
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 15 September 2023
Sec. Terrestrial Microbiology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1220901
This article is part of the Research TopicInsights in Terrestrial Microbiology: 2022View all 7 articles
 Chakriya Sansupa1†
Chakriya Sansupa1† Nattaphon Suphaphimol1†
Nattaphon Suphaphimol1† Paradha Nonthijun1
Paradha Nonthijun1 Teewararat Ronsuek1
Teewararat Ronsuek1 Saranphong Yimklan2
Saranphong Yimklan2 Natthawat Semakul2
Natthawat Semakul2 Thapakorn Khrueraya3
Thapakorn Khrueraya3 Nakarin Suwannarach1,4
Nakarin Suwannarach1,4 Witoon Purahong5*
Witoon Purahong5* Terd Disayathanoowat1,4*
Terd Disayathanoowat1,4*Diverse microorganisms from the three domains of life (Archaea, Bacteria, and Eukaryota) cause deterioration in mural paintings worldwide; however, few studies have simultaneously targeted these three domains. This study aims to survey the microbiome and its potential for biodeterioration on unpreserved Lan Na mural paintings in Sean Khan temple, Chiang Mai, Thailand. The overview of the archaeal, bacterial, and fungal communities was reported by Illumina sequencing, whereas the potential for deterioration was revealed by culturable techniques and a literature search. The abundant microbes reported in this study were also found in other ancient mural paintings worldwide. Halococcus, a salt-tolerant archaeon, as well as the eubacterial genus Crossiella dominated the prokaryotic community. On the other hand, the main fungal group was the genus Candida (Ascomycota). However, a low number of fungi and bacteria were isolated. Most of the isolates showed the ability to survive in the drought conditions of mural paintings but could not perform discoloration activities. The deterioration activity mainly affected calcium compounds, which are the main components of painting substrates. Aspergillus and several bacterial isolates could dissolve calcium compounds, but only Trichaptum species could induce crystal formation. These results suggest that deterioration of painting substrate should be taken into consideration in addition to deterioration of color in mural paintings. For the Lan Na painting in Sean Khan temple, the plaster is the prime target for biodeterioration, and thus we suggest that the preservation effort should focus on this component of the mural painting.
Paint is one of the world’s oldest manmade products, with roots dating back to prehistoric times (Ravikumar, 2012). Paintings, like murals in a historical building heritage, are crucial landmarks that reflect and contribute to the long-term development of a region or country. In addition, it is linked to a sense of self-identity and is frequently exploited by the tourism industry. As past studies have demonstrated that the decline of mural paintings can impact social and economic systems, it is crucial to conserve these artworks for historical research as well as to ensure the continued social and economic stability of these systems (Silverman and Ruggles, 2007).
The deterioration of mural paintings can be influenced by several threats. These include intrinsic changes in the substrates’ physicochemical properties as well as extrinsic factors like atmospheric conditions and biological activities (Tiano, 2002; Dahmani et al., 2020). Atmospheric conditions and environmental factors, such as humidity, temperature, pH, and light, may cause structural or aesthetic damage to the paintings (Sterflinger, 2000; Warscheid and Braams, 2000; Rosling et al., 2009). Furthermore, as paintings contain a wide variety of organic and inorganic materials, they can be exploited by a wide range of microbial species. If the environmental conditions are favorable, many different types of microbes can colonize the paintings and cause damage to the artwork (Ciferri, 1999).
Diverse microorganisms, including archaea, bacteria and fungi, contribute to the destruction of cultural assets, especially those that contain elements required for their metabolism (Nuhoglu et al., 2006; Wiktor et al., 2009; Ettenauer et al., 2014; Karaca et al., 2015; Mihajlovski et al., 2017). Few studies have investigated simultaneously these three domains. Discoloration of paint and structural deconstruction of the painting layer are major issues caused by these organisms, leading to the deterioration of the art work (Ciferri, 1999). Microorganisms can utilize the organic compounds in the painting layers and produce enzymes or organic acids that decolorized and dissolved the compounds in the painting. Calcium compounds, for example, can be dissolved by many microorganisms (Rölleke et al., 1996; Rosado et al., 2013; Suphaphimol et al., 2022). Biomineralization is the process by which living organisms influence the precipitation of mineral materials (Skinner and Ehrlich, 2014). The formation of salt or calcium compound can also lead to the deconstruction of the painting’s layer, as it could increase the pressure on the mural materials and crack the artwork (Pérez-Alonso et al., 2004; Piñar and Sterflinger, 2009; Čukovska et al., 2017; Suphaphimol et al., 2022). Due to the ability of microorganisms to cause mural deterioration, as described above, it is of utmost importance to observe and investigate the possible threats that these organisms pose to a mural painting. As a result, we can determine the deterioration risk of the mural paintings and suggest promising ways to preserve them.
Lan Na culture is the unique culture of the northern Thailand and their neighborhood area because this kingdom is considered as the melting pot of big cultures especially Siamese (Central part of Thailand), Chinese and Burmese (Myanmar). Lan Na heritage is unique for its cuisine, architecture, clothing and mural paintings for over 700 years. One of the remained pieces of evidence of the last dynasty named Tipjakara (1732–1943) (Penth, 2001) and the biodeterioration of their mural painting was monitored (Suphaphimol et al., 2022). Only two mural painting sites from the first dynasty named Mungrai (1296–1578) (Penth, 2001) were found, one is a well preserved mural painting in the tunnel of Wat Umong, and another the unpreserved mural painting in Wat Sean Khan.
In this study, we investigated the microbial communities from the three domains of life associated with the mural painting in Sean Khan temple and determined the biodeterioration potentials of these microbes. Specifically, we identified archaea, bacteria and fungi using high-throughput sequencing and isolated bacteria and fungi using traditional culture methods. We then subjected the microbial isolates to deterioration tests, including discoloration, solubilization of calcium compounds, and crystal formation. Our study aimed to answer the following questions: (i) What microbial taxa dominate the mural paintings? (ii) Are any of these taxa capable of deteriorating the paintings? and (iii) How do they cause deterioration? To our knowledge, this study is among the few to reveal information about the microbes associated with mural paintings in Lan Na culture, and it is the first to contribute such information about mural art in the Mungrai dynasty period.
This study was carried out at Sean Khan temple, Chiang Mai, northern Thailand (18°47′ 48′′ N, 99° 59′ 4′′ E). The climatic data from 2008–2022 showed the minimum and maximum ranging from 9°C in December to 42.2°C in March, the rain regime average was ranging from 4.2–216.9 millimeters per month with a monsoon period from June to October and the relative humidity was ranging from 45% to 77% (data collected from weather station of the Thai Meteorological Department—Chiang Mai). The temple is abandoned and located in a residential area (Figures 1A,B). There is a Samanea saman (Jacq.) Merr. tree (Fabaceae) at the western side of the pagoda and below ground pagoda is surrounded by round eggplants [Solanum melongena L. (Solanaceae)] and Siam weeds [Chromolaena odorata (L.) R.M.King & H.Rob. (Asteraceae)]. The mural painting was discovered in 2019 in the inner part of a pagoda tunnel, 55 cm in width, 120 cm in height and 140 cm long. The mural was painted on walls at the end and on both sides of the tunnel (Figure 1C). It was painted using the fresco technique, water-based pigment on fresh, fine-grained plaster (Piovesan et al., 2012), which was commonly used during that time. Red and black were used as the primary colors of the painting (Figure 1C). The painting was fading, thus, to present the clear picture of the painting, we copy the mural painting line on transparent paper with 1:1 scale and then adjust it on Adobe Illustrator Cs6 (Figure 1D).
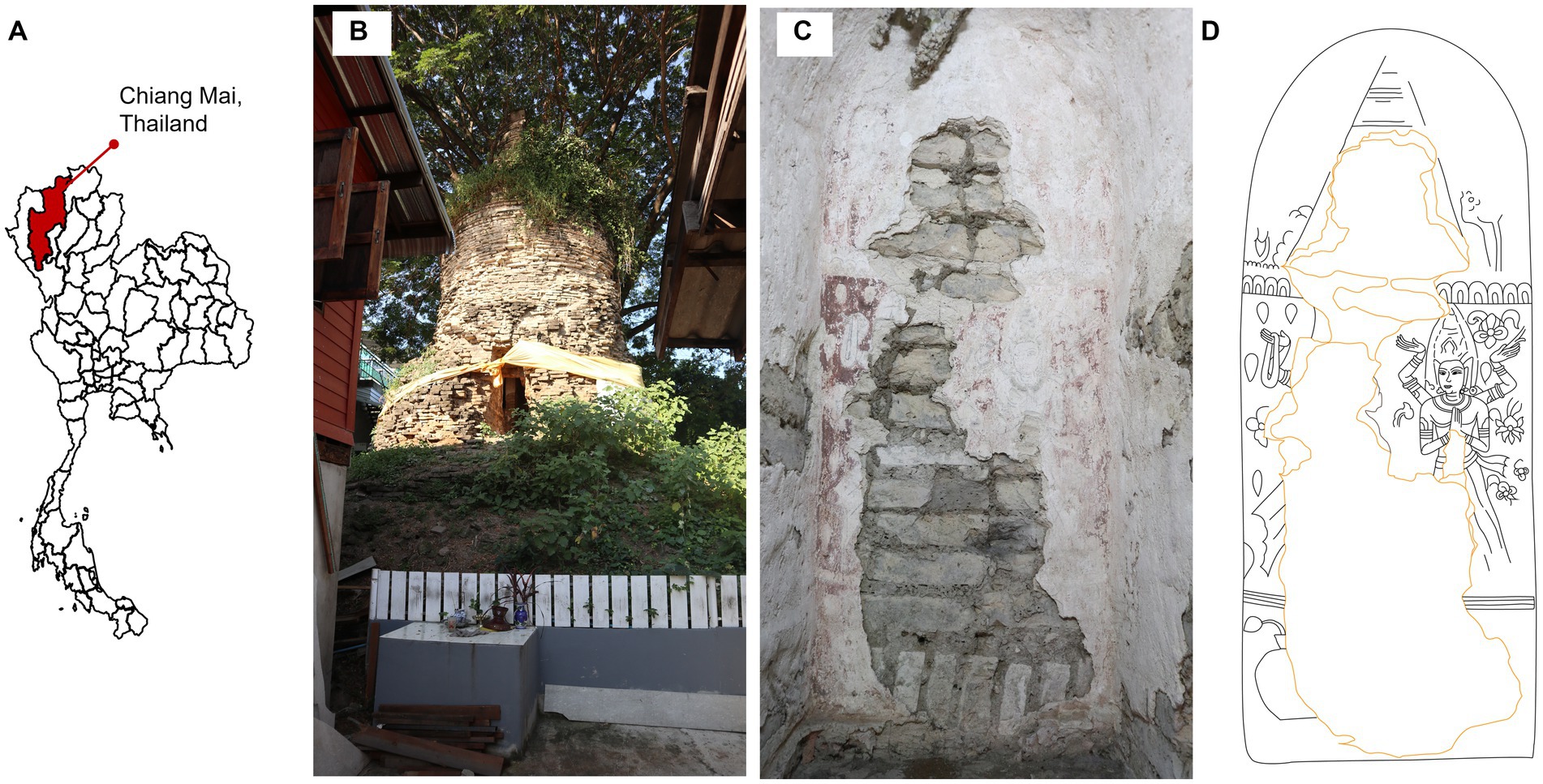
Figure 1. Study site. (A) Study site location, (B) landscape outside the pagoda, (C) overview of mural painting on the wall inside the pagoda tunnel and (D) skatch of the mural painting.The pagoda and mural photoes were taken by Thapakorn Khrueraya.
