- 1Department of Laboratory Medicine, Affiliated Hospital of Zunyi Medical University, Zunyi, Guizhou, China
- 2School of Laboratory Medicine, Zunyi Medical University, Zunyi, Guizhou, China
- 3Department of Blood Transfusion, Affiliated Hospital of Zunyi Medical University, Zunyi, Guizhou, China
In bacteria, DegS protease functions as an activating factor of the σE envelope stress response system, which ultimately activates the transcription of stress response genes in the cytoplasm. On the basis of high-throughput RNA sequencing, we have previously found that degS knockout inhibits the expression of flagellum synthesis- and chemotaxis-related genes, thereby indicating that DegS may be involved in the regulation of V. cholerae motility. In this study, we examined the relationships between DegS and motility in V. cholerae. Swimming motility and chemotaxis assays revealed that degS or rpoE deletion promotes a substantial reduction in the motility and chemotaxis of V. cholerae, whereas these activities were restored in ΔdegS::degS and ΔdegSΔrseA strains, indicating that DegS is partially dependent on σE to positively regulate V. cholerae activity. Gene-act network analysis revealed that the cAMP–CRP–RpoS signaling pathway, which plays an important role in flagellar synthesis, is significantly inhibited in ΔdegS mutants, whereas in response to the overexpression of cyaA/crp and rpoS in the ΔdegS strain, the motility and chemotaxis of the ΔdegS + cyaA/crp and ΔdegS + rpoS strains were partially restored compared with the ΔdegS strain. We further demonstrated that transcription levels of the flagellar regulatory gene flhF are regulated by DegS via the cAMP–CRP–RpoS signaling pathway. Overexpression of the flhF gene in the ΔdegS strain partially restored motility and chemotaxis. In addition, suckling mouse intestinal colonization experiments indicated that the ΔdegS and ΔrpoE strains were characterized by the poor colonization of mouse intestines, whereas colonization efficacy was restored in the ΔdegSΔrseA, ΔdegS + cyaA/crp, ΔdegS + rpoS, and ΔdegS + flhF strains. Collectively, our findings indicate that DegS regulates the motility and chemotaxis of V. cholerae via the cAMP–CRP–RpoS–FlhF pathway, thereby influencing the colonization of suckling mouse intestines.
Introduction
Vibrio cholerae is classified into more than 200 serogroups based on surface O-antigens, among which the O1 and O139 serotypes are characterized as pathogenic strains responsible for cholera epidemics and pandemics, whereas non-O1/non-O139 V. cholerae (NOVC) produce no cholera toxins and consequently do not cause cholera (Lekshmi et al., 2018). However, NOVC are now recognized as pathogens responsible for sporadic and local infectious outbreaks (Xie et al., 2020), in which they can cause gastrointestinal infection (Lee et al., 2007), as well as extra-intestinal disease, such as bacteremia (Li et al., 2020b; Wang et al., 2022), meningitis (Hao et al., 2015), bacterial emphysema (Marinello et al., 2017), and have even been associated with significant mortality (Engel et al., 2016). Although in recent years, an increasing number of cases of NOVC infection have been reported, the pathogenic mechanisms of non-O1/non-O139 V. cholerae have yet to be sufficiently established. Bacterial motility, intestinal colonization, and cholera toxin secretion have been established to be the three most important factors associated with the pathophysiology of V. cholerae, and it is accordingly reasoned that reducing these virulence factors could make an important contribution to the prevention and treatment of cholera (Charles and Ryan, 2011). Bacterial motility has currently been identified as a virulence determinant in V. cholerae, which has a unipolar flagellum and is dependent on chemotaxis and motility to initiate appropriate virulence gene expression during the early stages of infection (Martinez et al., 2009; Matilla and Krell, 2018; Subramanian and Kearns, 2019). Within the host, V. cholerae typically utilizes its flagellum to cross the protective layer of the intestinal mucosa and enter the microvilli of intestinal epithelial cells, wherein it attaches to and colonizes the epithelial cells (Almagro-Moreno et al., 2015). When exposed to environmental selection pressure, V. cholerae can also escape from the site of colonization and enter the external environment by controlling its motility (Nielsen et al., 2006).Non-motile mutants of V. cholerae have been observed to be less virulent than the wild-type (WT) strain in young mouse model (Syed et al., 2009), whereas in humans, non-motile mutants of live attenuated V. cholerae vaccines are characterized by reduced reactivity (Coster et al., 1995). Consequently, motility plays essential roles in both the life cycle and pathogenicity of V. cholerae, and thus from the perspective of cholera prevention and treatment, it would be beneficial to study the molecular basis of V. cholerae motility and establish means whereby this motility could be impaired.
DegS is a serine protease that plays an important role in regulation of the σE (rpoE) stress response (de Regt et al., 2015). Interestingly, on the basis of RNA sequencing analysis, we previously found that degS knockout inhibited the expression of flagellum synthesis- and chemotaxis-related genes, and the findings of gene-act network analysis provided evidence to indicate that the cAMP–CRP–RpoS signaling pathway is significantly inhibited in the degS mutant (Huang et al., 2019a). Cyclic adenylate (cAMP), a second messenger involved in the regulation of cell function, binds to its signal transduction receptor (CRP) and plays important roles not only in a range of catabolic functions but also in flagellar synthesis, toxin production, and other non-catabolic processes (Manneh-Roussel et al., 2018). There are two predicted CRP binding sites in the vicinity of the rpoS promoter, and the cAMP–CRP complex can bidirectionally regulate rpoS expression at different stages of development (Cheng and Sun, 2009). The findings of previous research have revealed that deletion of rpoS markedly reduces the expression of several flagellar synthesis and chemotactic genes, which accordingly impairs bacterial motility and chemotaxis (Hengge, 2009). In the present study, we found that trends in the expression of nine flagellum synthesis- and chemotaxis-related genes are affected by degS knockout in a similar manner to seven genes affected by rpoS knockout. Furthermore, we established that the motility of a ΔdegS mutant on motility plates is less pronounced than that of the WT strain, thereby indicating that DegS may regulate the motility of V. cholerae. In this study, we systematically investigate the effects of DegS on the motility of V. cholerae.
Regulation of the expression of genes associated with flagellar synthesis is currently a particularly active area of focus in dynamics research, and also a critical link in dynamic regulation (Echazarreta and Klose, 2019; Khan et al., 2020). In this regard, the flagellar genes of V. cholerae have been established to be transcribed in a four-tiered transcriptional hierarchy (Prouty et al., 2001). Among these genes, flhF, which is transcribed in a class II operon and positively regulates class III gene transcription (Kazmierczak and Hendrixson, 2013; Arroyo-Pérez and Ringgaard, 2021), encodes a membrane-associated signal recognition particle family GTP-binding protein (FlhF), which restricts flagellum assembly to the cell pole (Green et al., 2009). In V. cholerae, a deficiency in flhF can result in flagellum loss (Correa et al., 2005). In this study, we established that DegS modulates the expression of the flagellum regulatory gene flhF via the cAMP–CRP–RpoS signaling pathway, thereby influencing the motility, chemotaxis, and colonization of V. cholerae.
Materials and methods
Bacterial strains and growth conditions
As a wild-type strain in this study, we used non-O1/non-O139 V. cholerae HN375, which was obtained from the China Center for Type Culture Collection (accession number CCTCC AB209168; Luo et al., 2011). Escherichia coli DH5α and DH5α-λpir were used for cloning and WM3064 was used as the donor strain in sexual pilus conjugation assay. All strains were grown in Luria–Bertani (LB) medium at 37°C until reaching the exponential stage of growth stage. The medium was supplemented with the following antibiotic or arabinose concentrations as needed: 100 μg/ml ampicillin, 50 μg/ml chloramphenicol, and 0.1% arabinose. Supplementary Table 1 lists all strains used in this study.
DNA manipulation and genetic techniques
All deletion mutants were constructed from the HN375 wild-type strain using the suicide plasmid pWM91 (Wu et al., 2015). The primers used for amplification are listed in Supplementary Table 2. To facilitate complementation, the coding region of crp or cyaA was cloned into plasmid pBAD24 and then sexually pilus conjugated into Δcrp or ΔcyaA, to generate the complemented strains Δcrp::crp and ΔcyaA::cyaA, respectively. Similar methods were used to construct the ΔdegSΔrseA::rseA, ΔrpoE::rpoE, and ΔrpoS::rpoS strains. To produce overexpressing strains, the recombinant plasmid pBAD24-crp or pBAD24-cyaA was used to transform ΔdegS strains via electroporation to yield the ΔdegS + crp- or ΔdegS + cyaA-overexpressing strain, respectively. Similar methods were used to construct the ΔdegS + rpoS, ΔdegS + mcp, and ΔdegS + flhF strains. To generate double overexpression strains, the crp coding sequence was cloned into a pBAD33 plasmid and used the transform the ΔdegS + cyaA overexpressing strain via electroporation. To induce gene expression, all complementary and overexpressing strains were cultured in LB medium supplemented with 0.1% arabinose.
Quantitative real-time PCR assay
Having reached the exponential stage of growth (OD600 = 0.6), bacterial cultures were harvested by centrifugation at 8,000 rpm for 5 min. Total RNA was extracted from the pelleted cells using TRIzol reagent and reverse transcribed to generate cDNA using PrimeScript™ RT reagent Kit with gDNA Eraser (Takara, China). The concentration and purity of RNA and cDNA were determined. qRT-PCR was performed using TB Green Premix Ex Taq™II (Huang et al., 2019a). For each experimental group, analyses were performed in triplicate.
Swimming motility assay
After bacterial cultures had reached the exponential stage of growth (OD600 = 0.6), cell suspensions were used to inoculate motility plates containing 0.25% agar, 1% tryptone, and 0.5% NaCl. After culturing for 14 h at 37°C, the diameters of the swimming zones around the sites of inoculation were measured at 2-h intervals. All experiments were performed in triplicate.
