
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 04 April 2023
Sec. Microbe and Virus Interactions with Plants
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1139456
This article is part of the Research Topic Actinobacteria Plant Interaction: Recent Molecular Tools and Biology View all 6 articles
Anthracnose disease caused by Colletotrichum gloeosporioides is one of the devastating diseases of yams (Dioscorea sp.) worldwide. In this study, we aimed to isolate endophytic actinobacteria from yam plants and to evaluate their potential for the control of yam anthracnose based on bioassays and genomic analyses. A total of 116 endophytic actinomycete strains were isolated from the surface-sterilized yam tissues from a yam orchard in Hainan Province, China. In total, 23 isolates showed antagonistic activity against C. gloeosporioides. An endophytic actinomycete, designated HNM0140T, which exhibited strong antifungal activities, multiple biocontrol, and plant growth-promoting (PGP) traits was subsequently selected to colonize in the tissue-cultured seedlings of yam and was tested for its in vivo biocontrol potential on yam anthracnose. The results showed that treatment with strain HNM0140T markedly reduced the severity and incidence of yam anthracnose under greenhouse conditions. Morphological and chemotaxonomic analyses showed that strain HNM0140T was assigned to the genus Streptomyces. Phylogenetic analysis based on the 16S rRNA gene sequences indicated that strain HNM0140T formed a separate cluster together with Streptomyces lydicus ATCC 25470T (99.45%), Streptomyces chattanoogensis NRRL ISP-5002T (99.45%), and Streptomyces kronopolitis NEAU-ML8T (98.97%). The phylogenomic tree also showed that strain HNM0140T stably clustered with Streptomyces lydicus ATCC 25470T. The ANI and dDDH between strain HNM0140T and its closest related-type species were well below the recommended thresholds for species demarcation. Hence, based on the phylogenetic, genomic, and phenotypic analyses, strain HNM0140T should represent a new streptomycete species named Streptomyces endophytica sp. nov. Genomic analysis revealed that strain HNM0140T harbored 18 putative BGCs for secondary metabolites, some PGP-related genes, and several genes coding for antifungal enzymes. The presented results indicated that strain HNM0140T was a promising biocontrol agent for yam anthracnose.
Yam (Dioscorea spp.) is a very important tuber crop that is widely cultivated in both tropical and sub-tropical regions around the world. Yam tubers are widely consumed as a valuable source of food across the globe (Egesi et al., 2007). Chinese yam (Dioscorea opposita Thunb.) is among the most widely cultivated yam species in China both as a traditional medicine and as a nutritious food source (Li et al., 2014). However, anthracnose disease, caused by Colletotrichum gloeosporioides, is one of the devastating diseases of yam and seriously threatens yam production worldwide (Abang et al., 2003). The pathogenic fungus can not only infect all parts of yam plants but also affect all growth stages of yam, which results in serious yield losses, especially in susceptible genotypes (Nwadili et al., 2017). Currently, the anthracnose disease of yam has been controlled mainly using chemical methods (Onyeka et al., 2006). However, chemical fungicides have been reported to not always be effective against anthracnose disease, especially during the rainy season (Ntui et al., 2021). Meanwhile, their frequent use has caused deleterious effects, including the development of fungicide-resistant pathogens and damage to the environment (Onyeka et al., 2006). Thereby, it is important to call for investigating alternative control measures including biological control, which is favored as an ecofriendly alternative to chemical fungicides for its decreasing reliance on the dangerous use of chemicals and its lesser cost (Okigbo, 2005).
As an effective tool of biological control, microbial antagonism has been attracting the attention of researchers in recent years, some of whom have found a novel method for selecting antagonists against anthracnose diseases in a variety of crops, such as strawberry, chili, and rubber (Thilagam and Hemalatha, 2019; Gu et al., 2020; Marian et al., 2020). Actinomycetes, as prolific producers of antibiotics, have been studied extensively as promising antagonists to control plant pathogens (Doumbou et al., 2001; Solanki et al., 2016; Zhang et al., 2020). Many previous studies have suggested that actinomycetes inhabit inside plant tissues as endophytes (Govindasamy et al., 2014), some of which have been used for effective biocontrol and as PGP agents (Shimizu, 2011; Singh and Dubey, 2018; Gao et al., 2021). However, only a few studies have reported the utilization of actinomycetes to manage anthracnose diseases of yam (Soares et al., 2006; Palaniyandi et al., 2011, 2013, 2016).
Thus, the present study was designed to isolate endophytic actinobacteria from yam plants and to evaluate their potential for the biocontrol of yam anthracnose. A novel endophytic actinobacterial strain of HNM0140T isolated from the root of yam displayed a strong antifungal activity against C. gloeosporioides, which was isolated from naturally infected yam leaves, and its taxonomic status was established through a polyphasic taxonomic approach. We identified strain HNM0140T as a novel actinobacterial species named Streptomyces endophytica sp. nov. and evaluated its biocontrol ability against yam anthracnose. Moreover, the complete genome of strain HNM0140T was also sequenced and analyzed for further understanding of its biocontrol potential at genomic levels.
Plant materials (leaf, stem, and root samples) of healthy Dioscorea opposita Thunb. plants were collected from a yam orchard in Danzhou City (19°31′N, 109°35′E) of Hainan Province, China. The surface-sterilized procedures of plant materials were performed according to the method previously described (Huang et al., 2012). A total of 1 g of surface-sterilized plant materials was then cut into small fragments and homogenized in 10 ml of sterile phosphate-buffered saline (PBS). The resultant homogenates (0.1 ml) were plated onto three isolation media: humic acid–vitamin agar (Hayakawa and Nonomura, 1987), tap water–yeast extract agar (Crawford et al., 1993), and IMA-2 agar (Shimizu et al., 2000) supplemented with 50 mg/L of actidione, 50 mg/L of nystatin, and 100 mg/L of potassium dichromate (K2Cr2O7). The plates were incubated at 28°C for 28 days. Presumptive actinobacterial colonies that appeared on the isolated plates were picked and streaked onto ISP2 agar (Shirling and Gottlieb, 1966) and cultured at 28°C for 1–2 weeks.
Antagonistic activities of the endophytic isolates against C. gloeosporioides, a major pathogen of yam, were evaluated on potato dextrose agar (PDA) plates using the dual culture in vitro assay as described previously by Palaniyandi et al. (2011). All isolates of endophytic actinobacteria were streaked onto PDA plates and incubated at 28°C for 3 days. Mycelial disks of 5-mm diameter of C. gloeosporioides were placed onto the described PDA plates 5 cm away from each actinobacterial colony. The phytopathogenic fungi were separately incubated on non-inoculated PDA plates as a control. After culturing at 28°C for 7 days, the plates were examined for the inhibition zone between the pathogen and the actinobacterial isolate according to the method described previously (Crawford et al., 1993). Endophytic actinobacteria, designated as strain HNM0140T, were obtained from the root of D. opposita Thunb., which was selected for further studies on the basis of its strong antifungal activity against C. gloeosporioides. Strain HNM0140T was also tested for its antifungal activities against other phytopathogenic fungi, namely, Fusarium oxysporum f. sp. cubense race 4 from banana, Magnaporthe oryzae from rice, Lasiodiplodia theobromae from mango, Neoscytalidium dimidiatum from pitaya, Colletotrichum capsici from chili, and Stagonosporopsis cucurbitacearum from watermelon. These aforementioned pathogens were provided by the College of Life Sciences, Hainan University, Haikou, China.
The production of siderophore and protease by strain HNM0140T was detected as described in a previous study (Naik et al., 2008). The β-1,4-glucanase activity of strain HNM0140T was screened according to a previously described method (Palaniyandi et al., 2011). The chitinase activity of strain HNM0140T was assessed using colloidal chitin agar medium as described in a previous study (Hsu and Lockwood, 1975). The nitrogen-fixing ability of strain HNM0140T was assessed using N-free semisolid JNFb medium (Döbereiner et al., 1995). The phosphate solubilization activity of strain HNM0140T was detected as described in a previous study (Hamdali et al., 2008). The ACC deaminase activity and indole acetic acid production (IAA) were evaluated as described previously (El-Tarabily, 2008). The ability of the PGP of strain HNM0140T was assessed by a plate-based bioassay according to the modified method described previously (Palaniyandi et al., 2011). An agar block (6-mm diameter) containing mycelia of the strain HNM0140T was placed in the center of plates containing MS medium (Murashige and Skoog, 1962) with 0.8% (w/v) agar and grown at 28°C for 3 days. Twenty stratified Arabidopsis Col-0 seeds were placed 3 cm away from the strain HNM0140T culture on the MS plate and incubated in a growth chamber at 25°C for 7 days. The MS plate inoculated with Arabidopsis seeds only was used as the control. After 7 days, the root lengths and total fresh weight of the plants were measured.
