- 1Department of Plant and Microbial Biology, University of Zurich, Zurich, Switzerland
- 2School of Biosciences, University of Nottingham, Loughborough, United Kingdom
Since its discovery six decades ago, the predatory bacterium Bdellovibrio bacteriovorus has sparked recent interest as a potential remedy to the antibiotic resistance crisis. Here we give a comprehensive historical overview from discovery to progressive developments in microscopy and molecular mechanisms. Research on B. bacteriovorus has moved from curiosity to a new model organism, revealing over time more details on its physiology and fascinating predatory life cycle with the help of a variety of methods. Based on recent findings in cryo-electron tomography, we recapitulate on the intricate molecular details known in the predatory life cycle including how this predator searches for its prey bacterium, to how it attaches, grows, and divides all from within the prey cell. Finally, the newly developed B. bacteriovorus progeny leave the prey cell remnants in the exit phase. While we end with some unanswered questions remaining in the field, new imaging technologies and quantitative, systematic advances will likely help to unravel them in the next decades.
From curiosity to new model organism
What started as a serendipity-based discovery from a soil sample (Stolp and Petzold, 1962) was regarded for a long time as a curiosity in the microbial world. Recently, the research field of predatory bacteria and Bdellovibrio in particular, is gaining interest as a novel way to fight antimicrobial resistant pathogens and is growing rapidly due to methodological and technical advances over the last six decades. The number of published articles on Bdellovibrio per year, as indicated in Figure 1, has steadily increased in recent years—a trend coinciding with the rise of antimicrobial resistance studies.
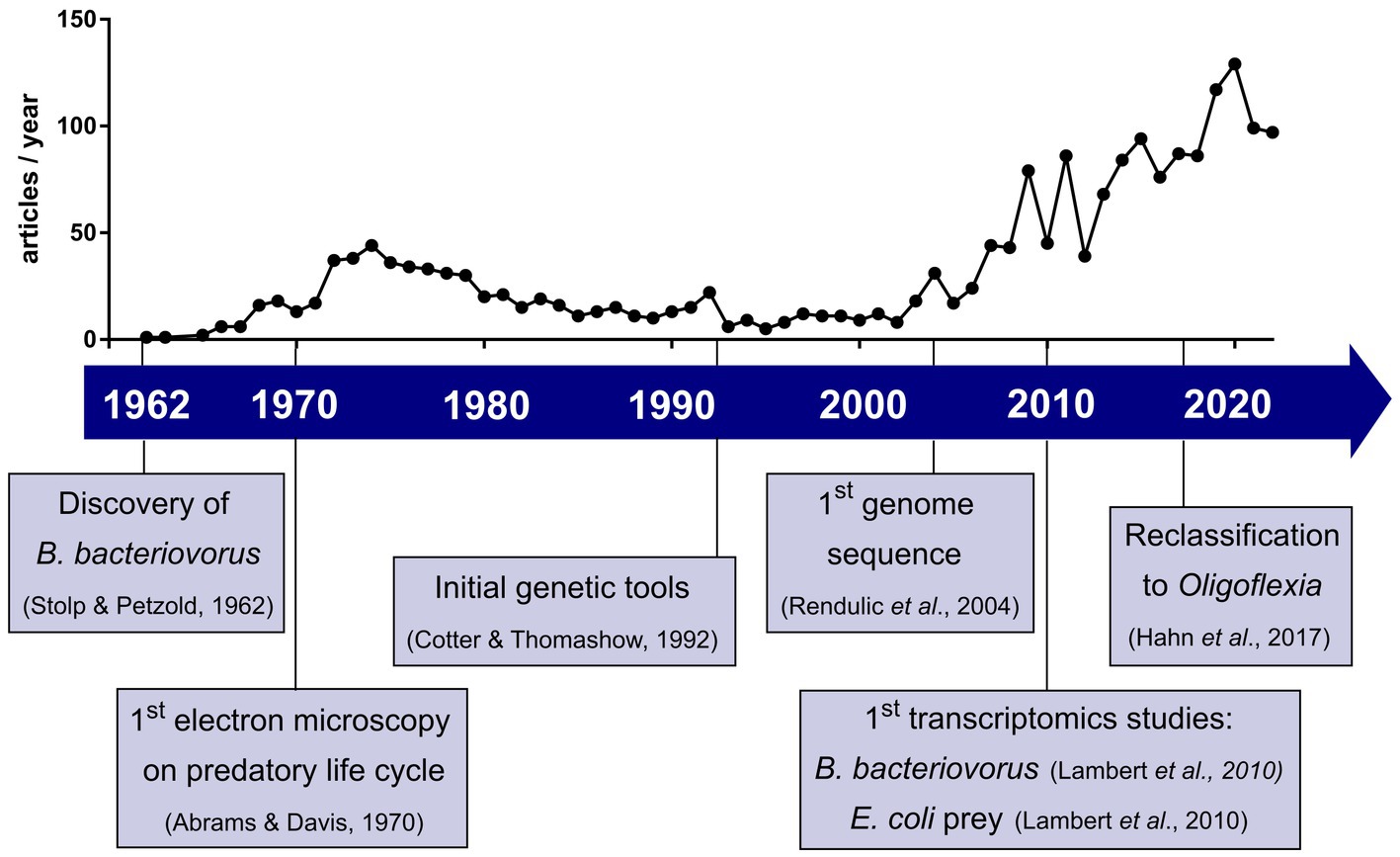
Figure 1. Development of Bdellovibrio research field in the last six decades with selected key events. Plot shows the number of articles published per year when searching Bdellovibrio on Web of Science™ until 2022.
In 1962, whilst attempting to isolate bacteriophages, Stolp and Petzold discovered a small ‘parasitic’ organism which showed lytic activity against phytopathogenic Pseudomonas (Stolp and Petzold, 1962). But unlike bacteriophages, this new organism, named Bdellovibrio bacteriovorus, formed plaques after a longer incubation period and which increased in size (Stolp and Starr, 1963). Visualization using phase contrast microscopy showed that only Gram-negative species were susceptible to B. bacteriovorus (Starr and Baigent, 1966). Interestingly, a subset of B. bacteriovorus was shown to be able to grow independently of prey as host-independent (HI) strains (Shilo and Bruff, 1965). Further studies on how the predator invades bacterial prey cells led to an initial characterization of attachment and invasion (Varon and Shilo, 1968, 1969; Abram et al., 1974). Due to the lack of genetic tools and methods available, the mechanistic hypotheses proposed could not be investigated further. Early electron microscopy identified the morphology of B. bacteriovorus (Abram and Davis, 1970) as well as the predator–prey interactions in invasion and intracellular growth (Burnham et al., 1968). These studies set the foundations of the predatory life cycle we know today (Figure 2).
