
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Microbiol., 09 May 2023
Sec. Infectious Agents and Disease
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1182612
Clostridioides difficile is a gram-positive, spore-forming, obligate anaerobe that infects the colon. C. difficile is estimated to cause nearly half a million cases in the United States annually, with about 29,000 associated deaths. Unfortunately, the current antibiotic treatment is not ideal. While antibiotics can treat the infections, they also disrupt the gut microbiota that mediates colonization resistance against enteric pathogens, including C. difficile; disrupted gut microbiota provides a window of opportunity for recurrent infections. Therefore, therapeutics that restore the gut microbiota and suppress C. difficile are being evaluated for safety and efficacy. This review will start with mechanisms by which gut bacteria affect C. difficile pathogenesis, followed by a discussion on biotherapeutics for recurrent C. difficile infections.
Clostridioides difficile is a spore-forming bacterium that infects the large intestine. Contact with C. difficile results in various outcomes: from no colonization to asymptomatic carriages, from mild diarrhea to life-threatening complications (Rupnik et al., 2009; Crobach et al., 2018). Clostridioides difficile infection (CDI) is one of the most common causes of healthcare-associated infections in the United States: the estimated burden of CDI was 462,100 cases in 2017, of which roughly half were healthcare-associated cases (Guh et al., 2020). The cumulative incidence of CDI ranges from 1.12 to 631.80 cases per 100,000 population per year based on a meta-analysis of the global CDI burden (Balsells et al., 2019).
One of the most challenging tasks in treating CDI is managing recurrent infections. Vancomycin and metronidazole have been used as first-line CDI treatments for decades. However, they also disrupt the commensal gut microbes; CDI recurrence occurs in at least 20–30% of cases within 60 days of either treatment, potentially involving failure to promptly restore the gut microbial community that defends against enteric pathogens (McFarland et al., 2002; Pépin et al., 2006; DuPont, 2011; Louie et al., 2011). In 2011, fidaxomicin, a narrow-spectrum antibiotic, was approved for the treatment of CDI. It selectively eradicates C. difficile while affecting the rest of the microbiota to a lesser extent than vancomycin (Tannock et al., 2010; Krutova et al., 2022). As a result, fidaxomicin is associated with reduced recurrence, but about 1 in 7 patients still relapse following treatment (Louie et al., 2011). Patients with recurrent CDI may benefit from tapered/pulsed antibiotic regimens or monoclonal antibodies (Wilcox et al., 2017; Johnson et al., 2021). However, neither therapies target “dysbiosis,” the root cause of the problem. Since the key to curing the infection lies in the intact gut microbiota, researchers and clinicians are implementing a two-pronged approach, involving antibiotics to remove the pathogen, followed by biotherapeutics to replenish the gut microbiota.
Microbiota protects against C. difficile by competing for nutrients, activating immunity, producing antibiotics, or modulating the gut metabolome (Horvat and Rupnik, 2018; Rosa et al., 2018). Additionally, the composition of the gut community influences disease severity. For example, bacteria capable of fiber degradation and bile acid metabolism were linked to less severe diseases. Meanwhile, some bacterial groups, including Escherichia, Streptococcus, Enterococcus, Helicobacter, and Klebsiella, were associated with worse infection outcomes (Schubert et al., 2015; Lesniak et al., 2022). Much is known about the colonization resistance conferred by the gut microbiota; however, only recent studies have started to reveal the interactions between specific gut bacterial species and C. difficile. The microbe-microbe interactions will be the topic of the first section below. In the second section of this review, I will discuss live biotherapeutics for CDI.
C. difficile spores must germinate into vegetative cells to colonize the gut and cause disease. Both processes: spore germination and vegetative growth, are influenced by bile acids (Schäffler and Breitrück, 2018). Primary bile acids are those synthesized by the liver and secreted into the intestinal lumen, where they are metabolized into secondary bile acids by the gut microbiota. While primary bile acids, such as cholate and taurocholate, stimulate C. difficile spore germination, secondary bile acids, such as lithocholate (LCA) and deoxycholate (DCA), inhibit the vegetative growth of C. difficile (Sorg and Sonenshein, 2008, 2010; Heeg et al., 2012; Thanissery et al., 2017).
The transformation from primary to secondary bile acids requires 7α-dehydroxylation. Only a few gut bacterial species have such activity; one of the best-studied is Clostridium scindens (Studer et al., 2016; Solbach et al., 2018). By combining metagenomic analyses and mathematical modeling, Buffie and colleagues identified a positive correlation between C. scindens and C. difficile resistance in clinical samples and mice (Buffie et al., 2015). In addition, adoptively transferring C. scindens to C. difficile-susceptible mice led to reduced pathogen burden, milder weight loss, and improved survival. C. scindens-mediated protection depends on bile acids since pretreatment of C. scindens-spiked intestinal content with a bile acid sequestrant abolished its C. difficile inhibitory capacity, while engraftment of C. scindens in gnotobiotic mice deficient in 7α-dehydroxylation restored DCA and LCA and delayed C. difficile expansion (Buffie et al., 2015; Studer et al., 2016). In agreement with these results, the baiCD gene cluster, which encodes a key enzyme in bile acid 7α-dehydroxylation, is less prevalent in fecal samples from CDI patients than samples from C. difficile negative individuals (Solbach et al., 2018).
