
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 31 July 2023
Sec. Virology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1205297
This article is part of the Research Topic Advances and Insights in the Diagnosis of Viral Infections and Vaccines Development in Animals View all 24 articles
 Yingyu Liu1†
Yingyu Liu1† Chenyan Sheng1†
Chenyan Sheng1† Yu Zhou2
Yu Zhou2 Jianming Li3
Jianming Li3 Qinglong Gong1
Qinglong Gong1 Kun Shi3
Kun Shi3 Fei Liu1
Fei Liu1 Lihui Xu1
Lihui Xu1 Zhenzhen Cui3
Zhenzhen Cui3 Xue Leng3*
Xue Leng3* Rui Du3*
Rui Du3*Mink circovirus disease caused by Mink Circovirus (MiCV) is a serious infectious disease of mink that has become prevalent in recent years in China, severely affecting the reproductive performance of mink and causing significant economic losses to farms. To date, there have been few studies on MiCV, its pathogenic mechanism is not clear, and there is no effective vaccine or drug to prevent and control the disease. Therefore, it is necessary to establish a rapid and reliable molecular diagnostic method, which would aid future studies of this novel virus. In our study, we developed a sensitive and specific TaqMan-based quantitative real-time PCR assay targeting the MiCV Cap gene. The assay showed no cross-reaction with other tested animal viruses. The assay is highly sensitive, with a detection limit of as low as 10 plasmid DNA copies and 2.38 × 10−2 pg of viral DNA. The intra and inter--assay coefficients of variation were both low. The positive detection rate of MiCV in clinical samples from minks, foxes, and raccoon dogs were 58.8% (133/226), 50.7% (72/142), and 42.2% (54/128), respectively, giving a total positive detection rate of 52.2% (259/496). Higher contamination levels were observed in samples from the environment in direct or indirect contact with animals, with a total positive detection rate of 75.1% (220/293). These epidemiological results showed that minks, foxes, and raccoon dogs had high infection rates of MiCV. This was also the first study to detect MiCV on the ground and equipment of fur-bearing animal farms. Our assay is highly sensitive and specific for the diagnosis and quantification of MiCV, and should provide a reliable real-time tool for epidemiological and pathogenetic study of MiCV infection.
Circoviruses have various hosts among mammals and birds (Li et al., 2010; Decaro et al., 2014). In recent years, in addition to the well-known Porcine Circovirus, Circoviruses that can infect canines, penguins, bats, and other animals have been reported (Piewbang et al., 2018; Matsumoto et al., 2019; Morandini et al., 2019; Canuti et al., 2022). Mink circovirus disease, caused by a new Circovirus member, Mink Circovirus (MiCV), has only been discovered in China and is reported to be able to infect foxes and raccoon dogs (Yang et al., 2018). Taxonomically, MiCV belongs to the genus Circovirus in the family Circoviridae. Circoviruses are small envelope-free icosahedral viruses, with a diameter of 15–25 nm, and a circular single-stranded DNA genome, which is the smallest known auto replicating viral genome (Breitbart et al., 2017; Rosario et al., 2017). The genome of MiCV comprises about 1700 nucleotides, including two main open reading frames (ORFs), one encoding the replication-related protein (Rep) and the other encoding viral capsid protein (Cap; Ge et al., 2018a). In Circovirus, the ORF encoding Rep is on the sense strand of the genome, while the Rep of Cyclovirus is encoded by the complementary strand of double-stranded DNA (Breitbart et al., 2017; Rosario et al., 2017; Shulman and Davidson, 2017; Zhao et al., 2019). The Rep protein (35.8 kDa) is mainly involved in virus replication and plays an important role in virus proliferation, while Cap, is the main structural protein (27.8 kDa) and has good immunogenicity (Rosario et al., 2017).
Mink circovirus disease has a wide epidemic range in China. Existing epidemiological research results showed that the positive rate of MiCV was 30.30% in Heilongjiang Province, 38.46% in Jilin Province, 52.88% in Shandong Province, 58.46% in Liaoning Province and 67.90% in Hebei Province (Ge et al., 2018a). MiCV has caused great economic losses to the mink breeding industry. Minks infected with the virus showed poor appetite, mental fatigue, rough fur, diarrhea, and even death, and the virus easily causes mixed infection with Aleutian mink disease virus, Canine distemper virus and other viruses (Ge et al., 2018b). Most minks recovered spontaneously, however, their growth was poor, their fur quality was significantly reduced, and the reproductive capacity of females was lower than before infection. MiCV can be transmitted between minks of different ages.
