- 1College of Food Science and Nutritional Engineering, China Agricultural University, Beijing, China
- 2The Supervision, Inspection and Testing Center of Genetically Modified Organisms, Ministry of Agriculture, Beijing, China
- 3Beijing Laboratory for Food Quality and Safety, College of Food Science and Nutritional Engineering, China Agricultural University, Beijing, China
Aspergillus is widely distributed in nature and occupies a crucial ecological niche, which has complex and diverse metabolic pathways and can produce a variety of metabolites. With the deepening of genomics exploration, more Aspergillus genomic informations have been elucidated, which not only help us understand the basic mechanism of various life activities, but also further realize the ideal functional transformation. Available genetic engineering tools include homologous recombinant systems, specific nuclease based systems, and RNA techniques, combined with transformation methods, and screening based on selective labeling. Precise editing of target genes can not only prevent and control the production of mycotoxin pollutants, but also realize the construction of economical and efficient fungal cell factories. This paper reviewed the establishment and optimization process of genome technologies, hoping to provide the theoretical basis of experiments, and summarized the recent progress and application in genetic technology, analyzes the challenges and the possibility of future development with regard to Aspergillus.
1. Introduction
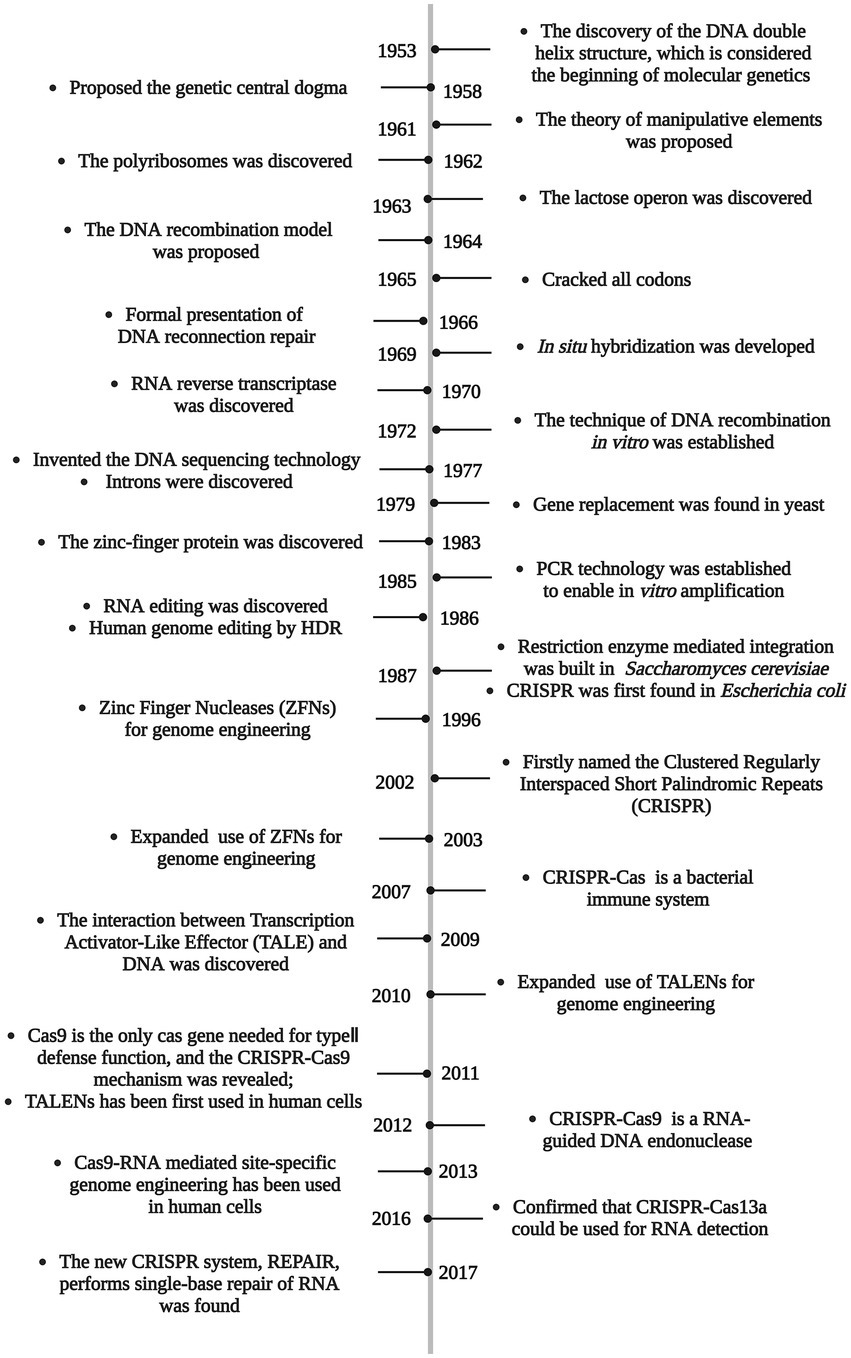
The discovery of the DNA double helix in 1953 is considered the beginning of molecular genetics (Wilkins et al., 1953). Since Sanger (Sanger et al., 1977) invented the dideoxy termination method (also named chain termination method) as well as Maxam and Gilbert (1977) invented the chemical degradation method to sequence genes, humans have obtained the key to the genome. At present, genome sequencing technology has developed to the third generation of high-throughput single-molecule sequencing, which facilitates us to quickly obtain genomic data of different species (Chiara et al., 2021). Subsequently, the principles of gene damage and repair have been clarified that provided a theoretical basis for artificially modified genes. Finally, the discovery of various restriction enzymes and ligases, which are known as “molecular scissors and glues,” provides powerful tools. Gene editing technologies came into being, realized the replacement, insertion, deletion, and modification of specific nucleic acid segments (Pant et al., 2022).
Scientists have long attempted to acquire new traits that do not exist in nature but meet human expectation by influencing genes of organisms. The simplest and most crude way to induce mutations is through physicochemical methods, including physical mutagens (such as UV light, ionizing radiation, and radioactive material) and chemical mutagens (such as benzene and alkaloid; Rivera et al., 2014), but extreme uncertainty and tedious laborious works limit their systematic-scale use. Molecular tools are then used to create random genetic mutations through the insertion of foreign nucleic acid segments or the movement of mobile genetic elements. Since random mutation has the potential risk to disrupt or alter other genes, new strategies have been explored to solve this uncontrollability. Finally, specific and targeted technologies combined with the homologous recombination mechanism realize the current arbitrary study of target genes (Li et al., 2020). From the initial simple gene replacement in yeast in 1979 (Struhl et al., 1979) to the advent of multifunctional Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)-Cas9 technology that is currently popular around the world (Jinek et al., 2012), gene editing technologies have been explored for more than 40 years (Figure 1). Genetic engineering can break through species limits, which is expected to achieve unprecedented scientific breakthroughs, but in order to avoid genetic contamination in nature and to ensure ethical compliance, some legal framework on genetically modified organism or strain (GMO) have been enacted, such as the Directive 2001/18/EC of the European Parliament, that genetic engineering must be carried out under strict laws (Garrigues et al., 2021). Recent advances in genome biology have expanded the field of genetic engineering, various methods can be used alone or in combination to achieve unprecedented collaboration and for interdisciplinary applications.
Aspergillus is a class of saprophytic filamentous fungi that reproduce by asexual conidiospores and sexual ascospores, this genus contains four subgenera and 339 species, and are distributed ubiquitously in natural habitats (Houbraken et al., 2020). Some Aspergillus species, such as A. nidulans, A. niger, and A. oryzae are Generally Recognized As Safe (GRAS) by the Food and Drug Administration (FDA), playing significant roles in industrial biotechnology and fermentation technology, and as extraordinary eukaryotic host for microbial cell factories for the production of enzymes, antibiotics, nutrilite, and so on (Li et al., 2022). However, some Aspergillus species are terrible food spoilage fungus, which widely contaminating raw materials and processed foods, spatiotemporally involving planting, harvesting, processing, storage and sales, causing foods sensory quality reduction, physical damage, and chemical composition destruction (Luo et al., 2018). These fungi can also produce harmful secondary metabolites mycotoxins. The Food and Agriculture Organization (FAO) has reported 25% contamination of food by mycotoxins worldwide (Eskola et al., 2020), and most mycotoxins are certified by the International Agency for Research on Cancer (IARC) as Group I or Group II carcinogens, which pose a serious threat to animals and humans health (Ostry et al., 2017).
In recent years, with the popularization of gene sequencing technology, genome sequences of filamentous fungi have been established (http://www.genomesonline.org/; http://genome.jgi.doe.gov/), and the explosive growth of Aspergillus whole genomes data provide the possibility to change fungal characters (Robey et al., 2021). The gene expression can be controlled during both transcription and post transcription levels through gene editing technologies, then combined with omics analysis and high-throughput screening technologies has achieved industrial application. In this review, we systematically summarized the genetic modifications in Aspergillus, from the development process, gene editing techniques, to efficient transformation methods, and hoping to facilitate the background supplement of experimenters. Furthermore, we discuss the major challenges and solutions of genetic engineering, and finally, we propose potential directions in the development of the Aspergillus system, which would be helpful for the research and development of Aspergillus expression cell factories or mycotoxin prevention and control, and then draw on advantages and avoid disadvantages.
2. Genome editing technologies
2.1. Homologous recombination based systems
Homologous Recombination (HR) based systems are relatively early gene editing technologies and a breakthrough in eukaryotic gene editing (Capecchi, 1989). The principle is to introduce the foreign gene into the recipient cell, and the exogenous DNA fragment replaces the target gene in situ through site-specific recombinases mediating the recombination of target sites to achieve specific gene modification. Typical examples include the Lambda-red system, which is only for Escherichia coli, as well as the Cre-loxP system in Saccharomyces cerevisiae and the similar FLP-FRT system that are widely applicable to fungi.
2.1.1. Lambda-red system
In 1998, Murphy reported the use of λ bacteriophage Red system (Lambda-red system) for gene modification in E. coli, which was an early implementation of sequence-specific recombinases (Murphy, 1998). It does not rely on the recombinant system of E. coli own, but uses three proteins (Exo, Gam, and Beta) from bacteriophages through vector plasmids transformation to modify genes. Exo exonuclease degrades linear double-stranded DNA (dsDNA) starting from the 5′ ends to produce single-stranded DNA (ssDNA) fragments with a 3′ sticky end, Gam protein prevents the E. coli endogenous RecBCD and SbcCD nucleases from cutting linear DNA, Beta annealing protein protects the ssDNA created by Exo (Dillingham and Kowalczykowski, 2008). The Lambda-red system is independent of restriction sites and has applied to gene editing flexibly, but it is only suitable for E. coli (Zeng et al., 2022). However, this system in E. coli can be used to achieve efficient expression of fusion PCR fragments and based on homologous recombination to achieve gene editing in Aspergillus (Chaveroche et al., 2000; Langfelder et al., 2002). In addition, with the development of new technologies, the Lambda-red system can already be more widely used by combining with such as CRISPR technology.
2.1.2. Cre-loxP and FLP-FRT system
In 1981 (Sternberg and Hamilton, 1981), the DNA sequence specific Cyclization recombination enzyme (Cre) was discovered in P1 bacteriophages, and its C-terminal domain contains catalytic active sites that can catalyze recombination between pairs of locus of X-overP1 (loxP) sites. The loxP site consists of two reverse palindromic sequence domains and the intermediate spacer sequence, the former identifies and binds the Cre and the latter determines the direction of loxP sites. Cre recombinase can act on DNA substrates of various structures (such as linear, circular, and supercoiled DNA) independent of cofactors. Depending on the location and orientation of loxP sites in the genome, Cre recombinase can initiate deletions, inversions, translocations, and cassette exchange of the two loxP sites that flank a genomic segment of interest (named “floxed” locus; Nagy, 2000; Figure 2).
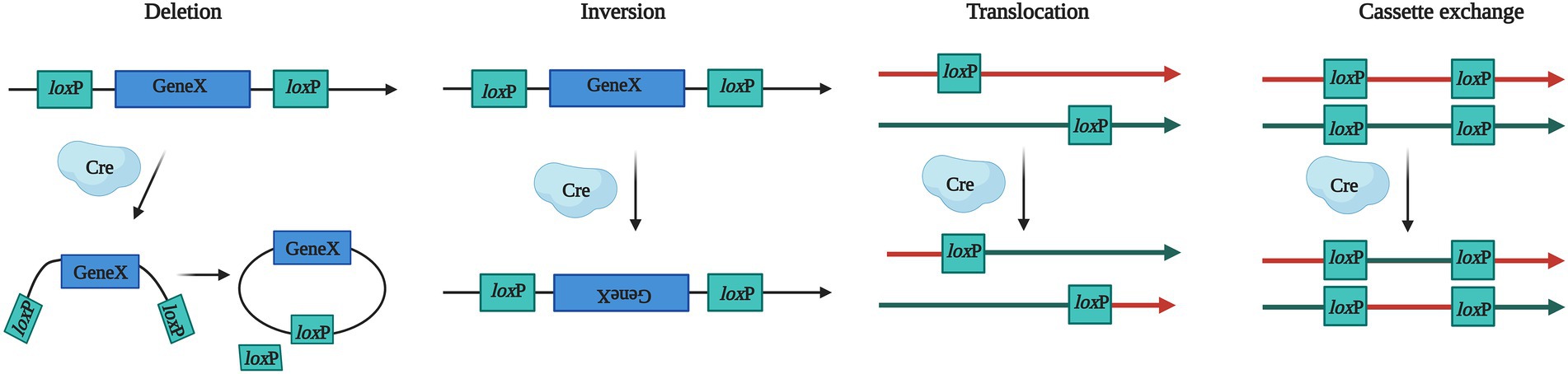
Figure 2. Biological mechanisms for Cre–loxP system. Cre recombinase can recognize the loxP site and promote gene recombination. LoxP is a unique gene sequence from the P1 phage, which guides the Cre to target sites of the genome. Gene X stands for target gene. The Cre–loxP system can produce four types of gene modification, including deletion, inversion, translocation, and cassette exchange.
The Cre–loxP system provides the possibility for multigene manipulation that need two steps to achieve gene recycling, loxP sites and a marker gene need to be integrated into the host genome firstly, then the Cre was expressed to complete the gene modification process. The Cre–loxP system has been successfully applied in Aspergillus spp. Modified Cre-loxP system is a powerful tool for producing high levels of various organic acids in A. niger (Xu et al., 2019), and this site-specific recombinase implemented the integration of a heterologous DNA fragment as large as 21 kb in A. nidulans (Roux and Chooi, 2022). In addition, Cre recombinase has been used in Saccharomyces (Moon et al., 2022), Lecanicillium (Nguyen et al., 2018), Trichoderma (Steiger et al., 2011), Neurospora (Honda and Selker, 2009), Neotyphodium (Florea et al., 2012) and so on, setting the foundations for the further development of fungal cell factories.
The FLP-FRT system derived from S. cerevisiae is similar to the Cre-loxP system (Sadowski, 1995), using flippase (FLP) recognizes a pair of FLP recombinase target (FRT) sequences that flank a genomic region of interest, and also rely on HR with 30–50 base-pairs of homologous arms for non-specificity gene knockin or knockout. The FLP-FRT system was first successfully applied in Penicillium chrysogenum and Sordaria macrospora (Kopke et al., 2010). At present, these techniques have developed into recombination-based genetic engineering (recombineering), which widely applied in yeast, filamentous fungi, plant, and mammal (Zeng et al., 2022), However, it can only act on specific sequences in the genome that are recognized by recombinases, and there is a high probability of misintegration.
