM APH(3′)-Id and aminoglycosides (5–400 μM) in a total volume of 250 μL. The steady-state velocities were calculated from the linear period of the reaction process curve and applied to plot the substrate concentration-velocity curve. The data were fit by nonlinear regression with the velocity as a function of substrate using the Michaelis–Menten equation. Curve fitting and calculation of the dynamic parameters Km and kcat were completed in GraphPad Prism 8.0.2 (GraphPad Software, Inc.).
Bioinformatics analysis
Prokka v1.14.6 (Seemann, 2014) was used to predict the potential open reading frames (ORFs) of the assembled genome sequence, and the deduced proteins were annotated using DIAMOND v2.0.11 (Buchfink et al., 2021) according to the Comprehensive Antibiotic Resistance Database (CARD) (McArthur et al., 2013) to identify antimicrobial resistance genes. The annotation parameters were set as follows: E-value <1E-9, identity >95% and coverage >85%. Multiple sequence alignment and visualization were conducted using MAFFT v7.490 (Katoh and Standley, 2013) and GeneDoc v2.7 (Nicholas, 1997). Construction of phylogenetic trees and their visualization were conducted using IQ-TREE v1.6.9 (Nguyen et al., 2015) and iTOL v6.7 (Letunic and Bork, 2021). The crystal structure of the aminoglycoside phosphotransferase APH(3′)-Ia (SMTL ID: 3r78.1), which shared the highest amino acid identity of 77.49% with APH(3′)-Id, was used as a template for homology modeling of APH(3′)-Id in SWISS-MODEL (Waterhouse et al., 2018) to predict the tertiary structure. The tertiary structure alignment of the APH(3′)-Id model and kanamycin-bound APH(3′)-Ia (PDB 4FEU chain B) was performed by PyMOL (Schrödinger, 2023). A map of the genetic environment surrounding aph(3′)-Id was generated by clinker v0.0.24 (Gilchrist and Chooi, 2021). The Expasy ProtParam Tool (Gasteiger et al., 2003) was used for prediction of the molecular weight and pI value of the protein APH(3′)-Id.
Nucleotide sequence accession numbers
The following accession numbers have been assigned for the complete genome sequence of Kluyvera intermedia DW18 in GenBank: CP123488 for chromosome, CP123489 for plasmid pDW18-1 and CP123490 for plasmid pDW18-2. The nucleotide sequence of the novel AME gene aph(3′)-Id has been assigned GenBank accession number OQ819314.
Results and discussion
Identification and characteristics of Kluyvera intermedia DW18
The 16S rRNA gene of DW18 shared the highest similarity of 100% (100% identity and 100% coverage) with that of K. intermedia MAG 4195 (GCA_029202345.1). The genome sequence of DW18 shared 98.95% ANI with that of K. intermedia N2-1 (GCA_009649915.1), which exceeded the threshold of 95% to define a bacterial species. Currently, the ANI between genomes is considered the best alternative to DDH and 16S rRNA gene homology analyses as the gold standard for prokaryotic classification, and the ANI threshold for species membership was empirically defined as 95% (Konstantinidis and Tiedje, 2005; Richter and Rosselló-Móra, 2009). Therefore, the isolate DW18 in this study was classified into the species K. intermedia and designated K. intermedia DW18.
General features of the Kluyvera intermedia DW18 genome
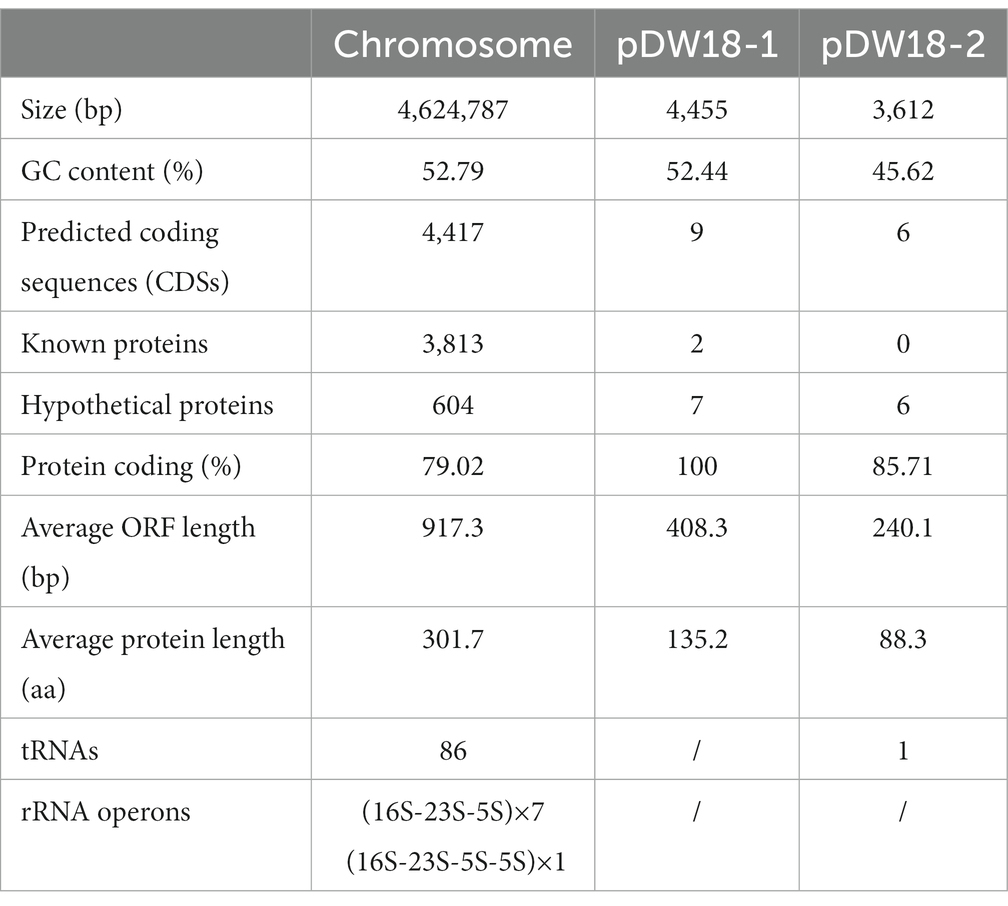
The complete genome of K. intermedia DW18 was assembled, and it was found to consist of a chromosome and two circular plasmids. The chromosome is approximately 4.62 Mb in length and encodes 4,417 ORFs with an average GC content of 52.79%. The two circular plasmids were designated pDW18-1 and pDW18-2 (Table 4). pDW18-1 is 4,455 bp in length and encodes 9 ORFs, while pDW18-2 is 3,612 bp in length and encodes 6 ORFs. In the NCBI database, a total of 23 K. intermedia genomes were present, and only four (K. intermedia N2-1, GCA_009649915.1, K. intermedia HR2, GCA_009650415.1, K. intermedia NCTC12125, GCA_900635475.1, and K. intermedia MAG 4159, GCA_029202345.1) of them had complete genome sequences. Among the four strains with complete genome sequences, two (K. intermedia N2-1 and K. intermedia HR2) carried a large plasmid each (pN2-1, 245,785 bp in length, and pHR2-1, 245,785 bp in length). The other strains, NCTC12125 and MAG 4159, did not carry any plasmid. DW18, however, carried two small plasmids, pDW18-1 (4,455 bp) and pDW18-2 (3,612 bp). The chromosome size of the five K. intermedia strains with complete genomes (including DW18 in this work) varied slightly, ranging from 4.62 Mb (DW18) to 4.74 Mb (NCTC12125).
