
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 31 March 2023
Sec. Infectious Agents and Disease
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1158086
This article is part of the Research Topic Insights in Infectious Agents and Disease: 2022 View all 41 articles
The anaerobic gut bacteria and opportunistic pathogen Bacteroides fragilis can cause life-threatening infections when leaving its niche and reaching body sites outside of the gut. The antimicrobial metronidazole is a mainstay in the treatment of anaerobic infections and also highly effective against Bacteroides spp. Although resistance rates have remained low in general, metronidazole resistance does occur in B. fragilis and can favor fatal disease outcomes. Most metronidazole-resistant Bacteroides isolates harbor nim genes, commonly believed to encode for nitroreductases which deactivate metronidazole. Recent research, however, suggests that the mode of resistance mediated by Nim proteins might be more complex than anticipated because they affect the cellular metabolism, e.g., by increasing the activity of pyruvate:ferredoxin oxidoreductase (PFOR). Moreover, although nim genes confer only low-level metronidazole resistance to Bacteroides, high-level resistance can be much easier induced in the laboratory in the presence of a nim gene than without. Due to these observations, we hypothesized that nim genes might induce changes in the B. fragilis proteome and performed comparative mass-spectrometric analyses with B. fragilis 638R, either with or without the nimA gene. Further, we compared protein expression profiles in both strains after induction of high-level metronidazole resistance. Interestingly, only few proteins were repeatedly found to be differentially expressed in strain 638R with the nimA gene, one of them being the flavodiiron protein FprA, an enzyme involved in oxygen scavenging. After induction of metronidazole resistance, a far higher number of proteins were found to be differentially expressed in 638R without nimA than in 638R with nimA. In the former, factors for the import of hemin were strongly downregulated, indicating impaired iron import, whereas in the latter, the observed changes were not only less numerous but also less specific. Both resistant strains, however, displayed a reduced capability of scavenging oxygen. Susceptibility to metronidazole could be widely restored in resistant 638R without nimA by supplementing growth media with ferrous iron sulfate, but not so in resistant 638R with the nimA gene. Finally, based on the results of this study, we present a novel hypothetic model of metronidazole resistance and NimA function.
The genus Bacteroides comprises several of the most common anaerobic bacteria in the human intestine and accounts for 30% of the human fecal isolates (Kuwahara et al., 2004). Their role is predominantly beneficial as they break down carbohydrates which would otherwise be inaccessible to the human host. Thereby they produce volatile short-chain fatty acids such as succinic acid which are absorbed in the gut (Wexler, 2007). Bacteroides spp., however can also cause severe disease when the integrity of the intestinal wall is compromised, e.g., through cancer or injuries, and they can leave their intestinal niche to cause abscesses in other body sites or bacteremia (Aldridge and Sanders, 2002; Wexler, 2007). This is especially true for Bacteroides fragilis which accounts only for 1% of the intestinal microbiome but for as much 30%–60% of all anaerobic clinical isolates in total (Wexler, 2007; Tan et al., 2017). Importantly, B. fragilis secretes fragilysin, a metalloprotease which digests E-cadherin and can disrupt the tight junctions of the intestinal epithelium (Remacle et al., 2014).
Bacteroides infections are commonly treated with carbapenems and metronidazole. Metronidazole is a 5-nitroimidazole drug specifically targeting anaerobic and microaerophilic microbes (reviewed in Leitsch, 2019). The specificity of metronidazole for anaerobes and microaerophiles is due to a requirement for the reduction of the nitro group, which only takes place quantitatively in low oxygen environments. The resulting reactive intermediates damage the cell through forming adducts with proteins, inducing DNA strand breaks and by depleting cellular redox buffers (Leitsch, 2019). Arguably, either the nitroimidazole anion, resulting from a single electron transfer to the nitro group, or the nitroso radical, resulting from a double electron transfer to the nitro group, are causing most of the damage. In contrast to many other antibiotics, metronidazole has retained usability due to overall low resistance rates, especially in true anaerobes such as Bacteroides spp. (Snydman et al., 2017). However, metronidazole resistance in Bacteroides spp. does occur and can be found in a sizeable proportion of isolates in certain areas (Vieira et al., 2006; Yehya et al., 2014; Sheikh et al., 2015).
Metronidazole resistance in the field is mainly but not exclusively observed in isolates carrying nim genes. To date, 12 different homologs of nim were detected in Bacteroidetes (nimA-nimL), all of which are believed to encode nitroreductases capable of transferring six electrons to the nitro group of metronidazole, resulting in a non-toxic amine (reviewed in Alauzet et al., 2019). Although there are data supporting this hypothesis (Carlier et al., 1997), a direct mode of Nim proteins as nitroreductases has been questioned by other findings (Leitsch et al., 2014; Paunkov et al., 2022a). Importantly, Nim expression levels are widely independent of the degree of metronidazole resistance (Leitsch et al., 2014; Paunkov et al., 2022a) although the proposed role of Nim proteins as nitroreductases would logically imply a positive correlation of expression levels and the degree of metronidazole resistance. Such positive correlation, however, could be only observed in the initial stage of resistance as conferred by nim genes (Kupc et al., 2022), but not after adaptation to higher, medically relevant, doses of the drug (4 μg mL−1 according to EUCAST, see https://www.eucast.org/clinical_breakpoints). Furthermore, nim genes are positively correlated to a greatly increased activity of pyruvate:ferredoxin oxidoreductase (PFOR; Paunkov et al., 2022a), a central metabolic enzyme in most anaerobes, including B. fragilis (Narikawa, 1986). This is surprising given that PFOR is commonly believed to be a major metronidazole-reducing enzyme (Narikawa, 1986) and therefore a source of reactive metronidazole intermediates. After induction of high-level metronidazole resistance in the laboratory, i.e., adaptation to high concentrations of metronidazole, PFOR activity is retained in B. fragilis 638R with the nimA gene, but lost without it (Paunkov et al., 2022a). Moreover, B. fragilis 638R carrying the nimA gene also retains good tolerance to oxygen after induction of high-level metronidazole resistance, whereas as metronidazole-resistant 638R without the nimA gene becomes highly sensitive to oxygen (Paunkov et al., 2022a).
These observations suggest that the development of high-level metronidazole resistance is strongly facilitated in the presence of the nimA gene and might be caused by a different mechanism than observed in strains without the nimA gene. In order to address this question and to identify differentially expressed proteins we analyzed and compared the proteomes of B. fragilis 638R without a nim gene and of B. fragilis 638R harboring nimA by quantitative mass spectrometry before and after induction of high-level metronidazole resistance.
Vitamin K1, brain-heart-infusion broth (BHI), and agar were purchased from Carl Roth (Karlsruhe, Germany). Wilkins-Chalgren anaerobe agar (WC) was purchased from Oxoid (Basingstoke, England). Hemin was purchased from Sigma-Aldrich (St. Luis, United States) and Anaerocult A was purchased from Merck (Darmstadt, Germany). Metronidazole was purchased from Sigma-Aldrich (St. Luis, United States), and Etests were purchased from bioMérieux (Marcy-l’Étoile, France).
The experiments were performed with B. fragilis 638R (division I, cfiA-) and a transconjugant daughter strain with plasmid pI417 harboring the nimA gene (Breuil et al., 1989). The MICs for metronidazole according to Etests were: 638R, 0.25 μg mL−1 (Paunkov et al., 2022b); and 638R nimA, 1.5 μg μg mL−1 (Paunkov et al., 2022b). Importantly, the breakpoint concentration for metronidazole resistance amounts to 4 μg mL−1 according to EUCAST, 2023. The nimA gene is positioned behind insertion element IS1168 (Haggoud et al., 1994) whose sequence can be accessed via GenBank number X71444. Both strains were obtained from the strain repository of the Institute of Medical Microbiology in Szeged, Hungary. The genome sequence of 638R can be accessed in the NCBI database via accession number FQ312004. Induction of high-level resistance was performed as described previously (Paunkov et al., 2022a) by passaging 638R and 638R nimA with increasing metronidazole concentrations in the agar (factor 2 with every passage). The induction process was terminated after having achieved growth at a metronidazole concentration of 64 μg mL−1. The four strains, i.e., 638R and its three derivatives, are designated as 638R, 638RR, 638R nimA, and 638R nimAR, respectively, throughout the manuscript, “R” indicating “resistant.”
Cells were grown on either WC agar plates or BHI agar plates with hemin and vitamin K1 supplementation. When cultures were prepared for mass spectrometry, cells were grown anaerobically at 37°C in BHI broth (Carl Roth, Germany) supplemented with 1 μg mL−1 vitamin K1 and 5 μg mL−1 hemin (BHI-S) inside 2.5 L anaerobic jars (Merck, Darmstadt, Germany) conditioned with the Anaerocult A system (0% O2 and 18% CO2).
RT-qPCR was performed exactly as described recently (Paunkov et al., 2022a) with rpoD and gapdh mRNA as internal standards for normalization. The primers for the amplification of a 176 nt fragment within the flavodiiron protein A (fprA) cDNA were 5′-CAATGTCAGCAAAAGTAATCCC-3′ (forward) and 5′-GTGAACGAACCGAAATACCC-3′ (reverse).