2.2. Sequence specific nuclease based technologies
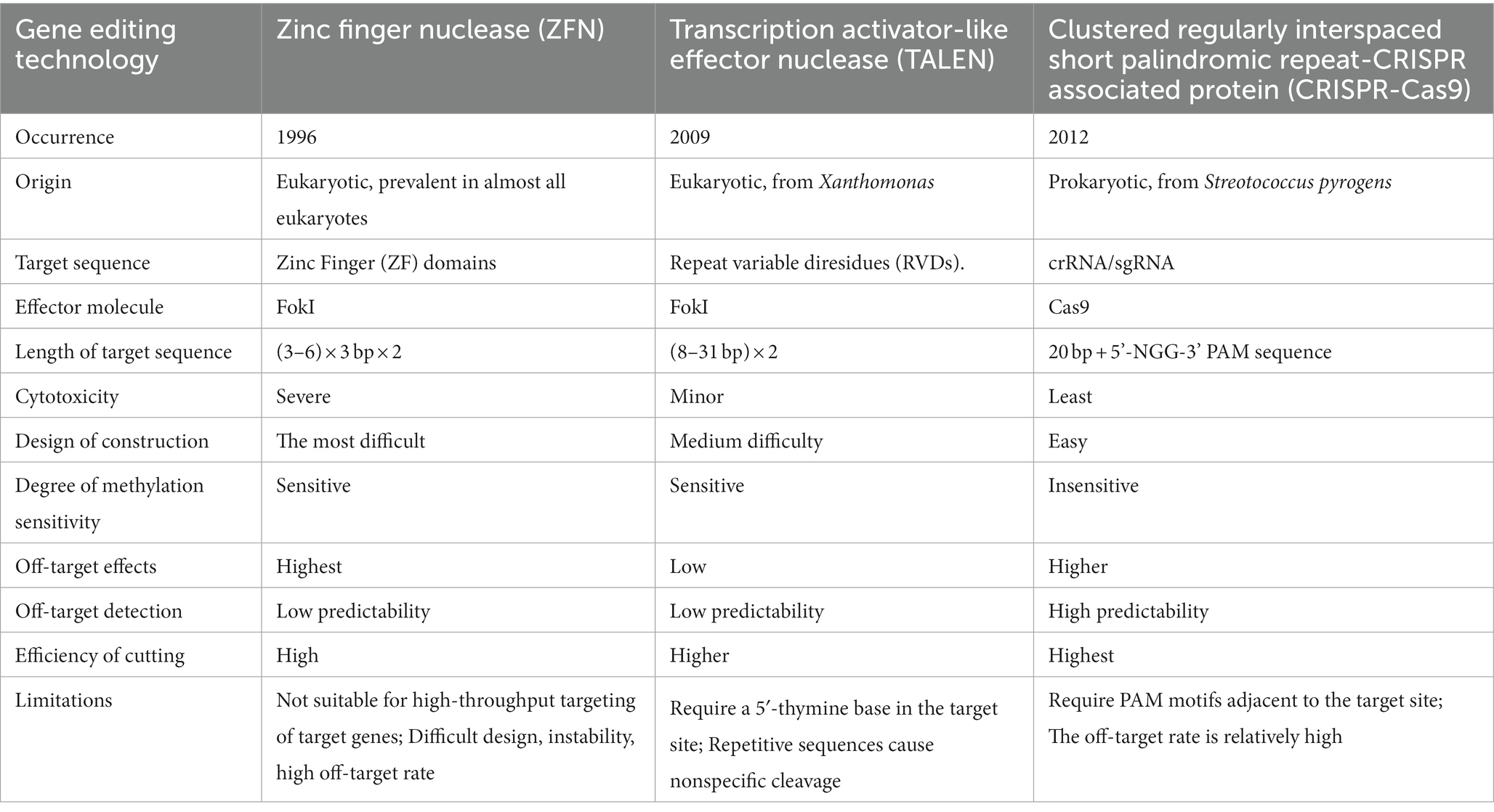
Compared with traditional HR gene editing technologies, nuclease-based gene editing techniques are also rely on homologous sequences. However, designed nucleases can more accurately act on target sequences to reduce random insertion, and more importantly, enable realize the recognition and modification of arbitrary gene sequences. Currently, manipulation nuclease-based gene editing technologies include Zinc Finger Nuclease (ZFN) system, Transcription Activator-Like Effector Nuclease (TALEN) system, Clustered Regulatory Interspaced Short Palindromic Repeat-CRISPR associated protein (CRISPR-Cas) system and single Base Editor (BE), etc. The first three systems are based on the genomic target site causing Double-Strand Breaks (DSBs) of DNA, which in turn activates the internal repair mechanism of cells. Among them, CRISPR has emerged as a powerful instrument for exploring genomics (Pant et al., 2022). The principles and comparison of these three techniques are shown in charts (Figure 3; Table 1). In 2016, the development of BE technology realized single base conversion without causing DSBs of DNA and no more need for homologous templates, effectively avoiding the off-target effect (Komor et al., 2016).
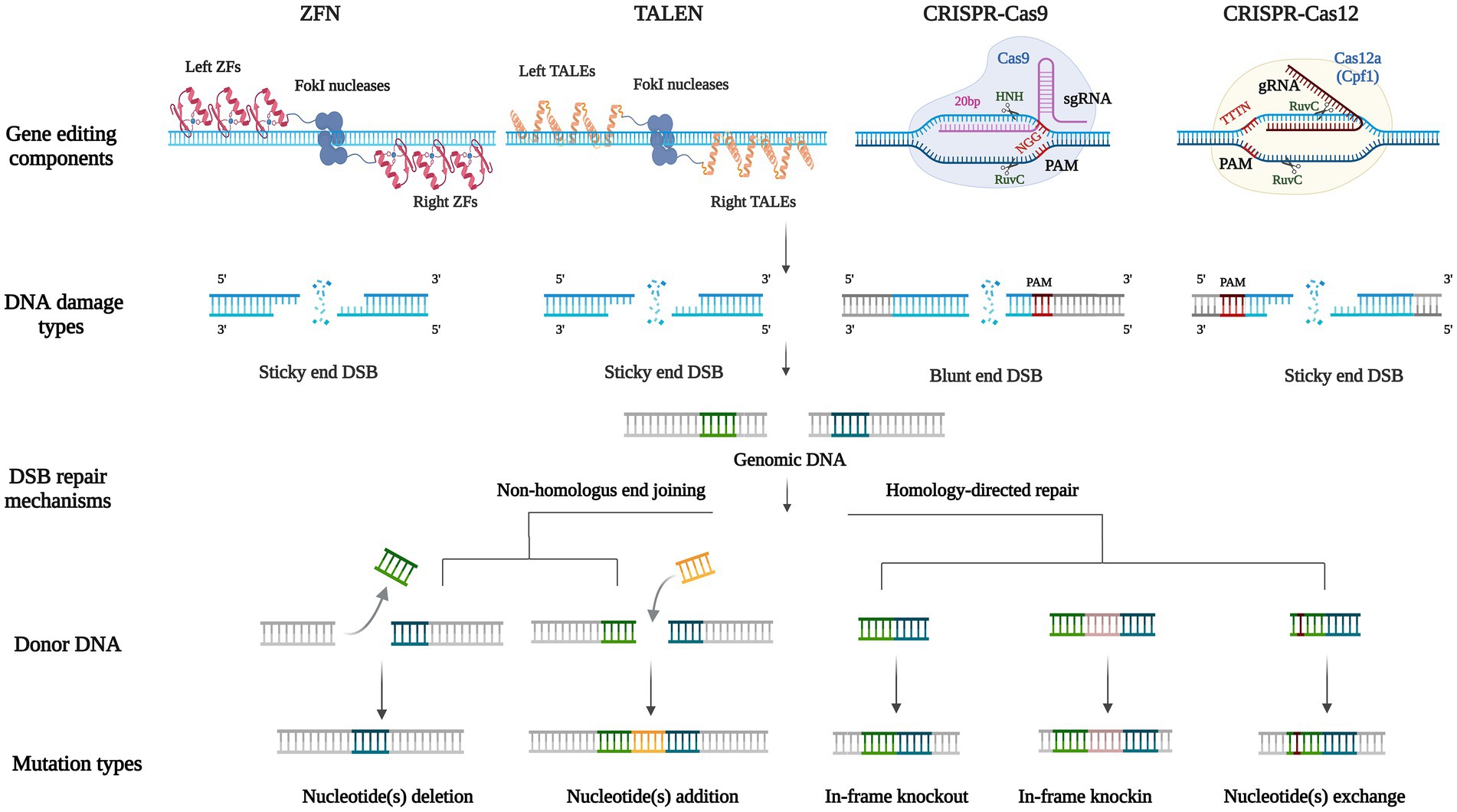
Figure 3. Biological mechanisms for nuclease-based gene editing technologies. Including gene editing components, the type of DNA damage, double-strand break repair mechanisms, donor DNA, and mutation types of ZFN, TALEN, CRISPR-Cas9, and CRISPR-Cas12 systems. Both ZFN and TALEN systems locate similar FokI endonucleases (the dark blue combined sphere) by DNA-binding domains on the left and right of the target site (the red zinc finger domains and the yellow transcription activator-like effectors domains), producing sticky ends double-strand breaks. The CRISPR systems consist of guiding RNA to locate target fragments containing PAM (the bases highlighted in red), and Cas nuclease domains cleave DNA to produce double-strand breaks. Cas9 includes HNH and RuvC catalytic nuclease domains, each cleaving one strand of the target DNA to produce a blunt end. Cas12 contains a RuvC nuclease domain that acts on both single strands of the target DNA to produce a sticky end. NHEJ and HDR are two DSB repair mechanisms, based on the complementarity of homologous fragments between genomic DNA and donor DNA (base pairs of the same color), to produce different mutation types including nucleotide(s) deletion, nucleotide(s) addition, in-frame deletion, in-frame knockout, and nucleotide(s) exchange.
2.2.1. Zinc finger nuclease
In 1996, scientists introduced the Zinc Finger Nuclease (ZFN) technology (Kim et al., 1996). ZFN monomer is a protein fused with a non-specific FokI endonuclease catalytic domain and 3–6 Cys2-His2 Zinc Finger (ZF) DNA-binding domains, each of which targets a DNA triplet base pairs. The dimerization of two FokI nuclease domains, by which designing ZF domains in both directions of 5′–3′ and 3′–5′ for target sequences and connecting them to different Fok1 respectively, then the middle target DNA sequence could be cleaved into DSBs (Carroll, 2011; Figure 3). The application of ZFN in A. nidulans is relatively mature (Fayyaz et al., 2020), but still not been widely used due to the cumbersome process of constructing ZF domains and verifying their specificity, but also because this system has some cytotoxicity, and thus there have few studies in fungi.
2.2.2. Transcription activator-like effector nuclease
Transcription Activator-Like Effector (TALE) was first found in the plant pathogen Xanthomonas in 2009 (Boch et al., 2009). Each TALE contains a central region of tandem direct repeats composed of 33–35 amino acids (mostly 34), this sequence is conserved except for the 12 and 13th amino acid residues, which are named Repeat Variable Diresidue (RVD). The 12th amino acid residue stabilizes the RVD loop, whereas the 13th amino acid residue makes a base-specific contact in the target DNA bases (NI recognizes A, NG recognizes T, HD recognizes C, and NN recognizes G) and therefore TALENs are able to identify Single Nucleotide Polymorphisms (SNPs; Bedell et al., 2012; Figure 3). The composition of TALENs is similar to ZFNs in that their carboxylate terminal are both fused with a catalytic FokI endonuclease to produce a specific DSB. However, compared with the triplet base pairs recognized by each ZFN monomer, one TALEN monomer recognizes only one base and therefore a specific TALEN nuclease can theoretically be designed and constructed for any DNA sequence, to improve its specificity and flexibility (Cermak et al., 2011; Joung and Sander, 2013). At present, TALEN technology has been successfully applied to generate relatively stable genetic modification of human cells and model organisms including Drosophila, Caenorhabditis elegans, zebrafish, mice, and Arabidopsis thaliana (Fayyaz et al., 2020). In A. oryzae, it has been verified that the TALEN can achieve the deletion of large fragments of the target sequence (Mizutani et al., 2017). However, each base of the target sequence requires a TALE recognition module, so the construction process of TALENs still laborious.
2.2.3. Clustered regularly interspaced short palindromic repeats-CRISPR associated protein system
In 1987, Ishino et al. discovered the repeated sequences of palindromic repeats interspaced by a 32 bp sequence in E. coli (Ishino et al., 1987), and Dutch scientists Jansen et al. first named it the CRISPR and CRISPR-associated protein (Cas; Jansen et al., 2002). Subsequently, Barrangou et al. demonstrated that CRISPR-Cas is an adaptive immunity system present in bacteria and archaea (Barrangou et al., 2007). The in vitro remodeling of CRISPR-Cas9 (Jinek et al., 2012) and the demonstration of its gene editing function in human cells (Cong et al., 2013) marked the beginning of a new era of gene engineering. In 2015, following the successful genome editing of the CRISPR-Cas9 system in the model fungus N. crassa, Mortensen’s team established a universal CRISPR-Cas9 system for the first time in six species of Aspergillus (A. nidulans, A. niger, A. aculeatus, A. brasiliensis, A. carbonarius, and A. luchuensis; Nodvig et al., 2015).
2.2.3.1. Mechanism of CRISPR-Cas system
According to the complex of Cas proteins and the domains organization of the effectors, CRISPR-Cas systems are divided into two classes including six different types. Class 1 includes Type I, Type III, and Type IV, and need the Cas complex participating in the recognition and degradation of foreign DNA. Class 2 contains three types (II, V, and VI) and divided into 10 subtypes, which only need one Cas protein mediated by RNA to perform interference function, has been greatly developed in gene editing (Haft et al., 2005; Koonin et al., 2017).
The Class 2 Type II CRISPR-Cas9 systems have been studied relatively thoroughly. In the natural bacterial CRISPR-Cas9 system, the Cas9 contains two single stranded nuclease domains, RuvC (the crossover junction endodeoxyribonuclease) and HNH (named for characteristic histidine and asparagine residues). The guide RNA (gRNA, i.e., protospacer) exists as a CRISPR array, transcribed to form a primary transcript, and then processed into mature CRISPR-RNA (crRNA) with the participation of RNase III. Parts of infective viruses or phages DNA sequence would be integrated into the bacterial CRISPR array. When the foreign DNA invades again the mature crRNA combined with a trans-activating RNA (tracrRNA) to form crRNA: tracrRNA dimers, which can combine Cas9 protein and lead nucleases to degrade this foreign DNA based on complementary sequences (Barrangou et al., 2007; Jiang and Doudna, 2017; Figure 4A).
