
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Microbiol., 16 March 2023
Sec. Microbiotechnology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1146650
This article is part of the Research TopicFungi for Food and Environmental SustainabilityView all 5 articles
 Vinita Verma1
Vinita Verma1 Arpita Batta2
Arpita Batta2 Harikesh B. Singh3
Harikesh B. Singh3 Alok Srivastava4
Alok Srivastava4 Sanjay Kumar Garg4
Sanjay Kumar Garg4 Vijay Pal Singh4
Vijay Pal Singh4 Pankaj Kumar Arora1*
Pankaj Kumar Arora1*The CRISPR/Cas9 system is a genome-editing tool that allows for precise and efficient modifications to the DNA of a cell. This technology can be used in endophytic fungi, which live within plants and can have beneficial effects on their host, making them important for agriculture. Using CRISPR/Cas9, researchers can introduce specific genetic changes into endophytic fungal genomes, allowing them to study the function of genes, improve their plant-growth-promoting properties, and create new, more beneficial endophytes. This system works by using the Cas9 protein, which acts as a pair of molecular scissors, to cut DNA at specific locations determined by a guide RNA. Once the DNA is cut, the cell’s natural repair mechanisms can be used to insert or delete specific genes, allowing for precise editing of the fungal genome. This article discusses the mechanism and applications of CRISPR/Cas9 to fungal endophytes.
Fungal endophytes are fungal species living within plant tissues that have no harmful effect on the plant but are useful to the host for a number of reasons (Collinge et al., 2019; Jørgensen et al., 2020). They are abundant within the intercellular spaces of the roots, stems, and leaves of plants. There is no visible evidence of fungal endophytes on host plants (Corrêa et al., 2014). The symbiotic relationship between endophytic fungal species and their hosts has been reported to possess interchangeable advantages. The plant provides essential nutrients to the endophytic species, which, in turn, confer tolerance and resistance against phytopathogen attack to their host.
Fungal endophytes produce a wide range of biologically active compounds, for example, anthraquinone, chromone, sesquiterpenoid, phenols, xanthone, quinolone, quinones, cyclic peptides, coumarins, piperazine, flavonoids, lignans, glycosides, terpenoids, saponins, and phenylpropanoids (Singh et al., 2021; Deshmukh et al., 2022). To date, a number of bioactive compounds from various endophytic fungi have been reported (Deshmukh et al., 2022). These bioactive compounds shield their host plants from adverse environmental circumstances and bio-agents (Verma et al., 2009). However, biological and non-biological factors affecting the host plant’s growth will also affect the fungal endophyte community.
With the development of clustered regularly interspaced short palindromic repeats–CRISPR-associated protein 9 (CRISPR-Cas9) genome-editing techniques, researchers have been able to change genomic sequences more precisely (Muñoz et al., 2019; Sarma et al., 2021; Rajan et al., 2022). The CRISPR-Cas system is an effective tool that can be applied to replacing, deleting, or inserting genes into the genomes of both eukaryotic and prokaryotic organisms (Jinek et al., 2012; Cong et al., 2013; Qi et al., 2013; Hsu et al., 2014). Crisper-Cas technology has been used to edit the genomes of several filamentous fungi; however, there is a limited study on endophytic fungi genome editing (Salazar-Cerezo et al., 2020; Satish et al., 2020). Although endophytic fungi produce many bioactive compounds, and CRISPR/Cas9 technology may increase their bioactive compound production, there are no review articles available on this topic. The purpose of this mini-review is to summarize the role of CRISPR/Cas9 technology in editing fungal endophyte genomes.
CRISPR/Cas systems have been discovered in bacteria and archaea and can be classified into three groups based on their Cas effectors (Cas9, Cas13, and Cas12), which are further subdivided into six types and more than 20 subtypes (Makarova et al., 2018; Li et al., 2019; Jiang et al., 2021). There is an increased use of the CRISPR-Cas Type II system because it is more efficient and simple to use. The RNA guide (sgRNA) and the Cas9 endonuclease are the main components of the CRISPR-Cas9 Type II system for gene targeting and cleavage. The short guide RNA (sgRNA) contains a simple chimeric strand of RNA that leads Cas9 to the target gene’s location in the genome, where it needs to be blocked from expressing. The Cas9 enzyme can bind to DNA and cause a double-strand break (DSB) in the target gene. It is important to note that Cas9 needs a short protospacer adjacent motif (PAM) that is located adjacent to a non-target DNA strand to achieve complementary target-DNA binding and cleavage (Mojica et al., 2009; Sternberg et al., 2014; Jiang et al., 2021). There are two methods to repair DSBs: homologous recombination (HR) and non-homologous end joining (NHEJ), achieving the target sequence editing (Figure 1). The NHEJ system is the dominant method for repairing DSB, which can re-join the ends of the DSB, causing nucleotide insertions, substitutions, or deletions. Due to these indels, non-sense sequences were produced, or premature stop codons appeared, blocking the transcription of the target gene (Muñoz et al., 2019). The HR system requires constructing homology-directed DNA repair templates, which are then introduced together with CRISPR-Cas components to change target sequences exactly.