PCR amplification of fprA gene was performed using forward 5′-GAG GAT CCA TGG AAC AGA AAA CAA GAA TTA AAG G-3′ and reverse 5′-GAG GTA CCT TAT ACT CTG TCT TTT TTT AAA CGG C-3′ primers containing BamHI and KpnI restriction sites, respectively. The gene was inserted into the pFD340 vector with the same restriction sites. The resulting pFDfprA plasmid was introduced into E. coli 10-beta cells using the manufacturer’s instructions. Transformed cells were selected on LB plates with 100 μg ml−1 ampicillin. An established tri-parental filter mating protocol (Mays et al., 1982), using E. coli RK231 as a helper strain, was applied to transfer plasmids from E. coli 10-beta to B. fragilis 638R. Briefly, E. coli 10-beta and E. coli RK231 cells were grown on LB plates with 100 μg mL−1 ampicillin and 150 μg mL−1 streptomycin and spectinomycin, respectively. Bacteroides fragilis 638R cells were grown on BHI-S plates with 20 μg mL−1 rifampicin. A single colony of each strain was grown with antibiotics overnight at 37°C then diluted 1:100 (E. coli strains) or 1:40 (B. fragilis 638R) and further grown in antibiotic-free medium until an OD600 of 0.5 was reached. 1 mL of B. fragilis 638R and 0.25 mL of each E. coli culture were united in sterile centrifuge tubes, and centrifuged for 30 s at maximum speed. The resulting pellet was resuspended in 100 μL of BHIS medium and transferred onto sterile filters placed on mating plates which had been previously incubated overnight inside an anaerobic chamber at 37°C. Plates with filters carrying a mixture of all three strains were then incubated overnight at 37°C aerobically. Cells were resuspended by vortexing the filters for 1 min inside 15 mL falcon tubes in 1 ml of BHIS medium. Conjugants were grown and selected on BHIS plates with 10 μg mL−1 erythromycin, 20 μg mL−1 rifampicin, and 100 μg mL−1 gentamycin after 48 h of anaerobic incubation at 37°C.
2 × 109 cells were harvested from an overnight culture (3,000 × g for 5 min) and suspended in 14 mL of BHI medium which had previously been saturated with oxygen through vigorous shaking. Cells were then transferred into a 15 ml glass tube and an oxygen microsensor (OX-500, Unisense, Denmark) of an oximeter (OXY-Meter, Unisense, Denmark) was inserted. In order to prevent inflow of air, the glass tube was sealed with Parafilm M. Oxygen concentrations were measured for 60 min with automatic recording of values every 30 s. Oxygen removal by the components of the medium was also measured in the same setup in the absence of cells. These background values were subtracted from the values obtained in the presence of cells.
Susceptibility tests were performed by picking colonies with a sterile swab and resuspending them in 1× PBS before application on BHI agar plates (either with or without supplementation with ferrous iron sulfate). Plates were left for 15 min to dry and Etests were applied. Plates were transferred to anaerobic jars and incubated for 48 h at 37°C. The MICs were read at the intersection of the border of growth inhibition and the test strip. Etest images were obtained using BioRad GelDoc XR.
For protein isolation, 4 × 109 cells from an overnight culture grown in BHI-S broth were harvested by centrifugation at 3,000 × g for 10 min. After centrifugation, the medium was removed and cells were washed with 1 ml ddH2O and centrifuged once again at 3,000 × g for 10 min. The supernatant was removed and cells were resuspended in 120 μL denaturing protein sample buffer [7 M urea, 2 M thiourea, 1% DTT, 1% Biolyte® ampholytes (BioRad), 4% CHAPS] and 1 μL of Turbonuclease® from Serratia marcescens (Sigma-Aldrich) was added for the removal of genomic DNA in the samples. Cells were then sonicated at 4°C (10 cycles 30 s pulse on, and 30 s pulse off) and centrifuged for 20 min at 4°C and 20,000 × g. The resulting supernatant was transferred into fresh tubes, and protein concentrations were determined using Bradford assay. For protein digestion, sample extracts were prepared with a filter-aided sample preparation protocol based on Wisniewski et al. (2009) and Wisniewski (2016). In brief, 30 μg protein solved in lysis buffer was filled up to 500 μL with 8 M Urea in 50 mM Tris buffer (both Roth, Karlsruhe, Germany) and loaded onto an Pall 10 kDa ultrafiltration device (Pall Corporation, Port Washington, NY, United States). After centrifugation (20 min at 10,000 × g), cysteine residues were reduced with 200 mM DTT (37°C, 30 min) and alkylated with 500 mM iodoacetamide (37°C, 30 min) on the filter (both Sigma-Aldrich, St. Louis, MO, United States). This was followed by two washes with 100 μL 50 mM Tris before digestion with trypsin/LysC Mix for 14 h (Promega Technical Manual, Madison, WI, United States). Digested peptides were recovered with 3 times 50 μL of 50 mM Tris buffer. Before LC-MS analysis, acidified peptide extracts were purified using C18 spin columns (Pierce, Thermo Fisher Scientific) according to the manufacturer’s protocol. Dried peptides were redissolved in 300 μL 0.1% trifluoroacetic acid (Thermo Fisher Scientific, Waltham, MA, United States) and 300 ng digested protein was injected into the nanoRSLC-nESI-QExactive-Orbitrap HF MS/MS system (Thermo Fisher Scientific). Mass spectrometry analysis was performed as described in Gutiérrez et al. (2019).
For the label-free quantitative shotgun proteomic approach, mass spectrometry analysis was done in two technical replicates per biological replicate, i.e., six replicates in total per isolate. Evaluation of raw data was accomplished with the software Proteome Discoverer version 2.4.1.15. (Thermo Fisher Scientific). A combination of the Bacteroides fragilis database downloaded from RefSeq (taxonomy 817, https://www.ncbi.nlm.nih.gov/refseq/, accessed on 24 October 2022) and a common contaminant database (https://www.thegpm.org/crap/, accessed on 25 June 2019) was used. Furthermore, the sequence of the nitroimidazole resistance protein NimA (WP_032488596.1 from Phocaeicola vulgatus) was added. Search parameters were applied as follows: trypsin as an enzyme; maximally 2 missed cleavages; 10 ppm precursor mass tolerance and 0.02 Da fragment mass tolerance; dynamic modifications allowed were oxidation/+15.995 Da (M)/deamidation/+0.984 Da (N, Q)/Gln->pyro-Glu/−17.027 Da (Q)/acetylation/+42.011 Da (N-Terminus)/methionine loss/−131.040 Da (M) and static modification carbamidomethylation/+57.021 Da (C).
For intensity-based label-free quantification (LFQ), protein abundance raw values were generated in Proteome Discoverer software including normalization to total area sums. Further ANOVA analysis was performed in R version 4.0.5 (R Core Team, 2021). Prior to data import into R, protein abundances of technical replicates were aggregated by the mean. Furthermore, proteins with at least one missing value per group were excluded from the quantification analysis, in order to keep so-called “ON/OFF” proteins, but maintain high data quality. Statistical analysis by ANOVA depending on the number of groups was performed in R Studio. Proteins recognized with more than two tryptic peptides and displaying a fold change higher/lower than +/−2-fold with an adjusted value of p for controlling the false discovery rate according to Benjamini-Hochberg lower than 0.05, were considered to be upregulated/downregulated.
Statistical tests were performed with GraphPad Prism 9 software. More details are given in the respective table and figure legends.
In a previous study, we had shown that strain B. fragilis 638R displayed elevated PFOR activity (Paunkov et al., 2022a) when the nimA gene was present. The nimA gene confers reduced susceptibility to metronidazole, with an MIC of approximately 1.5 μg mL−1 as compared to an MIC of 0.25 μg mL−1 in 638R wildtype (Paunkov et al., 2022b). We had also observed that the development of high-level metronidazole resistance (≥ 64 μg μg mL−1) proceeds much faster and does not entail grave physiological impairments such as the loss of PFOR activity or high sensitivity to oxygen when nimA was present (Paunkov et al., 2022a). Hence, we speculated that high-level metronidazole resistance was caused by different mechanisms in 638R wildtype (henceforth designated as 638R) and in 638R with a nimA gene (henceforth designated as 638R nimA), and that this would be mirrored by differential expression of relevant proteins. In order to address this hypothesis, we subjected cell extracts of strain 638R and of 638R nimA to proteomic analyses using mass spectrometry. The analysis was performed twice at two different time points in order to ensure reproducibility of the results. Further, also highly metronidazole-resistant derivatives of 638R (henceforth designated as 638RR) and of 638R nimA (henceforth designated as 638R nimAR), both resistant to at least 64 μg mL−1 of metronidazole, were analyzed by mass spectrometry and compared to the original strains. The highly metronidazole-resistant cell lines had been generated in a recent study (Paunkov et al., 2022a) by growing cells on increasing concentrations of metronidazole with each subculture. The mass spectrometric data obtained were deposited at the PRIDE database (Perez-Riverol et al., 2022).
First, it was assessed if the presence of nimA induces changes in the protein expression profile as compared to the 638R wildtype. Protein numbers in all samples (biological triplicates in technical duplicates) ranged from about 1,000 to 1,500. Only changes in expression of at least twofold and at a significance level of p < 0.01 were considered. Further, only proteins of which peptides were found in all samples were considered. The first analysis with 638R nimA gave 24 differentially expressed proteins as compared to 638R (Table 1; Supplementary Table 1), whereas the second analysis gave 46 differentially expressed proteins (Table 1; Supplementary Table 2). However, although the number of differentially expressed proteins was similar, only five proteins were shared in both data sets (Table 2). Of these, only one protein was downregulated, i.e., exo-alpha-sialidase (WP_005794995), the other four were upregulated. One of the upregulated proteins was galactokinase (8.6-fold upregulated in the first run, and 12.5-fold in the second) and another was type A flavoprotein (FprA; WP_005785280), a flavodiiron protein commonly found in bacteria (Wasserfallen et al., 1998) which reduces either molecular oxygen to water or nitric oxide (NO) to nitrous oxide (N2O; Martins et al., 2019). FprA was upregulated 3.7-fold in the first run, and 12.9-fold in the second.
