- 1African Genome Center, Mohammed VI Polytechnic University, Ben Guerir, Morocco
- 2Department of Biochemistry, Ahmadu Bello University, Zaria, Nigeria
Background: Metaproteomics is a subfield in meta-omics that is used to characterize the proteome of a microbial community. Despite its importance and the plethora of publications in different research area, scientists struggle to fully comprehend its functional impact on the study of microbiomes. In this study, bibliometric analyses are used to evaluate the current state of metaproteomic research globally as well as evaluate the specific contribution of Africa to this burgeoning research area. In this study, we use bibliometric analyses to evaluate the current state of metaproteomic research globally, identify research frontiers and hotspots, and further predict future trends in metaproteomics. The specific contribution of Africa to this research area was evaluated.
Methods: Relevant documents from 2004 to 2022 were extracted from the Scopus database. The documents were subjected to bibliometric analyses and visualization using VOS viewer and Biblioshiny package in R. Factors such as the trends in publication, country and institutional cooperation networks, leading scientific journals, author’s productivity, and keywords analyses were conducted. The African publications were ranked using Field-Weighted Citation Impact (FWCI) scores.
Results: A total of 1,138 documents were included and the number of publications increased drastically from 2004 to 2022 with more publications (170) reported in 2021. In terms of publishers, Frontiers in Microbiology had the highest number of total publications (62). The United States of America (USA), Germany, China, and Canada, together with other European countries were the most productive. Institution-wise, the Helmholtz Zentrum für Umweltforschung, Germany had more publications while Max Plank Institute had the highest total collaborative link strength. Jehmlich N. was the most productive author whereas Hettich RL had the highest h-index of 63. Regarding Africa, only 2.2% of the overall publications were from the continent with more publication outputs from South Africa. More than half of the publications from the continent had an FWCI score ≥ 1.
Conclusion: The scientific outputs of metaproteomics are rapidly evolving with developed countries leading the way. Although Africa showed prospects for future progress, this could only be accelerated by providing funding, increased collaborations, and mentorship programs.
1. Introduction
The development of “omics” technologies has revolutionized the field of molecular biology by enabling the analysis of biological systems at various molecular levels. The omics approaches which include genomics, transcriptomics, proteomics, and metabolomics focus on a specific aspect of molecular biology, allowing researchers to gain a comprehensive understanding of biological systems. Of course, these have contributed to numerous technological advancements across a wide range of fields (Seyhan and Carini, 2019).
One special field of omics named “meta-omics” has enabled the understanding of the microbial world. It involves the use of high-throughput sequencing technologies to investigate the collective genomic and functional potential of microbial communities. Using meta-omics, complex microbial communities (which cannot be cultured and studied in isolation) can be explored (Zhang et al., 2019; Mauger et al., 2022).
The different subfields of meta-omics (metagenomics, metatranscriptomics, metaproteomics, and metabolomics) work hand-in-hand to fully characterize the microbial population. In brief, metagenomics involves sequencing of DNA fragments in a sample, without the need for isolation and cultivation of individual microbes while metatranscriptomics focuses on the study of the RNA transcripts expressed by the microbial community, providing insights into the active functional pathways and metabolic processes of the community (Shakya et al., 2019; Maghini et al., 2021). Metaproteomics and metabolomics, on the other hand, involve the study of proteins and small molecules produced by microbial communities, providing insights into the metabolic, functional, and biochemical pathways of the community (Nephali et al., 2020; Armengaud, 2023). Hence, new biological molecules of significant importance can be identified, thereby unlocking new opportunities for biotechnology and environmental sciences (Zhang et al., 2019; Tiwari and Taj, 2020).
While metagenomics provides information on the potential functional capabilities of microbial communities, metaproteomics can confirm which functions are actively being performed (Schiebenhoefer et al., 2019). As many studies are focusing on metagenomics, coupling these studies with metaproteomics can address some of the limitations of metagenomics (Issa Isaac et al., 2019; Zheng et al., 2020). For example, metagenomics can be limited by the quality and completeness of the genomic data obtained from environmental samples, while metaproteomics can provide direct evidence of the proteins that are being expressed by microbial communities in the sample (Nowrotek et al., 2019; Renu et al., 2019).
Promising metaproteomics research has been published in recent years. The increase in the publication output could be associated with the technological advancements made in instrumentation (Bailey et al., 2019). One of the advancements is the development of Nano-Liquid chromatography (LC) techniques that are more advantageous than the conventional LC techniques, as well as the advent of high-resolution mass spectrometry (MS) that enables the identification and quantification of tens of thousands of peptides and proteins per sample (Vargas Medina et al., 2020). These have contributed to the understanding of proteins and their significance in different conditions (Hardouin et al., 2021).
However, despite the remarkable advancements, metaproteomics is still in its infancy, and its full potential has not been realized, particularly in developing nations (Hamdi et al., 2021). For instance, few studies in omics were conducted in Africa, most of which are collaborative with the developed world. Although these could be attributed to the lack of funding from the government and private sectors, the limited expertise in use of the advanced technologies, bioinformatics analysis, and interpretation of the generated data could be a limiting factor (El Jaddaoui et al., 2020; Iskandar et al., 2021).
Interestingly, the field of metaproteomics has existed for so long, and its impact on the functional characterization of the microbial population is far-reaching. Many publications have widely used the technology to determine the abundance, diversity, and activity of proteins in different research domains (Chandran et al., 2020; Rane et al., 2022). To mention a few, metaproteomics has been used in studies such as examining the microbiome composition and function (Kumar et al., 2021), comparative proteomics in health and disease states (Calabrese et al., 2022), evaluating the effects of environmental toxins (Jiao et al., 2022), and insights into metabolic pathways (Armengaud, 2023). Furthermore, the technique has been applied in the development of novel biomarkers for medical diagnostics as well as a better understanding of the function proteins play in host-pathogen interactions (Moreira et al., 2021). It is important to note that several reviews have discussed improvements made in metaproteomics (Schiebenhoefer et al., 2019; Andersen et al., 2021; Hardouin et al., 2021; Salvato et al., 2021; Bahule et al., 2022). However, there is scanty information concerning the state-of-art in the field of metaproteomics compared to other omics.
In this article, we conducted a bibliometric analysis in the field of metaproteomics. This kind of analysis has been conducted in other fields to have a full grasp of the achievements made (Md Khudzari et al., 2018; Yu et al., 2018; Kushairi and Ahmi, 2021; Wang et al., 2021). The specific objectives are to assess the current state of metaproteomic research on a global scale, evaluate its significance and potential impact, and specifically examine the contribution of Africa in the research field. The study aims to provide insights into the overall trends, collaboration patterns, and key research areas in metaproteomics globally while highlighting Africa’s research output and its potential role in advancing the field. By understanding the global landscape and Africa’s contributions, this study can inform future research directions, identify potential collaborations, and promote the growth of metaproteomics research in Africa.
2. Materials and methods
2.1. Data source and search strategy
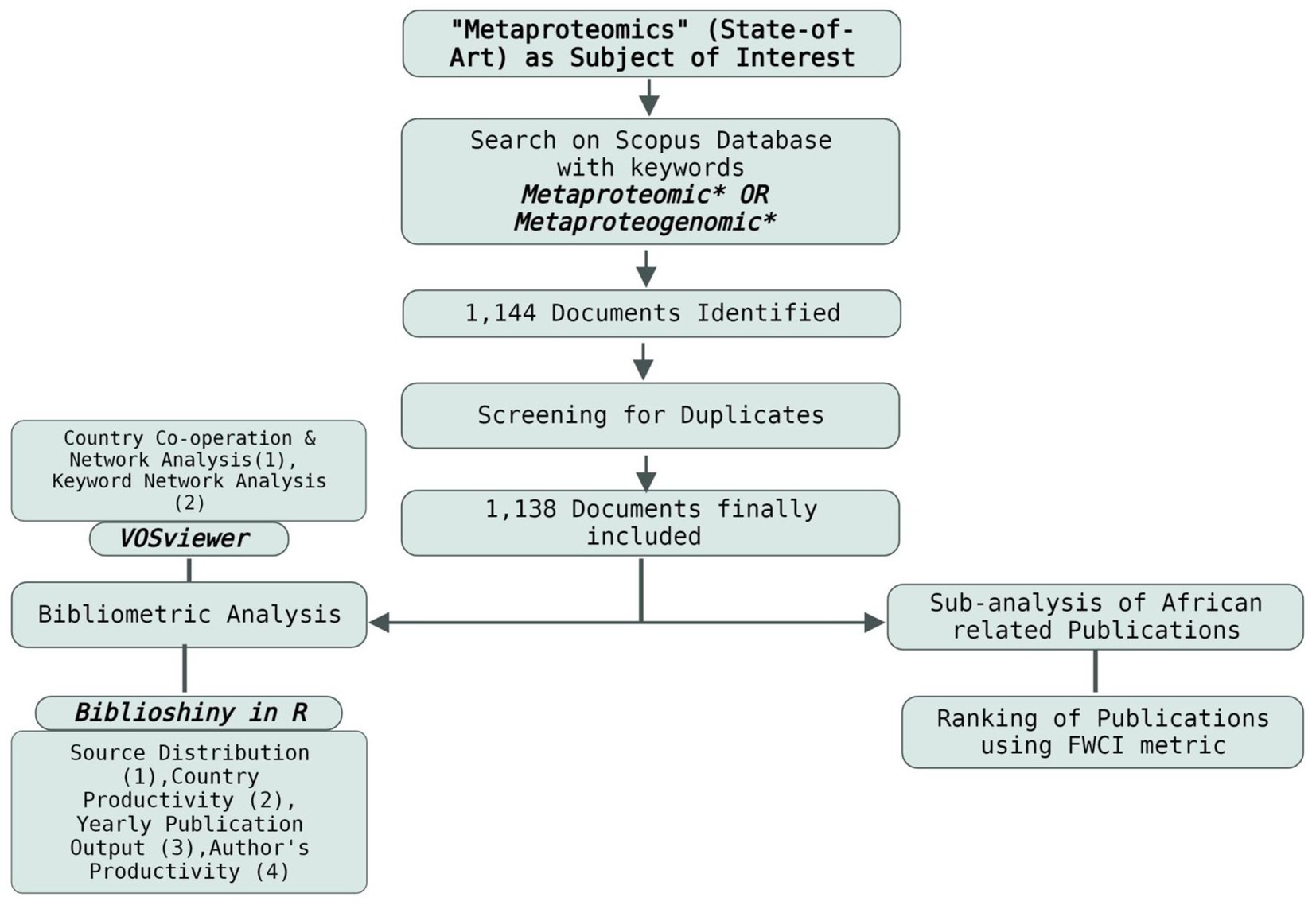
The bibliometric analysis was conducted by searching for publications on metaproteomics in the Scopus database. Initially, a search was conducted on the Web of Science and Scopus databases to draw a comparison between their outputs. It was discovered that the Scopus database contained the majority of publications. The Scopus database is a comprehensive abstract and indexing database, offering extensive coverage across a wide range of subjects. It provides access to a significant number of international journals, ensuring global coverage and facilitating the inclusion of research from different geographic regions (Baas et al., 2020; Zhang et al., 2020). Keywords were selected by initially conducting a review of the relevant literature to identify commonly used terms related to our research topic. The MeSH (Medical Subject Headings) term of “Metaproteomics,” thus “Metaproteogenomic” was identified and the keywords “Metaproteomic” and “Metaproteogenomic” were used to search the titles, abstracts, and keywords of the Scopus database in combination with the Boolean operator “OR.” The wild card “*” was used at the end of the search term to cater to all other variants of the term, that is, “metaproteomic, metaproteomics, metaproteogenomic, and metaproteogenomics.” Only English publications were considered for this work. Hence, the query string used is as follows: TITLE-ABS-KEY (metaproteomic* OR metaproteogenomic*) AND [LIMIT-TO (LANGUAGE, “English”)] AND [EXCLUDE (PUBYEAR, 2023)]. Consequently, the results starting from 2004 to 2022 were collected. Altogether, 1,144 publications of all literature types were retrieved and included in order to ensure the overall comprehensiveness of the metaproteomics research. The completeness of the bibliographic metadata was determined using Biblioshiny (Supplementary Figure 1). In the database, information on journals, publication date, authors’ institutions, countries, publication sources, abstract, citation frequency, keywords, and bibliographies was selected and subsequently downloaded in a CSV file. In order to clean the data, duplicates were searched and removed, and the remaining literature was used for subsequent analysis. To obtain more information, the author’s h-index and Scopus ID, in addition to the journal cite scores, were retrieved from the database. It is important to mention that, in order to eliminate possible bias caused by database updates, data searching and gathering were conducted on the same day (January 17, 2023).
2.2. Bibliometric analyses and data visualization
The bibliometric analysis was conducted according to previous studies (Md Khudzari et al., 2018; Ejaz et al., 2022). VOSviewer (due to its user-friendly interface, ability to handle large datasets, and intuitive visualization options) and the Biblioshiny package in R (Aria and Cuccurullo, 2017) were used for the analysis. The VOSviewer has been known to be useful in developing more sophisticated bibliometric maps (Yu et al., 2018, 2020). The combination of the software and the R package enables in-depth analyses that aid in a more holistic understanding of scholarly collaboration, research impact, and citation dynamics in specific studies (Ragazou et al., 2022).
Using VOSviewer (Version 1.6.18), Country cooperation network, Institutional collaboration network, and Keyword analysis were conducted. Briefly, after importing the CSV file into the software, the co-authorship option was chosen, and the unit of analysis was designated as “countries.” The maximum number of writers per document and the counting method were left as defaults, while threshold criteria were set to a minimum of five (5) papers per country and a minimum of one (1) citation. For the Institutional collaboration network, a thesaurus file was developed and imported into VOSviewer in order to harmonize and re-label synonymic institutional names. Thereafter, co-authorship was selected, and organizations were chosen as the unit of analysis, while the counting method and the maximum number of organizations per document were left as defaults. The threshold criteria were set at a minimum of three (3) documents and zero (0) citations per organization. The type of analysis chosen for the keyword analysis was co-occurrence, and the unit of analysis was Author keywords, with a minimum number of keyword occurrences of five (5). The default counting method was used. A thesaurus file was developed and imported into VOSviewer to re-label synonymic single words and congeneric phrases. Overall, VOSviewer was used to construct network maps for keyword analysis, as well as collaboration between countries and institutions.
