
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 22 May 2023
Sec. Infectious Agents and Disease
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1192832
This article is part of the Research Topic Insights in Infectious Agents and Disease: 2022 View all 41 articles
 Santiago Maffia-Bizzozero1†
Santiago Maffia-Bizzozero1† Cintia Cevallos2†
Cintia Cevallos2† Federico Remes Lenicov2
Federico Remes Lenicov2 Rosa Nicole Freiberger2
Rosa Nicole Freiberger2 Cinthya Alicia Marcela Lopez2
Cinthya Alicia Marcela Lopez2 Alex Guano Toaquiza2
Alex Guano Toaquiza2 Franco Sviercz2
Franco Sviercz2 Patricio Jarmoluk2
Patricio Jarmoluk2 Cristina Bustos1
Cristina Bustos1 Adriana Claudia D’Addario1
Adriana Claudia D’Addario1 Jorge Quarleri2*‡
Jorge Quarleri2*‡ M. Victoria Delpino2*‡
M. Victoria Delpino2*‡Introduction: Pulmonary and extrapulmonary manifestations have been described after infection with SARS-CoV-2, the causative agent of coronavirus disease 2019 (COVID-19). The virus is known to persist in multiple organs due to its tropism for several tissues. However, previous reports were unable to provide definitive information about whether the virus is viable and transmissible. It has been hypothesized that the persisting reservoirs of SARS-CoV-2 in tissues could be one of the multiple potentially overlapping causes of long COVID.
Methods: In the present study, we investigated autopsy materials obtained from 21 cadaveric donors with documented first infection or reinfection at the time of death. The cases studied included recipients of different formulations of COVID-19 vaccines. The aim was to find the presence of SARS-CoV-2 in the lungs, heart, liver, kidneys, and intestines. We used two technical approaches: the detection and quantification of viral genomic RNA using RT-qPCR, and virus infectivity using permissive in vitro Vero E6 culture.
Results: All tissues analyzed showed the presence of SARS-CoV-2 genomic RNA but at dissimilar levels ranging from 1.01 × 102 copies/mL to 1.14 × 108 copies/mL, even among those cases who had been COVID-19 vaccinated. Importantly, different amounts of replication-competent virus were detected in the culture media from the studied tissues. The highest viral load were measured in the lung (≈1.4 × 106 copies/mL) and heart (≈1.9 × 106 copies/mL) samples. Additionally, based on partial Spike gene sequences, SARS-CoV-2 characterization revealed the presence of multiple Omicron sub-variants exhibiting a high level of nucleotide and amino acid identity among them.
Discussion: These findings highlight that SARS-CoV-2 can spread to multiple tissue locations such as the lungs, heart, liver, kidneys, and intestines, both after primary infection and after reinfections with the Omicron variant, contributing to extending knowledge about the pathogenesis of acute infection and understanding the sequelae of clinical manifestations that are observed during post-acute COVID-19.
SARS-CoV-2 has caused millions of deaths worldwide. Pulmonary and extrapulmonary manifestations have been described after infection with SARS-CoV-2, the causative agent of COVID-19 (Elrobaa and New, 2021). Until now, most efforts have been made to understand acute COVID-19 in the early stages. Most COVID-19 patients recover within a few days to a few weeks after infection. However, people who have been exposed to SARS-CoV-2 may have a variety of new, recurring, or ongoing health sequelae. These disorders are known as post-acute COVID-19, also known as long-COVID, that cannot be explained by other diagnoses and take at least 4 weeks after infection to appear. Post-acute COVID-19 symptoms can affect anyone who contracted the infection, but some groups might be more at risk than others for developing such sequelae, such as those who have experienced more severe COVID-19 illness, especially those who were hospitalized or needed intensive care (Nalbandian et al., 2021; Sanyaolu et al., 2022; Soriano et al., 2022). Nevertheless, post-acute COVID-19 has been observed even among young and middle-aged adults after six to 12 months after mild infection (Peter et al., 2022). Some studies have reported that COVID-19 vaccines had a protective effect against long-COVID even among patients vaccinated before or after SARS-CoV-2 infection/COVID-19 (Al-Aly et al., 2022; Gao et al., 2022; Nascimento et al., 2023). Additionally, the evidence shows that a SARS-CoV-2 reinfection further increases risks of death, hospitalization, and sequelae in multiple organ systems in the acute and post-acute phase, particularly in those who were unvaccinated and had one vaccination or two or more vaccinations before reinfection (Bowe et al., 2022). The underlying mechanisms of post-acute COVID-19 pathogenesis are not well understood, but an association with SARS-CoV-2 persistence in some tissues has been proposed, particularly those with high expression of the viral receptor ACE2, thus acting as a viral reservoir for SARS-CoV-2 (Neurath et al., 2021; Chen et al., 2022). Several reports have described the persistence of viral RNA and/or antigens in gastrointestinal tissues, cardiovascular system, reproductive tissues, brain, muscles, breast tissue, lymph nodes, stool, and urine (Lamers et al., 2020; Trypsteen et al., 2020; Martin-Cardona et al., 2021; Neurath et al., 2021; Chen et al., 2022; Cheung et al., 2022; Goh et al., 2022; Stein et al., 2022; Tejerina et al., 2022). Fundamentally, the underlying significance of established risk factors, including age, obesity, and diabetes, as well as why some people advance to severe forms of COVID-19 after a two- to three-week period, is still unknown. Only autopsy investigations can provide an answer to disease pathophysiology and death queries (Layne et al., 2022). These autopsy studies have provided evidence of the ability of SARS-CoV-2 to infect multiple organs (Edler et al., 2020; Deinhardt-Emmer et al., 2021; Fitzek et al., 2021; Maiese et al., 2021). Moreover, autopsy material from different tissues revealed the persistence of SARS-CoV-2 genomic RNA for a time ranging from 22 to 27 days between death and autopsy (Stein et al., 2022). Although SARS-CoV-2 genomic RNA and even subgenomic mRNA have been found in several organs, this does not necessarily indicate that the virus can replicate and be infectious (Casagrande et al., 2020; Wolfel et al., 2020; Aquila et al., 2021; Beltempo et al., 2021; Sablone et al., 2021). Currently, host (infection time, SARSCoV-2 infection-induced and/or vaccine-induced immunity) and virus-related (variant, viral load) factors can modify the course of COVID-19 and even the probability of post-COVID occurrence. Understanding the mechanism of acute COVID-19 as well as its long-term consequences is crucial because it causes multi-organ damage (including the lungs, heart, liver, kidney, and intestine). However, comparative research between the many implicated organs is still scarce. The objectives of this study are to describe the involvement of the aforementioned organs after viral spreading, assess the SARS-CoV-2 viral load and genomic characterization, and elucidate the SARS-CoV-2 infectivity between decedents with first infection and those with SARS-CoV-2 reinfection and clinical records of COVID sequelae.
