- 1College of Life Sciences, Liaocheng University, Liaocheng, China
- 2Jiangxi Zymerck Biotechnology Co., Ltd., Nanchang, China
Ursodeoxycholic acid (UDCA) is a fundamental treatment drug for numerous hepatobiliary diseases that also has adjuvant therapeutic effects on certain cancers and neurological diseases. Chemical UDCA synthesis is environmentally unfriendly with low yields. Biological UDCA synthesis by free-enzyme catalysis or whole-cell synthesis using inexpensive and readily available chenodeoxycholic acid (CDCA), cholic acid (CA), or lithocholic acid (LCA) as substrates is being developed. The free enzyme-catalyzed one-pot, one-step/two-step method uses hydroxysteroid dehydrogenase (HSDH); whole-cell synthesis, mainly uses engineered bacteria (mainly Escherichia coli) expressing the relevant HSDHs. To further develop these methods, HSDHs with specific coenzyme dependence, high enzyme activity, good stability, and high substrate loading concentration, P450 monooxygenase with C-7 hydroxylation activity and engineered strain harboring HSDHs must be exploited.
Introduction
Ursodeoxycholic acid (UDCA) is an endogenous bile acid that has been shown to dissolve gallstone (Park et al., 2020; Haal et al., 2021); treat cholestasis (Leoni et al., 2018; Marschall, 2019; Razori et al., 2019; Chappell et al., 2020; Linhares et al., 2020; Haal et al., 2021; Kato et al., 2022), biliary pancreatitis (Rose et al., 1991; Testoni et al., 2000; Byelyayeva, 2015), primary cholangitis (Carbone et al., 2018; Deneau et al., 2019; Cacciato et al., 2020; Qian et al., 2020; Yao et al., 2020; Yu et al., 2021), colitis (colon cancer) (Eun-Kyung et al., 2017; He et al., 2022), (non)alcoholic hepatitis (Khachidze, 2019; Wu et al., 2020; Xie et al., 2022), primary biliary cirrhosis (van der Feen et al., 2016; Ye et al., 2019; Colombo et al., 2021), and drug-induced hepatitis (Karim and Yesmin, 2020); and improve liver transplantation outcomes (Harms et al., 2019; Kato et al., 2022), while gut microbiota-derived UDCA also attenuates colon inflammation in low-birth-weight infants by enhancing macrophage M2 polarization (Pi et al., 2022). In recent years, it has been found that UDCA can also mediate and influence the regulation of oncogenic signaling pathways in the cancer genome for cancer therapy (Goossens and Bailly, 2019; Lugini et al., 2022); it can specifically regulate the threshold of apoptosis, inhibit cancer cell growth (Eun-Kyung et al., 2017; Chen et al., 2022), and induce autophagy and apoptosis (Lim and Han, 2015; Lee et al., 2017). It has also been found that UDCA can improve damaged mitochondrial function and exert neuroprotective effects in the treatment of progressive neurological diseases (Bell et al., 2018; Abdelkader, 2020; Qi et al., 2020; Sathe et al., 2020; West et al., 2020; Yang et al., 2020; Huang, 2021).
Originally discovered as a natural active product from polar bear bile and named by Hammarsten in 1902, UDCA has gradually been shown to exhibit better efficacy in the treatment of gallbladder and liver-related diseases than other endogenous bile acids (Beuers et al., 1992; Combes et al., 1995; Im and Martinez, 2004). To date, UDCA is the only drug approved by the U.S. Food and Drug Administration (FDA) for the treatment of primary biliary cirrhosis (Shah and Kowdley, 2020). UDCA is now prepared by chemical or biosynthetic routes: the chemical route produces UDCA by a seven-step synthesis using cholic acid (CA) or chenodeoxycholic acid (CDCA) as starting substrates (Hofmann et al., 1963), with CA and CDCA isolated from inexpensive and readily available bovine bile and chicken, duck, and goose bile, respectively. Unfortunately, this route requires the use of toxic and dangerous hydrazine, CrO3 and pyridine reagents and generates a large amount of waste, in addition to an overall yield of only approximately 30%. To improve the efficiency of UDCA synthesis, alternative routes were developed, and Dangate et al. (2011) used o-iodobenzoic acid, hydrazine and sodium metal dissolved in n-propanol in the reaction to circumvent the group protection and deprotection steps of the original process and increase the yield of CA to UDCA to 53%; however, this preparation process is still not optimized in terms of cost and environmental protection. UDCA produced by chemical synthesis is still far from meeting market demand in terms of both quantity and quality (Tonin and Arends, 2018). Compared with chemical synthesis, biosynthesis is greener, efficient and safe, and UDCA biosynthesis using bile acid substrates is gradually being developed.
Biological UDCA synthesis is mainly free enzyme-catalyzed synthesis or whole-cell synthesis (Eggert et al., 2014). Free enzyme-catalyzed synthesis uses CDCA or CA as a substrate, and UDCA is synthesized through a multienzyme cascade reaction; whole-cell synthesis involves the addition of CDCA or lithocholic acid (LCA) substrate during microbial culture, which is converted into UDCA by microbial cells.
UDCA synthesis catalyzed by free enzymes
Compared with whole-cell UDCA synthesis, free enzyme-catalyzed UDCA synthesis has been more extensively studied. Free enzyme-catalyzed UDCA synthesis uses CDCA, CA or LCA as starting substrates, with most of the studies focusing on CDCA and CA substrates. On the one hand, CDCA and CA are isolated from poultry (chicken, duck, and goose) and bovine bile, and the raw materials are inexpensive and easily available; on the other hand, the catalytic efficiency of hydroxysteroid dehydrogenase (HSDH) for the conversion of CDCA and CA into UDCA is relatively high. Currently, only CDCA substrate is used for free enzyme-catalyzed UDCA synthesis, and UDCA synthesis can also be catalyzed by substrate-specific HSDH using CA as substrate, but the key step of this synthetic route still requires a chemical reaction, i.e., the reduction of the intermediate product carbonyl (C=O) to methylene using the Wolff–Kishner reaction (CH2). Using LCA as a substrate, the conversion to UDCA requires a C-7 carbonyl oxygenation reaction via P450 monooxygenase.
CDCA as a substrate for UDCA synthesis
Chenodeoxycholic acid is converted into UDCA upon the isomerization of the C-7 hydroxyl group. UDCA synthesis using CDCA as a substrate involves two key enzymes: 7α-hydroxysteroid dehydrogenase (7α-HSDH) and 7β-hydroxysteroid dehydrogenase (7β-HSDH). 7α-HSDH first catalyzes the oxidative dehydrogenation of CDCA to produce 7-ketolithocholic acid (7-KLCA), and 7β-HSDH then catalyzes the hydrogenation and reduction of 7-KLCA to produce UDCA (Figure 1). 7α-HSDH specifically oxidizes the α-hydroxyl group at C7 and reduces NAD+/NADP+, while 7β-HSDH specifically reduces the hydroxyl group at C7 to generate the β-hydroxyl group and oxidizes NADPH or NADH (Figure 1).
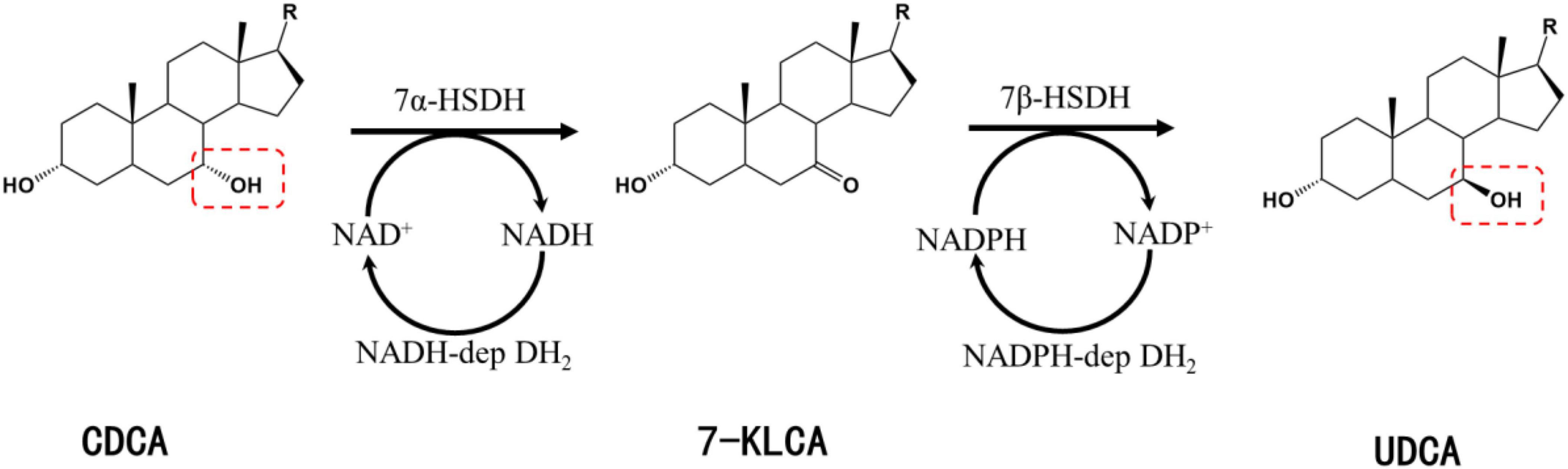
Figure 1. Synthesis of UDCA from CDCA catalyzed by 7α-HSDH and 7β-HSDH. R, 4-pentynoic acid; red dashed box, C-7 hydroxyl group.
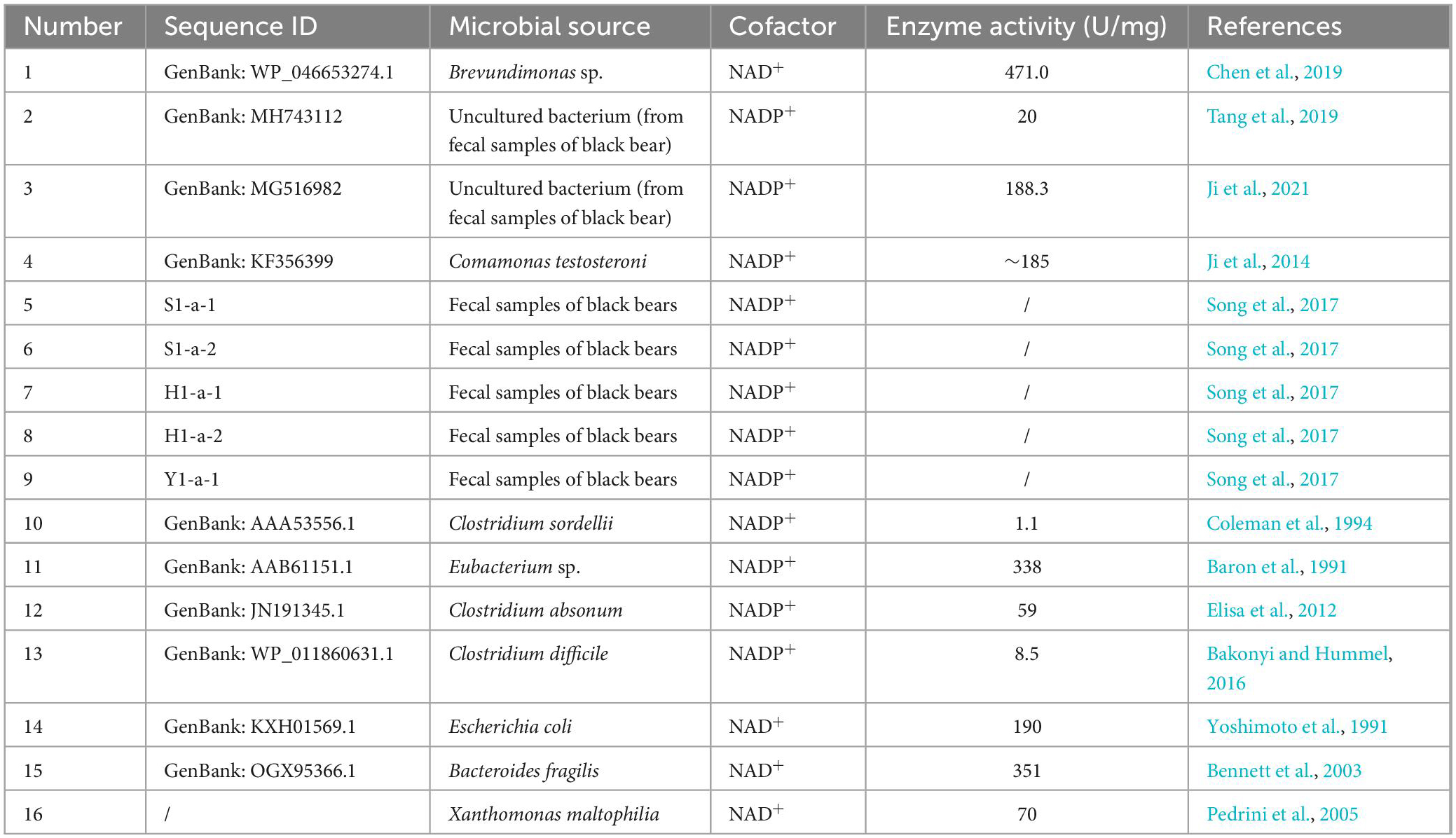
7α-Hydroxysteroid dehydrogenase and 7β-HSDH belong to the same short-chain dehydrogenase/reductase (SDR) family with a molecular mass of approximately 30 kDa and generally exist in homodimeric or homotetrameric structures (Oppermann et al., 2003; Kavanagh et al., 2008). The first 7α-HSDHs were isolated from aerobic or anaerobic bacteria, and in recent years, a larger number of 7α-HSDHs have been identified from black bear feces by homologous alignment (Ji et al., 2014, 2021; Tang et al., 2019) or metagenomics methods (Song et al., 2017). The cofactor-dependent and enzymatic activities of the reported 7α-HSDHs are shown in Table 1.