For the Biblioshiny, the CSV file was uploaded for analysis of trends in publication, major document types, peer-reviewed scientific journals, country productivity, and distributions of the author’s productivity. The analyses were performed by clicking the tabs on the web interface as follows:
Trends in publication: To access information in relation to the trend in publication, the Biblioshiny→ Overview→ Annual Scientific Production→ Table tabs were clicked. The annual scientific production is based on the total number of publications within each year.
Major document types: The Biblioshiny → Overview→ Main information tabs were clicked to have the information on the document types and their quantity.
Peer-reviewed scientific journals: Information related to the Leading Scientific Journals was accessed by clicking Biblioshiny → Sources → Most relevant Sources tabs.
Country Productivity: For the Country’s Productivity, Biblioshiny → Authors → Countries’ Scientific Production tabs were selected. Information on the total citation count and the average article citations of the countries was obtained by clicking Biblioshiny → Authors → Most Cited Countries tabs. The map of international collaboration between the countries was obtained by clicking Biblioshiny → Social Structure → Countries’ Collaboration World Map tabs.
Distribution of Author’s Productivity: Information concerning the leading Authors in Metaproteomics research was accessed by clicking Biblioshiny → Authors → Most Relevant Authors tabs. The author’s productivity period was ascertained by clicking Biblioshiny → Authors → Author’s Production Over-Time → Plots tabs. The analysis of the Author’s productivity using Lotka’s law was performed by clicking the tabs Biblioshiny → Authors → Lotka’s law.
Trending topics: To analyze trending terms based on the Keywords, the trend topics plot was generated by clicking the Biblioshiny → Documents → Trend topics → Plot tabs.
2.3. Sub-analysis of African publication trends and ranking using FWCI parameter
The emergence of African contributions to the field of metaproteomics was investigated by assessing the distribution and the overall trends in publication activities. To achieve this, data from publications related to Africa were extracted and analyzed using Excel. The ranking of the publications was conducted using the FWCI parameter from Scopus (Zanotto and Carvalho, 2021).
3. Results
3.1. Publications retrieval and screening process
From the Scopus database, 1,144 publications were collected. After screening for duplicates, the number of articles was reduced to 1,138. A bibliometric analysis of the 1,138 publications on metaproteomics obtained from the database was conducted to understand the global impacts of metaproteomics research. The publications included research articles, reviews, conference papers, editorials, book chapters, and others. The workflow of the retrieval process is indicated in Figure 1.
3.2. The trend in publication output and major document types
The analysis revealed that the number of publications on metaproteomics has been increasing steadily since the early 2000s. Initially, studies on the topic started in 2004 with a single study. Afterwards, a slow increase in the number of publications was observed from 2006 to 2009. A significant rise in the number of publications was observed from 2010 onwards (Figure 2A). From 2010 onwards, there was a steady rise in the number of publications. Although a high number of publications (164; 14.4%) were recorded in 2022, there was more publication output in 2021 (170; 14.9%). Moreover, there was no publication record found in 2005. Overall, the estimated annual growth rate (EAGR) of the publications computed from the Biblioshiny package was 37.25%.
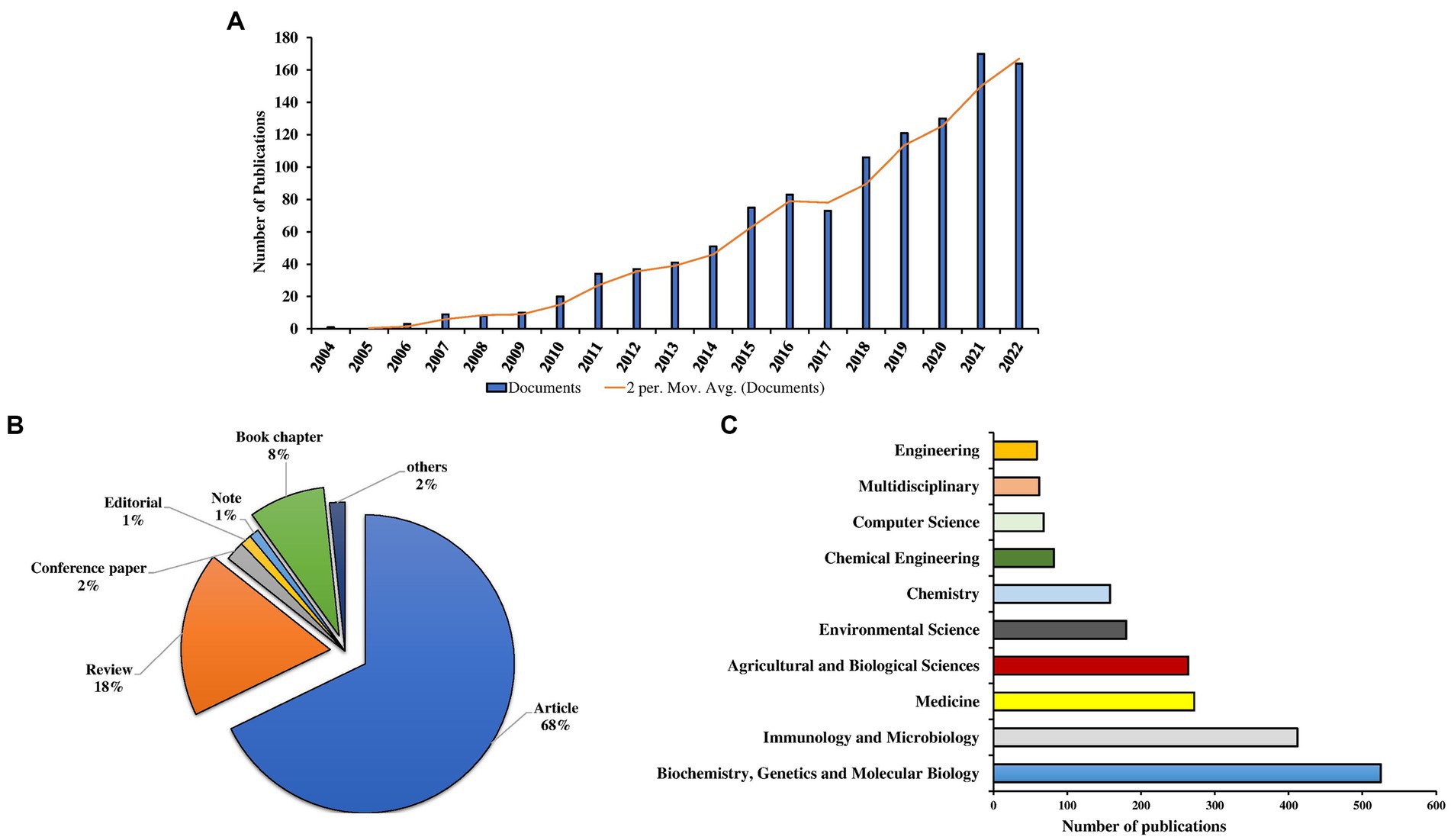
Figure 2. Global publication trend (A), document types (B), and global leading subject areas (C) in metaproteomics research.
In terms of the document types (Figure 2B), the majority of the publications were research articles (772; 68%), followed by reviews (203; 18%) and book chapters (94; 8%). Others, including conference papers, editorials, and notes, account for 3.95%. Information gathered from the Scopus database revealed that the majority of the publications are all open access (678; 59.6%).
3.3. Leading subject areas and peer-reviewed scientific journals
The publications on metaproteomics included in the Scopus database cover 23 subject areas. The Major areas with the most densely distributed publications are Biochemistry, Genetics, and Molecular Biology with 525 published documents representing 46.13% of the total number of scholarly works (Figure 2C). Other subject areas with publications include Immunology and Microbiology (412; 36.20%), Medicine (272; 23.90%), Agricultural and Biological science (264; 23.19%), Environmental science (180; 15.82%), Chemistry (158; 13.88%), Chemical Engineering (82; 7.21%), Computer Science (68; 5.98%), Multidisciplinary (62; 5.45%), and Engineering (59; 5.19%) (Figure 2C).
The present finding investigated the major Journal houses that published the metaproteomic research, and among the top 10, Springer Nature (86; 7.56%) and Wiley-Blackwell (77; 6.77%) were the top publishers (Table 1). The American Chemical Society (ACS), Elsevier, Frontiers Media S.A., and the Public Library of Science were also among them (Table 1).
In terms of the top 10 journals, Frontiers in Microbiology has the highest number of publications (62; 5.45%), followed by the Journal of Proteome Research (50; 4.39%), Proteomics (49; 4.31%), Microbiome (37; 3.25%), and ISME Journal (32; 2.88%). Conversely, while assessing the Journal’s CiteScore from the Scopus database (2021 report), Microbiome has the highest score of 24.5, followed by Nature Communications (23.2), and ISME Journal (18.2) (Table 1).
3.4. Country productivity and cooperation network
Looking at the country’s productivity, the top 20 most productive countries (in terms of publications) are the USA (359), Germany (257), China (172), and Canada (90). These countries have the highest total citations (>1,504), in addition to other countries like Spain (1,728), Sweden (1,620), and the United Kingdom (1,414). Contrariwise, Switzerland, Finland, and the UK have high average article citations of 147.3, 58.0, and 56.6, respectively (Table 2).
The map and network of international and country cooperation (Figures 3A,B) demonstrate close collaboration among countries or regions involved in metaproteomics research. In the first cluster, the USA is the most affiliated country, linked to 38 countries or territories with a total link strength of 330, while China, as the second, is affiliated with 28 countries with a link strength of 119. Germany also has a total of 30 links and a total link strength of 334, followed by the UK in the second cluster. The final cluster includes Italy and Canada with more collaboration networks.
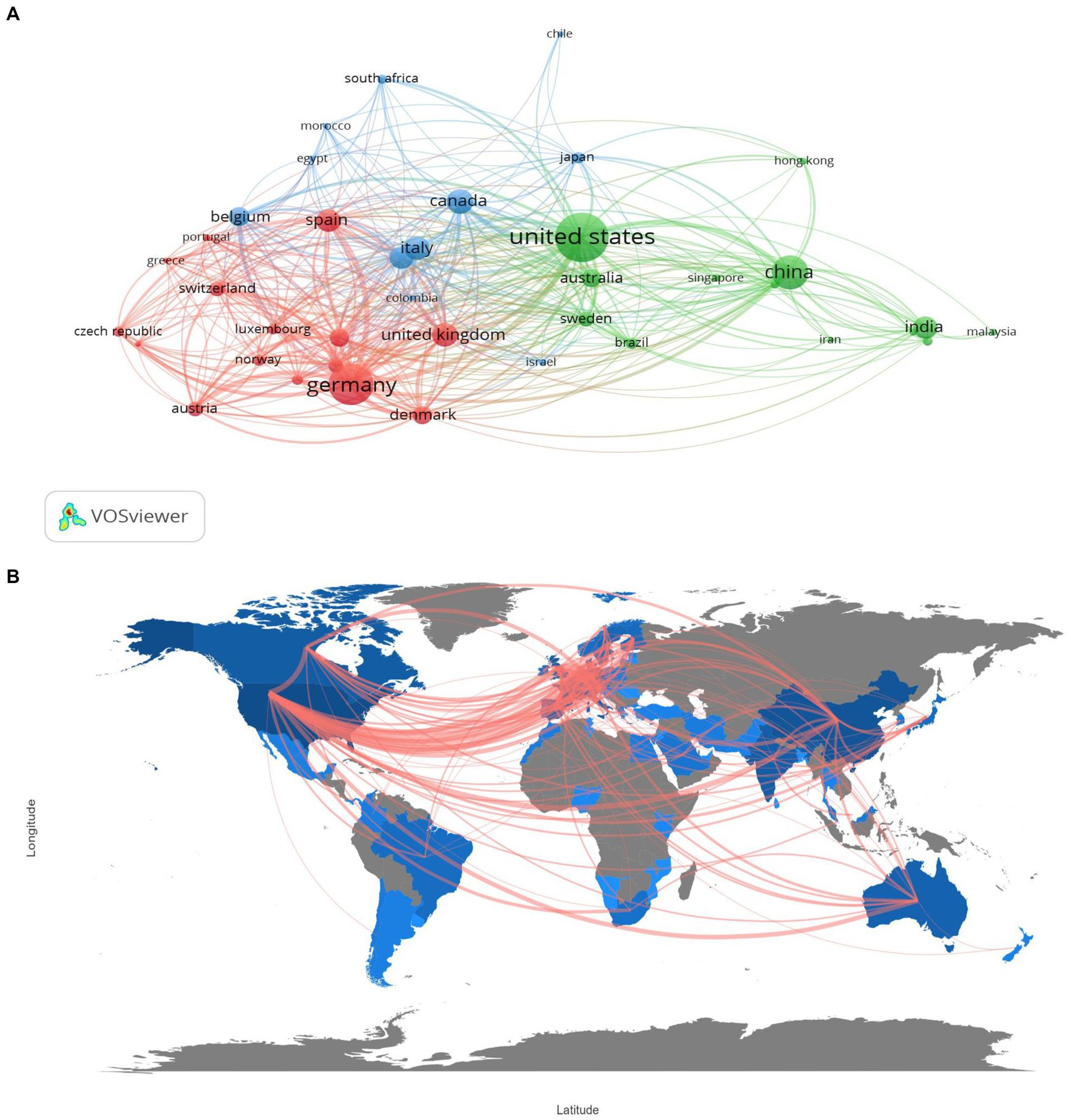
Figure 3. Network of country co-operation (A), and worldwide map indicating international collaborations (B) in metaproteomics research.
In terms of African countries, Morocco had the highest number of collaborations (14 links), followed by Egypt (13 links) and South Africa (11 links), with total link strengths of 20, 18, and 28, respectively.
