
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 15 March 2023
Sec. Food Microbiology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1150217
This article is part of the Research Topic Postharvest Disease Management in Fruits and Vegetables: Recent Advances and Mechanisms View all 6 articles
The use of synthetic fungicides against postharvest Alternaria rot adversely affects human health and the environment. In this study, as a safe alternative to fungicides, Bacillus subtilis strain Y17B isolated from soil exhibited significant antifungal activity against Alternaria alternata. Y17B was identified as B. subtilis based on phenotypic identification and 16S rRNA sequence analysis. To reveal the antimicrobial activity of this strain, a PCR-based study detected the presence of antifungal lipopeptide (LP) biosynthetic genes from genomic DNA. UPLC Q TOF mass spectrometry analysis detected the LPs surfactin (m/z 994.64, 1022.68, and 1026.62), iturin (m/z 1043.56), and fengycin (m/z 1491.85) in the extracted LP crude of B. subtilis Y17B. In vitro antagonistic study demonstrated the efficiency of LPs in inhibiting A. alternata growth. Microscopy (SEM and TEM) studies showed the alteration of the morphology of A. alternata in the interaction with LPs. In vivo test results revealed the efficiency of LPs in reducing the growth of the A. alternata pathogen. The overall results highlight the biocontrol potential of LPs produced by B. subtilis Y17B as an effective biological control agent against A. alternata fruit rot of cherry.
Alternaria alternata is a common storage pathogen that causes considerable postharvest contamination and rot of fruit crops. It can grow rapidly at low temperatures and causes food spoilage during transport and storage (Ostry, 2008; Xu et al., 2018). Alternaria species are reported to produce more than 60 different secondary metabolites, including mutagenic Alternaria mycotoxins, during their growth on the substrate, which leads to a high risk of human and animal health hazards (Pruß et al., 2014; Andersen et al., 2015). Every year, a large amount of food and feedstuff is contaminated and spoiled due to the growth of Alternaria species and their toxins. Additionally, previous studies have reported that A. alternata is associated with asthma (Sanchez and Bush, 2001). Owing to the great damage caused by Alternaria species, it is necessary to study and develop an effective management strategy for this fungus. Currently, the use of fungicides is the most widely used method to control A. alternata (Soylu and Kose, 2015) but their application has a long-term residual effect, which poses a high potential risk to health, food safety, and environmental pollution (Özkara et al., 2016).
Biological control is regarded as a reliable and safe alternative to chemicals (Li et al., 2016). Some bacterial species have been reported as promising biological control agents due to their strong antagonistic activity. They possess multiple modes of action and produce a wide variety of biologically active compounds with antifungal potential against different preharvest and postharvest diseases (Gond et al., 2015; Mnif et al., 2016). Among these antagonistic bacteria, Bacillus is well known for its production of broad-spectrum antimicrobial compounds, plant growth promotion, enhancement of plant biomass, and induction of systemic resistance in plants against plant pathogenic fungi (Ongena and Jacques, 2008; Alina et al., 2015; Fatima et al., 2023). Bacillus species have recently emerged as an interesting source of biocontrol agents for postharvest fungal disease management. These species showed different antifungal mechanisms, including the production of antifungal volatile organic compounds (VOCs) and lipopeptides (LPs), the induction of disease resistance, and nutrient competition (Wang et al., 2022b). B. amyloliquefaciens, B. subtilis, B. licheniformis, and B. cereus have been reported to produce LPs with a high antifungal activity (Lawrance et al., 2014; Zhao et al., 2017; Basit et al., 2018; de Souza et al., 2018). Three families of LPs, iturin, surfactin, and fengycin, have been reported to be inhibitory against pathogenic fungi and oomycetes (Stein, 2005; Ongena and Jacques, 2008). Based on their structures, iturin is a cyclopeptide consisting of C14 to C17, β-amino fatty acids, and heptapeptides. Surfactin comprises cyclic lactone rings (C13 to C16), β-hydroxy fatty acids, and heptapeptides. Fengycin is a decapeptide and is composed of β-hydroxy fatty acid chains, which form cyclic lactone rings (Ahmad et al., 2020). The antimicrobial potential of LPs is closely related to the peptide cyclization, amino acid residual sequence, and branching length of the fatty acid chain (Zhao et al., 2018). LPs are low molecular weight compounds that are synthesized by a complex of multi-enzymes, called non-ribosomal peptide synthetases (NRPSs). Most of the short sequence peptides, generally less than 50 amino acids, have been mostly identified as antimicrobial peptides. Therefore, NRPSs can generate a range of variants based on their amino acid sequence and fatty acid chain length. Bacillus species can simultaneously produce different families of LPs and their isoforms, i.e., surfactin, fengycin, and iturin (Finking and Marahiel, 2004; Montesinos, 2007; Marahiel, 2009; Han et al., 2015; Yang et al., 2015; Deng et al., 2017).
The antifungal potential of LPs has been reported against several postharvest fungal pathogens (Arrebola et al., 2010; Arroyave-Toro et al., 2017; Cozzolino et al., 2020). However, the antifungal potential of LPs produced by B. subtilis against A. alternata has not yet been systematically studied. To understand the mechanism of LPs against fruit decay of postharvest cherry caused by Alternaria, we were prompted to evaluate its antifungal potential. The primary objective of the present study was (a) to isolate a Bacillus strain with promising antifungal potential against A. alternata, (b) to identify genes in the B. subtilis Y17B genome that are involved in synthesizing antifungal LPs, (c) to identify antifungal LPs in the crude extract of Y17B on the basis of molecular weight using ultra-high-performance liquid chromatography-quadrupole time-of-flight mass spectrometry (UPLC Q TOF-MS) and test their combined antifungal activity in vitro, and (d) to examine alterations of the morphology of A. alternata upon interaction with these LPs using scanning electron microscopy (SEM) and transmission electron microscopy (TEM). Furthermore, the biocontrol potential of LPs was tested on cherry fruit to demonstrate the ability of LPs to protect it against A. alternata infection. To the best of our knowledge, this is the first report to explain the efficiency of LP activity in enhancing the shelf life of cherry fruit against A. alternata.
The soil samples were collected from Tengchong (Thermal Sea area), Baoshan City, Yunnan Province, China in 2019. Isolation was carried out using a method described previously (Chen et al., 2017) with slight modification. Each soil sample (5 g) was placed in a 100-ml flask containing 50 ml of sterile water mixed with 0.1% NaCl to make the sample suspensions a concentration of 10−1, which were then incubated overnight at room temperature. The sample suspensions were adjusted to concentrations of 10−4 and 10−6 through serial dilution, and 10 μl of each concentration was streaked on Luria–Bertani (LB) agar medium and incubated at 37°C for 4 days. Single purified colonies were obtained by re-streaking two times and preserved in 25% glycerol at −18°C. The single colonies were further screened for their ability to produce antifungal agents. Alternaria alternata ACT-1, isolated by Ahmad et al. (2020) and associated with postharvest fruit rot of cherry, was preserved in 25% glycerol solution and occasionally retrieved and grown on potato dextrose agar (PDA) medium prior to conducting experiments.
The antifungal effect of bacterial strains was evaluated in a dual culture assay described previously (Wu et al., 2019) with slight modification. A mycelium plug (5 mm) was taken from a 7-day-old culture of A. alternata and placed at the center of the PDA plates. Then, 10 μl of bacterial culture was inoculated 30 mm away from the mycelium plug and incubated at 28°C for 7 days post inoculation (dpi). To assess antifungal activity, the percentage of growth inhibition was calculated using the following formula described by Vincent (1947) at 7 dpi:
I = [(C−T)/C] × 100
where I is the percentage of growth inhibition, C is growth in control, and T is growth in treatment.
For morphological characterization, B. subtilis Y17B culture (10 μl) was streaked on an LB agar plate and incubated at 37°C for 4 days. The colony patterns were observed under a stereomicroscope. For SEM study, the specimen was washed twice with phosphate buffer, then fixed with 2.5% glutaraldehyde buffer and stored at 4°C for 24 h. Then, the specimen was rinsed three times with 100 mM phosphate buffer sequentially for 5 min and postfixed in 1% osmium tetroxide fixation solution for 2 h. Next, the specimen was subjected to gradient dehydration with 30, 50, 60, 70, 80, 90, 95, and 100% ethanol solution sequentially for 15 min. The specimen was dried with a Leica EM CPD030 automated dryer (Leica Microsystems Inc., Germany), attached to a sample stage, and sputtered with gold coating by ion sputtering (HITACHI MC1000). Morphology was observed using a HITACHI-SU8010 SEM (Hitachi High-Technologies Corporation, Tokyo, Japan). Biochemical and physiological identification of strain Y17B was carried out following the instructions featured in Bergey’s manual of systematic bacteriology (Vos et al., 2011).
