- Department of Physiology of Industrial Microorganisms, Danylo Zabolotny Institute of Microbiology and Virology, National Academy of Science of Ukraine, Kyiv, Ukraine
This study was carried out to select the bacteriocinogenic strains among Enterococcus strains isolated from Ukrainian traditional dairy products using a low-cost media for screening, that containing molasses and steep corn liquor. A total of 475 Enterococcus spp. strains were screened for antagonistic activity against Pseudomonas aeruginosa, Escherichia coli, Staphylococcus aureus, and Listeria monocytogenes indicator strains. The initial screening revealed that 34 Enterococcus strains during growth in low-cost medium containing corn steep liquor, peptone, yeast extract, and sucrose produced metabolites with inhibition activity against at least of the indicator strains used. Enterocin genes entA, entP, and entB were detected in 5 Enterococcus strains by PCR assay. Genes of enterocins A and P were found in E. faecalis 58 and Enterococcus sp. 226 strains, enterocins B and P – in Enterococcus sp. 423, enterocin A – in E. faecalis 888 and E. durans 248 strains. Bacteriocin-like inhibitory substances (BLIS) produced by these Enterococcus strains were thermostable and sensitive to proteolytic enzymes. To our knowledge, this is the first report on the isolation of enterocin-producing wild Enterococcus strains from traditional Ukrainian dairy products using a low-cost media for screening bacteriocinogenic strains. Strains E. faecalis 58, Enterococcus sp. 423, and Enterococcus sp. 226 are promising candidates for practical use as producers of bacteriocins with inhibitory activity against L. monocytogenes using molasses and steep corn liquor as cheap sources of carbon and nitrogen, that can significantly reduce the cost of industrial bacteriocin production. Further studies will be required to determine the dynamic of bacteriocin production, its structure, and mechanisms of antibacterial action.
1. Introduction
Enterococci are widely distributed in nature and one of the main niches of their habitat are fermented milk products obtained by spontaneous fermentation of raw milk. Natural resistance to a wide range of temperatures, pH, and high NaCl concentrations allows them to survive in cheeses but they are also commonly found in other fermented milk products, for example artisanal fermented milks such as Kumis and Dahi (Giraffa, 2003; Ogier and Serror, 2008; Chaves-López et al., 2011; Farah et al., 2016). Due to their enzymatic activities, Enterococcus strains can impart special organoleptic characteristics to cheeses and are often included in starter cultures. Enterococci can also improve the microbiological quality and extend the shelf life of end-products due to their antagonistic properties against pathogens (Giraffa, 2003; Foulquié Moreno et al., 2006). According to the literature, one of the mechanisms of the antagonistic action of enterococci is their ability to produce bacteriocins, which allows the use of bacteriocinogenic Enterococcus strains as protective starter cultures, as well as producers of bacteriocins that can be used as purified preparations (Ng et al., 2020). Bacteriocins of lactic acid bacteria (LAB) have been intensively studied over the past decades and there is still more interest in screening for new producer strains (Nami et al., 2019; Lauková et al., 2020; Choeisoongnern et al., 2021; Sosa et al., 2021). According to a definition, bacteriocins are ribosomally synthesized peptides that inhibit the growth of phylogenetically closely related microorganisms (Darbandi et al., 2022). As was shown in numerous studies, LAB bacteriocins can also inhibit the growth of many opportunistic and foodborne pathogens, and are of great practical interest as natural preservatives in the food industry (Sameli et al., 2021; Kasimin et al., 2022). Bacteriocins of enterococci, so-called enterocins, mainly belong to subclass IIa, which are small, heat-stable peptides with antagonistic activity against the Listeria monocytogenes and other pathogenic bacteria (Darbandi et al., 2022; Kasimin et al., 2022). Also, anticancer, antiviral, and antibiofilm activities of enterocins were reported (Ermolenko et al., 2019; Lee et al., 2021; Sharma et al., 2021). Such a wide range of biological activity of enterocins significantly expands the potential field of the practical application of Enterococcus strains, including as probiotics (Nami et al., 2019; Ng et al., 2020; Choeisoongnern et al., 2021).
The composition and the concentration of nutrients in the medium greatly influence LAB bacteriocin production (López et al., 2007). Most studies on the production of enterocins have been conducted using commercial laboratory media such as MRS, BHI, and M17. Commonly, after the initial screening of bacteriocinogenic LAB strains, the next stage is the optimization of culture conditions for large-scale biomass or bacteriocin production using cheaper cultural media (Metsoviti et al., 2011; Schirru et al., 2014; Sosa et al., 2021). Data concerning enterocin production by Enterococcus strains using inexpensive media are very rare and mainly concern certain strains (Coetzee et al., 2007; López et al., 2007; Bali et al., 2014).
However, there is evidence that strains selected as bacteriocinogenic in the MRS medium do not produce bacteriocin when grown on media based on cheaper substrates, since the synthesis of bacteriocins strongly depends on growth conditions (Audisio et al., 2001; Todorov and Dicks, 2005; Schirru et al., 2014; Vandera et al., 2019). Thus, it is more reasonable to initially carry out primary screening on cheap media suitable for industrial production.
The aim of this study was the screening of potentially bacteriocinogenic Enterococcus strains, using low-cost by-product-based media among dairy-derived Enterococcus strains. To the best of our knowledge, this is the first report about the presence of bactericonogenic strains among enterococci isolated from artisan Ukrainian traditional dairy products prepared by spontaneous fermentation.
2. Materials and methods
2.1. Bacterial strains
The 475 Enterococcus strains from the Lactic Acid Bacteria Culture Collection of the Department of Physiology of Industrial Microorganisms, Zabolotny Institute of Microbiology and Virology NAS, (Kyiv, Ukraine) were used in this study. The E. faecalis (n = 350), E. durans (n = 92), E. faecium (n = 15). Enterococcus spp. (n = 17) and 1 E. hirae strains were previously isolated from Ukrainian traditional dairy products such as fermented milk, cottage cheese, bryndza, and sour cream (Garmasheva et al., 2018). The strains were stored in MRS broth (HiMedia, India) supplemented with 30% glycerol at −50°C. Enterococcus strains were subcultured in MRS broth at 37°C for 24 h. The cultures were activated by two successive transfers in the MRS broth before use.
2.2. Initial screening for antagonistic activity
Production of antimicrobial metabolites was tested using four media having the following composition (w/v): medium I – 1.00% peptone, 0.05% yeast extract, 0.25% corn steep liquor, 2.00% sucrose; medium II – 1.00% peptone, 0.05% yeast extract, 0.25% corn steep liquor, 2.00% sugar beet molasses; medium III – 1.00% peptone, 0.05% yeast extract, 0.25% corn steep liquor; medium IV – 2.00% sugar beet molasses (pH 7.0 ± 0.2). Corn steep liquor and sugar-beet molasses were procured from local companies Interstarch Ukraine (Kyiv, Ukraine) and Radekhivskyi Sugar LLC (Ternopil region, Ukraine), respectively. The MRS broth also was used. All media were sterilized by autoclaving at 121°C for 15 min. Samples of 24 h-old Enterococcus cultures were serially diluted in sterile saline solution, plated on MRS agar and viable cell counts were determined for each medium used.
The antagonistic activity was determined by the well-diffusion assay as described previously (Schillinger and Lücke, 1989). The 6 mm-diameter stainless steel cylinders were laid on Petri dishes with Nutrient agar (NA), and the dishes were overlaid with 15 mL of molten NA (0.75%) inoculated with 100 μL of an overnight culture of the indicator strain. The Pseudomonas aeruginosa АТСС 9027, Escherichia coli ATCC 25922, Staphylococcus aureus ATCC 25923, and Listeria monocytogenes NCTC 5105 were used as indicator strains. After solidification, the cylinders were removed, and aliquots of 24 h-old Enterococcus cultures (50 μL) were poured into wells and kept at 4°C for 2 h. Then the Petri dishes were incubated for 24 h at 37°C. A clear zone around the wells was accepted as a positive result. Each determination was carried out in duplicates.
