
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 19 May 2023
Sec. Aquatic Microbiology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1164937
This article is part of the Research TopicMicrobial Diversity and Resources in Tidal FlatsView all 17 articles
The booming mudflat aquaculture poses an accumulation of organic matter and a certain environmental threat. Protease-producing bacteria are key players in regulating the nitrogen content in ecosystems. However, knowledge of the diversity of protease-producing bacteria in coastal mudflats is limited. This study investigated the bacterial diversity in the coastal mudflat, especially protease-producing bacteria and their extracellular proteases, by using culture-independent methods and culture-dependent methods. The clam aquaculture area exhibited a higher concentration of carbon, nitrogen, and phosphorus when compared with the non-clam area, and a lower richness and diversity of bacterial community when compared with the clam naturally growing area. The major classes in the coastal mud samples were Bacteroidia, Gammaproteobacteria, and Alphaproteobacteria. The Bacillus-like bacterial community was the dominant cultivated protease-producing group, accounting for 52.94% in the non-clam area, 30.77% in the clam naturally growing area, and 50% in the clam aquaculture area, respectively. Additionally, serine protease and metalloprotease were the principal extracellular protease of the isolated coastal bacteria. These findings shed light on the understanding of the microbes involved in organic nitrogen degradation in coastal mudflats and lays a foundation for the development of novel protease-producing bacterial agents for coastal mudflat purification.
The coastal mudflat is a complex and dynamic ecological system, which is deeply influenced by the geological, physicochemical, and biological factors of marine and terrestrial land. Previous studies have reported that the spatiotemporal dynamics of bacterial abundance, diversity, and activity in tidal flat sediments were significantly affected by the biogeochemical heterogeneity of salinity, pH, carbon, nitrogen, sulfur, and phosphorus (Taylor et al., 2014; Soares et al., 2018; Zhang G. et al., 2021; Niu et al., 2022; Zhang et al., 2022). The seasonality and sediment depth also affected the bacterial communities in the mudflat (Böer et al., 2009; Gobet et al., 2012). Moreover, bacteria were found to be deeply involved in carbon fixation and carbon-containing compound degradation, as well as nitrogen cycling processes including nitrogen fixation, ammonium oxidation, and nitrite and nitrous oxide reduction in the coastal mudflats (Hou et al., 2013; Zhou et al., 2018; Li et al., 2021; Li Q. et al., 2022).
Coastal mudflats are of great significance in regulating regional climate and sustaining ecological balance. In recent years, mudflat planting, mudflat aquaculture, and eco-tourism are flourishing in coastal areas, which have promoted economic growth, but also posed serious environmental threats (Long et al., 2016). With the expansion of human activities, the contents of organic carbon and nitrogen in coastal mudflats were significantly increased, resulting in a large amount of organic matter entering the sea and the atmosphere (Hu et al., 2019; Li T. et al., 2022). The nitrogen cycle is a main component of global biochemical cycles, and the nitrogen budget plays an important role in keeping ecological balance (Zhang X. et al., 2020; Hutchins and Capone, 2022). Current studies of bacterial functions in the nitrogen cycle mainly focused on nitrogen fixation, anaerobic ammonia oxidation, nitrification, and denitrification (Zhou et al., 2018; Li et al., 2021; Li Q. et al., 2022).
Protease is an important participant in the degradation of organic nitrogen, and protease-producing bacteria have been reported as the main microflora that regulate the nitrogen content in ecosystems (Zhou et al., 2013; Zhang et al., 2015; Tornkvist et al., 2019; Zhang Y. Z. et al., 2020). Related studies based on culture-dependent methods have been conducted in a tropical aquaculture environment (Wei et al., 2021), in the coastal sediments and soils of Antarctica (Zhou et al., 2013; Liu et al., 2021), the sediments of the Bohai Sea, Yellow Sea, and South China Sea (Zhou et al., 2009; Zhang et al., 2019), and the sediments of Jiaozhou Bay (Zhang et al., 2015), showing the diversity of protease-producing bacteria and their extracellular protease in various environments. In addition, a variety of microorganisms with enzyme-producing capacities have been isolated from coastal mudflats (Das, 2012; Suthindhiran et al., 2013; Gaonkar and Furtado, 2020). To the best of our knowledge, reports on the diversity of protease-producing bacteria in coastal mudflats are relatively rare. We speculated that an abundant and diverse protease-producing bacteria community existed in the coastal mudflats, especially in mudflat aquaculture with a high content of organic nitrogen. Therefore, it is necessary to study the diversity of protease-producing bacteria in coastal mudflats that are heavily influenced by anthropogenic activity.
Jiaozhou Bay is a representative semi-enclosed bay of China's Yellow Sea. Under the influence of dense and long-time human activities, the ecological factors of Jiaozhou Bay have changed significantly (Zhang L. et al., 2021; Lin et al., 2022). Eutrophication and high organic nitrogen content occurred in the coastal sediments of the bay due to terrestrial inputs (Liu et al., 2010; Li H. et al., 2019; Zhang L. et al., 2021), all of which make it an ideal area for studying the organic nitrogen biodegradation in the coastal mudflats. In this study, mud samples were collected from three stations in the coastal mudflat of Jiaozhou Bay, including the non-clam area, the clam naturally growing area, and the clam aquaculture area. The variation of bacterial taxonomy composition of different loci was investigated by culture-independent methods. In addition, protease-producing bacteria were isolated from these samples by culture-dependent methods, and the diversity of both the bacteria and the extracellular proteases as well as their distribution in different stations were further analyzed. Our research provides experimental evidence to elucidate the ecological role of microorganisms in organic nitrogen degradation by decomposing protein in coastal mudflats and lays a foundation for the development of measures to protect the coastal mudflat.
Mud samples were collected from the coastal mudflat of Jiaozhou Bay, including three stations: the non-clam area (sample D1-3, hereafter collectively referred to as sample D), the clam naturally growing area (sample F1-3, hereafter collectively referred to as sample F), and the clam aquaculture area (sample H1-3, hereafter collectively referred to as sample H) on 18 March 2021. The geographic location of sampling sites is shown in Supplementary Figures S1–S4. The surface mud of each station was removed, and then, a deeper mud sample (5–10 cm) was collected aseptically using stainless-steel sterile shovels. Three replicates of all samples were taken from each station. The collected mud samples were stored in airtight sterile plastic bags at 4°C for subsequent geochemical characteristics detection and microbiological analysis. The salinity of the mud samples was measured using a soil salt meter PNT3000 (STEPS, Germany). The pH, total carbon content (TC), total nitrogen content (TN), and total phosphorus content (TP) of each sample were measured according to NY/T 1121.2-2006, NY/T1121.6-2006, NY/T1121.24-2012, and NY/T88-1988, respectively. The paired sample t-test was applied to evaluate the statistical significance between different variables of this study. The level of significance was set to a p-value of < 0.05.
The total genomic DNA of the mud samples was extracted with OMEGA-soil DNA Kit (Omega Bio-Tek, USA). The V3–V4 regions of bacterial 16S rRNA gene were amplified using TransStart FastPfu Polymerase (TransGen Biotech, China), with Barcoded sequencing primers 338F (5′-ACTCCTACGGGAGGCAGCA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′). The generated PCR mixtures were then sequenced on an Illumina Miseq PE300 sequencing platform (Shanghai Majorbio, China). The obtained large short-read libraries were merged and trimmed, and the standard quality control was performed with Usearch 8.0.161 by the sequencing company. The remaining reads of the three samples were then pooled, dereplicated, and finally assigned to each OTU using a 97% identity. OTU clustering and taxonomy assignment were processed by mapping the reads of each sample to representative OTU sequences using a Perl script, followed by comparing with a 16S rRNA gene database (RDP Release 11.1 http://rdp.cme.msu.edu/) at an 80% confidence threshold. The data were analyzed on the online Majorbio Cloud Platform (www.majorbio.com) developed by Shanghai Majorbio Bio-pharm Technology Co. Ltd. (Ren et al., 2022). Statistical analysis of alpha diversity was conducted via t-test at a significance level of 5% (p < 0.05).
