
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 17 April 2023
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1149418
This article is part of the Research Topic The role of regulatory networks in virulence and antimicrobial resistance of microbial pathogens View all 7 articles
Efflux pumps function as an advanced defense system against antimicrobials by reducing the concentration of drugs inside the bacteria and extruding the substances outside. Various extraneous substances, including antimicrobials, toxic heavy metals, dyes, and detergents, have been removed by this protective barrier composed of diverse transporter proteins found in between the cell membrane and the periplasm within the bacterial cell. In this review, multiple efflux pump families have been analytically and widely outlined, and their potential applications have been discussed in detail. Additionally, this review also discusses a variety of biological functions of efflux pumps, including their role in the formation of biofilms, quorum sensing, their survivability, and the virulence in bacteria, and the genes/proteins associated with efflux pumps have also been explored for their potential relevance to antimicrobial resistance and antibiotic residue detection. A final discussion centers around efflux pump inhibitors, particularly those derived from plants.
Bacteria are already well known for their ability to persist in extreme environments via their evolving numerous defense mechanisms against various substances that are lethal for their survival. Currently, antimicrobial resistance, the most complicated health concern across the world, which is primarily associated with an extended hospital stay, expensive treatment, and high mortality and morbidity rates, needs undeniable attention and multiple operative approaches involving the “One Health” concept. As the concept specifies, the interrelatedness among food, animals, and human health with the environment and the intentional collaboration between different sectors' efforts to confront the standing challenge (Taneja and Sharma, 2019), but regrettably, of antimicrobial resistance (AMR) development due to environmental influences have received less care as compared to human and animal health in spite of the significant deviations in the geographical dissemination of AMR associated with environment (O'Neill, 2015). The ultimate result of the increasing incidences of AMR is the development of multidrug-resistant phenotypes, and the multidrug-resistant (MDR) bacteria is a serious problem worldwide (Cooper and Shlaes, 2011; Blair et al., 2014; O'Neill, 2015; Venter, 2019). Moreover, unexpectedly, this problem occurs at the same time when several pharmaceutical companies stopped investing in research to find new drugs to treat MDR due to a lack of high financial returns. In the bacterial cell membrane, the presence of transporters helps in the extrusion of noxious compounds, ultimately decreasing the concentration of external and unwanted or irrelevant substances and enhancing growth. However, the role of some transporters in the biodegradation of various deadly substances present in our environment is also well known (Ganas et al., 2007). As per the previous reports available, there are various resistance mechanisms contributing toward the development of resistant phenotypes—both extrinsic and intrinsic (Ganas et al., 2007). However, the role of intrinsic mechanisms such as the overexpression of multidrug-resistant efflux pump systems in evolving MDR phenotypes has gained partial attention (Nikaido and Pagès, 2012). The role of these efflux pumps is not only limited to functions as an efficient transporter but also limited to combat the stress from external environmental for bacterial survivability. However, it should be noted that some efflux pumps are substrate specific while others have a broad range of substrate specificity, which means that they can pump out multiple substrates including different classes of antibiotics, thus prompting multidrug resistance in bacteria (Nikaido and Pagès, 2012). Therefore, the efflux pump systems which form an important antibiotic resistance mechanism are widely considered due to their unique ability to extrude a variety of noxious compounds including dyes, detergents, different classes of antibiotics, and disinfectants (Nikaido and Pagès, 2012; Venter et al., 2015) outside the cell and thus are intensely associated with MDR expansion. Moreover, these specific features of the efflux pump systems have made them more prominent as multidrug efflux pumps (Webber and Piddock, 2003; Piddock, 2006a).
For the survivability of bacteria, these transporters located in the bacterial cell membrane play a crucial role in facilitating the reduction of external and irrelevant substance concentrations and promoting bacterial growth. However, evidence indicating the role of these transporters in the biodegradation of deadly compounds that are present in the environment has previously been reported (Ganas et al., 2007), suggesting the role of these efflux pump systems not only as a transporter but also in defending bacterial growth and survivability under various environmental stresses. Substrate specificity of these efflux pumps against various substrates, such as different antimicrobial classes, induces the pathogens to become multidrug resistant (Hernando-Amado et al., 2016). However, in some cases it has also been observed that the ejection of the substrates or organic solvents results into overexpression of transporters, which further, triggers the co-selection of the altered features of antimicrobial resistance in bacteria (Blair and Piddock, 2016). As a consequence of overexpression of the efflux pump systems, biofilm formation and quorum sensing (QS) will also be impacted by bacterial pathogenicity (Alcalde-Rico et al., 2016; Kong et al., 2017). In addition to antimicrobials, efflux pumps also have the potential to export virulence determinants, such as adhesins, toxins, and other crucial proteins, which are very critical for colonization within host cells (Piddock, 2006b). Several studies have already been reported on unfolding the development of antibiotic resistance associated with efflux pump systems along with numerous novel efflux pump transporters and simultaneous proteins. Additionally, in recent years, several other functions of an efflux pump system besides antibiotic resistance, such as virulence and bacterial self-protection against various environmental pollutants, were investigated. However, the particular mechanisms of regulation of these efflux pump systems and the active domains of the transporters are still not clearly understood. There are several inducing factors located on the inner and outer membranes of bacteria that stimulate the activity of efflux pump systems and promote their structural alterations within the fluid membrane environment. A brief overview of the role of efflux pumps in Gram-positive and Gram-negative bacteria is presented in this review article. Furthermore, the applications of efflux pump regulators to identify antimicrobial resistance and antibiotic residues, as well as the discovery of new efflux pump inhibitors derived from plants, have been discussed in this study.
Efflux pump systems are predominantly classified into six major superfamilies based on their energy source utilized to export the substrates: ATP-binding cassette (ABC) superfamily, multidrug and toxic compound extrusion (MATE) superfamily, major facilitator superfamily (MFS), resistance nodulation and cell division (RND) superfamily, small multidrug resistance (SMR) superfamily, and proteobacterial antimicrobial compound efflux (PACE) (Hassan et al., 2018) (Figure 1). One of the chief differentiating features of these efflux pumps is the source of energy they utilize; for example, the ABC superfamily draws energy via ATP hydrolysis (Verchère et al., 2012); however, other EP families such as RND, MFS, SMR, and MATE utilize either proton motive force (PMF) derived from H+ or electrochemical gradient of Na+ for exporting their substrates and for extruding several compounds (Kim and Hummer, 2012). Additionally, there are some other features as well for differentiating these efflux pumps based upon their composition or structure of the efflux pump transporters; for example, in an RND efflux pump system, three components, namely, an outer membrane protein (OMP) channel, an inner membrane transporter protein, and two modules that were connected via a membrane fusion protein (MFP) combine to form a composite known as a tripartite complex, which ultimately works together for the extrusion of noxious agents (Daury et al., 2016; Neuberger et al., 2018). However, research on novel multidrug-resistant (MDR) efflux pump systems in bacteria is still underway.
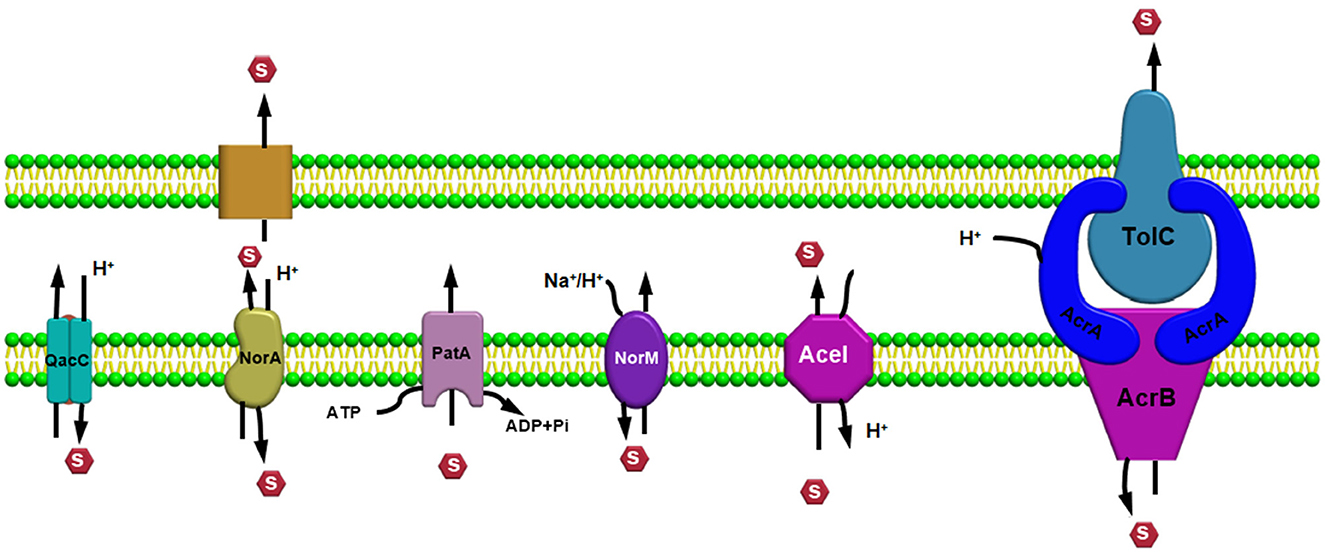
Figure 1. Efflux pump major super-families based on their energy source utilized to export the substrates.
The MacAB-TolC efflux pump system is a tripartite complex belonging to the ABC and is the most comprehensively studied EP system among gram negative bacteria which is actively associated with the extrusion of macrolides and polypeptides virulence factors driven via ATPase MacB and further participates in enterotoxin heat stable enterotoxins (TII) secretion in Escherichia coli (Lu and Zgurskaya, 2013; Fitzpatrick et al., 2017; Jo et al., 2017). Additionally, lipopolysaccharides (LPS) and analogous glycolipids can be considered as organic substrates of the MacAB-TolC pump systems (Lu and Zgurskaya, 2013). As the MacAB-TolC (pump system) is a tripartite complex with three components, including MacB, which is an inner membrane protein that functions as a homodimer complex and consists of two domains, namely, the (i) N-terminal, a nucleotide-binding domain that enables power generation via ATP hydrolysis and the (ii) C-terminal cytoplasmic tail (Lu and Zgurskaya, 2013). Other important components of the EP system are MacA, a periplasmic adaptor protein (PAP) that gets stimulated when ATPase binds specifically with the lipopolysaccharide core, and an outer membrane channel protein TolC that functions as an exit duct for substrate transport (Lu and Zgurskaya, 2013; Fitzpatrick et al., 2017). We observed from previous literature that, in Serratia marcescens, the absence of the MacAB (efflux pump system) resulted in increased susceptibility to polymixins and aminoglycosides, with a significant decrease in swimming motility and biofilm formation ability, and even caused impairment in defense capability to combat superoxide stress (Shirshikova et al., 2021). In addition, in their research study, Shi et al. (2019) observed that, in Agrobacterium tumefaciens 5A {a strain that is arsenite [As (III)] resistant}, MacAB efflux pump confers resistance against a variety of macrolide and penicillin-like antibiotics along with arsenite [As (III)].
However, Gram-positive bacteria harboring the ABC efflux pump systems, containing single component or transmembrane protein, have also been observed: for example, EfrAB transporter in Enterococcus faecalis, Msr protein in Streptococcus, LmrA in Lactococcus lactis (Hellmich et al., 2015), and PatA/B in Streptococcus pneumoniae (Garvey et al., 2011a; Iannelli et al., 2014). EfrAB, an MDR efflux pump system, is a heterodimeric protein conferring resistance toward different antibiotics such as gentamicin, chloramphenicol, and streptomycin (Lerma et al., 2014). In addition, the expression of the EfrAB pump was found to be highly induced when exposed to these antibiotics in sub-inhibitory concentration, and a decline in the expression was observed in the presence of a sub-inhibitory concentration of EDTA (3 mM) (Lerma et al., 2014). The LmrA transporter protein in L. lactis is a homodimer protein having one nucleotide-binding domain and six α-helix domains, which initially identifies and then proceeds toward the extrusion of macrolides and lincosamides (Hellmich et al., 2012). In Streptococcus, the Msr protein, comprising only two nucleotide-binding domains [without any transmembrane domain (TMD)], was found to be responsible for conferring resistance against macrolides and to collaborate with the Mef transport family (Zhang et al., 2016; Tatsuno et al., 2018). Further, in the case of the PatA/B efflux pump system, nearly the entire hydrophilic fluoroquinolone family members, such as ciprofloxacin, function as a substrate for this pump (Baylay and Piddock, 2015).
Members of the multidrug and toxic compound extrusion (MATE) family are broadly disseminated among the three domains, namely, archaea, bacteria, and eukarya (Kuroda and Tsuchiya, 2009). This class of transporters belongs to the multidrug/oligosaccharide-lipid/polysaccharide (MOP) family, which is further categorized into two subfamilies, namely, (i) NorM and DinF found in prokaryotes and (ii) the eMATE subfamily in eukaryotes based on their sequence similarity (Brown et al., 1999; Omote et al., 2006). MATE transporter proteins in bacteria are mainly involved in the extrusion process of amphiphilic cationic drugs, such as norfloxacin, outside the cell. However, in plants, these transporters are involved in physiological roles like developing resistance toward herbicides, leaf senescence, auxin synthesis, tolerance against aluminum, homeostasis, and sequestration of organic compounds (which are plant-based) inside the vacuoles (Remy and Duque, 2014). Further, the MATE transporters were found to be located in both the proximal (convoluted and straight) tubules in the kidney in the case of mammals and in the canalicular domain inside hepatocytes, where the organic compounds are transported across this membrane domain during the final round of drug elimination process (Omote et al., 2006; Terada and Inui, 2008).
All life forms on earth need a balanced extrusion of exogenous noxious substances outside to sustain homeostasis. Therefore, the extrusion of lethal compounds such as xenobiotics requires the efficient involvement of the transporters. First, MATE family transporters were known for their ability to develop antibiotic resistance in Vibrio parahaemolyticus (Morita et al., 1998) but they later became well-known in other domains as well. Moreover, MATE is abundant throughout the human body, but its expression is highly concentrated in the bile canaliculi of the liver and within the brush border membrane of the kidney (Otsuka et al., 2005). Metformin, cimetidine, procainamide, and acyclovir (Masuda et al., 2006; Tanihara et al., 2007) are the cationic drugs that are recognized by the MATE transporter, and therefore, they act as influential factors in controlling plasma concentrations. However, previous literature suggests the significant role of MATE in inducing drug-associated nephrotoxicity during the last phase of pharmacodynamics of cationic drugs. An example is cisplatin, a clinically vital anticancer agent which has not been recognized by the MATE efflux transporter and has ultimately resulted in nephrotoxicity due to renal accumulation (Yokoo et al., 2007). Moreover, cimetidine, which is a histamine (H2) receptor antagonist that is a well-known MATE inhibitor functioning as a gastric acid inhibitor (Ito et al., 2012; Wagner et al., 2016), must be carefully handled and administered. Therefore, a deep understanding of the MATE transporter's working mechanisms is necessary for refining the efficiencies of cationic drugs. The MATE transporters are energetically involved in several physiological functions, which makes them attractive pharmaceutical targets. The MATE transporter protein structure is composed of 12-transmembrane helices arranged in two six-transmembrane (TM) domains, namely, the N- and C-lobes.
However, the positioning of these helices is arranged in an altered manner so as to make it different from the transporters belonging to the major facilitator superfamily (MFS) (He et al., 2010; Van Veen, 2010), supporting the previous assumptions that these two transporters should be considered as distinct superfamilies (Brown et al., 1999). Previous studies on the effective changes in the mechanisms, which are required either for the survivability of a particular microbe or for the development of a multidrug-resistant variant, have been observed in the case of a variety of transporter proteins. As a result of consecutive conformational variations, the transporter protein substitutes within two main conformations, one of which functions via exposing the substrate binding pocket (located centrally) to the inside and the other disclosures the pocket toward the outside of the cell. However, the structure and catalytic reactions mediated desired conformational changes are still not properly understood and one of the main reason behind this is the evidence indicating two crystal structures of the MATE transporter protein having an outward fronting conformation. Additionally, transporter proteins belonging to the MATE family were primarily well-known for mediating drugs or ions antiport via utilizing Na+ or H+ energy sources. The difference in the utilization of different energy sources such as H+ or Na+ gradient in different family members within the MATE superfamily has been suggested according to previous x-ray crystallographic studies concerning the NorM subfamily depending upon Na+ coupling in the C-lobe (He et al., 2010; Lu et al., 2013a), and a quite similar pattern was noticed in case of the eMATE subfamily (Miyauchi et al., 2017; Tanaka et al., 2017) but the DinF subfamily members was found to utilize H+ in the N-lobe (Lu et al., 2013b; Tanaka et al., 2013; Radchenko et al., 2015; Mousa et al., 2017; Kusakizako et al., 2019).
Several secondary carrier families having similar topological features were identified before 1993, but no information was available to prove or to relate these families by a common origin. Among these families, a sugar porter's family including glucose facilitators found in mammals, two families of specific and multidrug efflux pump transporters, a metabolic uptake transporter family, and an oligosaccharide transporting family, including lactose permease present in E. coli, have been well studied. In addition, the knowledge and effort of bioinformatics helped us to understand the mechanisms and provided evidence to relate these families, and further, the name “major facilitator superfamily” (MFS) was coined (Marger and Saier, 1993). The major facilitator superfamily (MFS) transporters can be categorized into three main groups based on the mode of transportation of their substrates: (i) uniporters, which are responsible for the transference of a single substrate, (ii) symporters, which transport a substrate along with the simultaneous transportation of a coupling ion (e.g., H+), and (iii) antiporters, which are responsible for the simultaneous transportation of two substrates in opposite directions in such a way that the transportation of one substrate is dependent on prior discharge of the co-substrate (Forrest et al., 2011) (Figures 2A–C). In addition, it has been observed that the uniporters are able to transport their substrates without utilizing any energy source below their concentration gradient, while in the case of antiporters and symporters, the stored energy of the coupling ion concentration gradient is utilized for substrate transportation against their concentration gradient. However, irrespective of the differences in the mode of transportation of the substrates, all the members belonging to the major facilitator superfamily have the identical core structure comprising 12 TM helices, which are organized on two domains, namely, the N-domain and the C-domain (having similar structure) (Hirai et al., 2002; Yan, 2013), and are further partitioned into two inverted repeats of three helices.
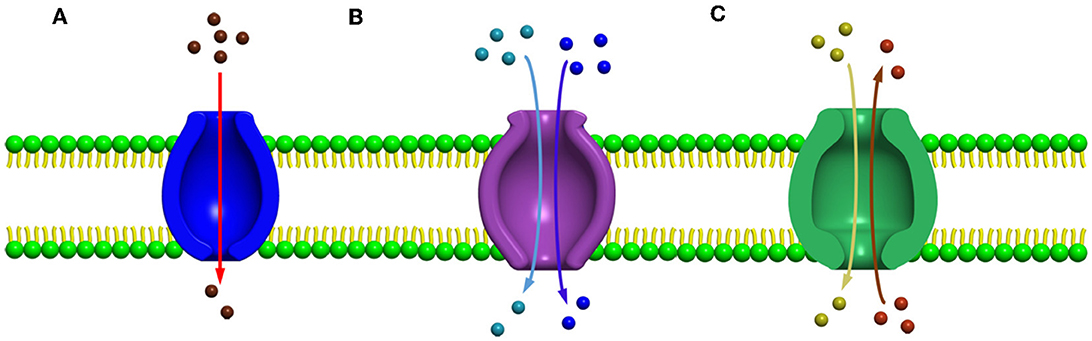
Figure 2. Major facilitator super family (MFS) transporters can be categorized into three main groups based upon their mode of transportation of their substrates (A) uniport, (B) symport, and (C) antiport.
