- College of Plant Protection, Shandong Agricultural University, Tai'an, China
Edible fungi are not only delicious but are also rich in nutritional and medicinal value, which is highly sought after by consumers. As the edible fungi industry continues to rapidly advance worldwide, particularly in China, the cultivation of superior and innovative edible fungi strains has become increasingly pivotal. Nevertheless, conventional breeding techniques for edible fungi can be arduous and time-consuming. CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats/CRISPR-associated nuclease 9) is a powerful tool for molecular breeding due to its ability to mediate high-efficiency and high-precision genome modification, which has been successfully applied to many kinds of edible fungi. In this review, we briefly summarized the working mechanism of the CRISPR/Cas9 system and highlighted the application progress of CRISPR/Cas9-mediated genome-editing technology in edible fungi, including Agaricus bisporus, Ganoderma lucidum, Flammulina filiformis, Ustilago maydis, Pleurotus eryngii, Pleurotus ostreatus, Coprinopsis cinerea, Schizophyllum commune, Cordyceps militaris, and Shiraia bambusicola. Additionally, we discussed the limitations and challenges encountered using CRISPR/Cas9 technology in edible fungi and provided potential solutions. Finally, the applications of CRISPR/Cas9 system for molecular breeding of edible fungi in the future are explored.
1. Introduction
Edible fungi have long been valued worldwide for their rich nutritional content, medicinal benefits, and pleasant taste (Pérez et al., 2021). With the rapid expansion of the edible fungi market, the need for high-quality and high-yield strains has been rapidly increasing. At present, there are various defects in the existing edible fungi production, such as strain degeneration (Zhu et al., 2020; Pérez et al., 2021; Chen et al., 2022b), unstable quality, poor resistance to diseases and insects, and easily becoming soft and brown during storage (Zhang et al., 2021). However, the traditional breeding method is inefficient, time-consuming, and labor-intensive (Boontawon et al., 2021b), and the breeding direction is uncertain, which limited molecular breeding for the improvement of cultivars. Consequently, molecular breeding techniques, such as genome editing, have emerged as a promising alternative.
Genome editing is a novel technology for manipulating genomic DNA sequences at specific sites (Osakabe et al., 2016; Tehrani et al., 2019). Zinc-finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs) were the first generation of genome-editing techniques but were complex, inefficient, and costly (Hsu et al., 2014; Juillerat et al., 2014), which limited their widespread application in edible fungi. In contrast, the CRISPR/Cas9 system is simple, flexible, stable, and cost-effective, making it a more practical option for genome editing in edible fungi (Xiao-tian et al., 2021). In this review, we provided a comprehensive summary of recent applications of CRISPR/Cas9 in the genetic improvement of edible fungi. We also highlighted potential issues and future prospects of this technology in the field of molecular breeding. Overall, our review aimed to provide a serviceable reference for researchers and practitioners interested in the molecular breeding of edible fungi.
2. Features of the application of CRISPR/Cas9 in edible fungi
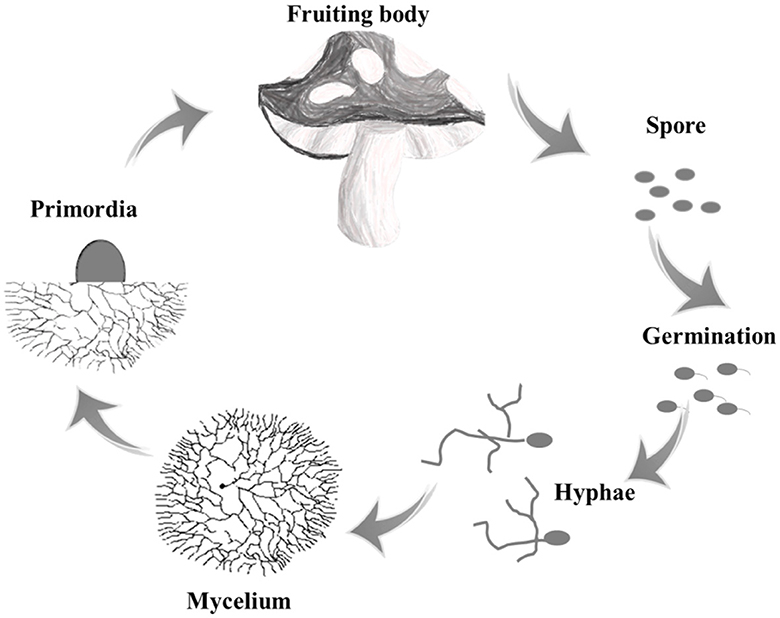
Edible fungi are a group of macro-fungus that can form large fleshy or colloidal fruit bodies or sclerotia tissues, which mainly included basidiomycetes and ascomycetes. Taking basidiomycetes as an example, the lifecycle of a mushroom begins with the spores, which are released from the basidium and dispersed by wind. When the spores land in a suitable environment, they germinate and grow into threadlike structures called hyphae. These hyphae then fuse together to form mycelium, which is the vegetative part of the fungus that grows underground or within a substrate. As the mycelium matures, it produces fruiting bodies, which are the mushrooms that we see above ground (Figure 1) (Gupta et al., 2018). The initial stage of a sub-entity that lacks organizational differentiation in appearance is usually referred to as a primordium or abbreviated as primordia.
Edible fungi are rich in nutrients, such as protein, fiber, vitamins, and minerals, and have been found to have anti-inflammatory and immune-boosting properties. In recent years, CRISPR/Cas9 genome-editing technology was applied in the molecular breeding of edible fungi to improve the yield and quality of edible fungi. The working principle of the CRISPR/Cas9 system has been explained in detail in many studies (Jinek et al., 2012; Barrangou and Marraffini, 2014; Sander and Joung, 2014; Wang and Coleman, 2019; Ullah et al., 2020), which will not be elaborated on here. The CRISPR/Cas9 system works in edible fungi similar to the manner described in the abovementioned literature but with specific application features. CRISPR-related research in edible fungi is relatively lagging behind (Xiao-tian et al., 2021) compared with that in plants and mammals, due to the fact that most edible fungi are not model fungi in the traditional sense, coupled with complex genetic background and immature genetic manipulation methods.
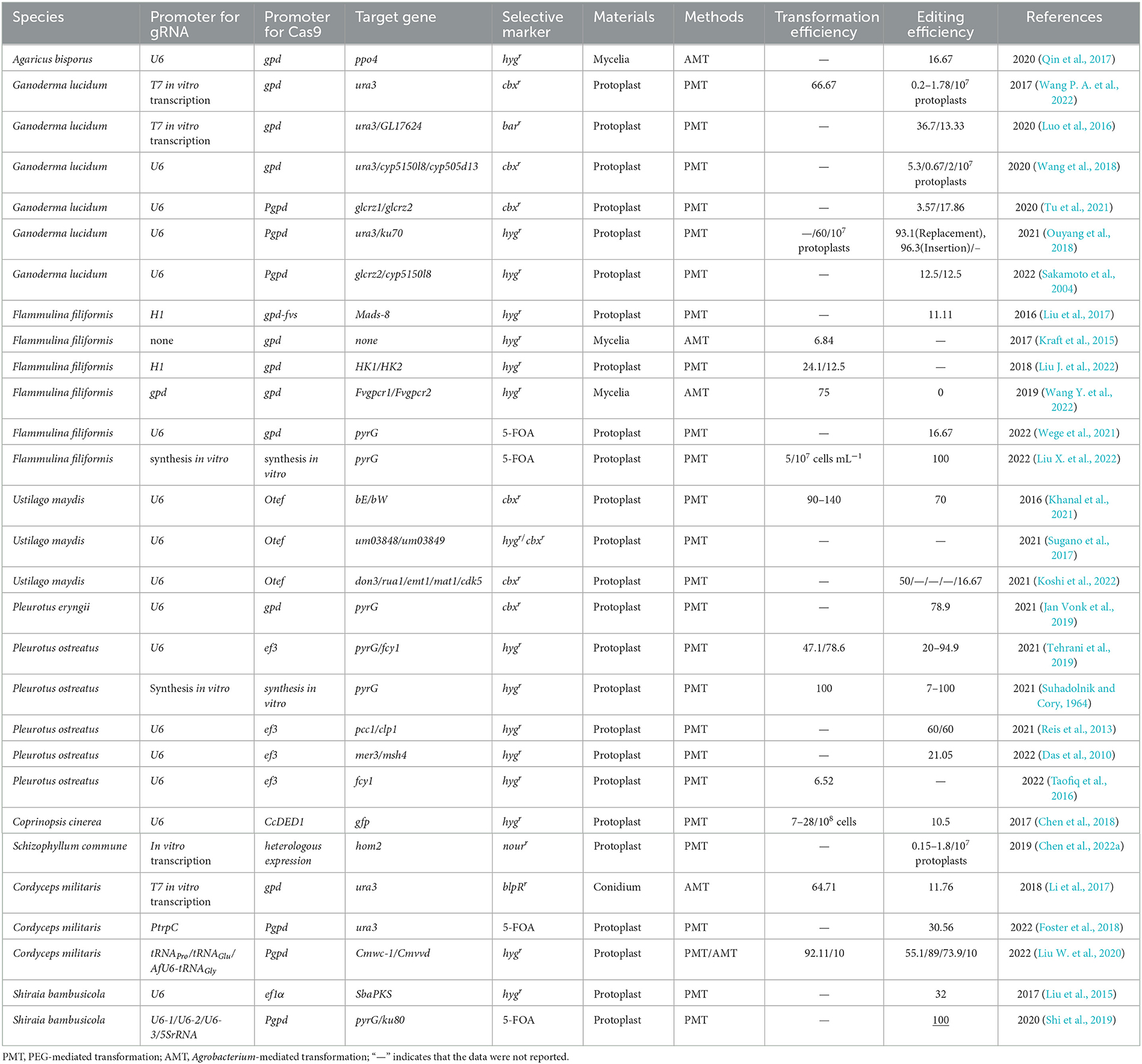
In the CRISPR system vector of edible fungi, constitutive promoters, such as trpC, gpdA, and tef1 promoters from Aspergillus nidulan (Svahn et al., 2015; Zhang et al., 2016; Song et al., 2019), have been employed to activate the expression of the Cas9 protein. The sgRNA typically undergoes transcription in vitro driven by the T7 promoter and in vivo driven by the U6 promoter (Gao and Zhao, 2014; Shi et al., 2017). The typical CRISPR/Cas9 system vector for fungal genome editing involves the connection of the Cas9 gene and sgRNA to either a single vector or a double vector. The complex is then delivered to the target cells and integrated into the target genome via either polyethylene glycol (PEG) transformation or Agrobacterium-mediated transformation (AMT) (Song et al., 2019). Furthermore, a few studies have successfully achieved transformants of edible fungi using pre-assembled Cas9-RNPs (Cas9-guide RNA ribonucleoproteins). There are a variety of options available for the transformation of host receptor materials in edible fungi due to their unique characteristics. These options include protoplasts, spores, or mycelia. However, the selection of screening markers in edible fungi is limited. Available markers include genes that mediate drug resistance, autotrophic markers, nutritional genes, and visual distinction reporters (Deng et al., 2017a), such as carboxin and hygromycin resistance genes (hph) and uracil nutrient deficiency markers. The use of CRISPR/Cas9 technology for molecular breeding of edible fungi is not only more convenient but also more efficient than traditional breeding methods (Figure 2). Despite the late adoption of the CRISPR/Cas9 technology in the field of edible fungi, a significant number of studies have been conducted in recent years. Here, we highlight the progress of the CRISPR/Cas9 system in the application of edible fungi (Table 1).
