
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 03 August 2023
Sec. Microbiotechnology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1138979
This article is part of the Research Topic Genetically Modified Organisms: Tricks or Threats, Volume II View all 4 articles
 Fang Zhang1†
Fang Zhang1† Liying Wang2†
Liying Wang2† Jiayu Jin3†
Jiayu Jin3† Yulu Pang4
Yulu Pang4 Hao Shi5
Hao Shi5 Ziyi Fang5
Ziyi Fang5 Han Wang5
Han Wang5 Yujie Du3
Yujie Du3 Yufan Hu5
Yufan Hu5 Yingchun Zhang2
Yingchun Zhang2 Xiaoyue Ding2*
Xiaoyue Ding2* Zuobin Zhu2*
Zuobin Zhu2*Escherichia coli (E. coli) mutant strains have been reported to extend the life span of Caenorhabditis elegans (C. elegans). However, the specific mechanisms through which the genes and pathways affect aging are not yet clear. In this study, we fed Drosophila melanogaster (fruit fly) various E. coli single-gene knockout strains to screen mutant strains with an extended lifespan. The results showed that D. melanogaster fed with E. coli purE had the longest mean lifespan, which was verified by C. elegans. We conducted RNA-sequencing and analysis of C. elegans fed with E. coli purE (a single-gene knockout mutant) to further explore the underlying molecular mechanism. We used differential gene expression (DGE) analysis, enrichment analysis, and gene set enrichment analysis (GSEA) to screen vital genes and modules with significant changes in overall expression. Our results suggest that E. coli mutant strains may affect the host lifespan by regulating the protein synthesis rate (cfz-2) and ATP level (catp-4). To conclude, our study could provide new insights into the genetic influences of the microbiota on the life span of a host and a basis for developing anti-aging probiotics and drugs.
With the rapid development of modern medical technology, society is increasingly aging. Improving healthy aging and searching for longevity substances have become current biomedical research priorities. In a report published in Cell in 2017, researchers screened 3,983 Escherichia coli (E. coli) mutants obtained from an E. coli single-gene knockout library using Caenorhabditis elegans (C. elegans) as a model. They investigated 29 mutants that significantly prolonged the lifespan of C. elegans. The results showed that colanic acid (CA) significantly prolonged the lifespan of C. elegans (Han et al., 2018). In 2019, based on the previously mentioned report, it was found that E. coli mutants can affect the growth and development of C. elegans through the balance between bacterial iron and reactive oxygen species (ROS) (Zhang et al., 2019). Hence, intestinal bacteria have provided a new direction in anti-aging research.
In humans, food digestion and absorption processes occur in the gut, where a considerable number of microorganisms exist. Being the largest and most complex micro-ecosystem in the human body, gut microbes and their metabolites regulate human health and play an essential role in bridging the diet and host gap. Recent studies have shown that fecal transplantation improves physiological functions and prolongs the life span in mice, suggesting that gut bacteria can influence the aging process (Barcena et al., 2019). However, gut bacteria are too complex and difficult to standardize. In this regard, it has been suggested that the genetic variation in gut microbes can influence the host’s viability and drive the host’s evolution. A study by Guo et al. published in the December 2019 issue of science suggested that to study the effects of gut flora metabolites on the host, it is essential to simplify the study population by starting with a single gut bacterium and subjecting it to multiple knockouts (Guo et al., 2019). E. coli, constituting about 0.1% of the human gut flora, is the most widely studied prokaryotic model organism and an important species in biotechnology and microbiology (Gao et al., 2014). Therefore, we screened for E. coli mutants with putative anti-aging effects on the host from essential gut bacteria (E. coli) using hosts and intestinal bacteria. We also explored how the bacterial mutant strains regulate aging, using the transcriptional changes in the intestinal bacteria that affect the host’s aging as an entry point.