Besides its role in secondary bile acid biosynthesis, C. scindens secretes 1-acetyl-β-carboline, a tryptophan-derived antibiotic; its antimicrobial activity against C. difficile is enhanced in the presence of DCA or LCA (Kang et al., 2019). The dual C. difficile-inhibitory mechanism and its effectiveness in animal models make this 7α-dehydroxylating bacterium and related Clostridium species promising probiotic candidates for CDI. However, Amrane and colleagues detected C. scindens in C. difficile-positive stool samples, indicating C. scindens, on its own, may not inhibit C. difficile in patients and need additional microbial components to protect against CDI (Amrane et al., 2018).
The gut microbiota also influences C. difficile expansion and pathogenesis via microbial-derived nutrients, especially following antibiotic treatments. Gut microbes, such as Bacteroides thetaiotaomicron, can cleave sialic acids from the mucosal glycoconjugates but lack the catabolic enzymes to consume the sugar, thereby supplying nutrients for others in the gut lumen (Martens et al., 2008). While the gut microbiota efficiently consumes sialic acids in healthy individuals, antibiotic treatments disrupt this equilibrium, resulting in a transient excess of sialic acids (Ng et al., 2013). C. difficile, by upregulating the sialic acid catabolic pathway, can utilize the now available sialic acids for growth and expansion. Similarly, perturbing the gut microbiota chemically or by antibiotics leads to a transient spike in microbiota-derived succinate levels in mice. C. difficile adapts to the metabolic shift, induces a pathway to metabolize succinate to butyrate, and gains a competitive advantage (Ferreyra et al., 2014).
In adult and pediatric patients, Enterococci positively correlate with C. difficile burden and susceptibility (Ozaki et al., 2004; Auchtung et al., 2020; Smith et al., 2022). Consistent with the clinical observations, Enterococci are associated with more severe CDI in mouse models. Mice colonized with vancomycin-resistant Enterococcus prior to C. difficile infection showed worse pathology of colonic tissues and increased toxin levels in the cecal content (Keith et al., 2020). In contrast, C. difficile colonization is delayed in mice receiving antibiotics that deplete Enterococci (Smith et al., 2022). How do Enterococci influence CDI susceptibility? Enterococcus species, such as Enterococcus faecalis, convert arginine to ornithine using the arginine deiminase pathway. The resulting gut lumen featuring high ornithine and low arginine favors C. difficile pathogenesis: C. difficile ferments ornithine for energy; meanwhile, arginine limitation may provide an environment cue for C. difficile to increase toxin production (Pruss et al., 2022; Smith et al., 2022). Other than Enterococci, commensal gut bacteria such as Clostridium sardiniense and Paraclostridium bifermentans can modulate C. difficile infection via the same pathway (Girinathan et al., 2021). C. sardiniense supplies ornithine to C. difficile and is associated with worsened infection outcomes in mice. In contrast, P. bifermentans competes for ornithine, protecting mice against lethal C. difficile infection.
Dietary sources also influence intestinal amino acid levels and C. difficile susceptibility. For example, a soy protein diet increased the abundance of Lactobacillus bacteria, which digest soy proteins into available amino acids for C. difficile; therefore, mice on a soy protein diet were more susceptible to CDI than mice given a diet that contains casein as a protein source (Yakabe et al., 2022). Future research exploring nutrient crossfeed in CDI patients and their influence on infection outcomes can help identify avenues for new therapeutic interventions.
Bacteriocins are antimicrobial peptides produced by bacteria that can have narrow- or broad-spectrum activities. Several gut bacterial species produce bacteriocins that can directly inhibit C. difficile. For example, Bacillus thuringiensis DPC 6431, a bacterial strain derived from human feces, produces bacteriocin thuricin CD, which has a narrow-spectrum of activity that kills C. difficile while sparing many intestinal commensals (Rea et al., 2010). The lytic activity of thuricin CD involves permeabilization and depolarization of the target cell membrane, likely due to pore formation (Mathur et al., 2017). Similarly, a human gut symbiont, Ruminococcus gnavus E1, synthesizes an antimicrobial sactipeptide, Ruminococcin C1 (RumC1). RumC1 demonstrated bactericidal activities against a panel of Gram-positive bacteria, including C. difficile, by possibly inhibiting nucleic acid synthesis (Chiumento et al., 2019).
Bacteriocin-mediated killing also occurs among different strains of C. difficile. Diffocins are phage tail-like R-type bacteriocins synthesized by C. difficile to kill non-self C. difficile strains. They act as molecular syringes to puncture cell membranes after binding to the target cell surface receptors, disrupt the membrane potential, and result in bacterial death (Gebhart et al., 2012; Schwemmlein et al., 2018). This potent inter-strain killing mechanism may explain how colonization with non-toxigenic C. difficile protects animals against subsequent C. difficile challenges (Borriello and Barclay, 1985). Whether these antibacterial peptides are produced in vivo and how they contribute to resistance against C. difficile in the host remains to be uncovered.
