- Department of Chemistry and Biochemistry, University of Northern British Columbia, Prince George, BC, Canada
This mini-review covers the use of nitrogen-15 in bacterial and fungal natural product discovery and biosynthetic characterization from 1970 to 2022. Nitrogen is an important element in a number of bioactive and structurally intriguing natural products including alkaloids, non-ribosomal peptides, and hybrid natural products. Nitrogen-15 can be detected at natural abundance utilizing two-dimensional nuclear magnetic resonance and mass spectrometry. Additionally, it is a stable isotope that can be added to growth media for both filamentous fungi and bacteria. With stable isotope feeding, additional two-dimensional nuclear magnetic resonance and mass spectrometry strategies have become available, and there is a growing trend to use nitrogen-15 stable isotope feeding for the biosynthetic characterization of natural products. This mini-review will catalog the use of these strategies, analyze the strengths and weaknesses of the different approaches, and suggest future directions for the use of nitrogen-15 in natural product discovery and biosynthetic characterization.
1. Nitrogen-15 in natural products
Nitrogen is often found within structurally interesting and bioactive microbial natural products in varying frequencies and moieties. Natural product classes with significant nitrogen representation include alkaloids, non-ribosomal peptides, polyketide macrolactams, and ribosomally synthesized post-translationally modified peptides (RiPPs). Due to this prevalence, tools and methodologies that can detect nitrogen are of great use to those who study microbial natural products. Important tools are nuclear magnetic resonance (NMR) and mass spectrometry (MS). 15N-NMR can be employed strategically for structure elucidation and discovery, while MS methodologies are pivotal for biosynthetic characterization and discovery particularly by detecting nitrogen-15 isotope ratio shifts.
2. Nitrogen-15 NMR
2.1. Nitrogen-15 NMR background
Nitrogen-15 nuclear magnetic resonance (NMR) spectroscopy is a powerful tool for elucidating natural product structures which contain nitrogen atoms. With steady instrument improvements and new techniques, valuable NMR data can be gathered with increasingly less material (Breton and Reynolds, 2013). Of the commonly found elements in natural products—hydrogens (i.e., protons), carbon, oxygen, nitrogen, sulfur and phosphorous– experiments can be run to directly probe small variations in structural environments for 1H, 13C,14N, 15N, and 31P nuclei (Parella and Sánchez-Ferrando, 2004; Martin, 2011). Nitrogen has two NMR active isotopes: 14N and 15N. 14N is the more abundant isotope; however, 14N is a spin-1 nucleus, and the signal suffers from significant line broadening due to quadrupolar relaxation (Witanowski and Webb, 1972). Alternately, nitrogen-15 is a spin-1/2 nucleus that does not experience significant line broadening, and is thus the more commonly observed nitrogen isotope (Witanowski and Webb, 1972). However, 15N has a natural abundance of only 0.36%, making it less sensitive than other more abundant nuclei such as carbon-13 or proton. The 15N isotope's negative gyromagnetic ratio (−2.7126) (Martin and Williams, 2010) also results in decreased sensitivity of the 15N isotope in NMR experiments in comparison to isotopes with a positive gyromagnetic ratio.
Methods for overcoming both 15N and 13C signal insensitivities incorporate a suite of two-dimensional (2D) NMR experiments in which the sensitivity of the proton is transferred to the more insensitive nuclei, increasing the signal-to-noise ratio and permitting data analysis of lower-abundance metabolites (Marek and Lycka, 2002; Martin and Williams, 2010). In general, two phenomena are exploited to transfer sensitivity from the proton to insensitive nuclei, such as 13C and 15N: the nuclear Overhauser effect, and polarization transfer for scalar coupled nuclei (Morris and Freeman, 1979; Freeman, 1997). Sensitivity transfer through the nuclear Overhauser effect is directly related to the gyromagnetic ratio of the observed nuclei. Because of this relationship, the nuclear Overhauser effect is not of value for sensitivity enhancement of the 15N nuclei with its negative gyromagnetic ratio (Freeman, 1997). The base experiment for non-selective sensitivity enhancement for scalar coupled nuclei is called the Insensitive Nuclei Enhanced by Polarization Transfer (INEPT) experiment (Morris and Freeman, 1979). Polarization transfer in scalar coupled nuclei is independent of the sign of the gyromagnetic ratio and, as such, is very useful for 15N experiments (Morris and Freeman, 1979). One caveat is that because polarization transfer relies on scalar coupling, through-bond connectivity must occur for this enhancement. Usefully, in inverse detected 2D experiments, the 15N isotope experiences significant increases in detection sensitivity (Freeman, 1997) due to the large population disparity between 1H and 15N, which arises from the sizeable gyromagnetic ratio difference between 1H (26.7522) and 15N (−2.7126) (Marek and Lycka, 2002). With experiments built off the INEPT foundation (Morris and Freeman, 1979), 15N is routinely used in NMR methods for elucidating amino acids for protein structure determination (Mulder and Filatov, 2010; Williamson et al., 2014), while usage in small molecule structure determination has varied in the literature (Marek and Lycka, 2002; Martin and Williams, 2010). The low natural abundance can be circumvented by incorporating stable isotope (or labeled) precursors into natural products to increase the 15N signal (Bax, 1989; Ohki and Kainosho, 2008; Chokkathukalam et al., 2014; Deev et al., 2017). However, with 2D experiments, natural abundance is usually enough to collect data from pure samples (Martin and Hadden, 2000; Marek and Lycka, 2002). Another barrier to its use has been the historical tendency to report nitrogen referenced to various internal and external references (Witanowski and Webb, 1972; Wishart et al., 1995), which has made it difficult to look to the literature for typical chemical shifts that are widely applicable. Fortunately, over the last several decades, there has been a shift to reference 15N NMR spectra to either nitromethane or liquid ammonia set equal to 0 ppm (IUPAC recommendation Harris et al., 2002; Martin and Williams, 2010). Additionally, experimental work has been done to compare the shifts of nitromethane and liquid ammonia, meaning chemical shift values can be easily converted between these two 15N references (Witanowski and Webb, 1972; Martin et al., 2007; Martin and Williams, 2010; Martin, 2011). Nitrogen chemical shifts referenced to liquid ammonia tend to have positive signs. Nitromethane is more deshielded than most nitrogens in natural products; this has historically been represented with shielded nitrogens possessing negative values in relation to nitromethane (IUPAC recommendation (Harris et al., 2002). Since about 2015, chemists are reversing that convention in relation to nitromethane (Martin and Williams, 2015). However, because most natural product nitrogens are more shielded than nitromethane, if a negative nitrogen chemical shift is reported, it can be generally assumed the historical conventions are being followed.
