
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 17 May 2023
Sec. Extreme Microbiology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1150224
This article is part of the Research Topic The Impact of the Space Environment on Microbial Growth and Behavior View all 13 articles
 Claudia Fagliarone1
Claudia Fagliarone1 Claudia Mosca1
Claudia Mosca1 Giorgia Di Stefano1,2
Giorgia Di Stefano1,2 Stefan Leuko3
Stefan Leuko3 Ralf Moeller3,4
Ralf Moeller3,4 Elke Rabbow5
Elke Rabbow5 Petra Rettberg5
Petra Rettberg5 Daniela Billi1*
Daniela Billi1*Cyanobacteria are gaining considerable interest as a method of supporting the long-term presence of humans on the Moon and settlements on Mars due to their ability to produce oxygen and their potential as bio-factories for space biotechnology/synthetic biology and other applications. Since many unknowns remain in our knowledge to bridge the gap and move cyanobacterial bioprocesses from Earth to space, we investigated cell division resumption on the rehydration of dried Chroococcidiopsis sp. CCMEE 029 accumulated DNA damage while exposed to space vacuum, Mars-like conditions, and Fe-ion radiation. Upon rehydration, the monitoring of the ftsZ gene showed that cell division was arrested until DNA damage was repaired, which took 48 h under laboratory conditions. During the recovery, a progressive DNA repair lasting 48 h of rehydration was revealed by PCR-stop assay. This was followed by overexpression of the ftsZ gene, ranging from 7.5- to 9-fold compared to the non-hydrated samples. Knowing the time required for DNA repair and cell division resumption is mandatory for deep-space experiments that are designed to unravel the effects of reduced/microgravity on this process. It is also necessary to meet mission requirements for dried-sample implementation and real-time monitoring upon recovery. Future experiments as part of the lunar exploration mission Artemis and the lunar gateway station will undoubtedly help to move cyanobacterial bioprocesses beyond low Earth orbit. From an astrobiological perspective, these experiments will further our understanding of microbial responses to deep-space conditions.
Long-term human presence on the Moon and future settlements on Mars will depend on the development of self-sufficient life support systems that will minimize dependence on resupply from Earth. Microorganisms offer great promise to support deep-space exploration: A so-called biomanufactory was proposed for human outposts on Mars, based on in situ resource utilization and biologically driven subunits for needed bioprocesses (Berliner et al., 2021). In such a scenario, cyanobacteria are gaining considerable interest due to their ability to produce oxygen and their potential as bio-factories for space biotechnology/synthetic biology applications (for a review see Mapstone et al., 2022). The “PowerCell” concept proposed cultivating cyanobacteria with raw materials available on the Moon and Mars to feed the bacteria used in biotechnology/synthetic biology (Rothschild, 2016). Recently, it was shown that the biomass produced by the desert cyanobacterium Chroococcidiopsis sp. 029 can be used as feedstock for bacteria (Billi et al., 2021).
Future lunar exploration mission Artemis and the lunar Gateway station (Crusan et al., 2019; Smith et al., 2020) will undoubtedly offer a unique opportunity to advance the use of cyanobacterial bioprocesses in space. Microorganisms on board the International Space Station (ISS) have exhibited global alterations in metabolic functions and gene expression, causing variations in biofilm formation, sporulation, and virulence (for a review see Milojevic and Weckwerth, 2020). Many unknowns remain in bridging the gaps in knowledge required to move cyanobacterial bioprocesses from Earth to space, and, for realistic microbial bioprocesses in space, a deeper understanding of the recovery process after traveling in space and subsequent reactivation is mandatory. In NASA's BioSentinel CubeSat, launched as a secondary payload on Artemis-1, dried cells of Saccharomyces cerevisiae are scheduled to be reactivated during an 18-month permanence in deep space, meaning that DNA damage repair will be monitored in deep space (Massaro Tieze et al., 2020; Santa Maria et al., 2020).
The investigations performed using ground-based simulations or facilities installed outside the ISS offer a baseline testbed for deep-space experiments by providing insights into microbial survivability after they have traveled through space and undergone subsequent reactivation. Post-flight analyses of the rehydration of the cyanobacterium Chroococcidiopsis sp. 029 exposed to space and Mars-like conditions during the BOSS (Biofilms Organisms Surfing Space) space mission showed: i) the upregulation of DNA repair genes in cells exposed to space vacuum (Mosca et al., 2021) and ii) the absence of increased genome variants in cells exposed to Mars-like conditions (Napoli et al., 2022). An upregulation of DNA repair genes was also reported during the rehydration of dried cells irradiated with Fe ions (Mosca et al., 2022) during the STARLIFE irradiation campaign (Moeller et al., 2017).
These results on the recovery of dried Chroococcidiopsis sp. 029 with accumulated DNA damage laid the foundation for its suitability for deep-space experimentation, which is necessary to unravel the effect of reduced/microgravity on the recovery process. However, even though the role of DNA repair genes was reported, it is fundamental to assess the timeframe needed for DNA repair and cell division resumption upon rehydration. This knowledge might be vital to meet mission requirements for dried-sample implementation and real-time monitoring upon recovery, in case of extensive pre-flight periods and unguaranteed sample return.
