
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 14 June 2023
Sec. Evolutionary and Genomic Microbiology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1150070
This article is part of the Research Topic Antimicrobial Resistance Genomics in Bacterial Zoonotic Pathogens View all 5 articles
 Magdalena Nüesch-Inderbinen1*
Magdalena Nüesch-Inderbinen1* Michael Biggel1
Michael Biggel1 Adrian Haussmann1
Adrian Haussmann1 Andrea Treier1
Andrea Treier1 Lore Heyvaert2
Lore Heyvaert2 Nicole Cernela1
Nicole Cernela1 Roger Stephan1
Roger Stephan1Background: Linezolid is a critically important oxazolidinone antibiotic used in human medicine. Although linezolid is not licensed for use in food-producing animals, the use of florfenicol in veterinary medicine co-selects for oxazolidinone resistance genes.
Objective: This study aimed to assess the occurrence of cfr, optrA, and poxtA in florfenicol-resistant isolates from beef cattle and veal calves from different herds in Switzerland.
Methods: A total of 618 cecal samples taken from beef cattle and veal calves at slaughter originating from 199 herds were cultured after an enrichment step on a selective medium containing 10 mg/L florfenicol. Isolates were screened by PCR for cfr, optrA, and poxtA which are genes known to confer resistance to oxazolidinones and phenicols. One isolate per PCR-positive species and herd was selected for antimicrobial susceptibility testing (AST) and whole-genome sequencing (WGS).
Results: Overall, 105 florfenicol-resistant isolates were obtained from 99 (16%) of the samples, corresponding to 4% of the beef cattle herds and 24% of the veal calf herds. Screening by PCR revealed the presence of optrA in 95 (90%) and poxtA in 22 (21%) of the isolates. None of the isolates contained cfr. Isolates included for AST and WGS analysis were Enterococcus (E.) faecalis (n = 14), E. faecium (n = 12), E. dispar (n = 1), E. durans (n = 2), E. gallinarum (n = 1), Vagococcus (V.) lutrae (n = 2), Aerococcus (A.) urinaeequi (n = 1), and Companilactobacillus (C.) farciminis (n = 1). Thirteen isolates exhibited phenotypic linezolid resistance. Three novel OptrA variants were identified. Multilocus sequence typing identified four E. faecium ST18 belonging to hospital-associated clade A1. There was a difference in the replicon profile among optrA- and poxtA-harboring plasmids, with rep9 (RepA_N) plasmids dominating in optrA-harboring E. faecalis and rep2 (Inc18) and rep29 (Rep_3) plasmids in poxtA-carrying E. faecium.
Conclusion: Beef cattle and veal calves are reservoirs for enterococci with acquired linezolid resistance genes optrA and poxtA. The presence of E. faecium ST18 highlights the zoonotic potential of some bovine isolates. The dispersal of clinically relevant oxazolidinone resistance genes throughout a wide variety of species including Enterococcus spp., V. lutrae, A. urinaeequi, and the probiotic C. farciminis in food-producing animals is a public health concern.
The gastrointestinal tract of beef cattle and veal calves is reportedly a reservoir of antibiotic-resistant bacteria and clinically important antimicrobial resistance genes (Cameron and McAllister, 2016). Antimicrobial resistance (AMR) in food animals is thought to be promoted by antimicrobial use which is particularly high in the veal production sector (Rell et al., 2020; Becker et al., 2022). In beef cattle and veal calves, antimicrobial treatment consists of standard antimicrobial agents (aminoglycosides, β-lactams, diaminopyrimidines, phenicols, sulfonamides, and tetracyclines) depending on symptoms and body site of infection and restricted compounds which are classified as critically important for human medicine (fluoroquinolones, macrolides, and third- or fourth-generation cephalosporins) (European Union, 2019; World Health Organization, 2019; Swiss Veterinary Society, 2022). Florfenicol is a fluorinated derivative of chloramphenicol that is exclusively used in veterinary medicine and was licensed in Europe in 1995 for the treatment of bovine respiratory diseases (Schwarz et al., 2004). In Switzerland, florfenicol is recommended by the Swiss Veterinary Society (SVS) and the Federal Food Safety and Veterinary Office (FSVO) as a first-line antibiotic for the treatment of bovine respiratory disease in calves, calf meningitis, infectious bovine keratoconjunctivitis (IBK; also known as pinkeye) in cattle, and as a second-line antibiotic (after tetracycline) for metaphylaxis in calves and for the treatment of otitis in calves (Swiss Veterinary Society, 2022). The use of florfenicol in food-producing animals has selected for florfenicol-resistant bacteria, for example, enterococci harboring the phenicol exporter genes fexA or fexB, the 23S rRNA methyltransferase gene cfr, and genes encoding ribosomal protection proteins optrA and poxtA (Schwarz et al., 2004). Importantly, cfr, optrA, and poxtA may confer resistance not only to phenicols but also to oxazolidinones, potentially resulting in cross-resistance between florfenicol which is used exclusively in animals, and linezolid which is a critically important antibiotic to treat severe infections in humans (Long et al., 2006; Antonelli et al., 2018; Wang et al., 2018). Thus, due to the possibility of zoonotic transmission of AMR bacteria and horizontal transfer of resistance determinants to pathogenic strains within the human gut, the use of florfenicol in food-producing animals, including beef cattle and veal calves, may have serious consequences for public health. For beef, in particular, a possible transmission route may be through the consumption of undercooked or raw meat (rare steak or beef tartare). Linezolid-resistant enterococci from food-producing cattle have been described in Europe recently (de Jong et al., 2018; Timmermans et al., 2022), but data from beef cattle and veal calves in Switzerland are lacking. Therefore, this study was designed to (i) assess the occurrence of florfenicol resistance in presumptive enterococci from beef cattle and veal calves at slaughter in Switzerland using microbiological screening methodologies combined with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI–TOF MS) for species identification, (ii) identify any oxazolidinone resistance genes among the isolates using conventional polymerase chain reaction (PCR) protocols, and (iii) characterize a representative subset of the isolates by evaluating phenotypic AMR and applying whole-genome sequencing (WGS) analyses.
A total of 618 cecal samples (360 from beef cattle and 258 from veal calves) from 199 herds (106 cattle beef herds and 93 calf veal herds) were collected between September and October 2021 at a major abattoir in Zürich, Switzerland. Given that herd size varied widely (1–19 animals per herd), samples were obtained from all animals from each herd processed during the sampling period, excluding any milk-producing cows or breeding bulls within a herd. Cecal contents were obtained from each animal immediately after slaughter by cutting through the cecal wall with sterile scissors. Each cecum was swabbed once to retrieve content, and each swab was placed in a sterile blender bag (Seward, Worthing, UK). Samples were transported to the laboratory in cooler boxes and processed on the same day. For each swab specimen, 50-ml brain heart infusion (BHI) broth with 6% NaCl (Oxoid, Pratteln, Switzerland) was added directly to each bag. The samples were homogenized using a Stomacher® laboratory blender (Seward, Worthing, UK) and incubated for 18–24 h at 37°C. One loopful of the enriched culture was streaked on Bile Esculin Azide (BAA) agar (Merck, Darmstadt, Germany), supplemented with 10 mg/L florfenicol (Sigma-Aldrich, Buchs, Switzerland), and incubated under aerobic conditions for 48 h at 37°C. All bacterial colonies with morphological characteristics similar to Enterococcus spp. (small transparent colonies with brown-black halos) were subcultured on BAA agar with 10 mg/L florfenicol for 48 h at 37°C, and single colonies were subcultured on sheep blood agar (Difco, Becton Dickinson, Allschwil, Switzerland) for 24 h at 37°C. When morphologically different colonies were observed, both observed representative colonies were subcultured separately.
