- Key Laboratory of New Animal Drug Project, Gansu Province/Key Laboratory of Veterinary Pharmaceutical Development, Ministry of Agriculture and Rural Affairs, Lanzhou Institute of Husbandry and Pharmaceutical Sciences of CAAS, Lanzhou, China
Escherichia coli is a common inhabitant of the intestinal microbiota and is responsible for udder infection in dairy cattle and gastro-urinary tract infections in humans. We isolated E. coli strains from a dairy farm environment in Xinjiang, China, and investigated their epidemiological characteristics, phenotypic and genotypic resistance to antimicrobials, virulence-associated genes, and phylogenetic relationship. A total of 209 samples were collected from different sources (feces, slurry, water, milk, soil) and cultured on differential and selective agar media (MAC and EMB). The presumptive identification was done by the VITEK2 system and confirmed by 16S rRNA gene amplification by PCR. Antimicrobial susceptibility testing was done by micro-dilution assay, and genomic characterization was done by simple and multiplex polymerase chain reaction (PCR). A total of 338 E. coli strains were identified from 141/209 (67.5%) of the samples. Most of the E. coli strains were resistant to sulfamethoxazole/trimethoprim (62.43%), followed by cefotaxime (44.08%), ampicillin (33.73%), ciprofloxacin (31.36%), tetracycline (28.99%), and a lesser extent to florfenicol (7.99%), gentamicin (4.44%), amikacin (1.77%), and fosfomycin (1.18%). All of the strains were susceptible to meropenem, tigecycline, and colistin sulfate. Among the resistant strains, 44.4% were identified as multi-drug resistant (MDR) showing resistance to at least one antibiotic from ≥3 classes of antibiotics. Eighteen out of 20 antibiotic-resistance genes (ARGs) were detected with sul2 (67.3%), blaTEM (56.3%), gyrA (73.6%), tet(B) (70.4%), aph(3)-I (85.7%), floR (44.4%), and fosA3 (100%, 1/1) being the predominant genes among different classes of antibiotics. Among the virulence-associated genes (VAGs), ompA was the most prevalent (86.69%) followed by ibeB (85.0%), traT (84.91%), ompT (73.96%), fyuA (23.1%), iroN (23.1%), and irp2 gene (21.9%). Most of the E. coli strains were classified under phylogenetic group B1 (75.45%), followed by A (18.34%), C (2.96%), D (1.18%), E (1.18%), and F (0.30%). The present study identified MDR E. coli strains carrying widely distributed ARGs and VAGs from the dairy environment. The findings suggested that the dairy farm environment may serve as a source of mastitis-causing pathogens in animals and horizontal transfer of antibiotic resistance and virulence genes carrying bacterial strains to humans via contaminated milk and meat, surface water and agricultural crops.
1. Introduction
Escherichia coli (E. coli) is an opportunistic and common inhabitant of the intestinal microbiota of animals as well as humans (He et al., 2019). Moreover, E. coli is also the most common organism responsible for causing udder infection in animals (Cheng et al., 2019) and bloodstream infections in humans (Jara et al., 2021). The use of antibiotics to prevent disease and promote the health of growing animals remains an integral part of livestock farming. It has been 50 years since antibiotic-supplemented feeds were first approved for livestock to improve overall health and increase the productivity of animals (Afema et al., 2018). However, the emergence and spread of pathogens resistance to multiple antibiotics has become a growing problem for veterinary medicine and public health (Murray et al., 2022). It was estimated that antimicrobial resistance results in $55 billion annual economic loss in USA (Dadgostar, 2019). The China also rank high in consumption of antibiotics for food producing animals especially in dairy sector.
Transmission of antimicrobial resistance (AMR) may occur by multiple ways, but contact with human and animal feces is the most common pathway (Graham et al., 2019). Dairy cattle act as a potential source of spread of antibiotic-resistant and zoonotic bacterial strains, especially Shiga-toxin producing E. coli (STEC) through the contamination of the farm environment and food products such as milk and meat, and direct contact with animals (Amézquita-López et al., 2018; Sobur et al., 2019). Animal farming, especially intensive livestock farming, plays a major role in AMR transmission between humans, animals, and the environment (Manyi-Loh et al., 2018). Due to the widespread use of antimicrobials in livestock production, livestock manure is considered a hotspot for the spread and transmission of AMR genes. Genetically diverse E. coli strains exist in animal manures, and their ability to survive in various ecological niches (Beattie et al., 2020). E. coli strains carrying blaCTX-M and blaCMY genes confer resistance to β-lactam antibiotics are frequently found in animal manure (Cookson et al., 2022). Therefore, animal manure is thought to be harming to animals via udder infections by environmental pathogens such as E. coli, humans via contaminated food products, and environment by using manure as fertilizer in soil or waste water (Sarowska et al., 2019). This increases the potential of antibiotic resistance genes (ARGs) to integrate into human intestinal microbiota by horizontal gene transfer mechanism (Lima et al., 2020). A better understanding of the transmission and spread of AMR, especially in areas with intensive livestock production, is important to understand. Therefore, the present study investigated the prevalence of E. coli in the dairy farm environment and their drug resistance characteristics. We also investigated the diversity of virulence associated genes (VAGs) responsible for pathogenicity and their distribution within phylogenetic groups.
2. Materials and methods
2.1. Sample sources and collection strategy
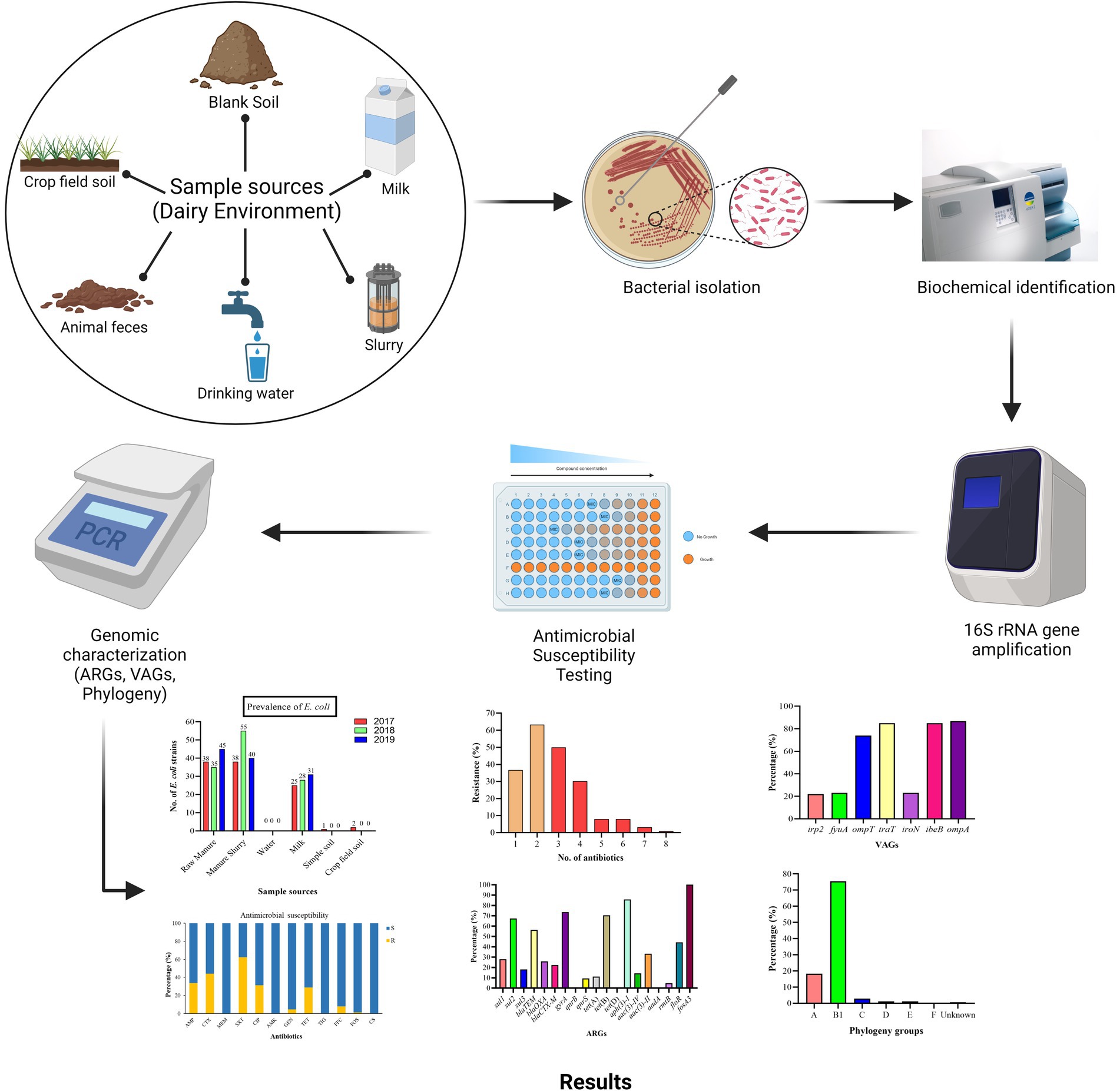
A total of 209 environment samples were collected from 2017–2019 from a large dairy farm (herd size = 25,000 animals) in Xinjiang province, China. The environmental samples included were fecal samples (n = 50), manure slurry from a storage tank (n = 36), raw milk (n = 90), water samples from the residential area (n = 9), soil samples (n = 12), and crop field soil (n = 12) based on random sampling technique (Figure 1). A 50 g of manure sample was collected from the animal living area and storage tank from five different sites using a five-point mixed sampling method (Sharp et al., 2012) and stored in sterile zipper bags. Raw milk samples (10 mL) were collected and transferred to sterile falcon tubes according to the guidelines of the National Mastitis Council (Hogan and Smith, 1992). The water samples (50 mL) were collected in sterile water bottles from the residential area by randomly selecting three different water outlets. The blank and crop field soil samples were collected from different sites on farm and fodder growing fields, respectively. All the collected samples were kept at 4°C and transferred to the laboratory within 24 h for further processing.