C. elegans is a unique experimental system owing to its small size, rapid life cycle, transparency, and well-annotated genome. Most importantly, an estimated 60%–80% of human homologous genes exist in the nematode genome (Kaletta and Hengartner, 2006). The fruit fly, Drosophila melanogaster, is another vital model organism for studying aging (Piper and Partridge, 2018). Seventy-seven percent of human aging-related genes are also expressed in Drosophila, and their lifespan experiments date back to 1913. The following aging features make Drosophila a good model organism for studying aging: increased sleep fragment, impaired negative geotaxis, reduced autonomic flight and crawl function, reduced resting metabolic rate, and neurological and cardiac decline (Tamura et al., 2003; Gargano et al., 2005; Iliadi et al., 2012). Moreover, its simple culture and short lifespan facilitate high-throughput screening experiments.
Using high-throughput screening, we selected 25 E. coli mutant strains that have been shown to have anti-aging ability in C. elegans (Han et al., 2018), which were subjected to longevity and climbing experiments. Active and UV-inactivated bacteria were used in an attempt to detect any differences between the two. Combining the phenotypic and bioinformatic analyses, we further investigated the pathways related to lifespan extension, including the evolutionarily conserved insulin/insulin-like growth factor (IGF), the target of rapamycin (TOR), and germline signaling pathways (Guarente and Kenyon, 2000; Pletcher et al., 2002; Hahn and Denlinger, 2011; Partridge et al., 2011). We tried to lay the foundation for subsequent molecular mechanistic studies and provide a research basis for developing anti-aging probiotics and subsequent drugs.
D. melanogaster was selected from the Canton Special (CS) strain of wild-type Drosophila from the Drosophila Resource and Technology Platform, Center of Excellence in Molecular Cell Science, Chinese Academy of Sciences. E. coli single gene knockout strains were obtained from the single knockout library (E. coli Keio Knockout Collection). C. elegans strains were obtained from Caenorhabditis Genetics Center (CGC), University of Minnesota, United States. C. elegans strains were grown on a nematode growth medium (NGM) at 20°C. All flies were reared on standard cornmeal-yeast-agar medium at 25°C with a photoperiod of 12 h:12hLD (light:dark). E. coli was incubated in the Luria-Bertani Culture and used for the experiment when it reached OD600 = 1.
Ten-day post-adult male Drosophila were transferred to tubes containing an experimental growth medium grown (standard cornmeal-yeast-agar medium without yeast extract) at 25°C. Every 20 individuals were fed in one tube, in groups of four, with activated and inactivated E. coli of different mutant strains, focusing on mutant strains purE, aroG, and wild type BW25113. The experiment was repeated three times. During this process, each tube contains 200 μL experimental bacteria (OD600 = 1), which is replaced every 10 days. In addition, inactivated strains are obtained by UV irradiation of mutant strains for 30 min.
C. elegans and Drosophila lifespan assays are briefly described as follows. For the C. elegans lifespan assay, young adult worms were transferred to NGM containing E. coli BW25113 and purE. Then, worm survival was measured daily by observing the pharyngeal movement and the touch-provoked movement using a platinum wire. Worms were considered dead if there was no pharyngeal and no touch-provoked movement. For the Drosophila lifespan assay, the second day of Drosophila tube entry was considered day one. The number of Drosophila deaths in each strain tube was recorded at 13:00 daily until they all died. Regarding the survival index, the mean lifespan is the mean of the number of days that all fruit flies in each tube survived; the maximum lifespan of each group is the lifespan of the fruit fly that last died; Survival was assessed every day based on the number of dead and alive flies; LT50 was calculated as the number of days it takes for 50% of the flies to die (Charalambous et al., 2022). The mean lifespan of Drosophila is calculated as follows. Let the number of days the fruit flies survived be “d,” and the number of flies that die on the same day be “xd.” Then, the mean lifespan of Drosophila per tube is
Drosophila climbing experiments for health span assays were performed as follows. The tubes of days 10 and 15 were placed vertically upside down at 18:00 daily. The number of fruit flies that reached the top of the tube in 10 s was examined after they had adapted, and the climbing process was recorded using a video camera for observation. The climbing index was calculated as follows: the number of fruit flies reaching the top of the tube within 10 s divided by the total number of alive fruit flies. Each tube was examined at least three times with at least 1 min apart. The average of the climbing index of each tube was recorded for each group.