In addition to the above functionally validated 7α-HSDHs, a search for “7α-hydroxysteroid dehydrogenase” in the NCBI RefSeq database yielded more than 1,200 entries (O’Leary et al., 2016). In addition, some processes related to 7α-HSDH mutants or selective oxidation, derivative synthesis, and salt formation reactions have been patented (Gupta et al., 2009; Monti et al., 2012; Schmid et al., 2013; Bakonyi et al., 2020).
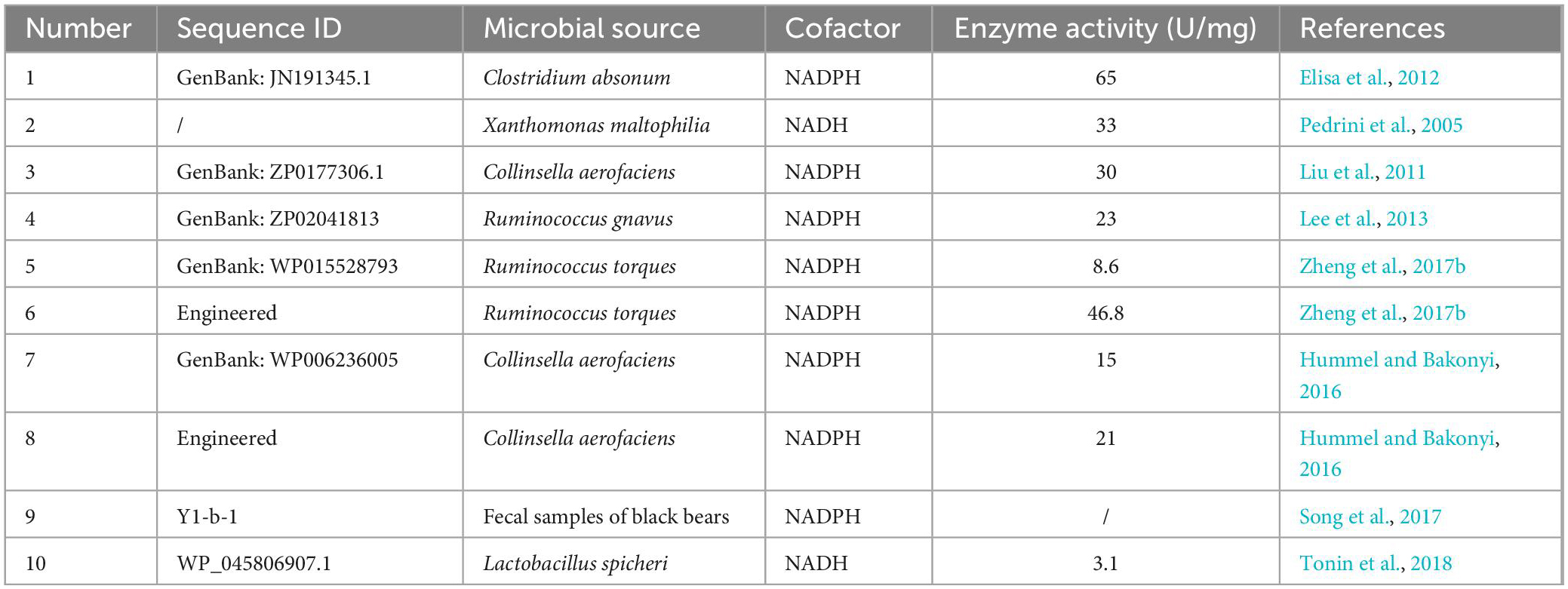
Unlike 7α-HSDH, 7β-HSDH has not been used as frequently for biotransformation (Table 2). Moreover, the specific enzymatic activity and enzymatic stability of 7β-HSDH are poor (Hummel and Bakonyi, 2016; Zheng et al., 2017b), and only one wild-type 7β-HSDH derived from Clostridium absonum was used in two different UDCA synthesis processes (Bovara et al., 1996; Monti et al., 2011). 7β-HSDH almost always requires NADPH as a coenzyme, and thus far, only two NAD+-dependent 7β-HSDHs have been identified: one from Lactobacillus spicheri (Tonin et al., 2018) and one from Xanthomonas maltophilia with a specific enzymatic activity of 33 U/mg, but the protein sequences have not been reported (Pedrini et al., 2005). To improve the activity and stability of 7β-HSDHs, numerous studies have focused on the protein engineering of 7β-HSDH to obtain mutants that meet industrial application requirements (Hummel and Bakonyi, 2016; Liu et al., 2016, 2018, 2019; Zheng et al., 2017b).
CA as a substrate for UDCA synthesis
Cholic acid is currently the most abundant and inexpensive bile acid. It exists in the bile of cattle, sheep and pigs and is most abundant in bovine bile, with the bile acid content reaching 44 g in 1,000 ml of bovine bile (Scalia et al., 1998). There are four synthetic pathways for the synthesis of UDCA from CA, as shown in Figure 2, but each pathway requires the reduction of the intermediate product C-12 carbonyl to methylene with the help of the Wolff–Kishner reaction (Newman et al., 2014), so the full UDCA biosynthesis process is not yet possible using bile acid as the substrate.
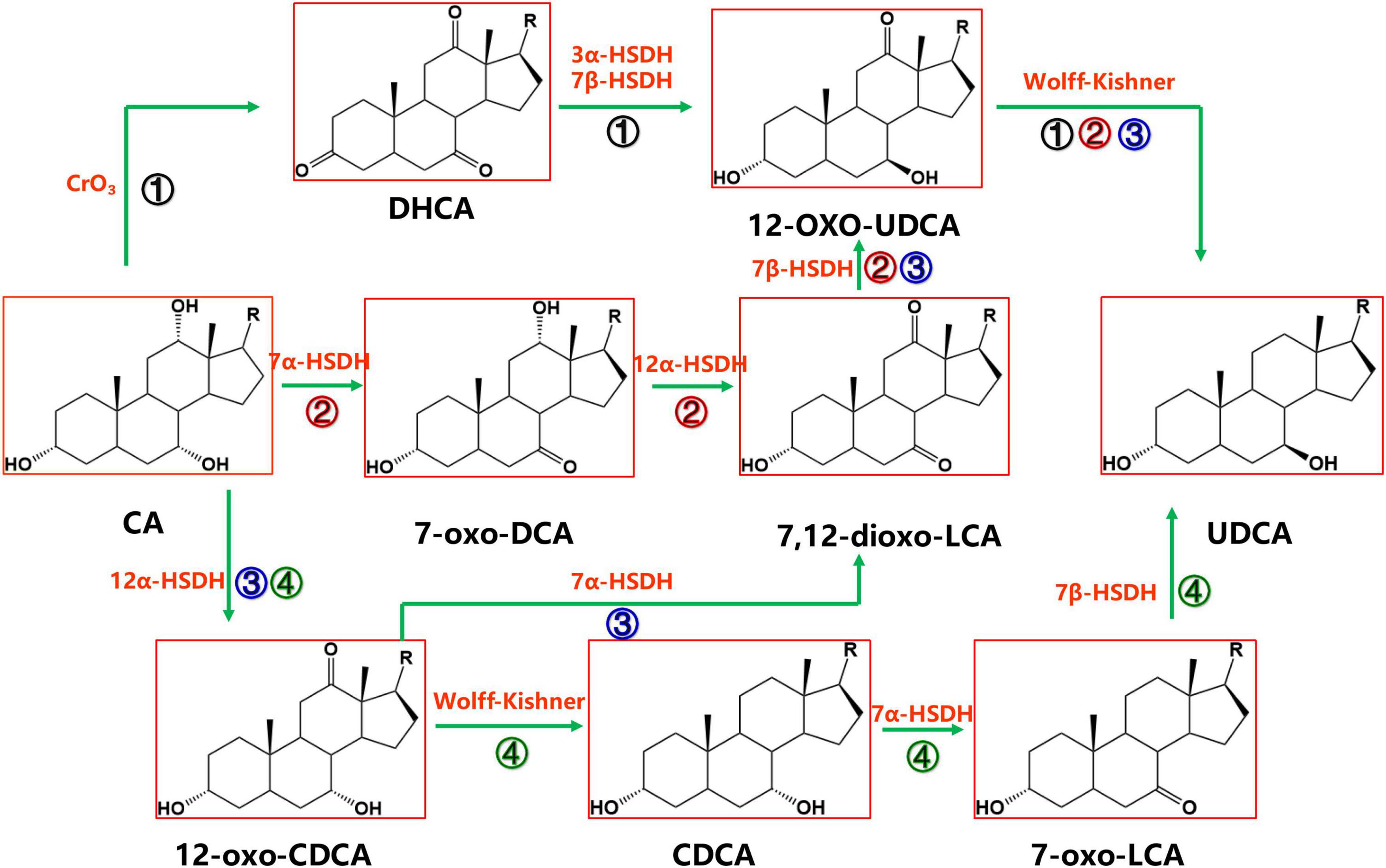
Figure 2. Chemoenzymatic routes for the production of UDCA from CA. DHCA, dehydrocholic acid; 12-oxo-UDCA, 12-oxo-ursodeoxycholic acid; CA, cholic acid; 7-oxo-DCA, 7-oxo-cholic acid; 7,12-dioxo-LCA, 7,12-dioxo-lithocholic acid; UDCA, ursodeoxycholic acid; 12-oxo-CDCA, 12-oxo-chenodeoxycholic acid; CDCA, chenodeoxycholic acid; R, 4-pentynoic acid. ➀, route 1; ➁, route 2; ➂, route 3; ➃, route 4.
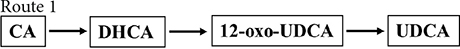
Cholic acid was dehydrogenated (CrO3) to produce dehydrocholic acid (DHCA) (Carrea et al., 1992), the C-3 and C-7 carbonyl groups of DHCA were reduced by 3α-HSDH and 7β-HSDH, respectively, to produce 12-oxo-UDCA (Carrea et al., 1992; Bakonyi et al., 2012; Braun et al., 2012; Liu et al., 2012; Sun et al., 2012), and finally 12-oxo-UDCA was synthesized into UDCA by the Wolff–Kishner reaction (Carrea et al., 1992; Bovara et al., 1996; Bakonyi et al., 2012; Braun et al., 2012; Liu et al., 2012; Sun et al., 2012).
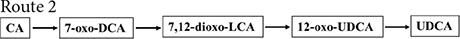
7α-Hydroxysteroid dehydrogenase catalyzed the dehydrogenation of the CA C-7 hydroxyl group to generate 7-oxo-DCA, 12α-HSDH continued to oxidize the C-12 hydroxyl group to a carbonyl group to generate 7,12-dioxo-LCA, 7β-HSDH then reduced the C-7 carbonyl group to a β hydroxyl group to synthesize 12-oxo-UDCA (Bovara et al., 1996; Monti et al., 2009), and 12-oxo-UDCA was finally synthesized into UDCA by the Wolff–Kishner reaction.