One of the most well-studied MFS transporters in bacteria is the lactose permease (LacY) present in E. coli, which has worked as an appropriate protein model, for understanding the diverse features of the mechanisms involved in transportation associated with MFS transporters (Guan and Kaback, 2006; Smirnova et al., 2011; Kaback, 2015). However, in humans, the most studied MFS efflux pump (EP) system is the glucose transporter which plays a very crucial role in maintaining glucose homeostasis in the human body, which is also referred to as the SLC2 protein. Moreover, the deterioration or faulty regulation of these proteins may lead to important diseases such as diabetes (Type II) (Augustin, 2010; Cura and Carruthers, 2012). Further, the roles of the Msr transporter belonging to the ABC superfamily and the Mef system of the MFS family were found to increase the macrolides extrusion co-actively, which further enhanced the resistance toward 14–15-member ring macrolides (Nunez-Samudio and Chesneau, 2013). Besides their active participation in the extrusion process, these MFS transporters are also linked with other biological pathways, such as the role of MdrT and MdrM in promoting the immune response in hosts by activating the production of interferon-β (IFN-B, which is a type I interferon response) and in maintaining the stability of the cell wall (Pasqua et al., 2021). In the case of Staphylococcus aureus, the presence of a constitutively expressing MFS efflux pump system Tet38 has been reported to influence the progression of host cell invasion comprising internalization, adhesion, and relocation within epithelial cells and the consequent step involved in epithelial cell infection due to Staphylococcus aureus, which includes the viability of bacteria and trafficking within phagolysosomes (Pasqua et al., 2021).
However, in Acinetobacter baumannii, loss of the MFS transporter protein AbaQ resulted in a significant decrease in bacterial virulence and motility capability (Pasqua et al., 2021), and furthermore, a substantial reduction in bacterial motility and virulence was detected when the efflux pumps encoding genes (such as RND, MATE, SMR, and ABC) are inactivated (Pérez-Varela et al., 2019). The working mechanisms of MFS transporters still require more explanation. However, an updated form of the classical rocker–switch model for understanding the changes emerging in the conformation of the MFS transporter associated with a form of substrate transportation known as the “clamp-and-switch” model has been previously proposed (Quistgaard et al., 2016), which also discussed the function of gating residues, binding of substrates, and the conformational change observed in the updated model (Quistgaard et al., 2016). In the past decades, many molecular biology, biochemical, and biophysical experiments have been conducted to understand the function of MFS transporters due to their physiological and pharmacological significance.
Resistance nodulation division (RND) superfamily members are the most well-known efflux pump systems because of their astonishing capability of extruding a wide range of substrates, which includes various classes of antibiotics, such as tetracyclines, chloramphenicol, different β-lactam antibiotics, and besides antibiotics, several other compounds such as dyes, detergents, metals, bacterial metabolites are also utilized as a substrate by this pump (Munita and Arias, 2016). The only feature which is common among these compounds is the amphiphilic nature of the substrates utilized by this pump (Yamaguchi et al., 2015). RND family members are part of multiprotein complexes which are arranged within the bacteria in a way that connects the outer membrane, periplasmic space, and the inner membrane to form a tripartite complex; thus, the RND transporters can facilitate the transport of compounds into these compartments by allowing them to work in conjunction with other transmembrane transporters. RND transporters are capable of capturing substrates not only from the cytoplasm but also from the inner membrane's outer leaflets or from the periplasm (Du et al., 2015). Consequently, it has been observed that these protein complexes are larger than several other transmembrane proteins of bacteria, which usually range over only a single membrane. Resistance nodulation division (RND) family members is comprised of three components, including (i) a periplasmic adaptor protein (PAP) (ii) an outer membrane protein (OMP) channel, and (iii) an MFP, which connects IMP and OMP, and are referred as a tripartite system for having three components together to form a complex (Figure 3). The main function of this tripartite complex is to identify and bind the substrates found in the periplasmic space, outer leaflet of the inner membrane, or the cytoplasm and lastly extrude or expel outside the cell through the outer membrane channel. However, other transporter protein functions as a single unit within the inner membrane for transporting substrates via the membrane bilayer (Nikaido, 2011; Du et al., 2018). Besides the participation of efflux pumps in the development of antibiotic resistance, other mechanisms such as alteration in drug target or inactivation or modification of antibiotics also have a vital role in AMR (Walsh, 2003; Blair et al., 2015; Colque et al., 2020). However, it has been noticed that comparative to the single factor that contributes to the resistance development against a particular class or group of antibiotics, multidrug resistance (MDR) can progress due to a reduction in bacterial membrane permeability (Nikaido, 2009) and via overexpression of the MDR efflux pump system in both Gram-positive and Gram-negative bacteria (Nikaido, 2009; Allen et al., 2010; Li et al., 2015). Besides being intrinsically expressed or present, bacteria also acquire resistance by RND efflux pumps via plasmids, etc. (Levy and Marshall, 2004; Allen et al., 2010), and mutation is intensely associated with the significant rise in the expression level of both intrinsic and extrinsic efflux pump systems in the clinical strains (Blair et al., 2015). This resulted in an increase in the expression level, which is one of the chief reasons for MDR development (Blair et al., 2014). In the current scenario, resistance nodulation division (RND) superfamily members are considered among the core contributors to MDR progress in clinical isolates (Nikaido, 1996; Blair et al., 2014).
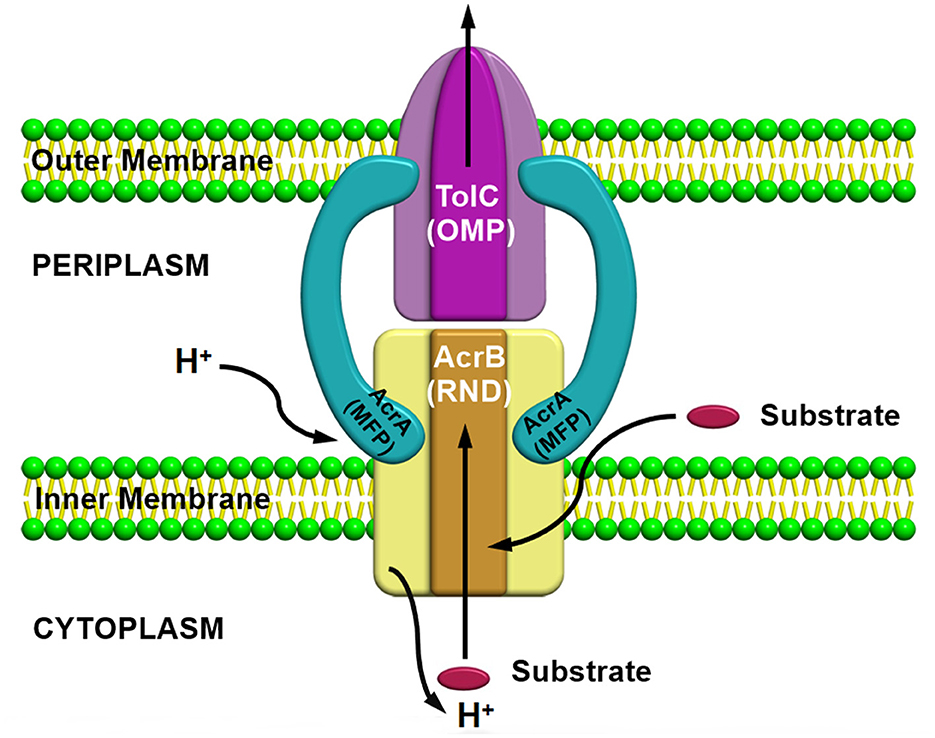
Figure 3. AcrAB-TolC a tripartite efflux pump system containing three components together to form a complex.
Besides, the ability of these RND transporters to work as a tripartite pump in the extrusion of a wide variety of substrates such as different classes of antibiotics, detergents (Zwama and Yamaguchi, 2018), dyes, and various other noxious compounds outside the cell (Du et al., 2014), the overexpression of the efflux pump genes as a result of an increase in the transcriptional level of the operonic genes due to mutations in their regulatory proteins and genes (Nishino et al., 2008, 2009) has already been well studied and established in pathogenic bacteria within clinical settings (Oethinger et al., 1998; Nishino et al., 2008, 2009; Blair et al., 2015; Yu et al., 2020; Salehi et al., 2021). Examples of some of the well-studied RND efflux pump systems observed in Gram-negative bacteria are AcrAB-TolC in E. coli and Salmonella enterica, MexAB-OprM in Psedomonas aeruginosa, and the AdeABC system in A. baumannii (Bhakdi et al., 1988; Nikaido, 1996; Xu et al., 2020; Lv et al., 2021) are intensely associated with the extrusion process of a wide range of lethal compounds. Other examples such as the CmeABC system in Campylobacter jejuni, SmeABC in S. enterica, and the MtrCDE efflux pump system in Neisseria gonorrhoeae, have also been reported previously (Gilson et al., 1990; Singh et al., 2006; Wehmeier et al., 2009; Guérin et al., 2014; Wright and Tate, 2015; Lv et al., 2021). An important role of the RND transporter proteins, such as MtrD, MexB, AcrB, and AdeB, within the system, is in the substrate binding step (where the substrate may be specific) and in the drug transportation process, which is crucial for resistance development clinically (Delmar et al., 2014). An example is the mutation in AcrB gene which resulted in ciprofloxacin treatment failure (B). According to the previous literature, the transcriptional level of the regulatory proteins belonging to the TetR family such as AcrR (Blanco et al., 2016; Chetri et al., 2019a), MtrR (Lin et al., 2017), CmeR (Zwama and Nishino, 2021), TtgR (Bay and Turner, 2009), MexR (Srinivasan and Rajamohan, 2013), NalC/NalD (Bay et al., 2008), SmeT (Lytvynenko et al., 2016), and RarA belonging to the AraC family (Chetri et al., 2018b) influences the expression of these efflux pumps. The amino acid (AA) residues within the efflux pump systems are very crucial and actively participate in the substrate binding course and the substitution, or any changes in the AA residues probably can modify the substrate affinity (Banigan et al., 2015). Further, alteration in prominent AA residues was demonstrated to associate with efflux pump-facilitated resistance toward various drugs (Padariya et al., 2018). AcrAB-TolC, MexAB-OprM, and CmeABC are among the most well-characterized multidrug resistant efflux pump systems belonging to the RND superfamily and are responsible for survivability and pathogenicity in bacteria. There are reports indicating the co-existence of different defense mechanisms including porin deficiencies and β-lactamase production with the overexpression of efflux pumps in E. coli (Chetri et al., 2019a,c), but rare evidence of insertions into penicillin-binding protein 3 (PBP3) of E. coli affecting their activity against monobactams and cephalosporins has already been reported (Alm et al., 2015). In carbapenem-resistant E. coli isolates, the overexpression of the AcrAB-TolC efflux pump simultaneously with the downregulation of OmpF due to regulation via the MarA gene was observed (Chetri et al., 2020). Further, for a comparative analysis study in carbapenem-resistant E. coli isolates, the β-lactamase gene (blaNDM-1, blaCTX-M-14, and blaCTX-M-3) were targeted and amplified (Shimizu et al., 2017; Chetri et al., 2019b). However, a study showing the capability of the AcrAB-TolC efflux pump system in influencing bacterial attachment, then penetration within the host, and further proliferation and persistence within animals has been reported (Piddock, 2006b) previously. Besides, the role of the AcrAB-TolC efflux pump in E. coli, AcrEF-TolC has also been reported to have a significant role in developing resistance toward different antibiotics (Chetri et al., 2018a).
Extensive usage of heavy metals (HME) in antimicrobials is not an infrequent event, for example, in disinfectants, and as a result, the RND efflux pump superfamily has developed the ability to resist these compounds for their survivability. Further, these RND superfamily is divided into two main categories: (i) hydrophobic and amphiphilic (HAE)-RND family, for example, AcrB, AcrD, AcrF, MdtC, MdtD, and YhiV, (ii) HME-RND family, for example, CusA which utilizes Cu(I) and Ag(I) as the most important substrate (Delmar et al., 2014), where CusA functions as a membrane fusion protein, CusC as an outer membrane channel protein to form a tripartite complex, the CusCBA efflux pump system (Long et al., 2012). However, the mechanical pathways required for the exportation of metal ions have previously been predicted (Long et al., 2012). Further, for survival, bacteria adapts in different ways such as efflux pump-associated biodegradation to get rid of the harmful effects of the lethal chemical compounds resulting from organic pollutants, for example, in Pseudomonas Putida, the occurrence of the TtgABC efflux pump system is responsible for conferring tolerance toward toluene (Blanco et al., 2016). Consequently, due to pressure exerted via overuse of antimicrobials, mutations in functional genes occurs, which may influence various changes in the AA residues located in the binding sites, which affect the substrate specificity of an efflux pump, and as a result, bacteria become less receptive toward antimicrobials, i.e., antimicrobial-resistant phenotype significantly altering the scenario in the environmental, clinical, and laboratory settings (Zwama and Nishino, 2021). The genetic changes occurring in an efflux pump denote an evolutionary adaptation within a microorganism against antimicrobials, and it is undoubtedly complexing the entire therapeutic option management system of pathogenic bacteria (Zwama and Nishino, 2021).
Small multidrug resistance (SMR) family members utilize proton gradient to export a wide range of antibiotics, hence contributing toward MDR development. On the basis of the sequence, the SMR family can be divided into two main physiological subtypes (Kermani et al., 2020): (i) guanidinium exporter (representative of “Gdx”) which is responsible for the allocation of bacterial metabolites such as guanidinium ion in exchange of two protons (Nelson et al., 2017; Kermani et al., 2018) and (ii) quaternary ammonium compound (Qac) representative subtype, which is responsible for the export of certain hydrophobic, cationic, and quaternary ammonium compounds (Nelson et al., 2017; Kermani et al., 2018). After the discovery of the first antiseptics containing quaternary ammonium around 100 years ago, Qac cluster proteins were found to be nearly associated with the propagation of multidrug resistance elements (Russell, 2002; Pal et al., 2015; Gillings, 2017; Zhu et al., 2017). Several bacteria harboring both subtypes of SMR transporter proteins were detected. Additionally, it was observed that the two subtypes do not interfere with each other's physiological role, for example, Qac protein will not participate in the transportation of guanidinium ion (Gdm+) and likewise, the guanidinium exporter will not involve in the export of any cationic or quaternary ammonium compounds but will require a substrate that is having guanidine moiety (Kermani et al., 2020). In E. coli, an example of the most well-studied Qac transporter protein is EmrE, and also an EmrE mutant, i.e., S64V that is having diminished conformational exchange dynamics based on extensive NMR measurements has been recently reported (Shcherbakov et al., 2021). SMR family members comprise short polypeptide chains of 100–150 amino acids and can stretch the cytoplasmic membrane as 4 TM α-helices (Bay et al., 2008). Proteins with short hydrophilic loops allow solubilization of a broad range of drugs, noxious lipophilic compounds that comprise DNA intercalating agents, lethal metabolites such as nicotine, and polyamine compounds such as spermidine (Bay et al., 2008; Bay and Turner, 2009). There are several reports indicating the active role of these family members in developing resistance toward different substrates—like in A. baumannii—AbeS transporter is found to be responsible for the transportation of ethidium, benzalkonium, and acriflavine, which simultaneously increases resistance toward amikacin (Lytvynenko et al., 2016; Lin et al., 2017). Similarly, in Klebsiella pneumoniae activation of the efflux pump system, KpnEF confers resistance against chlorhexidine, other antiseptics, benzalkonium chloride, etc. (Srinivasan and Rajamohan, 2013). In both P. aeruginosa and E. coli, the role of EmrE transporter protein in recognizing the substrates and mediating the elimination of polyaromatic compounds has already been reported (Banigan et al., 2015; Chetri et al., 2018b), but the Qac proteins are particularly associated in resistance development against different antibiotics and antiseptics (Jaglic and Cervinkova, 2012). Frequent incidences of QacA or QacB genes in E. faecalis and S. aureus have been observed, whereas in the case of Enterobacteriaceae and Pseudomonas spp., the dissemination of qacE gene was more prevalent (Jaglic and Cervinkova, 2012). SMR family members are plasmid-encoded and can often occur on integrons (the mobile genetic elements [MGEs]), which influences the menace of horizontal spread of resistance (Bay and Turner, 2009). Incidences of QAC exposure influencing the overexpression of the efflux pump supports the horizontal transfer of integrons or MGEs harboring fluoroquinolone resistance determinants within Class I intergron (QacED1) (Anandapadamanaban et al., 2016; Buffet-Bataillon et al., 2016). The transfer of both the antibiotic-resistant and disinfectant-resistant genes between diverse species regulates the bactericidal effect of these lethal compounds.
The abundance of these efflux pump systems in proteobacteria has provided a purpose for the naming of the family as the proteobacterial antimicrobial compound efflux (PACE) family (Hassan et al., 2015a). One of the well-studied transporter proteins of the PACE family, Acinetobacter chlorhexidine efflux (AceI) found to be responsible for developing resistance toward different compounds and additionally, numerous homologs of AceI conferring resistance against various biocides and mediating transportation of fluorescent dyes, such as proflavine and acriflavine, has been observed which differentiates this family from other families of efflux pump (Hassan et al., 2015a). Presently, PACE family members responsible for the extrusion of around 10 substrates have been reported and the transporter proteins belonging to this family are included in the Database of Transporter Classification (Saier et al., 2016), which comes under proteobacterial chlorhexidine efflux (CHX) family (Elbourne et al., 2017). Transporter protein of the PACE superfamily has been reported for the export of biocides such as acriflavine and chlorhexidine; however, transporter AbgT having a role in the extrusion of sulphonamides was observed. Regulators play an important role in modulation the efflux pump systems for drug extrusion, and the factors influencing drug resistance such as under tetracycline exposure, TetR regulates TetB expressional level and in case of cationic antimicrobials pressure, QacR controls qacA expression (Grkovic et al., 2002). In bacterial membranes, the regulators control the systems in such a way that, when there is a requirement of efflux, and only the expression of the efflux pump will proceed; thus, avoiding lethal effect of overexpressed efflux pumps that are constitutively expressing within the bacteria and saving the cellular resources (Andersson and Levin, 1999). From a research viewpoint, the identification of the efflux pump (EP) system recognizing the substrates or novel factors associated with resistance or tolerance against drugs can be analyzed through observation of the changes occurring in their transcriptional level, and it is possible because of the constricted regulatory control of EP genes. The analysis of the transcriptional response of A. baumannii toward chlorhexidine (a membrane-active biocide) revealed the role of Acinetobacter chlorhexidine efflux protein (AceI) system in A. baumannii (Hassan et al., 2013, 2015b). Antibiotic-resistant strains of A. baumannii are becoming increasingly prevalent in hospitals because they have emerged as a major cause of Gram-negative infections.
