
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 13 April 2023
Sec. Microbial Physiology and Metabolism
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1146210
This article is part of the Research Topic Fungal Green Biotechnology and Biomanufacturing View all 6 articles
In the control of plant diseases, biocontrol has the advantages of being efficient and safe for human health and the environment. The filamentous fungus Trichoderma harzianum and its closely related species can inhibit the growth of many phytopathogenic fungi, and have been developed as commercial biocontrol agents for decades. In this review, we summarize studies on T. harzianum species complex from the perspective of strain improvement. To elevate the biocontrol ability, the production of extracellular proteins and compounds with antimicrobial or plant immunity-eliciting activities need to be enhanced. In addition, resistance to various environmental stressors should be strengthened. Engineering the gene regulatory system has the potential to modulate a variety of biological processes related to biocontrol. With the rapidly developing technologies for fungal genetic engineering, T. harzianum strains with increased biocontrol activities are expected to be constructed to promote the sustainable development of agriculture.
Biotic stresses in plants are caused by diverse organisms such as fungi, bacteria, viruses, weeds, and insects (Redondo-Gómez, 2013). A recent study reassessed the figures for five staple crop losses associated with biotic stresses, showing that global crop loss estimates per crop were 21.5, 30.0, 22.6, 17.2, and 21.4% for wheat, rice, maize, potato, and soybean, respectively (Savary et al., 2019). Consequently, chemical pesticides are commonly used in agricultural systems. However, the excessive and irrational use of chemical pesticides can lead to non-target effects, potential environmental and public health risks, and the generation of resistance among pests (Jasuja, 2015; Goswami et al., 2018). In comparison, biocontrol methods employing the natural enemies of pests have the advantage of being safe with lower risks of pest resistance, resulting in them being widely used in agricultural production.
Trichoderma are well-known beneficial microorganisms in agriculture because of their ability to kill pathogenic fungi and promote plant growth (Verma et al., 2007). As biofungicides, Trichoderma species can inhibit the growth of many phytopathogenic fungi and oomycetes, e.g., Fusarium solani, Sclerotinia sclerotiorum, Botrytis cinerea, Macrophomina phaseolina, Cordana musae, Rhizoctonia solani, and Pythium ultimum (Anees et al., 2010; Samuelian, 2016; Zhang et al., 2016; Hewedy et al., 2020; Erazo et al., 2021). Inhibition is believed to involve three main mechanisms (Figure 1): (1) competition for nutrients (e.g., carbon, nitrogen, and iron) or infection spots with pathogenic fungi (Sivan, 1989; Güçlü and Özer, 2022); (2) mycoparasitism (Mukherjee et al., 2022); and (3) antibiosis through the synthesis of secondary metabolites with inhibitory or lethal effects on pathogenic fungi (El-Debaiky, 2017; Mironenka et al., 2021). In addition, Trichoderma species can indirectly prevent pathogen infection by inducing plant resistance responses (Harman et al., 2004; Woo et al., 2022).
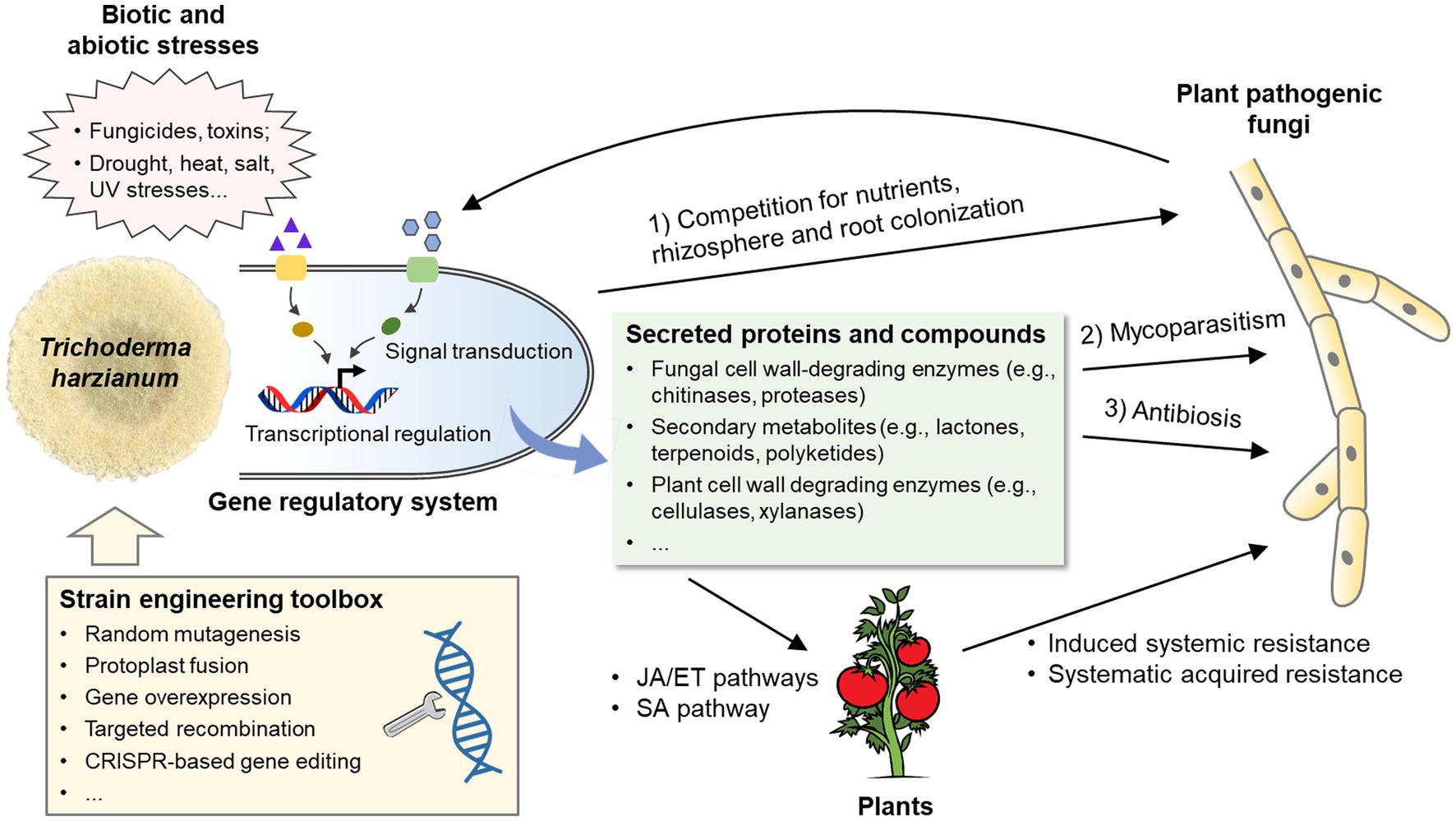
Figure 1. Biological processes involved in the biocontrol capacity of Trichoderma harzianum. Members in the T. harzianum species complex inhibit the growth of plant pathogenic fungi through competition, mycoparasitism and antibiosis. Meanwhile, T. harzianum activates defensive reactions in plants, which include induced systemic resistance and systemic acquired resistance. A set of secret proteins and secondary metabolites produced by T. harzianum play important roles in the above processes. In addition, T. harzianum is subjected to a combination of different biotic and abiotic stresses in the field. Signaling pathways and the downstream transcriptional regulation system are responsible for the regulation of responses to fungal pathogens and environmental stresses. With the genetic engineering toolbox, the biocontrol capacity of T. harzianum can be significantly improved. JA, jasmonic acid; ET, ethylene; SA, salicylic acid.
Trichoderma harzianum is one of the most frequently used Trichoderma species in the management of plant diseases (Meher et al., 2020; Rush et al., 2021). It has been used for the production of more than twenty commercial biocontrol agents all over the world (Woo et al., 2014), of which eight are listed in Table 1. T. harzianum not only has mycoparasitic properties but also the ability to promote plant growth by adjusting the balance of hormones and acting as a biofertilizer to promote the uptake of mineral ions and carbon dioxide (Stewart and Hill, 2014; Marra et al., 2021). In the comparison of 27 Trichoderma species, T. harzianum/T. afroharzianum was found to produce the highest number of known biopesticides and plant growth-promoting compounds (Rush et al., 2021). Classical random mutagenesis (Szekeres et al., 2007; Marzano et al., 2013) and protoplast fusion (Prabavathy et al., 2006) have been successfully used to generate T. harzianum strains with improved performance. Along with a deeper understanding of the molecular mechanisms of biocontrol (Daguerre et al., 2014; Sood et al., 2020; Abbas et al., 2022; Chen et al., 2022), rational genetic engineering has become a feasible strategy for improving the strains of T. harzianum (Chen et al., 2021). Nevertheless, most of the commercial strains are reported to be wild-type isolates and no information on genetic improvement was reported. This phenomenon can be related to the restrictions and public concerns about genetically modified organisms (GMOs) (Chen et al., 2022).
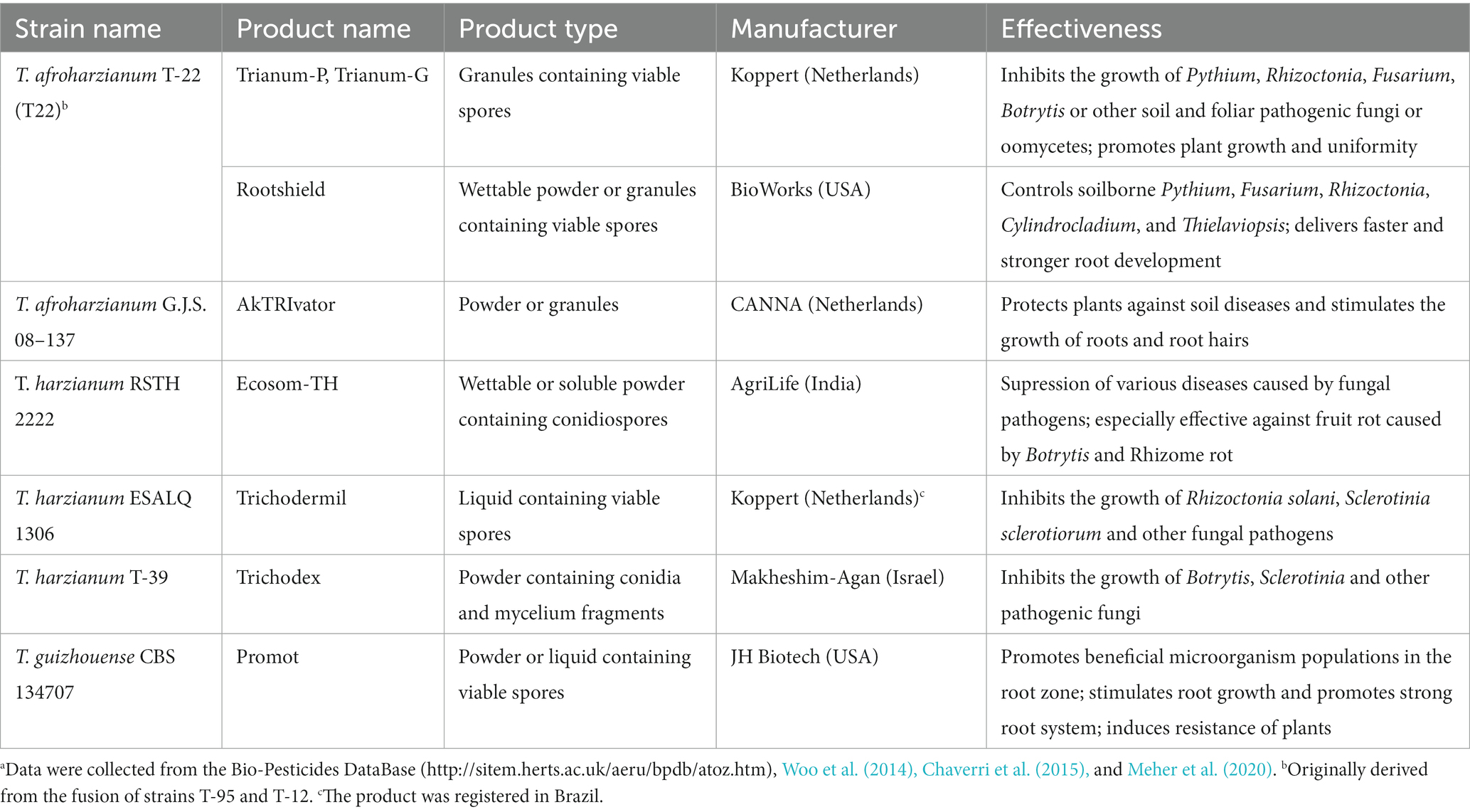
Table 1. Selected Trichoderma harzianum species complex strains used for the manufacture of biocontrol products.a
In this article, we review studies on the development of T. harzianum strains with enhanced biocontrol activity in laboratory level. These include the strengthening of protein and chemical effectors for biocontrol, enhancing the robustness of strains, and modulation of the gene regulatory system controlling these processes. It should be noted that with the development of systematic taxonomy in the fungal community, many previously described “T. harzianum” strains have been identified as other Trichoderma species (Mach et al., 1999; Chaverri et al., 2015; Fanelli et al., 2018; Cai and Druzhinina, 2021). For example, the strain T22, widely used as commercial biocontrol agents, was re-identified to be T. afroharzianum belonging to the T. harzianum species complex (Chaverri et al., 2015; Kubicek et al., 2019). Therefore, the review covers the research progresses in the T. harzianum complex (Figure 2), considering that many mechanisms for biocontrol are conserved among the members in this species complex.
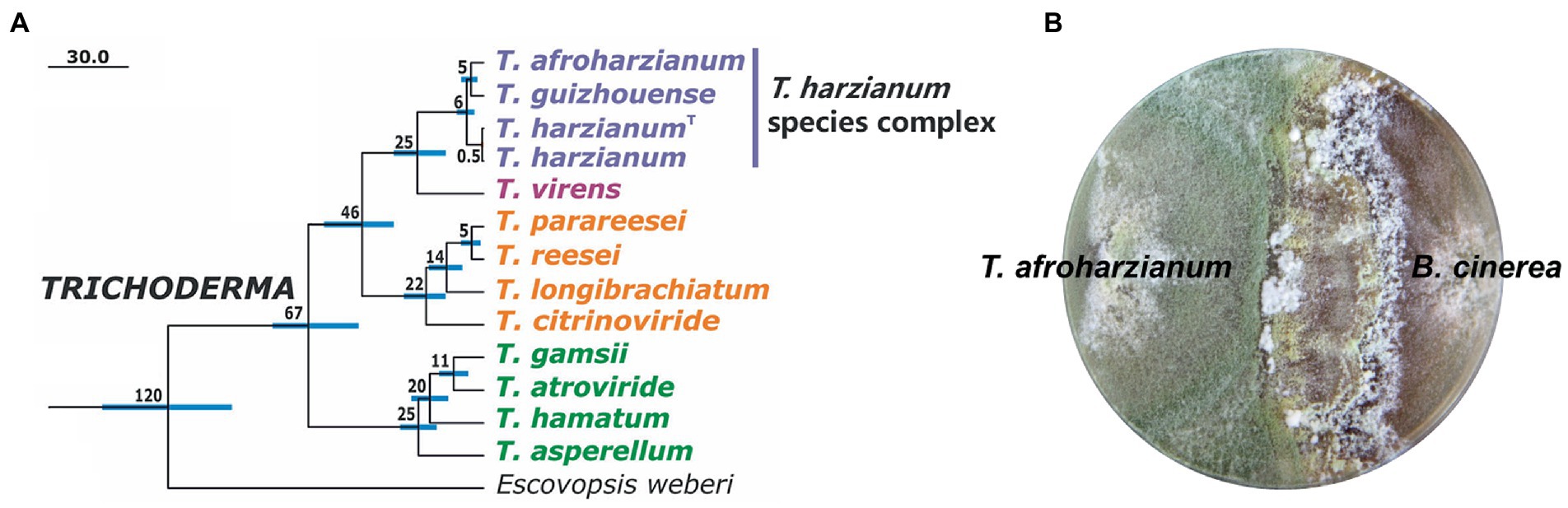
Figure 2. Trichoderma harzianum species complex for biocontrol. (A) Phylogenetic relationship of three species in T. harzianum complex and other Trichoderma species. The chronogram was adapted from Kubicek et al. (2019). The numbers represent chronological ages of the nodes in Mya. The NCBI GenBank accession numbers of the genomes are: T. afroharzianum T6776, JOKZ00000000; T. guizhouense NJAU 4742, LVVK00000000; T. harzianum CBS 226.95 (type culture, indicated withT), MBGI00000000; T. harzianum TR274, NQLC00000000; T. virens Gv29-8, ABDF00000000; T. parareesei CBS 125925, LFMI00000000; T. reesei QM6a, AAIL00000000; T. longibrachiatum ATCC 18648, MBDJ00000000; T. citrinoviride TUCIM 6016, MBDI00000000; T. gamsii T6085, JPDN00000000; T. atroviride IMI 206040, ABDG00000000; T. hamatum GD12, ANCB00000000; T. asperellum CBS 433.97, MBGH00000000; Escovopsis weberi CC031208-10, LGSR00000000. Some other species in the T. harzianum complex, such as T. atrobrunneum and T. simmonsii, also show good biocontrol potentials. (B) Overgrowth of T. afroharzianum T22 against plant pathogen B. cinerea on agar plate.