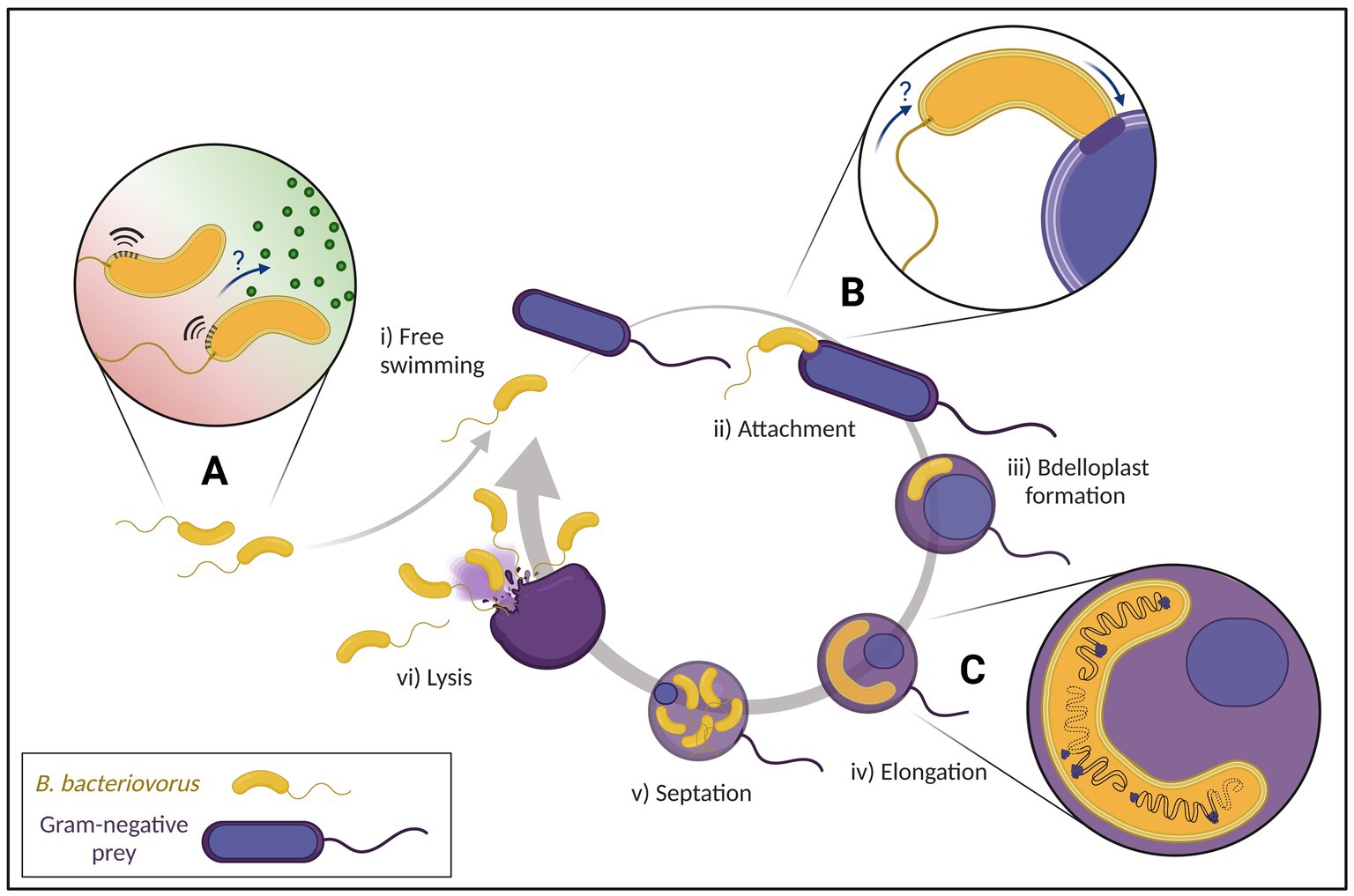
Figure 2. Predatory life cycle of Bdellovibrio bacteriovorus with focus on current advances of research. (i) Free swimming attack phase B. bacteriovorus move to regions with high prey density (A) by fast movement and possible chemotaxis (‘?’). (ii) After an initial reversible attachment to the prey, an irreversible attachment on suitable prey is formed (B) before entering the prey periplasm with suggested flagellar reabsorption (‘?’). (iii) After sealing the entry site behind itself, B. bacteriovorus establishes within the prey cell turning into an osmotically stable bdelloplast. (iv) The predator breaks down the prey cell contents to elongate while multiple asynchronous chromosomal replications take place (C). (v) Once nutrient availability is depleted, B. bacteriovorus undergoes synchronous septation. (vi) Prey cell lysis releases progeny cells into the environment to repeat the predatory life cycle.
Based on this initial work, various aspects of the predator’s biology were explored deeper in the 1970s and 80s, prominently by three research groups: Samuel Conti’s group performed early chemotaxis experiments (LaMarre et al., 1977; Straley and Conti, 1977; Straley et al., 1979), and Sydney Rittenberg’s group investigated the biochemistry and metabolism of B. bacteriovorus—its breakdown and uptake of prey nutrients as well as the synthesis of its own macromolecules (Hespell et al., 1973, 1975; Kuenen and Rittenberg, 1975; Nelson and Rittenberg, 1981; Rosson and Rittenberg, 1981; Rayner et al., 1985). Furthermore, B. bacteriovorus population studies by Mazel Varon spearheaded research on the effect of predator–prey and predator-environment interactions on predatory ability (Varon and Zeigler, 1978; Varon, 1981; Varon and Shilo, 1981). After the initial boom of publications in the first two decades of Bdellovibrio research, the following years of the 1980s and 90s saw a decline in the number of publications in this field. It was not until the advent of the genomics era that re-sparked the interest in Bdellovibrio research. Cotter and Thomashow pioneered the first genetic work, successfully conjugating DNA from E. coli into B. bacteriovorus and proving B. bacteriovorus to be a genetically tractable model (Cotter and Thomashow, 1992). Undoubtedly, one of the most important milestones was in 2004, where Rendulic and colleagues published the first whole genome sequence of B. bacteriovorus HD100 (Rendulic et al., 2004). While most findings are based on B. bacteriovorus strains HD100 and 109J, numerous B. bacteriovorus genomes have been sequenced, isolated from a variety of different environments (Wurtzel et al., 2010; Hobley et al., 2012b; Oyedara et al., 2016). Recently, the order Bdellovibrionales was reclassified from the Deltaproteobacteria into the new class of Oligoflexia (Hahn et al., 2017). In the past 60 years, other genera and species of obligate predatory prokaryotes were discovered, generally referred to as BALOs (Bdellovibrio and like organisms). These BALOs have also been shown to be efficient against pathogenic bacteria and biofilms, but will not be discussed further in this mini review (Chanyi et al., 2016; Bratanis et al., 2020).
In 2010, the first transcriptomics study in B. bacteriovorus identified differentially expressed predatory and HI genes (Lambert et al., 2010a). Complementing this was the profiling of differentially regulated E. coli genes in response to the predator invasion (Lambert et al., 2010b). These genetic and transcriptomic approaches allowed a much broader view, thus supporting a deeper understanding of all stages of the predatory life cycle. Genetic manipulation coupled with early “omics” data and more recent structural determination studies of B. bacteriovorus proteins have revealed important functions and molecular mechanisms in different aspects of the predator’s biology (Lovering et al., 2011; Lerner et al., 2012; Lambert et al., 2015; Meek et al., 2019; Harding et al., 2020).
In the last decade, the therapeutic applications of B. bacteriovorus have become a prominent field of research including its use against antibiotic-resistant pathogens. The unique lifestyle of B. bacteriovorus has garnered interest as a predator against an impressive variety of pathogens (Dashiff and Kadouri, 2011; Iebba et al., 2014) and efficacy against Gram-negative biofilms (Kadouri and O’Toole, 2005; Núñez et al., 2005). Multiple animal models have been used to assess predatory ability alongside host safety in vivo (Atterbury et al., 2011; Romanowski et al., 2016; Shatzkes et al., 2016, 2017; Willis et al., 2016; Findlay et al., 2019). Concurrently, in vitro testing in sera and human cell lines has deepened the understanding of the human immune response towards B. bacteriovorus (Gupta et al., 2016; Monnappa et al., 2016; Baker et al., 2017; Raghunathan et al., 2019), a necessary step in developing B. bacteriovorus as a potential human therapeutic.