Efflux pump genes are present in all species of bacteria, and the efflux pump encoding genes are located either on a chromosome or a plasmid, especially in clinical cases (Piddock, 2006b; Poole, 2007). The understanding of the regulation of these efflux pumps, finding their function, their participation in developing resistance toward specific or multiple substrates, and their response under different environmental conditions, all of which were very crucial for the implementation of an accurate action plan. For a variety of membrane transporters, alternating access mechanisms have shown evolutionary persistence over the past few decades (Jardetzky, 1966). During sequential conformational modifications, a substrate-binding pocket in the middle of the transporter is exposed to the inner cellular space, while the other exposes the pocket to the outer cellular space through multiple conformational changes (Raturi et al., 2021). However, the structure and catalysis of MATE transporters underlying conformational changes are still not clearly understood. Partly, this is due to the fact that, particularly, two previously reported crystal structures of MATE transporters were with an outward-facing orientation (Raturi et al., 2021). Therefore, the MATE transporters facilitating drug/ion coupled transportation via Na+ or H+ coupling (Lu et al., 2013a) have already been described. The functionally diverse ABC family members utilize ATP to export their substrates and some act as ion channel modulators (Li et al., 2017). The structure of the ABC family transporter consists of two domains—one with substrate binding pocket (TMDs) and another with nucleotide-binding domain (NBDs)—which binds and initiates hydrolysis of ATP to stimulate the transport cycle (Dawson and Locher, 2006; Jin et al., 2012; Choudhury et al., 2014; Kodan et al., 2014; Johnson and Chen, 2017; Verhalen et al., 2017). An “alternating access” mechanism involved in the transportation of the substrate has been observed based on the functional and structural data available, in which the transportation triggers the switch in the conformation between inward-open, occluded, and outward-open states required for substrate transportation, and additionally, these conformational alterations are associated with the dimerization and dissociation of the nucleotide-binding domain-mediated via binding and hydrolysis of ATP (Khare et al., 2009; Korkhov et al., 2012; Woo et al., 2012). Consequently, in E. coli, the presence of ABC transporter protein MsbA utilizing the ATPase activity to transfer lipopolysaccharide precursor lipid A from the inner-cytoplasmic membrane to the outer-periplasmic membrane has been observed (Woebking et al., 2005), and as a result of its catalytic action, this enzyme is referred to as a flippase. Further, the cryogenic electron microscopy (cryo-EM) studies of the structural aspects of MsbA protein helped to understand the functional states and the visualization of the transportation route (Mi et al., 2017). However, some ABC transporters have shown that the MacB exhibits only outward-directed transport mechanism for the substrate entering through the periplasmic side, and as a result, the state of the substrate binding pocket remains in the outward direction. The transporters have the ability to interfere with their substrates from the outside facing binding pocket followed by a conformational alteration coupled with ATP hydrolysis, which enables the reduction in the affinity toward the ligand inducing its shifting into the exterior part (Perez et al., 2015; Locher, 2016). Similar to the ABC transporter family, the mechanism of alternating access was also reported in MFS transporters as well where in the course of a transport cycle the conformational switch of the two domains within inward to outward open states has been reported (Jardetzky, 1966). According to the data obtained from lactose permease symporter (LacY), the conformational switch of the alternating access mechanism is activated via induced fit from the ligand binding and further, and the transport rate is governed by the electrochemical proton gradients (Kaback, 2015). The fundamental component of the model is the formation of a ternary complex when the proton and substrate bind in an ordered manner. However, various MFS transporters bind in different sequences as well, such as initiated with binding with proton, then substrate, or with a substrate first and then proton. Additionally, it has been observed that the transportation cycle of MFS protein generally involves the binding (as an initial step) followed by the release of substrate and proton, however, variations in the stoichiometric drug–proton interconversion within family members have been reported (Schaedler and Van Veen, 2010; Fluman et al., 2014). Small multidrug resistance (SMR) efflux pump system utilizes proton motive force as an energy source was previously reported via an experiment with S. aureus Smr (Sau-Smr) conducted first by Grinius and Goldberg in 1994 (Littlejohn et al., 1992; Grinius and Goldberg, 1994). Moreover, this experiment revealed that an electrochemical proton gradient was utilized as the energy source for tetraphenylphosphonium (TPP) efflux due to the reconstitution of Sau-Smr efflux within proteoliposomes. Further, under similar experimental conditions, similar results were obtained for Eco-EmrE, the SMR homolog (Yerushalmi et al., 1995, 1996). Therefore, SMR proteins are considered a proton-dependent MDR efflux pump system (Paulsen et al., 1996). Despite QACs, SMR proteins are able to transport neutral and negatively charged compounds (Jack et al., 2000; Nishino and Yamaguchi, 2001). However, previous studies revealed that the nature of the compound has a significant effect on the energy required for substrate transportation observed in Eco-EmrE studies, for example, the transportation of TPP requires movement of the charge for an active efflux besides the transportation of methyl viologen (MV) (a lipophilic divalent cation) (Yerushalmi et al., 1995; Rotem and Schuldiner, 2004). Further, as a result of several structural studies of the AcrB protein of E. coli by utilizing X-ray crystallography (Murakami et al., 2002; Nakashima et al., 2011; Eicher et al., 2012) and cryo-EM techniques (Du et al., 2014), the transporter protein was well-studied and became the most expansively characterized member of RND family (Zgurskaya, 2009; Zwama et al., 2019). Additionally, the interaction of AcrB with a wide range of substrates, which are chemically distinct, was analyzed to understand the poly-specificity feature of the transporter protein (Murakami et al., 2006; Nakashima et al., 2011; Eicher et al., 2012, 2014; Hung et al., 2013; Mousa et al., 2016; Sjuts et al., 2016; Zwama et al., 2018). Further, the movement of the substrates via the transporter is driven by the conformational changes taking place within the system, which is also revealed by the previous studies (Du et al., 2014; Blair et al., 2015). Moreover, the understanding of the function of these EP systems and the inhibition mediated via small molecules was simplified with the help of molecular simulation studies (Vargiu and Nikaido, 2012; Vargiu et al., 2018). The entire protein domains present were defined by the initial crystal structure developed for AcrB, which revealed that the region of transmembrane consists of 12 α-helices that anchor the inner membrane protein and supports proton movement and is accompanied by the transportation of the substrates and energy supply to drive translocation (Murakami et al., 2006; Seeger et al., 2006). Further, the entry of the substrates into the multiprotein complex is facilitated by the groove located between 8 and 9 transmembrane helices, which provide one out of two distinct access sites for the substrates, and the remaining protein residue projects into periplasm which is required for binding of the substrates and to link up with AcrA (periplasmic protein) as well as with the outer membrane channel protein TolC that is rooted in the outer membrane. The carrier domain of AcrB having the ability to span periplasm consist of four sub-domains, namely, PN1, PN2, PC1, and PC2, each of which comprises a common motif pattern, namely, two β-strand–α-helix–β-strand motif. The carrier sub-domains of all three protomers are arranged in such a way that a central pore along with a long channel extending through the membrane has been formed for the translocation of the substrate (Zwama and Yamaguchi, 2018).
Further, it has been reported that a second access site for substrate was found to be located in a cleft that formed in between PC1 and PC2 domains. The binding of AcrA and TolC is facilitated by the docking domains (DN and DC), which are located on the top of the porter domain. Additionally, tight binding of the top of AcrB with the TolC base is very essential for preventing the substrate leakage into periplasm aided by β-strands, which are placed antiparallel to generate a funnel-like construction. The crystal structure initially reported by Murakami et al. (2002) revealed the crystallized symmetrical homotrimeric structure without any bound ligand and explained the entire structural design of AcrB. According to three consequent studies published in 2006, it has been first reported that the establishment of an AcrB complex with various ligands that demonstrated the adoption of diverse conformations by each of the three protomers resulted in an asymmetrical homotrimeric structure assembly (Murakami et al., 2006; Seeger et al., 2006; Su et al., 2006), which has been indicated to be an active state. Therefore, under these circumstances, three different conformations have been well-defined as follows: (i) loose (L) that is mainly associated with the access for the substrates, (ii) tight (T) that is linked with the binding course, and (iii) open (O) state coupled with the extrusion conformation (Seeger et al., 2009; Nikaido, 2011; Eicher et al., 2012). As the substrates move over the protein, the monomer rotates between L, T, and O and returns back to L, and this “L conformer” which is accessible for substrate binding is also recognized as the resting state (Murakami et al., 2006; Seeger et al., 2006; Nakashima et al., 2011). First, the substrate enters into AcrB during the unliganded L state either from the periplasm or inner membrane envelope via one of the two defined entry sites, which are then released into a proximal binding pocket (PBP). Additionally, as a result of the movement of substrates via AcrB in the direction of the distal binding pocket (DBP), which is situated deep in the carrier domain, a conformational alteration toward the T state assisting the transfer of the substrate from the central pore of AcrB toward TolC entrance has been observed. Subsequently, the ejection of the substrate from AcrB into TolC occurs via the rotating functional mechanism, which involves the protein transitional step, i.e., from state T to state O (Murakami et al., 2006; Seeger et al., 2006). Under that situation, state O which indicates an open state, a channel which is generated for substrate extrusion from AcrB to TolC and during the transportation of substrate, and several compressions and hindrances occurred inside the porter domain which facilitated the transportation of substrate in a unidirectional way toward central funnel referred as “peristaltic pump mechanism” (Seeger et al., 2006, 2008; Pos, 2009). In AcrB, the substrate entry via the two primary sites, primarily, via the proximal binding pocket (PBP) and then toward the distal binding pocket (DBP), plays an important role in defining the poly-specificity feature of the transporter.
However, in the case of hydrophobic substrates which are separated in the inner membrane's outer leaflet can enter through the TMD, containing hydrophobic grooves and are well defined as TM8 and TM9 (Murakami et al., 2006; Pos, 2009). Thereafter, tunnel 1 is formed which then elongates from the periplasmic membrane plane at the end of the TM8/TM9 groove till it links the proximal binding pocket (PBP) 2. However, comparatively, the entry of compounds confined to periplasm occurs via another site placed within the porter domains PC1 and PC2, ~15 Å on top of the membrane plane, leads toward tunnel 2, which further elongates in the direction of the protein center (Eicher et al., 2012). Finally, after linking tunnels 1 and 2, it leads toward a distal binding pocket located deep within AcrB protein, where the presence of a glycine-rich switch loop that is proposed to contribute to the unidirectional movement of the substrates further facilitates the separation of distal and proximal binding pockets (Eicher et al., 2012; Zwama et al., 2018; Tam et al., 2020). As per the previous AcrB structural studies, the high molecular mass drugs, such as erythromycin and rifampicin, co-crystallize within the proximal binding pocket; however, the distal binding pockets are found to be responsible for holding both the low and high molecular mass drugs such as minocycline, doxorubicin, and neopentyl glycol derivative C7NG (Nakashima et al., 2011; Ababou and Koronakis, 2016; Sakurai et al., 2019). AcrB in E. coli and MexB homolog in P. aeruginosa and have similar structures and substrate-binding sites; therefore, both have similarities in a broad substrate specificity as well. Distal binding pocket (DBP) is a huge chamber surrounded by hydrophobic phenylalanine (Phe) residues and polar residues (Vargiu et al., 2018); further, the side chains present on these hydrophobic residues interact with the substrates through Pi–Pi stacking and van der Waals interaction, and on the contrary, polar residues interact via H-bonding. There are several pieces of evidence supporting the manifestation of broad substrate specificity of the pump as demonstrated by the different binding positions adopted by minocycline and doxorubicin. Moreover, according to previous structural studies, the role of a switch loop containing Gln 616–Gly 619 in regulating substrate's entrance into the distal binding pocket (Vargiu et al., 2018) and also in functioning as a barrier to separate the distal binding pocket from proximal pocket has been observed. Additionally, it has been suggested that physical contact between the substrate with the switch loop is essential for the translocation of the substrate before leaving the proximal binding pocket, and the movement instigated via the flexible loop facilitated the transportation of the substrates deep inside the distal binding site; however, without passing through the loop, the extrusion of the substrates via TolC is not possible (Nakashima et al., 2011; Eicher et al., 2012; Vargiu and Nikaido, 2012; Jamshidi et al., 2018). Immediately, after the binding of the substrate into the distal binding pocket occurs, it leads to a conformational change in the protein from state T to O and subsequently, the disintegration of tunnels 1 and 2 because of a transition of the coil to helix at N-terminal end of TM8 and extensive reorientation of carrier domains has been noticed (Zwama et al., 2017). Furthermore, for substrate transportation, a third tunnel that directs the substrates from AcrB on the way to TolC for extrusion is created in the state O (Eicher et al., 2012). Besides AcrB, the role of AcrA is also very crucial as repacking of AcrA helps to inhibit the leaking of the substrate into the periplasm via closing the cavities that exist inside the pump, and further, the changes arising in AcrA which result into the opening of the closed base of TolC are obviously initiated via the ligand binding phase due to the conformational changes occurring in AcrB. In addition, if the assemblage is exposed to the periplasm, it will restrict the opening of the TolC channel and the absence of any substrate inside a pump or extrusion of a substrate facilitates the conformational changes in AcrB from the drug transportation state to the resting state L (Wang Y. et al., 2017; Wang Z. et al., 2017). According to the aforementioned discussion regarding the homotrimeric symmetrical structure of AcrB which crystallizes immediately and forms substrate or inhibitor bound protein co-complexes because of their flexibility and multiple binding site mechanisms of AcrB, the movement for transportation of the substrates driven via multiple structural changes has been observed (Sakurai et al., 2019). A well-organized protein crystal required for the investigation of x-ray diffraction patterns can be difficult to grow when either of these processes is involved. However, currently, a number of compound-bounded AcrB structural studies have been carried out, which facilitated our understanding of the molecular foundation of inhibition (Murakami et al., 2006; Sennhauser et al., 2009; Nakashima et al., 2011, 2013; Eicher et al., 2012; Sjuts et al., 2016; Wang Y. et al., 2017; Wang Z. et al., 2017). Distal binding pockets (DBP) contain two characteristics crucial to inhibitor binding: (i) channels that translocate hydrophilic substrates and bind multiple drugs and (ii) a hydrophobic catcher that is located proximate to the distal binding pocket that divides this channel, which gives more attention to AcrB targeted small molecule inhibitors designing.
Besides the contribution of efflux pumps in the development of antibiotic resistance, some other mechanisms such as biofilm formation, which is clearly a community of microbes affixed to a surface, also contribute to the progress of bacterial resistance and tolerance; additionally, it has been suggested that the efflux pump system has the capability to influence the function of biofilm either directly or indirectly (Hall and Mah, 2017; Alav et al., 2018). In A. baumannii, biofilm formation is inhibited due to tigecycline exposure in sub-inhibitory concentration via the downregulation of the adeG gene encoding for efflux pumps (Chen et al., 2017). Quorum sensing (QS) is a network that is generated within cells to encourage mutual communication at the cellular level, and the utilization of signal transduction for participation in activities occurring inside bacteria has already been observed. The association between biofilm and QS improves bacterial viability in response to different variations in environmental signals. As an example, the ejection of acylated homoserine lactone (AHL) via the efflux pump system MexAB-OprM in P. aeruginosa is maintained by the QS contribution and overexpression of the efflux pump, which ultimately resulted in the release of QS signals (Piddock, 2006b). According to the available literature, it has been assumed that the bacterial QS mechanism is restricted if inhibitors hinder the efflux pump activity (Gupta et al., 2017). Similarly, in S. aureus, the inhibition of the NorA efflux pump activity due to the presence of an appropriate concentration of efflux pump inhibitor has also been described in a previous study (Sabatini et al., 2017). Additionally, the role of efflux pump systems in facilitating the pathogens in adapting to the harsh environment present within a mature biofilm, which involves response against envelope stress in maintaining the membrane rigidity and in conferring resistance toward high osmotic stress (Robin et al., 2022), such as the essential role played by the MacAB-TolC efflux pump in biofilm formation (Robin et al., 2022), has been reported. Pmt and AbaF are the two well-known MFS transporter proteins found in A. baumannii, which play an important role in the formation of biofilms such as Pmt in extracellular DNA extrusion and active participation of AbaF in emancipating biofilm materials (Pasqua et al., 2021). There is evidence indicating variations in bacterial membrane influencing the functioning of biofilm formation mediated via different MDR efflux pump systems belonging to the RND family (Yoon et al., 2016), as shown in A. baumannii, where the inhibition of adeAB gene expression or deletion resulting in reduced expression or hindrance of biofilm and QS systems have been described earlier (Richmond et al., 2016; Dou et al., 2017). A previous study proposed the incidence of a positive association between mRNA levels of efflux pump genes and biofilm formation, which can be influenced by the MIC levels of antibiotics such as polymyxin B or colistin (Sato et al., 2018). For example, MexGHI, an efflux pump system recognized in P. aerugiosa responsible for phenazine transportation, plays a crucial role in biofilm morphogenesis (Sakhtah et al., 2016). Similarly, the QS system also has a similar correlation with the efflux pump system (Liang et al., 2016). Additionally, a study evidenced the mutation in mexI, an efflux pump gene of P. aeruginosa, mediating the loss and reduced expression of quorum sensing molecules or virulence factors (Wolloscheck et al., 2018). The role of ATP-binding cassette (ABC) type efflux pump systems in conferring resistance toward antifungal agents that are used against fungi, mostly Candida species, is well known and their involvement in QS molecules secretion and influential capability toward biofilm formation is found to be similar to that of RND family members (Cannon and Holmes, 2015).