3.5. Top institutions and distribution of author’s productivity
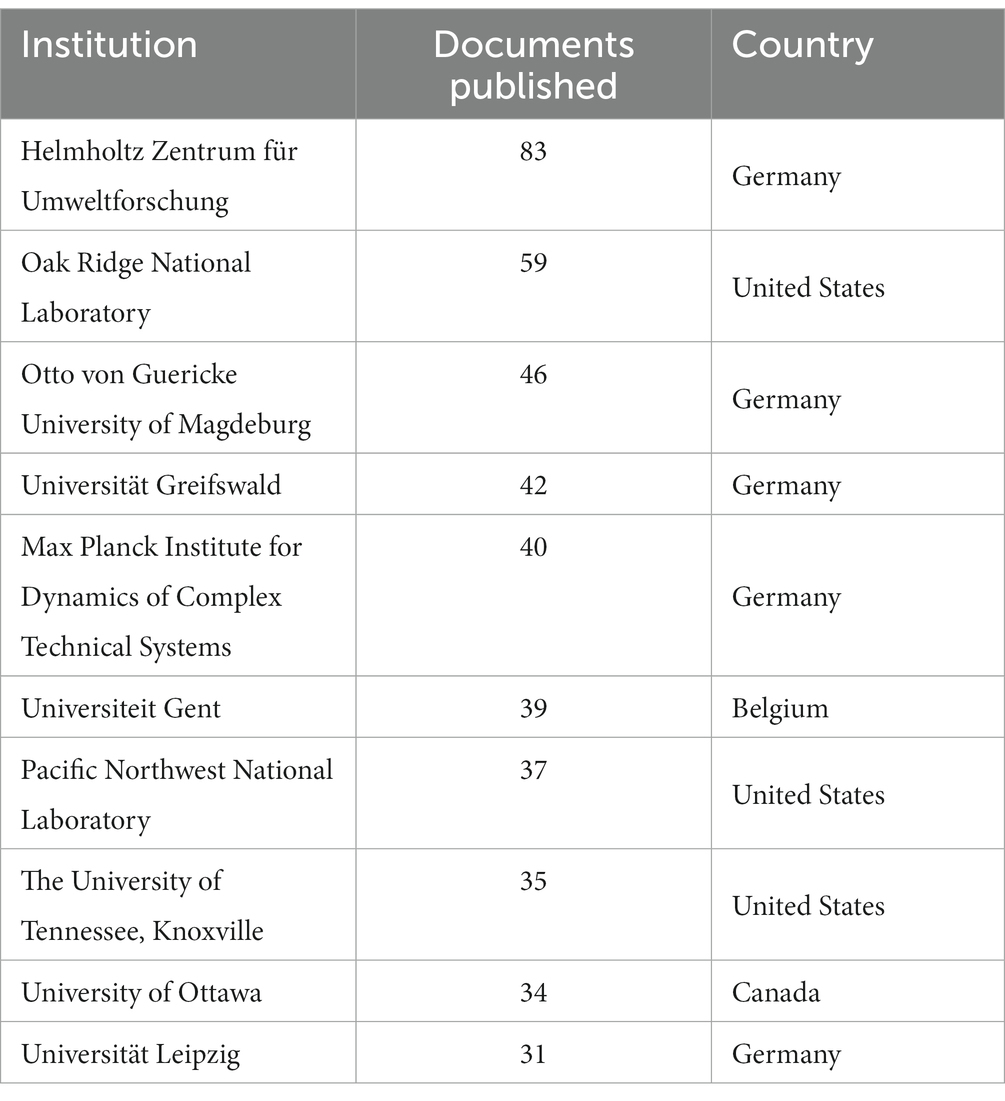
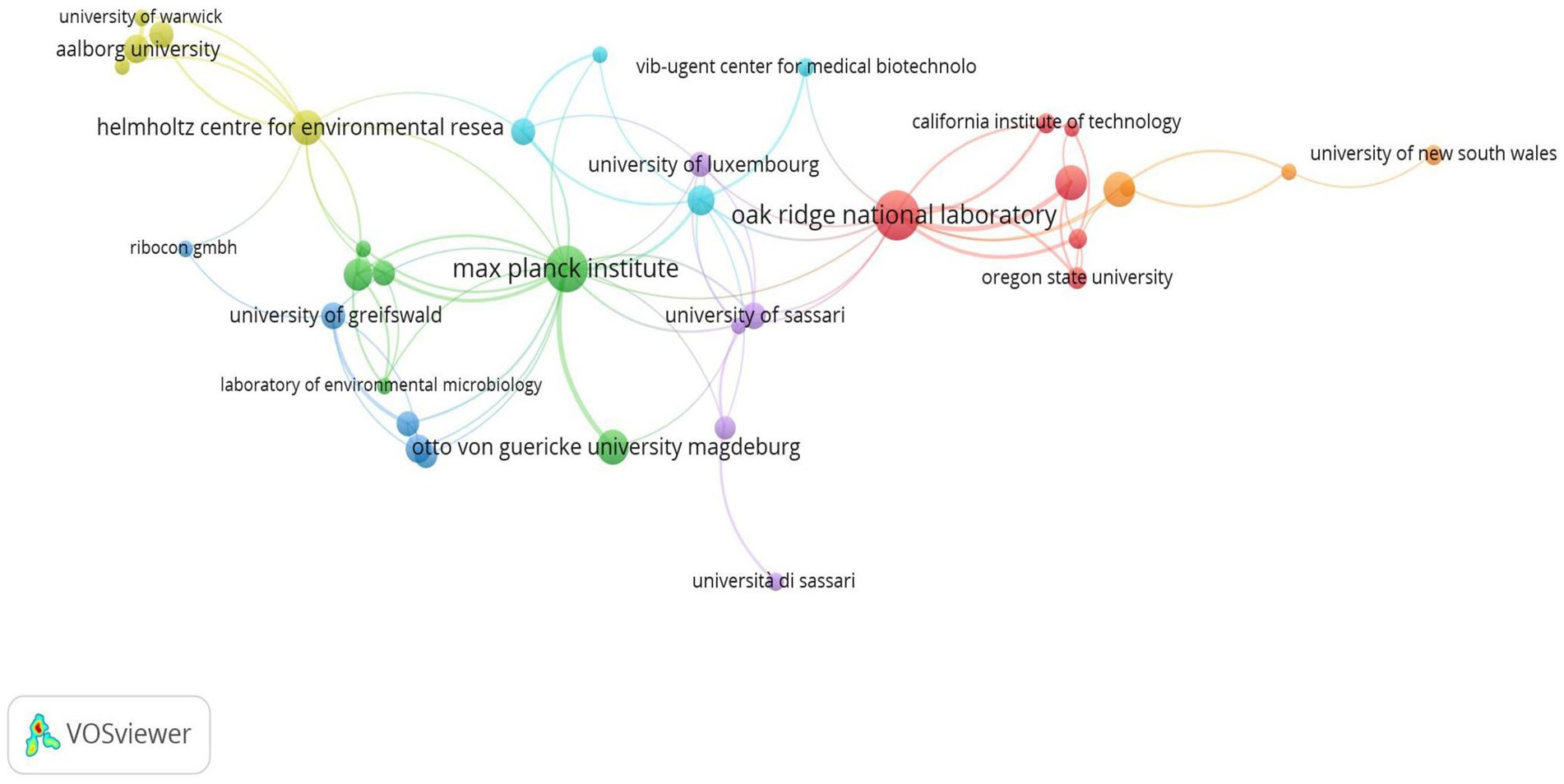
Among the institutions, Helmholtz Zentrum für Umweltforschung, Germany (83; 7.29%), Oak Ridge National Laboratory, USA (59; 5.18%), Otto von Guericke University of Magdeburg, Germany (46; 4.04%), and Universität Greifswald, Germany (42; 3.69%) are the top institutions (Table 3). In terms of institutional collaborations, the Max Planck Institute, Oak Ridge National Laboratory, and Helmholtz Centre for Environmental Research have the highest total link strength of collaborations (Supplementary Figure 2). Figure 4 depicts the collaboration network among these institutions.
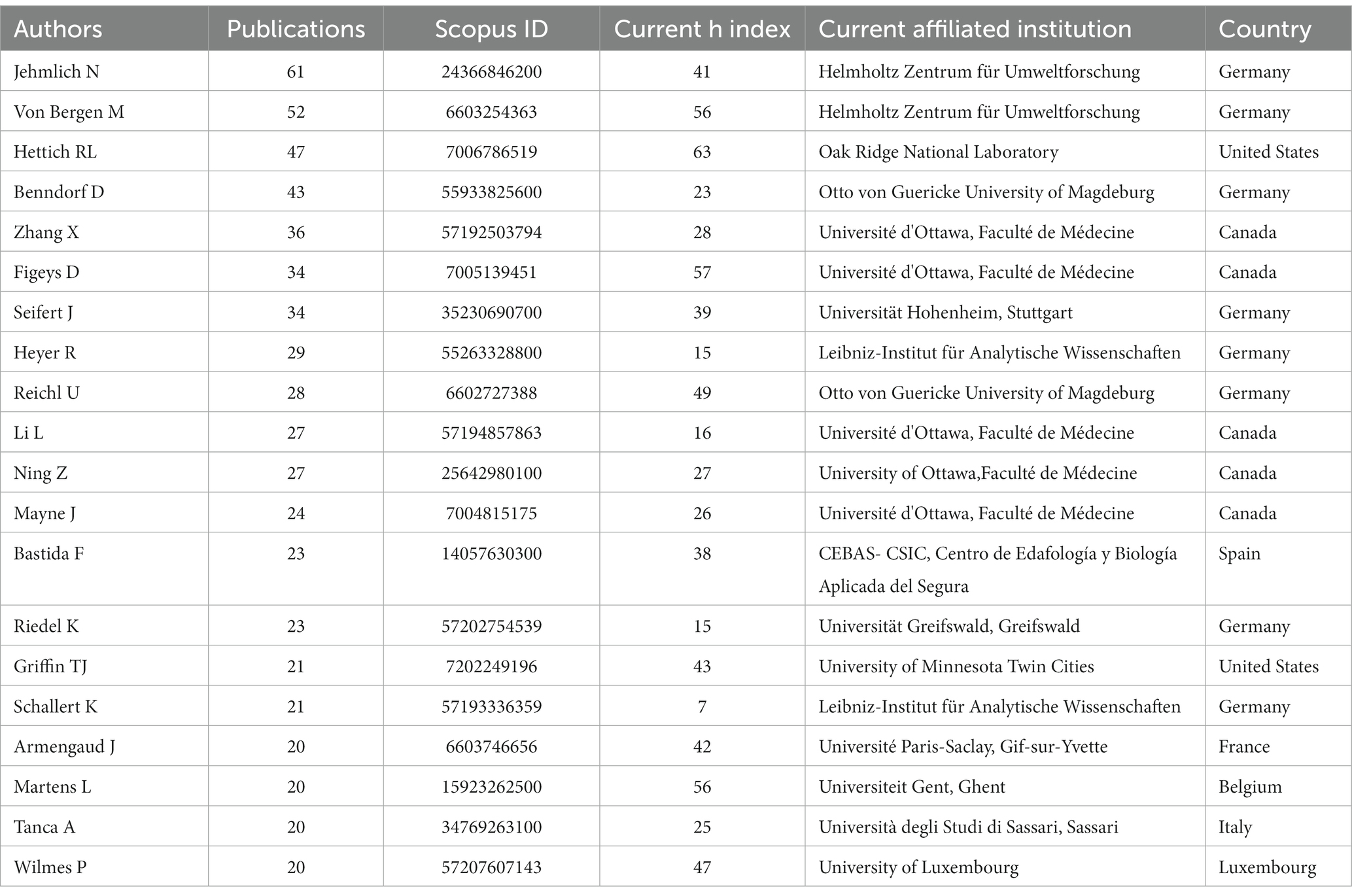
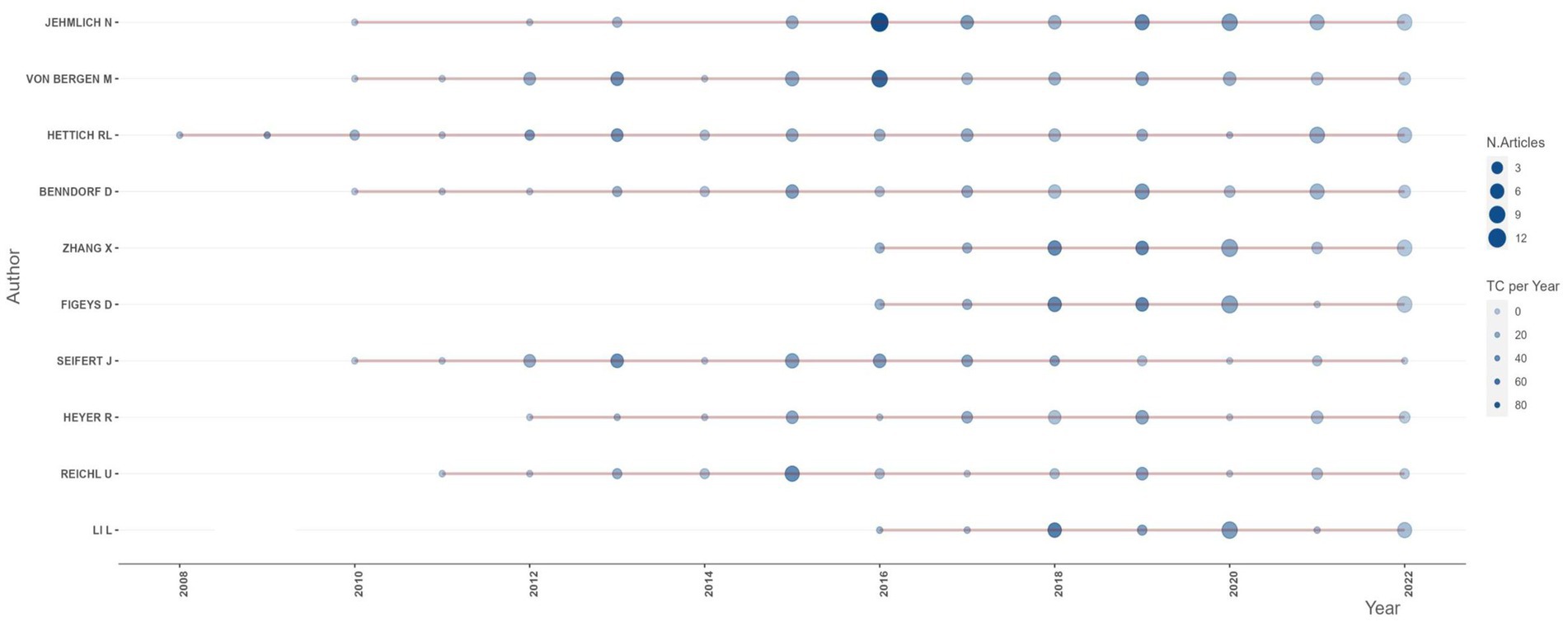
In terms of authorship, the major authors (top 20) are from Germany, Canada, the United States of America (USA), France, Spain, Belgium, and Luxembourg, in that order. Among them, the author “Jehmlich N” from Helmholtz Centre for Environmental Research, Germany, has the highest publication record with 61 articles. His productive period spans from 2010, and he currently holds an h-index of 41 (Table 4; Figure 5). “Von Bergen M.” and “Hettich R.L.”, affiliated with the Helmholtz Centre for Environmental Research, Germany, and the Oak Ridge National Laboratory, USA, respectively, are ranked second and third. Similar to Jehmlich N., “Von Bergen M.” started his productive period in 2010 and currently holds an h-index of 56, while “Hettich R.L.” has an index of 63 with his first publication in 2008. Other authors with their h-index and productivity period are shown in Table 4 and Figure 5, respectively. An analysis of Lotka’s law reveals a skewed distribution of productivity among the authors contributing to metaproteomics research (Supplementary Figure 3).
