- 1College of Veterinary Medicine, Northwest A&F University, Yangling, Shaanxi, China
- 2Lanzhou Institute of Husbandry and Pharmaceutical Sciences of Chinese Academy of Agricultural Science, Lanzhou, Gansu, China
Staphylococci, mainly including Staphylococcus aureus and coagulase-negative staphylococci (CNS), are one of the most common pathogens causing bovine mastitis worldwide. In this study, we investigated the antimicrobial resistance and virulence profiles of staphylococci from clinical bovine mastitis in Ningxia Hui Autonomous Region of China. Antimicrobial resistance was determined by disc diffusion combined with E-test method. Genes of antimicrobial resistance and virulence factors were determined by PCR. A total of 332 staphylococcal isolates were confirmed from 1,519 mastitic milk samples, including 172 S. aureus and 160 CNS isolates. Fifteen CNS species were identified, with S. chromogenes being the most frequent found (49.4%), followed by S. equorum (13.8%). Noticeably, 2 S. agnetis isolates were found among the CNS isolates. To our knowledge, this is the first report documenting the presence of S. agnetis from bovine mastitis in China. The S. aureus and CNS isolates showed high resistance against penicillin, followed by erythromycin and tetracycline. Multidrug resistance was found in 11.6 and 16.3% of the S. aureus and CNS isolates, respectively. Resistance to penicillin was attributed to the presence of blaZ, erythromycin resistance to ermC (alone or combined with ermB) and tetracycline resistance to tetK (alone or combined with tetM). Notably, one S. equorum isolate and one S. saprophyticus isolate were both methicillin-resistant and mecA positive. Additionally, all S. aureus isolates carried the adhesin genes fnbpA, clfA, clfB, and sdrC, and most of them contained cna and sdrE. Conversely, only a few of the CNS isolates carried clfA, cna, and fnbA. Regarding toxin genes, all S. aureus isolates harbored hlb, and most of them were hlg positive. The lukE-lukD, lukM, sec, sed, sei, sen, seo, tst, seg, seh, and sej were also detected with low frequencies. However, no toxin genes were observed in CNS isolates. This study reveals high species diversity of staphylococci from clinical bovine mastitis in Ningxia Hui Autonomous Region of China. The findings for the genetic determinants of antimicrobial resistance and virulence factor provide valuable information for control and prevention of staphylococcal bovine mastitis.
1. Introduction
Bovine mastitis remain the most frequent and costly disease affecting dairy cattle due to its effects on health, welfare, and productivity. Staphylococci, mainly including Staphylococcus aureus and coagulase-negative staphylococci (CNS), are one of the most common etiological agents causing bovine mastitis worldwide. S. aureus is generally considered major mastitis pathogen and mainly induce clinical mastitis, while CNS have traditionally considered minor mastitis-causing pathogen and usually cause subclinical mastitis (Naranjo-Lucena and Slowey, 2023). Currently, however, reports of subclinical and clinical mastitis cases caused by different CNS species have surfaced largely and they have emerged as an important pathogen (Li et al., 2015; De Visscher et al., 2017; Mahato et al., 2017; Ferreira et al., 2022). Among the group of CNS commonly isolated from bovine milk samples, S. chromogenes, S. epidermidis, S. haemolyticus, S. simulans, and S. xylosus have been identified as the CNS species most likely to cause mastitis (Leroy et al., 2015).
Mastitis is the most common reason for antimicrobials use to control or prevent staphylococcal infections in dairy cattle. Unfortunately, the selective pressure from antimicrobial agents significantly contributes to the dissemination of resistant strains, which greatly attenuate the therapeutic effectiveness of antimicrobial therapy (Isaac et al., 2017; Ahmed et al., 2020). Antimicrobial resistance of staphylococci are mainly attributed to various resistant determinants, such as genes blaZ and mecA for β-lactams resistance, tets for tetracyclines resistance, and erms for macrolides resistance. The reduced susceptibility of staphylococci against these commonly used antimicrobials in veterinary medicine might promote their persistence in the dairy herd (Kot et al., 2012; Piessens et al., 2012). Therefore, surveillance of antimicrobial resistance is important to ensure optimal results of antimicrobial use and minimize the risk for development and spread of antimicrobial resistance (Waller et al., 2011).
Staphylococci possess a wide variety of virulence factors, including different cell wall-associated adhesins and toxins, that facilitate the bacteria to avoid the immune system and contribute to increased severity of infections. Although most of these factors are originally identified in S. aureus, they have also been detected in CNS, including the isolates from bovine origin (González-Martín et al., 2020). In the last decades, the virulence factors in S. aureus isolates from bovine mastitis had been frequently reported worldwide. However, despite the emergence of CNS as pathogens, the knowledge regarding their virulence as well antimicrobial resistance in CNS is still poorly understood and is not usually identified at species level, especially the isolates from bovine mastitis in China, which makes it difficult to control infection because a great diversity of species have their own characteristics. Thus, this study was designed to investigate the antimicrobial resistance and virulence profiles of staphylococci isolated from clinical bovine mastitis cases in Ningxia Hui Autonomous Region of China.
2. Materials and methods
2.1. Bacterial isolation and identification
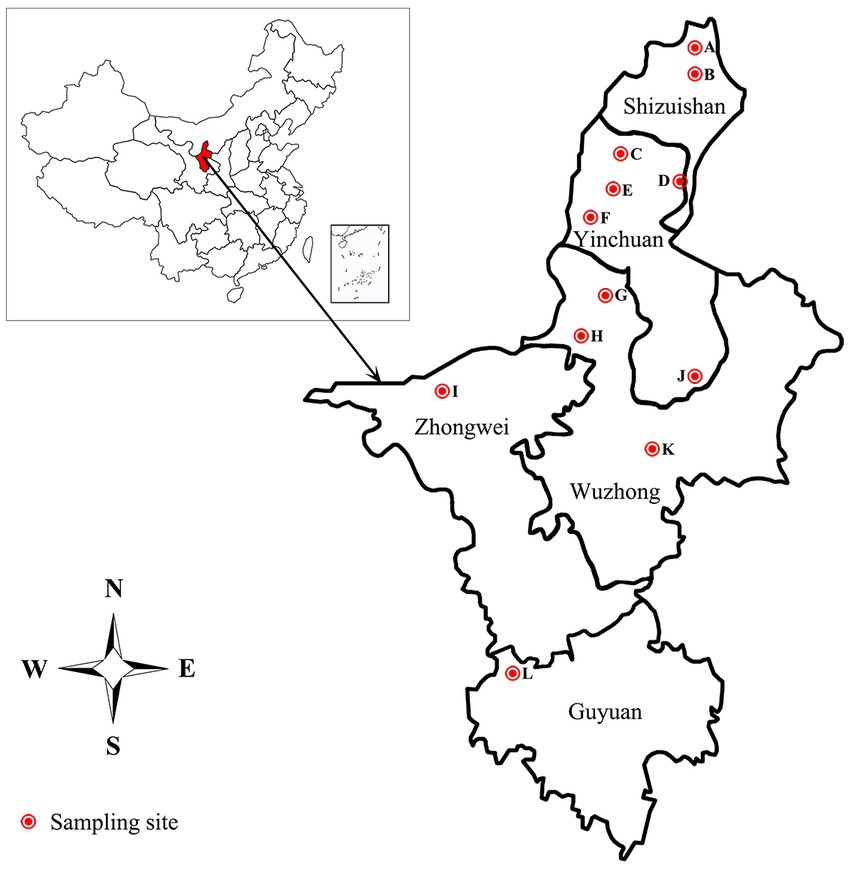
The 332 staphylococcal isolates tested in this study were isolated from 1,519 clinical mastitic milk samples from cows from 12 commercial dairy herds located in Ningxia Hui Autonomous Region in China during July 2021 to Aug 2022 (Figure 1; Supplementary Table S1). Bovine udder showing obvious signs, such as oedema, lumps, increase in temperature, hardening or pain, and milk samples showing any visual evidence of abnormality, such as the presence of clots, flakes or blood, were classified as clinical mastitis (Schmidt et al., 2015; Pérez et al., 2020). Before sampling, teats were disinfected using hydrophilic cotton saturated with 70% ethanol. The first milk squirts were discarded, and 5–10 mL of milk was collected in sterile tubes and transported to the laboratory under refrigeration in cool boxes with ice packs. After transportation to the laboratory, 100 μL of milk was inoculated onto blood agar plates supplemented with 5% defibrinated sheep blood and incubated at 37°C for 48 h. Colonies were initially identified as staphylococcal isolates by appearance (shape, color, and size), Gram staining, catalase and coagulase testing. The suspected isolates were further confirmed by PCR and sequencing as described in our previous study (Yang et al., 2019). Briefly, the genomic DNA was extracted through the Bacterial DNA Kit (Omega Bio-Tek, Norcross, GA) according to the manufacturer’s instructions.1 The 16S rRNA gene was amplified by the 16S rDNA Bacterial Identification PCR Kit (Takara, Shiga, Japan) in accordance with the manufacturer’s recommendation.2 The PCR products were purified and sequenced by Sanger sequencing by Sangon Biotech (Shanghai) Co., Ltd. in China. Nucleotide sequences were analyzed with the program NCBI-BLAST.3 Sequences with 99 to 100% identity to sequences deposited in public domain databases were considered to be positive identification. Confirmed isolates were kept into tryptic soy broth with 20% glycerol at −70°C for molecular testing.