The isolates were subjected to species identification using MALDI-TOF-MS (Bruker Daltonics, Billerica, MA, USA) and the Compass FlexControl version 3.4 software with the Compass database version 4.1.80. The presence of optrA and poxtA was established by singleplex PCR using custom synthesized primers (Microsynth, Balgach, Switzerland) and conditions described previously (Wang et al., 2015; Egan et al., 2020b). The cfr gene was searched using primer and conditions described earlier (Kehrenberg and Schwarz, 2006; Nüesch-Inderbinen et al., 2022a). All the isolates were stored in 20% glycerol at −20°C until further analysis.
One isolate per PCR-positive species and herd was selected for antimicrobial susceptibility testing (AST) and whole-genome sequencing (WGS). The isolates were selected based on the following criteria: (i) They represented various bacterial species in proportion to their occurrence [Aerococcus urinaeequi, Companilactobacillus farciminis, Enterococcus (E.) dispar, E. faecalis, E. faecium, E. gallinarum, and Vagococcus lutrae] and (ii) they were collected from animals originating from different herds.
Antimicrobial susceptibility tests were performed using the agar disk diffusion method according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI, 2023). Antimicrobial substances included chloramphenicol and linezolid (Becton, Dickinson, Heidelberg, Germany). E. faecalis ATCC 29212 was used as a quality control strain. Zone diameters were interpreted according to the CLSI (2023). Zone diameter breakpoints were ≤ 12 mm for chloramphenicol and ≤ 20 mm for linezolid, respectively. For bacterial genera where interpretive criteria for susceptibility testing are not available, the CLSI classification was not applied.
Whole-genome sequences were determined using short-read sequencing (Illumina MiniSeq, Illumina, San Diego, CA, USA). Additionally, isolates containing plasmidal optrA or poxtA genes were long-read sequenced on a MinION Mk1B device (Oxford Nanopore Technologies, Oxford, UK). All isolates were first grown on sheep blood agar (Difco, Becton Dickinson, Allschwil, Switzerland), and genomic DNA was extracted using the DNeasy Blood and Tissue Kit (Qiagen, Hombrechtikon, Switzerland). Short-read sequencing libraries were prepared using the Nextera DNA Flex Library Preparation Kit (Illumina, San Diego, CA, USA) and sequenced on an Illumina MiniSeq instrument with 2 × 150 bp paired-end chemistries. For long-read sequencing, DNA was isolated using the MasterPure Complete DNA and RNA Purification Kit (Lucigen), and libraries were prepared using an SQK-LSK109 Ligation Sequencing Kit (Oxford Nanopore Technologies, Oxford, UK). The sequencing was carried out on a MinION Mk1B device using the FLO-MIN106 (R9) flow cell (Oxford Nanopore Technologies, Oxford, UK). Basecalling, demultiplexing, and barcode trimming were performed with guppy v4.2.2 (Oxford Nanopore Technologies, Oxford, UK) and quality-assessed with LongQC v1.2.0 (Fukasawa et al., 2020).
Short-reads from Illumina underwent assembly using SPAdes v.3.14.1 (Bankevich et al., 2012) implemented in Shovill v1.1.0 (Seemann, 2019). Hybrid assemblies were generated with Unicycler v.0.5.0 (Wick et al., 2017) using default settings. Genes were annotated using Bakta v1.6.1 (Schwengers et al., 2021), and isolates were typed in silico using mlst v.2.22.0 (Seemann, 2021b). For isolates that could not be definitively identified by MALDI-TOF MS, WGS was conducted to confirm the bacterial species. For species that could not be definitively identified by MALDI-TOF MS, genome assemblies were typed using the Ribosomal Multilocus Sequence Typing (rMLST) database (https://pubmlst.org/species-id) (Jolley et al., 2012).
Clade affiliation of E. faecium was determined using tools for the phylogenetic analysis described by Gouliouris et al. (2021) and phylogenetic clustering with 50 publicly available genomes for which clades were previously assigned (Lebreton et al., 2013). Following core genome alignment using Snippy v.4.6.0 (Seemann, 2015) with the chromosome of E. faecium SRR24 (GCF_009734005.1) as reference and masking of recombinant regions detected with Gubbins v.2.4.1 (Croucher et al., 2015), clusters were defined from the SNP alignment using fastbaps v.1.0.5 (Tonkin-Hill et al., 2019).
Antimicrobial resistance genes and plasmid replicons were identified using Abricate 1.0.1 (Seemann, 2021a) (70% coverage, 90% identity) in combination with the ResFinder 4.1 (Zankari et al., 2012) and PlasmidFinder databases (Carattoli et al., 2014), respectively. Plasmid replicon families were determined based on conserved domains identified in replicon sequences using BLAST (Madden, 2002). The optrA nucleotide sequences were compared with the wild-type (WT) optrAE349 (GenBank accession number KP399637) (Wang et al., 2015), and allele numbers of optrA were assigned according to the scheme from Freitas et al. (2020). OptrA variants were defined based on alterations in the deduced amino acid sequences. The nucleotide sequences of poxtA genes were compared to that described in S. aureus AOUC-0915 (GenBank accession no. MF095097) (Antonelli et al., 2018). Reads were also queried for mutations in the 23S rRNA genes associated with linezolid resistance (G2505A and G2576T) using LRE-finder 1.0 (Hasman et al., 2019).
Sequencing data and genome assemblies are available under BioProject no. PRJNA783265.
Overall, 99 (16%) of the 618 cecal samples exhibited bacterial growth on BAA-florfenicol plates after 48 h, corresponding to 26 (13%) of the 199 herds. Among the 99 samples, six were from beef cattle (2% of the 360 beef cattle samples) and 93 from veal calves (36% of the 258 veal calf samples), corresponding to four (4%) of the 106 beef cattle herds and 22 (24%) of 93 the veal calf herds (Table 1). Six cecum samples yielded more than one distinct isolate, resulting in a total of 105 florfenicol-resistant strains (Table 1; Supplementary Table S1).
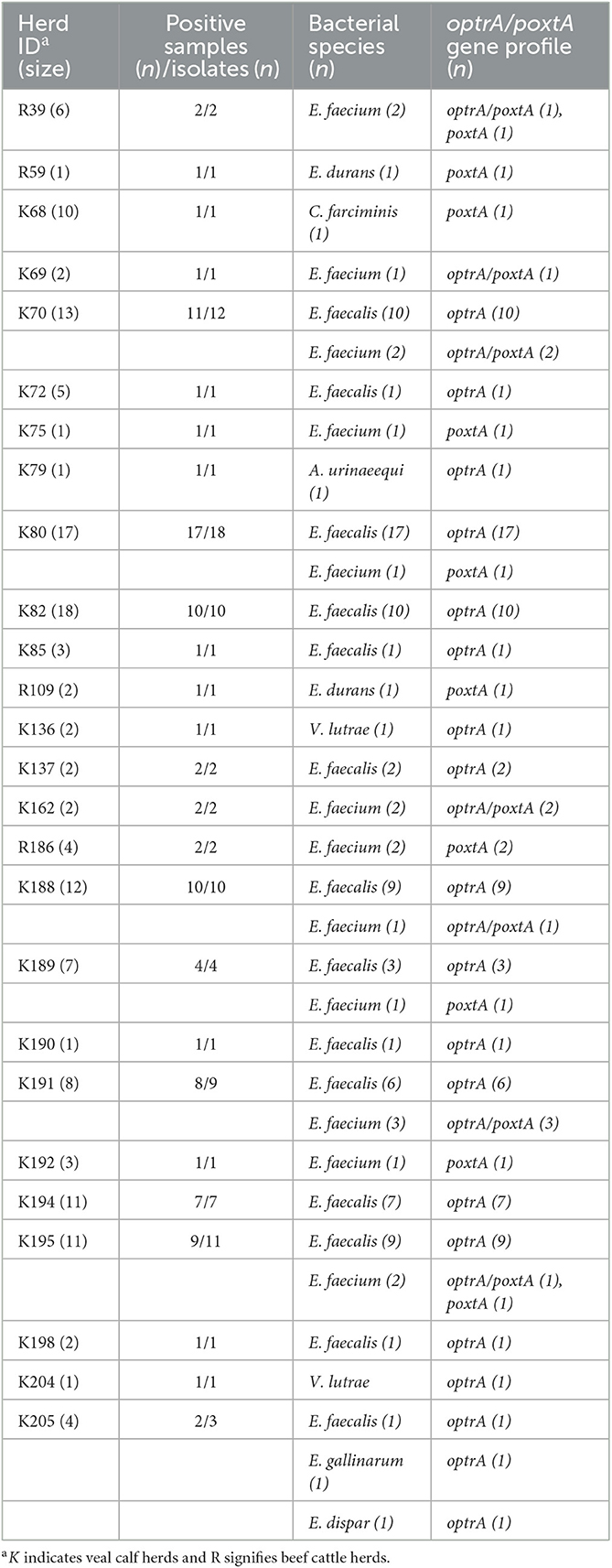
Table 1. Distribution of florfenicol-resistant isolates and oxazolidinone resistance genes among cecal samples from beef cattle and veal calves from 26 herds in Switzerland.