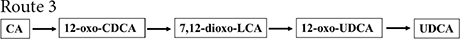
12α-Hydroxysteroid dehydrogenase catalyzed the dehydrogenation of the CA C-12 hydroxyl group to generate 12-oxo-CDCA (Giovannini et al., 2008). 7α-HSDH oxidized the 12-oxo-CDCA C-7 hydroxyl group to a carbonyl group to generate 7,12-dioxo-LCA (Bovara et al., 1996; Monti et al., 2009) and reduced the C-7 carbonyl group to a β hydroxyl group by 7β-HSDH to generate 12-oxo-UDCA (Bovara et al., 1996; Monti et al., 2009). Then, UDCA was finally synthesized by the Wolff–Kishner reaction.

12α-Hydroxysteroid dehydrogenase (Giovannini et al., 2008) catalyzed the dehydrogenation of the CA C-12 hydroxyl group to generate 12-oxo-CDCA, and then the carbonyl group was removed by the Wolff–Kishner reaction to generate CDCA, which entered the CDCA-to-UDCA enzymatic synthesis pathway.
Among the four routes, Route 4 is preferable. In Route 4, UDCA can be synthesized using both CA and CDCA, which are abundant and inexpensive substrates; compared with Route 1, Route 4 cuts one chemical reaction step, offering a greener and more efficient synthesis; compared with Route 2 and Route 3, the 7α-HSDH and 7β-HSDH combination in Route 4 provides a higher substrate conversion rate (Line 127–131).
LCA as a substrate for UDCA synthesis
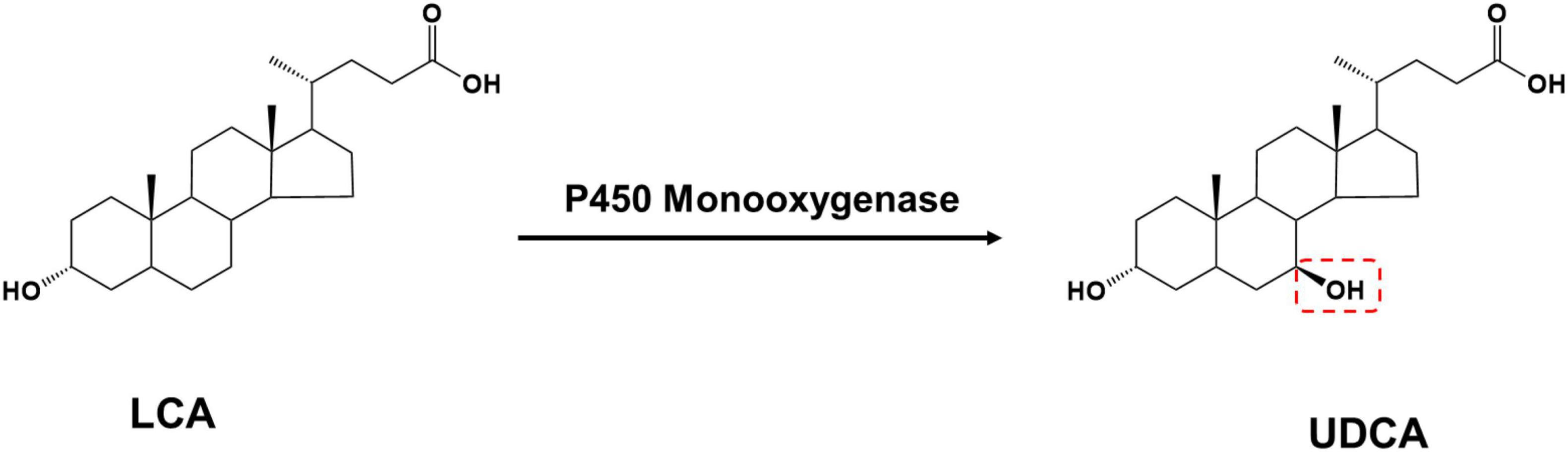
Lithocholic acid is found in the bile of farm animals such as cattle, sheep, and pigs and is an abundant and inexpensive waste product from meat processing. Based on the structural formula, UDCA synthesis using LCA as a substrate only requires LCA C-7 hydroxylation (Figure 3).
P450 monooxygenases catalyze this reaction; however, P450 enzymes face many challenges in practical applications: their redox catalytic process using O2 is highly dependent on the electron-providing cofactor NAD(P)H and the electron transporting reducing chaperone protein, and the complex molecular configuration of this protein reduces P450 enzyme stability and catalytic efficiency. The use of H2O2 instead of O2 eliminates the need for cofactors and their complex electron transport chains, thus simplifying the P450 enzyme catalytic pathway. However, the vast majority of natural P450 enzymes have low or no activity in the presence of H2O2 (Ma et al., 2018). Currently, only a few microorganisms can convert LCA into UDCA, and the only free enzyme that catalyzes UDCA synthesis from LCA is an engineered cytochrome P450 monooxygenase OleP (Grobe et al., 2020), which was initially found to be involved in the epoxidation reaction in the oleandomycin biosynthetic pathway and was later found to also recognize 12-membered macrolides as substrates. Using protein engineering, the regioselectivity of OleP was further modified to catalyze the conversion of LCA into UDCA: the researchers first docked LCA as a substrate to the OleP protein, analyzed the interaction of the substrate with amino acid residues, and obtained five mutants capable of producing UDCA. Furthermore, a small mutant library containing 4,480 mutants was constructed using the 3DM database (Kuipers et al., 2010), from which a mutant with substantially increased selectivity in the C-7 hydroxylated region was screened for UDCA synthesis.
Mode of UDCA synthesis catalyzed by free enzymes
Ursodeoxycholic acid synthesis using CDCA as a substrate and catalyzed by free enzymes is currently the most promising biosynthesis method, and CDCA has obvious advantages over the other two substrates in terms of cost, conversion rate and technical maturity. UDCA synthesis using CDCA as a substrate is generally carried out with one-pot, two-step or one-pot, one-step methods. The former method uses a step-by-step reaction, in which 7α-HSDH catalyzes CDCA to produce 7-KLCA in the first step. After that reaction is completed, the enzyme is inactivated (or spatially isolated from the second step), and then 7β-HSDH is added for the second step. If a high temperature was used to inactivate the enzyme after the first step, the temperature needs to be lowered before the second step (Zheng et al., 2015, 2017a). This method has a high conversion rate but a low substrate concentration, low efficiency, and high time consumption. The latter method involves the addition of 7α-HSDH, 7β-HSDH, the substrate and all the auxiliary materials to the reactor together, and the reaction is simultaneously completed in one step. This method has a high substrate concentration, efficiency and energy savings because there is no intermediate step required to raise and lower the temperature, but it is limited by the reaction equilibrium of the enzyme and the low substrate conversion rate, resulting in a low product yield and complicated subsequent purification process (Zheng et al., 2015, 2017a).
A new 7β-HSDH from Ruminococcus torques was identified, heterologously expressed in Escherichia coli and used together with NADPH-dependent 7α-HSDH from C. absonum in a one-pot, one-step reaction (Zheng et al., 2015). The final conversion rate was limited to 73% due to chemical equilibrium, without additional coenzyme regeneration. Later, a one-pot, two-step reaction strategy was used, in which the enzyme involved in the first step was thermally inactivated after the first step (dehydrogenation) to prevent the reduction of 7-KLCA to CDCA in the second step, and the UDCA yield was significantly increased to more than 98% at a substrate loading of 10 mM, and no residual 7-KLCA intermediate was detected.
7α-Hydroxysteroid dehydrogenase from C. absonum DSM 599 and 7β-HSDH from R. torques ATCC 359157 were used to produce several tens of kilograms of UDCA by the one-pot, one-step method with a conversion rate of more than 90% and a product purity of 98.5%, achieving large-scale UDCA production with CDCA (Zhang et al., 2019).
Almost all of the discovered 7β-HSDHs are NADPH-dependent enzymes, and a wild-type NADH-dependent 7β-HSDH from L. spicheri was identified, recombinantly expressed and purified by Tonin et al. (2018). This enzyme, in combination with the NAD+-dependent type 7α-HSDH from Stenotrophomonas maltophilia, converted CA and CDCA to UCA or UDCA with a >90% yield and only NAD+ addition required for catalysis.
To improve the efficiency of HSDHs, enzymes can be immobilized before use, regardless of whether a one-pot, two-step or one-pot, one-step method is used. Yang et al. (2017) used the enzyme immobilization technique to improve the activity and thermal stability of 7α-HSDH and 7β-HSDH, and the catalytic efficiency and reusability of 7α-HSDH and 7β-HSDH coimmobilized at EP-0.5-C were improved: the conversion of the coimmobilized enzymes was increased by approximately 45%, and the conversion remained at 83.7% after seven consecutive cycles.
7α-Hydroxysteroid dehydrogenase/lactate dehydrogenase (LDH) and 7β-HSDH/glucose dehydrogenase (GDH) were immobilized on epoxy resin ES-103 to produce immobilized enzymes LDH-7αHSDH@ES-103 and 7βHSDH-GDH@ES-103, respectively, for catalyzing UDCA synthesis with CDCA (Zheng et al., 2017a). By optimizing the immobilization pH, time, and enzyme–resin loading ratio, the specific activities of LDH-7αHSDH@ES-103 and 7βHSDH-GDH@ES-103 were increased by 12-fold and 516-fold, respectively. Subsequently, continuous UDCA production from CDCA was achieved by using the immobilized enzyme in two continuous packed-bed reactors. In the packed-bed reactor, the UDCA yield was close to 100%, and the reaction duration was at least 12 h, which was superior to that of the batch reaction. The continuous immobilized enzyme method developed in this study may provide a reference for UDCA synthesis by large-scale CDCA bioconversion.
A novel method for in vitro tauroursodeoxycholic acid (TUDCA) preparation was taken (Ji et al., 2016): a pair of 7α-HSDH and 7β-HSDH from C. absonum DSM599 were coimmobilized using modified chitosan microspheres as a carrier. In the batch reaction catalyzed by the dual-enzyme coupling system, more than 72% of taurochenodeoxycholic acid (TCDCA) was converted. The TUDCA yield achieved by coimmobilized enzyme microsphere catalysis was over 62%, and this method has potential applications in the in vitro synthesis of TUDCA and other high-value bile acid derivatives.
Protein engineering and cofactor regeneration strategies for enzymes used in UDCA synthesis
Protein engineering of hydroxysteroid dehydrogenase
To improve the efficiency and conversion rate of enzymatic UDCA synthesis, protein engineering technology has been applied to modify UDCA synthesis enzymes to improve their stability and activity or enhance the tolerance of HSDHs toward high substrate concentrations and even change the coenzyme specificity of HSDHs to reduce the production cost.
A combination of directed evolution and high-throughput screening was used for improving the catalytic efficiency and substrate-concentration tolerance of 7α-HSDH in C. absonum (Huang et al., 2019). Compared with the wild type, the best mutant 7α-3 increased the enzymatic activity by 5.5-fold and the catalytic efficiency for CDCA and NADP+ by 10-fold and 14-fold, respectively. In addition, the tolerance of 7α-3 to high substrate concentrations was significantly enhanced compared with that of the wild type. Due to the improved catalytic efficiency and enhanced substrate-concentration tolerance, 7α-3 converted 100 mM CDCA to 7-KLCA with a 99% conversion rate in 2 h. The conversion rate of the wild type under the same conditions was only 85% in 16 h.
Error-prone PCR, DNA shuffling, and site mutation were used to target and modify 7β-HSDH from R. torques, improve the activity, thermal stability, and optimal pH of 7β-HSDH (Zheng et al., 2017b). The best mutant, V3-1, had a 5.5-fold higher specific activity and a 28-fold longer half-life than the wild type. In addition, the optimal reaction pH of the mutant changed from acidic to weakly basic. In the cascade reaction, the yield of UDCA synthesized by the V3-1 mutant increased to 942 g L–1 d–1 compared with 141 g L–1 d–1 for the wild type. This study provides a useful strategy for the modification of HSDHs to promote UDCA biosynthesis.
Almost all existing reported 7β-HSDH are strictly dependent on NADPH; however, NADH is more economical than NADPH and is a preferred cofactor for HSDH synthesis in UDCA applications. You et al. (2018) used a strategy called “Cofactor specificity reversal: small-and-smart library design (CSR-SaSLiD) strategy” to coengineer recombinant 7β-HSDH derived from R. torques to alter its cofactor dependence. They designed a CSR-SaSLiD library consisting of only five mutants, resulting in the successful discovery of two ideal 7β-HSDH mutants, G39D and G39D/T17A. G39D showed a 953,000-fold reversal in NADH/NADPH cofactor specificity, while G39D/T17A showed a 223-fold increase in activity against NADH. Through molecular dynamics simulations, they elucidated the structural mechanism by which mutations reverse cofactor preference and increase catalytic activity and further demonstrated that the CSR-SaSLiD strategy can be extended to other 7β-HSDHs. This work provides an efficient method for altering the cofactor preference of HSDHs and subsequently restoring enzyme activity and can reduce the cost of transformation.
