- 1Institute of Environmental Biotechnology, Graz University of Technology, Graz, Austria
- 2KWS SAAT SE & Co. KGaA, Einbeck, Germany
- 3Microbiome Biotechnology Department, Leibniz-Institute for Agricultural Engineering and Bioeconomy (ATB), Potsdam, Germany
- 4Institute for Biochemistry and Biology, University of Potsdam, Potsdam, Germany
The importance of crop-associated microbiomes for the health and field performance of plants has been demonstrated in the last decades. Sugar beet is the most important source of sucrose in temperate climates, and—as a root crop—yield heavily depends on genetics as well as on the soil and rhizosphere microbiomes. Bacteria, fungi, and archaea are found in all organs and life stages of the plant, and research on sugar beet microbiomes contributed to our understanding of the plant microbiome in general, especially of microbiome-based control strategies against phytopathogens. Attempts to make sugar beet cultivation more sustainable are increasing, raising the interest in biocontrol of plant pathogens and pests, biofertilization and –stimulation as well as microbiome-assisted breeding. This review first summarizes already achieved results on sugar beet-associated microbiomes and their unique traits, correlating to their physical, chemical, and biological peculiarities. Temporal and spatial microbiome dynamics during sugar beet ontogenesis are discussed, emphasizing the rhizosphere formation and highlighting knowledge gaps. Secondly, potential or already tested biocontrol agents and application strategies are discussed, providing an overview of how microbiome-based sugar beet farming could be performed in the future. Thus, this review is intended as a reference and baseline for further sugar beet-microbiome research, aiming to promote investigations in rhizosphere modulation-based biocontrol options.
1. Introduction
The holobiont concept (Zilber-Rosenberg and Rosenberg, 2008) changed the view on microbes in many scientific disciplines. It states that practically all multicellular lifeforms are inhabited, depending on—or at least are affected by—the interplay with microbial life. The collective genome of plant-associated microbiota exceeds the host genome in both size and number of functions by far and is thus referred to as its second genome (Berendsen et al., 2012; de la Fuente Cant et al., 2020). Given the importance of plant-associated microbes for the health, vigor, and resilience of their host, the microbiome of plants and its modulation is a potential key factor for crop management and crop development in the future (Berg et al., 2015, 2021; Mendes and Raaijmakers, 2015).
Sugar beet (Beta vulgaris ssp. vulgaris, L.) is the most important regional source of sucrose in moderate climates of the northern hemisphere. Its biomass production is ranked eighth amongst the most produced field crops worldwide (FAOSTAT, 2022). Sugar beets are biennial, meaning that flowers and seeds are produced in the second year. Since flowering detracts sucrose from taproots, sugar beets are harvested annually. The wild ancestor of all beet crops is the sea beet (Beta maritima L.), a native plant still frequently found on European coastlines. Sugar beet thrives on most soil types, as long as pH is near neutral, easing its geographically widespread cultivation (Draycott, 2003). In contrast to many other crops, the breeding of sugar beet out of the Silesian Beet happened in times when the basics of genetics were understood. Therefore, its development and breeding trends over the decades are comparably well documented (Panella and Lewellen, 2007). Early sugar beet cultivars were bred in Northern Europe, a region with a non-humid, temperate climate and low pest and disease pressure. When these cultivars were planted in other regions, the yield was severely decimated by pests and pathogens (Panella and Lewellen, 2007). Sugar beet was intensively studied regarding physiology, anatomy, chemical, biochemical constitution, genomic traits, nutrient requirements, and convenient agricultural practices to optimize yield in the last 150 years, and was first genome sequenced in 2014 (Dohm et al., 2014). Still, leaf pathogens, root and storage rots, and microbes interfering with sucrose extractions illustrate the importance of sugar beet-associated microbial communities for both plant health and yield. All these mentioned facts make sugar beet an interesting model plant for microbiome research.
Despite the widespread cultivation of sugar beet, our knowledge in sugar beet microbiomes and microbiome-based strategies in future agricultural systems have not reached their full potential thus far. To fully exploit this potential for crop protection and plant growth promotion (PGP), a deep and holistic understanding of both the plant itself and the environment-plant interactions is crucial. Since the rhizosphere is the primary soil-plant interface, we have to especially emphasize the establishment, formation, and dynamics of its microbiome in this context. We hereby try to connect current knowledge about sugar beet-associated microbial communities to their physical, chemical, and biological context, namely the specific traits of the host plant. We aim to describe the sugar beet holobiont as defined by Berg et al. (2020), as the entirety of the microbial community members and its “theater of activity”. In the first section of this review, we will provide an overview of the current knowledge on sugar beet microbiome to be considered in experimental setups of future studies, highlight knowledge gaps, and discuss the sugar beet holobiont following its ontology from seed to postharvest roots. The second section summarizes potential or already tested biocontrol agents and their natural occurrence in the plant host and presents the current application strategies for microbiome-based agricultural practices.
2. Part I: Microbial journeys on and in sugar beets: from seed to beet
2.1. Seed endophytes as source for rhizosphere microbes
Plant holobionts represent dynamic systems where the plant and microbiota influence each other and both develop over time. During their life cycle, starting with seed germination, plants provide several microhabitats (root: rhizosphere and root endosphere; leaves and stems: phyllosphere; flowers: anthosphere; fruits: carposphere; seeds: spermosphere) with very different physicochemical properties. Consequently, these plant microhabitats harbor specific microbiota connected by endophytic communities (Hardoim et al., 2015), that may differ on very small spatial scales (Ottesen et al., 2013; Wassermann et al., 2019b; Kusstatscher et al., 2020). Endophytes may either migrate through the vascular system of plants (James et al., 2002; Compant et al., 2005, 2021) or move through the apoplast (Gasser et al., 2011; Compant et al., 2021), the latter requiring cell wall-degrading enzymes (Dong et al., 1994; James et al., 2002; Liu et al., 2017). In homologous organs, endophytic microbiomes partially differ according to phylogeny (Abdelfattah et al., 2021), even up to cultivar-specific differences in seeds (Rybakova et al., 2017; Wolfgang et al., 2020).
Seed tissues usually undergo serious physiological changes during maturation, including enrichment of starch and dehydration. Therefore, seed endophytes often require additional characteristics and adaptions, e.g., tolerance toward high osmotic pressure, amongst other typical traits (Truyens et al., 2015). Seed endophytes are difficult to discover because the majority are in a dormant, non-cultivable stage. Using microbial community fingerprinting methods developed in the 1990's, mainly based on PCRs amplifying 16Sr DNA genes, seed microbiota could be analyzed and were found to be surprisingly diverse (Berg and Raaijmakers, 2018). Nevertheless, the origin of seed endophytes is still a matter of debate, since it appears that plant taxa differ in to what extent endophytes are either horizontally acquired or vertically transmitted. Vertically transmitted seed endophytes represent a core community in seeds (Truyens et al., 2015; Nelson, 2018; Shahzad et al., 2018). However, seed endo- and ectophytes play an important role in early plant development, including rhizosphere development (Berg and Raaijmakers, 2018).
In sugar beet, an influence of both the genotype and the environment on microbial seed communities was reported, although a core community is conserved to some extent (Wolfgang et al., 2020). This microbial inheritance was equally observed in other crops, e.g., Arabidopsis (Truyens et al., 2013), maize (Johnston-Monje et al., 2016), oak (Abdelfattah et al., 2021; Fort et al., 2021), beans (Klaedtke et al., 2016) and tomato (Bergna et al., 2018). Microbial inheritance represents the basis of the phylosymbiosis concept (Lim and Bordenstein, 2020). Dent et al. (2004) were the first to use culture-independent PCR-DGGE for seed community profiling in sugar beet. They discovered both bacterial and fungal DNA in the seed coat (=fruit) that showed specific band patterns depending on seed acreage, which was correlated with differing germination rates under field conditions. Bacterial seed communities appeared to be complex, while fungal communities seemed to be species-poor (Dent et al., 2004; Spanner et al., 2021). A commonality in bacterial sugar beet seed communities with other plant species is the high abundance of Pseudomonas and Pantoea (Truyens et al., 2015; Nelson, 2018; Wassermann et al., 2019a). Other genera contribute to the bacterial seed community to various extents, including Paenibacillus, Sphingomonas, Curtobacterium, Massilia, Methylobacterium, Saccharibacillus, and Kosakonia (Wolfgang et al., 2020). Some fungal taxa known for phytopathogenic traits, e.g., Cercospora, Fusarium, and Alternaria, can establish in seeds via the xylem sap flow (Spanner et al., 2021), with negative implications for the next sugar beet generation. Archaea represent 0–1.1% relative abundance and mainly comprise Woesearchaeia, indicating a minor role of Archaea in seeds (Wolfgang et al., 2020). In addition to genetic and epigenetic information of the plant itself, seed endophyte communities can thus be interpreted as a third layer of inheritable information about the direct environment of the host plant, which is directly or indirectly provided and modified by the parental plant to the next generation of plants. This further indicates that the sugar beet seed community is modifiable by the substrate of the mother plant, which may have implications for sugar beet breeding or plant pathogen control.
The process of germination in sugar beet is epigeal (Milford, 2006). Germination is generally considered the most vulnerable life stage, where plants are highly susceptible to abiotic and biotic stressors (Hegarty, 1977). Interaction dynamics between seedling, seed-associated microbiomes, and soil microbiomes during seed germination, including the establishment and development of rhizosphere communities, may constitute the most decisive life stage for further plant health (Nelson, 2018). Dominating taxa in seeds also dominate in soilless germinated sugar beet seedling roots, namely Kosakonia, Methylobacterium, Pantoea, and Pseudomonas (Wolfgang et al., 2020). According to the mentioned study, ~63–83% of seed endophytes survive germination and colonize developing roots. In vitro roots show lower alpha diversity, a community shift, and a less balanced microbiome composition than seeds (Wolfgang et al., 2020), an effect as well observed in Brassica plants (Barret et al., 2015), maize (Johnston-Monje et al., 2016) and wheat (Huang et al., 2016). This indicates a first selection of vertically transmitted endophytes in germinating roots, although it is yet unassessed whether this process is stochastic or specific. However, the instability of the seedling community indicates a “reshuffling” of the seed community, potentially creating new niches for soil-derived microbes (Figure 1A). In this life stage, seed endophytes have several advantages compared to soil-derived microbiota to further colonize the seedling. Firstly, colonization of the newly germinated seedlings is more rapid, since they already inhabit the seed and do not have to compete for niches and nutrients yet. Secondly, they are already adapted to live inside plant tissue, and thirdly, they are protected to an extent from environmental stressors by host tissue (Hallmann and Berg, 2007; Kaga et al., 2009; Hardoim et al., 2012). Altogether, seed microbes provide ideal starter cultures for seedlings, while microorganisms acquired from soil allow an adaptation to local conditions (Bergna et al., 2018); both sources ensure an adapted microbiota assembly for the whole plant life cycle.
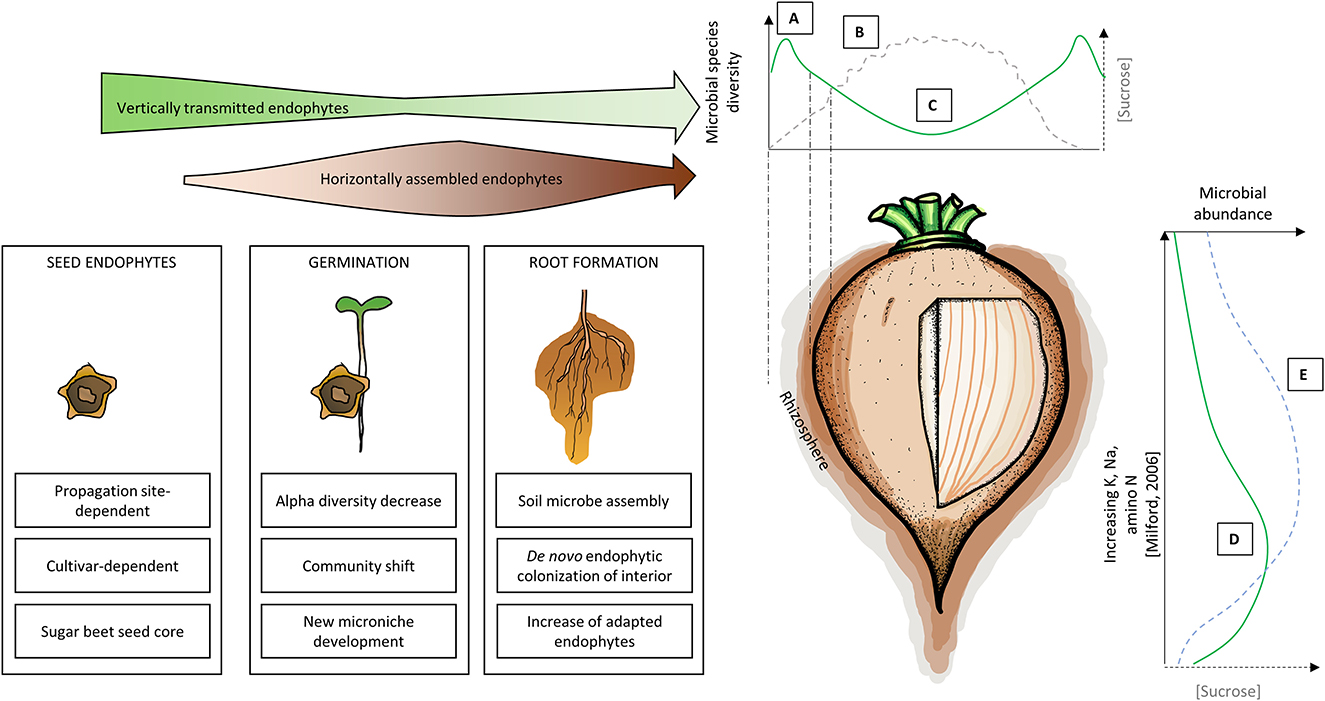
Figure 1. Simplified temporal (Left) and spatial (Right) holobiont model of sugar beet taproot. The arrow width indicates the relative importance of vertically and horizontally assembled endophytes (Top Left). A: Root exudation and/or endophyte release leads to an increase in measured diversity in taproot-associated rhizosphere communities (Zachow et al., 2014; Cardinale et al., 2020; Wolfgang et al., 2020). CFU number in the peel can exceed the CFU number in the rhizosphere (Okazaki et al., 2014). B: Relative sugar content increases toward the center, higher in proximity to vascular bundles. Sucrose further decreases with increasing distance to the secondary cambia (Milford, 2006; Hoffmann and Kenter, 2018). C: Diversity decreases toward the center, while the relative abundance of copiotrophic bacteria increases (Lilley et al., 1996; Okazaki et al., 2014). D: Microbial abundance is highest in the root elongation zone near the root tip, with a high relative abundance of exudate responders, e.g., Variovorax and Pseudomonas (Jacobs et al., 1985; Lübeck et al., 2000; Shi et al., 2009a). E: The sugar content of beet tissue is highest in lower taproot (Milford, 2006).