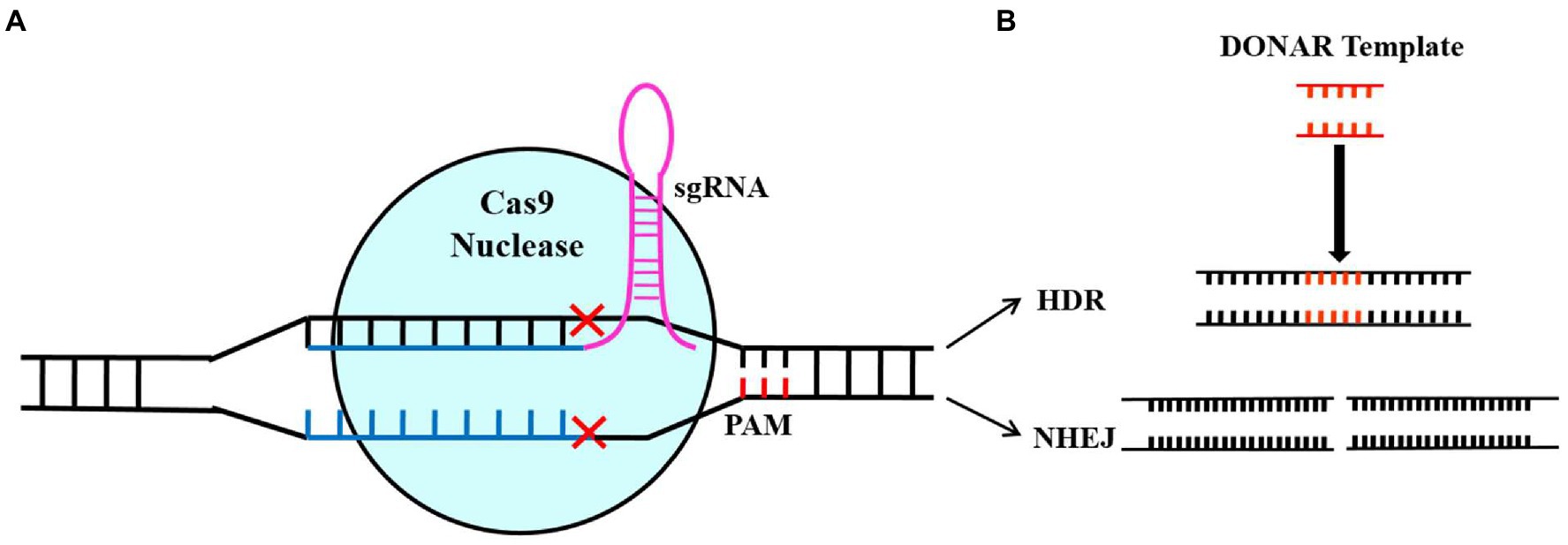
Figure 1. Illustration of Cas9/sgRNA-based genome editing. The diagram showing the sgRNA-mediated Cas9 protein binding to the target sequence and knocking out the double strand of DNA (A) and the self-repair mechanism of the cell after DNA double-strand breakage (B). A non-homologous end-joining repair pathway results in random loss, introduction, and replacement of bases at damaged points, thus resulting in gene mutation. By using the homology repair pathway (HR), the gene of interest will be precisely edited based on fragments of donor DNA (Song et al., 2019).
Cas9/guide RNA complexes are in vivo generated by delivering the Cas9 gene and guide DNA in expression cassettes. A DNA shuttle, selective markers, and promoter and terminator sequences that are compatible with the native host are required for this method, which is often unavailable for new isolates (Sagita et al., 2021). To overcome this problem, it is now possible to prepare Cas9-gRNA ribonucleoprotein complex in vitro and transform them into fungal protoplasts. CRISPR/Cas9 systems in endophytic fungi must follow the following steps.
Cas9 protein has approximately 1,400 amino acids in total and performs endonuclease functions as part of the CRISPR/Cas9 system. Because the CRISPR/Cas9 system originated in bacteria or archaea, Cas9 proteins are usually optimized for fungal codons when CRISPR/Cas9 system was used in fungi (Song et al., 2019). The fusion of nuclear localization signal to either of C-terminal (Nødvig et al., 2015; Song et al., 2018) or N-terminal (Hu et al., 2018)/both termini (Katayama et al., 2016) of Cas9 protein is the way to translocate the Cas9 protein into the nucleus. The nucleoplasmin (Wang et al., 2016), Human c-Myc (Chen et al., 2018), Simian virus 40 (Wang et al., 2016; Song et al., 2018), and Myceliophthora hac-1 (Liu et al., 2017) are reported nuclear localization signals (NLS) that are used for the localization of Cas9 in the cells of fungal strains. Cas9 gene expression depends on the type of promoter used. Cas9 exogenous genes’ expression efficiency is greatly influenced by the strength of the promoter driving transcription, thus selecting a suitable promoter is vital for its function (Ouedraogo and Tsang, 2020). There are many types of promoters, but the most common ones are constitutive promoters. It occurs continuously in all tissues and organs of an organism. This type of promoter-initiated gene has relatively stable levels of transcription and expression. Examples of constitutive promoters are gpdA (Nødvig et al., 2015), tef1 (DiCarlo et al., 2013; Nødvig et al., 2015), and pkiA (Song et al., 2018). Inducible promoters can be applied to control the expression of the Cas9 gene while toxicity due to DNA scission activity of Cas9 is perceived. In fungi, no reports on the toxicity of Cas9 gene expression have been declared to date (Ouedraogo and Tsang, 2020).