2.2. Antimicrobial susceptibility testing
Antimicrobial susceptibility was determined by disc diffusion method on Mueller-Hinton agar (Oxoid, United Kingdom) according to the protocol of Clinical and Laboratory Standards Institute (CLSI, 2018). The panel of antimicrobial agents (Oxoid) included penicillin (10 U), cefoxitin (30 μg), gentamicin (10 μg), erythromycin (15 μg), tetracycline (30 μg), ciprofloxacin (5 μg), nitrofurantoin (300 μg), trimethoprim-sulfamethoxazole (1.25/23.75 μg), chloramphenicol (30 μg), quinupristin/dalfopristin (15 μg), and linezolid (30 μg). Susceptibility to cefoxitin was used to detect the methicillin-resistance phenotype. The E-test strips (Liofilchem, Roseto, Italy) were used to detect the vancomycin (0.016 to 256 μg/mL) susceptibility of the staphylococcal isolates. S. aureus ATCC 25923 was used as quality control strain. Multidrug-resistant (MDR) isolates were defined as an isolate being resistant to at least 3 antimicrobial agents belonging to different antimicrobial categories (Magiorakos et al., 2012).
2.3. Detection of antimicrobial resistance and virulence genes
The resistance genes for penicillin (blaZ), methicillin (mecA and mecC), tetracycline (tetK and tetM), and erythromycin (ermA, ermB, and ermC) were tested by PCR as described previously using specific primer sets in Supplementary Table S2 (Paterson et al., 2012; Yang et al., 2016). Similarly, the adhesins encoding genes fnbpA (fibronectin bind protein), clfA and clfB (clumping factor), cna (collagen binding protein), sdrC, sdrD and sdrE (serine-aspartic acid repeat proteins), bbp (bone sialoprotein-binding protein), ebpS (elastin-binding protein) and map/eap (major histocompatibility complex class II analogous protein/extracellular adherence protein), as well as toxins encoding genes sea, seb, sec, sed, see, seg, seh, sei, sej, sen, seo, and sem (staphylococcal enterotoxins), tst (toxic shock syndrome toxin-1), eta and etb (exfoliative toxins), lukS/lukF-PV, lukE-lukD, and lukM (leukocidins), hla, hlb, hld, and hlg (hemolysins) and edin (epidermal cell differentiation inhibitor) were evaluated through PCR (Supplementary Table S2; Jarraud et al., 2002; Peacock et al., 2002). The PCR products were analyzed using 1.0% agarose gel electrophoresis.
3. Results
3.1. Bacterial identification
Overall, 172 S. aureus and 160 CNS isolates were identified from the 332 staphylococcal isolates. Among the CNS isolates, a total of 15 species were identified. The predominant species were S. chromogenes (49.4%), followed by S. equorum (13.8%), S. succinus (9.4%), S. xylosus (6.3%), S. simulans (5.0%), S. haemolyticus (4.4%), S. hominis (2.5%), S. saprophyticus (1.9%), S. lugdunensis (1.9%), S. gallinarum (1.9%), S. agnetis (1.3%), S. auricularis (0.6%), S. cohnii (0.6%), S. epidermidis (0.6%), and S. hyicus (0.6%) (Table 1).
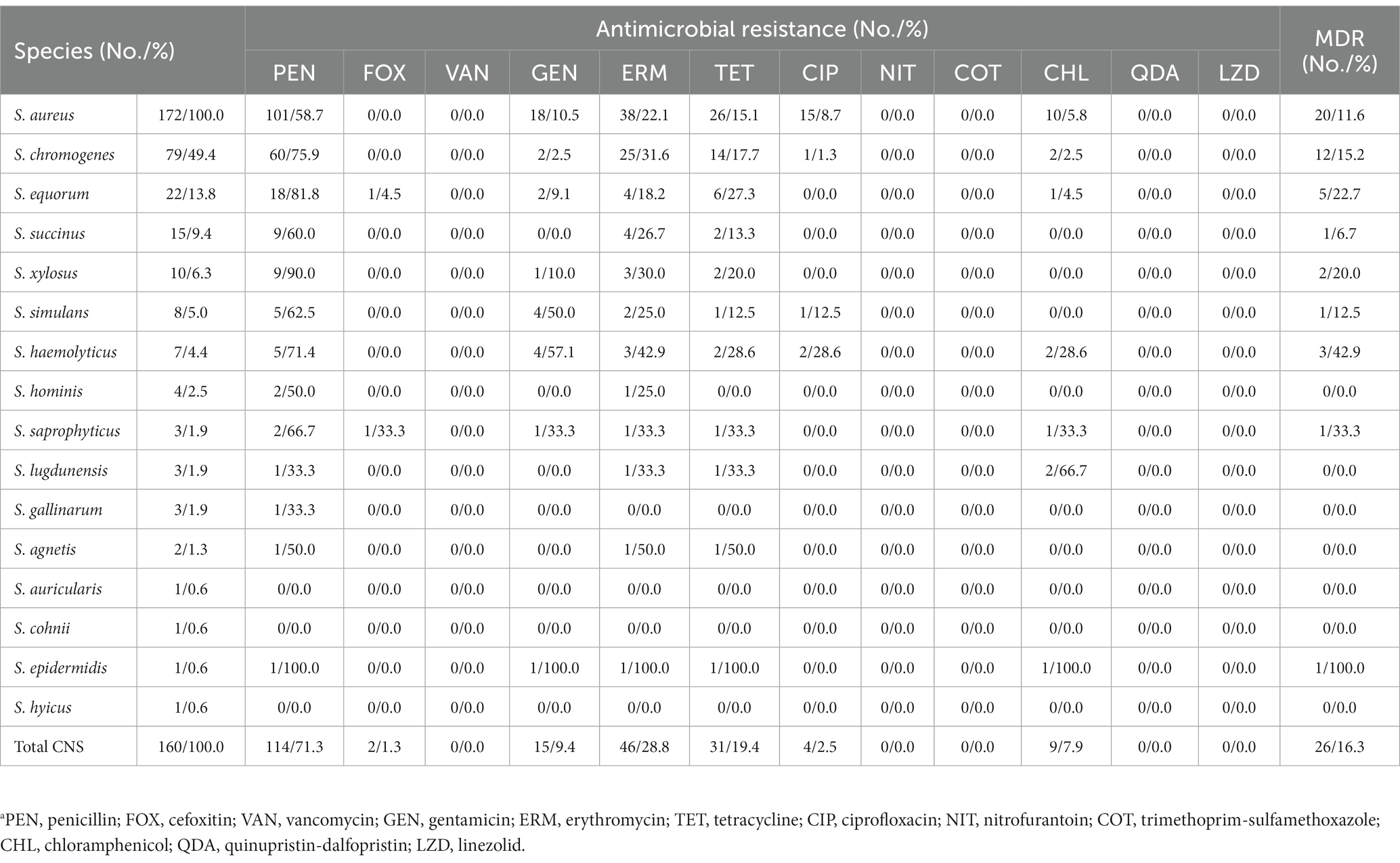
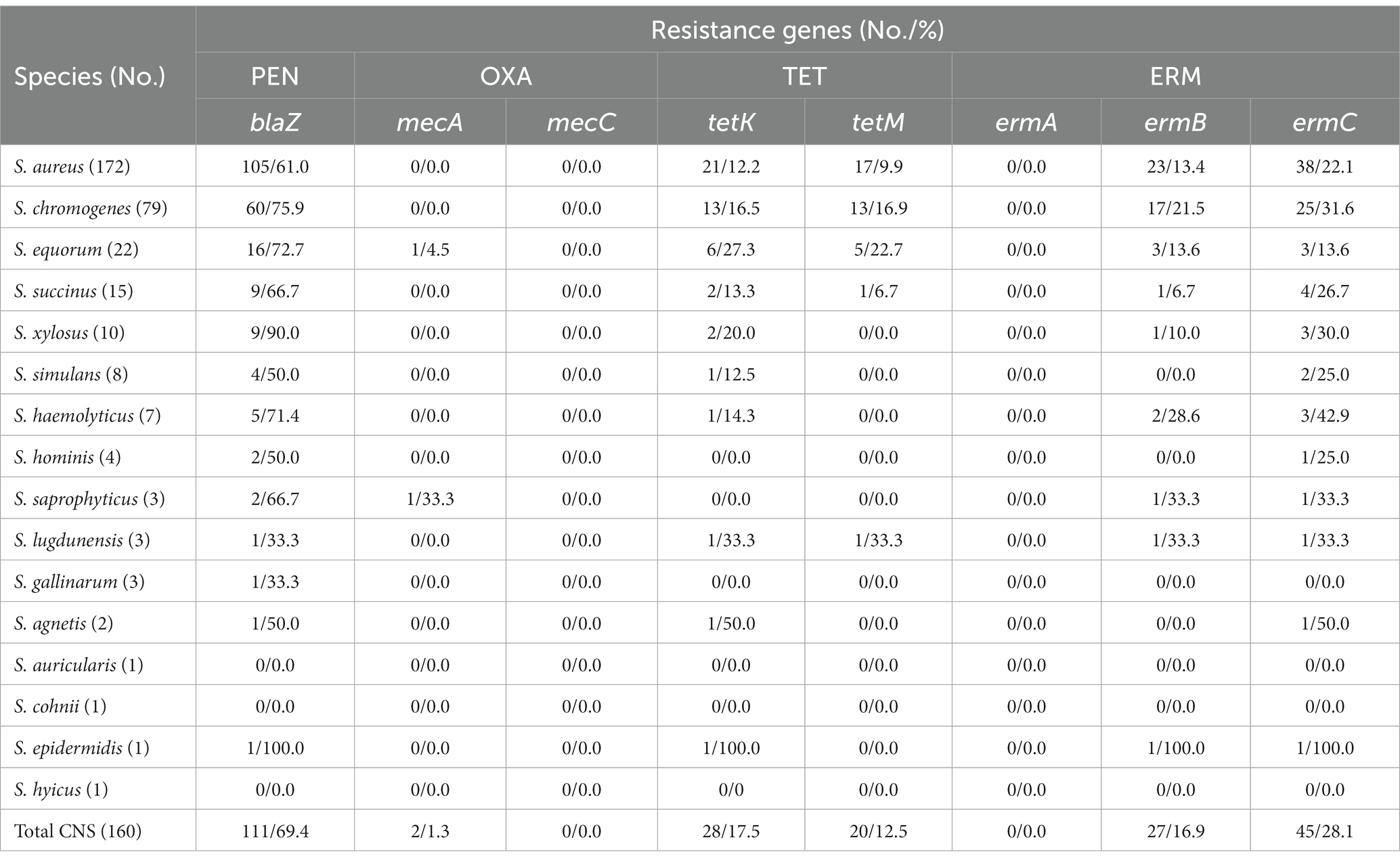
Table 1. Distribution and the antimicrobial resistance of staphylococci isolated from clinical bovine mastitisa.