Chemotaxis assay
The capillary chemotactic assay performed in this study was based on a modified version of that previously described (Gordillo et al., 2007; Elgamoudi and Korolik, 2022). After bacteria had reached the exponential stage of growth, cell cultures were centrifuged at 1,200 × g for 2 min. The resulting cell pellets were washed twice with chemotaxis buffer (PH = 7.0) consisting of 10.0 ml 1 M KPO4, 0.2 ml 0.5 M EDTA, 3.915 g NaCl, 0.1 ml 10 mM methionine, 1.0 ml 10 M lactic acid, 1.0 L ddH2O, and adjusted to OD600 = 0.3. A 300-μl aliquot of the bacterial suspension was drawn into the needle cap of a 1-ml syringe, and 200 μl of a chemotaxis solution [chemotaxis buffer was used as a blank control (Partridge et al., 2019)] was aspirated into the syringe. After laying on a horizontal surface for 1 h, the movement of bacteria into the syringe was measured by quantifying the number of bacteria that accumulated by random motility or chemotaxis. The relative chemotaxis index (RCI) was calculated as the ratio between the numbers of bacteria entering the test syringe and those in the control syringe (Gordillo et al., 2007). All experiments were performed in triplicate.
Suckling mouse colonization assay
An intestinal colonization model was generated using 6-day-old CD1 suckling mice. All animal experiments performed in this study were approved by the Ethics Committee of Zunyi Medical University. Suckling mice were assigned randomly to each experimental group (n = 8 animals per group) and maintained in a specific pathogen-free environment. After the bacteria had reached the exponential stage of growth, the cells were collected by centrifugation at 1,200 × g for 2 min. The pellets thus obtained were washed twice with 1× phosphate-buffered saline, and the bacterial suspension was adjusted to OD600 = 0.5 and then diluted 100 times to a concentration of 1 × 107 CFU per ml. Each suckling mouse received 50 μl (~5 × 105 CFU) of cell suspensions via gavage, with the same volume of 1× phosphate-buffered saline serving as a baseline control (Sarkar et al., 2018; Wang et al., 2018; Cho et al., 2022). The inoculation dose was determined through several pre-experimental explorations, and we found that higher concentration including 5 × 107 CFU per 50 μl and 5 × 106 CFU per 50 μl of bacterial suspension resulted in massive death of the suckling mice in 18 h, while 5 × 105 CFU per 50 μl did not. After 18 h of observation, the intestines of sacrificed mice were dissected out, weighed, and ground. Having diluted gut preparation 100-fold, 50-μL aliquots were spread on LB plates supplemented with 0.01 mg/ml streptomycin, and colonies were counted after incubation at 37°C for 18 h. The results were expressed as the logarithm of CFUs/g intestine (cfu/g).
Statistical analysis
Data are presented as the means ± standard deviation. The data were analyzed using a two-way analyses of variance (ANOVA) and two-tailed unpaired t-tests. Two-way ANOVA was performed in conjunction with Šídák’s multiple comparison test, where the independent variable was time (h) and the dependent variable was diameter (mm).
Results
The positive regulation of Vibrio cholerae motility by DegS is partially dependent on σE
Our transcriptome sequencing data revealed that degS knockdown resulted in a significant down-regulation of nine motility-related gene (Figure 1A; Supplementary Table 3), implying that DegS may influence V. cholerae motility. To verify this conjecture, we carried out swimming motility assays, and found that the diameter of the swimming zone of the degS mutant on motility plates was significantly smaller than that of the WT strain (Figure 1B; Supplementary Figure 1). Subsequently, we performed a chemotaxis assay using three widely utilized attractants, namely 50 mM fructose (Liu et al., 2020b), 100 mM serine (L-ser; Roggo et al., 2018), and 100 μM aspartic acid (L-Asp; Long et al., 2017). Either with or without attractant, the motility of the ΔdegS strain was found to be lower than that of the WT strain. However, motility was partially restored in the ΔdegS::degS strain, although not in the empty vector strain (ΔdegS + pBAD24; Figure 1C). These observations thus provided evidence that DegS positively regulates the motility of V. cholerae.
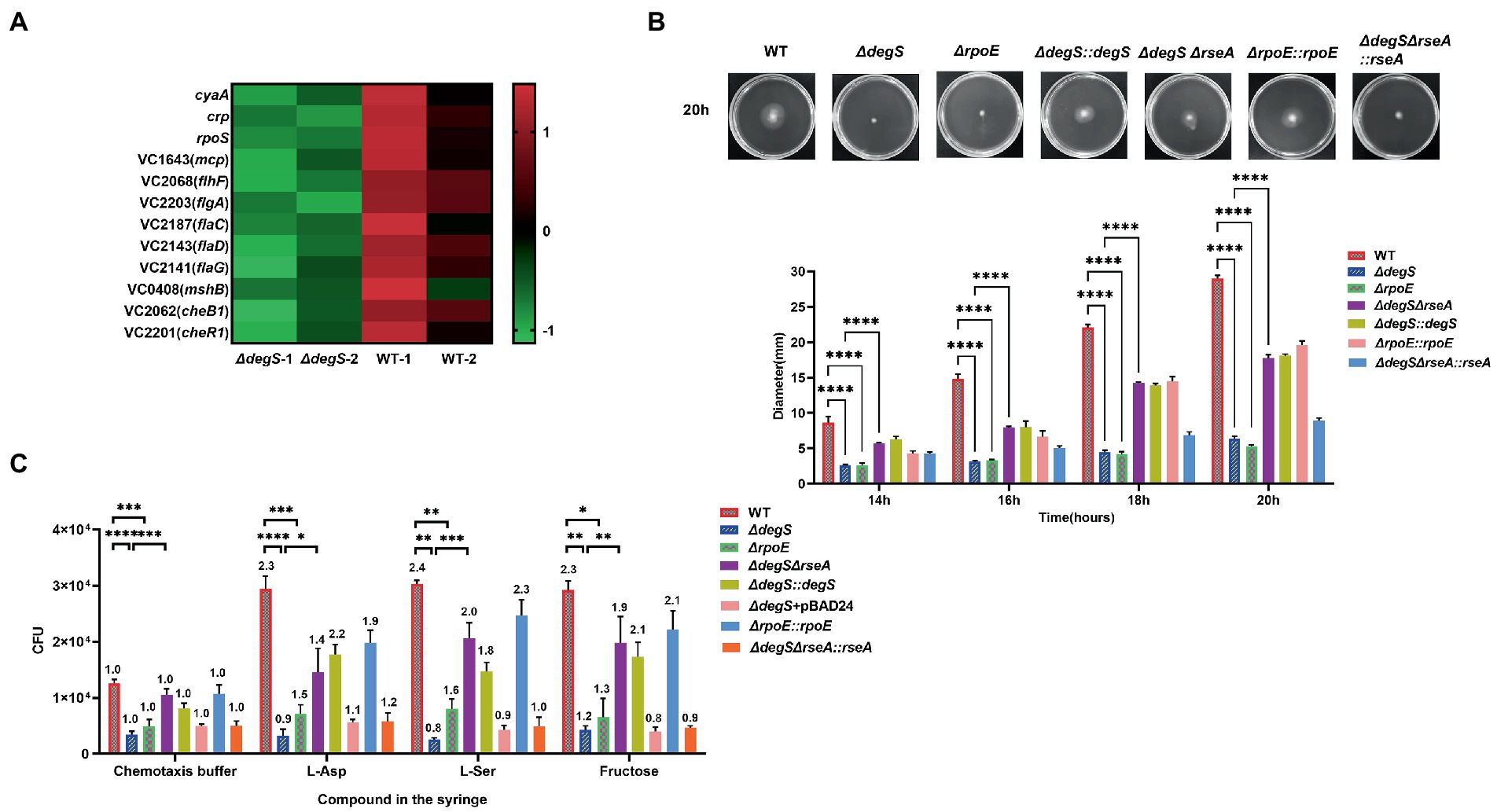
Figure 1. The positive regulation of Vibrio cholerae motility by DegS is partially dependent on σE. (A) RNA-Seq analysis of differentially expressed genes in ΔdegS and wild-type (WT) strains reveled that crp, cyaA, rpoS, and nine motility-related genes were downregulated in ΔdegS strains. (B) The swimming motility of the WT, ΔdegS, ΔdegS::degS, ΔrpoE, ΔrpoE::rpoE, ΔdegSΔrseA, and ΔdegSΔrseA::rseA strains were assessed on soft agar plates. The representative pictures for the swimming zones are obtained at 20 h. Data were analyzed using a two-way ANOVA. ****, p < 0.0001 between two strains. (C) Analysis of the chemotactic responses to aspartate, serine, and fructose by the WT, ΔdegS, ΔdegS::degS, ΔdegS + pBAD24, ΔrpoE, ΔrpoE::rpoE, ΔdegSΔrseA, and ΔdegSΔrseA::rseA strains. Error bars indicate the SDs based on three replicated experimental values (n = 3). *, p < 0.05; **, p < 0.01; ***, p < 0.001; and ****, p < 0.0001 between two strains (two-tailed unpaired t-tests). Numbers on top of each bar indicate the relative chemotactic indexes (RCI).
It has previously been established that the essential function of DegS is to provide active σE for cells by degrading RseA (Alba et al., 2001), and the absence of DegS has been demonstrated to result in a significant reduction in σE activity (Ades et al., 1999). In contrast, the absence of RseA coincides with a high constitutive activity σE (De Las Peñas et al., 1997; Missiakas et al., 1997). To investigate whether RpoE is involved in the DegS-regulated motility of V. cholerae, we constructed a rpoE mutant (ΔrpoE), a degS and rseA double-knockout strain (ΔdegSΔrseA) and the corresponding complemented strains (ΔrpoE::rpoE and ΔdegSΔrseA::rseA). qRT-PCR results confirmed that expression of the rpoE gene in these strains was consistent with the aforementioned results (Supplementary Figure 2). The ΔrpoE strain showed a significant decrease in swimming motility and chemotaxis compared to the WT strain, and the ΔrpoE::rpoE strain could revert to near WT strain levels. Whereas the ΔdegSΔrseA strain showed swimming motility and chemotaxis close to WT strain levels, the ΔdegSΔrseA::rseA strain showed phenotypic characteristics similar to the ΔdegS strain (Figures 1B,C; Supplementary Figure 1). These findings accordingly indicate that the positive regulation of V. cholerae motility via DegS is partially dependent on σE.