To process a fully developed guide RNA, CRISPR/Cas9 tool requires an 80-nucleotide trans-activating CRISPR RNA (tracrRNA; Carroll, 2012; Chylinski et al., 2013). For the simplification of heterologous editing of the genome, the trans-activating CRISPR RNA is built for fusing it at the 5′ end of crRNA (20-nucleotide target sequence) to produce a single guide RNA (sgRNA; Chylinski et al., 2013). Moreover, adequate expression and the processing of sgRNA consider important means for developing an editing system in CRISPR/Cas. Most often, in vivo sgRNA expression results from pol III promoters including 5S RNA, snr52, tRNA, and U6. While using pol II promoters to express, sgRNA demands a ribosomal splicing sequence for the processing of pre-crRNA (Ouedraogo and Tsang, 2020).
The Cas9 and sgRNA expression cassettes can be delivered to fungal cells by using single-vector or dual-vector systems (Pohl et al., 2016; Zhang et al., 2016). Single-vector expression systems are significantly more accurate and efficient than dual-vector expression systems. CRISPR/Cas9 systems are mainly transformed into fungal cells using polyethylene glycol (PEG) and Agrobacterium-mediated transformation (AMT). PEG-mediated transformation is one of the easiest methods for transforming Cas9 and gRNA expression cassettes into fungi, thus most studies use this method (Song et al., 2019). Nevertheless, some researchers have found that Agrobacterium-mediated CRISPR/Cas9 systems can also be used for efficient genome editing in fungi.
The direct delivery of the ribonucleoprotein (RNP) complex composed of Cas9 protein and gRNA is an alternative method. The implementation of the RNP complex for genome editing, via the CRISPR tool, is quite advantageous as it allows for reducing difficulties in selecting promoters for Cas9 expression processing of single guide RNA (sgRNA). Moreover, in vitro assembly of sgRNA using the RNP complex has been reported to minimize the off-target reactions due to the transient feature of the Cas9-sgRNA-RNP complex (Foster et al., 2018).
The CRISPR-Cas9 system has been extensively used in model filamentous fungi such as Sporormiella minima (Zheng et al., 2017), Aspergillus oryzae (Katayama et al., 2016), Aspergillus niger (Kuivanen et al., 2016), Aspergillus nidulans (Nødvig et al., 2015), Talaromyces atroroseus (Nielsen et al., 2017), Neurospora crassa (Matsu-Ura et al., 2015), Alternaria alternata (Wenderoth et al., 2017), and Penicillium chrysogenum (Pohl et al., 2016). In contrast, its application to non-model endophytic fungi is less common (Table 1). This section discusses CRISPR/Cas9 applications to the genome editing of a few endophytic fungi.
Epichloë species are fungal endophytes of cold-season grasses. These endophytes are responsible for increasing durability, productivity, and plant health by providing the ability to withstand harsh conditions and tolerate drought (Malinowski and Belesky, 2000; Malinowski and Belesky, 2019); however, some Epichloë species have been reported to produce toxic alkaloids that harm livestock (Florea et al., 2021). Furthermore, four different classes of alkaloids are synthesized by the Epichloë species to protect the host plant from invertebrates (Clay and Schardl, 2002; Schardl et al., 2013). However, introducing non-toxic fungal endophytes to different forage varieties may improve livestock performance (Florea et al., 2021). Strains of Epichloë coenophiala, which has been unintentionally co-propagated with a member of grass called tall fescue, have been reported to produce an extremely injurious ergopeptine-type ergot alkaloid known as ergovaline (Thompson et al., 2001; Aiken et al., 2013; Klotz, 2015). Only a small amount of ergovaline has a detrimental effect on plant reproduction, resulting in a loss of plant productivity and ultimately livestock health. The dimethylallyltryptophan synthase gene (dmaW) was found to be essential for the biosynthesis of the ergot alkaloid (Wang et al., 2004), which was determined by deleting the dmaW gene from the Epichlo hybrida strain Lp1 and subsequent complementation with the ortholog from the plant pathogen Claviceps fusiformis (Tsai et al., 1995). A study based on long-read sequencing of the genome revealed the presence of ergot alkaloid biosynthesis (EAS) genes clusters in Epichloë hybrida Lp1 and Epichloë coenophiala strain e19, which are fungal endophytes of Lolium perenne and Lolium arundinaceum, respectively. Moreover, Epichloë coenophiala has been reported with two homologous copies of ergot alkaloid biosynthesis gene clusters EAS1 (196 kb) and EAS2 (75 kb); however; Epichloë hybrida has a single EAS (83 kb) gene cluster (Florea et al., 2021).