The link between DNA damage repair and cell division arrest is provided by the regulation of the ftsZ gene expression. In bacteria such as Escherichia coli, when RecA binds to single-stranded DNA, it stimulates the autocatalytic cleavage of LexA. This is then followed by the expression of the SOS repair genes, including the sulA gene that causes cell division delay through the inhibition of the FtsZ polymerization (Kreuzer, 2013). A comparative genome analysis suggested that the E. coli-like SOS model might not be valid for all cyanobacteria: The lexA gene, which regulates the SOS system, is unevenly distributed among the analyzed genomes, while the sulA gene, which stalls cell division until DNA reparation, is ubiquitous (Cassier-Chauvat et al., 2016). Thus far, it is unknown whether the E. coli-like SOS model is present in Chroococcidiopsis sp. 029, although it was reported that FtsZ is a critical component of its cell division machinery (Billi, 2009).
The present study aimed to determine the timeframe between DNA damage repair and cell division resumption upon the rehydration of dried Chroococcidiopsis sp. 029 that has undergone DNA damage. Lesions were accumulated during the exposure to space and Mars-like conditions during the BOSS space experiment and Fe-ion radiation during the STARLIFE project.
The presence of genome lesions and their repair upon rehydration was determined by using a PCR-stop assay by amplifying a 4-kbp genome fragment. Cell division resumption was determined by monitoring the expression of the cell division ftsZ gene by quantitative polymerase chain reaction (RT-qPCR) during 48 h of rehydration.
Chroococcidiopsis sp. 029 was isolated by Roseli Ocampo-Friedmann from cryptoendolithic growth in sandstone in the Negev Desert (Israel) and maintained at the University of Rome Tor Vergata, as part of the Culture Collection of Microorganisms from Extreme Environments established by E. Imre Friedmann. Cultures were grown under routine conditions at 25°C in BG-11 liquid medium (Rippka et al., 1979) under a photon flux density of 40 μmol/m2/s provided by fluorescent cool-white bulbs. Dried samples for the BOSS space experiment of the EXPOSE-R2 space mission were prepared as previously reported (Billi et al., 2019). Biofilms were obtained by growing cyanobacterial cells on top of BG-11 agarized medium for about 2 months, following which they were allowed to air-dry for about 15 days under routine growth conditions and finally stored in the dark under laboratory conditions. Dried cells for the STARLIFE experiment were prepared as previously reported (Verseux et al., 2017). Briefly, cells from cultures in the early stationary phase were immobilized on Millipore filters, air-dried overnight in a sterile hood, and stored in the dark under laboratory conditions.
In the context of the BOSS space experiment, which aimed to investigate biofilm resistance under space and Mars-like conditions in LEO (Cottin and Rettberg, 2019), dried biofilms of Chroococcidiopsis sp. 029 were accommodated in the compartment C4 of the tray 1 “space” and of the tray 2 “Mars” of the EXPOSE-R2 hardware (Figure 1). In the present study, we analyzed samples from the middle-layer carrier that were exposed for 672 days to a space vacuum and 0.5 Gy of cosmic ionizing radiation and temperature values ranging from −20.9°C to 57.98°C (Dachev et al., 2017; Rabbow et al., 2017). After retrieval to Earth, 2.5-year-old samples (considering 900 days from launch to space and sample shipping to the laboratory) were stored for an additional 4.5 years in an air-dried state under laboratory conditions. Control biofilms were air-dried and stored in the dark at room temperature for 7 years.