2.3. Screening of the enterocin-encoding genes by PCR
The total DNA was extracted from overnight cultures as described previously (Yost and Nattress, 2000). Briefly, 1.5 mL of overnight culture was centrifugated at 5000 rpm for 5 min, and the cell pellet was resuspended in 1 mL of sterile distilled water and centrifugated at the same condition. The resulting bacterial pellet was resuspended in 50 μL of sterile distilled water, heated at 95°C for 5 min, and centrifuged at 13000 rpm for 15 min. Then the supernatant was used as a template DNA in the PCR reaction. To investigate the occurrence of enterocin structural genes among Enterococcus strains, we performed a screening using primers specific to three well-known enterocin genes: for enterocin A – the forward primer 5´-GGTACCACTCATAGTGGAAA-3′ and reverse primer 5´-CCCTGGAATTGCTCCACCTAA-3′; for enterocin B – the forward primer 5´-СAAAATGTAAAAGAATTAAGATCG-3′ and reverse primer 5´-AGAGTATACATTTGCTAACCC-3′; for enterocin P – the forward primer 5´-GCTACGCGTTCATATGGTAAT-3′ and reverse primer 5´-TCCTGCAATATTCTCTTTAGC-3′. PCR amplification conditions were as follows: initial denaturation for 5 min at 95°C, 30 cycles of denaturation for 30 s at 95°C, annealing at 58°C (for the primers of entA) or 56°C (for the primers of entB and entP) for 30 s and extension at 72°C for 30 s, and a final elongation step of 5 min at 72°C (De Vuyst et al., 2003). The amplified PRC products were examined using 1.5% (w/v) agarose gel in a TBE buffer containing ethidium bromide with a DNA ladder GeneRuler 100 bp (Fermentas, United States). Fragments with the specific expected sizes were recorded as positive results for each bacteriocin-encoding gene for each Enterococcus strain. Strain E. durans 140D UCM B-2530 (UCM – Ukrainian Collection of Microorganisms) harboring entA, entB, and entP genes (GenBank accession numbers OP740237, OP740236, and OP690158, respectively) was used as a positive control.
2.4. Effect of enzyme and heat treatment on antagonistic activity of cell-free supernatants
Cell-free culture supernatants (CFS) were obtained by centrifugation of 250 mL of the overnight cultures at 5,000 rpm for 10 min and sterilized through 0.45 μm pore-size filters. Antagonistic activity of CFS was determined using L. monocytogenes and Enterococcus spp. strains as indicators. The inhibition activity of CFS was expressed in arbitrary units per milliliter (AU/mL), which was defined as the highest reciprocal dilution of CFS that gave inhibition of indicator strains. For exclusion of antibacterial action of organic acids, CFSs were adjusted to pH 6.5 ± 0.1 using 1 N NaOH and sterilized. The neutralized and filtered supernatant was assayed for antagonistic activity against L. monocytogenes and E. faecalis 888 strains by agar well diffusion assay.
CFSs were treated with the α-amylase, lipase, α-chymotrypsin, proteinase K, pepsin, and trypsin, each at a final concentration of 1 mg/mL. After 1 h of incubation at 37°C, enzymes were inhibited by heating at 100°C for 5 min. Antimicrobial activity against L. monocytogenes and E. faecalis 888 was examined using the agar well diffusion assay. The untreated cell-free supernatants were used as controls. The CFSs were heated at 100°C for 10 and 20 min and in an autoclave at 121°C, for 20 min, and the antagonistic activity was determined. Unheated samples were used as controls. Each determination was carried out in duplicates.
2.5. Partial purification of BLIS
Enterococcus strains were grown in Medium I at 37°C for 24 h. The bacterial culture was centrifuged at 8000 rpm for 15 min and the cell pellet was discarded. The supernatant was adjusted to pH 6.5 and filtered through a 0.45 μm filter. Solid ammonium sulfate was gently added to the supernatant (to a final saturation of 20, 40, 60, 80%), stirred, and held overnight at 4°C. Precipitates were collected by centrifugation at 8000 rpm for 30 min at 4°C and resuspended in 2 mL of 20 mmol sodium phosphate buffer (pH 7.0). The inhibitory activity of protein precipitates against E. faecalis 888 and L. monocytogenes was evaluated. Each determination was carried out in triplicates. The dissolved partially purified bacteriocins were desalted and freeze dried for further investigation.
2.6. Statistical analysis
Cluster analysis, a one-way analysis of variance (ANOVA), multiple mean comparisons using LSD test, and graphing procedures were carried out with the Statistika 7.0 software (Systat Inc., United States). The correlation between antagonistic action and viable cell count during growth in different media was analyzed by using Pearson’s correlation coefficient tests. All data are presented as the mean ± standard deviation of means. Statistical significance was defined as p < 0.05.
3. Results
3.1. Initial antagonistic activity screening
Using the agar well diffusion assay the antagonistic activity of 475 Enterococcus strains against P. aeruginosa, E. coli, S. aureus, and L. monocytogenes strains was determined. The presence of a clear zone around the well was assessed as a positive result. The medium I with 2% sucrose was used for initial screening. Only 34 (7%) Enterococcus strains produced metabolites with antagonistic action against at least one indicator strain used. Thus, these Enterococcus strains and 3 strains that had no antagonistic activity were selected for further investigation. Among them, 21 Enterococcus strains inhibited the growth of S. aureus, 13 strains – the growth of P. aeruginosa, 9 Enterococcus strains showed activity against E. coli, and only 3 strains – against L. monocytogenes. It should be noted, that zones of growth inhibition were larger for the L. monocytogenes strain (16–18 mm) compared to other indicator strains (8–10 mm). The antibacterial action against S. aureus and E. coli strains exhibited during growth in the medium I was lost after replacement of sucrose with molasses (Medium II) or their exclusion from the medium (Medium III). The strains E. faecalis 266 and E. durans 300a remained active against P. aeruginosa, and three strains – against L. monocytogenes. At the same time, 5 strains remained active against E. coli, 3 strains – against S. aureus, and 4 – against P. aeruginosa when grown in 2% molasses. We also used the MRS medium because MRS is the most commonly used medium for LAB antagonistic activity detection. All 37 Enterococcus strains, selected during the initial screening, showed antibacterial action against P. aeruginosa, 14 – against E. coli, 5 – against L. monocytogenes, and none – against S. aureus. In addition, when grown in media II, III IV, and MRS broth some strains showed antibacterial action that was not observed during the initial screening.
Cluster analysis was applied for analysis of the antagonistic activity spectra of the selected Enterococcus strains during growth on different media (Figure 1). Analysis of antibacterial activity in five media used revealed, that some strains had the same spectra of growth inhibition during growth in different media. Four Enterococcus strains showed only anti-P. aeruginosa activity.
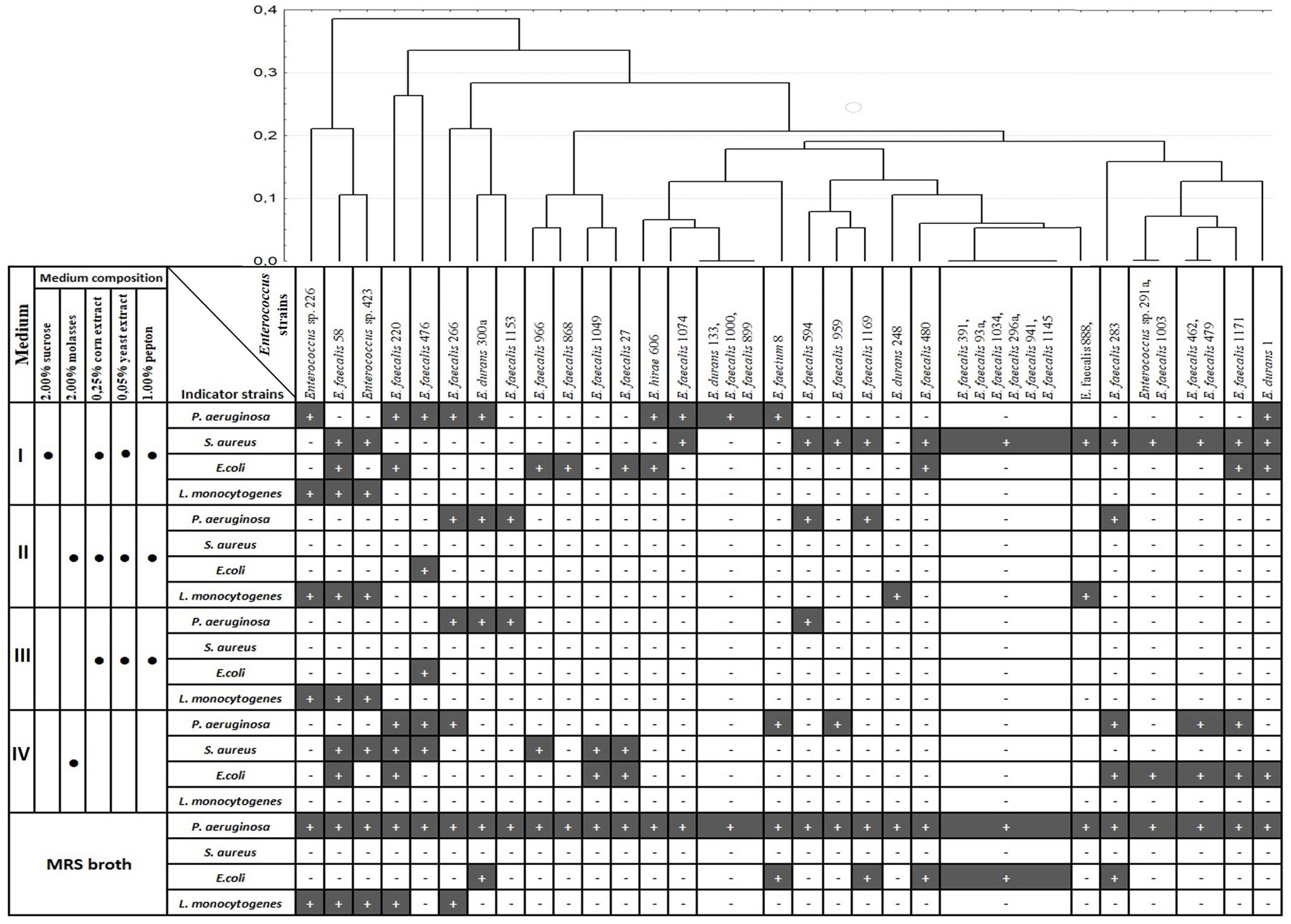
Figure 1. Dendrogram of antagonistic activity spectra of 37 Enterococcus strains depending on growth medium. Clustering was performed with Percent disagreement and UPGMA. Black bar (+) indicates the growth inhibition of indicator strain; medium component is included (•).