Genomic DNA of B. subtilis Y17B was extracted using a bacterial DNA kit D3350-01 (Omega Bio-Tek) according to the manufacturer’s protocol. The concentration and purity of the DNA was measured using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Massachusetts, United States). The 16S rRNA gene was amplified using the primers 27F (AGAGTTTGATCCTGGCTCAG) and 1492R (CTACGGCTACCTTGTTACGA; Hou et al., 2018). The PCR (BIO-RAD T100™) conditions were as follows; initial denaturation at 94°C for 3 min, followed by 34 cycles of denaturation at 94°C for 30 s, annealing at 58°C for 30 s, and extension at 72°C for 30 s/kb, and a final extension at 72°C for 5 min. The PCR product was sequenced by Personal bio (Shanghai Personal Biotechnology Co., Ltd., China). The obtained sequences were subjected to an NCBI BLAST similarity search. For phylogenetic analysis, the published reference sequences already available in the NCBI database that were most homologous to the DNA sequences of B. subtilis Y17B strain were used for alignments. GenBank accession numbers and the complete record of isolates from NCBI are listed in the caption of Figure 1. The concatenated sequences and the outgroup were analyzed using Molecular Evolutionary Genetics Analysis (MEGA Version X; Kumar et al., 2018). The sequence alignment of genes was performed using Clustal W. The evolutionary history was inferred by constructing a maximum likelihood tree (ML; Saitou and Nei, 1987) of 16S rRNA gene sequences based on the Tamura Nei model (Tamura and Nei, 1993). The confidence values for clade stability within the phylogenetic tree were determined using bootstrap analysis with 1,000 replicates of the aligned sequences.

Figure 1. Maximum likelihood phylogenetic tree of the tested bacteria based on the Taimura Nei model. The scale bar represents 0.02 nucleotide substitutions per nucleotide position. The numbers on the nodes are bootstrap values inferred from 1,000 replicates. The reference isolates used in this study were as follows: B. aerophilus 28 K (GenBank ID. AJ831844), B. altitudinis 41KF2b (AJ831842), B. stratosphericus 41KF2a (AJ831841), B. licheniformis (X68416), B. aerius 24 K (AJ831843), B. subtilis HZ-72 (MF136610), B. safensis FO-036b (AF234854), B. subtilis Rhizo-02 (KF015515), B. subtilis CSL2 (KX281166), B. flexus (AB021185), B. marisflavi TF-11 (AF483624), B. lentus IAM12466 (D16272), B. subtilis NRS-744 (NR116192), B. licheniformis (X68416), and Pseudomonas aeruginosa ATCC10145 (NR_114471).
Genomic DNA extraction was carried out using a bacterial DNA kit D3350-01 (Omega Bio-Tek) according to the manufacturer’s protocol. DNA amplification was carried out using specifically designed primers for LP biosynthetic genes: surfactin, fengycin, and iturin (Table 1). Primer specificity was checked using the NCBI nucleotide BLAST tool. PCR amplification of genomic DNA was carried using a TRAN kit (Trans-Gen Biotech, China) as per the manufacturer’s protocol. The PCR conditions were as follows; initial denaturation at 94°C for 3 min, followed by 34 cycles of denaturation at 94°C for 30 s, annealing at 50°C for 30 s, and extension at 72°C for 30 s/kb, and a final extension at 72°C for 5 min. A negative control was also included in the PCR amplification reaction. The amplified PCR product from each reaction was visualized on a 1% agarose gel stained with ethidium bromide.
The crude extract of LPs were extracted using a method described previously (Gu et al., 2017) with slight modification. B. subtilis Y17B was inoculated into 50 ml of LB liquid medium and incubated for 24 h at 37°C with shaking at 180 rpm. Y17B culture (50 ml) was taken and inoculated into 3 l of Landy medium and incubated at 37°C with shaking at 180 rpm for 2 days. The supernatant was collected by centrifugation (Eppendorf centrifuge 5804R, Germany) at 12,000 × g for 20 min at 4°C. LPs in the supernatant were precipitated by adjusting the pH to 2 and kept for 6 h at room temperature. The precipitates were collected by centrifugation at 12,000 × g for 20 min at 4°C and dissolved in HPLC grade methanol (Fisher Chemicals, United Kingdom). The pH of the solution was adjusted to 7 by adding 1 M NaOH and the mixture was vortexed for 1 min followed by centrifugation at 4,000 × g for 5 min. The supernatant was passed through a silica gel column and eluted with a mixed solution of HPC grade methanol and methylene chloride at a 5:1 ratio, respectively, as a mobile phase with a 1 ml/min flow rate. The eluted solution was dried under a gentle stream of pure nitrogen at 45°C and the LPs were harvested.
The extracted LPs were dissolved in HPLC grade acetonitrile (ACN) to obtain 50 μg/ml concentrated crude LP extract solution, which was analyzed by UPLC Q TOF-MS to identify the antifungal agents and their isomers. UPLC Q TOF-MS was performed using a Waters ACQUITY UPLC® system (Waters Co., United States) equipped with a binary solvent manager FTN delivery system, auto-sampler manager, and detector. Chromatographical separation was achieved using an ACQUITY UPLC® HSS T3 1.8 μm (2.1 × 100 mm) Column (Part No. 186003539; Serial No. 02163904225158). The mobile phase contained (A) 0.1% aqueous formic acid (v/v) and (B) HPLC grade acetonitrile (ACN). The gradient elution conditions of UPLC were adjusted as follows; 0–7% B (0–11 min), 7–12% B (11–13.9 min), 12–15% B (13.9–14 min), 15–34% B (14–29 min), 34–70% B (29–32 min), 70–100% B (32–35 min), 100% B (35–42 min), 100–0% B (42–45 min), and 0% B (45–60 min). The flow rate was 0.3 ml/min. The pressure range was set at 0 psi (minimum) to 13,000 psi (maximum). The temperature of the column and auto-sampler was maintained at 35°C and 10°C, respectively. An aliquot of 5 μl of the sample was injected and mass spectrometry (MS) analysis was performed using a Waters Xevo-G2QTOF-MS. The MS (Waters Co., United States) was equipped with an electrospray ionization (ESI) source operating in positive ionization mode. The collision energy and ramp collision energy were set at 6 V and 20 to 30 V, respectively. The capillary voltage and cone voltage were set at 2.23 kV and 40 V, respectively. The source and desolvation temperatures were set at 100°C and 500°C, respectively. The desolvation gas flow was set at 995 l/h at a temperature of 450°C. The cone gas was set at 50 l/h. The Q_TOF acquisition rate was 0.2 s and the inter-scan delay was 0.1 s. A mass range was scanned from 200 to 2000 Da. All data were collected using Mass-Lynx V4.1. software (Waters Co., United States).
A. alternata mycelial plugs (5 mm) were taken from 7-day-old culture and placed in the center of fresh PDA plates. Subsequently, 10 μl of 300 μg/ml LP solution was prepared with 0.01 M PBS buffer and inoculated 30 mm away from the fungal mycelial block on a sterilized filter paper block. For the control treatment, only 10 μl of PBS was inoculated 30 mm away from the fungal block. The Petri plates were sealed and incubated at 28°C for 7 days. The percentage of growth inhibition was measured using the following formula described by Vincent (1947) at 7 dpi:
I = [(C−T)/C] × 100
where I is the percentage of growth inhibition, C is growth in control, and T is growth in treatment.
PDA was autoclaved and precooled to 40–45°C and poured onto 90 mm plates. LP solution (1.5 ml) at different concentrations (50, 100, 200, 300, and 500 μg/ml) was mixed into the PDA plates. The control treatment contained 1.5 ml of PBS only. After solidification of the PDA, a mycelial plug (5 mm) from a 7-day-old culture of A. alternata was placed in the middle of each Petri plate and incubated at 28°C for 7 days. The percentage of growth inhibition was calculated using the following formula described by Vincent (1947) at 7 dpi:
I = [(C−T)/C] × 100
where I is the percentage of growth inhibition, C is growth in control, and T is growth in treatment. The experiment was repeated three times.
The ultrastructural changes in the pathogen were studied using a protocol described previously (Wu et al., 2019) with modification. An aliquot of 10 μl of 300 μg/ml LPs was inoculated on sterilized filter paper (5 mm) and placed 30 mm away from the fungal mycelial block and incubated for 7 dpi. Fungal mycelium was taken from the edge of the inhibition zone produced by the LPs. For the control treatment, sample was taken from the edge of mycelial growth, which was inoculated with only 10 μl of PBS. Samples were fixed into 2.5% glutaraldehyde and stored at 4°C for 48 h. For SEM analysis, the samples were prepared using the same procedure as described above in the ‘Morphological, Biochemical, and Physiological Characteristics of B. subtilis Y17B’ section. The samples were observed using a HITACHI SU8010 SEM (Hitachi High-Technologies Corporation, Tokyo, Japan). For TEM analysis, the samples were embedded in Epon 812 and then kept in an oven for 7 days for polymerization. The ultrathin sections were collected and analyzed using a HITACHI-7500 Bio-Transmission electron microscope (Hitachi High-Technologies Corporation, Tokyo, Japan).
