- 1National Key Laboratory for Germplasm Innovation and Utilization of Horticultural Crops, Zhengzhou Fruit Research Institute, Chinese Academy of Agricultural Sciences, Zhengzhou, China
- 2National Key Laboratory for Germplasm Innovation and Utilization for Fruit and Vegetable Horticultural Crops, College of Plant Science and Technology, Huazhong Agricultural University, Wuhan, China
Fruit rot caused by Colletotrichum magnum is a crucial watermelon disease threatening the production and quality. To understand the pathogenic mechanism of C. magnum, we optimized the Agrobacterium tumefaciens-mediated transformation system (ATMT) for genetic transformation of C. magnum. The transformation efficiency of ATMT was an average of around 245 transformants per 100 million conidia. Southern blot analysis indicated that approximately 75% of the mutants contained a single copy of T-DNA. Pathogenicity test revealed that three mutants completely lost pathogenicity. The T-DNA integration sites (TISs) of three mutants were Identified. In mutant Cm699, the TISs were found in the intron region of the gene, which encoded a protein containing AP-2 complex subunit σ, and simultaneous gene deletions were observed. Two deleted genes encoded the transcription initiation protein SPT3 and a hypothetical protein, respectively. In mutant Cm854, the TISs were found in the 5′-flanking regions of a gene that was similar to the MYO5 encoding Myosin I of Pyricularia oryzae (78%). In mutant Cm1078, the T-DNA was integrated into the exon regions of two adjacent genes. One was 5′-3′ exoribonuclease 1 encoding gene while the other encoded a WD-repeat protein retinoblastoma binding protein 4, the homolog of the MSl1 of Saccharomyces cerevisiae.
Introduction
Colletotrichum magnum, is one of the major pathogens causing watermelon anthracnose (Rossman et al., 2016; Damm et al., 2019; Guo et al., 2022). Recently, it was reported on Cucumis sativus, Capsicum spp., Lagenaria siceraria, Lobelia chinensis, and Luffa cylindrica in China (Tsay et al., 2010; Li et al., 2013; Liu F. et al., 2022). C. magum, whoes conidia were larger than those of other Colletotrichum species, belongs to C. magum species complex (Damm et al., 2019). When Colletotrichum species infect the host, they establish biotrophic and necrotrophic lifestyles (Munch et al., 2008). To colonize host tissues, pathogens of Colletotrichum species generally form appressoria which penetrate the epidermis of the host using penetration pegs. The formation of appressoria usually begins with the germination of conidia (Tucker and Talbot, 2001). In the biotrophic phase of the pathogen, the primary hyphae infect the epidermal and mesophyll cells and do not cause macroscopically visible damage to the host (De Silva et al., 2017). Subsequently, when differentiation of the primary hyphae produces secondary hyphae that spread throughout host tissue, the necrotrophic infection phase occurs (Wharton et al., 2001; Moraes et al., 2015).
The taxonomy, population structure, and disease epidemiology of C. magnum have been extensively studied (Damm et al., 2019). However, little is known about the pathogenicity of C. magnum. Agrobacterium tumefaciens-mediated transformation (ATMT) allows the study of numerous fungi to discover candidate pathogenic genes (Tsuji et al., 2003; Lawrence et al., 2010; Zhang et al., 2013; Martinez-Cruz et al., 2017; Li et al., 2019; Binh et al., 2021; Chauhan et al., 2021). Compared to conventional transformation methods, this approach provides a high percentage of T-DNA single-copy integration (De Groot et al., 1998; Münch et al., 2011) and is suitable for random insertional mutagenesis. The T-DNA integration sites (TISs) of transformants labeled with T-DNA can be easily identified (Liu et al., 1995; Sun et al., 2019).
The molecular determinants of fungal pathogenicity remain to be clarified and require more attention. Depending on the method of plant infection and the genome size of the fungus, fungi presumably containing 60–360 virulence genes (Idnurm and Howlett, 2001). Some genes that play important roles in signal cascades, cell wall degradation, the formation of infection structures, respond to the host environment, participat in the synthesis of toxins, and avoid and overcom plant defenses, have been identified as virulence or pathogenicity genes (Idnurm and Howlett, 2001; Werner et al., 2007; Gong et al., 2022; Jiao et al., 2022). To form an infection structure, fungi secrete effector molecules, such as the mutant of C. higginsianum is deficient in dihydroxynaphthalene melanin metabolism and is unable to form mature and infectious appressoria, thereby exhibiting reduced pathogenicity (Liu et al., 2013). In C. gloeosporioides and Magnaporthe grisea, the protein encoded by the protein kinase A is necessary for the mature appressorium to mediated plant infection (Gilbert et al., 2006; Cai et al., 2013). In C. lindemuthianum, the transcriptional activator gene (CLTA1) is disrupted, and mutant H433 cannot form necrotrophic secondary hyphae (Dufresne et al., 2000). Furthermore, allantoicase genes may participate in appressorium formation in C. graminicola (Münch et al., 2011).
In the present study, we established a stable ATMT protocol for C. magnum and generated a library of hygromycin B transformants. In the virulence assays on leaves, three mutants with defective pathogenicity were generated. The TISs of the three mutants were identified. Six putative pathogenic genes were identified.
Materials and methods
Fungus and plasmid preparation
The C. magnum wild-type (WT) strain CAASZK4 was isolated from watermelons in Kaifeng City, Henan Province, China (Guo et al., 2022). The strain AGL-1 of A tumefaciens, containing plasmid pATMT1 which carries a hygromycin B resistance cassette, was used as a T-DNA donor for fungal transformation (Zheng et al., 2011; Li et al., 2019).
ATMT of Colletotrichum magnum
The WT strain (CAASZK4) were grown on potato dextrose agar (PDA) medium complemented with a range of hygromycin B concentrations (0, 20, 40, 60, 80, 100, 150, 200, and 250 mg/L) and were incubated at 27°C in darkness for 7 d.
ATMT-based transformation of C. magnum was performed according to the protocol described by Cai et al. with slight modifications (Cai et al., 2013). The AGL-1 strain grew at 28°C in 5 mL of Luria–Bertani (LB) medium supplemented 25 mg/L rifampicin and 50 mg/L kanamycin. Cultures were centrifuged again. Finally, the cells were resuspended in induction medium (IM, Bundock et al., 1995) supplemented with acetosyringone (AS, 200 μM), and incubated for 6 h to achieve OD600 nm values (0.6–0.8).
C. magnum grew on synthetic nutrient-poor agar medium (SNA, Nirenberg, 1976) for 25 d at 27°C in the dark. Subsequently, the conidia of C. magnum were cleaned thrice sterile water and diluted in IM to achieve different concentrations (1 × 106–1 × 108 spores/ml). Aliquots of 100 μL of bacterial culture and 100 μL of conidial suspension were plated onto the 0.45 μm pore nitrocellulose filter of the co-cultivation medium (CM) containing 200 μM AS. After cocultivation for 84 h at 23°C in the dark, the membranes were transferred to PDA containing 200 mg/L cefotaxime, 100 mg/L timentin, and 80 mg/L hygromycin B, and cultured for 18 d at 27°C in the dark. Mycelial plugs from the edges where fresh mycelia were cultured on PDA containing 80 mg/L hygromycin B were used to confirm resistance. To determine the mitotic stability of the transformants, 40 randomly selected transformants were cultured on PDA without hygromycin B for four generations and then transferred back to PDA with hygromycin B.
DNA extraction and sequence analysis
The transformants and the WT strains of C. magnum were transferred to 100 mL fresh liquid PDA and cultured for 20 d at 27°C in the dark. Genomic DNA of 40 transformants and the WT strains of C. magnum were extracted by CTAB (Sangon Biotech, China).