Synthetic sea salt was purchased from Qingdao Sea-Salt Aquarium Technology Co. Ltd. and then dissolved at a concentration of 3% to make artificial seawater. The screening solid medium was prepared by adding 1% casein, 2% gelatin, 0.2% yeast extract, and 1.5% agar into artificial seawater with a final pH of 8.0. Protease-producing bacteria were screened using the dilution-plate method according to previous studies, with minor modifications (Zhang et al., 2015; Liu et al., 2021). In brief, approximately 1 g of mud sample was diluted serially 10 times to 10−6 using artificial seawater. Aliquots of 100 μl of each diluted sample were spread on the screening plates for subsequent incubation at 20°C until visible hydrolytic zones were formed. Colonies of different morphology with hydrolytic zone were further repeatedly streaked on the same medium at least three times to obtain a pure strain.
The 16S rRNA gene of culturable bacteria was amplified by colony PCR technique using a Colony PCR kit (Mei5 Biotechnology, China) with the universal primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTTC-3′), and then, the PCR products were sequenced by Shanghai Tsingke Biotechnology Co. Ltd., China. Isolates with at least two different nucleotides in their 16S rRNA gene sequences were identified as different strains. Phylogenetic trees were constructed using MEGA 11 with the neighbor-joining method. The GenBank accession numbers of 16S rRNA gene sequences in this study were OQ625902, OQ625903, OQ625905–OQ625912, OQ625916, OQ625917, OQ625921, OQ625938, OQ625939, OQ625941, OQ625993–OQ625996, OQ626000–OQ626019, OQ626205-OQ626214, OQ626220, OQ626221, OQ626228, OQ629890, OQ629902, OQ629992–OQ629994, OQ642131, OQ651245, and OQ690495. The strains' corresponding accession numbers are shown in Supplementary Table 1.
The protease hydrolysis ability of the pure strains toward different substrates, including milk powder, casein, and gelatin, was detected using the method of Zhou et al. (2009), with minor modifications. Protein substrate solid media were prepared by mixing each protein substrate (1.0% milk powder, 0.5% casein, or 0.5% gelatin) with 0.2% yeast extract, 1.5% agar, and artificial seawater and adjusting the final pH to 8.0. The bacterial strains were streaked on the substrate plates and then incubated at 20°C for 4 days. The specific value of the diameter of the formed hydrolytic zone divided by the diameter of the colony, and the H/C ratio was measured and calculated.
The bacterial strain was cultured in a liquid screening medium at 20°C and 200 rpm for 4 days. The supernatant of the bacterial culture was collected after centrifugation at 12,000 rpm at 4°C for 10 min and then used for subsequent protease inhibitor assay. The inhibitors contained phenylmethylsulfonyl fluoride (PMSF, Sigma) at a concentration of 1.0 mM, 1,10-phenanthroline (OP, Sigma) at a concentration of 1.0 mM, E64 (Merck) at a concentration of 0.1 mM, and pepstatin A (PA, Merck) at a concentration of 0.1 mM. The supernatant was properly diluted with 50 mM Tris-HCl (pH 8.0), followed by incubating with each inhibitor at 4°C for 60 min, and then, the residue protease activity was measured by the digestion of casein as formerly described (Chen et al., 2003). In brief, 100 μl of diluted solution and 100 μl of 2% casein in 50 mM Tris-HCl (pH 8.0) were mixed and incubated at 25°C for 20 min. To stop the reaction, 200 μl of 0.4 M TCA was added to the reaction mixture. After centrifugation at 12,000 rpm and 4°C for 5 min, the supernatant was collected and every 100 μl of supernatant was incubated with 500 μl of 0.4 M Na2CO3 and 100 μl of Folin–Ciocalteu's phenol reagent at 40°C for 10 min, and then, the absorbance was measured at 660 nm. One unit of protease activity was defined as the amount of enzyme that released 1 μg of tyrosine per milliliter of reaction mixture per minute. A sample without the addition of an inhibitor was set as the negative control. The difference between the relative residue activity of each sample and the negative control was taken as the inhibition ratio (%).
The mud samples were collected from three stations in a coastal mudflat of Jiaozhou Bay, including the non-clam area (sample D), the clam naturally growing area (sample F), and the clam aquaculture area (sample H). The results of the geochemical analysis are shown in Table 1. All mud samples exhibited slightly alkaline pH (8.21–8.74). The salinity ranged from 4.75 to 6.25 g/kg, with the highest observed in sample H and the lowest in sample D. The contents of TC, TN, and TP in the samples ranged from 9.64–22.42 g/kg (TC), 0.52–0.79 g/kg (TN), and 0.31–0.62 g/kg (TP), respectively. The highest values were found in sample H (TC and TP) and sample F (TN), while the lowest were found in sample D (TC, TN, and TP). The carbon content, nitrogen content, and phosphorus content in the clam naturally growing area and the clam aquaculture area were significantly higher than the corresponding values in the non-clam area, indicating that the artificial culture and growth of clams may be relevant to eutrophication in a tidal area.
The microbial community of the mud samples was investigated by using culture-independent methods based on Illumina high-throughput sequencing. A total of 113,717 effective sequence reads were obtained, and the sequencing coverage rate exceeded 97%. A total of 5,135 OTUs were assigned, which were arranged from high to low as sample F (2,090), sample D (1,574), and sample H (1,471). The alpha diversity indexes are shown in Table 2. The estimators Ace and Chao reveal the richness of the bacterial community. Sample F showed higher Ace and Chao values than samples D and H, indicating higher bacterial richness in sample F. The estimators Shannon and Simpson express the diversity of the microbial community, with a positive correlation between Shannon and a negative correlation between Simpson. Sample F, again, exhibited the highest Shannon value and the lowest Simpson value, implying that the microbial diversity of Sample F was the highest of all the samples analyzed. The richness and diversity of the bacterial community in the clam naturally growing area were significantly higher than that of the clam aquaculture area and the non-clam area. However, the results of statistical analysis indicated that there are no statistically significant differences in microbial richness and diversity between the clam aquaculture area and the non-clam area.
In total, 52 phyla, 136 classes, 304 orders, 468 families, and 833 genera were identified in the three mud samples. Figure 1 shows the top phyla and classes of the mud samples. It seems that the bacterial diversity of sample F was the highest and that of H was the lowest (Figure 1), which was consistent with the above alpha diversity estimators. The phylum with more than 5% was considered the dominant phylum. Proteobacteria (44.47%) and Bacteroidota (27.26%) were definitively dominant in sample D, with a sum of 71.73% (Figure 1A), while in sample F, the major phyla were Proteobacteria (22.76%), Desulfobacterota (17.92%), Bacteroidota (13.44%), Chloroflexi (10.14%), and Acidobacteriota (7.45%), with a sum of 71.71% (Figure 1A). The dominant phyla constituted 89.76% of the bacterial community in sample H, which were Proteobacteria (29.13%), Bacteroidota (23.40%), Firmicutes (21.03%), Desulfobacterota (9.93%), and Campilobacterota (6.27%) (Figure 1A). On class level, Bacteroidia, Gammaproteobacteria, and Alphaproteobacteria were dominant in the mud samples, contributing to a sum of 70.92, 36.40, and 52.35% of the bacterial community in samples D, F, and H, respectively. It is noted that Bacilli with an abundance of 11.38% and Clostridia with an abundance of 9.34% were two major classes in sample H but were barely found in sample D and sample F (Figure 1B). On the genus level, Woeseia and Loktanella were the major bacterial genera in the non-clam area, Woeseia was dominant in the clam naturally growing area, and Trichococcus, Lutibacter, and Psychrobacter were dominant in the clam aquaculture area (Supplementary Figure 5).