2.2. Isolation and identification of Escherichia coli
The 25 g of fecal, manure, and soil samples were first mixed in 225 mL of phosphate-buffered saline (PBS) to solubilize them. After mixing, 1 mL of the liquid was transferred to a 10 mL LB broth tube for bacterial enrichment by incubation at 37°C with continuous mixing at 160 rpm. From each tube, 100 μL of the enrichment culture was sub-cultured on MacConkey (MAC) agar under prior mentioned incubation conditions. However, the water and milk samples were swabbed directly onto MacConkey agar and incubated at 37°C overnight. Based on colony shape and color, large, smooth, and pink colonies were picked and further streaked onto Eosin Methylene Blue (EMB) agar and incubated overnight at 37°C. The appearance of a metallic green sheen with dark center colonies on EMB agar was indicative of E. coli growth (Peng et al., 2022). Further, presumptive identification was done by the VITEK2 system (BioMerieux, France) (Alfinete et al., 2022) and confirmation by 16S rRNA gene amplification by PCR using primers reported previously (Liu et al., 2021). The PCR-amplified product was visualized on 1% agarose gel under the GelDoc XR system (Supplementary Figure S1). The confirmed isolates were preserved in 20% glycerol at −80°C for further analysis.
2.3. Detection of virulence-associated genes
The 16S rRNA-confirmed E. coli isolates were subjected to the identification of seven VAGs by the previously described method (Hu et al., 2022). The genomic DNA was extracted using a DNA extraction kit (Tiangen Biotech Beijing, Co., Ltd.) following the manufacturer’s guidelines. The virulence genes were identified by PCR amplification of target gene primers mentioned in Supplementary Table S1. The PCR reaction mixture (25 μL) consisted of 12.5 μL PreTaq Mix (Vazyme Biotech, China), 1 μL forward primer, 1 μL reverse primer, 1 μL genomic DNA, and 9.5 μL of deionized water under the following conditions; prior denaturation at 94°C for 5 min followed by 35 cycles of denaturation at 94°C for the 30s, annealing at varying temperatures mentioned in Supplementary Table S1 for 30s, initial extension at 72°C for 30s followed by a final extension at 72°C for 5 min. After amplification, the PCR product was run on 1% agarose gel electrophoresis at 180 V/200 mA followed by ethidium bromide staining for visualization, and images were taken under the GelDoc XR system (Supplementary Figure S2).
2.4. Antimicrobial susceptibility testing
The AST was done by broth micro-dilution assay following the EUCAST guidelines.1 Briefly, the preserved isolates were thawed at room temperature and re-suspended in LH broth by vigorous mixing (120 rpm) at 37°C for 12 h. The loopful enriched broth was streaked on MacConkey agar following the overnight incubation. The bacterial inoculum was prepared by adjusting the cell density at 5 × 105 CFU/mL. The 96-well round bottom plate was used for broth dilution assay and 100 μL of Mueller Hinton (MH) broth was added from the 1st well to the 12th well with a micropipette. Next, 50 μL of prepared bacterial inoculum was added from the 1st to 11th well by keeping the 12th well as a negative control. The antibiotics were selected based on medical and veterinary use which includes trimethoprim-sulfamethoxazole (SXT), ampicillin (AMP), cefotaxime (CTX), tetracycline (TET), ciprofloxacin (CIP), gentamicin (GEN), amikacin (AMK), colistin sulfate (CS), florfenicol (FFC), meropenem (MEM), and tigecycline (TIG) were added from 1st to 10th well by keeping 11th well as a positive control. The reference strain E. coli ATCC 25922 was used as a quality control. The MIC (minimum concentration that inhibits visible growth of bacteria) of fosfomycin was calculated by the agar dilution method, recommended by EUCAST. The MIC of all antibiotics was evaluated by visualizing the growth in the bottom of the plate well as tinny buttons/turbidity. The MIC values were compared with standard EUCAST MIC breakpoints (Supplementary Table S2). The strains showing resistance to at least one antibiotic from ≥3 classes were classified as MDR.
2.5. Detection of antibiotic resistance genes
Phenotypically resistant E. coli strains were subjected to the detection of 20 ARGs from eight antibiotic classes (Supplementary Table S3) according to the method described previously (Yu et al., 2020). The bacterial DNA was extracted using a DNA extraction kit (Tiangen Biotech Beijing, Co., Ltd.) and used as a template for PCR amplification of 20 ARGs (listed in Supplementary Table S3). The PCR reaction mixture (25 μL) consisted of 12.5 μL PreTaq Mix (Vazyme Biotech, China), 1 μL of forward and reverse primer each, 1 μL of bacterial DNA, and 9.5 μL of deionized water. The reactions were performed under the following conditions: denaturation at 94°C for 5 min followed by 35 cycles of denaturation at 94°C for 30s, annealing at varying temperatures (see Supplementary Table S3) for 30s, and extension at 72°C for 30s, followed by a final extension at 72°C for 5 min. After amplification, the PCR product was separated on 1% agarose gel at 180 V/200 mA and stained with ethidium bromide for visualization using the GelDoc XR system.
2.6. Phylogenetic analysis
The phylogenetic grouping of E. coli strains was carried out by 2 sets of PCR using primers listed in Supplementary Table S4. The quadruple PCR reaction mixture (25 μL) consists of Premix Taq TM 12.5 μL, 1 μL of each forward and reverse primers (chuA, yjaA, tspE4C2), 2 μL of each arpA forward and reverse primers, 1.5 μL DNA template, and 1 μL dd H2O. PCR was carried out under the following conditions; pre-denaturation at 94°C for 4 min, 30 cycles of denaturation at 94°C for 5 s followed by annealing at 59°C for 20 s, and extension at 72°C for 5 min. The PCR reaction for group E and C identification consisted of Premix Taq TM 12.5 μL, 0.6 μL of trpBA forward and reverse primers each, 1 μL of each group-specific primer (Supplementary Table S4), 1.5 μL DNA template, and 2.8 μL dd H2O. In the PCR reaction solution, trpBA primers were added as an internal control. The PCR amplifications conditions were pre-denaturation at 94°C for 4 min, 30 cycles of denaturation at 94°C for 5 s, annealing at 57°C (group E) or 59°C (group C) for 20 s, and final extension at 72°C for 5 min. After the PCR amplification, the PCR product was run on 1% agarose gel and visualized under the GelDoc XR system (Supplementary Figure S3), and the phylogenetic group was identified by comparing the results with Supplementary Table S4.
2.7. Data analysis
The prevalence was calculated using the formula described by Thrusfield (2018).
The antimicrobial susceptibility data were analyzed by descriptive statistics using Microsoft Excel. Moreover, the data for various factors such as sampling source and sampling year affecting the prevalence, AMR and virulence rates were analyzed using the Pearson’s Chi-Squared test keeping the level of significance, α = 5% (Zhao et al., 2021; Ma et al., 2022). p-value < 0.05 was considered statistically significant and vice versa. The graphical representation of data was done by GraphPad Prism version 8.2.1 and Microsoft Excel.
3. Results
3.1. Isolation of Escherichia coli strain from different sources
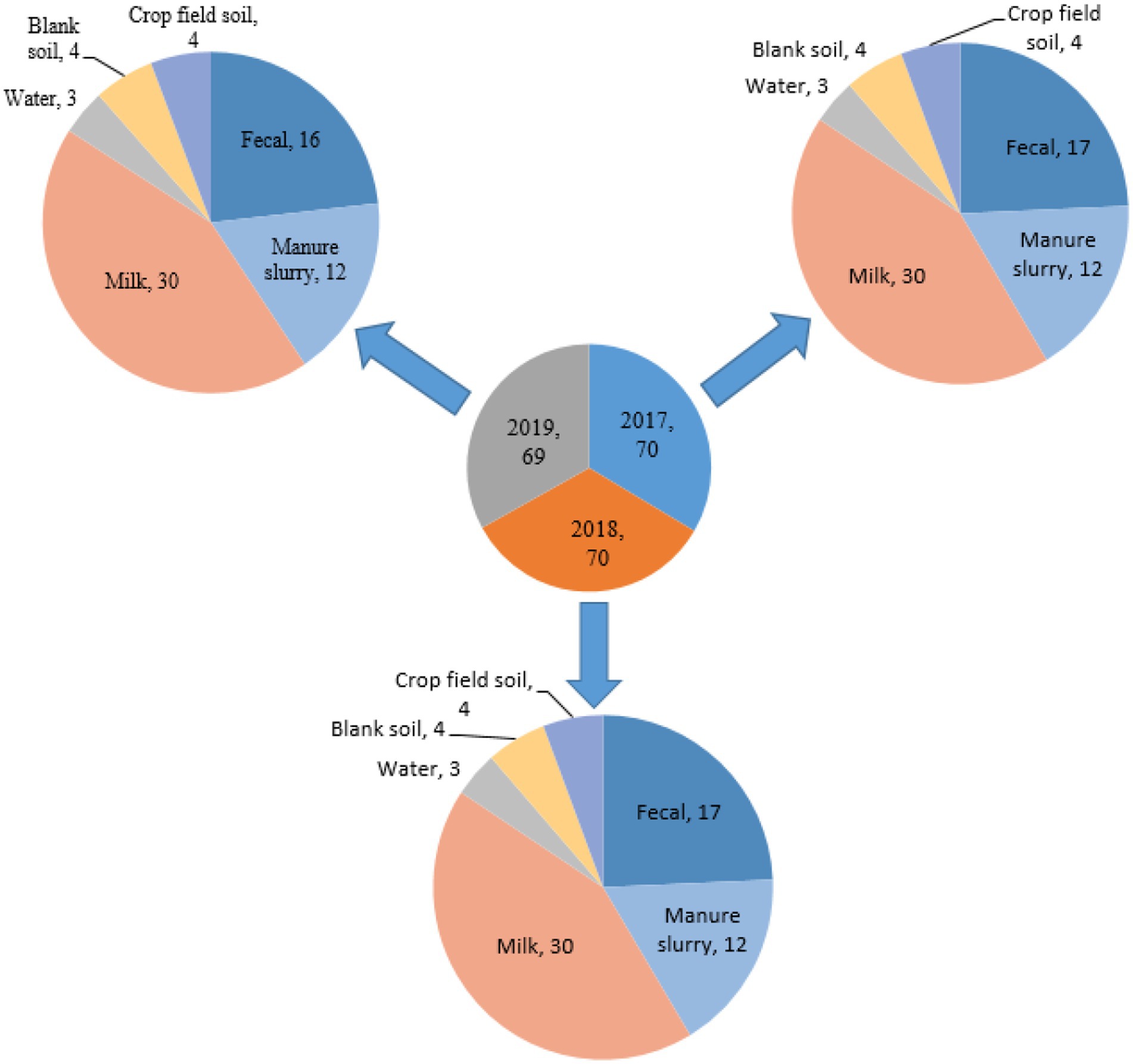
A total of 209 samples were collected from different sites of dairy environment including fecal samples (n = 50), manure slurry from the storage tank (n = 36), raw milk (n = 90), water samples (n = 9), soil samples (n = 12), and crop field soil (n = 12) samples. In total, 534 suspected E. coli strains were isolated from 141/209 (67.5%) samples based on colony characteristics. Subsequently, 338 E. coli strains were confirmed by 16S rRNA gene amplification. The isolation rates were comparable over the years, with 30.8% (104/338) in 2017, 34.9% (118/338) in 2018, and 34.3% (116/338) in 2019 (Figure 2A). Overall, most of the E. coli strains were isolated from manure slurry (39.3%, 133/338), followed by fecal samples (34.9%, 118/338), raw milk (24.8%, 84/338), crop field soil (0.59%, 2/338), and least from blank soil (0.29%, 1/338). However, none of the E. coli strains was isolated from water samples (Figure 2B). In 2019, a higher number of E. coli strains were isolated from fecal and milk samples compared to other sampling years while more E. coli strains were isolated from slurry samples in 2018. Moreover, only 1 and 2 strains were isolated in 2017 from blank and crop field soil samples, respectively, while none in 2018 and 2019 (Figure 2B).
