
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 03 August 2023
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1208314
This article is part of the Research Topic Antimicrobial Resistance in Zoonotic Bacteria View all 12 articles
The advent of antimicrobials-resistant (AMR), including colistin-resistant bacteria, poses a significant challenge to animal and human health, food safety, socio-economic growth, and the global environment. This study aimed to ascertain the colistin resistance prevalence and molecular mechanisms of colistin resistance in Enterobacteriaceae. The colistin resistance was determined using broth microdilution assay, PCR; and Sanger sequencing of mcr genes responsible for colistin resistance in Enterobacteriaceae (n = 627), including Escherichia coli (436), Salmonella spp. (n = 140), and Klebsiella pneumoniae (n = 51), obtained from chicken and chicken meats. Out of 627 Enterobacteriaceae, 8.6% of isolates exhibited colistin resistance phenotypically. Among these colistin resistant isolates, 9.3% (n = 37) were isolated from chicken meat, 7.2% (n = 11) from the cloacal swab of chicken and 7.9% (n = 6) from the litter samples. Overall, 12.96% of colistin-resistant isolates were positive with mcr genes, in which mcr-1 and mcr-5 genes were determined in 11.11% and 1.85% of colistin-resistant isolates, respectively. The E. coli isolates obtained from chicken meats, cloacal swabs and litter samples were found positive for mcr-1, and Salmonella spp. originated from the chicken meat sample was observed with mcr-5, whereas no mcr genes were observed in K. pneumoniae strains isolated from any of the collected samples. The other colistin resistance genes, including mcr-2, mcr-3, mcr-4, mcr-6, mcr-7, mcr-8, mcr-9, and mcr-10 were not detected in the studied samples. The mcr-1 and mcr-5 genes were sequenced and found to be 100% identical to the mcr-1 and mcr-5 gene sequences available in the NCBI database. This is the first report of colistin resistance mcr-5 gene in Malaysia which could portend the emergence of mcr-5 harboring bacterial strains for infection. Further studies are needed to characterize the mr-5 harbouring bacteria for the determination of plasmid associated with mcr-5 gene.
Antimicrobial agents are essential medicines in animals and humans to curb infections. Owing to the overuse and abuse of antimicrobial agents, the globe is being faced with the rapid proliferation of resistant microbes. Currently, the advent of antimicrobial-resistant (AMR) bacteria poses a significant challenge to animal and human health, food safety, socio-economic growth, and the global environment (Theuretzbacher, 2017). The proliferation of Gram-negative bacterial strains which are resistant to multiple drugs, and the absence of new drugs to combat such microbes, reintroduced colistin as a last-line therapy (Ezadi et al., 2019).
The resistance to colistin is a crucial problem to be tackled today. Numerous studies have demonstrated that this colistin resistance was present in various bacterial strains around the world. The plasmid-mediated mcr genes are accountable for exceptional colistin resistance, as it is a conduit that spreads via horizontal transmission from one bacterial strain to another and through food chain or direct contact to humans, animals, and the environments (Gharaibeh and Shatnawi, 2019). Before 2015, all documented colistin-resistance was chromosomally regulated, involving modification of a two-component regulatory structure, phoPQ and pmrAB with the negative regulator, mgrB gene alteration (Cannatelli et al., 2013; Liu et al., 2016). In 2016, the Escherichia coli strains isolated from humans, retail chicken meat and pork, and Klebsiella pneumoniae strains isolated from humans were reported with the plasmid-encoded mcr-1 gene in China (Liu et al., 2016). The plasmid-mediated new mcr genes have rapidly emerged. The therapeutic effectiveness of colistin has been compromised by the advent of the plasmid-encoded mcr genes, including mcr-1 to mcr-10, which were reported during the last four years (Ling et al., 2020). The E. coli isolates recovered from chicken liver, and chicken feed in the trough (Yu et al., 2016), and poultry meat in Malaysia were found positive for mcr-1 gene (Aklilu and Raman, 2020).
Poultry meat is an important source of protein for humans, it could also be a significant conduit for spreading multidrug-resistant bacterial species from food-producing animals to humans. Previous study has shown that retail chicken meat plays a role in disseminating multiple antibiotic-resistant strains among humans and their environment, posing a severe threat to environmental health and food safety (Aidara-Kane et al., 2013). The colistin resistance-producing gene, mcr-1, was present in 52.1 percent of the E. coli isolates from raw chicken meat (Aklilu and Raman, 2020). Colistin resistance was found in more in 36.4% of bacteria from poultry chicken and 20% of strains isolated from native chicken in Bangladesh (Islam et al., 2020). In Nepal, it was reported that 27 (22.8%) of colistin-resistant E. coli in broiler farms carried the mcr-1 gene (Joshi et al., 2019). Aside from that, the horizontal transmission is thought to be the main mechanism for the spread of colistin resistance mcr genes in Enterobacteriaceae worldwide (Gharaibeh and Shatnawi, 2019).
With potent in vitro transfer rates and frequently harboured alongside other resistance determinants like β -lactamases, mcr genes have been identified on a variety of conjugative plasmids (IncI2, IncHI2, IncX4, and pHNSHP45) (EMA (European Medicine Agencey), 2016). As the mobilized colistin resistance (mcr) genes driven by plasmids are quickly emerging, appropriate information on colistin resistance, including the incidence and epidemiological studies of mcr-positive cases, is required to apply steps to prevent and manage its dissemination. Therefore, the aim of the study was to determine the prevalence and molecular determinants underlying colistin resistance in Enterobacteriaceae (E. coli, Salmonella spp., and Klebsiella pneumoniae) isolates recovered from poultry and poultry meats in Malaysia.
The ethical board of Universiti Putra Malaysia (UPM), Institutional Animal Care and Use Committee (IACUC) approved the research study protocol for collecting cloacal swabs from live poultry (UPM/IACUC/AUP-R091/2019).
The research study was performed in which chicken meat samples from supermarkets and cloacal swabs, and litter samples from chicken farms within Selangor, Malaysia (Figure 1) were collected from July 2019 to February 2021. Selangor is the densely populated area in Malaysia and most of the poultry farms are located in this state. Sterile plastic bags were used to collect the meat samples. The cloacal swab samples were collected aseptically from the healthy chicken and kept in sterile transport media, Stuart media. Litter samples were collected from the farms’ floors using a sterile spoon, and placed in a sterile plastic bag. In total, 543 samples, including 350 chicken meats, 144 cloacal swabs, and 49 litter samples (Table 1), were collected from supermarkets and poultry farms in different areas of Selangor in Malaysia (Figure 1). All collected samples were immediately transported in a sealed icebox to the Bacteriology Laboratory, Faculty of Veterinary Medicine at Universiti Putra Malaysia (UPM), Serdang, Selangor, Malaysia.