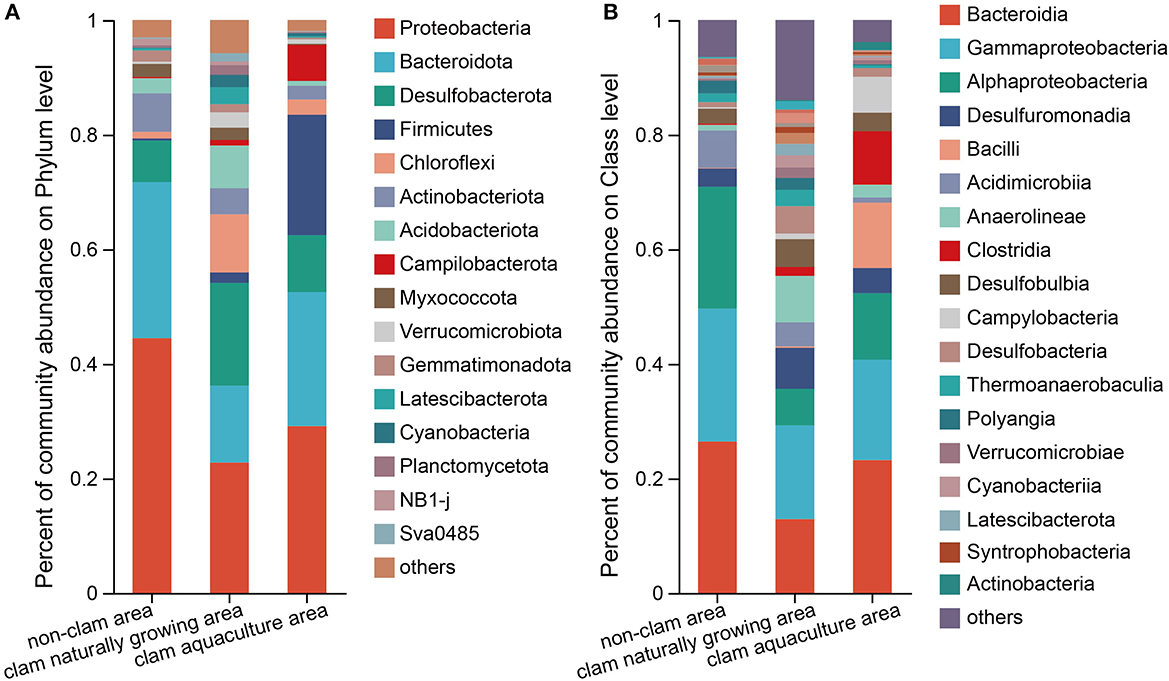
Figure 1. Composition of the bacterial community of the mud samples on phylum level (A) and class level (B). The mud samples were taken from the non-clam area (sample D), the clam naturally growing area (sample F), and the clam aquaculture area (sample H). The microbial community was investigated by culture-independent methods. The column stands for different mud samples, and the row represents the relative percentage of each bacterial type, which is depicted by different colors.
Protease is a key factor involved in nitrogen recycling (Zaman et al., 1999; Kamimura and Hayano, 2000). To further study the microbe-associated organic nitrogen degradation in coastal mudflats, we examined the cultivable protease-producing bacterial communities in mud samples by using culture-dependent methods. Moreover, culture-dependent methods have been commonly used for the discovery of novel species and bacterial products, which may further develop as protease-producing microbial agents for depollution of the environment. In general, the cultivability of the microbial community in the coastal area ranges from 0.1 to 0.01% (Al-Mailem et al., 2014; Rajeev et al., 2019). A number of colonies appeared on the screening plates of the 10−1-10−4 diluted mud samples after cultivation. The bacterial abundance reached 104 cells/g mud in the non-clam area (sample D) and the clam aquaculture area (sample H) and 105 cells/g mud in the clam naturally growing area (sample F). Clear hydrolytic zones were found around approximately 60% of the colonies on the screening plates. These results indicated that a large quantity of protease-producing bacteria inhabited the studied coastal muds.
The pure strain of the above colonies was obtained by repeated streaking cultivation. The nearly full-length 16S rRNA genes of the isolates were amplified and sequenced, and isolates with two or more different nucleotides in their 16S rRNA gene were considered as different strains. Finally, a total of 61 different protein hydrolyzing bacterial strains were isolated; 17 strains from sample D, 26 strains from sample F, and 18 strains from sample H (Supplementary Table 2). The 61 strains were classified into three phyla, four classes, nine families, and 18 genera (Figure 2). On the phylum and class level of the culturable fraction, the phyla Firmicutes and Proteobacteria and the classes Bacilli and Gammaproteobacteria were the largest groups in the three studied areas (Figure 2A). On the family level of the culturable fraction, Bacillaceae was the major family in all the studied areas and the dominance of Pseudoalteromonadaceae exists only in the clam naturally growing area (Figure 2B). The family Bacillaceae contains Bacillus-like species, including the five genera Bacillus, Alkalihalobacillus, Rossellomorea, Halobacillus, and Cytobacillus, with an abundance of 52.94% in sample D, 30.77% in sample F, and 50% in sample H, implying a dominance of Bacillus-like bacteria in the analyzed culturable fraction from coastal mudflats (Figures 2B, C). Pseudoalteromonas dominated in sample F (34.62%), meanwhile only a minor genus dominated in sample D (5.88%) and sample H (5.6%) (Figure 2C). In addition, the isolated protease-producing bacteria of sample F affiliated with 13 genera were apparently more diverse than that of sample D with seven genera and sample H with eight genera (Figure 2C), which was consistent with the findings of bacterial diversity by the above culture-independent methods.
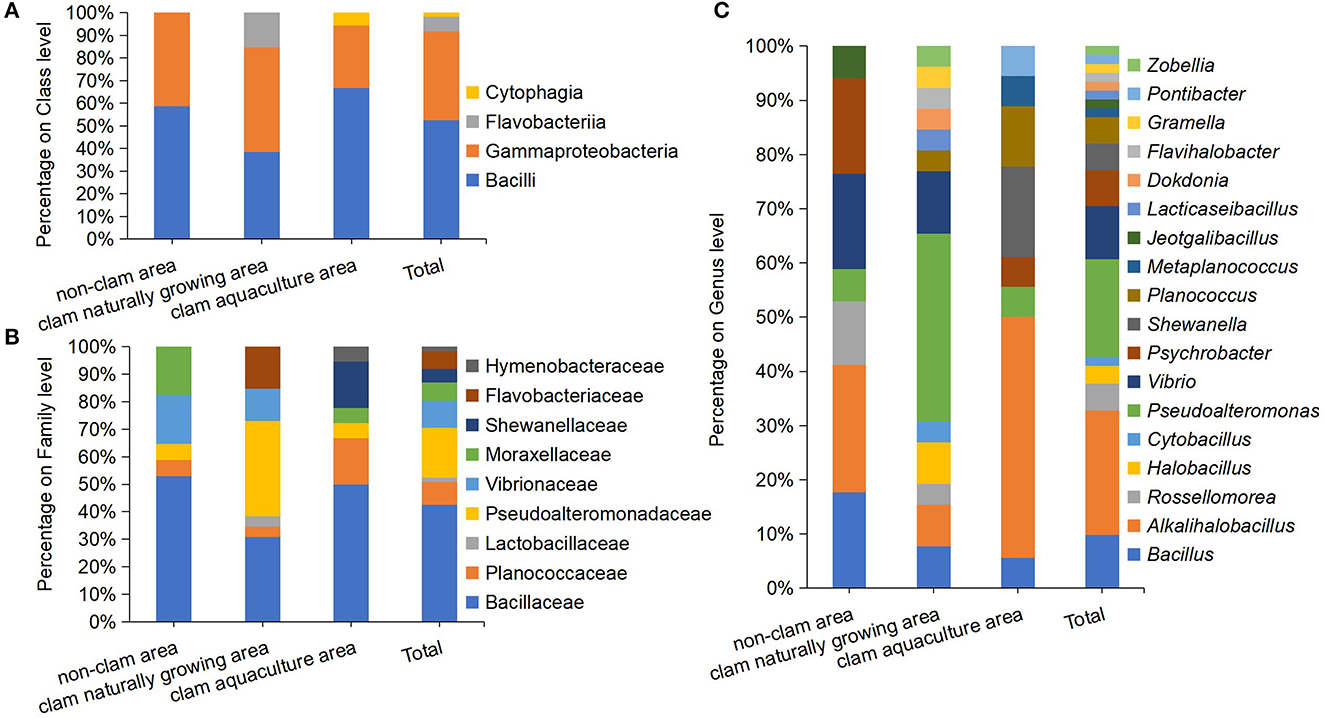
Figure 2. Diversity and distribution of culturable protease-producing bacteria of mud samples on class level (A), family level (B), and genus level (C). The mud samples were taken from the non-clam area (sample D), the clam naturally growing area (sample F), and the clam aquaculture area (sample H), and the microbial community was investigated by culture-dependent methods. Each color represents the percentage of the taxon in the total isolates.
In addition, neighbor-joining phylogenetic trees of the protease-producing strains with different genera based on 16S rRNA gene sequences were constructed (Figure 3). The relationship between the strains isolated from the non-clam area, the clam naturally growing area, and the clam aquaculture area is shown in Figures 3A–C, respectively. Alkalihalobacillus strain H1-7 illustrated a distant relationship with other recognized species and may represent a new taxonomic unit worthy of further research and investigation.