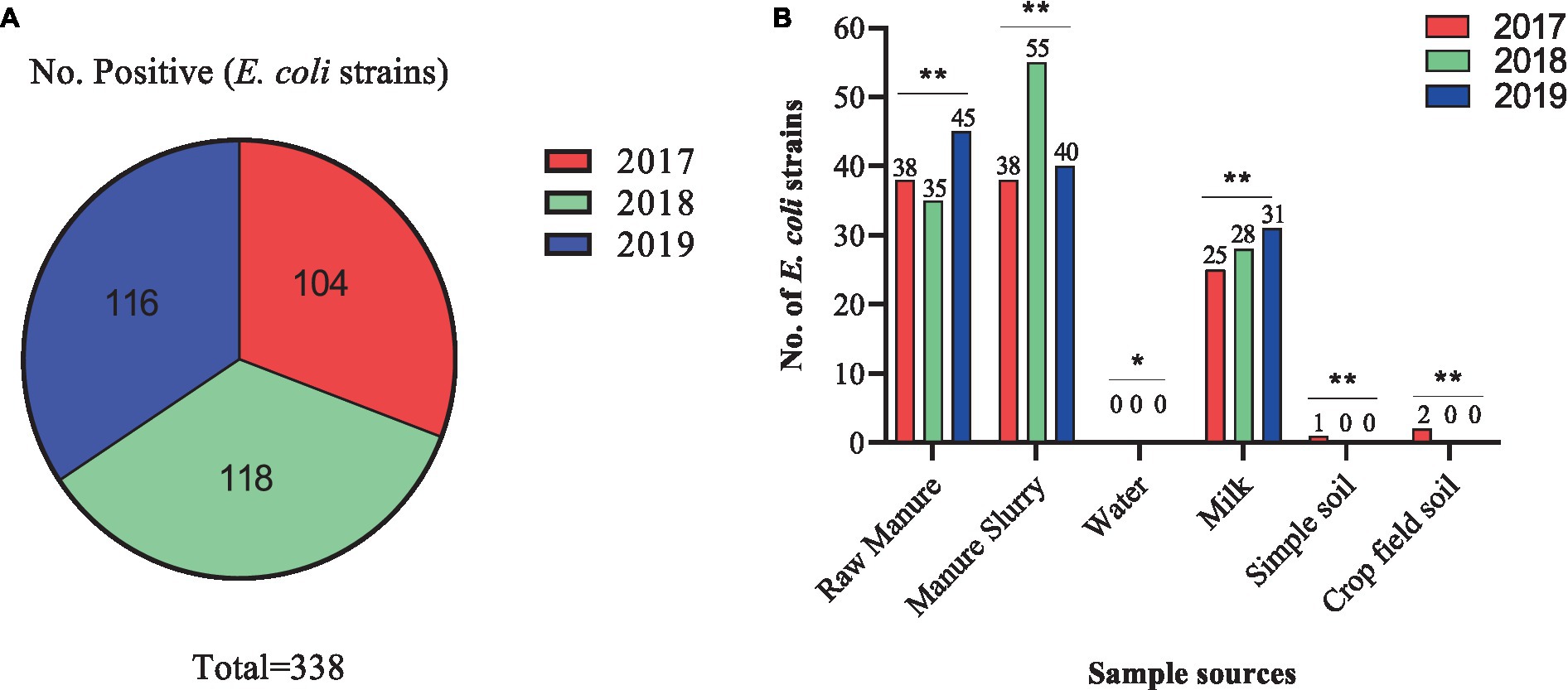
Figure 2. Isolation of Escherichia coli strains in different sampling years and sources. (A) Total number of E. coli strains isolated in different years. (B) Distribution of E. coli strains isolated from different sources in different years; **, indicate non-significant difference (p > 0.05); *, indicate not applicable; ***, indicate significant difference (p < 0.05).
3.2. Antimicrobial susceptibility of Escherichia coli strains
The AST of 338 E. coli strains showed that 284/338 (84.0%) were resistant to at least one antibiotic and 54/338 (26.0%) were susceptible strains (Figure 3A). Most of the strains were resistant to trimethoprim/ sulfamethoxazole (62.43%, 211/338), followed by cefotaxime (44.08%, 149/338), ampicillin (33.73%, 114/338), ciprofloxacin (31.36%, 106/338), tetracycline (28.99%, 98/338), and less to florfenicol (7.99, 27/338), gentamicin (4.44%, 15/338), amikacin (1.77%, 6/338), and fosfomycin (1.18%, 4/338). All of the E. coli strains were susceptible to meropenem, tigecycline, and colistin sulfate (Figure 3B). All E. coli strains from 2017–2019 were found 100% susceptible to meropenem, tigecycline, and colistin sulfate. Additionally, the AMR rate of AMP (44.23%), CIP (40.38%), TET (44.23%), and GEN (4.81%) was noted higher with a significant difference (p < 0.05) in 2017 than in other sampling years. Moreover, E. coli strains showed higher AMR to CTX (55.93%), SXT (68.64%), and AMK (3.39%) in 2018 with a significant difference (p < 0.05). However, none of the E. coli strain from 2017 and 2019 exhibited resistance to AMK and FOS (Figure 3C).
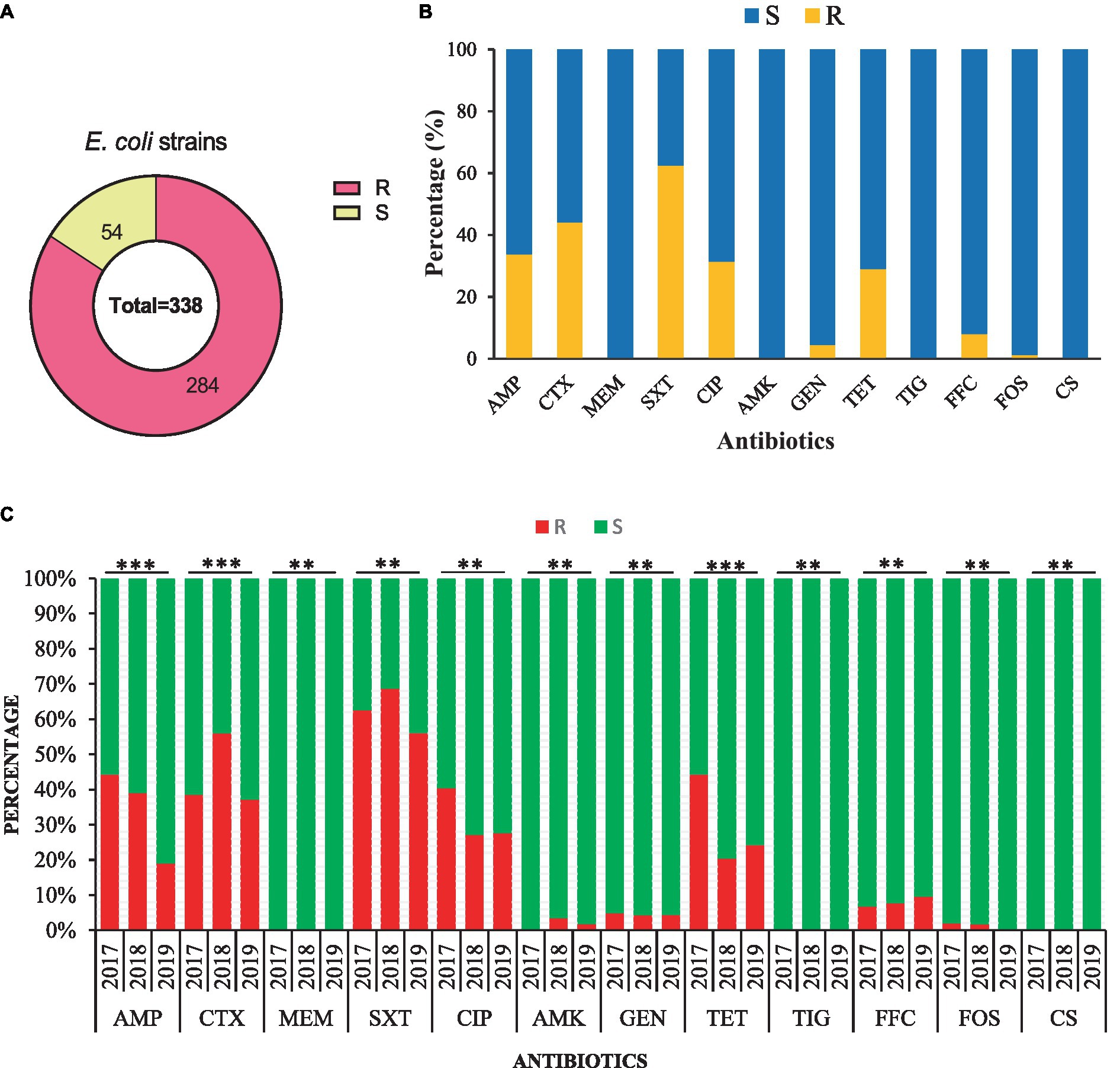
Figure 3. Antimicrobial susceptibility of the 338 E. coli strains isolated from the dairy environment. (A) The overall resistant (R) and susceptible (S) E. coli strains. (B) The overall antimicrobial susceptibility of 338 E. coli strains against individual antibiotic tested. (C) The comparison of antimicrobial susceptibility of E. coli strains from different sampling years. AMP, ampicillin; CTX, cefotaxime; MEM, meropenem; SXT, trimethoprim-sulfamethoxazole; CIP, ciprofloxacin; AMK, amikacin; GEN, gentamicin; TET, tetracycline; TIG, tigecycline; FFC, florfenicol; FOS, fosfomycin; CS, colistin sulfate; **, indicate non-significant difference (p > 0.05); *, indicate not applicable; ***, indicate significant difference (p < 0.05).