Resistance profiles of Kluyvera intermedia DW18
Of the 16 antimicrobials from two classes (aminoglycosides and β-lactams) tested, DW18 showed higher MIC levels for eight, including four aminoglycosides (ribostamycin 64 μg/mL, kanamycin 32 μg/mL, paromomycin 32 μg/mL, and spectinomycin 16 μg/mL) and four β-lactams (penicillin 2048 μg/mL, cefoxitin 512 μg/mL, ampicillin 256 μg/mL, and cefazolin 64 μg/mL) (Table 2). On the one hand, for β-lactams, the resistance profile of K. intermedia DW18 was generally consistent with previous reports, but on the other hand, for aminoglycosides, it showed different resistance phenotypes. Previous studies showed that K. intermedia was resistant to a variety of β-lactam antibiotics, such as penicillin, amoxycillin, and cefoxitin, but was susceptible to aminoglycosides (Stock, 2002). Some isolates also showed resistance to carbapenems and other cephalosporins such as imipenem, meropenem, cefepime, and ceftriaxone (Ribeiro et al., 2014; Paskova et al., 2018).
In our present project studying the resistance status of animal bacteria, we sequenced the draft genomes of several strains isolated from animal specimens. When analyzing the resistance genotype of the DW18 genome based on the CARD with thresholds of identity >95% and coverage >85%, we found that no functionally characterized aminoglycoside and β-lactam resistance gene was annotated in the genome even though it showed high MIC levels for some aminoglycosides and β-lactams.
To determine the potential resistance mechanism of DW18 against aminoglycosides, we examined the annotation data of the genome sequence and found that among the predicted genes, the gene showing the highest identity with the functionally characterized aminoglycoside resistance gene was an aph(3′)-Ia-like gene (finally designated aph(3′)-Id in this work), which showed amino acid similarity (100% coverage and 77.49% identity) with APH(3′)-Ia (CAE51638.1). To determine whether the predicted resistance gene was indeed functional, we cloned the ORF of the gene with its promoter region into the T-vector pMD19, and the resistance function of this gene was confirmed. Compared with the control strains (DH5α or DH5α carrying pMD19), the recombinant strain (pMD19-aph(3′)-Id/DH5α) showed 1,024-, 512-, 128-, and 16-fold increased MICs for kanamycin, ribostamycin, paromomycin and neomycin, respectively. No significant increases in the MICs for the other 6 aminoglycosides tested were observed (Table 2). There are only three other aph(3′)-I genes [aph(3′)-Ia, aph(3′)-Ib and aphA15] present in CARD, and only aph(3′)-Ia and aphA15 had the resistance profile documented (Siregar et al., 1995; Riccio et al., 2001). aph(3′)-Id, aph(3′)-Ia and aphA15 all showed higher MIC values for kanamycin and neomycin. The aph(3′)-Id gene in this work did not confer resistance to gentamicin or amikacin; however, the aph(3′)-Ia gene showed high MICs for gentamicin (>1,024 μg/mL) and amikacin (8 μg/mL). According to the evolutionary relationship, substrate spectrum and nomenclature proposed for genes encoding AMEs (Shaw et al., 1993), the novel resistance gene in this work was thus designated aph(3′)-Id.
Homologs of the novel aminoglycoside 3’-O-phosphotransferase APH(3′)-Id
When analyzing the distribution of aph(3′)-Id and aph(3′)-Id-like genes, only 4 genes with amino acid identities >80% were found according to the BLAST search against the NCBI nonredundant protein database. These genes were derived from K. intermedia (WEJ84261.1, WP_153742217.1, WP_062773153.1) and Klebsiella sp. (WP_181482651.1). The deduced amino acid sequence of APH(3′)-Id shared the highest identity of 99.26% with a hypothetical APH(3′)-I family aminoglycoside O-phosphotransferase (WEJ84261.1) from K. intermedia. The other two proteins from K. intermedia also shared high identities of 98.89% (WP_153742217.1) and 97.05% (WP_062773153.1) with APH(3′)-Id. However, the protein from Klebsiella sp. shared the lowest identity of 80.07% (WP_181482651.1) with APH(3′)-Id. This indicated that APH(3′)-Id was relatively conserved within the species K. intermedia.
The novel aminoglycoside 3′-O-phosphotransferase gene aph(3′)-Id was 816 bp in length and encoded a protein of 271 amino acids (30.69 kDa) with a pI value of 4.90. APH(3′)-Id shared the highest amino acid identities of 77.49, 58.67 and 39.85% with the functionally characterized APH(3′) proteins APH(3′)-Ia (CAE51638.1), APH(3′)-Ib (AAA26412.1) and APH(3′)-IIc (ADQ43421.1), respectively. Furthermore, a phylogenetic tree that included all 19 functionally characterized APH(3′) enzymes collected from the CARD database showed that APH(3′)-Id clustered closest to APH(3′)-I enzymes (Figure 1). Currently, the aph(3′)-Id genes were mainly found in the chromosomes of Kluyvera intermedia. The aph(3′)-Ib and aph(3′)-IIc genes were only found in Pseudomonas aeruginosa and Stenotrophomonas maltophilia, respectively. The aph(3′)-Ia gene, however, was initially identified in the translocator element Tn903 of Escherichia coli (Oka et al., 1981), but now is widely distributed in a variety of bacterial genera, such as Acinetobacter, Aeromonas, Avibacterium, Brucella, and Enterobacter. This suggested that aph(3′)-Id may not spread as rapidly and widely across multiple strains as aph(3′)-Ia.