Regulation of Vibrio cholerae motility by DegS may involve the co-regulation of cAMP and CRP
Previous studies have shown that the cAMP–CRP complex regulates flagellum biosynthesis (Liu et al., 2020a). Our transcriptome data revealed that the levels of cyaA and crp transcripts were significantly lower in the ΔdegS strain (Figure 1A). We further verified these observations by qRT-PCR and found that DegS and RpoE positively regulate the expression of cyaA and crp (Figure 2A). CyaA encodes adenylyl cyclase, which can synthesize cAMP from ATP, and crp encodes the cyclic adenylyl receptor protein (CRP; Zhang et al., 2013). To gain a better understanding of the role of cAMP and CRP in the DegS-mediated regulation of V. cholerae motility, we overexpressed cyaA or crp in ΔdegS mutants (ΔdegS + cyaA and ΔdegS + crp) and performed swimming and chemotaxis assays, the results of which revealed that overexpression of neither cyaA nor crp could compensate for the lost motility and chemotaxis of ΔdegS strains (Figures 2B,C; Supplementary Figure 3). In most cases, cAMP and CRP are mutually dependent on each other for functional activity (Kolb et al., 1993; Busby and Ebright, 1999), and when both cyaA and crp were overexpressed in the ΔdegS strains (ΔdegS + cyaA/crp), we found that the motility and chemotaxis of the ΔdegS + cyaA/crp strain could be partially restored (Figures 2B,C). These findings indicate that the regulation of V. cholerae motility by DegS may involve the co-regulation of cAMP and CRP.
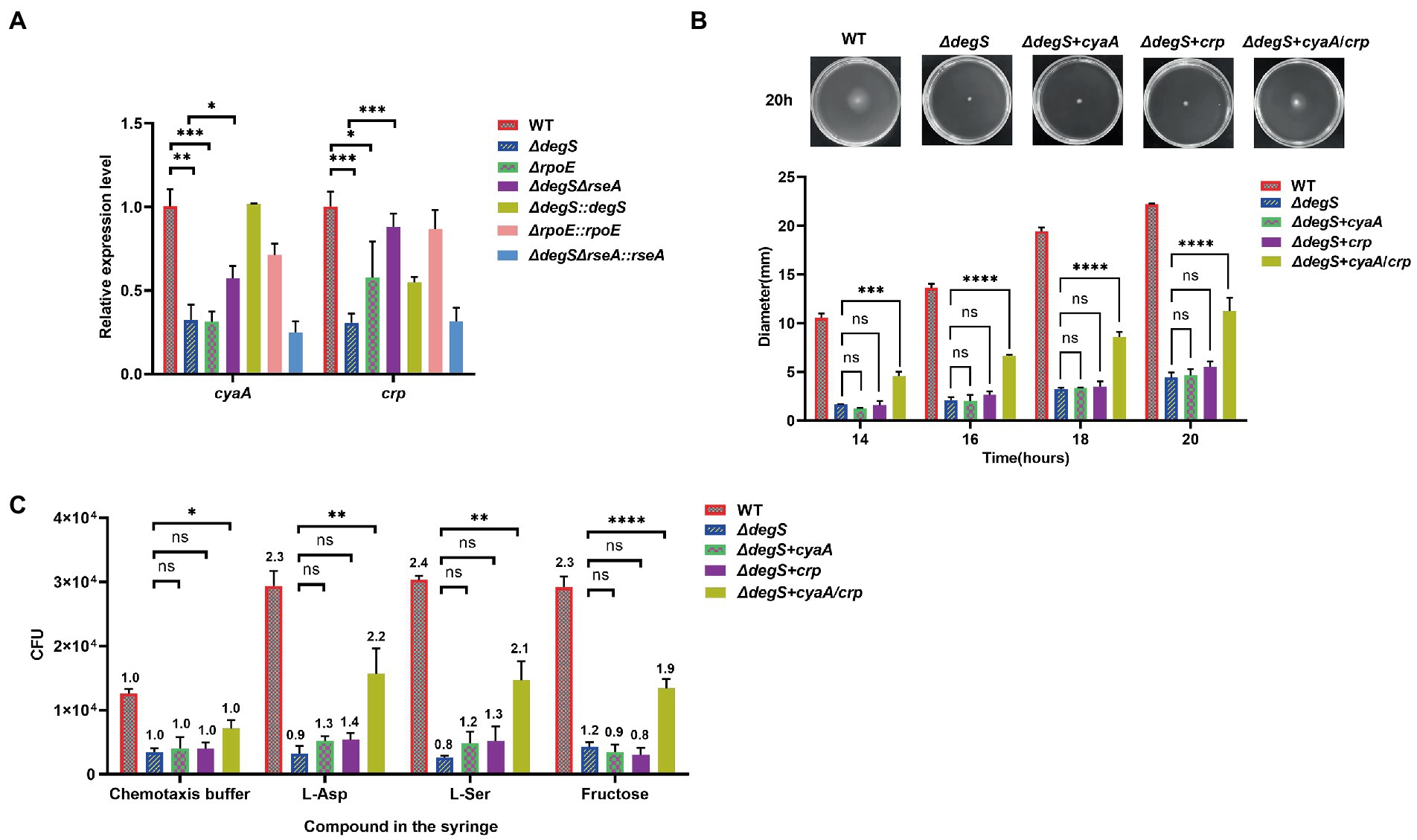
Figure 2. The regulation of Vibrio cholerae motility by DegS may involve the co-regulation of cAMP and CRP. (A) The mRNA levels of cyaA and crp in the WT, ΔdegS, ΔdegS::degS, ΔrpoE, ΔrpoE::rpoE, ΔdegSΔrseA, and ΔdegSΔrseA::rseA strains. Statistical analyses were performed using an unpaired t-test. *, p < 0.05; **, p < 0.01; and ***, p < 0.001 between two strains. (B) The swimming motility of WT, ΔdegS, ΔdegS + crp, ΔdegS + cyaA and ΔdegS + cyaA/crp strains was determined. The representative pictures for the swimming zones are obtained at 20 h. Statistical analyses were performed using a two-way ANOVA. ***, p < 0.001 and ****, p < 0.0001 between two strains; ns, not significant. (C) Analysis of the chemotactic responses to aspartate, serine, and fructose for the WT, ΔdegS, ΔdegS + crp, ΔdegS + cyaA, and ΔdegS + cyaA/crp strains. Error bars indicate the SDs based on three replicated experimental values (n = 3). *, p < 0.05; **, p < 0.01; and ****, p < 0.0001 between two strains; ns, not significant (unpaired t-tests). Numbers on top of each bar indicate the relative chemotactic indexes (RCI).
RpoS participates in the DegS-mediated regulation of Vibrio cholerae motility
Knockout of the rpoS gene has been demonstrated to markedly reduce the expression of several flagellum synthesis and chemotactic genes, which impairs bacterial motility and chemotaxis (Hengge, 2009). The cAMP–CRP complex has been characterized as a transcriptional activator of rpoS (Guo et al., 2015), and the results of our qRT-PCR analyses indicated that degS positively affects the transcriptional levels of rpoS via cyaA and crp (Figure 3A). To examine the role of RpoS in the DegS-mediated regulation of motility in V. cholerae, we generated rpoS knockout, rpoS compensation and rpoS overexpression strains (ΔrpoS, ΔrpoS::rpoS and ΔdegS + rpoS). Compared with WT strain, the motility and chemotaxis of ΔrpoS mutant were significantly reduced, whereas these activities were partially compensated in the ΔrpoS::rpoS and ΔdegS + rpoS strains (Figures 3B,C; Supplementary Figure 4). These results thus tend to indicate that RpoS participates in the DegS-mediated regulation of V. cholerae motility.