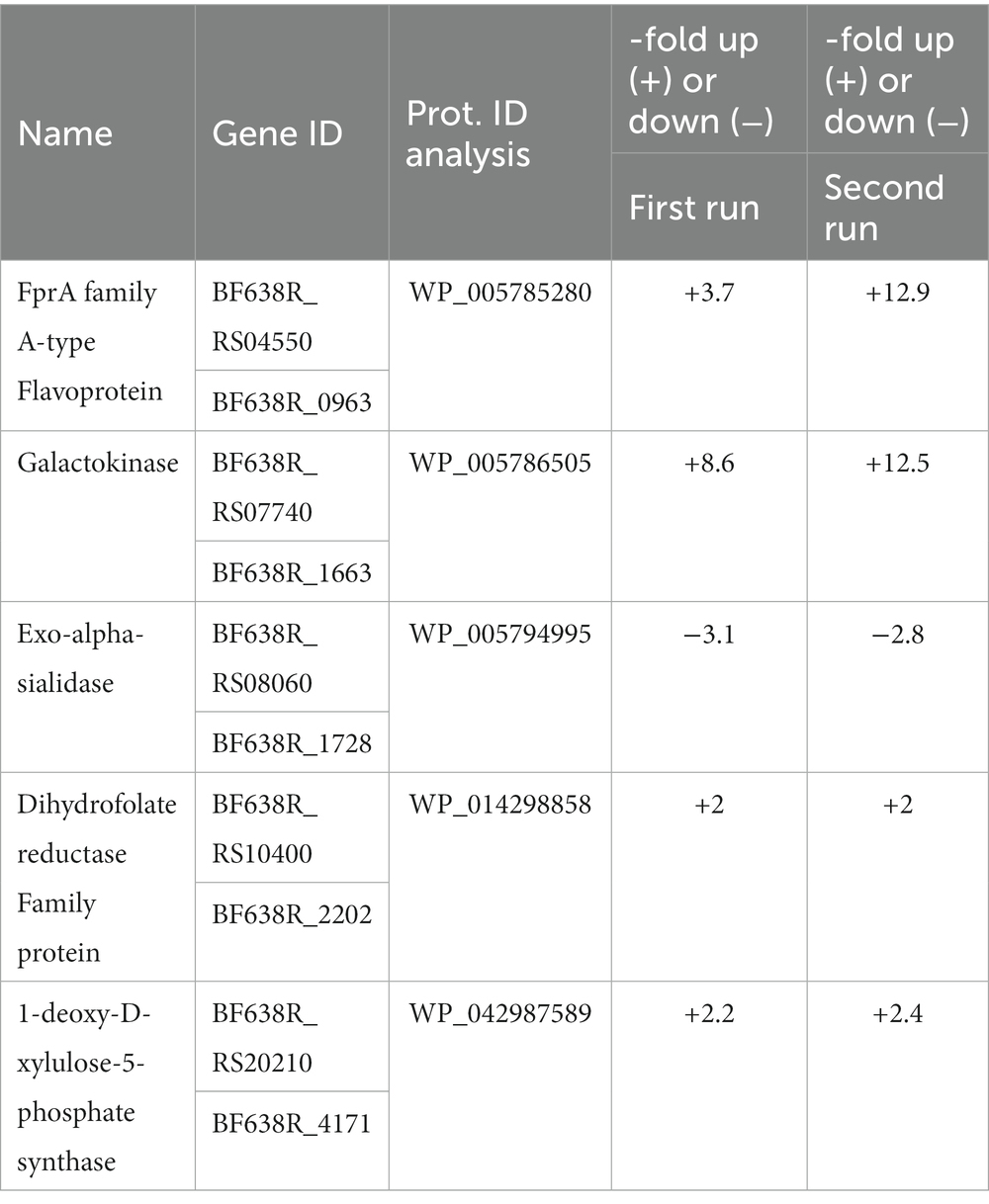
Table 2. Proteins consistently upregulated or downregulated in 638R nimA vs. 638R in two independent mass-spectrometric analyses.
Next, we analyzed the proteomes of 638RR and 638R nimAR and compared them with their respective non-resistant parent strains. Applying the same selection criteria as stated above, 41 proteins were found to be differentially expressed in 638R nimAR as compared to the non-resistant parent 638R nimA (Table 1; Supplementary Table 3). Only eight of the 41 proteins were upregulated in expression, seven of which are obviously organized in an operon for nucleotide sugar synthesis (WP_005801381, WP_032600526, WP_100766060, WP_005784923, WP_005784927, WP_005784941, and WP_014298264). Downregulated enzymes included aminotransferases, N-acetyl transferases, enzymes involved in capsule synthesis (Zhang et al., 2003; WP_005817136, WP_014298686), and numerous other factors whose correlation with metronidazole resistance remained unclear. None of the five proteins found to be differentially upregulated in 638R nimA as compared to 638R were further upregulated or downregulated, respectively, in 638R nimAR.
In 638RR, a comparably large number of proteins, i.e., 237, were found to be differentially expressed (Table 1; Supplementary Table 4) as compared to wildtype 638R. The levels of most differentially expressed proteins were increased (189) rather than decreased (48), and a very large proportion of the proteins, i.e., at least 84 of 237, are likely or known to be surface-associated, 41 being transporters or pores (Figure 1; Supplementary Table 5). When comparing 638RR to 638R nimA, three of the five proteins previously identified to be consistently differentially expressed in 638R nimA as compared to 638R were also found to be differentially expressed in 638RR (Supplementary Table 4). Two of these, however, exo-alpha-sialidase and 1-deoxy-D-xylulose-5-phosphate, were inversely expressed in the two strains, i.e., upregulated in one and downregulated in the other. Only FprA was upregulated in 638R nimA and 638RR alike. When comparing 638RR to 638R nimAR, 19 proteins were found differentially expressed in both strains (Supplementary Table 4). Eleven of these proteins, however, were upregulated in one strain but downregulated in the other, suggesting that they are not inherently related to metronidazole resistance. Of the other eight proteins, all were consistently downregulated in both strains. Five of these are putatively involved in carbohydrate metabolism (WP_005801178, WP_005795861, WP_008768174, WP_005794845, and WP_005817136), one is a hypothetical protein (WP_032542280), another an acetyl transferase (WP_032531450), and finally, HemY (WP_005791059) is a protoporphyrinogen oxidase which is tentatively involved in heme synthesis.
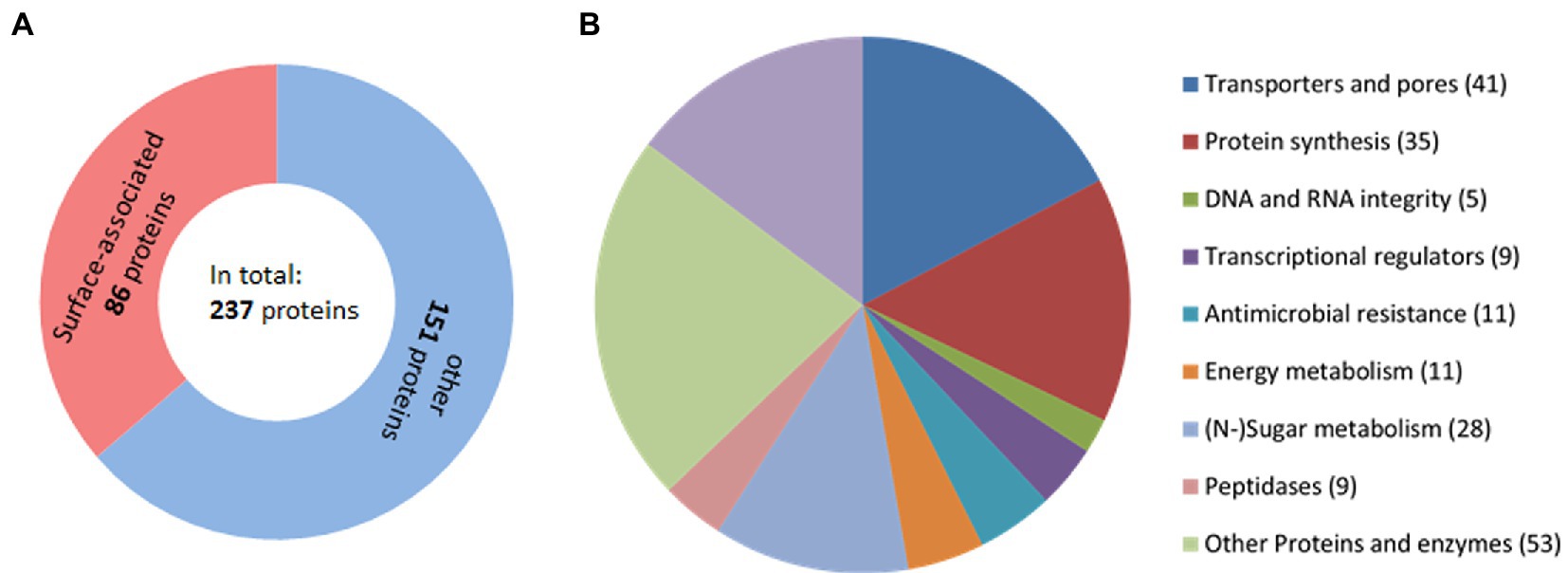
Figure 1. Proteins identified to be differentially expressed in Bacteroides fragilis 638RR as compared to 638R. (A) Total number of proteins differentially expressed as compared to wildtype 638R and their cellular localization. (B) Pie chart indicating the functions of the proteins differentially expressed in 638RR.