Latest methods and technology on imaging and quantification
In the last couple of decades, many new methods and technologies, especially microscopy, have been essential for the analysis of B. bacteriovorus. Combining epifluorescence microscopy with genetic manipulations has revealed interesting morphological phenotypes alongside molecular mechanisms (Fenton et al., 2010a; Lambert et al., 2016; Banks et al., 2022). Further development of fluorescently labelled D-amino acids proved to be a useful tool in investigating peptidoglycan (PG) modifications by the predator using super-resolution microscopy (Kuru et al., 2017).
Electron microscopy has been used to resolve key structures like the flagellar motor in B. bacteriovorus (Morehouse et al., 2011; Chaban et al., 2018). Further advancements in electron microscopy techniques such as cryo-electron tomography (cryo-ET) supported the elucidation of novel structures within predator cells, to a resolution of individual proteins (Xu et al., 2019; Kaplan et al., 2022, Preprint). Alongside visualizing protein structures, the use of X-ray crystallography has generated models of individual B. bacteriovorus proteins, allowing elucidation of protein functions by evaluating interactions (Lambert et al., 2015; Cho et al., 2019; Meek et al., 2019).
Until recently, the enumeration of Bdellovibrio were based on double-layer overlay plates and plaque counting requiring three to four days (Rittenberg, 1982; Lambert and Sockett, 2008). Two new methods for Bdellovibrio spp. enumeration in under one hour have been reported in 2022. The resazurin assay relies on the detection of NADH produced by B. bacteriovorus, which transforms weakly fluorescent resazurin into the highly fluorescent resorufin (Jang et al., 2022). Alternatively, the SYBR Green assay quantifies total DNA content to determine predator concentration (Remy et al., 2022). The same authors developed an independent R-package, CuRveR, to analyze the decrease of prey OD600nm opposite to the increase of fluorescence in the predator (when expressing a fluorescent cytoplasmic protein).
Searching for prey
One major unknown in the B. bacteriovorus life cycle is how exactly the predator finds its prey (Figure 2A). Attack phase (AP) predator cells move up to 160 μm/s using a single polar flagellum (Lambert et al., 2006b), searching for suitable prey. B. bacteriovorus can pause swimming in prey limited environments and subsequently resume when prey bacteria are reintroduced (Sathyamoorthy et al., 2021), in a fine balance of energy conservation and prey detection for optimal predation.
Fast swimming speeds and altered motion make finding prey appear random, with limited evidence of any active sensing towards specific prey. A study of hydrodynamic forces from B. bacteriovorus movement showed that the self-generated disruption to the local liquid environment results in movement away from open spaces towards surfaces or obstacles. Prey hydrodynamic forces cause localization at surfaces, increasing encounter rate with B. bacteriovorus (Jashnsaz et al., 2017).
Combining speed with weak chemotaxis to amino acids (LaMarre et al., 1977) and high bacterial populations (regardless of prey suitability; Straley and Conti, 1977), B. bacteriovorus increases the chances of colliding with other bacteria. If the prey found is suitable, the initial reversible attachment becomes irreversible. Sequencing of the B. bacteriovorus HD100 genome (Rendulic et al., 2004) has revealed 20 methyl-accepting chemotaxis proteins (mcps) where multiple copies of genes encode the chemotaxis machinery alongside an absence of quorum sensing systems (Pasternak et al., 2013; Sester et al., 2020). Transposon mutants of mcps took longer to reduce prey cell numbers by half in liquid culture (Lambert et al., 2003), were unable to produce lytic plaques on prey cell lawns, and also had a significantly reduced ability to clear preformed biofilms (Medina et al., 2008). Cryo-ET data identifies chemosensory arrays at the flagellated pole of B. bacteriovorus in the attack and attachment phases (Borgnia et al., 2008; Butan et al., 2011; Kaplan et al., 2022, Preprint), which seem to be degraded and therefore absent in later stages (Kaplan et al., 2022, Preprint). While there clearly is a role for chemotaxis, it is not yet fully understood which exact part of the predatory cycle it impacts.
Attaching to and entering the prey
During the attachment and invasion stages, several unique structures at the invasive pole of B. bacteriovorus are described. Abundantly found were fimbriae (Evans et al., 2007; Mahmoud and Koval, 2010), thought to be involved in the adhesion to facilitate invasion (Avidan et al., 2017). In nearly all AP cells, novel structures dubbed “rose-like complexes” were visualized with piliated and non-piliated Type IVa pili (T4aP) basal bodies often neighboring these structures (Kaplan et al., 2022, Preprint). It has been predicted that these T4aP are used to pull the prey cell closer (Evans et al., 2007; Mahmoud and Koval, 2010). Structural comparison (at macromolecular resolution) suggested similarities of the rose-like complexes to a tripartite efflux pump, possibly a Type I secretion system (Alav et al., 2021) and the non-piliated T4aP basal bodies to a Type II secretion system (Ghosal et al., 2019). Both structures may be implicated in effector secretion that modify the prey during invasion into an osmotically stable structure known as the bdelloplast (Rittenberg and Langley, 1975).
Kaplan et al., 2022 (Preprint) seems to detect by cryo-ET, that a majority of B. bacteriovorus HD100 cells seem to internalize the flagellum via a resorption mechanism once the predator is stably attached to the prey (Figure 2B). This differs from mechanisms found in other bacteria, which release their flagella under starvation, mechanical stress or by programmed ejection (Ferreira et al., 2019; Kaplan et al., 2019, 2021; Zhu et al., 2019; Zhuang et al., 2020). Why this occurs can only be speculated, perhaps as an energy conservation strategy employed by the predator (Smith and Chapman, 2010). However, this newest flagellar resorption report is opposed to a study with B. bacteriovorus 109 J, where the majority of predatory flagella were unshed (Lambert et al., 2006a).
Development within the prey
Within the bdelloplast B. bacteriovorus transitions into a growth phase whereupon the prey contents are used for replication with prey-derived chemical cues driving the transcriptional changes required (Rotem et al., 2015). The genomic DNA of B. bacteriovorus has been shown to be tightly packed in a nucleoid, with ribosomes arranged at the edge (Borgnia et al., 2008; Butan et al., 2011; Kaplan et al., 2022, Preprint). Interestingly, the nucleoid area of 0.25 ± 0.03 μm2 is so dense during attack phase, it excludes freely diffusing monomeric proteins (Kaljević et al., 2021). Compaction, however, varies throughout the life cycle with relaxation of the nucleoid occurring between prey cell entry and DNA replication, resulting in multiple highly condensed nucleoids.