3.3. AMR characteristics of Escherichia coli strains isolated from different sources
Most of the E. coli strains from all samples were resistant to trimethoprim/sulfamethoxazole (SXT) and 100% susceptible to meropenem (MEM), tigecycline (TIG), and colistin sulfate (CS). Moreover, E. coli strains from fecal samples exhibited higher resistance to ampicillin (AMP), ciprofloxacin (CIP), and tetracycline (TET) in 2017 than other sampling years with a significant difference (Figure 4A). A similar trend was observed for E. coli strains isolated from milk and manure slurry (Figures 4B,C). Furthermore, E. coli strains isolated from blank and crop field soil in 2017 exhibited 100% resistant to CTX, CIP, TET, and SXT, while none of the strains isolated from 2018 and 2019 was resistant (Figures 4D,E). In addition, E. coli strains from crop field showed 50% resistance to AMP and florfenicol (FFC).
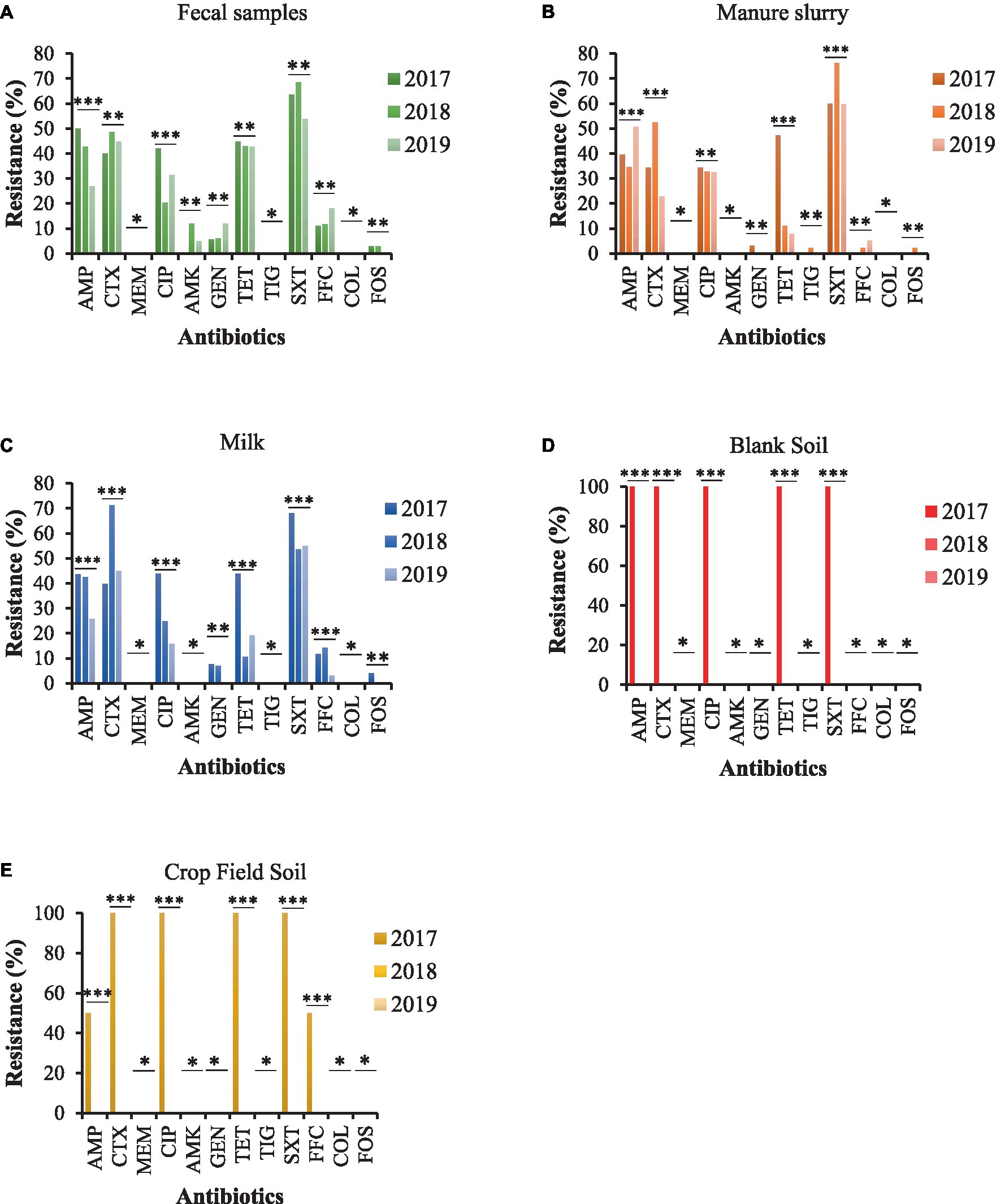
Figure 4. AMR rates of E. coli strains isolated from different sources. (A) fecal sample. (B) manure slurry from the storage tank. (C) raw milk. (D) blank soil. (E) crop field soil. AMP, ampicillin; CTX, cefotaxime; MEM, meropenem; CIP, ciprofloxacin; AMK, amikacin; GEN, gentamicin; TET, tetracycline; TIG, tigecycline; SXT, trimethoprim/sulfamethoxazole; FOS, fosfomycin; COL, colistin; FFC, florfenicol; **, indicate non-significant difference (p > 0.05); *, indicate not applicable; ***, indicate significant difference (p < 0.05).
3.4. Drug resistance spectrum
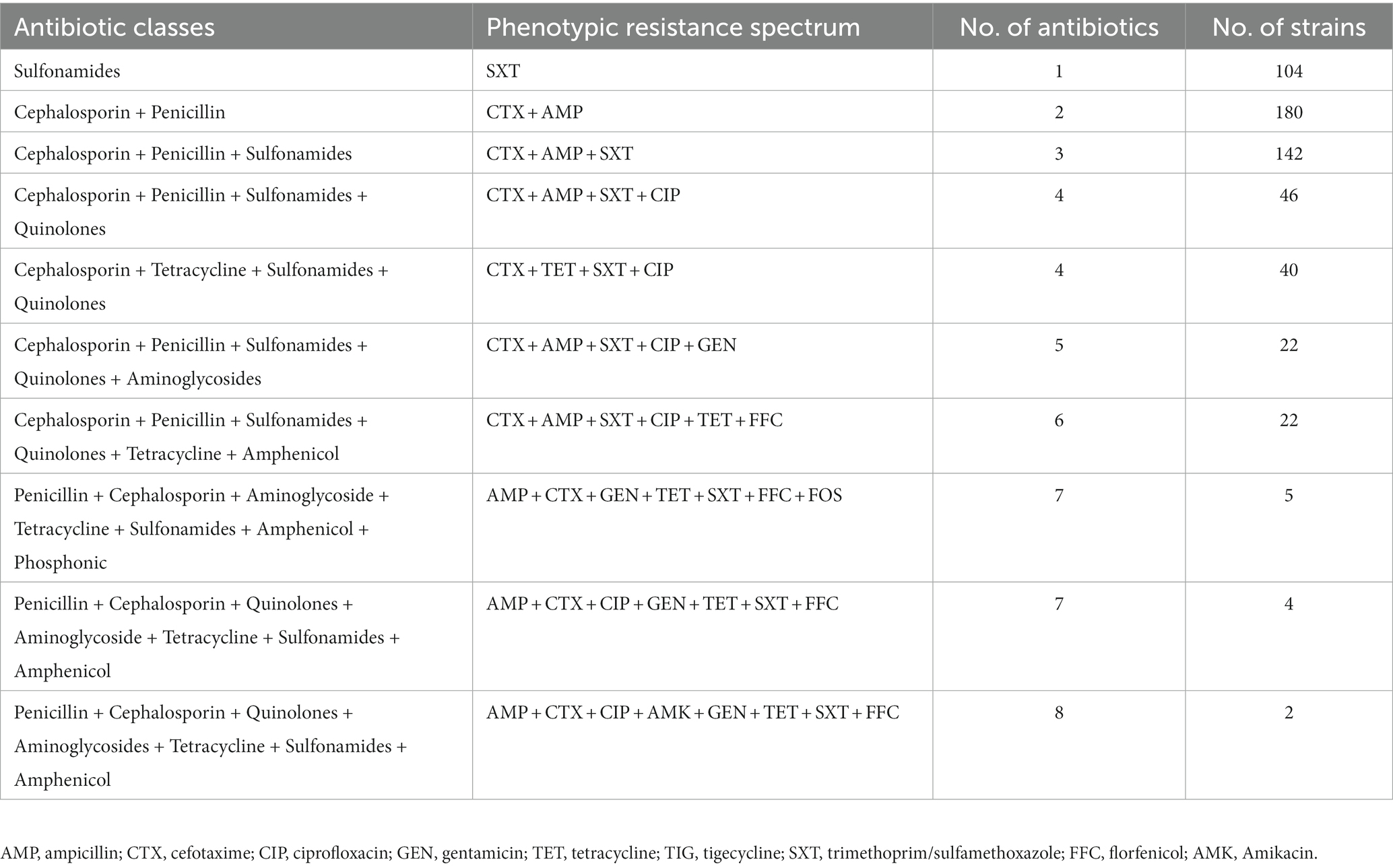
Among the resistant strains, 126/284 (44.4%) were identified as multi-drug resistant (MDR) and 158/284 (55.6%) were recognized as non-MDR (Figure 5A). Most of the strains showed resistance to 2 antibiotics (63.38%), followed by 3 (50.0%), 1 (36.61%), 4 (30.28%), 5 and 6 (7.75% each), 7 (3.17%), and 8 (0.70%) antibiotics (Figure 5B). Moreover, diverse AMR patterns were recognized such as CTX + AMP, CTX + AMP + SXT, CTX + AMP + SXT + CIP, CTX + TET + SXT + CIP, CTX + AMP + SXT + CIP + GEN, CTX +AMP + SXT + CIP + TET + FFC, AMP + CTX + GEN + TET + SXT + FFC + FOS, and AMP + CTX + CIP + GEN + TET + SXT + FFC. Only 1 strain showed resistance to 8 antibiotics, AMP + CTX + CIP + AMK + GEN + TET + SXT + FFC (Table 1).