Species identification revealed that E. faecalis (n = 78/105, 74%) was the predominant species, followed by E. faecium (n = 19/105, 18%), E. durans (n = 2/105, 2%), E. dispar (n = 1/105, 1%), and E. gallinarum (n = 1/105, 1%) (Table 1). Non-enterococcal species included Vagococcus lutrae (n = 2/105, 2%), Aerococcus urinaeequi (n = 1/105, 1%), and Companilactobacillus farciminis (n = 1/105, 1%) (Table 1).
PCR screening demonstrated the presence of oxazolidinone resistance genes in all 105 isolates (Table 1; Supplementary Table S1). The optrA gene was identified in 95 (90%) of the isolates, including E. faecalis (n = 78), E. faecium (n = 12), E. dispar (n = 1), E. gallinarum (n = 1), V. lutrae (n = 2), and A. urinaeequi (n = 1) (Table 1). The poxtA gene was present in a total of 22 (21%) of the isolates consisting of E. faecium (n = 19), E. durans (n = 2), and C. farciminis (n = 1). In 12 of the isolates, poxtA was co-harbored with optrA (Table 1). The cfr gene was not detected in any of the isolates (Supplementary Table S1).
For 19 of the 26 positive herds (herds R39, R59, K68, K69, K72, K75, K79, K82, K85, R109, K136, K137, K162, R186, K190, K192, K194, K198, and K204), all positive samples within each herd yielded the same bacterial species (Supplementary Table S1). Of these, one randomly selected isolate per herd was used for further analysis. For six herds (herds K70, K80, K188, K189, K191, and K195), two distinct bacterial species were isolated within each herd, and, in each case, one isolate of each species was included for subsequent analysis (Supplementary Table S1). Finally, one herd (K205) yielded samples containing three different species, all of which were included for further analysis (Supplementary Table S1). Thus, a total of 34 isolates from four cattle beef and from 30 veal calves were available for further characterization, including E. faecalis (n = 14), E. faecium (n = 12), E. durans (n = 2), E. dispar (n = 1), E. gallinarum (n = 1), V. lutrae (n = 2), A. urinaeequi (n = 1), and C. farciminis (n = 1). Thereof, optrA only was present in 19 isolates, poxtA only in 10, and optrA/poxtA in five isolates, respectively.
All 34 isolates were tested for their susceptibility to chloramphenicol and linezolid (Table 2). Resistance to chloramphenicol was noted for 23 (77%) of the enterococcal isolates, including E. faecalis (n = 12), E. faecium (n = 10), and E. gallinarum (n = 1). Resistance to linezolid was detected in 13 (43%) of the enterococcal isolates, including E. faecalis (n = 9), E. faecium (n = 3), and E. gallinarum (n = 1) (Table 2).
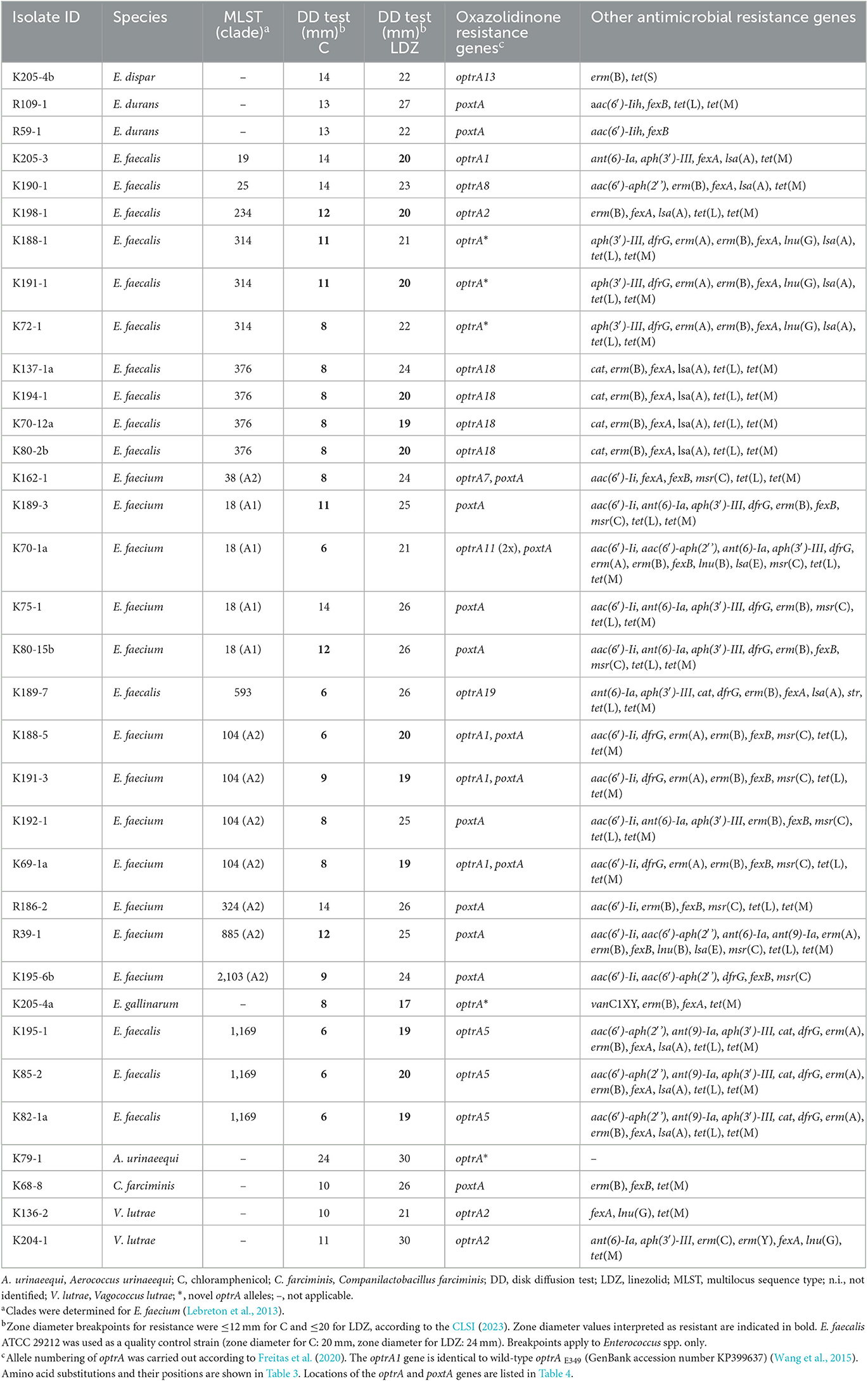
Table 2. WGS-based identification of resistance genes including linezolid-resistant genes in isolates from beef cattle and veal calves.
Multilocus sequence typing analysis assigned the 14 E. faecalis to seven different sequence types (STs), namely, ST19 (n = 1), ST25 (n = 1), ST234 (n = 1), ST314 (n = 3), ST376 (n = 4), ST593 (n = 1), and ST1169 (n = 3) (Table 2). The 12 E. faecium belonged to six different STs, namely ST38 (n = 1), ST18 (n = 4), ST104 (n = 4), ST324 (n = 1), ST885 (n = 1), and ST2013 (n = 1). E. faecium ST18 was assigned to hospital-associated clade A1 and the remaining E. faecium STs to clade A2 (Table 2).