Cell wall-degrading enzymes (mainly chitinases, glucanases, and proteases) play an important role in the antagonistic effect of Trichoderma species toward fungal pathogens. The fungal inhibitory activity of Trichoderma isolates was reported to be positively correlated with the production of extracellular lytic enzymes (Rai et al., 2016). As summarized below, increasing the expression of fungal cell wall-degrading enzymes is an effective strategy for enhancing the biocontrol capacity of T. harzianum (Table 2). Additionally, the expression of these enzymes in transgenic plants resulted in increased resistance to fungal pathogens (Distefano et al., 2008; Mercado et al., 2015).
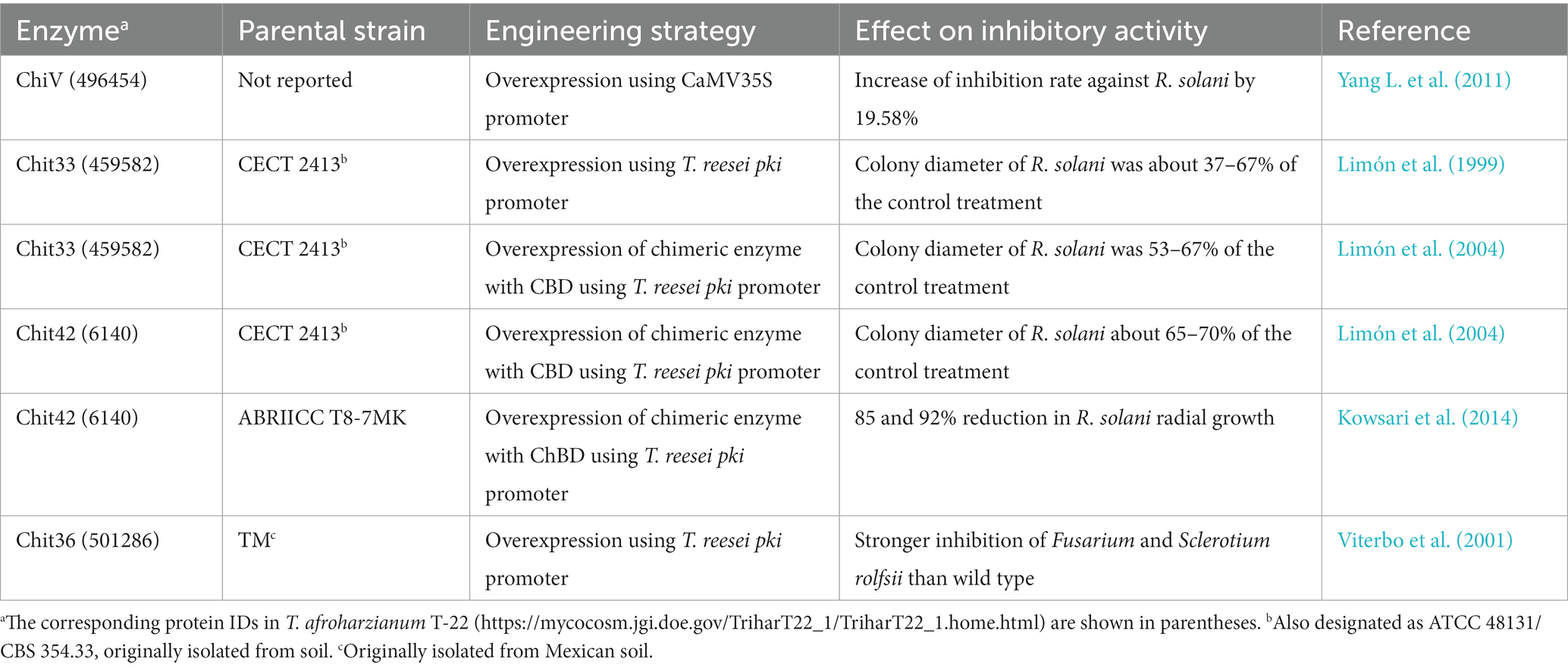
Table 2. Studies on improving the biocontrol ability of T. harzianum strains by overexpressing fungal cell wall-degrading enzymes.
Chitin is a major component of the cell wall in most fungi (Brown et al., 2020). The chitinolytic system of Trichoderma species includes several chitinases and β-1,4-N-acetylglucosaminidases (Ghasemi et al., 2020). The most frequently studied chitinases in T. harzianum are Chit42/Ech42 (García et al., 1994; Carsolio et al., 1999; Woo et al., 1999), Chit33 (Limón et al., 1999; de las Mercedes Dana et al., 2001), and Chit46 (Deng et al., 2019), which are named by their molecular mass. Purified or heterologously expressed chitinases effectively inhibit the growth of phytopathogenic fungi (Wu et al., 2013). Correspondingly, introduction of the chit42 gene to plants increased their resistances to fungal pathogens (Lorito et al., 1998).
Interspecific and intraspecific protoplasmic fusions were reported to enhance chitinase activity and antagonistic activity in T. harzianum (Balasubramanian et al., 2012; Hassan, 2014). On the other hand, rational genetic engineering has also been used to improve the chitinase activity of T. harzianum strains. Both overexpression and enzyme engineering strategies were applied to this end. Overexpression of the chit33 gene using a constitutive promoter resulted in an approximately 200-fold increase in extracellular chitinase activity, and the inhibitory ability against R. solani was effectively improved (Limón et al., 1999). Moreover, the main chitinases produced by T. harzianum lack a specific chitin-binding domain (ChBD), which affects their affinity for insoluble chitin in the fungal cell wall. The transformants with the overexpression of a chimeric chitinase carrying ChBD from a T. atroviride chitinase showed higher chitinase activities and stronger inhibition against R. solani, compared with those without ChBD (Kowsari et al., 2014; Eslahi et al., 2021). Similarly, the addition of cellulose binding domains (CBDs) with binding ability to the chitin surface to chitinases led to not only increased chitinase activity but also more effective inhibition against R. solani, B. cinerea, and Phytophthora citrophthora than the wild-type strain (Limón et al., 2004).
β- and α-linked glucans are also major components of the scaffold and matrix of the fungal cell wall (Kang et al., 2018). β-1,3-exoglucanase, β-1,3-endoglucanase, and β-1,6-endoglucanase have been reported to be associated with the biological control ability of T. harzianum (de la Cruz et al., 1996; Cohen-Kupiec et al., 1999; de la Cruz and Llobell, 1999; Donzelli et al., 2001). After contact with F. solani, the expression level of β-1,3-endoglucanase in T. harzianum was significantly upregulated compared with that before contact (Vieira et al., 2013). Furthermore, endo-β-1,3-glucanase, cellulase (β-1,4-glucanase), and α-1,3-glucanase purified from T. harzianum were shown to inhibit the growth of several pathogenic fungi (Thrane et al., 1997; Ait-Lahsen et al., 2001). Although gene knockout has been used to study the function of β-1,3-endoglucanase in biocontrol (Suriani Ribeiro et al., 2019), overexpression of glucanase-encoding genes for enhanced biocontrol performance has rarely been reported. An endo-β-1,6-glucanase BGN16.2 was successfully overexpressed using the T. reesei pki promoter; however, its effect on biocontrol ability remains to be studied (Delgado-Jarana et al., 2000).
In addition to chitinases and glucanases, proteases play an important role in the degradation of fungal cell walls. A proteomic study found that an aspartic protease was highly expressed in T. harzianum in the presence of the cell walls of P. ultimum and B. cinerea (Suárez et al., 2005). Multiple proteases from T. harzianum, such as serine proteases (Yan and Qian, 2009; Liu and Yang, 2013; Fan et al., 2014) and aspartic proteases (Delgado-Jarana et al., 2002; Liu and Yang, 2007; Deng et al., 2018), showed significant inhibitory activities against pathogenic fungi. After ultraviolet light (UV) irradiation, the extracellular protease activities of some T. harzianum mutants were 6 to 12.5 times higher than that of the wild-type strain, and certain mutants were proven to be more effective against fungal pathogens during in vitro plate antagonism experiments (Szekeres et al., 2004). Overexpression of the serine protease-encoding gene prb1 was reported to increase protease production and enhance antagonistic activity against R. solani (Flores et al., 1997).
In addition to fungal cell wall-degrading enzymes, T. harzianum produces other proteins, such as plant cell wall-degrading enzymes, L-amino acid oxidase, cerato-platanins and hydrophobins, to inhibit pathogens and/or induce plant resistances.
Trichoderma spp. can secrete plant cell wall-degrading enzymes as elicitors to induce plant resistance to pathogens. For example, cellulases and xylanases from Trichoderma have been reported to induce plant defense responses via the ethylene/H2O2/calcium/jasmonic acid signaling pathways (Saravanakumar et al., 2016; Guo et al., 2021). By constructing a gene-silenced mutant and investigating its effect on the transcriptome of Arabidopsis, Thpg1 (encoding an endopolygalacturonase) was found to be required for active root colonization and plant defense induction by T. harzianum T34 (Morán-Diez et al., 2009). Finally, a swollenin from T. guizhouense can promote the growth of cucumber by altering the root cell wall architecture (Meng et al., 2019). According to the evolutionary analysis of genes, 41% of plant cell wall-degrading enzymes and auxiliary proteins in Trichoderma were obtained via lateral gene transfer from other classes of Ascomycota (Druzhinina et al., 2018).
Proteomic analysis revealed that the expression of L-amino acid oxidase (LAAO) was induced in media containing deactivated B. cinerea mycelia as the sole carbon source (Yang et al., 2009). LAAO has inhibitory effects on pathogenic bacteria and fungi. For the inhibition of R. solani, T. harzianum LAAO physically interacts with the cell wall proteins of the pathogen and triggers the mitochondria-mediated apoptosis pathway, including cytochrome c release and the activation of apoptosis factors, caspases 3 and 9 (Yang C. A. et al., 2011; Yang et al., 2012).
Cerato-platanins are small, secreted cysteine-rich proteins that act as effectors and elicitors in fungus-plant interactions. Although the cerato-platanin family protein Epl1 is not necessary for the biocontrol ability of T. harzianum, the absence of epl1 was found to affect the expression level of mycoparasitic genes (Gomes et al., 2015; Gao et al., 2020). Furthermore, removal of epl1 from T. harzianum not only reduced the jasmonic acid-mediated defense response in tomato, but also lost its ability to downregulate the expression of B. cinerea virulence genes (Gomes et al., 2017; Gao et al., 2020). Another type of surface-active small protein, hydrophobin, is also involved in interactions between Trichoderma and plants (Viterbo and Chet, 2006). Thhdy1, a class II hydrophobin from T. harzianum, acts as an elicitor to activate the expression of jasmonic acid/ethylene defense-related and brassinosteroid-associated genes that are involved in plant systemic resistance (Yu et al., 2020). Therefore, the construction of Thhdy1-overexpressing T. harzianum strains is expected to enhance their biocontrol activity.
Reactive oxygen species (ROS) act as signals to regulate diverse biological processes. The production of ROS has been suggested to be one of the mechanisms of induced systemic resistance in plants by T. harzianum (Lara-Ortíz et al., 2003; Zhang et al., 2017). NADPH oxidases, although not extracellular proteins, are involved in the formation of ROS and are therefore indirectly associated with the biocontrol ability of T. harzianum. Transformants overexpressing the NADPH oxidase-encoding gene nox1 showed higher inhibitory activity against P. ultimum than the wild-type. According to the result of transcriptomic analysis, the nox1-overexpressing transformant had upregulated expression of genes linked to protease, cellulase, and chitinase activities in the interaction with P. ultimum compared to the wild-type strain (Montero-Barrientos et al., 2011).
The antibiosis activity of T. harzianum is generally mediated by the production of low-molecular-weight compounds, which can directly or indirectly inhibit the growth of pathogens. These include a variety of classes of compounds, such as peptides (McMullin et al., 2017; Kai et al., 2018; van Bohemen et al., 2021), polyketides (Zhao et al., 2020), and terpenes (Song et al., 2018; Figure 3). Various methods have been developed for the discovery of new metabolites with antimicrobial activity in T. harzianum. First, the one strain-many compounds (OSMAC) method was used to activate secondary metabolic gene clusters, which in turn altered their metabolic pathways to synthesize new metabolites (Yu et al., 2021). Using this method, eleven compounds were obtained from a T. harzianum strain, of which triharzianin B, triharzianin C, trichoharin A, triharzin C, 5-hydroxy-3-hydroxymethyl-2-methyl-7-methoxychromone, trichoacorenol B and harzianone exhibited antifungal activity against Aspergillus fumigatus and Trichoderma sp. (Wang X.-Y. et al., 2021). Second, the mutant strains of T. harzianum may produce new compounds. For example, several mutants obtained by UV mutagenesis exhibited increased Fusarium-inhibiting activity and produced two new compounds, including an isonitrile compound with broad antibiotic activity against fungi and bacteria (Graeme-Cook and Faull, 1991; Faull et al., 1994). Third, mining of new isolates of T. harzianum from soil, plant root systems, and rhizomes allowed for the identification of new chemical derivatives with inhibitory activities, such as α-pyrone and decalin derivatives (Nuansri et al., 2022), pentadecaibins (van Bohemen et al., 2021), azaphilone D and E (Zhang et al., 2020), harzianopyridone (Ahluwalia et al., 2015), tandyukisin (Yamada et al., 2014), trichosordarin A (Liang et al., 2020), trichoharzianol (Jeerapong et al., 2015), harzianic acid (Vinale et al., 2009), and nafuredin C (Zhao et al., 2020).
