
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 06 April 2023
Sec. Microbial Physiology and Metabolism
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1160736
This article is part of the Research TopicChromosome Architecture and DNA Topology in ProkaryotesView all 8 articles
Topoisomerase IV (Topo IV) is the main decatenation enzyme in Escherichia coli; it removes catenation links that are formed during DNA replication. Topo IV binding and cleavage sites were previously identified in the E. coli genome with ChIP-Seq and NorfIP. Here, we used a more sensitive, single-nucleotide resolution Topo-Seq procedure to identify Topo IV cleavage sites (TCSs) genome-wide. We detected thousands of TCSs scattered in the bacterial genome. The determined cleavage motif of Topo IV contained previously known cleavage determinants (−4G/+8C, −2A/+6 T, −1 T/+5A) and additional, not observed previously, positions −7C/+11G and −6C/+10G. TCSs were depleted in the Ter macrodomain except for two exceptionally strong non-canonical cleavage sites located in 33 and 38 bp from the XerC-box of the dif-site. Topo IV cleavage activity was increased in Left and Right macrodomains flanking the Ter macrodomain and was especially high in the 50–60 kb region containing the oriC origin of replication. Topo IV enrichment was also increased downstream of highly active transcription units, indicating that the enzyme is involved in relaxation of transcription-induced positive supercoiling.
Replication of long DNA molecules, especially, circular chromosomes, leads to accumulation of positive DNA supercoiling (Postow et al., 2001), which, through rotation of the DNA polymerase complex, is thought to lead to formation of precatenanes (overtwisted sister chromosomes) (Peter et al., 1998; Cebrián et al., 2015). If left unresolved, precatenanes interfere with bulk DNA segregation during replication and lead to sister chromosomes catenation after the replication is completed (Adams et al., 1992; Lucas et al., 2001). Specialized enzymes, topoisomerases, resolve such topological challenges and support proper DNA replication and segregation processes (Adams et al., 1992; Khodursky et al., 2000; Schvartzman et al., 2019).
In Escherichia coli, topoisomerase III, topoisomerase IV, and DNA gyrase are thought to contribute to the resolution of catenanes and segregation of replicating molecules (Adams et al., 1992; Hiasa et al., 1994; Khodursky et al., 2000; Seol et al., 2013; Lee et al., 2019). While the functions of these enzymes may be partially redundant, topoisomerase IV (Topo IV) is considered to be the main decatenase (Zechiedrich et al., 1997). Topo IV is encoded by the parC and parE genes and functions as a heterotetrameric complex ParC2ParE2 (Kato et al., 1990; Peng and Marians, 1993). Topo IV is a classical type-IIA topoisomerase: it introduces a transient double-stranded break in one segment of DNA (called G-segment, for “Gate”) and transfers another DNA segment (called T-segment, for “Transport”) through the break. To maintain the break during the transfer, the enzyme produces 4-nt 5′-overhangs with 5′-ends covalently linked to catalytic ParC Tyr residues located in the DNA-gate of the enzyme (Peng and Marians, 1993). In vitro and in vivo, Topo IV efficiently decatenates circular DNA molecules and resolves precatenanes (Adams et al., 1992; Zechiedrich and Cozzarelli, 1995; Seol et al., 2013). Topo IV specificity toward DNA crossovers is linked to the structure of the ParC C-terminal domain (CTD) (Hirsch and Klostermeier, 2021). Topo IV relaxes positive supercoiling more efficiently than negative supercoiling and thus may partially substitute for DNA gyrase, another type-IIA topoisomerase that also relieves positive supercoiling and, uniquely, introduces negative supercoils in an ATP-dependent process (Crisona et al., 2000; Khodursky et al., 2000; Ashley et al., 2017).
In E. coli, depletion of Topo IV results in a par phenotype (Kato et al., 1990): DNA replication rate is mildly inhibited (Khodursky et al., 2000), segregation of replicated DNA is impaired (Wang et al., 2008), genome structure is reorganized (Conin et al., 2022), and the rate of formation of “ghost” cells lacking DNA is increased (Grainge et al., 2007; Wang et al., 2008). Conversely, increased intracellular amounts of Topo IV accelerate segregation of replicated DNA (Wang et al., 2008).
Escherichia coli Topo IV has several known protein partners which modulate its activity via direct protein–protein interactions with CTDs. The condensin complex MukBEF interacts through its hinge domain with Topo IV (Hayama and Marians, 2010; Li et al., 2010; Vos et al., 2013). In vitro, this interaction was reported to stimulate the relaxation and decatenation activities of Topo IV (Hayama and Marians, 2010; Li et al., 2010), though this finding was later disputed (Kumar et al., 2022). In vivo, the interaction between MukBEF and Topo IV is necessary for timely segregation of newly replicated regions containing the origin of replication (Nicolas et al., 2014; Zawadzki et al., 2015) and chromosomal DNA (Kumar et al., 2017). At the origin, Topo IV and MukBEF were shown to form clusters consisting of 15–16 Topo IV and MukBEF complexes (Nicolas et al., 2014; Zawadzki et al., 2015; Nolivos et al., 2016). These clusters may act as a scaffold to initiate chromosomal DNA condensation (Vos et al., 2013). In vitro, decatenation and relaxation of negative supercoiling by Topo IV were reported to be stimulated by low concentrations of SeqA. The effect appears to be specific, since topoisomerase I or DNA gyrase activities were unaffected. However, high concentrations of SeqA caused non-specific inhibition of activities of all topoisomerases (Kang et al., 2003). SeqA binds hemimethylated DNA and therefore effectively follows the replisome during replication. It is required for temporary cohesion of newly replicated regions, which may be achieved by inhibition of Topo IV activity on SeqA-bound hemimethylated chromosomal regions behind the replisome (Joshi et al., 2013; Helgesen et al., 2021).
The Ter macrodomain (MD) of the E. coli chromosome is organized by the MatP protein, which binds to matS sites scattered through Ter (Mercier et al., 2008). MatP forms dimers and dimers of dimers, binds MukBEF, and promotes its unloading from DNA (Nolivos et al., 2016). The interaction of MatP and Topo IV with MukBEF was shown to be competitive (Fisher et al., 2021) and it was proposed that MatP-mediated displacement of MukBEF-Topo IV complexes from Ter promotes the turnover of complexes and their loading at the origin (Nolivos et al., 2016). In the middle of Ter, a dif-site is located. It is recognized by heterotetrameric resolvase complex XerC/XerD that resolves chromosome dimers to monomers (Blakely et al., 1993). A Topo IV cleavage hot-spot was found in the vicinity of the dif-site (Hojgaard et al., 1999; El Sayyed et al., 2016). XerC is required for Topo IV binding and cleavage at this site implying a direct interaction between Topo IV and XerC, which, however, has not yet been demonstrated (El Sayyed et al., 2016). Another essential component of resolvase machinery, an FtsK DNA translocase, also binds Topo IV and stimulates its decatenation and relaxation activities (Espeli et al., 2003; Bigot and Marians, 2010). Considering the FtsK interaction with XerD and activation of recombination by XerC/XerD (Grainge et al., 2011; Keller et al., 2016), a complex multiprotein machine must be operating at the dif-site to secure the resolution of chromosome dimers and catenanes.
The Topo IV cleavage specificity was extensively studied in vitro. Several symmetrical nucleotide determinants at positions −4G/+8C, −2A/+6 T, −1 T/+5A relative to the left cleavage site located at position +1 were identified as important for cleavage (Leo et al., 2005; Richter et al., 2007; Arnoldi et al., 2013). On a whole-genome level and in vivo, Topo IV binding and cleavage were studied by, respectively, ChIP-Seq and NorfIP. The latter method utilizes stabilization of the intermediate covalent cleavage complex between Topo IV and DNA by a topoisomerase poison norfloxacin. With NorfIP, the strong cleavage at dif was validated and hundreds of cleavage sites scattered throughout the E. coli genome were revealed (El Sayyed et al., 2016). Since NorfIP has a limited resolution, we here decided to map Topo IV cleavage sites with Topo-Seq, a method previously developed in our laboratory. Topo-Seq also employs stabilization properties of topoisomerase poisons to trap covalent complexes of type-II topoisomerases with DNA, but relies on a specific protocol for preparation of sequencing libraries from single-stranded DNA, which allows to map cleavage sites strand specifically and with a single-nucleotide resolution (Sutormin et al., 2019). Using Topo-Seq, we identified several thousand Topo IV cleavage sites in the E. coli genome and derived a Topo IV cleavage motif. The motif contains several determinants that were not reported previously and lacks the signs of DNA wrapping observed in the DNA gyrase cleavage motif (Sutormin et al., 2019). On a whole-genome level, Topo IV cleavage sites were significantly depleted in the Ter MD and highly enriched in a 50–60 kb region containing the origin of replication oriC. We found two strong non-canonical Topo IV cleavage signals, 33 and 38 bp from the XerC-box of the dif-site. Finally, we found that Topo IV enrichment was increased downstream of active transcription units, where positive supercoiling is accumulated. These results significantly expand the knowledge about Topo IV activity in the E. coli genome. With minimal modification, the Topo-Seq approach can be extended for studies of Topo IV in other bacteria.
Escherichia coli DY330 parC-SPA strain (W3110 lacU169 gal490 cI857 Δ(cro-bioA) parC-SPA) with parC gene fused with the sequence encoding the SPA tag (purchased from Horizon Discovery Biosciences) was used in Topo IV Topo-Seq experiments. E. coli DY330 parC-SPA gyrA-S83L was constructed from the original strain by recombineering. pKD46 plasmid was used for recombineering.