In this study, three C. elegans fed with E. coli purE mutation samples and three BW25113 control samples were sequenced on the DNBSEQ platform, averagely generating about 1.19 Gb per sample (BioProject: F21FTSECWLJ1283_NEMyjzwN). Raw reads with rRNA, low quality, joint contamination, and high content of unknown base nucleotides were filtered out. Then, clean reads were matched to the reference genome (NCBI: GCF_000002985.6_WBcel235) using HISAT and assembled using StringTie. Clean reads were compared to the reference sequence using the Bowtie 2 software. Differential gene expression between the C. elegans fed with purE and BW25113 were analyzed by the Huada Online Analysis Software (Dr. Tom) using read count. The fold change >2 and Q-value <0.05 indicated significantly different gene expressions. The differentially expressed genes were mapped for gene function evaluation using the gene ontology (GO) database and the online analysis platform of Dr. Tom. They were also analyzed using the Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis and Gene Set Enrichment Analysis (GSEA).
Statistical analyses were performed using student’s t-test (SPSS 19.0). Data were presented as mean ± standard deviation. Asterisks denote significant differences (*p < 0.05) as determined by student’s t-test. The anti-aging ability of the mutant strains was analyzed together with lifespan and health span (climbing index) assays. The survival curves of the Drosophila and C. elegans were analyzed with the Kaplan–Meyer method and the log-rank test.
To test whether the different strains of E. coli extend the lifespan, we measured the mean lifespan of D. melanogaster fed with different E. coli mutant strains. The results showed that Drosophila fed with E. coli purE and aroG mutant strains had a significantly longer lifespan than those fed with E. coli BW25113 (Supplementary Table S1, Table 1, and Figure 1A). Drosophila fed with E. coli