A WGS analysis identified 12 different optrA allelic variants (including the WT optrAE349, referred to in this study as optrA1) and three novel alleles in a total of 24 isolates. The corresponding amino acid sequences of the OptrA variants are summarized in Table 3. Among the isolates, E. faecalis ST19 (n = 1) and E. faecium ST104 (n = 3) harbored optrA1 (GenBank acc. no. KP399637), E. faecalis ST234 (n = 1) and V. lutrae (n = 2) carried optrA2 (GenBank acc. no. KT862781), E. faecalis ST1169 (n = 3) contained optrA5 (GenBank acc. no. KT862783), E. faecium ST38 (n = 1) carried optrA7 (GenBank acc. no. KT862784), E. faecalis ST25 (n = 1) contained optrA8 (GenBank acc. no. KT862775), E. faecium ST18 (n = 1) harbored two copies of optrA11 (GenBank acc. no. KT862782), E. dispar (n = 1) contained an optrA13 (GenBank acc. no. KT892063), E. faecalis ST376 (n = 4) harbored optrA18 (GenBank acc. no. KX620942), and E. faecalis ST593 (n = 1) contained optrA19 (GenBank acc. no.KX620939). Three novel OptrA variants were found: E. faecalis ST314 (n = 3) harbored an optrA allelic variant encoding OptrA EK (Table 3). E. gallinarum (n = 1) and A. urinaeequi (n = 1) contained novel optrA alleles corresponding to variants OptrA EYDKWVDASKELYNKQLEIG, and OptrA EYKCLKVDASKELYNKQLEIG, respectively (Table 3).
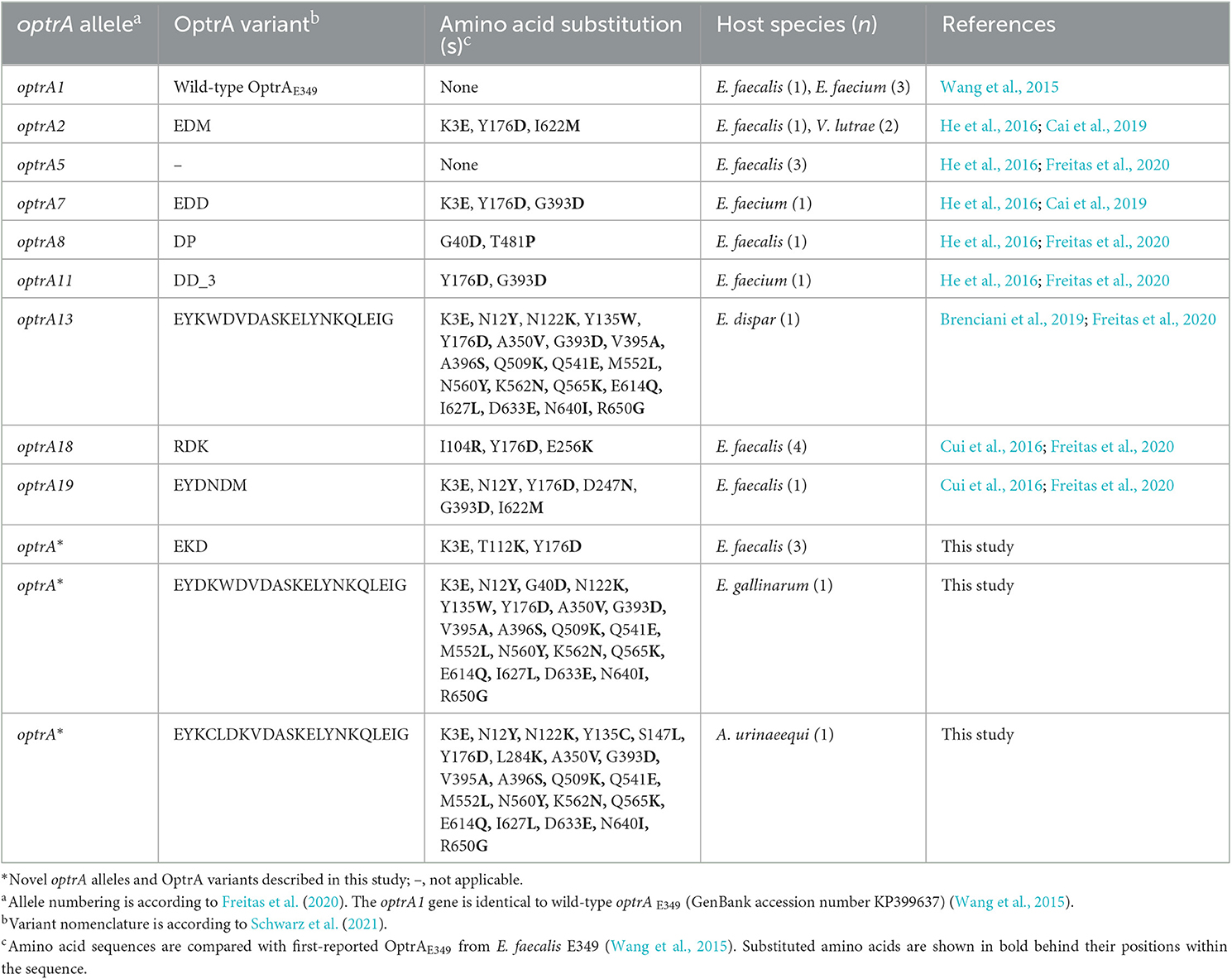
Table 3. OptrA variants were detected among florfenicol-resistant isolates from beef cattle and veal calves.
The WGS analysis identified the WT poxtA gene (GenBank accession no. MF095097) in E. faecium ST38 (n = 1), ST18 (n = 4), ST104 (n = 4), ST324 (n = 1), ST885 (n = 1), and ST2103 (n = 1), E. durans (n = 2), and C. farciminis (n = 1) (Table 2).
Almost all the isolates (n = 32, 94%) carried the phenicol resistance gene fexA (n = 17), fexB (n = 14), or fexA and fexB (n = 1) (Table 2). None of the isolates contained mutations in the 23S rRNA genes associated with linezolid resistance.
A number of additional resistance genes were identified, including genes conferring resistance to aminoglycosides [aac(6′)-aph(2′'), aac(6′)-Ii, aac(6′)-Iih, ant(6)-Ia, ant(9)-Ia, ant(6)-Ia, aph(3′)-III, and str], chloramphenicol (cat), lincosamides [lnu(G), lsa(A), and lsa(E)], macrolides [erm(A), erm(B), and msr(C)], tetracycline [tet(L) and tet(M)], trimethoprim (dfrG), and vancomycin (vanC1XY) (Table 2).
Among the AMR genes, lsa(A) was found in every E. faecalis and msr(C) in every E. faecium isolate, respectively. Furthermore, tet(L) and/or tet(M) was identified in 31 (91%) of the isolates, and E. gallinarum (n = 1) harbored the vancomycin resistance gene vanC1XY (Table 2). A. urinaeequi (isolate ID K79-1) did not carry any AMR genes apart from optrA (Table 2).
Overall, optrA was located within the chromosome (n = 5), on plasmids (n = 18), or both (n = 1) (Table 4). In E. faecalis, nine of the 10 identified optrA-encoding plasmids were categorized within the replicon type rep9a and rep9b belonging to the RepA_N family, with plasmid sizes ranging between 51.4 and 78.7 kb (Table 4). In E. faecalis ST376, optrA was associated with rep9a plasmids (n = 4), and in E. faecalis, ST25, ST234, and ST314, optrA were found on rep9b plasmids (n = 5). The remaining E. faecalis (ST19) harbored optrA on a 36.3 kb repUS40_1 Rep_3 family plasmid (Table 4).