Importantly, several proteins involved in the import or synthesis of heme were found differentially expressed in 638RR as compared to 638R (Table 3; Supplementary Tables 4, 5). Apart from HemY, two homologs of heme-binding protein HmuY (WP_032556399 and WP_010993105), their cognate TonB-dependent receptors (WP_032564333 and WP_005788361), and a ChaN-like lipoprotein, presumably involved in heme uptake (Chan et al., 2006), were all strongly downregulated. One of the HmuY homologs, WP_032556399, had also recently been found downregulated at the mRNA level in 638RR (Paunkov et al., 2022a). In contrast, several enzymes dependent on heme or iron were upregulated in 638RR (Table 3; Supplementary Tables 4, 5): catalase, cytochrome ubiquinol oxidase subunit I, and fumarate reductase cytochrome b subunit. Further, also NADH:ubiquinone reductase subunits B, C, and F were upregulated. NADH:ubiquinone reductase feeds electrons into the electron transport chain which are used downstream by fumarate reductase and cytochrome ubiquinol oxidase in the reduction of fumarate or oxygen, respectively (Baughn and Malamy, 2004; Franke and Deppenmeier, 2018; Butler et al., 2022). In addition to fumarate reductase, also three other metabolic enzymes, i.e., pyruvate phosphate dikinase (PPDK), acetyl-CoA hydrolase, and butyrate kinase, were upregulated (Table 3). Pyruvate formate-lyase (PFL), an important metabolic enzyme which breaks down pyruvate to acetyl-CoA and formate, however, was strongly downregulated (Table 3).
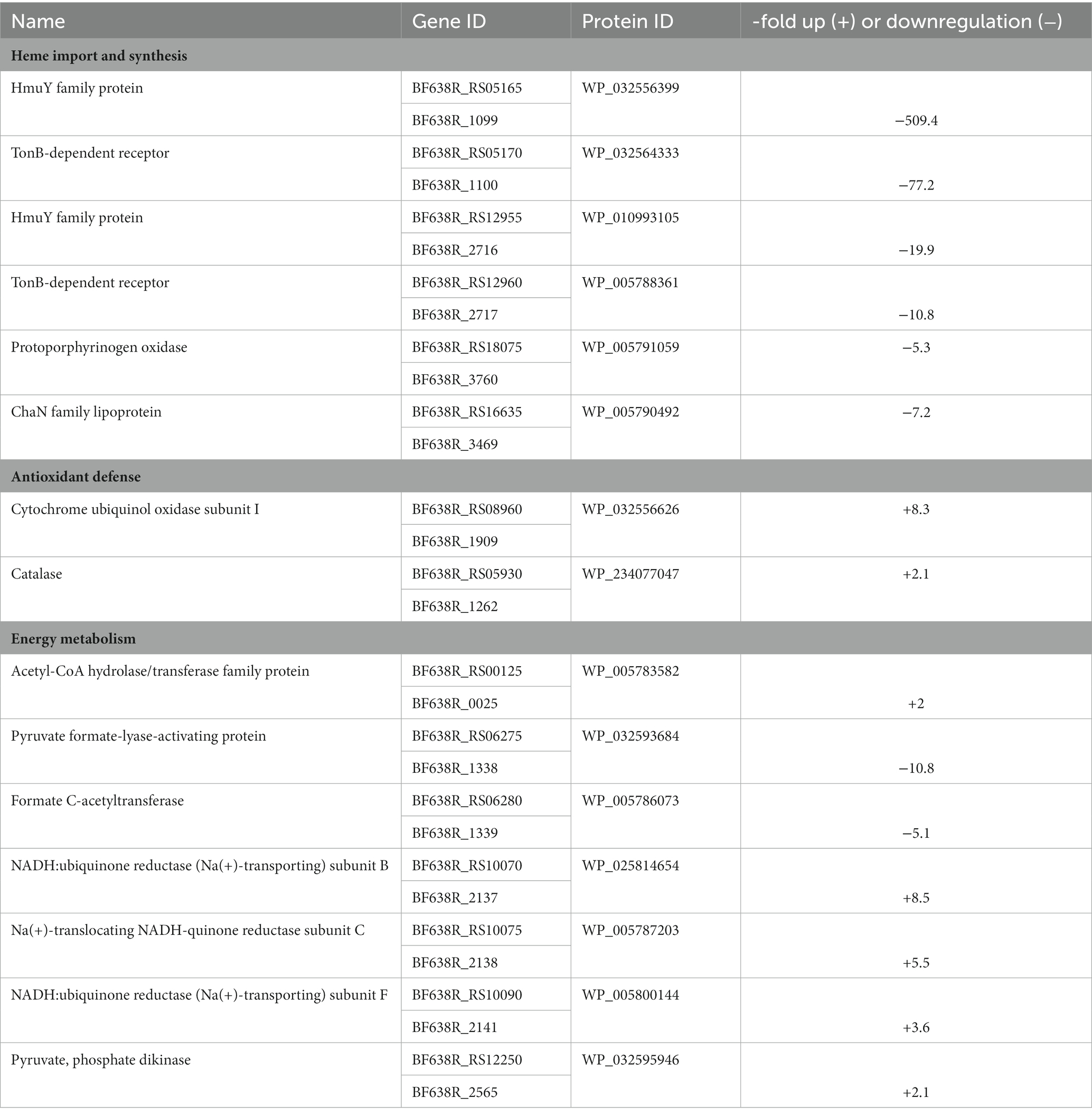
Table 3. Proteins involved in heme import, antioxidant defense, and energy metabolism differentially expressed in 638RR vs. 638R.
Thirty-five proteins involved in protein synthesis, i.e., 31 ribosomal proteins, translation initiation factor IF-3, RluA family pseudouridine synthase, elongation factor G, and pseudouridine synthase were upregulated (Supplementary Tables 4, 5), while none were downregulated in 638RR. Further, a subset of 11 surface-associated proteins which are assumed or known to be involved in antimicrobial resistance were upregulated (Table 4; Supplementary Tables 4, 5): an ABC transporter ATP-binding protein, an aminoglycoside phosphotransferase, an MBL fold metallo-hydrolase, a TolC protein, and nine components of RND (Resistance-Nodulation-Division) family transporters. The latter are of special significance as they had been reported before to be associated with metronidazole resistance (Pumbwe et al., 2006, 2007). Indeed, eight of the nine could be matched in the NCBI database to BmeA2 and BmeB2, BmeA5 and BmeB5, BmeA13 and BmeB13, and BmeA15 and BmeB15. Of these, especially BmeA5 and BmeB5 have been implied to be involved in metronidazole resistance as part of the BmeRABC5 multidrug efflux system (Pumbwe et al., 2007), but also BmeB2 had been found upregulated in B. fragilis strains after selection for metronidazole resistance (Pumbwe et al., 2006).
Finally, a large number of transporters, channels, peptidases, nucleases, transcriptional regulators, and enzymes involved in carbohydrate metabolism were differentially expressed in 638RR as compared to 638R (Supplementary Tables 4, 5).
The flavodiiron protein FprA was found to be more strongly expressed in 638R nimA and in 638RR as compared to 638R (Supplementary Tables 1, 2, 4). Flavodiiron proteins are a class of enzymes often found in anaerobes (Martins et al., 2019) and are important factors in the antioxidant and/or antinitrosative defense by reducing oxygen to water and/or nitric oxide (NO) to nitrous oxide (N2O), respectively. In order to test if overexpression of FprA conferred enhanced tolerance to metronidazole, we cloned the fprA gene into the pFD340 shuttle vector (Smith et al., 1992) and introduced the resulting plasmid pFDfprA into 638R by tri-parental mating. Subsequent analysis by RT-qPCR showed that the level of fprA was approximately 3-fold higher in the pFDfprA transconjugant than in 638R wildtype (Figure 2). Despite this elevated level of fprA mRNA, however, 638R pFDfprA displayed an even lower MIC of metronidazole as determined by Etest (Supplementary Figure 1) than the 638R parent, i.e., 0.125 μg mL−1 as compared to 0.25 μg mL−1 (Paunkov et al., 2022b). We asked if a 3-fold overexpression of fprA mRNA would be insufficient to obtain a measurable effect on the MLC of metronidazole and if fprA mRNA levels would be even higher in 638R nimA and 638RR. The levels of fprA mRNA in both strains, however, proved to be unchanged as compared to 638R wildtype (Figure 2), indicating that regulation of FprA expression occurs at the translational rather than the transcriptional level.