2.2. Microbial assembly and dynamics in the sugar beet rhizosphere
The rhizosphere is the critical soil-plant interface for resource exchange and the interplay between the plant host and soil microbiota (Weller et al., 2002; Philippot et al., 2013; Pantigoso et al., 2022). In the sugar beet rhizosphere, the number of cultivable bacteria and fungi is approximately 108 and 2*105 CFUs per gram fresh weight, respectively (Zachow et al., 2008). Proteobacteria, Acidobacteria, and Actinobacteria dominate the root community of 14d-old sugar beet cultivars. The archaeal family Nitrososphaeraceae is steadily represented but derived from soil (Wolfgang et al., 2020). Protists play a pivotal role in shaping sugar beet rhizosphere microbiomes (Bazany et al., 2022), but are rarely investigated on metagenomic levels. Interestingly, sugar beet rhizospheres were repeatedly reported to display a comparable (Mendes et al., 2011), or even higher bacterial alpha diversity than surrounding bulk soil in the early seedling stages (Zachow et al., 2014; Wolfgang et al., 2020), but also in the rhizosphere of pre-harvest taproots (Cardinale et al., 2020), while also significant lower rhizosphere alpha diversity compared to bulk soil was reported (Cui et al., 2022). Rhizosphere communities in vascular plants are usually less diverse than soil communities (Berendsen et al., 2012). This so-called “rhizosphere effect” is caused by specific root exudates that enrich or repel a specific fraction of the given soil community (Houlden et al., 2008; Berg and Smalla, 2009; de la Fuente Cant et al., 2020). There are several non-exclusive possible explanations for this deviant phenomenon in sugar beet that can be categorized into methodological and ecological-physiological explanations. Methodological explanations include (I) a higher concentration of clay in bulk soil compared to the rhizosphere soil, which could lead to a decreased DNA yield during DNA extraction (Novinscak and Filion, 2011). However, this should as well affect the rhizosphere of other crops grown in the same soil; (II) sugar beet root exudates enhance the growth of bacteria that are beneath the detection threshold in bulk soil or seeds. Ecological explanations for this phenomenon may include (III) soil bacteria and seed endophytes belonging to different genotypes both colonizing the rhizosphere, leading to an overall increased genetic diversity in the rhizosphere; (IV) a phylogenetic legacy. Sea beet (Beta maritima) as the progenitor of all beets is adapted to grow in sea drift lines, where abiotic selective forces like salinity, temperature, and UV exposure are more critical for plant survival than biotic stressors. Therefore, root exudates of beet seedlings may hardly contain compounds repellent to soil microbes and enrich microbes rather unspecifically. Given a temporal change in chemical root exudate composition, selective assembly of soil microbes may appear later in the life cycle once the seedling is established in its habitat; (V) the abundance of microbial responders toward sugar beet root exudates differ depending on the soil community. Since soils used in different studies differ in both biotic and abiotic properties, some soil microbiomes may be simply more responsive upon compounds exuded into the rhizosphere; (VI) It is a side effect of simple sugar content in root exudates, supporting trophic interactions between microbes. Sugar beet rhizospheres display a high abundance of polyhydroxyalkanoate (PHA) producing bacteria like Burkholderia, Ensifer, Erwinia, Lysobacter, Pantoea, Pseudomonas, and Variovorax (Gasser et al., 2009). This indicates an adaption of the bacterial community to excess carbon sources in combination with limiting other nutrient sources in the sugar beet rhizosphere (Lee, 1996). However, at the current stage, further meta-analyses or standardized longitudinal studies are needed to either support or falsify these hypotheses.
Appropriate nitrogen fertilization management in sugar beet is crucial for optimizing net sucrose yield. Available nitrogen is usually the plant growth-limiting nutrient. Although root biomass is positively correlated with N fertilization, sugar concentration in roots negatively correlates with N fertilization, resulting in an optimum curve regarding sucrose yield dependent on N application rate (Milford, 2006; Hergert, 2010). Consequently, fertilization regimes usually aim to let sugar beets become N-deficient several weeks before harvest to increase processing efficacy (Stevanato et al., 2016). Fertilization affects bacterial soil and rhizosphere microbiome community composition, including lowering the abundance of bacterial and archaeal nitrogen fixation-associated gene transcripts. Reduced nitrogen cycle-related gene abundance in sugar beet was observed when using chemical fertilization, alone or in combination with manuring (Cardinale et al., 2020). An indication for reduced nitrogen fixation with a concurrent increase of nitrification and aerobic ammonia oxidation in sugar beet rhizospheres was also found in a long-term crop rotation field (Du et al., 2022a). Archaeal nitrification in the rhizosphere further positively correlates with increasing soil depth (Stevanato et al., 2016).
Microbial rhizosphere communities shift during the maturation of the host plant. Sugar beet fibrous roots can reach up to 3 m depth (Stevanato et al., 2010), with the highest root length density at a depth of around 1 m (Stevanato et al., 2010, 2016). With the increasing length of the taproot, a vertical gradient of microbial diversity, increasing with depth, establishes alongside the fine roots under field conditions (Stevanato et al., 2016). Proteobacteria are enriched compared to bulk soil in pre-harvest taproot rhizospheres, Acidobacteria, Actinobacteria, Bacteroidetes, and Gemmatimonadetes were mentioned as codominant bacterial phyla (Houlden et al., 2008; Kusstatscher et al., 2019a; Cardinale et al., 2020; Huang et al., 2020; Du et al., 2022a). The fungal rhizosphere community of pre-harvest taproots is dominated by Guehomyces, Humicola, Mortierella, Plectosphaerella, and Vishniacozyma (Kusstatscher et al., 2019b; Du et al., 2022b), but is most likely less conserved than bacterial communities (Houlden et al., 2008).
Bacterial rhizosphere communities in sugar beet are dynamic across ontogenesis compared to other crops. Still, an increasingly crop-specific rhizosphere community establish over time (Houlden et al., 2008). As the physical, chemical, and biological activity of a plant itself shapes its surroundings even past its lifespan, continuous cropping of sugar beet severely affects soil traits. Continuous cropping usually leads to detrimental effects on yield and soil properties due to an imbalance in soil nutrients, autotoxicity of root exudates, and shifts in microbial community composition (Huang et al., 2020; Pervaiz et al., 2020; Cui et al., 2022). Effects in sugar beet include potassium depletion in soil (Samadi, 2012), rapid crop yield loss mainly due to plant disease intensification (Stevanato et al., 2019), and accumulation of potentially autotoxic root exudates (Huang et al., 2021). In fields where sugar beet was continuously grown for 0 (rotation cropping), 1, 5, and 30 years, bacterial and fungal alpha diversity was already lower after 1 year of sugar beet cultivation compared to planting in virgin soil but increased in year 30. This diversity increase however was attributable to potentially phytopathogenic species (Acidobacteria, Alternaria, Fusarium, Cladosporium) while beneficial genera (Bacillus, Actinobacteria, Pseudomonas) declined. The genera Sphingomonas, Haliangium, Gaiella, and Lysobacter were positively correlated, while Lysinibacillus and Sphingobacterium negatively correlated with sugar beet cropping time (Huang et al., 2020; Du et al., 2022a,b). It appears that long-time rotation with other crops does not fully restore the negative effects on sugar beet biomass, health parameters, and yield as well as on the community shift in bacterial (Cui et al., 2022; Du et al., 2022a) and fungal (Cui et al., 2022; Du et al., 2022b) rhizosphere microbiome compared to sugar beets grown in virgin soil. Strategies to re-establish the microbiome-related beneficial effects on plant growth found in virgin soil, while increasing the sustainability of the crop is a challenge of future sugar beet research.
2.2.1. Root exudates fine-tune microbial communities in sugar beet rhizosphere
Root exudates are the main mechanism plants use for altering the microbial communities in their proximity while surrounding soil is the main source of diversity (Bais et al., 2006; Pantigoso et al., 2022). Furthermore, plant root exudation is used for recruiting pathogen antagonists from the environment (Cook et al., 1995; Mendes et al., 2011; Carrión et al., 2019). This assembly however may be specific to host traits and phylogeny. Transcriptomic profiling of Ps. aeruginosa-exposed root exudates of different sugar beet cultivars showed even cultivar-dependent responses (Mark et al., 2005). Additionally, the colonization patterns of sugar beet rhizosphere are strain-specific (Zachow et al., 2010); chemical root exudate composition shows spatially distinct patterns and may change when the root matures (Badri and Vivanco, 2009; Sasse et al., 2018) or when the root is exposed to nutrient deficiency (Khorassani et al., 2011). Over 200 compounds were measured in sugar beet root exudates, with salicylic acid and especially citramalic acid playing important roles in phosphor solubilization from the soil. Citramalic acid is rarely observed in plants and may even be produced by sugar beet-associated microbes (Khorassani et al., 2011). Root exudate chemical composition in sugar beet further appears to prevent close associations with arbuscular mycorrhizal fungi (Steinkellner et al., 2007). The root zone directly behind the root tip is considered the most active section regarding root exudate secretion (Sasse et al., 2018). A remarkably high proportion of Variovorax paradoxus (45%) was found in the cultivable fraction of lower root sections, but not in other endosphere compartments (1–4% in peel and inner core) (Lilley et al., 1996). The community in lower root sections was distinctly different from the soil, rhizosphere, peel, and inner core. Variovorax increases sugar beet biomass and was found to preferentially colonize root hair and the rhizodermis (Natsagdorj et al., 2019). Lübeck et al. (2000) tracked a fluorescein-labeled Pseudomonas fluorescens strain over 20 days during the germination of sugar beet seedlings. Strain density was 10-fold decreased at the root base within 1–2 days, while it remained stable and active at lower root parts and the root tip, again indicating an important role of root exudates in the rhizosphere and subsequently root endosphere colonization.
To summarize: the sugar beet rhizosphere microbiome is dominated by Proteobacteria and a dynamic habitat that is heavily influenced by the substrate. Microbes that should act as biofertilizers, biostimulants, or biocontrol agents (BCA) have to establish themselves in the rhizosphere. Thus, they should be copiotrophic and competitive to establish in this nutrient-rich and contested microhabitat. However, with a better understanding of the ecology of keystone species, including trophic interactions and other synergisms with other present microbiota, this statement can be challenged.
2.3. Gradients inside of the sugar beet taproot determine endophytic communities
The taproot of sugar beet is the actual agricultural target organ, where the rhizosphere community dynamics have the biggest impact on further plant health. Historically, three phases of vegetative development of sugar beet are distinguished: shoot growth, tuberization (storage root growth), and “ripening” (accumulation of sucrose in the tuber) (Milford, 2006). Only recently this classification could be proved by a detailed investigation of key enzyme profiles for carbohydrate metabolism and phytohormones patterns (Jammer et al., 2020). Apart from dynamics over time, the taproot itself has several internal spatially related gradients that could partially explain the responses of microbial communities that were observed so far.
2.3.1. Root endophytes follow spatial and sucrose gradients
The sugar beet root simultaneously produces 6–7 concentric secondary cambia within the pericycle at an early seedling stage (Milford, 2006; Jammer et al., 2020). Sucrose is transported from leaves via vascular bundles to roots and then passively diffuses via the apoplast to the target root cell, which actively enriches sucrose in its vacuole (Wyse, 1979; Saftner et al., 1983). Therefore, sucrose concentration in the apoplast decreases with increasing distance to active secondary cambia, resulting in an increasing sucrose gradient from the outer to the inner core of taproots (Hoffmann and Kenter, 2018). This may explain why the cultivable fractions of endophytes are less diverse in the root core compared to the outer core, with a higher proportion of copiotrophs (Lilley et al., 1996; Okazaki et al., 2014). Okazaki et al. (2014) found Pseudoxanthomonas, Polaromonas, and Devosia in the outer core, and Devosia, Bosea, and Lysobacter to be dominant in the inner core. By contrast, another study (Lilley et al., 1996) found Bacillus and Flavobacterium to dominate the outer core, while Pseudomonas and Bacillus dominated the inner core. In the latter study, the number of cultivable endophytes of peel was even higher (1.09*108 g−1) than in rhizosphere (5.27*107 g−1), an effect that is not universal to root crops (Kõiv et al., 2019). However, a proximal-distal community shift was also observed using 16S rRNA amplicon sequencing of other root vegetables (Kõiv et al., 2019). Interestingly, the bacterial endosphere communities differ between taproots and secondary roots, with some taxa (e.g., Niastella spp.) being only found in secondary roots (Okazaki et al., 2021). In addition to a proximal-distal sucrose gradient, there is also a vertical gradient in the taproot: sucrose concentration is highest in the lower root part (16–20%) and decreases progressively from hypocotyl (ca. 15%) to lower (13%) and upper crown (7–9%). Decreasing sucrose concentration is accompanied by increased K, Na, and alpha-amino N (Milford, 2006). Interestingly, there is also a corresponding gradient of cultivable endophytes: the number of bacterial and fungal endophytes decrease from bottom to top (Jacobs et al., 1985; Shi et al., 2009a). That means the number of cultivable endophytes inside of taproots is correlated with apoplastic sucrose concentration as well as negatively correlated with spatial distance from soil (Figure 1B). However, since >90% of sugar beet-associated microbes appear to be non-cultivable (Lilley et al., 1996), these statements still need to be verified using cultivation-independent methods.
2.3.2. Root endophytes shift during host ontogenesis
At harvest, the taproot consists of ca. 77% water, 17–18% sucrose, 4–5% marc (=insoluble compounds), and 1–2% water-soluble but non-sucrose compounds. Both dry matter and sugar content change during plant development, especially in the first 100 days post-sowing (Hoffmann et al., 2005). Harvest date, genotype, fertilizer, plant density, soil type, and seasonal weather can cause the proportion of dry matter and sucrose to vary (Milford, 2006). Secondary roots may act as a possible entrance for root endophytes (Jacobs et al., 1985; Shi et al., 2009b), but secondary roots are not desired by farmers.
Comparable to rhizosphere communities (Houlden et al., 2008), endophytic communities respond to seasonal variations. The number of cultivable taproot endophytes has an optimum in early fall (Shi et al., 2009a). Furthermore, the inner core of taproots significantly changes toward a lower diversity and lower evenness during taproot maturation due to an increasing abundance of Bacillus (Lilley et al., 1996). Shi et al. investigated sugar beet endophytes in four different life stages using amplicon sequencing. Bacterial (Shi et al., 2014), fungal (Shi et al., 2016), and archaeal (Shi et al., 2015) microbiomes of two cultivars at two different locations differed mainly due to developmental stage, rather than to cultivar or location. Life stage-dependent differences in bacteria were attributable to Deltaproteobacteria, Nitrospirae, Acidobacteria, Gemmatimonadetes, Alphaproteobacteria, and Sphingobacteria (Shi et al., 2014). By investigating bacterial communities only in the secondary roots of sugar beet across a growing season, a seasonal shift between life stages (Ikeda et al., 2023). They found some taxa that decrease in abundance after the seedling stage (e.g., Janthinobacterium, Streptomyces), some taxa to have an optimum during tuber growth (e.g., Rhizobiaceae, Chitinophagaceae, Pseudomonas), while some taxa increase across the whole plant ontogenesis until reaching an optimum during sucrose accumulation stage (e.g., Steroidobacter). Bacterial alpha diversity in secondary root endophytes increased until the late tuber growth phase and subsequently remained relatively stable (Ikeda et al., 2023). Longitudinal changes in endophytic or rhizosphere microbiomes may be linked to pathogen tolerance. Susceptibility toward plant pathogens decreases with the increasing age of the plant, irrespective if the cultivar itself is regarded as tolerant or susceptible to the pathogen (Liu et al., 2019). Current results indicate domain-specific diversity optima in different plant life stages: while bacterial diversity was highest during tuber growth (Shi et al., 2014; Ikeda et al., 2023), fungal diversity was highest during rosette formation (Shi et al., 2016), and archaeal diversity (dominant: Methanococci, Crenarchaeota, and Thermoplasmata) was highest during sucrose accumulation (Shi et al., 2015). However, the results of the mentioned studies are based on comparable small numbers of biological replicates and were not compared to the natural microbiome dynamics in bulk soil. Therefore, it is yet unclear if the observed microbiome shifts are due to a change in plant physiology (e.g., root exudation) or the natural seasonal dynamics in the corresponding bulk soil microbiome. During sugar beet harvest time, studies reported a high abundance and diversity of fungi, which significantly decreased in decaying sugar beets (Kusstatscher et al., 2019a,b). Fungal taxa such as Candida and Penicillium together with the gram-positive bacterium Lactobacillus were the main storage rot disease indicators in the microbiome of decaying sugar beets (see also Section 3.2).
