
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 13 April 2023
Sec. Virology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1171500
Baculovirus expression vector system (BEVS) is a powerful and versatile platform for recombinant protein production in insect cells. As the most frequently used baculovirus, Autographa californica multiple nucleopolyhedrovirus (AcMNPV) encodes 155 open reading frames (ORFs), including a considerable number of non-essential genes for the virus replication in cell culture. Studies have shown that protein production in BEVS can be improved by removing some viral dispensable genes, and these AcMNPV vectors also offer the possibility of accommodating larger exogenous gene fragments. In this study, we, respectively, deleted 14 DNA fragments from AcMNPV genome, each of them containing at least two contiguous genes that were known nonessential for viral replication in cell culture or functionally unknown. The effects of these fragment-deletions on virus replication and exogenous protein production were examined. The results showed that 11 of the 14 fragments, containing 43 genes, were dispensable for the virus replication in cultured cells. By detecting the expression of intracellularly expressed and secreted reporter proteins, we demonstrated that nine of the fragment-deletions benefited protein production in Sf9 cells and/or in High Five cells. After combining the deletion of some dispensable fragments, we obtained two AcMNPV vectors shortened by more than 10 kb but displayed an improved capacity for recombinant protein production. The deletion strategies used in this study has the potential to further improve the BEVS.
Baculoviruses are a group of double-stranded DNA viruses with varied genome sizes from 80 to 180 kb. Among these viruses, the most well studied species is Autographa californica multiple nucleopolyhedrovirus (AcMNPV) (Rohrmann, 2019). Since the first successful utilization of AcMNPV in the expression of human β-interferon in insect cells (Smith et al., 1983), the baculovirus expression vector system (BEVS) has been used for the production of a large number of exogenous proteins. BEVS is a powerful platform for the production of functional eukaryotic proteins that require post-translational modifications such as glycosylation and phosphorylation. AcMNPV has a genome size of about 134 kb which can accommodate large foreign genes. The virus can only infect insects, so it is safe for mammals. As many types of cells can be transduced by AcMNPV, it is also a useful vector for gene delivery (Drugmand et al., 2012; Van Oers et al., 2015; Weissmann et al., 2016; Targovnik et al., 2021).
Over the past four decades, significant improvements to the original BEVS have been achieved. The establishment of bacmid system by incorporation of bacterial replicon into the viral genome has allowed quick and efficient generation of recombinant baculovirus (Luckow et al., 1993; Je et al., 2001a,b; Zhao et al., 2003). The production of multi-subunit protein complexes using a single baculovirus vector has been made possible by the development of MultiBac system (Berger et al., 2013). Alternatively, multiple foreign genes can be inserted at several loci within the baculovirus genome using multilocus baculovirus vectors (Kanai et al., 2013). These improvements have promoted the application potential of BEVS in the production of protein complexes in basic researches and virus like particles (VLPs) in the development of multivalent vaccines.
AcMNPV contains 155 predicted protein-coding genes (Chen T. et al., 2021). The functions of many of these genes are still unknown, and more than half of them are not required in the virus replication in cultured cells (Wang et al., 2007; Hitchman et al., 2010a; Chen T. et al., 2021; Yu et al., 2023). AcMNPV has two morphologically distinct virion types: budded virion (BV) and occlusion-derived virion (ODV). ODV is embedded in an alkaline-soluble protein matrix, which can maintain the stability of the virus particle in the environment and also be degradable in insect’s midgut, and therefore it is responsible for the primary infection or oral infection in insects. BVs produced from the primary infection will spread the infection to other cells and tissues, resulting in systemic infection of the host (Blissard and Theilmann, 2018). Commercially available BEVS usually lack the polyhedrin (polh) gene, as only BV is required for the infection of cultured cells. Besides polyhedrin, some other ODV-associated proteins are also nonessential for the production of BV and expression of recombinant proteins in cell culture (Rohrmann, 2019). In the development of baculoviruses as optimized vectors for gene delivery and expression, it has been found that removing some viral dispensable genes could not only generate smaller baculovirus genomes to accommodate larger exogenous gene fragments, but also improve the production of exogenous proteins. For example, deletion of chitinase (chiA) and cathepsin (cath) genes was found to be able to improve the stability of intracellular and secreted proteins and therefore increase the production of recombinant proteins (Kaba et al., 2004; Hitchman et al., 2010b). On this basis, a baculovirus vector that was deficient in chiA, cath, p26, p10 and p74 was constructed, and it was revealed that further deletion of p26, p10 and p74 had an additive beneficial effect on recombinant protein expression (Hitchman et al., 2010a). The bacmid with the deletion of all of the five nonessential genes is now commercially available (flashBACULTRA, Oxford Expression Technologies). In addition, knockout of ODV-E26 has been shown to be able to effectively increase recombinant protein production in insect larvae (Pazmiño-Ibarra et al., 2019).
The function of many AcMNPV genes are not well understood, and the asymmetric distribution of essential and nonessential genes in the baculovirus genome provides ample opportunity for further improvement of the baculovirus vector by engineering the viral genome (Vijayachandran et al., 2013). Most of the existing gene-knockout studies focus on one or two single genes, such as Ac18 (Wang et al., 2007), Ac124 (Fang et al., 2019), Ac68 and lef3 (Nie et al., 2012), Ac16 and Ac17 (Nie and Theilmann, 2010), Ac145 and Ac150 (Lapointe et al., 2004). In terms of viral replication and BV production, Ac18, Ac68, Ac124, Ac145 and Ac150 are not important, while lef3, Ac16 and Ac17 are also nonessential but the gene deletion will delay or reduce viral replication. In a recent study, 42 AcMNPV genes whose functions were not fully understood were, respectively, knocked out, and 36 of them were found to be dispensable for BV production (Chen T. et al., 2021). A most recent study found that 51 AcMNPVs containing deletions of in total 62 nonessential genes had similar infectivity as the parental virus, and a potential minimal AcMNPV with a genome of 90 kb was proposed (Yu et al., 2023).
In this study, we deleted 14 DNA fragments from AcMNPV genome, each of them containing at least two contiguous genes that were known nonessential for viral replication in cell culture or functionally unknown. The effects of the fragment deletion on viral proliferation and foreign gene expression were examined. These data provided evidence for further improvement of BEVS by engineering the viral genome.
Spodoptera frugiperda 9 (Sf9) cells (Invitrogen) were cultured in SFX insect medium (HyClone, GE healthcare life science) supplemented with 1% fetal bovine serum (HyClone). Trichoplusia ni (Tn) cells (High Five) were maintained in Serum-free insect cell culture medium at 27°C. Bacmid BAC10:KO1629 (Zhao et al., 2003) and Bac563-5 T (Zhang et al., 2021) were propagated in E. coli strain HS996. Plasmids pTriEx-GFP, pTriEx-OD-Fc and pTriEx-Fluc were stored in our laboratory (Zhang et al., 2021).
Baculovirus gene fragments were knocked out from the parental bacmid BAC10:KO1629 (named as bAc in this study) (Zhao et al., 2003) or Bac563-5 T (Zhang et al., 2021) using the Red/ET recombination system (the recombinase gene was introduced by pSC101 plasmid). RpsL-amp counter-selection cassette with homologous arms containing the target viral gene flanking sequences was amplified by PCR (Primer sequences are listed in Table 1). The target viral gene fragment was replaced with the amplified fragment that was transformed into HS996 competent cells, resulting in the corresponding fragment-knockout (KO) bacmid which was named as bAcΔX, where X represented the knockout genes in the DNA fragment.
To generate KO recombinant baculoviruses, a plasmid carrying a reporter gene was co-transfected with the linearized KO bacmid into Sf9 cells using FuGENE HD Transfection Reagent (Promega). Supernatants containing the recombinant baculoviruses were harvested at 5 days post transfection (dpt) by centrifugation at 300 × g for 5 min to remove cell debris. Viral titers were determined by 50% tissue culture infective doses (TCID50) assay after two passages. The approaches for the generation of KO Bacmids and recombinant viruses are illustrated in Figure 1.