To evaluate the antifungal effect of LPs against A. alternata infection on cherry fruit, an antagonism assay was conducted. For this experiment, medium-sized healthy cherry fruit (Lapins cultivar) were selected and dipped in 1% NaOCl for 60 s and later rinsed three times with sterilized water for surface disinfection. Fruits were punctured with a sterilized needle to approximately make 3 mm × 2 mm (depth × width) wounds. Each wound was inoculated with a 10-μl suspension of A. alternata (108 conidia/ml). Subsequently, 10 μl of 300 μg/ml LP solution was applied to the wounds. Inoculated fruits with only a 10-μl suspension of A. alternata were used as a disease control group. All fruits were placed in sterilized six-well culture cluster flat-bottom plates (Costar 3,516) and covered with a zipper bag (240 mm × 170 mm). The samples were incubated for 7 dpi at 28°C. Disease progress was evaluated by measuring the size (mm) of the lesions on the surface of the cherry fruit 7 dpi. The experiment consisted of a cherry fruit with three replicates per treatment.
The experiments were performed in a completely randomized design and subjected to one-way ANOVA, and the means were separated by Fisher’s least significant difference (LSD) test (p ≤ 0.05) using the statistical package Statistix (Ver. 8.1).
A total of 63 isolated bacterial strains from soil samples were evaluated for their ability to inhibit the fungal growth of A. alternata in a dual culture assay. Among all the strains, B. subtilis Y17B significantly inhibited the mycelial growth of A. alternata. Subsequently, 10 μl (cfu 107/ml) of fresh B. subtilis Y17B culture inhibited fungal mycelial growth by 61.3% at 7 dpi (Figure 2).
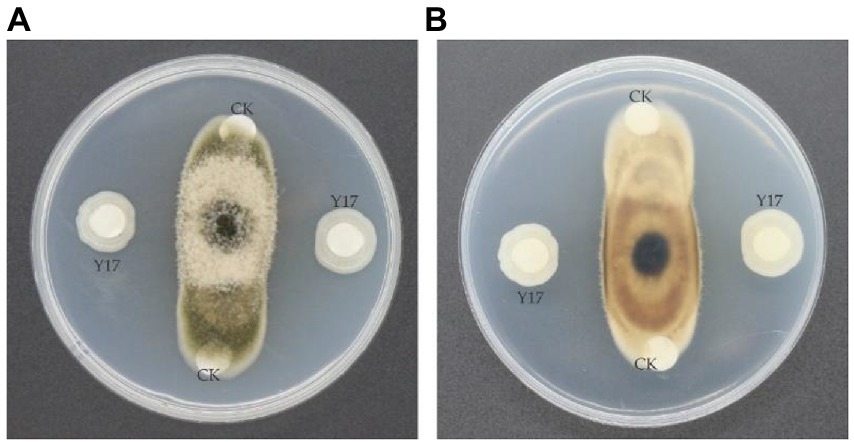
Figure 2. Antifungal activity of the Y17B strain against A. alternata. (A,B) Front (A) and reverse (B) side of the dual culture plate 7 dpi. CK, control (only LB medium); Y17, inoculated with Bacillus subtilis Y17B.
The colony of B. subtilis Y17B was off white to creamy with irregular margins, sticky and had a smooth surface on the LB agar. Under a stereomicroscope, the colonies were joined together and appeared as a chain. SEM revealed that Y17B cells were straight and rod-shaped with round ends, organized in chains, and motile (Figure 3). The bacterial cell was 1.47 to 1.76 μm in length and 765 to 920 nm in width. The results of the biochemical identification are presented in Table 2. B. subtilis Y17B showed a positive result in catalase, citrate, Voges–Proskauer (VP), and nitrate reduction tests, and a negative result in a Methyl red (MR) test. Additionally, B. subtilis Y17B was able to utilize carbon substrates, including D-glucose, D-xylose, and sucrose.
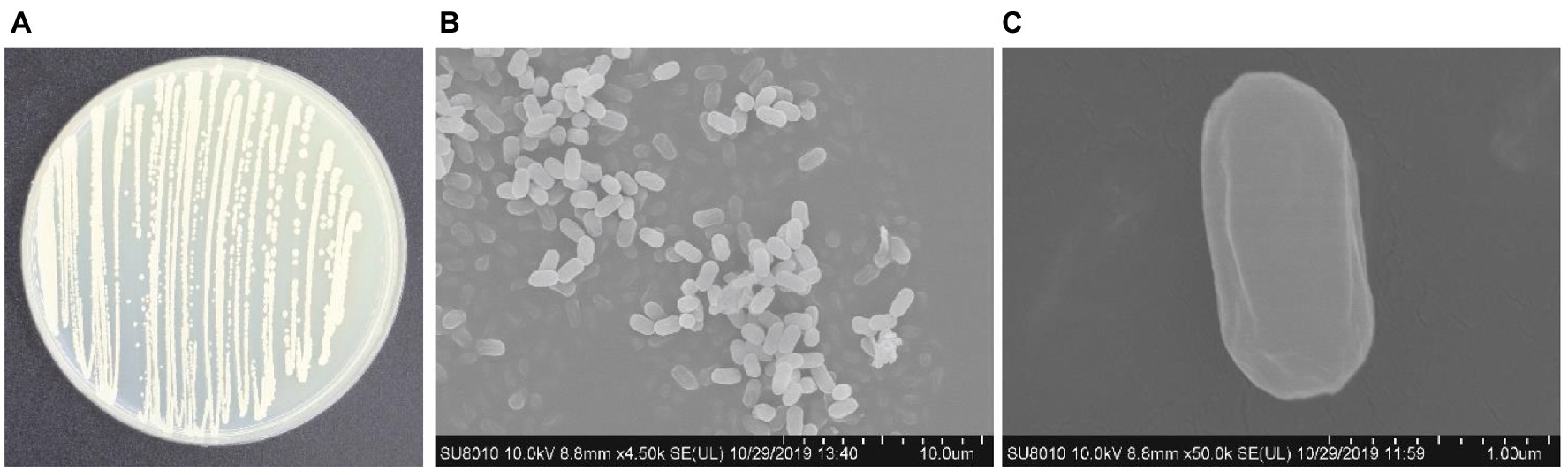
Figure 3. Morphological features of B. subtilis Y17B. (A) Colony morphology. (B) Bacterial cells in chain form. (C) Single bacterium. Scale bars: 10 μm (B); 1 μm (C).
Based on the 16S rRNA gene, the Y17B strain was identified as B. subtilis with >99.9% similarity with B. subtilis strain CSL2 (GenBank ID, KX281166). Phylogenetic analysis of concatenated sequences of the 16S rRNA gene resulted in an ML tree consisting of four distinct clades. Y17B was grouped with B. subtilis strains in one clade along with other isolates of Bacillus species. Pseudomonas aeruginosa ATCC10145 was used as an outgroup and formed a separate clade (Figure 1). Therefore, based on molecular and phylogenetic analysis, the tested bacterium was described as Bacillus subtilis strain Y17B (GenBank ID, MT571500).
PCR amplification of surfactin, iturin, and fengycin from the genome of B. subtilis Y17B indicated the presence of LP biosynthetic genes. The gel showed the bands of all antifungal LPs according to their primer fragment size (bps), in contrast to the negative control without template DNA, which did not produce any band (Figure 4). The amplified products of the surfactin (band size of approximately 1,298 bps), iturin (approximately 1,244 bps), and fengycin (approximately 1,447 bps) genes were detected.
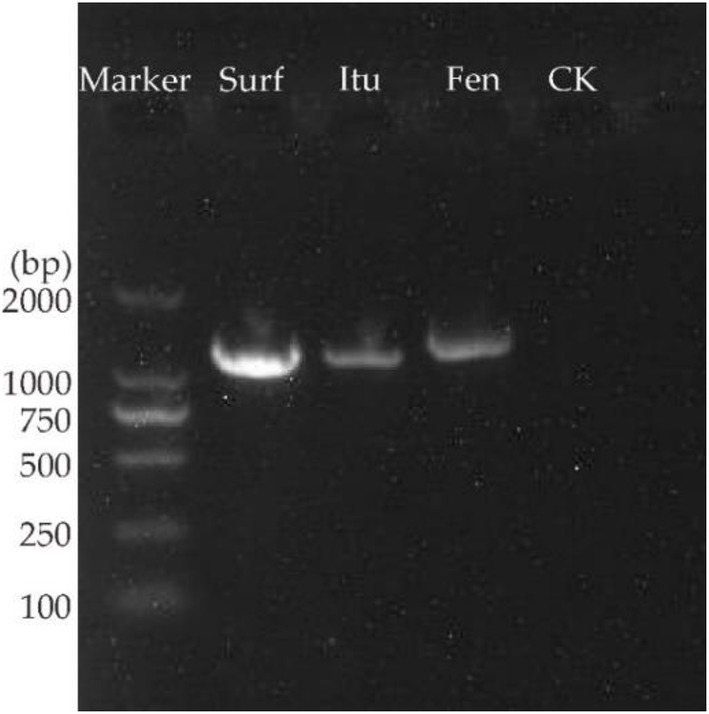
Figure 4. PCR amplification of antifungal LPs biosynthetic genes in B. subtilis Y17B. Surf, surfactin; Itu, iturin; Fen, fengycin; CK, control (without template DNA).