In many places in China, minks, foxes, and raccoon dogs are kept on the same fur-bearing animal farms. A study showed that MiCV can infect not only minks, but also foxes and raccoon dogs,accompanied by similar clinical symptoms such as drowsiness, anorexia, pale mouth and unkempt fur (Yang et al., 2018). Besides, the persistence of the virus in the environment is also an important factor in the spread of the disease. To date, no large-scale MiCV prevalence studies in minks, foxes and raccoon dogs have been carried out. Viral contamination in the environment is also unknown. There have been few studies on MiCV, its pathogenic mechanism is not clear, and there is no effective vaccine and drug to treat the disease. Therefore, it is important to establish a rapid and effective detection method for disease prevention and control. Quantitative real-time PCR (qPCR) detection has the advantages of strong specificity, high sensitivity and relatively low cost (Abdel-Moneim et al., 2017; Tong et al., 2020; de Oliveira Lopes et al., 2021; Gong et al., 2021). It can quickly determine viruses in different tissue samples and has become an important tool in veterinary virology and disease control (Varga and James, 2006; Vázquez et al., 2017; Cui et al., 2018).
In this study, a TaqMan-based qPCR method was established to detect MiCV DNA. Subsequently, we detected MiCV DNA in clinical samples from minks, foxes, raccoon dogs and farm environments in China.
MiCV strain HB3 (GenBank accession No. MK561562.1), Aleutian mink disease virus (AMDV) DL125 strain (Isolated by Dr. Liu of the Economic Animal Infectious Diseases Laboratory of Jilin Agricultural University in 2015 citation), Pseudorabies virus (PRV) JL03 strain, and Porcine circovirus 2 (PCV-2) were stored in the Economic Animal Infectious Diseases Laboratory of Jilin Agricultural University. The vaccine strains mink enteritis virus (MEV), canine distemper virus (CDV), and canine adenovirus type 2 (CAdV-2) were purchased from Teyan Biotechnology, Ltd., (Jilin, China) and QiLu Animal Health Products, Ltd., (Shandong, China), respectively.
Two hundred and twenty-six mink anal swab samples, 142 fox anal swab samples, 128 raccoon dog anal swab samples, and 293 environmental swab samples were collected from 6 farms in Jilin, Heilongjiang, and Shandong provinces in China from 2021 to 2022.
According to the MiCV genome sequences obtained in the laboratory in recent years and all 13 of MiCV genome sequences deposited in GenBank, DNAMAN (LynnonBiosoft, USA) was used to perform multiple comparisons to obtain a highly conserved region in the Cap region of the genome. Primer 5.0 (PREMIER Biosoft International, Palo Alto, California, USA) was used to design the required primers (real-time quantitative PCR primers and common PCR primers) and probes (Table 1). The primers and probes were synthesized by Comate Bioscience Co. Ltd., (Changchun, China).
All swab samples were repeatedly frozen and thawed three times and centrifuged. According to the manufacturer’s protocol, a Viral DNA Kit (Omega Biotek, Winooski, VT, USA) was used to extract DNA from 200 μl of the supernatant. After the DNA samples were eluted into 100 μl of Elution Buffer, they were stored at – 20°C until required.
The 684 bp fragment of the Cap gene was amplified from the DNA of MiCV strain HB3 using PCR with the MiCV-Cap-F and MiCV-Cap-R primers. A Midi Purification Kit (Tiangen Biotech, Beijing, China) was used to purify the amplification products from the agarose gel, which were then ligated into the vector pMD18-T (Takara Biotechnology, Dalian, China) to construct recombinant plasmid pMD-MiCV. pMD-MiCV was transformed into Escherichia coli DH5α cells (TransGen Biotech, Beijing, China), purified. Using a plasmid miniprep Kit (Axygen A Corning Brand, Suzhou, China), and verified by sequencing (carried out by Comate Bioscience, Changchun, China). The target plasmid pMD-MiCV had an original concentration of 210.47 ng/μl, as measured using a Nanodrop One spectrophotometer (Thermo Scientific, Wilmington, DE, USA). Ten-fold serial dilutions of plasmid DNA in elution buffer were created and stored at −20°C until further testing. According to the following formula: the copy number of the plasmid (copies/μl) = [concentration of plasmid (ng/μl) × (6.02 × 1023)]/[(plasmid length × 660 × 109)], the copy number of the plasmid was 5.68 × 1010 copies/μl. The original plasmid was diluted using elution buffer to 1.0 × 1010 copies/μl, stored as the standard plasmid-20°C until required.