In vitro plantlets of yam (cultivars: D. opposita Thunb.) were raised from nodal explants for 1 month in 75 × 105-mm glass flasks using MS medium supplemented with bacto agar (0.8%) and sucrose (3%). These explants were incubated in a growth chamber at 25°C under a 16-h photoperiod for another month. The resultant seedlings were prepared for the following colonization test. The colonization of strain HNM0140T on tissue-cultured seedlings of yam was examined using the method described previously by Shimizu et al. (2001). Strain HNM0140T was inoculated into 150 ml of ISP2 broth and cultured on a shaker (180 rpm) at 28°C for 3 days. Then, 2 ml of the mycelial suspension (5–6 × 106 CFU/ml) was added to the surface of MS medium in glass flasks supporting yam seedlings. For the control, 2 ml of ISP2 broth was also spread on the surface of the MS medium. These seedlings, inoculated and non-inoculated with strain HNM0140T, were grown for 2 weeks. Then, the seedlings were taken from the flasks and were cut into small fragments at each node. These fragments were placed on ISP2 agar and incubated at 28°C for ~14 days. For each seedling, the colonized strain was re-isolated and re-identified by comparing the morphological characteristics and the 16S rRNA gene sequence with those of strain HNM0140T. The detached leaves were also observed by scanning electron microscopy (S-3000N, Hitachi, Japan) according to the methods previously described (Shimizu et al., 2009).
To validate its suitability as a biocontrol agent, the seedlings of yam, treated and untreated with strain HNM0140T in flasks as previously, were transferred into a tray (50 × 25 × 7 cm3) containing autoclaved soil. Then, the trays were transferred into a large bioclimatic chamber, and the seedlings were acclimatized at 30/25°C day/night temperature and 12-h photoperiod for 2 weeks. At the end of the acclimatization period, the seedlings were then spray-inoculated with C. gloeosporioides spore suspension (2 × 106 spores/ml) using an atomizer at the rate of ~5 ml per seedling. Each experiment was replicated three times and eight plants were used for each replicate. These inoculated seedlings were transferred into a humid chamber (RH 95%, 28°C, dark) for 24 h and then placed in a greenhouse with 85% relative humidity under a 12-h photoperiod at 28°C for 2 weeks. The yam seedlings were monitored daily for anthracnose, and the disease incidence and severity were assessed after 2 weeks of inoculation of C. gloeosporioides. The disease incidence was calculated as the percentage of diseased leaves among the total leaves studied in each tray (Palaniyandi et al., 2011). The disease severity index was calculated according to the methods described previously (Simons and Green, 1994). The disease severity of each seedling was scored using a 0–6-point scale on the basis of the percentage of the whole seedling area affected by anthracnose, where 0 = 0%, 1 = 1%, 2 = 2%, 3 = 3–9%, 4 = 10–24%, 5 = 25–50%, and 6 >50% (Onyeka et al., 2006).
Genomic DNA was extracted from strain HNM0140T using the Wizard® Genomic DNA Purification Kit (Promega Corp.). PCR amplification and sequencing of the 16S rRNA gene of strain HNM0140T were performed as described by Huang et al. (2012). The 16S rRNA gene sequence of strain HNM0140T was aligned with those of the type strains by the EzTaxon-e server (Yoon et al., 2017). Phylogenetic trees were generated using neighbor-joining (Saitou and Nei, 1987), minimum-evolution (Rzhetsky and Nei, 1992), and maximum-likelihood (Felsenstein, 1981) methods from the MEGA 7.0 software (Kumar et al., 2016). Evolutionary distances were calculated using the Kimura 2-parameter model (Kimura, 1980). Confidence interval levels of the branch nodes were studied by a bootstrap analysis of 1,000 replications (Felsenstein, 1985).
The whole-genome sequencing and assembly of strain HNM0140T were carried out as described by Zhou et al. (2018). Protein-coding gene prediction was carried out by Glimmer v3.02 software (Delcher et al., 2007), whereas rRNA and tRNA were predicted using Infernal 1.1 (Nawrocki and Eddy, 2013) and tRNAscan-SEv1.31 (Lowe and Eddy, 1997) software, respectively. Their functions were annotated using the databases of clusters of orthologous genes (COGs), the Gene Ontology (GO), and the Kyoto Encyclopedia of Genes and Genomes (KEGG). The secondary metabolite gene cluster prediction was performed using the online antiSMASH 6.0 software (Blin et al., 2021).
The phylogenomic tree of the whole genome was constructed on the Type Strain Genome Server (TYGS) (Meier-Kolthoff et al., 2022). Digital DNA–DNA hybridizations (dDDHs) were calculated with a Genome-to-Genome Distance Calculator (GGDC) v3.0 (Meier-Kolthoff et al., 2022). A comparison of the average nucleotide identity (ANI) values was performed using the JSpeciesWS Online Service (Richter et al., 2016).
Morphological characteristics of strain HNM0140T were observed by scanning electron microscopy (S-3400N, Hitachi, Japan) after incubation for 14 days at 28°C on ISP2 agar. Cultural characteristics of strain HNM0140T were determined after 2 weeks at 28°C using standard ISP 1–ISP 7 media (Shirling and Gottlieb, 1966). Colors of the colony and diffusible pigments on various media were determined by a comparison with the ISCC–NBS color charts (Kelly, 1964). The effects of different temperatures, pH values, and sodium chloride (NaCl) concentrations on growth were observed using ISP2 medium as the basal medium after incubation at 28°C for 14 days as described in a previous study (Huang et al., 2019). The carbon substrate and nitrogen source utilization was examined following the method of Shirling and Gottlieb (1966) and the method of Williams et al. (1983), respectively. Hydrolysis of aesculin, starch, and Tween 40, peptonization and coagulation of milk, and production of urease were tested as described in a previous study (Yokota et al., 1993).
Cell biomass obtained after culturing strain HNM0140T in shake flasks of ISP2 broth for 4–6 days at 28°C was used for chemotaxonomic analyses. The analyses of amino acids and sugars in whole-cell hydrolysates of strain HNM0140T were performed as described previously by Lechevalier and Lechevalier (1980). Phospholipid and menaquinone analysis was performed according to the methods of Minnikin et al. (1984). Fatty acids of strain HNM0140T were extracted and analyzed using the method of Sasser (1990).
A total of 116 endophytic actinobacteria were obtained from the different tissue samples of D. opposita Thunb. Out of them, 23 isolates showed more or less antifungal activity against C. gloeosporioides (Supplementary Table S1). Especially, strain HNM0140T exhibited a strong antagonistic activity against C. gloeosporioides (Figures 1A, B). Consequently, strain HNM0140T was selected further for biocontrol of yam anthracnose diseases caused by the pathogen.