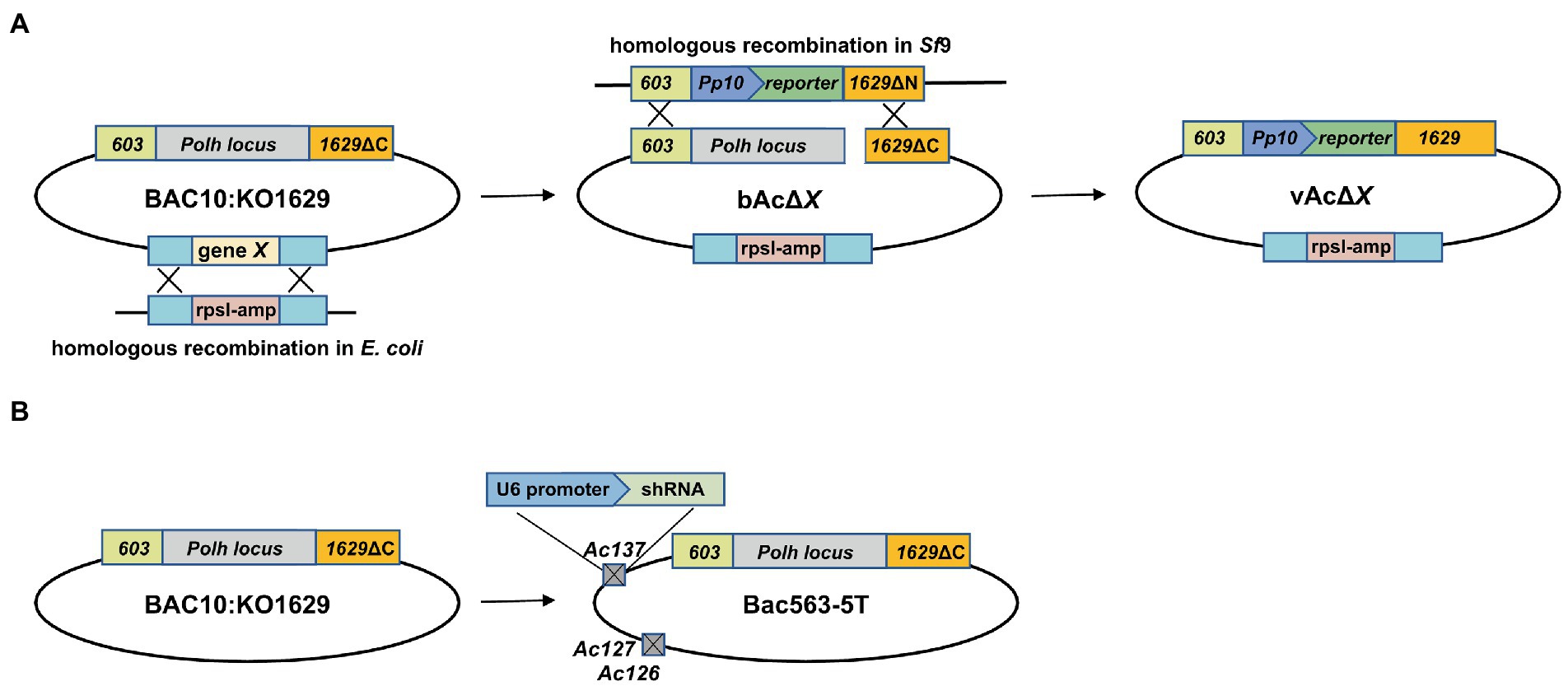
Figure 1. Schematic diagram of the construction of bacmids and generation of AcMNPV expressing a reporter gene. (A) Generation of fragment-knockout (KO) bacmid and AcMNPV expressing a reporter gene. DNA fragment containing predicted nonessential genes (gene X) was deleted from the AcMNPV bacmid BAC10:KO1629 by replacing the target fragment with a rpsl-Amp cassette via homologous recombination in E. coli strain HS996, and the KO bacmid was named as bAcΔX. The recombinant virus carrying the reporter gene expression cassette at the polyhedrin locus, vAcΔX, was generated in Sf9 cells by co-transfection with the linearized bAcΔX and pTriEx plasmid. The parental virus vAc generated from BAC10:KO1629 was used as a control in this study. (B) Schematic diagram of Bac563-5 T used in this study. Bac563-5 T was modified from BAC10:KO1629 by knocking out ChiA/v-cath (Ac126-127) and p10 (Ac137) and inserting an shRNA expression cassette at the Ac137 locus to inhibit the virus infection-induced apoptosis (Je et al., 2001a; Zhang et al., 2021).
To compare the growth kinetics of different KO viruses (vAcΔX) with their parental virus (vAc), Sf9 cells were infected with the recombinant viruses expressing GFP at a multiplicity of infection (MOI) of 0.5. The supernatants of the infected cell cultures were harvested at 24, 48, 72, 96 and 120 h post infection (hpi). Titers of BV were determined by a TCID50 endpoint dilution assay in triplicates and the growth curves were made using GraphPad Prism software.
Sf9 cells and High Five cells were infected with recombinant baculoviruses at an MOI of 3 and harvested for protein detection at 5 days post infection (dpi). Protein samples were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), followed by staining with Coomassie brilliant blue R-250. To calculate the expression level of target protein as a % of total cellular protein, protein bands were quantified by densitometry scanning using Image J software.
To monitor the expression of GFP in baculovirus infected cells, GFP fluorescence was observed by an inverted fluorescence microscope (Lecia DMI8) and the mean fluorescence intensity of the infected cells were analyzed by flow cytometry (NovoCyte 3000RYB).
Fc-tagged recombinant proteins were detected by Western blot using an HRP-conjugated goat anti-human antibody (CoWin Biotech). An enzyme-linked immunosorbent assay (ELISA) was used to quantify the secreted OD-Fc. Purified GP5-Fc (Wu et al., 2014), a recombinant protein containing the same Fc sequence, was used as a standard for the calibration of the protein concentration. Briefly, ELISA plates were individually coated with serial dilutions of GP5-Fc or cell culture medium containing OD-Fc, incubated at 4°C overnight, and then blocked with 5% skimmed milk in PBST at 37°C for 1 h. After three washes with PBST, HRP conjugated Goat Anti-Human IgG (CoWin Biotech) was added and incubated with the antigen for 1 h at 37°C. Unbound antibodies were washed away with PBST for three times. 3,3,5,5-tetramethylbenzidine (TMB) chromogenic substrate (Sangon Biotech) was added and incubated for 5 min at room temperature. The reaction was stopped by adding of H2SO4 and the absorbance was read at a wavelength of 450 nm using an Epoch Microplate Spectrophotometer (BioTek Epoch).
AcMNPV encodes 155 ORFs, and more than half of the genes are non-essential for the production of BV. To construct smaller baculovirus genome for better expression of foreign genes and insertion of larger exogenous DNA fragments, we generated 14 fragment-knockout (KO) bacmids based on BAC10:KO1629 (Table 2). In each of them, a fragment containing at least two predicted nonessential genes (Je et al., 2001b; Chen T. et al., 2021; Yu et al., 2023) was replaced with the rpsl-amp counter-selection cassette by using the Red/ET recombination systems (Figure 1A). All the knockout fragments were confirmed by PCR (data not shown). In total, the 14 KO bacmids contained 62 knockout ORFs.
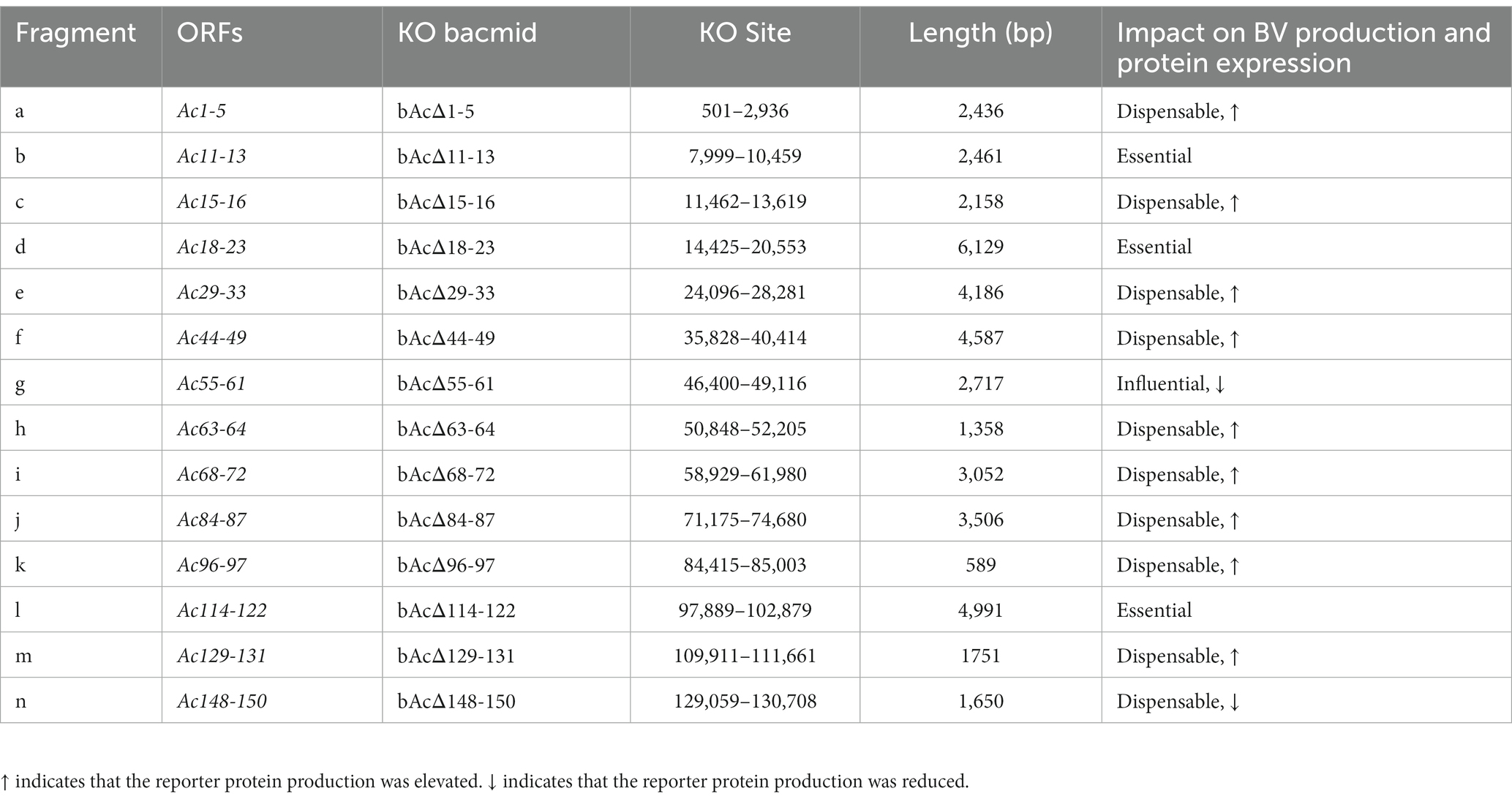
Table 2. Summary of the impact of the knockout of 14 DNA fragments on AcMNPV BV production and protein expression.
