- 1ICAR-Directorate of Onion and Garlic Research, Pune, India
- 2Department of Botany, S.N. Arts, D.J.M. Commerce and B.N.S. Science College, Sangamner, Maharashtra, India
- 3Department of Biotechnology, Centurion University of Technology and Management, Paralakhemundi, India
Plant-endophyte symbiosis influences plant defense and growth. Serendipita indica is a root endophyte that promotes growth and induces tolerance against biotic and abiotic stress in plants. In this study, we examined the effect of S. indica colonization on herbivore (Spodoptera exigua) resistance of onion (Allium cepa L.). We found that colonization of S. indica in the roots of onion significantly reduced the feeding damage of leaves by S. exigua larvae, and also resulted in a reduction in weight gain of the larvae when fed on S. indica plants. This enhanced resistance is a result of modulation of antioxidant and defense enzymes/genes in the host by S. indica mutualism. Specifically, the activities of enzymes such as Superoxide dismutase, peroxidase, polyphenol oxidase, phenylalanine ammonia-lyase, and H2O2 content were significantly higher in the early stages of S. exigua feeding in the S. indica colonized plants compared to the non-colonized counterparts. Similarly, defense genes also showed modulation in response to this tripartite interaction of onion -S. indica mutualism and S. exigua herbivory. The hierarchical cluster analysis and principal component analysis indicated a clear difference in the onion biochemical responses, which is due to the S. indica symbiosis. Our investigation demonstrates that onion-S. indica symbiosis significantly decreases chewing injury by efficiently modulating antioxidant and defense enzyme activities and gene expression in response to S. exigua herbivory. Therefore, S. indica can be used as a potential biocontrol agent for sustainable management of this important pest of Alliums.
Introduction
Onion (Allium cepa) is an important vegetable from the Allium genus in the Amaryllidaceae family. Onions are cultivated and consumed worldwide. Onions hold high economic and medicinal significance for their uses in culinary practices and traditional medicines. Quercetin, the major bioactive compound in onion acts as a lachrymatic agent and along with other secondary metabolites provides anti-bacterial, anti-fungal, anti-cancer, and antioxidant activities (Khandagale and Gawande, 2019). On the other hand, allicin, an organosulphur compound in garlic is used as a stimulant, diuretic, diaphoretic, and expectorant (Nanda et al., 2016). India is the second largest producer of onion in the world (FAOSTAT, 2021). However, India cultivates onions in the largest production area of 1.4 lakh million hectare with productivity of 16.4 t/ha (FAOSTAT, 2021). Thus, it is evident that the productivity of onions in India is low compared to the global productivity rates. Severe crop losses due to different pathogen attacks and insect infestations are the major causes of this lower productivity rate in India. Among the insect pests, the Lepidopteran insect Spodoptera spp. has been reported to be a notorious pest in onions causing huge crop losses (Fernandes et al., 2012; Soumia et al., 2020; Cokola et al., 2021).
Spodoptera is a genus of moths having mostly polyphagous feeding habits. In onions, the infestation of S. frugiperda (fall armyworms) and S. exigua (beet armyworm) have been reported. S. frugiperda is a major pest of some cereals and field crops, including maize, sugarcane, millets, sorghum, and cotton, and causes huge crop damage. However, both recent and earlier reports have confirmed the infestation of S. frugiperda on onions in different parts of the world (Fernandes et al., 2012; Cokola et al., 2021). Similarly, S. exigua infests vegetable crops and cereals, including cruciferous crops, okra, and wheat. The first report of S. exigua infestation in Allium spp. (A. fistulosum, welsh onion) came in 1993. Kawai et al. (1993) reported that S. exigua could invade the leaf blades and the infestation was contagious among the onion hills. Similarly, several subsequent studies reporting the infestation incidents of S. exigua on onions confirmed that onion is a host plant for this notorious pest (Zheng et al., 2000; Ueno, 2015; Ilmi et al., 2022). These reports of the host shifting suggest the strong polyphagous and well-diverse host adaptability of the Spodoptera spp. Moreover, in India it is emerging pest of onion, recorded a significant foliage damage of 70–80% in rabi onions when subjected to infestation by S. exigua (Soumia et al., 2020). Therefore, the management and control of Spodoptera spp. are crucial in achieving crop security. Currently, the use of synthetic pesticides is the most practiced method of controlling Spodoptera infestation. However, the exploitation of these broad-spectrum chemical pesticides has caused the elimination of natural enemies, insect resurgence, and insecticide resistance along with other environmental hazards. Therefore, alternative environment-friendly and sustainable solutions to control pest infestations are required.
Serendipita indica (formerly known as Piriformospora indica) is a widely-studied plant endosymbiont having phyto-promotional and stress-alleviating roles (Gill et al., 2016). Notably, S. indica is a cultivable symbiont with beneficial properties such as growth promotion, induction of stress tolerance etc. The role of S. indica in enhancing plant growth, development, and nutrient acquisition has already been reported (Gill et al., 2016). In recent times, the use of S. indica as a biocontrol agent in different crops against several biotic stressors is being focused on. Several reports have confirmed the use of S. indica as a biocontrol agent in controlling the infestation of both pathogens and insects. For instance, the use of S. indica as a biocontrol agent protected Arabidopsis plants from the soil-borne vascular pathogen Verticillium dahliae. S. indica restricted the growth of V. dahliae in Arabidopsis and regulated the Arabidopsis defense responses against the pathogen (Sun et al., 2014). Similarly, Atia et al. (2020) reported that S. indica improved tolerance of cucumber plants against the root-knot nematode infection by regulating photosynthesis and innate immune responsive genes. Interestingly, the use of S. indica has been explored to control insect infestations, including the Spodoptera spp. The colonization of S. indica in rice plants improved the resistance against Cnaphalocrocis medinalis (rice leaffolder) through the activation of antioxidant- and jasmonic acid (JA)-mediated defense responses (Chen et al., 2022). In sweet potato plants, the colonization of S. indica enhanced the JA levels, induced the expression of defense-responsive genes, and increased trypsin inhibitory activity resulting in a significant reduction in the performance of S. litura (Li et al., 2021). Therefore, considering these facts, the potential of S. indica as a biocontrol agent against S. exigua has been investigated in this study. Further, the mechanism of onion defense modulation against the S. exigua infestation is explored by analyzing the key defense-related gene expressions, including WRKY transcription factors, and JA and salicylic acid (SA) pathway-related genes. In addition, the antioxidant activities in onions are studied by analyzing the peroxidase (POD), superoxide dismutase (SOD), and hydrogen peroxide (H2O2) levels. Furthermore, defense enzymes like phenylalanine ammonia-lyase (PAL) and polyphenol oxidase (PPO) were also included in the study. Overall, the findings of this study provide valuable insights into the S. indica-mediated control of S. exigua infestations in onions.
Materials and methods
Plant material and fungal culture
The susceptible onion cultivar “Bhima Super” was used as plant material for this study. The culture of S. indica was obtained from Dr. J. Vadassery (National Institute for Plant Genome Research, New Delhi, India) and maintained on potato dextrose agar (PDA) medium.
Serendipita indica colonization of onion plant roots
The cultivation and harvesting of S. indica mycelium were done as described in Roylawar et al. (2021). Ten grams of minced mycelium per 100 g of autoclaved soil was mixed in a perforated plastic crate (20′ × 13′ × 11′). Seeds of Bhima Super were sown in these autoclaved soil crates with S. indica mixture while, in control, seeds were sown in autoclaved soil only. The colonization of onion roots was examined microscopically after 30 days of sowing in S. indica-treated pots. Roots of 10 randomly selected S. indica-treated plants were incubated at 90°C in 10% KOH for 5 min. Followed by 1 min. in 1 N HCl and then stained with trypan blue (0.02%) for 60 min. at room temperature. Colonization was observed under a light microscope in 40X magnification (LEICA DM2500).
Insect material and rearing
The egg masses and larvae of S. exigua (Lepidoptera: Noctuidae) collected from the experimental field of ICAR-Directorate of Onion and Garlic Research, Rajgurunagar (Pune, India) (18.8543°N, 73.8876°E) during the Kharif season were reared in laboratory under ambient conditions [25 ± 2°C and 65 ± 5% relative humidity (RH); 16:8 h light:dark photoperiod]. Larvae were raised on fresh castor leaves in transparent acrylic jars covered with a piece of muslin cloth as described by Saeed et al. (2017). When the pupae were ready, they were surface sterilized with 0.1% sodium hypochlorite solution and transferred into fresh acrylic jars for adult emergence and mating. Castor leaves were used as the oviposition substrate whereas cotton swabs with 50% honey and water as a diet in adult cages. Insects were sub-cultured consecutively for three generations. Subsequent progenies emerging from the sub-culture were used for the present experiment.
Onion-Spodoptera exigua interaction assays
To study the effect of induced systemic resistance of S. indica-treated onion against S. exigua, single onion plants potted in individual plastic pots (20 cm diameter) were taken with twelve replications each for S. indica-treated and control. For the in-planta experiments, artificial infestation with one third instar larvae per plant (eight-week-old S. indica treated onion plant) was released under glasshouse conditions. Before the feeding assay, larvae were starved for 6 h. After 24 h of infestation, total leaves and damaged leaves were counted and their overall damage rating is converted to a 0–3 scale (0 = No damage; 1 = up to 1/3 of leaf area scarped; 2 = 1/3 to ½ of leaf area scraped; 3 = more than ½ of leaf area scraped) (Kavitha and Reddy, 2012).