purE, especially, had the longest mean lifespan, which was verified by C. elegans (Supplementary Table S1 and Figure 2A). The survival curves and half-lives of Drosophila fed with E. coli purE mutant strain showed a significant increase in host lifespan compared to the control group (p = 0.0212), as well as a higher median lifespan (LT50 = 22) (Supplementary Table S1 and Figures 1C–E). Meanwhile, the effects of live bacteria were more pronounced than that of inactivated bacteria, except for the value of p, but still significant. To better assess the health of flies with increased lifespan, day 10 and day 15 Drosophila climbing experiment data showed a significant increase in the climbing ability of Drosophila fed with E. coli purE mutant strain compared with those fed with E. coli BW25113 (Figure 1B). The mean climbing index was higher than that of Drosophila fed with E. coli BW25113.
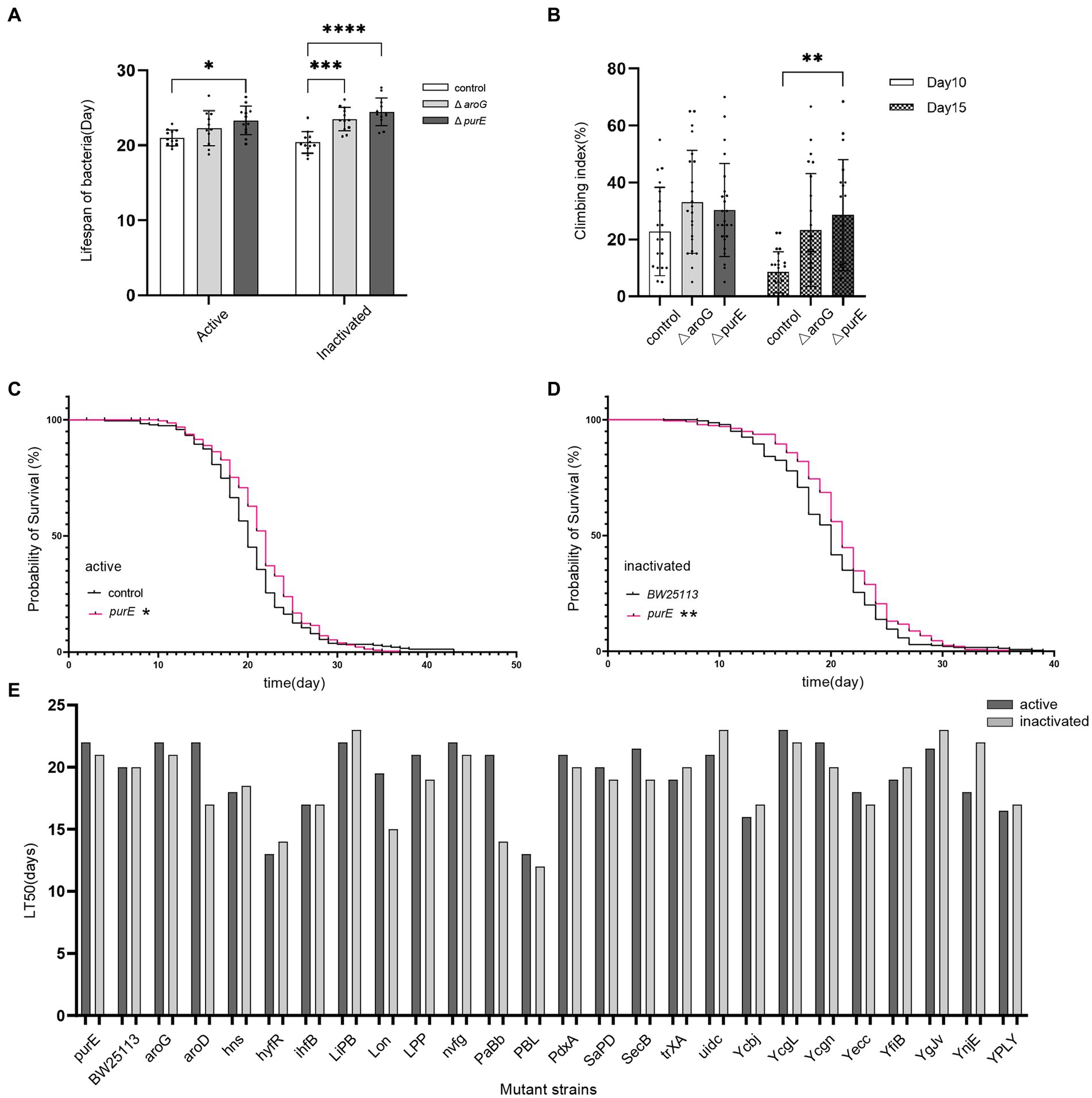
Figure 1. Comparison of the lifespan and climbing index of Drosophila fed with E. coli mutant strains purE, aroG, and BW25113. (A) Mean lifespan of Drosophila fed with E. coli mutant strains purE, aroG, and BW25113. (B) Days 10 and 15 of the climbing index of Drosophila fed E. coli purE compared with those fed with BW25113. * indicates significant differences, compared with BW25113 (student’s t-test value of p <0.05). **, ***, and **** indicate student’s t-test value of p < 0.01, p < 0.001, and p < 0.0001, respectively. (C) Survival curves of Drosophila fed with active E. coli purE (red lines) and BW25113 (black lines). (D) Survival curves of Drosophila fed with inactivated E. coli purE (red lines) and BW25113 (black lines). * indicates significant differences, compared with BW25113 (Log-rank test value of p < 0.05). **indicate Log-rank test value of p < 0.01.(E) The survival of Drosophila evaluated as the number of days it takes to reach 50% fly mortality (LT50) following feeding E. coli mutant strains.