2.3.3. Nitrogen fertilization affects root endophytes
Sugar beet growing requires a comparably high amount of fertilizer, mainly nitrogen. Choosing an adequate amount of N fertilizer is crucial since too high concentrations of alpha-amino N in storage roots decrease the extractability of sucrose. Furthermore, nitrogen fertilizing increases leaf area (=photosynthetic potential) and size of root cells, but not the overall sugar content in the cell. Thus, bigger roots have a lower relative sugar concentration (Milford and Watson, 1971; Wyse, 1979; Hergert, 2010; Stevanato et al., 2019). In addition to the sucrose gradient in the apoplast depending on the distance to cambial structures, there is a reverse pattern regarding nitrogenous compounds and potassium ions, hypothesized to be important for sucrose transport to parenchymal cells (Milford, 2006). Similar to rhizosphere communities (Cardinale et al., 2020; see section 2.2), fertilizing affects the root endosphere bacterial community: in a bacterial metagenome study with chemical ammonium sulfate fertilization (Tsurumaru et al., 2015), the authors found several genes coding for PGP effects, but were not able to amplify known N2-fixating genes. In a follow-up study, a positive correlation between Firmicutes and nitrogen fertilization was observed, while Sphingomonadaceae was negatively correlated with nitrogen fertilization in the taproot endosphere (Okazaki et al., 2021). However, the endophyte community in the mentioned studies was dominated by Rhizobiales, mainly Bradyrhizobium, and Mesorhizobium (Tsurumaru et al., 2015; Okazaki et al., 2021). Given the importance of nitrogen sources for sugar beet growing, the importance of microbes in the soil nitrogen cycle, and the importance of root-internal alpha-amino N for post-harvest sucrose extraction, linking the right strategy for sugar beet nitrogen fertilization with the effects on and in different soil microbiomes needs further evaluation.
To summarize, the temporal and spatial dynamics during sugar beet development as well as the consequent changes in microbial communities highlight the importance of good sampling strategies for microbiome research. An elaborate and precise description of the considered plant organ or position on the beet, sampling procedure, plant developmental stage, available raw data, and metadata is crucial for the comparability of future results. Studies of different origins often report contradicting results, maybe owing to non-standardized sampling procedures, and have to be further evaluated.
2.4. Microbial assembly and dynamics in the sugar beet phyllosphere
The epiphyllosphere is generally thought to be a nutrient-scarce environment (Lindow and Brandl, 2003; Koskella, 2020), while the leaf interior is the main tissue of photosynthetic activity. Thus, epi- and endophyllospheres comprise very different conditions regarding UV exposure, temperature, nutrient availability, etc. Distinguishing between epi- and endophyllosphere is difficult because stomata and substomatal chambers directly connect the leaf interior and leaf exterior. Fluctuations in phyllosphere physicochemical conditions are thought to favor bacteria with versatile metabolic capabilities, which are often non-cultivable in the lab. However, pigmented microbes usually dominate leaf microbiomes (Lindow and Brandl, 2003; Bashir et al., 2022; Gouka et al., 2022).
Sugar beet phyllosphere microbiomes are subject to permanent fluctuations due to canopy dynamics, including leaf formation, senescence, and available habitat size in the apoplast due to changes in plant cell size and leaf size (Milford, 2006). In general, sugar beet plants produce new leaves as long as they remain in a vegetative state, and leaves usually wilt in the order they are developed. Leaves produced later in ontogenesis harbor higher bacterial endophyte diversity than earlier leaves, and the abundance of antagonists toward fungal phytopathogens is highest in a 90% leaf cover stage (Zachow et al., 2008).
Pseudomonas is the most frequently reported genus in sugar beet leaves. Using cultivation-dependent methods, other bacterial genera included Arthrobacter, Clavibacter, Enterobacter, Erwinia, and Klebsiella (Thompson et al., 1993, 1995b). Culture-independent analyses of sugar beet phyllosphere revealed the phyllosphere to contain a strikingly similar taxonomic composition compared to seed microbiomes. Pseudomonas is dominant (15–48% relative abundance), while Sphingomonas, Pantoea, Methylobacterium, and Massilia appear as codominant taxa (Wolfgang et al., 2020; Bertoldo et al., 2021; Della Lucia et al., 2021; Okazaki et al., 2021). This pattern is similar in both hydroponically grown and field-grown seedlings (Della Lucia et al., 2021). This points toward strong selective pressure in phyllosphere tissue for vertically transmittable endophytes even before seed formation, resulting in relatively conserved bacterial communities. With currently available data, it appears that phyllosphere bacterial diversity shifted and increased during domestication, while rhizosphere diversity decreased (see Section 2.6); sea beet phyllospheres are strongly dominated by Sphingomonas and Methylobacterium, together accounting for ca. 84% relative abundance (Broccanello et al., 2022). This however could as well refer to fertilization regimes, since bacterial phyllosphere microbiomes in sugar beet respond stronger upon NPK fertilization than in bacterial endosphere communities (Okazaki et al., 2021). Bacterial phyllosphere communities differ between petioles and laminae: Phyllobacterium, Methylobacterium, and Sphingomonas are higher-abundant, while Pseudomonas and Enterobacteriaceae are lower-abundant in petioles (Okazaki et al., 2021). Phyllosphere abundance of Methylobacterium and Mucilaginibacter was further linked to tolerance toward leaf pathogen Cercospora beticola in both sea beets and sugar beets (Broccanello et al., 2022). In contrast to bacteria, fungi isolated from sugar beet leaves usually belong to opportunistic plant pathogens (Alternaria, Aspergillus, Cladosporium, Acremonium, Fusarium, Penicillium, Phoma, Plectosphaerella, Pleospora, Pythium) but also saprobionts (Cryptococcus, Epicoccum, Sporobolomyces, Stemphylium) (Thompson et al., 1993; Larran et al., 2000; Shi et al., 2009a; Pusenkova et al., 2016). The number of cultivable filamentous fungi and yeasts increase in senescing leaves (Thompson et al., 1993), especially Alternaria alternata and Pleospora herbarum gradually increase during senescence (Larran et al., 2000). Senescent leaves and leaf debris will be degraded and incorporated into the soil together with their respective microbes, which is especially important for phytopathogen management.
2.5. Sugar beet anthosphere is a yet unexploited microhabitat
The formation of sugar beet flowers and seeds starts with bolting. Bolting usually requires vernalization but can also be induced spontaneously under cold weather conditions in the first year of growing (Owen et al., 1940; Milford, 2006). Bolting detracts sucrose from taproots, thus lowering yield. Sugar beet flowers are inconspicuous in appearance, which is typical for anemophilic plants. In the wild type, flowers merge into clusters, producing multigerm seedballs, which are fused corky fruits with up to five seeds (Lange et al., 1999; Panella and Lewellen, 2007). Seedballs are hypothesized to be an adaptation to hydrochory (Francis, 2006). Modern sugar beet cultivars display monogermity (seedball with one single embryo), which is a recessive maternal trait in beets. Consequently, modern sugar beet cultivars are usually 2- or 3-way hybrids (Panella and Lewellen, 2007). Hybridization and breeding history influence plant-microbiome interactions (see Section 2.6). Counterintuitively, offspring microbiomes can resemble rather the microbiomes of the pollen-providing parent than the seed-bearing mother plant, which was recently found in specific oilseed rape (Brassica napus L.) cultivars (Wassermann et al., 2022), but the exact implications of parent sex and respective microbiomes on offspring microbiomes has yet to be elucidated in sugar beet. Therefore, anthosphere microbiome dynamics may have yet unknown potential for microbiome-based breeding approaches.
Flowering lasts 1 month or longer, and sugar beet pollen can be transported across large distances by wind (Darmency et al., 2009). Consequently, both stamen and pollen are exposed considerably long to environmental factors including horizontally transmitted microbiota. Sugar beet pollination does not require insects. Still, flowers produce nutrient-rich pollen and odorous nectar, attracting several insect taxa that transport pollen (Free et al., 1975) or may transmit microbes to flowers and subsequently seeds (Hardoim et al., 2015; Berg and Raaijmakers, 2018; Nelson, 2018). However, such microbes will not form close relationships with the host plant, unless they can adapt to the selective force of the spermosphere (see Section 2.1). Nevertheless, anthosphere microbiome modulation, e.g., by introducing beneficial seed endophytes during flowering before seed maturation, may as well have the potential to promote plant health of the offspring generation.
2.6. The impact of domestication on the sugar beet microbiome
Plant domestication changes the morphology, physiology, and genetic potential of a given species, but also unintentionally shapes associated microbiomes via host-mediated, multi-generational microbiome selection (Johnston-Monje and Raizada, 2011, 2013; Orozco-Mosqueda et al., 2018; Pérez-Jaramillo et al., 2018). Sea beet, as the wild ancestor of sugar beet, is frequently used for comparative studies and backcrossing approaches to increase tolerance of modern cultivars toward biotic (Whitney, 1989), and abiotic stresses (Monteiro et al., 2013; Stevanato et al., 2019). Sea beet however carries traits undesired by breeders, namely annual life cycle, red pigments in root tissues, fangy or sprangled roots, multigerm flowers, elongated crowns, multiple crowns, low sucrose concentration, and low sucrose extractability (Panella and Lewellen, 2007). The Silesian beet was the first sugar beet line and was bred out of fodder beet and chard in the late 18th century (Fischer, 1989). Soon breeding started to diverge, resulting in cultivars of the Z-type (high sugar content), the E-type (optimized for biomass), and the N-type (intermediate type) (Francis, 2006). Ongoing breeding activities led to an increase in sugar concentration in sugar beet tap roots from 12 to ca. 18% (Milford, 2006; Dohm et al., 2014). Sugar yield continuously increased, attributable to higher average temperatures in spring (Jaggard et al., 2007), improved management practices (Hoffmann and Kenter, 2018), and most importantly breeding progress (Hoffmann and Loel, 2015).
Crop breeding in general mainly focuses on genomic traits, leading to higher yields, but also genetic homogeneity and erosion of genetic diversity (Gopal and Gupta, 2016). Due to the historically recent development of sugar beet, some authors regard the genetic background of sugar beet as relatively narrow compared to several other crops (Bosemark, 1979; Fischer, 1989; Stevanato et al., 2019; Galewski et al., 2022). Still, intentional as well as unintentional admixture and introgression events in the course of domestication and breeding re-introduced considerable genetic variation in beet crops (Galewski and McGrath, 2020). Nevertheless, the process of sugar beet domestication is correlated with a community shift in bacteria: bacterial rhizosphere communities of sea beet have a significantly higher alpha diversity and evenness than modern sugar beet cultivars when grown in the same substrate (Zachow et al., 2014). Such domestication-related effects were observed in other plant species like wheat (Germida and Siciliano, 2001), and beans (Pérez-Jaramillo et al., 2017) as well. Decreasing Bacteroidetes abundance in the rhizosphere was observed in several crops, including sugar beet, as a consequence of domestication (Pérez-Jaramillo et al., 2018). Differences between root community structure and its corresponding soil are bigger in modern sugar beets compared to sea beets; modern sugar beet roots have lesser but stronger responders to root exudates already in the seedling state (namely Novosphingobium, Pseudomonas, Sphingomonas). These differences in rhizosphere bacteria may have implications for soil-borne disease pathogenesis: bacterial isolates extracted from sea beet roots contained on average fewer antagonists of fungal phytopathogens but displayed higher tolerance toward salt stress when compared to isolates from sugar beet (Zachow et al., 2014). Since bacterial alpha diversity is often positively correlated with pathogen tolerance (Berg et al., 2017; Du et al., 2022a), the microbiome itself may also contribute to the resilience of the sea beet. In addition, the coastline as natural habitat itself may contain low pathogen pressure compared to osmotic pressure, which again may emphasize the reciprocal adaption of the host and its microbiome within its natural habitat. For sugar beet farming, however, high microbial rhizosphere alpha diversity may not always increase yield under different field conditions or in different cultivars. Some microbiome-mediated services to plants, e.g., stress resilience, nutrient acquisition, or phytopathogen antagonism, may not increase plant performance if conditions are already optimal or the corresponding stress is absent. The advantage of high microbial alpha diversity in the rhizosphere is usually more apparent under suboptimal growing conditions. To re-introduce resilience to the sugar beet crop using bacterial diversity, the lower alpha diversity in modern cultivars indicate actually a potential for intentionally introduced beneficial microorganisms (e.g., via seed priming) to establish in the rhizosphere, since some micro-niches in roots may not be occupied. In general, implementing functional hologenomic in breeding programs holds the potential to further increase desired plant traits and field performances of a crop (Nogales et al., 2016).
3. Part II: Microbiome management and pathogen control in sugar beet
3.1. Modulating sugar beet-associated microbiomes in the field
Using microbes to improve plant performance and health is a promising approach for the agriculture of the 21st century, not only in sugar beet. Several management options are available to modulate microbial communities in the field, including physical, chemical, biological, and logistical practices. In general, biological options include single strains (Section 3.1.1) or consortia (Section 3.1.2) with bioactive properties, the use of microbiota-active metabolites, application of microbiome transplants, or changing the environmental setting to indirectly shift the structure and functions of microbiomes (Berg and Smalla, 2009; Orozco-Mosqueda et al., 2018; Berg et al., 2021). Seed endophytes are of special interest for agricultural applications and targeted microbiome modulations (Berg and Raaijmakers, 2018; Shahzad et al., 2018) because of their vertical transmission, but environmental microbiomes known for high resilience toward biotic and abiotic stress can be exploited as well (Zachow et al., 2013). Methods to apply microorganisms include drenching, soil amendments, spraying, seed or seedling inoculation, tissue atomization, and direct injection. The upcoming sections discuss several examples of microbiome-based control, ranging from the application of single BCAs (Section 3.1.1) and consortia (Section 3.1.2) to the potential of soil amendments (Section 3.1.3) and disease-suppressive soils (Section 3.1.4) to positively influence sugar beet vigor.
3.1.1. Sugar beet biocontrol and plant growth promotion are available
Applying microbes to sugar beet usually aims at suppressing pathogens in the field to increase yield, and antagonism toward pathogens is often correlated with plant growth promotion (PGP, Table 1). Organic farming of sugar beet is challenging at market prices, amongst others due to insufficient availability, and missing or scattered scientific knowledge on environmental-friendly alternatives for pathogen suppression (Stevanato et al., 2019). Important pathogens of sugar beet include mainly fungal pathogens (Table 1), especially different anastomosis groups of Rhizoctonia solani J.G.KÜHN [teleomorph: Thanatephorus cucumeris (A.B.Frank) Donk], Cercospora beticola SACC., and the oomycete Globisporangium ultimum (Trow) Uzuhashi, Tojo and Kakish. (syn.: Pythium ultimum). Diseases caused by these pathogens include crown rot, damping-off, and early and late root rot. As an example, late root rot, a disease caused by R. solani AG2-2IIIB, can lead locally to yield losses >50% on the field or during storage at sugar refineries. It is estimated to affect 24% of the acreage in the USA, and 5–10% in Europe (Büttner et al., 2004; Jacobsen, 2006; Windels et al., 2009). First sugar beet resistance to Rhizoctonia root rot was discovered in breeding lines developed in Fort Collins, USA (Panella et al., 2015), but these cultivars often insufficiently carried other desired traits like bolting resistance, yield or processing quality (Büttner et al., 2004). Nowadays, Rhizoctonia-tolerant varieties have been improved significantly thanks to breeding progress and additionally provide protection against other plant diseases. Since Rhizoctonia tolerance is a polygenic trait (Galewski et al., 2022) and due to its quantitative inheritance however, yield potential could not be improved as fast as for single gene-mediated tolerances, e.g., resistance against beet necrotic yellow vein virus (BNYVV) causing rhizomania (Scholten and Lange, 2000), or beet cyst nematodes (Heterodera schachtii A.Schmidt) (Cai et al., 1997). In seeds, the genus Kosakonia seems to be correlated with Rhizoctonia-tolerant sugar beet cultivars (Wolfgang et al., 2020), but further investigations are needed to generalize this correlation. Anyway, using biocontrol and plant growth-promoting microbes has considerable potential, especially in susceptible cultivars providing high yields.
