- 1College of Medicine and Dentistry, James Cook University, Townsville, QLD, Australia
- 2Centre for Molecular Therapeutics, Australian Institute of Tropical Health and Medicine, James Cook University, Cairns, QLD, Australia
Tuberculosis (TB) has remained at the forefront of the global infectious disease burden for centuries. Concerted global efforts to eliminate TB have been hindered by the complexity of Mycobacterium tuberculosis (Mtb), the emergence of antibiotic resistant Mtb strains and the recent impact of the ongoing pandemic of coronavirus disease 2019 (COVID19). Examination of the immunomodulatory role of gastrointestinal microbiota presents a new direction for TB research. The gut microbiome is well-established as a critical modulator of early immune development and inflammatory responses in humans. Recent studies in animal models have further substantiated the existence of the ‘gut-lung axis’, where distal gastrointestinal commensals modulate lung immune function. This gut microbiome-lung immune crosstalk is postulated to have an important correlation with the pathophysiology of TB. Further evaluation of this gut immunomodulation in TB may provide a novel avenue for the exploration of therapeutic targets. This mini-review assesses the proposed mechanisms by which the gut-lung axis impacts TB susceptibility and progression. It also examines the impact of current anti-TB therapy on the gut microbiome and the effects of gut dysbiosis on treatment outcomes. Finally, it investigates new therapeutic targets, particularly the use of probiotics in treatment of antibiotic resistant TB and informs future developments in the field.
Introduction
Tuberculosis
Tuberculosis (TB) remains at the forefront of the global disease burden with over 10 million new cases and 1.6 million deaths in 2021 (World Health Organization, 2023). Currently, it is the second highest infectious cause of death after COVID19 and has a disproportionate socioeconomic impact on low to middle income countries through catastrophic healthcare costs for individuals (World Health Organization, 2023). Current global initiatives to target TB are centered around addressing social determinants, early case identification and provision of effective antibiotics in the framework of universal healthcare (World Health Organization, 2022). Concerted global efforts led by the WHO through the END TB strategy and sustainable development goals have led to a gradual reduction in case incidence over the past decade (World Health Organization, 2022). However, the COVID19 pandemic is estimated to have reversed progress by 12 years through its disruption to case detection and treatment access, with consequent increases in mortality and morbidity (Wingfield et al., 2021; World Health Organization, 2022). Additionally, the emergence of drug resistant mycobacteria strains has rendered many first line antibiotic regimens ineffective, driving the resurgence of TB in low and middle income countries (Tiberi et al., 2022). Novel vaccine development remains hindered by an incomplete understanding of Mycobacterium tuberculosis (Mtb) pathogenesis and spreading of multidrug resistance threatens to further negate progress (McShane, 2019; Stephanie et al., 2021).
The gut microbiome
A novel avenue for TB treatment and management is modulation of the gut-lung axis – the bidirectional relationship between the composition and metabolism of the gastrointestinal microbiome and regulation of lung immune responses recently evidenced in animal studies (Mori et al., 2021). The gut microbiome has long been recognized for its multifaceted role in health through detoxification, protection against pathogens, regulation of metabolism and modulation of the immune system (Wu and Wu, 2012; Zheng et al., 2020). Germ free (GF) animal models have defective lymphoid tissue, increased likelihood of Th1/Th2 imbalances as well as reduced intraepithelial lymphocytes, IgA antibodies and Th17 immunoregulatory cells (Zheng et al., 2020). Microbiome development is influenced by a range of factors including maternal diet, infections, probiotic use, genetics, geography, delivery method, gestational age, diet, and antibiotic use (Vandenplas et al., 2020; Li P. et al., 2022; Wernroth et al., 2022). The infant microbiome composition is highly dynamic in response to these environmental factors (Li P. et al., 2022; Wernroth et al., 2022), and the diversity generated in this period modulates IgE homeostasis, determining allergic susceptibility (Méndez et al., 2021; Zhu et al., 2021).
Short chain fatty acids (SCFAs), such as butyrate, are produced by the gut microbiota and function as signaling molecules that modulate inflammatory responses, regulate macromolecule metabolism and reduce colorectal cancer risk (He et al., 2020). They modulate pH, regulate mucus production and act as colonic epithelial cell energy source, directly promoting gut integrity (Blaak et al., 2020). Butyrate reduces IL-12 and IFNγ production by inhibiting histone deacetylase (HDAC), mammalian target of rapamycin (mTOR) kinase and nuclear factor kappa B (NF-κB) signaling, preventing skewing to the inflammatory Th1/M1 phenotype (He et al., 2020; Kotlyarov, 2022). Furthermore, butyrate inhibits nitric oxide and LPS-mediated induction of proinflammatory cytokines such as IL-6, IL-12, IL-1β, and TNFα (He et al., 2020; Kotlyarov, 2022). SCFAs also inhibit IL-12 release from dendritic cells (DC), inhibiting antigen specific CD8+ T cell activity and increasing infection risk (Nastasi et al., 2017). While predominately acting anti-inflammatory, SCFAs can also produce pro-inflammatory states through G protein-coupled receptors (GPCRs), and promote CD8+ T cells memory potential (Bachem et al., 2019; He et al., 2020). High butyrate or propionate levels in children are also protective against the development of atopy (Roduit et al., 2019). Thus, microbiome composition and SCFA production has a significant impact on immune development and dysfunction with lasting implications for health in adult life (Zheng et al., 2020).
Gut-lung axis
Emerging evidence supports the role of gut microbiota in modulating immunity and inflammation at distal sites such as the lungs (Osei Sekyere et al., 2020). Changes in microbiota metabolites or composition correlate to defective immune responses in many respiratory diseases (Comberiati et al., 2021). Animal models demonstrated more severe Escherichia coli pneumonia in mice with gut commensal depletion due to decreased alveolar macrophages (AM) activity via reduced toll-like receptor (TLR) signaling, NF-κB DNA-binding activity, TNFα, CXCR2 and ICAM expression on intestinal mucosa (Chen et al., 2011). Similarly, a murine model of Streptococcus pneumoniae infection found that antibiotic treated mice given fecal suspensions by oral gavage showed enhanced AM function (Schuijt et al., 2016), however antibiotic effects and oral gavage use may have confounded results (Budden et al., 2017). Clostridium spp. may be an important regulator of allergic asthma through the induction of IL10+ CTLA4+ colonic T regulatory cells (Tregs) (Di Gangi et al., 2020). Animal models demonstrate antibiotics specific to Clostridium spp., such as vancomycin, reduce CD4+CD25+ Tregs (Di Gangi et al., 2020). Recent evidence suggests that Helicobacter pylori infection is inversely correlated with asthma severity (Chen et al., 2011; Lim et al., 2016; Kato et al., 2017; Tsigalou et al., 2019), however this is contradicted by other studies (Wang et al., 2012, 2013, 2017; Molina-Infante et al., 2018), suggesting its role remains unclear.
Polysaccharide A (PSA), produced by Bacteroides fragilis, suppresses adverse inflammatory responses, inhibiting asthma pathogenesis (Johnson et al., 2018). PSA signals through the TLR2/TLR1 heterodimer to activate multiple signaling pathways that promote immune tolerance such as activating IL-10 producing Tregs (Figure 1; Erturk-Hasdemir et al., 2019). Furthermore, PSA also induces dose dependent interferon beta (IFNβ) expression by colonic lamina propria DCs through TLR4 activation which has been shown to be protective in infection with vesicular stomatitis virus or influenza A virus (Stefan et al., 2020; Wirusanti et al., 2022). This supports the hypothesis that microbiota critically modulate homeostatic type 1 IFN expression essential for a rapid antiviral response and effective viral clearance (Van Winkle et al., 2022; Wirusanti et al., 2022).
