- 1State Key Laboratory of Tropical Oceanography (LTO), South China Sea Institute of Oceanology, Chinese Academy of Sciences, Guangzhou, China
- 2Key Laboratory for Water Quality and Conservation of the Pearl River Delta, Institute of Environmental Research at Greater Bay Area, Ministry of Education, Guangzhou University, Guangzhou, China
- 3University of Chinese Academy of Sciences, Beijing, China
Denitrification is vital to nitrogen removal and N2O release in ecosystems; in this regard, paddy soils exhibit strong denitrifying ability. However, the underlying mechanism of N2O emission from denitrification in paddy soils is yet to be elucidated. In this study, the potential N2O emission rate, enzymatic activity for N2O production and reduction, gene abundance, and community composition during denitrification were investigated using the 15N isotope tracer technique combined with slurry incubation, enzymatic activity detection, quantitative polymerase chain reaction (qPCR), and metagenomic sequencing. Results of incubation experiments showed that the average potential N2O emission rates were 0.51 ± 0.20 μmol⋅N⋅kg–1⋅h–1, which constituted 2.16 ± 0.85% of the denitrification end-products. The enzymatic activity for N2O production was 2.77–8.94 times than that for N2O reduction, indicating an imbalance between N2O production and reduction. The gene abundance ratio of nir to nosZ from qPCR results further supported the imbalance. Results of metagenomic analysis showed that, although Proteobacteria was the common phylum for denitrification genes, other dominant community compositions varied for different denitrification genes. Gammaproteobacteria and other phyla containing the norB gene without nosZ genes, including Actinobacteria, Planctomycetes, Desulfobacterota, Cyanobacteria, Acidobacteria, Bacteroidetes, and Myxococcus, may contribute to N2O emission from paddy soils. Our results suggest that denitrification is highly modular, with different microbial communities collaborating to complete the denitrification process, thus resulting in an emission estimation of 13.67 ± 5.44 g N2O⋅m–2⋅yr–1 in surface paddy soils.
1. Introduction
Nitrous oxide (N2O) is not only a significant ozone-depleting substance (Ravishankara et al., 2009), but is also a well-known greenhouse gas with strong radiative forcing effects (Zumft and Kroneck, 2007). Since the industrial revolution, the concentration of N2O has increased at an annual rate of 0.25%, and the atmospheric N2O concentration has reached 331 ppb (Solomon et al., 2007). Moreover, the greenhouse effect of N2O is 310 times higher than that of the equivalent carbon dioxide (Lashof and Ahuja, 1990). Thus, N2O has received increasing attention owing to the environmental problems that it may cause.
Agricultural fields that receive substantial amounts of nitrogen fertilizers are typically known as N2O emission hotspots (Syakila and Kroeze, 2011). According to predictions, by 2030, agricultural soils will become the primary source of N2O emission, contributing 59% of the total N2O emission released into the atmosphere (Hu et al., 2015). Although nitrification, denitrification, and nitrate dissimilation to ammonium can all generate N2O, denitrification is the primary pathway for N2O emission in terrestrial ecosystems (Sanford et al., 2012; Butterbach-Bahl et al., 2013; Harris et al., 2021). Furthermore, in the denitrification process, N2O exists as an intermediate product and can be both generated and consumed, leading to the emission of N2O being regulated by multiple functional genes (Zumft, 1997) During denitrification, NO3– is successively reduced to NO2–, NO, N2O, and finally to N2 (Zumft, 1997). Diverse phylogenetic denitrifying bacteria contain different functional genes, including napA, nirS, nirK, norB, nosZ I, and nosZ II genes, which encode enzymes that complete the denitrification process (Zumft, 1997). The napA gene encodes NO3– reductase, which catalyzes the reduction of NO3– to NO2– (Arnoux et al., 2003). NO2– and N2O reductions are considered the rate-limiting steps in denitrification (Zumft, 1997). NO2– reduction is catalyzed by NO2– reductases, including the nirS gene-encoded copper-containing NO2– reductase and the nirK gene-encoded cytochrome cd1-containing NO2– reductase (Zumft, 1997). Copper-containing or cytochrome cd1-containing NO2– reductases are functionally identical but have different structures and catalytic sites, and generally do not coexist in one bacterial species (Coyne et al., 1989). The NO reductase (NOR), encoded by the norB gene, is responsible for the reduction of NO to N2O (Braker and Tiedje, 2003). The N2O reductase (NOS) catalyzes the reduction of N2O, converting the greenhouse gas N2O into relatively harmless N2, thereby reducing its contribution to the greenhouse effect. NOS, encoded by either nosZ I or nosZ II genes, can complete denitrification or N2O reduction only (Zumft and Kroneck, 2007). The nosZ I and nosZ II genes generally do not coexist in the same bacteria, except in Thauera linaloolentis 47LolT (Semedo et al., 2020).
Nitrous oxide is an intermediate product of the denitrification process, and N2O emission is typically associated with enzymes encoded by functional genes and the bacteria that harbor these genes during denitrification (Black et al., 2019). Denitrification is a complex process affected by multiple factors, including environmental parameters, the microbial composition of functional genes, and key enzymatic activities (Groffman, 2012). Rich et al. (2003) showed the community composition as well as environmental factors lead to the variation in denitrification rates in meadow and forest soils. The analysis of denitrification genes abundance can establish a stronger correlation with potential N2O emissions (Morales et al., 2010). For instance, the ratio of nir to nosZ can partly determine the extent of N2O emission in soils (Domeignoz-Horta et al., 2015) and lakes (Saarenheimo et al., 2015). However, the comprehensive mechanisms of denitrification and N2O emissions are yet to be elucidated.
In this study, we first investigated the dissolved N2O concentration in paddy water and calculated the N2O flux at the water–air exchange to provide background information regarding N2O emissions in paddy fields. Focusing on paddy soils, we performed 15N isotope tracer experiments to determine the potential N2O emission rate and calculate the end-product ratio, i.e., N2O/(N2+N2O). The enzymatic activities of the NOR and NOS were measured using ELISA assay kits. Subsequently, qPCR and metagenomic analysis were performed to determine the abundance and community composition of key functional genes, respectively. The aim of this study is to demonstrate the contribution and underlying mechanism of denitrification to N2O emission in paddy soils and provide a more comprehensive understanding of the N2O emission process in paddy soils.
2. Materials and methods
2.1. Sample acquisition and physiochemical property determination
The soil to be tested was obtained from two paddy fields (22°55′16″N, 113°29′24″E; 22°54′24″N, 113°29′36″E) in May 2021 from Guangdong Province, China (Figure 1). Both paddy fields had been planted with rice for many years and the paddy fields were under water-logged conditions at the time of sampling. The dissolved oxygen (DO) in the paddy water was first determined using a portable multifunctional parameter meter equipped with a DO probe (HQ40D, HACH, Loveland, CO, USA). Quickly insert a 12.5 mL vial (Exetainer, Labco Limited, Lampeter, UK) below the surface of the paddy water until the vial was overflowing and there were no air bubbles inside. The cap was immediately sealed and 200 μL of saturated ZnCl2 solution was added to stop microbial activities. The sample was in triplicate and stored at −4°C. The dissolved N2O concentration of in situ water was determined within 24 h. The pH level was determined using a pH meter (Mettler Toledo S220, Greifensee, Switzerland). Salinity and temperature were measured using salinometers (ATAGO, Tokyo, Japan) and geothermometers (HG04-SYQX-2, Beijing, China), respectively.
Surface soil (0–10 cm) was sampled using a sterilized shovel, and each soil sample was mixed in triplicate. Paddy soil was divided into three parts: one for measuring potential rates of N2 and N2O emissions, which was stored at 4°C; another for DNA extraction and enzymatic activity measurement, which was kept at −80°C; and the last part was stored at 4°C for pH determination and analysis of dissolved inorganic nitrogen (DIN), including NO3–, NO2– and NH4+. DIN in the paddy soil was extracted using 2 M KCl with a soil extraction ratio of 1:5 (wt./vol.) before measurement (Bao, 2000). The DIN content was determined using the spectrophotometric method described by Wu et al. (2016) and Guan et al. (2017).
2.2. Dissolved N2O analysis
The concentration of N2O was measured using static headspace gas chromatography (Xu et al., 2005). The water in the vial was replaced with 5 mL of He to achieve headspace, agitated vigorously for 15 min, and stored in the dark to attain gas–liquid equilibrium. A gas chromatograph equipped with an electron capture detector (GC-2014C, Shimadzu, Tokyo, Japan) was used to measure the concentration of N2O (CG) in the headspace. The dissolved N2O concentration (CL) before gas replacement and the △N2O (i.e., the net increase in N2O) were calculated by the following equations (Lin et al., 2016):
where K0 is the equilibrium constant (Weiss and Price, 1980); R is the ideal gas constant; T is the temperature at equilibrium; VG and VL are the gas and liquid volumes after He replacement, respectively; and CN2Oeq is the N2O concentration in equilibrium with the atmospheric concentration calculated based on Weiss and Price (1980).
N2O saturation was calculated as follows:
The estimated N2O flux through the water–air interface was calculated as follows:
where K denotes the gas change rate calculated based on the method of Borges (2004).
2.3. Determination of potential N2 production rate and N2O emission rate
The procedures for slurry incubation were modified from Thamdrup and Dalsgaard (2002) and described in detail by Xiang et al. (2023). A mixture of fresh soil and water was pre-incubated in the dark for 72 h at a weight-to-volume ratio of 1:7. The soil slurry was then flushed with He to eliminate N2O and create an anaerobic environment before being transferred to vials using a syringe. The vials were separated into two groups, one group (three vials) was for measuring the concentration of remaining 14NO3– such that Fn (the proportion of 14NO3– in the total NO3– pool after adding 15NO3–) can be calculated, and the other group (12 vials) was injected with 15NO3– (15N 99.6%) to the final concentration of 100 μmol⋅L–1. The vials containing 15NO3– were incubated for T0 (T0 = 0 h) and T2 (T2 = 2 h) at in situ temperature in darkness, respectively, and 200 μL of saturated ZnCl2 solution was injected immediately.