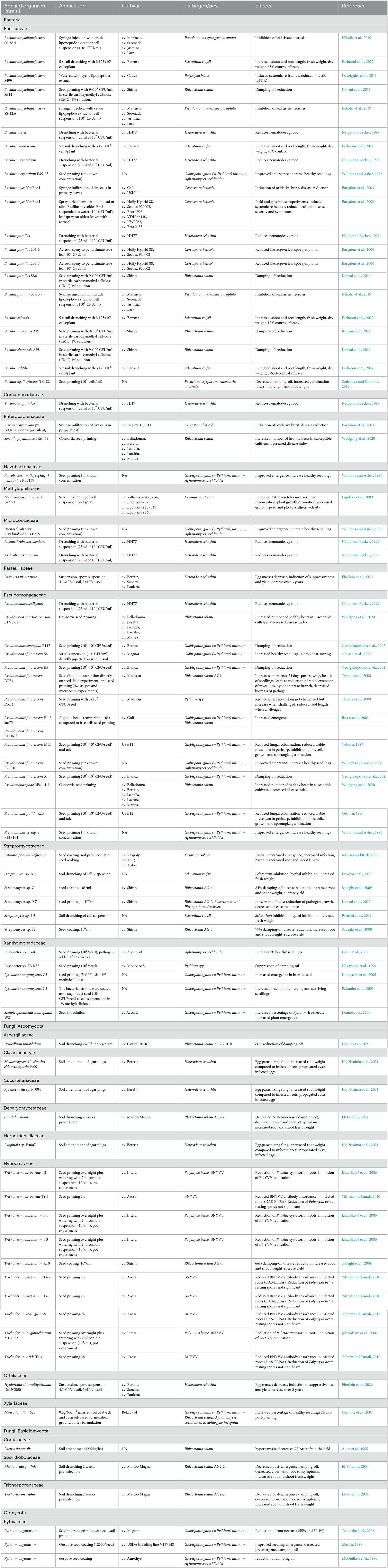
Table 1. Biocontrol of selected phytopathogens and plant growth promotion potential of different biocontrol agents (BCAs).
Microbes already adapted to an endophytic lifestyle or a specific host plant appear to be more effective in biocontrol and PGP. For instance, strains isolated from sugar beet rhizosphere were reported to outperform BCAs isolated from other plant hosts in Rhizoctonia-infested sugar beet (Karimi et al., 2016). On the other hand, sugar beet-adapted microbiota can also show positive effects in other crops (Natsagdorj et al., 2019). Endophytes were frequently successfully tested for sugar beet PGP, including strains of Acinetobacter, Achromobacter, Bacillus, Burkholderia, Chryseobacterium, and Pseudomonas. Positive effects include increased fresh weight, dry weight, number of leaves, absorption of K, N, Mg, and P, Vitamin B and C content, leave size, germination rate, plant height, shortened germination time, and lower tissue water content (Çakmakçi et al., 2001; Shi et al., 2009b, 2011, 2017; Gasser et al., 2011; Piernik et al., 2017). Nevertheless, it may be reasonable to isolate microbes from non-target hosts or environments adapted to given stressors. Zachow et al. (2013) used sugar beet plants to enrich bacterial taxa from substrate amended with either a pH-tolerant moss species, cold-adapted primroses, or drought-adapted lichens for targeted extraction of strains both mediating tolerance toward abiotic stress and compatible with the host plant. Such crop-adapted microbiome transplants hold the big potential to isolate strains that increase crop resilience toward the numerous biotic and abiotic challenges agriculture will face during the Anthropocene (Berg and Cernava, 2022).
Most biocontrol approaches to control phytopathogens use members of either Bacillus or Pseudomonas solely applied as seed priming (Table 1). Both genera carry several beneficial traits regarding agricultural applications, e.g., the ability to produce endospores. Furthermore, the importance of Pseudomonas in disease-suppressive soils (Mendes et al., 2011), as well as in sugar beet-associated bacterial communities, was frequently reported (Thompson et al., 1993; Zachow et al., 2008; Wolfgang et al., 2020; Okazaki et al., 2021), especially fluorescent pseudomonads (Thompson et al., 1993, 1995a,b; Rainey et al., 1994; Lilley et al., 1997; Ellis et al., 1999). Pseudomonas strains produce interesting secondary metabolites with antifungal properties, e.g., lipopeptides like amphisin, poaeamide, viscosin, and viscosinamide (Andersen et al., 2003; Zachow et al., 2015). There is evidence that some Pseudomonas adapt lipopeptide and endochitinase production when exposed to different sugar concentrations (Nielsen et al., 1998), which is advantageous regarding the variable sugar concentration in sugar beet.
Biocontrol of beet pathogens was even achieved with subcellular components of BCAs. For instance, reduced root necrosis caused by G. ultimum was observed when seedlings were primed with only cell wall proteins of Pythium oligandrum DRECHSLER, most likely triggering host defense response (Takenaka et al., 2003). To summarize, a high number of potential BCAs in sugar beet is readily available and scientifically investigated, but has yet to be established in current agricultural practices.
3.1.2. Consortia vs. single strains in seed priming: pros and cons
Combining and applying different BCA strains as consortiums can have additive or synergistic effects on plant health (Carrión et al., 2019). Verticillium wilt symptoms were reduced using Verticillium tricorpus and non-pathogenic Fusarium oxysporum as soil amendments under greenhouse conditions (Lopisso et al., 2017). Another study reported a yield increase, an increase in the numbers of healthy beets by up to 65%, and a 26% reduction of disease index in Rhizoctonia-susceptible cultivars under field conditions using seed-priming with a tripartite bacterial consortium consisting of Pseudomonas poae, Ps. brassicacearum, and Serratia plymuthica (Wolfgang et al., 2020). On the other hand, the combination of Pseudomonas and Bacillus BCAs did not increase the biocontrol of Globisporangium (Pythium) ultimum in sugar beet compared to the single usage of Pseudomonas strains (Georgakopoulos et al., 2002). However, approaches using consortia are confronted with the hurdle that every BCA has to be licensed separately, which poses high investment costs.
3.1.3. Amendments and the importance of C/N ratio for biocontrol
Indirectly managing microbial communities in field soils via amendments was practiced for millennia. With recent molecular methods, we can nowadays identify the specific responses of soil microbiomes upon different amendments on the microbial community level (Vida et al., 2020). Organic amendments can increase yield or decrease the severity of diseases caused by beet pathogens or pests (e.g., Postma and Schilder, 2015). Despite promising results, the main disadvantage of amendments scientifically is the lack of comparability due to multivariate differences, including age, composition, present microbiota, source of single components, etc. N-rich, easily decomposable amendments favor R. solani infection in the short term but suppress it with increasing decomposition time in lettuce (Lactuca sativa, L.). Furthermore, an amendment-dependent trade-off between plant growth promotion and microbial disease control capability was observed (Bonanomi et al., 2020). C/N-ratio of amendments may be an important factor for microbiome capability for pathogen control: sugar beets exude a comparably high amount of inorganic nitrogen (exudate C/N-ratio ≤ 12:1), and R. solani infection was controlled under laboratory conditions by locally applying 2% solutions of glucose, fructose, sorbose or xylose to germinating seedlings. This control effect was reversible by adding additional nitrogen sources to the soil (Shimizu et al., 2018). N content of the taproot may play a role in some pathogeneses: R. solani-tolerant cultivars often contain higher concentrations of alpha-amino N. The concentration of alpha-amino N within the root further increases during infection, irrespective of the Rhizoctonia tolerance of the sugar beet cultivar (Ogata et al., 2006). Still, soil microbiota species identity plays a role in disease onset: using N-rich vermicompost as an amendment, a high amount of Firmicutes and Streptomycetacae in the sugar beet root community was found (Wolfgang et al., 2020); these taxa are known to display antagonism toward Rhizoctonia (Table 1). Competition between pathogen and microbial communities for nitrogen sources is hereby suggested to influence biocontrol of damping-off caused by R. solani, but this needs further evaluation.
3.1.4. Suppressive soils: a treasure chest for future biocontrol options
Disease-suppressive soils harbor microbiomes where a given phytopathogen cannot establish or does not cause disease despite its prevalence and abundance in the soil. Soil suppressiveness is lost after soil sterilization and thus attributed to specific microbiome composition or members (Mendes et al., 2011; van der Voort et al., 2016). Soil suppressiveness to R. solani in sugar beet was investigated elaborately, broadening our understanding of both sugar beet microbiomes and plant microbiomes in general. Soil suppressiveness may evolve under continuous cropping conditions (Raaijmakers and Mazzola, 2016), but the exact prerequisites of soil microbiomes to develop suppressiveness are yet not identified. Still, slow-growing and heat-tolerant bacteria like Streptomycetaceae, Micrococcaceae, Mycobacteriaceae, and Solibacteraceae correlate with suppressiveness toward Rhizoctonia (van der Voort et al., 2016).
Several mechanisms for sugar beet protection through the activity of associated microbiota in suppressive soils were found, hereby categorized into the “four lines of defense” (Figure 2) with R. solani as the model pathogen. The first defensive line includes the biotic background suppressiveness of soil, for instance, mediated by microbes inhibiting pathogens via volatile organic compounds (VOC) as “long-range” antagonists, e.g., plant growth-promoting Streptomyces spp. and Paraburkholderia graminis isolates originating from sugar beet rhizosphere (Cordovez et al., 2015; Carrión et al., 2018). The second line of defense includes microbes activated upon Rhizoctonia infection in the rhizosphere: if R. solani is applied to sugar beet seedlings in suppressive soil, Oxalobacteraceae, Burkholderiaceae, Sphingobacteriaceae, and Sphingomonadaceae are significantly enriched compared to non-stressed rhizosphere (Chapelle et al., 2016). Transcriptomic analyses indicated R. solani to produce oxalic and phenylacetic acids, activating specific rhizobacteria. These activated bacteria show upregulated ppGpp pathways, leading to fungal growth inhibition, induced plant resistance responses, and co-activation of other microorganisms. The third line of defense includes root endophytes changing their transcriptomic profile upon Rhizoctonia infection, namely Pseudomonas, Chitinophaga, and Flavobacterium (Carrión et al., 2019). These bacteria may therefore be regarded as soterobionts (disease-preventing bacteria; Cernava and Berg, 2022) of the sugar beet-Rhizoctonia pathosystem. Lastly, the fourth line of defense contains the plant host-specific defense mechanisms and physiological traits, that as well affect the associated microbes and can be influenced via breeding.
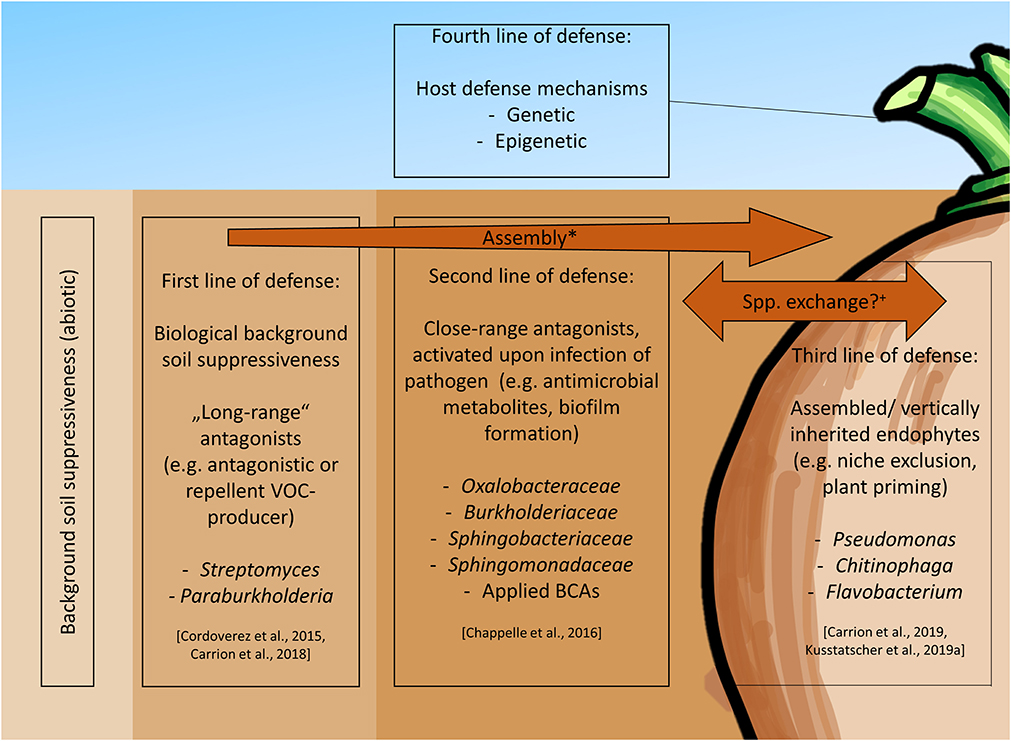
Figure 2. Microbiome-based defense mechanism toward Rhizoctonia solani in sugar beet. *Root exudate-mediated enrichment of rhizosphere-associated microbes. +The deposition of initial seed endophytes from taproot toward the rhizosphere is not proven in sugar beet so far.
Pathogen tolerance may occur in susceptible cultivars grown in conducive soils, and such plants are promising sources for future biocontrol candidates. Kusstatscher and colleagues (Kusstatscher et al., 2019a) investigated the bacterial and fungal rhizosphere community of pre-harvest sugar beet taproots in conducive soils, targeting R. solani-diseased beets and healthy beets surrounded by diseased beets. Lactobacillales, Candida, Fusarium, and Penicillium were identified as indicator species for diseased beets, while Flavobacteriales, Cyanobacteria, Plectosphaerella, Vishniacozyma, and Sordariomycetes were indicators for symptomless beets under pathogen pressure. Our current knowledge on microbiota active in disease-suppressive soils and on indicator taxa for disease tolerance in conducive soils provides the potential to predict realized pathogen pressure on a given field, as well as several disease-preventing taxa that can be applied as BCAs in future studies.
3.2. Controlling root rots in postharvest beets
After harvest, sugar beet taproots are usually stored in piles directly on the field or in the proximity of sugar refineries (Draycott, 2003; Kusstatscher et al., 2019b; Kohout et al., 2020). As storage time increases, autochthonous sucrose synthase of sugar beet cleaves sucrose to fructose and glucose to provide life-sustaining metabolism. In unwounded sugar beets, >90% of sucrose loss is due to the activity of autogenous sucrose synthase (Klotz and Finger, 2004). However, the combination of high water and sucrose content with wounds, cracks, or root tip breakage provides optimal conditions for microbial infections (Liebe and Varrelmann, 2016; Liebe et al., 2016; Kusstatscher et al., 2019b). Especially fungal infections with Penicillium spp. and Botrytis cinerea lead to an increase in fructose and glucose concentration, mainly attributable to fungal acid invertases (Klotz and Finger, 2004). Furthermore, oomycetes like Aphanomyces cochlioides DRECHSLER can cause severe sugar losses (Campbell and Klotz, 2006). Since fungi are the main group causing post-harvest sucrose losses, post-harvest microbiome studies focus—in contrast to all other sugar beet-related microbiome research—on fungal communities.
Damage to the root is the most important factor determining fungal rot severity, followed by genotype and environment (Liebe and Varrelmann, 2016). After harvest, the fungal microbiome of sugar beet taproots is dominated by Plectosphaerella cucumerina, Pyrenochaeta, and Leptosphaeria. The oomycete community is dominated by Aphanomyces and Globisporangium/Pythium. In addition to mechanical damage, storage temperature has severe consequences for the fungal community in stored roots: while lower temperatures (8°C) favor Botrytis, higher temperatures (20°C) favor Fusarium, Penicillium, and partially Pichia (Liebe and Varrelmann, 2016). During rot, the overall diversity of fungi decreases (Liebe et al., 2016; Kusstatscher et al., 2019b), but diversity within the genus Fusarium increases (Liebe et al., 2016). These fungal taxa however could also be targeted using biocontrol approaches, reducing fungal sucrose degradation in stored beets, to increase efficiency and thus sustainability of sugar beet production.
The microbiome in fields, where stored beets originate from, has a big influence on associated fungal taxa. Storage rots were long thought to be erratic, because visually healthy beets may develop rot symptoms (Campbell and Klotz, 2006; Christ et al., 2011). However, fungal indicator species for storability were identified in the rhizosphere and outer endosphere of harvested beets, namely Plectosphaerella and Vishniacozyma. These taxa were associated with symptomless stored beets next to rotten beets while decaying beets were characterized by a high abundance of Lactobacillus, Gluconobacter, Candida, and Penicillium (Kusstatscher et al., 2019b). Measuring such indicator taxa in field soils before storage would enable refineries to rank and process sugar beet lots based on their proposed storability, reducing losses due to storage rots.