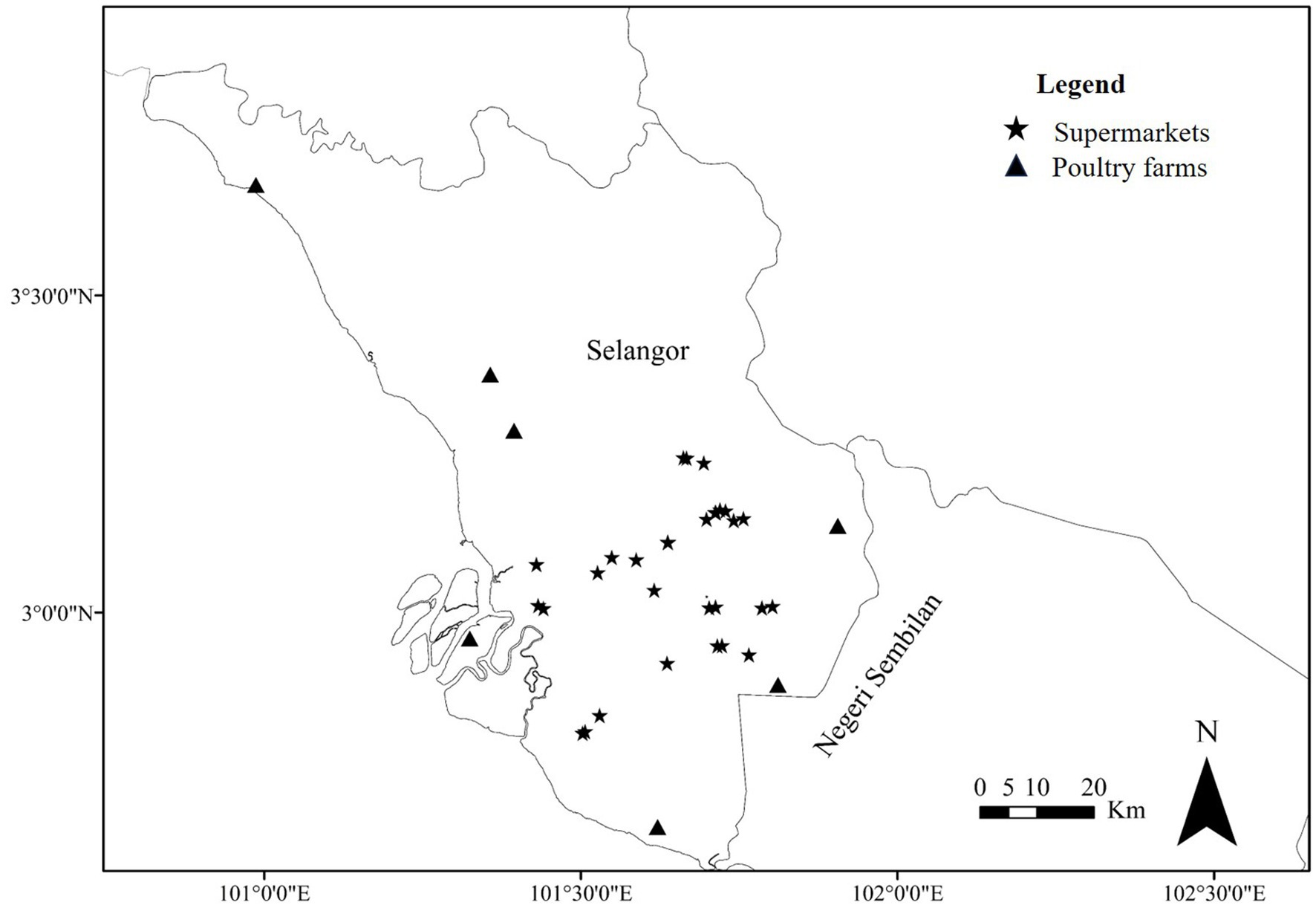
Figure 1. Map showing the sampling areas of Selangor, Malaysia. Star (*) and triangle (▲) marks indicating supermarkets and poultry farms, respectively. The geographic information software, ArcGIS v.10.4.1 for Windows was used to generate the sampling spot on the map.
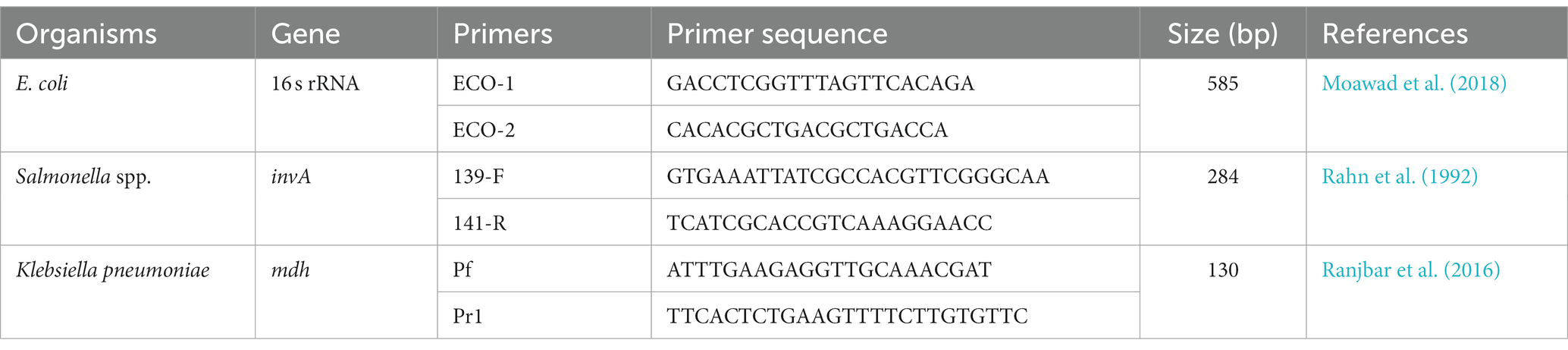
Table 1. List of the primers and sequences for the confirmation of E. coli, Salmonella spp., and Klebsiella pneumoniae.
In the Enterobacteriaceae family, Salmonella spp., E. coli, and K. pneumoniae have public health importance and are more prevalent with colistin resistance mcr genes (Elbediwi et al., 2019). For this reason, Salmonella spp., E. coli, and K. pneumoniae, were isolated and identified phenotypically using the standard protocol of traditional cultural and biochemical tests (Ghafur et al., 2019; Sharma et al., 2019). In the pre-enrichment step, the samples were cultured in buffered peptone water (BPW, Oxoid, United Kingdom), including 25 g of meat in 225 mL, 10 g litter in 90 mL, and cloacal swabs in 10 mL BPW medium, samples were incubated at 37°C for 24 h. The meat samples with BPW were homogenized for 2 min in a stomacher. For Salmonella spp. isolation, 100 μL of homogenized BPW was mixed with 10 mL of Rappaport-Vassiliadis (RVS; Oxoid, United Kingdom). Then the RVS mixtures were incubated at 42°C for 24 h. One loop-full RVS of each sample was sub-cultured onto Xylose Lysine Deoxycholate agar (XLD; Oxoid, United Kingdom) and kept at 37°C for 24 h for incubation. On XLD, typically, Salmonella colonies were red with a black centre. One pure colony from XLD was cultured onto nutrient agar (NA, Oxoid, United Kingdom). For presumptive identification, biochemical tests (TSI, Urease, Citrate, and SIM) were performed with pure cultures grown onto NA. Serological confirmation of Salmonella spp. was performed using a slide agglutination test using Poly ‘O’ and ‘H’ antisera (Remel, United Kingdom). For E. coli and K. pneumoniae isolation, a loopful of suspension of BPW was inoculated onto MacConkey (Oxoid, United Kingdom). One presumptive colony of E. coli and K. pneumoniae were then sub-cultured onto NA and subjected to a standard biochemical test for their presumptive confirmation.
The genomic DNA was extracted from the pure culture of the isolates cultured on nutrient agar using the boiling and snap chill method (Pui et al., 2011). The phenotypically positive E. coli, Salmonella spp., and Klebsiella pneumoniae species were then confirmed with conventional PCR with species-specific gene primers (Table 1), in which positive controls included E. coli ATCC 25922, Salmonella ATCC 14028 and Klebsiella pneumoniae ATCC 700603 (Rahn et al., 1992; Ranjbar et al., 2016; Moawad et al., 2018).
Colistin-susceptibility and MIC in isolates were assessed by the ISO-20776 standard broth microdilution technique (BMD) jointly recommended by the EUCAST (2016a) with few modifications. In brief, a two-fold dilution (0.125–128 μg/mL) of the colistin sulfate salt (Sigma-Aldrich) was prepared on a 96 well microtitre plate. The bacterial inoculum was inoculated in each well with a final concentration of 5 × 105 CFU/mL. Then the microtitre plate was incubated at 37°C for 16–20 h. After the incubation period, 30 μL of 0.015% resazurin solution was added to each well of the microtitre plate and incubated again at 37C for 1 h, during which the plate was routinely checked every 15 min. The colistin MICs were interpreted with the naked eye and recorded by observing the color change of resazurin (discoloration from blue to pink or purple indicates colistin resistance, while susceptibility is deduced when no color change- blue color). The test was performed in triplicates. During colistin-susceptibility testing, colistin-susceptible E. coli ATCC25922 and colistin-resistant (ColR) E. coli NCTC 13846 were utilized as negative and positive controls, respectively.