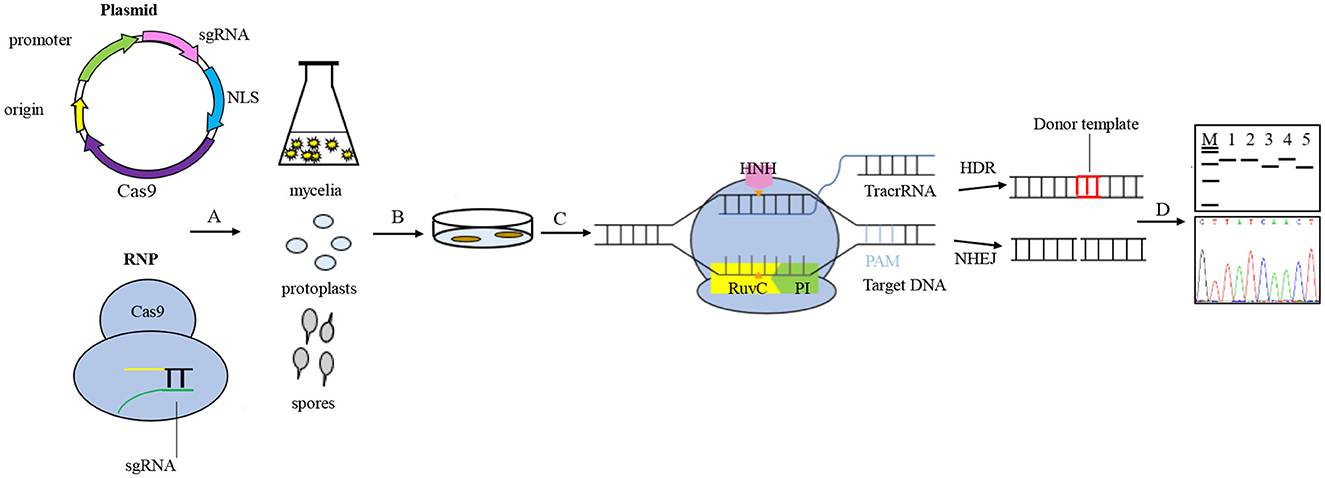
Figure 2. General procedures for genome editing in edible fungi using the CRISPR/Cas9 system. Vector construction generally connects the Cas9 gene and sgRNA to a single vector or a double vector, or using pre-assembled Cas9-RNPs. A. The vectors were transported to target cells and integrated into the target genome through either polyethylene glycol (PEG) transformation or Agrobacterium-mediated transformation (AMT). The host receptor materials for edible fungi transformation include protoplasts, spores, and mycelia. B. The transformants were screened based on the traits shown by the screening marker genes. C. The process of genome editing involves the Cas9 enzyme specifically recognizing and binding to genomic target sites, guided by sgRNA. The HNH and RuvC nuclease active domains of the Cas9 protein are responsible for cleaving 3- to 4-bp upstream of the protospacer adjacent motif (PAM) site of the target DNA, resulting in double-strand breaks (DSBs) and stimulating the genomic DNA to initiate a self-repair mechanism. The homologous recombination (HR) pathway precisely edits the target gene guided by the donor DNA fragment, while the non-homologous end joining (NHEJ)-dominated repair pathway ligates the break ends directly without using a homologous template, which can sometimes cause targeted mutations, such as random loss, insertion, or base substitutions. D. The target gene mutants are screened and verified using diagnostic PCR and Sanger sequencing.
To successfully apply CRISPR/Cas9 to edible fungi, several key requirements must be met: (1) Selecting the appropriate CRISPR/Cas9 system: The CRISPR/Cas9 system must be carefully selected based on the type of edible fungus being studied. The system must be efficient, accurate, and specific to ensure successful gene editing. (2) Design of sgRNA: The sgRNA must be designed to target specific genes of interest. They must also be specific enough to avoid off-target effects, which could lead to unintended consequences. (3) Transformation methods: The CRISPR/Cas9 components must be delivered into fungal cells using an appropriate transformation method. This can include protoplast transformation or Agrobacterium-mediated transformation. (4) Validation of gene editing: Gene editing must be validated to ensure that the desired changes have been made. This can be accomplished through PCR analysis, sequencing, or other molecular techniques. (5) Optimization of growth conditions: The growth conditions of edited fungi must be optimized to ensure proper growth and development. This may include factors, such as temperature, light, and nutrient availability.
Overall, the successful application of CRISPR/Cas9 in edible fungi requires careful selection of the CRISPR/Cas9 system, design of guide RNA, transformation method, validation of gene editing, and optimization of growth conditions. These requirements pave the way for using gene editing to enhance the yield and quality of edible fungi.
3. Application progress of CRISPR/Cas9 in edible fungi
Recent advancements in molecular biology, genomics, and bioinformatics, coupled with the relentless efforts of scientific researchers, have led to the widespread application of the CRISPR/Cas9 genome-editing system in many edible fungi, including Agaricus bisporus, Flammulina filiformis, Pleurotus eryngii, Pleurotus ostreatus, Cordyceps militaris, and so on.
3.1. Applications of CRISPR/Cas9 genome-editing technology in Agaricus bisporus
Agaricus bisporus, which belongs to the basidiomycota class, is one of the most extensively cultivated and consumed edible fungi in the world (Cai et al., 2022). Browning in A. bisporus caused by polyphenol oxidase (PPO) is a common and economically detrimental phenomenon, which inevitably influences the product quality, sensory acceptance, and nutritional value of the mushroom, resulting in certain economic losses (Sun et al., 2012; Wang et al., 2017). Various genetic manipulation experiments have been applied in the cultivation of anti-browning A. bisporus. In 2016, one of six PPO genes was knocked out by CRISPR/Cas9 in the A. bisporus, reducing the enzyme's activity by 30% and showing the ability to resist browning. It is the first successful application of the CRISPR/Cas9 system in an edible fungi (Waltz, 2016). The edited button mushroom received a green light from the United States Department of Agriculture (USDA), as it had not transferred foreign genes. The research marks a technical milestone. Similar research was conducted by Yang et al. (2020). AbPPO4 is a browning-related gene, which is highly expressed in fruiting bodies (Wu et al., 2010). To further clarify the browning mechanism of A. bisporus, the genome-editing vector pAbGEB1-AbPPO4 carrying the hph was successfully constructed and then transformed into A. bisporus mycelium cells by Agrobacterium-mediated transformation (Yang et al., 2020). The results showed that the designed CRISPR/Cas9 system successfully edited the AbPPO4 gene in A. bisporus and it was also found that the mutation of AbPPO4 gene had an adverse effect on the growth of its mycelium. In conclusion, the AbPPO4 gene is likely to not only participate in the browning-related phenotype of A. bisporus, but also play an important role in the growth and development of A. bisporus.
3.2. Applications of CRISPR/Cas9 genome-editing technology in Ganoderma lucidum
Ganoderma lucidum is an esteemed traditional Chinese medicinal material that is known for its health-boosting properties. It can synthesize a variety of biologically active substances with anti-tumor and anti-bacterial effects (Zhong and Xiao, 2009; Xiao and Zhong, 2016), such as highly oxygenated lanostane-type triterpenoids–ganoderic acids (GAs). However, the synthesis of GAs in G. lucidum is hindered by the orotidine 5′-monophosphate decarboxylase gene (ura3) (Pei et al., 2018). In 2017, Qin et al. (2017) established the CRISPR/Cas9 genome-editing system in both G. lucidum and Ganoderma lingzhi for the first time and further promoted ura3 gene disruption. First, the CRISPR/Cas9 expression vector pMD18-Glcas9 carrying the carboxin resistance gene was constructed, which was driven by the Pgpd promoter and then transformed into the protoplasts of G. lucidum using a modified PEG-mediated transformation method. The G. lucidum strain Gl-Cas9 capable of expressing Cas9 protein was successfully obtained with a conversion efficiency of 66.6%. The sgRNAs transcribed in vitro and driven by the T7 promoter were transferred into the G. lucidum strain that successfully expressed Cas9 protein. The two designed sgRNAs targeting different sites of ura3 finally yielded 3 and 5 uracil nutrition-deficient strains. The researchers hoped to improve the editing efficiency by a one-time transformation of sgRNA and Cas9 gene expression elements but unfortunately failed in the end. Despite this setback, this study accelerated strain improvement through rational genetic methods, helping to provide a broadly applicable gene-knockout methodology in higher fungi.
Previous studies demonstrated that adding calcium ions during G. lucidum mycelial fermentation could significantly increase the yield of GA (Xu and Zhong, 2012). Calcineurin-responsive zinc finger transcription factor CRZ1 is a key transcription factor for calcium signal transduction. In 2020, Li and Zhong (2020) utilized a bioinformatic analysis to identify two proteins, GlCRZ1 and GlCRZ2, in the G. lucidum proteome that displayed the highest similarity to CRZ1. The crz1 homologous genes glcrz1 and glcrz2 were individually knocked out using the CRISPR/Cas9 genome-editing system with carboxin resistance and PEG-mediated protoplast transformation methodology. The mutants and wild-type strains were cultured under different calcium ion conditions, including the absence of calcium ions, a high concentration of calcium ions (100 mM), and a low concentration of calcium ions (10 mM), to observe the growth of mycelia and the yield of GA. Compared with the wild-type strain, the addition of Ca2+ enhanced GA production, and the disruption of glcrz1 did not impede cell growth, but abolished the effect of calcium signal on stimulating GA biosynthesis. For glcrz2-knockout strains, even with Ca2+ treatment, their biomass and GA production were hampered severely. These results indicated that glcrz1 was required to transmit calcium signals to regulate GA synthesis, and glcrz2 was essential for cell growth and GA secondary metabolism. This study provides valuable insights into the role of CRZ1 in regulating secondary metabolism in basidiomycetes mushrooms. In 2020, Wang et al. (2020) constructed the CRISPR/Cas9 genome-editing system in G. lucidum to study the functional genes involved in GA biosynthesis. The researchers evaluated the efficiency of ura3 disruption using various sgRNA constructs containing an endogenous promoter and self-cleaving ribozyme HDV. They also identified a U6 promoter suitable for in vivo expression of sgRNA. Then, the ura3 disruption plasmid was constructed and transformed into the protoplasts of G. lucidum by PEG-mediated transformation. The highlight is that HDV ribozymes were introduced at the 3′ end of the sgRNA to achieve precise transcriptional termination to improve gene-editing efficiency. This newly developed system was subsequently applied to precisely edit the functional gene cyp5150l8, which is responsible for transforming lanosterol to GA. Compared with the wild-type strain, the titers of the four identified GAs in the cyp5150l8 disruptant were significantly reduced. To test the editing efficiency of this system in targeting other functional genes in G. lucidum, another cyp505d13 gene involved in the bioproduction of squalene-type triterpenoid 2,3; 22,23-squalene dioxide was also successfully disrupted with an editing efficiency of 22.2%. This study is of great significance for strain molecular breeding and metabolic engineering of G. lucidum and other basidiomycetes mushrooms.
To further clarify the biosynthetic pathway of metabolites in G. lucidum, Wang P. A. et al. (2022) applied CRISPR/Cas9-assisted in situ complementation in G. lucidum, which is the first report on the application of CRISPR/Cas9 to complement mutants in situ in basidiomycetes. The results showed that the content of four GAs was restored in the in situ complemented strains of glcrz2 and cyp5150l8. This study is of great value for the identification of various genes of other basidiomycetes. In 2020, Liu K. et al. (2020) increased the frequency of CRISPR/Cas9-mediated gene disruption in G. lucidum by adding an intron upstream of the Cas9 gene, which is indispensable for the efficient expression of heterologous phosphinothricin-resistance gene (bar) and green fluorescent protein gene (gfp) in G. lucidum. The four sgRNA cassettes driven by T7 promoter, including two ura3 and two GL26016 gene targeting sequences, were transformed into the protoplasts of G. lucidum by PEG-mediated genetic transformation, and then, the transformants were screened on specific selection plates. The dual sgRNA-mediated CRISPR/Cas9 system established in this study successfully achieved the deletion of ura3 and GL17624 genes in G. lucidum. The deletion frequency was significantly improved, making this system a valuable tool for creating gene deletion mutants and studying gene function in G. lucidum and other basidiomycetes. In 2021, Tu et al. (2021) no longer focused on the knockout of the target gene ura3 in G. lucidum, but broadened their horizons to study the insertion and replacement of ura3, which remains a serious challenge due to the low efficiency of homologous recombination in G. lucidum. To eliminate the interference of the main repair pathway of DSBs induced by the Cas9-NHEJ repair pathway, the sgRNA-directed gene deletion method was first used to inactivate the NHEJ repair mechanism by disrupting the Ku70 gene (Ku70) using the CRISPR/Cas9 system. The plasmid carrying the codon-optimized hph was constructed and introduced into G. lucidum protoplasts by PEG-mediated transformation, and ku70 mutants were successfully obtained. When the target DNA with ura3 gene and 1.5-kb homologous 5′- and 3′-flanking sequences were used as donor templates, the insertion and replacement frequencies of the target gene in ku70 mutant were 96.3 and 93.1%, respectively, while those in the control strain (Cas9) were 3.3 and 0%, respectively. The results demonstrated that the ku70 mutant was an effective receptor for gene insertion and replacement. This study provided a reference for the targeted insertion and replacement of genes in G. lucidum, which will promote our in-depth study of the gene function of G. lucidum.
3.3. Applications of CRISPR/Cas9 genome-editing technology in Flammulina filiformis
Flammulina filiformis, previously referred to as F. velutipes, is a globally important edible basidiomycetes for its nutritional value and medicinal properties worldwide (Wang et al., 2018). However, the hyphae of F. velutipes must be stimulated at low temperatures (10–15°C) to promote the formation of primordium and further develop into fruit bodies (Sakamoto et al., 2004), which leads to some problems in the large-scale production, such as high energy consumption, high cost, and heavy pollution. Therefore, it is of great scientific significance to explore the growth and development mechanism of F. velutipes fruit body by molecular biology methods.