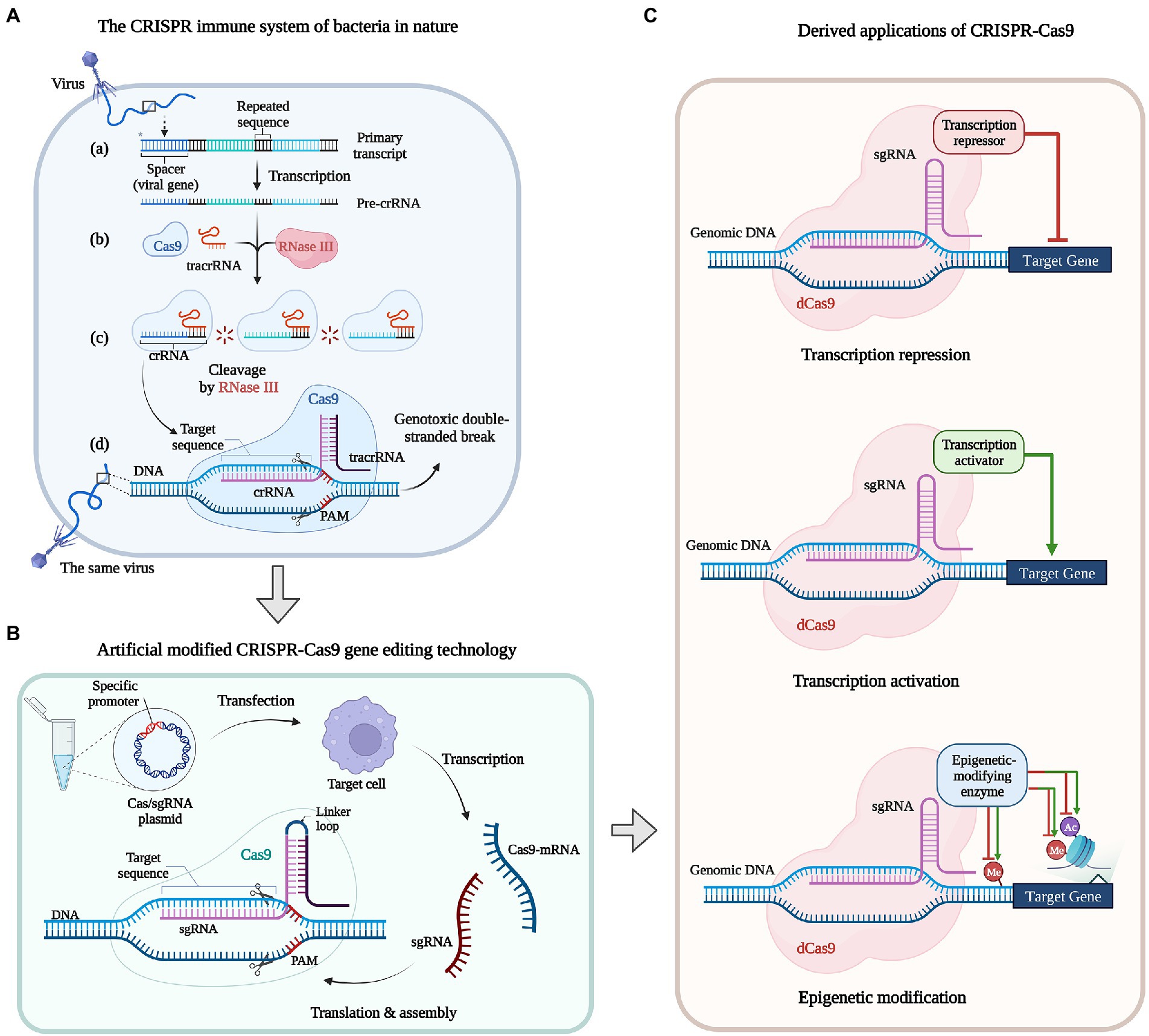
Figure 4. Development of the CRISPR-Cas system: natural, modified, and derived applications. (A) The CRISPR immune system of bacteria in nature. (a) Viral DNA is inserted into the bacterial genome as a spacer in between the repeated CRISPR sequences. (b) Trans-activating RNA (tracrRNA) recognize CRISPR-RNA (crRNA) sequences and target Cas9 enzymes to the crRNA. (c) The crRNA: tracrRNA duplex cleave the longer pre-crRNA by RNaseIII and then produce mature crRNA. (d) When the same virus infect bacterium again, the Cas9: crRNA generated during the first infection recognizes Protospacer Adjacent Motif (PAM) motifs in the viral genome, enabling target-DNA recognition and Cas9-mediated cleavage to prevent the re-infection. (B) Artificial modified CRISPR-Cas9 gene editing technology. In vitro designed Cas/ single guide RNA (sgRNA) expression plasmid(s) with specific promoters are transduced into target cells, Cas/sgRNA mRNAs are transcribed in vivo, which are translated, modified, and assembled to form a CRISPR-Cas9 system. (C) Derived applications of CRISPR-Cas9. dead Cas9(dCas9) can be used to mediate transcriptional repression, activation and epigenetic modification of target genes when fused with a small-molecule repressor, activator and epigenetic-modifying enzymes.
Currently, the modified CRISPR-Cas9 system derived from Streptococcus pyogenesis is the most mature type of CRISPR-Cas (Cong et al., 2013), and S. cerevisiae is the first fungal species that had implemented CRISPR-Cas9 for genome editing (DiCarlo et al., 2013). Adding a linker loop between crRNA and tracrRNA fuses the crRNA: tracrRNA dimers into a more refined single guide RNA (sgRNA), which composed of two parts (Pant et al., 2022). The 20-bp gRNA part responsible for identifying genomic DNA regions with a short Protospacer Adjacent Motif (PAM) composed of three bases of NGG (N stands for any base), and the gRNA scaffold part binding to Cas9 and activate its ability of cleavage (Jinek et al., 2012). The target strand hybridizing to gRNA and the complementary strand is, respectively, cleaved by HNH and RuvC then produce a blunt end DSB at the target region which is located 3–5 bases upstream of the PAM (Hsu et al., 2014; Figure 4B).
The Class 2 Type V effector Cas12a, also called Cpf1 (Cas in Prevotella and Francisella 1), requires no tracrRNA, which uses the guide crRNA alone to target dsDNA and activates RuvC to cut both strands of the target DNA then produce a sticky end DSB (Zetsche et al., 2015). Vanegas et al. first realized the application of Cpf1 in A. nidulans, which catalyzed oligonucleotide-mediated genomic site directed mutagenesis and marker-free gene targeting (Vanegas et al., 2019). The crRNA of Cas12a is significantly shorter and more flexible than Cas9 sgRNA, thus thought that Cas12a has the ability to target multiple genome loci simultaneously, as well as possess RNase activity in addition to its DNA cleavage activity. In addition, the new Class 2 Type VI effector Cas13 that can edit RNA, has considerable research value (Li et al., 2019; Safari et al., 2019).
2.2.3.2. Application of CRISPR-Cas system
Clustered Regularly Interspaced Short Palindromic Repeats-Cas systems provide diversified sequence-specific gene regulation tools that have led to various gene editing techniques. Cas9 nuclease becomes the Cas9 nickase (Cas9n) when inactivate the HNH or RuvC nuclease domain that induces single-strand break (also called nick) instead of DSB, but do not interfere with the binding and interaction of gRNA with the target DNA double strands. Thus CRISPR-Cas systems can be modified for diverse functions based on the nuclease deactivated Cas9 (dCas9) combined with regulatory parts (Figure 4C; Kundert et al., 2019; Uygun et al., 2020). For example, the CRISPR activation (CRISPRa) is fusing a transcriptional activator or multiple copies of the activation domain, while CRISPR interference (CRISPRi) is fusing repressor domains to the Cas9n enzyme that can block the elongation of transcripts (Qi et al., 2013). Epigenetic modifications at specific genomic locations can be achieved when fusing dCas9 with epigenetic-modifying enzymes, for example, the modification of methyl or acetyl groups for the histidine of target gene can affect gene function to a certain extent (Zhuo et al., 2021). Moreover, multiple sgRNA expression plasmids can be transferred into one cell to edit multiple genes synchronous, such as A. oryzae (Li et al., 2023) and even have the potential function of genome screening (Cong et al., 2013; Zetsche et al., 2017; Zeng et al., 2022). At present, a considerable part of common Aspergillus gene function studies have applied the CRISPR technology, also have applied in fungal factories, such as glucoamylase-hyperproducing industrial in A. niger (Liu D. et al., 2022) and kojic acid production in A. oryzae (Chen Z. et al., 2022).
2.2.3.3. Improvement of CRISPR-Cas system
Compared with ZFN and TALEN, CRISPR-Cas improved the accuracy of gene editing by gRNA matched to the target DNA region. In addition, unlike the FokI endonuclease must be dimerized to cleave targeted DNA, Cas performs functions as a monomeric protein thus avoiding delicate and complex protein design and assembly. However, Cas nucleases may inherit the low sequence specificity from the prokaryotic innate defense system, which increases the chance of non-specific cleavage and off-target effect.
Many strategies for components efficient expression, normal operation and function expansion have been developed. The operation of the CRISPR-Cas system requires the presence of two key component elements in hosts, Cas protein and sgRNA, Zheng et al. summarized three types of transformation in A. niger: (1) Cas protein expression plasmid and sgRNA expression plasmid both in DNA form, (2) sgRNA in RNA form and Cas protein expression plasmid in DNA form, and (3) RNP complex (Cas/sgRNA ribonucleoprotein complex) formed in vitro, which provides a useful reference for the application of CRISPR technology in Aspergillus (Zheng et al., 2021). The construction of Cas protein and sgRNA expression cassette is crucial, and the selection of promoters is related to the efficient expression of component elements and the successful application in target organisms. The promoters used for Cas protein expression are mostly constitutive strong promoters such as PgpdA from A. nidulans (Zhang et al., 2016), Ptef1 (Nodvig et al., 2015), PcoxA (Sarkari et al., 2017), and pkiA (Song et al., 2018), or inducible promoters such as PglaA from A. niger (Zhang et al., 2020). Fusing a nucleus localization signal on the plasmid can precisely translocate the Cas9 protein to the nucleus (Goeckel et al., 2019). The gRNA expression cassettes are divided into promoters recognized by RNA polymerase II or RNA polymerase III. The use of pol II promoter, including gpdA, mbfA, etc., requires the presence of ribosomal splicing sequence for pre-crRNA processing. The pol III promoter such as U6 (Zhang et al., 2016), the S. cerevisiae SNR52 promoter (Fuller et al., 2015), and the bacteriophage T7 promoter (Kuivanen et al., 2016), have been optimized for high transcription. Various tRNA promoters can also drive the sgRNA expression but the expression level is low (Song et al., 2018), and the endogenous 5S rRNA promoter proposed by Zheng et al. can significantly improve the expression level of sgRNA and achieve 100% gene targeting inactivation efficiency (Zheng et al., 2019). More professional and detailed description has been reviewed by Zheng et al. (2021) and references therein. Adding a cell-specific promoter to the Cas or sgRNA expression vector can achieve the spatiotemporal control of gene editing, and counteracts undesired side effects in non-target cells (Zhou et al., 2018; Maggio et al., 2020). Other studies showed that both Cas9 and Cas12a proteins can be engineered to recognize different PAM sequences thus expanded the application scope (Corsi et al., 2022; Zhu et al., 2022). Kleinstiver et al. broadened the targeting range of Staphylococcus aureus Cas9 by modifying PAM sites in human cells, although the specificity and off-target effect remains not very ideal (Kleinstiver et al., 2015a,b).
Improvements in the high off-target rate of CRISPR can be achieved by modifying the components. One approach is to design truncated guide sequences, which minimize mismatches and secondary structures of sgRNA that reduce off-target binding (Fu et al., 2014). Modifications directed to control active Cas amount can reduce unnecessary and non-specific cleavage, some physicochemical methods such as light (Nihongaki et al., 2015) or exogenous small molecules (Dow et al., 2015) can partly inhibit the Cas9 expression. Another way is to use the Cas nickase instead of the Cas nuclease, producing the single-strand break instead of the double-strand break, which does not induce NHEJ repair and still activates precise HDR repair (Kuo et al., 2020). Single-strand breaks can reduce off-target effects, but also reduce repair efficiency, so it has been proposed to use double nickases, such as two Cas9ns (Ran et al., 2013), two cpf1n (Suzuki et al., 2016; Zetsche et al., 2017), and dCas9-FokI dimers (Tsai et al., 2014), to improve the specificity and efficiency of gene editing.
Due to its simplicity, operability, and efficiency, CRISPR has been widely used in a variety of advanced eukaryotes that are difficult to operate with traditional gene editing strategies. Meanwhile, several laboratories around the world have developed various CRISPR-Cas systems for Aspergillus (Nakamura et al., 2021). Recently studied in A. flavus, the CRISPR-Cas9 utilized the autonomous maintenance in Aspergillus (AMA1) autonomously replicating sequence has achieved relatively efficient multiple-gene knockouts, satisfactory gene-targeting efficiencies (>90%) have also been obtained in A. nidulans, A. fumigatus, A. terreus, and A. niger (Chang, 2023). It likely have a broad application in aspergilli.
2.2.4. Base editor
Zinc Finger Nuclease, TALEN, and CRISPR-Cas all rely on Non-Homologous End Joining (NHEJ) and Homology Directed Repair (HDR), which induce breaks at target sites to activate DNA repair. NHEJ is prone to cause frameshift mutations, which in turn affect the function of target genes. Although the accuracy of HDR is higher than NHEJ, its efficiency in cells is very low, only about 0.1–5%. The Base Editor (BE) technology does not require DSBs or homologous templates to perform single base conversion, which has effectively improved the above problems (Komor et al., 2016). BE technique combines nuclease-free dCas9 or Cas9n, cytosine deaminase, DNA Uracil Glycosylase Inhibitor (UGI), and sgRNA. It is possible to directly deaminates cytosine (C) to uracil (U) at the targeted site without DSBs, and the U is not excised due to the presence of uracil glycosylase inhibitor. As DNA replicates, U is replaced by thymine (T), and guanine (G) that was originally complementary to C will be replaced with adenine (A), finally achieving single-base precise editing of C to T and G to A within a certain active frame (Gaudelli et al., 2017; Chen L. et al., 2022). The advent of BE technology has promoted the effectiveness and scope of use of point mutation gene editing. If the termination codon (TAA, TGA, and TAG) appears in advance, gene knockout can be achieved. For multi-copy sites, Cas9 will produce multiple DSBs, causing DNA damage and even apoptosis, so under the same conditions, BE-induced terminator generation to achieve gene knockout is safer than Cas, and can be used as a safe and efficient genome-wide screening system (Chen L. et al., 2022; Liu N. et al., 2022).