3.6. Keyword analysis and current trending topics
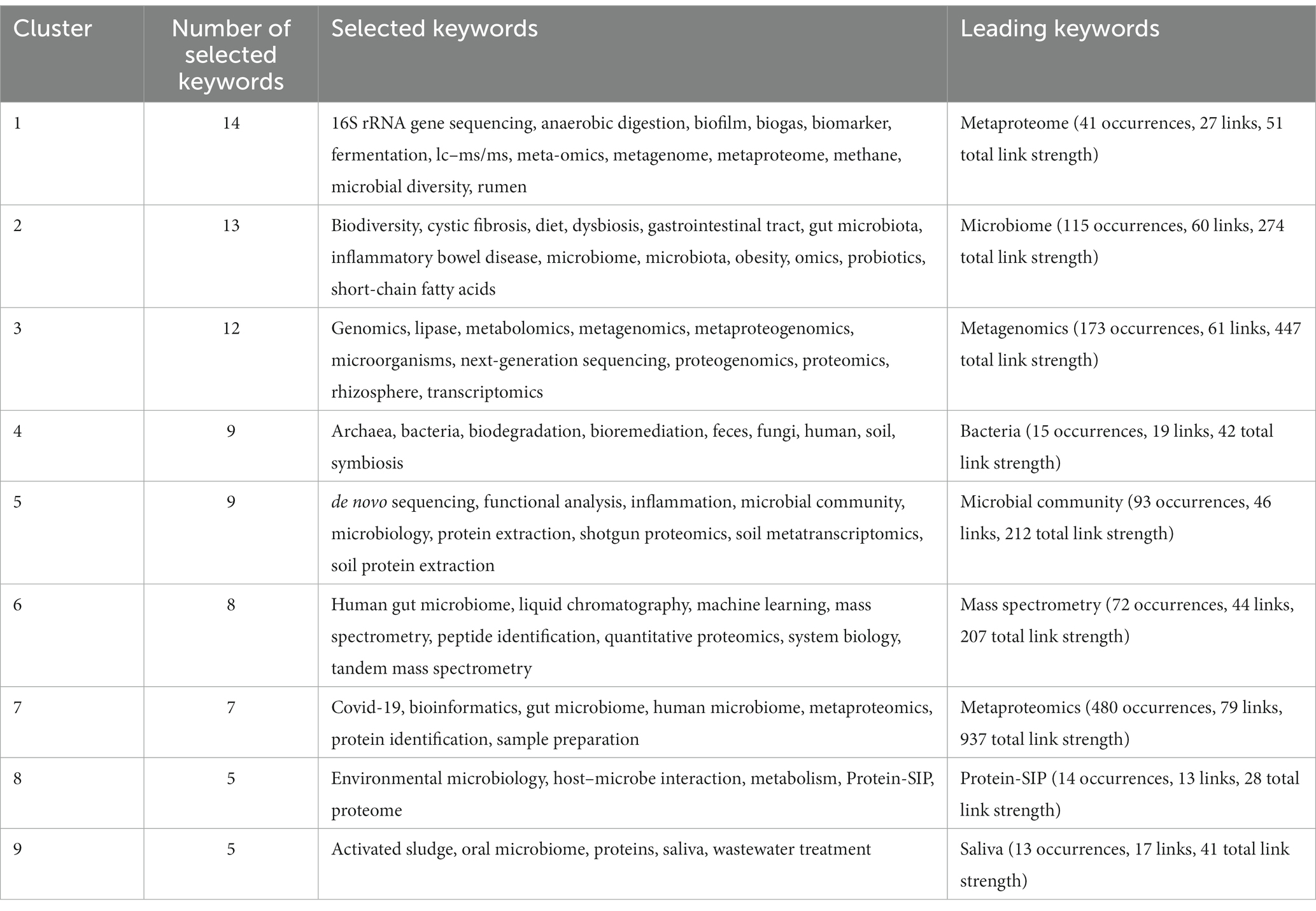
Among 1,138 publications, a total of 2,459 keywords were identified. After re-labeling synonymous single words and consolidating similar phrases, 82 keywords met the threshold of a minimum of 5 occurrences for mapping in VOS viewer (Version 1.6.18). As shown in Figures 6A,B, nine clusters were formed and categorized in Table 5 to clearly indicate the selected and leading keywords in each cluster. Clusters 1 to 3 contained more than 10 keywords, while clusters 4 to 9 had fewer than 10 keywords. Cluster 1 had the highest number of selected keywords (14), followed by Cluster 2 (13) and Cluster 3 (12).
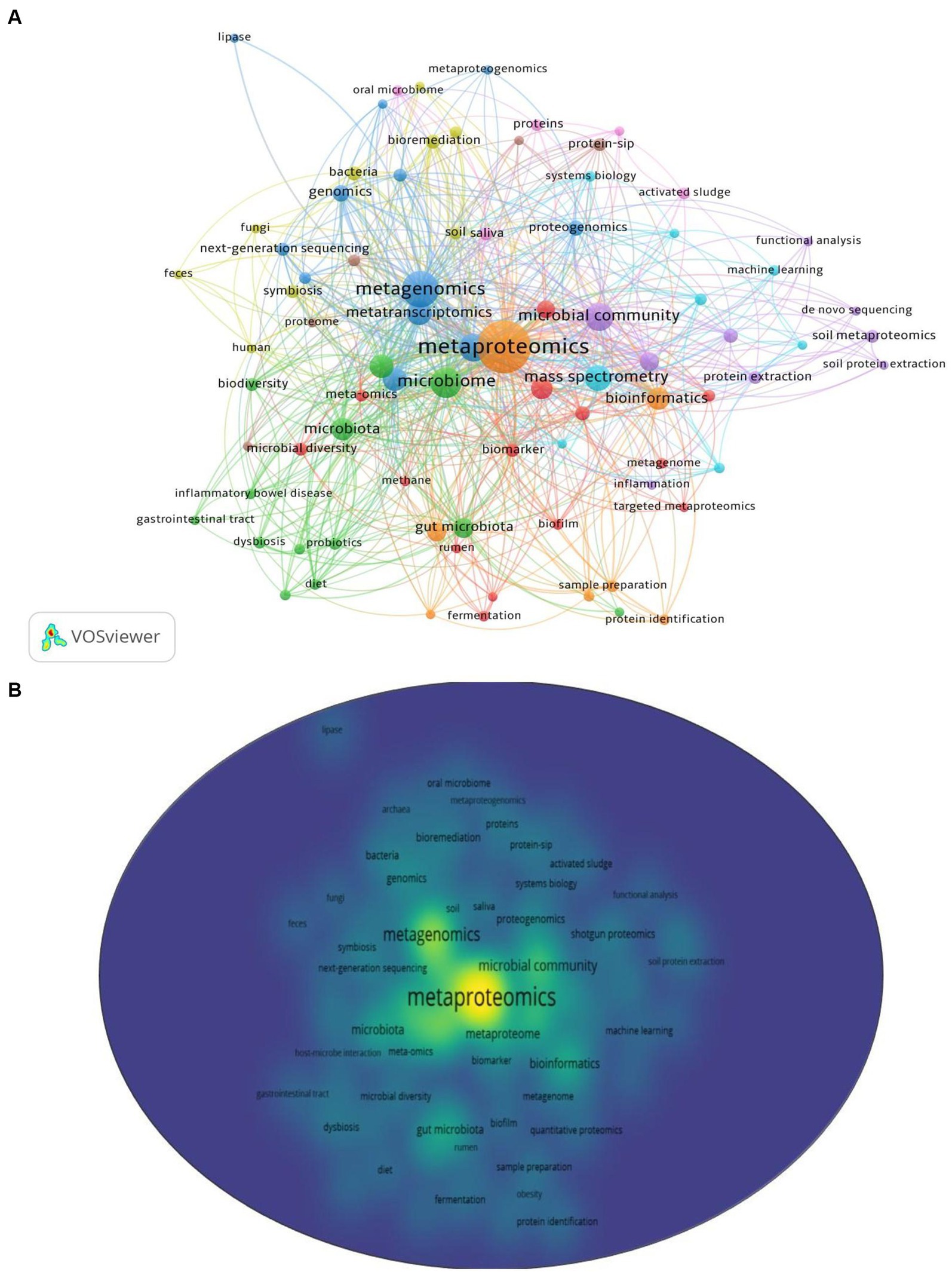
Figure 6. Analyses of keywords used in metaproteomics research (A) with their density visualization (B).
Some leading keywords encountered in each cluster, based on their degree of occurrence, included metaproteome (Cluster 1; 41 occurrences; 27 links; 51 total link strength), microbiome (Cluster 2; 115 occurrences; 60 links; total link strength 274), metagenomics (Cluster 3; 173 occurrences; 61 links; 447 total link strength), bacteria (Cluster 4; 15 occurrences; 19 links; 42 total link strength), microbial community (Cluster 5; 93 occurrences; 46 links; 212 total link strength), mass spectrometry (Cluster 6; 72 occurrences; 44 links; 207 total link strength), metaproteomics (Cluster 7; 480 occurrences; 79 links; 937 total link strength), Protein-SIP (Cluster 8; 14 occurrences; 13 links; 28 total link strength), and saliva (Cluster 9; 13 occurrences; 17 links; 41 total link strength).
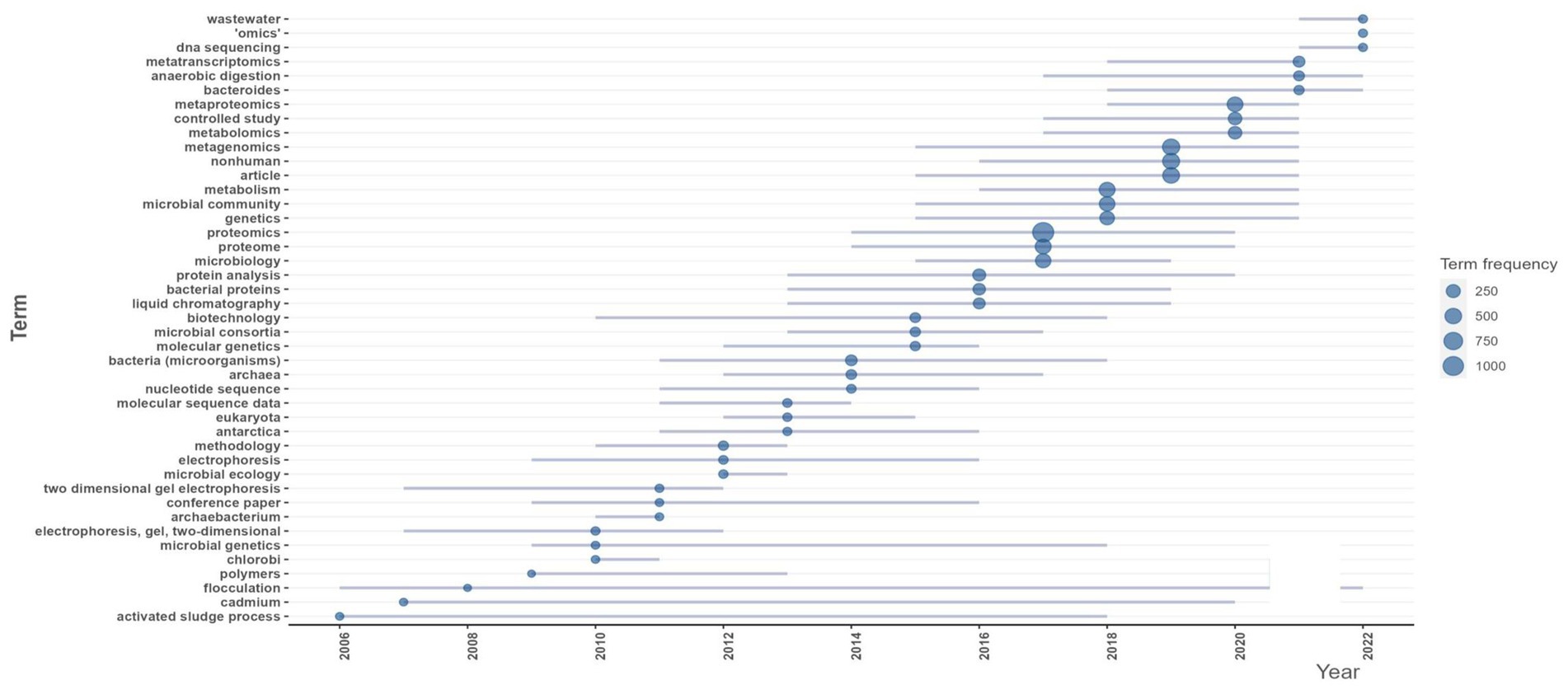
In order to analyze the trending terms based on the keywords, the Trend topic plot was constructed (Figure 7). The size of the circles shows the frequency of the term, and the length of the lines shows how long it has been studied (Abafe et al., 2022). From the plot, wastewater, omics, and DNA sequencing are the trending topics in recent years. The three most commonly used terms are proteomics (f = 1110), metagenomics (f = 564), and metaproteomics (f = 412) (Figure 7).
3.7. Publication analyses from Africa
The Scopus database retrieved 13 articles, 9 reviews, 1 book chapter, 1 editorial, and 1 short survey related to metaproteomics research in Africa (Figure 8A). Accordingly, the major research areas that utilized metaproteomics include Immunology and Microbiology (10 publications), Biochemistry, Genetics, and Molecular Biology (9 publications), as well as Medicine (8 publications) (Figure 8B).