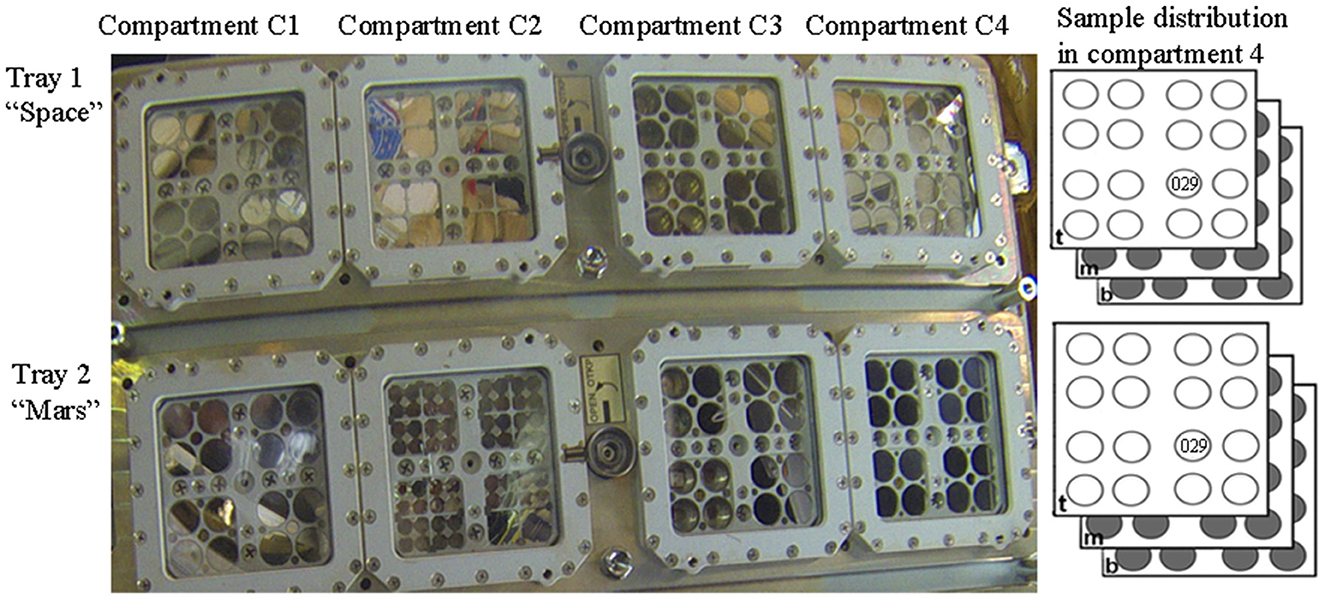
Figure 1. Sample distribution in the EXPOSE-R2 hardware. In compartment 4 of Tray 1 “Space” and Try 2 “Mars”, samples were allocated in top-layer (t), middle (m), and bottom-layer carrier (b). Dried biofilms of Chroococcidiopsis sp. 029 used in this study were exposed in the position 11 middle-layer of the carrier. Photo credits: Roscosmos and ESA.
Cells were first immobilized on Millipore filters and shipped to the Heavy Ion Medical Accelerator in Chiba (HIMAC, Gunma University Heavy Ion Medical Center, Japan) for irradiation, which was performed in June 2018 during the STARLIFE project, as previously reported (Mosca et al., 2022). After irradiation with 2 kGy of Fe ions, the air dried samples were stored in the dark at room temperature for 2 years.
Aliquots of dried cells and dried-exposed cells (about 25 mm2) were resuspended in 1 ml of BG-11 and allowed to recover under standard growth conditions as reported above.
Genomic DNA was extracted from dried samples and from samples at different time points of rehydration by modifying a method previously reported (Billi et al., 1998). Briefly, the lysis of Chroococcidiopsis cells was achieved by adding hot phenol and glass beads. Then the pooled aqueous phases were first extracted with phenol–chloroform–isoamyl alcohol and followed by chloroform–isoamyl alcohol as described (Mosca et al., 2019). The PCR-stop assay was performed by amplifying a 4-kbp genome fragment using 6 ng of genomic DNA in 12-μl reaction mixtures containing 0.5 μM each of primers Chroo-4 K-2-F (5′-GCTACTCGTTGCTTTGCGTC-3′) and Chroo-4 K-2-R (5′-TTCCCCATACTTTGCTTCCCA-3′) and 6 μl of High-Fidelity Master Mix (Thermo Fisher Scientific, Waltham, MA, USA) as described (Mosca et al., 2019). A no-template control was included. Each one of the 12-μl PCR reaction mixtures was loaded onto a 1.5% agarose gel containing 0.5 mg/ml of ethidium bromide, subjected to electrophoresis for about 1 h at 90 V, and visualized with a trans-illuminator.
At different time points of rehydration, total RNA was extracted as described (Fagliarone et al., 2020). Real-time reactions were performed in 12 μl, including 1 μg of cDNA template, 6 μl of iTaqTM universal SYBR® Green Supermix (Bio-Rad Laboratories, Hercules, CA, USA), and 400 nM of forward primer chrftsZ-F (5′-GTTGACTTTGCCGATGTCCG-3′) and reverse primer chrftsZ-R (5′-CTTCTCTGGCACGGGACTTT-3′) that were designed based on a 473-bp fragment previously identified (Billi, 2009). PCR cycling conditions were as follows: a cycle of 95°C for 10 min, then 40 cycles of 95°C for 15 s, and 60°C for 1 min, followed by a ramp from 60 to 95°C for the melting curve stage. Quantification was performed using the Ct comparative method (2−ΔΔCt method) and using the 16S rRNA gene as the housekeeping gene (GenBank accession number AF279107) and employing the primers chr16S-F (5′-TACTACAATGCTACGGACAA-3′) and chr16S-R (5′-CCTGCAATCTGAACTGAG-3′). All cDNA quantities were normalized to 16S rRNA quantities, and fold changes of gene transcripts were compared to the values of non-rehydrated samples set as 1. Values >1 were considered upregulated, and values <1 were considered downregulated. Five rehydration time points (1, 6, 12, 24, and 48 h) were evaluated for dried cells exposed to Fe ions and 7 years of dried storage. Three rehydration time points (1, 6, and 48 h) were tested for dried biofilms exposed during the BOSS space mission, due to the limited availability of samples. In each PCR protocol, three replicates were performed, and no-template control was included.