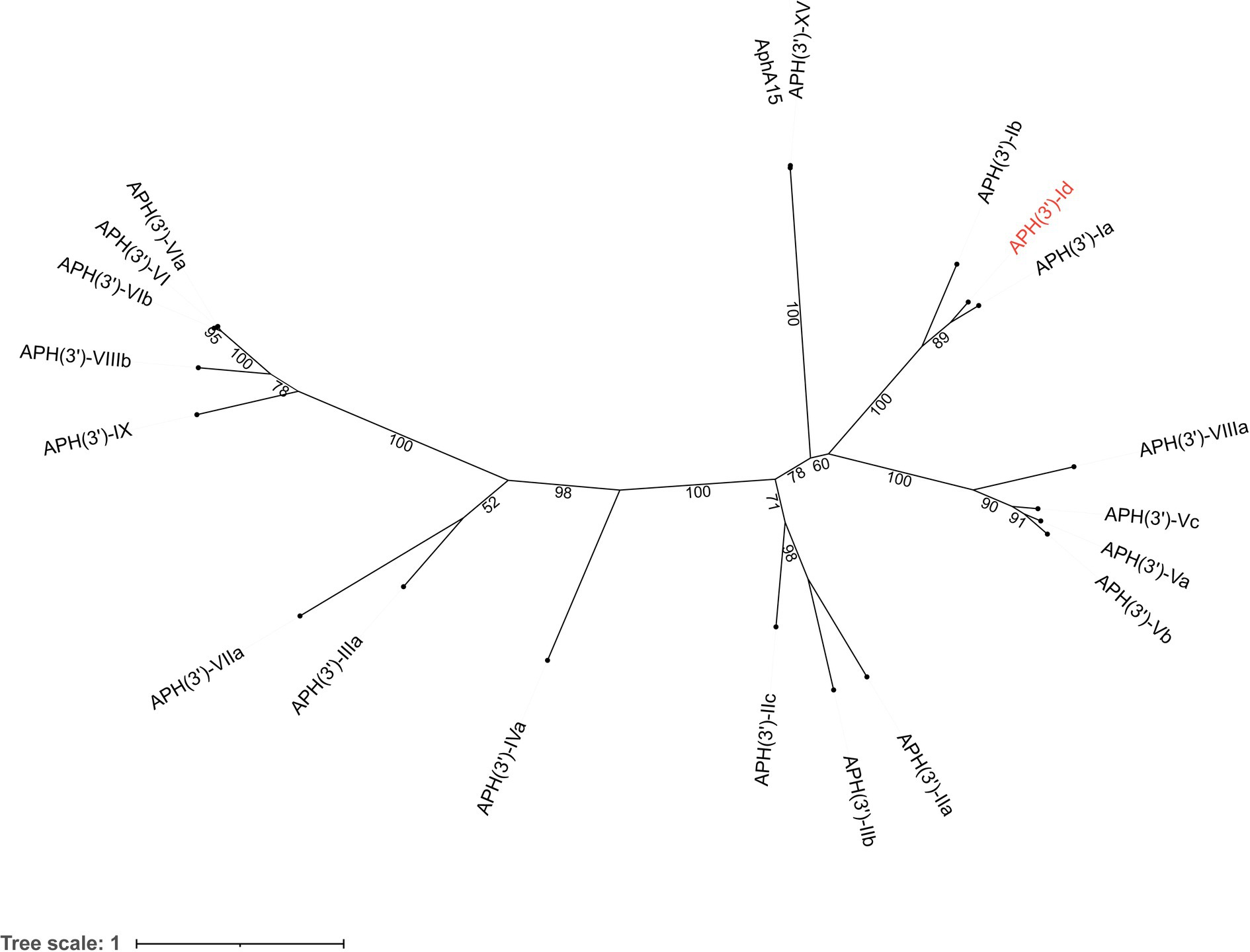
Figure 1. A phylogenetic tree showing the relationship of APH(3′)-Id with other APH(3′) enzymes. APH(3′)-Id is colored in red. The numbers on the branches are bootstrapping values. The proteins and their accession numbers: APH(3′)-Ia (CAE51638.1), APH(3′)-Ib (AAA26412.1), APH(3′)-XV (ABY48974.1), AphA15 (CAD91341.1), APH(3′)-VIIIa (AAG11411.2), APH(3′)-Vc (AAB21326.1), APH(3′)-Va (AAA26699.1), APH(3′)-Vb (AAC32025.1), APH(3′)-IIa (CAA23892.1), APH(3′)-IIb (CAA62365.1′), APH(3′)-IIc (ADQ43421.1), APH(3′)-IVa (CAA27061.1), APH(3′)-IIIa (AGV10830.1), APH(3′)-VIIa (AAA76822.1), APH(3′)-IX (ENV34035.1), APH(3′)-VIIIb (EPF73263.1), APH(3′)-VIb (CAF29483.1), APH(3′)-VI (AGI04227.1), and APH(3′)-VIa (CAA30578.1).
To analyze the structure related to the resistance function of APH(3′)-Id, multiple sequence alignment of APH(3′)-Id and other functionally characterized APH(3′) proteins was performed (Figure 2). The results showed that APH(3′)-Id contained three conserved functional domains of the aminoglycoside phosphotransferase family, including Motif 1 (V--HGD----N), Motif 2 (G--D-GR-G) and Motif 3 (D--R/K--F/Y---LDE) (Shaw et al., 1993) (Figure 2). Six of eight active residues responsible for the phosphorylation of kanamycin in APH(3′)-IIa were identified in APH(3′)-Id (D167, D198, R219, E238, E269 and F271) (Nurizzo et al., 2003) (Figure 2). All nine kanamycin binding sites of APH(3′)-Ia were also identified in APH(3′)-Id (D165, F166, D167, D198, N234, E238, D268, E269, and F271) (Stogios et al., 2013) (Figures 2, 3). The tertiary structure alignment of the APH(3′)-Id model and kanamycin-bound APH(3′)-Ia showed