To define the effect of the fragment deletions on baculovirus replication, linearized KO bacmids and the parental bacmid were, respectively, co-transfected with pTriEx-GFP into Sf9 cells. The expression of GFP in the transfected Sf9 cells was visualized by using a fluorescence microscope at 5dpi (Figure 2A). GFP producing cells were observed in most of the wells, except that the cells transfected with bacmid bAcΔ114-122 had no fluorescence signal, suggesting that the deletion of Ac114-122 eliminated the very late gene expression under the control of p10 promoter. The subsequent infection results showed that deletions of Ac11-13 and Ac18-23, in addition to Ac114-122, abolished the production of infectious BVs, indicating that essential genes were included in these three fragments. Infection of vAcΔ55-61 exhibited much less fluorescent cells than the parental virus vAc, hinting that BV production might be affected by the knockout of Ac55-61. The deletion of the rest 10 fragments did not damage the production of progeny viruses (Figure 2B).
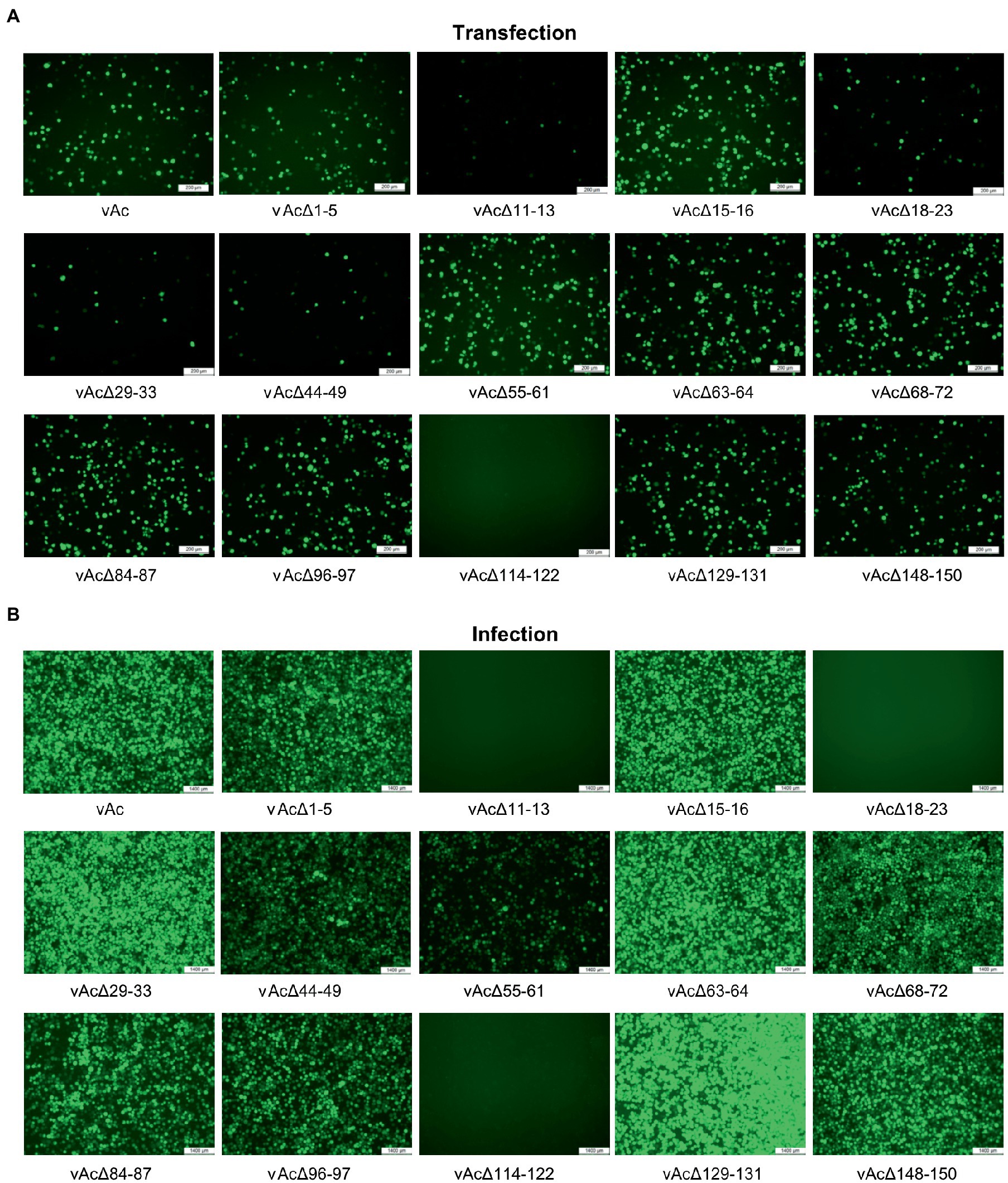
Figure 2. Characterization of KO AcMNPV expressing GFP by fluorescence microscopy. (A) The influence of the DNA fragment deletion on GFP expression after transfection. Sf9 cells were co-transfected with the KO bacmids and pTriEx-GFP, and the images were taken at 5 days post transfection (dpt). Bar, 200 μm. (B) The cells infected with the KO AcMNPV. The cell culture supernatants were harvested at 5 dpt and used to infect Sf9 cells, and the images were captured at 5 days post infection (dpi). Bar, 200 μm.
To characterize the replication kinetics of the KO viruses which can produce infectious BVs, supernatants from Sf9 cells infected with the recombinant viruses were titrated and used to infect cells at an MOI of 0.5, and then the titers of the viruses released in the cell culture supernatants were measured at intervals of 24 h until 120 hpi for the generation of growth kinetics curves (Figure 3). The results showed that vAcΔ1-5, vAcΔ15-16, vAcΔ29-33, vAcΔ44-49, vAcΔ63-64, vAcΔ84-87, vAcΔ96-97, vAcΔ129-131 and vAcΔ148-150 had similar growth curves as vAc. Compared with the parental virus, Further research is needed vAcΔ68-72 had higher titers of BVs and vAcΔ55-61 produced obviously lower levels of progeny BVs in late infection. Although the deletion of Ac55-61 reduced the virus proliferation in late infection, the growth kinetics curves confirmed that the 43 genes included in the above 11 deleted fragments were nonessential for AcMNPV replication.
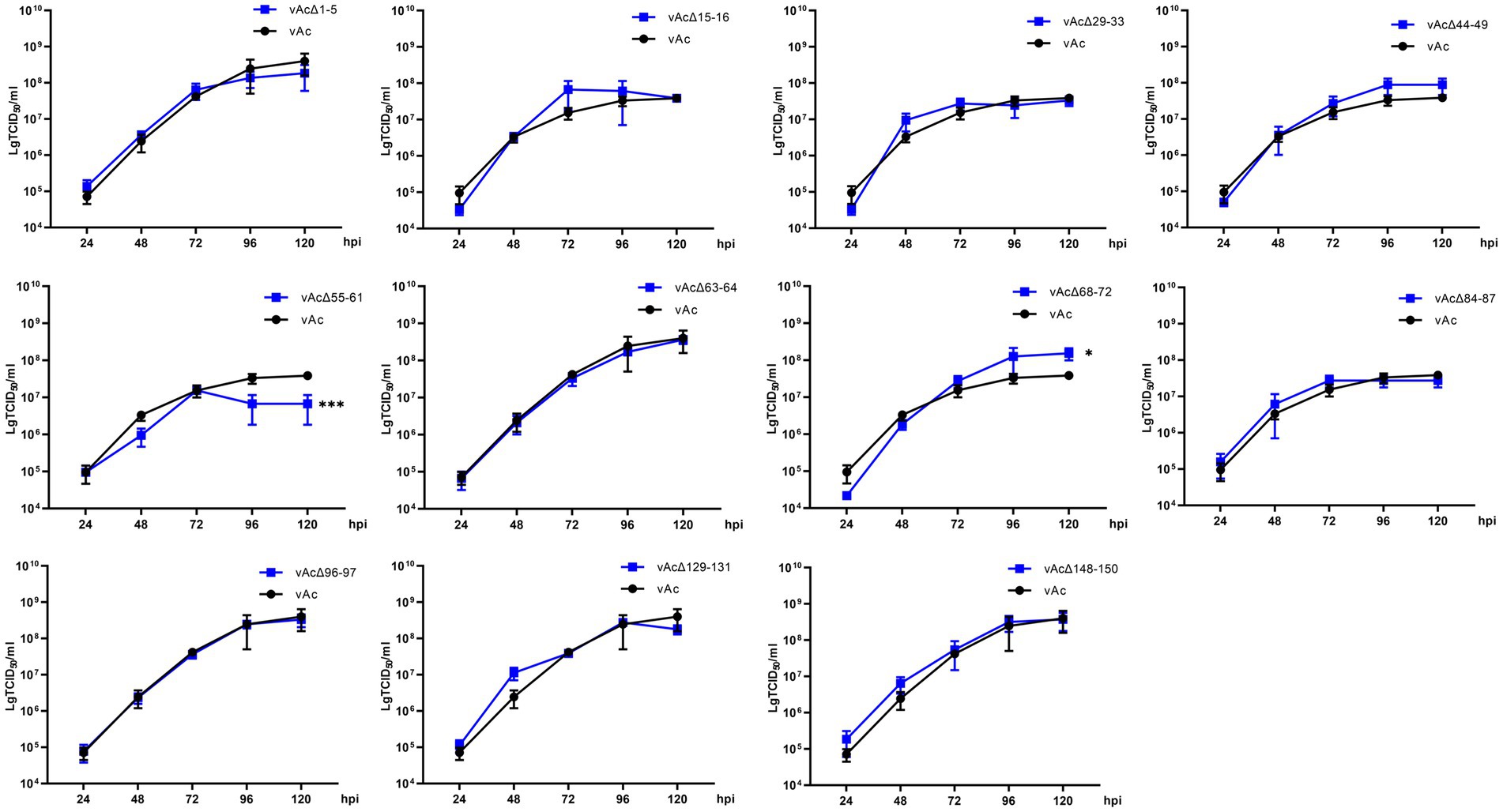
Figure 3. Growth kinetics curves. Sf9 Cells were infected by the virus at an MOI of 0.5. The cell culture supernatants from the infected cells were collected at intervals of 24 h, and BV titers were determined by an endpoint dilution assay in triplicate. *p < 0.05, ***p < 0.001 vs. vAc.