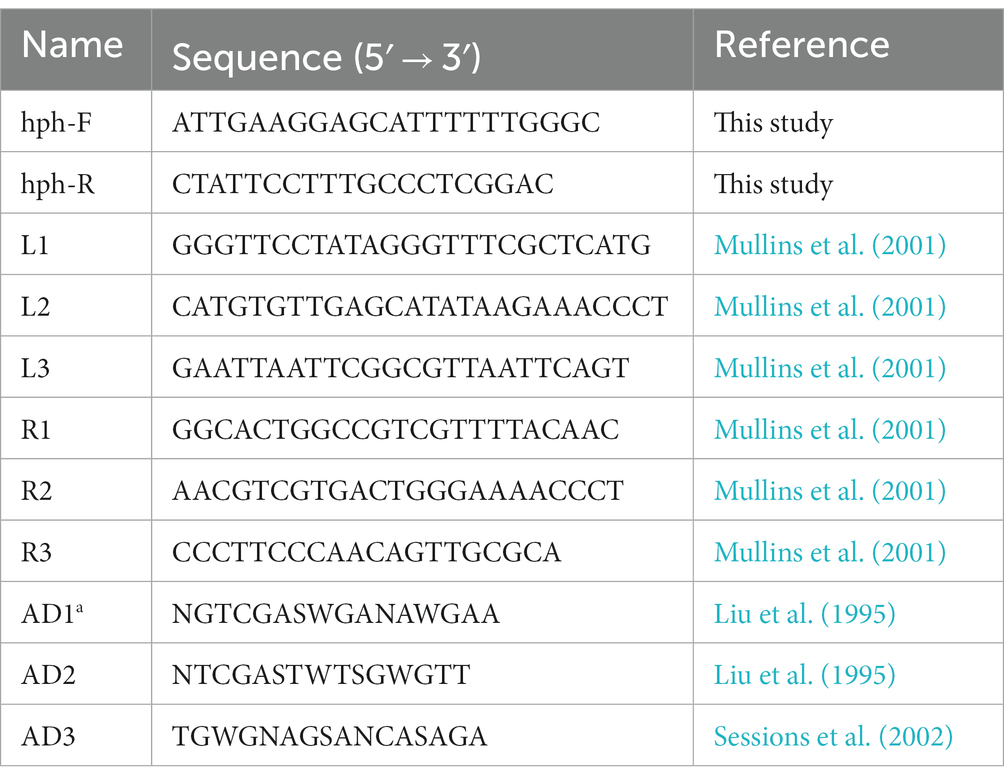
To analyze genome integration of the transferred genes, the hph gene was amplified using the primer pair hph-F + hph-R (Table 1) and sequenced. PCR was were performed according to the protocol described by Guo et al. with slight modifications (Guo et al., 2022). The annealing temperature of hph was 60°C. PCR amplicons were purified and sequenced by Sangon Biotech Company (Shanghai, China).
The digestion of genomic DNA from 40 transformants or WT strains with EcoRI and HindIII (NEB, Ipswich, United States), electrophoresised on 0.75% agarose, were depurated, and transferred to nylon membranes (Hybond-N+; General Electric Company, Boston, United States). In the hybridization experiments, the digoxigenin (DIG) labeled probe corresponded to 1,380 bp hph of T-DNA from plasmid pATMT1 (Figure 1), and the hph gene was amplified using the primers hph-F and hph-R. Hybridization and chemiluminescence detection of the hybridized Dig-labeled probes were performed using the DIG High Prime DNA Labeling and Detection Starter Kit (Roche, Germany).
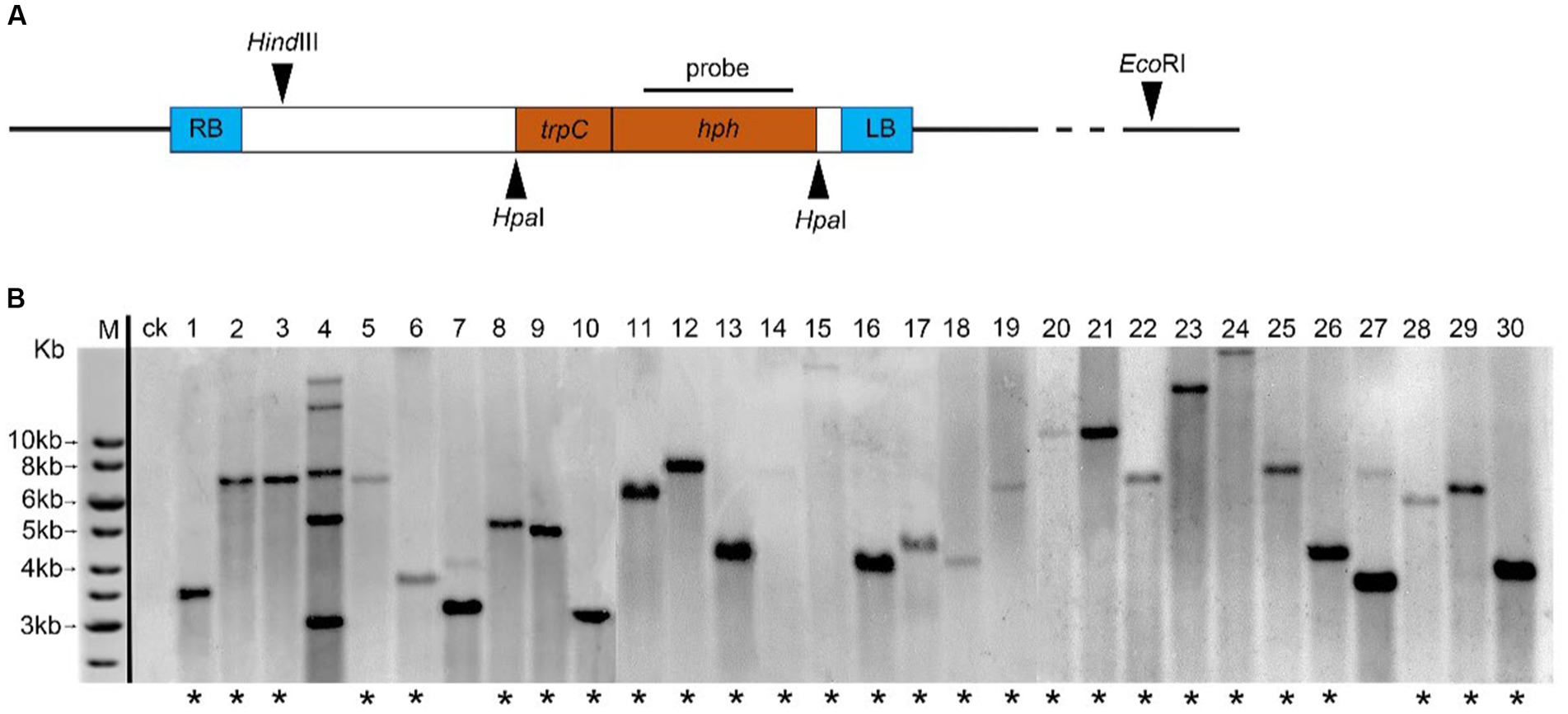
Figure 1. Analysis of T-DNA copy number in C. magnum transformants generated by Agrobacterium tumefaciens-mediated transformation (ATMT). (A) Probe used for hybridization, HindIII and EcoRI restriction sites are indicated; (B) Genomic Southern blot analysis of 30 transformants of C. magnum (1–30) generated by ATMT. Genomic DNA was restricted with HindIII and EcoRI. One band (lanes marked with an asterisk) represents a single T-DNA integration event 20, 22, and 28 represent genomic Southern blot analysis of Cm699, Cm854, and Cm1078, respectively.
Pathogenicity assay and morphological characterization of transformants
Mycelial plugs (5 mm) from 1,460 transformants and WT strains of C. magnum were cultured in PDA at 27°C in the dark for 5–15 d. The pathogenicity of these transformants and WT strain was evaluated on healthy detached watermelon leaves (Citrullus lanatus cv. Hongheping, 5–6 true leaves), which were previously washed with tap water, surface-sterilized with 75% ethanol for 30 s, and rinsed thrice with sterile water. Mycelial discs (5 mm in diameter) were individually placed on the right half of the leaves. PDA blocks (5 mm in diameter) were placed on the left half of the same leaves as controls. All leaves were incubated in sealed sterile plastic plates at 27°C in the dark. Leaf symptoms were assessed at 4 d post-inoculation. All transformants and WT strains were tested in triplicates.
Mycelial plugs (5 mm) from transformants and WT strains were transferred on 60 mm Petri dishes containing SNA medium, and incubated in the dark at 27°C for 25 d. The mycelia, conidia, and appressoria were observed under a light microscopy (Olympus BX51, Japan). The conidia formed in 60-mm Petri dishes were cleaned thrice with sterile water and resuspended in 1 mL sterile water. The number of conidia was calculated using a hemocytometer. To induce the formation of appressoria, the conidia were suspended in 1% glucose and incubated at 27°C in the dark. After 48 h, the germination rate of the conidia was measured. All transformants and WT strains were tested in triplicates.