Colistin-resistant Enterobacteriaceae isolates were identified with MIC values greater than 2 μg/mL colistin (EUCAST, 2016b). The genomic DNA of the colistin-resistant (col-R) isolates was assessed with conventional PCR to detect colistin resistance (mcr) gene variants (mcr-1 to mcr-10). According to previous studies, the detection of mcr-1 to mcr-5 (Rebelo et al., 2018) and mcr-6 to mcr-9 (Borowiak et al., 2020) was performed with multiplex PCR. The uniplex PCR was performed for mcr-10 (Xu et al., 2021) by the previously designed oligonucleotide primers and the protocol (Table 2).
Amplified PCR products were examined on 1.5% agarose (Conda, Madrid, Spain) prepared in 100 mL of 0.5 × TBE Buffer stained with 4 μL of Nucleic Acid stain (ETB “out” Nucleic Acid, Cat. No. FYD007-200P, Yestern Biotech Co. ltd, Taiwan) for mcr gene. The expected bands for mcr-1 to mcr-10 (Table 2) were visualized and photographed under UV light using AlphaImager 2,200 (AlphaImager, United States).
The amplified PCR products for the mcr-1 gene of E. coli strains, E. coli E172, and mcr-5 gene from one Salmonella spp. strain S283 were sent to commercial company for DNA sequencing with the same primers for Sanger sequencing (Table 2) and compared to previously reported mcr-1 and mcr-5 sequences in the NCBI database. The consensus sequences of mcr-1 and mcr-5 were obtained based on the alignment of the forward and reverse sequences using BioEdit v. 7.2 program (Hall, 1999).
The mcr gene sequences were used in the phylogenetic tree construction, and analysis was carried out according to Li et al. (2019). The sequences from this study and those from GenBank for the mcr-1 and mcr-5 were aligned separately using the MEGA X software (Kumar et al., 2018) to compare their similarities. There are four different types of mcr-5 gene variants such as mcr-5.1, mcr-5.2, mcr-5.3 and mcr-5.4 has been identified in the world. We have selected partial sequences of these four mcr-5 variants from GenBank in the NCBI database and analysed with our mcr-5 sequence data. The phylogenetic tree for mcr-1 and mcr-5 was constructed with aligned sequences by the neighbor-joining method using the Kimura 2-parameter model, and Bootstrap values were calculated using 500 replicates.
Microsoft Excel sheets (MS-2019) were used to input data, which were then uploaded into the SPSS program v. 25.0 (IBM, Armonk, NY, United States). Descriptive analysis was used to quantify the prevalence, in which level of significance was assessed using the χ2 test. For statistical significance, p- values less than 0.05 (p < 0.05) was taken into consideration.
In total, 627 Enterobacteriaceae (E. coli, Salmonella spp., and K. pneumoniae) isolates, including 398 isolates from meat samples from supermarkets, 153 isolates from cloacal swabs, and 76 isolates from litter samples from poultry farms, were isolated and identified (Table 3). The 398 isolates from meat samples from supermarkets were classified as E. coli (n = 258, 73.7%%), Salmonella spp. (n = 122, 34.9%) and K. pneumoniae (n = 18, 5.1%). The poultry cloacal swabs from poultry farms yielded E. coli (n = 134, 93.1%), Salmonella spp. (n = 4, 2.8%) and K. pneumoniae (n = 15, 10.4%), and litter samples from poultry farms generated E. coli (n = 44, 89.8%), Salmonella spp. (n = 14, 28.6%) and K. pneumoniae (n = 18, 36.7%). Salmonella spp. was highly prevalent in collected chicken meat samples (p = 0.000), E. coli in cloacal swab (p = 0.000), and K. pneumoniae was in litter samples (p = 0.000) (Table 3). E. coli, Salmonella spp., and K. pneumoniae isolates were confirmed by PCR showing a band size of 585 bp, 284 bp, and 130 bp, respectively.
Out of 627 Enterobacteriaceae isolates, 8.6% (n = 54) of isolates exhibited colistin resistance using the broth microdilution assay. Among these, 9.3% (n = 37) were isolated from chicken meat, 7.2% (n = 11) from the cloacal swab of chicken and 7.9% (n = 6) from the litter samples (Table 4). Overall, the phenotypic colistin resistance of the isolates from chicken meat, cloacal swabs, and litter samples was indifferent (Tables 4, p = 0.712). On the other hand, 54 colistin-resistant Enterobacteriaceae isolates were comprised of E. coli (n = 32, 7.34%), Salmonella spp. (n = 16, 11.4%), and K. pneumoniae (n = 6, 11.76%) (Figure 2).
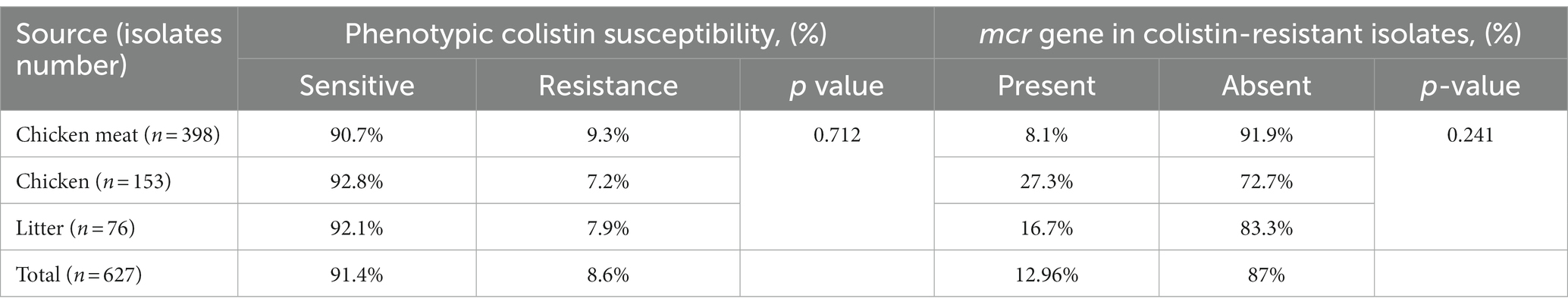
Table 4. Phenotypic colistin resistance and mechanism of colistin resistance with mcr genes in isolates from chicken meat, chicken and litter.
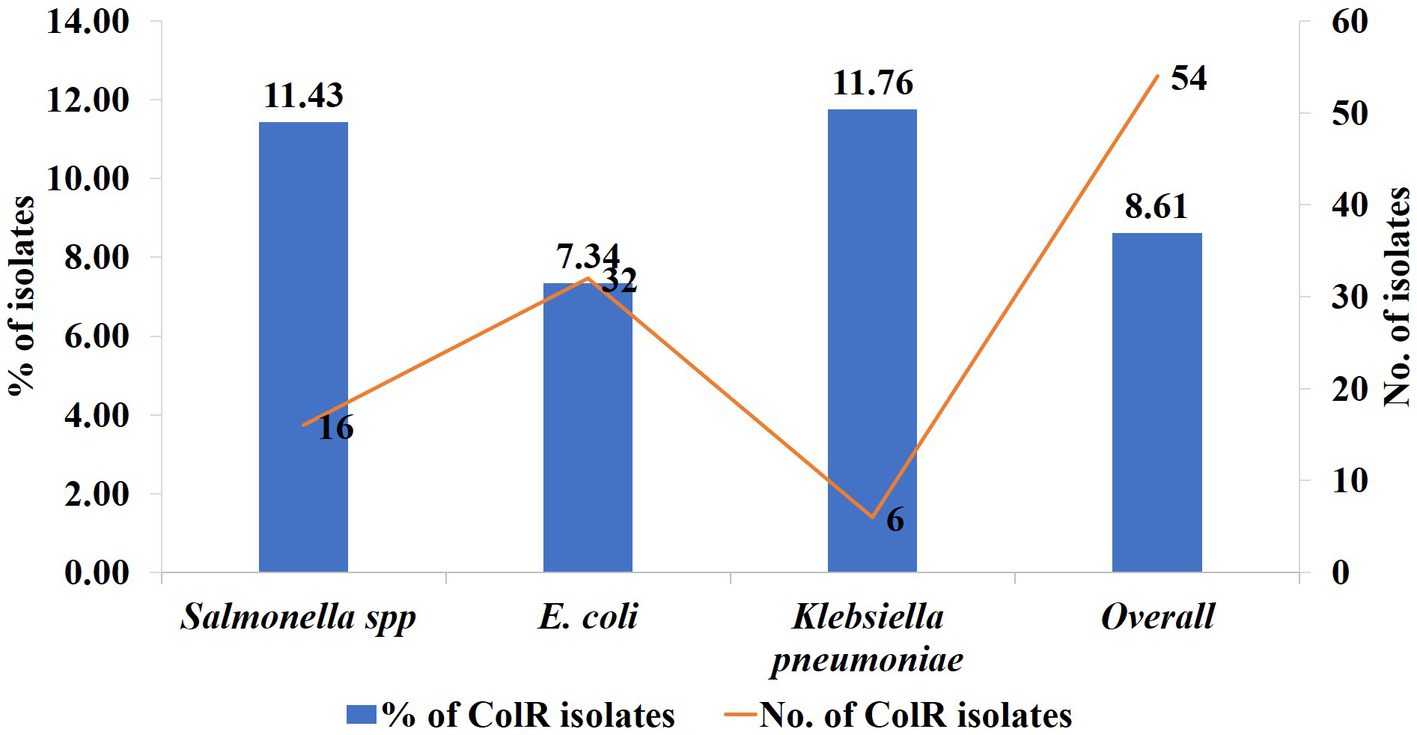
Figure 2. The rate trend and number of phenotypically colistin-resistant Enterobacteriaceae isolates, E. coli, Salmonella spp., and K. pneumoniae. The blue column = rate trend and red line = number of isolates, left y axis = % of colR isolates, right y axis = number of colR isolates.