that the overall structures of the two were similar, especially the kanamycin binding position. Similar to APH(3′)-Ia, the ATP binding sites of APH(3′)-Id, located in the hinge region between the N- and the C-terminal lobe of the bilobal fold, also had a similar spatial structure to that of eukaryotic protein kinases. The difference was that APH(3′)-Ia had a β-sheet (R6, E7, T8, C9), while APH(3′)-Id had an α-helix (L19, D20, T21, E22, L23), at the N terminus (Figure 3).
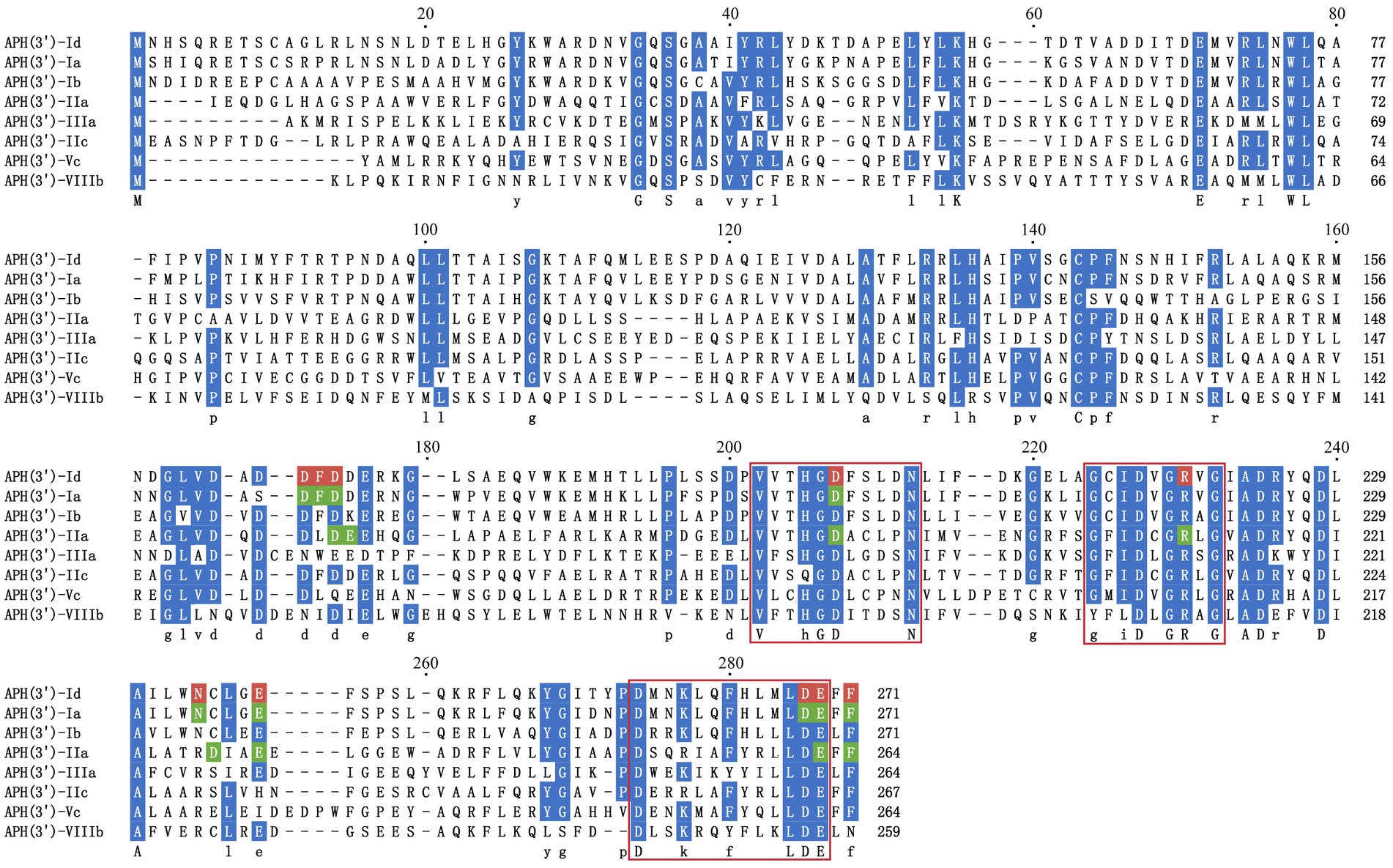
Figure 2. Multiple sequence alignment of APH(3′)-Id with other functionally characterized relatives. Dark blue indicates highly similar residues; red indicates residues related to kanamycin binding sites in APH(3′)-Id; green indicates residues representing kanamycin binding sites in APH(3′)-Ia and APH(3′)-IIa; three conserved functional domains of the aminoglycoside phosphotransferase family are framed in red squares; gaps are represented using hyphens. The numbers on the right represent the length of the corresponding sequence. Accession numbers of APH(3′) proteins: APH(3′)-Ia (CAE51638.1), APH(3′)-Ib (AAA26412.1), APH(3′)-IIa (CAA23892.1), APH(3′)-IIIa (AGV10830.1), APH(3′)-IIc (ADQ43421.1), APH(3′)-Vc (AAB21326.1) and APH(3′)-VIIIb (EPF73263.1).
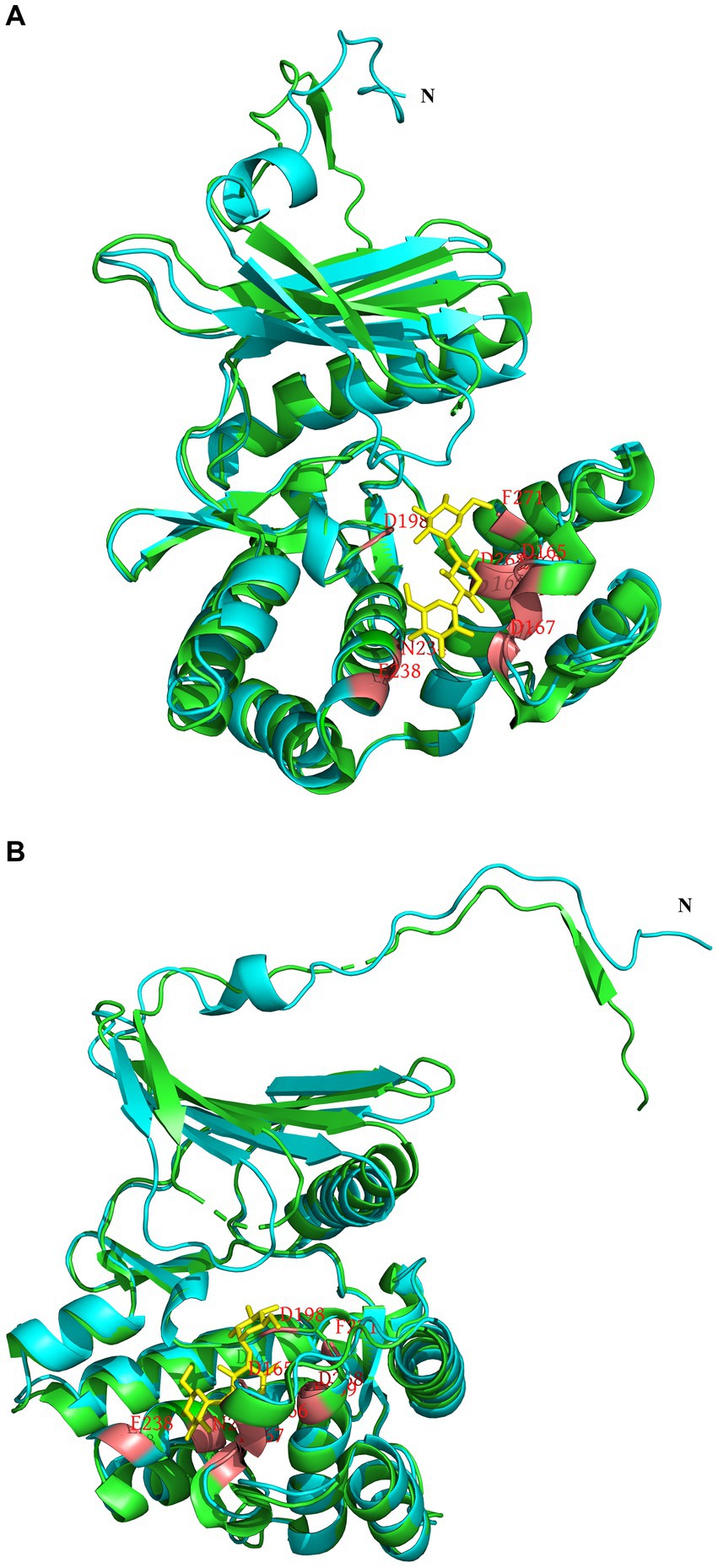
Figure 3. Tertiary structure alignment of the APH(3′)-Id model and kanamycin-bound APH(3′)-Ia. (A,B) are different illustrations of the same alignment. The APH(3′)-Id model is colored blue; APH(3′)-Ia is colored green; the position of amino acid residues bound with kanamycin is colored red in the APH(3′)-Id model and APH(3′)-Ia; kanamycin is colored yellow.