This study involved 21 decedents with antemortem SARS-CoV-2 infection (named from “A” to “U”). They were subjected to minimally invasive forensic autopsy between 5 January and 6 August 2022 at the Judicial Morgue of Argentina (Table 1). Image-guided tissue sampling was performed in the right lung, right kidney, small intestine, right lobe of the liver, and left heart ventricle in regions with edema, congestive hemorrhage, and organ consolidation in the case of the right lung.
Samples were collected by forensic medical personnel when a cadaver was admitted, following international recommendations for sample collection in the context of the COVID-19 pandemic regarding the use of personal protective equipment (Centers for Disease Control and Prevention, 2020). This involved wearing a surgical jacket and trousers, goggles, an N95/FFP3 face mask, and high rubber boots, along with disposable equipment such as an impermeable surgical gown, a surgical cap, and gloves. To prevent contamination, samples were taken from cadavers in suspicious cases in an isolated environment that included restricted access to the facilities, closed rooms with negative air pressure, and appropriate decontamination and waste inactivation (Centers for Disease Control and Prevention, 2020).
The decedents ranged in age from 3 months to 94 years (mean: 61; median: 67). Eleven cadavers were female and nine were male. Small tissue samples were obtained from the lung, heart, liver, and kidney of each cadaver, as well as 16 from the small intestine, and stored in RNAlater® (Sigma-Aldrich) at −80°C until use. On average, tissue samples were collected 1.9 days postmortem while the corpses were maintained at 4°C.
SARS-CoV-2 RNA was posthumously confirmed in all cases by routinely performing real-time reverse transcription-polymerase chain reaction (RT-qPCR) on nasopharyngeal swab samples. Ten of the deaths occurred after hospitalization (H) while the remaining 11 were non-hospitalized (NH). COVID-19 vaccine varied among the 13 cases representing different formulations available. Seven cases had not been vaccinated and no records were obtained for the remaining one. As a rule, nasopharyngeal swabs and radiological examinations were performed on all suspected cases to distinguish between cases where SARS-CoV-2 infection was the sole cause of death and those where COVID-19 was merely a contributing factor. However, the specific chain of events leading to their deaths is only available in clinical records that we were unable to access due to Argentine legal regulations, limiting the scope of this study.
Pre-autopsy nasopharyngeal swabs can reveal whether a corpse is infected or not. In the initial hours following death, thoracic radiological examinations (RE) and macroscopic autopsy records can provide insights into the extent and severity of the illness.
Five cases (E, F, K, T, U) were categorized as SARS-CoV-2 reinfections because SARS-CoV-2 was diagnosed for them in the past 6–12 months. This time frame reduces the probability that the positive post-mortem test was related to the first infection (Table 1). A wide range of symptoms, including fever, exhaustion, headache, shortness of breath, coughing, chest discomfort, heart palpitation, digestive issues (diarrhea, stomach pain), or neurological issues (anxiety, disturbed sleep, ageusia, anosmia), was recorded from such diagnoses until death. These symptoms lasted several weeks or even months after infection, while some disappeared or recurred. This study was approved by the Cuerpo Médico Forense (Corte Suprema de Justicia de la Nación Argentina).
Frozen tissues (0.5 cm3) were mechanically disaggregated using fine-point scissors and scalpels in 0.6 mL of Dulbecco’s Modified Eagle Medium (DMEM). Homogenates were centrifuged at 12,000 × g for 10 min at 4°C, and supernatants were stored at −70°C until use.
RNA was extracted using chemagic™ Viral DNA/RNA kit special H96 (PerkinElmer, Germany) on the automated chemagic™ 360 instrument (PerkinElmer, Germany). The efficiency of RNA extraction was independent of organ origin. RNA was quantified using a NanoDrop™ (Thermo Scientific™) and its load was normalized before SARS-CoV-2 RNA detection was performed using RT-qPCR (DisCoVery SARS-CoV-2 RT-PCR Detection Kit Rox) amplifying ORF1ab and N viral genes following the manufacturer’s instructions. The internal control was successfully amplified in all samples with a mean cycle threshold (Ct) value of 24.74 ± 3.19. Samples were defined as positive when Ct values were below 36 for both ORF1ab and N genes. The limit of detection (LOD) of this qualitative assay is 10 viral genomic RNA copies/mL.
Viral ORF1ab and N genomic RNA quantification were performed using a standard curve performed with quantified SARS-CoV-2 positive RNA control (GISAID EPI_ISL_420600) isolated from local viral strain cultured in Vero E6 cells and quantified against a WHO SARS-CoV-2 standard.
The viral load in samples was calculated by interpolation of the corresponding Ct value with a standard curve, which had been built with the Ct values obtained following PCR amplification of samples containing serial dilutions of quantified SARS-CoV-2 positive RNA control. The LOD for the quantitative assay is 100 SARS-CoV-2 genomic RNA copies/mL.
RT-PCR was done using a SuperScript III One-Step RT-PCR System with Platinum Taq High Fidelity DNA (Invitrogen). The pairs of primers used were the following: S-RBD Fw: 5´-CTTGTGCCCTTTTGGTGAAGT-3′ and Rv: 5´-GTGGATCACGGACAGCATCA-3′. The amplification cycle was 52°C for 30 min, 94°C for 2 min followed by 40 cycles: 94°C for 15 s, 58°C for 30s, 68°C for 90 s. Primer sets yielded a single product of the correct size.
The purified template DNA was bidirectionally sequenced using a BigDye Terminator Ready Reaction Cycle Sequencing Kit (Applied Biosystems) using the same primers pairs on an ABI Prism 3,130 Genetic Analyzer (Applied Biosystems). The obtained overlapping SARS-CoV-2 Spike sequences (nucleotide position 22,614 to 23,215 numbered according to GenBank accession number NC_045512) were then assembled using BLAST multiple sequences software by comparison with SARS-CoV-2 reference sequences. The sample information and corresponding GenBank accession number for each sample are listed in Table 1.
A distance-based phylogenetic tree based on S protein consensus sequences was constructed to infer the phylogenetic relationships between SARS-CoV-2 isolates obtained from each/tissue. Individual S gene nucleotide consensus sequences were first constructed using multiple sequence alignment via the ClustalW approach implemented in MEGA11. All ambiguous positions were removed for each sequence pair (pairwise deletion option). The evolutionary history among SARS-CoV-2 isolates from autopsy tissues and reference sequences from viral variants and sub-variants was inferred using the Neighbor-Joining method (Saitou and Nei, 1987). The distance-based neighbor-joining (NJ) phylogenetic tree was then constructed based on nucleotide sequences in MEGA 11 using bootstrap values of 1,000 replicates and a 70% threshold score (Tamura et al., 2021).
The tree is drawn to scale involving 69 nucleotide sequences, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the p-distance method (Nei, 2000) and are in the units of the number of base differences per site. The rate variation among sites was modeled with a gamma distribution (shape parameter = 1). Codon positions included were 1st + 2nd + 3rd + Noncoding. All ambiguous positions were removed for each sequence pair (pairwise deletion option). There were a total of 602 positions in the final dataset. Evolutionary analyses were conducted in MEGA11 (Tamura et al., 2021).