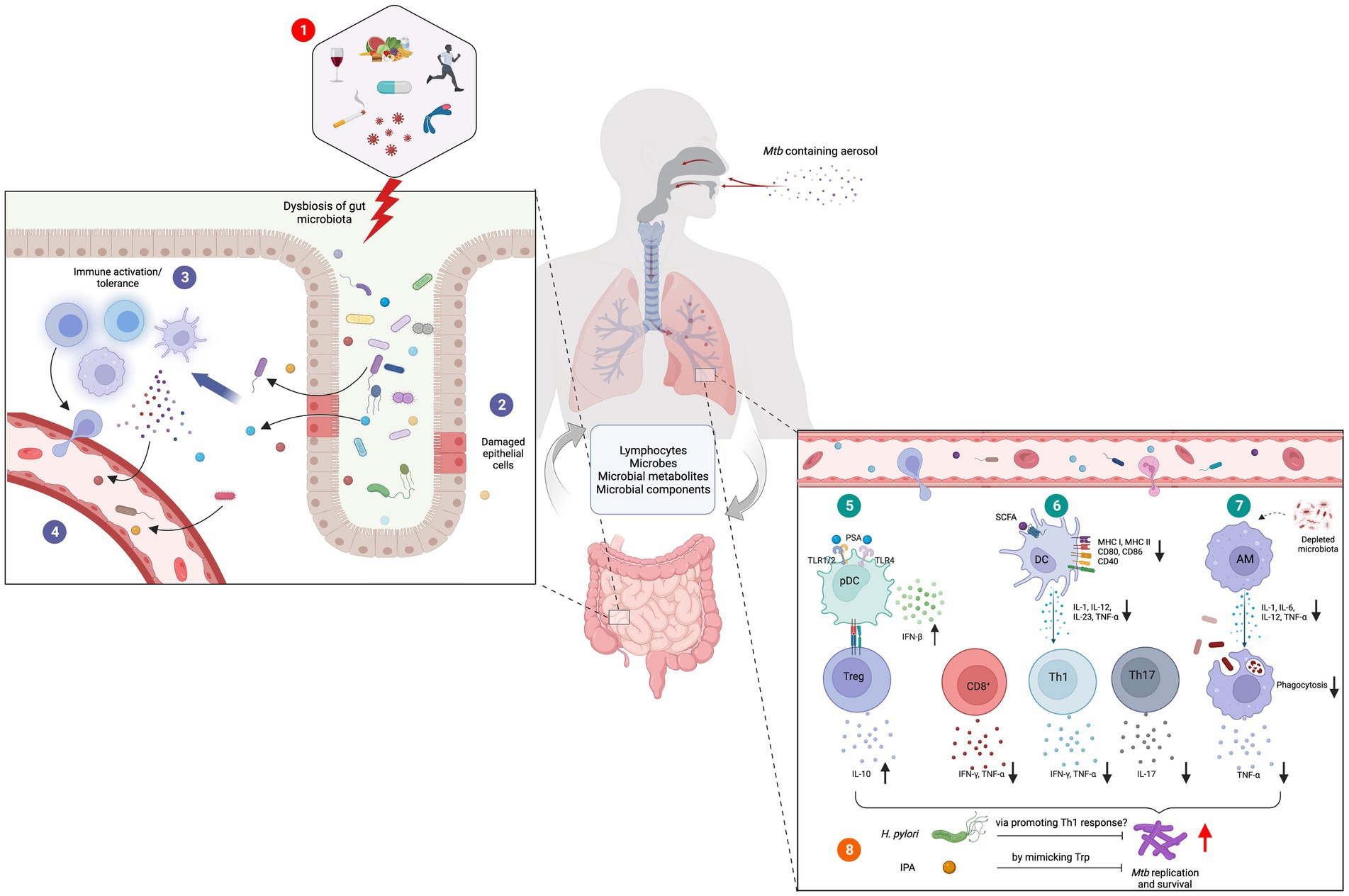
Figure 1. Role of gut-lung axis in Mtb infections. (1) Diet, alcohol, smoking, infectious diseases, lifestyle habits, antibiotics and genetic conditions are among the major causes of gut dysbiosis. (2) Altered gut microbiota cause damages to the gut epithelial layer facilitating the translocation of microbes, microbial components, and metabolites into the tissue (3) followed by immune activation. (4) Some of these activated immune cells, immune molecules, microbes, and their metabolites migrate to distal sites through blood circulation. (5) In the lung tissue, bacterial components such as PSA induce the secretion of IFNβ from plasmacytoid DC (pDCs) and activation and expansion of Tregs. (6) Microbial metabolites such as SCFA downregulate the expression and secretion of co-stimulatory molecules and cytokines in DCs. This results in defective Th1 and Th2 responses and diminished antigen specific CD8+ T cell activation. (7) Depleted gut microbiota hinders the phagocytic capacity of AMs supporting the survival and growth of Mtb. Certain organisms and their metabolites have demonstrated anti-tubercular activities. (8) H. pylori and IPA are believed to restrict Mtb survival by promoting pro-inflammatory Th1 response and mimicking Trp, respectively. Figure was created with Biorender.com.
Gut dysbiosis results in systemic inflammation and bacterial translocation, eventuating in lung dysbiosis (Donovan et al., 2020). This process may activate the inflammasome NOD-like receptor protein 3 (NLRP3) triggering sterile inflammation and neutrophil recruitment that could cause gut epithelial damage and increased permeability (Donovan et al., 2020; Liu Q. et al., 2021). Dysbiosis is also strongly correlated with inflammatory bowel disease (IBD), with 60% of IBD patients also having subclinical lung disease (Raftery et al., 2020). SCFAs regulate lung immune tone by binding to free fatty acid receptors (FFAR), a classic GPCR found on AMs, causing basal IL-1β expression and modulating type 1 IFN responses to respiratory syncytial virus (RSV) infections (Liu Q. et al., 2021). Additionally, patients with severe chronic obstructive pulmonary disease (COPD) were found to have lower SCFAs levels, possibly due to their role in epithelial barrier integrity via increased ZO-1 dense contact proteins expression (Kotlyarov, 2022). Microbiota have also shown to be protective against Streptococcus pneumoniae infection through enhancement of primary AM phagocytosis and responsiveness to pathogenic associated molecular patterns (PAMPs) (Schuijt et al., 2016).
The gut-lung axis in Mycobacterium tuberculosis infection
Millions of individuals acquire a latent or active TB infection annually without obvious immune deficiency, indicating the existence of previously unidentified risk factors (Namasivayam et al., 2018). Alterations in the gut-lung axis is hypothesized to be a contributing factor in Mtb infection pathogenesis and its clinical presentation (Comberiati et al., 2021). Early GF models initially suggested no difference in Mtb tissue load between GF and conventional mice (Suter and Kirsanow, 1962; Huempfner and Deuschle, 1966), however recent developments in the understanding of gut microbiome-mediated immunomodulation has renewed interest in the area. Human immunodeficiency virus (HIV) infection, malnutrition, diabetes, alcohol, smoking and air pollution which are important risk factors for TB have all also been shown to cause changes in the gut microbiome (Naidoo et al., 2019). These factors and others can cause gut dysbiosis (Figure 1), resulting in alterations in the microbiota’s biosynthetic pathways, changes in the lung microbiome and downstream immunomodulatory effects (Shah et al., 2021). This results in reduced resistance to colonization by external pathogens, escape of a contained pathogen or loss of benign lung commensals, leading to lung disease (Naidoo et al., 2019).