Cloning and sequencing of flanking T-DNA sequences
The T-DNA flanking sequences inserted into the genome were cloned using thermal asymmetric interlaced PCR (TAIL-PCR) protocol with minor modification (Liu et al., 1995; Mullins et al., 2001). Right border primers (R1, R2, and R3) and left border primers (L1, L2, and L3) and arbitrary degenerate primers (AD1, AD2, and AD-3) were utilized for TAIL-PCR (Table 1). PCR conditions were set as described by Mullins and Sessions (Mullins et al., 2001; Sessions et al., 2002), with minor modifications (Supplementary Table S1). The third TAIL-PCR products of all transformants displaying the highest brightness were purified using the FastPure Gel DNA Extraction Mini Kit (Vazyme, China). This fragment was ligated to the vector pTOPO-Blunt, which was transferred into Escherichia coli strain Top10 using a CV16-Zero Background pTOPO-Blunt Cloning Kit (Aidlab, China). E. coli harboring the pTOPO-Blunt vector was sequenced by Sangon Biotech Company (Shanghai, China). To isolate the tagged genes, the sequences flanking the T-DNA of each transformant were used to search the local genome database of WT C. magnum by BLAST+.1 Gene structure was predicted using the C. fioriniae, C. gloeosporioides, C. graminicola, C. higginsianum, C. orbiculare, and C. sublineola by the FGENESH program.2
Whole-genome resequencing and analyses
Four micrograms of high-quality genomic DNA from three transformants (Cm699, Cm854, and Cm1078) were extracted from fresh mycelia using CTAB (Sangon Biotech, China) and used to construct a sequencing library, following the manufacturer’s instructions (Illumina Inc.). Paired-end sequencing libraries with an insert size of approximately 200 or 400 bp were sequenced using an Illumina HiSeq 2,500, NovaSeq 6,000, or MiSeq sequencer, with a reading length of 150 bp. In total, more than 6 GB of sequence data were generated for each transformant. The upstream and downstream genome sequences (500 bp) of the TISs were obtained using a comprehensive approach called TDNAscan3 searches against the genome database of WT C. magnum CAASZK4 (Sun et al., 2019). Using the GFF3 file of WT C. magnum as a reference genome organism, TDNAscan annotated all identified T-DNA insertions. Sequence homology searches were performed using NCBI protein database.4
Primers were designed to validate the TISs, T-DNA integration patterns, and deleted genes of Cm699, (Supplementary Table S2).
Results
Establishing a transformant library of Colletotrichum magnum using ATMT
To determine the hygromycin B sensitivity of C. magnum, the CAASZK4 strain was incubated on PDA supplemented with various concentrations of hygromycin B. These results indicate that the growth of C. magnum was inhibited by 60 mg/L hygromycin (Supplementary Figure S1). To exclude the possibility of false-positive transformants, 70 mg/L was chosen for the subsequent selection of resistant transformants.
Using the modified ATMT protocol, we obtained a library containing 1,460 transformants of the watermelon pathogen C. magnum. On an average, 245 transformants were generated from 108 conidia. Analysis of 40 randomly selected transformants harboring hygromycin B resistance revealed that all 40 transformants showed mitotically stable integrated T-DNA. These transformants, which were successively cultured for four generations on PDA medium without the selection marker hygromycin B, did not result in the loss of integrated T-DNA, as shown by the ability of all 40 transformants to grow on the screening medium containing hygromycin B and hph gene amplification analysis (Supplementary Figures S2, S3).
T-DNA copy number variation of transformants
To effectively identify the T-DNA integration events in the mutant library, 40 randomly selected transformants and WT strains were subjected to genomic Southern blot hybridization. Genomic DNA digested using EcoRI and HindIII was detected with the digoxigenin (Dig)-labeled probe harboring 1,380 bp hph gene (Figure 1A). Of the tested transformants, 30 displayed single-site TISs, nine harbored two TISs, and one harbored more than two TISs. Representative selections of 30 transformants are shown in Figure 1B.
Identification of transformants with reduced pathogenicity
In total 1,460 transformants were analyzed for pathogenicity on detached leaves. Fourteen transformants were screened out for their strongly reduced virulence and further identified and analyzed on watermelon leaves. The three transformants of the 14 strains exhibited strongly reduced virulence in leaves assays. Interestingly, the three transformants (Cm699, Cm854, and Cm1078) caused no visible disease symptoms in watermelon leaves (Figure 2).
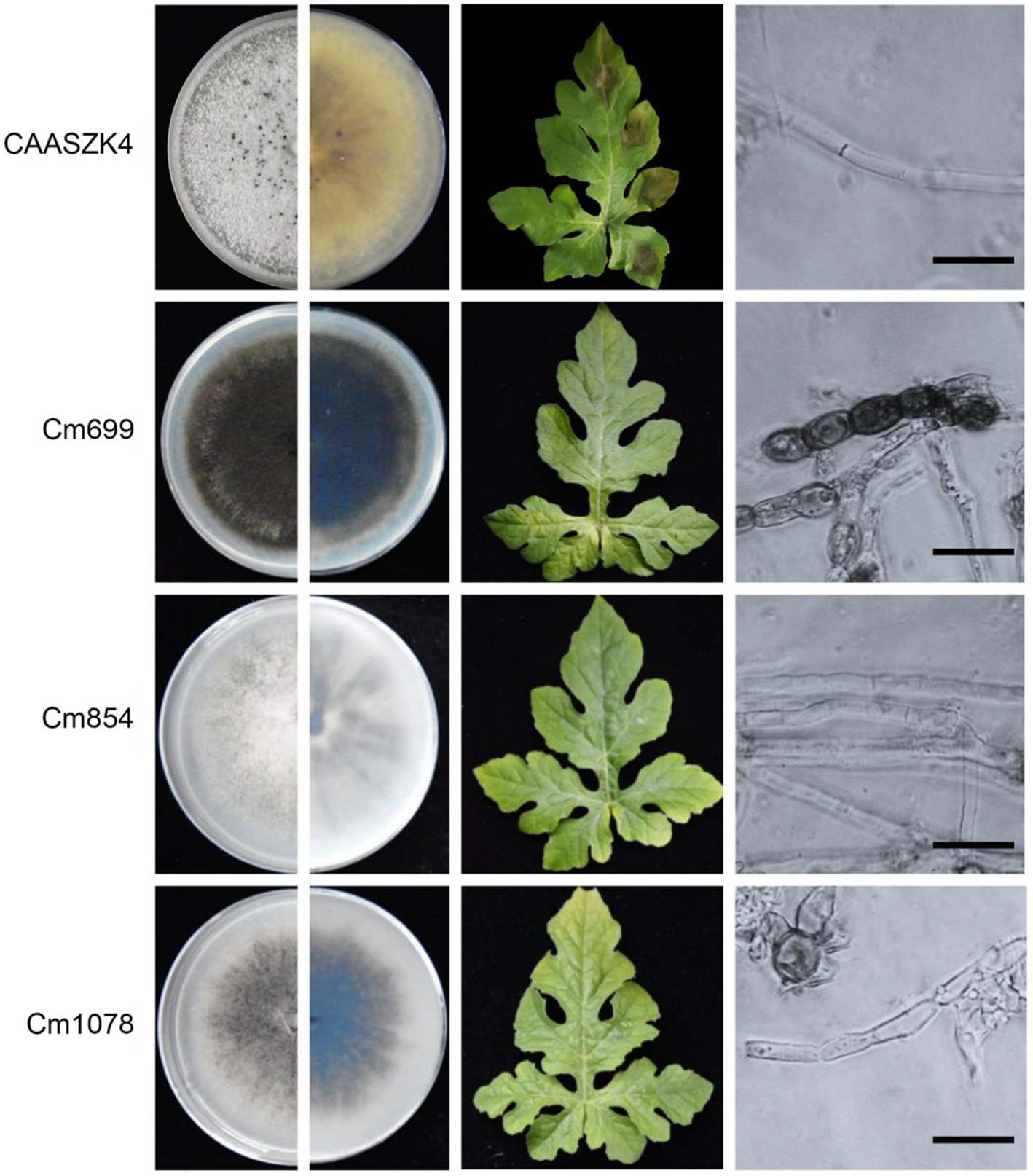
Figure 2. Phenotype and pathogenic development of C. magnum transformants produced by Agrobacterium tumefaciens-mediated transformation (ATMT). The phenotype during axenic growth on PDA is shown in the left panel. Pathogenicity assays on detached watermelon leaves (4 days post-inoculation (dpi)). Structure of C. magnum transformants mycelia. Bars represent 50 μm.
Morphological and growth characterization of transformants with reduced pathogenicity
Three transformants (Cm699, Cm854, and Cm1078) exhibited significant morphology alterations compared to the WT strain (Figure 2). Of the three transformants, the hyphae of Cm699 showned severe deformity and pigment deposition compared to those of the WT strain (Figure 2); Cm699 and Cm1078 did not produce conidia, whereas Cm854 produced significantly fewer conidia than the WT strain (Figure 3A). In addition, the conidia germination rates of Cm854 were significantly lower than those of the WT strain (Figure 3B).