For further biochemical and gene expression analyses, leaf tissue was also collected at 6, 12, and 24 h-post infestations (hpi), powdered in liquid nitrogen, and stored at −80° C.
Insect feeding assays
The S. indica-colonized onion plants were used to conduct the S. exigua feeding bioassays (Li et al., 2021). The 3rd instar S. exigua larvae were placed on the onion leaves for the feeding evaluations. Both initial and final larval weights (n = 15) were determined using an analytical balance after 3 days of feeding and the weight gain was measured.
Evaluation of H2O2 content
The level of hydrogen peroxide (H2O2) was estimated according to the method described by Roylawar et al. (2021). In 4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES) buffer (50 mM; pH 7.5) 50 mg of frozen leaf sample was mixed and centrifuged at 12,000 rpm (4°C) for 5 min. The supernatant (100 μL) was mixed with 1 mL reaction volume containing HEPES buffer (50 mM, pH 7.5) and potassium titanium oxalate (2.5% in 20% H2SO4). The absorbance was recorded at 410 nm and the H2O2 content was deduced from the standard calibration curve.
SOD activity
200 mg leaf tissue from each treatment was homogenized in an ice-cold potassium phosphate buffer (0.1 M, pH 7.2) with ethylenediaminetetraacetic acid disodium salt (Na-EDTA) (0.1 M) and polyvinylpyrrolidone (0.5%). Homogenate was centrifuged for 15 min. (20,000 rpm at 4°C) and the supernatant was used for the estimation of SOD and POD enzymatic activities. Superoxide dismutase activity was measured according to Beauchamp and Fridovich (1971). The assay was performed in a 3 mL reaction volume containing phosphate buffer (50 mM, pH 7.8), riboflavin (50 μM), methionine (20 mM), EDTA (1 mM), nitro blue tetrazolium chloride (NBT) (1 mM), and 70 μL enzyme extract. The reaction mixtures were prepared in two replicate sets for each treatment. One set was kept in the dark and the other was incubated in the light for 20 min. Under 100 μE m−2 s−1 light intensity at 26 ± 2° C. Absorbance was measured at 560 nm, and the dark-incubated reaction mixtures of each sample were used as a blank.
POD activity
The POD activity was measured as described by Xu et al. (2011). The assay was performed in a 3 mL reaction mixture containing phosphate buffer (0.1 M, pH 7.2), guaiacol (30 mM), H2O2 (20 mM), and enzyme extract. The POD activity was determined using the molar extinction coefficient of 26.6 mM−1 cm−1 and expressed in Ug−1 fresh wt.
PPO activity
For PPO activity measurement, 400 mg frozen leaves were homogenized in ice-cold 1 mL of 50 mM phosphate buffer (pH 7.1) containing 0.1% (w/v) SDS. The homogenate was centrifuged at 10,000 g for 15 min. at 4° C and the supernatant was collected. The protein concentration of the enzyme extract was determined by the Bradford method (Bradford, 1976). The activity of PPO was determined spectrophotometrically at a wavelength of 400 nm in a reaction mixture (1 mL) containing 50 mM phosphate buffer, pH 5.5, 10 mM 3-methyl-catechol, and 150 μL enzyme extract. The assay buffer was aerated for 5 min before use. Catalase (280 Units ml−1; Sigma) was added to remove any H2O2 present and prevent interference by peroxidases (Sherman et al., 1991). In addition, the specificity of the assay was verified by performing parallel assays with 1 mM kojic acid, a specific PPO inhibitor. Enzyme activity was determined according to Cheema and Sommerhalter (2015). One unit of PPO activity was defined as the amount of enzyme producing 1 μmol of quinone per minute at 30° C. Specific activity was expressed in U/mg protein.
PAL activity
The activity of PAL was measured as reported by Roylawar et al. (2021). For enzyme extraction, 1 g of each leaf sample was ground in liquid nitrogen and homogenized in 3 mL of cold Tris–HCl buffer (50 mM, pH 8.0), containing 2-mercaptoethanol (1.5 mM) and 0.5% polyvinylpyrrolidone (PVP), then centrifuged at 12,000 rpm for 10 min. at 4° C. The resultant supernatant was used as an enzyme extract for the PAL assay. The assay was performed in a 3 mL reaction mixture containing 1.5 mL of Tris–HCl buffer (50 mM, pH 8.0), 200 μL of L-phenylalanine (1.5 mM), 0.9 mL of distilled water, and 0.3 mL of enzyme extract. The reaction mixture was incubated at 30 ± 1° C for 90 min., and then 1 mL of HCl (2 N) was added to stop the reaction. Further, 3 mL of toluene was added to the mixture before centrifugation at 14,000 rpm for 10 min. at 4° C. The upper layer (enzyme-toluene mixture) was removed carefully and read at 290 nm. Toluene (in the absence of the enzyme) was used as a blank. A standard curve for trans-cinnamic acid (0–100 μM) was plotted to deduce the quantity of cinnamic acid formed in the sample.
Defense gene expression analyses
RNA isolation and cDNA synthesis
The leaf samples were collected at 12 and 24 hpi from all treatments. Three plants pooled as one biological replicate and two such biological replicates were incorporated into the study. Total RNA was isolated from the leaves of two biological replicates using the RNeasy Plant Mini Kit (Qiagen, Germany). The potential genomic DNA contamination was eliminated with Dnase I (Fermentas, Lithuania) treatment. RNAs were quantified using NanoDrop (ND1000 Spectrophotometer, ThermoFisher Scientific, United States). First-strand cDNA was synthesized by reverse transcription from 1 μg RNA using a revert aid first-strand cDNA synthesis kit (Fermenats, Lithuania). cDNAs were stored at −80°C until used in further study.
RT-qPCR analyses
Gene sequences of onion available at NCBI were used for primer designing by Gene Runner software. Expression analysis of defense genes was performed in LightCylcer 480 II instrument (Roche, Germany). A 10 μL PCR reaction was carried out in triplicate for each treatment with Light cycler 480 SYBR Green l master mix (Roche, Germany). The PCR cycling was programmed as follows: initial denaturation at 95°C for 5 min, 40 cycles of 95°C for 15 s, 55–60°C for 45 s, and 72°C for 30 s. The melting curve analysis was done to confirm PCR specificity. The onion Actin gene (AcActin) was used as the internal control for the normalization of cycle threshold (CT) value and relative transcript abundance was calculated according to the 2−∆∆Ct method (Schmittgen and Livak, 2008). The details of primers used in the present qPCR analyses are depicted in Table 1.
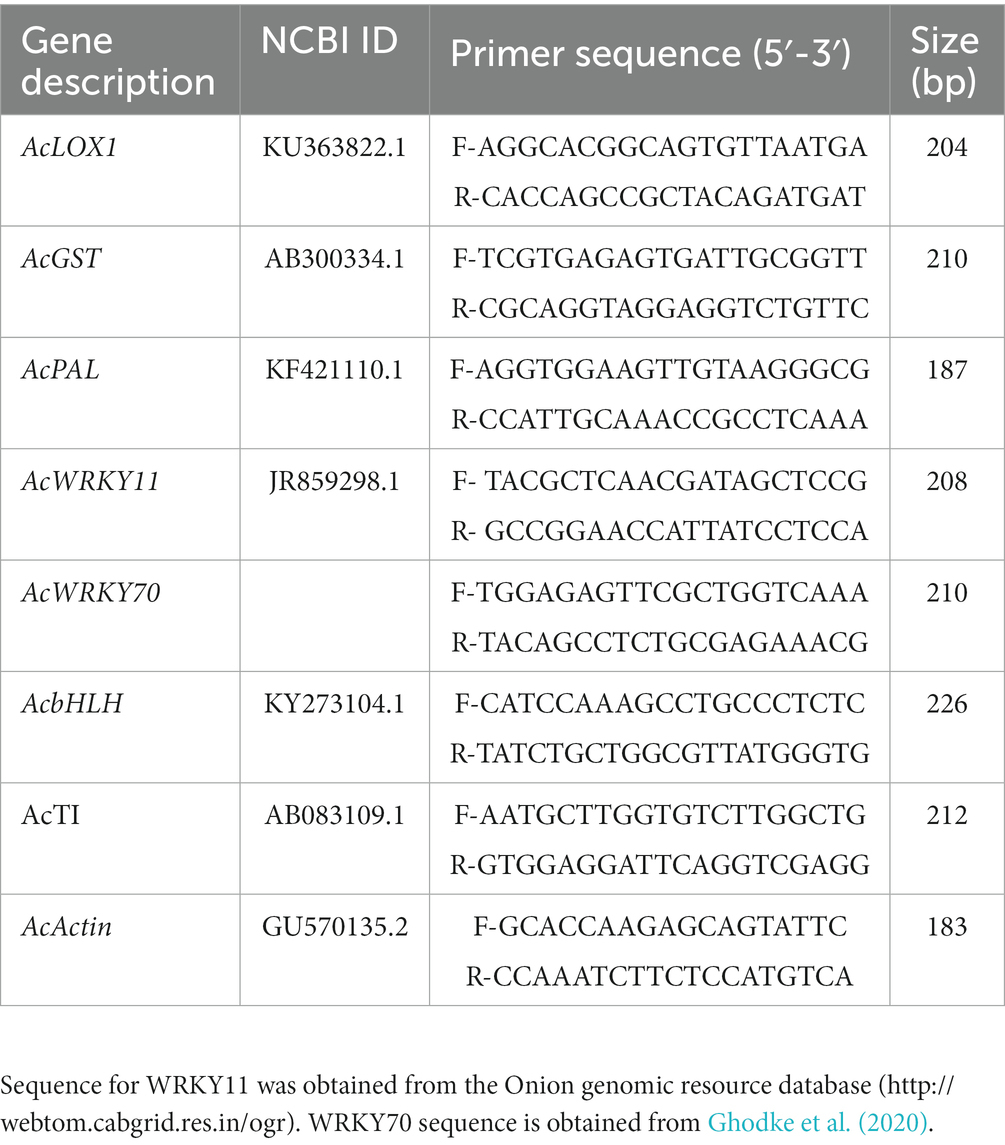
Table 1. Selected genes and respective primer set used for gene expression analysis from onion leaves.