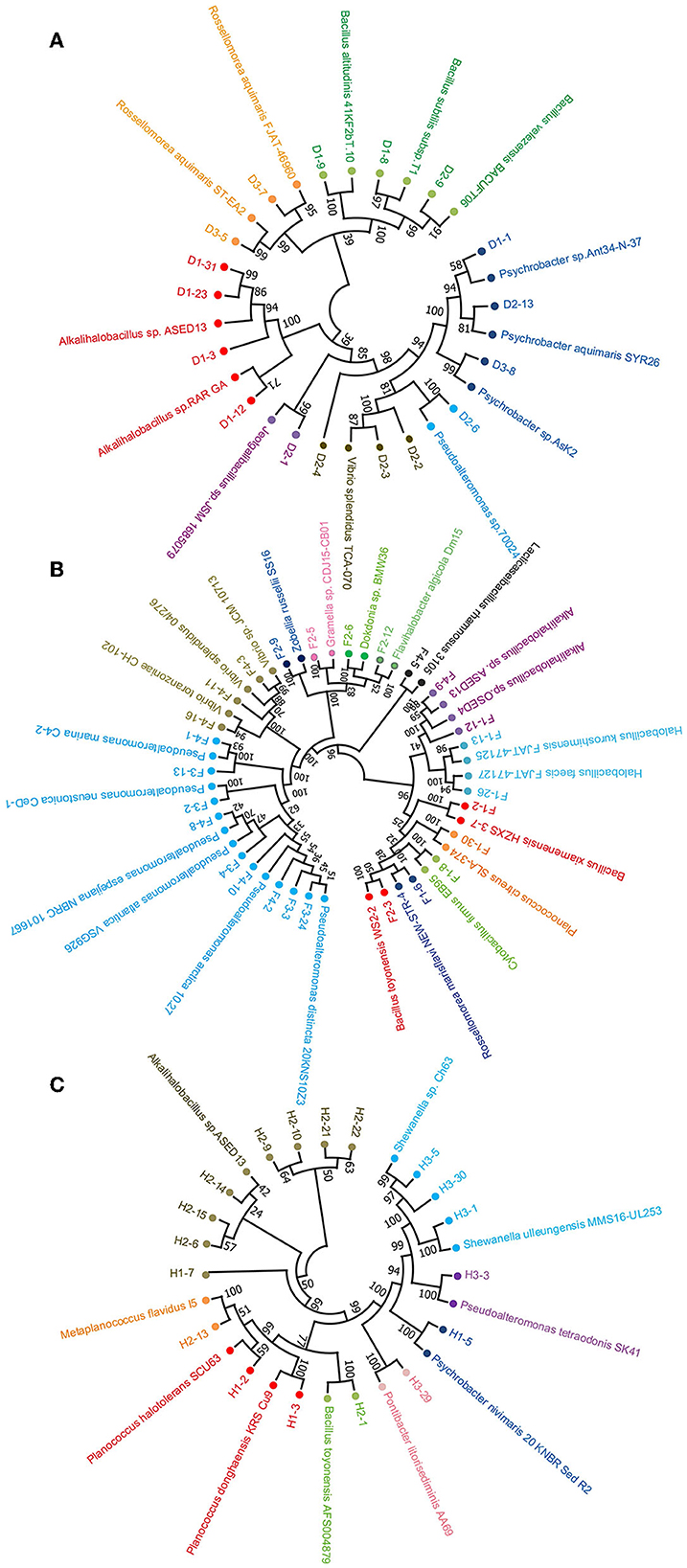
Figure 3. Neighbor-joining phylogenetic tree based on 16S rRNA sequencing of protease-producing bacteria isolated from three stations of coastal mudflat in Jiaozhou Bay using MEGA 11. The stations were the non-clam area (A), the clam naturally growing area (B), and the clam aquaculture area (C). Strains affiliated with different genera are shown in different colors.
Furthermore, the diversity of the bacterial extracellular protease was investigated by protein substrate specificity testing and protease inhibitor assay. Among the total 61 isolated strains, the extracellular proteases from 59, 55, and 55 strains could hydrolyze milk powder, casein, and gelatin, respectively, forming clear hydrolytic zones (Supplementary Table 2). The difference in the H/C ratios of the bacterial strains reflected their variation in substrate specificity. There was a certain variety in the activity of extracellular protease toward different protein substrates. The extracellular proteases secreted from Alkalihalobacillus strains F4-9 and H1-7, Bacillus strains D1-8, F1-2, and F2-3, Halobacillus strain F1-13, Jeotgalibacillus strain D2-1, and Psychrobacter strains H1-5 had high hydrolytic activity toward milk powder, with H/C ratios higher than five. The proteases from the Alkalihalobacillus strain D1-12, Bacillus strains D1-8 and F1-2, and Planococcus strain H1-3 showed high hydrolytic activity toward casein. The proteases from the Bacillus strain D2-9, Halobacillus strain F1-13, and Psychrobacter strains D2-13, D1-1, and H1-5 exhibited high hydrolytic activity toward gelatin.
In addition, the diversity of bacterial extracellular proteases was further investigated by detecting the inhibition ratios of PMSF (serine protease inhibitor), OP (metalloprotease inhibitor), Pepstatin A (aspartic protease inhibitor), and E64 (cysteine protease inhibitor). The 61 strains were cultivated in the liquid screening medium, and only 20 strains affiliated with the genera Alkalihalobacillus, Bacillus, Planococcus, Rossellomorea, Halobacillus, Jeotgalibacillus, Psychrobacter, Pseudoalteromonas, and Shewanella were able to produce enough extracellular proteases for protease inhibitor assay (Table 3). The extracellular protease activities of all 20 strains were inhibited by PMSF at the degree of 8.77–68.97%, indicating that all the coastal mudflat strains produced serine proteases. OP inhibited the protease activities of 16 strains by more than 10%, indicating that a majority of the isolated strains produced extracellular metalloproteases. In particular, an inhibition ratio as high as 84.97% was observed in the Alkalihalobacillus strain F1-12, suggesting that the Alkalihalobacillus strain F1-12 mainly produced extracellular metalloproteases. In contrast, Pepstatin A inhibited extracellular protease activity by 20% or less, indicating that these strains secrete very little aspartic or cysteine proteases. Moreover, E64 exhibited little inhibitory effect on the extracellular protease activities, with the exception of the Alkalihalobacillus strain F1-12, Bacillus strains D1-8 and D1-9, and Halobacillus strain F1-13, demonstrating that only a minority of the strains produced extracellular cysteine protease.
Investigating microbial diversity and functionality is of crucial importance in demonstrating “who's there and what do they do.” The culture-dependent method is the traditional way to explore and discover novel bacterial strains and products (Chen et al., 2003; Zhang et al., 2015). In recent years, the microbial diversity of environmental samples has been commonly investigated by culture-independent methods based on the rapid development of Illumina sequencing (Li Q. et al., 2022; Zhang et al., 2022). In this study, culture-dependent methods and culture-independent methods were interrelated for a better understanding of the bacterial diversity in the coastal mudflat, especially the protease-producing bacteria and their extracellular proteases.
The coastal mudflats are the transition zone of marine and terrestrial land, serving as an indispensable member of the global biogeochemical system. Excessive anthropogenic nitrogen input is a key factor influencing the coastal ecosystem (Li K. et al., 2019). Nitrogen loading caused eutrophication of the tidal flat environment, leading to hypoxia, red tides, and sediment loss (Cui et al., 2013; Cheng et al., 2020). In addition to inorganic nitrogen, environmental organic matters contain nitrogen mostly as protein. The organic feed and fertilizers applied in farming and aquaculture, animal feces and carcasses, as well as plant debris, contained a large amount of protein, an excess of which is an organic pollutant in coastal areas (Hodge et al., 2000; Davies et al., 2022). Soil enzymatic activity, especially soil hydrolase enzymes including protease, serves as a biological indicator to examine soil health (Maliang et al., 2020; Farooq et al., 2021). Microbes that inhabit coastal sediments play an important part in coastal biochemical cycling (Jiao et al., 2018). The protease-producing bacteria participated in the protection of coastal mudflats by secreting protease and degrading the protein in coastal mud.