An NADP+-dependent 7α-HSDH gene from Clostridium difficile was cloned and expressed in E. coli (Bakonyi and Hummel, 2016). It was found that unlike other 7α-HSDHs, this enzyme did not exhibit substrate inhibition. By site mutation based on structural analysis, they transformed the cofactor specificity of this enzyme to accept NAD(H), thereby reducing the cost of using cofactors in UDCA synthesis.
Lou et al. (2017) identified the cofactor-specific determining sites (csss) of 7α-HSDH from C. absonum, namely T15, R16, R38, and R194, and then performed alanine scanning mutagenesis and molecular dynamics (MD) simulations. The results showed that the csss of wild-type 7α-HSDH had high affinity for NADP(H) but low catalytic efficiency (kcat) for NADP+. However, the mutant R194A had a higher catalytic efficiency (kcat/Km) for NADP+ than the wild type and other mutants, which increased the catalytic efficiency of R194A toward CDCA oxidation to more than three times that of the wild type.
In addition, several patents have reported numerous evolutionary studies and related applications of HSDH enzymes, also focusing on enzyme activity enhancement, stability enhancement, high-substrate-concentration tolerance improvement or cofactor-specific transformation (Weuster-botz et al., 2017, 2021; Liu et al., 2018).
Cofactor regeneration strategy
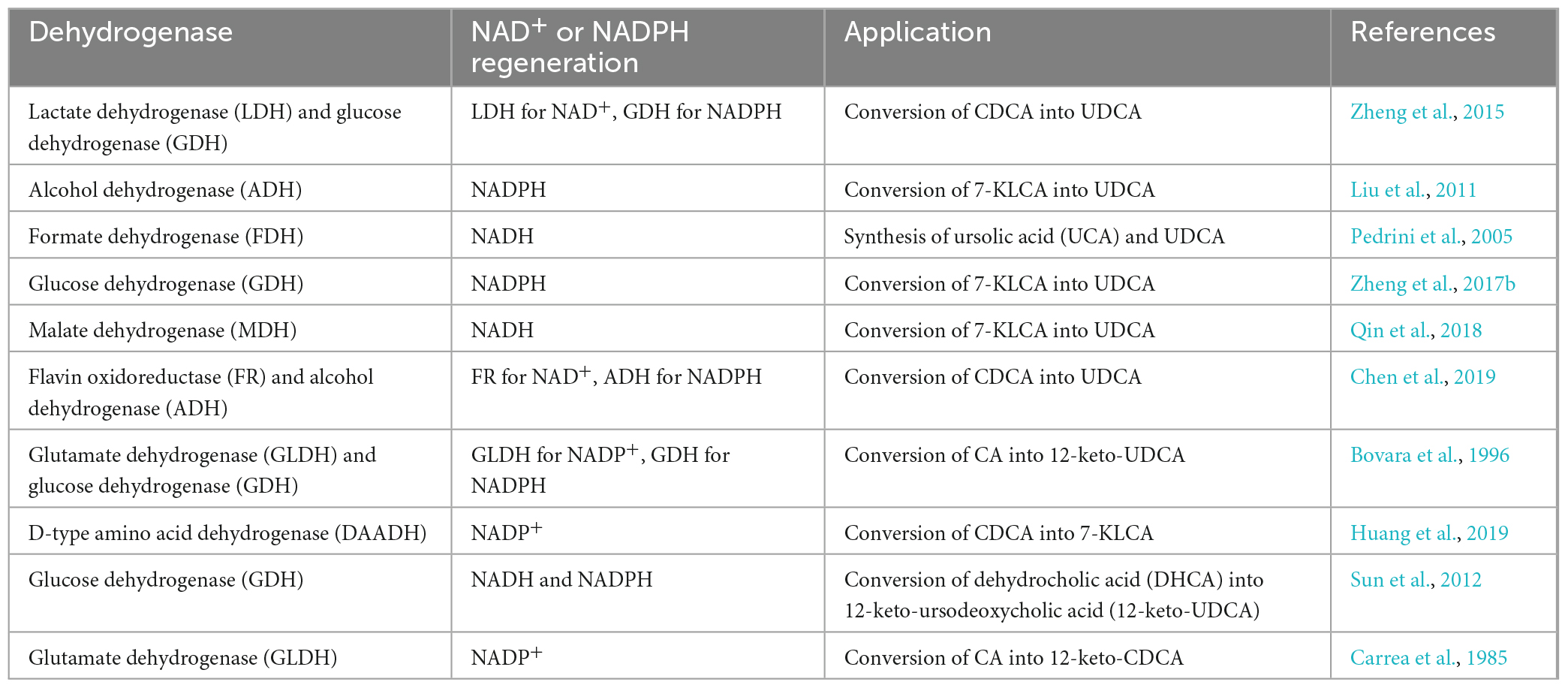
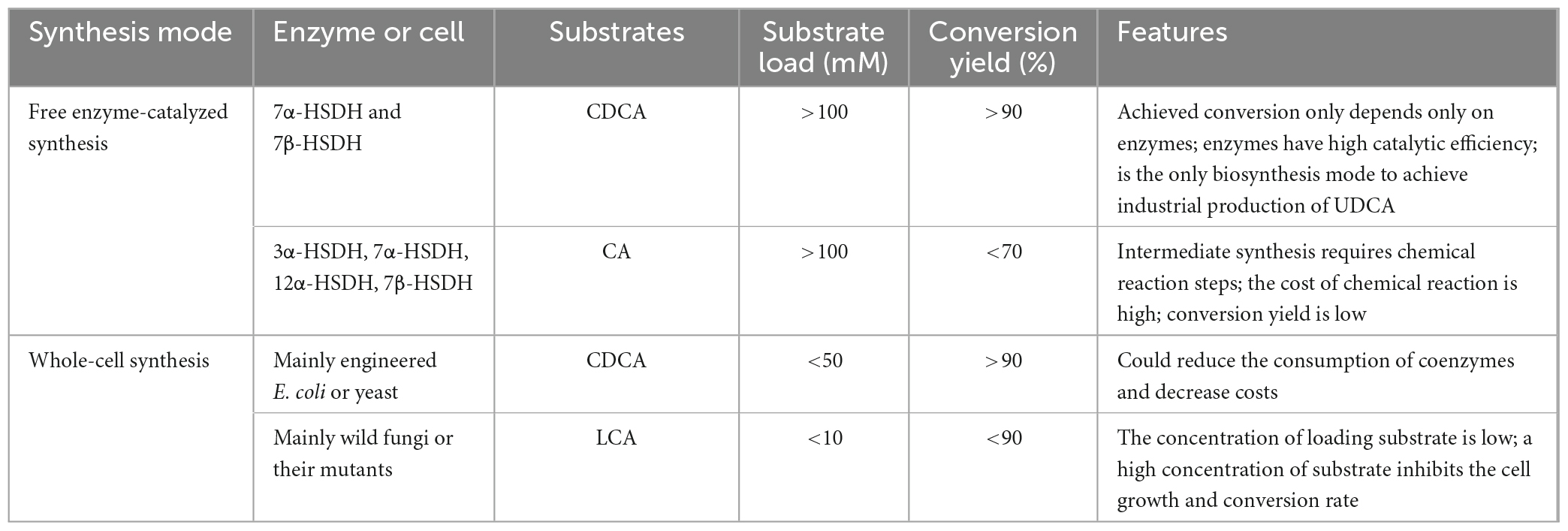
The enzymes that catalyze the biosynthesis of UDCA from CDCA or CA are HSDHs, and they all belong to the family. One of the typical features of this family of enzymes is that they have an NAD(P)(H) binding region (TGXXX[AG]XG), and their reactions all require NAD(H) or NADP(H) coenzymes (Figure 1; Kavanagh et al., 2008; Savino et al., 2016; Wang et al., 2017). Theoretically, equimolar amounts of cofactors are required in relation to the amount of substrate that is consumed or product that is generated, and these cofactors are often expensive, making it infeasible to increase the cofactor amount from an economic point of view; on the other hand, excess cofactors also inhibit HSDH activity, which technically limits cofactor application. Therefore, according to the process economics of biocatalytic reactions and the feasibility of industrial applications, HSDH systems must meet the requirements of efficient and low-cost cofactor regeneration systems in addition to having suitable enzymes and compatible reaction engineering technologies in their applications. Cofactor regeneration refers to the regeneration of the cofactor from the oxidation state to the reduction state or vice versa, which allows the cofactor to be maintained at a certain catalyst amount level. A number of methods have been proposed to address this cofactor regeneration issue, including chemical, photochemical, enzymatic, and electrochemical regeneration systems (Zhang, 2001; Hummel and Gröger, 2014). Enzymatic UDCA synthesis must also take advantage of a suitable cofactor regeneration system for industrial UDCA production. The efficiency of a cofactor regeneration system is usually measured by the cofactor conversion number (turnover number, TN) and the total conversion number (total turnover number, TTN) (Weckbecker et al., 2010; Kara et al., 2014; Mordhorst and Andexer, 2020). Bioconversion processes with TTNs of 103–105 are usually considered economically feasible. Coenzyme I (NAD+) and coenzyme II (NADPH) in enzymatic UDCA synthesis are almost always regenerated enzymatically, and dehydrogenases are used (Table 3).
As listed in Table 3, there are a variety of dehydrogenases available for NAD(H) and NADP(H) regeneration in the biological synthesis of UDCA. However, some commonly used dehydrogenases have notable characteristics (Table 4): the most commonly used are LDH for NAD+ regeneration and GDH for NADPH regeneration. One of the reasons for this is that coenzyme NAD+ regeneration using the LDH/pyruvate system is more advantageous in terms of cost compared to other coenzyme regeneration systems; LDH is more active and more stable in the reaction system, and the reaction substrate pyruvate is relatively inexpensive. In conclusion, the LDH/pyruvate system is advantageous for NAD+ regeneration because of its low cost, high stability and easy post-processing. Regarding NADPH regeneration, the GDH/glucose system has very outstanding features, including being extremely stable to oxygen and having a highly specific activity for NADP+. Moreover, glucose is inexpensive, easy to obtain, stable, highly reducible, harmless to enzymes and coenzymes, and increases enzyme stability. These advantages make GDH/glucose the best dehydrogenase system for NADPH regeneration. Although the byproduct gluconate produced by the GDH/glucose coenzyme regeneration system may complicate the post-reaction treatment, this system still has significant advantages over other coenzyme regeneration systems in terms of cost and stability in large-scale applications. The GDH/glucose regeneration system is currently the best method for regenerating NADPH.
Whole-cell UDCA synthesis
Research on whole-cell UDCA synthesis began with the discovery that wild microbial strains convert CDCA or LCA to UDCA, and researchers found that some microorganisms convert CDCA to UDCA when cultured alone (Macdonald et al., 1981; Kole and Altosaar, 1985; Lepercq, 2004) or in coculture (Hirano and Masuda, 1981; MacDonald et al., 1982); however, wild microorganisms can only carry very low substrate concentrations (micromolar or a few millimolar CDCA), and once the substrate concentration increases, microbial growth is inhibited. Moreover, their conversion rate is also very low, with gram-level substrate concentrations being converted into only milligram-level product concentrations. It was later confirmed that these microorganisms self-express 7α-HSDH and/or 7β-HSDH.
For the whole-cell catalytic study of converting LCA into UDCA, Sawada et al. (1982) isolated 609 fungal strains with hydroxylation abilities from soil samples, from which a strain of Fusarium spp. Fusarium equiseti M41 with good C-7 hydroxylation ability was selected, and this strain converted 1 g/L LCA into 350 mg/L UDCA after 112 h of incubation. Previously, Carlström et al. (1981) also reported that Absidia coerulea and Rhizoctonia solani also had the ability to hydroxylate C-7.
Kollerov et al. (2013) further screened different species of actinomycetes and filamentous fungi for their ability to convert LCA into UDCA and identified for the first time that Bipolaris, Gibberella, Cunninghamella, Curvularia, Pseudonocardia, Saccharothrix, Amycolatopsis, Lentzea, Saccharopolyspora, and Nocardia genera directly converted LCA into UDCA. Moreover, they screened a Gibberella zeae VKM F-2600 strain with high β-hydroxylation activity at C-7 against LCA. After optimizing the conditions, close to 90% of 1 g/L LCA was converted into UDCA. Recently, a mutant G. zeae M23 was obtained by UV mutagenesis (Kollerov and Donova, 2022). The mutant strain offered better conversion ability and could convert 88% of 4 g/L LCA into UDCA. This yield of UDCA is superior to previously reported yields.