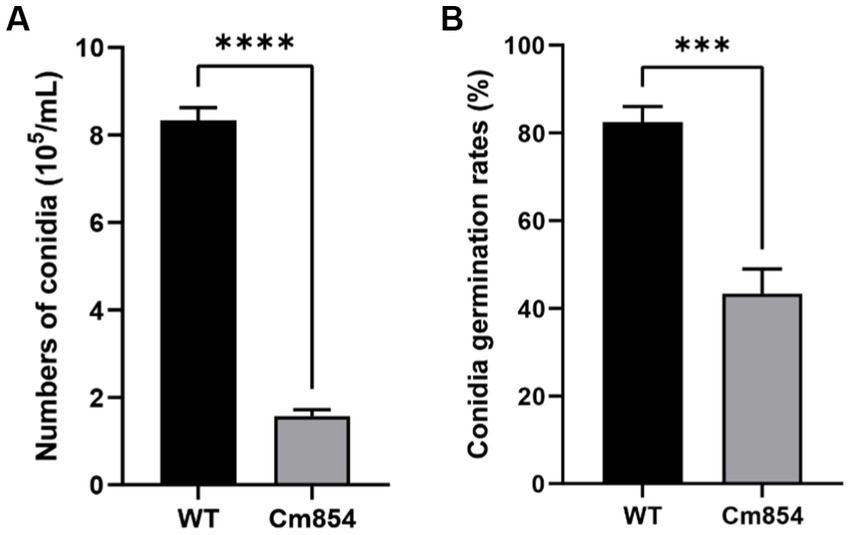
Figure 3. Numbers of conidia and the conidia germination rates of the wild-type (WT) strain CAASZK4 and of mutant Cm854. (A) Numbers of conidia of the wild-type (WT) strain and of mutant Cm854; (B) Conidia germination rates of the wild-type (WT) strain CAASZK4 and of mutant Cm854. Data were analysed with GraphPad Prism 9.0 (https://www.graphpad.com/). Asterisks over the error bars indicate the significant difference at the p = 0.05 level.
The growth rates of Cm699 (2.5) and Cm854 (10.8) were significantly lower than those of the WT strain (11.9), and there was no significant difference between the Cm1078 (11.6) and WT strains (Figure 4).
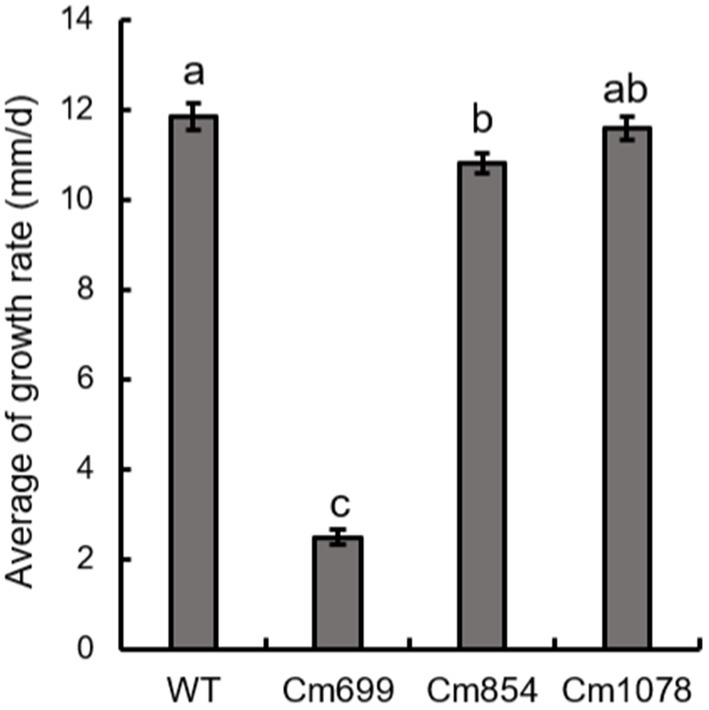
Figure 4. The growth rate of the three pathogenicity mutants and WT strains obtained in this study. Data were analysed with SPSS Statistics 19.0 (WinWrap Basic; http://www.winwrap.com) by one-way analysis of variance, and means were compared using Duncan’s test at a significance level of p = 0.05. Letters over the error bars indicate the significant difference at the p = 0.05 level.
Identification of T-DNA insertion sites
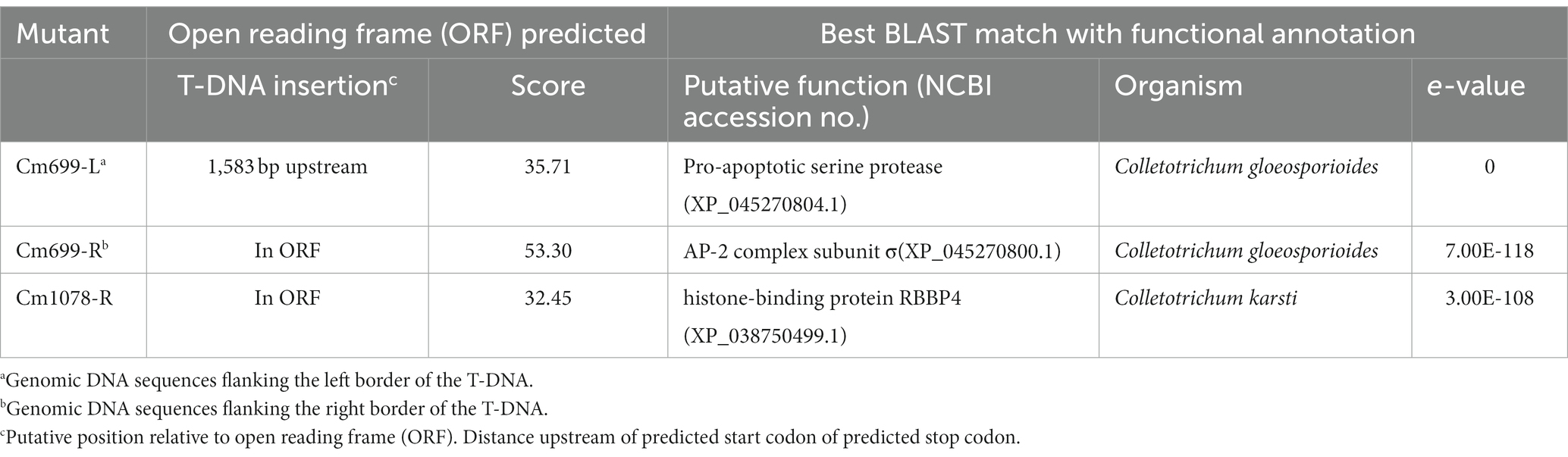
According to genomic Southern blot experiments, three transformants (Cm699, Cm854, and Cm1078) harbored single T-DNA integrations (Figure 1B). To identify the genomic loci of the transformants where T-DNA was inserted, TAIL-PCR was performed on three pathogenic transformants. PCR fragments displayed more specific and higher intensity bands in the third round than those in the first and second of TAIL-PCRs (Supplementary Figure S5). Thus, PCR amplicons resulting from the third round of TAIL-PCR were cloned and sequenced. Three genomic DNA sequences flanking the left and right border of the T-DNA ranging from 0.7 to 3.0 kb were obtained, respectively. In two transformants (Cm699 and Cm1078), comparison of the sequences flanking the left and right border of the T-DNA with the genome sequence of WT C. magnum revealed significant similarities (Table 2). However, the sequence flanking the T-DNA of Cm854 showed no significant similarity with the genomic sequence of WT C. magnum. A analysis of T-DNA insertions demonstrated that two transformants (Cm699 and Cm1078) had interrupted protein-coding genes (Table 3).
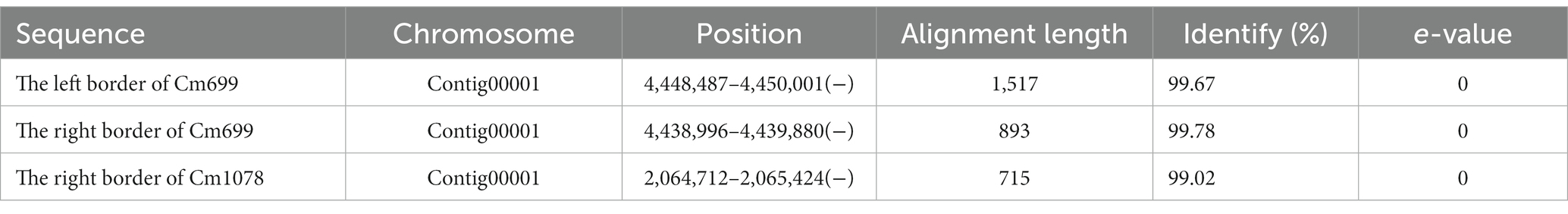
Table 2. The sequences flanking left border and the right border of the T-DNA were aligned to the genome sequence of WT C. magnum by BLAST+.