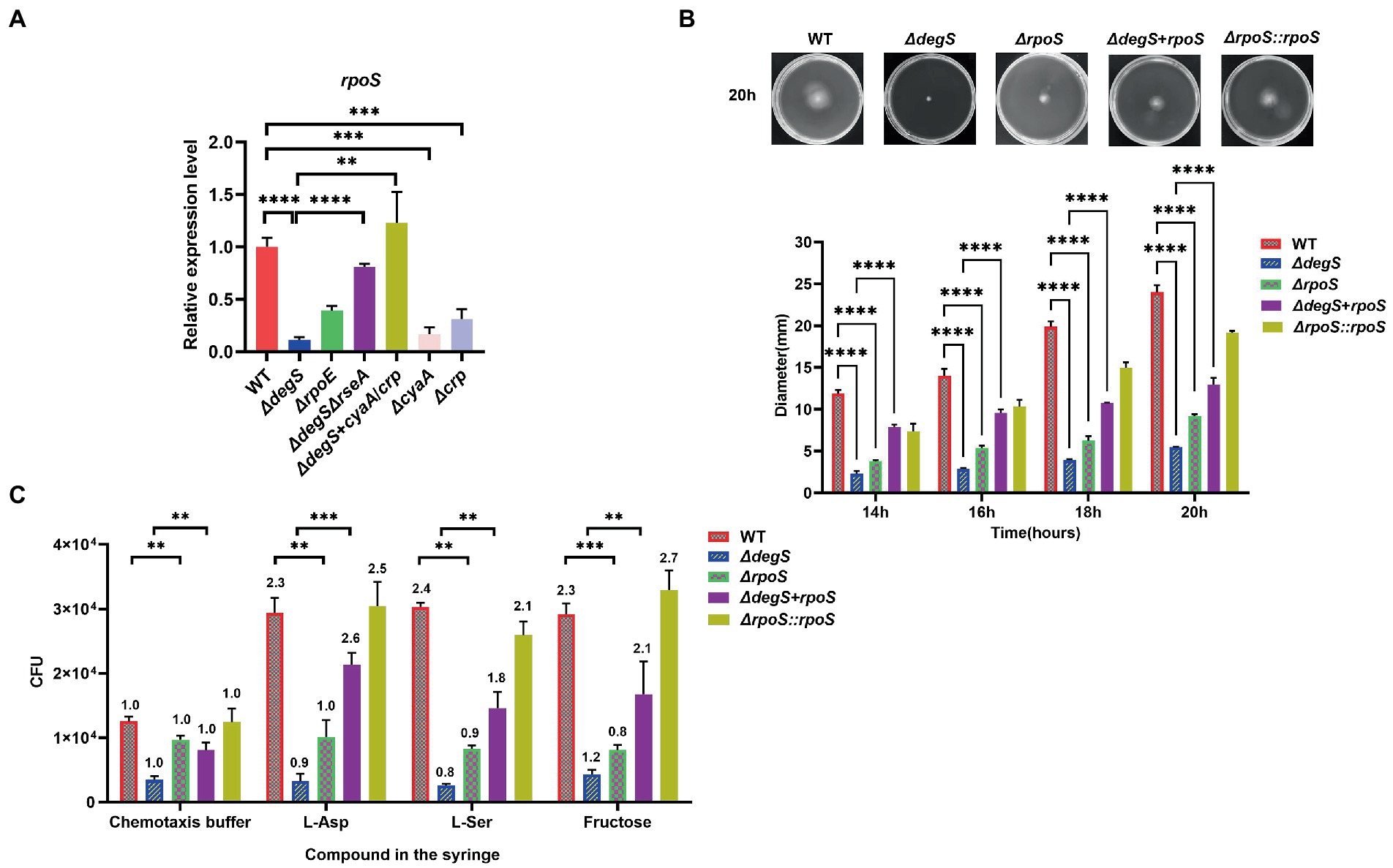
Figure 3. RpoS participates in the DegS-mediated regulation of Vibrio cholerae motility. (A) The mRNA levels of rpoS in WT, ΔdegS, ΔrpoE, ΔdegSΔrseA, ΔcyaA, Δcrp, and ΔdegS + cyaA/crp strains (unpaired t-tests). (B) The swimming motility of WT, ΔdegS, ΔrpoS, ΔrpoS::rpoS, and ΔdegS + rpoS strains was assessed. The representative pictures for the swimming zones are obtained at 20 h. Statistical analyses were performed using a two-way ANOVA. ***, p < 0.001 and ****, p < 0.0001 between two strains. (C) Analysis of the chemotactic responses to aspartate, serine, and fructose for WT, ΔdegS, ΔrpoS, ΔrpoS::rpoS, and ΔdegS + rpoS strains. Error bars indicate the SDs based on three replicated experimental values (n = 3). **, p < 0.01; ***, p < 0.001 (unpaired t-tests). Numbers on top of each bar indicate the relative chemotactic indexes (RCI).
The effect of DegS on Vibrio cholerae motility may be involved in the expression of FlhF
Our transcriptome data revealed that compared with the WT strain, nine flagellar synthesis and chemotaxis genes were significantly repressed in ΔdegS strains, namely, VC1643 (mcp), VC2068 (flhF), VC2203 (flgA), VC2187 (flaC), VC2143 (flaD), VC2141 (flaG), VC0408 (mshB), VC2062 (cheB1), and VC2201 (cheR1; Figure 1A). Furthermore, our qRT-PCR results confirmed that the deletion of either degS or rpoE significantly inhibited the expression of these nine genes, and the compensation of degS or rpoE could restore the transcription levels of these genes (Figure 4A). To further establish which of these genes might participate in the DegS regulation of V. cholerae motility, we determined the transcriptional levels of the nine dynamic-related genes in ΔcyaA, Δcrp, and other strains. Compared with the WT strain, the transcription levels of the mcp, flhF, flaC, and flaD genes were substantially reduced in both the ΔcyaA and Δcrp strains, whereas levels were found to be restored in the ΔcyaA::cyaA and Δcrp::crp strains (Figure 4B). In addition, the transcription of these four genes was partially restored in the ΔdegAΔrseA strain, whereas the transcription of these four genes was suppressed by the compensation of rseA (Figure 4A). In V. cholerae, FlaC, and FlaD have been identified as components of the flagellum filament, although appear to be non-essential for filament synthesis and motility (Rui et al., 2010). Contrastingly, MCP and FlhF play important roles in the motility of bacterial flagella (Huang et al., 2019b; Arroyo-Pérez and Ringgaard, 2021). Moreover, RpoS has been shown to be required for the expression of both mcp and flhF (Nielsen et al., 2006). Our qRT-PCR analysis in the present study revealed that the transcription levels of mcp and flhF indeed positively regulated by RpoS, and the levels were partially recovered in the ΔdegS + cyaA/crp and ΔdegS + rpoS strains (Figure 4C), thereby indicating that DegS may control the transcription of mcp and flhF via the cAMP–CRP–RpoS pathway. To further assess whether FlhF and MCP play roles in the DegS regulation of V. cholerae motility, we, respectively, overexpressed proteins in the ΔdegS mutant and performed swimming and chemotaxis assays. The results revealed that motility and chemotaxis were partially restored in the ΔdegS + flhF strain, although not in the ΔdegS + mcp strain (Figures 4D,E; Supplementary Figure 5). Taken together, these observations indicate that the DegS-mediated regulation of V. cholerae motility may be involved in the regulation of FlhF expression.
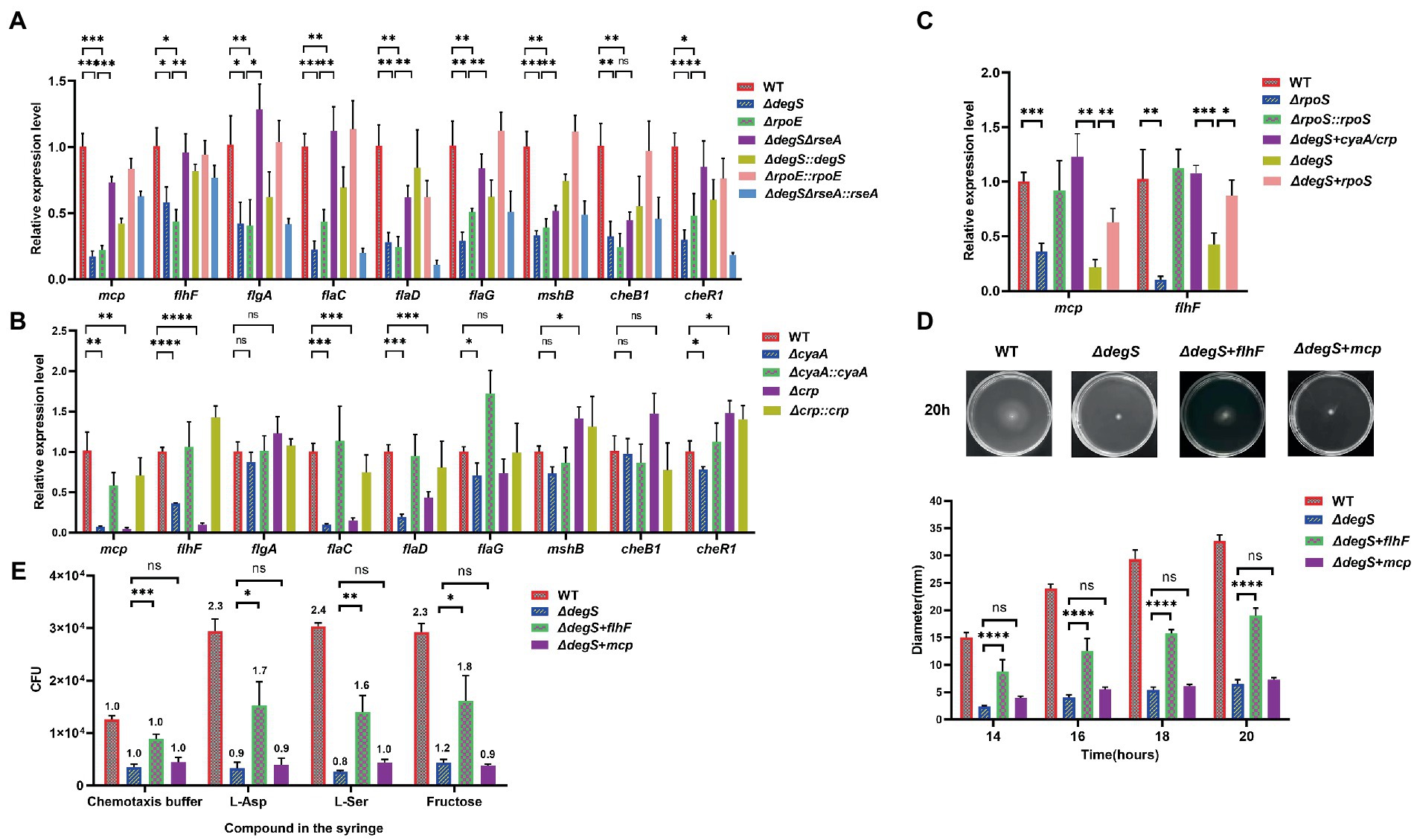
Figure 4. The effect of DegS on Vibrio cholerae motility may involve the expression of FlhF. (A,B) The mRNA levels of nine flagellum synthesis- and chemotaxis-related genes in WT, ΔdegS, ΔrpoE, ΔrpoE::rpoE, ΔdegS ΔrseA, ΔdegS ΔrseA::rseA, ΔdegS::degS, ΔcyaA, ΔcyaA::cyaA, Δcrp, and Δcrp::crp strains (unpaired t-tests). (C) The mRNA levels of mcp and flhF in WT, ΔdegS, ΔrpoS, ΔrpoS::rpoS, ΔdegS + cyaA/crp, and ΔdegS + rpoS strains (unpaired t-tests). (D) The swimming motility of WT, ΔdegS, ΔdegS + flhF, and ΔdegS + mcp strains was assessed. The representative pictures for the swimming zones are obtained at 20 h. Statistical analyses were performed using a two-way ANOVA. ****, p < 0.0001 between two strains; ns, not significant. (E) The analysis of chemotactic responses to aspartate, serine, and fructose for WT, ΔdegS, ΔdegS + flhF, and ΔdegS + mcp strains. Error bars indicate the SDs based on three replicated experimental values (n = 3). *, p < 0.05; **, p < 0.01; ***, p < 0.001; ns, not significant (unpaired t-tests). Numbers on top of each bar indicate the relative chemotactic indexes (RCI).