It has been reported that nitrogen and phosphorus were released from an aquaculture farm and introduced pollution and eutrophication to the surrounding environments (Kawasaki et al., 2016). Consistently, in our study, the total concentrations of carbon, nitrogen, and phosphorus in the clam aquaculture area were 22.4 g/kg, 0.76 g/kg, and 0.62 g/kg, respectively, all of which were significantly higher than the corresponding values in the non-clam area of 9.64 g/kg (TC), 0.52 g/kg (TN), and 0.31 g/kg (TP). Recent studies have reported that 57% of nitrogen and 76% of phosphorus in aquatic feed were lost to the aquaculture water environment (Kong et al., 2020), and that the total nitrogen concentrations in the contaminated seawater of fishing harbors were two times higher than those of uncontaminated samples (Cheffi et al., 2022). All the related studies demonstrated that the fast-growing aquaculture industry had a great influence on the biogeochemistry factors of the surrounding areas.
We found that Proteobacteria was the most abundant phylum (44.47%) of the non-clam area in the coastal mudflat of Jiaozhou Bay, and the dominant class Gammaproteobacteria constituted 23.23% of the bacterial community. The dominance of Proteobacteria in the microbiota of various coastal environments has been reported in previous studies. Proteobacteria was found to be the most abundant phylum in two Atlantic coastal areas of France and Portugal, representing 89.3 and 82.3% of the total microbiota, respectively (Leite et al., 2017), as well as in the surrounding seawater of cultivation farms along coastal areas of the Yellow Sea (35.25%) (Ahmed et al., 2021), and in three Kerkennian fishing harbors (51.02–66.7%) (Cheffi et al., 2022). The bacterial community of Jiaozhou Bay sediments was also investigated, and the results showed that Proteobacteria was the most dominant phylum (61.3%), among which Gammaproteobacteria constituted the most abundant class (32.8%) (Liu et al., 2014). In addition, Proteobacteria and Gammaproteobacteria predominantly existed in the clam naturally growing area and the clam aquaculture area, with the composition of Proteobacteria accounting for 22.76 and 29.13%, as well as Gammaproteobacteria accounting for 16.37 and 17.51%. This was consistent with previous reports on the prevalence of Proteobacteria in the microbiota of marine invertebrates, such as oysters and hydroid (Fernández et al., 2013; Guo et al., 2017). On the other hand, we investigated the cultivable protease-producing bacteria in the studied areas and found an abundant bacterial community in the phylum Proteobacteria, especially in the class Gammaproteobacteria, which was affiliated with four genera Pseudoalteromonas, Vibrio, Psychrobacter, and Shewanella. Proteobacteria accounted for 41.18% in the non-clam area, 46.15% in the clam naturally growing area, and 27.78% in the clam aquaculture area of the total culturable protease-producing bacteria, which was consistent with the findings of the culture-independent approaches.
Members of Firmicutes were found as a minor phylum in the sediments of Jiaozhou Bay (Liu et al., 2014), which was consistent with our findings that Firmicutes only account for 0.37% of the microbial biomass in the non-clam area. However, the amount of Firmicutes increased to 1.85% in the clam naturally growing area and even to 21.03% in the clam aquaculture area. These findings are in line with the reports of Zhao et al. (2022), who observed Firmicutes as the most abundant phylum of the core bacterial communities in clams, with a composition of 26.2%. The prevalence of Firmicutes has also been shown in the microbiota of several marine invertebrates and aquatic environments of different geographic locations, such as the marine ark shell in the Japan Sea (Romanenko et al., 2009), the Easter oyster in the coastal Bay of USA (King et al., 2012), and the sponge in the Western Mediterranean Sea (Bauvais et al., 2015).
On the genus level, we found that Woeseia, Loktanella, Trichococcus, Lutibacter, and Psychrobacter were dominant genera in the studied mudflat areas. Woeseia genus was considered a core member of the microbial community in the marine ecosystem, which is characterized by the ability to assimilate inorganic carbon (Dyksma et al., 2016). Woeseia strains have been frequently isolated from coastal sediments and coastal seawaters (Du et al., 2016; Xu et al., 2022). Loktanella was reported to be a dominant genus in the coastal microbial community (Cardoso et al., 2019), and large numbers of Loktanella strains have been isolated from tidal flat sediments (Park et al., 2013, 2014; Tanaka et al., 2014). Trichococcus strains were characterized by their psychrotolerant ability, which was commonly isolated from environments at low temperatures such as cold spring (Zakharyuk et al., 2021). Lutibacter was reported to be predominant in shallow water marine sediment (Kerfahi et al., 2014), and some strains have been isolated from tidal flat sediments (Choi and Cho, 2006). However, except for a Trichococcus pasteurii strain that was isolated from freshwater crawfish and has been reported to produce alkaline proteases to degrade the myofibrillar protein (Qiu et al., 2022), no other studies report on the protease-producing ability of these four genera. What is different is the Psychrobacter genus. Psychrobacter strains were also known for psychrophilic characteristics, and some strains have been reported to produce cold-active protease (Amato and Christner, 2009; Perfumo et al., 2020). In this study, the Psychrobacter genus was found to be the third in abundance in sample H and the 35th in sample D based on the results of culture-independent methods. Correspondingly, as a result of culture-dependent methods, Psychrobacter was found to be a predominant genus of the analyzed culturable fraction in coastal mudflats, with an abundance of 17.65% in sample D, 5.6% in sample H, and 6.56% in total.
Bacillus genus, as a member of the phylum Firmicutes, has been frequently reported because of their bioremediation potentials of organic and inorganic compounds, which are also essential in the clam aquaculture area (Hsieh et al., 2020; Kumar et al., 2022; Liu et al., 2023). Zhang et al. (2015) isolated sixty-six protease-producing bacteria from Jiaozhou Bay sediments. Of all these cultivable protease-producing bacteria, Bacillus was found to be a major group (25.8%) (Zhang et al., 2015). Correspondingly, in our study, Bacillus-like bacterial communities in the class Bacilli and family Bacillaceae that were affiliated with five genera, including Bacillus, Alkalihalobacillus, Rossellomorea, Halobacillus, and Cytobacillus, accounted for 52.94% in the non-clam area, 30.77% in the clam naturally growing area, and 50% in the clam aquaculture area. Analogously, Bacillus-like species were also found as one of the dominant groups in other marine habits, e.g., sediments of mangrove wetlands in Hainan, China (Liu et al., 2017), the coast of South India (Sinimol et al., 2016), and the continental slope of eastern Arabian Sea (Farha et al., 2019). In addition, members of the genus Bacillus and related genera were widespread in terrestrial environments and exhibited rapid growth in high protein media (Manktelow et al., 2020), which may also cause the high composition of Bacillus-like species isolated from the studied areas, originating from surrounding terrestrial habits.
Additionally, according to the inhibitor assay on protease, we found that serine protease and metalloprotease were secreted by the isolated bacteria of the studied area as the principal types of proteases, generally similar to previous studies on sediments of the Antarctic Sea, sediments of the South China Sea, soils of the Antarctica South Shetland Islands, and sediments of Jiaozhou Bay (Zhou et al., 2009; Hou et al., 2013; Zhang et al., 2015; Liu et al., 2021). Relatively few strains isolated from Jiaozhou Bay sediments were found to secrete cysteine proteases (Zhang et al., 2015), and a minority of bacterial strains isolated from the soils of the Antarctica South Shetland Islands were reported to secrete aspartic and/or cysteine proteases (Liu et al., 2021). In this study, we also found that a small quantity of aspartic protease and cysteine protease was produced by a few strains isolated from the non-clam area and the clam naturally growing area. These bacterial extracellular protease findings may shed light on the understanding of organic nitrogen degradation in coastal mudflat areas and the development of microbial protease agents to prevent pollution and eutrophication.
In summary, this study analyzed the bacterial taxonomy composition of different clam-growing areas in the coastal mudflats of Jiaozhou Bay, especially protease-producing bacteria and their extracellular protease. The results showed that the diversity of the bacterial community and the protease-producing bacteria of the clam naturally growing area was higher than that of the non-clam area and the clam aquaculture area. Bacillus-like species, including the genera Bacillus, Alkalihalobacillus, Rossellomorea, Halobacillus, and Cytobacillus, were the dominant cultivated protease-producing groups in the Jiaozhou Bay coastal mudflats, and serine proteases and metalloproteases were the principal types of proteases produced by the bacteria. These findings contribute to a better understanding of the function of protease-producing bacteria in organic nitrogen degradation in coastal mudflat areas and the development of bacterial protease agents to improve the coastal aquaculture quality.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
JY designed the research and wrote the manuscript. ZL and XG conducted the research. GL collected samples, carried out bioinformatic analyzes, and revised the manuscript. YL, NJ, XX, and QS contributed ideas to the study. All authors contributed to the article and approved the submitted version.