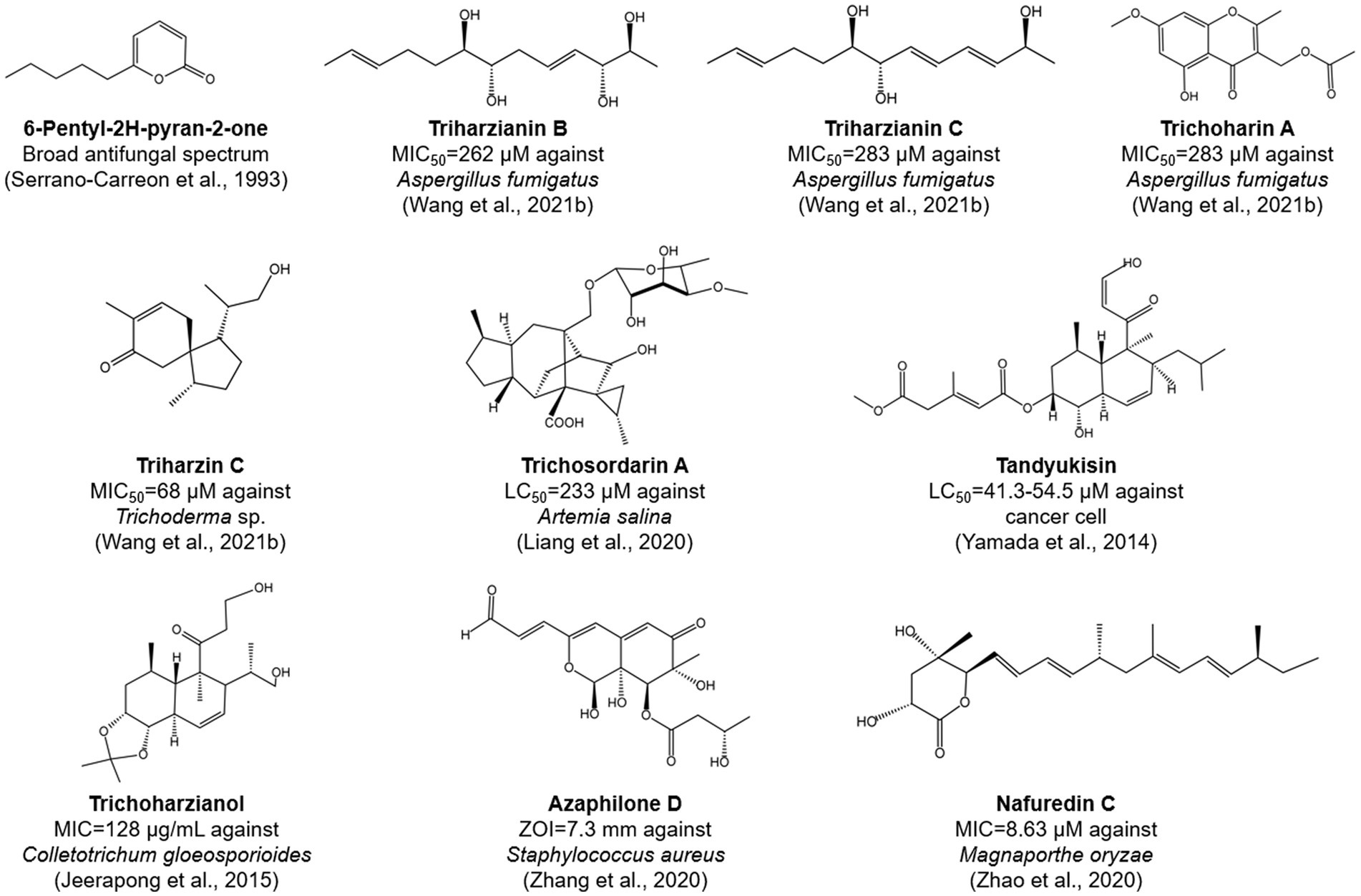
Figure 3. Chemical structures of selected bioactive compounds produced by T. harzianum species complex strains. MIC, minimum inhibitory concentration; LC, lethal concentration; ZOI, zone of inhibition.
Understanding of the synthetic pathway is important for improving the production level and modifying the structures of natural products, which help to improve the inhibitory ability of T. harzianum against pathogens. Despite the extensive reports on bioactive compounds from T. harzianum, the biosynthetic pathways of most of these molecules remain unresolved so far.
Lactone compounds such as butenolides (e.g., harzianolide) and pyrones are commonly isolated from T. harzianum. Harzianolide could significantly promote tomato seedling growth and activate plant systemic resistance (Cai et al., 2013), and its biosynthesis pathway was shown to involve the rearrangements and decarboxylation of a heptaketide (Avent et al., 1992).
6-Pentyl-2H-pyran-2-one (6-PP) is an unsaturated volatile lactone with a coconut aroma, and is commonly detected in the secondary metabolites produced by T. harzianum and other Trichoderma species (Keswani et al., 2014; Vinale et al., 2014). 6-PP can inhibit the growth of a broad spectrum of fungal pathogens such as Fusarium moniliforme and R. solani (Scarselletti and Faull, 1994; El-Hasan et al., 2007). Furthermore, it can promote plant growth and induce plant defenses against pathogenic fungi (Garnica-Vergara et al., 2016; Lazazzara et al., 2021). Deciphering the 6-PP biosynthetic pathway is yet to be accomplished, and most of the clues from T. atroviride isotopic labeling experiments have suggested that the oxidation of linoleic acid by lipoxygenase might be a major step in the biosynthesis of 6-PP by Trichoderma (Serrano-Carreon et al., 1993). However, a gene deletion study showed that the single lipoxygenase-encoding gene lox1 is dispensable for the production of 6-PP and for the antagonistic capacity of T. atroviride against the plant pathogen B. cinerea (Speckbacher et al., 2020). The authors proposed that the synthesis of 6-PP may involve the action of polyketide synthase. In addition, 6-PP can be degraded or converted into the intracellular microsomal fraction of T. atroviride, which decreases its concentration in culture (Flores et al., 2019).
Ergosterol, a component of the fungal cell membrane, can upregulate the expression of plant defense-related genes and elicit responses through induction of the oxidative burst by inhibition of H+-ATPase activity on the plasma membrane (Rossard et al., 2010; Khoza et al., 2019). Hydroxy-methylglutaryl-CoA reductase (encoded by hmgR) is a rate-limiting enzyme involved in the synthesis of farnesyl diphosphate (FPP), an important intermediate in sterol synthesis (Figure 4). Partial silencing of the hmgR gene in T. harzianum led to a reduction in antifungal activity against the plant pathogens R. solani and Fusarium oxysporum and a 15.8-fold increase in the expression of erg7 in the sterol pathway (Cardoza et al., 2007).
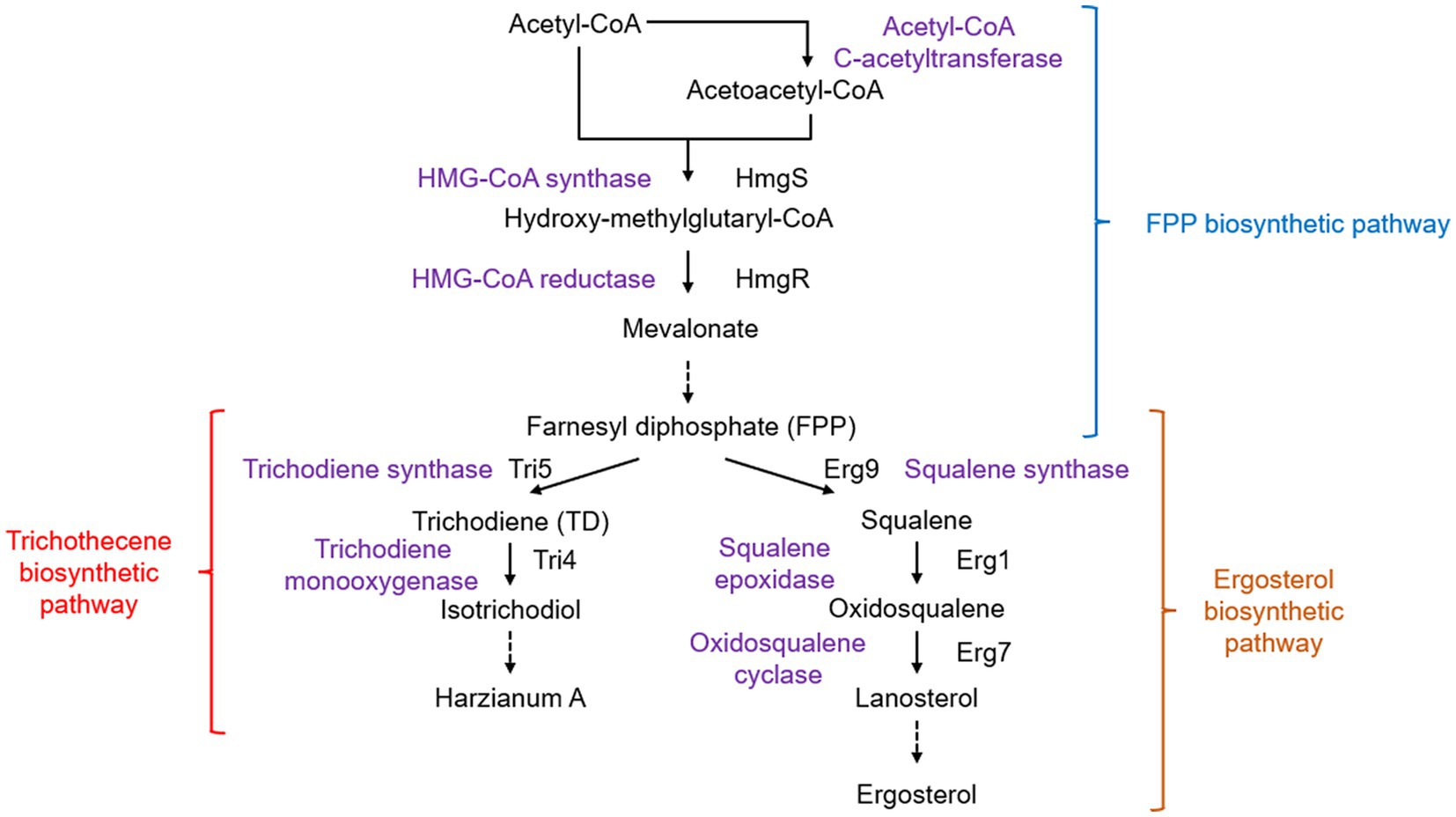
Figure 4. Biosynthetic pathways of sterol and trichothecenes. Solid arrows show direct chemical reactions, while dashed arrows represent a series of chemical reactions.
Silencing of the squalene epoxidase-encoding gene erg1 led to lower ergosterol production and increased erg7 expression (Cardoza et al., 2006b). In addition, silencing erg1 was found to increase the production of squalene, which can induce the expression of tomato defense-related genes in a concentration-dependent manner. The ability of T. harzianum to colonize tomato roots has also been enhanced (Malmierca et al., 2015b). However, overexpression of erg1, although it did not show any effect on ergosterol levels, led to a substantial decrease in the amount of squalene and also reduced the priming ability of some defense-related genes in the salicylic acid and jasmonic acid pathways (Cardoza et al., 2014).
The synthesis of sesquiterpene compounds, including trichothecenes in fungi, also uses FPP as a precursor (McCormick et al., 2011). Many trichothecenes are fungal toxins with some showing good antifungal activity. Harzianum A, a non-phytotoxic trichothecene produced by Trichoderma arundinaceum, was found to have antagonistic activity against fungal pathogens and induce plant defense response genes (Malmierca et al., 2012). In trichothecene biosynthesis, the first step is to cyclize FPP to form trichodiene (TD) using trichodiene synthase encoded by tri5 (Fekete et al., 1997). Although a tri5 homologous gene has been isolated from T. harzianum ATCC 90237 (Gallo et al., 2004), this strain was later identified as T. arundinaceum (Degenkolb et al., 2008). Currently, T. harzianum is thought to be unable to synthesize trichothecenes. When T. harzianum was transformed with tri5 from T. arundinaceum, the production of TD resulted in the upregulation of plant defense-related genes in tomatoes (Malmierca et al., 2015a). This TD-producing strain showed enhanced biocontrol activity against F. graminearum and reduced mycotoxin deoxynivalenol contamination (Taylor et al., 2022). Transgenic T. harzianum with both tri5 and tri4 produced 12,13-epoxytrichothec-9-ene and downregulated tomato genes involved in fungal root colonization and pathogen defense (Cardoza et al., 2015). These findings highlight the intricate interactions between host plants, fungal pathogens, and antagonists mediated by trichothecene compounds.
Azaphilones as a family of polyketide-based secondary metabolites were isolated from the T. harzianum species complex. These compounds were shown to have antifungal, antiviral or radical scavenging activities (Vinale et al., 2006; Pang et al., 2020; Xie et al., 2022). The gene cluster for the biosynthesis of trigazaphilones in T. guizhouense has been identified (Pang et al., 2020). Another gene cluster hac is responsible for the biosynthesis of harzianic acid in T. afroharzianum and T. guizhouense, with two transcriptional activators identified to be involved in its regulation (Xie et al., 2021; Pang et al., 2022).
The products of many gene clusters containing polyketide synthase (PKS)-encoding genes in T. harzianum remain unknown. In vitro plate confrontation experiments against S. sclerotiorum, R. solani, and F. oxysporum revealed that the PKS-encoding genes pksT-1 and pksT-2 are differentially regulated in T. harzianum in response to fungal pathogens. The pksT-2 knockout mutant showed a significant change in the color of the conidia, but the biocontrol activity of the mutant was not tested (Yao et al., 2016). Additionally, heterologous expression of a polyketide synthase-nonribosomal peptide synthetase gene cluster from T. harzianum in Aspergillus nidulans has led to the discovery of new tetronate compounds with potential antimicrobial activities (Zhu et al., 2021).
In addition to the production of the biocontrol effectors mentioned above, it is also important to improve the resistance of T. harzianum to various stresses in practical applications. The survival characteristics of these strains may be significantly influenced by physical and chemical environmental factors such as pH, temperature, and fungicides in the soil (Lo et al., 1998). Therefore, the ecology of T. harzianum should be better understood to deploy biocontrol agents for disease control.
The synergistic application of fungicide and T. harzianum can reduce the amount of fungicide used while ensuring the same inhibition rate (Wang et al., 2019; Becker et al., 2021); however, this is based on a situation where T. harzianum shows resistance to fungicides. After exposure to UV light, mutant strains obtained by screening on specific plates supplemented with fungicides showed cross-resistance to prochloraz and bromuconazole (Figueras-Roca et al., 1996) or to benomyl and thiabendazole (Hatvani et al., 2006). Thmfs1, a major facilitator superfamily transporter gene, is partially responsible for trichodermin secretion in T. harzianum. A strain overexpressing Thmfs1 displayed increased resistance to a wide range of antimicrobial compounds (Liu et al., 2012; Table 3). In addition to chemical fungicides, the tolerance to metabolites secreted by pathogenic fungi should be taken into the consideration. For example, the metabolite fusaric acid produced by F. oxysporum inhibits the growth of T. harzianum. A UV-C mutant was not only more tolerant to fusaric acid but also more effective against Fusarium wilt in tomatoes than the wild-type (Marzano et al., 2013).
Besides the resistance to antifungal chemicals, the tolerance to other abiotic stresses needs to be taken into account when applying T. harzianum in specific environments. The response to heat stress is a highly conserved system by inducing the synthesis of heat-shock proteins (Lindquist and Craig, 1988). When T. harzianum conidia were heat-shocked at 45°C for 2 h, the hsp70-overexpressed strains showed better growth than the wild-type under various oxidative, osmotic, and salt stresses (Montero-Barrientos et al., 2008). Transformants with the superoxide dismutase (SOD)-encoding gene showed a significantly higher resistance to heat and salt stress. Although the wild-type strain could not grow at 40°C or in the presence of 2 mol/l NaCl, the sod transformant maintained its inhibitory activity against S. sclerotiorum under these conditions (Yang et al., 2010). In addition, hydrophobins play important roles in the resistance of Trichoderma spores to several kinds of abiotic stresses (e.g., UV radiation), and the perturbation of hydrophobin-encoding genes can result in species-specific changes of phenotypes (Cai et al., 2020).
Mycoviruses are widely observed among fungal species, some of which are harmful to their hosts. Recently, T. harzianum hypovirus 1 (ThHV1) was identified in a T. harzianum isolate, and strains carrying both ThHV1 and its defective RNA were found to have a decreased mycoparasitism ability (You et al., 2019). Therefore, the viruses in T. harzianum may also be related to their stable performance.
The synthesis of protein and chemical effectors, as well as the responses to environmental stresses, are tightly regulated in Trichoderma species for biocontrol. In eukaryotes, gene regulatory systems respond to external signals and typically undergo multiple signal transitions to regulate downstream gene expression. Modification of the gene regulatory system can often alter the expression levels of multiple genes simultaneously, making it an efficient strategy for strain engineering (Pang et al., 2022).
The sensing of pathogenic fungi and the consequent responses in Trichoderma involve the combinatorial action of different signaling pathways (Howell, 2003; Zeilinger and Omann, 2007). Several classical signal transduction pathways in fungi have been linked to their ability to combat phytopathogens in Trichoderma spp., including G protein signaling, mitogen-activated protein kinase (MAPK) cascades, and cAMP pathways (Mendoza-Mendoza et al., 2003; Omann and Zeilinger, 2010).