C3HeB/FeJ mice fed with a high fat diet showed proinflammatory responses that increased the risk for developing active TB and impaired the immune protection from BCG vaccination in obese mice (Arias et al., 2019). The authors hypothesized that this was due to a reduction in the Firmicutes/Bacteroidetes phyla ratio and decreased Porphyromonadaceae family abundance in the gut microbiota (Arias et al., 2019). Additionally, increases in the genera associated with dysbiosis such as Alistipes, Parasuterella, Mucispirillum, and Akkermansia were observed (Arias et al., 2019). However, an increased Firmicutes/Bacteroidetes ratio was found in a murine model of type 2 diabetes (T2D) generated through the administration of an energy dense diet (Sathkumara et al., 2021). The study also suggested that these changes in the gut microbiome may increase susceptibility to TB through alterations in SCFA metabolism (Sathkumara et al., 2021). Additionally, rhesus macaque monkeys that developed severe disease from Mtb infection had distinct intestinal microbiota compared to those with less severe disease (Namasivayam et al., 2019). In particular, animals with more severe disease had enriched Lachonospiraceae and Clostridiaceae and depleted Streptococcaceae bacterial families (Namasivayam et al., 2019). These findings, support the importance of factors that impact gut dysbiosis due to crosstalk on the gut-lung axis and implications for TB pathogenesis and susceptibility.
Aerosol Mtb infection in mice can lead to rapid changes in intestinal bacterial composition, particularly in the orders Closteridiales and Bacteroidetales (Winglee et al., 2014). A low dose Mtb aerosol challenge caused rapid intestinal dysbiosis and loss of diversity in both diabetic and nondiabetic mice in a murine model of T2D (Sathkumara et al., 2021). A similar study of Mtb infection in a murine model found similar trends but these were not significant over the duration of the study (Namasivayam et al., 2017). Patients with a Mtb respiratory infection have been shown to have reduced gut microbiome diversity compared to healthy controls, evidencing the role of the gut-lung axis in susceptibility to TB (Hu et al., 2019; Comberiati et al., 2021). Pulmonary Mtb infection was also shown to decrease the α diversity of the gut microbiome, particularly through alterations in the populations of the genus Bacteroides in recent human studies (Hu et al., 2019; Wang Y. et al., 2022; Wang S. et al., 2022). One cross-sectional study contradicted these results by finding no significant alterations in gut microbiota in latent TB patients, but the same patients were not sampled before and after infection (Wipperman et al., 2017; Naidoo et al., 2019).
Gut bacteria from the phylum Bacteroidetes, which include many beneficial commensals, were also found to be reduced in patients with recurrent TB (Table 1; Luo et al., 2017). Species from the genera Lachnospira and Roseburia (phylum Firmicutes) were also depleted in TB patients, with possible negative ramifications for the production of SCFAs and their consequent downstream regulatory effects (Luo et al., 2017). Species from the more pathogenic phyla Actinobacteria and Proteobacteria including Escherichia coli were increased in recurrent TB patients’ feces samples, consistent with previous studies analyzing sputum (Luo et al., 2017). Both the genera Prevotella and Lachnospira were shown to inversely correlate with recurrent TB, positively correlate with peripheral CD4+ cell counts in new cases and decrease in both new and recurrent TB patient groups (Luo et al., 2017). However, in a HIV+ positive population on anti-retroviral treatment with high TB incidence, increased oral anaerobes including species of the genus Prevotella, were associated with higher levels of pulmonary SCFA that positively correlated with risk of developing active TB (Segal et al., 2017).
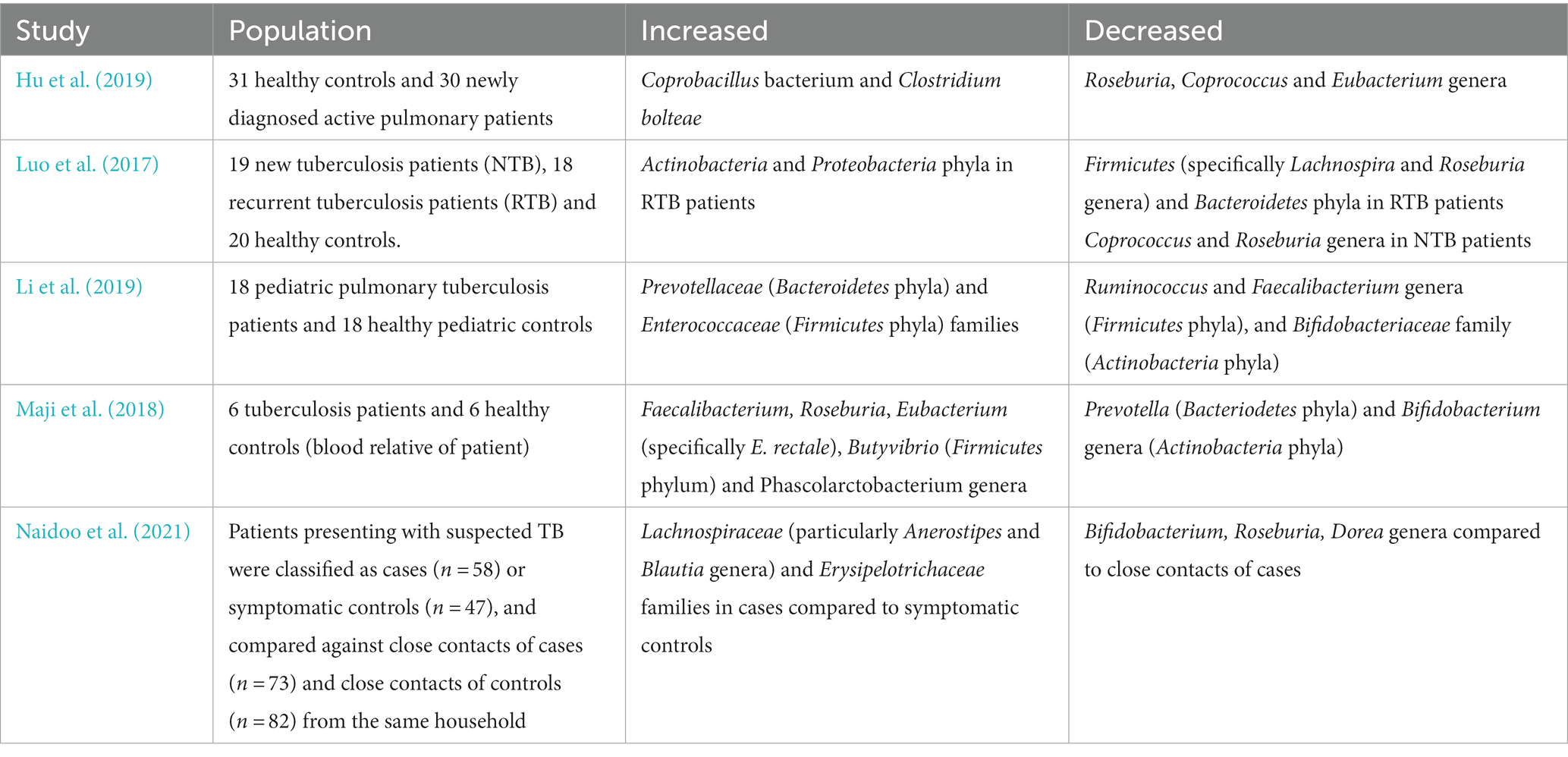
Table 1. Summary of the gut microbiome changes in patients infected with Mtb relative to healthy controls in key human studies.
A case–control study of pulmonary TB in pediatric patients also found significant gut dysbiosis in comparison to healthy controls with enriched Prevotellaceae and Enterococcaceae, but decreased beneficial Oscillospiraceae and Bifidobacteriaceae bacterial families (Table 1; Li et al., 2019). The authors speculated that the increased populations of Prevotella may induce a proinflammatory cytokine production that worsens TB, though this remains to be mechanistically confirmed (Li et al., 2019). Faecalibacterium ruminococcaceae and Faecalibacterium prausnitzii species depletion (part of the Oscillospiraceae family) may have negative effects through reduced immunomodulatory SCFAs while reductions in the bacterial family Bifidobacteriacae have been associated with many other respiratory diseases (Li et al., 2019).