All 54 isolated colistin-resistant (Col-R) Enterobacteriaceae were analyzed to observe the presence of mcr-1 to mcr-10. Overall, 12.96% (n = 7) of colistin-resistant Enterobacteriaceae isolates were found possessing colistin resistance mcr genes comprising 8.1% (n = 3), 27.3% (n = 3), and 16.67% (n = 1) of Col-R isolates from the chicken meat, chicken and litter samples, respectively. Variations of colistin resistance from different sources were not statistically significant (value of p > 0.05) as shown in Table 4. Out of seven mcr harboring Col-R isolates, 11.11% (n = 6) and 1.85% (n = 1) were found with mcr-1 and mcr-5, respectively. The E. coli isolates obtained from chicken meats, cloacal swabs and litter samples were found positive for mcr-1, and Salmonella spp. originated from the chicken meat sample was observed with mcr-5, whereas no mcr genes were observed in K. pneumoniae strains isolated from any of the collected samples (Figure 3). The other colistin resistance genes, including mcr-2, mcr-3, mcr-4, mcr-6, mcr-7, mcr-8, mcr-9, and mcr-10 were not detected in the studied samples.
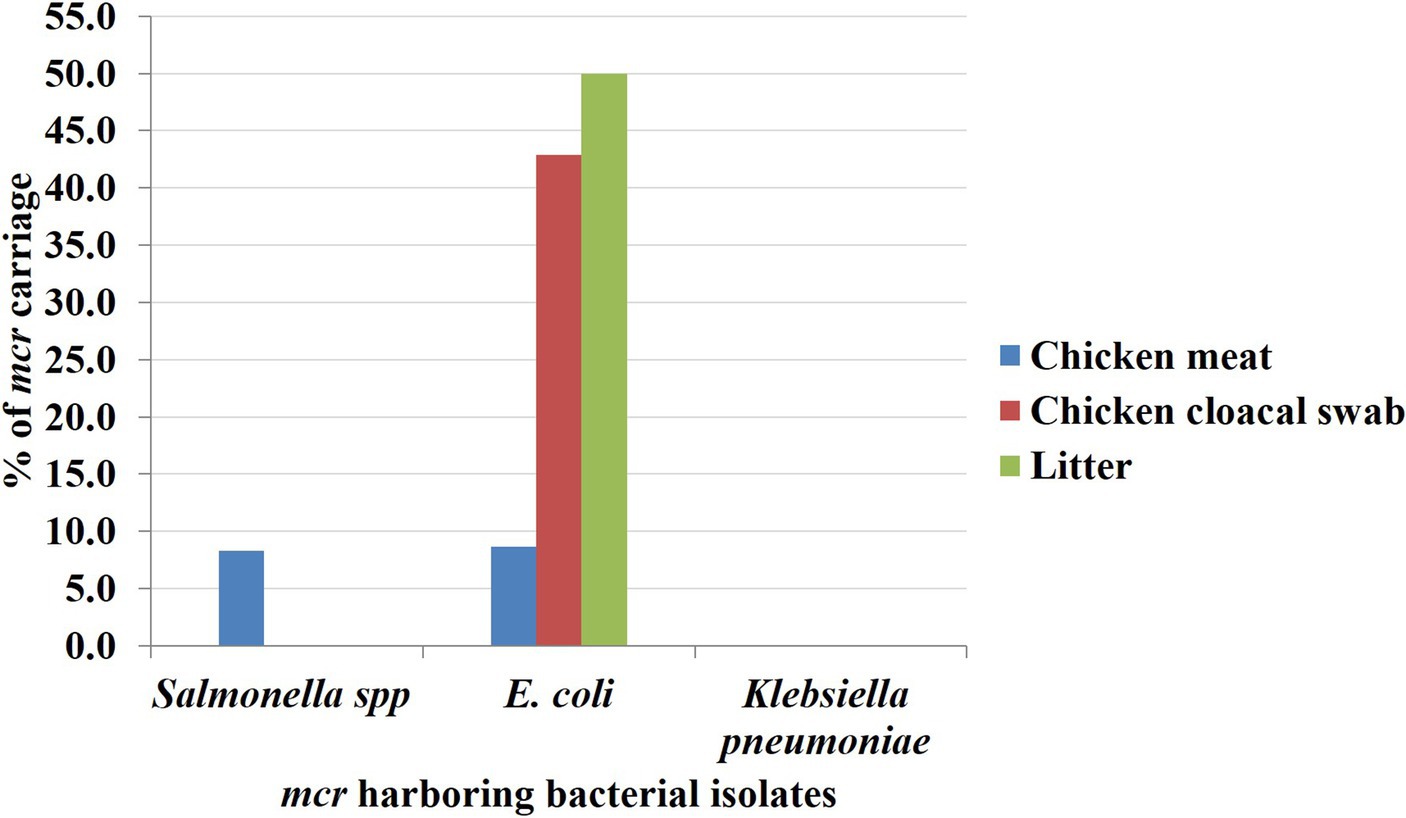
Figure 3. The rate of Enterobacteriaceae isolates harboring mcr gene based on sample sources. The rate of mcr genes was marked by blue, red and green bars to the Y-axis for chicken meat, chicken cloacal swab and litter samples, respectively.
In BLAST analysis, our studied ¬mcr-1 sequence was found to be 100% identical to the mcr-1 sequence (Genbank: NG_050417.1) in the NCBI database with 99% query coverage. On the other hand, our studied ¬mcr-5 sequence was found to be 100% identical to the mcr-5 sequence (Genbank: NG_055658) in the NCBI database with 98% query coverage.
The broth microdilution test was conducted to assess the MIC value of all isolates following EUCAST guidelines, with the epidemiological cutoff value >2 μg/mL for colistin resistance. Control strains, colistin-resistant isolates Escherichia coli NTCC 13846 and susceptible isolate Escherichia coli ATCC 25922, showed growth up to 4 μg/mL and 0.5 μg/mL colistin concentration, respectively. The MIC value of isolated colistin-resistant bacteria exhibited 4 to 128 μg/mL of colistin. The mcr-carrying isolates were observed with MIC values of 4 and 8 μg/mL colistin (Figure 4). In contrast, mcr negative colistin-resistant isolates had extremely high MIC levels, 128 μg/mL colistin (Figure 4). Most of the Col-R isolates from three sources exhibited MIC values from 4 to 8 μg/mL of colistin (Supplementary Figure S1).