In 2016, Luo et al. (2016) first explored the editing effect of a CRISPR/Cas9 editing system on F. velutipes genome. Based on the gene transcriptome data of F. velutipes mycelium and primordium stages, the knockout vector pgFvs-cas9-sgRNA of Mads-8 gene related to growth and development was successfully constructed, and the vector was transformed into protoplast using the PEG-mediated method. In 2018, Ouyang et al. (2018) constructed a CRISPR/Cas9 system of F. velutipes that targets genes involved in cold-induced fruiting body formation. The researchers established CRISPR/Cas9 vectors pgfvs-Cas9-HK1-sgRNA and pgfvs-Cas9-HK2-sgRNA for cold-inducible genes HK 1 and HK 2, respectively, which are related to primordium formation in F. velutipes. These vectors were transformed into F. velutipes by PEG-mediated protoplast transformation with transformation rates of 24.1 and 12.5%, respectively. The abovementioned studies showed that the recombinant vector was successfully transformed into the genome of F. velutipes, providing a theoretical foundation for further exploring the growth mechanism of F. velutipes and cultivating high-quality F. velutipes strains.
In 2019, Lin et al. (2019) reported a study on the knockout of functional genes in F. velutipes using the CRISPR/Cas9 system. Two putative G protein-coupled receptor genes Fvgpcr1 and Fvgpcr2 in F. velutipes were obtained by amino acid homology alignment (Blast P) and transcriptome data (Cabrera et al., 2015). To target these genes, four expression vectors were constructed: pCAMBIA0390-hph-Fvcas9-Fvgpcr1-sgRNA1/sgRNA2; pCAMBIA0390-hph-Fvcas9-Fvgpcr2-sgRNA1/sgRNA2. The vectors were successfully integrated into the genome of F. velutipes by Agrobacterium-mediated transformation of F. velutipes mycelium. Unfortunately, the FvCas9 protein was normally expressed, but no Fvgpcr gene-knockout mutant was obtained. In 2017, Lin et al. optimized the Streptococcus pyogenes Cas9 (SpCas9) codon to synthesize the full-length sequence of FvCas9 and constructed the binary expression vector FpiC. The mycelia of F. velutipes monokaryon “Dan3” transformed by the Agrobacterium-mediated method were used to obtain transformants. However, the transformation rate was low, only 6.84% (Liu et al., 2017); 5 years later, Liu J. et al. (2022) first established a CRISPR/Cas9 genome-editing system in the mushroom F. velutipes using a fast, accurate, and transgene-free RNP complex delivery technology (Wang P. A. et al., 2022), greatly improving the targeting efficiency to 100%. In this study, a CRISPR/Cas9 genome-editing method based on in vitro assembled Cas9 and RNPs was developed, which avoids the use of any exogenous DNA, thus saving time and manpower in plasmid construction.
The dual sgRNA-mediated fragment deletion strategy has attracted increasing attention (Zhou et al., 2014; Kraft et al., 2015), possibly due to its high efficiency and divergent visualization (Liu X. et al., 2022). In 2022, Liu X. et al. (2022) successfully established a CRISPR/Cas9 genome-editing system based on double sgRNAs in F. velutipes for the first time. Under the control of the gpd promoter and FfU6 promoter, plasmids harboring basidiomycetes codon-optimized Cas9 expression cassette and dual sgRNAs targeting pyrG (orotidine 5′ phosphate decarboxylase gene) were transferred into the protoplasts of the F. velutipes strain Dan3 by PEG-mediated transformation. Finally, several pyrG mutants with fragment deletions or small insertions and deletions (indels) were identified, resulting in genetic alterations in genomic information. This study showed a good example of the application of advanced tools in cultivated mushrooms.
3.4. Applications of CRISPR/Cas9 genome-editing technology in Ustilago maydis
Ustilago maydis is a kind of basidiomycetes; often eating it can not only protect the liver system but also help digestion and defecation, and is the pathogen of corn smut disease as well. In 2016, Schuster et al. (2016) reported the establishment of the CRISPR/Cas9 system for generating targeted alterations in the genome of the U. maydis. bE and bW form an active homeodomain complex in non-allelic combinations triggering filamentation and pathogenic development (Kämper et al., 1995). Due to the presence of an active bE-bW complex and autocrine pheromone stimulation, these strains can form filamentous colonies (Fuz+ phenotype) on solid media containing charcoal (Banuett and Herskowitz, 1989). Thus, bE and bW are used as marker genes to screen mutants in U. maydis. The plasmids pCas9_sgRNA_bW2 and pCas9_sgRNA_bE1 were generated and introduced into the U. maydis SG200 strain by PEG-mediated protoplast transformation. Colony screening and sequencing showed that the mutation efficiency was up to 70%. This technology greatly improves the genetic efficiency of U. maydis. In 2021, Wege et al. (2021) created several different types of mutants in U. maydis using the CRISPR/Cas9 technology. These include targeted gene deletion through homologous recombination of short double-stranded oligonucleotides, introduction of point mutations, heterologous complementation at genomic sites, and endogenous N-terminal labeling using the fluorescent protein mCherry. All applications do not rely on permanent selection markers, and only transient expression of endonuclease Cas9hf and sgRNA is required. The techniques demonstrated in this study have the potential to accelerate the study of maize smut communities but could also serve as a template for genome editing in other important fungi.
U. maydis can use nitrate as the sole nitrogen source. Khanal et al. (2021) characterized the role of nitrate assimilation genes in the virulence of U. maydis, including two genes, um03848 and um03849, which serve as nitrite reductase and nitrate transporter, respectively. The deletion mutants for um03848, um03849, or both genes were obtained by homologous recombination. Additionally, the pCas9-um03849 plasmid carrying carboxin resistance gene was constructed and used to transform protoplasts of compatible U. maydis mating strains to produce INDEL mutation. All mutants obtained by CRISPR-mediated and homologous recombination were examined, and it was inferred that genes um03849 and um03848 were used for nitrate utilization alone or together. This is the first study to verify the role of nitrate assimilation genes in maize and opens up a possible avenue for further study of nitrogen assimilation pathways using U. maydis as a model organism.
3.5. Applications of CRISPR/Cas9 genome-editing technology in Pleurotus eryngii
Pleurotus eryngii, classified as basidiomycetes, has been widely cultivated in many countries, especially in China (Wang et al., 2021). P. eryngii contains a plethora of unique and versatile bioactive compounds. Nevertheless, studies on functional genes involved in the synthesis of these compounds have been severely impeded by the absence of a mature and effective gene-editing system for P. eryngii. In 2021, Wang et al. (2021) innovatively established the CRISPR/Cas9 system in P. eryngii and improved the genetic transformation system. At first, they determined that the endogenous sdhB gene point-mutated variant (cbxr) was a more stable selective marker than hph. In addition, the codon of the Cas9 protein was further optimized, and a native efficient U6 promoter was selected to guide sgRNA transcription, thereby improving the genome-editing efficiency. A more efficient and stable pyrG gene-editing system was established in P. eryngii by optimizing the PEG-mediated protoplast transformation system. Different insertions and deletions were incorporated through NHEJ and HDR to realize the genome-editing function of the pyrG system. Although this system has not yet been used for gene editing of functional genes in P. eryngii, it is just around the corner based on the establishment of the CRISPR/Cas9 system of this study. This study paves the way for gene function research and molecular breeding of P. eryngii.
3.6. Applications of CRISPR/Cas9 genome-editing technology in Pleurotus ostreatus
Pleurotus ostreatus is one of the most economically important mushroom species and possesses both edible and medicinal value (Boontawon et al., 2021c). Extensive breeding of P. ostreatus has improved its cultivation and food qualities (Sonnenberg et al., 2017; Barh et al., 2019). However, these classical breeding methods are laborious and time-consuming. Thus, methodologies for more time- and cost-effective molecular breeding are needed (Yamasaki et al., 2022). The implementation of the CRISPR/Cas9 system in P. ostreatus will undoubtedly facilitate its molecular breeding. An efficient genome-editing system for non-genetically modified (non-GM) in P. ostreatus using plasmid-based CRISPR/Cas9 was constructed (Boontawon et al., 2021b). Plasmids harboring Cas9 expression cassettes and different sgRNAs targeting fcy1 and pyrG were individually transferred into protoplasts of the PC9 strain. Mutants with resistance to 5-fluorocytosine and 5-fluoroorotic acid (5-FOA) were obtained. Small insertions/deletions or insertion of plasmid fragments at the target site were found in some resistant strains, which proved that CRISPR/Cas9-assisted genome editing of P. ostreatus is available. It is the first step toward using CRISPR/Cas9 for modern molecular breeding of non-GM cultivated strains in the future. Soon after, Boontawon et al. (2021c) realized plasmid-free genome editing in P. ostreatus using Cas9 RNP, and strains resistant to 5-FOA were generated using this method. The results demonstrated Cas9 RNP-assisted gene mutation could be used for the molecular breeding of P. ostreatus as well as other edible fungi.
The formation of dikaryons in Agaricomycetes is mainly controlled by A and B mating-type loci. Based on the results of previous studies, Boontawon et al. (2021a) speculated the molecular mechanism of dikaryon formation was related to pcc1 and clp1. Such plasmids harboring the Cas9 expression cassette and different sgRNAs targeting pcc1 or clp1 were introduced into strain PC9 to obtain mutants. The dikaryosis of the clp1 mutant was damaged, suggesting that the function of clp1 may be necessary for clamp cell formation. This study not only provides a successful example of the application of CRISPR/Cas9 in functional studies of cultivated mushrooms but also provides a reference for further revealing the molecular mechanism of sexual development in Agaricomycetes. In addition to the monokaryotic P. ostreatus strain PC9, CRISPR/Cas9 has also been successfully performed on the dikaryotic P. ostreatus. In 2022, Yamasaki et al. (2022) reported a plasmid-based CRISPR/Cas9 technique for transforming both nuclei of dikaryotic P. ostreatus, PC9 × #64, to generate strains with low basidiospore production ability. Meiosis-related genes mer3 or msh4 were selected as target genes to design two different sgRNAs. These four different plasmids were introduced into PC9 × # 64 to obtain mutants with low basidiospore production ability, so as to develop an efficient and rapid molecular breeding method for cultivated mushrooms. Koshi et al. (2022) achieved marker-free genome editing for the first time by transiently expressing Cas9, sgRNA, and hph in P. ostreatus. The gene of fcy1 was edited by transiently expressing Cas9, sgRNA, and hph, and strains with 5-fluorocytosine resistance and hyg sensitivity were isolated. The genome-editing effect of fcy1 in the mutant strains was confirmed through Sanger sequencing. This significant achievement marks a new chapter in fungal repetitive genome editing.
3.7. Applications of CRISPR/Cas9 genome-editing technology in Coprinopsis cinerea
Coprinopsis cinerea is an excellent model organism for studying the growth, development, and reproductive mechanism of other edible and medicinal fungi that are challenging to cultivate in laboratory conditions. With the completion of its whole-genome sequencing, it has been revealed to possess numerous crucial genes for synthesizing terpenoids and enzymes (Pei et al., 2018). Therefore, it is crucial to develop gene-editing technologies that are suitable for C. cinerea. In 2017, Sugano et al. (2017) constructed the CRISPR/Cas9 genome-editing system in C. cinerea, and it was optimized by a high-throughput transformation system. First, they developed a new method for protoplast preservation to reduce the preparation time of protoplasts. In addition, a novel stronger promoter-CcDED1pro was screened to drive the Cas9 expression, and sgRNA was designed to target the gfp gene, which was constructed in the CRISPR/Cas9 vector and transformed into the protoplasts of a stable gfp expression line using the PEG-mediated method. Finally, the loss of gfp function was successfully detected. The cryopreserved protoplast is a powerful tool for high-throughput CRISPR/Cas9, thereby accelerating gene function studies and molecular breeding in edible fungi.