The microorganisms that convert CDCA to UDCA produce 7α-HSDH and 7β-HSDH, and the microorganisms that convert LCA to UDCA produce P450 monooxygenases. These microorganisms became an important source of late HSDH genes and protein sequences. However, wild microorganisms, because of their low HSDH expression, have low enzyme activities, and slightly higher concentrations of substrate also inhibit their growth and conversion. Therefore, relevant whole-cell catalytic studies were little explored by researchers. It was not until the heterologous expression of related HSDHs using engineered strains, especially E. coli, and the more successful synthesis of UDCA from CDCA with these engineered bacteria that whole-cell catalysis research came back into the limelight. Compared with free-enzyme catalysis, whole-cell catalysis is advantageous because it uses the coenzyme regeneration systems of microbial cells and eliminates the steps of bacterial crushing and enzyme extraction. Regarding the synthesis of UDCA from LCA, progress has not yet been made in whole-cell catalysis research because of the lack of P450 enzyme resources and harsh reaction conditions.
An engineered E. coli integrating 7α-HSDH and 7β-HSDH had been constructed (Shi et al., 2017) to convert TCDCA into TUDCA and CDCA into UDCA. The recombinant E. coli integrating Clostridium sardiniense-derived 7α-HSDH and Ruminococcus gnavus-derived 7β-HSDH was able to convert approximately 50% of TCDCA to TUDCA, and this study provided a potential pathway for producing a bear bile substitute using inexpensive and readily available chicken bile. Later, they developed a deep tank static process through fermentation optimization and balanced bidirectional reactions to control the ratio of converted TUDCA to TCDCA similar to that of medicinal bear bile powder, providing a practical yet environmentally friendly industrial production method for producing artificial bear bile powder substitutes from chicken bile powder using microbial cell factories (Xu et al., 2019). Recently, the group constructed a Saccharomyces cerevisiae host expressing these two enzymes and optimized the gene combination and whole-cell transformation conditions. From the perspective of food and drug safety, S. cerevisiae is a GRAS strain and has a higher safety profile than E. coli (Jin et al., 2022).
The whole-cell synthesis of 12-keto-UDCA, a key intermediate in the chemical-enzymatic synthesis of UDCA, had been achieved (Sun et al., 2012). 7β-HSDH mutant from Collinsella aerofaciens and 3α-HSDH from Comamonas testosteroni were used to reduce DHCA to 12-keto-UDCA by a two-step reaction. Researchers first coexpressed 3α-HSDH, 7β-HSDH, and a formate dehydrogenase (FDH) mutant from Mycobacterium vaccae N10 in the same strain of E. coli BL21(DE3) cells, thus constructing a single-strain whole-cell catalytic system; then, they coexpressed 3α-HSDH, 7β-HSDH and a GDH from Bacillus subtilis in the same strain of E. coli BL21(DE3) cells to construct another single-strain whole-cell catalytic system. After comparing the two whole-cell catalytic systems, the single-strain expression system constructed by 3α-HSDH, β-HSDH, and GDH achieved the best results, and the system was able to almost completely convert 100 mM DHCA into 12-keto-UDCA (>99.5 mM) within 4.5 h.
Braun et al. (2012) similarly achieved the whole-cell synthesis of 12-keto-UDCA. They expressed NADH-dependent 3α-HSDH from C. testosteroni in an E. coli strain already overexpressing 7β-HSDH (from C. aerofaciens) and an NAD(P) bispecific FDH mutant (from M. vaccae). This engineered cell was able to directly convert 50 mM DHCA to 12-keto-UDCA with only a small amount of intermediate product remaining.
The complete conversion of 30 mM CDCA into UDCA in one pot had been achieved, using different coenzyme-specific 7α-HSDH and 7β-HSDH and an independent coenzyme cycling system (Chen et al., 2019). In the study, an NAD+ regeneration system was developed by using flavin oxidoreductase (FR) and flavin mononucleotide (FMN). The FR/FMN system subsequently coupled NAD+-dependent 7α-HSDH from Brevundimonas sp. to oxidize CDCA to 7-KLCA. When whole cells of E. coli coexpressed 7α-HSDH and FR, a complete conversion of 50 mM CDCA into 7-keto-LCA was achieved. For the reduction of 7-KLCA, NADPH-dependent 7β-HSDH from Clostridium sp. was used, and alcohol dehydrogenase from Thermoanaerobacter brockii (TbADH) and isopropanol were used as cofactor regeneration systems. When catalyzed by this E. coli-engineered bacterial whole-cell system, 40 mM 7-KLCA was reduced to UDCA with a conversion rate of 26.8%. Then, 12.5 mM CDCA was completely converted into UDCA when both cells were reacted simultaneously in a one-pot, one-step method. Unfortunately, bile acids, especially UDCA, inhibit the activity of these enzymes, so the high concentration of substrate and intermediate products prevented the complete conversion of 7α-OH, and the substrate inhibition problem still needs to be solved by enzyme engineering.
Regarding whole-cell catalysis, several patented technologies have been granted. Weuster-botz et al. (2017) reported the recombinant whole-cell biocatalytic production of UDCA with a 7β-HSDH enzyme mutant in patent US 20170191104; Hummel and Bakonyi (2022), in turn, provided a novel 3α-HSDH mutant and a method for UDCA production using a recombinant microorganism containing this mutant.
The biological synthesis modes of UDCA are summarized in Table 5. In short, each of these synthesis methods has different characteristics. In terms of synthesis efficiency, CDCA is the most efficient substrate and has the highest conversion rate in both enzymatic and whole-cell synthesis methods, which is the focus of current research.
Conclusion and future perspectives
With the continuous application development of UDCA and its derivatives in the fields of hepatobiliary diseases and tumor therapy in recent years, UDCA synthesis research has made great strides. Currently, chemical synthesis is still the main method for large-scale UDCA production, but this method is not sufficient for meeting the market demand due to factors such as low yield, cumbersome steps and environmental unfriendliness. Biosynthesis, on the other hand, has gradually become the mainstream research direction for UDCA preparation because of its advantages of a high conversion rate, low byproduct levels and environmental safety.
Ursodeoxycholic acid biosynthesis has demonstrated its superiority over chemical UDCA synthesis, and the industrial UDCA production process using CDCA as substrate has been realized. However, there are still some limitations of this process in actual production. First, the enzymes used (especially 7β-HSDH) are unstable, and their enzyme activities are rapidly reduced in the reaction system. Second, the substrate-concentration tolerance of enzyme reaction systems is still low, and some enzymes are seriously inhibited by the products or intermediates. A one-pot, one-step method can allow the substrate concentration to be increased, but the conversion rate is low, and the residual substrate and intermediate products put more pressure on the downstream purification; a one-pot, two-step method can increase the conversion rate, but it also requires the substrate concentration to be lowered, resulting in a low production efficiency. It also generates a large amount of wastewater, which results in the risk of environmental harm. Although the expression of existing HSDHs by E. coli heterologous expression systems is considerable and the enzyme source is no longer a major limiting factor, mechanical shear stress, and physical damage during the production process can easily cause enzyme loss and lead to a decrease in enzyme recovery. In addition, the potential of coenzyme dosage optimization and whole-cell catalysis in industrial applications still needs to be further explored; the biotransformation of LCA to UDCA is still far from being industrially relevant because of the actual catalytic capacity limitation of the P450 enzyme monooxygenase.
The abovementioned problems are the main factors limiting industrial UDCA production by various biotransformation methods. In addition, the biosynthetic pathway sometimes involves the issue of microbial pathogenicity assessment, which requires the development of efficient heterologous expression systems other than E. coli systems. There is no doubt that enzymes are the most central component of the biosynthetic UDCA process, so the further expansion of enzyme sources, the development of screening procedures for new enzymes, and the enhancement of efforts to modify enzymes in the field of protein engineering should result in biocatalysts that can be applied at high substrate concentrations of >100 mM. In contrast, the application of immobilized enzyme technology and the use of recombinant whole-cell catalysis should greatly reduce the cost of catalysis and facilitate product isolation and purification.
The problems to be solved in order to achieve UDCA biosynthesis are as follows: 1. developing one-pot, one-step and whole-cell catalytic methods by mining or modifying cofactor-specific HSDH and cofactor regeneration enzymes while also taking into account the enzyme activity and substrate high-carrying-capacity requirements; and 2. screening wild strains from bacteria and actinomycetes that can transform LCA into UDCA, selecting the P450 monooxygenases with C-7 hydroxylation functions, and further modifying them by protein engineering technology to obtain enzymes that can meet industrial application requirements and make full use of LCA raw material resources.
Author contributions
PS and RF: conceptualization. XZ, WF, WX, CW, SX, and SY: literature search. PS and XZ: writing—original draft preparation. PS, WX, and SY: writing—review and editing. SX and RF: funding acquisition. All authors read and approved the manuscript.
Funding
This work was supported by the Natural Science Foundation of Shandong Province (grant number ZR2022MC159).
Acknowledgments
We would like to thank AJE (https://www.aje.cn) for English language editing.
Conflict of interest
PS, CW, SX, SY, and RF were employed by Jiangxi Zymerck Biotechnology Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdelkader, N. F. (2020). Ursodeoxycholic and tauroursodeoxycholic acids as antiapoptotic agents: modulation of Parkinson’s disease. Diagn. Manage. Parkinsons Dis. 1, 653–664. doi: 10.1016/B978-0-12-815946-0.00037-5
Allen, S. J., and Holbrook, J. J. (1995). Isolation, sequence and overexpression of the gene encoding NAD-dependent formate dehydrogenase from the methylotrophic yeast Candida methylica. Gene 162, 99–104. doi: 10.1016/0378-1119(95)00347-9
Bakonyi, D., and Hummel, W. (2016). Cloning, expression, and biochemical characterization of a novel NADP+-dependent 7α-hydroxysteroid dehydrogenase from Clostridium difficile and its application for the oxidation of bile acids. Enzyme Microb. Technol. 99, 16–24. doi: 10.1016/j.enzmictec.2016.12.006
Bakonyi, D., Hummel, W., and Gross, R. (2020). Coupled, self-sufficient biotransformation of chenodeoxcholic acid to ursodeoxycholic acid and novel enzyme mutants applicable in said process. U.S. Patent No 20200407766. Washington, DC: U.S. Patent and Trademark Office.