Eradicating C. difficile requires two actions: killing the vegetative cells and inhibiting spores. While antibiotics, such as vancomycin, are effective at the former, they fail to keep spores at bay: antibiotic-resistant spores can germinate into vegetative cells, produce toxins, and cause colonic inflammation again once antibiotic treatment discontinues (Rupnik et al., 2009). Furthermore, antibiotic treatments also disrupt the normal gut microbiota, alter the gut metabolic state, and leave an opportunity for pathogenic bacteria to thrive (Theriot et al., 2014; Vincent and Manges, 2015; Theriot et al., 2016; Staley et al., 2017; Contijoch et al., 2019). Therefore, probiotic-based therapies that restore the gut microbiota composition and provide sustained protection became attractive alternatives. Figure 1 summarizes live biotherapeutic products (LBPs) for recurrent CDI at various stages of clinical development. They represent two broad categories: donor-derived microbiota products and defined microbial components.
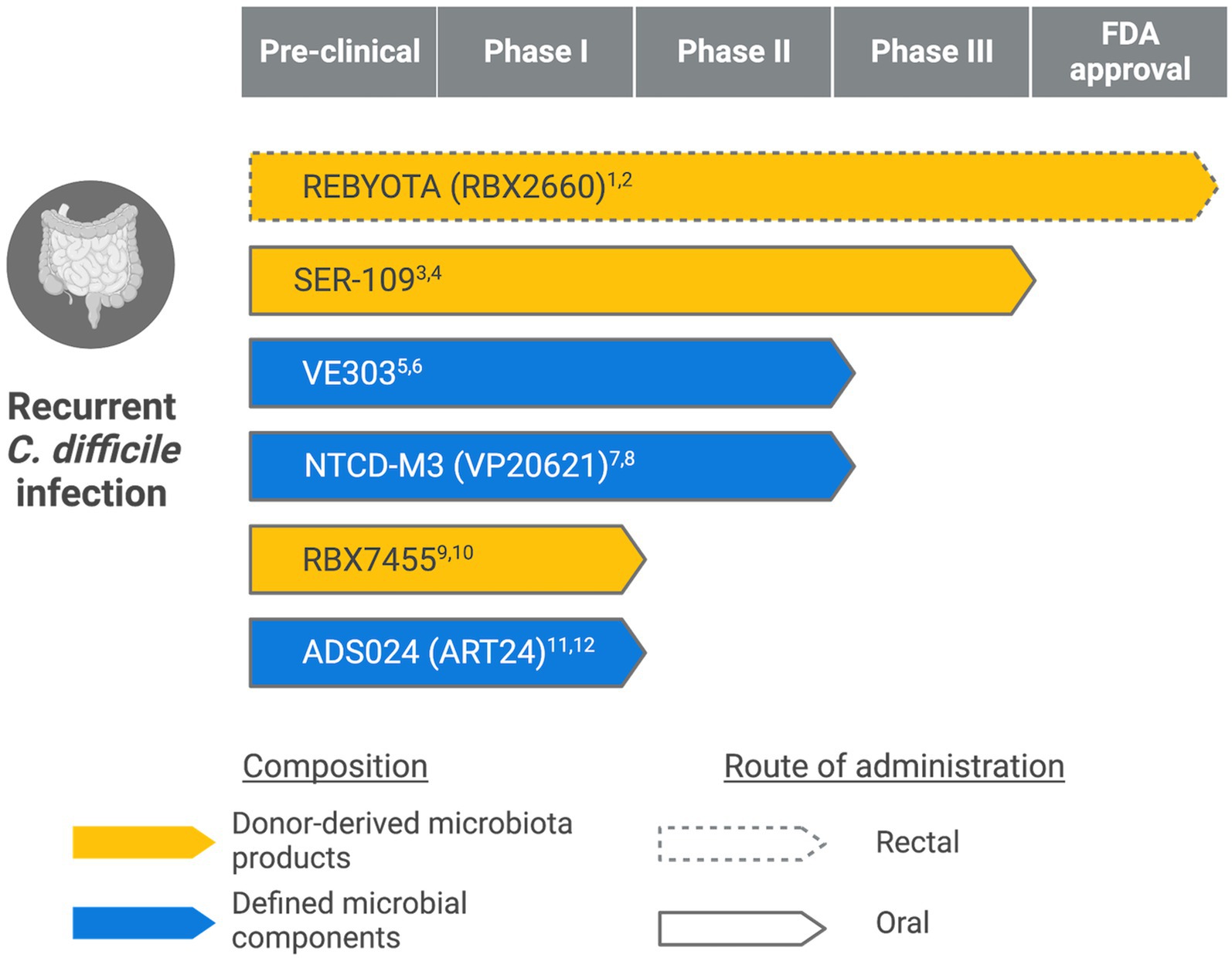
Figure 1. Live biotherapeutic products in different stages of clinical development. (Created with BioRender.com) 1ClinicalTrials.gov Identifier: NCT03244644. 2https://www.fda.gov/news-events/press-announcements/fda-approves-first-fecal-microbiota-product. 3ClinicalTrials.gov Identifier: NCT03183128. 4https://www.serestherapeutics.com/our-programs/. 5ClinicalTrials.gov Identifier: NCT03788434. 6https://www.vedantabio.com/pipeline/ve303. 7ClinicalTrials.gov Identifier: NCT01259726. 8https://www.destinypharma.com/pipeline/clostridioides-difficile-infections/. 9ClinicalTrials.gov Identifier: NCT02981316. 10https://www.rebiotix.com/clinical-trials/rbx7455-oral-c-diff-prevention/. 11ClinicalTrials.gov Identifier: NCT04891965. 12https://adisotx.com/.