The 29N2 amounts (D29) and 30N2 amounts (P30) in the vials above were measured via membrane inlet mass spectrometry (HPR40, Hiden, Warrington, UK), and the potential N2 production rate (RN2) was calculated as follows (Xiao et al., 2018; Wu et al., 2021):
The N2O concentration in each vial was determined via a procedure similar to that used for determining the N2O concentration in the water sample (Xu et al., 2005). The potential N2O emission rates (RN2O) was calculated based on the dissolved N2O concentrations in the vials at T0 and T2 (i.e., CL0 and CL2, respectively) (Xiang et al., 2023).
The ratio of denitrification end-products was calculated as follows:
2.4. Measurement of enzymatic activity of NOR and NOS
The enzymatic activities of NOR (NO→N2O) and NOS (N2O→N2) in the soils were determined using microbial NOR and NOS ELISA kits (Yilaisa Biotechnology Co., Ltd., Jiangsu, China). Briefly, 1 g of fresh soil, standards, and HRP-labeled detection antibodies were added based on the manufacturer’s protocol. A microplate reader (BioTek Elx800, Winooski, VT, USA) was used to measure the absorbance at 450 nm. Enzymatic activity was calculated from the absorbance based on the standard curve.
2.5. DNA extraction and qPCR
Soil DNA was extracted using a Fast DNA Spin Kit for Soil (MP Biomedical, Irvine, CA, USA) based on the manufacturer’s protocol. The concentration and purity of the extracted DNA were verified using a NanoDrop spectrophotometer (Thermo Scientific, Wilmington, DE, USA) and agarose gel electrophoresis, respectively. Primer sets 515F/806R (Caporaso et al., 2011), cd3af/R3cd (Throback et al., 2004), F1aCu/R3Cu (Hallin and Lindgern, 1999), nosZ 2F/nosZ 2R (Henry et al., 2006), nosZ IIF/nosZ IIR (Jones et al., 2013) were used to quantify the abundance of bacterial 16S, nirS, nirK, nosZ I, and nosZ II genes, respectively. qPCR was performed using the iQ5 Real-Time PCR system (Bio-Rad, Hercules, CA, USA). The amplification procedures are present in Supplementary Table 1 and have been described previously (Xiang et al., 2022). Plasmids, samples, and negative controls were prepared in triplicate and quantified simultaneously. The specificity of the amplified products was verified via melting curve analysis and agarose gel electrophoresis. Results of the amplification efficiency (90–110%) and correlation coefficient (R2 > 97%) are shown.
2.6. Metagenomic sequencing and gene annotation
The extracted DNA was sent to Genewiz (Suzhou, China) for library construction and shotgun sequencing. The raw sequencing reads of each sample exceeded 10 Gb and were trimmed using Trimmomatic (v 0.38) (Bolger et al., 2014) to obtain high-quality clean reads and ensure the accuracy of subsequent analysis. The clean reads were assembled to contigs using metaSPAdes (v 3.13.2) (Bankevich et al., 2012) with k-mer sizes of 21, 33, 55, 77, 99, and 127. The open reading frames (ORFs) of the contigs were predicted using Prodigal (v 2.6.3) (Hyatt et al., 2010) and were searched against KEGG (Aramaki et al., 2019) to obtain the corresponding KO numbers. The denitrification genes and corresponding KO numbers were as follows: napA, K02567; nirS, K15864; nirK, K00368; nosZ, K00376. The nosZ gene has only one KO number but includes nosZ I and nosZ II genes. We first obtained the protein sequence of nosZ gene and searched against the NCBI database.1 The nosZ I and nosZ II genes were distinguished based on their top-10 hits. The protein sequences of the ORFs were annotated using Kraken 2 (v 2.0.8b) (Wood et al., 2019) and GTDB combined (Parks et al., 2018; Chaumeil et al., 2019). The normalized abundance of ORF for all samples, i.e., the expression levels measured on the transcripts per million (TPM) scale, were estimated using Salmon (v 1.1.0) (Patro et al., 2017). Raw metagenomic sequencing data were deposited in the NCBI under BioProject PRJNA957066.
2.7. Statistical analysis
Redundancy analysis (RDA) was performed using Canoco 5 (v 5.0) and linear regression analysis, and graphs were generated using Graphism (v 8.0). The spearman correlation analysis between physiochemical properties and the abundance of denitrification genes was conducted by SPSS (v 26.0). P-value less than 0.05 (p < 0.05) was considered statistically significant.
3. Results
3.1. Physiochemical properties of paddy fields
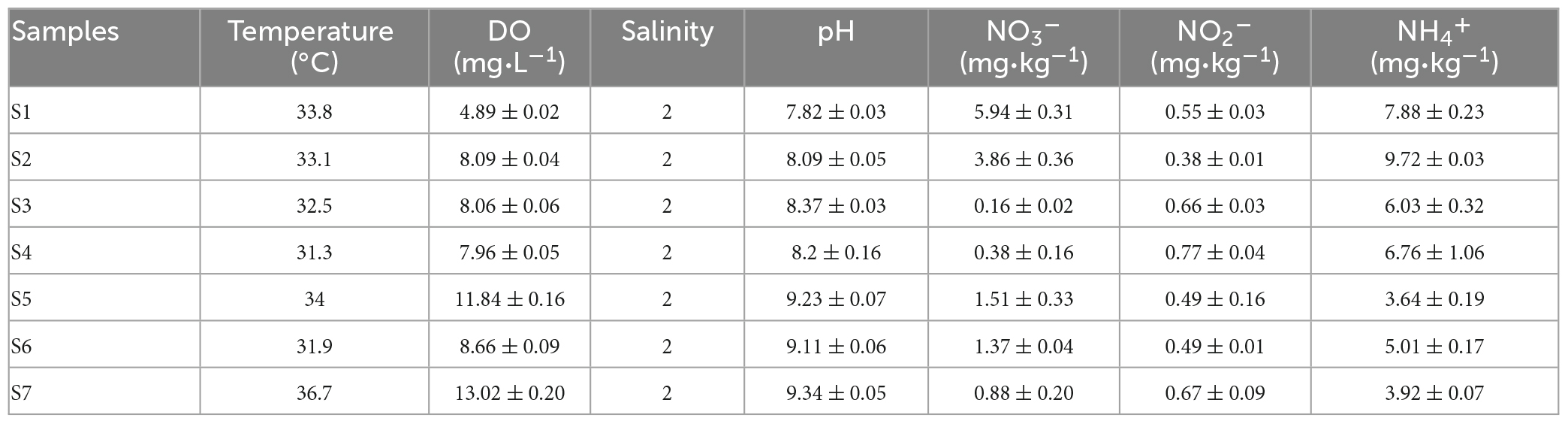
The physiochemical properties of the paddy soils are shown in Table 1. The temperature at the time of sampling was relatively high, i.e., 31.3–36.7°C. The DO of paddy water was low and fluctuated in the range of 4.89 ± 0.02 to 13.02 ± 0.20 mg⋅L–1. The salinity value was two among all paddy water samples. The soil pH was weakly alkaline and its value ranged from 7.82 ± 0.03 to 9.34 ± 0.05. NH4+ was the main existing form of soil DIN, followed by NO3–. The NH4+ and NO3– concentrations varied from 3.64 ± 0.19 to 9.72 ± 0.03 and 0.16 ± 0.02 to 5.94 ± 0.31 mg⋅kg–1, respectively. The NO2– concentrations were low in soil samples, with a minimum value of 0.38 ± 0.01 mg⋅kg–1.
3.2. N2O concentration, ΔN2O concentration, N2O saturation, and N2O flux of air–water exchange
Paddy water showed high N2O concentrations and saturations (Figure 2). The dissolved N2O concentration ranged from 123.65 ± 16.09 to 235.59 ± 5.59 nmol⋅L–1, with minimum and maximum values indicated by samples S2 and S1, respectively (Figure 2A). Excluding dissolved N2O at the air–water equilibrium, the ΔN2O concentration fluctuated between 117.56 ± 16.09 and 229.62 ± 5.59 nmol⋅L–1 (Figure 2B). The N2O dissolved in paddy water was supersaturated, with an average N2O saturation of 2588.13 ± 659.94% (Figure 2C). The paddy fields were a net source of atmospheric N2O, and the maximum N2O flux from the paddy water was estimated to be between 367.28 ± 50.26 and 747.03 ± 18.19 μmol⋅m–2⋅d–1 (Figure 2D).

Figure 2. Dissolved nitrous oxide (N2O) concentration (A), △N2O concentration (B), N2O saturation (C), and N2O emission flux (D) of paddy water.
3.3. Potential denitrification activities in surface paddy soils
The potential N2 production and N2O emission rates were measured via slurry incubation combined with 15N isotope tracer experiments (Figure 3). The paddy soils showed comparable N2 production rates, with an average rate of 24.63 ± 6.76 μmol⋅N⋅kg–1⋅h–1 (Figure 3A). The potential N2O emission rates fluctuated between 0.26 ± 0.15 and 0.90 ± 0.20 μmol⋅N⋅k⋅g–1⋅h–1, with an average value of 0.51 ± 0.20 μmol⋅N⋅kg–1⋅h–1 (Figure 3B). On average, the N2O emitted constituted 2.16 ± 0.85% of the denitrification end-products (Figure 3C), and the potential denitrification rates ranged from 10.92 ± 1.19 to 30.03 ± 1.33 μmol⋅N⋅kg–1⋅h–1 in paddy soils (Figure 3D).