3.8. Applications of CRISPR/Cas9 genome-editing technology in Schizophyllum commune
Schizophyllum commune is a basidiomycetes fungus that provides a protein source of both high quality and quantity. However, the main challenge in gene deletion in S. commune is the relatively low incidence of homologous recombination, coupled with a relatively high incidence of NHEJ. In 2019, Jan Vonk et al. (2019) used pre-assembled Cas9 RNPs for high-throughput targeted gene deletion in S. commune. First, pre-assembled Cas9-sgRNA RNPs were used to delete the homeodomain transcription factor hom2 in S. commune. However, it was found that Cas9 RNPs alone were not efficient enough for gene disruption in the absence of positive selection markers. Consequently, a repair template containing a selection marker was utilized in combination with Cas9-sgRNA RNPs to increase gene deletion rates. Sequencing analysis confirmed that the hom2 gene was successfully replaced by the nourseothricin resistance cassette in all cases. Furthermore, an Δku80 background was used to increase the rate of homologous recombination and reduce the number of ectopically integrated transformants, resulting in a higher gene deletion rate than in the wild-type strain. Finally, homology arms of 250 bp were identified to be sufficient for consistent recruitment of homology-directed repair. This report provides a new possibility for optimizing the CRISPR/Cas9 system to improve the genetic accessibility of non-model species.
3.9. Applications of CRISPR/Cas9 genome-editing technology in Cordyceps militaris
Cordyceps militaris is an Ascomycete species known for its bioactive constituents, such as cordycepin (Suhadolnik and Cory, 1964), cordyceps polysaccharide (Yu et al., 2004), and ergosterol (Bok et al., 1999). Studies have reported a diverse range of biological properties associated with these constituents, including anti-oxidant, anti-tumor, anti-aging, and anti-inflammatory (Das et al., 2010; Reis et al., 2013; Taofiq et al., 2016). It is meaningful to explore the synthetic mechanism of these substances and establish an efficient genome-editing method. However, there are still few reports on CRISPR/Cas9 genome editing in C. militaris, due to its complex genetic background and high proportion of fruiting body gemmated degeneration.
The first application of the CRISPR system in C. militaris was reported by Chen et al. (2018) in 2018, who successfully established an efficient CRISPR/Cas9 gene destruction system in the species. With the help of the newly discovered promoter Pcmlsm3 and terminator Tcmura3, a vector containing three functional domains of the codon-optimized Cas9, sgRNA, and phosphinothricin acetyltransferase gene blpR was successfully constructed and transformed into wild-type C. militaris by PEG-mediated protoplast transformation. The stable expression of Cas9 was proved by fluorescent gfp tag and Western blot analyses. Regrettably, no ura3 mutant was obtained, as the transcription level of sgRNA was extremely low. To circumvent the above problem, the strategy of in vitro introduction of sgRNA was adopted. The sgRNA and donor single-stranded DNA (ssDNA) synthesized in vitro were transformed into the C. militaris strains expressing cmcas9 and screened by 5-FOA. The CRISPR/Cas9 system effectively generated mutants with site-specific deletions and insertions. As mentioned earlier, the application of the CRISPR system in the C. militaris is reported for the first time, which can greatly accelerate molecular breeding in C. militaris.
To develop powerful genome engineering tools, Chen et al. (2022a) successfully achieved multiplex gene precise editing and large DNA fragment deletion by the CRISPR/Cas9-TRAMA system in C. militaris. Leveraging the multiplex RNA-processing function of endogenous tRNA elements, the vector C9TRAMA was constructed by introducing the extrachromosomal plasmid and combining it with the homologous template of target sites. The marker-free CRISPR/Cas9-TRAMA multiplex gene-editing system, which can efficiently edit multiple targets at any position of the C. militaris genome to delete large synthetic clusters, was developed by transforming the vector into C. militaris via PEG-mediated protoplast transformation. Moreover, the greatest advantage of the Cas9-TRAMA system is that it could easily be removed without any accidental foreign DNA residue. The researchers constructed and transformed the vectors C9TRAMA-Cns1B, -Cns123, and-Egt1 to further edit the synthetases of cordycepin and ergothioneine, which proved that this system can be used for protein modification, promoter strength evaluation, and 10-kb metabolic synthetic cluster deletion. The Cas9-TRAMA system developed in this study provides a feasible approach for extracting valuable metabolic resources from medicinal mushrooms.
Because the AMA1 plasmid is difficult to insert into the fungal genome, it is easy to lose after being cultured under non-selective conditions, allowing for the reuse of dominant selection markers in the next transformation (Wang and Coleman, 2019). Therefore, in theory, the AMA1-based CRISPR/Cas9 genome-editing technology can achieve unlimited rounds of genetic engineering. In 2022, Meng et al. (2022) constructed an efficient CRISPR/Cas9 genome-editing system based on AMA1-based plasmids for the first time in C. militaris. The photoreceptor genes Cmwc-1 and Cmvvd were selected as the target genes, as the appearance of albinism or deepened color of mycelium could confirm their disruption. The plasmids pAMA1-Cas9-sgRNACmwc − 1 and pAMA1-Cas9-sgRNACmvvd were constructed and transformed into protoplasts by PEG-mediated transformation. The transformants were identified on a selective medium after light exposure, with efficiencies of 55.1 and 88.89%. To achieve a more precise target modification, mutants based on the vector harboring a sgRNA supplementation with an HDR template were obtained at an efficiency of 73.9%. Double-gene disruption of Cmwc-1 and Cmvvd was also achieved with an efficiency of 10%. The CRISPR/Cas9 system with endogenous tRNAPro of C. militaris and the heterologous chimeric RNA Pol III promoter AfU6-t RNA Gly exhibited an editing efficiency exceeding 80%, making them more suitable promoters. Finally, the transformation efficiency of PEG-mediated protoplast transformation and Agrobacterium-mediated transformation in C. militaris was compared. The results showed that the former had a notably higher efficiency than the latter. These findings hold tremendous potential for the utilization of CRISPR/Cas9 technology in characterizing gene function and molecular breeding of edible fungi in the future.
3.10. Applications of CRISPR/Cas9 genome-editing technology in Shiraia bambusicola
Shiraia bambusicola, a member of the Ascomycete family, thrives on the twigs of bamboo. Its fruiting bodies showcase a superior ability to produce high-value pharmacological drugs such as hypocrellin. This compound has been widely applied in many pharmaceutical fields, including the treatment of sciatica, pertussis, tracheitis, and rheumatic arthritis (Zhao et al., 2016). Unfortunately, inadequate molecular tools have impeded research and utilization of this promising compound. In 2017, Deng et al. (2017b) constructed the CRISPR/Cas9 system in Shiraia sp. SUPER-H168 and successfully deleted the key gene in the hypocrellin biosynthesis pathway-polyketide synthase (SbaPKS). First, the PlentiU plasmid carrying hph was used as the starting vector. The human U6 promoter was used to activate the transcription of sgRNA, and the constitutive promoter ef1α was used to activate the expression of the Cas9 protein. The plasmids, harboring solely sgRNA and Cas9-hph, were transformed into the wild-type protoplast by PEG–CaCl2 transformation. There was no significant change in the morphology of the transformants compared with that of the wild-type strain, indicating that the Cas9 protein or sgRNA had no deleterious effect on the growth of the wild-type strain. Then, the PlentiU vector loaded with both sgRNA and Cas9-hph was transformed into Shiraia sp. SUPER-H168 to obtain the SbaPKS-edited mutant. The relative expression levels of SbaPKS and its adjacent genes in the ΔSbaPKS mutant were extremely downregulated compared with those of the wild-type strain, underscoring the involvement of SbaPKS in hypocrellin biosynthesis. The successful implementation of the CRISPR/Cas9 system in S. bambusicola will open a new window for deciphering the mechanism of hypocrellin biosynthesis and metabolites of medicinal fungi.
In 2020, Deng et al. (2020) constructed a more efficient CRISPR/Cas9 system in S. bambusicola for enhancing hypocrellin production by optimizing sgRNA transcription elements and interfering with the endogenous NHEJ pathway. 5SrRNA was selected for transcribing sgRNA, owing to the fact that the genome-editing efficiency of 5SrRNA CRISPR plasmid was higher than that of other U6 mediating systems. In addition, the ku80 gene was destroyed by the CRISPR system, and the homologous donor with pyrG cassette was integrated into the targeting genomic locus of the ku80 gene via the DSBs repair system. The transformants were then screened on a regeneration medium containing 5-FOA. The results indicated that Δku80 mutant was no damage to S. bambusicola and the homologous recombination efficiency was significantly increased to 100%. Subsequently, the optimized system was applied to improve hypocrellin production via gene modification and integration, increasing ~12-fold than that of the wild-type strain. This study lays the foundation for the regulation of metabolic pathways using the efficient CRISPR/Cas9 system in edible fungi.
4. Challenges and optimization of the CRISPR/Cas9 system in edible fungi
4.1. Low editing efficiency
Editing efficiency refers to the proportion of strains that achieve successful editing of target genes (Ruiping et al., 2019). Low editing efficiency is a common problem in the CRISPR/Cas9 system for edible fungi, which can be attributed to various reasons, including low transformation efficiency and regeneration efficiency of host material (Li et al., 2017), low expression level of Cas9 or sgRNA (Foster et al., 2018; Wang and Coleman, 2019), low targeting efficiency of sgRNA (Schuster et al., 2016; Xiao-tian et al., 2021) and low precision repair ratio (Yuan et al., 2017).
The optimization scheme can be considered from the following aspects: (1) Optimizing the genetic transformation system. It is necessary to establish stable and efficient genetic transformation systems (Li et al., 2017) given the complex cell wall structures of edible fungi. Combining multiple wall lytic enzymes under different digestion conditions may result in higher quality protoplasts (Liu X. et al., 2022). (2) Optimizing Cas9 expression and the sgRNA transcription strategy (Xuping et al., 2019). The insufficient expression of Cas9 is usually caused by the presence of rare codons. Therefore, codon optimization for target species is necessary (Liu et al., 2015; Ullah et al., 2020). Numerous facts prove that codon optimization makes the Cas9 gene more efficiently expressed in specific species (Schuster et al., 2016; Qin et al., 2017; Chen et al., 2018; Wang et al., 2021; Liu X. et al., 2022). In addition, incorporating a suitable nuclear localization sequence to Cas9 protein is conducive to the specific expression of Cas9 protein in the nucleus of edible fungi, thereby improving the editing efficiency to a certain extent (Shi et al., 2019; Ullah et al., 2020). (3) The targeting efficiency of sgRNA affects the editing efficiency of CRISPR. Currently, there are many online tools to assist in the design of sgRNA for reference to select the optimal sgRNA. The related websites are shown in Table 2. (4) Selecting the appropriate promoter. Generally, endogenous gpdA, TrpC, and other promoters are selected to express Cas9 gene, while the U6 promoter is used to express sgRNA (Ruiping et al., 2019). (5) Improving repair accuracy. Some studies have shown that the efficiency of precise repair can be improved by inhibiting NHEJ repair or enhancing HDR repair (Yan and Finnigan, 2018; Tu et al., 2021).
4.2. Off-target effects
Off-target effects may exist in all species, and edible fungi are no exception. During the process of genome editing, base mismatches can occur between the sgRNA and the target DNA sequence (Fu et al., 2013) or sgRNA sequences, forming a DNA bulge that differs from the off-target DNA sequence (Lin et al., 2014) which can lead the sgRNA to pair with other bases. This can result in the cutting of non-target sequences and unnecessary mutations, which represents a major challenge of the CRISPR/Cas9 system. The off-target effect can be reduced via the following aspects: First, detecting the off-target effect in time. At present, many methods to detect the off-target effect have been developed. Chromatin immunoprecipitation sequencing (ChIP-seq) and genomewide, unbiased identification of DSBs enabled by sequencing (GUIDE-seq) are two such methods that could accurately identify off-target sites for CRISPR/Cas9 (Kuscu et al., 2014; Tsai et al., 2015; Lin et al., 2018; Rodriguez et al., 2021). Second, optimizing the sgRNA sequence. This can be achieved through the following steps: (1) Minimizing the number of base pairs between sgRNA and predicted off-target site sequences; sgRNA and predicted off-target sites should avoid four consecutive or interval base pairing; 1–5 bases near the PAM sequence have the greatest influence on the correct binding of sgRNA to the target DNA, so these five bases should be as specific as possible when designing sgRNA (Pei et al., 2018). (2) Modifying the length of sgRNA. Shortening the length of sgRNA from 20 nucleotides to 17–18 nucleotides (Fu et al., 2014) or adding two guanine nucleotides at the 5′ end of sgRNA (Cho et al., 2014) can significantly reduce undesired mutagenesis at some off-target sites. (3) Controlling sgRNA concentration. The off-target effect can be minimized by the reasonable concentration of the plasmid harboring sgRNA (Hsu et al., 2013; Pattanayak et al., 2013). Furthermore, using the RNA polymerase II transcription system to express sgRNA can better regulate the content of sgRNA, thereby minimizing off-target effects (Kiani et al., 2014).