The African green monkey kidney cell line Vero E6 (ATCC, Rockville, MD) was cultured as monolayers in a 5% CO2 atmosphere at 37°C in DMEM (Sigma-Aldrich, Argentina) supplemented with 2 mM L-glutamine, 10% heat-inactivated fetal bovine serum (FBS) (Sigma-Aldrich, Argentina), 100 U/mL penicillin and 100 μg/mL streptomycin.
Vero E6 cells were seeded at a concentration of 8 × 104 cells/well in 24-well plates and incubated with supernatants from frozen tissues that were mechanically disaggregated as described in point 2.2. The supernatants were diluted 1/4 in DMEM and incubated with the cells for 4 h at 37°C under a 5% CO2 atmosphere. The cells were then washed six times with DMEM, and the last wash was stored at −70°C to detect any remaining SARS-CoV-2 genomic RNA. The cells were incubated in DMEM supplemented with 2 mM L-glutamine, 2% heat-inactivated FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin. Cytopathic effect, evidenced by the destruction of the cell monolayer observed under a light microscope at 20× magnification, was monitored every 24 h. After 7 days, the supernatants were inspected for the presence of SARS-CoV-2 RNA using RT-qPCR, as described in point 2.3.
Live SARS-CoV-2 manipulation was performed in biosafety level 3 facilities at the INBIRS.
Twenty-one cadavers referred to the Judicial Morgue of Argentina between January and August 2022 were studied. They involved 100 small tissue samples collected from five different organs (lung, heart, liver, kidney, and small intestine).
Twenty-one corpses sent to the Argentine Judicial Morgue between January and August 2022 were studied. From them, 100 small tissue samples were collected from five different organs: lung, heart, liver, kidney, and small intestine. The most frequent causes of death included adult respiratory distress syndrome with bilateral lung compromise during COVID-19 as well as exacerbations of preexisting comorbidities and COVID-19.
Qualitatively, SARS-CoV-2 RNA was found in all tissues analyzed for both viral target genes studied (ORF1ab, N), in 47 out of 100 tissue samples collected but showing dissimilar frequencies as follows: 13/21 in the lung, 7/21 in the heart, 9/21 in the liver, 8/21 in the kidney, as well as 10/16 in small intestine tissue.
Among the studied cases, the presence of SARS-CoV-2 RNA was verified in at least one tissue sample in all cases.
Quantitatively, the highest viral load (expressed as median values in SARS-CoV-2 RNA copies/mL, for ORF1ab and N gene) was measured in lung samples (4.4 × 103 and 6.2 × 103). In this tissue, viral loads ranged from 1.01 × 102 copies/mL to 1.14 × 108 copies/mL. Comparatively, such median viral loads were lower in the liver (5.2 × 102 and 2.9 × 102), heart (6.1 × 103 and 8.3 × 102), kidney (4.7 × 102 and 6.0 × 102), and small intestine (9.8 × 102 and 3.2 × 102) (Figures 1A,B).
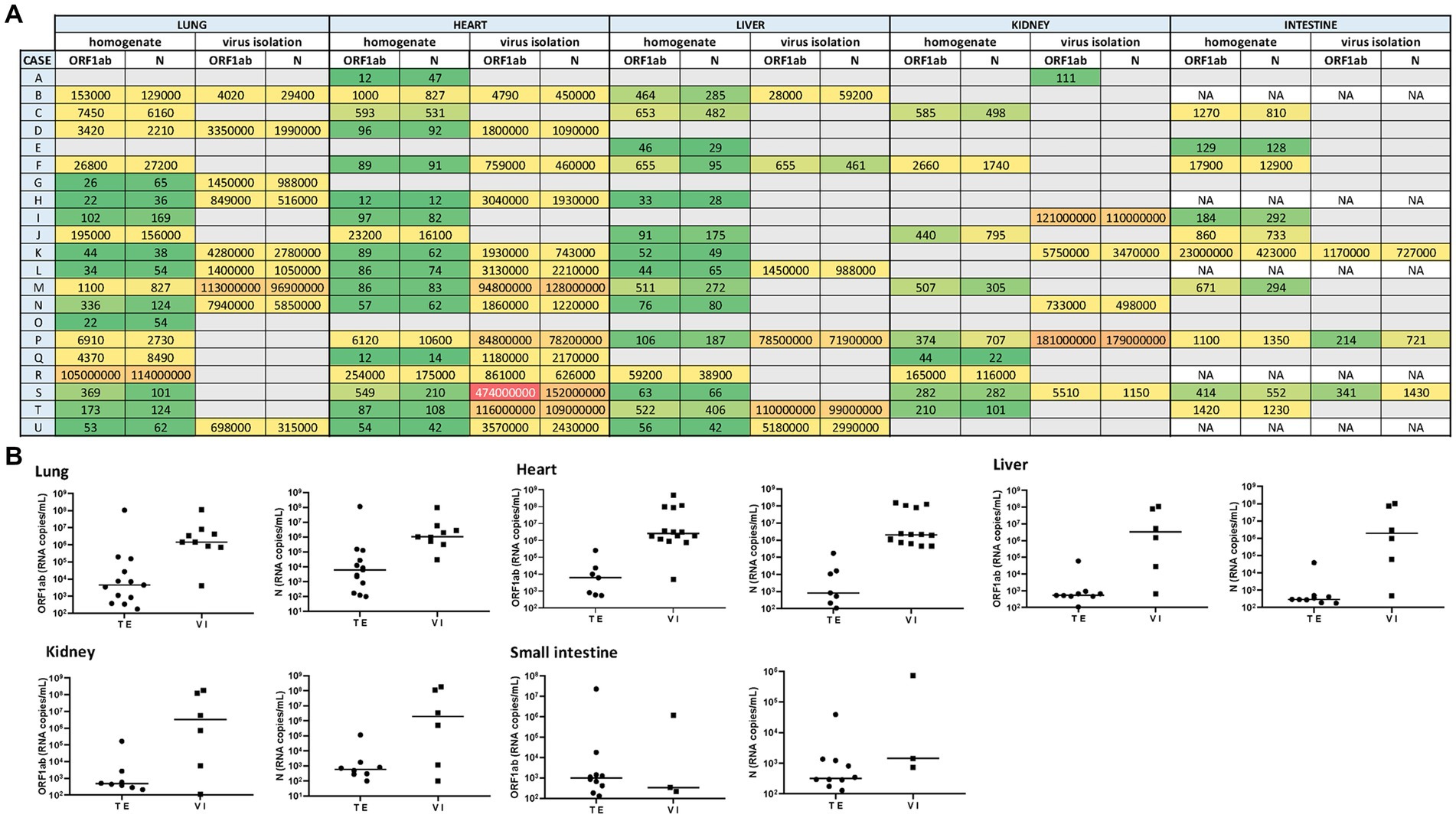
Figure 1. (A) Heat map representing the SARS-CoV-2 viral load recorded for each autopsy case from different tissues (including its homogenate and infectivity measurement in cell culture). A grid of colored squares ranging from green (minimum) to red (maximum) is used to represent the values of the viral load (expressed as copies/mL). The gray cells indicate an undetectable viral load, while “NA” indicates that the sample was not available. (B) The SARS-CoV-2 viral load (RNA copies/mL) was measured directly from tissue extracts (TE) and after 7-day-cell culture virus isolation (VI). The ORF1ab and N viral genes were detected in different tissues (lung, heart, liver, kidney, and small intestine) extract and culture supernatants from cell culture virus isolation.