The viable cell count for the five media used is presented in Figure 2. The majority of the Enterococcus strains (54%) had higher viable cell counts in the MRS medium, and 62% of the strains had the lowest number of CFU ml−1 in 2% molasses (medium IV). Correlation analysis between antagonistic action and viable cell counts (CFU/ml) during growth in different media revealed that only one weak positive correlation (r = 0.34) was observed between CFU/ml value and ability to inhibit P. aeruginosa during growth on 2% molasses (Medium IV) (Table 1). A strong positive correlation was observed between anti-listerial activity during growth on different media (r = 0.54–1.00). Activities against P. aeruginosa and E. coli correlated between Medium II and Medium III (r = 0.79 and 0.70 respectively). Antagonistic action against S. aureus during growth in 2% molasses (Medium IV) weakly correlated (r = 0.3) with antagonistic action against E. coli during growth on Media I and II and L. monocytogenes during growth on Medium II and III (Table 1).
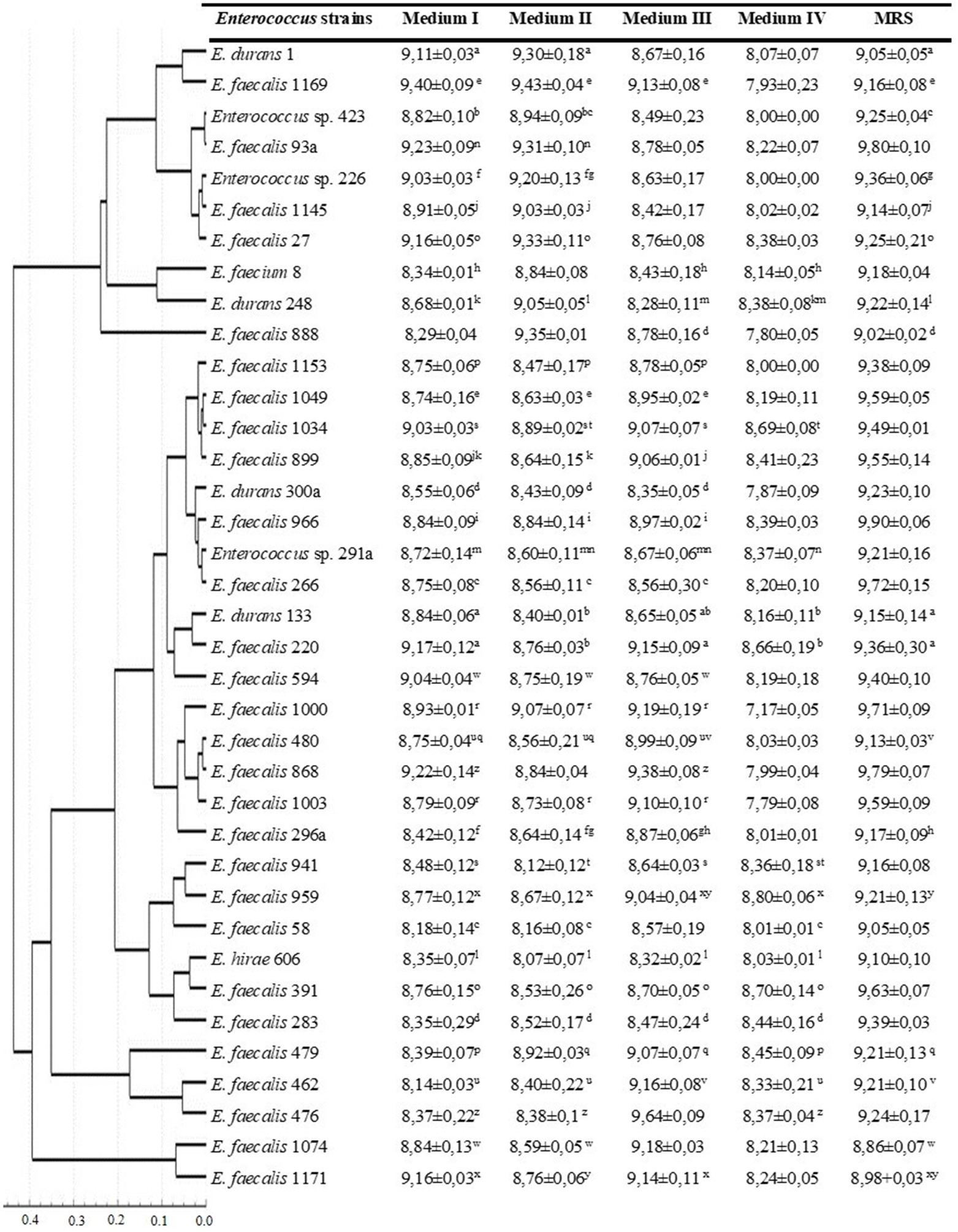
Figure 2. Viable cell counts of Enterococcus strains during growth in different media after 24 h of cultivation. Dendrogram based on viable cell counts (CFU ml−1) of Enterococcus strains after 24 h of cultivation in different media. Clustering was performed with Statistika 7.0, Pearson’s correlation coefficient and UPGMA. Mean values with different superscript letters within each row are significantly different (p < 0.05).
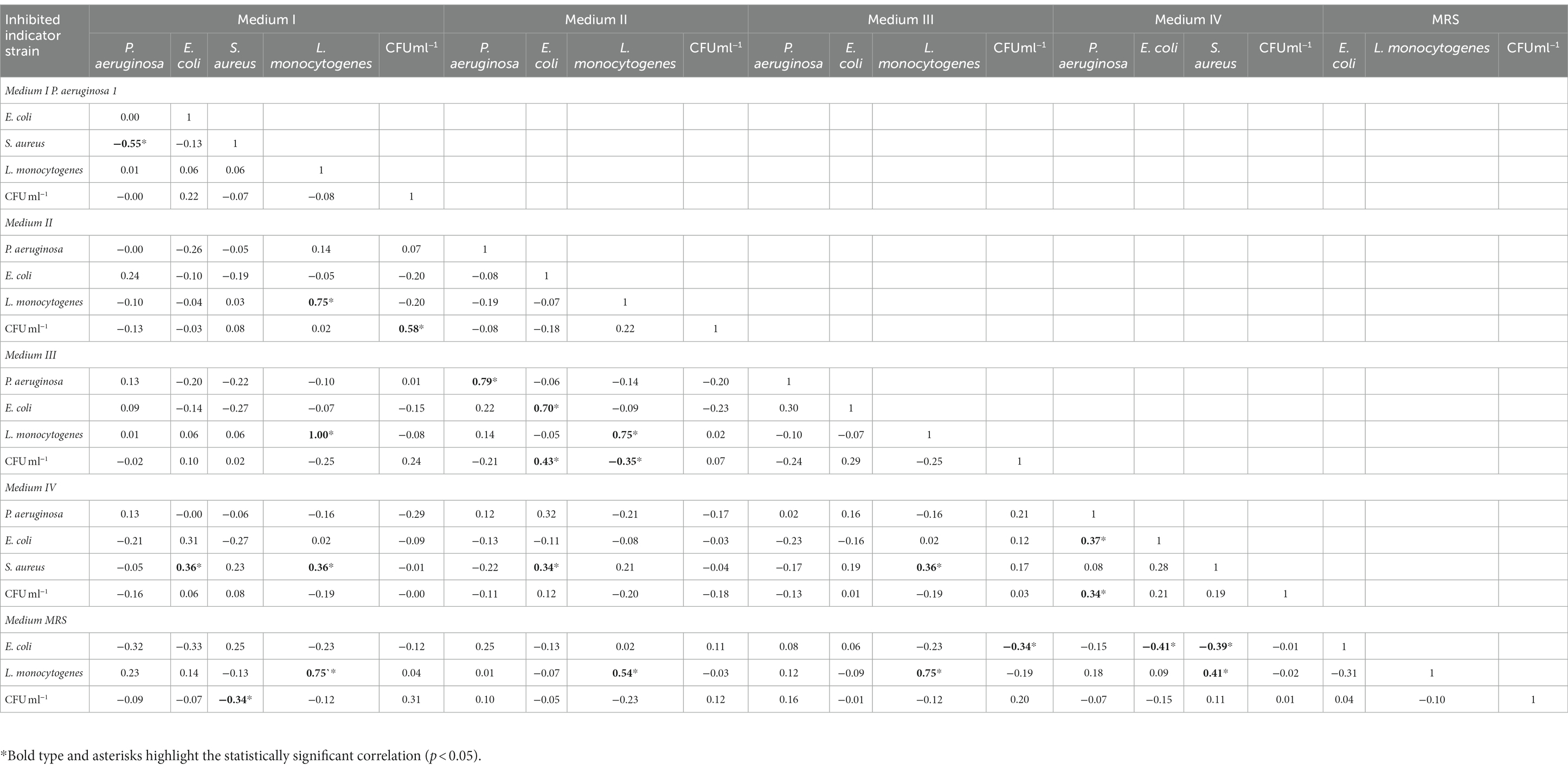
Table 1. Correlation coefficients between viable cell count of Enterococcus strains (CFU ml−1) and ability of enterococci to inhibit growth of indicator strains depending on growth medium.