The CRISPR-based study targeted these EAS gene clusters through the transformation of ribonucleoprotein (RNP) complexes of Cas9-2NLS (edited Cas9 nuclease), pairs of sgRNAs, and transiently opted plasmids into Epichloe spp. This method led to deleting EAS1 and EAS2 clusters (Figure 2A) separately and both of these clusters simultaneously in Epichloë coenophiala. In addition, the tool was able to delete both the EAS cluster as well as alkaloid biosynthesis genes, including dmaW and lolC, which had previously been quite hard to delete in Epichloë coenophiala. For these reasons, CRISPR-RNP is an excellent way to provide non-transgenic endophytes free of toxic genes, so forage cultivars can be improved and further research can be carried out (Florea et al., 2021).
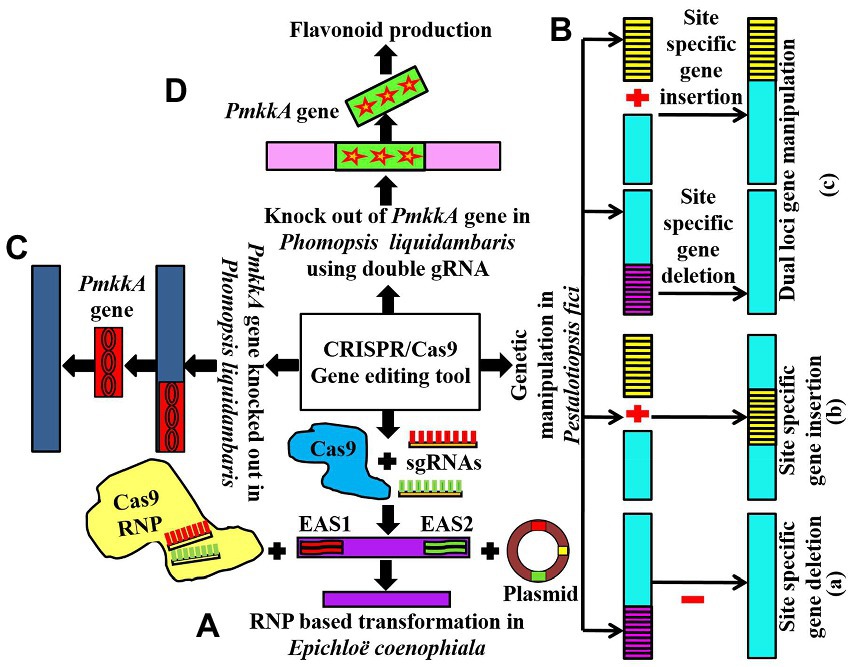
Figure 2. (A) Showing RNP-based transformation and disruption of EAS1 and EAS2 gene clusters in Epichloë coenophiala using the CRISPR/Cas9 technique; (Ba,b) communicating CRISPR/Cas9-based single locus gene manipulation in Pestalotiopsis fici; (c) showing dual loci gene editing in Pestalotiopsis fici using CRISPR/Cas9 technology; (C) showing knocking out of PmkkA gene in Phomopsis liquidambaris using CRISPR/Cas9 tool (D) conveys the role of CRISPR/Cas9 to knockout PmkkA gene using double gRNA in Phomopsis liquidambaris resulting in the production of flavonoids.
The CRISPR/Cas9 system was introduced into the endophytic fungus Pestalotiopsis fici using polyethylene glycol (PEG)-mediated protoplast transformation (Xu et al., 2021). The researchers used this approach to efficiently insert new genes into P. fici, mutate two loci simultaneously, and delete long DNA fragments (Figure 2B). For two-site gene editing and site-specific gene insertion, the efficiency was up to 44.4% and 48.0%, respectively. This study shows that using the CRISPR/Cas9 system to edit genes in Pestalotiopsis fici improves the efficiency of editing genes and reduces the number of rounds of transformation required (half the time needed in the traditional method of Agrobacterium-mediated transformation). As a result, the CRISPR/Cas9-dependent genome-manipulating tool increases the efficiency of homologous recombination and multi-loci gene editing, which reduces the number of genetic engineering attempts. For further research on Pestalotiopsis fici, the development of a well-organized CRISPR/Cas9 genome-editing tool will be beneficial, but it will also be beneficial for model fungal strains in the same genus to extract additional potent secondary metabolic compounds (Xu et al., 2021).