Overall, the median viral load measured in the tissue samples from non-hospitalized cases was lower than those hospitalized (1.4 × 103 vs. 1.2 × 102 copies/mL, respectively). The correlation was observed between the viral load and the levels of damage observed in the heart and lungs at the time of death.
Specimens (tissue samples) with a cycle-threshold (Ct) value of ≤25 in the SARS-CoV-2 RNA detection were considered appropriate for obtaining good-quality viral genome sequences (World Health Organization, 2022). Following this criterion and based on Spike-gene sequences, we were able to characterize 27 SARS-CoV-2 genomic RNA from 17 cases (excluding F, N, P, and U cases). The phylogenetic relatedness after NJ analysis showed that all S-gene nucleotide sequences clustered into the Omicron variant clade and were distributed into three distinguishable sub-variants, including 20 closely related to BA.1.1, two related to BA.5.2, and five sequences segregated in the BA.2 clade (Figure 2).
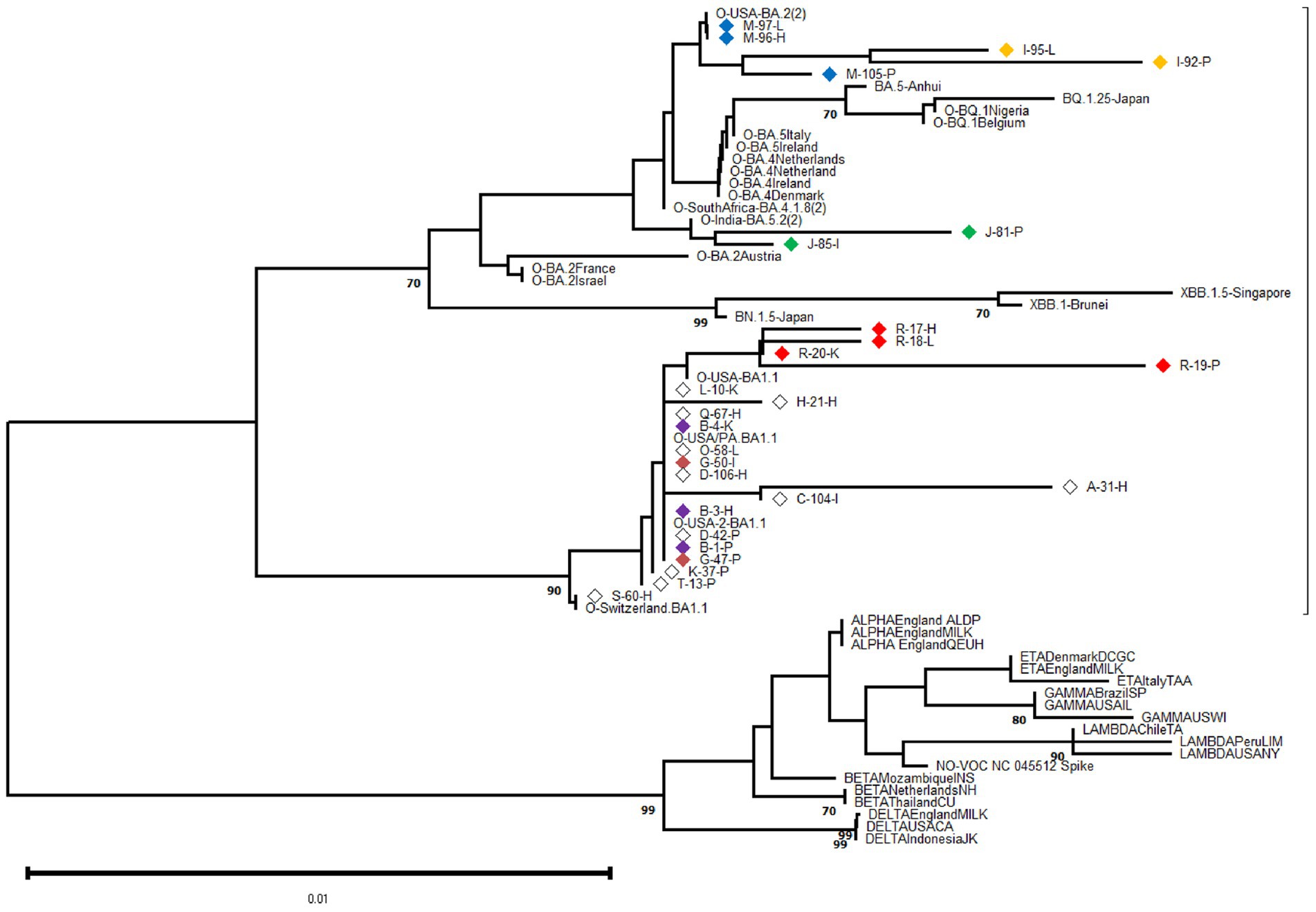
Figure 2. Phylogenetic analysis was performed on 602-base-pair-long nucleotide sequences of the SARS-CoV-2 Spike gene using the Neighbor Joining method with 1,000 bootstrap replicates. The lateral branches displayed the Omicron variant (as well as its sub-variants and recombinants represented by dashed lines), while the initially black branches represented the remaining non-Omicron variants. Each SARS-CoV-2 variant was accompanied by its reference sequence. The nucleotide sequences of SARS-CoV-2 isolates identified from autopsy materials were named according to the code provided in Table 1. Viral isolates from different tissues/cases were identified with a particular diamond color, while white diamonds represented unique viral isolates/cases. The numbers marked along the branches represented the bootstrap values higher than 0.7 out of 1,000 bootstrap resamplings. The tree scale depicted the number of substitutions/site.
At the intra-host level, we characterized viral isolates from different tissues in six cases (B, G, I, J, M, R) at the genomic level. For each case, the viral isolates were closely related even when depicting minor Spike-gene nucleotide differences when comparing genomic RNA obtained from different tissues. The sequence analysis revealed that viral isolation from different tissues from one donor was highly related, revealing only minor Spike-gene nucleotide differences.
At the inter-host level, there was no clustering among S-gene sequences according to any particular tissue of origin (Figure 2). Similarly, when comparing the Spike amino acid sequences (particularly in the Receptor Binding Domain or RBD) obtained from pulmonary tissues (n = 9), heart (n = 8), and other organs including kidney, liver, and intestine (n = 10), minor contributions in the frequency of amino acids were observed (Figure 3).
