
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 29 March 2023
Sec. Extreme Microbiology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1147162
This article is part of the Research TopicInsights in Extreme Microbiology: 2022View all 11 articles
 Maryam Yavari-Bafghi1
Maryam Yavari-Bafghi1 Maryam Rezaei Somee2
Maryam Rezaei Somee2 Mohammad Ali Amoozegar1*
Mohammad Ali Amoozegar1* Seyed Mohammad Mehdi Dastgheib3
Seyed Mohammad Mehdi Dastgheib3 Mahmoud Shavandi3*
Mahmoud Shavandi3*Groundwater pollution is one of the major environmental concerns. The entrance of pollutants into the oligotrophic groundwater ecosystems alters native microbial community structure and metabolism. This study investigated the application of innovative Small Bioreactor Chambers and CaO2 nanoparticles for phenol removal within continuous-flow sand-packed columns for 6 months. Scanning electron microscopy and confocal laser scanning microscopy analysis were conducted to indicate the impact of attached biofilm on sand surfaces in bioremediation columns. Then, the influence of each method on the microbial biodiversity of the column’s groundwater was investigated by next-generation sequencing of the 16S rRNA gene. The results indicated that the simultaneous application of biostimulation and bioaugmentation completely eliminated phenol during the first 42 days. However, 80.2% of phenol remained in the natural bioremediation column at the end of the experiment. Microbial diversity was decreased by CaO2 injection while order-level groups known for phenol degradation such as Rhodobacterales and Xanthomonadales dominated in biostimulation columns. Genome-resolved comparative analyses of oligotrophic groundwater prokaryotic communities revealed that Burkholderiales, Micrococcales, and Cytophagales were the dominant members of the pristine groundwater. Six-month exposure of groundwater to phenol shifted the microbial population towards increasing the heterotrophic members of Desulfobacterales, Pseudomonadales, and Xanthomonadales with the degradation potential of phenol and other hydrocarbons.
In recent decades, increasing demand for petroleum hydrocarbon, leakage from storage tanks and pipelines, and unsafe waste transportation have become severe global environmental issues. The pollutants are easily transported by the natural water flow and contaminate the downstream groundwater (Koshlaf et al., 2016; Wang et al., 2016; Lu et al., 2017). Phenols are the major organic compounds present in the effluents of various industries (e.g., petroleum), which contaminate groundwater by infiltration through the soil of the polluted area (Mohammadi et al., 2015; Sun et al., 2015). They are considered the main pollutants having high solubility in water (84.2 g L–1) and hence carcinogenic impacts on humans, animals, and plants (Villegas et al., 2016).
Groundwater, as a most critical water resource, is widely used in washing and irrigation processes for about 40% of the world’s agricultural products and also influences the rates and types of biogeochemical cycles in water networks (Lall et al., 2020). Although all domains of life are present and active in groundwater, archaea and bacteria showed a clear dominance over eukarya (Lopez-Fernandez et al., 2018). Prokaryotes, with estimated total abundances of 102–106 cells, have a vital role in the biogeochemical cycles (Clark et al., 2018). The entrance of hydrocarbon pollutants into the oligotrophic groundwater ecosystems will increase microbial metabolism, change their community structure, and affect their role in cycling elements (Flynn et al., 2008). Therefore, pollutant removal from groundwater has great importance.
Among the numerous physical, chemical, and biological groundwater treatment approaches, bioremediation has been widely used as a safe, economical, and eco-friendly method in recent years (Dong et al., 2019; Cecconet et al., 2020). However, it is a time-consuming process due to the limited amount of dissolved oxygen (DO) and low microbial count of the groundwater (Lu et al., 2017). To overcome this problem, biostimulation [by injection of oxygen-releasing compounds (ORCs)] and bioaugmentation (using exogenous microbial consortia inoculation) were applied in several studies (Pickup et al., 2001; Mikkonen et al., 2018).
Oxygen-releasing compounds, like calcium peroxide (CaO2), decompose to oxygen and hydroxyl radicals after exposure to water and accelerate the contaminant degradation (Mosmeri et al., 2018, 2019). According to previous studies, the generated O2 stimulates the intrinsic groundwater microorganisms, and OH● radicals destroy the aromatic structure of pollutants (Yeh et al., 2010; Gholami et al., 2019). CaO2 nanoparticles have been applied for groundwater treatment in a lot of research (Cassidy and Irvine, 1999; Xue et al., 2018; Mosmeri et al., 2019). For instance, in Qian et al. (2016) study, 2,4-dichlorophenol was removed by nanoscale CaO2 from groundwater. On the other side, the application of bioaugmentation to treat groundwater samples polluted with phenol, arsenic, and total petroleum-hydrocarbons (TPH) has recently received increasing interest (Dey et al., 2016; Hedbavna et al., 2016; Poi et al., 2018; Zhuang and Fang, 2020). However, achieving and maintaining a sufficient microbial mass over time is the main challenge for implementing bioaugmentation. In our previous study, Small Bioreactor Chambers (SBCs) were developed that enabled high concentrations of external specific microbial consortiums to stay active (alive) and grow normally inside the 3D chambers in a suspended state for phenol removal of groundwater. The innovative design of SBCs allowed diffusion of phenol, DO, and nutrients into the encapsulated culture, however, confined the inner bacteria due to the presence of cellulose acetate membranes with pores of up to 0.22 μm in diameter. In addition, the SBCs contained a lyophilized specific consortium which was in a dry state and not active. The penetration of water through the membrane activated the freeze-dried culture and as long as the nutrients and DO were present in the water, the consortium was active and stable (Yavari-Bafghi et al., 2022). Due to the specific SBCs structure, indigenous or consortium bacteria, cannot pass through the 0.22 microfiltration (MF) membrane and as a result, SBCs are more environmentally friendly than polymer beads (González, 2001; Duan et al., 2016).
Despite the vast number of publications on the ecological characteristics of groundwater, the microbial community dynamics and the metabolic context in response to pollutant exposure during the bioremediation process, especially in Iran, have been less noticed.
Current work aims to (i) investigate the performance and impact of simultaneous use of CaO2 nanoparticles and SBCs on the intrinsic microorganisms of oligotrophic groundwater within phenol bioremediation during continuous-flow experiments (pilot level), (ii) survey the microbial community dynamics of the groundwater along the phenol pollution, and (iii) study the microbial potential metabolic capabilities in hydrocarbon degradation and biogeochemical cycles. To achieve these objectives, we conducted a 6-month biostimulation and bioaugmentation investigation through the sand-packed columns with a continuous flow of phenol-polluted groundwater. The genome-resolved comparative analyses of the pristine and phenol-contaminated groundwater were performed afterward.
Phenol (99.99%), calcium chloride (99.99%), sodium hydroxide (99.99%), sodium azide, and glutaraldehyde (25% in H2O, grade l) were prepared from Sigma-Aldrich, R2A agar, and H2O2 (30%) were purchased from Merck. All other chemicals were of analytical grade and obtained from commercial sources. The sand and groundwater used in this study were prepared as described in our previous work (Yavari-Bafghi et al., 2022).
Calcium peroxide nanoparticles were synthesized based on calcium chloride (CaCl2) and hydrogen peroxide reaction conforming to the procedure described by Khodaveisi et al. (2011).
According to the results of batch experiments in our previous work (Yavari-Bafghi et al., 2022), column experiments were conducted to investigate the CaO2 nanoparticles and SBCs performance in phenol removal from oligotrophic groundwater through the permeable reactive barrier (PRB). Four continuous up-flow Plexiglas reactors (100 cm length and 9 cm inner diameter) were packed with underground originated acid-washed and autoclaved sands with particle size in the 2–4 mm range. The phenol (100 mg/L) contaminated groundwater was passed through the columns with a 20 cm/day flow rate. To simulate the underground conditions, all the experiments were carried out in a cold room with a temperature of 15 ± 0.5°C.
As illustrated in Figure 1, column I was considered the abiotic column in which the filter-sterilized phenol-contaminated groundwater containing 1 g/L sodium azide was used as the inlet flow to study the chemical phenol removal efficiency by calcium peroxide nanoparticles. Column II was used to simulate the natural bioremediation process as the blank column. Columns III (CaO2 injected) and (IV) (CaO2 injected + SBCs placed) were applied to investigate the efficiency of biostimulation and bioaugmentation processes, respectively in the phenol removal of groundwater.