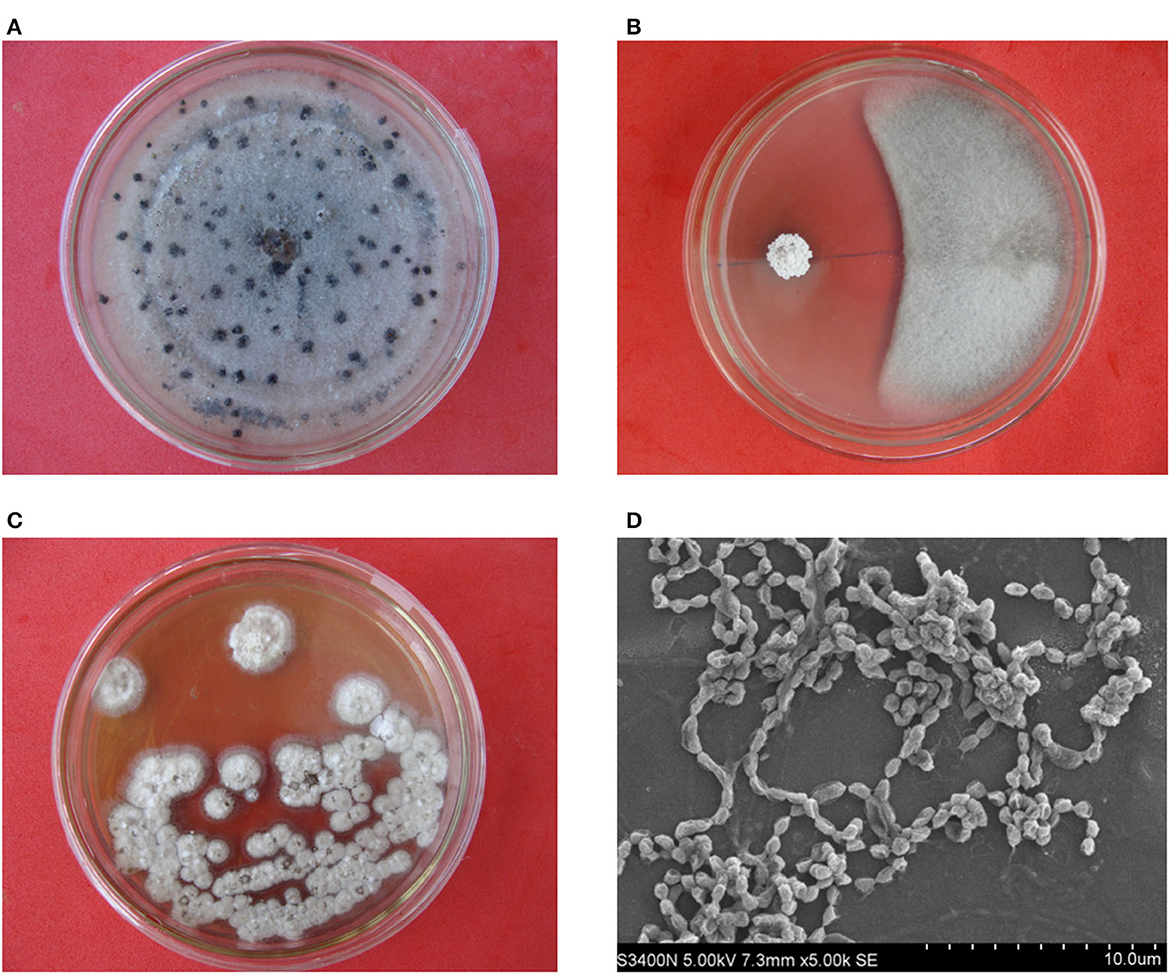
Figure 1. Antifungal activity of strain HNM0140T against C. gloeosporioides on a PDA plate and morphological features of HNM0140T colonies and spore chains. (A) C. gloeosporioides growth without antagonist challenge. (B) Inhibition of C. gloeosporioides growth by strain HNM0140T. (C) Morphological features of strain HNM0140T colonies. (D) Scanning electron micrograph showing spore chains of strain HNM0140T.
The various biocontrol and PGP traits of strain HNM0140T are summarized in Table 1. Strain HNM0140T showed a strong antagonistic activity on C. gloeosporioides, C. capsici, and F. oxysporum f. sp. cubense race 4, only a moderate activity on M. oryzae and S. cucurbitacearum, and a weaker activity on L. theobromae and N. dimidiatum (Table 1). Strain HNM0140T exhibited positive results for the production of protease, β-1,4-glucanase, chitinase, and siderophore. The strain also showed activity to growth in N-deficient media, phosphate solubilization, IAA synthesis, and ACC deaminase (Table 1). The strain was also capable of inducing multiple lateral root development of Arabidopsis seedlings with more root hairs, whereas the control seedlings exhibited longer roots with few lateral roots and fewer root hairs (Supplementary Figure 1).
The results for the colonization test of strain HNM0140T showed that the strain was successfully able to grow on MS medium (Figure 2A), but only one type of mycelial colony that had the same morphological characteristics with strain HNM0140T was expanding from the roots, stems, and leaves of inoculated tissue-culture seedlings of yam within 2 weeks after placing their fragments on ISP2 agar (Figures 2B, C). The re-isolated strains from fragments were analyzed using 16S rRNA gene sequencing, and the results suggested that these strains were 100% identical to the original strain HNM0140T. At the same time, abundant mycelia were observed by SEM on the leaves from seedlings treated with strain HNM0140T (Figure 2D). The results indicated that strain HNM0140T was endophytic.
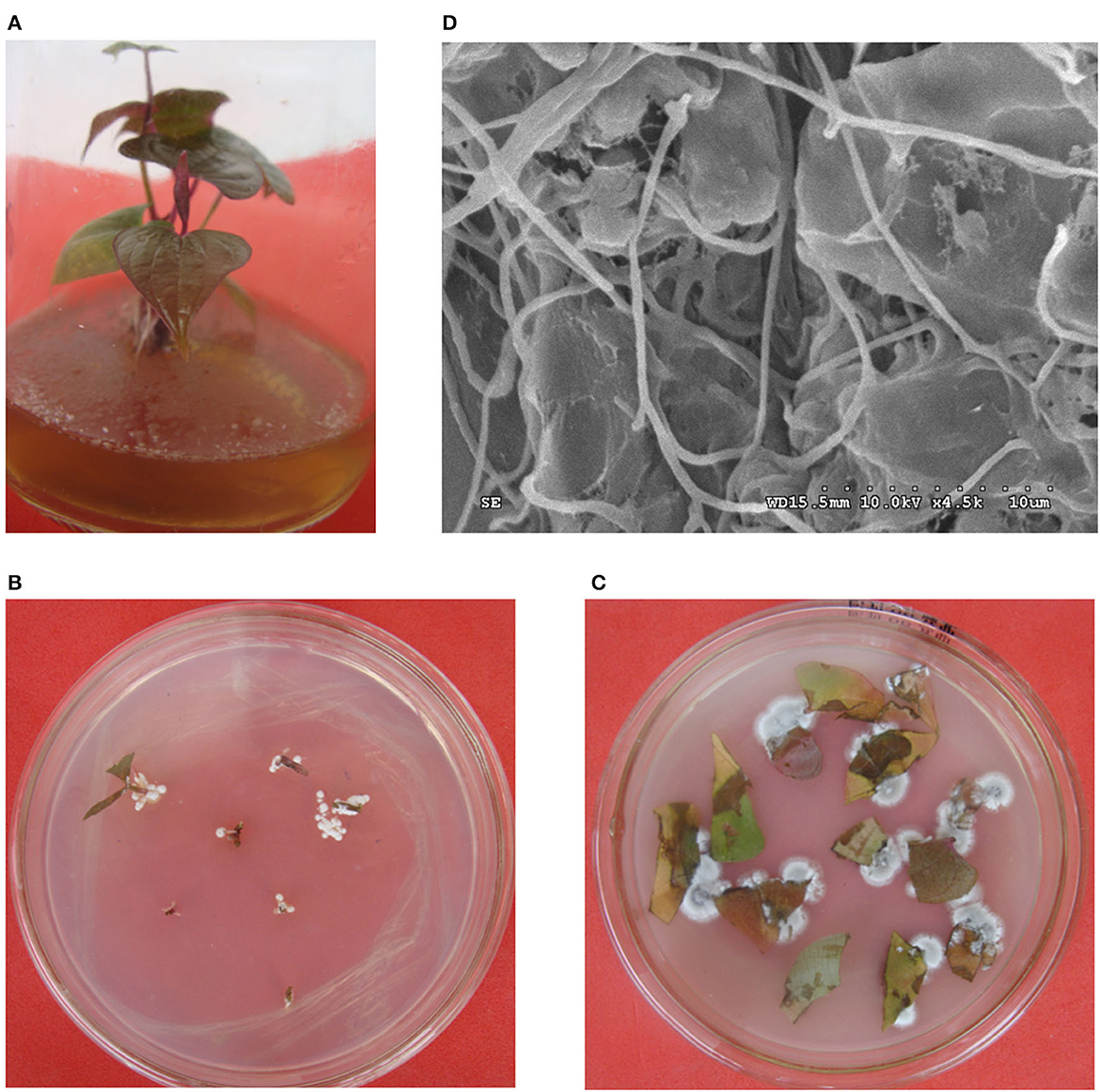
Figure 2. Colonization of strain HNM0140T on tissue-cultured seedlings of yam. (A) Strain HNM0140T-colonized seedling. (B) Re-isolation of strain HNM0140T from segments of stems of tissue-cultured seedlings of yam. (C) Re-isolation of strain HNM0140T from segments of leaves of tissue-cultured seedlings of yam. (D) Scanning electron micrograph of the surface of leaves of tissue-cultured seedlings of yam after colonization of strain HNM0140T.