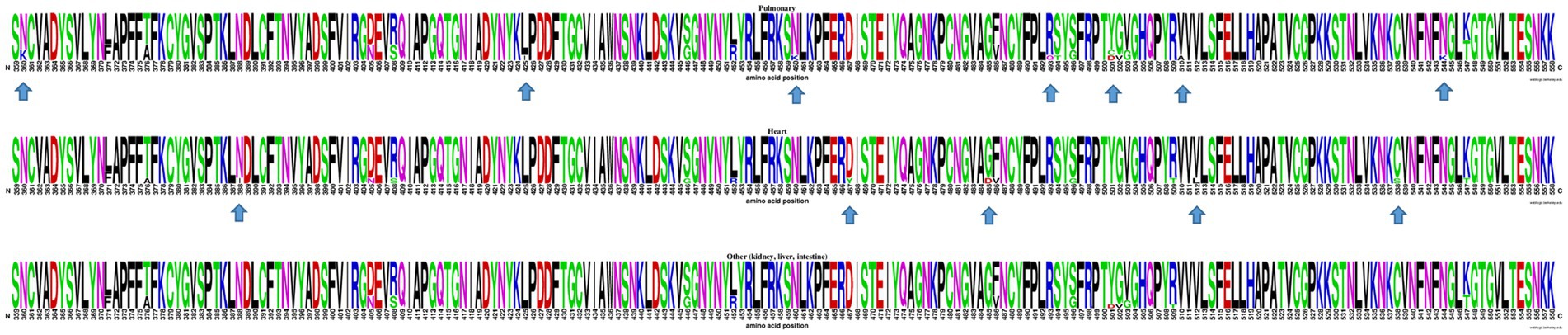
Figure 3. WebLogo profiles of SARS-CoV-2 receptor binding domain partial amino acid sequences (aa 359–558) characterized in viral isolates from different tissues are shown above. The upper profile represents sequences from the pulmonary tissue, the center profile represents sequences from the heart tissue, and the lower profile represents sequences from other tissues, including kidney, liver, and intestine. The size of the letter (bits; Y axis) indicates the frequency of the amino acid substitutions at a certain residue position (X axis). Different residues at the same position are scaled according to their frequency and colored based on their amino acid characteristics. An arrow is used to indicate RBD amino acid positions with a distinctive but infrequent residue.
Virus isolation from the five tissue samples that were studied (lung, heart, liver, kidney, and small intestine) was performed using supernatants from disaggregated tissues. After 7 days, the presence of infectious SARS-CoV-2 was evidenced by the cytopathic effect on Vero E6 monolayers.
Thus, infectious SARS-CoV-2 was confirmed after supernatants from tissue extract added on Vero E6 cells in 9 out of 21 lung samples (2 from reinfection cases), as well as in extrapulmonary organs, including 14 out of 21 hearts (4 from reinfection cases), 6 out of 21 livers (3 from reinfection cases), 6 out of 21 kidneys (1 from reinfection cases), and 3 out of 16 small intestine samples (1 from reinfection cases) (Figures 1A,B).
After 7 days, to determine the presence of infectious (that is, replication-competent) SARS-CoV-2 in tissue specimens, we carried out its recovery in a Vero cell monolayer. We have assessed the plaque formation as the lytic cytopathic effect. Thus, infectious SARS-CoV-2 was confirmed after supernatants from tissue extract added on Vero E6 cells identified in 9 out of 21 lung extracts samples (3 from reinfection cases), as well as in extrapulmonary organs, including 14 out of 21 hearts (5 from reinfection cases), 6 out of 21 livers (2 from reinfection cases), 6 out of 21 kidneys (2 from reinfection cases), and 3 out of 16 small intestine samples (2 from reinfection cases) (Figures 1A,B).
Tissue samples from 6 out of the 8 COVID-19 vaccine recipients were included among those with confirmed viral infectivity. Moreover, infectious virus was detected in tissue samples from 5 out of 9 lungs, 6 out of 14 hearts, 2 out of 6 livers, and 3 out of 6 kidneys, in which genomic RNA was undetectable upon direct analysis. Because no viable virus or viral RNA was detected in the cell culture media used for the last wash, it can be concluded that the isolated virus came from viral replication.
The highest viral load measured by RT-qPCR in 7-day/cell culture supernatants (expressed as median values in copies/mL, for ORF1ab and N gene) were found in the lung (1.4 × 106 and 1.06 × 106) and heart (1.9 × 106 and 1.6 × 106) samples. Such viral load decreased slightly in the liver (3.3 × 106 and 2 × 106), small intestine (3.4 × 102 and 1.4 × 103), and kidney (3.2 × 106 and 2 × 106) samples.
Autopsy material originating from those with an antemortem COVID-19 diagnosis is contagious thanks to the presence of SARS-CoV-2. Based on previous autopsy series (Desai et al., 2020; Trypsteen et al., 2020; Vincent and Taccone, 2020; Wolfel et al., 2020; Damiani et al., 2021), SARS-CoV-2 primarily infects the lungs and then disseminates through the blood, infecting cells of the different permissive organs and tissues. Several studies report the persistence of viral RNA shedding for an extended period after the onset of acute symptoms, as well as the presence of viral RNA and/or antigen in the gastrointestinal tissues of convalescent patients (Lamers et al., 2020; Trypsteen et al., 2020; Martin-Cardona et al., 2021; Cheung et al., 2022; Zollner et al., 2022).
The presence of replicating SARS-CoV-2 in samples from cadavers has been previously suggested by the detection of subgenomic SARS-CoV-2 RNA in post-mortem nasopharyngeal swabs (Schroder et al., 2021; Grassi et al., 2022). However, this finding is limited by the fact that the detection of SARS-CoV-2 subgenomic RNAs is not a definitive indicator of active replication (Alexandersen et al., 2020). The presence of viable SARS-CoV-2 in different autopsy tissues was recently reported (Plenzig et al., 2021a; Stein et al., 2022) but it was not possible to observe the same in samples obtained from two exhumed corpses (Plenzig et al., 2021b).
This study includes 21 cases (10 of them with previous hospitalization) with post-mortem nasopharyngeal swabs that tested positive for the presence of SARS-CoV-2 RNA. The autopsies were carried out from which 100 tissue samples were collected including lung, heart, liver, kidney, and small intestine.
We have demonstrated the presence of SARS-CoV-2 genomic RNA, as well as the presence of the replication-competent virus, providing certain proof of the viability of the virus regardless of COVID-19 vaccination status. However, we do not have data available on the presence of neutralizing antibodies against the virus. Five cases of SARS-CoV-2 reinfection were found, and considering the clinical records they presented several clinical signs and symptoms compatible with post-acute COVID. Importantly, viral particles were isolated from tissue samples from these cases, and the genomic characterization of SARS-CoV-2 did not offer any evidence of prevalent viral variants, such as Delta when their first infection diagnosis was carried out, thus offering additional support for the reinfection event.