Kinetic parameters of APH(3′)-Id
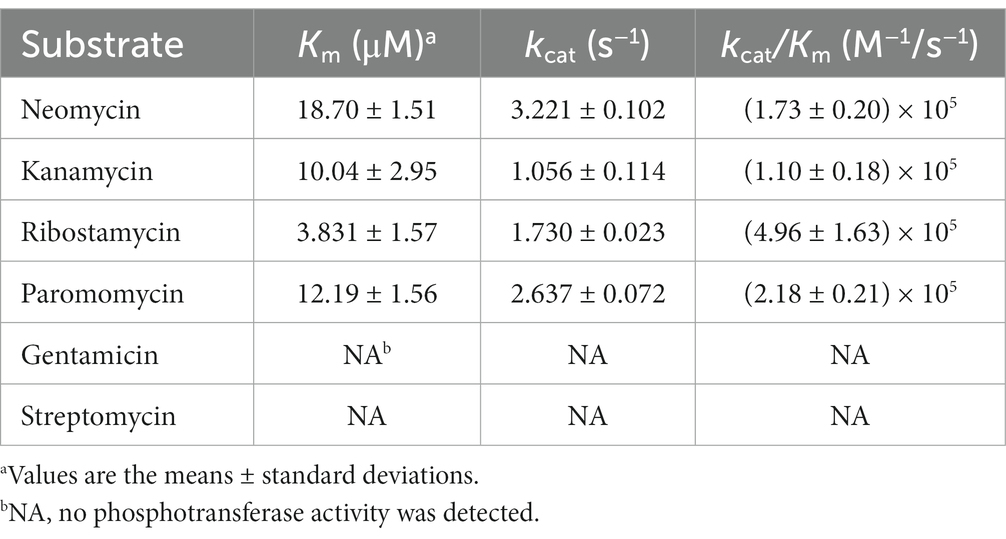
The induced expressed protein APH(3′)-Id was soluble (Supplementary Figure S1). After nickel column purification, His-tag removal and ultrafiltration concentration, 2 mL of 1.01 mg/mL (33.06 μM) APH(3′)-Id protein was obtained (Supplementary Figures S2, S3). The kinetic analysis of the phosphotransferase APH(3′)-Id showed that the purified APH(3′)-Id could phosphorylate four substrates, including ribostamycin, kanamycin, neomycin and paromomycin, which was consistent with the results for APH(3′)-I enzymes reported previously (Ramirez and Tolmasky, 2010). In the APH(3′)-I subclass, the enzyme with the closest evolutionary relationship to APH(3′)-Id was APH(3′)-Ia. By comparing the kinetic parameters between APH(3′)-Id and APH(3′)-Ia (Siregar et al., 1995), it was found that the affinity and catalytic efficiencies of APH(3′)-Id for the substrates were slightly weaker than those of APH(3′)-Ia. For example, when kanamycin was the substrate, the Km of APH(3′)-Ia was lower than that of APH(3′)-Id (1.2 ± 0.2 μM vs. 10.04 ± 2.95 μM), while the kcat/Km of APH(3′)-Ia was higher than that of APH(3′)-Id (8.5 × 107 M−1/s−1 vs. 1.10 × 105 M−1/s−1). A similar result was observed when neomycin was the substrate. The Km of APH(3′)-Ia for neomycin was lower than that of APH(3′)-Id (3.6 ± 0.3 μM vs. 18.70 ± 1.51 μM), while the kcat/Km of APH(3′)-Ia for neomycin was higher than that of APH(3′)-Id (5.1 × 107 M−1/s−1 vs. 1.73 × 105 M−1/s−1).
The substrate spectrum of APH(3′)-Id was also consistent with the resistance profile shown by the recombinant strain (pMD19-aph(3′)-Id/DH5α). Consistent with the MIC results, no phosphotransferase activity was found for gentamicin or streptomycin (Table 5). The catalytic efficiencies of APH(3′)-Id varied from the MIC values of the cloned aph(3′)-Id. APH(3′)-Id showed the highest catalytic efficiency for ribostamycin [kcat/Km of (4.96 ± 1.63) × 105 M−1/s−1], followed by paromomycin [kcat/Km of (2.18 ± 0.21) × 105 M−1/s−1], neomycin [kcat/Km of (1.73 ± 0.20) × 105 M−1/s−1], and kanamycin [kcat/Km of (1.10 ± 0.18) × 105 M−1/s−1]. However, the cloned aph(3′)-Id showed the highest MIC increase for kanamycin (1024-fold), followed by ribostamycin (512-fold), paromomycin (128-fold) and neomycin (16-fold). Neomycin had a high enzymatic catalytic efficiency but a low MIC value. It is possible that it was poorly transferred into the cell and could not reach the saturation concentration of the enzyme, so the enzyme could not fully perform its catalytic function (Siregar et al., 1995). In addition, APH(3′)-Id showed a high MIC level for kanamycin, while the catalytic efficiency was not very high. Different phosphate donors may result in different catalytic efficiencies of enzymes. ATP was used as the phosphate donor in this study. In a previous kinetic assay, the kcat of kanamycin phosphorylated by APH(3′)-Ia with GTP as a donor was 10-fold higher than that with ATP as the donor (Stogios et al., 2013).
As mentioned above, the substrate spectrum of APH(3′)-Id appeared to be similar to that of the other APH(3′)-I enzymes, which showed modification activity against several aminoglycosides, including kanamycin, ribostamycin, neomycin, paromomycin, lividomycin, gentamicin, butirosin and amikacin (Shaw et al., 1993; Siregar et al., 1995; Ramirez and Tolmasky, 2010). The substrate spectrum of the APH(3′)-II enzymes was the same with APH(3′)-I subclass (Shaw et al., 1993; Lu et al., 2021). APH(3′)-III enzymes had a broader substrate spectrum than APH(3′)-II, showing modification activity for isepamicin (Shaw et al., 1993; Ramirez and Tolmasky, 2010). In general, the substrate profile of APH(3′)-Id was consistent with that of the APH(3′)-I enzymes, and it further supported that this novel enzyme can be concluded into the APH(3′)-I subclass.
Genetic environment and distribution of aph(3′)-Id
The aph(3′)-Id gene is located in the chromosome of K. intermedia DW18. To explore its genetic context, the structure of an approximately 20 kb sequence with aph(3′)-Id at the center was analyzed (Figure 4). When using the nucleotide sequence of aph(3′)-Id as a query to search the NCBI nonredundant nucleotide database, a total of six aph(3′)-Id-like genes with identities of more than 80.0% were found. Of these six aph(3′)-Id-like genes, four were from the same species, K. intermedia, and each shared particularly high similarity (100% coverage and > 98% identity) with aph(3′)-Id, while the other two aph(3′)-Id-like genes were from Klebsiella sp. (CP056483.1 and CP055481.1) and shared a lower similarity (both with 100% coverage and 81.13% identity) with aph(3′)-Id. The genetic context of aph(3′)-Id with the four aph(3′)-Id-like genes that shared higher identities with aph(3′)-Id was then analyzed.