A study examining gut microbiome alterations in TB patients through 16S rRNA gene and whole-genome shotgun sequencing also found significant depletion of Provetalla and increased Bacteroides intestinal bacterial genera (Table 1; Maji et al., 2018). Considerable increases in butyrate and propionate producing intestinal bacteria in Faecalibacterium, Roseburia, Eubacterium and Phascolarctobacterium genera were detected among TB patients (Maji et al., 2018). Pre-treatment TB patients also showed increased butyrate-producing anaerobic species from the bacterial families Lachnospiraceae and Erysipelotrichaeceae in stool samples compared to healthy controls (Table 1; Naidoo et al., 2021). Butyrate inhibits IFNγ and IL-17A, strongly reducing Th17 proliferation and causing a detrimental dysregulation of the immune response against Mtb (Lachmandas et al., 2016; Segal et al., 2017; Naidoo et al., 2021). Additionally, through induction of immunosuppressive Tregs in the gut and consequent IL-10 release, butyrate may suppress critical pro-inflammatory T cell responses in TB patients facilitating immune evasion and chronic infection (Figure 1; Maji et al., 2018). Furthermore, this study showed a positive correlation between species from the Lachnospiraceae and Erysipelotrichaceae family and interferon regulation, inflammasome activation, cell death signaling and antibacterial activity in TB patients (Naidoo et al., 2021).
In contrast, a small cohort study found that healthy controls have higher levels of SCFA producing bacteria in their gut microbiota, such as the butyrate-producing Roseburia, Coprococcus and Eubacterium genera, compared to TB patients (Table 1; Hu et al., 2019). TB patients had altered activity of microbiome metabolic pathways through decreased production of precursor metabolites and energy, decreased degradation/utilization/assimilation capacities and increased vitamin synthesis (Hu et al., 2019). The study authors found that active TB patients had a unique microbiota signature through strain level single nucleotide polymorphisms and species patterns that differentiates them from healthy controls (Hu et al., 2019).
Colonization by Helicobacter spp. has diverse effects on the pathogenesis of TB. Murine models of Helicobacter hepaticus show increased inflammation, severe lung pathology, increased Mtb burden and worsened mortality and morbidity after aerosol challenge (Arnold et al., 2015; Majlessi et al., 2017). This is postulated to result from increased IL-10 (Cervantes and Hong, 2017), which suppresses macrophage activation and DC function in the early immune response to Mtb (Redford et al., 2011). Helicobacter pylori in contrast may induce a protective immunomodulatory response that reduces the TB risk in humans (Perry et al., 2010). Individuals with H. pylori infections were found to have heightened IFNγ and Th1-like responses to Mtb antigens (Figure 1), more likely to maintain latency and less likely to develop active disease (Perry et al., 2010, 2013). However, these results contradict the results of three studies which found no association between Mtb and H. pylori (Sanaka et al., 1998; Tsang et al., 1998; Torres et al., 2003), and three studies that found that H. pylori increased the incidence of Mtb infections (Mitchell et al., 1992; Woeltje et al., 1997; Philippou et al., 2003). H. pylori may worsen TB through its ability to increase Treg populations, interfere with DC maturation and prevent T lymphocyte maturation (Bustamante-Rengifo et al., 2021). However, the exact relationship between H. pylori and Mtb remains to be fully elucidated through larger studies with appropriate controls.
TB, antibiotics, and BCG
Murine models have consistently demonstrated that antibiotics can cause dramatic alterations in the gut microbiome,(Khan et al., 2016; Namasivayam et al., 2017; Dumas et al., 2018) potentially contributing to TB pathogenesis and affecting treatment efficacy. A mouse model of dysbiosis induced by the administration of wide-spectrum antibiotics showed increased colonization of the lungs by Mtb and reduced mucosal associated invariant T (MAIT) cells during the first week of infection (Dumas et al., 2018). Inoculation with a gavage of microbiota from non-treated mice reversed these changes and regenerated the MAIT cell population (Dumas et al., 2018). Another study of antibiotic therapy in a murine model of TB also demonstrated significant alterations in the microbiome (Khan et al., 2016). The antibiotic regimen was chosen to specifically not change Mtb viability but solely cause gut dysbiosis (Khan et al., 2016). Mice on the antibiotic therapy had higher Mtb burden both in the lungs and extra-pulmonary sites (Khan et al., 2016). This likely resulted from the suppression of Th1 immunity and enhancement of Tregs due to the alterations in gut microbiota (Khan et al., 2016). Furthermore, fecal transplants were effective to rebuild the gut microbiota, improve Th1 immunity, inhibit Treg populations and reduce Mtb burden (Khan et al., 2016). Similarly, treatment of mice with widely used anti-TB drugs isoniazid and pyrazinamide prior to Mtb infection resulted in significant gut dysbiosis and increased Mtb burden (Khan et al., 2019). This was associated with impaired AM metabolism and defective bactericidal activity and was reversible with fecal transplantation form untreated animals (Khan et al., 2019). These antibiotics are associated with more selective alterations in the gut microbiome including decreases in the Closteridia genera,(Khan et al., 2019) associated with Treg induction (Atarashi et al., 2013).
The use of broad-spectrum antibiotics on Mtb infected mice showed that the antibiotics caused significant gut dysbiosis that resulted in the deregulation of approximately 7,592 long noncoding RNA (lncRNA) sequences (Yang et al., 2022). One especially depleted highly conserved lncRNA, temporarily named lncRNA-CGB (commensal gut bacteria associated lncRNA), was especially downregulated in both mouse models and humans with active TB, correlating with poor outcomes in Mtb infections (Yang et al., 2022). Both murine lncRNA-CGB Genomic knock-out (KO) models and a human lncRNA-CGB knockdown CD3+ T cell model showed impaired ability to control Mtb replication and infections (Yang et al., 2022). B. fragilis, typically depleted in Mtb infections, was found to be a key up-regulator of lnRNA-CGB in mouse models and patients with active TB (Yang et al., 2022) LncRNA-CGB was found to epigenetically regulate IFNγ through interaction with enhancer of zeste homolog 2 (EZH2) (Yang et al., 2022). These findings provide a direct link between microbiota and the immune protection against TB conferred through the gut-lung axis.
A recent study also indicated that parenteral BCG vaccination in murine models causes time-dependent development of gut dysbiosis associated with increased production of butyrate (Jeyanathan et al., 2022). Additionally, BCG induces mild self-limiting, time-dependent intestinal inflammation causing significantly increased intestinal permeability, enabling the leakage of luminal molecules such as butyrate through the epithelium (Jeyanathan et al., 2022). Naïve mice treated with antibiotics and then transplanted with microbiota from BCG-immunized hosts were found to have alveolar macrophages (AM) with elevated MHC II, IL-6, and TNF production at base line and upon stimulation (Jeyanathan et al., 2022). These results indicate that a component of the protection conferred by BCG vaccination occurs through modulation of the lung-gut axis (Jeyanathan et al., 2022).