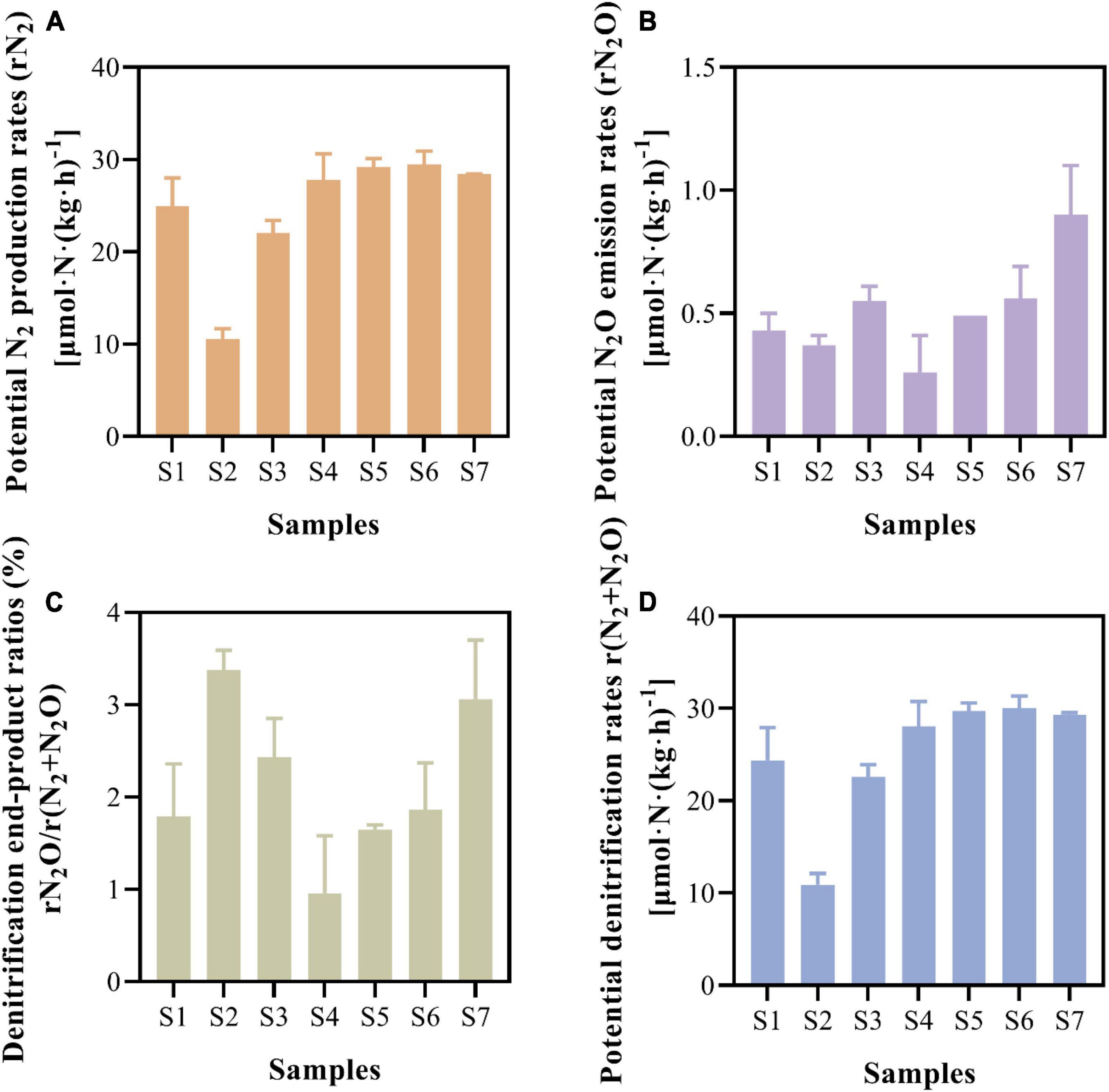
Figure 3. Potential N2 production rates (A), potential N2O emission rates (B), ratios of denitrification end-products (C), and potential denitrification rates (D) of paddy soils.
3.4. Enzymatic activities of NOR and NOS and enzymatic activity ratios of NOR to NOS
The enzymatic activities related to N2O production and N2O reduction were measured (Figure 4). The results showed that the activity of NOR was relatively high, ranging from 413.49 ± 23.84 to 829.52 ± 18.34 U⋅g–1 (Figure 4A). The activity of NOS fluctuated between 85.19 ± 0.32 and 148.92 ± 3.16 U⋅g–1 in the paddy soils (Figure 4B). The activity of NOR was 2.77 to 9.42 times than that of NOS, indicating that the production rate of N2O was higher than the N2O reduction rate at the enzymatic activity level (Figure 4C).
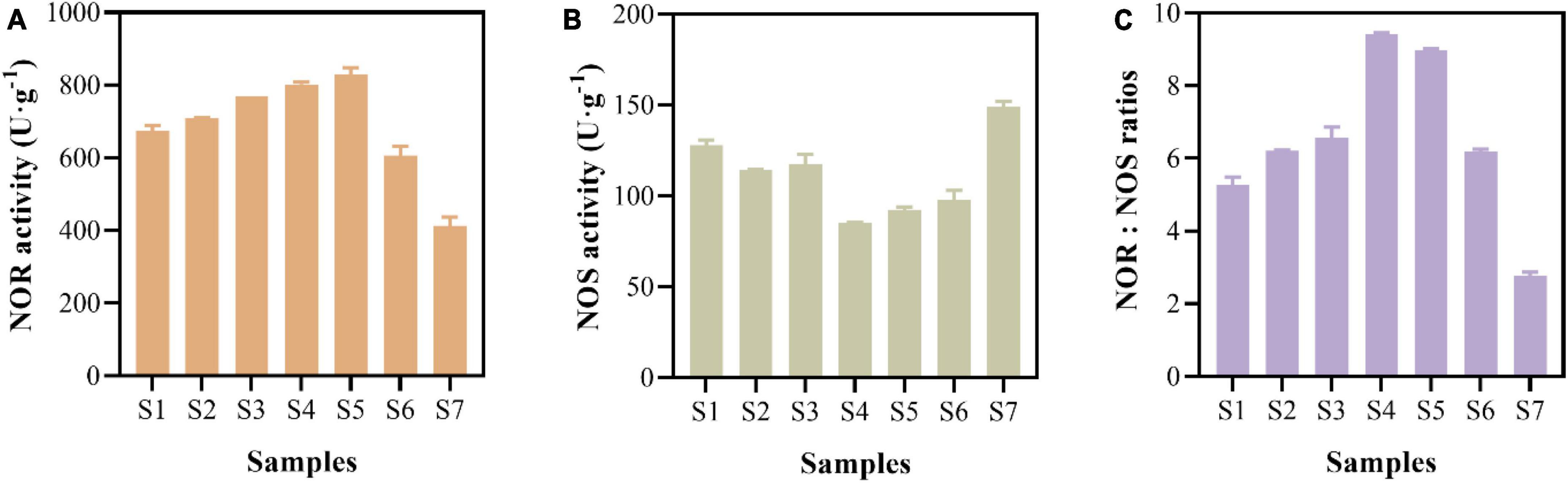
Figure 4. NO reductase (NOR) activity (A), N2O reductase (NOS) activity (B), and activity ratios of NOR to NOS (C) of paddy soils.
3.5. Abundance and abundance ratios of denitrification genes
The abundance of denitrification functional genes was successfully quantified in all paddy soils (Figure 5). The nir and nosZ genes include the nirS and nirK genes and nosZ I and nosZ II genes, respectively. The paddy soil showed higher abundance of the nir gene, with the nirS gene abundance ranging from (1.96 ± 0.20) × 108 to (1.45 ± 0.09) × 109 copies⋅g–1, and the nirK gene abundance ranging from (5.22 ± 0.02) × 107 to (1.68 ± 0.35) × 108 copies⋅g–1, respectively (Figure 5A). The abundance of the nirK gene was positive correlated with the NH4+ concentration (r = 0.786, p < 0.05) by the spearman correlation analysis. Moreover, the abundance of nir genes was positively correlated with the potential N2O emission rate (r = 0.786, p < 0.05). The abundance of the nosZ I and nosZ II genes fluctuated between (7.18 ± 0.29) × 106 and (2.68 ± 0.13) × 107 copies⋅g–1 and between (2.09 ± 0.19) × 108 and (2.86 ± 0.13) × 108 copies⋅g–1, respectively (Figure 5B). Based on the gene abundance ratios of nirS/nirK (average: 9.12 ± 9.00) and nosZ II/nosZ I (average: 21.27 ± 10.28), the nirS and nosZ II genes were the most abundant in paddy soils (Figure 5C). The abundances of nosZ I and nosZ II genes ranged from 108 to 109 and 108, respectively (Figures 5A, B). The gene abundance ratios of nir/nosZ [which ranged from 0.94 to 5.17 (Figure 5C)] exceeded 1, except in sample S4.