Heterotrimeric G-protein complexes consist of α, β, and γ subunits, and most filamentous fungi have three Gα subunits: Gα1, Gα2, and Gα3. Knockout of the Thga1 gene, which encodes the GαI protein, led to reduced growth rate, decreased 6-PP and chitinase production, and complete loss of the capacity to overgrow and lyse R. solani, B. cinerea, and S. sclerotiorum during in vitro plate confrontation (Sun et al., 2016). Knockout of another Gα-encoding gene, Thga3, results in an 80% reduction in hydrophobin expression and a 23% reduction in chitinase activity (Ding et al., 2018, 2020). Despite the demonstrated role of G proteins, the function of G protein-coupled receptors (GPCRs) has not yet been studied in T. harzianum. In T. atroviride, silencing of Gpr1, a cAMP-receptor-like family GPCR, results in the loss of capacity to activate the expression of chitinase and protease genes and to attach to host hyphae (Omann et al., 2012).
Highly conserved MAPK cascades play a crucial role in the transmission of extracellular and intracellular signals in fungi by controlling transcription factors through a phosphorylation cascade (Martínez-Soto and Ruiz-Herrera, 2017). hog1 is a homolog of the MAPK-encoding gene HOG1, controlling the hyperosmotic stress response in Saccharomyces cerevisiae. A mutant strain containing hyperactive point-mutated hog1 and another with hog1 silencing was constructed in T. harzianum. Both mutant strains showed strongly reduced antagonistic activity against the plant pathogens Phoma betae and Colletotrichum acutatum (Delgado-Jarana et al., 2006).
The significant changes in the transcriptome of T. harzianum during interactions with fungal pathogens involve the action of a set of transcription factors (Figure 5). CRE1, a conserved carbon catabolite repressor in fungi, is the first demonstrated transcription factor in the biocontrol with T. harzianum. Before contact with the plant pathogen, CRE1 can bind to two single sites in the promoter of chitinase gene chit42 to inactivate its expression. Confrontation with B. cinerea relieved the binding of Cre1 to the chit42 promoter (Lorito et al., 1996). In contrast, the expression of chit42 is triggered by soluble chitooligosaccharides which can be produced by constitutive chitinolytic enzymes (Zeilinger et al., 1999). In addition, a BrlA-like binding motif in the chit42 promoter was found to be related to the regulation of its expression in T. atroviride (Brunner et al., 2003).
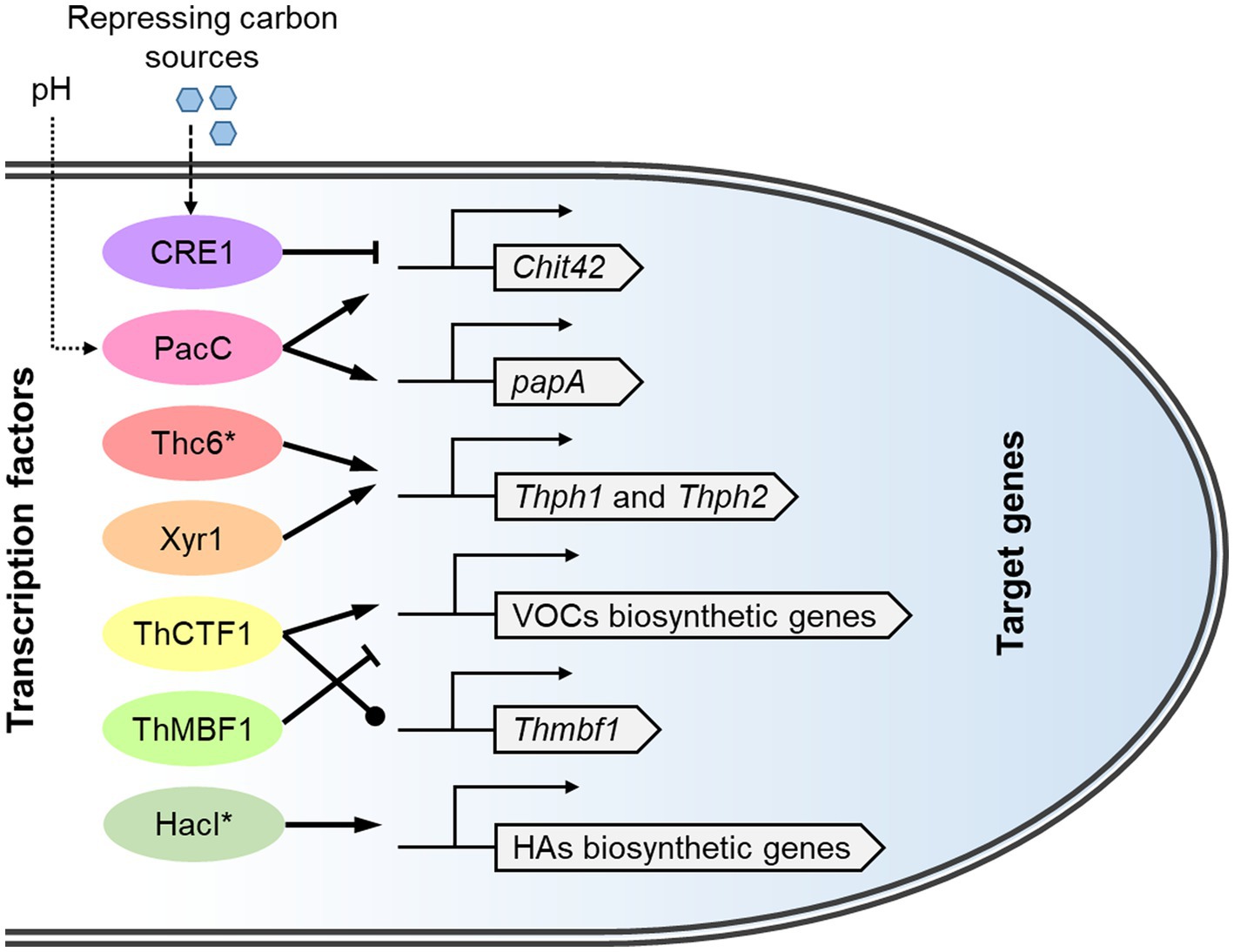
Figure 5. The transcriptional regulatory system of biocontrol-related genes in T. harzianum species complex. The transcription factors respond to upstream signals and regulate the expression of target genes involved in biocontrol. The corresponding protein IDs in T. afroharzianum T-22 (https://mycocosm.jgi.doe.gov/TriharT22_1/TriharT22_1.home.html) are: CRE1, 298,239; PacC, 516,100; Thc6, 503,211; Xyr1, 455,911; ThCTF1, 348,498; ThMBF1, 493,750; HacI, 207,786; chit42, 6,140; papA, 315,275; Thph1, 627,343; Thph2, 555,537. VOCs, Volatile organic compounds; HAs, harzianic acids. Transcription factors reported to be engineering targets for strain improvement are marked with asterisks.
The zinc cluster family transcription factor Thc6 is involved in the induction of systemic plant resistance by T. harzianum. Overexpression mutants of Thc6 could activate the expression of the jasmonic acid pathway genes and reduce the disease index of maize treated with Curvularia lunata (Fan et al., 2015). As a homolog of the cellulase transactivator ACE2 in T. reesei, Thc6 can bind to the promoters of cellulase genes Thph1 and Thph2. Knockout mutants of these two genes resulted in the loss of the ability to activate the expression of immune defense-related genes in plants (Saravanakumar et al., 2016, 2018).
Another zinc cluster transcription factor, ThCTF1, is involved in regulating the synthesis of 6-PP in T. harzianum. The Thctf1 deletion mutant did not produce two secondary metabolites derived from 6-PP and showed reduced antimicrobial capacity (Rubio et al., 2009). Through suppression subtractive hybridization between the wild-type strain T34 and a Thctf1-null mutant, a helix-turn-helix family regulator, ThMBF1, was identified to be differentially expressed. Overexpression of Thmbf1 exacerbated the incidence of fungal diseases in tomato plants, suggesting that this gene has a negative role in the biocontrol process (Rubio et al., 2017).
The transcription factor PacC/Rim101 plays an important role in adaptation to ambient pH in fungi (Denison, 2000; Franco-Frías et al., 2014). Pac1/ThPacC (homologous to PacC/Rim101) regulates many genes involved in T. harzianum antagonism, such as chit42 and protease papA. The silencing of pac1 seems to promote the production of certain metabolites that inhibit some plant pathogenic fungi, but it negatively affects the parasitic capacity of T. harzianum (Moreno-Mateos et al., 2007). Another study revealed that the ThpacC knockout strain did not produce the antifungal molecules homodimericin A and 8-epi-homodimericin A and showed reduced inhibition against S. sclerotiorum (Wu et al., 2021). However, neither constitutive activation nor overexpression of Pac1/ThPacC increased biocontrol ability in the above two studies.
The biocontrol capacity of T. harzianum involves complex interactions between the pathogens and plants. To date, the molecular mechanisms underlying the action of many effector proteins and compounds against phytopathogens have not been fully elucidated. The activities of these effector molecules and their combinatorial effects on different types of pathogens need to be investigated in detail to guide strain engineering. In particular, attention should be paid to the effects of the molecules or strains on the defense response and growth of plants, in addition to the results of traditional plate confrontation experiments.
The sequencing and annotation of the T. hazianum genome enabled the discovery of effector proteins and metabolites connected to biocontrol activity on a large scale (Rush et al., 2021). According to annotations from the JGI MycoCosm portal,1 there are approximately 60 secondary metabolic gene clusters in most sequenced strains in the T. harzianum species complex. For silent biosynthetic gene clusters, their products are expected to be identified and increased for production using molecular biology tools, such as promoter exchange, overexpression of pathway-specific transcription factors, and heterologous expression (Brakhage and Schroeckh, 2011). The genome-driven approach has been used to mine bioactive natural products from T. harzianum, which resulted in the discovery of several unique compounds and widened the knowledge of their biosynthetic pathways (Chen et al., 2019; Zhu et al., 2021).
Transcriptomic and secretomic analyses have suggested that hundreds of genes in T. harzianum are significantly differentially expressed during interaction with fungal pathogens (Vieira et al., 2013; Steindorff et al., 2014; Ramada et al., 2016). Systematic investigation of the functions of these genes can provide more targets for engineering strains with enhanced biocontrol capacities. For example, aquaglyceroporin, which facilitates the transport of water and solutes across the membrane, was found to be significantly upregulated in T. harzianum during its interaction with F. solani. A transformant overexpressing its encoding gene was capable of significantly reducing Fusarium sp. growth compared to the wild-type (Vieira et al., 2018).
Additionally, although the defective RNA of ThHV1 decreases the pathogen-inhibitory ability of T. harzianum, some other viruses enhance mycoparasitic ability by regulating the activity of cell wall-degrading enzymes. Compared to ThPV1-cured strains, β-1,3-glucanase activity and the ability to combat P. ostreatus and R. solani were increased in ThPV1-containing strains (Chun et al., 2018). In the future, dsRNA in T. harzianum strains can be mined from their genomes to identify beneficial viruses for improving their biocontrol abilities. Nevertheless, the effects of virus-infected T. harzianum strains on the physiological characteristics of plants and the plant root microbiome need to be studied.
Overexpression, mutagenesis, and domain-swapping strategies have been successfully used to engineer regulatory proteins in filamentous fungi (e.g., T. reesei) to improve the production of plant biomass-degrading enzymes (Liu and Qu, 2021; Zhao et al., 2022). However, understanding of the roles of regulatory proteins in biocontrol is still limited. Through the construction of gene disruption mutants, MAPKs, adenylate cyclase, protein kinase A, and GTPase activators have been linked to the inhibition of pathogens and production of secondary metabolites in Trichoderma species (Mukherjee et al., 2003; Hinterdobler et al., 2019; Segreto et al., 2021). These signaling proteins might have similar functions in T. harzianum and need to be studied and tested as potential targets for strain engineering in the future.
Despite the studies summarized in Figure 5, knowledge of the transcriptional regulation of biocontrol-related genes in T. harzianum is fragmented. Transcriptional activator(s) binding to the promoters of chitinase-encoding genes have yet to be identified (Lorito et al., 1996). Overexpression or improvement in the activity of such activators is expected to increase the expression of a set of fungal cell wall-degrading enzymes. In T. atroviride, the xylanase transcriptional regulator XYR1 positively regulates the expression of lignocellulolytic enzyme genes and activation of plant defense responses (Reithner et al., 2014). Overexpression of xyr1 has been shown to increase the production of cellulases and xylanases in T. harzianum (da Silva Delabona et al., 2017), but its effect on biocontrol ability needs to be studied.
In addition to transcriptional factors, proteins that regulate chromatin structure can significantly affect the expression levels of targeted genes. The lae1 (encoding putative methyltransferase) and tgf-1 (encoding histone acetyltransferase) genes were proven to be related to mycoparasitism in T. atroviride (Karimi Aghcheh et al., 2013; Gómez-Rodríguez et al., 2018). Overexpression of lae1 in T. harzianum results in a significant increase in cellulolytic gene expression (Delabona et al., 2020), and its function in secondary metabolite synthesis and biocontrol is worth investigating.
Based on the understanding of the molecular mechanisms for biocontrol, systems metabolic engineering strategies could be employed to construct T. harzianum strains with increased pathogen-inhibiting capacity and enhanced robustness (Ko et al., 2020). For the identified effector proteins and compounds, cutting-edge technologies for protein engineering and combinatorial biosynthesis are expected to be used to modify their structures for higher activities toward pathogens (Staunton and Wilkinson, 2001). The introduction of heterologous genes related to biocontrol is another approach to improve the ability of T. harzianum to combat pathogens. An insect-specific neurotoxin gene from the scorpion Androctonus australis was heterologously expressed in Metarhizium anisopliae, which significantly increased its ability to kill pest insects (Wang and St Leger, 2007). Similarly, heterologous genes (e.g., peptaibol synthetic gene clusters from other Trichoderma species) may be introduced into T. harzianum to expand the range of its action. The safety of the engineered strains, however, should be carefully evaluated, and the transfer of transgenic genes should be well controlled (Stirling and Silver, 2020).
The multiplex genetic engineering of strains requires the development of highly efficient gene manipulation tools. Traditionally, polyethylene glycol-mediated and Agrobacterium-mediated transformation methods have been used to construct mutants in T. harzianum (Cai et al., 2021), which allowed gene overexpression and targeted genetic recombination (e.g., gene knock-out). New methods for strain engineering, for example, CRISPR/Cas9-based genome editing, have offered straightforward platforms to carry out multiplex genetic modifications in filamentous fungi (Kun et al., 2019; Wang Q. et al., 2021). The first application of CRISPR/Cas9-based genome editing in Trichoderma was reported in T. reesei (Liu et al., 2015). Through recycling of selection marker genes, consecutive rounds of gene deletion were achieved in T. reesei (Chai et al., 2022). Recently, this technique was used in T. harzianum to inactivate the pyr4 gene to construct an uracil-deficient strain (Vieira et al., 2021). The genome editing methods also have the advantage of being easy to achieve genetic modifications without introduce foreign DNA, which can overcome some restrictions on the use of GMOs. For heterologous expression of biosynthetic gene clusters, the assembly of large DNA fragments has been reported based on homologous recombination in yeast or directly in filamentous fungus (Chiang et al., 2021). These methods are expected to aid in the systematic genetic modification of T. harzianum for the development of next-generation biocontrol agents.
To be used under field conditions, the strains in biocontrol agents are required to be genetically stable and eco-friendly. Exogenous DNA is usually integrated to the chromosome to ensure stability in strain engineering of Trichoderma (Cardoza et al., 2006a; Yang L. et al., 2011). So far, the only element reported for autonomous replication of plasmids in Trichoderma is AMA1 from A. nidulans (Kubodera et al., 2002). Such plasmids are easy to lose and not suitable for the construction of improved strains for practical application. On the other hand, the use of antibiotic-resistance genes as selectable markers in strain engineering could pose a threat to environment and public health. Therefore, it is better to use selection markers other than antibiotic-resistant genes (e.g., auxotrophic markers) or to remove antibiotic-resistance genes in the final strains (Zhao et al., 2016). With the use of advanced genetic manipulation technologies and well-implemented risk assessments, engineered biocontrol strains have the potential to step out of laboratories to increase agricultural production in the near future.