2.2. Nitrogen-15 NMR in natural product structure elucidation and discovery
Two-dimensional (2D) NMR experiments that indirectly detect nitrogen-15 improve the sensitivity limitations due to nitrogen's natural abundance. These experiments can detect one-bond couplings between nitrogen-15 and their attached protons (the sensitive 1H-15N HSQC/HMQC), detecting two-to-three bond couplings between tertiary nitrogen and protons in the molecule (long-range 1H-15N HMBC) (Martin and Hadden, 2000; Martin et al., 2007; Martin and Williams, 2010); and even detect carbon-13-nitrogen-15 correlations (13C-15N HSQC) with appropriate labeling. These 2D 15N NMR experiments are invaluable in solving complex bioactive natural product structures (Figure 1). Examples include the taumycins A (1) and B (Bishara et al., 2008), hyrtioreticulin F (2) (Imada et al., 2013) and coibamide A (Medina et al., 2008). The structure of the polyketide, forazoline A (3), was solved (after stable isotope feeding) utilizing the first example of carbon-13-nitrogen-15 direct detection via 13C-15N HSQC (Wyche et al., 2014). This same experiment proved vital in the recent structure elucidation of the structurally-unprecedented ecteinamine after discovery using an integrated genomic-metabolomic strategy by the Bugni group (Wu et al., 2022).
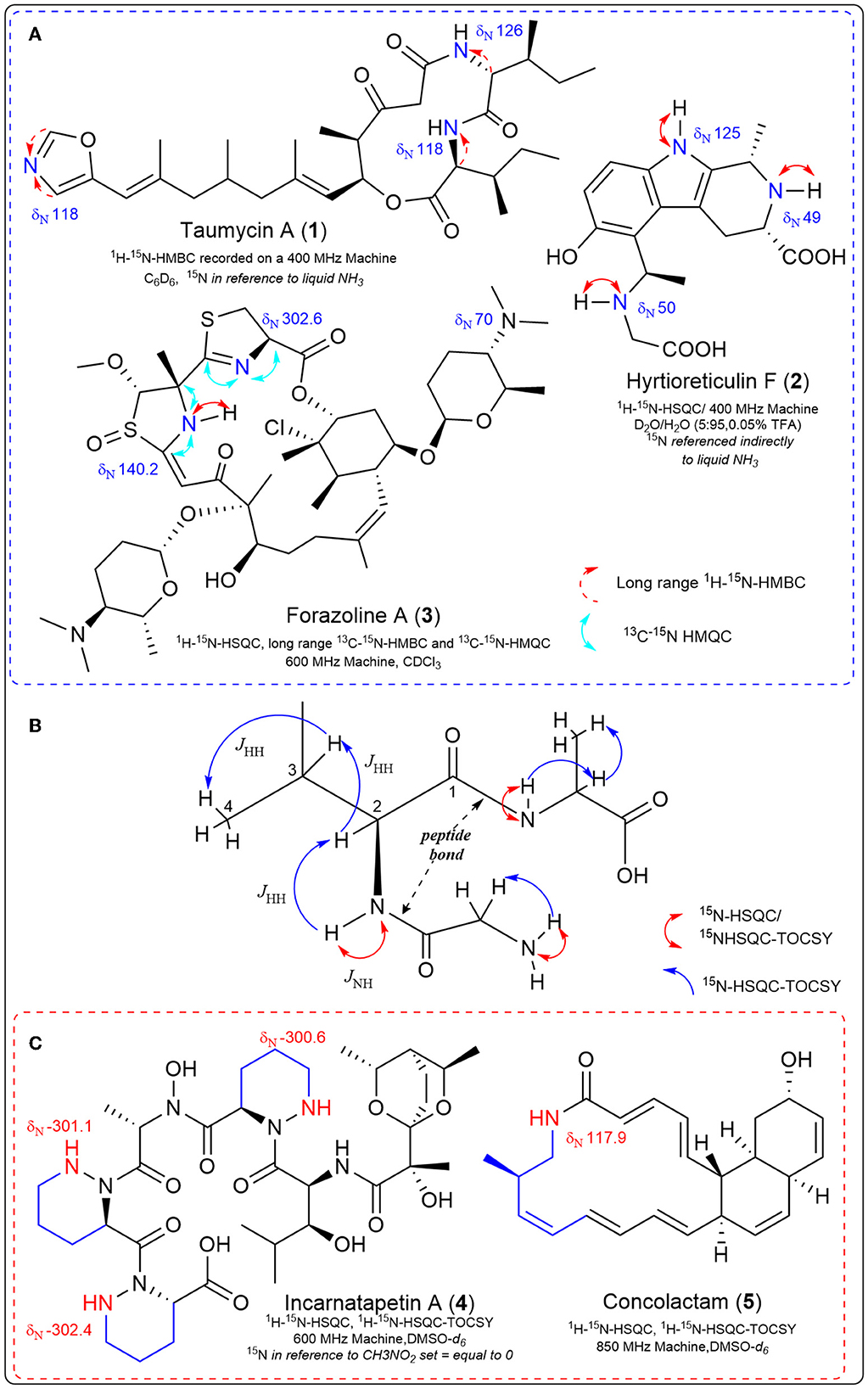
Figure 1. Experimentally determined nitrogen chemical shifts are provided, along with key correlations observed from various nitrogen-15 NMR experiments. (A) Select natural products structures that required 15N NMR 2D experiments during structure elucidation. (B) 1H-15N HSQC-TOCSY is an NMR experiment that allows us to observe the proton spin system of a single amino acid embedded in a peptide, beginning with the N-H correlation. The carbonyl of the peptide bond provides an insulator functionality that constrains the observation of single amino acid spin systems in the TOCSY spectrum. (C) Key examples where 15N NMR spectroscopy was integrated with genomic data for natural product discovery.