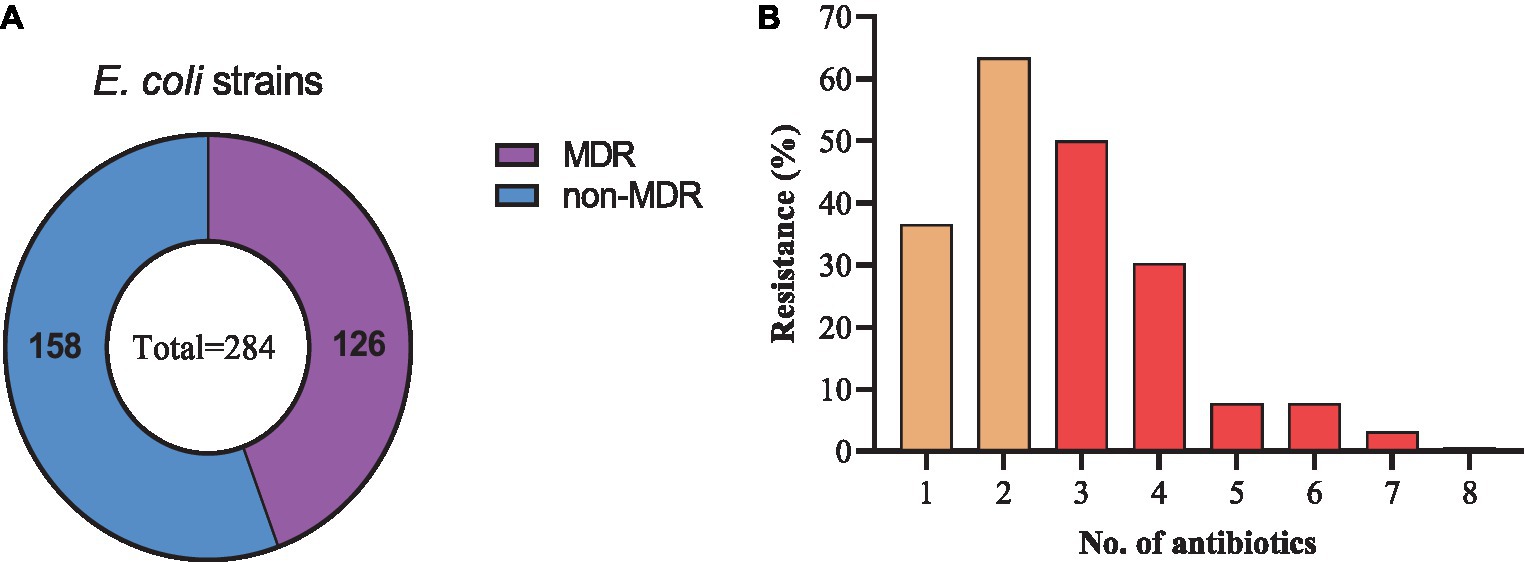
Figure 5. Drug-resistance spectrum of E. coli strains isolated from the dairy environment. (A) proportions of MDR and non-MDR strains. (B) Percentage resistance spectrum of 284 E. coli strains to 1 ~ 8 antibiotics.
3.5. Detection of ARGs and correlation with phenotypic resistance
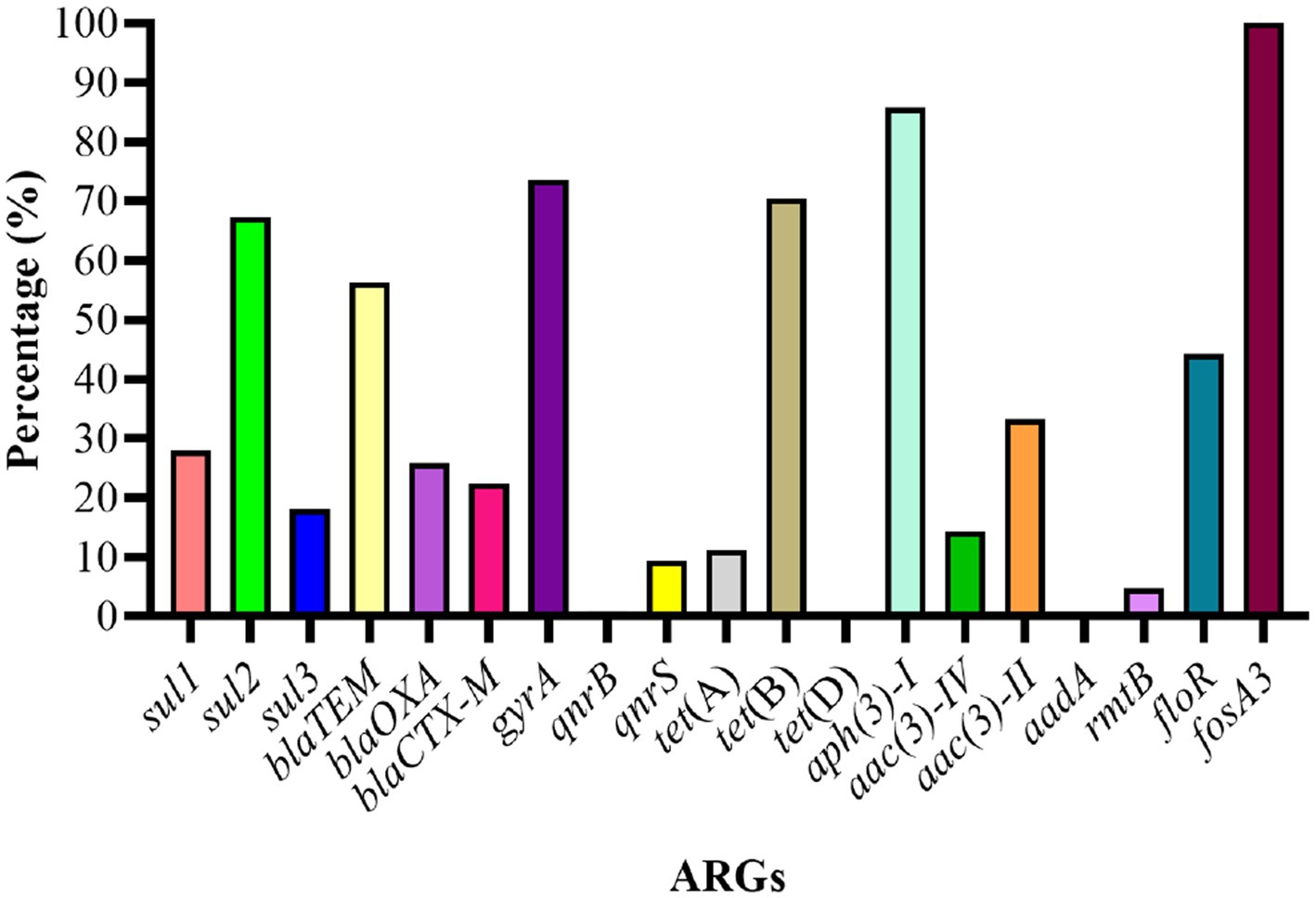
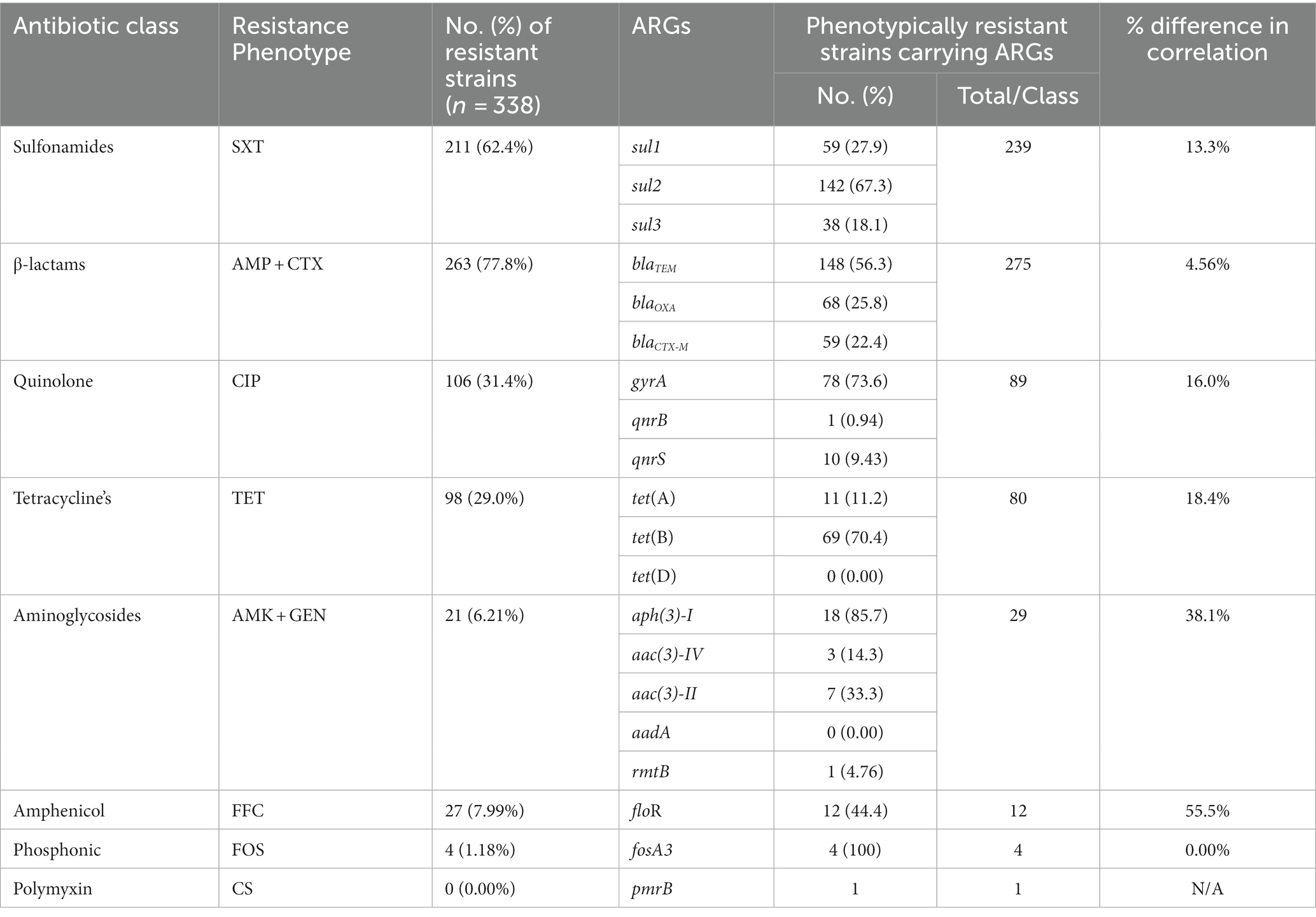
The genotypic analysis was done by targeting 20 ARGs among 8 classes of antibiotics (mentioned in Table 1). Eighteen out of 20 ARGs were identified, and the prevalent genotypes included sul2 (67.3%, sulfonamides), blaTEM (56.3%, beta-lactams), gyrA (73.6%, quinolones), tet(B) (70.4%, tetracycline’s), aph(3)-I (85.7%, aminoglycosides), floR (44.4%, amphenicol), and fosA3 (100%, phosphonic). The percentage distribution of other ARGs identified was as follows: sulfonamides (sul1, 27.9%; sul3, 18.1%), β-lactams (blaOXA, 25.8%; blaCTX-M, 22.4%), aminoglycosides (aac(3)-IV, 14.3%; aac(3)-II, 33.3%; aadA, 0.00%; rmtB, 4.76%), quinolones (qnrB, 0.94%; qnrS, 9.43%), polymyxin (pmrB, 0.35%) and tetracycline’s (tet(A), 11.2%; tet(D), 0.00%) (Figure 6).
The correlation between phenotypic resistance and genotypic detection of ARGs was noted variable. For example, no strains showed resistance to colistin sulfate phenotypically but one strain was carrying the ARG upon genotypic analysis. Moreover, the number of strains carrying ARGs was noted higher as compared to phenotypic resistance among sulfonamides, beta-lactams, and aminoglycosides-resistant strains while the inverse was noted among quinolone, tetracycline, and amphenicol-resistant strains. However, the phenotypic and genotypic expression was observed 100% correlated for fosfomycin-resistant strains (Table 2).