UPLC Q TOF-MS analysis was performed to identify the metabolites produced by B. subtilis Y17B. Figure 5 presents the total ion chromatogram (TIC) of LPs detected from crude extract of B. subtilis Y17B, which confirms the presence of three major and dominant antifungal LP families, surfactin, iturin, and fengycin, and their homologs. Three known surfactins with an acyl fatty chain length ranging from C12 to C14 were detected, along with one known iturin (C14). There were two peaks corresponding to fengycin (C16), and bacillomycin was not observed. Two homologs of surfactin [M + H] + were detected at 994.64 and 1022.681 m/z (retention time, 35.47 and 43.51 min, respectively). Iturin was identified [M + H] + at 1043.56 m/z (retention time, 31.16 min) and fengycin was observed [M + H] + at 1491.84 m/z (retention time, 32.12 min). LPs were identified by comparing their masses with those previously reported in the literature for respective compounds (Table 3).
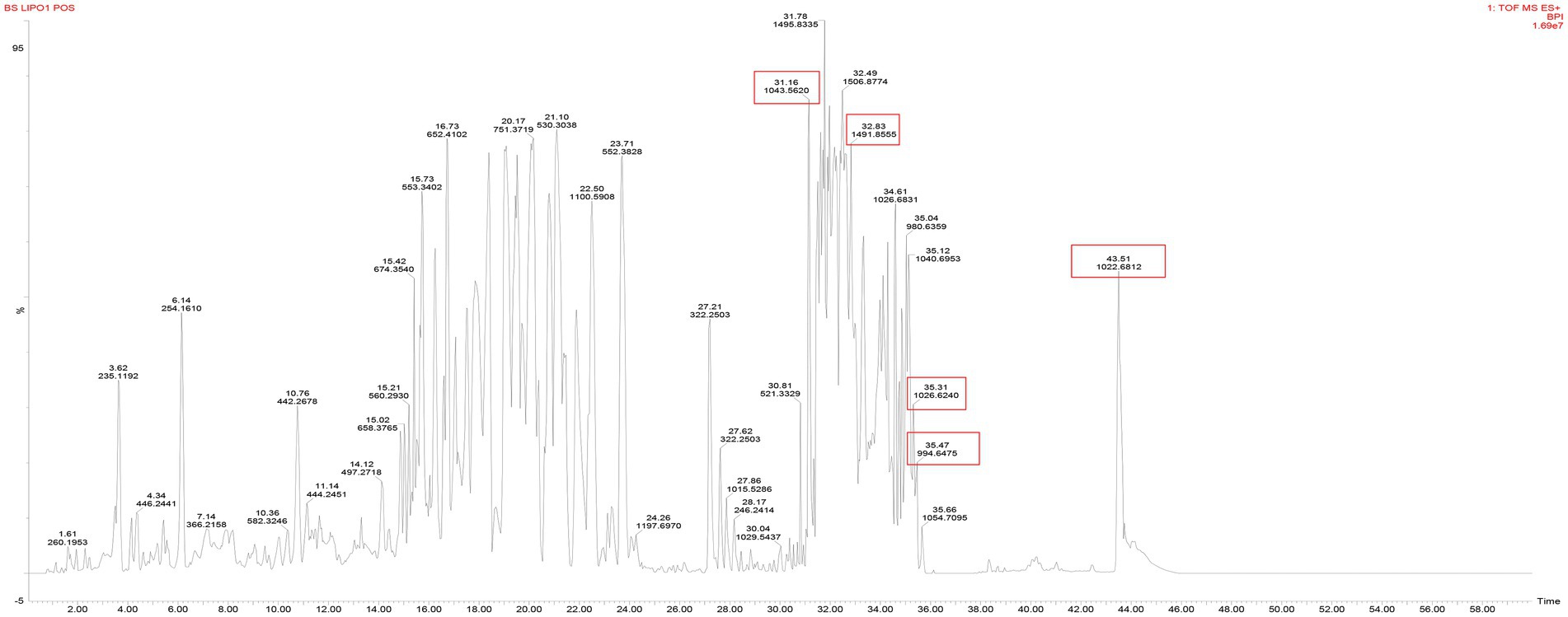
Figure 5. Total ion chromatogram of Y17B LPs with their molecular weight (m/z) and retention time (RT).
The antifungal LPs significantly inhibited A. alternata growth. The fungal growth of A. alternata was reduced by 54% in the dual culture assay by treatment with 10 μl of 300 μg/ml of LPs of B. subtilis Y17B. The growth of A. alternata was gradually reduced as the concentration of LPs of B. subtilis Y17B was increased (Figure 6).

Figure 6. Antagonistic interaction of LPs with the pathogen. (A,B) Front (A) and reverse side (B) of the dual culture plate.
Antifungal activity was evaluated by amending the PDA medium with the crude extract of LPs. The LPs produced by B. subtilis Y17B significantly inhibited the mycelial growth of A. alternata. Fungal mycelial growth was inhibited 76, 58, 52, 37, 25, and 10% at 500, 300, 200, 150, 100, and 50 μg/ml concentrations of LPs, respectively, 7 dpi (Figure 7). Therefore, the crude extract of LPs from B. subtilis Y17B inhibited A. alternata in a dose-dependent manner.
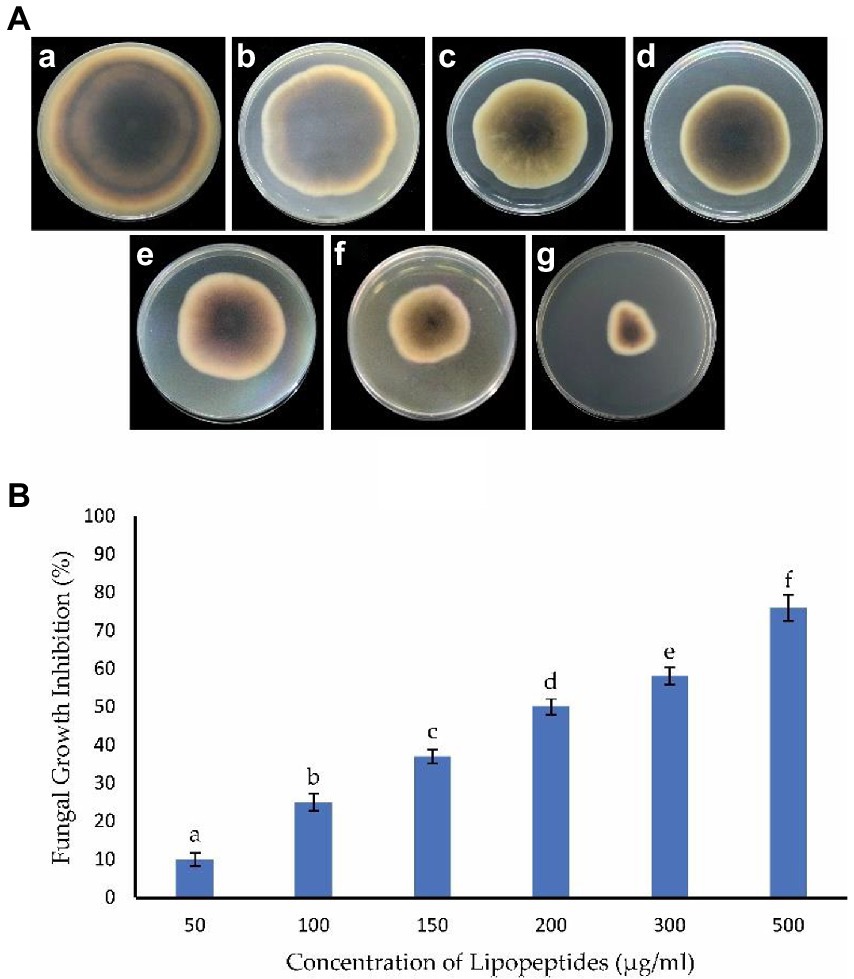
Figure 7. Antagonistic effect of B. subtilis Y17B LPs against A. alternata. (A) Assessment of LPs at different concentrations against A. alternata. The reverse side of the plate is shown. (A) Control, (B) 50 μg/mL of LPs, (C) 100 μg/mL of LPs, (D) 150 μg/mL of LPs, (E) 200 μg/mL of LPs, (F) 300 μg/mL of LPs, and (G) 500 μg/mL of LPs. (B) Graph representing the antagonistic effect of LPs on A. alternata mycelium inhibition (%). The same letters are not significantly different from each other. Analyzed using a LSD test (p ≤ 0.05).
SEM analysis revealed that the untreated control hyphae of A. alternata seemed intact, regular, long, dense, and plump with a column-like trunk, and conidia were smooth, normal, and flask-shaped. However, the LP-treated hyphae and conidia showed alterations in structure, exhibiting irregular shapes, loose cell walls, ruptured cell wall surfaces, surface subsidence, and shriveled trunks. The results demonstrate the leakage of cellular contents due to LP treatment (Figure 8). TEM analysis was performed to observe internal structural alterations of the LP-treated pathogen. The untreated control hyphae and conidia showed distinct cell walls, intact plasma membranes, smooth surfaces, septa, and uniformly distributed and clearly visible cytoplasm. By contrast, the cell walls of the treated hyphae and conidia were thinner, representing the cytoplasm leakage. Cell walls showed irregular structure, such as a lack of discernible layers, uneven thickness, and gapped structures. Additionally, LP treatment caused plasmolysis and fragmentation of the plasma membranes. The cell membranes were dissolved and ruptured. Furthermore, the damage to the cell walls and membranes finally resulted in serious leakage of the cytoplasm, resulting in cell death (Figure 9).