3.2. Antimicrobial susceptibility testing
The antimicrobial susceptibility of the staphylococcal isolates against 12 antimicrobial agents were evaluated. The S. aureus isolates showed highest resistance rate to penicillin (101, 58.7%), followed by erythromycin (38, 22.1%), tetracycline (26, 15.1%), gentamicin (18, 10.5%), ciprofloxacin (15, 8.7%), and chloramphenicol (10, 5.8%). In addition, 20 (11.6%) S. aureus isolates exhibited MDR. Similar to the antimicrobial resistance profile of S. aureus, the CNS isolates displayed high resistance to penicillin (114, 71.3%), followed by erythromycin (46, 28.8%), tetracycline (31, 19.4%), gentamicin (15, 9.4%), chloramphenicol (9, 7.9%), ciprofloxacin (4, 2.5%), and cefoxitin (2, 1.3%). Methicillin-resistant phenotype was detected in 1 S. equorum and 1 S. saprophyticus isolates based on their susceptibility to cefoxitin. Antimicrobial resistance rates varied by CNS species. Multidrug resistance was found in 26 (16.3%) CNS isolates, including S. chromogenes (12, 15.2%), S. equorum (5, 22.7%), S. succinus (1, 6.7%), S. xylosus (2, 20.0%), S. simulans (1, 12.5%), S. haemolyticus (3, 42.9%), S. saprophyticus (1, 33.3%), and S. epidermidis (1, 100.0%). None of the staphylococcal isolates showed resistance to nitrofurantoin, trimethoprim-sulfamethoxazole, quinupristin/dalfopristin, linezolid or vancomycin in this study (Table 1, Supplementary Table S3).
3.3. Genetic determinants for antimicrobial resistance
The staphylococcal isolates showed higher resistance to penicillin, erythromycin and tetracycline compared to other tested antimicrobial agents in this study. Hence, the resistance encoding genes for these antimicrobial agents as well as methicillin resistant genes mecA and mecC were tested and shown in Table 2 and Supplementary Table S3. In S. aureus isolates, the blaZ was detected in 105 (61.0%) isolates. All penicillin-resistant S. aureus isolates carried blaZ. Besides, 4 penicillin-susceptible isolates also contained this gene. The tetK and tetM were determined in 21 (12.2%) and 17 (9.9%) isolates, respectively. All tetK positive (alone or combined with tetM) isolates showed resistance to tetracycline. Five tetracycline-resistant S. aureus isolates were negative for tetK or tetM. Additionally, genes ermC and ermB were found in 38 (22.1%) and 23 (13.4%) S. aureus isolates, respectively. And all erythromycin-resistant isolates harbored ermC alone or in combination with ermB. None of the isolates were positive for the mecA, mecC or ermA.
Among the 160 CNS isolates evaluated, the blaZ was found in 111 (69.4%) isolates and all of them showed resistance to penicillin. Two S. equorum and 1 S. simulans that were resistant against penicillin were negative for blaZ. Importantly, both of the methicillin-resistant isolates, 1 S. equorum and 1 S. saprophyticus, carried mecA. The tetK and tetM were determined in 28 (17.5%) and 20 (12.5%) CNS isolates, respectively. All tetK-carrying (alone or combined with tetM) isolates showed resistance to tetracycline. Three tetracycline-resistant isolates, including 1 S. chromogenes, 1 S. haemolyticus and 1 S. saprophyticus, did not harbored tetK or tetM. Moreover, ermC and ermB were detected in 45 (28.1%) and 27 (16.9%) CNS isolates, respectively. All ermC-carrying (alone or combined with ermB) isolates displayed resistance to erythromycin. One erythromycin-resistant S. equorum was negative for ermC or ermB. None of the CNS isolates were positive for the mecC or ermA (Table 2, Supplementary Table S3).
3.4. Genetic determinants for virulence factors
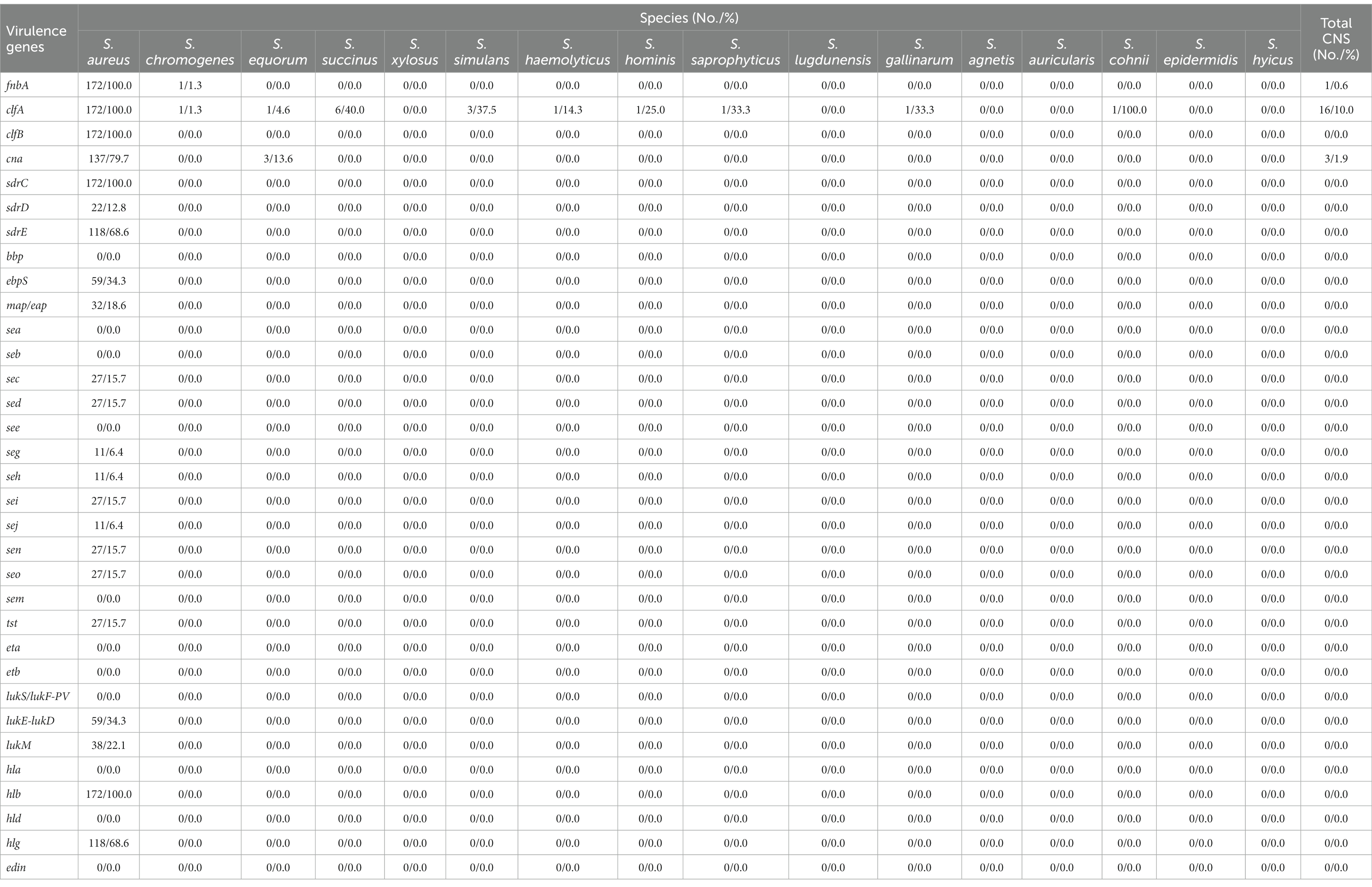
The presence and distribution of adhesin and toxin genes in staphylococcal isolates were presented in Table 3 and Supplementary Table S3. All S. aureus isolates harbored the adhesin genes fnbpA, clfA, clfB, and sdrC. Most of them contained cna (137, 79.7%) and sdrE (118, 68.6%), while genes ebpS, sdrD and map/eap were only found in 34.3% (59), 12.8% (22), and 18.6% (32) of the isolates, respectively. For the toxin genes, hlb was present in all S. aureus isolates, followed by hlg (118, 68.6%), lukE-lukD (59, 34.3%), and lukM (38, 22.1%). Genes sec, sed, seg, seh, sei, sej, sen, seo and tst were only observed in 15.7% (27), 15.7% (27), 6.4% (11), 6.4% (11), 15.7% (27), 6.4% (11), 15.7% (27), 15.7% (27), and 15.7% (27) of the isolates, respectively. None of the S. aureus isolates were positive for bbp, sea, seb, see, sem, eta, etb, lukS/lukF-PV, hla, hld, and edin. In contrast to S. aureus, virulence genes were detected in a small number of the CNS isolates. Only 10.0% (16) of the CNS isolates were positive for clfA, 1.9% (3) for cna and 0.6% (1) for fnbA. The toxin-encoding genes were not observed in any of the CNS isolates.