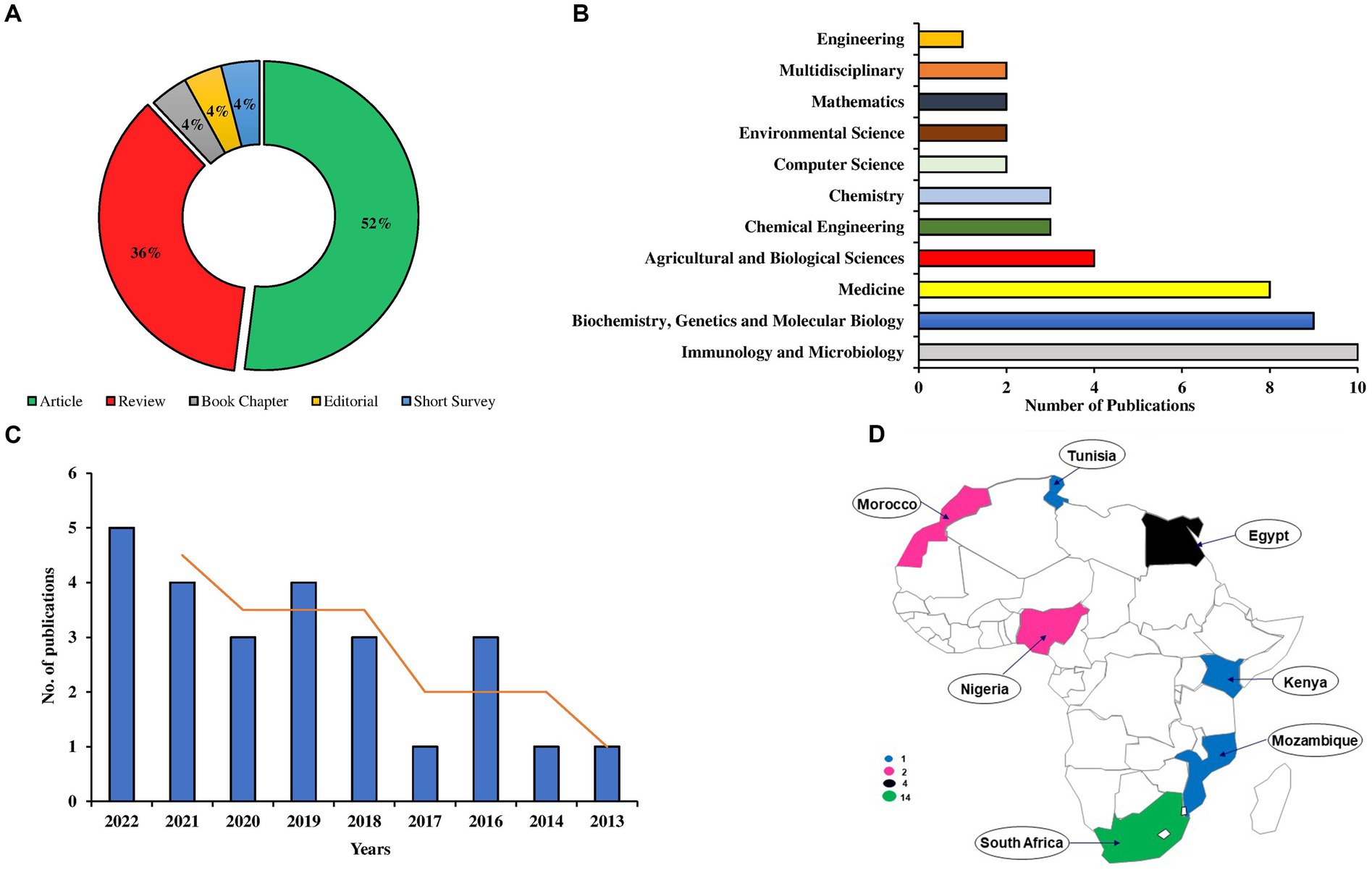
Figure 8. Analyses of the African metaproteomics publication including the document types (A), leading subject areas (B), yearly publication trend (C), and geographical distribution of publication output (D).
Looking at the trend of publication, it was observed that metaproteomics research in Africa began in 2013 (Figure 8C). From 2013 onwards, there has been an exponential growth in the publication outputs, with an EAGR of 19.58% (Supplementary Figure 4).
The countries in Africa, including South Africa (14; 56%) and Egypt (4; 16%), have the highest number of publications, while Nigeria and Morocco have 2 publications, accounting for 8% each. Other countries with a single publication in metaproteomics are Kenya, Mozambique, and Tunisia (Figure 8D). Supplementary Figure 4 provides additional information about the general characteristics of the African publications. Furthermore, we previously highlighted the country cooperation networks of three African countries: Morocco, Egypt, and South Africa, in Figure 3A. The top 10 journals for African publications are presented in Supplementary Table 1, with Frontiers in Cellular and Infection Microbiology and Microbiome emerging as the leading journals.
For the keyword analyses, a minimum threshold of two (2) occurrences was set, resulting in the identification of 10 keywords that met the criteria. The majority of the keywords are metaproteomics, metagenomics, metatranscriptomics, and microbiome. In terms of the network, the leading keywords are metabolites (Cluster 1), metaproteomics (Cluster 2), and metagenomics (Cluster 3). According to the Trend topic plot, it is evident that non-human and human research, along with review writing, are currently trending topics in the field. In concordance with the global trend, proteomics (f = 17) and metagenomics (f = 17) are the most commonly used terms (Figure 9).
3.8. Ranking publications from Africa using the field-weighted citation impact (FWCI) scores
The African publications were ranked using FWCI (Zanotto and Carvalho, 2021). About 52% of publications had an FWCI score of 1 or higher, indicating a positive impact (Table 6). For instance, most of the publications from South Africa, Egypt, Morocco, Kenya, Tunisia, and Nigeria have FWCI scores ≥1. One study from Morocco and a study from Mozambique have scores <1, respectively.
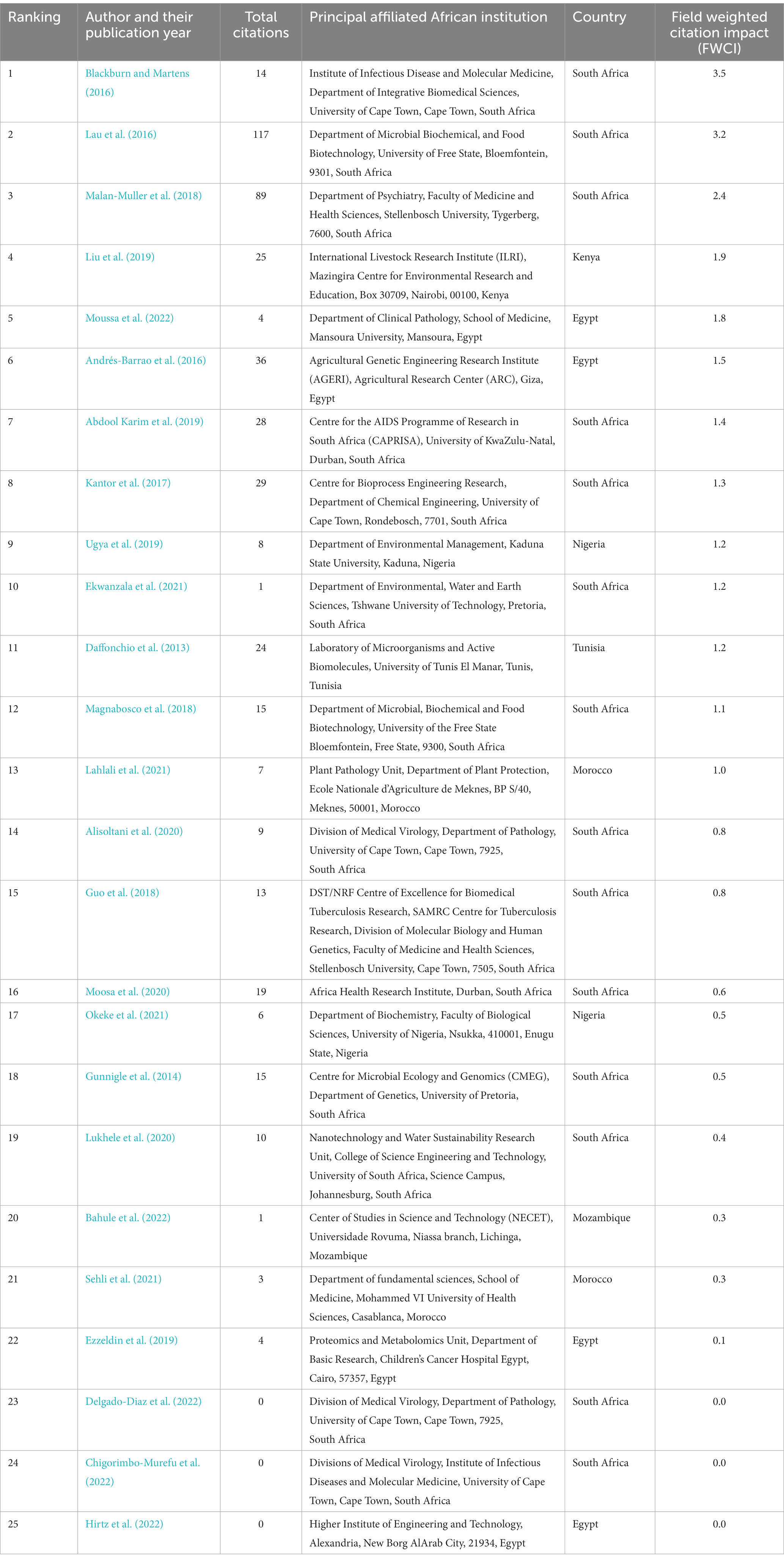
Table 6. Field weighted citation impact (FWCI) of African-affiliated publications in metaproteomics.
4. Discussion
Metaproteomics is a rapidly growing field of research that seeks to understand the complex microbial communities that inhabit the world (Kumar et al., 2021). To better understand the impact of this research area, tracking and analyzing its scientific outputs will be paramount in identifying emerging trends and key contributors in the field. To achieve this goal, we conducted a bibliometric analysis of more than 1,100 publications on metaproteomics obtained from the Scopus database. As the Web of Science is also commonly utilized for this type of study, we started our study with a comparative analysis, which revealed that 97.3% of the total extracted publications from the Scopus database are shared with the Web of Science. Only 2.7% of the publications (31 in total) are unique to the Web of Science database (statistics of these 31 publications have been included in the Supplementary material). From the analysis, information on the publication patterns and collaboration structures, among others was studied. A similar statistical analysis was adopted recently in order to understand the trends and the focus of the link between gut microbiota and type 1 diabetes (Guo et al., 2023).
Since 2004, the yearly trend in publication outputs on metaproteomics has been increasing with a steady growth rate starting from 2010. This increase, in part, could be attributed to the increased interest of scientists in identifying novel proteins for several biotechnological applications from microbial communities (Purohit et al., 2020; Mishra et al., 2021). Although there is not much difference in publication output between 2021 and 2022, the higher numbers in 2021 could be attributed to several notable advancements in metaproteomics tools within the year. For instance, the use of single-cell proteomics to study microbial communities and the development of methods for analyzing post-translational modifications in metaproteomics samples (Taylor et al., 2021; Armengaud, 2023), in addition to COVID-19 pandemic which spurred interest in using the technique to study the human microbiome and its role in infectious diseases (He et al., 2021; Sun et al., 2022). Overall, the global EAGR suggests that, under normal circumstances, the number of publications in metaproteomics is expected to increase by 37% over time. The abundance of research articles in this field suggests that many authors are actively engaged in experimental research and eager to advance our understanding by conducting thorough analyses of their findings. Their contributions are likely to shed new light on the intricate workings of protein chemistry in a particular phenomenon, potentially leading to significant breakthroughs and progress.
Based on the leading subject areas, most of the publications are related to Biochemistry, Genetics, and Molecular Biology as well as Immunology and Microbiology. This indicates that metaproteomics is a promising field that helps scientists gain a more comprehensive understanding of biological systems. Using the techniques of metaproteomics, an enhanced understanding of genetics, as well as possible changes that might occur in disease states, could be achieved (Henry et al., 2022). Also, some publications were on Agriculture and Environmental sciences, enabling the design of strategies that ensures agricultural productivity and environmental sustainability (Chandran et al., 2020; Bahule et al., 2022).
Analysis of the top 10 journals reveals that all of them are of high quality, with both Q1 or Q2 rankings and impact factors above 4. When examining the journals’ CiteScore, it becomes apparent that both Microbiome and Nature Communications have a substantial impact on the metaproteomics research community. The CiteScore could influence the decision of some authors when selecting a journal for their work (Roldan-Valadez et al., 2019). Overall, based on the findings, it could be asserted that researchers in the field of metaproteomics will be inclined to publish their findings in these top 10 journals.
In terms of country productivity and cooperation network, Europe and America have the highest total number of publications. Individually, the USA has the highest number of total citations followed by Germany and China, indicating that these countries have a high overall impact on the research output. Contrary to the total citation counts, Switzerland, Finland, and the UK exhibit high average article citations, indicating the production of high-quality research by these countries (Roldan-Valadez et al., 2019). As the total citation measures quantity (Tahamtan and Bornmann, 2019), the average citation measures quality and determines scientific contribution as well as plays a role in advancing a specific field. For collaboration, it appears that the USA, Germany, China, Italy, and Canada are the most connected countries in addition to some other European countries. The collaboration networks observed could be attributed to scientific advancements and investment in research and development (R&D) in these countries (El Jaddaoui et al., 2020; Guo et al., 2023).
Consistent with the above analysis, the institutions from Europe and America have the highest publication outputs, with German institutions taking the lead. Generally, German institutions have a long history of scientific excellence, and they have been at the forefront of many scientific breakthroughs. Specifically, in the case of metaproteomics research, German institutions have been able to leverage their expertise in proteomics and microbiology to develop methods and techniques in metaproteomic analyses (Schiebenhoefer et al., 2019). In terms of collaboration networks, there are more links among German institutions, with the Max Planck Institute having the highest collaboration. This observation is not surprising, as the institution has been observed to be frequently involved with metaproteomic research. To sum it up, the high publication outputs in the aforementioned top 10 institutions from Europe (Germany) and America may be due to a variety of factors, including access to funding, a high level of expertise and specialized knowledge, successful collaborations with other researchers and institutions, and strategic research priorities that are in line with the institution’s goals.
