- 1Department of Microbiology and Immunology, Faculty of Biology, Research Institute of the University of Bucharest, University of Bucharest, Bucharest, Romania
- 2Earth, Environmental and Life Sciences Section, Research Institute of the University of Bucharest, University of Bucharest, Bucharest, Romania
- 3Institute and Clinic for Occupational, Social and Environmental Medicine, University Hospital, LMU Munich, Munich, Germany
- 4Institute for Medical Information Processing, Biometry, and Epidemiology – IBE, LMU Munich, Munich, Germany
- 5Pettenkofer School of Public Health, Munich, Germany
- 6Department of Infectious Diseases, Institute of Biomedicine, University of Gothenburg, Gothenburg, Sweden
- 7Centre for Antibiotic Resistance Research in Gothenburg (CARe), University of Gothenburg, Gothenburg, Sweden
- 8Centre for Infectious Disease Control, National Institute for Public Health and the Environment, Bilthoven, Netherlands
- 9German Centre for Infection Research (DZIF), Partner Site Munich, Munich, Germany
- 10Max von Pettenkofer Institute, Faculty of Medicine, LMU Munich, Munich, Germany
- 11Department of Infectious Diseases and Tropical Medicine, LMU University Hospital Munich, Munich, Germany
- 12Comprehensive Pneumology Center Munich (CPC-M), German Center for Lung Research (DZL), Munich, Germany
- 13Romanian Academy of Sciences, Bucharest, Romania
- 14The Romanian Academy, Bucharest, Romania
Antibiotics are an essential tool of modern medicine, contributing to significantly decreasing mortality and morbidity rates from infectious diseases. However, persistent misuse of these drugs has accelerated the evolution of antibiotic resistance, negatively impacting clinical practice. The environment contributes to both the evolution and transmission of resistance. From all anthropically polluted aquatic environments, wastewater treatment plants (WWTPs) are probably the main reservoirs of resistant pathogens. They should be regarded as critical control points for preventing or reducing the release of antibiotics, antibiotic-resistant bacteria (ARB), and antibiotic-resistance genes (ARGs) into the natural environment. This review focuses on the fate of the pathogens Enterococcus faecium, Staphylococcus aureus, Clostridium difficile, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacteriaceae spp. (ESCAPE) in WWTPs. All ESCAPE pathogen species, including high-risk clones and resistance determinants to last-resort antibiotics such as carbapenems, colistin, and multi-drug resistance platforms, were detected in wastewater. The whole genome sequencing studies demonstrate the clonal relationships and dissemination of Gram-negative ESCAPE species into the wastewater via hospital effluents and the enrichment of virulence and resistance determinants of S. aureus and enterococci in WWTPs. Therefore, the efficiency of different wastewater treatment processes regarding the removal of clinically relevant ARB species and ARGs, as well as the influence of water quality factors on their performance, should be explored and monitored, along with the development of more effective treatments and appropriate indicators (ESCAPE bacteria and/or ARGs). This knowledge will allow the development of quality standards for point sources and effluents to consolidate the WWTP barrier role against the environmental and public health AR threats.
1. Introduction
The discovery of antibiotics was one of the most significant medical achievements of the 20th century, saving many lives and contributing to the control of numerous infectious diseases. The majority of the antibiotics that are used today were discovered in the period referred to as the golden age, i.e., 1940–1960, when at least 20 clinically relevant classes were developed (Katz and Baltz, 2016; O’Neill, 2016; Hutchings et al., 2019). However, the existing classes of antibiotics act selectively against only a few microbial cell targets: cell wall, plasma membrane, synthesis of proteins and metabolites, or DNA transcription and replication, allowing the bacterial species to develop resistance mechanisms during the millions of years of coevolution. Although many international authorities advocate for incentives to encourage the development of new antibiotics,1 (The Joint Programming Initiative on Antimicrobial Resistance, 2018) the interest of pharmaceutical companies in developing novel antibiotics has dramatically declined. Thus, in the last decades, only two new antibiotic classes (lipopeptides and oxazolidinones) providing coverage only against Gram-positive bacteria have been developed and approved, the other new molecules being analogs of the existing classes, drugs repurposed for the treatment of infectious diseases, non-antibiotics or antibiotic adjuvants (Xie et al., 2022).
The natural ability of bacteria to develop resistance has been accelerated by the selective pressure exerted by the improper use of antibiotics in human therapy, animal husbandry, and agriculture. Between 2000 and 2010, global antibiotic use increased by 36%, reaching 45% in the case of carbapenems, and the amount of all antibiotics used in animal food was estimated to be 200,235 tons in 2030 (Dhingra et al., 2020; Boyd et al., 2021). The acute limitation of currently available therapeutic options leads to increased morbidity and mortality rates, longer treatment duration, and higher hospitalization costs, questioning the efficacy of modern medical practices, which will become very risky because of common infections. Worldwide, it is estimated that 4.95 million deaths were associated with and 1.2 million attributable to bacterial AMR in 2019, mainly from lower respiratory infections and involving six leading pathogens (E. coli, S. aureus, K. pneumoniae, Streptococcus pneumoniae, A. baumannii, and P. aeruginosa) (Murray et al., 2022).
Clinical surveillance programs indicate that the prevalence of human pathogens exhibiting multidrug (MDR), extended drug (XDR), or pan-drug (PDR) resistance is rising to dangerously high levels in all parts of the world (Cheesman et al., 2017). The most threatening resistant pathogens are known under different acronyms, such as ESKAPE (E) [Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, Enterobacter sp. (Escherichia coli), ESCAPE or AmpC-producing SPICE (Serratia, Providencia, indole-positive Proteus, Morganella, Providencia species/Acinetobacter, Citrobacter, Enterobacter species) or are included in different “black” lists (Rice, 2008; Moy and Sharma, 2017)]. In addition, the WHO published the critical/high/medium priority pathogens list for R&D of new antibiotics, including carbapenem-resistant A. baumannii and P. aeruginosa, carbapenem-resistant and extended-spectrum beta-lactamase (ESBL)-producing Enterobacteriaceae, clarithromycin-resistant Helicobacter pylori, fluoroquinolone-resistant Campylobacter spp., Salmonella and Shigella, cephalosporin-resistant, fluoroquinolone-resistant Neisseria gonorrhoeae, ampicillin-resistant Haemophilus influenzae, vancomycin-resistant E. faecium, methicillin-resistant, vancomycin-intermediate, and resistant S. aureus, and penicillin-non-susceptible Streptococcus pneumoniae (WHO, 2017; Asokan et al., 2019). Furthermore, with the increasing use of whole-genome sequencing to analyze antibiotic-resistant pathogens, it has become clear that many of the significant resistance problems are associated with a few successful bacterial clones within a species, with worldwide dissemination in hospitals and, possibly, in the community and the natural environment (Oliver et al., 2015; Andersson and Hughes, 2017; MacLean and San Millan, 2019).
Numerous scientific publications reported the presence in the natural environment of clinically relevant antibiotic-resistant bacteria (ARB) and antibiotic-resistance genes (ARG) (Nnadozie and Odume, 2019; Pazda et al., 2019). Thus, the environment can serve as a source/reservoir of already resistant pathogens or may acquire ARGs from other human / animal-associated bacteria or the environmental resistome. Further, these ARGs could be transferred to the clinic and vice versa (Figure 1).
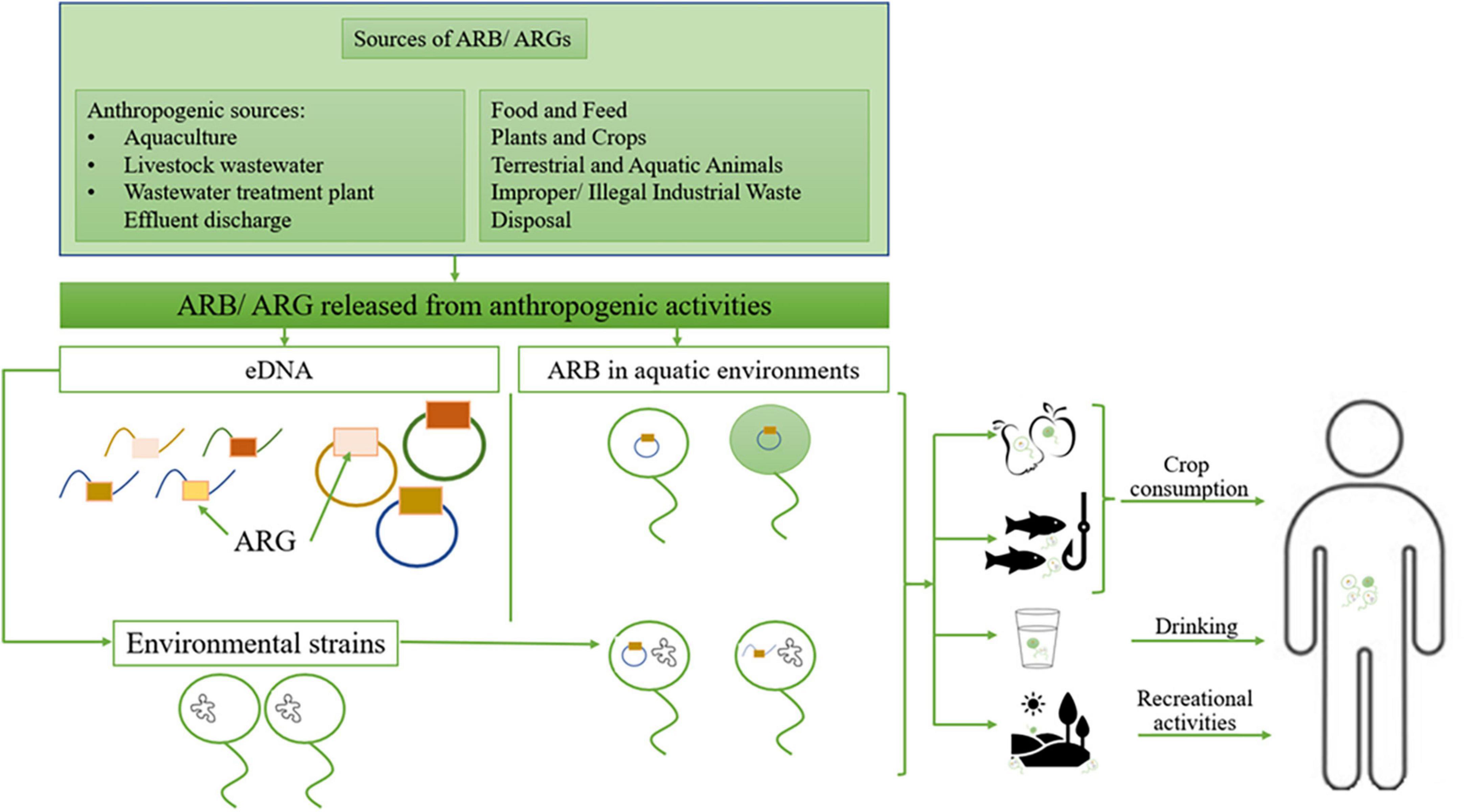
Figure 1. The environment role as a source of resistant pathogens; ARB, antibiotic-resistant bacteria; ARG, antibiotic resistance genes; eDNA, environmental DNA. Adapted from Denissen et al. (2022).
The potential gene exchange between environmental and clinical resistomes through horizontal gene transfer (HGT) involves mobile genetic elements (MGE) such as plasmids, integrative and conjugative elements (ICE), transposons, and integrons (Singer et al., 2016; Bengtsson-Palme et al., 2018; Larsson and Flach, 2022). The HGT occurs mainly via three mechanisms: conjugation (transfer of DNA between bacteria by direct cell-to-cell contact, considered the main AMR dissemination strategy), natural transformation (uptake of the free DNA from the environment), and transduction (transfer of DNA between bacteria via a bacteriophage) (von Wintersdorff et al., 2016; Chen et al., 2018). The MGE, such as integrons, are frequently associated with multi-drug resistance (MDR), increasing the risk of co-selection and persistence of multiple resistance determinants under the selective pressure of antimicrobial agents (Deng et al., 2015).
Many environmental ARGs have been shown to possess a high sequence similarity to those in human / domesticated animal fecal microbes (Nesme et al., 2014) and human/animal pathogens (Forsberg et al., 2012). For example, the quinolone resistance gene qnrA was first detected in clinical isolates of K. pneumoniae in the USA and then found in the aquatic Gram-negative bacteria Shewanella algae and in Aeromonas spp. isolated from the Seine River (France) (Cattoir et al., 2008; Perry and Wright, 2013). Most of the ARGs with known origins were likely mobilized from their native context by MGEs, such as IS/ISCR elements (Ebmeyer et al., 2021). However, their recent origin is still unknown for the great majority of the ARGs, probably because the original environmental bacterial hosts have not yet been sequenced (Larsson and Flach, 2022).
The “One Health” approach published in 2017 reinforced the importance of addressing the AMR issue outside the clinical sector by encompassing the environmental contribution to the emergence, accumulation, and spread of AMR (Fletcher, 2015; Baker et al., 2018; MacLean and San Millan, 2019). In order to map and understand the transmission routes of ARGs among clinical and environmental bacteria, research on the distribution patterns of ARGs and their temporal dynamics in different microbiomes and studies tracking the acquisition of ARGs in complex communities are required (Andersson and Hughes, 2017; MacLean and San Millan, 2019; Surleac et al., 2020).
There are numerous reports on the detection of wastewater effluents and impacted rivers of a wide variety of clinically significant ARB (Zhang et al., 2009; Giebułtowicz et al., 2018; Limayem et al., 2019).
