- 1Department of Pathology, Microbiology and Immunology, New York Medical College, Valhalla, NY, United States
- 2Department of Basic and Clinical Sciences, Albany College of Pharmacy and Health Sciences, Albany, NY, United States
- 3Electron Microscopy Laboratory, Department of Pathology, Westchester Medical Center, Valhalla, NY, United States
Francisella tularensis is a highly virulent Gram-negative bacterium that causes the fatal zoonotic disease tularemia. The mechanisms and signaling pathways leading to the absent in melanoma 2 (Aim2) inflammasome activation have been elegantly elucidated using Francisella novicida as a model. Although not pathogenic for humans, F. novicida can cause tularemia in mice, and the inflammatory response it triggers is the polar opposite to that observed in mice infected with F. tularensis strains. This study aimed to understand the mechanisms of Aim2 inflammasome activation in F. tularensis-infected macrophages. The results reveal that macrophages infected with the F. tularensis live vaccine strain (LVS) induce lower levels of Aim2-dependent IL-1β than those infected with F. novicida. The suppression/weak activation of Aim2 in F. tularensis LVS-infected macrophages is due to the suppression of the cGAS-STING DNA-sensing pathway. Furthermore, the introduction of exogenous F. tularensis LVS DNA into the cytosol of the F. tularensis LVS-infected macrophages, alone or in conjunction with a priming signal, failed to restore IL-1β levels similar to those observed for F. novicida-infected macrophages. These results indicated that, in addition to the bacterial DNA, DNA from some other sources, specifically from the damaged mitochondria, might contribute to the robust Aim2-dependent IL-1β levels observed in F. novicida-infected macrophages. The results indicate that F. tularensis LVS induces mitophagy that may potentially prevent the leakage of mitochondrial DNA and the subsequent activation of the Aim2 inflammasome. Collectively, this study demonstrates that the mechanisms of Aim2 inflammasome activation established for F. novicida are not operative in F. tularensis.
Introduction
Francisella tularensis (F. tularensis) is a non-motile, facultative intracellular, Gram-negative bacterium that causes a fatal zoonotic disease, tularemia. Francisella tularensis is classified into F. tularensis subspecies tularensis, F. tularensis subspecies holarctica, and F. tularensis subspecies mediasiatica. Francisella novicida is classified as a separate species because it is genetically and phenotypically different from F. tularensis (Kingry and Petersen, 2014). While F. tularensis subspecies tularensis and F. tularensis subspecies holarctica cause tularemia in humans, F. novicida is associated with disease in immunocompromised individuals only (Pechous et al., 2009; Sjödin et al., 2012). Francisella tularensis subspecies tularensis (also known as Type A strains) has a very high virulence in humans, with a lethal dose that could be lower than 10 colony colony-forming (CFUs) and is responsible for the vast majority of tularemia cases in North America. Due to its extremely high virulence, high rate of morbidity and mortality, and ease of aerosolization, F. tularensis subspecies tularensis SchuS4 strain is classified by the CDC as a Tier 1 Category A Select Agent (Ellis et al., 2002). Francisella tularensis subspecies holarctica (Type B strains) cause tularemia in healthy individuals. However, they are less virulent compared to F. tularensis subspecies tularensis strains, with a lethal dose of 10,000 CFUs or less (Pechous et al., 2009). The live vaccine strain (LVS) was developed by Russian scientists through serial in vivo and in vitro passages of F. tularensis subspecies holarctica, resulting in attenuation of its virulence in humans. The F. tularensis LVS was later gifted to the United States by the former Soviet Union in the 1950s (Eigelsbach and Downs, 1961). Francisella tularensis LVS shares a high DNA sequence homology and undergoes an identical intramacrophage replication cycle with highly virulent F. tularensis strains such as SchuS4. Hence, in research laboratories, F. tularensis LVS is used to study the virulence factors of Francisella and the pathogenesis of tularemia (Jones et al., 2014).
Francisella tularensis is transmitted to humans or animals by contact with infected materials, consumption of contaminated food and water, inhalation of aerosols caused by the disruption of materials containing the organism, and through bites of infected ticks, mosquitoes, and flies (Dennis et al., 2001). Depending on the route of bacterial entry, the clinical manifestations range from ulceroglandular, oculoglandular, and respiratory forms of tularemia (Dennis et al., 2001). The extreme virulence of F. tularensis ultimately depends on its ability to enter, persist, and replicate within the host cells without evoking an immediate innate immune response. Francisella enters macrophages through phagocytosis by forming asymmetric pseudopod loops (Clemens et al., 2005). Following internalization, bacteria initially reside in the phagosome and prevent its fusion with lysosomes (Golovliov et al., 2003; Clemens et al., 2004; Santic et al., 2005). Within 30 min to several hours, phagosomal membrane disruption and bacterial escape to the cytosol occur. Once in the cytosol, Francisella undergoes several rounds of replication that end with the death of the host cell (Lai et al., 2001; Oyston et al., 2004; Mariathasan et al., 2005; Santic et al., 2010).
In cytosol, the sensing of foreign nucleic acids is a major anti-microbial immune mechanism. The nucleic acid sensors include the retinoic acid-inducible gene (RIG)-I-like receptors (RLRs), NOD-like receptors (NLRs), and absent in melanoma 2-like receptors (ALRs). Additionally, DNA sensors that induce the production of Type I interferons (Type-I IFNs) include cyclic-GMP-AMP synthase (cGAS), the stimulator of interferon genes (STING), DNA-dependent activator of IFN-regulatory factors (DAI), and leucine-rich repeat flightless-interacting protein 1 (LRRFIP1; Takaoka et al., 2007; Burdette et al., 2011). All these sensors recognize pathogen-associated molecular patterns (PAMPs) and/or sterile endogenous molecules produced by the host cell tissue injury that are called damage-associated molecular patterns (DAMPs). Recognition of PAMPs or DAMPs initiates several signaling pathways, leading to the induction of proinflammatory cytokines and interferons via MyD88-dependent NF-κB and interferon-regulatory factors (IRFs) pathways, respectively (Kaiser et al., 2008). Absent in Melanoma 2 (AIM2) is an interferon inducible ALR family protein. AIM2 contains a C-terminal HIN-200 domain, which binds directly to cytosolic double-stranded DNA and an N-terminal pyrin motif, which recruits and interacts with ASC (Fernandes-Alnemri et al., 2009). AIM2 detects bacterial and viral DNA and senses mislocalized self-DNA released into the cytosol during inflammatory and autoimmune diseases (Hornung et al., 2009).
The majority of the studies related to cytosolic recognition of Francisella and activation of the AIM2 inflammasome have been done using F. novicida. Once the host cell is infected with F. novicida, a robust AIM2-dependent inflammasome response is induced, culminating in the production of a potent proinflammatory cytokine response and pyroptosis to clear the infection. In agreement with cell-based assays, Aim2, Caspase-1, and Asc are required for survival during F. novicida infection in mice (Mariathasan et al., 2005; Fernandes-Alnemri et al., 2010; Jones et al., 2010; Rathinam et al., 2010). Mice lacking either caspase-1 or Asc have a higher bacterial burden in the lung, liver, and spleen after 2 days, and 100% of them die within 3–5 days post-infection. In contrast, 25–30% of wild-type mice survive the infection(Mariathasan et al., 2005; Fernandes-Alnemri et al., 2010; Jones et al., 2010). Additionally, several studies using F. novicida have shown the critical protective role of inflammasome-induced IL-1β cytokine (Mariathasan et al., 2005; Li et al., 2006; Henry and Monack, 2007).
It has been shown that there is a delay in the activation of the inflammasome and significantly lower levels of IL-1β are produced by macrophages infected with F. tularensis LVS, as compared to those infected with F. novicida (Dotson et al., 2013). Similar to F. tularensis LVS, F. tularensis SchuS4-infected macrophages also fail to induce early inflammasome activation and IL-1β production. On the other hand, F. novicida-infected macrophages strongly activate the inflammasome. It has been established that infection of murine macrophages with F. novicida leads to the induction of IFN-β in a cGAS-STING-dependent manner, which subsequently binds to interferon-alpha/beta receptor in an autocrine fashion. This binding leads to the induction of multiple transcription factors that drive the expression of interferon-stimulated genes (ISGs) and guanylate binding proteins (GBPs), causing bacterial cell lysis by damaging the bacterial cell wall and the release of bacterial DNA in the host cell cytosol (Henry et al., 2007; Man et al., 2015; Storek et al., 2015). The released DNA is sensed by AIM2, which leads to the recruitment of the adaptor ASC and the cleavage of pro-caspase1 into its active form. The active caspase1 acts as a biological scissor and cleaves immature pro-IL-1β and pro-IL-18 into their active forms. Caspase1 also cleaves gasdermin D (GSDMD) into N and C terminal domains. The cleaved N-terminal domain of GSDMD forms pores in the cell membrane, that allows the secretion of bioactive forms of IL-1β and IL-18. The potassium efflux caused by the perforation of cell membrane eventually leads to the lytic form of cell death known as pyroptosis (Shi et al., 2015). Thus, the mechanisms and signaling pathways involved in AIM2 activation are known for the human avirulent F. novicida strain. However, these mechanisms remain unknown for the human virulent F. tularensis strains. This study aimed to investigate the innate immune evasion mechanisms of F. tularensis LVS, specifically those that lead to suppression/weak activation of AIM2 inflammasome and contribute to restricting early inflammation, and how these mechanisms differ between F. tularensis LVS and F. novicida.
Materials and methods
Animals
Six to eight-week-old mice were used in experiments. Aim2−/− [B6. 129P2-Aim2Gt(CSG445)Byg/J; Stock number:013144, Jackson Laboratory] and the corresponding wild-type C57BL/6 mice (000664 C57BL/6 J, Jackson Laboratory) were housed in a pathogen-free environment in the Animal Resource Facility of New York Medical College. All experiments were conducted according to the protocols approved by the institutional animal care and use committee (IACUC).
Bacterial strains
All work with F. tularensis subspecies holarctica LVS and F. novicida (BEI Resources, Manassas, VA, United States) was carried out in a Bio-Safety Level-2 laboratory. Bacterial stocks were prepared by growing bacteria on Mueller-Hinton (MH) agar plates at 37°C with 5% CO2. The individual colonies were picked from MH-agar plates and grown overnight in MH-broth (containing 10% glucose, 0.021% anhydrous calcium chloride, 0.000138% hydrous magnesium chloride, 2.5% ferric pyrophosphate, and 2.5% Isovitalex). Aliquots of mid-log phase bacteria (OD600 of 0.2) were snap-frozen and stored at −80°C. A vial of the frozen stock was thawed at 37°C and then plated on MH-agar and grown at 37°C with 5% CO2 for 48 h for use in the experiments.
