- 1School of Agricultural Engineering and Food Science, Shandong University of Technology, Zibo, China
- 2School of Resources and Environmental Engineering, Shandong University of Technology, Zibo, China
- 3School of Life Sciences and Medicine, Shandong University of Technology, Zibo, Shandong, China
- 4School of Environment and Safety Engineering, Qingdao University of Science and Technology, Qingdao, China
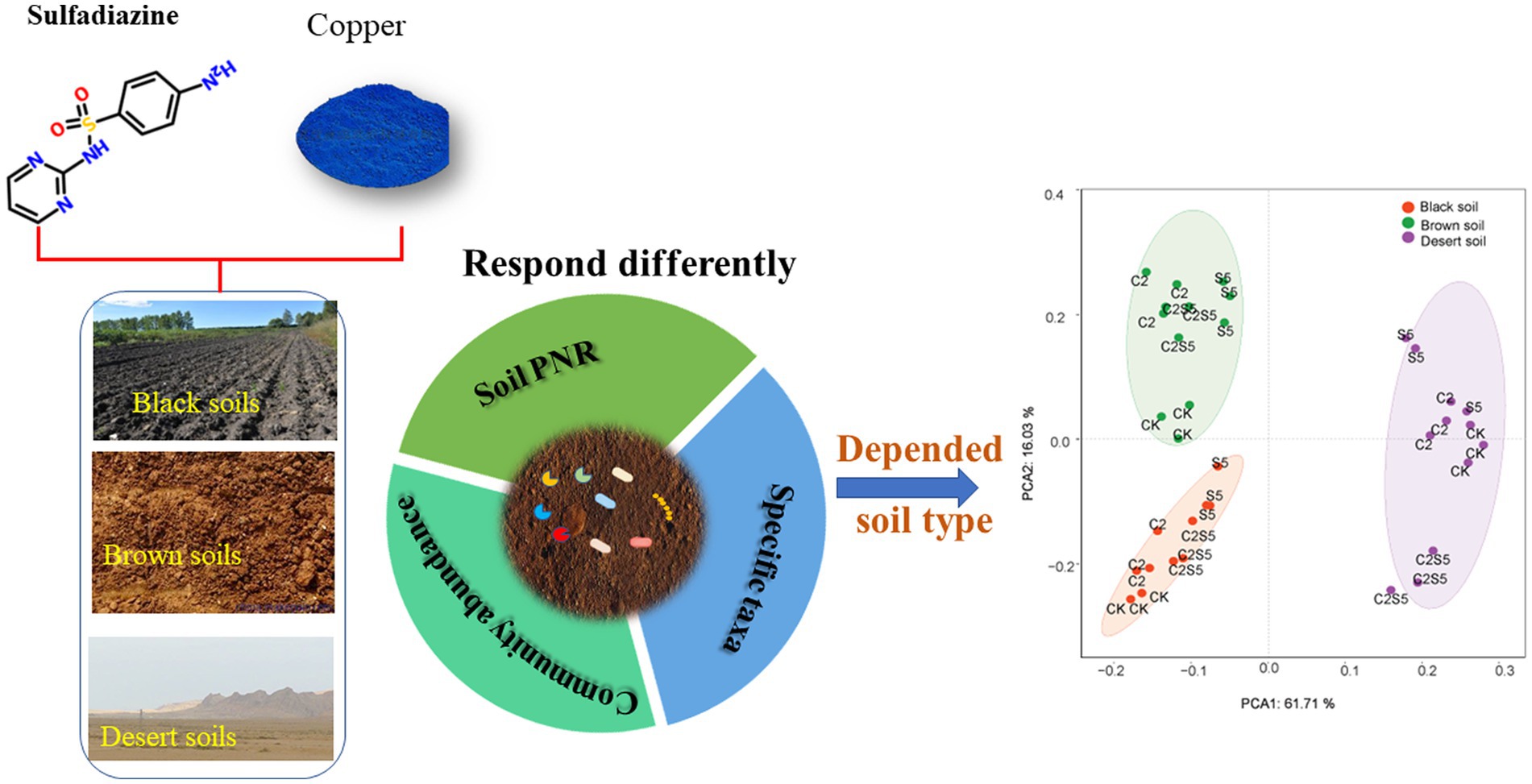
Introduction: Sulfadiazine (SDZ) and copper (Cu) are frequently detected in agricultural soils, but little is known on their single or combined impact on ammonia oxidizing microbial community and function across different soils.
Methods: In this study, a microcosm was conducted to distinguish the microbial ecotoxicity of SDZ and Cu across different soils by analyzing soil potential nitrification rate (PNR) and the amoA gene sequences.
Results: The results showed that the single spiking of SDZ caused a consistent decrease of soil PNR among three tested soils, but no consistent synergistic inhibition of SDZ and Cu was observed across these soils. Moreover, across three tested soils, the distinct responses to the single or joint exposure of SDZ and Cu were found in amoA gene abundance, and diversity as well as the identified genus taxa of ammonia-oxidizing archaea (AOA) and bacteria (AOB). Meanwhile, only the specific genus taxa of AOA or AOB consistently corresponded to the variation of soil PNR across different treated soils. The further principal component analysis (PCA) exhibited that the variable influence of SDZ and Cu on ammonia oxidizing microbial community and function was greatly dependent on soil type.
Discussion: Therefore, in addition to ecological functionality and the specific prokaryotic taxa, soil microbial ecotoxicity of SDZ and Cu also was dependent on edaphic factors derived from soil types. This study proposes an integrative assessment of soil properties and multiple microbial targets to soil contamination management.
Highlights
• PNR and amoA genes responded differently in soils exposed to SDZ and Cu.
• The impacts of SDZ and Cu on AOA and AOB communities depended on soil types.
• The specific phylotype corresponded to the PNR variation under the stress of SDZ and Cu.
1. Introduction
Recently, increasing attention is paid to soil contamination by veterinary antibiotics and heavy metals (Guo et al., 2018), especially their combined toxic effects on soil microbial ecology (Xu et al., 2016; Grenni et al., 2018; Wei et al., 2018; Wang L. et al., 2019). Moreover, a few investigations found that combined treatments of antibiotics and heavy metals produced higher toxicity on soil microbial ecology than the single one (Xu et al., 2016; Wang L. et al., 2019), which were strongly dependent on the added ratio, exposure time (Yang et al., 2020), and the specific biological indicator (Liu et al., 2015). It was reported that the co-existence of heavy metals and antibiotics could change soil ecological function by shifting microbial community structure (Cycoń et al., 2019). In light of the pivotal roles in the nitrogen cycling of ecosystems (Falkowski et al., 2008; Ke et al., 2013; Wang et al., 2015), ammonia-oxidizing microorganisms and the compounding function were widely chosen as biomarkers of soil environmental contamination (Katipoglu-Yazan et al., 2016; Zhang et al., 2017; Wei et al., 2018). It was found that combined exposure to antibiotics and heavy metals caused stronger toxicity than the single one on ammonia-oxidizers’ abundance and nitrification activity (Liao et al., 2019a,b; Wang L. et al., 2019). The abundant data also indicated that the entered toxicant could alter soil microbial activity (Gil-Sotres et al., 2005; Hammesfahr et al., 2011; Cycoń et al., 2016) and then shift their community structure (Wang L. et al., 2019). However, it was reported that the functional redundancy and the specific phylotypes modulate the contribution of soil microbial diversity and composition to multifunctionality (Li et al., 2021). Thus, it still could be debated whether or how soil contaminants caused real toxic effects on soil nitrification. This calls for further investigation to differentiate the responses of soil nitrification function and ammonia oxidizing microorganisms under contaminated stress; more importantly, the ecological mechanisms in mediating these responses need to be investigated.
Moreover, many investigations have proved that soil properties modulated soil prokaryotic diversity, composition, and ecological function (Tourna et al., 2008; Griffiths et al., 2011; Xu et al., 2020; Yang et al., 2021), which should regulate the response of microbes to the soil contamination stress (Bernier and Surette, 2013; Xian et al., 2015; Grenni et al., 2018). In comparison, ammonia oxidizers are more susceptible to changes in farming practices, such as tillage, fertilization, and planting (Zhou Z. et al., 2014; Azziz et al., 2016; Yan et al., 2018; Sun et al., 2019). It is also found that the distribution pattern of ammonia-oxidizers’ communities is different across distinct soils (Azziz et al., 2016; Xia et al., 2016; Liu et al., 2019). Thus, there are grounds to assume that the influence of antibiotics and heavy metals on ammonia-oxidizers’ community and function would be greatly dependent on individual soil types. Sulfonamides and copper, two common food additives largely applied in livestock farming, are frequently detected co-existence in agricultural soils as the wide application of manures (Grenni et al., 2018; Cycoń et al., 2019; Quaik et al., 2020; Yang et al., 2020), but few studies compared their toxic influence on ammonia-oxidizing microbial community and function across various soils.