Third, modifying the Cas9 protein. (1) Reducing or depriving the endonuclease activity of Cas9 protein. The Cas9 protein can be activated by non-targeting sequences within a certain mismatch range, resulting in mis-cleavage. To prevent this, mutants, such as H841A or D10A, which cleave the function of HNH or RuvC domains, can be used (Chiang et al., 2016). Alternatively, single-notch enzyme activity mutants, such as dCas9 and FokI-dCas9, can also be employed (Gao et al., 2016; Pan et al., 2016; Terao et al., 2016). These two methods can increase the degree of sequence matching required to activate Cas9 to a certain extent which significantly enhances the specificity of the system. (2) Reducing the ability of Cas9 protein to bind to DNA. Site-directed mutagenesis of Cas9 protein can be generated to form enhanced specificity Cas9 (eSpCas9) (Slaymaker et al., 2016) and high-fidelity-HF1 (SpCas9-HF1) (Kleinstiver et al., 2016). The mutants weakened the non-specific binding ability of Cas9 protein to DNA, thereby reducing off-target effects. (3) Enhancing the recognition ability of Cas9 protein to standard PAM sequence. This can be achieved by using Mutazyme II to randomly mutagenize the residues of SpCas9 and construct mutagenized PAM-interacting domain libraries. Positive selection and site-depletion assay of SpCas9 variants can then be performed to obtain variants with improved Cas9 PAM specificities (Kleinstiver et al., 2015). For example, the D1135E SpCas9 variant possesses improved discrimination against the off-targets with non-canonical NAG/NGA PAMs while retaining robust on-target activity on canonical NGG PAM sites (Kang et al., 2022). Fourth, controlling the action time of functional elements, such as Cas9 and sgRNA, in fungal cells. (1) Cas9 mRNA and sgRNA can be prepared by in vitro transcription and transferred into the cells (Nagy et al., 2017). Due to their susceptibility to degradation, the Cas9 and sgRNA will be eliminated rapidly, resulting in a shorter retention time of the CRISPR components in the cells. This approach can help reduce off-target effects. (2) The inducible promoter is used to express Cas9, so that Cas9 can only be expressed when the gene needs to be edited, such as alcohol dehydrogenase A (alcA) from A.nidulans (Waring et al., 1989) and benzoate p-hydrolase A (bphA) or catalase R (catR) from Aspergillus niger (Sharma et al., 2012; Antunes et al., 2016). (3) Using self-replicating plasmids. Cas9 in the plasmid will be lost from the cell during passage without screening pressure. (4) Deleting the functional elements, such as Cas9 and sgRNA, after editing by the CRISPR/Cas9 system using the FLP/FRT site-specific recombination system (Xuping et al., 2019), thus reducing the action time of CRISPR components in cells to reduce the off-target effect.
4.3. Difficulty in screening mutants
The difficulty in screening mutants in edible fungi arises from non-homozygous editing and fewer available selection markers (Deng et al., 2017a; Xiao-tian et al., 2021). Due to the heterokaryosis of edible fungi, many unedited nuclei in protoplasts are mixed in the screening of mutants, increasing the difficulty of screening. Therefore, many studies have utilized monokaryons of edible fungi for genome editing. It has been reported that the addition of chemical reagents, such as inositol and benomyl, increased the proportion of monokaryotic protoplasts in Aspergillus oryzae. This can greatly improve the obtaining efficiency of homozygous transformants and can avoid the multi-round single-spore isolation process in the late stage of transformation, thereby reducing the difficulty of screening (Zou et al., 2021). In addition, spore isolation to obtain mononuclear, haploid mutants, as the starting strain for gene editing, may be a solution (Han and Yixin, 2021). However, differences in morphological, physiological, and biochemical characteristics between monokaryons and dikaryons may need to be examined and circumvented.
Another limiting factor for mutant screening in edible fungi stems from the limited number of selective markers. Several commonly used markers have been used for screening positive transformants, including resistance genes (carboxin resistance gene and hph), auxotrophic markers (uridine auxotrophy), and mutagenic markers (pyrG and ura3). However, there are some limitations and potential problems with the above markers. For example, hph could not be stably expressed in P. eryngii. The transformants lost the hph in the third passage (Wang et al., 2021). The uracil auxotroph mutants can grow normally in a uridine-deleted medium only when the fragment containing the ura3 expression element is correctly inserted (Chen et al., 2017). Recently, some scientists have proposed the design of recyclable cassettes that can be removed from the targeted genome after use for the reuse of selectable markers (Fuller et al., 2015). For example, Solis-Escalante et al. (2013) reported a novel recyclable dominant marker cassette amdSYM consisting of an Ashbya gossypii TEF2 promoter and terminator and a codon-optimized acetamidase gene (amdS) from A. nidulans as the first dominant recyclable marker, which opens the door to rapid and simple genetic manipulation in industrial strains.
5. Future prospects of CRISPR/Cas9 system in edible fungi
5.1. Extending the PAM range
The PAM is located at the 3′-terminus of the target DNA, which is a short nucleotide motif of ~2–6 bp and is responsible for recognition by the Cas9 protein (Sternberg et al., 2014). Although the CRISPR/Cas9 genome-editing system has been widely used in edible fungi genome editing, its target selection is often restricted to DNA sequences with the NGG PAM motif, which severely hampers the scope of CRISPR/Cas9 genome editing in edible fungi. Researchers have been working to develop novel Cas9 variants to expand the scope of genome-targeted editing. SpCas9 variants, namely xCas9 and Cas9-NG, have been found to show great potential in improving targeting specificity and expanding targeting range. xCas9, for instance, can efficiently induce target site mutations of both NG and GAT PAM sequences (Hua et al., 2019). Cas9-NG can edit all NGG, NGA, NGT, and NGC sites, although with slightly reduced activity (Ren et al., 2019). Moreover, Cas9-NG can not only recognize NAC, NTG, NTT, and NCG apart from NG PAM (Ren et al., 2019) but can also achieve efficient editing at AT-rich PAM sites, such as GAT, GAA, and CAA (Zhong et al., 2019). Furthermore, researchers are actively exploring the discovery of new orthologous enzymes of Cas9 through sequence alignment, which is expected to significantly expand the application range of the CRISPR/Cas9 system. This development holds great promise for the genetic manipulation of edible fungi in the future.
5.2. Realization of multi-gene editing
Being a multicellular eukaryotic microorganism, edible fungi possess a more complex genetic background and tend to exhibit slower progress in genetic research as compared to bacteria and other prokaryotes. Consequently, most of the current reports on CRISPR/Cas9 in edible fungi mainly focus on the level of system construction and single gene editing (Song et al., 2019; Xuping et al., 2019). Traditional genetic approaches for continuous manipulating of multiple genes in edible fungi are neither efficient nor functional because of low homologous recombination rate and limited selection markers (Wang and Coleman, 2019). The CRISPR/Cas9 system could realize multiple gene editing by introducing multiple sgRNAs for different sites, or a single sgRNA for a conserved sequence of multiple sites at one time (Mali et al., 2013), which greatly facilitates study of the function of multiple genes (Xuping et al., 2019). Up to now, the following methods can be referred to achieve multiple gene editing: Multiple mature sgRNA synthesized in vitro could be introduced into Cas9-positive cells using the protoplast-mediated method. Co-transformation of sgRNA and donor DNA for different targets could realize multiple gene editing (Liu et al., 2015). In addition, the multiple RNA-processing functions of tRNA elements are expected to achieve multiple gene editing (Chen et al., 2022a). These methods are expected to provide a reference for multi-gene editing of edible fungi.
5.3. Application of base editors
In recent years, base editing technology based on CRISPR/Cas9 has gradually developed into a powerful genome-editing tool because of its advantages of no DSBs, no need for exogenous DNA templates, and no dependence on host homologous recombination repair. It has been developed and applied in animals (Komor et al., 2016; Kim et al., 2017; Zhang et al., 2017; Liu et al., 2018), plants (Shimatani et al., 2017; Zong et al., 2017; Tian et al., 2018), yeasts (Satomura et al., 2017; Pan et al., 2021), and bacteria (Zheng et al., 2018; Xia et al., 2020; Heo et al., 2022), which also provides a reference for the establishment and application of base editing systems in edible fungi. Base editors include cytosine base editor (CBE) and adenine base editor (ABE). CBE fuses catalytically impaired CRISPR/Cas9 mutant, cytidine deaminase enzyme, and uracil glycosylase inhibitor (UGI) to produce a base substitution from C-G to T-A near the sgRNA binding site (Komor et al., 2016). Similar to CBE, the difference is that ABE uses adenosine deaminase, which results in a base conversion from A-T to G-C (Gaudelli et al., 2017). As base conversion can be achieved without the introduction of DSBs, the base editor should be widely used for early termination of codon introduction and repair (Yannan and Yuhui, 2022). For instance, CBE can convert codons, such as CAA, CAG, and CGA to termination codons TAA, TAG, and TGA, respectively, thereby terminating gene expression prematurely. Similarly, ABE can convert the initiation codon ATG to ACG, which disrupts the initiation of gene translation. These base editors can be utilized for gene inactivation, investigating gene function, and directed modification of biological metabolic (Yannan and Yuhui, 2022). The establishment of base editors in edible fungi will greatly facilitate the study of gene function and the creation of high-quality germplasm resources.
6. Conclusion
The CRISPR/Cas9 genome-editing technology has already gained immense potential for a vast range of applications following its rapid development in recent years. Compared with ZFNs and TALENs systems, the CRISPR/Cas9 genome-editing technology system has shown great superiority with the advantages of simplicity, efficiency, and versatility. Meanwhile, it also shows irreplaceable application value in strain improvement, molecular breeding, and metabolite regulation of edible fungi. However, at present, there are still deficiencies in the application of this technology in edible fungi, such as off-target effect, low editing efficiency, and difficulty in mutant screening, which need more radical solutions. The advancement of the modern edible fungi industry is of great significance to ensure food safety, promote the development of ecological agriculture, and achieve the goal of “dual-carbon”. With the advent of the post-genome era and the in-depth utilization of the CRISPR/Cas9 technology, genome editing of edible fungi will step onto the right track, which will bring breakthroughs in gene mining of target traits, targeted improvement of varieties, and creation of new germplasm (Figure 3).
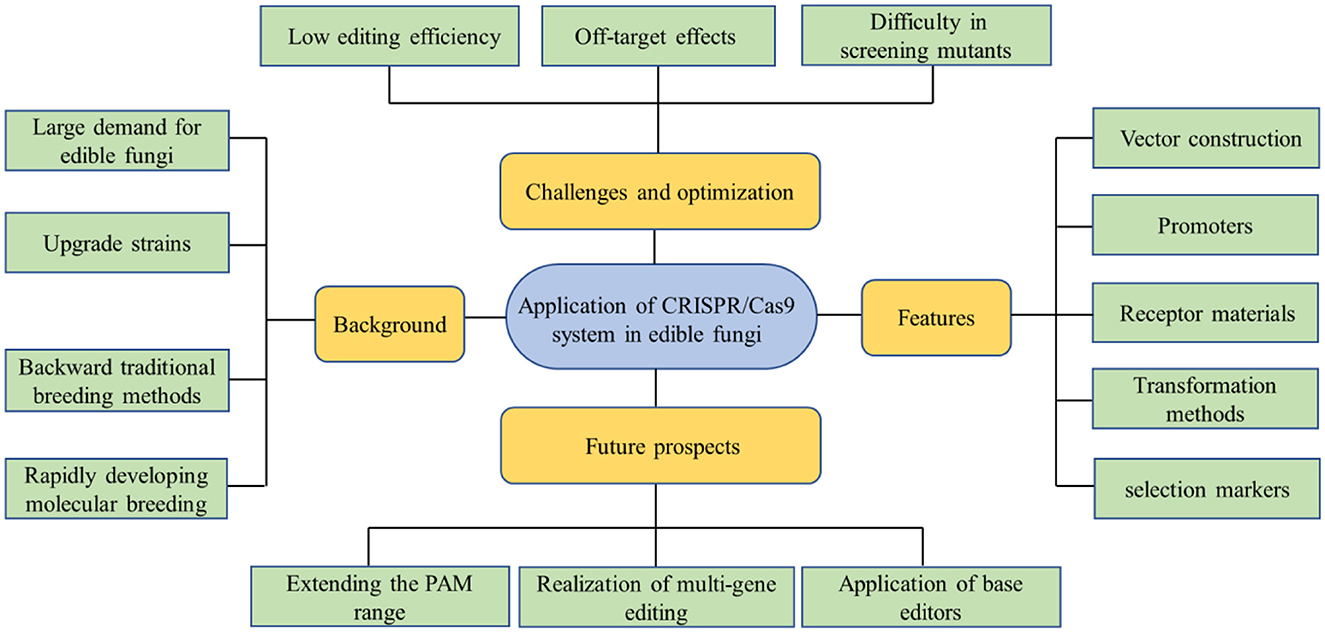
Figure 3. This concept map is used to summarize the entire article. Due to the intricate genetic background of edible fungi and the challenging nature of genetic manipulation, advancements in genetic research have been comparatively sluggish. However, with the swift progress of molecular biology, the CRISPR/Cas9 system is expected to have a significant impact on gene function research and molecular breeding of edible fungi.