ZX and GL drafted the manuscript. GL, LG, and WL revised the manuscript. All authors collected literature information and discussed about the organization of the manuscript, read, and approved the final manuscript.
This work was supported by National Natural Science Foundation of China (32100088), Shandong Natural Science Foundation (ZR2021QC073), Young Scholars Program of Shandong University (YSPSDU to GL), and Science and Technology Project of Shanghai Tobacco Group Beijing Cigarette Factory Co., Ltd. (TP2021-T2). The authors declare that the funder Shanghai Tobacco Group Beijing Cigarette Factory Co., Ltd., had the following involvement in the study: collection of literature and the writing of this article.
The authors thank Zhongfeng Zhang and Peng Zhang for helpful discussions.
WL was employed by Shanghai Tobacco Group Beijing Cigarette Factory Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbas, A., Mubeen, M., Zheng, H., Sohail, M. A., Shakeel, Q., Solanki, M. K., et al. (2022). Trichoderma spp. genes involved in the biocontrol activity against Rhizoctonia solani. Front. Microbiol. 13:884469. doi: 10.3389/fmicb.2022.884469
Ahluwalia, V., Kumar, J., Rana, V. S., Sati, O. P., and Walia, S. (2015). Comparative evaluation of two Trichoderma harzianum strains for major secondary metabolite production and antifungal activity. Nat. Prod. Res. 29, 914–920. doi: 10.1080/14786419.2014.958739
Ait-Lahsen, H., Soler, A., Rey, M., de la Cruz, J., Monte, E., and Llobell, A. (2001). An antifungal exo-alpha-1,3-glucanase (AGN13.1) from the biocontrol fungus Trichoderma harzianum. Appl. Environ. Microbiol. 67, 5833–5839. doi: 10.1128/AEM.67.12.5833-5839.2001
Anees, M., Tronsmo, A., Edel-Hermann, V., Hjeljord, L. G., Héraud, C., and Steinberg, C. (2010). Characterization of field isolates of Trichoderma antagonistic against Rhizoctonia solani. Fungal Biol. 114, 691–701. doi: 10.1016/j.funbio.2010.05.007
Avent, A. G., Hanson, J. R., and Truneh, A. (1992). The biosynthesis of harzianolide by Trichoderma harzianum. Phytochemistry 31, 791–793. doi: 10.1016/0031-9422(92)80016-8
Balasubramanian, N., Thamil Priya, V., Gomathinayagam, S., and Lalithakumari, D. (2012). Fusant Trichoderma HF9 with enhanced extracellular chitinase and protein content. Appl. Biochem. Microbiol. 48, 409–415. doi: 10.1134/S0003683812040035
Becker, P., Esker, P., and Umaña, G. (2021). Incorporation of microorganisms to reduce chemical fungicide usage in black Sigatoka control programs in Costa Rica by use of biological fungicides. Crop Prot. 146:105657. doi: 10.1016/j.cropro.2021.105657
Besoain, X., Perez, L., Araya, A., Lefever, L., Sanguinetti, M., and Montealegre, J. (2007). New strains obtained after UV treatment and protoplast fusion of native Trichoderma harzianum: their biocontrol activity on Pyrenochaeta lycopersici. Electron. J. Biotechnol. 10, 01–617. doi: 10.2225/vol10-issue4-fulltext-16
Brakhage, A. A., and Schroeckh, V. (2011). Fungal secondary metabolites – strategies to activate silent gene clusters. Fungal Genet. Biol. 48, 15–22. doi: 10.1016/j.fgb.2010.04.004
Brown, H. E., Esher, S. K., and Alspaugh, J. A. (2020). Chitin: A "hidden figure" in the fungal cell wall. Curr. Top. Microbiol. Immunol. 425, 83–111. doi: 10.1007/82_2019_184
Brunner, K., Montero, M., Mach, R. L., Peterbauer, C. K., and Kubicek, C. P. (2003). Expression of the ech42 (endochitinase) gene of Trichoderma atroviride under carbon starvation is antagonized via a BrlA-like cis-acting element. FEMS Microbiol. Lett. 218, 259–264. doi: 10.1111/j.1574-6968.2003.tb11526.x
Cai, F., and Druzhinina, I. (2021). In honor of John Bissett: authoritative guidelines on molecular identification of Trichoderma. Fungal Divers. 107, 1–69. doi: 10.1007/s13225-020-00464-4
Cai, F., Gao, R., Zhao, Z., Ding, M., Jiang, S., Yagtu, C., et al. (2020). Evolutionary compromises in fungal fitness: hydrophobins can hinder the adverse dispersal of conidiospores and challenge their survival. ISME J. 14, 2610–2624. doi: 10.1038/s41396-020-0709-0
Cai, F., Kubicek, C. P., and Druzhinina, I. S. (2021). Genetic transformation of Trichoderma spp. Methods Mol. Biol. 2290, 171–185. doi: 10.1007/978-1-0716-1323-8_12
Cai, F., Yu, G., Wang, P., Wei, Z., Fu, L., Shen, Q., et al. (2013). Harzianolide, a novel plant growth regulator and systemic resistance elicitor from Trichoderma harzianum. Plant Physiol. Biochem. 73, 106–113. doi: 10.1016/j.plaphy.2013.08.011
Cardoza, R., Hermosa, R., Vizcaino, J., González, F., Llobell, A., Monte, E., et al. (2007). Partial silencing of a hydroxy-methylglutaryl-CoA reductase-encoding gene in Trichoderma harzianum CECT 2413 results in a lower level of resistance to lovastatin and lower antifungal activity. Fungal Genet. Biol. 44, 269–283. doi: 10.1016/j.fgb.2006.11.013
Cardoza, R. E., Malmierca, M. G., and Gutiérrez, S. (2014). Overexpression of erg1 gene in Trichoderma harzianum CECT 2413: effect on the induction of tomato defence-related genes. J. Appl. Microbiol. 117, 812–823. doi: 10.1111/jam.12574
Cardoza, R., Malmierca, M., Olivera, E., Alexander, N., Monte, E., and Gutierrez, S. (2015). Effects of Trichothecene production on the plant defense response and fungal physiology: overexpression of the Trichoderma arundinaceum tri4 gene in T. harzianum. Appl. Environ. Microb. 81, 6355–6366. doi: 10.1128/AEM.01626-15
Cardoza, R. E., Vizcaino, J. A., Hermosa, M. R., Monte, E., and Gutiérrez, S. (2006a). A comparison of the phenotypic and genetic stability of recombinant Trichoderma spp. generated by protoplast- and Agrobacterium-mediated transformation. J. Microbiol. 44, 383–395.
Cardoza, R. E., Vizcaíno, J. A., Hermosa, M. R., Sousa, S., González, F. J., Llobell, A., et al. (2006b). Cloning and characterization of the erg1 gene of Trichoderma harzianum: effect of the erg1 silencing on ergosterol biosynthesis and resistance to terbinafine. Fungal Genet. Biol. 43, 164–178. doi: 10.1016/j.fgb.2005.11.002
Carsolio, C., Benhamou, N., Haran, S., Cortés, C., Gutiérrez, A., Chet, I., et al. (1999). Role of the Trichoderma harzianum endochitinase gene, ech42, in mycoparasitism. Appl. Environ. Microbiol. 65, 929–935. doi: 10.1128/AEM.65.3.929-935.1999
Chai, S., Zhu, Z., Tian, E., Xiao, M., Wang, Y., Zou, G., et al. (2022). Building a versatile protein production platform using engineered Trichoderma reesei. ACS Synth. Biol. 11, 486–496. doi: 10.1021/acssynbio.1c00570
Chaverri, P., Branco-Rocha, F., Jaklitsch, W., Gazis, R., Degenkolb, T., and Samuels, G. J. (2015). Systematics of the Trichoderma harzianum species complex and the re-identification of commercial biocontrol strains. Mycologia 107, 558–590. doi: 10.3852/14-147
Chen, S., Daly, P., Zhou, D., Li, J., Wang, X., Deng, S., et al. (2022). The use of mutant and engineered microbial agents for biological control of plant diseases caused by Pythium: achievements versus challenges. Fungal Biol. Rev. 40, 76–90. doi: 10.1016/j.fbr.2022.03.001
Chen, M., Liu, Q., Gao, S. S., Young, A. E., Jacobsen, S. E., and Tang, Y. (2019). Genome mining and biosynthesis of a polyketide from a biofertilizer fungus that can facilitate reductive iron assimilation in plant. Proc. Natl. Acad. Sci. U. S. A. 116, 5499–5504. doi: 10.1073/pnas.1819998116
Chen, P., Pang, G., Cai, F., and Druzhinina, I. S. (2021). “Strain improvement and genetic engineering of Trichoderma for industrial applications” in Encyclopedia of Mycology. eds. Ó. Zaragoza and A. Casadevall (Oxford: Elsevier), 505–517.
Chiang, Y.-M., Lin, T.-S., Chang, S.-L., Ahn, G., and Wang, C. C. C. (2021). An Aspergillus nidulans platform for the complete cluster refactoring and total biosynthesis of fungal natural products. ACS Synth. Biol. 10, 173–182. doi: 10.1021/acssynbio.0c00536
Chun, J., Yang, H. E., and Kim, D. H. (2018). Identification of a novel partitivirus of Trichoderma harzianum NFCF319 and evidence for the related antifungal activity. Front. Plant Sci. 9:1699. doi: 10.3389/fpls.2018.01699
Cohen-Kupiec, R., Broglie, K. E., Friesem, D., Broglie, R. M., and Chet, I. (1999). Molecular characterization of a novel beta-1,3-exoglucanase related to mycoparasitism of Trichoderma harzianum. Gene 226, 147–154. doi: 10.1016/S0378-1119(98)00583-6
da Silva Delabona, P., Rodrigues, G. N., Zubieta, M. P., Ramoni, J., Codima, C. A., Lima, D. J., et al. (2017). The relation between xyr1 overexpression in Trichoderma harzianum and sugarcane bagasse saccharification performance. J. Biotechnol. 246, 24–32. doi: 10.1016/j.jbiotec.2017.02.002
Daguerre, Y., Siegel, K., Edel-Hermann, V., and Steinberg, C. (2014). Fungal proteins and genes associated with biocontrol mechanisms of soil-borne pathogens: a review. Fungal Biol. Rev. 28, 97–125. doi: 10.1016/j.fbr.2014.11.001
de la Cruz, J., and Llobell, A. (1999). Purification and properties of a basic endo-beta-1,6-glucanase (BGN16.1) from the antagonistic fungus Trichoderma harzianum. Eur. J. Biochem. 265, 145–151. doi: 10.1046/j.1432-1327.1999.00698.x
de la Cruz, J., Pintor, J., Benítez, T., Llobell, A., and Romero, L. (1996). A novel endo-beta-1,3-glucanase, BGN13.1, involved in the mycoparasitism of Trichoderma harzianum. J. Bacteriol. 177, 6937–6945. doi: 10.1128/jb.177.23.6937-6945.1995
de Las Mercedes Dana, M., Limón, M. C., Mejías, R., Mach, R. L., Benítez, T., Pintor-Toro, J. A., et al. (2001). Regulation of chitinase 33 (chit33) gene expression in Trichoderma harzianum. Curr. Genet. 38, 335–342. doi: 10.1007/s002940000169
Degenkolb, T., Dieckmann, R., Nielsen, K. F., Gräfenhan, T., Theis, C., Zafari, D., et al. (2008). The Trichoderma brevicompactum clade: a separate lineage with new species, new peptaibiotics, and mycotoxins. Mycol. Prog. 7, 177–219. doi: 10.1007/s11557-008-0563-3
Delabona, P. D. S., Codima, C. A., Ramoni, J., Zubieta, M. P., de Araújo, B. M., Farinas, C. S., et al. (2020). The impact of putative methyltransferase overexpression on the Trichoderma harzianum cellulolytic system for biomass conversion. Bioresour. Technol. 313:123616. doi: 10.1016/j.biortech.2020.123616
Delgado-Jarana, J., Pintor-Toro, J. A., and Benítez, T. (2000). Overproduction of β-1,6-glucanase in Trichoderma harzianum is controlled by extracellular acidic proteases and pH. Biochim. Biophys. Acta. Protein Proteom. 1481, 289–296. doi: 10.1016/S0167-4838(00)00172-2
Delgado-Jarana, J., Rincón, A. M., and Benı́Tez, T. A. (2002). Aspartyl protease from Trichoderma harzianum CECT 2413: cloning and characterization. Microbiology 148, 1305–1315. doi: 10.1099/00221287-148-5-1305
Delgado-Jarana, J., Sousa, S., González, F., Rey, M., and Llobell, A. (2006). ThHog1 controls the hyperosmotic stress response in Trichoderma harzianum. Microbiology 152, 1687–1700. doi: 10.1099/mic.0.28729-0
Deng, J. J., Huang, W. Q., Li, Z. W., Lu, D. L., Zhang, Y., and Luo, X. C. (2018). Biocontrol activity of recombinant aspartic protease from Trichoderma harzianum against pathogenic fungi. Enzym. Microb. Technol. 112, 35–42. doi: 10.1016/j.enzmictec.2018.02.002
Deng, J. J., Shi, D., Mao, H. H., Li, Z. W., Liang, S., Ke, Y., et al. (2019). Heterologous expression and characterization of an antifungal chitinase (Chit46) from Trichoderma harzianum GIM 3.442 and its application in colloidal chitin conversion. Int. J. Biol. Macromol. 134, 113–121. doi: 10.1016/j.ijbiomac.2019.04.177
Denison, S. H. (2000). pH regulation of gene expression in fungi. Fungal Genet. Biol. 29, 61–71. doi: 10.1006/fgbi.2000.1188
Ding, J., Jiang, X., Mei, J., Sun, Q., and Li, M. (2018). Functions of Thga3 gene in Trichoderma harzianum based on transcriptome analysis. Chin. J. Biol. Control. 34, 124–132. doi: 10.16409/j.cnki.2095-039x.2018.01.015
Ding, J., Mei, J., Huang, P., Tian, Y., Liang, Y., Jiang, X., et al. (2020). Gα3 subunit Thga3 positively regulates conidiation, mycoparasitism, chitinase activity, and hydrophobicity of Trichoderma harzianum. AMB Express 10:221. doi: 10.1186/s13568-020-01162-9
Distefano, G., La Malfa, S., Vitale, A., Lorito, M., Deng, Z., and Gentile, A. (2008). Defence-related gene expression in transgenic lemon plants producing an antimicrobial Trichoderma harzianum endochitinase during fungal infection. Transgenic Res. 17, 873–879. doi: 10.1007/s11248-008-9172-9
Donzelli, B. G., Lorito, M., Scala, F., and Harman, G. E. (2001). Cloning, sequence and structure of a gene encoding an antifungal glucan 1,3-beta-glucosidase from Trichoderma atroviride (T. harzianum). Gene 277, 199–208. doi: 10.1016/S0378-1119(01)00681-3
Druzhinina, I. S., Chenthamara, K., Zhang, J., Atanasova, L., Yang, D., Miao, Y., et al. (2018). Massive lateral transfer of genes encoding plant cell wall-degrading enzymes to the mycoparasitic fungus Trichoderma from its plant-associated hosts. PLoS Genet. 14:e1007322. doi: 10.1371/journal.pgen.1007322
El-Debaiky, S. A. (2017). Antagonistic studies and hyphal interactions of the new antagonist Aspergillus piperis against some phytopathogenic fungi in vitro in comparison with Trichoderma harzianum. Microb. Pathog. 113, 135–143. doi: 10.1016/j.micpath.2017.10.041
El-Hasan, A., Walker, F., Schöne, J., and Buchenauer, H. (2007). Antagonistic effect of 6-pentyl-alpha-pyrone produced by Trichoderma harzianum toward Fusarium moniliforme. J. Plant Dis. Prot. 114, 62–68. doi: 10.1007/BF03356205
Erazo, J. G., Palacios, S. A., Pastor, N., Giordano, F. D., Rovera, M., Reynoso, M. M., et al. (2021). Biocontrol mechanisms of Trichoderma harzianum ITEM 3636 against peanut brown root rot caused by Fusarium solani RC 386. Biol. Control 164:104774. doi: 10.1016/j.biocontrol.2021.104774
Eslahi, N., Kowsari, M., Zamani, M. R., and Motallebi, M. (2021). The profile change of defense pathways in Phaseouls vulgaris L. by biochemical and molecular interactions of Trichoderma harzianum transformants overexpressing a chimeric chitinase. Biol. Control 152:104304. doi: 10.1016/j.biocontrol.2020.104304
Fan, L., Fu, K., Yu, C., Li, Y., Li, Y., and Chen, J. (2015). Thc6 protein, isolated from Trichoderma harzianum, can induce maize defense response against Curvularia lunata. J. Basic Microbiol. 55, 591–600. doi: 10.1002/jobm.201300814
Fan, H., Liu, Z., Zhang, R., Wang, N., Dou, K., Mijiti, G., et al. (2014). Functional analysis of a subtilisin-like serine protease gene from biocontrol fungus Trichoderma harzianum. J. Microbiol. 52, 129–138. doi: 10.1007/s12275-014-3308-9
Fanelli, F., Liuzzi, V. C., Logrieco, A. F., and Altomare, C. (2018). Genomic characterization of Trichoderma atrobrunneum (T. harzianum species complex) ITEM 908: insight into the genetic endowment of a multi-target biocontrol strain. BMC Genomics 19:662. doi: 10.1186/s12864-018-5049-3
Faull, J. L., Graeme-Cook, K. A., and Pilkington, B. L. (1994). Production of an isonitrile antibiotic by an UV-induced mutant of Trichoderma harzianum. Phytochemistry 36, 1273–1276. doi: 10.1016/S0031-9422(00)89649-1