Anthracnose symptoms in control yam seedlings were first observed after 3 days of challenge inoculation with C. gloeosporioides. After that, the disease aggravated with increased incidence and severity in subsequent days. On day 14, high disease incidence and severity (95% and 90%, respectively) were detected in untreated yam seedlings, and most of the seedlings died rapidly (Figure 3A). However, yam seedlings treated with strain HNM0140T showed a low disease incidence and severity (22% and 20%, respectively) after 14 days (Figures 3B, C). These results indicated that yam seedlings colonized by strain HNM0140T displayed consistent resistance against the pathogen and reduced markedly the anthracnose incidence and severity of yam seedlings. Consequently, strain HNM0140T can be considered as a candidate for the biocontrol of yam anthracnose caused by C. gloeosporioides.
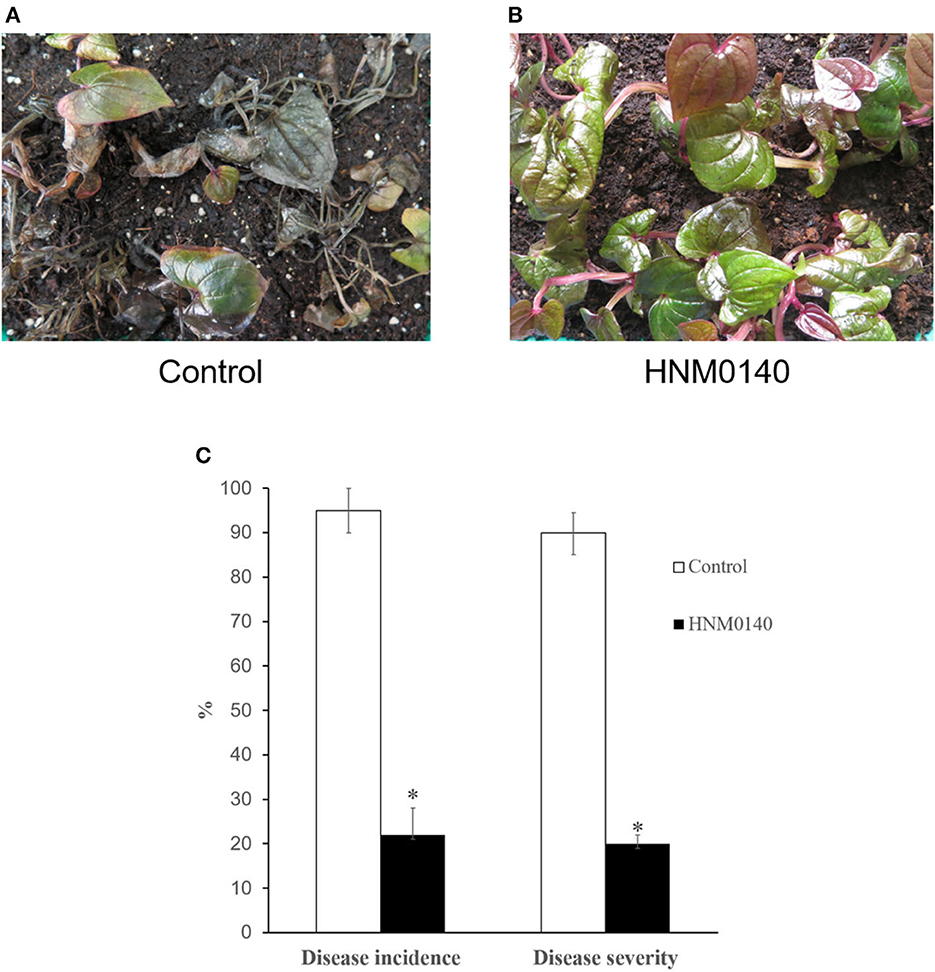
Figure 3. Effect of strain HNM0140Ton yam anthracnose under greenhouse conditions. (A) Anthracnose symptoms on non-treated yam plants 14 days after inoculation with C. gloeosporioides. (B) Anthracnose symptoms on treated yam plants 14 days after inoculation with C. gloeosporioides. (C) Differences in disease incidence and severity of anthracnose of the treatment relative to the non-treated plants. Asterisk indicates significance in treated than in the non-treated category according to Fisher's protected LSD test at P = 0.05.
The almost complete 16S rRNA gene sequence (1492 nt, GenBank Accession Number NT365784) of strain HNM0140T was obtained. Based on an EzBioCloud analysis, strain HNM0140T showed the highest similarity to Streptomyces lydicus ATCC 25470T (99.45%), Streptomyces chattanoogensis NRRL ISP-5002T (99.45%), and Streptomyces kronopolitis NEAU-ML8T (98.97%), and <99% similarity to other type species of the genus Streptomyces. Phylogenetic analysis (Figure 4; Supplementary Figures 2, 3) indicated that strain HNM0140T formed a separate cluster together with Streptomyces lydicus ATCC 25470T, Streptomyces chattanoogensis NRRL ISP-5002T, and Streptomyces kronopolitis NEAU-ML8T. The phylogenomic tree showed that strain HNM0140T stably clustered with Streptomyces lydicus ATCC 25470T (Figure 5). Strain HNM0140T also showed the highest ANI value of 87.05% and the highest dDDH value of 51% with Streptomyces lydicus ATCC 25470T, while the values of ANI and dDDH between strain HNM0140T and other related type species of the genus Streptomyces are shown in Table 2. It is apparent that all values are well below the recommended thresholds (ANI < 95–96% and dDDH < 70%) for species demarcation (Richter and Rosselló-Móra, 2009; Meier-Kolthoff et al., 2013). Therefore, the present genotypic data indicated that strain HNM0140T represented a novel species of the genus Streptomyces.
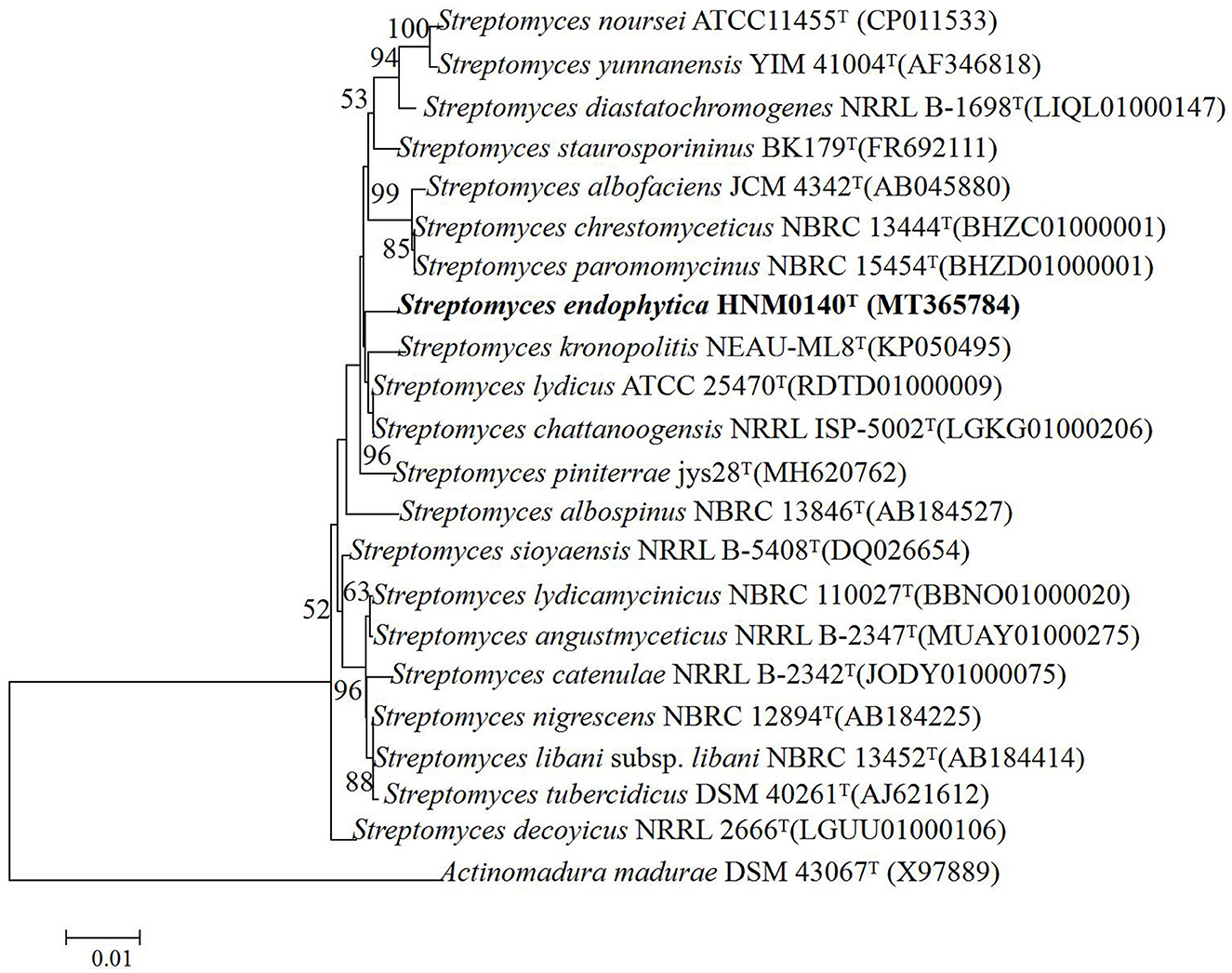
Figure 4. Neighbor-joining phylogenetic tree of strain HNM0140T based on 16S rRNA gene sequences. The bootstrap values (%) presented at the branches were calculated from 1,000 replications, only values >50% are given. Scale bar indicates 0.01 substitutions per nucleotide position.