Fungal endophyte Phomopsis liquidambaris has been reported to promote growth and nitrogen content in peanuts and rice. However, further investigation of the interaction mechanisms between Phomopsis liquidambaris and its host plants is still limited due to the lack of advanced genetic tools and systematic studies. Nonetheless, Huang et al. (2020) used the CRISPR/Cas9 technique to evaluate the impact of disrupting the PmkkA gene in Phomopsis liquidambaris, which encodes mitogen activated protein kinase kinase (MAPKK) in the cell wall integrity MAPK pathway (Figure 2C). It has been revealed that the CWI MAPK pathway plays a key role in cell wall integrity which defends the cells from extreme adverse conditions such as hypo-osmotic stress and heat shock (Bermejo et al., 2008). In rice seedlings, the PmkkA mutant strain induces a significant increase (reactive oxygen species) in ROS production, glucanase, and chitinase activities in comparison with the wild type strain, which resulted in robust resistance and growth suppression on rice. These results show that the PmkkA gene is important in interacting with its host rice plant, as well as inhibiting the plant’s immune system (Huang et al., 2020). The investigation resulted in the successful exploitation of the CRISPR/Cas9 technique which would be beneficial for a deep study on the mutualistic interaction of Phomopsis liquidambaris with its host plants system.
Yang et al. (2021) reported flavonoid production (naringenin, kaempferol, and quercetin) from the endophytic fungus P. liquidambaris by disrupting the MAPKK gene using the modified CRISPR-Cas9 system. To disturb the MAPKK gene, they introduced double sgRNA into the CRISPR-Cas9 system (Figure 2D). This modified CRISPR-Cas9 system is simple and beneficial to reduce the toxicity of high Cas9/gRNA concentration to cells and the off-target effects.
Based on the aforementioned examples, we can conclude that gene editing is more convenient for endophytic fungal strains. CRISPR/Cas9 systems have been developed for few endophytic fungi today. It can be expanded to more endophytic fungi to enhance their properties. Furthermore, since only a few selective markers are available for endophytic fungal species, re-processing these markers and allowing further genetic engineering will serve as vital support for genetic research (Meng et al., 2022). It will be advantageous for endophytic fungi to overexpress genes encoding secondary metabolites using CRISPR/CAS9.
Overall, CRISPR/Cas9 is an advanced gene manipulation technique that plays an important role in genetically manipulating fungal endophytes. Furthermore, removing or inserting the functional genes of interest provides researchers with an opportunity to discover novel genes in endophytic fungal strains for genetic research. By implementing CRISPR/Cas9 in molecular biology, inventors will also be able to overcome the biological challenges they face. The development of efficient, feasible, and versatile CRISPR/Cas9 gene editing tools will enable leftfield outcomes instead of tedious and time-consuming traditional methods.
By using the CRISPR/Cas9 system, it is possible to edit multiple genes simultaneously and obtain mutants that have multiple mutations in a single transformation. This improves the efficiency of genome editing as well as the production of secondary metabolites in endophytic fungi. CRISPR/Cas9 has, however, only been applied to a limited number of real applications in endophytic fungi for the production of secondary metabolites. Therefore, CRISPR/Cas9 systems in endophytic fungi are still in the early stages of development, with most of the research focusing on assessing their feasibility.
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
PA acknowledges the Department of Biotechnology, India, for awarding him Ramalingaswami Re-entry Fellowship.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Aiken, G. E., Klotz, J. L, Johnson, J. M., Strickland, J. R., and Schrick, F. N. (2013). Postgraze assessment of toxicosis symptoms for steers grazed on toxic endophyte-infected tall fescue pasture. J. Anim. Sci. 91, 15878–5884.
Bermejo, C., Rodríguez, E., García, R., Rodríguez-Peña, J. M., Rodríguez de la Concepción, M. L., Rivas, C., et al. (2008). The sequential activation of the yeast HOG and SLT2 pathways is required for cell survival to cell wall stress. Mol. Biol. Cell 19, 1113–1124. doi: 10.1091/mbc.e07-08-0742
Carroll, D. (2012). Genome engineering with zinc-finger nucleases. Genetics 188, 773–782. doi: 10.1534/genetics.111.131433
Chen, B. X., Wei, T., Ye, Z. W., Yun, F., Kang, L. Z., Tang, H. B., et al. (2018). Efficient CRISPR-Cas9 gene disruption system in edible-medicinal mushroom Cordyceps militaris. Front. Microbiol. 9:1157. doi: 10.3389/fmicb.2018.01157
Chylinski, K., Le Rhun, A., and Charpentier, E. (2013). The tracrRNA and Cas9 families of type II CRISPR-Cas immunity systems. RNA Biol. 10, 726–737. doi: 10.4161/rna.24321
Clay, K., and Schardl, C. (2002). Evolutionary origins and ecological consequences of endophyte symbiosis with grasses. Am. Nat. 160, S99–S127. doi: 10.1086/342161
Collinge, D. B., Jørgensen, H. J., Latz, M. A., Manzotti, A., Ntana, F., Rojas Tayo, E. C., et al. (2019). “Searching for novel fungal biological control agents for plant disease control among endophytes” in Endophytes for a growing world. eds. T. Hodkinson, F. Doohan, M. Saunders, and B. Murphy (Cambridge: Cambridge University Press), 25–51.