DegS affects intestinal colonization by Vibrio cholerae via the cAMP–CRP–RpoS–FlhF pathway
Flagellum-driven motility plays an essential role in bacterial colonization (Baban et al., 2013; Tamar et al., 2016; Kajikawa et al., 2018), and our in vitro experiments revealed that DegS affects V. cholerae motility via the cAMP–CRP–RpoS–FlhF signaling pathway. To establish whether the regulatory mechanism is equally important in intestinal colonization by V. cholerae, we performed analyses using an intestinal colonization model in suckling mice. The results revealed the poor colonization ability of the ΔdegS and ΔrpoE strains, whereas the colonization efficacy of the ΔdegSΔrseA strain was comparable to that of the WT strain (Figure 5A), which tends to indicate that the effect of DegS on the intestinal colonization of V. cholerae is partially dependent on σE. The colonization ability of ΔcyaA, Δcrp and ΔrpoS was also observed to be lower than that of the WT strain, whereas colonization efficacy was effectively restored in the ΔdegS + cyaA/crp, ΔdegS + rpoS, and ΔdegS + flhF strains (Figures 5B,C). These findings indicate that cAMP–CRP, RpoS, and FlhF are involved in the DegS-mediated regulation of V. cholerae intestinal colonization.
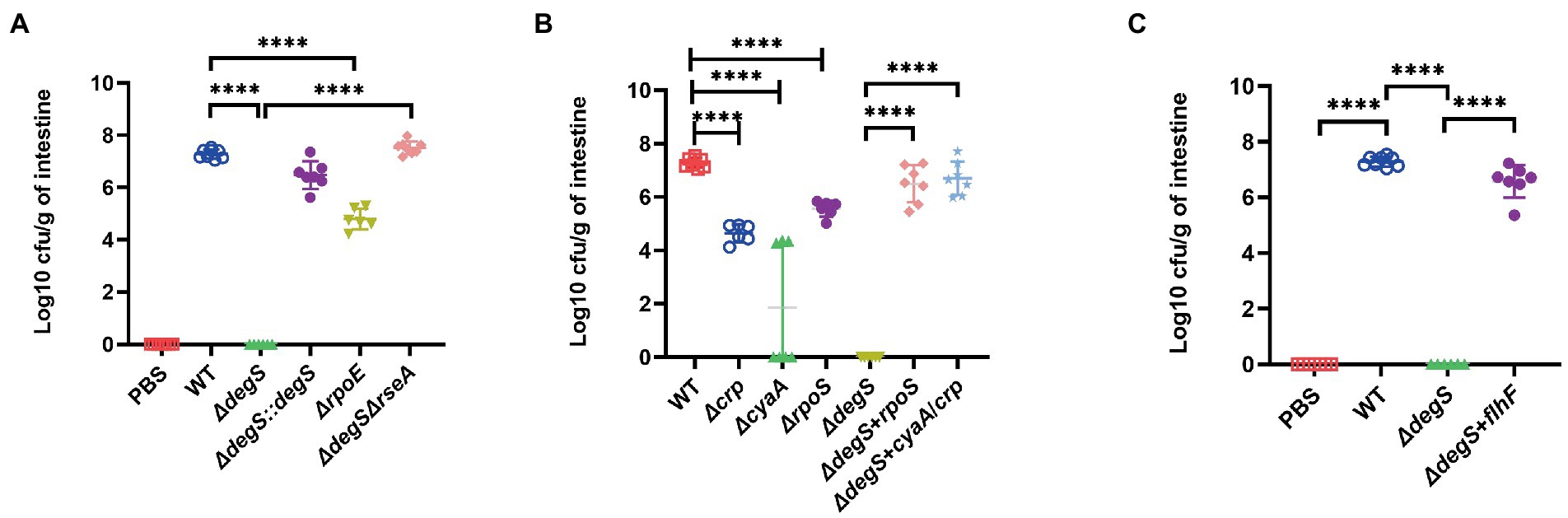
Figure 5. DegS influences Vibrio cholerae intestinal colonization via the cAMP–CRP–RpoS–FlhF pathway. (A–C) The numbers of bacteria in the intestine of suckling mice after infection with 1 × 107 colony-forming units of the WT, ΔdegS, ΔdegS::degS, ΔrpoE, ΔdegSΔrseA, ΔcyaA, Δcrp, ΔrpoS, ΔdegS + rpoS, Δdegs + cyaA/crp, and ΔdegS + flhF strains for 18 h. The results are expressed as the logarithm of colony-forming units/g intestine (cfu/g; ±SD, n = 8). ****, p < 0.0001 between two strains (unpaired t-tests).
Discussion
In this study, we showed that DegS protease plays essential roles in V. cholerae motility, chemotaxis, and colonization, and proposed a model in which DegS protease regulates expression of the flagellum regulatory gene flhF via the cAMP–CRP–RpoS signaling pathway, thereby regulating V. cholerae motility and influencing its intestinal colonization capacity (Figure 6). DegS, a membrane-anchored periplasmic protease, plays an important role in the σE-mediated stress response pathway (Sohn et al., 2007; Chaba et al., 2011), and as a pressure sensor protein of the σE-mediated stress response, active DegS promotes RseA cleavage and the release active σE, thereby inducing the expression of σE-dependent genes (Walsh et al., 2003). Multiple transcriptional regulatory systems that are directly controlled by σE are involved in a wide range of biological processes, including stress responses, virulence, motility, biofilm formation, and quorum sensing (Liang et al., 2021), and previous studies have established that RpoE can promote the motility of Salmonella enterica serovar typhi (S. typhi; Zhang et al., 2015; Spöring et al., 2018). In the present study, we showed that V. cholerae motility is significantly inhibited by the deletion of degS or rpoE, and that this motility could be partially restored in ΔdegSΔrseA strains (Figures 1B,C). On the basis of these observations, we thus speculate that the σE-mediated stress response could be the process linking the deletion of degS with impaired motility.
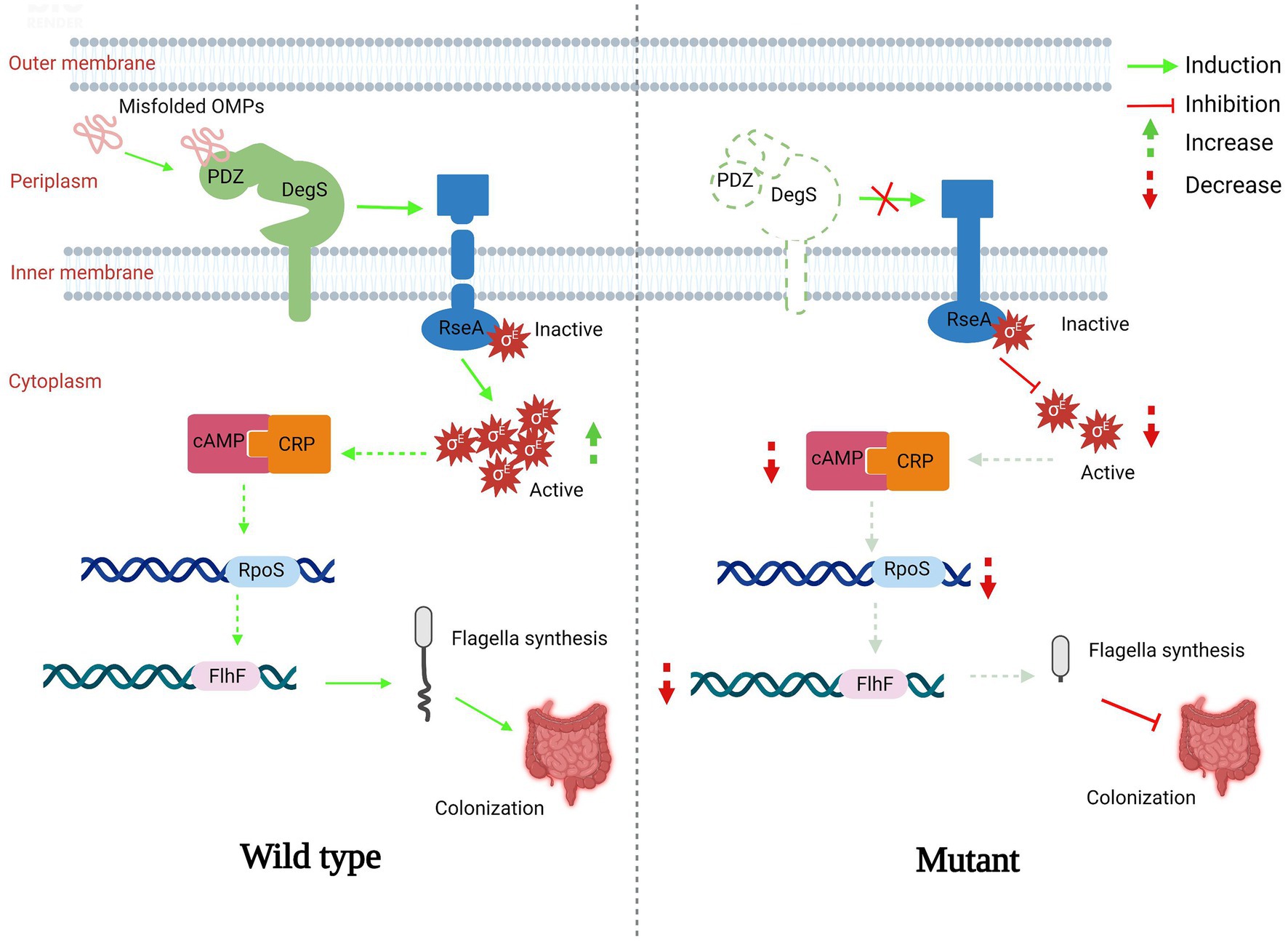
Figure 6. A proposed model showing the mechanisms whereby DegS positively regulates Vibrio cholerae motility. In the normal state (wild type), the PDZ domain of DegS binds to a large number of misfolded outer membrane proteins and promotes RseA cleavage to release active σE. Subsequently, the expression of flhF is induced via the cAMP–CRP–RpoS signaling pathway, resulting in the synthesis of bacterial flagella. In the degS knockout state (mutant), RseA cleavage is suppressed, amounts of σE and cAMP–CRP complex are reduced, and the transcription of rpoS and flhF is downregulated, resulting in impaired bacterial flagella development and poor colonization ability. A red arrow represents inhibition and a downward-pointing dashed red arrow represents a reduction. A green arrow represents induction and an upward-pointing green dashed arrow represents an increase.