2.3. RNA technologies
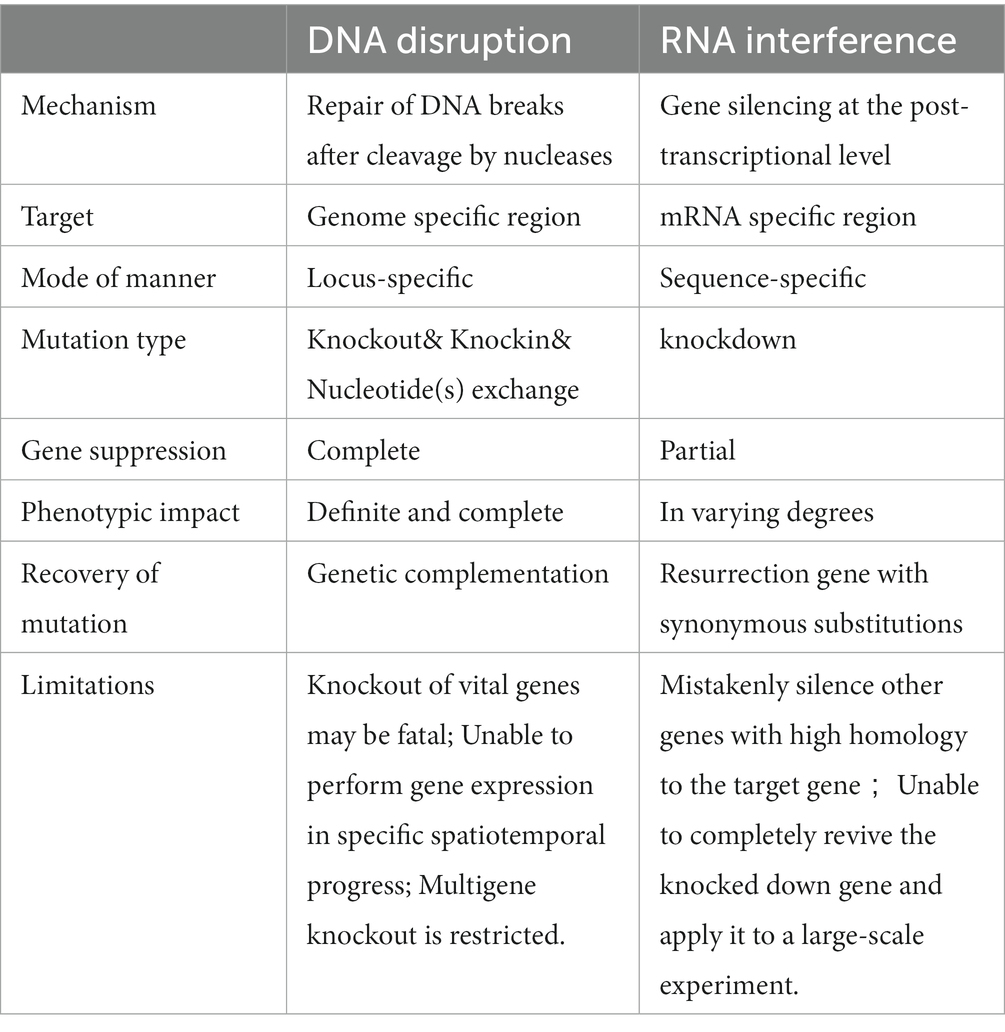
Some RNA technologies, including RNA interference (RNAi) and ribozymes, can verify gene function through disturbing mRNA translation have also applied in fungi.
RNA interference (RNAi) evolved by eukaryotes is a conserved transcriptional and post-transcriptional levels gene silencing mechanism, which is involved in multiple biological processes, particularly in host defense and gene regulation (Fire et al., 1998). The filamentous fungus Neurospora crassa is one of the first organisms used for RNAi studies in 1992 (Romano and Macino, 1992). RNAi is adopted by inactivating RNA translation via the RNase III enzyme (Dicer) in an ATP-dependent manner without affecting the normal gene expression (Fire et al., 1998; Borges and Martienssen, 2015; Nargesi et al., 2021; Figure 5). It should be noted that due to the high specificity and potential efficiency of RNAi inhibitory gene expression, a relatively less amount of dsRNA molecules (far less than the amount of endogenous mRNA) can completely inhibit the expression of the corresponding gene, which carried out in a catalytic scale-up and may cause needless and even harmful gene silencing. Therefore, the effective expression of various genes in normal body has a set of mechanisms to strictly prevent the formation of dsRNA (Liu et al., 2021; Wang et al., 2021). The RNAi technology has been used in many higher eukaryotes by transferring designed siRNA into the host for gene knockdown. Methods for siRNA preparation include chemical synthesis, in vitro transcription, degradation of long dsRNA by RNase III, expression of siRNA in cells through siRNA expression vector or viral vector, and siRNA expression frame prepared by PCR (Gebremichael et al., 2021). In summary, RNA silence has the potential to offer efficient tools that gene disruption methods cannot provide in the exploration of gene function, but there are also shortcomings (Table 2). Detailed analyses were carried out by Abo-Al-Ela (2021) and Gebremichael et al. (2021).
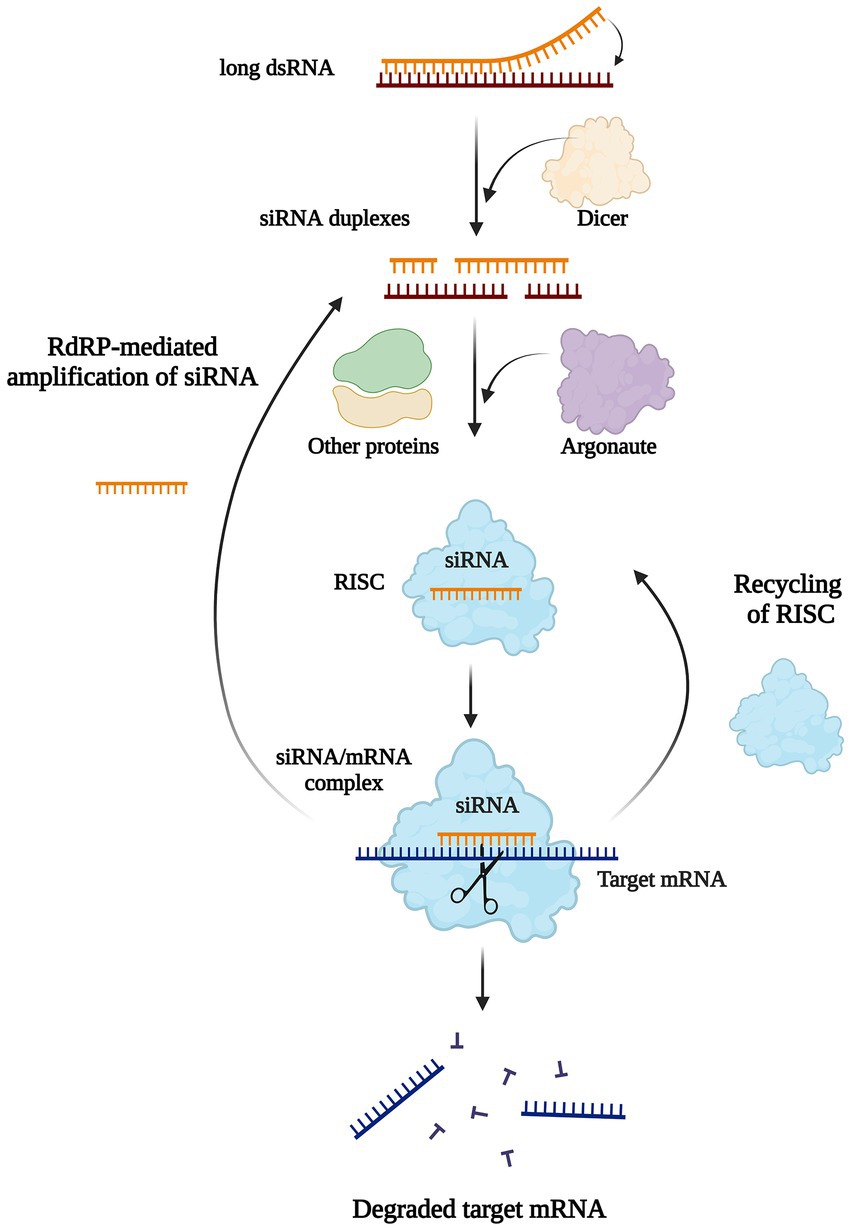
Figure 5. Biological mechanisms of RNAi-mediated gene silencing. Long double-stranded RNAs or hairpin RNAs bind to Dicer are cleaved, and then generate multiple 21–23 bp double-stranded small interfering RNAs (siRNAs) duplexes. Each siRNA is unchained into a passenger (sense) strand and a guide (antisense) strand by intracellular RNA helicase. The guide strand binds with Argonaute (AGO) and other proteins to form an RNA Induced Silencing Complex (RISC), while the passenger strand is degraded by subsequent cellular events in the cytoplasm. The siRNA/RISC complex then binds the complementary mRNA resulting in the degradation of the target transcript or inhibition of translation. The components of siRNA/mRNA complex can be recycled to the RISC complex or generate siRNA duplexes by the action of RNA-dependent RNA Polymerase (RdRP).
In 1980, the ribozyme (also known as catalytic RNA) was found to have the specific catalytic function similar to enzyme, which expands the range of enzymes from proteins to nucleic acids. Ribozymes can specifically recognize and cut target mRNAs then detach from degraded mRNAs and bind to other target mRNAs without affecting the host RNA. Ribozymes are widely exist in viruses, bacteria, and lower eukaryotes. At present, seven types have been recognized, which are Type I introns, Type II introns, RnaseP, hammerhead ribozymes (HH), hairpin ribozymes, Neurospora varkud Satellite (VS) ribozymes, and hepatitis delta virus (HDV) ribozymes (Deng et al., 2022). Among them, the HH consists of only 30–50 nt and requires divalent metal ions such as Mg2+ to participate in the catalytic process, and the specific cleavage site is at the 3′ end of triplet NUX (N stands for any base; X stands for C, U, A, but not G; Teplova et al., 2020). It is the only ribozyme that has been successfully applied to filamentous fungi and A. giganteus is the first reported case, with seven different designed hammerhead ribozymes that could reduce the beta-glucuronidase (uidA) gene expression by up to 100% (Mueller et al., 2006). Compared with other gene silencing strategies, ribozymes possess unique properties such as smaller in molecular weight, more flexible rational design and relative stability in organisms. Therefore, ribozymes can also be used in combination with other techniques such as CRISPR, where Gao et al. fused a hammerhead-type ribozyme and a hepatitis delta virus ribozyme sequences flanking the sgRNA expression plasmid, and the resulting sgRNA then was released by activities of these two ribozyme more efficient and accurate (Gao and Zhao, 2014). Moreover, the heritability of ribozyme is not stable, and the necessary cofactors should meet the need for intracellular catalysis without imposing a harmful burden on cells. In addition, the efficiency of biological action on target mRNA requires to improve and appropriately modify ribozymes to prevent degradation (Torres-Martinez and Ruiz-Vazquez, 2017; Wang et al., 2017).
3. Transformation methods
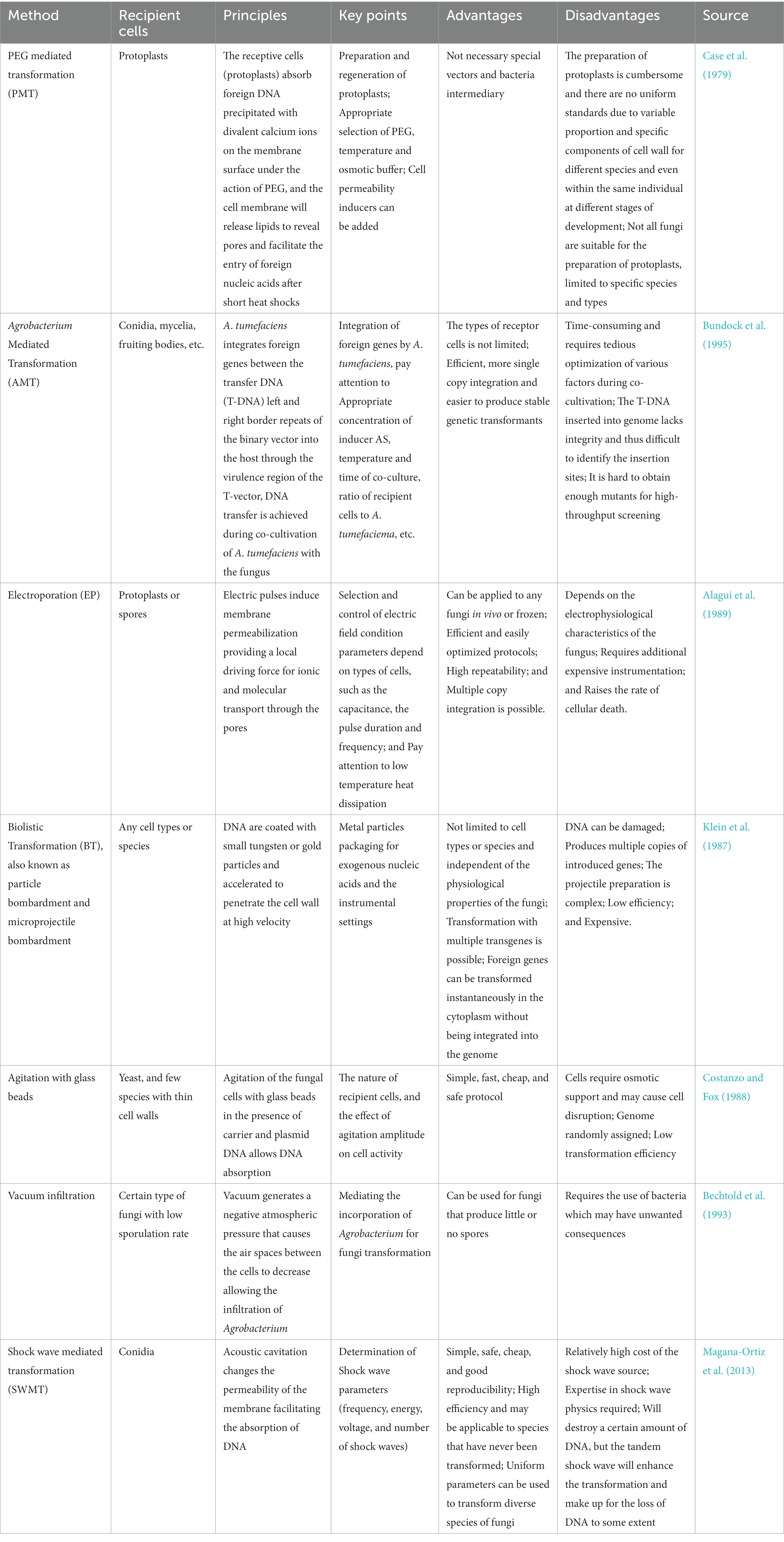
Transformation refers to the entry of foreign genetic biomolecules carrying modification information into the recipient cell, which may hampered by varying extents of the rigid cell wall and the multicellular structure of fungi. Allowing exogenous biomolecules (including DNA/RNA fragments, circular or linearized plasmids, and ribonucleoproteins) to pass through the cell wall and into as many as recipient cells are keys to breaking the bottleneck of low transformation efficiency of fungi. Currently, several proven transformation methods have been developed for Aspergillus spp., including Polyethylene glycol (PEG) Mediated Transformation (PMT) of protoplasts, Agrobacterium tumefaciens Mediated Transformation (AMT), Electroporation (EP), and Biolistic Transformation (BT). For a specialized review that focuses on transformation methods with filamentous fungi, the readers are directed to Yue et al. (2021). In addition, there are some physical methods, such as agitation with glass beads, vacuum infiltration, Shock Wave Mediated Transformation (SWMT), etc., which are relatively inefficient but can improve the shortcomings of the universal methods and play an auxiliary role (Table 3; Rivera et al., 2014; Li et al., 2017). The rate of these transformation approaches depends on the technique and the target species. However, some effective and simple methods that established in mammalian cell researches are not suitable for fungi, such as viral vector mediated transformation, nanomaterial mediated transformation, and liposome mediated transformation (Wang, 2021). Combining transformation technologies and genome editing technologies is expected to realize the best modification of fungal genomes.