Fecal microbiota transplantation (FMT) involves the transfer of donor microbiota to replenish a recipient’s gut microbial composition. Despite being generally well-tolerated and effective against recurrent CDI, the possible transmission of infectious agents poses safety concerns, especially in the immunocompromised population (DeFilipp et al., 2019; Tariq et al., 2019). Therefore, standardized screening and manufacturing processes were urgently needed. Recently, FDA approved REBYOTA (also known as RBX2660), the first fecal microbiota product for recurrent CDI. REBYOTA is manufactured under a standardized process that includes rigorous pathogen testing to minimize the health risks of FMT. While effective – 70.6% of patients treated with REBYOTA remain free of CDI recurrence within 8 weeks (Khanna et al., 2022); it requires storage at −80°C and is delivered to patients via enema. The same company is developing an oral capsule-based therapy called RBX7455 that contains lyophilized bacteria stable at room temperature (Khanna et al., 2021). If it succeeds in clinical trials and gains regulatory approval, RBX7455 may offer a more convenient option for recurrent CDI treatment.
SER-109 is another donor-derived therapeutic. The manufacturing of SER109 enriches Firmicute spores while inactivating potential bacterial, viral, and fungal pathogens (McGovern et al., 2021; Feuerstadt et al., 2022; McChalicher et al., 2022). In a phase III clinical trial (ClinicalTrials.gov Identifier: NCT03183128), subjects dosed with SER-109 were less likely to have recurrent infections following standard-of-care antibiotic treatment than patients in the placebo group (Feuerstadt et al., 2022). These spore-forming Firmicutes may mediate protection against C. difficile by competing for essential nutrients and modifying bile acid profiles in the gut (Ridlon et al., 2014; Theriot et al., 2016).
Donor-derived microbiota products contain a mixture of microbes and vary in composition (Khoruts et al., 2021); it is, therefore, challenging to correlate clinical efficacy with biological components. In contrast, defined microbial components have standardized compositions and can be rationally designed based on biological functions.
VE303 is a defined bacterial consortium consisting of eight commensal strains of Clostridium (Dsouza et al., 2022). In a phase 1a/b study, healthy volunteers dosed with VE303 after vancomycin pretreatment showed accelerated recovery of diverse microbial communities and increased levels of secondary bile acids and short-chain fatty acids associated with colonization resistance against C. difficile. VE303 is also safe and well-tolerated in the study subjects. Subsequent clinical trials will evaluate the safety and efficacy of VE303 in preventing recurrent CDI (ClinicalTrials.gov Identifier: NCT03788434).
Researchers have also focused on non-toxigenic C. difficile (NTCD), which lacks genes for toxin production and frequently colonizes hospitalized patients (Shim et al., 1998). The initial colonization with NTCD could prevent subsequent toxigenic C. difficile infections in animal models and patients (Wilson and Sheagren, 1983; Borriello and Barclay, 1985; Seal et al., 1987). One NTCD strain, NTCD-M3 (previously known as VP20621), has demonstrated safety and efficacy in a phase II clinical trial (Gerding et al., 2015): oral administration of NTCD-M3 spores was safe and well-tolerated and reduced CDI recurrence in patients clinically cured with antibiotics (Villano et al., 2012; Gerding et al., 2015). However, the concern with NTCD is the possibility of gaining toxin-producing genes via horizontal gene transfer (Brouwer et al., 2013); whether this occurs in vivo remains unclear and requires close monitoring.
Besides restoring the host microbiota, LBP could act directly on C. difficile. ADS024 (formerly ART24) is a single strain LBP of Bacillus velezensis isolated from a fecal sample of a healthy donor. ADS024 exhibits dual actions on clinically relevant C. difficile strains, including direct inhibition and toxin degradation (O’Donnell et al., 2022; Xie et al., 2023). The product has recently completed a phase I study, which evaluates the safety of ADS024 in recently cured CDI patients (ClinicalTrials.gov Identifier: NCT04891965).
Given the complex interactions between C. difficile and the gut microbes, combined treatments targeting the pathogen and the microbiota may yield better clinical outcomes than antibiotic treatments alone. Live biotherapeutics can protect against recurrent CDI by expediting the microbiota recovery, restoring the metabolic profile, mediating colonization resistance, or directly inhibiting C. difficile (Gerding et al., 2015; Khanna et al., 2021, 2022; Dsouza et al., 2022; Feuerstadt et al., 2022; O’Donnell et al., 2022). After colonization, the live ingredients in the LBP remain viable and potentially provide sustained protection against enteric pathogens. However, LBP’s non-traditional features also require distinct approaches and considerations in regulatory approval, manufacturing, and prescription (Dreher-Lesnick et al., 2017). Further research to understand the interactions among the introduced microbes, the microbiota, and host immunity will be crucial as these results will inform treatments for enteric infections and other conditions involving the gut microbiota.