To determine whether the KO bacmids can be used for improved expression of foreign gene, Sf9 cells and High Five cells were, respectively, infected by the recombinant viruses carrying GFP expression cassette driven by p10 promoter at the polh locus at an MOI of 3. GFP fluorescence of the cells were monitored by an inverted fluorescence microscope (Figures 4A,B), and the mean fluorescence intensity was measured by flow cytometry at 5 dpi (Figure 4C). The results showed that the GFP fluorescence was significantly enhanced both in Sf9 and High Five cells infected by vAcΔ1-5, vAcΔ15-16, vAcΔ29-33, vAcΔ44-49, vAcΔ68-72, vAcΔ84-87, vAcΔ96-97 and vAcΔ129-131, compared to the control AcMNPV. Although the rise of GFP fluorescence was not significant in Sf9 cells infected with vAcΔ63-64, the fluorescence signal was significantly elevated in High Five cells. In cells infected with vAcΔ55-61 and vAcΔ148-150, the GFP fluorescence was obviously weakened, suggesting the severe suppression of GFP expression by the deletion of Ac55-61 and Ac148-150.
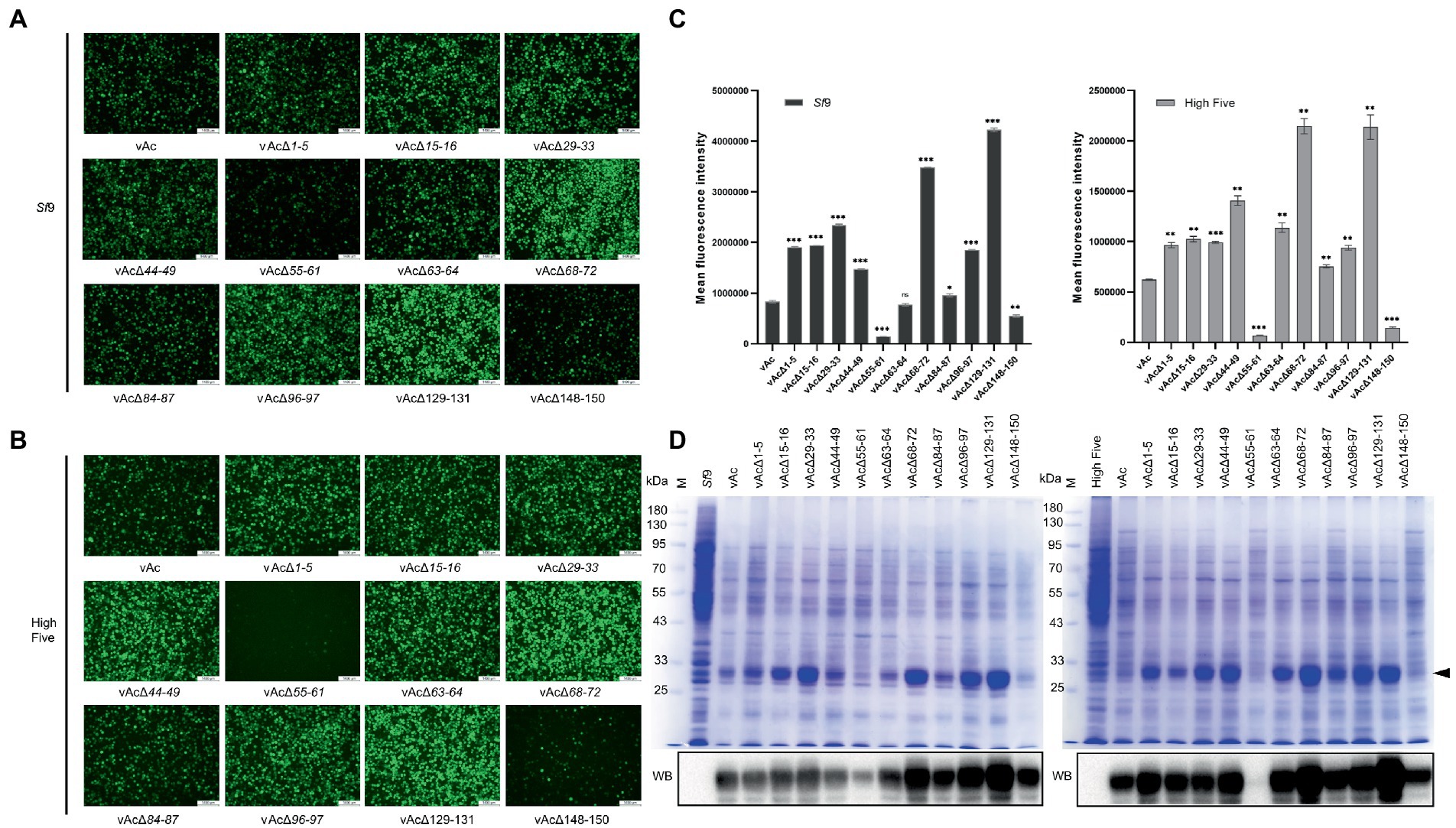
Figure 4. Production of fluorescence protein GFP in Sf9 and High Five cells. (A) Observation of reporter GFP expression in transfected Sf9 cells under fluorescence microscope. The cells were infected by the virus at an MOI of 3. (B) Observation of reporter GFP expression in High Five cells infected by the indicated viruses under fluorescence microscope. (C) Mean fluorescence intensity of GFP measured by flow cytometry. Data were collected from 3×104 cells for each sample, and presented as the means ±SD of two replicates. ns: not significant, *p < 0.05, **p < 0.01, ***p < 0.001 vs. vAc. (D) Detection of the GFP protein levels by SDS-PAGE (upper panel) and Western blot (lower panel). The total proteins of the indicated cell lysates were separated by SDS-PAGE and visualized by coomassie brilliant blue R250 staining, and the GFP protein band is indicated by a black arrow. In Western blot analysis, the His-tagged recombinant GFP was probed by an anti-His monoclonal antibody.
SDS-PAGE and Western blot analyzes of the infected cell lysates showed that the GFP protein level was reduced more severely in vAcΔ55-61 infected cells than in vAcΔ148-150 infected cells. The results also confirmed that the protein production was elevated by most of the KO recombinant viruses (Figure 4D). The data demonstrated that the GFP production in BEVS could be obviously improved by deletion of some baculovirus fragments containing multiple non-essential genes.
To explore whether the knockout of non-essential genes could enhance extracellular production of secreted proteins, OD-Fc, a highly glycosylated recombinant protein containing the outer domain of HIV-1 gp120 (Chen et al., 2007; Zhang et al., 2021), was expressed using KO bacmids bAcΔ29-33, bAcΔ68-72 and bAcΔ129-131 that resulted in the best expression of GFP, and bAcΔ84-87 which only moderately improved the GFP production in High Five cells. The expression and secretion of OD-Fc was examined by Western blot at 5 dpi. The analysis detected both of the predicted molecular mass of unglycosylated OD-Fc (indicated by arrows in Figure 5A) and glycosylated OD-Fc (indicated by right square brackets in Figure 5A) in the infected cell lysates, and only the modified form of the protein appeared in the cell culture supernatant. The blotting results showed that the production and secretion of OD-Fc were generally elevated in Sf9 and High Five cells infected with the KO baculoviruses, except in the vAcΔ29-33 infected High Five cells. No obvious change in the molecular weight of the glycosylated form of protein was observed, suggesting that the glycosylation modification process was not affected by the viral gene deletions.