Author contributions
QZ and LY designed the study and incorporated all the necessary modifications. YZ and SC drafted the manuscript and arranged the references. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the China Tobacco Shandong Industrial Corporation major project (202102004).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Antunes, M. S., Hodges, T. K., and Carpita, N. C. (2016). A benzoate-activated promoter from Aspergillus niger and regulation of its activity. Appl. Microbiol. Biotechnol. 100, 5479–5489. doi: 10.1007/s00253-016-7373-3
Banuett, F., and Herskowitz, I. (1989). Different a alleles of Ustilago maydis are necessary for maintenance of filamentous growth but not for meiosis. Proc. Nat. Acad. Sci. 86, 5878–5882. doi: 10.1073/pnas.86.15.5878
Barh, A., Sharma, V. P., Annepu, S. K., Kamal, S., Sharma, S., and Bhatt, P. (2019). Genetic improvement in Pleurotus (oyster mushroom): a review. 3 Biotech 9, 322. doi: 10.1007/s13205-019-1854-x
Barrangou, R., and Marraffini, L. A. (2014). CRISPR-Cas systems: prokaryotes upgrade to adaptive immunity. Mol. Cell 54, 234–244. doi: 10.1016/j.molcel.2014.03.011
Bok, J. W., Lermer, L., Chilton, J., Klingeman, H. G., and Towers, G. H. N. (1999). Antitumor sterols from the mycelia of Cordyceps sinensis. Phytochemistry 51, 891–898. doi: 10.1016/S0031-9422(99)00128-4
Boontawon, T., Nakazawa, T., Horii, M., Tsuzuki, M., Kawauchi, M., Sakamoto, M., et al. (2021a). Functional analyses of Pleurotus ostreatus pcc1 and clp1 using CRISPR/Cas9. Fungal Genet. Biol. 154, 103599. doi: 10.1016/j.fgb.2021.103599
Boontawon, T., Nakazawa, T., Inoue, C., Osakabe, K., Kawauchi, M., Sakamoto, M., et al. (2021b). Efficient genome editing with CRISPR/Cas9 in Pleurotus ostreatus. AMB Express 11, 30. doi: 10.1186/s13568-021-01193-w
Boontawon, T., Nakazawa, T., Xu, H., Kawauchi, M., Sakamoto, M., and Honda, Y. (2021c). Gene targeting using pre-assembled Cas9 ribonucleoprotein and split-marker recombination in Pleurotus ostreatus. FEMS Microbiol. Lett. 368, fnab080. doi: 10.1093/femsle/fnab080
Cabrera, I. E., Pacentine, I. V., Lim, A., Guerrero, N., Krystofova, S., Li, L., et al. (2015). Global analysis of predicted G protein-coupled receptor genes in the filamentous fungus, Neurospora crassa. G3 Genes Genomes Genetics 5, 2729–2743. doi: 10.1534/g3.115.020974
Cai, M., Zhong, H., Ma, Q., Yang, K., and Sun, P. (2022). Physicochemical and microbial quality of Agaricus bisporus packaged in nano-SiO2/TiO2 loaded polyvinyl alcohol films. Food Control 131, 108452. doi: 10.1016/j.foodcont.2021.108452
Chen, B.-X., Wei, T., Ye, Z.-W., Yun, F., Kang, L.-Z., Tang, H.-B., et al. (2018). Efficient CRISPR-Cas9 gene disruption system in edible-medicinal mushroom Cordyceps militaris. Front. Microbiol. 9, 1157. doi: 10.3389/fmicb.2018.01157
Chen, B.-X., Xue, L.-N., Wei, T., Wang, N., Zhong, J.-R., Ye, Z.-W., et al. (2022a). Multiplex gene precise editing and large DNA fragment deletion by the CRISPR-Cas9-TRAMA system in edible mushroom Cordyceps militaris. Microb. Biotechnol. 15, 2982–2991. doi: 10.1111/1751-7915.14147
Chen, B.-X., Xue, L.-N., Wei, T., Ye, Z.-W., Li, X.-H., Guo, L.-Q., et al. (2022b). Enhancement of ergothioneine production by discovering and regulating its metabolic pathway in Cordyceps militaris. Microb. Cell Fact. 21, 169. doi: 10.1186/s12934-022-01891-5
Chen, J., Lai, Y., Wang, L., Zhai, S., Zou, G., Zhou, Z., et al. (2017). CRISPR/Cas9-mediated efficient genome editing via blastospore-based transformation in entomopathogenic fungus Beauveria bassiana. Sci. Rep. 7, 45763. doi: 10.1038/srep45763
Chiang, T. W. W., le Sage, C., Larrieu, D., Demir, M., and Jackson, S. P. (2016). CRISPR-Cas9(D10A) nickase-based genotypic and phenotypic screening to enhance genome editing. Sci. Rep. 6, 24356. doi: 10.1038/srep24356
Cho, S. W., Kim, S., Kim, Y., Kweon, J., Kim, H. S., Bae, S., et al. (2014). Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 24, 132–141. doi: 10.1101/gr.162339.113
Das, S. K., Masuda, M., Sakurai, A., and Sakakibara, M. (2010). Medicinal uses of the mushroom Cordyceps militaris: current state and prospects. Fitoterapia 81, 961–968. doi: 10.1016/j.fitote.2010.07.010
Deng, H., Gao, R., Liao, X., and Cai, Y. (2017a). CRISPR system in filamentous fungi: current achievements and future directions. Gene 627, 212–221. doi: 10.1016/j.gene.2017.06.019
Deng, H., Gao, R., Liao, X., and Cai, Y. (2017b). Genome editing in Shiraia bambusicola using CRISPR-Cas9 system. J. Biotechnol. 259, 228–234. doi: 10.1016/j.jbiotec.2017.06.1204
Deng, H., Liang, W., Fan, T. P., Zheng, X., and Cai, Y. (2020). Modular engineering of Shiraia bambusicola for hypocrellin production through an efficient CRISPR system. Int. J. Biol. Macromol. 165, 796–803. doi: 10.1016/j.ijbiomac.2020.09.208
Foster, A. J., Martin-Urdiroz, M., Yan, X., Wright, H. S., Soanes, D. M., and Talbot, N. J. (2018). CRISPR-Cas9 ribonucleoprotein-mediated co-editing and counterselection in the rice blast fungus. Sci. Rep. 8, 14355. doi: 10.1038/s41598-018-32702-w
Fu, Y., Foden, J. A., Khayter, C., Maeder, M. L., Reyon, D., Joung, J. K., et al. (2013). High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 31, 822. doi: 10.1038/nbt.2623
Fu, Y. F., Sander, J. D., Reyon, D., Cascio, V. M., and Joung, J. K. (2014). Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 32, 279–284. doi: 10.1038/nbt.2808
Fuller, K. K., Chen, S., Loros, J. J., and Dunlap, J. C. (2015). Development of the CRISPR/Cas9 system for targeted gene disruption in Aspergillus fumigatus. Eukaryotic Cell 14, 1073–1080. doi: 10.1128/EC.00107-15
Gao, Y., Xiong, X., Wong, S., Charles, E. J., Lim, W. A., and Qi, L. S. (2016). Complex transcriptional modulation with orthogonal and inducible dCas9 regulators. Nat. Methods 13, 1043. doi: 10.1038/nmeth.4042
Gao, Y., and Zhao, Y. (2014). Self-processing of ribozyme-flanked RNAs into guide RNAs in vitro and in vivo for CRISPR-mediated genome editing. J. Integr. Plant Biol. 56, 343–349. doi: 10.1111/jipb.12152
Gaudelli, N. M., Komor, A. C., Rees, H. A., Packer, M. S., Badran, A. H., Bryson, D. I., et al. (2017). Programmable base editing of A.T to G.C in genomic DNA without DNA cleavage. Nature 551, 464. doi: 10.1038/nature24644
Gupta, S., Summuna, B., Gupta, M., and Annepu, S. K. (2018). “Edible mushrooms: cultivation, bioactive molecules, and health benefits,” in Bioactive Molecules in Food,” eds J.-M. Mérillon and K. G. Ramawat (Cham: Springer International Publishing), 1–33.
Han, X., and Yixin, L. (2021). Progress and challenge of the CRISPR-Cas system in gene editing for filamentous fungi. Synthetic Biol. J. 2, 274–286. doi: 10.12211/2096-8280.2020-078
Heo, Y. B., Hwang, G. H., Kang, S. W., Bae, S., and Woo, H. M. (2022). High-fidelity cytosine base editing in a GC-rich Corynebacterium glutamicum with reduced DNA off-target editing effects. Microbiol. Spectr. 10, e0376022 doi: 10.1128/spectrum.03760-22
Hsu, P. D., Lander, E. S., and Zhang, F. (2014). Development and applications of CRISPR-Cas9 for genome engineering. Cell 157, 1262–1278. doi: 10.1016/j.cell.2014.05.010
Hsu, P. D., Scott, D. A., Weinstein, J. A., Ran, F. A., Konermann, S., Agarwala, V., et al. (2013). DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31, 827. doi: 10.1038/nbt.2647
Hua, K., Tao, X. P., Han, P. J., Wang, R., and Zhu, J. K. (2019). Genome engineering in rice using Cas9 variants that recognize NG PAM sequences. Mol. Plant 12, 1003–1014. doi: 10.1016/j.molp.2019.03.009
Jan Vonk, P., Escobar, N., Wosten, H. A. B., Lugones, L. G., and Ohm, R. A. (2019). High-throughput targeted gene deletion in the model mushroom Schizophyllum commune using pre-assembled Cas9 ribonucleoproteins. Sci. Rep. 9, 7632. doi: 10.1101/563759
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., and Charpentier, E. (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. doi: 10.1126/science.1225829
Juillerat, A., Dubois, G., Valton, J., Thomas, S., Stella, S., Maréchal, A., et al. (2014). Comprehensive analysis of the specificity of transcription activator-like effector nucleases. Nucleic Acids Res. 42, 5390–5402. doi: 10.1093/nar/gku155
Kämper, J., Reichmann, M., Romeis, T., Bölker, M., and Kahmann, R. (1995). Multiallelic recognition: nonself-dependent dimerization of the bE and bW homeodomain proteins in ustilago maydis. Cell 81, 73–83. doi: 10.1016/0092-8674(95)90372-0
Kang, M. J., Zuo, Z. C., Yin, Z. X., and Gu, J. R. (2022). Molecular mechanism of D1135E-induced discriminated CRISPR-Cas9 PAM recognition. J. Chem. Inf. Model. 62, 3057–3066. doi: 10.1021/acs.jcim.1c01562
Khanal, S., Schroeder, L., Nava-Mercado, O. A., Mendoza, H., and Perlin, M. H. (2021). Role for nitrate assimilatory genes in virulence of Ustilago maydis. Fungal Biol. 125, 764–775. doi: 10.1016/j.funbio.2021.04.010
Kiani, S., Beal, J., Ebrahimkhani, M. R., Huh, J., Hall, R. N., Xie, Z., et al. (2014). CRISPR transcriptional repression devices and layered circuits in mammalian cells. Nat. Methods 11, 723–U155. doi: 10.1038/nmeth.2969
Kim, K., Ryu, S.-M., Kim, S.-T., Baek, G., Kim, D., Lim, K., et al. (2017). Highly efficient RNA-guided base editing in mouse embryos. Nat. Biotechnol. 35, 435. doi: 10.1038/nbt.3816
Kleinstiver, B. P., Pattanayak, V., Prew, M. S., Tsai, S. Q., Nguyen, N. T., Zheng, Z., et al. (2016). High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529, 490. doi: 10.1038/nature16526
Kleinstiver, B. P., Prew, M. S., Tsai, S. Q., Topkar, V. V., Nguyen, N. T., Zheng, Z., et al. (2015). Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 523, 481–U249. doi: 10.1038/nature14592
Komor, A. C., Kim, Y. B., Packer, M. S., Zuris, J. A., and Liu, D. R. (2016). Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420. doi: 10.1038/nature17946
Koshi, D., Ueshima, H., Kawauchi, M., Nakazawa, T., Sakamoto, M., and Hirata, M. (2022). Marker-free genome editing in the edible mushroom, Pleurotus ostreatus, using transient expression of genes required for CRISPR/Cas9 and for selection. J. Wood Sci. 68, 1–8. doi: 10.1186/s10086-022-02033-6
Kraft, K., Geuer, S., Will, A. J., Chan, W. L., Paliou, C., Borschiwer, M., et al. (2015). Deletions, inversions, duplications: engineering of structural variants using CRISPR/Cas in mice. Cell Rep. 10, 833–839. doi: 10.1016/j.celrep.2015.01.016
Kuscu, C., Arslan, S., Singh, R., Thorpe, J., and Adli, M. (2014). Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat. Biotechnol. 32, 677. doi: 10.1038/nbt.2916
Li, D., Tang, Y., Lin, J., and Cai, W. (2017). Methods for genetic transformation of filamentous fungi. Microb. Cell Fact. 16, 168. doi: 10.1186/s12934-017-0785-7
Li, H., and Zhong, J.-J. (2020). Role of calcineurin-responsive transcription factor CRZ1 in ganoderic acid biosynthesis by Ganoderma lucidum. Process Biochem. 95, 166–173. doi: 10.1016/j.procbio.2020.05.027
Lin, J., Yang, X., Wei, T., Guo, L., Lin, J., Chen, Y., et al. (2019). Construction and transformation of CRISPR/Cas9 genome editing vector of Flammulina filiformis G protein-coupled receptor gene. Mycosystema 38, 349–361. doi: 10.13346/j.mycosystema.180280
Lin, L., Liu, Y., Xu, F., Huang, J., Daugaard, T. F., Petersen, T. S., et al. (2018). Genome-wide determination of on-target and off-target characteristics for RNA-guided DNA methylation by dCas9 methyltransferases. Gigascience 7, 1–19. doi: 10.1093/gigascience/giy011
Lin, Y., Cradick, T. J., Brown, M. T., Deshmukh, H., Ranjan, P., Sarode, N., et al. (2014). CRISPR/Cas9 systems have off-target activity with insertions or deletions between target DNA and guide RNA sequences. Nucleic Acids Res. 42, 7473–7485. doi: 10.1093/nar/gku402
Liu, J., Cui, H., Wang, R., Xu, Z., Yu, H., Song, C., et al. (2022). A simple and efficient CRISPR/Cas9 system using a ribonucleoprotein method for Flammulina filiformis. J. Fungi 8, 1000. doi: 10.3390/jof8101000
Liu, J., Liu, J., Zhang, D., Zhang, M., Xu, Z., Wang, R., et al. (2017). Agrobacterium-mediated Gene Transformation of Cas9 into Flammulina velutipes. Acta Edulis Fungi 24, 25–29.
