
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 06 October 2023
Sec. Systems Microbiology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1190765
Background: Growing evidence has well documented the close association between the gut microbiome and allergic respiratory disease, which has been notably represented by allergic asthma. However, it is unclear whether this association is a causal link. Therefore, we investigated the potential causal associations between the gut microbiome and allergic asthma or other allergic diseases.
Methods: In this study, we performed two-sample Mendelian randomization (MR) analyses by using the publicly available genome-wide association study (GWAS) summary data. Single-nucleotide polymorphisms (SNPs) that significantly correlated were selected as instrumental variables. The inverse variance weighted (IVW) method was used to examine the potential causal gut microbial genera for allergic asthma and other allergic diseases. The robustness of the primary findings of the MR analyses was ensured by using different sensitivity analyses.
Results: Combining the findings from multiple analyses, the host genetic-driven increases in Butyricimonas at the genus level were positively correlated with the risk of allergic asthma. In addition, phylum Bacteroidetes and class Bacteroidia were also found to have negative associations with the risk of allergic asthma; genus Slackia was identified as having potential causal effects with allergic asthma. No clear evidence of pleiotropy and heterogeneity was observed in genus Butyricimonas. Butyricimonas was also found to have an association with allergic rhinitis, but not with other allergic diseases.
Conclusion: Our findings indicate that there are new gut microbial genera that were causally associated with the risk of allergic asthma and other allergic diseases, and offer novel insights into the pathogenesis of allergic respiratory diseases.
Allergic asthma is the most common phenotype of asthma. These patients are usually allergic to aeroallergens from the environment. After repeatedly inhale allergens, leading to acute bronchoconstriction, mediating the immune response produced by IgE and cytokines, promoting mast cells release mediators and recruitment of other cell types, which cause a late asthmatic response (Lotvall et al., 2011; Schatz and Rosenwasser, 2014; Akar-Ghibril et al., 2020). Allergic asthma is often accompanied by allergic rhinitis and conjunctivitis (Verschoor and von Gunten, 2019). Genetic, environmental and stress factors interact in susceptible individuals, resulting in changes in their immune system and enhanced airway responsiveness (Gaffin and Phipatanakul, 2009; Haczku and Panettieri, 2010; Thomsen, 2015). Presently, the treatments for allergic asthma mainly include environmental control measures, allergen immunotherapy, and anti-inflammatory inhaled corticosteroids (ICS). There are still some patients with allergic asthma may require sustained systemic corticosteroids to control their symptoms. These patients will be away from work or school for several days during the onset of allergic asthma and are at risk from the many potential serious side effects of long-term corticosteroid treatment. Therefore, it is imperative to explore the etiology of allergic asthma in order to promote the development of treatment strategies with low damage or even no side effects.
Recently, the causal relationship between the gut microbiome and allergic asthma risk has attracted widespread attention (Smith et al., 2013; Stiemsma and Turvey, 2017; Barcik et al., 2020). Many recent studies had showed that dysbiosis of gut microbiome may lead to the development of allergic disease, as gut microbiome has a significant impact on the maturation of immune cells and resistance to pathogens early (Azad et al., 2013). The gut microbiome seems to be a potential therapeutic target for preventing allergic asthma and other allergic diseases (Atarashi et al., 2011; Cukrowska et al., 2020; Wu et al., 2022). Several studies had shown that allergic asthma patients have dysbiosis of gut microbiome, with a significantly decreased in the relative abundance of some gut microbiome (Fujimura et al., 2016; Arrieta et al., 2018). Metabolites of gut microbiome can also have an impact on allergic asthma. Short-chain fatty acids (SCFAs) are metabolites produced by symbiotic bacteria in the gastrointestinal tract, maintained by fermentation of indigestible digestive products and intestinal amino acids, and they can be absorbed into the circulatory system (Le Poul et al., 2003; Trompette et al., 2014). In both ovalbumin (OVA)- and house dust mite (HDM)-induced models of allergic asthma, SCFAs have been documented to reduce inflammation (Trompette et al., 2014; Cait et al., 2018). However, the human gut microbiome can also produce pro-inflammatory metabolites, such as histamine (Pugin et al., 2017). The number of histamine-secreting bacteria in fecal samples of patients with allergic asthma is significantly higher, which correlates with the severity of the disease (Barcik et al., 2019). The guidelines issued by the World Allergy Organization (WAO) in 2015 indicated that use gut probiotics in not-exclusively breastfed infants places a relatively higher value on possible prevention of allergies (Cuello-Garcia et al., 2016), it can also reduce the risk of developing recurrent wheezing, which has been widely explored in many randomized controlled trials (RCT) (Ivakhnenko and Nyankovskyy, 2013; Moossavi et al., 2018). A meta-analysis involving 19 RCTs found that probiotics could significantly reduce wheeze incidence among infants with allergic asthma (Wei et al., 2020). Another meta-analysis involving 17 RCTs found that gut probiotics could significantly reduce the risk of eczema (Chen et al., 2022). Eczema is the earliest allergic reaction that occurs in childhood and is considered an early sign of a series of allergic diseases, such as allergic rhinitis and allergic asthma (Silverberg and Simpson, 2013; Chen et al., 2022). Although this study has not yet found that supplementation of gut microbiome in infants has a significant preventive effect on allergic asthma and allergic rhinitis, it demonstrates that the gut microbiome is important for forming the immune system and establishing the cell balance. Even so, these evidences are contradictory, they all prove that the gut microbiome is indeed involved in the immune regulation of allergic asthma, but the mechanism is still unclear. The causal association between gut microbiome and allergic asthma has not been fully confirmed.
Mendelian randomization (MR) uses instrumental variables (IVs) and is a common method that is used to examine whether there is a causal association between exposure and complex outcomes (Liu et al., 2022). The results of MR analysis mainly depend on the aggregated data of the genome-wide association studies (GWAS) database, thus greatly reducing the impact of confounding factors. Therefore, in this study we used the most up-to-date GWAS database to conduct an MR analysis to investigate the potential causal associations between gut microbial genera and the risk of allergic diseases including allergic asthma, allergic rhinitis, allergic purpura, allergic conjunctivitis, allergic contact dermatitis, and allergic urticaria (Bernstein et al., 2014; Gaudinski and Milner, 2017).