Conventional anti-TB regimens of isoniazid-rifampicin-pyrazinamide-ethambutol (HRZE) induce significant long-term gut dysbiosis that persist well beyond treatment cessation both in companion murine and human studies (Namasivayam et al., 2017; Wipperman et al., 2017). Subjects showed increased populations of Erysipelayoclotridium, Prevotella and Fusobacterium, and decreased population of Blautia, Lactobacillus, Coprococcus, Bifidobacterium and Bacteroides genera compared to latent TB controls (Wipperman et al., 2017). Polysaccharides produced by Bacteroides have been demonstrated to induce IL-10 producing Treg cells in mice (Johnson et al., 2018; Erturk-Hasdemir et al., 2019). Similarly, some Lactobaccillus spp. also can increase Treg cell populations and contribute to immune tolerance (Ding et al., 2017), Coprococcus can modulate IL-1β, IFNγ, and other cytokines in response to fungal stimuli (Schirmer et al., 2016), and Bifidobacterium is capable of inducing intestinal Th17 cells in animal models (Tan et al., 2016). This supports the notion that gut dysbiosis drives changes in immune signaling that potentially contribute to the variable efficacy of HRZE treatment (Wipperman et al., 2017). Additionally, the persistence of gut dysbiosis beyond treatment cessation may increase the likelihood of reinfection or other infections in TB patients (Osei Sekyere et al., 2020).
Probiotics and gut metabolites
Due to the adverse dysbiotic effects, expense and adherence challenge of conventional anti-TB therapy and growing evidence of the role of microbiota in TB, probiotics are now being explored as novel therapeutic avenue (Wipperman et al., 2017; Rahim et al., 2022). Probiotics have been previously shown to be effective in suppressing antibiotic resistant organisms which otherwise require more intensive and expensive treatments (Rahim et al., 2022). Lacticaseibacillus rhamnosus isolated from the vagina of healthy women, inhibited Mtb growth in culture medium and showed intracellular killing activity against drug sensitive and resistant strains of Mtb in a murine macrophage cell line without cytotoxicity (Rahim et al., 2022). A murine experiment involving the intragastric administration of Lactobacillus casei indicated a protective role for probiotics against the gut-based adverse reactions of isoniazid and rifampicin through modulation of SCFAs (Li Y. et al., 2022). This was consistent with a larger randomized open label dose–response clinical trial of Lactobacillus casei that found reduced anti-TB associated gastrointestinal adverse effects (Lin et al., 2020).
As evidence strongly supports that microbiome immunomodulation is primarily modulated through their secreted metabolites, the use of inactivated microbial cells or their components, termed as postbiotics, is also being investigated (Liu Y. et al., 2021). In a whole-cell screen of a fragment library, a recent study found that the gut microbial metabolite indole propionic acid (IPA) showed significant dose-dependent anti-tubercular in vitro potency comparable to first line antibiotics (Negatu et al., 2018). In vivo experiments confirmed the effect of IPA through 7-fold reductions in bacterial load in the spleen of Mtb-infected mice (Negatu et al., 2018). The antimycobacterial-specific activity in vitro of IPA has been shown to occur through mimicking the allosteric inhibitory effect of tryptophan (Trp) on anthranilate synthase TrpE as part of the physiological negative feedback loop on tryptophan synthesis (Kaufmann, 2018; Negatu et al., 2019). Thus, while evidence supports the beneficial role of probiotics and microbial metabolites in TB, it remains limited and should be explored through larger scale animal experiments.
Conclusion
The existence of the gut-lung axis has been consistently supported through recent literature. Gut microbiota have been shown to influence lung immunity through production of signaling molecules or SCFAs that modulate inflammatory responses in disease states. Significant evidence also supports a role for the gut-lung axis in TB pathogenesis and recent studies have demonstrated direct links between bacterial species and immunity against TB. However, evidence around the changes in microbiota composition in Mtb infection and its significance is contradictory and needs to be evaluated in more detail through large scale human studies. The use of antibiotics in TB therapy has been linked to significant gut dysbiosis which may be resolved through supplementation with probiotics. The limited research in the use of probiotics in TB is favorable to its use, but this hypothesis needs to be tested more thoroughly in animal and human studies. The strength of association between TB pathogenesis and microbiota alterations holds promise for the development of new therapeutics particularly through probiotic agents or purified bacterial compounds to be used against TB.
Author contributions
AE and AK conceived the manuscript. AE wrote the first draft. HS provided editorial and intellectual input and designed the Figure. All authors contributed to manuscript revision and approved the submitted version.
Funding
AE was supported by an Amuthan Medical Research Grant from James Cook University. AK was supported by an NHMRC Ideas (APP2001262) and Investigator Grant (APP2008715).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Arias, L., Goig, G. A., Cardona, P., Torres-Puente, M., Díaz, J., Rosales, Y., et al. (2019). Influence of gut microbiota on progression to tuberculosis generated by high fat diet-induced obesity in C3HeB/FeJ mice. Front. Immunol. 10:2464. doi: 10.3389/fimmu.2019.02464
Arnold, I. C., Hutchings, C., Kondova, I., Hey, A., Powrie, F., Beverley, P., et al. (2015). Helicobacter hepaticus infection in BALB/c mice abolishes subunit-vaccine-induced protection against M. tuberculosis. Vaccine 33, 1808–1814. doi: 10.1016/j.vaccine.2015.02.041
Atarashi, K., Tanoue, T., Oshima, K., Suda, W., Nagano, Y., Nishikawa, H., et al. (2013). Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 500, 232–236. doi: 10.1038/nature12331
Bachem, A., Makhlouf, C., Binger, K. J., de Souza, D. P., Tull, D., Hochheiser, K., et al. (2019). Microbiota-derived short-chain fatty acids promote the memory potential of antigen-activated CD8(+) T cells. Immunity 51, 285–297.e5. doi: 10.1016/j.immuni.2019.06.002
Blaak, E. E., Canfora, E. E., Theis, S., Frost, G., Groen, A. K., Mithieux, G., et al. (2020). Short chain fatty acids in human gut and metabolic health. Benef. Microbes. 11, 411–455. doi: 10.3920/BM2020.0057
Budden, K. F., Gellatly, S. L., Wood, D. L. A., Cooper, M. A., Morrison, M., Hugenholtz, P., et al. (2017). Emerging pathogenic links between microbiota and the gut–lung axis. Nat. Rev. Microbiol. 15, 55–63. doi: 10.1038/nrmicro.2016.142