The diversity of research partners, a high proportion of foreign postgraduates and visiting academics, and robust research funding are all possible contributors to the dynamism of international collaboration and increased publication outputs (Roldan-Valadez et al., 2019). The top 20 authors with the highest publication records are primarily from Europe and America. The foremost author among them serves as the group leader for Microbiome Biology at the Helmholtz-Center for Environmental Research in Germany. The key areas of interest for this author’s research are the study of microbiome biology in terrestrial microbial communities, metaproteomic studies, and both qualitative and quantitative microbial proteomics. In particular, the author is known for their work in identifying key microbial players using protein-SIP, a cutting-edge technique in the field of microbiology. The author’s first publication, titled “Phylogenetic and proteomic analysis of an anaerobic toluene-degrading community,” examined, for the first time, a sulfate-reducing community grown with toluene as a carbon source by a combination of molecular genetics and proteomic techniques in order to uncover the physiological interplay in this microbial anaerobic community (Jehmlich et al., 2010). The author achieved the highest publication output in 2016, resulting in a total citation count of 87.5, and subsequently, in 2020, the author’s publication output amassed a total citation count of 30.75. As well, it is worth highlighting that one of the top 20 authors is Wilmes P., who is a notable figure in the field. In collaboration with Bond P.L., Wilmes P. proposed the term “metaproteomics” in their 2004 publication entitled “The application of two-dimensional polyacrylamide gel electrophoresis and downstream analysis to a mixed community of prokaryotic microorganisms” (Wilmes and Bond, 2004). This contribution to the scientific community has had a significant impact on the field of metaproteomics and serves as a testament to Wilmes P.’s expertise and innovative thinking. In terms of h-index, Hettich R.L. (the leader of the Bioanalytical Mass Spectrometry group at Oak Ridge National Laboratory) has the highest value of 63 among the top 20. The h-index used here measures the productivity and impact of a researcher’s publications (Roldan-Valadez et al., 2019). It also takes into account engagements in secondary research, which could influence its value. An assessment of the author’s productivity in this field reveals that it adheres to Lotka’s law, which claims that a small percentage of authors produce the majority of output in a given field (Kushairi and Ahmi, 2021). This information could be useful for developing strategies to support and encourage the productivity of all authors in the field, regardless of their current level of output.
Keyword analyses offer a comprehensive overview of the research area’s trajectory and themes (Donthu et al., 2021). Based on the keyword analyses, Cluster 1 had the highest number of selected keywords, implying that it contains keywords frequently used in the context of metaproteomic research (Donthu et al., 2021). In terms of leading keywords, metaproteomics had the highest occurrence, followed by metagenomics and other omics. This is unsurprising as the omics fields are often coupled together in order to have a thorough understanding of the microbial world (Kumar et al., 2021; Jiang et al., 2022).
Further the keyword clusters generated give an overview of the thematic research hotspots. For instance Cluster 1 sheds light on the interest in understanding microbial processes and diversity in biotechnology and the environment while the terms in Cluster 2 are related to metaproteomics research aimed at understanding the importance of gut microbiota in human health. Clusters 3 and 4 on the other hand indicate interest in the concept of molecular components and interactions within biological systems as well as understanding the role of metaproteomics in investigating the soil microbiome and biodegradation and bioremediation processes. Further the other clusters concentrated on techniques for analyzing the protein expressions interactions and functional roles of microorganisms in the soil and gut microbiome in diseases such as COVID-19 as well as researching microbial communities and their functional proteins in activated sludge and wastewater systems. Some important diseases that have been researched using the metaproteomics approach in humans include obesity (Zhong et al., 2019; Biemann et al., 2021; Calabrese et al., 2022) cystic fibrosis (Debyser et al., 2019; Saralegui et al., 2022) inflammatory bowel disease (Moon et al., 2018; Zhang et al., 2018; Lehmann et al., 2019) and the oral microbiome (Jagtap et al., 2012; Bostanci et al., 2021). In addition to the research conducted on human diseases metaproteomics has also been extensively applied in the field of soil ecosystems – soil metaproteomics (Bastida et al., 2016b, 2018; Chourey and Hettich, 2018; Liu et al., 2019) – with specific focus on the rhizosphere (Renu et al., 2019; White et al., 2021). Other environmental phenomena such as bioremediation (Bastida et al., 2016a; Aishwarya et al., 2022) and biodegradation (Chang et al., 2018; Gunasekaran et al., 2022; Xie et al., 2022) has been explored
To further determine the metabolically active players in microbial communities, Protein SIP experiments are increasingly being conducted in the area of environmental microbiology. In addition, studies concerning fermentation or anaerobic digestion of samples from the environment such as biofilms or from wastewater treatment plants which leads to the production of biogas such as methane have been investigated (Joyce et al., 2018; Heyer et al., 2019; Lam et al., 2021; Chen et al., 2022).
Overall, the main instrumentation used in the analysis is tandem mass spectrometry. The peptides are initially separated using liquid chromatography, particularly in the LC–MS/MS procedure. Shotgun proteomics and quantitative proteomics are popular approaches employed to identify and quantify proteins (Balotf et al., 2022). The generated data are analyzed using bioinformatics tools in order to identify the proteomes in the studies.
Sub-analysis of African contributions in metaproteomics revealed that the continent contributed only 25 publications of the total 1,138. Among the 54 countries in Africa, only 7 countries engage in metaproteomic research. As previously mentioned, a lack of infrastructure, funding, and expertise (El Jaddaoui et al., 2020) could contribute to these low statistics. Therefore, considering the significant scientific impact of metaproteomics and omics in general, it is essential to redirect more research efforts toward Africa to characterize the microbial diversity on the continent (Cowan et al., 2022). Remarkably, the positive EAGR observed suggests that research efforts in the continent are increasing, although more has to be done in order to accelerate the growth. In terms of country collaboration, the collaboration network in the continent is relatively weak compared to global networking. Among the countries, only South Africa was noticed to establish research partnerships with the advanced countries.
Interestingly, in comparison with the major publishers, the publications from the African continent are also found in high-impact journals. Likewise, the top keywords used in Africa are comparable to those used globally, with metaproteomics ranking first. This word has links with the other omics terms. The trend plot portrays trending research topics related to non-human studies as well as human studies. Additionally, it is worth noting that review writing was highly predominant. To increase research output beyond review writing, it is recommended to establish more laboratories and research centers specifically dedicated to metaproteomics.
The FWCI calculates how frequently a given publication is cited relative to others in the same field. Given its normalization, it can be used to directly compare an article’s performance against those of other articles (even those in different subject areas). The FWCI ≥1 in some of the African publications indicates that the output performs exactly as expected by the global average, while the values <1 in some of the publications indicate underperformance in comparison to the global average (Zanotto and Carvalho, 2021). Hence, there is a need for African authors to develop means to improve their performance in order to increase the impact and visibility of their work.
To further emphasize, the insights gained from metaproteomics research have wide-ranging applications in fields such as medicine, agriculture, and environmental science. For instance, metaproteomics enables the taxonomic and functional analysis of complex microbial communities in various samples, allowing the quantification of proteins and providing a detailed understanding of cellular phenotypes at the molecular level. By leveraging metaproteomics, researchers can gain comprehensive insights into these complex biological systems and pave the way for advancements in various disciplines. Fernanda Salvato recently discussed, in detail, the extensive examples of metaproteomics-based approaches used to address a wide range of questions in diverse areas of biological research (Salvato et al., 2021)
5. Conclusion
In conclusion, the bibliometric analyses of more than 1,100 publications on metaproteomics from the Scopus database indicated an increasing trend of publications with a high percentage of open-access research articles. The majority of these publications fall within the field of Biochemistry, Genetics, and Molecular Biology, with Microbiome and Nature Communications emerging as the top publishers based on their CiteScore rankings. With respect to the author’s contribution and country productivity, Jehmlich N. was the most productive author, while the United States, Germany, and China were the most productive countries, collectively contributing to 69% of the total publications. Keyword analyses revealed metaproteomics and metagenomics as the highest co-occurring keywords in addition to other omics. Zooming in on Africa, only 2.2% of total publications came from the continent, with South Africa producing more publications. Based on the EAGR and the FWCI, more studies are required in the region in order to meet the global scale and contribute to the advancement of metaproteomics. To encourage this research area in Africa, some initiatives have begun to emerge. For instance, in Morocco, the Sharifian Phosphate Office (OCP) has funded a new proteomic platform at Mohammed VI Polytechnic University (RD is the principal investigator), which will help to advance the field of proteomics.
5.1. Recommendations
With the strides made by this emerging field, its global challenges cannot be overlooked. Some of the drawbacks in this field are in the areas of sample complexity and preparation, accurate protein database construction for microbial communities, false discovery rate assessments, annotation, and software integration (Issa Isaac et al., 2019; Rechenberger et al., 2019; Saito et al., 2019; Duong and Lee, 2023). Each of these challenges can affect the quality and reproducibility of metaproteomic results and addressing them will require collaborative efforts between researchers and software developers. By doing so, standardized protocols and tools that will improve data quality, accuracy, and reproducibility could be developed. In addition, the current bibliometric analysis has revealed a significant merit of metaproteomics in studying the spatiotemporal characterization of microbial communities at the functional level. Recently, Manuel Kleiner published a very interesting and detailed review that illustrates the diversity of questions that can be addressed solely through metaproteomics in the study of microbial communities, including those associated with plants and animals (Kleiner, 2019). This emerging area holds significant potential for advancing scientific knowledge, addressing biotechnological challenges, improving human health, and informing environmental management strategies. Further, to advance the field of metaproteomics in Africa, several steps can be taken. This includes prioritizing metaproteomics research by providing funding, equipment, and training opportunities. Also, encouraging collaboration between African and international researchers through joint projects, exchange programs, and workshops is essential. For instance, the International Metaproteomics Initiative (Van Den Bossche et al., 2021) is a commendable step toward advancing the field and promoting knowledge sharing. Lastly, support should be provided for publications in high-impact journals, travel grants, and mentoring programs to enhance the quality of research on the continent.
Overall, the findings of this study provide valuable insights that can be used to advocate for increased support, funding, and resources for metaproteomics research on a global scale, as well as specifically in Africa. These findings serve as a basis for informed decision-making by policymakers, enabling them to drive policies, allocate funds, and distribute resources in a manner that promotes the growth and impact of metaproteomics research. By harnessing the potential of metaproteomics, policymakers can address complex societal challenges by studying the intricate dynamics of microbial communities and their functions in various fields. Further, the application of metaproteomics in studying soil microbiomes, bioremediation, and biodegradation, for instance, can contribute to the development of policies related to sustainable agriculture, effective waste management, and environmental conservation.
5.2. Limitations of the study
Although the Scopus and Web of Science databases have been recognized as the largest searchable collection of citations and abstracts in the field of literature research, it would be interesting to incorporate other databases such as PubMed, and Cochrane into the present analysis subsequently. Also, language bias introduced in the selection of documents may inadvertently exclude contributions from non-English-published articles. Moreover, refining and expanding the search strategy to include a broader range of relevant terms, synonyms, and variations could help in capturing more documents for the bibliometric analysis.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
RD, AE, and AA: conceptualization. AA and SA: methodology. AA, SA, NS, AE, and RD: formal analysis, investigation, and writing—review and editing. AA, SA, and RD: writing—original draft preparation. RD, AA, AE, and SA: resources. RD: supervision. All authors contributed to the article and approved the submitted version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1217727/full#supplementary-material
References
Abafe, E. A., Bahta, Y. T., and Jordaan, H. (2022). Exploring biblioshiny for historical assessment of global research on sustainable use of water in agriculture. Sustainability 14:10651. doi: 10.3390/su141710651
Abdool Karim, S. S., Baxter, C., Passmore, J. S., McKinnon, L. R., and Williams, B. L. (2019). The genital tract and rectal microbiomes: their role in HIV susceptibility and prevention in women. J. Int. AIDS Soc. 22:e25300. doi: 10.1002/jia2.25300
Aishwarya, S., Rajalakshmi, S., and Veena Gayathri, K. (2022). “The proteome mapping—metabolic modeling, and functional elucidation of the microbiome in the remediation of dyes and treating industrial effluents” in Metagenomics to bioremediation: applications, cutting edge tools, and future outlook, eds Vineet, K, Muhammad, B, Sushil Kumar, S, and Vinod Kumar, G (Academic Press), 311–328.