3.3. Microbiome management for healthy sugar beet production and storage
Microbiome-based management or modulation options for all life stages of the sugar beet were already suggested and partially commercialized (Figure 3). Sugar beet-adapted microbes can be isolated to test antagonism toward pathogens and plant growth-promoting traits. Microbes derived from extreme environments can be investigated for tolerance-mediating effects to mitigate abiotic stress. Protection from biotic and abiotic stressors is especially important in the vulnerable seedling state during rhizosphere establishment, therefore seed priming technologies are specifically important to assist in the assembly of beneficial rhizosphere microbiomes. With microbial abiotic and biotic stress protection, a reduction of fertilizers and pesticides is possible. Investigating suppressive soils provides the opportunity to directly (application of transplants or suppressiveness-mediating microbiota) or indirectly (modulation via amendments) modulate soil microbiota to the benefit of crop vigor. Phylosymbioses studies could be used for microbiome-assisted breeding, potentially further increasing yield or desired crop traits. Finally, considering disease indicator taxa and biocontrol approaches in postharvest beets may increase storability, consequently efficiency and ecological sustainability of sugar beet farming. In this way, microbiome-based management systems provide tools to meet the upcoming agricultural challenges we will face in the Anthropocene (Berg and Cernava, 2022).
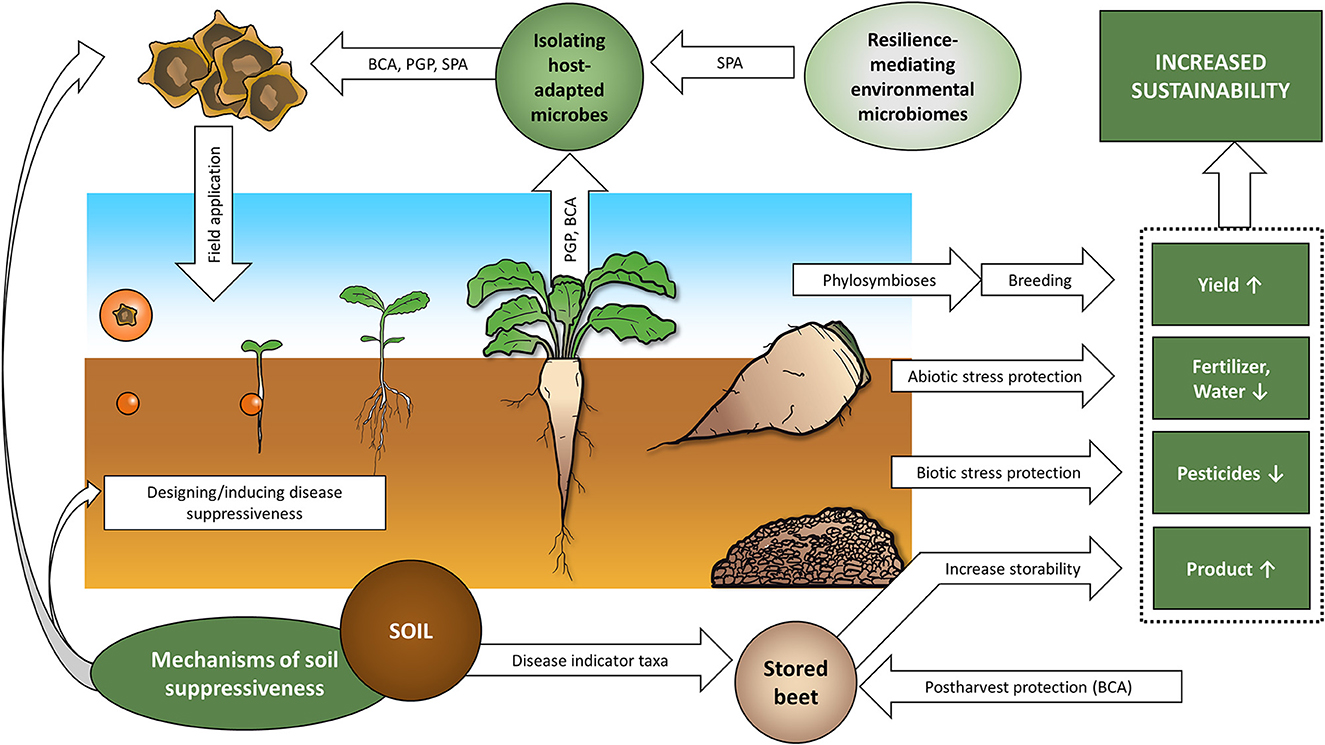
Figure 3. Summary of microbiome-based targets to increase sugar beet farming sustainability. BCA, Biological control agents; PGP, plant growth promoter; SPA, stress protecting agents. For a description see Section 3.3.
4. Conclusions and outlook
1. Sugar beet is a very interesting model plant for microbiome research due to its recent and documented breeding history, narrow genetic background, economic importance, well-studied relatives, susceptibility toward fungal pathogens, and also its long history of microbiological research.
2. The well-studied sugar beet microbiome is associated with some breakthroughs in plant microbiome research. However, additional holistic and especially mechanistic studies using comprehensive combinations of advanced microbiome research methods are required. Further research should specifically consider the known spatial and temporal scales realized in the sugar beet holobiont, being summarized in this review. Therefore, especially sampling design and procedures should be described in as much detail as possible (age, life stage, cultivar, plant organ, position on plant organ, etc.) to properly contextualize observations and to increase comparability between different studies.
3. Future research should as well address not only bacteria, but all microbiome members, including protists and viruses to provide a more holistic view of potential autochthonous regulatory mechanisms and interactions in a given microbiome. Combined studies of the plant genome and microbiome by multi-omics suggest further ground-breaking results.
4. Sugar beet microbiomes were strongly shaped by breeding, and early evidence for plant-microbe coevolution was found. Phylosymbiosis studies will promote microbiome-assisted breeding efforts, which is essential for further sustainable sugar beet production.
5. A system-immanent challenge for scientific work is the constant success and development in sugar beet breeding, together with the characteristic mating system. Modern sugar beet cultivars are hybrids, new cultivars are bred and licensed every year and usually have to be licensed on a national level. Consequently, most scientific studies use different cultivars, because of availability in a given country or shutdown of production. In this way, sugar beet microbiome research always has to consider cultivar-dependent effects as an additional layer of variability when comparing different studies.
6. The sugar beet microbiome is highly diverse, microhabitat- and cultivar-specific. Although several successful microbiome management strategies were already developed, understanding plant-microbiome interactions novel microbiome management strategies. In particular, the connection between leaf microbiomes and susceptibility toward air-borne foliage diseases (e.g., diseases caused by Ramularia beticola, Erysiphe betae, or Cercospora beticola) or insect pests are potential targets for future biocontrol strategies. Moreover, adapting sugar beet cultivation to abiotic stress conditions under climate change, and reducing greenhouse gas emissions are important challenges of the future.
Author contributions
AW and GB wrote the manuscript and created figures. All authors contributed to the article and approved the submitted version.
Funding
This work was performed as part of the BIOINSECTICIDES research project (F42422) at Graz University of Technology. Open access funding provided by Graz University of Technology Open Access Publishing Fund.
Acknowledgments
The authors would like to thank Birgit Wassermann for assistance in drawing figure elements and Alejandra Ordonez for comments on the final manuscript draft.
Conflict of interest
NT and RT are employed by KWS SAAT SE & Co. KGaA.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdelfattah, A., Wisniewski, M., Schena, L., and Tack, A. J. M. (2021). Experimental evidence of microbial inheritance in plants and transmission routes from seed to phyllosphere and root. Environ. Microbiol. 23, 2199–2214. doi: 10.1111/1462-2920.15392
Allen, M. F., Boosalis, M. G., Kerr, E. D., Muldoon, A. E., and Larsen, H. J. (1985). Population dynamics of sugar beets, Rhizoctonia solani, and Laetisaria arvalis: Responses of a host, plant pathogen, and hyperparasite to perturbation in the field. Appl. Environ. Microbiol. 50, 1123–1127. doi: 10.1128/aem.50.5.1123-1127.1985
Andersen, J. B., Koch, B., Nielsen, T. H., Sørensen, D., Hansen, M., Nybroe, O., et al. (2003). Surface motility in Pseudomonas sp. DSS73 is required for efficient biological containment of the root-pathogenic microfungi Rhizoctonia solani and Pythium ultimum. Microbiology. 149, 37–46. doi: 10.1099/mic.0.25859-0
Badri, D. V., and Vivanco, J. M. (2009). Regulation and function of root exudates. Plant, Cell Environ. 32, 666–681. doi: 10.1111/j.1365-3040.2009.01926.x
Bais, H. P., Weir, T. L., Perry, L. G., Gilroy, S., and Vivanco, J. M. (2006). The role of root exudates in rhizosphere interactions with plants and other organisms. Annu. Rev. Plant Biol. 57, 233–266. doi: 10.1146/annurev.arplant.57.032905.105159
Bargabus, R. L., Zidack, N. K., Sherwood, J. E., and Jacobsen, B. J. (2002). Characterisation of systemic resistance in sugar beet elicited by a non-pathogenic, phyllosphere-colonizing Bacillus mycoides, biological control agent. Physiol. Mol. Plant Pathol. 61, 289–298. doi: 10.1006/pmpp.2003.0443
Bargabus, R. L., Zidack, N. K., Sherwood, J. E., and Jacobsen, B. J. (2003). Oxidative burst elicited by Bacillus mycoides isolate Bac J, a biological control agent, occurs independently of hypersensitive cell death in sugar beet. Mol. Plant-Microbe Interact. 16, 1145–1153. doi: 10.1094/MPMI.2003.16.12.1145
Bargabus, R. L., Zidack, N. K., Sherwood, J. E., and Jacobsen, B. J. (2004). Screening for the identification of potential biological control agents that induce systemic acquired resistance in sugar beet. Biol. Control. 30, 342–350. doi: 10.1016/j.biocontrol.2003.11.005
Barret, M., Briand, M., Bonneau, S., Préveaux, A., Valière, S., Bouchez, O., et al. (2015). Emergence shapes the structure of the seed microbiota. Appl. Environ. Microbiol. 81, 1257–1266. doi: 10.1128/AEM.03722-14
Bashir, I., War, A. F., Rafiq, I., Reshi, Z. A., Rashid, I., and Shouche, Y. S. (2022). Phyllosphere microbiome: Diversity and functions. Microbiol. Res. 254, 126888. doi: 10.1016/j.micres.2021.126888
Bazany, K. E., Wang, J. T., Delgado-Baquerizo, M., Singh, B. K., and Trivedi, P. (2022). Water deficit affects inter-kingdom microbial connections in plant rhizosphere. Environ. Microbiol. 24, 3722–3734. doi: 10.1111/1462-2920.16031
Berendsen, R. L., Pieterse, C. M. J., and Bakker, P. A. H. M. (2012). The rhizosphere microbiome and plant health. Trends Plant Sci. 17, 478–486. doi: 10.1016/j.tplants.2012.04.001
Berg, G., and Cernava, T. (2022). The plant microbiota signature of the Anthropocene as a challenge for microbiome research. Microbiome 10, 1–12. doi: 10.1186/s40168-021-01224-5
Berg, G., Köberl, M., Rybakova, D., Müller, H., Grosch, R., and Smalla, K. (2017). Plant microbial diversity is suggested as the key to future biocontrol and health trends. FEMS Microbiol. Ecol. 93, 1–9. doi: 10.1093/femsec/fix050
Berg, G., Krause, R., and Mendes, R. (2015). Cross-kingdom similarities in microbiome ecology and biocontrol of pathogens. Front. Microbiol. 6, 1–5. doi: 10.3389/fmicb.2015.01311
Berg, G., Kusstatscher, P., Abdelfattah, A., Cernava, T., and Smalla, K. (2021). Microbiome modulation—toward a better understanding of plant microbiome response to microbial inoculants. Front. Microbiol. 12, 1–12. doi: 10.3389/fmicb.2021.650610
Berg, G., and Raaijmakers, J. M. (2018). Saving seed microbiomes. ISME J. 12, 1167–1170. doi: 10.1038/s41396-017-0028-2
Berg, G., Rybakova, D., Fischer, D., Cernava, T., Vergès, M. C. C., Charles, T., et al. (2020). Microbiome definition re-visited: old concepts and new challenges. Microbiome 8, 1–22. doi: 10.1186/s40168-020-00875-0
Berg, G., and Smalla, K. (2009). Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol. Ecol. 68, 1–13. doi: 10.1111/j.1574-6941.2009.00654.x
Bergna, A., Cernava, T., Rändler, M., Grosch, R., Zachow, C., and Berg, G. (2018). Tomato seeds preferably transmit plant beneficial endophytes. Phytobiomes. 2, 183–193. doi: 10.1094/PBIOMES-06-18-0029-R
Bertoldo, G., Della Lucia, M. C., Squartini, A., Concheri, G., Broccanello, C., Romano, A., et al. (2021). Endophytic microbiome responses to sulfur availability in Beta vulgaris (L.). Int. J. Mol. Sci. 22. doi: 10.3390/ijms22137184
Bonanomi, G., Zotti, M., Idbella, M., Di Silverio, N., Carrino, L., Cesarano, G., et al. (2020). Decomposition and organic amendments chemistry explain contrasting effects on plant growth promotion and suppression of Rhizoctonia solani damping off. PLoS ONE. 15, 1–20. doi: 10.1371/journal.pone.0230925
Bosemark, N. O. (1979). “Genetic poverty of the sugarbeet in Europe,” in Broadening the Genetic Base of Crops, Zeven, A., and van Harten, A. M. (eds). (Wageningen: Centre for Agricultural Publishing and Documentation) p. 28–35.