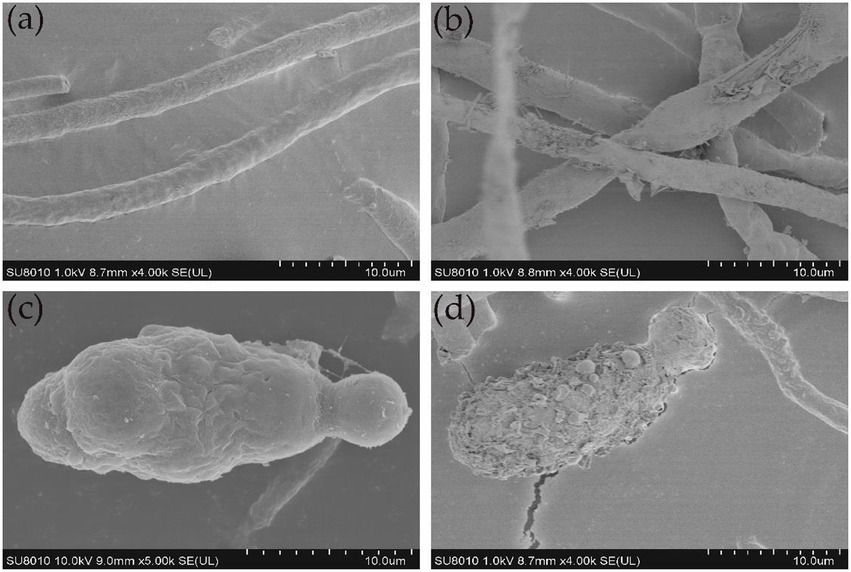
Figure 8. Ultrastructural changes in the hyphae and conidia of A. alternata. (A) Control group; healthy hyphae. (B) Effect of 10 μL of 300 μg/mL of LPs on hyphae. (C) Control group; healthy conidia. (D) Effect of 10 μL of 300 μg/ml of LPs on conidia.
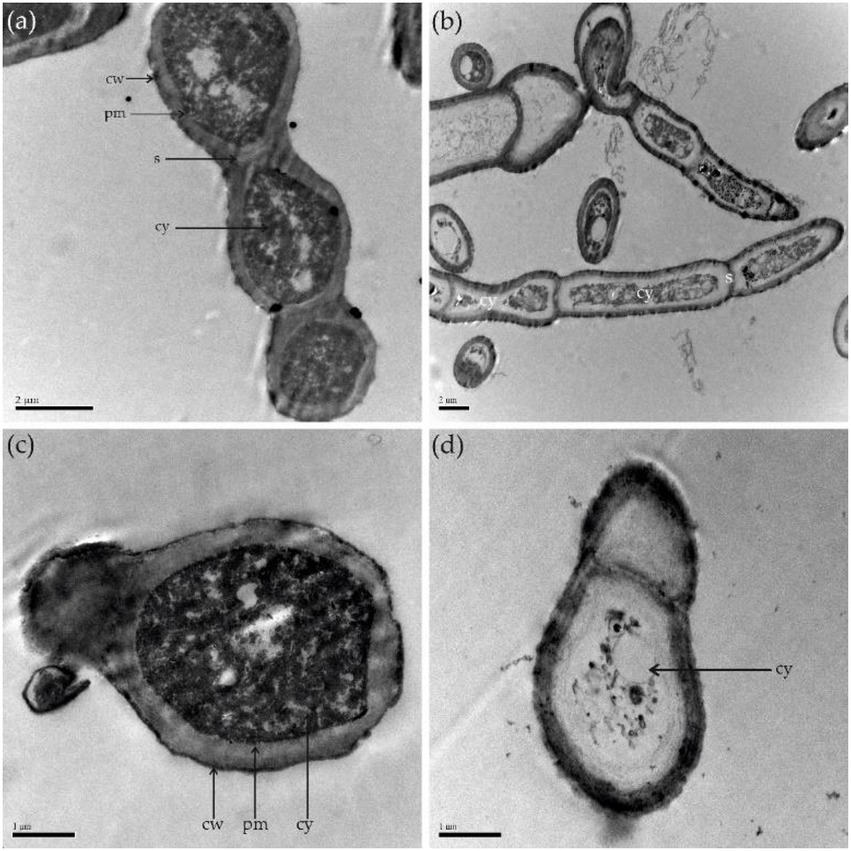
Figure 9. Transmission electron microscopy. (A) Control group; healthy hyphae. (B) Hyphae treated with 10 μl of 300 μg/ml of LPs. (C) Healthy conidia. (D) Conidia treated with 10 μL of 300 μg/mL of LPs. cw, cell wall; cy, cytoplasm; pm, plasma membrane; s, septum.
Alternaria alternata caused typical fruit rot symptoms in all inoculated cherry fruit, with the formation of dark olive-green to whitish aerial mycelia around the wounds. Conidia production was also observed at the site of infection. The rot lesions expanded toward the surrounding area and had an average diameter of 20.4 ± 1.0 mm in untreated fruit. The application of LPs reduced the lesion size induced by A. alternata compared with non-treated fruit. A significant effect of 10 μl of LPs with a concentration of 300 μg/ml was observed in this treatment and lesion diameter was reduced to 7.5 ± 1.1 mm (more than 63%), as shown in Figure 10.
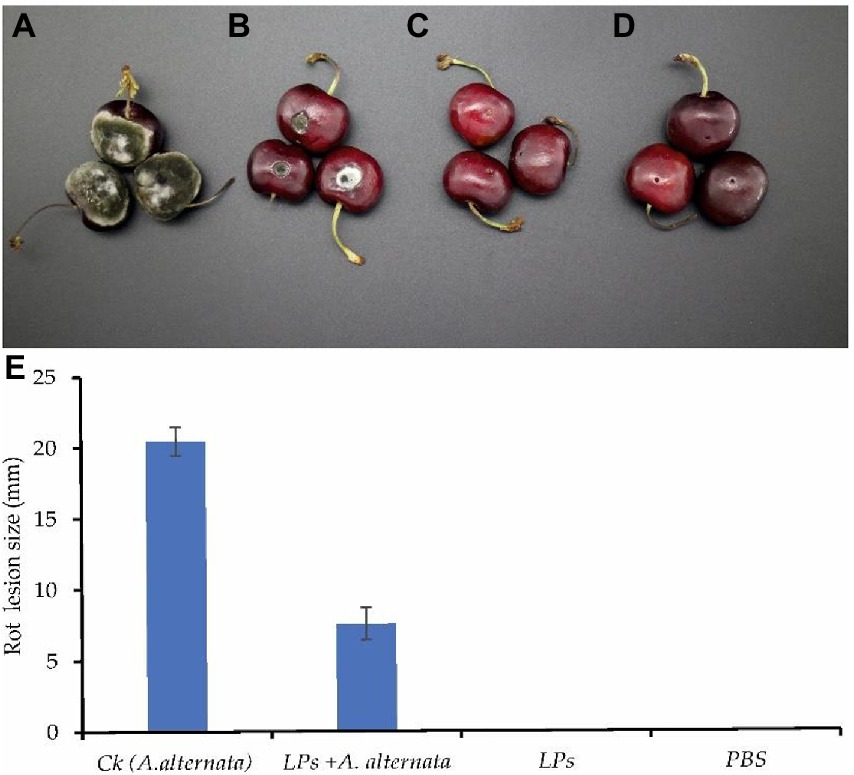
Figure 10. LPs from Y17B suppressed fruit rot development in cherry fruit. (A) Ck: cherry fruit infected with A. alternata only. (B) Treatment: cherry fruit infected with A. alternata and treated with LPs. (C) Cherry fruit treated with LPs only. (D) Cherry fruit treated with PBS only. (E) Mean values of rot lesion diameter ± SD.
Cherry is the most important fruit crop in the world, and postharvest rot of cherry fruit caused by the fungus A. alternata is one of the most devastating postharvest diseases, reducing the quality and quantity of fruit (Ahmad et al., 2020). Synthetic chemicals are commonly used to control fungal pathogens but excessive and regular use of these chemical agents causes environmental contamination due to their long-term residual effects. LPs produced by the Bacillus genus possess a strong antifungal action, which reduces the risk of postharvest fungal rots. The added advantage of LPs is that they are safe as they do not pose any health and environmental hazard (Ongena and Jacques, 2008; Ali et al., 2016).