The standard plasmid pMD-MiCV was serially diluted 10 times with elution buffer, 1.0 × 106 copies/μl to 1.0 × 101 copies/μl as templates to establish the standard curve, created by plotting the logarithm of the plasmid copy number against the measured cycle threshold (Ct) values. The standard curve, the correlation coefficient of the standard curve, and the qPCR efficiencies were calculated using the GraphPad Prism 7 software (GraphPad Inc., La Jolla, CA, USA). The amount of DNA quantified in each sample was expressed as number of copies per reaction. The final volume was 20 μl including 10 μl of TB Green® Premix Ex Taq™ II (Takara Biomedical Technology, Beijing, China), 0.8 μl each of the MiCV-F and MiCV-R primers (10 μmol/L), 0.4 μl of the probe MiCV-P (10 μmol/L), 1 μl of template, and 6 μl of ddH2O. The qPCR reaction was carried out in eight tubes by fluorescence quantitative PCR in qTOWER3 G instrument (Analytik Jena AG, Jena, Germany). The procedure comprised incubation at 95°C for 3 min, followed by 45 cycles of 95°C for 10 s and 57°C for 40 s.
To exclude qPCR cross-reactivities between MiCV and other pathogens, six viruses, namely, AMDV, PRV (PRV has occurred in mink in recent years, and might be related to eating pigs’ viscera), PCV-2, MEV, CDV and CAdV-2 were subjected to qPCR to confirm the specificity of the technique. MiCV strain HB3 was used as the positive control and ddH2O was used as the negative control, and each sample was repeated 3 times.
To compare the sensitivity of qPCR and conventional PCR, dilutions of plasmid pMD-MiCV (1.0 × 105 copies/μl to1.0 × 100 copies/μl) and DNA of MiCV strain HB3 (2.38 × 102 pg/μl to 2.38 × 10−3 pg/μl) were used as templates for qPCR and conventional PCR. The conventional PCR reaction was performed using the primer pair MiCV-cap-F and MiCV-cap-R. Besides, the diagnostic sensitivity of the qPCR was tested by the positive clinical samples verified by sequencing.
Three different dilutions of standard plasmid (1.0 × 102 copies/μl, 1.0 × 104 copies/μl, and 1.0 × 106 copies/μl) were used for amplification in three parallel assays under the same conditions. The coefficient of variation (CV) was calculated according to the formula CV = (SD [Ct value]/overall average [Ct value]) × 100, to evaluate the intra-and inter-assay repeatability and stability in the qPCR method.
A total of 789 clinical samples were collected from different fur-bearing animal farms in China. The samples included anal swab samples and environmental swab samples. The anal swab samples were collected by inserting sterile cotton swab into the anus about 2 to 3 cm, gently rotating it, and pulling it out. The anal swab samples were then placed into sampling tubes that contained 2 ml of sterile phosphate-buffered saline. The environmental swab samples were collected using a sterile cotton swab to clean each sampling area including cages, troughs, soil under the cages, sewage under the cages, aisle floors, breeder’s clothes, and equipment, for 20 to 30 s, then placing it into the sample tubes holding 2 ml of sterile phosphate-buffered saline. All samples were stored at −80°C immediately after being transported back to the laboratory at low temperature. The qPCR assays were performed on these samples for MiCV detection.
Ten-fold serial dilutions of plasmids were used to construct a standard curve by plotting the logarithm of the plasmid copy number against the measured Ct values (Figure 1). The standard curve had a wide dynamic range of 101 to106 copies/μl with a linear correlation (R2) of 0.996 between the Ct value and the logarithm of the plasmid copy number. The amplification efficiency of the obtained qPCR was 93%.
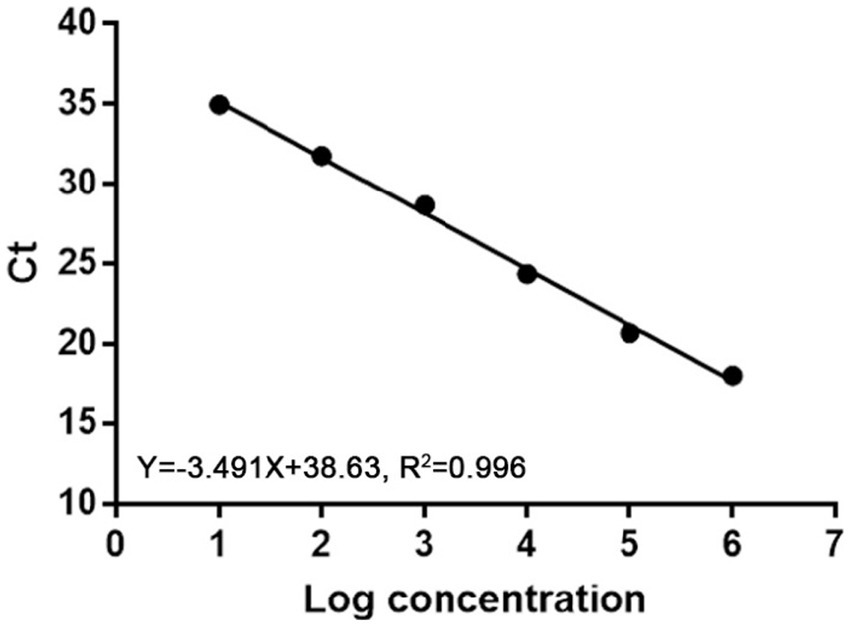
Figure 1. Standard curve of real-time PCR to detect MiCV. The correlation coefficient (R2) was 0.996 and the slope was −3.491.