Bakonyi, D., Wirtz, A., and Hummel, W. (2012). Large-scale enzymatic synthesis of 12-ketoursodeoxycholic acid from dehydrocholic acid by simultaneous combination of 3α-hydroxysteroid dehydrogenase from Pseudomonas testosteroni and 7β-hydroxysteroid dehydrogenase from Collinsella aerofaciens. Z. Naturforsch. B. 67, 1037–1044. doi: 10.5560/znb.2012-0165
Baron, S. F., Franklund, C. V., and Hylemon, P. B. (1991). Cloning, sequencing, and expression of the gene coding for bile acid 7 alpha-hydroxysteroid dehydrogenase from Eubacterium sp. strain VPI 12708. J. Bacteriol. 173, 4558–4569. doi: 10.1128/jb.173.15.4558-4569.1991
Bell, S. M., Barnes, K., Clemmens, H., Al-Rafiah, A. R., Al-Ofi, E. A., Leech, V., et al. (2018). Ursodeoxycholic acid improves mitochondrial function and redistributes drp1 in fibroblasts from patients with either sporadic or familial alzheimer’s disease. J. Mol. Biol. 430, 3942–3953. doi: 10.1016/j.jmb.2018.08.019
Bennett, M. J., McKnight, S. L., and Coleman, J. P. (2003). Cloning and characterization of the NAD-dependent 7 alpha-hydroxysteroid dehydrogenase from Bacteroides fragilis. Curr. Microbiol. 47, 475–484. doi: 10.1007/s00284-003-4079-4
Beuers, U., Spengler, U., Kruis, W., Aydemir, U., Wiebecke, B., Heldwein, W., et al. (1992). Ursodeoxycholic acid for treatment of primary sclerosing cholangitis: a placebo-controlled trial. Hepatology 16, 707–714. doi: 10.1002/hep.1840160315
Bovara, R., Carrea, G., Riva, S., and Secundo, F. (1996). A new enzymatic route to the synthesis of 12-ketoursodeoxycholic acid. Biotechnol. Lett. 18, 305–308. doi: 10.1007/BF00142949
Braun, M., Sun, B., Anselment, B., and Weuster-Botz, D. (2012). Novel whole-cell biocatalysts with recombinant hydroxysteroid dehydrogenases for the asymmetric reduction of dehydrocholic acid. Appl. Microbiol. Biotechnol. 95, 1457–1468. doi: 10.1007/s00253-012-4072-6
Byelyayeva, N. (2015). Effectiveness of ursodeoxycholic acid (UDCA) in patients with chronic pancreatitis (CP) and biliary sludge. Pancreatology 15:69. doi: 10.1016/j.pan.2015.05.260
Cacciato, V., Benizzelli, G., Grillo, F., Borro, P., Marenco, S., Pasta, A., et al. (2020). Primary biliary cholangitis: histological and clinical liver progression in non responders to ursodeoxycholic acid. Digest. Liver Dis. 52, 35–36. doi: 10.1016/j.dld.2019.12.124
Carbone, M., Nardi, A., Flack, S., Carpino, G., Varvaropoulou, N., Gavrila, C., et al. (2018). Pretreatment prediction of response to ursodeoxycholic acid in primary biliary cholangitis: development and validation of the UDCA response score. Lancet Gastroenterol. 9, 626–634. doi: 10.1016/S2468-1253(18)30163-8
Carlström, K., Kirk, D. N., and Sjövall, J. (1981). Microbial synthesis of 1 beta- and 15 beta-hydroxylated bile acids. J. Lipid Res. 22, 1225–1234. doi: 10.1016/S0022-2275(20)37315-6
Carrea, G., Bovara, R., Longhi, R., and Riva, S. (1985). Preparation of 12-ketochenodeoxycholic acid from cholic acid using coimmobilized 12α-hydroxysteroid dehydrogenase and glutamate dehydrogenase with NADP+ cycling at high efficiency. Enzyme Microb. Technol. 7, 597–600. doi: 10.1016/0141-0229(85)90027-4
Carrea, G., Pilotti, A., Riva, S., Canzi, E., and Ferrari, A. (1992). Enzymatic synthesis of 12-ketoursodeoxycholic acid from dehydrocholic acid in a membrane reactor. Biotechnol. Lett. 14, 1131–1134. doi: 10.1007/bf01027015
Chappell, L. C., Bell, J. L., Smith, A., Linsell, L., Juszczak, E., Dixon, P. H., et al. (2020). Ursodeoxycholic acid versus placebo in women with intrahepatic cholestasis of pregnancy (PITCHES): a randomized controlled trial. Obstetr. Anesth. Digest. 3, 151–152. doi: 10.1097/01.aoa.0000693804.53035.f5
Chen, W., Feng, Z., and Sun, Q. (2022). A novel ursodeoxycholic acid-chitosan-folate conjugates for the delivery of calcitriol for cancer therapy. J. Drug Deliv. Sci. Technol. 73:103410. doi: 10.1016/J.JDDST.2022.103410
Chen, X., Cui, Y., Feng, J., Wang, Y., Liu, X., Wu, Q., et al. (2019). Flavin oxidoreductase-mediated regeneration of nicotinamide adenine dinucleotide with dioxygen and catalytic amount of flavin mononucleotide for one-pot multi-enzymatic preparation of ursodeoxycholic acid. Adv Synth. Catal. 361, 2497–2504. doi: 10.1002/adsc.201900111
Coleman, J. P., Hudson, L. L., and Adams, M. J. (1994). Characterization and regulation of the NADP-linked 7 alpha-hydroxysteroid dehydrogenase gene from Clostridium sordellii. J. Bacteriol. 176, 4865–4874. doi: 10.1128/jb.176.16.4865-4874.1994
Colombo, C., Alicandro, G., Oliver, M., Lewindon, P. J., Ramm, G. A., Ooi, C. Y., et al. (2021). Ursodeoxycholic acid and liver disease associated with cystic fibrosis: a multicenter cohort study. J. Cyst. Fibros. 21, 220–226. doi: 10.1016/j.jcf.2021.03.014
Combes, B., Carithers, R. L., Maddrey, W. C., Lin, D., Mcdonald, M. F., Wheeler, D. E., et al. (1995). A randomized, double-blind, placebo-controlled trial of ursodeoxycholic acid in primary biliary cirrhosis. Hepatology 22, 759–766. doi: 10.1016/0270-9139(95)90294-5
Dangate, P. S., Salunke, C. L., and Akamanchi, K. G. (2011). Regioselective oxidation of cholic acid and its 7β epimer by using o-iodoxybenzoic acid. Steroids 76, 1397–1399. doi: 10.1016/j.steroids.2011.07.009
Deneau, M., Perito, E., Ricciuto, A., Gupta, N., Kamath, B. M., Palle, S., et al. (2019). Ursodeoxycholic acid therapy in pediatric primary sclerosing cholangitis: predictors of gamma glutamyltransferase normalization and favorable clinical course. J. Pediatr. 209, 92–96. doi: 10.1016/j.jpeds.2019.01.039
Eggert, T., Bakonyi, D., and Hummel, W. (2014). Enzymatic routes forthe synthesis of ursodeoxycholic acid. J. Biotechnol. 191, 11–21. doi: 10.1016/j.jbiotec.2014.08.006
Elisa, F. E., Maria, B. G., Fausto, P., Armando, N., Sergio, R., and Daniela, M. (2012). In search of sustainable chemical processes: cloning, recombinant expression, and functional characterization of the 7 alpha- and 7 beta-hydroxysteroid dehydrogenases from Clostridium absonum. Appl. Microbiol. Biotechnol. 95, 1221–1233. doi: 10.1007/s00253-011-3798-x
Eun-Kyung, K., Hee, C. J., Euijoo, K., and Jae, K. Y. (2017). Ursodeoxycholic acid inhibits the proliferation of colon cancer cells by regulating oxidative stress and cancer stem-like cell growth. PLoS One 12:e0181183. doi: 10.1371/journal.pone.0181183
Giovannini, P. P., Grandini, A., Perrone, D., Pedrini, P., Fantin, G., and Fogagnolo, M. (2008). 7α- and 12α-Hydroxysteroid dehydrogenases from Acinetobacter calcoaceticus lwoffii: a new integrated chemo-enzymatic route to ursodeoxycholic acid. Steroids 73, 1385–1390. doi: 10.1016/j.steroids.2008.06.013
Goossens, J. F., and Bailly, C. (2019). Ursodeoxycholic acid and cancer: from chemoprevention to chemotherapy. Pharmacol. Therapeut. 203:107396. doi: 10.1016/j.pharmthera.2019.107396
Grobe, S., Badenhorst, C. P. S., Bayer, T., Hamnevik, E., Wu, S., Grathwol, C. W., et al. (2020). Engineering regioselectivity of a P450 monooxygenase enables the synthesis of ursodeoxycholic acid via 7ß-hydroxylation of lithocholic acid. Angew. Chem. Int. Edit. 60, 753–757. doi: 10.1002/anie.202012675
Gupta, A., Tschentscher, A., and Bobkova, M. (2009). Process for the enantioselective reduction and oxidation, respectively, of steroids. U.S. Patent No 20090280525. Washington, DC: U.S. Patent and Trademark Office.
Haal, S., Guman, M. S. S., Boerlage, T. C. C., Acherman, Y. I. Z., Brauw, D. L. M., Bruin, S., et al. (2021). Ursodeoxycholic acid for the prevention of symptomatic gallstone disease after bariatric surgery (UPGRADE): a multicentre, double-blind, randomised, placebo-controlled superiority trial. Lancet Gastroenterol. 12, 993–1001. doi: 10.1016/S2468-1253(21)00301-0
Harms, M. H., van Buuren, H. R., Corpechot, C., Thorburn, D., Janssen, H., Lindor, K. D., et al. (2019). Ursodeoxycholic acid therapy and liver transplant-free survival in patients with primary biliary cholangitis. J. Hepatol. 71, 357–365. doi: 10.1016/j.jhep.2019.04.001
He, Q., Wu, J., Ke, J., Zhang, Q., Zeng, W., Luo, Z., et al. (2022). Therapeutic role of ursodeoxycholic acid in colitis-associated cancer via gut microbiota modulation. Mol. Ther. 109, 2126–2140. doi: 10.1016/j.ymthe.2022.10.014
Hirano, S., and Masuda, N. (1981). Epimerization of the 7-hydroxy group of bile acids by the combination of two kinds of microorganisms with 7 alpha- and 7 beta-hydroxysteroid dehydrogenase activity, respectively. J. Lipid Res. 22, 1060–1068. doi: 10.1016/S0022-2275(20)40663-7
Hofmann, A. F., Lundgren, G., Theander, O., Brimacombe, J. S., and Cook, M. C. (1963). The preparation of chenodeoxycholic acid and its glycine and taurine conjugates. Acta Chem. Scand. 17, 173–186. doi: 10.3891/acta.chem.scand.17-0173
Hollmann, F., Kleeb, A., Otto, K., and Schmid, A. (2005). Coupled chemoenzymatic transfer hydrogenation catalysis for enantioselective reduction and oxidation reactions. Tetrahedron 16, 3512–3519. doi: 10.1016/j.tetasy.2005.09.026
Höllrigl, V., Hollmann, F., Kleeb, A. C., Buehler, K., and Schmid, A. (2008). TADH, the thermostable alcohol dehydrogenase from Thermus sp. ATN1: a versatile new biocatalyst for organic synthesis. Appl. Microbiol. Biotechnol. 81, 263–273. doi: 10.1007/s00253-008-1606-z
Huang, B., Zhao, Q., Zhou, J., and Xu, G. (2019). Enhanced activity and substrate tolerance of 7α-hydroxysteroid dehydrogenase by directed evolution for 7-ketolithocholic acid production. Appl. Microbiol. Biotechnol. 103, 2665–2674. doi: 10.1007/s00253-019-09668-4
Huang, F. (2021). Ursodeoxycholic acid as a potential alternative therapeutic approach for neurodegenerative disorders: effects on cell apoptosis, oxidative stress and inflammation in the brain. Brain Behav. Immun. 18:100348. doi: 10.1016/J.BBIH.2021.100348
Hummel, W., and Bakonyi, D. (2016). 7-beta-hydroxysteroid dehydrogenase mutants and process for the preparation of ursodeoxycholic acid. WIPO Patent Application No WO/2016/016213 A1. Geneva: World Intellectual Property Organization.
Hummel, W., and Bakonyi, D. (2022). 3α-hydroxysteroid dehydrogenase mutants and process for the preparation of ursodeoxycholic acid. U.S. Patent No 11306343. Washington, DC: U.S. Patent and Trademark Office.