Fekete, C., Logrieco, A., Giczey, G., and Hornok, L. (1997). Screening of fungi for the presence of the trichodiene synthase encoding sequence by hybridization to the Tri5 gene cloned from Fusarium poae. Mycopathologia 138, 91–97. doi: 10.1023/A:1006882704594
Figueras-Roca, M., Cristani, C., and Vannacci, G. (1996). Sensitivity of Trichoderma isolates and selected resistant mutants to DMI fungicides. Crop Prot. 15, 615–620. doi: 10.1016/0261-2194(96)00005-1
Flores, A., Chet, I., and Herrera-Estrella, A. (1997). Improved biocontrol activity of Trichoderma harzianum by over-expression of the proteinase-encoding gene prb1. Curr. Genet. 31, 30–37. doi: 10.1007/s002940050173
Flores, C., Nieto, M., Millán-Gómez, D., Caro, M., Galindo, E., and Serrano-Carreón, L. (2019). Elicitation and biotransformation of 6-pentyl-α-pyrone in Trichoderma atroviride cultures. Process Biochem. 82, 68–74. doi: 10.1016/j.procbio.2019.04.019
Franco-Frías, E., Ruiz-Herrera, J., and Aréchiga-Carvajal, E. T. (2014). Transcriptomic analysis of the role of Rim101/PacC in the adaptation of Ustilago maydis to an alkaline environment. Microbiology 160, 1985–1998. doi: 10.1099/mic.0.076216-0
Gallo, A., Mulè, G., Favilla, M., and Altomare, C. (2004). Isolation and characterisation of a trichodiene synthase homologous gene in Trichoderma harzianum. Physiol. Mol. Plant Pathol. 65, 11–20. doi: 10.1016/j.pmpp.2004.11.005
Gao, R., Ding, M., Jiang, S., Zhao, Z., Chenthamara, K., Shen, Q., et al. (2020). The evolutionary and functional paradox of cerato-platanins in fungi. Appl. Environ. Microbiol. 86, e00696–e00620. doi: 10.1128/AEM.00696-20
García, I., Lora, J. M., de la Cruz, J., Benítez, T., Llobell, A., and Pintor-Toro, J. A. (1994). Cloning and characterization of a chitinase (CHIT42) cDNA from the mycoparasitic fungus Trichoderma harzianum. Curr. Genet. 27, 83–89. doi: 10.1007/BF00326583
Garnica-Vergara, A., Barrera-Ortiz, S., Muñoz-Parra, E., Raya-González, J., Méndez-Bravo, A., Macías-Rodríguez, L., et al. (2016). The volatile 6-pentyl-2H-pyran-2-one from Trichoderma atroviride regulates Arabidopsis thaliana root morphogenesis via auxin signaling and ethylene insensitive 2 functioning. New Phytol. 209, 1496–1512. doi: 10.1111/nph.13725
Ghasemi, S., Safaie, N., Shahbazi, S., Shams-Bakhsh, M., and Askari, H. (2020). The role of cell wall degrading enzymes in antagonistic traits of Trichoderma virens against Rhizoctonia solani. Iran. J. Biotechnol. 18:e2333. doi: 10.30498/IJB.2020.2333
Gomes, E. V., Costa, M. D. N., de Paula, R. G., de Azevedo, R. R., da Silva, F. L., Noronha, E. F., et al. (2015). The cerato-platanin protein Epl-1 from Trichoderma harzianum is involved in mycoparasitism, plant resistance induction and self cell wall protection. Sci. Rep. 5:17998. doi: 10.1038/srep17998
Gomes, E. V., Ulhoa, C. J., Cardoza, R. E., Silva, R. N., and Gutiérrez, S. (2017). Involvement of Trichoderma harzianum Epl-1 protein in the regulation of Botrytis virulence- and tomato defense-related genes. Front. Plant Sci. 8:880. doi: 10.3389/fpls.2017.00880
Gómez-Rodríguez, E. Y., Uresti-Rivera, E. E., Patrón-Soberano, O. A., Islas-Osuna, M. A., Flores-Martínez, A., Riego-Ruiz, L., et al. (2018). Histone acetyltransferase TGF-1 regulates Trichoderma atroviride secondary metabolism and mycoparasitism. PLoS One 13:e0193872. doi: 10.1371/journal.pone.0193872
Goswami, S., Singh, V., Chakdar, H., and Choudhary, P. (2018). Harmful effects of fungicides: current status. Int. J. Agric. Environ. Biotech. 1025–1033.
Graeme-Cook, K. A., and Faull, J. L. (1991). Effect of ultraviolet-induced mutants of Trichoderma harzianum with altered antibiotic production on selected pathogens in vitro. Can. J. Microbiol. 37, 659–664. doi: 10.1139/m91-112
Güçlü, T., and Özer, N. (2022). Trichoderma harzianum antagonistic activity and competition for seed colonization against seedborne pathogenic fungi of sunflower. Lett. Appl. Microbiol. 74, 1027–1035. doi: 10.1111/lam.13698
Guo, R., Ji, S., Wang, Z., Zhang, H., Wang, Y., and Liu, Z. (2021). Trichoderma asperellum xylanases promote growth and induce resistance in poplar. Microbiol. Res. 248:126767. doi: 10.1016/j.micres.2021.126767
Harman, G. E., Howell, C. R., Viterbo, A., Chet, I., and Lorito, M. (2004). Trichoderma species — opportunistic, avirulent plant symbionts. Nat. Rev. Microbiol. 2, 43–56. doi: 10.1038/nrmicro797
Hassan, M. M. (2014). Influence of protoplast fusion between two Trichoderma spp. on extracellular enzymes production and antagonistic activity. Biotechnol. Biotechnol. Equip. 28, 1014–1023. doi: 10.1080/13102818.2014.978206
Hatvani, L., Manczinger, L., Kredics, L., Szekeres, A., Antal, Z., and Vágvölgyi, C. (2006). Production of Trichoderma strains with pesticide-polyresistance by mutagenesis and protoplast fusion. Anton. Leeuw. 89, 387–393. doi: 10.1007/s10482-005-9042-x
Herrera, R., Nunez, D., Romero, N., Besoain, X., Perez, L., and Montealegre, J. (2012). Sensitivity of wild-type and mutant Trichoderma harzianum strains to fungicides. Cienc. Investig. Agrar. 39, 569–576. doi: 10.4067/S0718-16202012000300016
Hewedy, O. A., Abdel Lateif, K. S., Seleiman, M. F., Shami, A., Albarakaty, F. M., and El-Meihy, R. M. (2020). Phylogenetic diversity of Trichoderma strains and their antagonistic potential against soil-borne pathogens under stress conditions. Biology (Basel) 9:189. doi: 10.3390/biology9080189
Hinterdobler, W., Schuster, A., Tisch, D., Özkan, E., Bazafkan, H., Schinnerl, J., et al. (2019). The role of PKAc1 in gene regulation and trichodimerol production in Trichoderma reesei. Fungal Biol. Biotechnol. 6:12. doi: 10.1186/s40694-019-0075-8
Howell, C. R. (2003). Mechanisms employed by Trichoderma species in the biological control of plant diseases: the history and evolution of current concepts. Plant Dis. 87, 4–10. doi: 10.1094/PDIS.2003.87.1.4
Jasuja, D. N. (2015). A review on toxicological effects of fungicides. Res. J. Pharm. Biol. Chem. Sci. 6, 348–360.
Jeerapong, C., Phupong, W., Bangrak, P., Intana, W., and Tuchinda, P. (2015). Trichoharzianol, a new antifungal from Trichoderma harzianum F031. J. Agric. Food Chem. 63, 3704–3708. doi: 10.1021/acs.jafc.5b01258
Kai, K., Mine, K., Akiyama, K., Ohki, S., and Hayashi, H. (2018). Anti-plant viral activity of peptaibols, trichorzins HA II, HA V, and HA VI, isolated from Trichoderma harzianum HK-61. J. Pestic. Sci. 43, 283–286. doi: 10.1584/jpestics.D18-039
Kang, X., Kirui, A., Muszyński, A., Widanage, M. C. D., Chen, A., Azadi, P., et al. (2018). Molecular architecture of fungal cell walls revealed by solid-state NMR. Nat. Commun. 9:2747. doi: 10.1038/s41467-018-05199-0
Karimi Aghcheh, R., Druzhinina, I. S., and Kubicek, C. P. (2013). The putative protein methyltransferase LAE1 of Trichoderma atroviride is a key regulator of asexual development and mycoparasitism. PLoS One 8:e67144. doi: 10.1371/journal.pone.0067144
Keswani, C., Mishra, S., Sarma, B. K., Singh, S. P., and Singh, H. B. (2014). Unraveling the efficient applications of secondary metabolites of various Trichoderma spp. Appl. Microbiol. Biotechnol. 98, 533–544. doi: 10.1007/s00253-013-5344-5
Khoza, T. G., Dubery, I. A., and Piater, L. A. (2019). Identification of candidate ergosterol-responsive proteins associated with the plasma membrane of Arabidopsis thaliana. Int. J. Mol. Sci. 20:1302. doi: 10.3390/ijms20061302
Ko, Y.-S., Kim, J. W., Lee, J. A., Han, T., Kim, G. B., Park, J. E., et al. (2020). Tools and strategies of systems metabolic engineering for the development of microbial cell factories for chemical production. Chem. Soc. Rev. 49, 4615–4636. doi: 10.1039/D0CS00155D
Kowsari, M., Motallebi, M., and Zamani, M. (2014). Protein engineering of Chit42 towards improvement of chitinase and antifungal activities. Curr. Microbiol. 68, 495–502. doi: 10.1007/s00284-013-0494-3
Kubicek, C. P., Steindorff, A. S., Chenthamara, K., Manganiello, G., Henrissat, B., Zhang, J., et al. (2019). Evolution and comparative genomics of the most common Trichoderma species. BMC Genomics 20:485. doi: 10.1186/s12864-019-5680-7
Kubodera, T., Yamashita, N., and Nishimura, A. (2002). Transformation of Aspergillus sp. and Trichoderma reesei using the pyrithiamine resistance gene (ptrA) of Aspergillus oryzae. Biosci. Biotechnol. Biochem. 66, 404–406. doi: 10.1271/bbb.66.404
Kun, R. S., Gomes, A. C. S., Hildén, K. S., Salazar Cerezo, S., Mäkelä, M. R., and de Vries, R. P. (2019). Developments and opportunities in fungal strain engineering for the production of novel enzymes and enzyme cocktails for plant biomass degradation. Biotechnol. Adv. 37:107361. doi: 10.1016/j.biotechadv.2019.02.017
Lara-Ortíz, T., Riveros-Rosas, H., and Aguirre, J. (2003). Reactive oxygen species generated by microbial NADPH oxidase NoxA regulate sexual development in Aspergillus nidulans. Mol. Microbiol. 50, 1241–1255. doi: 10.1046/j.1365-2958.2003.03800.x
Lazazzara, V., Vicelli, B., Bueschl, C., Parich, A., Pertot, I., Schuhmacher, R., et al. (2021). Trichoderma spp. volatile organic compounds protect grapevine plants by activating defense-related processes against downy mildew. Physiol. Plant. 172, 1950–1965. doi: 10.1111/ppl.13406
Liang, X.-R., Ma, X.-Y., and Ji, N.-Y. (2020). Trichosordarin A, a norditerpene glycoside from the marine-derived fungus Trichoderma harzianum R5. Nat. Prod. Res. 34, 2037–2042. doi: 10.1080/14786419.2019.1574782
Limón, M. C., Chacón, M. R., Mejías, R., Delgado-Jarana, J., Rincón, A. M., Codón, A. C., et al. (2004). Increased antifungal and chitinase specific activities of Trichoderma harzianum CECT 2413 by addition of a cellulose binding domain. Appl. Microbiol. Biotechnol. 64, 675–685. doi: 10.1007/s00253-003-1538-6
Limón, M. C., Pintor-Toro, J. A., and Benítez, T. (1999). Increased antifungal activity of Trichoderma harzianum transformants that overexpress a 33-kDa chitinase. Phytopathology 89, 254–261. doi: 10.1094/PHYTO.1999.89.3.254
Lindquist, S., and Craig, E. A. (1988). The heat-shock proteins. Annu. Rev. Genet. 22, 631–677. doi: 10.1146/annurev.ge.22.120188.003215
Liu, R., Chen, L., Jiang, Y., Zhou, Z., and Zou, G. (2015). Efficient genome editing in filamentous fungus Trichoderma reesei using the CRISPR/Cas9 system. Cell Discov. 1:15007. doi: 10.1038/celldisc.2015.7
Liu, M., Liu, J., and Wang, W. M. (2012). Isolation and functional analysis of Thmfs1, the first major facilitator superfamily transporter from the biocontrol fungus Trichoderma harzianum. Biotechnol. Lett. 34, 1857–1862. doi: 10.1007/s10529-012-0972-x
Liu, G., and Qu, Y. (2021). Integrated engineering of enzymes and microorganisms for improving the efficiency of industrial lignocellulose deconstruction. Eng. Microbiol. 1:100005. doi: 10.1016/j.engmic.2021.100005
Liu, Y., and Yang, Q. (2007). Cloning and heterologous expression of aspartic protease SA76 related to biocontrol in Trichoderma harzianum. FEMS Microbiol. Lett. 277, 173–181. doi: 10.1111/j.1574-6968.2007.00952.x
Liu, Y., and Yang, Q. (2013). Cloning and heterologous expression of serine protease SL41 related to biocontrol in Trichoderma harzianum. J. Mol. Microbiol. Biotechnol. 23, 431–439. doi: 10.1159/000346830
Lo, C. T., Nelson, E. B., Hayes, C. K., and Harman, G. E. (1998). Ecological studies of transformed Trichoderma harzianum strain 1295-22 in the rhizosphere and on the phylloplane of creeping bentgrass. Phytopathology 88, 129–136. doi: 10.1094/PHYTO.1998.88.2.129
Lorito, M., Mach, R. L., Sposato, P., Strauss, J., Peterbauer, C. K., and Kubicek, C. P. (1996). Mycoparasitic interaction relieves binding of the Cre1 carbon catabolite repressor protein to promoter sequences of the ech42 (endochitinase-encoding) gene in Trichoderma harzianum. Proc. Natl. Acad. Sci. U. S. A. 93, 14868–14872. doi: 10.1073/pnas.93.25.14868
Lorito, M., Woo, S. L., Fernandez, I. G., Colucci, G., Harman, G. E., Pintor-Toro, J. A., et al. (1998). Genes from mycoparasitic fungi as a source for improving plant resistance to fungal pathogens. Proc. Natl. Acad. Sci. U. S. A. 95, 7860–7865. doi: 10.1073/pnas.95.14.7860
Mach, R. L., Peterbauer, C. K., Payer, K., Jaksits, S., Woo, S. L., Zeilinger, S., et al. (1999). Expression of two major chitinase genes of Trichoderma atroviride (T. harzianum P1) is triggered by different regulatory signals. Appl. Environ. Microbiol. 65, 1858–1863. doi: 10.1128/AEM.65.5.1858-1863.1999
Malmierca, M. G., Cardoza, R. E., Alexander, N. J., Mccormick, S. P., Hermosa, R., Monte, E., et al. (2012). Involvement of Trichoderma trichothecenes in the biocontrol activity and induction of plant defense-related genes. Appl. Environ. Microbiol. 78, 4856–4868. doi: 10.1128/AEM.00385-12
Malmierca, M. G., Mccormick, S. P., Cardoza, R. E., Alexander, N. J., Monte, E., and Gutiérrez, S. (2015a). Production of trichodiene by Trichoderma harzianum alters the perception of this biocontrol strain by plants and antagonized fungi. Environ. Microbiol. 17, 2628–2646. doi: 10.1111/1462-2920.12506
Malmierca, M. G., Mccormick, S. P., Cardoza, R. E., Monte, E., Alexander, N. J., and Gutiérrez, S. (2015b). Trichodiene production in a Trichoderma harzianum erg1-silenced strain provides evidence of the importance of the sterol biosynthetic pathway in inducing plant defense-related gene expression. Mol. Plant-Microbe Interact. 28, 1181–1197. doi: 10.1094/MPMI-06-15-0127-R
Marra, R., Lombardi, N., Piccolo, A., Bazghaleh, N., Prashar, P., Vandenberg, A., et al. (2021). Mineral biofortification and growth stimulation of lentil plants inoculated with Trichoderma strains and metabolites. Microorganisms 10:87. doi: 10.3390/microorganisms10010087
Martínez-Soto, D., and Ruiz-Herrera, J. (2017). Functional analysis of the MAPK pathways in fungi. Rev. Iberoam. Micol. 34, 192–202. doi: 10.1016/j.riam.2017.02.006
Marzano, M., Gallo, A., and Altomare, C. (2013). Improvement of biocontrol efficacy of Trichoderma harzianum vs. Fusarium oxysporum f. sp. lycopersici through UV-induced tolerance to fusaric acid. Biol. Control 67, 397–408. doi: 10.1016/j.biocontrol.2013.09.008
Mccormick, S. P., Stanley, A. M., Stover, N. A., and Alexander, N. J. (2011). Trichothecenes: from simple to complex mycotoxins. Toxins (Basel) 3, 802–814. doi: 10.3390/toxins3070802
Mcmullin, D. R., Renaud, J. B., Barasubiye, T., Sumarah, M. W., and Miller, J. D. (2017). Metabolites of Trichoderma species isolated from damp building materials. Can. J. Microbiol. 63, 621–632. doi: 10.1139/cjm-2017-0083
Meher, J., Rajput, R. S., Bajpai, R., Teli, B., and Sarma, B. K. (2020). “Trichoderma: A globally dominant commercial biofungicide” in Trichoderma: Agricultural applications and beyond. eds. C. Manoharachary, H. B. Singh, and A. Varma (New York: Cham Springer International Publishing), 195–208.