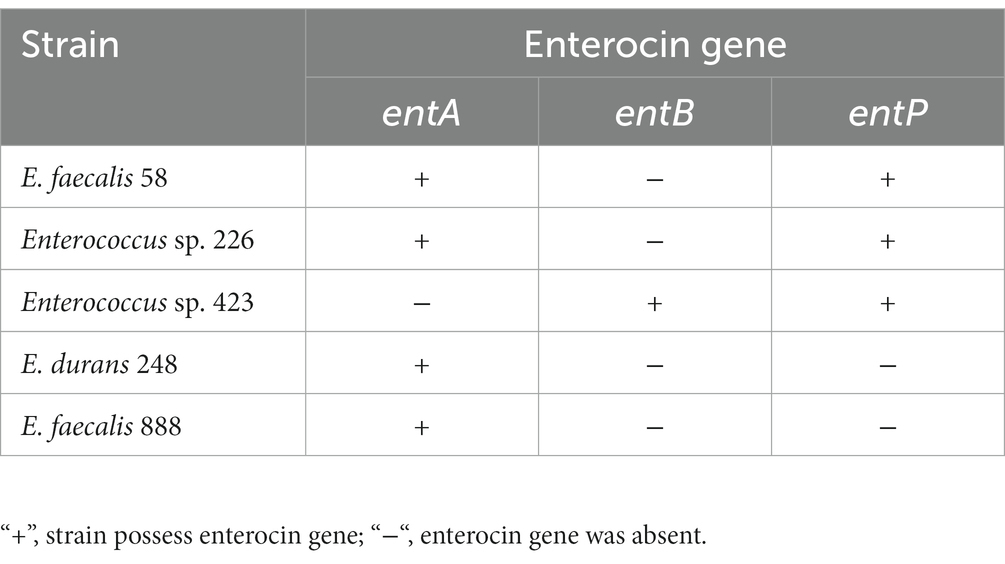
3.2. Detection of enterocin genes
Among 37 tested, enterocin genes were observed in 5 Enterococcus strains (Table 2). The structural genes of enterocins A and P were found in E. faecalis 58 and Enterococcus sp. 226 strains, enterocins B and P – in Enterococcus sp. 423, and enterocin A – in E. faecalis 888 and E. durans 248 strains.
3.3. Antagonistic activity against Enterococcus spp. strains
Each of the five Enterococcus strains harboring entA, entB, or entP genes was tested for antagonistic activity against 37 Enterococcus strains, that were selected after initial screening. E. faecalis 58, Enterococcus sp. 226, and Enterococcus sp. 423 showed antibacterial action against 27 E. faecalis strains (including E. faecalis 888 with entA gene detected by PCR) and 1 E. hirae strain. Four E. durans, 1 E. faecium, and 1 Enterococcus sp. strains were not sensitive. Furthermore, E. faecalis 58, Enterococcus sp. 226 strains, and Enterococcus sp. 423 did not inhibit the growth of each other and were resistant to their antimicrobial metabolites. These three strains were selected for further investigation of the nature of their antibacterial metabolites.
3.4. Antagonistic activity of cell-free supernatants
Neutralized cell-free supernatants (CFS) of E. faecalis 58, Enterococcus sp. 226, and Enterococcus sp. 423 strains expressed no inhibition of the growth of P. aeruginosa, S. aureus, or E. coli strains, but showed antagonistic activity (the same as overnight cultures with bacterial cells) against L. monocytogenes and E. faecalis 888 strains, that also was chosen as the indicator strain. Antagonistic activity of CFSs was observed to be at 51200 AU/mL against L. monocytogenes strain and 1,600–3,200 AU/mL against E. faecalis 888 strain.
3.5. Nature of bacteriocin-like substances
The antibacterial metabolites produced by E. faecalis 58 Enterococcus sp. 226 and Enterococcus sp. 423 strains were found to be heat stable as they could resist heat treatment at 100°C for 20 min and at 121°C for 20 min, although the inhibitory activity of the cell-free supernatants was lower after autoclaving (Table 3). The inhibitory activity was completely lost after treatments of the cell-free supernatants with proteolytic enzymes proteinase K, trypsin, pepsin, and pancreatin. The activity was not affected by treatment with α-amylase and lipase.
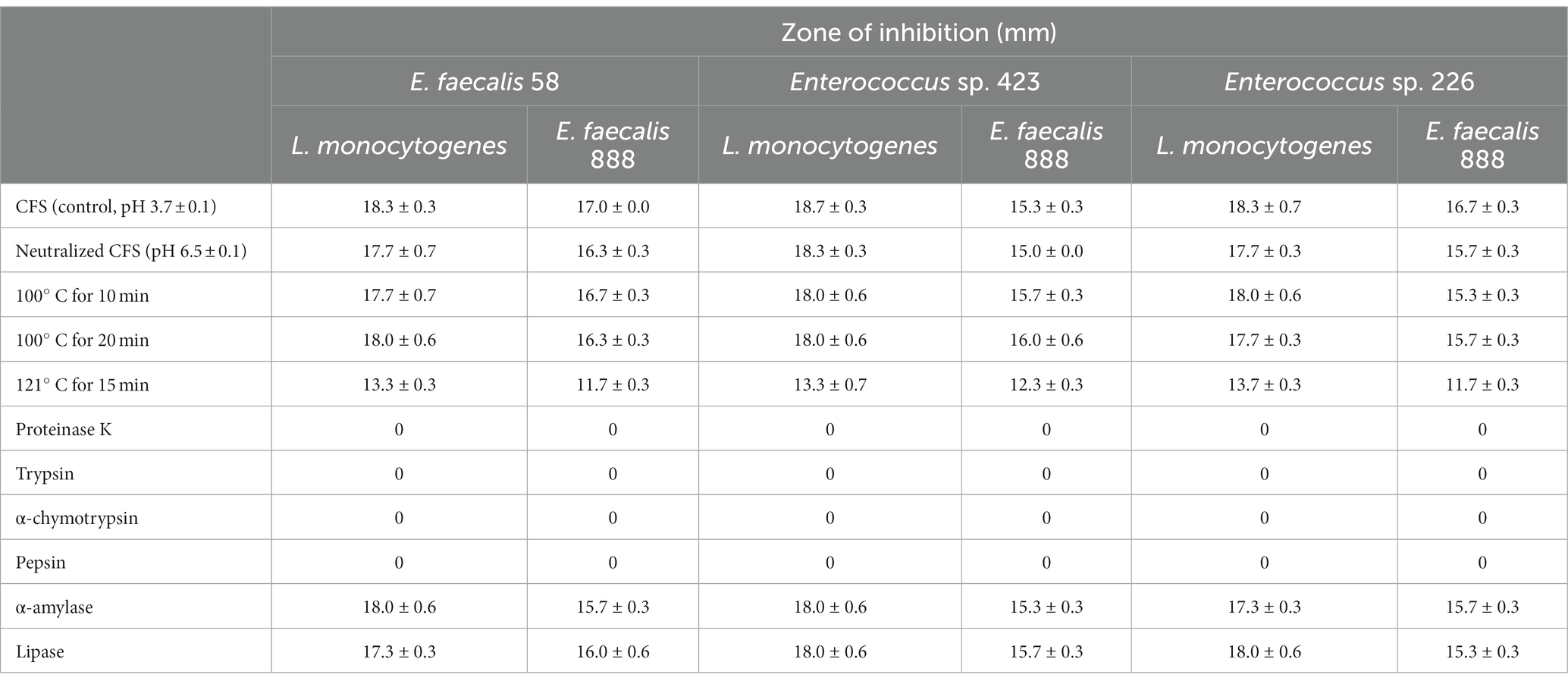
Table 3. Effect of neutralization, heat and enzyme treatments on inhibitory activity of cell-free supernatants, obtained after growth of Enterococcus strains in Medium I, against E. faecalis 888 and L. monocytogenes.