This study was supported by the National Natural Science Foundation of China (Grant No. 32000004), the Research Fund of Qingdao Special Food Research Institute (Grant No. 6602422203), the Qingdao Agricultural University Research Fund for High-level Professionals (Grant No. 1120088), and the Breeding Plan of Shandong Provincial Qingchuang Research Team-Innovation Team of Functional Plant Protein-Based Food (2021).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1164937/full#supplementary-material
Ahmed, A., Khurshid, A., Tang, X., Wang, J., Khan, T. U., Mao, Y., et al. (2021). Structural and Functional Impacts of Microbiota on Pyropia yezoensis and Surrounding Seawater in Cultivation Farms along Coastal Areas of the Yellow Sea. Microorganisms. 9, 1291. doi: 10.3390/microorganisms9061291
Al-Mailem, D., Eliyas, M., Khanafer, M., and Radwan, S. (2014). Culture-dependent and culture-independent analysis of hydrocarbonoclastic microorganisms indigenous to hypersaline environments in Kuwait. Microb. Ecol. 67, 857–865. doi: 10.1007/s00248-014-0386-5
Amato, P., and Christner, B. C. (2009). Energy metabolism response to low-temperature and frozen conditions in Psychrobacter cryohalolentis. Appl. Environ. Microbiol. 75, 711–718. doi: 10.1128/AEM.02193-08
Bauvais, C., Zirah, S., Piette, L., Chaspoul, F., Domart-Coulon, I., Chapon, V., et al. (2015). Sponging up metals: bacteria associated with the marine sponge Spongia officinalis. Marine Environ. Res. 104, 20–30. doi: 10.1016/j.marenvres.2014.12.005
Böer, S. I., Hedtkamp, S. I. C., van Beusekom, J. E. E., Fuhrman, J. A., Boetius, A., and Ramette, A. (2009). Time- and sediment depth-related variations in bacterial diversity and community structure in subtidal sands. ISME J. 3, 780–791. doi: 10.1038/ismej.2009.29
Cardoso, D. C., Cretoiu, M. S., Stal, L. J., and Bolhuis, H. (2019). Seasonal development of a coastal microbial mat. Scient. Rep. 9, 490. doi: 10.1038/s41598-019-45490-8
Cheffi, M., Belmabrouk, S., Karray, F., Hentati, D., Bru-Adan, V., and Godon, J.-J. (2022). Study of microbial communities and environmental parameters of seawater collected from three Tunisian fishing harbors in Kerkennah Islands: Statistical analysis of the temporal and spatial dynamics. Marine Pollut. Bull. 185, 114350. doi: 10.1016/j.marpolbul.2022.114350
Chen, X.-L., Zhang, Y.-Z., Gao, P.-J., and Luan, X.-W. (2003). Two different proteases produced by a deep-sea psychrotrophic bacterial strain, Pseudoaltermonas sp. SM9913. Marine Biol. 143, 989–993. doi: 10.1007/s00227-003-1128-2
Cheng, F. Y., Van Meter, K. J., Byrnes, D. K., and Basu, N. B. (2020). Maximizing US nitrate removal through wetland protection and restoration. Nature. 588, 625–630. doi: 10.1038/s41586-020-03042-5
Choi, D. H., and Cho, B. C. (2006). Lutibacter litoralis gen. nov., sp. nov., a marine bacterium of the family Flavobacteriaceae isolated from tidal flat sediment. Int. J. Syst. Evol. Microbiol. 56, 771–776. doi: 10.1099/ijs.0.64146-0
Cui, S., Shi, Y., Groffman, P. M., Schlesinger, W. H., and Zhu, Y. G. (2013). Centennial-scale analysis of the creation and fate of reactive nitrogen in China (1910–2010). Proc. Nat. Acad. Sci. USA. 110, 2052–2057. doi: 10.1073/pnas.1221638110
Das, n. (2012). Bacterial isolates of marine coast as commercial producer of protease. OnLine J. Biol. Sci. 12, 96–107. doi: 10.3844/ojbsci.2012.96.107
Davies, B., Coulter, J. A., and Pagliari, P. H. (2022). Soil enzyme activity behavior after urea nitrogen application. Plants. 11, 2247. doi: 10.3390/plants11172247
Du, Z. J., Wang, Z. J., Zhao, J. X., and Chen, G. J. (2016). Woeseia oceani gen. nov., sp. nov., a chemoheterotrophic member of the order Chromatiales, and proposal of Woeseiaceae fam. nov. Int. J. Syst. Evol. Microbiol. 66, 107–112. doi: 10.1099/ijsem.0.000683
Dyksma, S., Bischof, K., Fuchs, B. M., Hoffmann, K., and Mumann, M.arc. (2016). Ubiquitous Gammaproteobacteria dominate dark carbon fixation in coastal sediments. ISME J. 10, 1939–1953. doi: 10.1038/ismej.2015.257
Farha, A. K., Tr, T., Purushothaman, A., Salam, J. A., and Hatha, A. M. (2019). Phylogenetic diversity and biotechnological potentials of marine bacteria from continental slope of eastern Arabian Sea. J. Genetic Eng. Biotechnol. 16, 253–258. doi: 10.1016/j.jgeb.2018.06.002
Farooq, T., Kumar, U., Mo, J., Shakoor, A., Wang, J., Rashid, M., et al. (2021). Intercropping of peanut-tea enhances soil enzymatic activity and soil nutrient status at different soil profiles in subtropical southern China. Plants (Basel, Switzerland). 10, 881. doi: 10.3390/plants10050881
Fernández, T. N., Mazón-Suástegui, J. M., Vázquez-Juárez, R., Ascencio-Valle, F., and Romero, J. (2013). Changes in the composition and diversity of the bacterial microbiota associated with oysters (Crassostrea corteziensis, Crassostrea gigas and Crassostrea sikamea) during commercial production. FEMS Microbiol. Ecol. 88, 69–83. doi: 10.1111/1574-6941.12270
Gaonkar, S. K., and Furtado, I. J. (2020). Characterization of Extracellular Protease from the Haloarcheon Halococcus sp. Strain GUGFAWS-3 (MF425611). Current Microbiol. 77, 1024–1034. doi: 10.1007/s00284-020-01896-6
Gobet, A., Böer, S. I., Huse, S. M., van Beusekom, J. E. E., Quince, C., Sogin, M. L., et al. (2012). Diversity and dynamics of rare and of resident bacterial populations in coastal sands. ISME J. 6, 542–553. doi: 10.1038/ismej.2011.132
Guo, H., Rischer, M., Sperfeld, M., Weigel, C., Menzel, K. D., Clardy, J., et al. (2017). Natural products and morphogenic activity of γ-Proteobacteria associated with the marine hydroid polyp Hydractinia echinata. Bioorg. Med. Chem. 25, 6088–6097. doi: 10.1016/j.bmc.2017.06.053
Hodge, A., Stewart, J., Robinson, D., Griffiths, B. S., and Fitter, A. H. (2000). Plant N capture and microfaunal dynamics from decomposing grass and earthworm residues in soil. Soil. Biol. Biochem. 32, 1763–1772. doi: 10.1016/S0038-0717(00)00095-X
Hou, L., Zheng, Y., Liu, M., Gong, J., Zhang, X., Yin, G., et al. (2013). Anaerobic ammonium oxidation (anammox) bacterial diversity, abundance, and activity in marsh sediments of the Yangtze Estuary. J. Geophys. Res. 118, 1237–1246 doi: 10.1002/jgrg.20108
Hsieh, H.-Y., Lin, C.-H., Hsu, S.-Y., and Stewart, G. C. (2020). A Bacillus spore-based display system for bioremediation of atrazine. Appl. Environ. Microbiol. 86, e01230–e01220. doi: 10.1128/AEM.01230-20
Hu, W., Zhang, W., Zhang, L., Tong, C., Sun, Z., Chen, Y., et al. (2019). Nitrogen along the hydrological gradient of marsh sediments in a subtropical estuary: pools, processes, and fluxes. Int. J. Environ. Res. Public Health. 16, 2043. doi: 10.3390/ijerph16112043
Hutchins, D. A., and Capone, D. G. (2022). The marine nitrogen cycle: new developments and global change. Nat. Rev. Microbiol. 20, 401–414. doi: 10.1038/s41579-022-00687-z