In the phylogeny analysis, it is observed that mcr-1 genes in the obtained isolates were divided into two clades, named clade A, including subclade I and subclade II, and clade B. The colistin resistance mcr-1 gene sequence from E. coli strains E48, E13, E278, E297, E331 recovered from cloacal swabs and chicken meat samples were clustered in subclade I. These isolates were closely related to subclade II, grouped with E. coli and Salmonella spp. strains recovered from humans and animals obtained from the NCBI GenBank database. The nucleotide sequence of the mcr-1 gene of E. coli E172 obtained from chicken litter samples was very close to the previously identified isolate in Malaysia and human isolate in China. On the other hand, mcr-1 sequences of E. coli isolates recovered from chicken meats, and chicken cloacal swab samples were grouped in one cluster and were found close to isolates from China and Brazil (Figure 5).
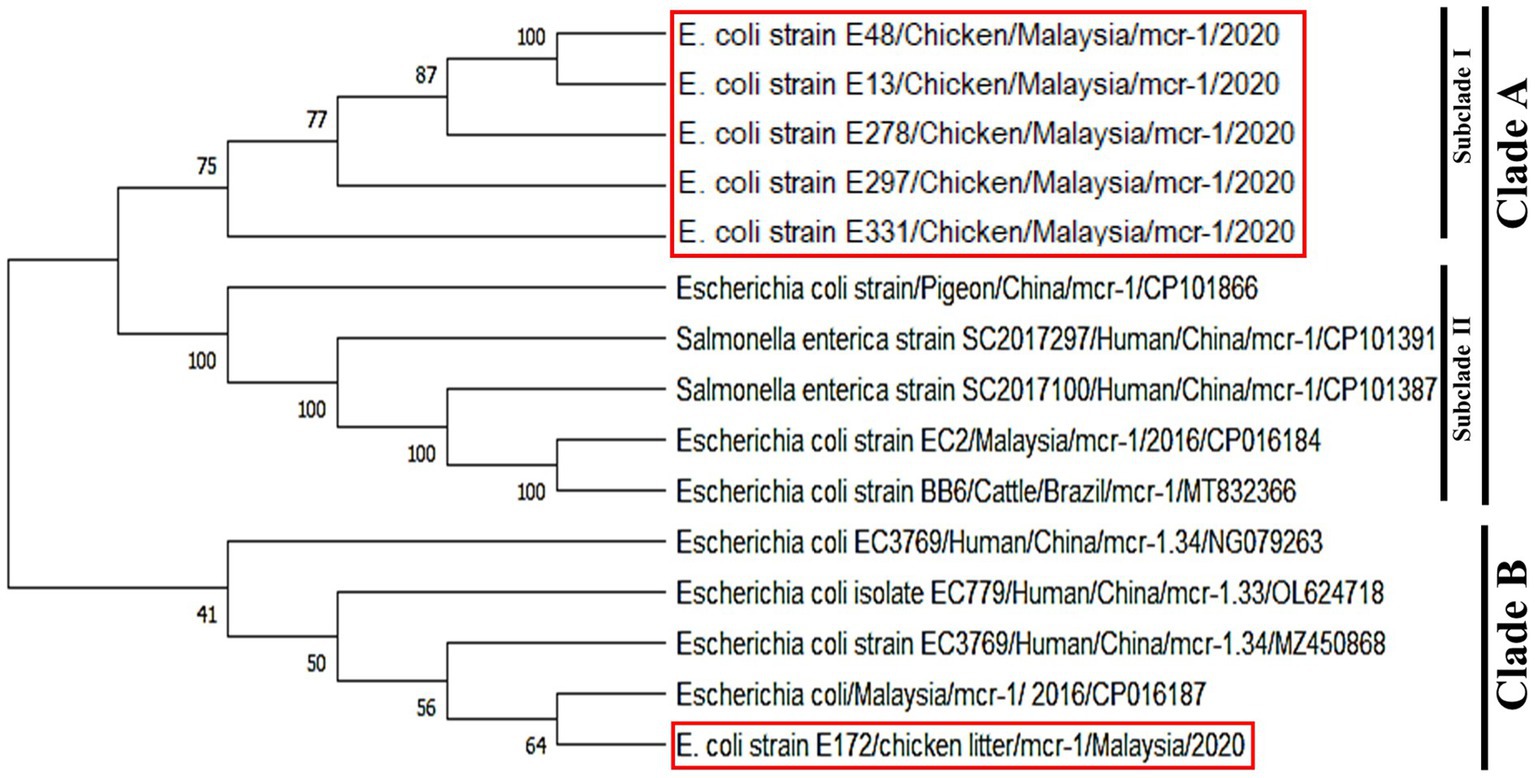
Figure 5. Phylogenetic tree of mcr-1 gene showing the relationship of the partial coding regions of mcr-1 gene sequences. The studied sequences were marked by red boxes. MEGA X was used to create the phylogenetic tree.
In the phylogeny analysis of the mcr-5 gene, it was revealed that the nucleotide sequence of the mcr-5 gene of Salmonella spp. obtained from chicken meat has a close relation to the mcr-5.3 gene in the E. coli isolate obtained from the horse in Brazil (Figure 6).
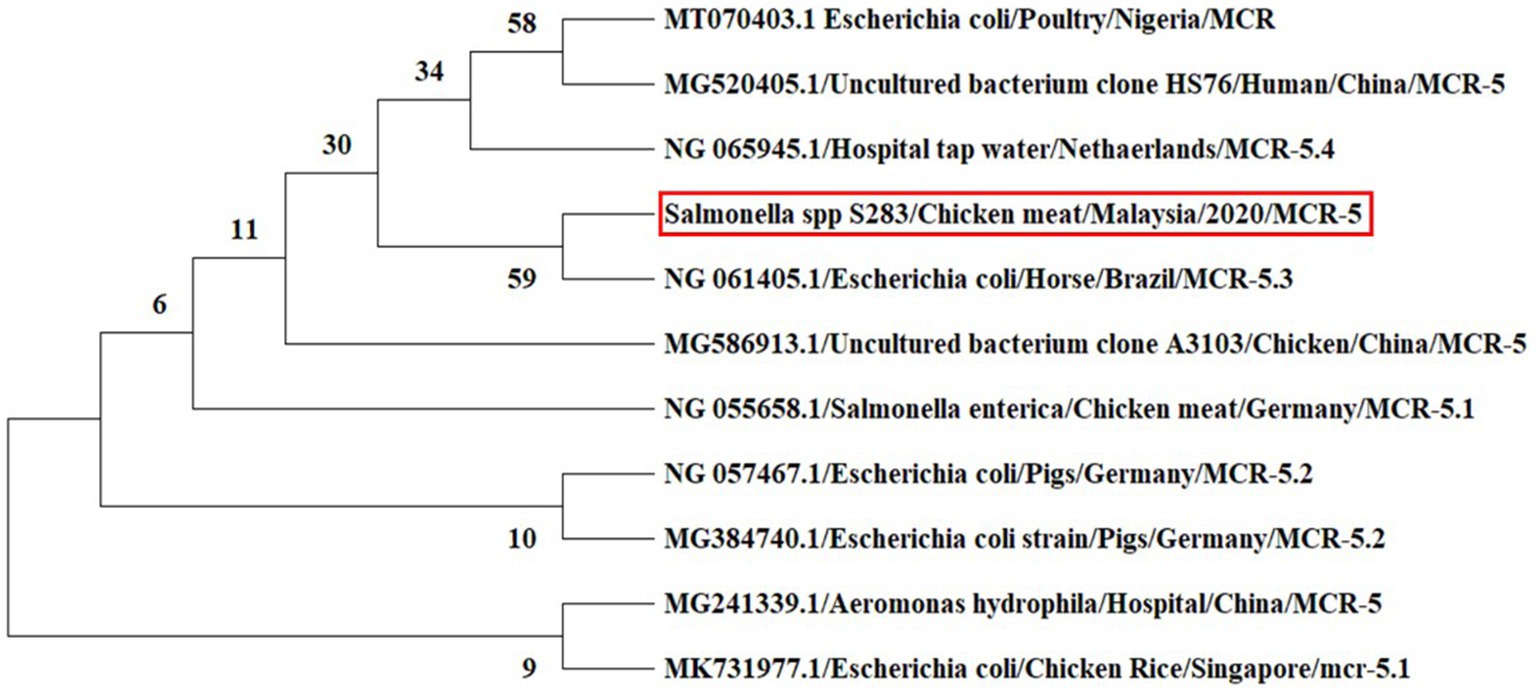
Figure 6. Phylogenetic tree inferred from the nucleotide sequences of mcr-5 gene using the maximum likelihood method for distance calculations using MEGA X software. Bootstrap percentages retrieved in 500 replications are shown at the nodes. The studied sequence is marked by the red box.