In this study, the antifungal activity of B. subtilis Y17B and its LPs against A. alternata has been elucidated. Previous research reports state that the major reason for the antifungal activity of Bacillus species is the production of LPs, i.e., surfactin, iturin, and fengycin, which are synthesized by NRPSs (Romero et al., 2007; Pramudito et al., 2018). In our study, the PCR amplification of the biosynthetic genes surfactin, iturin, and fengycin from the genome of B. subtilis Y17B revealed the presence of gene clusters for non-ribosomal lipopeptide synthetases for surfactin, iturin, and fengycin. Several recent studies have reported that the genomes of B.subtilus and B. amyloliquefaciens contain biosynthetic genes that encode for antifungal LPs, including fengycin, surfactin, bacillibactin, iturin, subtilosin, and bacilysin (Farzand et al., 2019; Su et al., 2020). The presence of antifungal LPs in the crude extract of B. subtilis Y17B was further confirmed by UPLC Q TOF-MS analysis. The LPs were detected based on their molecular weight (m/z). Previous studies have reported the potential of Bacillus species for producing several antifungal LPs in response to different fungal infections, which were detected by mass spectrometry (MS) based on their molecular weight (Frikha-Gargouri et al., 2017; Farzand et al., 2019). In another instance, several antifungal LPs, including surfactin, bacillomycin, and fengycin, were detected with molecular masses of 994.64, 1031.54, and 1491.84, respectively, in Bacillus XT1 culture through Q TOF-MS analysis (Toral et al., 2018).
LPs produced by B. subtilis Y17B showed an antagonistic effect against A. alternata in both dual culture and on PDA amended with LPs. A recent study reported that LPs from B. altitudinis Q7 and B. subtilis BS-01 show stable antifungal activity against A. alternata and A. solani, respectively, in extreme environments and are good biological antagonists (Awan et al., 2022; Guo et al., 2022). Additionally, several studies have previously reported the antagonistic effect of LPs and VOCs of Bacillus species on different fungal pathogens, especially A. alternata (Chowdhury et al., 2015; Kaur et al., 2015; Wang et al., 2022a; Jia et al., 2023). Bacillus strains B. subtilis 6,051, B. amyloliquefaciens FZB42, and Bacillus XT1 CECT have been reported as biocontrol agents due to their production of antifungal LPs, which inhibit different fungi, such as Aspergillus species, Fusarium graminearum, and Botrytis cinerea (Bais et al., 2004; Toral et al., 2018; Hanif et al., 2019). Similar results were reported by Ali et al. (2016) who showed that the culture filtrates of B. subtilis and B. amyloliquefaciens exert strong antagonistic activity against Alternaria species due to the presence of the antimicrobial cyclic LP families fengycin-A and fengycin-B. In another study, LCMS analysis of the crude mixture of B. subtilis revealed the presence of antifungal LPs. These LPs inhibited the growth of Verticillium dahliae by causing cell lysis, hyphal swelling, and the downregulation of melanin-related genes. LPs inhibited fungal gene expression associated with the signaling pathway of protein catabolism and secondary metabolism (Yu et al., 2019). Another study reported a similar result in that the enhanced transcription of genes involved in flavonoid biosynthesis and accumulation of flavonoids was induced by B. subtilis Y2 to fight against the pathogen A. brassicicola, which causes black spot disease of pear fruit (Wang et al., 2023).
In this study, ultrastructural changes were observed in A. alternata. The LPs produced by YB17 inhibited the germination of conidia and caused alterations in the hyphal structures of A. alternata. SEM analysis revealed damage to the fungal structures of A. alternata, which appeared irregular in shape, with loose and ruptured cell walls, surface subsidence, and shriveled trunks. In agreement with our results, a previous study revealed that LPs produced by B. subtilis YM 10–20 cause the inhibition of conidia germination in Penicillium roqueforti (Chitarra et al., 2003), and LPs of B. subtilis BS-99-H cause alterations in the morphology of Pestalotiopsis eugeniae, which is associated with wax apple fruit rot (Lin et al., 2010). Several studies have shown that LPs from Bacillus species alter and severely damage fungal hyphae and spores (Souto et al., 2004; Etchegaray et al., 2008). Additionally, in other studies, the LPs produced by B. amyloliquefaciens were reported to be strong antifungal agents against F. graminearum and F. solani (Torres et al., 2017; Hanif et al., 2019). TEM studies showed that the cell walls of the treated hyphae were thinner than the hyphae in the untreated controls, with uneven thickness of conidia and plasmolysis and fragmentation of the plasma membranes, which represents cytoplasm leakage. Similarly, TEM images of B. cinerea after treatment with LPs of XT1 Lps showed damage and alteration to the ultrastructure of the fungal hyphae and ultimately cytoplasmic leakage (Toral et al., 2018). A similar study, using SEM and TEM analysis, showed that fengycin BS155 extracted from marine B. subtilis causes morphological changes in the plasma membrane and cell wall of Magnaporthe grisea. Biochemical and proteomic assays revealed that fengycin BS155 possesses the ability to reduce mitochondrial membrane potential (MMP), induce oxidative stress, and downregulate the expression level of oxidative stress-scavenging enzymes. Additionally, fengycin BS155 causes chromatin condensation in fungal hyphal cells, resulting in cell death (Zhang and Sun, 2018).
An antagonism assay in cherry fruit was also conducted to evaluate the antifungal activity of the LPs produced by B. subtilis Y17B against postharvest fruit rot of cherry caused by A. alternata. The results showed that rot disease in cherry fruit was significantly reduced by the application of LPs due to their antifungal action against A. alternata. A previous study suggested that the LPs from B. subtilis ABS-S14 act as defense-related genes to inhibit pathogen growth (Waewthongrak et al., 2014). Another study produced similar results showing that LPs produced by B. mojavensis P1709 are an effective tool for controlling postharvest decay and mycotoxin contamination in cherry tomatoes (Galitskaya et al., 2022). In conclusion, we report that LPs produced by B. subtilis strain Y17B potentially exert strong antifungal activity against A. alternata, causing ultrastructural damage to the pathogen and reducing the disease intensity of A. alternata on cherry fruit. B. subtilis Y17B can be formulated as a biopesticide against the postharvest fruit rot pathogen A. alternata. For future studies, the antifungal potential of B. subtilis Y17B against A. alternata and the degradation of the Alternaria mycotoxins tenuazonic acid and alternariol could be studied under the influence of purified surfactin, iturin, and fengycin. However, the exact mechanism by which purified LPs of B. subtilis Y17B cause fungal cell death still needs to be investigated.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
TA, FX, and YL contributed to the conception and designed the experiments. TA performed the isolation, antifungal, antagonism assays, SEM and TEM analysis, and wrote the first draft of manuscript. FX, TA, and CN contributed to the identification of the strain and phylogenetic analysis. TA and YL contributed to the identification of biosynthetic genes and UPLC Q TOF-MS analysis. CC and XY analyzed the data. AM, CC, CN, XY, and YX contributed to the writing and editing of the manuscript. All authors contributed to the article and approved the submitted version.
This research was funded by National Key Research and Development Program of China (No. 2022YFE0139500) and National Key Research and Development Program of China (No. 2019YFC1604502).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Ahmad, T., Liu, Y., Shujian, H., and Moosa, A. (2020). First record of Alternaria alternata causing postharvest fruit rot of sweet cherry (Prunus avium) in China. Plant Dis. 104:2030. doi: 10.1094/PDIS-11-19-2322-PDN
Ali, G. S., El-Sayed, A. S., Patel, J. S., Green, K. B., Ali, M., Brennan, M., et al. (2016). Ex vivo application of secreted metabolites produced by soil-inhabiting bacillus spp. efficiently controls foliar diseases caused by Alternaria spp. Appl. Environ. Microbiol. 82, 478–490. doi: 10.1128/AEM.02662-15
Alina, S. O., Constantinscu, F., and Petruta, C. C. (2015). Biodiversity of Bacillus subtilis group and beneficial traits of bacillus species useful in plant protection. Rom. Biotechnol. Lett 20, 10737–10750.
Andersen, B., Nielsen, K. F., Pinto, V. F., and Patriarca, A. (2015). Characterization of Alternaria strains from Argentinean blueberry, tomato, walnut and wheat. Int. J. Food Microbiol. 196, 1–10. doi: 10.1016/j.ijfoodmicro.2014.11.029
Arrebola, E., Jacobs, R., and Korsten, L. J. J. O. A. M. (2010). Iturin a is the principal inhibitor in the biocontrol activity of bacillus amyloliquefaciens PPCB004 against postharvest fungal pathogens. Journal of Applied Microbiology, 108, 386–395. doi: 10.1111/j.1365-2672.2009.04438.x
Arroyave-Toro, J. J., Mosquera, S., and Villegas-Escobar, V. J. B. C. (2017). Biocontrol activity of Bacillus subtilis EA-CB0015 cells and lipopeptides against postharvest fungal pathogens. Biol. Control 114, 195–200. doi: 10.1016/j.biocontrol.2017.08.014
Awan, Z. A., Shoaib, A., Schenk, P. M., Ahmad, A., Alansi, S., and Paray, B. A. (2022). Antifungal potential of volatiles produced by Bacillus subtilis BS-01 against Alternaria solani in Solanum lycopersicum. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1089562
Bais, H. P., Fall, R., and Vivanco, J. M. (2004). Biocontrol of Bacillus subtilis against infection of Arabidopsis roots by pseudomonas syringae is facilitated by biofilm formation and surfactin production. Plant Physiol. 134, 307–319. doi: 10.1104/pp.103.028712
Basit, M., Rasool, M. H., Naqvi, S. A. R., Waseem, M., and Aslam, B. (2018). Biosurfactants production potential of native strains of Bacillus cereus and their antimicrobial, cytotoxic and antioxidant activities. Pak. J. Pharm. Sci. 31, 251–256.