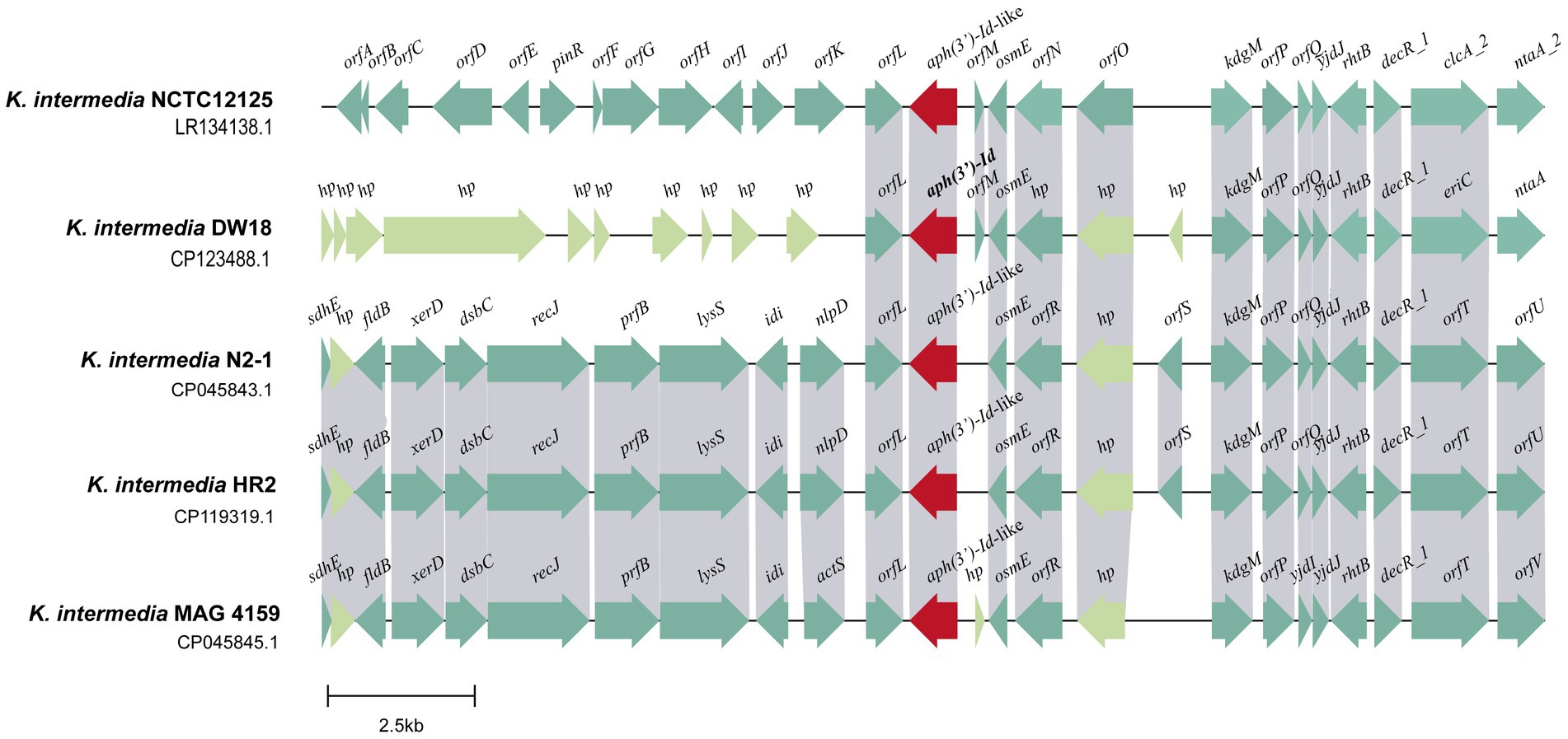
Figure 4. Genetic environment of the aph(3′)-Id and aph(3′)-Id-like genes. ORFs are shown as arrows drawn to scale to indicate the direction of transcription and colored based on gene function classification. Regions with ≥80% nucleotide identity are colored gray. hp: hypothetical protein.
Multisequence alignment of the five 20 kb sequences from K. intermedia (including one from this work) demonstrated that the downstream regions of the aph(3′)-Id(−like) genes were similar among them. This region of DW18 is mainly composed of genes related to transcription (osmE and decR_1) and transport (eriC, rhtB and kdgM). Nitrilotriacetate is an industrial chelating agent extensively used in textile and wastewater treatment processes. The product of the ntaA gene, nitrilotriacetate monooxygenase component A, is the core component of the nitrilotriacetate biodegradation process and may help prevent the harm caused by chelating agents in sewage (Kim K. J. et al., 2006). However, the upstream fragments were completely different between the gene from this work and any of the other four, except the orfL gene next to the aph(3′)-Id(−like) genes. The product of the orfL gene belongs to the multifunctional glutathionine S-transferase family, members of which can detoxify several chemical pollutants in the environment, such as 2,4,6-trinitrotoluen (TNT), toxic chlorinated organic compounds and anthracene (Mcguinness et al., 2007; Dixit et al., 2011; Gunning et al., 2014). No MGE was found in the flanking regions of the aph(3′)-Id(−like) genes (Figure 4). These results suggest that aph(3′)-Id may be an intrinsic gene of Kluyvera, and modification of aminoglycosides may be one of its normal physiological functions to protect the organism from environmental drugs. Although no MGE was detected near aph(3′)-Id, the possibility of horizontal transfer of this resistance gene between bacteria of different species or genera in the wild in the long run cannot be ruled out.
Conclusion
In this study, we reported the complete genome sequence of K. intermedia DW18 isolated from sewage in an animal farm and identified and characterized a novel aminoglycoside phosphotransferase gene, designated aph(3′)-Id, encoded in its chromosome. APH(3′)-Id shared the highest amino acid identity of 77.49% with the aminoglycoside 3′-O-phosphotransferase APH(3′)-Ia. APH(3′)-Id, consistent with the other enzymes of the APH(3′)-I subclass, conferred strong modification activity for specific aminoglycosides. These findings could help clinicians choose effective antimicrobials to treat infections caused by this bacterial species.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
HZ, KL, QB, and DL: conceived and designed the experiments. YS, NL, GZ, YZ, JZ, JL, and TZ: performed the experiments. YS, XZ, QL, and XL: data analysis and interpretation. YS, QB, and DL: drafting of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the Science and Technology Project of Wenzhou City, China (N20210001), the Science and Technology Project of Jinhua City, China (2022–2-013, 2022–4-017) and the Zhejiang Provincial Natural Science Foundation of China (LY19C060002 and LQ17H190001).