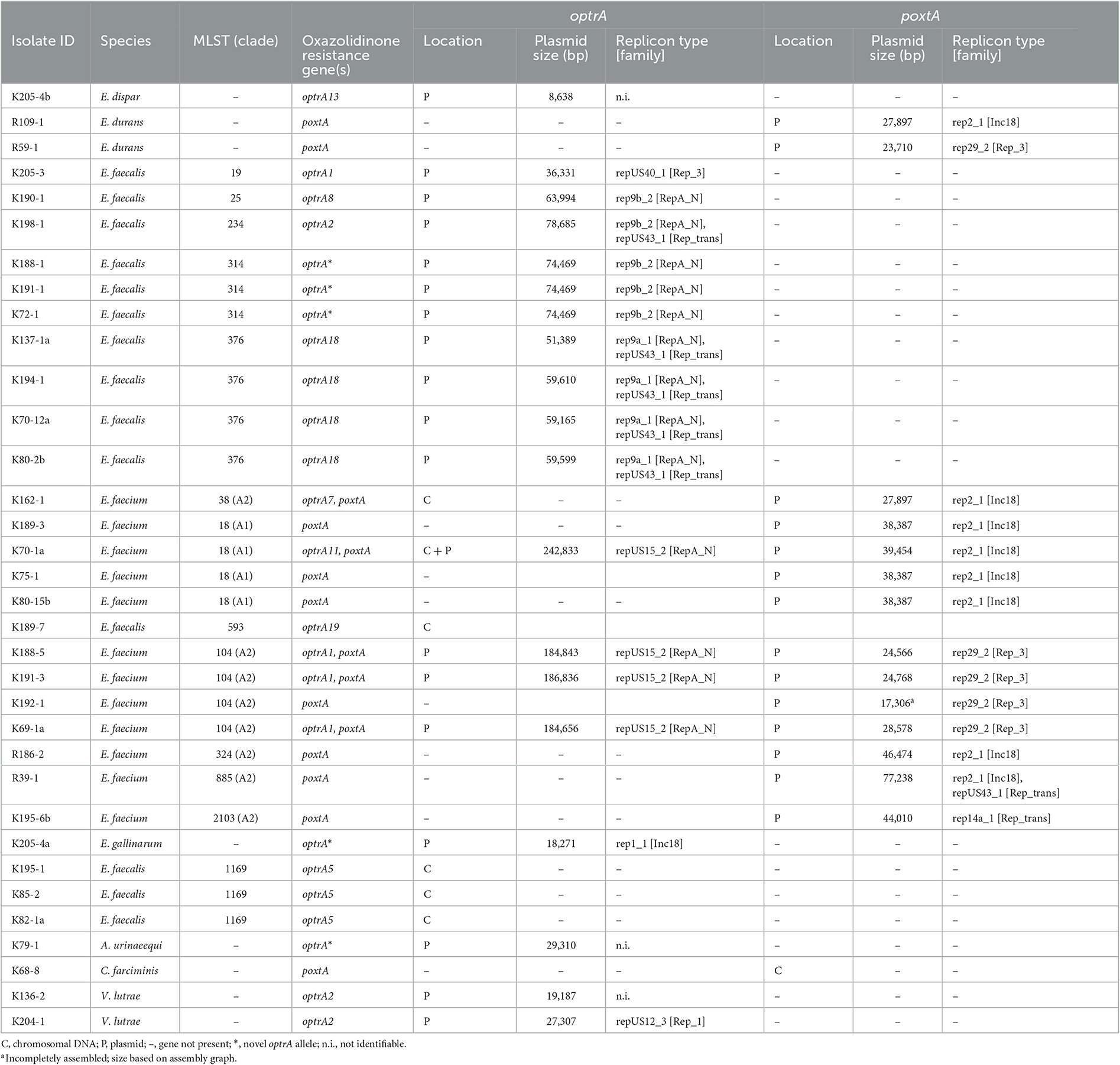
Table 4. Location of optrA and poxtA genes among florfenicol-resistant isolates from beef cattle and veal calves.
Among the E. faecium isolates, all optrA-carrying plasmids (n = 4) belonged to repUS15_2 (RepA_N) (Table 4). In E. faecium ST104, plasmid sizes (18.5 kb) were identical (n = 3). In E. faecium ST18, optrA was carried on a 24.3 kb repUS15_2 (RepA_N) plasmid (Table 4). In E. gallinarum, optrA was identified on an 18.3 kb rep1_1 (Inc18) plasmid, and V. lutrae carried optrA on a 27.3 kb repUS12_3 (Rep_1) plasmid (Table 4).
PlasmidFinder (Carattoli et al., 2014) did not identify rep genes in a total of three optrA-encoding plasmids from A. urinaeequi, E. dispar, and V. lutrae (Table 4).
The poxtA genes were located within the chromosome (n = 1) and on plasmids (n = 14).
Among E. faecium ST18, ST38, ST324, and ST885, poxtA was identified on rep2_1 plasmids (n = 7) belonging to the Inc18 family, ranging between 27.9 and 77.2 kb in size (Table 4). In E. faecium ST104, poxtA was found on 17.3–12.5 kb rep29_2 (Rep_3) plasmids (n = 4), and in E. faecium ST 2103, poxtA was located on a 44 kb rep14a_1 (Rep_trans) plasmid (n = 1) (Table 4). In E. durans, poxtA was identified on a 27.9 kb rep2_1 (Inc18) (n = 1) and a 23.7kb rep29_2 (Rep_3) plasmid (n = 1), respectively (Table 4).
In this study, we found the presence of florfenicol-resistant Enterococcus-like bacteria in 16% of cecal samples of healthy beef cattle and veal calves in Switzerland. Our data show that the occurrence of florfenicol-resistant isolates differed widely between beef cattle (2%) and veal calves (36%), corresponding to a herd prevalence of 4% for beef cattle and 24% for veal calves, respectively. These results may reflect the high usage of antimicrobials in Swiss calf fattening operations (Becker et al., 2022), which, for its part, is mainly due to the predisposition to infectious diseases of young calves challenged by transportation, co-mingling, and inadequate housing (Becker et al., 2020). In addition, most isolates analyzed in this study were associated with the presence of tetracycline resistance genes. Tetracyclines are widely used for the treatment of bovine infectious diseases and metaphylaxis (Swiss Veterinary Society, 2022) and are, therefore, similar to florfenicol, likely to further promote the spread and persistence of bacteria that harbor oxazolidinone resistance genes. In this study, all 105 florfenicol-resistant isolates carried oxazolidinone resistance genes. Notably, for many of the isolates, the presence of these genes alone did not result in phenotypic resistance. This observation corroborates previous studies that describe varying linezolid MICs among Enterococcus spp., their STs, and their optrA or poxtA genes (Wang et al., 2015; Cai et al., 2019; Nüesch-Inderbinen et al., 2022a). While bacterial host factors are suggested to influence the expression of linezolid resistance genes (Cai et al., 2019), the mechanisms that determine the resistance phenotype of Gram-positive bacteria harboring linezolid resistance determinants remain unclear. The lack of a linezolid resistance phenotype among many isolates in this study indicates that our selective approach using florfenicol to detect linezolid resistance mechanisms in enterococci and other bacterial genera may be favorable for screening samples from food-producing animals.
The majority (76%) of the florfenicol-resistant isolates were represented by E. faecalis or E. faecium. These two enterococcal species are part of the normal gastrointestinal flora in animals and humans, but they also belong to the most important nosocomial pathogens worldwide (Aarestrup, 2015; Zaheer et al., 2020). Among the isolates collected during this study, WGS analysis identified E. faecalis ST19 (isolate ID K205-3)-harboring plasmid-borne optrA1. Isolate K205-3 represented one of the 13 phenotypically linezolid-resistant isolates identified in this study according to CLSI definitions. The optrA1 gene belongs to one of the most widespread variants identified among optrA-positive enterococci from food-producing animals and humans worldwide (Wang et al., 2015). E. faecalis ST19-harboring optrA1 has also been isolated from a hospitalized patient in Ireland (Egan et al., 2020b). Similarly, E. faecalis ST593 carrying optrA19 (Optra EYDNDM) identified in one veal calf in this study has been associated with urinary tract infection in humans (Cai et al., 2019), highlighting the potential of optrA-harboring E. faecalis occurring in food-producing animals to cause disease in humans.