Bone marrow-derived macrophages and cell culture assays
Bone marrow cells isolated from 6- to 8-week-old wild-type and Aim2−/− knockout mice were differentiated into bone marrow-derived macrophages (BMDMs) as previously described (Toda et al., 2021). Briefly, bone marrow cells were cultured in Dulbecco’s modified Eagle medium (DMEM) containing 10% heat-inactivated fetal bovine serum (FBS), 2% L-glutamine, 1% sodium pyruvate, 1% HEPES, and 20% medium conditioned with L929 cells. The differentiated BMDMs were plated in 12- or 24-well plates and incubated overnight at 37°C in the presence of 5% CO2 prior to use in experiments. Immortalized Gasdermin D (Gsdmd−/−) BMDMs and their corresponding wild-type immortalized BMDMs were kindly provided by Dr. Vijay Rathinam, UCONN Health. BMDMs from Sting−/− mice were kindly provided by Dr. Penghua Wang, UCONN Health.
The BMDMs were infected with Francisella cultures at a multiplicity of infection (MOI) of 100:1 (Bacteria: cell ratio). Infection was synchronized by centrifuging the plate at 1,000 rpm at 4°C for 10 min, followed by incubation for 2 h at 37°C with 5% CO2. After 2 h, the medium was replaced with medium containing 250 μg/mL of gentamicin to kill all the extracellular and adherent bacteria. The plate was incubated again for 1 h at 37°C in the presence of 5% CO2. After 1 h of incubation, the medium was replaced with medium without any antibiotic. Culture supernatants were collected at 6-, 12-, and 24-h post-infection and stored at −80°C for cytokines analysis. Cells were lysed at 4- and 24-h post-infection or as otherwise indicated, with 0.1% sodium deoxycholate, diluted 10-fold serially, plated on MH-chocolate agar plates, and incubated at 37°C in the presence of 5% CO2 for 48 h. The colonies were counted, and the results were expressed as Log10 colony forming units (CFU)/mL.
Cell death assays
Bone marrow-derived macrophages were seeded in a 24-well plate and incubated at 37°C in the presence of 5% CO2 overnight. The following day, the BMDMs were infected with bacteria at an MOI of 100. Cell death was measured by lactate dehydrogenase (LDH) release in the cell culture supernatants using the CyQUANT LDH Cytotoxicity Assay Kit (Invitrogen) according to the manufacturer’s instructions.
Isolation of Francisella tularensis genomic DNA
The genomic DNA from F. tularensis LVS was purified using the Genomic DNA Mini Kit (Invitrogen) according to the manufacturer’s instructions. DNA concentration and purity were measured with a spectrophotometer and stored at −20°C for future use.
DNA transfection
Wild-type BMDMs (1 × 106 cells/well) were infected with F. tularensis LVS or F. novicida for 2 h and then transfected with 1, 4, and 8 μg of F. tularensis LVS DNA using DOTAP liposomal transfection reagent. One microgram of F. tularensis LVS DNA is approximately equivalent to 1.6 million copies of the F. tularensis LVS genome. BMDMs infected with F. tularensis LVS but not transfected with DOTAP: DNA and uninfected BMDMs were used as controls. Cell culture supernatants were collected at various time intervals post-infection to determine levels of IL-1β, IFN-β, and TNF-α by ELISA.
Cytokines analysis
Cell culture supernatants were collected from the infected and/or DNA-transfected BMDMs after 6, 12, and 24 h. Interferon-beta (IFN-β), IL-1β, and TNF-α levels were measured using LEGEND MAX™ Mouse ELISA Kits (Biolegend) according to the manufacturer’s instructions. The results were expressed as pg./mL.
Western blot analysis
Bone marrow-derived macrophages were infected with F. tularensis LVS or F. novicida for 6 h and then lysed using RIPA lysis buffer containing a protease and phosphatase inhibitor cocktail. The protein concentrations were determined by the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific, United States). An equal amount of protein from each sample were loaded onto a 10% Sodium dodecyl-sulfate polyacrylamide gel, and separated by electrophoresis. The proteins were then transferred to a 0.22 μM polyvinylidene difluoride (PVDF) membrane. The membrane was blocked for 1 h in 5% nonfat milk. The membrane was incubated overnight at 4°C with primary antibodies diluted in 5% Bovine Serum Albumin (BSA) against PINK1 (Novus Biologicals, United States), LC3A/B, and β-actin (Cell Signaling Technology, United States). The following day, the membrane was washed and incubated with 1:20,000 HRP-conjugated secondary antibodies at room temperature for 1 h. The membrane was washed and then incubated with Pierce ECL Plus Substrate. The protein bands were viewed using an X-ray Imager. The band intensities on the blots were quantified using the Bio-Rad Image Lab™ Software Version 3.0.
Transmission electron microscopy
The BMDMs were infected with either F. tularensis LVS or F. novicida at an MOI of 100, as described previously. The uninfected BMDMs were used as controls. At 6- and 12-h post-infection, the cells were prepared for TEM as described earlier (Steele et al., 2019) and then examined under the transmission electron microscope (Hitachi HT7700). The identification of F. tularensis LVS and F. novicida was made based on the size, shape, and location of the bacteria inside the macrophages. Detection of autophagic vacuoles characterized by the presence of double-membrane vacuoles; the extent of mitophagy characterized by engulfment of mitochondria by autophagic vacuoles, and mitochondrial dynamics defined by the presence of elongated and fragmented mitochondria was performed. Quantification of the number of autophagic vacuoles, engulfed mitochondria, and elongated and fragmented mitochondria was performed by counting these structures in 10 micrographs per group.
Statistical analysis
All data were statistically analyzed by one-way ANOVA followed by Tukey–Kramer multiple comparison test. The data were presented as Mean ± Standard error of the mean (SEM), and the differences between the experimental groups were considered statistically significant at a p < 0.05 level.
Results
Francisella tularensis LVS-infected macrophages produce low levels of IL-1β
Bone marrow-derived macrophages from wild-type and Aim2−/− mice and immortalized Gsdmd−/− BMDMs and their wild-type counterparts were infected with 100 MOI of F. tularensis LVS and F. novicida. Uninfected macrophages served as negative controls. The culture supernatants from the infected and control BMDMs were analyzed 24 h post-infection for secreted levels of IL-1β. F. tularensis LVS-infected wild-type BMDMs induced significantly low levels of IL-1β as compared to F. novicida-infected BMDMs. No detectable levels of IL-1β were observed in the culture supernatants of F. tularensis LVS-infected Aim2−/− BMDMs, while in those from F. novicida-infected Aim2−/− BMDMs, these levels were significantly lower than those observed for the wild-type BMDMs (Figure 1A). A trend similar to that observed for the wild-type and Aim2−/− BMDMs was also observed for the immortalized wild-type and Gsdmd−/− macrophages. However, the magnitude of IL-1β response was much lower than that observed for the primary BMDMs (Figure 1B). Overall, these results demonstrate that F. tularensis LVS-infected primary or immortalized BMDMs produce very low levels of Aim2-dependent IL-1β. On the other hand, very high levels of IL-1β observed in F. novicida-infected BMDMs were dependent on Aim2 and GsdmD.
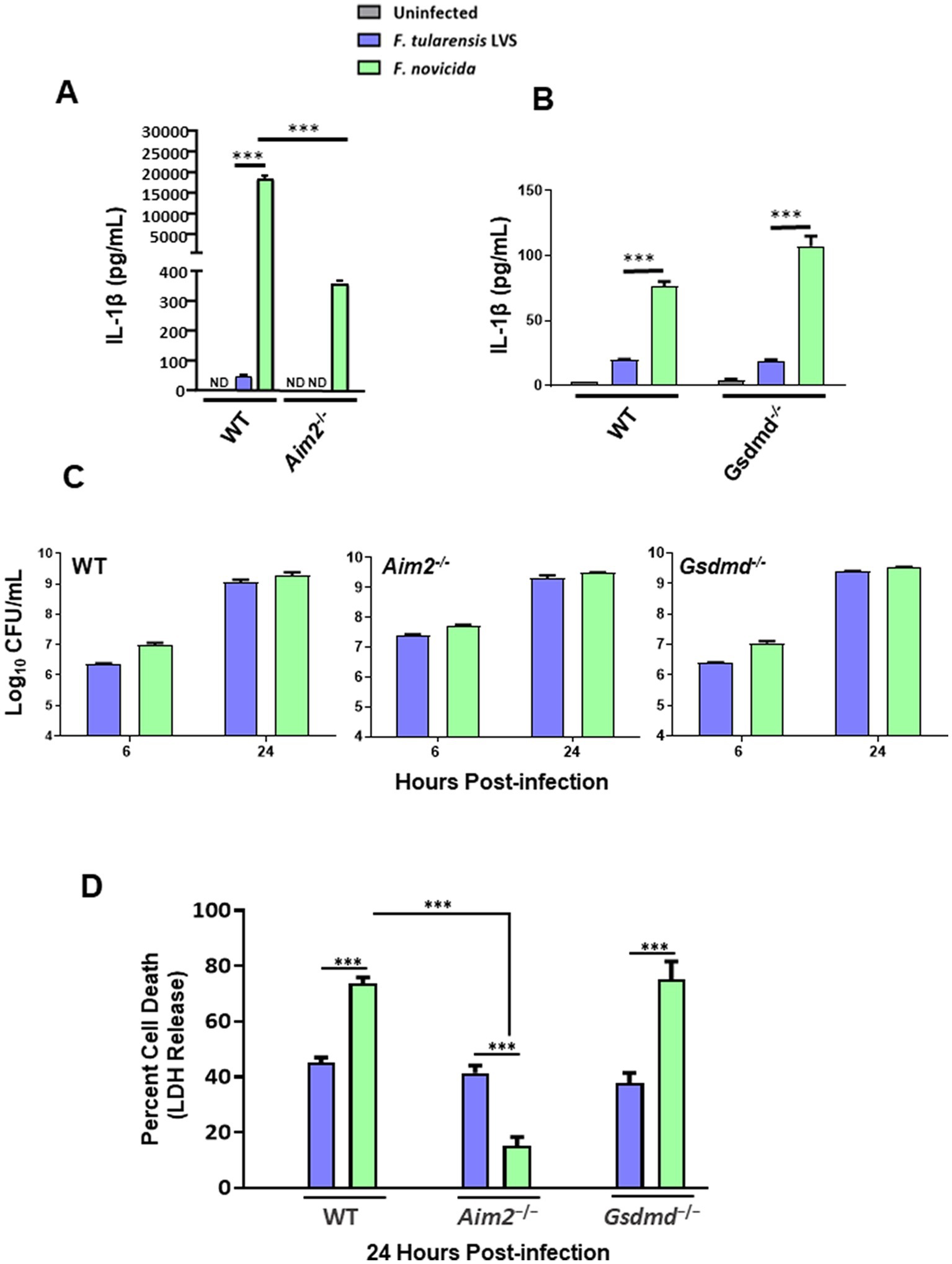
Figure 1. Francisella tularensis live vaccine strain (LVS) infection in macrophages leads to low levels of IL-1β production. Wild-type (WT), and Aim2−/− bone marrow-derived macrophages (BMDMs) (A) and immortalized WT and Gsdmd−/− BMDMs (B) were infected with F. tularensis LVS or Francisella novicida (MOI 100). ELISA was used to analyze the secreted levels of IL-1β in the culture supernatants 24 h post-infection. Results are expressed as pg./mL (mean ± SEM) and are cumulative of three independent experiments with up to six biological replicates. The p values were determined using one-way ANOVA. ***p < 0.001. (C) The same cell lines were infected with F. tularensis LVS or F. novicida (MOI 100) for 2 h, followed by gentamycin treatment for an hour to kill extracellular bacteria. Intracellular bacteria were enumerated at 6 and 24 h post-infection. Results are expressed as Log10 CFU/mL (mean ± SEM) and are cumulative from two independent experiments, each with three biological replicates. (D) The cell culture supernatants collected at 24 h from WT and Aim2−/− BMDMs and immortalized Gsdmd−/-BMDMs infected with F. tularensis LVS, or F. novicida (MOI 100) were analyzed for LDH release. Results are expressed as a percentage of LDH release and are cumulative from three independent experiments with up to six biological replicates. The p-values were determined using one-way ANOVA. ***p < 0.001. ND, not detected.