Powder X-ray diffraction (PXRD) (Rigaku SmartLab, Cu Kα, λ = 1.541 84 Å) was used to identify the crystalline phases in the well-grounded samples. The solid samples were collected and grounded in an agate mortar. The XRD patterns were collected in the range of 2θ = 10–60°, and then matched and indexed carefully using X’pert High Score Plus program. The PXRD patterns were drawn by Origin program.
Samples were taken in August 2020 using aseptic procedures. Six square plots (10 cm2) were randomly selected as representative samples from the painting. Three subsamples were collected from each plot using sterile swabs. The swabs from the same sampling points were collected in the same tube to make a composite sample. The composite samples were stored in DNA/RNA Shield (Zymo Research, Irvine, CA, United States) for microbial community analysis using a culture-independent method, and in 0.1% Tween 80 for microbial isolation using culture plates (Suphaphimol et al., 2022). At the sampling time, the temperature in the main hall was 29.8 ± 0.2°C with a relative humidity of 46.2% ± 2.4% and a light concentration of 8.4 ± 0.3 Lux. However, the painting surface was dry, due to the location of the painting deep in the tunnel, and the tunnel faces to the eastern side (Raining wind in Thailand normally flows from the southern part). Since algae did not appear on the painting, we omit algae from the investigation.
DNA was extracted from all samples collected from the mural paintings, using Quick-DNA Fecal/soil Microbe Microprep kit (Zymo Research, Irvine, CA, United States) following the manufacturer’s protocols. The DNA was then amplified on V3 – V4 region of 16S rRNA gene, using primer 341F (5’-CCTACGGGNGGCWGCAG-3′) and 805R (5’-GACTACHVGGGTA-TCTAATCC-3′), for both archaeal and bacterial study (Klindworth et al., 2013; Wasimuddin Schlaeppi et al., 2020) and ITS1 regions, using ITS1F (5’-CTTGGTCATTTAGGAAGTAA-3′) and ITS2R (5’-GCTGCGTTCTTCATCGATGC-3′), for fungal study (White et al., 1990). The amplicon was then sequenced using Illumina MiSeq platform (2 × 300 bp). The amplification and sequencing analysis were processed by Macrogen (Seoul, Korea). The raw sequence data of 16S rRNA and ITS gene were deposited in the National Center for Biotechnology Information (NCBI) under the accession number PRJNA838707.
The sequencing data were analyzed on QIIME2 software (Bolyen et al., 2019). Firstly, prokaryotic and fungal primers were removed using Cutadapt (Martin, 2011). The trimmed sequences were then quality-filtered, merged, and chimeras removed using DADA2 (Callahan et al., 2016). Amplicon sequence variants (ASVs) that contained less than 2 sequences were removed from the dataset to eliminate potential sequencing errors. The remaining data were taxonomically classified with the Silva database version 138 (Pruesse et al., 2007; Quast et al., 2013) for bacteria and UNITE database version 8.0 (Abarenkov et al., 2021) for fungi. Bacterial and fungal diversity indexes, including observed richness, Shannon and Simpson, were analyzed with “phyloseq” as implemented in R software version 4.2.2 (R Development Core, 2019).
The community data of microorganisms were subjected to a correlation analysis using the “Hmisc” package (Harrell and Dupont, 2023) in R software version 4.2.2., employing a correlation coefficient threshold of 0.8 and a significance level of 0.05. The resulting correlation coefficient data were visualized using Cytoscape version 3.9.1.
Samples collected for the culture-dependent method were shaken thoroughly and serially diluted by 0.85% (v/v) of NaCl to get the concentration ranging from 10−1 to 10−3. After that, 100 μL of each concentration was inoculated on tryptic soy agar (TSA) plates and potato dextrose agar (PDA) plates, for bacterial and fungal isolation, respectively. The culture plates were incubated at 30°C, under aseptic condition, for 24–72 h for bacteria and 3–5 d for fungi. Colonies showing different morphology were selected and transferred to fresh plates to obtain pure isolates.
Genomic DNA was extracted from bacterial and fungal pure isolates using PureLink Genomic DNA mini Kit (Invitrogen, Walthan, MA, United States), following the manufacture protocol. Specifically, the fungal pure cultures were prepared following the thermolysis method (Zhang et al., 2010) before subjected to DNA extraction kits. Briefly, fungal mycelia on a culture media were transferred into pure water in microcentrifuge tube, vortexed thoroughly, and centrifuged. The supernatant was then discarded and the remaining part was used for DNA extraction (Zhang et al., 2010). Bacterial and fungal DNA were amplified through PCR using the 27F (5’-AGAGTTTGATCMTGGCTCAG-3′) and 1492R (5’-TACGGYTACCTTGTTACGACT-3′) primer set for bacteria and the ITS5 (5′-GGAAGTAAAAGTCGT AACAAGG-3′) and ITS4 (5’-TCCTCCGCTTATTGATATGC-3′) primer set for fungi (Suphaphimol et al., 2022). The PCR conditions were used as previously described by Suphaphimol et al. (2022). All PCR products were sequenced with dideoxy method by Macrogen (Macrogen Inc., Seoul, Korea). The obtained sequences were then edited and aligned using Bioedit (version 7.1.1) (Hall, 1999). Taxonomic identification of each isolate was performed by comparing the sequences to those in the NCBI database using BLASTn. All sequences derived in this study were submitted to NCBI under the accession number as presented in Supplementary Table S1.
The ability of the isolates to survive under dry/drought conditions, which are the prevailing conditions in the pagoda tunnel, was tested using the sorbitol method (Hallsworth et al., 1998; Lasudee et al., 2018). Briefly, TSA and PDA media were prepared by mixing the media components with 0, 175, 285, 405, 520, 660, and 780 g/L of sorbitol to adjust the water activity of the media to 0.998, 0.976, 0.957, 0.919, 0.844, and 0.807, respectively. The microbial isolates were then inoculated onto the media. Isolates that grew on media with 0.919 aw or lower were considered drought-tolerant (Lasudee et al., 2018).
In this study, we used ferric oxide calcined and charcoal activated as the representatives of red and black colors found in the study site. The ability of microbial isolates to potentially decolorate colors on the mural painting were investigated using a modified method from Suphaphimol et al. (2022). In detail, 0.8% (w/v) of the colors were separately suspended in culture media. For bacteria, discoloration activities were tested by the well diffusion approach. Bacterial isolates were incubated in tryptic soy broth (TSB) for 24 h, and then the isolates were adjusted to 1.0 McFarland standard and placed in the agar well of TSA mixed with the tested colors. On the other hand, fungal isolates were incubated on PDA for 24 h, then harvested using a 7 mm cock borer and placed on a PDA plate mixed with the tested colors. Both bacterial and fungal isolates were incubated for about 3 and 5 d, respectively. An isolate producing a halo zone was considered as a potential isolate to decolorize the painting.
Since the main component of the mural wall is calcium compounds, in this study we test the ability of microbial isolates to solubilize insoluble calcium compounds. The solubilizations were determined using methods described by Fomina et al. (2005) and Kumla et al. (2014). Briefly, basal media were individually mixed with 0.3% (w/v) of calcium carbonate (CaCO3), calcium phosphate [Ca3(PO4)2], zinc carbonate (ZnCO3), feldspar, and kaolinite. Bacterial and fungal isolates were placed on the prepared media as described in 2.5.1 and incubated in darkness for 3 d (bacteria) and 5 d (fungi). Isolate producing a solubilization zone (halo zone) is considered as a potential isolate to degrade the painting layers.
Each of the isolates underwent testing to determine its capacity to induce the precipitation of calcium carbonate using the procedure described by Ma et al. (2020). Briefly, microbial isolates were incubated in a modified B4 medium, which included calcium acetate [(CH3COO)2Ca]. The incubated substrates were fragmented into smaller specimens and subsequently subjected to desiccation within a heated incubation apparatus for 72 h, prior to being coated with a layer of gold for investigating the formation of calcium crystals. Calcium precipitates were observed using a scanning electron microscope, JSM 5910 (JEOL, Akishima, Tokyo, Japan).