Bustamante-Rengifo, J. A., Astudillo-Hernandez, M., and Del Pilar, C.-O. M. (2021). Effect of Helicobacter pylori and Helminth Coinfection on the immune response to Mycobacterium tuberculosis. Curr. Microbiol. 78, 3351–3371. doi: 10.1007/s00284-021-02604-8
Cervantes, J., and Hong, B.-y. (2017). The gut–lung axis in tuberculosis. Pathog. Dis. 75:97. doi: 10.1093/femspd/ftx097
Chen, L. W., Chen, P. H., and Hsu, C. M. (2011). Commensal microflora contribute to host defense against Escherichia coli pneumonia through toll-like receptors. Shock 36, 67–75. doi: 10.1097/SHK.0b013e3182184ee7
Comberiati, P., Di Cicco, M., Paravati, F., Pelosi, U., Di Gangi, A., Arasi, S., et al. (2021). The role of gut and lung microbiota in susceptibility to tuberculosis. Int. J. Environ. Res. Public Health 18:2220. doi: 10.3390/ijerph182212220
Di Gangi, A., Di Cicco, M. E., Comberiati, P., and Peroni, D. G. (2020). Go with your gut: the shaping of T-cell response by gut microbiota in allergic asthma. Front. Immunol. 11:1485. doi: 10.3389/fimmu.2020.01485
Ding, Y. H., Qian, L. Y., Pang, J., Lin, J. Y., Xu, Q., Wang, L. H., et al. (2017). The regulation of immune cells by lactobacilli: a potential therapeutic target for anti-atherosclerosis therapy. Oncotarget 8, 59915–59928. doi: 10.18632/oncotarget.18346
Donovan, C., Liu, G., Shen, S., Marshall, J. E., Kim, R. Y., Alemao, C. A., et al. (2020). The role of the microbiome and the NLRP3 inflammasome in the gut and lung. J. Leukoc. Biol. 108, 925–935. doi: 10.1002/JLB.3MR0720-472RR
Dumas, A., Corral, D., Colom, A., Levillain, F., Peixoto, A., Hudrisier, D., et al. (2018). The host microbiota contributes to early protection against lung colonization by Mycobacterium tuberculosis. Front. Immunol. 9:2656. doi: 10.3389/fimmu.2018.02656
Erturk-Hasdemir, D., Oh, S. F., Okan, N. A., Stefanetti, G., Gazzaniga, F. S., Seeberger, P. H., et al. (2019). Symbionts exploit complex signaling to educate the immune system. Proc. Natl. Acad. Sci. U. S. A. 116, 26157–26166. doi: 10.1073/pnas.1915978116
He, J., Zhang, P., Shen, L., Niu, L., Tan, Y., Chen, L., et al. (2020). Short-chain fatty acids and their association with signalling pathways in inflammation, glucose and lipid metabolism. Int. J. Mol. Sci. 21:6356. doi: 10.3390/ijms21176356
Hu, Y., Feng, Y., Wu, J., Liu, F., Zhang, Z., Hao, Y., et al. (2019). The gut microbiome signatures discriminate healthy from pulmonary tuberculosis patients. Front. Cell. Infect. Microbiol. 9:90. doi: 10.3389/fcimb.2019.00090
Hu, Y., Yang, Q., Liu, B., Dong, J., Sun, L., Zhu, Y., et al. (2019). Gut microbiota associated with pulmonary tuberculosis and dysbiosis caused by anti-tuberculosis drugs. J. Infect. 78, 317–322. doi: 10.1016/j.jinf.2018.08.006
Huempfner, H. R., and Deuschle, K. W. (1966). Experimental tuberculosis in germ-free and conventional mice. Am. Rev. Respir. Dis. 93, 465–467. doi: 10.1164/arrd.1966.93.3P1.465
Jeyanathan, M., Vaseghi-Shanjani, M., Afkhami, S., Grondin, J. A., Kang, A., D’Agostino, M. R., et al. (2022). Parenteral BCG vaccine induces lung-resident memory macrophages and trained immunity via the gut-lung axis. Nat. Immunol. 23, 1687–1702. doi: 10.1038/s41590-022-01354-4
Johnson, J. L., Jones, M. B., and Cobb, B. A. (2018). Polysaccharide-experienced effector T cells induce IL-10 in FoxP3+ regulatory T cells to prevent pulmonary inflammation. Glycobiology 28, 50–58. doi: 10.1093/glycob/cwx093
Kato, H., Pattison, R., and Bhandari, S. (2017). Inverse relationship between Helicobacter pylori infection and asthma in US adults with peptic ulcer disease. Indian J. Gastroenterol. 36, 158–159. doi: 10.1007/s12664-017-0736-6
Kaufmann, S. H. E. (2018). Indole propionic acid: a small molecule links between gut microbiota and tuberculosis. Antimicrob. Agents Chemother. 62:18. doi: 10.1128/aac.00389-18
Khan, N., Mendonca, L., Dhariwal, A., Fontes, G., Menzies, D., Xia, J., et al. (2019). Intestinal dysbiosis compromises alveolar macrophage immunity to Mycobacterium tuberculosis. Mucosal Immunol. 12, 772–783. doi: 10.1038/s41385-019-0147-3
Khan, N., Vidyarthi, A., Nadeem, S., Negi, S., Nair, G., and Agrewala, J. N. (2016). Alteration in the gut microbiota provokes susceptibility to tuberculosis. Front. Immunol. 7:529. doi: 10.3389/fimmu.2016.00529
Kotlyarov, S. (2022). Role of short-chain fatty acids produced by gut microbiota in innate lung immunity and pathogenesis of the heterogeneous course of chronic obstructive pulmonary disease. Int. J. Mol. Sci. 23:4768. doi: 10.3390/ijms23094768
Lachmandas, E., van den Heuvel, C. N., Damen, M. S., Cleophas, M. C., Netea, M. G., and van Crevel, R. (2016). Diabetes mellitus and increased tuberculosis susceptibility: the role of short-chain fatty acids. J. Diabetes Res. 2016, 6014631–6014615. doi: 10.1155/2016/6014631
Li, P., Chang, X., Chen, X., Tang, T., Liu, Y., Shang, Y., et al. (2022). Dynamic colonization of gut microbiota and its influencing factors among the breast-feeding infants during the first two years of life. J. Microbiol. 60, 780–794. doi: 10.1007/s12275-022-1641-y
Li, Y., Zhao, L., Hou, M., Gao, T., Sun, J., Luo, H., et al. (2022). Lactobacillus casei improve anti-tuberculosis drugs-induced intestinal adverse reactions in rat by modulating gut microbiota and short-chain fatty acids. Nutrients 14:1668. doi: 10.3390/nu14081668
Li, W., Zhu, Y., Liao, Q., Wang, Z., and Wan, C. (2019). Characterization of gut microbiota in children with pulmonary tuberculosis. BMC Pediatr. 19:445. doi: 10.1186/s12887-019-1782-2
Lim, J. H., Kim, N., Lim, S. H., Kwon, J. W., Shin, C. M., Chang, Y. S., et al. (2016). Inverse relationship between Helicobacter Pylori infection and asthma among adults younger than 40 years: a cross-sectional study. Medicine (Baltimore) 95:e2609. doi: 10.1097/md.0000000000002609
Lin, S., Zhao, S., Liu, J., Zhang, J., Zhang, C., Hao, H., et al. (2020). Efficacy of proprietary Lactobacillus casei for anti-tuberculosis associated gastrointestinal adverse reactions in adult patients: a randomized, open-label, dose-response trial. Food Funct. 11, 370–377. doi: 10.1039/c9fo01583c
Liu, Q., Tian, X., Maruyama, D., Arjomandi, M., and Prakash, A. (2021). Lung immune tone via gut-lung axis: gut-derived LPS and short-chain fatty acids’ immunometabolic regulation of lung IL-1beta, FFAR2, and FFAR3 expression. Am. J. Physiol. Lung Cell. Mol. Physiol. 321, L65–L78. doi: 10.1152/ajplung.00421.2020
Liu, Y., Wang, J., and Wu, C. (2021). Microbiota and tuberculosis: a potential role of probiotics, and Postbiotics. Front. Nutr. 8:626254. doi: 10.3389/fnut.2021.626254
Luo, M., Liu, Y., Wu, P., Luo, D. X., Sun, Q., Zheng, H., et al. (2017). Alternation of gut microbiota in patients with pulmonary tuberculosis. Front. Physiol. 8:822. doi: 10.3389/fphys.2017.00822