Differences in IL-1β levels in Francisella tularensis LVS and Francisella novicida-infected BMDMs are not due to differences in intracellular bacterial replication
After observing that IL-1β levels were much lower following infection with F. tularensis LVS than with F. novicida, we investigated whether this discrepancy was due to differences in intracellular bacterial replication. To determine this, wild-type and Aim2−/− BMDMs, and immortalized wild-type as well as Gsdmd−/− BMDMs were infected with F. tularensis LVS or F. novicida at 100 MOI. The BMDMs were lysed at 6- and 24-h post-infection to enumerate intracellularly replicated bacteria. It was found that equal numbers of F. tularensis LVS and F. novicida were recovered from infected wild-type, Aim2−/− and Gsdmd−/− macrophages at 6- and 24-h post-infection (Figure 1C). These results demonstrate that the differences in IL-1β levels in F. tularensis LVS- and F. novicida-infected BMDMs are not due to differences in the numbers of intracellular bacteria.
Francisella tularensis-infected BMDMs exhibit lower cell death than Francisella novicida-infected BMDMs
Previous studies have shown that F. novicida infection induces pyroptotic cell death, as indicated by elevated LDH release in the culture supernatants (Fernandes-Alnemri et al., 2010; Man et al., 2015, 2016). To investigate this further, we measured LDH release as a readout for pyroptosis in wild-type, Aim2−/− and immortalized Gsdmd−/− BMDMs infected with F. tularensis LVS or F. novicida at 24 h post-infection. In wild-type BMDMs, nearly 40% of F. tularensis LVS-infected BMDMs underwent cell death, as compared to almost 70% of F. novicida-infected BMDMs. Additionally, unlike F. tularensis LVS-infected BMDMs, the cell death of F. novicida-infected BMDMs was dependent on Aim2. Furthermore, the cell death observed in F. novicida was independent of GsdmD, as the extent of cell death in the immortalized Gsdmd−/− BMDMs was similar to that observed for wild-type BMDMs infected with F. novicida (Figure 1D). Overall, these results indicate that significantly higher F. novicida-infected BMDMs undergo Aim2-dependent but GsdmD-independent cell death than F. tularensis LVS-infected BMDMs.
Francisella tularensis LVS-infected BMDMs produce low levels of IFN-β
Our previous results demonstrated that F. tularensis LVS weakly activates Aim2 inflammasome, which may impair the downstream events such as cleavage of GsdmD, pyroptotic cell death, and the release of the secreted active form of IL-1β. We next examined the impact of F. tularensis LVS infection on the cGAS-STING pathway upstream of Aim2 by determining the levels of IFN-β in wild-type, Aim2−/−, Sting−/− and immortalized Gsdmd−/− BMDMs. The results show that similar to IL-1β, F. tularensis LVS induces small quantities of IFN-β in the wild-type BMDMs, and these levels were similar to those observed in Aim2−/− BMDMs. However, no detectable levels of IFN-β were observed in Sting−/− BMDMs infected with F. tularensis LVS. In contrast, F. novicida-infected wild-type BMDMs produced very high levels of IFN-β in a STING-dependent manner, and these levels remained significantly elevated in Aim2−/− BMDMs at 12 and 24 post-infection (Figures 2A,B) and in immortalized Gsdmd−/− BMDMs 24 h post-infection (Figure 2C). Overall, these results demonstrate that BMDMs infected with F. tularensis LVS produce very low levels of IFN-β compared to F. novicida-infected cells in a STING-dependent manner. These results suggest that suppression or low activation of the Aim2-inflammasome in F. tularensis LVS-infected BMDMs could result from a weak activation of the cGAS-STING pathway.
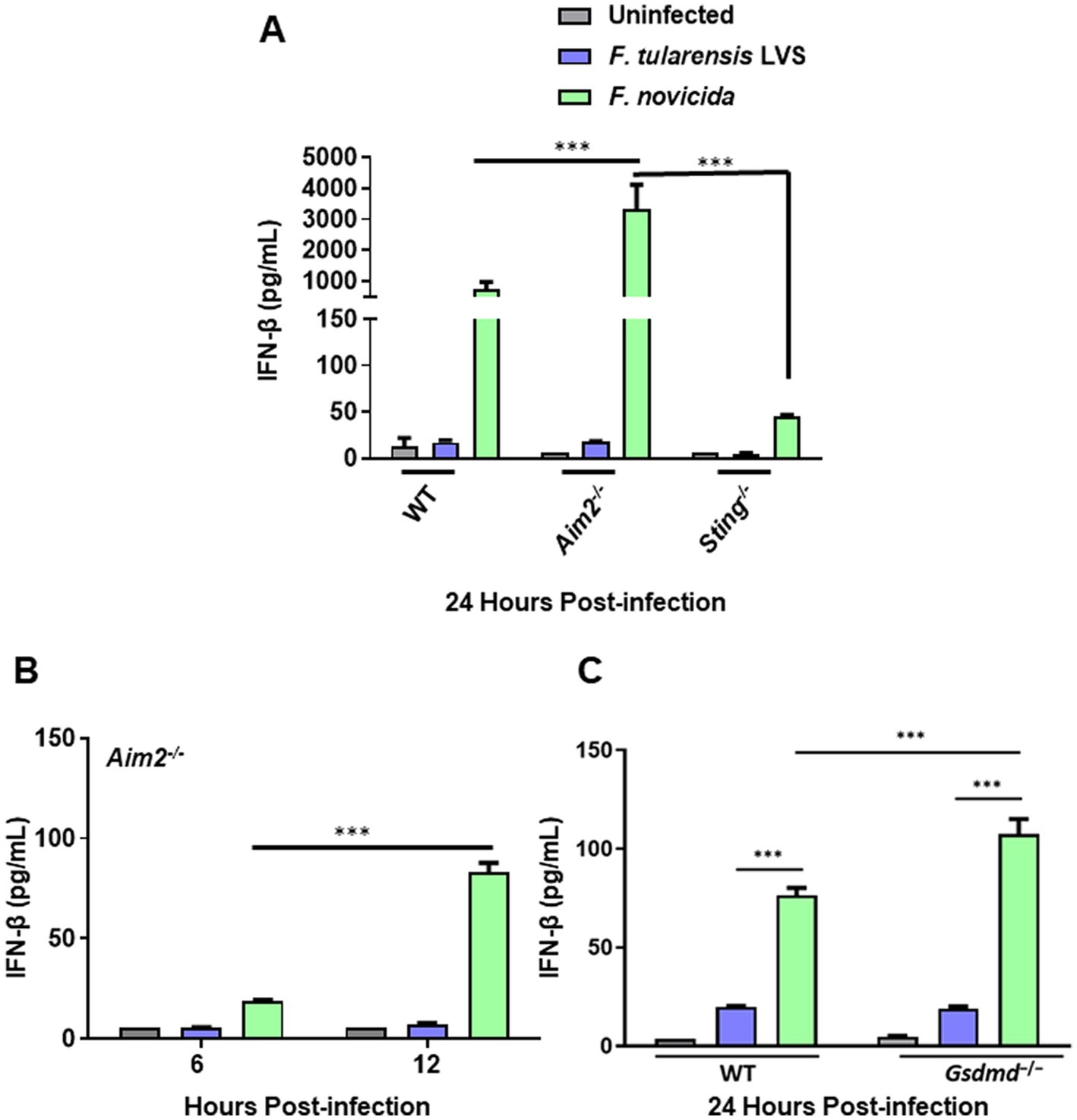
Figure 2. Francisella tularensis LVS-infected macrophages produce low levels of IFN-β. Wild-type (WT), Aim2−/− and Sting−/− BMDMs (A,B) and immortalized WT and Gsdmd−/− BMDMs (C) were infected with F. tularensis LVS, or F. novicida (MOI 100). The culture supernatants were analyzed at 6 and 12 h (B) and 24 h post-infection (A,C) for secreted levels of IFN-β by ELISA. Results are expressed as pg./mL (mean ± SEM) and are cumulative from three independent experiments with up to six biological replicates. The p values were determined using one-way ANOVA. ***p < 0.001.
The introduction of exogenous Francisella tularensis LVS DNA in Francisella tularensis LVS-infected BMDMs does not lead to enhanced IL-1β levels
DNA isolated from F. tularensis LVS (1 or 4 μg; 1 μg DNA = ~ 1 × 106 genomic copies of F. tularensis LVS) was introduced into the wild-type and Aim2−/− BMDMs infected with F. tularensis LVS. Francisella novicida infected, uninfected, and non-DNA transfected BMDMs were used as additional controls. The activation of Aim2 inflammasome was determined by measuring the secretion of IL-1β in the culture supernatants of these BMDMs. The results showed that both wild-type and Aim2−/− BMDMs either untransfected or transfected with F. tularensis LVS DNA and infected with F. tularensis LVS produced similarly low levels of IL-1β as compared to untransfected and F. novicida-infected BMDMs. As observed earlier, the production of IL-1β in F. novicida-infected BMDMs was Aim2-dependent (Figures 3A,B).