Statistical analysis
The gene expression data were analyzed by two-way ANOVA at p ≤ 0.05 to evaluate the statistical significance. Similarly, the principal component analysis (PCA) and hierarchical clustering analysis (HCA) of H2O2, SOD, POD, PAL, and PPO activity were performed on the MetaboAnalyst 5.0 platform (Pang et al., 2021). The PCA was carried out to compare onion plant response to S. exigua infestation under control conditions, with or without S. indica treatments. The HCA was employed to evaluate the similarities and dissimilarities between the response variables. The clustering was done by using the Euclidean distance and Ward clustering algorithm. All data sets were normalized before they were used for PCA and HCA.
Results
Serendipita indica colonization in the onion root and efficacy against Spodoptera exigua
Colonization of onion root by S. indica was confirmed after 30 days post inoculation (DPI) by microscopy. Pear-shaped chlamydospores were observed in the onion root cortex (Figure 1A). In control plants there was no colonization observed. The efficacy of the S. indica treatment in controlling S. exigua was evaluated by checking the performance of S. exigua larvae (Figures 1B,C). The results suggested that in the treated plants, the insect larvae performance was reduced as compared to the control plants (non-treated). The larval weights were significantly less on the treated plants as compared to the larvae fed on the non-treated plants (Figure 1D). The chewing injury by S. exigua larvae in S. indica colonized plants was significantly reduced compared to non-colonized plants (Supplementary Table S1).
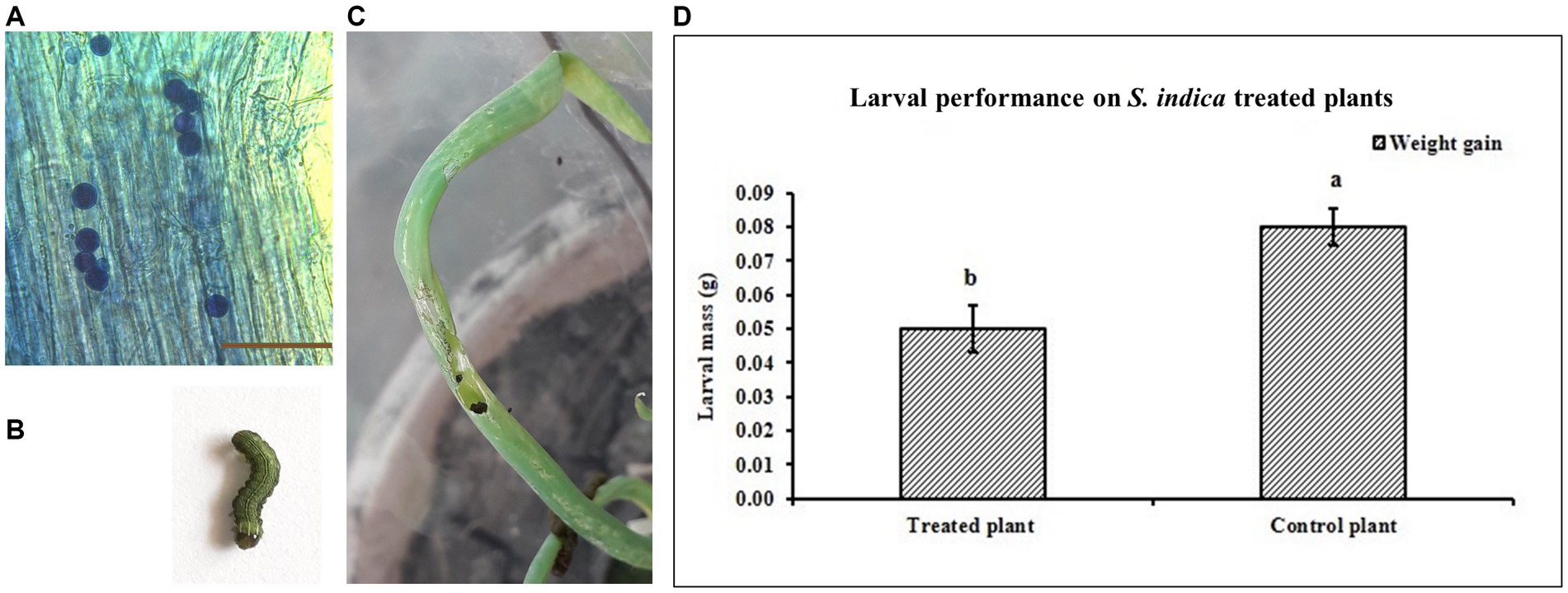
Figure 1. (A) Colonization of onion root by S. indica. (B) 3rd instar larva of S. exigua. (C) Leaf damaged due to feeding of S. exigua. (D) Performance of the S. exigua larvae on the S. indica-treated and untreated (control) plants. Data are represented as mean ± SE and the alphabets indicate the statistical significance computed at p < 0.05.
Quantification of H2O2
The H2O2 levels were measured at 6, 12, and 24 h post infestation (hpi). No significant increase in the H2O2 levels was observed at 6 hpi in control plants and S. indica colonized plants in response to S. exigua infestation. Whereas, at 12 hpi S. indica-colonized plants showed an 18.55% increase in the H2O2 level as compared to non-colonized plants in response to S. exigua (Figure 2). Furthermore, in the late stage of infestation, the level of H2O2 was highly reduced in S. indica-colonized plants, while the non-colonized plants showed a 55.73% increase in H2O2 accumulation. There was no significant difference observed in control and S. indica-colonized uninfested plants.
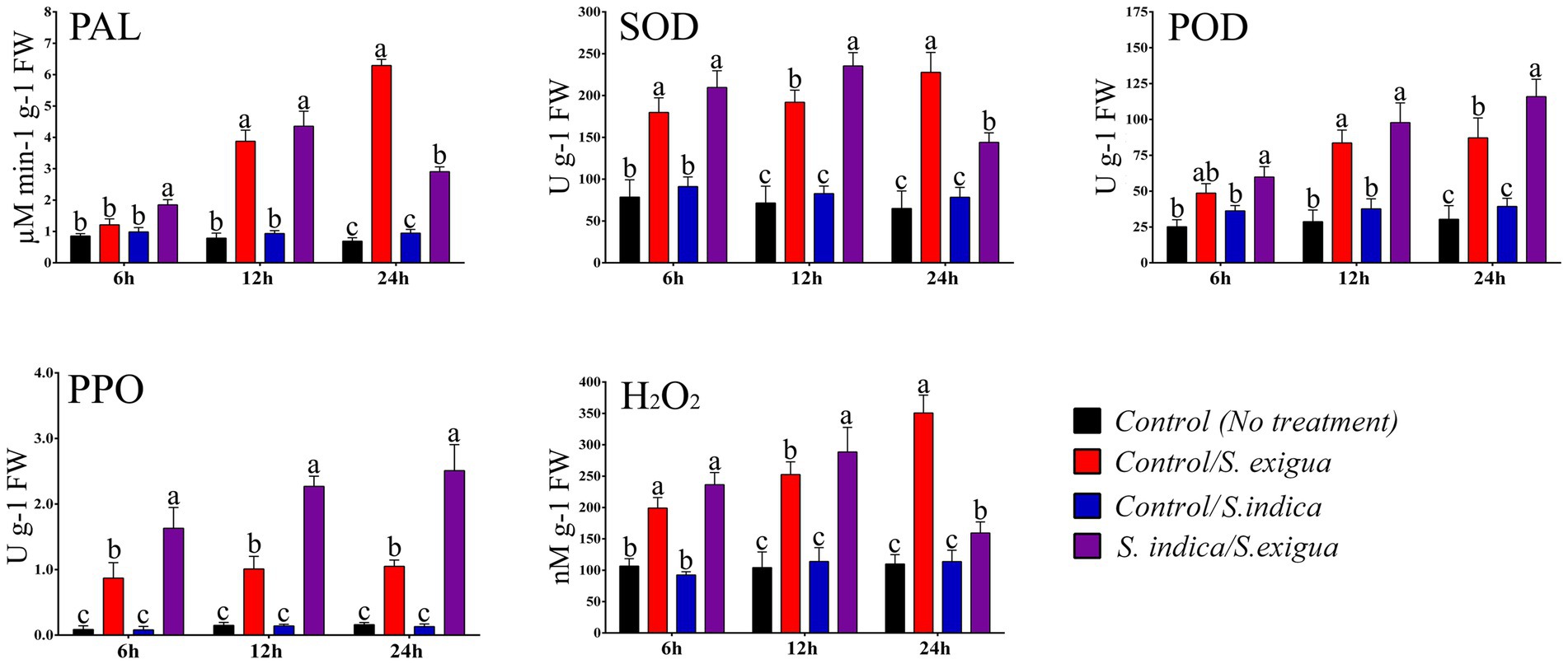
Figure 2. The dynamics of different stress-responsive enzyme activities and H2O2 levels in different onion plant groups. Data are represented as mean ± SE and the alphabets indicate the statistical significance computed at p < 0.05.