Hummel, W., and Gröger, H. (2014). Strategies for regeneration of nicotinamide coenzymes emphasizing self-sufficient closed-loop recycling systems. J. Biotechnol. 191, 22–31. doi: 10.1016/j.jbiotec.2014.07.449
Im, E., and Martinez, J. D. (2004). Ursodeoxycholic acid (UDCA) can inhibit deoxycholic acid (DCA)-induced apoptosis via modulation of EGFR/Raf-1/ERK signaling in human colon cancer cells. J. Nutr. 134, 483–486. doi: 10.1093/jn/134.2.483
Ji, Q., Tan, J., Zhu, L., Lou, D., and Wang, B. (2016). Preparing tauroursodeoxycholic acid (TUDCA) using a double-enzyme-coupled system. Biochem. Eng. J. 105, 1–9. doi: 10.1016/j.bej.2015.08.005
Ji, S., Pan, Y., Zhu, L., Tan, J., Tang, S., Yang, Q., et al. (2021). A novel 7α-hydroxysteroid dehydrogenase: magnesium ion significantly enhances its activity and thermostability. Int. J. Biol. Macromol. 177, 111–118. doi: 10.1016/J.IJBIOMAC.2021.02.082
Ji, W., Chen, Y., Zhang, H., Zhang, X., Li, Z., and Yu, Y. (2014). Cloning, expression and characterization of a putative 7alpha-hydroxysteroid dehydrogenase in Comamonas testosterone. Microbiol. Res. 169, 148–154. doi: 10.1016/j.micres.2013.07.009
Jin, L., Yang, L., Zhao, S., and Wang, Z. T. (2022). A green strategy to produce potential substitute resource for bear bile using engineered Saccharomyces cerevisiae. Bioresour. Bioprocess. 9, 1–10. doi: 10.1186/s40643-022-00517-3
Kara, S., Schrittwieser, J. H., Hollmann, F., and Ansorge-Schumacher, M. B. (2014). Recent trends and novel concepts in cofactor-dependent biotransformations. Appl. Microbiol. Biotechnol. 98, 1517–1529. doi: 10.1007/s00253-013-5441-5
Karim, M. A. U., and Yesmin, N. (2020). Hepatoprotective efficacy of ursodeoxycholic acid in pediatric acute lymphoblastic leukemia. J. Pediat. Hematol Oncol. 5:S4. doi: 10.1016/j.phoj.2021.04.011
Kato, S., Konuma, T., Monna-Oiwa, M., Isobe, M., Takahashi, S., and Nannya, Y. (2022). Higher cryopreserved CD34+cell dose is associated with decreased hepatic veno-occlusive disease/sinusoidal obstruction syndrome after single-unit cord blood transplantation in adults given prophylactic ursodeoxycholic acid and intravenous heparin. Transpl. Cell Ther. 28, 779.e1–e9. doi: 10.1016/j.jtct.2022.08.013
Kavanagh, K. L., Jörnvall, H., Persson, B., and Oppermann, U. (2008). Medium- and short-chain dehydrogenase/reductase gene and protein families: the SDR superfamily: functional and structural diversity within a family of metabolic and regulatory enzymes. Cell. Mol. Life Sci. 65, 3895–3906. doi: 10.1007/s00018-008-8588-y
Khachidze, T. (2019). Comparison of the efficacy of ursodeoxycholic acid (UDCA) versus vitamin E plus vitamin C in non-diabetic patients with nonalcoholic Steatohepatitis (NASH). HPB 2, 398–399. doi: 10.1016/j.hpb.2019.10.2084
Kole, M. M., and Altosaar, I. (1985). Conversion of chenodeoxycholic acid to ursodeoxycholic acid by Clostridium absonumin culture and by immobilized cells. FEMS Microbiol. Lett. 28, 69–72. doi: 10.1111/j.1574-6968.1985.tb00766.x
Kollerov, V., and Donova, M. (2022). Ursodeoxycholic acid production by Gibberella zeae mutants. AMB Expr. 12:105. doi: 10.1186/s13568-022-01446-2
Kollerov, V. V., Monti, D., Deshcherevskaya, N. O., Lobastova, T. G., Ferrandi, E. E., Larovere, A., et al. (2013). Hydroxylation of lithocholic acid by selected Actinobacteria and Filamentous fungi. Steroids 78, 370–378. doi: 10.1016/j.steroids.2012.12.010
Kuipers, R. K., Joosten, H. J., van Berkel, W. J., Leferink, N. G., Rooijen, E., Ittmann, E., et al. (2010). 3DM: systematic analysis of heterogeneous superfamily data to discover protein functionalities. Proteins 78, 2101–2113. doi: 10.1002/prot.22725
Lee, J. Y., Arai, H., Nakamura, Y., Fukiya, S., Wada, M., and Yokota, A. (2013). Contribution of the 7β-hydroxysteroid dehydrogenase from Ruminococcus gnavus N53 to ursodeoxycholic acid formation in the human colon. J. Lipid Res. 54, 3062–3069. doi: 10.1194/jlr.M039834
Lee, W. S., Jung, J. H., Panchanathan, R., Yun, J. W., Kim, D. H., Kim, H. J., et al. (2017). Ursodeoxycholic acid induces death receptor-mediated apoptosis in prostate cancer cells. J. Cancer Prev. 22, 16–21. doi: 10.15430/JCP.2017.22.1.16
Leoni, M. C., Amelung, L., Lieveld, F. I., van den Brink, J., de Bruijne, J., Arends, J. E., et al. (2018). Adherence to ursodeoxycholic acid therapy in patients with cholestatic and autoimmune liver disease. Clin. Res. Hepatol Gastroenterol. 1, 37–44. doi: 10.1016/j.clinre.2018.08.006
Lepercq, P. (2004). Epimerization of chenodeoxycholic acid to ursodeoxycholic acid by Clostridium baratii isolated from human feces. FEMS Microbiol. Lett. 235, 65–72. doi: 10.1016/j.femsle.2004.04.011
Lim, S. C., and Han, S. I. (2015). Ursodeoxycholic acid effectively kills drug-resistant gastric cancer cells through induction of autophagic death. Oncol. Rep. 34, 1261–1268. doi: 10.3892/or.2015.4076
Linhares, F. S., Cardoso, A., Terrabuio, D., Harriz, M., Guedes, L., Carrilho, F. J., et al. (2020). Ursodeoxycholic acid and/or ciprofibrate for treating patients with presumptive diagnosis of low phospholipid cholelithiasis, a clinical spectrum of progressive familial intrahepatic cholestasis type 3. Hepatology 1:460. doi: 10.1016/S0168-8278(20)31403-3
Liu, L., Aigner, A., and Schmid, R. D. (2011). Identification, cloning, heterologous expression, and characterization of a NADPH-dependent 7 beta-hydroxysteroid dehydrogenase from Collinsella aerofaciens. Appl. Microbiol. Biotechnol. 90, 127–135. doi: 10.1007/s00253-010-3052-y
Liu, L., Braun, M., Gebhardt, G., Weuster-Botz, D., Gross, R., and Schmid, R. D. (2012). One-step synthesis of 12-ketoursodeoxycholic acid from dehydrocholic acid using a multienzymatic system. Appl. Microbiol. Biotechnol. 97, 633–639. doi: 10.1007/s00253-012-4340-5
Liu, L., Schmid, R., and Aigner, A. (2016). Novel 7beta-hydroxysteroid dehydrogenases and their use. U.S. Patent No 20160194615. Washington, DC: U.S. Patent and Trademark Office.
Liu, L., Schmid, R., and Aigner, A. (2018). Novel 7 beta-hydroxysteroid dehydrogenases and their use. U.S. Patent No 20180273916. Washington, DC: U.S. Patent and Trademark Office.
Liu, L., Schmid, R., and Aigner, A. (2019). 7β-hydroxysteroid dehydrogenases and their use. U.S. Patent No 10465171. Washington, DC: U.S. Patent and Trademark Office.
Lou, D., Wang, Y., Tan, J., Zhu, L., Ji, S., and Wang, B. (2017). Functional contribution of coenzyme specificity-determining sites of 7α-hydroxysteroid dehydrogenase from Clostridium absonum. Comput. Biol. Chem. 70, 89–95. doi: 10.1016/j.compbiolchem.2017.08.004
Lugini, A., Verna, S., Buzzacchino, F., Minelli, M., and Cristofani, L. (2022). 1134P Prevention of hepatic toxicities associated with anaplastic lymphoma kinase inhibitors in the treatment of non-small cell lung cancer by administration of ursodeoxycholic acid: Analysis from the monoinstitutional analysis. Ann. Oncol. 33:1069. doi: 10.1016/J.ANNONC.2022.07.1258
Ma, N., Chen, Z., Chen, J., Chen, J., Wang, C., Zhou, H., et al. (2018). Dual-functional small molecules for generating an efficient cytochrome P450BM3 peroxygenase. Angew. Chem. 130, 7754–7759. doi: 10.1002/anie.201801592
Macdonald, I. A., Hutchison, D. M., and Forrest, T. P. (1981). Formation of urso- and ursodeoxy-cholic acids from primary bile acids by Clostridium absonum. J. Lipid Res. 22, 458–466. doi: 10.1016/s0022-2275(20)34960-9
MacDonald, I. A., Rochon, Y. P., Hutchison, D. M., and Holdeman, L. V. (1982). Formation of ursodeoxycholic acid from chenodeoxycholic acid by a 7 beta-hydroxysteroid dehydrogenase-elaborating Eubacterium aerofaciens strain cocultured with 7 alpha-hydroxysteroid dehydrogenase-elaborating organisms. Appl. Environ. Microbiol. 44, 1187–1195. doi: 10.1128/aem.44.5.1187-1195.1982
Marschall, H. U. (2019). Ursodeoxycholic acid for intrahepatic cholestasis in pregnancy. Lancet 394, 810–812. doi: 10.1016/S0140-6736(19)31607-1
Monti, D., Ferrandi, E. E., Riva, S., and Polentini, F. (2012). New process for the selective oxidation of bile acids, their salts or derivatives. WO Patent No 2012131591. Geneva: World Intellectual Property Organization.
Monti, D., Ferrandi, E. E., Zanellato, I., Hua, L., Polentini, F., Carrea, G., et al. (2009). One-pot multienzymatic synthesis of 12-ketoursodeoxycholic acid: subtle cofactor specificities rule the reaction equilibria of five biocatalysts working in a row. Adv. Synth. Catal. 351, 1303–1311. doi: 10.1002/adsc.200800727
Monti, D., Ottolina, G., Carrea, G., and Riva, S. (2011). Redox reactions catalyzed by isolated enzymes. Chem. Rev. 111, 4111–4140. doi: 10.1021/cr100334x
Mordhorst, S., and Andexer, J. N. (2020). Round, round we go-strategies for enzymatic cofactor regeneration. Nat. Prod. Rep. 37, 1316–1333. doi: 10.1039/d0np00004c
Nagao, T., Mitamura, T., Wang, X., Negoro, S., Yomo, T., Urabe, I., et al. (1992). Cloning, nucleotide sequences, and enzymatic properties of glucose dehydrogenase isozymes from Bacillus megaterium IAM1030. J. Bacteriol. 174, 5013–5020. doi: 10.1128/jb.174.15.5013-5020.1992
Nathalie, B., Thierry, F., Dominique, G., Pascal, H., and Jean, D. (1991). Cloning of the D-lactate dehydrogenase gene from lactobacillus Delbrueckii subsp. bulgaricus by complementation in Escherichia coli. FFBS Lett. 290, 61–64. doi: 10.1016/0014-5793(91)81226-X
Newman, S. G., Gu, L., Lesniak, C., Victor, G., Meschke, F., Abahmane, L., et al. (2014). Rapid Wolff-Kishner reductions in a silicon carbide microreactor. Green Chem. 16, 176–180. doi: 10.1039/C3GC41942H
O’Leary, N. A., Wright, M. W., Brister, J. R., Ciufo, S., Haddad, D., McVeigh, R., et al. (2016). Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44, 733–745. doi: 10.1093/nar/gkv1189
Oppermann, U., Filling, C., Hult, M., Shafqat, N., Wu, X. Q., Lindh, M., et al. (2003). Short-chain dehydrogenases/reductases (SDR): the 2002 update. Chem-Biol Interact. 143, 247–253. doi: 10.1016/S0009-2797(02)00164-3
Park, D. J., Park, Y. S., Yoo, M. W., Hwang, S. H., Seong-Yeob, R., Kyoung Kwon, O., et al. (2020). Efficacy and safety of ursodeoxycholic acid in the prevention of gallstone formation after gastrectomy in patients with gastric cancer: a randomized, double-blind, placebo-controlled study (Pegasus-D). Surg. Oncol. 46, e147. doi: 10.1016/j.ejso.2019.11.385
Pedrini, P., Andreotti, E., Guerrini, A., Dean, M., Fantin, G., and GIovannini, P. P. (2005). Xanthomonas maltophilia CBS 897.97 as a source of new 7α- and 7β-hydroxysteroid dehydrogenases and cholylglycine hydrolase: improved biotransformations of bile acids. Steroids 71, 189–198. doi: 10.1016/j.steroids.2005.10.002
Pennacchio, A., Pucci, B., Secundo, F., La Cara, F., Rossi, M., and Raia, C. A. (2008). Purification and characterization of a novel recombinant highly enantioselective short-chain NAD(H)-dependent alcohol dehydrogenase from Thermus thermophilus. Appl. Environ. Microbiol. 74, 3949–3958. doi: 10.1128/aem.00217-08
Pi, Y., Wu, Y., Han, D., Zou, X., Ni, D., and Wang, J. (2022). O13. Gut microbiota-derived ursodeoxycholic acid alleviates low birth weight-induced colonic inflammation by enhancing M2 macrophage polarization. Anim. Sci. J. 13:149. doi: 10.1016/J.ANSCIP.2022.04.014
Qi, H., Shen, D., Jiang, C., Wang, H., and Chang, M. (2020). Ursodeoxycholic acid protects dopaminergic neurons from oxidative stress via regulating mitochondrial function, autophagy, and apoptosis in MPTP/MPP+-induced Parkinson’s disease. Neurosci. Lett. 741:135493. doi: 10.1016/j.neulet.2020.135493
Qian, J., Yao, T., Wang, Y., and Wang, G. (2020). Treatment of primary biliary cholangitis with ursodeoxycholic acid, prednisolone and immunosuppressants in patients not responding to ursodeoxycholic acid alone and the prognostic indicators. Clin. Res. Hepatol. Gastroenterol. 44, 874–884. doi: 10.1016/j.clinre.2020.03.026
Qin, H. P., Zhang, H. P., Zhong, Y. H., et al. (2018). A method for enzymatic synthesis of ursodeoxycholicAcid. CN Patent No 108251491A. Beijing: State Administration for Market Regulation.