The overall flowchart for this study is illustrated in Figure 1. We conducted a two-sample MR study to investigate the causal relationships between host genetic-driven gut microbiome and allergic asthma. For significant results, we then explored whether associations exist among the gut microbiome, lung functions (FEV1, FVC, and FEV1/FVC ratio), and five allergic diseases, namely, allergic rhinitis, allergic purpura, allergic conjunctivitis, allergic contact dermatitis, and allergic urticaria.
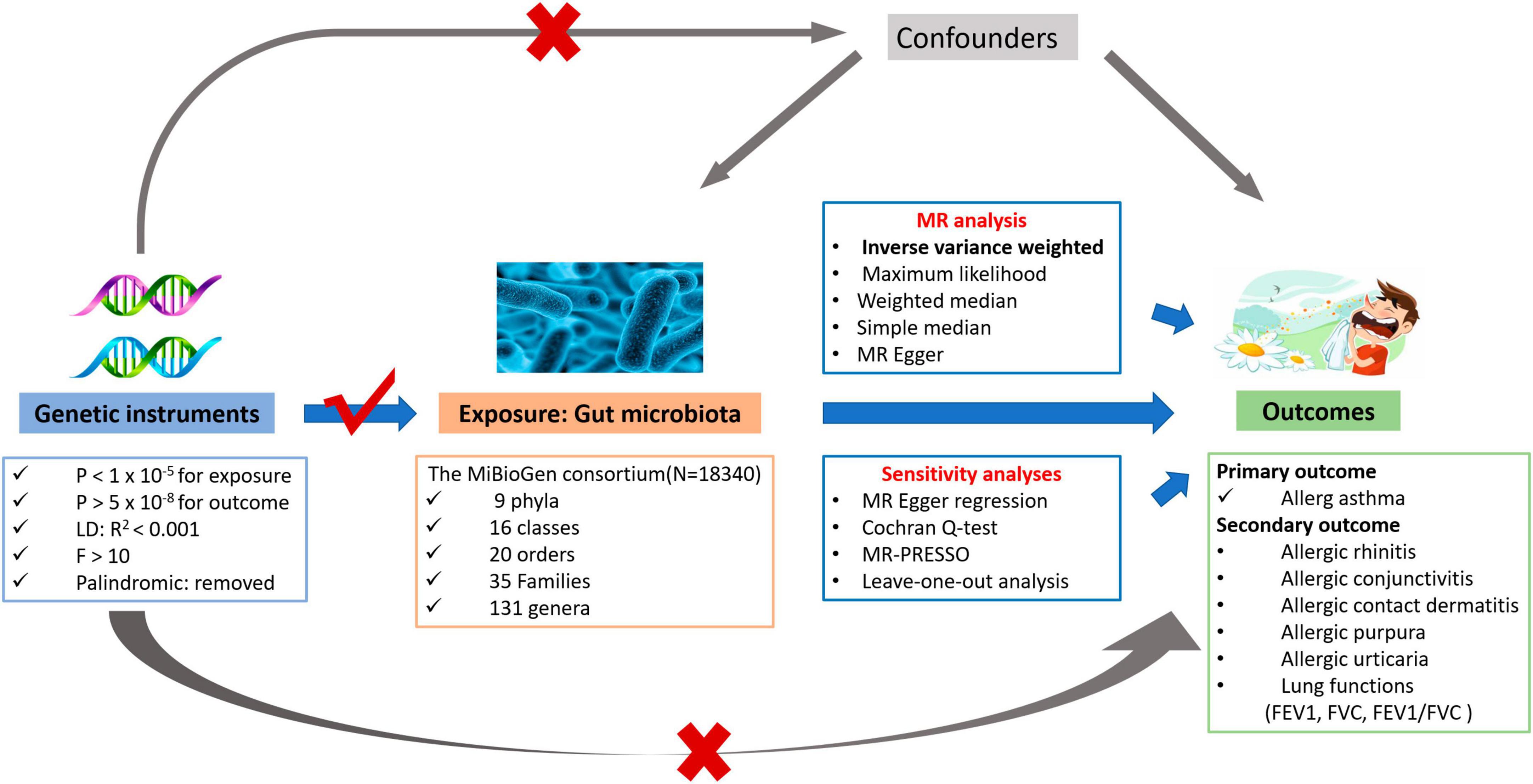
Figure 1. The overview flowchart of hypothesis and study design. This study was a two-sample Mendelian randomization analysis testing the causal effects between gut microbiome and allergic asthma. There are three assumptions required for a valid genetic instrument: the SNPs were significantly correlated with gut microbiome, they were independent of confounders, and the allergic asthma was only related through gut microbiome.
Summary genetic association estimates for the human gut microbiome were obtained from the largest, genome-wide meta-analysis, the MiBioGen consortium study (Kurilshikov et al., 2021), which consisted of 18,340 individuals from 24 population-based cohorts of European (n = 13266), Hispanic, Middle Eastern, Asian, and African ancestries. In addition, the GWAS study was adjusted for sex, age, genetic principal components, and cohort-specific potential microbiome batch effects.
Genome-wide association study summary statistics for allergic asthma and five other allergic diseases were drawn from the FinnGen consortium round 8 databases (Supplementary Table 1) after adjusting for sex, age, 10 principal components, and genotyping batch. FinnGen is a large public-private partnership aiming to collect and analyze genome and health data from 500,000 Finnish biobank participants. The diagnoses were based on ICD-10 and the following study IDs were used to obtain the outcome data: allergic asthma, allergic rhinitis, allergic purpura, allergic conjunctivitis, allergic contact dermatitis and allergic urticaria.
For lung function, we focused on forced expiratory volume in 1-second (FEV1), forced vital capacity (FVC), and FEV1/FVC ratio (Supplementary Table 1). The related summary statistics were extracted from UK Biobank, comprising a sample size of 344,267 participants of European ancestry, which were then adjusted for age, age*age, gender, height, and smoking status.