The PXRD is non-destructive and useful tool for phase identification and analyzing functional groups and chemical components in materials. The well-indexed PXRD patterns of the three different colored samples (Figure 2); (i) the white (background), (ii) the red, and (iii) the black; unambiguously reveal that the common and major construction materials are sand (Quartz, SiO2, 00–046-1,045) and limestone (Calcite, CaCO3, 00–005-0586). The colorless (white) SiO2 and CaCO3 were fine and commonly used as painting background materials. The characteristic peaks of Hematite (Fe2O3), the red painting materials, and some unidentified crystalline phases can be observed in PXRD patterns of the red and black samples. Note that the significantly high background in the PXRD pattern of the black sample indicates the amorphous phase composition, which could be both the organic and other carbonous painting materials.
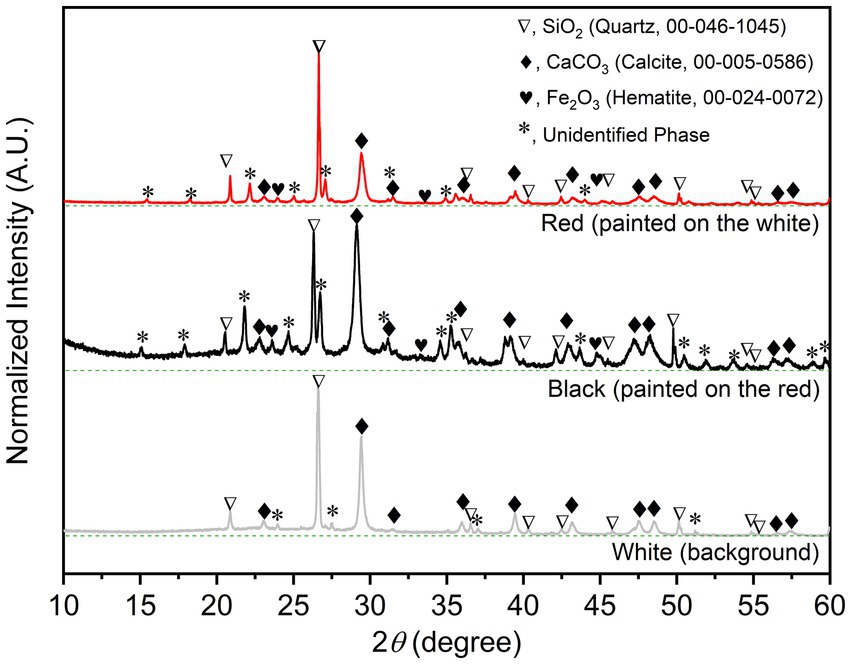
Figure 2. Indexed PXRD patterns with baselines (dashed green line) of the bulk solid samples of the wall-painting materials sampled from the three different areas.
Overall, a total of 417,264 reads, assigned to 205 prokaryotic ASVs (35 archaeal ASVs and 270 bacterial ASVs) and 631,710 reads, assigned 140 fungal ASVs, were obtained in this study. At the analysis sequencing depth, the rarefaction curves were gradually flattened, indicating that the obtained reads were sufficient to represent all microbes in the communities (Supplementary Figure S1). As shown in Table 1, bacteria are more diverse than fungi and archaea, accounting for two and three times higher than archaea and fungi, respectively.
For prokaryotic taxa, the relative abundance of archaea was much higher than that of bacteria, accounting for 84.58% (Figure 3A). Here, we detected two archaeal genera which are Halococcus and Halomarina. These two genera belonged to the phylum Euryarchaeota, Class Halobacteria, Order Halobacteriales, and Family Halococcaceae. The relative abundance and occurrence percentage of Halococcus are much higher than those of Halomarina, with more than 98% of relative abundance and about 74% of occurrence (Figures 3B,C). More diverse taxa were found in bacteria, compared to archaea. Fourteen phyla, 23 Classes, 50 orders and 106 genera were derived in this study. The most abundant phyla belonged to Actinomycetota (Actinobacteria), followed by Bacillota (Firmicutes), and Pseudomonadota (Proteobacteria). At the genus level, Crossiella obtained about 50% of total relative abundance and the occurrence percentage of this genus was 11.51%. Whilst Bacillus and Virgibacillus were found in a very low abundance (<1%), their occurrences were 6.23% and 5.86%, respectively (Figures 3D,E).
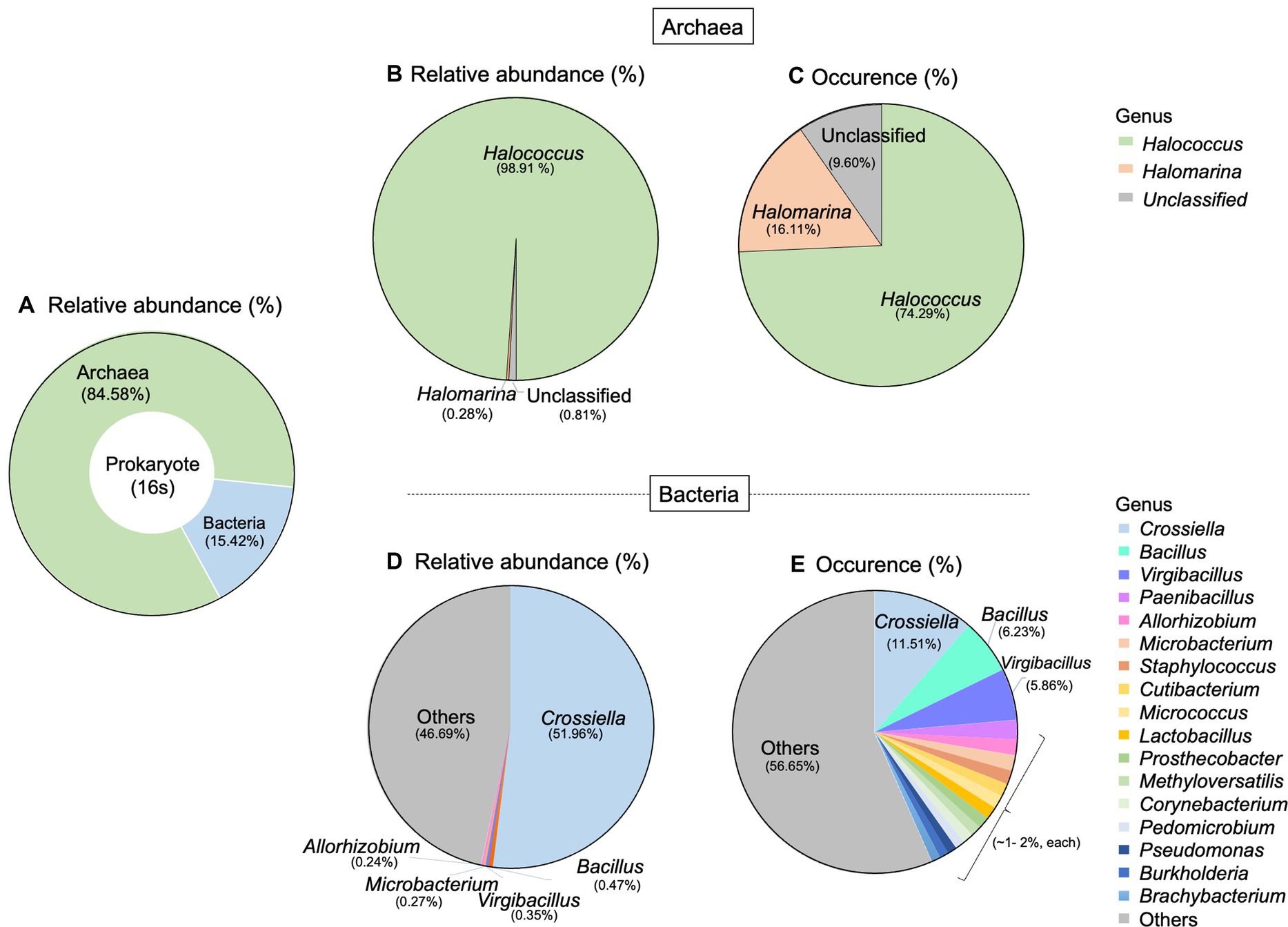
Figure 3. Taxonomic distribution of prokaryote associated with Lan Na’s mural painting. (A) The proportion of archaea and bacteria found in this study. (B) Relative abundance and (C) occurrence of archaea genera. (D) Relative abundance and (E) occurrence of bacterial genera. These data are average proportions across all samples.
Regarding fungi, 2 phyla, 13 classes, 25 orders, and 55 genera were detected. The most abundant phylum was Ascomycota (99.94%), and the other phylum was Basidiomycota (0.05%). At the genus level, Candida was the dominant genus in terms of both abundance (95.66%) and occurrence (34.13%). Daldinia had an abundance of 4.17% and an occurrence of 3.17%. Aspergillus, on the other hand, had a very low abundance, but it occurred in about 10.04% of the total fungal community (Figures 4A,B).