These 2D nitrogen-15 experiments can also be used in the discovery process, as first exemplified in 2007 in the genomisotopic approach developed by the Gerwick lab. The genomisotopic approach used 15N isotopically labeled precursors as predicted by in silico adenylation domain specificity of a genetically encoded non-ribosomal peptide to link orphan secondary metabolite gene clusters to the putatively produced natural product (Gross et al., 2007). In this method, use of the 1H-15N HMBC experiment to identify a 15N labeled isoleucine in tandem with bioassay-guided fractionation led to the successful isolation and structure elucidation of orfamide A representative of a new peptide class.
In fact, one advantage of 15N NMR is the large spectral range (~900 ppm), such that different nitrogen-containing functional groups can have widely dispersed 15N chemical shifts (Williamson et al., 1999). Due to fewer nitrogen atoms (compared to protons or carbons) in small molecule natural products, 15N spectra tend to be less crowded. Specific experiments designed to use this larger spectral dispersion to provide a means of separating crowded proton and carbon signals include the 1H-15N-HSQC-TOCSY and 1H-15N-HSQC-NOESY (Williamson et al., 1999). The 1H-15N-HSQC-TOCSY experiment allows observations of proton spin systems which are ultimately connected to a 15N resonance, that is, the set of scalar-coupled protons adjacent to a nitrogen-attached proton (Figure 1). 1H-15N HSQC experiments can be used to link a primary or secondary nitrogen with its attached proton, which, due to the large dispersion of nitrogen signals, allows observation of spin systems outside the crowded proton spectral window (Williamson et al., 1999). The TOCSY portion of the 1H-15N HSQC-TOCSY allows observation of magnetization transfer through scalar-coupled protons to observe a complete amino acid 1H spin system in peptide natural products (Figure 1). Recently these spectroscopic experiments have been integrated with genomic information to discover piperazic acid (Piz)-containing polyketide-nonribosomal peptides (PK-NRP) (Morgan et al., 2020, 2022a,b). and macrolactams (Shin et al., 2023) (Figure 1). In both cases, the dispersion of the nitrogen signals allowed detection of the predicted genetically encoded chemical structures in crude microbial extracts, significantly speeding up the discovery process. 15N NMR was the ideal tool to uncover genomically encoded Piz-containing natural products because Piz contains a unique nitrogen chemical shift and N-H correlation (Williams et al., 2011) distinct from other amides within peptides. Thus, we utilized the sensitive 15N-HSQC experiment to quickly screen microbial extracts obtained from small amounts of bacterial cultures fed with the stable isotope Piz-precursor 15N2 L-Orn (Morgan, 2021). After quickly screening through dozens of 15N2 L-Orn labeled microbial extracts, the less sensitive 1H-15N-HSQC-TOCSY was applied for unequivocal detection of piperazic acid-containing extracts that aligned with genetically encoded chemical structures of interest (Morgan et al., 2020). We then conducted microbial scale-up, with efficient isolation guided by the 1H-15N-HSQC-TOCSY Piz signal leading to the cytotoxic incarnatapeptin B, the related incarnatapeptin A (4) and the piperazic-acid containing dentigerumycins G and H (Morgan et al., 2020, 2022a,b). For the discovery of the novel and cytotoxic muanlactam and antibacterial concolactam (5) by the Oh group, the presence and expression of genetically encoded macrolactams was detected through PCR analysis of an in-house gDNA library utilizing macrolactam-specific primers (Shin et al., 2023). Then, bacterial strains identified in the PCR experiments were fed the stable isotope 15NH4Cl to enrich all amide bonds in crude extracts and permit detection of the macrolactams utilizing 1H-15N-HSQC-TOCSY NMR experiments. After this detection, scale-up provided efficient discovery of the muanlactam, 5 and structural reassignment of salinilactam (Shin et al., 2023). The application of 1H-15N-HSQC-TOCSY to the discovery of polyketide macrolactams by the Oh group provides strong support for the power of this method as the novel macrolactams contain but one nitrogen each.
3. Nitrogen mass spectrometry
3.1. Nitrogen mass spectrometry background
As discussed above, nitrogen has two stable isotopes, nitrogen-14 and nitrogen-15 (15N), with 99.635% and 0.365% natural abundance, respectively (Wieser and Brand, 2017). Due to the low natural abundance of 15N, precursors enriched in 15N have been used in biosynthetic characterization to study the metabolic pathways of living organisms for a century (Rinkel and Dickschat, 2015). The most common application of 15N is in metabolic labeling experiments, in which the organism is grown in a medium containing 15N-labeled compounds, such as ammonium or nitrate. These labeled compounds are then incorporated into the organism's biomass and metabolic intermediates, and the distribution of 15N can be used to trace the metabolic pathways of the organism. Originally, isotope ratio mass spectrometry (IRMS) and degradation prior to mass spectrometric analysis were used to assess distribution of incorporated nitrogen-15 in metabolic pathways (Kahn et al., 2002). As mass spectrometers technology developed with increased sensitivity, techniques like ESI-MS began to be utilized for detecting isotopic ratios patterns between 14N and 15N to detect 15N enrichment in metabolic labeling (Kahn et al., 2002; Wieser and Brand, 2017). The advantage of utilizing mass spectrometry (MS) combined with stable isotopes for biosynthetic characterization is the higher sensitivity of the MS instrumentation (Chokkathukalam et al., 2014).