To gain a further understanding of the mechanisms underlying the regulation of V. cholerae motility by DegS, we performed RNA-seq analysis to investigate the genes that are differentially expressed between ΔdegS and WT strains (Figure 1A), and gene-act network analysis indicated that the cAMP–CRP–RpoS signaling pathway may participate in the DegS-mediated regulation of V. cholerae motility (Huang et al., 2019a). cAMP–CRP serves as an important global regulator of the expression of numerous regulators and operons. It has been established that more than 7% of the genes in E. coli are regulated by the cAMP–CRP system (Zheng et al., 2004). For example, it has been shown that the cAMP–CRP complex bind directly to a region upstream of FlhDC, the main operon associated flagellar production in E. coli (Soutourina et al., 1999; Fahrner and Berg, 2015). In the present study, we found that that cAMP and CRP influence the transcription of certain flagellum biosynthesis-related genes in V. cholerae (Figure 4B), and that the impaired motility and chemotaxis of a degS mutant could be partly reversed by the overexpression of cyaA and crp (Figures 2B,C). We accordingly speculate that the cAMP–CRP complex participates in the DegS-mediated regulation of V. cholerae motility.
The cAMP–CRP complex and RpoS collaborate to regulate multiple cellular processes (Franchini et al., 2015), and as early as 2002, a close association between cAMP–CRP and RpoS was proposed (Hengge-Aronis, 2002b). Two potential cAMP–CRP binding sites are located upstream and downstream of the rpoS promoter (Hengge-Aronis, 2002a), and direct binding of the cAMP–CRP complex to the rpoS promoter has been demonstrated in S. typhi (Cheng and Sun, 2009). In the present study, qRT-PCR was performed to confirm that the cAMP–CRP system also positively regulates transcription of the rpoS gene in V. cholerae (Figure 3A). We established that only simultaneous overexpression of cyaA and crp could restore the levels of rpoS transcription in the degS knockout strain (Figure 3A), which accordingly leads us to speculate that DegS may regulate the expression of RpoS by regulating the cAMP–CRP complex. RpoS has been identified as an important regulator of flagellum biosynthesis in Yersinia (Guan et al., 2015), and in V. cholerae, the interaction between RpoS and its antiregulator RssB has been shown to influence motility and colonization ability (Wurm et al., 2017; Wölflingseder et al., 2022). We also established that the motility and colonization of V. cholerae are suppressed by the deletion of rpoS (Figures 3B,C, 5B). A previous comparison of the genes differentially expressed between the WT and ΔrpoS strains of V. cholerae has revealed that the transcription of flhF is dependent on rpoS (Nielsen et al., 2006), and in the present study, we confirmed this association based on qRT-PCR analysis and suggest that RpoS is involved in the transcriptional regulation of flhF by DegS (Figure 4C).
It has been established that the flagellar genes in V. cholerae are expressed on the basis of a four-tiered transcriptional hierarchy, and that flhF is transcribed within a class II operon (Prouty et al., 2001). The findings of previous studies have indicated that the flhF mutant of V. cholerae lacks a polar flagellum, which results in a significant reduction in bacterial motility (Correa et al., 2005), and we demonstrated that transformation with the pBAD24-flhF plasmid could partially compensate for the deficient motility and chemotaxis of the degS mutant (Figures 4D,E). FlhF plays roles in the polar targeting of the V. cholerae flagellum and promotes flagellum assembly by recruiting the earliest flagellar structural component, the inner-membrane MS-ring protein FliF, to the cell pole (Green et al., 2009). FlhF functions as a positive regulator of class III flagellar promoters involved in the synthesis of the basal bodyhook, motor component (MotY), and “core” flagellin (FlaA; Echazarreta and Klose, 2019). In Campylobacter jejuni, FlhF has been shown to bind directly to the flgI promoter, which encodes a component of the flagellar P-ring, thereby controlling flagellar biosynthesis (Li et al., 2020a). On the basis of these previous finding, we propose that DegS may influence the motility and chemotaxis of V. cholerae by regulating FlhF expression via the cAMP–CRP–RpoS signaling pathway. Given that the respective overexpression of cyaA/crp, rpoS, and flhF in ΔdegS strains only partially restored the normal phenotypes of the degS mutant, we speculate that there may be additional factors involved in the DegS-mediated regulation of motility and chemotaxis in V. cholerae. Accordingly, the specific mechanisms whereby DegS regulates the motility of V. cholerae via fhlF warrant further analyses.
In order to successfully infect a host, V. cholerae is dependent on flagellar motility to facilitate penetration of the host intestinal mucosa, and thereby effectively colonize the host intestines (Butler and Camilli, 2005). Our observations in the present study indicate that the DegS-mediated regulation of V. cholerae flagellar motility via the cAMP–CRP–RpoS–FlhF pathway may contribute to intestinal colonization (Figure 5). In this regard, it has previously been demonstrated that Salmonella might use the σE-dependent cell envelope stress response as a cue to determine the spatiotemporal stage of infection during host colonization (Spöring et al., 2018). Colonization of the intestines of suckling mice by V. cholerae has been shown to be reduced by mutation of the rpoE gene (Kovacikova and Skorupski, 2002), which is consistent with our findings in this study, in which we showed that the deletion of degS or rpoE had the effect of suppressing the intestinal colonization capacity of V. cholerae, whereas this lost function was partially restored in the ΔdegSΔrseA strain (Figure 5A). We therefore speculate that V. cholerae may be dependent on a DegS-mediated stress response pathway for effective colonization. Furthermore, the cAMP–CRP signaling pathway has been established to modulates the expression of numerous virulence genes and colonization factors during bacterial host colonization (Kariisa et al., 2015; Manneh-Roussel et al., 2018). In this regard, our findings indicate that the DegS-mediated regulation of V. cholerae colonization involves regulation of the flagellum regulatory gene flhF via the cAMP–CRP–RpoS signaling pathway.
Conclusion
In this study, we demonstrate that deletion of the degS gene attenuates the motility and chemotaxis of V. cholerae, which in turn influences the ability of V. cholerae to colonize the small intestine of suckling mice. Moreover, we established that the underlying regulatory process is dependent on rpoE. Our findings also indicate that DegS plays important regulatory roles in the motility and chemotaxis V. cholerae and that the underlying mechanisms may involve the regulation of FlhF expression via the cAMP–CRP–RpoS signaling pathway. Collectively, the findings of this study enhance our current understanding of the biological function of DegS and provide new insights into the regulation of V. cholerae motility. However, further details are required regarding the regulation of cAMP–CRP by DegS-σE and the mechanisms whereby FlhF plays a role in the DegS-mediated regulation of V. cholerae motility.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
JH and XM designed the study. MZ, KW, JZ, HL, HY, and LW performed the experiments. MZ, KW, MH, and GW analyzed the data. MZ and JH wrote the paper. XM and JH conceived and supervised the project. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the National Natural Science Foundation of China (No. 32060035) and the Science and Technology Project of Guizhou [Qiankehejichu-ZK[2021]470].