Cong, L., Ran, F. A., Cox, D., Lin, S., Barretto, R., Habib, N., et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. doi: 10.1126/science.1231143
Corrêa, R. C. G., Rhoden, S. A., Mota, T. R., Azevedo, J. L., Pamphile, J. A., de Souza, C. G. M., et al. (2014). Endophytic fungi: expanding the arsenal of industrial enzyme producers. J. Ind. Microbiol. Biotechnol. 41, 1467–1478. doi: 10.1007/s10295-014-1496-2
Deshmukh, S. K., Dufossé, L., Chhipa, H., Saxena, S., Mahajan, G. B., and Gupta, M. K. (2022). Fungal endophytes: a potential source of antibacterial compounds. J. Fungi 8:164. doi: 10.3390/jof8020164
DiCarlo, J. E., Norville, J. E., Mali, P., Rios, X., Aach, J., and Church, G. M. (2013). Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 41, 4336–4343. doi: 10.1093/nar/gkt135
Florea, S., Jaromczyk, J., and Schardl, C. L. (2021). Non-transgenic CRISPR-mediated knockout of entire ergot alkaloid gene clusters in slow-growing asexual polyploid fungi. Toxins 13:153. doi: 10.3390/toxins13020153
Foster, A. J., Martin-Urdiroz, M., Yan, X., Wright, H. S., Soanes, D. M., and Talbot, N. J. (2018). CRISPR-Cas9 ribonucleoprotein-mediated co-editing and counter selection in the rice blast fungus. Sci. Rep. 8, 1–12. doi: 10.1038/s41598-018-32702-w
Hsu, P. D., Lander, E. S., and Zhang, F. (2014). Development and applications of CRISPR-Cas9 for genome engineering. Cells 157, 1262–1278. doi: 10.1016/j.cell.2014.05.010
Hu, P., Zhao, X., Zhang, Q., Li, W., and Zu, Y. (2018). Comparison of various nuclear localization signal-fused Cas9 proteins and Cas9 mRNA for genome editing in Zebrafish. G3 8, 823–831. doi: 10.1534/g3.117.300359
Huang, P. W., Yang, Q., Zhu, Y. L., Zhou, J., Sun, K., Mei, Y. Z., et al. (2020). The construction of CRISPR-Cas9 system for endophytic Phomopsis liquidambaris and its PmkkA-deficient mutant revealing the effect on rice. Fungal Genet. Biol. 136:103301. doi: 10.1016/j.fgb.2019.103301
Jiang, C., Lv, G., Tu, Y., Cheng, X., Duan, Y., Zeng, B., et al. (2021). Applications of CRISPR/Cas9 in the synthesis of secondary metabolites in filamentous fungi. Front. Microbiol. 12:638096. doi: 10.3389/fmicb.2021.638096
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., and Charpentier, E. (2012). A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. doi: 10.1126/science.1225829
Jørgensen, H. J. L., Collinge, D. B., Rojas, E. C., Latz, M. A. C., Manzotti, A., Ntana, F., et al. (2020). Plant endophytes. Encycl. Life Sci. 0028893. doi: 10.1002/9780470015902.a0028893
Katayama, T., Tanaka, Y., Okabe, T., Nakamura, H., Fujii, W., Kitamoto, K., et al. (2016). Development of a genome editing technique using the CRISPR/Cas9 system in the industrial filamentous fungus Aspergillus oryzae. Biotechnol. Lett. 38, 637–642. doi: 10.1007/s10529-015-2015-x
Klotz, J. L. (2015). Activities and effects of ergot alkaloids on livestock physiology and production. Toxins 7, 2801–2821. doi: 10.3390/toxins7082801
Kuivanen, J., Wang, Y. M. J., and Richard, P. (2016). Engineering Aspergillus Niger for galactaric acid production: elimination of galactaric acid catabolism by using RNA sequencing and CRISPR/Cas9. Microb. Cell Fact. 15, 1–9. doi: 10.1186/s12934-016-0613-5
Li, Y., Li, S. Y., Wang, J., and Liu, G. (2019). CRISPR/Cas systems towards next-generation biosensing. Trends Biotechnol. 37, 730–743. doi: 10.1016/j.tibtech.2018.12.005
Liu, Q., Gao, R., Li, J., Lin, L., Zhao, J., Sun, W., et al. (2017). Development of a genome-editing CRISPR/Cas9 system in thermophilic fungal Myceliophthora species and its application to hyper-cellulase production strain engineering. Biotechnol. Biofuels 10, 1–14. doi: 10.1186/s13068-016-0693-9
Makarova, K. S., Wolf, Y. I., and Koonin, E. V. (2018). Classification and nomenclature of CRISPR-Cas systems: where from here? CRISPR J. 1, 325–336. doi: 10.1089/crispr.2018.0033