RW conceptualized the manuscript topic, conducted literature research, created the figure, and wrote the manuscript.
This publication was supported by the Princeton University Library Open Access Fund.
The author would like to thank Uday S. Ganapathy (Center for Discovery and Innovation, Hackensack Meridian Health) for carefully reading and editing the manuscript.
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Amrane, S., Bachar, D., Lagier, J. C., and Raoult, D. (2018). Clostridium scindens is present in the gut microbiota during Clostridium difficile infection: a metagenomic and Culturomic analysis. J. Clin. Microbiol. 56, e01663–e01617. doi: 10.1128/JCM.01663-17
Auchtung, J. M., Preisner, E. C., Collins, J., Lerma, A. I., and Britton, R. A. (2020). Identification of simplified microbial communities that inhibit Clostridioides difficile infection through dilution/extinction. mSphere 5, e00387–e00320. doi: 10.1128/mSphere.00387-20
Balsells, E., Shi, T., Leese, C., Lyell, I., Burrows, J., Wiuff, C., et al. (2019). Global burden of Clostridium difficile infections: a systematic review and meta-analysis. J. Global Health 9:010407. doi: 10.7189/jogh.09.010407
Borriello, S. P., and Barclay, F. E. (1985). Protection of hamsters against Clostridium difficile ileocaecitis by prior colonisation with non-pathogenic strains. J. Med. Microbiol. 19, 339–350. doi: 10.1099/00222615-19-3-339
Brouwer, M. S. M., Roberts, A. P., Hussain, H., Williams, R. J., Allan, E., and Mullany, P. (2013). Horizontal gene transfer converts non-toxigenic Clostridium difficile strains into toxin producers. Nat. Commun. 4:2601. doi: 10.1038/ncomms3601
Buffie, C. G., Bucci, V., Stein, R. R., McKenney, P., Ling, L., Gobourne, A., et al. (2015). Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517, 205–208. doi: 10.1038/nature13828
Chiumento, S., Roblin, C., Kieffer-Jaquinod, S., Tachon, S., Leprètre, C., Basset, C., et al. (2019). Ruminococcin C, a promising antibiotic produced by a human gut symbiont. Sci. Adv. 5:eaaw9969. doi: 10.1126/sciadv.aaw9969
Contijoch, E. J., Britton, G. J., Yang, C., Mogno, I., Li, Z., Ng, R., et al. (2019). Gut microbiota density influences host physiology and is shaped by host and microbial factors. eLife 8:e40553. doi: 10.7554/eLife.40553
Crobach, M. J. T., Vernon, J. J., Loo, V. G., Kong, L. Y., Péchiné, S., Wilcox, M. H., et al. (2018). Understanding Clostridium difficile Colonization. Clin. Microbiol. Rev. 31, e00021–e00017. doi: 10.1128/CMR.00021-17
DeFilipp, Z., Bloom, P. P., Torres Soto, M., Mansour, M. K., MRA, S., Huntley, M. H., et al. (2019). Drug-Resistant E. coli bacteremia transmitted by fecal microbiota transplant. N. Engl. J. Med. 381, 2043–2050. doi: 10.1056/NEJMoa1910437
Dreher-Lesnick, S. M., Stibitz, S., and Carlson, P. E. Jr. (2017). U.S. regulatory considerations for development of live biotherapeutic products as drugs. Microbiol. Spectr. 5, BAD-0017-2017. doi: 10.1128/microbiolspec.BAD-0017-2017
Dsouza, M., Menon, R., Crossette, E., Bhattarai, S. K., Schneider, J., Kim, Y. G., et al. (2022). Colonization of the live biotherapeutic product VE303 and modulation of the microbiota and metabolites in healthy volunteers. Cell Host Microbe 30, 583–598.e8. doi: 10.1016/j.chom.2022.03.016
DuPont, H. L. (2011). The search for effective treatment of Clostridium difficile infection. N. Engl. J. Med. 364, 473–475. doi: 10.1056/NEJMe1013236
Ferreyra, J. A., Wu, K. J., Hryckowian, A. J., Bouley, D. M., Weimer, B. C., and Sonnenburg, J. L. (2014). Gut microbiota-produced succinate Promotes C. difficile infection after antibiotic treatment or motility disturbance. Cell Host Microbe 16, 770–777. doi: 10.1016/j.chom.2014.11.003
Feuerstadt, P., Louie, T. J., Lashner, B., Wang, E. E. L., Diao, L., Bryant, J. A., et al. (2022). SER-109, an Oral microbiome therapy for recurrent Clostridioides difficile infection. N. Engl. J. Med. 386, 220–229. doi: 10.1056/NEJMoa2106516
Gebhart, D., Williams, S. R., Bishop-Lilly, K. A., Govoni, G. R., Willner, K. M., Butani, A., et al. (2012). Novel high-molecular-weight, R-type Bacteriocins of Clostridium difficile. J. Bacteriol. 194, 6240–6247. doi: 10.1128/JB.01272-12