This review aims to present an update regarding the contribution of WWTPs to the antibiotic resistance reservoir, focusing on ESCAPE pathogens and associated ARGs. We performed a literature search in Pubmed, Scopus, and Google Scholar databases using the combinations of the following keywords: ESCAPE pathogens (searched both as abbreviation and individually for each pathogen) + antibiotic resistance / wastewater treatment plants / antibiotic resistance genes / wastewater / high-risk clones / multidrug resistance. Consequently, a title and abstract screening was performed, and the papers published in the last 10 years, as well as some significant studies published previously, have been considered. The articles duplicated, irrelevant, and without full text were excluded. The remaining studies have been grouped into six categories, corresponding to each ESCAPE pathogen species and one last category, referring to the abundance of ARGs in WWTPs environments and the efficiency of different wastewater treatments in decreasing or removing ARGs.
2. WWTPs—An escape gate for ESCAPE pathogens and associated ARGs
Wastewater treatment plants receive a continuous discharge of human-associated ARB, ARG, antibiotics, disinfectants, and metals, which are co-locating inside the WWTP, together with the commensal microbiota, generating the perfect premises for antibiotic resistance emergence, accumulation, and dissemination (Bougnom and Piddock, 2017; Karkman et al., 2019; Kraemer et al., 2019; Paulshus et al., 2019; Pärnänen et al., 2019; Reichert et al., 2019).
Although WWTPs reduce the number of total bacteria released into the environment, the current wastewater treatment technologies do not altogether remove and/or biodegrade ARB, ARG, and antibiotics (Jäger et al., 2018; Müller et al., 2018). The sewage’ inner surfaces favor the colonization with wastewater-derived microorganisms that develop biofilms, which are continuously exposed to antibiotic residues and ARB from wastewater (Auguet et al., 2017). An analysis of the biofilm-embedded bacteria from influent and effluent sewage showed a lower susceptibility to antibiotics than bacteria from wastewater (Lépesová et al., 2018). Moreover, biofilms could facilitate the HGT, as shown by an in vitro study, demonstrating an increased transfer rate of the plasmidial CTX-M-15 gene in a K. pneumoniae biofilm compared to planktonic growth conditions (Hennequin et al., 2012). The transfer of conjugative transposons carrying ARG, such as Tn916, might also be responsible for acquiring resistance mechanisms in biofilm bacteria (Hannan et al., 2010). Microfluidic systems combined with laser confocal microscopy, fluorescent labeling of bacteria, and flow cytometry have been developed to investigate the HGT of ARGs in activated sludge biofilm (Qiu et al., 2018). WWTP discharges might also impact the river-streambed biofilm communities that often prevail in natural environments compared to free, planktonic bacteria (Proia et al., 2016). The released WWTPs effluents could impact the river biofilms through the fixation of pharmaceuticals and the persistence of the discharged microorganisms in the streambed biofilms (Aubertheau et al., 2017; Subirats et al., 2017). It has been shown that some ESCAPE pathogens (enterococci, A. baumannii, and P. aeruginosa), as well as associated ARGs (ermB, blaTEM, tetM, and qnrS), are associated with the particulate fraction of the WWTP effluent and are more probably remaining in the riverbed of the receiving water due to sedimentation (Brown et al., 2019). However, little information is available regarding the ARG distribution in the river streambed and how wastewater-associated microorganisms, including potential pathogens, contribute to maintaining the streambed resistome.
Wastewater systems, especially those receiving hospital or livestock wastewater, were demonstrated to be significant sources of significant epidemic pathogens belonging to the so-called high-risk clones: E. coli sequence type (ST) 131 (Finn et al., 2020), E. faecium HiRECC (Sadowy and Luczkiewicz, 2014), Acinetobacter IC2 carrying OXA-23, and IC1 carrying OXA-72 (Goic-Barisic et al., 2017; Higgins et al., 2018), K. pneumoniae ST11, and ST258 (Surleac et al., 2020), and P. aeruginosa ST235, ST111, and ST175. Furthermore, increased correlations of ESCAPE pathogens with clinically relevant ARGs (e.g., blaNDM-1, vanA) were found in WWTP effluents influenced by hospital wastewaters. Furthermore, isolates carrying the mcr-1 gene, providing resistance to the last resort antibiotic colistin, have been reported in WWTP sewage, probably originating from food-producing animals (Ovejero et al., 2017). Additionally, the selective pressure within the wastewater treatment plants (WWTP)/sewer systems, an ecosystem with consistent exposure to antibiotics and other chemical pollutants such as biocides, heavy metals, etc., could contribute to the emergence of new antibiotic-resistant variants, with possible epidemic risk (Flach et al., 2018; Danner et al., 2019; Kraupner et al., 2021). In a study aimed to assess the presence of ESKAPE strains in process water from delivery and unclean areas as well as wastewater from the in-house WWTPs of German poultry slaughterhouses, at least one of the target species was detected in 87.5% of the wastewater samples and 86% of the process water samples, with the following decreasing order of their prevalence: E. coli > A. calcoaceticus-A. baumannii (ACB) complex > S. aureus > K. pneumoniae > Enterobacter spp., Enterococcus spp., and P. aeruginosa (Savin et al., 2020).
In the next subchapters we will present an update of the current knowledge on the ESCAPE pathogens to WWTP relationships.
2.1. Enterococcus faecium
Enterococci are ubiquitous in nature, found in soil, plants, surface water, wastewater, and food, as well as in the gastrointestinal tract of animals and humans (Sanderson et al., 2020). E. faecium and E. faecalis are the predominant species in wastewater, likely due to the continuous input of fecal waste into these systems. Comparative genomics indicated a lower diversity of E. faecium isolates in the wastewaters than E. faecalis, suggesting that E. faecium isolates may be more adapted specifically to clinical environments from which they are released in the natural environment (Sanderson et al., 2020).
Łuczkiewicz et al. (2010) reported a significant increase in resistance prevalence, after water treatment, for both E. faecalis and E. faecium, to fluoroquinolones (ciprofloxacin, levofloxacin, and moxifloxacin), tetracycline, and erythromycin. The E. faecium and E. faecalis isolates were expressing high-level resistance to aminoglycosides (up to 4.5% for gentamicin) and glycopeptides (for teicoplanin up to 2.7% and for vancomycin up to 6.8%) (Łuczkiewicz et al., 2010). E. faecium, with high prevalence of resistance to ciprofloxacin, was reported to be positively selected during WWTP treatment (Ferreira da Silva et al., 2006).
Enterococcus faecium belonging to a high-risk clonal group named clonal complex-17 (CC17), is associated with clinical infections and hospital outbreaks worldwide, exhibiting increased levels of ciprofloxacin resistance and carrying the esp gene associated with adhesion to cells and pathogenicity (van Schaik et al., 2010). This nosocomial clone may survive and pass through the wastewater system reaching the environmental reservoirs (Leclercq et al., 2013; Sadowy and Luczkiewicz, 2014; Sanderson et al., 2020). Leclercq et al. (2013) reported the identification of a large number of E. faecium strains belonging to the CC17 complex resistant to ciprofloxacin and carrying acquired macrolide resistance genes in hospital and retirement home effluents. In Gauteng, South Africa, isolates of E. faecium CC17 clonal complexes were recovered only from hospital sewage but were not detected further in WWTP and surface waters (Hamiwe et al., 2019).
Enterococci were demonstrated to survive in beach sediments, thus their release from untreated wastewater into the sea could pose a potential threat to the health of recreational users (Anderson et al., 2015); thus, the release of treated and into the sea. In marine outfalls directly impacted by treated wastewater, a lower number of enterococci by four orders of magnitude (<100 CFU/100 mL) was reported in comparison to corresponding treated wastewater, which is below the value described by the New Bathing Water Directive 2006/7/EC for coastal water of “excellent quality.” However, in WWTP effluents and marine outfalls, the isolates belonging to the significant nosocomial HiRECC (formerly CC17) constituted 24.6% of all E. faecium. Furthermore, these isolates were resistant to ciprofloxacin and ampicillin and showed an MDR phenotype in the vast majority (Sadowy and Luczkiewicz, 2014).
It has been shown that planktonic Enterococcus spp. isolated from influent wastewater exhibited low resistance to ampicillin and ciprofloxacin, while those recovered from the influent biofilm were resistant to vancomycin, and most were MDR (Lépesová et al., 2018). In the Netherlands, ampicillin-resistant E. faecium strains with intermediate vancomycin resistance have been isolated from WWTP-treated effluent but not from the receiving surface water (Taučer-Kapteijn et al., 2016).
Multidrug vancomycin-resistant enterococci (VRE) were detected in unchlorinated effluent samples, suggesting an exposure risk for WWTP workers (Rosenberg Goldstein et al., 2014). The majority of E. faecium isolates (86%) from the WWTP effluent in the Czech Republic harbored the vanA gene and belonged to ST17, ST18, and ST78, while those isolated from the WWTP effluent and downstream in Poland (57%) harbored vanC1 (27.6%) (Oravcova et al., 2017; Gotkowska-Płachta, 2021).
Vancomycin-resistant enterococci have been used as indicators of antimicrobial resistance in two WWTPs from the same municipality, one with a biologically aerated filter (BAF) and the other with conventional activated sludge (CAS). The BAF system assured a better removal rate of total enterococci, VRE, and levofloxacin resistance, while CAS was selected for nitrofurantoin resistance and reduced quinupristin/dalfopristin and streptomycin-resistant enterococci (Sanderson et al., 2019).
The results of the current studies indicate an incomplete removal of resistant enterococci, including VRE strains belonging to clinically significant sequence types, during wastewater treatment.
2.2. Staphylococcus aureus
Drug-susceptible and drug-resistant S. aureus have been identified in the US’s influent and effluent samples collected from four WWTPs (Goldstein et al., 2012). From the very few studies documenting the release of methicillin-resistant S. aureus (MRSA) through wastewater systems into the environment (Börjesson et al., 2009; Boopathy, 2017; Kozajda and Jeżak, 2020), it appears that hospital wastewaters add to the load of MDR S. aureus entering WWTP. In a study performed in Poland, among 149 S. aureus isolates (2 from the air and 147 from wastewater), only two were MRSA, and over 60% were resistant to penicillin. In contrast, ∼20% showed MDR phenotypes (Kozajda and Jeżak, 2020). Thompson et al. (2013) reported detecting one MRSA isolate resistant to nine antibiotics in hospital effluent. MRSA ST398 carrying the mecA gene and exhibiting an MDR phenotype (resistance to clindamycin, ciprofloxacin, tetracycline, and aminoglycosides) was detected in an effluent water sample in Spain (Gómez et al., 2016). A comparative study of the MRSA prevalence and the genotypic and phenotypic characteristics of MRSA isolates from a municipal (M) and a swine slaughterhouse (S) WWTP revealed a different profile of the SCCmec types in the two types of WWTP and higher abundance of mecA gene in the S-WWTP (Wan and Chou, 2015).
In one study, although wastewater treatment reduced the number of MRSA isolates and diversity, the remaining strains exhibited a higher resistance level and virulence (the presence of the PVL gene) (Börjesson et al., 2009). Both hospital and community-related clonal complexes have been detected in fully treated WWTP effluents. The genetic analysis of the S. aureus isolates obtained after treatment showed diversity in the spa type and carriage of MGEs, suggesting that treatment could facilitate persistence, evolution, and genetic shifts (Amirsoleimani et al., 2019). A recent study evaluated the occurrence and environmental health risk of S. aureus and MRSA from hospital effluent to sewage treatment plant (STP) and finally to river water at the basin level; a high abundance of over 90% has been detected in the sewage treatment plant, and the contribution of the pollution load derived from the target STP effluent to river water ranged from 2 to 25% (Azuma et al., 2022).
The few available studies report the presence of multidrug-resistant S. aureus and MRSA belonging to different clonal complexes in the WWTP effluents and surface water, suggesting the importance of reducing or inactivating S. aureus and MRSA before the effluent is discharged into rivers.
2.3. Clostridioides difficile
More than 25% of the hospital- and community-associated Clostridioides difficile infections originate in community sources such as asymptomatic carriers, animals, food, and WWTP (Hensgens et al., 2012; Warriner et al., 2017). In addition, the resistance genes to metronidazole and vancomycin antibiotics used for treating C. difficile infections, i.e., the nim and vanA genes, were detected in wastewater decades ago (Trinh and Reysset, 1996).