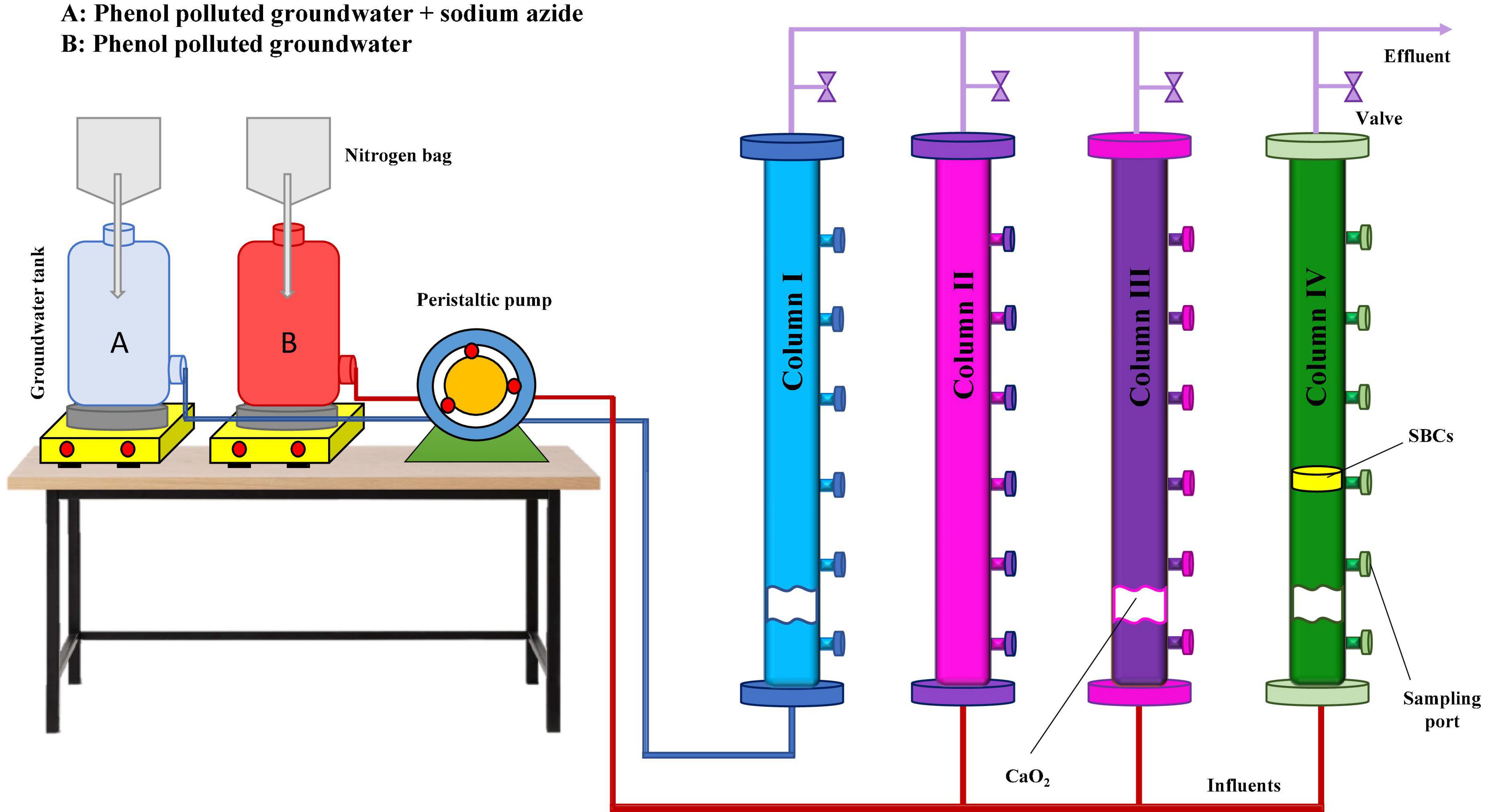
Figure 1. Schematic representation of column experiments. Column I: phenol removal in the abiotic condition by CaO2 nanoparticles (chemical), column II: natural bioremediation of phenol (blank), column III: phenol remediation by CaO2 nanoparticles and native groundwater microbial population (biostimulation), and column IV: phenol remediation by CaO2 nanoparticles and SBCs (biostimulation and bioaugmentation).
Before the experiments, groundwater from the feed tank (pH 7.4 ± 0.01 and DO of 4 mg/L) flowed through the packed columns for 4 weeks to perform the adaptation phase. Initially, 15 g of CaO2 nanoparticles were injected into columns I, III, and IV. Then, to maintain the desired conditions in the columns, 15 and 5 g of CaO2 were re-injected after 8 and 16 weeks, respectively. SBCs were placed in column IV simultaneously with the first CaO2 injection. DO, pH, phenol concentration, and microbial count were measured within 6 months.
The DO and pH of the water samples of each column effluent were measured using HQ40d multimeter (HACH). Phenol concentration in groundwater was analyzed by high-performance liquid chromatography (HPLC) (SPD-M10 A, Shimadzu, Japan). The HPLC analysis conditions were as previously presented by Yavari-Bafghi et al. (2022). Moreover, the total culturable microbial count of the effluent samples was measured by the standard plate count (SPC) method on R2A agar, and the colonies were counted by a standard colony counter (SC6 Plus, Stuart). At the end of the experiments, the physicochemical parameters of the effluent of each column were analyzed through standard methods (Supplementary Table 1) and compared with groundwater parameters reported in the authors’ previous study (Yavari-Bafghi et al., 2022).
Additionally, scanning electron microscopy (SEM) (VEGA\\TESCAN-XMU) and confocal laser scanning microscopy (CLSM) (Leica TCS SPE) were applied according to Gholami et al. (2018) and Inoué (2006), respectively, to study the microbial attachment and biofilm formation on the sand surfaces.
To study the effects of CaO2 and SBCs application on the structure and diversity of the oligotrophic groundwater microbial community by 16S rRNA gene amplicon sequencing, total microbial DNA was extracted from column’s sand surfaces using DNeasy® PowerSoil® (QIAGEN) according to the manufacturer’s protocol. In addition, the genomic DNA of the SBC’s consortium was extracted through the NucleoSpin® Microbial DNA isolation kit (Macherey-Nagel) according to the manufacturer’s protocol to investigate the consortium dynamics during the experiment in comparison to our previous study (Yavari-Bafghi et al., 2022). The quality and concentration of the extracted DNA were analyzed by NanoDrop UV–Vis spectrophotometer (ND-1000, USA). Additionally, to ensure the structural integrity of the extracted DNA, 5 μl DNA was loaded on 1.0% agarose gel and visualized under UV in a gel documentation system. The V3–V4 region of the 16S rRNA gene was analyzed using 341F and 806R universal primers and sequenced on the Illumina MiSeq platform (Novogene company, Hong Kong).
Paired-end reads were merged to tags based on overlaps using FLASH (V1.2.7) (Magoc and Salzberg, 2011). To obtain high-quality clean tags, quality filtering on the raw tags was performed according to the Qiime (V1.7.0) (Caporaso et al., 2010). Operational taxonomic units (OTUs) were generated using Uparse software (V7.0.1090) with a 97% similarity threshold for the tags (Edgar, 2013). SSUrRNA database (SILVA138.1) was applied for species annotation of each representative sequence at each taxonomic rank, using Mothur software. The phylogenetic relationship of all sequences was obtained by MUSCLE (V3.8.31) which can compare multiple sequences rapidly (Edgar, 2004; Quast et al., 2013). Histograms of the relative abundance (%) of taxa in different samples were plotted in RStudio (V3.1.1).
Comparative metagenomic analyses were performed during 6 months of continuous phenol pollution (100 mg/L) to investigate the dynamics of oligotrophic groundwater microbial communities due to phenol contamination. Sampling from the deep groundwater well located in the research institute of the petroleum industry, Tehran, Iran (35°44′18.7″ N 51°15′34.0″ E) was done in May 2020 using a water pump. This water sample was considered a control point without any pollution (GW). The sands used in the second column in the PRB studies were collected after 6 months of experiments. As mentioned before, the phenol (100 mg/L) contaminated groundwater was passed through the column for 6 months and the sands, which were autoclaved before applying into the column, were considered a suitable substrate for the formation of microbial biofilms during the experiment (second point, R2).
For the first sampling point, 80 L of water sample were collected and pre-filtered through 20 μm (Albet DP5891150, Germany), and 5 μm pore-size (Albet DP5895150, Germany) filters (15 cm in diameter). Microbial biomass was finally concentrated on 0.22 μm pore-size cellulose acetate filters (Sartorius 11107-142-N, Germany) using a peristaltic pump. Sand samples were collected from the sampling port of the column using sterile laboratory pincers. Water filters and sand samples were stored at −80°C until DNA extraction.
A standard phenol-chloroform protocol (Martín-Cuadrado et al., 2007) was used for extracting community DNA from the water sample. DNeasy PowerMax Soil DNA Extraction Kit (QIAGEN 12988-10, Germany) was applied for DNA extraction of the sand samples, according to the manufacturer’s instructions. Extracted DNA samples were sequenced using Illumina Novaseq 6000 platform (PE150) (Novogene, Hong Kong).
A subset of 5 million reads was separated from each dataset, and the reads affiliated with ribosomal RNA genes were detected using SSU-ALIGN (Nawrocki, 2009). Then, the putative prokaryotic 16S rRNA gene sequences were blasted against the SILVA reference database (release 138.1 SSUParc) using BLAST, and their taxonomic affiliation was assigned based on their closest hit if the read was ≥90 bp at the similarity threshold of ≥90.
Paired-end reads of the sequenced datasets were interleaved and quality trimmed using reformat.sh and bbduck.sh scripts of the BBMap toolkit, respectively (Bushnell, 2014). All trimmed sequences of each dataset were assembled separately using MEGAHIT (k-mer list 49, 69, 89, 109, 129, and 149) (Li et al., 2015). MetaBat2 software binned contigs ≥1 kb into metagenome-assembled genomes (MAGs) based on their different mapping depth and tetranucleotide frequency (Kang et al., 2019). The quality and completeness of the reconstructed MAGs were evaluated with CheckM (Parks et al., 2015). The taxonomy of MAGs with ≥40% completeness and ≤5% contamination was assigned using GTDB-tk (release 202) (Chaumeil et al., 2020). Putative genes were predicted with Prodigal (Hyatt et al., 2010) and preliminarily annotated using Prokka in the metagenomics mood (Seemann, 2014). Finally, the eggNOG-mapper was used to annotate further each MAG’s predicted protein sequences (Huerta-Cepas et al., 2019).
Four continuous-flow columns were set up to investigate the phenol removal rate from oligotrophic groundwater by CaO2 nanoparticles and SBCs. The experimental columns were as follows: (I) abiotic control column (CaO2 + sodium azide), (II) natural remediation column (without CaO2 and SBCs), (III) biostimulation column (CaO2), and (IV) simultaneous biostimulation and bioaugmentation column (CaO2 + SBCs).
As presented in Figure 2, the DO in the chemical column I effluent increased notably after the first injection and peaked on the 21st day, reaching 14.14 mg/L. Production of Ca(OH)2 from calcium peroxide decomposition increased the pH in the injection zone (Figure 2B), resulting in the higher stability of solid peroxide (Gholami et al., 2018). According to Northup and Cassidy (2008), CaO2 in an alkaline environment tends to produce O2 rather than OH●. Therefore, most CaO2 nanoparticles in column I were converted to oxygen, which completely remained in the abiotic column.