Based on the previously published studies and getting more acceptable instrumental variables (Sanna et al., 2019), the SNPs association threshold was set to be p < 1.0 × 10–5, but it was not associated with the outcome (p > 5.0 × 10–8). A total of five levels, namely, phylum, class, order, family, and genus, of bacterial taxa were analyzed in our study. Then, to retain the independent instrumental variables, SNPs were clumped and discarded at linkage disequilibrium r2 < 0.001 within a 10,000 kilobase pairs window, which was based on reference data of European ancestry from the 1000 Genomes Project (Genomes Project et al., 2015). Subsequently, weak instrumental variables were removed using the F statistic (F < 10). Finally, after harmonizing the exposure and the outcome data, the palindromic SNPs (A/T, G/C) were also excluded from the MR analysis.
A two-sample MR analysis was conducted using the inverse-variance weighted (IVW) method as the primary analysis. IVW is classically used in combination with the Wald ratio estimates to obtain an unbiased estimate (Burgess et al., 2013). To provide robust evidence of results, four additional methods were employed to validate the findings, namely, the maximum likelihood method (Burgess et al., 2015), weighted median (Bowden et al., 2016), simple median (Burgess et al., 2016), and MR–Egger method (Bowden et al., 2015). The MR–Egger regression analysis allows free evaluation of the non-zero intercept value as an estimated value of average pleiotropic bias, and it gives a consistent estimate even if all of the instrumental variables are invalid. If some instrumental variables were valid, an unbiased estimate of causality could be provided by the weighted median method and maximum likelihood method.
To ensure the robustness of MR findings, The MR–Egger intercept test and Cochrane’s Q heterogeneity method were conducted to detect any potential horizontal pleiotropy (Higgins and Thompson, 2002). We also performed the Pleiotropy Residual Sum and Outlier (MR–PRESSO) test to find and correct the outlier instrumental variables reflecting pleiotropic biases (Verbanck et al., 2018). Additionally, we applied the leave-one-out method to rule out the possibility of the influence of a single SNP on the causality.
All of the analyses were conducted using the TwoSampleMR package and MR–PRESSO package in R software (version 4.1.2). The MR results are presented as odds ratios (ORs) with 95% confidence intervals (CIs). The significant threshold for each level was corrected based on the Bonferroni’s method (p < 0.05/n). A value of p < 0.05 but above the Bonferroni’s–corrected significance threshold was considered suggestive of evidence for a potential causal effect.
Under the suggestive significance threshold of p < 1.0 × 10–5, a total of 1,958 independent SNPs from 175 bacterial taxa (excluding the unknown taxa) were identified after a series of quality control. The 175 bacterial taxa consisted of 11 classes, 26 families, 117 genera, 12 orders, and 9 phyla. To account for multiple testing, we implemented the Bonferroni’s-corrected method to examine for associations with allergic asthma risks using the IVW model, and the results are listed as follows: class p < 4.5 × 10–3 (0.05/11), family p < 1.9 × 10–3 (0.05/26), genus p < 4.2 × 10–4 (0.05/117), order p < 4.2 × 10–3 (0.05/12), and phylum p < 5.6 × 10–3 (0.05/9).
Based on the IVW model, the host genetic-driven increases in Butyricimonas at the genus level were positively associated with a higher risk of allergic asthma (OR = 1.279, 95% CI = 1.12–1.46, p = 2.87 × 10–4, Table 1). The conclusion was confirmed by the results of the maximum likelihood method (OR = 1.283, 95% CI = 1.12–1.47, p = 3.24 × 10–4), weighted median method (OR = 1.31, 95% CI = 1.098–1.563, p = 2.74 × 10–3), and simple median method (OR = 1.27, 95% CI = 1.053–1.532, p = 1.24 × 10–2). In addition, four suggestive causal effects were identified, within the phylum Bacteroidetes (OR = 0.8, 95% CI = 0.673–0.951, p = 1.14 × 10–2), class Bacteroidia (OR = 0.819 95% CI = 0.695–0.965, p = 1.69 × 10–2), and genus Slackia (OR = 1.169, 95% CI = 1.021–1.34, p = 2.42 × 10–2). The details on instrumental variables are summarized in Supplementary Table 2.
In the sensitivity analysis, no clear evidence of pleiotropy (MR–Egger intercept = 0.009, MR–Egger p = 0.654, and MR–PRESSO Global Test p = 0.952) and heterogeneity (Cochran Q = 6.485, p = 0.927) was observed in genus Butyricimonas (Table 1). The results of leave-one-out sensitivity analysis revealed that no single SNP drives the causal associations between the genus Butyricimonas and allergic asthma (Figure 2). Furthermore, no evidence of heterogeneity or pleiotropy bias was noted among the phylum Bacteroidetes, class Bacteroidia, and genus Slackia as well (Figure 2). Thus, the two-sample MR estimates found that the genus Butyricimonas was positively related to allergic asthma risk, phylum Bacteroidetes and class Bacteroidia were negatively related to allergic asthma risk, and genus Slackia was identified as having potential causal effects with allergic asthma (Table 1).
