- 1Department of Environmental Microbiology, Babasaheb Bhimrao Ambedkar University, Lucknow, India
- 2College of Agriculture Sciences, Teerthanker Mahaveer University, Moradabad, Uttar Pradesh, India
- 3Department of Plant Science, Faculty of Applied Sciences, MJP Rohilkhand University, Bareilly, India
Fungal lipases (triacylglycerol acyl hydrolases EC 3.1.1.3) are significant industrial enzymes and have several applications in a number of industries and fields. Fungal lipases are found in several species of fungi and yeast. These enzymes are carboxylic acid esterases, categorized under the serine hydrolase family, and do not require any cofactor during the catalyzing of the reactions. It was also noticed that processes including the extraction and purification of lipases from fungi are comparatively easier and cheaper than other sources of lipases. In addition, fungal lipases have been classified into three chief classes, namely, GX, GGGX, and Y. Fungal lipases have applications not only in the hydrolysis of fats and oils (triglycerides) but are also involved in synthetic reactions such as esterification, acidolysis, alcoholysis, interesterification, and aminolysis. The production and activity of fungal lipases are highly affected by the carbon source, nitrogen source, temperature, pH, metal ions, surfactants, and moisture content. Therefore, fungal lipases have several industrial and biotechnological applications in many fields such as biodiesel production, ester synthesis, production of biodegradable biopolymers, formulations of cosmetics and personal care products, detergent manufacturing, degreasing of leather, pulp and paper production, textile industry, biosensor development, and drug formulations and as a diagnostic tool in the medical sector, biodegradation of esters, and bioremediation of wastewater. The immobilization of fungal lipases onto different carriers also helps in improving the catalytic activities and efficiencies of lipases by increasing thermal and ionic stability (in organic solvents, high pH, and temperature), being easy to recycle, and inducing the volume-specific loading of the enzyme onto the support, and thus, these features have proved to be appropriate for use as biocatalysts in different sectors.
1. Introduction
Triacylglycerol acyl hydrolases (EC 3.1.1.3) are also known as lipases and are recognized as a group of potential industrial enzymes, responsible for catalyzing the hydrolysis or breakdown of insoluble fats and oils (triglycerides), and they can release monoglycerides, diglycerides, glycerol, and free fatty acids over an oil–water interface (Geoffry and Achur, 2018; Patel and Shah, 2020). Moreover, lipases are carboxylic acid esterases that belong to the serine hydrolase family and do not require any cofactor to catalyze the reactions (Basheer et al., 2011). Lipases constitute the third biggest family of digestive enzymes after proteases and carbohydrates. They are a chief group of biocatalysts in the field of biotechnology (Demera et al., 2019; Lima et al., 2019). Furthermore, these enzymes in an organic medium are also able to catalyze synthetic (formation) reactions such as esterification, interesterification, alcoholysis, aminolysis, and acidolysis in addition to hydrolysis of triglycerides (Anobom et al., 2014; Lima et al., 2019). In both aqueous and non-aqueous mediums, lipases have high efficiency to catalyze reactions as they contain high stability against a high range of temperatures, pH, and even organic solvents. It is also known that lipases have a hydrophobic lid essential for their interfacial activity (Khan et al., 2017; Mehta et al., 2017; Tan et al., 2018; Bharathi and Rajalakshmi, 2019). Firstly, Clade Bernad, in 1856, had observed the lipase enzyme in pancreatic juice, where lipase performed the function of hydrolysis of oil and fats droplets and was capable of converting them into soluble digestible products (Jamilu et al., 2022). Lipases have been reported in animals, insects, and plants, as well as microorganisms such as bacteria, fungi, yeasts, and algae (Mehta et al., 2017; Sarmah et al., 2018; Bharathi and Rajalakshmi, 2019).
Lipases are highly diverse and mainly ubiquitous in animals, plants, and microorganisms (Bharathi and Rajalakshmi, 2019). Lipases that are specially derived from microbial sources have gained increasing attention in the industrial fields rather than those that are derived from animals and plants due to their suitable characteristic features and functional ability under highly difficult conditions and remain stable in organic solvents, chemical selectivity, enantio-selectivity, and do not need any cofactor to increase their catalytic efficiency during reactions (Bharathi and Rajalakshmi, 2019; Thapa et al., 2019).
Among microbial-origin sources of lipases, fungi have been recognized as good producers of extracellular lipases, and processes including extraction and purification are comparatively easier than other sources of lipases (Treichel et al., 2010; Geoffry and Achur, 2018). Because of their versatility, fungal lipases are recognized as Potential biocatalysts in both the industrial and biotechnological sectors. Fungal lipases have applications in several industries such as leather, textile, cosmetics, biodiesel production, detergent manufacturing, medicine and pharmaceutical, pulp and paper production, dairy product formation, beverages, medical and diagnostics, biosurfactant formation, fatty acid production, and the oleochemical industry (Kaur et al., 2016; Geoffry and Achur, 2018; Avhad and Marchetti, 2019; Jamilu et al., 2022).
The production of fungal lipases is largely affected by the composition of the medium, temperature, pH, inoculum volume, aeration, agitation, and several other factors. These other factors have been observed that affect the number of microbes, and several strategies have been applied that optimize the different parameters of the fermentation process by using statistical experimental designs (Kishan et al., 2013; Lima et al., 2019).
The review article mainly focuses on fungal lipases and their catalytic potential, sources of fungal lipases, classes of fungal lipases, different methods of fungal lipase immobilization, factors affecting the activity and production of fungal lipases, molecular techniques in fungal lipase production, and their industrial and biotechnological applications in various industries and sectors.
2. Bio-catalytic potential of fungal lipases
Fungi and yeast are distinguished as potential sources of fungal lipase among the different kinds of microorganisms (Tan et al., 2003). Since fungal lipases are substrate-specific and stable under a wide range of chemical and physical conditions, they have received significant attention in industries and other sectors, as fungi produce extracellular lipolytic enzymes that are easily extracted and purified, thus lowering production costs and making these preferred sources over bacterial lipases (Mehta et al., 2017).
Now days, the chief fungal strains, such as Candida rugosa, Rhizopus oryzae, Mucor miehei, Rhizopus japonicus, Rhizopus arrhizus, Rhizopus delemar, Rhizopus niveus, Aspergillus niger, and Humicola lanuginosa, are produced commercial lipases in their culture medium (Chandra et al., 2020). Fungal-originated commercial lipases have applications in several industrial sectors such as detergent formation, food and dairy products production, pharmaceutical and medicine, biodiesel production, oleochemical industries, bioremediation, the leather industry, bioremediation of wastewater, cosmetics and perfumeries, the medical and pharmaceutical sectors, ester synthesis, and the paper industry. Furthermore, some fungal lipases also have applications as diagnostic tools in the area of the medical field. Fungal lipases have tremendous potential as biocatalysts for biomolecule production due to their specificity and benefits for future development (Mehta et al., 2017; Geoffry and Achur, 2018; Mahfoudhi et al., 2022).
The major important advantages are as follows:
• They have high efficacy under mild reaction conditions.
• Easier to practice in the natural reaction medium and products.
• Capable to decrease contamination from the environment.
• Accessibility of lipases from diverse fungal sources.
• Enhancement of catalytic power of lipases through genetic engineering (Mahfoudhi et al., 2022).
3. Classes of fungal lipases
According to their protein topology, lipases were originally classified as bacterial lipases because of their high sequence diversity (Arpigny and Jaeger, 1999). There are eight families of lipases. The classification of lipase has been modified many times, and recently, there is a modified classification that contains 15 families, all of which are part of the ESTHER database available at http://bio0web.ensam.inra.fr/esther (Fu et al., 2013; Mahfoudhi et al., 2022). An ESTHER is an extensive database covering all the information on a huge variety of the α/β hydrolases fold superfamily that comprises lipases (Lenfant et al., 2012). An additional classification created on comparatively simplified lipase data has been recently published in the Lipase Engineering Database (LED), in addition to that by Arpigny and Jaeger (1999; http://www.led.uni-stuttgart.de). This classification now comprises not only bacterial lipases but also mammalian, fungal, and yeast lipases.
Based on the oxyanion hole pattern, this classification (shown in Figure 1) categorized these enzymes into three classes: GX, GGGX, and Y (Fischer and Pleiss, 2003). The backbone N–H of an amino acid (X) in a GX motif forms the first part of the oxyanion hole, whereas in GGGX types, the backbone N–H of the third glycine forms the first part, and later, a third type of oxyanion hole (Y-type) was discovered. A bulky amino acid, mainly tyrosine or aspartate, forms the oxyanion hole in Y-type hydrolases (Gupta et al., 2015). According to this classification and based on amino acid sequence similarity, yeasts and fungal lipases are classified into five subclasses. The GX class contains two subclasses, the GGGX class also contains two subclasses, and the Y class only contains one subclass (Borrelli and Trono, 2015; Gupta et al., 2015).
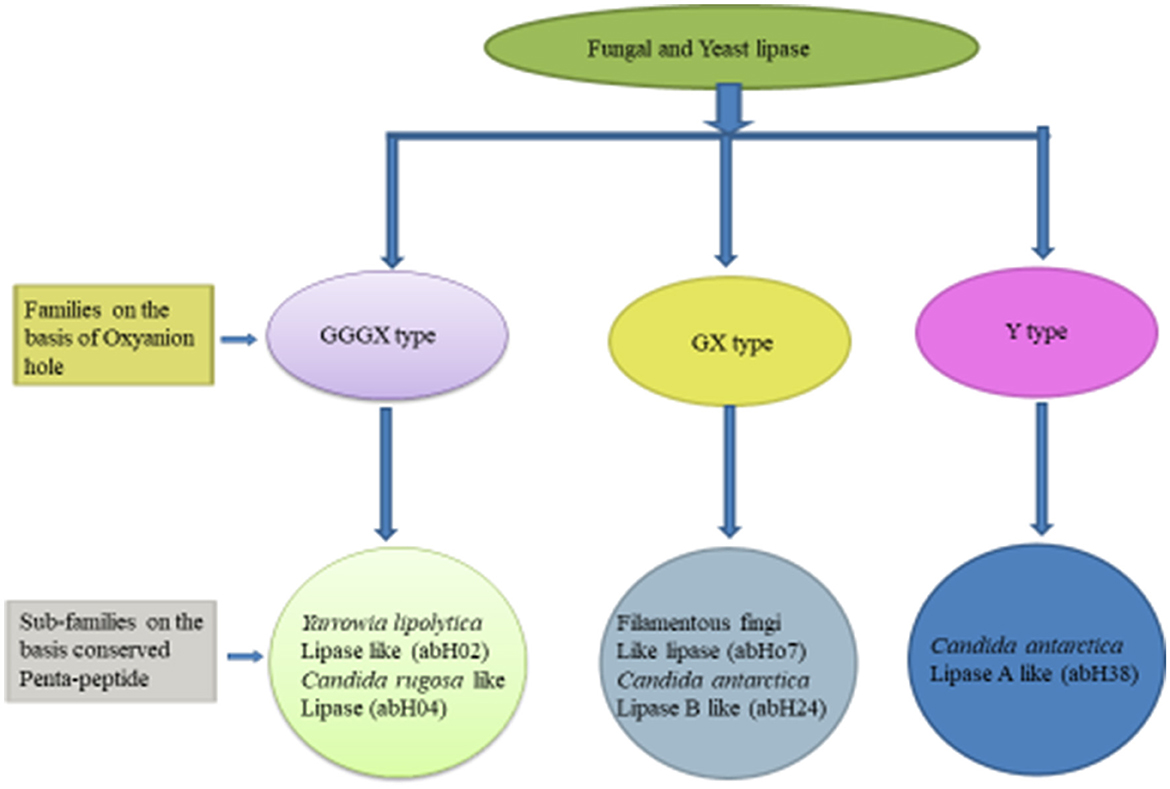
Figure 1. Classification of fungal and yeast lipase (Mehta et al., 2017; Mahfoudhi et al., 2022).
4. Sources of lipases
Among the various sources of lipases, the microbial sources of lipases have gained greater industrial attention because of their selectivity, stability, and broad substrate specificity (Kumar et al., 2016; Mehta et al., 2017; Mahfoudhi et al., 2022). Several microbes are recognized as potential sources of lipases, and these microbes are grouped as fungi, yeast, and bacteria. In this review, we discussed only the fungal and yeast sources of lipases.
4.1. Fungal sources of lipases
A large number of fungal strains are known to be capable of producing lipases that have distinctive catalytic properties that are crucial to various commercial applications (Pandey et al., 2016). In the commercial and industrial world, the most important fungal species that produce lipases are Aspergillus sp., Penicillium sp., Rhizopus sp., Fusarium sp., Geotrichum sp., Trichoderma sp., and Mucor sp. (Mohanasrinivasan et al., 2009; Bharathi and Rajalakshmi, 2019; Joshi et al., 2019). Moreover, some recently isolated fungi are also documented as they can produce lipase enzymes such as Rhizopus oryzae R1 (Helal et al., 2021), Stemphylium lycopersici, Sordida sp. (Rocha et al., 2020), Aspergillus niger 13 F, Fusarium solani 7 F (Patel and Shah, 2020), Aspergillus flavus (Ezema et al., 2022), Aspergillus terreus AH-F2 (Shabbir and Mukhtar, 2018), and Thermomyces lanuginosus (Tišma et al., 2019). The production of lipases by fungi is varied according to the fungal organism and composition of the growth medium and physical conditions such as the nitrogen source, carbon source, pH, and temperature (Pandey et al., 2016). Among the fungal sources, filamentous fungi are recognized as prominent lipase producers, and methods including extraction, purification, and processing are comparatively simple and easy when compared to other sources. Rocha et al. (2020) isolated two endophytic fungi, namely, Stemphylium lycopersici and Sordida sp., from the leaves of Humiria balsamifera and Tocoyena bullata, and both fungi have been observed to be capable of yielding the lipase enzyme of 397 U/ml/min and 286 U/ml/min, respectively. Shabbir and Mukhtar (2018) also isolated a fungus Aspergillus terreus AH-F2 from soil and oily samples that shows a maximum lipase production of 5.0 U/ml/min in a defined medium at pH 6.0. In another study, Patel and Shah (2020) isolated two fungi, namely, Fusarium solani 7 F and Aspergillus niger 13 F, and both the fungi are capable of producing 5.95 U/mL/min of crude lipase after 4 days of incubation at 30°C. Some fungal strains, their isolation sites, culture conditions, screening substrate, and specific enzyme activities are summarized in Table 1.