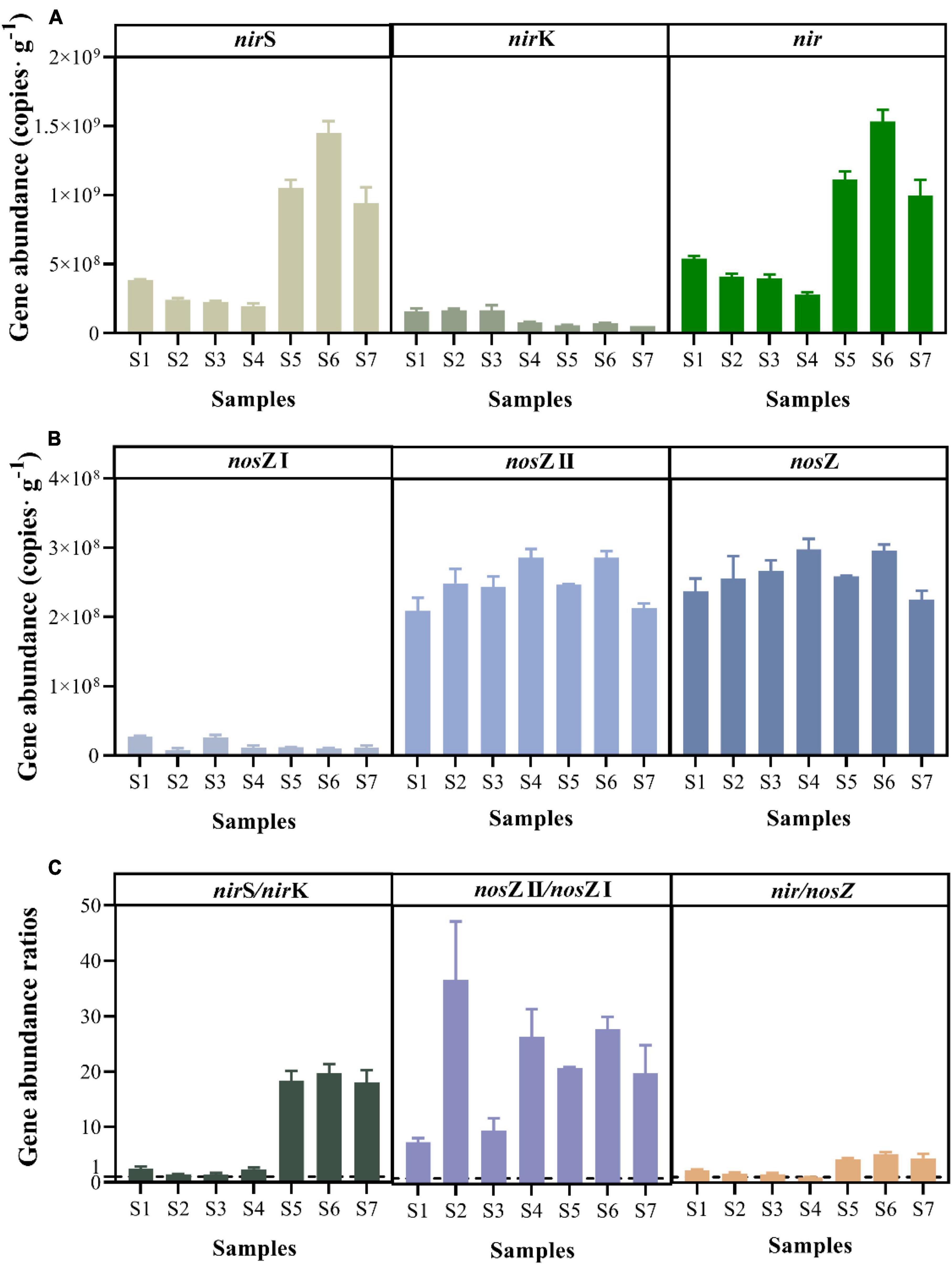
Figure 5. Abundance of key functional genes for denitrification, including nir (nirS and nirK) (A), nosZ (nosZ I and nosZ II) (B), and gene abundance ratios (C) of paddy soils.
3.6. Community composition of denitrification genes
In total, 395,500,000 high-quality sequences were obtained after quality control (Supplementary Table 2). At the phylum level, the prokaryotic communities mainly belonged to Proteobacteria (27.87–41.8%), Actinobacteria (19.14–29.43%), unclassified bacteria (13.63–15.77%), Chloroflexi (2.73–5.06%), and Acidobacteria (2.37–4.32%) (Supplementary Figure 1). Community composition and diversity based on denitrification genes (including napA, nirS, nirK, norB, nosZ I, and nosZ II) were analyzed (Figure 6).
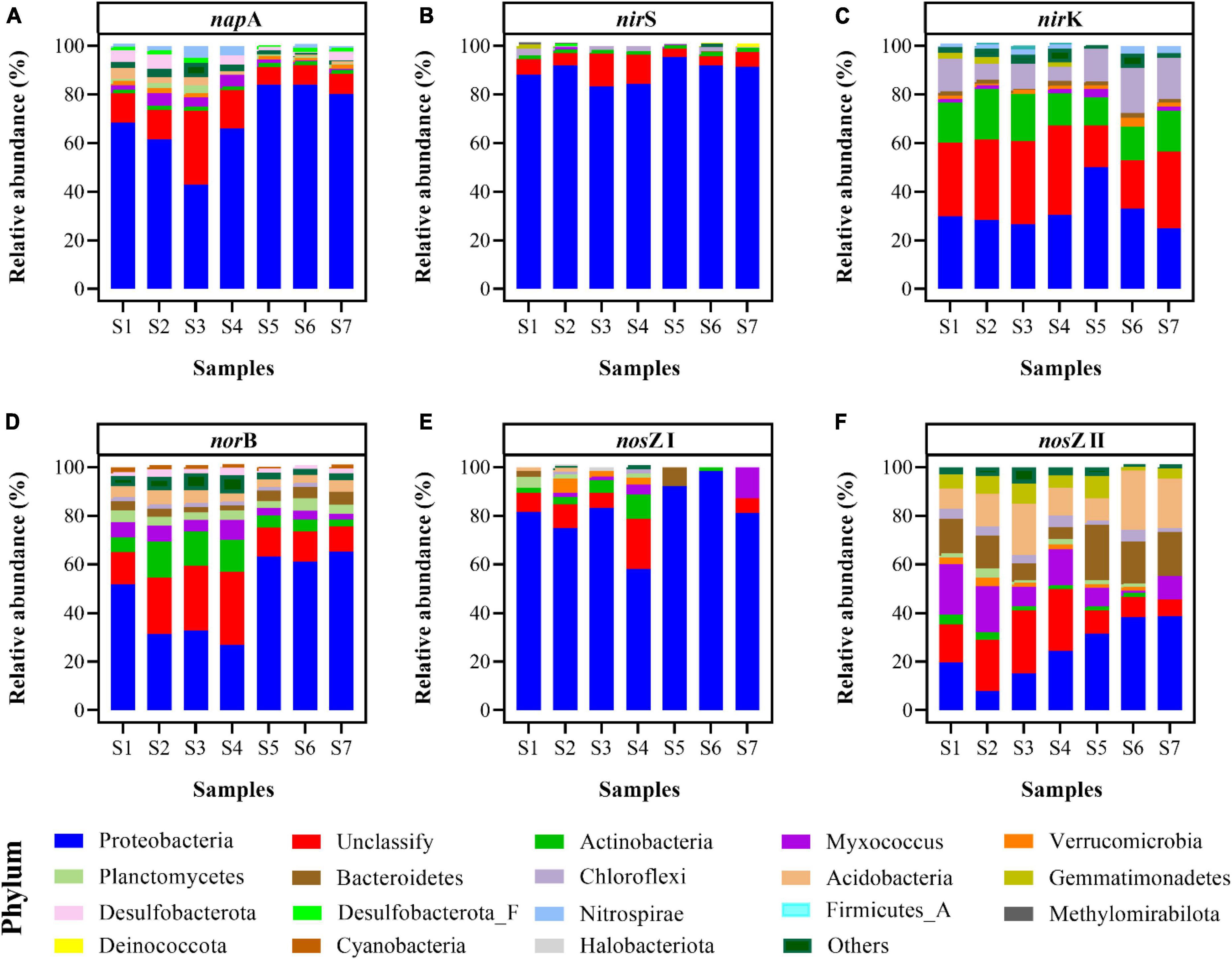
Figure 6. Community compositions of key functional genes for denitrification, including napA (A), nirS (B), nirK (C), norB (D), nosZ I (E), and nosZ II (F) at the phylum level of paddy soils.
The napA gene, catalyzing the reduction of NO3– to NO2–, was abundant in Proteobacteria. The mean abundance of Proteobacteria of the napA gene was 21.22 ± 15.38 TPM (Supplementary Table 3), which constituted 69.58 ± 14.78% of the napA gene community (Figure 6A). Additionally, unclassified bacteria constituted 13.39 ± 8.06% of the napA gene sequences, and the napA gene distributed in Desulfobacterota and Myxococcus for some samples (Figure 6A).
The enzymes encoded by the nirS and nirK genes contributed to the reduction of NO2– to NO. Similar to the composition of the napA gene, a large proportion of the nirS gene appeared in Proteobacteria (83.33–95.39%), and a small proportion of unclassified bacteria (3.34–13.31%) was detected in all paddy soils. The abundances of Proteobacteria and unclassified bacteria in the nirS gene sequences ranged from 9.85 to 59.86 TPM and from 0.71 to 3.47 TPM, respectively (Supplementary Table 4). Additionally, Chloroflexi (0.14–1.89 TPM) and Actinobacteria (0.06–0.33 TPM) were observed in all samples (Figure 6B). The nirK gene was mainly distributed in unclassified bacteria (17.23–34.27%), Proteobacteria (25.05–49.96%), Actinobacteria (11.82–20.80%), and Chloroflexi (7.60–18.42%) (Figure 6C), and their abundances were 9.12 ± 5.40, 8.90 ± 3.40, 4.89 ± 2.73, and 3.27 ± 1.12 TPM, respectively (Supplementary Table 5). Besides, the community composition of the nirK gene was positively correlated with the potential N2O emission (r = 0.821, p < 0.05).
The norB gene, catalyzing the reduction of NO to N2O, was abundant in Proteobacteria, unclassified bacteria, and Actinobacteria. Their mean abundances were 21.77 ± 46.32, 7.24 ± 24.72, 2.14 ± 12.61 TPM, respectively (Supplementary Table 6), and they constituted 26.74–65.35, 10.22–30.36, and 3.03–14.77% of the norB gene sequences, respectively (Figure 6D). In addition, Myxococcus and Acidobacteria, which contained the norB gene, were detected in all soil samples.