The most common bacteria associated with bacterial infections in chickens are E. coli, Salmonella spp., and K. pneumoniae. These microorganisms are known to cause serious health issues, which can result in increased mortality, decreased productivity, and higher costs for both disease prevention and treatment (Ibrahim et al., 2021). Escherichia coli isolates were the most prevalent in the collected samples, followed by Salmonella spp. and K. pneumoniae isolates. Escherichia coli isolates were high prevalence in chicken cloacal swabs (93.1%), Salmonella spp. were in chicken meats (34.9%), and K. pneumoniae isolates were in litter samples (36.7%). Previous studies have also reported 83 and 53.04% of cloacal swabs were found positive for E. coli in Malaysia and China, respectively (Suryadevara et al., 2020; Li et al., 2022). In Indonesia, 13.75% of cloacal swabs obtained from broiler chicken farms were found positive for K. pneumoniae (Permatasari et al., 2020). Another study in Malaysia showed that the prevalence of Salmonella spp. and E. coli from cloacal swabs obtained from broilers were 6.5 and 51.8%, respectively (Ibrahim et al., 2021). Backyard chickens in Malaysia were documented with 2.5% of Salmonella spp. (Jajere et al., 2019). In contrast, a much higher prevalence of Salmonella spp. (48%) in chicken meat samples was observed in Bangladesh (Islam et al., 2016). In Nepal, 33.33% of chicken meat samples were positive for E. coli (Joshi et al., 2019) which is lower than our findings, indicating that the frequency of E. coli in chicken meat might vary widely. Different levels of hygienic practices in various geographic regions and environmental factors, such as exposure to poor sanitation, could be the source of the variations in prevalence (Dawadi et al., 2021).
Colistin is being prescribed for the therapeutic purposes of bacterial infection in humans that are resistant to multiple drugs despite its side effects. The introduction of the colistin resistance (mcr) gene in bacterial strains that are already resistant to many antibiotics, creates the ineffectiveness of colistin, the last resort drug. In this study, the Enterobacteriaceae isolates recovered from various sources, comprising chicken meat from supermarkets and cloacal swabs and litter samples from poultry farms, were prevalent with colistin resistance and mcr genes. The MIC value of colistin-resistant isolates ranged from 4 μg/mL to 128 μg/mL of colistin, however, low MIC values were found in mcr positive isolates. In previous studies, the mcr-1 had been documented from Klebsiella pneumoniae, and E. coli isolates in Malaysia (Yu et al., 2016; Mobasseri et al., 2019; Aklilu and Raman, 2020). The colistin resistance mcr-1 gene is also observed in E. coli isolates from poultry in many countries in Asia (Trung et al., 2017; Joshi et al., 2019; Amin et al., 2020). A greater percentage of mcr-1 (>94%) was depicted in bacterial strains from turkey and broiler feces in Germany (Irrgang et al., 2016). The colistin resistance gene (mcr-1) was found in 25.8% of poultry (turkey and chicken) meats from Italy (5 samples) and Germany (28 samples) but not in any samples (turkey and chicken meats) from Switzerland, Denmark, Austria, or Hungary, according to a Swiss study (Zurfluh et al., 2016). In Brazil, 19.5% of chicken meat and liver samples were positive for E. coli harboring mcr-1 (Monte et al., 2017). All the colistin-resistant E. coli obtained from raw beef and beef products in Egypt were found positive for mcr-1 (Sabala et al., 2021). In comparison with previous studies, a low prevalence of mcr-1 gene in colistin-resistant isolates was found in the current research. The explanation for the low incidence in Malaysia isolates is unknown, although other resistance mechanisms, such as chromosomal alteration and modulation in the mgrB gene, could be involved (Chen et al., 2017).
The colistin resistance gene, mcr-5 was detected in Salmonella spp. for the first time in Malaysia. There were no reports about this gene in any bacterial strains from animals or humans in Malaysia to the best of our knowledge. In this study, the mcr-5 gene in Salmonella spp. was found at a very low rate. This finding is consistent with a previous report in which 2.5% (8/315) of colistin-resistant Salmonella spp. originated from pigs and meats (pork) in Germany (Borowiak et al., 2019), and 0.7% of human vaginal swab samples in Yangzhou in China (Zhang et al., 2018) were found to be positive for the mcr-5 gene. The mcr-5 gene was also observed in colistin-resistant E. coli strains obtained from cloacal swabs of poultry in Nigeria (Ngbede et al., 2020), veal in Belgium (Timmermans et al., 2021), pork samples in Cambodia (Pungpian et al., 2021) and poultry and pigs samples in Spain and China (García-Meniño et al., 2019). The situation is concerning because these resistant pathogens could be spread to humans through the food chain or close contact with animals (Dawadi et al., 2021).
The mcr-1 gene was found among the isolates showing MIC values of 4 μg/mL and 8 μg/mL of colistin in this study which is consistent with the previous studies in Bangladesh and China, which showed that colistin-resistant isolates with mcr-1 had a MIC value of 4 μg/mL, and 8 to 16 μg/mL of colistin, respectively (Islam et al., 2017; Amin et al., 2020).
In nucleotide BLAST, the sequences of the mcr-1 gene isolated from E. coli and the mcr-5 gene obtained from Salmonella spp. were found to be 100% identical to previously reported mcr-1 and mcr-5 gene sequences in the NCBI database. In the phylogenetic analysis, mcr-1 gene sequences were closely related to previous reports, mcr-1 gene sequence of E. coli isolates recovered from chicken in Malaysia (Yu et al., 2016) and E. coli strain isolated from human in China (Genbank_MZ450868). This suggests that mcr-1 gene in E. coli has been circulating in Malaysia, which is a threat to animals and public health. The colistin resistance mcr-5 gene was detected in Salmonella spp. isolate for the first time in Malaysia. The various types of mcr-5 gene variants, including mcr-5.1 to mcr-5.4, were identified in the world (Fleres et al., 2019; Ling et al., 2020). In the phylogeny, the mcr-5 gene sequence of the current study was closely related to mcr-5.3 gene recovered from E. coli isolates obtained from the horse in Brazil (GenBank database, NG_061405).
The introduction and dissemination of colistin resistance with mcr genes in Enterobacteriaceae is a major worldwide issue. Colistin-resistant Enterobacteriaceae were observed in poultry meats and poultry farms in the present study. The mcr-1 and mcr-5 genes were found in colistin-resistant E. coli and Salmonella spp., respectively. The mcr-5 has been identified in Malaysia for the first time, which could signal the advent of mcr-5 harboring bacterial strains for infection. The existence of mcr-positive Enterobacteriaceae in poultry and poultry meat in Malaysia emphasizes the importance of proper poultry waste disposal and good hygiene practices among people who are exposed to poultry and poultry meats.
The nucleotide sequences of mcr-1 and mcr-5 have been submitted to the GenBank of the NCBI (accession no. OR333822, OR333835). The raw data supporting the findings of this article will be made available by the authors, without undue reservation.
The animal study was reviewed and approved by the ethical board of Universiti Putra Malaysia (UPM), Institutional Animal Care and Use Committee (IACUC) approved the research study protocol for collecting cloacal swabs from live poultry (UPM/IACUC/AUP-R091/2019).
MK and ZZ: conceptualization, software, investigation, data curation, writing - original draft preparation, visualization, and acquisition. MK, ZZ, LH, NF, and NA: methodology, validation, and writing - review and editing. RK: formal analysis. ZZ: resources, supervision, and project administration. All authors contributed to the article and approved the submitted version.
This research was funded by the UPM trust fund (grant number: 6282525) in collaboration with Bangladesh Agricultural Research Council (BARC), Dhaka, Bangladesh.