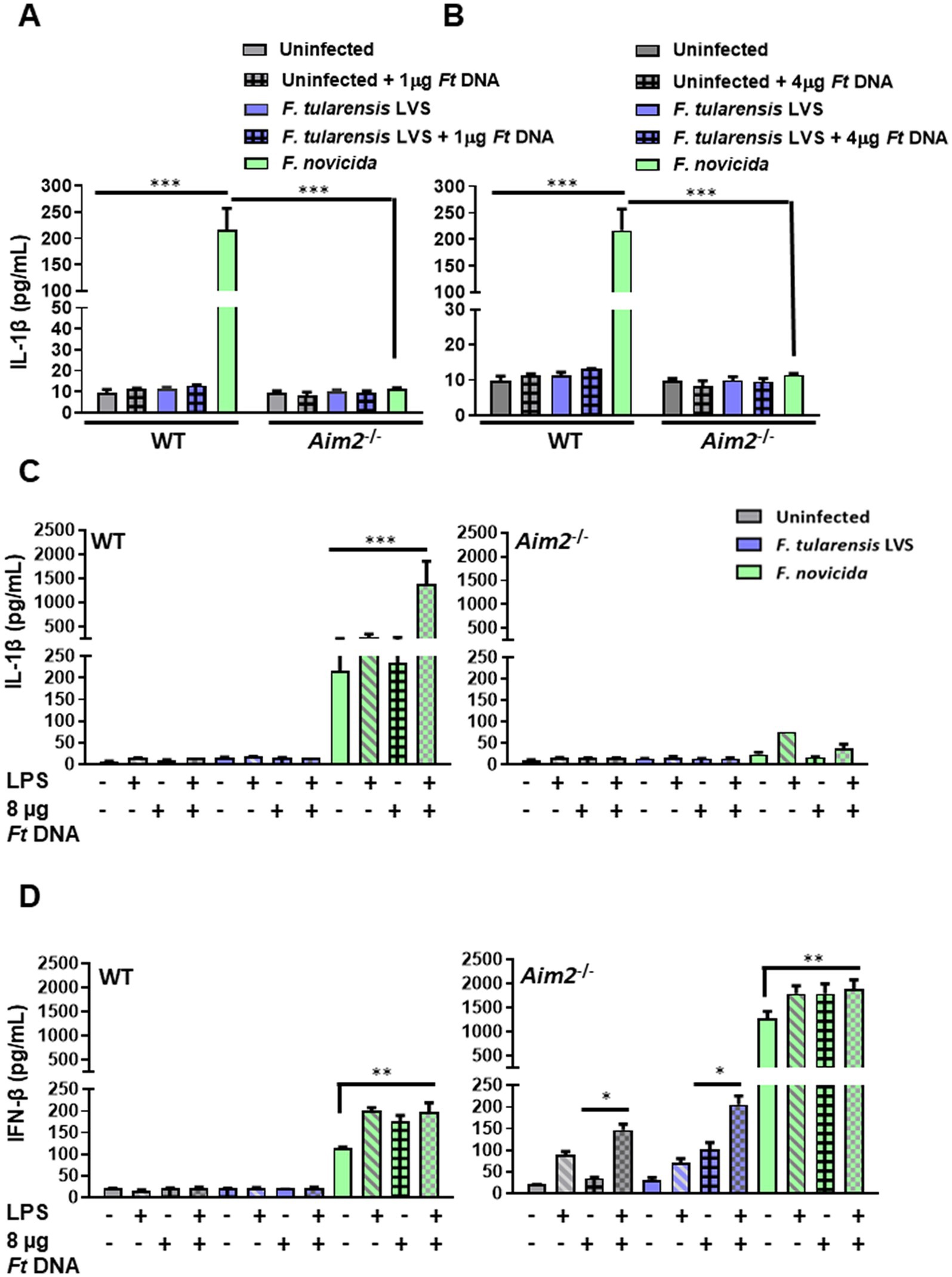
Figure 3. The introduction of exogenous Francisella tularensis LVS DNA and/or stimulation with a potent TLR4 agonist does not lead to enhanced IL-1β levels in F. tularensis LVS-infected macrophages. Wild-type (WT) and Aim2−/− BMDMs were infected (MOI 100) with F. tularensis LVS or Francisella novicida for 2 h, then transfected with (A) 1 μg and (B) 4 μg of F. tularensis LVS DNA. The culture supernatants from these cells were analyzed 24 h post-infection for the levels of secreted IL-1β by ELISA. Results are expressed as pg./mL (mean ± SEM) and are cumulative of two independent experiments with a total of up to six biological replicates. The p values were determined using one-way ANOVA. ***p < 0.001. LPS-primed or unprimed WT and Aim2−/− BMDMs were infected with F. tularensis LVS or F. novicida (MOI 100) for 2 h, then transfected with 8 μg/mL of F. tularensis LVS DNA. Culture supernatants were analyzed for the levels of secreted IL-1β (C) and IFN-β (D) at 24 h post-infection by ELISA. Results are expressed as pg/mL (mean ± SEM) and are cumulative of two independent experiments with up to six replicates. The p values were determined using one-way ANOVA. *p < 0.05; **p < 0.01; ***p < 0.001.
We conducted further experiments with a higher DNA concentration (8 μg) and with or without prior priming with lipopolysaccharide (LPS) derived from Escherichia coli, which is a potent TLR4 agonist. As an additional control, F. tularensis LVS DNA was also added to F. novicida-infected cells. The culture supernatants were then analyzed for the secreted levels of IL-1β at 24 h post-infection. The results showed that introducing 8 μg of F. tularensis LVS DNA with or without the LPS priming did not affect the levels of secreted IL-1β in uninfected and F. tularensis LVS-infected wild-type or the Aim2−/− BMDMs. Moreover, only the introduction of F. tularensis LVS DNA into LPS-primed F. novicida-infected BMDMs promoted robust inflammasome activation as indicated by higher IL-1β secretion as compared to the untransfected F. novicida controls (217.047 ± 40.3 vs. 1392.64 ± 969 pg./mL, respectively). This significant increase in IL-1β levels also confirmed that DNA transfection was successful, which was enhanced further upon priming the BMDMs with E. coli LPS in F. novicida-infected BMDMs (Figure 3C). These results demonstrate that suppression or weak activation of the Aim2-inflammasome by F. tularensis LVS was not relieved even after the introduction of exogenous F. tularensis LVS DNA or by LPS priming.
The addition of exogenous Francisella tularensis LVS DNA leads to enhanced IFN-β levels only in Aim2−/− BMDMs
To examine whether the introduction of F. tularensis LVS DNA results in cGAS-STING-dependent production of IFN-β, we determined the levels of secreted IFN-β. The results showed that the levels of IFN-β in untransfected or LPS-primed wild-type BMDMs, infected with F. tularensis LVS and treated with F. tularensis LVS DNA, were similar to those observed for the uninfected controls (Figure 3D). However, the LPS-primed, DNA-treated, or LPS-primed and DNA-treated, and F. novicida-infected BMDMs all induced significantly higher levels of IFN-β as compared to the untreated F. novicida-infected BMDMs. These results indicated that F. tularensis LVS possesses suppressive mechanisms, which, even after E. coli LPS priming or the introduction of F. tularensis LVS DNA, do not result in the production of IFN-β except in the Aim2−/− BMDMs.
We next examined the impact of IFN-β production in untreated, LPS-primed, F. tularensis LVS DNA-treated, and F. tularensis LVS or F. novicida infected Aim2−/− BMDMs. It was observed that LPS priming and introduction of F. tularensis LVS DNA in uninfected Aim2−/− BMDMs induced significantly higher levels of IFN-β as compared to untreated, LPS-primed or DNA-treated alone counterparts. A similar trend with significantly higher IFN-β levels were observed in LPS-primed, DNA-treated, and F. tularensis LVS or the F. novicida-infected Aim2−/− BMDMs (Figure 3D). These results indicate F. tularensis LVS-infected Aim2−/− BMDMs do respond to LPS priming and introduction of F. tularensis LVS DNA. However, these responses are muted in the wild-type BMDMs.
Stimulation with Escherichia coli LPS and/or the addition of exogenous Francisella tularensis LVS DNA does not lead to enhanced TNF-α response in Francisella tularensis LVS-infected BMDMs
We next determined if F. tularensis LVS exerts its suppressive effect only on cGAS-STING-dependent Aim2-inflammasome signaling or if other signaling pathways are similarly impacted. We specifically looked at the levels of TNF-α, a readout for the activation of the NF-κB signaling pathway. Very high levels of TNF-α were observed in BMDMs primed with E. coli LPS compared to the uninfected or BMDMs transfected with F. tularensis LVS DNA. The TNF-α levels did not increase when the BMDMs were treated with both E. coli LPS plus F. tularensis LVS DNA and infected with F. tularensis LVS. Surprisingly, the levels of TNF-α in LPS primed alone or those treated with both LPS and DNA and infected with F. tularensis LVS were significantly lower than those observed for their respective uninfected controls. Significantly high levels of TNF-α were observed in all groups of F. novicida-infected BMDMs than those observed for uninfected or F. tularensis LVS-infected BMDMs. Also, more or less similar levels were observed in F. novicida-infected BMDMs, irrespective of the treatment (Figure 4). Together, these results again highlight the differences between F. tularensis LVS and F. novicida in the type of innate immune response induced by the infected macrophages. Additionally, these results also confirm that F. tularensis is capable of suppressing an ongoing proinflammatory immune response.
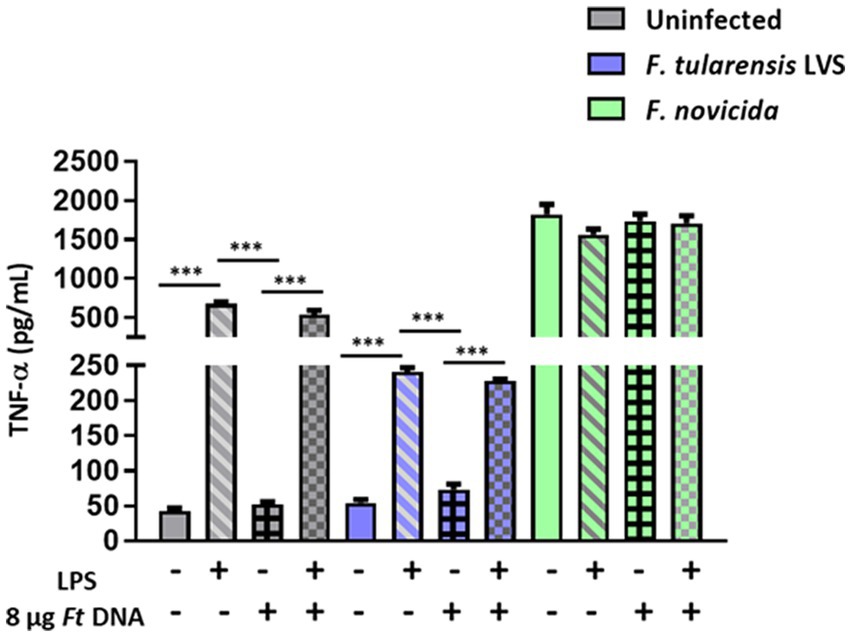
Figure 4. Stimulation with Escherichia coli LPS and/or the introduction of exogenous Francisella tularensis LVS DNA does not lead to enhanced TNF-α response in F. tularensis LVS-infected BMDMs. Escherichia coli LPS-primed or unprimed wild-type (WT) BMDMs were infected with F. tularensis LVS, or F. novicida at (MOI 100) for 2 h, then transfected with 8 μg/mL of F. tularensis LVS (Ft) DNA. Culture supernatants were analyzed for the levels of TNF-α at 24 h post-infection by ELISA. Results are expressed as pg./mL (mean ± SEM) and are cumulative from two independent experiments with up to six biological replicates. The p values were determined using one-way ANOVA. ***p < 0.001.