N2O reductase, encoded by the nosZ I and nosZ II genes, reduces N2O to N2. Proteobacteria (58.14–98.14%) was the main phylum of the nosZ I gene (Figure 6E), and its abundance fluctuated between 2.54 and 7.93 TPM (Supplementary Table 7). Only a few unclassified bacteria (0.00–20.57%) and Actinobacteria (0.00–10.05%) were indicated in the nosZ I gene sequence (Figure 6E). Meanwhile, for the nosZ II gene, the community composition was more diverse (Figure 6F). Proteobacteria was the dominant phylum; its abundance ranged from 2.04 to 13.21 TPM (Supplementary Table 8) and it constituted 7.97–38.67% of the relative abundance (Figure 6F). Other phyla, including Acidobacteria (8.01–24.36%), unclassified bacteria (7.00–25.95%), Bacteroidetes (4.92–22.84%), Myxococcus (2.07–20.99%), Gemmatimonadetes (1.05–9.28%), and Chloroflexi (0.75–4.78%), were observed in all soil samples (Figure 6F).
In general, Proteobacteria was the most typical group among the denitrification genes, and the relative abundance of classes in Proteobacteria based on the denitrification genes was analyzed (Supplementary Figure 2). Gammaproteobacteria was the dominant Proteobacteria in napA (Supplementary Figure 2A), nirS (Supplementary Figure 2B), and norB genes (Supplementary Figure 2D). Proteobacteria containing nirK (Supplementary Figure 2C), nosZ I (Supplementary Figure 2E), and nosZ II genes (Supplementary Figure 2F) were mainly composed of Alphaproteobacteria and Gammaproteobacteria.
The results of RDA showed that pH of the paddy soils significantly affected the community composition of most denitrification genes, including napA, nirS, nirK, nosZ I, and nosZ II genes (p < 0.05) (Supplementary Figure 3). In addition, the community composition of the nirS gene was affected by the NO3– concentration (p < 0.05) (Supplementary Figure 3A).
4. Discussion
4.1. Contribution of denitrification to N2O emission
Soil and water in paddy fields are closely related but rarely investigated simultaneously. In this study, we determined the N2O emission capacity of both paddy soil and paddy water. The potential N2O emission rates fluctuated between 0.26 ± 0.15 and 0.90 ± 0.20 μmol⋅N⋅kg–1⋅h–1 (Figure 3B), which was higher than those of soils from Hebei Province (0.029 μmol⋅N⋅kg–1⋅h–1) (Zhao et al., 2019) and similar to those of sediments from the Chongming Dongtan wetland (0.21–0.84 μmol⋅N⋅kg–1⋅h–1) (Gao et al., 2022). Based on the potential N2O emission rate in surface paddy soils, the potential N2O emission by denitrification reached 0.31 ± 0.12 mol N2O⋅m–2⋅yr–1, generating an emission estimation of 13.67 ± 5.44 g N2O⋅m–2⋅yr–1 in surface paddy soils (estimation method supplied in Supplementary material).
The concentration of N2O in the paddy water was high, i.e., 123.54 to 235.59 nmol⋅L–1 (Figure 2A). The concentration of N2O in the paddy water was approximately 18.17 times that in Indian estuaries (Rao and Sarma, 2013) and 7.72–14.72 times that in the Jinshui River (Zhao and Zhang, 2021). Excessive application of nitrogen fertilizer in paddy fields can promote the release of a significant amount of N2O (Gupta et al., 2021). As expected, supersaturation was observed in the paddy water, and its average N2O saturation (2588.13 ± 659.94%) was significantly higher than that of water from the Jinshui and Qi Rivers (Zhao and Zhang, 2021) and from the Shanghai River (770%) (Yu et al., 2013). The mean N2O flux (489.59 ± 147.28 μmol⋅m–2⋅d–1) in the paddy water were significantly higher than that in the upper Pearl River estuarine water (313 ± 150 μmol⋅m–2⋅d–1) (Lin et al., 2016) and the Shanghai city river (140 μmol⋅m–2⋅d–1) (Yu et al., 2013). Based on the N2O flux, the average N2O release reached 0.18 ± 0.05 mol⋅N2O m–2⋅yr–1, which generated an emission estimation of 7.86 ± 2.37 g N2O⋅m–2⋅yr–1 in the paddy water (estimation method supplied in Supplementary material). The N2O released from the surface paddy soils contributed to the N2O dissolved in the paddy water and was further converted, and the remaining N2O was released from the paddy water into the atmosphere.
4.2. Abundance of denitrification genes and enzymatic activities related to N2O production and reduction in paddy soil
Microbial denitrification is a four-step reduction process catalyzed by different enzymes, which are mainly encoded by the napA, nirK/S, norB, and nosZI/II genes (Zumft, 1997). Both functional genes and enzymatic activities are vital to denitrification. N2O is an intermediate product of the denitrification process, and the relative contributions of N2O production and reduction determine whether N2O is released into the atmosphere as well as the amount released (Saarenheimo et al., 2015).
Regarding the abundance of the denitrification genes, previous studies pertaining to paddy soils (Zhao et al., 2019; Zhang et al., 2021) and wetland soils (Jiang et al., 2020) showed higher abundance of the nirS gene than the nirK gene. Similar to these studies, the abundance of nirS gene in the paddy soils was 1.39–19.71 times higher than that of nirK gene (Figures 5A, C). Furthermore, it was consistent with the better adaptation of the nirS gene to stable and watery environments compared with the nirK gene (Petersen et al., 2012). In addition, the abundance ratio of nirS to the 16S rRNA gene was positively correlated with the potential N2O emission rate (Supplementary Figure 4), as similarly observed in soils (Assémien et al., 2019; Zhao et al., 2019). Among the genes contributing to N2O reduction, the abundance of nosZ II gene was higher than that of nosZ I gene, and the abundance ratios of nosZ II to nosZ I genes ranged from 2.67 to 10.91 (Figures 5B, C). The ratios were consistent with the typically reported value ranging between 1.5 and 10 in different environments (Jones et al., 2013; Frame et al., 2014; Tsiknia et al., 2015). Additionally, previous studies demonstrated that the abundance of the nosZ II gene contributed significantly to the soil N2O sink capacity (Jones et al., 2014). Thus, nosZ II gene may be vital to N2O reduction. Considering genes related to N2O production and reduction, the nir/nosZ abundance ratios ranged from 0.94 to 5.17 (Figure 5C), which were similar to those in farmland soil (Zhao et al., 2019) and lake sediments (Saarenheimo et al., 2015). The ratios revealed an imbalance between N2O production and reduction, indicating that the potential of paddy soils to produce N2O is greater than reduce it at the genetic level (Domeignoz-Horta et al., 2015).
Microbial denitrification is an enzyme-mediated biochemical process; however, the activities of denitrifying enzymes have been disregarded in most researches (Morales et al., 2010; Petersen et al., 2012). Some studies showed that enzymatic activity affected sediment N2O emissions (Zheng et al., 2014; Su et al., 2019). NOR and NOS are enzymes that catalyze the production and reduction processes of N2O, respectively (Braker and Tiedje, 2003; Zumft and Kroneck, 2007). In our study, the NOR activity in paddy soil (413.19–829.52 U⋅g–1) was higher than that in Donghu sediment (274.70 U⋅g–1), while the NOS activity (85.19–148.92 U⋅g–1) was lower than that in Donghu sediment (188.73 U⋅g–1) (Zhang et al., 2022). NOR activity was higher than NOS activity, with the ratio of NOR to NOS ranging from 2.77 to 9.42 (Figure 4). This ratio was higher than the ratio detected in Donghu Lake sediments, i.e., 1.46 (Zhang et al., 2022), and that determined from riparian sediments, i.e., 0.33 (Su et al., 2019). These enzymatic activities agreed well with the genetic potential of N2O emissions (Figure 5C) and potential N2O emission rates (Figure 3C) in the paddy soils. The enzymatic activity ratios suggest that paddy soils have a higher potential to produce N2O than to reduce it at the protein level. Enzyme are proteins encoded by functional genes, in view of the fact that the gene abundance of nir was higher than that of nosZ gene (Figure 5C), the higher activity of NOR than NOS was more likely to be the result of gene expression.
4.3. Modularity of denitrification process and taxonomy groups of different denitrifying bacteria
By performing metagenomic sequencing, we achieved a more comprehensive understanding of the bacterial community composition in paddy soils, particularly that of denitrifying bacteria. Bacteria in the paddy soils investigated were mainly Proteobacteria, Actinobacteria, Chloroflexi, and Acidobacteria at the phylum level (Supplementary Figure 1); such microbial community composition has been similarly detected in paddy soils from Jiangxi Province (Zhang et al., 2021). However, the dominant community composition varied for different denitrification genes (Figure 6).