The gyrA-S83L mutation conferring resistance to fluoroquinolone antibiotics (Bagel et al., 1999), was introduced to the E. coli DY330 parC-SPA strain by recombineering using the template oligonucleotide complementary to the lagging DNA strand (Sawitzke et al., 2011). A culture of E. coli DY330 parC-SPA harboring the pKD46 plasmid was prepared for recombineering as described in Datsenko and Wanner (2000) and 50 μl aliquot of cells (~6*108 cells) was electroporated with 0.2 μl of 50 μM chemically synthesized 70-mer oligonucleotide (~6*1012 molecules) (GCGCCATGCGGACGATCGTGTCATAGACCGCCAGATCACCATGGGGATGG-TATTTACCGATTACGTCACC). Fluoroquinolone-resistant mutants were selected on LB plates containing 500 nM ciprofloxacin. To verify the mutation acquisition, a fragment of gyrA gene was PCR amplified using gyrA_F (ATGAGCGACCTTGCGAGAGAAATTACACCG) and gyrA_R (CCGGAATTTTTTCCG-TGCCGTC) primers and subjected to Sanger sequencing.
Minimal inhibitory concentrations (MIC) for ciprofloxacin and tetracycline were measured for E. coli DY330 parC-SPA and E. coli DY330 parC-SPA gyrA-S83L liquid cultures by a serial dilution method in 200-μL 96-well plates in LB. The plates were incubated at 37°C for 24 h and MIC was defined as a minimal concentration suppressing the growth of a liquid culture.
Purification and sequencing of DNA fragments covalently attached to the poisoned Topo IV (Topo-Seq) were performed with E. coli DY330 parC-SPA and E. coli DY330 parC-SPA gyrA-S83L as described previously for E. coli DNA gyrase (Sutormin et al., 2019). Briefly, ciprofloxacin (final concentration 10 μM) was added to the 50 ml of exponentially growing cell culture in LB medium when OD600 ~ 0.6, and the culture was incubated for 15 min with rotation at 37°C. Cells were harvested by centrifugation and washed twice with TES buffer (10 mM Tris–HCl pH 7.5, 1 mM EDTA, 250 mM NaCl). Then, cells were resuspended in 1 ml of TES and disrupted by sonication (DNA fragmentation range 200–800 bp). Lysate was cleared by centrifugation and incubated with 100 μl of ANTI-FLAG M2 affinity gel (Sigma-Aldrich). After immunoprecipitation, the resin was treated with Proteinase K (Sigma-Aldrich) in 200 μl of TES and DNA fragments were purified with AMPure XP magnetic beads from the supernatant (Beckman Coulter). Non-treated control was processed identically, except that ciprofloxacin was not added. For mock control, DNA was extracted from 100 μl of cleared lysate using the GeneJET DNA purification kit (Thermo Scientific).
NGS libraries were prepared using Accel NGS 1S kit (Swift Bioscience). DNA sequencing was performed with Illumina HiSeq 4000 in a 150 + 150 bp paired-end mode. Library preparation and sequencing were performed at Skoltech Genomics Core Facility. Topo-Seq was performed in triplicate.
Raw reads were filtered with Trimmomatic (Bolger et al., 2014) and then aligned to the E. coli W3110 MuSGS genome (the genome may be obtained from GEO: GSE95567) using BWA-MEM (Li and Durbin, 2010). SAM, BAM, and bed files were prepared with Samtools (Li et al., 2009). The number of DNA fragments’ 3′-ends (N3E) was calculated per position, based on the read alignments stored in SAM files. Normalization of N3E tracks and identification of Topoisomerase Cleavage Sites (TCSs) were performed as described previously (Sutormin et al., 2019). The resultant tracks were further analyzed using custom Python scripts.1 E. coli reference genome annotation with transcription units was obtained from the EcoCyc database (Karp et al., 2018).
Topo IV ChIP-Seq and NorfIP data were taken from publicly available GEO dataset GSE75641 (El Sayyed et al., 2016). Raw reads were aligned to the E. coli W3110 MuSGS reference genome, and alignment was converted to coverage depth tracks using Samtools (Li et al., 2009). Fold enrichment tracks were obtained by position-wise division of IP samples by corresponding mock samples. MatP and MukB ChIP-Seq datasets were taken from publicly available GEO dataset GSE67221 (Nolivos et al., 2016) and processed as described before. 3C data for wt E. coli were taken from GSE107301 (Lioy et al., 2018). Contact maps were generated using the pipelines2,3 (Lioy et al., 2018). E. coli W3110 MuSGS reference genome was used for reads alignment. Contact data were filtered with thresholds: 3 restriction fragments for uncuts and 2 restriction fragments for loops. Resultant contact maps were binned at 10 kb resolution. RNA-Seq data were taken from GEO dataset GSE181687 (Sutormin et al., 2022). Full list of NGS datasets used in this study can be found in Supplementary Table 2.
Plasmid DNA was extracted with GeneJet Plasmid Miniprep Kit (Thermo Fisher) from stationary phase (overnight growth, 5 ml) and exponential phase (OD600 = 0.6, 50 ml) cultures of E. coli DY330 parC-SPA and E. coli DY330 parC-SPA gyrA-S83L strains cultivated in LB at 37°C. 1.2 μg of plasmid was separated by electrophoresis (120 V) in 1% agarose gel in ice-cold TAE buffer supplemented with 2.5 μg/ml chloroquine and visualized by ethidium bromide staining.
Escherichia coli DY330 parC-SPA or E. coli DY330 parC-SPA gyrA-S83L cells were grown and treated with Cfx as described in the Topo-Seq section. 50-μL aliquots of the cells were stained for 5 min in 20 ng/ml 4′,6-diamidino-2-phenylindole (DAPI). Cells were spotted on agarose pads (1.2% agarose in PBS) and imaged at 100× magnification using a Nikon Eclipse Ti microscope controlled by NIS-Elements BR 4.51.01 and equipped with the Nikon Plan Apo VC 100 × 1.40 oil objective and Nikon DS-Qi2 digital monochrome camera. Images were processed using ImageJ2 v2.35 software (Schneider et al., 2012).
To map the sites of E. coli Topo IV activity genome-wide with single-nucleotide resolution, Topo-Seq, a protocol previously developed to monitor genome-wide distribution of E. coli DNA gyrase activity (Sutormin et al., 2019), was applied to exponentially growing cultures of E. coli DY330 parC-SPA strain. This strain encodes an affinity-tagged ParC subunit of Topo IV and behaves as wild-type, i.e., shows no par phenotype. We reasoned that the affinity tag should allow us to purify covalent ParC-DNA adducts formed in the presence of Topo IV poison ciprofloxacin (Cfx). While Cfx also targets the DNA gyrase, the adducts of this enzyme shall not be recovered during affinity purification. Three parallel DY330 parC-SPA cultures were treated with 10 μM Cfx (~300× minimal inhibitory concentration – MIC), SPA-tagged ParC subunits were affinity purified (Supplementary Figure 1A, verified by MS), followed by strand-specific sequencing of covalently attached DNA fragments and their mapping on a reference E. coli genome. The detected Topo IV cleavage sites (TCSs) had an enrichment patterns observed previously for DNA gyrase cleavage sites (GCSs) (Sutormin et al., 2019) and expected for type-IIA topoisomerases. Enrichment at TCSs has a bimodal shape with a sharp 4-bp gap in the middle. As explained elsewhere, the numbers of 3′-ends (N3Es) of enriched and sequenced DNA fragments comprise the gap ‘walls’ and precisely mark the cleavage site (Figure 1A). In total, 835 unique TCSs were identified in the reference genome. Among them, 356 TCSs were observed in at least two out of three biological replicates (Supplementary Figure 1B and Supplementary Table 1). Typically, around a primary strong TCS several weaker satellite TCSs were observed (Figure 1A, red and black arrows).
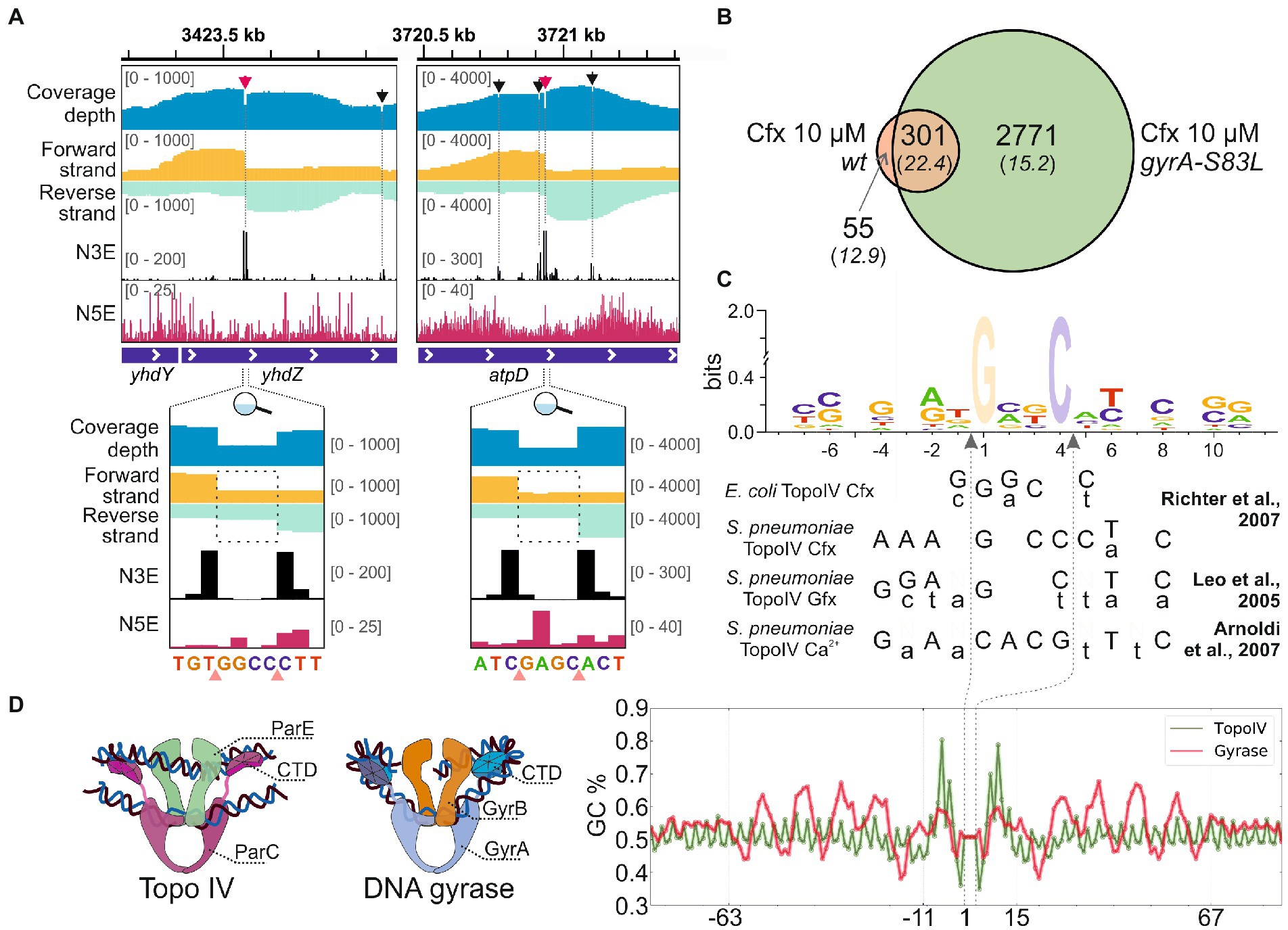
Figure 1. Detection of Topo IV TCSs with Topo-Seq. (A) Representative TCSs and cleavage patterns. The total sequencing coverage depth is shown in blue at the top. Coverage depths for forward and reverse strands are shown in gold and turquoise, respectively. Profiles of the number of 5′- (N5E) and 3′-ends (N3E) are shown in black and red, correspondingly. Tracks depths are shown in brackets. TCSs are shown with arrows and dashed lines. Primary and satellite TCSs (see text) are shown with red and black arrows, correspondingly. Zoom-in views of primary cleavage sites are shown below. Cleavage sites are shown with pink triangles. Dashed rectangles mark the gaps between coverage depths on forward and reverse strands. The data were visualized in IGV (Thorvaldsdóttir et al., 2013). (B) A Venn diagram representing an overlap between TCSs sets identified for wt and gyrA-S83L strains. (C) Logo representation of Topo IV cleavage motif determined with Topo-Seq. Cfx-biased positions at positions +1 and + 4 are translucent. Below the logo, cleavage consensus sequences identified previously for E. coli and S. pneumonia Topo IV in vitro are shown (Leo et al., 2005; Richter et al., 2007; Arnoldi et al., 2013). Preferred bases are shown in capital letters and non-preferred bases are shown in lowercase letters. Gfx—gemifloxacin. Vertical dashed arrows indicate positions of cleavage sites. (D) The Topo IV motif (green) and the DNA gyrase ‘combined’ motif (red) are shown as plots of GC content (right). Cartoon representations of Topo IV and DNA gyrase complexes with G- and T-segment DNAs are shown on the left.