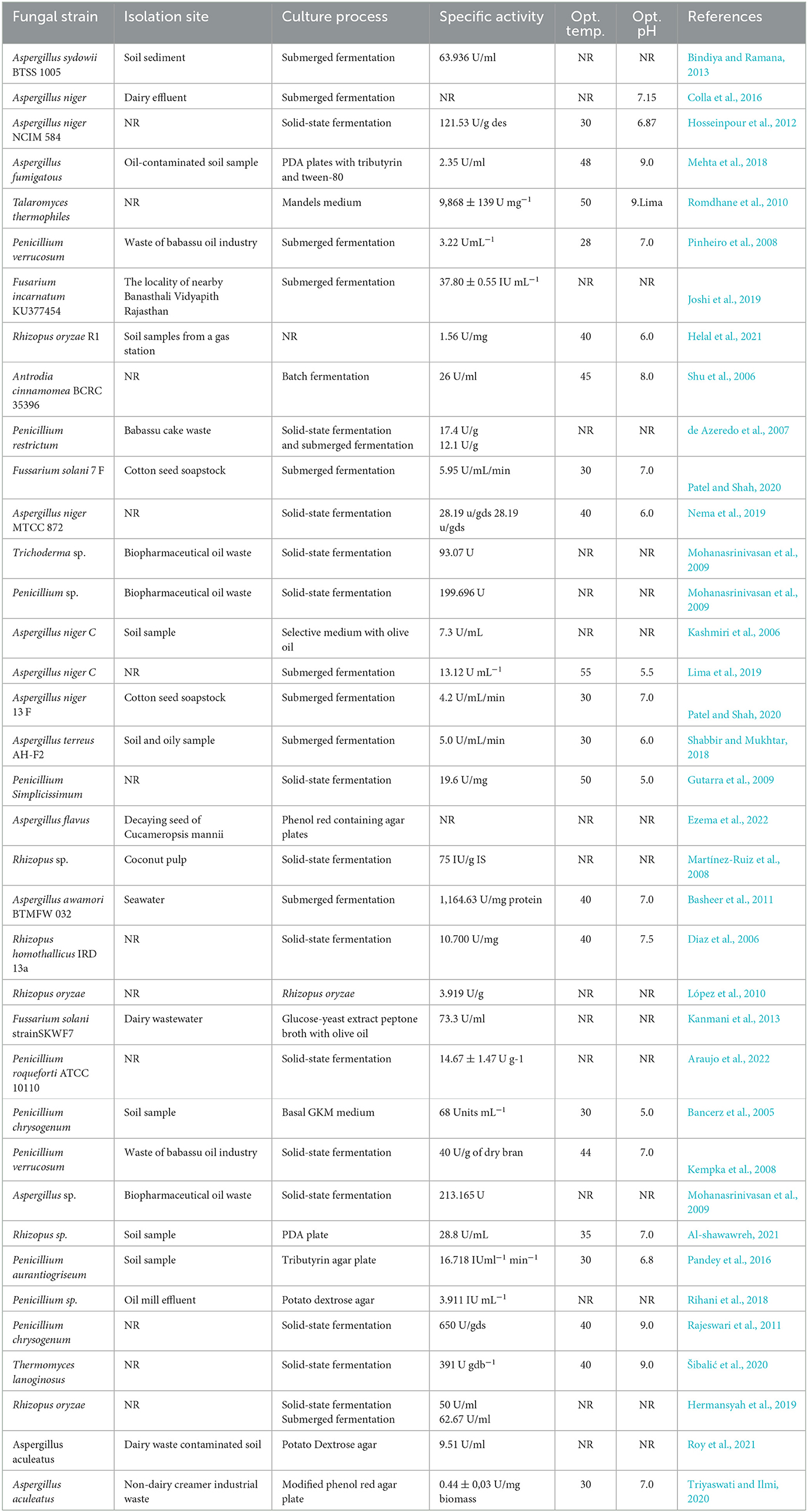
Table 1. The names of some fungi, their isolation sites, culture processes, and enzymatic activities.
4.2. Yeast sources of lipases
Yeast-derived lipases have distinctive applications in the chemical and pharmaceutical industries and in biodiesel production (Singh and Mukhopadhyay, 2012). Recently, the reviewed literature survey demonstrated that Yarrowia sp., Cryptococcus sp., Candida 99–125, Meyerozyma guilliermondii, Candida utilis, Magnusiomyces capitatus JF5, Pichia sp. Rhodotorula sp., Candida rugosa, and Candida utilis are very good primary producers of lipases (Dalmau et al., 2000; Dominguez et al., 2003; Tan et al., 2003; Thirunavukarasu et al., 2016; Bharathi and Rajalakshmi, 2019; Knob et al., 2020; Salgado et al., 2020). According to the literature review, Candida sp. is the lipase-producing category of yeasts with the most potential. The lipase from Candida sp. has been widely recognized for its structural, biochemical, and catalytic properties.
Thirunavukarasu et al. (2016) reported that Cryptococcus sp. can produce a maximum lipase activity of 753 ± 19 U/g dry substrate (gds) after growing on agro-industrial residues. Recently, Knob et al. (2020) observed that after culture conditions using statistical design, Meyerozyma guilliermondii showed lipase activity of 285.82 U ml−1 when grown in a 2% of cheese whey at 4.0 pH for 24 h. Some names of yeasts, isolation sites, culture processes, and enzymatic activities are summarized in Table 2.
5. Immobilization of fungal lipase
The term immobilization of enzyme designates an enzyme that is either physically or chemically confined or localized in a certain space or region with a plenteous holding of its catalytic activity in several cycles to follow (Mohamad et al., 2015). The immobilization of enzymes can enhance enzyme stability and catalytic power (Liese and Hilterhaus, 2013; Thangaraj and Solomon, 2019). There are several methods or techniques for the immobilization of enzymes by using different supports, and these techniques are either reversible (physical adsorption) or irreversible (membrane confinement or encapsulation, cross-linking, entrapment, and covalent-binding; Bilal et al., 2021).
Physical adsorption is an old, simple, and less costly technique involved in the binding of atoms, ions, or molecules of a dissolved solid, liquid, or gas to the surface region of a carrier by weak interactions such as Landon–van der Waals interactions, hydrogen bonding, ionic bonding, and hydrophobic interactions (Ahmad and Sardar, 2015). In contrast, covalent bonding is an irreversible method that involves the binding of the enzyme to a carrier through strong interactions during catalytic reactions and improves the stability of the enzyme automatically (Zheng et al., 2012), while the membrane-confining or encapsulation technique entraps the enzyme within a semi-permeable spherical membrane with a selectively controlled permeability (Nedovic et al., 2011; Thangaraj and Solomon, 2019). In the entrapment technique, the enzyme is enclosed within the lattice of a polymer matrix or membrane without a chemical reaction. Furthermore, the cross-linking process involves the formation of physical or chemical intermolecular cross-linkages between a protein and other protein molecules or a functional group of an insoluble matrix to form a three-dimensional structure. These techniques have made great advancements in fungal lipases, such as thermal or ionic stability, easy recycling, and volume-specific loading of the enzyme onto the support, and thus immobilization can improve the catalytic activity of lipases to apply them in different areas.
Asmat et al. (2019) immobilized the lipase of Candida rugosa on multi-walled carbon nanotubes (MWCNTs) as a carrier material by covalent bonding through glutaraldehyde, and the immobilized lipase showed high catalytic activity of about 3-fold of free lipase, exhibited high thermal stability in a broad range of pH, and was used to synthesize isoamyl and ethyl butyrate, which have characteristic banana and pineapple flavors, respectively. Similarly, Bayramoglu et al. (2022) also immobilized the Candida rugosa lipase by using magnetic chitosan beads as a carrier by Schiff base reaction, and the immobilized lipase enzyme showed better thermal and storage stability at various temperatures than free lipase and was capable of synthesizing isopentyl acetate and isoamyl acetate from isopentyl alcohol and isoamyl alcohol through the esterification reaction in the hexane medium. In another similar study, Aghaei et al. (2021) also immobilized the Candida rugosa lipase by cross-bonding on epoxy-activated Cloisite 30 B as a carrier and successively used it in the olive oil hydrolysis and synthesis of the isoamyl acetate (banana flavor) and biodiesel production at ~95.4%. Moreover, Gricajeva et al. (2018) immobilized the lipase (Resinase A 2x) of Aspergillus sp. by using cross-linking on the magnetic particles as a support and found that it has applications in the synthesis of 2-phenylethyl butanoate (fragrance compound and flavor). Lira et al. (2021) also immobilized the lipase of Thermomyces lanuginosus on agro-industrial wastes as a support and found that it has applications in the production of hexyl laurate.
6. Factors that affect the production and activity of fungal lipases
Several factors strongly affect the production and catalytic activity of lipases, and the presence of several inducers, such as carbon sources, nitrogen sources, pH, and temperature, are the major factors considered.
6.1. Carbon source
The production of lipases from fungi largely depends on the induction of lipase enzyme-encoding genes in microbes. Therefore, carbon sources play an important function in the stimulation of lipase enzyme-encoding genes in all kinds of microbial lipase-producing sources (microorganisms). Carbon sources help in the improvement of the fermentation process, which, in turn, increases cellular metabolism and results in increased fungal lipase activity (Alabdalall et al., 2020).
However, several oily carbon sources, such as olive oil, palm oil, and other vegetable oils, have been used as inducers for lipase enzyme production in microbes. Sethi et al. (2013) observed that when mustard seed oil was used as a carbon source in the culture medium of Aspergillus terreus, it showed a significant yield of lipase. Fatima et al. (2021) also observed that when an amalgam of olive oil cake and sugar cane bagasse is used as a carbon source, an increased production of lipase is attained by fungal strains. Olive oil cake has been found to be a good inducer of the lipase enzyme when compared to other carbon sources (Fatima et al., 2021). Different lipid sources, such as mustard oil, neem oil, coconut oil, olive oil, sunflower oil, palm oil, and cucumber oil, and many non-lipid sources of carbon such as sucrose, lactose, maltose, glucose, galactose, xylose, fructose, and mannitol, have been used in the media of Aspergillus sydowii to check the effect of these sources for lipase production by Bindiya and Ramana (2013). Carbohydrates act as monovalent carbon sources for lipase production, due to which, in the presence of starch and sucrose, a low yield of lipase was achieved by Aspergillus niger, but when fructose was used as a carbon source, it showed high lipase production of 777.44 U/ml (Alabdalall et al., 2020). Tween 80 has been used to assist in improving the recovery of lipase in Acinetobacter sp. (Li et al., 2001; Mahfoudhi et al., 2022).
6.2. Nitrogen source
In the synthesis of lipase enzymes, nitrogen sources also have an important role because these are necessary for the growth of microbes and the stimulation of lipase production in microbes. Different organic and inorganic nitrogen sources, such as peptone, tryptone, sodium nitrate, ammonium salts, urea, and yeast extract, have a major role in the production of lipase in various microorganisms (Oliveira et al., 2016; Bharathi and Rajalakshmi, 2019; Priyanka et al., 2019; Fatima et al., 2021). Higher lipolytic activity was demonstrated by Rhizopus sp. showed higher lipolytic activity, when added urea in their growing medium (Rodriguez et al., 2006; Mehta et al., 2017). Similarly, an amalgam of peptone and another nitrogen extract has been utilized for the production of lipase from Aspergillus sp. (Bharathi and Rajalakshmi, 2019; Colonia et al., 2019). It was also observed that a mixture of nitrogen sources influences lipase enzyme activity. Trichoderma harzianum achieved maximum lipase activity when glucose and peptone acted as sources of carbon and nitrogen in their culture medium, whereas minimum lipase activity was observed with glucose and yeast extracts as sources of carbon and nitrogen, respectively. Furthermore, Bindiya and Ramana (2013) used different nitrogen sources with a concentration of 1% w/v for investigating their effects on lipase production in Aspergillus sydowii. The nitrogen sources used in the study were (NH4)2SO4, yeast extract, NaNO3, tryptone, KNO3, NH2CONH2, beef extract, and NH4Cl. The highest activity of lipase output was achieved when using NH4Cl.
6.3. Temperature
Temperature also has a significant effect on microbial lipase production and can change the physical properties of cell membranes, resulting in the influencing of the secretion of the extracellular enzyme. An optimum temperature is crucial and has a vital role in the secretion of enzymes in the shake-flask method. A higher biomass output of lipase production was achieved at a temperature of 37°C (Mehta et al., 2017; Bharathi and Rajalakshmi, 2019). Researchers recorded that a small increase in temperature at 38°C stimulates the output of lipase (Bharathi and Rajalakshmi, 2019; de Souza et al., 2019). It has been observed that a lower temperature decreases the output of the lipase enzyme, while a higher temperature also affects its activity. In a research study, Mukhtar et al. (2016) studied the effect of different incubation temperatures ranging from 25 to 55°C on the production of lipase enzyme from Aspergillus niger. The highest output of lipase was obtained at 30°C followed by 35, 40, 25, 50, and 55°C. In another study, Mahmoud et al. (2015) investigated the effect of various incubating temperatures, viz., 10, 20, 30, and 45°C, on the production of lipase from Aspergillus terreus. The maximum activity of lipase was achieved at 45°C (15 U/mL), which was decreased with the decrease in temperatures, viz., 30°C (12 U/ml), 20°C (9.5 U/ml), and 10°C (3.0 U/ml).
Comménil et al. (1995) mentioned that the Botrytis cinerea strain produced a temperature-sensitive fungal lipase that achieved the highest activity at 38°C and was completely inactivated above 60°C. Furthermore, the Botrytis cinerea lipase can also be stable at room temperature, with 98% of its initial activity observed after an incubation period of 48 h. Nomuraea rileyi and Rhizopus oryzae showed their highest enzymatic activities at 60°C, and great stabilities were noted at 50°C (Supakdamrongkul et al., 2010; Saranya and Ramachandra, 2020). Recently, a lipase was isolated and purified from Cladosporium tenuissimum, and the lipase achieved its maximum activity at 60°C (Saranya and Ramachandra, 2020).
6.4. The pH of the medium
In general, bacterial-origin lipases have either an alkaline pH or neutral pH. Recent research reported that conditions with an alkaline pH or a slightly neutral pH stimulated the production of lipase in both bacteria and fungi (Ramakrishnan et al., 2016; Bharathi and Rajalakshmi, 2019). According to Taskin et al. (2016), Rhodotorula glutinis HL 25 was observed with fair lipase activity when the pH of their lipase-producing medium was kept slightly neutral. On the other hand, acidic pH stimulates the production of fungal lipases. Turati et al. (2019) recorded the enhanced output and activity of fungal lipase when the pH of the reaction process was kept at 4.0. Mahmoud et al. (2015) investigated the effect of different pH ranges from 2.0 to 12 on the production of lipase from Aspergillus terreus; it showed maximum enzyme activity at pH 8.0.