To bridge the aforementioned knowledge gap, two typical groups of ammonia-oxidizing microorganisms, namely ammonia-oxidizing archaea (AOA) and ammonia-oxidizing bacteria (AOB), and the involved soil potential nitrification were chosen as biomarkers to elucidate the toxicity difference of SDZ and Cu across distinctive soils. The present study would offer an implication for the integrative management of soil contamination.
2. Materials and methods
2.1. Soil samples and materials
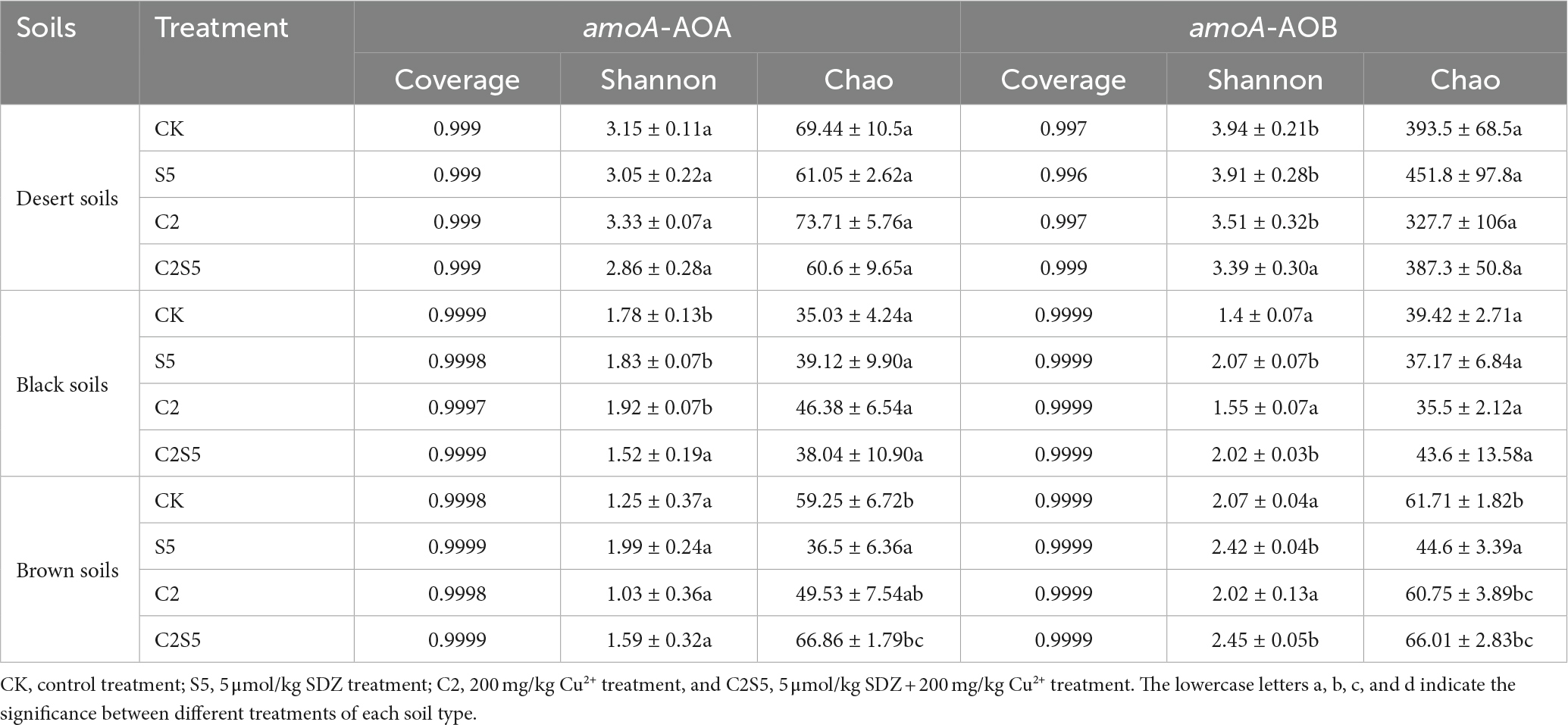
Black soils, Brown soils, and Desert soils, three typical soil types of China, were sampled from three farmland zones along distinct climatic gradients (Table 1). Surface soil samples (0–20 cm) were collected from a fallow plot without plants to minizine the influence of crop growing. Three sampling plots were collected from each farmland zone. Five random sites were sampled from each plot, and about 2 kg of soil was homogenized as one soil sample after picking out large stones, plant litter, and animal debris. The collected soil samples were screened (~2 mm) and then divided into two subsamples: One subsample was stored at ~4°C for microbial analysis in the next 2 weeks, and the other one was air-dried for the analysis of soil properties. Soil type and the selected soil physicochemical properties are listed in Table 1.
Sulfadiazine (AR, ≥ 98.6%) was purchased from Shanghai Yuanye Biological Technology Co. Ltd. (Shanghai, China) and prepared in a stock solution (40 mmol/l) with ultra-pure water after being dissolved by methanol (HPLC grade). A total of 1,000 mg/l of Cu2+ stock solution was prepared with CuSO4 in ultra-pure water. The reagents, except methanol, are all of the analytical grade and purchased from Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China).
2.2. Soil property analysis
Multiple soil abiotic variables were determined according to the protocols described in the reference (Lu, 1999; Carter and Gregorich, 2007): soil pH was measured using a suspension of 1:2 soil/water with a glass electrode (FE22, Mettler Toledo, Shanghai, China); soil organic carbon (SOC) was determined by acid dichromate wet oxidation method; total N was determined by the Kjeldahl method; soil inorganic nitrogen (NH4-N and NO3-N) was extracted with 1 M KCl and analyzed by a flow injection analyzer (SAN++, Skalar, Netherlands). The clays were analyzed by a laser particle size analyzer (Rise 2008, Rise, China) after being scattered with sodium hexametaphosphate. The cation exchange capacity (CEC) was measured by analyzing the concentration of exchangeable cations in ammonium acetate extract of soils with an inductively coupled plasma-atomic emission spectrometry (ICP-AES, Optima 5300DV, Perkin Elmer). Cu in soils was determined with an inductively coupled plasma atomic emission spectrometer (ICP-MS, Vista MPX, United States, Varian) after digested by HCl, HNO3, and HF (8 ml, 3:9:4 v1/v2/v3) in a microwave digestion system (ETHOS 1, Italy, Milestone; Hu et al., 2011; Liao et al., 2019a,b). For the analysis of SDZ in soil, the method of Zhou A. et al. (2014) was employed with a modification, and the detailed procedure was described in the previous report (Liao et al., 2019a,b).
2.3. Incubation experiment
A soil sample of 50 g was added to a plastic bottle (250 ml) and then pre-incubated in dark at 25 ± 2°C for soil microbial recovery after adjusting water content to 50% of the field water-holding capacity [WHC, determined by Keen’s box method (Kumar et al., 2018)]. At the end of 7 days’ pre-incubation, the soils were spiked with SDZ and CuSO4, respectively, to their designed concentrations. Then, all soils were thoroughly mixed, and each soil treatment was split into two subsamples: a subsample amended with 100 mg NH4-N/kg fresh soil using a stock solution of (NH4)2SO4 and additional deionized water to reach soil moisture of 60% of WHC for soil potential nitrification rates (PNRs) analysis according to the ISO method (Rusk et al., 2004) with slight modification, seen in our prior publications (Liu et al., 2014; Liao et al., 2019a,b); another one, used for the amoA gene analysis, was humidified directly with deionized water to 60% of WHC and incubated for another 14 days to keep the same incubating days with soil PNR. During the whole incubation, soil moisture was maintained at 60% of WHC by adding regularly sterilized ultra-pure water.