Because SARS-CoV-2 has numerous distinctive mutations, it can be recognized from partial S-gene direct sequences, including the RBD (European Centre for Disease Prevention and Control, 2021; World Health Organization, 2021). Therefore, we used this approach to detect the presence of three different Omicron sub-variants, including BA.1.1 (20 out of 27 isolates), BA.2 (5 out of 27), and BA.5.2 (2 out of 27). Such distribution matches the Argentinean epidemiology map during the sampling time (Torres et al., 2023). However, further research is warranted, as whole-genome sequencing (WGS) of SARS-CoV-2 using next-generation sequencing (NGS) technology remains the gold standard for tracking and identifying new variants, especially for characterizing minority variants, including preexistent ones (Berno et al., 2022).
Among the studied cases, there were COVID-19 vaccine recipients with completed or unscheduled vaccinations, and even with hybrid immunity. Breakthrough infections with Omicron sub-variants are frequent, despite the strong protection provided by COVID-19 mRNA vaccines and even hybrid immunity against SARS-CoV-2 variants of concern (Van Zelm, 2022; Pradenas et al., 2023).
Interestingly, the tissues studied revealed the presence of SARS-CoV-2 by detecting its genomic RNA and/or its infectivity in cell culture. Thus, autopsies of people who have died from COVID-19 are associated with a reliable biological risk because the virus remains viable in several organs. Although the quantities were widely varied in most cases, viral loads in lung tissue samples appeared higher than in other organs, which probably implies host-related undetermined factors, such as increased affinity to the receptor of human cells and less efficient viral clearance between different tissues (Beyer and Forero, 2022). Likewise, the antemortem clinical condition was diverse among the cases studied, suggesting that the SARS-CoV-2 viral load in lung may vary greatly according to the proliferative and exudative phases of diffuse alveolar damage, which itself drives the multiple organ dissemination of SARS-CoV-2 (Delorey et al., 2021; Odilov et al., 2021). Because tissue samples were collected within a short postmortem interval (a mean of 1.9 days), even between hospitalized and non-hospitalized cases, we have minimized the bias due to cellular autolysis, associated with postmortem changes that alter the extracellular space by releasing intracellular content and influencing the postmortem viral load. Nevertheless, other tissue and cell-related factors may be involved such as their differential cellular permissiveness and the relative abundance of super-permissive cells (Lee et al., 2022). Furthermore, the absence of SARS-CoV-2 RNA tested in tissue homogenate did not necessarily correlate with the absence of viable SARS-CoV-2. Additionally, the presence of viral RNA did not necessarily implicate the presence of infective viral particles.
It could reflect the persistence of RNA in tissues even when the infectious virus has been eliminated (Griffin, 2022). Lung, heart, and kidney samples revealed their infectiousness only when SARS-CoV-2 was isolated in cell culture.
The infectivity time for the SARS-CoV-2 in fresh cadaveric tissue and exhumed corpses even after months in an earth grave warrants further investigation. Furthermore, as the number of individuals recovered from COVID-19 is increasing, it is mandatory to define whether and when organs from these potential donors can be safely used. Few available reports among donors with prior COVID-19 infection indicate low rates of transmission to the recipient and transplanted organ dysfunction, depending on the infection status (Kute et al., 2021; Saharia et al., 2023). However, few studies have been conducted until now, indicating that viral transmission by organ donation could not be completely ruled out.
In conclusion, this study reinforces that autopsies of COVID-19 cases are associated with a consistent biological risk because the virus remains viable and with a high load, necessitating utmost caution in the handling of all corpses.
Thus, the studies involving potentially infected corpses should be performed in adequate biosafety facilities only by expert professionals and technical operators wearing the correct protective equipment following the international biosafety normative (Centers for Disease Control and Prevention, 2020).
Negative clinical records of SARS-CoV-2 do not guarantee microbiologically negative yields, highlighting the importance of carrying out additional clinical tests and examinations in organ donors who have recovered from SARS-CoV-2 infection. In the end, this research makes a strong argument for continuing research of this kind in the future to characterize the mechanisms underlying post-acute COVID-19 sequelae in the pulmonary and wide range of extrapulmonary organ systems. It also adds to our understanding of the virus’s multiple tissue tropisms even after reinfection and plausible persistence of SARS-CoV-2.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
MD and JQ contributed equally to the conception and design of this study, funding acquisition, project administration, acquisition, analysis, interpretation of data, and writing of the manuscript. SM-B, AD’A, and CB were responsible for the autopsy and sample collection at Morgue. CC focused on bioinformatics analysis of data. FR SARS-CoV-2 viral load data curation. RF, CL, AG, FS, CC, and PJ contributed to molecular biology experiments. All authors contributed to the article and approved the submitted version.
This study was supported by the Argentinean National Agency of Scientific and Technological Promotion (grant number PICTO 2021-00005 to JQ). The funder had no role in the study design, data collection, interpretation, or the decision to submit the work for publication.
The authors thank Cecilia Monzani for her technical assistance with nucleotide sequencing.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Al-Aly, Z., Bowe, B., and Xie, Y. (2022). Long COVID after breakthrough SARS-CoV-2 infection. Nat. Med. 28, 1461–1467. doi: 10.1038/s41591-022-01840-0
Alexandersen, S., Chamings, A., and Bhatta, T. R. (2020). SARS-CoV-2 genomic and subgenomic RNAs in diagnostic samples are not an indicator of active replication. Nat. Commun. 11:6059. doi: 10.1038/s41467-020-19883-7
Aquila, I., Ricci, P., Bonetta, C. F., Sacco, M. A., Longhini, F., Torti, C., et al. (2021). Analysis of the persistence time of the SARS-CoV-2 virus in the cadaver and the risk of passing infection to autopsy staff. Med. Leg. J. 89, 40–53. doi: 10.1177/0025817220980601
Beltempo, P., Curti, S. M., Maserati, R., Gherardi, M., and Castelli, M. (2021). Persistence of SARS-CoV-2 RNA in post-mortem swab 35 days after death: a case report. Forensic Sci. Int. 319:110653. doi: 10.1016/j.forsciint.2020.110653
Berno, G., Fabeni, L., Matusali, G., Gruber, C. E. M., Rueca, M., Giombini, E., et al. (2022). SARS-CoV-2 variants identification: overview of molecular existing methods. Pathogens 11:1058. doi: 10.3390/pathogens11091058
Beyer, D. K., and Forero, A. (2022). Mechanisms of antiviral immune evasion of SARS-CoV-2. J. Mol. Biol. 434:167265. doi: 10.1016/j.jmb.2021.167265
Bowe, B., Xie, Y., and Al-Aly, Z. (2022). Acute and postacute sequelae associated with SARS-CoV-2 reinfection. Nat. Med. 28, 2398–2405. doi: 10.1038/s41591-022-02051-3
Casagrande, M., Fitzek, A., Puschel, K., Aleshcheva, G., Schultheiss, H. P., Berneking, L., et al. (2020). Detection of SARS-CoV-2 in human retinal biopsies of deceased COVID-19 patients. Ocul. Immunol. Inflamm. 28, 721–725. doi: 10.1080/09273948.2020.1770301
Centers for Disease Control and Prevention. (2020). Centers for Disease Control and Prevention collection and submission of postmortem specimens from deceased persons with known or Supected COVID-19, march 2020. Available at: https://www.cdc.gov/coronavirus/2019-ncov/hcp/guidance-postmortem-specimens.html (Accessed December 23, 2020).