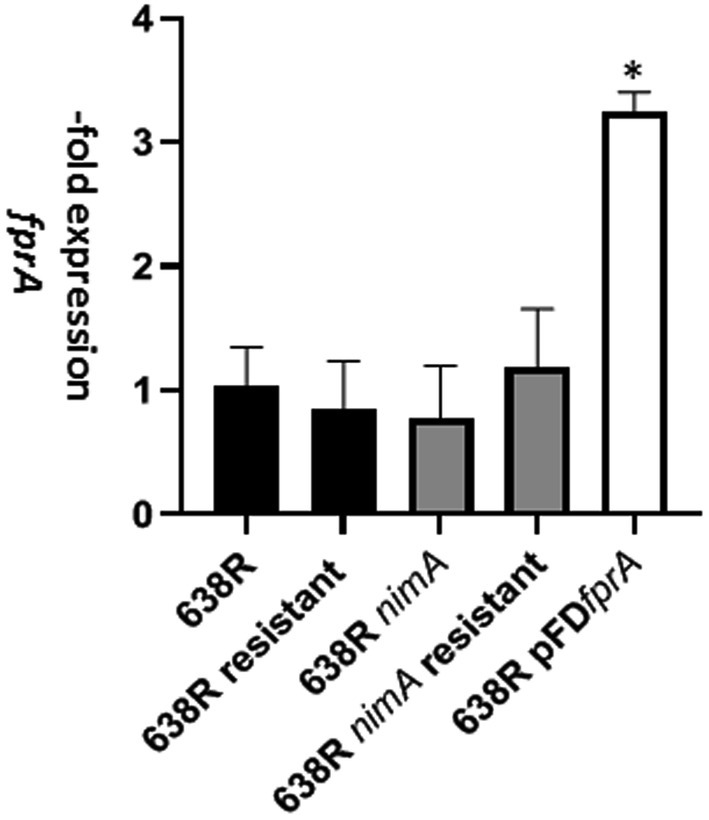
Figure 2. Levels of fprA mRNA as determined by RT-qPCR in 638R, 638RR, 638R nimA, 638R nimAR, and 638R pFDfprA which carries an additional copy of the fprA gene on shuttle plasmid pFD340 under transcriptional control of insertion element 4351 (IS4351). Measurements were performed in three independent experiments each. The asterisk indicates 638R pFDfprA > 638R with p < 0.001 according to Brown-Forsythe and Welch ANOVA tests with a post-hoc two stage linear step-up procedure of Benjamini, Krieger, and Yekutieli.
FprA had already been previously described in B. fragilis (Meehan et al., 2012) and in Bacteroides thetaiotaomicron (Mishra and Imlay, 2013), albeit under different names, i.e., Oxe and Roo, respectively. In B. thetaiotaomicron, the deletion of the roo gene strongly reduces oxygen scavenging capability (Lu and Imlay, 2017), suggesting that also B. fragilis FprA acts as an oxygen reducing enzyme. As such, it has the same function as cytochrome ubiquinol oxidase, also commonly termed cytochrome bd oxidase (Baughn and Malamy, 2004). The latter, however, is membrane-localized and harnesses electrons from the quinone pool, whereas flavodiiron proteins are cytoplasmic and derive electrons from reductants such as rubredoxin (Martins et al., 2019). Ultimately, both oxygen scavenging enzymes depend on NAD(P)H oxidation and have similar turnovers. In a previous study, we found that oxygen scavenging capacity in 638R nimA is slightly but significantly enhanced as compared to 638R (see Supplementary Figure 1 in Paunkov et al., 2022b) which is in accordance with the higher levels of FprA in the former (Table 2). We asked how oxygen scavenging rates would develop in the course of development of metronidazole resistance and measured oxygen scavenging rates in 638RR and 638R nimAR. These proved to be approximately halved in both strains and were not significantly different from each other (Table 5). At least for 638R nimAR, this result suggests that either less reductant, i.e., NADH or NADPH, is available for oxygen scavenging or that cytochrome ubiquinol oxidase is less active. An impairment of FprA-mediated oxygen scavenging can be ruled out because the FprA expression level is equal to that in 638R nimA and good supply with iron is indicated by the retention of high PFOR activity (Paunkov et al., 2022a). In the case of 638RR, the data do not allow a firm conclusion to be drawn because low intracellular iron levels brought about by reduced import of hemin, and therefore also of iron, can negatively affect both, FprA and cytochrome ubiquinol oxidase. It is important to note, however, that oxygen scavenging is totally abrogated when growth media are not supplemented with hemin (Paunkov et al., 2022b). Thus, oxygen scavenging is specifically retained in 638RR, albeit at a slower rate, whereas other enzyme activities such as PFOR are also lost as in hemin-deprived 638R (Paunkov et al., 2022a).
Based on the results of the proteomic analysis of 638RR, we argued that metronidazole resistance could be caused by three different strategies: (1) inactivation of enzymes which depend on iron and/or heme for the reduction of metronidazole by depleting intracellular iron pools; (2) upregulation of RND transporters which pump metronidazole out of the cell; and (3) overexpression of ribosomal proteins and other proteins involved in protein synthesis which increases the tolerance to metronidazole by enabling the quick replacement of proteins damaged by reactive metronidazole intermediates (Leitsch, 2019). In order to assess the relative contribution of these three potential strategies, we exposed 638RR to an iron source alternative to hemin, i.e., 100 μM ferrous iron sulfate, for one subculture in order to replenish the cells with iron. Ferrous iron sulfate had been described before to abrogate metronidazole resistance in a feoAB mutant (Veeranagouda et al., 2014) which is defective in importing ferrous iron released from heme (Rocha et al., 2019). After exposure to ferrous iron sulfate for one subculture, MICs to metronidazole were determined by Etest, likewise on plates with added 100 μM ferrous sulfate. Importantly, 638RR did not show any inhibition by metronidazole according to Etest when plates were not supplemented with ferrous iron (Figure 3). However, when grown with ferrous iron sulfate, 638RR displayed an MIC of 4 μg mL−1 (Figure 3A). This result indicates that the depletion of intracellular iron levels is the major strategy to achieve high-level metronidazole resistance in 638R and that other changes observed in the proteome of 638RR confer protection against metronidazole to a smaller degree only. In contrast, the addition of 100 μM ferrous iron sulfate did only slightly if at all reduce the level of resistance in 638R nimAR (Figure 3B). Further, the addition of ferrous iron sulfate had practically no effect on the susceptibility to metronidazole in 638R (Figure 3A). In 638R nimA, the addition of 100 μM ferrous iron even enhanced protection against metronidazole (Figure 3B) about twofold.
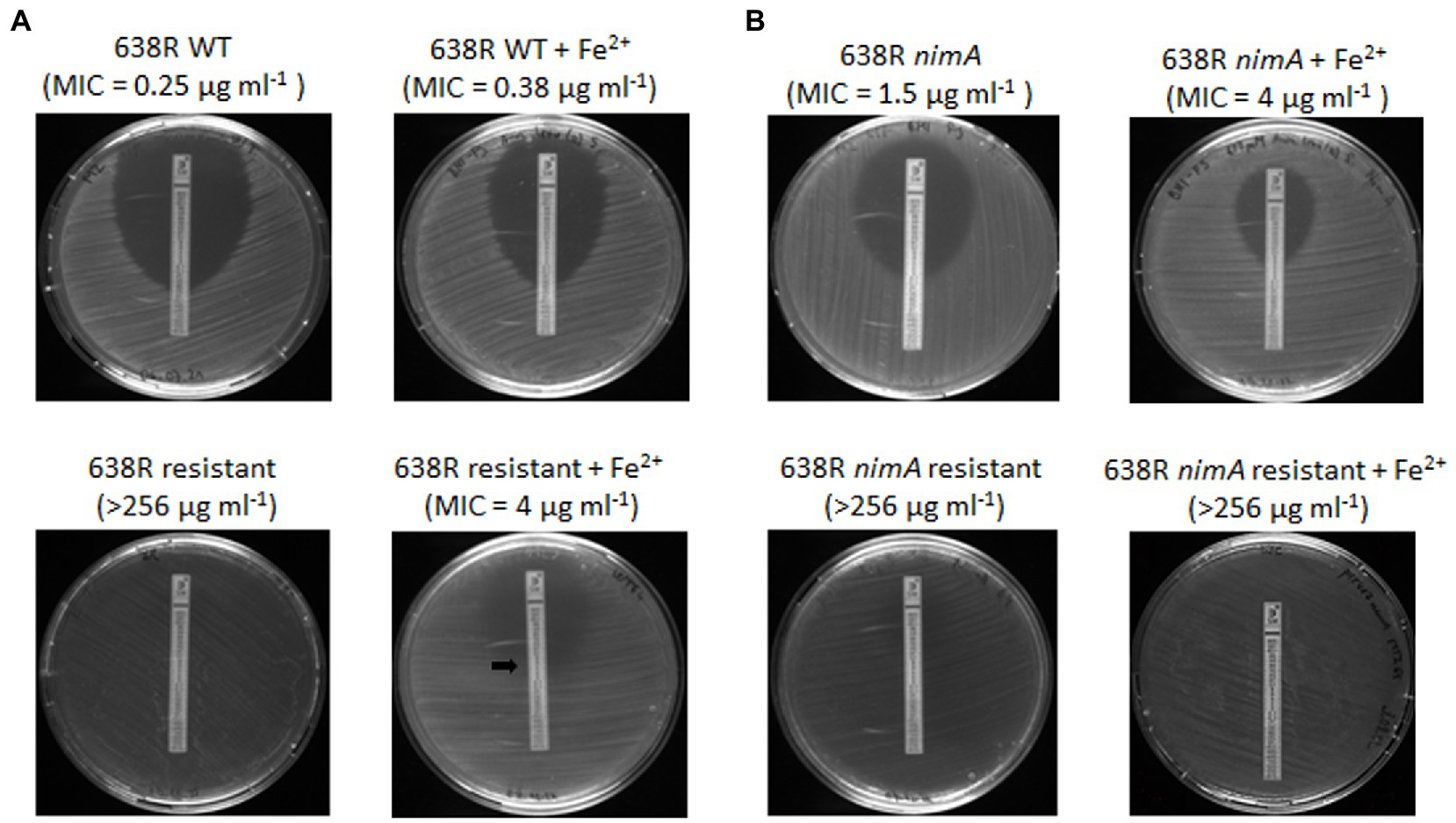
Figure 3. Susceptibilities of (A) 638R and 638RR, and (B) 638R nimA and 638R nimAR as determined by Etest, either with or without supplementation of 100 μM ferrous iron sulfate. Due to the fainter appearance of 638RR as compared to 638R, the point of intersection with the Etest strip is indicated by a black arrow.