3.2. Nitrogen-15 MS for biosynthetic characterization and discovery
Feeding specific 15N-enriched precursors has been used in conjunction with mass spectrometry for natural product biosynthetic characterization (Feng et al., 2009; Rinkel and Dickschat, 2015). By feeding these specific stable isotope precursors enriched with 15N into a natural product and then analyzing the resulting isotopomers and isotopic ratios with mass spectrometry, it is possible to determine the biosynthetic pathway of the natural product (Figure 2). Enrichment with 15N typically involves the incorporation of a precursor labeled with the stable isotope. In mass spectrometry, the incorporation of a 15N-enriched precursor into a molecule can be detected through the observation of isotopic peaks in the mass spectrum. When a molecule enriched with 15N isotopes is ionized in the mass spectrometer, it produces a series of isotopic peaks with increased m/z values compared to the peaks of the unlabeled molecule. The number of 15N atoms in the molecule determines the spacing between the isotopic peaks. For example, if a molecule with three nitrogen atoms incorporates 15N into all three positions, the isotopic peaks will show an increase of three mass units higher than the corresponding peaks of the unlabeled molecule. The increased spacing between the isotopic peaks provides evidence that the molecule has incorporated 15N. Additionally, the relative abundance of the isotopic peaks can provide information about the extent of incorporation of the 15N-labeled precursor. The higher the relative abundance of the isotopic peaks, the greater the extent of incorporation of the 15N-labeled precursor into the molecule.
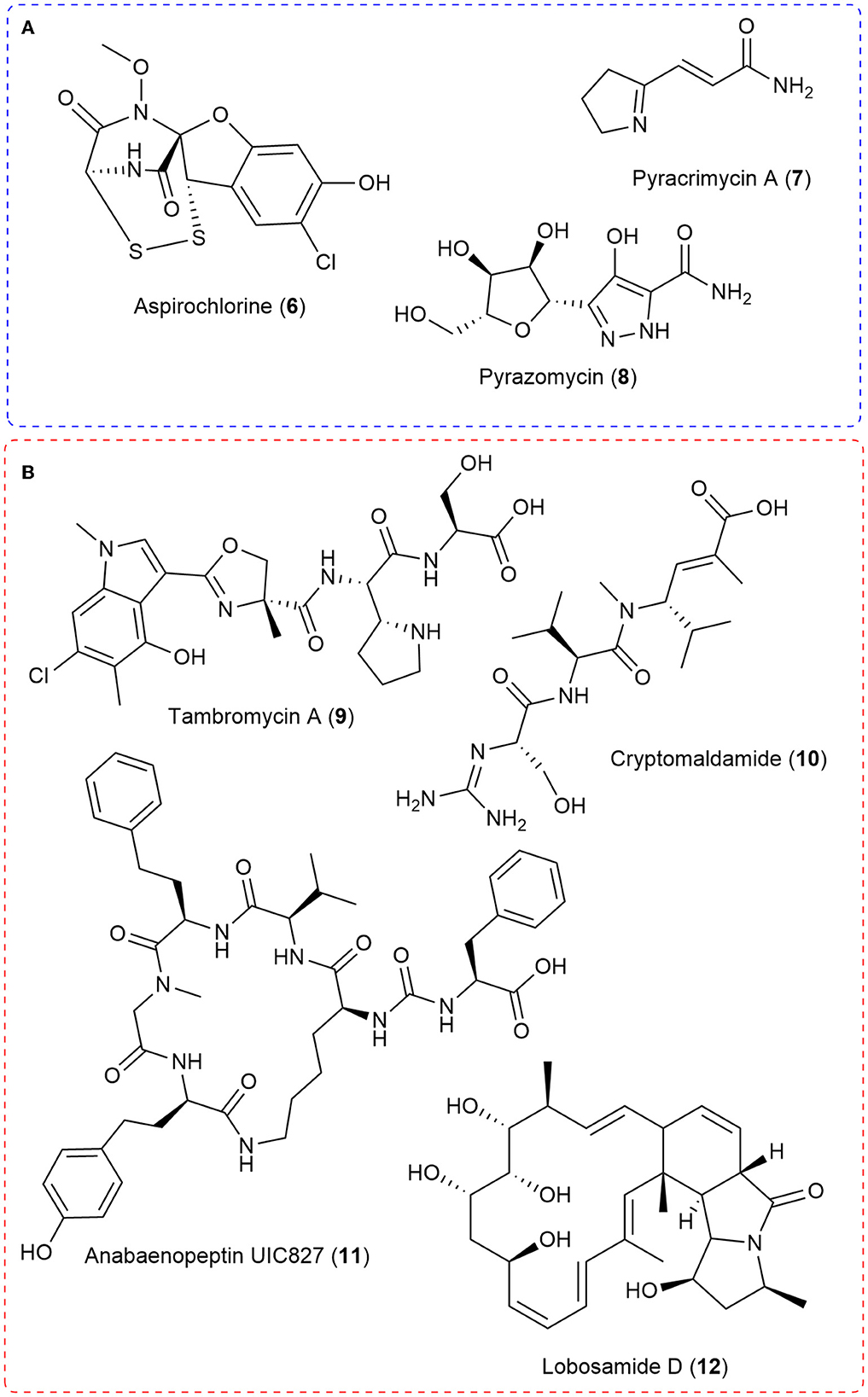
Figure 2. (A) Select natural products examples where stable isotope feeding of 15N-labeled precursors allowed the use of MS isotopic ratios to characterize all or key components of the biosynthetic pathway. (B) A selection of natural products discovered through integrated genomics and MS metabolomic strategies in which 15N played a crucial role.