Chen, Y., Liu, S. A., Mou, H., Ma, Y., Li, M., and Hu, X. (2017). Characterization of Lipopeptide biosurfactants produced by bacillus licheniformis MB01 from marine sediments. Front. Microbiol. 8:871. doi: 10.3389/fmicb.2017.00871
Chitarra, G., Breeuwer, P., Nout, M., Van Aelst, A., Rombouts, F., and Abee, T. (2003). An antifungal compound produced by Bacillus subtilis YM 10–20 inhibits germination of Penicillium roqueforti conidiospores. J. Appl. Microbiol. 94, 159–166. doi: 10.1046/j.1365-2672.2003.01819.x
Chowdhury, S. P., Hartmann, A., Gao, X., and Borriss, R. (2015). Biocontrol mechanism by root-associated bacillus amyloliquefaciens FZB42–a review. Front. Microbiol. 6:780. doi: 10.3389/fmicb.2015.00780
Cozzolino, M. E., Distel, J. S., García, P. A., Mascotti, M. L., Ayub, M. J., Benazzi, L. M., et al. (2020). Control of postharvest fungal pathogens in pome fruits by lipopeptides from a bacillus sp. SL-6. 261:108957. doi: 10.1016/j.scienta.2019.108957
De Souza, C. G., Martins, F. I. C. C., Zocolo, G. J., Figueiredo, J. E. F., Canuto, K. M., and De Brito, E. S. (2018). Simultaneous quantification of lipopeptide isoforms by UPLC-MS in the fermentation broth from Bacillus subtilis CNPMS22. Anal. Bioanal. Chem. 410, 6827–6836. doi: 10.1007/s00216-018-1281-6
Deng, Q., Wang, W., Sun, L., Wang, Y., Liao, J., Xu, D., et al. (2017). A sensitive method for simultaneous quantitative determination of surfactin and iturin by LC-MS/MS. Anal. Bioanal. Chem. 409, 179–191. doi: 10.1007/s00216-016-9984-z
Etchegaray, A., De Castro Bueno, C., De Melo, I. S., Tsai, S. M., De Fátima Fiore, M., Silva-Stenico, M. E., et al. (2008). Effect of a highly concentrated lipopeptide extract of Bacillus subtilis on fungal and bacterial cells. Arch. Microbiol. 190, 611–622. doi: 10.1007/s00203-008-0409-z
Farzand, A., Moosa, A., Zubair, M., Khan, A. R., Ayaz, M., Massawe, V. C., et al. (2019). Transcriptional profiling of diffusible Lipopeptides and fungal virulence genes during bacillus amyloliquefaciens EZ1509-mediated suppression of Sclerotinia sclerotiorum. Phytopathology, 110, 317–326. doi: 10.1094/PHYTO-05-19-0156-R
Fatima, R., Mahmood, T., Moosa, A., Aslam, M. N., Shakeel, M. T., Maqsood, A., et al. (2023). Bacillus thuringiensis CHGP12 uses a multifaceted approach for the suppression of fusarium oxysporum f. sp. ciceris and to enhance the biomass of chickpea plants. Pest Manag. Sci. 79, 336–348. doi: 10.1002/ps.7203
Finking, R., and Marahiel, M. A. (2004). Biosynthesis of nonribosomal peptides. Annu. Rev. Microbiol. 58, 453–488. doi: 10.1146/annurev.micro.58.030603.123615
Frikha-Gargouri, O., Ben Abdallah, D., Ghorbel, I., Charfeddine, I., Jlaiel, L., Triki, M. A., et al. (2017). Lipopeptides from a novel bacillus methylotrophicus 39b strain suppress agrobacterium crown gall tumours on tomato plants. Pest Manag. Sci. 73, 568–574. doi: 10.1002/ps.4331
Galitskaya, P., Karamova, K., Biktasheva, L., Galieva, G., Gordeev, A., and Selivanovskaya, S. (2022). Lipopeptides produced by Bacillus mojavensis P1709 as an efficient tool to maintain postharvest cherry tomato quality and quantity. Agriculture 12:609. doi: 10.3390/agriculture12050609
Gond, S. K., Bergen, M. S., Torres, M. S., and White, J. F. Jr. (2015). Endophytic bacillus spp. produce antifungal lipopeptides and induce host defence gene expression in maize. Microbiol. Res. 172, 79–87. doi: 10.1016/j.micres.2014.11.004
Gu, Q., Yang, Y., Yuan, Q., Shi, G., Wu, L., Lou, Z., et al. (2017). Bacillomycin D produced by bacillus amyloliquefaciens is involved in the antagonistic interaction with the plant-pathogenic fungus fusarium graminearum. Appl. Environ. Microbiol. 83, e01075–e01017. doi: 10.1128/AEM.01075-17
Guo, P., Yang, F., Ye, S., Li, J., Shen, F., and Ding, Y. J. J. O. B. M. (2022). Characterization of lipopeptide produced by Bacillus altitudinis Q7 and inhibitory effect on Alternaria alternata, Journal of Basic Microbiology. 63, 26–38 doi: 10.1002/jobm.202200530.
Han, Q., Wu, F., Wang, X., Qi, H., Shi, L., Ren, A., et al. (2015). The bacterial lipopeptide iturins induce V erticillium dahliae cell death by affecting fungal signalling pathways and mediate plant defence responses involved in pathogen-associated molecular pattern-triggered immunity. Environ. Microbiol. 17, 1166–1188. doi: 10.1111/1462-2920.12538
Hanif, A., Zhang, F., Li, P., Li, C., Xu, Y., Zubair, M., et al. (2019). Fengycin produced by bacillus amyloliquefaciens FZB42 inhibits fusarium graminearum growth and mycotoxins biosynthesis. Toxins 11:295. doi: 10.3390/toxins11050295
Hou, Q., Bai, X., Li, W., Gao, X., Zhang, F., Sun, Z., et al. (2018). Design of primers for evaluation of lactic acid bacteria populations in complex biological samples. Front. Microbiol. 9:2045. doi: 10.3389/fmicb.2018.02045
Jia, Q., Fan, Y., Duan, S., Qin, Q., Ding, Y., Yang, M., et al. (2023). Effects of bacillus amyloliquefaciens XJ-BV2007 on growth of Alternaria alternata and production of Tenuazonic acid. Toxins 15:53. doi: 10.3390/toxins15010053
Kaur, P. K., Kaur, J., and Saini, H. S. (2015). Antifungal potential of Bacillus vallismortis R2 against different phytopathogenic fungi. Span. J. Agric. Res. 13:e1004. doi: 10.5424/sjar/2015132-6620
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lawrance, A., Balakrishnan, M., Joseph, T. C., Sukumaran, D. P., Valsalan, V. N., Gopal, D., et al. (2014). Functional and molecular characterization of a lipopeptide surfactant from the marine sponge-associated eubacteria bacillus licheniformis NIOT-AMKV06 of Andaman and Nicobar Islands, India. Mar. Pollut. Bull. 82, 76–85. doi: 10.1016/j.marpolbul.2014.03.018
Li, X., Zhang, Y., Wei, Z., Guan, Z., Cai, Y., and Liao, X. (2016). Antifungal activity of isolated bacillus amyloliquefaciens SYBC H47 for the biocontrol of peach gummosis. PLoS One 11:2125. doi: 10.1371/journal.pone.0162125
Lin, H., Chen, T., and Liu, S. (2010). Bioactivity of antifungal substance iturin a produced by Bacillus subtilis strain BS-99-H against Pestalotiopsis eugeniae, a causal pathogen of wax apple fruit rot. Plant Pathol. Bull. 19, 225–233.