Only one sample for each condition was allocated in the EXPOSE-R2 facility. Therefore, three replicates for biologically distinct samples (different cultures) exposed to air-drying under laboratory conditions and Fe-ion irradiation were used, as well as three replicates of a single sample exposed to space vacuum and Mars-like conditions. Data are shown with standard deviation, and significance was assessed by using Student's t-test.
The presence of DNA damage was qualitatively determined by using a PCR-stop assay that evaluates the genome suitability as a template for PCR amplifications since lesions impede DNA polymerase progression (Kumar et al., 2004). When a 4-kbp genome fragment was amplified as a target, a reduction of the amplicon intensity was observed in dried biofilms exposed to Mars-like conditions (Figure 2, lane 4) compared to control liquid culture (Figure 2, lane 2) and dried control biofilms maintained in the dark at room temperature for 7 years (Figure 2, lane 3). No amplicons occurred in the PCR-stop assay performed with no genomic DNA (not shown). DNA damage in dried cells exposed to a space vacuum and to 2 kGy of Fe ions was previously reported (Mosca et al., 2021, 2022).
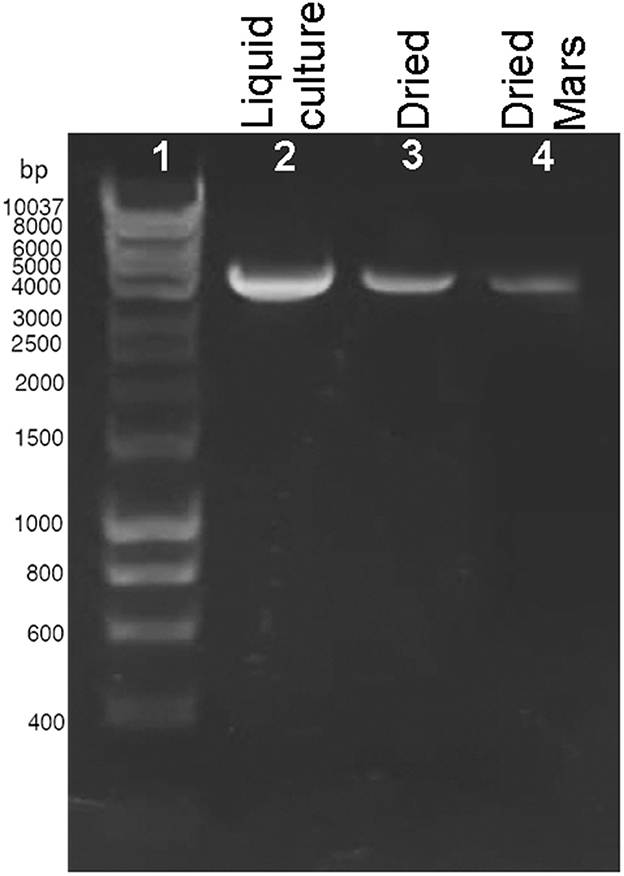
Figure 2. DNA damage accumulation in Chroococcidiopsis sp. 029 evaluated by PCR-stop assay with a 4-kbp amplification target. Control liquid culture (lane 2), dried biofilms maintained for 7 years in the dark, at room temperature (lane 3), and dried biofilms exposed to Mars-like conditions (lane 4). Lane 1: Hyperladder 1 kbp (Bioline Meridian Life Science, Memphis, TN, USA).
During 48 h of rehydration, the 4-kbp amplicon was gradually restored in both dried cells exposed to 2 kGy of Fe ions (Figure 3A) and dried biofilms maintained in the dark at room temperature for 7 years (Figure 3B). For both samples, the intensity of the amplicon band continued to increase after 6 h of rehydration (Figures 3A, B), and no amplicons were observed in the PCR-stop assay performed with no genomic DNA (not shown). Due to the limited sample availability of the dried samples employed in the BOSS space mission, the DNA recovery during rehydration was not evaluated.