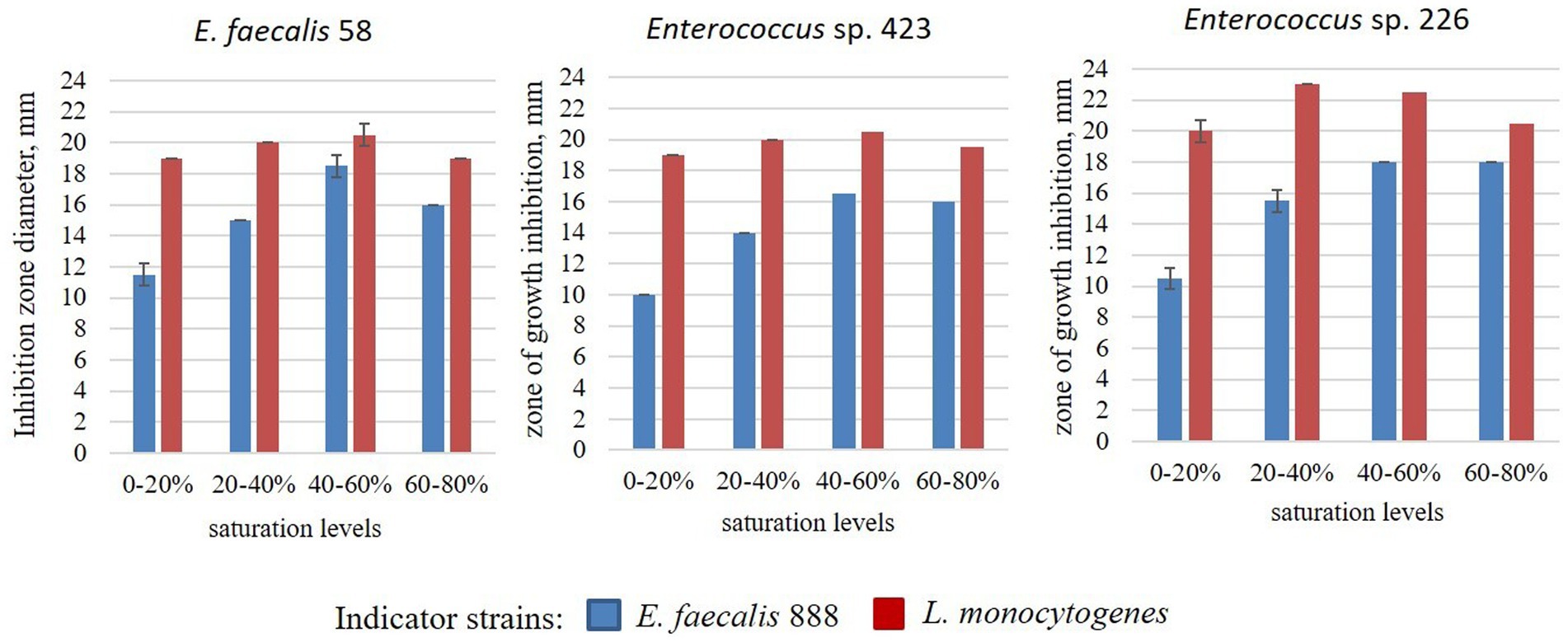
3.6. Partial purification of BLIS
The size of the inhibition zones of the crude extracts of enterocins obtained by ammonium sulfate precipitation is presented in Figure 3. The antagonistic action of the crude extracts against E. faecalis 888 increased with increasing of ammonium sulfate concentration to 40–60%. At the same time, antagonistic action against L. monocytogenes was stronger compared to E. faecalis 888, it reached the maximum at 20–40% ammonium sulfate and did not change dramatically up to 80%.
4. Discussion
In this work, screening of potentially bacteriocinogenic strains among dairy-derived enterococci was carried out. At the initial screening stage, we evaluated the antagonistic activity of bacterial cultures that were grown in a medium containing sucrose, CSL, peptone, and yeast extract. Only 34 (from 475) Enterococcus strains were selected because they exhibited antagonistic action against at least one of the indicator strains. To our knowledge, this is the first report about using a low-cost medium for screening of bacteriocinogenic enterococci, because commonly MRS or other commercial media are used for initial screening. As was shown in many studies, MRS broth is the optimal medium for bacteriocin production by Enterococcus strains compared to other complex media, such as BHI broth or M17 broth (De Kwaadsteniet et al., 2005; Yang et al., 2018). But commercial growth media are too costly for industrial production. We can assume that the use of cheap media suitable for large-scale cultivation instead of MRS for screening will make it possible to avoid the stage of medium composition optimization to reduce the cost of bacteriocin production. In our work, we use four media containing sucrose, molasses, and CSL in different combinations. All these components were used for bacteriocin production by different Enterococcus strains and other LAB (Coetzee et al., 2007; Bali et al., 2014). The main carbohydrate in molasses is sucrose but its concentration may vary (Audisio et al., 2001), so for initial screening sucrose was used as a carbon source (Medium I). As was shown, sucrose is one of the well-preferred carbon sources for Nisin production by L. lactis strains (Parente and Ricciardi, 1999). Yeast extract and peptone also were added to the media, because these components are required for bacteriocin production, as previously shown (Mirhosseini and Emtiazi, 2011; Bali et al., 2014).
During growth in media containing sucrose or molasses as the carbon source, most enterococci strains had compatible viable cell counts. The absence in the culture medium of the main source of carbohydrates (sucrose or molasses) did not affect the growth of 75% of strains. During growth in 2% molasses, only 40,5% of strains had the same viable cell counts as in at least one of the other media used, with exception of MRS. Overall, the growth rate did not correlate with the growth inhibition of indicator strains.
Detection of genes encoding bacteriocin production by PCR is a commonly used method for screening of bacteriocin-producing LAB strains. The analysis of literature data showed that the presence of enterocin genes entA, entB, entP, entL50A, entL50B, entAS-48, cyl, or bac31 is most often determined in enterococci by PCR analysis (Vandera et al., 2018, 2019; Furlaneto-Maia et al., 2020; Lauková et al., 2020a). According to the results obtained by the authors, genes of enterocin A, B, and P are most often detected in enterococci isolated from different econiches (Ogaki et al., 2016; Vandera et al., 2018; Kubašová et al., 2020). None of the Enterococcus strains analyzed by Vandera et al. (2018) possessed entL50A, entL50B, entAS-48, or bac31 genes. Only the gene of enterocin P was detected in strain E. lactis Q1 using PCR amplifications with primers specific for genes entA, entB, entP, LB50A, and LB50B (Braïek et al., 2017). The structural enterocin genes entA, entB, entP, and entX were the most frequently observed in Enterococcus spp. isolated from food and environment, but genes encoding for enterocin L50A/B, Q, K, 31, and AS48 were not found (Furlaneto-Maia et al., 2020). Thus, in the present study, we focused on screening the most frequently occurring enterocin genes entA, entB, and entP. Enterocin typing by specific PCR revealed that only 5 from 37 antagonistic Enterococcus strains harbored the most widespread enterocin structural genes entA, ent B, and entP. According to data of literature, enterocins A (Aymerich et al., 1996), B (Casaus et al., 1997), and P (Cintas et al., 1997) are produced by Enterococcus strains isolated from different sources (Cocolin et al., 2007; Ogaki et al., 2016; Vandera et al., 2018; Furlaneto-Maia et al., 2020). Enterocins A, B, and P had a broad and very similar inhibitory spectrum against Gram-positive bacteria, such as Listeria spp., Clostridium spp., Enterococcus spp., and Staphylococcus aureus, but did not inhibit the growth of Gram-negative indicator strains (Aymerich et al., 1996; Casaus et al., 1997; Cintas et al., 1997). Multiple-enterocin-producing enterococci are also frequently found in raw milk and artisanal dairy products (Furlaneto-Maia et al., 2020). Strains Enterococcus sp. 423, 226, and E. faecalis 58 harbor a combination of two enterocin genes (entB/entP and entA/entP, respectively), and exhibit anti-listerial and anti-enterococcal activity, CFSs of these stains retained antagonistic activity after neutralization. Thus, the activity spectra of E. faecalis 58, Enterococcus sp. 226 and 423 correspond to the data previously presented by other authors regarding the activity of enterocins A, B, and P (Aymerich et al., 1996; Casaus et al., 1997; Cintas et al., 1997). But, neutralized CFSs did not inhibit the growth of S. aureus indicator strain used in our study. These results are in agreement with previous studies, which show that the growth of 38 Staphylococcus spp. strains was not inhibited by crude bacteriocin of E. faecium EF9a, which possessed genes for enterocin A, B, P, and LB50 (Lauková et al., 2020). Antimicrobial metabolites produced by E. faecalis 58, Enterococcus sp. 226 and 423 strains are thermostable as they remained active after heating at 100°C for 20 min and lost activity after autoclaving (121°C, 15 min) only on 25%. This is in agreement with the results of other authors, who also reported about enterocins, that withstood heat treatment at 100°C and autoclaving (Kumar et al., 2010). At the same time, resistance to autoclaving may vary among described enterocins, some of them lost activity after autoclaving at 121°C (Alvarado et al., 2005). The loss of antibacterial activity of CFS after treatment with proteinase K, α-chymotrypsin, and trypsin was observed. Lipase and α-amylase did not affect the antimicrobial activity of CFS, indicating that the antimicrobial metabolites are proteinaceous in nature and do not depend on the lipid or carbohydrate moiety. These results agreed with previously reported data concerning enzymatic treatment sensitivity of studied enterocins (Du Toit et al., 2000; Cocolin et al., 2007; Al-Seraih et al., 2017), so we can assume that these strains are potential producers of bacteriocin-like substances (BLIS). We did not determine the effect of pH on CFS antagonistic activity in this study. However, we can assume that the antimicrobial metabolites are active at least in the pH range of 3.7–6.5 since there was no difference in the activity of the CFSs before and after neutralization. Enterococcus sp. 423, 226, and E. faecalis 58 strains were insensitive to their own CFS. Resistance in the bacteriocin-producing cells may indicate the expression of immunity peptides along with bacteriocin (Bastos et al., 2015; Teso-Pérez et al., 2021). Cross-resistance in the E. faecalis 58, Enterococcus sp. 423, and 226 strains may indicate a strong similarity of the BLIS produced by these strains.