6.5. Surfactants
Surfactants are ionic, long-chain organic molecules with a hydrophobic part and a hydrophilic part, and they can increase the permeability of the cell membrane, thus facilitating the export of many other molecules across the membrane and also stimulating the secretion of the protein. Surfactants are capable of changing the internal charges of lipase from cationic to anionic and vice versa. Due to this, the physicochemical properties of lipases are changed (Essamri et al., 1998). Surfactants are important gradients for emulsion preparations during the lipase assay at each step of enzyme production, purification, and their characterization (Supakdamrongkul et al., 2010). According to Saranya and Ramachandra (2020), several lipases revealed deviation in their affinities to the substrate when surfactants, such as Triton X-100, Tween 20, SDS, and Tween 80, were used in different concentrations. Lipase produced by the fungus Nomuraea rileyi showed enhanced activity in the presence of Tween 80 and SDS, whereas Rhizopus oryzae, a thermophilic fungus, showed less activity when Tween 80 and SDS were added (Supakdamrongkul et al., 2010).
In general, surfactants, such as Tween 80, Tween 20, Triton X-100, and cetyltrimethylammonium bromide (CTAB), enhance the activity of lipases when they are incorporated into the fermentation medium, but their enhancing effects may be varied from surfactants to strains (Silva et al., 2005; Niaz et al., 2013; Das et al., 2017; Geoffry and Achur, 2018). Thus, the selection of an appropriate surfactant must be important for obtaining high lipase activity.
6.6. Moisture content
Moisture content plays a major role in fungus growth and the production of lipases, especially in solid-state fermentation processes (Singhania et al., 2009). Moisture affects the physical properties of the solid substrate for several reasons, such as decreasing substrate porosity, altering the particle structure of the substrate, promoting the development of stickiness, reducing gas volume and aeration, and limiting the diffusion of oxygen in the substrate layer (Mukhtar et al., 2016; Oliveira et al., 2016).
High moisture content ultimately decreases the filamentous growth of fungi and thus results in a decrease in the activity and production of lipases. Optimum moisture content enhances lipase production and hydrolytic activity.
6.7. Metal ions
Metal ions are a significant ingredient and play different roles in influencing the structure and working of enzymes including lipases (Mahfoudhi et al., 2022). Different fungal lipases showed different behaviors toward different metal ions. Lipase produced by Aspergillus japonicas is inhibited when incubated with Mn2+ and Hg2+, whereas it remains stable when incubated with Ca2+ (Jayaprakash and Ebenezer, 2012). Similarly, Katiyar and Ali (2013) noticed that increased activity of lipase has been shown by Candida rugosa in the presence of Ca2+ ions. Interestingly, Saranya and Ramachandra (2020) reported that there is no effect of Ca2+, Mg2+, Na+, and K+ on the activity of lipase produced from Cladosporium tenuissimum. Toida et al. (1995) also noticed that metal ions, such as Cu2+, Fe3+, Hg2+, Zn2+, and Ag+, inhibited the lipase activity isolated from Aspergillus oryzae.
7. Molecular techniques in lipase production
The increased industrial application of lipases is responsible for the scale-up in the productivity of lipases, and several approaches have been practiced to fulfill this aim. In this section, we resume several processes and metabolic engineering techniques to carry out fungal lipase production. Industrial lipases are chiefly produced from fungi extracellularly. There have been several studies conducted on the production of free-type lipases, focusing primarily on the parameters related to optimizing the medium and operating conditions in batch cultures. Fed-batch fermentation has recently resulted in a significant increase in lipase productivity. Low productivity and high cost are generally associated with wild-type lipases. They naturally lose the optimal specificities and required properties for industrial feedstock. Hence, to obtain the standards of quantity and manipulation of industrial processes, we try to opt for other substitute methods, such as cloning and expression of the recombinant genes of lipases, to find an appropriate quantity of pure biocatalysts (Mahfoudhi et al., 2022).
Codon optimization in the lip3 gene of Candida rugosa can improve lipase production by ~50- to 70-fold (Chang et al., 2006). In a study, Hwang et al. (2014) indicated that promoter optimization is a generally used technique, which significantly advances the production of lipases. Techniques such as gene modification can make genes adjustable for expression within the recombinant host cells. Additionally, Yaver et al. (2000) have engendered insertion mutant libraries in a recombinant Aspergillus oryzae strain capable of expressing the Thermomyces lanuginosus lipase using restriction enzyme-mediated integration as a mutagen, and the results have proved that genetic modification can be used to modulate the expression of heterologous proteins. It is possible to improve lipase production using recombinant lipase, but its yield is limited by many post-translational events, such as disulfide bond formation, solubility, miss-folding, secretion, proteolysis, and even toxicity to host cells (Makrides, 1996; Mahfoudhi et al., 2022). To overcome these limitations, genetic and metabolic engineering can play an important role in increasing the production of recombinant lipase. Scaling up fermentation through efficient and convenient techniques can enhance lipase production. By optimizing fermentation conditions, Zhao et al. (2008) significantly enhanced the production of the Candida rugosa lipase in constitutive recombinant Pichia pastoris in both laboratory and pilot scales. In this experiment, a pH-stat and two-stage fermentation strategy combined with exponential feeding was used to scale up fermentation from 5 to 800 L, which achieved an excellent balance between the expression of recombinant lipase and the growth of the host cells. In the 800 L scale, an optimal lipase activity of ~14,000 IU/ml and a cell wet weight of 500 g/L have been determined. It is possible to effectively control the cell growth rate in large-scale fermentations of recombinant lipases by tuning their cell lyse and proteolytic sensitivity of lipase (Narayanan and Chou, 2009). Significant interest has been shown in improving biofuel production by upgrading lipase production through metabolic engineering (Atsumi and Liao, 2008; Lee et al., 2008). Zhou et al. (2019) cloned the Rhizomucor miehei lipase gene (rml) in Yarrowia lipolytica Polg and obtained 19.5 U/ml lipase activity; after codon optimization of the rml gene, an increased lipase activity (26.9 U/ml) was obtained. Subsequently, a method was developed for constructing hybrid promoters harboring different copy numbers of the upstream activation sequences fragment (UAS1B), and the recombinant strain Po1g/hp12d-rml 25# reached 38.9 U/ml. Wang et al. (2018) increased the 48.7% recombinant lipase activity of Galactomyces geotrichum in Pichia pastoris through codon optimization of the lipase gene.
8. Industrial and biotechnological applications of fungal lipases
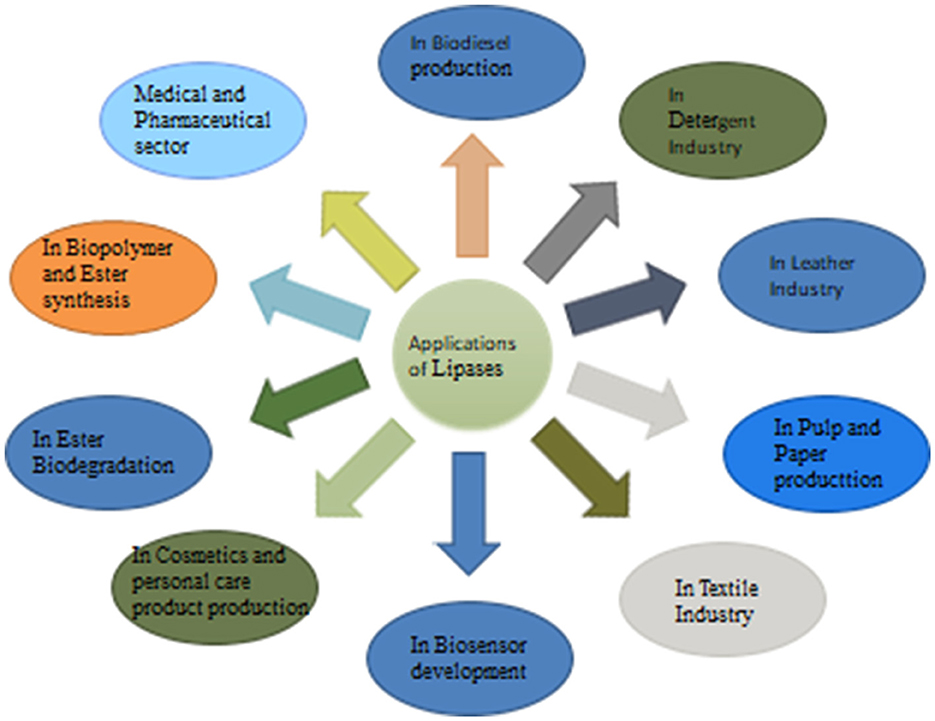
Fungal lipases are widely known for their enzymatic properties and substrate specificity, which make them versatile tools for industrial applications. They have become a chief group of biotechnological enzymes because of the versatility of their properties and easy mass production. Figure 2 represents the systematic applications of fungal lipases.
8.1. In biodiesel production
Continuous steady growth in population, transportation, and industrialization in the world results in the origin of increased demand for energy for industrial and domestic uses (Santos et al., 2020). To fulfill this increased demand for energy, we depend mainly on fossil fuels (petroleum-derived diesel), but the reservoir of fossil fuels (petroleum-derived diesel) is limited in quantity and will be completely exhausted in the future. A high amount of CO2, CO, and NOx gases is released into the environment by the burning of diesel, which in turn increases pollution and produces a greenhouse gas effect on the Earth (Quayson et al., 2020; Almeida et al., 2021). Therefore, we should try to search for alternative renewable sources of energy that could fulfill our needs for energy in the upcoming future. Biodiesel could be an effective alternative resource for our energy needs and has many advantages over fossil fuels (Santos et al., 2020). Biodiesel is a mixture of fatty acid alkyl esters and is non-toxic, biodegradable, and has low carbon, sulfur, and high hexadecane contents. Therefore, biodiesel produces low carbon dioxide (CO2), carbon monoxide (CO), and nitrogen oxide (NOx) emissions in comparison to petroleum-derived diesel (Almeida et al., 2021). Moreover, in this way, we can reduce pollution and greenhouse effects by using increased amounts of biodiesel. Many methods of biodiesel manufacturing, such as dilution, pyrolysis, micro-emulsion, and transesterification, are used. The substrates, such as animal fats (bovine tallow and lard), greases (trap grease and float grease), used cooking oils, and vegetable oils (rapeseed, cotton seed, palm, soybean, corn, jatropha, peanut, sunflower, and canola), are used for biodiesel production by transesterification. Transesterification can be either catalytic or non-catalytic, and hence, the production of biodiesel by using enzymes has become popular now due to its low energy consumption and mild reaction conditions (Vilar et al., 2018; Pérez et al., 2019; Carvalho et al., 2020). Transesterification reaction requires a small chain alcohol (methanol, ethanol, and propanol), a catalyst (acid, base, and catalyst), and either vegetable oil or animal fats (Almeida et al., 2021).
Enzymes, especially lipases, are used as a catalyst for biodiesel production in the transesterification process. Lipases have high catalytic activity, regioselectivity, and stereoselectivity in mild physical conditions such as pH, temperature, and pressure.
Several yeast and fungal lipases have catalyzed the production of biodiesel, including Aspergillus niger, Rhizomucor miehei, Ralstonia sp, Candida rugosa, Candida antarctica, Rhizopus oryzae, Thermomyces lanuginosa, and Magnusiomyces capitatus (Fan et al., 2012). Winayanuwattikun et al. (2011) immobilized the lipase of Candida rugosa B on seabeds EC-OD and used it as a biocatalyst for the synthesis of enzyme-catalyzed biodiesel. Similarly, Tian et al. (2021) reported that Rhizomucor miehei lipase (RML), with only a single chain of α/β type protein, is largely used in the preparation of biodiesel, due to its high catalytic activity and tolerance against methanol.
8.2. In detergent manufacturing
A detergent is either a sodium salt of long-chain benzene sulfonic acid or a sodium salt of long-chain alkyl hydrogen sulfate that possesses cleansing properties in water, as is the case with soaps. Both anionic groups (sulfate or sulfonic) and non-ionic groups (long-chain hydrocarbon) are present in a detergent; these groups make a detergent more soluble in water. Detergent is a water-soluble cleansing agent that combines dirt and impurities and makes them more soluble in water without forming scum with the salt of hard water. The use of detergents in washing is increasing the popularity of laundry and household detergents, because they are anti-static, dispersible in water, and give softness to fabric. Several brands of laundry and household detergents with different properties are now available in the market (Verma et al., 2012). Higher use of detergents increases the detergent load in the environment and becomes a pollutant. Moreover, these detergents also have a higher washing temperature. The use of hydrolytic fungal lipases as additives in the preparation of household and laundry detergents is commercially important, because lipases can decrease the detergent product load from the environment by saving energy by lowering the washing temperature of the detergent that is to be used (Mehta et al., 2017). Navo Nordisk, in 1994, first introduced the first commercial lipase. Lipolase was produced by the fungus Thermomyces lanuginosus and has been expressed in Aspergillus oryzae. Thermomyces sp. produces the most significant detergent by using the lipases that are primarily used in Lipolase and Novozymes (Gillis, 1988; Mehta et al., 2017). Furthermore, these lipases, including Humicola lanuginosa, Talaromyces thermophilus, Rhizopus oryzae, and candida sp., are also used in the preparation of detergents that promise better washing performance and energy savings (Nishioka et al., 1990; Salleh et al., 1993; Jaeger et al., 1994; Romdhane et al., 2010; Mahfoudhi et al., 2022). Wang et al. (2018) reported an alkaline, low-temperature, and strong surfactant tolerance recombinant lipase of Trichoderma lentiform ACCC30425 in Pichia pastoris GS115; this lipase is suitable for use in the detergent industry. Xiao et al. (2021) also reported a recombinant lipase Lip486 without clear homology alao to know lipase, Lip486 was discovered by using functional metagenomics technology and belongs to a new sub-familycalled lipolytic enzyme family II. The recombinant Lip486 expresses great activity and stability even in alkaline pH and medium–low temperature environments; these properties will make the lipase most important for use in detergent and other industries in the future.
8.3. In the leather industry
Leather industries are involved in the manufacturing of goods, such as bags, shoes, footwear, garments, belts, and purses, and have a considerable role in the world's economy and foreign exchange earnings and employment (Dixit et al., 2015; Khambhaty, 2020). However, these industries also harm the environment by releasing several toxic chemicals that are used during various steps of the tanning process. Leather industries have been established in a large number over the past few years, because leather and leather products have significant importance in the world's fashion industry. Leather production involves the conversion of raw animal skin and hides into leather by a series of different mechanical and chemical processes, such as soaking, dehairing, bating, degreasing, and post-tanning. Various chemicals are used in different processes of tanning that are released as waste, contributing to the significant increase in chemical oxygen demand (COD), total dissolved solids (TDSs), sulfates, chlorides, and heavy metals to the wastewater to be released into water bodies such as rivers, lakes, and the sea, thus polluting them enormously (Dayanandan et al., 2013; Dixit et al., 2015; Khambhaty, 2020). In order to reduce the application of these toxic chemicals, enzymes have been recognized as the best practical alternative to use during tanning, and these enzymes can help in waste management (Khambhaty, 2020). Lipases are an important group of enzymes in the various processes of leather production, because animal skin and hides contain protein and fat in collagen fibers. These skin and hides must be partially and totally removed before they are tanned. Lipases especially break down the lipids without damaging the leather and show the degreasing process with lower environmental impact (Choudhary et al., 2004; Dayanandan et al., 2013; Mehta et al., 2017).