Alisoltani, A., Manhanzva, M. T., Potgieter, M., Balle, C., Bell, L., Ross, E., et al. (2020). Microbial function and genital inflammation in young South African women at high risk of HIV infection. Microbiome 8:165. doi: 10.1186/s40168-020-00932-8
Andersen, T. O., Kunath, B. J., Hagen, L. H., Arntzen, M. Ø., and Pope, P. B. (2021). Rumen metaproteomics: closer to linking rumen microbial function to animal productivity traits. Methods 186, 42–51. doi: 10.1016/j.ymeth.2020.07.011
Andrés-Barrao, C., Saad, M. M., Cabello Ferrete, E., Bravo, D., Chappuis, M.-L., Ortega Pérez, R., et al. (2016). Metaproteomics and ultrastructure characterization of Komagataeibacter spp. involved in high-acid spirit vinegar production. Food Microbiol. 55, 112–122. doi: 10.1016/j.fm.2015.10.012
Aria, M., and Cuccurullo, C. (2017). Bibliometrix: an R-tool for comprehensive science mapping analysis. J. Informet. 11, 959–975. doi: 10.1016/j.joi.2017.08.007
Armengaud, J. (2023). Metaproteomics to understand how microbiota function: the crystal ball predicts a promising future. Environ. Microbiol. 25, 115–125. doi: 10.1111/1462-2920.16238
Baas, J., Schotten, M., Plume, A., Côté, G., and Karimi, R. (2020). Scopus as a curated, high-quality bibliometric data source for academic research in quantitative science studies. Quant. Sci. Studies 1, 377–386. doi: 10.1162/qss_a_00019
Bahule, C. E., Da Silva Martins, L. H., Chaúque, B. J. M., and Lopes, A. S. (2022). Metaproteomics as a tool to optimize the maize fermentation process. Trends Food Sci. Technol. 129, 258–265. doi: 10.1016/j.tifs.2022.09.017
Bailey, M., Thomas, A., Francis, O., Stokes, C., and Smidt, H. (2019). The dark side of technological advances in analysis of microbial ecosystems. J. Anim. Sci. Biotechnol. 10:49. doi: 10.1186/s40104-019-0357-2
Balotf, S., Wilson, R., Tegg, R. S., Nichols, D. S., and Wilson, C. R. (2022). Shotgun proteomics as a powerful tool for the study of the proteomes of plants, their pathogens, and plant-pathogen interactions. Proteomes 10:5. doi: 10.3390/proteomes10010005
Bastida, F., Jehmlich, N., Lima, K., Morris, B. E. L., Richnow, H. H., Hernández, T., et al. (2016a). The ecological and physiological responses of the microbial community from a semiarid soil to hydrocarbon contamination and its bioremediation using compost amendment. J. Proteome 135, 162–169. doi: 10.1016/j.jprot.2015.07.023
Bastida, F., Jehmlich, N., Torres, I. F., and García, C. (2018). The extracellular metaproteome of soils under semiarid climate: a methodological comparison of extraction buffers. Sci. Total Environ. 619-620, 707–711. doi: 10.1016/j.scitotenv.2017.11.134
Bastida, F., Torres, I. F., Moreno, J. L., Baldrian, P., Ondoño, S., Ruiz-Navarro, A., et al. (2016b). The active microbial diversity drives ecosystem multifunctionality and is physiologically related to carbon availability in Mediterranean semi-arid soils. Mol. Ecol. 25, 4660–4673. doi: 10.1111/mec.13783
Biemann, R., Buß, E., Benndorf, D., Lehmann, T., Schallert, K., Püttker, S., et al. (2021). Fecal metaproteomics reveals reduced gut inflammation and changed microbial metabolism following lifestyle-induced weight loss. Biomol. Ther. 11:726. doi: 10.3390/biom11050726
Blackburn, J. M., and Martens, L. (2016). The challenge of metaproteomic analysis in human samples. Expert Rev. Proteomics 13, 135–138. doi: 10.1586/14789450.2016.1135058
Bostanci, N., Grant, M., Bao, K., Silbereisen, A., Hetrodt, F., Manoil, D., et al. (2021). Metaproteome and metabolome of oral microbial communities. Periodontol. 85, 46–81. doi: 10.1111/prd.12351
Calabrese, F. M., Porrelli, A., Vacca, M., Comte, B., Nimptsch, K., Pinart, M., et al. (2022). Metaproteomics approach and pathway modulation in obesity and diabetes: a narrative review. Nutrients 14:47. doi: 10.3390/nu14010047
Chandran, H., Meena, M., and Sharma, K. (2020). Microbial biodiversity and bioremediation assessment through omics approaches. Front. Environ. Chem. 1:570326. doi: 10.3389/fenvc.2020.570326
Chang, B.-V., Fan, S.-N., Tsai, Y.-C., Chung, Y.-L., Tu, P.-X., and Yang, C.-W. (2018). Removal of emerging contaminants using spent mushroom compost. Sci. Total Environ. 634, 922–933. doi: 10.1016/j.scitotenv.2018.03.366
Chen, Y., Ping, Q., Li, D., Dai, X., and Li, Y. (2022). Comprehensive insights into the impact of pretreatment on anaerobic digestion of waste active sludge from perspectives of organic matter composition, thermodynamics, and multi-omics. Water Res. 226:119240. doi: 10.1016/j.watres.2022.119240
Chigorimbo-Murefu, N. T. L., Potgieter, M., Dzanibe, S., Gabazana, Z., Buri, G., Chawla, A., et al. (2022). A pilot study to show that asymptomatic sexually transmitted infections alter the foreskin epithelial proteome. Front. Microbiol. 13:928317. doi: 10.3389/fmicb.2022.928317
Chourey, K., and Hettich, R. L. (2018). “Utilization of a detergent-based method for direct microbial cellular lysis/proteome extraction from soil samples for metaproteomics studies” in Microbial proteomics: methods and protocols. ed. D. Becher (New York, NY: Springer), 293–302.
Cowan, D., Lebre, P., Amon, C., Becker, R., Boga, H., Boulangé, A., et al. (2022). Biogeographical survey of soil microbiomes across sub-Saharan Africa: structure, drivers, and predicted climate-driven changes. Microbiome 10:131. doi: 10.1186/s40168-022-01297-w
Daffonchio, D., Ferrer, M., Mapelli, F., Cherif, A., Lafraya, Á., Malkawi, H. I., et al. (2013). Bioremediation of southern Mediterranean oil polluted sites comes of age. New Biotechnol. 30, 743–748. doi: 10.1016/j.nbt.2013.05.006
Debyser, G., Aerts, M., Van Hecke, P., Mesuere, B., Duytschaever, G., Dawyndt, P., et al. (2019). “A method for comprehensive proteomic analysis of human faecal samples to investigate gut dysbiosis in patients with cystic fibrosis” in Emerging sample treatments in proteomics. ed. J.-L. Capelo-Martínez (Cham: Springer International Publishing), 137–160.
Delgado-Diaz, D. J., Jesaveluk, B., Hayward, J. A., Tyssen, D., Alisoltani, A., Potgieter, M., et al. (2022). Lactic acid from vaginal microbiota enhances cervicovaginal epithelial barrier integrity by promoting tight junction protein expression. Microbiome 10:141. doi: 10.1186/s40168-022-01337-5
Donthu, N., Kumar, S., Mukherjee, D., Pandey, N., and Lim, W. M. (2021). How to conduct a bibliometric analysis: an overview and guidelines. J. Bus. Res. 133, 285–296. doi: 10.1016/j.jbusres.2021.04.070
Duong, V.-A., and Lee, H. (2023). Bottom-up proteomics: advancements in sample preparation. Int. J. Mol. Sci. 24:5350. doi: 10.3390/ijms24065350
Ejaz, H., Zeeshan, H. M., Ahmad, F., Bukhari, S. N. A., Anwar, N., Alanazi, A., et al. (2022). Bibliometric analysis of publications on the omicron variant from 2020 to 2022 in the Scopus database using R and VOSviewer. Int. J. Environ. Res. Public Health 19:12407. doi: 10.3390/ijerph191912407
Ekwanzala, M. D., Budeli, P., and Unuofin, J. O. (2021). “Chapter 8—application of metatranscriptomics in wastewater treatment processes” in Wastewater treatment. eds. M. P. Shah, A. Sarkar, and S. Mandal (Cham: Elsevier), 187–204.
El Jaddaoui, I., Allali, I., Sehli, S., Ouldim, K., Hamdi, S., Al Idrissi, N., et al. (2020). Cancer omics in Africa: present and prospects. Front. Oncol. 10:606428. doi: 10.3389/fonc.2020.606428
Ezzeldin, S., El-Wazir, A., Enany, S., Muhammad, A., Johar, D., Osama, A., et al. (2019). Current understanding of human metaproteome association and modulation. J. Proteome Res. 18, 3539–3554. doi: 10.1021/acs.jproteome.9b00301
Graf, A. C., Striesow, J., Pané-Farré, J., Sura, T., Wurster, M., Lalk, M., et al. (2021). An innovative protocol for metaproteomic analyses of microbial pathogens in cystic fibrosis sputum. Front. Cell. Infect. Microbiol. 11:724569. doi: 10.3389/fcimb.2021.724569
Gunasekaran, V., Canela, N., and Constantí, M. (2022). Comparative proteomic analysis of an ethyl tert-butyl ether-degrading bacterial consortium. Microorganisms 10, 10:2331. doi: 10.3390/microorganisms10122331
Gunnigle, E., Ramond, J.-B., Frossard, A., Seeley, M., and Cowan, D. (2014). A sequential co-extraction method for DNA, RNA and protein recovery from soil for future system-based approaches. J. Microbiol. Methods 103, 118–123. doi: 10.1016/j.mimet.2014.06.004
Guo, K., Li, J., Li, X., Huang, J., and Zhou, Z. (2023). Emerging trends and focus on the link between gut microbiota and type 1 diabetes: A bibliometric and visualization analysis. Front. Microbiol. 14:1137595. doi: 10.3389/fmicb.2023.1137595
Guo, X., Li, Z., Yao, Q., Mueller, R. S., Eng, J. K., Tabb, D. L., et al. (2018). Sipros ensemble improves database searching and filtering for complex metaproteomics. Bioinformatics 34, 795–802. doi: 10.1093/bioinformatics/btx601
Hamdi, Y., Zass, L., Othman, H., Radouani, F., Allali, I., Hanachi, M., et al. (2021). Human OMICs and computational biology research in Africa: current challenges and prospects. OMICS 25, 213–233. doi: 10.1089/omi.2021.0004
Hardouin, P., Chiron, R., Marchandin, H., Armengaud, J., and Grenga, L. (2021). Metaproteomics to decipher CF host-microbiota interactions: overview, challenges and future perspectives. Genes 12:892. doi: 10.3390/genes12060892
He, F., Zhang, T., Xue, K., Fang, Z., Jiang, G., Huang, S., et al. (2021). Fecal multi-omics analysis reveals diverse molecular alterations of gut ecosystem in COVID-19 patients. Anal. Chim. Acta 1180:338881. doi: 10.1016/j.aca.2021.338881
Henry, C., Bassignani, A., Berland, M., Langella, O., Sokol, H., and Juste, C. (2022). Modern metaproteomics: a unique tool to characterize the active microbiome in health and diseases, and pave the road towards new biomarkers—example of Crohn’s disease and ulcerative colitis flare-ups. Cells 11:1340. doi: 10.3390/cells11081340
Heyer, R., Schallert, K., Siewert, C., Kohrs, F., Greve, J., Maus, I., et al. (2019). Metaproteome analysis reveals that syntrophy, competition, and phage-host interaction shape microbial communities in biogas plants. Microbiome 7:69. doi: 10.1186/s40168-019-0673-y
Hirtz, C., Mannaa, A. M., Moulis, E., Pible, O., O’Flynn, R., Armengaud, J., et al. (2022). Deciphering black extrinsic tooth stain composition in children using metaproteomics. ACS Omega 7, 8258–8267. doi: 10.1021/acsomega.1c04770
Iskandar, K., Molinier, L., Hallit, S., Sartelli, M., Hardcastle, T. C., Haque, M., et al. (2021). Surveillance of antimicrobial resistance in low- and middle-income countries: a scattered picture. Antimicrobial Resistance Infection Control 10:63. doi: 10.1186/s13756-021-00931-w
Issa Isaac, N., Philippe, D., Nicholas, A., Raoult, D., and Eric, C. (2019). Metaproteomics of the human gut microbiota: challenges and contributions to other OMICS. Clin. Mass Spectr. 14, 18–30. doi: 10.1016/j.clinms.2019.06.001
Jagtap, P., McGowan, T., Bandhakavi, S., Tu, Z. J., Seymour, S., Griffin, T. J., et al. (2012). Deep metaproteomic analysis of human salivary supernatant. Proteomics 12, 992–1001. doi: 10.1002/pmic.201100503
Jehmlich, N., Kleinsteuber, S., Vogt, C., Benndorf, D., Harms, H., Schmidt, F., et al. (2010). Phylogenetic and proteomic analysis of an anaerobic toluene-degrading community. J. Appl. Microbiol. 109, 1937–1945. doi: 10.1111/j.1365-2672.2010.04823.x
Jiang, N., Wu, R., Wu, C., Wang, R., Wu, J., and Shi, H. (2022). Multi-omics approaches to elucidate the role of interactions between microbial communities in cheese flavor and quality. Food Rev. Intl., 1–13. doi: 10.1080/87559129.2022.2070199
Jiao, H., Huang, Z., Chen, Z., Wang, H., Liu, H., and Wei, Z. (2022). Lead removal in flue gas from sludge incineration by denitrification: insights from metagenomics and metaproteomics. Ecotoxicol. Environ. Saf. 244:114059. doi: 10.1016/j.ecoenv.2022.114059
Joyce, A., Ijaz, U. Z., Nzeteu, C., Vaughan, A., Shirran, S. L., Botting, C. H., et al. (2018). Linking microbial community structure and function during the acidified anaerobic digestion of grass. Front. Microbiol. 9:540. doi: 10.3389/fmicb.2018.00540
Kantor, R. S., Huddy, R. J., Iyer, R., Thomas, B. C., Brown, C. T., Anantharaman, K., et al. (2017). Genome-resolved meta-omics ties microbial dynamics to process performance in biotechnology for thiocyanate degradation. Environ. Sci. Technol. 51, 2944–2953. doi: 10.1021/acs.est.6b04477
Kleiner, M. (2019). Metaproteomics: much more than measuring gene expression in microbial communities. mSystems 4, 1–6. doi: 10.1128/msystems.00115-19
Kumar, V., Singh, K., Shah, M. P., Singh, A. K., Kumar, A., and Kumar, Y. (2021). “Chapter 1—application of Omics Technologies for Microbial Community Structure and Function Analysis in contaminated environment” in Wastewater treatment. eds. M. P. Shah, A. Sarkar, and S. Mandal (Cham: Elsevier), 1–40.