Mendoza-Mendoza, A., Pozo, M. J., Grzegorski, D., Martínez, P., García, J. M., Olmedo-Monfil, V., et al. (2003). Enhanced biocontrol activity of Trichoderma through inactivation of a mitogen-activated protein kinase. Proc. Natl. Acad. Sci. U. S. A. 100, 15965–15970. doi: 10.1073/pnas.2136716100
Meng, X., Miao, Y., Liu, Q., Ma, L., Guo, K., Liu, D., et al. (2019). TgSWO from Trichoderma guizhouense NJAU4742 promotes growth in cucumber plants by modifying the root morphology and the cell wall architecture. Microb. Cell Factories 18:148. doi: 10.1186/s12934-019-1196-8
Mercado, J. A., Barceló, M., Pliego, C., Rey, M., Caballero, J. L., Muñoz-Blanco, J., et al. (2015). Expression of the β-1,3-glucanase gene bgn13.1 from Trichoderma harzianum in strawberry increases tolerance to crown rot diseases but interferes with plant growth. Transgenic Res. 24, 979–989. doi: 10.1007/s11248-015-9895-3
Mironenka, J., Różalska, S., Soboń, A., and Bernat, P. (2021). Trichoderma harzianum metabolites disturb Fusarium culmorum metabolism: Metabolomic and proteomic studies. Microbiol. Res. 249:126770. doi: 10.1016/j.micres.2021.126770
Montero-Barrientos, M., Hermosa, R., Cardoza, R. E., Gutiérrez, S., and Monte, E. (2011). Functional analysis of the Trichoderma harzianum nox1 gene, encoding an NADPH oxidase, relates production of reactive oxygen species to specific biocontrol activity against Pythium ultimum. Appl. Environ. Microbiol. 77, 3009–3016. doi: 10.1128/AEM.02486-10
Montero-Barrientos, M., Hermosa, R., Nicolás, C., Cardoza, R. E., Gutiérrez, S., and Monte, E. (2008). Overexpression of a Trichoderma HSP70 gene increases fungal resistance to heat and other abiotic stresses. Fungal Genet. Biol. 45, 1506–1513. doi: 10.1016/j.fgb.2008.09.003
Morán-Diez, E., Hermosa, R., Ambrosino, P., Cardoza, R. E., Gutiérrez, S., Lorito, M., et al. (2009). The ThPG1 endopolygalacturonase is required for the Trichoderma harzianum–plant beneficial interaction. Mol. Plant-Microbe Interact. 22, 1021–1031. doi: 10.1094/MPMI-22-8-1021
Moreno-Mateos, M. A., Delgado-Jarana, J., Codón, A. C., and Benítez, T. (2007). pH and Pac1 control development and antifungal activity in Trichoderma harzianum. Fungal Genet. Biol. 44, 1355–1367. doi: 10.1016/j.fgb.2007.07.012
Mukherjee, P. K., Latha, J., Hadar, R., and Horwitz, B. A. (2003). TmkA, a mitogen-activated protein kinase of Trichoderma virens, is involved in biocontrol properties and repression of conidiation in the dark. Eukaryot. Cell 2, 446–455. doi: 10.1128/EC.2.3.446-455.2003
Mukherjee, P. K., Mendoza-Mendoza, A., Zeilinger, S., and Horwitz, B. A. (2022). Mycoparasitism as a mechanism of Trichoderma-mediated suppression of plant diseases. Fungal Biol. Rev. 39, 15–33. doi: 10.1016/j.fbr.2021.11.004
Nuansri, S., Rukachaisirikul, V., Rungwirain, N., Kaewin, S., Yimnual, C., Phongpaichit, S., et al. (2022). α-Pyrone and decalin derivatives from the marine-derived fungus Trichoderma harzianum PSU-MF79. Nat. Prod. Res. 36, 5462–5469. doi: 10.1080/14786419.2021.2015593
Omann, M., Lehner, S. M., Escobar Rodriguez, C., Brunner, K., and Zeilinger, S. J. M. (2012). The seven-transmembrane receptor Gpr1 governs processes relevant for the antagonistic interaction of Trichoderma atroviride with its host. Microbiology 158, 107–118. doi: 10.1099/mic.0.052035-0
Omann, M., and Zeilinger, S. (2010). How a mycoparasite employs G-protein signaling: using the example of Trichoderma. J Signal Transduct. 2010:123126, 1–8. doi: 10.1155/2010/123126
Pang, G., Sun, T., Ding, M., Li, J., Zhao, Z., Shen, Q., et al. (2022). Characterization of an exceptional fungal mutant enables the discovery of the specific regulator of a silent PKS-NRPS hybrid biosynthetic pathway. J. Agric. Food Chem. 70, 11769–11781. doi: 10.1021/acs.jafc.2c03550
Pang, G., Sun, T., Yu, Z., Yuan, T., Liu, W., Zhu, H., et al. (2020). Azaphilones biosynthesis complements the defence mechanism of Trichoderma guizhouense against oxidative stress. Environ. Microbiol. 22, 4808–4824. doi: 10.1111/1462-2920.15246
Perez, L., Polanco, R., Rios, J. C., Montealegre, J., Valderrama, L., Herrera, R., et al. (2007). The increase in endochitinases and β-1,3-glucanases in the mutant Th650-NG7 of the Trichoderma harzianum Th650, improves the biocontrol activity on Rhizoctonia solani infecting tomato. IOBC WPRS Bull. 30, 135–138.
Prabavathy, V. R., Mathivanan, N., Sagadevan, E., Murugesan, K., and Lalithakumari, D. (2006). Self-fusion of protoplasts enhances chitinase production and biocontrol activity in Trichoderma harzianum. Bioresour. Technol. 97, 2330–2334. doi: 10.1016/j.biortech.2005.10.031
Rai, S., Kashyap, P. L., Kumar, S., Srivastava, A. K., and Ramteke, P. W. (2016). Identification, characterization and phylogenetic analysis of antifungal Trichoderma from tomato rhizosphere. Springerplus 5:1939. doi: 10.1186/s40064-016-3657-4
Ramada, M. H. S., Steindorff, A. S., Bloch, C. Jr., and Ulhoa, C. J. (2016). Secretome analysis of the mycoparasitic fungus Trichoderma harzianum ALL 42 cultivated in different media supplemented with Fusarium solani cell wall or glucose. Proteomics 16, 477–490. doi: 10.1002/pmic.201400546
Redondo-Gómez, S. (2013). “Abiotic and biotic stress tolerance in plants” in Molecular Stress Physiology of Plants. eds. G. R. Rout and A. B. Das (India: Springer India), 1–20.
Reithner, B., Mach-Aigner, A. R., Herrera-Estrella, A., and Mach, R. L. (2014). Trichoderma atroviride transcriptional regulator Xyr1 supports the induction of systemic resistance in plants. Appl. Environ. Microbiol. 80, 5274–5281. doi: 10.1128/AEM.00930-14
Rossard, S., Roblin, G., and Atanassova, R. (2010). Ergosterol triggers characteristic elicitation steps in Beta vulgaris leaf tissues. J. Exp. Bot. 61, 1807–1816. doi: 10.1093/jxb/erq047
Rubio, M. B., Hermosa, R., Reino, J. L., Collado, I. G., and Monte, E. (2009). Thctf1 transcription factor of Trichoderma harzianum is involved in 6-pentyl-2H-pyran-2-one production and antifungal activity. Fungal Genet. Biol. 46, 17–27. doi: 10.1016/j.fgb.2008.10.008
Rubio, M. B., Pardal, A. J., Cardoza, R. E., Gutiérrez, S., Monte, E., and Hermosa, R. (2017). Involvement of the transcriptional coactivator ThMBF1 in the biocontrol activity of Trichoderma harzianum. Front. Microbiol. 8:2273. doi: 10.3389/fmicb.2017.02273
Rush, T. A., Shrestha, H. K., Gopalakrishnan Meena, M., Spangler, M. K., Ellis, J. C., Labbé, J. L., et al. (2021). Bioprospecting Trichoderma: A systematic roadmap to screen genomes and natural products for biocontrol applications. Front. Fungal. Biol. 2:716511. doi: 10.3389/ffunb.2021.716511
Samuelian, S. (2016). Potential of Trichoderma harzianum for control of banana leaf fungal pathogens when applied with a food source and an organic adjuvant. 3 Biotech 6:8. doi: 10.1007/s13205-015-0327-0
Saravanakumar, K., Fan, L., Fu, K., Yu, C., Wang, M., Xia, H., et al. (2016). Cellulase from Trichoderma harzianum interacts with roots and triggers induced systemic resistance to foliar disease in maize. Sci. Rep. 6:35543. doi: 10.1038/srep35543
Saravanakumar, K., Wang, S., Dou, K., Lu, Z., and Chen, J. (2018). Yeast two-hybrid and label-free proteomics based screening of maize root receptor to cellulase of Trichoderma harzianum. Physiol. Mol. Plant Pathol. 104, 86–94. doi: 10.1016/j.pmpp.2018.10.002
Savary, S., Willocquet, L., Pethybridge, S. J., Esker, P., Mcroberts, N., and Nelson, A. (2019). The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 3, 430–439. doi: 10.1038/s41559-018-0793-y
Scarselletti, R., and Faull, J. L. (1994). In vitro activity of 6-pentyl-alpha-pyrone, a metabolite of Trichoderma harzianum, in the inhibition of Rhizoctonia solani and Fusarium oxysporum f. sp. lycopersici. Mycol. Prog. 98, 1207–1209. doi: 10.1016/S0953-7562(09)80206-2
Segreto, R., Bazafkan, H., Millinger, J., Schenk, M., Atanasova, L., Doppler, M., et al. (2021). The TOR kinase pathway is relevant for nitrogen signaling and antagonism of the mycoparasite Trichoderma atroviride. PLoS One 16:e0262180. doi: 10.1371/journal.pone.0262180
Serrano-Carreon, L., Hathout, Y., Bensoussan, M., and Belin, J. M. (1993). Metabolism of linoleic acid or mevalonate and 6-pentyl-alpha-pyrone biosynthesis by Trichoderma species. Appl. Environ. Microbiol. 59, 2945–2950. doi: 10.1128/aem.59.9.2945-2950.1993
Sivan, A. (1989). The possible role of competition between Trichoderma harzianum and Fusarium oxysporum on rhizosphere colonization. Phytopathology 79, 198–203. doi: 10.1094/Phyto-79-198
Song, Y. P., Fang, S. T., Miao, F. P., Yin, X. L., and Ji, N. Y. (2018). Diterpenes and sesquiterpenes from the marine algicolous fungus Trichoderma harzianum X-5. J. Nat. Prod. 81, 2553–2559. doi: 10.1021/acs.jnatprod.8b00714
Sood, M., Kapoor, D., Kumar, V., Sheteiwy, M. S., Ramakrishnan, M., Landi, M., et al. (2020). Trichoderma: the "secrets" of a multitalented biocontrol agent. Plants (Basel) 9:762. doi: 10.3390/plants9060762
Speckbacher, V., Ruzsanyi, V., Martinez-Medina, A., Hinterdobler, W., Doppler, M., Schreiner, U., et al. (2020). The lipoxygenase lox1 is involved in light- and injury-response, conidiation, and volatile organic compound biosynthesis in the mycoparasitic fungus Trichoderma atroviride. Front. Microbiol. 11:2004. doi: 10.3389/fmicb.2020.02004
Staunton, J., and Wilkinson, B. (2001). Combinatorial biosynthesis of polyketides and nonribosomal peptides. Curr. Opin. Chem. Biol. 5, 159–164. doi: 10.1016/S1367-5931(00)00185-X
Steindorff, A. S., Ramada, M. H., Coelho, A. S., Miller, R. N., Pappas, G. J. Jr., Ulhoa, C. J., et al. (2014). Identification of mycoparasitism-related genes against the phytopathogen Sclerotinia sclerotiorum through transcriptome and expression profile analysis in Trichoderma harzianum. BMC Genomics 15:204. doi: 10.1186/1471-2164-15-204
Stewart, A., and Hill, R. (2014). “Chapter 31 - applications of Trichoderma in plant growth promotion” in Biotechnology and Biology of Trichoderma. eds. V. K. Gupta, M. Schmoll, A. Herrera-Estrella, R. S. Upadhyay, I. Druzhinina, and M. G. Tuohy (Amsterdam: Elsevier), 415–428.