4. Discussion
A variety of bacteria have been implicated in bovine mastitis, with staphylococci being considered one of the most significant and prevalent causative agents in China and other countries (Gao et al., 2017). Understanding the pathogen profile for mastitis is critical to management (Dyson et al., 2022). In routine mastitis diagnostic laboratories, CNS are usually not identified to the species level but are reported as a single group. Consequently, limited knowledge is available regarding the epidemiology and relative importance of different species in this group (Ruegg et al., 2015; Schmidt et al., 2015; Dyson et al., 2022; Zigo et al., 2022). Although a protective effect against clinical mastitis has been postulated (Addis et al., 2020), ascribing the beneficial effect to the CNS as a group is probably inaccurate and still a topic of debate; such effect will rather be situated at the species or even strain level (Vanderhaeghen et al., 2014). The CNS group isolated from bovine milk samples consists of more than 50 different species and subspecies (Locatelli et al., 2013), and the distribution of CNS species change over time and vary between different regions (Dyson et al., 2022). In our study, 172 S. aureus and 160 CNS isolates were identified from the 332 staphylococcal isolates through 16S rRNA gene sequencing. A total of 15 species were confirmed among the CNS isolates. These species were frequently observed in both clinical and subclinical mastitis with slight differences among herds worldwide (Frey et al., 2013; Condas et al., 2017; El-Razik et al., 2017; Lianou et al., 2021), but the proportion of different Staphylococcus species varied between studies carried out in different countries (Schmidt et al., 2015; Xu et al., 2015). In accordance with the previous reports (Rall et al., 2014; Dos Santos et al., 2016; Valckenier et al., 2021), the predominant CNS species analyzed in this study was S. chromogenes. Normally, S. equorum was a less frequently detected species among CNS from dairy cattle (Adkins et al., 2018; Mahmmod et al., 2018; Jenkins et al., 2019; Valckenier et al., 2021). However, the S. equorum was the second most prevalent CNS species in our study, similar to the report that high proportion of this species was found in bulk milk (De Visscher et al., 2017), suggesting that this species might be relevant for udder health in the sampling site. Additionally, although S. epidermidis and S. haemolyticus commonly presents a high prevalence among CNS from bovine origin (Lee and Lee, 2022), the S. epidermidis and S. haemolyticus in our study were only observed in few isolates. This may indicate that these two species are not significant causative agent of mastitis in our studied area. Management practices, origin and strategy of samples, housing systems, climate and herd size used in the studies could probably explain some of the differences. Moreover, the distribution of the most common species has been shown to change over time (Koop et al., 2012; Nyman et al., 2018). Notably, although the prevalence was low, S. agnetis isolates were identified among the CNS isolates in the current study. This Staphylococcus species, an emerging pathogen, was described as a separate species in 2012 and frequently isolated from mastitic milk samples in other countries (Condas et al., 2017; Mahato et al., 2017; Poulsen et al., 2017; Rahmdel et al., 2018; Szafraniec et al., 2020). To our knowledge, this is the first report documenting the occurrence of S. agnetis from bovine mastitis in China. Further sampling is required to ascertain the true prevalence and significance of this species in local dairy herds.
Antimicrobial therapy has been used as a successful strategy for controlling staphylococcal mastitis. β-Lactams, tetracyclines and macrolides were commonly used to treat staphylococcal mastitis. But the therapeutic effects are hampered by the increasing number of drug-resistant strains (Kim et al., 2019; Achek et al., 2020). In the present study, the most resistance was observed against penicillin in both S. aureus and CNS isolates, followed by erythromycin and tetracycline. Meanwhile, low resistance rates of gentamicin, ciprofloxacin and chloramphenicol were also found in the staphylococcal isolates tested in this study. Resistance to these antimicrobials was also frequently reported by other authors (Cheng et al., 2019; Vasileiou et al., 2019; Francisco et al., 2021; Lianou et al., 2021; Mostafa Abdalhamed et al., 2022). In agreement with other recent studies (Fernandes Dos Santos et al., 2016; Taponen et al., 2016), resistance to the tested antimicrobials was higher in CNS than that in S. aureus with the exception of gentamicin and ciprofloxacin in the present study. Nevertheless, our results were similar to previous studies reporting low-level resistance to gentamicin and ciprofloxacin in both S. aureus and CNS isolates from bovine mastitis (Frey et al., 2013; Mahato et al., 2017; Martins et al., 2017; Klibi et al., 2018), probably due to the low frequent use of these antimicrobials in dairy farm in comparison with penicillin, erythromycin and tetracycline. Notably, 1 S. equorum and 1 S. saprophyticus isolates were resistant to methicillin in the current study. The occurrence of methicillin resistance in these 2 CNS species isolated from humans, livestock and farm environment has been previously described (Cicconi-Hogan et al., 2014; Teeraputon et al., 2017; Lu et al., 2020; Bonvegna et al., 2021; Garbacz et al., 2021). However, to the best of the available knowledge, there are no reports of the methicillin resistance in S. equorum and S. saprophyticus causing bovine mastitis in China. These resistant bacteria have been reported as an emerging problem in veterinary medicine and pose a threat to public health due to their transfer from animals to the humans caring for them (Kim et al., 2019). Moreover, our findings were in accord with previous study found that CNS often exhibit greater tendency to develop multidrug resistance (MDR) than S. aureus (Schmidt et al., 2015). The high phenotypic resistance could be explained by the frequent use of these antimicrobials for the treatment of mastitis or other diseases such as lameness, respiratory, or reproductive problems. An augmented exposure to antimicrobials can lead to an increase in resistant strains and consequently to the diversity we observed in the resistance profile of the isolates (Fernandes Dos Santos et al., 2016; Osman et al., 2016). Furthermore, we found very large differences in antimicrobial resistance between different CNS species, possibly due to the limited number of isolates at the species level.
In this study, the most commonly antimicrobial resistance determined was against penicillin, erythromycin and tetracycline. Thus, the genes conferring resistance to these antimicrobials as well as methicillin were detected. Corresponding to the phenotypic resistance, blaZ showed high prevalence and was found in all penicillin-resistant S. aureus isolates in this study. However, 4 blaZ-containing isolates were susceptible to penicillin. Previous studies also found the phenomenon that some blaZ-positive S. aureus isolates were susceptible to penicillin (Ruegg et al., 2015; Andrade et al., 2021). The discrepancy may be attributable to the lack of blaZ expression (Hammad et al., 2014). In CNS, all blaZ-positive isolates were resistant to penicillin. But 3 penicillin-resistant CNS isolates were negative for this gene. This may be attributed to the fact that mechanisms such as efflux pump or biofilm other than expression of the blaZ gene can cause penicillin resistance because multiple mechanisms of resistance often exist in these isolates (Osman et al., 2015; Addetia et al., 2019; Francisco et al., 2021). Moreover, the mecA was observed in both of the methicillin-resistant isolates (one isolate each of S. equorum and S. saprophyticus), which confirmed the phenotypic resistance to methicillin. To date, at least 38 tetracycline resistance genes have been found, and the genes tetK and tetM has been commonly found in species of staphylococci (Ruegg et al., 2015). In this study, despite the low occurrence of tetM, the tetK was determined in most of the tetracycline-resistant staphylococcal isolates. This may not be surprising because tetK is very frequent in staphylococci species from cows with clinical mastitis (Klibi et al., 2018). Furthermore, all tetK-carrying (alone or combined with tetM) isolates showed resistance to tetracycline. A few staphylococcal isolates showed phenotypic resistance to tetracycline but were negative for tetM or tetK. Additionally, 10 genes have been identified encoding resistance to erythromycin until now, being ermA, ermB, and ermC the major mechanism in staphylococci for erythromycin resistance (Sun et al., 2018). But in our study, aside from 1 erythromycin-resistant S. equorum that was negative for ermC or ermB, the ermC alone or in combination with ermB were detected in all erythromycin-resistant staphylococcal isolates, which is supported by previous research indicating that ermC is the most prevalent erm gene recovered from cases of staphylococcal bovine mastitis and most of the isolates exhibited phenotypic resistance to erythromycin (Ruegg et al., 2015). The coexistence of these tetracyclines and macrolides resistance genes has been frequently reported in S. aureus or CNS isolates from bovine mastitis in China and other countries (Klibi et al., 2018; Naranjo-Lucena and Slowey, 2023). The discrepancies observed between the phenotypic susceptibility and resistance genes could be due to the presence of other resistance-encoding genes, such as tetL or tetO for tetracyclin and ermE, ermT, mefA, or mefE for erythromycin, or due to a mutation in the primer-annealing site (DiPersio et al., 2008; Schmidt et al., 2015).