Several genes including napA, nirS, nirK, and norB are closely related with the production of N2O (Zumft, 1997). Our results showed that microbial groups with different denitrification genes exhibited distinct taxonomic characteristics. Proteobacteria was the most abundant phylum for the napA gene in paddy soils, which was consistent with the results of sediments from the Pearl River Estuary (Wang et al., 2021). In addition, other napA-harboring bacteria were affiliated with Myxococcus and Actinobacteria (Figure 6A and Supplementary Table 3). Previous studies based on high-throughput amplicon sequencing or clone libraries demonstrated that the nirS gene belonged to Proteobacteria in sediments (Liu et al., 2020), red soil (Ye et al., 2022) and paddy soil (Yoshida et al., 2009). We performed metagenomic analysis, which avoided primer preference, and discovered that the nirS gene predominantly existed in Proteobacteria, but also in a small proportion of unclassified bacteria, Chloroflexi, and Actinobacteria in all the paddy soils (Figure 6A and Supplementary Table 4). The community diversity of nirS gene in this study was higher than that reported in other studies (Jiao et al., 2018; Liu et al., 2020; Ye et al., 2022). However, the community composition of the nirK gene was more diverse than that of nirS gene. The sequences of nirK gene were assigned to unclassified bacteria, Proteobacteria, Actinobacteria, and Chloroflexi (Figure 6C and Supplementary Table 5). Similarly, Proteobacteria and Chloroflexi were the dominant phyla of the nirK gene in sediments from the Pearl River Estuary (Wang et al., 2021). Owing to primer limitations, information regarding groups containing the norB gene is limited. In this study, norB gene was widespread in Proteobacteria, unclassified bacteria, Actinobacteria, Myxococcus, Acidobacteria, Planctomycetes, and Bacteroidetes (Figure 6 and Supplementary Table 6). Moreover, Gammaproteobacteria contained high levels of both the nirS gene (9.22–57.61 TPM) and norB gene (16.58–42.58 TPM) (Figure 7A). This result was similar to a previous finding where a high percentage of organisms with the nirS gene was discovered among bacteria that also contained the norB gene (Graf et al., 2014). However, the other groups of microorganisms did not contain the nirS gene but contained the norB gene (Figure 7). Many dominant phyla of the norB gene (including Actinobacteria, Planctomycetes, Desulfobacterota, Cyanobacteria, Acidobacteria, Bacteroidetes, and Myxococcus) do not have or have a relatively low abundance of the nirS gene because of the predominant distribution of the nirS gene in the Proteobacteria phylum (Figure 7).
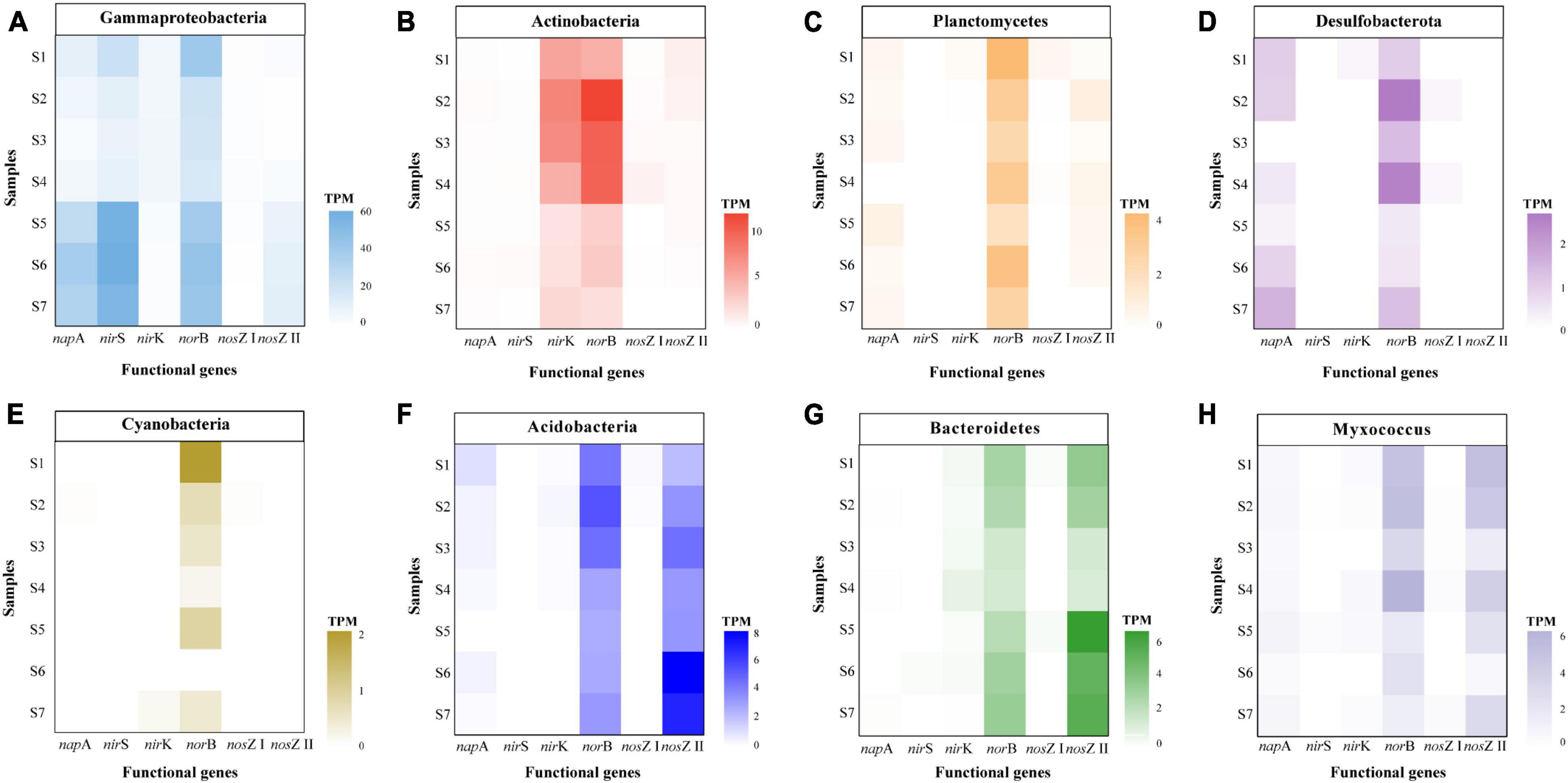
Figure 7. Abundance of key functional genes for denitrification in dominant phyla of norB gene, including Gammaproteobacteria (A), Actinobacteria (B), Planctomycetes (C), Desulfobacterota (D), Cyanobacteria (E), Acidobacteria (F), Bacteroidetes (G), and Myxococcus (H).
The reduction of N2O, which is the only pathway for the N2O sink, is solely related to nosZ I and nosZ II genes (Jones et al., 2013). Although the nosZ I gene was predominately related to Proteobacteria (Green et al., 2010; Xiang et al., 2022), many previously overlooked phyla were identified in all paddy soils, including Actinobacteria and Myxococcus (Figure 6C and Supplementary Table 7). Thus, previous community studies based on high-throughput amplicon sequencing of nirS and nosZ I genes inevitably overlooked some denitrifying bacterial diversity. The nosZ II gene appeared in Proteobacteria, Acidobacteria, unclassified bacteria, Bacteroidetes, Myxococcus, Gemmatimonadetes, and Chloroflexi in all the paddy soils investigated (Figure 6D and Supplementary Table 8); similarly, these phyla of the nosZ II gene have been reported in sediments from Bohai Sea and Jiulong River (Dai et al., 2022). For the nosZ I and nosZ II genes, Gemmatimonadetes appeared only in the nosZ II gene (Supplementary Tables 7, 8), as detected in soils from Hebei Province (Zhao et al., 2019). In addition, the abundance of nosZ II was higher than that of nosZ I in many phyla, including Acidobacteria, Bacteroidetes, Myxococcus, and Chloroflexi (Supplementary Tables 7, 8), indicating that these phyla are more likely to be NosZ-II-type N2O-reducing bacteria.
NapA, nirS, and nosZ I genes were mainly affiliated with Proteobacteria, whereas nirK, norB, nosZ II genes were composed of different phyla (Figure 6). As the norB gene contributes to N2O production (Zumft, 1997), we further analyzed the abundance of denitrification genes in the dominant community of the norB gene. Gammaproteobacteria and many phyla, including Actinobacteria, Planctomycetes, Desulfobacterota, Cyanobacteria, Acidobacteria, Bacteroidetes, and Myxococcus, harbored the norB gene and did not have or had a relatively lower abundance of the nosZ gene (Figures 7A–E). This imbalance between N2O production and the reduction in these phyla may have contributed to the release of N2O from paddy soils. In addition, the abundance of the norB and nosZ II genes was relatively high in Acidobacteria, Bacteroidetes, and Myxococcus, whereas the abundance of napA, nirK, and nosZ I genes was low, and that of nirS gene was absent (Figures 7F–H). This suggests that Acidobacteria, Bacteroidetes, and Myxococcus ware more likely to be NosZ-II-type N2O-reducing bacteria that contain the norB gene but lack the nir gene. This was consistent with the discovery of bacteria that contain the nosZ gene and lack the nir gene, mainly in Bacteroidetes (Graf et al., 2014), and further extended the understanding that Bacteroidetes did not contain the nir gene but harbored the norB and nosZ II genes. For Alphaproteobacteria, the similar abundance distribution of the norB gene and the nosZ I gene indicate that they possess both N2O production and reduction potential (Supplementary Figure 5). It is inferred that their contribution to N2O emissions is likely smaller than that of Gammaproteobacteria. Results of co-occurrence analysis between different denitrification genes showed that denitrification in paddy soil was a highly modular and truncated process that caused an imbalance in the N2O production and reduction processes, thus resulting in N2O emission. However, more data regarding the distribution of denitrification genes in diverse microbial taxa are required to obtain a more comprehensive understanding of the modularity in denitrification.
5. Conclusion
This study provides valuable insights into N2O emission during paddy soil denitrification. The high potential for N2O emission was facilitated by the imbalance between N2O production and reduction processes in terms of gene abundance (nir/nosZ) and enzymatic activity (NOR/NOS). The nirS and nosZ II genes were abundant in paddy soils. Furthermore, the composition of denitrification genes demonstrated a highly modularized denitrification process in paddy fields. Gammaproteobacteria and other phyla, including Actinobacteria, Planctomycetes, Desulfobacterota, Cyanobacteria, Acidobacteria, Bacteroidetes, and Myxococcus, containing the norB gene without nosZ genes, may contribute to N2O emission from paddy soils. These results enhance our understanding of N2O emission during denitrification and provide a theoretical basis for mitigating greenhouse gas emissions in agricultural ecosystems.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in below: https://www.ncbi.nlm.nih.gov/, PRJNA957066.