Serendipita indica primed temporal modulation of activities of stress-responsive enzymes
We have assessed the enzymatic modulation of selected stress-responsive enzymes at 6, 12, and 24 hpi in uninfested and infested plants. As observed in the plots (Figure 2) S. indica-primed S. exigua-infested plants showed an enhanced temporal modulation of the enzymes. In the case of SOD, the enzymatic activity can be very well correlated with the accumulation of H2O2. In the early stage of S. exigua infestation comparatively, there was no significant change in the level of SOD and POD in control plants and S. indica colonized plants. Compared to the uninfested plants, S. exigua infestation significantly triggered the enzymatic response in both primed and unprimed plants at all-time points. At 12 hpi S. indica-primed plants showed 18.41% more expression of SOD upon S. exigua infestation compared to control-infested plants. However, the level of SOD was significantly decreased by 17.69% at 24 hpi in S. indica primed plants compared to control plants upon S. exigua infestation. The enzymatic expression of POD was insignificant at 12 hpi however, the expression significantly enhanced by 24.77% at 24 hpi in S. indica-primed plants compared to control plants upon S. exigua infestation.
The other stress-responsive enzymes like PAL and PPO showed 31.53 and 46.62% significantly enhanced expression at 6 hpi in S. indica-primed plants compared to control plants upon S. exigua infestation. Moreover, at 12 hpi PAL did not show any significant difference however, PPO showed 55.50% enhanced expression in S. indica-primed plants compared to control plants upon S. exigua infestation. Furthermore, the level of PAL has significantly decreased at 24 hpi by 53.8% while, PPO expression was significantly enhanced by 58.16% in S. indica primed plants compared to control plants upon S. exigua infestation. We have noticed that level of PAL was significant in an early phase of infestation while PPO was significantly higher at all studied time points. In uninfested plants, we did not observe any significant changes throughout the study indicating that together with antioxidants enzymes PAL and PPO are actively triggered in response to S. exigua infestation. It is worth mentioning that the major significant changes were observed at 12 and 24 hpi after S. exigua infestation.
PCA and HCA analysis
The H2O2, SOD, POD, PAL, and PPO activities in the control (without S. indica treatment) and test (with S. indica treatment) were compared by PCA under S. exigua infestation. The data from three time points, including 6, 12, and 24 hpi were collected and used in the PCA. A score plot was plotted between PC1 and PC2, the principal components accounting for 99.8% of the total variance (Figure 3). On the score plot, the S. indica-treated samples and the non-treated samples were well-separated.
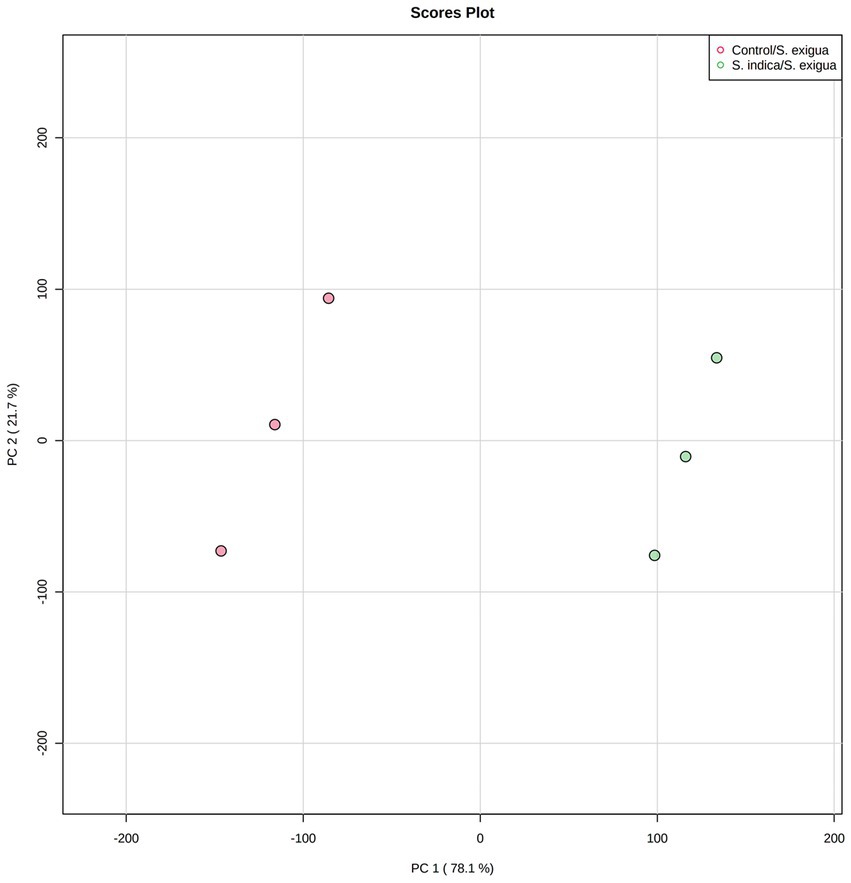
Figure 3. PCA analysis of the onion responses variable to S. indica treatment and S. exigua infestation.
The same data set was used to generate an HCA heatmap (Figure 4). The results revealed that the S. indica-treated samples clustered together (Pi_1, Pi_2, and Pi_3), whereas the control samples formed a separate cluster (Ctrl_1, Ctrl_2, and Ctrl_3). The HCA result matched with the PCA results, suggesting the obvious difference in the onion biochemical responses to the S. indica treatments under the insect infestation. Out of the fifteen divergent features, 12 clustered together, showing overall higher values in the treated samples and lower in the control. Three variables (PAL_24 hpi, H2O2_24 hpi, and SOD_24 hpi) exhibited overall higher values in the control than the treated plants and formed a separate cluster. Collectively, these data suggested that in the S. indica-treated plants, the overall biochemical activities of H2O2, POD, SOD, PPO, and PAL were upregulated as compared to the untreated control under S. exigua infestation. However, at 24 hpi, the activity of PAL, H2O2, and SOD was found to be higher in the control than in the treated.
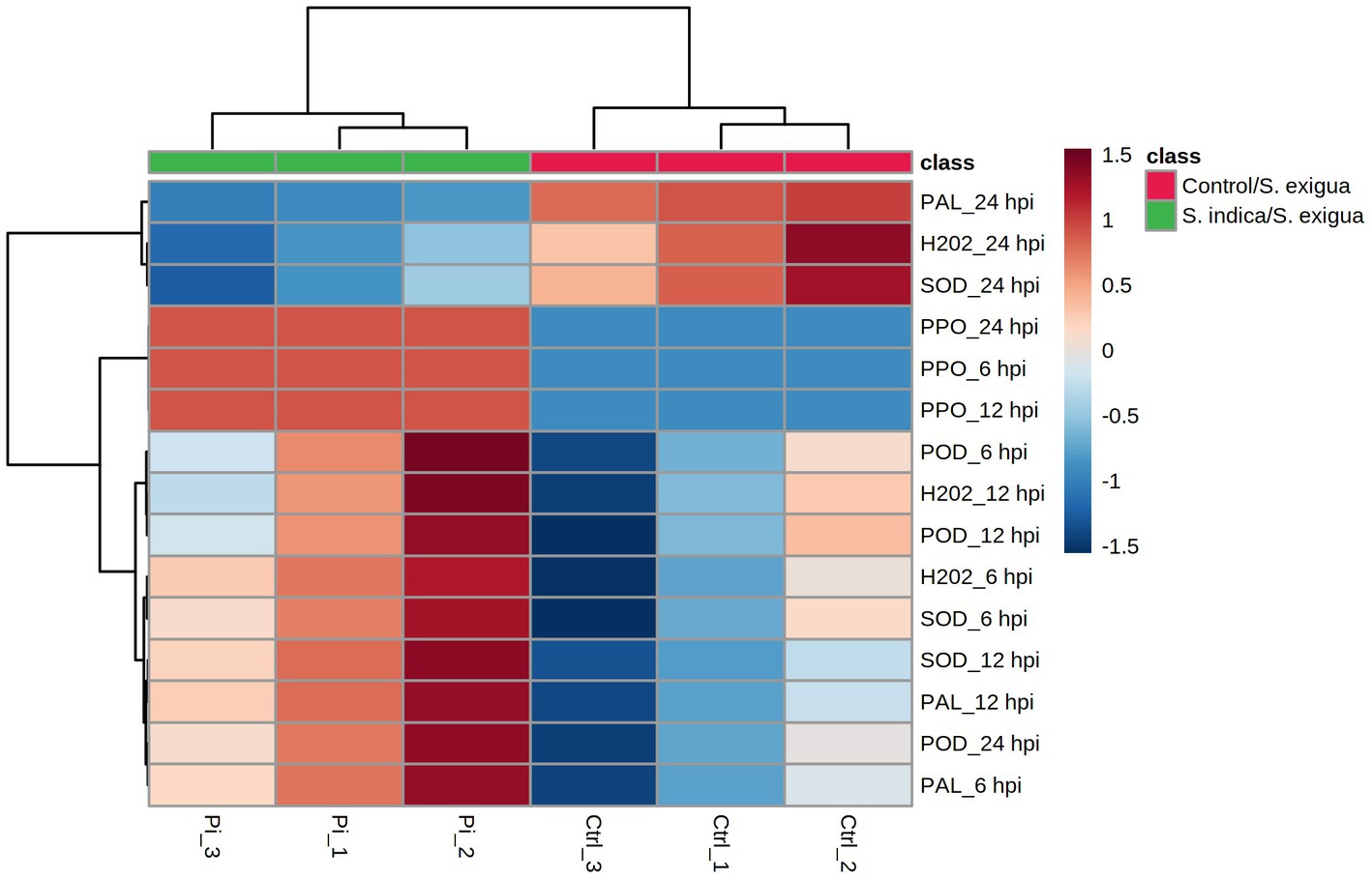
Figure 4. HCA analysis of the onion responses variable to S. indica treatment and S. exigua infestation represented as a heatmap. Clustering was done by using the Euclidean distance and Ward’s clustering algorithm.