4. Challenges and solutions to gene editing technology
The main processes of gene editing technology include the transference of foreign target DNA fragments into the recipient cell nucleus, correct integration into the genome and efficient expression of target genes. Currently, target fragments are usually generated in the form of linear PCR cassettes, which are available mainly through two systems. One is the split-marker PCR system that requires two DNA products containing overlapping fragments of a selective marker. Another is the fusion PCR system that requires fusing three PCR products, two for the HR in the genomic DNA flaking a selection region and the other for a selective marker and/or the replaceable promoter (Yon and Fried, 1989; Szewczyk et al., 2006). It is also possible to use plasmids to carry fragments of interest, and both linear and circular cassettes are suitable for transformation. But more importantly, there are still great challenges for the correct homologous recombination of foreign DNA sequences into the genome and the efficient expression of target genes.
4.1. Improve the efficiency of homologous recombination
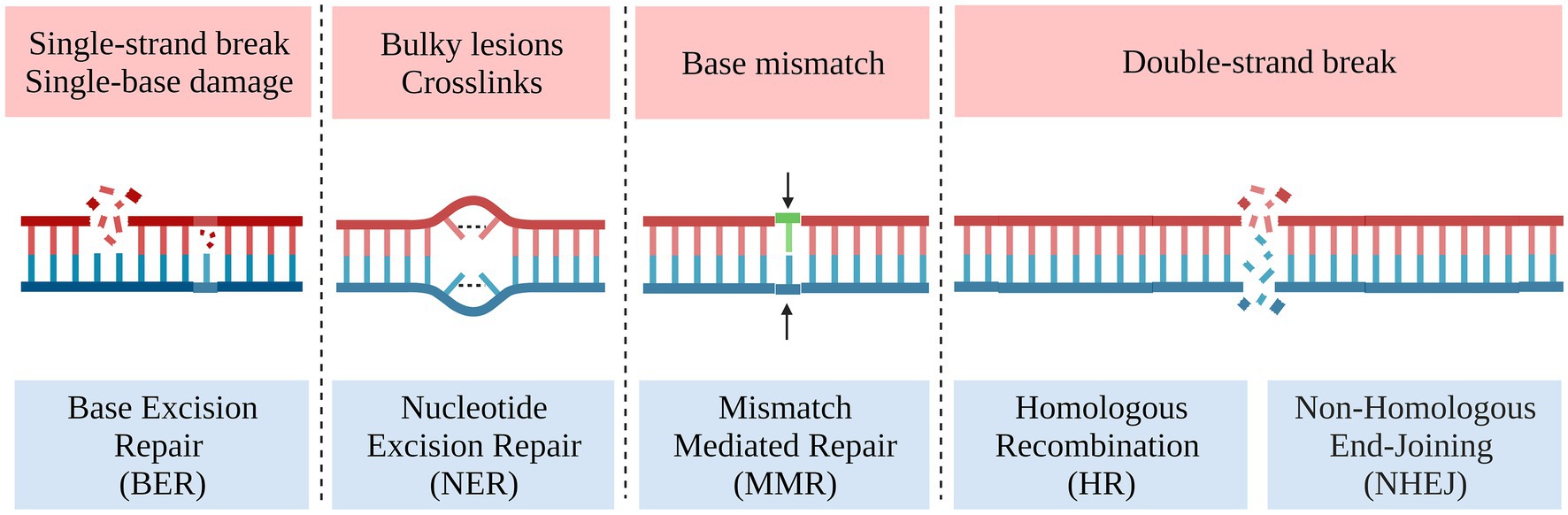
Genetic damage and repair occur in cells at any time, accordingly, there are a variety of repair systems for different DNA damage (Figure 6; Ciccia and Elledge, 2010). The Double-Strand Break Repair (DSBR) systems in eukaryotic cells include the Non-Homologous End Joining (NHEJ), which is simpler but prone to random insertions and deletions, and Homology Directed Repair (HDR) that requires the presence of a homologous template to activate, which is necessary for the precise integration of DNA fragments into specific genomic loci (Szostak et al., 1983; Scully et al., 2019). However, eukaryotic microorganisms have a complex genetic background and inherently less HDR efficiency, and at least two generations are required to obtain stable homozygotes. All these are important reasons limiting the application of gene editing technology and solutions always from the perspective of improving HDR efficiency.
4.1.1. Physicochemical methods
Firstly, synchronize the cell cycle. In wild-type cells, the HDR functions only in the S and G2 phases of the cycle, while NHEJ can occur throughout all the cell cycle phases (Deriano and Roth, 2013). Usually, the G1/G0 phase of cells has the DNA content of diploid cells (2 N), while the G2/M phase has the DNA content of tetraploid cells (4 N), while the DNA content of the S phase is between diploid and tetraploid. The detection of intracellular DNA content by insertive nucleic acid fluorescent dye combined with flow cytometry can screen out S phase cells. Furthermore, some small molecules, such as hydroxyurea (HU) and nocodazole, can prevent the reduction of nucleotides to deoxynucleotides and reversibly inhibit DNA synthesis in the S phase without affecting other cell cycle operations (Rosenkranz and Levy, 1965; Sinclair and Morton, 1965). But these cell cycle synchronization methods inevitably produce abnormal cell division and growth.
Alternatively, adding enhancers or inhibitors targeting DSBR components. The main components involved in NHEJ including KU70-KU80 heterodimer that binds to the cleaved double-strand DNA end to recruit other proteins and pull the two separated DNA segments together. DNA-PKCS and its substrate Artemis, which are responsible for hydrolyzing the terminal single-stranded region at the end of DNA to create an effective substrate for ligase. DNA ligase IV together with XRCC4 catalyzes the formation of a new linkage between the DNA termini that have been processed by Artemis (Scully et al., 2019). Previous studies suggested that various small molecule compounds that repress the NHEJ pathway can effectively enhance the efficiency of HDR, such as the W7 and chlorpromazine (inhibit the production of KU co-factor), the vanillin, Nu7026 and Nu7441 (inhibit DNA-PKCS activity), and the most commonly used inhibitor Scr7 (inhibit DNA ligase IV activity; Ying et al., 2019). On the other hand, it has been reported that RS-1 acts as an enhancer and promotes the HDR efficiency of mammalian cells, but there have been no reports in fungi (Jayathilaka et al., 2008). However, it is worth noting that some molecules are cytotoxic and need to determine the threshold and range of toxic effects.
4.1.2. Biotechnological methods
Successful HDR requires single or double homologous fragments between the donor and acceptor DNA, but base mismatches will seriously reduce its frequency, and thus the length of the flanking sequences is an important decisive factor of HDR efficiency. Different organisms require different homologous arm lengths, and the upper limit of homologous recombination efficiency can be achieved varies greatly (Fujitani et al., 1995). In yeast, 50–100 bp homologous sequences can accomplish gene replacement and achieve 50–100% homologous recombination efficiency (Takita et al., 1997), while filamentous fungi require at least 1 kb homologous region but the efficiency of correct homologous recombination is still below 30%. It is necessary to determine the optimal length based on the species of Aspergillus, the size of the inserted DNA fragment, and similar properties of the system.
Non-Homologous End Joining and HR are two different repair pathways but NHEJ dominants DSB repair, so inhibiting the function of essential factors (including KU70/KU80, DNA ligaseIV, and their homologs) in the NHEJ pathway would improve the efficiency of HDR. Ninomiya Y et al. reported that inactivation of the NHEJ pathway led to 100% homologous recombination efficiency in N. crassa, and then the approach was rapidly developed in other filamentous fungi (Ninomiya et al., 2004). Currently, it is widely used to construct engineered Aspergillus with convenient NHEJ defect background to achieve efficient further gene editing, such as ΔKu70/Ku80 (including A. nidulans, A. fumigatus, A. sojae, A. oryzae, A. niger, A. parasiticus, A. flavus, A. chevalieri var. intermedius, and A. westerdijkiae), and ΔligD (including A. oryzae and A. luchuensis; Ying et al., 2019; Madhavan et al., 2022). In addition, in strains loss of NHEJ ability, the integration efficiency is highly dependent on the targeted gene locus.
4.2. Efficient expression of foreign genes
The expression of heterologous genes can be limited in transcription, post-transcription, and translation levels. Several genetic strategies have been applied to reduce the expression constraints and enhance gene functionality.
Theoretically, increasing the copy number of the target gene can improve its transcription level, but it also depends largely on the integration place of the expression cassette in the genome (Graessle et al., 1997). AT-rich sequences in the coding regions act as internal polyadenylation sequences and produce incomplete transcripts that will limit heterologous gene expression at the transcriptional level, replacing an AT-rich sequence with a more GC-rich sequence can overcome the premature termination of transcription (Roberts et al., 1992). In addition, the presence of introns in eukaryotic systems can effectively improve mRNA stability. Deletion of introns in A. niger almost completely inhibited endogenous acid lipase expression (Zhu et al., 2020), and artificially adding introns to heterologous protein genes may be another way to improve mRNA stability (Deckers et al., 2020).
A more feasible method is the modification of the pre-transcriptional level, using strong promoters or optimized codons in desired gene cassettes for integration. Since the original pathway regulation is interactive and the expression of a gene is coordinated by both global and pathway specific multiple systems, selecting the appropriate promoter can be difficult but necessary. So far, a series of strong promoters are divided into constituent promoters and inducible promoters, which were developed to induce the expression of heterologous genes. The constituent promoters mainly include translation-elongation factor 1α (tef1), glyceraldehyde-3-phosphate dehydrogenase (gpdA), pyruvate kinase (pkiA), phosphoglycerate kinase (pgkA), etc. The inducible promoters mainly include xylose (xlnA), glucoamylase alpha (glaA), alpha-amylase (amyA), xylanase (exlA), cellobiohydrolase I (cbh1), heat shock protein (hsp70), etc. (Cazier and Blazeck, 2021; Garrigues et al., 2021; Zhuo et al., 2021). In addition, the alterations in 5’UTR can be used in combination with various strong promoters to enhance the expression of heterologous genes (Cao et al., 2021).
Genes show a nonrandom usage of synonymous codons and well-expressed genes are highly biased toward a subset of the present synonymous codons, significant heterogeneity in the codon usage exists among genes within species and the grade of codon bias is positively correlated with gene expression (Karlin and Mrazek, 2000). Therefore, codon optimization is also an effective method for attaining faster translation rates and higher accuracy, genes can be manipulated with rare codons replaced by their optimal synonyms and then reconstructed by overlap extension of synthetic oligonucleotides, improving the expression levels of heterologous genes (Koushki et al., 2011; Tanaka et al., 2014).
4.3. Screening for correct transformants
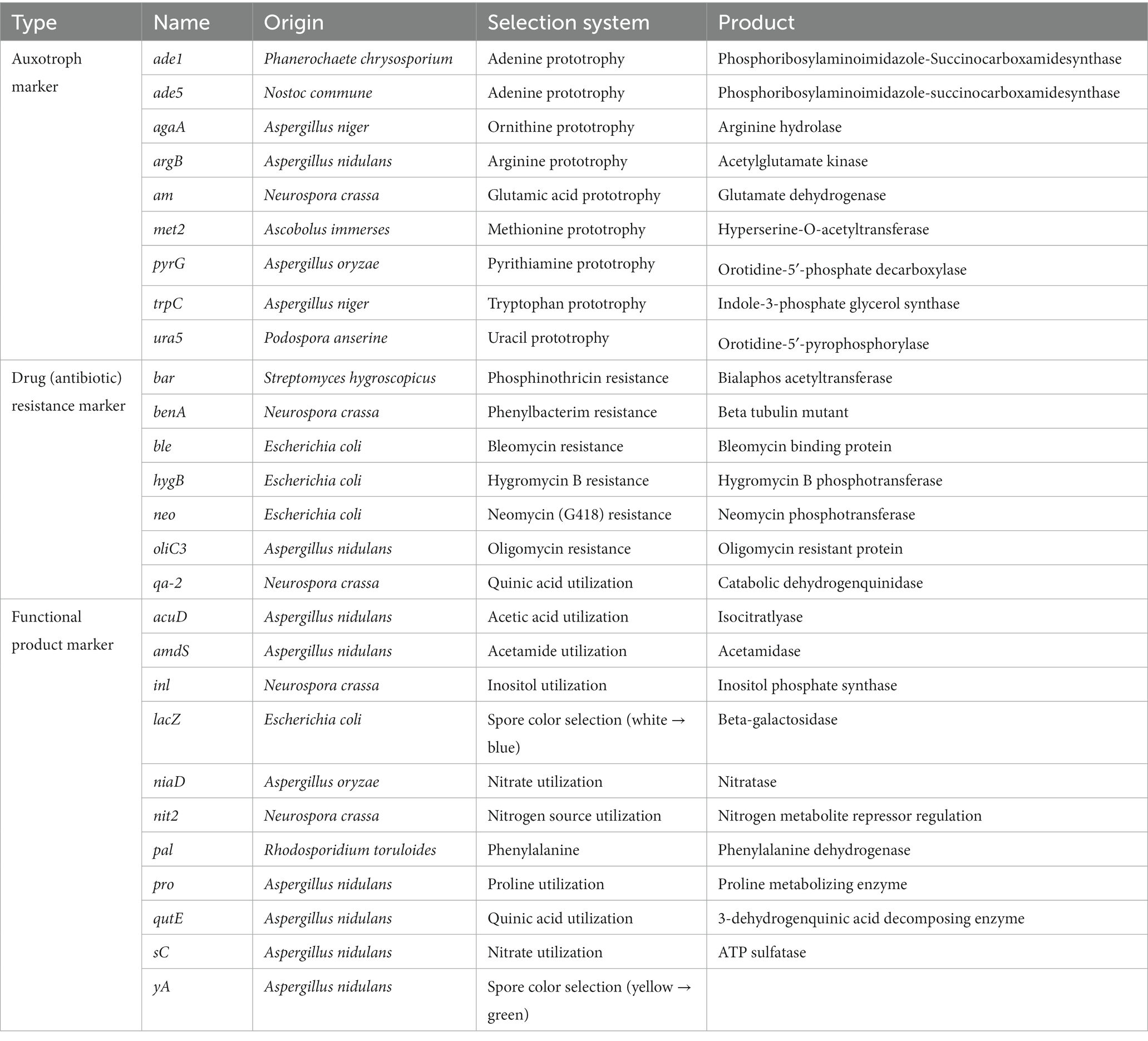
Effective selective markers are critical for molecular manipulation, including drug resistance genes, autotrophic or nutritional genes, and visual distinction reporters, which are all used for screening positive filamentous fungi (Table 4; Jin et al., 2021).