Razori, M. V., Maidagan, P. M., Ciriaci, N., Andermatten, R. B., Barosso, I. R., Martín, P. L., et al. (2019). Anticholestatic mechanisms of ursodeoxycholic acid in lipopolysaccharide-induced cholestasis. Biochem. Pharmacol. 168, 48–56. doi: 10.1016/j.bcp.2019.06.009
Rose, E., Navvrro, S., Bru, C., Garcia-Pugés, A., and Valderrama, R. (1991). Occult microlithiasis in ‘idiopathic’ acute pancreatitis: Prevention of relapses by cholecystectomy or ursodeoxycholic acid therapy. Gastroenterology 6, 1701–1709. doi: 10.1016/0016-5085(91)90410-M
Sathe, A. G., Tuite, P., Chen, C., Ma, Y., Chen, W., Cloyd, J., et al. (2020). Pharmacokinetics, safety, and tolerability of orally administered ursodeoxycholic acid in patients with Parkinson’s diseasea-a pilot study. J. Clin. Pharmacol. 60, 744–750. doi: 10.1002/jcph.1575
Savino, S., Ferrandi, E. E., Forneris, F., Rovida, S., Riva, S., Monti, D., et al. (2016). Structural and biochemical insights into 7β-hydroxysteroid dehydrogenase stereoselectivity. Proteins 84, 859–865. doi: 10.1002/prot.25036
Sawada, H., Kulprecha, S., Nilubol, N., Yoshida, T., Kinoshita, S., and Taguchi, H. (1982). Microbial production of ursodeoxycholic acid from lithocholic acid by Fusarium equiseti M41. Appl. Environ. Microbiol. 44, 1249–1252. doi: 10.1128/aem.44.6.1249-1252.1982
Scalia, S., Williams, J. R., Shim, J. H., Law, B., and Morgan, E. D. (1998). Supercritical fluid extraction of bile acids from bovine bile raw materials. Chromatographia 48, 785–789. doi: 10.1007/BF02467648
Schmid, R., Braun, M., Liu, L., Aigner, A., and Weuster-botz, D. (2013). Novel 7 alpha-hydroxysteroid dehydrogenase knockout mutants and use therefor. U.S. Patent No 20130224792. Washington, DC: U.S. Patent and Trademark Office.
Shah, R. A., and Kowdley, K. V. (2020). Current and potential treatments for primary biliary cholangitis. Lancet Gastroenterol. 5, 306–315. doi: 10.1016/s2468-1253(19)30343-7
Shi, J., Wang, J., Yu, L., Yang, L., Zhao, S., and Wang, Z. (2017). Rapidly directional biotransformation of tauroursodeoxycholic acid through engineered Escherichia coli. J. Ind. Microbiol. Biotechnol. 44, 1073–1082. doi: 10.1007/s10295-017-1935-y
Song, C., Wang, B., Tan, J., Zhu, L., and Luo, D. (2017). Discovery of tauroursodeoxycholic acid biotransformation enzymes from the gut microbiome of black bears using metagenomics. Sci. Rep. 7:45495. doi: 10.1038/srep45495
Sun, B., Kantzow, C., Bresch, S., Castiglione, K., and Weuster-Botz, D. (2012). Multi-enzymatic one-pot reduction of dehydrocholic acid to 12-keto-ursodeoxycholic acid with whole-cell biocatalysts. Biotechnol. Bioeng. 110, 68–77. doi: 10.1002/bit.24606
Tang, S., Pan, Y., Luo, D., Ji, S., Zhu, L., Tan, J., et al. (2019). Structural and functional characterization of a novel acidophilic 7α-hydroxysteroid dehydrogenase. Protein Sci. 28, 910–919. doi: 10.1002/pro.3599
Testoni, P., Caporuscio, S., Bagnolo, F., and Lella, F. (2000). Idiopathic recurrent pancreatitis: long-term results after ERCP, endoscopic sphincterotomy, or ursodeoxycholic acid treatment. Am. J. Gastroenterol. 7, 1702–1707. doi: 10.1016/S0002-9270(00)01084-4
Tian, G., Wang, Q., Wei, X., Ma, X., and Chen, S. (2017). Glutamate dehydrogenase (RocG) in Bacillus licheniformis WX-02: Enzymatic properties and specific functions in glutamic acid synthesisfor poly-γ-glutamic acid production. Enzyme Microbiol. Technol. 99, 9–15. doi: 10.1016/j.enzmictec.2017.01.002
Tonin, F., and Arends, I. W. C. E. (2018). Latest development in the synthesis of ursodeoxycholic acid (UDCA): a critical review. Beilstein J. Org. Chem. 14, 470–483. doi: 10.3762/bjoc.14.33
Tonin, F., Otten, L. G., and Arends, I. W. C. E. (2018). NAD+-dependent enzymatic route for the epimerization of hydroxysteroids. ChemSusChem 12, 3192–3203. doi: 10.1002/cssc.201801862
van der Feen, C., van der Doef, H. P., van der Ent, C. K., and Houwen, R. H. (2016). Ursodeoxycholic acid treatment is associated with improvement of liver stiffness in cystic fibrosis patients. J. Cyst. Fibros. 6, 834–838. doi: 10.1016/j.jcf.2016.07.009
Wang, R., Wu, J., Jin, D. K., Chen, Y., Lv, Z., Chen, Q., et al. (2017). Structure of NADP+-bound 7β-hydroxysteroid dehydrogenase reveals two cofactor-binding modes. Acta Crystallogr. F. 73, 246–252. doi: 10.1107/s2053230x17004460
Weckbecker, A., Gröger, H., and Hummel, W. (2010). Regeneration of nicotinamide coenzymes: principles and applications for the synthesis of chiral compounds. Adv. Biochem. Eng. Biotechnol. 120, 195–242. doi: 10.1007/10_2009_55
West, R. J. H., Ugbode, C., Fort-Aznar, L., and Sweeney, S. T. (2020). Neuroprotective activity of ursodeoxycholic acid in CHMP2BIntron5 models of frontotemporal dementia. Neurobiol. Dis. 144:105047. doi: 10.1016/j.nbd.2020.105047
Weuster-botz, D., Braun, M., Aigner, A., Sun, B., Kantzow, C., Bresch, S., et al. (2021). Novel 7beta-hydroxysteroid dehydrogenase mutants and process for the preparation of ursodeoxycholic acid. U.S. Patent No 20210139862. Washington, DC: U.S. Patent and Trademark Office.
Weuster-botz, D., Sun, B., and Quirin, C. (2017). Method for biocatalytic whole cell reduction of dehydrocholic acid compounds, and 7-beta-hydroxysteroid dehydrogenase mutants. U.S. Patent No 20170191104. Washington, DC: U.S. Patent and Trademark Office.
Wu, P., Zhao, J., Guo, Y., Yu, Y., Wu, X., Xiao, H., et al. (2020). Ursodeoxycholic acid alleviates nonalcoholic fatty liver disease by inhibiting apoptosis and improving autophagy via activating AMPK. Biochem. Biophys. Res. Commun. 529, 834–838. doi: 10.1016/j.bbrc.2020.05.128
Xie, P., Peng, Y., and Qiu, L. (2022). Responsive oligochitosan nano-vesicles with ursodeoxycholic acid and exenatide for NAFLD synergistic therapy via SIRT1. Carbohyd. Polym. 288:119388. doi: 10.1016/J.CARBPOL.2022.119388
Xu, Y., Yang, L., Zhao, S., and Wang, Z. (2019). Large-scale production of tauroursodeoxycholic acid products through fermentation optimization of engineered Escherichia coli cell factory. Microbiol. Cell Fact. 18:34. doi: 10.1186/s12934-019-1076-2
Xu, Z., Jing, K., Liu, Y., and Cen, P. (2006). High-level expression of recombinant glucose dehydrogenase and its application in NADPH regeneration. J. Ind. Microbiol. Biotechnol. 34, 83–90. doi: 10.1007/s10295-006-0168-2
Yang, Q., Wang, B., Zhang, Z., Lou, D., Tan, J., and Zhu, L. (2017). The effects of macromolecular crowding and surface charge on the properties of an immobilized enzyme: activity, thermal stability, catalytic efficiency and reusability. RSC Adv. 7, 38028–38036. doi: 10.1039/c7ra06544b
Yang, Y., Liu, S., Gao, H., Wang, P., Zhang, Y., Zhang, A., et al. (2020). Ursodeoxycholic acid protects against cisplatin-induced acute kidney injury and mitochondrial dysfunction through acting on ALDH1L2. Free Rad. 152, 821–837. doi: 10.1016/j.freeradbiomed.2020.01.182
Yao, T., Qian, J., and Wang, G. (2020). Efficacy of ursodeoxycholic acid combined with prednisolone and immunosuppressant triple therapy in the treatment of refractory primary biliary cholangitis. Med. Clin-barcelona 4, 165–170. doi: 10.1016/j.medcli.2020.03.013
Ye, H., Zhang, J., Chen, X., Wu, P., Chen, L., and Zhang, G. (2019). Ursodeoxycholic acid alleviates experimental liver fibrosis involving inhibition of autophagy. Life Sci. 242:117175. doi: 10.1016/j.lfs.2019.117175
Yoshimoto, T., Higashi, H., Kanatani, A., Lin, X. S., Nagai, H., Oyama, H., et al. (1991). Cloning and sequencing of the 7 alpha-hydroxysteroid dehydrogenase gene from Escherichia coli HB101 and characterization of the expressed enzyme. J. Bacteriol. 173, 2173–2179. doi: 10.1128/jb.173.7.2173-2179.1991
You, Z., Chen, Q., Shi, S., Zheng, M., Pan, J., Qian, X., et al. (2018). Switching cofactor dependence of 7β-hydroxysteroid dehydrogenase for cost-effective production of ursodeoxycholic acid. ACS Catal. 9, 466–473. doi: 10.1021/acscatal.8b03561
Yu, K., Li, P., Xu, T., Xu, J., Wang, K., Chai, J., et al. (2021). Decreased infiltration of CD4+ Th1 cells indicates a good response to ursodeoxycholic acid (UDCA) in primary biliary cholangitis. Pathol. Res. Pract. 217:153291. doi: 10.1016/j.prp.2020.153291
Zhang, X., Fan, D., Hua, X., and Zhang, T. (2019). Large-scale production of ursodeoxycholic acid from chenodeoxycholic acid by engineering 7α- and 7β-hydroxysteroid dehydrogenase. Bioproc. Biosyst. Eng. 42, 1537–1545. doi: 10.1007/s00449-019-02151-4
Zheng, M., Chen, K., Wang, R., Li, H., Li, C., and Xu, J. (2017b). Engineering 7β-hydroxysteroid dehydrogenase for enhanced ursodeoxycholic acid production by multiobjective directed evolution. J. Agr. Food Chem. 65, 1178–1185. doi: 10.1021/acs.jafc.6b05428
Zheng, M., Chen, F., Li, H., Li, C., and Xu, J. (2017a). Continuous production of ursodeoxycholic acid by using two cascade reactors with co-immobilized enzymes. ChemBioChem 19, 347–353. doi: 10.1002/cbic.201700415
Zheng, M., Wang, R., Li, C., and Xu, J. (2015). Two-step enzymatic synthesis of ursodeoxycholic acid with a new 7β-hydroxysteroid dehydrogenase from Ruminococcus torques. Process. Biochem. 50, 598–604. doi: 10.1016/j.procbio.2014.12.026
Keywords: ursodeoxycholic acid, chenodeoxycholic acid, biological synthesis, free-enzyme catalysis, whole-cell synthesis
Citation: Song P, Zhang X, Feng W, Xu W, Wu C, Xie S, Yu S and Fu R (2023) Biological synthesis of ursodeoxycholic acid. Front. Microbiol. 14:1140662. doi: 10.3389/fmicb.2023.1140662
Received: 09 January 2023; Accepted: 13 February 2023;
Published: 24 February 2023.
Edited by:
Cormac Murphy, University College Dublin, IrelandReviewed by:
Marina Donova, Institute of Biochemistry and Physiology of Microorganisms (RAS), RussiaFouzia Sadiq, Shifa Tameer-e-Millat University, Pakistan
Copyright © 2023 Song, Zhang, Feng, Xu, Wu, Xie, Yu and Fu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peng Song,  c29uZ3BlbmdAbGN1LmVkdS5jbg==,
c29uZ3BlbmdAbGN1LmVkdS5jbg==,  c29uZ3BlbmdAenltZXJjay5jb20=
c29uZ3BlbmdAenltZXJjay5jb20=
 Peng Song
Peng Song Xue Zhang1
Xue Zhang1