Subsequent cell division of B. bacteriovorus is remarkably different from typical bacterial cell division. Unlike many bacteria, B. bacteriovorus can follow non-binary division by elongating and dividing into a variable number of progenies correlating with prey cell size (Thomashow and Rittenberg, 1979; Fenton et al., 2010b; Kaljević et al., 2021). This filamentous cell division is controlled in part by DivIVA, responsible for progeny cell morphology with cell division coordinated by ParAB chromosomal partitioning systems (Milner et al., 2020). A recent study investigated B. bacteriovorus cell division by fluorescently labelling key components of the chromosome partitioning system ParABS (Kaljević et al., 2021). Imaging showed multiple asynchronous rounds of replication (Figure 2C) first initiated at the invasive pole, which is again different compared to the polarity of other monoflagellated bacteria like Caulobacter crescentus (Jensen and Shapiro, 1999) or Vibrio cholerae (Fogel and Waldor, 2004). The specific cues for staggered initiation of DNA replication and subsequent synchronous division are unknown.
Exit phase
The mechanisms behind the exit phase by which the predator progeny leave the prey cell are not yet well understood, partially due to the technical difficulty of synchronizing B. bacteriovorus that exit the prey at a similar time. Initial hints on the exit mechanisms came from the study of secondary messenger signaling where cyclic GMP-AMP (cGAMP) has been shown to be critical in controlling gliding motility, the impairment of which leaves B. bacteriovorus HD100 stranded inside of the empty prey cell (Hobley et al., 2012a; Lowry et al., 2022). Furthermore, the deletion of two N-acetyl glucosamine (GlcNAc) deacetylase genes (bd0468 and bd3279) left a “ghost” structure after predator exit, consisting mainly of PG and outer membrane porins (Lambert et al., 2016). Moreover, this predator mutant was delayed in escape time from prey cell remnants compared to wild type. While GlcNAc deacetylation of the prey PG happens during invasion/establishment by the predator (Lambert et al., 2016), this prey PG modification was shown to be important for DslA, a lysozyme that specifically acts on the GlcNAc deacetylated prey PG facilitating prey cell exit (Harding et al., 2020). B. bacteriovorus ΔdslA showed an increased time from division until leaving the prey remnants but did not abolish the exit process fully. Therefore DslA is likely part of a multifactorial system using different enzymes to break through the cell wall and outer membrane for exit from the prey cell. Fenton et al. observed that B. bacteriovorus progeny cells that escape through pores of the bdelloplast are consistently shorter than AP cells before infection. Further maturation/elongation occurs after release into the environment (Fenton et al., 2010a). While it was observed that gliding motility in B. bacteriovorus HD100 plays an integral part in exit from the prey cell, it is yet unknown how this mechanism interacts in the network of other established processes like enzyme secretion.
Prospects for Bdellovibrio bacteriovorus research
Six decades after the discovery of B. bacteriovorus, researchers from 17 countries came together in virtual space for the first anniversary symposium ‘Celebrating sixty years of Bdellovibrio research’ to reflect on Bdellovibrio history and exchange the newest insights. Reflected by the modern way of meeting in an online format, the last six decades have transformed knowledge in many ways.
There are still many key questions on the cellular and molecular level to be solved with regards to the fascinating and useful predatory life cycle that B. bacteriovorus undergoes: (1) What structures and mechanisms gauge the suitability/specificity of the prey? (2) How can B. bacteriovorus possess such condensed genomic DNA, yet still be highly transcriptionally active and tightly regulated? (3) How does B. bacteriovorus gain access to the valuable prey cytoplasmatic contents? (4) What is the ultimate key signal that concludes B. bacteriovorus growth inside the prey and initiates division? (5) What are the multifactorial steps facilitating the exit of predator progeny cells from the prey cell remnants?
The expansion of novel technologies and improved methodologies has helped to gather data rapidly, expediting the sharing of new insights. Moving from light microscopy to fluorescence-assisted methods enabled more precise localization of structures, whilst the newest developments in cryo-ET enabled imaging at astonishing nm-resolution. While the latter resulted in new findings that we review here from Kaplan et al. (2022, Preprint), we expect that the coming decade will shed even more light into the precise mechanisms of how the predator interacts with its bacterial prey. Molecular structures determined by X-ray crystallography, fluorescence microscopy and bioassays have allowed a detailed view into the mechanistic side. Alphafold (Jumper et al., 2021; Varadi et al., 2022) has given researchers an additional powerful tool to help build hypotheses to elucidate the function of many uncharacterized predatory proteins. New developments in super-resolution microscopy and automated intelligence-assisted software help to analyze big microscopy data and gain a more in-depth understanding not only at the single cell level, but also on the level of different bacterial (prey/predator) populations. The newest trend in quantitative research will provide more knowledge on B. bacteriovorus behavior and (predator/prey) population dynamics with the help of mathematical modelling (de Dios Caballero et al., 2017; Hobley et al., 2020; Summers and Kreft, 2022), where models that dovetail nicely with wet lab experiments are of special value (Hobley et al., 2020). With these new tools in hand, the research community will certainly gain a better understanding of the complex interactions within microbial communities. The technological advances on different ‘omics’ technologies in the last two decades help us to understand the complex interactions of bacterial predator and prey. With a multitude of transcriptomics studies (Lambert et al., 2010a; Karunker et al., 2013; Dwidar et al., 2017), there is certainly much to be gained from applying further holistic technologies.
While research on B. bacteriovorus has sometimes learned from advances made in the related predator Myxococcus xanthus (Seef et al., 2021), it has since developed into a growing research field on its own. The newest technologies equip us with valuable tools to resolve the many remaining questions in the future, which is an important prerequisite to leverage B. bacteriovorus in the fight against antimicrobial resistance.
Author contributions
TL, RF, and SH wrote the manuscript. SH made Figure 1 (with Inkscape). RF made Figure 2 (Created with BioRender.com). All authors contributed to the article and approved the submitted version.
Funding
This work was funded by the Swiss National Science Foundation Ambizione Grant PZ00P3_193401 to TL and SH. RF was supported by the Wellcome Trust Antimicrobials and Antimicrobial Resistance DTP studentship 108876/B/15/Z.