Gerding, D. N., Meyer, T., Lee, C., Cohen, S. H., Murthy, U. K., Poirier, A., et al. (2015). Administration of Spores of nontoxigenic Clostridium difficile strain M3 for prevention of recurrent C. difficile infection: a randomized clinical trial. JAMA 313:1719. doi: 10.1001/jama.2015.3725
Girinathan, B. P., DiBenedetto, N., Worley, J. N., Peltier, J., Arrieta-Ortiz, M. L., Immanuel, S. R. C., et al. (2021). In vivo commensal control of Clostridioides difficile virulence. Cell Host Microbe 29, 1693–1708.e7. doi: 10.1016/j.chom.2021.09.007
Guh, A. Y., Mu, Y., Winston, L. G., Johnston, H., Olson, D., Farley, M. M., et al. (2020). Trends in U.S. burden of Clostridioides difficile infection and outcomes. N. Engl. J. Med. 382, 1320–1330. doi: 10.1056/NEJMoa1910215
Heeg, D., Burns, D. A., Cartman, S. T., and Minton, N. P. (2012). Spores of Clostridium difficile clinical isolates display a diverse germination response to bile salts. PLoS One 7:e32381. doi: 10.1371/journal.pone.0032381
Horvat, S., and Rupnik, M. (2018). Interactions between Clostridioides difficile and fecal microbiota in in vitro batch model: growth, sporulation, and microbiota changes. Front. Microbiol. 9:1633. doi: 10.3389/fmicb.2018.01633
Johnson, S., Lavergne, V., Skinner, A. M., Gonzales-Luna, A. J., Garey, K. W., Kelly, C. P., et al. (2021). Clinical practice guideline by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA): 2021 focused update guidelines on Management of Clostridioides difficile infection in adults. Clin. Infect. Dis. 73, e1029–e1044. doi: 10.1093/cid/ciab549
Kang, J. D., Myers, C. J., Harris, S. C., Kakiyama, G., Lee, I. K., Yun, B. S., et al. (2019). Bile acid 7α-Dehydroxylating gut bacteria secrete antibiotics that inhibit Clostridium difficile: role of secondary bile acids. Cell Chem. Biol. 26, 27–34.e4. doi: 10.1016/j.chembiol.2018.10.003
Keith, J. W., Dong, Q., Sorbara, M. T., Becattini, S., Sia, J. K., Gjonbalaj, M., et al. (2020). Impact of antibiotic-resistant bacteria on immune activation and Clostridioides difficile infection in the mouse intestine. Infect. Immun. 88, e00362–e00319. doi: 10.1128/IAI.00362-19
Khanna, S., Assi, M., Lee, C., Yoho, D., Louie, T., Knapple, W., et al. (2022). Efficacy and safety of RBX2660 in PUNCH CD3, a phase III, randomized, double-blind, placebo-controlled trial with a Bayesian primary analysis for the prevention of recurrent Clostridioides difficile infection. Drugs 82, 1527–1538. doi: 10.1007/s40265-022-01797-x
Khanna, S., Pardi, D. S., Jones, C., Shannon, W. D., Gonzalez, C., and Blount, K. (2021). RBX7455, a non-frozen, orally administered investigational live biotherapeutic, is safe, effective, and shifts patients microbiomes in a phase 1 study for recurrent Clostridioides difficile infections. Clin. Infect. Dis. 73, e1613–e1620. doi: 10.1093/cid/ciaa1430
Khoruts, A., Staley, C., and Sadowsky, M. J. (2021). ‘Faecal microbiota transplantation for Clostridioides difficile: mechanisms and pharmacology’, nature reviews. Gastroenterol. Hepatol. 18, 67–80. doi: 10.1038/s41575-020-0350-4
Krutova, M., Wilcox, M., and Kuijper, E. (2022). Clostridioides difficile infection: are the three currently used antibiotic treatment options equal from pharmacological and microbiological points of view? Int. J. Infect. Dis. 124, 118–123. doi: 10.1016/j.ijid.2022.09.013
Lesniak, N. A., Schubert, A. M., Flynn, K. J., Leslie, J. L., Sinani, H., Bergin, I. L., et al. (2022). The gut bacterial community potentiates Clostridioides difficile infection severity. MBio 13, e01183–e01122. doi: 10.1128/mbio.01183-22
Louie, T. J., Miller, M. A., Mullane, K. M., Weiss, K., Lentnek, A., Golan, Y., et al. (2011). Fidaxomicin versus vancomycin for Clostridium difficile infection. N. Engl. J. Med. 364, 422–431. doi: 10.1056/NEJMoa0910812
Martens, E. C., Chiang, H. C., and Gordon, J. I. (2008). Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe 4, 447–457. doi: 10.1016/j.chom.2008.09.007
Mathur, H., Fallico, V., O’Connor, P. M., Rea, M. C., Cotter, P. D., Hill, C., et al. (2017). Insights into the mode of action of the Sactibiotic Thuricin CD. Front. Microbiol. 8:696. doi: 10.3389/fmicb.2017.00696
McChalicher, C., Abdulaziz, A., Zhou, S. S., Lombardo, M. J., Hasson, B., Auniņš, J. G., et al. (2022). ‘Manufacturing process of SER-109, a purified investigational microbiome therapeutic, reduces risk of coronavirus transmission from donor stool’, open forum. Infect. Dis. 9:ofac448. doi: 10.1093/ofid/ofac448