Defense gene expression analysis
To provide further molecular insight for S. indica primed induced resistance against S. exigua we have performed the qRT-PCR analysis of seven genes at 12 and 24 hpi (Figure 5). The relevant expression of AcWRKY11 and AcWRKY70 were observed to be interconnected to each other. At 12 hpi in control S. exigua infested plants, the expression of AcWRKY11 was insignificant (−1.64 fold) which further drastically down-regulated at 24 hpi (−4.28 fold) as compared to S. indica primed S. exigua infested plants. While the expression of AcWRKY70 was opposite of AcWRKY11 which was highly up-regulated from 12 hpi (3.61fold) to 24 hpi (5.29 fold) in control S. exigua infested plants as compared to S. indica primed plant, where it got downregulated at 12 hpi (0.15 fold) after S. exigua infestation. This trend indicated that the expression of AcWRKY11 and AcWRKY70 was modulated more likely by the treatment of S. indica in response to S. exigua infestation. Furthermore, the expression of AcbHLH at 12 hpi (2.57 fold) was significantly up-regulated in the primed plant after S. exigua infestation while at 24 hpi there was no significant difference in both the control and primed plant in response to S. exigua infestation. Another pair of genes AcLOX1 and AcGST which are the marker genes for JA- and SA- mediated pathways respectively, showed an opposite trend where in primed plants LOX1was upregulated at 12 hpi (4.26 fold) and 24 hpi (6.43 fold) while AcGSTwas significant in the unprimed plant at 12 hpi (3.97 fold) and 24 hpi (7.13 fold) in response to S. exigua infestation. We have noticed that both AcWRKY11 and AcLOX1 were down-regulated in the late stage of infestation in unprimed plants. AcPAL showed a significant change in the early stage of infestation (2.07 fold) in control S. exigua infested plants, however, there was no significant change in the late stage of infestation compared to S. indica primed plants (3.04 fold). A relevant increase in the relative expression level of the onion gene coding for Trypsin inhibitor (AcTI) is significantly up at the early stage of infestation in control S. exigua infested plants (2.84 fold) while, it was strongly up-regulated in the late stage of infestation in S. indica primed plants (3.75 fold). In this temporal expression of the gene in the absence of S. exigua infestation, S. indica primed plants showed a relative expression in the range of 0.29 to 1.42 folds indicating the defense response might have been triggered by the insect, while the S. indica priming attuned the intensity of response in defense pathway-specific manner.
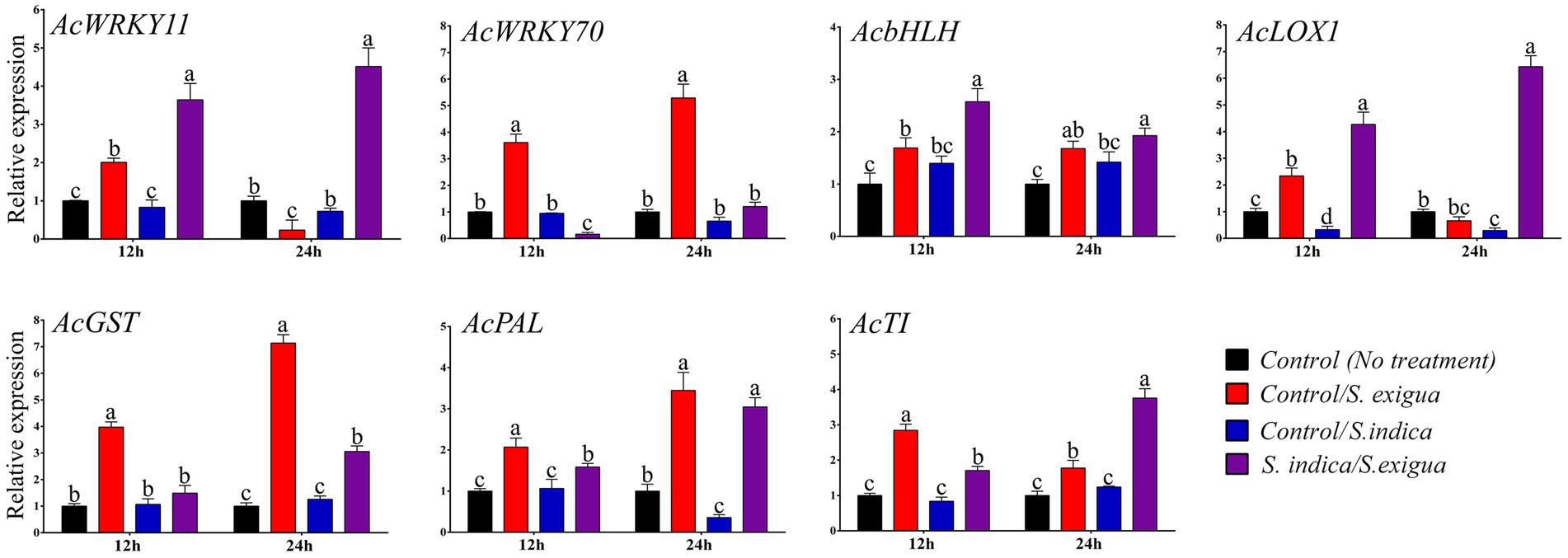
Figure 5. Expression analysis of the selected onion genes in different onion plant groups. Data are represented as mean ± SE and the alphabets indicate the statistical significance computed at p < 0.05.
Discussion
Growth promotion and induction of tolerance to biotic and abiotic stress after S. indica symbiosis have been evidenced from several studies in several plant species including onion (Fakhro et al., 2010; Tsai et al., 2020; Roylawar et al., 2021). Such mutualism or symbiosis of plants and microbial communities is an important factor in a natural ecosystem as well as in agriculture. In the present study, we reported that leaf damage by caterpillar S. exigua and its weight gain was significantly reduced when fed upon S. indica-colonized onion plants. Earlier studies also investigated the colonization of S. indica and its impact on insect herbivory. Li et al. (2021) demonstrated that symbiosis of sweet potato and S. indica enhanced growth and resistance against S. litura which was evidenced by reduced weight gain and less feeding injury. Similarly, S. indica colonization of rice roots resulted in increased resistance against rice leaffolder and rice water weevil which showed that S. indica enhanced the resistance of plants to leaf and root herbivory by insects (Cosme et al., 2016; Chen et al., 2022). This enhanced resistance is by modulation of antioxidant and defense enzymes/genes in the host as a result of mutual interaction between S. indica and the host (Gill et al., 2016).
Biotic and abiotic stresses led to the production of reactive oxygen species in plants. The H2O2 accumulation was higher at the initial stages and reduced in the late stage in S. indica colonized plants in response to herbivory by S. exigua than the non-colonized counterpart. It has been shown that the treatment with JA has a positive role in the accumulation of H2O2 in wounded tomato seedlings (Orozco-Cárdenas et al., 2001). Furthermore, H2O2 acts as a key signal molecule in oxidative stress signaling and has an adverse effect on insects (Zhu-Salzman et al., 2008; Petrov and Van Breusegem, 2012). This observation suggests that colonization of roots by S. indica assisted the host to limit the oxidative stress caused due to insect herbivory. The plant system must scavenge these excess ROS to protect cells from oxidative damage. An earlier study reported a significantly lower level of malondialdehyde (MDA) content in S. indica-colonized plants than in non-colonized rice plants after leaffolder infestation (Chen et al., 2022). Similarly, S. indica colonization also showed such modulation of ROS after infection by pathogenic fungi in crops like wheat and chickpea (Serfling et al., 2007; Narayan et al., 2017). AM fungi, R. irregularis increased resistance against root-knot nematode with a significant reduction in ROS content in tomatoes (Sharma and Sharma, 2017). There is a close relationship between the perception of H2O2 and the stimulation of defense response to insect herbivory in plants which interferes with feeding, digestion, and nutrient absorption by insects (Maffei et al., 2006; Wu and Baldwin, 2010).