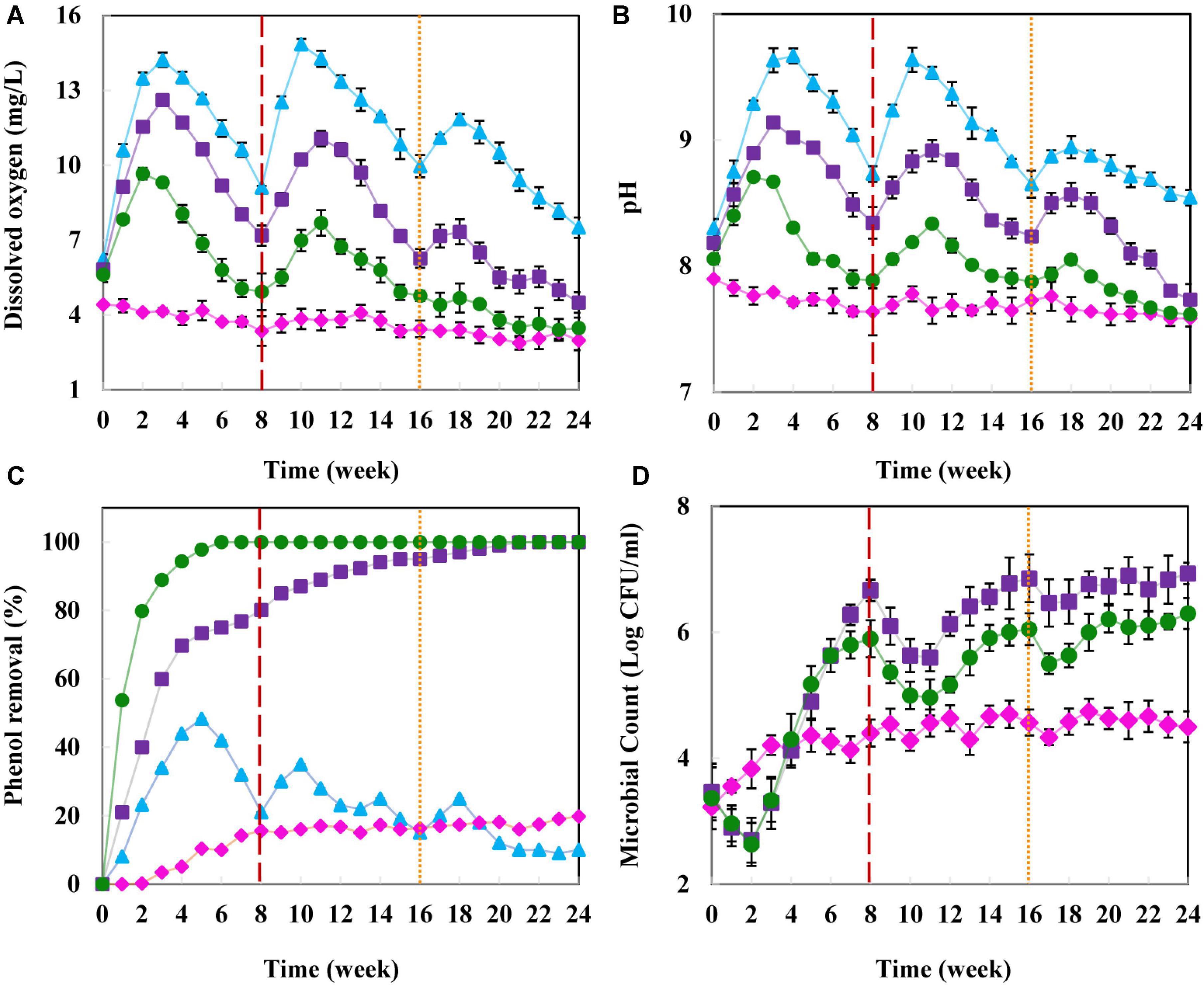
Figure 2. Remediation of phenol from groundwater in the column experiments. (A) Dissolved oxygen concentration (mg/L), (B) pH, (C) phenol removal (%), and (D) intrinsic microbial count (log CFU/mL). Abiotic column (▲), natural remediation column II (◆), biostimulation column III (■), and simultaneous biostimulation and bioaugmentation column IV (●). The red and orange dashed lines, respectively indicate 15 and 5 g CaO2 nanoparticles re-injection to provide the proper contaminant removal condition.
By injection of nanoparticles into columns III and IV, the DO increased in comparison to the blank column II (Figure 2A). Within 21 days, it reached 12.62 and 9.31 mg/L in columns III and IV, respectively. However, due to the high microbial count (intrinsic and SBCs’ communities) (Figure 2D) and oxygen consumption in column IV, DO dropped sharply (Figure 2A). To overcome this problem, the oxidative agents were re-injected into the groundwater after 8 and 16 weeks of the experiment.
After the first nanoparticle injection, the pH of column I effluent significantly increased and reached to 9.17 within 4 weeks (Figure 2B). In the three other columns, the pH slowly decreased, consistent with previous studies (Gholami et al., 2018; Mosmeri et al., 2019). According to Padhi and Gokhale (2017), it seems possible that these results are due to the production of acidic metabolites from the degradation of contaminants by microorganisms.
Only 19.8% of the phenol was naturally removed from groundwater in column II within 6 months (Figure 2C). Meanwhile, the addition of CaO2 and SBCs to column IV resulted in the complete phenol remediation after 42 days. The contaminant removal efficiency in column III containing CaO2 increased to 90% after 12 weeks and reached 100% in the 22nd week. In the abiotic column I, 48.3% of the phenol was removed within 5 weeks; however, a remarkable reduction in the contaminant removal efficiency was observed afterward.
Further CaO2 injection into column I at weeks 8 and 16 could not improve the phenol removal by more than 48.3%. The majority of H2O2 generated from CaO2 was converted to oxygen in the abiotic column I, and OH● content in water was not enough to degrade 100 mg/L of phenol (Figure 2C). These results were consistent with previous studies (Northup and Cassidy, 2008; Mosmeri et al., 2017; Gholami et al., 2018). Moreover, in accordance to prior surveys, our results indicated that by increasing the groundwater DO, microbial growth and activity gradually increased (Mosmeri et al., 2017; Gholami et al., 2019; Yavari-Bafghi et al., 2022).
In the natural bioremediation column I, the culturable microbial count was increased logarithmically to more than 3.2 × 104 CFU/ml, then significantly limited due to the lack of enough available oxygen for microorganisms. After injecting CaO2 nanoparticles into the biotic columns III and IV, the microbial count dropped initially due to the negative impacts of OH radicals generated from nanoparticles on the intrinsic microorganisms. Then, the count increased by adapting the microbial population to the oxidative condition (Figure 2D).
In summary, the results showed that the phenol remediation of oligotrophic groundwater was entirely successful in the presence of CaO2 and SBCs. Furthermore, the simultaneous application of biostimulation and bioaugmentation methods significantly increased the contaminant removal efficiency. Our results agree with Liu N. et al. (2018), which used a PRB containing CaO2 to remediate nitrobenzene-contaminated groundwater and observed the highest removal rate in the presence of calcium peroxide and nitrobenzene-degrading bacteria after 20 days. Additionally, a comparison of the phenol removal in the natural remediation column II and the abiotic column I indicates the crucial role of microorganisms in the remediation process. The innovative SBCs concept of macro-encapsulation allowed the possibility of selective contaminant removal in the water using the encapsulation of a specific bacterial culture. This study proved the beneficial effects of native bacterial biostimulation and bioaugmentation with external bacteria in oligotrophic environments through continuous-flow reactors. Besides, this bioremediation procedure is considered in-situ cost-effective, environmentally friendly, and performs at high efficiencies compared to conventional ex situ pump and treat technologies.
Apart from the planktonic groundwater microorganisms, the attached microbial population (biofilm) on the sand surfaces also plays a key role in contaminant remediation. Therefore, bacterial adhesion to the sand surface and biofilm thickness was respectively examined by SEM and CLSM.
As represented in Figure 3 and Supplementary Figure 1, no recognizable microorganism was observed on the sand surface in the abiotic column I (Figure 3A and Supplementary Figure 1A). It proved that no microbial degradation was involved in phenol removal from the column I effluent, and remediation was conducted by hydroxyl radicals (OH●). The groundwater microorganisms were attached to the sand surfaces and formed a thin biofilm layer (Figure 3B and Supplementary Figure 1B). However, by stimulating the microbial activity, the attached microbial communities with a thick biofilm layer can be seen on the sand surfaces (Figures 3C, D and Supplementary Figures 1C, D). It has resulted from the direct effect of oxygen-releasing nanoparticles on microbial growth. Present findings seem to be consistent with the results of Mosmeri et al. (2019) and Gholami et al. (2018), which investigated the impact of CaO2 on groundwater microbial communities. It can also be indicated from Figure 3C that the attached microbial communities, due to more desirable oxygen conditions in column III, formed a thicker biofilm compared to others.
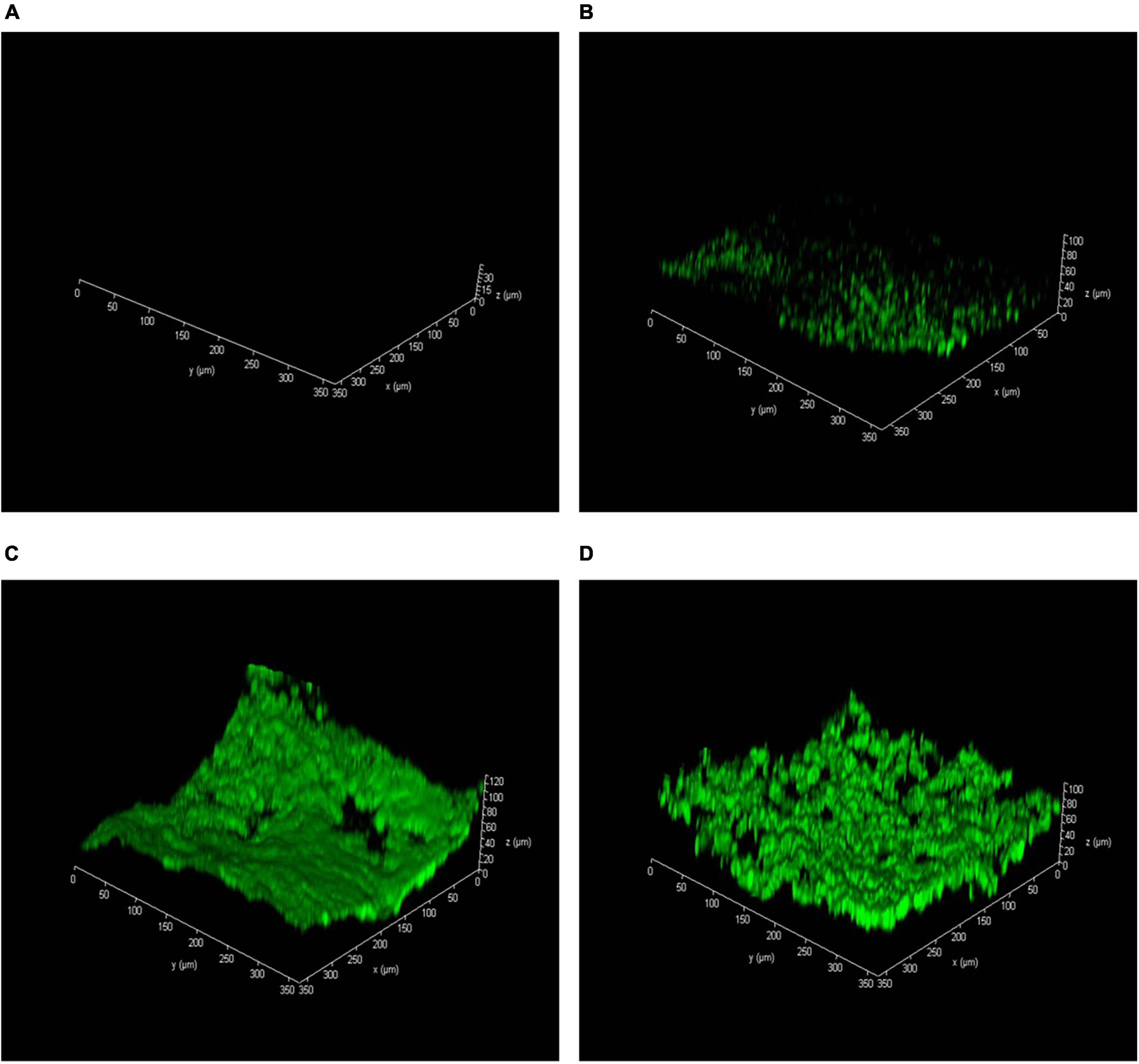
Figure 3. Acridine orange-stained confocal laser scanning microscopy (CLSM) images of biofilm thickness on the sand surface after 6 months. (A) Abiotic column I, (B) natural bioremediation column II, (C) biostimulation column III, and (D) biostimulation and bioaugmentation column IV.