Drug (antibiotic) resistance genes are generally used as dominant selection markers in species that are no endogenous drug resistance. For example, the A. flavus NRRL 3357 is resistant to hygromycin, so hygB gene cannot be used as the marker gene, while the ble and pyr genes have been successfully used for A. flavus transformation. The other limitation is that antibiotics are usually expensive, and there are concerns about issues with the contamination of genetically modified organisms. Auxotrophic genes encode essential proteins for the biosynthesis of necessary nutrients, among them, the pyrG gene is the most widely used marker. PyrG is involved in uridine biosynthesis and is also a target for the antimetabolite 5-fluoroorotic acid (5-FOA). The pyrG deficient mutants are resistant to 5-FOA, meanwhile, it could not grow in media without uridine or uracil because of the loss of ability to synthesize uracil autologously. Therefore, it can show a two-way screening effect and distinguish the wild type and mutant from two perspectives (Wang et al., 2022). Fluorescent signal genes encode fluorescent proteins that can be visually distinguished, such as enhanced Green Fluorescent Protein (eGFP) and Discosoma sp. Red fluorescent protein (DsRed; Chen et al., 2019). It is relatively simple to operate but requires special observation instruments. In addition, phenotypic differences, such as pigment production, colony color or traits between wild-type strains and mutant strains can be used to achieve marker free screening of mutants, albeit only in individual samples. For A. fumigatus, the mutants of expected genomic alteration can be screened by the colorless (albino; Fuller et al., 2015). In A. carbonarius, the yellow conidia mutants can also be clearly distinguished from the wild type of the black conidia (Weyda et al., 2017).
Notably, the combination of multiple selection markers can greatly reduce false positives and enable the construction of multi-gene mutants or complemented strains conveniently. However, there are still fewer available selection markers for continuous genetic replacement, and may not be suitable for any strain. The solution is to combine recombinase and reuse marker genes, which allows the same selection gene for screening the positive clones. Zhang et al. constructed a one-step unmarked genetic modification by the Cre-loxP based CRISPR-Cas9 system, the target gene is knocked out through the CRISPR-Cas9 system, and Cre recombinase is induced by light and then deletes selection markers, which ensures that the same marker can be used for future genetic manipulation (Zhang et al., 2016). This one-step unmarked genetic modification system has been successfully applied in Hypocrea jecorina Qm6a, N. crassa, A. niger, Fusarium graminearum, Metarhizium anisopliae, etc. (Connolly et al., 2018; Enghiad et al., 2021; Rozhkova and Kislitsin, 2021).
5. Conclusion and future directions
Aspergillus with powerful and attractive ability to synthesize, modified, and secrete various metabolites. Filamentous fungi express more than 50% of industrial enzymes, in which Aspergillus plays a vital role in expressing these industrial enzymes (Arnau et al., 2020). Compared to bacteria and yeasts, Aspergillus performs polarized cell growth, where hyphal tip secretes proteins intensively and efficiently, reaching 10–1,000-fold of bacterial, yeast, or mammalian cells. On the other hand, Aspergillus has a variety of enzymes that can decompose and utilize a variety of carbon and nitrogen sources, as cell factories which have more diverse fermentation substrates, and extracellular proteins are easier to purify. But the development of filamentous fungal expression platforms is much more complex, time-consuming, and needs extensive development. At present, A. niger and A. oryzae have been used as mature cell factories to produce enzymes, recombinant proteins, antibiotics, organic acids, polyunsaturated fatty acids, nutrients, etc. in industrialization, species of Aspergillus can decompose plastics and pollutants, it has been widely used in the pharmaceutical, agricultural and food industries, as well as the environmental field, safely, efficiently, and economically (Ying et al., 2019; Li et al., 2022). Even mycotoxins synthesis, which is considered harmful and regulated comprehensively through complex pathways such as anabolism and catabolism, can be controlled in the cascade of external environmental factors (Gao et al., 2021), the knockdown of key genes in mycotoxin biosynthesis gene clusters or cross-regulatory pathways can prevent and control harmful mycotoxin synthesis from the source. Overexpression of specific genes can activate silencing pathways and may discover more useful secondary metabolites (Yuan et al., 2022). Therefore, the study of Aspergillus inevitably needs to go deep into the genetic level, the exploration of mechanisms and modification of gene functions can be realized through gene editing technologies by pathways reconstruction or activating silent pathways. Gene manipulation systems require vector construction, transformation, and genome editing, with the ultimate goal of achieving the expression of desirable traits and stable inheritance. But opportunities and challenges coexist, firstly, Aspergillus genome background is limited, and up to now the available gene information is still lacking except for model strains. For transformation, compared with other microorganisms such as yeast and bacteria, the transfer of exogenous vectors is blocked by rigid cell wall and complex mycelial structure. From the perspective of gene fragment integration, due to the intrinsic ploidy, propagation, and inefficient homologous recombination, it has not been as efficient as desired, and so far, no industrial strains built using the CRISPR technology have been used commercially. In conclusion, the search for a versatile, effective and stable genetic tool suitable for Aspergillus still confronted with many challenges.
Traditional gene-level gene editing strategies have certain technological barriers, such as the design process being complex and relatively costly and time-consuming, the off-target rate being high, only a single gene can be modified at a time, and only simple gene knockout or knockin can be performed. Further transcription-level editing can enable genome-scale knockout screening, genome architecture engineering, and RNA editing. The advent of new technologies, such as CRISPR-Cas, provides brand new ideas and greater operable space. The selective uniting of multiple gene editing technologies and the combination with efficient expression systems have significantly improved the precision and accuracy of biotechnology and applications, and the scope of function is evolving from single to multi-gene and from low to high throughput (Yilmaz, 2021; Antony et al., 2022). In the future, other types of genetic modifications may be achieved, such as gene motivation and gene interference with a controlled degree of expression, exploring more orthologues of functional enzymes within organisms, as well as cross-species gene mapping. Under the premise of ethics, gene editing technology is bound to become a powerful tool for humans.
Author contributions
JG and ZL: conceptualization and formal analysis. JG and ZZ: writing—original draft preparation. JG and HL: writing—review and editing. ZL and HK: supervision and validation. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (No. 32172170) and Graduate Independent Innovation Research Foundation of China Agricultural University (No. 2022TC160).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abo-Al-Ela, H. G. (2021). RNA interference in aquaculture: a small tool for big potential. J. Agric. Food Chem. 69, 4343–4355. doi: 10.1021/acs.jafc.1c00268
Alagui, A., Apparu, M., Dhardemare, A. D., Riche, F., and Vidal, M. (1989). Stability of the bond in TC-99M=N as a function of pH—demonstration of the transformation of TC-99M NCl4 - to TC-99M 04. Appl. Radiat. Isot. 40, 813–817. doi: 10.1016/0883-2889(89)90102-0
Antony, J. S., Hinz, J. M., and Wyrick, J. J. (2022). Tips, tricks, and potential pitfalls of CRISPR genome editing in Saccharomyces cerevisiae. Front. Bioeng. Biotechnol. 10:924914. doi: 10.3389/fbioe.2022.924914
Arnau, J., Yaver, D., and Hjort, C. M. (2020). “Strategies and challenges for the development of industrial enzymes using fungal cell factories” in Grand Challenges in Fungal Biotechnology. ed. H. Nevalainen (Cham: Springer International Publishing), 179–210.
Barrangou, R., Fremaux, C., Deveau, H., Richards, M., Boyaval, P., Moineau, S., et al. (2007). CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712. doi: 10.1126/science.1138140
Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. 82, 259–266. doi: 10.1385/0-89603-391-0:259
Bedell, V. M., Wang, Y., Campbell, J. M., Poshusta, T. L., Starker, C. G., Krug, R. G. II, et al. (2012). In vivo genome editing using a high-efficiency TALEN system. Nature 491, 114–118. doi: 10.1038/nature11537
Boch, J., Scholze, H., Schornack, S., Landgraf, A., Hahn, S., Kay, S., et al. (2009). Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326, 1509–1512. doi: 10.1126/science.1178811
Borges, F., and Martienssen, R. A. (2015). The expanding world of small RNAs in plants. Nat. Rev. Mol. Cell Biol. 16, 727–741. doi: 10.1038/nrm4085
Bundock, P., Dendulkras, A., Beijersbergen, A., and Hooykaas, P. J. J. (1995). Transkingdom T-DNA transfer from Agrobacterium tumefaciens to Saccharomyces cerevisiae. EMBO J. 14, 3206–3214. doi: 10.1002/j.1460-2075.1995.tb07323.x
Cao, J., Novoa, E. M., Zhang, Z., Chen, W. C. W., Liu, D., Choi, G. C. G., et al. (2021). High-throughput 5 ' UTR engineering for enhanced protein production in non-viral gene therapies. Nature. Communications 12, 4138–4147. doi: 10.1038/s41467-021-24436-7
Capecchi, M. R. (1989). Altering the genome by homologous recombination. Science 244, 1288–1292. doi: 10.1126/science.2660260
Carroll, D. (2011). Genome engineering with zinc-finger nucleases. Genetics 188, 773–782. doi: 10.1534/genetics.111.131433
Case, M. E., Schweizer, M., Kushner, S. R., and Giles, N. H. (1979). Efficient transformation of Neurospora crassa by utilizing hybrid plasmid DNA. Proc. Natl. Acad. Sci. U. S. A. 76, 5259–5263. doi: 10.1073/pnas.76.10.5259
Cazier, A. P., and Blazeck, J. (2021). Advances in promoter engineering: novel applications and predefined transcriptional control. Biotechnol. J. 16:e2100239. doi: 10.1002/biot.202100239
Cermak, T., Doyle, E. L., Christian, M., Wang, L., Zhang, Y., Schmidt, C., et al. (2011). Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 39:e82. doi: 10.1093/nar/gkr218
Chang, P.-K. (2023). A simple CRISPR/Cas9 system for efficiently targeting genes of Aspergillus section Flavi species, Aspergillus nidulans, Aspergillus fumigatus, Aspergillus terreus, and Aspergillus Niger. Microbiol. Spectr. 11:e0464822. doi: 10.1128/spectrum.04648-22
Chaveroche, M. K., Ghigo, J. M., and d’Enfert, C. (2000). A rapid method for efficient gene replacement in the filamentous fungus Aspergillus nidulans. Nucleic Acids Res. 28:97e. doi: 10.1093/nar/28.22.e97
Chen, Z., Chen, T., Wang, H., Jiang, C., Liu, Y., Wu, X., et al. (2022). Disruption of Aokap6 near the kojic acid gene cluster affects the growth and kojic acid production in Aspergillus oryzae. World J. Microbiol. Biotechnol. 38:175. doi: 10.1007/s11274-022-03361-x
Chen, L., Zhang, S., Xue, N., Hong, M., Zhang, X., Zhang, D., et al. (2022). Engineering a precise adenine base editor with minimal bystander editing. Nat. Chem. Biol. 1, 101–110. doi: 10.1038/s41589-022-01163-8
Chen, Z., Zheng, W., Chen, L., Li, C., Liang, T., Chen, Z., et al. (2019). Green fluorescent protein and Discosoma sp. red fluorescent protein tagged organelle marker lines for protein subcellular localization in rice. Front. Plant Sci. 10:1421. doi: 10.3389/fpls.2019.01421
Chiara, M., D'Erchia, A. M., Gissi, C., Manzari, C., Parisi, A., Resta, N., et al. (2021). Next generation sequencing of SARS-CoV-2 genomes: challenges, applications and opportunities. Brief. Bioinform. 22, 616–630. doi: 10.1093/bib/bbaa297
Ciccia, A., and Elledge, S. J. J. M. C. (2010). The DNA damage response: making it safe to play with knives. Mol. Cell 40, 179–204. doi: 10.1016/j.molcel.2010.09.019
Cong, L., Ran, F. A., Cox, D., Lin, S., Barretto, R., Habib, N., et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. doi: 10.1126/science.1231143
Connolly, L. R., Erlendson, A. A., Fargo, C. M., Jackson, K. K., Pelker, M. M. G., Mazzola, J. W., et al. (2018). “Application of the Cre/lox system to construct auxotrophic markers for quantitative genetic analyses in Fusarium graminearum” in Plant Pathogenic Fungi and Oomycetes: Methods and Protocols Vol. 1848. eds. W. Ma and T. Wolpert, 235–263.
Corsi, G. I., Qu, K., Alkan, F., Pan, X., Luo, Y., and Gorodkin, J. (2022). CRISPR/Cas9 gRNA activity depends on free energy changes and on the target PAM context. Nature. Communications 13, 1–14. doi: 10.1038/s41467-022-30515-0
Costanzo, M. C., and Fox, T. D. (1988). Transformation of yeast by agitation with glass beads. Genetics 120, 667–670. doi: 10.1093/genetics/120.3.667
Deckers, M., Deforce, D., Fraiture, M.-A., and Roosens, N. H. C. (2020). Genetically Modified Micro-Organisms for Industrial Food Enzyme Production: An Overview. Foods 9, 326–346. doi: 10.3390/foods9030326
Deng, J., Wilson, T. J., Wang, J., Peng, X., Li, M., Lin, X., et al. (2022). Structure and mechanism of a methyltransferase ribozyme. Nat. Chem. Biol. 18, 556–564. doi: 10.1038/s41589-022-00982-z
Deriano, L., and Roth, D. B. (2013). Modernizing the nonhomologous end-joining repertoire: alternative and classical NHEJ share the stage. Annu. Rev. Genet. 47, 433–455.
DiCarlo, J. E., Norville, J. E., Mali, P., Rios, X., Aach, J., and Church, G. M. (2013). Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 41, 4336–4343. doi: 10.1093/nar/gkt135
Dillingham, M. S., and Kowalczykowski, S. C. (2008). RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiol. Mol. Biol. Rev. 72, 642–671. doi: 10.1128/MMBR.00020-08
Dow, L. E., Fisher, J., O'Rourke, K. P., Muley, A., Kastenhuber, E. R., Livshits, G., et al. (2015). Inducible in vivo genome editing with CRISPR-Cas9. Nat. Biotechnol. 33, 390–398. doi: 10.1038/nbt.3155
Enghiad, B., Huang, C., Guo, F., Jiang, G., Wang, B., Tabatabaei, S. K., et al. (2021). Cas12a-assisted precise targeted cloning using in vivo Cre-lox recombination. Nat. Commun. 12, 1171–1181. doi: 10.1038/s41467-021-21275-4
Eskola, M., Kos, G., Elliott, C. T., Hajslova, J., Mayar, S., and Krska, R. (2020). Worldwide contamination of food-crops with mycotoxins: validity of the widely cited 'FAO estimate' of 25. Crit. Rev. Food Sci. Nutr. 60, 2773–2789. doi: 10.1080/10408398.2019.1658570
Fayyaz, M., Chew, K. W., Show, P. L., Ling, T. C., Ng, I. S., and Chang, J.-S. (2020). Genetic engineering of microalgae for enhanced biorefinery capabilities. Biotechnol. Adv. 43:107554. doi: 10.1016/j.biotechadv.2020.107554
Fire, A., Xu, S. Q., Montgomery, M. K., Kostas, S. A., Driver, S. E., and Mello, C. C. (1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811. doi: 10.1038/35888
Florea, S., Machado, C., Andreeva, K., and Schardl, C.L., (2012). Cre/lox system: A practical tool to efficiently eliminate selectable markers in fungal endophytes.