Kushairi, N., and Ahmi, A. (2021). Flipped classroom in the second decade of the Millenia: A Bibliometrics analysis with Lotka’s law. Educ. Inf. Technol. 26, 4401–4431. doi: 10.1007/s10639-021-10457-8
Lahlali, R., Ibrahim, D. S. S., Belabess, Z., Kadir Roni, M. Z., Radouane, N., Vicente, C. S. L., et al. (2021). High-throughput molecular technologies for unraveling the mystery of soil microbial community: challenges and future prospects. Heliyon 7:e08142. doi: 10.1016/j.heliyon.2021.e08142
Lam, T.-K., Yang, J.-T., Lai, S.-J., Liang, S.-Y., and Wu, S.-H. (2021). Meta-proteomics analysis of microbial ecosystem during the anaerobic digestion of chicken manure in biogas production farm. Bioresour. Technol. Rep. 13:100643. doi: 10.1016/j.biteb.2021.100643
Lau, M. C. Y., Kieft, T. L., Kuloyo, O., Linage-Alvarez, B., van Heerden, E., Lindsay, M. R., et al. (2016). An oligotrophic deep-subsurface community dependent on syntrophy is dominated by sulfur-driven autotrophic denitrifiers. Proc. Natl. Acad. Sci. 113, E7927–E7936. doi: 10.1073/pnas.1612244113
Lehmann, T., Schallert, K., Vilchez-Vargas, R., Benndorf, D., Püttker, S., Sydor, S., et al. (2019). Metaproteomics of fecal samples of Crohn’s disease and ulcerative colitis. J. Proteome 201, 93–103. doi: 10.1016/j.jprot.2019.04.009
Liu, D., Keiblinger, K. M., Leitner, S., Wegner, U., Zimmermann, M., Fuchs, S., et al. (2019). Response of microbial communities and their metabolic functions to drying–rewetting stress in a temperate forest soil. Microorganisms 7:5. doi: 10.3390/microorganisms7050129
Lukhele, T., Selvarajan, R., Nyoni, H., Mamba, B. B., and Msagati, T. A. M. (2020). Acid mine drainage as habitats for distinct microbiomes: current knowledge in the era of molecular and omic technologies. Curr. Microbiol. 77, 657–674. doi: 10.1007/s00284-019-01771-z
Maghini, D. G., Moss, E. L., Vance, S. E., and Bhatt, A. S. (2021). Improved high-molecular-weight DNA extraction, nanopore sequencing and metagenomic assembly from the human gut microbiome. Nat. Protoc. 16, 458–471. doi: 10.1038/s41596-020-00424-x
Magnabosco, C., Timmers, P. H. A., Lau, M. C. Y., Borgonie, G., Linage-Alvarez, B., Kuloyo, O., et al. (2018). Fluctuations in populations of subsurface methane oxidizers in coordination with changes in electron acceptor availability. FEMS Microbiol. Ecol. 94:fiy089. doi: 10.1093/femsec/fiy089
Malan-Muller, S., Valles-Colomer, M., Raes, J., Lowry, C. A., Seedat, S., and Hemmings, S. M. J. (2018). The gut microbiome and mental health: implications for anxiety- and trauma-related disorders. OMICS 22, 90–107. doi: 10.1089/omi.2017.0077
Mauger, S., Monard, C., Thion, C., and Vandenkoornhuyse, P. (2022). Contribution of single-cell omics to microbial ecology. Trends Ecol. Evol. 37, 67–78. doi: 10.1016/j.tree.2021.09.002
Md Khudzari, J., Kurian, J., Tartakovsky, B., and Raghavan, G. S. V. (2018). Bibliometric analysis of global research trends on microbial fuel cells using Scopus database. Biochem. Eng. J. 136, 51–60. doi: 10.1016/j.bej.2018.05.002
Mishra, S., Lin, Z., Pang, S., Zhang, W., Bhatt, P., and Chen, S. (2021). Recent advanced technologies for the characterization of xenobiotic-degrading microorganisms and microbial communities. Front. Bioeng. Biotechnol. 9:632059. doi: 10.3389/fbioe.2021.632059
Moon, C., Stupp, G. S., Su, A. I., and Wolan, D. W. (2018). Metaproteomics of colonic microbiota unveils discrete protein functions among colitic mice and control groups. Proteomics 18:1700391. doi: 10.1002/pmic.201700391
Moosa, Y., Kwon, D., de Oliveira, T., and Wong, E. B. (2020). Determinants of vaginal microbiota composition. Front. Cell. Infect. Microbiol. 10:467. doi: 10.3389/fcimb.2020.00467
Moreira, M., Schrama, D., Farinha, A. P., Cerqueira, M., Raposo de Magalhães, C., Carrilho, R., et al. (2021). Fish pathology research and diagnosis in aquaculture of farmed fish; a proteomics perspective. Animals 11:125. doi: 10.3390/ani11010125
Moussa, D. G., Ahmad, P., Mansour, T. A., and Siqueira, W. L. (2022). Current state and challenges of the global outcomes of dental caries research in the meta-omics era. Front. Cell. Infect. Microbiol. 12:887907. doi: 10.3389/fcimb.2022.887907
Nephali, L., Piater, L. A., Dubery, I. A., Patterson, V., Huyser, J., Burgess, K., et al. (2020). Biostimulants for plant growth and mitigation of abiotic stresses: a metabolomics perspective. Meta 10:505. doi: 10.3390/metabo10120505
Nowrotek, M., Jałowiecki, Ł., Harnisz, M., and Płaza, G. A. (2019). Culturomics and metagenomics: in understanding of environmental resistome. Front. Environ. Sci. Eng. 13:40. doi: 10.1007/s11783-019-1121-8
Okeke, E. S., Ita, R. E., Egong, E. J., Udofia, L. E., Mgbechidinma, C. L., and Akan, O. D. (2021). Metaproteomics insights into fermented fish and vegetable products and associated microbes. Food Chem. 3:100045. doi: 10.1016/j.fochms.2021.100045
Purohit, J., Chattopadhyay, A., and Teli, B. (2020). Metagenomic exploration of plastic degrading microbes for biotechnological application. Curr. Genomics 21, 253–270. doi: 10.2174/1389202921999200525155711
Ragazou, K., Passas, I., Garefalakis, A., and Dimou, I. (2022). Investigating the research trends on strategic ambidexterity, agility, and open innovation in SMEs: perceptions from Bibliometric analysis. J. Open Innov. Technol. Market Complexity 8:118. doi: 10.3390/joitmc8030118
Rane, N. R., Tapase, S., Kanojia, A., Watharkar, A., Salama, E.-S., Jang, M., et al. (2022). Molecular insights into plant–microbe interactions for sustainable remediation of contaminated environment. Bioresour. Technol. 344:126246. doi: 10.1016/j.biortech.2021.126246
Rechenberger, J., Samaras, P., Jarzab, A., Behr, J., Frejno, M., Djukovic, A., et al. (2019). Challenges in clinical metaproteomics highlighted by the analysis of acute leukemia patients with gut colonization by multidrug-resistant enterobacteriaceae. Proteomes 7:2. doi: 10.3390/proteomes7010002
Renu,, Gupta, S. K., Rai, A. K., Sarim, K. M., Sharma, A., Budhlakoti, N., et al. (2019). Metaproteomic data of maize rhizosphere for deciphering functional diversity. Data Brief 27:104574. doi: 10.1016/j.dib.2019.104574
Roldan-Valadez, E., Salazar-Ruiz, S. Y., Ibarra-Contreras, R., and Rios, C. (2019). Current concepts on bibliometrics: a brief review about impact factor, Eigenfactor score, CiteScore, SCImago journal rank, source-normalised impact per paper, H-index, and alternative metrics. Irish J. Med. Sci. 188, 939–951. doi: 10.1007/s11845-018-1936-5
Saito, M. A., Bertrand, E. M., Duffy, M. E., Gaylord, D. A., Held, N. A., Hervey, W. J. I., et al. (2019). Progress and challenges in ocean metaproteomics and proposed best practices for data sharing. J. Proteome Res. 18, 1461–1476. doi: 10.1021/acs.jproteome.8b00761
Salvato, F., Hettich, R. L., and Kleiner, M. (2021). Five key aspects of metaproteomics as a tool to understand functional interactions in host-associated microbiomes. PLoS Pathog. 17:e1009245. doi: 10.1371/journal.ppat.1009245
Saralegui, C., García-Durán, C., Romeu, E., Hernáez-Sánchez, M. L., Maruri, A., Bastón-Paz, N., et al. (2022). Statistical evaluation of metaproteomics and 16S rRNA amplicon sequencing techniques for study of gut microbiota establishment in infants with cystic fibrosis. Microbiol. Spectr. 10:e0146622. doi: 10.1128/spectrum.01466-22
Schiebenhoefer, H., Van Den Bossche, T., Fuchs, S., Renard, B. Y., Muth, T., and Martens, L. (2019). Challenges and promise at the interface of metaproteomics and genomics: an overview of recent progress in metaproteogenomic data analysis. Expert Rev. Proteomics 16, 375–390. doi: 10.1080/14789450.2019.1609944
Sehli, S., Allali, I., Chahboune, R., Bakri, Y., Al Idrissi, N., Hamdi, S., et al. (2021). Metagenomics approaches to investigate the gut microbiome of COVID-19 patients. Bioinform. Biol. Insights 15:1177932221999428. doi: 10.1177/1177932221999428
Seyhan, A. A., and Carini, C. (2019). Are innovation and new technologies in precision medicine paving a new era in patients centric care? J. Transl. Med. 17:114. doi: 10.1186/s12967-019-1864-9
Shakya, M., Lo, C.-C., and Chain, P. S. G. (2019). Advances and challenges in metatranscriptomic analysis. Front. Genet. 10:904. doi: 10.3389/fgene.2019.00904
Sun, Z., Song, Z.-G., Liu, C., Tan, S., Lin, S., Zhu, J., et al. (2022). Gut microbiome alterations and gut barrier dysfunction are associated with host immune homeostasis in COVID-19 patients. BMC Med. 20:24. doi: 10.1186/s12916-021-02212-0
Tahamtan, I., and Bornmann, L. (2019). What do citation counts measure? An updated review of studies on citations in scientific documents published between 2006 and 2018. Scientometrics 121, 1635–1684. doi: 10.1007/s11192-019-03243-4
Taylor, M. J., Lukowski, J. K., and Anderton, C. R. (2021). Spatially resolved mass spectrometry at the single cell: recent innovations in proteomics and metabolomics. J. Am. Soc. Mass Spectrom. 32, 872–894. doi: 10.1021/jasms.0c00439
Tiwari, A., and Taj, G. (2020). “Applications of advanced omics technology for harnessing the high altitude agriculture production” in Microbiological advancements for higher altitude agro-ecosystems & sustainability. eds. R. Goel, R. Soni, and D. C. Suyal (New York, NY: Springer), 447–463.
Ugya, A. Y., Hua, X., Agamuthu, P., and Ma, J. (2019). Molecular approach to uncover the function of bacteria in petrochemical refining wastewater: a mini review. Appl. Ecol. Environ. Res. 17, 3645–3665. doi: 10.15666/aeer/1702_36453665
Van Den Bossche, T., Arntzen, M. Ø., Becher, D., Benndorf, D., Eijsink, V. G. H., Henry, C., et al. (2021). The metaproteomics initiative: a coordinated approach for propelling the functional characterization of microbiomes. Microbiome 9:243. doi: 10.1186/s40168-021-01176-w
Vargas Medina, D. A., Maciel, E. V. S., de Toffoli, A. L., and Lanças, F. M. (2020). Miniaturization of liquid chromatography coupled to mass spectrometry: 2. Achievements on modern instrumentation for miniaturized liquid chromatography coupled to mass spectrometry. TrAC Trends Anal. Chem. 128:115910. doi: 10.1016/j.trac.2020.115910
Wang, Q., Li, R., and Zhan, L. (2021). Blockchain technology in the energy sector: from basic research to real world applications. Comput. Sci. Rev. 39:100362. doi: 10.1016/j.cosrev.2021.100362
White, R. A., Rosnow, J., Piehowski, P. D., Brislawn, C. J., and Moran, J. J. (2021). In situ non-destructive temporal measurements of the rhizosphere microbiome ‘hot-spots’ using metaproteomics. Agronomy 11:2248. doi: 10.3390/agronomy11112248
Wilmes, P., and Bond, P. L. (2004). The application of two-dimensional polyacrylamide gel electrophoresis and downstream analyses to a mixed community of prokaryotic microorganisms. Environ. Microbiol. 6, 911–920. doi: 10.1111/j.1462-2920.2004.00687.x
Xie, X., Zheng, H., Zhang, Q., Fan, J., Liu, N., and Song, X. (2022). Co-metabolic biodegradation of structurally discrepant dyestuffs by Klebsiella sp. KL-1: a molecular mechanism with regards to the differential responsiveness. Chemosphere 303:135028. doi: 10.1016/j.chemosphere.2022.135028
Yu, D., Wang, W., Zhang, W., and Zhang, S. (2018). A bibliometric analysis of research on multiple criteria decision making. Curr. Sci. 114, 747–758. doi: 10.18520/cs/v114/i04/747-758
Yu, D., Xu, Z., and Pedrycz, W. (2020). Bibliometric analysis of rough sets research. Appl. Soft Comput. 94:106467. doi: 10.1016/j.asoc.2020.106467
Yu, D., Xu, Z., and Wang, W. (2018). Bibliometric analysis of fuzzy theory research in China. Knowl. Based Syst. 141, 188–199. doi: 10.1016/j.knosys.2017.11.018
Zanotto, E. D., and Carvalho, V. (2021). Article age- and field-normalized tools to evaluate scientific impact and momentum. Scientometrics 126, 2865–2883. doi: 10.1007/s11192-021-03877-3
Zhang, X., Deeke, S. A., Ning, Z., Starr, A. E., Butcher, J., Li, J., et al. (2018). Metaproteomics reveals associations between microbiome and intestinal extracellular vesicle proteins in pediatric inflammatory bowel disease. Nat. Commun. 9:2873. doi: 10.1038/s41467-018-05357-4
Zhang, X., Li, L., Butcher, J., Stintzi, A., and Figeys, D. (2019). Advancing functional and translational microbiome research using meta-omics approaches. Microbiome 7:154. doi: 10.1186/s40168-019-0767-6
Zhang, Y., Liu, H., Kang, S.-C., and Al-Hussein, M. (2020). Virtual reality applications for the built environment: research trends and opportunities. Autom. Constr. 118:103311. doi: 10.1016/j.autcon.2020.103311
Zheng, Q., Wang, Y., Lu, J., Lin, W., Chen, F., and Jiao, N. (2020). Metagenomic and metaproteomic insights into photoautotrophic and heterotrophic interactions in a Synechococcus culture. mBio 11:e03261-19. doi: 10.1128/mBio.03261-19
Keywords: metaproteomics, metagenomics, microbiome, bibliometric analyses, field weighted citation impact
Citation: Ascandari A, Aminu S, Safdi NEH, El Allali A and Daoud R (2023) A bibliometric analysis of the global impact of metaproteomics research. Front. Microbiol. 14:1217727. doi: 10.3389/fmicb.2023.1217727
Edited by:
Utkarsh Sood, University of Delhi, IndiaCopyright © 2023 Ascandari, Aminu, Safdi, El Allali and Daoud. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rachid Daoud, cmFjaGlkLmRhb3VkQHVtNnAubWE=; Achraf El Allali, YWNocmFmLmVsYWxsYWxpQHVtNnAubWE=
 AbdulAziz Ascandari
AbdulAziz Ascandari Suleiman Aminu
Suleiman Aminu Nour El Houda Safdi
Nour El Houda Safdi Achraf El Allali
Achraf El Allali Rachid Daoud
Rachid Daoud