Acknowledgments
The authors would like to acknowledge all study participants and individuals who contributed to this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1224464/full#supplementary-material
References
Alcock, B. P., Huynh, W., Chalil, R., Smith, K. W., Raphenya, A. R., Wlodarski, M. A., et al. (2023). CARD 2023: expanded curation, support for machine learning, and resistome prediction at the comprehensive antibiotic resistance database. Nucleic Acids Res. 51, D690–d699. doi: 10.1093/nar/gkac920
Azucena, E., and Mobashery, S. (2001). Aminoglycoside-modifying enzymes: mechanisms of catalytic processes and inhibition. Drug Resist. Updat. 4, 106–117. doi: 10.1054/drup.2001.0197
Bengtsson-Palme, J., Boulund, F., Fick, J., Kristiansson, E., and Larsson, D. G. (2014). Shotgun metagenomics reveals a wide array of antibiotic resistance genes and mobile elements in a polluted lake in India. Front. Microbiol. 5:648. doi: 10.3389/fmicb.2014.00648
Buchfink, B., Reuter, K., and Drost, H. G. (2021). Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat. Methods 18, 366–368. doi: 10.1038/s41592-021-01101-x
Caminero, J. A., and Scardigli, A. (2015). Classification of antituberculosis drugs: a new proposal based on the most recent evidence. Eur. Respir. J. 46, 887–893. doi: 10.1183/13993003.00432-2015
Carter, J. E., and Evans, T. N. (2005). Clinically significant Kluyvera infections: a report of seven cases. Am. J. Clin. Pathol. 123, 334–338. doi: 10.1309/61xp-4ktl-jywm-5h35
Costa, B. O., Cardoso, M. H., and Franco, O. L. (2020). Development of peptides that inhibit aminoglycoside-modifying enzymes and β-lactamases for control of resistant Bacteria. Curr. Protein Pept. Sci. 21, 1011–1026. doi: 10.2174/1389203721666200915113630
D’Costa, V. M., King, C. E., Kalan, L., Morar, M., Sung, W. W., Schwarz, C., et al. (2011). Antibiotic resistance is ancient. Nature 477, 457–461. doi: 10.1038/nature10388
Dellinger, R. P., Levy, M. M., Rhodes, A., Annane, D., Gerlach, H., Opal, S. M., et al. (2013). Surviving sepsis campaign: international guidelines for management of severe sepsis and septic shock: 2012. Crit. Care Med. 41, 580–637. doi: 10.1097/CCM.0b013e31827e83af
Dixit, P., Mukherjee, P. K., Sherkhane, P. D., Kale, S. P., and Eapen, S. (2011). Enhanced tolerance and remediation of anthracene by transgenic tobacco plants expressing a fungal glutathione transferase gene. J. Hazard. Mater. 192 -, 270–276. doi: 10.1016/j.jhazmat.2011.05.018
Djordjevic, S., Stokes, H., and Roy Chowdhury, P. (2013). Mobile elements, zoonotic pathogens and commensal bacteria: conduits for the delivery of resistance genes into humans, production animals and soil microbiota. Front. Microbiol. 4:e00086. doi: 10.3389/fmicb.2013.00086
Doi, Y., and Arakawa, Y. (2007). 16S ribosomal RNA methylation: emerging resistance mechanism against aminoglycosides. Clin. Infect. Dis. 45, 88–94. doi: 10.1086/518605
Farmer, J. J., Fanning, G. R., Huntley-Carter, G. P., Holmes, B., Hickman, F. W., Richard, C., et al. (1981). Kluyvera, a new (redefined) genus in the family Enterobacteriaceae: identification of Kluyvera ascorbata sp. nov. and Kluyvera cryocrescens sp. nov. in clinical specimens. J. Clin. Microbiol. 13, 919–933. doi: 10.1128/jcm.13.5.919-933.1981
Forsberg, K. J., Reyes, A., Wang, B., Selleck, E. M., Sommer, M. O., and Dantas, G. (2012). The shared antibiotic resistome of soil bacteria and human pathogens. Science 337, 1107–1111. doi: 10.1126/science.1220761
Gasteiger, E., Gattiker, A., Hoogland, C., Ivanyi, I., Appel, R. D., and Bairoch, A. (2003). ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788. doi: 10.1093/nar/gkg563
Gilchrist, C. L. M., and Chooi, Y. H. (2021). Clinker & clustermap.Js: automatic generation of gene cluster comparison figures. Bioinformatics 37, 2473–2475. doi: 10.1093/bioinformatics/btab007
Grimont, F., and Grimont, P. A. D. (2006). “The genus Enterobacter” in The prokaryotes: A handbook on the biology of Bacteria volume 6: Proteobacteria: Gamma subclass. eds. M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (New York, NY: Springer New York), 197–214.
Gunning, V., Tzafestas, K., Sparrow, H., Johnston, E. J., Brentnall, A. S., Potts, J. R., et al. (2014). Arabidopsis glutathione transferases U24 and U25 exhibit a range of detoxification activities with the environmental pollutant and explosive, 2,4,6-trinitrotoluene. Plant Physiol. 165, 854–865. doi: 10.1104/pp.114.237180
Hon, W. C., McKay, G. A., Thompson, P. R., Sweet, R. M., Yang, D. S., Wright, G. D., et al. (1997). Structure of an enzyme required for aminoglycoside antibiotic resistance reveals homology to eukaryotic protein kinases. Cells 89, 887–895. doi: 10.1016/s0092-8674(00)80274-3
Jain, C., Rodriguez, R. L., Phillippy, A. M., Konstantinidis, K. T., and Aluru, S. (2018). High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 9:5114. doi: 10.1038/s41467-018-07641-9
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kim, C., Cha, J. Y., Yan, H., Vakulenko, S. B., and Mobashery, S. (2006). Hydrolysis of ATP by aminoglycoside 3′-phosphotransferases: an unexpected cost to bacteria for harboring an antibiotic resistance enzyme. J. Biol. Chem. 281, 6964–6969. doi: 10.1074/jbc.M513257200
Kim, K. J., Kim, S., Lee, S., Kang, B. S., Lee, H. S., Oh, T. K., et al. (2006). Crystallization and initial crystallographic characterization of the Corynebacterium glutamicum nitrilotriacetate monooxygenase component a. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 62, 1141–1143. doi: 10.1107/s1744309106042072
Konstantinidis, K. T., and Tiedje, J. M. (2005). Genomic insights that advance the species definition for prokaryotes. Proc. Natl. Acad. Sci. U. S. A. 102, 2567–2572. doi: 10.1073/pnas.0409727102
Krause, K. M., Serio, A. W., Kane, T. R., and Connolly, L. E. (2016). Aminoglycosides: an overview. Cold Spring Harb. Perspect. Med. 6, 1–2. doi: 10.1101/cshperspect.a027029
Letunic, I., and Bork, P. (2021). Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–w296. doi: 10.1093/nar/gkab301
Lu, W., Li, K., Huang, J., Sun, Z., Li, A., Liu, H., et al. (2021). Identification and characteristics of a novel aminoglycoside phosphotransferase, APH(3′)-IId, from an MDR clinical isolate of Brucella intermedia. J. Antimicrob. Chemother. 76, 2787–2794. doi: 10.1093/jac/dkab272
Magnet, S., Courvalin, P., and Lambert, T. (2001). Resistance-nodulation-cell division-type efflux pump involved in aminoglycoside resistance in Acinetobacter baumannii strain BM4454. Antimicrob. Agents Chemother. 45, 3375–3380. doi: 10.1128/aac.45.12.3375-3380.2001
Martínez, J. L. (2008). Antibiotics and antibiotic resistance genes in natural environments. Science 321, 365–367. doi: 10.1126/science.1159483
McArthur, A. G., Waglechner, N., Nizam, F., Yan, A., Azad, M. A., Baylay, A. J., et al. (2013). The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother. 57, 3348–3357. doi: 10.1128/aac.00419-13
Mcguinness, M. C., Mazurkiewicz, V., Brennan, E., and Dowling, D. N. (2007). Dechlorination of pesticides by a specific bacterial glutathione S-transferase, BphKLB400: potential for bioremediation. Eng. Life Sci. 7, 611–615. doi: 10.1002/elsc.200720218
Nguyen, L. T., Schmidt, H. A., von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Nicholas, K.B. (1997). GeneDoc: Analysis and visualization of genetic variation, EMBNEW. Embnew News 4.