To increase the specificity of Cfx treatment, a mutation in the gyrA gene leading to the S83L single amino acid substitution conferring resistance of DNA gyrase to fluoroquinolones was introduced by recombineering. Phenotypically, the mutant strain behaves as the parental strain, with the average cell length and DNA content statistically indistinguishable from it (Supplementary Figures 2A–C, raw quantification data in Supplementary Table 3), and similar supercoiling level (Supplementary Figure 2D). In agreement with published data (Bagel et al., 1999), compared to parental DY330 parC-SPA the mutation led to specific 32-fold increase of MIC to Cfx (Supplementary Figure 2E). A Topo-Seq experiment revealed that in the presence of 10 μM Cfx the amount of Topo IV TCSs in the gyrA-S83L mutant was ~8-times higher than in the parental wt strain. A total of 6,768 unique TCSs were identified, of which 3,072 were recovered in at least two biological replicates (Supplementary Figure 1C and Supplementary Table 1). 301 (85%) out of 356 TCSs detected in the wild-type gyrA strain were also observed in the gyrA-S83L mutant, indicating that the gyrase mutation did not significantly alter the basal pattern of Topo IV activity. These shared TCSs had an increased average enrichment and, likely, comprise a core set of strong sites of Topo IV activity in the E. coli genome (Figure 1B). 3,127 TCSs were replicated at least in two independent experiments, either for wt or a derivative mutant strain, formed a dataset for further analyses (Supplementary Table 1).
Positions of identified TCSs were compared with positions of GCSs identified with Cfx earlier (Sutormin et al., 2019). Only 178 cleavage sites were shared between Topo IV and DNA gyrase (5.7% of TCSs and 3.8% of GCSs). Low fraction of coincide cleavage sites indicates that Cfx-induced cleavage bias was not dominating the individual cleavage preferences of the studied topoisomerases.
The single-nucleotide resolution of Topo-Seq allowed us to identify the Topo IV cleavage motif. In addition to fluoroquinolone-specific nucleotide bias previously observed with Topo-Seq for DNA gyrase [more frequent cleavage before G nucleotides (Sutormin et al., 2019)], sequences flanking TCSs had several symmetrically enriched positions (−4G/+8C, −2A/+6 T, −1 T/+5A) previously reported as important cleavage determinants for Topo IV in vitro (Leo et al., 2005; Richter et al., 2007; Arnoldi et al., 2013; Figure 1C). Interestingly, these positions were also conserved for DNA gyrase (Supplementary Figure 3). The Topo-Seq data extended the Topo IV cleavage motif by revealing additional enriched positions −7C/+11G and −6C/+10G (Figure 1C). These positions were not conserved in the DNA gyrase motif (Supplementary Figure 3). It should be noted that the revealed Topo IV motif is rather weak and allows alternative nucleotides in most positions. On a broader scale, the Topo IV motif lacked any signs of periodic regions characteristic for the DNA gyrase motif (Sutormin et al., 2019) or any other motifs within 2 kb from the cleavage site, indicating that Topo IV is unlikely to wrap the immediate extensions of a G-segment DNA around its CTDs in vivo (Figure 1D).
The sites of Topo IV cleavage activity revealed by Topo-Seq are non-uniformly distributed over the E. coli genome (Figure 2A). First, the Topo IV enrichment was depleted in the Ter MD (1.2–1.9 Mb marked with a gray rectangle in Figure 2A and zoomed-in in Figure 2B) with only 250 TCSs observed vs. 475 expected for this region (p-value 6.0e-34, binomial test). This depletion is in line with previous observations made with NorfIP (El Sayyed et al., 2016) and is likely mediated by the MatP protein which unloads the MukBEF-Topo IV complexes from the chromosome and is thought to coordinate sister chromosomes decatenation with cell division (Nolivos et al., 2016; Fisher et al., 2021). Indeed, the decrease in the Topo IV activity matched well with the region occupied by MatP (Figure 2B). This region also lacks high-range chromosomal contacts (Figure 2B; Nolivos et al., 2016; Lioy et al., 2018). Topo IV enrichment was gradually and symmetrically increased left and right of Ter MD, in so-called Left and Right MDs, respectively, and then decreased again (Valens et al., 2004). Specifically, average enrichments of TCSs in the 500-kb regions of Right and Left MDs adjacent to Ter MD were significantly higher than the enrichments of TCSs in the neighboring 500-kb regions (p-values 0.03 and 0.02, respectively; two-tailed t-test). This may indicate that active resolution of precatenanes occurs in the Left and Right MDs at the borders of the Ter MD. Gyrase cleavage enrichment was also depleted in Ter MD and increased in Left and Right MDs. However, unlike with Topo IV, gyrase enrichment did not show local maxima in these MDs and extended further toward the origin (Supplementary Figure 4; Sutormin et al., 2019).