Jiao, S., Chen, W., Wang, J., Du, N., Li, Q., Wei, G., et al. (2018). Soil microbiomes with distinct assemblies through vertical soil profiles drive the cycling of multiple nutrients in reforested ecosystems. Microbiome. 6, 146. doi: 10.1186/s40168-018-0526-0
Kamimura, Y., and Hayano, K. (2000). Properties of protease extracted from tea-field soil. Biol. Fertil. Soils. 30, 351–355. doi: 10.1007/s003740050015
Kawasaki, N., Kushairi, M. R. M., Nagao, N., Yusoff, F., Imai, A., Kohzu, A., et al. (2016). Release of Nitrogen and Phosphorus from Aquaculture Farms to Selangor River, Malaysia. Int. J. Environ. Sci. Develop. 7, 113–116. doi: 10.7763/IJESD.2016.V7.751
Kerfahi, D., Hall-Spencer, J. M., Tripathi, B. M., Milazzo, M., Lee, J., Adams, J., et al. (2014). Shallow water marine sediment bacterial community shifts along a natural CO2 gradient in the mediterranean sea off Vulcano, Italy. Microbial. Ecol. 67, 819–828. doi: 10.1007/s00248-014-0368-7
King, G. M., Judd, C., Kuske, C. R., and Smith, C. (2012). Analysis of stomach and gut microbiomes of the eastern oyster (Crassostrea virginica) from coastal Louisiana, USA. PLoS ONE. 7, e51475. doi: 10.1371/journal.pone.0051475
Kong, W., Huang, S., Yang, Z., Shi, F., Feng, Y., Khatoon, Z., et al. (2020). Fish feed quality is a key factor in impacting aquaculture water environment: evidence from incubator experiments. Scient. Rep. 10, 187. doi: 10.1038/s41598-019-57063-w
Kumar, R., Singh, A., Maurya, A., Yadav, P., Yadav, A., Chowdhary, P., et al. (2022). Effective bioremediation of pulp and paper mill wastewater using Bacillus cereus as a possible kraft lignin-degrading bacterium. Bioresour. Technol. 352, 127076. doi: 10.1016/j.biortech.2022.127076
Leite, L., Jude-Lemeilleur, F., Raymond, N., Henriques, I., Garabetian, F., and Alves, A. (2017). Phylogenetic diversity and functional characterization of the Manila clam microbiota: a culture-based approach. Environ. Sci. Pollut. Res. 24, 21721–21732. doi: 10.1007/s11356-017-9838-z
Li, H., Chi, Z., Li, J., Wu, H., and Yan, B. (2019). Bacterial community structure and function in soils from tidal freshwater wetlands in a Chinese delta: Potential impacts of salinity and nutrient. Sci. Total Environ. 696, 134029.1–134029.9. doi: 10.1016/j.scitotenv.2019.134029
Li, K., Liu, C., Ma, Y., and Wang, X. (2019). Land-based dissolved organic nitrogen dynamics and bioavailability in Jiaozhou Bay, China. Estuarine, Coastal Shelf Sci. 220, 13–24. doi: 10.1016/j.ecss.2019.02.045
Li, Q., Zhang, Y., Hu, J., and Dai, Q. (2022). Response of bacterial communities and nitrogen-cycling genes in newly reclaimed mudflat paddy soils to nitrogen fertilizer gradients. Environ. Sci. Pollut. Res. 29, 71113–71123. doi: 10.1007/s11356-022-20770-5
Li, T., Zhang, G., Wang, S., Mao, C., Tang, Z., Rao, W., et al. (2022). The isotopic composition of organic carbon, nitrogen and provenance of organic matter in surface sediment from the Jiangsu tidal flat, southwestern Yellow Sea. Marine Pollut. Bull. 182, 114010. doi: 10.1016/j.marpolbul.2022.114010
Li, Y., Wang, Y., Shen, C., Xu, L., Yi, S., Zhao, Y., et al. (2021). Structural and predicted functional diversities of bacterial microbiome in response to sewage sludge amendment in coastal mudflat soil. Biology. 10, 1302. doi: 10.3390/biology10121302
Lin, J., Zhang, L., Zhang, M., Zhang, H., Guo, C., Feng, S., et al. (2022). Distribution, sources, and ecological risk of organophosphate esters in the urbanized Jiaozhou Bay, East China. Environ. Sci. Pollut. Res. 29, 70167–70178. doi: 10.1007/s11356-022-20367-y
Liu, C., Ma, S., Wang, X., Ou, Y., and Du, H. (2023). Biodegradation of organic compounds in the coal gangue by Bacillus sp. into humic acid. Biodegradation. 34, 125–138. doi: 10.1007/s10532-022-10007-0
Liu, J., Liu, W., Xing, S., Zhang, X., He, H., Chen, J., et al. (2021). Diversity of protease-producing bacteria in the soils of the South Shetland Islands, Antarctica. Antonie van Leeuwenhoek 114, 457–464. doi: 10.1007/s10482-021-01533-7
Liu, M., Cui, Y., Chen, Y., Lin, X., Huang, H., Bao, S., et al. (2017). Diversity of Bacillus-like bacterial community in the sediments of the Bamenwan mangrove wetland in Hainan, China. Can. J. Microbiol. 63, 238–245. doi: 10.1139/cjm-2016-0449
Liu, S. M., Zhu, B. D., Zhang, J., Wu, Y., Liu, G. S., Deng, B., et al. (2010). Environmental change in Jiaozhou Bay recorded by nutrient components in sediments. Marine Pollut. Bull. 60, 1591–1599. doi: 10.1016/j.marpolbul.2010.04.003
Liu, X., Hu, H.-W., Liu, Y.-R., Xiao, K.-Q., Cheng, F.-S., Li, J., et al. (2014). Bacterial composition and spatiotemporal variation in sediments of Jiaozhou Bay, China. J. Soils Sed. 15, 732–744. doi: 10.1007/s11368-014-1045-7
Long, X. H., Liu, L. P., Shao, T. Y., Shao, H. B., and Liu, Z. P. (2016). Developing and sustainably utilize the coastal mudflat areas in China. Sci. Total Environ. 569–570, 1077–1086. doi: 10.1016/j.scitotenv.2016.06.170
Maliang, H., Tang, L., Lin, H., Chen, A., and Ma, J (2020). Influence of high-dose continuous applications of pyroligneous acids on soil health assessed based on pH, moisture content and three hydrolases. Environ. Sci. Pollut. Res. 27, 15426–15439. doi: 10.1007/s11356-020-08075-x
Manktelow, C. J., White, H., Crickmore, N., and Raymond, B. (2020). Divergence in environmental adaptation between terrestrial clades of the Bacillus cereus group. FEMS Microbiol. Ecol. 97, 228. doi: 10.1093/femsec/fiaa228
Niu, Y., Zheng, Y., Hou, L., Gao, D., Chen, F., Pei, C., et al. (2022). Microbial dynamics and activity of denitrifying anaerobic methane oxidizers in China's estuarine and coastal wetlands. Sci. Total Environ. 806, 150425. doi: 10.1016/j.scitotenv.2021.150425
Park, S., Jung, Y. T., and Yoon, J. H. (2013). Loktanella sediminilitoris sp. nov., isolated from tidal flat sediment. Int. J. System. Evolut. Microbiol. 63, 4118–4123. doi: 10.1099/ijs.0.049841-0
Park, S.ooyeon., Jung, Yong-Taek., Won, Sung-Min., Park, Ja-Min., and Yoon, Jung-Hoon. (2014). Loktanella aestuariicola sp. nov., an alphaproteobacterium isolated from a tidal flat. Antonie van Leeuwenhoek. 106, 707–714. doi: 10.1007/s10482-014-0240-2
Perfumo, A., Freiherr von Sass, G. J., Nordmann, E. L., Budisa, N., and Wagner, D. (2020). Discovery and characterization of a new cold-active protease from an extremophilic bacterium via comparative genome analysis and in vitro expression. Front. Microbiol. 11, 881. doi: 10.3389/fmicb.2020.00881
Qiu, L., Zhao, Y., Ma, H., Tian, X., Bai, C., Liao, T., et al. (2022). The quality and bacterial community changes in freshwater crawfish stored at 4°C in vacuum packaging. Molecules. 27, 8618. doi: 10.3390/molecules27238618