Clostridium difficile is a chlorine-resistant, spore-forming bacterium that can survive in contaminated aquatic environments such as wastewater (Baghani et al., 2020). C. difficile was detected in 11.8% of untreated human wastewater samples in a study performed in Texas (Norman et al., 2011) and in 96% of anaerobically digested sludge samples, 92% of raw sludge, and 73% of dewatered biosolids and effluent discharges from two Southern Ontario WWTPs (Xu et al., 2014). The analysis of a conventional activated sludge treatment plant and a waste stabilization pond system in Iran revealed the presence of C. difficile in 13.6% of digested sludge samples and 5% of the waste stabilization ponds samples, all strains being toxigenic (positive for the tcdB gene) (Nikaeen et al., 2015). Resistant hypervirulent C. difficile (ribotype 078 or toxinotype V) strains have been detected in the WWTPs influents and effluents in China, and the raw sewage, digested sludge, and biosolids from Southern Ontario WWTPs (Xu et al., 2014). The C. difficile isolates from raw sewage influents and treated effluents of WWTPs from southern Switzerland exhibited a large diversity, belonging to 13 different known ribotypes (009, 010, 014, 015, 039, 052, 053, 066, 070, 078, 101, 106, and 117), to which non-typeable strains were added. Eight ribotypes (010, 014, 015, 039, 066, 078, 101, and 106) were also detected in hospitalized symptomatic patients in the respective region. The most frequently isolated ribotype (40%) was the hypervirulent ribotype 078, present in more than 50% of the sampled WWTPs (6/9). In contrast, the toxigenic emerging ribotype 066, associated with hospital infection, was isolated from the effluent of one plant. Most isolated strains (85%) were toxigenic, with 49% harboring the profile A + B + CDT + and 51% the profile A + B + CDT–(Romano et al., 2012). In another study performed in Slovenia, C. difficile was detected in all analyzed WWTP samples, the recovered isolates belonging to 32 different ribotypes, of which 014/020 and 010 were the most prevalent (Steyer et al., 2015). The analysis of 18 WWTPs from across the East of England, half of which were located downstream of hospitals, has revealed the presence of C. difficile in the effluent of all WWTPs, belonging to 38 STs, out of which 13 were common to clinical isolates analyzed in the same temporo-spatial sequence (Moradigaravand et al., 2018). In New Zealand, toxigenic C. difficile has also been detected with high frequency in wastewater (10 out of 13 toxigenic isolates), belonging to eight PCR-ribotypes (RTs), including two novel RTs (878 and 879). However, all C. difficile isolates were susceptible to the first-line human antimicrobials used to treat C. difficile infection (Rivas et al., 2020).
These studies confirm the extensive escape of toxigenic and resistant C. difficile from WWTPs into surface waters, raising the need to monitor these bacteria in treated wastewater.
2.4. Acinetobacter baumannii
Acinetobacter baumannii, a leading cause of nosocomial infections, hospital outbreaks, and sporadic acute community-acquired infections with severe evolution in critically ill patients, is known for its ability to develop resistance to multiple antibiotics (Yakkala et al., 2019). MDR A. baumannii was reported in untreated hospital wastewater in India, Brazil, China, and Croatia (Ferreira et al., 2011; Zhang et al., 2013; Seruga Music et al., 2017; Marathe et al., 2019). In a study performed in China, Acinetobacter spp. isolates from urban WWTPs effluents were found to express high resistance to rifampin (72.4%), chloramphenicol (69%), and amoxicillin plus clavulanic acid (37.9%), and 84.5% of the tested isolates were MDR (Zhang et al., 2009).
Goic-Barisic et al. (2017) reported the release of hospital wastewaters containing clinical carbapenem-resistant A. baumannii to the Adriatic Sea without any pre-treatment, as this is not legally required in Croatia. Carbapenem-resistant isolates belonging to international clonal lineage IC2 carrying OXA-23, IC1 carrying OXA-72, and even pan-drug resistant isolates were detected in the WWTP effluent (Goic-Barisic et al., 2017; Higgins et al., 2018). In addition, carbapenem-resistant A. baumannii isolates were detected in the urban sewage receiving hospital wastewater and the river (Seruga Music et al., 2017; Higgins et al., 2018). The prevalence of carbapenem-resistant A. baumannii and ARGs in untreated and treated wastewater has been analyzed for three consecutive seasons; during 2019, in Poland, the highest prevalence of target bacteria was recorded in the wastewater collected in June and September, as compared to February. The ISAba1/blaOXA-51 complex associated with carbapenem resistance was identified in 13 isolates. The number of resistant Acinetobacter isolates increased in river water samples collected downstream compared to upstream from the WWTP (Hubeny et al., 2022). A study performed on A. baumannii isolated in 2018 and 2019 from hospital settings, hospital collecting sewage tanks, and the receiving WWTPs located in the central geographical regions of Romania has shown that the strains isolated from hospital effluents belonged to epidemic clones, such as ST2 and exhibited high MDR rates. The WGS analysis revealed the relatedness between clinical and hospital wastewater strains and the possible dissemination of clinical A. baumannii belonging to ST2 in the wastewater (Gheorghe-Barbu et al., 2022).
The current evidence shows that WWTP might be a source of dissemination in the environment of Acinetobacter strains carrying clinically significant ARGs, including carbapenem-resistance genes.
2.5. Pseudomonas aeruginosa
Pseudomonas aeruginosa is an environmental bacterium that can cause human infections, particularly in patients with compromised host defense mechanisms. It is associated with urinary, gastrointestinal, soft tissue, bone, joint, and surgical site infections (European Centre for Disease Prevention and Control, 2022). The MDR/XDR P. aeruginosa high-risk clones such as ST235, ST111, and ST175, associated with chronic and hospital-acquired infections with significant morbidity and mortality, are disseminated in hospitals worldwide (Del Barrio-Tofiño et al., 2020). They are characterized by the increasing prevalence of transferable ARGs, particularly those encoding carbapenemases (such as blaIMP or blaVIM), ESBLs, and defective outer membrane porins.
Antibiotic-resistant P. aeruginosa isolates are highly concentrated in hospital effluents (Kerr and Snelling, 2009; Slekovec et al., 2012; Hocquet et al., 2016) and are continuously discharged into natural water basins mainly through sewage and further possibly spread to the soil through natural fertilizers (Topp et al., 2018; Manaia et al., 2022; Marutescu et al., 2022). Slekovec et al. (2012) reported high levels of P. aeruginosa in WWTP sludge (2.95 × 106 CFU/kg), within the general range found in hospital wastewater. The treated effluent of three hospital WWTPs was demonstrated to discharge MDR P. aeruginosa strains into a city river from Brazil, the major tributary of the Amazon river. The P. aeruginosa isolates from the respective river exhibited resistance profiles matching some of the resistant P. aeruginosa in the discharged effluent, suggesting that the hospital WWTPs pose a public health risk to residents that live in contact with this stream (Magalhães et al., 2016). The high-risk P. aeruginosa clones ST235, ST111, and ST395 were identified throughout the wastewater network of Besançon (France) and recovered from treated water and in the river downstream (Slekovec et al., 2012). Müller et al. (2018) reported MDR and XDR P. aeruginosa isolates in the clinical/urban system in Germany belonging to the epidemic outbreak clones ST235, mainly isolated from the undiluted clinical wastewater, with only one strain detected in the effluent of the WWTP, with ST111. These clones can also produce a strong biofilm, likely increasing their capacity to colonize sewer and drinking water plumbing systems (Müller et al., 2018).
Wastewater treatment plants receiving hospital wastewater have also been shown to be an essential source of carbapenem-resistant P. aeruginosa. They may therefore contribute to the environmental dissemination of resistance to this important class of antibiotics. In a longitudinal study, carbapenem-resistant bacteria were isolated from the wastewater of a maximum-care hospital for 2 years. The largest ST235 P. aeruginosa cluster contained WWTP effluent strains, suggesting the dissemination of this high-risk clone associated with severe infections into the environment (Kehl et al., 2022). Investigation of genetic relatedness between P. aeruginosa strains from wastewater treatment (WWT) lagoons and hospital-associated P. aeruginosa, as well as community-acquired clones collected in the same geographic area, confirmed the ability of some of these clones to survive in and disseminate from WWT lagoons ponds to connected streams. The WWT lagoons were colonized by highly diverse P. aeruginosa, with most genotypes harboring virulence genes involved in human colonization and infection (Petit et al., 2013). By contrast, Golle et al. (2017) have shown little overlap between a diverse population of carbapenem-resistant P. aeruginosa from WWTP influents and clinical genotypes (Golle et al., 2017). Also, Fuentefria et al. (2011) have found differences in resistance patterns, and frequency of MDR strains among P. aeruginosa from the hospital wastewater (Rio Grande do Sul, RS, Brazil) and surface water, the MDR strains being more frequent in the hospital (Fuentefria et al., 2011). In a Polish study, the cultivable Pseudomonas strains sampled from raw and treated wastewater and the receiving coastal waters of the Puck Bay, Baltic Sea, exhibited a low prevalence of carbapenems (meropenem and imipenem) resistance and of clinically relevant ESBLs (Luczkiewicz et al., 2015). The analysis of MDR P. aeruginosa strains isolated during two consecutive years (2018 and 2019) from hospital settings and hospital collecting sewage tanks. WWTPs located in the central geographical regions of Romania have revealed the presence of epidemic clones ST233, and ST357 in the wastewater and the release of P. aeruginosa strains belonging to ST357, ST640c, and ST621 from hospitals into the wastewaters (Gheorghe-Barbu et al., 2022).
Multidrug and carbapenem-resistant P. aeruginosa strains have been detected in wastewater in different geographical regions, with contrasting results regarding the relatedness between carbapenem-resistant strains found in wastewater and clinical strains. The variations in the local biodiversity of the aquatic microbial communities, pollution sources, and sampling and analysis methods could explain these discrepancies.
2.6. Enterobacteriaceae
Different studies report that Enterobacteriaceae strains collected downstream of WWTP discharge points can be resistant to different classes of antibiotics, the most frequently reported for E. coli isolates being resistance to penicillin (ampicillin, piperacillin, and amoxicillin plus clavulanate) associated with AmpC or ESBLs production, fluoroquinolones (ciprofloxacin and levofloxacin) as well as to trimethoprim/sulfamethoxazole and tetracycline (Amador et al., 2015; Kotlarska et al., 2015; Voigt et al., 2019; Gumede et al., 2021).
The ESBL and quinolone resistance genes found in E. coli and K. pneumoniae isolates from WWTP raw and treated water are frequently located on class 1 integrons with various gene cassette arrays as well as on IncP-1 and IncFIB plasmids, proving their high risk of spread to human and natural environments (Alouache et al., 2014; Hassen et al., 2021). The NDM-5 carbapenemase producer E. coli ST617 was reported in the effluent of a WWTP discharged into the river Rhine (Zurfluh et al., 2017), suggesting that E. coli resistant to carbapenems may be present in the community and are released into the aquatic environment with WWTP effluents.
Many studies identify hospital sewage as a more critical source of MDR enterobacterial strains, including carbapenem-resistant isolates, as compared to municipal wastewater (likely due to the selection pressure of antibiotic residues and biocides and higher fecal carriage of ARB in hospitals), underscoring the necessity of an appropriate treatment of the hospitals and other clinical settings wastewater prior to its discharge (Hutinel et al., 2019; King et al., 2020; Addae-Nuku et al., 2022; Mutuku et al., 2022).
Within the past 20 years, a clone of the sequence type ST131 has become the predominant MDR extraintestinal E. coli human pathogen (ExPEC) globally due to its ability to acquire antibiotic resistance (Croxen and Finlay, 2010). Thus, ExPEC strains show resistance to fluoroquinolones, extended-spectrum cephalosporins, primarily associated with CTX-M-15 (Coque et al., 2008) and carbapenems (O’Hara et al., 2014; Ortega et al., 2016; Stoesser et al., 2016). In addition, ST131 isolates were identified in an urban WWTP in the Czech Republic (Jamborova et al., 2018), in Norway (Paulshus et al., 2019), in Canada (Finn et al., 2020), in Nigeria (Ben Said et al., 2016), in the surface waters of the rural catchment area in Germany (Müller et al., 2018), hospital wastewater from Japan (Gomi et al., 2017), and in a wastewater treatment network in France (Bréchet et al., 2014).
Due to its capacity to maintain and transmit MGEs, K. pneumoniae is positioned as a critical trafficker for amplifying and spreading ARGs among the different environmental niches (Lai et al., 2019), including those encoding for last-resort antibiotics, such as carbapenems or colistin (Galarde-López et al., 2022). Colistin- and/or tigecycline-resistant K. pneumoniae ST29 strains, harboring the Tn21-like mercury resistance operon transposons and silver, copper, and arsenic resistance were detected in the WWTP influents from Japan (Hayashi et al., 2021). Lepuschitz et al. (2019) reported that water samples from rivers upstream of Australian cities were negative for ESBL and carbapenemase-producing K. pneumoniae. In contrast, all samples taken one to three kilometers downstream of the same cities’ WWTP release points were positive, demonstrating the impact of wastewater effluents and anthropogenic pollution on the aquatic environment (Amos et al., 2014).
Resistance to carbapenems is conferred mainly by the carbapenemase genes blaKPC, blaNDM, and blaOXA, carried by plasmids and transposons. Over 100 different K. pneumoniae STs clones have been described to carry blaKPC genes (Munoz-Price et al., 2013). The world’s widespread KPC-producing K. pneumoniae ST258 emerged in the 2000s as an important human pathogen in urinary and respiratory tract infections. The KPC-2-producing K. pneumoniae belonging to the international CC258 has been detected in the treated effluent of an urban WWTP receiving hospital wastewater (Zurfluh et al., 2017). The MDR, KPC-2-producing K. pneumoniae ST11, has been isolated from a sampling site in Tokyo Bay, Japan, near a WWTP (Sekizuka et al., 2018). A study in Puerto Rico 6 months after Hurricane Maria, a category V storm, revealed various clinically significant mobile β-lactam ARGs downstream of WWTP discharge, including KPC-2 within an ISKpn6-like transposase (Davis et al., 2020).
An NDM-1-producing K. pneumoniae ST147 clone was reported in the wastewater of a WWTP in Basel (Switzerland), most probably originating in the wastewater from clinical settings (Nüesch-Inderbinen et al., 2018). In addition, Surleac et al. (2020) described carbapenemases-producing K. pneumoniae ST35, ST219, ST364, ST395, ST485, and ST1878 in wastewaters collected from WWTPs in Southern Romania, of which ST395 has clinical importance, while ST35 and ST485 are sporadically related to clinical cases.