The specificity of the qPCR assay was evaluated using eight different reactions, which included, MiCV, AMDV, PRV, PCV-2, MEV, CDV, CAdV2 and a negative water control. Strong fluorescent signals were obtained from reactions with MiCV in three replications; while no signals were obtained from the other six virus samples and the water control (Figure 2). The results demonstrated that the MiCV assay specifically detected the target virus without cross detection of any non-target pathogens.
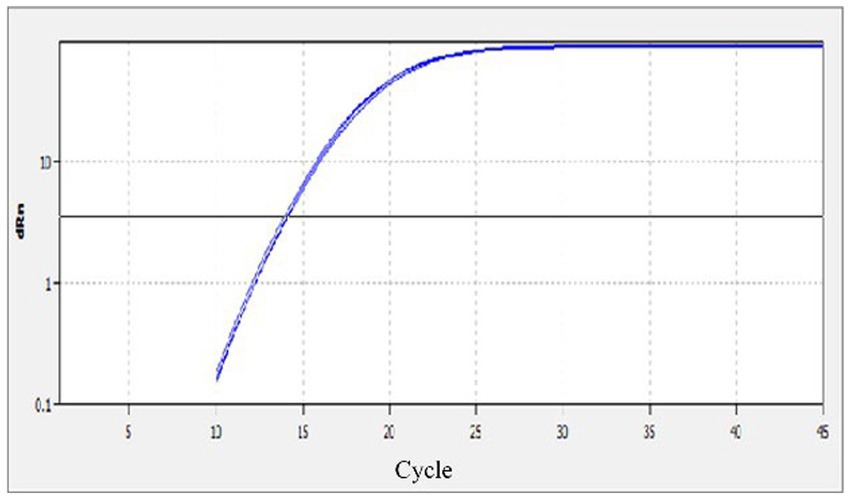
Figure 2. Specificity of the qPCR assay. The templates included DNA or cDNA of MiCV, AMDV, PRV, PCV-2, MEV, CDV, CAdV2 and the water control.
The sensitivity of the qPCR assays were evaluated by testing 10-fold serial dilutions of the DNA standards and the positive clinical samples. For the standard plasmid DNA of pMD-MiCV, the detection limit of qPCR was 1.0 × 101 copies/μl, while the detection limit of conventional PCR was 1.0 × 103 copies/μl (Figures 3A,B). For the virus DNA of MiCV strain HB3, the detection limit of qPCR was 2.38 × 10−2 pg/μl, while the detection limit of conventional PCR was 2.38 × 101 pg/μl (Figures 4A,B). For the positive clinical samples, 35 of 35 positive samples were verified as positive by qPCR assay. Thus, the diagnostic sensitivity of the qPCR was 100%.
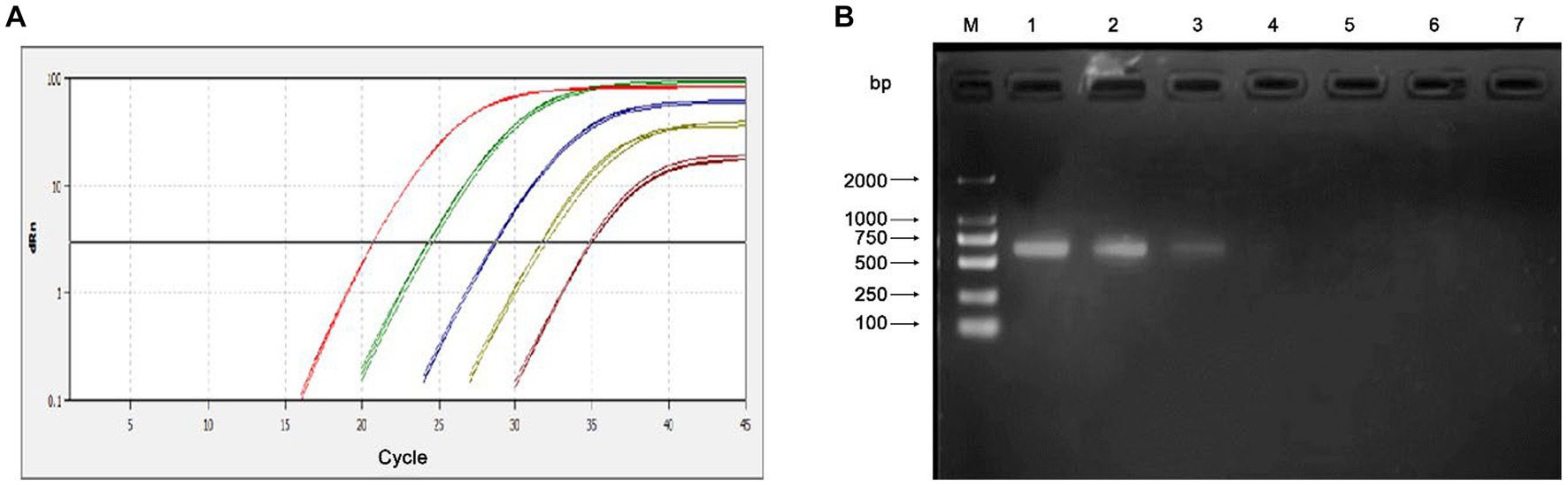
Figure 3. Sensitivity of the qPCR assay for the plasmid DNA of pMD-MiCV. (A) The qPCR amplification curve. (B) Electrophoresis of conventional PCR reactions. The template concentrations of plasmid DNA ranged from 1.0 × 105 copies/μl to 1.0 × 100 copies/μl. A negative control was included.