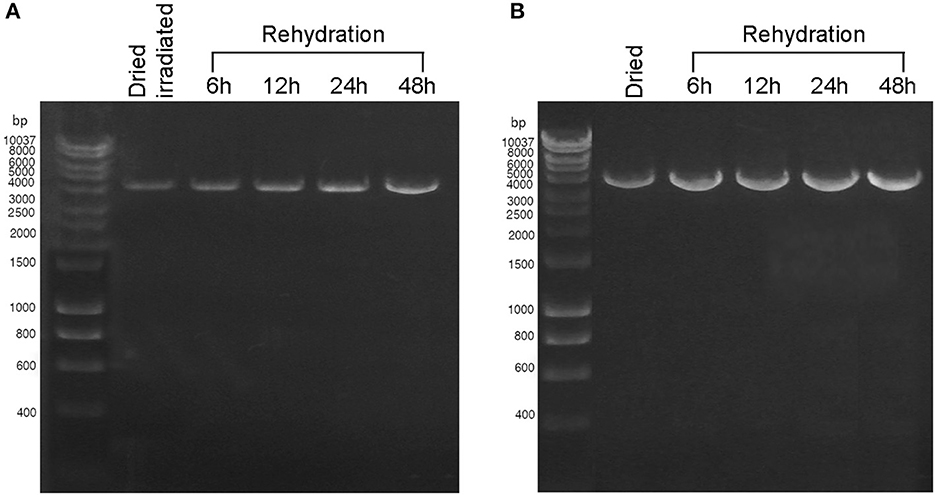
Figure 3. DNA damage restoration upon the rehydration of Chroococcidiopsis sp. 029 evaluated by PCR-stop assay with a 4-kbp amplification target. (A) Dried cells were exposed to 2 kGy of Fe ions (lane 2) and after 6, 12, 24, and 48-h recovery (lanes 3–6). (B) Dried cells were stored for 7 years in the dark at room temperature (lane 2) and after 6, 12, 24, and 48 h of recovery (lanes 3–6). Lane 1: Hyperladder 1 kbp (Bioline Meridian Life Science, Memphis, TN, USA).
The cell division resumption was monitored during the 48 h of rehydration of dried cells with accumulated DNA damage, by determining the expression of the ftsZ gene (Figure 4). Dried biofilms exposed to a space vacuum did not show any upregulation of the ftsZ gene during 1 h and 6 h of rehydration, while this gene was 7.5-fold upregulated after 48 h of rehydration, compared to the non-rehydrated sample (Figure 4A). In comparison to the non-rehydrated sample, dried biofilms exposed to Mars-like conditions showed a 2.5-fold increase in the transcriptional level of the ftsZ gene after 6 h and a 9-fold increase after 48 h recovery (Figure 4B). During the rehydration of dried cells irradiated with 2 kGy of Fe ions, the ftsZ gene showed a 3-fold upregulation after 48 h of rehydration (Figure 4C). The ftsZ gene was not upregulated during 1, 6, 12, and 24 h of rehydration in biofilms that had been air-dried in the dark for 7 years at room temperature. However, the gene was 10.4-fold over-expressed after 48 h of rehydration compared to the non-rehydrated sample (Figure 4D).
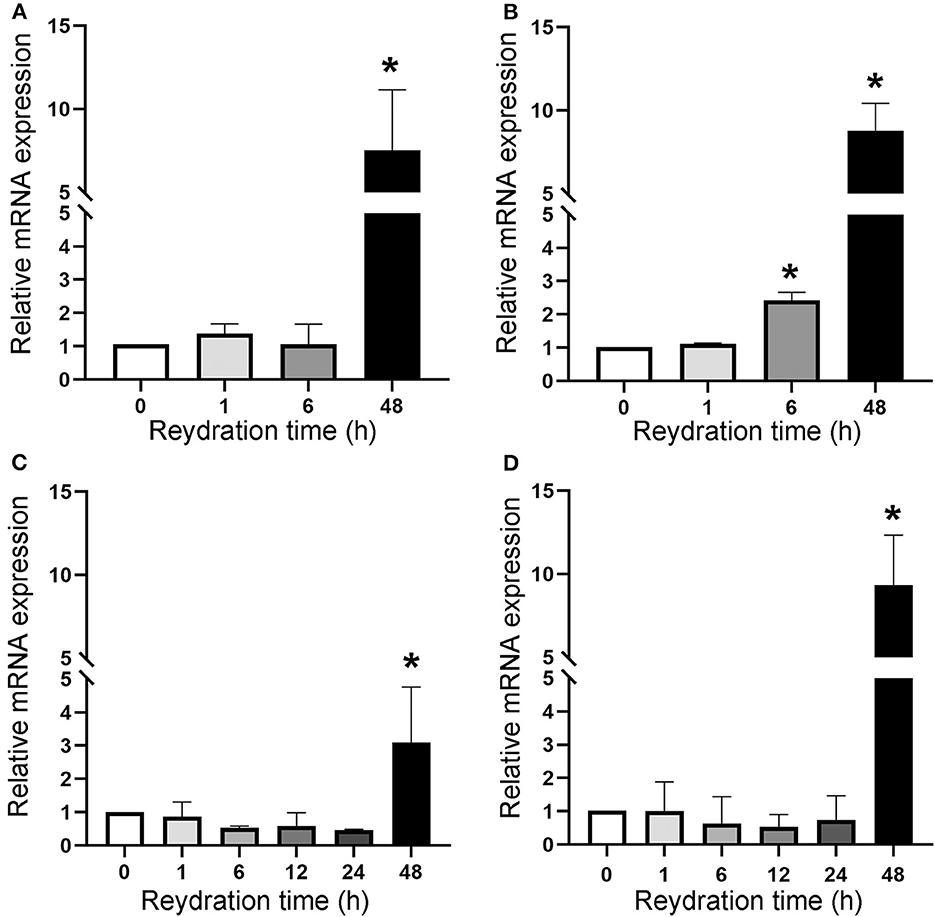
Figure 4. Expression of ftsZ gene upon the recovery of dried biofilms of Chroococcidiopsis sp. 029 exposed to space vacuum (A), Mars-like conditions (B), irradiation with Fe ions (C), and in dried biofilms maintained for 7 years in the dark at room temperature (D). Values from dried samples were considered control and set as 1. Data represent mean ± SD (n = 3), *p < 0.05.