Efflux pumps largely contribute toward multidrug resistance, both intrinsic and extrinsic in bacteria; therefore, the active participation of efflux pump inhibitors (EPIs) have a potential impact in preventing antimicrobial resistance globally. A significant challenge in the discovery of EPI has been its highly hydrophobic nature and the substrate-binding poly-specific site for the target. However, the primary studies on the x-ray crystal structures of different compounds such as pyranopyridine (Sjuts et al., 2016) and pyridopyrimidine (Nakashima et al., 2013) (Table 1) revealed that the EPIs bound to different efflux pumps of RND family of P. aeruginosa and E. coli have helped to facilitate the rapid advancement in our understanding of different mechanisms involved and the binding interactions of these EPIs. Moreover, a strong binding interface between the Phe-abundant hydrophobic trap and the inhibitor D13-9001 participates in inhibiting the conformational change from state L to T and further hinders the functional rotation mechanism. Further, it has been suggested that the entry of the substrates into the binding cleft situated between the domains PC1 and PC2 is blocked by the hydrophilic moiety existing on the inhibitor D13-9001. Phe 178 is an imperative amino acid that exists in the hydrophobic trap, which plays a vital role in the binding of inhibitors with the aromatic substrates via utilizing π-π stacking (Nakashima et al., 2013; Sjuts et al., 2016), and it has been reported that, by applying a structure-guided design for an effective inhibitors development, the hydrophobic trap discovery has a major role (Aron and Opperman, 2018). As these efflux pumps are considered excellent targets for antimicrobial combination treatment, facilitating the usage of synthetic (Table 1) or plant-based (Table 2) efflux pump inhibitors (EPIs) to support antimicrobial therapeutic options toward bacterial infections (Opperman and Nguyen, 2015; Alibert et al., 2017). There are several effective approaches to restrict or avoid the activity of an active efflux pump system found in bacteria such as decreasing the antibiotic binding affinity toward transporters via chemical structural modification of the drug, escalating the OM permeability to increase the drug concentration within cell, knocking out or impeding the efflux pump associated genes, and disrupting the supply of ATP or via designing compounds which have the capability to compete with the antimicrobial agents for active sites for their action to obstruct the efflux pump activity (Jamshidi et al., 2016). Additionally, approaches such as computational analysis or artificial abstraction from plants were utilized for the detection of numerous inhibitors active against both Gram-positive and Gram-negative bacteria. Researchers working on screening different natural EP inhibitor compounds via utilizing high-throughput methods have observed a natural compound referred to as Terminalia chebula, which is isolated from an Indian medicinal plant, having the ability to abolish the binding of ligands with Na+, which resulted in a change in the conformation of the transporter protein NorM found in N. gonorrhea to closed state (Kesherwani et al., 2017). In E. coli, P. aeruginosa, and A. baumannii, one of the most widely studied efflux pump inhibitors of AcrB, phenylalanyl-arginine-β-naphthylamide (PAβN), is recognized for its ability to bind with the hydrophobic pocket located within AcrB, further inhibiting the drug extrusion activity (Lomovskaya et al., 2001; Ribera et al., 2002; Kinana et al., 2016). However, few other EPIs (MBX inhibitors) having the ability to bind strongly to AcrB have also been reported (Sjuts et al., 2016). Additionally, this inhibitor can be combined with a secondary metabolite, i.e., carolacton produced by mycobacteria, which can be utilized as a potential therapeutic option against pathogens over-expressing the AcrAB-TolC efflux pump system (Donner et al., 2017). Another example of EPIs is tannic acid, which inhibits the multidrug efflux pump systems, such as Tet and Msr (Tintino et al., 2017), and is well studied in S. aureus. Tannic acid has also been reported to reduce the MIC levels of different antibiotics such as erythromycin and tetracycline significantly (Tintino et al., 2017). However, in clinical settings, the usage of an antibiotic along with an efflux pump inhibitor (EPI) as combination therapy is a potential challenge that depends upon the internal permeability characteristics of the bacterial outer membrane. Further, in a study done by Yang et al. (2017) the use of combination therapy against multidrug-resistant P. aeruginosa comprising tobramycin and EPI (NMP, paroxetine, or DBP) encourages the binding of EPI with tetracycline; additionally, the response of the combination of tobramycin plus EPI along with rifampicin, fluoroquinolone, and fosfomycin was also analyzed, which revealed that they exert a robust effect cooperatively upon the bacteria, which resulted into reduction in the MIC80 level against these antibiotics (Yang et al., 2018). Another example of combination therapy containing levofloxacin and EPI (trimethoprim and sertraline) used against P. aeruginosa harboring the MexAB-OprM efflux pump system showed a progressive advantage over monotherapy (using levofloxacin alone) (Adamson et al., 2015). Similar research conducted by Prasch and Bucar (2015) has also validated that a reduction in the antibiotic dose was observed when the combination therapy comprising EPI inhibitor and antibiotics was co-administered. Besides the aforementioned EPIs, several plant-based EPIs, have also been investigated, and above 20 different EPIs derived from plants have been reported based on their extraction mechanisms (Garvey et al., 2011b; Shiu et al., 2013). Additionally, vegetable-derived compounds, such as artesunate, berberine (Table 2), and curcumin, have also been reported to have the ability to inhibit EP activity in E. coli and P. aeruginosa, besides their anti-inflammatory and antibacterial activities (Li et al., 2011; Negi et al., 2014; Laudadio et al., 2019). Different food items such as vegetables, including Momoedica balsamina, spices, such as cumin and pepper, and oils extracted from aromatic plants are known for being an exceptional source of EPIs (Karumathil et al., 2018; Li et al., 2019; Tariq et al., 2019; Solnier et al., 2020; Muniz et al., 2021); further, the application of flavonoids such as flavonolignans in combating MDR via inhibiting efflux pump systems have been described previously (Chambers et al., 2019).
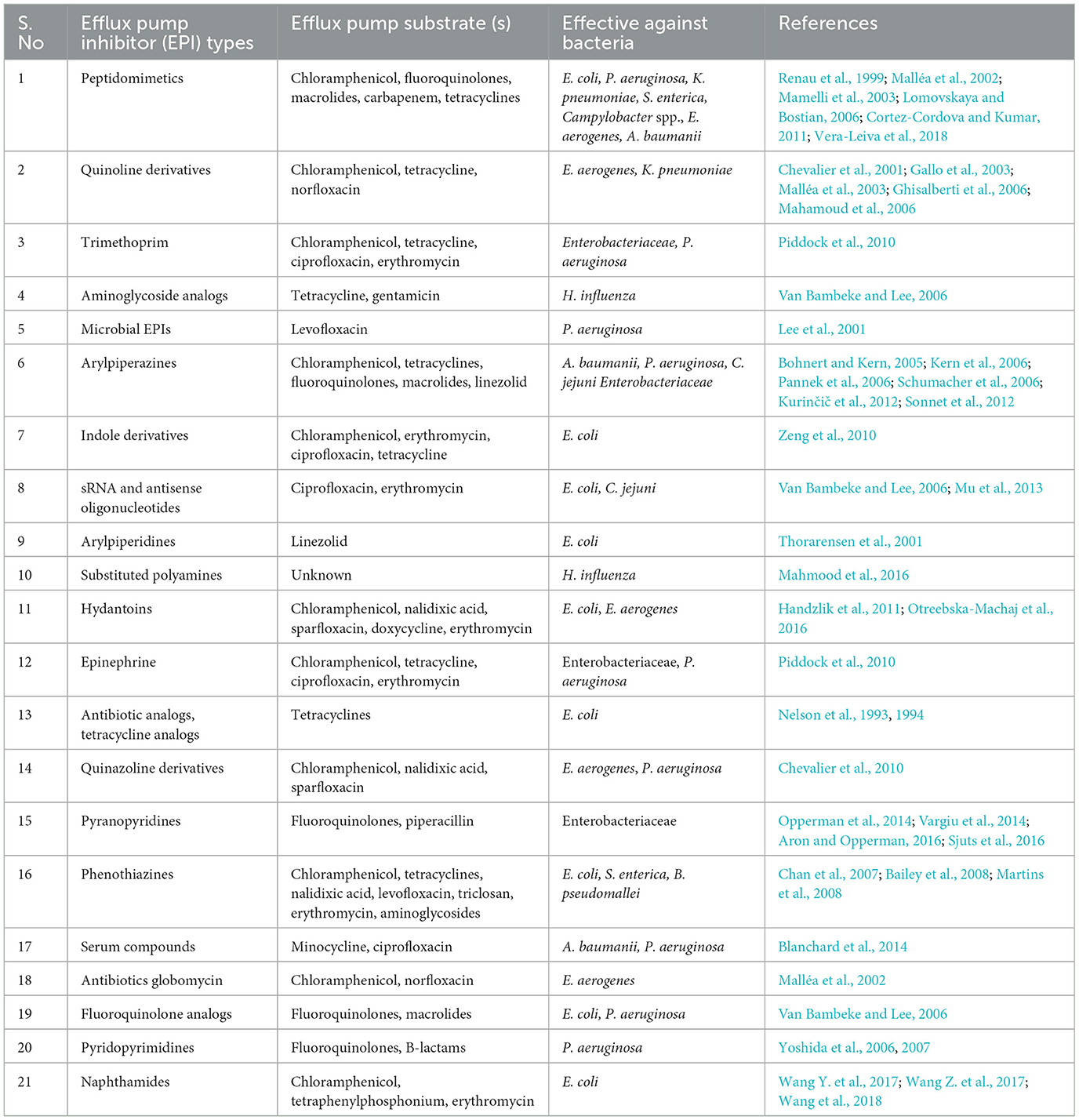
Table 1. Examples of synthetic efflux pump inhibitors (EPIs) having inhibitory action against Gram-negative bacteria.
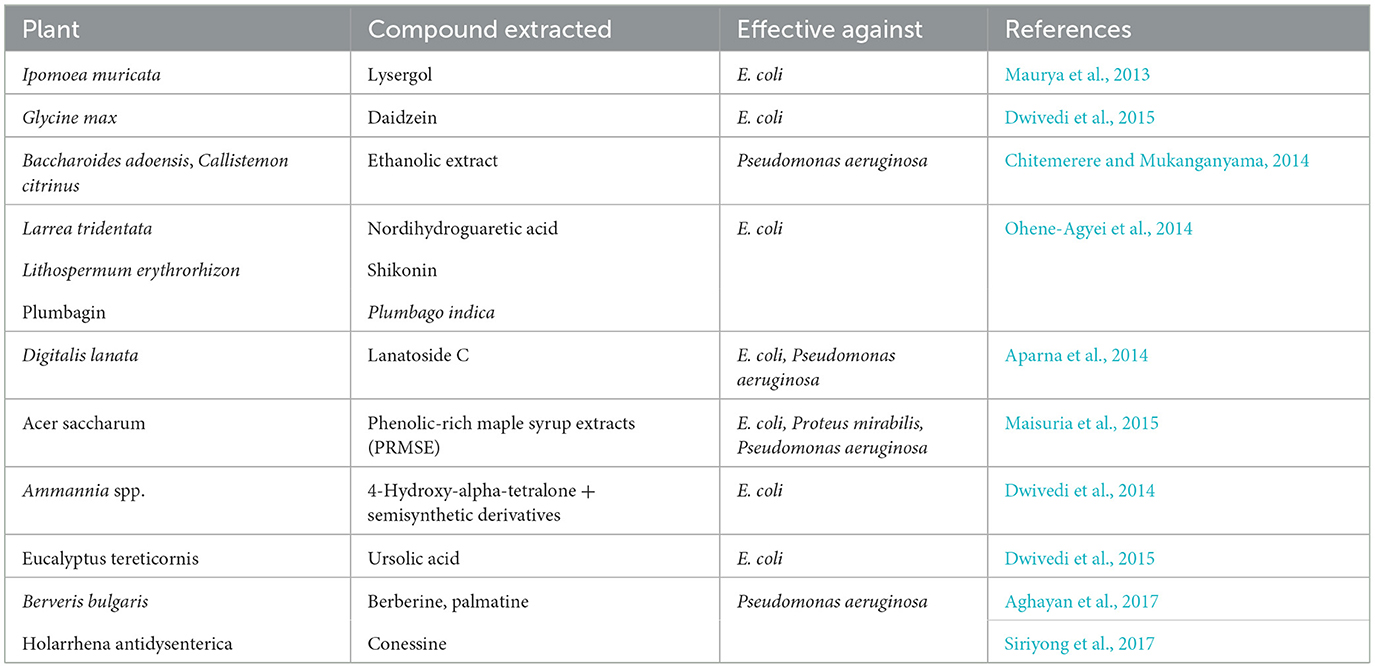
Table 2. List of plant-derived efflux pump inhibitors (EPIs) having inhibitory action toward Gram-negative bacteria.
However, incidences of another resistance mechanism associated with membrane-embedded MDR efflux pumps which can extrude a wide range of noxious compounds such as antibiotics, dyes, and biocides have been reported (Mitchell et al., 2019; Jesin et al., 2020), which can be a promising novel approach for tackling the increasing risk of antibiotic-resistant bacteria. Based on previous studies, all the bacterial species possess MDR efflux pumps, conferring intrinsic resistance against antibiotics (Bay and Turner, 2009), and six different superfamilies of efflux pumps have already been reported, but mostly, the small multidrug resistance efflux pump family was studied extensively (Sun et al., 2014). In hospitals, the commonly used disinfectants, the quaternary ammonium compounds (QACs), toxic compounds such as ethidium bromide, and many other compounds such as biocides, cetyltrimethylammonium bromide (CTAB), and benzalkonium chloride (BZK) are utilized as a substrate for acquiring energy by the SMRs family members (Bay et al., 2008). A study by Kadry et al. (2017) revealed the existence of a strong correlation between the gene encoding SMR protein with the enhanced level of resistance toward compounds such as benzalkonium chloride in clinical isolates of P. aeruginosa.
In this review study, we have discussed the important role played by efflux pump family members in developing resistance against single or multiple antibiotics as well as against various compounds other than the antimicrobial agents including heavy metals, preservatives, and toxins. Apart from different classes of antibiotics, efflux pumps also provide resistance against disinfectants and virulence factors and even develop cross-resistance against other different noxious compounds, which further leads to the development of a multidrug-resistant phenotype. The working mechanism along with the regulation of an efflux pump system is an intricate process that actively participates in different biological processes in bacteria such as biofilm formation, quorum sensing, and adhesion of bacteria to host, and their progressive multiplication have been highlighted in this review. This review further highlights the importance of the discovery of novel EPIs, which should be plant-based, safe, and non-toxic, and many efflux pump inhibitors that are under clinical trial as well. Efflux pump inhibitors derived from plants are of great importance and are studied widely presently. However, the variations in function and regulatory mechanisms of the efflux pump systems along with the progress and utilization of a variety of efflux pump inhibitors still need further exploration. According to the progressive research work, it has been suggested that TetR family members can be studied further for the discovery of antibiotic residues associated with multidrug resistance. In addition to the role of the aforementioned active regulatory proteins, some other proteins with an ability to identify single or multiple substrates and specific substrate-binding domains that can be utilized to identify antibiotic residues involved in substrate binding have been reported earlier. Therefore, other than the TetR regulatory protein, the detection of residues of antibiotics and transcriptional regulatory protein requires the utilization of efflux pump concomitant genes. Additionally, the proteins with a high substrate binding specificity toward diverse antimicrobials with more appropriate features are very demanding. Moreover, this review discussed the association of the expressional levels of efflux pump systems with various other defense mechanisms such as biofilm formation and quorum sensing, which provides novel insights for expanding the extent of fundamental research. In this article, we provided an overview of efflux genes and significant transporters indicating an area which requires persistent exploration to provide supervision for clinical practices.
SC was involved in the literature review, writing of the original draft, figure illustrations, conceptualization, preparing the draft of the manuscript, reviewing, editing, revising the manuscript, has contributed to the article, and approved the submitted version.
SC would like to acknowledge the support of management, Dr. S. Sumaya (Principal, Thassim Beevi College, Kilakarai, Tamil Nadu, India) and Dr. M. S. Irfan Ahmed (Director—Research and Industry–Institute Relations) for providing a stimulating environment.
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Ababou, A., and Koronakis, V. (2016). Structures of gate loop variants of the AcrB drug efflux pump bound by erythromycin substrate. PLoS ONE 11, e159154. doi: 10.1371/journal.pone.0159154
Adamson, D. H., Krikstopaityte, V., and Coote, P. J. (2015). Enhanced efficacy of putative efflux pump inhibitor/antibiotic combination treatments versus MDR strains of Pseudomonas aeruginosa in a Galleria mellonella in vivo infection model. J. Antimicrob. Chemother. 70, 2271–2278. doi: 10.1093/jac/dkv111
Aghayan, S. S., Mogadam, H. K., Fazli, M., Darban-Sarokhalil, D., Khoramrooz, S. S., Jabalameli, F., et al. (2017). The effects of berberine and palmatine on efflux pumps inhibition with different gene patterns in Pseudomonas aeruginosa isolated from burn infections. Avicenna J. Med. Biotechnol. 9, 2.