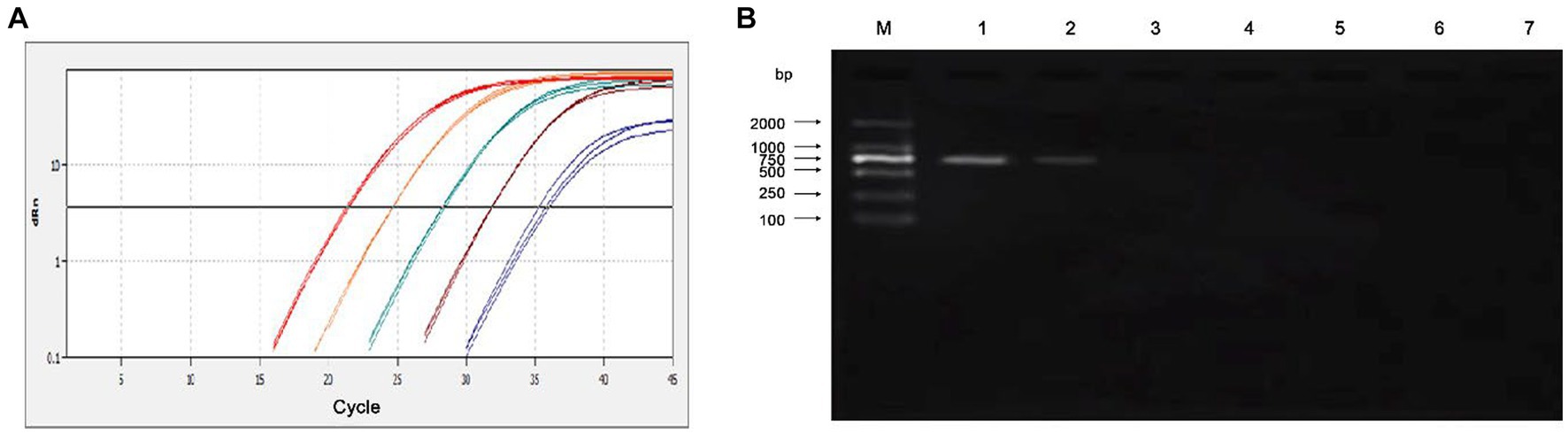
Figure 4. Sensitivity of the qPCR assay for the virus DNA of MiCV strain HB3. (A) The qPCR amplification curve. (B) Electrophoresis of conventional PCR reactions. The template concentrations of virus DNA ranged from 2.38 × 102 pg/μl to 2.38 × 10−3 pg/μl. A negative control was included.
The intra-and inter-assay reproducibility was assessed using three dilutions of the standard plasmid pMD-MiCV DNA (1.0 × 102 copies/μl, 1.0 × 104 copies/μl, and 1.0 × 106 copies/μl). The values of the intra-assay co-efficient of variation (CV) ranged from 0.46 to 0.96%, and the values of the inter-assay CV ranged from 1.37 to 1.98%, indicating that the qPCR method was highly reproducible.
The application of qPCR was evaluated by detecting 226 mink anal swabs, 142 fox anal swabs and 128 raccoon dog anal swabs. The positive detection rates of MiCV in minks, foxes and raccoon dogs were 58.8, 50.7, and 42.2%, respectively (Table 2). The total positive detection rate was 52.2%. The positive detection rate of MiCV in minks was higher than that in foxes and raccoon dogs. The positive detection rate of MiCV in fur-bearing animals ≥1 year old was 55.1%, and was 49.6% in fur-bearing animals <1 year old. The positive detection rate in male fur-bearing animals was 51.0%, and was 53.5% in female fur-bearing animals (Table 3). Environmental samples from fur-bearing animal farms had a higher positive detection rate of 75.1%. Among them, the positive rates were higher in samples from cages, troughs, soil under the cages and sewage under the cages than in samples from floors, breeder’s clothes, and equipment (Table 4).