The antioxidant defense is a vital element of the basic metabolism and enables the plant to deal immediately with rapid environmental stress. Anti-oxidative and defense enzymes were induced in onion plants in response to feeding injury by S. exigua to protect cells from oxidative damage as well as insect herbivory. Colonization of root by S. indica was reported to elevate the levels of these enzymes in several plants in response to biotic or abiotic stress (Sun et al., 2010; Lin et al., 2019; Roylawar et al., 2021). In this study, we demonstrated the elevation of antioxidative and defense enzyme activities in S. indica-colonized onion plants. At the early stage of infection higher activities of H2O2-metabolizing enzymes might have helped in imparting better protection against chewing injury by S. exigua in onion plants colonized by S. indica. From the obtained results, we assume that S. indica-colonized plants were characteristically able to early percept insect herbivory signals leading to triggering of early defense response in the plant. SOD plays a primary role in scavenging ROS by the dismutation of superoxide radicals into O2 and H2O2. Chen et al. (2022) reported the elevation of SOD activity in S. indica colonized rice and increased resistance to leaffolder. Similarly, POD enzymes are ubiquitous and are one of the important components in the ROS scavenging system. In contrast, Li et al. (2021) demonstrated that S. indica could not increase the SOD in sweet potatoes. It suggests that the response of S. indica to anti-oxidative enzymes is host-dependent (Lin et al., 2019; Tsai et al., 2020). Similarly, higher SOD activities were reported in S. indica-primed other crops in response to pathogens (Kumar et al., 2009; Roylawar et al., 2021). Its key role in immediate response against insect pests in various plants was reported such as turfgrasses against western chinch bug (Gulsen et al., 2010), tomato, cowpea, and cotton against S. litura (Singh et al., 2013), rice against brown planthopper (Wei et al., 2009), groundnut against S. litura (War et al., 2012). Further, PAL is an important enzyme in the phenylpropanoid pathway for SA biosynthesis and is known to involve in plant defense against disease and insects. Here, we observed that in the early stage of S. exigua feeding PAL activity was higher in S. indica-treated plants and then declined in an advanced stage. It indicates that S. indica modulated this enzyme response in such a way that plant exhibits immediate higher defense against stress. PAL activity was reported to increase upon insect attack in poplar, maize, Brachypodium, cotton, etc. (Hu et al., 2009; Cass et al., 2015; Sha et al., 2015; Lv et al., 2017). Moreover, different biostatistical methods, including PCA and HCA have been used in several studies to screen and analyze the difference between experimental sample sets (test) and controls (Zhang et al., 2020; Sarfraz et al., 2021). In this study, the PCA and HCA results corroborated each other. There was an obvious difference observed in the onion biochemical responses to the S. indica treatments under the insect infestation. In the S. indica-treated plants, the overall activities of H2O2, POD, SOD, PPO, and PAL were upregulated as compared to the untreated control under S. exigua infestation. In addition, there was an insignificant change in the activities of all the enzymes in control and S. indica control treatment while higher activity was in the early stage of feeding in S. indica/S. exigua treatment suggesting effective priming of defense response in onion against herbivory by S. indica.
In the present study, the expression of genes involved in SA and JA-signalling, like PAL and LOX was studied. PAL is an important gene in the phenylpropanoid pathway for SA biosynthesis, a crucial phytohormone involved in plant defense (Rivas-San Vicente and Plasencia, 2011). AcPAL expression was seen to be induced upon S. exigua herbivory in onion. Recently, Dixit et al. (2020) reported the higher expression of PAL gene is associated with the resistant cotton varieties under H. armigera and S. litura infestation. Similarly, JA is an important phytohormone involved in plant defense signaling against insects. LOX gene, a well-studied JA-pathway gene encoding lipoxygenase enzyme, is involved in the catalyzation of the hydroperoxidation of linolenic acid, an important step in JA synthesis (Bell et al., 1995). Here, AcLOX1 expression was found to be highly induced in S. indica/ S. exigua treatment at both early and late stages of herbivory in onion. Earlier reports also mentioned that root colonization by S. indica induces the expression of genes for JA biosynthesis (Lin et al., 2019; Li et al., 2021; Roylawar et al., 2021). It ultimately enhanced resistance to both wound/herbivory and necrotrophic pathogens for diseases. LOX1 interacts with the other LOX genes for the synthesis of oxylipins and regulates the resistance to insect herbivory (Zhou et al., 2014). It was also seen in Maize, that 9-lox are more prominently expressed upon herbivory by larvae of S. exigua than 13-lox (Woldemariam et al., 2018).
In the present study, we have studied the expression of two WRKY and one basic-helix–loop–helix (bHLH) transcription factor (TF). WRKY TFs regulate many metabolic processes and are well studied for their role in plants’ defense response against abiotic and biotic stress. Expression of WRKY11 was found to be induced in S. indica/S. exigua treatment in onion. Earlier studies reported that WRKY11 is involved in bacterial pathogen defense, drought, and salt stress in rice and soybean (Lee et al., 2018; Wang et al., 2018). It was proposed that WRKY11 induces JA-responsive genes by repressing SA-responsive genes together with WRKY70 (Journot-Catalino et al., 2006). The higher level of WRKY11 in our studies may have positively regulated the expression of LOX1. This is also evident from the suppression of WRKY70 in primed plants. We have observed significantly higher expression of WRKY70 in control/S. exigua treatment over others. It suggests that the colonization of S. indica fine-tuned the defense response more efficiently. WRKY70 is an inducer of SA-related genes and a suppressor of JA-related genes (Li et al., 2004). Higher jasmonate level increases plant endurance against different insect herbivore in natural habitat (Machado et al., 2016). WRKY TFs are also reported to impart resistance to plants against insects by regulating the mitogen-activated protein kinase (MAPK) pathways (Yao et al., 2020). Our previous study on onion and S. indica interaction also hinted at such modulation of WRKYs to enhance the resistance of the host plant (Roylawar et al., 2021). Similarly, bHLH TFs are involved in JA-mediated signaling in plants (Goossens et al., 2017). The significantly higher-level expression of the AcbHLH gene in S. indica/S. exigua treatment also assures that the JA-mediated defense pathway is actively getting regulated in S. indica-primed plants against insect herbivory. An earlier study in sweet potatoes also reported similar findings where S. indica colonization induced the expression of IbbHLH3 upon wounding by S. litura (Li et al., 2021). In tomatoes, the jasmonate-induced bHLH gene homolog was found to be associated with terpene biosynthesis ultimately increasing resistance to the H. armigera (Cao et al., 2022).
Protease inhibitors affect the availability of nutrients and weaken the development of insect pests. Among them, trypsin inhibitors are well-known and studied protease inhibitors involved in pest resistance (Fürstenberg-Hägg et al., 2013). Here, in onion, we observed induced expression of AcTI upon infestation by S. exigua. This expression was more in control/S. exigua treatment at an earlier stage and later stages expression were higher in S. indica/ S. exigua treatment. It was earlier reported that in S. indica colonized sweet potato, the activity of trypsin inhibitor was 60% higher than in non-colonized plants upon S. litura herbivory (Li et al., 2021). Trypsin proteinase inhibitors’ role in defense response against S. exigua was also reported in Nicotiana attenuate (Steppuhn and Baldwin, 2007).
Conclusion
Onion plants colonized with S. indica significantly reduced the chewing injury by larvae of S. exigua and indeed retarded the growth of larvae. The elevated enzyme activities and defense genes expression at an early stage of chewing damage suggest efficient priming of the defense response in onion by S. indica. The primed plants have shown early perception of insect feeding by higher activity of H2O2. The higher level of activity of PPO and expression of WRKY11, LOX, and bHLH gene in all the time points indicated the prominent role of JA signaling in defense against S. exigua. A more targeted approach is needed to dissect the mechanism of this induced resistance where the role of ROS and WRKY11 will be studied in detail. Our results indicate the potential use of S. indica as a biocontrol agent against chewing insects in onion.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
SG and PR: conceptualization and supervision. PR, KK, and PS: methodology. PR and SN: formal analysis, data curation, and visualization. PR, KK, SJ, and PS: investigation. SG and SJ: resources. PR, SN, and KK: writing – original draft preparation. PR, KK, SJ, SN, and SG: writing – review and editing. SG: project administration and funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by the Indian Council of Agricultural Research, New Delhi, India (HORTDOGRSIL201800400084). The Article Processing Charge (APC) was funded by ICAR-DOGR, Rajgurunagar (Pune, India).