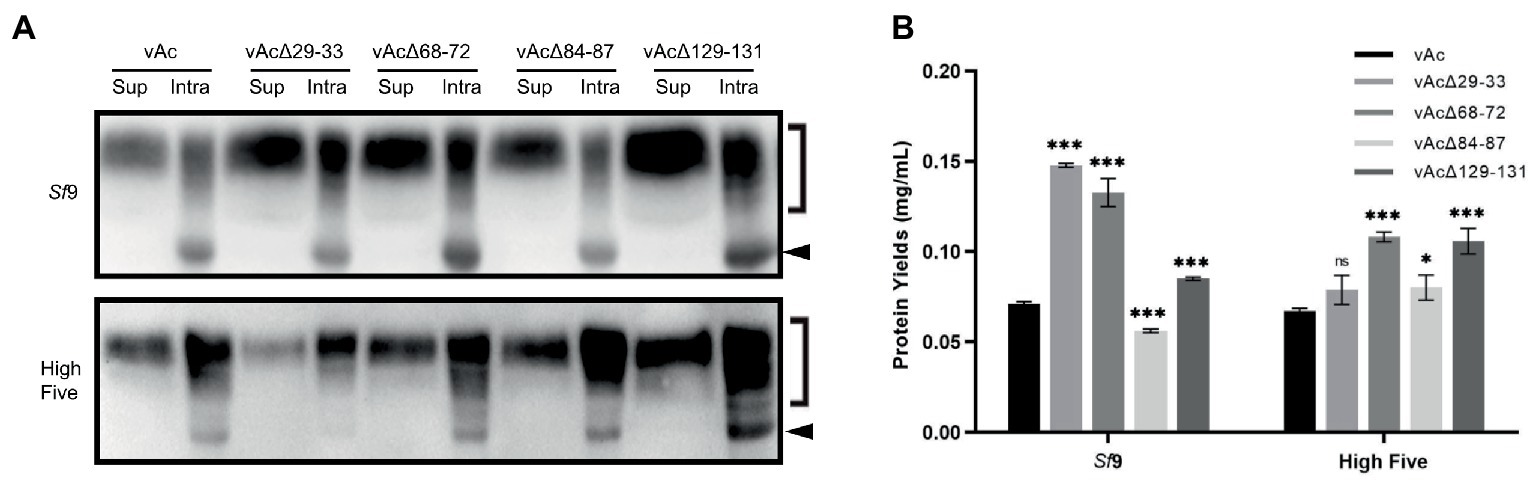
Figure 5. Expression of glycoprotein OD-Fc in Sf9 and High Five cells. (A) Detection of the intracellular and secreted OD-Fc by Western blot at 5 dpi. The unmodified target protein band is indicated by a black arrow, and the glycosylated form is indicated by a right square bracket. The OD-Fc protein was probed by HRP-conjugated anti-Human antibody in Western blot analysis. (B) Quantification of the level of OD-Fc secreted in the cell culture supernatant by ELISA at 5 dpi. Data were presented as the means ±SD of three replicates. ns: not significant, *p < 0.05, **p < 0.01, ***p < 0.001 vs. vAc.
To better quantify the highly glycosylated and heterogeneous OD-Fc secreted in the cell culture media, ELISA analyzes were performed using purified GP5-Fc with the same Fc tag as a standard. Supernatant of the cell cultures infected with the recombinant baculoviruses was harvested for protein detection at 5 dpi. As shown in Figure 5B, the data confirmed that the production of secreted OD-Fc was significantly improved in both Sf9 and High Five cells infected with vAcΔ68-72 and vAcΔ129-131. The deletion of 29–33 resulted in two-fold increase of the yield of secreted OD-Fc in Sf9 cells but no significant improvement in High Five cells. In contrast, the production of secreted OD-Fc by vAcΔ84-87 was only significantly increased in High Five cells. These data demonstrate that knockout of some multiple genes which are nonessential for viral survival or infectivity are beneficial for the production of secreted glycoprotein in BEVS.
In our previous report, we have developed a baculovirus vector Bac563-5 T carrying an shRNA expression cassette to inhibit virus infection-induced apoptosis and improve protein production in both Sf9 and High Five cells. In this vector, the nonessential ChiA/v-cath (Ac126-127) and p10 (Ac137) genes have been deleted (Je et al., 2001a; Zhang et al., 2021). Based on Bac563-5 T, we investigated whether the dispensable fragment deletion could be combined to further enhance protein production in BEVS.
By knocking out of Ac15-16 and Ac29-33, respectively, from Bac563-5 T, we obtained Bac563-5 T-Δc and Bac563-5 T-Δe (Table 3). After generation of baculoviruses expressing firefly luciferase (Fluc) as a reporter protein, it revealed that neither the deletion of Ac15-16 nor Ac29-33 impacted the virus replication (Table 3), and both the fragment deletion benefited Fluc expression in Sf9 and High Five cells (Figures 6A,B).
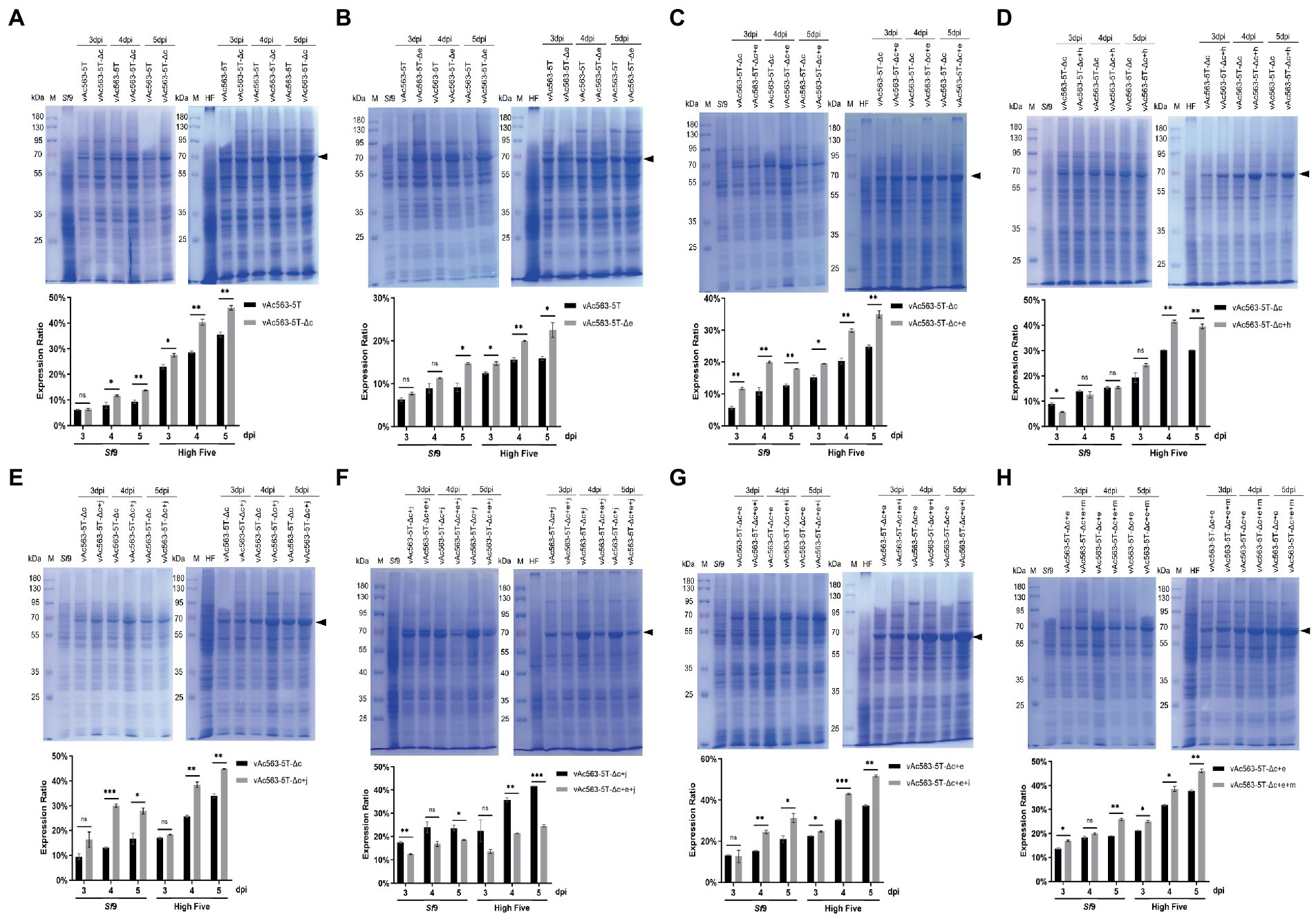
Figure 6. Expression of firefly luciferase (Fluc) using the indicated bacmid containing superimposed deletion of multiple nonessential fragments. (A-H) Impact of combination deletion of the indicated fragment on the Fluc expression. Cell lysates were harvested at 3, 4, and 5 dpi, separated by SDS-PAGE and visualized by coomassie brilliant blue R250 staining. The Fluc protein band is indicated by a black arrow. The protein bands were quantified by densitometry scanning using Image J software, and the expression levels of Fluc as a % of total cellular protein were calculated and the data were presented in the bar charts. *p < 0.05, **p < 0.01, ***p < 0.001.
Based on Bac563-5 T-Δc, the deletion of Ac29-33, Ac63-64 and Ac84-87 was, respectively, combined with the deletions of Ac15-16, Ac126-127 and Ac137. The results showed that the virus titer decreased approximately one order of magnitude after the knockout of Ac63-64 (Bac563-5 T-Δc + h), suggesting that combined deletion of Ac63-64 with Ac15-16, Ac126-127 and Ac137 affected the virus replication ability, although the protein production was not reduced (Figure 6D). In contrast, the virus propagation ability was maintained (Table 3) and Fluc protein production was improved in the other two combinations (Bac563-5 T-Δc + e and Bac563-5 T-Δc + j) (Figures 6C,E). However, when both the deletions of Ac29-33 and Ac84-87 were combined on Bac563-5 T-Δc, the resulting Bac563-5 T-Δc + e + j had a lower protein yield than Bac563-5 T-Δc + j (Figure 6F).
