
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 14 April 2023
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1184238
This article is part of the Research Topic Transmission Mechanisms and Control Strategies of Antimicrobial Resistome in Livestock Farms and Farmland Ecosystems View all articles
Agricultural fertilization caused the dissemination of antibiotic resistance genes (ARGs) in agro-ecological environment, which poses a global threat to crop-food safety and human health. However, few studies are known about the influence of different agricultural fertilization modes on antibiotic resistome in the paddy-upland rotation soils. Therefore, we conducted a field experiment to compare the effect of different fertilization (chemical fertilizer, slow release fertilizer and commercial organic fertilizer replacement at various rates) on soil antibiotic resistome in paddy-upland rotation fields. Results revealed that a total of 100 ARG subtypes and 9 mobile genetic elements (MGEs) occurred in paddy-upland rotation soil, among which MDR-ARGs, MLSB-ARGs and tet-ARGs were the dominant resistance determinants. Long-term agricultural fertilization remarkably facilitated the vertical accumulation of ARGs, in particular that blaampC and tetO in relative abundance showed significant enrichment with increasing depth. It’s worth noting that slow release fertilizer significantly increased soil ARGs, when comparable to manure with 20% replacing amount, but chemical fertilizer had only slight impact on soil ARGs. Fertilization modes affected soil microbial communities, mainly concentrated in the surface layer, while the proportion of Proteobacteria with the highest abundance decreased gradually with increasing depth. Furthermore, microbial community and MGEs were further proved to be essential factors in regulating the variability of ARGs of different fertilization modes by structural equation model, and had strong direct influence (λ = 0.61, p < 0.05; λ = 0. 55, p < 0.01). The results provided scientific guidance for reducing the spreading risk of ARGs and control ARG dissemination in agricultural fertilization.
The overuse of antibiotics accelerates the spread of antibiotic resistance genes (ARGs) among bacteria, thereby reducing the effectiveness of antibiotic therapy (Cohen et al., 2017). The proliferation of ARGs has been a major problem of antibiotic contamination in the environment, which may lead to the emergence of multidrug-resistant human pathogens and ultimately reduce the efficiency of antibiotic therapy (Mahnert et al., 2019; Tiedje et al., 2019). With the rapid development of large-scale breeding industry, the problem of ARGs contamination caused by antibiotic abuse is becoming increasingly prominent (Rahman et al., 2018). The farmland application of livestock manure can amend the soil fertility and ensure sustainable soil productivity, which has a long agricultural history, and is being vigorously promoted due to the rising cost of fertilizer as well as the necessity of unwanted wastes disposal (Zhen et al., 2014). Currently, returning manure to the field is an effective way of livestock manure resource utilization, which could realize the circulation of agricultural planting and breeding resources (Xie et al., 2018; Zhi et al., 2022).
However, ARGs inevitably transfers horizontally into the farmland along with fertilization, causing serious pollution to soil environment (Zhu D. et al., 2019; Han et al., 2021). Several studies have examined the effect of manure amendment on the fate and migration of ARGs in soil (Duan et al., 2019; Yang et al., 2020; Liu et al., 2021), where the use of manure containing residual antibiotics generally increased soil ARGs. Gu et al. (2020) found that application of pig manure, chicken manure and cow manure increased soil ARG abundances in soil by 36, 30 and 11 times, respectively, compared with the control soil. Liu et al. (2021) found that the abundance and diversity of ARGs showed an upward trend in farmland soils with repeated and continuous application of livestock manure. Obviously, manure application significantly increased the diversity and abundance of ARGs in soil, and also markedly changed the composition of bacteria related to ARGs profiles (Pu et al., 2020; Barrios et al., 2021; Guo et al., 2022). ARGs-resistant bacteria contained in the soil environment may enter the plant through horizontal transfer from endophytes, and transfer exogenous ARGs to the plant endophytic system (Guron et al., 2019; Chen et al., 2019b). A large number of ARGs and a variety of related integrons have been found in fresh fruits and vegetables, increasing the risk of transmission from ARGs to humans through the food chain (Zhang et al., 2019).
The accumulation and propagation of ARGs in soil were significantly affected by different fertilization patterns. Studies have shown that the application of chemical fertilizer and organic fertilizer can increase the level of ARGs compared with the soil without fertilization (Xu et al., 2019; Wang F. H. et al., 2020). In particular, the application of livestock manure can significantly increase the diversity and abundance of ARGs in soil, resulting in the improvement of microbial resistance level in soil environment (Gu et al., 2020; Yang et al., 2020; Han et al., 2021). In order to achieve “reducing fertilizer and controlling pollution,” it is necessary to optimize fertilization mode and structure to improve the use efficiency of agricultural fertilizer and accelerate the construction of ecological circular agriculture (Zhou et al., 2019a; Chen et al., 2021; Sanz et al., 2021; Xu F. et al., 2021). However, there are few studies on the distribution pattern and vertical migration of ARGs in paddy-upland rotation fields with unique habitat, especially under different fertilization modes (Lin et al., 2016; Rahman et al., 2018; Lin et al., 2022). The results are helpful to clarify the occurrence characteristics of soil ARGs pollution spectrum under different fertilization modes, further understand the effects of different fertilization modes on farmland ecosystem, and explore the optimal fertilization mode.
In this study, the paddy-upland rotation fields in Erhai River basin of Yunnan Province were selected for research, where were set up a plot experiment (chemical fertilizer, substitution model of different amount of livestock manure, slow release fertilizer). High throughput-quantitative PCR (HT-qPCR) combined with real-time quantification PCR (qPCR) were used to explore the occurrence profiles and vertical migration patterns of ARGs in paddy-upland rotation fields under different fertilization modes. Meanwhile, we also further studied microbial communities and environmental factors, and their contribution on ARG shift in paddy-upland rotation fields, further aiming to find an appropriate agricultural fertilization to mitigate and control the spread of ARGs in farmland. This work would provide a reference for the pollution control and risk assessment of ARGs in paddy-upland rotation soil.
The field experiment 8 years’ fertilization was conducted at an experimental station located in Dali City (100°10′E, 25°53′N), Yunnan Province, China. The experimental plot has maintained a fixed paddy-upland rotation mode of the rice-garlic and rice-broad bean rotation cycles. The rice used was Oryza sativa L. (japonica hybrid early-maturing varieties), the garlic planted was Allium sativum L (medium-maturing varieties), and the broad bean is Vicia faba L (medium-maturing varieties), which are highly suitable for planting in Yunnan Province. Initial physical and chemical properties (mean values) of soil in experimental area were as follows: total nitrogen (TN), 3.3 g/kg; total phosphorus (TP), 0.97 g/kg; total potassium, 19.3 g/kg; pH, 7.1; organic matter (OM), 57.3 g/kg.
The experimental field was designed in random block design with 8 treatments: no fertilization (CK); conventional fertilizer treatment (CF); conventional fertilizer reduction 20% treatment (T1); conventional fertilization replaced by 100% organic fertilizer replacement in terms of nitrogen (T2); conventional fertilization replaced by 100% organic fertilizer replacement in terms of phosphorus (T3); conventional fertilization replaced by 100% organic fertilizer replacement in terms of nitrogen under considering 25% organic fertilizer mineralization rate (T4); conventional fertilization replaced by 100% organic fertilizer replacement in terms of phosphorus under considering 25% organic fertilizer mineralization rate (T5); slow release fertilizer instead of chemical fertilizer (T6). Fertilizer application amount of each treatment is shown in Supplementary Table S1. The commercial organic fertilizer used in this experiment consists of 80% cow manure and 20% other animal manure. There were three plots for each treatment, and a total of 24 plots for 8 fertilization modes. The area of each experimental plot is 30 m2 (6 m × 5 m), and the ridge of field between plots was built with cement (the width is 0.24 m, the height is 0.20 m, and 1 m is built below the ground surface). Soil samples were collected from different depths of 0–20 cm, 20–50 cm and 50–80 cm after garlic harvest in May. Each plot was sampled at 5 points and mixed evenly, and then about 1 kg of the samples were placed in the refrigerator at −20°C by quartering method for DNA extraction.
Soil samples DNA was extracted from 0.5 g (dry weight) soil samples collected at different soil depths using FastDNA® Soil Rotation kit (MP Biomedicals, LLC, Santa Ana, United States) according to the manufacturer’s protocol. The concentration and purity of DNA extracted from soil samples were measured on a NanoVue Plus spectrophotometer (GE Healthcare, United States) and the concentration of ARGs were recalculated based on the weight of the soil used for DNA extraction. Finally, DNA samples extracted from soils with different fertilization modes were stored at−20°C for subsequent measurement.
High-throughput quantitative PCR (HT-qPCR) reactions adopted Wafergen SmartChip Real-time PCR system (MicroAnaly Genetech Co., Ltd., China), which qualified 134 ARGs, 7 transpoase genes, 2 integrase genes and 16S rRNA. Details of the primers used in this study are listed in Supplementary Table S2, and the design of the primers used followed previous studies (Zhu et al., 2013; Wang et al., 2016). The reaction system contained 2 ng/μL DNA template, 500 nM each primer, 1 × LightCycler 480 SYBR Green I Master mix and 0.1 mg/ml albumin from bovine serum. The procedure steps were set as follows: initial denaturation at 95°C for 10 min, followed by 40 cycles, including denaturation at 95°C for 30 s, annealing at 60°C for 30 s (Chen et al., 2017; Han et al., 2018).
Real-time quantification PCR (qPCR) reactions were conducted on a 7,500 Real-Time PCR System (Applied Biological) using the DNA/RNA/RNase-free 96-well reaction plates. The total abundance of bacteria was evaluated by analyzing 16S rRNA genes of bacteria, and the variation of ARGs abundance caused by differences in background bacterial levels and DNA extraction efficiency was corrected. The qPCR reaction mixtures (20 μl) consists of 6.8 μl RNAase-free water, 0.4 μl ROX Reference DyeII, 10 μl SYBR® Premix Ex Taq TM II (Tli RNase H Plus, Takara), 0.4 μl each primer and 2 μl DNA extract with a suitable concentration or standard plasmid. The qPCR amplification conditions were set as follows: initial denaturation (95°C, 30 s), then 40 cycles of 5 s at 95°C and 34 s at 60°C. Melting curves were analyzed at temperatures ranging from 60°C to 95°C, with 1°C/read to ensure specificity, and the method of calibration curves for each target gene was shown in previous studies (Yang et al., 2019). Each DNA sample was amplified in three replicates, and RNAase-free water was used as a negative control, a threshold cycle of 31 was used as the detection limit (Ouyang et al., 2015; Xie et al., 2016). The specificity was detected by melting curve, while the fold change, relative abundance of each target gene and the normalized copy numbers of ARGs per bacterial cell (normalized abundance) were calculated according to previous studies (Su et al., 2015; Chen et al., 2016; Wang et al., 2017).