3.6. Virulome gene analysis
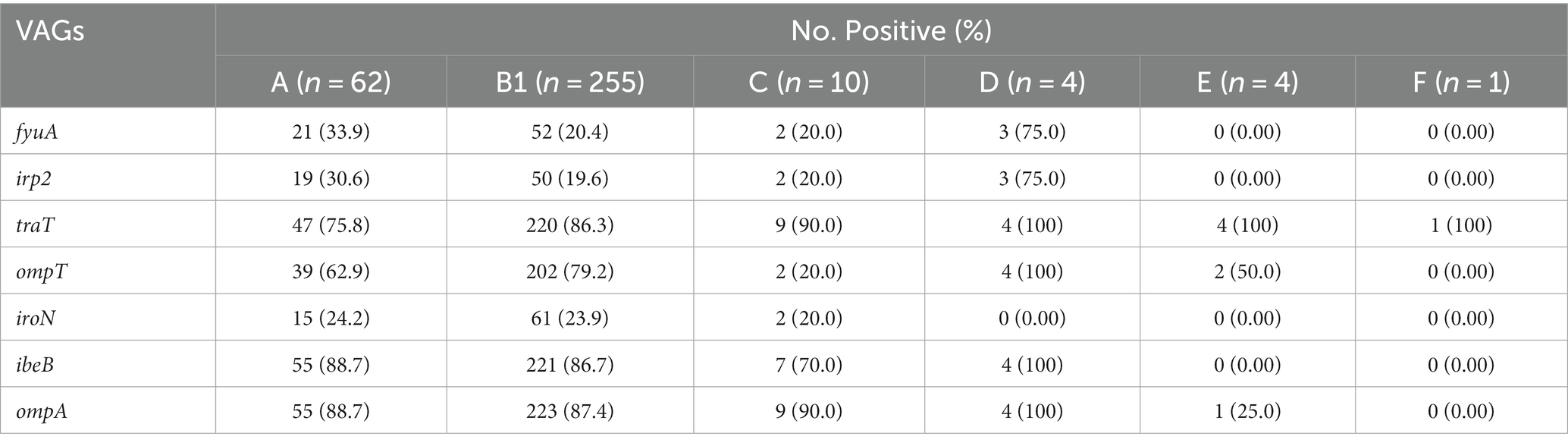
Among the VAGs, ompA was most prevalent (86.69%), followed by ibeB (85.0%), traT (84.91%), ompT (73.96%), fyuA (23.1%), iroN (23.1%), and irp2 (21.9%) (Figure 7A). All of the E. coli strains carrying the fyaA gene were also carrying the iroN gene. Moreover, 93.59% of E. coli strains carrying irp2 were also harboring the fyuA gene. VAGs such as ompT, traT, iroN, ibeB, and ompA were detected in E. coli strains from all sources while irp2 and fyuA genes were not observed from manure slurry and fecal samples, respectively. However, both irp2 and fyuA genes (36.9%, 31/84) were identified in strains of milk origin. Collectively, a higher percentage of VAGs was identified in strains of milk origin as compared to feces and slurry (Figure 7B).
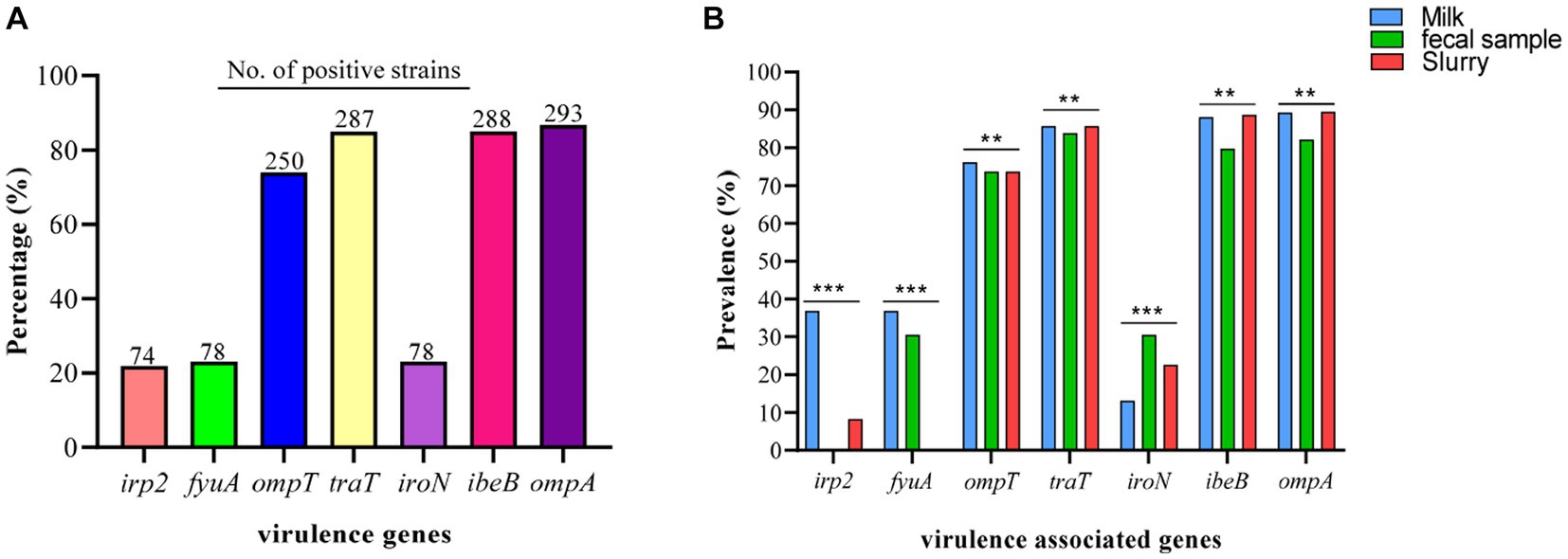
Figure 7. Escherichia coli strains carrying virulence-associated genes (VAGs). (A) Overall percentage and number of positive E. coli strains to carry VAGs. (B) Percentage distribution of VAGs among the E. coli strains isolated from different sources.
3.7. Distribution of VAGs among phylogenetic groups
The phylogenetic analysis of 338 E. coli strains showed most of the strains belong to the B1 group (75.45%, 255/338), followed by A (18.34%, 62/338), C (2.96%, 10/338), D (1.18%, 4/338), E (1.18%, 4/338), and F (0.30%, 1/338). However, the phylogenetic group for 2 of the strains was not identified. The most prevalent VAGs among the various phylogenetic groups were as follows; B1 (ompA, 87.4%), A (ibeB and ompA, 88.7%), C (traT and ompA, 90.0%), D (traT, ibeB, and ompA, 100%), E (traT, 100%), and F (traT, 100%). Moreover, the percentage distribution of other VAGs among the phylogenetic groups is presented in Table 3.