Rajeev, M., Sushmitha, T. J., Toleti, S. R., and Pandian, S. K. (2019). Culture dependent and independent analysis and appraisal of early stage biofilm-forming bacterial community composition in the Southern coastal seawater of India. Sci. Total Environ. 666, 308–320. doi: 10.1016/j.scitotenv.2019.02.171
Ren, Y., Yu, G., Shi, C., Liu, L., Guo, Q., Han, C., et al. (2022). Majorbio Cloud: A one-stop, comprehensive bioinformatic platform for multiomics analyses. iMeta. 1, e12. doi: 10.1002/imt2.12
Romanenko, L. A., Uchino, M., Kalinovskaya, N. I., and Mikhailov, V. V. (2009). Isolation, phylogenetic analysis and screening of marine mollusc-associated bacteria for antimicrobial, hemolytic and surface activities. Microbiol. Res. 163, 633–644. doi: 10.1016/j.micres.2006.10.001
Sinimol, S., Sarika, A. R., and Jayakumaran Nair, A. (2016). Diversity and antagonistic potential of marine microbes collected from south-west coast of India. 3 Biotech. 6, 7. doi: 10.1007/s13205-015-0318-1
Soares, A. R. A., Kritzberg, E. S., Custelcean, I., and Berggren, M. (2018). Bacterioplankton responses to increased organic carbon and nutrient loading in a boreal estuary—separate and interactive effects on growth and respiration. Microbial. Ecol.76, 144–155. doi: 10.1007/s00248-017-1115-7
Suthindhiran, K., Jayasri, M. A., Dipali, D., and Prasar, A. (2013). Screening and characterization of protease producing actinomycetes from marine saltern. J. Basic Microbiol. 54, 1098–1109. doi: 10.1002/jobm.201300563
Tanaka, N., Romanenko, L. A., Kurilenko, V. V., Svetashev, V. I., Kalinovskaya, N. I., Mikhailov, V. V., et al. (2014). Loktanella maritima sp. nov. isolated from shallow marine sediments. Int. J. Syst. Evol. Microbiol. 64, 2370–2375. doi: 10.1099/ijs.0.061747-0
Taylor, J. D., Ellis, R., Milazzo, M., Hall-Spencer, J. M., and Cunliffe, M. (2014). Intertidal epilithic bacteria diversity changes along a naturally occurring carbon dioxide and pH gradient. FEMS Microbiol. Ecol. 89, 670–678. doi: 10.1111/1574-6941.12368
Tornkvist, A., Liu, C., and Moschou, P. N. (2019). Proteolysis and nitrogen: emerging insights. J. Exper. Botany. 70, 2009–2019 doi: 10.1093/jxb/erz024
Wei, Y., Bu, J., Long, H., Zhang, X., Cai, X., and Huang, A. (2021). Community structure of protease-producing bacteria cultivated from aquaculture systems: potential impact of a tropical environment. Front. Microbiol. 12, 638129. doi: 10.3389/fmicb.2021.638129
Xu, N., Wang, W., Xu, K., Xu, Y., Ji, D., Chen, C., et al. (2022). Cultivation of different seaweed species and seasonal changes cause divergence of the microbial community in coastal seawaters. Front. Microbiol. 13, 988743. doi: 10.3389/fmicb.2022.988743
Zakharyuk, A., Valyshev, A., Plotnikov, A., Kopitsyn, D., Suzina, N., Shcherbakova, et al. (2021). Trichococcus shcherbakoviae subsp. psychrophilus subsp. nov., a psychrotolerant facultative anaerobe isolated from a cold spring. Int. J. Syst. Evol. Microbiol. 71, 5068. doi: 10.1099/ijsem.0.005068
Zaman, M., Di, H. J., Cameron, K. C., and Frampton, C. M. (1999). Gross nitrogen mineralization and nitrification rates and their relationships to enzyme activities and the soil microbial biomass in soils treated with dairy shed effluent and ammonium fertilizer at different water potentials. Biol. Fertil. Soils. 29, 178–186. doi: 10.1007/s003740050542
Zhang, G., Bai, J., Tebbe, C. C., Zhao, Q., Jia, J., Wang, W., et al. (2021). Salinity controls soil microbial community structure and function in coastal estuarine wetlands. Environ. Microbiol. 23, 1020–1037. doi: 10.1111/1462-2920.15281
Zhang, J., Chen, M., Huang, J., Guo, X., Zhang, Y., Liu, D., et al. (2019). Diversity of the microbial community and cultivable protease-producing bacteria in the sediments of the Bohai Sea, Yellow Sea and South China Sea. PLoS ONE. 14, e0215328. doi: 10.1371/journal.pone.0215328
Zhang, L., Xiong, L., Li, J., and Huang, X. (2021). Long-term changes of nutrients and biocenoses indicating the anthropogenic influences on ecosystem in Jiaozhou Bay and Daya Bay, China. Marine Pollut Bull. 168, 112406. doi: 10.1016/j.marpolbul.2021.112406
Zhang, X., Ward, B. B., and Sigman, D. M. (2020). Global nitrogen cycle: critical enzymes, organisms, and processes for nitrogen budgets and dynamics. Chem. Rev. 120, 5308–5351. doi: 10.1021/acs.chemrev.9b00613
Zhang, X. Y., Han, X. X., Chen, X. L., Dang, H. Y., Xie, B. B., Qin, Q. L., et al. (2015). Diversity of cultivable protease-producing bacteria in sediments of Jiaozhou Bay, China. Front. Microbiol. 6, 1021. doi: 10.3389/fmicb.2015.01021
Zhang, Y. Z., Zhang, W. X., and Chen, X. L. (2020). Mechanisms for induction of microbial extracellular proteases in response to exterior proteins. Appl. Environ. Microbiol. 86, e01036–e01020. doi: 10.1128/AEM.01036-20
Zhang, Z., Han, P., Zheng, Y., Jiao, S., Dong, H., Liang, X., et al. (2022). Spatiotemporal Dynamics of Bacterial Taxonomic and Functional Profiles in Estuarine Intertidal Soils of China Coastal Zone. Microb. Ecol. 85, 383–399. doi: 10.21203/rs.3.rs-1155844/v1
Zhao, Z., Mao, Z., Xing, P., Tao, Y., and Wu, Q. (2022). Intrahabitat differences in bacterial communities associated with Corbicula fluminea in the large shallow eutrophic Lake Taihu. Appl. Environ. Microbiol. 88, e02328–e02321. doi: 10.1128/aem.02328-21
Zhou, M. Y., Chen, X. L., Zhao, H. L., Dang, H. Y., Luan, X. W., Zhang, X. Y., et al. (2009). Diversity of both the cultivable protease-producing bacteria and their extracellular proteases in the sediments of the South China sea. Microb. Ecol. 58, 582–590. doi: 10.1007/s00248-009-9506-z
Zhou, M. Y., Wang, G. L., Li, D., Zhao, D. L., Qin, Q. L., Chen, X. L., et al. (2013). Diversity of both the cultivable protease-producing bacteria and bacterial extracellular proteases in the coastal sediments of King George Island, Antarctica. PLoS ONE. 8, e79668. doi: 10.1371/journal.pone.0079668
Keywords: bacterial community, protease-producing bacteria, diversity, clam naturally growing area, clam aquaculture area, coastal mudflats
Citation: Liu Z, Liu G, Guo X, Li Y, Ji N, Xu X, Sun Q and Yang J (2023) Diversity of the protease-producing bacteria and their extracellular protease in the coastal mudflat of Jiaozhou Bay, China: in response to clam naturally growing and aquaculture. Front. Microbiol. 14:1164937. doi: 10.3389/fmicb.2023.1164937
Received: 13 February 2023; Accepted: 24 April 2023;
Published: 19 May 2023.
Edited by:
Xue-Wei Xu, Ministry of Natural Resources, ChinaReviewed by:
Xuwang Zhang, Dalian University of Technology, ChinaCopyright © 2023 Liu, Liu, Guo, Li, Ji, Xu, Sun and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jie Yang, cGhkeWFuZzEzMTRAMTYzLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.