Figure 5. Phylogenomic tree of strain HNM0140T reconstructed on the TYGS (https://tygs.dsmz.de/). Tree inferred with FastME 2.1.6.1 from genome blast distance phylogeny (GBDP) distances calculated from genome sequences. The branch lengths are scaled in terms of the GBDP distance formula d5. The numbers above branches are GBDP pseudo-bootstrap support values >60% from 100 replications.
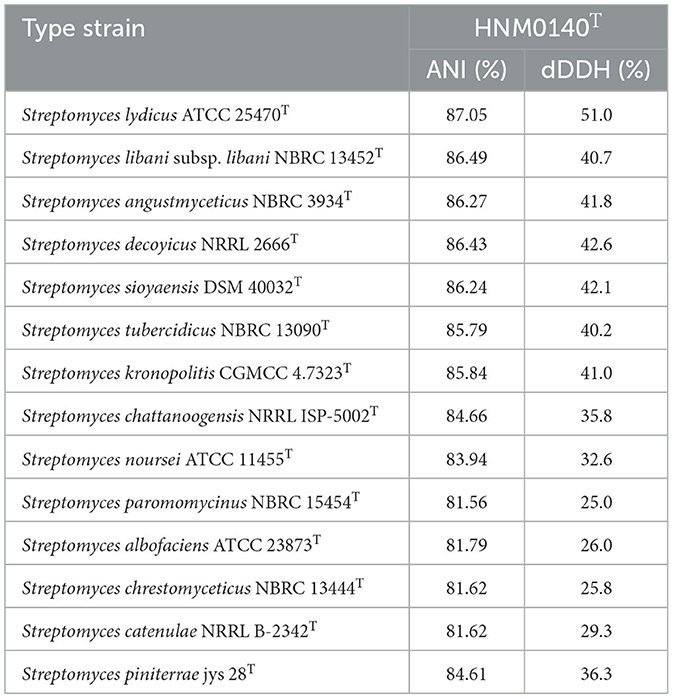
Table 2. Average nucleotide identity (ANI) and dDDH values between strain HNM0140T and its closest Streptomyces species.
Strain HNM0140T grew well on ISP 1–ISP 6 medium, except for ISP 7 (Supplementary Table S2). The color of aerial hyphae was either white or white gray, while that of substrate mycelia tended to be media dependent. Aerial hyphae further differentiated into long or curl spore chains, showing rugose spore ornamentation (Figures 1C, D). The organism produced no pigment on all test media. Strain HNM0140T was found to survive at pH 3.0–10.0 (optimum pH 7.0) and at 15–40°C (optimum 28°C) and with 0–12% (w/v) NaCl (optimum for 0–5%). The difference in phenotypic properties shown in Table 3 revealed that strain HNM0140T can be clearly distinct from the type strains of its closely related neighbors.
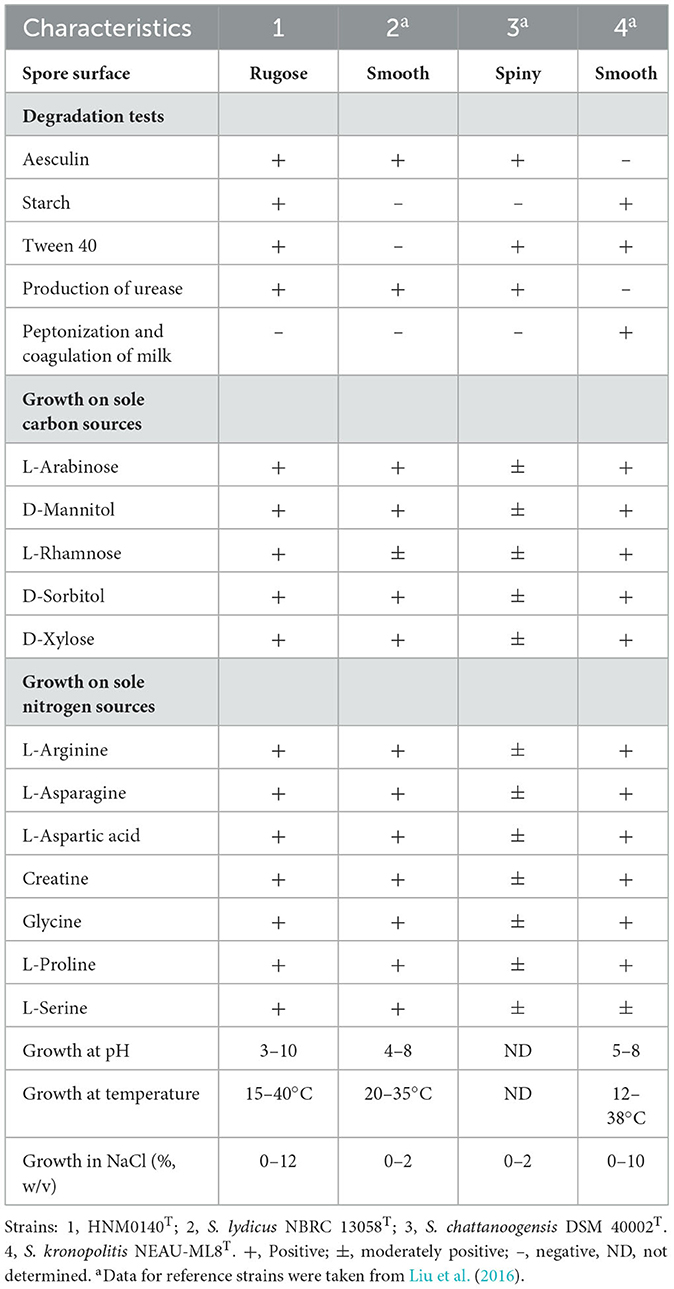
Table 3. Differential physiological characteristics between strain HNM0140T and its closely related strains.