Fu, Y., Sander, J. D., Reyon, D., Cascio, V. M., and Joung, J. K. (2014). Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 32, 279–284. doi: 10.1038/nbt.2808
Fujitani, Y., Yamamoto, K., and Kobayashi, I. (1995). Dependence of frequency of homologous recombination on the homology length. Genetics 141:797. doi: 10.1093/genetics/141.2.797
Fuller, K. K., Chen, S., Loros, J. J., and Dunlap, J. C. (2015). Development of the CRISPR/Cas9 system for targeted gene disruption in Aspergillus fumigatus. Eukaryot. Cell 14, 1073–1080. doi: 10.1128/EC.00107-15
Gao, J., Xu, X. G., Huang, K. L., and Liang, Z. H. (2021). Fungal G-protein-coupled receptors: a promising mediator of the impact of extracellular signals on biosynthesis of Ochratoxin a. Front. Microbiol. 12:631392. doi: 10.3389/fmicb.2021.631392
Gao, Y., and Zhao, Y. (2014). Self-processing of ribozyme-flanked RNAs into guide RNAs in vitro and in vivo for CRISPR-mediated genome editing. J. Integr. Plant Biol. 56, 343–349. doi: 10.1111/jipb.12152
Garrigues, S., Martínez-Reyes, N., and de Vries, R. P. (2021). “Genetic engineering for strain improvement in filamentous fungi” in Encyclopedia of Mycology. eds. Ó. Zaragoza and A. Casadevall (Oxford: Elsevier), 489–504.
Gaudelli, N. M., Komor, A. C., Rees, H. A., Packer, M. S., Badran, A. H., Bryson, D. I., et al. (2017). Programmable base editing of a.T to G.C in genomic DNA without DNA cleavage. Nature 551, 464–471. doi: 10.1038/nature24644
Gebremichael, D. E., Haile, Z. M., Negrini, F., Sabbadini, S., Capriotti, L., Mezzetti, B., et al. (2021). RNA interference strategies for future management of plant pathogenic fungi: prospects and challenges. Plan. Theory 10, 650–670. doi: 10.3390/plants10040650
Goeckel, M. E., Basgall, E. M., Lewis, I. C., Goetting, S. C., Yan, Y., Halloran, M., et al. (2019). Modulating CRISPR gene drive activity through nucleocytoplasmic localization of Cas9 in S. cerevisiae. Fung. Biol. Biotechnol. 6:2. doi: 10.1186/s40694-019-0065-x
Graessle, S., Haas, H., Friedlin, E., Kurnsteiner, H., Stoffler, G., and Redl, B. (1997). Regulated system for heterologous gene expression in Penicillium chrysogenum. Appl. Environ. Microbiol. 63, 753–756. doi: 10.1128/aem.63.2.753-756.1997
Haft, D. H., Selengut, J., Mongodin, E. F., and Nelson, K. E. (2005). A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput. Biol. 1, 474–483. doi: 10.1371/journal.pcbi.0010060
Honda, S., and Selker, E. U. (2009). Tools for fungal proteomics: multifunctional Neurospora vectors for gene replacement, protein expression and protein purification. Genetics 182, 11–23. doi: 10.1534/genetics.108.098707
Houbraken, J., Kocsube, S., Visagie, C. M., Yilmaz, N., Wang, X. C., Meijer, M., et al. (2020). Classification of Aspergillus, Penicillium, Talaromyces and related genera (Eurotiales): an overview of families, genera, subgenera, sections, series and species. Stud. Mycol. 95, 5–169. doi: 10.1016/j.simyco.2020.05.002
Hsu, P. D., Lander, E. S., and Zhang, F. (2014). Development and applications of CRISPR-Cas9 for genome engineering. Cells 157, 1262–1278. doi: 10.1016/j.cell.2014.05.010
Ishino, Y., Shinagawa, H., Makino, K., Amemura, M., and Nakata, A. (1987). Nucleotide sequence of the IAP gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 169, 5429–5433. doi: 10.1128/jb.169.12.5429-5433.1987
Jansen, R., van Embden, J. D. A., Gaastra, W., and Schouls, L. M. (2002). Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microbiol. 43, 1565–1575. doi: 10.1046/j.1365-2958.2002.02839.x
Jayathilaka, K., Sheridan, S. D., Bold, T. D., Bochenska, K., Logan, H. L., Weichselbaum, R. R., et al. (2008). A chemical compound that stimulates the human homologous recombination protein RAD51. Proc. Natl. Acad. Sci. U. S. A. 105, 15848–15853. doi: 10.1073/pnas.0808046105
Jiang, F., and Doudna, J. A. (2017). CRISPR-Cas9 structures and mechanisms. Annu. Rev. Biophys. 46, 505–529.
Jin, F. J., Hu, S., Wang, B. T., and Jin, L. (2021). Advances in genetic engineering technology and its application in the industrial fungus Aspergillus oryzae. Front. Microbiol. 12:644404. doi: 10.3389/fmicb.2021.644404
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., and Charpentier, E. (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. doi: 10.1126/science.1225829
Joung, J. K., and Sander, J. D. (2013). TALENs: a widely applicable technology for targeted genome editing. Nat. Rev. Mol. Cell Biol. 14, 49–55. doi: 10.1038/nrm3486
Karlin, S., and Mrazek, J. (2000). Predicted highly expressed genes of diverse prokaryotic genomes. J. Bacteriol. 182, 5238–5250. doi: 10.1128/JB.182.18.5238-5250.2000
Kim, Y. G., Cha, J., and Chandrasegaran, S. (1996). Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc. Natl. Acad. Sci. U. S. A. 93, 1156–1160. doi: 10.1073/pnas.93.3.1156
Klein, T. M., Wolf, E. D., Wu, R., and Sanford, J. C. (1987). High velocity microprojectiles for delivering nucleic acids into living cells. Nature 327, 70–73. doi: 10.1038/327070a0
Kleinstiver, B. P., Prew, M. S., Tsai, S. Q., Nguyen, N. T., Topkar, V. V., Zheng, Z., et al. (2015a). Broadening the targeting range of Staphylococcus aureus CRISPR-Cas9 by modifying PAM recognition. Nat. Biotechnol. 33, 1293–1298. doi: 10.1038/nbt.3404
Kleinstiver, B. P., Prew, M. S., Tsai, S. Q., Topkar, V. V., Nguyen, N. T., Zheng, Z., et al. (2015b). Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 523:481. doi: 10.1038/nature14592
Komor, A. C., Kim, Y. B., Packer, M. S., Zuris, J. A., and Liu, D. R. (2016). Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424. doi: 10.1038/nature17946
Koonin, E. V., Makarova, K. S., and Zhang, F. (2017). Diversity, classification and evolution of CRISPR-Cas systems. Curr. Opin. Microbiol. 37, 67–78. doi: 10.1016/j.mib.2017.05.008
Kopke, K., Hoff, B., and Kueck, U. (2010). Application of the Saccharomyces cerevisiae FLP/FRT recombination system in filamentous fungi for marker recycling and construction of knockout strains devoid of heterologous genes. Appl. Environ. Microbiol. 76, 4664–4674. doi: 10.1128/AEM.00670-10
Koushki, M. M., Rouhani, H., and Farsi, M. (2011). Genetic manipulation of fungal strains for the improvement of heterologous genes expression (a mini-review). Afr. J. Biotechnol. 10, 7939–7948. doi: 10.5897/AJB11.041
Kuivanen, J., Wang, Y. J., and Richard, P. (2016). Engineering Aspergillus Niger for galactaric acid production: elimination of galactaric acid catabolism by using RNA sequencing and CRISPR/Cas9. Microb. Cell Factories 15:210. doi: 10.1186/s12934-016-0613-5
Kundert, K., Lucas, J. E., Watters, K. E., Fellmann, C., Ng, A. H., Heineike, B. M., et al. (2019). Controlling CRISPR-Cas9 with ligand-activated and ligand-deactivated sgRNAs. Nat. Commun. 10, 2127–2137. doi: 10.1038/s41467-019-09985-2
Kuo, J., Yuan, R., Sanchez, C., Paulsson, J., and Silver, P. A. (2020). Toward a translationally independent RNA-based synthetic oscillator using deactivated CRISPR-Cas. Nucleic Acids Res. 48, 8165–8177. doi: 10.1093/nar/gkaa557
Langfelder, K., Gattung, S., and Brakhage, A. A. (2002). A novel method used to delete a new Aspergillus fumigatus ABC transporter-encoding gene. Curr. Genet. 41, 268–274. doi: 10.1007/s00294-002-0313-z
Li, Q. H., Lu, J. C., Zhang, G. Q., Liu, S., Zhou, J. W., Du, G. C., et al. (2022). Recent advances in the development of Aspergillus for protein production. Bioresour. Technol. 348. doi: 10.1016/j.biortech.2022.126768
Li, Q., Lu, J., Zhang, G., Zhou, J., Li, J., Du, G., et al. (2023). CRISPR/Cas9-mediated multiplexed genome editing in Aspergillus oryzae. J. Fungi 9, 109–120. doi: 10.3390/jof9010109
Li, D. D., Tang, Y., Lin, J., and Cai, W. W. (2017). Methods for genetic transformation of filamentous fungi. Microb. Cell Factories 16, 168–180. doi: 10.1186/s12934-017-0785-7
Li, C., Zhou, J., Du, G., Chen, J., Takahashi, S., and Liu, S. (2020). Developing Aspergillus Niger as a cell factory for food enzyme production. Biotechnol. Adv. 44:107630. doi: 10.1016/j.biotechadv.2020.107630
Li, T., Zhu, L., Xiao, B., Gong, Z., Liao, Q., and Guo, J. (2019). CRISPR-Cpf1-mediated genome editing and gene regulation in human cells. Biotechnol. Adv. 37, 21–27. doi: 10.1016/j.biotechadv.2018.10.013
Liu, D., Liu, Q., Guo, W., Liu, Y., Wu, M., Zhang, Y., et al. (2022). Development of genetic tools in Glucoamylase-Hyperproducing industrial Aspergillus Niger strains. Biology 11, 1396–1496. doi: 10.3390/biology11101396
Liu, Z., Wang, S., Tapeinos, C., Torrieri, G., Kankanen, V., El-Sayed, N., et al. (2021). Non-viral nanoparticles for RNA interference: principles of design and practical guidelines. Adv. Drug Deliv. Rev. 174, 576–612. doi: 10.1016/j.addr.2021.05.018
Liu, N., Zhou, L., Lin, G., Hu, Y., Jiao, Y., Wang, Y., et al. (2022). HDAC inhibitors improve CRISPR-Cas9 mediated prime editing and base editing. Mol. Therap. Nucleic Acids 29, 36–46. doi: 10.1016/j.omtn.2022.05.036
Luo, Y., Liu, X., and Li, J. (2018). Updating techniques on controlling mycotoxins—A review. Food Control 89, 123–132. doi: 10.1016/j.foodcont.2018.01.016
Madhavan, A., Arun, K. B., Sindhu, R., Binod, P., Awasthi, M. K., and Pandey, A. (2022). Aspergillus spp., a versatile cell factory for enzymes and metabolites: interventions through genome editing. Indian J. Exp. Biol. 60, 647–658. doi: 10.56042/ijeb.v60i09.65635
Magana-Ortiz, D., Coconi-Linares, N., Ortiz-Vazquez, E., Fernandez, F., Loske, A. M., and Gomez-Lim, M. A. (2013). A novel and highly efficient method for genetic transformation of fungi employing shock waves. Fungal Genet. Biol. 56, 9–16. doi: 10.1016/j.fgb.2013.03.008
Maggio, I., Zittersteijn, H. A., Wang, Q., Liu, J., Janssen, J. M., Ojeda, I. T., et al. (2020). Integrating gene delivery and gene-editing technologies by adenoviral vector transfer of optimized CRISPR-Cas9 components. Gene Ther. 27, 209–225. doi: 10.1038/s41434-019-0119-y
Maxam, A. M., and Gilbert, W. (1977). New method for sequencing DNA. Proc. Natl. Acad. Sci. U. S. A. 74, 560–564. doi: 10.1073/pnas.74.2.560
Mizutani, O., Arazoe, T., Toshida, K., Hayashi, R., Ohsato, S., Sakuma, T., et al. (2017). Detailed analysis of targeted gene mutations caused by the platinum fungal TALENs in Aspergillus oryzae RIB40 strain and a ligD disruptant. J. Biosci. Bioeng. 123, 287–293. doi: 10.1016/j.jbiosc.2016.09.014
Moon, H. Y., Sim, G. H., Kim, H. J., Kim, K., and Kang, H. A. (2022). Assessment of Cre-lox and CRISPR-Cas9 as tools for recycling of multiple-integrated selection markers in Saccharomyces cerevisiae. J. Microbiol. 60, 18–30. doi: 10.1007/s12275-022-1580-7
Mueller, D., Stahl, U., and Meyer, V. (2006). Application of hammerhead ribozymes in filamentous fungi. J. Microbiol. Methods 65, 585–595. doi: 10.1016/j.mimet.2005.10.003
Murphy, K. C. (1998). Use of bacteriophage lambda recombination functions to promote gene replacement in Escherichia coli. J. Bacteriol. 180, 2063–2071. doi: 10.1128/JB.180.8.2063-2071.1998
Nagy, A. (2000). Cre recombinase: the universal reagent for genome tailoring. Genesis 26, 99–109. doi: 10.1002/(SICI)1526-968X(200002)26:2<99::AID-GENE1>3.0.CO;2-B
Nakamura, M., Gao, Y., Dominguez, A. A., and Qi, L. S. (2021). CRISPR technologies for precise epigenome editing. Nat. Cell Biol. 23, 11–22. doi: 10.1038/s41556-020-00620-7
Nargesi, S., Kaboli, S., Thekkiniath, J., Heidari, S., Keramati, F., Seyedmousavi, S., et al. (2021). Recent advances in genome editing tools in medical mycology research. J. Fungi 7, 257–271. doi: 10.3390/jof7040257
Nguyen, H. N., Ishidoh, K.-I., Kinoshita, H., and Nihira, T. (2018). Targeted cloning of a large gene cluster from Lecanicillium genome by Cre/loxP based method. J. Microbiol. Methods 150, 47–54. doi: 10.1016/j.mimet.2018.05.017
Nihongaki, Y., Kawano, F., Nakajima, T., and Sato, M. (2015). Photoactivatable CRISPR-Cas9 for optogenetic genome editing. Nat. Biotechnol. 33, 755–760. doi: 10.1038/nbt.3245
Ninomiya, Y., Suzuki, K., Ishii, C., and Inoue, H. (2004). Highly efficient gene replacements in Neurospora strains deficient for non-homologous end-joining. Proc. Natl. Acad. Sci. U. S. A. 101, 12248–12253. doi: 10.1073/pnas.0402780101
Nodvig, C. S., Nielsen, J. B., Kogle, M. E., and Mortensen, U. H. (2015). A CRISPR-Cas9 system for genetic engineering of filamentous fungi. PLoS One 10:e133085. doi: 10.1371/journal.pone.0133085
Ostry, V., Malir, F., Toman, J., and Grosse, Y. (2017). Mycotoxins as human carcinogens—the IARC monographs classification. Mycotoxin Res. 33, 65–73. doi: 10.1007/s12550-016-0265-7
Pant, S., Ritika, P., Nag, P., Ghati, A., Chakraborty, D., Maximiano, M. R., et al. (2022). Employment of the CRISPR/Cas9 system to improve cellulase production in Trichoderma reesei. Biotechnol. Adv. 60:108022. doi: 10.1016/j.biotechadv.2022.108022
Qi, L. S., Larson, M. H., Gilbert, L. A., Doudna, J. A., Weissman, J. S., Arkin, A. P., et al. (2013). Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cells 152, 1173–1183. doi: 10.1016/j.cell.2013.02.022
Ran, F. A., Hsu, P. D., Lin, C.-Y., Gootenberg, J. S., Konermann, S., Trevino, A. E., et al. (2013). Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cells 154, 1380–1389. doi: 10.1016/j.cell.2013.08.021
Rivera, A. L., Magana-Ortiz, D., Gomez-Lim, M., Fernandez, F., and Loske, A. M. (2014). Physical methods for genetic transformation of fungi and yeast. Phys Life Rev 11, 184–203. doi: 10.1016/j.plrev.2014.01.007
Roberts, I. N., Jeenes, D. J., Mackenzie, D. A., Wilkinson, A. P., Sumner, I. G., and Archer, D. B. (1992). Heterologous gene expression in Aspergillus Niger a glucoamylase-porcine pancreatic prophospholipase-a(2) fusion protein is secreted and processed to yield mature enzyme. Gene 122, 155–161. doi: 10.1016/0378-1119(92)90043-O
Robey, M. T., Caesar, L. K., Drott, M. T., Keller, N. P., and Kelleher, N. L. (2021). An interpreted atlas of biosynthetic gene clusters from 1,000 fungal genomes. Proc. Natl. Acad. Sci. U. S. A. 118:e2020230118. doi: 10.1073/pnas.2020230118
Romano, N., and Macino, G. (1992). Quelling transient inactivation of gene expression in Neurospora crassa by transformation with homologous sequences. Mol. Microbiol. 6, 3343–3353. doi: 10.1111/j.1365-2958.1992.tb02202.x
Rosenkranz, H. S., and Levy, J. A. (1965). Hydroxyurea: a specific inhibitor of deoxyribonucleic acid synthesis. Biochim. Biophys. Acta 95, 181–183. doi: 10.1016/0005-2787(65)90225-X
Roux, I., and Chooi, Y.-H. (2022). Cre/lox-mediated chromosomal integration of biosynthetic gene clusters for heterologous expression in Aspergillus nidulans. ACS Synth. Biol. 11, 1186–1195. doi: 10.1021/acssynbio.1c00458
Rozhkova, A. M., and Kislitsin, V. Y. (2021). CRISPR/Cas genome editing in filamentous fungi. Biochemistry 86, S120–S139. doi: 10.1134/s0006297921140091
Sadowski, P. D. (1995). “The FLP recombinase of the 2-MU-M plasmid of Saccharomyces cerevisiae” in Progress in Nucleic Acid Research and Molecular Biology. eds. W. E. Cohn and K. Moldave (Journal of molecular biology), 53–91.