The concentration of NO3–, NO2–, CaCO3, Cl–, Fe (total), PO4–, color, and turbidity of the effluents from each column were examined at the end of the experiment and were compared with oligotrophic groundwater parameters (Supplementary Table 2). Among mentioned factors, PO4–, Fe (total), Cl–, and color showed no significant changes. However, there was a substantial decrease in the NO3– and NO2– concentrations of the biotic columns II, III, and IV effluent (Supplementary Table 2). These results indicate that groundwater microorganisms consume nitrate as a nitrogen source during phenol biodegradation. In line with the study of Ye et al. (2017), nitrate in groundwater can also be removed by the denitrification process.
CaCO3 concentration was increased from 184 mg/L in the groundwater to 253 mg/L by flowing through the abiotic column I, which contained sodium azide. However, in columns II, III, and IV effluents, the concentration of CaCO3 trend was decreased. According to previous reports, the coincidence of the denitrification and high pH leads to the precipitation of bicarbonates in groundwater (Dupraz et al., 2004; Hamdan et al., 2017). Meanwhile, microbial growth in columns III, IV, and II, raised the water turbidity from 1.8 to 2.5, 2.2, and 1.9, respectively. In contrast, abiotic column I effluent, which lacked microbial growth, was the most transparent sample.
Microbial community compositions of columns at the order level are visualized in Supplementary Figure 2. Although Rhodobacterales with more than 70% relative abundance were dominated in samples from columns III and IV, they were present in lower than one percent in another column. In accordance with the current results, Jehmlich et al. (2010) demonstrated that the order Rhodobacterales abundance increased along the chronic hydrocarbon pollution in groundwater. Therefore, the presence of this order in columns III and IV indicates its ability to remove phenol under aerobic conditions. The order Burkholderiales showed a high abundance (45.01%) in column II, while it had a percentage of around three in other columns. The reduction in the prevalence of order Burkholderiales in biostimulation columns III and IV was probably due to the toxic effects of hydroxyl radical generation, which was previously mentioned in section “3.1.1. Phenol removal experiments.”
The order Xanthomonadales had relatively similar frequency in all three columns. Despite the presence of Syntrophobacterales in the natural bioremediation column II, its frequency reached zero in the third and fourth columns. Orders Desulfobacterales, Bacteriovoracales, Pseudomonadales, and Bdellovibrionales, which were identified in column II, were not detected in columns III and IV. Representatives of Syntrophobacterales, Desulfobacterales, and Bacteriovoracales generally have anaerobic respiration or fermentation metabolism (Koval et al., 2015; Dyksma et al., 2018; Liu P. et al., 2018); thus, their abundance was higher in the oxygen-deficient column II than in the biostimulation columns (Supplementary Figure 2).
According to Supplementary Figure 2, column II (natural bioremediation) exhibited the highest species richness among the samples collected at the end of the experiment. It seems that the application of ORCs had a remarkable impact on the microbial diversity of two other columns (III and IV). The same results were obtained in the bioremediation of benzene-contaminated groundwater by ORC (Mosmeri et al., 2019). Although the composition of the microbial community was changed in the presence of CaO2 nanoparticles, it significantly increased the remediation efficiency.
Considering the environmental concerns about using the bioaugmentation method, the microbial community dynamics of the ph100 consortium were investigated at the end of the experiment. This microbial community was compared with the initial consortium’s composition which was indicated in our previous study (Yavari-Bafghi et al., 2022). According to the results, the microbial composition of the ph100 consortium was constant during the continuous-flow experiment. Representatives of Burkholderiales from Gammaproteobacteria were the dominant order with almost the same relative abundance in both samples (Supplementary Figure 3). Based on the stability of the consortium’s composition, it can be concluded that the SBCs have no microbial leakage, and only the contaminated water penetrates through the SBCs microfiltration structure membrane. As a result, the SBCs can solve a significant part of the environmental concern of the bioaugmentation method.
Comparative metagenomic analyses demonstrated that continuous exposure to phenol pollution in oligotrophic groundwater samples causes a shift in the microbial community composition. Hydrogen and CO2 are frequently found in deep groundwater ecosystems since they are formed in the Earth’s crust or mantle. Chemolithotrophs in these environments obtain their metabolic energy and carbon from the oxidation of reduced inorganic compounds and fixation of CO2, respectively (Purkamo, 2017). As demonstrated in Figure 4, the microbial community of pristine groundwater (GW sample) was dominated by Burkholderiales (Burkholderiaceae), Micrococcales, Cytophagales, and Sphingomonadales. In a previous study, it has been proved that the family Burkholderiaceae belongs to the order Burkholderiales including obligate and facultative chemolithotroph members (Pérez-Pantoja et al., 2012).
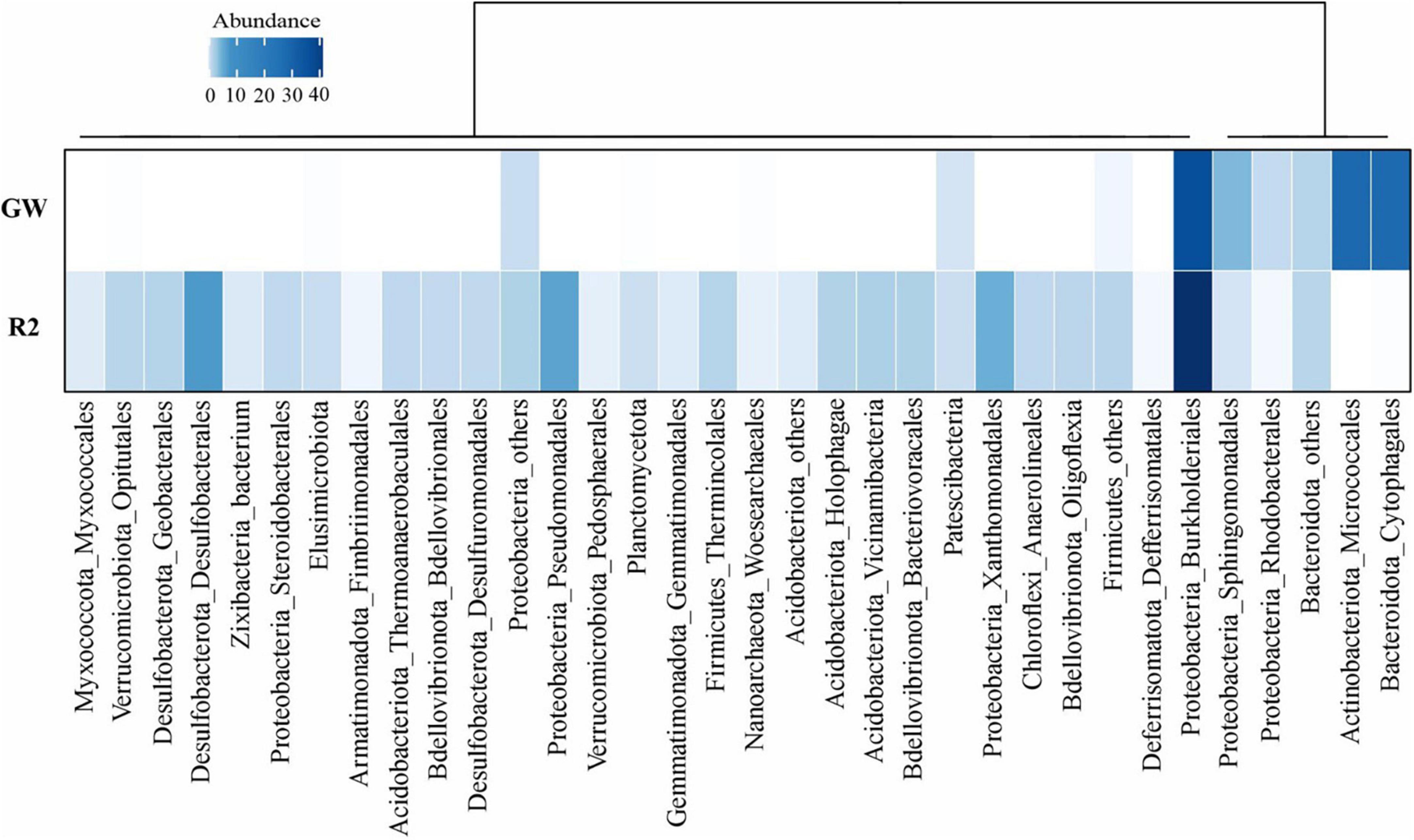
Figure 4. Prokaryotic community composition of the groundwater samples according to the abundance of 16S rDNA gene reads in unassembled metagenomes. Rows are the name of samples (GW: unpolluted and R2: phenol polluted groundwater). Column names are microbial taxa at the phylum and order level. For some taxa with lower frequency, the sum of orders is displayed as others or in their corresponding higher taxonomic level. There are a total number of 34 taxa for both samples. Dendrograms represent the clustering of columns based on Pearson correlation. The figure was plotted using “circlize” and “ComplexHeatmap” packages in R.