Malinowski, D. P., and Belesky, D. P. (2000). Adaptations of endophyte-infected cool-season grasses to environmental stresses: mechanisms of drought and mineral stress tolerance. Crop. Sci. 40, 923–940. doi: 10.2135/cropsci2000.404923x
Malinowski, D. P., and Belesky, D. P. (2019). Epichloë (formerly Neotyphodium) fungal endophytes increase adaptation of cool-season perennial grasses to environmental stresses. Acta Agrobot. 72:1767. doi: 10.5586/aa.1767
Matsu-Ura, T., Baek, M., Kwon, J., and Hong, C. (2015). Efficient gene editing in Neurospora crassa with CRISPR technology. Fungal Biol. Biotechnol. 2, 1–7. doi: 10.1186/s40694-015-0015-1
Meng, G., Wang, X., Liu, M., Wang, F., Liu, Q., and Dong, C. (2022). Efficient CRISPR/Cas9 system based on autonomously replicating plasmid with an AMA1 sequence and precisely targeted gene deletion in the edible fungus, Cordyceps militaris. J. Microbial. Biotechnol. 15, 2594–2606. doi: 10.1111/1751-7915.14107
Mojica, F. J. M., Diez-Villasenor, C., Garcia-Martinez, J., and Almendros, C. (2009). Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 155, 733–740. doi: 10.1099/mic.0.023960-0
Muñoz, I. V., Sarrocco, S., Malfatti, L., Baroncelli, R., and Vannacci, G. (2019). CRISPR-Cas for fungal genome editing: a new tool for the management of plant diseases. Front. Plant Sci. 10:135. doi: 10.3389/fpls.2019.00135
Nielsen, M. L., Isbrandt, T., Rasmussen, K. B., Thrane, U., Hoof, J. B., Larsen, T. O., et al. (2017). Genes linked to production of secondary metabolites in Talaromyces atroroseus revealed using CRISPR-Cas9. PLoS One 12:0169712. doi: 10.1371/journal.pone.0169712
Nødvig, C. S., Nielsen, J. B., Kogle, M. E., and Mortensen, U. H. (2015). A CRISPR-Cas9 system for genetic engineering of filamentous fungi. PLoS One 10:0133085. doi: 10.1371/journal.pone.0133085
Ouedraogo, J. P., and Tsang, A. (2020). CRISPR_Cas systems for fungal research. Fungal Biol. Rev. 34, 189–201. doi: 10.1016/j.fbr.2020.10.002
Pohl, C., Kiel, J. A., Driessen, A. J., Bovenberg, R. A., and Nygard, Y. (2016). CRISPR/Cas9 based genome editing of Penicillium chrysogenum. ACS Synth. Biol. 5, 754–764. doi: 10.1021/acssynbio.6b00082
Qi, L. S., Larson, M. H., Gilbert, L. A., Doudna, J. A., Weissman, J. S., Arkin, A. P., et al. (2013). Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cells 152, 1173–1183. doi: 10.1016/j.cell.2013.02.022
Rajan, A., Shrivastava, S., Kumar, A., Singh, A. K., and Arora, P. K. (2022). CRISPR-Cas system: from diagnostic tool to potential antiviral treatment. Appl. Microbiol. Biotechnol. 106, 5863–5877. doi: 10.1007/s00253-022-12135-2
Sagita, R., Quax, W. J., and Haslinger, K. (2021). Current state and future directions of genetics and genomics of Endophytic fungi for bioprospecting efforts. Front. Bioeng. Biotechnol. 9:649906. doi: 10.3389/fbioe.2021.649906
Salazar-Cerezo, S., Kun, R. S., De Vries, R. P., and Garrigues, S. (2020). CRISPR/Cas9 technology enables the development of the filamentous ascomycete fungus Penicillium subrubescens as a new industrial enzyme producer. Enzyme Microb. Technol. 133:109463. doi: 10.1016/j.enzmictec.2019.109463
Sarma, H., Islam, N. F., Prasad, R., Prasad, M. N. V., Ma, L. Q., and Rinklebe, J. (2021). Enhancing phytoremediation of hazardous metal (loid) s using genome engineering CRISPR–Cas9 technology. J. Hazard. Mater. 414:125493. doi: 10.1016/j.jhazmat.2021.125493
Satish, L., Shamili, S., Muthubharathi, B. C., Ceasar, S. A., Kushmaro, A., Singh, V., et al. (2020). CRISPR-Cas9 system for fungi genome engineering toward industrial applications. Genome Engineer. via CRISPR-Cas9 Syst. 69–81. doi: 10.1016/B978-0-12-818140-9.00006-4
Schardl, C. L., Young, C. A., Pan, J., Florea, S., Takach, J. E., Panaccione, D., et al. (2013). Currencies of mutualisms: sources of alkaloid genes in vertically transmitted Epichloae. Toxins 5, 1064–1088. doi: 10.3390/toxins5061064