As Bac563-5 T-Δc + e gave a higher average virus titer than Bac563-5 T-Δc + h, we further knocked out Ac68-72 and Ac129-131 from Bac563-5 T-Δc + e, generated Bac563-5 T-Δc + e + i and Bac563-5 T-Δc + e + m. Both of the bacmids maintained good virus propagation ability (Table 3). Notably, the yields of Fluc protein were close to 50% of total proteins in High Five cells and 30% in Sf9 cells (Figures 6G,H). Taken together, these data demonstrated that foreign protein production in BEVS could be noticeably increased by deletion of some large fragments containing multiple non-essential genes from AcMNPV genome.
AcMNPV encodes about 150 open reading frames (ORFs) and the functions of many genes in viral replication and foreign protein expression are still unclear. Previous studies have suggested that more than half of the genes in AcMNPV and BmNPV baculovirus genomes are not required for the production of infectious BV when they are individually deleted (Ono et al., 2012; Chen T. et al., 2021; Yu et al., 2023). It has been proposed that the removal of redundant viral proteins may facilitate the virus to make better use of cell machineries and substrates to synthesize more target proteins, and it has been found that single and multiple deletions of some viral nonessential genes such as polh, chiA, cath, p26, p10 and p74 can increase recombinant protein yield without affecting the virus viability (Hitchman et al., 2010a). Whether deletion of large fragments containing other multiple nonessential genes could benefit the production of exogenous proteins remains to be investigated.
In this study, we systematically deleted 14 DNA fragments from AcMNPV genome to determine the effects on BEVS properties such as viral proliferation and foreign gene expression and secretion. The results revealed that deletion of Ac1-5, Ac15-16, Ac29-33, Ac44-49, Ac55-61, Ac63-64, Ac68-72, Ac84-87, Ac96-97, Ac129-131 or Ac148-150 was not lethal, demonstrating that the 43 genes included in the 11 fragments are nonessential for virus replication in cultured cells. AcMNPV mutants deleted for these fragments had normal replication ability in Sf9 cells, except that vAcΔ55-61 significantly produced less BVs than its parental virus. In AcMNPV and BmNPV, it has been shown that individual deletion of Ac55 (Bm44), Ac56 (Bm45), Ac57 (Bm46), Ac58/59 (Bm47) and Ac60 (Bm48) does not affect the virus reproduction (Ono et al., 2012; Chen T. et al., 2021). For Ac61, its gene product FP25K is involved in nuclear trafficking of occlusion-associated proteins (Garretson et al., 2016). Studies have demonstrated that the gene knockout does not reduce BV production in BmNPV and AcMNPV (Ono et al., 2012; Garretson et al., 2016; Yu et al., 2023), but results in a ‘few polyhedra phenotype’ which are influenced by the host insect cells (Cheng et al., 2012). As virus mutants with single gene deletion in Ac55-61 are normal for BV production, it is still unclear why vAcΔ55-61 has reduced BV productivity. It has been found that knockout of either ie-1 or ie-0 is not lethal for AcMNPV, but the mutant lacking the two genes lost the ability to produce BV (Stewart et al., 2005). It remains to be investigated whether there are functional compensation genes in Ac55-61.
For the other three AcMNPV mutants, the deletion of Ac11-13, Ac18-23 and Ac114-122 abolished the production of infectious BVs, indicating that these three fragments may contain essential genes for the virus replication. Among Ac11-13, Ac11 is a highly conserved gene. Its homolog in BmNPV (bm4) is not associated with BV or ODV (Ono et al., 2012), and deletion of Ac11 does not affect viral DNA replication, but it has been shown essential for infectious BV production and ODV envelopment in AcMNPV (Tao et al., 2015). The gene product can interact with components of the ESCRT-III complex and may be involved in the release of nucleocapsid at the nuclear membrane (Yue et al., 2018). Ac12 has been identified dispensable for BV production (Chen T. et al., 2021), but a more recent study found that deletion of Ac12 had a significant negative effect on the production of infectious viruses (Yu et al., 2023). Ac13 is a conserved gene in all sequenced alphabaculoviruses and it encodes a late expression protein with a putative nuclear localization signal motif. Its gene product contains coiled-coil regions and has some structural similarity to some membrane proteins. Deletion of Ac13 did not affect viral genome replication, nucleocapsid assembly or occlusion body (OB) formation but caused lower titers of BV due to lack of efficient nuclear egress from nucleus to cytoplasm (Chen X. et al., 2021). Its homolog in BmNPV (bm5) was found to be present in both the inner-and outer nuclear membranes (Nagamine et al., 2019). Bm5 disruption resulted in lower titers of BV, fewer numbers of ODV and aberrant expression of various viral genes at the very late stage of infection (Kokusho et al., 2016). We tried to introduce Ac11 back into vAcΔ11-13 and failed in the rescue of infectious virus particles (data not shown), which indicated that Ac12 and Ac13 cannot be simultaneously deleted from AcMNPV genome.
For Ac18-23, all the homolog genes have been demonstrated dispensable for BmNPV production (Ono et al., 2012), but a recent study shows that deletion of Bm14 (homolog of Ac23) causes reduced BV and ODV production and also delayed death of infected larvae (Xu et al., 2019). Ac23 encodes a fusion protein (F), which is involved in the formation of the BV envelope structure (Lung et al., 2003) and enhancing the infectivity of the budded virus (Wang et al., 2008). Reintroducing Ac23 into vAcΔ18-23 in our study was not able to restore the virus infectivity (data not shown), suggesting that other genes in Ac18-22 may also affect the production of infectious virus particles.
The design of Ac114-122 deletion was based on the data that these genes were dispensable for BmNPV (Ono et al., 2012). However, our data here revealed that deletion of this fragment was lethal for AcMNPV. Consistent with our results, Ac120 (Chen T. et al., 2021) and Ac115 (Yu et al., 2023) were proven important for BV production in recent studies. It remains to be investigated whether re-introduction of Ac115 and Ac120 can restore the infectivity of vAcΔ114-122.
To test if the AcMNPV mutants deficient in multiple nonessential genes were appropriate vectors for the expression of exogenous proteins, we constructed recombinant baculoviruses for the intracellular expression of GFP and secretory production of glycoprotein OD-Fc. Among the 10 mutants with normal replication ability, most of them resulted in higher protein productivity, and only vAcΔ148-150 obviously gave a lower protein yield than the parental vector. Interestingly, some KO mutants exhibited cell type-dependent effects on protein production. Deletion of Ac63-64 and Ac84-87 resulted in more significant improvement of GFP production in High Five than in Sf9 cells. In contrast, lack of Ac15-16, Ac29-33 and Ac96-97 benefited the protein production more prominently in Sf9 cells than in High Five cells. The yield of secreted glycoprotein OD-Fc showed similar cell type-dependent tendency as the GFP production. Previous studies have discovered that Sf9 cells have a higher capacity for production of infectious virus particles, while High Five cells have higher susceptibility to baculovirus infection and a number of metabolic pathways can be hijacked to support mass production, therefore they exhibit higher protein productivity than Sf9 cells (Monteiro et al., 2014; Wilde et al., 2014). Our data here suggested that the effect of deletion of genes with different functions on the protein production in BEVS varied in different host cells. Consequently, the impacts of different cell lines should also be considered for developing the baculovirus expression vector system.
Based on our previously constructed Bac563-5 T with the knockout of ChiA/v-cath (Ac126-127) and p10 (Ac137) (Je et al., 2001a; Zhang et al., 2021), we tried to delete more dispensable fragments from this Bacmid. Despite the fact that individual deletion of these nonessential fragments increased protein expression, simultaneous knockout of them may have an adverse impact on protein expression, and distinct combinations of deletions may have diverse effects on protein expression. In the end, we successfully generated three bacmid vectors each containing three more dispensable fragments removed. Compared with BAC10:KO1629 (Zhao et al., 2003), all these three bacmids are more than 10 kb shorter in length but the viruses have comparable replication capabilities in Sf9. Among them, Bac563-5 T-Δc + e + j had reduced protein production than Bac563-5 T-Δc + e after the deletion of Ac84-87, but Bac563-5 T-Δc + e + i (with combined deletion of Ac15-16, Ac29-33, Ac68-72, Ac126-127 and Ac137) and Bac563-5 T-Δc + e + m (with combined deletion of Ac15-16, Ac29-33, Ac126-127, Ac137 and Ac129-131) showed improved protein expression performance (maps illustrating the deletions in the genome are shown in Figure 7). In Sf9 cells, the amount of expressed Fluc occupied around 30% of the total intracellular proteins at 5dpi, while in High five cells, the ratio was close to 50% of the total proteins. More efforts are needed to investigate whether more dispensable fragments can be removed from the AcMNPV genome to maintain or further improve the protein productivity in BEVS.