Mitophagy is induced upon Francisella tularensis LVS infection
Mitophagy is a process in which the damaged mitochondria are selectively removed by autophagy. We investigated the levels of PINK1 protein and LC3A/B in wild-type BMDMs infected with F. tularensis LVS or F. novicida (MOI 100) 6 h post-infection by western blot analysis. The western blot results showed higher levels of full-length and cleaved PINK1 in F. tularensis LVS-infected BMDMs compared to uninfected or F. novicida-infected BMDMs (Figure 5A). This increase was associated with an increase in LC3-II to LC3-I ratio in F. tularensis LVS-infected macrophages compared to the uninfected control or the F. novicida- infected cells (Figure 5B). These results suggest that PINK1-mediated autophagy may be involved in removing stressed/damaged mitochondria during F. tularensis LVS infection.
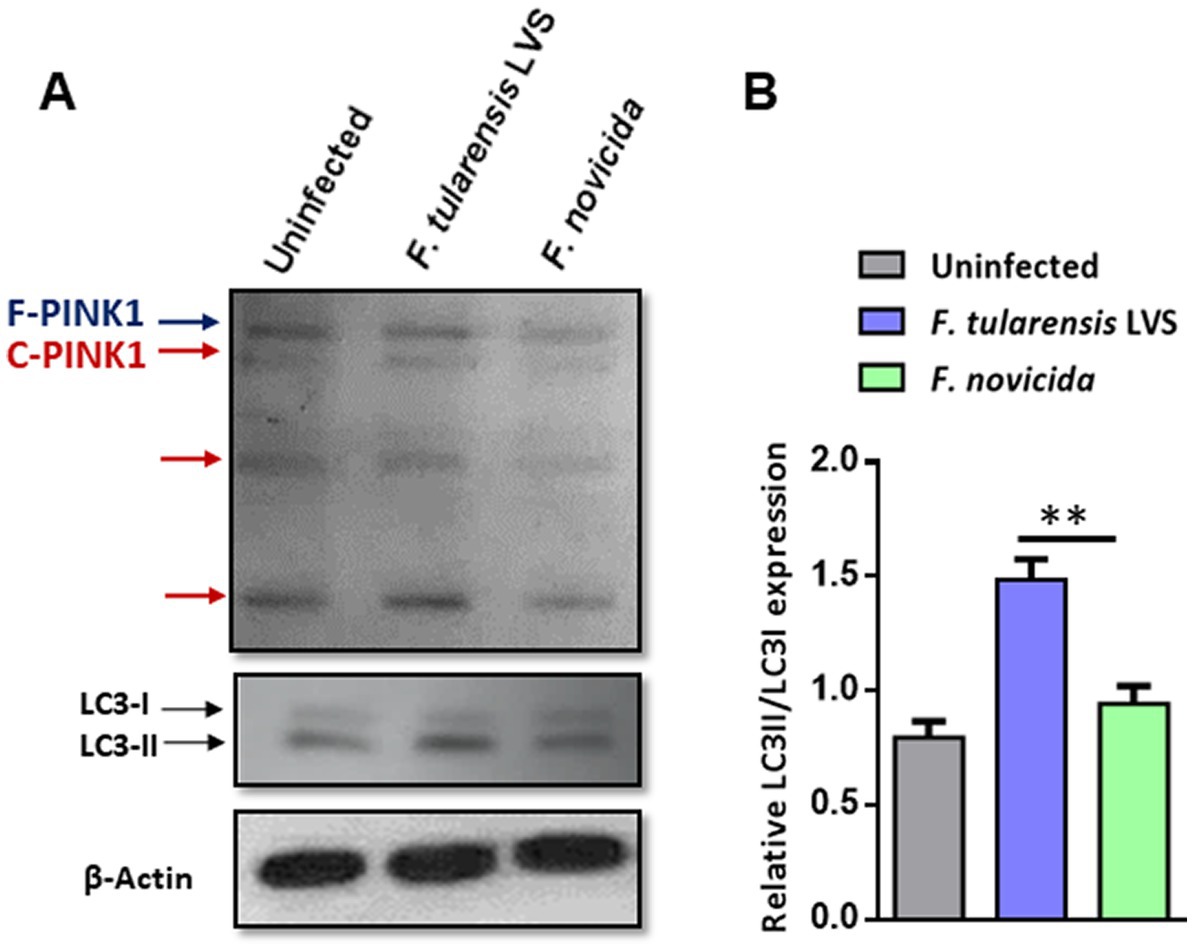
Figure 5. Mitophagy is induced upon Francisella tularensis LVS infection. (A) Western blot analysis of uninfected and BMDMs infected with F. tularensis LVS or Francisella novicida (MOI 100). Lysates were collected 6 h post-infection and subjected to SDS-PAGE, then blotted for PINK1, LC3 I/II, and β-Actin. F-PINK1, full-length PINK1, and C-PINK1, cleaved PINK1. A representative blot out of three is shown. (B) Relative LC3II/LC3I expression was quantitated from three blots. The p values were determined using one-way ANOVA. **p < 0.01.
We investigated further by examining BMDMs infected with F. tularensis LVS or F. novicida at 6- and 12-h post-infection for the signs of mitochondrial damage or the induction of mitophagy by TEM. Assessment of mitochondrial morphology in the uninfected macrophages revealed tubular networks, while F. tularensis LVS-infected cells possessed fragmented and round mitochondria as early as one-hour post-infection (not shown). Also, F. tularensis LVS-infected macrophages showed an increase in autophagic vacuoles, some of which contained the engulfed mitochondria. Conversely, macrophages infected with F. novicida showed mostly tubular and some round, swollen mitochondria, but no engulfed mitochondria were observed (not shown). At 6 h post-infection, mitochondrial fragmentation increased significantly in F. tularensis LVS-infected cells. Low levels of mitochondrial fragmentation were also observed in both uninfected and F. novicida-infected BMDMs. Furthermore, F. tularensis LVS infection was associated with the presence of higher numbers of autophagic vacuoles, while none were detected in uninfected and F. novicida-infected BMDMs (Figures 6A,B). At 12 h post-infection, the number of fragmented mitochondria per cell decreased from that observed at 6 h, and this was associated with higher numbers of autophagic vacuoles containing damaged mitochondria and autophagosomes in F. tularensis LVS compared to the F. novicida infected BMDMs. Additionally, the numbers of autophagosomes were significantly higher in F. tularensis LVS-infected BMDMs at 12 h post-infection than those observed at 6 h (Figures 7A,B). These results indicate that enhanced mitophagy is induced during F. tularensis LVS-infected than F. novicida-infected macrophages.
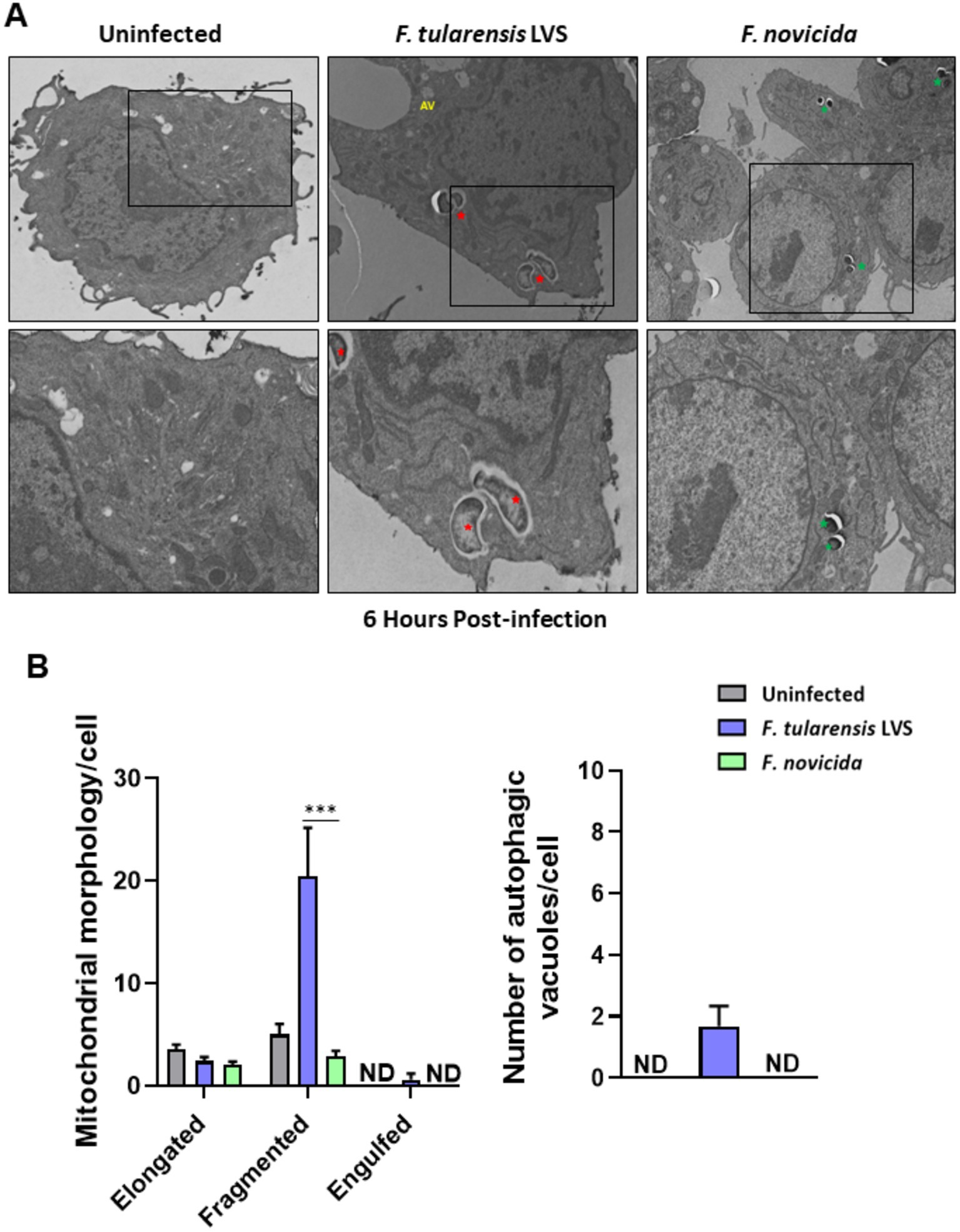
Figure 6. Increased mitophagy is observed in BMDMs infected with Francisella tularensis LVS 6 h post-infection. (A) Representative transmission electron microscopy images randomly selected from uninfected, F. tularensis LVS, and Francisella novicida infected BMDMs 6 h post-infection are shown. The identification of F. tularensis LVS and F. novicida was done on the basis of the size, shape, and location of the bacteria within macrophages. Francisella tularensis is represented by red stars and F. novicida is represented by green stars. The black boxes indicate the higher magnification of the areas shown in lower panels. (B) Quantitation of mitochondrial morphology and autophagic vacuoles per cell. Representative results from three independent experiments are shown. The p values were determined using one-way ANOVA. ***p < 0.001. ND, not detected; AV, autophagic vacuoles.