For example, the biosynthesis of aspirochlorine (6) was solved by feeding the stable isotope enriched 13C2-15N-Gly to the producing Aspergillus oryzae strain and demonstrating via liquid chromatography-mass spectrometry that only the carbon-13 was incorporated into aspirocholine. This result helped demonstrate that phenylalanine was being transformed into a glycine-like moiety and that glycine was only utilized during N-methylation (Chankhamjon et al., 2014). The Charusanti group recently established the biosynthesis of the antibiotic pyracrimycin A (7) in a Streptomyces strain utilizing the isotopic ratios in MS analysis after feeding with 15N-proline and 15N-13C3-serine, together and individually (Nielsen et al., 2022). 15N-enriched precursors were utilized to demonstrate that αNH2 nitrogen atom of glutamic acid was involved in forming the nitrogen-nitrogen bond into the rare C-nucleoside antibiotic pyrazomycin (8) and point to a new nitrogen-nitrogen bond forming enzyme PyrN (Zhao et al., 2020). More recently, detailed bioinformatic analysis in tandem with feeding synthesized 15N-labeled Piz to the 4 producing Streptomyces incarnatus NRRL8089 strain followed by high resolution mass spectrometry isotopic ratio analysis demonstrated that free piperazic acid residues are most commonly incorporated into growing non-ribosomal peptide chains (Wei et al., 2022).
Another strategy that has been recently developed is utilizing 15N-labeled stable isotope precursors in integrated genomic and mass spectrometry metabolomic approaches (Chokkathukalam et al., 2014). This integration of 15N with genomic and metabolomic approaches has allowed the discovery of novel compounds and importantly, the linking of natural product structures with their producing biosynthetic gene clusters (Figure 2). When coupled with metabolomic data analysis methods such as multivariate statistical analyses or molecular networking, strains or extracts with potentially novel components can be prioritized for investigation (Hou et al., 2012; Covington et al., 2017). This type of approach has seen the discovery of molecules such as tambromycin (9) (Goering et al., 2016) and columbamides (Kleigrewe et al., 2015) and often linked them to their gene cluster using 15N-labeled precursors and MS analysis. The structure of 9 was discovered using the metabologenomics approach whereby statistical analysis of the data from simultaneous bioinformatic analysis of 172 draft genomes and MS analysis of extracts from the corresponding bacteria in four media led to an m/z value for an unknown molecule that correlated with several bacteria carrying the same gene cluster. Scale-up allowed isolation and structure elucidation of the novel cytotoxic non-ribosomal peptide, tambromycin (9). Stable isotope feeding of 15N1-Lysine (as compared to unlabeled media) was followed by MS isotopic analysis to link tambromycin with its' gene cluster. Next, the Gerwick Lab discovered both columbamides and cryptomaldamide (10) integrating genomic and MS analysis. The columbamides, a mild cannabinomimetic, were also discovered by matching molecular networks of MS data from cyanobacterial extracts with putative gene clusters obtained from their draft genomes (Kleigrewe et al., 2015). In the case of 10, they used MALDI-MS after 15N-Nitrate feeding and noticed a 5 dalton shift in the m/z value of interest after 4 days of growth. In both cases, they used MS to guide isolation and 1D and 2D NMR to solve the final structures (Kinnel et al., 2017). Building from these ideas, the Orjala lab utilized shifts in MS isotopic ratio after comparing with data from unlabeled media extracts with 99% 15N-nitrate labeled media extracts to match the m/z value with isotopic shifts indicative of 15N incorporation with predicted nitrogen rich gene clusters from sequenced cyanobacteria. Through this approach, they isolated four peptides, two known and two unknown. They named the unknown peptides anabaenopeptin UIC827 (11) and nostopyrrolidonamide after solving their structures with NMR and MS/MS data (May et al., 2020). Finally, the Medema and Linington labs have recently developed IsoAnalyst which can analyze MS data of extracts arising from isotope labeling, including 15N-labeled precursors, and connect them with their predicted biosynthetic gene clusters. The developers used IsoAnalyst to match multiple gene clusters with their chemical phenotype and discovered lobosamide D (12) after structure elucidation with 1D and 2D NMR (McCaughey et al., 2022). In conclusion, using 15N-labeled stable isotope precursors in integrated genomic and MS metabolomic approaches has proven to be a successful strategy for discovering novel compounds and linking natural product structures with their producing biosynthetic gene clusters.
4. Discussion
Due to their utility, both 15N-NMR and MS have been incorporated into various methodologies and strategies to solve natural product structures, to uncover novel natural product structures, to reveal the biogenesis of the natural products, and to link biosynthetic gene clusters to specific natural products. Indeed, nitrogen-15 NMR is a premier tool due to nitrogen's presence in many of the most structurally interesting and bioactive natural products. Nitrogen-15 NMR experiments are inconsistently used for structure elucidation purposes, perhaps because of the historical challenges and the low natural abundance. However, experiments by Gary Martin of Merck (amongst others) demonstrate that inversely detected nitrogen-15 2D data can be collected at natural abundance for even around 1 mg of pure sample (Martin and Hadden, 2000). 15N NMR as a discovery tool has produced structurally interesting and bioactive products from its first use by Gross et al. (2007), to the discovery of the incarnatapeptins (Morgan et al., 2020), to the most recent use by Oh et al. combining genomics and spectroscopic tools (Shin et al., 2023). The application of the 15N-HSQC-TOCSY experiments for targeted and efficient discovery approaches for both polyketide-nonribosomal peptide hybrids and macrolactams demonstrates that these approaches are worth considering for a wide range of nitrogen-containing natural products. Moreover, the structural revision of salinilactam during the discovery process by Oh et al., demonstrates that these 2D nitrogen-15 NMR experiments are a critical tool for structure elucidation of natural products. NMR instrumentation now routinely come equipped with the ability to detect nitrogen-15. With these modern enhancements, obtaining 15N NMR 2D experiments during dataset collection for all small molecule natural products should be habitual. Furthermore, if novel structures report nitrogen, journals and editors could require 2D nitrogen-15 NMR experiments alongside the proton and carbon spectra whenever not prohibited by older equipment.