With the upcoming long-term space explorations, it is becoming increasingly important to broaden our understanding of microbial bioprocesses toward moving the same from Earth to space. Cyanobacteria are expected to play a key role in supporting long-term human presence on the Moon and future settlements on Mars (Mapstone et al., 2022). Thus, a deeper understanding of their recovery process after traveling in space and subsequent reactivation is very relevant.
Here, the timeframe required for DNA repair and cell division resumption under laboratory growth conditions was assessed during the rehydration of dried cells of the cyanobacterium Chroococcidiopsis sp. 029 that accumulated DNA damage during the BOSS space experiment (Mosca et al., 2021) and Fe-ion during the STARLIFE irradiation campaign (Mosca et al., 2022). Upon rehydration, the expression of the cell division gene ftsZ gene and the progression of the DNA damage restoration indicated that the cell division resumption required 48 h from the rehydration onset. Although the key role of FtsZ in the cell division machinery of Chroococcidiopsis sp. 029 was previously reported (Billi, 2009), the molecular mechanism involved in delaying cell division until DNA damage is repaired remains to be elucidated. However, a possible involvement of the sulA gene is anticipated since, in agreement with its ubiquity in cyanobacteria (Cassier-Chauvat et al., 2016), it is present in the genome of Chroococcidiopsis sp. 029 (Billi D. personal communication).
Due to the limited sample availability of the flight samples, the recovery of DNA damage was monitored by PCR-stop assay during the rehydration of air-dried cells and dried cells exposed to Fe-ion radiation, but not of dried cells exposed to Mars-like conditions and space vacuum. Although the PCR-stop assay did not determine the lesion type, the presence of DNA lesions in dried cells exposed to space vacuum and Fe-ion irradiation is known to be largely represented by single- and double-strand breaks and oxidative damage. Previous research has shown that genes of the homologous recombination RecF pathway and base excision are upregulated during the rehydration of dried cells exposed to space vacuum and Fe-ion irradiation (Mosca et al., 2021, 2022).
In the BOSS space mission, dried Chroococcidiopsis sp. 029 allocated in the middle-layer carrier of tray 1 “Space” was exposed to a space vacuum combined with 0.5 Gy of cosmic ionizing radiation (Dachev et al., 2017). Although this is a non-lethal dose for a cyanobacterium that can tolerate up to 12 kGy in the dried state (Verseux et al., 2017), it simulates the dose accumulated during a trip to Mars. Hence, it is anticipated that a 48-h recovery should be allowed in future space experiments aimed to investigate the effects of reduced gravity/microgravity on the DNA repair process upon the rehydration of dried cells that accumulated DNA damage.
In the BOSS space mission, dried Chroococcidiopsis sp. 029 allocated in the middle-layer carrier of tray 2 “Mars” experienced about 0.5 Gy of cosmic ionizing radiation in a Mars-like atmosphere (Rabbow et al., 2017). The exposure to 980 Pa of a gas mixture composed mainly of CO2 might explain reduced DNA damage as suggested by a 2.5-fold expression of the ftsZ gene after 6 h of rehydration, whereas a 9-fold expression occurred after 48 h of rehydration. The absence of oxygen in the tray 2 “Mars” might have caused reduced DNA damage as previously reported (Schulte-Frohlinde et al., 1994). In the context of microbial-based technologies for space settlements on Mars, a scenario of accidental failures causing exposure to a combination of low-pressure and CO2 should be taken into account (Cockell, 2022). Thus, this result is relevant for biomanufactory on Mars.
The suitability of Chroococcidiopsis sp. 029 for deep-space investigations was further supported by DNA repair and cell division resumption of dried cells exposed to Fe ions, which are the more lethal components of the galactic cosmic radiation since they induce complex DNA damage (Horneck et al., 2010). The DNA repair and cell division resumption of dried biofilms stored for 7 years in air-dried state in the dark at room temperature further highlighted the suitability of this cyanobacterium for space missions that require extensive pre-flight periods.
In conclusion, the monitoring of the ftsZ gene expression determined the timeframe for DNA repair and cell division resumption. Data from this study have important implications for further studies on the effects of reduced gravity/microgravity on this process, as well as, for deep-space missions requiring the implementation of dried samples and real-time monitoring upon reactivation. Within the upcoming lunar exploration mission Artemis and lunar Gateway station, deep-space experiments will facilitate further understanding toward moving microbial bioprocesses beyond Earth. From an astrobiological perspective, such experimentation will further our understanding of microbial response to deep-space conditions.