LL-Diaminopimelic acid was detected in the cell wall of strain HNM0140T and the cell sugars, namely, glucose and ribose were detected in its whole-organism hydrolysates. Polar lipid analysis revealed that the polar lipid profiles of strain HNM0140T contained phosphatidylmethylethanolamine, diphosphatidylglycerol, phosphatidylinositol mannoside, phosphatidylethanolamine, and one unidentified phospholipid (Supplementary Figure 4). The menaquinone profiles of strain HNM0140T were identified as MK-9(H6) (46.3%), MK-9(H4) (40.3%), MK-9(H8) (8.8%), and MK-9(H11) (4.6%). The fatty-acid profiles of strain HNM0140T comprised iso-C16:0 (32.2%), C16:0 (16.9%), and anteiso-C15:0 (12.4%) as its major compositions, and minor amounts (<10%) of anteiso-C17:0, iso-C15:0, iso-C14:0, iso-C18:0, and C17:0 were also detected. All the aforementioned chemotaxonomic characteristics were consistent with its assignment to the genus Streptomyces.
In conclusion, a combination of phylogenetic, genomic, phenotypic, and chemotaxonomic analyses mentioned previously clearly indicated that strain HNM0140T should represent a new streptomycete species, for which the name proposed is Streptomyces endophytica sp. nov.
The complete genome of strain HNM0140T consisted of one linear chromosome (6,901,780 bp) with G+C content of 72.2%, 6,056 protein-coding genes (CDS), 66 tRNA genes, 18 rRNA genes, and 3 ncRNA genes. Among all CDS, the number of genes annotated into the COG, GO, and KEGG functional categories was 3,916, 1,562, and 2,126, respectively (Table 4). Based on the antiSMASH 6.0 analysis, strain HNM0140T was found to possess 18 putative biosynthetic gene clusters (BGCs) for secondary metabolites, which were distributed across 13 different types, including three butyrolactone, two RiPP-like, two siderophore, two terpene, one ectoine, one lanthipeptide-class-iii, one lassopeptide, one linaridin, one other, and four hybrid gene clusters (Supplementary Table S3). Seven putative BGCs showed at least 50% similarity to known BGCs, including three BGCs encoding for ectoine, desferrioxamine E, and althiomycin with 100% similarity. Among them, ectoine was an osmotic and cold stress protectant produced by Streptomyces (Prabhu et al., 2004). Desferrioxamine E is a cyclic trihydroxamate siderophore that exhibits antimicrobial activity against Mycobacterium species by inhibiting biofilm formation (Ishida et al., 2011). Althiomycin is a broad-spectrum antibiotic that displays antibacterial activity against both Gram-negative and -positive bacteria by inhibiting protein synthesis (Fujimoto et al., 1970). Other putative BGCs showed a low similarity (<30%) to known BGCs, including two BGCs with no similarity, which highlighted the potential of strain HNM0140T to produce novel antimicrobial metabolites.
Genome annotation and analysis uncovered that strain HNM0140T possessed a series of genes related to PGP, such as N2 fixation, production of IAA, phosphate solubilization, siderophore, and production of ACC deaminase (Supplementary Table S4). The genome of strain HNM0140T harbors a nifU gene, which encodes a protein (NifU) involved in the synthesis of iron–sulfur metalloclusters that are thought to be required for nitrogen fixation (Hwang et al., 1996). Strain HNM0140T also contains an iaaM gene that potentially participated in the indole-3-acetamide pathway of the indole-3-acetic acid biosynthesis (Qin et al., 2017). The genome of strain HNM0140T carries multiple genes that were related to the transport and solubilization of inorganic polyphosphates, including two gdh genes and a pqqD gene that participated in the synthesis of gluconic acid, which was believed to be responsible for the degradation of mineral phosphates (Farhat et al., 2015), and two pit genes, five phn genes, and three pst genes that participated in the transport of phosphonates. Strain HNM0140T also harbors plenty of rhbCDEF genes involved in the synthesis of the siderophores. Moreover, two BGCs encoding for siderophores, such as desferrioxamine E and ficellomycin, were predicted to be in the genome of strain HNM0140T (Supplementary Table S3). The acdS gene coding for ACC deaminase was also found in the genome, which has been reported to promote plant growth in stress conditions by decreasing ethylene concentrations (Santoyo et al., 2016). In addition, several gene coding for antifungal enzymes, such as β-glucanase and chitinase, were also present in the genome, indicating the potential inhibitory mechanism of strain HNM0140T against C. gloeosporioides.
Streptomyces endophytica (en.do.phy′ti.ca. Gr. pref. endo within; Gr. neut. n. phyton plant; L. fem. suff. -ica adjectival suffix used with the sense of belonging to; N.L. fem. adj. endophytica within plant, endophytic, pertaining to the original isolation from plant tissues).
Gram-stain-positive, aerobic and non-motile actinomycete, which forms an extensively branched substrate and aerial hyphae that differentiate into long or curl spore chains, consisting of rugose spores at maturity, grows well on ISP1–ISP6 media. No diffusible pigments were observed on ISP1–ISP7 agars. The growth occured at pH 3.0–10.0 (optimum pH 7.0), at 15–40°C (optimum 28°C), and up to 12% (w/v) NaCl tolerance (optimum 0–5%). It was positive for hydrolysis of starch, aesculin, Tween 40 and production of urease but negative for milk peptonization and coagulation. D-Xylose, L-arabinose, raffinose, D-mannitol, L-rhamnose, myo-inositol, fructose, raffinose, sucrose, D-galactose, and D-glucose are utilized as sole carbon sources. L-Aspartic acid, glycine, L-arginine, creatine, L-asparagine, L-proline, and L-serine are utilized as sole nitrogen sources.
The cell wall of strain HNM0140T consists of LL-diaminopimelic acid. MK-9(H6) and MK-9(H4) are its major menaquinones, and minor amounts of MK-9(H8) and MK-9(H11) were also detected. Iso-C16:0, C16:0, and anteiso-C15:0 are its major fatty-acid compositions, and minor amounts of anteiso-C17:0, iso-C15:0, iso-C14:0, iso-C18:0, and C17:0 were also detected. The major phospholipids comprise phosphatidylmethylethanolamine, diphosphatidylglycerol, phosphatidylinositol mannoside, and phosphatidylethanolamine.
The type strain, HNM0140T (=CCTCC AA2019073T = LMG 31910T), was isolated from the root of yam collected from Danzhou City of Hainan Province, China. The 16S rRNA gene sequence of strain HNM0140T was deposited in the GenBank/EMBL/DDBJ (accession no. MT365784). The complete genome of HNM0140T consists of 6,901,780 bp and the DNA G+C content of the type strain is 72.2% mol%. The complete genome sequence of HNM0140T is available in GenBank under Accession Number CP110636.
A new endophytic actinobacterium strain HNM0140T was isolated from the root of yam (Dioscorea opposita Thunb.) and selected for its strong antifungal activity against C. gloeosporioides. Strain HNM0140T was identified as a novel Streptomyces species through polyphasic taxonomic analyses, and its name was proposed as Streptomyces endophytica sp. nov. The organism could colonize in the tissue-cultured seedlings of yam and markedly reduce the severity and incidence of yam anthracnose under greenhouse conditions. The whole genome of strain HNM0140T harbored 18 BGCs for secondary metabolites, PGP-related genes, and several genes coding for antifungal enzymes. Therefore, strain HNM0140T will be a promising candidate for the biocontrol of yam anthracnose.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
SZ and XH conceived and designed the study. SZ and YZ carried out all the experiments. CL, WW, YX, WX, and DH conducted the data analysis. SZ prepared the manuscript. XH revised it. All authors reviewed and approved the manuscript.
This study received funding from the newly introduced doctoral research project in Guilin Medical University (Grant No. 20501020044) and the Natural Science Foundation of Hainan Province (Grant No. 311036).