McFarland, L. V., Elmer, G. W., and Surawicz, C. M. (2002). Breaking the cycle: treatment strategies for 163 cases of recurrent Clostridium difficile disease. Am. J. Gastroenterol. 97, 1769–1775. doi: 10.1111/j.1572-0241.2002.05839.x
McGovern, B. H., Ford, C. B., Henn, M. R., Pardi, D. S., Khanna, S., Hohmann, E. L., et al. (2021). SER-109, an investigational microbiome drug to reduce recurrence after Clostridioides difficile infection: lessons learned from a phase 2 trial. Clin. Infect. Dis. 72, 2132–2140. doi: 10.1093/cid/ciaa387
Ng, K. M., Ferreyra, J. A., Higginbottom, S. K., Lynch, J. B., Kashyap, P. C., Gopinath, S., et al. (2013). Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature 502, 96–99. doi: 10.1038/nature12503
O’Donnell, M. M., Hegarty, J. W., Healy, B., Schulz, S., Walsh, C. J., Hill, C., et al. (2022). Identification of ADS024, a newly characterized strain of bacillus velezensis with direct Clostridiodes difficile killing and toxin degradation bio-activities. Sci. Rep. 12:9283. doi: 10.1038/s41598-022-13248-4
Ozaki, E., Kato, H., Kita, H., Karasawa, T., Maegawa, T., Koino, Y., et al. (2004). Clostridium difficile colonization in healthy adults: transient colonization and correlation with enterococcal colonization. J. Med. Microbiol. 53, 167–172. doi: 10.1099/jmm.0.05376-0
Pépin, J., Routhier, S., Gagnon, S., and Brazeau, I. (2006). Management and outcomes of a first recurrence of Clostridium difficile-associated disease in Quebec, Canada. Clin. Infect. Dis. 42, 758–764. doi: 10.1086/501126
Pruss, K. M., Enam, F., Battaglioli, E., DeFeo, M., Diaz, O. R., Higginbottom, S. K., et al. (2022). ‘Oxidative ornithine metabolism supports non-inflammatory C. difficile colonization’, nature. Metabolism 4, 19–28. doi: 10.1038/s42255-021-00506-4
Rea, M. C., Sit, C. S., Clayton, E., O'Connor, P. M., Whittal, R. M., Zheng, J., et al. (2010). Thuricin CD, a posttranslationally modified bacteriocin with a narrow spectrum of activity against Clostridium difficile. Proc. Natl. Acad. Sci. U. S. A. 107, 9352–9357. doi: 10.1073/pnas.0913554107
Ridlon, J. M., Kang, D. J., Hylemon, P. B., and Bajaj, J. S. (2014). Bile acids and the gut microbiome. Curr. Opin. Gastroenterol. 30, 332–338. doi: 10.1097/MOG.0000000000000057
Rosa, R., Donskey, C. J., and Munoz-Price, L. S. (2018). The intersection between colonization resistance, antimicrobial stewardship, and Clostridium difficile. Curr. Infect. Dis. Rep. 20:27. doi: 10.1007/s11908-018-0631-z
Rupnik, M., Wilcox, M. H., and Gerding, D. N. (2009). Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat. Rev. Microbiol. 7, 526–536. doi: 10.1038/nrmicro2164
Schäffler, H., and Breitrück, A. (2018). Clostridium difficile – from colonization to infection. Front. Microbiol. 9:646. doi: 10.3389/fmicb.2018.00646
Schubert, A. M., Sinani, H., and Schloss, P. D. (2015). Antibiotic-induced alterations of the murine gut microbiota and subsequent effects on colonization resistance against Clostridium difficile. MBio 6, e00974–e00915. doi: 10.1128/mBio.00974-15
Schwemmlein, N., Pippel, J., Gazdag, E. M., and Blankenfeldt, W. (2018). Crystal structures of R-type Bacteriocin sheath and tube proteins CD1363 and CD1364 from Clostridium difficile in the pre-assembled state. Front. Microbiol. 9:1750. doi: 10.3389/fmicb.2018.01750
Seal, D., Borriello, S. P., Barclay, F., Welch, A., Piper, M., and Bonnycastle, M. (1987). Treatment of relapsing Clostridium difficile diarrhoea by administration of a non-toxigenic strain. Eur. J. Clin. Microbiol. 6, 51–53. doi: 10.1007/BF02097191
Shim, J. K., Johnson, S., Samore, M. H., Bliss, D. Z., and Gerding, D. N. (1998). Primary symptomless colonisation by Clostridium difficile and decreased risk of subsequent diarrhoea. Lancet 351, 633–636. doi: 10.1016/S0140-6736(97)08062-8
Smith, A. B., Jenior, M. L., Keenan, O., Hart, J. L., Specker, J., Abbas, A., et al. (2022). Enterococci enhance Clostridioides difficile pathogenesis. Nature 611, 780–786. doi: 10.1038/s41586-022-05438-x
Solbach, P., Chhatwal, P., Woltemate, S., Tacconelli, E., Buhl, M., Gerhard, M., et al. (2018). BaiCD gene cluster abundance is negatively correlated with Clostridium difficile infection. PLoS One 13:e0196977. doi: 10.1371/journal.pone.0196977