The authors would like to thank the authorities of the Bangladesh Agriculture Research Council, Bangladesh, and Universiti Putra Malaysia for their financial support during this research.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1208314/full#supplementary-material
Aidara-Kane, A., Andremont, A., and Collignon, P. (2013). Antimicrobial resistance in the food chain and the AGISAR initiative. J. Infect. Public Health 6, 162–165. doi: 10.1016/j.jiph.2013.04.001
Aklilu, E., and Raman, K. (2020). MCR-1 gene encoded Colistin-resistant Escherichia coli in raw chicken meat and bean sprouts in Malaysia. Int. J. Microbiol. 2020:8853582. doi: 10.1155/2020/8853582
Amin, M. B., Sraboni, A. S., Hossain, M. I., Roy, S., Mozmader, T. A. U., Unicomb, L., et al. (2020). Occurrence and genetic characteristics of mcr-1-positive colistin-resistant E. coli from poultry environments in Bangladesh. J. Glob. Antimicrob. Resist. 22, 546–552. doi: 10.1016/j.jgar.2020.03.028
Borowiak, M., Baumann, B., Fischer, J., Thomas, K., Deneke, C., Hammerl, J. A., et al. (2020). Development of a novel mcr-6 to mcr-9 multiplex PCR and assessment of mcr-1 to mcr-9 occurrence in Colistin-resistant Salmonella enterica isolates from environment, feed, animals and food (2011–2018) in Germany. Front. Microbiol. 11:80. doi: 10.3389/fmicb.2020.00080
Borowiak, M., Hammerl, J. A., Deneke, C., Fischer, J., Szabo, I., and Malorny, B. (2019). Characterization of mcr-5-Harboring Salmonella enterica subsp. Enterica Serovar typhimurium isolates from animal and food origin in Germany. Antimicrob. Agents Chemother. 63:e00063-19. doi: 10.1128/AAC.00063-19
Cannatelli, A., D’Andrea, M. M., Giani, T., Di Pilato, V., Arena, F., Ambretti, S., et al. (2013). In vivo emergence of colistin resistance in Klebsiella pneumoniae producing KPC-type carbapenemases mediated by insertional inactivation of the PhoQ/PhoP mgrB regulator. Antimicrob. Agents Chemother. 57, 5521–5526. doi: 10.1128/AAC.01480-13
Chen, Y., Luo, Y., Pettengill, J., Timme, R., Melka, D., Doyle, M., et al. (2017). Singleton sequence type 382, an emerging clonal Group of Listeria monocytogenes associated with three multistate outbreaks linked to contaminated stone fruit, caramel apples, and leafy green salad. J. Clin. Microbiol. 55, 931–941. doi: 10.1128/JCM.02140-16
Dawadi, P., Bista, S., and Bista, S. (2021). Prevalence of Colistin-resistant Escherichia coli from poultry in south Asian developing countries. Vet. Med. Int. 2021, 1–5. doi: 10.1155/2021/6398838
Elbediwi, M., Li, Y., Paudyal, N., Pan, H., Li, X., Xie, S., et al. (2019). Global burden of Colistin-resistant bacteria: mobilized Colistin resistance genes study. Microorganisms 7, 1–18. doi: 10.3390/microorganisms7100461
EMA (European Medicine Agency) (2016). Updated Advice on the Use of Colistin Products in Animals within the European Union: Development of Resistance and Possible Impact on Human and Animal Health. European Medicines Agency, Committee for Medicinal Products for Veterinary Use (CVMP); London, UK.
EUCAST (2016a) Recommendations for MIC determination of colistin (polymyxin E) As recommended by the joint CLSI-EUCAST Polymyxin Breakpoints Working Group. Available at: https://Www.Eucast.Org
EUCAST (2016b). EUCAST breakpoint tables for interpretation of MICs and zone diameters,version 1.0. Basel: European Committee on Antimicrobial Susceptibility Testing. Version 6.
Ezadi, F., Ardebili, A., and Mirnejad, R. (2019). Antimicrobial susceptibility testing for Polymyxins: challenges, issues, and recommendations. J. Clin. Microbiol. 57, 1–20. doi: 10.1128/JCM.01390-18
Fleres, G., Couto, N., Schuele, L., Chlebowicz, M. A., Mendes, C. I., Van Der Sluis, L. W. M., et al. (2019). Detection of a novel mcr-5.4 gene variant in hospital tap water by shotgun metagenomic sequencing. J. Antimicrob. Chemother. 74, 3626–3628. doi: 10.1093/jac/dkz363
García-Meniño, I., Díaz-Jiménez, D., García, V., de Toro, M., Flament-Simon, S. C., Blanco, J., et al. (2019). Genomic characterization of prevalent mcr-1, mcr-4, and mcr-5 Escherichia coli within swine enteric Colibacillosis in Spain. Front. Microbiol. 10:2469. doi: 10.3389/fmicb.2019.02469
Ghafur, A., Shankar, C., GnanaSoundari, P., Venkatesan, M., Mani, D., Thirunarayanan, M. A., et al. (2019). Detection of chromosomal and plasmid-mediated mechanisms of colistin resistance in Escherichia coli and Klebsiella pneumoniae from Indian food samples. J. Glob. Antimicrob. Resist. 16, 48–52. doi: 10.1016/j.jgar.2018.09.005
Gharaibeh, M. H., and Shatnawi, S. Q. (2019). An overview of colistin resistance, mobilized colistin resistance genes dissemination, global responses, and the alternatives to colistin: a review. Vet. World 12, 1735–1746. doi: 10.14202/vetworld.2019.1735-1746
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucl. Acids. Symp. Ser. 41, 95–98.
Ibrahim, S., Wei Hoong, L., Lai Siong, Y., Mustapha, Z., CS, C. W. Z., Aklilu, E., et al. (2021). Prevalence of antimicrobial resistance (AMR) salmonella spp. and Escherichia coli isolated from broilers in the East Coast of peninsular Malaysia. Antibiotics 10. doi: 10.3390/antibiotics10050579
Irrgang, A., Roschanski, N., Tenhagen, B. A., Grobbel, M., Skladnikiewicz-Ziemer, T., Thomas, K., et al. (2016). Prevalence of mcr-1 in E. coli from livestock and food in Germany, 2010-2015. PLoS One 11:e0159863. doi: 10.1371/journal.pone.0159863
Islam, M. J., Mahbub-E-Elahi, A. T. M., Ahmed, T., and Hasan, M. K. (2016). Isolation and identification of salmonella spp. from broiler and their antibiogram study in Sylhet, Bangladesh. J. Appl. Biol. Biotechnol. 4, 046–051. doi: 10.7324/jabb.2016.40308
Islam, A., Rahman, Z., Monira, S., Rahman, M. A., Camilli, A., George, C. M., et al. (2017). Colistin resistant Escherichia coli carrying mcr-1 in urban sludge samples: Dhaka, Bangladesh. Gut Pathog. 9:77. doi: 10.1186/s13099-017-0227-4
Islam, S., Urmi, U. L., Rana, M., Sultana, F., Jahan, N., Hossain, B., et al. (2020). High abundance of the colistin resistance gene mcr-1 in chicken gut-bacteria in Bangladesh. Sci. Rep. 10:17292. doi: 10.1038/s41598-020-74402-4
Jajere, S. M., Hassan, L., Abdul Aziz, S., Zakaria, Z., Abu, J., Nordin, F., et al. (2019). Salmonella in native “village” chickens (Gallus domesticus): prevalence and risk factors from farms in south-central peninsular Malaysia. Poult. Sci. 98, 5961–5970. doi: 10.3382/ps/pez392
Joshi, P. R., Thummeepak, R., Paudel, S., Acharya, M., Pradhan, S., Banjara, M. R., et al. (2019). Molecular characterization of Colistin-resistant Escherichia coli isolated from chickens: first report from Nepal. Microb. Drug Resist. 25, 846–854. doi: 10.1089/mdr.2018.0326
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Li, J., Liu, S., Fu, J., Yin, J., Zhao, J., Zhong, C., et al. (2019). Co-occurrence of Colistin and Meropenem resistance determinants in a Stenotrophomonas strain isolated from sewage water. Microb. Drug Resist. 25, 317–325. doi: 10.1089/mdr.2018.0418