Author contributions
HX conducted the experiments and wrote the original draft. HX, YH, and AL reviewed and edited the manuscript. HX, JW, YW, FY, JY, and JL performed field sampling. YH and AL funded this study. All authors have read and approved the final version of the manuscript.
Funding
This study was funded by the National Natural Science Foundation of China (Nos. 42276130 and 42006122), the Innovation Team Project of Guangdong Provincial Department of Education (No. 2021KCXTD016), and the Basic and Applied Basic Research Foundation for Guangdong Province (Nos. 2019B1515120066, 2020A1515110597, and 2021A1515011548).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1218207/full#supplementary-material
Footnotes
References
Aramaki, T., Blanc-Mathieu, R., Endo, H., Ohkubo, K., Kanehisa, M., Goto, S., et al. (2019). KofamKOALA: KEGG ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics 36, 2251–2252. doi: 10.1093/bioinformatics/btz859
Arnoux, P., Sabaty, M., Alric, J., Frangioni, B., Guigliarelli, B., Adriano, J.-M., et al. (2003). Structural and redox plasticity in the heterodimeric periplasmic nitrate reductase. Nat. Struct. Biol. 10, 928–934. doi: 10.1038/nsb994
Assémien, F. L., Cantarel, A. A. M., Florio, A., Lerondelle, C., Pommier, T., Gonnety, J. T., et al. (2019). Different groups of nitrite-reducers and N2O-reducers have distinct ecological niches and functional roles in West African cultivated soils. Soil Biol. Biochem. 129, 39–47. doi: 10.1016/j.soilbio.2018.11.003
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Black, A., Wakelin, S., Hamonts, K., Gerard, E., and Condron, L. M. (2019). Impacts of long term copper exposure on abundance of nitrogen cycling genes and denitrification activity in pasture soils. Appl. Soil Ecol. 138, 253–261. doi: 10.1016/j.apsoil.2019.03.009
Bolger, A., Lohse, M., and Usadel, B. (2014). Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–20. doi: 10.1093/bioinformatics/btu170
Borges, A. V. (2004). Gas transfer velocities of CO2 in three European estuaries (Randers Fjord. Scheldt, and Thames). Limnol. Oceanogr. 49, 1630–1641. doi: 10.4319/lo.2004.49.5.1630
Braker, G., and Tiedje, J. (2003). Nitric oxide reductase (norB) genes from pure cultures and environmental samples. Appl. Environ. Microbiol. 69, 3476–3483. doi: 10.1128/AEM.69.6.3476-3483.2003
Butterbach-Bahl, K., Baggs, E. M., Dannenmann, M., Kiese, R., and Zechmeister-Boltenstern, S. (2013). Nitrous oxide emissions from soils: How well do we understand the processes and their controls? Philos. Trans. R. Soc. Lond. B Biol. Sci. 368:20130122. doi: 10.1098/rstb.2013.0122
Caporaso, J., Lauber, C., Walters, W., Berg-Lyons, D., Lozupone, C., Turnbaugh, P., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U.S.A. 108, 4516–4522. doi: 10.1073/pnas.1000080107
Chaumeil, P.-A., Mussig, A., Philip, H., and Parks, D. (2019). GTDB-Tk: A toolkit to classify genomes with the genome taxonomy database. Bioinformatics 36, 1925–1927. doi: 10.1093/bioinformatics/btz848
Coyne, M. S., Arunakumari, A., Averill, B. A., and Tiedje, J. M. (1989). Immunological identification and distribution of dissimilatory heme cd1 and nonheme copper nitrite reductases in denitrifying bacteria. Appl. Environ. Microbiol. 55, 2924–2931. doi: 10.1128/aem.55.11.2924-2931.1989
Dai, X., Chen, M., Wan, X., Tan, E., Zeng, J., Chen, N., et al. (2022). Potential contributions of nitrifiers and denitrifiers to nitrous oxide sources and sinks in China’s estuarine and coastal areas. Biogeosciences 19, 3757–3773. doi: 10.5194/bg-19-3757-2022
Domeignoz-Horta, L. A., Aymé, S., Bizouard, F., Léonard, J., Bru, D., Philippot, L., et al. (2015). The diversity of the N2O reducers matters for the N2O:N2 denitrification end-product ratio across an annual and a perennial cropping stystem. Front. Microbiol. 6:971. doi: 10.3389/fmicb.2015.00971
Frame, C., Deal, E., Nevison, C., and Casciotti, K. (2014). N2O production in the eastern South Atlantic: Analysis of N2O stable isotopic and concentration data. Glob. Biogeochem. Cycles 28, 1262–1278. doi: 10.1002/2013GB004790
Gao, D., Hou, L., Liu, M., Zheng, Y., Yin, G., and Niu, Y. (2022). N2O emission dynamics along an intertidal elevation gradient in a subtropical estuary: Importance of N2O consumption. Environ. Res. 205:112432. doi: 10.1016/j.envres.2021.112432
Graf, D., Jones, C., and Hallin, S. (2014). Intergenomic comparisons highlight modularity of the denitrification pathway and underpin the importance of community structure for N2O emissions. PLoS One 9:e114118. doi: 10.1371/journal.pone.0114118
Green, S., Prakash, O., Gihring, T., Akob, D., Jasrotia, P., Jardine, P., et al. (2010). Denitrifying bacteria isolated from terrestrial subsurface sediments exposed to mixed-waste contamination. Appl. Environ. Microbiol. 76, 3244–3254. doi: 10.1128/AEM.03069-09
Groffman, P. M. (2012). Terrestrial denitrification: Challenges and opportunities. Ecol. Process. 1:11. doi: 10.1186/2192-1709-1-11
Guan, F., Hong, Y., Wu, J., Wang, Y., Bin, L., Tang, B., et al. (2017). A fast sodium hypobromite oxidation method for the sequential determination of ammonia nitrogen in small volume. J. Ecol. Sci. 36, 42–48. doi: 10.14108/j.cnki.1008-8873.2017.02.006
Gupta, K., Kumar, R., Baruah, K. K., Hazarika, S., Karmakar, S., and Bordoloi, N. (2021). Greenhouse gas emission from rice fields: A review from Indian context. Environ. Sci. Pollut. Res. Int. 28, 30551–30572. doi: 10.1007/s11356-021-13935-1
Hallin, S., and Lindgern, P.-E. (1999). PCR detection of genes encoding nitrite reductase in denitrifying bacteria. Appl. Environ. Microbiol. 65, 1652–1657. doi: 10.1128/AEM.65.4.1652-1657.1999
Harris, E., Diaz-Pines, E., Stoll, E., Schloter, M., Schulz, S., Duffner, C., et al. (2021). Denitrifying pathways dominate nitrous oxide emissions from managed grassland during drought and rewetting. Sci. Adv. 7:eabb7118. doi: 10.1126/sciadv.abb7118
Henry, S., Bru, D., Stres, B., Hallet, S., and Philippot, L. (2006). Quantitative detection of the nosZ gene, encoding nitrous oxide reductase, and comparison of the abundances of 16S rRNA, narG, nirK, and nosZ genes in soils. Appl. Environ. Microbiol. 72, 5181–5189. doi: 10.1128/AEM.00231-06
Hu, H., Chen, D., and He, J.-Z. (2015). Microbial regulation of terrestrial nitrous oxide formation: Understanding the biological pathways for prediction of emission rates. FEMS Microbiol. Rev. 39, 729–749. doi: 10.1093/femsre/fuv021
Hyatt, D., Chen, G.-L., Locascio, P., Land, M., Larimer, F., and Hauser, L. (2010). Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119
Jiang, X., Liu, W., Yao, L., Liu, G., and Yang, Y. (2020). The roles of environmental variation and spatial distance in explaining diversity and biogeography of soil denitrifying communities in remote Tibetan wetlands. FEMS Microbiol. Ecol. 96:fiaa063. doi: 10.1093/femsec/fiaa063
Jiao, L., Wu, J., He, X., Wen, X., Li, Y., and Hong, Y. (2018). Significant microbial nitrogen loss from denitrification and anammox in the land-sea interface of low permeable sediments. Int. Biodeterior. Biodegrad. 135, 80–89. doi: 10.1016/j.ibiod.2018.10.002
Jones, C. M., Graf, D. R., Bru, D., Philippot, L., and Hallin, S. (2013). The unaccounted yet abundant nitrous oxide-reducing microbial community: A potential nitrous oxide sink. ISME J. 7, 417–426. doi: 10.1038/ismej.2012.125
Jones, C. M., Spor, A., Brennan, F. P., Breuil, M.-C., Bru, D., Lemanceau, P., et al. (2014). Recently identified microbial guild mediates soil N2O sink capacity. Nat. Clim. Change 4, 801–805. doi: 10.1038/nclimate2301
Lashof, D., and Ahuja, D. (1990). Relative contributions of greenhouse gas emissions to global warming. Nature 344, 529–531. doi: 10.1038/344529a0
Lin, H., Dai, M., Kao, S.-J., Wang, L., Roberts, E., Yang, J.-Y. T., et al. (2016). Spatiotemporal variability of nitrous oxide in a large eutrophic estuarine system: The Pearl River Estuary, China. Mar. Chem. 182, 14–24. doi: 10.1016/j.marchem.2016.03.005
Liu, X., Wu, J., Hong, Y., Jiao, L., Li, Y., Wang, L., et al. (2020). Nitrogen loss by nirS-type denitrifying bacterial communities in eutrophic coastal sediments. Int. Biodeterior. Biodegrad. 150:104955. doi: 10.1016/j.ibiod.2020.104955
Morales, S. E., Cosart, T., and Holben, W. E. (2010). Bacterial gene abundances as indicators of greenhouse gas emission in soils. ISME J. 4, 799–808. doi: 10.1038/ismej.2010.8
Parks, D. H., Chuvochina, M., Waite, D. W., Rinke, C., Skarshewski, A., Chaumeil, P. A., et al. (2018). A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 36, 996–1004. doi: 10.1038/nbt.4229
Patro, R., Duggal, G., Love, M., Irizarry, R., and Kingsford, C. (2017). Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419. doi: 10.1038/nmeth.4197
Petersen, D., Blazewicz, S., Firestone, M., Herman, D., Turetsky, M., and Waldrop, M. (2012). Abundance of microbial genes associated with nitrogen cycling as indices of biogeochemical process rates across a vegetation gradient in Alaska. Environ. Microbiol. 14, 993–1008. doi: 10.1111/j.1462-2920.2011.02679.x
Rao, G. D., and Sarma, V. V. S. S. (2013). Contribution of N2O emissions to the atmosphere from Indian monsoonal estuaries. Tellus B 65, 1–8. doi: 10.3402/tellusb.v65i0.19660
Ravishankara, A. R., Daniel, J. S., and Portmann, R. W. (2009). Nitrous oxide (N2O): The dominant ozone-depleting substance emitted in the 21st century. Science 326, 123–125. doi: 10.1126/science.1176985
Rich, J., Heichen, R. S., Bottomley, P. J., Cromack, K., and Myrold, D. D. (2003). Community composition and functioning of denitrifying bacteria from adjacent meadow and forest soils. Appl. Environ. Microbiol. 69, 5974–5982. doi: 10.1128/AEM.69.10.5974-5982.2003
Saarenheimo, J., Rissanen, A. J., Arvola, L., Nykänen, H., Lehmann, M. F., and Tiirola, M. (2015). Genetic and environmental controls on nitrous oxide accumulation in lakes. PLoS One 10:e0121201. doi: 10.1371/journal.pone.0121201
Sanford, R. A., Wagner, D. D., Wu, Q., Chee-Sanford, J. C., Thomas, S. H., Cruz-Garcia, C., et al. (2012). Unexpected nondenitrifier nitrous oxide reductase gene diversity and abundance in soils. Proc. Natl. Acad. Sci. U.S.A. 109, 19709–19714. doi: 10.1073/pnas.1211238109
Semedo, M., Wittorf, L., Hallin, S., and Song, B. (2020). Differential expression of clade I and II N2O reductase genes in denitrifying Thauera linaloolentis 47LolT under different nitrogen conditions. FEMS Microbiol. Lett. 367:fnaa205. doi: 10.1093/femsle/fnaa205
Solomon, S., Qin, D., Manning, M., Chen, Z., Marquis, M., and Averyt, K., et al. (eds) (2007). “Summary for policymakers climate change 2007: The physical science basis,” in Contribution of working group I to the fourth assessment report of the intergovernmental panel on climate change, (Cambridge: Cambridge University Press), 1–18.