Metronidazole resistance is a complex phenomenon in most anaerobes (reviewed in Leitsch, 2019), which is also underscored by the numerous changes in the proteome of B. fragilis 638RR in this study. Our data suggest that the impairment of hemin import, and therefore also of iron import in general, is mainly responsible for the development of metronidazole resistance in vitro in the absence of the nimA gene. Although a large subset of proteins is differentially expressed in 638RR, supplementation with high concentrations of ferrous iron widely abrogates metronidazole resistance. This result mirrors observations by others (Veeranagouda et al., 2014) that metronidazole resistance as caused by impeded iron uptake via heme can be widely abolished through the addition of ferrous iron. The low-level protection against metronidazole remaining after the addition of ferrous iron as seen herein might be due to other changes such as the upregulation of RND transporters and of ribosomal proteins.
The depletion of heme and iron has severe consequences for the activities of iron and heme-dependent enzymes which, consequently, cannot be equipped anymore with functional cofactors. We assume that the enzymes involved in energy metabolism which were found upregulated in this study, e.g., fumarate reductase or NADH:ubiquinone reductase are upregulated in the—probably futile—attempt to compensate for the loss of activity of iron and heme-dependent enzymes. This response, however, is not stereotypical because PFOR expression was not found altered in 638RR although PFOR activity is lost early during the development of resistance (Narikawa et al., 1991; Paunkov et al., 2022a). Moreover, PFOR levels are also unaltered in 638R nimA although this strain displays a more than twofold higher PFOR activity as compared to 638R (Paunkov et al., 2022a). These observations indicate that gene expression studies alone are insufficient to resolve the mechanism behind metronidazole resistance. However, when combining the findings of our present study, of our previous study (Paunkov et al., 2022a), and of earlier studies on the effect of heme deprivation on metabolic end products in B. fragilis (Macy et al., 1975; Chen and Wolin, 1981), a scenario of the energy metabolism in 638RR can be proposed (Figure 4). Three major end metabolites of the B. fragilis metabolism (Frantz and McCallum, 1979) are likely to be formed at much lower quantities or not at all: acetate, succinate, and propionate. Acetate is formed from acetyl-CoA, previously generated by PFOR and pyruvate formate-lyase (PFL), but PFOR is inactive due to the lack of iron–sulfur clusters which function as prosthetic groups, and PFL is downregulated approximately 11-fold (Table 4). Succinate is normally formed from fumarate by fumarate reductase and partly further processed to propionate, but fumarate reductase is dependent on the heme cofactor which is in short supply due to impaired heme import, resulting in strongly diminished activity (Paunkov et al., 2022a). Thus, the only major fermentative options remaining for the quantitative restoration of NAD+ are the reduction of pyruvate to lactate by lactate dehydrogenase (LDH) and the reduction of oxaloacetate to malate by malate dehydrogenase (MDH), resulting in far less ATP generated per mol of glucose (Figure 4). This is consistent with the impaired growth observed in 638RR (Paunkov et al., 2022a), indicating that these changes come at a very high cost and are probably only tolerable under the optimal growth conditions of in vitro cell culture. In return, the decreased activities of iron-dependent enzymes or factors can be expected to confer resistance to metronidazole, arguably by slowing down metronidazole reduction, i.e., activation.
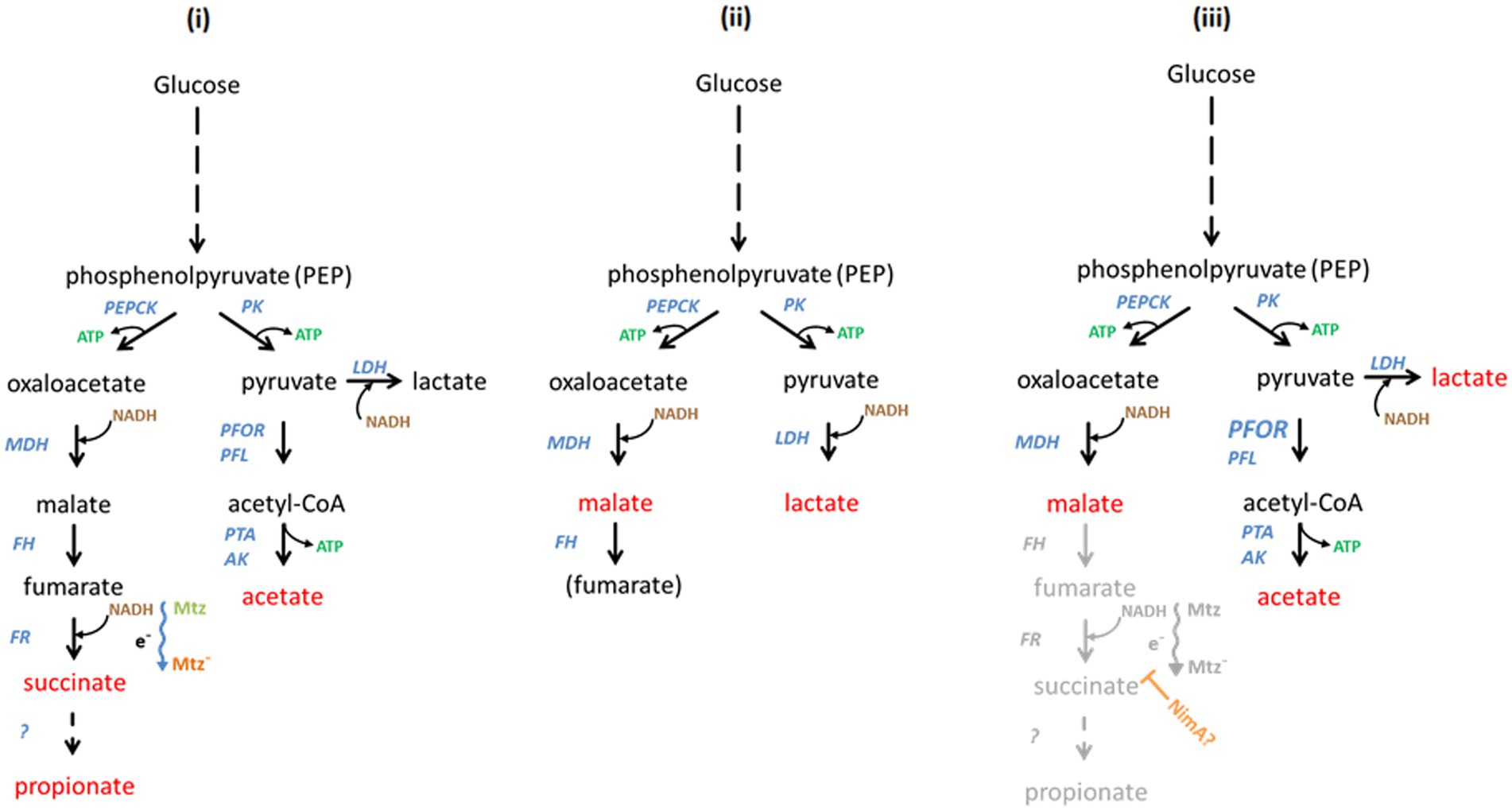
Figure 4. Hypothetical scheme of the interdependence of Bacteroides fragilis metabolism, induced metronidazole resistance, and of NimA-mediated protection from metronidazole, respectively. The main metabolic end products are indicated in red, the enzymes involved in blue, ATP in green, and NADH in brown. In the left panel (i) the pathways in B. fragilis leading to the formation of the metabolic end products (Frantz and McCallum, 1979) are depicted. Glucose is broken down to phosphoenolpyruvate (PEP) which can be either converted to oxaloacetate by phosphoenolpyruvate carboxykinase (PEPCK) or to pyruvate by pyruvate kinase (PK). Oxaloacetate is reduced to malate by malate dehydrogenase (MDH). Fumarate hydratase (FH), alternatively termed fumarase, dehydrates malate to fumarate which is then converted by fumarate reductase (FR) to succinate. Finally, succinate is converted to propionate by as yet unknown enzymes. The conversion of fumarate to succinate depends on an electron chain (indicated by e− and a wavy arrow) which might be involved in metronidazole reduction (Mtz to Mtz−). Importantly, also cytochrome ubiquinol oxidase depends on this electron chain. Pyruvate is converted to acetyl-CoA by pyruvate:ferredoxin oxidoreductase (PFOR) and/or pyruvate formate-lyase (PFL). Ultimately, acetyl-CoA is converted to acetate by phosphotransacetylase (PTA) and acetate kinase (AK). Alternatively, pyruvate can be reduced to lactate by lactate dehydrogenase (LDH). Lactate, however, is not a major end product under normal conditions. In the middle panel (ii) the altered metabolism of 638RR is shown. Due to the lack of iron and/or heme, PFOR and FR are not functional. Further, PFL is strongly downregulated. Consequently, NAD+ must be recycled through the reduction of pyruvate and oxaloacetate, rendering lactate and malate the major metabolic end products. Pathways for the reduction of metronidazole are shut-down. The loss of cytochrome ubiquinol oxidase activity is compensated by upregulation of FprA. The presumptive metabolism of 638R nimA is shown in the right panel (iii). The formation of acetate is up due to increased activity of PFOR (indicated by enlarged letters). This is might be necessitated by a hypothetical interference of NimA (orange) with the electron chain which involved in the formation of succinate and could also lead to less metronidazole being reduced (inhibited pathway shown in gray). Importantly, this would also lead to a decreased activity of cytochrome ubiquinol oxidase, necessitating a compensatory upregulation of FprA. Finally, in order to ensure efficient regeneration of NAD+, more lactate could be formed in order to compensate for the decreased formation of succinate. Also malate might accumulate when formation of succinate is down.