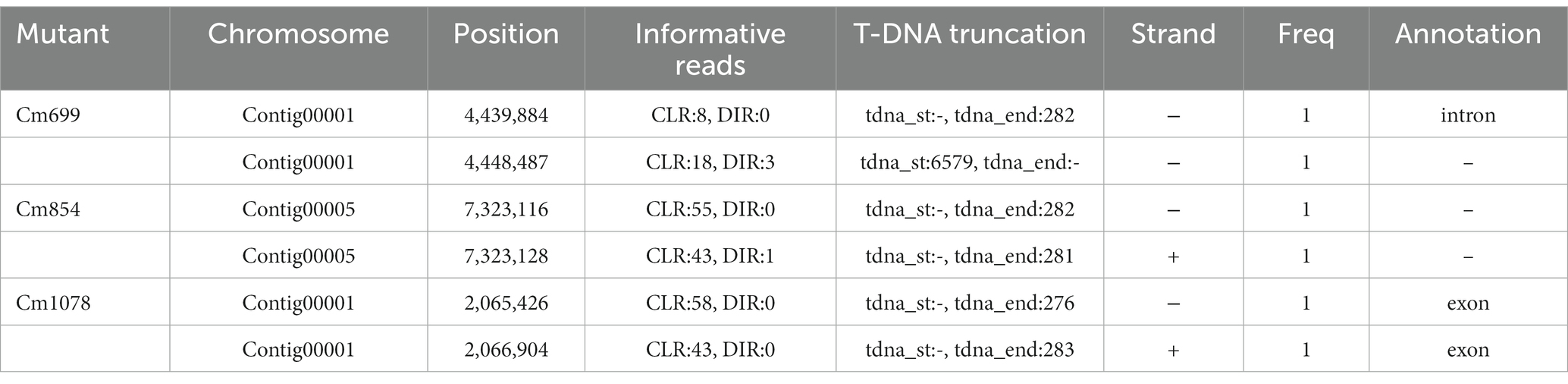
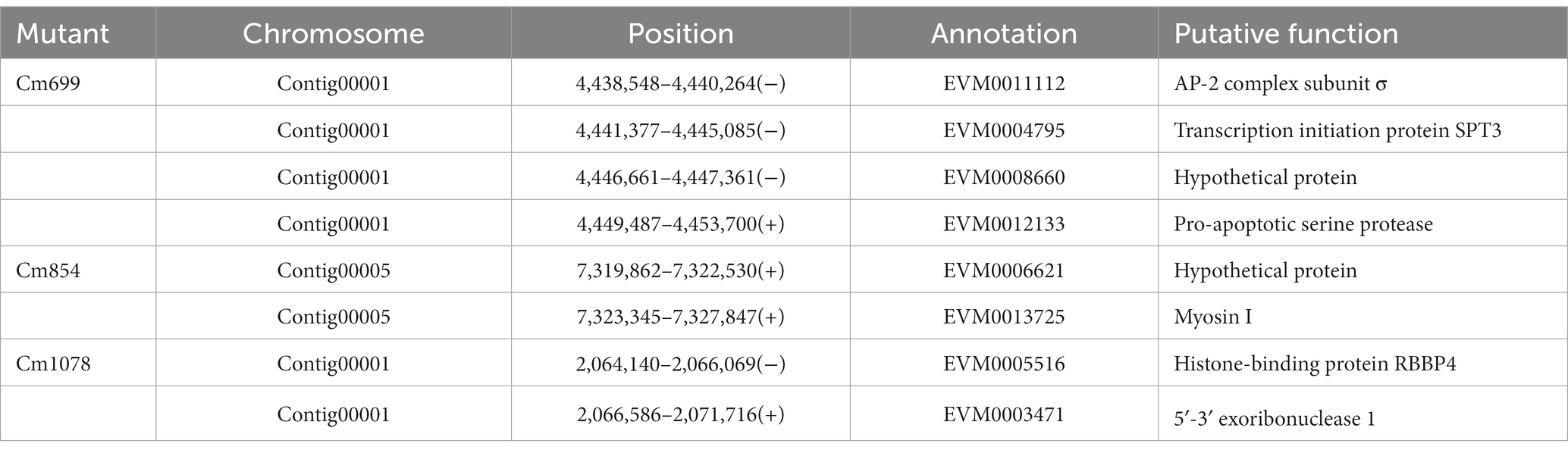
To further localize and characterize the TISs, whole-genome resequencing data from three transformants (Cm699, Cm854, and Cm1078) were utilized. Six T-DNA insertions in the three transformants were identified using TDNAscan (Table 4). However, all T-DNA insertions in the two transformants (Cm854 and Cm1078) were truncated (Table 5; Supplementary Figure S5). Furthermore, two T-DNA integration patterns, single-copy T-DNA with complete LB and RB and truncated tandem T-DNA repeats of LB or RB, were found in C. magnum (Figure 5). Using the local genome database of WT C. magnum as a reference genome, we found that T-DNA integration of transformant Cm699 occurred in the intron region starting 1,336-bp downstream of the start codon of the AP-2 complex subunit σ gene, and two genes were deleted, in the integration process. (Figure 5A). Based on whole-genome resequencing data, the two deleted genes were predicted to be the transcription initiation protein gene and hypothetical gene, respectively (Table 5). Furthermore, integration occurred within the promoter region 217-bp upstream of the start codon of the Myosin I gene in the transformant Cm854 (Figure 5B). In the Cm1078 transformant, the integration had in the exon region of two adjacent genes, one exon region starting 893-bp downstream of the start codon of WD-repeat protein retinoblastoma binding protein 4 gene (RBBP4), while the other exon region starting 4,812-bp upstream of the termination codon of 5′-3′exoribonuclease 1 gene (XRN1) (Figure 5C).
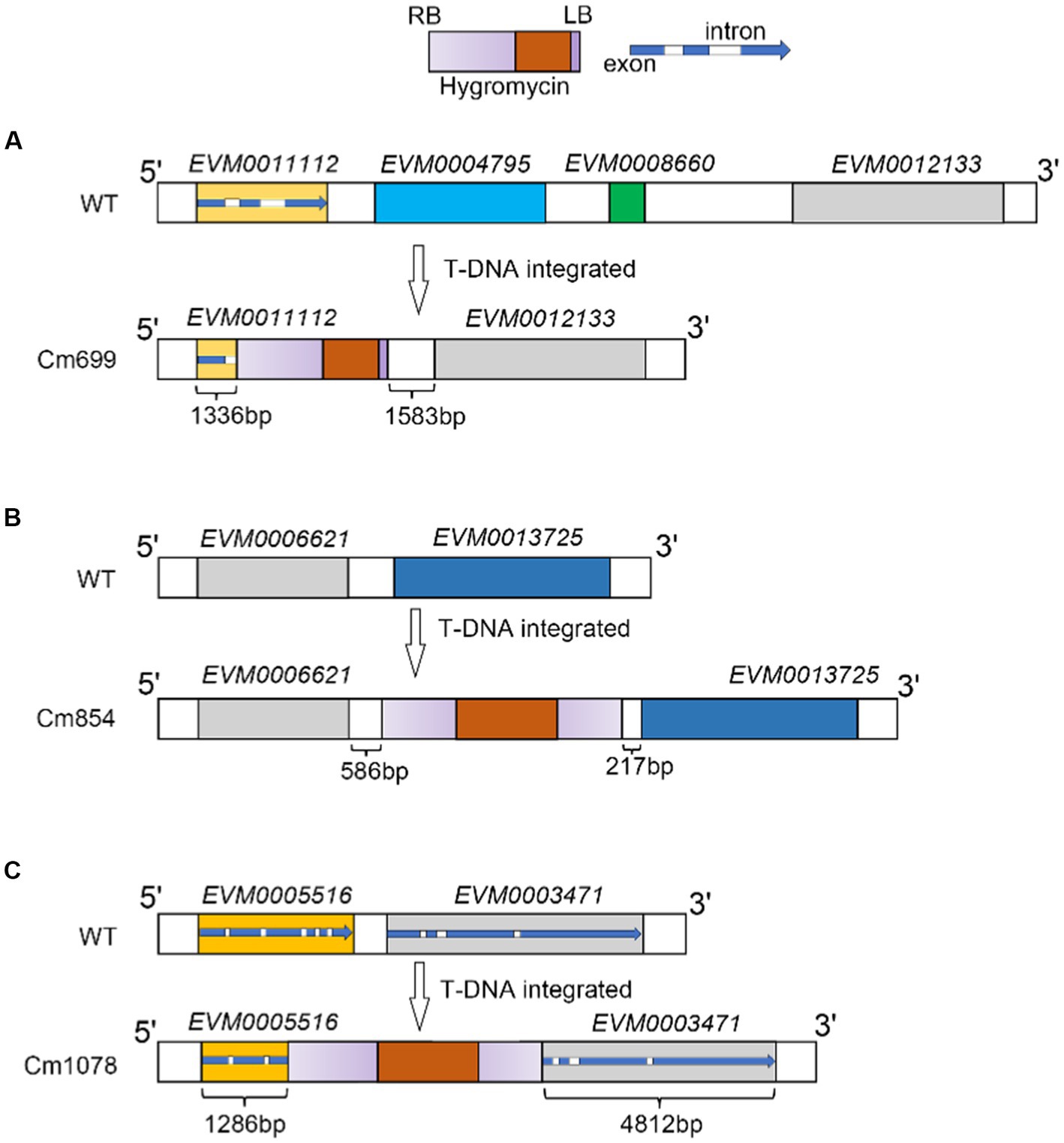
Figure 5. T-DNA integration sites patterns of T-DNA integration and in transformants Cm699, Cm854, and Cm1078. (A), (B), (C) Illustration of T-DNA integration into Cm699, Cm854, and Cm1078, respectively. The rectangle in gradient purple represents the ideal T-DNA between LB and RB. The rectangle in orange represents the T-DNA fragment coding the hygromycin resistance gene. In the blue arrow, the blue represents the exon as well as the white represents the intron. The integration sites were identified by whole genome re-sequencing in the text.