Moreover, fungal-origin lipases are more efficient when compared to those originating from animals and plants as they are characterized by high yield, the rapid growth of fungi on cheaper media, and more stable, easy, and safer production methods. In a study, Aspergillus tamarii MTCC5152 was recognized for its high-level production of lipase (758 ± 3.61 u/g) through solid-state fermentation, the crude lipase (3%) used for tanning fleshing, and its 92% fat solubility (Dayanandan et al., 2013). Similarly, in another study, Moujehed et al. (2022) reported that Lip2 lipase is produced from the yeast Yarrowia lipolytica. It is effective in degreasing sheepskins, and Lip2 uses only 6 mg/kg of raw skin and is capable of decreasing even in 15 min at pH 8.0 and 30°C successfully.
8.4. In pulp and paper production
The pulp and paper industries are growing rapidly around the world and have a crucial role in the economic, social, and environmental development of any country. The increase in population and economic development will also increase the demand for paper. The paper-making process involves raw materials that can be categorized into three groups, namely, wood, non-wood, and recycled waste paper (Abd El-Sayed et al., 2020). In paper industries, some chemical compounds, such as hydrogen peroxidase, sodium carbonate, sodium hydroxide, sodium silicate, diethylenetriaminepenta-acetic acid, and surfactants, are used in high amounts in different steps of conventional methods; these chemicals are very toxic and become highly pollutant in the environment after releasing in wastewater (Yakubu et al., 2019). Several enzymes, such as lipase, amylase, esterase, cellulase, xylanase, hemicellulase, and pectinase, are used as a substitute for these chemicals to reduce toxic waste. Wood is the brassy reservoir of paper pulp and pitch that contains hydrophobic matter, such as triglycerides and waxes, and the presence of this matter creates a very difficult problem in the processing of paper pulp. These triglycerides and waxes might be present as holes or spots and sticky deposits in the finished product (Yakubu et al., 2019).
Therefore, the use of lipases can remove the pitch from the pulpy matter during the paper manufacturing process, and ~90% of present glycerides in the pitch get dissolved into diglycerides, monoglycerides, free fatty acids, and glycerol by lipases, which are less sticky and more hydrophilic (Jaeger and Reetz, 1998). Nippon paper industries in Japan introduced a method to sort out the pitch problem of wood by using the Candida rugosa lipase, which can reduce about 90% of triglyceride from the pitch. In Jujo's paper company, Hata and Coworkers in1990 noticed that lipases could reduce pitch problems by reducing the content of triglycerides in ground wood pulp. In another study, Candida cylindracea produced lipase that when added to the groundwood stock chest could reduce pitch and talc consumption substantially (de María et al., 2005; Mehta et al., 2017).
8.5. In the textile industry
Textile industries are the oldest industries that have existed for many centuries, and they contribute considerably to the economy of several developing countries. Textile industries also help to fulfill one of the introductory demands of humankind (Rahman et al., 2020; Kumar et al., 2021). These industries are facing remarkable environmental and resource challenges, because they are using an intensive amount of chemicals and are thus the most polluting industries. Some chemicals are carcinogenic and have an allergic effect that affects human health. These industries produce waste with chemicals, such as formaldehyde, chlorine, dyes, detergents, and heavy metals (lead and mercury), which cause complex environmental problems (Hooda, 2020; Kumar et al., 2021). To replace these chemicals and overcome these environmental problems, textile industries are now shifting from conventional methods to biological methods that involve the application of enzymes, as they are reliable, stable, safe, and biodegradable; thus, they are widely used in place of toxic chemicals, creating an eco-friendly environment, and they also help in improving the quality of manufacturing products (Kumar et al., 2021). The use of fungal lipases has become crucial in textile industries, and fungal lipases are also used to help in the removal of size lubricants to provide the fabric with better absorbency for raised levelness in dyeing. Commercial preparations also contain lipase enzymes that are used for the desizing of denim and other cotton fabric (Hasan et al., 2006; Mehta et al., 2017). In the textile industry, polyester is a synthetic fiber with certain distinct advantages, such as softness, high strength, strain, wash ability, stretch machine abrasion, and wrinkled resistance; when the polyester fiber is treated enzymatically by lipase, it assists in improving its ability to absorb the dyes and cationic chemicals that support tissue preservation. Lipase produced from Aspergillus oryzae is used to modify the polyethylene terephthalate (PET) fabric by increasing its polarity and anti-static ability (Mehta et al., 2017; Kumar and Kumar, 2020). In another study, El Menoufy et al. (2022) reported that the fungus Aspergillus niger NRRL-599 produced a lipase through solid-state fermentation and produced an immobilized lipase by using gelatin-coated titanium nanoparticles. The immobilized lipase is used in the treatment of wool after dyeing to improve color and strength.
8.6. In biosensor development
Quantitative determination of triglycerides, mainly fats and oils, in the food industry and clinical diagnosis are of high importance, and this function can be performed by the biosensor, which is a lipid sensing device that is cheaper and faster working compared to the chemical methods of determination of triglycerides (Mehta et al., 2017). A biosensor device based on the enzyme-catalyzed dissolution of biodegradable polymer films has been formed (Singh and Mukhopadhyay, 2012). Sumner et al. (2001) have investigated three polymerase-enzyme systems, namely, a dextran hydrogel, degraded by the enzyme dextranase; a poly (ester amide), degraded by the protein degrading α-chymotrypsin; and a poly (tri-methylene) succinate, degraded by a lipase enzyme. The basic concept behind the lipase biosensor is to quantify the glycerol released from triglycerides in an analytical sample by the use of enzymatic, colorimetric, and chemical methods (Hasan et al., 2006; Pérez et al., 2019). Moreover, fungus and yeast lipases along with glucose oxidase immobilized on pH/oxygen electrodes can be used as lipid biosensors and for the estimation of triglycerides and blood cholesterol (Imamura et al., 1989). In addition, fungus lipases as biosensors are also used to estimate the level of lipids in clinical diagnosis. For instance, Candida rugosa produced a lipase that has been developed as a DNA probe (Benjamin and Pandey, 2001). The lipase from Candida rugosa was immobilized on the aluminosilicate, which was used for the detection of organophosphate insecticides such as diazinon in a liquid medium (Zehani et al., 2014). In another study, the lipase of Candida rugosa acts as a biocatalyst for the hydrolysis of triglycerides into glycerol and fatty acid and is used as a biosensor for the detection of esters (β-hydro acid) and triglycerides in blood serum (Califano et al., 2014). Lipase, when immobilized at the Nafion membrane onto a graphite–epoxy transducer, can be used for the quantitative determination of glycerides in food samples (Escamilla-Mejía et al., 2015).
8.7. In cosmetics and personal care products production
Lipases have played a significant role in the cosmetic sector and personal care products such as softening, cleaning, aroma, and coloring. This sector has a considerable market value after the food and pharma sectors (Marion and Oliver, 2013; Mehta et al., 2017). Lipases have high potential uses in perfumeries and cosmetics, as they show activities in surfactants and aroma production (Mehta et al., 2017). The esterification of glycerol produces mono- and diglycerol that are used as surfactants in the cosmetics and perfume industries. Lipases catalyze the transesterification of 3,7-dimethyl-4,7-octadien-1-ol, which produces rose oxide, and it is a very important fragrance ingredient in the perfume industry (Izumi et al., 1997). Moreover, Mouad et al. (2016) have reported that immobilized Rhizomucor miehei lipase is used as a biocatalyst in making personal care products such as skin creams and bath oils. In addition, Candida antarctica B yields a lipase, which synthesizes amphiphilic compounds that attain great attention in the cosmetic industry because they have a range of beneficial characteristics for the skin (Mouad et al., 2016).
Unichem International (Spain) has produced isopropyl palmitate, isopropyl myristate, and 2-ethylhexyl palmitate for use as an emollient in personal care products such as skin and sun tan creams and bath oils, and the company also claims that the use of lipases as a substitute for conventional acid as a catalyst can improve the quality of the product with minimum downstream refining (Hasan et al., 2006). Lipases are used in the production of hair waving and also as an ingredient of topical anti-obese creams or in oral administration (Mehta et al., 2017). Vitamin A (retinol) and its derivatives have been used in large amounts in pharmaceuticals and cosmetics, such as skin care products, as water-soluble derivatives of retinol are prepared with the catalytic reaction of the immobilized lipase (Maugard et al., 2002; Mahfoudhi et al., 2022). Nippon Oil and Fats Co. Ltd. was granted a patent for the formulation of propylene glycerol mono-fatty acid ester in the presence of lipase enzyme; this ester is used as an emulsifier and pearling agent in the cosmetic and food industries (Kim et al., 1997). In other research, Khan and Rathod (2020) synthesized n-butyl palmitate (a significant valuable ingredient of cosmetics) from Candida antarctica B lipase after covalent immobilization on hydrophobic and macroporous polyacrylate beads in a solvent-free medium.
8.8. In medical and pharmaceutical applications
The application of lipases as diagnostic tools is growing rapidly, as their increased levels show the indication of several diseases and also act as important drug targets or marker enzymes in the medical field. The levels of lipase in the blood act as a diagnostic tool for the detection of some pathological concerns such as pancreatic injury and acute pancreatitis (Mahfoudhi et al., 2022). Lipase is the preliminary enzyme for fat metabolism, and its deficiency causes dangerous health problems. Lipase acts as an activator of tumor necrosis factor and assists in the treatment of malignant tumors, and the determination of the lipase level is also significant in the diagnosis of heart ailments (Singh and Mukhopadhyay, 2012). Moreover, lipases isolated from bacteria, fungi, yeast, and some protozoa have been used in the medical and pharmaceutical fields. Candida rugosa produces a lipase that is immobilized on nylon supports in the presence of organic solvents; the immobilized lipase is used in synthesizing lovastatin, a widely used drug in the treatment of serum cholesterol reduction (Yang et al., 1997; Mahfoudhi et al., 2022). Similarly, Sharma and Kanwar (2014) reported that the fungus Serratia marcescens produces a lipase that can yield a chiral 3-phenyl glycosidic acid by enantioselective hydrolysis, and this intermediate compound is used in the synthesis of diltiazem hydrochloride, a drug used as a coronary vasodilator in many countries (Pérez et al., 2019). Similarly, lipase can emulsify fats together with proteases and could be used in the treatment of digestive tract diseases (Hasan et al., 2006). In another study, it has been noticed that lipases isolated from yeast and fungi can be used as a therapeutic agent for the treatment of gastrointestinal disturbances, dyspepsia, and cutaneous manifestations of digestive allergies (Mehta et al., 2017).
8.9. In the synthesis of biodegradable biopolymer and esters
Biopolymers, such as polysaccharides, polyphenols, and polyesters, indicate a great degree of diversity and complexity, and these polymers have attained the focus of many researchers, as they are synthesized from natural renewable resources and are also biodegradable (Pérez et al., 2019). Interestingly, fungal lipases are exploited as biocatalysts in the production of biodegradable compounds. The esterification of oleic acid and butanol results in the synthesis of 1-butyl oleate, which reduces the viscosity of biodiesel during its use in the winter (Hasan et al., 2006). Similarly, a mixture of 2-ethyl-1-hexyl esters is achieved in an appropriate amount by enzymatic transesterification of rapeseed oil fatty acids and functions as a solvent (Singh and Mukhopadhyay, 2012).
In addition, lipases not only act as biocatalysts in biodegradable biopolymer synthesis but also in ester synthesis in transesterification reactions in organic solvents. The esters of short-chain fatty acids are used as flavoring agents in the food industry (Mehta et al., 2017). The Candida rugosa lipase catalyzes the esterification reaction between fatty acids with sulcatol in toluene (Janssen et al., 1999). In a study, after physical adsorption of the lipases of Mucor javanicus (MJL), Candida rugosa (CRL), and Candida sp. (CALA) on a Diaion HP-20 (hydrophobic and mesoporous support), these lipases were used as biocatalysts in the synthesis of the aromatic esters geranyl butyrate, hexyl butyrate, and propyl butyrate, respectively (Dos Santos et al., 2021). The esterification reaction between lauryl alcohol and palmitic acid has also used the lipase from Candida antarctica (Novozym 435) as a catalyst, and thus it is capable of producing more than 90% of lauryl palmitate under optimized conditions (Syamsul et al., 2010). Moreover, the lipase isolated from the Aspergillus ibericus has applications as a catalyst in esterification reactions and aroma ester production (Oliveira et al., 2016).
In another study, Mehta et al. (2020) reported that the esterification of ethanol and acetic acid produces ethyl acetate, while the esterification of ethanol and lactic acid gives ethyl lactate; both reactions are catalyzed by the lipase of Aspergillus fumigatus.
8.10. In ester biodegradation
The application of lipases in the biodegradation of esters is a new developing approach in biotechnology. Lipases can biodegrade aliphatic and aromatic esters through their catalytic actions. Lipases also act as catalysts for transesterification reactions in an organic solvent, through which they catalyze the biodegradation of polyesters (Pérez et al., 2019). The lipase-mediated degradation of a polymer is a robust substitute approach to conventional methods because of its biocompatibility and mild conditions. Lipase produced from Penicillium fellutanum can degrade polyester Nylon-200 by ~80% by weight in 7 days (Amin et al., 2021). Another study was carried out by da Costa et al. (2020) in which they observed that Yarrowia lipolytica produces a lipase that can degrade aromatic ester polyethylene terephthalate (PET).
8.11. In the bioremediation of wastewater
The lipase-mediated bioremediation of wastewater is a novel, robust, and extensively used waste management technique in lipase biotechnology. Lipases have applications in activated sludge and other aerobic waste processes where thin layers of fats are continuously removed from the surface of aerated tanks to maintain the oxygen supply; this skimmed fast rich liquid is digested by the lipases (Cruz et al., 2020). Fungal sp. can be applied for degrading oil spills in the coastal environment, which may enhance ecorestoration and also help in the enzymatic processing of oil in industries (Gopinath et al., 1998). Species that belong to the genera Aspergillus, Fusarium, Cladosporium, Trichoderma, Mortierella, Penicillium, Beauveria, and Engyodontium are examples of fungi that have recently been designated as potential bioremediation agents in soil (Islam and Datta, 2015). The lipase of Candida rugosa has applications as an anaerobic digester (Singh and Mukhopadhyay, 2012). Lipases are involved in the effective breakdown of solids and the removal and prevention of fat blockages or filming in the waste system in several industrial processes, such as the treatment of sewage, the cleaning of holding tanks, septic tanks, and grease traps, and P (Singh and Mukhopadhyay, 2012). Aspergillus terreus and Aspergillus niger produce lipases that are used in the bioremediation of polluted soil and the degradation of polyvinyl alcohol films, respectively (Mahmoud et al., 2015; Mehta et al., 2017). Lipases of Aspergillus uvarum and Aspergillus ibericus also have applications in bioremediation processes (Salgado et al., 2016). Leal et al. (2006) noticed that synthetic mobile dairy wastewater with or without pre-hydrolysis, is treated anaerobically by solid peprations of Penicillium restrictum and Aspergillus niger, both of which were isolated from oil-polluted soil and tested for lipase production. The obtained lipase was found to biodegrade polyaromatic hydrocarbons present in petroleum-contaminated soil (Mauti et al., 2016). Basheer et al. (2011) isolated a fungus Aspergillus awamori BTMFW032 from seawater, which was able to produce lipase and could reduce ~92% of the fat and oil contents from the mill effluent containing waste oil. A lipase isolated from Geotrichum candidum has applications in bioremediation and controlling the decolorization of olive mill wastewater (Kumar et al., 2020).