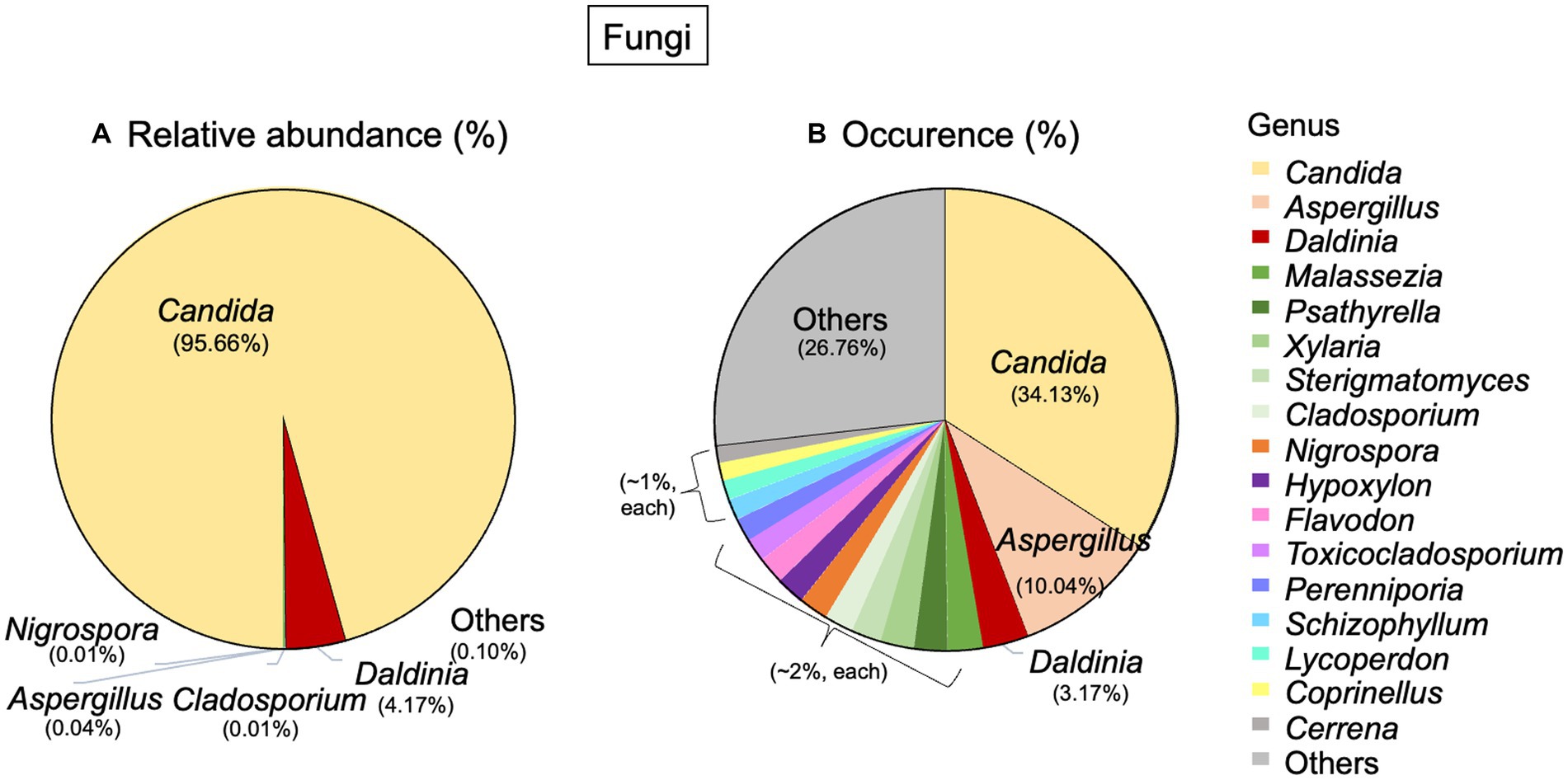
Figure 4. Taxonomic distribution of fungi associated with Lan Na’s mural painting. (A) Relative abundance and (B) occurrence of fungal genera. These data are average proportions across all samples.
The co-occurrence network analysis pattern of the microbial community present on the mural painting within the temple was examined by assessing its statistically significant correlations (Figure 5). The network exhibits both positive and negative correlations among distinct microorganism groups. The network encompasses 30 nodes and 103 edges, portraying the interactions between these microbial entities. Notably, Candida sp. and Halococcus sp. emerge as the predominant groups, displaying a noteworthy negative association with other bacterial groups. Furthermore, the network delineates the presence of three distinct sub-networks, as depicted in the accompanying figure.
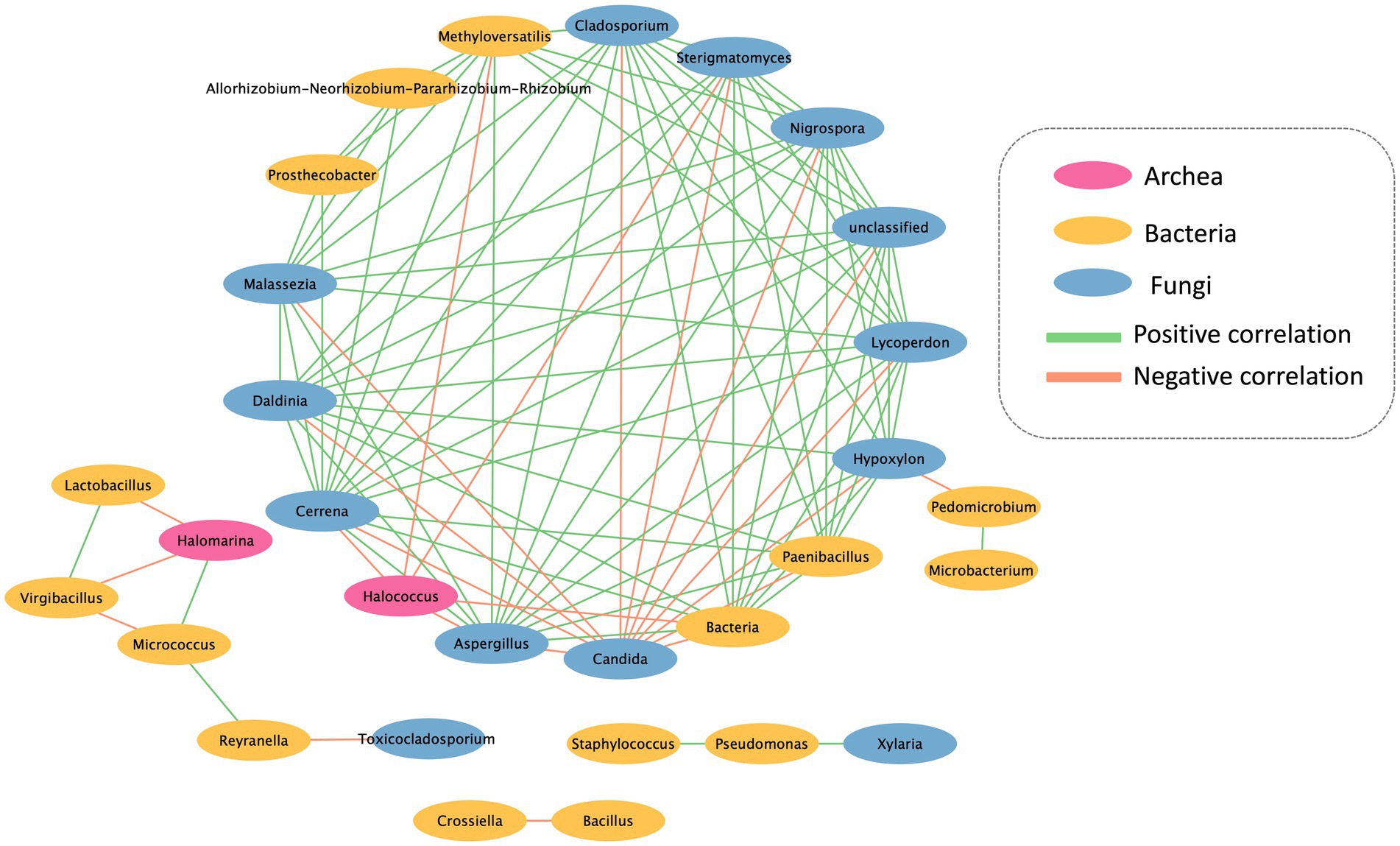
Figure 5. Interkingdom network between archaea, bacteria and fungi found in Lan Na’s mural painting.
Six bacterial and two fungal isolates were obtained from the mural painting samples in Sean Khan temple. As shown in Table 2, the bacterial isolates were identified as Staphylococcus epidermidis (SKB1, SKB2, and SKB5), Bacillus licheniformis (SKB3), Bacillus amyloliquefaciens (SKB4), and Burkholderia cepacia (SKB6), while the fungal isolates were identified as Trichaptum sp. (SKF1) and Aspergillus sp. (SKF2). When subjected to the isolated dry condition using the sorbital method, we found that almost all bacterial and fungal isolates, except for isolate Trichaptum sp. (SKF1), were able to survive in drought conditions (aw = 0.957 or higher) (Table 2). Sequences of all isolates were submitted to NCBI and the accession number of each isolate was presented in Supplementary Table S1.
Here, we tested the ability of microbial isolates to discolor calcined ferric oxide and activated charcoal. However, no isolates showed the ability to discolor them. In contrast, several isolates were capable of solubilizing calcium compounds. Bacterial isolates Staphylococcus epidermidis (SKB1, SKB5), Burkholderia cepacia (SKB6), and fungal isolate Trichaptum sp. (SKF1) and Aspergillus sp. (SKF2) were able to solubilize calcium carbonate, while only fungal isolate Trichaptum sp. (SKF1) and Aspergillus sp. (SKF2) was able to solubilize calcium phosphate. Almost all isolates, except for Burkholderia cepacian) (SKB6) and Trichaptum sp. (SKF1), were able to solubilize zinc carbonate (Table 2).
All microbial isolates were subjected to a culture medium with calcium compound to test the ability to participate in calcium precipitation or form crystals. As shown in Table 2 and Figure 6, no bacterial isolate was able to form the crystal structure, whereas fungal isolates Trichaptum sp. (SKF1) could form the crystal structure.
