
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
CORRECTION article
Front. Microbiol. , 07 August 2023
Sec. Evolutionary and Genomic Microbiology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1222851
This article is a correction to:
Out of the Qinghai-Tibetan plateau: Origin, evolution and historical biogeography of Morchella (both Elata and Esculenta clades)
 Qing Meng1,2
Qing Meng1,2 Zhanling Xie1,2*
Zhanling Xie1,2* Hongyan Xu1,3
Hongyan Xu1,3 Jing Guo1,2
Jing Guo1,2 Yongpeng Tang4
Yongpeng Tang4 Ting Ma1
Ting Ma1 Qingqing Peng1
Qingqing Peng1 Bao Wang1,2
Bao Wang1,2 Yujing Mao1,2
Yujing Mao1,2 Shangjin Yan1
Shangjin Yan1 Jiabao Yang1,2
Jiabao Yang1,2 Deyu Dong1,2
Deyu Dong1,2 Yingzhu Duan4
Yingzhu Duan4 Fan Zhang5
Fan Zhang5 Taizhen Gao4
Taizhen Gao4A corrigendum on
Out of the Qinghai-Tibetan plateau: origin, evolution and historical biogeography of Morchella (both Elata and Esculenta clades)
by Meng, Q., Xie, Z., Xu, H., Guo, J., Tang, Y., Ma, T., Peng, Q., Wang, B., Mao, Y,. Yan, S., Yang, J., Dong, D., Duan, Y., Zhang, F., and Gao, T. (2022). Front. Microbiol. 13:1078663. doi: 10.3389/fmicb.2022.1078663
In the published article, there was an error in the legend for Figure 2C and Figure 3C as published. The names of various Morchella phylospecies were incorrectly presented. The corrected parts of the legends appear below.