We are grateful to Jinlan Xiao, Ji Zhu, Yuxin Yang, Yu Li, Yuhui Xie, and Yaxian Feng for their help in this study.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1139456/full#supplementary-material
Abang, M. M., Winter, S., Mignouna, H. D., Green, K. R., and Asiedu, R. (2003). Molecular taxonomic, epidemiological and population genetic approaches to understanding yam anthracnose disease. Afr. J. Biotechnol. 2, 486–496. doi: 10.5897/AJB2003.000-1098
Blin, K., Shaw, S., Kloosterman, A. M., Charlop-Powers, Z., Van Wezel, G. P., Medema, M. H., et al. (2021). AntiSMASH 6.0: improving cluster detection and comparison capabilities. Nucleic. Acids Res. 49, W29–W35. doi: 10.1093/nar/gkab335
Crawford, D. L., Lynch, J. M., Whipps, J. M., and Ousley, M. A. (1993). Isolation and characterization of actinomycete antagonists of a fungal root pathogen. Appl. Environ. Microb. 59, 3899–3905. doi: 10.1128/aem.59.11.3899-3905.1993
Delcher, A. L., Bratke, K. A., Powers, E. C., and Salzberg, S. L. (2007). Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics. 23, 673–679. doi: 10.1093/bioinformatics/btm009
Döbereiner, H. G., Evans, E., Seifert, U., and Wortis, M. (1995). Spinodal fluctuations of budding vesicles. Phys. Rev. Lett. 75, 3360–3363. doi: 10.1103/PhysRevLett.75.3360
Doumbou, C. L., Hamby Salove, M. K., Crawford, D. L., and Beaulieu, C. (2001). Actinomycetes, promising tools to control plant diseases and to promote plant growth. Phytoprotection 82, 85–102. doi: 10.7202/706219ar
Egesi, C. N., Onyeka, T. J., and Asiedu, R. (2007). Severity of anthracnose and virus diseases of water yam (Dioscorea alata L.) in Nigeria I: effects of yam genotype and date of planting. Crop Prot. 26, 1259–1265. doi: 10.1016/j.cropro.2006.10.025
El-Tarabily, K. A. (2008). Promotion of tomato (Lycopersicon esculentum Mill.) plant growth by rhizosphere competent 1-aminocyclopropane-1-carboxylic acid deaminase-producing streptomycetes actinomycetes. Plant Soil. 308, 161–174. doi: 10.1007/s11104-008-9616-2
Farhat, M. B, Boukhris, I., and Chouayekh, H. (2015). Mineral phosphate solubilization by Streptomyces sp. CTM396 involves the excretion of gluconic acid and is stimulated by humic acids. FEMS Microbiol. Lett. 362, 1–7. doi: 10.1093/femsle/fnv008
Felsenstein, J. (1981). Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17, 368–376. doi: 10.1007/BF01734359
Felsenstein, J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x
Fujimoto, H., Kinoshita, T., Suzuki, H., and Umezawa, H. (1970). Studies on the mode of action of althiomycin. J. Antibiot. 23, 271–275. doi: 10.7164/antibiotics.23.271
Gao, Y., Ning, Q., Yang, Y., Liu, Y., Niu, S., Hu, X., et al. (2021). Endophytic Streptomyces hygroscopicus OsiSh-2-mediated balancing between growth and disease resistance in host rice. Mbio 12, e01566–e01521. doi: 10.1128/mBio.01566-21
Govindasamy, V., Franco, C. M. M., and Gupta, V. V. S. R. (2014). “Endophytic actinobacteria: diversity and ecology,” in Advances in Endophytic Research, eds V. Verma, and A. Gange (New Delhi: Springer), 27–59. doi: 10.1007/978-81-322-1575-2_2
Gu, L., Zhang, K., Zhang, N., Li, X., and Liu, Z. (2020). Control of the rubber anthracnose fungus Colletotrichum gloeosporioides using culture filtrate extract from Streptomyces deccanensis QY-3. Antonie van Leeuwenhoek. 113, 1573–1585. doi: 10.1007/s10482-020-01465-8
Hamdali, H., Bouizgarne, B., Hafidi, M., Lebrihi, A., Virolle, M. J., and Ouhdouch, Y. (2008). Screening for rock phosphate solubilizing Actinomycetes from Moroccan phosphate mines. Appl. Soil Ecol.38, 12–19. doi: 10.1016/j.apsoil.2007.08.007
Hayakawa, M., and Nonomura, H. (1987). Humic acid-vitamin agar, a new medium for the selective isolation of soil actinomycetes. J. Ferment. Technol. 65, 501–509. doi: 10.1016/0385-6380(87)90108-7
Hsu, S. C., and Lockwood, J. L. (1975). Powdered chitin agar as a selective medium for enumeration of actinomycetes in water and soil. Appl. Microbiol. 29, 422–426. doi: 10.1128/am.29.3.422-426.1975
Huang, X., Kong, F., Zhou, S., Huang, D., Zheng, J., and Zhu, W. (2019). Streptomyces tirandamycinicus sp. nov., a novel marine sponge-derived actinobacterium with potential against Streptococcus agalactiae. Front. Microbiol. 10, 482. doi: 10.3389/fmicb.2019.00482
Huang, X., Zhuang, L., Lin, H. P., Li, J., Goodfellow, M., and Hong, K. (2012). Isolation and bioactivity of endophytic filamentous actinobacteria from tropical medicinal plants. Afr. J. Biotechnol. 11, 9855–9864. doi: 10.5897/AJB11.3839
Hwang, D. M., Dempsey, A., Tan, K. T., and Liew, C. C. (1996). A modular domain of NifU, a nitrogen fixation cluster protein, is highly conserved in evolution. J. Mol. Evol. 43, 536–540. doi: 10.1007/BF02337525
Ishida, S., Arai, M., Niikawa, H., and Kobayashi, M. (2011). Inhibitory effect of cyclic trihydroxamate siderophore, desferrioxamine E, on the biofilm formation of Mycobacterium species. Biol. Pharm. Bull. 34, 917–920. doi: 10.1248/bpb.34.917
Kelly, K. L. (1964). Inter-Society Color Council—National Bureau of Standards Color Name Charts Illustrated with Centroid Colors. Washington, DC: US Government Printing Office.
Kimura, M. (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120. doi: 10.1007/BF01731581
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lechevalier, M. P., and Lechevalier, H. A. (1980). “The chemotaxonomy of actinomycetes,” in Actinomycete Taxonomy, Special Publication 6, eds A. Dietz and D. W. Thayer (Arlington, VA: Society of Industrial Microbiology), 227–291.
Li, M., Li, J., Liu, W., Liu, L., Lu, J., Niu, J., et al. (2014). A protocol for in vitro production of microtubers in Chinese yam (Dioscorea opposita). Biosci. Biotechnol. Biochem. 78, 1005–1009. doi: 10.1080/09168451.2014.912119
Liu, C., Ye, L., Li, Y., Jiang, S., Liu, H., Yan, K., et al. (2016). Streptomyces kronopolitis sp. nov., an actinomycete that produces phoslactomycins isolated from a millipede (Kronopolites svenhedind Verhoeff). Int. J. Syst. Evol. Microbiol. 66, 5352–5357. doi: 10.1099/ijsem.0.001520
Lowe, T. M., and Eddy, S. R. (1997). tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955–964. doi: 10.1093/nar/25.5.955
Marian, M., Ohno, T., Suzuki, H., Kitamura, H., Kuroda, K., and Shimizu, M. (2020). A novel strain of endophytic Streptomyces for the biocontrol of strawberry anthracnose caused by Glomerella cingulata. Microbiol. Res. 234, 126428. doi: 10.1016/j.micres.2020.126428
Meier-Kolthoff, J. P., Auch, A. F., Klenk, H. P., and Göker, M. (2013). Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 14, 60. doi: 10.1186/1471-2105-14-60
Meier-Kolthoff, J. P., Sardà Carbasse, J., Peinado-Olarte, R. L., and Göker, M. (2022). TYGS and LPSN: a database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acid Res. 50, D801–D807. doi: 10.1093/nar/gkab902
Minnikin, D. E., O'Donnell, A. G., Goodfellow, M., Alderson, G., Athalye, M., Schaal, A., et al. (1984). An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J. Microbiol. Methods. 2, 233–241. doi: 10.1016/0167-7012(84)90018-6
Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15, 473–497.