The entrance of phenol as a carbon source into the oligotrophic groundwater altered the microbial community towards a higher relative abundance of Desulfobacterales, Burkholderiales (Rhodocyclaceae), Pseudomonadales, and Xanthomonadales in the R2 sample.
The order Burkholderiales of the phylum Proteobacteria was the most abundant member of the community in both R2 and GW samples. Its relative abundance increased from 35.57% (Burkholderiaceae) in GW to 45.01% (Rhodocyclaceae) in R2. According to Silva et al. (2013), the most effective bacteria that harbor the metabolic pathways for phenol degradation are represented by genera from the order Burkholderiales.
Furthermore, sulfate-reducing bacteria (SRB) in the R2 sample comprised up to 11% of the community (Desulfobacterales, Geobacterales, Desulfuromonadales, and Myxococcales). The nitrate- and sulfate-reducing bacteria were presented in almost all samples of marine oil-polluted sediments and are considered the most crucial hydrocarbon (HC) degrading communities in aquatic ecosystems with low levels of DO (Paissé et al., 2008; Stagars et al., 2017; Rezaei Somee et al., 2021). Additionally, the orders Pseudomonadales and Xanthomonadales were prevalent in the diesel-contaminated soils and have caused the efficient removal of pollutants (Koshlaf et al., 2016). It indicated that the bioremediation process successfully raised the population of the phenol-degrading species.
Among microbial members of groundwater samples, the representatives of Woesearchaeales from the phylum Nanoarchaeota were the only archaea observed in both metagenomic samples with 1% frequency.
In accordance with the previous study that reported the negative response of Cytophagales to oil pollution in marine ecosystems (Rezaei Somee et al., 2021), Figure 4 represents that phenol pollution also significantly reduced the relative abundance of orders Micrococcales and Cytophagales. However, the relative abundance of Firmicutes, Chloroflexi, Acidobacteriota, and Verrucomicrobiota consistently increased from GW to R2 in response to phenol pollution. The entrance of high concentrations of HC pollutant (phenol) into the oligotrophic groundwater environment introduces new sources of carbon and more taxonomic groups were able to consume these carbon sources. As a result, the frequency of microbial groups increased and the amount of DO showed a significant reduction. In the absence of oxygen, alternative final electron acceptors such as nitrate and sulfate increased and consequently, a higher relative abundance of nitrate- and sulfate-reducing bacteria besides phenol-degrading members became dominant in the microbial community of the R2 sample. Furthermore, the alpha diversity of samples has been calculated based on the Shannon–Wiener index in R using the “vegan” package. As could be seen in Supplementary Figure 4, the R2 sample has a higher alpha index (2.3) than the pristine groundwater sample (1.4), indicating increased microbial diversity after phenol entrance.
A total of 47 metagenome-assembled genomes (MAGs) with completeness ≥40% and contamination ≤5% were recovered from two sequenced metagenomes, among which 46 belonged to domain bacteria and one to domain Archaea. Reconstructed bacterial MAGs were mainly affiliated with Proteobacteria (46.8%), Bdellovibrionota (10.63%), Bacteroidota (8.51%), and Desulfobacterota (6.38%), along with some representatives of other phyla.
Metabolic pathways in each MAG have been recovered based on the KEGG orthologous (KO) list of corresponding MAG via the KEGG Mapper Reconstruct website tool1 (Supplementary Table 3). The distribution of energy-related modules (e.g., carbon, nitrogen, and sulfur) present in at least one MAG is represented in Figure 5.
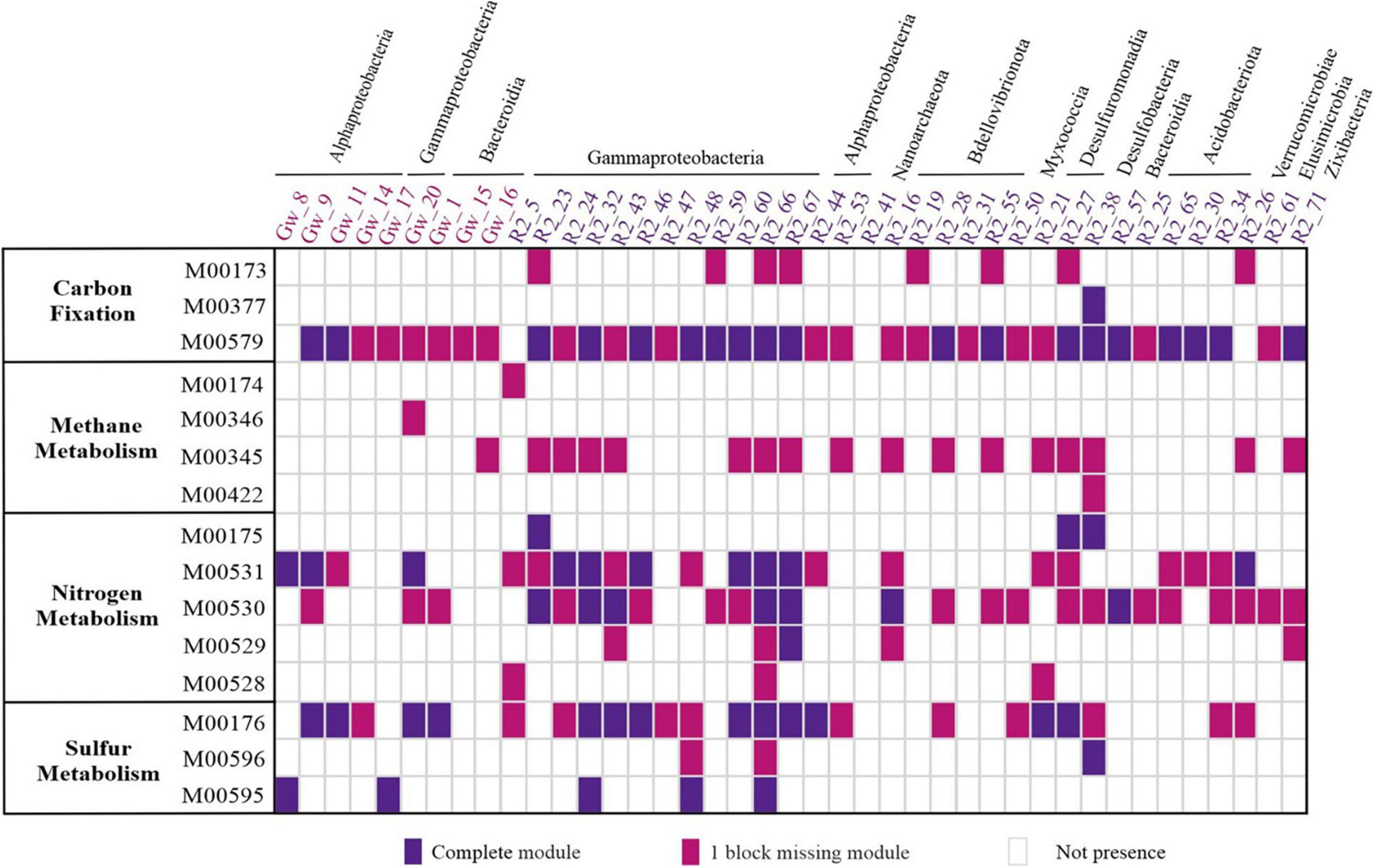
Figure 5. Energy metabolism pathways modules present in recovered MAGs from the groundwater metagenomes. The columns represent the taxonomy recovered of MAGs. The color of each MAG indicates the MAG origin. The row shows the cluster of metabolic pathways. Reductive citrate cycle (Arnon–Buchanan cycle) (M00173), reductive acetyl-CoA pathway (Wood–Ljungdahl pathway) (M00377), and phosphate acetyltransferase-acetate kinase pathway (M00579). Methane oxidation, methanotroph (M00174), formaldehyde assimilation, serine pathway (M00346), formaldehyde assimilation, ribulose monophosphate pathway (M00345), and Acetyl-CoA pathway (M00422). Nitrogen fixation (M00175), assimilatory nitrate reduction (M00531), dissimilatory nitrate reduction (M00530), nitrification (M00528), and denitrification (M00529). Assimilatory sulfate reduction (M00176), dissimilatory sulfate reduction (M00596), and thiosulfate oxidation by SOX complex (M00595). For the presence of a module that has all the enzymes of a pathway the value 2 and for the module that lacks one of the enzymes of a pathway the value 1 is considered.
The KEGG orthologous accession numbers (KOs) of a collection of reported enzymes involved in the degradation of different aromatic and aliphatic hydrocarbons (HCs) under aerobic and anaerobic conditions were collected and surveyed in the annotated MAGs (Abbasian et al., 2015, 2016; Rabus et al., 2016). Figure 6 represents the distribution of KEGG orthologues detected at least in one MAG (n = 93 genes). Mono/dioxygenases triggering the degradation of alkane, cyclododecane, biphenyl, phenol, toluene, xylene, and naphthalene/phenanthrene were detected in 43 recovered MAGs of the groundwater samples. Furthermore, the key enzymes responsible for initiating the degradation of alkane, ethylbenzene, phenol, and toluene exclusively under anaerobic conditions were detected in four reconstructed MAGs.