Alav, I., Sutton, J. M., and Rahman, K. M. (2018). Role of bacterial efflux pumps in biofilm formation. J. Antimicrob. Chemother. 73, 2003–2020. doi: 10.1093/jac/dky042
Alcalde-Rico, M., Hernando-Amado, S., Blanco, P., and Martínez, J. L. (2016). Multidrug efflux pumps at the crossroad between antibiotic resistance and bacterial virulence. Front. Microbiol. 7, 1483. doi: 10.3389/fmicb.2016.01483
Alibert, S., N'gompaza Diarra, J., Hernandez, J., Stutzmann, A., Fouad, M., Boyer, G., et al. (2017). Multidrug efflux pumps and their role in antibiotic and antiseptic resistance: a pharmacodynamic perspective. Expert Opin. Drug Metab. Toxicol. 13, 301–309 doi: 10.1080/17425255.2017.1251581
Allen, H. K., Donato, J., Wang, H. H., Cloud-Hansen, K. A., Davies, J., and Handelsman, J. (2010). Call of the wild: antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 8, 251–259. doi: 10.1038/nrmicro2312
Alm, R. A., Johnstone, M. R., and Lahiri, S. D. (2015). Characterization of Escherichia coli NDM isolates with decreased susceptibility to aztreonam/avibactam: role of a novel insertion in PBP3. J. Antimicrob. Chemother. 70, 1420–1428. doi: 10.1093/jac/dku568
Anandapadamanaban, M., Pilstål, R., Andresen, C., Trewhella, J., Moche, M., Wallner, B., et al. (2016). Mutation-induced population shift in the MexR conformational ensemble disengages DNA binding: a novel mechanism for MarR family derepression. Structure 24, 1311–1321. doi: 10.1016/j.str.2016.06.008
Andersson, D. I., and Levin, B. R. (1999). The biological cost of antibiotic resistance. Curr. Opin. Microbiol. 2, 489–493. doi: 10.1016/s1369-5274(99)00005-3
Aparna, V., Dineshkumar, K., Mohanalakshmi, N., Velmurugan, D., and Hopper, W. (2014). Identification of natural compound inhibitors for multidrug efflux pumps of Escherichia coli and Pseudomonas aeruginosa using in silico high-throughput virtual screening and in vitro validation. PLoS ONE 9, e101840. doi: 10.1371/journal.pone.0101840
Aron, Z., and Opperman, T. J. (2016). Optimization of a novel series of pyranopyridine RND efflux pump inhibitors. Curr. Opin. Microbiol. 33, 1–6. doi: 10.1016/j.mib.2016.05.007
Aron, Z., and Opperman, T. J. (2018). The hydrophobic trap—the Achilles heel of RND efflux pumps. Res. Microbiol. 169, 393–400. doi: 10.1016/j.resmic.2017.11.001
Augustin, R. (2010). The protein family of glucose transport facilitators: it's not only about glucose after all. IUBMB Life 62, 315–333. doi: 10.1002/iub.315
Bailey, A. M., Paulsen, I. T., and Piddock, L. J. (2008). RamA confers multidrug resistance in Salmonella enterica via increased expression of acrB, which is inhibited by chlorpromazine. Antimicrob. Agents Chemother. 52, 3604–3611. doi: 10.1128/AAC.00661-08
Banigan, J. R., Gayen, A., Cho, M. K., and Traaseth, N. J. (2015). A structured loop modulates coupling between the substrate-binding and dimerization domains in the multidrug resistance transporter EmrE. J. Biol. Chem. 290, 805–814. doi: 10.1074/jbc.M114.601963
Bay, D. C., Rommens, K. L., and Turner, R. J. (2008). Small multidrug resistance proteins: a multidrug transporter family that continues to grow. Biochim. Biophys. Acta. Biomembr. 1778, 1814–1838. doi: 10.1016/j.bbamem.2007.08.015
Bay, D. C., and Turner, R. J. (2009). Diversity and evolution of the small multidrug resistance protein family. BMC Evol. Biol. 9, 1–27. doi: 10.1186/1471-2148-9-140
Baylay, A. J., and Piddock, L. J. (2015). Clinically relevant fluoroquinolone resistance due to constitutive overexpression of the PatAB ABC transporter in Streptococcus pneumoniae is conferred by disruption of a transcriptional attenuator. J. Antimicrob. Chemother. 70, 670–679. doi: 10.1093/jac/dku449
Bhakdi, S., Mackman, N., Menestrina, G., Gray, L., Hugo, F., Seeger, W., et al. (1988). The hemolysin of Escherichia coli. Eur. J. Epidemiol. 4, 135–143. doi: 10.1007/BF00144740
Blair, J., Webber, M. A., Baylay, A. J., Ogbolu, D. O., and Piddock, L. J. (2015). Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 13, 42–51. doi: 10.1038/nrmicro3380
Blair, J. M., and Piddock, L. J. (2016). How to measure export via bacterial multidrug resistance efflux pumps. MBio 7, e00840-16. doi: 10.1128/mBio.00840-16
Blair, J. M., Richmond, G. E., and Piddock, L. J. (2014). Multidrug efflux pumps in Gram-negative bacteria and their role in antibiotic resistance. Future Microbiol. 9, 1165–1177. doi: 10.2217/fmb.14.66
Blanchard, C., Barnett, P., Perlmutter, J., and Dunman, P. M. (2014). Identification of Acinetobacter baumannii serum-associated antibiotic efflux pump inhibitors. Antimicrob. Agents Chemother. 58, 6360–6370. doi: 10.1128/AAC.03535-14
Blanco, P., Hernando-Amado, S., Reales-Calderon, J. A., Corona, F., Lira, F., Alcalde-Rico, M., et al. (2016). Bacterial multidrug efflux pumps: much more than antibiotic resistance determinants. Microorganisms 4, 14. doi: 10.3390/microorganisms4010014
Bohnert, J. A., and Kern, W. V. (2005). Selected arylpiperazines are capable of reversing multidrug resistance in Escherichia coli overexpressing RND efflux pumps. Antimicrob. Agents Chemother. 49, 849–852. doi: 10.1128/AAC.49.2.849-852.2005
Brown, M. H., Paulsen, I. T., and Skurray, R. A. (1999). The multidrug efflux protein NorM is a prototype of a new family of transporters. Mol. Microbiol. 31, 394–395. doi: 10.1046/j.1365-2958.1999.01162.x
Buffet-Bataillon, S., Tattevin, P., Maillard, J. Y., Bonnaure-Mallet, M., and Jolivet-Gougeon, A. (2016). Efflux pump induction by quaternary ammonium compounds and fluoroquinolone resistance in bacteria. Future Microbiol. 11, 81–92. doi: 10.2217/fmb.15.131
Cannon, R. D., and Holmes, A. R. (2015). Learning the ABC of oral fungal drug resistance. Mol. Oral Microbiol. 30, 425–437. doi: 10.1111/omi.12109
Chambers, C. S., Viktorová, J., Rehorová, K., Biedermann, D., Turková, L., Macek, T., et al. (2019). Defying multidrug resistance! Modulation of related transporters by flavonoids and flavonolignans. J. Agric. Food Chem. 68, 1763–1779. doi: 10.1021/acs.jafc.9b00694
Chan, Y. Y., Ong, Y. M., and Chua, K. L. (2007). Synergistic interaction between phenothiazines and antimicrobial agents against Burkholderia pseudomallei. Antimicrob. Agents Chemother. 51, 623–630. doi: 10.1128/AAC.01033-06
Chen, H., Cao, J., Zhou, C., Liu, H., Zhang, X., and Zhou, T. (2017). Biofilm formation restrained by subinhibitory concentrations of tigecyclin in Acinetobacter baumannii is associated with downregulation of efflux pumps. Chemotherapy 62, 128–133. doi: 10.1159/000450537
Chetri, S., Bhowmik, D., Dhar, D., Chakravarty, A., and Bhattacharjee, A. (2019b). Effect of concentration gradient carbapenem exposure on expression of blaNDM-1 and acrA in carbapenem resistant Escherichia coli. Infect. Genet. Evol. 73, 332–336. doi: 10.1016/j.meegid.2019.05.024
Chetri, S., Bhowmik, D., Paul, D., Pandey, P., Chanda, D. D., Chakravarty, A., et al. (2019a). AcrAB-TolC efflux pump system plays a role in carbapenem non-susceptibility in Escherichia coli. BMC. microbial. 19, 1–7. doi: 10.1186/s12866-019-1589-1
Chetri, S., Das, B. J., Bhowmik, D., Chanda, D. D., Chakravarty, A., and Bhattacharjee, A. (2020). Transcriptional response of mar, sox and rob regulon against concentration gradient carbapenem stress within Escherichia coli isolated from hospital acquired infection. BMC Res. Notes. 13, 1–7. doi: 10.1186/s13104-020-04999-2
Chetri, S., Dolley, A., Bhowmik, D., Chanda, D. D., Chakravarty, A., and Bhattacharjee, A. (2018a). Transcriptional response of AcrEF-TolC against fluoroquinolone and carbapenem in Escherichia coli of clinical origin. Indian J. Med. Microbiol. 36, 537–540. doi: 10.4103/ijmm.IJMM_18_308
Chetri, S., Singha, K., Bhowmik, D., Chanda, D. D., Chakravarty, A., and Bhattacharjee, A. (2018b). Sub-inhibitory concentration of ertapenem induces overexpression of regulator of antibiotic resistance A in Escherichia coli. Indian J. Med. Microbiol. 36, 569–572. doi: 10.4103/ijmm.IJMM_18_436
Chetri, S., Singha, M., Bhowmik, D., Nath, K., Chanda, D. D., Chakravarty, A., et al. (2019c). Transcriptional response of OmpC and OmpF in Escherichia coli against differential gradient of carbapenem stress. BMC Res. Notes. 12, 1–6. doi: 10.1186/s13104-019-4177-4
Chevalier, J., Atifi, S., Eyraud, A., Mahamoud, A., Barbe, J., and Pagès, J. M. (2001). New pyridoquinoline derivatives as potential inhibitors of the fluoroquinolone efflux pump in resistant enterobacter a erogenes strains. J. Med. Chem. 44, 4023–4026. doi: 10.1021/jm010911z
Chevalier, J., Mahamoud, A., Baitiche, M., Adam, E., Viveiros, M., Smarandache, A., et al. (2010). Quinazoline derivatives are efficient chemosensitizers of antibiotic activity in Enterobacter aerogenes, Klebsiella pneumoniae and Pseudomonas aeruginosa resistant strains. Int. J. Antimicrob. Agents. 36, 164–168. doi: 10.1016/j.ijantimicag.2010.03.027
Chitemerere, T. A., and Mukanganyama, S. (2014). Evaluation of cell membrane integrity as a potential antimicrobial target for plant products. BMC Complement. Altern. Med. 14, 1–8. doi: 10.1186/1472-6882-14-278
Choudhury, H. G., Tong, Z., Mathavan, I., Li, Y., Iwata, S., Zirah, S., et al. (2014). Structure of an antibacterial peptide ATP-binding cassette transporter in a novel outward occluded state. Proc. Natl. Acad. Sci. USA 111, 9145–9150. doi: 10.1073/pnas.1320506111
Colque, C. A., Albarracín Orio, A. G., Feliziani, S., Marvig, R. L., Tobares, A. R., Johansen, H. K., et al. (2020). Hypermutator Pseudomonas aeruginosa exploits multiple genetic pathways to develop multidrug resistance during long-term infections in the airways of cystic fibrosis patients. Antimicrob. Agents Chemother. 64, e02142-19. doi: 10.1128/AAC.02142-19
Cooper, M. A., and Shlaes, D. (2011). Fix the antibiotics pipeline. Nature 472, 32–32. doi: 10.1038/472032a
Cortez-Cordova, J., and Kumar, A. (2011). Activity of the efflux pump inhibitor phenylalanine-arginine β-naphthylamide against the AdeFGH pump of Acinetobacter baumannii. Int. J. Antimicrob. Agents. 37, 420–424. doi: 10.1016/j.ijantimicag.2011.01.006
Cura, A. J., and Carruthers, A. (2012). The role of monosaccharide transport proteins in carbohydrate assimilation, distribution, metabolism and homeostasis. Compr. Physiol. 2, 863. doi: 10.1002/cphy.c110024
Daury, L., Orange, F., Taveau, J. C., Verchere, A., Monlezun, L., Gounou, C., et al. (2016). Tripartite assembly of RND multidrug efflux pumps. Nat. Commun. 7, 1–8. doi: 10.1038/ncomms10731
Dawson, R. J., and Locher, K. P. (2006). Structure of a bacterial multidrug ABC transporter. Nature. 443, 180–185. doi: 10.1038/nature05155
Delmar, J. A., Su, C. C., and Yu, E. W. (2014). Bacterial multidrug efflux transporters. Annu. Rev. Biophys. 43, 93–117. doi: 10.1146/annurev-biophys-051013-022855
Donner, J., Reck, M., Bunk, B., Jarek, M., App, C. B., Meier-Kolthoff, J. P., et al. (2017). The biofilm inhibitor carolacton enters gram-negative cells: studies using a TolC-deficient strain of Escherichia coli. Msphere 2, e00375-17. doi: 10.1128/mSphereDirect.00375-17
Dou, Y., Song, F., Guo, F., Zhou, Z., Zhu, C., Xiang, J., et al. (2017). Acinetobacter baumannii quorum-sensing signalling molecule induces the expression of drug-resistance genes. Mol. Med. Rep. 15, 4061–4068. doi: 10.3892/mmr.2017.6528
Du, D., van Veen, H. W., and Luisi, B. F. (2015). Assembly and operation of bacterial tripartite multidrug efflux pumps. Trends Microbiol. 23, 311–319. doi: 10.1016/j.tim.2015.01.010
Du, D., Wang, Z., James, N. R., Voss, J. E., Klimont, E., Ohene-Agyei, T., et al. (2014). Structure of the AcrAB–TolC multidrug efflux pump. Nature 509, 512–515. doi: 10.1038/nature13205
Du, D., Wang-Kan, X., Neuberger, A., Van Veen, H. W., Pos, K. M., Piddock, L. J., et al. (2018). Multidrug efflux pumps: structure, function and regulation. Nat. Rev. Microbiol. 16, 523–539. doi: 10.1038/s41579-018-0048-6
Dwivedi, G. R., Maurya, A., Yadav, D. K., Khan, F., Darokar, M. P., and Srivastava, S. K. (2015). Drug resistance reversal potential of ursolic acid derivatives against nalidixic acid-and multidrug-resistant Escherichia coli. Chem. Biol. Drug Des. 86, 272–283. doi: 10.1111/cbdd.12491
Dwivedi, G. R., Upadhyay, H. C., Yadav, D. K., Singh, V., Srivastava, S. K., Khan, F., et al. (2014). 4-Hydroxy-α-tetralone and its derivative as drug resistance reversal agents in multi drug resistant Escherichia coli. Chem. Biol. Drug Des. 83, 482–492. doi: 10.1111/cbdd.12263
Eicher, T., Cha, H. J., Seeger, M. A., Brandstätter, L., El-Delik, J., Bohnert, J. A., et al. (2012). Transport of drugs by the multidrug transporter AcrB involves an access and a deep binding pocket that are separated by a switch-loop. Proc. Natl. Acad. Sci. USA 109, 5687–5692. doi: 10.1073/pnas.1114944109
Eicher, T., Seeger, M. A., Anselmi, C., Zhou, W., Brandstätter, L., Verrey, F., et al. (2014). Coupling of remote alternating-access transport mechanisms for protons and substrates in the multidrug efflux pump AcrB. Elife 3, e03145. doi: 10.7554/eLife.03145
Elbourne, L. D., Tetu, S. G., Hassan, K. A., and Paulsen, I. T. (2017). TransportDB 2.0: a database for exploring membrane transporters in sequenced genomes from all domains of life. Nucleic. Acids. Res. 45, D320–D324. doi: 10.1093/nar/gkw1068
Fitzpatrick, A. W., Llabrés, S., Neuberger, A., Blaza, J. N., Bai, X. C., Okada, U., et al. (2017). Structure of the MacAB–TolC ABC-type tripartite multidrug efflux pump. Nat. Microbiol. 2, 1–8. doi: 10.1038/nmicrobiol.2017.70
Fluman, N., Adler, J., Rotenberg, S. A., Brown, M. H., and Bibi, E. (2014). Export of a single drug molecule in two transport cycles by a multidrug efflux pump. Nat. Commun. 5, 4615. doi: 10.1038/ncomms5615
Forrest, L. R., Krämer, R., and Ziegler, C. (2011). The structural basis of secondary active transport mechanisms. Biochim. Biophys. Acta. Bioenerg. 1807, 167–188. doi: 10.1016/j.bbabio.2010.10.014
Gallo, S., Chevalier, J., Mahamoud, A., Eyraud, A., Pagès, J. M., and Barbe, J. (2003). 4-alkoxy and 4-thioalkoxyquinoline derivatives as chemosensitizers for the chloramphenicol-resistant clinical Enterobacter aerogenes 27 strain. Int. J. Antimicrob. Agents. 22, 270–273. doi: 10.1016/S0924-8579(03)00215-2
Ganas, P., Mihasan, M., Igloi, G. L., and Brandsch, R. (2007). A two-component small multidrug resistance pump functions as a metabolic valve during nicotine catabolism by Arthrobacter nicotinovorans. Microbiology 153, 1546–1555. doi: 10.1099/mic.0.2006/004234-0
Garvey, M. I., Baylay, A. J., Wong, R. L., and Piddock, L. J. (2011a). Overexpression of patA and patB, which encode ABC transporters, is associated with fluoroquinolone resistance in clinical isolates of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 55, 190–196. doi: 10.1128/AAC.00672-10
Garvey, M. I., Rahman, M. M., Gibbons, S., and Piddock, L. J. (2011b). Medicinal plant extracts with efflux inhibitory activity against Gram-negative bacteria. Int. J. Antimicrob. Agents. 37, 145–151.doi: 10.1016/j.ijantimicag.2010.10.027
Ghisalberti, D., Mahamoud, A., Chevalier, J., Baitiche, M., Martino, M., Pagès, J. M., et al. (2006). Chloroquinolines block antibiotic efflux pumps in antibiotic-resistant Enterobacter aerogenes isolates. Int. J. Antimicrob. Agents. 27, 565–569. doi: 10.1016/j.ijantimicag.2006.03.010
Gillings, M. R. (2017). Class 1 integrons as invasive species. Curr. Opin. Microbiol. 38, 10–15. doi: 10.1016/j.mib.2017.03.002
Gilson, L., Mahanty, H. K., and Kolter, R. (1990). Genetic analysis of an MDR-like export system: the secretion of colicin V. EMBO J. 9, 3875–3884. doi: 10.1002/j.1460-2075.1990.tb07606.x
Grinius, L. L., and Goldberg, E. B. (1994). Bacterial multidrug resistance is due to a single membrane protein which functions as a drug pump. J. Biol. Chem. 269, 29998–30004. doi: 10.1016/S0021-9258(18)43980-4
Grkovic, S., Brown, M. H., and Skurray, R. A. (2002). Regulation of bacterial drug export systems. Microbiol. Mol. Biol. Rev. 66, 671–701. doi: 10.1128/MMBR.66.4.671-701.2002
Guan, L., and Kaback, H. R. (2006). Lessons from lactose permease. Annu. Rev. Biophys. Biomol. Struct. 35, 67. doi: 10.1146/annurev.biophys.35.040405.102005
Guérin, F., Galimand, M., Tuambilangana, F., Courvalin, P., and Cattoir, V. (2014). Overexpression of the novel MATE fluoroquinolone efflux pump FepA in Listeria monocytogenes is driven by inactivation of its local repressor FepR. PLoS ONE 9, e106340. doi: 10.1371/journal.pone.0106340