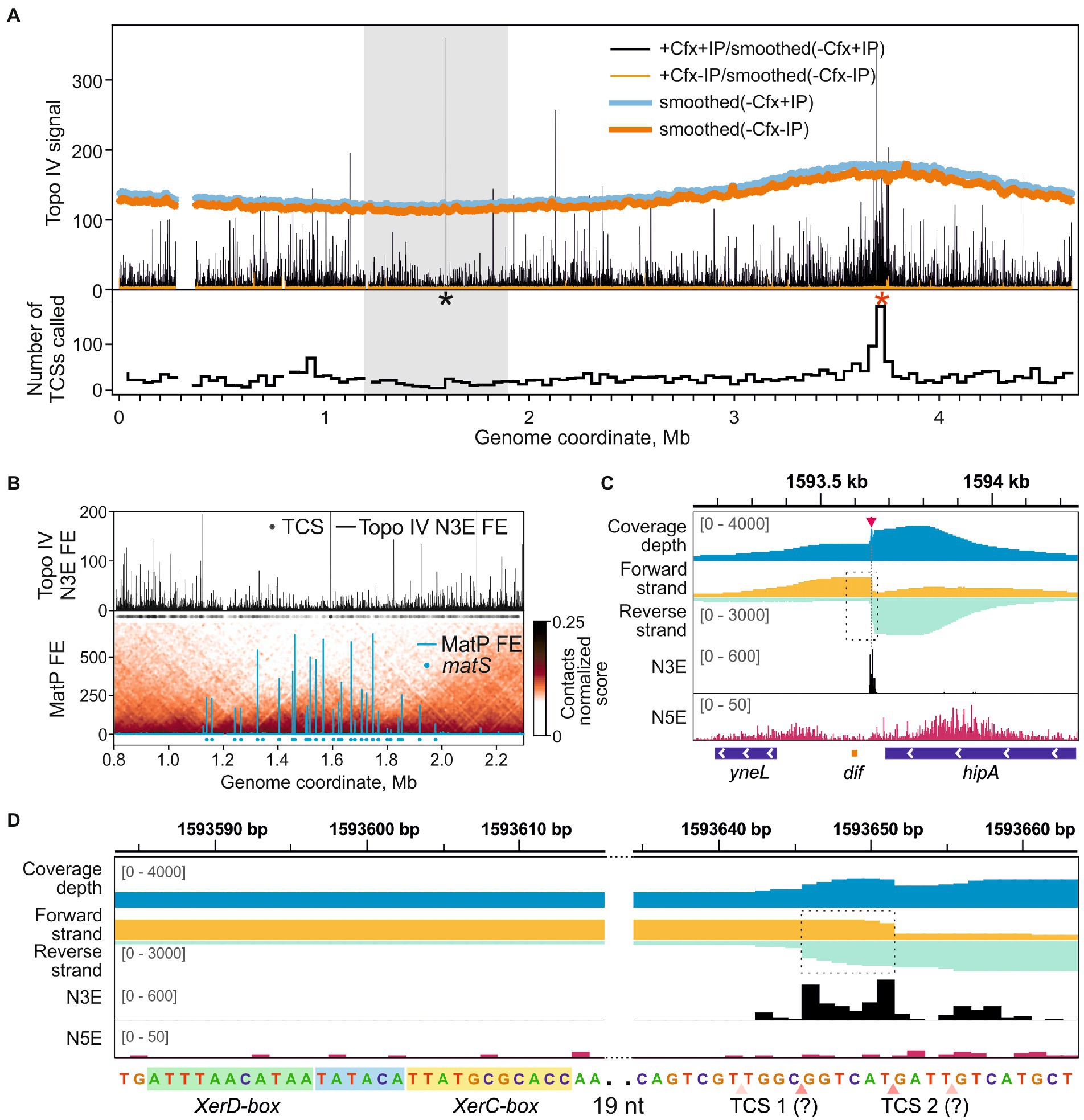
Figure 2. Topo IV cleavage activity at the dif-site. (A) Topo IV enrichment over the E. coli genome. Topo IV N3E fold enrichment (FE) for the +Cfx/+IP sample and the +Cfx/-IP control is shown in black and orange, respectively. Smoothed coverage depth tracks for -Cfx/+IP and -Cfx/-IP controls are shown in thick blue and dark orange curves, respectively. A black asterisk marks the position of the dif-site; red asterisk marks the position of oriC. A gray rectangle indicates the E. coli chromosome Ter MD. A plot representing the number of TCSs in 40 kb-wide genomic bins is shown below. (B) Topo IV N3E FE in the Ter macrodomain (upper plot). Black circles represent the positions of detected TCSs. MatP FE in Ter is shown below. Positions of matS-sites are shown with blue circles. Normalized contact map in Ter is shown in the underlying layer (resolution 10 kb). MatP data are taken from Nolivos et al. (2016), 3C contact data are taken from Lioy et al. (2018). (C) Topo IV enrichment and cleavage in the vicinity of the dif-site (orange rectangle below). N5E, N3E, and total coverage depth tracks are shown as in Figure 1A. Coverage depths for forward and reverse strands are shown in gold and turquoise, respectively. TCS is marked with a red arrow and a dashed line. A dashed rectangle indicates a region, containing the dif-site and the adjacent TCS, which is zoomed in (D). The data are visualized in IGV (Thorvaldsdóttir et al., 2013). (D) Zoom-in of the dif-site region. Genomic sequence with marked XerD and XerC boxes is given below. Hypothetical asymmetric adjacent TCSs are shown with pink triangles below. Translucent triangles indicate the absent or depleted cleavage in the second DNA strand. Dashed rectangle marks the anomalously enriched region.
One of the strongest Topo IV cleavage sites in the E. coli genome detected with Topo-Seq was located near the dif-site inside the Ter MD (Figures 2A,C). The site is recognized by the XerC/XerD recombinase. Previously, a TCS was mapped with low resolution in vitro in the vicinity of the dif-site (Hojgaard et al., 1999). Later, a strong cleavage at dif was detected in vivo using NorfIP (El Sayyed et al., 2016). However, the limited precision of the method did not allow to identify the exact position of the cleavage site. With Topo-Seq we detected two strong cleavage signals 33 and 38 bp away from the XerC-box (Figure 2D). The two N3E spikes were separated by 6 bp as observed in conventional cleavage sites (Figure 1A). Yet, sequenced fragments were not separated by a 4-bp gap but instead overlapped by 6 bp, which resulted in a local coverage peak in the center of a broader peak (Figure 2D). Such a pattern cannot be explained by a conventional double-stranded DNA cleavage by a single type-IIA topoisomerase complex or by extension of 3′-ends formed by Topo IV by DNA repair enzymes (the extension should have resulted in a 4-bp overlap with the two cleavage signals separated by 2 bp). We speculate that the observed pattern is formed by a superposition of two separate single-stranded cleavage events. Presumably, only reverse DNA strand is cleaved at one site, while the opposite, forward, DNA strand is cleaved at the other site, presumably by a different Topo IV complex. On a sequence level, the two cleavage sites did not resemble the derived Topo IV cleavage consensus, indicating that the enzyme was either attracted by the adjacent resolvase complex or that a different sequence recognition mode is realized at this site. There were no gyrase cleavage hot-spots either at dif or in the entire Ter MD in the presence of Cfx or other gyrase poisons (Supplementary Figure 4), indicating that the dif TCS is a highly specific target of Topo IV. However, visual analysis of gyrase enrichment profiles revealed a weak cleavage signal matching the position of Topo IV TCS at dif in either enriched or mock sequencing data for Cfx and oxalinic acid treated samples, but not for MccB17 treated samples (Supplementary Figure 5). The former two drugs, but not the latter one, inhibit Topo IV. This, and the fact that cleavage is observed in the absence of affinity purification suggests that the Topo IV induced cleavage at dif in the presence of fluoroquinolone poisons is so massive that it can be detected by sequencing genomic DNA without any enrichment.
Topo IV had an increased number of highly enriched cleavage sites near the oriC origin of replication (Figure 2A, a red asterisk). Overall, 950 TCSs were detected in the Ori MD vs. 622 TCSs expected (p-value 1.1e-16, binomial test). A particularly enriched region (251 TCSs vs. 42 expected, ~6-fold enrichment, p-value 1.1e-16, binomial test) spans 50–60 kb with oriC roughly in the middle. This enrichment was not observed by either ChIP-Seq or NorfIP earlier (El Sayyed et al., 2016). Interestingly, in the wt background the increased density of sites was less pronounced. Gyrase also had an increased number of GCSs both in the Ori MD and in the vicinity of oriC. This enrichment was less pronounced than that for Topo IV (Supplementary Figure 4) and is transcription-dependent (Sutormin et al., 2019).
To investigate the role of Topo IV in the relaxation of transcription-induced supercoiling, we analyzed the average enrichment of Topo IV in and around transcription units (TUs) stratified by their transcription level. Topo IV enrichment was decreased for least-expressed TUs (LETUs) and increased for highly expressed TUs (HETUs) (Supplementary Figures 6A,B). In addition, Topo IV enrichment was increased in HETUs downstream regions (Figure 3 and Supplementary Figure 6B), where positive supercoiling is accumulated (Sutormin et al., 2019; Guo et al., 2021; Visser et al., 2022). Especially high enrichment was observed in downstream regions of rRNA operons, which are highly transcribed in exponentially growing cells (Supplementary Figure 6C). Similar Topo IV enrichment was detected for both wt and gyrase mutant strains (Supplementary Figure 6D). Topo IV is known to interact with the MukBEF condensin and could have been expected to follow the MukBEF enrichment pattern if MukBEF enrichment was somehow affected by transcription or supercoiling (e.g., if elongating RNAPs move condensin to downstream regions). Indeed, on the level of enrichment sites, significantly more than expected TCSs were observed in MukB [a subunit of the MukBEF complex; MukB ChIP-Seq data from Nolivos et al. (2016)] enriched regions (938 TCSs vs. 648 TCSs expected, p-value 1.1e-16, binomial test). However, when we analyzed the MukB enrichment pattern relative to TUs, no comparable level of enrichment was observed downstream of HETUs (Supplementary Figures 6B,E). Therefore, the observed Topo IV enrichment pattern likely reflects the distinct properties of this topoisomerase and is unrelated to MukB. Overall, the Topo IV enrichment at TUs resembles enrichment pattern for DNA gyrase (Supplementary Figure 6F) indicating that Topo IV is also involved in relaxation of transcription-induced positive supercoiling. Correspondingly, correlations between Topo IV and DNA gyrase enrichments in 5 kb regions downstream of TUs were positive and significant (Pearson correlation coefficients 0.56 and 0.46 for wt and mutant strains, respectively), though Topo IV enrichment was less prominent and spanned over shorter distance compared to DNA gyrase enrichment (Supplementary Figures 6B,C). Interestingly, in contrast to DNA gyrase, Topo IV was not depleted in intergenic regions (compare Supplementary Figures 6D,F).
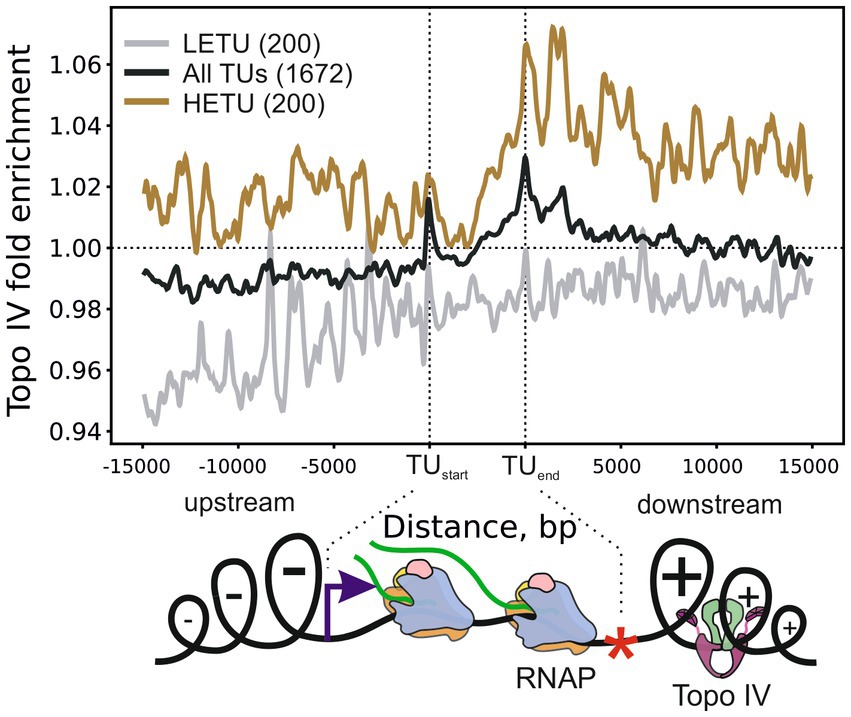
Figure 3. Metagene plot showing Topo IV enrichment in TUs, their upstream, and downstream regions. Enrichment is shown for all TUs (black curve), highly expressed (HETU, orange curve), and least-expressed (LETU, gray curve) sets. The number of TUs in each group is indicated in parentheses. Topo IV fold enrichment is given relative to the input sample (+Cfx + IP/-Cfx-IP). Bottom, graphical representation of the Liu and Wang twin-domain model (Liu and Wang, 1987) showing localization of RNAP and Topo IV. Data are shown for the E. coli DY330 parC-SPA strain.