Hypervirulent K. pneumoniae strains are recognized as an urgent threat to human health, producing community-associated invasive diseases that can affect young, healthy people and hospital-acquired infections. In clinical settings, hypervirulent K. pneumoniae has a marked propensity to acquire antibiotic resistance and rarely carries virulence plasmids (Turton et al., 2019). The ST147 K. pneumoniae high-risk clones (blaNDM/blaOXA-48) have been identified in the wastewater of a full-care hospital, and the effluent of the WWTP receiving this wastewater, suggesting the persistence of this clone during the wastewater treatment in a study performed in Germany (Kehl et al., 2022). Also, the transmission of an MDR K. pneumoniae ST101 clone from hospital to wastewater and its persistence after chlorine treatment was demonstrated in Romania. The strains belonging to this clone harbored multiple acquired ARGs (incl. blaCTX-M- 15, blaOXA- 48) and chromosomal mutations involved in antibiotic resistance. Twenty-nine virulence genes were identified in iron acquisition, biofilm, pili formation, adherence, and the type six secretion system–T6SS-III (Popa et al., 2021). The persistence of carbapenemase-producing K. pneumoniae from hospital to environment via municipal WWTP has also been found in a study performed in the US (Loudermilk et al., 2022). A recent study has demonstrated that carbapenemase-producing, hyper-virulent K. pneumoniae strains could be transmitted from wastewater via bioaerosols to the upper respiratory tract of WWTP employees (Rolbiecki et al., 2021). All these studies demonstrate that the WWTPs contribute to the transmission and spreading of MDR Enterobacteriaceae in the aquatic environment, posing a risk for the communities living in proximity and, eventually, for the WWTP workers.
3. WWTPs effluents–A source of ESCAPE-related ARGs into the aquatic environment
ESCAPE-related ARGs, encoding every known type of mechanism (target protection, target modification, drug modification, reduced permeability or efflux), have been widely reported in both wastewater and downstream environments across continents (Freeman et al., 2018; Pazda et al., 2019; Surleac et al., 2020; Popa et al., 2021; Gheorghe-Barbu et al., 2022). These ARGs have been detected in bacterial isolates or total genomic DNA samples (Table 1).
Dutch WWTPs were shown to discharge on average 106 copies of ARGs per liter of effluent to the receiving water bodies, the dominant ones being the ARGs to sulphonamides (sul1) and macrolides (ermB), their high occurrence being possibly associated with MGEs or might be due to the prolonged use of these antibiotics in humans and animals (Pallares-Vega et al., 2019). Some ARGs, like the resistance to sulfonamides (sul1, sul2), glycopeptides (vanA), and tetracycline (tetM), were shown to be more abundant in treated wastewater, a fact that might also be explained by a long history of usage of sulphonamide and tetracycline antibiotics (Heuer et al., 2011; Caucci et al., 2016; Pärnänen et al., 2019). Several other ARGs conferring resistance to different classes of antibiotics, including macrolide-lincosamide-streptogramin B (MLSB) (ermA, ermB), beta-lactams (blaSHV-02, blaVEB, blaOXA-10, blaCTX-M-32, blaTEM, and blaCMY-2), glycopeptides (vanA), aminoglycosides (aadA, aadA1, and aadA2) and also multidrug resistance (qacEdelta1) were significantly enriched in effluent wastewater from Chinese and European WWTPs (Hembach et al., 2017). It has been hypothesized that a positive selection or community change during wastewater treatment could enrich vanA (Pärnänen et al., 2019). Worrisome is the detection of the colistin resistance gene mcr-1 in influent samples of seven WWTPs investigated in Germany and its release into the aquatic environment (Hembach et al., 2017).
The absolute abundance of nine ARGs and a class 1 integron-associated integrase gene in the wastewater effluents of 16 WWTPs from ten different European countries could be ranked along the following order: int1 > sul1 > tetM > blaOXA–58 > blaTEM > blaOXA–48 > blaCTX–M–32 > mcr-1 > blaCTX–M–15 > blaKPC–3 (Cacace et al., 2019). Downstream of the plants, these ARGs were quantifiable in many cases, suggesting an impact of the effluent on the receiving waterbody (Cacace et al., 2019). The aph(III)a, blaOXA, ermB, ermF, sul1, and tetM resistance genes had a wide-spread occurrence in effluents discharged from 12 WWTPs in the Danube River Basin (Romania, Serbia, Hungary, Slovenia, Croatia, Slovakia, Czechia, Austria, Germany); in this study, the highest absolute concentrations (gene copies/mL) were observed for four ARGs (aph(III)a, tetM, vanA, and mecA) at the WWTP Bucharest (Romania), four ARGs (blaOXA, blaSHV, qnrS, and tetB) at the WWTP Sabac (Serbia), two ARGs (ermB, sul1) at the WWTP Varazdin (Croatia) and one ARG (ermF) at the WWTP Brno (Czechia) (Alygizakis et al., 2019).
The analysis of the abundance of resistance and virulence genes in Staphylococcus spp. strains from untreated and treated wastewater, an activated sludge (AS) bioreactor, and from surface water collected upstream and downstream the WWTP, revealed that S. aureus was present in 63% of the samples, with 20% of the strains carrying the vanA gene. The hla virulence gene was present in most of the isolates (80%), followed by the PVL gene (Zieliński et al., 2020). The highest abundance of virulence and resistance genes among S. aureus strains was observed in the untreated wastewater, gradually decreasing in treated water and downstream river water, probably due to the synergic effect of wastewater treatment, dilution, as well as influence location-specific abiotic and biotic factors, such as temperature, pH, solar radiation, mineral and organic content, microbial community structure, seasonal influence, competition, temporal variations in the prescription level of antibiotics, etc. (Caucci et al., 2016; Auguet et al., 2017; Zieliński et al., 2020).
The analysis of the water metagenomes sampled immediately downstream, and at 6.4 km from a WWTP effluent released into the surface water of a small river located in the rural area of Knox County, Ohio, showed that the metagenome just downstream of the WWTP effluent was substantially enriched in 15 different ARGs relative to the remote location. Among these enriched ARGs were 6 ARGs commonly associated with A. baumannii, such as msrE, mphE (macrolide resistance), and tet (tetracycline resistance), that persisted 6.4 km downriver. In addition, the samples from directly downstream of the WWTP were enriched in Acinetobacter spp. and gut-associated taxa: Bacteroides and Firmicutes. The ARG levels, taxa composition, and prevalence were independent of the seasonal effluent chlorination and nitrogen and phosphorus concentrations (Murphy et al., 2021).
Using a hiseq-based metagenomic sequencing approach to investigate the core resistome of urban WWTPs persisting through wastewater treatment processes, the influents and effluents of 12 urban WWTPs in Republic of Korea were analyzed. The abundance of some core ARGs such as macrolide-lincosamide-streptogramin–and tetracycline-resistance genes was higher in the influent samples, while others such as sul1, APH(3″)-lb, and RbpA were more abundant in the effluent. Most core ARGs were carried by ESCAPE pathogens, such as A. baumannii, E. cloacae, and P. aeruginosa. Also, the abundance of core ARGs in the effluent was correlated with an abundance of phages, suggesting a role of transduction in disseminating ARGs from WWTP effluent to surface water (Raza et al., 2022).
Generally, conventional WWTP treatments can reduce ARGs’ overall richness, concentration, and relative abundance in variable proportions (Yang et al., 2014; Pallares-Vega et al., 2019; Li et al., 2022). The primary treatment is usually not efficient in removing pathogens and ARB, requiring the use of advanced treatments (e.g., sand and membrane filtration) or disinfection methodologies (e.g., chlorination, UV radiation, and ozonation) after conventional activated sludge treatment (Hazra and Durso, 2022). Many WWTPs only use conventional activated sludge for wastewater treatment, which, however, has been shown to have better removed fecal coliforms, enterococci, and E. coli in comparison to septic tank systems; but, despite this reduction, an increase in the MDR profiles has been observed during wastewater treatment (Barrios-Hernández et al., 2019; Hazra and Durso, 2022). From an analysis of the influent concentrations and the removal of ARGs in a large number of WWTPs in the Netherlands, the researchers concluded that WWTP design parameters such as size, presence of primary clarification, type of phosphorus removal, and operational parameters such as hydraulic retention time, sludge retention time, anaerobic contact time and effluent total suspended solids did not affect the removal of the studied ARGs and MGEs significantly (Pallares-Vega et al., 2019). Disinfection steps, i.e., chlorination (Mao et al., 2015; Yuan et al., 2015), UV irradiation (Sousa et al., 2017), and ozone (Alexander et al., 2016), alone or in combination (UV and chlorination) (Zhang et al., 2015), were applied in WWTPs to assess their efficiency in removing ARB and ARGs. The results showed that they were not able to altogether remove the ARGs. Moreover, some of the cleaning methods could increase the proportion of ARB through the alteration of bacterial cell permeability by the activation of efflux pumps, accumulation of intermediate disinfection by-products, and promotion of HGT, particularly in case of prolonged contact with low chlorine concentrations (Huang et al., 2011). Chlorination could result in the survival of the ARB in more significant percentages due to their different susceptibility to disinfection (Mao et al., 2015). The ozone treatment causes DNA damage but has a reduced effect on guanine-rich DNA (Alexander et al., 2016). UV light is partially absorbed by organic and inorganic matter present in wastewater; thus, the DNA damage depends on the UV dosage; additionally, different sensitivity of ARB to UV radiation was noticed (Hazra and Durso, 2022). A membrane bioreactor has been shown to increase the municipal WWTP efficiency for decreasing the abundance of ARGs and loads of E. coli, enterococci, and P. aeruginosa strains (Ng et al., 2019). Still, the ARGs of 12 antibiotic classes, including aminoglycosides, beta-lactams, macrolides-lincosamides-streptogramins, polymyxins, quinolones, rifamycin, tetracyclines, and multidrug resistance, persisted during treatment. Constructed wetlands (CWs) seem to be a sustainable “green” wastewater treatment option that can be used as an alternative to WWTP or as a component of conventional WWTP, having the ability to remove antibiotics, ARB, and ARGs (Dires et al., 2018; de Oliveira et al., 2019; Riaz et al., 2020; Hazra and Durso, 2022). CWs can be used alone or even after primary treatment, contributing to 2–4 log pathogens reduction (Sleytr et al., 2007; Karimi et al., 2014) and increasing the performance of WWTP when coupled as secondary/tertiary units to primary treatment (Chen et al., 2014).
A recent comprehensive review on the performance of primary, secondary and tertiary treatments of wastewater in removing antibiotics, bacterial pathogens, ARB and ARGs concludes that combinations of different methods are more efficient in the elimination of a large number of ARB (Hazra and Durso, 2022).
4. Conclusion and perspectives
In this review, we have focused on the relationships between ESCAPE pathogens and the WWTPs. Different studies performed in different geographical areas report that ESCAPE pathogens and their associated ARGs, including high-risk clones and resistance determinants to last resort antibiotics such as carbapenems and colistin, as well as MDR platforms are present in wastewater. The sources for the enrichment of ESCAPE pathogens are diverse, including clinical settings, animal husbandry facilities, urban and municipal wastewater systems, etc. Some studies are demonstrating the clonal relationships and dissemination of ESCAPE clinical strains, such as A. baumannii, P. aeruginosa and K. pneumoniae into the wastewater via hospital effluents. Also, WWTPs seem to provide a suitable environment for the enrichment of resistance and virulence determinants in ESCAPE bacteria, such as enterococci and S. aureus. Therefore, the efficiency of different wastewater treatment processes and combinations thereof regarding the removal of clinically relevant ARB species and ARGs, as well as the influence of water quality factors on their performance should be explored and monitored. Also, the development of more effective disinfection and treatment methods could contribute to prevent or reduce the release of antibiotics, ARB and ARGs into the natural environment via WWTP effluents. In order to achieve these goals, research is needed to select the most appropriate indicators (ESCAPE bacteria and/or ARGs) and standard methods for assessing them, to facilitate the comparison of resistance levels between countries and different WWTPs (Uluseker et al., 2021). The quantification of known ARGs along the wastewater treatment chain need to be complemented with information regarding the origin of these genes, their genetic context, the type and level of selective pressure, potential for HGT and compatibility with putative pathogenic hosts (Bengtsson-Palme and Larsson, 2015). Also, the possible use of current microbiological indicator species such as enterococci and E. coli in managing the risks associated with the environmental resistome, depending on the total counts as well as the resistance level in these strains, needs to be explored in the future. This knowledge will allow to establish quality threshold limits regarding the AR for point sources and effluents that are being directly discharged to water bodies and agricultural land for irrigation, and ultimately to reinforce the barrier role of WWTPs against environmental and public health threats.