The original contributions presented in the study are included in the article, further inquiries can be directed to the corresponding author.
DB: supervision, conceptualization, and writing—review and editing. PR: conceived and coordinated the BOSS experiment. RM: conceived and coordinated the STARLIFE experiment. ER and SL: contributed to sample exposure. CF, CM, and GDS: contributed to methodology, validation, and data analysis. All authors discussed the results and commented on the manuscript. All authors contributed to the article and approved the submitted version.
This research was supported by the Italian Space Agency (ASI, DC-VUM-2017-034, Grant No. 2019-3 U.0 Life in Space to DB). RM and SL were supported by the Grant DLR-FuE-Projekt ISS LIFE (Programm RF-FuW, Teilprogramm 475).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Berliner, A. J., Hilzinger, J. M., Abel, A. J., McNulty, M. J., Makrygiorgos, G., Averesch, N. J. H., et al. (2021). Towards a biomanufactory on Mars. Front. Astron. Space Sci. 8. doi: 10.3389/fspas.2021.711550
Billi, D. (2009). Loss of topological relationships in a Pleurocapsalean cyanobacterium (Chroococcidiopsis sp.) with partially inactivated ftsZ. Ann. Microbiol. 59, 1–4. doi: 10.1007/BF03178322
Billi, D., Gallego Fernandez, B., Fagliarone, C., Chiavarini, S., and Rothschild, L. J. (2021). Exploiting a perchlorate-tolerant desert cyanobacterium to support bacterial growth for in situ resource utilization on Mars. Int. J. Astrobiol. 20, 29–35. doi: 10.1017/S1473550420000300
Billi, D., Grilli Caiola, M., Paolozzi, L., and Ghelardini, P. (1998). A method for DNA extraction from the desert cyanobacterium Chroococcidiopsis and its application to identification of ftsZ. Appl. Environ. Microbiol. 64, 4053–4056. doi: 10.1128/AEM.64.10.4053-4056.1998
Billi, D., Staibano, C., Verseux, C., Fagliarone, C., Mosca, C., Baqué, M., et al. (2019). Dried biofilms of desert strains of Chroococcidiopsis survived prolonged exposure to space and Mars-like conditions in low Earth orbit. Astrobiology 19, 1008–1017. doi: 10.1089/ast.2018.1900
Cassier-Chauvat, C., Veaudor, T., and Chauvat, F. (2016). Comparative genomics of DNA recombination and repair in cyanobacteria: biotechnological implications. Front. Microbiol. 7, 1809. doi: 10.3389/fmicb.2016.01809
Cockell, C. S. (2022). Bridging the gap between microbial limits and extremes in space: Space microbial biotechnology in the next 15years. Microb. Biotechnol. 15, 29–41. doi: 10.1111/1751-7915.13927
Cottin, H., and Rettberg, P. (2019). EXPOSE-R2 on the International Space Station (2014-2016): Results from the PSS and BOSS astrobiology experiments. Astrobiology 19, 975–978. doi: 10.1089/ast.2019.0625
Crusan, J., Bleacher, J., Caram, J., Craig, D., Goodliff, K., Herrmann, N., et al. (2019). NASA's Gateway: An Update on Progress and Plans for Extending Human Presence to Cislunar Space. Manhattan, NY: IEEE Aerospace Conference (IEEE). p. 1–19.
Dachev, T. P., Bankov, N. G., Tomov, B. T., Matviichuk, Y. N., Dimitrov, P. G., Häder, D.-P., et al. (2017). Overview of the ISS radiation environment observed during the ESA EXPOSE-R2 mission in 2014–2016. Space Weather 15, 1475–1489. doi: 10.1002/2016SW001580
Fagliarone, C., Napoli, A., Chiavarini, S., Baqué, M., de Vera, J.-P., and Billi, D. (2020). Biomarker preservation and survivability under and Mars-like UV flux of a desert cyanobacterium capable of trehalose and sucrose accumulation. Front. Astron. Space Sci. 7, 31. doi: 10.3389/fspas.2020.00031
Horneck, G., Klaus, D. M., and Mancinelli, R. L. (2010). Space microbiology. Microbiol. Mol. Biol. Rev. 74, 121–156. doi: 10.1128/MMBR.00016-09
Kreuzer, K. N. (2013). DNA damage responses in prokaryotes: Regulating gene expression, modulating growth patterns, and manipulating replication fork. Cold Spring Harb. Perspect. Biol. 5, a012674. doi: 10.1101/cshperspect.a012674
Kumar, A., Tyagi, M. B., and Jha, P. N. (2004). Evidences showing ultraviolet-B radiation-induced damage of DNA in cyanobacteria and its detection by PCR assay. Biochem. Biophys. Res. Commun. 318, 1025–1030. doi: 10.1016/j.bbrc.2004.04.129