Chen, Y., Gu, S., Chen, Y., Lu, H., Shi, D., Guo, J., et al. (2022). Six-month follow-up of gut microbiota richness in patients with COVID-19. Gut 71, 222–225. doi: 10.1136/gutjnl-2021-324090
Cheung, C. C. L., Goh, D., Lim, X., Tien, T. Z., Lim, J. C. T., Lee, J. N., et al. (2022). Residual SARS-CoV-2 viral antigens detected in GI and hepatic tissues from five recovered patients with COVID-19. Gut 71, 226–229. doi: 10.1136/gutjnl-2021-324280
Damiani, S., Fiorentino, M., De Palma, A., Foschini, M. P., Lazzarotto, T., Gabrielli, L., et al. (2021). Pathological post-mortem findings in lungs infected with SARS-CoV-2. J. Pathol. 253, 31–40. doi: 10.1002/path.5549
Deinhardt-Emmer, S., Wittschieber, D., Sanft, J., Kleemann, S., Elschner, S., Haupt, K. F., et al. (2021). Early postmortem mapping of SARS-CoV-2 RNA in patients with COVID-19 and the correlation with tissue damage. elife 10:60361. doi: 10.7554/eLife.60361
Delorey, T. M., Ziegler, C. G. K., Heimberg, G., Normand, R., Yang, Y., Segerstolpe, A., et al. (2021). COVID-19 tissue atlases reveal SARS-CoV-2 pathology and cellular targets. Nature 595, 107–113. doi: 10.1038/s41586-021-03570-8
Desai, N., Neyaz, A., Szabolcs, A., Shih, A. R., Chen, J. H., Thapar, V., et al. (2020). Temporal and spatial heterogeneity of host response to SARS-CoV-2 pulmonary infection. Nat. Commun. 11:6319. doi: 10.1038/s41467-020-20139-7
Edler, C., Schroder, A. S., Aepfelbacher, M., Fitzek, A., Heinemann, A., Heinrich, F., et al. (2020). Dying with SARS-CoV-2 infection-an autopsy study of the first consecutive 80 cases in Hamburg, Germany. Int. J. Legal Med. 134, 1275–1284. doi: 10.1007/s00414-020-02317-w
Elrobaa, I. H., and New, K. J. (2021). COVID-19: pulmonary and extra pulmonary manifestations. Front. Public Health 9:711616. doi: 10.3389/fpubh.2021.711616
European Centre for Disease Prevention and Control. (2021). Europe WHORO for methods for the detection and characterisation of SARS-CoV-2 variants–first update what is New in this update: Key messages. Available at: https://www.ecdc.europa.eu/sites/default/files/documents/Methods-for-the-detection-char-SARS-CoV-2-variants_2nd%20update_final.pdf (Accessed August 25, 2022).
Fitzek, A., Schadler, J., Dietz, E., Ron, A., Gerling, M., Kammal, A. L., et al. (2021). Prospective postmortem evaluation of 735 consecutive SARS-CoV-2-associated death cases. Sci. Rep. 11:19342. doi: 10.1038/s41598-021-98499-3
Gao, P., Liu, J., and Liu, M. (2022). Effect of COVID-19 vaccines on reducing the risk of long COVID in the real world: a systematic review and Meta-analysis. Int. J. Environ. Res. Public Health 19:422. doi: 10.3390/ijerph191912422
Goh, D., Lim, J. C. T., Fernaindez, S. B., Joseph, C. R., Edwards, S. G., Neo, Z. W., et al. (2022). Case report: persistence of residual antigen and RNA of the SARS-CoV-2 virus in tissues of two patients with long COVID. Front. Immunol. 13:939989. doi: 10.3389/fimmu.2022.939989
Grassi, S., Arena, V., Cattani, P., Dell'aquila, M., Liotti, F. M., Sanguinetti, M., et al. (2022). SARS-CoV-2 viral load and replication in postmortem examinations. Int. J. Legal Med. 136, 935–939. doi: 10.1007/s00414-021-02753-2
Griffin, D. E. (2022). Why does viral RNA sometimes persist after recovery from acute infections? PLoS Biol. 20:e3001687. doi: 10.1371/journal.pbio.3001687
Kute, V. B., Fleetwood, V. A., Meshram, H. S., Guenette, A., and Lentine, K. L. (2021). Use of organs from SARS-CoV-2 infected donors: is it safe? A contemporary review. Curr. Transplant. Rep. 8, 281–292. doi: 10.1007/s40472-021-00343-0
Lamers, M. M., Beumer, J., Van Der Vaart, J., Knoops, K., Puschhof, J., Breugem, T. I., et al. (2020). SARS-CoV-2 productively infects human gut enterocytes. Science 369, 50–54. doi: 10.1126/science.abc1669
Layne, S. P., Walters, K. A., Kash, J. C., and Taubenberger, J. K. (2022). More autopsy studies are needed to understand the pathogenesis of severe COVID-19. Nat. Med. 28, 427–428. doi: 10.1038/s41591-022-01684-8
Lee, J. Y., Wing, P. A. C., Gala, D. S., Noerenberg, M., Jarvelin, A. I., Titlow, J., et al. (2022). Absolute quantitation of individual SARS-CoV-2 RNA molecules provides a new paradigm for infection dynamics and variant differences. elife 11:74153. doi: 10.7554/eLife.74153
Maiese, A., Manetti, A. C., La Russa, R., Di Paolo, M., Turillazzi, E., Frati, P., et al. (2021). Autopsy findings in COVID-19-related deaths: a literature review. Forensic Sci. Med. Pathol. 17, 279–296. doi: 10.1007/s12024-020-00310-8
Martin-Cardona, A., Lloreta Trull, J., Albero-Gonzalez, R., Paraira Beser, M., Andujar, X., Ruiz-Ramirez, P., et al. (2021). SARS-CoV-2 identified by transmission electron microscopy in lymphoproliferative and ischaemic intestinal lesions of COVID-19 patients with acute abdominal pain: two case reports. BMC Gastroenterol. 21:334. doi: 10.1186/s12876-021-01905-3
Nalbandian, A., Sehgal, K., Gupta, A., Madhavan, M. V., Mcgroder, C., Stevens, J. S., et al. (2021). Post-acute COVID-19 syndrome. Nat. Med. 27, 601–615. doi: 10.1038/s41591-021-01283-z
Nascimento, T., do Valle Costa, L., Ruiz, A. D., Ledo, C. B., Fernandes, V. P. L., Cardoso, L. F., et al. (2023). Vaccination status and long COVID symptoms in patients discharged from hospital. Sci. Rep. 13:2481. doi: 10.1038/s41598-023-28839-y
Neurath, M. F., Uberla, K., and Ng, S. C. (2021). Gut as viral reservoir: lessons from gut viromes, HIV and COVID-19. Gut 70, 1605–1608. doi: 10.1136/gutjnl-2021-324622