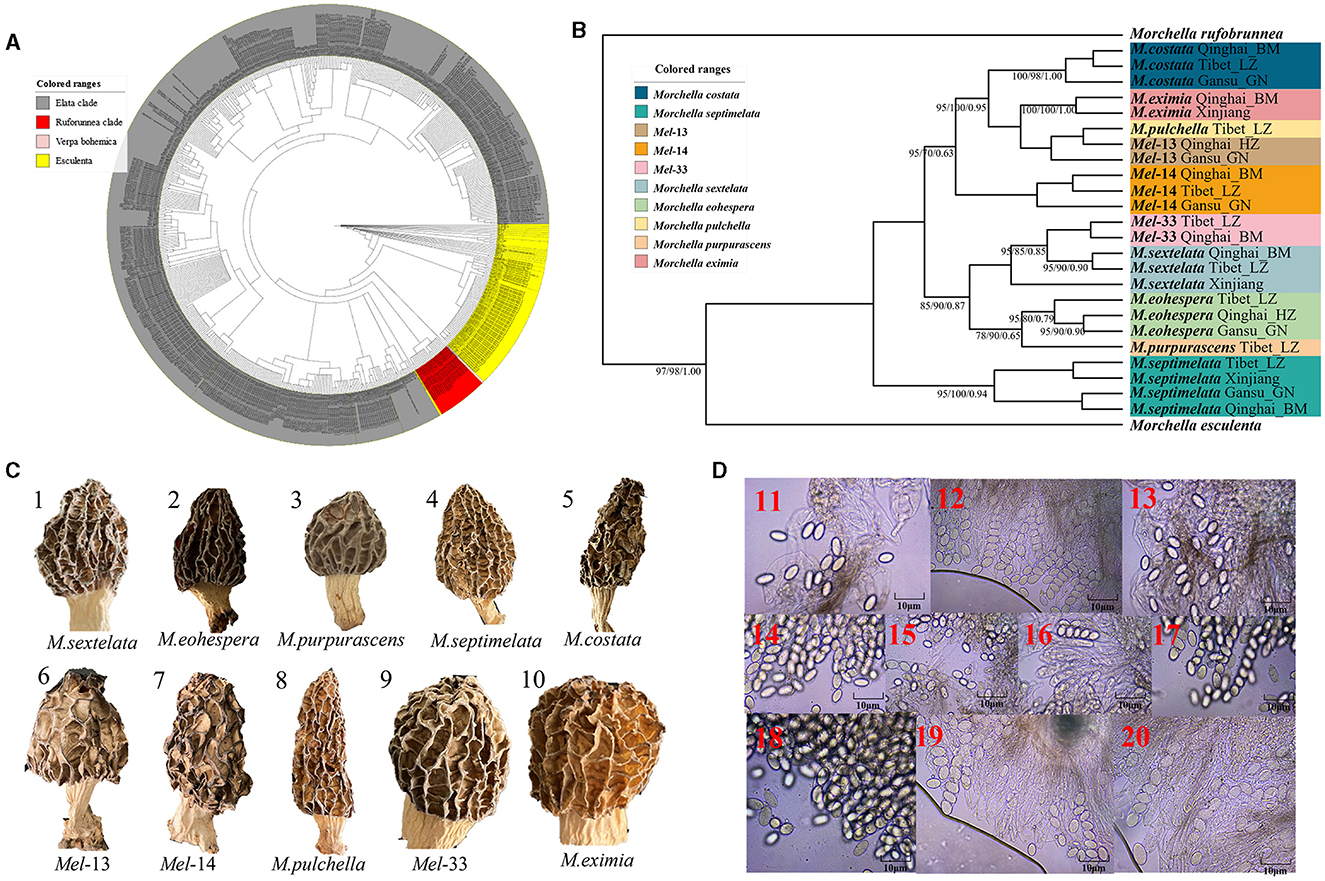
Figure 2. Species recognition of the Morchella in Elata clade from the QTPs. (A) Bayesian inference phylogenetic analyses of the Elata clade were inferred from 115 internal transcribed spacer (ITS) sequences representing a total of 10 phylospecies. (B) Phylogenetic analyses of the Elata clade were inferred from 120 (24*5) multi-genes (ITS + LSU + EF1-α + RPB1 + RPB2) sequences representing a total of ten phylospecies. Branches are labeled where MP/ML support is greater than 60% and collapsed below that support threshold. BPP is labeled were greater than 0.95. (C) Morphological diversity of the 10 Elata clades' ascocarps from the QTPs: M. sextelata/Mel-6 (1), M. norvegiensis = M. eohespera/Mel-19 (2), M. purpurascens/Mel-20 (3), M. septimelata/Mel-7 (4), M. costata (5), M. deliciosa/Mel-13 (6), Mel-14 (7), M. pulchella/Mel-31 (8), Mel-33 (9), M. eximia/Mel-5 (10). (D) Micromorphological ascospores of the 10 Elata clades.
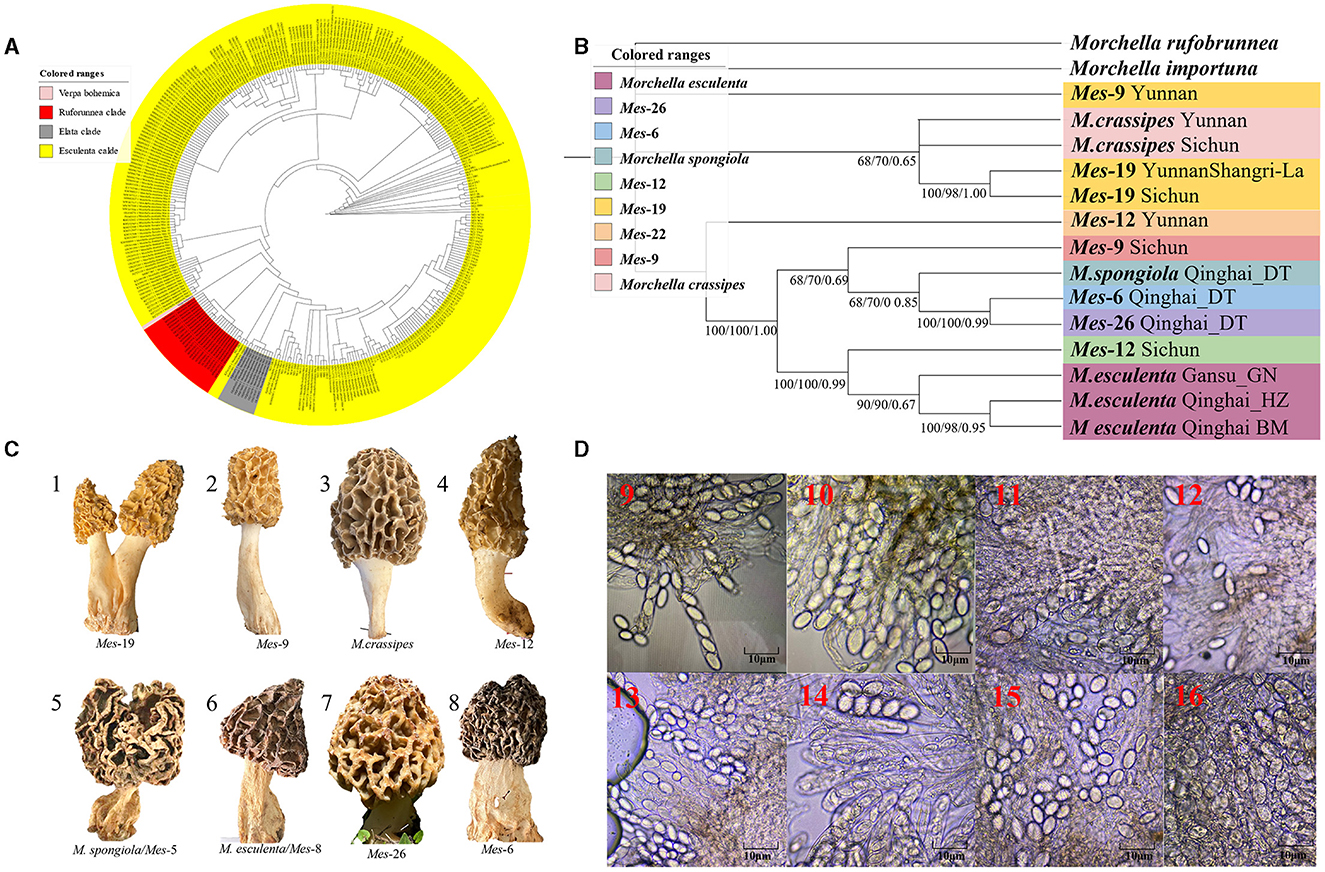
Figure 3. Species recognition of the Morchella in Esculenta clade from the QTPs. (A) Bayesian inference phylogenetic analyses of the Esculenta clade were inferred from 101 ITS sequences representing a total of 8 phylospecies. (B) Phylogenetic analyses of the Esculenta clade were inferred from 70 (14*5) multi-genes (ITS + LSU + EF1-α + RPB1 + RPB2) sequences