Mapstone, L. J., Leite, M. N., Purton, S., Crawford, I. A., and Dartnell, L. (2022). Cyanobacteria and microalgae in supporting human habitation on Mars. Biotechnol. Adv. 59, 107946. doi: 10.1016/j.biotechadv.2022.107946
Massaro Tieze, S., Liddell, L. C., Santa Maria, S. R., and Bhattacharya, S. (2020). BioSentinel: a biological cubesat for deep space exploration. Astrobiology Apr 13. doi: 10.1089/ast.2019.2068
Milojevic, T., and Weckwerth, W. (2020). Molecular mechanisms of microbial survivability in outer space: a systems biology approach. Front. Microbiol. 11, 923. doi: 10.3389/fmicb.2020.00923
Moeller, R., Raguse, M., Leuko, S., Berger, T., Hellweg, C., Fujimori, A., et al. (2017). STARLIFE—An international campaign to study the role of galactic cosmic radiation in astrobiological model systems. Astrobiology 17, 101–109. doi: 10.1089/ast.2016.1571
Mosca, C., Fagliarone, C., Napoli, A., Rabbow, E., Rettberg, P., and Billi, D. (2021). Revival of anhydrobiotic cyanobacterium biofilms exposed to space vacuum and prolonged dryness: Implications for future missions beyond low Earth orbit. Astrobiology 21, 541–550. doi: 10.1089/ast.2020.2359
Mosca, C., Napoli, A., Fagliarone, C., Fujimori, A., Moeller, R., and Billi, D. (2022). Role of DNA repair pathways in the recovery of a dried, radioresistant cyanobacterium exposed to high-LET radiation: implications for the habitability of Mars. Int J Astrobiol 1–12. doi: 10.1017/S1473550422000131
Mosca, C., Rothschild, L. J., Napoli, A., Ferr,é, F., Pietrosanto, M., Fagliarone, C., et al. (2019). Over-expression of UV-damage DNA repair genes and ribonucleic acid persistence contribute to the resilience of dried biofilms of the desert cyanobacetrium Chroococcidiopsis exposed to Mars-like UV flux and long-term desiccation. Front. Microbiol. 10, 2312. doi: 10.3389/fmicb.2019.02312
Napoli, A., Micheletti, D., Pindo, M., Larger, S., Cestaro, A., de Vera, J. P., et al. (2022). Absence of increased genomic variants in the cyanobacterium Chroococcidiopsis exposed to Mars-like conditions outside the space station. Sci. Rep. 12, 8437. doi: 10.1038/s41598-022-12631-5
Rabbow, E., Rettberg, P., Parpart, A., Panitz, C., Schulte, W., Molter, F., et al. (2017). EXPOSE-R2: the astrobiological ESA mission on board of the International Space Station. Front. Microbiol. 8, 1533. doi: 10.3389/fmicb.2017.01533
Rippka, R., Deruelles, J., Waterbury, J. B., Herdman, M., and Stanier, R. Y. (1979). Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 111, l−61. doi: 10.1099/00221287-111-1-1
Rothschild, L. J. (2016). Synthetic biology meets bioprinting: enabling technologies for humans on Mars (and Earth). Biochem. Soc. Trans. 44, 1158–1164. doi: 10.1042/BST20160067
Santa Maria, S. R., Marina, D. B., Massaro Tieze, S., Liddell, L. C., and Bhattacharya, S. (2020). BioSentinel: long-term Saccharomyces cerevisiae preservation for a deep space biosensor mission. Astrobiology. doi: 10.1089/ast.2019.2073 [Epub ahead of print].
Schulte-Frohlinde, D., Ludwig, D. C., and Rettberg, P. (1994). Influence of thiols and oxygen on the survival of gamma-irradiated plasmid DNA and cells. Adv. Space Res. 14, 277–284. doi: 10.1016/0273-1177(94)90478-2
Smith, M., Craig, D., Herrmann, N., Mahoney, E., Krezel, J., McIntyre, N., et al. (2020). The Artemis Program: An Overview of NASA's Activities to Return Humans to the Moon. IEEE Aerospace Conference, Big Sky, MT.
Keywords: outer space, DNA damage, cell division, Fe-ion radiation, desert cyanobacteria
Citation: Fagliarone C, Mosca C, Di Stefano G, Leuko S, Moeller R, Rabbow E, Rettberg P and Billi D (2023) Enabling deep-space experimentations on cyanobacteria by monitoring cell division resumption in dried Chroococcidiopsis sp. 029 with accumulated DNA damage. Front. Microbiol. 14:1150224. doi: 10.3389/fmicb.2023.1150224
Received: 23 January 2023; Accepted: 21 April 2023;
Published: 17 May 2023.
Edited by:
Donatella Tesei, University of Natural Resources and Life Sciences Vienna, AustriaReviewed by:
Rajeshwar P. Sinha, Banaras Hindu University, IndiaCopyright © 2023 Fagliarone, Mosca, Di Stefano, Leuko, Moeller, Rabbow, Rettberg and Billi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daniela Billi, YmlsbGlAdW5pcm9tYTIuaXQ=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.