No anti-listerial activity was observed only during the growth of ent-genes possessing Enterococcus sp. 423, 226, and E. faecalis 58 strains in 2% molasses, although in some cases viable cell counts were compatible with other media used. The absence of antagonistic action upon growth in 2% molasses was presumably attributable to the insufficiency of nutrients for enterocin production. This result is in agreement with other studies, that reported the absence of enterocins production when Enterococcus strains were grown in molasses (Todorov and Dicks, 2005; Coetzee et al., 2007). Similar to this result, authors have shown that E. faecium strains grew in cheese whey with no enterocin A production and produced maximum bacteriocin in cheese whey with supplemented of yeast extract (Mirhosseini and Emtiazi, 2011). At the same time, selected Enterococcus strains showed anti-listerial activity during growth in the medium that contains only CSL, peptone, and yeast extract. Although the сorn steep liquor is mainly used as a source of nitrogen, it also contains carbohydrates and other nutrients, in an amount sufficient for the growth and synthesis of bacteriocins (Coetzee et al., 2007). Thus, the production at least of enterocin P by strains E. faecalis 58, Enterococcus sp. 423, and Enterococcus sp. 226 when growing in four of five media used may be confirmed since the anti-listerial activity was detected in their cell-free supernatants. Furthermore, anti-enterococcal activity also was detected, CFSs inhibited the growth of E. faecalis indicator strains, but not E. faecium and E. durans strains. At the same time, enterocins B3A-B3B and B20A-B20B produced by E. faecalis strains were not active against the E. faecalis ATCC 29212, as was shown by authors (Al-Seraih et al., 2017).
Strains E. faecalis 888 and E. durans 248 possess only the entA gene and have anti-listerial activity only when grown in Medium II. Therefore, the entA gene in these strains was expressed only when growing on this medium. It is also possible that the expression also occurred in other media but in insufficient quantity for detection by well diffusion assay. As was shown by authors, E. faecium strains that possessed only the entA gene showed remarkably weaker anti-listerial action compared with isolates that possessed entA-entB-entP or entA-entB genes (Vandera et al., 2018). Strain E. faecalis 888 was susceptible to CFSs of Enterococcus sp. 423, 226, and E. faecalis 58, but strain E. durance 248 was not. Therefore, we cannot rule out the possibility that the antilisterial activity could be mediated by other bacteriocins, whose genes were not determined in this study. Furthermore, anti-listerial activity was detected in E. faecalis 220 and 266 strains when growing in MRS broth, but entA, entB, or entP genes were not detected in these strains. As was shown by the authors, E. lactis strains that harboured the entP gene, not showed antibacterial activity against L. monocytogenes (Morandi et al., 2013). Thus, the results of our study once again corroborate the data of other authors, that the medium composition has a great influence on LAB bacteriocin production (Audisio et al., 2001), and the most effective approach is to use both microbiological methods and screening for the presence of enterocin genes using PCR analysis.
It should be noted, that no antibacterial activity was observed against P. aeruginosa, E. coli, and S. aureus after centrifugation and removal of cells from bacterial cultures. The loss of antilisterial activity by filter-sterilized supernatants of Enterococcus strains the authors explained by the removal of all viable cells from broth supernatants (Vandera et al., 2019). The same observation about the need for cell-to-cell contact between the producer strain and the indicator strain was reported by other authors (Saraoui et al., 2016). This fact is of interest and could be attributed to the presence of other antimicrobial metabolites strongly associated with the bacterial cell surface. This may also be evidenced by smaller growth inhibition zones of these indicator strains compared to L. monocytogenes during initial screening. The levels of acid production (titrable acidity) were comparable in different strains (data not shown), so the antagonistic action is unlikely to be mediated by organic acid. The fact that some Enterococcus strains showed different spectra of activity when grown on five media may also indicate the production of specific antimicrobial metabolites. The strains E. faecalis 266 and 220 were active against L. monocytogenes when grown on MRS broth. At the same time, strain E. faecalis 266 showed activity only against P. aeruginosa on four other media, while strain E. faecalis 220 on the 2% molasses was active against P. aeruginosa, E. coli, and S. aureus. The absence of antibacterial activity in cell-free neutralized supernatants also may indicate the sensitivity of the antimicrobial metabolites to pH, as was shown by authors for some bacteriocins (Vandera et al., 2018). For example, the strong activity of Nisin at acidic pH is considerably reduced at a neutral pH range (Noutsopoulos et al., 2017). Thus, further research is needed to clarify this issue.
Although enterococci are widespread in fermented foods and used as a starter or protective cultures, some Enterococcus strains may harbor recognized virulence determinants and have caused nosocomial infections in humans (Ogier and Serror, 2008). Our previous preliminary study showed that autochthonous enterococci isolated from Ukrainian traditional dairy products can produce tyramine (95,4% of strains), 20% of strains exhibited β-hemolytic activity, and 56,76% were resistant to 5–11 antibiotics (Garmasheva et al., 2018). In particular, among seven potential bacteriocin producers selected in the present study, three strains (Enterococcus sp. 226, Enterococcus sp. 423, and E. durans 248) were β-hemolytic, and all strains were able to produce tyramine. Overall, seven Enterococcus strains exhibited susceptibility to 13 of 17 of the most clinically relevant antibiotics tested, including benzylpenicillin, vancomycin, and teicoplanin, with the exclusion of E. faecalis 266 and E. faecalis 220 that were resistant to benzylpenicillin and vancomycin, respectively. Further in-depth analysis of other putative virulence factors must be conducted to evaluate the potential for practical use of these bacteriocinogenic Enterococcus strains. The presence of virulence factors completely excludes the use of the bacteriocinogenic Enterococcus strains as a starter or protective culture for inoculation of raw materials or products, respectively. But such Enterococcus strains can be used as producers of bacteriocins followed by their isolation from the culture fluid and purification. It was shown, that CFS and semi-purified BLIS produced by Enterococcus hirae ST57ACC have no cytotoxic effect against HT-29 cells (Cavicchioli et al., 2019). Cell-free supernatant of bacteriocinogenic E. faecium TJUQ1 strain was used for the preparation of composite film with antibacterial activity (Feng et al., 2021). But the safety of each purified bacteriocin needs to be assessed before use in the food industry.