Acknowledgments
The authors gratefully acknowledge J. Vadassery, National Institute for Plant Genome Research (NIPGR), New Delhi, India for providing Serendipita indica culture.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1190942/full#supplementary-material
References
Atia, M., Abdeldaym, E. A., Abdelsattar, M., Ibrahim, D., Saleh, I., Elwahab, M. A., et al. (2020). Piriformospora indica promotes cucumber tolerance against root-knot nematode by modulating photosynthesis and innate responsive genes. Saudi J. Biol. Sci. 27, 279–287. doi: 10.1016/j.sjbs.2019.09.007
Beauchamp, C., and Fridovich, I. (1971). Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Anal. Biochem. 44, 276–287. doi: 10.1016/0003-2697(71)90370-8
Bell, E., Creelman, R. A., and Mullet, J. E. (1995). (1995) a chloroplast lipoxygenase is required for wound-induced jasmonic acid accumulation in Arabidopsis. Proc. Natl. Acad. Sci. 92, 8675–8679. doi: 10.1073/pnas.92.19.8675
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Cao, Y., Liu, L., Ma, K., Wang, W., Lv, H., Gao, M., et al. (2022). The Jasmonate-induced bHLH gene SlJIG functions in terpene biosynthesis and resistance to insects and fungus. J. Integr. Plant Biol. 64, 1102–1115. doi: 10.1111/jipb.13248
Cass, C. L., Peraldi, A., Dowd, P. F., Mottiar, Y., Santoro, N., Karlen, S. D., et al. (2015). Effects of phenylalanine ammonia lyase (PAL) knockdown on cell wall composition, biomass digestibility, and biotic and abiotic stress responses in Brachypodium. J. Exp. Bot. 66, 4317–4335. doi: 10.1093/jxb/erv269
Cheema, S., and Sommerhalter, M. (2015). Characterization of polyphenol oxidase activity in Ataulfo mango. Food Chem. 171, 382–387. doi: 10.1016/j.foodchem.2014.09.011
Chen, C. Y., Huang, P. H., Yeh, K. W., and Wang, S. J. (2022). Colonization of Piriformospora indica enhances insect herbivore resistance of rice plants through jasmonic acid-and antioxidant-mediated defense mechanisms. J. Plant Interact. 17, 9–18. doi: 10.1080/17429145.2021.2008031
Cokola, M. C., Ndjadi, S. S., Bisimwa, E. B., Ahoton, L. E., and Francis, F. (2021). First report of Spodoptera frugiperda (lepidoptera: Noctuidae) on onion (Allium cepa L.) in south Kivu, eastern DR Congo. Rev. Bras. Entomol. 65:e20200083. doi: 10.1590/1806-9665-RBENT-2020-0083
Cosme, M., Lu, J., Erb, M., Stout, M. J., Franken, P., and Wurst, S. (2016). A fungal endophyte helps plants to tolerate root herbivory through changes in gibberellin and jasmonate signaling. New Phytol. 211, 1065–1076. doi: 10.1111/nph.13957
Dixit, G., Srivastava, A., Rai, K. M., Dubey, R. S., Srivastava, R., and Verma, P. C. (2020). Distinct defensive activity of phenolics and phenylpropanoid pathway genes in different cotton varieties toward chewing pests. Plant Signal. Behav. 15:1747689. doi: 10.1080/15592324.2020.1747689
Fakhro, A., Andrade-Linares, D. R., von Bargen, S., Bandte, M., Büttner, C., Grosch, R., et al. (2010). Impact of Piriformospora indica on tomato growth and on interaction with fungal and viral pathogens. Mycorrhiza 20, 191–200. doi: 10.1007/s00572-009-0279-5
Fernandes, F. L., Diniz, J. F. S., Alves, F. M., and Silva, L. O. D. (2012). Injury and spatial distribution of Spodoptera frugiperda (J. E. Smith) (lepidoptera: noctuidae) in onion Allium cepa (Alliaceae) in alto Paranaíba, Minas Gerais, Brazil. Entomol. News 122, 257–260. doi: 10.3157/021.122.0307
Fürstenberg-Hägg, J., Zagrobelny, M., and Bak, S. (2013). Plant defense against insect herbivores. Int. J. Mol. Sci. 14, 10242–10297. doi: 10.3390/ijms140510242
Ghodke, P., Khandagale, K., Thangasamy, A., Kulkarni, A., Narwade, N., Shirsat, D., et al. (2020). Comparative transcriptome analyses in contrasting onion (Allium cepa L.) genotypes for drought stress. PLoS One 15:e0237457. doi: 10.1371/journal.pone.0237457
Gill, S. S., Gill, R., Trivedi, D. K., Anjum, N. A., Sharma, K. K., Ansari, M. W., et al. (2016). Piriformospora indica: potential and significance in plant stress tolerance. Front. Microbiol. 7:332. doi: 10.3389/fmicb.2016.00332
Goossens, J., Mertens, J., and Goossens, A. (2017). Role and functioning of bHLH transcription factors in jasmonate signalling. J. Exp. Bot. 68, 1333–1347. doi: 10.1093/jxb/erw440
Gulsen, O., Eickhoff, T., Heng-Moss, T., Shearman, R., Baxendale, F., Sarath, G., et al. (2010). Characterization of peroxidase changes in resistant and susceptible warm season turfgrasses challenged by Blissus occiduus. Arthropod Plant Interact. 4, 45–55. doi: 10.1007/s11829-010-9086-3
Hu, Z. H., Zhang, W., Shen, Y. B., Fu, H. J., Su, X. H., and Zhang, Z. Y. (2009). Activities of lipoxygenase and phenylalanine ammonia lyase in poplar leaves induced by insect herbivory and volatiles. J. Forest. Res. 20, 372–376. doi: 10.1007/s11676-009-0063-6
Ilmi, Z., Muhibbudin, M., Abdullah, A., Ali, S. M., and Artika, W. (2022). Distribution patterns and larval prevalence of Spodoptera exigua Hubner PEST on onion (Allium cepa) in Pidie District Lambideng. J. Pharm. Biol. Sci. 17, 18–21. doi: 10.9790/3008-1701011821
Journot-Catalino, N., Somssich, I. E., Roby, D., and Kroj, T. (2006). The transcription factors WRKY11 and WRKY17 act as negative regulators of basal resistance in Arabidopsis thaliana. Plant Cell 18, 3289–3302. doi: 10.1105/tpc.106.044149
Kavitha, K., and Reddy, K. D. (2012). Screening techniques for different insect pests in crop plants. Int. J. Bioresour. Stress Manag. 3, 188–195.
Kawai, A., Kashio, T., and Kawamoto, K. (1993). Distribution pattern and analysis of the damage caused by Spodoptera exigua (Hubner) to welsh onion [Allium fistulosum]. Bulletin of the National Research Institute of vegetables. Ornamental Plants and Tea. Series A. (Japan). 6, 137–148.
Khandagale, K., and Gawande, S. (2019). Genetics of bulb colour variation and flavonoids in onion. J. Hort. Sci. Biotech. 94, 522–532. doi: 10.1080/14620316.2018.1543558
Kumar, M., Yadav, V., Tuteja, N., and Johri, A. K. (2009). Antioxidant enzyme activities in maize plants colonized with Piriformospora indica. Microbiology 155, 780–790. doi: 10.1099/mic.0.019869-0
Lee, H., Cha, J., Choi, C., Choi, N., Ji, H. S., Park, S. R., et al. (2018). Rice WRKY11 plays a role in pathogen defense and drought tolerance. Rice 11, 1–12. doi: 10.1186/s12284-018-0199-0
Li, J., Brader, G., and Palva, E. T. (2004). The WRKY70 transcription factor: a node of convergence for Jasmonate-mediated and salicylate- mediated signals in plant Defense. Plant Cell 16, 319–331. doi: 10.1105/tpc.016980
Li, Q., Kuo, Y. W., Lin, K. H., Huang, W., Deng, C., Yeh, K. W., et al. (2021). Piriformospora indica colonization increases the growth, development, and herbivory resistance of sweet potato (Ipomoea batatas L.). Plant Cell Rep. 40, 339–350. doi: 10.1007/s00299-020-02636-7
Lin, H. F., Xiong, J., Zhou, H. M., Chen, C. M., Lin, F. Z., Xu, X. M., et al. (2019). Growth promotion and disease resistance induced in anthurium colonized by the beneficial root endophyte Piriformospora indica. BMC Plant Biol. 19, 1–10. doi: 10.1186/s12870-019-1649-6
Lv, M., Kong, H., Liu, H., Lu, Y., Zhang, C., Liu, J., et al. (2017). Induction of phenylalanine ammonia-lyase (PAL) in insect damaged and neighboring undamaged cotton and maize seedlings. Int. J. Pest Manag. 63, 166–171. doi: 10.1080/09670874.2016.1255804
Machado, R. A., McClure, M., Herve, M. R., Baldwin, I. T., and Erb, M. (2016). Benefits of jasmonate-dependent defenses against vertebrate herbivores in nature. Elife 5:e13720. doi: 10.7554/eLife.13720
Maffei, M. E., Mithofer, A., Arimura, G. I., Uchtenhagen, H., Bossi, S., BerteaCM, C. L. S., et al. (2006). Effects of feeding Spodoptera littoralis on Lima beanleaves. III. Membrane depolarization and involvement of hydrogen peroxide. Plant Physiol. 140, 1022–1035. doi: 10.1104/pp.105.071993
Nanda, S., Chand, S. K., Mandal, P., Tripathy, P., and Joshi, R. K. (2016). Identification of novel source of resistance and differential response of allium genotypes to purple blotch pathogen, Alternaria porri (Ellis) Ciferri. Plant Pathol. J. 32, 519–527. doi: 10.5423/PPJ.OA.02.2016.0034