9. Application of CRISPR/Cas genome editing technology to modify lipase encoding genes
Genome editing is a form of genetic engineering in which DNA is purposefully added, subtracted, or altered in living cells. The acronym CRISPR (clustered regularly interspaced short palindromic repeat) refers to the special arrangement of brief, partially repeated DNA sequences present in the prokaryotic genomes. Prokaryotes use CRISPR and its associated protein (Cas9) as a form of adaptive immunity to protect themselves against viruses or bacteriophages (Hille and Charpentier, 2016; Asmamaw and Zawdie, 2021).
The three fundamental steps of the CRISPR defensive system, namely, adaptation (spacer acquisition), crRNA synthesis (expression), and target interference, protect bacteria from recurrent viral infections. CRISPR loci are a collection of brief repetitive sequences that are present in prokaryotic chromosomal or plasmid DNA. The Cas gene, which produces the nuclease protein (Cas protein) necessary to break or destroy viral nucleic acid, is typically found adjacent to the CRISPR gene (Rath et al., 2015).
Nearly, 50% of bacteria, about all the archaea, and even some bacteriophages hold the genes essential for CRISPR-Cas adaptive immunity, which supplies a memory of previous infections through encoding short sequences of DNA at clustered regularly interspaced short palindromic repeat (CRISPR) loci within their genome. These previous infections are stored as spacer sequences, each of which is flanked by repeat sequences. Firstly, these spacer sequence repeats are transcribed into pre-crRNA and then processed into functional crRNAs (Jiang and Doudna, 2015; McCarty et al., 2020). Inherently, the native CRISPR-Cas system is multiplexed; an organism can encode one or several CRISPR arrays and also express many Cas (CRISPR-associated) proteins that help the acquisition of new spacers and further processing of the CRISPR arrays (McCarty et al., 2020). By using Cas proteins and a library of CRISPR RNAs (crRNAs) from CRISPR loci, the CRISPR-Cas system can detect and silence foreign nucleic acids through sequencing and RNA editing. The invasive (non-self) DNA is targeted by CRISPR-Cas via base pairing with the crRNA guide sequence, leading to Cas protein-mediated DNA cleavage (Barrangou et al., 2007; Garneau et al., 2010).
Fungal research has recently been revolutionized by CRISPR/Cas9 genome-editing technology. The first CRISPR/Cas9 genome editing system was introduced to Saccharomyces cerevisiae by DiCarlo et al. (2013). Consequently, Liu et al. (2015) applied the CRISPR/Cas9 genome-editing system to Trichoderma reesei; Matsu-Ura et al. (2015) and Nodvig et al. (2015) applied this system to the model fungi N. crassa and A. nidulans, respectively. Since then, the CRISPR/Cas9 genome-editing system has found widespread applications in the genetic alteration of many filamentous fungi, including numerous vital genera such as Neurospora, Trichoderma, Penicillium, and Aspergillus (Jin et al., 2022). This genome-editing strategy and its application have been well-established, particularly in Aspergilli.
Liu et al. (2017) successfully engineered the genome (gene level) of Myceliophthora thermophila and Myceliophthora heterothallic with the CRISPR/Cas9 technique and found that engineered fungal sp. is able to produce significantly increased (5- to 13-fold) cellulase. Similarly, in this way, we can engineer the genomes (by adding high-potential lipase genes) of lipase-producing fungi with the CRISPR/Cas9 system to increase the lipase production ability of these fungi. Li et al. (2023) modified the genome of Aspergillus oryzae with CRISPR/Cas9 technology, and the heterologous lipase gene (TLL) achieved the precise integration of different genetic loci in one step.
10. Conclusion and future prospects
Lipases have been found and characterized in a wide range of fungi and yeasts and act as biocatalysts in the hydrolysis of oils and fats (triglycerides) into fatty acids and glycerol. Lipases are also recognized as having applications in different industries and sectors such as bioethanol production, pulp and paper production, biosensor development, the production of cosmetics and personal care products, food processing, the textile industry, the medical and pharmaceutical sector, bioremediation of wastewater, biodegradable biopolymer synthesis, ester synthesis, and ester biodegradation. Lipase immobilization could improve the thermal or ionic stability, easy recycling, and volume-specific loading of enzymes onto the support, all of which lead to better activity of lipases. Advanced techniques such as genetic engineering also enhance the increased production of lipases in fungi.
Therefore, encouraging the isolation of new fungal species with a high yield of lipase is needed in order to meet the industrial and domestic requirements of lipases in the near future. New optimization techniques to induce high levels of fungal lipase production with better quality must also be sought after. To improve the immobilization techniques for better performance, lipases as diagnostic tools in the medical sector should be further investigated. The application of fungal lipases in bioremediation should also be studied more and can be carried out on a large scale. Moreover, advanced techniques, such as genetic engineering and molecular techniques, should be used profusely to improve production and quality. Genome editing in fungal strains through the CRISPR/Cas9 technique may be used for improving the production and application of lipases in various fields.
Author contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abd El-Sayed, E. S., El-Sakhawy, M., and El-Sakhawy, M. A. M. (2020). Non-wood fibers as raw material for pulp and paper industry. Nord. Pulp Pap. Res. J. 35, 215–230. doi: 10.1515/npprj-2019-0064
Aghaei, H., Yasinian, A., and Taghizadeh, A. (2021). Covalent immobilization of lipase from Candida rugosa on epoxy-activated cloisite 30B as a new heterofunctional carrier and its application in the synthesis of banana flavor and production of biodiesel. Int. J. Biol. Macromol. 178, 569–579. doi: 10.1016/j.ijbiomac.2021.02.146
Ahmad, R., and Sardar, M. (2015). Enzyme immobilization: An overview on nanoparticles as immobilization matrix. Biochem. Analyt. Biochem. 4, 1. doi: 10.4172/2161-1009.1000178
Alabdalall, A. H., ALanazi, N. A., Aldakeel, S. A., AbdulAzeez, S., and Borgio, J. F. (2020). Molecular, physiological, and biochemical characterization of extracellular lipase production by Aspergillus niger using submerged fermentation. PeerJ. 8, e9425. doi: 10.7717/peerj.9425
Alami, N. H., Nasihah, L., Umar, R. L. A., Kuswytasari, N. D., Zulaika, E., and Shovitri, M. (2017). “Lipase production in lipolytic yeast from Wonorejo mangrove area,” in AIP Conference Proceedings, Vol. 1854 (AIP Publishing LLC). doi: 10.1063/1.4985392
Almeida, F. L., Travalia, B. M., Goncalves, I. S., and Forte, M. B. S. (2021). Biodiesel production by lipase-catalyzed reactions: Bibliometric analysis and study of trends. Biofuel Bioprod. Biorefin. 15, 1141–1159. doi: 10.1002/bbb.2183
Al-shawawreh, M. F. M. (2021). Isolation of endogenous fungal isolate producing extracellular lipase with potential in olive oil hydrolysis. Rev. Int. Geogr. Educ. Online. 11, 1–9.
Amin, M., Bhatti, H. N., Sadaf, S., and Bilal, M. (2021). Optimization of lipase production by response surface methodology and its application for efficient biodegradation of polyester vylon-200. Catal. Lett. 21, 1–14. doi: 10.1007/s10562-021-03603-x
Anobom, C. D., Pinheiro, A. S., De-Andrade, R. A., Aguieiras, E. C., Andrade, G. C., Moura, M. V., et al. (2014). From structure to catalysis: Recent developments in the biotechnological applications of lipases. Biomed Res. Int. 2014, 684506. doi: 10.1155/2014/684506
Araujo, S. C., Ramos, M. R. M. F., do Espírito Santo, E. L., de Menezes, L. H. S., de Carvalho, M. S., Tavares, I. M. D. C., et al. (2022). Optimization of lipase production by Penicillium roqueforti ATCC 10110 through solid-state fermentation using agro-industrial residue based on a univariate analysis. Prep. Biochem. Biotechnol. 52, 325–330. doi: 10.1080/10826068.2021.1944203
Arpigny, J. L., and Jaeger, K. E. (1999). Bacterial lipolytic enzymes: Classification and properties. Biochem. J. 343, 177–183. doi: 10.1042/bj3430177
Asmamaw, M., and Zawdie, B. (2021). Mechanism and applications of CRISPR/Cas-9-mediated genome editing. Biol.: Targets Ther. 15, 353–361. doi: 10.2147/BTT.S326422
Asmat, S., Anwer, A. H., and Husain, Q. (2019). Immobilization of lipase onto novel constructed polydopamine grafted multiwalled carbon nanotube impregnated with magnetic cobalt and its application in synthesis of fruit flavours. Int. J. Biol. Macromol. 140, 484–495. doi: 10.1016/j.ijbiomac.2019.08.086
Atsumi, S., and Liao, J. C. (2008). Metabolic engineering for advanced biofuels production from Escherichia coli. Curr. Opin. Biotechnol. 19, 414–419. doi: 10.1016/j.copbio.2008.08.008
Avhad, M. R., and Marchetti, J. M. (2019). “Uses of enzymes for biodiesel production,” in Advanced Bioprocessing for Alternative Fuels, Biobased Chemicals, and Bioproducts (Sawston: Woodhead Publishing), 135–152. doi: 10.1016/B978-0-12-817941-3.00007-3
Bancerz, R., Ginalska, G., Fiedurek, J., and Gromada, A. (2005). Cultivation conditions and properties of extracellular crude lipase from the psychrotrophic fungus Penicillium chrysogenum 9′. J. Ind. Microbiol. 32, 253–260. doi: 10.1007/s10295-005-0235-0
Barrangou, R., Fremaux, C., Deveau, H., Richards, M., Boyaval, P., Moineau, S., et al. (2007). CRISPR provides acquired resistance against viruses in prokaryotes. Science. 315, 1709–1712. doi: 10.1126/science.1138140
Basheer, S. M., Chellappan, S., Beena, P. S., Sukumaran, R. K., Elyas, K. K., and Chandrasekaran, M. (2011). Lipase from marine Aspergillus awamori BTMFW032: Production, partial purification and application in oil effluent treatment. New Biotechnol. 28, 627–638. doi: 10.1016/j.nbt.2011.04.007
Bayramoglu, G., Celikbicak, O., Kilic, M., and Arica, M. Y. (2022). Immobilization of Candida rugosa lipase on magnetic chitosan beads and application in flavor esters synthesis. Food Chem. 366, 130699. doi: 10.1016/j.foodchem.2021.130699
Benjamin, S., and Pandey, A. (2001). Isolation and characterization of three distinct forms of lipases from Candida rugosa produced in solid state fermentation. Braz. Arch. Biol. Technol. 44, 213–221. doi: 10.1590/S1516-89132001000200016
Bharathi, D., and Rajalakshmi, G. (2019). Microbial lipases: An overview of screening, production and purification. Biocatal. Agric. Biotechnol. 22, 101368. doi: 10.1016/j.bcab.2019.101368
Bilal, M., Hussain, N., Américo-Pinheiro, J. H. P., Almulaiky, Y. Q., and Iqbal, H. M. (2021). Multi-enzyme co-immobilized nano-assemblies: Bringing enzymes together for expanding bio-catalysis scope to meet biotechnological challenges. Int. J. Biol. Macromol. 186, 735–749. doi: 10.1016/j.ijbiomac.2021.07.064
Bindiya, P., and Ramana, T. (2013). Production of lipase by repeated batch fermentation using immobilized Aspergillus sydowii. Int. J. Chem. Biochem. Sci. 4, 72–78.
Borrelli, G. M., and Trono, D. (2015). Recombinant lipases and phospholipases and their use as biocatalysts for industrial applications. Int. J. Mol. Sci. 16, 20774–20840. doi: 10.3390/ijms160920774
Califano, V., Bloisi, F., Aronne, A., Federici, S., Nasti, L., Depero, L. E., et al. (2014). Biosensor applications of MAPLE deposited lipase. Biosensors 4, 329–339. doi: 10.3390/bios4040329
Carvalho, R. S., Cruz, I. A., Américo-Pinheiro, J. H. P., Soriano, R. N., de Souza, R. L., Bilal, M., et al. (2020). Interaction between Saccharomyces cerevisiae and Lactobacillus fermentum during co-culture fermentation. Biocatal. Agric. Biotechnol. 29, 101756. doi: 10.1016/j.bcab.2020.101756
Chandra, P., Singh, R., and Arora, P. K. (2020). Microbial lipases and their industrial applications: A comprehensive review. Microb. Cell Factories 19, 1–42. doi: 10.1186/s12934-020-01428-8
Chang, S. W., Lee, G. C., and Shaw, J. F. (2006). Efficient production of active recombinant Candida rugosa LIP3 lipase in Pichia pastoris and biochemical characterization of the purified enzyme. J. Agric. Food Chem. 54, 5831–5838. doi: 10.1021/jf060835e
Choudhary, R. B., Jana, A. K., and Jha, M. K. (2004). Enzyme technology applications in leather processing. Ind. J. Chem. Technol. 11, 659–671.