4. Discussion
Antimicrobial resistance particularly in the Enterobacteriaceae family possesses a major threat to global public health. The present study isolated E. coli from the dairy environment, which serves as a reservoir of bacterial pathogens and ARGs and a source of spread of ARGs between the bacterial species via horizontal gene transfer and to humans via fecal contamination of drinking water and milk. The isolation rates of E. coli in this study were found similar to the findings of Sobur et al. (2019) who reported 75% prevalence of E. coli from dairy cattle and farm environment. Other studies conducted by Li et al. (2022) reported 84.6% E. coli isolation from fecal samples of cattle, chicken, and pigs while 34.4% was noted by Liu et al. (2021) from raw milk samples and 81.1% from raw cheese (Imre et al., 2022). Beattie et al. (2020) also reported a similar isolation rate of E. coli from dairy manure in the USA. It is also reported that the presence of E. coli in the dairy environment may be the cause of clinical mastitis in dairy cows (Su et al., 2016).
The antimicrobial susceptibility results showed higher resistance to SXT, followed by CTX, AMP, CIP, TET, and the least resistance to FFC, GEN, AMK, and FOS. These results are consistent with the findings of Peng et al. (2022) who isolated E. coli strains from pigs that were highly resistant to SXT (80.38%), AMP (92.86%), and TET (96.26%). A similar study conducted by Beattie et al. (2020) reported that E. coli strains from manure isolates showed higher resistance to AMP and CTX. However, a lower resistance rate to AMK and GEN was also noted by Lu et al. (2022) and Liu et al. (2021) respectively. We noted the E. coli strains were susceptible to the “last resort” antimicrobials such as MEM, TIG, and CS, which is consistent with the findings of a study conducted by Hu et al. (2019) in the human setting. Moreover, other studies conducted by Wang et al. (2021), Zou et al. (2021), and Ma et al. (2022) in animal settings also reported 100% susceptibility of MEM and TIG against E. coli strains. We noted no resistance to CS, which is consistent with a previous report that colistin resistance is decreasing in animal and human settings because of the CS ban in China (Wang et al., 2020). The percentage of MDR E. coli was noted at 44.4% in the current study, which is comparable with what was previously reported (54.4%) by Su et al. (2016) and lower than what was reported by Yu et al. (2020) in dairy milk. Another study conducted by Salinas et al. (2019) reported higher resistance to SXT, CIP, AMP, CTX, and TET by E. coli isolated from child and domestic animal origin, which is also consistent with current findings.
We identified 18 ARGs out of 20 belonging to different classes of antibiotics. The most prevalent ARGs were sul2 (67.3%, sulfonamides), blaTEM (56.3%, beta-lactam), gyrA (73.6%, quinolones), tet(B) (70.4%, tetracycline’s), aph(3)-I (85.7%, aminoglycosides), floR (44.4%, amphenicol), and fosA3 (100%, fosfomycin). Previous studies reported AMR in humans is linked to food animals raised for milk and meat purposes because of environmental contamination and drug residues (Bacanlı and Başaran, 2019; Pormohammad et al., 2019; Ma et al., 2021). The use of antimicrobial drugs in food animals also enhances the percentage of MDR bacteria and ARGs in human microbiota (Ma et al., 2022). Moreover, E. coli is also known to serve as donor bacteria for horizontal gene transfer within and between species (Oladeinde et al., 2019). Lima et al. (2020) highlighted the importance of animal manure and manure-substituted agriculture lands as a major source of antibiotic residues, ARGs, and AMR bacteria in the environment posing a potential threat to public health via horizontal gene transfer mechanisms with the help of mobile genetic elements such as plasmids, transposons, and integrons. Qian et al. (2018) detected 109 ARGs from the fresh manure of chicken, cattle, and pigs responsible for AMR to a class of antibiotics widely used in human and animal settings.
We investigated multiple VAGs in the isolated E. coli and most of the investigated VAGs (ompA, ibeB, traT, ompT, fyuA, iroN, irp2) belong to ExPEC, which may cause urinary tract infections in humans. VAGs are responsible for the production of virulence factors which play an important role in the pathogenicity of bacteria through multiple mechanisms such as adhesion, invasion, toxin production, and immune evasion (Kudva et al., 2020). Virulence genes investigated in the present study have various functions. ompA encodes for outer membrane protein A, ibeB is an invasion protein gene, traT encodes for complement resistance protein, ompT encodes for outer membrane protease protein, fyuA encodes for yersiniabactin receptor, iroN encodes for aerobactin receptor, and irp2 gene encodes for iron-responsive element binding protein 2. Zhang et al. (2021) investigated similar VAGs in E. coli strains isolated from healthy waterfowls in Hainan, China. A study carried out by Raimondi et al. (2019) also identified similar VAGs in E. coli isolated from the feces of healthy individuals in Italy. Another study conducted by Khalifeh and Obaidat (2022) identified the iroN gene in E. coli strains from milk and fecal origin similar to the present study. In the current study, most of the E. coli strains were classified under phylogenetic group B1, which is consistent with the findings of Raimondi et al. (2019). These results suggest that the occurrence of ARGs and VAGs may vary by antimicrobial use and other unknown factors. This study also suggests regular monitoring of antimicrobial usage on dairy farms and proper manure treatment before disposal be ensured.
5. Conclusion
The present study identified multi-drug resistant E. coli strains carrying various ARGs and VAGs in the dairy environment, which may pose a potential threat to human, animal, and environmental health. Moreover, all of the E. coli strains were susceptible to meropenem, tigecycline, and colistin sulfate, which may be considered as critical antibiotics for therapeutic purposes in human and animal settings. Given the widespread distribution of AMR in the dairy environment, it is a potential reservoir of transferring ARGs genes to humans via various direct and indirect gene transfer mechanisms. This prudent the use of antibiotics on dairy farms, proper manure treatment, and enhancement of sanitation, especially in milk processing and transportation, are necessary to reduce the risk to food safety, public health, and environmental health.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
MS: writing–original draft. ZH and XG: data curation and formal analysis. MT and RH: graphical representation of data. SW and RS: review and editing. XW and HZ: project administration. WP: conceptualization, supervision, project visualization, and funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Key Research and Development Program (2016YFD0501305) and the Agricultural Science and Technology Innovation Program of the Chinese Academy of Agricultural Sciences (25-LZIHPS-03).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1197579/full#supplementary-material
Footnotes
References
Afema, J. A., Ahmed, S., Besser, T. E., Jones, L. P., Sischo, W. M., and Davis, M. A. (2018). Molecular epidemiology of dairy cattle-associated Escherichia coli carrying Bla CTX-M genes in Washington state. Appl. Environ. Microbiol. 84, e02430–e02417. doi: 10.1128/AEM.02430-17
Alfinete, N. W., Bolukaoto, J. Y., Heine, L., Potgieter, N., and Barnard, T. G. (2022). Virulence and phylogenetic analysis of enteric pathogenic Escherichia coli isolated from children with diarrhoea in South Africa. Int. J. Infect. Dis. 114, 226–232. doi: 10.1016/j.ijid.2021.11.017
Amézquita-López, B. A., Soto-Beltrán, M., Lee, B. G., Yambao, J. C., and Quiñones, B. (2018). Isolation, genotyping and antimicrobial resistance of Shiga toxin-producing Escherichia coli. J. Microbiol. Immunol. Infect. 51, 425–434. doi: 10.1016/j.jmii.2017.07.004
Bacanlı, M., and Başaran, N. (2019). Importance of antibiotic residues in animal food. Food Chem. Toxicol. 125, 462–466. doi: 10.1016/j.fct.2019.01.033
Beattie, R. E., Bakke, E., Konopek, N., Thill, R., Munson, E., and Hristova, K. R. (2020). Antimicrobial resistance traits of Escherichia coli isolated from dairy manure and freshwater ecosystems are similar to one another but differ from associated clinical isolates. Microorganisms 8:747. doi: 10.3390/microorganisms8050747
Cheng, J., Qu, W., Barkema, H. W., Nobrega, D. B., Gao, J., Liu, G., et al. (2019). Antimicrobial resistance profiles of 5 common bovine mastitis pathogens in large Chinese dairy herds. J. Dairy Sci. 102, 2416–2426. doi: 10.3168/jds.2018-15135
Cookson, A. L., Marshall, J. C., Biggs, P. J., Rogers, L. E., Collis, R. M., Devane, M., et al. (2022). Whole-genome sequencing and virulome analysis of Escherichia coli isolated from New Zealand environments of contrasting observed land use. Appl. Environ. Microbiol. 88, e00277–e00222. doi: 10.1128/aem.00277-22
Dadgostar, P. (2019). Antimicrobial resistance: implications and costs. Infect. Drug Resist. 12, 3903–3910. doi: 10.2147/IDR.S234610
Graham, D. W., Bergeron, G., Bourassa, M. W., Dickson, J., Gomes, F., Howe, A., et al. (2019). Complexities in understanding antimicrobial resistance across domesticated animal, human, and environmental systems. Ann. N. Y. Acad. Sci. 1441, 17–30. doi: 10.1111/nyas.14036
He, T., Wang, R., Liu, D., Walsh, T. R., Zhang, R., Lv, Y., et al. (2019). Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat. Microbiol. 4, 1450–1456. doi: 10.1038/s41564-019-0445-2
Hogan, J., and Smith, K. (1992). Using bulk tank milk cultures in a dairy practice. Workshop mastitis microbiology diagnostics, National Mastitis Council, Arlington.