Stable isotope 15N labeling combined with MS is a powerful tool for biosynthetic characterization and integrated MS-metabolomic and genomic approaches. These strategies have been useful in uncovering new chemistry with a multitude of MS instrumentation. Perhaps more importantly, 15N MS is an invaluable tool in biosynthetic characterization, from how molecules like aspirocholorine are made to how the intriguing non-ribosomal piperazic acid residue is incorporated in the building of natural products. Tools such as IsoAnalyst will surely become indispensable for linking genotype to phenotype. However, there are still some significant limitations when using mass spectrometry as a discovery tool, such as varying tendencies of molecules to ionize, media interference in crude samples (Watrous et al., 2012), unpredictable fragmentation patterns of some natural product classes (Liu et al., 2009; Novák et al., 2015; Townsend et al., 2018), and the need to use NMR in order to determine the final structure usually. At the same time, NMR usually suffers from sensitivity limitations. Therefore, just as MS and NMR data are utilized together to solve natural product structure de novo, so could the discovery process be amplified utilizing approaches that integrate the ability of 15N NMR to detect precise chemical environments with the sensitivity of MS data and the clarity of isotopic ratio shifts. As stable isotopes are increasingly included for structure elucidation, discovery, and gene cluster linking, the community could consider other tools to exploit the presence of precisely included 15N and 13C isotopes, such as increased usage of 2D 15N NMR experiments and analysis of carbon-nitrogen NMR splitting patterns.
Author contributions
The author confirms being the sole contributor of this work and has approved it for publication.
Conflict of interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Bax, A. (1989). Two-dimensional NMR and protein structure. Annu. Rev. Biochem. 58, 223–256. doi: 10.1146/annurev.bi.58.070189.001255
Bishara, A., Rudi, A., Aknin, M., Neumann, D., Ben-Califa, N., and Kashman, Y. (2008). Taumycins A and B, two bioactive lipodepsipeptides from the Madagascar sponge Fascaplysinopsis sp. Org. Lett. 10, 4307–4309. doi: 10.1021/ol801750y
Breton, R. C., and Reynolds, W. F. (2013). Using NMR to identify and characterize natural products. Nat. Prod. Rep. 30, 501. doi: 10.1039/c2np20104f
Chankhamjon, P., Boettger-Schmidt, D., Scherlach, K., Urbansky, B., Lackner, G., Kalb, D., et al. (2014). Biosynthesis of the halogenated mycotoxin aspirochlorine in koji mold involves a cryptic amino acid conversion. Angewandte Chemie Int. Ed. 53, 13409–13413. doi: 10.1002/anie.201407624
Chokkathukalam, A., Kim, D.-H., Barrett, M. P., Breitling, R., and Creek, D. J. (2014). Stable isotope-labeling studies in metabolomics: new insights into structure and dynamics of metabolic networks. Bioanalysis 6, 511–524. doi: 10.4155/bio.13.348
Covington, B. C., McLean, J. A., and Bachmann, B. O. (2017). Comparative mass spectrometry-based metabolomics strategies for the investigation of microbial secondary metabolites. Nat. Prod. Rep. 34, 6–24. doi: 10.1039/C6NP00048G
Deev, S. L., Paramonov, A. S., Shestakova, T. S., Khalymbadzha, I. A., Chupakhin, O. N., Subbotina, J. O., et al. (2017). 15 N-labelling and structure determination of adamantylated azolo-azines in solution. Beilstein J. Org. Chem. 13, 2535–2548. doi: 10.3762/bjoc.13.250
Feng, X., Ratnayake, A. S., Charan, R. D., Janso, J. E., Bernan, V. S., Schlingmann, G., et al. (2009). Probing natural product biosynthetic pathways using Fourier transform ion cyclotron resonance mass spectrometry. Bioorganic Med. Chem. 17, 2154–2161. doi: 10.1016/j.bmc.2008.10.073
Freeman, R. (1997). Spin Choreography: Basic Steps in High Resolution NMR. Oxford, Sausalito, CA: Spektrum; University Science Books.