In 638R nimA, only five changes in the proteome were consistently observed as compared to 638R, one of them being the upregulation of flavodiiron protein FprA. Interestingly, the higher expression of FprA is a trait shared with 638RR, although it cannot be ruled out at present that in the latter this change is an attempt to compensate for low iron levels and does not lead to more actual FprA activity. Further, the induction of high-level resistance resulted in fewer changes in 638R nimA than in 638R and the significance of these changes is presently unclear. For financial reasons, the comparative analysis of 638R nimA and its resistant daughter strain was not repeated so it cannot be ruled out that many of the changes found would prove irreproducible in a second analysis, as was the case after repeating the analysis of 638R nimA vs. 638R. Still, the comparable paucity of changes in 638R nimAR as compared to 638RR (41 vs. 237) could be interpreted as compatible with the notion of Nim proteins acting as nitroreductases in the sense that direct detoxification of metronidazole by Nim would render extensive changes in protein expression unnecessary for survival. However, Nim levels are not further enhanced after induction of high-level resistance as has been repeatedly shown (Leitsch et al., 2014; Paunkov et al., 2022a; this study). This is hard to reconcile with a potential role of Nim proteins as nitroreductases which implies that more drug requires more enzyme to be metabolized. Further, said role would also not explain elevated PFOR activity as observed in 638R nimA and 638R nimE (Paunkov et al., 2022a). A clue to the solution of this conundrum might be the function of FprA. In Bacteroides, oxygen is reduced to water by cytochrome ubiquinol oxidase (Baughn and Malamy, 2004) and by FprA (Lu and Imlay, 2017). However, cytochrome ubiquinol oxidase is a heme-dependent enzyme and its function must be compromised in 638RR which imports little to no hemin. A direct interaction of FprA with metronidazole is rather unlikely because overexpression of fprA in 638R did not confer any resistance to metronidazole, but upregulation of FprA might well be a strategy to compensate for impaired cytochrome ubiquinol oxidase activity. A potential interdependence of FprA overexpression and NimA is not obvious. However, the increased PFOR activity in 638R nimA (Paunkov et al., 2022a) indicates that metabolic flux might be redirected from succinate formation by fumarate reductase to acetate formation by the PFOR pathway (Figure 4). Fumarate reductase and cytochrome ubiquinol oxidase depend on electrons fueled into a quinone-based electron transport chain through the oxidation of NADH by NADH:ubiquinone reductase and NADH dehydrogenase (Ito et al., 2020). Any interference with this process would result in less reducing power being available for the formation of succinate by fumarate reductase and for the reduction of oxygen by cytochrome ubiquinol oxidase. An inhibition of cytochrome ubiquinol oxidase function, in turn, might necessitate the upregulation of FprA expression to ensure efficient oxygen scavenging. Importantly, this proposed interference might also have implications for metronidazole reduction. It is conceivable that leakage of electrons along the electron transport chain could lead to the formation of toxic metronidazole intermediates, and that less metronidazole would be reduced via this route if the flux of electrons was restricted. This might give other routes of reduction enough time to act, eventually resulting in the formation of less toxic products such as the corresponding aminoimidazole (Carlier et al., 1997). It is presently unclear how NimA could achieve this proposed interference mechanistically, but a metabolomic study targeting the metabolic end products in 638R with and without the nimA gene should provide an answer if our proposed model does indeed apply. If it applied, then 638R nimA should produce more acetate and malate and less succinate and propionate than 638R. This shift could even be more pronounced in 638R nimAR. Thus, further studies should focus on the concentrations of metabolic end products and of cofactors such as NAD+/NADH, NADP+/NADPH, FAD, FMN and quinones, in order to obtain a more complete picture of the effect of nimA or other nim genes on B. fragilis physiology. This would be a timely undertaking because metronidazole resistance in Bacteroides spp. is increasingly becoming a problem in clinical settings in certain countries (Vieira et al., 2006; Yehya et al., 2014; Sheikh et al., 2015).
The raw mass spectrometric data files presented in the study are deposited in the PRIDE repository, accession number PXD039809 (http://www.ebi.ac.uk/pride/archive/projects/PXD039809).
AP performed the experiments, conceived the experiments, and analyzed the data. KH performed the mass-spectrometric analyses and analyzed the data. DS performed the experiments. JS conceived the experiments and analyzed the data. DL conceived the experiments, analyzed the data, and wrote the manuscript. All authors contributed to the article and approved the submitted version.
This research was funded by the Austrian Science Fund (FWF) [grant number I 4234]. For the purpose of open access, the author has applied a CC BY public copyright license to any Author Accepted Manuscript version arising from this submission. Further, JS was funded by grant ANN_130760 from the National Research, Development and Innovation Office of Hungary (NKFIH).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1158086/full#supplementary-material
Alauzet, C., Lozniewski, A., and Marchandin, H. (2019). Metronidazole resistance and nim genes in anaerobes: a review. Anaerobe 55, 40–53. doi: 10.1016/j.anaerobe.2018.10.004
Aldridge, K. E., and Sanders, C. V. (2002). Susceptibility trending of blood isolates of the Bacteroides fragilis group over a 12-year period to clindamycin, ampicillin-sulbactam, cefoxitin, imipenem, and metronidazole. Anaerobe 8, 301–305. doi: 10.1016/S1075-9964(03)00028-3
Baughn, A. D., and Malamy, M. H. (2004). The strict anaerobe Bacteroides fragilis grows in and benefits from nanomolar concentrations of oxygen. Nature 427, 441–444. doi: 10.1038/nature02285
Breuil, J., Dublanchet, A., Truffaut, N., and Sebald, M. (1989). Transferable 5-nitroimidazole resistance in the Bacteroides fragilis group. Plasmid 21, 151–154. doi: 10.1016/0147-619X(89)90060-7
Butler, N. L., Ito, T., Foreman, S., Morgan, J. E., Zagorevsky, D., Malamy, M. H., et al. (2022). Bacteroides fragilis maintains concurrent capability for anaerobic and nanaerobic respiration. J. Bacteriol. 2022:e0038922. doi: 10.1128/jb.00389-22
Carlier, J. P., Sellier, N., Rager, M. N., and Reysset, G. (1997). Metabolism of a 5-nitroimidazole in susceptible and resistant isogenic of Bacteroides fragilis. Antimicrob. Agents Chemother. 41, 1495–1499. doi: 10.1128/AAC.41.7.1495
Chan, A. C., Lelj-Garolla, B., Rosell, F. I., Pedersen, K. A., Mauk, A. G., and Murphy, M. E. P. (2006). Cofacial heme binding is linked to dimerization by a bacterial heme transport protein. J. Mol. Biol. 362, 1108–1119. doi: 10.1016/j.jmb.2006.08.001
Chen, M., and Wolin, M. J. (1981). Influence of heme and vitamin B12 on growth and fermentations of Bacteroides species. J. Bacteriol. 145, 466–471. doi: 10.1128/jb.145.1.466-471.1981
Franke, T., and Deppenmeier, U. (2018). Physiology and central carbon metabolism of the gut bacterium Prevotella copri. Mol. Microbiol. 109, 528–540. doi: 10.1111/mmi.14058
Frantz, J. C., and McCallum, R. E. (1979). Growth yields and fermentation balance of Bacteroides fragilis cultured in glucose-enriched medium. J. Bacteriol. 137, 1263–1270. doi: 10.1128/jb.137.3.1263-1270.1979
Gutiérrez, A. M., Sotillo, J., Schlosser, S., Hummel, K., and Miller, I. (2019). Towards understanding non-infectious growth-rate retardation in growing pigs. Proteomes 7:31. doi: 10.3390/proteomes7030031