representing a total of 8 phylospecies. Branches are labeled where MP/ML support is greater than 60% and collapsed below that support threshold. BPP is labeled were greater than 0.95. (C) Morphological diversity of the 8 Esculenta clades' ascocarps from the QTPs: Mes-19 (1); Mes-9 (2), M. crassipes (3), Mes-12 (4), M. vulgaris = M. spongiola/Mes-5 (5), M. esculenta/Mes-8 (6), Mes-26 (7), Mes-6 (8). (D) Micromorphological ascospores of the 8 Esculenta clades.
Figure 2. Species recognition of the Morchella in Elata clade from the QTPs. (C) Morphological diversity of the 10 Elata clades' ascocarps from the QTPs: M. sextelata/Mel-6 (1), M. norvegiensis = M. eohespera/Mel-19 (2), M. purpurascens/Mel-20 (3), M. septimelata/Mel-7 (4), M. costata (5), M. deliciosa/Mel-13 (6), Mel-14 (7), M. pulchella/Mel-31 (8), Mel-33 (9), M. eximia/Mel-5 (10).
Figure 3. Species recognition of the Morchella in Esculenta clade from the QTPs. (C) Morphological diversity of the 8 Esculenta clades' ascocarps from the QTPs: Mes-19 (1); Mes-9 (2), M. crassipes (3), Mes-12 (4), M. vulgaris = M. spongiola/Mes-5 (5), M. esculenta/Mes-8 (6), Mes-26 (7), Mes-6 (8).
In the published article, in Figures 2–4, 6 and Table 1 the names of several Morchella phylospecies were incorrectly presented.
The corrected Figures 2–4, Figure 6 and Table 1 and their captions appear below.