The pathogenicity of staphylococci is mainly related to its capacity to encode and produce a multitude of virulence factors, facilitating their adhesion and invasion of the host cells and establishment of infection (Klibi et al., 2018). The initial attachment of staphylococci to epithelial cells of the teat canal depends on the interaction of bacterial adhesins with host surface proteins, peptides and molecules located in the basement membrane (Stutz et al., 2011). In the current study, the S. aureus isolates exhibited high prevalence of fnbpA, clfA, clfB, sdrC, cna, and sdrE. Similar observations have also been reported in our previous study and by other reports (Yang et al., 2020; Avila-Novoa et al., 2022; Ibrahim et al., 2022). However, the frequencies of ebpS, sdrD, and map/eap found in our study was lower than those reported by other authors (Cheraghi et al., 2017; Kot et al., 2022). Our findings indicated that a diversity of adhesins were involved in the initial attachment of host cells and colonization of the mammary gland by S. aureus in Ningxia Hui Autonomous Region. In addition, this group of isolates was also evaluated for toxin genes related to the invasion of host cells and the evasion of immune response. Hemolysins are pore-forming toxins that attack cell membranes and cause platelet damage, lysosome destruction, ischemia, and necrosis (Abril et al., 2020). Most S. aureus isolates from bovine and human origins have been reported to primarily possess the hla, which causes incomplete or partial hemolysis (Zhang et al., 2018; Khan et al., 2021). However, the hlb and hlg were the predominant hemolysin genes in this study, and none of the isolates contained hla. Leukocidins are also pore-forming two-component toxins that specifically attack immune cells (Abril et al., 2020). Similar to previous studies (Haveri et al., 2007, 2008; Thomas et al., 2021), lukE-lukD was the most prevalent leukocidin-encoding gene in our study, followed by lukM. Moreover, enterotoxins and toxic shock syndrome toxin-1 are pyrogenic toxins known as staphylococcal superantigens causing staphylococcal food poisoning and are able to interrupt host immune responses (Podkowik et al., 2013; Abril et al., 2020). In the present study, enterotoxin-encoding genes sec, sed, seg, seh, sei, sej, sen, and seo as well as toxic shock syndrome toxin-1-encoding gene tst were detected with low frequencies. These findings were in accordance with those of other reports involved in bovine S. aureus (Hummerjohann et al., 2014; Rall et al., 2014; Mello et al., 2016 Vaughn et al., 2020). The variation in the prevalence of the tested virulence factors could be associated with the genetic diversity of strains, the source and sizes of samples or their geographic locations (Avila-Novoa et al., 2022). Given that certain virulence genes are overrepresented in some clonal lineages and that some combinations are correlated with high pathogenic potential (Achek et al., 2020), further investigations need to be performed to explore the diversity of virulence factors combination in S. aureus pathogenesis.
Consistent with other studies (Supré et al., 2011; Xu et al., 2015; França et al., 2021), the virulence genes in CNS were significantly less prevalent than that in the S. aureus in our study. Previous study indicated that collagen binding protein (cna) and fibronectin binding protein (fnbA) were often associated with CNS attachment in bovines (Pizauro et al., 2019). However, in the current study, only few of the CNS isolates carried cna and fnbA. Similar results were obtained from subclinical mastitis milk in China (Xu et al., 2015). Additionally, a few of the CNS isolates were positive for clfA in this study, in line with the report by Felipe et al. that 12.2% of the CNS isolates contained this gene (Felipe et al., 2017). The capacity to adhere to bovine mammary epithelial cells strongly differs among the different CNS isolates and potentially reflects intra-species diversity in ecology and epidemiological behavior (Souza et al., 2016). Recent studies have provided strong evidence for the presence of toxin genes and production of the corresponding toxins in CNS in China and other countries, especially the enterotoxins (Rall et al., 2014; Salaberry et al., 2015; Mahato et al., 2017; Martins et al., 2017; Pizauro et al., 2019). However, the toxin-encoding genes were not observed in any of the CNS isolates in our study. Our results were similar to those presented by other authors that all CNS species were negative for the toxin genes, even though a wide range of genes were tested (Nemati et al., 2008; Klempt et al., 2022). This may be attributed to the fact that the low prevalence of toxin-producing CNS isolates in the studied area, or the presence of other toxin genes which were not tested (Xu et al., 2015). Another possible reason is the requirement of different primer sets to detect the target genes (Vanderhaeghen et al., 2014). Furthermore, previous studies demonstrated that the use of antimicrobial drugs influenced the expression of virulence genes in staphylococci. The connection between genetic elements conferring resistance to antimicrobials and expression of virulence factors is intricately linked to the ability of bacteria to communicate through two-component system and quorum sensing system and has not yet been fully elucidated (Pérez et al., 2020). In further research, large sample size and sufficient numbers of isolates of each species are needed to explore the species-specific association between antimicrobial resistance and virulence factors in staphylococci.
5. Conclusion
This study provides high species diversity of staphylococci from clinical bovine mastitis in Ningxia Hui Autonomous Region in China. Noticeably, to our knowledge, we first describe the occurrence of S. agnetis from bovine mastitis in China. The S. aureus and CNS isolates displayed high frequencies of phenotypic and genotypic resistance to penicillin, erythromycin and tetracycline, which remind the government to pay continuous attention to the commonly used antimicrobial agents in dairy industry. Moreover, the high occurrence of the adhesin genes fnbpA, clfA, clfB, sdrC, cna, and sdrE tested in this study as well as the toxin genes hlb and hlg in S. aureus indicate their pathogenic potential causing bovine mastitis in the studied area. Further investigation is necessary to explore the diversity of virulence factors combination in S. aureus pathogenesis. Furthermore, despite the absence of toxin genes in CNS in this study, a more extensive examination is needed to demonstrate the true toxigenic potential of this organisms group in mastitis.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Ethics statement
The animal study was reviewed and approved by Animal Welfare and Ethics Committee of Northwest A&F University. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
FY conceptualized the study, designed the methodology, conducted the tests, and wrote the original paper. WS, NM, and YZ helped to conduct the experimental tests. XD and QL provided resources, made the review, as well as were the leadership and responsible for funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
This work was financially supported by the Key Research and Development Program of Gansu Province (Grant No. 21YF5NA141) and the National Key R&D Program of China during the 14th Five-year Plan Period (Grant No. 2022YFD1302101).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1190790/full#supplementary-material
Footnotes
1. ^https://www.omegabiotek.com/product/e-z-n-a-bacterial-dna-kit/
References
Achek, R., El-Adawy, H., Hotzel, H., Tomaso, H., Ehricht, R., Hamdi, T. M., et al. (2020). Short communication: diversity of staphylococci isolated from sheep mastitis in northern Algeria. J. Dairy Sci. 103, 890–897. doi: 10.3168/jds.2019-16583
Addetia, A., Greninger, A. L., Adler, A., Yuan, S., Makhsous, N., Qin, X., et al. (2019). A novel, widespread qacA allele results in reduced chlorhexidine susceptibility in Staphylococcus epidermidis. Antimicrob. Agents Chemother. 63, e02607–e02618. doi: 10.1128/AAC.02607-18
Addis, M. F., Maffioli, E. M., Ceciliani, F., Tedeschi, G., Zamarian, V., Tangorra, F., et al. (2020). Influence of subclinical mastitis and intramammary infection by coagulase-negative staphylococci on the cow milk peptidome. J. Proteome 226:103885. doi: 10.1016/j.jprot.2020.103885
Adkins, P. R. F., Dufour, S., Spain, J. N., Calcutt, M. J., Reilly, T. J., Stewart, G. C., et al. (2018). Cross-sectional study to identify staphylococcal species isolated from teat and inguinal skin of different-aged dairy heifers. J. Dairy Sci. 101, 3213–3225. doi: 10.3168/jds.2017-13974
Ahmed, W., Neubauer, H., Tomaso, H., El Hofy, F. I., Monecke, S., Abdeltawab, A. A., et al. (2020). Characterization of staphylococci and streptococci isolated from milk of bovides with mastitis in Egypt. Pathogens 9:381. doi: 10.3390/pathogens9050381
Andrade, N. C., Laranjo, M., Costa, M. M., and Queiroga, M. C. (2021). Virulence factors in Staphylococcus associated with small ruminant mastitis: biofilm production and antimicrobial resistance genes. Antibiotics (Basel) 10:633. doi: 10.3390/antibiotics10060633
Avila-Novoa, M. G., Solis-Velazquez, O. A., Guerrero-Medina, P. J., González-Gómez, J. P., González-Torres, B., Velázquez-Suárez, N. Y., et al. (2022). Genetic and compositional analysis of biofilm formed by Staphylococcus aureus isolated from food contact surfaces. Front. Microbiol. 13:1001700. doi: 10.3389/fmicb.2022.1001700
Bonvegna, M., Grego, E., Sona, B., Stella, M. C., Nebbia, P., Mannelli, A., et al. (2021). Occurrence of methicillin-resistant coagulase-negative staphylococci (MRCoNS) and methicillin-resistant Staphylococcus aureus (MRSA) from pigs and farm environment in northwestern Italy. Antibiotics (Basel) 10:676. doi: 10.3390/antibiotics10060676
Cheng, J., Qu, W., Barkema, H. W., Nobrega, D. B., Gao, J., Liu, G., et al. (2019). Antimicrobial resistance profiles of 5 common bovine mastitis pathogens in large Chinese dairy herds. J. Dairy Sci. 102, 2416–2426. doi: 10.3168/jds.2018-15135
Cheraghi, S., Pourgholi, L., Shafaati, M., Fesharaki, S. H., Jalali, A., Nosrati, R., et al. (2017). Analysis of virulence genes and accessory gene regulator (agr) types among methicillin-resistant Staphylococcus aureus strains in Iran. J. Glob. Antimicrob. Resist. 10, 315–320. doi: 10.1016/j.jgar.2017.06.009
Cicconi-Hogan, K. M., Belomestnykh, N., Gamroth, M., Ruegg, P. L., Tikofsky, L., and Schukken, Y. H. (2014). Short communication: prevalence of methicillin resistance in coagulase-negative staphylococci and Staphylococcus aureus isolated from bulk milk on organic and conventional dairy farms in the United States. J. Dairy Sci. 97, 2959–2964. doi: 10.3168/jds.2013-7523
CLSI (2018). Performance standards for antimicrobial susceptibility testing. 29 (Wayne, PA: CLSI), M100–MS29.