Safari, F., Zare, K., Negahdaripour, M., Barekati-Mowahed, M., and Ghasemi, Y. (2019). CRISPR Cpf1 proteins: structure, function and implications for genome editing. Cell Biosci. 9:36. doi: 10.1186/s13578-019-0298-7
Sanger, F., Nicklen, S., and Coulson, A. R. (1977). DNA sequencing with chain terminating inhibitors. Proc. Natl. Acad. Sci. U. S. A. 74, 5463–5467. doi: 10.1073/pnas.74.12.5463
Sarkari, P., Marx, H., Blumhoff, M. L., Mattanovich, D., Sauer, M., and Steiger, M. G. (2017). An efficient tool for metabolic pathway construction and gene integration for Aspergillus Niger. Bioresour. Technol. 245, 1327–1333. doi: 10.1016/j.biortech.2017.05.004
Scully, R., Panday, A., Elango, R., and Willis, N. A. (2019). DNA double-strand break repair-pathway choice in somatic mammalian cells. Nat. Rev. Mol. Cell Biol. 20:1. doi: 10.1038/s41580-019-0152-0
Sinclair, W. K., and Morton, R. A. (1965). X-ray and ultraviolet sensitivity of synchronized Chinese hamster cells at various stages of cell cycle. Biophys. J. 5, 1–25. doi: 10.1016/S0006-3495(65)86700-5
Song, L., Ouedraogo, J. P., Kolbusz, M., Nguyen, T. T. M., and Tsang, A. (2018). Efficient genome editing using tRNA promoter-driven CRISPR/Cas9 gRNA in Aspergillus Niger. PLoS One 13:e0202868. doi: 10.1371/journal.pone.0202868
Steiger, M. G., Vitikainen, M., Uskonen, P., Brunner, K., Adam, G., Pakula, T., et al. (2011). Transformation system for Hypocrea jecorina (Trichoderma reesei) that favors homologous integration and employs reusable bidirectionally selectable markers. Appl. Environ. Microbiol. 77, 114–121. doi: 10.1128/AEM.02100-10
Sternberg, N., and Hamilton, D. (1981). Bacteriophage-P1 site-specific recombination: recombination between loxP sites. J. Mol. Biol. 150, 467–486. doi: 10.1016/0022-2836(81)90375-2
Struhl, K., Stinchcomb, D. T., Scherer, S., and Davis, R. W. (1979). High frequency transformation of yeast: autonomous replication of hybrid DNA molecules. Proc. Natl. Acad. Sci. U. S. A. 76, 1035–1039. doi: 10.1073/pnas.76.3.1035
Suzuki, K., Tsunekawa, Y., Hernandez-Benitez, R., Wu, J., Zhu, J., Kim, E. J., et al. (2016). In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature 540, 144–149. doi: 10.1038/nature20565
Szewczyk, E., Nayak, T., Oakley, C. E., Edgerton, H., Xiong, Y., Taheri-Talesh, N., et al. (2006). Fusion PCR and gene targeting in Aspergillus nidulans. Nat. Protoc. 1, 3111–3120. doi: 10.1038/nprot.2006.405
Szostak, J. W., Orrweaver, T. L., Rothstein, R. J., and Stahl, F. W. (1983). The double-strand break repair model for recombination. Cells 33, 25–35. doi: 10.1016/0092-8674(83)90331-8
Takita, Y., Takahara, M., Nogami, S., Anraku, Y., and Ohya, Y. (1997). Applications of the long and accurate polymerase chain reaction method in yeast molecular biology: direct sequencing of the amplified DNA and its introduction into yeast. Yeast 13, 763–768. doi: 10.1002/(SICI)1097-0061(19970630)13:8<763::AID-YEA135>3.0.CO;2-0
Tanaka, M., Tokuoka, M., and Gomi, K. (2014). Effects of codon optimization on the mRNA levels of heterologous genes in filamentous fungi. Appl. Microbiol. Biotechnol. 98, 3859–3867. doi: 10.1007/s00253-014-5609-7
Teplova, M., Falschlunger, C., Krasheninina, O., Egger, M., Ren, A., Patel, D. J., et al. (2020). Crucial roles of two hydrated Mg2+ ions in reaction catalysis of the pistol ribozyme. Angew. Chem. Int. Edn. 59, 2837–2843. doi: 10.1002/anie.201912522
Torres-Martinez, S., and Ruiz-Vazquez, R. M. (2017). The RNAi universe in fungi: a varied landscape of small RNAs and biological functions. Annu. Rev. Microbiol. 71, 371–391. doi: 10.1146/annurev-micro-090816-093352
Tsai, S. Q., Wyvekens, N., Khayter, C., Foden, J. A., Thapar, V., Reyon, D., et al. (2014). Dimeric CRISPR RNA-guided Fokl nucleases for highly specific genome editing. Nat. Biotechnol. 32:569. doi: 10.1038/nbt.2908
Uygun, Z. O., Yeniay, L., and Sagin, F. G. (2020). CRISPR-dCas9 powered impedimetric biosensor for label-free detection of circulating tumor DNAs. Anal. Chim. Acta 1121, 35–41. doi: 10.1016/j.aca.2020.04.009
Vanegas, K. G., Jarczynska, Z. D., Strucko, T., and Mortensen, U. H. (2019). Cpf1 enables fast and efficient genome editing in Aspergilli. Fungal Biol. Biotechnol. 6:6. doi: 10.1186/s40694-019-0069-6
Wang, P. (2021). Genetic transformation in Cryptococcus species. J. Fungi 7, 56–65. doi: 10.3390/jof7010056
Wang, Y., Chen, H. Y., Ma, L., Gong, M., Wu, Y. Y., Bao, D. P., et al. (2022). Use of CRISPR-Cas tools to engineer Trichoderma species. Microb. Biotechnol. 15, 2521–2532. doi: 10.1111/1751-7915.14126
Wang, S. X., Chen, H. Q., Tang, X., Zhang, H., Chen, W., and Chen, Y. Q. (2017). Molecular tools for gene manipulation in filamentous fungi. Appl. Microbiol. Biotechnol. 101, 8063–8075. doi: 10.1007/s00253-017-8486-z
Wang, P., Zhou, Y., and Richards, A. M. (2021). Effective tools for RNA-derived therapeutics: siRNA interference or miRNA mimicry. Theranostics 11, 8771–8796. doi: 10.7150/thno.62642
Weyda, I., Yang, L., Vang, J., Ahring, B. K., Lubeck, M., and Lubeck, P. S. (2017). A comparison of agrobacterium-mediated transformation and protoplast-mediated transformation with CRISPR-Cas9 and bipartite gene targeting substrates, as effective gene targeting tools for Aspergillus carbonarius. J. Microbiol. Methods 135, 26–34. doi: 10.1016/j.mimet.2017.01.015
Wilkins, M. H. F., Stokes, A. R., and Wilson, H. R. (1953). Molecular structure of deoxypentose nucleic acids. Nature 171, 738–740. doi: 10.1038/171738a0
Xu, Y., Shan, L., Zhou, Y., Xie, Z., Ball, N. S., Cao, W., et al. (2019). Development of a Cre-loxP-based genetic system in Aspergillus Niger ATCC1015 and its application to construction of efficient organic acid-producing cell factories. Appl. Microbiol. Biotechnol. 103, 8105–8114. doi: 10.1007/s00253-019-10054-3
Yilmaz, S. G. (2021). Genome editing technologies: CRISPR, LEAPER, RESTORE, ARCUT, SATI, and RESCUE. EXCLI J. 20, 19–45. doi: 10.17179/excli2020-3070
Ying, D., Kai-Feng, W., Wei-Jian, W., Yi-Rong, M., Tian-Qiong, S., He, H., et al. (2019). Increasing the homologous recombination efficiency of eukaryotic microorganisms for enhanced genome engineering. Appl. Microbiol. Biotechnol. 103, 4313–4324. doi: 10.1007/s00253-019-09802-2
Yon, J., and Fried, M. (1989). Precise gene fusion by PCR. Nucleic Acids Res. 17:4895. doi: 10.1093/nar/17.12.4895
Yuan, B., Keller, N. P., Oakley, B. R., Stajich, J. E., and Wang, C. C. C. (2022). Manipulation of the global regulator mcrA Upregulates secondary metabolite production in Aspergillus wentii using CRISPR-Cas9 with in vitro assembled Ribonucleoproteins. ACS Chem. Biol. 17, 2828–2835. doi: 10.1021/acschembio.2c00456
Yue, J.-J., Yuan, J.-L., Wu, F.-H., Yuan, Y.-H., Cheng, Q.-W., Hsu, C.-T., et al. (2021). Protoplasts: from isolation to CRISPR/Cas genome editing application. Front. Genome Edit. 3:717017. doi: 10.3389/fgeed.2021.717017
Zeng, Y., Hong, Y. X., Azi, F., Liu, Y. G., Chen, Y. S., Guo, C. C., et al. (2022). Advanced genome-editing technologies enable rapid and large-scale generation of genetic variants for strain engineering and synthetic biology. Curr. Opin. Microbiol. 69:102175. doi: 10.1016/j.mib.2022.102175
Zetsche, B., Gootenberg, J. S., Abudayyeh, O. O., Slaymaker, I. M., Makarova, K. S., Essletzbichler, P., et al. (2015). Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cells 163, 759–771. doi: 10.1016/j.cell.2015.09.038
Zetsche, B., Heidenreich, M., Mohanraju, P., Fedorova, I., Kneppers, J., DeGennaro, E. M., et al. (2017). Multiplex gene editing by CRISPR-Cpf1 using a single crRNA array. Nat. Biotechnol. 35, 31–34. doi: 10.1038/nbt.3737
Zhang, L., Zhao, X., Zhang, G., Zhang, J., Wang, X., Zhang, S., et al. (2016). Light-inducible genetic engineering and control of non-homologous end-joining in industrial eukaryotic microorganisms: LML 3.0 and OFN 1.0. Sci. Rep. 6. doi: 10.1038/srep20761
Zhang, L., Zheng, X., Cairns, T. C., Zhang, Z., Wang, D., Zheng, P., et al. (2020). Disruption or reduced expression of the orotidine-5′-decarboxylase gene pyrG increases citric acid production: a new discovery during recyclable genome editing in Aspergillus Niger. Microb. Cell Factories 19:76. doi: 10.1186/s12934-020-01334-z
Zheng, X., Zheng, P., and Sun, J. (2021). CRISPR/Cas-based genome editing in Aspergillus Niger. Chin. J. Biotechnol. 37, 980–990. doi: 10.13345/j.cjb.200613
Zheng, X., Zheng, P., Zhang, K., Cairns, T. C., Meyer, V., Sun, J., et al. (2019). 5S rRNA promoter for guide RNA expression enabled highly efficient CRISPR/Cas9 genome editing in Aspergillus Niger. ACS Synth. Biol. 8, 1568–1574. doi: 10.1021/acssynbio.7b00456
Zhou, W., Cui, H., Ying, L., and Yu, X.-F. (2018). Enhanced cytosolic delivery and release of CRISPR/Cas9 by black phosphorus nanosheets for genome editing. Angew. Chem. Int. Edn. 57, 10268–10272. doi: 10.1002/anie.201806941
Zhu, Z., Li, R., Zhang, H., Wang, J., Lu, Y., Zhang, D., et al. (2022). PAM-free loop-mediated isothermal amplification coupled with CRISPR/Cas12a cleavage (Cas-PfLAMP) for rapid detection of rice pathogens. Biosens. Bioelectron. 204:114076. doi: 10.1016/j.bios.2022.114076
Zhu, S. Y., Xu, Y., and Yu, X. W. (2020). Improved homologous expression of the acidic lipase from Aspergillus Niger. J. Microbiol. Biotechnol. 30, 196–205. doi: 10.4014/jmb.1906.06028
Keywords: genetic engineering, homologous recombination, CRISPR, RNA interference, transformation, selective marker, cell factory
Citation: Gao J, Liu H, Zhang Z and Liang Z (2023) Establishment, optimization, and application of genetic technology in Aspergillus spp.. Front. Microbiol. 14:1141869. doi: 10.3389/fmicb.2023.1141869
Edited by:
Xiao-Jun Ji, Nanjing Tech University, ChinaReviewed by:
Wei Cao, Tianjin University of Science and Technology, ChinaXiaomei Zheng, Tianjin Institute of Industrial Biotechnology (CAS), China
Copyright © 2023 Gao, Liu, Zhang and Liang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhihong Liang, bHpoMTA1QGNhdS5lZHUuY24=
 Jing Gao
Jing Gao Huiqing Liu1
Huiqing Liu1 Zhihong Liang
Zhihong Liang