Previously, using NorfIP, a total of 571 Topo IV unique cleavage sites in the E. coli genome were identified in three independent experiments, with 234 common sites detected in at least two experiments, and 84 common sites detected in three experiments [(El Sayyed et al., 2016) and also see Supplementary Figure 7]. With Topo-Seq, the numbers of detected unique and doubly replicated TCSs were ~ 10 times higher (6,872 and 3,127, respectively), and the number of TCSs observed in at least four experiments (301) was ~3.5 times higher. The majority (84%) of Topo IV TCSs detected with NorfIP (190 out of 226 TCSs mapped to the reference genome used in this work) were colocalized with TCSs detected with Topo-Seq. The higher number of detected TCSs and a large fraction of NorfIP TCSs replicated with Topo-Seq confirms superior sensitivity of cleavage sites detection by our method (Figure 4A). In addition, while with NorfIP a typical binding/cleavage signal spanned several hundreds of bp, Topo-Seq identified TCSs with single-nucleotide resolution (Figure 4B). The increased sensitivity of Topo-Seq might be caused by differences in treatment (15 min with 10 μM Cfx for Topo-Seq vs. 10 min with 2 μM norfloxacin for NorfIP) which should lead to more efficient trapping of Topo IV at our conditions, and by a more efficient method for sequencing library preparation from modified or damaged DNA (Accel NGS 1S for Topo-Seq vs. standard Illumina TruSeq for NorfIP).
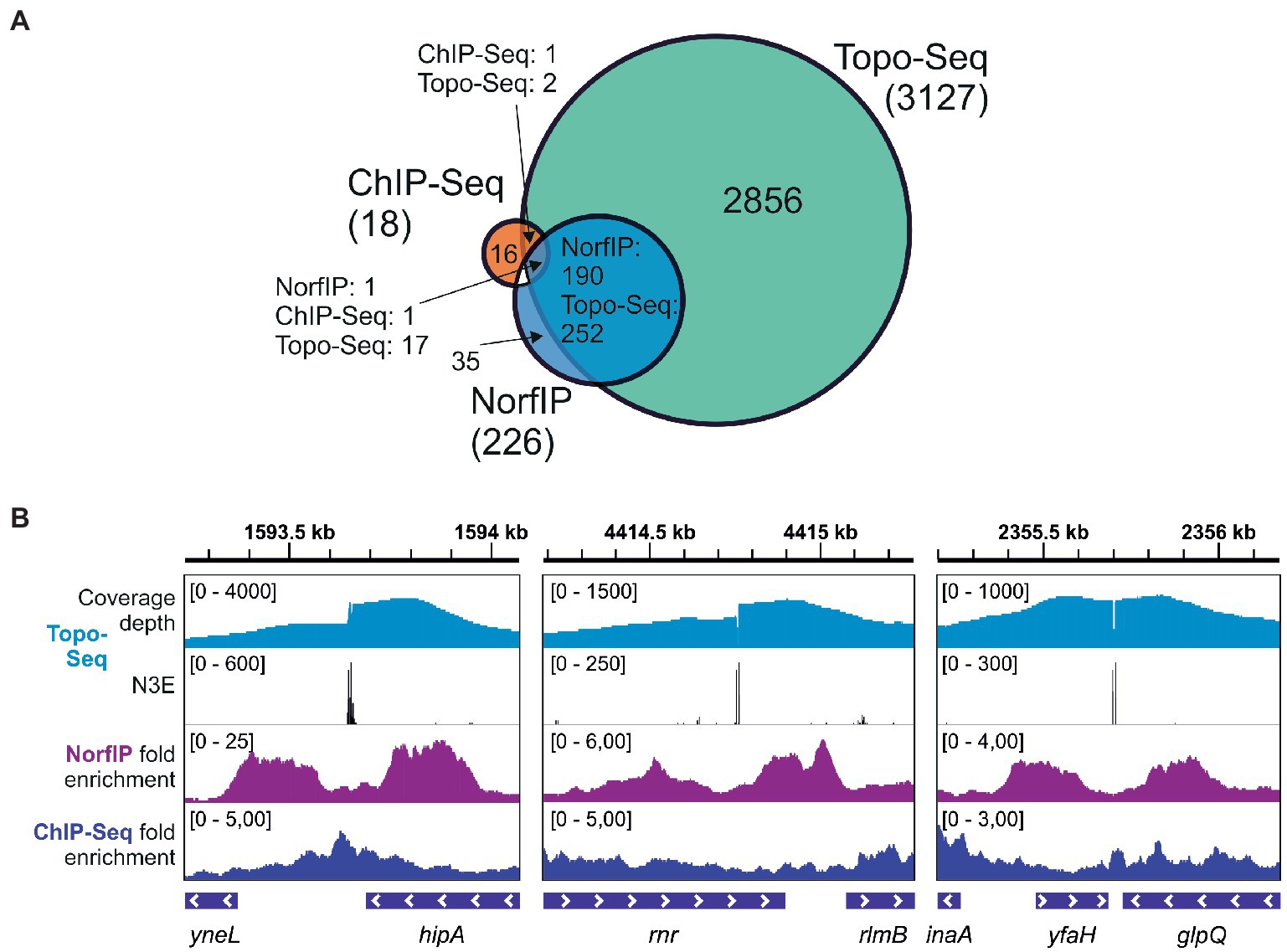
Figure 4. Sensitivity and precision of Topo-Seq. (A) A Venn diagram representing colocalization of Topo IV binding sites identified with ChIP-Seq and TCSs identified with NorfIP and Topo-Seq. Numbers of binding/cleavage sites are given for each subset, the total number of sites is shown in parentheses. (B) Representative genomic regions with binding/cleavage sites detected with ChIP-Seq, NorfIP, and Topo-Seq. ChIP-Seq and NorfIP data are taken from El Sayyed et al. (2016) and mapped to the E. coli reference genome used.
Only 19 regions were found to be significantly enriched with Topo IV by ChIP-Seq (El Sayyed et al., 2016), a method that does not distinguish between the binding and cleavage. Colocalization between the binding regions identified by ChIP-Seq and TCSs was limited (Figure 4). This result indicates that Topo IV cleavage occurs in only some binding regions (El Sayyed et al., 2016).
Using Topo-Seq, we identified several thousand Topo IV TCSs in the E. coli genome, which expands the repertoire of previously detected TCSs by almost an order of magnitude. We found that Topo IV enrichment pattern in the E. coli genome differs significantly from that of another type-II topoisomerase, the DNA gyrase. Topo IV cleavage is depleted in the Ter MD (except for strong and highly atypical cleavage at the dif-site), and gradually increases toward the Left and Right MDs. Additionally, Topo IV was strongly enriched in the Ori region.
We applied Topo-Seq to wt E. coli strain and for a derivative containing a gyrA-S83L mutation that confers resistance for fluoroquinolones. Surprisingly, sensitivity of Topo-Seq was much higher in the derivative strain, resulting in ~8-times more TCSs detected. Several independent observations suggest that the gyrA-S83L mutation by itself does not affect supercoiling and has a neutral effect on cell fitness (Aleixandre et al., 1989; Hallett and Maxwell, 1991; Marcusson et al., 2009). Likewise, our gyrA-S83L strain was phenotypically similar to wt and did not alter genome topology, DNA segregation, and plasmid supercoiling levels in our experimental conditions (Supplementary Figure 2). However, other reports suggest that substitutions of the GyrA S83 residue slightly reduce the supercoiling activity of the enzyme in vitro (a 10% decrease, Webber et al., 2017) and plasmid supercoiling in vivo, as judged by activity of a reporter gene controlled by supercoiling-sensitive promoter (a 14–22% decrease, Bagel et al., 1999). Such a decrease in gyrase activity, if it happened in our gyrA-S83L strain, could have been compensated by increased Topo IV activity. This effect, and increased availability of Cfx molecules unable to bind the mutant DNA gyrase might be jointly responsible for the appearance of additional Topo IV TCSs in the gyrase mutant background.
The single-nucleotide resolution of Topo-Seq and the large number of detected TCSs allowed us to identify the cleavage motif of E. coli Topo IV. As expected from observations made in vitro for fluoroquinolone-induced DNA cleavage by purified E. coli and S. pneumoniae Topo IV enzymes (Leo et al., 2005; Richter et al., 2007), Cfx induced strong preference toward cleavage before G nucleotides (Figure 1C). Consensus sequences of cleavage sites determined in vitro for S. pneumoniae Topo IV in the presence of Cfx and gemifloxacin (Gfx) (Leo et al., 2005; Richter et al., 2007) share significant similarity with the Topo-Seq-derived motif for the E. coli enzyme. Moreover, sites of Ca2+-induced cleavage (a method considered more native) had similar cleavage determinants (−4G/+8C, −2A/+6 T) suggesting intrinsic properties of the detected motif (Arnoldi et al., 2013). It should be noted that a consensus sequence obtained previously in vitro for E. coli Topo IV and the Topo-Seq-derived motif, seem to lack similarity between flanking sequences. The in vitro motif is shorter and lacks specificity determinants at positions −4/+8, −2/+6, probably, due to a lower sensitivity of the approach (Figure 1C). However, if one considers weaker nucleotide associations revealed in vitro, the two motifs become similar (Richter et al., 2007). With Topo-Seq, we further extended the Topo IV cleavage motif with −7C/+11G, −6C/+10G positions. The E. coli DNA gyrase cleavage motif determined in vitro and in vivo, by Topo-Seq, has similar cleavage determinants (−4G/+8C, −2A/+6 T, −1 T/+5A) indicating the conserved mode of G-segment recognition by type-IIA topoisomerases. −7C/+11G and −6C/+10G positions enriched in the Topo IV motif and absent in the gyrase motif, may thus determine Topo IV cleavage specificity. The overall motif of Topo IV is much shorter than DNA gyrase motif, lacks the periodic flanking regions, and generally resembles the eukaryotic Top2 motif (Gittens et al., 2019). The absence of periodic regions confirms that the ParC CTD does not interact with proximal extensions of G-segment DNA (Peng and Marians, 1995) but rather binds segments of crossing over DNA, which should favor resolution of precatenanes and catenanes. Differences between identified Topo IV and DNA gyrase motifs might contribute to the low fraction of cleavage sites that coincide for Topo IV and DNA gyrase.