Goering, A. W., McClure, R. A., Doroghazi, J. R., Albright, J. C., Haverland, N. A., Zhang, Y., et al. (2016). Metabologenomics: correlation of microbial gene clusters with metabolites drives discovery of a nonribosomal peptide with an unusual amino acid monomer. ACS Cent. Sci. 2, 99–108. doi: 10.1021/acscentsci.5b00331
Gross, H., Stockwell, V. O., Henkels, M. D., Nowak-Thompson, B., Loper, J. E., and Gerwick, W. H. (2007). The genomisotopic approach: a systematic method to isolate products of orphan biosynthetic gene clusters. Chem. Biol. 14, 53–63. doi: 10.1016/j.chembiol.2006.11.007
Harris, R. K., Becker, E. D., Cabral De Menezes, S. M., Goodfellow, R., and Granger, P. (2002). NMR nomenclature: Nuclear spin properties and conventions for chemical shifts (IUPAC recommendations 2001). Concepts Magnetic Resonance 14, 326–346. doi: 10.1002/cmr.10035
Hou, Y., Braun, D. R., Michel, C. R., Klassen, J. L., Adnani, N., Wyche, T. P., et al. (2012). Microbial strain prioritization using metabolomics tools for the discovery of natural products. Anal. Chem. 84, 4277–4283. doi: 10.1021/ac202623g
Imada, K., Sakai, E., Kato, H., Kawabata, T., Yoshinaga, S., Nehira, T., et al. (2013). Reticulatins A and B and hyrtioreticulin F from the marine sponge Hyrtios reticulatus. Tetrahedron 69, 7051–7055. doi: 10.1016/j.tet.2013.06.043
Kahn, M. L., Parra-Colmenares, A., Ford, C. L., Kaser, F., McCaskill, D., and Ketchum, R. E. (2002). A mass spectrometry method for measuring 15N incorporation into pheophytin. Anal. Biochem. 307, 219–225. doi: 10.1016/S0003-2697(02)00046-5
Kinnel, R. B., Esquenazi, E., Leao, T., Moss, N., Mevers, E., Pereira, A. R., et al. (2017). A Maldiisotopic approach to discover natural products: cryptomaldamide, a hybrid tripeptide from the marine cyanobacterium moorea producens. J. Nat. Prod. 80, 1514–1521. doi: 10.1021/acs.jnatprod.7b00019
Kleigrewe, K., Almaliti, J., Tian, I. Y., Kinnel, R. B., Korobeynikov, A., Monroe, E. A., et al. (2015). Combining mass spectrometric metabolic profiling with genomic analysis: a powerful approach for discovering natural products from cyanobacteria. J. Nat. Prod. 78, 1671–1682. doi: 10.1021/acs.jnatprod.5b00301
Liu, W.-T., Ng, J., Meluzzi, D., Bandeira, N., Gutierrez, M., Simmons, T. L., et al. (2009). Interpretation of tandem mass spectra obtained from cyclic nonribosomal peptides. Anal. Chem. 81, 4200–4209. doi: 10.1021/ac900114t
Marek, R., and Lycka, A. (2002). 15N NMR spectroscopy in structural analysis. COC 6, 35–66. doi: 10.2174/1385272023374643
Martin, G. E. (2011). “Chapter 5 - using 1,1- and 1,n-ADEQUATE 2D NMR data in structure elucidation protocols,” in Annual Reports on NMR Spectroscopy, editor G. A. Webb (Academic Press), 215–291.
Martin, G. E., and Hadden, C. E. (2000). Long-range 1 H– 15 N heteronuclear shift correlation at natural abundance. J. Nat. Prod. 63, 543–585. doi: 10.1021/np9903191
Martin, G. E., Solntseva, M., and Williams, A. J. (2007). “Applications of 15N NMR spectroscopy in alkaloid chemistry,” in Modern Alkaloids, eds. E. Fattorusso and O. Taglialatela-Scafati (Weinheim: John Wiley & Sons. Ltd.), 409–471.
Martin, G. E., and Williams, A. J. (2010). “Utilizing long-range 1 H−15 N 2-D NMR spectroscopy for chemical structure elucidation and confirmation,” in eMagRes, eds. R. K. Harris and R. L. (Wasylishen: American Cancer Society).
Martin, G. E., and Williams, A. J. (2015). “Applications of 1H−15N long-range heteronuclear shift correlation and 15N NMR in alkaloid chemistry,” in Annual Reports on NMR Spectroscopy (Elsevier), 1–76.
May, D. S., Crnkovic, C. M., Krunic, A., Wilson, T. A., Fuchs, J. R., and Orjala, J. E. (2020). 15N stable isotope labeling and comparative metabolomics facilitates genome mining in cultured cyanobacteria. ACS Chem. Biol. 15, 758–765. doi: 10.1021/acschembio.9b00993
McCaughey, C. S., van Santen, J. A., van der Hooft, J. J. J., Medema, M. H., and Linington, R. G. (2022). An isotopic labeling approach linking natural products with biosynthetic gene clusters. Nat. Chem. Biol. 18, 295–304. doi: 10.1038/s41589-021-00949-6
Medina, R. A., Goeger, D. E., Hills, P., Mooberry, S. L., Huang, N., Romero, L. I., et al. (2008). Coibamide A, a potent antiproliferative cyclic depsipeptide from the panamanian marine cyanobacterium Leptolyngbya sp. J. Am. Chem. Soc. 130, 6324–6325. doi: 10.1021/ja801383f
Morgan, K. D. (2021). Chemical and Genetic Investigations of Marine and Terrestrial Bacteria Towards Bioactive Natural Product Discovery. doi: 10.14288/1.0397473
Morgan, K. D., Williams, D. E., Patrick, B. O., Remigy, M., Banuelos, C. A., Sadar, M. D., et al. (2020). Incarnatapeptins A and B, nonribosomal peptides discovered using genome mining and 1H/15N HSQC-TOCSY. Org. Lett. 22, 4053–4057. doi: 10.1021/acs.orglett.0c00818
Morgan, K. D., Williams, D. E., Ryan, K. S., and Andersen, R. J. (2022a). Corrigendum to “dentigerumycin F and G: dynamic structures retrieved through a genome-mining/nitrogen NMR methodology” [Tetrahedron Lett. 94C (2022) 153688]. Tetrahedron Lett. 105, 154030. doi: 10.1016/j.tetlet.2022.154030