Colla, L. M., Primaz, A. L., Benedetti, S., Loss, R. A., Lima, M. D., Reinehr, C. O., et al. (2016). Surface response methodology for the optimization of lipase production under submerged fermentation by filamentous fungi. Braz. J. Microbiol. 47, 461–467. doi: 10.1016/j.bjm.2016.01.028
Colonia, B. S. O., Woiciechowski, A. L., Malanski, R., Letti, L. A. J., and Soccol, C. R. (2019). Pulp improvement of oil palm empty fruit bunches associated to solid-state biopulping and biobleaching with xylanase and lignin peroxidase cocktail produced by Aspergillus sp. LPB-5. Bioresour. Technol. 285, 121361. doi: 10.1016/j.biortech.2019.121361
Comménil, P., Belingheri, L., Sancholle, M., and Dehorter, B. (1995). Purification and properties of an extracellular lipase from the fungus Botrytis cinerea. Lipids 30, 351–356. doi: 10.1007/BF02536044
Cruz, Y. W., Vieira, Y. A., Vilar, D. S., Torres, N. H., Aguiar, M. M., Cavalcanti, E. B., et al. (2020). Pulp wash: a new source for production of ligninolytic enzymes and biomass and its toxicological evaluation after biological treatment. Environ. Technol. 41, 1837–1847. doi: 10.1080/09593330.2018.1551428
da Costa, A. M., de Oliveira Lopes, V. R., Vidal, L., Nicaud, J. M., de Castro, A. M., and Coelho, M. A. Z. (2020). Poly (ethylene terephthalate) (PET) degradation by Yarrowia lipolytica: Investigations on cell growth, enzyme production and monomers consumption. Process Biochem. 95, 81–90. doi: 10.1016/j.procbio.2020.04.001
Dalmau, E. J. L. M., Montesinos, J. L., Lotti, M., and Casas, C. (2000). Effect of different carbon sources on lipase production by Candida rugosa. Enzyme Microb. Technol. 26, 657–663. doi: 10.1016/S0141-0229(00)00156-3
Das, A., Bhattacharya, S., Shivakumar, S., Shakya, S., and Sogane, S. S. (2017). Coconut oil induced production of a surfactant-compatible lipase from Aspergillus tamarii under submerged fermentation. J. Basic Microbiol. 57, 114–120. doi: 10.1002/jobm.201600478
Dayanandan, A., Rani, S., Shanmugavel, M., Gnanamani, A., and Rajakumar, G. S. (2013). Enhanced production of Aspergillus tamarii lipase for recovery of fat from tannery fleshings. Braz. J. Microbiol. 44, 1089–1095. doi: 10.1590/S1517-83822013000400010
de Azeredo, L. A., Gomes, P. M., Sant'Anna, G. L., Castilho, L. R., and Freire, D. M. (2007). Production and regulation of lipase activity from Penicillium restrictum in submerged and solid-state fermentations. Curr. Microbiol. 54, 361–365. doi: 10.1007/s00284-006-0425-7
de María, P. D., Carboni-Oerlemans, C., Tuin, B., Bargeman, G., and van Gemert, R. (2005). Biotechnological applications of Candida antarctica lipase A: State-of-the-art. J. Mol. Catal., 37, 36–46. doi: 10.1016/j.molcatb.2005.09.001
de Souza, C. E. C., Ribeiro, B. D., and Coelho, M. A. Z. (2019). Characterization and application of Yarrowia lipolytica lipase obtained by solid-state fermentation in the synthesis of different esters used in the food industry. Appl. Biochem. Biotechnol. 189, 933–959. doi: 10.1007/s12010-019-03047-5
Demera, L. L., Barahona, P. P., and Barriga, E. J. C. (2019). Production, extraction and characterization of lipases from the antarctic yeast Guehomyces pullulans. Am. J. Biochem. Biotechnol. 15, 75–82. doi: 10.3844/ajbbsp.2019.75.82
Diaz, J. M., Rodríguez, J. A., Roussos, S., Cordova, J., Abousalham, A., Carriere, F., et al. (2006). Lipase from the thermotolerant fungus Rhizopus homothallicus is more thermostable when produced using solid state fermentation than liquid fermentation procedures. Enzyme Microb. Technol. 39, 1042–1050. doi: 10.1016/j.enzmictec.2006.02.005
DiCarlo, J. E., Norville, J. E., Mali, P., Rios, X., Aach, J., and Church, G. M. (2013). Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 41, 4336–4343. doi: 10.1093/nar/gkt135
Dixit, S., Yadav, A., Dwivedi, P. D., and Das, M. (2015). Toxic hazards of leather industry and technologies to combat threat: A review. J. Clean. Prod. 87, 39–49. doi: 10.1016/j.jclepro.2014.10.017
Dominguez, A., Costas, M., Longo, M. A., and Sanromán, A. (2003). A novel application of solid state culture: Production of lipases by Yarrowia lipolytica. Biotechnol. Lett. 25, 1225–1229. doi: 10.1023/A:1025068205961
Dos Santos, M. M. O., Gama, R. S., de Carvalho Tavares, I. M., Santos, P. H., Gonçalves, M. S., de Carvalho, M. S., et al. (2021). Application of lipase immobilized on a hydrophobic support for the synthesis of aromatic esters. Biotechnol. Appl. Biochem. 68, 538–546. doi: 10.1002/bab.1959
El Menoufy, H. A., Gomaa, S. K., Haroun, A. A., Farag, A. N., Shafei, M. S., Shetaia, Y. M., et al. (2022). Comparative studies of free and immobilized partially purified lipase from Aspergillus niger NRRL-599 produced from solid-state fermentation using gelatin-coated titanium nanoparticles and its application in textile industry. Egypt. Pharm. J. 21, 143. doi: 10.4103/epj.epj_90_21
Escamilla-Mejía, J. C., Rodríguez, J. A., Álvarez-Romero, G. A., and Galán-Vidal, C. A. (2015). Monoenzymatic lipase potentiometric biosensor for the food analysis based on a pH sensitive graphite-epoxy composite as transducer. J. Mex. Chem. Soc. 59, 19–23.
Essamri, M., Deyris, V., and Comeau, L. (1998). Optimization of lipase production by Rhizopus oryzae and study on the stability of lipase activity in organic solvents. J. Biotechnol., 60, 97–103. doi: 10.1016/S0168-1656(97)00193-4
Ezema, B. O., Omeje, K. O., Bill, R. M., Goddard, A. D. O., Eze, S. O., and Fernandez-Castane, A. (2022). Bioinformatic characterization of a triacylglycerol lipase produced by Aspergillus flavus isolated from the decaying seed of Cucumeropsis mannii. J. Biomol. Struct. Dyn. 2022, 1–15. doi: 10.1080/07391102.2022.2035821
Fan, M., Zhang, P., and Ma, Q. (2012). Enhancement of biodiesel synthesis from soybean oil by potassium fluoride modification of a calcium magnesium oxides catalyst. Bioresour. Technol. 104, 447–450. doi: 10.1016/j.biortech.2011.11.082
Fatima, S., Faryad, A., Ataa, A., Joyia, F. A., and Parvaiz, A. (2021). Microbial lipase production: A deep insight into the recent advances of lipase production and purification techniques. Biotechnol. Appl. Biochem. 68, 445–458. doi: 10.1002/bab.2019
Fischer, M., and Pleiss, J. (2003). The Lipase Engineering Database: A navigation and analysis tool for protein families. Nucleic Acids Res. 31, 319–321. doi: 10.1093/nar/gkg015
Fu, J., Leiros, H. K. S., de Pascale, D., Johnson, K. A., Blencke, H. M., and Landfald, B. (2013). Functional and structural studies of a novel cold-adapted esterase from an Arctic intertidal metagenomic library. Appl. Microbiol. Biotechnol. 97, 3965–3978. doi: 10.1007/s00253-012-4276-9
Garneau, J. E., Dupuis, M. É., Villion, M., Romero, D. A., Barrangou, R., Boyaval, P., et al. (2010). The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468, 67–71. doi: 10.1038/nature09523
Geoffry, K., and Achur, R. N. (2018). Screening and production of lipase from fungal organisms. Biocatal. Agric. Biotechnol. 14, 241–253. doi: 10.1016/j.bcab.2018.03.009
Gillis, A. (1988). Research discovers new roles for lipases. J. Am. Oil Chem. Soc. 65, 846–852. doi: 10.1007/BF02544495
Gopinath, S., Hilda, A., and Ramesh, V. M. (1998). Detection of biodegradability of oils and related substances. J. Environ. Biol. 19, 157–165.
Gricajeva, A., Kazlauskas, S., Kalediene, L., and Bendikiene, V. (2018). Analysis of Aspergillus sp. lipase immobilization for the application in organic synthesis. Int. J. Biol. Macromol. 108, 1165–1175. doi: 10.1016/j.ijbiomac.2017.11.010
Gupta, R., Kumari, A., Syal, P., and Singh, Y. (2015). Molecular and functional diversity of yeast and fungal lipases: Their role in biotechnology and cellular physiology. Prog. Lipid. Res. 57, 40–54. doi: 10.1016/j.plipres.2014.12.001
Gutarra, M. L., Godoy, M. G., Maugeri, F., Rodrigues, M. I., Freire, D. M., and Castilho, L. R. (2009). Production of an acidic and thermostable lipase of the mesophilic fungus Penicillium simplicissimum by solid-state fermentation. Bioresour. Technol. 100, 5249–5254. doi: 10.1016/j.biortech.2008.08.050
Hasan, F., Shah, A. A., and Hameed, A. (2006). Industrial applications of microbial lipases. Enzyme Microb. Technol. 39, 235–251. doi: 10.1016/j.enzmictec.2005.10.016
Helal, S. E., Abdelhady, H. M., Abou-Taleb, K. A., Hassan, M. G., and Amer, M. M. (2021). Lipase from Rhizopus oryzae R1: in-depth characterization, immobilization, and evaluation in biodiesel production. J. Genet. Eng. Biotechnol. 19, 1–13. doi: 10.1186/s43141-020-00094-y
Hermansyah, H., Andikoputro, M. I., and Alatas, A. (2019). “Production of lipase enzyme from Rhizopus oryzae by solid state fermentation and submerged fermentation using wheat bran as substrate,” in AIP Conference Proceedings, Vol. 2085 (AIP Publishing LLC). doi: 10.1063/1.5094991
Hille, F., and Charpentier, E. (2016). CRISPR-Cas: biology, mechanisms and relevance. Philos. Trans. R. Soc. Lond., B, Biol. Sci. 371, 20150496. doi: 10.1098/rstb.2015.0496
Hooda, S. (2020). Bio-processing and herbal treatment on textile: A route to sustainability. Int. J. Home Sci. 6, 285–288.
Hosseinpour, M. N., Najafpour, G. D., Younesi, H., Khorrami, M., and Vaseghi, Z. (2012). Lipase production in solid state fermentation using Aspergillus niger: Response Surf. Methodol. 25, 3b. doi: 10.5829/idosi.ije.2012.25.03b.01
Hwang, H. T., Qi, F., Yuan, C., Zhao, X., Ramkrishna, D., Liu, D., et al. (2014). Lipase-catalyzed process for biodiesel production: Protein engineering and lipase production. Biotechnol. Bioeng. 111, 639–653. doi: 10.1002/bit.25162
Imamura, S., Takahashi, M., Misaki, H., and Matsuura, K. (1989). Method and reagent containing lipases for enzymatic determination of triglycerides. West Germany Patent 3, 912, 226.
Islam, R., and Datta, B. (2015). Fungal diversity and its potential in environmental cleanup. Int. J. Res. 2, 815–825.
Izumi, T., Tamura, F., Akutsu, M., Katou, R., and Murakami, S. (1997). Enzymatic transesterification of 3, 7-dimethyl-4, 7-octadien-1-ol using lipases. J. Chem. Technol. Biotechnol. 68, 57–64. doi: 10.1002/(SICI)1097-4660(199701)68:1<57::AID-JCTB607>3.0.CO;2-A
Jaeger, K. E., Ransac, S., Dijkstra, B. W., Colson, C., van Heuvel, M., and Misset, O. (1994). Bacterial lipases. FEMS Microbiol. Rev. 15, 29–63. doi: 10.1111/j.1574-6976.1994.tb00121.x
Jaeger, K. E., and Reetz, M. T. (1998). Microbial lipases form versatile tools for biotechnology. Trends Biotechnol. 16, 396–403. doi: 10.1016/S0167-7799(98)01195-0
Jamilu, H., Ibrahim, A. H., and Abdullahi, S. Z. (2022). Isolation, optimization and characterization of lipase producing bacteria from abbatoir soil. Int. J. Adv. Sci. Technol. 3, 2708–7972 doi: 10.51542/ijscia.v3i1.9
Janssen, A. E., Sjursnes, B. J., Vakurov, A. V., and Halling, P. J. (1999). Kinetics of lipase-catalyzed esterification in organic media: Correct model and solvent effects on parameters. Enzyme Microb. Technol. 24, 463–470. doi: 10.1016/S0141-0229(98)00134-3
Jayaprakash, A., and Ebenezer, P. (2012). Purification and characterization of Aspergillus japonicus lipase from a pig fat production medium. J. Acad. Ind. Res. 1, 1–7.
Jiang, F., and Doudna, J. A. (2015). The structural biology of CRISPR-Cas systems. Curr. Opin. Struct. Biol. 30, 100–111. doi: 10.1016/j.sbi.2015.02.002
Jin, F. J., Wang, B. T., Wang, Z. D., Jin, L., and Han, P. (2022). CRISPR/Cas9-based genome editing and its application in Aspergillus species. J. Fungi. 8, 467. doi: 10.3390/jof8050467
Joshi, R., Sharma, R., and Kuila, A. (2019). Lipase production from Fusarium incarnatum KU377454 and its immobilization using Fe3O4 NPs for application in waste cooking oil degradation. Bioresour. Technol. Rep. 5, 134–140. doi: 10.1016/j.biteb.2019.01.005
Kanmani, P., Karthik, S., Aravind, J., and Kumaresan, K. (2013). The use of response surface methodology as a statistical tool for media optimization in lipase production from the dairy effluent isolate Fusarium solani. Int. Sch. Res. Notices 2013, 528708. doi: 10.5402/2013/528708
Kashmiri, M. A., Adnan, A., and Butt, B. W. (2006). Production, purification and partial characterization of lipase from Trichoderma viride. Afr. J. Biotechnol. 5.