Soil bacterial communities were analyzed by next-generation amplicon sequencing of 16S rRNA genes. PCR was used to amplify the V4 hypervariable region of 16S rRNA gene in bacteria and archaea with primer 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and the reverse primer 806R (5′-GGACTACHVGGGTWTCTAAT-3′). PCR mixture (50 μl) consists of 10 μl DNA template, 3 μl each primer, 6 μl nuclease-free PCR-grade water, 3 μl DMSO and 25 μl Phusion High-Fidelity PCR Master Mix, and the PCR amplification process was as follows: initial denaturation at 98°C for 30 s, followed by 30 cycles consisting of denaturation at 98°C for 15 s, annealing at 58°C for 15 s, and extension at 72°C for 15 s. The PCR system was finally kept at 72°C for 1 min. PCR products were purified and subjected on Illumina HiSeq4000 platform of GUHE INFO (Hangzhou, China). High-quality clean reads were obtained by qualitative filtering of the original reads, with sequences with a similarity of 97% clustered into the same Operational Taxonomic Units (OTUs) (Edgar, 2013).
The content of organic matter (OM), total nitrogen (TN), total phosphorus (TP) and pH of the soil samples were determined according to the previous studies and the national standard methods (Han et al., 2018; Cheng et al., 2019; Mu et al., 2022). The OM in paddy soil was determined by dichromate oxidation and titration with ferrous sulfate solution. The TN and TP in soil were determined by Semi-micro Kjeldahl Method and spectrophotometry. pH was determined with a pH meter (in a 1:5 soil-water ratio suspension).
The abundance and standard deviation of all genes were calculated by Microsoft Excel 2010. SPSS 22.0 was used for correlation analysis and ANOVA analysis, and p < 0.05 was taken as the significant difference level. Histogram, heat map and boxplot plots were drawn using Origin 2021 software, and RDA analysis was performed using Canoco5. Structural equation models (SEMs) was established using R 4.2.2 with the “piecewiseSEM” package to describe the relationship among fertilization treatment, soil properties, bacterial community, mobile genetic elements (MGEs) and the abundance of ARGs in paddy-upland soil (Lefcheck, 2016). We used the Fisher’s C test to judge the goodness of the modeling results. The models were modified stepwise according to the pathway significance (p < 0.05) and the goodness of the model (0 ≤ Fisher’s C/df ≤ 2 and 0.05 < p ≤ 1.00).
A total of 100 ARGs subtypes and 9 MGEs (7 transposon genes and 2 integron genes) were detected in paddy-upland rotation soils by HT-qPCR assay (Figure 1A). From the perspective of different categories and mechanisms of action, the ARGs detected in paddy-upland rotation soils included the various types of ARGs (acc-ARGs, bla-ARGs, sul-ARGs, MLSB-ARGs, MDR-ARGs, tet-ARGs, van-ARGs and other ARGs; Figure 1B). Among them, tet-ARGs and MLSB-ARGs were more detected in paddy-upland rotation soil (22.9 and 21.1%, respectively). MDR-ARGs and bla-ARGs account for 17.0 and 13.6%, which were high risk for the treatment of human diseases because they could endow bacteria with multidrug resistance, and have been reported in soil by many studies (Chen et al., 2016; Xie et al., 2018; Li et al., 2019). acc-ARGs accounted for 11.1%, sul-ARGs and van-ARGs accounted for 6.1 and 5.1%, respectively. Detected ARGs were classed into four resistance mechanisms in all treatments, including antibiotic deactivation, efflux pump, cellular protection and others, in which antibiotic deactivation and efflux pump (accounting for 36.7 and 37.5%, respectively) were the two dominant mechanisms (Figure 1C). As for the abundance of ARGs in paddy-upland rotation soils, the abundance of MDR-ARGs was the highest, and the sum of all samples was 3.34 × 105 copies, among which the mexF abundance of each subtype was the highest. Secondly, sul-ARGs, tet-ARGs and bla-ARGs was more abundant. The total abundance of all treatments with fertilizer application ranged from 6.8 × 104 to 9.6 × 104 copies/g, organic fertilizer application from 1.9 × 105 to 9.8 × 105 copies/g, and slow release fertilizer application from 1.6 × 105 copies/g. Besides, the total abundance of ARGs in organic fertilizer and slow-release fertilizer treatments was also significantly higher than that in unfertilized soil ((4.5 ± 0.6) × 104 copies/g). The above results indicated that agricultural fertilization could increase the pollution load of ARGs in the farmland soil to different degrees, other students have made similar findings (Rahman et al., 2018; Xu Y. et al., 2021).
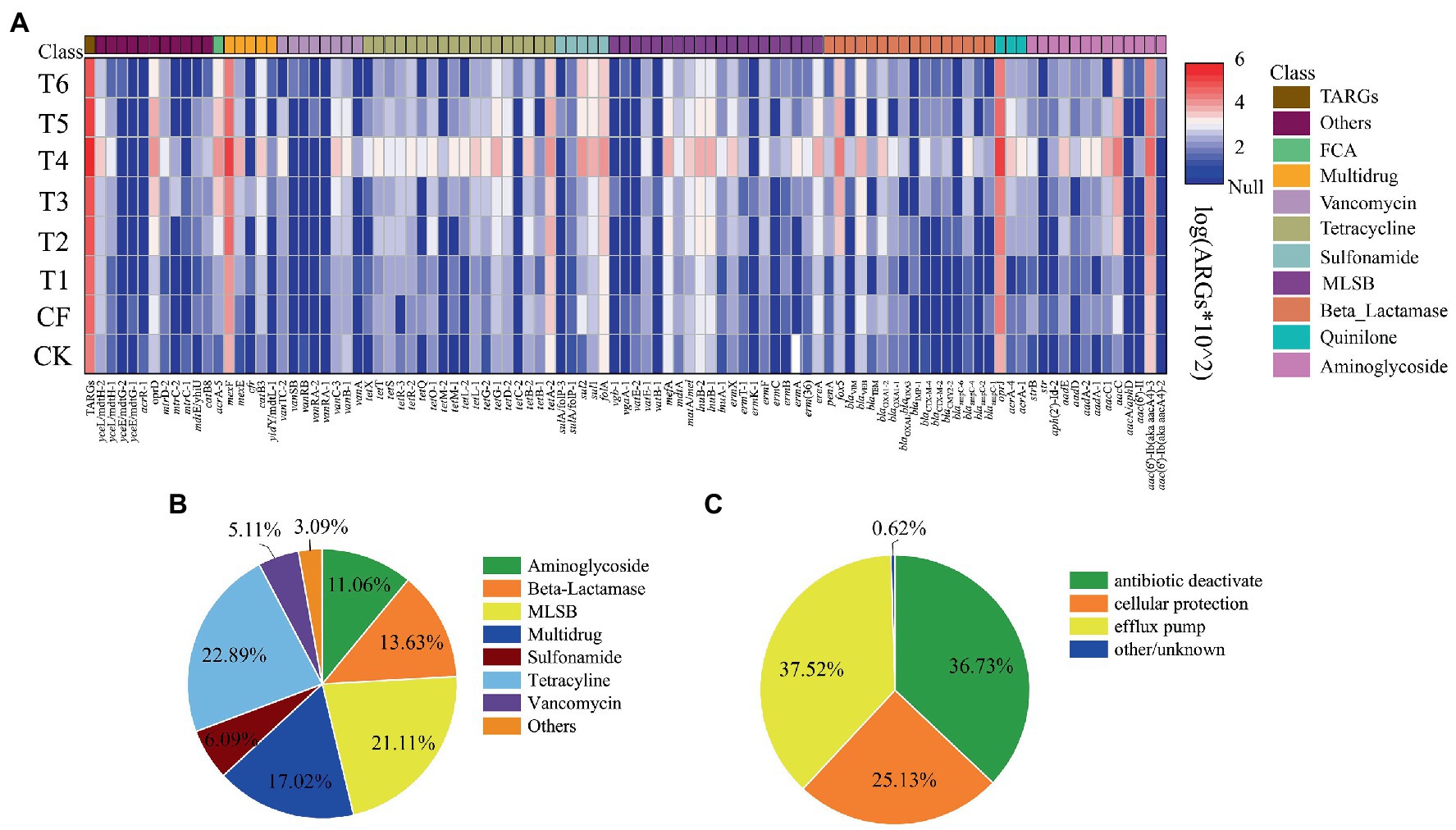
Figure 1. The content of detected ARGs under different fertilization modes in high-throughput sequencing technology (A) the abundance of all genes detected, (B) proportion of different subtypes of ARGs, (C) proportion of antibiotic resistance mechanisms. CK, no fertilization; CF, conventional fertilization; T1, conventional fertilization was reduced by 20%; T2, organic fertilizer replaced T1 (in terms of nitrogen); T3, organic fertilizer replaced T1 and urea supplemented nitrogen (in terms of phosphorus); T4, considering the mineralization rate of organic fertilizer is 25%, organic fertilizer replaced T1 (in terms of nitrogen); T5, considering the mineralization rate of organic fertilizer is 25%, organic fertilizer replaces T1, and urea supplemented nitrogen (in terms of phosphorus); T6, slow release fertilizer.
It is worth noting that the diversity of ARGs in paddy-upland rotation soil varied with different fertilization modes (Figure 2). There were 68 ARGs subtypes in the paddy-upland rotation soil without fertilization (CK), and 83 ARG subtypes in the T6 soil with slow and controlled release fertilizer. The gene subtypes of ARGs were the most abundant in manured soils of T2, T3, T4 and T5, which was consistent with a previous study, in which the diversity of ARGs increased in the soil environment after application of organic fertilizer (Nolvak et al., 2016; Pu et al., 2020). Moreover, similar to the diversity characteristics of ARGs pollution, MGEs were detected at the largest number in T4 with the largest amount of manure application, which was significantly higher than that in other fertilization types (p < 0.05). According to the Venn diagram (Supplementary Figure S1), 53 gene subtypes (48.6%) of ARGs were core ARGs (i.e., co-occurring ARGs) in paddy-upland rotation soils of six different fertilization modes. Among them, the diversity and number of ARGs were the lowest in CK, and the number of ARGs increased in other treatments after fertilization. Notably, mineral fertilizer application also affected ARG abundance to a certain extent, which may be related to changes in soil microbial structure (Sun et al., 2019). The total detected number of ARGs was the highest in the treatment of manure application, followed by the treatment of slow release fertilizer and chemical fertilizer, which were 1.4, 1.3 and 1.1 times higher than that in unfertilized soil, respectively. The diversity of ARGs in soil with organic fertilizer application was more complex than that in unfertilized soil, which significantly changed the composition of ARGs (p < 0. 05) and was consistent with previous studies (Zhang et al., 2017; Han et al., 2018; Guo et al., 2022).