5. Conclusion
The present work is, to our knowledge, the first report on the screening of bacteriocinogenic Enterococcus strains isolated from Ukrainian traditional dairy products using low-cost by-product-based media. In this study, three Enterococcus strains that harbor genes of enterocins and produce BLIS with anti-listerial and anti-enterococcal activity were selected. We can assume that the use of the method described here will make it possible to exclude the stage of selecting and optimizing the medium for the large-scale production of bacteriocins. Strains E. faecalis 58, Enterococcus sp. 423, and Enterococcus sp. 226 are promising candidates for practical use as producers of bacteriocins with inhibitory activity against L. monocytogenes using molasses and steep corn liquor as cheap sources of carbon and nitrogen, that can significantly reduce the cost of industrial bacteriocin production. Further studies will be required to determine the dynamic of bacteriocin production, its structure, and mechanisms of antibacterial action.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
IG designed and performed the experiments, analyzed the results, and wrote the manuscript. LO performed the experiments. All authors contributed to the article and approved the submitted version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Al-Seraih, A., Belguesmia, Y., Baah, J., Szunerits, S., Boukherroub, R., and Drider, D. (2017). Enterocin B3A-B3B produced by LAB collected from infant faeces: potential utilization in the food industry for Listeria monocytogenes biofilm management. Antonie Van Leeuwenhoek 110, 205–219. doi: 10.1007/s10482-016-0791-5
Alvarado, C., García-Almendárez, B. E., Martin, S. E., and Regalado, C. (2005). Anti-Listeria monocytogenes bacteriocin-like inhibitory substances from Enterococcus faecium UQ31 isolated from artisan Mexican-style cheese. Curr. Microbiol. 51, 110–115. doi: 10.1007/s00284-005-4549-y
Audisio, M. C., Oliver, G., and Apella, M. C. (2001). Effect of different complex carbon sources on growth and bacteriocin synthesis of Enterococcus faecium. Int. J. Food Microbiol. 63, 235–241. doi: 10.1016/s0168-1605(00)00429-3
Aymerich, T., Holo, H., Håvarstein, L. S., Hugas, M., Garriga, M., and Nes, I. F. (1996). Biochemical and genetic characterization of enterocin a from E. faecium, a new antilisterial bacteriocin in the pediocin family of bacteriocins. Appl. Environ. Microbiol. 62, 1676–1682. doi: 10.1128/aem.62.5.1676-1682
Bali, V., Bera, M. B., and Panesar, P. S. (2014). Utilization of agro-industrial byproducts for bacteriocin production using newly isolated Enterococcus faecium BS13. Int. J. Agricul. Biosyst. Eng. 8, 562–566. doi: 10.5281/zenodo.1092922
Bastos, M. C. F., Coelho, M. L. V., and Santos, O. C. S. (2015). Resistance to bacteriocins produced by gram-positive bacteria. Microbiology 161, 683–700. doi: 10.1099/mic.0.082289-0
Braïek, O. B., Ghomrassi, H., Cremonesi, P., Morandi, S., Fleury, Y., Chevalier, P. L., et al. (2017). Isolation and characterisation of an enterocin P-producing Enterococcus lactis strain from a fresh shrimp (Penaeus vannamei). Antonie Van Leeuwenhoek 110, 771–786. doi: 10.1007/s10482-017-0847-1
Casaus, P., Nilsen, T., Cintas, L. M., Nes, I. F., Hernández, P. E., and Holo, H. (1997). Enterocin B, a new bacteriocin from Enterococcus faecium T136 which can act synergistically with enterocin a. Microbiology 143, 2287–2294. doi: 10.1099/00221287-143-7-2287
Cavicchioli, V. Q., Todorov, S. D., Iliev, I., Ivanova, I., Drider, D., and Nero, L. A. (2019). Physiological and molecular insights of bacteriocin production by Enterococcus hirae ST57ACC from Brazilian artisanal cheese. Braz. J. Microbiol. 50, 369–377. doi: 10.1007/s42770-019-00068-4
Chaves-López, C., Serio, A., Martuscelli, M., Paparella, A., Osorio-Cadavid, E., and Suzzi, G. (2011). Microbiological characteristics of kumis, a raditional fermented Colombian milk, with particular emphasis on enterococci population. Food Microbiol. 28, 1041–1047. doi: 10.1016/j.fm.2011.02.006
Choeisoongnern, T., Sirilun, S., Waditee-Sirisattha, R., Pintha, K., Peerajan, S., and Chaiyasu, C. (2021). Potential probiotic Enterococcus faecium OV3-6 and its bioactive peptide as alternative bio-preservation. Foods 10:2264. doi: 10.3390/foods10102264
Cintas, L. M., Casaus, P., Håvarstein, L. S., Hernández, P. E., and Nes, I. F. (1997). Biochemical and genetic characterization of enterocin P, a novel sec-depend bacteriocin from Enterococcus faecium P13 with a broad antimicrobial spectrum. Appl. Environ. Microbiol. 63, 4321–4330. doi: 10.1128/aem.63.11.4321-4330.1997
Cocolin, L., Foschino, R., Comi, G. M., and Fortina, G. (2007). Description of the bacteriocins produced by two strains of Enterococcus faecium isolated from Italian goat milk. Food Microbiol. 24, 752–758. doi: 10.1016/j.fm.2007.03.001
Coetzee, J. C., Todorov, S. D., Gorgens, J. F., and Dicks, L. M. (2007). Increased production of bacteriocin ST4SA by Enterococcus mundtii ST4SA in modified corn steep liquor. Ann. Microbiol. 57, 617–622. doi: 10.1007/BF03175363
Darbandi, A., Asadi, A., Ari, M. M., Ohadi, E., Talebi, M., Zadeh, M. H., et al. (2022). Bacteriocins: properties and potential use as antimicrobials. J. Clin. Lab. Anal. 36:e24093. doi: 10.1002/jcla.24093
De Kwaadsteniet, M., Todorov, S. D., Knoetze, H., and Diks, L. M. (2005). Characterization of a 3944 Da bacteriocin produced by Enterococcus mundtii ST15, with activity against gram positive and gram negative bacteria. Int. J. Food Microbiol. 105, 433–444. doi: 10.1016/j.ijfoodmicro.2005.03.021
De Vuyst, L., Foulquie Moreno, M. R., and Revets, H. (2003). Screening for enterocins and detection of hemolysin and vancomycin resistance in enterococci of different origins. Int. J. Food Microbiol. 84, 299–318. doi: 10.1016/S0168-1605(02)00425-7
Du Toit, M., Franz, C. M. A. P., Dicks, L. M. T., and Holzapfel, W. H. (2000). Preliminary characterization of bacteriocins produced by Enterococcus faecium and Enterococcus faecalis isolated from pig faeces. J. Appl. Microbiol. 88, 482–494. doi: 10.1046/j.1365-2672.2000.00986.x
Ermolenko, E. I., Desheva, Y. A., Kolobov, A. A., Kotyleva, M. P., Sychev, I. A., and Suvorov, A. N. (2019). Anti-influenza activity of enterocin B in vitro and protective effect of bacteriocinogenic enterococcal probiotic strain on influenza infection in mouse model. Probiotics Antimicrob. Proteins 11, 705–712. doi: 10.1007/s12602-018-9457-0
Farah, N., Mehdi, A., Soomro, S. I., Soomro, N. I., Tareb, R., Desmasures, N., et al. (2016). Draft Genome Sequence of Enterococcus mundtii QAUEM2808, Isolated from Dahi, a Fermented Milk Product. Genome Announc. 4, e00995–e00916. doi: 10.1128/genomeA.00995-16
Feng, Y., Yin, N., Zhou, Z., and Han, Y. (2021). Physical and antibacterial properties of bacterial cellulose films supplemented with cell-free supernatant enterocin-producing Enterococcus faecium TJUQ1. Food Microbiol. 99:103828. doi: 10.1016/j.fm.2021.103828
Foulquié Moreno, M. R., Sarantinopoulos, P., Tsakalidou, E., and De Vuyst, L. (2006). The role and application of enterococci in food and health. Int. J. Food Microbiol. 106, 1–24. doi: 10.1016/j.foodcont.2020.107425
Furlaneto-Maia, L., Ramalho, R., Rocha, K. R., and Furlaneto, M. C. (2020). Antimicrobial activity of enterocins against Listeria sp. and other food spoilage bacteria. Biotechnol. Lett. 2, 797–806. doi: 10.1007/s10529-020-02810-7
Garmasheva, I. L., Kovalenko, N. K., and Oleschenko, L. T. (2018). Resistance to antibiotics, decarboxylase and hemolytic activities of enterococci isolated from traditional dairy products. Mikrobiol. Z. 80, 3–14. doi: 10.15407/microbiolj80.01.003