Stirling, F., and Silver, P. A. (2020). Controlling the implementation of transgenic microbes: are we ready for what synthetic biology has to offer? Mol. Cell 78, 614–623. doi: 10.1016/j.molcel.2020.03.034
Suárez, M. B., Sanz, L., Chamorro, M. I., Rey, M., González, F. J., Llobell, A., et al. (2005). Proteomic analysis of secreted proteins from Trichoderma harzianum. Identification of a fungal cell wall-induced aspartic protease. Fungal Genet. Biol. 42, 924–934. doi: 10.1016/j.fgb.2005.08.002
Sun, Q., Jiang, X., Pang, L., Wang, L., and Li, M. (2016). Functions of thga1 gene in Trichoderma harzianum based on transcriptome analysis. Biomed. Res. Int. 2016, 8329513–8329519. doi: 10.1155/2016/8329513
Suriani Ribeiro, M., Graciano De Paula, R., Raquel Voltan, A., de Castro, R. G., Carraro, C. B., José De Assis, L., et al. (2019). Endo-β-1,3-glucanase (GH16 family) from Trichoderma harzianum participates in cell wall biogenesis but is not essential for antagonism against plant pathogens. Biomol. Ther. 9:781. doi: 10.3390/biom9120781
Szekeres, A., Hatvani, L., and Leitgeb, B. (2007). Colony morphology mutants of Trichoderma harzianum with increased beta-1,4-N-acetyl-glucosaminidase production. Acta Microbiol. Immunol. Hung. 54, 23–34. doi: 10.1556/amicr.54.2007.1.3
Szekeres, A., Kredics, L., Antal, Z., Kevei, F., and Manczinger, L. (2004). Isolation and characterization of protease overproducing mutants of Trichoderma harzianum. FEMS Microbiol. Lett. 233, 215–222. doi: 10.1111/j.1574-6968.2004.tb09485.x
Taylor, L., Gutierrez, S., Mccormick, S. P., Bakker, M. G., Proctor, R. H., Teresi, J., et al. (2022). Use of the volatile trichodiene to reduce Fusarium head blight and trichothecene contamination in wheat. Microb. Biotechnol. 15, 513–527. doi: 10.1111/1751-7915.13742
Thrane, C., Tronsmo, A., and Jensen, D. F. (1997). Endo-1,3-β-glucanase and cellulase from Trichoderma harzianum: purification and partial characterization, induction of and biological activity against plant pathogenic Pythium spp. Eur. J. Plant Pathol. 103, 331–344. doi: 10.1023/A:1008679319544
van Bohemen, A. I., Ruiz, N., Zalouk-Vergnoux, A., Michaud, A., Robiou Du Pont, T., Druzhinina, I., et al. (2021). Pentadecaibins I-V: 15-residue peptaibols produced by a marine-derived Trichoderma sp. of the harzianum clade. J. Nat. Prod. 84, 1271–1282. doi: 10.1021/acs.jnatprod.0c01355
Verma, M., Brar, S. K., Tyagi, R. D., Surampalli, R. Y., and Valéro, J. R. (2007). Antagonistic fungi, Trichoderma spp.: panoply of biological control. Biochem. Eng. J. 37, 1–20. doi: 10.1016/j.bej.2007.05.012
Vieira, P. M., Coelho, A. S., Steindorff, A. S., de Siqueira, S. J., Silva Rdo, N., and Ulhoa, C. J. (2013). Identification of differentially expressed genes from Trichoderma harzianum during growth on cell wall of Fusarium solani as a tool for biotechnological application. BMC Genomics 14:177. doi: 10.1186/1471-2164-14-177
Vieira, A. A., Vianna, G. R., Carrijo, J., Aragão, F. J. L., and Vieira, P. M. (2021). Generation of Trichoderma harzianum with pyr4 auxotrophic marker by using the CRISPR/Cas9 system. Sci. Rep. 11:1085. doi: 10.1038/s41598-020-80186-4
Vieira, P. M., Zeilinger, S., Brandão, R. S., Vianna, G. R., Georg, R. C., Gruber, S., et al. (2018). Overexpression of an aquaglyceroporin gene in Trichoderma harzianum affects stress tolerance, pathogen antagonism and Phaseolus vulgaris development. Biol. Control 126, 185–191. doi: 10.1016/j.biocontrol.2018.08.012
Vinale, F., Flematti, G., Sivasithamparam, K., Lorito, M., Marra, R., Skelton, B., et al. (2009). Harzianic acid, an antifungal and plant growth promoting metabolite from Trichoderma harzianum. J. Nat. Prod. 72, 2032–2035. doi: 10.1021/np900548p
Vinale, F., Marra, R., Scala, F., Ghisalberti, E. L., Lorito, M., and Sivasithamparam, K. (2006). Major secondary metabolites produced by two commercial Trichoderma strains active against different phytopathogens. Lett. Appl. Microbiol. 43, 143–148. doi: 10.1111/j.1472-765X.2006.01939.x
Vinale, F., Sivasithamparam, K., Ghisalberti, E., Woo, S., Nigro, M., Marra, R., et al. (2014). Trichoderma secondary metabolites active on plants and fungal pathogens. Open Mycol. J. 8, 127–139. doi: 10.2174/1874437001408010127
Viterbo, A. D. A., and Chet, I. (2006). TasHyd1, a new hydrophobin gene from the biocontrol agent Trichoderma asperellum, is involved in plant root colonization. Mol. Plant Pathol. 7, 249–258. doi: 10.1111/j.1364-3703.2006.00335.x
Viterbo, A., Haran, S., Friesem, D., Ramot, O., and Chet, I. (2001). Antifungal activity of a novel endochitinase gene (chit36) from Trichoderma harzianum Rifai TM. FEMS Microbiol. Lett. 200, 169–174. doi: 10.1111/j.1574-6968.2001.tb10710.x
Wang, S.-Q., Ma, J., Wang, M., Wang, X.-H., Li, Y.-Q., and Chen, J. (2019). Combined application of Trichoderma harzianum SH2303 and difenoconazole-propiconazolein controlling southern corn leaf blight disease caused by Cochliobolus heterostrophus in maize. J. Integr. Agric. 18, 2063–2071. doi: 10.1016/S2095-3119(19)62603-1
Wang, C., and St Leger, R. J. (2007). A scorpion neurotoxin increases the potency of a fungal insecticide. Nat. Biotechnol. 25, 1455–1456. doi: 10.1038/nbt1357
Wang, X.-Y., Xu, T.-T., Sun, L.-J., Cen, R.-H., Su, S., Yang, X.-Q., et al. (2021). The chemical diversity, the attractant, anti-acetylcholinesterase, and antifungal activities of metabolites from biocontrol Trichoderma harzianum uncovered by OSMAC strategy. Bioorg. Chem. 114:105148. doi: 10.1016/j.bioorg.2021.105148
Wang, Q., Zhao, Q., Liu, Q., He, X., Zhong, Y., Qin, Y., et al. (2021). CRISPR/Cas9-mediated genome editing in Penicillium oxalicum and Trichoderma reesei using 5S rRNA promoter-driven guide RNAs. Biotechnol. Lett. 43, 495–502. doi: 10.1007/s10529-020-03024-7
Woo, S., Donzelli, B., Scala, F., Mach, R., Harman, G., Kubicek, C., et al. (1999). Disruption of the ech42 (endochitinase-encoding) gene affects biocontrol activity in Trichoderma harzianum P1. Mol. Plant-Microbe Interact. 12, 419–429. doi: 10.1094/MPMI.1999.12.5.419
Woo, S. L., Hermosa, R., Lorito, M., and Monte, E. (2022). Trichoderma: a multipurpose, plant-beneficial microorganism for eco-sustainable agriculture. Nat. Rev. Microbiol. doi: 10.1038/s41579-022-00819-5
Woo, S. L., Ruocco, M., Vinale, F., Nigro, M., Marra, R., Lombardi, N., et al. (2014). Trichoderma-based products and their widespread use in agriculture. Open Mycol. J. 8, 71–126. doi: 10.2174/1874437001408010071
Wu, Q., Bai, L., Liu, W., Li, Y., Lu, C., Li, Y., et al. (2013). Construction of a Streptomyces lydicus A01 transformant with a chit42 gene from Trichoderma harzianum P1 and evaluation of its biocontrol activity against Botrytis cinerea. J. Microbiol. 51, 166–173. doi: 10.1007/s12275-013-2321-8
Wu, M., Wei, H., Ma, K., Cui, P., Zhu, S., Lai, D., et al. (2021). ThpacC acts as a positive regulator of homodimericin A biosynthesis and antifungal activities of Trichoderma harzianum 3.9236. J. Agric. Food Chem. 69, 12695–12704. doi: 10.1021/acs.jafc.1c04330
Xie, L., Zang, X., Cheng, W., Zhang, Z., Zhou, J., Chen, M., et al. (2021). Harzianic acid from Trichoderma afroharzianum is a natural product inhibitor of acetohydroxyacid synthase. J. Am. Chem. Soc. 143, 9575–9584. doi: 10.1021/jacs.1c03988
Xie, X., Zhao, Z., Yang, H., Pan, H., Zhu, C., Hu, J., et al. (2022). Nigirpexin E, a new azaphilone derivative with anti-tobacco mosaic virus activity from soil-derived fungus Trichoderma afroharzianum LTR-2. J. Antibiot. (Tokyo) 75, 117–121. doi: 10.1038/s41429-021-00485-4
Yamada, T., Mizutani, Y., Umebayashi, Y., Inno, N., Kawashima, M., Kikuchi, T., et al. (2014). Tandyukisin, a novel ketoaldehyde decalin derivative, produced by a marine sponge-derived Trichoderma harzianum. Tetrahedron Lett. 55, 662–664. doi: 10.1016/j.tetlet.2013.11.107
Yan, L., and Qian, Y. (2009). Cloning and heterologous expression of SS10, a subtilisin-like protease displaying antifungal activity from Trichoderma harzianum. FEMS Microbiol. Lett. 290, 54–61. doi: 10.1111/j.1574-6968.2008.01403.x
Yang, C. A., Cheng, C. H., Lee, J. W., Lo, C. T., Liu, S. Y., and Peng, K. C. (2012). Monomeric L-amino acid oxidase-induced mitochondrial dysfunction in Rhizoctonia solani reveals a novel antagonistic mechanism of Trichoderma harzianum ETS 323. J. Agric. Food Chem. 60, 2464–2471. doi: 10.1021/jf203883u
Yang, C. A., Cheng, C. H., Liu, S. Y., Lo, C. T., Lee, J. W., and Peng, K. C. (2011). Identification of antibacterial mechanism of L-amino acid oxidase derived from Trichoderma harzianum ETS 323. FEBS J. 278, 3381–3394. doi: 10.1111/j.1742-4658.2011.08262.x
Yang, H. H., Yang, S. L., Peng, K. C., Lo, C. T., and Liu, S. Y. (2009). Induced proteome of Trichoderma harzianum by Botrytis cinerea. Mycol. Res. 113, 924–932. doi: 10.1016/j.mycres.2009.04.004
Yang, L., Yang, Q., Sun, K., Tian, Y., and Li, H. (2010). Agrobacterium tumefaciens-mediated transformation of SOD gene to Trichoderma harzianum. World J. Microbiol. Biotechnol. 26, 353–358. doi: 10.1007/s11274-009-0182-4
Yang, L., Yang, Q., Sun, K., Tian, Y., and Li, H. (2011). Agrobacterium tumefaciens mediated transformation of ChiV gene to Trichoderma harzianum. Appl. Biochem. Biotechnol. 163, 937–945. doi: 10.1007/s12010-010-9097-7
Yao, L., Tan, C., Song, J., Yang, Q., Yu, L., and Li, X. (2016). Isolation and expression of two polyketide synthase genes from Trichoderma harzianum 88 during mycoparasitism. Braz. J. Microbiol. 47, 468–479. doi: 10.1016/j.bjm.2016.01.004
You, J., Zhou, K., Liu, X., Wu, M., Yang, L., Zhang, J., et al. (2019). Defective RNA of a novel mycovirus with high transmissibility detrimental to biocontrol properties of Trichoderma spp. Microorganisms 7:507. doi: 10.3390/microorganisms7110507
Yu, C., Dou, K., Wang, S., Wu, Q., Ni, M., Zhang, T., et al. (2020). Elicitor hydrophobin Hyd1 interacts with Ubiquilin1-like to induce maize systemic resistance. J. Integr. Plant Biol. 62, 509–526. doi: 10.1111/jipb.12796
Yu, J. Y., Shi, T., Zhou, Y., Xu, Y., Zhao, D. L., and Wang, C. Y. (2021). Naphthalene derivatives and halogenate quinoline from the coral-derived fungus Trichoderma harzianum (XS-20090075) through OSMAC approach. J. Asian Nat. Prod. Res. 23, 250–257. doi: 10.1080/10286020.2020.1729752
Zeilinger, S., Galhaup, C., Payer, K., Woo, S. L., Mach, R. L., Fekete, C., et al. (1999). Chitinase gene expression during mycoparasitic interaction of Trichoderma harzianum with its host. Fungal Genet. Biol. 26, 131–140. doi: 10.1006/fgbi.1998.1111
Zeilinger, S., and Omann, M. (2007). Trichoderma biocontrol: signal transduction pathways involved in host sensing and mycoparasitism. Gene Regul. Syst. Bio. 1, 227–234. doi: 10.4137/GRSB.S397
Zhang, F., Chen, C., Zhang, F., Gao, L., Liu, J., Chen, L., et al. (2017). Trichoderma harzianum containing 1-aminocyclopropane-1-carboxylate deaminase and chitinase improved growth and diminished adverse effect caused by Fusarium oxysporum in soybean. J. Plant Physiol. 210, 84–94. doi: 10.1016/j.jplph.2016.10.012
Zhang, F., Ge, H., Zhang, F., Guo, N., Wang, Y., Chen, L., et al. (2016). Biocontrol potential of Trichoderma harzianum isolate T-aloe against Sclerotinia sclerotiorum in soybean. Plant Physiol. Biochem. 100, 64–74. doi: 10.1016/j.plaphy.2015.12.017
Zhang, S., Sun, F., Liu, L., Bao, L., Fang, W., Yin, C., et al. (2020). Dragonfly-associated Trichoderma harzianum QTYC77 is not only a potential biological control agent of Fusarium oxysporum f. sp. cucumerinum but also a source of new antibacterial agents. J. Agric. Food Chem. 68, 14161–14167. doi: 10.1021/acs.jafc.0c05760
Zhao, H., Lovett, B., and Fang, W. (2016). “Chapter five - genetically engineering entomopathogenic fungi” in Advances in Genetics. eds. B. Lovett and R. J. St. Leger (Cambridge, MA: Academic Press), 137–163.
Zhao, S., Xiang, B., Yang, L., Chen, J., Zhu, C., Chen, Y., et al. (2022). Genetic modifications of critical regulators provide new insights into regulation modes of raw-starch-digesting enzyme expression in Penicillium. Biotechnol. Biofuels Bioprod. 15:62. doi: 10.1186/s13068-022-02162-6
Zhao, D. L., Zhang, X. F., Huang, R. H., Wang, D., Wang, X. Q., Li, Y. Q., et al. (2020). Antifungal nafuredin and epithiodiketopiperazine derivatives from the mangrove-derived fungus Trichoderma harzianum D13. Front. Microbiol. 11:1495. doi: 10.3389/fmicb.2020.01495
Keywords: Trichoderma harzianum , biocontrol, mycoparasitism, strain improvement, fungal engineering
Citation: Xiao Z, Zhao Q, Li W, Gao L and Liu G and (2023) Strain improvement of Trichoderma harzianum for enhanced biocontrol capacity: Strategies and prospects. Front. Microbiol. 14:1146210. doi: 10.3389/fmicb.2023.1146210
Received: 17 January 2023; Accepted: 20 March 2023;
Published: 13 April 2023.
Edited by:
Gen Zou, Shanghai Academy of Agricultural Sciences, ChinaReviewed by:
Feng Marc Cai, Sun Yat-sen University, ChinaCopyright © 2023 Xiao, Zhao, Li, Gao and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guodong Liu, Z2RsaXVAc2R1LmVkdS5jbg==; Liwei Gao, Z2FvbGl3ZWkwMUBjYWFzLmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.