Acknowledgments
This mini review was inspired by the first global Symposium on Bdellovibrio research (6th and 7th September 2022, online) ‘Celebrating 60 years of Bdellovibrio research’ organized by SH (University of Zurich, United States), Laura Hobley (University of Nottingham, United Kingdom) and Andrew L. Lovering (University of Birmingham, United Kingdom). We are very thankful to Andrew L. Lovering (University of Birmingham, United Kingdom), Mohammed Kaplan (California Institute of technology, United States), and Florian Lindner (University of Zurich, United States) for useful comments on the manuscript. Sincere apologies to all the authors whose important work could not be mentioned in this brief review.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abram, D., and Davis, B. K. (1970). Structural properties and features of parasitic Bdellovibrio bacteriovorus. J. Bacteriol. 104, 948–965. doi: 10.1128/jb.104.2.948-965.1970
Abram, D., e Melo, J. C., and Chou, D. (1974). Penetration of Bdellovibrio bacteriovorus into host cells. J. Bacteriol. 118, 663–680. doi: 10.1128/jb.118.2.663-680.1974
Alav, I., Bavro, V. N., and Blair, J. M. A. (2021). Interchangeability of periplasmic adaptor proteins AcrA and AcrE in forming functional efflux pumps with AcrD in Salmonella enterica serovar Typhimurium. J. Antimicrob. Chemother. 76, 2558–2564. doi: 10.1093/jac/dkab237
Atterbury, R. J., Hobley, L., Till, R., Lambert, C., Capeness, M. J., Lerner, T. R., et al. (2011). Effects of orally administered Bdellovibrio bacteriovorus on the well-being and Salmonella colonization of young chicks. Appl. Environ. Microbiol. 77, 5794–5803. doi: 10.1128/AEM.00426-11
Avidan, O., Petrenko, M., Becker, R., Beck, S., Linscheid, M., Pietrokovski, S., et al. (2017). Identification and characterization of differentially-regulated type IVb Pilin genes necessary for predation in obligate bacterial predators. Sci. Rep. 7:1013. doi: 10.1038/s41598-017-00951-w
Baker, M., Negus, D., Raghunathan, D., Radford, P., Moore, C., Clark, G., et al. (2017). Measuring and modelling the response of Klebsiella pneumoniae KPC prey to Bdellovibrio bacteriovorus predation, in human serum and defined buffer. Sci. Rep. 7:8329. doi: 10.1038/s41598-017-08060-4
Banks, E. J., Valdivia-Delgado, M., Biboy, J., Wilson, A., Cadby, I. T., Vollmer, W., et al. (2022). Asymmetric peptidoglycan editing generates cell curvature in Bdellovibrio predatory bacteria. Nat. Commun. 13:1509. doi: 10.1038/s41467-022-29007-y
Borgnia, M. J., Subramaniam, S., and Milne, J. L. S. (2008). Three-dimensional imaging of the highly bent architecture of Bdellovibrio bacteriovorus by using Cryo-Electron tomography. J. Bacteriol. 190, 2588–2596. doi: 10.1128/JB.01538-07
Bratanis, E., Andersson, T., Lood, R., and Bukowska-Faniband, E. (2020). Biotechnological potential of Bdellovibrio and like organisms and their secreted enzymes. Front. Microbiol. 11:662. doi: 10.3389/fmicb.2020.00662
Burnham, J. C., Hashimoto, T., and Conti, S. F. (1968). Electron microscopic observations on the penetration of Bdellovibrio bacteriovorus into gram-negative bacterial hosts. J. Bacteriol. 96, 1366–1381. doi: 10.1128/jb.96.4.1366-1381.1968
Butan, C., Hartnell, L. M., Fenton, A. K., Bliss, D., Sockett, R. E., Subramaniam, S., et al. (2011). Spiral architecture of the nucleoid in Bdellovibrio bacteriovorus. J. Bacteriol. 193, 1341–1350. doi: 10.1128/JB.01061-10
Chaban, B., Coleman, I., and Beeby, M. (2018). Evolution of higher torque in Campylobacter-type bacterial flagellar motors. Sci. Rep. 8:97. doi: 10.1038/s41598-017-18115-1
Chanyi, R. M., Koval, S. F., and Brooke, J. S. (2016). Stenotrophomonas maltophilia biofilm reduction by Bdellovibrio exovorus: S. maltophilia biofilm reduction by Bdellovibrio. Environ. Microbiol. Rep. 8, 343–351. doi: 10.1111/1758-2229.12384
Cho, S. Y., Song, W. S., and Yoon, S. (2019). Crystal structure of the flagellar cap protein FliD from Bdellovibrio bacteriovorus. Biochem. Biophys. Res. Commun. 519, 652–658. doi: 10.1016/j.bbrc.2019.09.024
Cotter, T. W., and Thomashow, M. F. (1992). A conjugation procedure for Bdellovibrio bacteriovorus and its use to identify DNA sequences that enhance the plaque-forming ability of a spontaneous host-independent mutant. J. Bacteriol. 174, 6011–6017. doi: 10.1128/jb.174.19.6011-6017.1992
Dashiff, A., and Kadouri, D. E. (2011). Predation of Oral pathogens by Bdellovibrio bacteriovorus 109J: Oral pathogens and B. bacteriovorus. Mol Oral Microbiol 26, 19–34. doi: 10.1111/j.2041-1014.2010.00592.x
de Dios Caballero, J., Vida, R., Cobo, M., Máiz, L., Suárez, L., Galeano, J., et al. (2017). Individual patterns of complexity in cystic fibrosis lung microbiota, including predator Bacteria, over a 1-year period. MBio 8, e00959–e00917. doi: 10.1128/mBio.00959-17
Dwidar, M., Im, H., Seo, J. K., and Mitchell, R. J. (2017). Attack-phase Bdellovibrio bacteriovorus responses to extracellular nutrients are analogous to those seen during late Intraperiplasmic growth. Microb. Ecol. 74, 937–946. doi: 10.1007/s00248-017-1003-1
Evans, K. J., Lambert, C., and Sockett, R. E. (2007). Predation by Bdellovibrio bacteriovorus HD100 requires type IV Pili. J. Bacteriol. 189, 4850–4859. doi: 10.1128/JB.01942-06
Fenton, A. K., Kanna, M., Woods, R. D., Aizawa, S.-I., and Sockett, R. E. (2010a). Shadowing the actions of a predator: backlit fluorescent microscopy reveals synchronous nonbinary Septation of predatory Bdellovibrio inside prey and exit through discrete Bdelloplast pores. J. Bacteriol. 192, 6329–6335. doi: 10.1128/JB.00914-10
Fenton, A. K., Lambert, C., Wagstaff, P. C., and Sockett, R. E. (2010b). Manipulating each MreB of Bdellovibrio bacteriovorus gives diverse morphological and predatory phenotypes. J. Bacteriol. 192, 1299–1311. doi: 10.1128/JB.01157-09
Ferreira, J. L., Gao, F. Z., Rossmann, F. M., Nans, A., Brenzinger, S., Hosseini, R., et al. (2019). γ-proteobacteria eject their polar flagella under nutrient depletion, retaining flagellar motor relic structures. PLoS Biol. 17:e3000165. doi: 10.1371/journal.pbio.3000165
Findlay, J. S., Flick-Smith, H. C., Keyser, E., Cooper, I. A., Williamson, E. D., and Oyston, P. C. F. (2019). Predatory bacteria can protect SKH-1 mice from a lethal plague challenge. Sci. Rep. 9:7225. doi: 10.1038/s41598-019-43467-1