Broccanello, C., Ravi, S., Deb, S., Bolton, M., Secor, G., Richards, C., et al. (2022). Bacterial endophytes as indicators of susceptibility to Cercospora leaf spot (CLS) disease in Beta vulgaris L. Sci. Rep. 12, 1–13. doi: 10.1038/s41598-022-14769-8
Büttner, G., Pfähler, B., and Märländer, B. (2004). Greenhouse and field techniques for testing sugar beet for resistance to Rhizoctonia root and crown rot. Plant Breed. 123, 158–166. doi: 10.1046/j.1439-0523.2003.00967.x
Cai, D., Kleine, M., Kifle, S., Harloff, H. J., Sandal, N. N., Marcker, K. A., et al. (1997). Positional cloning of a gene for nematode resistance in sugar beet. Science (80-.). 275, 832–834. doi: 10.1126/science.275.5301.832
Çakmakçi, R., Kantar, F., and Sahin, F. (2001). Effect of N2-fixing bacterial inoculations on yield of sugar beet and barley. J. Plant Nutr. Soil Sci. 164, 527–531. doi: 10.1002/1522-2624(200110)164:5<527::AID-JPLN527>3.0.CO;2-1
Campbell, L. G., and Klotz, K. (2006). “Storage,” in Sugar Beet, Draycott, A. P. (ed). (Oxford: Blackwell Publishing Ltd) p. 387–408. doi: 10.1002/9780470751114.ch15
Cardinale, M., Ratering, S., Sadeghi, A., Pokhrel, S., Honermeier, B., and Schnell, S. (2020). The response of the soil microbiota to long-term mineral and organic nitrogen fertilization is stronger in the bulk soil than in the rhizosphere. Genes (Basel). 11, 456. doi: 10.3390/genes11040456
Carrión, V. J., Cordovez, V., Tyc, O., Etalo, D. W., de Bruijn, I., de Jager, V. C. L., et al. (2018). Involvement of Burkholderiaceae and sulfurous volatiles in disease-suppressive soils. ISME J. 12, 2307–2321. doi: 10.1038/s41396-018-0186-x
Carrión, V. J., Perez-Jaramillo, J., Cordovez, V., Tracanna, V., De Hollander, M., Ruiz-Buck, D., et al. (2019). Pathogen-induced activation of disease-suppressive functions in the endophytic root microbiome. Science (80-.). 366, 606–612. doi: 10.1126/science.aaw9285
Cernava, T., and Berg, G. (2022). The emergence of disease-preventing bacteria within the plant microbiota. Environ. Microbiol. 24, 3259–3263. doi: 10.1111/1462-2920.15896
Chapelle, E., Mendes, R., Bakker, P. A. H., and Raaijmakers, J. M. (2016). Fungal invasion of the rhizosphere microbiome. ISME J. 10, 265–268. doi: 10.1038/ismej.2015.82
Christ, D. S., Märländer, B., and Varrelmann, M. (2011). Characterization and mycotoxigenic potential of Fusarium species in freshly harvested and stored sugar beet in Europe. Phytopathology. 101, 1330–1337. doi: 10.1094/PHYTO-01-11-0002
Compant, S., Cambon, M. C., Vacher, C., Mitter, B., Samad, A., and Sessitsch, A. (2021). The plant endosphere world–bacterial life within plants. Environ. Microbiol. 23, 1812–1829. doi: 10.1111/1462-2920.15240
Compant, S., Reiter, B., Sessitsch, A., Nowak, J., Clément, C., and Barka, E. A. (2005). Endophytic colonization of Vitis vinifera L. by plant growth-promoting bacterium Burkholderia sp. strain PsJN. Appl. Environ. Microbiol. 71, 1685–1693. doi: 10.1128/AEM.71.4.1685-1693.2005
Cook, R. J., Thomashow, L. S., Weller, D. M., Fujimoto, D., Mazzola, M., Bangera, G., et al. (1995). Molecular mechanisms of defense by rhizobacteria against root disease. Proc. Natl. Acad. Sci. U. S. A. 92, 4197–4201. doi: 10.1073/pnas.92.10.4197
Cordovez, V., Carrion, V. J., Etalo, D. W., Mumm, R., Zhu, H., van Wezel, G. P., et al. (2015). Diversity and functions of volatile organic compounds produced by Streptomyces from a disease-suppressive soil. Front. Microbiol. 6, 1–13. doi: 10.3389/fmicb.2015.01081
Cui, R., Geng, G., Wang, G., Stevanato, P., Dong, Y., Li, T., et al. (2022). The response of sugar beet rhizosphere micro-ecological environment to continuous cropping. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.956785
Darmency, H., Klein, E. K., Gestat De Garanb,é, T., Gouyon, P. H., Richard-Molard, M., and Muchembled, C. (2009). Pollen dispersal in sugar beet production fields. Theor. Appl. Genet. 118, 1083–1092. doi: 10.1007/s00122-009-0964-y
de la Fuente Cant,ó, C., Simonin, M., King, E., Moulin, L., Bennett, M. J., Castrillo, G., et al. (2020). An extended root phenotype: the rhizosphere, its formation and impacts on plant fitness. Plant J. 103, 951–964. doi: 10.1111/tpj.14781
Della Lucia, M. C., Bertoldo, G., Broccanello, C., Maretto, L., Ravi, S., Marinello, F., et al. (2021). Novel effects of leonardite-based applications on sugar beet. Front. Plant Sci. 12, 1–10. doi: 10.3389/fpls.2021.646025
Dent, K. C., Stephen, J. R., and Finch-Savage, W. E. (2004). Molecular profiling of microbial communities associated with seeds of Beta vulgaris subsp. vulgaris (sugar beet). J. Microbiol. Methods 56, 17–26. doi: 10.1016/j.mimet.2003.09.001
Desoignies, N., Schramme, F., Ongena, M., and Legrève, A. (2013). Systemic resistance induced by Bacillus lipopeptides in beta vulgaris reduces infection by the rhizomania disease vector Polymyxa betae. Mol. Plant Pathol. 14, 416–421. doi: 10.1111/mpp.12008
Dohm, J. C., Minoche, A. E., Holtgräwe, D., Capella-Gutiérrez, S., Zakrzewski, F., Tafer, H., et al. (2014). The genome of the recently domesticated crop plant sugar beet (Beta vulgaris). Nature. 505, 546–549. doi: 10.1038/nature12817
Dong, Z., Canny, M. J., McCully, M. E., Roboredo, M. R., Cabadilla, C. F., Ortega, E., et al. (1994). A nitrogen-fixing endophyte of sugarcane stems: a new role for the apoplast. Plant Physiol. 105, 1139–1147. doi: 10.1104/pp.105.4.1139
Draycott, A. P. (2003). “Introduction,” in Nutrients for sugarbeet production: soil-plant relationships, Draycott, A. P., and Christenson, D. R. (Wallingford, UK: CAB International) p. 1–8. doi: 10.1079/9780851996233.0001
Du, J., Song, B., Jia, Q., Liu, S., Li, X., Liu, H., et al. (2022a). Effect of long-term sugar beet cultivation on rhizosphere bacterial diversity, community structure and sugar yield of sugar beet. Rhizosphere. 22, 100507. doi: 10.1016/j.rhisph.2022.100507
Du, J., Song, B., Li, X., and Huang, W. (2022b). Long-term cultivation of sugar beet: Effect on rhizosphere micro-flora, soil fertility and beet productivity. Sugar Tech. 24, 1821–1831. doi: 10.1007/s12355-022-01124-4
Dunne, C., Moenne-Loccoz, Y., de Bruijn, F. J., and O'Gara, F. (2000). Overproduction of an inducible extracellular serine protease improves biological control of Pythium ultimum by Stenotrophomonas maltophilia strain W81. Microbiology. 146, 2069–2078. doi: 10.1099/00221287-146-8-2069
Eberlein, C., Heuer, H., and Westphal, A. (2020). Biological suppression of populations of Heterodera schachtii adapted to different host genotypes of sugar beet. Front. Plant Sci. 11, 1–13. doi: 10.3389/fpls.2020.00812
Ellis, R. J., Thompson, I. P., and Bailey, M. J. (1999). Temporal fluctuations in the pseudomonad population associated with sugar beet leaves. FEMS Microbiol. Ecol. 28, 345–356. doi: 10.1111/j.1574-6941.1999.tb00589.x
El-Tarabily, K. A. (2004). Suppression of Rhizoctonia solani diseases of sugar beet by antagonistic and plant growth-promoting yeasts. J. Appl. Microbiol. 96, 69–75. doi: 10.1046/j.1365-2672.2003.02043.x
Errakhi, R., Lebrihi, A., and Barakate, M. (2009). In vitro and in vivo antagonism of actinomycetes isolated from Moroccan rhizospherical soils against Sclerotium rolfsii: A causal agent of root rot on sugar beet (Beta vulgaris L.). J. Appl. Microbiol. 107, 672–681. doi: 10.1111/j.1365-2672.2009.04232.x
FAOSTAT (2022). Available online at: https://www.fao.org/faostat/en/#data (accessed June 24, 2022).
Farhaoui, A., Adadi, A., Tahiri, A., El Alami, N., Khayi, S., Mentag, R., et al. (2022). Biocontrol potential of plant growth-promoting rhizobacteria (PGPR) against Sclerotiorum rolfsii diseases on sugar beet (Beta vulgaris L.). Physiol. Mol. Plant Pathol. 119, 101829. doi: 10.1016/j.pmpp.2022.101829
Fischer, H. E. (1989). Origin of the “Weisse Schlesische Rübe” (white Silesian beet) and resynthesis of sugar beet. Euphytica. 41, 75–80. doi: 10.1007/BF00022414
Fort, T., Pauvert, C., Zanne, A. E., Ovaskainen, O., Caignard, T., Barret, M., et al. (2021). Maternal effects shape the seed mycobiome in Quercus petraea. New Phytol. 230, 1594–1608. doi: 10.1111/nph.17153
Francis, S. A. (2006). “Development of sugar beet,” in Sugar Beet, Draycott, A. P. (Oxford: Blackwell Publishing Ltd) p. 9–29. doi: 10.1002/9780470751114.ch2
Free, J. B., Williams, I., Longden, P. C., and Johnson, M. G. (1975). Insect pollination of sugar-beet (Beta vulgaris) seed crops. Ann. Appl. Biol. 81, 121–134. doi: 10.1111/j.1744-7348.1975.tb00529.x
Galewski, P., Funk, A., and McGrath, J. M. (2022). Select and sequence of a segregating sugar beet population provides genomic perspective of host resistance to seedling Rhizoctonia solani Infection. Front. Plant Sci. 12, 785267. doi: 10.3389/fpls.2021.785267
Galewski, P., and McGrath, J. M. (2020). Genetic diversity among cultivated beets (Beta vulgaris) assessed via population-based whole genome sequences. BMC Genomics. 21, 1–14. doi: 10.1186/s12864-020-6451-1
Gasser, I., Cardinale, M., Müller, H., Heller, S., Eberl, L., Lindenkamp, N., et al. (2011). Analysis of the endophytic lifestyle and plant growth promotion of Burkholderia terricola ZR2-12. Plant Soil. 347, 125–136. doi: 10.1007/s11104-011-0833-8
Gasser, I., Müller, H., and Berg, G. (2009). Ecology and characterization of polyhydroxyalkanoate-producing microorganisms on and in plants. FEMS Microbiol. Ecol. 70, 142–150. doi: 10.1111/j.1574-6941.2009.00734.x
Georgakopoulos, D. G., Fiddaman, P., Leifert, C., and Malathrakis, N. E. (2002). Biological control of cucumber and sugar beet damping-off caused by Pythium ultimum with bacterial and fungal antagonists. J. Appl. Microbiol. 92, 1078–1086. doi: 10.1046/j.1365-2672.2002.01658.x
Germida, J. J., and Siciliano, S. D. (2001). Taxonomic diversity of bacteria associated with the roots of modern, recent and ancient wheat cultivars. Biol. Fertil. Soils 33, 410–415. doi: 10.1007/s003740100343
Gopal, M., and Gupta, A. (2016). Microbiome selection could spur next-generation plant breeding strategies. Front. Microbiol. 7, 1–10. doi: 10.3389/fmicb.2016.01971
Gouka, L., Raaijmakers, J. M., and Cordovez, V. (2022). Ecology and functional potential of phyllosphere yeasts. Trends Plant Sci. 27, 1109–1123. doi: 10.1016/j.tplants.2022.06.007
Grimme, E., Zidack, N. K., Sikora, R. A., Strobel, G. A., and Jacobsen, B. J. (2007). Comparison of Muscodor albus volatiles with a biorational mixture for control of seedling diseases of sugar beet and root-knot nematode on tomato. Plant Dis. 91, 220–225. doi: 10.1094/PDIS-91-2-0220
Haj Nuaima, R., Ashrafi, S., Maier, W., and Heuer, H. (2021). Fungi isolated from cysts of the beet cyst nematode parasitized its eggs and counterbalanced root damages. J. Pest Sci. 94, 563–572. doi: 10.1007/s10340-020-01254-2
Hallmann, J., and Berg, G. (2007). Spectrum and population dynamics of bacterial root endophytes. Microb. Root Endophytes 9, 15–31. doi: 10.1007/3-540-33526-9_2
Haque, M. E., Lakshman, D. K., Qi, A., and Khan, M. F. R. (2021). Penicillium pinophilum has the potential to reduce damping-off caused by Rhizoctonia solani in sugar beet. Sugar Tech. 23, 872–880. doi: 10.1007/s12355-021-00958-8
Hardoim, P. R., Hardoim, C. C. P., van Overbeek, L. S., and van Elsas, J. D. (2012). Dynamics of seed-borne rice endophytes on early plant growth stages. PLoS ONE. 7, e30438. doi: 10.1371/journal.pone.0030438
Hardoim, P. R., van Overbeek, L. S., Berg, G., Pirttil,ä, A. M., Compant, S., Campisano, A., et al. (2015). The hidden world within plants: ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol. Mol. Biol. Rev. 79, 293–320. doi: 10.1128/MMBR.00050-14
Hegarty, T. W. (1977). Seed and seedling susceptibility to phased moisture stress in soil. J. Exp. Bot. 28, 659–668. doi: 10.1093/jxb/28.3.659
Hergert, G. W. (2010). Sugar beet fertilization. Sugar Tech. 12, 256–266. doi: 10.1007/s12355-010-0037-1
Hoffmann, C., and Loel, J. (2015). Bedeutung der Züchtung für den Ertragsanstieg von Zuckerrüben. Sugar Ind. 140, 48–56. doi: 10.36961/si16195
Hoffmann, C. M., and Kenter, C. (2018). Yield potential of sugar beet–have we hit the ceiling? Front. Plant Sci. 9, 1–6. doi: 10.3389/fpls.2018.00289
Hoffmann, C. M., Kenter, C., and Bloch, D. (2005). Marc concentration of sugar beet (Beta vulgaris L) in relation to sucrose storage. J. Sci. Food Agric. 85, 459–465. doi: 10.1002/jsfa.2002
Houlden, A., Timms-Wilson, T. M., Day, M. J., and Bailey, M. J. (2008). Influence of plant developmental stage on microbial community structure and activity in the rhizosphere of three field crops. FEMS Microbiol. Ecol. 65, 193–201. doi: 10.1111/j.1574-6941.2008.00535.x
Huang, W., Sun, D., Fu, J., Zhao, H., Wang, R., and An, Y. (2020). Effects of continuous sugar beet cropping on rhizospheric microbial communities. Genes (Basel). 11, 13. doi: 10.3390/genes11010013
Huang, W., Sun, D., Wang, R., and An, Y. (2021). Integration of transcriptomics and metabolomics reveals the responses of sugar beet to continuous cropping obstacle. Front. Plant Sci. 12, 1–11. doi: 10.3389/fpls.2021.711333
Huang, Y., Kuang, Z., Wang, W., and Cao, L. (2016). Exploring potential bacterial and fungal biocontrol agents transmitted from seeds to sprouts of wheat. Biol. Control. 98, 27–33. doi: 10.1016/j.biocontrol.2016.02.013
Ikeda, S., Okazaki, K., Takahashi, H., Tsurumaru, H., and Minamisawa, K. (2023). Seasonal shifts in bacterial community structures in the lateral root of sugar beet grown in an andosol field in Japan. Microbes Environ. 38, 1–13. doi: 10.1264/jsme2.ME22071