Giraffa, G. (2003). Functionality of enterococci in dairy products. Int. J. Food Microbiol. 88, 215–222. doi: 10.1016/s0168-1605(03)00183-1
Kasimin, M. E., Shamsuddin, S., Molujin, A. M., Sabullah, M. K., Gansau, J. A., and Jawan, R. (2022). Enterocin: promising biopreservative produced by Enterococcus sp. Microorganisms 10:684. doi: 10.3390/microorganisms10040684
Kubašová, I., Diep, D. B., Ovchinnikov, K. V., Lauková, A., and Strompfová, V. (2020). Bacteriocin production and distribution of bacteriocin-encoding genes in enterococci from dogs. Int. J. Antimicrob. Agents 55:105859. doi: 10.1016/j.ijantimicag.2019.11.016
Kumar, M., Tiwari, S. K., and Srivastava, S. (2010). Purification and characterization of enterocin LR/6, a bacteriocin from Enterococcus faecium LR/6. Appl. Biochem. Biotechnol. 160, 40–49. doi: 10.1007/s12010-009-8586-z
Lauková, A., Focková, V., and Simonová, M. P. (2020). Enterococcus mundtii isolated from Slovak raw goat milk and its bacteriocinogenic potential. Int. J. Environ. Res. Public Health 17:9504. doi: 10.3390/ijerph17249504
Lauková, A., Strompfová, V., and Simonová, M. P. (2020a). Enterocin structural gene screening in enterococci from Pannon white breed rabbits. Probiotics. Antimicrob. Proteins. 12, 1246–1252. doi: 10.1007/s12602-020-09641-y
Lee, J. S., Park, S. W., Lee, H. B., and Kang, S. S. (2021). Bacteriocin-like inhibitory substance (BLIS) activity of Enterococcus faecium DB1 against biofilm formation by Clostridium perfringens. Probiotics Antimicrob. Proteins 13, 1452–1457. doi: 10.1007/s12602-021-09813-4
López, R. L., García, M. T., Abriouel, H., Omar, N. B., Grande, M. J., Martínez-Cañamero, M., et al. (2007). Semi-preparative scale purification of enterococcal bacteriocin enterocin EJ97, and evaluation of substrates for its production. J. Ind. Microbiol. Biotechnol. 34, 779–785. doi: 10.1007/s10295-007-0254-0
Metsoviti, M., Paramithiotis, S., Drosinos, E. H., Skandamis, P. N., Galiotou-Panayotou, M., and Papanikolaou, S. (2011). Biotechnological valorization of low-cost sugar-based media for bacteriocin production by Leuconostoc mesenteroides E131. New Biotechnol. 28, 600–609. doi: 10.1016/j.nbt.2011.03.004
Mirhosseini, M., and Emtiazi, G. (2011). Optimisation of enterocin a production on a whey-based substrate. World Appl. Sci. J. 14, 1493–1499. doi: 10.1016/j.idairyj.2008.02.001
Morandi, S., Silvetti, T., and Brasca, M. (2013). Biotechnological and safety characterization of Enterococcus lactis, a recently described species of dairy origin. Antonie Van Leeuwenhoek 103, 239–249. doi: 10.1007/s10482-012-9806-z
Nami, Y., Bakhshayesh, R. V., Jalaly, H. M., Lotfi, H., Eslami, S., and Hejazi, M. A. (2019). Probiotic properties of Enterococcus isolated from artisanal dairy products. Front. Microbiol. 10:300. doi: 10.3389/fmicb.2019.00300
Ng, Z. J., Zarin, M. A., Lee, C. K., and Tan, J. S. (2020). Application of bacteriocins in food preservation and infectious disease treatment for humans and livestock: a review. RSC Adv. 10, 38937–38964. doi: 10.1039/d0ra06161a
Noutsopoulos, D., Kakouri, A., Kartezini, E., Pappas, D., Hatziloukas, E., and Samelis, J. (2017). Growth, nisA gene expression and in situ Nisin a activity of novel Lactococcus lactis subsp. cremoris co-starter culture in commercial hard cheese production. J. Food Prot. 80, 2137–2146. doi: 10.4315/0362-028X.JFP-17-245
Ogaki, M. B., Rocha, K. R., Terra, M. R., Furlaneto, M. C., and Furlaneto-Maia, L. (2016). Screening of the enterocin-encoding genes and antimicrobial activity in Enterococcus species. J. Microbiol. Biotechnol. 26, 1026–1034. doi: 10.4014/jmb.1509.09020
Ogier, J. C., and Serror, P. (2008). Safety assessment of dairy microorganisms: the Enterococcus genus. Int. J. Food Microbiol. 126, 291–301. doi: 10.1016/j.ijfoodmicro.2007.08.017
Parente, E., and Ricciardi, A. (1999). Production, recovery and purification of bacteriocins from lactic acid bacteria. Appl. Microbiol. Biotechnol. 52, 628–638. doi: 10.1007/s002530051570
Sameli, N., Skandamis, P. N., and Samelis, J. (2021). Application of Enterococcus faecium KE82, an enterocin A-B-P-producing strain, as an adjunct culture enhances inactivation of Listeria monocytogenes during traditional protected designation of origin Galotyri processing. J. Food Prot. 84, 87–98. doi: 10.4315/JFP-20-278
Saraoui, T., Fall, P. A., Leroi, F., Antignac, J.-P., Chéreau, S., and Pilet, M. F. (2016). Inhibition mechanism of Listeria monocytogenes by a bioprotective bacteria Lactococcus piscium CNCM I-4031. Food Microbiol. 53, 70–78. doi: 10.1016/j.fm.2015.01.002
Schillinger, U., and Lücke, F. K. (1989). Antibacterial activity of Lactobacillus sake isolated from meat. Appl. Environ. Microbiol. 55, 1901–1906. doi: 10.1128/aem.55.8.1901-1906.1989
Schirru, S., Favaro, L., Mangia, N. P., Basaglia, M., Casella, S., Comunian, R., et al. (2014). Comparison of bacteriocins production from Enterococcus faecium strains in cheese whey and optimised commercial MRS medium. Ann. Microbiol. 64, 321–331. doi: 10.1007/s13213-013-0667-0
Sharma, P., Kaur, S., Chadha, B. S., Kaur, R., Kaur, M., and Kaur, S. (2021). Anticancer and antimicrobial potential of enterocin 12a from Enterococcus faecium. BMC Microbiol. 21:39. doi: 10.1186/s12866-021-02086-5
Sosa, F. M., Parada, R. B., Marguet, E. R., and Vallejo, M. (2021). Utilization of agro-industrial byproducts for bacteriocin production using Enterococcus spp. strains isolated from Patagonian marine invertebrates. Curr. Microbiol. 79:16. doi: 10.1007/s00284-021-02712-5
Teso-Pérez, C., Martínez-Bueno, M., Peralta-Sánchez, J. M., Valdivia, E., Maqueda, M., Fárez-Vidal, M. E., et al. (2021). Enterocin cross-resistance mediated by ABC transport systems. Microorganisms 9:1411. doi: 10.3390/microorganisms9071411
Todorov, S. D., and Dicks, L. M. (2005). Optimization of bacteriocin ST311LD production by Enterococcus faecium ST311LD, isolated from spoiled black olives. J. Microbiol. 43, 370–374.
Vandera, E., Parapouli, M., Kakouri, A., Koukkou, A.-I., Hatziloukas, E., and Samelis, J. (2019). Structural enterocin gene profiles and mode of antilisterial activity in synthetic liquid media and skim milk of autochthonous Enterococcus spp. isolates from artisan Greek Graviera and Galotyri cheeses. Food Microbiol. 86:103335. doi: 10.1016/j.fm.2019.103335
Vandera, E., Tsirka, G., Kakouri, A., Koukkou, A., and Samelis, J. (2018). Approaches for enhancing in situ detection of enterocin genes in thermized milk, and selective isolation of enterocin-producing Enterococcus faecium from Baird-Parker agar. Int. J. Food Microbiol. 281, 23–31. doi: 10.1016/j.ijfoodmicro.2018.05.020
Yang, E., Fan, L., Yan, J., Jiang, Y., Doucette, C., Fillmore, S., et al. (2018). Influence of culture media, pH and temperature on growth and bacteriocin production of bacteriocinogenic lactic acid bacteria. AMB Expr. 8:10. doi: 10.1186/s13568-018-0536-0
Keywords: Enterococcus , bacteriocin, Listeria monocytogenes , screening, low-cost media
Citation: Garmasheva IL and Oleschenko LT (2023) Screening of bacteriocin-producing dairy Enterococcus strains using low-cost culture media. Front. Microbiol. 14:1168835. doi: 10.3389/fmicb.2023.1168835
Edited by:
Celia Costa Silva, Universidade dos Açores, PortugalReviewed by:
Franca Rossi, Experimental Zooprophylactic Institute of Abruzzo and Molise G. Caporale, ItalyTakeshi Zendo, Kyushu University, Japan
Copyright © 2023 Garmasheva and Oleschenko. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Inna L. Garmasheva, Z2FybWFzaGV2YS5pbEBnbWFpbC5jb20=
 Inna L. Garmasheva
Inna L. Garmasheva Ljubov T. Oleschenko
Ljubov T. Oleschenko