Morgan, K. D., Williams, D. E., Ryan, K. S., and Andersen, R. J. (2022b). Dentigerumycin F and G: dynamic structures retrieved through a genome-mining/nitrogen-NMR methodology. Tetrahedron Lett. 94, 153688. doi: 10.1016/j.tetlet.2022.153688
Morris, G. A., and Freeman, R. (1979). Enhancement of nuclear magnetic resonance signals by polarization transfer. J. Am. Chem. Soc. 101, 760–762. doi: 10.1021/ja00497a058
Mulder, F. A. A., and Filatov, M. (2010). NMR chemical shift data and ab initio shielding calculations: emerging tools for protein structure determination. Chem. Soc. Rev. 39, 578–590. doi: 10.1039/B811366C
Nielsen, J. B., Gren, T., Mohite, O. S., Jørgensen, T. S., Klitgaard, A. K., Mourched, A.-S., et al. (2022). Identification of the biosynthetic gene cluster for pyracrimycin A, an antibiotic produced by Streptomyces sp. ACS Chem. Biol. 17, 2411–2417. doi: 10.1021/acschembio.2c00480
Novák, J., Lemr, K., Schug, K. A., and Havlíček, V. (2015). CycloBranch: de novo sequencing of nonribosomal peptides from accurate product ion mass spectra. J. Am. Soc. Mass Spectrom. 26, 1780–1786. doi: 10.1007/s13361-015-1211-1
Ohki, S., and Kainosho, M. (2008). Stable isotope labeling methods for protein NMR spectroscopy. Progress Nuclear Magnetic Resonance Spectrosc. 53, 208–226. doi: 10.1016/j.pnmrs.2008.01.003
Parella, T., and Sánchez-Ferrando, F. (2004). Improved multiplicity-edited ADEQUATE experiments. J. Magnetic Resonance 166, 123–128. doi: 10.1016/j.jmr.2003.10.010
Rinkel, J., and Dickschat, J. S. (2015). Recent highlights in biosynthesis research using stable isotopes. Beilstein J. Org. Chem. 11, 2493–2508. doi: 10.3762/bjoc.11.271
Shin, Y.-H., Im, J. H., Kang, I., Kim, E., Jang, S. C., Cho, E., et al. (2023). Genomic and spectroscopic signature-based discovery of natural macrolactams. J. Am. Chem. Soc. 145, 1886–1896. doi: 10.1021/jacs.2c11527
Townsend, C., Furukawa, A., Schwochert, J., Pye, C. R., Edmondson, Q., and Lokey, R. S. (2018). CycLS: accurate, whole-library sequencing of cyclic peptides using tandem mass spectrometry. Bioorganic Medicinal Chem. 26, 1232–1238. doi: 10.1016/j.bmc.2018.01.027
Watrous, J., Roach, P., Alexandrov, T., Heath, B. S., Yang, J. Y., Kersten, R. D., et al. (2012). Mass spectral molecular networking of living microbial colonies. Proc. Natl. Acad. Sci. 109, E1743–E1752. doi: 10.1073/pnas.1203689109
Wei, Z.-W., Niikura, H., Morgan, K. D., Vacariu, C. M., Andersen, R. J., and Ryan, K. S. (2022). Free piperazic acid as a precursor to nonribosomal peptides. J. Am. Chem. Soc. 144, 13556–13564. doi: 10.1021/jacs.2c03660
Wieser, M. E., and Brand, W. A. (2017). “Isotope ratio studies using mass spectrometry,” in Encyclopedia of Spectroscopy and Spectrometry, eds. J. C. Lindon, G. E. Tranter, and D. W. Koppenaal (Oxford: Elsevier), 488–500.
Williams, David. E., Dalisay, D. S., Patrick, B. O., Matainaho, T., Andrusiak, K., Deshpande, R., et al. (2011). Padanamides A and B, Highly Modified Linear Tetrapeptides Produced in Culture by a Streptomyces sp. Isolated from a Marine Sediment. Org. Lett. 13, 3936–3939. doi: 10.1021/ol2014494
Williamson, R. T., Buevich, A. V., Martin, G. E., and Parella, T. (2014). LR-HSQMBC: a sensitive NMR technique to probe very long-range heteronuclear coupling pathways. J. Org. Chem. 79, 3887–3894. doi: 10.1021/jo500333u
Williamson, R. T., Márquez, B. L., and Gerwick, W. H. (1999). Use of 1H-15N PEP-HSQC-TOCSY at natural abundance to facilitate the structure elucidation of naturally occurring peptides. Tetrahedron 55, 2881–2888. doi: 10.1016/S0040-4020(99)00081-2
Wishart, D. S., Bigam, C. G., Yao, J., Abildgaard, F., Dyson, H. J., Oldfield, E., et al. (1995). 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J. Biomol. NMR 6, 135–140. doi: 10.1007/BF00211777
Witanowski, M., and Webb, G. A. (1972). “Nitrogen NMR spectroscopy,” in Annual Reports on NMR Spectroscopy, ed. E. F. Mooney (Elsevier), 395–464.
Wu, Q., Bell, B. A., Yan, J.-X., Chevrette, M. G., Brittin, N. J., Zhu, Y., et al. (2022). Metabolomics and genomics enable the discovery of a new class of nonribosomal peptidic metallophores from a marine micromonospora. J. Am. Chem. Soc. 145, 58–69. doi: 10.1021/jacs.2c06410
Wyche, T. P., Piotrowski, J. S., Hou, Y., Braun, D., Deshpande, R., McIlwain, S., et al. (2014). Forazoline A, a novel marine-derived polyketide with antifungal in vivo efficacy. Angew. Chem. Int. Ed. Engl. 53, 11583–11586. doi: 10.1002/anie.201405990
Keywords: natural products, nitrogen-15, NMR-guided discovery, mass spectrometry, structure elucidation, biosynthetic characterization, biosynthetic gene clusters
Citation: Morgan KD (2023) The use of nitrogen-15 in microbial natural product discovery and biosynthetic characterization. Front. Microbiol. 14:1174591. doi: 10.3389/fmicb.2023.1174591
Received: 26 February 2023; Accepted: 17 April 2023;
Published: 10 May 2023.
Edited by:
Sylvie Lautru, Centre National de la Recherche Scientifique (CNRS), FranceReviewed by:
Sonia Ilaria Maffioli, Naicons Srl, ItalyChristophe Corre, University of Warwick, United Kingdom
Copyright © 2023 Morgan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kalindi D. Morgan, a2FsaW5kaS5tb3JnYW5AdW5iYy5jYQ==
 Kalindi D. Morgan
Kalindi D. Morgan