Islam, M. T., Hashidoko, Y., Deora, A., Ito, T., and Tahara, S. (2005). Suppression of damping-off disease in host plants by the rhizoplane bacterium Lysobacter sp. strain SB-K88 is linked to plant colonization and antibiosis against soilborne Peronosporomycetes. Appl. Environ. Microbiol. 71, 3786–3796. doi: 10.1128/AEM.71.7.3786-3796.2005
Jacobs, M. J., Bugbee, W. M., and Gabrielson, D. A. (1985). Enumeration, location, and characterization of endophytic bacteria within sugar beet roots. Can. J. Bot. 63, 1262–1265. doi: 10.1139/b85-174
Jacobsen, B. J. (2006). Root rot diseases of sugar beet. Proc. Nat. Sci. Matica Srp. Novi Sad. 2006, 9–19. doi: 10.2298/ZMSPN0610009J
Jaggard, K. W., Qi, A., and Semenov, M. A. (2007). The impact of climate change on sugarbeet yield in the UK: 1976–2004. J. Agric. Sci. 145, 367–375. doi: 10.1017/S0021859607006922
Jakubíková, L., Šubíková, V., Nemčovi,č, M., and Farkaš, V. (2006). Selection of natural isolates of Trichoderma spp. for biocontrol of Polymyxa betae as a vector of virus causing rhizomania in sugar beet. Biologia (Bratisl). 61, 347–351. doi: 10.2478/s11756-006-0063-3
James, E. K., Gyaneshwar, P., Mathan, N., Barraquio, W. L., Reddy, P. M., Iannetta, P. P. M., et al. (2002). Infection and colonization of rice seedlings by the plant growth-promoting bacterium Herbaspirillum seropedicae Z67. Mol. Plant-Microbe Interact. 15, 894–906. doi: 10.1094/MPMI.2002.15.9.894
Jammer, A., Albacete, A., Schulz, B., Koch, W., Weltmeier, F., van der Graaff, E., et al. (2020). Early-stage sugar beet taproot development is characterized by three distinct physiological phases. Plant Direct. 4, 1–29. doi: 10.1002/pld3.221
Johnston-Monje, D., Lundberg, D. S., Lazarovits, G., Reis, V. M., and Raizada, M. N. (2016). Bacterial populations in juvenile maize rhizospheres originate from both seed and soil. Plant Soil. 405, 337–355. doi: 10.1007/s11104-016-2826-0
Johnston-Monje, D., and Raizada, M. N. (2011). Conservation and diversity of seed associated endophytes in Zea across boundaries of evolution, ethnography and ecology. PLoS ONE. 6, e20396. doi: 10.1371/journal.pone.0020396
Johnston-Monje, D., and Raizada, M. N. (2013). Surveying diverse zea seed for populations of bacterial endophytes. Mol. Microb. Ecol. Rhizosph. 1, 445–455. doi: 10.1002/9781118297674.ch42
Kaga, H., Mano, H., Tanaka, F., Watanabe, A., Kaneko, S., and Morisaki, H. (2009). Rice seeds as sources of endophytic bacteria. Microbes Environ. 24, 154–162. doi: 10.1264/jsme2.ME09113
Karimi, E., Sadeghi, A., Dehaji, P. A., Dalvand, Y., Omidvari, M., and Kakuei Nezhad, M. (2012). Biocontrol activity of salt tolerant Streptomyces isolates against phytopathogens causing root rot of sugar beet. Biocontrol Sci. Technol. 22, 333–349. doi: 10.1080/09583157.2012.658552
Karimi, E., Safaie, N., Shams-Baksh, M., and Mahmoudi, B. (2016). Bacillus amyloliquefaciens SB14 from rhizosphere alleviates Rhizoctonia damping-off disease on sugar beet. Microbiol. Res. 192, 221–230. doi: 10.1016/j.micres.2016.06.011
Khorassani, R., Hettwer, U., Ratzinger, A., Steingrobe, B., Karlovsky, P., and Claassen, N. (2011). Citramalic acid and salicylic acid in sugar beet root exudates solubilize soil phosphorus. BMC Plant Biol. 11. doi: 10.1186/1471-2229-11-121
Klaedtke, S., Jacques, M. A., Raggi, L., Préveaux, A., Bonneau, S., Negri, V., et al. (2016). Terroir is a key driver of seed-associated microbial assemblages. Environ. Microbiol. 18, 1792–1804. doi: 10.1111/1462-2920.12977
Klotz, K. L., and Finger, F. L. (2004). Impact of temperature, length of storage and postharvest disease on sucrose catabolism in sugarbeet. Postharvest Biol. Technol. 34, 1–9. doi: 10.1016/j.postharvbio.2004.05.016
Kobayashi, D. Y., Reedy, R. M., Palumbo, J. D., Zhou, J. M., and Yuen, G. Y. (2005). A clp gene homologue belonging to the Crp gene family globally regulates lytic enzyme production, antimicrobial activity, and biological control activity expressed by Lysobacter enzymogenes strain C3. Appl. Environ. Microbiol. 71, 261–269. doi: 10.1128/AEM.71.1.261-269.2005
Kohout, C. K., Ukowitz, C., Reiter, D., Zitz, U., Moder, K., Emerstorfer, F., et al. (2020). Bacterial growth dynamics and corresponding metabolite levels in the extraction area of an Austrian sugar beet factory using antimicrobial treatment. J. Sci. Food Agric. 100, 2713–2721. doi: 10.1002/jsfa.10303
Kõiv, V., Arbo, K., Maiväli, Ü., Kisand, V., Roosaare, M., Remm, M., et al. (2019). Endophytic bacterial communities in peels and pulp of five root vegetables. PLoS ONE. 14, 1–17. doi: 10.1371/journal.pone.0210542
Kusstatscher, P., Cernava, T., Harms, K., Maier, J., Eigner, H., Berg, G., et al. (2019a). Disease incidence in sugar beet fields is correlated with microbial diversity and distinct biological markers. Phytobiomes J. 3, 22–30. doi: 10.1094/PBIOMES-01-19-0008-R
Kusstatscher, P., Wicaksono, W. A., Bergna, A., Cernava, T., Bergau, N., Tissier, A., et al. (2020). Trichomes form genotype-specific microbial hotspots in the phyllosphere of tomato. Environ. Microbiomes 15, 1–10. doi: 10.1186/s40793-020-00364-9
Kusstatscher, P., Zachow, C., Harms, K., Maier, J., Eigner, H., Berg, G., et al. (2019b). Microbiome-driven identification of microbial indicators for postharvest diseases of sugar beets. Microbiome 7, 1–12. doi: 10.1186/s40168-019-0728-0
Lange, W., Brandenburg, W. A., and De Bock, T. S. M. (1999). Taxonomy and cultonomy of beet (Beta vulgaris L.). Bot. J. Linn. Soc. 130, 81–96. doi: 10.1111/j.1095-8339.1999.tb00785.x
Larran, S., Mónaco, C., and Alippi, H. E. (2000). Endophytic fungi in beet (Beta vulgaris var. esculenta L.) leaves. Adv. Hort. Sci. 14, 193–196. doi: 10.1400/14064
Lee, S. Y. (1996). Bacterial polyhydroxyalkanoates. Biotechnol. Bioeng. 49, 1–14.3. doi: 10.1002/(SICI)1097-0290(19960105)49:1<1::AID-BIT1>3.0.CO;2-P
Liebe, S., and Varrelmann, M. (2016). Effect of environment and sugar beet genotype on root rot development and pathogen profile during storage. Phytopathology. 106, 65–75. doi: 10.1094/PHYTO-07-15-0172-R
Liebe, S., Wibberg, D., Winkler, A., Pühler, A., Schlüter, A., and Varrelmann, M. (2016). Taxonomic analysis of the microbial community in stored sugar beets using high-throughput sequencing of different marker genes. FEMS Microbiol. Ecol. 92, 1–12. doi: 10.1093/femsec/fiw004
Lilley, A. K., Fry, J. C., Bailey, M. J., and Day, M. J. (1996). Comparison of aerobic heterotrophic taxa isolated from four root domains of mature sugar beet (Beta vulgaris). FEMS Microbiol. Ecol. 21, 231–242. doi: 10.1111/j.1574-6941.1996.tb00350.x
Lilley, A. K., Hails, R. S., Cory, J. S., and Bailey, M. J. (1997). The dispersal and establishment of pseudomonad populations in the phyllosphere of sugar beet by phytophagous caterpillars. FEMS Microbiol. Ecol. 24, 151–157. doi: 10.1111/j.1574-6941.1997.tb00431.x
Lim, S. J., and Bordenstein, S. R. (2020). An introduction to phylosymbiosis. Proc. R. Soc. B Biol. Sci. 287. doi: 10.1098/rspb.2019.2900
Lindow, S. E., and Brandl, M. T. (2003). Microbiology of the phyllosphere. Appl. Environ. Microbiol. 69, 1875–1883. doi: 10.1128/AEM.69.4.1875-1883.2003
Liu, H., Carvalhais, L. C., Crawford, M., Singh, E., Dennis, P. G., Pieterse, C. M. J., et al. (2017). Inner plant values: diversity, colonization and benefits from endophytic bacteria. Front. Microbiol. 8, 1–17. doi: 10.3389/fmicb.2017.02552
Liu, Y., Qi, A., and Khan, M. F. R. (2019). Age-dependent resistance to Rhizoctonia solani in sugar beet. Plant Dis. 103, 2322–2329. doi: 10.1094/PDIS-11-18-2001-RE
Lopisso, D. T., Kühlmann, V., and Siebold, M. (2017). Potential of soil-derived fungal biocontrol agents applied as a soil amendment and a seed coating to control Verticillium wilt of sugar beet. Biocontrol Sci. Technol. 27, 1019–1037. doi: 10.1080/09583157.2017.1357800
Lübeck, P. S., Hansen, M., and Sørensen, J. (2000). Simultaneous detection of the establishment of seed-inoculated Pseudomonas fluorescens strain DR54 and native soil bacteria on sugar beet root surfaces using fluorescence antibody and in situ hybridization techniques. FEMS Microbiol. Ecol. 33, 11–19. doi: 10.1016/S0168-6496(00)00038-6
Mark, G. L., Dow, J. M., Kiely, P. D., Higgins, H., Haynes, J., Baysse, C., et al. (2005). Transcriptome profiling of bacterial responses to root exudates identifies genes involved in microbe-plant interactions. Proc. Natl. Acad. Sci. U. S. A. 102, 17454–17459. doi: 10.1073/pnas.0506407102
Martin, F. N. (1987). The use of Pythium oligandrum for biological control of preemergence damping-off caused by P. ultimum. Phytopathology. 77, 1013. doi: 10.1094/Phyto-77-1013
McQuilken, M. P., Whipps, J. M., and Cooke, R. C. (1990). Control of damping-off in cress and sugar-beet by commercial seed-coating with Pythium oligandrum. Plant Pathol. 39, 452–462. doi: 10.1111/j.1365-3059.1990.tb02521.x
Mendes, R., Kruijt, M., De Bruijn, I., Dekkers, E., Van Der Voort, M., Schneider, J. H. M., et al. (2011). Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science (80-.). 332, 1097–1100. doi: 10.1126/science.1203980
Mendes, R., and Raaijmakers, J. M. (2015). Cross-kingdom similarities in microbiome functions. ISME J. 9, 1905–1907. doi: 10.1038/ismej.2015.7
Milford, G. F., and Watson, D. J. (1971). The effect of nitrogen on the growth and sugar content of sugar-beet. Ann. Bot. 35, 287–300. doi: 10.1093/oxfordjournals.aob.a084478
Milford, G. F. J. (2006). “Plant structure and crop physiology,” in Sugar Beet, Draycott, A. P. (Oxford: Blackwell Publishing Ltd) p. 30–49. doi: 10.1002/9780470751114.ch3
Monteiro, F., Romeiras, M. M., Batista, D., and Duarte, M. C. (2013). Biodiversity assessment of sugar beet species and its wild relatives: Linking ecological data with new genetic approaches. Am. J. Plant Sci. 04, 21–34. doi: 10.4236/ajps.2013.48A003
Moussa, T. A. A., and Rizk, M. (2002). Biocontrol of sugarbeet pathogen Fusarium solani (Mart.) Sacc. by Streptomyces aureofaciens. Pakistan J. Biol. Sci. 5, 556–559. doi: 10.3923/pjbs.2002.556.559
Nakayama, T., Homma, Y., Hashidoko, Y., Mizutani, J., and Tahara, S. (1999). Possible role of xanthobaccins produced by Stenotrophomonas sp. strain SB-K88 in suppression of sugar beet damping-off disease. Appl. Environ. Microbiol. 65, 4334–4339. doi: 10.1128/AEM.65.10.4334-4339.1999
Natsagdorj, O., Sakamoto, H., Santiago, D. M. O., Santiago, C. D., Orikasa, Y., Okazaki, K., et al. (2019). Variovorax sp. has an optimum cell density to fully function as a plant growth promoter. Microorganisms. 7, 82. doi: 10.3390/microorganisms7030082
Neipp, P. W., and Becker, J. O. (1999). Evaluation of biocontrol activity of rhizobacteria from Beta vulgaris against Heterodera schachtii. J. Nematol. 31, 54−61.
Nelson, E. B. (2018). The seed microbiome: origins, interactions, and impacts. Plant Soil. 422, 7–34. doi: 10.1007/s11104-017-3289-7
Nielsen, M. N., Sørensen, J., Fels, J., and Pedersen, H. C. (1998). Secondary metabolite- and endochitinase-dependent antagonism toward plant-pathogenic microfungi of Pseudomonas fluorescens isolates from sugar beet rhizosphere. Appl. Environ. Microbiol. 64, 3563–3569. doi: 10.1128/AEM.64.10.3563-3569.1998
Nikolić, I., Berić, T., Dimkić, I., Popović, T., Lozo, J., Fira, D., et al. (2019). Biological control of Pseudomonas syringae pv. aptata on sugar beet with Bacillus pumilus SS-10.7 and Bacillus amyloliquefaciens (SS-12.6 and SS-38.4) strains. J. Appl. Microbiol. 126, 165–176. doi: 10.1111/jam.14070
Nogales, A., Nobre, T., Valadas, V., Ragonezi, C., Döring, M., Polidoros, A., et al. (2016). Can functional hologenomics aid tackling current challenges in plant breeding? Brief. Funct. Genomics 15, 288–297. doi: 10.1093/bfgp/elv030
Novinscak, A., and Filion, M. (2011). Effect of soil clay content on RNA isolation and on detection and quantification of bacterial gene transcripts in soil by quantitative reverse transcription-PCR. Appl. Environ. Microbiol. 77, 6249–6252. doi: 10.1128/AEM.00055-11
Ogata, N., Takahashi, H., Taguchi, K., Okazaki, K., and Nakatsuka, K. (2006). Change in the amount of amino nitrogen in crown root of sugar beet (Beta vulgaris L.) infected with Rhizoctonia solani. Japanese J. Crop Sci. 74, 357–363. doi: 10.1626/jcs.74.357
Okazaki, K., Iino, T., Kuroda, Y., Taguchi, K., Takahashi, H., Ohwada, T., et al. (2014). An assessment of the diversity of culturable bacteria from main root of sugar beet. Microbes Environ. 29, 220–223. doi: 10.1264/jsme2.ME13182
Okazaki, K., Tsurumaru, H., Hashimoto, M., Takahashi, H., Okubo, T., Ohwada, T., et al. (2021). Community analysis-based screening of plant growth-promoting bacteria for sugar beet. Microbes Environ. 36, 1–11. doi: 10.1264/jsme2.ME20137
Orozco-Mosqueda, M., del, C., Rocha-Granados, M., del, C., Glick, B. R., and Santoyo, G. (2018). Microbiome engineering to improve biocontrol and plant growth-promoting mechanisms. Microbiol. Res. 208, 25–31. doi: 10.1016/j.micres.2018.01.005
Osburn, R. M. (1989). Dynamics of sugar beet seed colonization by Pythium ultimum and Pseudomonas species: effects on seed rot and damping-off. Phytopathology. 79, 709. doi: 10.1094/Phyto-79-709
Ottesen, A. R., González Peña, A., White, J. R., Pettengill, J. B., Li, C., Allard, S., et al. (2013). Baseline survey of the anatomical microbial ecology of an important food plant: Solanum lycopersicum (tomato). BMC Microbiol. 13, 114. doi: 10.1186/1471-2180-13-114
Owen, F. W., Carsner, E., and Stout, M. (1940). Photothermal induction of flowering in sugar beet. Am. Soc. Sugar Beet Technol. 61, 101–124.