Discussion
C. magnum is a crcucial fungus that induces watermelon anthracnose (Damm et al., 2019). Understanding the mechanisms of its pathogenesis will facilitate disease management. ATMT is an effective method for exploring pathogenicity associated genes in pathogenic fungi (Fitzgerald et al., 2003; Leclerque et al., 2004; White and Chen, 2006; Zhang et al., 2013; Li et al., 2019; Villena et al., 2020; Binh et al., 2021; Casado-Del Castillo et al., 2021; Chauhan et al., 2021; Liu et al., 2021). Howerever, this method has not been applied to C. magnum. In this study, we optimized the ATMT protocol for the stable transformation of C. magnum, yielding130–360 (average = 245) transformants per 108 spores.
The mode and frequency of T-DNA insertions into the pathogenic genome are vital for the identification of disrupted genes. In Magnaporthe oryzae, not only single-copy T-DNA integration into genome, but multi-copy integration of the entire Ti-plasmid was found (Li et al., 2007). In this study, 75% of the transfromants had a single copy of T-DNA randomly located in the genome, whereas only approximately 2% multi-copy integration was found, which was consistent with previous reports (Münch et al., 2011). The transformants generated using the ATMT protocol, in combination with the identification of TISs, were used to determine the virulence factors of C. magnum, which can be applied in forward genetics.
Pathogenically defective transformants were predicted through the pathogenicity assay of the transformants, which were obtained using ATMT protocol for C. magnum. The penetration barriers (cuticle and epidermis) of the host are the main structures that defend against pathogenic infections. If both penetration barriers are destroyed and the quiescent infection is dispruted, the transformants are capable of colonizing the host tissues (Takano et al., 2000; Tsuji et al., 2003). Beacuse the defense reaction of young leaves is incomplete compared to that of old leaves when the host is infected by fungi, young leaves are more suitable for screening and assessing the pathogenicity of transformants (Cai et al., 2013). In this study, pathogenicity assay for all transformants was conducted on healthy and wounded watermelon leaves (5–6 true leaves), and the mycelium of the pathogen was used to inoculate watermelon leaves. Of these, compared with the WT strain, three transformants revealed no pathogenicity and 11 transformants showed impaired pathogenicity on healthy detached watermelon leaves (Figure 2; Supplementary Figure S6). It may be that potential pathogenic genes of three transformants affected the ability of the mycelium to infect the host.
As the integration of T-DNA into the genome of the pathogen affected the expression of pathogenic genes of pathogen, defective transformants were obtained. Hence, the TISs analysis was necessary. To identify the location and mode of the T-DNA insertions into the fungal genome, flanking sequence analysis was performed via TAIL-PCR (Huser et al., 2009; Liu et al., 2013). However, the application of TAIL-PCR for the identification of TISs was limited because the left and/or right ends of T-DNA can be truncated before insertion into the genome (Schouten et al., 2017). In this study, no LB-or RB-flanking sequences were cloned from the Cm854 transformants. For the Cm1078 transformants, LB flanking sequences were not cloned. This result may be explained by the truncation of LB, similar to observations in other fungi (Maruthachalam et al., 2008; Cai et al., 2013). Next-generation sequencing technologies have complemented the deficiency of TAIL-PCR for T-DNA site identification. The TISs of three transformants (Cm699, Cm854, and Cm1078) were successfully identified using whole-genome resequencing data (Table 5). Moreover, the sequences of 500 bp flanking TISs and the position of TISs in the genome of C. magnum were obtained using TDNAscan. Notably, the TISs of Cm699 and Cm1078 examined using TDNAscan were consistent with those obtained using TAIL-PCR (Tables 3, 5).
Some genes regulate infection-related morphogenesis during pathogenesis (Idnurm and Howlett, 2001). In the present study, six potentially pathogenic genes were identified. One potential pathogenicity gene encoded a protein containing the AP-2 complex subunit σ. AP-2 complexes were composed of four subunits: two large subunits (β2 and α), a medium subunit μ, and a small subunit σ (Liu C. et al., 2022). In S. cerevisiae, AP2 has been shown to facilitate the formation of the major class of endocytic vesicles (Myers and Payne, 2013; Lu et al., 2016). Moreover, we found that two genes were deleted in the transformant Cm699 (Figure 5A), which was probably a result of T-DNA integration employing double-strand break repair (Kleinboelting et al., 2015). These two genes may also be potential pathogenic genes that are not similar to known genes in fungi, and may be novel fungal pathogenic factors. T-DNA insertion into the promoter region of the potential pathogenicity gene was probably the main element responsible for the observed pathogenic defect in the transformant (Lee et al., 1990). In the transformant Cm854, the observed pathogenicity defect was presumably the reason for the disruption of the promoter region of a gene encoding Myosin I, which shares sequence homology with MYO5 of Pyricularia oryzae (78%) by T-DNA integration. In S. cerevisiae, although the phenotype of MYO5 deletion mutants did not change, growth and actin cytoskeletal organization were affected (Goodson et al., 1996). The observed pathogenicity defect in the transformant Cm1078 was possibly a result of a T-DNA insertion into the region of between two adjacent genes, which were simultaneously disrupted. One gene encoded a WD-repeat protein RBBP4, which also known as chromatin-remodeling factor RBAP48, shares sequence homology with MSl1 of S. cerevisiae (Ruggieri et al., 1989; Qian et al., 1993; Miao et al., 2020). Furthermore, the MSl1 gene could suppress the defects in S. cerevisiae sporulation (Ruggieri et al., 1989). The other gene encoded a protein, which exhibited homology with the XRN1 of S. cerevisiae, was not only involved in the termination of DNA transcription, but also in RNA degradation (Hsu and Stevens, 1993; Kim et al., 2004; Sharma et al., 2022).
In summary, the modified ATMT protocol is suitable for identifying novel genes required for the pathogenicity of the watermelon pathogen C. magnum. Six potential virulence genes were identified in C. magnum. These findings provide foundation for the pathogenic mechanism of C. magnum.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
QG and ZG conceptualized the study and designed the experiments. ZG performed the experiments, acquired and analyzed the data, and drafted the original manuscript. HW, CL, and QG reviewed and edited the manuscript. BP, BK, LL, HW, and QG supervised the work. All authors contributed to the article and approved the submitted version.