Author contributions
LGM and MCC conceived the review. LGM, MP, IG-B, ICB, GPG, and COV wrote the draft and drown the figure. DR-M, FB, HB, C-FF, MAK, BS, LW, DGJL, DN, KR, AMRH, AW, and HS completed and corrected the drafts. MCC integrated all the comments from the authors and assembled the final form of the review. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by the ERANET-JPI-EC-AMR-AWARE-WWTP, Contract nos. 26/1.06.2017, C1.2.PFE-CDI.2021-587, and 41PFE/30.12.2021, and the Swedish Research Council Formas, Grant no. 2021-00922. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
Addae-Nuku, D., Kotey, F., Dayie, N., Osei, M., Tette, E., Debrah, P., et al. (2022). Multidrug-resistant bacteria in hospital wastewater of the Korle Bu teaching hospital in Accra, Ghana. Environ. Health Insights 16:11786302221130613. doi: 10.1177/11786302221130613
Alexander, J., Knopp, G., Dötsch, A., Wieland, A., and Schwartz, T. (2016). Ozone treatment of conditioned wastewater selects antibiotic resistance genes, opportunistic bacteria, and induces strong population shifts. Sci. Totaal Environ. 559, 103–112. doi: 10.1016/j.scitotenv.2016.03.154
Alouache, S., Estepa, V., Messai, Y., Ruiz, E., Torres, C., and Bakour, R. (2014). Characterization of ESBLs and associated quinolone resistance in Escherichia coli and Klebsiella pneumoniae isolates from an urban wastewater treatment plant in Algeria. Microb. Drug Resist. 20, 30–38. doi: 10.1089/mdr.2012.0264
Alygizakis, N., Besselink, H., Paulus, G., Oswald, P., Hornstra, L., et al. (2019). Characterization of wastewater effluents in the Danube river basin with chemical screening, in vitro bioassays and antibiotic resistant genes analysis. Environ. Int. 127, 420–429. doi: 10.1016/j.envint.2019.03.060
Amador, P., Fernandes, R., Prudêncio, M., Barreto, M., and Duarte, I. (2015). Antibiotic resistance in wastewater: occurrence and fate of Enterobacteriaceae producers of class a and class C β-lactamases. J. Environ. Sci. Health A Tox Hazard. Subst. Environ. Eng. 50, 26–39.
Amirsoleimani, A., Brion, G., Diene, S., François, P., and Richard, E. (2019). Prevalence and characterization of Staphylococcus aureus in wastewater treatment plants by whole genomic sequencing. Water Res. 158, 193–202. doi: 10.1016/j.watres.2019.04.035
Amos, G., Hawkey, P., Gaze, W., and Wellington, E. (2014). Waste water effluent contributes to the dissemination of CTX-M-15 in the natural environment. J. Antimicrob. Chemother. 69, 1785–1791. doi: 10.1093/jac/dku079
Anderson, K., Whitlock, J., and Harwood, V. (2015). Persistence and differential survival of fecal indicator bacteria in subtropical waters and sediments. Appl. Environ. Microbiol. 71, 3041–3048. doi: 10.1128/AEM.71.6.3041-3048.2005
Andersson, D., and Hughes, D. (2017). Selection and transmission of antibiotic-resistant bacteria. Microbiol. Spectr. 5:MTBP0013-2016. doi: 10.1128/microbiolspec.MTBP-0013-2016
Asokan, G., Ramadhan, T., Ahmed, E., and Sanad, H. (2019). WHO global priority pathogens list: a bibliometric analysis of medline-pubmed for knowledge mobilization to infection prevention and control practices in Bahrain. Oman Med. J. 34, 184–193. doi: 10.5001/omj.2019.37
Aubertheau, E., Stalder, T., Mondamert, L., Ploy, M., Dagot, C., and Labanowski, J. (2017). Impact of wastewater treatment plant discharge on the contamination of river biofilms by pharmaceuticals and antibiotic resistance. Sci. Total Environ. 579, 1387–1398. doi: 10.1016/j.scitotenv.2016.11.136
Auguet, O., Pijuan, M., Borrego, C., Rodriguez-Mozaz, S., Triadó-Margarit, X., Giustina, S., et al. (2017). Sewers as potential reservoirs of antibiotic resistance. Sci. Total Environ. 605-606, 1047–1054.
Azuma, T., Murakami, M., Sonoda, Y., Ozaki, A., and Hayashi, T. (2022). Occurrence and quantitative microbial risk assessment of methicillin-resistant Staphylococcus aureus (MRSA). in a sub-catchment of the Yodo river, Basin, Japan. Antibiotics 11:1355. doi: 10.3390/antibiotics11101355
Baghani, A., Alimohammadi, M., Aliramezani, A., Talebi, M., Mesdaghinia, A., and Douraghi, M. (2020). Isolation and characterization of a multidrug-resistant Clostridioides difficile toxinotype V from municipal wastewater treatment plant. J. Environ. Health Sci. Eng. 18, 1281–1288. doi: 10.1007/s40201-020-00546-0
Baker, S., Thomson, N., Weill, F., and Holt, K. (2018). Genomic insights into the emergence and spread of antimicrobial-resistant bacterial pathogens. Science 360, 733–738.
Barrios-Hernández, M., Pronk, M., Garcia, H., Boersma, A., Brdjanovic, D., van Loosdrecht, M., et al. (2019). Removal of bacterial and viral indicator organisms in full-scale aerobic granular sludge and conventional activated sludge systems. Water Res. X 6, 100040. doi: 10.1016/j.wroa.2019.100040
Ben Said, L., Jouini, A., Alonso, C., Klibi, N., Dziri, R., Boudabous, A., et al. (2016). Characteristics of extended-spectrum β-lactamase (ESBL)- and pAmpC beta-lactamase-producing Enterobacteriaceae of water samples in Tunisia. Sci. Total Environ. 550, 1103–1109.
Bengtsson-Palme, J., and Larsson, D. G. J. (2015). Antibiotic resistance genes in the environment: prioritizing risks. Nat. Rev. Microbiol. 13:396.
Bengtsson-Palme, J., Kristiansson, E., and Larsson, D. G. J. (2018). Environmental factors influencing the development and spread of antibiotic resistance. FEMS Microbiol Rev. 42:fux053.
Boopathy, R. (2017). Presence of methicillin-resistant Staphylococcus aureus (MRSA). in sewage treatment plant. Bioresour. Technol. 240, 144–148. doi: 10.1016/j.biortech.2017.02.093
Börjesson, S., Melin, S., Matussek, A., and Lindgren, P. (2009). A seasonal study of the mecA gene and Staphylococcus aureus including methicillin-resistant S. aureus in a municipal wastewater treatment plant. Water Res. 43, 925–932. doi: 10.1016/j.watres.2008.11.036
Bougnom, B., and Piddock, L. (2017). Wastewater for urban agriculture: a significant factor in dissemination of antibiotic resistance. Environ. Sci. Technol. 51, 5863–5864. doi: 10.1021/acs.est.7b01852
Boyd, N., Teng, C., and Frei, C. (2021). Brief overview of approaches and challenges in new antibiotic development: a focus on drug repurposing. Front. Cell Infect. Microbiol. 11:684515. doi: 10.3389/fcimb.2021.684515
Bréchet, C., Plantin, J., Sauget, M., Thouverez, M., Talon, D., Cholley, P., et al. (2014). Wastewater treatment plants release large amounts of extended-spectrum -lactamase-producing Escherichia coli into the environment. Clin. Infect. Dis. 58, 1658–1665. doi: 10.1093/cid/ciu190
Brown, P., Borowska, E., Schwartz, T., and Horn, H. (2019). Impact of the particulate matter from wastewater discharge on the abundance of antibiotic resistance genes and facultative pathogenic bacteria in downstream river sediments. Sci. Total Environ. 649, 1171–1178. doi: 10.1016/j.scitotenv.2018.08.394
Cacace, D., Fatta-Kassinos, D., Manaia, C., Cytryn, E., Kreuzinger, N., and Rizzo, L. (2019). Antibiotic resistance genes in treated wastewater and in the receiving water bodies: a pan-European survey of urban settings. Water Res. 162, 320–330. doi: 10.1016/j.watres.2019.06.039
Cattoir, V., Poirel, C., Aubert, C., and Nordmann, P. (2008). Unexpected occurrence of plasmid-mediated quinolone resistance determinants in environmental Aeromonas spp. Emerg. Infect. Dis. 14, 231–237. doi: 10.3201/eid1402.070677
Caucci, S., Karkman, A., Cacace, D., Rybicki, M., Timpel, P., and Veiko. (2016). Berendonk, seasonality of antibiotic prescriptions for outpatients and resistance genes in sewers and wastewater treatment plant outflow. FEMS Microbiol. Ecol. 5:fiw060. doi: 10.1093/femsec/fiw060
Cheesman, M., Ilanko, A., Blonk, B., and Cock, I. (2017). Pharmacognosy Reviews Developing New antimicrobial therapies: are synergistic combinations of plant extracts/compounds with conventional antibiotics the solution? Pharmacognosy Rev. 11, 57–72. doi: 10.4103/phrev.phrev_21_17
Chen, J., Liu, Y.-S., Su, H.-C., Ying, G.-G., Liu, F., Liu, S.-S., et al. (2014). Removal of antibiotics and antibiotic resistance genes in rural wastewater by an integrated constructed wetland. Environ. Sci. Pollut. Res. 22, 1794–1803.
Chen, J., Quiles-Puchalt, N., Chiang, Y., Bacigalupe, R., Fillol-Salom, A., Chee, M., et al. (2018). Genome hypermobility by lateral transduction. Science 362, 207–212. doi: 10.1126/science.aat5867
Coque, T., Novais, A., Carattoli, A., Poirel, L., Pitout, J., Peixe, L., et al. (2008). Dissemination of clonally related Escherichia coli strains expressing extended-spectrum β-lactamase CTX-M-15. Emerg. Infect. Dis. 14, 195–200.
Croxen, M., and Finlay, B. (2010). Molecular mechanisms of Escherichia coli pathogenicity. Nat. Rev. Microbiol. 8, 26–38.
Danner, M., Robertson, A., Behrends, V., and Reiss, J. (2019). Antibiotic pollution in surface fresh waters: occurrence and effects. Sci. Total Environ. 664, 793–804.
Davis, B., Riquelme, M., Ramirez-Toro, G., Bandaragoda, C., Garner, E., Rhoads, W., et al. (2020). Demonstrating an integrated antibiotic resistance gene surveillance approach in Puerto rican watersheds post-hurricane Maria. Environ. Sci. Technol. 54, 15108–15119. doi: 10.1021/acs.est.0c05567
de Oliveira, M., Frihling, B., Velasques, J., Filho, F., Cavalheri, P., and Migliolo, L. (2019). Pharmaceuticals residues and xenobiotics contaminants: occurrence, analytical techniques and sustainable alternatives for wastewater treatment. Sci. Total Environ. 705:135568. doi: 10.1016/j.scitotenv.2019.135568
Del Barrio-Tofiño, E., López-Causapé, C., and Oliver, A. (2020). Pseudomonas aeruginosa epidemic high-risk clones and their association with horizontally-acquired β-lactamases: 2020 update. Int. J. Antimicrob. Agents 56:106196.
Deng, Y., Bao, X., Ji, L., Chen, L., Liu, J., Chen, D., et al. (2015). Resistance integrons: class 1, 2 and 3 integrons. Ann. Clin. Microbiol. Antimicrob. 14:45.
Denissen, J., Reyneke, B., Waso-Reyneke, M., Havenga, B., Barnard, T., Khan, S., and Khan, W. (2022). Prevalence of ESKAPE pathogens in the environment: Antibiotic resistance status, community-acquired infection and risk to human health. Int. J. Hyg. Environ. Health 244:114006. doi: 10.1016/j.ijheh.2022.114006
Dhingra, S., Rahman, N., Peile, E., Rahman, M., Sartelli, M., Islam, T., et al. (2020). Microbial resistance movements: an overview of global public health threats posed by antimicrobial resistance, and how best to counter. Front. Public Health 8:535668.
Dires, S., Birhanu, T., Ambelu, A., and Sahilu, G. (2018). Antibiotic resistant bacteria removal of subsurface flow constructed wetlands from hospital wastewater. J. Environ. Chem. Eng. 6, 4265–4272.
Ebmeyer, S., Kristiansson, E., and Larsson, D. G. J. (2021). A framework for identifying the recent origins of mobile antibiotic resistance genes. Commun. Biol. 4:8. doi: 10.1038/s42003-020-01545-5
European Centre for Disease Prevention and Control (2022). Antimicrobial Resistance in the EU/EEA (EARS-Net). - Annual Epidemiological Report 2021. Stockholm: ECDC.
Ferreira, A., Marchetti, D., De Oliveira, L., Gusatti, C., Fuentefria, D., and Corção, G. (2011). Presence of OXA-23-producing isolates of Acinetobacter baumannii in wastewater from hospitals in southern Brazil. Microb. Drug. Resist. 17, 221–227. doi: 10.1089/mdr.2010.0013
Ferreira, da Silva, M., Tiago, I., Verıssımo, A., Boaventura, R., Nunes, O., et al. (2006). Antibiotic resistance of enterococci and related bacteria in an urban wastewater treatment plant. FEMS Microbiol. Ecol. 55, 322–329. doi: 10.1111/j.1574-6941.2005.00032.x
Finn, T., Scriver, L., Lam, L., Duong, M., Peirano, G., Lynch, T., et al. (2020). A comprehensive account of Escherichia coli sequence type 131 in wastewater reveals an abundance of fluoroquinolone-resistant clade a strains. Appl. Environ. Microbiol. 86:e01913-19. doi: 10.1128/AEM.01913-19
Flach, C.-F., Genheden, M., Fick, J., and Larsson, D. G. J. (2018). A comprehensive screening of E. coli isolates from Scandinavia’s largest sewage treatment plant indicates no selection for antibiotic resistance. Environ. Sci. Technol. 52:11419. doi: 10.1021/acs.est.8b03354
Fletcher, S. (2015). Understanding the contribution of environmental factors in the spread of antimicrobial resistance. Environ. Health Prev. Med. 20, 243–252.
Forsberg, K., Reyes, A., Wang, B., Selleck, E., Sommer, M., and Dantas, G. (2012). The shared antibiotic resistome of soil bacteria and human pathogens. Science 337, 1107–1111.