WGS analysis further identified four E. faecalis ST376 (isolates K137-1a, K194-1, K70-12a, and K80-2b, recovered from calves across different herds) harboring plasmid-encoded optrA18 (OptrA RDK), a globally widespread optrA variant originally described in human clinical E. faecalis (Cui et al., 2016). E. faecalis ST376 carrying optrA18 has been found in fecal samples of healthy adults and children in China, in fresh commercial flowers in China, and in a cecal isolate from a fattening pig in Switzerland (Cai et al., 2019; Yu et al., 2021; Nüesch-Inderbinen et al., 2022a). The detection of optrA18-positive E. faecalis ST376 among bovine enterococci across different herds in the present study provides further evidence for its worldwide dissemination throughout multiple ecological niches.
WGS showed that the bovine E. faecium from this study included four E. faecium ST18 belonging to the epidemic hospital-associated lineage referred to as clade A1 (isolates K189-3, K70-1a, K75-1, and K80-15b, respectively) (Lebreton et al., 2013). Three of these isolates carried plasmid-mediated poxtA with the fourth additionally harboring a chromosomal optrA. Similar linezolid-resistant E. faecium ST18 strains carrying plasmid-mediated poxtA with or without chromosomal optrA have been recovered from hospitalized patients in Ireland between 2016 and 2019 (Egan et al., 2020a). In addition, optrA-harboring E. faecalis ST18 has been found in fecal samples from veal calves in Belgium in 2019 (Timmermans et al., 2022), emphasizing the dispersal of poxtA and/or optrA-positive E. faecium ST18 across different hosts and regions. The actual risk of zoonotic transmission of Enterococcus spp. from livestock to humans is probably quite low (Gouliouris et al., 2018; Cattoir, 2022). Furthermore, our data are not discriminative enough to infer a strong relatedness between clinical and livestock isolates; nevertheless, it cannot be excluded that the occurrence of E. faecium ST18 in veal calves may represent a potential zoonotic risk. Additional screening of the employees in the slaughterhouse may have provided indications of possible transmission events; however, this was not performed in this study.
The remaining E. faecium analyzed in this study were part of clade A2, which is mostly represented by strains from animal commensals (Lebreton et al., 2013). Isolates from clade A2 included optrA1/poxtA-carrying E. faecium ST104 collected across four veal calf herds, which has previously been found in fecal samples from veal calves in Belgium (Timmermans et al., 2022).
Apart from E. faecalis and E. faecium, we detected plasmid-encoded optrA in E. dispar, E. durans, and E. gallinarum, demonstrating a high diversity among optrA and/or poxtA-carrying enterococci circulating among beef cattle and veal calf herds in Switzerland. E. gallinarum harboring vanC1XY and optrA2 (OptrA EDM) was previously recovered from a porcine fecal swab in China in 2018 (Yao et al., 2020), and E. gallinarum co-harboring vanC1XY and OptrA (KLDP) was found recently in healthy humans in Switzerland in 2021 (Nüesch-Inderbinen et al., 2022b), but this is to our knowledge the first description of a bovine E. gallinarum carrying vanC1XY and a novel optrA (OptrA EYDKWDVDASKELYNKQLEIG) variant. Although E. gallinarum harboring vanC1XY has been isolated from a patient with sepsis in Malaysia in 2019 (Mastor et al., 2020), this species is generally not considered pathogenic to humans, and its zoonotic potential is probably low (Cattoir, 2022).
In this study, optrA and poxtA were distributed unevenly among the enterococcal species and plasmid families, with most optrA found in E. faecalis and identified on rep9 variants of RepA_N plasmids which are narrow host range plasmids considered specific for E. faecalis (Mikalsen et al., 2015). By contrast, in optrA-harboring E. faecium, the repUS15_2 was the predominant replicon type but was absent in E. faecalis. Furthermore, plasmid-encoded poxtA was detected predominantly among E. faecium and found on broad host range rep2 (Inc18) and on narrow host range rep29 (Rep_3) plasmids which were lacking in E. faecalis, comparable to previous observations (Mikalsen et al., 2015). Taken together, our findings indicate that the dissemination of oxazolidinone resistance genes among bovine isolates is driven by different plasmid families in E. faecalis and E. faecium, similar to the development observed in clinically relevant enterococcal lineages (Mikalsen et al., 2015).
Finally, this study revealed the presence of optrA and poxtA in several non-enterococcal isolates, including V. lutrae, A. urinaeequi, and C. farciminis. While optrA-positive V. lutrae and optrA-harboring A. urinaeequi have previously been isolated from porcine samples in China (Yang et al., 2021; Zhu et al., 2022a), this is only the second report of optrA in A. urinaeequi and the first description of a novel, plasmid-encoded optrA variant encoding OptrA EYKCLKVDASKELYNKQLEIG.
Furthermore, while poxtA has been identified in Lactobacillaceae, e.g., in L. salivarius recovered from human and porcine samples in China (Shen et al., 2022; Zhu et al., 2022b), to the best of our knowledge, this is the first report of poxtA in C. farciminis [formerly L. farciminis (Zheng et al., 2020)]. C. farciminis is authorized and presumed safe as a probiotic feed additive for animals in the EU (Directive 70/524/EEC, JLO 297:15/11/2001) (Coeuret et al., 2004; EFSA, 2021). The identification in this study of poxtA-carrying C. farciminis notably co-harboring erm(B), fexB, and tet(M) points to the possibility of transmission of AMR genes within the bovine intestine and raises questions regarding the role of probiotics as reservoirs of transferable AMR genes.
Our study shows that oxazolidinone resistance determinants occur in a variety of florfenicol-resistant Enterococcus spp., in V. lutrae, A. urinaeequi, and the probiotic C. farciminis, within the intestinal tract of beef cattle and veal calves. A potential health hazard is highlighted by the presence of poxtA and poxtA/optrA-harboring E. faecium ST18, which is associated with nosocomial infections. Co-selection of oxazolidinone resistance genes through the use of florfenicol represents a serious threat to the efficacy of linezolid and represents an issue that should be considered using a one-health approach.
The data presented in this study are deposited in the NCBI repository, accession number PRJNA783265: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA783265.
No animal was killed for the purpose of providing samples. Ethics approval was not required for this study. All information was treated anonymously.
The study was designed by RS. Sampling was accomplished by AH. Microbiological analyses were performed by AH, AT, and LH. WGS was done by MB and NC. Bioinformatics analyses were conducted by MB. Data analyses were conducted by AH, MB, MN-I, and RS. The manuscript was written by MN-I. RS and MB contributed to writing and revising the manuscript. All authors agreed on the final version. All authors contributed to the article and approved the submitted version.
This study was supported in part by the Swiss Federal Office of Public Health, Division of Communicable Diseases.