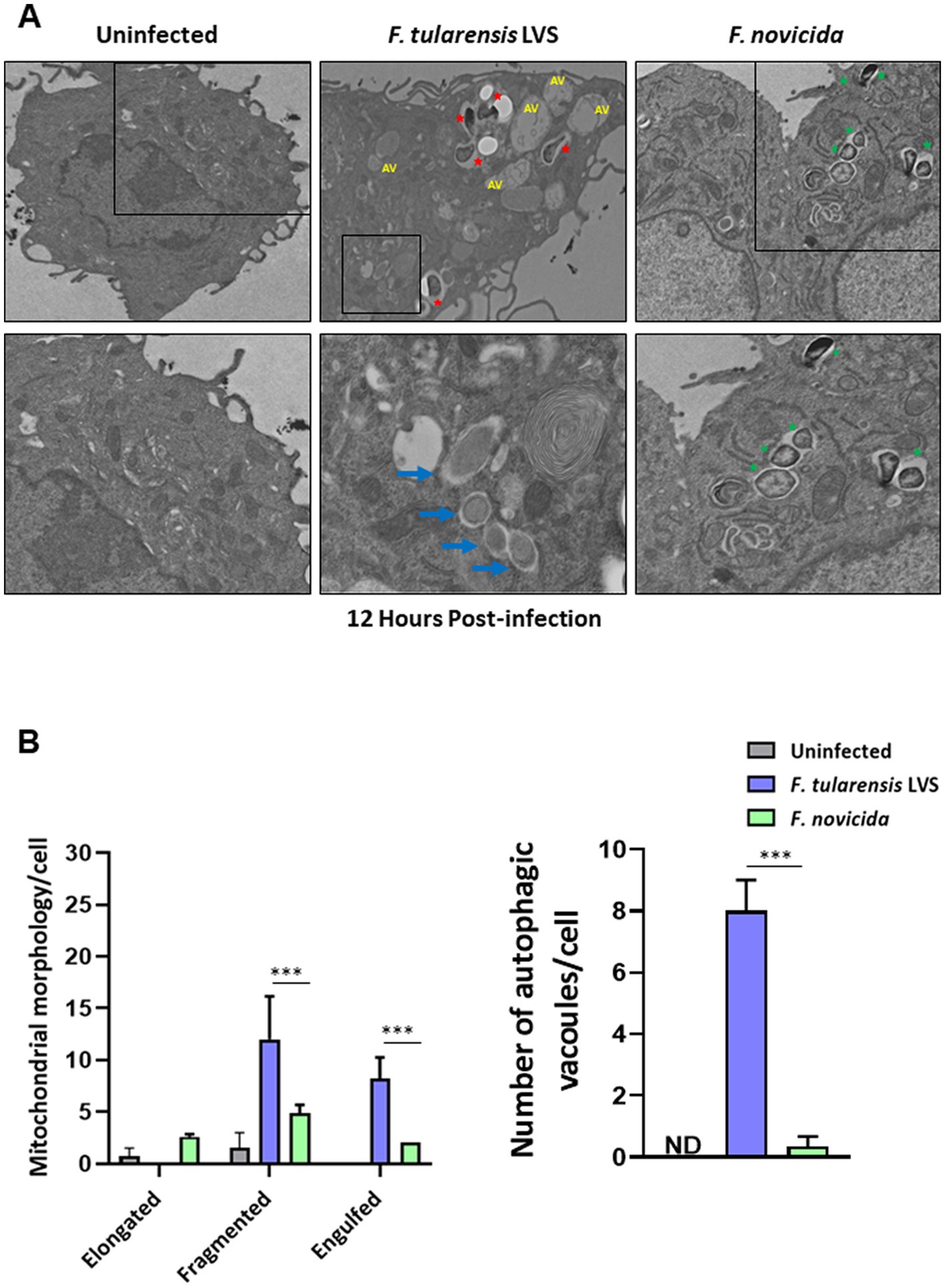
Figure 7. Increased mitophagy is observed in BMDMs infected with Francisella tularensis LVS 12 h post-infection. (A) Representative transmission electron microscopy images randomly selected from uninfected, F. tularensis LVS, and F. novicida infected BMDMs 12 h post-infection are shown. Francisella tularensis is represented by red stars and F. novicida is represented by green stars. Autophagic vacuoles are indicated by blue arrows. The black boxes indicate the higher magnification of the areas shown in lower panels. (B) Quantitation of mitochondrial morphology and autophagic vacuoles per cell. Representative results from three independent experiments are shown. The p values were determined using one-way ANOVA. ***p < 0.001. ND, not detected; AV, autophagic vacuoles.
Discussion
The mechanisms and signaling pathways leading to the activation of Aim2 inflammasome have been elegantly elucidated using the F. novicida strain as a model. However, these mechanisms remain unknown for F. tularensis strains. This study aimed to understand the mechanisms of suppression of Aim2 inflammasome by F. tularensis LVS and to investigate if the mechanisms known for F. novicida are also operative in F. tularensis strains.
Our results showed that in F. tularensis LVS-infected BMDMs, very low levels of IL-1β were produced in an Aim2-dependent manner than in F. novicida-infected BMDMs. The differences in IL-1β levels in F. tularensis LVS and F. novicida-infected BMDMs were not due to differences in their growth kinetics since both strains grew similarly in wild-type BMDMs or BMDMs lacking inflammasome sensors. However, there were differences in the extent of cell death, as indicated by the LDH release in macrophages infected with F. tularensis LVS and F. novicida. In an in vivo study (Hall et al., 2008) comparing the pathogenesis of F. tularensis LVS and F. novicida side-by-side, it was reported that there are differences in the propensity of these two Francisella subspecies to infect cells. The number of cells infected by F. novicida at the initiation of infection is higher than those infected by F. tularensis LVS. Furthermore, no progressive increase in the number of F. novicida-infected cells is observed. On the other hand, despite the number of cells initially infected by F. tularensis LVS are low, an increasing number of cells continue to become infected as the infection progresses. Therefore, the number of both F. novicida and F. tularensis observed during the later stages of infection are identical. Moreover, F. novicida causes a more rapid killing of the cells than F. tularensis LVS, as observed by the rapid depletion of macrophages and dendritic cells. Our in vitro results mirror this in vivo profile and show recovery of identical numbers of both F. tularensis LVS and F. novicida from the infected macrophages, however, they reveal significant differences in cell death between F. tularensis and F. novicida-infected macrophages.
Gasdermin D plays a crucial role in pyroptosis, which is characterized by the formation of pores in the cell membrane. In addition to IL-1β and IL-18, these pores facilitate the rapid release of intracellular contents, including the lytic cell death marker, LDH, into the extracellular environment (Banerjee et al., 2018). Interestingly, in Gsdmd−/− macrophages infected with F. novicida, the levels of LDH were similar to those observed in wild-type macrophages (Figure 1D). These results corroborate a previous study that has shown that Gsdmd-deficient macrophages, similar to wild-type macrophages, exhibit increased LDH release despite their inability to undergo GsdmD-dependent pyroptosis. This robust GsdmD-independent LDH release has been reported to be due to cell lysis caused by necrotic rather than pyroptotic cell death in Gsdmd−/− macrophages (Schneider et al., 2017).
Previous studies have shown that IFN-β production by activated cGAS-STING pathway is a prerequisite for optimal Aim2 inflammasome activation in F. novicida-infected macrophages (Henry et al., 2007; Storek et al., 2015), and that IFN-β production is negatively regulated via Aim2 inflammasome (Corrales et al., 2016; Nakaya et al., 2017; Banerjee et al., 2018; Yan et al., 2018). Our results revealed significantly lower levels of IFN-β in F. tularensis LVS-infected BMDMs than F. novicida-infected BMDMs in a STING-dependent manner, suggesting that suppression/weak activation of cGAS-STING pathway by F. tularensis LVS may result in suppression/weak activation of the Aim2 inflammasome. However, the levels of IFN-β in F. tularensis LVS-infected BMDMs did not change irrespective of the presence or absence of Aim2 or GsdmD and remained significantly lower than those observed for F. novicida-infected BMDMs. On the other hand, higher levels of IFN-β in Aim2−/− and Gsdmd−/− BMDMs infected with F. novicida were observed as compared to their wild-type counterparts. These results indicated that even trace amounts of DNA that can be sensed by the c-GAS-STING pathway might not be released during the replication of F. tularensis, which may serve as a potential mechanism to prevent activation of the Aim2 inflammasome. Furthermore, these results also indicate potential differences that could exist in the ability of F. novicida and F. tularensis LVS DNA to stimulate or be recognized by the cGAS-STING pathway. Previous studies have shown that AT-rich motifs in the genome of Plasmodium falciparum are sensed by the STING pathway, leading to a potent type-1 interferon response (Sharma et al., 2011). It would be of great interest to investigate if the existence of similar AT-rich motifs in higher abundance in F. novicida DNA than in F. tularensis LVS DNA contributes to differential activation of cGAS-STING pathway resulting in higher levels of IFN-β.
We next sought to elucidate the mechanisms behind suppression/weak activation of the Aim2 inflammasome in F. tularensis LVS-infected BMDMs. We hypothesized that the low levels of IFN-β and IL-1β observed upon F. tularensis LVS infection were due to insufficient release of bacterial DNA. To investigate this, we introduced exogenous F. tularensis DNA into the cytosol of BMDMs infected with F. tularensis to activate the cGAS-STING signaling pathway and prime the Aim2 inflammasome for IFN-β and IL-1β, respectively. We anticipated that this approach would bring the levels of IL-1β in F. tularensis LVS-infected macrophages similar to those observed for F. novicida-infected counterparts. However, even with the introduction of exogenous F. tularensis LVS DNA, the levels of secreted IL-1β remained similar to untransfected and F. tularensis LVS-infected BMDMs, indicating that additional exogenous DNA failed to activate Aim2-dependent IL-1β production in F. tularensis LVS-infected BMDMs. These intriguing results prompted us to investigate further why exogenous DNA did not activate the Aim2-inflammasome in F. tularensis LVS-infected BMDMs.
Activation of the inflammasome is a complex process that requires two signals. Signal-one or the priming signal is provided by a TLR ligand that activates the NF-κB pathway for the transcriptional upregulation of certain inflammasome components, including pro-IL-1β. Signal-two, or the activation signal, is provided by microbial components, endogenous molecules, and/or stress signals that are recognized by the inflammasome sensors such as Aim2 or Nlrp3 to activate the inflammasome (Broz and Dixit, 2016). Since F. tularensis also suppress TLR-dependent signaling pathways (Dotson et al., 2013), we reasoned that low levels of IL-1β in F. tularensis LVS-infected BMDMs could be due to a lack of signal-one or that the amount of introduced DNA is not sufficient for the activation of Aim2 as signal-two. We next primed the cells by treating them with a potent TLR4 ligand, E. coli LPS, and introduced a higher DNA concentration (8 μg) of exogenous F. tularensis LVS DNA. However, neither priming with E. coli LPS nor a higher quantity of exogenous F. tularensis LVS DNA improved the levels of IL-1β. Contradictorily, providing both signal-one and -two further elevated the levels of IL-1β in F. novicida-infected BMDMs in an Aim2-dependent fashion. Overall, these results demonstrate that suppression/weak activation of Aim2 inflammasome by F. tularensis LVS was not relieved even after the introduction of exogenous F. tularensis LVS DNA or boosting the signal-one by LPS priming. However, LPS priming resulted in the production of TNF-α in F. tularensis LVS-infected BMDMs at lower levels compared to the uninfected cells, while TNF-α levels were produced at extremely high levels during F. novicida infection irrespective of LPS stimulation. These results confirm previous findings about the ability of F. tularensis to suppress activated macrophages (Dueñas et al., 2006; Gillette et al., 2014).