Freeman, C., Scriver, L., Neudorf, K., Truelstrup, H., Jamieson, R., and Yost, C. (2018). Antimicrobial resistance gene surveillance in the receiving waters of an upgraded wastewater treatment plant. Facets 3, 128–138.
Fuentefria, D., Ferreira, A., and Corção, G. (2011). Antibiotic-resistant Pseudomonas aeruginosa from hospital wastewater and superficial water: are they genetically related? J. Environ. Manag. 92, 250–255. doi: 10.1016/j.jenvman.2010.09.001
Galarde-López, M., Velazquez-Meza, M., Bobadilla-Del-Valle, M., Carrillo-Quiroz, B., Cornejo-Juárez, P., Ponce-de-León, A., et al. (2022). Surveillance of antimicrobial resistance in hospital wastewater: identification of carbapenemase-producing Klebsiella spp. Antibiotics 11:288. doi: 10.3390/antibiotics11030288
Gheorghe-Barbu, I., Barbu, I. C., Popa, L. I., Pîrcălăbioru, G. G., Popa, M., Măruţescu, L., et al. (2022). Temporo-spatial variations in resistance determinants and clonality of Acinetobacter baumannii and Pseudomonas aeruginosa strains from Romanian hospitals and wastewaters. Antimicrob. Resist. Infect. Control 11:115. doi: 10.1186/s13756-022-01156-1
Giebułtowicz, J., Tyski, S., Wolinowska, R., Grzybowska, W., Zaręba, T., Drobniewska, A., et al. (2018). Occurrence of antimicrobial agents, drug-resistant bacteria, and genes in the sewage-impacted Vistula river (Poland). Environ. Sci. Pollut. Res. Int. 25, 5788–5807. doi: 10.1007/s11356-017-0861-x
Goic-Barisic, I., Seruga Music, M., Kovacic, A., Tonkic, M., and Hrenovic, J. (2017). Pan drug-resistant environmental isolate of Acinetobacter baumannii from croatia. Microb. Drug Resist. 23, 494–496. doi: 10.1089/mdr.2016.0229
Goldstein, R., Micallef, S., Gibbs, S., Davis, J., He, X., George, A., et al. (2012). Methicillin-resistant Staphylococcus aureus (MRSA). detected at four US wastewater treatment plants. Environ. Health Perspect. 120:1551. doi: 10.1289/ehp.1205436
Golle, A., Janezic, S., and Rupnik, M. (2017). Low overlap between carbapenem resistant Pseudomonas aeruginosa genotypes isolated from hospitalized patients and wastewater treatment plants. PLoS One 12:e0186736. doi: 10.1371/journal.pone.0186736
Gómez, P., Lozano, C., Benito, D., Estepa, V., Tenorio, C., Zarazaga, M., et al. (2016). Characterization of staphylococci in urban wastewater treatment plants in Spain, with detection of methicillin resistant Staphylococcus aureus ST398. Environ. Pollut. 212, 71–76. doi: 10.1016/j.envpol.2016.01.038
Gomi, R., Matsuda, T., Matsumura, Y., Yamamoto, M., Tanaka, M., Ichiyama, S., et al. (2017). Occurrence of clinically important lineages, including the sequence type 131 C1-M27 subclone, among extended-spectrum- -lactamase-producing Escherichia coli in wastewater. Antimicrob. Agents Chemother. 61:e00564-17. doi: 10.1128/AAC.00564-17
Gotkowska-Płachta, A. (2021). The prevalence of virulent and multidrug-resistant enterococci in river water and in treated and untreated municipal and hospital wastewater. Int. J. Environ. Res. Public Health 18:563. doi: 10.3390/ijerph18020563
Gumede, S., Abia, A., Amoako, D., and Essack, S. (2021). Analysis of wastewater reveals the spread of diverse extended-spectrum β-lactamase-producing E. coli strains in uMgungundlovu district, South Africa. Antibiotics 10:860.
Hamiwe, T., Kock, M., Magwira, C., Antiabong, J., and Ehlers, M. (2019). Occurrence of enterococci harbouring clinically important antibiotic resistance genes in the aquatic environment in Gauteng, South Africa. Environ. Pollut. 245, 1041–1049. doi: 10.1016/j.envpol.2018.11.040
Hannan, S., Ready, D., Jasni, A., Rogers, M., Pratten, J., and Roberts, A. (2010). Transfer of antibiotic resistance by transformation with eDNA within oral biofilms. FEMS Immunol. Med. Microbiol. 59, 345–349. doi: 10.1111/j.1574-695X.2010.00661.x
Hassen, B., Abbassi, M., Ruiz-Ripa, L., Mama, O., Ibrahim, C., Benlabidi, S., et al. (2021). Genetic characterization of extended-spectrum β-lactamase-producing Enterobacteriaceae from a biological industrial wastewater treatment plant in Tunisia with detection of the colistin-resistance mcr-1 gene. FEMS Microbiol. Ecol. 97:fiaa231.
Hayashi, W., Iimura, M., Soga, E., Koide, S., Izumi, K., Yoshida, S., et al. (2021). Presence of colistin- and tigecycline-resistant Klebsiella pneumoniae ST29 in municipal wastewater influents in Japan. Microb. Drug Resist. 27, 1433–1442. doi: 10.1089/mdr.2020.0514
Hazra, M., and Durso, L. (2022). Performance efficiency of conventional treatment plants and constructed wetlands towards reduction of antibiotic resistance. Antibiotics 11:114. doi: 10.3390/antibiotics11010114
Hembach, N., Schmid, F., Alexander, J., Hiller, C., Rogall, E., and Schwartz, T. (2017). Occurrence of the mcr-1 colistin resistance gene and other clinically relevant antibiotic resistance genes in microbial populations at different municipal wastewater treatment plants in Germany. Front. Microbiol. 8:1282. doi: 10.3389/fmicb.2017.01282
Hennequin, C., Aumeran, C., Robin, F., Traore, O., and Forestier, C. (2012). Antibiotic resistance and plasmid transfer capacity in biofilm formed with a CTX-M-15-producing Klebsiella pneumoniae isolate. J. Antimicrob. Chemother. 67, 2123–2130. doi: 10.1093/jac/dks169
Hensgens, M., Keessen, E., Squire, M., Riley, T., Koene, M., and de Boer, E. (2012). Clostridium difficile infection in the community: a zoonotic disease? Clin. Microbiol. Infect. 18, 635–645. doi: 10.1111/j.1469-0691.2012.03853.x
Heuer, H., Schmitt, H., and Smalla, K. (2011). Antibiotic resistance gene spread due to manure application on agricultural fields. Curr. Opin. Microbiol. 14, 236–243.
Higgins, P., Hrenovic, J., Seifert, H., and Dekic, S. (2018). Characterization of Acinetobacter baumannii from water and sludge line of secondary wastewater treatment plant. Water Res. 140, 261–267. doi: 10.1016/j.watres.2018.04.057
Hocquet, D., Muller, A., and Bertrand, X. (2016). What happens in hospitals does not stay in hospitals: antibiotic-resistant bacteria in hospital wastewater systems. J. Hosp. Infect. 93, 395–402.
Huang, J., Hu, H., Tang, F., Li, Y., Lu, S., and Lu, Y. (2011). Inactivation and reactivation of antibiotic-resistant bacteria by chlorination in secondary effluents of a municipal wastewater treatment plant. Water Res. 45, 2775–2781. doi: 10.1016/j.watres.2011.02.026
Hubeny, J., Korzeniewska, E., Buta-Hubeny, M., Zieliński, W., Rolbiecki, D., and Harnisz, M. (2022). Characterization of carbapenem resistance in environmental samples and Acinetobacter spp. isolates from wastewater and river water in Poland. Sci. Total Environ. 822:153437. doi: 10.1016/j.scitotenv.2022.153437
Hutchings, M., Truman, A., and Barrie Wilkinson, B. (2019). Antibiotics: past, present and future. Curr. Opin. Microbiol. 51, 72–80.
Hutinel, M., Huijbers, P., Fick, J., Åhrén, C., Larsson, D. G. J., and Flach, C.-F. (2019). Population-level surveillance of antibiotic resistance in Escherichia coli through sewage analysis. Euro Surveillance 24:1800497. doi: 10.2807/1560-7917.ES.2019.24.37.1800497
Jäger, T., Hembach, N., Elpers, C., Wieland, A., Alexander, J., Hiller, C., et al. (2018). Reduction of antibiotic resistant bacteria during conventional and advanced wastewater treatment, and the disseminated loads released to the environment. Front. Microbiol. 9:2599. doi: 10.3389/fmicb.2018.02599
Jamborova, I., Johnston, B., Papousek, I., Kachlikova, K., Micenkova, L., Clabots, C., et al. (2018). Extensive genetic commonality among wildlife, wastewater, community, and nosocomial isolates of Escherichia coli sequence type 131 (H30R1 and H30Rx subclones). that carry blaCTX–M–27 or blaCTX–M–15. Antimicrob. Agents Chemother. 62:e00519-18. doi: 10.1128/AAC.00519-18
Karimi, B., Ehrampoush, M., and Jabary, H. (2014). Indicator pathogens, organic matter and LAS detergent removal from wastewater by constructed subsurface wetlands. J. Environ. Health Sci. Eng. 12:52. doi: 10.1186/2052-336X-12-52
Karkman, A., Pärnänen, K., and Larsson, D. G. J. (2019). Fecal pollution can explain antibiotic resistance gene abundances in anthropogenically impacted environments. Nat. Commun. 10:80.
Katz, L., and Baltz, R. (2016). Natural product discovery: past, present, and future. J. Ind. Microbiol. Biotechnol. 43, 155–176.
Kehl, K., Schallenberg, A., Szekat, C., Albert, C., Sib, E., Exner, M., et al. (2022). Dissemination of carbapenem resistant bacteria from hospital wastewater into the environment. Sci. Total Environ. 806:151339.
Kerr, K., and Snelling, A. (2009). Pseudomonas aeruginosa: a formidable and ever-present adversary. J. Hosp Infect. 73, 338–344. doi: 10.1016/j.jhin.2009.04.020
King, T., Schmidt, S., and Essack, S. (2020). Antibiotic resistant Klebsiella spp. from a hospital, hospital effluents and wastewater treatment plants in the uMgungundlovu district, KwaZulu-Natal, South Africa. Sci. Total Environ. 712:135550. doi: 10.1016/j.scitotenv.2019.135550
Kotlarska, E., Łuczkiewicz, A., Pisowacka, M., and Burzyński, A. (2015). Antibiotic resistance and prevalence of class 1 and 2 integrons in Escherichia coli isolated from two wastewater treatment plants, and their receiving waters (Gulf of Gdansk, Baltic sea, Poland). Environ. Sci. Pollut. Res. Int. 22, 2018–2030. doi: 10.1007/s11356-014-3474-7
Kozajda, A., and Jeżak, K. (2020). Occupational exposure to Staphylococcus aureus in the wastewater treatment plants environment. Med. Pr. 71, 265–278. doi: 10.13075/mp.5893.00946
Kraemer, S., Ramachandran, A., and Perron, G. (2019). Antibiotic pollution in the environment: from microbial ecology to public policy. Microorganisms 7:180. doi: 10.3390/microorganisms7060180
Kraupner, N., Hutinel, M., Schumacher, K., Gray, D., Genheden, M., Fick, J., et al. (2021). Evidence for selection of multi-resistant E. coli by hospital effluent. Environ. Int. 150:106436. doi: 10.1016/j.envint.2021.106436
Lai, Y., Lu, M., and Hsueh, P. (2019). Hypervirulence and carbapenem resistance: two distinct evolutionary directions that led high-risk Klebsiella pneumoniae clones to epidemic success. Expert Rev. Mol. Diagnostics 19, 825–837. doi: 10.1080/14737159.2019.1649145
Larsson, D. G. J., and Flach, C.-F. (2022). Antibiotic resistance in the environment. Nat. Rev. Microbiol. 20, 257–269.
Leclercq, R., Oberle, K., Galopin, S., Cattoir, V., Budzinski, H., and Petit, F. (2013). Changes in enterococcal populations and related antibiotic resistance along a medical center-wastewater treatment plant-river continuum. Appl. Environ. Microbiol. 79, 2428–2434. doi: 10.1128/AEM.03586-12
Lépesová, K., Kraková, L., Pangallo, D., Medved’ová, A., Olejníková, P., Mackul’ak, T., et al. (2018). Prevalence of antibiotic-resistant coliform bacteria, Enterococcus spp. and Staphylococcus spp. in wastewater sewerage biofilm. J. Glob. Antimicrob. Resist. 14, 145–151. doi: 10.1016/j.jgar.2018.03.008
Lepuschitz, S., Schill, S., Stoeger, A., Pekard-Amenitsch, S., Huhulescu, S., Inreiter, N., et al. (2019). Whole genome sequencing reveals resemblance between ESBL-producing and carbapenem resistant Klebsiella pneumoniae isolates from Austrian rivers and clinical isolates from hospitals. Sci. Total Environ. 662, 227–235. doi: 10.1016/j.scitotenv.2019.01.179
Li, S., Ondon, B., Ho, S., Jiang, J., and Li, F. (2022). Antibiotic resistant bacteria and genes in wastewater treatment plants: from occurrence to treatment strategies. Sci. Total Environ. 838(Pt 4):156544.