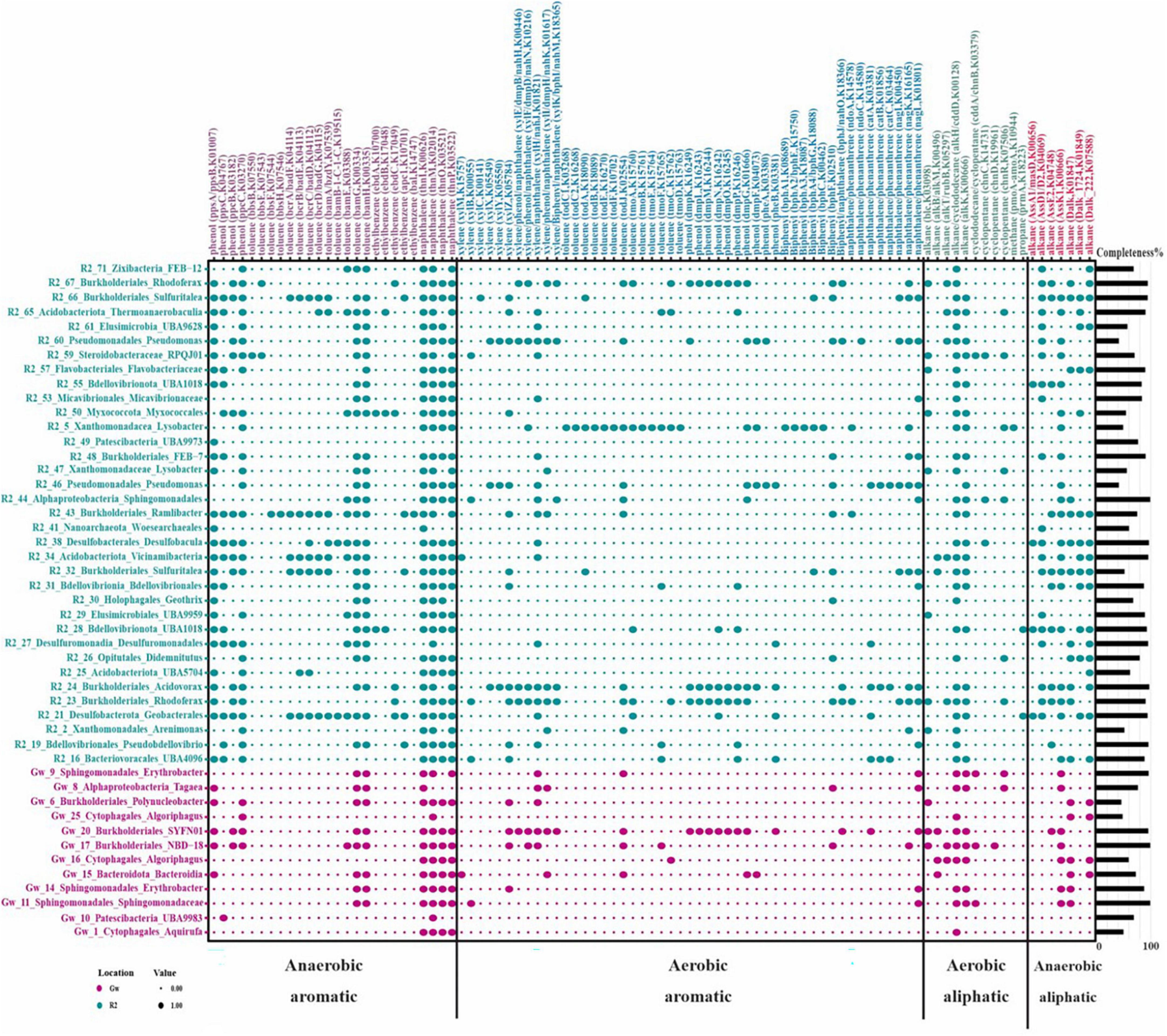
Figure 6. Hydrocarbon-degrading enzymes present in recovered MAGs from the groundwater metagenomes. Row names represent the taxonomy of recovered MAGs and their completeness is provided as a bar plot on the right side. The color indicates the MAG origin. Columns indicate the type of hydrocarbon and in the parenthesis is the name of the enzyme hydrolyzing this compound, followed by its corresponding KEGG orthologous accession number. The size of the dots indicates the presence or absence of each enzyme in each recovered MAG. The figure was plotted using the “reshape2” and “ggplot2” packages in RStudio.
The metabolic context of pristine groundwater (GW sample) reconstructed MAGs suggested a primarily heterotrophic and autotrophic lifestyle. In oligotrophic groundwater, chemolithotrophs are responsible for organic matter production through CO2 fixation (Purkamo, 2017). Representatives of Alphaproteobacteria (GW_9 and GW_11) in the GW sample were autotrophs capable of fixing carbon dioxide and producing acetate by using the phosphate acetyltransferase-acetate kinase pathway (Figure 5). Acetate is a key intermediate for heterotrophic bacteria in anaerobic conditions and is also considered as a carbon source for SRBs (Dyksma et al., 2018). Furthermore, sulfur-oxidizing chemolithoautotrophic bacteria such as NBD.18 (GW_17) and Alphaproteobacteria member (GW_8) are probably among the primary producers in GW sample by converting thiosulfate to sulfate through sulfate oxidation with SOX complex.
According to Figure 6, the distribution of HC-degrading enzymes is less frequent in reconstructed MAGs of the GW sample. However, sulfur-oxidizing MAGs, including GW_17, GW_20 (Burkholderiales), as well as GW_15 and GW_16 (Algoriphagus), potentially have enzymes involved in the degradation of phenol, toluene, xylene, ethylbenzene, alkanes, and naphthalene under both aerobic and anaerobic conditions. They were among microorganisms with alkB/M genes and were initiating the degradation of HCs under aerobic conditions in waters (Rezaei Somee et al., 2021).
In the R2 sample, despite recovering chemolithotrophic MAGs that potentially were capable of carbon fixation (Figure 5), MAGs with HC degradation ability were more prevalent due to the high concentrations of phenol as a carbon source (Figure 6).
R2_23 and R2_24 MAGs, affiliated to Rhodoferax and Acidovorax, respectively, had the genomic potential to degrade a diverse range of HCs, including phenol, alkane, biphenyl, toluene, ethylbenzene, and xylene under both aerobic and anaerobic conditions (Figure 6). Members of Acidovorax have been reported to potentially degrade various organic compounds like chlorobenzene (Monferrán et al., 2005), phenanthrene (Singleton et al., 2009), trichloroethylene (Futamata et al., 2005) and polyhydroxyalkanoates (Masood, 2017). Therefore, the genus Acidovorax can be one of the main genera that play a crucial role in the bioremediation of a wide range of oil derivatives in polluted ecosystems.
According to previous studies, the entrance of a carbon source into groundwater causes the activation of denitrification and increases nitrate reduction (Lee et al., 2021). Sulfuritalea (R2_32/66), Rhodoferax (R2_23), Ramlibacter (R2_43), and Zixibacteria (R2_71) had denitrification potential in the R2 sample. Sulfuritalea members have been identified in HC-polluted environments and reported to have conserved genes of the anaerobic benzoyl-CoA pathway, such as bamA, to initiate the degradation process under anaerobic conditions (Sperfeld et al., 2018). In the present study, the existence of aromatic and aliphatic compounds degradation genes under anaerobic conditions in the Sulfuritalea genome was confirmed and the presence of genes related to pollutant degradation in aerobic conditions was also reported. It seems that SRB members of Burkholderiales and Desulfobacteraceae, such as Sulfuritalea and FEB-7, have substrate specificity in degrading hydrocarbon compounds. According to a recent study, they often degrade HCs in samples that have aliphatic compounds (Shin et al., 2019).
Representatives of Zixibacteria (R2_71) had genes for the degradation of phenol, toluene, ethylbenzene, alkanes, and naphthalene under anaerobic conditions. The phylum Zixibacteria has no cultured representative, and its genome was first introduced in 2013 by metagenomics studies of sediment samples of a reservoir near the Colorado River (USA) (Castelle et al., 2013).
Phenol, naphthalene, and phenanthrene degradation genes have also been reported for representatives of Lysobacter (Maeda et al., 2009; Brescia et al., 2020). According to Figure 6, the Lysobacter (R2_5) genome also had toluene degradation genes under aerobic conditions. However, there is no report on this ability in previous studies.
Woesearchaeales from Nanoarchaeota that have been reported in metagenomics analyses of groundwater samples (Mehrshad et al., 2021) were recovered from both GW and R2 samples and contained several enzymes contributing to phenol (ppsA/ppsB), naphthalene (thnl), and alkane (AssD1/D2) degradation under aerobic conditions.
A nitrate-reducing member of Flavobacteriaceae (R2_57) was recovered from the R2 sample and contained aromatic compounds’ degrading enzymes under anaerobic conditions. They mostly have enzymes that participate in the degradation of PAHs under anaerobic conditions (Rezaei Somee et al., 2021) and are potent aquatic indigenous degraders that bloom in response to HC pollution (Momper et al., 2017).
The present study investigated the application of SBCs and CaO2 nanoparticles in phenol-contaminated groundwater remediation through the continuous flow of sand-packed plexiglass columns. In addition, the influence of each treatment approach on the diversity and richness of the oligotrophic groundwater microbial communities was evaluated using next-generation sequencing (NGS) technology. The results indicated that applying SBCs into the simulated groundwater substrate remarkably affected the phenol removal rate. During the first 42 days of experiment, simultaneous use of CaO2 nanoparticles and SBCs showed great potential in the complete phenol removal efficiency. Meanwhile, the natural bioremediation process was able to remove only 19.8% of the contaminant of the column’s effluent within 6 months.
The attached microbial communities on the sand surfaces were observed by SEM and CLSM, where their crucial role in the remediation process was specified. Furthermore, studying the biodiversity of the attached biofilm in each remediation process by NGS demonstrated that the addition of CaO2 nanoparticles into the groundwater stimulates the contaminant biodegrading microorganisms without any adverse effect on the groundwater. Consequently, the biostimulation and bioaugmentation process is suggested for phenol-contaminated groundwater bioremediation.
Moreover, our understanding of microbial dynamics in response to phenol pollution in oligotrophic groundwater enrolled as a valuable model for advancing knowledge of managing organic and hydrocarbon spill accidents, especially in Iran. In this work, the extensive analysis of groundwater metagenome along phenol pollution illustrated the 6-month exposure to 100 mg/L phenol altered the microbial community, and microbes with the capability of pollutant degradation became the dominant population in the pollution zone. Higher-resolution analysis of the microbial community of this ecosystem in future studies can reveal critical ecological adaptations to different pollutants.
The data presented in the study are deposited in the DDBJ/EMBL/GenBank repository, accession number PRJNA741716. All data have been released and are available at https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA741716.