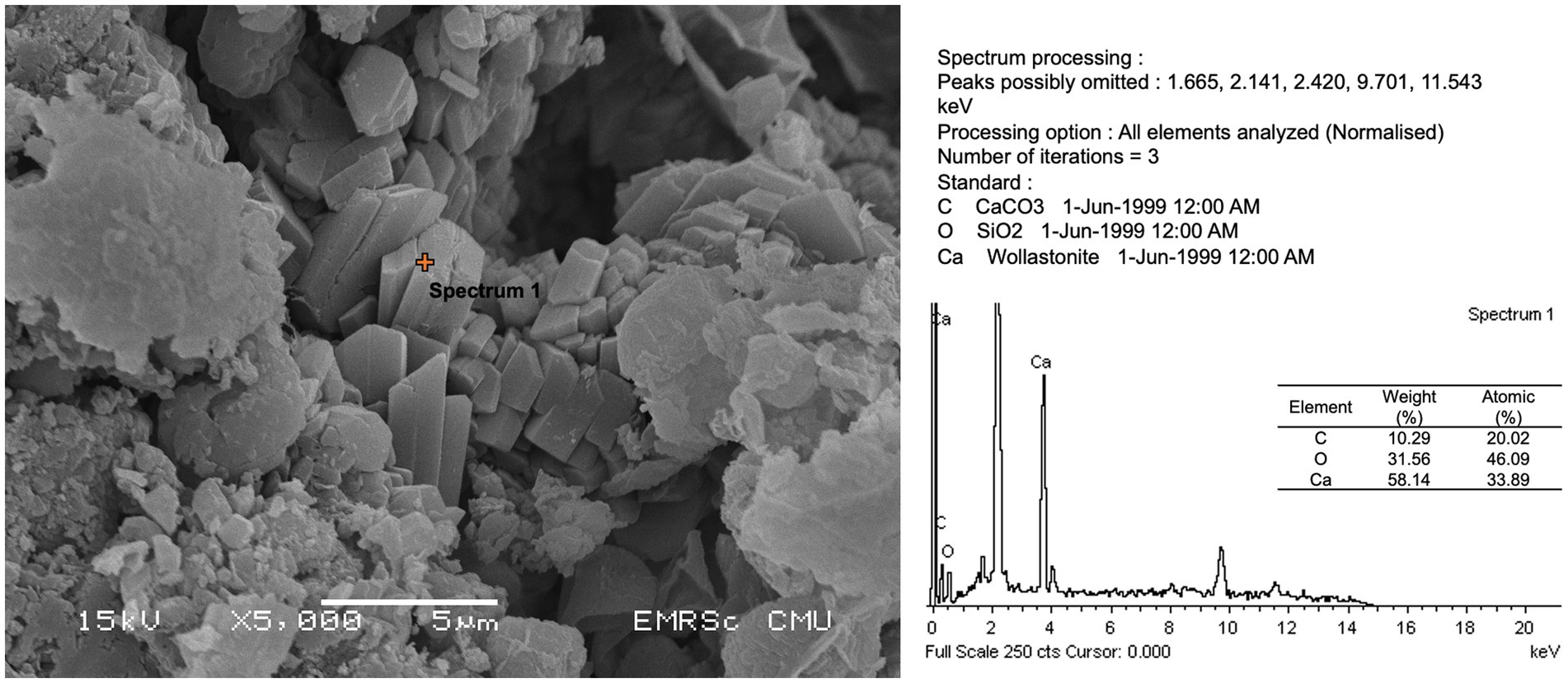
Figure 6. Calcium compound formation and energy-dispersive X-ray spectroscopy (EDS) analysis of isolate SKF1 (Trichaptum sp.).
The study of biodeterioration activities on mural paintings has been the central focus for decades. However, in northern Thailand, where a unique cultural style called “Lan Na” developed, little research has been conducted. This study was carried out at the Sean Khan temple in Chiang Mai, Thailand and was the first study on 13th-century Lan Na mural painting. We identified microbial communities, including archaea, bacteria, and fungi, associated with the paintings and determined the possible deterioration capacity of these microbes.
For prokaryotes, more than 80% of the total prokaryotic community belonged to archaea and about 15% were bacteria. This result was contradicted to the study on limestone walls of the old cathedral of Coimbra which found more diverse bacteria than archaea (Coelho et al., 2021). In the case of our study site, a neglected mural painting, water seepage into the painting substrate can result in the formation of salt efflorescence as the water evaporates (Amoroso and Fassina, 1983; Laiz et al., 2000). The accumulation of salts creates a high salt concentration environment, which in turn selects for halophiles. Our results demonstrate a significant proportion of haloarchaea within the archaeal communities. The most prevalent taxa were Halococcus, with a small portion of Halomarina, both of which are halophilic archaea. Archaea has been identified in several deteriorated ancient wall paintings (Rölleke et al., 1998; Piñar et al., 2001). Halococcus is typically found colonizing ancient wall paintings and is known to be a part of bacterioruberin-producing groups, which could potentially alter the color of frescoes (Piñar et al., 2001 Ettenauer et al., 2010, 2014; Coelho et al., 2021) (Table 3). Moreover, we found that Crossiella dominated the bacterial community. Crossiella has been identified as the predominant actinobacteria associated with salt efflorescence (Lepinay et al., 2018). This finding suggests that the prokaryotic communities inhabiting this Lanna painting have adapted to the specific conditions associated with salt efflorescence. Bacillus and Virgibacillus were the major members of the bacterial community. Bacillus and related genera (i.e., Paenibacillus) were also reported on mural paintings in Germany (Gorbushina et al., 2004), Spain (Jurado et al., 2021), and also in other Lan Na’s mural paintings in Thailand (Suphaphimol et al., 2022). However, the abundance of Bacillus is not new, as they are ubiquitous bacteria which can be found in any environment. On the other hand, the prevalent taxa dominated fungal communities was Candida, followed by Daldinia and Aspergillus. Candida was also reported on mural painting in Japan (Sugiyama et al., 2017) and discolored walls in Nigeria (Obidi and Okekunjo, 2017). Aspergillus, on the other hand, was generally found in mural paintings (Suphaphimol et al., 2022).
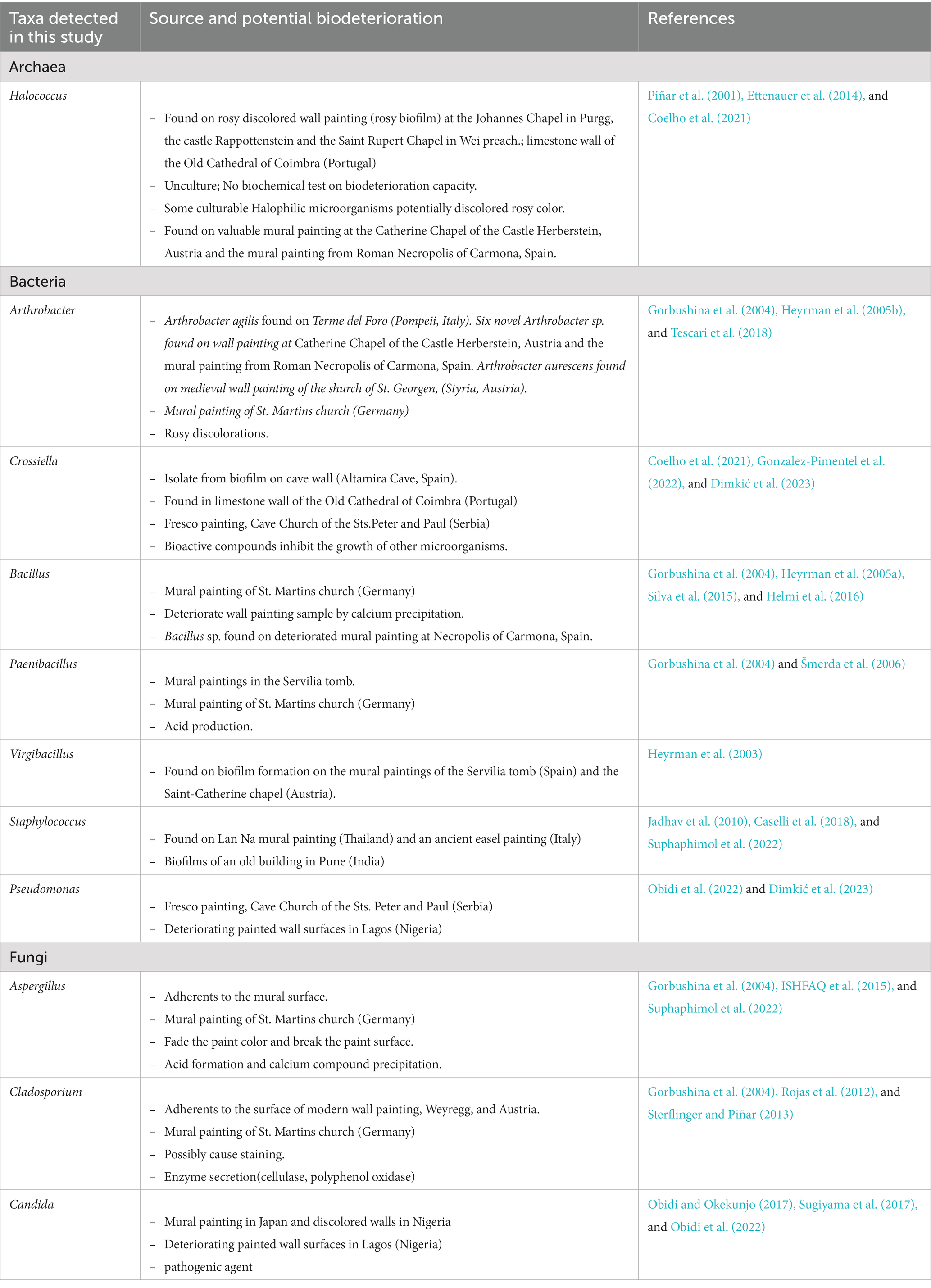
Table 3. Potential biodeterioration capacity of microbial taxa detected by the culture-independent method.
The interrelationships among microorganisms were investigated using network analysis. A majority of microorganisms displayed a positive correlation, indicating their potential to provide a favorable substrate and environment for the proliferation of other microorganisms. However, Candida sp. and Halococcus sp. exhibited a negative correlation with the other microorganisms. The colonization of Candida sp. was found to inhibit the growth of other microorganisms due to the production and secretion of proteinase or phospholipase enzymes, which can impact other organisms negatively (Pannanusorn et al., 2020). Furthermore, Halococcus sp. demonstrated the ability to generate halocins, which are protein-based antimicrobial substances. These halocins function by inhibiting the Na+ and H+ antiporter, leading to cell lysis and ultimately preventing the growth of other microorganisms (Kumar et al., 2021). However, Halomarina sp. exhibits a positive correlation with Micrococcus sp., while displaying a negative correlation with the remaining bacterial groups. Previous research indicates that both Microoccus sp. and Halomarina sp. possess the capability to synthesize carotenoids (Galiana et al., 2022), which may serve as antimicrobial agents against the other bacterial groups. Consequently, this leads to a negative correlation with Lactobacillus sp. and Virgibacillus sp. (Vargas-Sinisterra and Ramírez-Castrillón, 2021).