Sorg, J. A., and Sonenshein, A. L. (2008). Bile salts and glycine as Cogerminants for Clostridium difficile spores. J. Bacteriol. 190, 2505–2512. doi: 10.1128/JB.01765-07
Sorg, J. A., and Sonenshein, A. L. (2010). Inhibiting the initiation of Clostridium difficile spore germination using analogs of Chenodeoxycholic acid, a bile acid. J. Bacteriol. 192, 4983–4990. doi: 10.1128/JB.00610-10
Staley, C., Kaiser, T., Beura, L. K., Hamilton, M. J., Weingarden, A. R., Bobr, A., et al. (2017). Stable engraftment of human microbiota into mice with a single oral gavage following antibiotic conditioning. Microbiome 5:87. doi: 10.1186/s40168-017-0306-2
Studer, N., Desharnais, L., Beutler, M., Brugiroux, S., Terrazos, M. A., Menin, L., et al. (2016). Functional intestinal bile acid 7α-Dehydroxylation by Clostridium scindens associated with protection from Clostridium difficile infection in a Gnotobiotic mouse model. Front. Cell. Infect. Microbiol. 6:191. doi: 10.3389/fcimb.2016.00191
Tannock, G. W., Munro, K., Taylor, C., Lawley, B., Young, W., Byrne, B., et al. (2010). A new macrocyclic antibiotic, fidaxomicin (OPT-80), causes less alteration to the bowel microbiota of Clostridium difficile-infected patients than does vancomycin. Microbiology 156, 3354–3359. doi: 10.1099/mic.0.042010-0
Tariq, R., Pardi, D. S., Bartlett, M. G., and Khanna, S. (2019). Low cure rates in controlled trials of fecal microbiota transplantation for recurrent Clostridium difficile infection: a systematic review and meta-analysis. Clin. Infect. Dis. 68, 1351–1358. doi: 10.1093/cid/ciy721
Thanissery, R., Winston, J. A., and Theriot, C. M. (2017). Inhibition of spore germination, growth, and toxin activity of clinically relevant C. difficile strains by gut microbiota derived secondary bile acids. Anaerobe 45, 86–100. doi: 10.1016/j.anaerobe.2017.03.004
Theriot, C. M., Bowman, A. A., and Young, V. B. (2016). Antibiotic-induced alterations of the gut microbiota Alter secondary bile acid production and allow for Clostridium difficile spore germination and outgrowth in the large intestine. mSphere 1, e00045–e00015. doi: 10.1128/mSphere.00045-15
Theriot, C. M., Koenigsknecht, M. J., Carlson, P. E. Jr., Hatton, G. E., Nelson, A. M., Li, B., et al. (2014). Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat. Commun. 5:3114. doi: 10.1038/ncomms4114
Villano, S. A., Seiberling, M., Tatarowicz, W., Monnot-Chase, E., and Gerding, D. N. (2012). Evaluation of an Oral suspension of VP20621, spores of nontoxigenic Clostridium difficile strain M3, in healthy subjects. Antimicrob. Agents Chemother. 56, 5224–5229. doi: 10.1128/AAC.00913-12
Vincent, C., and Manges, A. R. (2015). Antimicrobial use, human gut microbiota and Clostridium difficile colonization and infection. Antibiotics (Basel) 4, 230–253. doi: 10.3390/antibiotics4030230
Wilcox, M. H., Gerding, D. N., Poxton, I. R., Kelly, C., Nathan, R., Birch, T., et al. (2017). Bezlotoxumab for prevention of recurrent Clostridium difficile infection. N. Engl. J. Med. 376, 305–317. doi: 10.1056/NEJMoa1602615
Wilson, K. H., and Sheagren, J. N. (1983). Antagonism of toxigenic Clostridium difficile by nontoxigenic C. difficile. J. Infect. Dis. 147, 733–736. doi: 10.1093/infdis/147.4.733
Xie, Y., Chupina, E. A., Nelson, B., Feng, H., Pothoulakis, C., Chesnel, L., et al. (2023). ADS024, a bacillus velezensis strain, protects human colonic epithelial cells against C. difficile toxin-mediated apoptosis. Front. Microbiol. 13:1072534. doi: 10.3389/fmicb.2022.1072534
Keywords: Clostridioides difficile , Clostridioides difficile infections (CDI), gut microbiota, microbe-microbe interactions, live biotherapeutic products (LBP), fecal microbiota transplantation (FMT)
Citation: Wang R (2023) Clostridioides difficile infection: microbe-microbe interactions and live biotherapeutics. Front. Microbiol. 14:1182612. doi: 10.3389/fmicb.2023.1182612
Received: 09 March 2023; Accepted: 03 April 2023;
Published: 09 May 2023.
Edited by:
Dieter Jahn, Technical University of Braunschweig, GermanyReviewed by:
Meina Neumann-Schaal, German Collection of Microorganisms and Cell Cultures GmbH (DSMZ), GermanyCopyright © 2023 Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ruojun Wang, cnc2Njg2QHByaW5jZXRvbi5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.