Singh, A., Singh, D. K., Kharwar, R. N., White, J. F., and Gond, S. K. (2021). Fungal endophytes as efficient sources of plant-derived bioactive compounds and their prospective applications in natural product drug discovery: insights, avenues, and challenges. Microorganisms 9:197. doi: 10.3390/microorganisms9010197
Song, L., Ouedraogo, J. P., Kolbusz, M., Nguyen, T. T. M., and Tsang, A. (2018). Efficient genome editing using tRNA promoter-driven CRISPR/Cas9 gRNA in Aspergillus Niger. PLoS One 13:0202868. doi: 10.1371/journal.pone.0202868
Song, R., Zhai, Q., Sun, L., Huang, E., Zhang, Y., Zhu, Y., et al. (2019). CRISPR/Cas9 genome editing technology in filamentous fungi: progress and perspective. Appl. Microbiol. Biotechnol. 103, 6919–6932. doi: 10.1007/s00253-019-10007-w
Sternberg, S. H., Redding, S., Jinek, M., Greene, E. C., and Doudna, J. A. (2014). DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 507, 62–67. doi: 10.1038/nature13011
Thompson, F. N., Stuedemann, J. A., and Hill, N. S. (2001). Anti-quality factors associated with alkaloids in eastern temperate pasture. Rangel. Ecol. Manage. 54, 474–489. doi: 10.2307/4003119
Tsai, H. F., Wang, H., Gebler, J. C., Poulter, C. D., and Schardl, C. L. (1995). The Claviceps purpurea gene encoding dimethylallyltryptophan synthase, the committed step for ergot alkaloid biosynthesis. Biochem. Biophys. Res. Commun. 216, 119–125. doi: 10.1006/bbrc.1995.2599
Verma, V. C., Kharwar, R. N., and Strobel, G. A. (2009). Chemical and functional diversity of natural products from plant associated endophytic fungi. Nat. Prod. Commun. 4, 1511–1532. doi: 10.1177/1934578X0900401114
Wang, J., Machado, C., Panaccione, D. G., Tsai, H. F., and Schardl, C. L. (2004). The determinant step in ergot alkaloid biosynthesis by an endophyte of perennial ryegrass. Fungal Genet. Biol. 41, 189–198. doi: 10.1016/j.fgb.2003.10.002
Wang, Y., Wei, D., Zhu, X., Pan, J., Zhang, P., and Huo, L. (2016). A ‘suicide’ CRISPR-Cas9 system to promote gene deletion and restoration by electroporation in Cryptococcus neoformans. Sci. Rep. 6:31145. doi: 10.1038/srep31145
Wenderoth, M., Pinecker, C., Voss, B., and Fischer, R. (2017). Establishment of CRISPR/Cas9 in Alternaria alternata. Fungal Genet. Biol. 101, 55–60. doi: 10.1016/j.fgb.2017.03.001
Xu, X., Huang, R., and Yin, W. B. (2021). An optimized and efficient CRISPR/Cas9 system for the endophytic fungus Pestalotiopsis fici. J. Fungi 7:809. doi: 10.3390/jof7100809
Yang, Q., Wu, M., Zhu, Y. L., Yang, Y. Q., Mei, Y. Z., and Dai, C. C. (2021). The disruption of the MAPKK gene triggering the synthesis of flavonoids in endophytic fungus Phomopsis liquidambaris. Biotechnol. Lett. 43, 119–132. doi: 10.1007/s10529-020-03042-5
Zhang, C., Meng, X., Wei, X., and Lu, L. (2016). Highly efficient CRISPR mutagenesis by microhomology-mediated end joining in Aspergillus fumigatus. Fungal Genet. Biol. 86, 47–57. doi: 10.1016/j.fgb.2015.12.007
Keywords: fungal endophytes, bioactive compounds, CRISPR/Cas9 technology, bioengineering, metabolite
Citation: Verma V, Batta A, Singh HB, Srivastava A, Garg SK, Singh VP and Arora PK (2023) Bioengineering of fungal endophytes through the CRISPR/Cas9 system. Front. Microbiol. 14:1146650. doi: 10.3389/fmicb.2023.1146650
Received: 17 January 2023; Accepted: 14 February 2023;
Published: 16 March 2023.
Edited by:
Ram Prasad, Mahatma Gandhi Central University, Motihari, IndiaReviewed by:
Gaurav Saxena, Shoolini University of Biotechnology and Management Sciences, IndiaCopyright © 2023 Verma, Batta, Singh, Srivastava, Garg, Singh and Arora. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pankaj Kumar Arora, YXJvcmE0ODRAZ21haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.