In this study, 5 μmol/kg of SDZ was chosen as the added rate according to the previous reports (An et al., 2015; Qiao et al., 2018), but a 200 mg/kg added rate of Cu was used based on risk control standards for soil contamination of agricultural land (GB15618-2018, China). Both single and combined treatments of SDZ and Cu were performed for each soil, which was designed as S5 for the single spiking of 5 μmol/kg SDZ, C2 for the single spiking of 200 mg/kg Cu2+, and C2S5 for the combined spiking of 5 μmol/kg SDZ and 200 mg/kg Cu2+. The soil treatments without any addition of Cu or SDZ were used as the control treatment designed as CK for each soil. Each soil treatment was conducted in triplicates.
2.4. Soil DNA extraction and amoA genes analysis
The fresh soil samples were transported in iced boxes to Lc-Bio Technology Co., Ltd. (Hangzhou, China) for DNA extraction, PCR amplification, sequencing, and taxonomic assignment (Liao et al., 2019a,b).
In brief, the total DNA was extracted from 0.25 g of each soil sample with the PowerSoil™ DNA isolation kit (TianGen Biotech, Beijing, China). The fluorescence quantitative PCR (qPCR, SYBR Green I) was conducted in triplicate on a real-time ABI7500 (Applied Biosystems, Warrington, United Kingdom) using the primer amoA-1F/amoA-2R for ammonia-oxidizing bacteria (AOB) and CrenamoA23F/A616R for ammonia-oxidizing archaea (AOA) (Supplementary Table S1). The standard curves of both AOA-amoA and AOB-amoA were drawn as described previously (Liao et al., 2019a,b). Quantification of amoA genes of AOA and AOB was conducted by comparing the values of cycle threshold (Ct) with the standard curves, respectively.
The primers used for PCR amplification were the same as the qPCR as well as the amplification systems and conditions. After purification using the SanPrep® quick PCR purification kit, pyrosequencing working of the target PCR products was accomplished on the Illumina Miseq PE300 sequencing platform (Illumina, Inc., CA, United States), and each treatment was performed in triplicate. The raw sequences were quality-filtered and processed using QIIME v2.0. Obtained sequences from different soil samples were, respectively, clustered into the operational taxonomic unit (OTU) at 97% sequence similarity using UPARSE (Version 7.1) after chimera detection using USTARCH7.0. The archaeal and bacterial OTUs were classified by using the ribosomal database project (RDP) classifier against the functional genomics resource (FGR) functional gene database.
2.5. Data analysis
The software package SPSS 18.0 (SPSS, Inc., United States) was used for data statistical analysis. The significant differences between soil treatments were identified by the one-way variance analysis (ANOVA) with Tukey’s HSD post hoc test at a value of p of <0.05 significant level. The principal component analysis (PCA) and the canonical correlation analysis (CCA) were used to account for the influence of soil properties on the variation of soil PNR and ammonia-oxidizers’ community in soils after being treated with SDZ and Cu.
3. Results
3.1. Variation of soil PNR and Amo genes’ abundance in three soils under the stress of SDZ and Cu
As shown in Figure 1, soil PNRs were all significantly (df = 3, F = 1031.84, p ≤ 0.05) inhibited in soils treated with SDZ and Cu singly or simultaneously; moreover, this inhibition was different across three tested soils. In detail, for the single exposure of SDZ, the highest decrease in soil PNR was observed in Brown soils, while the highest decrease of soil PNR was observed in Black soils when exposed to Cu. The combination of SDZ and Cu showed an antagonistic inhibition on soil PNR in Desert and Black soils, but there was a synergetic inhibition in Brown soils.
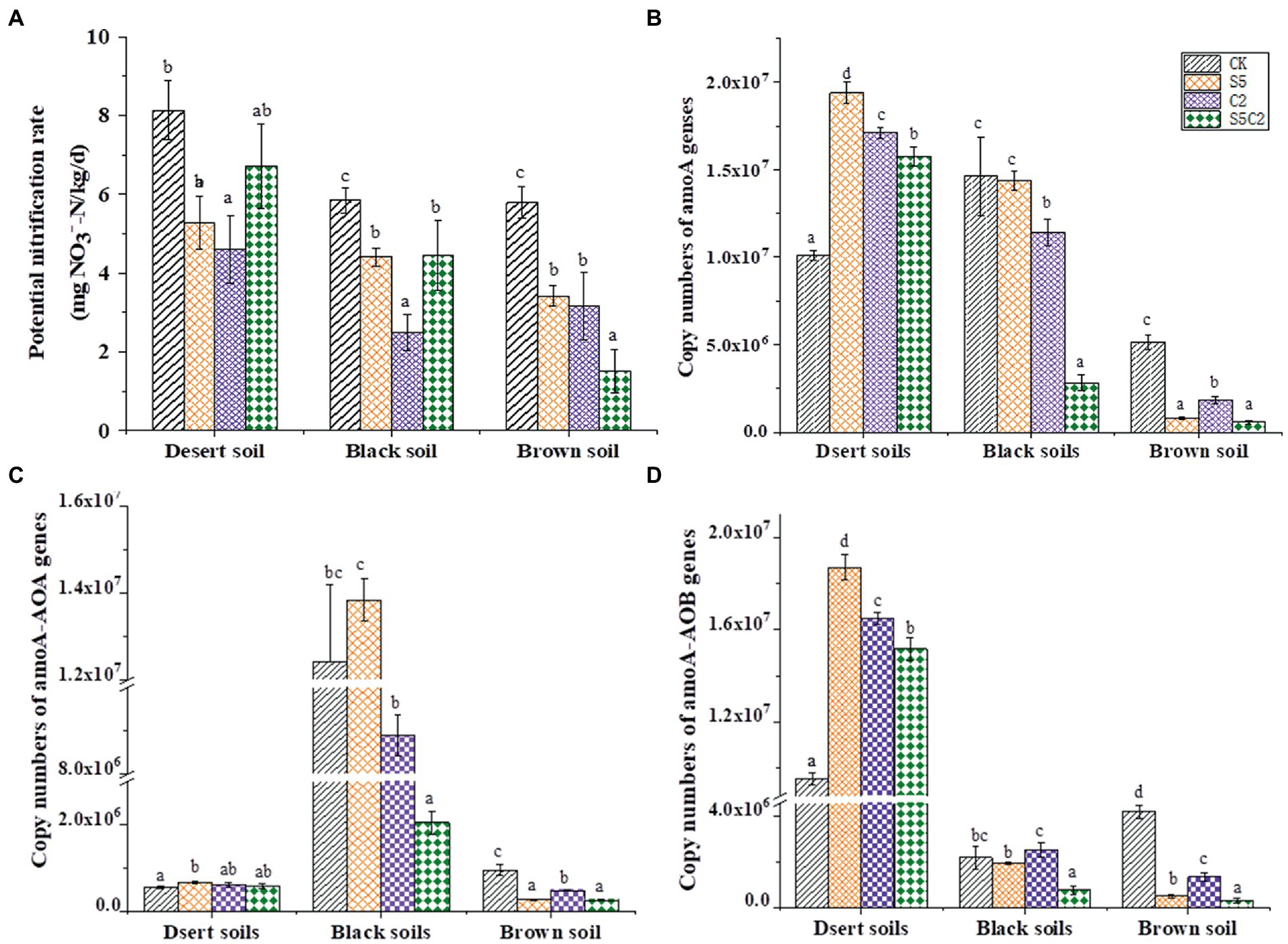
Figure 1. Variation of soil PNR (A), the abundance of the total amoA genes (B), AOA groups (C), and AOB groups (D) in different soil treatments (Error bar is standard deviation; CK, control treatment; S5, 5 μmol/kg SDZ treatment; C2, 200 mg/kg Cu2+ treatment, and C2S5, 5 μmol/kg SDZ + 200 mg/kg Cu2+ treatment).
In view of amoA gene abundance (Figure 1B), it was a significant proliferation (df = 3, F = 10.45, p ≤ 0.05) in Desert soils, while a significant inhibition (df = 3, F = 12.36, p ≤ 0.05) in Brown soils was observed when exposed to SDZ and Cu singly or simultaneously. In Black soils, amoA gene abundance was little changed with the single application of SDZ, while it was a synergetic inhibition to the combined exposure of SDZ and Cu. Similar results were also found for the individual AOA or AOB groups (Figures 1C,D). These results indicated that the abundance of ammonia-oxidizing microorganisms might not always agree with the variation of soil PNR in distinct soils under the single or combined stress of SDZ and Cu.