We further investigated the mechanism responsible for suppression/weak activation of Aim2 inflammasome that remained elusive from the preceding studies. We hypothesized that DNA coming from some other source might also contribute to the higher levels of inflammasome-dependent cytokines in F. novicida-infected BMDMs and that F. tularensis may have evolved mechanisms to cause immunosuppression. This notion was investigated next.
Studies have shown that during stress, there is a loss of membrane potential, increased release of mitochondrial DNA (mtDNA), decreased production of ATP, and increased fragmentation of dysfunctional mitochondria (Murphy, 2009; Larsen et al., 2012; Youle and Van Der Bliek, 2012; Friedman and Nunnari, 2014; Garza-Lombó et al., 2020). A previous study has demonstrated the ability of F. tularensis SchuS4 to manipulate mitochondrial activity early during infection, enhancing mitochondrial functions by impairing the metabolic shift from oxidative phosphorylation to glycolysis (Jessop et al., 2018). Francisella tularensis LVS is also capable of maintaining mitochondrial integrity during neutrophil infection in vivo to delay apoptosis (McCracken et al., 2016). Moreover, mtDNA was not detected in isolated mice leukocytes 2 days post-infection with F. tularensis LVS but started to appear after the third day of infection, reaching significantly higher levels on day six post-infection (Singh et al., 2017). The appearance of mtDNA is associated with an increase in proinflammatory cytokine response. Additionally, the enhanced inflammasome activation during F. novicida infection in vitro is found to be mediated by mitochondrial reactive oxygen species (mROS), suggesting the accumulation of stressed mitochondria (Crane et al., 2014). Collectively, these studies demonstrated that mtDNA released from stressed mitochondria might play a key role in activating the innate immune responses during Francisella infection. Based on these observations, we postulated that dysfunctional mitochondria might be removed early during F. tularensis LVS infection to avoid stimulating Aim2-mediated responses by the leaked mtDNA.
Our results indicate that mitophagy is induced during F. tularensis LVS, but not F. novicida infection. The results suggest a role for the PINK1/Parkin pathway, which converts LC3-I to LC3-II to induce mitophagy in F. tularensis LVS-infected macrophages. Furthermore, our electron microscopy results indicate mitochondrial fragmentation and mitophagy in F. tularensis LVS-infected BMDMs compared to their F. novicida-infected counterparts. However, a detailed investigation is warranted to understand the pathways and mechanisms involved in the induction of mitophagy and its direct association with the suppression of the Aim2 inflammasome in F. tularensis LVS-infected macrophages.
However, the question remains: why is there enhanced mitophagy in F. tularensis-infected BMDMs compared to F. novicida-infected ones? One potential explanation could be that several TCA cycle components are missing in F. tularensis, and these strains rely extensively on the host cell metabolism, which stresses out the mitochondria. Francisella novicida possesses all TCA cycle components and may impose less stress on mitochondria (Rohmer et al., 2007; Gesbert et al., 2015; Ziveri et al., 2017). Another question is why dysfunctional mitochondria are removed from F. tularensis LVS-infected BMDMs but not from those infected with F. novicida. M1 macrophages and activated DCs have an incomplete TCA cycle, and a pathogen with a deficiency in nine amino acids biosynthetic pathways may induce additional stress on mitochondria and the infected cell, resulting in the induction of autophagic pathways (Dieppedale et al., 2013; Williams and O’Neill, 2018; Breda et al., 2019). However, more detailed work is needed to be done to address these probable explanations.
Conclusion
This study reveals that F. tularensis LVS induces low levels of Aim2-dependent IL-1β than those infected with F. novicida. The suppression/weak activation of the Aim2 inflammasome is due to the suppression of the cGAS-STING DNA-sensing pathway upstream of Aim2 in F. tularensis LVS-infected BMDMs. The introduction of exogenous F. tularensis LVS DNA into the cytosol of the infected BMDMs alone or in conjunction with a priming signal provided by a potent TLR4-agonist failed to restore IL-1β levels similar to those observed for F. novicida. Collectively, the results from this study demonstrate that F. tularensis suppresses the activation of Aim2 inflammasome, and the mechanisms established for F. novicida are not operative in F. tularensis LVS.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Ethics statement
The animal study was reviewed and approved by IACUC New York Medical College.
Author contributions
MA, ZM, and JM carried out the experiments and data analysis. JY conducted the transmission electron microscopy. MM and CB conceived, coordinated, and supervised the research and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by National Institutes of Health Grants R56AI101109 and R21AI51277 (CB) and R15AI107698 (MM).
Acknowledgments
We acknowledge Vijay Rathinam and Penghua Wang of UCONN Health for providing immortalized wild type, Gasdermin D knockout macrophage cell lines, and sting knockout macrophages, respectively.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Banerjee, I., Behl, B., Mendonca, M., Shrivastava, G., Russo, A. J., Menoret, A., et al. (2018). Gasdermin D restrains type I interferon response to cytosolic DNA by disrupting ionic homeostasis. Immunity 49, 413–426.e5. doi: 10.1016/j.immuni.2018.07.006
Breda, D. C. N. S., Davanzo, G. G., Basso, P. J., Saraiva Câmara, N. O., and Moraes-Vieira, P. M. M. (2019). Mitochondria as central hub of the immune system. Redox Biol. 26:101255. doi: 10.1016/j.redox.2019.101255
Broz, P., and Dixit, V. M. (2016). Inflammasomes: mechanism of assembly, regulation and signalling. Nat. Rev. Immunol. 16, 407–420. doi: 10.1038/nri.2016.58
Burdette, D. L., Monroe, K. M., Sotelo-Troha, K., Iwig, J. S., Eckert, B., Hyodo, M., et al. (2011). STING is a direct innate immune sensor of cyclic di-GMP. Nature 478, 515–518. doi: 10.1038/nature10429
Clemens, D. L., Lee, B. Y., and Horwitz, M. A. (2004). Virulent and avirulent strains of Francisella tularensis prevent acidification and maturation of their phagosomes and escape into the cytoplasm in human macrophages. Infect. Immun. 72, 3204–3217. doi: 10.1128/IAI.72.6.3204-3217.2004
Clemens, D. L., Lee, B. Y., and Horwitz, M. A. (2005). Francisella tularensis enters macrophages via a novel process involving pseudopod loops. Infect. Immun. 73, 5892–5902. doi: 10.1128/IAI.73.9.5892-5902.2005
Corrales, L., Woo, S.-R., Williams, J. B., McWhirter, S. M., Dubensky, T. W., and Gajewski, T. F. (2016). Antagonism of the STING pathway via activation of the AIM2 Inflammasome by intracellular DNA. J. Immunol. 196, 3191–3198. doi: 10.4049/jimmunol.1502538
Crane, D. D., Bauler, T. J., Wehrly, T. D., and Bosio, C. M. (2014). Mitochondrial ROS potentiates indirect activation of the AIM2 inflammasome. Front. Microbiol. 5:438. doi: 10.3389/fmicb.2014.00438
Dennis, D. T., Inglesby, T. V., Henderson, D. A., Bartlett, J. G., Ascher, M. S., Eitzen, E., et al. (2001). Tularemia as a biological weapon: medical and public health management. JAMA 285, 2763–2773. doi: 10.1001/jama.285.21.2763
Dieppedale, J., Gesbert, G., Ramond, E., Chhuon, C., Dubail, I., Dupuis, M., et al. (2013). Possible links between stress defense and the tricarboxylic acid (TCA) cycle in FrancisellaPathogenesis. Mol. Cell. Proteomics 12, 2278–2292. doi: 10.1074/mcp.M112.024794
Dotson, R. J., Rabadi, S. M., Westcott, E. L., Bradley, S., Catlett, S. V., Banik, S., et al. (2013). Repression of Inflammasome by Francisella tularensis during early stages of infection. J. Biol. Chem. 288, 23844–23857. doi: 10.1074/jbc.M113.490086
Dueñas, A. I., Aceves, M., Orduña, A., Díaz, R., Sánchez Crespo, M., and García-Rodríguez, C. (2006). Francisella tularensis LPS induces the production of cytokines in human monocytes and signals via toll-like receptor 4 with much lower potency than E. coli LPS. Int. Immunol. 18, 785–795. doi: 10.1093/intimm/dxl015
Eigelsbach, H. T., and Downs, C. M. (1961). Prophylactic effectiveness of live and killed tularemia vaccines. I. Production of vaccine and evaluation in the white mouse and guinea pig. J. Immunol. 87, 415–425. doi: 10.4049/jimmunol.87.4.415
Ellis, J., Oyston, P. C. F., Green, M., and Titball, R. W. (2002). Tularemia. Clin. Microbiol. Rev. 15, 631–646. doi: 10.1128/CMR.15.4.631
Fernandes-Alnemri, T., Yu, J. W., Datta, P., Wu, J., and Alnemri, E. S. (2009). AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature 458, 509–513. doi: 10.1038/nature07710
Fernandes-Alnemri, T., Yu, J. W., Juliana, C., Solorzano, L., Kang, S., Wu, J., et al. (2010). The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nat. Immunol. 11, 385–393. doi: 10.1038/ni.1859
Friedman, J. R., and Nunnari, J. (2014). Mitochondrial form and function. Nature 505, 335–343. doi: 10.1038/nature12985
Garza-Lombó, C., Pappa, A., Panayiotidis, M. I., and Franco, R. (2020). Redox homeostasis, oxidative stress and mitophagy. Mitochondrion 51, 105–117. doi: 10.1016/j.mito.2020.01.002
Gesbert, G., Ramond, E., Tros, F., Dairou, J., Frapy, E., Barel, M., et al. (2015). Importance of branched-chain amino acid utilization in Francisella intracellular adaptation. Infect. Immun. 83, 173–183. doi: 10.1128/IAI.02579-14
Gillette, D. D., Curry, H. M., Cremer, T., Ravneberg, D., Fatehchand, K., Shah, P. A., et al. (2014). Virulent type a Francisella tularensis actively suppresses cytokine responses in human monocytes. Front. Cell. Infect. Microbiol. 4:45. doi: 10.3389/fcimb.2014.00045