Li, Z., Xin, L., Peng, C., Liu, C., Wang, P., Yu, L., et al. (2022). Prevalence and antimicrobial susceptibility profiles of ESBL-producing Klebsiella pneumoniae from broiler chicken farms in Shandong Province, China. Poult. Sci. 101:102002. doi: 10.1016/j.psj.2022.102002
Ling, Z., Yin, W., Shen, Z., Wang, Y., Shen, J., and Walsh, T. R. (2020). Epidemiology of mobile colistin resistance genes mcr-1 to mcr-9. J. Antimicrob. Chemother. 75, 3087–3095. doi: 10.1093/jac/dkaa205
Liu, Y.-Y., Ang, Y., Walsh, T. R., Yi, L.-X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/S1473-3099(15)00424-7
Moawad, A. A., Hotzel, H., Neubauer, H., Ehricht, R., Monecke, S., Tomaso, H., et al. (2018). Antimicrobial resistance in Enterobacteriaceae from healthy broilers in Egypt: emergence of colistin-resistant and extended-spectrum β-lactamase-producing Escherichia coli. Gut Pathog. 10, 1–12. doi: 10.1186/s13099-018-0266-5
Mobasseri, G., Teh, C. S. J., Ooi, P. T., and Thong, K. L. (2019). The emergence of colistin-resistant Klebsiella pneumoniae strains from swine in Malaysia. J. Glob. Antimicrob. Resist. 17, 227–232. doi: 10.1016/j.jgar.2018.12.015
Monte, D. F., Mem, A., Fernandes, M. R., Cerdeira, L., Esposito, F., Galvão, J. A., et al. (2017). Chicken meat as a reservoir of colistin-resistant Escherichia coli strains carrying mcr-1 genes in South America. Antimicrob. Agents Chemother. 61, 2–5. doi: 10.1128/AAC.02718-16
Ngbede, E. O., Poudel, A., Kalalah, A., Yang, Y., Adekanmbi, F., Adikwu, A. A., et al. (2020). Identification of mobile colistin resistance genes (mcr-1.1, mcr-5 and mcr-8.1) in Enterobacteriaceae and Alcaligenes faecalis of human and animal origin, Nigeria. Int. J. Antimicrob. Agents 56:106108. doi: 10.1016/j.ijantimicag.2020.106108
Permatasari, D. A., Witaningrum, A. M., Wibisono, F. J., and Effendi, M. H. (2020). Detection and prevalence of multidrug-resistant Klebsiella pneumoniae strains isolated from poultry farms in Blitar, Indonesia. Biodiversitas 21, 4642–4647. doi: 10.13057/biodiv/d211024
Pui, C. F., Wong, W. C., Chai, L. C., Nillian, E., Ghazali, F. M., Cheah, Y. K., et al. (2011). Salmonella Typhi and Salmonella Typhimurium in sliced fruits using multiplex PCR. Food Control 22, 337–342. doi: 10.1016/j.foodcont.2010.05.021
Pungpian, C., Lee, S., Trongjit, S., Sinwat, N., Angkititrakul, S., Prathan, R., et al. (2021). Colistin resistance and plasmid-mediated mcr genes in Escherichia coli and salmonella isolated from pigs, pig carcass and pork in Thailand, Lao PDR and Cambodia border provinces. J. Vet. Sci. 22, 1–15. doi: 10.4142/jvs.2021.22.e68
Rahn, K., De Grandis, S. A. A., Clarke, R. C. C., McEwen, S. A. A., Galán, J. E. E., Ginocchio, C., et al. (1992). Amplification of an invA gene sequence of Salmonella typhimurium by polymerase chain reaction as a specific method of detection of salmonella. Mol. Cell. Probes 6, 271–279. doi: 10.1016/0890-8508(92)90002-F
Ranjbar, R., Izadi, M., Hafshejani, T. T., and Khamesipour, F. (2016). Molecular detection and antimicrobial resistance of Klebsiella pneumoniae from house flies (Musca domestica) in kitchens, farms, hospitals and slaughterhouses. J. Infect. Public Health 9, 499–505. doi: 10.1016/j.jiph.2015.12.012
Rebelo, A. R., Bortolaia, V., Kjeldgaard, J. S., Pedersen, S. K., Leekitcharoenphon, P., Hansen, I. M., et al. (2018). Multiplex PCR for detection of plasmid-mediated colistin resistance determinants, mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 for surveillance purposes. Eur. Secur. 23:17-00672. doi: 10.2807/1560-7917.ES.2018.23.6.17-00672
Sabala, R. F., Usui, M., Tamura, Y., Abd-Elghany, S. M., Sallam, K. I., and Elgazzar, M. M. (2021). Prevalence of colistin-resistant Escherichia coli harbouring mcr-1 in raw beef and ready-to-eat beef products in Egypt. Food Control 119:107436. doi: 10.1016/j.foodcont.2020.107436
Sharma, J., Kumar, D., Hussain, S., Pathak, A., Shukla, M., Kumar, V. P., et al. (2019). Prevalence, antimicrobial resistance and virulence genes characterization of nontyphoidal salmonella isolated from retail chicken meat shops in northern India. Food Control 102, 104–111. doi: 10.1016/j.foodcont.2019.01.021
Suryadevara, N., Yong, K. B., Ganapathy, B., Subramonie, S., Ragavan, N. D., Ramachandiran, M., et al. (2020). Molecular characterization of Escherichia coli from chickens in poultry farms of Malaysia. Res. J. Biotechnol. 15, 1–10.
Theuretzbacher, U. (2017). Global antimicrobial resistance in gram-negative pathogens and clinical need. Curr. Opin. Microbiol. 39, 106–112. doi: 10.1016/j.mib.2017.10.028
Timmermans, M., Wattiau, P., Denis, O., and Boland, C. (2021). Colistin resistance genes mcr-1 to mcr-5, including a case of triple occurrence (mcr-1, −3 and −5), in Escherichia coli isolates from faeces of healthy pigs, cattle and poultry in Belgium, 2012–2016. Int. J. Antimicrob. Agents 57:106350. doi: 10.1016/j.ijantimicag.2021.106350
Trung, N. V., Matamoros, S., Carrique-Mas, J. J., Nghia, N. H., Nhung, N. T., Chieu, T. T. B., et al. (2017). Zoonotic transmission of mcr-1 Colistin resistance gene from small-scale poultry farms, Vietnam. Emerg. Infect. Dis. 23, 529–532. doi: 10.3201/eid2303.161553
Xu, T., Zhang, C., Ji, Y., Song, J., Liu, Y., Guo, Y., et al. (2021). Identification of mcr-10 carried by self-transmissible plasmids and chromosome in Enterobacter roggenkampii strains isolated from hospital sewage water. Environ. Pollut. 268:115706. doi: 10.1016/j.envpol.2020.115706
Yu, C. Y., Ang, G. Y., Chin, P. S., Ngeow, Y. F., Yin, W.-F., and Chan, K.-G. (2016). Emergence of mcr-1-mediated colistin resistance in Escherichia coli in Malaysia. Int. J. Antimicrob. Agents 47, 504–505. doi: 10.1016/j.ijantimicag.2016.04.004
Zhang, J., Chen, L., Wang, J., Butaye, P., Huang, K., Qiu, H., et al. (2018). Molecular detection of colistin resistance genes (mcr-1 to mcr-5) in human vaginal swabs. BMC. Res. Notes 11:143. doi: 10.1186/s13104-018-3255-3
Keywords: Enterobacteriaceae , MCR, colistin resistance, poultry, Malaysia
Citation: Karim MR, Zakaria Z, Hassan L, Faiz NM and Ahmad NI (2023) The occurrence and molecular detection of mcr-1 and mcr-5 genes in Enterobacteriaceae isolated from poultry and poultry meats in Malaysia. Front. Microbiol. 14:1208314. doi: 10.3389/fmicb.2023.1208314
Received: 18 April 2023; Accepted: 12 July 2023;
Published: 03 August 2023.
Edited by:
Zuowei Wu, Iowa State University, United StatesReviewed by:
Nikolaos Strepis, Erasmus Medical Center, NetherlandsCopyright © 2023 Karim, Zakaria, Hassan, Faiz and Ahmad. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zunita Zakaria, enVuaXRhQHVwbS5lZHUubXk=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.