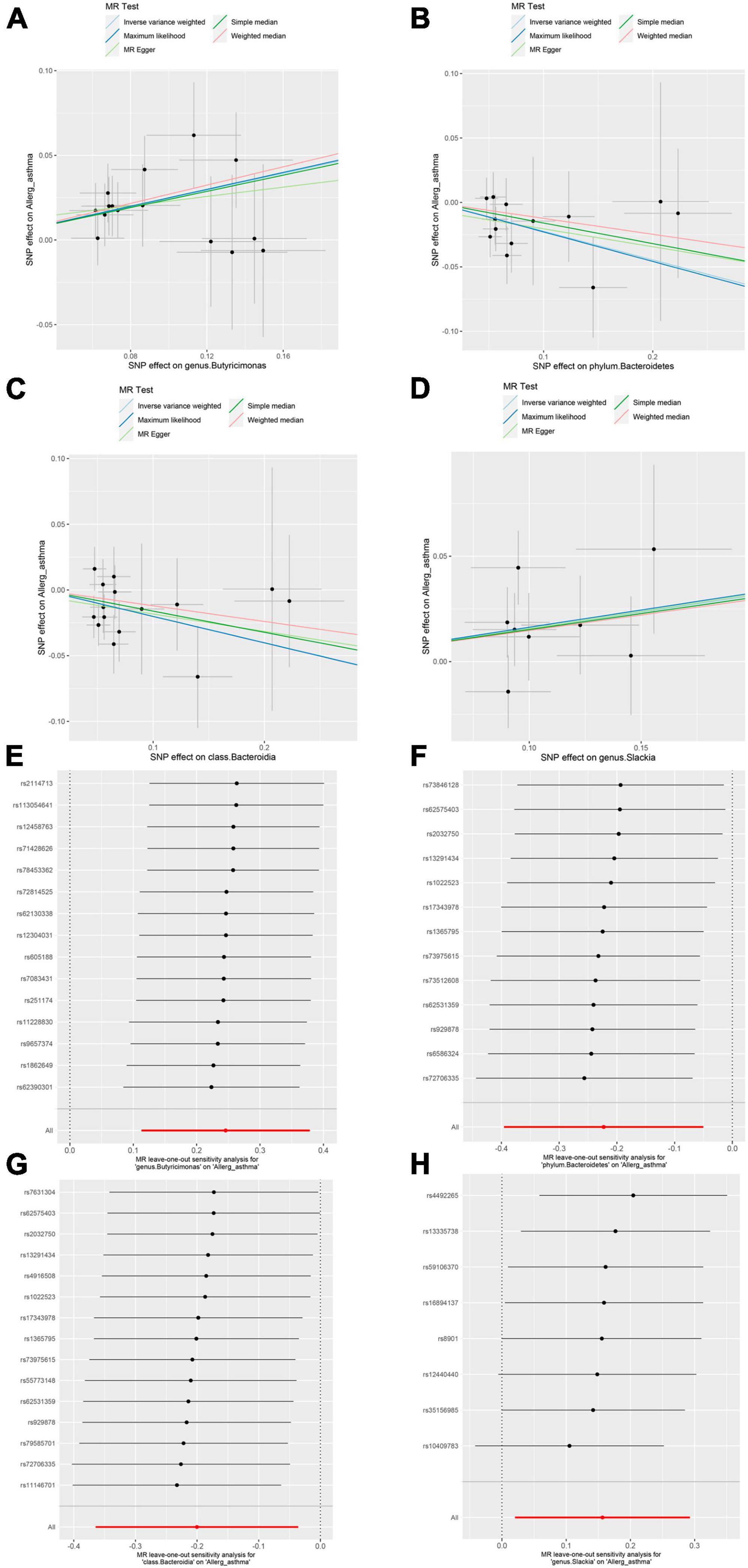
Figure 2. Mendelian randomization (MR) sensitivity analysis for gut microbiome on allergic asthma. Scatterplot of five MR tests between the genus Butyricimonas (A), phylum Bacteroidetes (B), class Bacteroidia (C) and genus Slackia (D) and allergic asthma. MR leave-one-out sensitivity analysis are included for the genus Butyricimonas (E), phylum Bacteroidetes (F), class Bacteroidia (G), and genus Slackia (H) on allergic asthma.
Lung function is one of the important characteristics for asthma diagnosis, with the FEV1, FVC, and FEV1/FVC ratio being particularly important (Kappen et al., 2023). GINA 2021 (Reddel et al., 2022) highlights the relevance of lung function testing for asthma diagnosis. Therefore, we assessed the associations between the gut microbiome and lung function. The five methods of MR analysis suggested that the genus Butyricimonas has no causal effect on FEV1 (OR = 0.981, 95% CI = 0.954–1.008, PIVW = 0.174), FVC (OR = 0.986, 95% CI = 0.961–1.011, PIVW = 0.278) or FEV1/FVC ratio (OR = 0.982, 95% CI = 0.954–1.012, PIVW = 0.234). No evidence was detected indicating a causal relationship between the other four potential gut microbiome and lung function (Figure 3 and Supplementary Table 3). In addition, the sensitivity analysis results supported the robustness of the MR analysis (Supplementary Tables 4, 5).
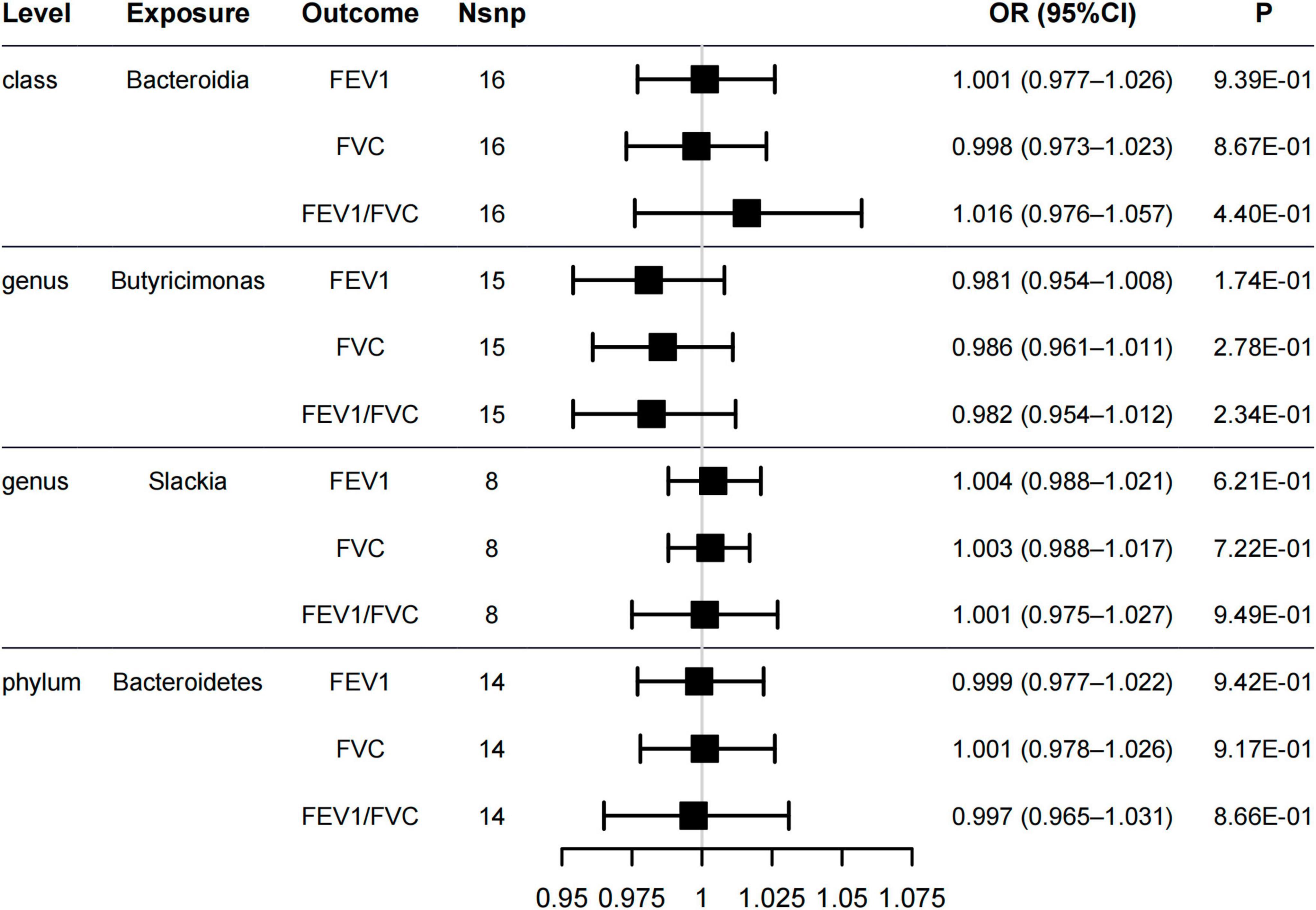
Figure 3. Forest plot of causal relationships estimated for five gut microbiome and lung functions [forced expiratory volume in 1-second (FEV1), forced vital capacity (FVC), and FEV1/FVC ratio] using the inverse variance weighted method.