Golovliov, I., Baranov, V., Krocova, Z., Kovarova, H., and Sjöstedt, A. (2003). An attenuated strain of the facultative intracellular bacterium Francisella tularensis can escape the phagosome of monocytic cells. Infect. Immun. 71, 5940–5950. doi: 10.1128/IAI.71.10.5940-5950.2003
Hall, J. D., Woolard, M. D., Gunn, B. M., Craven, R. R., Taft-Benz, S., Frelinger, J. A., et al. (2008). Infected-host-cell repertoire and cellular response in the lung following inhalation of Francisella tularensis Schu S4, LVS, or U112. Infect. Immun. 76, 5843–5852. doi: 10.1128/IAI.01176-08
Henry, T., Brotcke, A., Weiss, D. S., Thompson, L. J., and Monack, D. M. (2007). Type I interferon signaling is required for activation of the inflammasome during Francisellainfection. J. Exp. Med. 204, 987–994. doi: 10.1084/jem.20062665
Henry, T., and Monack, D. M. (2007). Activation of the inflammasome upon Francisella tularensis infection: interplay of innate immune pathways and virulence factors. Cell. Microbiol. 9, 2543–2551. doi: 10.1111/j.1462-5822.2007.01022.x
Hornung, V., Ablasser, A., Charrel-Dennis, M., Bauernfeind, F., Horvath, G., Caffrey, D. R., et al. (2009). AIM2 recognizes cytosolic dsDNA and forms a caspase-1 activating inflammasome with ASC. Nature 458, 514–518. doi: 10.1038/NATURE07725
Jessop, F., Schwarz, B., Heitmann, E., Buntyn, R., Wehrly, T., and Bosio, C. M. (2018). Temporal manipulation of mitochondrial function by virulent Francisella tularensis to limit inflammation and control cell death. Infect. Immun. 86, e00044–e00058. doi: 10.1128/IAI.00044-18
Jones, B. D., Faron, M., Rasmussen, J. A., and Fletcher, J. R. (2014). Uncovering the components of the Francisella tularensis virulence stealth strategy. Front. Cell. Infect. Microbiol. 4:32. doi: 10.3389/fcimb.2014.00032
Jones, J. W., Kayagaki, N., Broz, P., Henry, T., Newton, K., O’Rourke, K., et al. (2010). Absent in melanoma 2 is required for innate immune recognition of Francisella tularensis. Proc. Natl. Acad. Sci. 107, 9771–9776. doi: 10.1073/pnas.1003738107
Kaiser, W. J., Upton, J. W., and Mocarski, E. S. (2008). Receptor-interacting protein homotypic interaction motif-dependent control of NF-κB activation via the DNA-dependent activator of IFN regulatory factors. J. Immunol. 181, 6427–6434. doi: 10.4049/jimmunol.181.9.6427
Kingry, L. C., and Petersen, J. M. (2014). Comparative review of Francisella tularensis and Francisella novicida. Front. Cell. Infect. Microbiol. 4:35. doi: 10.3389/fcimb.2014.00035
Lai, X. H., Golovliov, I., and Sjöstedt, A. (2001). Francisella tularensis induces cytopathogenicity and apoptosis in murine macrophages via a mechanism that requires intracellular bacterial multiplication. Infect. Immun. 69, 4691–4694. doi: 10.1128/IAI.69.7.4691-4694.2001
Larsen, S., Nielsen, J., Hansen, C. N., Nielsen, L. B., Wibrand, F., Stride, N., et al. (2012). Biomarkers of mitochondrial content in skeletal muscle of healthy young human subjects. J. Physiol. 590, 3349–3360. doi: 10.1113/jphysiol.2012.230185
Li, H., Nookala, S., Bina, X. R., Bina, J. E., and Re, F. (2006). Innate immune response to Francisella tularensis is mediated by TLR2 and caspase-1 activation. J. Leukoc. Biol. 80, 766–773. doi: 10.1189/jlb.0406294
Man, S. M., Karki, R., Malireddi, R. K. S., Neale, G., Vogel, P., Yamamoto, M., et al. (2015). The transcription factor IRF1 and guanylate-binding proteins target activation of the AIM2 inflammasome by Francisella infection. Nat. Immunol. 16, 467–475. doi: 10.1038/ni.3118
Man, S. M., Karki, R., Sasai, M., Place, D. E., Kesavardhana, S., Temirov, J., et al. (2016). IRGB10 liberates bacterial ligands for sensing by the AIM2 and Caspase-11-NLRP3 Inflammasomes. Cells 167, 382–396.e17. doi: 10.1016/j.cell.2016.09.012
Mariathasan, S., Weiss, D. S., Dixit, V. M., and Monack, D. M. (2005). Innate immunity against Francisella tularensisis dependent on the ASC/caspase-1 axis. J. Exp. Med. 202, 1043–1049. doi: 10.1084/jem.20050977
McCracken, J. M., Kinkead, L. C., McCaffrey, R. L., and Allen, L.-A. H. (2016). Francisella tularensis modulates a distinct subset of regulatory factors and sustains mitochondrial integrity to impair human neutrophil apoptosis. J. Innate Immun. 8, 299–313. doi: 10.1159/000443882
Murphy, M. P. (2009). How mitochondria produce reactive oxygen species. Biochem. J. 417, 1–13. doi: 10.1042/BJ20081386
Nakaya, Y., Lilue, J., Stavrou, S., Moran, E. A., and Ross, S. R. (2017). AIM2-like receptors positively and negatively regulate the interferon response induced by cytosolic DNA. MBio 8, 1–17. doi: 10.1128/mBio.00944-17
Oyston, P. C. F., Sjostedt, A., and Titball, R. W. (2004). Tularaemia: bioterrorism defence renews interest in Francisella tularensis. Nat. Rev. Microbiol. 2, 967–978. doi: 10.1038/nrmicro1045
Pechous, R. D., McCarthy, T. R., and Zahrt, T. C. (2009). Working toward the future: insights into Francisella tularensis pathogenesis and vaccine development. Microbiol. Mol. Biol. Rev. 73, 684–711. doi: 10.1128/MMBR.00028-09
Rathinam, V. A. K., Jiang, Z., Waggoner, S. N., Sharma, S., Cole, L. E., Waggoner, L., et al. (2010). The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat. Immunol. 11, 395–402. doi: 10.1038/ni.1864
Rohmer, L., Fong, C., Abmayr, S., Wasnick, M., Larson Freeman, T. J., Radey, M., et al. (2007). Comparison of Francisella tularensis genomes reveals evolutionary events associated with the emergence of human pathogenic strains. Genome Biol. 8:R102. doi: 10.1186/gb-2007-8-6-r102
Santic, M., Molmeret, M., and Abu Kwaik, Y. (2005). Modulation of biogenesis of the Francisella tularensis subsp. novicida-containing phagosome in quiescent human macrophages and its maturation into a phagolysosome upon activation by IFN-?? Cell. Microbiol. 7, 957–967. doi: 10.1111/j.1462-5822.2005.00529.x
Santic, M., Pavokovic, G., Jones, S., Asare, R., and Kwaik, Y. A. (2010). Regulation of apoptosis and anti-apoptosis signalling by Francisella tularensis. Microbes Infect. 12, 126–134. doi: 10.1016/j.micinf.2009.11.003
Schneider, K. S., Groß, C. J., Dreier, R. F., Saller, B. S., Mishra, R., Gorka, O., et al. (2017). The Inflammasome drives GSDMD-independent secondary Pyroptosis and IL-1 release in the absence of Caspase-1 protease activity. Cell Rep. 21, 3846–3859. doi: 10.1016/j.celrep.2017.12.018
Sharma, S., DeOliveira, R. B., Kalantari, P., Parroche, P., Goutagny, N., Jiang, Z., et al. (2011). Innate immune recognition of an AT-rich stem-loop DNA motif in the plasmodium falciparum genome. Immunity 35, 194–207. doi: 10.1016/j.immuni.2011.05.016
Shi, J., Zhao, Y., Wang, K., Shi, X., Wang, Y., Huang, H., et al. (2015). Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 526, 660–665. doi: 10.1038/nature15514
Singh, A., Periasamy, S., Malik, M., Bakshi, C. S., Stephen, L., Ault, J. G., et al. (2017). Necroptotic debris including damaged mitochondria elicits sepsis-like syndrome during late-phase tularemia. Cell Death Dis. 3:17056. doi: 10.1038/cddiscovery.2017.56
Sjödin, A., Svensson, K., Öhrman, C., Ahlinder, J., Lindgren, P., Duodu, S., et al. (2012). Genome characterisation of the genus Francisella reveals insight into similar evolutionary paths in pathogens of mammals and fish. BMC Genomics 13:268. doi: 10.1186/1471-2164-13-268
Steele, S. P., Chamberlain, Z., Park, J., and Kawula, T. H. (2019). Francisella tularensis enters a double membraned compartment following cell-cell transfer. elife 8, 1–17. doi: 10.7554/eLife.45252
Storek, K. M., Gertsvolf, N. A., Ohlson, M. B., and Monack, D. M. (2015). cGAS and Ifi204 cooperate to produce type I IFNs in response to Francisella infection. J. Immunol. 194, 3236–3245. doi: 10.4049/jimmunol.1402764
Takaoka, A., Wang, Z., Choi, M. K., Yanai, H., Negishi, H., Ban, T., et al. (2007). DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature 448, 501–505. doi: 10.1038/nature06013
Toda, G., Yamauchi, T., Kadowaki, T., and Ueki, K. (2021). Preparation and culture of bone marrow-derived macrophages from mice for functional analysis. STAR Protoc. 2:100246. doi: 10.1016/j.xpro.2020.100246
Williams, N. C., and O’Neill, L. A. J. (2018). A role for the Krebs cycle intermediate citrate in metabolic reprogramming in innate immunity and inflammation. Front. Immunol. 9:141. doi: 10.3389/fimmu.2018.00141
Yan, S., Shen, H., Lian, Q., Jin, W., Zhang, R., Lin, X., et al. (2018). Deficiency of the AIM2–ASC signal uncovers the STING-driven Overreactive response of type I IFN and reciprocal depression of protective IFN-γ immunity in mycobacterial infection. J. Immunol. 200, 1016–1026. doi: 10.4049/jimmunol.1701177
Youle, R. J., and Van Der Bliek, A. M. (2012). Mitochondrial fission, fusion, and stress. Science 337, 1062–1065. doi: 10.1126/science.1219855
Keywords: Francisella tularensis, novicida, inflammasome, Aim2, immune suppression
Citation: Alqahtani M, Ma Z, Miller J, Yu J, Malik M and Bakshi CS (2023) Comparative analysis of absent in melanoma 2-inflammasome activation in Francisella tularensis and Francisella novicida. Front. Microbiol. 14:1188112. doi: 10.3389/fmicb.2023.1188112
Edited by:
Jason F. Huntley, University of Toledo, United StatesReviewed by:
Luke Kingry, Centers for Disease Control and Prevention (CDC), United StatesRoberto De Pascalis, United States Food and Drug Administration, United States
Kenta Watanabe, Yamaguchi University, Japan
Copyright © 2023 Alqahtani, Ma, Miller, Yu, Malik and Bakshi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Meenakshi Malik, TWVlbmFrc2hpLk1hbGlrQGFjcGhzLmVkdQ==; Chandra Shekhar Bakshi, U2hla2hhcl9CYWtzaGlAbnltYy5lZHU=
†These authors share senior authorship
 Maha Alqahtani1
Maha Alqahtani1 Zhuo Ma
Zhuo Ma Jen Yu
Jen Yu Meenakshi Malik
Meenakshi Malik Chandra Shekhar Bakshi
Chandra Shekhar Bakshi