As shown in Table 3, Crossiella promotes calcite formation in caves and is very abundant in moonmilk (Martin-Pozas et al., 2022). Furthermore, it was reported that this genus potentially inhibits the growth of several bacteria and fungi such as Bacillus cereus, Staphylococcus aureus, Aspergillus versicolor, Penicillium chrysogenum, and Fusarium solani (Gonzalez-Pimentel et al., 2022). The abundance of this taxa may inhibit the growth of several taxa on the painting surface. Although the biodeterioration potentials of Candida has not been studied, several members of this genus were presented as pathogenic agent (Turner and Butler, 2014; Obidi and Okekunjo, 2017). It was suggested that the presence of this group with a large proportion might be from animals, insects and their fecal pellets and may be influenced by the presence of visitors, which contribute to the movement of airborne particles (Sugiyama et al., 2017; Zucconi et al., 2022). Aspergillus, on the other hand, showed biodeterioration activities. For example, Aspergillus fumigatus and Aspergillus piperis were able to produce acid and calcium compounds which potentially deteriorate the mural paintings (Suphaphimol et al., 2022). However, it should be noted that these are the predictive metabolic potential of microbial communities found in this study. These data provided only a general assessment. Empirical activity measurements are necessary for a more accurate representation. Conducting activity assessments through enrichment or co-culture approaches would allow for a closer capture of the true metabolic potential of microbial communities.
Culture dependent method could provide the information on biodeterioration potentials. The isolated cultures can be used for further investigation of the processes and mechanisms causing the biodeterioration of mural paintings. In this study, six bacteria and two fungal isolates were obtained and were identified as Staphylococcus epidermidis, Bacillus licheniformis, B. amyloliquefaciens, Burkholderia cepacia, Trichaptum sp., and Aspergillus sp. Staphylococcus sp. and Aspergillus sp. were found in the mural painting in many studies (Jadhav et al., 2010; Caselli et al., 2018; Suphaphimol et al., 2022; Zucconi et al., 2022). Burkholderia cepacia was reported in soil or water samples (Miller et al., 2002), while Trichaptum sp. was found in the environment as a saprophytic fungus (Gafforov et al., 2020). As compared to previous study, some isolates found in this study were different from those on another Lan Na mural painting in the same region (Suphaphimol et al., 2022). These may be caused by different abiotic factors, such as temperature, relative humidity and light radiation, and different mural materials. However, notice should be taken that small number of microbes were isolated and no halophilic archaea was cultured. This may be caused by the fact that only two culture media were used in this study. To captured more diverse microorganisms, diverse type of culture media should be used in further study. Specifically, studies on murals that have undergone severe deterioration should incorporate selective culture media, particularly those with high salt content, to enhance the isolation efforts.
The mural painting in this study was created using the fresco technique. The based materials are composed of SiO2 and CaCO3, whereas red color and black colors are hematite (Fe2O3) and carbonous painting materials, respectively. Although, almost all isolates were identified as drought stress tolerant microbes which were able to growth and survive in dry condition of the temple, no discoloration activities were found in all cultured isolates. Discoloration of this painting may be caused by the uncultured microbes, such as the halophilic archaea, other abiotic factors or the deterioration of the based materials. Here, we found that five out of eight isolates, including SKB1 (Staphylococcus epidermidis), SKB5 (S. epidermidis), SKB6 (Burkholderia cepacia), SKF1 (Trichaptum sp.), and SKF2 (Aspergillus sp.), were able to dissolve CaCO3, which found to be the base material of the Lan Na’s mural painting in this study. The dissolution of this calcium compound may result from secondary metabolite secreting from the microorganism which could cause severe damage to the paint layers (Čukovska et al., 2017). We further found that isolate SK1, Trichaptum sp., was able to form calcium crystals which could further crack the paint layer and deteriorate the painting. The calcium crystal formation is one of the factors that generally damage or stain to the mural painting (Pérez-Alonso et al., 2004; Nevin et al., 2008). Although, previous studies showed that Bacillus licheniformis can precipitate the calcium compound and several Bacillus species capable of carbonatogenesis which is useful for stone restoration (Vahabi et al., 2013; Jroundi et al., 2017). Our results show that Bacillus licheniformis and B. amyloliquefaciens did not have the ability to precipitate calcium carbonate and form calcium compound crystals. This may be cause by the fact that our isolation strategy may not have specifically targeted the strains of Bacillus capable of carbonatogenesis, which is why we were unable to find any isolates with such ability. In addition, we noticed that several microorganisms, especially bacteria (i.e., Staphylococcus spp. and Burkholderia sp.) and fungi (i.e., Trichaptum sp.), detected as biodeterioration agents in this study were found in low abundance (rare taxa). However, when the environmental factors changes, these rare taxa could be highly active and destroy the painting (Ciferri, 1999; Martin-Pozas et al., 2022). Thus far, avoiding the favorable conditions of these rare taxa would be recommended in order to preserve the painting. We suggest that future study should include the effects of environmental changes to monitor changes in the microbial community composition.
Here, we showed that the substrate components deterioration should be taken into consideration in addition to color deterioration. However, we noted that this study investigated some biodeterioration capacity and did not cover all possible biodeterioration capacity. Further study is needed to investigate on other aspects, especially those involved in biodegradation of binders, as Wu et al. (2022) presented that biodegradation of organic binders that had been used in paintings can be easily attacked by microorganisms and may lead to loss of pigment layers. Thus, binders’s deterioration should be taken into account for the study of biodeterioration of painting in future work.
This study is the first to report on the microbiome associated with a 13th-century Lan Na mural painting in Sean Khan Temple. Halophilic archaea, especially those belonging to Halococcus dominated the prokaryotic community whereas Candida dominated the fungal community. Based on the literature, these dominant species may play an important role in discoloration and suppression of other microorganisms. When it comes to cultural collections, no cultured isolates were found as discoloration agents. However, some of them were able to dissolve calcium carbonate, the base material of the painting (Staphylococcus epidermidis, Burkholderia cepacia, and Aspergillus sp.), and form the crystal of calcium compound (Trichaptum sp.) which could impact the deterioration of the painting. To preserve this mural painting, further study is needed to find a possible way to eliminate those microbial taxa reported as biodeterioration agents. Once the biodeterioration agents have been identified, it is necessary to evaluate their in vitro inhibition to control these microbes. This evaluation will then enable suggestions for future large-scale applications.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, PRJNA838707.
TD substantial contributions to the conception or design of the work. TD and WP supervised and received funding. TD and NSuw collected the samples. NSup, PN, TR, SY, and NSe performed the laboratory work. CS and NSup analyzed the results and wrote the original draft preparation. CS visualized data and performed bioinformatic analysis. SY, NSe, TK, NSuw, WP, and TD reviewed and edited the manuscript. All authors contributed to the article and approved the submitted version.
Open Access funding was enabled and organized by the Department of Soil Ecology, UFZ-Helmholtz Centre for Environmental Research.
TD and CS are supported by the post-doctoral fellowship 2022, office of research administration, Chiang Mai University, Thailand. The Department of Biology, Faculty of Science, Chiang Mai University is also acknowledged. We are grateful for the great assistance provided by Nuttapol Noirungsee and Sahutchai Inwongwan with the article.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1220901/full#supplementary-material
Abarenkov, K., Zirk, A., Piirmann, R., Ivanov, F., Nilsson, H., and Kõljalg, U. (2021). UNITE QIIME release for Fungi. UNITE Comm. doi: 10.15156/BIO/1264708
Amoroso, G. G., and Fassina, V. (1983). Stone Decay and Conservation: Atmospheric Pollution, Cleaning, Consolidation and Protection. New York: Amsterdam.
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Caselli, E., Pancaldi, S., Baldisserotto, C., Petrucci, F., Impallaria, A., Volpe, L., et al. (2018). Characterization of biodegradation in a 17th century easel painting and potential for a biological approach. PLoS One 13:e0207630. doi: 10.1371/journal.pone.0207630
Ciferri, O. (1999). Microbial degradation of paintings. Appl. Environ. Microbiol. 65, 879–885. doi: 10.1128/AEM.65.3.879-885.1999
Coelho, C., Mesquita, N., Costa, I., Soares, F., Trovão, J., Freitas, H., et al. (2021). Bacterial and archaeal structural diversity in several biodeterioration patterns on the limestone walls of the old cathedral of Coimbra. Microorganisms 9:709. doi: 10.3390/microorganisms9040709
Čukovska, L. R., Ivanova, T. Š., and Kokolanski, Ž. (2017). Analytical study of the XIV century wall painting and lime mortars in the “St. George” church in Staro Nagoričane, republic of Macedonia. Maced. J. Chem. Chem. Eng. 36, 41–58. doi: 10.20450/mjcce.2017.1070
Dahmani, J., Benharbit, M., Fassar, M., Hajila, R., Zidane, L., Magri, N., et al. (2020). Vascular plants census linked to the biodeterioration process of the Portuguese city of Mazagan in El Jadida, Morocco. J. King Saud. Univ. Sci. 32, 682–689. doi: 10.1016/j.jksus.2018.10.015
Dimkić, I., Ćopić, M., Petrović, M., Stupar, M., Savković, Ž., Knežević, A., et al. (2023). Bacteriobiota of the cave Church of Sts. Peter and Paul in Serbia—culturable and non-culturable communities’ assessment in the bioconservation potential of a peculiar Fresco painting. Int. J. Mol. Sci. 24:1016. doi: 10.3390/ijms24021016
Ettenauer, J., Sterflinger, K., and Piñar, G. (2010). Cultivation and molecular monitoring of halophilic microorganisms inhabiting an extreme environment presented by a salt-attacked monument. International Journal of Astrobiology 9, 59–72. doi: 10.1017/S1473550409990383
Ettenauer, J., Jurado, V., Pinar, G., Miller, A., Santner, M., Saiz-Jimenez, C., et al. (2014). Halophilic microorganisms are responsible for the rosy discolouration of saline environments in three historical buildings with mural paintings. PLoS One 9:e103844. doi: 10.1371/journal.pone.0103844
Fomina, M. A., Alexander, I. J., Colpaert, J. V., and Gadd, G. M. (2005). Solubilization of toxic metal minerals and metal tolerance of mycorrhizal fungi. Soil Biol. Biochem. 37, 851–866. doi: 10.1016/j.soilbio.2004.10.013
Gafforov, Y., Ordynets, A., Langer, E., Yarasheva, M., de Mello Gugliotta, A., Schigel, D., et al. (2020). Species diversity with comprehensive annotations of wood-inhabiting poroid and corticioid fungi in Uzbekistan. Front. Microbiol. 11:598321. doi: 10.3389/fmicb.2020.598321
Galiana, C. P., Miralles-Robledillo, J. M., Bernabeu, E., Harfi, N., and Martínez-Espinosa, R. M. (2022). “Chapter 9 – genetic and protein engineering of halophilic enzymes” in Extremozymes and their industrial applications. eds. N. K. Arora, S. Agnihotri, and J. Mishra (London, United Kingdom: Academic Press), 249–278.