Condas, L. A. Z., De Buck, J., Nobrega, D. B., Carson, D. A., Roy, J. P., Keefe, G. P., et al. (2017). Distribution of non-aureus staphylococci species in udder quarters with low and high somatic cell count, and clinical mastitis. J. Dairy Sci. 100, 5613–5627. doi: 10.3168/jds.2016-12479
De Visscher, A., Piepers, S., Haesebrouck, F., Supré, K., and De Vliegher, S. (2017). Coagulase-negative Staphylococcus species in bulk milk: prevalence, distribution, and associated subgroup- and species-specific risk factors. J. Dairy Sci. 100, 629–642. doi: 10.3168/jds.2016-11476
DiPersio, L. P., DiPersio, J. R., Frey, K. C., and Beach, J. A. (2008). Prevalence of the erm(T) gene in clinical isolates of erythromycin-resistant group D Streptococcus and Enterococcus. Antimicrob. Agents Chemother. 52, 1567–1569. doi: 10.1128/AAC.01325-07
Dos Santos, D. C., Lange, C. C., Avellar-Costa, P., Dos Santos, K. R., Brito, M. A., and Giambiagi-deMarval, M. (2016). Staphylococcus chromogenes, a coagulase-negative Staphylococcus species that can clot plasma. J. Clin. Microbiol. 54, 1372–1375. doi: 10.1128/JCM.03139-15
Dyson, R., Charman, N., Hodge, A., Rowe, S. M., and Taylor, L. F. (2022). A survey of mastitis pathogens including antimicrobial susceptibility in southeastern Australian dairy herds. J. Dairy Sci. 105, 1504–1518. doi: 10.3168/jds.2021-20955
El-Razik, K. A. A., Arafa, A. A., Hedia, R. H., and Ibrahim, E. S. (2017). Tetracycline resistance phenotypes and genotypes of coagulase-negative staphylococcal isolates from bubaline mastitis in Egypt. Vet. World. 10, 702–710. doi: 10.14202/vetworld.2017.702-710
Felipe, V., Morgante, C. A., Somale, P. S., Varroni, F., Zingaretti, M. L., Bachetti, R. A., et al. (2017). Evaluation of the biofilm forming ability and its associated genes in Staphylococcus species isolates from bovine mastitis in Argentinean dairy farms. Microb. Pathog. 104, 278–286. doi: 10.1016/j.micpath.2017.01.047
Fernandes Dos Santos, F., Mendonça, L. C., Reis, D. R. L., Guimarães, A. S., Lange, C. C., Ribeiro, J. B., et al. (2016). Presence of mecA-positive multidrug-resistant Staphylococcus epidermidis in bovine milk samples in Brazil. J. Dairy Sci. 99, 1374–1382. doi: 10.3168/jds.2015-9931
Ferreira, E. M., Romero, L. C., Cunha, M. L. R. S. D., Malagó Junior, W., Camargo, C. H., Barioni Júnior, W., et al. (2022). Persistence of Staphylococcus spp. in milk from cows undergoing homeopathy to control subclinical mastitis. BMC Vet. Res. 18:273. doi: 10.1186/s12917-022-03364-8
França, A., Gaio, V., Lopes, N., and Melo, L. D. R. (2021). Virulence factors in coagulase-negative staphylococci. Pathogens 10:170. doi: 10.3390/pathogens10020170
Francisco, M. S., Rossi, C. C., Brito, M. A. V. P., Laport, M. S., Barros, E. M., and Giambiagi-deMarval, M. (2021). Characterization of biofilms and antimicrobial resistance of coagulase-negative Staphylococcus species involved with subclinical mastitis. J. Dairy Res. 88, 179–184. doi: 10.1017/S0022029921000285
Frey, Y., Rodriguez, J. P., Thomann, A., Schwendener, S., and Perreten, V. (2013). Genetic characterization of antimicrobial resistance in coagulase-negative staphylococci from bovine mastitis milk. J. Dairy Sci. 96, 2247–2257. doi: 10.3168/jds.2012-6091
Abril, A. G., Villa, T. G., Barros-Velázquez, J., Cañas, B., Sánchez-Pérez, A., Calo-Mata, P., et al. (2020). Staphylococcus aureus exotoxins and their detection in the dairy industry and mastitis. Toxins (Basel) 12:537. doi: 10.3390/toxins12090537
Gao, J., Barkema, H. W., Zhang, L., Liu, G., Deng, Z., Cai, L., et al. (2017). Incidence of clinical mastitis and distribution of pathogens on large Chinese dairy farms. J. Dairy Sci. 100, 4797–4806. doi: 10.3168/jds.2016-12334
Garbacz, K., Wierzbowska, M., Kwapisz, E., Kosecka-Strojek, M., Bronk, M., Saki, M., et al. (2021). Distribution and antibiotic-resistance of different Staphylococcus species identified by matrix assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) isolated from the oral cavity. J. Oral Microbiol. 13:1983322. doi: 10.1080/20002297.2021.1983322
González-Martín, M., Corbera, J. A., Suárez-Bonnet, A., and Tejedor-Junco, M. T. (2020). Virulence factors in coagulase-positive staphylococci of veterinary interest other than Staphylococcus aureus. Vet. Q. 40, 118–131. doi: 10.1080/01652176.2020.1748253
Hammad, A. M., Shimamoto, T., and Shimamoto, T. (2014). Genetic characterization of antibiotic resistance and virulence factors in Enterococcus spp. from Japanese retail ready-to-eat raw fish. Food Microbiol. 38, 62–66. doi: 10.1016/j.fm.2013.08.010
Haveri, M., Hovinen, M., Roslöf, A., and Pyörälä, S. (2008). Molecular types and genetic profiles of Staphylococcus aureus strains isolated from bovine intramammary infections and extramammary sites. J. Clin. Microbiol. 46, 3728–3735. doi: 10.1128/JCM.00769-08
Haveri, M., Roslöf, A., Rantala, L., and Pyörälä, S. (2007). Virulence genes of bovine Staphylococcus aureus from persistent and nonpersistent intramammary infections with different clinical characteristics. J. Appl. Microbiol. 103, 993–1000. doi: 10.1111/j.1365-2672.2007.03356.x
Hummerjohann, J., Naskova, J., Baumgartner, A., and Graber, H. U. (2014). Enterotoxin-producing Staphylococcus aureus genotype B as a major contaminant in Swiss raw milk cheese. J. Dairy Sci. 97, 1305–1312. doi: 10.3168/jds.2013-7643
Ibrahim, E. S., Arafa, A. A., Dorgam, S. M., Eid, R. H., Atta, N. S., El-Dabae, W. H., et al. (2022). Molecular characterization of genes responsible for biofilm formation in Staphylococcus aureus isolated from mastitic cows. Vet. World 15, 205–212. doi: 10.14202/vetworld.2022.205-212
Isaac, P., Bohl, L. P., Breser, M. L., Orellano, M. S., Conesa, A., Ferrero, M. A., et al. (2017). Commensal coagulase-negative Staphylococcus from the udder of healthy cows inhibits biofilm formation of mastitis-related pathogens. Vet. Microbiol. 207, 259–266. doi: 10.1016/j.vetmic.2017.05.025
Jarraud, S., Mougel, C., Thioulouse, J., Lina, G., Meugnier, H., Forey, F., et al. (2002). Relationships between Staphylococcus aureus genetic background, virulence factors, agr groups (alleles), and human disease. Infect. immun. 70, 631–641. doi: 10.1128/IAI.70.2.631-641.2002
Jenkins, S. N., Okello, E., Rossitto, P. V., Lehenbauer, T. W., Champagne, J., Penedo, M. C. T., et al. (2019). Molecular epidemiology of coagulase-negative Staphylococcus species isolated at different lactation stages from dairy cattle in the United States. PeerJ. 7:e6749. doi: 10.7717/peerj.6749
Khan, S., Marasa, B. S., Sung, K., and Nawaz, M. (2021). Genotypic characterization of clinical isolates of Staphylococcus aureus from Pakistan. Pathogens 10:918. doi: 10.3390/pathogens10080918
Kim, S. J., Moon, D. C., Park, S. C., Kang, H. Y., Na, S. H., and Lim, S. K. (2019). Antimicrobial resistance and genetic characterization of coagulase-negative staphylococci from bovine mastitis milk samples in Korea. J. Dairy Sci. 102, 11439–11448. doi: 10.3168/jds.2019-17028
Klempt, M., Franz, C. M. A. P., and Hammer, P. (2022). Characterization of coagulase-negative staphylococci and macrococci isolated from cheese in Germany. J. Dairy Sci. 105, 7951–7958. doi: 10.3168/jds.2022-21941
Klibi, A., Maaroufi, A., Torres, C., and Jouini, A. (2018). Detection and characterization of methicillin-resistant and susceptible coagulase-negative staphylococci in milk from cows with clinical mastitis in Tunisia. Int. J. Antimicrob. Agents 52, 930–935. doi: 10.1016/j.ijantimicag.2018.07.026