Gupta, D., Singh, A., and Khan, A. U. (2017). Nanoparticles as efflux pump and biofilm inhibitor to rejuvenate bactericidal effect of conventional antibiotics. Nanoscale Res. Lett. 12, 1–6. doi: 10.1186/s11671-017-2222-6
Hall, C. W., and Mah, T. F. (2017). Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol. Rev. 41, 276–301. doi: 10.1093/femsre/fux010
Handzlik, J., Szymańska, E., Chevalier, J., Otrebska, E., Kieć-Kononowicz, K., Pagès, J. M., et al. (2011). Amine–alkyl derivatives of hydantoin: new tool to combat resistant bacteria. Eur. J. Med. Chem. 46, 5807–5816. doi: 10.1016/j.ejmech.2011.09.032
Hassan, K. A., Elbourne, L. D., Li, L., Gamage, H. K., Liu, Q., Jackson, S. M., et al. (2015b). An ace up their sleeve: a transcriptomic approach exposes the AceI efflux protein of Acinetobacter baumannii and reveals the drug efflux potential hidden in many microbial pathogens. Front. Microbiol. 6, 333. doi: 10.3389/fmicb.2015.00333
Hassan, K. A., Jackson, S. M., Penesyan, A., Patching, S. G., Tetu, S. G., Eijkelkamp, B. A., et al. (2013). Transcriptomic and biochemical analyses identify a family of chlorhexidine efflux proteins. Proc. Natl. Acad. Sci. USA 110, 20254–20259. doi: 10.1073/pnas.1317052110
Hassan, K. A., Liu, Q., Elbourne, L. D., Ahmad, I., Sharples, D., Naidu, V., et al. (2018). Pacing across the membrane: the novel PACE family of efflux pumps is widespread in Gram-negative pathogens. Res. Microbiol. 169, 450–454. doi: 10.1016/j.resmic.2018.01.001
Hassan, K. A., Liu, Q., Henderson, P. J., and Paulsen, I. T. (2015a). Homologs of the Acinetobacter baumannii AceI transporter represent a new family of bacterial multidrug efflux systems. MBio 6, e01982–14. doi: 10.1128/mBio.01982-14
He, X., Szewczyk, P., Karyakin, A., Evin, M., Hong, W. X., Zhang, Q., et al. (2010). Structure of a cation-bound multidrug and toxic compound extrusion transporter. Nature 467, 991–994. doi: 10.1038/nature09408
Hellmich, U. A., Lyubenova, S., Kaltenborn, E., Doshi, R., van Veen, H. W., Prisner, T. F., et al. (2012). Probing the ATP hydrolysis cycle of the ABC multidrug transporter LmrA by pulsed EPR spectroscopy. J. Am. Chem. Soc. 134, 5857–5862. doi: 10.1021/ja211007t
Hellmich, U. A., Mönkemeyer, L., Velamakanni, S., van Veen, H. W., and Glaubitz, C. (2015). Effects of nucleotide binding to LmrA: a combined MAS-NMR and solution NMR study. Biochim. Biophys. Acta- Biomembr. 1848, 3158–3165. doi: 10.1016/j.bbamem.2015.10.003
Hernando-Amado, S., Blanco, P., Alcalde-Rico, M., Corona, F., Reales-Calderón, J. A., Sánchez, M. B., et al. (2016). Multidrug efflux pumps as main players in intrinsic and acquired resistance to antimicrobials. Drug Resist. Updat. 28, 13–27. doi: 10.1016/j.drup.2016.06.007
Hirai, T., Heymann, J. A., Shi, D., Sarker, R., Maloney, P. C., and Subramaniam, S. (2002). Three-dimensional structure of a bacterial oxalate transporter. Nat. Struct. Biol. 9, 597–600. doi: 10.1038/nsb821
Hung, L. W., Kim, H. B., Murakami, S., Gupta, G., Kim, C. Y., and Terwilliger, T. C. (2013). Crystal structure of AcrB complexed with linezolid at 3.5 Å resolution. J. Struct. Funct. Genomics. 14, 71–75. doi: 10.1007/s10969-013-9154-x
Iannelli, F., Santagati, M., Santoro, F., Oggioni, M. R., Stefani, S., and Pozzi, G. (2014). Nucleotide sequence of conjugative prophage Φ1207. 3 (formerly Tn 1207.3) carrying the mef (A)/msr (D) genes for efflux resistance to macrolides in Streptococcus pyogenes. Front. Microbiol. 5, 687. doi: 10.3389/fmicb.2014.00687
Ito, S., Kusuhara, H., Yokochi, M., Toyoshima, J., Inoue, K., Yuasa, H., et al. (2012). Competitive inhibition of the luminal efflux by multidrug and toxin extrusions, but not basolateral uptake by organic cation transporter 2, is the likely mechanism underlying the pharmacokinetic drug-drug interactions caused by cimetidine in the kidney. J. Pharmacol. Exp. Ther. 340, 393–403. doi: 10.1124/jpet.111.184986
Jack, D. L., Storms, M. L., Tchieu, J. H., Paulsen, I. T., and Saier Jr, M. H. (2000). A broad-specificity multidrug efflux pump requiring a pair of homologous SMR-type proteins. J. Bacteriol. 182, 2311–2313. doi: 10.1128/JB.182.8.2311-2313.2000
Jaglic, Z., and Cervinkova, D. (2012). Genetic basis of resistance to quaternary ammonium compounds–the qac genes and their role: a review. Vet. Med. 57, 275–281. doi: 10.17221/6013-VETMED
Jamshidi, S., Sutton, J. M., and Rahman, K. M. (2016). An overview of bacterial efflux pumps and computational approaches to study efflux pump inhibitors. Future Med. Chem. 8, 195–210. doi: 10.4155/fmc.15.173
Jamshidi, S., Sutton, J. M., and Rahman, K. M. (2018). Mapping the dynamic functions and structural features of acrb efflux pump transporter using accelerated molecular dynamics simulations. Sci. Rep. 8, 1–13. doi: 10.1038/s41598-018-28531-6
Jardetzky, O. (1966). Simple allosteric model for membrane pumps. Nature 211, 969–970. doi: 10.1038/211969a0
Jesin, J. A., Stone, T. A., Mitchell, C. J., Reading, E., and Deber, C. M. (2020). Peptide-based approach to inhibition of the multidrug resistance efflux pump AcrB. Biochemistry 59, 3973–3981. doi: 10.1021/acs.biochem.0c00417
Jin, M. S., Oldham, M. L., Zhang, Q., and Chen, J. (2012). Crystal structure of the multidrug transporter P-glycoprotein from Caenorhabditis elegans. Nature 490, 566–569. doi: 10.1038/nature11448
Jo, I., Hong, S., Lee, M., Song, S., Kim, J. S., Mitra, A. K., et al. (2017). Stoichiometry and mechanistic implications of the MacAB-TolC tripartite efflux pump. Biochem. Biophys. Res. Commun. 494, 668–673. doi: 10.1016/j.bbrc.2017.10.102
Johnson, Z. L., and Chen, J. (2017). Structural basis of substrate recognition by the multidrug resistance protein MRP1. Cell. 168, 1075–1085. doi: 10.1016/j.cell.2017.01.041
Kaback, H. R. (2015). A chemiosmotic mechanism of symport. Proc. Natl. Acad. Sci. USA 112, 1259–1264. doi: 10.1073/pnas.1419325112
Kadry, A. A., Serry, F. M., El-Ganiny, A. M., and El-Baz, A. M. (2017). Integron occurrence is linked to reduced biocide susceptibility in multidrug resistant Pseudomonas aeruginosa. Br. J. Biomed. Sci. 74, 78–84. doi: 10.1080/09674845.2017.1278884
Karumathil, D. P., Nair, M. S., Gaffney, J., Kollanoor-Johny, A., and Venkitanarayanan, K. (2018). Trans-cinnamaldehyde and eugenol increase Acinetobacter baumannii sensitivity to beta-lactam antibiotics. Front. Microbiol. 9, 1011. doi: 10.3389/fmicb.2018.01011
Kermani, A. A., Macdonald, C. B., Burata, O. E., Ben Koff, B., Koide, A., Denbaum, E., et al. (2020). The structural basis of promiscuity in small multidrug resistance transporters. Nat. Commun. 11, 1–9. doi: 10.1038/s41467-020-19820-8
Kermani, A. A., Macdonald, C. B., Gundepudi, R., and Stockbridge, R. B. (2018). Guanidinium export is the primal function of SMR family transporters. Proc. Natl. Acad. Sci. USA 115, 3060–3065. doi: 10.1073/pnas.1719187115
Kern, W. V., Steinke, P., Schumacher, A., Schuster, S., Baum, H. V., and Bohnert, J. A. (2006). Effect of 1-(1-naphthylmethyl)-piperazine, a novel putative efflux pump inhibitor, on antimicrobial drug susceptibility in clinical isolates of Escherichia coli. J. Antimicrob. Chemother. 57, 339–343. doi: 10.1093/jac/dki445
Kesherwani, M., Michael Gromiha, M., Fukui, K., and Velmurugan, D. (2017). Identification of novel natural inhibitor for NorM—a multidrug and toxic compound extrusion transporter—an insilico molecular modeling and simulation studies. J. Biomol. Struct. Dyn. 35, 58–77. doi: 10.1080/07391102.2015.1132391
Khare, D., Oldham, M. L., Orelle, C., Davidson, A. L., and Chen, J. (2009). Alternating access in maltose transporter mediated by rigid-body rotations. Mol. Cell. 33, 528–536. doi: 10.1016/j.molcel.2009.01.035
Kim, Y. C., and Hummer, G. (2012). Proton-pumping mechanism of cytochrome c oxidase: a kinetic master-equation approach. Biochim. Biophys. Acta. Bioenerg. 1817, 526–536. doi: 10.1016/j.bbabio.2011.09.004
Kinana, A. D., Vargiu, A. V., May, T., and Nikaido, H. (2016). Aminoacyl β-naphthylamides as substrates and modulators of AcrB multidrug efflux pump. Proc. Natl. Acad. Sci. USA 113, 1405–1410. doi: 10.1073/pnas.1525143113
Kodan, A., Yamaguchi, T., Nakatsu, T., Sakiyama, K., Hipolito, C. J., Fujioka, A., et al. (2014). Structural basis for gating mechanisms of a eukaryotic P-glycoprotein homolog. Proc. Natl. Acad. Sci. USA 111, 4049–4054. doi: 10.1073/pnas.1321562111
Kong, E. F., Tsui, C., Kucharíková, S., Van Dijck, P., and Jabra-Rizk, M. A. (2017). Modulation of Staphylococcus aureus response to antimicrobials by the Candida albicans quorum sensing molecule farnesol. Antimicrob. Agents. Chemother. 61, e01573-17. doi: 10.1128/AAC.01573-17
Korkhov, V. M., Mireku, S. A., and Locher, K. P. (2012). Structure of AMP-PNP-bound vitamin B12 transporter BtuCD–F. Nature. 490, 367–372. doi: 10.1038/nature11442
Kurinčič, M., Klančnik, A., and Smole Možina, S. (2012). Effects of efflux pump inhibitors on erythromycin, ciprofloxacin, and tetracycline resistance in Campylobacter spp. isolates. Microb. Drug Resist. 18, 492–501. doi: 10.1089/mdr.2012.0017
Kuroda, T., and Tsuchiya, T. (2009). Multidrug efflux transporters in the MATE family. Biochim. Biophys. Acta. Proteins Proteom. 1794, 763–768. doi: 10.1016/j.bbapap.2008.11.012
Kusakizako, T., Claxton, D. P., Tanaka, Y., Maturana, A. D., Kuroda, T., Ishitani, R., et al. (2019). Structural basis of H+-dependent conformational change in a bacterial MATE transporter. Structure 27, 293–301. doi: 10.1016/j.str.2018.10.004
Laudadio, E., Cedraro, N., Mangiaterra, G., Citterio, B., Mobbili, G., Minnelli, C., et al. (2019). Natural alkaloid berberine activity against Pseudomonas aeruginosa MexXY-mediated aminoglycoside resistance: in silico and in vitro studies. J. Nat. Prod. 82, 1935–1944. doi: 10.1021/acs.jnatprod.9b00317
Lee, M. D., Galazzo, J. L., Staley, A. L., Lee, J. C., Warren, M. S., Fuernkranz, H., et al. (2001). Microbial fermentation-derived inhibitors of efflux-pump-mediated drug resistance. IL Farmaco 56, 81–85. doi: 10.1016/S0014-827X(01)01002-3
Lerma, L. L., Benomar, N., Valenzuela, A. S., Muñoz, M. D. C. C., Gálvez, A., and Abriouel, H. (2014). Role of EfrAB efflux pump in biocide tolerance and antibiotic resistance of Enterococcus faecalis and Enterococcus faecium isolated from traditional fermented foods and the effect of EDTA as EfrAB inhibitor. Food Microbiol. 44, 249–257. doi: 10.1016/j.fm.2014.06.009
Levy, S. B., and Marshall, B. (2004). Antibacterial resistance worldwide: causes, challenges and responses. Nat. Med. 10, S122–S129. doi: 10.1038/nm1145
Li, B., Yao, Q., Pan, X. C., Wang, N., Zhang, R., Li, J., et al. (2011). Artesunate enhances the antibacterial effect of β-lactam antibiotics against Escherichia coli by increasing antibiotic accumulation via inhibition of the multidrug efflux pump system AcrAB-TolC. J. Antimicrob. Chemother. 66, 769–777. doi: 10.1093/jac/dkr017
Li, J., Liu, D., Tian, X., Koseki, S., Chen, S., Ye, X., et al. (2019). Novel antibacterial modalities against methicillin resistant Staphylococcus aureus derived from plants. Crit. Rev. Food Sci. Nutr. 59, S153–S161. doi: 10.1080/10408398.2018.1541865
Li, N., Wu, J. X., Ding, D., Cheng, J., Gao, N., and Chen, L. (2017). Structure of a pancreatic ATP-sensitive potassium channel. Cell. 168, 101–110. doi: 10.1016/j.cell.2016.12.028
Li, X. Z., Plésiat, P., and Nikaido, H. (2015). The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin. Microbiol. Rev. 28, 337–418. doi: 10.1128/CMR.00117-14
Liang, Z. B., Chen, Y. M., Chen, Y. F., Cheng, Y. Y., and Zhang, L. H. (2016). RND efflux pump and its interrelationship with quorum sensing system. FEMS Microbiol. Lett. 38, 894–901. doi: 10.16288/j.yczz.16-139
Lin, M. F., Lin, Y. Y., Tu, C. C., and Lan, C. Y. (2017). Distribution of different efflux pump genes in clinical isolates of multidrug-resistant Acinetobacter baumannii and their correlation with antimicrobial resistance. J. Microbiol. Immunol. Infect. 50, 224–231. doi: 10.1016/j.jmii.2015.04.004
Littlejohn, T. G., Paulsen, I. T., Gillespie, M. T., Tennent, J. M., Midgley, M., Jones, I. G., et al. (1992). Substrate specificity and energetics of antiseptic and disinfectant resistance in Staphylococcus aureus. FEMS Microbiol. Lett. 95, 259–265. doi: 10.1111/j.1574-6968.1992.tb05376.x
Locher, K. P. (2016). Mechanistic diversity in ATP-binding cassette (ABC) transporters. Nat. Struct. Mol. Biol. 23, 487–493. doi: 10.1038/nsmb.3216
Lomovskaya, O., and Bostian, K. A. (2006). Practical applications and feasibility of efflux pump inhibitors in the clinic—a vision for applied use. Biochem. Pharmacol. 71, 910–918. doi: 10.1016/j.bcp.2005.12.008
Lomovskaya, O., Warren, M. S., Lee, A., Galazzo, J., Fronko, R., Lee, M. A. Y., et al. (2001). Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: novel agents for combination therapy. Antimicrob. Agents Chemother. 45, 105–116. doi: 10.1128/AAC.45.1.105-116.2001
Long, F., Su, C. C., Lei, H. T., Bolla, J. R., Do, S. V., and Yu, E. W. (2012). Structure and mechanism of the tripartite CusCBA heavy-metal efflux complex. Philos. Trans. R. Soc. B Biol. Sci. 367, 1047–1058. doi: 10.1098/rstb.2011.0203
Lu, M., Radchenko, M., Symersky, J., Nie, R., and Guo, Y. (2013b). Structural insights into H+-coupled multidrug extrusion by a MATE transporter. Nat. Struct. Mol. Biol. 20, 1310–1317. doi: 10.1038/nsmb.2687
Lu, M., Symersky, J., Radchenko, M., Koide, A., Guo, Y., Nie, R., et al. (2013a). Structures of a Na+-coupled, substrate-bound MATE multidrug transporter. Proc. Natl. Acad. Sci. USA 110, 2099–2104. doi: 10.1073/pnas.1219901110
Lu, S., and Zgurskaya, H. I. (2013). MacA, a periplasmic membrane fusion protein of the macrolide transporter MacAB-TolC, binds lipopolysaccharide core specifically and with high affinity. J. Bacteriol. 195, 4865–4872. doi: 10.1128/JB.00756-13
Lv, Z., Zhao, M., Wang, W., Wang, Q., Huang, M., Li, C., et al. (2021). Changing Gly311 to acidic amino acid in the MATE family protein DTX6 leads to enhanced resistance of Arabidopsis to the dihydropyridine herbicides. Mol. Plant. 14, 2115–2125. doi: 10.1016/j.molp.2021.09.002
Lytvynenko, I., Brill, S., Oswald, C., and Pos, K. M. (2016). Molecular basis of polyspecificity of the small multidrug resistance efflux pump AbeS from Acinetobacter baumannii. J. Mol. Biol. 428, 644–657. doi: 10.1016/j.jmb.2015.12.006
Mahamoud, A., Chevalier, J., Davin-Regli, A., and Barbe, J. (2006). Quinoline derivatives as promising inhibitors of antibiotic efflux pump in multidrug resistant Enterobacter aerogenes isolates. Curr. Drug Targets. 7, 843–847. doi: 10.2174/138945006777709557
Mahmood, Y. H., Jamshidi, S., Mark Sutton, J. M., and Rahman, K. (2016). Current advances in developing inhibitors of bacterial multidrug efflux pumps. Curr. Med. Chem. 23, 1062–1081. doi: 10.2174/0929867323666160304150522
Maisuria, V. B., Hosseinidoust, Z., and Tufenkji, N. (2015). Polyphenolic extract from maple syrup potentiates antibiotic susceptibility and reduces biofilm formation of pathogenic bacteria. Appl. Environ. Microbiol. 81, 3782–3792. doi: 10.1128/AEM.00239-15
Malléa, M., Chevalier, J., Eyraud, A., and Pagès, J. M. (2002). Inhibitors of antibiotic efflux pump in resistant Enterobacter aerogenes strains. Biochem. Biophys. Res. Commun. 293, 1370–1373. doi: 10.1016/S0006-291X(02)00404-7
Malléa, M., Mahamoud, A., Chevalier, J., Alibert-Franco, S., Brouant, P., Barbe, J., et al. (2003). Alkylaminoquinolines inhibit the bacterial antibiotic efflux pump in multidrug-resistant clinical isolates. Biochem. J. 376, 801–805. doi: 10.1042/BJ20030963
Mamelli, L., Amoros, J. P., Pagès, J. M., and Bolla, J. M. (2003). A phenylalanine–arginine β-naphthylamide sensitive multidrug efflux pump involved in intrinsic and acquired resistance of Campylobacter to macrolides. Int. J. Antimicrob. Agents. 22, 237–241. doi: 10.1016/S0924-8579(03)00199-7
Marger, M. D., and Saier Jr, M. H. (1993). A major superfamily of transmembrane facilitators that catalyse uniport, symport and antiport. Trends Biochem. Sci. 18, 13–20. doi: 10.1016/0968-0004(93)90081-w
Martins, M., Dastidar, S. G., Fanning, S., Kristiansen, J. E., Molnar, J., Pages, J. M., et al. (2008). Potential role of non-antibiotics (helper compounds) in the treatment of multidrug-resistant Gram-negative infections: mechanisms for their direct and indirect activities. Int. J. Antimicrob. Agents. 31, 198–208. doi: 10.1016/j.ijantimicag.2007.10.025