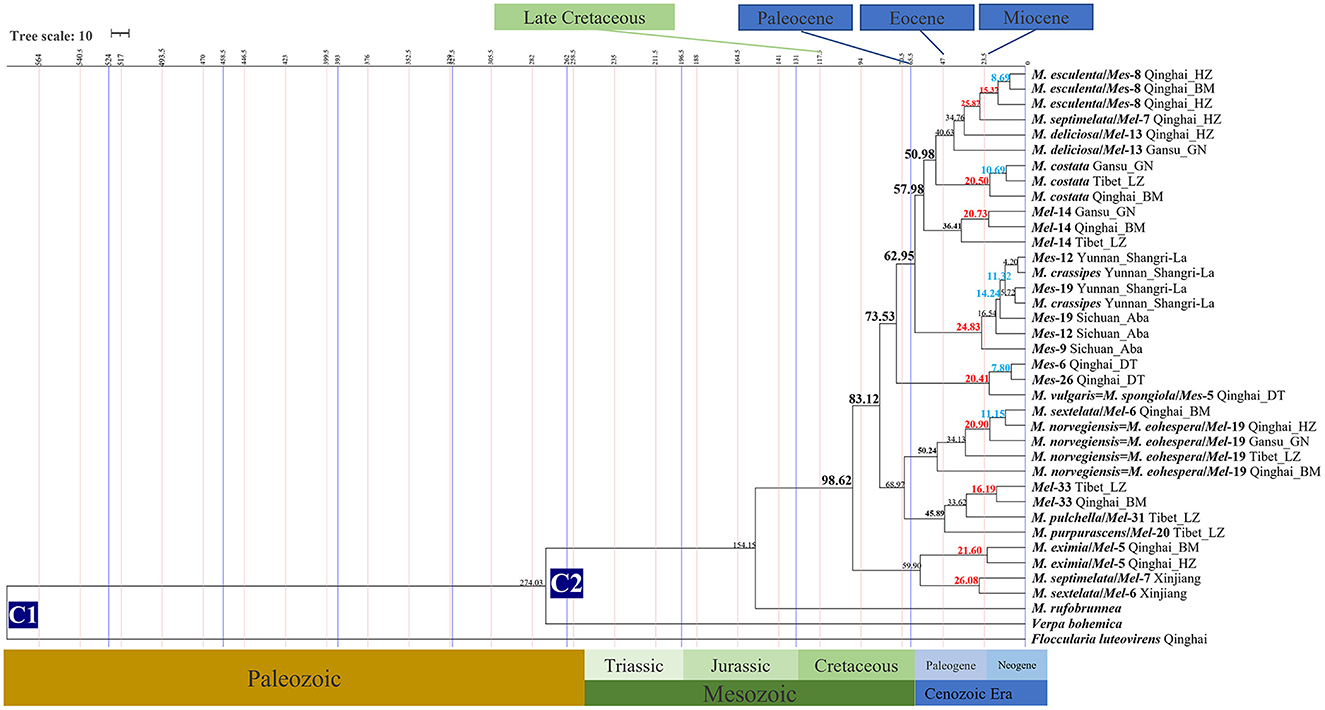
Figure 4. Chronogram and estimated divergence times of Morchella in QTPs generated by molecular clock analysis using the two concatenated datasets (ITS + LSU and EF1-α + RPB1 + RPB2) dataset. The chronogram was obtained using the Ascomycota-Basidiomycota divergence time of 582.08 Mya as the calibration point 1. The Morchella-Verpa bohenica divergence time of 274.06 Mya as the calibration point 2. The calibration point and objects of this study are marked in the chronogram. The geological time scale is millions of years ago (Mya). The red font is defined as the first uplift of the QTPs, and the blue font is defined as the second uplift of the QTPs.