Fogel, M. A., and Waldor, M. K. (2004). Distinct segregation dynamics of the two V. cholerae chromosomes. Mol. Microbiol. 55, 125–136. doi: 10.1111/j.1365-2958.2004.04379.x
Ghosal, D., Kim, K. W., Zheng, H., Kaplan, M., Truchan, H. K., Lopez, A. E., et al. (2019). In vivo structure of the Legionella type II secretion system by electron cryotomography. Nat. Microbiol. 4, 2101–2108. doi: 10.1038/s41564-019-0603-6
Gupta, S., Tang, C., Tran, M., and Kadouri, D. E. (2016). Effect of predatory Bacteria on human cell lines. PLoS One 11:e0161242. doi: 10.1371/journal.pone.0161242
Hahn, M. W., Schmidt, J., Koll, U., Rohde, M., Verbarg, S., Pitt, A., et al. (2017). Silvanigrella aquatica gen. Nov., sp. nov., isolated from a freshwater lake, description of Silvanigrellaceae fam. Nov. and Silvanigrellales Ord. Nov., reclassification of the order Bdellovibrionales in the class Oligoflexia, reclassification of the families Bacteriovoracaceae and Halobacteriovoraceae in the new order Bacteriovoracales Ord. Nov., and reclassification of the family Pseudobacteriovoracaceae in the order Oligoflexales. Int. J. Syst. Evol. Microbiol. 67, 2555–2568. doi: 10.1099/ijsem.0.001965
Harding, C. J., Huwiler, S. G., Somers, H., Lambert, C., Ray, L. J., Till, R., et al. (2020). A lysozyme with altered substrate specificity facilitates prey cell exit by the periplasmic predator Bdellovibrio bacteriovorus. Nat. Commun. 11:4817. doi: 10.1038/s41467-020-18139-8
Hespell, R. B., Miozzari, G. F., and Rittenberg, S. C. (1975). Ribonucleic acid destruction and synthesis during intraperiplasmic growth of Bdellovibrio bacteriovorus. J. Bacteriol. 123, 481–491. doi: 10.1128/jb.123.2.481-491.1975
Hespell, R. B., Rosson, R. A., Thomashow, M. F., and Rittenberg, S. C. (1973). Respiration of Bdellovibrio bacteriovorus strain 109J and its energy substrates for Intraperiplasmic growth. J. Bacteriol. 113, 1280–1288. doi: 10.1128/jb.113.3.1280-1288.1973
Hobley, L., Fung, R. K. Y., Lambert, C., Harris, M. A. T. S., Dabhi, J. M., King, S. S., et al. (2012a). Discrete cyclic di-GMP-dependent control of bacterial predation versus axenic growth in Bdellovibrio bacteriovorus. PLoS Pathog. 8:e1002493. doi: 10.1371/journal.ppat.1002493
Hobley, L., Lerner, T. R., Williams, L. E., Lambert, C., Till, R., Milner, D. S., et al. (2012b). Genome analysis of a simultaneously predatory and prey-independent, novel Bdellovibrio bacteriovorus from the river Tiber, supports in silico predictions of both ancient and recent lateral gene transfer from diverse bacteria. BMC Genomics 13:670. doi: 10.1186/1471-2164-13-670
Hobley, L., Summers, J. K., Till, R., Milner, D. S., Atterbury, R. J., Stroud, A., et al. (2020). Dual predation by bacteriophage and Bdellovibrio bacteriovorus can eradicate Escherichia coli prey in situations where single predation cannot. J. Bacteriol. 202:e00629-19. doi: 10.1128/JB.00629-19
Iebba, V., Totino, V., Santangelo, F., Gagliardi, A., Ciotoli, L., Virga, A., et al. (2014). Bdellovibrio bacteriovorus directly attacks Pseudomonas aeruginosa and Staphylococcus aureus cystic fibrosis isolates. Front. Microbiol. 5:280. doi: 10.3389/fmicb.2014.00280
Jang, H., Mun, W., Choi, S. Y., and Mitchell, R. J. (2022). Use of Resazurin to rapidly enumerate Bdellovibrio and like organisms and evaluate their activities. Microbiol. Spectr. 10, e00825–e00822. doi: 10.1128/spectrum.00825-22
Jashnsaz, H., al Juboori, M., Weistuch, C., Miller, N., Nguyen, T., Meyerhoff, V., et al. (2017). Hydrodynamic hunters. Biophys. J. 112, 1282–1289. doi: 10.1016/j.bpj.2017.02.011
Jensen, R. B., and Shapiro, L. (1999). The Caulobacter crescentus smc gene is required for cell cycle progression and chromosome segregation. Proc. Natl. Acad. Sci. 96, 10661–10666. doi: 10.1073/pnas.96.19.10661
Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. doi: 10.1038/s41586-021-03819-2
Kadouri, D., and O’Toole, G. A. (2005). Susceptibility of biofilms to Bdellovibrio bacteriovorus attack. Appl. Environ. Microbiol. 71, 4044–4051. doi: 10.1128/AEM.71.7.4044-4051.2005
Kaljević, J., Saaki, T. N. V., Govers, S. K., Remy, O., van Raaphorst, R., Lamot, T., et al. (2021). Chromosome choreography during the non-binary cell cycle of a predatory bacterium. Curr. Biol. 31, 3707–3720.e5. doi: 10.1016/j.cub.2021.06.024
Kaplan, M., Chang, Y.-W., Oikonomou, C. M., Nicolas, W. J., Jewett, A. I., Kreida, S., et al. (2022). Dynamic structural adaptations enable the endobiotic predation of Bdellovibrio bacteriovorus. bioRxiv [Preprint]. doi: 10.1101/2022.06.13.496000 (Accessed April 21, 2023).
Kaplan, M., Subramanian, P., Ghosal, D., Oikonomou, C. M., Pirbadian, S., Starwalt-Lee, R., et al. (2019). In situ imaging of the bacterial flagellar motor disassembly and assembly processes. EMBO J. 38:e100957. doi: 10.15252/embj.2018100957
Kaplan, M., Wang, Y., Chreifi, G., Zhang, L., Chang, Y.-W., and Jensen, G. J. (2021). Programmed Flagellar ejection in Caulobacter crescentus leaves PL-subcomplexes. J. Mol. Biol. 433:167004. doi: 10.1016/j.jmb.2021.167004
Karunker, I., Rotem, O., Dori-Bachash, M., Jurkevitch, E., and Sorek, R. (2013). A global transcriptional switch between the attack and growth forms of Bdellovibrio bacteriovorus. PLoS One 8:e61850. doi: 10.1371/journal.pone.0061850
Kuenen, J. G., and Rittenberg, S. C. (1975). Incorporation of long-chain fatty acids of the substrate organism by Bdellovibrio bacteriovorus during intraperiplasmic growth. J. Bacteriol. 121, 1145–1157. doi: 10.1128/jb.121.3.1145-1157.1975
Kuru, E., Lambert, C., Rittichier, J., Till, R., Ducret, A., Derouaux, A., et al. (2017). Fluorescent D-amino-acids reveal bi-cellular cell wall modifications important for Bdellovibrio bacteriovorus predation. Nat. Microbiol. 2, 1648–1657. doi: 10.1038/s41564-017-0029-y
LaMarre, A., Straley, S. C., and Conti, S. F. (1977). Chemotaxis toward amino acids by Bdellovibrio bacteriovorus. J. Bacteriol. 131, 201–207. doi: 10.1128/jb.131.1.201-207.1977
Lambert, C., Cadby, I. T., Till, R., Bui, N. K., Lerner, T. R., Hughes, W. S., et al. (2015). Ankyrin-mediated self-protection during cell invasion by the bacterial predator Bdellovibrio bacteriovorus. Nat. Commun. 6:8884. doi: 10.1038/ncomms9884