Hu, F., Guo, Y., Yang, Y., Zheng, Y., Wu, S., Jiang, X., et al. (2019). Resistance reported from China antimicrobial surveillance network (CHINET) in 2018. Eur. J. Clin. Microbiol. Infect. Dis. 38, 2275–2281. doi: 10.1007/s10096-019-03673-1
Hu, B., Yang, X., Liu, Q., Zhang, Y., Jiang, D., Jiao, H., et al. (2022). High prevalence and pathogenic potential of Shiga toxin-producing Escherichia coli strains in raw mutton and beef in Shandong, China. Curr. Res. Food Sci. 5, 1596–1602. doi: 10.1016/j.crfs.2022.08.021
Imre, K., Ban-Cucerzan, A., Herman, V., Sallam, K. I., Cristina, R. T., Abd-Elghany, S. M., et al. (2022). Occurrence, pathogenic potential and antimicrobial resistance of Escherichia coli isolated from raw Milk cheese commercialized in Banat region, Romania. Antibiotics 11:721. doi: 10.3390/antibiotics11060721
Jara, M. C., Frediani, A. V., Zehetmeyer, F. K., Bruhn, F. R. P., Müller, M. R., Miller, R. G., et al. (2021). Multidrug-resistant hospital bacteria: epidemiological factors and susceptibility profile. Microb. Drug Resist. 27, 433–440. doi: 10.1089/mdr.2019.0209
Khalifeh, O. M., and Obaidat, M. M. (2022). Urinary tract virulence genes in extended-spectrum beta-lactamase E. coli from dairy cows, beef cattle, and small ruminants. Acta Trop. 234:106611. doi: 10.1016/j.actatropica.2022.106611
Kudva, I. T., Cornick, N. A., Plummer, P. J., Zhang, Q., Nicholson, T. L., Bannantine, J. P., et al. (2020). Virulence mechanisms of bacterial pathogens. Hoboken, NJ: John Wiley & Sons.
Li, F., Cheng, P., Li, X., Liu, R., Liu, H., and Zhang, X. (2022). Molecular epidemiology and colistin-resistant mechanism of mcr-positive and mcr-negative Escherichia coli isolated from animal in Sichuan province, China. Front. Microbiol. 13:8548. doi: 10.3389/fmicb.2022.818548
Lima, T., Domingues, S., and Da Silva, G. J. (2020). Manure as a potential hotspot for antibiotic resistance dissemination by horizontal gene transfer events. Vet. Sci. 7:110. doi: 10.3390/vetsci7030110
Liu, H., Meng, L., Dong, L., Zhang, Y., Wang, J., and Zheng, N. (2021). Prevalence, antimicrobial susceptibility, and molecular characterization of Escherichia coli isolated from raw milk in dairy herds in northern China. Front. Microbiol. 12:656. doi: 10.3389/fmicb.2021.730656
Lu, Q., Zhang, W., Luo, L., Wang, H., Shao, H., Zhang, T., et al. (2022). Genetic diversity and multidrug resistance of phylogenic groups B2 and D in InPEC and ExPEC isolated from chickens in Central China. BMC Microbiol. 22, 1–12. doi: 10.1186/s12866-022-02469-2
Ma, F., Xu, S., Tang, Z., Li, Z., and Zhang, L. (2021). Use of antimicrobials in food animals and impact of transmission of antimicrobial resistance on humans. Biosafety Health 3, 32–38. doi: 10.1016/j.bsheal.2020.09.004
Ma, J., Zhou, W., Wu, J., Liu, X., Lin, J., Ji, X., et al. (2022). Large-scale studies on antimicrobial resistance and molecular characterization of Escherichia coli from food animals in developed areas of eastern China. Microbiol. Spectrum 10, e02015–e02022. doi: 10.1128/spectrum.02015-22
Manyi-Loh, C., Mamphweli, S., Meyer, E., and Okoh, A. (2018). Antibiotic use in agriculture and its consequential resistance in environmental sources: potential public health implications. Molecules 23:795. doi: 10.3390/molecules23040795
Murray, C. J., Ikuta, K. S., Sharara, F., Swetschinski, L., Aguilar, G. R., Gray, A., et al. (2022). Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399, 629–655. doi: 10.1016/S0140-6736(21)02724-0
Oladeinde, A., Cook, K., Lakin, S. M., Woyda, R., Abdo, Z., Looft, T., et al. (2019). Horizontal gene transfer and acquired antibiotic resistance in Salmonella enterica serovar Heidelberg following in vitro incubation in broiler ceca. Appl. Environ. Microbiol. 85, e01903–e01919. doi: 10.1128/AEM.01903-19
Peng, Z., Hu, Z., Li, Z., Zhang, X., Jia, C., Li, T., et al. (2022). Antimicrobial resistance and population genomics of multidrug-resistant Escherichia coli in pig farms in mainland China. Nat. Commun. 13:1116. doi: 10.1038/s41467-022-28750-6
Pormohammad, A., Nasiri, M. J., and Azimi, T. (2019). Prevalence of antibiotic resistance in Escherichia coli strains simultaneously isolated from humans, animals, food, and the environment: a systematic review and meta-analysis. Infect. Drug Resist. 12, 1181–1197. doi: 10.2147/IDR.S201324
Qian, X., Gu, J., Sun, W., Wang, X.-J., Su, J.-Q., and Stedfeld, R. (2018). Diversity, abundance, and persistence of antibiotic resistance genes in various types of animal manure following industrial composting. J. Hazard. Mater. 344, 716–722. doi: 10.1016/j.jhazmat.2017.11.020
Raimondi, S., Righini, L., Candeliere, F., Musmeci, E., Bonvicini, F., Gentilomi, G., et al. (2019). Antibiotic resistance, virulence factors, phenotyping, and genotyping of E. coli isolated from the feces of healthy subjects. Microorganisms 7:251. doi: 10.3390/microorganisms7080251
Salinas, L., Cárdenas, P., Johnson, T. J., Vasco, K., Graham, J., and Trueba, G. (2019). Diverse commensal Escherichia coli clones and plasmids disseminate antimicrobial resistance genes in domestic animals and children in a semirural community in Ecuador. Msphere 4, e00316–e00319. doi: 10.1128/mSphere.00316-19
Sarowska, J., Futoma-Koloch, B., Jama-Kmiecik, A., Frej-Madrzak, M., Ksiazczyk, M., Bugla-Ploskonska, G., et al. (2019). Virulence factors, prevalence and potential transmission of extraintestinal pathogenic Escherichia coli isolated from different sources: recent reports. Gut Pathogens 11, 1–16. doi: 10.1186/s13099-019-0290-0
Sharp, J. L., Mobley, C., Hammond, C., Withington, C., Drew, S., Stringfield, S., et al. (2012). A mixed methods sampling methodology for a multisite case study. J. Mixed Methods Res. 6, 34–54. doi: 10.1177/1558689811417133
Sobur, M. A., Sabuj, A. A. M., Sarker, R., Rahman, A. T., Kabir, S. L., and Rahman, M. T. (2019). Antibiotic-resistant Escherichia coli and Salmonella spp. associated with dairy cattle and farm environment having public health significance. Veterinary world 12, 984–993. doi: 10.14202/vetworld.2019.984-993
Su, Y., Yu, C.-Y., Tsai, Y., Wang, S.-H., Lee, C., and Chu, C. (2016). Fluoroquinolone-resistant and extended-spectrum β-lactamase-producing Escherichia coli from the milk of cows with clinical mastitis in southern Taiwan. J. Microbiol. Immunol. Infect. 49, 892–901. doi: 10.1016/j.jmii.2014.10.003
Wang, Y., Xu, C., Zhang, R., Chen, Y., Shen, Y., Hu, F., et al. (2020). Changes in colistin resistance and mcr-1 abundance in Escherichia coli of animal and human origins following the ban of colistin-positive additives in China: an epidemiological comparative study. Lancet Infect. Dis. 20, 1161–1171. doi: 10.1016/S1473-3099(20)30149-3
Wang, W., Yu, L., Hao, W., Zhang, F., Jiang, M., Zhao, S., et al. (2021). Multi-locus sequence typing and drug resistance analysis of swine origin Escherichia coli in Shandong of China and its potential risk on public health. Front. Public Health 9:780700. doi: 10.3389/fpubh.2021.780700
Yu, Z., Wang, J., Ho, H., Wang, Y., Huang, S., and Han, R. (2020). Prevalence and antimicrobial-resistance phenotypes and genotypes of Escherichia coli isolated from raw milk samples from mastitis cases in four regions of China. J. Global Antimicrobial Resist. 22, 94–101. doi: 10.1016/j.jgar.2019.12.016
Zhang, S., Chen, S., Rehman, M. U., Yang, H., Yang, Z., Wang, M., et al. (2021). Distribution and association of antimicrobial resistance and virulence traits in Escherichia coli isolates from healthy waterfowls in Hainan, China. Ecotoxicol. Environ. Safety 220:112317. doi: 10.1016/j.ecoenv.2021.112317
Zhao, X., Lv, Y., Adam, F. E. A., Xie, Q., Wang, B., Bai, X., et al. (2021). Comparison of antimicrobial resistance, virulence genes, phylogroups, and biofilm formation of Escherichia coli isolated from intensive farming and free-range sheep. Front. Microbiol. 12:699927. doi: 10.3389/fmicb.2021.699927
Keywords: antibiotic resistance genes (ARGs), antimicrobial resistance, multi-drug resistance, virulence associated genes, Escherichia coli , dairy environment
Citation: Shoaib M, He Z, Geng X, Tang M, Hao R, Wang S, Shang R, Wang X, Zhang H and Pu W (2023) The emergence of multi-drug resistant and virulence gene carrying Escherichia coli strains in the dairy environment: a rising threat to the environment, animal, and public health. Front. Microbiol. 14:1197579. doi: 10.3389/fmicb.2023.1197579
Edited by:
Xiaodan Huang, Lanzhou University, ChinaReviewed by:
Adriana Morar, Banat University of Agricultural Sciences and Veterinary Medicine, RomaniaMagdalena Zalewska, University of Warsaw, Poland
Copyright © 2023 Shoaib, He, Geng, Tang, Hao, Wang, Shang, Wang, Zhang and Pu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wanxia Pu, cHV3YW54aWFAY2Fhcy5jbg==
 Muhammad Shoaib
Muhammad Shoaib Zhoulin He
Zhoulin He Xiang Geng
Xiang Geng Shengyi Wang
Shengyi Wang Wanxia Pu
Wanxia Pu