3.2. Shift of ammoxidation community in three soils to the exposure of SDZ and Cu
The values of Chao and Shannon indexes of the AOA and the AOB groups were originally different among the three tested soils (Table 2). Moreover, the single or combined application of SDZ and Cu did not cause an obvious change in the richness and diversity of the AOA and the AOB groups in most of the cases, as based on Chao and Shannon indexes that had no significant variation in most of the soil treatments, except in Brown soils treated with SDZ alone (Table 2). However, various alterations in communities’ composition of the AOA and AOB groups were found across three tested soils to the same exposure of SDZ and Cu singly or simultaneously (Figure 2). In detail, among the different treatments of SDZ and Cu, there were only 7% of the AOA communities in common, while it was zero for the AOB community in Desert soils, but there were remained at least 25% in common for the AOA and AOB communities in Brown and Black soils (Figure 2).
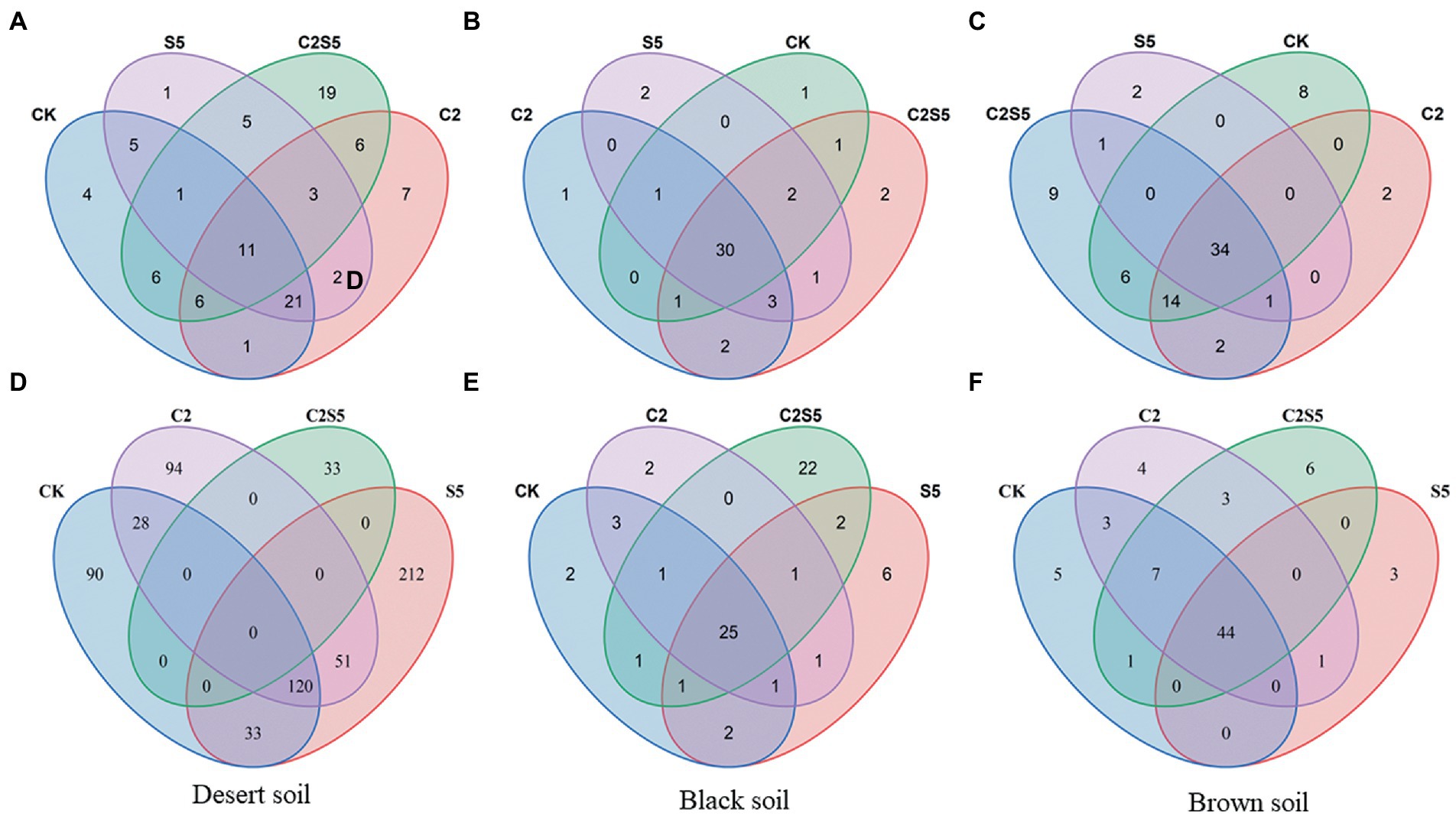
Figure 2. Venn diagram of overlapping AOA (A–C) and AOB (D–F) communities between different treatments of each investigated soil (CK, control treatment; S5, 5 μmol/kg SDZ treatment; C2, 200 mg/kg Cu2+ treatment, and C2S5, 5 μmol/kg SDZ + 200 mg/kg Cu2+ treatment).
Moreover, in view of the individual genus, various responses were also found among three tested soils to the exposure of SDZ and Cu singly or simultaneously, that is, the genus Nitrososphaera of the AOA group was significantly (df = 3, F = 12.90, p ≤ 0.05) proliferated in Desert soil, but it was inhibited in Black and Brown soils with the single application of SDZ or Cu, although the synergistic inhibition was all found in these three soils to the combined exposure of SDZ and Cu. In addition, both norank_Thaumarchaeota and norank_Crenarchaeota of the AOA group were mostly bolstered in three tested soils under the single or combined stress of SDZ and Cu (Figure 3A).
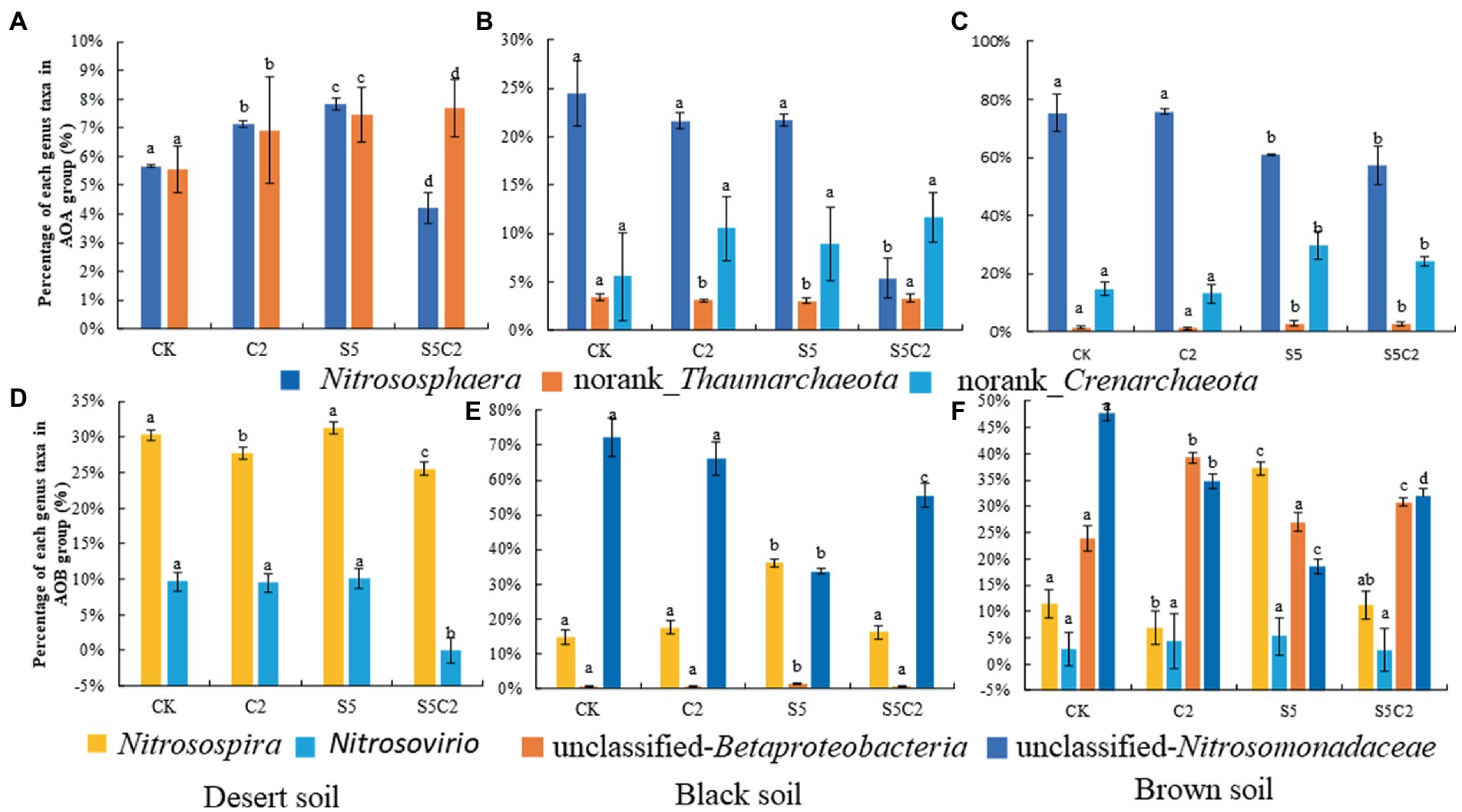
Figure 3. Relative abundance of the dominant AOA genera (A–C) and AOB (D–F) genera in different soil treatments (Error bar is standard deviation; CK, control treatment; S5, 5 μmol/kg SDZ treatment; C2, 200 mg/kg Cu2+ treatment, and C2S5, 5 μmol/kg SDZ + 200 mg/kg Cu2+ treatment).