Odilov, A., Volkov, A., Abdullaev, A., Gasanova, T., Lipina, T., and Babichenko, I. (2021). COVID-19: multiorgan dissemination of SARS-CoV-2 is driven by pulmonary factors. Viruses 14:39. doi: 10.3390/v14010039
Peter, R. S., Nieters, A., Krausslich, H. G., Brockmann, S. O., Gopel, S., Kindle, G., et al. (2022). Post-acute sequelae of covid-19 six to 12 months after infection: population based study. BMJ 379:e071050. doi: 10.1136/bmj-2022-071050
Plenzig, S., Bojkova, D., Held, H., Berger, A., Holz, F., Cinatl, J., et al. (2021a). Infectivity of deceased COVID-19 patients. Int. J. Legal Med. 135, 2055–2060. doi: 10.1007/s00414-021-02546-7
Plenzig, S., Holz, F., Bojkova, D., Kettner, M., Cinatl, J., Verhoff, M. A., et al. (2021b). Detection and infectivity of SARS-CoV-2 in exhumated corpses. Int. J. Legal Med. 135, 2531–2536. doi: 10.1007/s00414-021-02670-4
Pradenas, E., Marfil, S., Urrea, V., Trigueros, V., Pidkova, T., Pons-Grífols, A., et al. (2023). Impact of hybrid immunity booster vaccination and omicron breakthrough infection on SARS-CoV-2 VOCs cross-neutralization. iScience 26:106457. doi: 10.1016/j.isci.2023.106457
Sablone, S., Solarino, B., Ferorelli, D., Benevento, M., Chironna, M., Loconsole, D., et al. (2021). Post-mortem persistence of SARS-CoV-2: a preliminary study. Forensic Sci. Med. Pathol. 17, 403–410. doi: 10.1007/s12024-021-00375-z
Saharia, K. K., Ramelli, S. C., Stein, S. R., Roder, A. E., Kreitman, A., Banakis, S., et al. (2023). Successful lung transplantation using an allograft from a COVID-19-recovered donor: a potential role for subgenomic RNA to guide organ utilization. Am. J. Transplant. 23, 101–107. doi: 10.1016/j.ajt.2022.09.001
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Sanyaolu, A., Marinkovic, A., Prakash, S., Zhao, A., Balendra, V., Haider, N., et al. (2022). Post-acute Sequelae in COVID-19 survivors: an overview. SN Compr. Clin. Med. 4:91. doi: 10.1007/s42399-022-01172-7
Schroder, A. S., Edler, C., Ondruschka, B., Puschel, K., Schadler, J., Heinemann, A., et al. (2021). The handling of SARS-CoV-2 associated deaths - infectivity of the body. Forensic Sci. Med. Pathol. 17, 411–418. doi: 10.1007/s12024-021-00379-9
Soriano, J. B., Murthy, S., Marshall, J. C., Relan, P., and Diaz, J. V., WHO Clinical Case Definition Working Group on Post-COVID-19 Condition (2022). A clinical case definition of post-COVID-19 condition by a Delphi consensus. Lancet Infect. Dis. 22, e102–e107. doi: 10.1016/S1473-3099(21)00703-9
Stein, S. R., Ramelli, S. C., Grazioli, A., Chung, J. Y., Singh, M., Yinda, C. K., et al. (2022). SARS-CoV-2 infection and persistence in the human body and brain at autopsy. Nature 612, 758–763. doi: 10.1038/s41586-022-05542-y
Tamura, K. S. G., Stecher, G., and Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Tejerina, F., Catalan, P., Rodriguez-Grande, C., Adan, J., Rodriguez-Gonzalez, C., Muñoz, P., et al. (2022). Post-COVID-19 syndrome. SARS-CoV-2 RNA detection in plasma, stool, and urine in patients with persistent symptoms after COVID-19. BMC Infect. Dis. 22:211. doi: 10.1186/s12879-022-07153-4
Torres, C., Nabaes Jodar, M., Acuña, D., Montaño, R. M. Z., Culasso, A. C. A., Amadio, A. F., et al. (2023). Omicron waves in Argentina: dynamics of SARS-CoV-2 lineages BA.1, BA.2 and the emerging BA.2.12.1 and BA.4/BA.5. Viruses 15:312. doi: 10.3390/v15020312
Trypsteen, W., Van Cleemput, J., Snippenberg, W. V., Gerlo, S., and Vandekerckhove, L. (2020). On the whereabouts of SARS-CoV-2 in the human body: a systematic review. PLoS Pathog. 16:e1009037. doi: 10.1371/journal.ppat.1009037
Van Zelm, M. C. (2022). Immune memory to SARS-CoV-2 omicron BA.1 breakthrough infections: to change the vaccine or not? Sci. Immunol. 7:eabq5901. doi: 10.1126/sciimmunol.abq5901
Vincent, J. L., and Taccone, F. S. (2020). Understanding pathways to death in patients with COVID-19. Lancet Respir. Med. 8, 430–432. doi: 10.1016/S2213-2600(20)30165-X
Wolfel, R., Corman, V. M., Guggemos, W., Seilmaier, M., Zange, S., Muller, M. A., et al. (2020). Virological assessment of hospitalized patients with COVID-2019. Nature 581, 465–469. doi: 10.1038/s41586-020-2196-x
World Health Organization. (2021). WHO genomic sequencing of SARS-CoV-2 p. 94. Available at: https://www.who.int/publications/i/item/9789240018440 (Accessed July 27, 2022).
World Health Organization. (2022). End-to-end integration of SARS-CoV-2 and influenza sentinel surveillance: Revised interim guidance, 31 January 2022. Available at: https://apps.who.int/iris/handle/10665/351409 (Accessed 31 January 2022).
Keywords: SARS-CoV-2, virus isolation, postmortem tissue, Vero E6 cell line, omicron variants
Citation: Maffia-Bizzozero S, Cevallos C, Remes Lenicov F, Freiberger RN, Lopez CAM, Guano Toaquiza A, Sviercz F, Jarmoluk P, Bustos C, D’Addario AC, Quarleri J and Delpino MV (2023) Viable SARS-CoV-2 Omicron sub-variants isolated from autopsy tissues. Front. Microbiol. 14:1192832. doi: 10.3389/fmicb.2023.1192832
Received: 27 March 2023; Accepted: 08 May 2023;
Published: 22 May 2023.
Edited by:
Axel Cloeckaert, Institut National de recherche pour l’agriculture, l’alimentation et l’environnement (INRAE), FranceReviewed by:
Farid Rahimi, Australian National University, AustraliaCopyright © 2023 Maffia-Bizzozero, Cevallos, Remes Lenicov, Freiberger, Lopez, Guano Toaquiza, Sviercz, Jarmoluk, Bustos, D’Addario, Quarleri and Delpino. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jorge Quarleri, cXVhcmxlcmlAZm1lZC51YmEuYXI=; M. Victoria Delpino, bWRlbHBpbm9AZmZ5Yi51YmEuYXI=;
†These authors share first authorship
‡These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.