We thank Katrin Zurfluh for her technical assistance.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1150070/full#supplementary-material
Aarestrup, F. M. (2015). The livestock reservoir for antimicrobial resistance: a personal view on changing patterns of risks, effects of interventions and the way forward. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 370, 20140085. doi: 10.1098/rstb.2014.0085
Antonelli, A., D'Andrea, M. M., Brenciani, A., Galeotti, C. L., Morroni, G., Pollini, S., et al. (2018). Characterization of poxtA, a novel phenicol-oxazolidinone-tetracycline resistance gene from an MRSA of clinical origin. J. Antimicrob. Chemother. 73, 1763–1769. doi: 10.1093/jac/dky088
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Becker, J., Perreten, V., Schüpbach-Regula, G., Stucki, D., Steiner, A., and Meylan, M. (2022). Associations of antimicrobial use with antimicrobial susceptibility at the calf level in bacteria isolated from the respiratory and digestive tracts of veal calves before slaughter. J. Antimicrob. Chemother. 77, 2859–2866. doi: 10.1093/jac/dkac246
Becker, J., Schüpbach-Regula, G., Steiner, A., Perreten, V., Wüthrich, D., Hausherr, A., et al. (2020). Effects of the novel concept ‘outdoor veal calf' on antimicrobial use, mortality and weight gain in Switzerland. Prev. Vet. Med. 176, 104907. doi: 10.1016/j.prevetmed.2020.104907
Brenciani, A., Fioriti, S., Morroni, G., Cucco, L., Morelli, A., Pezzotti, G., et al. (2019). Detection in Italy of a porcine Enterococcus faecium isolate carrying the novel phenicol-oxazolidinone-tetracycline resistance gene poxtA. J. Antimicrob. Chemother. 74, 817–818. doi: 10.1093/jac/dky505
Cai, J., Schwarz, S., Chi, D., Wang, Z., Zhang, R., and Wang, Y. (2019). Faecal carriage of optrA-positive enterococci in asymptomatic healthy humans in Hangzhou, China. Clin. Microbio.l Infect. 25 630.e1–630.e6. doi: 10.1016/j.cmi.2018.07.025
Cameron, A., and McAllister, T. A. (2016). Antimicrobial usage and resistance in beef production. J. Anim. Sci. Biotechnol. 7, 68. doi: 10.1186/s40104-016-0127-3
Carattoli, A., Zankari, E., García-Fernández, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
Cattoir, V. (2022). The multifaceted lifestyle of enterococci: genetic diversity, ecology and risks for public health. Curr. Opin. Microbiol. 65, 73–80. doi: 10.1016/j.mib.2021.10.013
CLSI (2023). Performance Standards for Antimicrobial Susceptibility Testing. Supplement M100, 33rd Edn. Wayne, PE: CLSI.
Coeuret, V., Gueguen, M., and Vernoux, J. P. (2004). Numbers and strains of lactobacilli in some probiotic products. Int. J. Food Microbiol. 97, 147–156. doi: 10.1016/j.ijfoodmicro.2004.04.015
Croucher, N. J., Page, A. J., Connor, T. R., Delaney, A. J., Keane, J. A., Bentley, S. D., et al. (2015). Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 43, e15. doi: 10.1093/nar/gku1196
Cui, L., Wang, Y., Lv, Y., Wang, S., Song, Y., Li, Y., et al. (2016). Nationwide surveillance of novel oxazolidinone resistance gene optrA in Enterococcus isolates in China from 2004 to 2014. Antimicrob. Agents Chemother. 60, 7490–7493. doi: 10.1128/AAC.01256-16
de Jong, A., Simjee, S., Garch, F. E., Moyaert, H., Rose, M., Youala, M., et al. (2018). Antimicrobial susceptibility of enterococci recovered from healthy cattle, pigs and chickens in nine EU countries (EASSA Study) to critically important antibiotics. Vet. Microbiol. 216, 168–175. doi: 10.1016/j.vetmic.2018.02.010
EFSA (2021). Efficacy of the feed additive containing Companilactobacillus farciminis (formerly Lactobacillus farciminis) CNCM I-3740 (Biacton®) for chickens for fattening, turkeys for fattening and laying hens (ChemVet dk A/S). EFSA J. 19, e06627. doi: 10.2903/j.efsa.2021.6627
Egan, S. A., Corcoran, S., McDermott, H., Fitzpatrick, M., Hoyne, A., McCormack, O., et al. (2020a). Hospital outbreak of linezolid-resistant and vancomycin-resistant ST80 Enterococcus faecium harbouring an optrA-encoding conjugative plasmid investigated by whole-genome sequencing. J. Hosp. Infect. 105, 726–735. doi: 10.1016/j.jhin.2020.05.013
Egan, S. A., Shore, A. C., O'Connell, B., Brennan, G. I., and Coleman, D. C. (2020b). Linezolid resistance in Enterococcus faecium and Enterococcus faecalis from hospitalized patients in Ireland: high prevalence of the MDR genes optrA and poxtA in isolates with diverse genetic backgrounds. J. Antimicrob. Chemother. 75, 1704–1711. doi: 10.1093/jac/dkaa075
European Union (2019). Regulation 2019/6 of the European Parliament and of the Council of 11 December 2018 on veterinary medicinal products and repealing Directive 2001/82/EC. OJEU. 62, 4.
Freitas, A. R., Tedim, A. P., Novais, C., Lanza, V. F., and Peixe, L. (2020). Comparative genomics of global optrA-carrying Enterococcus faecalis uncovers a common chromosomal hotspot for optrA acquisition within a diversity of core and accessory genomes. Microb. Genom. 6, 350. doi: 10.1099/mgen.0.000350
Fukasawa, Y., Ermini, L., Wang, H., Carty, K., and Cheung, M. S. (2020). LongQC: A quality control tool for third generation sequencing long read data. G3 Genes GenomesGenet. 10, 1193–1196. doi: 10.1534/g3.119.400864
Gouliouris, T., Coll, F., Ludden, C., Blane, B., Raven, K. E., Naydenova, P., et al. (2021). Quantifying acquisition and transmission of Enterococcus faecium using genomic surveillance. Nat. Microbiol. 6, 103–111. doi: 10.1038/s41564-020-00806-7
Gouliouris, T., Raven, K. E., Ludden, C., Blane, B., Corander, J., Horner, C. S., et al. (2018). Genomic surveillance of Enterococcus faecium reveals limited sharing of strains and resistance genes between livestock and humans in the United Kingdom. MBio. 9, e01780–e01718. doi: 10.1128/mBio.01780-18
Hasman, H., Clausen, P. T. L. C., Kaya, H., Hansen, F., Knudsen, J. D., Wang, M., et al. (2019). LRE-Finder, a web tool for detection of the 23S rRNA mutations and the optrA, cfr, cfr(B) and poxtA genes encoding linezolid resistance in enterococci from whole-genome sequences. J. Antimicrob. Chemother. 74, 1473–1476. doi: 10.1093/jac/dkz092
He, T., Shen, Y., Schwarz, S., Cai, J., Lv, Y., Li, J., et al. (2016). Genetic environment of the transferable oxazolidinone/phenicol resistance gene optrA in Enterococcus faecalis isolates of human and animal origin. J. Antimicrob. Chemother. 71, 1466–1473. doi: 10.1093/jac/dkw016
Jolley, K. A., Bliss, C. M., Bennett, J. S., Bratcher, H. B., Brehony, C., Colles, F. M., et al. (2012). Ribosomal multilocus sequence typing: universal characterization of bacteria from domain to strain. Microbiol. 158, 1005–1015. doi: 10.1099/mic.0.055459-0
Kehrenberg, C., and Schwarz, S. (2006). Distribution of florfenicol resistance genes fexA and cfr among chloramphenicol-resistant Staphylococcus isolates. Antimicrob. Agents Chemother. 50, 1156–1163. doi: 10.1128/AAC.50.4.1156-1163.2006
Lebreton, F., van Schaik, W., McGuire, A. M., Godfrey, P., Griggs, A., Mazumdar, V., et al. (2013). Emergence of epidemic multidrug-resistant Enterococcus faecium from animal and commensal strains. MBio. 4, e00534-13. doi: 10.1128/mBio.00534-13
Long, K. S., Poehlsgaard, J., Kehrenberg, C., Schwarz, S., and Vester, B. (2006). The Cfr rRNA methyltransferase confers resistance to phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A antibiotics. Antimicrob. Agents Chemother. 50, 2500–2505. doi: 10.1128/AAC.00131-06
Mastor, N. N., Subbiah, V. K., Bakar, W. N. W. A., Begum, K., Alam, M. J., and Hoque, M. Z. (2020). Whole genome sequencing data of a clinical Enterococcus gallinarum strain EGR748 from Sabah, Malaysia. Data Brief 33, 106370. doi: 10.1016/j.dib.2020.106370
Mikalsen, T., Pedersen, T., Willems, R., Coque, T. M., Werner, G., Sadowy, E., et al. (2015). Investigating the mobilome in clinically important lineages of Enterococcus faecium and Enterococcus faecalis. BMC Genom. 16, 1–16. doi: 10.1186/s12864-015-1407-6
Nüesch-Inderbinen, M., Biggel, M., Zurfluh, K., Treier, A., and Stephan, R. (2022b). Faecal carriage of enterococci harbouring oxazolidinone resistance genes among healthy humans in the community in Switzerland. J. Antimicrob. Chemother. 77, 2779–2783. doi: 10.1093/jac/dkac260
Nüesch-Inderbinen, M., Haussmann, A., Treier, A., Zurfluh, K., Biggel, M., Stephan, R., et al. (2022a). Fattening pigs are a reservoir of florfenicol-resistant enterococci harboring oxazolidinone resistance genes. J. Food Prot. 85, 740–746. doi: 10.4315/JFP-21-431
Rell, J., Wunsch, N., Home, R., Kaske, M., Walkenhorst, M., Vaarst, M., et al. (2020). Stakeholders' perceptions of the challenges to improving calf health and reducing antimicrobial use in Swiss veal production. Prev. Vet. Med. 179, 104970. doi: 10.1016/j.prevetmed.2020.104970
Schwarz, S., Kehrenberg, C., Doublet, B., and Cloeckaert, A. (2004). Molecular basis of bacterial resistance to chloramphenicol and florfenicol. FEMS Microbiol. Rev. 28, 519–542. doi: 10.1016/j.femsre.2004.04.001
Schwarz, S., Zhang, W., Du, X. D., Krüger, H., Feßler, A. T., Ma, S., et al. (2021). Mobile oxazolidinone resistance genes in Gram-positive and Gram-negative bacteria. Clin. Microbiol. Rev. 34, e0018820. doi: 10.1128/CMR.00188-20
Schwengers, O., Jelonek, L., Dieckmann, M. A., Beyvers, S., Blom, J., and Goesmann, A. (2021). Bakta: rapid and standardized annotation of bacterial genomes via alignment-free sequence identification. Microb Genom 7, 000685. doi: 10.1099/mgen.0.000685
Seemann, T. (2015). Snippy. Available online at: https://github.com/tseemann/snippy (accessed January 9, 2023).