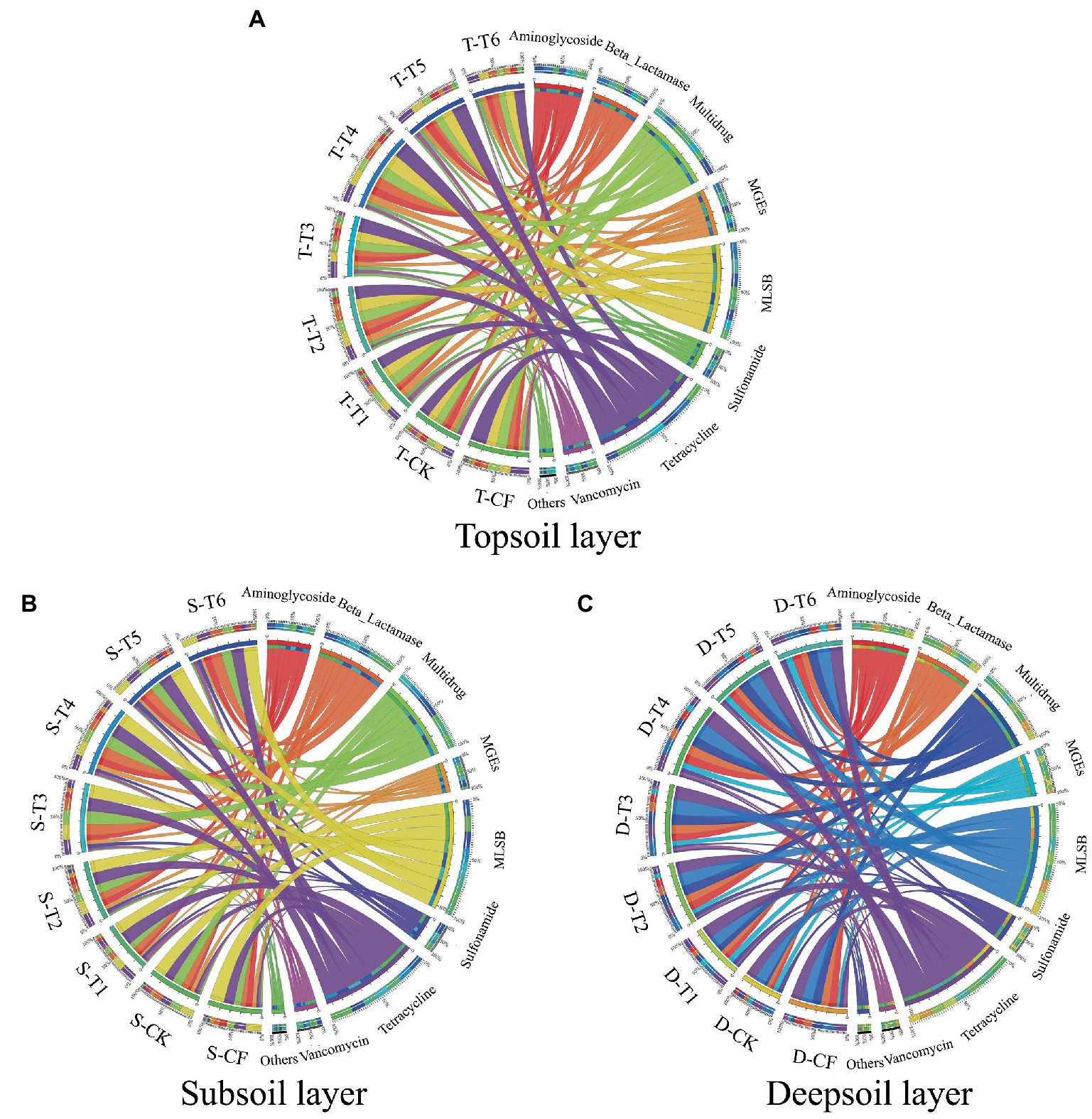
Figure 2. Distribution of each class of ARGs in paddy-upland rotation soil with different fertilization modes and different depths, (A) showing 0–20 cm in the topsoil, (B) showing 20–50 cm in the subsoil, and (C) showing 50–80 cm in the deepsoil. Please refer to Figure 1 for fertilization treatment abbreviations.
Based on the diversity of ARGs pollution in paddy-upland rotation soils under different fertilization methods, ARGs abundance was calculated to reveal the effect of fertilization modes on antibiotic resistome in soil. Figure 3 showed that the application of chemical fertilizer, organic fertilizer, slow release fertilizer all increased the abundance of ARGs. In paddy-upland rotation soils, the application of chemical fertilizer (T1, T2) increased str-ARGs and erm-ARGs slightly, while the application of slow-release fertilizer (T6) increased str-ARGs and sul-ARGs significantly. In addition, the application of commercial organic fertilizer in soil significantly increased the abundance of tet-ARGs, str-ARGs, erm-ARGs and sul-ARGs, which were greatly affected by the introduction of these genes into soil. Guo et al. (2018) also reported higher relative abundance of tet-ARGs and sul-ARGs in manure-amended soils. Additionally, consistent with previous studies, the level of ARG abundance in soil under fertilizer application was similar to that of CK, and the effect of chemical fertilizer on the ARGs abundance of paddy-upland rotation soil was not obvious (Liu et al., 2017; Wang F. H. et al., 2020). The total relative abundance was still the highest in T4, followed by T5 and T6, which were 5.5, 2.3 and 1.2 times of CK, respectively. The order of total relative abundance was T4 > T5 > T6 > T1 > T3 > T2 > CF > CK, the rule of ARGs abundance was slightly different from that of absolute abundance, but the effect of fertilizer application on ARGs abundance in paddy-upland rotation soil was basically that of commercial organic fertilizer > slow release fertilizer > chemical fertilizer.
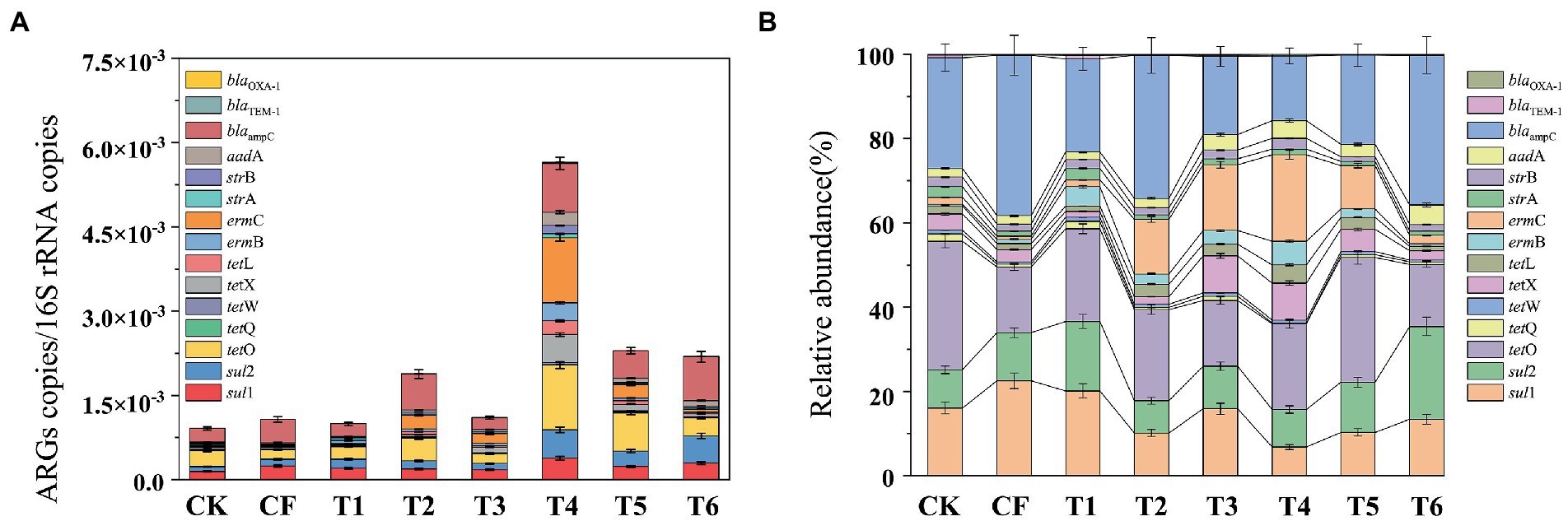
Figure 3. The level (A) and the proportion (B) in relative abundance of ARGs in paddy-upland rotation soil under different fertilization modes. Please refer to Figure 1 for fertilization treatment abbreviations.
The proportion of abundance of various ARGs in different fertilization modes were the same, blaampC occupied the highest proportion (the ratio of 15.30–60.01%), followed by sul1, sul2 and tetO, which is closely related to human health, was high in all paddy-upland rotation fields and its distribution was widespread, that should be paid attention to. Although the proportion of sul-ARGs (sul1 and sul2) decreased significantly in commercial organic fertilizer application, the abundance of sul-ARGs increased by 1.1–2.6 times compared with CK. In a previous study that applied cow manure to agricultural soils, sul-ARGs abundance was also significantly increased, with sul1 abundance increasing by 36% compared to background soil levels (Munir and Xagoraraki, 2011). This may be related to the fact that sulfonamides were commonly used as feed additives and antibiotics in the aquaculture industry, and since sulfonamides cannot be synthesized by microorganisms, sulfonamide resistance acquired by microorganisms was mainly due to the selective pressure (Guo et al., 2022). The distribution patterns of absolute and relative abundance of manure application treatments were slightly different, which might be caused by the difference of soil microbial biomass in different fertilization modes. In the treatment of chemical fertilizer, organic fertilizer and slow release fertilizer, the proportion of various ARGs changed, which may be due to the key role of host microorganisms in controlling the spread of ARGs in soil (Chen et al., 2019a; Li et al., 2021; Zhang et al., 2023). Overall, application of manure significantly increased the abundance of ARGs and changed the pollution distribution characteristics of different ARGs. The proportion of sul2 in T6 treatment with slow and controlled release fertilizer increased significantly compared with other treatments (p < 0.05), and the abundance was the highest in T6. Application of slow release fertilizer may have significant effects on sul2-related microbial community in soil to a large extent, which needs further study.
The absolute and relative abundances of ARGs showed different patterns in different soil layers under different fertilization treatments (p < 0.05). Supplementary Figure S2 showed that the five subtypes detected ARGs accumulated in the surface layer. In general, the absolute abundance of ARGs in paddy-upland rotation soils of different soil layers was the highest in topsoil, and decreased gradually with the increase of soil depth, with a distribution pattern of 0–20 cm > 20–50 cm > 50–80 cm. Moreover, the absolute abundance of sul-ARGs decreased most significantly in the subsoil, which decreased by about 1.5 logs compared with the topsoil. The absolute abundance of ARGs decreased in subsoil layer (20–50 cm), probably due to leaching from topsoil to deeper soil (Jechalke et al., 2014). For the relative abundance of ARGs in different soil layers, some categories accumulated in the surface layer, while others enriched in the deep layer, with inconsistent regularity. In terms of the average relative abundance of total ARGs, it generally showed a vertical accumulation to deepsoil, which was the same as the results of previous studies (Mckinney et al., 2018). This may be due to the fact that the paddy-upland rotation soil in this study was collected after harvest, and ARGs gradually migrated to the deeper layer after fertilization period of more than 1 month. In addition, after the flooding period of rice planting for a long time, the bacteria carrying ARGs accelerated their migration to the subsoil by the osmosis of water. In detail, sul-ARGs and tet-ARGs had the highest relative abundance in topsoil (average relative abundance of both was about 10−4), while erm-ARGs, str-ARGs and β-Lactam-ARGs had the highest relative abundance in subsoil (average relative abundance was about 10−4, 10−5 and 10−4). sul-ARGs was still higher in topsoil (8.18 × 10−5–9.98 × 10−4) than in subsoil and deepsoil (1.10 × 10−4–5.07 × 10−4 and 4.95 × 10−6–5.53 × 10−4), which may be related to the higher mobility of sulfa antibiotics and the lower soil absorption rate (Jechalke et al., 2014). Meanwhile, due to the uncertainty of soil properties, related MGEs and other factors, the distribution and accumulation of different types of ARGs in different soil layers may be different, with some enriched in 0–20 cm and others mainly in 50–80 cm.