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1159986/full#supplementary-material
References
Ades, S. E., Connolly, L. E., Alba, B. M., and Gross, C. A. (1999). The Escherichia coli sigma(E)-dependent extracytoplasmic stress response is controlled by the regulated proteolysis of an anti-sigma factor. Genes Dev. 13, 2449–2461. doi: 10.1101/gad.13.18.2449
Alba, B. M., Zhong, H. J., Pelayo, J. C., and Gross, C. A. (2001). degS (hhoB) is an essential Escherichia coli gene whose indispensable function is to provide sigma (E) activity. Mol. Microbiol. 40, 1323–1333. doi: 10.1046/j.1365-2958.2001.02475.x
Almagro-Moreno, S., Pruss, K., and Taylor, R. K. (2015). Intestinal colonization dynamics of Vibrio cholerae. PLoS Pathog. 11:e1004787. doi: 10.1371/journal.ppat.1004787
Arroyo-Pérez, E. E., and Ringgaard, S. (2021). Interdependent polar localization of FlhF and FlhG and their importance for flagellum formation of Vibrio parahaemolyticus. Front. Microbiol. 12:655239. doi: 10.3389/fmicb.2021.655239
Baban, S. T., Kuehne, S. A., Barketi-Klai, A., Cartman, S. T., Kelly, M. L., Hardie, K. R., et al. (2013). The role of flagella in Clostridium difficile pathogenesis: comparison between a non-epidemic and an epidemic strain. PLoS One 8:e73026. doi: 10.1371/journal.pone.0073026
Busby, S., and Ebright, R. H. (1999). Transcription activation by catabolite activator protein (CAP). J. Mol. Biol. 293, 199–213. doi: 10.1006/jmbi.1999.3161
Butler, S. M., and Camilli, A. (2005). Going against the grain: chemotaxis and infection in Vibrio cholerae. Nat. Rev. Microbiol. 3, 611–620. doi: 10.1038/nrmicro1207
Chaba, R., Alba, B. M., Guo, M. S., Sohn, J., Ahuja, N., Sauer, R. T., et al. (2011). Signal integration by DegS and RseB governs the σ E-mediated envelope stress response in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 108, 2106–2111. doi: 10.1073/pnas.1019277108
Charles, R. C., and Ryan, E. T. (2011). Cholera in the 21st century. Curr. Opin. Infect. Dis. 24, 472–477. doi: 10.1097/QCO.0b013e32834a88af
Cheng, Y., and Sun, B. (2009). Polyphosphate kinase affects oxidative stress response by modulating cAMP receptor protein and rpoS expression in Salmonella typhimurium. J. Microbiol. Biotechnol. 19, 1527–1535. doi: 10.4014/jmb.0903.03030
Cho, J. Y., Liu, R., and Hsiao, A. (2022). Microbiota-associated biofilm regulation leads to Vibrio cholerae resistance against intestinal environmental stress. Front. Cell. Infect. Microbiol. 12:861677. doi: 10.3389/fcimb.2022.861677
Correa, N. E., Peng, F., and Klose, K. E. (2005). Roles of the regulatory proteins FlhF and FlhG in the Vibrio cholerae flagellar transcription hierarchy. J. Bacteriol. 187, 6324–6332. doi: 10.1128/jb.187.18.6324-6332.2005
Coster, T. S., Killeen, K. P., Waldor, M. K., Beattie, D. T., Spriggs, D. R., Kenner, J. R., et al. (1995). Safety, immunogenicity, and efficacy of live attenuated Vibrio cholerae O139 vaccine prototype. Lancet 345, 949–952. doi: 10.1016/s0140-6736(95)90698-3
De Las Peñas, A., Connolly, L., and Gross, C. A. (1997). The sigmaE-mediated response to extracytoplasmic stress in Escherichia coli is transduced by RseA and RseB, two negative regulators of sigmaE. Mol. Microbiol. 24, 373–385. doi: 10.1046/j.1365-2958.1997.3611718.x
de Regt, A. K., Baker, T. A., and Sauer, R. T. (2015). Steric clashes with bound OMP peptides activate the DegS stress-response protease. Proc. Natl. Acad. Sci. U. S. A. 112, 3326–3331. doi: 10.1073/pnas.1502372112
Echazarreta, M. A., and Klose, K. E. (2019). Vibrio flagellar synthesis. Front. Cell. Infect. Microbiol. 9:131. doi: 10.3389/fcimb.2019.00131
Elgamoudi, B. A., and Korolik, V. (2022). A review of the advantages, disadvantages and limitations of chemotaxis assays for Campylobacter spp. Int. J. Mol. Sci. 23:1576. doi: 10.3390/ijms23031576
Engel, M. F., Muijsken, M. A., Mooi-Kokenberg, E., Kuijper, E. J., and van Westerloo, D. J. (2016). Vibrio cholerae non-O1 bacteraemia: description of three cases in the Netherlands and a literature review. Euro Surveill. 21:30197. doi: 10.2807/1560-7917.Es.2016.21.15.30197
Fahrner, K. A., and Berg, H. C. (2015). Mutations that stimulate flhDC expression in Escherichia coli K-12. J. Bacteriol. 197, 3087–3096. doi: 10.1128/jb.00455-15
Franchini, A. G., Ihssen, J., and Egli, T. (2015). Effect of global regulators RpoS and cyclic-AMP/CRP on the catabolome and transcriptome of Escherichia coli K12 during carbon- and energy-limited growth. PLoS One 10:e0133793. doi: 10.1371/journal.pone.0133793
Gordillo, F., Chávez, F. P., and Jerez, C. A. (2007). Motility and chemotaxis of Pseudomonas sp. B4 towards polychlorobiphenyls and chlorobenzoates. FEMS Microbiol. Ecol. 60, 322–328. doi: 10.1111/j.1574-6941.2007.00293.x
Green, J. C., Kahramanoglou, C., Rahman, A., Pender, A. M., Charbonnel, N., and Fraser, G. M. (2009). Recruitment of the earliest component of the bacterial flagellum to the old cell division pole by a membrane-associated signal recognition particle family GTP-binding protein. J. Mol. Biol. 391, 679–690. doi: 10.1016/j.jmb.2009.05.075
Guan, J., Xiao, X., Xu, S., Gao, F., Wang, J., Wang, T., et al. (2015). Roles of RpoS in Yersinia pseudotuberculosis stress survival, motility, biofilm formation and type VI secretion system expression. J. Microbiol. 53, 633–642. doi: 10.1007/s12275-015-0099-6
Guo, M., Wang, H., Xie, N., and Xie, Z. (2015). Positive effect of carbon sources on natural transformation in Escherichia coli: role of low-level cyclic AMP (cAMP)-cAMP receptor protein in the derepression of rpoS. J. Bacteriol. 197, 3317–3328. doi: 10.1128/jb.00291-15
Hao, Y., Wang, Y., Bi, Z., Sun, B., Jin, Y., Bai, Y., et al. (2015). A case of non-O1/non-O139 Vibrio cholerae septicemia and meningitis in a neonate. Int. J. Infect. Dis. 35, 117–119. doi: 10.1016/j.ijid.2015.05.004
Hengge, R. (2009). Proteolysis of sigmaS (RpoS) and the general stress response in Escherichia coli. Res. Microbiol. 160, 667–676. doi: 10.1016/j.resmic.2009.08.014
Hengge-Aronis, R. (2002a). Signal transduction and regulatory mechanisms involved in control of the sigma(S) (RpoS) subunit of RNA polymerase. Microbiol. Mol. Biol. Rev. 66, 373–395. doi: 10.1128/mmbr.66.3.373-395.2002
Hengge-Aronis, R. (2002b). Stationary phase gene regulation: what makes an Escherichia coli promoter sigmaS-selective? Curr. Opin. Microbiol. 5, 591–595. doi: 10.1016/s1369-5274(02)00372-7
Huang, J., Chen, Y., Chen, J., Liu, C., Zhang, T., Luo, S., et al. (2019a). Exploration of the effects of a degS mutant on the growth of Vibrio cholerae and the global regulatory function of degS by RNA sequencing. PeerJ 7:e7959. doi: 10.7717/peerj.7959
Huang, Z., Pan, X., Xu, N., and Guo, M. (2019b). Bacterial chemotaxis coupling protein: structure, function and diversity. Microbiol. Res. 219, 40–48. doi: 10.1016/j.micres.2018.11.001
Kajikawa, A., Suzuki, S., and Igimi, S. (2018). The impact of motility on the localization of Lactobacillus agilis in the murine gastrointestinal tract. BMC Microbiol. 18:68. doi: 10.1186/s12866-018-1219-3
Kariisa, A. T., Grube, A., and Tamayo, R. (2015). Two nucleotide second messengers regulate the production of the Vibrio cholerae colonization factor GbpA. BMC Microbiol. 15:166. doi: 10.1186/s12866-015-0506-5
Kazmierczak, B. I., and Hendrixson, D. R. (2013). Spatial and numerical regulation of flagellar biosynthesis in polarly flagellated bacteria. Mol. Microbiol. 88, 655–663. doi: 10.1111/mmi.12221
Khan, F., Tabassum, N., Anand, R., and Kim, Y. M. (2020). Motility of Vibrio spp.: regulation and controlling strategies. Appl. Microbiol. Biotechnol. 104, 8187–8208. doi: 10.1007/s00253-020-10794-7
Kolb, A., Busby, S., Buc, H., Garges, S., and Adhya, S. (1993). Transcriptional regulation by cAMP and its receptor protein. Annu. Rev. Biochem. 62, 749–797. doi: 10.1146/annurev.bi.62.070193.003533
Kovacikova, G., and Skorupski, K. (2002). The alternative sigma factor sigma(E) plays an important role in intestinal survival and virulence in Vibrio cholerae. Infect. Immun. 70, 5355–5362. doi: 10.1128/iai.70.10.5355-5362.2002
Lee, Y. L., Hung, P. P., Tsai, C. A., Lin, Y. H., Liu, C. E., and Shi, Z. Y. (2007). Clinical characteristics of non-O1/non-O139 Vibrio cholerae isolates and polymerase chain reaction analysis of their virulence factors. J. Microbiol. Immunol. Infect. 40, 474–480. doi: 10.37757/mr2017.v19.n4.6
Lekshmi, N., Joseph, I., Ramamurthy, T., and Thomas, S. (2018). Changing facades of Vibrio cholerae: an enigma in the epidemiology of cholera. Indian J. Med. Res. 147, 133–141. doi: 10.4103/ijmr.IJMR_280_17