Katiyar, M., and Ali, A. (2013). Effect of metal ions on the hydrolytic and transesterification activities of Candida rugosa lipase. J. Oleo Sci. 62, 919–924. doi: 10.5650/jos.62.919
Kaur, G., Singh, A., Sharma, R., Sharma, V., Verma, S., and Sharma, P. K. (2016). Cloning, expression, purification and characterization of lipase from Bacillus licheniformis, isolated from hot spring of Himachal Pradesh, India. 3 Biotech 6, 1–10. doi: 10.1007/s13205-016-0369-y
Kempka, A. P., Lipke, N. L., da Luz Fontoura Pinheiro, T., Menoncin, S., Treichel, H., Freire, D. M., et al. (2008). Response surface method to optimize the production and characterization of lipase from Penicillium verrucosum in solid-state fermentation. Bioprocess Biosyst Eng. 31, 119–125. doi: 10.1007/s00449-007-0154-8
Khambhaty, Y. (2020). Applications of enzymes in leather processing. Environ. Chem. Lett. 18, 747–769. doi: 10.1007/s10311-020-00971-5
Khan, F. I., Lan, D., Durrani, R., Huan, W., Zhao, Z., and Wang, Y. (2017). The lid domain in lipases: Structural and functional determinant of enzymatic properties. Front. Bioeng. Biotechnol. 5, 16. doi: 10.3389/fbioe.2017.00016
Khan, N. R., and Rathod, V. K. (2020). Microwave mediated lipase-catalyzed synthesis of n-butyl palmitate and thermodynamic studies. Biocatal. Agric. Biotechnol. 29, 101741. doi: 10.1016/j.bcab.2020.101741
Kim, K. K., Song, H. K., Shin, D. H., Hwang, K. Y., and Suh, S. W. (1997). The crystal structure of a triacylglycerol lipase from Pseudomonas cepacia reveals a highly open conformation in the absence of a bound inhibitor. Structure 5, 173–185. doi: 10.1016/S0969-2126(97)00177-9
Kishan, G., Gopalakannan, P., Muthukumaran, C., Muthukumaresan, K. T., Kumar, M. D., and Tamilarasan, K. (2013). Statistical optimization of critical medium components for lipase production from Yarrowia lipolytica (MTCC 35). J. Genet. Eng. Biotechnol. 11, 111–116. doi: 10.1016/j.jgeb.2013.06.001
Knob, A., Izidoro, S. C., Lacerda, L. T., Rodrigues, A., and de Lima, V. A. (2020). A novel lipolytic yeast Meyerozyma guilliermondii: efficient and low-cost production of acid and promising feed lipase using cheese whey. Biocatal. Agric. Biotechnol. 24, 101565. doi: 10.1016/j.bcab.2020.101565
Kumar, A., Dhar, K., Kanwar, S. S., and Arora, P. K. (2016). Lipase catalysis in organic solvents: Advantages and applications. Biol. Proced. Online 18, 1–11. doi: 10.1186/s12575-016-0033-2
Kumar, A., Gudiukaite, R., Gricajeva, A., Sadauskas, M., Malunavicius, V., Kamyab, H., et al. (2020). Microbial lipolytic enzymes–promising energy-efficient biocatalysts in bioremediation. Energy 192, 116674. doi: 10.1016/j.energy.2019.116674
Kumar, D., Bhardwaj, R., Jassal, S., Goyal, T., Khullar, A., and Gupta, N. (2021). Application of enzymes for an eco-friendly approach to textile processing. Environ. Sci. Pollut. Res. 4, 1–11. doi: 10.1007/s11356-021-16764-4
Kumar, J. A., and Kumar, M. S. (2020). A study on improving dyeability of polyester fabric using lipase enzyme. Autex Res. J. 20, 243–249. doi: 10.2478/aut-2019-0030
Kumar, S. S., and Gupta, R. (2008). An extracellular lipase from Trichosporon asahii MSR 54: Medium optimization and enantioselective deacetylation of phenyl ethyl acetate. Process Biochem. 43, 1054–1060. doi: 10.1016/j.procbio.2008.05.017
Leal, M. C., Freire, D. M., Cammarota, M. C., and Sant'Anna, G. L. Jr (2006). Effect of enzymatic hydrolysis on anaerobic treatment of dairy wastewater. Process Biochem. 41, 1173–1178. doi: 10.1016/j.procbio.2005.12.014
Lee, S. K., Chou, H., Ham, T. S., Lee, T. S., and Keasling, J. D. (2008). Metabolic engineering of microorganisms for biofuels production: From bugs to synthetic biology to fuels. Curr. Opin. Biotechnol. 19, 556–563. doi: 10.1016/j.copbio.2008.10.014
Lenfant, N., Hotelier, T., Velluet, E., Bourne, Y., Marchot, P., and Chatonnet, A. (2012). ESTHER, the database of the α/β-hydrolase fold superfamily of proteins: Tools to explore diversity of functions. Nucleic Acids Res. 41, D423–D429. doi: 10.1093/nar/gks1154
Li, C. Y., Cheng, C. Y., and Chen, T. L. (2001). Production of Acinetobacter radioresistens lipase using Tween 80 as the carbon source. Enzyme Microb. Technol. 29, 258–263. doi: 10.1016/S0141-0229(01)00396-9
Li, Q., Lu, J., Zhang, G., Zhou, J., Li, J., Du, G., et al. (2023). CRISPR/Cas9-mediated multiplexed genome editing in aspergillus oryzae. J. Fungi. 9, 109. doi: 10.3390/jof9010109
Liese, A., and Hilterhaus, L. (2013). Evaluation of immobilized enzymes for industrial applications. Chem. Soc. Rev. 42, 6236–6249. doi: 10.1039/c3cs35511j
Lima, L. G. R., Gonçalves, M. M. M., Couri, S., Melo, V. F., Sant'Ana, G. C. F., and Costa, A. C. A. D. (2019). Lipase production by Aspergillus niger C by submerged fermentation Braz Arch Biol Technol., 62. doi: 10.1590/1678-4324-2019180113
Lira, R. K. D. S., Zardini, R. T., de Carvalho, M. C. C., Wojcieszak, R., Leite, S. G. F., and Itabaiana, I. (2021). Agroindustrial wastes as a support for the immobilization of lipase from Thermomyces lanuginosus: Synthesis of hexyl laurate. Biomolecules 11, 445. doi: 10.3390/biom11030445
Liu, Q., Gao, R., Li, J., Lin, L., Zhao, J., Sun, W., et al. (2017). Development of a genome-editing CRISPR/Cas9 system in thermophilic fungal Myceliophthora species and its application to hyper-cellulase production strain engineering. Biotechnol. Biofuels. 10, 1–14. doi: 10.1186/s13068-016-0693-9
Liu, R., Chen, L., Jiang, Y., Zhou, Z., and Zou, G. (2015). Efficient genome editing in filamentous fungus Trichoderma reesei using the CRISPR/Cas9 system. Cell Discov. 1, 1–11. doi: 10.1038/celldisc.2015.7
López, E., Deive, F. J., Longo, M. A., and Sanroman, M. A. (2010). Strategies for utilisation of food-processing wastes to produce lipases in solid-state cultures of Rhizopus oryzae. Bioprocess Biosyst. Eng. 33, 929–935. doi: 10.1007/s00449-010-0416-8
Mahfoudhi, A., Benmabrouk, S., Fendri, A., and Sayari, A. (2022). Fungal lipases as biocatalysts: A promising platform in several industrial biotechnology applications. Biotechnol. Bioeng. 119, 3370–3392. doi: 10.1002/bit.28245
Mahmoud, G. A., Koutb, M. M., Morsy, F. M., and Bagy, M. M. (2015). Characterization of lipase enzyme produced by hydrocarbons utilizing fungus Aspergillus terreus. Eur. J. Biol. Res. 5, 70–77.
Makrides, S. C. (1996). Strategies for achieving high-level expression of genes in Escherichia coli. Microbiol. Rev. 60, 512–538. doi: 10.1128/mr.60.3.512-538.1996
Marion, B. A. S., and Oliver, T. (2013). Review article: Immobilized lipases in the cosmetics industry. Chem. Soc. Rev. 42, 6406–6442. doi: 10.1039/c3cs35484a
Martínez-Ruiz, A., García, H. S., Saucedo-Castañeda, G., and Favela-Torres, E. (2008). Organic phase synthesis of ethyl oleate using lipases produced by solid-state fermentation. Appl. Biochem. Biotechnol. 151, 393–401. doi: 10.1007/s12010-008-8207-2
Matsu-Ura, T., Baek, M., Kwon, J., and Hong, C. (2015). Efficient gene editing in Neurospora crassa with CRISPR technology. Fungal Biol. Biotechnol, 2, 1–7. doi: 10.1186/s40694-015-0015-1
Maugard, T., Rejasse, B., and Legoy, M. D. (2002). Synthesis of water-soluble retinol derivatives by enzymatic method. Biotechnol. Prog. 18, 424–428. doi: 10.1021/bp025508f
Mauti, G. O., Onguso, J., Kowanga, D. K., and Mauti, E. M. (2016). Biodegradation activity of Aspergillus niger lipase isolates from a tropical country garage. J. Sci. Innov. Res. 5, 15–18. doi: 10.31254/jsir.2016.5104
McCarty, N. S., Graham, A. E., Studená, L., and Ledesma-Amaro, R. (2020). Multiplexed CRISPR technologies for gene editing and transcriptional regulation. Nat. Commun. 11, 1281. doi: 10.1038/s41467-020-15053-x
Mehta, A., Bodh, U., and Gupta, R. (2018). Isolation of a novel lipase producing fungal isolate Aspergillus fumigatus and production optimization of enzyme. Biocatal. Biotransform. 36, 450–457. doi: 10.1080/10242422.2018.1447565
Mehta, A., Grover, C., Bhardwaj, K. K., and Gupta, R. (2020). Application of lipase purified from Aspergillus fumigatus in the syntheses of ethyl acetate and ethyl lactate. J. Oleo Sci. 69, 23–29. doi: 10.5650/jos.ess19202
Mohamad, N. R., Marzuki, N. H. C., Buang, N. A., Huyop, F., and Wahab, R. A. (2015). Review; agriculture and environmental biotechnology an overview of technologies for immobilization of enzymes and surface analysis techniques for immobilized enzymes. Biotechnol. Biotechnol. Equip. 29, 205–220. doi: 10.1080/13102818.2015.1008192
Mohanasrinivasan, V., Dhrisya, P., Dipinsha, K. P., Unnithan, C. M., Viswanath, K. M., and Devi, C. S. (2009). A comparative study of the lipase yield by solid state and submerged fermentations using fungal species from biopharmaceutical oil waste. Afr. J. Biotechnol. 8.
Mouad, A. M., Taupin, D., Lehr, L., Yvergnaux, F., and Porto, A. L. M. (2016). Aminolysis of linoleic and salicylic acid derivatives with Candida antarctica lipase B: A solvent-free process to obtain amphiphilic amides for cosmetic application. J. Mol. Catal. 126, 64–68. doi: 10.1016/j.molcatb.2016.01.002
Moujehed, E., Zarai, Z., Khemir, H., Miled, N., Bchir, M. S., Gablin, C., et al. (2022). Cleaner degreasing of sheepskins by the Yarrowia lipolytica LIP2 lipase as a chemical-free alternative in the leather industry. Colloids Surf. B 211, 112292. doi: 10.1016/j.colsurfb.2021.112292
Mukhtar, H., Khursheed, S., Mumtaz, M. W., Rashid, U., and Al-Resayes, S. I. (2016). Optimization of lipase biosynthesis from Rhizopus oryzae for biodiesel production using multiple oils. Chem. Eng. Technol. 39, 1707–1715. doi: 10.1002/ceat.201500584
Narayanan, N., and Chou, C. P. (2009). Alleviation of proteolytic sensitivity to enhance recombinant lipase production in Escherichia coli. Appl. Environ. Microbiol. 75, 5424–5427. doi: 10.1128/AEM.00740-09
Nedovic, V., Kalusevic, A., Manojlovic, V., Levic, S., and Bugarski, B. (2011). An overview of encapsulation technologies for food applications. Procedia Food Sci. 1, 1806–1815. doi: 10.1016/j.profoo.2011.09.265
Nema, A., Patnala, S. H., Mandari, V., Kota, S., and Devarai, S. K. (2019). Production and optimization of lipase using Aspergillus niger MTCC 872 by solid-state fermentation. Bull. Natl. Res. Cent. 43, 1–8. doi: 10.1186/s42269-019-0125-7
Niaz, M., Iftikhar, T., and Zia, M. A. (2013). Effect of nutritional factors on lipase biosynthesis by Aspergillus niger in solid state fermentation. Extensive J. Appl. Sci. 1, 9–16.
Nishioka, M., Joko, K., and Takama, M. (1990). Lipase manufacture with Candida for use in detergents. Japan. Pat. 1990, 292.
Nodvig, C. S., Nielsen, J. B., Kogle, M. E., and Mortensen, U. H. (2015). A CRISPR-Cas9 system for genetic engineering of filamentous fungi. PLoS ONE. 10, e0133085. doi: 10.1371/journal.pone.0133085
Oliveira, F., Moreira, C., Salgado, J. M., Abrunhosa, L., Venâncio, A., and Belo, I. (2016). Olive pomace valorization by Aspergillus species: Lipase production using solid-state fermentation. J. Sci. Food Agric. 96, 3583–3589. doi: 10.1002/jsfa.7544
Pandey, N., Dhakar, K., Jain, R., and Pandey, A. (2016). Temperature dependent lipase production from cold and pH tolerant species of Penicillium. Mycosphere 7, 1533–1545. doi: 10.5943/mycosphere/si/3b/5
Patel, G. B., and Shah, K. R. (2020). Isolation, screening and identification of Lipase producing fungi from cotton seed soapstock. Indian J. Sci. Technol. 13, 3762–3771. doi: 10.17485/IJST/v13i36.1099
Pérez, M. M., Gonçalves, E. C. S., Vici, A. C., Salgado, J. C. S., and de Moraes Polizeli, M. D. L. T. (2019). “Fungal lipases: Versatile tools for white biotechnology,” in Recent Advancement in White Biotechnology Through Fungi: Volume 1: Diversity and Enzymes Perspectives, eds A. Nath Yadav, S. Mishra, S. Singh and A. Gupta, 361–404. doi: 10.1007/978-3-030-10480-1_11
Pinheiro, T. D. L. F., Menoncin, S., Domingues, N. M., Oliveira, D. D., Treichel, H., Di Luccio, M., et al. (2008). Production and partial characterization of lipase from Penicillium verrucosum obtained by submerged fermentation of conventional and industrial media. Food Sci. Technol. 28, 444–450. doi: 10.1590/S0101-20612008000200028
Priyanka, P., Kinsella, G., Henehan, G. T., and Ryan, B. J. (2019). Isolation, purification and characterization of a novel solvent stable lipase from Pseudomonas reinekei. Protein Expr. Purif. 153, 121–130. doi: 10.1016/j.pep.2018.08.007
Quayson, E., Amoah, J., Hama, S., Kondo, A., and Ogino, C. (2020). Immobilized lipases for biodiesel production: Current and future greening opportunities. Renew. Sustain. Energy Rev. 134, 110355. doi: 10.1016/j.rser.2020.110355
Rahman, M., Hack-Polay, D., Billah, M. M., and Un Nabi, M. N. (2020). Bio-based textile processing through the application of enzymes for environmental sustainability. Int. J. Technol. Manag. Sustain. Dev. 19, 87–106. doi: 10.1386/tmsd_00017_1
Rajeswari, T., Palaniswamy, M., Raphy, P., and Padmapriya, B. (2011). Utilization of mixed agroindustrial by-products for alkaline lipase production from Penicillim chrysogenum. Int. J. Pharm. Biol. Sci. Arch. 2, 682–691.