The fertilization modes affected soil microbial community dramatically, and the soil microbial community change also occurred in the different dosage. Microorganisms were considered to be the main carriers of ARGs transmission, so the entry of host bacteria of ARGs in manure into the environment would change the local microbial community and promote the migration of ARGs (Li et al., 2020). Since fertilizer application and slow release fertilizer could not support plasmid transfer (Musovic et al., 2014), we supposed that the response of soil microbial community to fertilizer application might be a key factor in the changes of soil ARGs spectrum with inorganic fertilizer application. Over 100,000 sequences were obtained with over 40,000 sequences per sample on average. These sequences were assigned to 6,418 OTUs at 97% similarity. Rarefaction curves for OTUs indicated the sufficient depth of sequencing to account for most of the taxa amplified, ensuring the accuracy of sequencing. Figure 4A showed the compositions of the bacterial communities and the dynamic changes in the bacteria abundances at the top 20 phyla, respectively. The bacterial community compositions of the sample sites were similar at phylum level. Proteobacteria, Acidobacteria, Actinobacteria, Chloroflexi and Bacteriodetes were the dominant phyla, ranging from 69.6 to 87.6% of the total bacterial community. Proteobacteria was the most abundant phyla, accounting for 15.2 to 47.2%, dominated by the classes Alphaproteobacteria, Gammaproteobacteria and Betaproteobacteria. Acidobacteria was the second abundant phyla and dominated by Acidobacteria-Gp6 at class level.
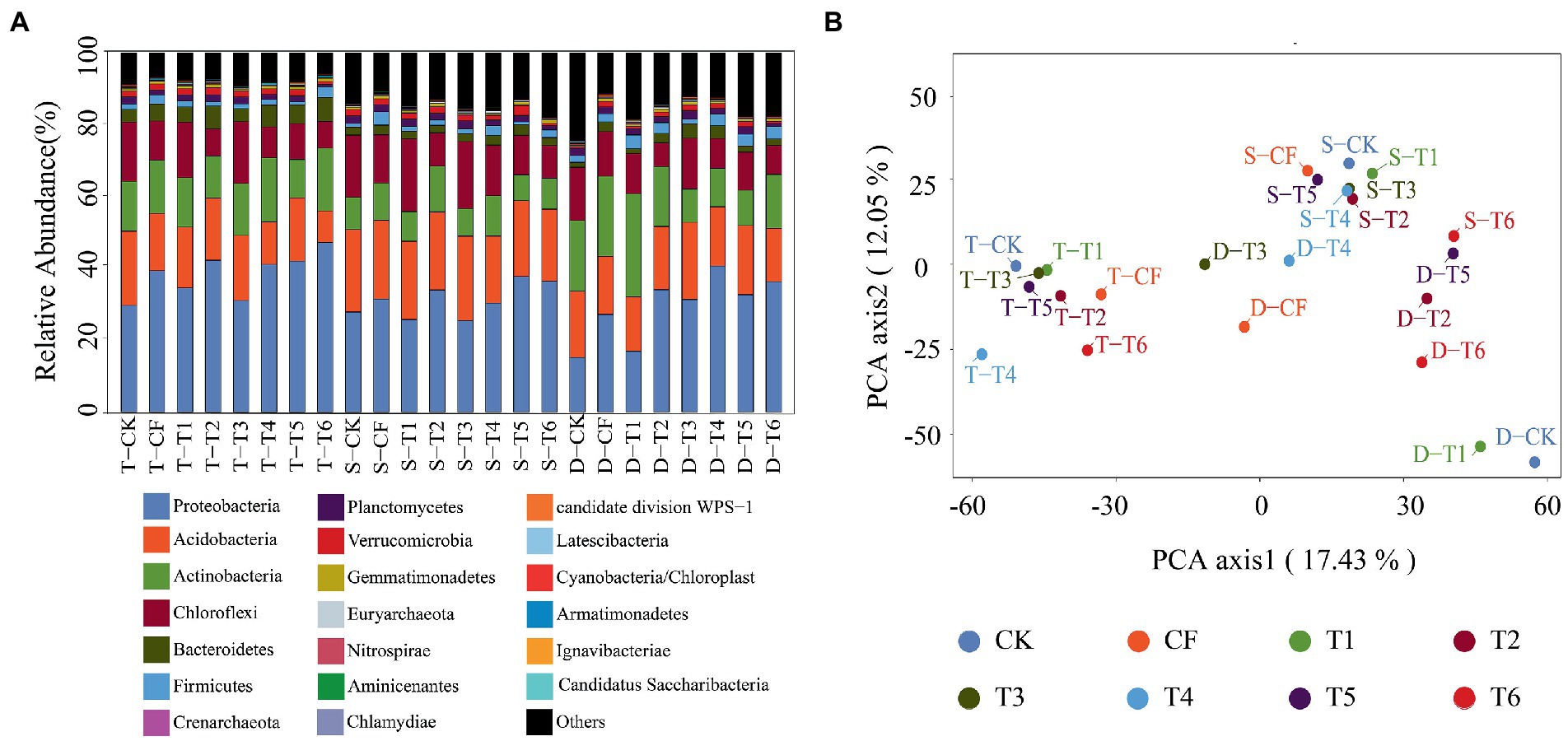
Figure 4. Distribution characteristics of soil microbial community under different fertilization modes, (A) showing the abundance of microbial community at each site across different soil layers; (B) showing PCA analysis of microbial communities. T. 0–20 cm in the topsoil; S. 20–50 cm in the subsoil, D. 50–80 cm in the deepsoil. Fertilization treatment abbreviation is shown in Figure 1.
Different fertilization modes changed the proportion of microbial abundance in microbial communities, and the abundance of microorganisms in different soil layers was also different due to the different soil properties. In the top five phylum levels, the proportion of Proteobacteria, Actinobacteria and Bacteroidetes in the soil with chemical fertilizer application (CF and T1) and slow controlled release fertilizer (T6) increased, while the proportion of Acidobacteria and Chloroflexi decreased. In T4 with the highest ARGs abundance, the proportion of Proteobacteria and Actinobacteria increased significantly (p < 0.05), 1.4 and 1.3 times of CK, respectively. Actinobacteria and Proteobacteria are considered to be important carriers of ARGs (Stegmann et al., 2015), and their increase could explain that the abundance of ARGs increases with the application of soil commercial organic fertilizer. The changes of ARGs in organic fertilizer application soil may be closely related to these two kinds of microorganisms. Consistent with previous studies (Chaudhry et al., 2012; Zhen et al., 2014), this work found an enhancement of manure and an inhibition of chemical fertilizer on the activity of soil microbial community; but the enhancement of manure was dosage-related. Therefore, the consequence of excessive use of manure involves not only a bloom of soil ARGs but also a suppression of soil microbial activity. The evolution of soil microbial community was significantly affected by the application of different fertilizers, and the composition of soil microbial community was closely related to ARGs migration.
Meanwhile, differences in the composition of microbial communities at different depths may also lead to changes in the abundance of ARGs. According to Principal component analysis (PCA) in Figure 4B, species diversity and microbial community composition of the samples in the same soil layer are similar, but there are differences in microbial community characteristics among different soil layers. The proportion of Proteobacteria with the highest abundance decreased significantly from 29.7–47.2% in topsoil to 15.2–40.7% in deepsoil. The effects of different fertilization methods on soil microbial communities at different depths were mainly concentrated in the surface layer, the evolution of microbial communities significantly affected the characteristics of ARGs (Zhang J. Y. et al., 2018).
The characteristics of ARGs in paddy-upland rotation soil under different fertilization modes were affected by many factors. In order to explore the potential factors responsible for the differences in the distribution of ARGs in different fertilizer modes, redundancy analysis was performed to determine the relationship between environmental factors, bacterial community and ARGs in this study, as shown in Supplementary Figure S3. Bacterial communities had an effect on the change of ARGs abundance, and Proteobacteria, which had the highest proportion in abundance and changed most obviously with the application of commercial organic fertilizer, was positively correlated with most changes of ARGs. Zhou et al. (2019b) also reported a high proportion of Proteobacteria in soil microorganisms applied with manure, and Proteobacteria pathogens may obtain ARGs from antibiotic-producing actinomycetes through horizontal transfer (Jiang et al., 2017). Soil microbial communities played a key role in controlling the spread of ARG in fertilized soils (Chen et al., 2019a).
In addition to microbes, changes of environmental factors (pH, TN, TP and OM) were also important indicators for tracking the variation of ARGs. The results showed that OM accounted for 48.1% of ARGs, which was significantly positively correlated with most of ARGs (p < 0.05). It was explained that the application of manure increased OM, which significantly affected the distribution characteristics of ARGs. Therefore, the amount of organic fertilizer applied has an impact on the abundance of ARGs, and reducing the application of manure can reduce the risk of ARGs to the soil environment to a certain extent. TN levels also explained the variations in ARGs between different fertilization modes samples, suggesting that fertilizer application may affect the distribution of ARGs in paddy-upland rotation soil, as some ARGs-carrying bacteria may increase due to increased nitrogen levels (Zhou et al., 2017; Qian et al., 2018). Recent studies have also confirmed that TN, TP and pH were important environmental factors driving the evolution of soil microbial community, possibly further leading to the change of ARGs in soil under different fertilization treatments (Cui et al., 2018; Wang F. H. et al., 2020). In addition, all samples were projected by Redundancy analysis (RDA), demonstrating a correlation between bacterial communities, environmental factors and the abundance of ARGs in soils with different fertilization modes. For different fertilization modes, Procrustes analysis (PC) (Figure 5) showed that changes in the abundance of ARGs in soil with fertilizer application were affected by both bacterial community and environmental factors, while different treatments in soil with organic fertilizer application were more concentrated in the environmental factors, and ARGs was mainly dominated by environmental factors.