Masuda, S., Terada, T., Yonezawa, A., Tanihara, Y., Kishimoto, K., Katsura, T., et al. (2006). Identification and functional characterization of a new human kidney–specific H+/organic cation antiporter, kidney-specific multidrug and toxin extrusion 2. J. Am. Soc. Nephrol. 17, 2127–2135. doi: 10.1681/ASN.2006030205
Maurya, A., Dwivedi, G. R., Darokar, M. P., and Srivastava, S. K. (2013). Antibacterial and synergy of clavine alkaloid lysergol and its derivatives against nalidixic acid-resistant Escherichia coli. Chem. Biol. Drug Des. 81, 484–490. doi: 10.1111/cbdd.12103
Mi, W., Li, Y., Yoon, S. H., Ernst, R. K., Walz, T., and Liao, M. (2017). Structural basis of MsbA-mediated lipopolysaccharide transport. Nature. 549, 233–237. doi. 10.1038/nature23649
Mitchell, C. J., Stone, T. A., and Deber, C. M. (2019). Peptide-based efflux pump inhibitors of the small multidrug resistance protein from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 63, e00730–e00719. doi: 10.1128/AAC.00730-19
Miyauchi, H., Moriyama, S., Kusakizako, T., Kumazaki, K., Nakane, T., Yamashita, K., et al. (2017). Structural basis for xenobiotic extrusion by eukaryotic MATE transporter. Nat. Commun. 8, 1–11. doi: 10.1038/s41467-017-01541-0
Morita, Y., Kodama, K., Shiota, S., Mine, T., Kataoka, A., and Mizushima, T. (1998). NorM, a putative multidrug efflux protein, of Vibrio parahaemolyticus and its homolog in Escherichia coli. Antimicrob. Agents Chemother. 42, 1778–1782. doi: 10.1128/aac.42.7.1778
Mousa, J. J., Newsome, R. C., Yang, Y., Jobin, C., and Bruner, S. D. (2017). ClbM is a versatile, cation-promiscuous MATE transporter found in the colibactin biosynthetic gene cluster. Biochem. Biophys. Res. Commun. 482, 1233–1239. doi: 10.1016/j.bbrc.2016.12.018
Mousa, J. J., Yang, Y., Tomkovich, S., Shima, A., Newsome, R. C., Tripathi, P., et al. (2016). MATE transport of the E. coli-derived genotoxin colibactin. Nat. Microbiol. 1, 1–7. doi: 10.1038/nmicrobiol.2015.9
Mu, Y., Shen, Z., Jeon, B., Dai, L., and Zhang, Q. (2013). Synergistic effects of anti-CmeA and anti-CmeB peptide nucleic acids on sensitizing Campylobacter jejuni to antibiotics. Antimicrob. Agents Chemother. 57, 4575–4577. doi: 10.1128/AAC.00605-13
Munita, J. M., and Arias, C. A. (2016). Mechanisms of antibiotic resistance. Microbiol. Spectr. 4, 4–2. doi: 10.1128/microbiolspec.VMBF-0016-2015
Muniz, D. F., dos Santos Barbosa, C. R., de Menezes, I. R. A., de Sousa, E. O., Pereira, R. L. S., Júnior, J. T. C., et al. (2021). In vitro and in silico inhibitory effects of synthetic and natural eugenol derivatives against the NorA efflux pump in Staphylococcus aureus. Food. Chem. 337, 127776. doi: 10.1016/j.foodchem.2020.127776
Murakami, S., Nakashima, R., Yamashita, E., Matsumoto, T., and Yamaguchi, A. (2006). Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature 443, 173–179. doi: 10.1038/nature05076
Murakami, S., Nakashima, R., Yamashita, E., and Yamaguchi, A. (2002). Crystal structure of bacterial multidrug efflux transporter AcrB. Nature 419, 587–593. doi: 10.1038/nature01050
Nakashima, R., Sakurai, K., Yamasaki, S., Hayashi, K., Nagata, C., Hoshino, K., et al. (2013). Structural basis for the inhibition of bacterial multidrug exporters. Nature 500, 102–106. doi: 10.1038/nature12300
Nakashima, R., Sakurai, K., Yamasaki, S., Nishino, K., and Yamaguchi, A. (2011). Structures of the multidrug exporter AcrB reveal a proximal multisite drug-binding pocket. Nature 480, 565–569. doi: 10.1038/nature10641
Negi, N., Prakash, P., Gupta, M. L., and Mohapatra, T. M. (2014). Possible role of curcumin as an efflux pump inhibitor in multi drug resistant clinical isolates of Pseudomonas aeruginosa. J. Clin. Diagnostic. Res. 8, DC04. doi: 10.7860/JCDR/2014/8329.4965
Nelson, J. W., Atilho, R. M., Sherlock, M. E., Stockbridge, R. B., and Breaker, R. R. (2017). Metabolism of free guanidine in bacteria is regulated by a widespread riboswitch class. Mol. Cell 65, 220–230. doi: 10.1016/j.molcel.2016.11.019
Nelson, M. L., Park, B. H., Andrews, J. S., Georgian, V. A., Thomas, R. C., and Levy, S. B. (1993). Inhibition of the tetracycline efflux antiport protein by 13-thio-substituted 5-hydroxy-6-deoxytetracyclines. J. Med. Chem. 36, 370–377. doi: 10.1021/jm00055a008
Nelson, M. L., Park, B. H., and Levy, S. B. (1994). Molecular requirements for the inhibition of the tetracycline antiport protein and the effect of potent inhibitors on the growth of tetracycline-resistant bacteria. J. Med. Chem. 37, 1355–1361. doi: 10.1021/jm00035a016
Neuberger, A., Du, D., and Luisi, B. F. (2018). Structure and mechanism of bacterial tripartite efflux pumps. Res. Microbiol. 169, 401–413. doi: 10.1016/j.resmic.2018.05.003
Nikaido, H. (1996). Multidrug efflux pumps of gram-negative bacteria. J. Bacteriol. 178, 5853–5859. doi: 10.1128/jb.178.20.5853-5859.1996
Nikaido, H. (2009). Multidrug resistance in bacteria Annu. Rev. Biochem. 78, 119–146. doi: 10.1146/annurev.biochem.78.082907.145923
Nikaido, H. (2011). Structure and mechanism of RND-type multidrug efflux pumps. Adv. Enzymol. Relat. Areas Mol. Biol. 77, 1. doi: 10.1002/9780470920541.ch1
Nikaido, H., and Pagès, J. M. (2012). Broad-specificity efflux pumps and their role in multidrug resistance of Gram-negative bacteria. FEMS Microbiol. Rev. 36, 340–363. doi: 10.1111/j.1574-6976.2011.00290.x
Nishino, K., Nikaido, E., and Yamaguchi, A. (2009). Regulation and physiological function of multidrug efflux pumps in Escherichia coli and Salmonella. Biochim. Biophys. Acta Proteins. Proteom. 1794, 834–843. doi: 10.1016/j.bbapap.2009.02.002
Nishino, K., Senda, Y., and Yamaguchi, A. (2008). The AraC-family regulator GadX enhances multidrug resistance in Escherichia coli by activating expression of mdtEF multidrug efflux genes. J. Infect. Chemother. 14, 23–29. doi: 10.1007/s10156-007-0575-y
Nishino, K., and Yamaguchi, A. (2001). Analysis of a complete library of putative drug transporter genes in Escherichia coli. J. Bacteriol. 183, 5803–5812. doi: 10.1128/JB.183.20.5803-5812.2001
Nunez-Samudio, V., and Chesneau, O. (2013). Functional interplay between the ATP binding cassette Msr (D) protein and the membrane facilitator superfamily Mef (E) transporter for macrolide resistance in Escherichia coli. Res. Microbiol. 164, 226–235. doi: 10.1016/j.resmic.2012.12.003
Oethinger, M., Podglajen, I., Kern, W. V., and Levy, S. B. (1998). Overexpression of the marA or soxS regulatory gene in clinical topoisomerase mutants of Escherichia coli. Antimicrob. Agents. Chemother. 42, 2089–2094. doi: 10.1128/aac.42.8.2089
Ohene-Agyei, T., Mowla, R., Rahman, T., and Venter, H. (2014). Phytochemicals increase the antibacterial activity of antibiotics by acting on a drug efflux pump. Microbiologyopen 3, 885–896. doi: 10.1002/mbo3.212
Omote, H., Hiasa, M., Matsumoto, T., Otsuka, M., and Moriyama, Y. (2006). The MATE proteins as fundamental transporters of metabolic and xenobiotic organic cations. Trends Pharmacol. Sci. 27, 587–593. doi: 10.1016/j.tips.2006.09.001
O'Neill, J. (2015). Antimicrobials in Agriculture and the Environment: Reducing Unnecessary Use and Waste. The Review on Antimicrobial Resistance. 1–41.
Opperman, T. J., Kwasny, S. M., Kim, H. S., Nguyen, S. T., Houseweart, C., D'Souza, S., et al. (2014). Characterization of a novel pyranopyridine inhibitor of the AcrAB efflux pump of Escherichia coli. Antimicrob. Agents Chemother. 58, 722–733. doi: 10.1128/AAC.01866-13
Opperman, T. J., and Nguyen, S. T. (2015). Recent advances toward a molecular mechanism of efflux pump inhibition. Front. Microbiol. 6, 421. doi: 10.3389/fmicb.2015.00421
Otreebska-Machaj, E., Chevalier, J., Handzlik, J., Szymańska, E., Schabikowski, J., Boyer, G., et al. (2016). Efflux pump blockers in Gram-negative bacteria: the new generation of hydantoin based-modulators to improve antibiotic activity. Front. Microbiol. 7, 622. doi: 10.3389/fmicb.2016.00622
Otsuka, M., Matsumoto, T., Morimoto, R., Arioka, S., Omote, H., and Moriyama, Y. (2005). A human transporter protein that mediates the final excretion step for toxic organic cations. Proc. Natl. Acad. Sci. USA 102, 17923–17928. doi: 10.1073/pnas.0506483102
Padariya, M., Kalathiya, U., and Baginski, M. (2018). Structural and dynamic insights on the EmrE protein with TPP+ and related substrates through molecular dynamics simulations. Chem. Phys. Lipids. 212, 1–11. doi: 10.1016/j.chemphyslip.2017.12.004
Pal, C., Bengtsson-Palme, J., Kristiansson, E., and Larsson, D. G. (2015). Co-occurrence of resistance genes to antibiotics, biocides and metals reveals novel insights into their co-selection potential. BMC Genom. 16, 1–14. doi: 10.1186/s12864-015-2153-5
Pannek, S., Higgins, P. G., Steinke, P., Jonas, D., Akova, M., Bohnert, J. A., et al. (2006). Multidrug efflux inhibition in Acinetobacter baumannii: comparison between 1-(1-naphthylmethyl)-piperazine and phenyl-arginine-β-naphthylamide. J. Antimicrob. Chemother. 57, 970–974. doi: 10.1093/jac/dkl081
Pasqua, M., Bonaccorsi di Patti, M. C., Fanelli, G., Utsumi, R., Eguchi, Y., Trirocco, R., et al. (2021). Host-Bacterial pathogen communication: the Wily role of the multidrug efflux pumps of the MFS family. Front. Mol. Biosci. 8, 723274. doi: 10.3389/fmolb.2021.723274
Paulsen, I. T., Brown, M. H., and Skurray, R. A. (1996). Proton-dependent multidrug efflux systems. Microbiol. Rev. 60, 575–608. doi: 10.1128/mr.60.4.575-608.1996
Perez, C., Gerber, S., Boilevin, J., Bucher, M., Darbre, T., Aebi, M., et al. (2015). Structure and mechanism of an active lipid-linked oligosaccharide flippase. Nature. 524, 433–438. doi: 10.1038/nature14953
Pérez-Varela, M., Corral, J., Aranda, J., and Barbé, J. (2019). Roles of efflux pumps from different superfamilies in the surface-associated motility and virulence of Acinetobacter baumannii ATCC 17978. Antimicrob. Agents Chemother. 63, e02190–e02118. doi: 10.1128/AAC.02190-18
Piddock, L. J. (2006a). Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin. Microbiol. Rev. 19, 382–402. doi: 10.1128/CMR.19.2.382-402.2006
Piddock, L. J. (2006b). Multidrug-resistance efflux pumps? Not just for resistance. Nat. Rev. Microbiol. 4, 629–636. doi: 10.1038/nrmicro1464
Piddock, L. J., Garvey, M. I., Rahman, M. M., and Gibbons, S. (2010). Natural and synthetic compounds such as trimethoprim behave as inhibitors of efflux in Gram-negative bacteria. J. Antimicrob. Chemother. 65, 1215–1223. doi: 10.1093/jac/dkq079
Poole, K. (2007). Efflux pumps as antimicrobial resistance mechanisms. Ann. Med. 39, 162–176. doi: 10.1080/07853890701195262
Pos, K. M. (2009). Drug transport mechanism of the AcrB efflux pump. Biochim. Biophys. Acta Proteins. Proteom. 1794, 782–793. doi: 10.1016/j.bbapap.2008.12.015
Prasch, S., and Bucar, F. (2015). Plant derived inhibitors of bacterial efflux pumps: an update. Phytochem. Rev. 14, 961–974. doi: 10.1007/s11101-015-9436-y
Quistgaard, E. M., Löw, C., Guettou, F., and Nordlund, P. (2016). Understanding transport by the major facilitator superfamily (MFS): structures pave the way. Nat. Rev. Mol. Cell. Biol. 17, 123–132. doi: 10.1038/nrm.2015.25
Radchenko, M., Symersky, J., Nie, R., and Lu, M. (2015). Structural basis for the blockade of MATE multidrug efflux pumps. Nat. Commun. 6, 1–11. doi: 10.1038/ncomms8995
Raturi, S., Nair, A. V., Shinoda, K., Singh, H., Bai, B., Murakami, S., et al. (2021). Engineered MATE multidrug transporters reveal two functionally distinct ion-coupling pathways in NorM from Vibrio cholerae. Commun. Biol. 4, 558. doi: 10.1038/s42003-021-02081-6
Remy, E., and Duque, P. (2014). Beyond cellular detoxification: a plethora of physiological roles for MDR transporter homologs in plants. Front. Physiol. 5, 201. doi: 10.3389/fphys.2014.00201
Renau, T. E., Léger, R., Flamme, E. M., Sangalang, J., She, M. W., Yen, R., et al. (1999). Inhibitors of efflux pumps in Pseudomonas aeruginosa potentiate the activity of the fluoroquinolone antibacterial levofloxacin. J. Med. Chem. 42, 4928–4931. doi: 10.1021/jm9904598
Ribera, A., Ruiz, J., Jiminez de Anta, M. T., and Vila, J. (2002). Effect of an efflux pump inhibitor on the MIC of nalidixic acid for Acinetobacter baumannii and Stenotrophomonas maltophilia clinical isolates. J. Antimicrob. Chemother. 49, 697–698. doi: 10.1093/jac/49.4.697
Richmond, G. E., Evans, L. P., Anderson, M. J., Wand, M. E., Bonney, L. C., Ivens, A., et al. (2016). The Acinetobacter baumannii two-component system AdeRS regulates genes required for multidrug efflux, biofilm formation, and virulence in a strain-specific manner. MBio 7, e00430–e00416. doi: 10.1128/mBio.00430-16
Robin, B., Nicol, M., Le, H., Tahrioui, A., Schaumann, A., Vuillemenot, J. B., et al. (2022). MacAB-TolC contributes to the development of Acinetobacter baumannii biofilm at the solid–liquid interface. Front. Microbiol. 12, 3878. doi: 10.3389/fmicb.2021.785161
Rotem, D., and Schuldiner, S. (2004). EmrE, a multidrug transporter from Escherichia coli, transports monovalent and divalent substrates with the same stoichiometry. J. Biol. Chem. 279, 48787–48793. doi: 10.1074/jbc.M408187200
Russell, A. D. (2002). Introduction of biocides into clinical practice and the impact on antibiotic-resistant bacteria. J. Appl. Microbiol. 92, 121S−135S. doi: 10.1046/j.1365-2672.92.5s1.12.x
Sabatini, S., Piccioni, M., Felicetti, T., De Marco, S., Manfroni, G., Pagiotti, R., et al. (2017). Investigation on the effect of known potent S. aureus NorA efflux pump inhibitors on the staphylococcal biofilm formation. RSC. Adv. 7, 37007–37014. doi: 10.1039/C7RA03859C
Saier Jr, M. H., Reddy, V. S., Tsu, B. V., Ahmed, M. S., Li, C., and Moreno-Hagelsieb, G. (2016). The transporter classification database (TCDB): recent advances. Nucleic Acids Res. 44, D372–D379. doi: 10.1093/nar/gkv1103
Sakhtah, H., Koyama, L., Zhang, Y., Morales, D. K., Fields, B. L., Price-Whelan, A., et al. (2016). The Pseudomonas aeruginosa efflux pump MexGHI-OpmD transports a natural phenazine that controls gene expression and biofilm development. Proc. Natl. Acad. Sci. USA 113, E3538–E3547. doi: 10.1073/pnas.1600424113
Sakurai, K., Yamasaki, S., Nakao, K., Nishino, K., Yamaguchi, A., and Nakashima, R. (2019). Crystal structures of multidrug efflux pump MexB bound with high-molecular-mass compounds. Sci. Rep. 9, 1–9. doi: 10.1038/s41598-019-40232-2
Salehi, B., Ghalavand, Z., Yadegar, A., and Eslami, G. (2021). Characteristics and diversity of mutations in regulatory genes of resistance-nodulation-cell division efflux pumps in association with drug-resistant clinical isolates of Acinetobacter baumannii. Antimicrob. Resist. Infect. Control. 10, 1–12. doi: 10.1186/s13756-021-00924-9
Sato, Y., Unno, Y., Ubagai, T., and Ono, Y. (2018). Sub-minimum inhibitory concentrations of colistin and polymyxin B promote Acinetobacter baumannii biofilm formation. PLoS ONE 13, e0194556. doi: 10.1371/journal.pone.0194556
Schaedler, T. A., and Van Veen, H. W. (2010). A flexible cation binding site in the multidrug major facilitator superfamily transporter LmrP is associated with variable proton coupling. FASEB J. 24, 3653–3661. doi: 10.1096/fj.10-156927
Schumacher, A., Steinke, P., Bohnert, J. A., Akova, M., Jonas, D., and Kern, W. V. (2006). Effect of 1-(1-naphthylmethyl)-piperazine, a novel putative efflux pump inhibitor, on antimicrobial drug susceptibility in clinical isolates of Enterobacteriaceae other than Escherichia coli. J. Antimicrob. Chemother. 57, 344–348. doi: 10.1093/jac/dki446
Seeger, M. A., Diederichs, K., Eicher, T., Brandstatter, L., Schiefner, A., Verrey, F., et al. (2008). The AcrB efflux pump: conformational cycling and peristalsis lead to multidrug resistance. Curr. Drug. Targets 9, 729–749. doi: 10.2174/138945008785747789
Seeger, M. A., Schiefner, A., Eicher, T., Verrey, F., Diederichs, K., and Pos, K. M. (2006). Structural asymmetry of AcrB trimer suggests a peristaltic pump mechanism. Science 313, 1295–1298. doi: 10.1126/science.1131542
Seeger, M. A., von Ballmoos, C., Verrey, F., and Pos, K. M. (2009). Crucial role of Asp408 in the proton translocation pathway of multidrug transporter AcrB: evidence from site-directed mutagenesis and carbodiimide labeling. Biochemistry 48, 5801–5812. doi: 10.1021/bi900446j
Sennhauser, G., Bukowska, M. A., Briand, C., and Grütter, M. G. (2009). Crystal structure of the multidrug exporter MexB from Pseudomonas aeruginosa. J. Mol. Biol. 389, 134–145. doi: 10.1016/j.jmb.2009.04.001