Gonzalez-Pimentel, J. L., Dominguez-Moñino, I., Jurado, V., Laiz, L., Caldeira, A. T., and Saiz-Jimenez, C. (2022). The rare actinobacterium Crossiella sp. is a potential source of new bioactive compounds with activity against bacteria and fungi. Microorganisms 10:1575. doi: 10.3390/microorganisms10081575
Gorbushina, A., Heyrman, J., Dornieden, T., González-delValle, M. A., Krumbein, W., Laiz, L., et al. (2004). Bacterial and fungal diversity and biodeterioration problems in mural painting environments of St. Martins church (Greene–Kreiensen, Germany). Int. Biodeterior. Biodegrad. 53, 13–24. doi: 10.1016/j.ibiod.2003.07.003
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98.
Hallsworth, J. E., Nomura, Y., and Iwahara, M. (1998). Ethanol-induced water stress and fungal growth. J. Biosci. Bioeng. 86, 451–456. doi: 10.1016/S0922-338X(98)80150-5
Harrell, F., and Dupont, C. (2023). Hmisc: Harrell Miscellaneous. Available at: https://cran.r-project.org/web/packages/Hmisc/index.html (Accessed June 23, 2023).
Helmi, F. M., Elmitwalli, H. R., Elnagdy, S. M., and El-hagrassy, A. F. (2016). Biomineralization consolidation of Fresco wall paintings samples by Bacillus sphaericus. Geomicrobiol J. 33, 625–629. doi: 10.1080/01490451.2015.1067654
Heyrman, J., Logan, N. A., Busse, H.-J., Balcaen, A., Lebbe, L., Rodriguez-Diaz, M., et al. (2003). Virgibacillus carmonensis sp. nov., Virgibacillus necropolis sp. nov. and Virgibacillus picturae sp. nov., three novel species isolated from deteriorated mural paintings, transfer of the species of the genus salibacillus to Virgibacillus, as Virgibacillus marismortui comb. nov. and Virgibacillus salexigens comb. nov., and emended description of the genus Virgibacillus. Int. J. Syst. Evol. Microbiol. 53, 501–511. doi: 10.1099/ijs.0.02371-0
Heyrman, J., Logan, N. A., Rodríguez-Díaz, M., Scheldeman, P., Lebbe, L., Swings, J., et al. (2005a). Study of mural painting isolates, leading to the transfer of “Bacillus maroccanus” and “Bacillus carotarum” to Bacillus simplex, emended description of Bacillus simplex, re-examination of the strains previously attributed to “Bacillus macroides” and description of Bacillus muralis sp. nov. Int. J. Syst. Evol. Microbiol. 55, 119–131. doi: 10.1099/ijs.0.63221-0
Heyrman, J., Verbeeren, J., Schumann, P., Swings, J., and De Vos, P. (2005b). Six novel Arthrobacter species isolated from deteriorated mural paintings. Int. J. Syst. Evol. Microbiol. 55, 1457–1464. doi: 10.1099/ijs.0.63358-0
Ishfaq, S., Ali, N., Tauseef, I., Khattak, M. N. K., Shinwari, Z., and Ali, M. (2015). Analysis of paint degradation by fungal and bacterial species. Pak. J. Bot. 47, 753–760.
Jadhav, G., Salunkhe, D., Nerkar, D., and Bhadekar, R. (2010). Novel Staphylococcus sp. isolated from wall scrapings of a historical building in India. Ann. Microbiol. 60, 197–201. doi: 10.1007/s13213-010-0027-2
Jroundi, F., Schiro, M., Ruiz-Agudo, E., Elert, K., Martín-Sánchez, I., González-Muñoz, M. T., et al. (2017). Protection and consolidation of stone heritage by self-inoculation with indigenous carbonatogenic bacterial communities. Nat. Commun. 8:279. doi: 10.1038/s41467-017-00372-3
Jurado, V., Gonzalez-Pimentel, J. L., Hermosin, B., and Saiz-Jimenez, C. (2021). Biodeterioration of Salón de Reinos, Museo Nacional del Prado, Madrid. Spain. Appl. Sci. 11:8858. doi: 10.3390/app11198858
Karaca, Z., Öztürk, A., and Çolak, E. (2015). Biofouling of marbles by oxygenic photosynthetic microorganisms. Environ. Sci. Pollut. Res. 22, 11285–11289. doi: 10.1007/s11356-015-4366-1
Klindworth, A., Pruesse, E., Schweer, T., Peplies, J., Quast, C., Horn, M., et al. (2013). Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 41:e1. doi: 10.1093/nar/gks808
Kumar, V., Singh, B., van Belkum, M. J., Diep, D. B., Chikindas, M. L., Ermakov, A. M., et al. (2021). Halocins, natural antimicrobials of Archaea: exotic or special or both? Biotechnol. Adv. 53:107834. doi: 10.1016/j.biotechadv.2021.107834
Kumla, J., Suwannarach, N., Bussaban, B., Matsui, K., and Lumyong, S. (2014). Indole-3-acetic acid production, solubilization of insoluble metal minerals and metal tolerance of some sclerodermatoid fungi collected from northern Thailand. Ann. Microbiol. 64, 707–720. doi: 10.1007/s13213-013-0706-x
Laiz, L., Recio, D., Hermosin, B., and Saiz-Jimenez, C. (2000). “Microbial Communities in Salt Efflorescences,”in Of Microbes and Art: The Role of Microbial Communities in the Degradation and Protection of Cultural Heritage. eds. O. Ciferri, P. Tiano, and G. Mastromei (Boston, MA, US: Springer), 77–88. doi: 10.1007/978-1-4615-4239-1_6
Lasudee, K., Tokuyama, S., Lumyong, S., and Pathom-aree, W. (2018). Actinobacteria associated with arbuscular mycorrhizal funneliformis mosseae spores, taxonomic characterization and their beneficial traits to plants: evidence obtained from mung bean (Vigna radiata) and Thai jasmine Rice (Oryza sativa). Front. Microbiol. 9:1247. doi: 10.3389/fmicb.2018.01247
Lepinay, C., Mihajlovski, A., Touron, S., Seyer, D., Bousta, F., and Di Martino, P. (2018). Bacterial diversity associated with saline efflorescences damaging the walls of a French decorated prehistoric cave registered as a world cultural heritage site. Int. Biodeterior. 130, 55–64. doi: 10.1016/j.ibiod.2018.03.016
Ma, W., Wu, F., Tian, T., He, D., Zhang, Q., Gu, J.-D., et al. (2020). Fungal diversity and its contribution to the biodeterioration of mural paintings in two 1700-year-old tombs of China. Int. Biodeterior. Biodegrad. 152:104972. doi: 10.1016/j.ibiod.2020.104972
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12. doi: 10.14806/ej.17.1.200
Martin-Pozas, T., Cuezva, S., Fernandez-Cortes, A., Cañaveras, J. C., Benavente, D., Jurado, V., et al. (2022). Role of subterranean microbiota in the carbon cycle and greenhouse gas dynamics. Sci. Total Environ. 831:154921. doi: 10.1016/j.scitotenv.2022.154921
Mihajlovski, A., Gabarre, A., Seyer, D., Bousta, F., and Di Martino, P. (2017). Bacterial diversity on rock surface of the ruined part of a French historic monument: the Chaalis abbey. Int. Biodeterior. Biodegrad. 120, 161–169. doi: 10.1016/j.ibiod.2017.02.019
Miller, S. C. M., LiPuma, J. J., and Parke, J. L. (2002). Culture-based and non-growth-dependent detection of the Burkholderia cepacia complex in soil environments. Appl. Environ. Microbiol. 68, 3750–3758. doi: 10.1128/AEM.68.8.3750-3758.2002
Nevin, A., Melia, J. L., Osticioli, I., Gautier, G., and Colombini, M. P. (2008). The identification of copper oxalates in a 16th century Cypriot exterior wall painting using micro FTIR, micro Raman spectroscopy and gas chromatography-mass spectrometry. J. Cult. Herit. 9, 154–161. doi: 10.1016/j.culher.2007.10.002
Nuhoglu, Y., Oguz, E., Uslu, H., Ozbek, A., Ipekoglu, B., Ocak, I., et al. (2006). The accelerating effects of the microorganisms on biodeterioration of stone monuments under air pollution and continental-cold climatic conditions in Erzurum, Turkey. Sci. Total Environ. 364, 272–283. doi: 10.1016/j.scitotenv.2005.06.034
Obidi, O. F., Awe, O. O., Igwo-Ezikpe, M. N., and Okekunjo, F. O. (2022). Empirical analysis of amylolytic and proteolytic activities of microbial isolates recovered from deteriorating painted wall surfaces in Lagos Nigeria. Bio-Res. 20, 1484–1496. doi: 10.4314/br.v20i1.9
Obidi, O., and Okekunjo, F. (2017). Bacterial and fungal biodeterioration of discolored building paints in Lagos, Nigeria. World J. Microbiol. Biotechnol. 33:196. doi: 10.1007/s11274-017-2362-y
Pannanusorn, S., Nakkao, N., and Cholnakasem, N. (2020). Biofilm formation, proteinase and phospholipase activities of Candida parapsilosis isolated from environment. J. Sci. Tech. 9, 129–141. doi: 10.14456/tjst.2020.11
Penth, H. (2001). A brief history of Lanna: Northern Thailand from past to present. (Chiang Mai, Thailand: Silkworm Books).