The prevalence of allergic diseases is increasing worldwide and affects respiratory, digestive, skin, and other systems. However, gut microbiome share a common central role in the risk factors, mechanism, and therapy of these allergic diseases. We evaluated the causal relationship between the five gut microbiome and other allergic diseases, namely, allergic rhinitis, allergic purpura, allergic conjunctivitis, allergic contact dermatitis, and allergic urticaria.
The genus Butyricimonas was associated with allergic rhinitis (OR = 1.192, 95% CI = 1.022–1.392, PIVW = 0.0256, Figure 4), but not with allergic purpura (OR = 1.247, 95% CI = 0.815–1.908, PIVW = 0.31), allergic conjunctivitis (OR = 1.109, 95% CI = 0.987–1.246, PIVW = 0.0806), allergic contact dermatitis (OR = 0.981, 95% CI = 0.782–1.23, PIVW = 0.867), and allergic urticaria (OR = 0.97, 95% CI = 0.748–1.257, PIVW = 0.816). Additionally, no significant causality between the other four gut microbiome and other allergic diseases was found in all of the models (Figure 4 and Supplementary Table 6). Neither pleiotropy bias (MR–Egger p > 0.1, Supplementary Table 7) or heterogeneity (Cochran’s Q p > 0.1, Supplementary Table 8) were found.
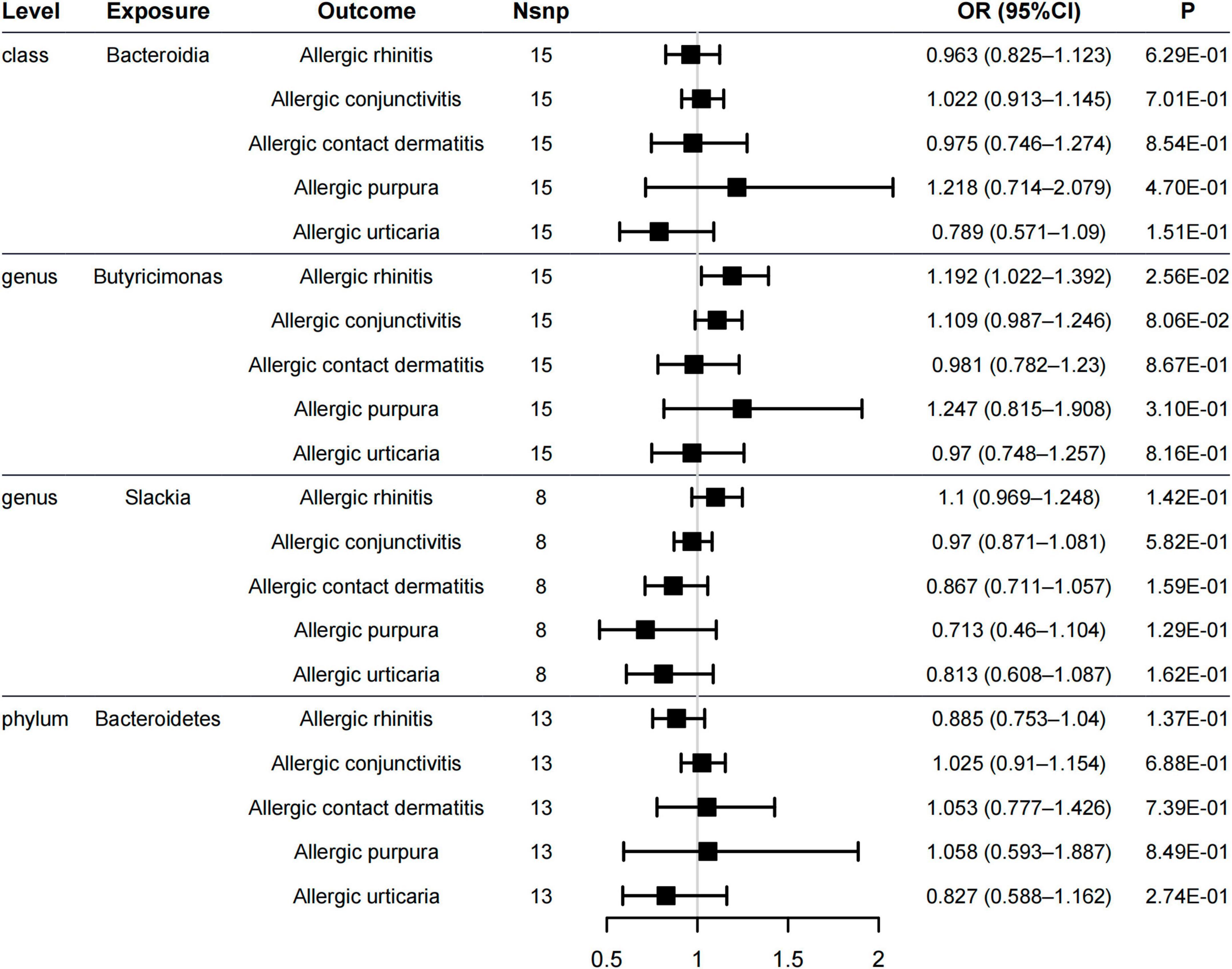
Figure 4. Forest plot of causal relationships estimated for five gut microbiome and other allergic diseases (allergic rhinitis, allergic purpura, allergic conjunctivitis, allergic contact dermatitis, and allergic urticaria) using the inverse variance weighted method.