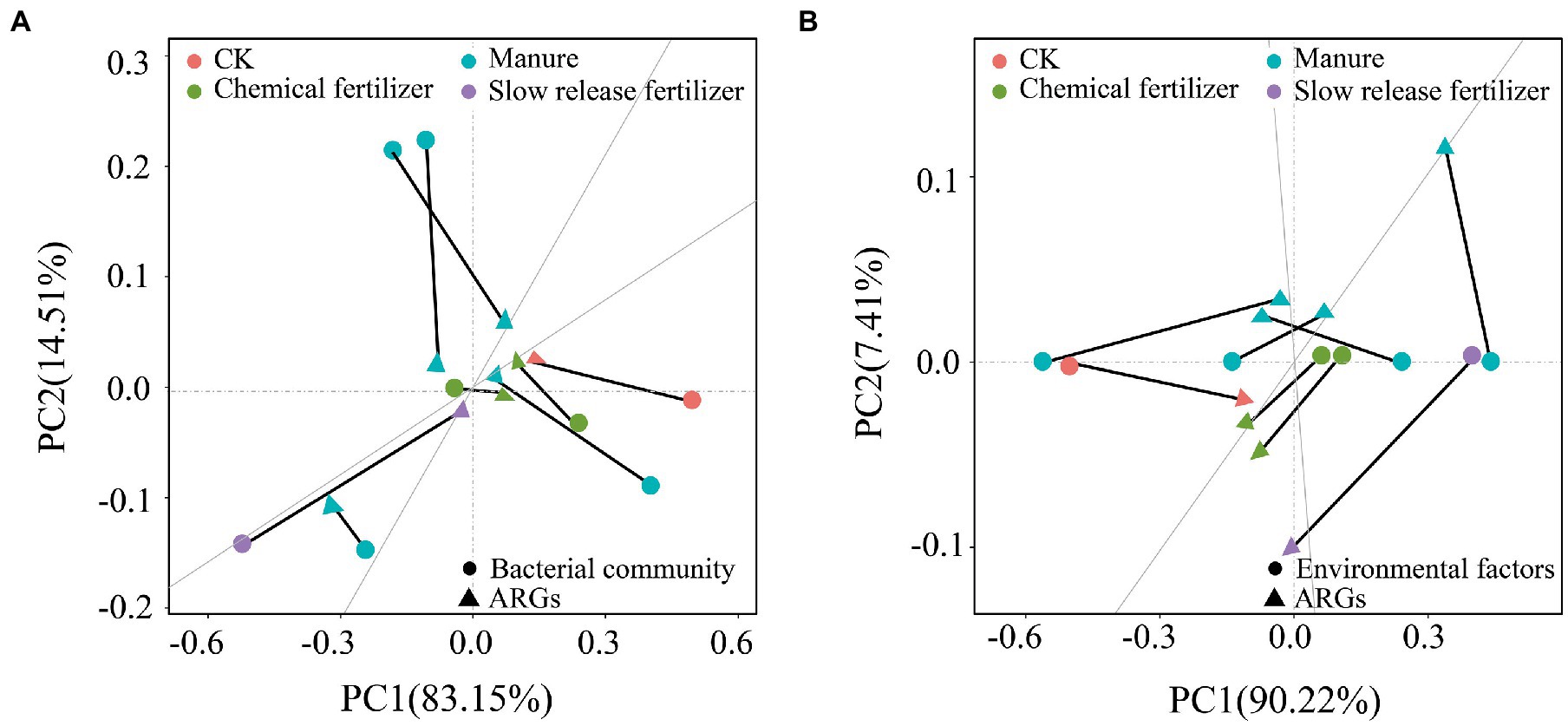
Figure 5. Procrustes analysis (PC) showing the correlation between microbial community (A) and environmental factors (B) with changes in ARGs in paddy soils under different fertilization modes.
The transmission and transfer of ARGs among environmental microorganisms are closely related to MEGs (Wang L. et al., 2020), so this study analyzed the correlation between ARGs and MGEs. The results showed that some ARGs were significantly correlated with intI1 and intI2, and MGEs promoted the migration and propagation of ARGs in paddy-upland rotation soil. The abundance of MGEs was also affected by different fertilization patterns, and its variation pattern was basically consistent with that of ARGs, showing change rule of T4 > T5 > T6 > T2 > T3 > T1 > CF > CK. Figure 6 showed the correlation coefficients between MGEs and the abundance of various ARGs. The results showed that intI1 was significantly positively correlated with tet-ARGs (tetO, tetQ, tetW, tetX and tetL), str-ARGs (strA, strB, aadA), ermB and blaOXA-1 (r > 0.95, p < 0.05), intI2 was also significantly positively correlated with str-ARGs (strA, strB, aadA), ermC, tetO and tetQ (r > 0.85, p < 0.05), which was consistent with previous findings that intI1 in environmental microorganisms is closely related to many ARGs (Guo et al., 2020; Zheng et al., 2020). The strong correlation may be due to the fact that the potential host bacteria of tet-ARGs and erm-ARGs share the same integrase gene of intI1 (Zhu et al., 2013, 2017b; Chen Z. et al., 2019). Compared with the non-fertilized soil, the abundance of intI1 in T4, T5 and T6 treatments increased by 14.8, 6.5 and 4.1 times, respectively, indicating that the application of organic fertilizer could promote the spread of ARGs in the soil (Nolvak et al., 2016; Fresia et al., 2019). MGEs were significantly enriched in paddy-upland rotation soil with organic fertilizer application and slow release fertilizer application. The higher abundance of MGEs and the faster horizontal gene transfer of ARGs promoted the further dissemination and evolution of ARGs in paddy-upland rotation soils, and the process interconnected the planetary microbiomes, thus forming a “high way” for the dispersal of ARGs (Zhu et al., 2017a; Zhu L. J. et al., 2019; Wang L. et al., 2020).
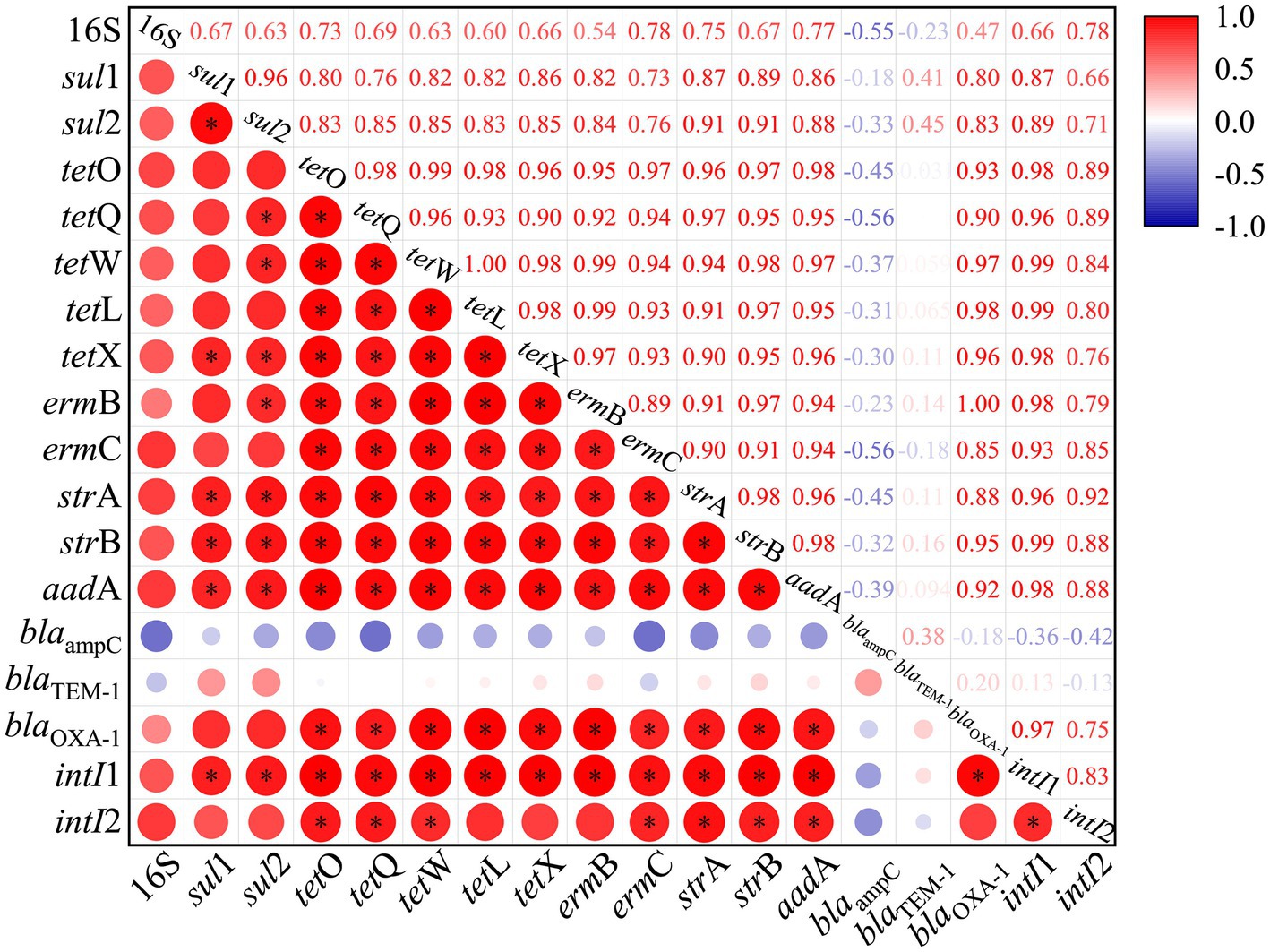
Figure 6. Correlation analysis between ARGs and MGEs in paddy soils, *Correlation is significant (p < 0.01).
The relationship between the abundance of ARGs and main driving factors was further depicted by Structural equation models (SEMs) (Figure 7). Different fertilization modes had significant direct effects on soil physicochemical properties (λ = −0. 85, p < 0.01) and bacterial communities (λ = 0. 20, p < 0.01), and indirectly affected ARGs abundance. Soil properties had a direct negative effect on the ARGs (λ = −0.19, p < 0.05) and can also indirectly affect the ARGs by altering bacterial community and MGEs, which both had a positive relationship with the ARGs. Previous studies have shown that TN is associated with efficient lateral transfer of ARGs, with more than 85% of sequenced ARGs and nitrogen processing genes present in the same host bacterial species (Zhang B. et al., 2018; Wang et al., 2021; Peng et al., 2022). Bacterial community directly impacted the abundance of ARGs (λ = 0.61, p < 0.05) also indirectly influenced MGEs (λ = −0.78, p < 0.05). MGEs can directly significantly alter the abundance of ARGs (λ = 0. 55, p < 0.01). intI1 gene is one of the most typical integrons and may carry a large number of ARGs fragments, which can be used as an ARGs indicator of influence (Gillings et al., 2015). Therefore, the increase of horizontal transfer potential of ARGs may be closely related to the high abundance of MGEs. Previous studies have shown that horizontal gene transfer plays an important role in ARG abundance under the condition of adequate bacterial community succession nutrition (Zhou et al., 2018; Yao et al., 2020), and in this study, microbial community and MGEs are also two factors that have a great influence on ARGs. Generally, the above analysis results showed that different fertilization modes had indirect effects on the distribution of ARGs by influencing the soil properties, microbial community and MGEs, among which the more significant effects on ARGs were related to the bacterial community of host bacteria and MGEs closely related to horizontal gene transfer.