Palumbo, J. D., Yuen, G. Y., Jochum, C. C., Tatum, K., and Kobayashi, D. Y. (2005). Mutagenesis of β-1,3-glucanase genes in Lysobacter enzymogenes strain C3 results in reduced biological control activity toward Bipolaris leaf spot of tall fescue and Pythium damping-off of sugar beet. Phytopathology. 95, 701–707. doi: 10.1094/PHYTO-95-0701
Panella, L., Campbell, L. G., Eujayl, I. A., Lewellen, R. T., and McGrath, J. M. (2015). USDA-ARS sugarbeet releases and breeding over the past 20 years. J. Sugarbeet Res. 52, 1–2. doi: 10.5274/jsbr.52.3.40
Panella, L., and Lewellen, R. T. (2007). Broadening the genetic base of sugar beet: introgression from wild relatives. Euphytica. 154, 383–400. doi: 10.1007/s10681-006-9209-1
Pantigoso, H. A., Newberger, D., and Vivanco, J. M. (2022). The rhizosphere microbiome: plant–microbial interactions for resource acquisition. J. Appl. Microbiol. 133, 2864–2876. doi: 10.1111/jam.15686
Pérez-Jaramillo, J. E., Carrión, V. J., Bosse, M., Ferrão, L. F. V., De Hollander, M., Garcia, A. A. F., et al. (2017). Linking rhizosphere microbiome composition of wild and domesticated Phaseolus vulgaris to genotypic and root phenotypic traits. ISME J. 11, 2244–2257. doi: 10.1038/ismej.2017.85
Pérez-Jaramillo, J. E., Carrión, V. J., de Hollander, M., and Raaijmakers, J. M. (2018). The wild side of plant microbiomes. Microbiome. 6, 4–9. doi: 10.1186/s40168-018-0519-z
Pervaiz, Z. H., Iqbal, J., Zhang, Q., Chen, D., Wei, H., and Saleem, M. (2020). Continuous cropping alters multiple biotic and abiotic indicators of soil health. Soil Syst. 4, 59. doi: 10.3390/soilsystems4040059
Philippot, L., Raaijmakers, J. M., Lemanceau, P., and van der Putten, W. H. (2013). Going back to the roots: the microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 11, 789–799. doi: 10.1038/nrmicro3109
Piernik, A., Hrynkiewicz, K., Wojciechowska, A., Szymańska, S., Lis, M. I., and Muscolo, A. (2017). Effect of halotolerant endophytic bacteria isolated from Salicornia europaea L. on the growth of fodder beet (Beta vulgaris L.) under salt stress. Arch. Agron. Soil Sci. 63, 1404–1418. doi: 10.1080/03650340.2017.1286329
Pigoleva, S. V., Zakharchenko, N. S., Pigolev, A. V., Trotsenko, Y. A., and Buryanov, Y. I. (2009). The influence of colonizing methylobacteria on morphogenesis and resistance of sugar beet and white cabbage plants to Erwinia carotovora. Appl. Biochem. Microbiol. 45, 604–609. doi: 10.1134/S0003683809060052
Postma, J., and Schilder, M. T. (2015). Enhancement of soil suppressiveness against Rhizoctonia solani in sugar beet by organic amendments. Appl. Soil Ecol. 94, 72–79. doi: 10.1016/j.apsoil.2015.05.002
Pusenkova, L. I., Il'yasova, E. Y., Lastochkina, O. V., Maksimov, I. V., and Leonova, S. A. (2016). Changes in the species composition of the rhizosphere and phyllosphere of sugar beet under the impact of biological preparations based on endophytic bacteria and their metabolites. Eurasian Soil Sci. 49, 1136–1144. doi: 10.1134/S1064229316100112
Raaijmakers, J. M., and Mazzola, M. (2016). Soil immune responses. Science (80-.). 352, 1392–1393. doi: 10.1126/science.aaf3252
Rainey, P. B., Bailey, M. J., and Thompson, I. P. (1994). Phenotypic and genotypic diversity of fluorescent pseudomonads isolated from field-grown sugar beet. Microbiology. 140, 2315–2331. doi: 10.1099/13500872-140-9-2315
Russo, A., Basaglia, M., Tola, E., and Casella, S. (2001). Survival, root colonisation and biocontrol capacities of Pseudomonas fluorescens F113 LacZY in dry alginate microbeads. J. Ind. Microbiol. Biotechnol. 27, 337–342. doi: 10.1038/sj.jim.7000154
Rybakova, D., Mancinelli, R., Wikström, M., Birch-Jensen, A. S., Postma, J., Ehlers, R. U., et al. (2017). The structure of the Brassica napus seed microbiome is cultivar-dependent and affects the interactions of symbionts and pathogens. Microbiome 5, 104. doi: 10.1186/s40168-017-0310-6
Sadeghi, A., Hesan, A. R., Askari, H., Qomi, D. N., Farsi, M., and Hervan, E. M. (2009). Biocontrol of Rhizoctonia solani damping-off of sugar beet with native Streptomyces strains under field conditions. Biocontrol Sci. Technol. 19, 985–991. doi: 10.1080/09583150902912665
Saftner, R. A., Daie, J., and Wyse, R. E. (1983). Sucrose uptake and compartmentation in sugar beet taproot tissue. Plant Physiol. 72, 1–6. doi: 10.1104/pp.72.1.1
Samadi, A. (2012). Impact of continuous sugar beet cropping on potassium quantity-intensity parameters in calcareous soils. J. Plant Nutr. 35, 1154–1167. doi: 10.1080/01904167.2012.676128
Sasse, J., Martinoia, E., and Northen, T. (2018). Feed your friends: do plant exudates shape the root microbiome? Trends Plant Sci. 23, 25–41. doi: 10.1016/j.tplants.2017.09.003
Scholten, O. E., and Lange, W. (2000). Breeding for resistance to rhizomania in sugar beet: a review. Euphytica. 112, 219–231. doi: 10.1023/A:1003988003165
Shahzad, R., Khan, A. L., Bilal, S., Asaf, S., and Lee, I. J. (2018). What is there in seeds? Vertically transmitted endophytic resources for sustainable improvement in plant growth. Front. Plant Sci. 9, 1–10. doi: 10.3389/fpls.2018.00024
Shi, Y., Lou, K., and Li, C. (2009a). Isolation, quantity distribution and characterization of endophytic microorganisms within sugar beet. African J. Biotechnol. 8, 835–840.
Shi, Y., Lou, K., and Li, C. (2009b). Promotion of plant growth by phytohormone-producing endophytic microbes of sugar beet. Biol. Fertil. Soils. 45, 645–653. doi: 10.1007/s00374-009-0376-9
Shi, Y., Lou, K., and Li, C. (2011). Growth promotion effects of the endophyte Acinetobacter johnsonii strain 3-1 on sugar beet. Symbiosis. 54, 159–166. doi: 10.1007/s13199-011-0139-x
Shi, Y., Yang, H., Zhang, T., Sun, J., and Lou, K. (2014). Illumina-based analysis of endophytic bacterial diversity and space-time dynamics in sugar beet on the north slope of Tianshan mountain. Appl. Microbiol. Biotechnol. 98, 6375–6385. doi: 10.1007/s00253-014-5720-9
Shi, Y. W., Li, C., Yang, H. M., Zhang, T., Gao, Y., Chu, M., et al. (2017). Colonization study of gfp-tagged Achromobacter marplatensis strain in sugar beet. J. Microbiol. 55, 267–272. doi: 10.1007/s12275-017-6371-1
Shi, Y. W., Li, C., Yang, H. M., Zhang, T., Gao, Y., Zeng, J., et al. (2016). Endophytic fungal diversity and space-time dynamics in sugar beet. Eur. J. Soil Biol. 77, 77–85. doi: 10.1016/j.ejsobi.2016.09.005
Shi, Y. W., TaPa, M. S., Li, C., Yang, H. M., Zhang, T., Gao, Y., et al. (2015). Diversity and space–time dynamics of endophytic archaea from sugar beet in the north slope of Tianshan Mountain revealed by 454 pyrosequencing and T-RFLP. World J. Microbiol. Biotechnol. 31, 1031–1039. doi: 10.1007/s11274-015-1853-y
Shimizu, Y., Sagiya, D., Matsui, M., and Fukui, R. (2018). Zonal soil amendment with simple sugars to elevate soil C/N ratios as an alternative disease management strategy for Rhizoctonia damping-off of sugar beet. Plant Dis. 102, 1434–1444. doi: 10.1094/PDIS-09-16-1279-RE
Smirnova, I., and Sadanov, A. (2019). Application of agriculturally important microorganisms for biocontrol of root rot infection of sugar beet. Arch. Phytopathol. Plant Prot. 52, 698–713. doi: 10.1080/03235408.2019.1588195
Spanner, R., Neubauer, J., Heick, T. M., Grusak, M. A., Hamilton, O., Rivera-Varas, V., et al. (2021). Seedborne Cercospora beticola can initiate Cercospora leaf spot from sugar beet (Beta vulgaris) fruit tissue. Phytopathology. 112, 1016–1028. doi: 10.1094/PHYTO-03-21-0113-R
Steinkellner, S., Lendzemo, V., Langer, I., Schweiger, P., Khaosaad, T., Toussaint, J. P., et al. (2007). Flavonoids and strigolactones in root exudates as signals in symbiotic and pathogenic plant-fungus interactions. Molecules. 12, 1290–1306. doi: 10.3390/12071290
Stevanato, P., Chiodi, C., Broccanello, C., and Concheri, G. (2019). Sustainability of the sugar beet crop. Sugar Tech. 21, 703–716. doi: 10.1007/s12355-019-00734-9
Stevanato, P., Squartini, A., Concheri, G., and Saccomani, M. (2016). Sugar beet yield and processing quality in relation to nitrogen content and microbiological diversity of deep soil layer. Sugar Tech 18, 67–74. doi: 10.1007/s12355-014-0365-7
Stevanato, P., Zavalloni, C., Marchetti, R., Bertaggia, M., Saccomani, M., McGrath, J. M., et al. (2010). Relationship between subsoil nitrogen availability and sugarbeet processing quality. Agron. J. 102, 17–22. doi: 10.2134/agronj2009.0041
Takenaka, S., Nakamura, Y., Kono, T., Sekiguchi, H., Masunaka, A., and Takahashi, H. (2006). Novel elicitin-like proteins isolated from the cell wall of the biocontrol agent Pythium oligandrum induce defence-related genes in sugar beet. Mol. Plant Pathol. 7, 325–339. doi: 10.1111/j.1364-3703.2006.00340.x
Takenaka, S., Nishio, Z., and Nakamura, Y. (2003). Induction of defense reactions in sugar beet and wheat by treatment with cell wall protein fractions from the mycoparasite Pythium oligandrum. Phytopathology. 93, 1228–1232. doi: 10.1094/PHYTO.2003.93.10.1228
Thompson, I. P., Bailey, M. J., Ellis, R. J., Lilley, A. K., McCormack, P. J., Purdy, K. J., et al. (1995a). Short-term community dynamics in the phyllosphere microbiology of field-grown sugar beet. FEMS Microbiol. Ecol. 16, 205–211. doi: 10.1111/j.1574-6941.1995.tb00284.x
Thompson, I. P., Bailey, M. J., Fenlon, J. S., Fermor, T. R., Lilley, A. K., Lynch, J. M., et al. (1993). Quantitative and qualitative seasonal changes in the microbial community from the phyllosphere of sugar beet (Beta vulgaris). Plant Soil. 150, 177–191. doi: 10.1007/BF00013015
Thompson, I. P., Ellis, R. J., and Bailey, M. J. (1995b). Autecology of a genetically modified fluorescent pseudomonad on sugar beet. FEMS Microbiol. Ecol. 17, 1–13. doi: 10.1111/j.1574-6941.1995.tb00122.x
Thrane, C., Harder Nielsen, T., Neiendam Nielsen, M., Sørensen, J., and Olsson, S. (2000). Viscosinamide-producing Pseudomonas fluorescens DR54 exerts a biocontrol effect on Pythium ultimum in sugar beet rhizosphere. FEMS Microbiol. Ecol. 33, 139–146. doi: 10.1111/j.1574-6941.2000.tb00736.x
Truyens, S., Weyens, N., Cuypers, A., and Vangronsveld, J. (2013). Changes in the population of seed bacteria of transgenerationally Cd-exposed Arabidopsis thaliana. Plant Biol. 15, 971–981. doi: 10.1111/j.1438-8677.2012.00711.x
Truyens, S., Weyens, N., Cuypers, A., and Vangronsveld, J. (2015). Bacterial seed endophytes: Genera, vertical transmission and interaction with plants. Environ. Microbiol. Rep. 7, 40–50. doi: 10.1111/1758-2229.12181
Tsurumaru, H., Okubo, T., Okazaki, K., Hashimoto, M., Kakizaki, K., Hanzawa, E., et al. (2015). Metagenomic analysis of the bacterial community associated with the taproot of sugar beet. Microbes Environ. 30, 63–69. doi: 10.1264/jsme2.ME14109
van der Voort, M., Kempenaar, M., van Driel, M., Raaijmakers, J. M., and Mendes, R. (2016). Impact of soil heat on reassembly of bacterial communities in the rhizosphere microbiome and plant disease suppression. Ecol. Lett. 19, 375–382. doi: 10.1111/ele.12567
Vida, C., de Vicente, A., and Cazorla, F. M. (2020). The role of organic amendments to soil for crop protection: induction of suppression of soilborne pathogens. Ann. Appl. Biol. 176, 1–15. doi: 10.1111/aab.12555
Wassermann, B., Abdelfattah, A., Wicaksono, W. A., Kusstatscher, P., Müller, H., Cernava, T., et al. (2022). The Brassica napus seed microbiota is cultivar-specific and transmitted via paternal breeding lines. Microb. Biotechnol. 15, 2379–2390. doi: 10.1111/1751-7915.14077
Wassermann, B., Cernava, T., Müller, H., Berg, C., and Berg, G. (2019a). Seeds of native alpine plants host unique microbial communities embedded in cross-kingdom networks. Microbiome. 7, 108. doi: 10.1186/s40168-019-0723-5
Wassermann, B., Müller, H., and Berg, G. (2019b). An apple a day: which bacteria do we eat with organic and conventional apples? Front. Microbiol. 10, 1–13. doi: 10.3389/fmicb.2019.01629
Weller, D. M., Raaijmakers, J. M., Gardener, B. B. M., and Thomashow, L. S. (2002). Microbial populations responsible for specific soil suppressiveness to plant pathogens. Annu. Rev. Phytopathol. 40, 309–348. doi: 10.1146/annurev.phyto.40.030402.110010
Whitney, E. D. (1989). Identification, distribution, and testing for resistance to rhizomania in Beta maritima. Plant Dis. 73, 287–290. doi: 10.1094/PD-73-0287
Williams, G. E., and Asher, M. J. C. (1996). Selection of rhizobacteria for the control of Pythium ultimum and Aphanomyces cochlioides on sugar-beet seedlings. Crop Prot. 15, 479–486. doi: 10.1016/0261-2194(96)00014-2
Windels, C. E., Jacobsen, B. J., and Harveson, R. M. (2009). “Rhizoctonia root and crown rot,” in Compendium of beet diseases and pests, eds. R. M. Harveson, L. E. Hanson, and G. L. Hein (St. Paul, MN: American Phytopathological Society Press), 33–36.
Wolfgang, A., Zachow, C., Müller, H., Grand, A., Temme, N., Tilcher, R., et al. (2020). Understanding the impact of cultivar, seed origin, and substrate on bacterial diversity of the sugar beet rhizosphere and suppression of soil-borne pathogens. Front. Plant Sci. 11, 1–15. doi: 10.3389/fpls.2020.560869
Wyse, R. (1979). Parameters controlling sucrose content and yield of sugarbeet roots. 20th Gen. Meet. Am. Soc. Sugarbeet Technol. 20, 368–385. doi: 10.5274/jsbr.20.4.368
Yilmaz, N. D. K., and Tunali, B. (2010). Evaluation of Trichoderma spp. from central and northern regions of Turkey for suppression of Polymyxa betae as a vector of rhizomania disease. Arch. Phytopathol. Plant Prot. 43, 1534–1542. doi: 10.1080/03235400902927154
Zachow, C., Fatehi, J., Cardinale, M., Tilcher, R., and Berg, G. (2010). Strain-specific colonization pattern of Rhizoctonia antagonists in the root system of sugar beet. FEMS Microbiol. Ecol. 74, 124–135. doi: 10.1111/j.1574-6941.2010.00930.x
Zachow, C., Jahanshah, G., Bruijn, I., De Song, C., Ianni, F., Gerhardt, H., et al. (2015). The novel lipopeptide poaeamide of the endophyte Pseudomonas poae RE * 1-1-14 is involved in pathogen suppression and root colonization. Mol. Plant-Microbe Interact. 28, 800–810. doi: 10.1094/MPMI-12-14-0406-R
Zachow, C., Müller, H., Tilcher, R., and Berg, G. (2014). Differences between the rhizosphere microbiome of Beta vulgaris ssp. maritima - ancestor of all beet crops–and modern sugar beets. Front. Microbiol. 5, 1–13. doi: 10.3389/fmicb.2014.00415
Zachow, C., Müller, H., Tilcher, R., Donat, C., and Berg, G. (2013). Catch the best: Novel screening strategy to select stress protecting agents for crop plants. Agronomy. 3, 794–815. doi: 10.3390/agronomy3040794
Zachow, C., Tilcher, R., and Berg, G. (2008). Sugar beet-associated bacterial and fungal communities show a high indigenous antagonistic potential against plant pathogens. Microb. Ecol. 55, 119–129. doi: 10.1007/s00248-007-9257-7
Keywords: biofertilization, Beta vulgaris, Rhizoctonia, phylosymbiosis, microbiome, biocontrol, soil-borne pathogens
Citation: Wolfgang A, Temme N, Tilcher R and Berg G (2023) Understanding the sugar beet holobiont for sustainable agriculture. Front. Microbiol. 14:1151052. doi: 10.3389/fmicb.2023.1151052
Received: 25 January 2023; Accepted: 31 March 2023;
Published: 17 April 2023.
Edited by:
Rouhallah Sharifi, Razi University, IranReviewed by:
Song Baiquan, Heilongjiang University, ChinaPiergiorgio Stevanato, University of Padua, Italy
Copyright © 2023 Wolfgang, Temme, Tilcher and Berg. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gabriele Berg, Z2FicmllbGUuYmVyZ0B0dWdyYXouYXQ=
 Adrian Wolfgang
Adrian Wolfgang Nora Temme2
Nora Temme2 Ralf Tilcher
Ralf Tilcher Gabriele Berg
Gabriele Berg