This MR study found that the genus Butyricimonas was significantly positively associated with a high risk of allergic asthma. In addition, the phylum Bacteroidetes and class Bacteroidia was also found to have negative associations with the risk of allergic asthma. Genus Slackia was identified as having suggestive causal effects with allergic asthma, but more genome-wide data are needed to confirm this result. Therefore, our findings further indicate that gut microbiome has a causal association with allergic asthma, and suggest the regulatory role of gut microbial genera in allergic asthma.
The main metabolite of the genus Butyricimonas is butyrate, which is an important energy source for colon cells. Most of the butyrate is metabolized into energy by those cells. Butyrate is believed to be beneficial to the gut, and it helps to promote the body’s anti-inflammatory and immune response, which can ameliorate allergen-induced airway and improve airway hyperresponsiveness in mice (Theiler et al., 2019; Yip et al., 2021). The severity of allergic asthma seems to be related to butyrate produced in the gut. The evidence showed that in the OVA-induced airway inflammation mouse model, supplementing butyrate bacteria from the feces of infants can reduce the airway inflammation (Arrieta et al., 2015). The phylum Bacteroidetes and class Bacteroidia are colonized bacteria present in the gut of healthy adults. The observational research suggested that the decrease of genus Butyricimonas is related to the development of allergic diseases and the increase of the risk of sensitization (Storro et al., 2011; Abrahamsson et al., 2012; Nylund et al., 2013). The phylum Bacteroidetes and class Bacteroidia has been previously shown to promote the cellular and physical maturation of the developing immune system through its ability to guide the development of CD4+T cells, thus inducing the differentiation of Th1 lineage and the correction of Th1/Th2 imbalance (Penders et al., 2007). Interestingly, contrary to the conclusions of observational studies and systematic review (Barcik et al., 2020; Yip et al., 2021) the results of this MR study show that there is a causal association between gut microbiome and allergic asthma or allergic rhinitis. In particular, genus Butyricimonas may increase the risk of allergic asthma and allergic rhinitis. The significant change in the proportion of gut microbiome and the increasing risk of allergic asthma seems to occur at the same time.
When establishing the causal effects, multi-levels of research evidence should be considered, but observational studies under different conditions are susceptible to confounding factors, thus reducing the accuracy of conclusions. Therefore, the relevance of observational studies cannot be equated with causal correlations. MR avoids the influence of confounding factors through genetic instrumental variables, and it can carry out accurate causal assessment. However, the development of allergic diseases is a long-term process, and more data are needed to support our findings.
Our study is the first MR analysis of gut microbiome, allergic asthma, and multiple allergic diseases. It has several advantages: First, compared with the inherent limitations of observational studies, MR studies are unlikely to be affected by reverse causality and confounding factors. Second, extensive GWAS sample data, two sets of independent IVs, and different methods are applied to causal association assessments to improve reliability. It must be acknowledged that this study also has the following limitations: First, this study is limited to individuals of European descent, and the difference in dietary habits lead to diversity of gut microbiome among different populations, so it may be limited to extrapolate the research findings to other ethnic groups. Second, this study is unable to determine whether overlapping participants were included in the exposure and outcome GWAS used in the two-sample MR analysis. Lastly, this study cannot further answer the different role the genus Butyricimonas played in this MR study and other observational studies.
In conclusion, this MR study shows there are causal associations between gut microbiome and allergic asthma in European populations, as well as a potential causal association between gut microbiome and allergic rhinitis. Several types of gut microbiome were found to be pathogenic or protective factors of allergic asthma, and may have the prospects for the prevention and treatment of allergic asthma. With this research we hope to strengthen the understanding of the characteristics of gut microflora.
The original contributions presented in this study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
WW, YQ, and CY designed the research. WW, YQ, XH, and YR collected and analyzed the data. YQ, XH, and YR performed the literature search. WW and CY drafted the manuscript. YQ and AP supervised the study. All authors were involved in writing the manuscript, contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (No. 82160211), the Jiangxi Provincial Natural Science Foundation (No. 20202ACBL206013), and the Key Research and Development Program of Jiangxi Science and Technology Department (No. 20202BBGL73018).
We want to acknowledge the participants and investigators of the FinnGen and the UK Biobank study. We also thank the members of the MiBioGen consortium for sharing summary-level data publicity available. We also thank Dr. Junhao Tu for his outstanding advice in this manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1190765/full#supplementary-material
Abrahamsson, T., Jakobsson, H., Andersson, A., Bjorksten, B., Engstrand, L., and Jenmalm, M. (2012). Low diversity of the gut microbiota in infants with atopic eczema. J. Allergy Clin. Immunol. 129, 434–40, 40e1–2.
Akar-Ghibril, N., Casale, T., Custovic, A., and Phipatanakul, W. (2020). Allergic endotypes and phenotypes of asthma. J. Allergy Clin. Immunol. Pract. 8, 429–440.