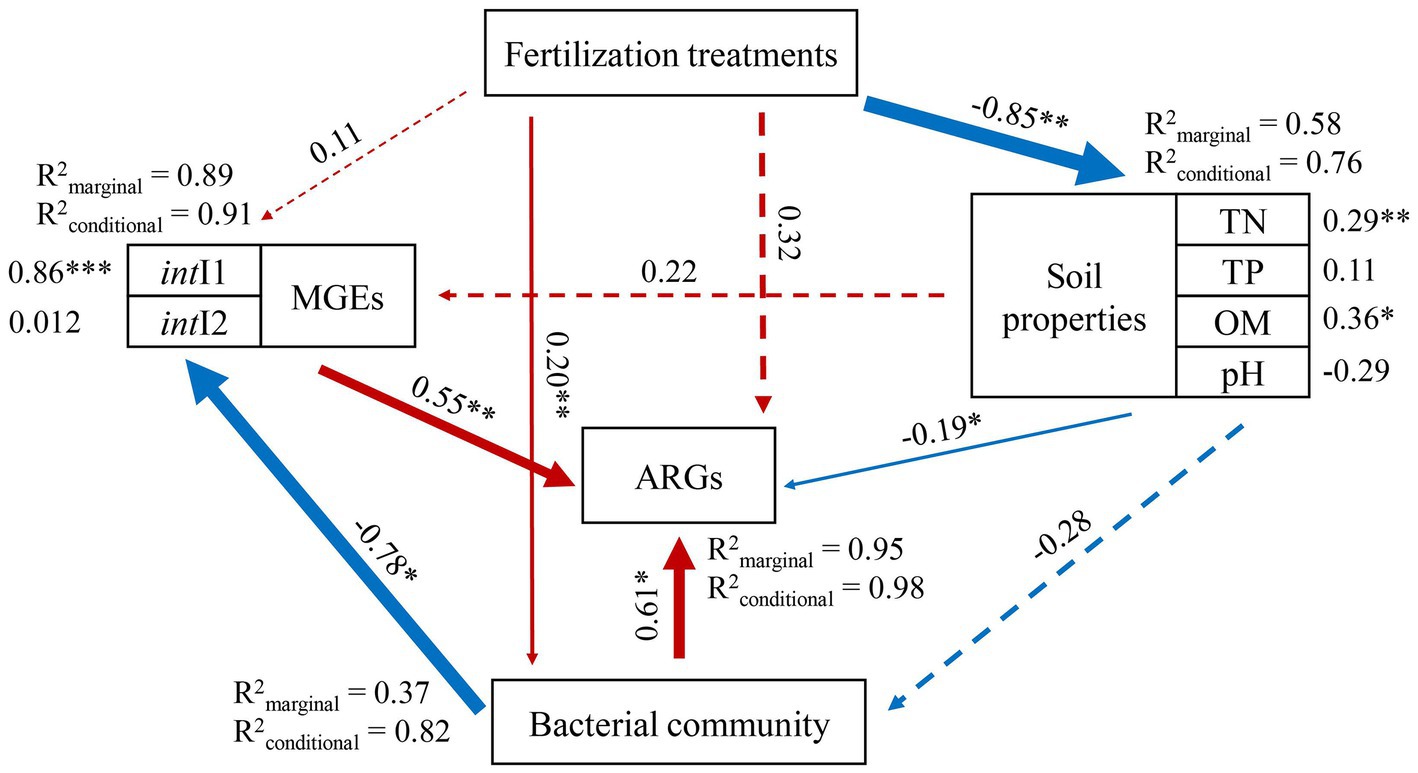
Figure 7. Structural equation models (SEMs) showing the effects of fertilization treatment, soil properties, bacterial community, MGEs and ARGs abundance in paddy-upland soil. Red and blue arrows represent positive and negative relationships, respectively. Solid and dashed arrows denote, respectively, significant and nonsignificant relationships. R2 represent the proportion of variance explained by all predictors. The hypothetical model fits the data well: Fisher’ C = 19.29, p = 0.37, df = 10, AIC = 59.29, BIC = 60.88. Significance levels of each predictor were *p < 0.05, **p < 0.01 and ***p < 0.001.
The present study, however, has some limitations. First, they fail to reproduce all the factors that might influence the fate of the ARGs. In particular, for paddy-upland rotation soil affected by weather and other natural factors, such as rainfall, drought and flood, previous studies observed soil ARGs migration in previous rainfall simulation experiments (Barrios et al., 2020; Hall et al., 2020). Secondly, the manure used in this study was commercial organic fertilizer refined from cow manure, and the effect of organic fertilizer made from other animal breeds on soil ARGs was not clear. There are some differences in the composition of manure of different animal breeds, and the results may have some influence.
Despite these limitations, ARGs in paddy-upland rotation soils with different fertilization modes had different pollution characteristics based on the results obtained from this study. The application of manure and slow release fertilizer had a greater burden on soil ARGs pollution, and changed the proportion of indigenous microbial community structure in the environment. Moreover, soil properties such as pH, nitrogen and phosphorus also fluctuated with the addition of fertilizers. We suggest to take on the fertilization modes to attention, thus minimizing the dispersal of resistance in soil and to human: (1) The application of manure should be appropriate, and it is best not to provide all the nutrients needed by crops with manure, which is easy to cause excessive nitrogen or phosphorus elements in the soil, and can promote the accumulation of ARGs to some extent; (2) In the same situation, when fertilizers are supplemented to crops, the application of slow release fertilizers should be reduced as far as possible, and simple chemical fertilizers can be used instead.
This study demonstrated that commercial organic and chemical/slow release fertilization resulted in redistribution of soil ARGs, which was closely related to the structural changes of soil microorganisms in different habitats. Meanwhile, MGEs and environmental factors, like OM and TN, also act as an important driving force in this process. Agricultural fertilization mainly increased the burden of ARGs pollution in topsoil, and some ARGs showed an enrichment trend with the increase of depth. Notably, there was no significant change in the abundance of total soil ARGs under chemical fertilizer at the current applied dose, while slow release fertilizer promoted the propagation of ARGs. These findings highlighted the potential role of fertilization patterns in influencing the fate of ARGs. The combined application of chemical fertilizer and organic fertilizer can provide enough crop nutrients and recycle waste resources to alleviate ARGs pollution and balance soil microbial community, so as to restore the health of soil environment and the stability of ecosystem. Therefore, we believe that the future agricultural fertilization needs to place more emphasis on precise positioning and matching, while the impact on soil ARGs also needs to be monitored and assessed over time.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
BH: data analysis and writing-original draft preparation. SS: sample pretreatment, DNA extraction, and MGEs detection. FY: supervision and writing-reviewing and editing. XW: ARGs detection and MGEs detection. WG: farm investigation and sample collection. KZ: project administration and supervision. All authors contributed to the article and approved the submitted version.
This study was financially supported by National Natural Science Foundation of China (42277033; 42077355; and 42107456), Fundamental Cutting-edge Projects of Research Institute (2022-jcqyrw-dyz), Central Public-interest Scientific Institution Basal Research Fund (No. Y2021PT01), Tianjin Dairy Cattle (mutton sheep) Industry Technology system innovation team construction project (ITTCRS2021000), and Transformation and Promotion Project of Agricultural Science and Technology Achievements in Tianjin (No. 202101040).
We would like to thank Yi Luo for her constructive suggestions on this manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1184238/full#supplementary-material
Barrios, R. E., Bartelt-Hunt, S. L., Li, Y., and Li, X. (2021). Modeling the vertical transport of antibiotic resistance genes in agricultural soils following manure application. Environ. Pollut. 285:117480. doi: 10.1016/j.envpol.2021.117480
Barrios, R. E., Khuntia, H. K., Bartelt-Hunt, S. L., Gilley, J. E., Schmidt, A. M., Snow, D. D., et al. (2020). Fate and transport of antibiotics and antibiotic resistance genes in runoff and soil as affected by the timing of swine manure slurry application. Sci. Total Environ. 712:136505. doi: 10.1016/j.scitotenv.2020.136505
Chaudhry, V., Rehman, A., Mishra, A., Chauhan, P. S., and Nautiyal, C. S. (2012). Changes in bacterial community structure of agricultural land due to long-term organic and chemical amendments. Microb. Ecol. 64, 450–460. doi: 10.1007/s00248-012-0025-y
Chen, Q. L., An, X. L., Li, H., Su, J. Q., Ma, Y. B., and Zhu, Y. G. (2016). Long-term field application of sewage sludge increases the abundance of antibiotic resistance genes in soil. Environ. Int. 92–93, 1–10. doi: 10.1016/j.envint.2016.03.026
Chen, Q. L., An, X. L., Li, H., Zhu, Y. G., Su, J. Q., and Cui, L. (2017). Do manure-borne or indigenous soil microorganisms influence the spread of antibiotic resistance genes in manured soil? Soil Biol. Biochem. 114, 229–237. doi: 10.1016/j.soilbio.2017.07.022
Chen, M. L., An, X. L., Liao, H., Yang, K., Su, J. Q., and Zhu, Y. G. (2021). Viral community and virus-associated antibiotic resistance genes in soils amended with organic fertilizers. Environ. Sci. Technol. 55, 13881–13890. doi: 10.1021/acs.est.1c03847
Chen, Q. L., An, X. L., Zheng, B. X., Gillings, M., Peñuelas, J., Cui, L., et al. (2019a). Loss of soil microbial diversity exacerbates spread of antibiotic resistance. Soil Ecol. Lett. 1, 3–13. doi: 10.1007/s42832-019-0011-0
Chen, Q. L., Cui, H. L., Su, J. Q., Penuelas, J., and Zhu, Y. G. (2019b). Antibiotic resistomes in plant microbiomes. Trends Plant Sci. 24, 530–541. doi: 10.1016/j.tplants.2019.02.010
Chen, Z., Zhang, W., Yang, L., Stedtfeld, R. D., Peng, A., Gu, C., et al. (2019). Antibiotic resistance genes and bacterial communities in cornfield and pasture soils receiving swine and dairy manures. Environ. Pollut. 248, 947–957. doi: 10.1016/j.envpol.2019.02.093
Cheng, J. H., Tang, X. Y., and Cui, J. F. (2019). Effect of long-term manure slurry application on the occurrence of antibiotic resistance genes in arable purple soil (entisol). Sci. Total Environ. 647, 853–861. doi: 10.1016/j.scitotenv.2018.08.028
Cohen, M. E., Salmasian, H., Li, J., Liu, J., Zachariah, P., Wright, J. D., et al. (2017). Surgical antibiotic prophylaxis and risk for postoperative antibiotic-resistant infections. J. Am. Coll. Surg. 225, 631–638e3. doi: 10.1016/j.jamcollsurg.2017.08.010
Cui, E. P., Gao, F., Liu, Y., Fan, X. Y., Li, Z. Y., Du, Z. J., et al. (2018). Amendment soil with biochar to control antibiotic resistance genes under unconventional water resources irrigation: proceed with caution. Environ. Pollut. 240, 475–484. doi: 10.1016/j.envpol.2018.04.143
Duan, M. L., Gu, J., Wang, X. J., Li, Y., Zhang, R. R., Hu, T., et al. (2019). Factors that affect the occurrence and distribution of antibiotic resistance genes in soils from livestock and poultry farms. Ecotoxicol. Environ. Safe 180, 114–122. doi: 10.1016/j.ecoenv.2019.05.005
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Fresia, P., Antelo, V., Salazar, C., Gimenez, M., D’Alessandro, B., Afshinnekoo, E., et al. (2019). Urban metagenomics uncover antibiotic resistance reservoirs in coastal beach and sewage waters. Microbiome 7, 35–39. doi: 10.1186/s40168-019-0648-z