Limayem, A., Wasson, S., Mehta, M., Pokhrel, A., Patil, S., Nguyen, M., et al. (2019). High-throughput detection of bacterial community and its drug-resistance profiling from local reclaimed wastewater plants. Front. Cell Infect. Microbiol. 9:303. doi: 10.3389/fcimb.2019.00303
Loudermilk, E., Kotay, S., Barry, K., Parikh, H., Colosi, L., and Mathers, A. (2022). Tracking Klebsiella pneumoniae carbapenemase gene as an indicator of antimicrobial resistance dissemination from a hospital to surface water via a municipal wastewater treatment plant. Water Res. 213:118151. doi: 10.1016/j.watres.2022.118151
Łuczkiewicz, A., Jankowska, K., Fudala-Ksia̧żek, S., and Olańczuk-Neyman, K. (2010). Antimicrobial resistance of fecal indicators in municipal wastewater treatment plant. Water Res. 44, 5089–5097.
Luczkiewicz, A., Kotlarska, E., Artichowicz, W., Tarasewicz, K., and Fudala-Ksiazek, S. (2015). Antimicrobial resistance of Pseudomonas spp. isolated from wastewater and wastewater-impacted marine coastal zone. Environ. Sci. Pollut. Res. Int. 22, 19823–19834. doi: 10.1007/s11356-015-5098-y
MacLean, R., and San Millan, A. (2019). The evolution of antibiotic resistance. Science 365, 1082–1083.
Magalhães, M., Pontes, G., Serra, P., Balieiro, A., Castro, D., Pieri, F., et al. (2016). Multidrug resistant Pseudomonas aeruginosa survey in a stream receiving effluents from ineffective wastewater hospital plants. BMC Microbiol. 16:193. doi: 10.1186/s12866-016-0798-0
Manaia, C., Aga, D., Cytryn, E., Gaze, W., Graham, D., Guo, J., et al. (2022). The Complex interplay between antibiotic resistance and pharmaceutical and personal care products in the environment. Environ Toxicol Chem. Online ahead of print. doi: 10.1002/etc.5555
Mao, D., Yu, S., Rysz, M., Luo, Y., Yang, F., Li, F., et al. (2015). Prevalence and proliferation of antibiotic resistance genes in two municipal wastewater treatment plants. Water Res. 85, 458–466. doi: 10.1016/j.watres.2015.09.010
Marathe, N. P., Berglund, F., Razavi, M., Pal, C., Dröge, J., Samant, S., et al. (2019). Sewage effluent from an Indian hospital harbors novel carbapenemases and integron-borne antibiotic resistance genes. Microbiome 7:97. doi: 10.1186/s40168-019-0710-x
Marutescu, L., Jaga, M., Postolache, C., Barbuceanu, F., Milita, N., and Romascu, L. (2022). Insights into the impact of manure on the environmental antibiotic residues and resistance pool. Front. Microbiol. 13:965132. doi: 10.3389/fmicb.2022.965132
Moradigaravand, D., Gouliouris, T., Ludden, C., Reuter, S., Jamrozy, D., and Blane, B. (2018). Genomic survey of Clostridium difficile reservoirs in the East of England implicates environmental contamination of wastewater treatment plants by clinical lineages. Microb. Genome 4:e000162. doi: 10.1099/mgen.0.000162
Moy, S., and Sharma, R. (2017). Treatment outcomes in infections caused by “SPICE” (Serratia, Pseudomonas, Indole-positive Proteus, Citrobacter, and Enterobacter). organisms: carbapenem versus noncarbapenem regimens. Clin. Ther. 39, 170–176.
Müller, H., Sib, E., Gajdiss, M., Klanke, U., Lenz-Plet, F., Barabasch, V., et al. (2018). Dissemination of multi-resistant gram-negative bacteria into German wastewater and surface waters. FEMS Microbiol. Ecol. 94:fiy057. doi: 10.1093/femsec/fiy057
Munoz-Price, L., Poirel, L., Bonomo, R., Schwaber, M., Daikos, G., Cormican, M., et al. (2013). Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect. Dis. 13, 785–796. doi: 10.1016/S1473-3099(13)70190-7
Murphy, A., Barich, D., Fennessy, M., and Slonczewski, J. (2021). An Ohio state scenic river shows elevated antibiotic resistance genes, including Acinetobacter tetracycline and macrolide resistance, downstream of wastewater treatment plant effluent. Microbiol. Spectr. 9:e0094121. doi: 10.1128/Spectrum.00941-21
Murray, C. J., Ikuta, K., Sharara, F., Swetschinski, L., Aguilar, G., Gray, A., et al. (2022). Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399, 629–655.
Mutuku, C., Melegh, S., Kovacs, K., Urban, P., Virág, E., Heninger, R., et al. (2022). Characterization of β-lactamases and multidrug resistance mechanisms in Enterobacterales from hospital effluents and wastewater treatment plant. Antibiotics 11:776.
Nesme, J., Cécillon, S., Delmont, T., Monier, J., Vogel, T., and Simonet, P. (2014). Large-scale metagenomic-based study of antibiotic resistance in the environment. Curr. Biol. 24, 1096–1100. doi: 10.1016/j.cub.2014.03.036
Ng, C., Tan, B., Jiang, X., Gu, X., Chen, H., Schmitz, B., et al. (2019). Metagenomic and resistome analysis of a full-scale municipal wastewater treatment plant in singapore containing membrane bioreactors. Front. Microbiol. 10:172. doi: 10.3389/fmicb.2019.00172
Nikaeen, M., Aghili Dehnavi, H., Hssanzadeh, A., and Jalali, M. (2015). Occurrence of Clostridium difficile in two types of wastewater treatment plants. J. Formos Med. Assoc. 114, 663–665.
Nnadozie, C., and Odume, O. (2019). Freshwater environments as reservoirs of antibiotic resistant bacteria and their role in the dissemination of antibiotic resistance genes. Environ. Pollut. 254(Pt B):113067.
Norman, K., Scott, H., Harvey, R., Norby, B., Hume, M., and Andrews, K. (2011). Prevalence and genotypic characteristics of Clostridium difficile in a closed and integrated human and swine population. Appl. Environ. Microbiol. 77, 5755–5760. doi: 10.1128/AEM.05007-11
Nüesch-Inderbinen, M., Zurfluh, K., Stevens, M., and Stephan, R. (2018). Complete and assembled genome sequence of an NDM-9- and CTX-M-15-producing Klebsiella pneumoniae ST147 wastewater isolate from Switzerland. J. Glob. Antimicrob. Resist. 13, 53–54. doi: 10.1016/j.jgar.2018.03.001
O’Hara, J., Hu, F., Ahn, C., Nelson, J., Rivera, J., Pasculle, A., et al. (2014). Molecular epidemiology of KPC-producing Escherichia coli: occurrence of ST131-fimH30 subclone harboring pKpQIL-like IncFIIk plasmid. Antimicrob. Agents Chemother. 58, 4234–4237. doi: 10.1128/AAC.02182-13
O’Neill, J. (2016). The review on antimicrobial resistance. Tackling drug-resistant infections globally: Final report and recommendations. Available online at: https://amr-review.org/sites/default/files/160525_Final%20paper_with%20cover.pdf (accessed March 15, 2023).
Oliver, A., Mulet, X., López-Causapé, C., and Juan, C. (2015). The increasing threat of Pseudomonas aeruginosa high-risk clones. Drug Resist. Updat. 21-22, 41–59. doi: 10.1016/j.drup.2015.08.002
Oravcova, V., Mihalcin, M., Zakova, J., Pospisilova, L., Masarikova, M., and Literak, I. (2017). Vancomycin-resistant enterococci with vanA gene in treated municipal wastewater and their association with human hospital strains. Sci Total Environ. 609, 633–643. doi: 10.1016/j.scitotenv.2017.07.121
Ortega, A., Sáez, D., Bautista, V., Fernández-Romero, S., Lara, N., and Aracil, B. (2016). Carbapenemase producing Escherichia coli is becoming more prevalent in Spain mainly because of the polyclonal dissemination of OXA-48. J. Antimicrob. Chemother. 71, 2131–2138. doi: 10.1021/es203399h
Ovejero, C., Delgado-Blas, J., Calero-Caceres, W., Muniesa, M., and Gonzalez-Zorn, B. (2017). Spread of mcr-1-carrying Enterobacteriaceae in sewage water from Spain. J. Antimicrob. Chemother. 72, 1050–1053. doi: 10.1093/jac/dkw533
Pallares-Vega, R., Blaak, H., van der Plaats, R., de Roda Husman, A., Leal, L., van Loosdrecht, M., et al. (2019). Determinants of presence and removal of antibiotic resistance genes during WWTP treatment: a cross-sectional study. Water Res. 161, 319–328. doi: 10.1016/j.watres.2019.05.100
Pärnänen, K., Narciso-da-Rocha, C., Kneis, D., Berendonk, T., Cacace, D., Do, T., et al. (2019). Antibiotic resistance in European wastewater treatment plants mirrors the pattern of clinical antibiotic resistance prevalence. Sci. Adv. 5:eaau9124. doi: 10.1126/sciadv.aau9124
Paulshus, E., Thorell, K., Guzman-Otazo, J., Joffre, E., Colque, P., Kühn, I., et al. (2019). Repeated isolation of extended-spectrum-β-lactamase-positive Escherichia coli sequence types 648 and 131 from community wastewater indicates that sewage systems are important sources of emerging clones of antibiotic-resistant bacteria. Antimicrob. Agents Chemother. 63:e00823-19.
Pazda, M., Kumirska, J., Stepnowski, P., and Mulkiewicz, E. (2019). Antibiotic resistance genes identified in wastewater treatment plant systems - a review. Sci. Total Environ. 697:134023.
Perry, J., and Wright, G. (2013). The antibiotic resistance “mobilome”: searching for the link between environment and clinic. Front. Microbiol. 4:138. doi: 10.3389/fmicb.2013.00138
Petit, S., Lavenir, R., Colinon-Dupuich, C., Boukerb, A., Cholley, P., Bertrand, X., et al. (2013). Lagooning of wastewaters favors dissemination of clinically relevant Pseudomonas aeruginosa. Res. Microbiol. 164, 856–866. doi: 10.1016/j.resmic.2013.06.007
Popa, L., Gheorghe, I., Barbu, I., Surleac, M., Paraschiv, S., and Măruţescu, L. (2021). Multidrug resistant Klebsiella pneumoniae ST101 clone survival chain from inpatients to hospital effluent after chlorine treatment. Front Microbiol. 11:610296. doi: 10.3389/fmicb.2020.610296
Proia, L., Schiller, D., Sànchez-Melsi, A., Sabater, S., Borrego, C. M., Rodríguez-Mozaz, S., et al. (2016). Occurrence and persistence of antibiotic resistance genes in river biofilms after wastewater inputs in small rivers. Environ. Pollut. 210, 121–128. doi: 10.1016/j.envpol.2015.11.035
Qiu, Y., Zhang, J., Li, B., Wen, X., Liang, P., Huang, X., et al. (2018). A novel microfluidic system enables visualization and analysis of antibiotic resistance gene transfer to activated sludge bacteria in biofilm. Sci. Total Environ. 642, 582–590. doi: 10.1016/j.scitotenv.2018.06.012
Raza, S., Shin, H., Hur, H., and Unno, T. (2022). Higher abundance of core antimicrobial resistant genes in effluent from wastewater treatment plants. Water Res. 208:117882.
Reichert, G., Hilgert, S., Fuchs, S., and Azevedo, J. (2019). Emerging contaminants and antibiotic resistance in the different environmental matrices of Latin America. Environ. Pollut. 250:113140. doi: 10.1016/j.envpol.2019.113140
Riaz, L., Anjum, M., Yang, Q., Safeer, R., Sikandar, A., Ullah, H., et al. (2020). “Treatment technologies and management options of antibiotics and AMR/ARGs,” in Antibiotics and Antimicrobial Resistance genes in the Environment, vol 1, ed. Z. M. Hashmi (Amsterdam: Elsevier).
Rice, L. (2008). Federal funding for the study of antimicrobial resistance in nosocomial pathogens: no ESKAPE. J. Infect. Dis. 197, 1079–1081. doi: 10.1086/533452
Rivas, L., Dupont, P., Gilpin, B., and Cornelius, A. (2020). Isolation and characterization of Clostridium difficile from a small survey of wastewater, food and animals in New Zealand. Lett. Appl. Microbiol. 70, 29–35. doi: 10.1111/lam.13238
Rolbiecki, D., Harnisz, M., Korzeniewska, E., Buta, M., Hubeny, J., and Zieliński, W. (2021). Detection of carbapenemase-producing, hypervirulent Klebsiella spp. in wastewater and their potential transmission to river water and WWTP employees. Int. J. Hyg. Environ. Health. 237:113831. doi: 10.1016/j.ijheh.2021.113831
Romano, V., Pasquale, V., Krovacek, K., Mauri, F., Demarta, A., and Dumontet, S. (2012). Toxigenic Clostridium difficile PCR ribotypes from wastewater treatment plants in southern Switzerland. Appl. Environ. Microbiol. 78, 6643–6646. doi: 10.1128/AEM.01379-12
Rosenberg Goldstein, R., Micallef, S., Gibbs, S., George, A., Claye, E., Sapkota, A., et al. (2014). Detection of vancomycin-resistant enterococci (VRE). at four U.S. wastewater treatment plants that provide effluent for reuse. Sci. Total Environ. 466-467, 404–411. doi: 10.1016/j.scitotenv.2013.07.039
Sadowy, E., and Luczkiewicz, A. (2014). Drug-resistant and hospital-associated Enterococcus faecium from wastewater, riverine estuary and anthropogenically impacted marine catchment basin. BMC Microbiol. 14:66. doi: 10.1186/1471-2180-14-66
Sanderson, H., Ortega-Polo, R., McDermott, K., Hall, G., Zaheer, R., Brown, R., et al. (2019). Quantification and multidrug resistance profiles of vancomycin-resistant enterococci isolated from two wastewater treatment plants in the same municipality. Microorganisms 7:626. doi: 10.3390/microorganisms7120626
Sanderson, H., Ortega-Polo, R., Zaheer, R., Goji, N., Amoako, K., Brown, R., et al. (2020). Comparative genomics of multidrug-resistant Enterococcus spp. isolated from wastewater treatment plants. BMC Microbiol. 20:20. doi: 10.1186/s12866-019-1683-4
Savin, M., Bierbaum, G., Hammerl, J., Heinemann, C., Parcina, M., Sib, E., et al. (2020). ESKAPE bacteria and extended-spectrum-β-lactamase-producing Escherichia coli isolated from wastewater and process water from german poultry slaughterhouses. Appl. Environ. Microbiol. 86:e02748-19.