Funding
This work was financially supported by Agricultural Science and Technology Innovation Program (CAAS-ASTIP-2022-ZFRI-09).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1220116/full#supplementary-material
Footnotes
1. ^https://blast.ncbi.nlm.nih.gov/Blast.cgi
2. ^http://linux1.softberry.com/berry.phtml
References
Binh, C. T., Thai, H. D., Ha, B. T. V., and Tran, V. T. (2021). Establishment of a new and efficient agrobacterium-mediated transformation system in the nematicidal fungus Purpureocillium lilacinum. Microbiol. Res. 249:126773. doi: 10.1016/j.micres.2021.126773
Bundock, P., Den Dulk-Ras, A., Beijersbergen, A., and Hooykaas, P. J. (1995). Trans-kingdom T-DNA transfer from Agrobacterium tumefaciens to Saccharomyces cerevisiae. EMBO J. 14, 3206–3214. doi: 10.1002/j.1460-2075.1995.tb07323.x
Cai, Z., Li, G., Lin, C., Shi, T., Zhai, L., Chen, Y., et al. (2013). Identifying pathogenicity genes in the rubber tree anthracnose fungus Colletotrichum gloeosporioides through random insertional mutagenesis. Microbiol. Res. 168, 340–350. doi: 10.1016/j.micres.2013.01.005
Casado-Del Castillo, V., Maccabe, A. P., and Orejas, M. (2021). Agrobacterium tumefaciens-mediated transformation of NHEJ mutant aspergillus nidulans conidia: an efficient tool for targeted gene recombination using selectable nutritional markers. J. Fungi 7:961. doi: 10.3390/jof7110961
Chauhan, A., Modgil, M., and Rajam, M. V. (2021). Establishment of Agrobacterium tumefaciens - mediated genetic transformation of apple pathogen Marssonina coronaria using marker genes under the control of CaMV 35S promoter. Microbiol. Res. 253:126878. doi: 10.1016/j.micres.2021.126878
Damm, U., Sato, T., Alizadeh, A., Groenewald, J. Z., and Crous, P. W. (2019). The Colletotrichum dracaenophilum, C. magnum and C. orchidearum species complexes. Stud. Mycol. 92, 1–46. doi: 10.1016/j.simyco.2018.04.001
De Groot, M. J. A., Bundock, P., Hooykaas, P. J. J., and Beijersbergen, A. G. M. (1998). Agrobacterium tumefaciens-mediated transformation of filamentous fungi. Nat. Biotechnol. 16, 839–842. doi: 10.1038/nbt0998-839
De Silva, D. D., Crous, P. W., Ades, P. K., Hyde, K. D., and Taylor, P. W. J. (2017). Life styles of Colletotrichum species and implications for plant biosecurity. Fungal Biol. Rev. 31, 155–168. doi: 10.1016/j.fbr.2017.05.001
Dufresne, M., Perfect, S., Pellier, A. L., Bailey, J. A., and Langin, T. (2000). A GAL4-like protein is involved in the switch between biotrophic and necrotrophic phases of the infection process of Colletotrichum lindemuthianum on common bean. Plant Cell 12, 1579–1589. doi: 10.1105/tpc.12.9.1579
Fitzgerald, A. M., Mudge, A. M., Gleave, A. P., and Plummer, K. M. (2003). Agrobacterium and PEG-mediated transformation of the phytopathogen Venturia inaequalis. Mycol. Res. 107, 803–810. doi: 10.1017/s0953756203008086
Gilbert, M. J., Thornton, C. R., Wakley, G. E., and Talbot, N. J. (2006). A P-type ATPase required for rice blast disease and induction of host resistance. Nature 440, 535–539. doi: 10.1038/nature04567
Gong, Z., Ning, N., Li, Z., Xie, X., Wilson, R. A., and Liu, W. (2022). Two Magnaporthe appressoria-specific (MAS) proteins, MoMas3 and MoMas5, are required for suppressing host innate immunity and promoting biotrophic growth in rice cells. Mol. Plant Pathol. 23, 1290–1302. doi: 10.1111/mpp.13226
Goodson, H. V., Anderson, B. L., Warrick, H. M., Pon, L. A., and Spudich, J. A. (1996). Synthetic lethality screen identifies a novel yeast myosin I gene (MYO5): myosin I proteins are required for polarization of the actin cytoskeleton. J. Cell Biol. 133, 1277–1291. doi: 10.1083/jcb.133.6.1277
Guo, Z., Luo, C. X., Wu, H. J., Peng, B., Kang, B. S., Liu, L. M., et al. (2022). Colletotrichum species associated with anthracnose disease of watermelon (Citrullus lanatus) in China. J. Fungi 8:790. doi: 10.3390/jof8080790
Hsu, C. L., and Stevens, A. (1993). Yeast cells lacking 5′-->3′ exoribonuclease 1 contain mRNA species that are poly (a) deficient and partially lack the 5′ cap structure. Mol. Cell. Biol. 13, 4826–4835. doi: 10.1128/mcb.13.8.4826-4835.1993
Huser, A., Takahara, H., Schmalenbach, W., and O'connell, R. (2009). Discovery of pathogenicity genes in the crucifer anthracnose fungus Colletotrichum higginsianum, using random insertional mutagenesis. Mol. Plant-Microbe Interact. 22, 143–156. doi: 10.1094/mpmi-22-2-0143
Idnurm, A., and Howlett, B. J. (2001). Pathogenicity genes of phytopathogenic fungi. Mol. Plant Pathol. 2, 241–255. doi: 10.1046/j.1464-6722.2001.00070.x
Jiao, W., Yu, H., Cong, J., Xiao, K., Zhang, X., Liu, J., et al. (2022). Transcription factor SsFoxE3 activating SsAtg8 is critical for sclerotia, compound appressoria formation, and pathogenicity in Sclerotinia sclerotiorum. Mol. Plant Pathol. 23, 204–217. doi: 10.1111/mpp.13154
Kim, M., Krogan, N. J., Vasiljeva, L., Rando, O. J., Nedea, E., Greenblatt, J. F., et al. (2004). The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature 432, 517–522. doi: 10.1038/nature03041
Kleinboelting, N., Huep, G., Appelhagen, I., Viehoever, P., Li, Y., and Weisshaar, B. (2015). The structural features of thousands of T-DNA insertion sites are consistent with a double-strand break repair-based insertion mechanism. Mol. Plant 8, 1651–1664. doi: 10.1016/j.molp.2015.08.011
Lawrence, G. J., Dodds, P. N., and Ellis, J. G. (2010). Transformation of the flax rust fungus, Melampsora lini: selection via silencing of an avirulence gene. Plant J. 61, 364–369. doi: 10.1111/j.1365-313X.2009.04052.x
Leclerque, A., Wan, H., Abschutz, A., Chen, S., Mitina, G. V., Zimmermann, G., et al. (2004). Agrobacterium-mediated insertional mutagenesis (AIM) of the entomopathogenic fungus Beauveria bassiana. Curr. Genet. 45, 111–119. doi: 10.1007/s00294-003-0468-2
Lee, H., Fu, Y. H., and Marzluf, G. A. (1990). Nucleotide sequence and DNA recognition elements of alc, the structural gene which encodes allantoicase, a purine catabolic enzyme of Neurospora crassa. Biochemistry 29, 8779–8787. doi: 10.1021/bi00489a039
Li, J. X., Hong, N., Peng, B., Wu, H. J., and Gu, Q. S. (2019). Transformation of Corynespora cassiicola by Agrobacterium tumefaciens. Fungal Biol. 123, 669–675. doi: 10.1016/j.funbio.2019.05.011
Li, Q. L., Mo, J. Y., Huang, S. P., Guo, T. X., and Hsiang, T. (2013). First report of leaf spot disease caused by Glomerella magna on Lobelia chinensis in China. Plant Dis. 97:1383. dio. doi: 10.1094/PDIS-03-13-0346-PDN
Li, G., Zhou, Z., Liu, G., Zheng, F., and He, C. (2007). Characterization of T-DNA insertion patterns in the genome of rice blast fungus Magnaporthe oryzae. Curr. Genet. 51, 233–243. doi: 10.1007/s00294-007-0122-5
Liu, R., Kim, W., Paguirigan, J. A., Jeong, M. H., and Hur, J. S. (2021). Establishment of Agrobacterium tumefaciens-mediated transformation of Cladonia macilenta, a odmel lichen-ofrming ufngus. J. Fungi 7:252. doi: 10.3390/jof7040252
Liu, C., Li, Z., Tian, D., Xu, M., Pan, J., Wu, H., et al. (2022). AP1/2beta-mediated exocytosis of tapetum-specific transporters is required for pollen development in Arabidopsis thaliana. Plant Cell 34, 3961–3982. doi: 10.1093/plcell/koac192
Liu, F., Ma, Z. Y., Hou, L. W., Diao, Y. Z., Wu, W. P., Damm, U., et al. (2022). Updating species diversity of Colletotrichum, with a phylogenomic overview. Stud. Mycol. 101, 1–56. doi: 10.3114/sim.2022.101.01