Haggoud, A., Reysset, G., Azeddoug, H., and Sebald, M. (1994). Nucleotide sequence analysis of two 5-nitroimidazole resistance determinants from Bacteroides strains and of a new insertion sequence upstream of the two genes. Antimicrob. Agents Chemoth. 38, 1047–1051. doi: 10.1128/AAC.38.5.1047
Ito, T., Gallegos, R., Matano, L. M., Butler, N. L., Hantman, N., Kaili, M., et al. (2020). Genetic and biochemical analysis of anaerobic respiration in Bacteroides fragilis and its importance in vivo. mBio 11, e03238–e03219. doi: 10.1128/mBio.03238-19
Kupc, M., Paunkov, A., Strasser, D., Sóki, J., and Leitsch, D. (2022). Initial expression levels of nimA are decisive for protection against metronidazole in Bacteroides fragilis. Anaerobe 77:102630. doi: 10.1016/j.anaerobe.2022.102630
Kuwahara, T., Yamashita, A., Hirakawa, H., Toh, H., Okada, N., Kuhara, S., et al. (2004). Genomic analysis of Bacteroides fragilis reveals extensive DNA inversions regulating cell surface adaption. Proc. Natl. Acac. Sci. USA 101, 14919–14924. doi: 10.1073/pnas.0404172101
Leitsch, D. (2019). A review on metronidazole: an old warhorse in antimicrobial chemotherapy. Parasitology 146, 1167–1178. doi: 10.1017/S0031182017002025
Leitsch, D., Sóki, J., Kolarich, D., Urbán, E., and Nagy, E. (2014). A study on Nim expression in Bacteroides fragilis. Microbiology 160, 616–622. doi: 10.1099/mic.0.074807-0
Lu, Z., and Imlay, J. A. (2017). The fumarate reductase of Bacteroides thetaiotaomicron, unlike that of Escherichia coli, is configured so that it does not generate reactive oxygen species. mBio 8, e01873–e01816. doi: 10.1128/mBio.01873-16
Macy, J., Probst, I., and Gottschalk, G. (1975). Evidence for cytochrome involvement in fumarate reduction and adenosine 5′-triphosphate synthesis by Bacteroides fragilis grown in the presence of hemin. J. Bacteriol. 123, 436–442. doi: 10.1128/jb.123.2.436-442.1975
Martins, M. C., Romão, C. V., Folgosa, F., Borges, P. T., Frazão, C., and Teixeira, M. (2019). How superoxide reductases and flavodiiron proteins combat oxidative stress in anaerobes. Free Radic. Biol. Med. 140, 36–60. doi: 10.1016/j.freeradbiomed.2019.01.051
Mays, T. D., Smith, C. J., Welch, R. A., Delfini, C., and Macrina, F. L. (1982). Novel antibiotic resistance transfer in Bacteroides. Antimicrob. Agents Chemother. 21, 110–118. doi: 10.1128/AAC.21.1.110
Meehan, B. M., Baughn, A. D., Gallegos, R., and Malamy, M. H. (2012). Inactivation of a single gene enables microaerobic growth of the obligate anaerobe Bacteroides fragilis. Proc Natl Acad. Sci. U S A. 109, 12153–12158. doi: 10.1073/pnas.1203796109
Mishra, S., and Imlay, J. A. (2013). An anaerobic bacterium, Bacteroides thetaiotaomicron, uses a consortium of enzymes to scavenge hydrogen peroxide. Mol. Microbiol. 90, 1356–1371. doi: 10.1111/mmi.12438
Narikawa, S. (1986). Distribution of metronidazole susceptibility factors in obligate anaerobes. J. Antimirob. Chemother. 18, 565–574. doi: 10.1093/jac/18.5.565
Narikawa, S., Suzuki, T., Yamamoto, M., and Nakamura, M. (1991). Lactate dehydrogenase activity as a cause of metronidazole resistance in Bacteroides fragilis NCTC 11295. J. Antimicrob. Chemother. 28, 47–53. doi: 10.1093/jac/28.1.47
Paunkov, A., Gutenbrunner, K., Sóki, J., and Leitsch, D. (2022b). Haemin deprivation renders Bacteroides fragilis hypersusceptible to metronidazole and cancels high-level metronidazole resistance. J. Antimicrob. Chemother. 77, 1027–1031. doi: 10.1093/jac/dkab485
Paunkov, A., Sóki, J., and Leitsch, D. (2022a). Modulation of iron import and metronidazole resistance in Bacteroides fragilis harboring a nimA gene. Front. Microbiol. 13:898453. doi: 10.3389/fmicb.2022.898453
Perez-Riverol, Y., Bai, J., Bandla, C., Hewapathirana, S., García-Seisdedos, D., Kamatchinathan, S., et al. (2022). The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 50, D543–D552. doi: 10.1093/nar/gkab1038
Pumbwe, L., Chang, A., Smith, R. L., and Wexler, H. M. (2007). BmeRABC5 is a multidrug efflux system that can confer metronidazole resistance in Bacteroides fragilis. Microb. Drug Resist. 13, 96–101. doi: 10.1089/mdr.2007.719
Pumbwe, L., Glass, D., and Wexler, H. M. (2006). Efflux pump overexpression in multiple-antibiotic-resistant mutants of Bacteroides fragilis. Antimicrob. Agents Chemother. 50, 3150–3153. doi: 10.1128/AAC.00141-06
R Core Team (2021). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria.
Remacle, A. G., Shiryaev, S. A., and Strongin, A. Y. (2014). Distinct interactions with cellular E-cadherin of the two virulent metalloproteinases encoded by a Bacteroides fragilis pathogenicity island. PLoS One 9:e113896. doi: 10.1371/journal.pone.0113896
Rocha, E. R., Bergonia, H. A., Gerdes, S., and Smith, J. C. (2019). Bacteroides fragilis requires the ferrous-iron transporter FeoAB and the CobN-like proteins BtuS1 and BtuS2 for assimilation of iron released from heme. Microbiology 8:e00669. doi: 10.1002/mbo3.669
Sheikh, S. O., Jabeen, K., Qaiser, S., Ahsan, S. T., Khan, E., and Zafar, A. (2015). High rate of non-susceptibility to metronidazole and clindamycin in anaerobic isolates: data from a clinical laboratory from Karachi, Pakistan. Anaerobe 33, 132–136. doi: 10.1016/j.anaerobe.2015.03.006
Smith, C. J., Rogers, M. B., and McKee, M. L. (1992). Heterologous gene expression in Bacteroides fragilis. Plasmid 27, 141–154. doi: 10.1016/0147-619X(92)90014-2
Snydman, D. R., Jacobus, N. V., McDermott, L. A., Goldstein, E. J., Harrell, L., Jenkins, S. G., et al. (2017). Trends in antimicrobial resistance among Bacteroides species and Parabacteroides species in the United States from 2010-2012 with comparison to 2008-2009. Anaerobe 43, 21–26. doi: 10.1016/j.anaerobe.2016.11.003
Tan, T. Y., Ng, L. S., Kwang, L. L., Rao, S., and Eng, L. C. (2017). Clinical characteristics and antimicrobial susceptibilities of anaerobic bacteremia in an acute care hospital. Anaerobe 43, 69–74. doi: 10.1016/j.anaerobe.2016.11.009
Veeranagouda, Y., Husain, F., Boente, R., Moore, J., Smith, C. J., Rocha, E. R., et al. (2014). Deficiency of the ferrous iron transporter FeoAB is linked with metronidazole resistance in Bacteroides fragilis. J. Antimicrob. Chemother. 69, 2634–2643. doi: 10.1093/jac/dku219
Vieira, J. M. B. D., Boente, R. F., Miranda, K. R., Avelar, K. E. S., Domingues, R. M. C. P., and Ferreira, M. C. (2006). Decreased susceptibility to nitroimidazoles among Bacteroides species in Brazil. Curr. Microbiol. 52, 27–32. doi: 10.1007/s00284-005-0068-0
Wasserfallen, A., Ragettli, S., Jouanneau, Y., and Leisinger, T. (1998). A family of flavoproteins in the domains archaea and bacteria. Eur. J. Biochem. 254, 325–332. doi: 10.1046/j.1432-1327.1998.2540325.x
Wexler, H. M. (2007). Bacteroides: the good, the bad, and the nitty-gritty. Clin. Microbiol. Rev. 20, 593–621. doi: 10.1128/CMR.00008-07
Wisniewski, J. R. (2016). Quantitative evaluation of filter aided sample preparation (FASP) and multienzyme digestion FASP protocols. Anal. Chem. 88, 5438–5443. doi: 10.1021/acs.analchem.6b00859
Wisniewski, J. R., Zougman, A., Nagaraj, N., and Mann, M. (2009). Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362. doi: 10.1038/nmeth.1322
Yehya, M., Hamze, M., Mallat, H., and Dabbousi, F. (2014). Prevalence and antibiotic susceptibility of Bacteroides fragilis group isolated from stool samples in North Lebanon. Braz. J. Microbiol. 44, 807–812. doi: 10.1590/S1517-83822013000300022
Keywords: Bacteroides fragilis, metronidazole, antimicrobial resistance, proteomics, nimA gene
Citation: Paunkov A, Hummel K, Strasser D, Sóki J and Leitsch D (2023) Proteomic analysis of metronidazole resistance in the human facultative pathogen Bacteroides fragilis. Front. Microbiol. 14:1158086. doi: 10.3389/fmicb.2023.1158086
Received: 03 February 2023; Accepted: 14 March 2023;
Published: 31 March 2023.
Edited by:
Axel Cloeckaert, Institut National de Recherche pour l’agriculture, l’alimentation et l’environnement (INRAE), FranceReviewed by:
Sylvia Valdezate, Carlos III Health Institute (ISCIII), SpainCopyright © 2023 Paunkov, Hummel, Strasser, Sóki and Leitsch. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: David Leitsch, ZGF2aWQubGVpdHNjaEBtZWR1bml3aWVuLmFjLmF0
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.