Nurizzo, D., Shewry, S. C., Perlin, M. H., Brown, S. A., Dholakia, J. N., Fuchs, R. L., et al. (2003). The crystal structure of aminoglycoside-3′-phosphotransferase-IIa, an enzyme responsible for antibiotic resistance. J. Mol. Biol. 327, 491–506. doi: 10.1016/s0022-2836(03)00121-9
Oka, A., Sugisaki, H., and Takanami, M. (1981). Nucleotide sequence of the kanamycin resistance transposon Tn903. J. Mol. Biol. 147, 217–226. doi: 10.1016/0022-2836(81)90438-1
Paskova, V., Medvecky, M., Skalova, A., Chudejova, K., Bitar, I., Jakubu, V., et al. (2018). Characterization of NDM-encoding plasmids from Enterobacteriaceae recovered from Czech hospitals. Front. Microbiol. 9:e01549. doi: 10.3389/fmicb.2018.01549
Pavan, M. E., Franco, R. J., Rodriguez, J. M., Gadaleta, P., Abbott, S. L., Janda, J. M., et al. (2005). Phylogenetic relationships of the genus Kluyvera: transfer of Enterobacter intermedius izard et al. 1980 to the genus Kluyvera as Kluyvera intermedia comb. nov. and reclassification of Kluyvera cochleae as a later synonym of K. intermedia. Int. J. Syst. Evol. Microbiol. 55, 437–442. doi: 10.1099/ijs.0.63071-0
Prats, G., Richard, C., Mirelis, B., and Lopez, P. (1987). Human isolates of Enterobacter intermedium. Zentralbl Bakteriol Mikrobiol Hyg A 266, 422–424. doi: 10.1016/s0176-6724(87)80222-5
Qing, G., Ma, L. C., Khorchid, A., Swapna, G. V., Mal, T. K., Takayama, M. M., et al. (2004). Cold-shock induced high-yield protein production in Escherichia coli. Nat. Biotechnol. 22, 877–882. doi: 10.1038/nbt984
Ramirez, M. S., and Tolmasky, M. E. (2010). Aminoglycoside modifying enzymes. Drug Resist. Updat. 13, 151–171. doi: 10.1016/j.drup.2010.08.003
Ribeiro, V. B., Zavascki, A. P., Rozales, F. P., Pagano, M., Magagnin, C. M., Nodari, C. S., et al. (2014). Detection of BlaGES-5 in Carbapenem-resistant Kluyvera intermedia isolates recovered from the hospital environment. Antimicrob. Agents Chemother. 58, 622–623. doi: 10.1128/aac.02271-13
Riccio, M. L., Pallecchi, L., Fontana, R., and Rossolini, G. M. (2001). In70 of plasmid pAX22, a Bla(VIM-1)-containing integron carrying a new aminoglycoside phosphotransferase gene cassette. Antimicrob. Agents Chemother. 45, 1249–1253. doi: 10.1128/aac.45.4.1249-1253.2001
Richter, M., and Rosselló-Móra, R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U. S. A. 106, 19126–19131. doi: 10.1073/pnas.0906412106
Schrödinger, L. (2023). PyMOL Available at: http://www.pymol.org.
Schulz, F., Eloe-Fadrosh, E. A., Bowers, R. M., Jarett, J., Nielsen, T., Ivanova, N. N., et al. (2017). Towards a balanced view of the bacterial tree of life. Microbiome 5:140. doi: 10.1186/s40168-017-0360-9
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Shaw, K. J., Rather, P. N., Hare, R. S., and Miller, G. H. (1993). Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol. Rev. 57, 138–163. doi: 10.1128/mr.57.1.138-163.1993
Siregar, J. J., Miroshnikov, K., and Mobashery, S. (1995). Purification, characterization, and investigation of the mechanism of aminoglycoside 3′-phosphotransferase type Ia. Biochemistry 34, 12681–12688. doi: 10.1021/bi00039a026
Smith, C. A., Toth, M., Stewart, N. K., Maltz, L., and Vakulenko, S. B. (2019). Structural basis for the diversity of the mechanism of nucleotide hydrolysis by the aminoglycoside-2-phosphotransferases. Acta Crystallogr D Struct Biol 75, 1129–1137. doi: 10.1107/s2059798319015079
Souvorov, A., Agarwala, R., and Lipman, D. J. (2018). SKESA: strategic k-mer extension for scrupulous assemblies. Genome Biol. 19:153. doi: 10.1186/s13059-018-1540-z
Stock, I. (2002). Natural antibiotic susceptibility of Enterobacter spp., with special reference to Enterobacter aerogenes and Enterobacter intermedius strains. J. Chemother. 14, 444–460. doi: 10.1179/joc.2002.14.5.444
Stogios, P. J., Spanogiannopoulos, P., Evdokimova, E., Egorova, O., Shakya, T., Todorovic, N., et al. (2013). Structure-guided optimization of protein kinase inhibitors reverses aminoglycoside antibiotic resistance. Biochem. J. 454, 191–200. doi: 10.1042/bj20130317
Suzuki, S., and Hoa, P. T. (2012). Distribution of quinolones, sulfonamides, tetracyclines in aquatic environment and antibiotic resistance in indochina. Front. Microbiol. 3:67. doi: 10.3389/fmicb.2012.00067
Toth, M., Frase, H., Antunes, N. T., and Vakulenko, S. B. (2013). Novel aminoglycoside 2-phosphotransferase identified in a gram-negative pathogen. Antimicrob. Agents Chemother. 57, 452–457. doi: 10.1128/aac.02049-12
Tsai, A., Uemura, S., Johansson, M., Puglisi, E. V., Marshall, R. A., Aitken, C. E., et al. (2013). The impact of aminoglycosides on the dynamics of translation elongation. Cell Rep. 3, 497–508. doi: 10.1016/j.celrep.2013.01.027
Vakulenko, S. B., and Mobashery, S. (2003). Versatility of aminoglycosides and prospects for their future. Clin. Microbiol. Rev. 16, 430–450. doi: 10.1128/cmr.16.3.430-450.2003
Walker, B. J., Abeel, T., Shea, T., Priest, M., Abouelliel, A., Sakthikumar, S., et al. (2014). Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963
Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., et al. (2018). SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–w303. doi: 10.1093/nar/gky427
Keywords: Kluyvera intermedia, resistance gene, APH(3′)-Id, aminoglycoside 3’-O-phosphotransferase, kinetic parameter
Citation: Sha Y, Lin N, Zhang G, Zhang Y, Zhao J, Lu J, Zhu T, Zhang X, Li Q, Zhang H, Lin X, Li K, Bao Q and Li D (2023) Identification and characterization of a novel chromosomal aminoglycoside 3’-O-phosphotransferase, APH(3′)-Id, from Kluyvera intermedia DW18 isolated from the sewage of an animal farm. Front. Microbiol. 14:1224464. doi: 10.3389/fmicb.2023.1224464
Edited by:
José Manuel Rodriguez-Martínez, Sevilla University, SpainReviewed by:
Ali H. A. Elbehery, University of Sadat City, EgyptElisa Binda, University of Insubria, Italy
Copyright © 2023 Sha, Lin, Zhang, Zhang, Zhao, Lu, Zhu, Zhang, Li, Zhang, Lin, Li, Bao and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiyu Bao, YmFvcXlAZ2Vub21pY3MuY24=; Dong Li, bGlkb25nQHdtdS5lZHUuY24=
 Yuning Sha
Yuning Sha Naru Lin
Naru Lin Guozhi Zhang
Guozhi Zhang Yuan Zhang
Yuan Zhang Jingxuan Zhao
Jingxuan Zhao Junwan Lu
Junwan Lu Tingting Zhu
Tingting Zhu Xueya Zhang
Xueya Zhang Qiaoling Li
Qiaoling Li Hailin Zhang
Hailin Zhang Xi Lin
Xi Lin Kewei Li
Kewei Li Qiyu Bao
Qiyu Bao Dong Li1,2*
Dong Li1,2*