Arrieta, M., Arevalo, A., Stiemsma, L., Dimitriu, P., Chico, M., Loor, S., et al. (2018). Associations between infant fungal and bacterial dysbiosis and childhood atopic wheeze in a nonindustrialized setting. J. Allergy Clin. Immunol. 142, 424–34.e10. doi: 10.1016/j.jaci.2017.08.041
Arrieta, M., Stiemsma, L., Dimitriu, P., Thorson, L., Russell, S., Yurist-Doutsch, S., et al. (2015). Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci. Transl. Med. 7:307ra152. doi: 10.1126/scitranslmed.aab2271
Atarashi, K., Tanoue, T., Shima, T., Imaoka, A., Kuwahara, T., Momose, Y., et al. (2011). Induction of colonic regulatory T cells by indigenous Clostridium species. Science 331, 337–341.
Azad, M., Konya, T., Maughan, H., Guttman, D., Field, C., Sears, M., et al. (2013). Infant gut microbiota and the hygiene hypothesis of allergic disease: Impact of household pets and siblings on microbiota composition and diversity. Allergy Asthma Clin. Immunol. 9:15. doi: 10.1186/1710-1492-9-15
Barcik, W., Boutin, R., Sokolowska, M., and Finlay, B. (2020). The role of lung and gut microbiota in the pathology of asthma. Immunity 52, 241–255.
Barcik, W., Pugin, B., Bresco, M., Westermann, P., Rinaldi, A., Groeger, D., et al. (2019). Bacterial secretion of histamine within the gut influences immune responses within the lung. Allergy 74, 899–909. doi: 10.1111/all.13709
Bernstein, J., Lang, D., Khan, D., Craig, T., Dreyfus, D., Hsieh, F., et al. (2014). The diagnosis and management of acute and chronic urticaria: 2014 update. J. Allergy Clin. Immunol. 133, 1270–1277.
Bowden, J., Davey Smith, G., and Burgess, S. (2015). Mendelian randomization with invalid instruments: Effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 44, 512–525.
Bowden, J., Davey Smith, G., Haycock, P., and Burgess, S. (2016). Consistent estimation in mendelian randomization with some invalid instruments using a weighted median estimator. Genet. Epidemiol. 40, 304–314.
Burgess, S., Bowden, J., Dudbridge, F., and Thompson, S. G. (2016). Assessing the effectiveness of robust instrumental variable methods using multiple candidate instruments with application to Mendelian randomization. arXiv [Preprint]. doi: 10.48550/arXiv.1606.03729
Burgess, S., Butterworth, A., and Thompson, S. (2013). Mendelian randomization analysis with multiple genetic variants using summarized data. Genet. Epidemiol. 37, 658–665.
Burgess, S., Scott, R., Timpson, N., Davey Smith, G., and Thompson, S. (2015). Using published data in Mendelian randomization: A blueprint for efficient identification of causal risk factors. Eur. J. Epidemiol. 30, 543–552. doi: 10.1007/s10654-015-0011-z
Cait, A., Hughes, M., Antignano, F., Cait, J., Dimitriu, P., Maas, K., et al. (2018). Microbiome-driven allergic lung inflammation is ameliorated by short-chain fatty acids. Mucosal Immunol. 11, 785–795. doi: 10.1038/mi.2017.75
Chen, L., Ni, Y., Wu, X., and Chen, G. (2022). Probiotics for the prevention of atopic dermatitis in infants from different geographic regions: A systematic review and Meta-analysis. J. Dermatol. Treatment 33, 2931–2939. doi: 10.1080/09546634.2022.2091101
Cuello-Garcia, C., Fiocchi, A., Pawankar, R., Yepes-Nunez, J., Morgano, G., Zhang, Y., et al. (2016). World Allergy Organization-McMaster University Guidelines for Allergic Disease Prevention (GLAD-P): Prebiotics. World Allergy Organ. J. 9:10. doi: 10.1186/s40413-016-0102-7
Cukrowska, B., Bierla, J., Zakrzewska, M., Klukowski, M., and Maciorkowska, E. (2020). The relationship between the infant gut microbiota and allergy. The role of bifidobacterium breve and prebiotic oligosaccharides in the activation of anti-allergic mechanisms in early life. Nutrients 12:946. doi: 10.3390/nu12040946
Fujimura, K., Sitarik, A., Havstad, S., Lin, D., Levan, S., Fadrosh, D., et al. (2016). Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat. Med. 22, 1187–1191. doi: 10.1038/nm.4176
Gaffin, J., and Phipatanakul, W. (2009). The role of indoor allergens in the development of asthma. Curr. Opin. Allergy Clin. Immunol. 9, 128–135.
Gaudinski, M., and Milner, J. (2017). Atopic dermatitis and allergic urticaria. Immunol. Allergy Clin. North Am. 37, 1–10.
Genomes Project, C., Auton, A., Brooks, L., Durbin, R., Garrison, E., Kang, H., et al. (2015). A global reference for human genetic variation. Nature 526, 68–74.
Haczku, A., and Panettieri, R. Jr. (2010). Social stress and asthma: The role of corticosteroid insensitivity. J. Allergy Clin. Immunol. 125, 550–558. doi: 10.1016/j.jaci.2009.11.005
Higgins, J., and Thompson, S. (2002). Quantifying heterogeneity in a meta-analysis. Stat. Med. 21, 1539–1558.
Ivakhnenko, O., and Nyankovskyy, S. (2013). Effect of the specific infant formula mixture of oligosaccharides on local immunity and development of allergic and infectious disease in young children: Randomized study. Pediatr. Polska 88, 398–404.
Kappen, J., Diamant, Z., Agache, I., Bonini, M., Bousquet, J., Canonica, G., et al. (2023). Standardization of clinical outcomes used in allergen immunotherapy in allergic asthma: An EAACI position paper. Allergy [Epub ahead of print].
Kurilshikov, A., Medina-Gomez, C., Bacigalupe, R., Radjabzadeh, D., Wang, J., Demirkan, A., et al. (2021). Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat. Genet. 53, 156–165.