Liu, K., Sun, B., You, H., Tu, J.-L., Yu, X., Zhao, P., et al. (2020). Dual sgRNA-directed gene deletion in basidiomycete Ganoderma lucidum using the CRISPR/Cas9 system. Microb. Biotechnol. 13, 386–396. doi: 10.1111/1751-7915.13534
Liu, R., Chen, L., Jiang, Y., Zhou, Z., and Zou, G. (2015). Efficient genome editing in filamentous fungus Trichoderma reesei using the CRISPR/Cas9 system. Cell Discov. 1, 15007. doi: 10.1038/celldisc.2015.7
Liu, W., An, C., Shu, X., Meng, X., Yao, Y., Zhang, J., et al. (2020). A dual-plasmid CRISPR/Cas system for mycotoxin elimination in polykaryotic industrial fungi. ACS Synth. Biol. 9, 2087–2095. doi: 10.1021/acssynbio.0c00178
Liu, X., Dong, J., Liao, J., Tian, L., Qiu, H., Wu, T., et al. (2022). Establishment of CRISPR/Cas9 genome-editing system based on dual sgRNAs in Flammulina filiformis. J. Fungi 8, 693. doi: 10.3390/jof8070693
Liu, Z., Chen, M., Chen, S., Deng, J., Song, Y., Lai, L., et al. (2018). Highly efficient RNA-guided base editing in rabbit. Nat. Commun. 9, 2717. doi: 10.1038/s41467-018-05232-2
Luo, R., Lin, J. F., Guo, L. Q., Ye, Z. W., Guo, T. F., and Yun, F. (2016). Construction of Flammulina velutipes genome editing vector by using CRISPR/Cas9 system. Sci. Technol. Food Industry 37, 230–234. doi: 10.13386/j.issn1002-0306.2016.20.037
Mali, P., Yang, L., Esvelt, K. M., Aach, J., Guell, M., DiCarlo, J. E., et al. (2013). RNA-guided human genome engineering via Cas9. Science 339, 823–826. doi: 10.1126/science.1232033
Meng, G., Wang, X., Liu, M., Wang, F., Liu, Q., and Dong, C. (2022). Efficient CRISPR/Cas9 system based on autonomously replicating plasmid with an AMA1 sequence and precisely targeted gene deletion in the edible fungus, Cordyceps militaris. Microb. Biotechnol. 15, 2594–2606. doi: 10.1111/1751-7915.14107
Nagy, G., Szebenyi, C., Csernetics, Á., Vaz, A. G., Tóth, E. J., Vágvölgyi, C., et al. (2017). Development of a plasmid free CRISPR-Cas9 system for the genetic modification of Mucor circinelloides. Sci. Rep. 7, 16800. doi: 10.1038/s41598-017-17118-2
Osakabe, Y., Sugano, S. S., and Osakabe, K. (2016). Genome engineering of woody plants: past, present and future. J. Wood Sci. 62, 217–225. doi: 10.1007/s10086-016-1548-5
Ouyang, P., Li, Q., Guo, L., Lin, J., Ye, Z., Wei, T., et al. (2018). Establishment of a CRISPR/Cas9 system for editing cold-induced gene HK1/HK2 in Flammulina velutipes. Acta Edulis Fungi 25, 1–7, 113. doi: 10.16488/j.cnki.1005-9873.2018.03.001
Pan, Y., Shen, N., Jung-Klawitter, S., Betzen, C., Hoffmann, G. F., Hoheisel, J. D., et al. (2016). CRISPR RNA-guided FokI nucleases repair a PAH variant in a phenylketonuria model. Scientific Reports 6, 35794. doi: 10.1038/srep35794
Pan, Y., Xia, S., Dong, C., Pan, H., Cai, J., Huang, L., et al. (2021). Random base editing for genome evolution in Saccharomyces cerevisiae. ACS Synth. Biol. 10, 2440–2446. doi: 10.1021/acssynbio.1c00217
Pattanayak, V., Lin, S., Guilinger, J. P., Ma, E., Doudna, J. A., and Liu, D. R. (2013). High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat. Biotechnol. 31, 839. doi: 10.1038/nbt.2673
Pei, W., Wen, L., and Xiang-ling, S. (2018). Research progress of CRISPR/Cas9 system in filamentous fungi. Biotechnol. Bull. 34, 1–8. doi: 10.13560/j.cnki.biotech.bull.1985.2017-1128
Pérez, G., Lopez-Moya, F., Chuina, E., Ibañez-Vea, M., Garde, E., López-Llorca, L. V., et al. (2021). Strain degeneration in pleurotus ostreatus: a genotype dependent oxidative stress process which triggers oxidative stress, cellular detoxifying and cell wall reshaping genes. J. Fungi 7, 26. doi: 10.3390/jof7100862
Qin, H., Xiao, H., Zou, G., Zhou, Z., and Zhong, J.-J. (2017). CRISPR-Cas9 assisted gene disruption in the higher fungus Ganoderma species. Process Biochem. 56, 57–61. doi: 10.1016/j.procbio.2017.02.012
Reis, F. S., Barros, L., Calhelha, R. C., Cirić, A., van Griensven, L. J. L. D., Soković, M., et al. (2013). The methanolic extract of Cordyceps militaris (L.) Link fruiting body shows antioxidant, antibacterial, antifungal and antihuman tumor cell lines properties. Food Chem. Toxicol. 62, 91–98. doi: 10.1016/j.fct.2013.08.033
Ren, B., Liu, L., Li, S., Kuang, Y., Wang, J., Zhang, D., et al. (2019). Cas9-NG greatly expands the targeting scope of the genome-editing toolkit by recognizing NG and other atypical PAMs in rice. Mol. Plant 12, 1015–1026. doi: 10.1016/j.molp.2019.03.010
Rodriguez, T. C., Dadafarin, S., Pratt, H. E., Liu, P. P., Amrani, N., Zhu, L. J., et al. (2021). Genome-wide detection and analysis of CRISPR-Cas off-targets. Reprogramming the Genome: Crispr-Cas-Based Human Disease Therapy 181, 31–43. doi: 10.1016/bs.pmbts.2021.01.012
Ruiping, X., Yan, Z., and Yinbing, B. (2019). Application of CRISPR/Cas9 technology in macrofungi research. Acta Edulis Fungi 26, 119–127. doi: 10.16488/j.cnki.1005-9873.2019.02.017
Sakamoto, Y., Tamai, Y., and Yajima, T. (2004). Influence of light on the morphological changes that take place during the development of the Flammulina velutipes fruit body. Mycoscience 45, 333–339. doi: 10.1007/S10267-004-0195-7
Sander, J. D., and Joung, J. K. (2014). CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 32, 347–355. doi: 10.1038/nbt.2842
Satomura, A., Nishioka, R., Mori, H., Sato, K., Kuroda, K., and Ueda, M. (2017). Precise genome-wide base editing by the CRISPR Nickase system in yeast. Sci. Rep. 7, 2095. doi: 10.1038/s41598-017-02013-7
Schuster, M., Schweizer, G., Reissmann, S., and Kahmann, R. (2016). Genome editing in Ustilago maydis using the CRISPR-Cas system. Fungal Genet. Biol. 89, 3–9. doi: 10.1016/j.fgb.2015.09.001
Sharma, R., Katoch, M., Govindappa, N., Srivastava, P. S., Sastry, K. N., and Qazi, G. N. (2012). Evaluation of the catalase promoter for expressing the alkaline xylanase gene (alx) in Aspergillus niger. FEMS Microbiol. Lett. 327, 33–40. doi: 10.1111/j.1574-6968.2011.02454.x
Shi, T.-Q., Gao, J., Wang, W.-J., Wang, K.-F., Xu, G.-Q., Huang, H., et al. (2019). CRISPR/Cas9-based genome editing in the filamentous fungus Fusarium fujikuroi and its application in strain engineering for gibberellic acid production. ACS Synth. Biol. 8, 445–454. doi: 10.1021/acssynbio.8b00478
Shi, T.-Q., Liu, G.-N., Ji, R.-Y., Shi, K., Song, P., Ren, L.-J., et al. (2017). CRISPR/Cas9-based genome editing of the filamentous fungi: the state of the art. Appl. Microbiol. Biotechnol. 101, 7435–7443. doi: 10.1007/s00253-017-8497-9
Shimatani, Z., Kashojiya, S., Takayama, M., Terada, R., Arazoe, T., Ishii, H., et al. (2017). Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat. Biotechnol. 35, 441. doi: 10.1038/nbt.3833
Slaymaker, I. M., Gao, L., Zetsche, B., Scott, D. A., Yan, W. X., and Zhang, F. (2016). Rationally engineered Cas9 nucleases with improved specificity. Science 351, 84–88. doi: 10.1126/science.aad5227
Solis-Escalante, D., Kuijpers, N. G. A., Bongaerts, N., Bolat, I., Bosman, L., Pronk, J. T., et al. (2013). amdSYM, a new dominant recyclable marker cassette for Saccharomyces cerevisiae. FEMS Yeast Res. 13, 126–139. doi: 10.1111/1567-1364.12024
Song, R., Zhai, Q., Sun, L., Huang, E., Zhang, Y., Zhu, Y., et al. (2019). CRISPR/Cas9 genome editing technology in filamentous fungi: progress and perspective. Appl. Microbiol. Biotechnol. 103, 6919–6932. doi: 10.1007/s00253-019-10007-w
Sonnenberg, A. S. M., Baars, J. J. P., Gao, W., and Visser, R. G. F. (2017). Developments in breeding of Agaricus bisporus var. bisporus: progress made and technical and legal hurdles to take. Appl. Microbiol. Biotechnol. 101, 1819–1829. doi: 10.1007/s00253-017-8102-2
Sternberg, S. H., Redding, S., Jinek, M., Greene, E. C., and Doudna, J. A. (2014). DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 507, 62–67. doi: 10.1038/nature13011
Sugano, S. S., Suzuki, H., Shimokita, E., Chiba, H., Noji, S., Osakabe, Y., et al. (2017). Genome editing in the mushroom-forming basidiomycete Coprinopsis cinerea, optimized by a high-throughput transformation system. Sci. Rep. 7, 1260. doi: 10.1038/s41598-017-00883-5
Suhadolnik, R. J., and Cory, J. G. (1964). Further evidence for the biosynthesis of cordycepin and proof of the structure of 3-deoxyribose. Biochim. Biophys. Acta 91, 661–662. doi: 10.1016/0926-6550(64)90021-0
Sun, H.-J., Wang, J., Tao, X.-M., Shi, J., Huang, M.-Y., and Chen, Z. (2012). Purification and characterization of polyphenol oxidase from rape flower. J. Agric. Food Chem. 60, 823–829. doi: 10.1021/jf2032999
Svahn, K. S., Chryssanthou, E., Olsen, B., Bohlin, L., and Göransson, U. (2015). Penicillium nalgiovense Laxa isolated from Antarctica is a new source of the antifungal metabolite amphotericin B. Fungal Biol. Biotechnol. 2, 1. doi: 10.1186/s40694-014-0011-x
Taofiq, O., Gonzalez-Paramas, A. M., Martins, A., Barreiro, M. F., and Ferreira, I. (2016). Mushrooms extracts and compounds in cosmetics, cosmeceuticals and nutricosmetics-A review. Ind. Crops Prod. 90, 38–48. doi: 10.1016/j.indcrop.2016.06.012
Tehrani, H. H., Becker, J., Bator, I., Saur, K., Meyer, S., Lóia, A. C. R., et al. (2019). Integrated strain- and process design enable production of 220 g L(-1) itaconic acid with Ustilago maydis. Biotechnol. Biofuels 12, 263. doi: 10.1186/s13068-019-1605-6