Gillings, M. R., Gaze, W. H., Pruden, A., Smalla, K., Tiedje, J. M., and Zhu, Y. G. (2015). Using the class 1 integron-integrase gene as a proxy for anthropogenic pollution. ISME J. 9, 1269–1279. doi: 10.1038/ismej.2014.226
Gu, Y. R., Shen, S. Z., Han, B. J., Tian, X. L., Yang, F. X., and Zhang, K. Q. (2020). Family livestock waste: An ignored pollutant resource of antibiotic resistance genes. Ecotoxicol. Environ. Saf. 197:110567. doi: 10.1016/j.ecoenv.2020.110567
Guo, T., Lou, C., Zhai, W., Tang, X., Hashmi, M. Z., Murtaza, R., et al. (2018). Increased occurrence of heavy metals, antibiotics and resistance genes in surface soil after long-term application of manure. Sci. Total Environ. 635, 995–1003. doi: 10.1016/j.scitotenv.2018.04.194
Guo, A. Y., Pan, C. R., Ma, J. Y., and Bao, Y. Y. (2020). Linkage of antibiotic resistance genes, associated bacteria communities and metabolites in the wheat rhizosphere from chlorpyrifos-contaminated soil. Sci. Total Environ. 741:140457. doi: 10.1016/j.scitotenv.2020.140457
Guo, Y. W., Xiao, X., Zhao, Y., Liu, J. G., Zhou, J. Z., Sun, B., et al. (2022). Antibiotic resistance genes in manure-amended paddy soils across eastern China: occurrence and influencing factors. Front. Environ. Sci. Eng. 16, 1–11. doi: 10.1007/s11783-021-1499-y
Guron, G., Arango-Argoty, G., Zhang, L., Pruden, A., and Ponder, M. A. (2019). Effects of dairy manure-based amendments and soil texture on lettuce-and radish-associated microbiota and resistomes. mSphere 4, e00239–e00219. doi: 10.1128/msphere.00239-19
Hall, M. C., Mware, N. A., Gilley, J. E., Bartelt-Hunt, S. L., Snow, D. D., Schmidt, A. M., et al. (2020). Influence of setback distance on antibiotics and antibiotic resistance genes in runoff and soil following the land application of swine manure slurry. Environ. Sci. Technol. 54, 4800–4809. doi: 10.1021/acs.est.9b04834
Han, X. M., Hu, H. W., Chen, Q. L., Yang, L. Y., Li, H. L., Zhu, Y. G., et al. (2018). Antibiotic resistance genes and associated bacterial communities in agricultural soils amended with different sources of animal manures. Soil Biol. Biochem. 126, 91–102. doi: 10.1016/j.soilbio.2018.08.018
Han, B. J., Yang, F. X., Tian, X. L., Mu, M. R., and Zhang, K. Q. (2021). Tracking antibiotic resistance gene transfer at all seasons from swine waste to receiving environments. Ecotoxicol. Environ. Saf. 219:112335. doi: 10.1016/j.ecoenv.2021.112335
Jechalke, S., Heuer, H., Siemens, J., Amelung, W., and Smalla, K. (2014). Fate and effects of veterinary antibiotics in soil. Trends Microbiol. 22, 536–545. doi: 10.1016/j.tim.2014.05.005
Jiang, X., Ellabaan, M. M. H., Charusanti, P., Munck, C., Blin, K., Tong, Y., et al. (2017). Dissemination of antibiotic resistance genes from antibiotic producers to pathogens. Nat. Commun. 8, 1–7. doi: 10.1038/ncomms15784
Lefcheck, J. S. (2016). Piecewise SEM: piecewise structural equation modelling in r for ecology, evolution, and systematics. Methods Ecol. Evol. 7, 573–579. doi: 10.1111/2041-210x.12512
Li, H., Cheng, W., Li, B., Xu, Y., and Zheng, X. (2020). The fate of antibiotic resistance genes during co-composting of swine manure with cauliflower and corn straw. Bioresour. Technol. 300:122669. doi: 10.1016/j.biortech.2019.122669
Li, S., Yao, Q., Wei, D., Zhou, B. K., Zhu, P., Cui, X. A., et al. (2019). Profiles of antibiotic resistome with animal manure application in black soils of Northeast China. J. Hazard. Mater. 384:121216. doi: 10.1016/j.jhazmat.2019.121216
Li, Z. H., Yuan, L., Shao, W., and Sheng, G. P. (2021). Evaluating the interaction of soil microorganisms and gut of soil fauna on the fate and spread of antibiotic resistance genes in digested sludge-amended soil ecosystem. J. Hazard. Mater. 420:126672. doi: 10.1016/j.jhazmat.2021.126672
Lin, D., Huang, D., Zhang, J. H., Yao, Y. L., Zhang, G. Q., Ju, F., et al. (2022). Reduction of antibiotic resistance genes (ARGs) in swine manure-fertilized soil via fermentation broth from fruit and vegetable waste. Environ. Res. 214:113835. doi: 10.1016/j.envres.2022.113835
Lin, H., Sun, W. C., Zhang, Z. L., Chapman, S. J., Freitag, T. E., Fu, J. R., et al. (2016). Effects of manure and mineral fertilization strategies on soil antibiotic resistance gene levels and microbial community in a paddy-upland rotation system. Environ. Pollut. 211, 332–337. doi: 10.1016/j.envpol.2016.01.007
Liu, P., Jia, S. Y., He, X. W., Zhang, X. X., and Ye, L. (2017). Different impacts of manure and chemical fertilizers on bacterial community structure and antibiotic resistance genes in arable soils. Chemosphere 188, 455–464. doi: 10.1016/j.chemosphere.2017.08.162
Liu, W. B., Ling, N., Guo, J. J., Ruan, Y., Wang, M., Shen, Q. R., et al. (2021). Dynamics of the antibiotic resistome in agricultural soils amended with different sources of animal manures over three consecutive years. J. Hazard. Mater. 401:123399. doi: 10.1016/j.jhazmat.2020.123399
Mahnert, A., Moissl-Eichinger, C., Zojer, M., Bogumil, D., Mizrahi, I., Rattei, T., et al. (2019). Man-made microbial resistances in built environments. Nat. Commun. 10, 968–912. doi: 10.1038/s41467-019-08864-0
McKinney, C. W., Dungan, R. S., Moore, A., and Leytem, A. B. (2018). Occurrence and abundance of antibiotic resistance genes in agricultural soil receiving dairy manure. FEMS Microbiol. Ecol. 94:fiy010. doi: 10.1093/femsec/fiy010
Mu, M., Yang, F., Han, B., Tian, X., and Zhang, K. (2022). Manure application: a trigger for vertical accumulation of antibiotic resistance genes in cropland soils. Ecotoxicol. Environ. Saf. 237:113555. doi: 10.1016/j.ecoenv.2022.113555
Munir, M., and Xagoraraki, I. (2011). Levels of antibiotic resistance genes in manure, biosolids, and fertilized soil. J. Environ. Qual. 40, 248–255. doi: 10.2134/jeq2010.0209
Musovic, S., Klumper, U., Dechesne, A., Magid, J., and Smets, B. F. (2014). Long-term manure exposure increases soil bacterial community potential for plasmid uptake. Environ. Microbiol. Rep. 6, 125–130. doi: 10.1111/1758-2229.12138
Nolvak, H., Truu, M., Kanger, K., Tampere, M., Espenberg, M., Loit, E., et al. (2016). Inorganic and organic fertilizers impact the abundance and proportion of antibiotic resistance and integron-integrase genes in agricultural grassland soil. Sci. Total Environ. 562, 678–689. doi: 10.1016/j.scitotenv.2016.04.035
Ouyang, W. Y., Huang, F. Y., Zhao, Y., Li, H., and Su, J. Q. (2015). Increased levels of antibiotic resistance in urban stream of Jiulongjiang River, China. Appl. Microbiol. Biotechnol. 99, 5697–5707. doi: 10.1007/s00253-015-6416-5
Peng, S., Song, D., Zhou, B., Hua, Q., Lin, X., and Wang, Y. (2022). Persistence of Salmonella typhimurium and antibiotic resistance genes in different types of soil influenced by flooding and soil properties. Ecotoxicol. Environ. Saf. 248:114330. doi: 10.1016/j.ecoenv.2022.114330
Pu, Q., Zhao, L. X., Li, Y. T., and Su, J. Q. (2020). Manure fertilization increase antibiotic resistance in soils from typical greenhouse vegetable production bases, China. J. Hazard. Mater. 391:122267. doi: 10.1016/j.jhazmat.2020.122267
Qian, X., Gu, J., Sun, W., Wang, X. J., Su, J. Q., and Stedfeld, R. (2018). Diversity, abundance, and persistence of antibiotic resistance genes in various types of animal manure following industrial composting. J. Hazard. Mater. 344, 716–722. doi: 10.1016/j.jhazmat.2017.11.020
Rahman, M. M., Shan, J., Yang, P. P., Shang, X. X., Xia, Y. Q., and Yan, X. Y. (2018). Effects of long-term pig manure application on antibiotics, abundance of antibiotic resistance genes (ARGs), anammox and denitrification rates in paddy soils. Environ. Pollut. 240, 368–377. doi: 10.1016/j.envpol.2018.04.135
Sanz, C., Casado, M., Navarro-Martin, L., Tadić, Đ., Parera, J., Tugues, J., et al. (2021). Antibiotic and antibiotic-resistant gene loads in swine slurries and their digestates: implications for their use as fertilizers in agriculture. Environ. Res. 194:110513. doi: 10.1016/j.envres.2020.110513
Stegmann, E., Frasch, H. J., Kilian, R., and Pozzi, R. (2015). Self-resistance mechanisms of actinomycetes producing lipid II-targeting antibiotics. Int. J. Med. Microbiol. 305, 190–195. doi: 10.1016/j.ijmm.2014.12.015
Su, J. Q., Wei, B., OuYang, W. Y., Huang, F. Y., Zhao, Y., Xu, H. J., et al. (2015). Antibiotic resistome and its association with bacterial communities during sewage sludge composting. Environ. Sci. Technol. 49, 7356–7363. doi: 10.1021/acs.est.5b01012
Sun, Y. M., Qiu, T. L., Gao, M., Shi, M. M., Zhang, H. F., and Wang, X. M. (2019). Inorganic and organic fertilizers application enhanced antibiotic resistome in greenhouse soils growing vegetables. Ecotoxicol. Environ. Saf. 179, 24–30. doi: 10.1016/j.ecoenv.2019.04.039