Marahiel, M. A. (2009). Working outside the protein-synthesis rules: insights into non-ribosomal peptide synthesis. J. Peptide Sci. 15, 799–807. doi: 10.1002/psc.1183
Mnif, I., Grau-Campistany, A., Coronel-León, J., Hammami, I., Triki, M. A., Manresa, A., et al. (2016). Purification and identification of Bacillus subtilis SPB1 lipopeptide biosurfactant exhibiting antifungal activity against Rhizoctonia bataticola and Rhizoctonia solani. Environ. Sci. Pollut. Res. 23, 6690–6699. doi: 10.1007/s11356-015-5826-3
Montesinos, E. (2007). Antimicrobial peptides and plant disease control. FEMS Microbiol. Lett. 270, 1–11. doi: 10.1111/j.1574-6968.2007.00683.x
Ongena, M., and Jacques, P. (2008). Bacillus lipopeptides: versatile weapons for plant disease biocontrol. Trends Microbiol. 16, 115–125. doi: 10.1016/j.tim.2007.12.009
Ostry, V. (2008). Alternaria mycotoxins: an overview of chemical characterization, producers, toxicity, analysis and occurrence in foodstuffs. World Mycotoxin J. 1, 175–188. doi: 10.3920/WMJ2008.x013
Özkara, A., Akyıl, D., and Konuk, M. (2016). “Pesticides, environmental pollution, and health” in Environmental health risk-hazardous factors to living species (London, UK: IntechOpen)
Pathak, K. V., and Keharia, H. (2014). Identification of surfactins and iturins produced by potent fungal antagonist, Bacillus subtilis K1 isolated from aerial roots of banyan (Ficus benghalensis) tree using mass spectrometry. Three biotech 4, 283–295. doi: 10.1007/s13205-013-0151-3
Pramudito, T. E., Agustina, D., Nguyen, T. K. N., and Suwanto, A. (2018). A novel variant of narrow-Spectrum antifungal bacterial Lipopeptides that strongly inhibit Ganoderma boninense. Prob. Antimicrob. Prot. 10, 110–117. doi: 10.1007/s12602-017-9334-2
Pruß, S., Fetzner, R., Seither, K., Herr, A., Pfeiffer, E., Metzler, M., et al. (2014). Role of the Alternaria alternata blue-light receptor LreA (white-collar 1) in spore formation and secondary metabolism. Appl. Environ. Microbiol. 80, 2582–2591. doi: 10.1128/AEM.00327-14
Romero, D., De Vicente, A., Olmos, J., Dávila, J., and Pérez-García, A. (2007). Effect of lipopeptides of antagonistic strains of Bacillus subtilis on the morphology and ultrastructure of the cucurbit fungal pathogen Podosphaera fusca. J. Appl. Microbiol. 103, 969–976. doi: 10.1111/j.1365-2672.2007.03323.x
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Sanchez, H., and Bush, R. K. (2001). A review of Alternaria alternata sensitivity. Revista iberoamericana de micologia 18, 56–59.
Sarwar, A., Hassan, M. N., Imran, M., Iqbal, M., Majeed, S., Brader, G., et al. (2018). Biocontrol activity of surfactin a purified from bacillus NH-100 and NH-217 against rice bakanae disease. Microbiol. Res. 209, 1–13. doi: 10.1016/j.micres.2018.01.006
Souto, G., Correa, O., Montecchia, M., Kerber, N., Pucheu, N., Bachur, M., et al. (2004). Genetic and functional characterization of a bacillus sp. strain excreting surfactin and antifungal metabolites partially identified as iturin-like compounds. J. Appl. Microbiol. 97, 1247–1256. doi: 10.1111/j.1365-2672.2004.02408.x
Soylu, E. M., and Kose, F. (2015). Antifungal activities of essential oils against citrus black rot disease agent Alternaria alternata. J. Essen. Oil Bear. Plants 18, 894–903. doi: 10.1080/0972060X.2014.895158
Stein, T. (2005). Bacillus subtilis antibiotics: structures, syntheses and specific functions. Mol. Microbiol. 56, 845–857. doi: 10.1111/j.1365-2958.2005.04587.x
Su, Z., Chen, X., Liu, X., Guo, Q., Li, S., Lu, X., et al. (2020). Genome mining and UHPLC–QTOF–MS/MS to identify the potential antimicrobial compounds and determine the specificity of biosynthetic gene clusters in Bacillus subtilis NCD-2. Genomics 21, 767–716. doi: 10.1186/s12864-020-07160-2
Tamura, K., and Nei, M. (1993). Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10, 512–526.
Toral, L., Rodríguez, M., Béjar, V., and Sampedro, I. (2018). Antifungal activity of lipopeptides from bacillus XT1 CECT 8661 against Botrytis cinerea. Front. Microbiol. 9:1315. doi: 10.3389/fmicb.2018.01315
Torres, M. J., Brandan, C. P., Sabaté, D. C., Petroselli, G., Erra-Balsells, R., and Audisio, M. C. (2017). Biological activity of the lipopeptide-producing bacillus amyloliquefaciens PGPBacCA1 on common bean Phaseolus vulgaris L. pathogens. Biol. Control 105, 93–99. doi: 10.1016/j.biocontrol.2016.12.001
Vincent, J. (1947). Distortion of fungal hyphae in the presence of certain inhibitors. Nature 159:850. doi: 10.1038/159850b0
Vos, P., Garrity, G., Jones, D., Krieg, N. R., Ludwig, W., Rainey, F. A., et al. (2011). Bergey's manual of systematic bacteriology, vol. 3 The Firmicutes, New York, USA: Springer Science & Business Media.
Waewthongrak, W., Leelasuphakul, W., and Mccollum, G. (2014). Cyclic lipopeptides from Bacillus subtilis ABS–S14 elicit defense-related gene expression in citrus fruit. PLoS One 9:e109386. doi: 10.1371/journal.pone.0109386
Wang, S.-Y., Herrera-Balandrano, D. D., Wang, Y.-X., Shi, X.-C., Chen, X., Jin, Y., et al. (2022b). Biocontrol ability of the Bacillus amyloliquefaciens group, B. amyloliquefaciens, B. velezensis, B. nakamurai, and B. siamensis, for the Management of Fungal Postharvest Diseases: a review. J. Agric. Food Chem. 70, 6591–6616. doi: 10.1021/acs.jafc.2c01745
Wang, D., Li, Y., Yuan, Y., Chu, D., Cao, J., Sun, G., et al. (2022a). Identification of non-volatile and volatile organic compounds produced by Bacillus siamensis LZ88 and their antifungal activity against Alternaria alternata. Biol. Control 169:104901. doi: 10.1016/j.biocontrol.2022.104901
Wang, X., Xie, S., Mu, X., Guan, B., Hu, Y., and Ni, Y. (2023). Investigating the resistance responses to Alternaria brassicicola in'Korla'fragrant pear fruit induced by a biocontrol strain Bacillus subtilis Y2. Postharvest Biol. Technol. 199:112293. doi: 10.1016/j.postharvbio.2023.112293
Wu, S., Liu, G., Zhou, S., Sha, Z., and Sun, C. (2019). Characterization of antifungal Lipopeptide biosurfactants produced by marine bacterium bacillus sp. CS30. Mar. Drugs 17:199. doi: 10.3390/md17040199
Xu, L., Tao, N., Yang, W., and Jing, G. (2018). Cinnamaldehyde damaged the cell membrane of Alternaria alternata and induced the degradation of mycotoxins in vivo. Ind. Crop. Prod. 112, 427–433. doi: 10.1016/j.indcrop.2017.12.038
Yang, H., Li, X., Li, X., Yu, H., and Shen, Z. (2015). Identification of lipopeptide isoforms by MALDI-TOF-MS/MS based on the simultaneous purification of iturin, fengycin, and surfactin by RP-HPLC. Anal. Bioanal. Chem. 407, 2529–2542. doi: 10.1007/s00216-015-8486-8
Yu, D., Fang, Y., Tang, C., Klosterman, S. J., Tian, C., and Wang, Y. J. M. P.-M. I. (2019). Genomewide transcriptome profiles reveal HowBacillus subtilis Lipopeptides inhibit Microsclerotia formation in Verticillium dahliae. MPMI 32, 622–634. doi: 10.1094/MPMI-08-18-0233-R
Zhang, L., and Sun, C. (2018). Fengycins, cyclic Lipopeptides from marine Bacillus subtilis strains, kill the plant-pathogenic fungus Magnaporthe grisea by inducing reactive oxygen species production and chromatin condensation. Applied and Environmental Microbiology, 84, e00445–e00418 doi: 10.1128/AEM.00445-18.
Zhao, H., Li, J., Zhang, Y., Lei, S., Zhao, X., Shao, D., et al. (2018). Potential of iturins as functional agents: safe, probiotic, and cytotoxic to cancer cells. Food Funct. 9, 5580–5587. doi: 10.1039/C8FO01523F
Zhao, H., Shao, D., Jiang, C., Shi, J., Li, Q., Huang, Q., et al. (2017). Biological activity of lipopeptides from bacillus. Appl. Microbiol. Biotechnol. 101, 5951–5960. doi: 10.1007/s00253-017-8396-0
Keywords: postharvest disease, fruit rot, Bacillus subtilis, biocontrol, antifungal, Alternaria alternata
Citation: Ahmad T, Xing F, Nie C, Cao C, Xiao Y, Yu X, Moosa A and Liu Y (2023) Biocontrol potential of lipopeptides produced by the novel Bacillus subtilis strain Y17B against postharvest Alternaria fruit rot of cherry. Front. Microbiol. 14:1150217. doi: 10.3389/fmicb.2023.1150217
Received: 23 January 2023; Accepted: 22 February 2023;
Published: 15 March 2023.
Edited by:
Junfeng Guan, Hebei Academy of Agriculture and Forestry Sciences (HAAFS), ChinaReviewed by:
Surekha K. Satpute, Savitribai Phule Pune University, IndiaCopyright © 2023 Ahmad, Xing, Nie, Cao, Xiao, Yu, Moosa and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yang Liu, bGl1eWFuZ0Bmb3N1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.