MS: conceptualization, supervision, review and editing, and funding acquisition. SD and MA: conceptualization, project administration, and review and editing. MY-B: methodology, investigation, software, validation, visualization, and writing—original draft. MR: software, validation, formal analysis, and review and editing. All authors contributed to the article and approved the submitted version.
This study was supported by a grant from the Research Institute of Petroleum Industry (RIPI), Tehran, Iran (grant number: 35670221).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1147162/full#supplementary-material
SBCs, Small Bioreactor Chambers; CaO2, calcium peroxide; NGS, next-generation sequencing; DO, dissolved oxygen; ORCs, oxygen-releasing compounds; PRB, permeable reactive barrier; SEM, scanning electron microscopy; CLSM, confocal laser scanning microscopy.
Abbasian, F., Lockington, R., Mallavarapu, M., and Naidu, R. (2015). A comprehensive review of aliphatic hydrocarbon biodegradation by bacteria. Appl. Biochem. Biotechnol. 176, 670–699.
Abbasian, F., Lockington, R., Megharaj, M., and Naidu, R. (2016). A review on the genetics of aliphatic and aromatic hydrocarbon degradation. Appl. Biochem. Biotechnol. 178, 224–250.
Brescia, F., Pertot, I., and Puopolo, G. (2020). “Lysobacter,” in Beneficial Microbes in Agro-Ecology, eds N. Amaresan, M. Senthil Kumar, and K. Annapurna (Amsterdam: Elsevier).
Bushnell, B. (2014). BBMap: a Fast, Accurate, Splice-Aware Aligner. Berkeley, CA: Lawrence Berkeley National Laboratory.
Caporaso, J., Stombaugh, J., Costello, E. K., Lozupone, C. A., McDonald, D., Pirrung, M., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336.
Cassidy, D. P., and Irvine, R. L. (1999). Use of calcium peroxide to provide oxygen for contaminant biodegradation in a saturated soil. J. Hazard. Mater. 69, 25–39. doi: 10.1016/s0304-3894(99)00051-5
Castelle, C. J., Hug, L. A., Wrighton, K. C., Thomas, B. C., Williams, K. H., Wu, D., et al. (2013). Extraordinary phylogenetic diversity and metabolic versatility in aquifer sediment. Nat. Commun. 4:2120. doi: 10.1038/ncomms3120
Cecconet, D., Sabba, F., Devecseri, M., Callegari, A., and Capodaglio, A. G. (2020). In situ groundwater remediation with bioelectrochemical systems: a critical review and future perspectives. Environ. Int. 137:105550. doi: 10.1016/j.envint.2020.105550
Chaumeil, P. A., Mussig, A. J., Hugenholtz, P., and Parks, D. H. (2020). GTDB-Tk: a toolkit to classify genomes with the genome taxonomy database. Bioinformatics 36, 1925–1927.
Clark, D. R., Ferguson, R. M. W., Harris, D. N., Matthews Nicholass, K. J., Prentice, H. J., Randall, K. C., et al. (2018). Streams of data from drops of water: 21st century molecular microbial ecology. WIREs Water 5:e1280.
Dey, U., Chatterjee, S., and Mondal, N. K. (2016). Isolation and characterization of arsenic-resistant bacteria and possible application in bioremediation. Biotechnol. Rep. 10, 1–7.
Dong, H., Li, L., Lu, Y., Cheng, Y., Wang, Y., Ning, Q., et al. (2019). Integration of nanoscale zero-valent iron and functional anaerobic bacteria for groundwater remediation: a review. Environ. Int. 124, 265–277. doi: 10.1016/j.envint.2019.01.030
Duan, L., Wang, H., Sun, Y., and Xie, X. (2016). Biodegradation of phenol from wastewater by microorganism immobilized in bentonite and carboxymethyl cellulose gel. Chem. Eng. Commun. 203, 948–956.
Dupraz, C., Visscher, P. T., Baumgartner, L. K., and Reid, R. P. (2004). Microbe-mineral interactions: early carbonate precipitation in a hypersaline lake (Eleuthera Island. Bahamas). Sedimentology 51, 745–765.
Dyksma, S., Lenk, S., Sawicka, J. E., and Mußmann, M. (2018). Uncultured gammaproteobacteria and desulfobacteraceae account for major acetate assimilation in a coastal marine sediment. Front. Microbiol. 9:3124. doi: 10.3389/fmicb.2018.03124
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797.
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Flynn, T. M., Sanford, R. A., and Bethke, C. M. (2008). Attached and suspended microbial communities in a pristine confined aquifer. Water Resour. Res. 44:W07425.
Futamata, H., Nagano, Y., Watanabe, K., and Hiraishi, A. (2005). Unique kinetic properties of phenol-degrading variovorax strains responsible for efficient trichloroethylene degradation in a chemostat enrichment culture. Appl. Environ. Microbiol. 71, 904–911. doi: 10.1128/AEM.71.2.904-911.2005
Gholami, F., Shavandi, M., Dastgheib, S. M. M., and Amoozegar, M. A. (2018). Naphthalene remediation from groundwater by calcium peroxide (CaO2) nanoparticles in permeable reactive barrier (PRB). Chemosphere 212, 105–113.
Gholami, F., Shavandi, M., Dastgheib, S. M. M., and Amoozegar, M. A. (2019). The impact of calcium peroxide on groundwater bacterial diversity during naphthalene removal by permeable reactive barrier (PRB). Environ. Sci. Pollut. Res. 26, 35218–35226. doi: 10.1007/s11356-019-06398-y
González, G. (2001). Biodegradation of phenolic industrial wastewater in a fluidized bed bioreactor with immobilized cells of Pseudomonas putida. Bioresour. Technol. 80, 137–142. doi: 10.1016/s0960-8524(01)00076-1
Hamdan, N., Kavazanjian, E., Rittmann, B. E., and Karatas, I. (2017). Carbonate mineral precipitation for soil improvement through microbial denitrification. Geomicrobiol. J. 34, 139–146.
Hedbavna, P., Rolfe, S. A., Huang, W. E., and Thornton, S. F. (2016). Biodegradation of phenolic compounds and their metabolites in contaminated groundwater using microbial fuel cells. Bioresour. Technol. 200, 426–434. doi: 10.1016/j.biortech.2015.09.092
Huerta-Cepas, J., Szklarczyk, D., Heller, D., Hernández-Plaza, A., Forslund, S. K., Cook, H., et al. (2019). eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 47, D309–D314. doi: 10.1093/nar/gky1085
Hyatt, D., Chen, G.-L., LoCascio, P. F., Land, M. L., Larimer, F. W., and Hauser, L. J. (2010). Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119
Inoué, S. (2006). “Foundations of confocal scanned imaging in light microscopy,” in Handbook Of Biological Confocal Microscopy, ed. J. Pawley (Boston, MA: Springer).
Jehmlich, N., Kleinsteuber, S., Vogt, C., Benndorf, D., Harms, H., Schmidt, F., et al. (2010). Phylogenetic and proteomic analysis of an anaerobic toluene-degrading community. J. Appl. Microbiol. 109, 1937–1945.
Kang, D. D., Li, F., Kirton, E., Thomas, A., Egan, R., An, H., et al. (2019). MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ 7:e7359. doi: 10.7717/peerj.7359
Khodaveisi, J., Banejad, H., Afkhami, A., Olyaie, E., Lashgari, S., and Dashti, R. (2011). Synthesis of calcium peroxide nanoparticles as an innovative reagent for in situ chemical oxidation. J. Hazard. Mater. 192, 1437–1440. doi: 10.1016/j.jhazmat.2011.06.060
Koshlaf, E., Shahsavari, E., Aburto-Medina, A., Taha, M., Haleyur, N., Makadia, T. H., et al. (2016). Bioremediation potential of diesel-contaminated Libyan soil. Ecotoxicol. Environ. Saf. 133, 297–305. doi: 10.1016/j.ecoenv.2016.07.027
Koval, S. F., Williams, H. N., and Stine, O. C. (2015). Reclassification of Bacteriovorax marinus as Halobacteriovorax marinus gen. nov., comb. nov. and Bacteriovorax litoralis as Halobacteriovorax litoralis comb. nov.; description of Halobacteriovoraceae fam. nov. in the class Deltaproteobacteria. Int. J. Syst. Evol. Microbiol. 65, 593–597. doi: 10.1099/ijs.0.070201-0
Lall, U., Josset, L., and Russo, T. (2020). A snapshot of the world’s groundwater challenges. Annu. Rev. Environ. Resour. 45, 171–194.
Lee, D., Ahn, Y., Pandi, K., Park, J., Yun, S.-T., Jang, M., et al. (2021). Evaluation of natural attenuation-potential and biogeochemical analysis in nitrate contaminated bedrock aquifers by carbon source injection. Sci. Total Environ. 780:146459. doi: 10.1016/j.scitotenv.2021.146459
Li, D., Liu, C. M., Luo, R., Sadakane, K., and Lam, T. W. (2015). MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31, 1674–1676. doi: 10.1093/bioinformatics/btv033
Liu, N., Zhang, Y., An, Y., and Wang, L. (2018). Preparation of integrative cubes as a novel biological permeable reactive barrier medium for the enhancement of in situ aerobic bioremediation of nitrobenzene-contaminated groundwater. Environ. Earth Sci. 77, 707.