Koop, G., De Vliegher, S., De Visscher, A., Supré, K., Haesebrouck, F., Nielen, M., et al. (2012). Differences between coagulase-negative Staphylococcus species in persistence and in effect on somatic cell count and milk yield in dairy goats. J. Dairy Sci. 95, 5075–5084. doi: 10.3168/jds.2012-5615
Kot, B., Piechota, M., Jakubczak, A., Gryzińska, M., Witeska, M., Grużewska, A., et al. (2022). The prevalence of virulence determinants in methicillin-resistant Staphylococcus aureus isolated from different infections in hospitalized patients in Poland. Sci. Rep. 12:5477. doi: 10.1038/s41598-022-09517-x
Kot, B., Piechota, M., Wolska, K. M., Frankowska, A., Zdunek, E., Binek, T., et al. (2012). Phenotypic and genotypic antimicrobial resistance of staphylococci from bovine milk. Pol. J. Vet. Sci. 15, 677–683. doi: 10.2478/v10181-012-0105-4
Lee, Y. J., and Lee, Y. J. (2022). Characterization of biofilm producing coagulase-negative staphylococci isolated from bulk tank milk. Vet. Sci. 9:430. doi: 10.3390/vetsci9080430
Leroy, F., Van Coillie, E., Braem, G., Piessens, V., Verbist, B., De Vuyst, L., et al. (2015). Short communication: subtyping of Staphylococcus haemolyticus isolates from milk and corresponding teat apices to verify the potential teat-skin origin of intramammary infections in dairy cows. J. Dairy Sci. 98, 7893–7898. doi: 10.3168/jds.2015-9415
Li, L., Feng, W., Zhang, Z., Xue, H., and Zhao, X. (2015). Macrolide-lincosamide-streptogramin resistance phenotypes and genotypes of coagulase-positive Staphylococcus aureus and coagulase-negative staphylococcal isolates from bovine mastitis. BMC Vet. Res. 11:168. doi: 10.1186/s12917-015-0492-8
Lianou, D. T., Petinaki, E., Cripps, P. J., Gougoulis, D. A., Michael, C. K., Tsilipounidaki, K., et al. (2021). Antibiotic resistance of staphylococci from bulk-tank milk of sheep flocks: prevalence, patterns, association with biofilm formation, effects on milk quality, and risk factors. Biology 10:1016. doi: 10.3390/biology10101016
Locatelli, C., Piepers, S., De Vliegher, S., Barberio, A., Supré, K., Scaccabarozzi, L., et al. (2013). Effect on quarter milk somatic cell count and antimicrobial susceptibility of Staphylococcus rostri causing intramammary infection in dairy water buffaloes. J. Dairy Sci. 96, 3799–3805. doi: 10.3168/jds.2012-6275
Lu, Y., Lu, Q., Cheng, Y., Wen, G., Luo, Q., Shao, H., et al. (2020). High concentration of coagulase-negative staphylococci carriage among bioaerosols of henhouses in Central China. BMC Microbiol. 20:21. doi: 10.1186/s12866-020-1709-y
Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Mahato, S., Mistry, H. U., Chakraborty, S., Sharma, P., Saravanan, R., and Bhandari, V. (2017). Identification of variable traits among the methicillin resistant and sensitive coagulase negative staphylococci in milk samples from mastitic cows in India. Front. Microbiol. 8:1446. doi: 10.3389/fmicb.2017.01446
Mahmmod, Y. S., Klaas, I. C., Svennesen, L., Pedersen, K., and Ingmer, H. (2018). Communications of Staphylococcus aureus and non-aureus Staphylococcus species from bovine intramammary infections and teat apex colonization. J. Dairy Sci. 101, 7322–7333. doi: 10.3168/jds.2017-14311
Martins, K. B., Faccioli, P. Y., Bonesso, M. F., Fernandes, S., Oliveira, A. A., Dantas, A., et al. (2017). Characteristics of resistance and virulence factors in different species of coagulase-negative staphylococci isolated from milk of healthy sheep and animals with subclinical mastitis. J. Dairy Sci. 100, 2184–2195. doi: 10.3168/jds.2016-11583
Mello, P. L., Moraes Riboli, D. F., Pinheiro, L., de Almeida Martins, L., Vasconcelos Paiva Brito, M., and Ribeiro de Souza da Cunha, M. (2016). Detection of enterotoxigenic potential and determination of clonal profile in Staphylococcus aureus and coagulase-negative staphylococci isolated from bovine subclinical mastitis in different Brazilian states. Toxins (Basel) 8:104. doi: 10.3390/toxins8040104
Mostafa Abdalhamed, A., Zeedan, G. S. G., Ahmed Arafa, A., Shafeek Ibrahim, E., Sedky, D., and Abdel Nabey Hafez, A. (2022). Detection of methicillin-resistant Staphylococcus aureus in clinical and subclinical mastitis in ruminants and studying the effect of novel green synthetized nanoparticles as one of the alternative treatments. Vet. Med. Int. 2022:6309984. doi: 10.1155/2022/6309984
Naranjo-Lucena, A., and Slowey, R. (2023). Invited review: antimicrobial resistance in bovine mastitis pathogens: a review of genetic determinants and prevalence of resistance in European countries. J. Dairy Sci. 106, 1–23. doi: 10.3168/jds.2022-22267
Nemati, M., Hermans, K., Vancraeynest, D., De Vliegher, S., Sampimon, O. C., Baele, M., et al. (2008). Screening of bovine coagulase-negative staphylococci from milk for superantigen-encoding genes. Vet. Rec. 163, 740–743. doi: 10.1016/j.zool.2007.04.002
Nyman, A. K., Fasth, C., and Waller, K. P. (2018). Intramammary infections with different non-aureus staphylococci in dairy cows. J. Dairy Sci. 101, 1403–1418. doi: 10.3168/jds.2017-13467
Osman, K. M., Abd El-Razik, K. A., Marie, H. S., and Arafa, A. (2015). Relevance of biofilm formation and virulence of different species of coagulase-negative staphylococci to public health. Eur. J. Clin. Microbiol. Infect. Dis. 34, 2009–2016. doi: 10.1007/s10096-015-2445-3
Osman, K. M., Abd El-Razik, K. A., Marie, H. S., and Arafa, A. (2016). Coagulase-negative staphylococci collected from bovine milk: species and antimicrobial gene diversity. J. Food Saf. 36, 89–99. doi: 10.1111/jfs.12216
Paterson, G. K., Larsen, A. R., Robb, A., Edwards, G. E., Pennycott, T. W., Foster, G., et al. (2012). The newly described mecA homologue, mecALGA251, is present in methicillin-resistant Staphylococcus aureus isolates from a diverse range of host species. J. Antimicrob. Chemother. 67, 2809–2813. doi: 10.1093/jac/dks329
Peacock, S. J., Moore, C. E., Justice, A., Kantzanou, M., Story, L., Mackie, K., et al. (2002). Virulent combinations of adhesin and toxin genes in natural populations of Staphylococcus aureus. Infect. immun. 70, 4987–4996. doi: 10.1128/IAI.70.9.4987-4996.2002
Pérez, V. K. C., Costa, G. M. D., Guimarães, A. S., Heinemann, M. B., Lage, A. P., and Dorneles, E. M. S. (2020). Relationship between virulence factors and antimicrobial resistance in Staphylococcus aureus from bovine mastitis. J. Glob. Antimicrob. Resist. 22, 792–802. doi: 10.1016/j.jgar.2020.06.010
Piessens, V., De Vliegher, S., Verbist, B., Braem, G., Van Nuffel, A., De Vuyst, L., et al. (2012). Characterization of coagulase-negative Staphylococcus species from cows’ milk and environment based on bap, icaA, and mecA genes and phenotypic susceptibility to antimicrobials and teat dips. J. Dairy Sci. 95, 7027–7038. doi: 10.3168/jds.2012-5400
Pizauro, L. J. L., de Almeida, C. C., Soltes, G. A., Slavic, D., de Ávila, F. A., Zafalon, L. F., et al. (2019). Short communication: detection of antibiotic resistance, mecA, and virulence genes in coagulase-negative Staphylococcus spp. from buffalo milk and the milking environment. J. Dairy Sci. 102, 11459–11464. doi: 10.3168/jds.2018-15920
Podkowik, M., Park, J. Y., Seo, K. S., Bystroń, J., and Bania, J. (2013). Enterotoxigenic potential of coagulase-negative staphylococci. Int. J. Food Microbiol. 163, 34–40. doi: 10.1016/j.ijfoodmicro.2013.02.005