For MiCV, there is no cell culture system for virus isolation and identification, and there is no effective vaccine or specific drug to prevent and control the disease. Some farmers used homemade inactivated vaccines (tissue supernatants of diseased mink inactivated using formalin) to prevent the disease and reduce morbidity; however, after a period of application, some minks developed symptoms of Aleutian disease. There are serious safety concerns with this type of vaccine; therefore, it has been discontinued (Wang et al., 2021). In view of this situation, the effective way to prevent the disease is to detect and eliminate the positive animals gradually in China. The recombinase polymerase amplification method had a minimum detection limit of 10 copies, but requires purification of amplification products and agarose gel electrophoresis to analyze the results (Ge et al., 2018c). The SYBR Green-based real-time PCR detection method for MiCV established by Cui could detect a minimum of 101 copies/μl, in which the specific primers referred to the Cap gene sequences of eight different MiCV strains (Cui et al., 2018).
In this study, a quantitative real-time PCR technology based on TaqMan was established. The standard curve of the qPCR was established as shown in Figure 1. The correlation coefficient (R2) was 0.996 and the amplification efficiency was 93%, which indicated that the method established in this study is highly efficient, and mighty be a suitable approach for MiCV diagnosis. The primers and probes were designed according to the whole genome sequences of MiCV obtained in the laboratory in recent years and all 13 of MiCV genome sequences deposited in GenBank. Besides, six viruses that could cause potential cross-reactivity in qPCR assays were assessed. The qPCR method could successfully detect MiCV and without any cross reaction with other viral pathogens, including AMDV, MEV, CDV, CAdV-2, PRV, and PCV-2, indicating the high specificity and reliability of this method for MiCV detection. For the plasmid DNA, the minimum detection limit of the established qPCR method was 1.0 × 101 copies/μl, whereas that for conventional PCR was 1.0 × 103 copies/μl. For the virus DNA of MiCV, the minimum detection limit of the established qPCR method was 2.38 × 10−2 pg/μl, the conventional PCR was 2.38 × 101 pg/μl. The results showed that the sensitivity of qPCR was higher than that of conventional PCR to detect MiCV. Moreover, the minimum detection limit of the qPCR method established in this study was lower than that of the previously established conventional PCR method and qPCR method (Wang et al., 2021). The values of the intra-assay CV ranged from 0.46 to 0.96%, and the values of the inter-assay CV ranged from 1.37 to 1.98%, which indicated that the qPCR method is highly reproducible.
MiCV infective rates in minks are high in some fur-bearing animal farms in China (Cui et al., 2018; Wang et al., 2021). In this study, we also detected a high positive detection rate of MiCV in minks, with positive detection rates exceeding 55% in Heilongjiang Province, Jilin Province, and Shandong Province. In clinical samples, the positive detection rate of MiCV in foxes was 52.4% in Heilongjiang Province, 50.9% in Jilin Province, and 48.9% in Shandong Province; in raccoon dogs it was 43.9% in Heilongjiang Province, 39.6% in Jilin Province, and 43.6% in Shandong Province. The total positive detection rate of MiCV in minks was higher (58.8%) than that in foxes (50.7%) and raccoon dogs (42.2%). However, the positive detection rates were lower than the 93% (40/43), 95.5% (21/22), and 69.2% (9/13) reported by Yang (Yang et al., 2018). We also found that the positive detection rate of MiCV in fur-bearing animals ≥ 1 year old was higher than that in animals <1 year old. The positive detection rate in male fur-bearing animals was lower than that in female fur-bearing animals. Usually when an infectious disease occurs on a farm, its pathogen can persist in the environment for a long time (Prieto et al., 2017). In this study we tested some environmental samples from cages, sinks, soil and sewage under cages. We found that the environmental samples of these fur-bearing animal farms have high positive detection rates. Among them, the samples from cages, troughs, soil under the cages, and sewage under cages had higher positive detection rates than the samples from aisle floors, breeder’s clothes and equipment. It is worth noting that these elements are frequently moved within the farm and in some exceptional cases are even transferred to other farms, which poses a significant risk to biosecurity. As far as we know, these results are the first application of the qPCR method to detect MiCV infection in environmental samples from fur-bearing animal farms. Unlike bacteria, there is no accepted standard culture method to quantify viruses in environmental samples; therefore, qPCR detection is a useful tool to study the epidemiology of viral diseases (Rodríguez-Lázaro et al., 2012). Although the presence of viral DNA cannot be interpreted as “infection” because its detection cannot determine infectivity, it might increase the risk of reinfection or transmission of the disease in the farms. These results focus attention on the role of environmental contamination in farms in the maintenance and transmission of this disease.