Liu, Y.-G., Mitsukawa, N., Oosumi, T., and Whittier, R. F. (1995). Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 8, 457–463. doi: 10.1046/j.1365-313X.1995.08030457.x
Liu, L., Zhao, D., Zheng, L., Hsiang, T., Wei, Y., Fu, Y., et al. (2013). Identification of virulence genes in the crucifer anthracnose fungus Colletotrichum higginsianum by insertional mutagenesis. Microb. Pathog. 64, 6–17. doi: 10.1016/j.micpath.2013.06.001
Lu, R., Drubin, D. G., and Sun, Y. (2016). Clathrin-mediated endocytosis in budding yeast at a glance. J. Cell Sci. 129, 1531–1536. doi: 10.1242/jcs.182303
Martinez-Cruz, J., Romero, D., De Vicente, A., and Perez-Garcia, A. (2017). Transformation of the cucurbit powdery mildew pathogen Podosphaera xanthii by Agrobacterium tumefaciens. New Phytol. 213, 1961–1973. doi: 10.1111/nph.14297
Maruthachalam, K., Nair, V., Rho, H. S., Choi, J., Kim, S., and Lee, Y. H. (2008). Agrobacterium tumefaciens-mediated transformation in Colletotrichum falcatum and C. acutatum. J. Microbiol. Biotechnol.. 18: 234–241. dio:
Miao, X., Sun, T., Barletta, H., Mager, J., and Cui, W. (2020). Loss of RBBP4 results in defective inner cell mass, severe apoptosis, hyperacetylated histones and preimplantation lethality in micedagger. Biol. Reprod. 103, 13–23. doi: 10.1093/biolre/ioaa046
Moraes, S. R. G., Escanferla, M. E., and Massola, N. S. Jr. (2015). Prepenetration and penetration of Colletotrichum gloeosporioides into guava fruit (Psidium guajava L.): effects of temperature, wetness period and fruit age. J. Phytopathol. 163, 149–159. doi: 10.1111/jph.12294
Mullins, E., Chen, X., Romaine, P., Raina, R., Geiser, D., and Kang, S. (2001). Agrobacterium-mediated transformation ofFusarium oxysporum: an efficient tool for insertional mutagenesis and gene transfer. Phytopathology 91, 173–180. doi: 10.1094/PHYTO.2001.91.2.173
Munch, S., Lingner, U., Floss, D. S., Ludwig, N., Sauer, N., and Deising, H. B. (2008). The hemibiotrophic lifestyle of Colletotrichum species. J. Plant Physiol. 165, 41–51. doi: 10.1016/j.jplph.2007.06.008
Münch, S., Ludwig, N., Floss, D. S., Sugui, J. A., Koszucka, A. M., Voll, L. M., et al. (2011). Identification of virulence genes in the corn pathogen Colletotrichum graminicola by Agrobacterium tumefaciens-mediated transformation. Mol. Plant Pathol. 12, 43–55. doi: 10.1111/j.1364-3703.2010.00651.x
Myers, M. D., and Payne, G. S. (2013). Clathrin, adaptors and disease: insights from the yeast Saccharomyces cerevisiae. Front. Biosci. 18, 862–891. doi: 10.2741/4149
Nirenberg, H. (1976). Untersuchungen über die morphologische und biologische differenzierung in der Fusarium-Sektion Liseola. Mitt. Biol. Bundesanst. für Land-und Forstwirtsch Berlin-Dahlem. 169, 1–117.
Qian, Y. W., Wang, Y. C., Hollingsworth, R. E. Jr., Jones, D., Ling, N., and Lee, E. Y. (1993). A retinoblastoma-binding protein related to a negative regulator of Ras in yeast. Nature 364, 648–652. doi: 10.1038/364648a0
Rossman, A. Y., Allen, W. C., and Castlebury, L. A. (2016). New combinations of plant-associated fungi resulting from the change to one name for fungi. IMA Fungus 7, 1–7. doi: 10.5598/imafungus.2016.07.01.01
Ruggieri, R., Tanaka, K., Nakafuku, M., Kaziro, Y., Toh-e, A., and Matsumoto, K. (1989). MSI1, a negative regulator of the RAS-cAMP pathway in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 86, 8778–8782. doi: 10.1073/pnas.86.22.8778
Schouten, H. J., Vande Geest, H., Papadimitriou, S., Bemer, M., Schaart, J. G., Smulders, M. J., et al. (2017). Re-sequencing transgenic plants revealed rearrangements at T-DNA inserts, and integration of a short T-DNA fragment, but no increase of small mutations elsewhere. Plant Cell Rep. 36, 493–504. doi: 10.1007/s00299-017-2098-z
Sessions, A., Burke, E., Presting, G., Aux, G., Mcelver, J., Patton, D., et al. (2002). A high-throughput Arabidopsis reverse genetics system. Plant Cell 14, 2985–2994. doi: 10.1105/tpc.004630
Sharma, S., Yang, J., Grudzien-Nogalska, E., Shivas, J., Kwan, K. Y., and Kiledjian, M. (2022). Xrn1 is a deNADding enzyme modulating mitochondrial NAD-capped RNA. Nat. Commun. 13:889. doi: 10.1038/s41467-022-28555-7
Sun, L., Ge, Y., Sparks, J. A., Robinson, Z. T., Cheng, X., Wen, J., et al. (2019). TDNAscan: a software to identify complete and truncated T-DNA insertions. Front. Genet. 10:685. doi: 10.3389/fgene.2019.00685
Takano, Y., Kikuchi, T., Kubo, Y., Hamer, J. E., Mise, K., and Furusawa, I. (2000). The Colletotrichum lagenarium MAP kinase gene CMK1 regulates diverse aspects of fungal pathogenesis. Mol. Plant-Microbe Interact. 13, 374–383. doi: 10.1094/MPMI.2000.13.4.374
Tsay, J. G., Chen, R. S., Wang, W. L., and Weng, B. C. (2010). First report of anthracnose on cucurbitaceous crops caused by Glomerella magna in Taiwan. Plant Dis. 94:787. doi: 10.1094/PDIS-94-6-0787A
Tsuji, G., Fujii, S., Fujihara, N., Hirose, C., Tsuge, S., Shiraishi, T., et al. (2003). Agrobacterium tumefaciens-mediated transformation for random insertional mutagenesis in Colletotrichum lagenarium. J. Gen. Plant Pathol. 69, 230–239. doi: 10.1007/s10327-003-0040-4
Tucker, S. L., and Talbot, N. J. (2001). Surface attachment and pre-penetration stage development by plant pathogenic fungi. Annu. Rev. Phytopathol. 39, 385–417. doi: 10.1146/annurev.phyto.39.1.385
Villena, C. I. F., Gomes, R. R., Fernandes, L., Florencio, C. S., Bombassaro, A., Grisolia, M. E., et al. (2020). Agrobacterium tumefaciens-mediated transformation of Fonsecaea monophora and Fonsecaea erecta for host-environment interaction studies. J. Fungi 6:325. doi: 10.3390/jof6040325
Werner, S., Sugui, J. A., Steinberg, G., and Deising, H. B. (2007). A chitin synthase with a myosin-like motor domain is essential for hyphal growth, appressorium differentiation, and pathogenicity of the maize anthracnose fungus Colletotrichum graminicola. Mol. Plant-Microbe Interact. 20, 1555–1567. doi: 10.1094/MPMI-20-12-1555
Wharton, P. S., Julian, A. M., and O'connell, R. J. (2001). Ultrastructure of the infection of sorghum bicolor by Colletotrichum sublineolum. Phytopathology 91, 149–158. doi: 10.1094/PHYTO.2001.91.2.149
White, D., and Chen, W. (2006). Genetic transformation of Ascochyta rabiei using agrobacterium-mediated transformation. Curr. Genet. 49, 272–280. doi: 10.1007/s00294-005-0048-8
Zhang, T., Qi, Z., Wang, Y., Zhang, F., Li, R., Yu, Q., et al. (2013). Agrobacterium tumefaciens-mediated transformation of Penicillium expansum PE-12 and its application in molecular breeding. Microbiol. Res. 168, 130–137. doi: 10.1016/j.micres.2012.11.001
Keywords: fungal transformation, ATMT, T-DNA integration, virulence gene, Colletotrichum magnum
Citation: Guo Z, Wu H, Peng B, Kang B, Liu L, Luo C and Gu Q (2023) Identifying pathogenicity-related genes in the pathogen Colletotrichum magnum causing watermelon anthracnose disease via T-DNA insertion mutagenesis. Front. Microbiol. 14:1220116. doi: 10.3389/fmicb.2023.1220116
Edited by:
Jian-Wei Guo, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Lian-Ming Liang, Yunnan University, ChinaRasappa Viswanathan, Indian Council of Agricultural Research (ICAR), India
Copyright © 2023 Guo, Wu, Peng, Kang, Liu, Luo and Gu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chaoxi Luo, Y3hsdW9AbWFpbC5oemF1LmVkdS5jbg==; Qinsheng Gu, Z3VxaW5zaGVuZ0BjYWFzLmNu
 Zhen Guo
Zhen Guo Huijie Wu
Huijie Wu Bin Peng
Bin Peng Baoshan Kang
Baoshan Kang Liming Liu
Liming Liu Chaoxi Luo
Chaoxi Luo Qinsheng Gu
Qinsheng Gu