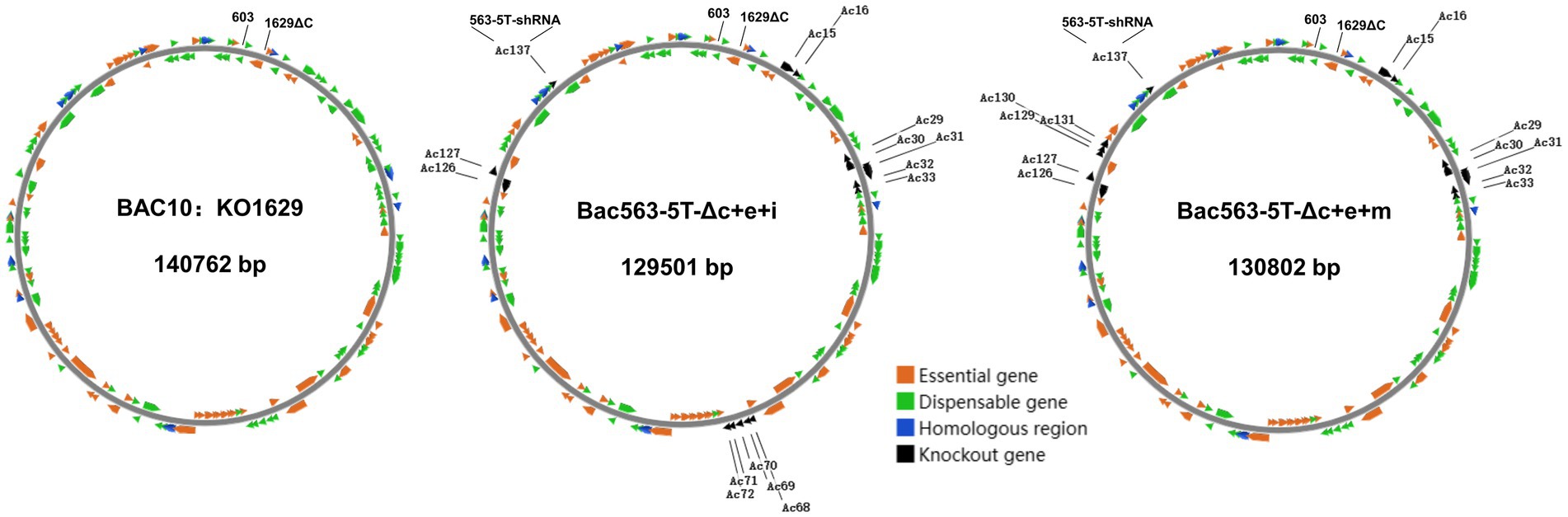
Figure 7. Schematic diagram of AcMNPV bacmid BAC10:KO1629 and the two shortened bacmids with improved capacity for recombinant protein production. BAC10:KO1629 (left) is 140,762 bp, containing 154 AcMNPV ORFs and a F replicon (Kanr) fragment inserted between ORF603 and truncated ORF1629. Compared with BAC10:KO1629, the shortened Bac563-5 T-Δc + e + i containing deletions of Ac15-16, Ac29-33, Ac68-72, Ac126-127 and Ac137 (middle) and Bac563-5 T-Δc + e + m containing deletions of Ac15-16, Ac29-33, Ac126-127, Ac129-131 and Ac137 (right) exhibit similar replication ability and better protein productivity.
Nowadays, the re-design and de novo synthesis of viral genome techniques have become mature. Since the first synthetic baculovirus was rescued in 2017, several re-designed AcMNPV have been constructed (Shang et al., 2017, 2021; Guo et al., 2022). In a recent report, a synthetic AcMNPV with 17 kb deletion in the C1 Region of the viral genome was generated, but the titer of infectious virus particles produced in Sf9 cells dropped two orders of magnitude (Guo et al., 2022). Synthetic virology provides a powerful platform for the understanding of baculovirus replication and the application of the virus as gene delivery and protein expression vectors. Compared to the traditional top-down strategy used in this study, it could be easier and quicker to generate a new viral genome containing multiple deletions or modifications using the bottom-up synthetic approach. However, the design a minimal baculovirus with comparable or better replication ability and protein productivity still requires more basic research on baculovirus biology for better understanding of the roles of viral proteins in the virus replication.
In this study, we constructed 14 fragment-knockout mutants of AcMNPV, each having at least two genes deleted, and identified 11 of the fragments containing 43 genes could be individually deleted without abolishing BV production. Using the AcMNPV mutants deficient in multiple nonessential genes as vector for exogenous protein expression, the results demonstrated that nine of the fragment-deletions benefited protein production. By combining the deletion of fragments dispensable for virus replication and recombinant protein expression, we obtained two AcMNPV vectors which were shortened more than 10 kb and simultaneously displayed improved capacity for recombinant protein production. This report could serve as a foundation for further improvement of the BEVS as a platform for protein production in biopharmaceutical industry and basic researches.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
HC, XX, and XZ conceived the study and designed the experiments. XZ, AH, YZ, HT, and KZ performed the experiments. XZ and ZZ analyzed the data. HC, XZ, and AH wrote the manuscript. All authors have read and approved the final manuscript.
This work was sponsored by Yangling science and technology project (2016SF-06), the Natural Science Basic Research Plan in Shaanxi Province of China (2021JM-098) and in part by Shaanxi Bacmid Biotechnology Co., Ltd. (K4050422140).
We thank the Teaching and Research Core Facility at College of Life Sciences, NWAFU for their support in this work.
XX is the founder and a shareholder of this company. Patents covering the AcMNPV vector described in this manuscript are as follows: patent ZL 2019 10650551.5 is owned by Northwest A&F University, and patents pending 2021103927781, 2021100308313, 2021103924181, 2021107427173, and 202110408273X are owned by Shaanxi Bacmid Biotechnology Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Berger, I., Garzoni, F., Chaillet, M., Haffke, M., Gupta, K., and Aubert, A. (2013). The Multibac protein complex production platform at the Embl. J. Vis. Exp. 77:E50159. doi: 10.3791/50159
Blissard, G. W., and Theilmann, D. A. (2018). Baculovirus entry and egress from insect cells. Annu. Rev. Virol. 5, 113–139. doi: 10.1146/Annurev-Virology-092917-043356
Chen, T., Duan, X., Hu, H., Shang, Y., Hu, Y., Deng, F., et al. (2021). Systematic analysis of 42 Autographa californica multiple nucleopolyhedrovirus genes identifies An additional six genes involved in the production of infectious budded virus. Virol. Sin. 36, 762–773. doi: 10.1007/S12250-021-00355-1
Chen, H., Xu, X., and Jones, I. M. (2007). Immunogenicity of the outer domain of a Hiv-1 clade C Gp120. Retrovirology 4:33. doi: 10.1186/1742-4690-4-33
Chen, X., Yang, X., Lei, C., Qin, F., Sun, X., and Hu, J. (2021). Autographa californica multiple nucleopolyhedrovirus Orf13 is required for efficient nuclear egress of nucleocapsids. Virol. Sin. 36, 968–980. doi: 10.1007/S12250-021-00353-3
Cheng, X., Kumar, S., Arif, B., Krell, P., Zhang, C., and Cheng, X. (2012). Cell-dependent polyhedra production and Virion occlusion of Acmnpv Fp25k mutants in vitro and in vivo. J. Gen. Virol. 94, 177–186. doi: 10.1099/Vir.0.045591-0
Drugmand, J.-C., Schneider, Y.-J., and Agathos, S. N. (2012). Insect cells as factories for biomanufacturing. Biotechnol. Adv. 30, 1140–1157. doi: 10.1016/J.Biotechadv.2011.09.014
Fang, Z., Que, Y., Li, J., and Zhang, Z. (2019). The deletion of the Acmnpv Ac124 gene resulted in a decrease in Chitinase transcription. Virus Res. 263, 151–158. doi: 10.1016/J.Virusres.2019.01.017
Garretson, T. A., Mccoy, J. C., and Cheng, X.-W. (2016). Baculovirus Fp25k localization: role of the coiled-coil domain. J. Virol. 90, 9582–9597. doi: 10.1128/Jvi.01241-16
Guo, Y., Hu, H., Xiao, H., Deng, F., Li, J., Wang, M., et al. (2022). Successful rescue of synthetic Acmnpv with a ~17 kb deletion in the C1 region of the genome. Viruses 14:2780. doi: 10.3390/V14122780
Hitchman, R. B., Possee, R. D., Crombie, A. T., Chambers, A., Ho, K., Siaterli, E., et al. (2010a). Genetic modification of a baculovirus vector for increased expression in insect cells. Cell Biol. Toxicol. 26, 57–68. doi: 10.1007/S10565-009-9133-Y
Hitchman, R. B., Possee, R. D., Siaterli, E., Richards, K. S., Clayton, A. J., Bird, L. E., et al. (2010b). Improved expression of secreted and membrane-targeted proteins in insect cells. Biotechnol. Appl. Biochem. 56, 85–93. doi: 10.1042/Ba20090130
Je, Y. H., Chang, J. H., Choi, J. Y., Roh, J. Y., Jin, B. R., O'reilly, D. R., et al. (2001a). A defective viral genome maintained in Escherichia Coli for the generation of baculovirus expression vectors. Biotechnol. Lett. 23, 575–582. doi: 10.1023/A:1010301404445
Je, Y. H., Chang, J. H., Roh, J. Y., and Jin, B. R. (2001b). Generation of Baculovirus expression vector using detective Autographa California nuclear Polyhedrosis virus genome maintained in Escherichia coli for Occ+ virus production. Int. J. Indust. Entomol 2, 155–160.