Su, X., Chen, Y., Wang, Y., Yang, X., and He, Q. (2019). Impacts of chlorothalonil on denitrification and N2O emission in riparian sediments: Microbial metabolism mechanism. Water Res. 148, 188–197. doi: 10.1016/j.watres.2018.10.052
Syakila, A., and Kroeze, C. (2011). The global nitrous oxide budget revisited. Greenhouse Gas Meas. Manag. 1, 17–26. doi: 10.3763/ghgmm.2010.0007
Thamdrup, B., and Dalsgaard, T. (2002). Production of N2 through anaerobic ammonium oxidation coupled to nitrate reduction in marine sediments. Appl. Environ. Microbiol. 68, 1312–1318. doi: 10.1128/AEM.68.3.1312-1318.2002
Throback, I. N., Enwall, K., Jarvis, A., and Hallin, S. (2004). Reassessing PCR primers targeting nirS, nirK and nosZ genes for community surveys of denitrifying bacteria with DGGE. FEMS Microbiol. Ecol. 49, 401–417. doi: 10.1016/j.femsec.2004.04.011
Tsiknia, M., Paranychianakis, N. V., Varouchakis, E. A., and Nikolaidis, N. P. (2015). Environmental drivers of the distribution of nitrogen functional genes at a watershed scale. J. FEMS Microbiol. Ecol. 91:fiv052. doi: 10.1093/femsec/fiv052
Wang, P., Li, J. L., Luo, X. Q., Ahmad, M., Duan, L., Yin, L. Z., et al. (2021). Biogeographical distributions of nitrogen-cycling functional genes in a subtropical estuary. Funct. Ecol. 36, 187–201. doi: 10.1111/1365-2435.13949
Weiss, R. F., and Price, B. A. (1980). Nitrous oxide solubility in water and seawater. Mar. Chem. 8, 347–359. doi: 10.1016/0304-4203(80)90024-9
Wood, D., Lu, J., and Langmead, B. (2019). Improved metagenomic analysis with Kraken 2. Genome Biol. 20:257. doi: 10.1186/s13059-019-1891-0
Wu, J., Hong, Y., Guan, F., Wang, Y., Tan, Y., Yue, W., et al. (2016). A rapid and high-throughput microplate spectrophotometric method for field measurement of nitrate in seawater and freshwater. Sci. Rep. 6:20165. doi: 10.1038/srep20165
Wu, J., Hong, Y., Liu, X., and Hu, Y. (2021). Variations in nitrogen removal rates and microbial communities over sediment depth in Daya Bay, China. Environ. Pollut. 286:117267. doi: 10.1016/j.envpol.2021.117267
Xiang, H., Hong, Y., Wu, J., and Long, A. (2022). Ecological distribution and diversity of key functional genes for denitrification in surface sediments of the northern South China Sea: Implications for potential N2O emissions. Front. Mar. Sci. 9:912402. doi: 10.3389/fmars.2022.912402
Xiang, H., Hong, Y., Wu, J., Wang, Y., Ye, F., Hu, Z., et al. (2023). NosZ–II–type N2O-reducing bacteria play dominant roles in determining the release potential of N2O from sediments in the Pearl River Estuary, China. Environ. Pollut. 329:121732. doi: 10.1016/j.envpol.2023.121732
Xiao, K., Wu, J., Li, H., Hong, Y., Wilson, A. M., Jiao, J. J., et al. (2018). Nitrogen fate in a subtropical mangrove swamp: Potential association with seawater-groundwater exchange. Sci. Total Environ. 635, 586–597. doi: 10.1016/j.scitotenv.2018.04.143
Xu, J., Wang, Y., Yin, J., and He, L. (2005). Determination of nitrous oxide dissolved in seawater by static headspace gas chromatographer. Mar. Environ. Sci. 24, 59–62. doi: 10.3969/j.issn.1007-6336.2005.04.016
Ye, J., Wu, J., and Hong, Y. (2022). Diverse nirS-type denitrifying bacteria contribute to vital nitrogen loss in natural acidic red soils. Curr. Microbiol. 79:289. doi: 10.1007/s00284-022-02982-7
Yoshida, M., Ishii, S., Otsuka, S., and Senoo, K. (2009). Temporal shifts in diversity and quantity of nirS and nirK in a rice paddy field soil. Soil Biol. Biochem. 41, 2044–2051. doi: 10.1016/j.soilbio.2009.07.012
Yu, Z., Deng, H., Wang, D., Ye, M., Tan, Y., Li, Y., et al. (2013). Nitrous oxide emissions in the Shanghai river network: Implications for the effects of urban sewage and IPCC methodology. Glob. Change Biol. 19, 2999–3010. doi: 10.1111/gcb.12290
Zhang, D., Li, M., Yang, Y., Yu, H., Xiao, F., Mao, C., et al. (2022). Nitrite and nitrate reduction drive sediment microbial nitrogen cycling in a eutrophic lake. Water Res. 220:118637. doi: 10.1016/j.watres.2022.118637
Zhang, X., Dang, D., Zheng, L., Wu, L., Wu, Y., Li, H., et al. (2021). Effect of Ag nanoparticles on denitrification and microbial community in a paddy soil. Front. Microbiol. 12:785439. doi: 10.3389/fmicb.2021.785439
Zhao, B., and Zhang, Q. (2021). N2O emission and its influencing factors in subtropical streams, China. Ecol. Process. 10:54. doi: 10.1186/s13717-021-00307-3
Zhao, S., Zhou, J., Yuan, D., Wang, W., Zhou, L., Pi, Y., et al. (2019). NirS-type N2O-producers and nosZ II-type N2O-reducers determine the N2O emission potential in farmland rhizosphere soils. J. Soils Sed. 20, 461–471. doi: 10.1007/s11368-019-02395-3
Zheng, X., Su, Y., Chen, Y., Wan, R., Liu, K., Li, M., et al. (2014). Zinc oxide nanoparticles cause inhibition of microbial denitrification by affecting transcriptional regulation and enzyme activity. Environ. Sci. Technol. 48, 13800–13807. doi: 10.1021/es504251v
Zumft, W. G. (1997). Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61, 533–536. doi: 10.1128/.61.4.533-616.1997
Keywords: denitrification, N2O emission, functional gene, microbial taxa, paddy soils
Citation: Xiang H, Hong Y, Wu J, Wang Y, Ye F, Ye J, Lu J and Long A (2023) Denitrification contributes to N2O emission in paddy soils. Front. Microbiol. 14:1218207. doi: 10.3389/fmicb.2023.1218207
Received: 06 May 2023; Accepted: 01 June 2023;
Published: 16 June 2023.
Edited by:
Xiaobo Liu, Nanjing University of Science and Technology, ChinaCopyright © 2023 Xiang, Hong, Wu, Wang, Ye, Ye, Lu and Long. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yiguo Hong, eWdob25nQGd6aHUuZWR1LmNu; Aimin Long, bG9uZ2FtQHNjc2lvLmFjLmNu
 Hua Xiang1,2,3
Hua Xiang1,2,3 Yiguo Hong
Yiguo Hong Jiapeng Wu
Jiapeng Wu Yu Wang
Yu Wang Fei Ye
Fei Ye