Maji, A., Misra, R., Dhakan, D. B., Gupta, V., Mahato, N. K., Saxena, R., et al. (2018). Gut microbiome contributes to impairment of immunity in pulmonary tuberculosis patients by alteration of butyrate and propionate producers. Environ. Microbiol. 20, 402–419. doi: 10.1111/1462-2920.14015
Majlessi, L., Sayes, F., Bureau, J. F., Pawlik, A., Michel, V., Jouvion, G., et al. (2017). Colonization with Helicobacter is concomitant with modified gut microbiota and drastic failure of the immune control of Mycobacterium tuberculosis. Mucosal Immunol. 10, 1178–1189. doi: 10.1038/mi.2016.140
McShane, H. (2019). Insights and challenges in tuberculosis vaccine development. Lancet Respir. Med. 7, 810–819. doi: 10.1016/S2213-2600(19)30274-7
Méndez, C. S., Bueno, S. M., and Kalergis, A. M. (2021). Contribution of gut microbiota to immune tolerance in infants. J Immunol Res 2021, 7823316–7823311. doi: 10.1155/2021/7823316
Mitchell, H. M., Li, Y. Y., Hu, P. J., Liu, Q., Chen, M., du, G. G., et al. (1992). Epidemiology of Helicobacter pylori in southern China: identification of early childhood as the critical period for acquisition. J. Infect. Dis. 166, 149–153. doi: 10.1093/infdis/166.1.149
Molina-Infante, J., Gutierrez-Junquera, C., Savarino, E., Penagini, R., Modolell, I., Bartolo, O., et al. (2018). Helicobacter pylori infection does not protect against eosinophilic esophagitis: results from a large multicenter case-control study. Am. J. Gastroenterol. 113, 972–979. doi: 10.1038/s41395-018-0035-6
Mori, G., Morrison, M., and Blumenthal, A. (2021). Microbiome-immune interactions in tuberculosis. PLoS Pathog. 17:e1009377. doi: 10.1371/journal.ppat.1009377
Naidoo, C. C., Nyawo, G. R., Sulaiman, I., Wu, B. G., Turner, C. T., Bu, K., et al. (2021). Anaerobe-enriched gut microbiota predicts pro-inflammatory responses in pulmonary tuberculosis. EBioMedicine 67:103374. doi: 10.1016/j.ebiom.2021.103374
Naidoo, C. C., Nyawo, G. R., Wu, B. G., Walzl, G., Warren, R. M., Segal, L. N., et al. (2019). The microbiome and tuberculosis: state of the art, potential applications, and defining the clinical research agenda. Lancet Respir. Med. 7, 892–906. doi: 10.1016/S2213-2600(18)30501-0
Namasivayam, S., Kauffman, K. D., McCulloch, J. A., et al. (2019). Correlation between disease severity and the intestinal microbiome in Mycobacterium tuberculosis-infected Rhesus macaques. MBio 10:19. doi: 10.1128/mBio.01018-19
Namasivayam, S., Maiga, M., Yuan, W., Thovarai, V., Costa, D. L., Mittereder, L. R., et al. (2017). Longitudinal profiling reveals a persistent intestinal dysbiosis triggered by conventional anti-tuberculosis therapy. Microbiome 5:71. doi: 10.1186/s40168-017-0286-2
Namasivayam, S., Sher, A., Glickman, M. S., and Wipperman, M. F. (2018). The microbiome and tuberculosis: early evidence for cross talk. MBio 9:18. doi: 10.1128/mBio.01420-18
Nastasi, C., Fredholm, S., Willerslev-Olsen, A., Hansen, M., Bonefeld, C. M., Geisler, C., et al. (2017). Butyrate and propionate inhibit antigen-specific CD8(+) T cell activation by suppressing IL-12 production by antigen-presenting cells. Sci. Rep. 7:14516. doi: 10.1038/s41598-017-15099-w
Negatu, D. A., Liu, J. J. J., Zimmerman, M., Kaya, F., Dartois, V., Aldrich, C. C., et al. (2018). Whole-cell screen of fragment library identifies gut microbiota metabolite Indole propionic acid as Antitubercular. Antimicrob. Agents Chemother. 62:17. doi: 10.1128/AAC.01571-17
Negatu, D. A., Yamada, Y., Xi, Y., Go, M. L., Zimmerman, M., Ganapathy, U., et al. (2019). Gut microbiota metabolite Indole propionic acid targets tryptophan biosynthesis in Mycobacterium tuberculosis. MBio 10:18. doi: 10.1128/mBio.02781-18
Osei Sekyere, J., Maningi, N. E., and Fourie, P. B. (2020). Mycobacterium tuberculosis, antimicrobials, immunity, and lung-gut microbiota crosstalk: current updates and emerging advances. Ann. N. Y. Acad. Sci. 1467, 21–47. doi: 10.1111/nyas.14300
Perry, S., Chang, A. H., Sanchez, L., Yang, S., Haggerty, T. D., and Parsonnet, J. (2013). The immune response to tuberculosis infection in the setting of Helicobacter pylori and helminth infections. Epidemiol. Infect. 141, 1232–1243. doi: 10.1017/S0950268812001823
Perry, S., de Jong, B. C., Solnick, J. V., Sanchez, M. L., Yang, S., Lin, P. L., et al. (2010). Infection with Helicobacter pylori is associated with protection against tuberculosis. PLoS One 5:e8804. doi: 10.1371/journal.pone.0008804
Philippou, N., Roussos, A., Tsimpoukas, F., Anastasakou, E., Mavrea, S., and Tsimogianni, A. (2003). Helicobacter pylori infection in patients with active pulmonarytuberculosis before the initiation of antituberculosis treatment. Ann. Gastroenterol. 16, 49–52.
Raftery, A. L., Tsantikos, E., Harris, N. L., and Hibbs, M. L. (2020). Links between inflammatory bowel disease and chronic obstructive pulmonary disease. Front. Immunol. 11:2144. doi: 10.3389/fimmu.2020.02144
Rahim, M. A., Seo, H., Kim, S., Tajdozian, H., Barman, I., Lee, Y., et al. (2022). In vitro anti-tuberculosis effect of probiotic Lacticaseibacillus rhamnosus PMC203 isolated from vaginal microbiota. Sci. Rep. 12:8290. doi: 10.1038/s41598-022-12413-z
Redford, P. S., Murray, P. J., and O’Garra, A. (2011). The role of IL-10 in immune regulation during M. tuberculosis infection. Mucosal Immunol. 4, 261–270. doi: 10.1038/mi.2011.7
Roduit, C., Frei, R., Ferstl, R., Loeliger, S., Westermann, P., Rhyner, C., et al. (2019). High levels of butyrate and propionate in early life are associated with protection against atopy. Allergy 74, 799–809. doi: 10.1111/all.13660
Sanaka, M., Kuyama, Y., Iwasaki, M., Hanada, Y., Tsuchiya, A., Haida, T., et al. (1998). No difference in seroprevalences of Helicobacter pylori infection between patients with pulmonary tuberculosis and those without. J. Clin. Gastroenterol. 27, 331–334. doi: 10.1097/00004836-199812000-00010
Sathkumara, H. D., Eaton, J. L., Field, M. A., Govan, B. L., Ketheesan, N., and Kupz, A. (2021). A murine model of tuberculosis/type 2 diabetes comorbidity for investigating the microbiome, metabolome and associated immune parameters. Anim. Model Exp. Med. 4, 181–188. doi: 10.1002/ame2.12159
Schirmer, M., Smeekens, S. P., Vlamakis, H., Jaeger, M., Oosting, M., Franzosa, E. A., et al. (2016). Linking the human gut microbiome to inflammatory cytokine production capacity. Cells 167:1897. doi: 10.1016/j.cell.2016.11.046
Schuijt, T. J., Lankelma, J. M., Scicluna, B. P., de Sousa e Melo, F., Roelofs, J. J. T. H., de Boer, J. D., et al. (2016). The gut microbiota plays a protective role in the host defence against pneumococcal pneumonia. Gut 65, 575–583. doi: 10.1136/gutjnl-2015-309728
Segal, L. N., Clemente, J. C., Li, Y., Ruan, C., Cao, J., Danckers, M., et al. (2017). Anaerobic bacterial fermentation products increase tuberculosis risk in antiretroviral-drug-treated HIV patients. Cell Host Microbe 21, 530–537.e4. doi: 10.1016/j.chom.2017.03.003