Terao, M., Tamano, M., Hara, S., Kato, T., Kinoshita, M., and Takada, S. (2016). Utilization of the CRISPR/Cas9 system for the efficient production of mutant mice using crRNA/tracrRNA with Cas9 nickase and Fokl-dCas9. Exp. Anim. 65, 275–283. doi: 10.1538/expanim.15-0116
Tian, S., Jiang, L., Cui, X., Zhang, J., Guo, S., Li, M., et al. (2018). Engineering herbicide-resistant watermelon variety through CRISPR/Cas9-mediated base-editing. Plant Cell Rep. 37, 1353–1356. doi: 10.1007/s00299-018-2299-0
Tsai, S. Q., Zheng, Z., Nguyen, N. T., Liebers, M., Topkar, V. V., Thapar, V., et al. (2015). GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 33, 187–197. doi: 10.1038/nbt.3117
Tu, J. L., Bai, X. Y., Xu, Y. L., Li, N., and Xu, J. W. (2021). Targeted gene insertion and replacement in the basidiomycete Ganoderma lucidum by inactivation of nonhomologous end joining using CRISPR/Cas9. Appl. Environ. Microbiol. 87, e01510–e01521. doi: 10.1128/AEM.01510-21
Ullah, M., Xia, L., Xie, S., and Sun, S. (2020). CRISPR/Cas9-based genome engineering: a new breakthrough in the genetic manipulation of filamentous fungi. Biotechnol. Appl. Biochem. 67, 835–851. doi: 10.1002/bab.2077
Waltz, E. (2016). Gene-edited CRISPR mushroom escapes US regulation. Nature 532, 293–293. doi: 10.1038/nature.2016.19754
Wang, P.-A., Zhang, J.-M., and Zhong, J.-J. (2022). CRISPR-Cas9 assisted in-situ complementation of functional genes in the basidiomycete Ganoderma lucidum. Process Biochem. 121, 689–697. doi: 10.1016/j.procbio.2022.08.015
Wang, P. A., Xiao, H., and Zhong, J. J. (2020). CRISPR-Cas9 assisted functional gene editing in the mushroom Ganoderma lucidum. Appl. Microbiol. Biotechnol. 104, 1661–1671. doi: 10.1007/s00253-019-10298-z
Wang, P. M., Liu, X. B., Dai, Y. C., Horak, E., Steffen, K., and Yang, Z. L. (2018). Phylogeny and species delimitation of Flammulina: taxonomic status of winter mushroom in East Asia and a new European species identified using an integrated approach. Mycol. Prog. 17, 1013–1030. doi: 10.1007/s11557-018-1409-2
Wang, Q., Chu, L., and Kou, L. (2017). UV-C Treatment maintains quality and delays senescence of oyster mushroom (Pleurotus ostreatus). Sci. Hortic. 225, 380–385. doi: 10.1016/j.scienta.2017.07.019
Wang, Q., and Coleman, J. J. (2019). Progress and challenges: development and implementation of CRISPR/Cas9 technology in filamentous fungi. Comput. Struct. Biotechnol. J. 17, 761–769. doi: 10.1016/j.csbj.2019.06.007
Wang, T., Yue, S., Jin, Y., Wei, H., and Lu, L. (2021). Advances allowing feasible pyrG gene editing by a CRISPR-Cas9 system for the edible mushroom Pleurotus eryngii. Fungal Genet. Biol. 147, 103509. doi: 10.1016/j.fgb.2020.103509
Wang, Y., Chen, H., Ma, L., Gong, M., Wu, Y., Bao, D., et al. (2022). Use of CRISPR-Cas tools to engineer Trichoderma species. Microb. Biotechnol. 15, 2521–2532. doi: 10.1111/1751-7915.14126
Waring, R. B., May, G. S., and Morris, N. R. (1989). Characterization of an inducible expression system in Aspergillus nidulans using alcA and tubulincoding genes. Gene 79, 119–130. doi: 10.1016/0378-1119(89)90097-8
Wege, S.-M., Gejer, K., Becker, F., Bölker, M., Freitag, J., and Sandrock, B. (2021). Versatile CRISPR/Cas9 systems for genome editing in Ustilago maydis. J. Fungi 7, 149. doi: 10.3390/jof7020149
Wu, J., Chen, H., Gao, J., Liu, X., Cheng, W., and Ma, X. (2010). Cloning, characterization and expression of two new polyphenol oxidase cDNAs from Agaricus bisporus. Biotechnol. Lett. 32, 1439–1447. doi: 10.1007/s10529-010-0329-2
Xia, P.-F., Casini, I., Schulz, S., Klask, C.-M., Angenent, L. T., and Molitor, B. (2020). Reprogramming acetogenic bacteria with CRISPR-targeted base editing via deamination. ACS Synth. Biol. 9, 2162–2171. doi: 10.1021/acssynbio.0c00226
Xiao, H., and Zhong, J. J. (2016). Production of useful terpenoids by higher-fungus cell factory and synthetic biology approaches. Trends Biotechnol. 34, 242–255. doi: 10.1016/j.tibtech.2015.12.007
Xiao-tian, L., Hao, Q., Li, T., Ang, R., and Ming-wen, Z. (2021). Research progress in CRISPR/Cas9 genome editing system in edible and medicinal fungi. Biotechnol. Bull. 37, 4–13. doi: 10.13560/j.cnki.biotech.bull.1985.2021-1055
Xu, Y. N., and Zhong, J. J. (2012). Impacts of calcium signal transduction on the fermentation production of antitumor ganoderic acids by medicinal mushroom Ganoderma lucidum. Biotechnol. Adv. 30, 1301–1308. doi: 10.1016/j.biotechadv.2011.10.001
Xuping, W., Jiaojiao, Z., and Caihong, D. (2019). Research progress of CRISPR/Cas9 mediated genome editing in edible and medicinal fungi. J. Fungal Res. 17, 215–223. doi: 10.13341/j.jfr.2019.8013
Yamasaki, F., Nakazawa, T., Oh, M., Bao, D., Kawauchi, M., Sakamoto, M., et al. (2022). Gene targeting of dikaryotic Pleurotus ostreatus nuclei using the CRISPR/Cas9 system. FEMS Microbiol. Lett. 369, fnac083. doi: 10.1093/femsle/fnac083
Yan, Y., and Finnigan, G. C. (2018). Development of a multi-locus CRISPR gene drive system in budding yeast. Sci. Rep. 8, 17277. doi: 10.1038/s41598-018-34909-3
Yang, L., Xiaofei, H., Pei, W., Shiling, L., and Shaopeng, H. (2020). Establishment of a CRISPR/Cas9 system in Agaricus bisporus. Acta Edulis Fungi 27, 16–22. doi: 10.16488/j.cnki.1005-9873.2020.03.003
Yannan, W., and Yuhui, S. (2022). Base editing technology and its application in microbial synthetic biology. Synthetic Biol. J. 3, 1–18. doi: 10.12211/2096-8280.2022-053
Yu, R., Song, L., Zhao, Y., Bin, W., Wang, L., Zhang, H., et al. (2004). Isolation and biological properties of polysaccharide CPS-1 from cultured Cordyceps militaris. Fitoterapia 75, 465–472. doi: 10.1016/j.fitote.2004.04.003
Yuan, W. X., Yu, Y. M., Hu, C. C., and & Zhao, Z. G. (2017). Current issues and progress in the application of CRISPR/Cas9 technique. Biotechnol. Bull. 33, 70–77. doi: 10.13560/j.cnki.biotech.bull.1985.2017.04.009
Zhang, C., Meng, X., Wei, X., and Lu, L. (2016). Highly efficient CRISPR mutagenesis by microhomology-mediated end joining in Aspergillus fumigatus. Fungal Genet. Biol. 86, 47–57. doi: 10.1016/j.fgb.2015.12.007
Zhang, Y., Qin, W., Lu, X., Xu, J., Huang, H., Bai, H., et al. (2017). Programmable base editing of zebrafish genome using a modified CRISPR-Cas9 system. Nat. Commun. 8, 118. doi: 10.1038/s41467-017-00175-6
Zhang, Y., Wang, D., Chen, Y., Liu, T., Zhang, S., Fan, H., et al. (2021). Healthy function and high valued utilization of edible fungi. Food Sci. Hum. Wellness 10, 408–420. doi: 10.1016/j.fshw.2021.04.003
Zhao, N., Lin, X., Qi, S.-S., Luo, Z.-M., Chen, S.-L., and Yan, S.-Z. (2016). De novo transcriptome assembly in Shiraia bambusicola to investigate putative genes involved in the biosynthesis of hypocrellin A. Int. J. Mol. Sci. 17, 311. doi: 10.3390/ijms17030311
Zheng, K., Wang, Y., Li, N., Jiang, F.-F., Wu, C.-X., Liu, F., et al. (2018). Highly efficient base editing in bacteria using a Cas9-cytidine deaminase fusion. Commun. Biol. 1, 32. doi: 10.1038/s42003-018-0035-5
Zhong, J. J., and Xiao, J. H. (2009). Secondary metabolites from higher fungi: discovery, bioactivity, and bioproduction. Biotechnology in China I: From Bioreaction To Bioseparation and Bioremediation 113, 79–150. doi: 10.1007/10_2008_26
Zhong, Z., Sretenovic, S., Ren, Q., Yang, L., Bao, Y., Qi, C., et al. (2019). Improving plant genome editing with high-fidelity xCas9 and non-canonical PAM-targeting Cas9-NG. Mol. Plant 12, 1027–1036. doi: 10.1016/j.molp.2019.03.011
Zhou, H. B., Liu, B., Weeks, D. P., Spalding, M. H., and Yang, B. (2014). Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice. Nucleic Acids Res. 42, 10903–10914. doi: 10.1093/nar/gku806
Zhu, W., Hu, J., Chi, J., Li, Y., Yang, B., Hu, W., et al. (2020). Label-free proteomics reveals the molecular mechanism of subculture induced strain degeneration and discovery of indicative index for degeneration in Pleurotus ostreatus. Molecules 25, 4920. doi: 10.3390/molecules25214920
Zong, Y., Wang, Y., Li, C., Zhang, R., Chen, K., Ran, Y., et al. (2017). Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat. Biotechnol. 35, 438. doi: 10.1038/nbt.3811
Keywords: edible fungi, CRISPR/Cas9, molecular breeding, application progress, genome editing
Citation: Zhang Y, Chen S, Yang L and Zhang Q (2023) Application progress of CRISPR/Cas9 genome-editing technology in edible fungi. Front. Microbiol. 14:1169884. doi: 10.3389/fmicb.2023.1169884
Received: 20 February 2023; Accepted: 26 April 2023;
Published: 25 May 2023.
Edited by:
Mingfeng Cao, Xiamen University, ChinaReviewed by:
Han Xiao, Shanghai Jiao Tong University, ChinaBibekananda Kar, Mayo Clinic, United States
Shekhar Mishra, University of Illinois at Urbana-Champaign, United States
Copyright © 2023 Zhang, Chen, Yang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Long Yang, bHlhbmdAc2RhdS5lZHUuY24=; Qiang Zhang, emhhbmdxaWFuZzE4NThAMTYzLmNvbQ==
†These authors have contributed equally to this work
 Yan Zhang†
Yan Zhang† Shutong Chen
Shutong Chen Long Yang
Long Yang Qiang Zhang
Qiang Zhang