In contrast to the AOA group, the individual genus response of the AOB group was more variable across three tested soils to the single or combined exposure of SDZ and Cu (Figure 3B), that is, Nitrosospira was bolstered up in three tested soils when exposed to SDZ, but it was significantly inhibited in Desert and Brown soils while increased in Black soils, to the single exposure of Cu. However, unclassified Betaproteobacterias and Nitrosomonadanceae were all significantly inhibited in Black and Brown soils to the single exposure of SDZ or Cu. However, for the combined exposure of SDZ and Cu, the synergistic effect was observed on a specific genus of AOB in Desert soils, while there was an antagonistic effect in Black and Brown soils.
3.3. The PCA results of different treatments for three tested soils
Principal components analysis (Figure 4) indicated that the separation of soil types by the first two PC derived from all measured biomarkers of the ammonia-oxidizing process (Figure 4B) corresponds to the distribution derived from basic soil properties (Figure 4A). Moreover, the similar soil distribution (Figures 4C,D) was also documented according to the individual AOA and AOB group in soils exposed to SDZ and Cu singly or simultaneously, which corresponds their variable variation across different soil treatments (Figures 1–3; Table 2). These results suggest that soil properties control the toxic effect of SDZ and Cu on soil ammonia-oxidizing microbial communities and function.
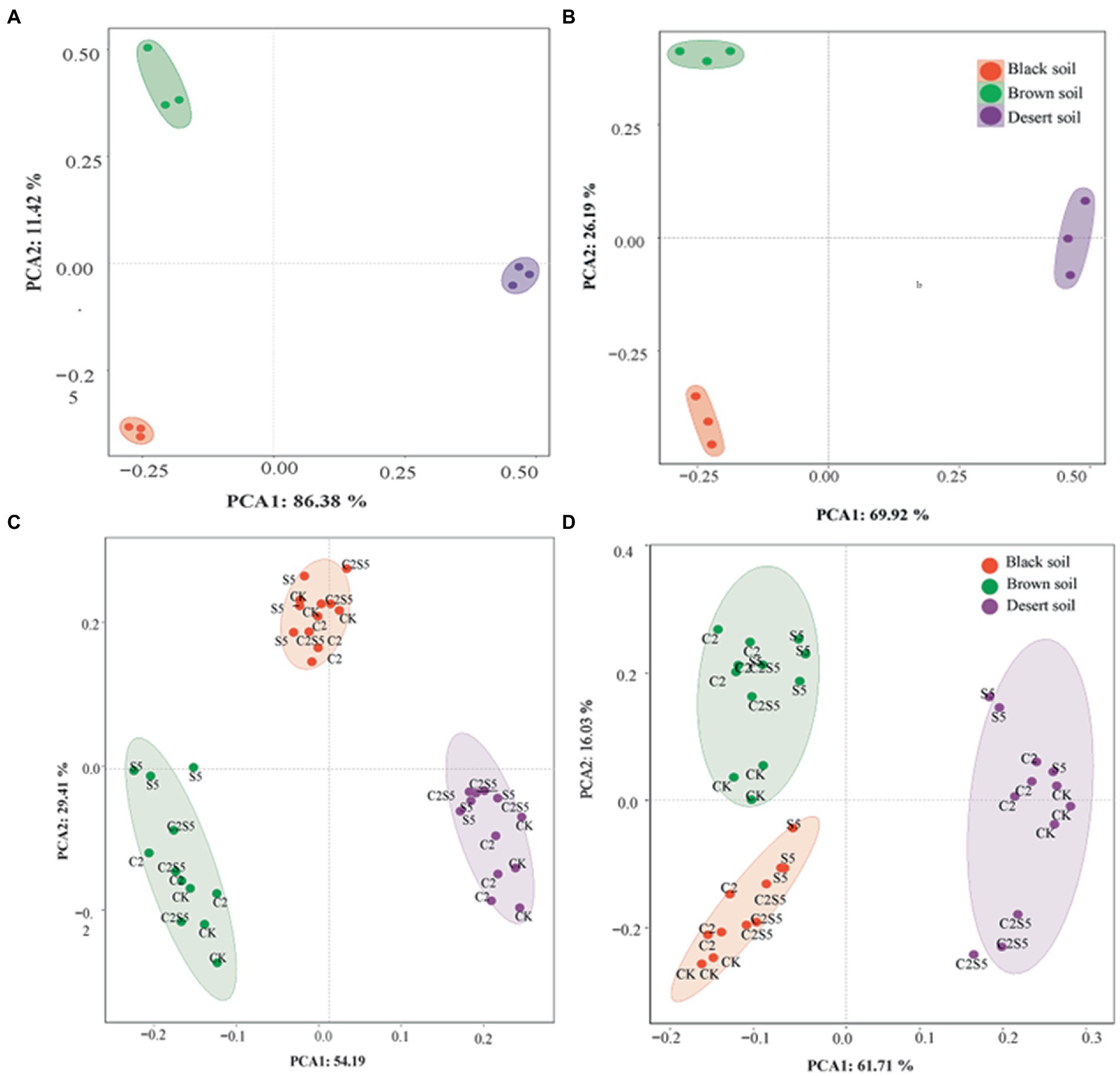
Figure 4. Grouping soils according to (A) basic soil properties from Table 1; (B) PNR and community abundance, diversity, and individual taxa of ammonia-oxidizing microorganism in control soil treatments from Figures 1–3 and Table 2; (C) PNR and community abundance, diversity and individual genus taxa of AOA in treated soils with SDZ and/Cu from Figures 1–3 and Table 2; (D) PNR and community abundance, diversity, and individual genus taxa of AOB in treated soils with SDZ and/Cu from Figures 1–3 and Table 2 (CK, control treatment; S5, 5 μmol/kg SDZ treatment; C2, 200 mg/kg Cu2+ treatment, and C2S5, 5 μmol/kg SDZ + 200 mg/kg Cu2+ treatment).
4. Discussion
4.1. Variable impact of SDZ on ammonia-oxidizing microorganisms across different soils
As the critical role in the turnover of soil nitrogen, ammonia-oxidizing archaea and bacteria as well as the related soil nitrification are widely chosen to evaluate the ecological risk of soil contamination (Xu et al., 2016; Liao et al., 2019a,b; Wang L. et al., 2019). Moreover, in addition to the exposure rate and time, soil properties are proved to be non-negligible in modulating the response of soil PNR to soil contaminants’ disturbance (Kotzerke et al., 2008; Toth et al., 2011; Ma et al., 2016; Wei et al., 2018). In the present study, the variable decrease of soil PNR was also found across different soils to the single exposure of SDZ, although there was a consistent inhibition in all the soil SDZ treatments (Figure 1A). The further CCA also exhibited a significantly dependent relationship between soil properties and PNR (Figure 5). No similar correspondence was found for the response of amoA gene abundance across three tested soils with the application of SDZ, even counting on the individual AOA or AOB group (Figures 1B–D). It proves that the single response of PNR could not be translated into the consistent effects of SDZ on the ammonia-oxidizing microbial community in soils. This result is different from the previous reports that SDZ had a similar influence on soil nitrification function and the involved ammonia-oxidizing microbial community (Ding and He, 2010; Xing and Jin, 2018). Moreover, along with a diverse variety of amoA gene abundance (Figure 1), variable responses in the community diversity, composition, as well as the specific taxa of AOA and AOB group were also found across three tested soils when exposed to SDZ (Figures 2 3; Table 2). These inconsistent findings might be related to soil properties controlling microbial composition and activity (Bernier and Surette, 2013; Grenni et al., 2018; Yang et al., 2020). Similar findings were also previously reported that microbial responses to pharmaceuticals in agricultural soils depended on soil types (Frková et al., 2020), as soil properties regulating their sorption and dissipation in soils, in addition to controlling microbial composition in soils (Kodešová et al., 2020). Moreover, the present study also found that the same separation of soil types by two first PC derived from all biomarkers of ammonia-oxidizing microorganisms corresponded to the distribution derived from basic soil properties (Figure 4), and the significant dependent relationship between soil properties and ammonia-oxidizing microbial community and function (Figure 5). Contrary to the stimulatory effects on the abundance and diversity of AOA and AOB groups, the single application of SDZ in three tested soils caused a well-consistent inhibition on soil PNR and the specific genus taxa of AOA or AOB (Figures 1A, 3). It might suggest that the specific genus taxa of ammonia-oxidizing microorganisms modulate the response of soil PNR to the exposure of antibiotics, in addition to soil properties (Toth et al., 2011; Ma et al., 2016; Wei et al., 2018).