Shcherbakov, A. A., Hisao, G., Mandala, V. S., Thomas, N. E., Soltani, M., Salter, E. A., et al. (2021). Structure and dynamics of the drug-bound bacterial transporter EmrE in lipid bilayers. Nat. Commun. 12, 1–13. doi: 10.1038/s41467-020-20468-7
Shi, K., Cao, M., Li, C., Huang, J., Zheng, S., and Wang, G. (2019). Efflux proteins MacAB confer resistance to arsenite and penicillin/macrolide-type antibiotics in Agrobacterium tumefaciens 5A. World J. Microbiol. Biotechnol. 35, 1–10. doi: 10.1007/s11274-019-2689-7
Shimizu, T., Harada, K., Tsuyuki, Y., Kimura, Y., Miyamoto, T., Hatoya, S., et al. (2017). In vitro efficacy of 16 antimicrobial drugs against a large collection of β-lactamase-producing isolates of extraintestinal pathogenic Escherichia coli from dogs and cats J. Med. Microbiol. 66, 1085–1091. doi: 10.1099/jmm.0.000535
Shirshikova, T. V., Sierra-Bakhshi, C. G., Kamaletdinova, L. K., Matrosova, L. E., Khabipova, N. N., Evtugyn, V. G., et al. (2021). The ABC-type efflux pump MacAB is involved in protection of Serratia marcescens against aminoglycoside antibiotics, polymyxins, and oxidative stress. Msphere 6, e00033–e00021. doi: 10.1128/mSphere.00033-21
Shiu, W. K., Malkinson, J. P., Rahman, M. M., Curry, J., Stapleton, P., Gunaratnam, M., et al. (2013). A new plant-derived antibacterial is an inhibitor of efflux pumps in Staphylococcus aureus. Int. J. Antimicrob. Agents. 42, 513–518. doi: 10.1016/j.ijantimicag.2013.08.007
Singh, A. K., Haldar, R., Mandal, D., and Kundu, M. (2006). Analysis of the topology of Vibrio cholerae NorM and identification of amino acid residues involved in norfloxacin resistance. Antimicrob. Agents. Chemother. 50, 3717–3723. doi: 10.1128/AAC.00460-06
Siriyong, T., Srimanote, P., Chusri, S., Yingyongnarongkul, B. E., Suaisom, C., Tipmanee, V., et al. (2017). Conessine as a novel inhibitor of multidrug efflux pump systems in Pseudomonas aeruginosa. BMC Complement Altern. Med. 17, 1–7. doi: 10.1186/s12906-017-1913-y
Sjuts, H., Vargiu, A. V., Kwasny, S. M., Nguyen, S. T., Kim, H. S., Ding, X., et al. (2016). Molecular basis for inhibition of AcrB multidrug efflux pump by novel and powerful pyranopyridine derivatives. Proc. Natl. Acad. Sci. USA. 113, 3509–3514. doi: 10.1073/pnas.1602472113
Smirnova, I., Kasho, V., and Kaback, H. R. (2011). Lactose permease and the alternating access mechanism. Biochemistry 50, 9684–9693. doi: 10.1021/bi2014294
Solnier, J., Martin, L., Bhakta, S., and Bucar, F. (2020). Flavonoids as novel efflux pump inhibitors and antimicrobials against both environmental and pathogenic intracellular mycobacterial species. Molecules 25, 734. doi: 10.3390/molecules25030734
Sonnet, P., Izard, D., and Mullié, C. (2012). Prevalence of efflux-mediated ciprofloxacin and levofloxacin resistance in recent clinical isolates of Pseudomonas aeruginosa and its reversal by the efflux pump inhibitors 1-(1-naphthylmethyl)-piperazine and phenylalanine-arginine-β-naphthylamide. Int. J. Antimicrob. Agents. 39, 77–80. doi: 10.1016/j.ijantimicag.2011.08.005
Srinivasan, V. B., and Rajamohan, G. (2013). KpnEF, a new member of the Klebsiella pneumoniae cell envelope stress response regulon, is an SMR-type efflux pump involved in broad-spectrum antimicrobial resistance. Antimicrob. Agents. Chemother. 57, 4449–4462. doi: 10.1128/AAC.02284-12
Su, C. C., Li, M., Gu, R., Takatsuka, Y., McDermott, G., Nikaido, H., et al. (2006). Conformation of the AcrB multidrug efflux pump in mutants of the putative proton relay pathway. J. Bacteriol. 188, 7290–7296. doi: 10.1128/JB.00684-06
Sun, J., Deng, Z., and Yan, A. (2014). Bacterial multidrug efflux pumps: mechanisms, physiology and pharmacological exploitations. Biochem. Biophys. Res. Commun. 453, 254–267. doi: 10.1016/j.bbrc.2014.05.090
Tam, H. K., Malviya, V. N., Foong, W. E., Herrmann, A., Malloci, G., Ruggerone, P., et al. (2020). Binding and transport of carboxylated drugs by the multidrug transporter AcrB. J. Mol. Biol. 432, 861–877. doi: 10.1016/j.jmb.2019.12.025
Tanaka, Y., Hipolito, C. J., Maturana, A. D., Ito, K., Kuroda, T., Higuchi, T., et al. (2013). Structural basis for the drug extrusion mechanism by a MATE multidrug transporter. Nature 496, 247–251. doi: 10.1038/nature12014
Tanaka, Y., Iwaki, S., and Tsukazaki, T. (2017). Crystal structure of a plant multidrug and toxic compound extrusion family protein. Structure 25, 1455–1460. doi: 10.1016/j.str.2017.07.009
Taneja, N., and Sharma, M. (2019). Antimicrobial resistance in the environment: the Indian scenario. Indian. J. Med. Res. 149, 119–128. doi: 10.4103/ijmr.IJMR_331_18
Tanihara, Y., Masuda, S., Sato, T., Katsura, T., Ogawa, O., and Inui, K. I. (2007). Substrate specificity of MATE1 and MATE2-K, human multidrug and toxin extrusions/H+-organic cation antiporters. Biochem. Pharmacol. 74, 359–371. doi: 10.1016/j.bcp.2007.04.010
Tariq, A., Sana, M., Shaheen, A., Ismat, F., Mahboob, S., Rauf, W., et al. (2019). Restraining the multidrug efflux transporter STY4874 of Salmonella Typhi by reserpine and plant extracts. Lett. Appl. Microbiol. 69, 161–167. doi: 10.1111/lam.13196
Tatsuno, I., Isaka, M., Masuno, K., Hata, N., Matsumoto, M., and Hasegawa, T. (2018). Functional predominance of msr (D), which is more effective as mef (A)-associated than mef (E)-associated, over mef (A)/mef (E) in macrolide resistance in Streptococcus pyogenes. Microb. Drug. Resist. 24, 1089–1097. doi: 10.1089/mdr.2017.0277
Terada, T., and Inui, K. I. (2008). Physiological and pharmacokinetic roles of H+/organic cation antiporters (MATE/SLC47A). Biochem. Pharmacol. 75, 1689–1696. doi: 10.1016/j.bcp.2007.12.008
Thorarensen, A., Presley-Bodnar, A. L., Marotti, K. R., Boyle, T. P., Heckaman, C. L., Bohanon, M. J., et al. (2001). 3-Arylpiperidines as potentiators of existing antibacterial agents. Bioorganic Med. Chem. Lett. 11, 1903–1906. doi: 10.1016/S0960-894X(01)00330-4
Tintino, S. R., Morais-Tintino, C. D., Campina, F. F., Costa, M. D. S., Menezes, I. R., de Matos, Y. M. L., et al. (2017). Tannic acid affects the phenotype of Staphylococcus aureus resistant to tetracycline and erythromycin by inhibition of efflux pumps. Bioorg. Chem. 74, 197–200. doi: 10.1016/j.bioorg.2017.08.004
Van Bambeke, F., and Lee, V. J. (2006). Inhibitors of bacterial efflux pumps as adjuvants in antibiotic treatments and diagnostic tools for detection of resistance by efflux. Recent Pat. Antiinfect. Drug Discov. 1, 157–175. doi: 10.2174/157489106777452692
Van Veen, H. W. (2010). Last of the multidrug transporters. Nature 467, 926–927. doi: 10.1038/467926a
Vargiu, A. V., and Nikaido, H. (2012). Multidrug binding properties of the AcrB efflux pump characterized by molecular dynamics simulations. Proc. Natl. Acad. Sci. USA 109, 20637–20642. doi: 10.1073/pnas.1218348109
Vargiu, A. V., Ramaswamy, V. K., Malloci, G., Malvacio, I., Atzori, A., and Ruggerone, P. (2018). Computer simulations of the activity of RND efflux pumps. Res. Microbiol. 169, 384–392. doi: 10.1016/j.resmic.2017.12.001
Vargiu, A. V., Ruggerone, P., Opperman, T. J., Nguyen, S. T., and Nikaido, H. (2014). Molecular mechanism of MBX2319 inhibition of Escherichia coli AcrB multidrug efflux pump and comparison with other inhibitors. Antimicrob. Agents Chemother. 58, 6224–6234. doi: 10.1128/AAC.03283-14
Venter, H. (2019). Reversing resistance to counter antimicrobial resistance in the World Health Organisation's critical priority of most dangerous pathogens. Biosci. Rep. 39:BSR20180474. doi: 10.1042/BSR20180474
Venter, H., Mowla, R., Ohene-Agyei, T., and Ma, S. (2015). RND-type drug efflux pumps from Gram-negative bacteria: molecular mechanism and inhibition. Front. Microbiol. 6, 377. doi: 10.3389/fmicb.2015.00377
Vera-Leiva, A., Carrasco-Anabalón, S., Lima, C. A., Villagra, N., Domínguez, M., Bello-Toledo, H., et al. (2018). The efflux pump inhibitor phenylalanine-arginine β-naphthylamide (PAβN) increases resistance to carbapenems in Chilean clinical isolates of KPC-producing Klebsiella pneumoniae. J. Glob. Antimicrob. Resist. 12, 73–76. doi: 10.1016/j.jgar.2017.12.003
Verchère, A., Broutin, I., and Picard, M. (2012). Photo-induced proton gradients for the in vitro investigation of bacterial efflux pumps. Sci. Rep. 2, 1–4. doi: 10.1038/srep00306
Verhalen, B., Dastvan, R., Thangapandian, S., Peskova, Y., Koteiche, H. A., Nakamoto, R. K., et al. (2017). Energy transduction and alternating access of the mammalian ABC transporter P-glycoprotein. Nature. 543, 738–741. doi: 10.1038/nature21414
Wagner, D. J., Hu, T., and Wang, J. (2016). Polyspecific organic cation transporters and their impact on drug intracellular levels and pharmacodynamics. Pharmacol. Res. 111, 237–246. doi: 10.1016/j.phrs.2016.06.002
Walsh, C. (2003). Where will new antibiotics come from? Nat. Rev. Microbiol. 1, 65–70. doi: 10.1038/nrmicro727
Wang, Y., Mowla, R., Guo, L., Ogunniyi, A. D., Rahman, T., Lopes, M. A. D. B., et al. (2017). Evaluation of a series of 2-napthamide derivatives as inhibitors of the drug efflux pump AcrB for the reversal of antimicrobial resistance. Bioorg. Med. Chem. Lett. 27, 733–739. doi: 10.1016/j.bmcl.2017.01.042
Wang, Y., Mowla, R., Ji, S., Guo, L., Lopes, M. A. D. B., Jin, C., et al. (2018). Design, synthesis and biological activity evaluation of novel 4-subtituted 2-naphthamide derivatives as AcrB inhibitors. Eur. J. Med. Chem. 143, 699–709. doi: 10.1016/j.ejmech.2017.11.102
Wang, Z., Fan, G., Hryc, C. F., Blaza, J. N., Serysheva, I. I., Schmid, M. F., et al. (2017). An allosteric transport mechanism for the AcrAB-TolC multidrug efflux pump. Elife 6, e24905. doi: 10.7554/eLife.24905
Webber, M. A., and Piddock, L. J. V. (2003). The importance of efflux pumps in bacterial antibiotic resistance. J. Antimicrob. Chemother. 51, 9–11. doi: 10.1093/jac/dkg050
Wehmeier, C., Schuster, S., Fähnrich, E., Kern, W. V., and Bohnert, J. A. (2009). Site-directed mutagenesis reveals amino acid residues in the Escherichia coli RND efflux pump AcrB that confer macrolide resistance. Antimicrob. Agents. Chemother. 53, 329–330. doi: 10.1128/AAC.00921-08
Woebking, B., Reuter, G., Shilling, R. A., Velamakanni, S., Shahi, S., Venter, H., et al. (2005). Drug-lipid A interactions on the Escherichia coli ABC transporter MsbA. J. Bacteriol. 187, 6363–6369. doi: 10.1128/JB.187.18.6363-6369.2005
Wolloscheck, D., Krishnamoorthy, G., Nguyen, J., and Zgurskaya, H. I. (2018). Kinetic control of quorum sensing in Pseudomonas aeruginosa by multidrug efflux pumps. ACS Infect. Dis. 4, 185–195. doi: 10.1021/acsinfecdis.7b00160
Woo, J. S., Zeltina, A., Goetz, B. A., and Locher, K. P. (2012). X-ray structure of the Yersinia pestis heme transporter HmuUV. Nat. Struct. Mol. Biol. 19, 1310–1315. doi: 10.1038/nsmb.2417
Wright, D. J., and Tate, C. G. (2015). Isolation and characterisation of transport-defective substrate-binding mutants of the tetracycline antiporter TetA (B). Biochim. Biophys. Acta. Biomembr. 1848, 2261–2270. doi: 10.1016/j.bbamem.2015.06.026
Xu, S., Chen, G., Liu, Z., Xu, D., Wu, Z., Li, Z., et al. (2020). Site-directed mutagenesis reveals crucial residues in Escherichia coli resistance-nodulation-division efflux pump OqxB. Microb. Drug. Resist. 26, 550–560. doi: 10.1089/mdr.2019.0165
Yamaguchi, A., Nakashima, R., and Sakurai, K. (2015). Structural basis of RND-type multidrug exporters. Front. Microbiol. 6, 327. doi: 10.3389/fmicb.2015.0032
Yan, N. (2013). Structural advances for the major facilitator superfamily (MFS) transporters. Trends. Biochem. Sci. 38, 151–159. doi: 10.1016/j.tibs.2013.01.003
Yang, X., Domalaon, R., Lyu, Y., Zhanel, G. G., and Schweizer, F. (2018). Tobramycin-linked efflux pump inhibitor conjugates synergize fluoroquinolones, rifampicin and fosfomycin against multidrug-resistant Pseudomonas aeruginosa. J. Clin. Med. 7, 158. doi: 10.3390/jcm7070158
Yang, X., Goswami, S., Gorityala, B. K., Domalaon, R., Lyu, Y., Kumar, A., et al. (2017). A tobramycin vector enhances synergy and efficacy of efflux pump inhibitors against multidrug-resistant Gram-negative bacteria. J. Med. Chem. 60, 3913–3932. doi: 10.1021/acs.jmedchem.7b00156
Yerushalmi, H., Lebendiker, M., and Schuldiner, S. (1995). EmrE, an Escherichia coli 12-kDa multidrug transporter, exchanges toxic cations and H+ and is soluble in organic solvents. J. Biol. Chem. 270, 6856–6863. doi: 10.1074/jbc.270.12.6856
Yerushalmi, H., Lebendiker, M., and Schuldiner, S. (1996). Negative dominance studies demonstrate the oligomeric structure of EmrE, a multidrug antiporter from Escherichia coli. J. Biol. Chem. 271, 31044–31048. doi: 10.1074/jbc.271.49.31044
Yokoo, S., Yonezawa, A., Masuda, S., Fukatsu, A., Katsura, T., and Inui, K. I. (2007). Differential contribution of organic cation transporters, OCT2 and MATE1, in platinum agent-induced nephrotoxicity. Biochem. Pharmacol. 74, 477–487. doi: 10.1016/j.bcp.2007.03.004
Yoon, E. J., Balloy, V., Fiette, L., Chignard, M., Courvalin, P., and Grillot-Courvalin, C. (2016). Contribution of the Ade resistance-nodulation-cell division-type efflux pumps to fitness and pathogenesis of Acinetobacter baumannii. MBio 7, e00697–e00616. doi: 10.1128/mBio.00697-16
Yoshida, K. I., Nakayama, K., Kuru, N., Kobayashi, S., Ohtsuka, M., Takemura, M., et al. (2006). MexAB-OprM specific efflux pump inhibitors in Pseudomonas aeruginosa. Part 5: Carbon-substituted analogues at the C-2 position. Bioorg. Med. Chem. Lett. 14, 1993–2004. doi: 10.1016/j.bmc.2005.10.043
Yoshida, K. I., Nakayama, K., Ohtsuka, M., Kuru, N., Yokomizo, Y., Sakamoto, A., et al. (2007). MexAB-OprM specific efflux pump inhibitors in Pseudomonas aeruginosa. Part 7: highly soluble and in vivo active quaternary ammonium analogue D13-9001, a potential preclinical candidate. Bioorg. Med. Chem. Lett. 15, 7087–7097. doi: 10.1016/j.bmc.2007.07.039
Yu, K., Zhang, Y., Xu, W., Zhang, X., Xu, Y., Sun, Y., et al. (2020). Hyper-expression of the efflux pump gene adeB was found in Acinetobacter baumannii with decreased triclosan susceptibility. J. Glob. Antimicrob. Resist. 22, 367–373. doi: 10.1016/j.jgar.2020.02.027
Zeng, B., Wang, H., Zou, L., Zhang, A., Yang, X., and Guan, Z. (2010). Evaluation and target validation of indole derivatives as inhibitors of the AcrAB-TolC efflux pump. Biosci. Biotechnol. Biochem. 74, 2237–2241. doi: 10.1271/bbb.100433
Zgurskaya, H. I. (2009). Covalently linked AcrB giant offers a new powerful tool for mechanistic analysis of multidrug efflux in bacteria. J. Bacteriol. 191, 1727–1728. doi: 10.1128/JB.01718-08
Zhang, Y., Tatsuno, I., Okada, R., Hata, N., Matsumoto, M., Isaka, M., et al. (2016). Predominant role of msr (D) over mef (A) in macrolide resistance in Streptococcus pyogenes. Microbiology 162, 46–52. doi: 10.1099/mic.0.000206
Zhu, Y. G., Zhao, Y. I., Li, B., Huang, C. L., Zhang, S. Y., Yu, S., et al. (2017). Continental-scale pollution of estuaries with antibiotic resistance genes. Nat. Microbiol. 2, 1–7. doi: 10.1038/nmicrobiol.2016.270
Zwama, M., Hayashi, K., Sakurai, K., Nakashima, R., Kitagawa, K., Nishino, K., et al. (2017). Hoisting-loop in bacterial multidrug exporter AcrB is a highly flexible hinge that enables the large motion of the subdomains. Front. Microbiol. 8, 2095. doi: 10.3389/fmicb.2017.02095
Zwama, M., and Nishino, K. (2021). Ever-adapting RND efflux pumps in gram-negative multidrug-resistant pathogens: a race against time. Antibiotics 10, 774. doi: 10.3390/antibiotics10070774
Zwama, M., and Yamaguchi, A. (2018). Molecular mechanisms of AcrB-mediated multidrug export. Res. Microbiol. 169, 372–383. doi: 10.1016/j.resmic.2018.05.005
Zwama, M., Yamaguchi, A., and Nishino, K. (2019). Phylogenetic and functional characterisation of the Haemophilus influenzae multidrug efflux pump AcrB. Commun. Biol. 2, 1–11. doi: 10.1038/s42003-019-0564-6
Keywords: efflux pump, MDR, antibiotic resistance, resistance nodulation division superfamily, efflux pump inhibitors (EPIs), Escherichia coli
Citation: Chetri S (2023) The culmination of multidrug-resistant efflux pumps vs. meager antibiotic arsenal era: Urgent need for an improved new generation of EPIs. Front. Microbiol. 14:1149418. doi: 10.3389/fmicb.2023.1149418
Received: 21 January 2023; Accepted: 13 March 2023;
Published: 17 April 2023.
Edited by:
Rami M. Elshazli, Horus University, EgyptReviewed by:
Padmani Sandhu, Institute of Microbial Technology (CSIR), IndiaCopyright © 2023 Chetri. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shiela Chetri, c29ueTI0MDY4OUBnbWFpbC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.