Ramakrishnan, V., Goveas, L. C., Suralikerimath, N., Jampani, C., Halami, P. M., and Narayan, B. (2016). Extraction and purification of lipase from Enterococcus faecium MTCC5695 by PEG/phosphate aqueous-two phase system (ATPS) and its biochemical characterization. Biocatal. Agric. Biotechnol. 6, 19–27. doi: 10.1016/j.bcab.2016.02.005
Rath, D., Amlinger, L., Rath, A., and Lundgren, M. (2015). The CRISPR-Cas immune system: biology, mechanisms and applications. Biochimie 117, 119–128. doi: 10.1016/j.biochi.2015.03.025
Rihani, A., Tichati, L., and Soumati, B. (2018). Isolation and identification of lipase producing fungi from local olive oil manufacture in east of algeria. Sci. Study Res. Chemistry Chem. Engg. Biotechnol. Food Industry. 19, 13–22.
Rocha, K. S., Queiroz, M. S., Gomes, B. S., Dallago, R., de Souza, R. O., Guimarães, D. O., et al. (2020). Lipases of endophytic fungi Stemphylium lycopersici and Sordaria sp.: Application in the synthesis of solketal derived Monoacylglycerols. Enzyme Microb. Technol. 142, 109664. doi: 10.1016/j.enzmictec.2020.109664
Rodriguez, J. A., Mateos, J. C., Nungaray, J., González, V., Bhagnagar, T., Roussos, S., et al. (2006). Improving lipase production by nutrient source modification using Rhizopus homothallicus cultured in solid state fermentation. Process Biochem. 41, 2264–2269. doi: 10.1016/j.procbio.2006.05.017
Romdhane, I. B. B., Fendri, A., Gargouri, Y., Gargouri, A., and Belghith, H. (2010). A novel thermoactive and alkaline lipase from Talaromyces thermophilus fungus for use in laundry detergents. Biochem. Eng. J. 53, 112–120. doi: 10.1016/j.bej.2010.10.002
Roy, M., Kumar, R., Ramteke, A., and Sit, N. (2021). Identification of lipase producing fungus isolated from dairy waste contaminated soil and optimization of culture conditions for lipase production by the isolated fungus. J. Microbiol. Biotechnol. Food Sci. 2021, 698–704. doi: 10.15414/jmbfs.2018.8.1.698-704
Salgado, J. M., Abrunhosa, L., Venâncio, A., Domínguez, J. M., and Belo, I. (2016). Combined bioremediation and enzyme production by Aspergillus sp. in olive mill and winery wastewaters. Int. Biodeterior. Biodegrad. 110, 16–23. doi: 10.1016/j.ibiod.2015.12.011
Salgado, V., Fonseca, C., Lopes da Silva, T., Roseiro, J. C., and Eusébio, A. (2020). Isolation and identification of Magnusiomyces capitatus as a lipase-producing yeast from olive mill wastewater. Waste Biomass Valorization 11, 3207–3221. doi: 10.1007/s12649-019-00725-7
Salleh, A. B., Musani, R., Basri, M., Ampon, K., Yunus, W. M. Z., and Razak, C. N. A. (1993). Extra-and intra-cellular lipases from a thermophilic Rhizopus oryzae and factors affecting their production. Can. J. Microbiol. 39, 978–981. doi: 10.1139/m93-147
Santos, S., Puna, J., and Gomes, J. (2020). A review on bio-based catalysts (immobilized enzymes) used for biodiesel production. Energies 13, 3013. doi: 10.3390/en13113013
Saranya, G., and Ramachandra, T. V. (2020). Novel biocatalyst for optimal biodiesel production from diatoms. Renew. Energ. 153, 919–934. doi: 10.1016/j.renene.2020.02.053
Sarmah, N., Revathi, D., Sheelu, G., Yamuna Rani, K., Sridhar, S., Mehtab, V., et al. (2018). Recent advances on sources and industrial applications of lipases. Biotechnol. Prog. 34, 5–28. doi: 10.1002/btpr.2581
Sethi, B. K., Rout, J. R., Das, R., Nanda, P. K., and Sahoo, S. L. (2013). Lipase production by Aspergillus terreus using mustard seed oil cake as a carbon source. Ann. Microbiol. 63, 241–252. doi: 10.1007/s13213-012-0467-y
Shabbir, A. R. O. O. S. H., and Mukhtar, H. (2018). Optimization process for enhanced extracellular lipases production from a new isolate of Aspergillus terreus ah-F2. Pak. J. Bot. 50, 1571–1578.
Sharma, S., and Kanwar, S. S. (2014). Organic solvent tolerant lipases and applications. Sci. World J. 2014, 625258. doi: 10.1155/2014/625258
Shu, C. H., Xu, C. J., and Lin, G. C. (2006). Purification and partial characterization of a lipase from Antrodia cinnamomea. Process Biochem. 41, 734–738. doi: 10.1016/j.procbio.2005.09.007
Šibalić, D., Šalić, A., Tušek, A. J., Sokač, T., Brekalo, K., Zelić, B., et al. (2020). Sustainable production of lipase from Thermomyces lanuginosus: process optimization and enzyme characterization. Ind. Eng. Chem. Res. 59, 21144–21154. doi: 10.1021/acs.iecr.0c04329
Silva, W. O. B., Mitidieri, S., Schrank, A., and Vainstein, M. H. (2005). Production and extraction of an extracellular lipase from the entomopathogenic fungus Metarhizium anisopliae. Process Biochem. 40, 321–326. doi: 10.1016/j.procbio.2004.01.005
Singh, A. K., and Mukhopadhyay, M. (2012). Overview of fungal lipase: A review. Appl. Biochem. Biotechnol. 166, 486–520. doi: 10.1007/s12010-011-9444-3
Singhania, R. R., Patel, A. K., Soccol, C. R., and Pandey, A. (2009). Recent advances in solid-state fermentation. Biochem. Eng. J. 44, 13–18. doi: 10.1016/j.bej.2008.10.019
Sumner, C., Krause, S., Sabot, A., Turner, K., and McNeil, C. J. (2001). Biosensor based on enzyme-catalysed degradation of thin polymer films. Biosens. Bioelectron 16, 709–714. doi: 10.1016/S0956-5663(01)00210-X
Supakdamrongkul, P., Bhumiratana, A., and Wiwat, C. (2010). Characterization of an extracellular lipase from the biocontrol fungus, Nomuraea rileyi MJ, and its toxicity toward Spodoptera litura. J. Invertebr. Pathol. 105, 228–235. doi: 10.1016/j.jip.2010.06.011
Syamsul, K. M. W., Salina, M. R., Siti, S. O., Hanina, M. N., Basyaruddin, M. A. R., and Jusoff, K. (2010). Green synthesis of lauryl palmitate via lipase-catalyzed reaction. World Appl. Sci. J. 11.
Tan, J. S., Abbasiliasi, S., Ariff, A. B., Ng, H. S., Bakar, M. H. A., and Chow, Y. H. (2018). Extractive purification of recombinant thermostable lipase from fermentation broth of Escherichia coli using an aqueous polyethylene glycol impregnated resin system. 3 Biotech 8, 1–7. doi: 10.1007/s13205-018-1295-y
Tan, T., Zhang, M., Wang, B., Ying, C., and Deng, L. (2003). Screening of high lipase producing Candida sp. and production of lipase by fermentation. Process Biochem. 39, 459–465. doi: 10.1016/S0032-9592(03)00091-8
Taskin, M., Ucar, M. H., Unver, Y., Kara, A. A., Ozdemir, M., and Ortucu, S. (2016). Lipase production with free and immobilized cells of cold-adapted yeast Rhodotorula glutinis HL25. Biocatal. Agric. Biotechnol. 8, 97–103. doi: 10.1016/j.bcab.2016.08.009
Thangaraj, B., and Solomon, P. R. (2019). Immobilization of lipases–a review. Part I: Enzyme immobilization. ChemBioEng Rev. 6, 157–166. doi: 10.1002/cben.201900016
Thapa, S., Li, H., OHair, J., Bhatti, S., Chen, F. C., Nasr, K. A., et al. (2019). Biochemical characteristics of microbial enzymes and their significance from industrial perspectives. Mol. Biotechnol. 61, 579–601. doi: 10.1007/s12033-019-00187-1
Thirunavukarasu, K., Purushothaman, S., Sridevi, J., Aarthy, M., Gowthaman, M. K., Nakajima-Kambe, T., et al. (2016). Degradation of poly (butylene succinate) and poly (butylene succinate-co-butylene adipate) by a lipase from yeast Cryptococcus sp. grown on agro-industrial residues. Int. Biodeterior. Biodegrad. 110, 99–107. doi: 10.1016/j.ibiod.2016.03.005
Tian, M., Fu, J., Wang, Z., Miao, C., Lv, P., He, D., et al. (2021). Enhanced activity and stability of Rhizomucor miehei lipase by mutating N-linked glycosylation site and its application in biodiesel production. Fuel 304, 121514. doi: 10.1016/j.fuel.2021.121514
Tišma, M., Tadić, T., BudŽaki, S., Ostojčić, M., Šalić, A., Zelić, B., et al. (2019). Lipase production by solid-state cultivation of Thermomyces lanuginosus on by-products from cold-pressing oil production. Processes 7, 465. doi: 10.3390/pr7070465
Toida, J., Kondoh, K., Fukuzawa, M., Ohnishi, K., and Sekoguchi, J. (1995). Purification and characterization of a lipase from Aspergillus oryzae. Biosci. Biotechnol. Biochem. 59, 1199–1203. doi: 10.1271/bbb.59.1199
Treichel, H., de Oliveira, D., Mazutti, M. A., Di Luccio, M., and Oliveira, J. V. (2010). A review on microbial lipases production. Food Bioproc. Tech. 3, 182–196. doi: 10.1007/s11947-009-0202-2
Triyaswati, D., and Ilmi, M. (2020). Lipase-producing filamentous fungi from non-dairy creamer industrial waste. Microbiol. Biotechnol. Lett. 48, 167–178. doi: 10.4014/mbl.1912.12018
Turati, D. F. M., Almeida, A. F., Terrone, C. C., Nascimento, J. M., Terrasan, C. R., Fernandez-Lorente, G., et al. (2019). Thermotolerant lipase from Penicillium sp. section Gracilenta CBMAI 1583: Effect of carbon sources on enzyme production, biochemical properties of crude and purified enzyme and substrate specificity. Biocatal. Agric. Biotechnol. 17, 15–24. doi: 10.1016/j.bcab.2018.10.002
Verma, N., Thakur, S., and Bhatt, A. K. (2012). Microbial lipases: Industrial applications and properties (a review). Int. J. Biol. Sci. 1, 88–92.
Vilar, D. S., Carvalho, G. O., Pupo, M. M., Aguiar, M. M., Torres, N. H., Américo, J. H., et al. (2018). Vinasse degradation using Pleurotus sajor-caju in a combined biological–Electrochemical oxidation treatment. Sep. Purif. Technol. 192, 287–296. doi: 10.1016/j.seppur.2017.10.017
Wang, Y., Ma, R., Li, S., Gong, M., Yao, B., Bai, Y., et al. (2018). An alkaline and surfactant-tolerant lipase from Trichoderma lentiforme ACCC30425 with high application potential in the detergent industry. AMB Express 8, 1–11. doi: 10.1186/s13568-018-0618-z
Winayanuwattikun, P., Kaewpiboon, C., Piriyakananon, K., Chulalaksananukul, W., Yongvanich, T., and Svasti, J. (2011). Immobilized lipase from potential lipolytic microbes for catalyzing biodiesel production using palm oil as feedstock. Afr. J. Biotechnol. 10, 1666–1673. doi: 10.5897/AJB10.1802
Xiao, Y., Liu, Y. D., Yuan, G., Mao, R. Q., and Li, G. (2021). An uncharacterized protein from the metagenome with no obvious homology to known lipases shows excellent alkaline lipase properties and potential applications in the detergent industry. Biotechnol. Lett. 43, 2311–2325. doi: 10.1007/s10529-021-03203-0
Yakubu, A., Saikia, U., and Vyas, A. (2019). “Microbial enzymes and their application in pulp and paper industry,” in Recent Advancement in White Biotechnology Through Fungi, eds A, Nath Yadav, S. Singh, S. Mishra and A. Gupta (Cham: Springer), 297–317. doi: 10.1007/978-3-030-25506-0_12
Yang, F., Weber, T. W., Gainer, J. L., and Carta, G. (1997). Synthesis of lovastatin with immobilized Candida rugosa lipase in organic solvents: Effects of reaction conditions on initial rates. Biotechnol. Bioeng. 56, 671–680. doi: 10.1002/(SICI)1097-0290(19971220)56:6<671::AID-BIT10>3.0.CO;2-B
Yaver, D. S., Lamsa, M., Munds, R., Brown, S. H., Otani, S., Franssen, L., et al. (2000). Using DNA-tagged mutagenesis to improve heterologous protein production in Aspergillus oryzae. Fungal Genet. Biol. 29, 28–37. doi: 10.1006/fgbi.1999.1179
Zehani, N., Kherrat, R., and Jaffrezic-Renault, N. (2014). Immobilization of Candida Rugosa lipase on Aluminosilicate incorporated in a polymeric membrane for the elaboration of an impedimetric biosensor. Sens. Transducers 27, 371.
Zhao, W., Wang, J., Deng, R., and Wang, X. (2008). Scale-up fermentation of recombinant Candida rugosa lipase expressed in Pichia pastoris using the GAP promoter. J. Ind. Microbiol 35, 189–195. doi: 10.1007/s10295-007-0283-8
Zheng, M. M., Dong, L., Lu, Y., Guo, P. M., Deng, Q. C., Li, W. L., et al. (2012). Immobilization of Candida rugosa lipase on magnetic poly (allyl glycidyl ether-co-ethylene glycol dimethacrylate) polymer microsphere for synthesis of phytosterol esters of unsaturated fatty acids. J. Mol. Catal. 74, 16–23. doi: 10.1016/j.molcatb.2011.08.008
Keywords: lipase, bioethanol, immobilization, biosensor, fungi
Citation: Kumar A, Verma V, Dubey VK, Srivastava A, Garg SK, Singh VP and Arora PK (2023) Industrial applications of fungal lipases: a review. Front. Microbiol. 14:1142536. doi: 10.3389/fmicb.2023.1142536
Received: 11 January 2023; Accepted: 21 March 2023;
Published: 28 April 2023.
Edited by:
Vineet Kumar, Central University of Rajasthan, IndiaReviewed by:
Sanjay Kumar Singh Patel, Konkuk University, Republic of KoreaJuliana Heloisa Pinê Américo-Pinheiro, São Paulo State University, Brazil
Aditi Singh, Amity University, Lucknow, India
Copyright © 2023 Kumar, Verma, Dubey, Srivastava, Garg, Singh and Arora. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pankaj Kumar Arora, YXJvcmE0ODRAZ21haWwuY29t
 Ashish Kumar
Ashish Kumar Vinita Verma
Vinita Verma Vimal Kumar Dubey2
Vimal Kumar Dubey2 Alok Srivastava
Alok Srivastava Pankaj Kumar Arora
Pankaj Kumar Arora