Lambert, C., Chang, C.-Y., Capeness, M. J., and Sockett, R. E. (2010a). The first bite— profiling the Predatosome in the bacterial pathogen Bdellovibrio. PLoS One 5:e8599. doi: 10.1371/journal.pone.0008599
Lambert, C., Evans, K. J., Till, R., Hobley, L., Capeness, M., Rendulic, S., et al. (2006a). Characterizing the flagellar filament and the role of motility in bacterial prey-penetration by Bdellovibrio bacteriovorus. Mol. Microbiol. 60, 274–286. doi: 10.1111/j.1365-2958.2006.05081.x
Lambert, C., Ivanov, P., and Sockett, R. E. (2010b). A transcriptional “scream” early response of E. coli prey to predatory invasion by Bdellovibrio. Curr. Microbiol. 60, 419–427. doi: 10.1007/s00284-009-9559-8
Lambert, C., Lerner, T. R., Bui, N. K., Somers, H., Aizawa, S.-I., Liddell, S., et al. (2016). Interrupting peptidoglycan deacetylation during Bdellovibrio predator-prey interaction prevents ultimate destruction of prey wall, liberating bacterial-ghosts. Sci. Rep. 6:26010. doi: 10.1038/srep26010
Lambert, C., Morehouse, K. A., Chang, C.-Y., and Sockett, R. E. (2006b). Bdellovibrio: growth and development during the predatory cycle. Curr. Opin. Microbiol. 9, 639–644. doi: 10.1016/j.mib.2006.10.002
Lambert, C., Smith, M. C. M., and Sockett, R. E. (2003). A novel assay to monitor predator-prey interactions for Bdellovibrio bacteriovorus 109 J reveals a role for methyl-accepting chemotaxis proteins in predation. Environ. Microbiol. 5, 127–132. doi: 10.1046/j.1462-2920.2003.00385.x
Lambert, C., and Sockett, R. E. (2008). Laboratory maintenance of Bdellovibrio. Curr. Protoc. Microbiol. 9, 7B.2.1–7B.2.13. doi: 10.1002/9780471729259.mc07b02s9
Lerner, T. R., Lovering, A. L., Bui, N. K., Uchida, K., Aizawa, S.-I., Vollmer, W., et al. (2012). Specialized peptidoglycan hydrolases sculpt the intra-bacterial niche of predatory Bdellovibrio and increase population fitness. PLoS Pathog. 8:e1002524. doi: 10.1371/journal.ppat.1002524
Lovering, A. L., Capeness, M. J., Lambert, C., Hobley, L., and Sockett, R. E. (2011). The structure of an unconventional HD-GYP protein from Bdellovibrio reveals the roles of conserved residues in this class of cyclic-di-GMP Phosphodiesterases. MBio 2, e00163–e00111. doi: 10.1128/mBio.00163-11
Lowry, R. C., Hallberg, Z. F., Till, R., Simons, T. J., Nottingham, R., Want, F., et al. (2022). Production of 3′,3′-cGAMP by a Bdellovibrio bacteriovorus promiscuous GGDEF enzyme, Bd0367, regulates exit from prey by gliding motility. PLoS Genet. 18:e1010164. doi: 10.1371/journal.pgen.1010164
Mahmoud, K. K., and Koval, S. F. (2010). Characterization of type IV pili in the life cycle of the predator bacterium Bdellovibrio. Microbiology 156, 1040–1051. doi: 10.1099/mic.0.036137-0
Medina, A. A., Shanks, R. M., and Kadouri, D. E. (2008). Development of a novel system for isolating genes involved in predator-prey interactions using host independent derivatives of Bdellovibrio bacteriovorus 109J. BMC Microbiol. 8:33. doi: 10.1186/1471-2180-8-33
Meek, R. W., Cadby, I. T., Moynihan, P. J., and Lovering, A. L. (2019). Structural basis for activation of a diguanylate cyclase required for bacterial predation in Bdellovibrio. Nat. Commun. 10:4086. doi: 10.1038/s41467-019-12051-6
Milner, D. S., Ray, L. J., Saxon, E. B., Lambert, C., Till, R., Fenton, A. K., et al. (2020). DivIVA controls progeny morphology and diverse ParA proteins regulate cell division or gliding motility in Bdellovibrio bacteriovorus. Front. Microbiol. 11:542. doi: 10.3389/fmicb.2020.00542
Monnappa, A. K., Bari, W., Choi, S. Y., and Mitchell, R. J. (2016). Investigating the responses of human epithelial cells to predatory Bacteria. Sci. Rep. 6:33485. doi: 10.1038/srep33485
Morehouse, K. A., Hobley, L., Capeness, M., and Sockett, R. E. (2011). Three motAB Stator gene products in Bdellovibrio bacteriovorus contribute to motility of a single flagellum during predatory and prey-independent growth. J. Bacteriol. 193, 932–943. doi: 10.1128/JB.00941-10
Nelson, D. R., and Rittenberg, S. C. (1981). Incorporation of substrate cell lipid a components into the lipopolysaccharide of Intraperiplasmically grown Bdellovibrio bacteriovorus. J. Bacteriol. 147, 860–868. doi: 10.1128/jb.147.3.860-868.1981
Núñez, M. E., Martin, M. O., Chan, P. H., and Spain, E. M. (2005). Predation, death, and survival in a biofilm: Bdellovibrio investigated by atomic force microscopy. Colloids Surf. B Biointerfaces 42, 263–271. doi: 10.1016/j.colsurfb.2005.03.003
Oyedara, O. O., de Luna-Santillana, E. J., Olguin-Rodriguez, O., Guo, X., Mendoza-Villa, M. A., Menchaca-Arredondo, J. L., et al. (2016). Isolation of Bdellovibrio sp. from soil samples in Mexico and their potential applications in control of pathogens. MicrobiologyOpen 5, 992–1002. doi: 10.1002/mbo3.382
Pasternak, Z., Pietrokovski, S., Rotem, O., Gophna, U., Lurie-Weinberger, M. N., and Jurkevitch, E. (2013). By their genes ye shall know them: genomic signatures of predatory bacteria. ISME J. 7, 756–769. doi: 10.1038/ismej.2012.149
Raghunathan, D., Radford, P. M., Gell, C., Negus, D., Moore, C., Till, R., et al. (2019). Engulfment, persistence and fate of Bdellovibrio bacteriovorus predators inside human phagocytic cells informs their future therapeutic potential. Sci. Rep. 9:4293. doi: 10.1038/s41598-019-40223-3
Rayner, J. R., Cover, W. H., Martinez, R. J., and Rittenberg, S. C. (1985). Bdellovibrio bacteriovorus synthesizes an OmpF-like outer membrane protein during both axenic and intraperiplasmic growth. J. Bacteriol. 163, 595–599. doi: 10.1128/jb.163.2.595-599.1985
Remy, O., Lamot, T., Santin, Y., Kaljević, J., de Pierpont, C., and Laloux, G. (2022). An optimized workflow to measure bacterial predation in microplates. STAR Protoc. 3:101104. doi: 10.1016/j.xpro.2021.101104
Rendulic, S., Jagtap, P., Rosinus, A., Eppinger, M., Baar, C., Lanz, C., et al. (2004). A predator unmasked: life cycle of Bdellovibrio bacteriovorus from a genomic perspective. Science 303, 689–692. doi: 10.1126/science.1093027
Rittenberg, S. C. (1982). “Bdellovibrios-interperiplasmic growth” in Experimental Microbial Ecology. eds. R. G. Burns and J. H. Slater (Oxford: Blackwell Scientific Publications), 379–394.