Liu, P., Pommerenke, B., and Conrad, R. (2018). Identification of syntrophobacteraceae as major acetate-degrading sulfate reducing bacteria in Italian paddy soil. Environ. Microbiol. 20, 337–354. doi: 10.1111/1462-2920.14001
Lopez-Fernandez, M., Simone, D., Wu, X., Soler, L., Nilsson, E., Holmfeldt, K., et al. (2018). Metatranscriptomes reveal that all three domains of life are active but are dominated by bacteria in the fennoscandian crystalline granitic continental deep biosphere. MBio 9:e1792-18. doi: 10.1128/mBio.01792-18
Lu, S., Zhang, X., and Xue, Y. (2017). Application of calcium peroxide in water and soil treatment: a review. J. Hazard. Mater. 337, 163–177. doi: 10.1016/j.jhazmat.2017.04.064
Maeda, R., Nagashima, H., Zulkharnain, A., Bin, Iwata, K., and Omori, T. (2009). Isolation and characterization of a car gene cluster from the naphthalene, phenanthrene, and carbazole-degrading marine isolate Lysobacter sp. strain OC7. Curr. Microbiol. 59, 154–159. doi: 10.1007/s00284-009-9414-y
Magoc, T., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Martín-Cuadrado, A.-B., López-García, P., Alba, J.-C., Moreira, D., Monticelli, L., Strittmatter, A., et al. (2007). Metagenomics of the deep mediterranean, a warm bathypelagic habitat. PLoS One 2:e914. doi: 10.1371/journal.pone.0000914
Masood, F. (2017). “Polyhydroxyalkanoates in the food packaging industry,” in Nanotechnology Applications in Food, 153–177. Available online at: https://doi.org/10.1016/B978-0-12-811942-6.00008-X
Mehrshad, M., Lopez-Fernandez, M., Sundh, J., Bell, E., Simone, D., Buck, M., et al. (2021). Energy efficiency and biological interactions define the core microbiome of deep oligotrophic groundwater. Nat. Commun. 12:4253. doi: 10.1038/s41467-021-24549-z
Mikkonen, A., Yläranta, K., Tiirola, M., Dutra, L. A. L., Salmi, P., Romantschuk, M., et al. (2018). Successful aerobic bioremediation of groundwater contaminated with higher chlorinated phenols by indigenous degrader bacteria. Water Res. 138, 118–128. doi: 10.1016/j.watres.2018.03.033
Mohammadi, S., Kargari, A., Sanaeepur, H., Abbassian, K., Najafi, A., and Mofarrah, E. (2015). Phenol removal from industrial wastewaters: a short review. Desalin. Water Treat. 53, 2215–2234.
Momper, L., Jungbluth, S. P., Lee, M. D., and Amend, J. P. (2017). Energy and carbon metabolisms in a deep terrestrial subsurface fluid microbial community. ISME J. 11, 2319–2333. doi: 10.1038/ismej.2017.94
Monferrán, M. V., Echenique, J. R., and Wunderlin, D. A. (2005). Degradation of chlorobenzenes by a strain of Acidovorax avenae isolated from a polluted aquifer. Chemosphere 61, 98–106. doi: 10.1016/j.chemosphere.2005.03.003
Mosmeri, H., Alaie, E., Shavandi, M., Dastgheib, S. M. M., and Tasharrofi, S. (2017). Bioremediation of benzene from groundwater by calcium peroxide (CaO2) nanoparticles encapsulated in sodium alginate. J. Taiwan Inst. Chem. Eng. 78, 299–306.
Mosmeri, H., Gholami, F., Shavandi, M., Dastgheib, S. M. M., and Alaie, E. (2019). Bioremediation of benzene-contaminated groundwater by calcium peroxide (CaO2) nanoparticles: continuous-flow and biodiversity studies. J. Hazard. Mater. 371, 183–190. doi: 10.1016/j.jhazmat.2019.02.071
Mosmeri, H., Tasharrofi, S., Alaie, E., and Sadegh Hassani, S. (2018). “Controlled-release oxygen nanocomposite for bioremediation of benzene contaminated groundwater,” in New Polymer Nanocomposites for Environmental Remediation, 601–622. Available online at: https://doi.org/10.1016/B978-0-12-811033-1.00023-8
Nawrocki, E. (2009). Structural RNA Homology Search and Alignment Using Covariance Models. St. Louis, MO: Washington University in St. Louis.
Northup, A., and Cassidy, D. (2008). Calcium peroxide (CaO2) for use in modified fenton chemistry. J. Hazard. Mater. 152, 1164–1170.
Padhi, S. K., and Gokhale, S. (2017). Benzene biodegradation by indigenous mixed microbial culture: kinetic modeling and process optimization. Int. Biodeterior. Biodegradation 119, 511–519.
Paissé, S., Coulon, F., Goñi-Urriza, M., Peperzak, L., McGenity, T. J., and Duran, R. (2008). Structure of bacterial communities along a hydrocarbon contamination gradient in a coastal sediment. FEMS Microbiol. Ecol. 66, 295–305. doi: 10.1111/j.1574-6941.2008.00589.x
Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P., and Tyson, G. W. (2015). CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055. doi: 10.1101/gr.186072.114
Pérez-Pantoja, D., Donoso, R., Agulló, L., Córdova, M., Seeger, M., Pieper, D. H., et al. (2012). Genomic analysis of the potential for aromatic compounds biodegradation in Burkholderiales. Environ. Microbiol. 14, 1091–1117.
Pickup, R., Rhodes, G., Alamillo, M., Mallinson, H. E., Thornton, S., and Lerner, D. (2001). Microbiological analysis of multi-level borehole samples from a contaminated groundwater system. J. Contam. Hydrol. 53, 269–284. doi: 10.1016/s0169-7722(01)00169-3
Poi, G., Shahsavari, E., Aburto-Medina, A., Mok, P. C., and Ball, A. S. (2018). Large scale treatment of total petroleum-hydrocarbon contaminated groundwater using bioaugmentation. J. Environ. Manage. 214, 157–163. doi: 10.1016/j.jenvman.2018.02.079
Purkamo, L. (2017). Thriving microbial life in ancient groundwater deep inside earth’s crust. Front. Young Minds 5:65. doi: 10.3389/FRYM.2017.00065/PDF
Qian, Y., Zhang, J., Zhang, Y., Chen, J., and Zhou, X. (2016). Degradation of 2,4-dichlorophenol by nanoscale calcium peroxide: implication for groundwater remediation. Sep. Purif. Technol. 166, 222–229.
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Rabus, R., Boll, M., Heider, J., Meckenstock, R. U., Buckel, W., Einsle, O., et al. (2016). Anaerobic microbial degradation of hydrocarbons: from enzymatic reactions to the environment. J. Mol. Microbiol. Biotechnol. 26, 5–28. doi: 10.1159/000443997
Rezaei Somee, M., Dastgheib, S. M. M., Shavandi, M., Ghanbari Maman, L., Kavousi, K., Amoozegar, M. A., et al. (2021). Distinct microbial community along the chronic oil pollution continuum of the Persian Gulf converge with oil spill accidents. Sci. Rep. 11:11316. doi: 10.1038/s41598-021-90735-0
Shin, B., Kim, M., Zengler, K., Chin, K.-J., Overholt, W. A., Gieg, L. M., et al. (2019). Anaerobic degradation of hexadecane and phenanthrene coupled to sulfate reduction by enriched consortia from northern Gulf of Mexico seafloor sediment. Sci. Rep. 9:1239. doi: 10.1038/s41598-018-36567-x
Silva, C. C., Hayden, H., Sawbridge, T., Mele, P., De Paula, S. O., Silva, L. C. F., et al. (2013). Identification of genes and pathways related to phenol degradation in metagenomic libraries from petroleum refinery wastewater. PLoS One 8:e61811. doi: 10.1371/journal.pone.0061811
Singleton, D. R., Guzmaìn Ramirez, L., and Aitken, M. D. (2009). Characterization of a polycyclic aromatic hydrocarbon degradation gene cluster in a phenanthrene-degrading acidovorax strain. Appl. Environ. Microbiol. 75, 2613–2620. doi: 10.1128/AEM.01955-08
Sperfeld, M., Diekert, G., and Studenik, S. (2018). Anaerobic aromatic compound degradation in Sulfuritalea hydrogenivorans sk43H. FEMS Microbiol. Ecol. 95:fiy199. doi: 10.1093/femsec/fiy199
Stagars, M. H., Mishra, S., Treude, T., Amann, R., and Knittel, K. (2017). Microbial community response to simulated petroleum seepage in Caspian sea sediments. Front. Microbiol. 8:764.
Sun, X., Wang, C., Li, Y., Wang, W., and Wei, J. (2015). Treatment of phenolic wastewater by combined UF and NF/RO processes. Desalination 355, 68–74.
Villegas, L. G. C., Mashhadi, N., Chen, M., Mukherjee, D., Taylor, K. E., and Biswas, N. (2016). A short review of techniques for phenol removal from wastewater. Curr. Pollut. Reports 2, 157–167.
Wang, H., Zhao, Y., Li, T., Chen, Z., Wang, Y., and Qin, C. (2016). Properties of calcium peroxide for release of hydrogen peroxide and oxygen: a kinetics study. Chem. Eng. J. 303, 450–457.
Xue, Y., Lu, S., Fu, X., Sharma, V. K., Mendoza-Sanchez, I., Qiu, Z., et al. (2018). Simultaneous removal of benzene, toluene, ethylbenzene and xylene (BTEX) by CaO2 based fenton system: enhanced degradation by chelating agents. Chem. Eng. J. 331, 255–264.
Yavari-Bafghi, M., Shavandi, M., Dastgheib, S. M. M., and Amoozegar, M. A. (2022). Simultaneous application of CaO2 nanoparticles and microbial consortium in Small bioreactor chambers (SBCs) for phenol removal from groundwater. Process Saf. Environ. Prot. 160, 465–477.
Ye, L., Yu, G., Zhou, S., Zuo, S., and Fang, C. (2017). Denitrification of nitrate-contaminated groundwater in columns packed with PHBV and ceramsites for application as a permeable reactive barrier. Water Supply 17, 1241–1248.
Yeh, C.-H., Lin, C.-W., and Wu, C.-H. (2010). A permeable reactive barrier for the bioremediation of BTEX-contaminated groundwater: microbial community distribution and removal efficiencies. J. Hazard. Mater. 178, 74–80. doi: 10.1016/j.jhazmat.2010.01.045
Keywords: oligotrophic groundwater, biodiversity, metagenome, phenol, bioremediation
Citation: Yavari-Bafghi M, Rezaei Somee M, Amoozegar MA, Dastgheib SMM and Shavandi M (2023) Genome-resolved analyses of oligotrophic groundwater microbial communities along phenol pollution in a continuous-flow biodegradation model system. Front. Microbiol. 14:1147162. doi: 10.3389/fmicb.2023.1147162
Received: 18 January 2023; Accepted: 13 March 2023;
Published: 29 March 2023.
Edited by:
Andreas Teske, University of North Carolina, Chapel Hill, United StatesReviewed by:
Anirban Chakraborty, Idaho State University, United StatesCopyright © 2023 Yavari-Bafghi, Rezaei Somee, Amoozegar, Dastgheib and Shavandi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mahmoud Shavandi, c2hhdmFuZGltQHJpcGkuaXI=; Mohammad Ali Amoozegar, YW1vb3plZ2FyQHV0LmFjLmly
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.