Sekizuka, T., Yatsu, K., Inamine, Y., Segawa, T., Nishio, M., Kishi, N., et al. (2018). Complete genome sequence of a blaKPC–2 -positive Klebsiella pneumoniae strain isolated from the effluent of an urban sewage treatment plant in Japan. mSphere 3:e00314-18. doi: 10.1128/mSphere.00314-18
Seruga Music, M., Hrenovic, J., Goic-Barisic, I., Hunjak, B., Skoric, D., and Ivankovic, T. (2017). Emission of extensively-drug resistant Acinetobacter baumannii from hospital settings to the natural environment. J. Hosp. Infect. 96, 232–237. doi: 10.1016/j.jhin.2017.04.005
Singer, A., Shaw, H., Rhodes, V., and Hart, A. (2016). Review of antimicrobial resistance in the environment and its relevance to environmental regulators. Front. Microbiol. 7:1728. doi: 10.3389/fmicb.2016.01728
Slekovec, C., Plantin, J., Cholley, P., Thouverez, M., Talon, D., Bertrand, X., et al. (2012). Tracking down antibiotic-resistant Pseudomonas aeruginosa isolates in a wastewater network. PLoS One 7:e49300. doi: 10.1371/journal.pone.0049300
Sleytr, K., Tietz, A., Langergraber, G., and Haberl, R. (2007). Investigation of bacterial removal during the filtration process in constructed wetlands. Sci. Total Environ. 380, 173–180.
Sousa, J., Macedo, G., Pedrosa, M., Becerra-Castro, C., Castro-Silva, S., Pereira, M., et al. (2017). Ozonation and UV254nm radiation for the removal of microorganisms and antibiotic resistance genes from urban wastewater. Hazard. Mater. 323, 434–441. doi: 10.1016/j.jhazmat.2016.03.096
Steyer, A., Gutiérrez-Aguirre, I., Rački, N., Beigot Glaser, S., Brajer Humar, B., Stražar, M., et al. (2015). The detection rate of enteric viruses and clostridium difficile in a waste water treatment plant effluent. Food Environ. Virol. Online ahead of print. doi: 10.1007/s12560-015-9183-7
Stoesser, N., Sheppard, A., Peirano, G., Sebra, R., Lynch, T., Anson, L., et al. (2016). First report of blaIMP–14 on a plasmid harboring multiple drug resistance genes in Escherichia coli sequence type 131. Antimicrob. Agents Chemother. 60, 5068–5071. doi: 10.1128/AAC.00840-16
Subirats, J., Triadó-Margarit, X., Mandaric, L., Acuña, V., Balcázar, J., Sabater, S., et al. (2017). Wastewater pollution differently affects the antibiotic resistance gene pool and biofilm bacterial communities across streambed compartments. Mol. Ecol. 26, 5567–5581. doi: 10.1111/mec.14288
Surleac, M., Czobor Barbu, I., Paraschiv, S., Popa, L., Gheorghe, I., Marutescu, L., et al. (2020). Whole genome sequencing snapshot of multi-drug resistant Klebsiella pneumoniae strains from hospitals and receiving wastewater treatment plants in Southern Romania. PLoS One 15:e0228079. doi: 10.1371/journal.pone.0228079
Taučer-Kapteijn, M., Hoogenboezem, W., Heiliegers, L., de Bolster, D., and Medema, G. (2016). Screening municipal wastewater effluent and surface water used for drinking water production for the presence of ampicillin and vancomycin resistant enterococci. Int. J. Hyg. Environ Health 219, 437–442. doi: 10.1016/j.ijheh.2016.04.007
The Joint Programming Initiative on Antimicrobial Resistance (2018). One health network on AMR. Available online at: https://health.ec.europa.eu/system/files/2020-01/amr_ev_20180205_co033_en_0.pdf (accessed May 13, 2023).
Thompson, J., Gündogdu, A., Stratton, H., and Katouli, M. (2013). Antibiotic resistant Staphylococcus aureus in hospital wastewaters and sewage treatment plants with special reference to methicillin-resistant Staphylococcus aureus (MRSA). J. Appl. Microbiol. 114, 44–54. doi: 10.1111/jam.12037
Topp, E., Larsson, D. G. J., Miller, D., Van den Eede, C., and Virta, M. (2018). Antimicrobial resistance and the environment: assessment of advances, gaps and recommendations for agriculture, aquaculture and pharmaceutical manufacturing. FEMS Microbiol. Ecol. 94:ix185. doi: 10.1093/femsec/fix185
Trinh, S., and Reysset, G. (1996). Detection by PCR of the nim genes encoding 5-nitroimidazole resistance in Bacteroides spp. J. Clin. Microbiol. 34, 2078–2084. doi: 10.1128/jcm.34.9.2078-2084.1996
Turton, J., Davies, F., Turton, J., Perry, C., Payne, Z., and Pike, R. (2019). Hybrid resistance and virulence plasmids in “high-risk” clones of Klebsiella pneumoniae, including those carrying blaNDM–5. Microorganisms 7:326. doi: 10.3390/microorganisms7090326
Uluseker, C., Kaster, K., Thorsen, K., Basiry, D., Shobana, S., Jain, M., et al. (2021). A review on occurrence and spread of antibiotic resistance in wastewaters and in wastewater treatment plants: mechanisms and perspectives. Front. Microbiol. 12:717809. doi: 10.3389/fmicb.2021.717809
van Schaik, W., Top, J., Riley, D., Boekhorst, J., Vrijenhoek, J., Schapendonk, C., et al. (2010). Pyrosequencing-based comparative genome analysis of the nosocomial pathogen Enterococcus faecium and identification of a large transferable pathogenicity island. BMC Genomics 11:239. doi: 10.1186/1471-2164-11-239
Voigt, A., Zacharias, N., Timm, C., Wasser, F., Sib, E., Skutlarek, D., et al. (2019). Association between antibiotic residues, antibiotic resistant bacteria and antibiotic resistance genes in anthropogenic wastewater – an evaluation of clinical influences. Chemosphere 241:125032. doi: 10.1016/j.chemosphere.2019.125032
von Wintersdorff, C. J., Penders, J., van Niekerk, J. M., Mills, N. D., Majumder, S., van Alphen, L. B., et al. (2016). Dissemination of antimicrobial resistance in microbial ecosystems through horizontal gene transfer. Front. Microbiol. 7:173. doi: 10.3389/fmicb.2016.00173
Wan, M., and Chou, C. (2015). Class 1 integrons and the antiseptic resistance gene (qaceδ1). in municipal and swine slaughterhouse wastewater treatment plants and wastewater—associated methicillin-resistant Staphylococcus aureus. Int. J. Environ. Res. Public Health 12, 6249–6260.
Warriner, K., Xu, C., Habash, M., Sultan, S., and Weese, S. (2017). Dissemination of Clostridium difficile in food and the environment: Significant sources of C. difficile community-acquired infection? J. Appl. Microbiol. 122, 542–553. doi: 10.1111/jam.13338
WHO (2017). Media Centre. News Release. WHO Publishes List of Bacteria for Which New Antibiotics are Urgently Needed. Geneva: WHO
Xie, Y., Sangaraiah, N., Meng, J., and Zhou, C. (2022). Unique carbazole-oxadiazole derivatives as new potential antibiotics for combating Gram-positive and -negative bacteria. J. Med. Chem. 65, 6171–6190. doi: 10.1021/acs.jmedchem.2c00001
Xu, C., Weese, J., Flemming, C., Odumeru, J., and Warriner, K. (2014). Fate of Clostridium difficile during wastewater treatment and incidence in Southern Ontario watersheds. J. Appl. Microbiol. 117, 891–904. doi: 10.1111/jam.12575
Yakkala, H., Samantarrai, D., Gribskov, M., and Siddavattam, D. (2019). Comparative genome analysis reveals niche-specific genome expansion in Acinetobacter baumannii strains. PLoS One 14:e0218204. doi: 10.1371/journal.pone.0218204
Yang, K., Pagaling, E., and Yan, T. (2014). Estimating the prevalence of potential enteropathogenic Escherichia coli and intimin gene diversity in a human community by monitoring sanitary sewage. Appl. Environ. Microbiol. 80, 119–127. doi: 10.1128/AEM.02747-13
Yuan, Q., Guo, M., and Yang, J. (2015). Fate of antibiotic resistant bacteria and genes during wastewater chlorination: implication for antibiotic resistance control. PLoS One 10:e0119403. doi: 10.1371/journal.pone.0119403
Zhang, C., Qiu, S., Wang, Y., Qi, L., Hao, R., and Liu, X. (2013). Higher isolation of NDM-1 producing Acinetobacter baumannii from the sewage of the hospitals in Beijing. PLoS One 8:e64857. doi: 10.1371/journal.pone.0064857
Zhang, Y., Marrs, C., Simon, C., and Xi, C. (2009). Wastewater treatment contributes to selective increase of antibiotic resistance among Acinetobacter spp. Sci. Total Environ. 407, 3702–3706. doi: 10.1016/j.scitotenv.2009.02.013
Zhang, Y., Zhuang, Y., Geng, J., Ren, H., Zhang, Y., Ding, L., et al. (2015). Inactivation of antibiotic resistance genes in municipal wastewater effluent by chlorination and sequential UV/chlorination disinfection. Sci. Total Environ. 512-513, 125–132. doi: 10.1016/j.scitotenv.2015.01.028
Zieliński, W., Korzeniewska, E., Harnisz, M., Hubeny, J., Buta, M., and Rolbiecki, D. (2020). The prevalence of drug-resistant and virulent Staphylococcus spp. in a municipal wastewater treatment plant and their spread in the environment. Environ Int. 143:105914. doi: 10.1016/j.envint.2020.105914
Keywords: ESCAPE pathogens, antibiotic resistance, wastewater treatment plants, antibiotic resistance genes, ESCAPE species dissemination, high-risk clones, multidrug resistance
Citation: Marutescu LG, Popa M, Gheorghe-Barbu I, Barbu IC, Rodríguez-Molina D, Berglund F, Blaak H, Flach C-F, Kemper MA, Spießberger B, Wengenroth L, Larsson DGJ, Nowak D, Radon K, de Roda Husman AM, Wieser A, Schmitt H, Pircalabioru Gradisteanu G, Vrancianu CO and Chifiriuc MC (2023) Wastewater treatment plants, an “escape gate” for ESCAPE pathogens. Front. Microbiol. 14:1193907. doi: 10.3389/fmicb.2023.1193907
Received: 25 March 2023; Accepted: 09 May 2023;
Published: 24 May 2023.
Edited by:
Eric N. Villegas, United States Environmental Protection Agency (EPA), United StatesReviewed by:
Scott P. Keely, United States Environmental Protection Agency (EPA), United StatesAbdennaceur Hassen, Centre de Recherches et des Technologies des Eaux, Tunisia
Copyright © 2023 Marutescu, Popa, Gheorghe-Barbu, Barbu, Rodríguez-Molina, Berglund, Blaak, Flach, Kemper, Spießberger, Wengenroth, Larsson, Nowak, Radon, de Roda Husman, Wieser, Schmitt, Pircalabioru Gradisteanu, Vrancianu and Chifiriuc. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mariana Carmen Chifiriuc, Y2FybWVuLmNoaWZpcml1Y0BiaW8udW5pYnVjLnJv; Marcela Popa, bWFyY2VsYS5wb3BhQGJpby51bmlidWMucm8=; Gratiela Pircalabioru Gradisteanu, Z3JhdGllbGEuZ3JhZGlzdGVhbnVAaWN1Yi51bmlidWMucm8=
 Luminita Gabriela Marutescu
Luminita Gabriela Marutescu Marcela Popa2*
Marcela Popa2* Irina Gheorghe-Barbu
Irina Gheorghe-Barbu Ilda Czobor Barbu
Ilda Czobor Barbu Carl-Fredrik Flach
Carl-Fredrik Flach Laura Wengenroth
Laura Wengenroth D. G. Joakim Larsson
D. G. Joakim Larsson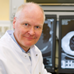 Dennis Nowak
Dennis Nowak Ana Maria de Roda Husman
Ana Maria de Roda Husman Heike Schmitt
Heike Schmitt Gratiela Pircalabioru Gradisteanu
Gratiela Pircalabioru Gradisteanu Corneliu Ovidiu Vrancianu
Corneliu Ovidiu Vrancianu Mariana Carmen Chifiriuc
Mariana Carmen Chifiriuc