Poulsen, L. L., Thøfner, I., Bisgaard, M., Olsen, R. H., Christensen, J. P., and Christensen, H. (2017). Staphylococcus agnetis, a potential pathogen in broiler breeders. Vet. Microbiol. 212, 1–6. doi: 10.1016/j.vetmic.2017.10.018
Rahmdel, S., Hosseinzadeh, S., Shekarforoush, S. S., Torriani, S., Gatto, V., and Pashangeh, S. (2018). Safety hazards in bacteriocinogenic Staphylococcus strains isolated from goat and sheep milk. Microb. Pathog. 116, 100–108. doi: 10.1016/j.micpath.2018.01.016
Rall, V. L., Miranda, E. S., Castilho, I. G., Camargo, C. H., Langoni, H., Guimarães, F. F., et al. (2014). Diversity of Staphylococcus species and prevalence of enterotoxin genes isolated from milk of healthy cows and cows with subclinical mastitis. J. Dairy Sci. 97, 829–837. doi: 10.3168/jds.2013-7226
Ruegg, P. L., Oliveira, L., Jin, W., and Okwumabua, O. (2015). Phenotypic antimicrobial susceptibility and occurrence of selected resistance genes in gram-positive mastitis pathogens isolated from Wisconsin dairy cows. J. Dairy Sci. 98, 4521–4534. doi: 10.3168/jds.2014-9137
Salaberry, S. R., Saidenberg, A. B., Zuniga, E., Melville, P. A., Santos, F. G., Guimarães, E. C., et al. (2015). Virulence factors genes of Staphylococcus spp. isolated from caprine subclinical mastitis. Microb. Pathog. 85, 35–39. doi: 10.1016/j.micpath.2015.05.007
Schmidt, T., Kock, M. M., and Ehlers, M. M. (2015). Diversity and antimicrobial susceptibility profiling of staphylococci isolated from bovine mastitis cases and close human contacts. J. Dairy Sci. 98, 6256–6269. doi: 10.3168/jds.2015-9715
Souza, F. N., Piepers, S., Della Libera, A. M. M. P., Heinemann, M. B., Cerqueira, M. M. O. P., and De Vliegher, S. (2016). Interaction between bovine-associated coagulase-negative staphylococci species and strains and bovine mammary epithelial cells reflects differences in ecology and epidemiological behavior. J. Dairy Sci. 99, 2867–2874. doi: 10.3168/jds.2015-10230
Stutz, K., Stephan, R., and Tasara, T. (2011). SpA, ClfA, and FnbA genetic variations lead to Staphaurex test-negative phenotypes in bovine mastitis Staphylococcus aureus isolates. J. Clin. Microbiol. 49, 638–646. doi: 10.1128/JCM.01148-10
Sun, X., Lin, Z. W., Hu, X. X., Yao, W. M., Bai, B., Wang, H. Y., et al. (2018). Biofilm formation in erythromycin-resistant Staphylococcus aureus and the relationship with antimicrobial susceptibility and molecular characteristics. Microb. Pathog. 124, 47–53. doi: 10.1016/j.micpath.2018.08.021
Supré, K., Haesebrouck, F., Zadoks, R. N., Vaneechoutte, M., Piepers, S., and De Vliegher, S. (2011). Some coagulase-negative Staphylococcus species affect udder health more than others. J. Dairy Sci. 94, 2329–2340. doi: 10.3168/jds.2010-3741
Szafraniec, G. M., Szeleszczuk, P., and Dolka, B. (2020). A review of current knowledge on Staphylococcus agnetis in poultry. Animals (Basel) 10:1421. doi: 10.3390/ani10081421
Taponen, S., Nykäsenoja, S., Pohjanvirta, T., Pitkälä, A., and Pyörälä, S. (2016). Species distribution and in vitro antimicrobial susceptibility of coagulase-negative staphylococci isolated from bovine mastitic milk. Acta Vet. Scand. 58:12. doi: 10.1186/s13028-016-0193-8
Teeraputon, S., Santanirand, P., Wongchai, T., Songjang, W., Lapsomthob, N., Jaikrasun, D., et al. (2017). Prevalence of methicillin resistance and macrolide-lincosamide-streptogramin B resistance in Staphylococcus haemolyticus among clinical strains at a tertiary-care hospital in Thailand. New Microbes. New Infect. 19, 28–33. doi: 10.1016/j.nmni.2017.05.007
Thomas, A., Chothe, S., Byukusenge, M., Mathews, T., Pierre, T., Kariyawasam, S., et al. (2021). Prevalence and distribution of multilocus sequence types of Staphylococcus aureus isolated from bulk tank milk and cows with mastitis in Pennsylvania. PLoS One 16:e0248528. doi: 10.1371/journal.pone.0248528
Valckenier, D., Piepers, S., Schukken, Y. H., De Visscher, A., Boyen, F., Haesebrouck, F., et al. (2021). Longitudinal study on the effects of intramammary infection with non-aureus staphylococci on udder health and milk production in dairy heifers. J. Dairy Sci. 104, 899–914. doi: 10.3168/jds.2020-18685
Vanderhaeghen, W., Piepers, S., Leroy, F., Van Coillie, E., Haesebrouck, F., and De Vliegher, S. (2014). Invited review: effect, persistence, and virulence of coagulase-negative Staphylococcus species associated with ruminant udder health. J. Dairy Sci. 97, 5275–5293. doi: 10.3168/jds.2013-7775
Vasileiou, N. G. C., Sarrou, S., Papagiannitsis, C., Chatzopoulos, D. C., Malli, E., Mavrogianni, V. S., et al. (2019). Antimicrobial agent susceptibility and typing of staphylococcal isolates from subclinical mastitis in ewes. Microb. Drug Resist. 25, 1099–1110. doi: 10.1089/mdr.2019.0009
Vaughn, J. M., Abdi, R. D., Gillespie, B. E., and Kerro Dego, O. (2020). Genetic diversity and virulence characteristics of Staphylococcus aureus isolates from cases of bovine mastitis. Microb. Pathog. 144:104171. doi: 10.1016/j.micpath.2020.104171
Waller, K. P., Aspán, A., Nyman, A., Persson, Y., and Andersson, U. G. (2011). CNS species and antimicrobial resistance in clinical and subclinical bovine mastitis. Vet. Microbiol. 152, 112–116. doi: 10.1016/j.vetmic.2011.04.006
Xu, J., Tan, X., Zhang, X., Xia, X., and Sun, H. (2015). The diversities of staphylococcal species, virulence and antibiotic resistance genes in the subclinical mastitis milk from a single Chinese cow herd. Microb. Pathog. 88, 29–38. doi: 10.1016/j.micpath.2015.08.004
Yang, F., Wang, Q., Wang, X., Wang, L., Li, X., Luo, J., et al. (2016). Genetic characterization of antimicrobial resistance in Staphylococcus aureus isolated from bovine mastitis cases in Northwest China. J. Integr. Agric. 15, 2842–2847. doi: 10.1016/S2095-3119(16)61368-0
Yang, F., Zhang, S., Shang, X., Li, H., Zhang, H., Cui, D., et al. (2020). Short communication: detection and molecular characterization of methicillin-resistant Staphylococcus aureus isolated from subclinical bovine mastitis cases in China. J. Dairy Sci. 103, 840–845. doi: 10.3168/jds.2019-16317
Yang, F., Zhang, S., Shang, X., Wang, X., Yan, Z., Li, H., et al. (2019). Short communication: antimicrobial resistance and virulence genes of Enterococcus faecalis isolated from subclinical bovine mastitis cases in China. J. Dairy Sci. 102, 140–144. doi: 10.3168/jds.2018-14576
Zhang, L., Gao, J., Barkema, H. W., Ali, T., Liu, G., Deng, Y., et al. (2018). Virulence gene profiles: alpha-hemolysin and clonal diversity in Staphylococcus aureus isolates from bovine clinical mastitis in China. BMC Vet. Res. 14:63. doi: 10.1186/s12917-018-1374-7
Keywords: Staphylococcus aureus , coagulase-negative staphylococci, antimicrobial resistance, virulence, bovine mastitis
Citation: Yang F, Shi W, Meng N, Zhao Y, Ding X and Li Q (2023) Antimicrobial resistance and virulence profiles of staphylococci isolated from clinical bovine mastitis. Front. Microbiol. 14:1190790. doi: 10.3389/fmicb.2023.1190790
Edited by:
Xiaodan Huang, Lanzhou University, ChinaReviewed by:
Manuela Oliveira, University of Lisbon, PortugalHuitian Gou, Gansu Agricultural University, China
Newton Valerio Verbisck, Brazilian Agricultural Research Corporation (EMBRAPA), Brazil
Amany Arafa, NRC, Egypt
Copyright © 2023 Yang, Shi, Meng, Zhao, Ding and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xuezhi Ding, ZGluZ3h1ZXpoaUBjYWFzLmNu; Qinfan Li, bGlxZjExMzFAMTYzLmNvbQ==
 Feng Yang
Feng Yang Wenli Shi
Wenli Shi Na Meng
Na Meng Yiyu Zhao1
Yiyu Zhao1 Xuezhi Ding
Xuezhi Ding