In this study, a highly sensitive, specific, repeatable and quantitative real-time PCR method for MiCV DNA detection was developed. The results showed that the positive detection rate of MiCV in minks was higher than that in foxes and raccoon dogs. The samples taken from the environment of the fur-bearing animal farms had high positive rates, and the positive detection rates in samples from cages, troughs, soil under the cages, and sewage under the cages were higher than those in the samples from the aisle floor, breeder clothes, and equipment. This study could contribute to control the spread of MiCV disease.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
The animal study was reviewed and approved by Animal Welffare and Research Ethics Committee of Jinlin Agricultural University (JLAU08201409).
YL and CS: conceived of the study, carried out the experiment and drafted the manuscript, conducting a research and investigation process, specifically performing the experiments, or data/evidence collection YZ and JL: provision of study materials, reagents, materials, patients, laboratory samples, animals, instrumentation, computing resources, or other analysis tools. QG, KS, and FL: management and coordination responsibility for the research activity and execution, including mentorship external to the core team. LX and ZC: application of statistical, mathematical, computational, or other formal techniques to analyze or synthesize study data, preparation, creation and/or presentation of the published work, specifically visualization/data presentation. XL: conceived of the study and revising the manuscript critically. RD: acquisition of the financial support for the project leading to this publication. All authors contributed to the article and approved the submitted version.
This research was supported by the Science and Technology Development Project of Jilin Province (grant numbers 20220101332JC and YDZJ202301ZYTS334) and the Science and Technology Research Project of Jilin Provincial Department of Education (grant number JJKH20210366KJ).
The authors grateful to all study participants for their contributions.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abdel-Moneim, A. S., Shehab, G. M., Alsulaimani, A. A., and Al-Malky, M. I. R. (2017). Development of Taq Man RT-qPCR for the detection of type a human respiratory syncytial virus. Mol. Cell. Probes 33, 16–19. doi: 10.1016/j.mcp.2017.02.006
Breitbart, M., Delwart, E., Rosario, K., Segalés, J., Varsani, A., and Ictv Report, C. (2017). ICTV virus taxonomy profile: Circoviridae. J. Gen. Virol. 98, 1997–1998. doi: 10.1099/jgv.0.000871
Canuti, M., Rodrigues, B., Bouchard, É., Whitney, H. G., Lang, A. S., Dufour, S. C., et al. (2022). Distinct epidemiological profiles of porcine circovirus 3 and fox circovirus in Canadian foxes (Vulpes spp.). Curr. Res. Micro. Sci. 3:100161. doi: 10.1016/j.crmicr.2022.100161
Cui, X., Shi, Y., Zhao, L., Gu, S., Wei, C., Yang, Y., et al. (2018). Application of real-time quantitative PCR to detect mink circovirus in naturally and experimentally infected minks. Front. Microbiol. 9:937. doi: 10.3389/fmicb.2018.00937
de Oliveira Lopes, G. A., Ferreira, L. R., de Souza Trindade, G., Fonseca, A. A. Jr., and Dos Reis, J. K. P. (2021). qPCR assay for the detection of Pseudocowpox virus. Arch. Virol. 166, 243–247. doi: 10.1007/s00705-020-04872-4
Decaro, N., Martella, V., Desario, C., Lanave, G., Circella, E., Cavalli, A., et al. (2014). Genomic characterization of a circovirus associated with fatal hemorrhagic enteritis in dog, Italy. PLoS One 9:e105909. doi: 10.1371/journal.pone.0105909
Ge, J., Cui, X., Shi, Y., Zhao, L., Wei, C., Wen, S., et al. (2018a). Development and application of an indirect enzyme-linked immunosorbent assay based on recombinant capsid protein for the detection of mink circovirus infection. BMC Vet. Res. 14:29. doi: 10.1186/s12917-018-1337-z
Ge, J., Gu, S., Cui, X., Zhao, L., Ma, D., Shi, Y., et al. (2018b). Genomic characterization of circoviruses associated with acute gastroenteritis in minks in northeastern China. Arch. Virol. 163, 2727–2735. doi: 10.1007/s00705-018-3908-5
Ge, J., Shi, Y., Cui, X., Gu, S., Zhao, L., and Chen, H. (2018c). Rapid and sensitive detection of mink circovirus by recombinase polymerase amplification. J. Virol. Methods 256, 1–5. doi: 10.1016/j.jviromet.2018.02.022