Le Poul, E., Loison, C., Struyf, S., Springael, J., Lannoy, V., Decobecq, M., et al. (2003). Functional characterization of human receptors for short chain fatty acids and their role in polymorphonuclear cell activation. J. Biol. Chem. 278, 25481–25489.
Liu, B., Ye, D., Yang, H., Song, J., Sun, X., Mao, Y., et al. (2022). Two-sample Mendelian randomization analysis investigates causal associations between gut microbial genera and inflammatory bowel disease, and specificity causal associations in ulcerative colitis or Crohn’s disease. Front. Immunol. 13:921546. doi: 10.3389/fimmu.2022.921546
Lotvall, J., Akdis, C., Bacharier, L., Bjermer, L., Casale, T., Custovic, A., et al. (2011). Asthma endotypes: A new approach to classification of disease entities within the asthma syndrome. J. Allergy Clin. Immunol. 127, 355–360. doi: 10.1016/j.jaci.2010.11.037
Moossavi, S., Miliku, K., Sepehri, S., Khafipour, E., and Azad, M. (2018). The prebiotic and probiotic properties of human milk: Implications for infant immune development and pediatric asthma. Front. Pediatr. 6:197. doi: 10.3389/fped.2018.00197
Nylund, L., Satokari, R., Nikkila, J., Rajilic-Stojanovic, M., Kalliomaki, M., Isolauri, E., et al. (2013). Microarray analysis reveals marked intestinal microbiota aberrancy in infants having eczema compared to healthy children in at-risk for atopic disease. BMC Microbiol. 13:12. doi: 10.1186/1471-2180-13-12
Penders, J., Thijs, C., van den Brandt, P., Kummeling, I., Snijders, B., Stelma, F., et al. (2007). Gut microbiota composition and development of atopic manifestations in infancy: The KOALA Birth Cohort Study. Gut 56, 661–667. doi: 10.1136/gut.2006.100164
Pugin, B., Barcik, W., Westermann, P., Heider, A., Wawrzyniak, M., Hellings, P., et al. (2017). A wide diversity of bacteria from the human gut produces and degrades biogenic amines. Microb. Ecol. Health Dis. 28:1353881. doi: 10.1080/16512235.2017.1353881
Reddel, H., Bacharier, L., Bateman, E., Brightling, C., Brusselle, G., Buhl, R., et al. (2022). Global initiative for asthma strategy 2021: Executive summary and rationale for key changes. Eur. Respir. J. 59: 2102730.
Sanna, S., van Zuydam, N., Mahajan, A., Kurilshikov, A., Vich Vila, A., Vosa, U., et al. (2019). Causal relationships among the gut microbiome, short-chain fatty acids and metabolic diseases. Nat. Genet. 51, 600–605.
Schatz, M., and Rosenwasser, L. (2014). The allergic asthma phenotype. J. Allergy Clin. Immunol. Pract. 2, 645–8; quiz9.
Silverberg, J., and Simpson, E. (2013). Association between severe eczema in children and multiple comorbid conditions and increased healthcare utilization. Pediatr. Allergy Immunol. 24, 476–486. doi: 10.1111/pai.12095
Smith, P., Howitt, M., Panikov, N., Michaud, M., Gallini, C., Bohlooly, Y., et al. (2013). The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341, 569–573.
Stiemsma, L., and Turvey, S. (2017). Asthma and the microbiome: Defining the critical window in early life. Allergy Asthma Clin. Immunol. 13:3.
Storro, O., Oien, T., Langsrud, O., Rudi, K., Dotterud, C., and Johnsen, R. (2011). Temporal variations in early gut microbial colonization are associated with allergen-specific immunoglobulin E but not atopic eczema at 2 years of age. Clin. Exp. Allergy 41, 1545–1554. doi: 10.1111/j.1365-2222.2011.03817.x
Theiler, A., Bärnthaler, T., Platzer, W., Richtig, G., Peinhaupt, M., Rittchen, S., et al. (2019). Butyrate ameliorates allergic airway inflammation by limiting eosinophil trafficking and survival. J. Allergy Clin. Immunol. 144, 764–776. doi: 10.1016/j.jaci.2019.05.002
Thomsen, S. F. (2015). Genetics of asthma: An introduction for the clinician. Eur. Clin. Respir. J. 2:24643. doi: 10.3402/ecrj.v2.24643
Trompette, A., Gollwitzer, E., Yadava, K., Sichelstiel, A., Sprenger, N., Ngom-Bru, C., et al. (2014). Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat. Med. 20, 159–166. doi: 10.1038/nm.3444
Verbanck, M., Chen, C., Neale, B., and Do, R. (2018). Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 50, 693–698.
Verschoor, D., and von Gunten, S. (2019). Allergy and atopic diseases: An update on experimental evidence. Int. Arch. Allergy Immunol. 180, 235–243.
Wei, X., Jiang, P., Liu, J., Sun, R., and Zhu, L. (2020). Association between probiotic supplementation and asthma incidence in infants: A meta-analysis of randomized controlled trials. J. Asthma 57, 167–178.
Wu, Z., Mehrabi Nasab, E., Arora, P., and Athari, S. (2022). Study effect of probiotics and prebiotics on treatment of OVA-LPS-induced of allergic asthma inflammation and pneumonia by regulating the TLR4/NF-kB signaling pathway. J. Transl. Med. 20:130. doi: 10.1186/s12967-022-03337-3
Keywords: allergic asthma, allergic diseases, causal relationship, gut microbial genera, Mendelian randomization
Citation: Wan W, Qiu Y, Huang X, Ren Y, Peng A and Yang C (2023) Causal relationship between Butyricimonas and allergic asthma: a two-sample Mendelian randomization study. Front. Microbiol. 14:1190765. doi: 10.3389/fmicb.2023.1190765
Received: 21 March 2023; Accepted: 25 September 2023;
Published: 06 October 2023.
Edited by:
George Tsiamis, University of Patras, GreeceReviewed by:
Niki Papapostolou, University General Hospital Attikon, GreeceCopyright © 2023 Wan, Qiu, Huang, Ren, Peng and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chunping Yang, NTI0Nzg5MEBxcS5jb20=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.