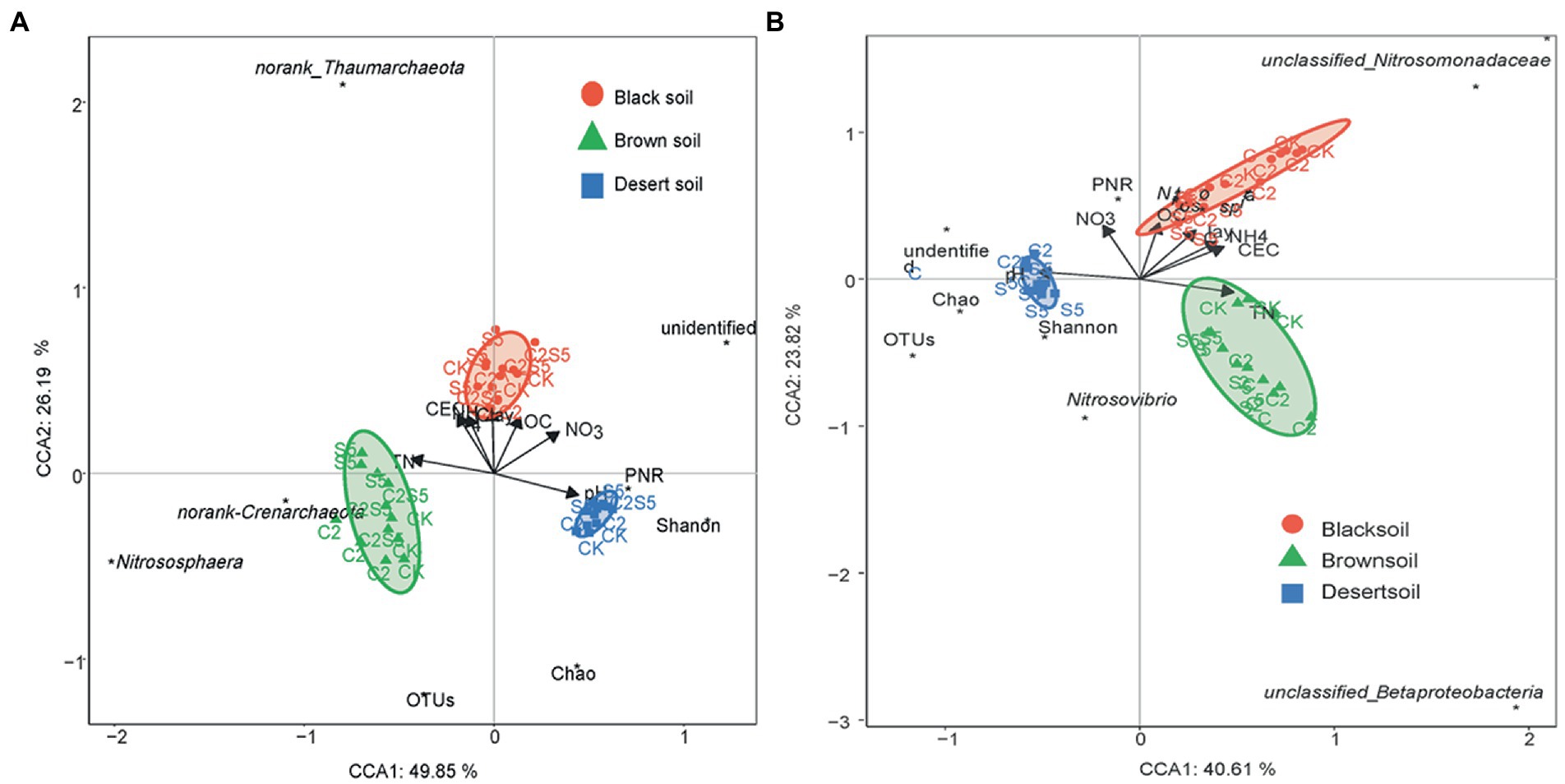
Figure 5. Canonical plots of discriminant analysis on soil parameters and all measured biomarkers of the ammonia-oxidizing process (A, for AOA groups; B, for AOB groups; CK, control treatment; S5, 5 μmol/kg SDZ treatment; C2, 200 mg/kg Cu2+ treatment, and C2S5, 5 μmol/kg SDZ + 200 mg/kg Cu2+ treatment).
4.2. Combined effect of SDZ and Cu on ammonia-oxidizing microorganisms across soils
Compared to the single exposure to the antibiotic, their combination with heavy metals could cause a higher inhibition of the abundance and function of ammonia-oxidizing microorganisms (Xu et al., 2016; Wang X. et al., 2019). In the present study, to the combined exposure of SDZ and Cu, a consistent inhibition on PNR was found in three tested soils, but a variable interaction was also found across three tested soils (Figure 1A). It was previously found that the functional redundancy of soil PNR also could overspread the toxicity of contaminants on ammonia-oxidizing community (Ruyters et al., 2013). To the combined exposure of SDZ and Cu in three tested soils, no consistent correspondence was also found considering the consistent inhibition of soil PNR (Figure 1A) versus the diverse variation of amoA genes abundance (Figures 1B–D) in the present study. In addition, no consistent correspondence was also found in community abundance, diversity, and the specific genus taxa of soil ammonia-oxidizing microorganisms across three tested soils to the combined exposure of SDZ and Cu (Figures 1 3; Table 2). The variable changes in the microbial community were previously investigated across different soils to antibiotic and heavy metal contamination (Grenni et al., 2018; Wang L. et al., 2019), and it was thought to be associated with the biogeographic distribution of soil ammonia-oxidizing microorganisms (Liu et al., 2019; Clark et al., 2021). However, the change observed in individual genus taxa of the AOA and AOB groups (Figure 3) was well corresponded to soil PNR (Figure 1A) across soils exposed to the combination of SDZ and Cu. It indicated that the combined effect of SDZ and Cu on soil nitrification might be dependent on specific genus taxa of ammonia-oxidizing microorganisms (Liu et al., 2015; Wei et al., 2018). In another word, it might be speculated that the functional redundancy of soil PNR and the corresponding specific taxa of AOA or AOB modulate the combined toxic effect of SDZ and Cu on the biological turnover of soil nitrogen (Li et al., 2021). The aforementioned diverse results also indicate that a multi-microbial targets analysis, including function, microbial communities, and the specific phylotype, is necessary to better understand the risk of joint pollution to the soil environment (Teng and Zhou, 2018). In addition, the division of soil treatments based on all the biological values under the combined stress of SDZ and Cu also closely corresponded to distribution derived from soil properties (Figure 4), especially dependent on soil pH and TN (Figure 5). It also indicates that the differentiations in the combined impact of SDZ and Cu on ammonia-oxidizing process are greatly attributed to variation in soil properties (Bernier and Surette, 2013; Xian et al., 2015; Grenni et al., 2018).