Li, X., Ren, F., Cai, G., Huang, P., Chai, Q., Gundogdu, O., et al. (2020a). Investigating the role of FlhF identifies novel interactions with genes involved in flagellar synthesis in Campylobacter jejuni. Front. Microbiol. 11:460. doi: 10.3389/fmicb.2020.00460
Li, X., Wu, Y., Sun, X., Ma, J., Li, X., Liu, C., et al. (2020b). Non-O1/non-O139 Vibrio cholerae bacteraemia in mainland China from 2005 to 2019: clinical, epidemiological and genetic characteristics. Epidemiol. Infect. 148:e186. doi: 10.1017/s0950268820001545
Liang, H., Zhang, Y., Wang, S., and Gao, H. (2021). Mutual interplay between ArcA and σ(E) orchestrates envelope stress response in Shewanella oneidensis. Environ. Microbiol. 23, 652–668. doi: 10.1111/1462-2920.15060
Liu, C., Sun, D., Zhu, J., Liu, J., and Liu, W. (2020a). The regulation of bacterial biofilm formation by cAMP-CRP: a mini-review. Front. Microbiol. 11:802. doi: 10.3389/fmicb.2020.00802
Liu, F., Wang, F., and Liu, J. (2020b). Characterization of chemotaxis and motility response towards fructose in Escherichia coli. Biochem. Biophys. Res. Commun. 527, 194–199. doi: 10.1016/j.bbrc.2020.04.087
Long, Z., Quaife, B., Salman, H., and Oltvai, Z. N. (2017). Cell-cell communication enhances bacterial chemotaxis toward external attractants. Sci. Rep. 7:12855. doi: 10.1038/s41598-017-13183-9
Luo, P., Su, T., Hu, C., and Ren, C. (2011). A novel and simple PCR walking method for rapid acquisition of long DNA sequence flanking a known site in microbial genome. Mol. Biotechnol. 47, 220–228. doi: 10.1007/s12033-010-9332-z
Manneh-Roussel, J., Haycocks, J. R. J., Magán, A., Perez-Soto, N., Voelz, K., Camilli, A., et al. (2018). cAMP receptor protein controls Vibrio cholerae gene expression in response to host colonization. MBio 9:e00966-18. doi: 10.1128/mBio.00966-18
Marinello, S., Marini, G., Parisi, G., Gottardello, L., Rossi, L., Besutti, V., et al. (2017). Vibrio cholerae non-O1, non-O139 bacteraemia associated with pneumonia, Italy 2016. Infection 45, 237–240. doi: 10.1007/s15010-016-0961-4
Martinez, R. M., Dharmasena, M. N., Kirn, T. J., and Taylor, R. K. (2009). Characterization of two outer membrane proteins, FlgO and FlgP, that influence Vibrio cholerae motility. J. Bacteriol. 191, 5669–5679. doi: 10.1128/jb.00632-09
Matilla, M. A., and Krell, T. (2018). The effect of bacterial chemotaxis on host infection and pathogenicity. FEMS Microbiol. Rev. 42, 40–67. doi: 10.1093/femsre/fux052
Missiakas, D., Mayer, M. P., Lemaire, M., Georgopoulos, C., and Raina, S. (1997). Modulation of the Escherichia coli sigmaE (RpoE) heat-shock transcription-factor activity by the RseA, RseB and RseC proteins. Mol. Microbiol. 24, 355–371. doi: 10.1046/j.1365-2958.1997.3601713.x
Nielsen, A. T., Dolganov, N. A., Otto, G., Miller, M. C., Wu, C. Y., and Schoolnik, G. K. (2006). RpoS controls the Vibrio cholerae mucosal escape response. PLoS Pathog. 2:e109. doi: 10.1371/journal.ppat.0020109
Partridge, J. D., Nhu, N. T. Q., Dufour, Y. S., and Harshey, R. M. (2019). Escherichia coli remodels the chemotaxis pathway for swarming. MBio 10:e00316-19. doi: 10.1128/mBio.00316-19
Prouty, M. G., Correa, N. E., and Klose, K. E. (2001). The novel sigma54- and sigma28-dependent flagellar gene transcription hierarchy of Vibrio cholerae. Mol. Microbiol. 39, 1595–1609. doi: 10.1046/j.1365-2958.2001.02348.x
Roggo, C., Picioreanu, C., Richard, X., Mazza, C., van Lintel, H., and van der Meer, J. R. (2018). Quantitative chemical biosensing by bacterial chemotaxis in microfluidic chips. Environ. Microbiol. 20, 241–258. doi: 10.1111/1462-2920.13982
Rui, H., Ritchie, J. M., Bronson, R. T., Mekalanos, J. J., Zhang, Y., and Waldor, M. K. (2010). Reactogenicity of live-attenuated Vibrio cholerae vaccines is dependent on flagellins. Proc. Natl. Acad. Sci. U. S. A. 107, 4359–4364. doi: 10.1073/pnas.0915164107
Sarkar, S., Hutton, M. L., Vagenas, D., Ruter, R., Schüller, S., Lyras, D., et al. (2018). Intestinal colonization traits of pandemic multidrug-resistant Escherichia coli ST131. J. Infect. Dis. 218, 979–990. doi: 10.1093/infdis/jiy031
Sohn, J., Grant, R. A., and Sauer, R. T. (2007). Allosteric activation of DegS, a stress sensor PDZ protease. Cells 131, 572–583. doi: 10.1016/j.cell.2007.08.044
Soutourina, O., Kolb, A., Krin, E., Laurent-Winter, C., Rimsky, S., Danchin, A., et al. (1999). Multiple control of flagellum biosynthesis in Escherichia coli: role of H-NS protein and the cyclic AMP-catabolite activator protein complex in transcription of the flhDC master operon. J. Bacteriol. 181, 7500–7508. doi: 10.1128/jb.181.24.7500-7508.1999
Spöring, I., Felgner, S., Preuße, M., Eckweiler, D., Rohde, M., Häussler, S., et al. (2018). Regulation of flagellum biosynthesis in response to cell envelope stress in Salmonella enterica Serovar typhimurium. MBio 9:e00736-17. doi: 10.1128/mBio.00736-17
Subramanian, S., and Kearns, D. B. (2019). Functional regulators of bacterial flagella. Annu. Rev. Microbiol. 73, 225–246. doi: 10.1146/annurev-micro-020518-115725
Syed, K. A., Beyhan, S., Correa, N., Queen, J., Liu, J., Peng, F., et al. (2009). The Vibrio cholerae flagellar regulatory hierarchy controls expression of virulence factors. J. Bacteriol. 191, 6555–6570. doi: 10.1128/jb.00949-09
Tamar, E., Koler, M., and Vaknin, A. (2016). The role of motility and chemotaxis in the bacterial colonization of protected surfaces. Sci. Rep. 6:19616. doi: 10.1038/srep19616
Walsh, N. P., Alba, B. M., Bose, B., Gross, C. A., and Sauer, R. T. (2003). OMP peptide signals initiate the envelope-stress response by activating DegS protease via relief of inhibition mediated by its PDZ domain. Cells 113, 61–71. doi: 10.1016/s0092-8674(03)00203-4
Wang, G., Fan, C., Wang, H., Jia, C., Li, X., Yang, J., et al. (2022). Type VI secretion system-associated FHA domain protein TagH regulates the hemolytic activity and virulence of Vibrio cholerae. Gut Microbes 14:2055440. doi: 10.1080/19490976.2022.2055440
Wang, J., Xing, X., Yang, X., Jung, I. J., Hao, G., Chen, Y., et al. (2018). Gluconeogenic growth of Vibrio cholerae is important for competing with host gut microbiota. J. Med. Microbiol. 67, 1628–1637. doi: 10.1099/jmm.0.000828
Wölflingseder, M., Tutz, S., Fengler, V. H., Schild, S., and Reidl, J. (2022). Regulatory interplay of RpoS and RssB controls motility and colonization in Vibrio cholerae. Int. J. Med. Microbiol. 312:151555. doi: 10.1016/j.ijmm.2022.151555
Wu, R., Zhao, M., Li, J., Gao, H., Kan, B., and Liang, W. (2015). Direct regulation of the natural competence regulator gene tfoX by cyclic AMP (cAMP) and cAMP receptor protein (CRP) in Vibrios. Sci. Rep. 5:14921. doi: 10.1038/srep14921
Wurm, P., Tutz, S., Mutsam, B., Vorkapic, D., Heyne, B., Grabner, C., et al. (2017). Stringent factor and proteolysis control of sigma factor RpoS expression in Vibrio cholerae. Int. J. Med. Microbiol. 307, 154–165. doi: 10.1016/j.ijmm.2017.01.006
Xie, H., Wu, Y., Liu, C., Guo, J., Ma, J., Li, X., et al. (2020). Oral infection caused by non-O1/non-O139 Vibrio cholerae in a patient with esophageal cancer undergoing esophagectomy and chemoradiotherapy: a case report. Infect. Drug Resist. 13, 3923–3927. doi: 10.2147/idr.S274077
Zhang, Y., Wang, L., Han, Y., Yan, Y., Tan, Y., Zhou, L., et al. (2013). Autoregulation of PhoP/PhoQ and positive regulation of the cyclic AMP receptor protein-cyclic AMP complex by PhoP in Yersinia pestis. J. Bacteriol. 195, 1022–1030. doi: 10.1128/jb.01530-12
Zhang, Q., Zhang, Y., Zhang, X., Zhan, L., Zhao, X., Xu, S., et al. (2015). The novel cis-encoded antisense RNA AsrC positively regulates the expression of rpoE-rseABC operon and thus enhances the motility of Salmonella enterica Serovar typhi. Front. Microbiol. 6:990. doi: 10.3389/fmicb.2015.00990
Keywords: Vibrio cholerae, DegS, motility, chemotaxis, colonization
Citation: Zou M, Wang K, Zhao J, Lu H, Yang H, Huang M, Wang L, Wang G, Huang J and Min X (2023) DegS protease regulates the motility, chemotaxis, and colonization of Vibrio cholerae. Front. Microbiol. 14:1159986. doi: 10.3389/fmicb.2023.1159986
Edited by:
Jeffrey H. Withey, Wayne State University, United StatesReviewed by:
Jyl S. Matson, University of Toledo, United StatesDhrubajyoti Nag, Wayne State University, United States
Copyright © 2023 Zou, Wang, Zhao, Lu, Yang, Huang, Wang, Wang, Huang and Min. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jian Huang, ODE1Mzc2NDhAcXEuY29t; Xun Min, bWlueHVuem11QDE2My5jb20=
†These authors have contributed equally to this work
 Mei Zou1,2†
Mei Zou1,2† Kaiying Wang
Kaiying Wang Jiajun Zhao
Jiajun Zhao Huifang Lu
Huifang Lu Jian Huang
Jian Huang