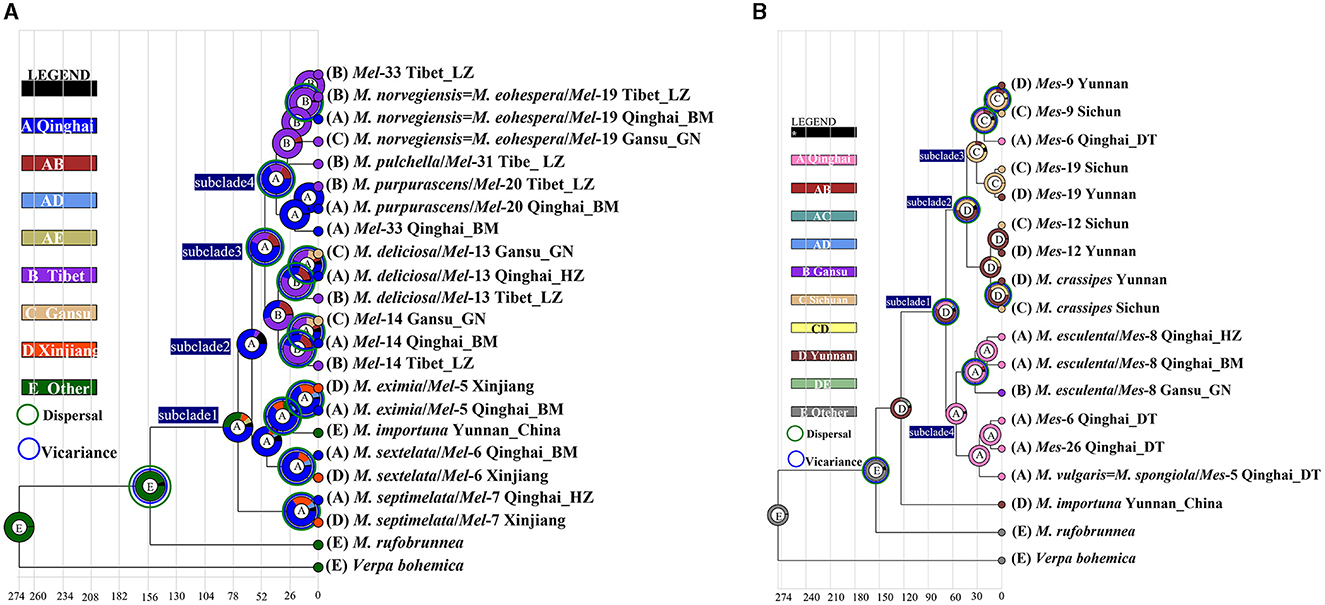
Figure 6. Ancestral area reconstruction of Morchella phylospecies in QTPs using the ITS dataset. The chronogram was obtained by molecular clock analysis using BEAST. The pie chart in each node indicates the possible ancestral distributions inferred from Bayesian Binary MCMC analysis (BBM) implemented in RASP. Bayesian credibility values (PP) over 0.85 are indicated near the pie chart of the tree. The green circle around the pie charts indicates possible dispersal events, the blue circle indicates possible vicariance events as suggested by BBM analysis. (A) Elata clade; (B) Esculenta clade.
In Supplementary Figures S1, S4, S5, and Table S1, the names of several Morchella phylospeices were incorrectly presented. The corrected Supplementary material accompanies this article.
A correction has been made to the Results section, paragraph 1. This sentence previously stated:
“A total of 101 individuals clustered with 10 phylogenetic species, including Mel-14, Mel-13, Morchella eohespera/Mel-19, Morchella eximia/Mel-5, Morchella costata, Morchella sextelata/Mel-6, Morchella septimelata/Mel-7, Morchella purpurascens/Mel-20, Mel-33, and Morchella pulchella/Mel-12 belongs to Elata clade (Figures 2A–C); and a total of 101 individuals clustered with 8 phylogenetic species, including Morchella spongiola, Mes-9, Mes-12, Mes-26, Morchella crassipes, Morchella esculenta, Mes-19, and Mes-6 belongs to Esculenta clade (Figures 3A–C).”
The corrected sentence appears below:
“A total of 101 individuals clustered with 10 phylogenetic species, including Mel-14, M. deliciosa/Mel-13, M. norvegiensis = M. eohespera/Mel-19, Morchella eximia/Mel-5, Morchella costata, Morchella sextelata/Mel-6, Morchella septimelata/Mel-7, Morchella purpurascens/Mel-20, Mel-33, and Morchella pulchella/Mel-31 belongs to Elata clade (Figures 2A–C); and a total of 101 individuals clustered with 8 phylogenetic species, including M. vulgaris = M. spongiola/Mes-5, Mes-9, Mes-12, Mes-26, Morchella crassipes, Morchella esculenta/Mes-8, Mes-19, and Mes-6 belongs to Esculenta clade (Figures 3A–C).”
A correction has been made to the Results section, paragraph 2. This sentence previously stated:
“The phylospecies of M. eohespera, Mel-13, Mel-14, M. eximia, M. costata, M. esculenta, M. crassipes, and….”