With metagene analysis, we demonstrated that Topo IV is enriched in downstream regions of highly active TUs. This observation indicates that similarly to DNA gyrase, Topo IV is involved in relaxation of transcription-induced positive supercoiling in vivo which is in line with its in vitro preferences (Crisona et al., 2000; Ashley et al., 2017). Enrichment of Topo IV in the downstream regions of active TUs was lower than the DNA gyrase enrichment, indicating a limited contribution of Topo IV to the resolution of transcription-induced supercoiling, which is in accordance with the previous observations (Zechiedrich et al., 2000). Topo IV may act as a partial backup of DNA gyrase in this process. We found no clear colocalization between MukB and Topo IV in the downstream regions of active TUs. We speculate that Topo IV may act independently of MukBEF in these regions. Likely, Topo IV interacts with a plectonemic DNA formed by excessive positive supercoiling, as no G-segment DNA wrapping was indicated by Topo IV motif. Correspondingly, inhibition of transcription with rifampicin was shown to abolish norfloxacin-induced DNA cleavage by Topo IV (El Sayyed et al., 2016). To test this model directly, a Topo-Seq may be performed when the MukBEF-Topo IV interaction is uncoupled or when transcription is inhibited by rifampicin. In contrast to DNA gyrase, Topo IV was not depleted in intergenic regions. Probably, this reflects the difference between the motifs recognized by the two enzymes: DNA gyrase has a long and GC-rich motif which is depleted in the AT-rich intergenic regions, while Topo IV has a less constrained DNA motif.
We observed a strong depletion of Topo IV activity in the Ter MD, likely reflecting the depletion of MukBEF-Topo IV complexes by MatP (Nolivos et al., 2016; Burmann et al., 2021; Conin et al., 2022) and overall reduced transcription activity of this region. Why should MukBEF-Topo IV be displaced from Ter, and Topo IV activity be limited to the dif-site? It was proposed that an unloading system based on matS-MatP is required to recycle MukBEF associated with DNA and promote its uploading at the replication origin (Nolivos et al., 2016). We speculate that Topo IV depletion may prevent knotting and catenating of over-replicated chromosomes when the two replisomes fuse and travel further, replicating opposite replichores (Syeda et al., 2020). If so, an increased fraction of abnormal cells in ΔmatP mutants might have signs of DNA over-replication in the Ter region. In line with this model, linearization of E. coli chromosome in Ter by tos/TelN corrects both the ΔmatP phenotype and DNA over-replication in the Ter region (El Sayyed et al., 2016; Midgley-Smith et al., 2019). To decipher how transcription and MatP affect the activity of Topo IV in the Ter MD, more Topo-Seq data are needed, e.g., with mutant cells lacking MatP and cells treated with RNA polymerase inhibitor rifampicin. Interestingly, the depletion of gyrase enrichment in Ter MD was not affected by rifampicin (Sutormin et al., 2019), indicating that MatP may play a role in this phenomenon directly or indirectly, e.g., by unloading of MukBEF or constraining DNA supercoiling diffusing into the MD from the adjacent regions.
Topo IV has a well-known cleavage hot-spot near the dif-site (Hojgaard et al., 1999; El Sayyed et al., 2016), which we here map precisely. The cleavage site has a non-conventional structure and, likely, is a composite of two single-strand cleavage sites separated by 6 bp. Sterically, it is unlikely that two Topo IV complexes can simultaneously attack such two closely adjacent DNA sites. The cleavage events may happen independently on separate DNA molecules (e.g., on sister chromosomes) or occur probabilistically, giving a mixed signal in a bulk method like Topo-Seq. If so, then, only a single-stranded break will be introduced into DNA duplex by a hypothetical structurally or functionally asymmetric complex. Such a break is unable to support the conventional strand passage mechanism to decatenate sister chromosomes but may efficiently resolve supercoiling via the nicking-closing mechanism demonstrated in vitro for DNA gyrase heterocomplex with a single catalytic tyrosine (Gubaev et al., 2016). Relaxation of excessive DNA supercoiling [generated by elongating DNA polymerase (Postow et al., 2001) and/or translocating FtsK (Saleh et al., 2005)] might be important in Ter MD for proper replication termination and segregation of the replicated chromosomes. Consistently, in cells with a linearized chromosome, Topo IV does not cleave at dif despite the presence of all components of the dif-XerC/XerD ensemble, which implies that a topological stress is required for Topo IV activity at this site (El Sayyed et al., 2016). The proposed single-strand cleavage model conflicts with evidence of norfloxacin-induced double-stranded DNA cleavage detected at dif by Southern blot (Hojgaard et al., 1999; El Sayyed et al., 2016). Probably, the constitutive single-strand break mediates increased fragility of the region which results in increased breakage during DNA extraction and processing steps. Additionally, our model implies the existence of a mechanism that controls the Topo IV cleavage activity and stops it after the first DNA strand is cleaved. Alternatively, an observed cleavage pattern may be introduced by a non-canonical Topo IV complex consisting of two Topo IV molecules, which, cleaving together, introduce a double-strand break. Overall, Topo IV cleavage at dif-site represents an unusual behavior that is likely specifically orchestrated by its protein partners—XerC and FtsK—and may play a specific biological function that remains to be defined.
With Topo-Seq we observed an increased density of highly enriched TCSs at the Ori region. The locally increased cleavage activity may reflect the action of supramolecular MukBEF-Topo IV complexes recruited to Ori and required for proper segregation of this region once replicated (Nicolas et al., 2014; Nolivos et al., 2016). We speculate that Topo IV may be involved in early steps of replication progression where its activity can be critical to resolve precatenanes when two replisomes are relatively close to each other. In line with this speculation, MukB interaction with Topo IV was shown to stimulate relaxation of only negatively supercoiled DNA (Vos et al., 2013). Likely, the interaction with MukBEF in vivo provides specificity for DNA precatenanes formed by replication, since they form of a right-handed plectoneme—a topological equivalent of negatively supercoiled DNA in a plectonemic form (Schvartzman et al., 2019). Increased transcriptional activity in the Ori region might also cause the accumulation of Topo IV activity, similarly to what has been observed for DNA gyrase (Sutormin et al., 2019). Topo-Seq experiments with cells treated with rifampicin may help to investigate the role of transcription in this phenomenon.
ChIP methods provide essential information on the genomic localization of DNA-binding proteins. The classical method is based on formaldehyde fixation of protein-DNA complexes, however, it suffers from low sensitivity, high background noise, and low precision (Waldminghaus and Skarstad, 2010; Chen et al., 2012). A plethora of methods such as Native ChIP, ChIP-Exo, CUT&RUN, CUT&TAG, etc., were developed to overcome these limitations (Ma et al., 2019; Leo and Romano, 2021). For topoisomerases, which naturally form covalent adducts with DNA during the catalytic cycle, using specific poisons or mutations, intermediate complexes can be stabilized and enriched without the crosslinking agent (McKie et al., 2020). Topoisomerase adducts, however, should be completely removed before sequencing, otherwise it may interfere with library preparation steps and reduce the precision of the method (El Sayyed et al., 2016). As we demonstrated here for E. coli Topo IV and previously for DNA gyrase and TopoI (Sutormin et al., 2019, 2022), Topo-Seq has a high sensitivity and a single-nucleotide resolution which makes it a method of choice for detection of topoisomerase cleavage sites in various model systems.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in Supplementary Table 2.
DS conceived the study, analyzed data, and prepared figures. DS, AGal, and AGaf performed the experiments. DS and KS wrote the draft and finalized the manuscript. All authors contributed to the article and approved the submitted version.
This work (experiments and bioinformatic analysis) was supported by a grant from the Ministry of Science and Higher Education of the Russian Federation (agreement No. 075-10-2021-114 from 11 October 2021). This work was also supported by Skoltech NGP Program (Skoltech-MIT joint project) and RFBR grant, project number 20-34-90069. Sequencing at Skoltech Genomics Core Facility was supported by the Skoltech Life Sciences Program grant.