Pérez-Alonso, M., Castro, K., Martinez-Arkarazo, I., Angulo, M., Olazabal, M. A., and Madariaga, J. M. (2004). Analysis of bulk and inorganic degradation products of stones, mortars and wall paintings by portable Raman microprobe spectroscopy. Anal. Bioanal. Chem. 379, 42–50. doi: 10.1007/s00216-004-2496-2
Piñar, G., Saiz-Jimenez, C., Schabereiter-Gurtner, C., Blanco-Varela, M. T., Lubitz, W., and Rölleke, S. (2001). Archaeal communities in two disparate deteriorated ancient wall paintings: detection, identification and temporal monitoring by denaturing gradient gel electrophoresis. FEMS Microbiol. Ecol. 37, 45–54. doi: 10.1111/j.1574-6941.2001.tb00852.x
Piñar, G., and Sterflinger, K. (2009). Microbes and building materials. Building Materials, Properties, Performance and Applications, Nova Science Publishers Inc. New York, 163–188.
Piovesan, R., Mazzoli, C., Maritan, L., and Cornale, P. (2012). Fresco and lime-paint: an experimental study and objective criteria for distinguishing between these painting techniques. Archaeometry 54, 723–736. doi: 10.1111/j.1475-4754.2011.00647.x
Pruesse, E., Quast, C., Knittel, K., Fuchs, B. M., Ludwig, W., Peplies, J., et al. (2007). SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35, 7188–7196. doi: 10.1093/nar/gkm864
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
R Development Core (2019). A language and environment for statistical computing. Computing 1. doi: 10.1890/0012-9658(2002)083[3097,CFHIWS]2.0.CO;2
Ravikumar, H. R. (2012). Biodegradation of paints: a current status. IJST 5, 1–11. doi: 10.17485/ijst/2012/v5i1.33
Rojas, T. I., Aira, M. J., Batista, A., Cruz, I. L., and González, S. (2012). Fungal biodeterioration in historic buildings of Havana (Cuba). Grana 51, 44–51. doi: 10.1080/00173134.2011.643920
Rölleke, S., Witte, A., Wanner, G., and Lubitz, Lubitz. (1998). Medieval wall paintings—a habitat for archaea: identification of archaea by denaturing gradient gel electrophoresis (DGGE) of PCR-amplified gene fragments coding for 16S rRNA in a medieval wall painting. International Biodeterioration & Biodegradation. 41, 85–92. doi: 10.1016/S0964-8305(98)80011-5
Rölleke, S., Muyzer, G., Wawer, C., Wanner, G., and Lubitz, W. (1996). Identification of bacteria in a biodegraded wall painting by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Appl. Environ. Microbiol. 62, 2059–2065. doi: 10.1128/aem.62.6.2059-2065.1996
Rosado, T., Martins, M. R., Pires, M., Mirao, J., Candeias, A., and Caldeira, A. (2013). Enzymatic monitorization of mural paintings biodegradation and biodeterioration. Int. J. Conserv. Sci. 4, 603–612.
Rosling, A., Roose, T., Herrmann, A. M., Davidson, F. A., Finlay, R. D., and Gadd, G. M. (2009). Approaches to modelling mineral weathering by fungi. Fungal Biol. Rev. 23, 138–144. doi: 10.1016/j.fbr.2009.09.003
Silva, M., Rosado, T., Teixeira, D., Candeias, A., and Caldeira, A. (2015). Production of green biocides for cultural heritage.Novel biotechnological solutions. Int. J. Conserv. Sci. 6, 519–530.
Silverman, H., and Ruggles, D. F. (2007). “Cultural heritage and human rights” in Cultural heritage and human rights. eds. H. Silverman and D. F. Ruggles (New York, NY: Springer New York), 3–29.
Skinner, H. C. W., and Ehrlich, H. (2014). “10.4 – Biomineralization” in Treatise on geochemistry. eds. H. D. Holland and K. K. Turekian. Second ed (Oxford: Elsevier), 105–162.
Šmerda, J., Sedláček, I., Páčová, Z., Krejčí, E., and Havel, L. (2006). Paenibacillus sepulcri sp. nov., isolated from biodeteriorated mural paintings in the Servilia tomb. Int. J. Syst. Evol. Microbiol. 56, 2341–2344. doi: 10.1099/ijs.0.64280-0
Sterflinger, K. (2000). Fungi as geologic agents. Geomicrobiol J. 17, 97–124. doi: 10.1080/01490450050023791
Sterflinger, K., and Piñar, G. (2013). Microbial deterioration of cultural heritage and works of art — tilting at windmills? Appl. Microbiol. Biotechnol. 97, 9637–9646. doi: 10.1007/s00253-013-5283-1
Sugiyama, J., Kiyuna, T., Nishijima, M., An, K.-D., Nagatsuka, Y., Tazato, N., et al. (2017). Polyphasic insights into the microbiomes of the Takamatsuzuka tumulus and Kitora tumulus. J. Gen. Appl. Microbiol. 63, 63–113. doi: 10.2323/jgam.2017.01.007
Suphaphimol, N., Suwannarach, N., Purahong, W., Jaikang, C., Pengpat, K., Semakul, N., et al. (2022). Identification of microorganisms dwelling on the 19th century lanna mural paintings from northern Thailand using culture-dependent and -independent approaches. Biology 11:228. doi: 10.3390/biology11020228
Tescari, M., Frangipani, E., Caneva, G., Casanova Municchia, A., Sodo, A., and Visca, P. (2018). Arthrobacter agilis and rosy discoloration in “Terme del Foro” (Pompeii, Italy). Int. Biodeterior. Biodegrad. 130, 48–54. doi: 10.1016/j.ibiod.2018.03.015
Tiano, P. (2002). “Biodegradation of cultural heritage: decay mechanisms and control methods” in Proceedings of the 9th ARIADNE Workshop “Historic Material and Their Diagnostic,(Prague, Czech Republic: ARCCHIP). 7–12.
Turner, S. A., and Butler, G. (2014). The Candida pathogenic species complex. Cold Spring Harb. Perspect. Med. 4:a019778. doi: 10.1101/cshperspect.a019778
Vahabi, A., Ramezanianpour, A., Sharafi, H., Zahiri, H., Vali, H., and Noghabi, K. (2013). Calcium carbonate precipitation by strain Bacillus licheniformis AK01, newly isolated from loamy soil: a promising alternative for sealing cement-based materials. J. Basic Microbiol. 55, 105–111. doi: 10.1002/jobm.201300560
Vargas-Sinisterra, A. F., and Ramírez-Castrillón, M. (2021). Yeast carotenoids: production and activity as antimicrobial biomolecule. Arch. Microbiol. 203, 873–888. doi: 10.1007/s00203-020-02111-7
Warscheid, T., and Braams, J. (2000). Biodeterioration of stone: a review. Int. Biodeterior. Biodegrad. 46, 343–368. doi: 10.1016/S0964-8305(00)00109-8
Wasimuddin Schlaeppi, K., Ronchi, F., Leib, S. L., Erb, M., and Ramette, A. (2020). Evaluation of primer pairs for microbiome profiling from soils to humans within the one health framework. Mol. Ecol. Resour. 20, 1558–1571. doi: 10.1111/1755-0998.13215
White, T., Bruns, T., Lee, S., Taylor, J., Innis, M., Gelfand, D., et al. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics” in Pcr Protocols: a Guide to Methods and Applications, eds. M. A Innis, J. J. Gelfand, D. H. Sninsky, and T. J. White (London, United Kingdom: Academic Press), 315–322.
Wiktor, V., De Leo, F., Urzì, C., Guyonnet, R., Grosseau, P., and Garcia-Diaz, E. (2009). Accelerated laboratory test to study fungal biodeterioration of cementitious matrix. Int. Biodeterior. Biodegrad. 63, 1061–1065. doi: 10.1016/j.ibiod.2009.09.004
Wu, F., Gu, J. D., Li, J., Feng, H., and Wang, W. (2022). “Microbial colonization and protective management of wall paintings” in Cultural heritage microbiology: recent developments (1 Birdcage Walk London SW1H 9JJ: Archetype Publications), 57–84.
Zhang, Y. J., Zhang, S., Liu, X. Z., Wen, H. A., and Wang, M. (2010). A simple method of genomic DNA extraction suitable for analysis of bulk fungal strains. Lett. Appl. Microbiol. 51, 114–118. doi: 10.1111/j.1472-765X.2010.02867.x
Keywords: biodeterioration of wall painting, deterioration of cultural heritage, microbiome associated with mural paintings, crystal formation on painting, biomineralization
Citation: Sansupa C, Suphaphimol N, Nonthijun P, Ronsuek T, Yimklan S, Semakul N, Khrueraya T, Suwannarach N, Purahong W and Disayathanoowat T (2023) Life on the wall: the diversity and activity of microbes on 13th – century AD. Lan Na mural painting. Front. Microbiol. 14:1220901. doi: 10.3389/fmicb.2023.1220901
Received: 11 May 2023; Accepted: 31 July 2023;
Published: 15 September 2023.
Edited by:
Luis Raul Comolli, Independent Researcher, Basel, SwitzerlandReviewed by:
Fadwa Jroundi, University of Granada, SpainCopyright © 2023 Sansupa, Suphaphimol, Nonthijun, Ronsuek, Yimklan, Semakul, Khrueraya, Suwannarach, Purahong and Disayathanoowat. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Witoon Purahong, d2l0b29uLnB1cmFob25nQHVmei5kZQ==; Terd Disayathanoowat, VGVyZC5kaXNAY211LmFjLnRo
†These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.