Tiedje, J. M., Wang, F., Manaia, C. M., Virta, M., Sheng, H. J., Ma, L. P., et al. (2019). Antibiotic resistance genes in the human-impacted environment: a one health perspective. Pedosphere 29, 273–282. doi: 10.1016/s1002-0160(18)60062-1
Wang, F. H., Han, W. X., Chen, S. M., Dong, W. X., Qiao, M., Hu, C. S., et al. (2020). Fifteen-year application of manure and chemical fertilizers differently impacts soil ARGs and microbial community structure. Front. Microbiol. 11:62. doi: 10.3389/fmicb.2020.00062
Wang, H., Qi, J. F., Xiao, D. R., Wang, Z. B., and Tian, K. (2017). A re-evaluation of dilution for eliminating PCR inhibition in soil DNA samples. Soil Biol. Biochem. 106, 109–118. doi: 10.1016/j.soilbio.2016.12.011
Wang, F., Stedtfeld, R. D., Kim, O. S., Chai, B., Yang, L., Stedtfeld, T. M., et al. (2016). Influence of soil characteristics and proximity to Antarctic research stations on abundance of antibiotic resistance genes in soils. Environ. Sci. Technol. 50, 12621–12629. doi: 10.1021/acs.est.6b02863
Wang, L., Wang, J., Wang, J., Zhu, L., Conkle, J. L., and Yang, R. (2020). Soil types influence the characteristic of antibiotic resistance genes in green-house soil with long-term manure application. J. Hazard. Mater. 392:122334. doi: 10.1016/j.jhazmat.2020.122334
Wang, P., Wu, D., You, X., Su, Y., and Xie, B. (2021). Antibiotic and metal resistance genes are closely linked with nitrogen-processing functions in municipal solid waste landfills. J. Hazard. Mater. 403:123689. doi: 10.1016/j.jhazmat.2020.123689
Xie, W. Y., McGrath, S. P., Su, J. Q., Hirsch, P. R., Clark, I. M., Shen, Q., et al. (2016). Long-term impact of field applications of sewage sludge on soil antibiotic resistome. Environ. Sci. Technol. 50, 12602–12611. doi: 10.1021/acs.est.6b02138
Xie, W. Y., Yuan, S. T., Xu, M. G., Yang, X. P., Shen, Q. R., Zhang, W. W., et al. (2018). Long-term effects of manure and chemical fertilizers on soil antibiotic resistome. Soil Biol. Biochem. 122, 111–119. doi: 10.1016/j.soilbio.2018.04.009
Xu, Y., Li, H. Y., Shao, Z. L., Li, X. C., Zheng, X. Q., and Xu, J. (2021). Fate of antibiotic resistance genes in farmland soil applied with three different fertilizers during the growth cycle of pakchoi and after harvesting. J. Environ. Manag. 289:112576. doi: 10.1016/j.jenvman.2021.112576
Xu, F., Liu, Y. L., Du, W. C., Li, C. L., Xu, M. L., Xie, T. C., et al. (2021). Response of soil bacterial communities, antibiotic residuals, and crop yields to organic fertilizer substitution in North China under wheat-maize rotation. Sci. Total Environ. 785:147248. doi: 10.1016/j.scitotenv.2021.147248
Xu, M., Stedtfeld, R. D., Wang, F., Hashsham, S. A., Song, Y., Chuang, Y. H., et al. (2019). Composting increased persistence of manure-borne antibiotic resistance genes in soils with different fertilization history. Sci. Total Environ. 689, 1172–1180. doi: 10.1016/j.scitotenv.2019.06.376
Yang, F. X., Han, B. J., Gu, Y. R., and Zhang, K. Q. (2020). Swine liquid manure: a hotspot of mobile genetic elements and antibiotic resistance genes. Sci. Rep. 10, 15037–15010. doi: 10.1038/s41598-020-72149-6
Yang, F. X., Zhang, K. Q., Zhi, S. L., Li, J. J., Tian, X. L., Gu, Y. R., et al. (2019). High prevalence and dissemination of beta-lactamase genes in swine farms in northern China. Sci. Total Environ. 651, 2507–2513. doi: 10.1016/j.scitotenv.2018.10.144
Yao, L., Li, Y., Li, Z., Shen, D., Feng, H., Zhou, H., et al. (2020). Prevalence of fluoroquinolone, macrolide and sulfonamide-related resistance genes in landfills from East China, mainly driven by MGEs. Ecotoxicol. Environ. Saf. 190:110131. doi: 10.1016/j.ecoenv.2019.110131
Zhang, Y., Hao, X., Thomas, B. W., McAllister, T. A., Workentine, M., Jin, L., et al. (2023). Soil antibiotic resistance genes accumulate at different rates over four decades of manure application. J. Hazard. Mater. 443:130136. doi: 10.1016/j.jhazmat.2022.130136
Zhang, Y. J., Hu, H. W., Chen, Q. L., Yan, H., and He, J. Z. (2019). Manure application did not enrich antibiotic resistance genes in root endophytic bacterial microbiota of cherry radish. Appl. Environ. Microbiol. 86, e02106–e02119. doi: 10.1128/aem.02106-19
Zhang, Y. J., Hu, H. W., Gou, M., Wang, J. T., Chen, D., and He, J. Z. (2017). Temporal succession of soil antibiotic resistance genes following application of swine, cattle and poultry manures spiked with or without antibiotics. Environ. Pollut. 231, 1621–1632. doi: 10.1016/j.envpol.2017.09.074
Zhang, J. Y., Sui, Q. W., Tong, J., Zhong, H., Wang, Y. W., Chen, M. X., et al. (2018). Soil types influence the fate of antibiotic-resistant bacteria and antibiotic resistance genes following the land application of sludge composts. Environ. Int. 118, 34–43. doi: 10.1016/j.envint.2018.05.029
Zhang, B., Wang, M. M., Wang, B., Xin, Y., Gao, J., and Liu, H. (2018). The effects of bioavailable copper on macrolide antibiotic resistance genes and mobile elements during tylosin fermentation dregs co-composting. Bioresour. Technol. 251, 230–237. doi: 10.1016/j.biortech.2017.12.051
Zhen, Z., Liu, H. T., Wang, N., Guo, L. Y., Meng, J., Ding, N., et al. (2014). Effects of manure compost application on soil microbial community diversity and soil microenvironments in a temperate cropland in China. PLoS One 9:e108555. doi: 10.1371/journal.pone.0108555
Zheng, W. L., Huyan, J. Q., Tian, Z., Zhang, Y., and Wen, X. H. (2020). Clinical class 1 integron-integrase gene - a promising indicator to monitor the abundance and elimination of antibiotic resistance genes in an urban wastewater treatment plant. Environ. Int. 135:105372. doi: 10.1016/j.envint.2019.105372
Zhi, B., Xiang, S., Wang, Y., Dai, Z., Du, P., Wang, R., et al. (2022). Redeploy manure resources to enhance the agro-pastoral cycle. Sci. Total Environ. 846:157439. doi: 10.1016/j.scitotenv.2022.157439
Zhou, Z. C., Feng, W. Q., Han, Y., Zheng, J., Chen, T., Wei, Y. Y., et al. (2018). Prevalence and transmission of antibiotic resistance and microbiota between humans and water environments. Environ. Int. 121, 1155–1161. doi: 10.1016/j.envint.2018.10.032
Zhou, X., Qiao, M., Su, J. Q., Wang, Y., Cao, Z. H., Cheng, W. D., et al. (2019a). Turning pig manure into biochar can effectively mitigate antibiotic resistance genes as organic fertilizer. Sci. Total Environ. 649, 902–908. doi: 10.1016/j.scitotenv.2018.08.368
Zhou, X., Qiao, M., Su, J. Q., and Zhu, Y. G. (2019b). High-throughput characterization of antibiotic resistome in soil amended with commercial organic fertilizers. J. Soils Sediments 19, 641–651. doi: 10.1007/s11368-018-2064-6
Zhou, Z. C., Zheng, J., Wei, Y. Y., Chen, T., Dahlgren, R. A., Shang, X., et al. (2017). Antibiotic resistance genes in an urban river as impacted by bacterial community and physicochemical parameters. Environ. Sci. Pollut. Res. 24, 23753–23762. doi: 10.1007/s11356-017-0032-0
Zhu, Y. G., Gillings, M., Simonet, P., Stekel, D., Banwart, S., and Penuelas, J. (2017a). Microbial mass movements. Science 357, 1099–1100. doi: 10.1126/science.aao3007
Zhu, Y. G., Johnson, T. A., Su, J. Q., Qiao, M., Guo, G. X., Stedtfeld, R. D., et al. (2013). Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc. Natl. Acad. Sci. U. S. A. 110, 3435–3440. doi: 10.1073/pnas.1222743110
Zhu, D., Xinag, Q., Yang, X. R., Ke, X., O'Connor, P., and Zhu, Y. G. (2019). Trophic transfer of antibiotic resistance genes in a soil detritus food chain. Environ. Sci. Technol. 53, 7770–7781. doi: 10.1021/acs.est.9b00214
Zhu, Y. G., Zhao, Y., Li, B., Huang, C. L., Zhang, S. Y., Yu, S., et al. (2017b). Continental-scale pollution of estuaries with antibiotic resistance genes. Nat. Microbiol. 2, 16270–16277. doi: 10.1038/nmicrobiol.2016.270
Keywords: antibiotic resistance genes, fertilization modes, distribution patterns, vertical profile, paddy-upland rotation soil
Citation: Han B, Shen S, Yang F, Wang X, Gao W and Zhang K (2023) Exploring antibiotic resistance load in paddy-upland rotation fields amended with commercial organic and chemical/slow release fertilizer. Front. Microbiol. 14:1184238. doi: 10.3389/fmicb.2023.1184238
Received: 11 March 2023; Accepted: 24 March 2023;
Published: 14 April 2023.
Edited by:
Xuming Wang, Beijing Academy of Agricultural and Forestry Sciences, ChinaReviewed by:
Qing Yuan, Nanjing University, ChinaCopyright © 2023 Han, Shen, Yang, Wang, Gao and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fengxia Yang, eWFuZ2Zlbmd4aWFjb21lb25AMTYzLmNvbQ==; Keqiang Zhang, a2VxaWFuZ3poYW5nNjhAMTYzLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.