Kaba, S. A., Salcedo, A. M., Wafula, P. O., Vlak, J. M., and Van Oers, M. M. (2004). Development of a Chitinase and V-Cathepsin negative Bacmid for improved integrity of secreted recombinant proteins. J. Virol. Methods 122, 113–118. doi: 10.1016/J.Jviromet.2004.07.006
Kanai, Y., Athmaram, T., Stewart, M., and Roy, P. (2013). Multiple large foreign protein expression by a single recombinant baculovirus: a system for production of multivalent vaccines. Protein Expr. Purif. 91, 77–84. doi: 10.1016/J.Pep.2013.07.005
Kokusho, R., Koh, Y., Fujimoto, M., Shimada, T., and Katsuma, S. (2016). Bombyx mori Nucleopolyhedrovirus Bm5 protein regulates progeny virus production and viral gene expression. Virology 498, 240–249. doi: 10.1016/J.Virol.2016.08.032
Lapointe, R., Popham, H. J., Straschil, U., Goulding, D., O'reilly, D. R., and Olszewski, J. A. (2004). Characterization of two Autographa californica nucleopolyhedrovirus proteins, Ac145 and Ac150, which affect Oral infectivity in a host-dependent manner. J. Virol. 78, 6439–6448. doi: 10.1128/Jvi.78.12.6439-6448.2004
Luckow, V. A., Lee, S. C., Barry, G. F., and Olins, P. O. (1993). Efficient generation of infectious recombinant baculoviruses by site-specific transposon-mediated insertion of foreign genes into a Baculovirus genome propagated in Escherichia coli. J. Virol. 67, 4566–4579. doi: 10.1128/Jvi.67.8.4566-4579.1993
Lung, O. Y., Cruz-Alvarez, M., and Blissard, G. W. (2003). Ac23, An envelope fusion protein homolog in the baculovirus Autographa californica multicapsid nucleopolyhedrovirus, is a viral pathogenicity factor. J. Virol. 77, 328–339. doi: 10.1128/Jvi.77.1.328-339.2003
Monteiro, F., Bernal, V., Saelens, X., Lozano, A. B., Bernal, C., Sevilla, A., et al. (2014). Metabolic profiling of insect cell lines: unveiling cell line determinants behind System's productivity. Biotechnol. Bioeng. 111, 816–828. doi: 10.1002/Bit.25142
Nagamine, T., Inaba, T., and Sako, Y. (2019). A nuclear envelop-associated Baculovirus protein promotes Intranuclear lipid accumulation during infection. Virology 532, 108–117. doi: 10.1016/J.Virol.2019.04.006
Nie, Y., Fang, M., Erlandson, M. A., and Theilmann, D. A. (2012). Analysis of the Autographa californica multiple nucleopolyhedrovirus overlapping gene pair Lef3 and Ac68 reveals that Ac68 is a per Os infectivity factor and that Lef3 is critical, but not essential, for virus replication. J. Virol. 86, 3985–3994. doi: 10.1128/Jvi.06849-11
Nie, Y., and Theilmann, D. A. (2010). Deletion of Acmnpv Ac16 and Ac17 results in delayed viral gene expression in budded virus infected cells but not transfected cells. Virology 404, 168–179. doi: 10.1016/J.Virol.2010.03.031
Ono, C., Kamagata, T., Taka, H., Sahara, K., Asano, S.-I., and Bando, H. (2012). Phenotypic grouping of 141 Bmnpvs lacking viral gene sequences. Virus Res. 165, 197–206. doi: 10.1016/J.Virusres.2012.02.016
Pazmiño-Ibarra, V., Mengual-Martí, A., Targovnik, A. M., and Herrero, S. (2019). Improvement of Baculovirus as protein expression vector and as biopesticide by Crispr/Cas9 editing. Biotechnol. Bioeng. 116, 2823–2833. doi: 10.1002/Bit.27139
Rohrmann, G. F.. (2019). Baculovirus Molecular Biology. 4th, Bethesda, MD: National Center For Biotechnology Information (Us).
Shang, Y., Hu, H., Wang, X., Wang, H., Deng, F., Wang, M., et al. (2021). Construction and characterization of a novel Bacmid Acbac-Syn based on a synthesized Baculovirus genome. Virol. Sin. 36, 1566–1574. doi: 10.1007/S12250-021-00449-W
Shang, Y., Wang, M., Xiao, G., Wang, X., Hou, D., Pan, K., et al. (2017). Construction and rescue of a functional synthetic Baculovirus. ACS Synth. Biol. 6, 1393–1402. doi: 10.1021/Acssynbio.7b00028
Smith, G. E., Summers, M. D., and Fraser, M. (1983). Production of human Beta interferon in insect cells infected with a Baculovirus expression vector. Mol. Cell. Biol. 3, 2156–2165. doi: 10.1128/Mcb.3.12.2156-2165.1983
Stewart, T. M., Huijskens, I., Willis, L. G., and Theilmann, D. A. (2005). The Autographa californica multiple nucleopolyhedrovirus Ie0-Ie1 gene complex is essential for wild-type virus replication, but either Ie0 or Ie1 can support virus growth. J. Virol. 79, 4619–4629. doi: 10.1128/Jvi.79.8.4619-4629.2005
Tao, X. Y., Choi, J. Y., Kim, W. J., An, S. B., Liu, Q., Kim, S. E., et al. (2015). Autographa californica multiple nucleopolyhedrovirus Orf11 is essential for budded-virus production and occlusion-derived-virus envelopment. J. Virol. 89, 373–383. doi: 10.1128/Jvi.01742-14
Targovnik, A. M., Simonin, J. A., Mc Callum, G. J., Smith, I., Cuccovia Warlet, F. U., Nugnes, M. V., et al. (2021). Solutions against emerging infectious and noninfectious human diseases through the application of Baculovirus technologies. Appl. Microbiol. Biotechnol. 105, 8195–8226. doi: 10.1007/S00253-021-11615-1
Van Oers, M. M., Pijlman, G. P., and Vlak, J. M. (2015). Thirty years of Baculovirus–insect cell protein expression: from dark horse to mainstream technology. J. Gen. Virol. 96, 6–23. doi: 10.1099/Vir.0.067108-0
Vijayachandran, L. S., Thimiri Govinda Raj, D. B., Edelweiss, E., Gupta, K., Maier, J., Gordeliy, V., et al. (2013). Gene gymnastics: synthetic biology for Baculovirus expression vector system engineering. Bioengineered 4, 279–287. doi: 10.4161/Bioe.22966
Wang, M., Tan, Y., Yin, F., Deng, F., Vlak, J. M., Hu, Z., et al. (2008). The F-like protein Ac23 enhances the infectivity of the budded virus of Gp64-null Autographa californica multinucleocapsid nucleopolyhedrovirus Pseudotyped with baculovirus envelope fusion protein F. J. Virol. 82, 9800–9804. doi: 10.1128/Jvi.00759-08
Wang, Y., Wu, W., Li, Z., Yuan, M., Feng, G., Yu, Q., et al. (2007). Ac18 is not essential for the propagation of Autographa californica multiple nucleopolyhedrovirus. Virology 367, 71–81. doi: 10.1016/J.Virol.2007.05.017
Weissmann, F., Petzold, G., VanderLinden, R., Huis in 't Veld, P. J., Brown, N. G., Lampert, F., et al. (2016). Bigbac enables rapid gene assembly for the expression of large multisubunit protein complexes. Proc. Natl. Acad. Sci. U. S. A. 113, E2564–E2569. doi: 10.1073/Pnas.1604935113
Wilde, M., Klausberger, M., Palmberger, D., Ernst, W., and Grabherr, R. (2014). Tnao38, high five and Sf9--evaluation of host-virus interactions in three different insect cell lines: baculovirus production and recombinant protein expression. Biotechnol. Lett. 36, 743–749. doi: 10.1007/S10529-013-1429-6
Wu, F., Peng, K., Tian, J., Xu, X., Zhou, E., and Chen, H. (2014). Immune response to fc tagged Gp5 glycoproteins of porcine reproductive and respiratory syndrome virus. Viral Immunol. 27, 343–349. doi: 10.1089/Vim.2014.0041
Xu, W., Fan, Y., Wang, H., Feng, M., and Wu, X. (2019). Bombyx mori nucleopolyhedrovirus F-like protein Bm14 affects the morphogenesis and production of occlusion bodies and the embedding of Odvs. Virology 526, 61–71. doi: 10.1016/J.Virol.2018.10.008
Yu, Y., Zhang, T., Lu, D., Wang, J., Xu, Z., Zhang, Y., et al. (2023). Genome-wide nonessential gene identification of Autographa californica multiple nucleopolyhedrovirus. Gene 863:147239. doi: 10.1016/J.Gene.2023.147239
Yue, Q., Yu, Q., Yang, Q., Xu, Y., Guo, Y., Blissard, G. W., et al. (2018). Distinct roles of cellular Escrt-I and Escrt-iii proteins in efficient entry and egress of budded Virions of Autographa californica multiple nucleopolyhedrovirus. J. Virol. 92, E01636–E01617. doi: 10.1128/Jvi.01636-17
Zhang, X., Zhao, K., Lan, L., Shi, N., Nan, H., Shi, Y., et al. (2021). Improvement of protein production by engineering a novel antiapoptotic baculovirus vector to suppress the expression of sf-Caspase-1 and Tn-Caspase-1. Biotechnol. Bioeng. 118, 2977–2989. doi: 10.1002/Bit.27807
Keywords: baculovirus expression vector system, Autographa californica multiple nucleopolyhedrovirus, non-essential genes, protein production, gene knockout
Citation: Zhang X, He A, Zong Y, Tian H, Zhang Z, Zhao K, Xu X and Chen H (2023) Improvement of protein production in baculovirus expression vector system by removing a total of 10 kb of nonessential fragments from Autographa californica multiple nucleopolyhedrovirus genome. Front. Microbiol. 14:1171500. doi: 10.3389/fmicb.2023.1171500
Received: 22 February 2023; Accepted: 24 March 2023;
Published: 13 April 2023.
Edited by:
Kohji Moriishi, University of Yamanashi, JapanReviewed by:
Jian Xu, East China Normal University, ChinaCopyright © 2023 Zhang, He, Zong, Tian, Zhang, Zhao, Xu and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongying Chen, Y2hlbmh5QG53c3VhZi5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.