We are grateful to Marina Serebryakova for mass spectrometry and to Svetlana Dubiley and Anthony Maxwell for extensive and fruitful discussions.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1160736/full#supplementary-material
1. ^ https://github.com/sutormin94/Topo IV_Topo-Seq_experiment
Adams, D. E., Shekhtman, E. M., Zechiedrich, E. L., Schmid, M. B., and Cozzarelli, N. R. (1992). The role of topoisomerase IV in partitioning bacterial replicons and the structure of catenated intermediates in DNA replication. Cells 71, 277–288. doi: 10.1016/0092-8674(92)90356-H
Aleixandre, V., Urios, A., Herrera, G., and Blanco, M. (1989). New Escherichia coli gyrA and gyrB mutations which have a graded effect on DNA supercoiling. Mol. Gen. Genet. 219, 306–312. doi: 10.1007/BF00261192
Arnoldi, E., Pan, X., and Fisher, M. (2013). Functional determinants of gate-DNA selection and cleavage by bacterial type II topoisomerases. Nucleic Acids Res. 41, 9411–9423. doi: 10.1093/nar/gkt696
Ashley, R. E., Dittmore, A., McPherson, S. A., Turnbough, C. L., Neuman, K. C., and Osheroff, N. (2017). Activities of gyrase and topoisomerase IV on positively supercoiled DNA. Nucleic Acids Res. 45, 9611–9624. doi: 10.1093/nar/gkx649
Bagel, S., Hullen, V., Wiedemann, B., and Heisig, P. (1999). Impact of gyrA and parC mutations on quinolone resistance, doubling time, and supercoiling degree of Escherichia coli. Antimicrob. Agents Chemother. 43, 868–875. doi: 10.1128/AAC.43.4.868
Bigot, S., and Marians, K. J. (2010). DNA chirality-dependent stimulation of topoisomerase IV activity by the C-terminal AAA+ domain of FtsK. Nucleic Acids Res. 38, 3031–3040. doi: 10.1093/nar/gkp1243
Blakely, G., May, G., McCulloch, R., Arciszewska, L. K., Burke, M., Lovett, S. T., et al. (1993). Two related recombinases are required for site-specific recombination at dif and cer in E. coli K12. Cells 75, 351–361. doi: 10.1016/0092-8674(93)80076-Q
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Burmann, F., Funke, L. F. H., Chin, J. W., and Lowe, J. (2021). Cryo-EM structure of MukBEF reveals DNA loop entrapment at chromosomal unloading sites. Mol. Cell 81, 4891–4906.e8. doi: 10.1016/j.molcel.2021.10.011
Cebrián, J., Castán, A., Martínez, V., Kadomatsu-Hermosa, M. J., Parra, C., Fernández-Nestosa, M. J., et al. (2015). Direct evidence for the formation of precatenanes during DNA replication. J. Biol. Chem. 290, 13725–13735. doi: 10.1074/jbc.M115.642272
Chen, Y., Negre, N., Li, Q., Mieczkowska, J. O., Slattery, M., Liu, T., et al. (2012). Systematic evaluation of factors influencing ChIP-seq fidelity. Nat. Methods 9, 609–614. doi: 10.1038/nmeth.1985.Systematic
Conin, B., Billault-Chaumartin, I., el Sayyed, H., Quenech’du, N., Cockram, C., Koszul, R., et al. (2022). Extended sister-chromosome catenation leads to massive reorganization of the E. coli genome. Nucleic Acids Res. 50, 2635–2650. doi: 10.1093/nar/gkac105
Crisona, N. J., Strick, T. R., Bensimon, D., Croquette, V., and Cozzarelli, N. R. (2000). Preferential relaxation of positively supercoiled DNA by E. coli topoisomerase IV in single-molecule and ensemble measurements. Genes Dev. 14, 2881–2892. doi: 10.1101/gad.838900
Datsenko, K. A., and Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97, 6640–6645. doi: 10.1073/pnas.120163297
el Sayyed, H., le Chat, L., Lebailly, E., Vickridge, E., Pages, C., Cornet, F., et al. (2016). Mapping topoisomerase IV binding and activity sites on the E. coli genome. PLoS Genet. 12:e1006025-22. doi: 10.1371/journal.pgen.1006025
Espeli, O., Lee, C., and Marians, K. J. (2003). A physical and functional interaction between Escherichia coli FtsK and topoisomerase IV. J. Biol. Chem. 278, 44639–44644. doi: 10.1074/jbc.M308926200
Fisher, G. L. M., Bolla, J. R., Rajasekar, K. V., Mäkelä, J., Baker, R., Zhou, M., et al. (2021). Competitive binding of MatP and topoisomerase IV to the MukB hinge domain. eLife 10:e70444. doi: 10.7554/eLife.70444
Gittens, W. H., Johnson, D. J., Allison, R. M., Cooper, T. J., Thomas, H., and Neale, M. J. (2019). A nucleotide resolution map of Top2-linked DNA breaks in the yeast and human genome. Nat. Commun. 10, 4846–4816. doi: 10.1038/s41467-019-12802-5
Grainge, I., Bregu, M., Vazquez, M., Sivanathan, V., Ip, S. C. Y., and Sherratt, D. J. (2007). Unlinking chromosome catenanes in vivo by site-specific recombination. EMBO J. 26, 4228–4238. doi: 10.1038/sj.emboj.7601849
Grainge, I., Lesterlin, C., and Sherratt, D. J. (2011). Activation of XerCD-dif recombination by the FtsK DNA translocase. Nucleic Acids Res. 39, 5140–5148. doi: 10.1093/nar/gkr078
Gubaev, A., Weidlich, D., and Klostermeier, D. (2016). DNA gyrase with a single catalytic tyrosine can catalyze DNA supercoiling by a nicking-closing mechanism. Nucleic Acids Res. 44, gkw740–gkw10366. doi: 10.1093/nar/gkw740
Guo, M. S., Kawamura, R., Littlehale, M., Marko, J. F., and Laub, M. T. (2021). High-resolution, genome-wide mapping of positive supercoiling in chromosomes. eLife 10:e67236. doi: 10.7554/eLife.67236
Hallett, P., and Maxwell, A. (1991). Novel quinolone resistance mutations of the Escherichia coli DNA Gyrase a protein: Enzymatic analysis of the mutant proteins. Antimicrob. Agents Chemother. 35, 335–340. doi: 10.1128/AAC.35.2.335
Hayama, R., and Marians, K. J. (2010). Physical and functional interaction between the condensin MukB and the decatenase topoisomerase IV in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 107, 18826–18831. doi: 10.1073/pnas.1008140107
Helgesen, E., Sætre, F., and Skarstad, K. (2021). Topoisomerase IV tracks behind the replication fork and the SeqA complex during DNA replication in Escherichia coli. Sci. Rep. 11, 474–478. doi: 10.1038/s41598-020-80043-4
Hiasa, H., DiGate, R. J., and Marians, K. J. (1994). Decatenating activity of Escherichia coli DNA gyrase and topoisomerases I and III during oriC and pBR322 DNA replication in vitro. J. Biol. Chem. 269, 2093–2099. doi: 10.1016/S0021-9258(17)42140-5
Hirsch, J., and Klostermeier, D. (2021). What makes a type IIA topoisomerase a gyrase or a topo IV? Nucleic Acids Res. 49, 6027–6042. doi: 10.1093/nar/gkab270
Hojgaard, A., Szerlong, H., Tabor, C., and Kuempel, P. (1999). Norfloxacin-induced DNA cleavage occurs at the dif resolvase locus in Escherichia coli and is the result of interaction with topoisomerase IV. Mol. Microbiol. 33, 1027–1036. doi: 10.1046/j.1365-2958.1999.01545.x
Joshi, M. C., Magnan, D., Montminy, T. P., Lies, M., Stepankiw, N., and Bates, D. (2013). Regulation of sister chromosome cohesion by the replication fork tracking protein SeqA. PLoS Genet. 9:e1003673. doi: 10.1371/journal.pgen.1003673
Kang, S., Han, J. S., Park, J. H., Skarstad, K., and Hwang, D. S. (2003). SeqA protein stimulates the relaxing and decatenating activities of topoisomerase IV. J. Biol. Chem. 278, 48779–48785. doi: 10.1074/jbc.M308843200
Karp, P. D., Billington, R., Caspi, R., Fulcher, C. A., Latendresse, M., Kothari, A., et al. (2018). The BioCyc collection of microbial genomes and metabolic pathways. Brief. Bioinform. 20, 1085–1093. doi: 10.1093/bib/bbx085
Kato, J., Nishimura, Y., Imamura, R., Niki, H., Hiraga, S., and Suzuki, H. (1990). New topoisomerase essential for chromosome segregation in E. coli. Cells 63, 393–404. doi: 10.1016/0092-8674(90)90172-B
Keller, A. N., Xin, Y., Boer, S., Reinhardt, J., Baker, R., Arciszewska, L. K., et al. (2016). Activation of Xer-recombination at dif: structural basis of the FtsKγ-XerD interaction. Sci. Rep. 6, 1–12. doi: 10.1038/srep33357
Khodursky, A. B., Peter, B. J., Schmid, M. B., DeRisi, J., Botstein, D., Brown, P. O., et al. (2000). Analysis of topoisomerase function in bacterial replication fork movement: use of DNA microarrays. Proc. Natl. Acad. Sci. U. S. A. 97, 9419–9424. doi: 10.1073/pnas.97.17.9419
Kumar, R., Bahng, S., and Marians, K. J. (2022). The MukB-topoisomerase IV interaction mutually suppresses their catalytic activities. Nucleic Acids Res. 50, 2621–2634. doi: 10.1093/nar/gkab1027
Kumar, R., Nurse, P., Bahng, S., Lee, C. M., and Marians, K. J. (2017). The MukB–topoisomerase IV interaction is required for proper chromosome compaction. J. Biol. Chem. 292, 16921–16932. doi: 10.1074/jbc.M117.803346
Lee, C. M., Wang, G., Pertsinidis, A., and Marians, K. J. (2019). Topoisomerase III acts at the replication fork to remove precatenanes. J. Bacteriol. 201, 1–13. doi: 10.1128/JB.00563-18
Leo, E., Gould, K. A., Pan, X., Capranico, G., Sanderson, M. R., Palumbo, M., et al. (2005). Novel symmetric and asymmetric DNA scission determinants for Streptococcus pneumoniae topoisomerase IV and Gyrase are clustered at the DNA breakage site. J. Biol. Chem. 280, 14252–14263. doi: 10.1074/jbc.M500156200