5. Conclusion
In summary, the single application of SDZ caused a consistent decrease in the soil nitrification rate of three tested soils, but it was more serious only in Brown soils when combined with Cu. In contrast, the single or combined impact of SDZ and Cu was variable in amoA gene abundance and community diversity of ammonia-oxidizing microorganisms across different soils. However, similar correspondences occurred between the PNR and the specific genus taxa of ammonia-oxidizing microorganisms among different soils exposed to SDZ and Cu singly or simultaneously. Finally, the specific phylotype of AOA or AOB plays an important role in modulating the response of soil nitrification to the contamination stress of SDZ and Cu. Thus, it should propose an integrative assessment of soil properties and multiple microbial targets in soil contamination management.
Data availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request. All the DNA sequence data in this manuscript are deposited in the GenBank databases, accession number KFXR00000000.
Author contributions
GH: data curation and original draft preparation. ZGW: English improvement and modification. JL, GW and FR: data curation and experiment investigation. YX, KL, and ML: visualization and editing. AL: conceptualization and methodology. HL and FW: reviewing and validation. All authors contributed to the article and approved the submitted version.
Funding
The study was supported by the National Natural Science Foundation of China (NSFC) under Grant Nos. 42077129, 41877122, 41671322, and 42177403 and the Shandong Natural Science Foundation under Grant Nos. ZR2020ZD19 and ZR2022YQ34.
Acknowledgments
The soil sampling was kindly helped by Mingjing Qu and her colleagues from the Provincial Academy of Agricultural Sciences of Jilin, and Xinjiang, China. The authors would like to thank Lianchuan BioTechnology Co., Ltd. for their help in high-throughput DNA sequencing data analysis.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpsyt.2023.1144873/full#supplementary-material
References
An, J., Chen, H., Wei, S., and Gu, J. (2015). Antibiotic contamination in animal manure, soil, and sewage sludge in Shenyang, Northeast China. Environ. Earth Sci. 74, 5077–5086. doi: 10.1007/s12665-015-4528-y
Azziz, G., Trasante, T., Monza, J., and Irisarri, P. (2016). The effect of soil type, rice cultivar and water management on ammonia-oxidizing archaea and bacteria populations. Appl. Soil Ecol. 100, 8–17. doi: 10.1016/j.apsoil.2015.11.009
Bernier, S. P., and Surette, M. G. (2013). Concentration-dependent activity of antibiotics in natural environments. Front. Microbiol. 4:20. doi: 10.3389/fmicb.2013.00020
Clark, I. M., Hughes, D. J., Fu, Q., Abadie, M., and Hirsch, P. R. (2021). Metagenomic approaches reveal differences in genetic diversity and relative abundance of nitrifying bacteria and archaea in contrasting soils. Sci. Rep. 11:15905. doi: 10.1038/s41598-021-95100-9
Cycoń, M., Borymski, S., Żołnierczyk, B., and Piotrowska-Seget, Z. (2016). Variable effects of non-steroidal anti-inflammatory drugs (nsaids) on selected biochemical processes mediated by soil microorganisms. Front. Microbiol. 7:1969. doi: 10.3389/fmicb.2016.01969
Cycoń, M., Mrozik, A., and Piotrowska-Seget, Z. (2019). Antibiotics in the soil environment–degradation and their impact on microbial activity and diversity. Front. Microbiol. 10:338. doi: 10.3389/fmicb.2019.00338
Ding, C., and He, J. (2010). Effect of antibiotics in the environment on microbial populations. Eur. J. Appl. Microbiol. Biotechnol. 87, 925–941. doi: 10.1007/s00253-010-2649-5
Falkowski, P. G., Fenchel, T., and Delong, E. (2008). The Microbial Engines That Drive Earth's Biogeochemical Cycles. Science 320, 1034–1039. doi: 10.1126/science.1153213
Frková, Z., Vystavna, Y., Koubová, A., Kotas, P., Grabicová, K., Grabic, R., et al. (2020). Microbial responses to selected pharmaceuticals in agricultural soils: microcosm study on the roles of soil, treatment and time. Soil Biol. Biochem. 149:107924. doi: 10.1016/j.soilbio.2020.107924
Gil-Sotres, F., Trasar-Cepeda, C., Leirós, M. C., and Seoane, S. (2005). Different approaches to evaluating soil quality using biochemical properties. Soil Biol. Biochem. 37, 877–887. doi: 10.1016/j.soilbio.2004.10.003
Grenni, P., Ancona, V., and Barra Caracciolo, A. (2018). Ecological effects of antibiotics on natural ecosystems: A review. Microchem. J. 136, 25–39. doi: 10.1016/j.microc.2017.02.006
Griffiths, R. I., Thomson, B. C., James, P., Bell, T., Bailey, M., and Whiteley, A. S. (2011). The bacterial biogeography of British soils. Environ. Microbiol. 13, 1642–1654. doi: 10.1111/j.1462-2920.2011.02480.x
Guo, T., Lou, C., Zhai, W., Tang, X., Hashmi, M. Z., Murtaza, R., et al. (2018). Increased occurrence of heavy metals, antibiotics and resistance genes in surface soil after long-term application of manure. Sci. Total Environ. 635, 995–1003. doi: 10.1016/j.scitotenv.2018.04.194
Hammesfahr, U., Bierl, R., and Thiele-Bruhn, S. (2011). Combined effects of the antibiotic sulfadiazine and liquid manure on the soil microbial-community structure and functions. J. Plant Nutr. Soil Sci. 174, 614–623. doi: 10.1002/jpln.201000322
Hu, X., Zhang, Y., Luo, J., Wang, T., Lian, H., and Ding, Z. (2011). Bioaccessibility and health risk of arsenic, mercury and other metals in urban street dusts from a mega-city, Nanjing, China. Environ. Pollut. 159, 1215–1221. doi: 10.1016/j.envpol.2011.01.037
Katipoglu-Yazan, T., Merlin, C., Pons, M. N., Ubay-Cokgor, E., and Orhon, D. (2016). Chronic impact of sulfamethoxazole on the metabolic activity and composition of enriched nitrifying microbial culture. Water Res. 100, 546–555. doi: 10.1016/j.watres.2016.05.043
Ke, X., Angel, R., Lu, Y., and Conrad, R. (2013). Niche differentiation of ammonia oxidizers and nitrite oxidizers in rice paddy soil. Environ. Microbiol. 15, 2275–2292. doi: 10.1111/1462-2920.12098
Kodešová, R., Chroňáková, A., Grabicová, K., Kočárek, M., Schmidtová, Z., Frková, Z., et al. (2020). How microbial community composition, sorption and simultaneous application of six pharmaceuticals affect their dissipation in soils. Sci. Total Environ. 746:141134. doi: 10.1016/j.scitotenv.2020.141134
Kotzerke, A., Sharma, S., Schauss, K., Heuer, H., Thiele-Bruhn, S., Smalla, K., et al. (2008). Alterations in soil microbial activity and N-transformation processes due to sulfadiazine loads in pig-manure. Environ. Pollut. 153, 315–322. doi: 10.1016/j.envpol.2007.08.020
Kumar, U., Mishra, V. N., and Kumar, N. (2018). Comparative study of soil quality indicators for rice-based cropping system in Vertisols of central plain of Chhattisgarh. Asian J. Soil Sci. 13, 136–147. doi: 10.15740/HAS/AJSS/13.2/136-147
Li, Y., Ge, Y., Wang, J., Shen, C., and Liu, Y. (2021). Functional redundancy and specific taxa modulate the contribution of prokaryotic diversity and composition to multifunctionality. Mol. Ecol. 30, 2915–2930. doi: 10.1111/mec.15935
Liao, Q., Li, M., Dong, Y., Shao, Y., Liu, X., Hu, X., et al. (2019b). Responses difference analysis of soil potential nitrification rates to Cu and sulfadiazine. Environ. Pollut. Bioavailab. 31, 219–225. doi: 10.1080/26395940.2019.1620132
Liao, Q., Li, M., Dong, Y., Wu, M., Meng, Z., Zhang, Q., et al. (2019a). Impacts of Cu and sulfadiazine on soil potential nitrification and diversity of ammonia-oxidizing archaea and bacteria. Environ. Pollut. Bioavailab. 31, 60–69. doi: 10.1080/26395940.2018.1564629
Liu, A., Cao, H., Yang, Y., Ma, X., and Liu, X. (2015). Combinational effects of sulfomethoxazole and copper on soil microbial community and function. Environ. Sci. Pollut. Res. 23, 4235–4241. doi: 10.1007/s11356-015-4892-x
Liu, A., Fang, D., Wang, C., Li, M., and Young, R. B. (2014). Recovery of soil nitrification after long-term zinc exposure and its co-tolerance to Cu in different soils. Environ. Sci. Pollut. Res. 22, 314–319. doi: 10.1007/s11356-014-3338-1
Liu, J., Yu, Z., Yao, Q., Sui, Y., Shi, Y., Chu, H., et al. (2019). Biogeographic distribution patterns of the archaeal communities across the black soil zone of Northeast China. Front. Microbiol. 10:23. doi: 10.3389/fmicb.2019.00023
Lu, R. K.. (1999). Analytical Methods of Soil Agrochemistry. China Agricultural Science and Technology Press, Beijing.