Seemann, T. (2019). Shovill. Available online at: https://github.com/tseemann/shovill (accessed January 9, 2023).
Seemann, T. (2021a). mlst. Available online at: https://github.com/tseemann/mlst (accessed January 9, 2023).
Seemann, T. (2021b). Abricate. Available online at: https://github.com/tseemann/abricate (accessed January 9, 2023).
Shen, W., Huang, Y., and Cai, J. (2022). An optimized screening approach for the oxazolidinone resistance gene optrA yielded a higher fecal carriage rate among healthy individuals in Hangzhou, China. Microbiol. Spectr. 10, e02974-22. doi: 10.1128/spectrum.02974-22
Swiss Veterinary Society (2022). Umsichtiger Einsatz von Antibiotika bei Rindern und Schweinen: Therapieleitfaden für Tierärztinnen und Tierärzte. Available online at: https://www.blv.admin.ch/dam/blv/de/dokumente/tiere/tierkrankheiten-und-arzneimittel/tierarzneimittel/therapieleitfaden.pdf.download.pdf/therapieleitfaden-de.pdf (accessed January 9, 2023).
Timmermans, M., Bogaerts, B., Vanneste, K., De Keersmaecker, S. C. J., Roosens, N. H. C., Kowalewicz, C., et al. (2022). Large diversity of linezolid-resistant isolates discovered in food-producing animals through linezolid selective monitoring in Belgium in 2019. J. Antimicrob. Chemother. 77, 49–57. doi: 10.1093/jac/dkab376
Tonkin-Hill, G., Lees, J. A., Bentley, S. D., Frost, S. D. W., and Corander, J. (2019). Fast hierarchical Bayesian analysis of population structure. Nucleic Acids Res. 47, 5539–5549. doi: 10.1093/nar/gkz361
Wang, X., Wang, Y., Zhou, Y., Li, J., Yin, W., Wang, S., et al. (2018). Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing Klebsiella pneumoniae. Emerg. Microbes Infect. 7, 122. doi: 10.1038/s41426-018-0124-z
Wang, Y., Lv, Y., Cai, J., Schwarz, S., Cui, L., Hu, Z., et al. (2015). A novel gene, optrA, that confers transferable resistance to oxazolidinones and phenicols and its presence in Enterococcus faecalis and Enterococcus faecium of human and animal origin. J. Antimicrob. Chemother. 70, 2182–2190. doi: 10.1093/jac/dkv116
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13, e1005595. doi: 10.1371/journal.pcbi.1005595
World Health Organization (2019). Critically Important Antimicrobials for Human Medicine. 6th Rev. Available online at: https://apps.who.int/iris/bitstream/handle/10665/312266/9789241515528-eng.pdf (accessed January 9, 2023).
Yang, Q., Zhu, Y., Schwarz, S., Wang, L., Liu, W., Yang, W., et al. (2021). A novel plasmid from Aerococcus urinaeequi of porcine origin co-harboring the tetracycline resistance genes tet(58) and tet(61). Vet. Microbiol. 257, 109065. doi: 10.1016/j.vetmic.2021.109065
Yao, T. G., Li, B. Y., Luan, R. D., Wang, H. N., and Lei, C. W. (2020). Whole genome sequence of Enterococcus gallinarum EG81, a porcine strain harbouring the oxazolidinone-phenicol resistance gene optrA with chromosomal and plasmid location. J. Glob. Antimicrob. Resist. 22, 598–600. doi: 10.1016/j.jgar.2020.06.012
Yu, Y., Ye, X. Q., Liang, H. Q., Zhong, Z. X., Cheng, K., Sun, J., et al. (2021). Lilium spp., as unnoticed environmental vector, spreading OptrA-carrying Enterococcus spp. Sci. Total Environ. 816, 151540. doi: 10.1016/j.scitotenv.2021.151540
Zaheer, R., Cook, S. R., Barbieri, R., Goji, N., Cameron, A., Petkau, A., et al. (2020). Surveillance of Enterococcus spp. reveals distinct species and antimicrobial resistance diversity across a One-Health continuum. Sci. Rep. 10, 1–16. doi: 10.1038/s41598-020-61002-5
Zankari, E., Hasman, H., Cosentino, S., Vestergaard, M., Rasmussen, S., Lund, O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644. doi: 10.1093/jac/dks261
Zheng, J., Wittouck, S., Salvetti, E., Franz, C. M. A. P., Harris, H. M. B., Mattarelli, P., et al. (2020). A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 70, 2782–2858. doi: 10.1099/ijsem.0.004107
Zhu, Y., Yang, W., Schwarz, S., Xu, Q., Yang, Q., Wang, L., et al. (2022a). Characterization of the novel optrA-carrying pseudo-compound transposon Tn7363 and an Inc18 plasmid carrying cfr(D) in Vagococcus lutrae. J. Antimicrob. Chemother. 77, 921–925. doi: 10.1093/jac/dkab478
Keywords: florfenicol, linezolid, optrA, poxtA, beef cattle, veal calves, E. faecalis, E. faecium clade A1
Citation: Nüesch-Inderbinen M, Biggel M, Haussmann A, Treier A, Heyvaert L, Cernela N and Stephan R (2023) Oxazolidinone resistance genes in florfenicol-resistant enterococci from beef cattle and veal calves at slaughter. Front. Microbiol. 14:1150070. doi: 10.3389/fmicb.2023.1150070
Received: 23 January 2023; Accepted: 18 May 2023;
Published: 14 June 2023.
Edited by:
Mariela E. Srednik, Iowa State University, United StatesReviewed by:
Dele Ogunremi, Canadian Food Inspection Agency (CFIA), CanadaCopyright © 2023 Nüesch-Inderbinen, Biggel, Haussmann, Treier, Heyvaert, Cernela and Stephan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Magdalena Nüesch-Inderbinen, bWFnZGFsZW5hLm51ZWVzY2gtaW5kZXJiaW5lbkB1emguY2g=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.