Leo, L., and Romano, N. C. (2021). Emerging single-cell technological approaches to investigate chromatin dynamics and centromere regulation in human health and disease. Int. J. Mol. Sci. 22, 1–22. doi: 10.3390/ijms22168809
Li, H., and Durbin, R. (2010). Fast and accurate long-read alignment with burrows-wheeler transform. Bioinformatics 26, 589–595. doi: 10.1093/bioinformatics/btp698
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Li, Y., Stewart, N. K., Berger, A. J., Vos, S., Schoeffler, A. J., Berger, J. M., et al. (2010). Escherichia coli condensin MukB stimulates topoisomerase IV activity by a direct physical interaction. Proc. Natl. Acad. Sci. U. S. A. 107, 18832–18837. doi: 10.1073/pnas.1008678107
Lioy, V. S., Cournac, A., Marbouty, M., Duigou, S., Mozziconacci, J., Espéli, O., et al. (2018). Multiscale structuring of the E. coli chromosome by nucleoid-associated and condensin proteins. Cells 172, 771–783.e18. doi: 10.1016/j.cell.2017.12.027
Liu, L. F., and Wang, J. C. (1987). Supercoiling of the DNA template during transcription. Proc. Natl. Acad. Sci. U. S. A. 84, 7024–7027. doi: 10.1073/pnas.84.20.7024
Lucas, I., Germe, T., Chevrier-Miller, M., and Hyrien, O. (2001). Topoisomerase II can unlink replicating DNA by precatenane removal. EMBO J. 20, 6509–6519. doi: 10.1093/emboj/20.22.6509
Ma, T., Ye, Z., and Wang, L. (2019). Genome wide approaches to identify protein-DNA interactions. Curr. Med. Chem. 26, 7641–7654. doi: 10.2174/0929867325666180530115711
Marcusson, L. L., Frimodt-Møller, N., and Hughes, D. (2009). Interplay in the selection of Fluoroquinolone resistance and bacterial fitness. PLoS Pathog. 5:e1000541. doi: 10.1371/journal.ppat.1000541
McKie, S. J., Maxwell, A., and Neuman, K. C. (2020). Mapping DNA topoisomerase binding and cleavage genome wide using next-generation sequencing techniques. Genes (Basel) 11, 1–16. doi: 10.3390/genes11010092
Mercier, R., Petit, M. A., Schbath, S., Robin, S., El Karoui, M., Boccard, F., et al. (2008). The MatP/matS site-specific system organizes the terminus region of the E. coli chromosome into a macrodomain. Cells 135, 475–485. doi: 10.1016/j.cell.2008.08.031
Midgley-Smith, S. L., Dimude, J. U., and Rudolph, C. J. (2019). A role for 3′ exonucleases at the final stages of chromosome duplication in Escherichia coli. Nucleic Acids Res. 47, 1847–1860. doi: 10.1093/nar/gky1253
Nicolas, E., Upton, A. L., Uphoff, S., Henry, O., Badrinarayanan, A., and Sherratt, D. (2014). The SMC complex MukBEF recruits topoisomerase IV to the origin of replication region in live Escherichia coli. mBio 5, e01001–e01013. doi: 10.1128/mBio.01001-13
Nolivos, S., Upton, A. L., Badrinarayanan, A., Müller, J., Zawadzka, K., Wiktor, J., et al. (2016). MatP regulates the coordinated action of topoisomerase IV and MukBEF in chromosome segregation. Nat. Commun. 7:10466. doi: 10.1038/ncomms10466
Peng, H., and Marians, K. J. (1993). Escherichia coli topoisomerase IV. Purification, characterization, subunit structure, and subunit interactions. J. Biol. Chem. 268, 24481–24490. doi: 10.1016/S0021-9258(20)80551-1
Peng, H., and Marians, K. J. (1995). The interaction of Escherichia coli topoisomerase IV with DNA. J. Biol. Chem. 270, 25286–25290. doi: 10.1074/jbc.270.42.25286
Peter, B. J., Ullsperger, C., Hiasa, H., Marians, K. J., and Cozzarelli, N. R. (1998). The structure of supercoiled intermediates in DNA replication. Cells 94, 819–827. doi: 10.1016/S0092-8674(00)81740-7
Postow, L., Crisona, N. J., Peter, B. J., Hardy, C. D., and Cozzarelli, N. R. (2001). Topological challenges to DNA replication: conformations at the fork. Proc. Natl. Acad. Sci. U. S. A. 98, 8219–8226. doi: 10.1073/pnas.111006998
Richter, S. N., Giaretta, G., Comuzzi, V., Leo, E., Mitchenall, L. A., Fisher, L. M., et al. (2007). Hot-spot consensus of fluoroquinolone-mediated DNA cleavage by gram-negative and gram-positive type II DNA topoisomerases. Nucleic Acids Res. 35, 6075–6085. doi: 10.1093/nar/gkm653
Saleh, O. A., Bigot, S., Barre, F.-X., and Allemand, J.-F. (2005). Analysis of DNA supercoil induction by FtsK indicates translocation without groove-tracking. Nat. Struct. Mol. Biol. 12, 436–440. doi: 10.1038/nsmb926
Sawitzke, J. A., Costantino, N., Li, X., Thomason, L. C., Bubunenko, M., Court, C., et al. (2011). Probing cellular processes with Oligo-mediated recombination and using the knowledge gained to optimize recombineering. J. Mol. Biol. 407, 45–59. doi: 10.1016/j.jmb.2011.01.030
Schneider, C. A., Rasband, W. S., and Eliceiri, K. W. (2012). NIH image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. doi: 10.1038/nmeth.2089
Schvartzman, J. B., Hernandez, P., Krimer, D. B., Dorier, J., and Stasiak, A. (2019). Closing the DNA replication cycle: from simple circular molecules to supercoiled and knotted DNA catenanes. Nucleic Acids Res. 47, 7182–7198. doi: 10.1093/nar/gkz586
Seol, Y., Hardin, A. H., Strub, M. P., Charvin, G., and Neuman, K. C. (2013). Comparison of DNA decatenation by Escherichia coli topoisomerase IV and topoisomerase III: Implications for non-equilibrium topology simplification. Nucleic Acids Res. 41, 4640–4649. doi: 10.1093/nar/gkt136
Sutormin, D., Galivondzhyan, A., Musharova, O., Travin, D., Rusanova, A., Obraztsova, K., et al. (2022). Interaction between transcribing RNA polymerase and topoisomerase I prevents R-loop formation in E. coli. Nat. Commun. 13, 1–19. doi: 10.1038/s41467-022-32106-5
Sutormin, D., Rubanova, N., Logacheva, M., Ghilarov, D., and Severinov, K. (2019). Single-nucleotide-resolution mapping of DNA gyrase cleavage sites across the Escherichia coli genome. Nucleic Acids Res. 47, 1373–1388. doi: 10.1093/nar/gky1222
Syeda, A. H., Dimude, J. U., Skovgaard, O., and Rudolph, C. J. (2020). Too much of a good thing: how ectopic DNA replication affects bacterial replication dynamics. Front. Microbiol. 11:534. doi: 10.3389/fmicb.2020.00534
Thorvaldsdóttir, H., Robinson, J. T., and Mesirov, J. P. (2013). Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 14, 178–192. doi: 10.1093/bib/bbs017
Valens, M., Penaud, S., Rossignol, M., Cornet, F., and Boccard, F. (2004). Macrodomain organization of the Escherichia coli. EMBO J. 23, 4330–4341. doi: 10.1038/sj.emboj.7600434
Visser, B. J., Sharma, S., Chen, P. J., Mcmullin, A. B., Bates, M. L., and Bates, D. (2022). Psoralen mapping reveals a bacterial genome supercoiling landscape dominated by transcription. Nucleic Acids Res. 50, 4436–4449. doi: 10.1093/nar/gkac244
Vos, S. M., Stewart, N. K., Oakley, M. G., and Berger, J. M. (2013). Structural basis for the MukB-topoisomerase IV interaction and its functional implications in vivo. EMBO J. 32, 2950–2962. doi: 10.1038/emboj.2013.218
Waldminghaus, T., and Skarstad, K. (2010). ChIP on Chip: surprising results are often artifacts. BMC Genomics 11, 414–415. doi: 10.1186/1471-2164-11-414
Wang, X., Reyes-Lamothe, R., and Sherratt, D. J. (2008). Modulation of Escherichia coli sister chromosome cohesion by topoisomerase IV. Genes Dev. 22, 2426–2433. doi: 10.1101/gad.487508
Webber, M. A., Buckner, M. M. C., Redgrave, L. S., Ifill, G., Mitchenall, L. A., Webb, C., et al. (2017). Quinolone-resistant gyrase mutants demonstrate decreased susceptibility to triclosan. J. Antimicrob. Chemother. 72, 2755–2763. doi: 10.1093/jac/dkx201
Zawadzki, P., Stracy, M., Ginda, K., Zawadzka, K., Lesterlin, C., Kapanidis, A. N., et al. (2015). The localization and action of topoisomerase IV in Escherichia coli chromosome segregation is coordinated by the SMC complex, MukBEF. Cell Rep. 13, 2587–2596. doi: 10.1016/j.celrep.2015.11.034
Zechiedrich, E. L., and Cozzarelli, N. R. (1995). Roles of topoisomerase IV and DNA gyrase in DNA unlinking during replication in Eschericia coli. Genes Dev. 9, 2859–2869. doi: 10.1101/gad.9.22.2859
Zechiedrich, E. L., Khodursky, A. B., Bachellier, S., Schneider, R., Chen, D., Lilley, D. M. J., et al. (2000). Roles of topoisomerases in maintaining steady-state DNA supercoiling in Escherichia coli. J. Biol. Chem. 275, 8103–8113. doi: 10.1074/jbc.275.11.8103
Keywords: E. coli , topoisomerase IV, genome topology, Topo-Seq, supercoiling, decatenation
Citation: Sutormin D, Galivondzhyan A, Gafurov A and Severinov K and (2023) Single-nucleotide resolution detection of Topo IV cleavage activity in the Escherichia coli genome with Topo-Seq. Front. Microbiol. 14:1160736. doi: 10.3389/fmicb.2023.1160736
Received: 07 February 2023; Accepted: 16 March 2023;
Published: 06 April 2023.
Edited by:
Tamara Basta, UMR9198 Institut de Biologie Intégrative de la Cellule (I2BC), FranceReviewed by:
Olivier Espeli, Centre National de la Recherche Scientifique (CNRS), FranceCopyright © 2023 Sutormin, Galivondzhyan, Gafurov and Severinov. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dmitry Sutormin, RC5BLlN1dG9ybWluQGdtYWlsLmNvbQ==; Konstantin Severinov, c2V2ZXJpa0B3YWtzbWFuLnJ1dGdlcnMuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.