Gong, H. Y., Li, Q. Y., Zhang, H., Ye, L., Shi, L., and Feng, Y. H. (2021). Development and comparison of qPCR and qLAMP for rapid detection of the decapod iridescent virus 1 (DIV1). J. Invertebr. Pathol. 182:107567. doi: 10.1016/j.jip.2021.107567
Li, L., Kapoor, A., Slikas, B., Bamidele, O. S., Wang, C., Shaukat, S., et al. (2010). Multiple diverse circoviruses infect farm animals and are commonly found in human and chimpanzee feces. J. Virol. 84, 1674–1682. doi: 10.1128/jvi.02109-09
Matsumoto, T., Sato, M., Nishizono, A., and Ahmed, K. (2019). A novel bat-associated circovirus identified in northern Hokkaido, Japan. Arch. Virol. 164, 2179–2182. doi: 10.1007/s00705-019-04286-x
Morandini, V., Dugger, K. M., Ballard, G., Elrod, M., Schmidt, A., Ruoppolo, V., et al. (2019). Identification of a novel Adélie penguin circovirus at cape crozier (Ross Island, Antarctica). Viruses 11:1088. doi: 10.3390/v11121088
Piewbang, C., Jo, W. K., Puff, C., van der Vries, E., Kesdangsakonwut, S., Rungsipipat, A., et al. (2018). Novel canine circovirus strains from Thailand: evidence for genetic recombination. Sci. Rep. 8:7524. doi: 10.1038/s41598-018-25936-1
Prieto, A., Fernández-Antonio, R., Díaz-Cao, J. M., López, G., Díaz, P., Alonso, J. M., et al. (2017). Distribution of Aleutian mink disease virus contamination in the environment of infected mink farms. Vet. Microbiol. 204, 59–63. doi: 10.1016/j.vetmic.2017.04.013
Rodríguez-Lázaro, D., Cook, N., Ruggeri, F. M., Sellwood, J., Nasser, A., Nascimento, M. S., et al. (2012). Virus hazards from food, water and other contaminated environments. FEMS Microbiol. Rev. 36, 786–814. doi: 10.1111/j.1574-6976.2011.00306.x
Rosario, K., Breitbart, M., Harrach, B., Segalés, J., Delwart, E., Biagini, P., et al. (2017). Revisiting the taxonomy of the family Circoviridae: establishment of the genus Cyclovirus and removal of the genus Gyrovirus. Arch. Virol. 162, 1447–1463. doi: 10.1007/s00705-017-3247-y
Shulman, L. M., and Davidson, I. (2017). Viruses with circular single-stranded DNA genomes are everywhere! Ann. Rev. Virol. 4, 159–180. doi: 10.1146/annurev-virology-101416-041953
Tong, M., Sun, N., Cao, Z., Cheng, Y., Zhang, M., Cheng, S., et al. (2020). Molecular epidemiology of Aleutian mink disease virus from fecal swab of mink in Northeast China. BMC Microbiol. 20:234. doi: 10.1186/s12866-020-01910-8
Varga, A., and James, D. (2006). Real-time RT-PCR and SYBR green I melting curve analysis for the identification of plum pox virus strains C, EA, and W: effect of amplicon size, melt rate, and dye translocation. J. Virol. Methods 132, 146–153. doi: 10.1016/j.jviromet.2005.10.004
Vázquez, L., Guadamuro, L., Giganto, F., Mayo, B., and Flórez, A. B. (2017). Development and use of a real-time quantitative PCR method for detecting and quantifying Equol-producing bacteria in human Faecal samples and slurry cultures. Front. Microbiol. 8:1155. doi: 10.3389/fmicb.2017.01155
Wang, L., Zhang, Y., Chen, T., Mi, L., Sun, X., Zhou, X., et al. (2021). The mink circovirus capsid subunit expressed by recombinant baculovirus protects minks against refractory diarrhea in field. Viruses 13:606. doi: 10.3390/v13040606
Yang, Y., Cheng, Y., Li, N., Cheng, S., Guo, L., Zhou, Y., et al. (2018). Mink circovirus can infect minks, foxes and raccoon dogs. Virol. Sin. 33, 561–564. doi: 10.1007/s12250-018-0059-5
Keywords: Mink Circovirus , quantitative real-time PCR, epidemiological investigation, detection, fur-bearing animals
Citation: Liu Y, Sheng C, Zhou Y, Li J, Gong Q, Shi K, Liu F, Xu L, Cui Z, Leng X and Du R (2023) Application of quantitative real-time PCR to detect Mink Circovirus in minks, foxes and raccoon dogs in northern China. Front. Microbiol. 14:1205297. doi: 10.3389/fmicb.2023.1205297
Received: 13 April 2023; Accepted: 10 July 2023;
Published: 31 July 2023.
Edited by:
Jingqiang Ren, Wenzhou University, ChinaReviewed by:
Xiaobo Wen, Hainan University, ChinaCopyright © 2023 Liu, Sheng, Zhou, Li, Gong, Shi, Liu, Xu, Cui, Leng and Du. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xue Leng, eHVlbEBqbGF1LmVkdS5jbg==; Rui Du, ZHVydWkxOTcxMDFAc2luYS5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.