Rittenberg, S. C., and Langley, D. (1975). Utilization of nucleoside monophosphates per se for intraperiplasmic growth of Bdellovibrio bacteriovorus. J. Bacteriol. 121, 1137–1144. doi: 10.1128/jb.121.3.1137-1144.1975
Romanowski, E. G., Stella, N. A., Brothers, K. M., Yates, K. A., Funderburgh, M. L., Funderburgh, J. L., et al. (2016). Predatory bacteria are nontoxic to the rabbit ocular surface. Sci. Rep. 6:30987. doi: 10.1038/srep30987
Rosson, R. A., and Rittenberg, S. C. (1981). Pyrimidine metabolism of Bdellovibrio bacteriovorus grown intraperiplasmically and axenically. J. Bacteriol. 146, 108–116. doi: 10.1128/jb.146.1.108-116.1981
Rotem, O., Pasternak, Z., Shimoni, E., Belausov, E., Porat, Z., Pietrokovski, S., et al. (2015). Cell-cycle progress in obligate predatory bacteria is dependent upon sequential sensing of prey recognition and prey quality cues. Proc. Natl. Acad. Sci. 112, E6028–E6037. doi: 10.1073/pnas.1515749112
Sathyamoorthy, R., Kushmaro, Y., Rotem, O., Matan, O., Kadouri, D. E., Huppert, A., et al. (2021). To hunt or to rest: prey depletion induces a novel starvation survival strategy in bacterial predators. ISME J. 15, 109–123. doi: 10.1038/s41396-020-00764-2
Seef, S., Herrou, J., de Boissier, P., My, L., Brasseur, G., Robert, D., et al. (2021). A tad-like apparatus is required for contact-dependent prey killing in predatory social bacteria. eLife 10:e72409. doi: 10.7554/eLife.72409
Sester, A., Korp, J., and Nett, M. (2020). “Secondary metabolism of predatory Bacteria” in The Ecology of Predation at the Microscale. eds. E. Jurkevitch and R. Mitchell (Cham: Springer), 127–153.
Shatzkes, K., Singleton, E., Tang, C., Zuena, M., Shukla, S., Gupta, S., et al. (2016). Predatory Bacteria attenuate Klebsiella pneumoniae burden in rat lungs. MBio 7, e01847–e01816. doi: 10.1128/mBio.01847-16
Shatzkes, K., Tang, C., Singleton, E., Shukla, S., Zuena, M., Gupta, S., et al. (2017). Effect of predatory bacteria on the gut bacterial microbiota in rats. Sci. Rep. 7:43483. doi: 10.1038/srep43483
Shilo, M., and Bruff, B. (1965). Lysis of gram-negative Bacteria by host-independent Ectoparasitic Bdellovibrio bacteriovorus isolates. J. Gen. Microbiol. 40, 317–328. doi: 10.1099/00221287-40-3-317
Smith, D. R., and Chapman, M. R. (2010). Economical evolution: microbes reduce the synthetic cost of extracellular proteins. MBio 1, e00131–e00110. doi: 10.1128/mBio.00131-10
Starr, M. P., and Baigent, N. L. (1966). Parasitic interaction of Bdellovibrio bacteriovorus with other Bacteria. J. Bacteriol. 91, 2006–2017. doi: 10.1128/jb.91.5.2006-2017.1966
Stolp, H., and Petzold, H. (1962). Untersuchungen über einen obligat parasitischen Mikroorganismus mit lytischer Aktivität für Pseudomonas-Bakterien. J. Phytopathol. 45, 364–390. doi: 10.1111/j.1439-0434.1962.tb02050.x
Stolp, H., and Starr, M. P. (1963). Bdellovibrio bacteriovorus gen. Et sp. n., a predatory, ectoparasitic, and bacteriolytic microorganism. Antonie Van Leeuwenhoek 29, 217–248. doi: 10.1007/BF02046064
Straley, S. C., and Conti, S. F. (1977). Chemotaxis by Bdellovibrio bacteriovorus toward prey. J. Bacteriol. 132, 628–640. doi: 10.1128/jb.132.2.628-640.1977
Straley, S. C., LaMarre, A. G., Lawrence, L. J., and Conti, S. F. (1979). Chemotaxis of Bdellovibrio bacteriovorus toward pure compounds. J. Bacteriol. 140, 634–642. doi: 10.1128/jb.140.2.634-642.1979
Summers, J. K., and Kreft, J.-U. (2022). Predation strategies of the bacterium Bdellovibrio bacteriovorus result in overexploitation and bottlenecks. Appl. Environ. Microbiol. 88:e0108221. doi: 10.1128/AEM.01082-21
Thomashow, M., and Rittenberg, S. (1979). “The Intraperiplasmic growth cycle–the life style of the Bdellovibrios", in Developmental Biology of Prokaryotes, ed. J. H. Parish Oxford, Blackwell Scientific, 115–138.
Varadi, M., Anyango, S., Deshpande, M., Nair, S., Natassia, C., Yordanova, G., et al. (2022). AlphaFold protein structure database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 50, D439–D444. doi: 10.1093/nar/gkab1061
Varon, M. (1981). Interaction of Bdellovibrio with its prey in mixed microbial populations. Microb. Ecol. 7, 97–105. doi: 10.1007/BF02032491
Varon, M., and Shilo, M. (1968). Interaction of Bdellovibrio bacteriovorus and host Bacteria I. kinetic studies of attachment and invasion of Escherichia coli B by Bdellovibrio bacteriovorus. J. Bacteriol. 95, 744–753. doi: 10.1128/jb.95.3.744-753.1968
Varon, M., and Shilo, M. (1969). Attachment of Bdellovibrio bacteriovorus to cell wall mutants of Salmonella spp. and Escherichia coli. J. Bacteriol. 97, 977–979. doi: 10.1128/jb.97.2.977-979.1969
Varon, M., and Shilo, M. (1981). Inhibition of the predatory activity of Bdellovibrio by various environmental pollutants. Microb. Ecol. 7, 107–111. doi: 10.1007/BF02032492
Varon, M., and Zeigler, B. P. (1978). Bacterial predator-prey interaction at low prey density. Appl. Environ. Microbiol. 36, 11–17. doi: 10.1128/aem.36.1.11-17.1978
Willis, A. R., Moore, C., Mazon-Moya, M., Krokowski, S., Lambert, C., Till, R., et al. (2016). Injections of predatory Bacteria work alongside host immune cells to treat Shigella infection in Zebrafish larvae. Curr. Biol. 26, 3343–3351. doi: 10.1016/j.cub.2016.09.067
Wurtzel, O., Dori-Bachash, M., Pietrokovski, S., Jurkevitch, E., and Sorek, R. (2010). Mutation detection with next-generation resequencing through a mediator genome. PLoS One 5:e15628. doi: 10.1371/journal.pone.0015628
Xu, M., Singla, J., Tocheva, E. I., Chang, Y.-W., Stevens, R. C., Jensen, G. J., et al. (2019). De novo structural pattern mining in cellular electron cryotomograms. Structure 27, 679–691.e14. doi: 10.1016/j.str.2019.01.005
Zhu, S., Schniederberend, M., Zhitnitsky, D., Jain, R., Galán, J. E., Kazmierczak, B. I., et al. (2019). In situ structures of polar and lateral flagella revealed by Cryo-Electron tomography. J. Bacteriol. 201:e00117-19. doi: 10.1128/JB.00117-19
Keywords: predatory bacteria, Bdellovibrio bacteriovorus , molecular mechanisms, history, methods, antimicrobial resistance, predatory life cycle, cryo-electron tomography
Citation: Lai TF, Ford RM and Huwiler SG (2023) Advances in cellular and molecular predatory biology of Bdellovibrio bacteriovorus six decades after discovery. Front. Microbiol. 14:1168709. doi: 10.3389/fmicb.2023.1168709
Edited by:
Robert J Mitchell, Ulsan National Institute of Science and Technology, Republic of KoreaReviewed by:
Mohammed Dwidar, Cleveland Clinic, United StatesSusan Fearn Koval, Western University, Canada
Copyright © 2023 Lai, Ford and Huwiler. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Simona G. Huwiler, c2ltb25hLmh1d2lsZXJAdXpoLmNo
 Ting F. Lai1
Ting F. Lai1 Rhian M. Ford
Rhian M. Ford Simona G. Huwiler
Simona G. Huwiler