Narayan, O. P., Verma, N., Singh, A. K., Oelmüller, R., Kumar, M., Prasad, D., et al. (2017). Antioxidant enzymes in chickpea colonised by Piriformospora indica participate in defense against the pathogen Botrytis cinerea. Sci. Rep. 7:13553. doi: 10.1038/s41598-017-12944-w
Orozco-Cárdenas, M. L., Narvaez-Vasquez, J., and Ryan, C. A. (2001). Hydrogen peroxide acts as a second messenger for the induction of defense genes in tomato plants in response to wounding, systemin, and methyl jasmonate. Plant Cell 13, 179–191. doi: 10.1105/tpc.13.1.179
Pang, Z., Chong, J., Zhou, G., de Lima Morais, D. A., Chang, L., Barrette, M., et al. (2021). MetaboAnalyst 5.0: narrowing the gap between raw spectra and functional insights. Nuc. Acids Res. 49, W388–W396. doi: 10.1093/nar/gkab382
Petrov, V. D., and Van Breusegem, F. (2012). Hydrogen peroxide – a central hub for information flow in plant cells. AoB Plants 2012:pls014. doi: 10.1093/aobpla/pls014
Rivas-San Vicente, M., and Plasencia, J. (2011). Salicylic acid beyond defence: its role in plant growth and development. J. Exp. Bot. 62, 3321–3338. doi: 10.1093/jxb/err031
Roylawar, P., Khandagale, K., Randive, P., Shinde, B., Murumkar, C., Ade, A., et al. (2021). Piriformospora indica primes onion response against stemphylium leaf blight disease. Pathogens 10:1085. doi: 10.3390/pathogens10091085
Saeed, Q., Ahmad, F., and Saeed, S. (2017). Development and survival of Spodoptera exigua (lepidoptera: Noctuidae) on alternate crops in cotton cropping pattern, with implications to integrated pest management. Environ. Entomol. 46, 595–601. doi: 10.1093/ee/nvx056
Sarfraz, Z., Shah, M. M., Iqbal, M. S., Nazir, M. F., Al-Ashkar, I., Rehmani, M. I. A., et al. (2021). Rendering multivariate statistical models for genetic diversity assessment in A-genome diploid wheat population. Agronomy 11:2339. doi: 10.3390/agronomy11112339
Schmittgen, T. D., and Livak, K. J. (2008). Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Serfling, A., Wirsel, S. G. R., Lind, V., and Deising, H. B. (2007). Performance of the biocontrol fungus Piriformospora indica on wheat under greenhouse and field conditions. Phytopathology 97, 523–531. doi: 10.1094/PHYTO-97-4-0523
Sha, P. J., Fan, Y. J., Wang, Z. C., and Shi, X. Y. (2015). Response dynamics of three defense related enzymes in cotton leaves to the interactive stress of Helicoverpa armigera (Hübner) herbivory and omethoate application. J Integ. Agric. 14, 355–364. doi: 10.1016/S2095-3119(14)60793-0
Sharma, I. P., and Sharma, A. K. (2017). Co-inoculation of tomato with an arbuscular mycorrhizal fungus improves plant immunity and reduces root-knot nematode infection. Rhizosphere 4, 25–28. doi: 10.1016/j.rhisph.2017.05.008
Sherman, T. D., Vaughn, K. C., and Duke, S. O. (1991). A limited survey of the phylogenetic distribution of polyphenol oxidase. Phytochemistry 30, 2499–2506. doi: 10.1016/0031-9422(91)85089-I
Singh, H., Dixit, S., Verma, P. C., and Singh, P. K. (2013). Differential peroxidase activities in three different crops upon insect feeding. Plant Signal. Behav. 8:e25615. doi: 10.4161/psb.25615
Soumia, P. S., Karuppaiah, V., Mahajan, V., and Singh, M. (2020). Beet armyworm Spodoptera exigua: emerging threat to onion production. Natl. Acad. Sci. Lett. 43, 473–476. doi: 10.1007/s40009-020-00892-5
Steppuhn, A., and Baldwin, I. T. (2007). Resistance management in a native plant: nicotine prevents herbivores from compensating for plant protease inhibitors. Ecol. Lett. 10, 499–511. doi: 10.1111/j.1461-0248.2007.01045.x
Sun, C., Johnson, J. M., Cai, D., Sherameti, I., Oelmüller, R., and Lou, B. (2010). Piriformospora indica confers drought tolerance in Chinese cabbage leaves by stimulating antioxidant enzymes, the expression of drought-related genes and the plastid-localized CAS protein. J. Plant Physiol. 167, 1009–1017. doi: 10.1016/j.jplph.2010.02.013
Sun, C., Shao, Y., Vahabi, K., Lu, J., Bhattacharya, S., Dong, S., et al. (2014). The beneficial fungus Piriformospora indica protects Arabidopsis from verticillium dahlia infection by downregulation plant defense responses. BMC Plant Biol. 14:268. doi: 10.1186/s12870-014-0268-5
Tsai, H. J., Shao, K. H., Chan, M. T., Cheng, C. P., Yeh, K. W., Oelmüller, R., et al. (2020). Piriformospora indica symbiosis improves water stress toleranceof rice through regulating stomata behavior and ROS scavenging systems. Plant Signal. Behav. 15:1722447. doi: 10.1080/15592324.2020.1722447
Ueno, T. (2015). Beet armyworm Spodoptera exigua (lepidoptera: Noctuidae): a major pest of welsh onion in Vietnam. J. Agric. Environ. Sci. 4, 181–185. doi: 10.15640/jaes.v4n2a21
Wang, Y., Jiang, L., Chen, J., Tao, L., An, Y., Cai, H., et al. (2018). Overexpression of the alfalfa WRKY11 gene enhances salt tolerance in soybean. PLoS One 13:e0192382. doi: 10.1371/journal.pone.0192382
War, A. R., Paulraj, M. G., War, M. Y., and Ignacimuthu, S. (2012). Herbivore-induced resistance in different groundnut germplasm lines to Asian armyworm, Spodoptera litura (fab.) (lepidoptera: Noctuidae). Acta Physiol. Plant. 34, 343–352. doi: 10.1007/s11738-011-0833-6
Wei, Z., Hu, W., Lin, Q., Cheng, X., Tong, M., Zhu, L., et al. (2009). Understanding rice plant resistance to the brown planthopper (Nilaparvata lugens): a proteomic approach. Proteomics 9, 2798–2808. doi: 10.1002/pmic.200800840
Woldemariam, M. G., Ahern, K., Jander, G., and Tzin, V. (2018). A role for 9-lipoxygenases in maize defense against insect herbivory. Plant Signal. Behav. 13, 4709–4723. doi: 10.1080/15592324.2017.1422462
Wu, J., and Baldwin, I. T. (2010). New insights into plant responses to attack from insect herbivores. Annu. Rev. Genet. 44, 1–24. doi: 10.1146/annurev-genet-102209-163500
Xu, H. W., Lu, Y., Tong, S. Y., and Song, F. B. (2011). Lipid peroxidation, antioxidant enzyme activity and osmotic adjustment changes in husk leaves of maize in black soils region of Northeast China. Afr. J. Agric. Res. 6, 3098–3102. doi: 10.5897/AJAR11.237
Yao, D. M., Zou, C., Shu, Y. N., and Li, S. S. (2020). WRKY transcription factors in Nicotiana tabacum modulate plant immunity against whitefly via interacting with MAPK cascade pathways. Insects 12:16. doi: 10.3390/insects12010016
Zhang, N., Zhou, S., Yang, D., and Fan, Z. (2020). Revealing shared and distinct genes responding to JA and SA Signaling in Arabidopsis by meta-analysis. Front. Plant Sci. 11:908. doi: 10.3389/fpls.2020.00908
Zheng, S., Henken, B., Wietsma, W., Sofiari, E., Jacobsen, E., Krens, F. A., et al. (2000). Development of bio-assays and screening for resistance to beet armyworm (Spodoptera Exigua Hübner) in Allium cepa L. and its wild relatives. Euphytica 114, 77–85. doi: 10.1023/A:1004089424419
Zhou, G., Ren, N., Qi, J., Lu, J., Xiang, C., Ju, H., et al. (2014). The 9-lipoxygenase Osr9-LOX1 interacts with the 13-lipoxygenase-mediated pathway to regulate resistance to chewing and piercing-sucking herbivores in rice. Physiol. Plant. 152, 59–69. doi: 10.1111/ppl.12148
Keywords: onion, Spodoptera exigua, Serendipita indica, insect herbivory, endophyte-mediated resistance
Citation: Roylawar P, Khandagale K, Nanda S, Soumia PS, Jadhav S, Mahajan V and Gawande S (2023) Colonization of Serendipita indica promotes resistance against Spodoptera exigua in onion (Allium cepa L.). Front. Microbiol. 14:1190942. doi: 10.3389/fmicb.2023.1190942
Edited by:
Tarun Belwal, Texas A&M University, United StatesReviewed by:
Saritha M., Central Arid Zone Research Institute (ICAR), IndiaMaruthadurai R., Central Coastal Agricultural Research Institute (ICAR), India
Copyright © 2023 Roylawar, Khandagale, Nanda, Soumia, Jadhav, Mahajan and Gawande. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Suresh Gawande, U3VyZXNoZ2F3YW5kZTc2QGdtYWlsLmNvbQ==
†These authors have contributed equally to this work
 Praveen Roylawar
Praveen Roylawar Kiran Khandagale
Kiran Khandagale Satyabrata Nanda
Satyabrata Nanda Parakkattu Sulochanan Soumia1
Parakkattu Sulochanan Soumia1 Suresh Gawande
Suresh Gawande