Naik, P. R., Sahoo, N., Goswami, D., Ayyadurai, N., and Sakthivel, N. (2008). Genetic and functional diversity among fluorescent pseudomonads isolated from the rhizo- sphere of banana. Microb. Ecol. 56, 492–504. doi: 10.1007/s00248-008-9368-9
Nawrocki, E. P., and Eddy, S. R. (2013). Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics. 29, 2933–2935. doi: 10.1093/bioinformatics/btt509
Ntui, V. O., Uyoh, E. A., Ita, E. E., Markson, A. A. A., Tripathi, J. N., Okon, N. I., et al. (2021). Strategies to combat the problem of yam anthracnose disease: status and prospects. Mol Plant Pathol. 22, 1302–1314. doi: 10.1111/mpp.13107
Nwadili, C. O., Augusto, J., Bhattacharjee, R., Atehnkeng, J., Lopez- Montes, A., Onyeka, T. J., et al. (2017). Comparative reliability of screening parameters for anthracnose resistance in water yam (Dioscorea alata). Plant Dis. 101, 209–216. doi: 10.1094/PDIS-06-16-0924-RE
Okigbo, R. N. (2005). Biological control of postharvest fungal rot of yam (Dioscorea spp.) with Bacillus subtilis. Mycopathologia. 159, 307–314. doi: 10.1007/s11046-004-2454-8
Onyeka, T. J., Pe'tro, D., Ano, G, Etienne, S., and Rubens, S. (2006). Resistance in water yam (Dioscorea alata) cultivars in the French West Indies to anthracnose disease based on tissue culture-derived whole-plant assay. Plant. Pathol. 55, 671–678. doi: 10.1111/j.1365-3059.2006.01436.x
Palaniyandi, S. A., Yang, S. H., Cheng, J. H., Meng, L., and Suh, J. W. (2011). Biological control of anthracnose (Colletotrichum gloeosporioides) in yam by Streptomyces sp. MJM5763. J. Appl. Microbiol. 111, 443–455. doi: 10.1111/j.1365-2672.2011.05048.x
Palaniyandi, S. A., Yang, S. H., Damodharan, K., and Suh, J. W. (2013). Genetic and functional characterization of culturable plant-beneficial actinobacteria associated with yam rhizosphere. J. Basic Microb. 53, 985–995. doi: 10.1002/jobm.201200531
Palaniyandi, S. A., Yang, S. H., and Suh, J. W. (2016). Foliar application of extract from an azalomycin-producing Streptomyces malaysiensis strain MJM1968 suppresses yam anthracnose caused by Colletotrichum gloeosporioides. J. Microbiol. Biotechn. 26, 1103–1108. doi: 10.4014/jmb.1601.01018
Prabhu, J., Schauwecker, F., Grammel, N., Keller, U., and Bernhard, M. (2004). Functional expression of the ectoine hydroxylase gene (thpD) from Streptomyces chrysomallus in Halomonas elongata. Appl. Environ. Microb.70, 3130–3132. doi: 10.1128/AEM.70.5.3130-3132.2004
Qin, S., Feng, W. W., Wang, T. T., Ding, P., Xing, K., and Jiang, J. H. (2017). Plant growth-promoting effect and genomic analysis of the beneficial endophyte Streptomyces sp. KLBMP 5084 isolated from halophyte Limonium sinense. Plant Soil 416, 117-132. doi: 10.1007/s11104-017-3192-2
Richter, M., and Rosselló-Móra, R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA. 106, 19126–19131. doi: 10.1073/pnas.0906412106
Richter, M., Rosselló-Móra, R., Glöckner, F. O., and Peplies, J. (2016). JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics. 32, 929–931. doi: 10.1093/bioinformatics/btv681
Rzhetsky, A., and Nei, M. (1992). A simple method for estimating and testing minimum evolution trees. Mol. Biol. Evol. 9, 945–967. doi: 10.1093/oxfordjournals.molbev.a040771
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Santoyo, G., Moreno-Hagelsieb, G., Orozco-Mosqueda Mdel, C., and Glick, B. R. (2016). Plant growth-promoting bacterial endophytes. Microbiol Res. 183, 92–99. doi: 10.1016/j.micres.2015.11.008
Sasser, M. (1990). Identification of Bacteria by Gas Chromatography of Cellular Fatty Acids. Newark, DE: MIDI Inc.
Shimizu, M. (2011). “Endophytic actinomycetes: biocontrol agents and growth promoters,” in Bacteria in Agrobiology: Plant Growth Responses, ed D. Maheshwari (Berlin, Heidelberg: Springer), 201–220. doi: 10.1007/978-3-642-20332-9_10
Shimizu, M., Fujita, N., Nakagawa, Y., Nishimura, T., Furumai, T., Igarashi, Y., et al. (2001). Disease resistance of tissue-cultured seedlings of rhododendron after treatment with Streptomyces sp. R-5. J. Gen. Plant Pathol. 6, 325–332. doi: 10.1007/PL00013040
Shimizu, M., Nakagawa, Y., Sato, Y., Furumai, T., Igarashi, Y., Onaka, H., et al. (2000). Studies on endophytic actinomycetes (I). Streptomyces sp. isolated from rhododendron and its antifungal activity. J. Gen. Plant. Pathol. 66, 360–366. doi: 10.1007/PL00012978
Shimizu, M., Yazawa, S., and Ushijima, Y. (2009). A promising strain of endophytic Streptomyces sp. for biological control of cucumber anthracnose. J. Gen. Plant. Pathol. 75, 27–36. doi: 10.1007/s10327-008-0138-9
Shirling, E. B., and Gottlieb, D. (1966). Methods for characterization of Streptomyces species. Int. J. Syst. Bacteriol. 16, 313–340. doi: 10.1099/00207713-16-3-313
Simons, S. A., and Green, K. R. (1994). Quantitative methods for assessing the severity of anthracnose on yam (Dioscorea alata). Trop Sci. 34, 216–224.
Singh, R., and Dubey, A. K. (2018). Diversity and applications of endophytic actinobacteria of plants in special and other ecological niches. Front. Microbiol. 9, 1767. doi: 10.3389/fmicb.2018.01767
Soares, A. C. F., Sousa, C. D. S., Garrido, M. D. S., Perez, J. O., and Almeida, N. S. D. (2006). Soil streptomycetes with in vitro activity against the yam pathogens Curvularia eragrostides and Colletotrichum gloeosporioides. Braz. J. Microbiol. 37, 456–461. doi: 10.1590/S1517-83822006000400010
Solanki, M. K., Malviya, M. K., and Wang, Z. (2016). “Actinomycetes bio-inoculants: a modern prospectus for plant disease management,” in Plant Growth Promoting Actinobacteria, eds G. Subramaniam, S. Arumugam, V. Rajendran (Singapore: Springer), 63–81. doi: 10.1007/978-981-10-0707-1_5
Thilagam, R., and Hemalatha, N. (2019). Plant growth promotion and chilli anthracnose disease suppression ability of rhizosphere soil actinobacteria. J. Appl. Microbiol. 126, 1835–1849. doi: 10.1111/jam.14259
Williams, S. T., Goodfellow, M., Alderson, G., Wellington, E. M. H., Sneath, P. H. A., and Sackin, M. J. (1983). Numerical classification of Streptomyces and related genera. J. Gen. Microbiol. 129, 1743–1813. doi: 10.1099/00221287-129-6-1743
Yokota, A., Tamura, T., Hasegawa, T., and Huang, L. H. (1993). Catenuloplanes japonicus gen. nov., sp. nov., nom. rev., a new genus of the order Actinomycetales. Int. J. Syst. Bacteriol. 43, 805–812. doi: 10.1099/00207713-43-4-805
Yoon, S. H., Ha, S. M., Kwon, S., Lim, J., Kim, Y., and Seo, H. (2017). Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 67, 1613–1617. doi: 10.1099/ijsem.0.001755
Zhang, D., Lu, Y., Chen, H., Wu, C., Zhang, H., Chen, L., et al. (2020). Antifungal peptides produced by actinomycetes and their biological activities against plant diseases. J. Antibiot. 73, 265–282. doi: 10.1038/s41429-020-0287-4
Keywords: Streptomyces endophytica sp. nov., anthracnose, endophytic actinobacteria, biological control, yam
Citation: Zhou S, Zhou Y, Li C, Wu W, Xu Y, Xia W, Huang D and Huang X (2023) Identification and genomic analyses of a novel endophytic actinobacterium Streptomyces endophytica sp. nov. with potential for biocontrol of yam anthracnose. Front. Microbiol. 14:1139456. doi: 10.3389/fmicb.2023.1139456
Received: 07 January 2023; Accepted: 06 March 2023;
Published: 04 April 2023.
Edited by:
Zhen Wang, Yulin Normal University, ChinaReviewed by:
Maria Carolina Quecine, University of São Paulo, BrazilCopyright © 2023 Zhou, Zhou, Li, Wu, Xu, Xia, Huang and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaolong Huang, aHVhbmd4aWFsb25nQGhhaW5hbnUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.