Shah, T., Shah, Z., Baloch, Z., and Cui, X. (2021). The role of microbiota in respiratory health and diseases, particularly in tuberculosis. Biomed. Pharmacother. 143:112108. doi: 10.1016/j.biopha.2021.112108
Stefan, K. L., Kim, M. V., Iwasaki, A., and Kasper, D. L. (2020). Commensal microbiota modulation of natural resistance to virus infection. Cells 183, 1312–1324.e10. doi: 10.1016/j.cell.2020.10.047
Stephanie, F., Saragih, M., and Tambunan, U. S. F. (2021). Recent Progress and challenges for drug-resistant tuberculosis treatment. Pharmaceutics 13:592. doi: 10.3390/pharmaceutics13050592
Suter, E., and Kirsanow, E. M. (1962). Fate of attenuated tubercle bacilli (BCG) in germ-free and conventional mice. Nature 195, 397–398. doi: 10.1038/195397b0
Tan, T. G., Sefik, E., Geva-Zatorsky, N., Kua, L., Naskar, D., Teng, F., et al. (2016). Identifying species of symbiont bacteria from the human gut that, alone, can induce intestinal Th17 cells in mice. Proc. Natl. Acad. Sci. U. S. A. 113, E8141–E8150. doi: 10.1073/pnas.1617460113
Tiberi, S., Utjesanovic, N., Galvin, J., Centis, R., D’Ambrosio, L., van den Boom, M., et al. (2022). Drug resistant TB – latest developments in epidemiology, diagnostics and management. Int. J. Infect. Dis. 124, S20–S25. doi: 10.1016/j.ijid.2022.03.026
Torres, M. A., Passaro, D. J., Watanabe, J., Parsonnet, J., Small, P., Miyagu, J., et al. (2003). No association between Helicobacter pylori and Mycobacterium tuberculosis infections among gastrointestinal clinic attendees in Lima. Peru. Epidemiol Infect. 130, 87–91. doi: 10.1017/s0950268802007653
Tsang, K. W., Lam, S. K. W. K., Lam, J., Karlberg, B. C, Wong, W. H., Hu,, et al. (1998). High seroprevalence of Helicobacter pylori in active bronchiectasis. Am. J. Respir. Crit. Care Med. 158, 1047–1051. doi: 10.1164/ajrccm.158.4.9712104
Tsigalou, C., Konstantinidis, T. G., Cassimos, D., Karvelas, A., Grapsa, A., Tsalkidis, A., et al. (2019). Inverse association between Helicobacter pylori infection and childhood asthma in Greece: a case-control study. Germs 9, 182–187. doi: 10.18683/germs.2019.1174
Van Winkle, J. A., Peterson, S. T., Kennedy, E. A., Wheadon, M. J., Ingle, H., Desai, C., et al. (2022). Homeostatic interferon-lambda response to bacterial microbiota stimulates preemptive antiviral defense within discrete pockets of intestinal epithelium. elife 11:74072. doi: 10.7554/eLife.74072
Vandenplas, Y., Carnielli, V. P., Ksiazyk, J., Luna, M. S., Migacheva, N., Mosselmans, J. M., et al. (2020). Factors affecting early-life intestinal microbiota development. Nutrition 78:110812. doi: 10.1016/j.nut.2020.110812
Wang, Y., Bi, Y., Zhang, L., and Wang, C. (2012). Is Helicobacter pylori infection associated with asthma risk? A meta-analysis based on 770 cases and 785 controls. Int. J. Med. Sci. 9, 603–610. doi: 10.7150/ijms.4970
Wang, Y., Deng, Y., Liu, N., Chen, Y., Jiang, Y., Teng, Z., et al. (2022). Alterations in the gut microbiome of individuals with tuberculosis of different disease states. Front. Cell. Infect. Microbiol. 12:836987. doi: 10.3389/fcimb.2022.836987
Wang, Y. C., Lin, T. Y., Shang, S. T., Chen, H. J., Kao, C. H., Wu, C. C., et al. (2017). Helicobacter pylori infection increases the risk of adult-onset asthma: a nationwide cohort study. Eur. J. Clin. Microbiol. Infect. Dis. 36, 1587–1594. doi: 10.1007/s10096-017-2972-1
Wang, S., Yang, L., Hu, H., Lv, L., Ji, Z., Zhao, Y., et al. (2022). Characteristic gut microbiota and metabolic changes in patients with pulmonary tuberculosis. Microb. Biotechnol. 15, 262–275. doi: 10.1111/1751-7915.13761
Wang, Q., Yu, C., and Sun, Y. (2013). The association between asthma and Helicobacter pylori: a meta-analysis. Helicobacter 18, 41–53. doi: 10.1111/hel.12012
Wernroth, M. L., Peura, S., Hedman, A. M., Hetty, S., Vicenzi, S., Kennedy, B., et al. (2022). Development of gut microbiota during the first 2 years of life. Sci. Rep. 12:9080. doi: 10.1038/s41598-022-13009-3
Wingfield, T., Karmadwala, F., MacPherson, P., Millington, K. A., Walker, N. F., Cuevas, L. E., et al. (2021). Challenges and opportunities to end tuberculosis in the COVID-19 era. Lancet Respir. Med. 9, 556–558. doi: 10.1016/S2213-2600(21)00161-2
Winglee, K., Eloe-Fadrosh, E., Gupta, S., Guo, H., Fraser, C., and Bishai, W. (2014). Aerosol Mycobacterium tuberculosis infection causes rapid loss of diversity in gut microbiota. PLoS One 9:e97048. doi: 10.1371/journal.pone.0097048
Wipperman, M. F., Fitzgerald, D. W., Juste, M. A. J., Taur, Y., Namasivayam, S., Sher, A., et al. (2017). Antibiotic treatment for tuberculosis induces a profound dysbiosis of the microbiome that persists long after therapy is completed. Sci. Rep. 7:10767. doi: 10.1038/s41598-017-10346-6
Wirusanti, N. I., Baldridge, M. T., and Harris, V. C. (2022). Microbiota regulation of viral infections through interferon signaling. Trends Microbiol. 30, 778–792. doi: 10.1016/j.tim.2022.01.007
Woeltje, K. F., Kilo, C. M., Johnson, K., Primack, J., and Fraser, V. J. (1997). Tuberculin skin testing of hospitalized patients. Infect. Control Hosp. Epidemiol. 18, 561–565. doi: 10.1086/647672
World Health Organization. Global tuberculosis report 2022. Geneva: World Health Organization. (2022).
World Health Organization. (2023) Tuberculosis: Key facts. Available at: https://www.who.int/news-room/fact-sheets/detail/tuberculosis (Accessed December 21, 2022)
Wu, H. J., and Wu, E. (2012). The role of gut microbiota in immune homeostasis and autoimmunity. Gut Microbes 3, 4–14. doi: 10.4161/gmic.19320
Yang, F., Yang, Y., Chen, L., Zhang, Z., Liu, L., Zhang, C., et al. (2022). The gut microbiota mediates protective immunity against tuberculosis via modulation of lnc RNA. Gut Microbes 14:2029997. doi: 10.1080/19490976.2022.2029997
Zheng, D., Liwinski, T., and Elinav, E. (2020). Interaction between microbiota and immunity in health and disease. Cell Res. 30, 492–506. doi: 10.1038/s41422-020-0332-7
Keywords: tuberculosis, microbiome, gut-lung-axis, immunity, dysbiosis, probiotics
Citation: Enjeti A, Sathkumara HD and Kupz A (2023) Impact of the gut-lung axis on tuberculosis susceptibility and progression. Front. Microbiol. 14:1209932. doi: 10.3389/fmicb.2023.1209932
Edited by:
Farzam Vaziri, University of California, Davis, United StatesReviewed by:
Aude Remot, Institut National de recherche pour l’agriculture, l’alimentation et l’environnement (INRAE), FranceAbbas Yadegar, Shahid Beheshti University of Medical Sciences, Iran
Copyright © 2023 Enjeti, Sathkumara and Kupz. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andreas Kupz, YW5kcmVhcy5rdXB6QGpjdS5lZHUuYXU=
 Aditya Enjeti1
Aditya Enjeti1 Harindra Darshana Sathkumara
Harindra Darshana Sathkumara Andreas Kupz
Andreas Kupz