Ma, T., Pan, X., Chen, L., Liu, W., Christie, P., Luo, Y., et al. (2016). Effects of different concentrations and application frequencies of oxytetracycline on soil enzyme activities and microbial community diversity. Eur. J. Soil Biol. 76, 53–60. doi: 10.1016/j.ejsobi.2016.07.004
Qiao, M., Ying, G.-G., Singer, A. C., and Zhu, Y.-G. (2018). Review of antibiotic resistance in China and its environment. Environ. Int. 110, 160–172. doi: 10.1016/j.envint.2017.10.016
Quaik, S., Embrandiri, A., Ravindran, B., Hossain, K., Al-Dhabi, N. A., Arasu, M. V., et al. (2020). Veterinary antibiotics in animal manure and manure laden soil: Scenario and challenges in Asian countries. J. King Saud Univ. Sci. 32, 1300–1305. doi: 10.1016/j.jksus.2019.11.015
Rusk, J., Hamon, R., Stevens, D., and McLaughliny, M. J. (2004). Adaptation of soil biological nitrification to heavy metals. Environ. Sci. Technol. 38, 3092–3097. doi: 10.1021/es035278g
Ruyters, S., Springael, D., and Smolders, E. (2013). Recovery of soil ammonia oxidation after long-term zinc exposure is not related to the richness of the bacterial nitrifying community. Microb. Ecol. 66, 312–321. doi: 10.1007/s00248-013-0210-7
Sun, R., Myrold, D. D., Wang, D., Guo, X., and Chu, H. (2019). AOA and AOB communities respond differently to changes of soil pH under long-term fertilization. Soil Ecol. Lett. (China) 1, 126–135. doi: 10.1007/s42832-019-0016-8
Teng, Y., and Zhou, Q. (2018). Response of soil enzymes, functional bacterial groups, and microbial communities exposed to sudan I-IV. Ecotoxicol. Environ. Saf. 166, 328–335. doi: 10.1016/j.ecoenv.2018.09.102
Toth, J. D., Feng, Y., and Dou, Z. (2011). Veterinary antibiotics at environmentally relevant concentrations inhibit soil iron reduction and nitrification. Soil Biol. Biochem. 43, 2470–2472. doi: 10.1016/j.soilbio.2011.09.004
Tourna, M., Freitag, T. E., Nicol, G. W., and Prosser, J. I. (2008). Growth, activity and temperature responses of ammonia-oxidizing archaea and bacteria in soil microcosms. Environ. Microbiol. 10, 1357–1364. doi: 10.1111/j.1462-2920.2007.01563.x
Wang, X., Gao, P., Li, D., Liu, J., Yang, N., Gu, W., et al. (2019). Risk assessment for and microbial community changes in Farmland soil contaminated with heavy metals and metalloids. Ecotoxicol. Environ. Saf. 185:109685. doi: 10.1016/j.ecoenv.2019.109685
Wang, L., Xia, X., Zhang, W., Wang, J., Zhu, L., Wang, J., et al. (2019). Separate and joint eco-toxicological effects of sulfadimidine and copper on soil microbial biomasses and ammoxidation microorganisms abundances. Chemosphere 228, 556–564. doi: 10.1016/j.chemosphere.2019.04.165
Wang, B., Zhao, J., Guo, Z., Ma, J., Xu, H., and Jia, Z. J. T. I. J. (2015). Differential contributions of ammonia oxidizers and nitrite oxidizers to nitrification in four paddy soils. ISME J. 9, 1062–1075. doi: 10.1038/ismej.2014.194
Wei, Z., Wang, J., Zhu, L., Wang, J., and Zhu, G. (2018). Toxicity of enrofloxacin, copper and their interactions on soil microbial populations and ammonia-oxidizing archaea and bacteria. Sci. Rep. 8:5828. doi: 10.1038/s41598-018-24016-8
Xia, Z., Bai, E., Wang, Q., Gao, D., Zhou, J., Jiang, P., et al. (2016). Biogeographic distribution patterns of bacteria in typical Chinese forest soils. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.01106
Xian, Y., Wang, M., and Chen, W. (2015). Quantitative assessment on soil enzyme activities of heavy metal contaminated soils with various soil properties. Chemosphere 139, 604–608. doi: 10.1016/j.chemosphere.2014.12.060
Xing, B. S., and Jin, R. C. (2018). Inhibitory effects of heavy metals and antibiotics on nitrifying bacterial activities in mature partial nitritation. Chemosphere 200, 437–445. doi: 10.1016/j.chemosphere.2018.02.132
Xu, Y., Yu, W., Ma, Q., Wang, J., Zhou, H., and Jiang, C. (2016). The combined effect of sulfadiazine and copper on soil microbial activity and community structure. Ecotoxicol. Environ. Saf. 134, 43–52. doi: 10.1016/j.ecoenv.2016.06.041
Xu, Z., Zhang, T., Wang, S., and Wang, Z. (2020). Soil ph and c/n ratio determines spatial variations in soil microbial communities and enzymatic activities of the agricultural ecosystems in northeast china: jilin province case. Appl. Soil Ecol. 155:103629. doi: 10.1016/j.apsoil.2020.103629
Yan, L., Wang, G., Ai, S., Huo, Z., Wang, Y., Gu, J. D., et al. (2018). Abundance of ammonia-oxidizing bacteria and archaea under different ventilation strategies during cattle manure composting. J. Environ. Manag. 212, 375–383. doi: 10.1016/j.jenvman.2018.02.032
Yang, T., Lupwayi, N., Marc, S. A., Siddique, K., and Bainard, L. D. (2021). Anthropogenic drivers of soil microbial communities and impacts on soil biological functions in agroecosystems. Glob. Ecol. Conserv. 27:e01521. doi: 10.1016/j.gecco.2021.e01521
Yang, R., Xia, X., Wang, J., Zhu, L., Wang, J., Ahmad, Z., et al. (2020). Dose and time-dependent response of single and combined artificial contamination of sulfamethazine and copper on soil enzymatic activities. Chemosphere 250:126161. doi: 10.1016/j.chemosphere.2020.126161
Zhang, X., Zhou, Y., Yu, B., Zhang, N., Wang, L., Fu, H., et al. (2017). Effect of copper oxide nanoparticles on the ammonia removal and microbial community of partial nitrification process. Chem. Eng. J. 328, 152–158. doi: 10.1016/j.cej.2017.07.028
Zhou, Z., Shi, X., Zheng, Y., Qin, Z., Xie, D., Li, Z., et al. (2014). Abundance and community structure of ammonia-oxidizing bacteria and archaea in purple soil under long-term fertilization. Eur. J. Soil Biol. 60, 24–33. doi: 10.1016/j.ejsobi.2013.10.003
Keywords: sulfadiazine, copper, toxicity, soil potential nitrification, AOA, AOB, soil type
Citation: Hou G, Wazir ZG, Liu J, Wang G, Rong F, Xu Y, Li M, Liu K, Liu A, Liu H and Wang F (2023) Effects of sulfadiazine and Cu on soil potential nitrification and ammonia-oxidizing archaea and bacteria communities across different soils. Front. Microbiol. 14:1153199. doi: 10.3389/fmicb.2023.1153199
Edited by:
Yaying Li, Institute of Urban Environment (CAS), ChinaReviewed by:
Peng Zhang, Nankai University, ChinaXinjian Zhang, Shandong Academy of Sciences, China
Copyright © 2023 Hou, Wazir, Liu, Wang, Rong, Xu, Li, Liu, Liu, Liu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aijv Liu, YWlqdmxpdUBzZHV0LmVkdS5jbg==; Hongliang Liu, ZW1pY3JvYmVAc2R1dC5lZHUuY24=
 Guoqin Hou
Guoqin Hou Zafran Gul Wazir1
Zafran Gul Wazir1 Jing Liu
Jing Liu Fangxu Rong
Fangxu Rong Aijv Liu
Aijv Liu Hongliang Liu
Hongliang Liu Fayuan Wang
Fayuan Wang