The corrected sentence appears below:
“The phylospecies of M. norvegiensis = M. eohespera/Mel-19, M. deliciosa/Mel-13, Mel-14, Morchella eximia, M. costata, M. esculenta/Mes-8, M. crassipes, and….”
A correction has been made to the Results section, paragraph 3. This sentence previously stated:
“such as the Mel-13, Mel-14, M. eohespera, suggests that not all of the Morchella species were narrowly distributed;”
The corrected sentence appears below:
“such as the M. deliciosa/Mel-13, Mel-14, M. norvegiensis = M. eohespera/Mel-19, suggests that not all of the Morchella species were narrowly distributed;”
A correction has been made to the Results section, paragraph 5. This sentence previously stated:
“(1) M. spongiola, there were…; (2) M. esculenta was widely…”
The corrected sentence appears below:
“(1) M. vulgaris = M. spongiola/Mes-5 there were…; (2) M. esculenta/Mes-8 was widely…”
A correction has been made to the Results section, paragraph 6. This sentence previously stated:
“(4) M. eohespera (Elata clade), were distributed in…; (5) Mel-13 and Mel-14 are widely distributed in Eurasia, especially in the QTPs. Mel-13 and…”
The corrected sentence appears below:
“(4) M. norvegiensis = M. eohespera/Mel-19 (Elata clade), were distributed in…; (5) M. deliciosa/Mel-13 and Mel-14 are widely distributed in Eurasia, especially in the QTPs. M. deliciosa/Mel-13 and…”
A correction has been made to the Discussion section, paragraph 3. This sentence previously stated:
“In our data, M. eohespera was differentiated at 50.24 Mya with the new uplift belts of Tengchong-Bango formatted and the uplift area of Songpan-Ganzi shrank to the east during Eocene; M. eohespera in the middle latitudes region were differentiated at 34.24 Mya with the further uplifted of Kunlun-Algin-Qilian during the Oligocene; ”
The corrected sentence appears below:
“In our data, M. norvegiensis = M. eohespera/Mel-19 was differentiated at 50.24 Mya with the new uplift belts of Tengchong-Bango formatted and the uplift area of Songpan-Ganzi shrank to the east during Eocene; M. norvegiensis=M. eohespera/Mel-19 in the middle latitudes region were differentiated at 34.24 Mya with the further uplifted of Kunlun-Algin-Qilian during the Oligocene;”
A correction has been made to the Discussion section paragraph 4. This sentence previously stated:
“(ii) provincialism in the QTPs: the specific local distributions of two species in the Elata clade (M. pulchella/Mel-12, M. purpurascens/Mel-20) were unique (only in the Tibet region).”
The corrected sentence appears below:
“(ii) provincialism in the QTPs: the specific local distributions of two species in the Elata clade (M. pulchella/Mel-31, M. purpurascens/Mel-20) were unique (only in the Tibet region).”
A correction has been made to the Discussion section, paragraph 6. This sentence previously stated:
“For example, in M. eohespera, the divergence time was estimated at 52.25 Mya, which was earlier than that of Europe (Supplementary Figure S4).”
The corrected sentence appears below:
“For example, in M. norvegiensis = M. eohespera/Mel-19, the divergence time was estimated at 52.25 Mya, which was earlier than that of Europe (Supplementary Figure S4).”
The authors apologize for these errors and state that this does not change the scientific conclusions of the article in any way. The original article has been updated.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Keywords: Morchella, Qinghai-Tibet plateau subkingdoms, multigene phylogenetics, age estimation, phylogeographic structure
Citation: Meng Q, Xie Z, Xu H, Guo J, Tang Y, Ma T, Peng Q, Wang B, Mao Y, Yan S, Yang J, Dong D, Duan Y, Zhang F and Gao T (2023) Corrigendum: Out of the Qinghai-Tibetan plateau: origin, evolution and historical biogeography of Morchella (both Elata and Esculenta clades). Front. Microbiol. 14:1222851. doi: 10.3389/fmicb.2023.1222851
Received: 15 May 2023; Accepted: 10 July 2023;
Published: 07 August 2023.
Edited and reviewed by: John R. Battista, Louisiana State University, United States
Copyright © 2023 Meng, Xie, Xu, Guo, Tang, Ma, Peng, Wang, Mao, Yan, Yang, Dong, Duan, Zhang and Gao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhanling Xie, eGllemhhbmxpbmcyMDEyQDEyNi5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.