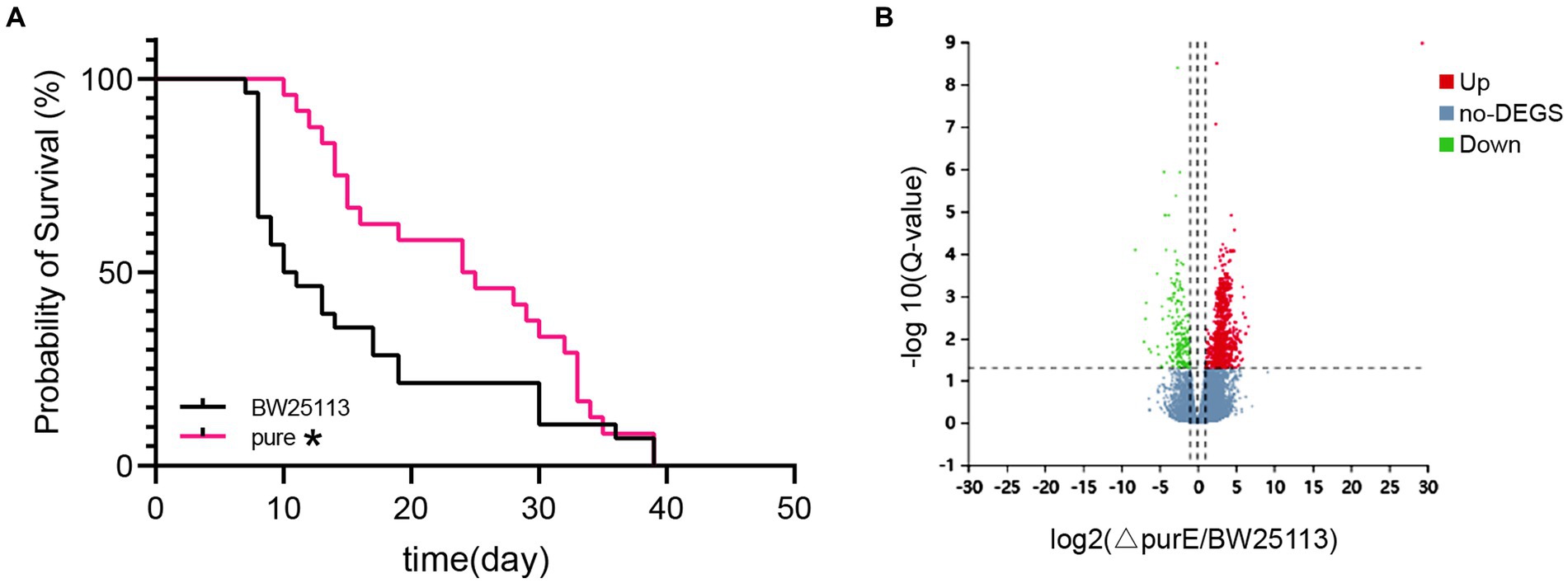
Figure 2. (A) Survival curve graph of C. elegans fed with E. coli purE and BW25113. (B) Volcano plot of the differential genes. * indicates significant differences, compared with BW25113 (Log-rank test value of p < 0.05).
The longevity of C. elegans fed with E. coli purE and BW25113 was also recorded, and survival curves were plotted (Figure 2A). There was a significant increase in longevity after feeding with E. coli purE.
Gene expression analysis of C. elegans fed with lifespan-extending E. coli mutant strains was performed. We used differential gene expression analysis, KEGG pathway enrichment analysis, and GO enrichment analysis to analyze the sequencing data. GSEA analysis was performed to find genes associated with an extended lifespan.
The samples were analyzed in three independent biological replicates (six RNA-Seq libraries). Pearson’s correlation coefficient analysis showed high reproducibility between the three replicates, ranging from 0.91 to 0.99 (Table 2).
The differentially expressed gene (DEG) analysis showed that there were 1,001 DEGs (Q-value <0.05) between E. coli purE and BW25113 obtained by the mean read count. Among them, there were 810 upregulated genes and 191 downregulated genes of more than twofold in E. coli purE, as shown in the volcano plot (Figure 2B and Supplementary Table S2).
To explore DEGs’ function and find whether they are associated with pathways known to extend the lifespan, we performed KEGG pathway and GO enrichment analyses. GO enrichment analysis showed that DEGs significantly enriched many biological processes, such as protein dephosphorylation, protein phosphorylation, and peptidyl-serine phosphorylation, and had significant effects on collagen trimer and the pseudopodium in the cellular component and the structural constituent of the cuticle in the molecular function (Figures 3A-C).
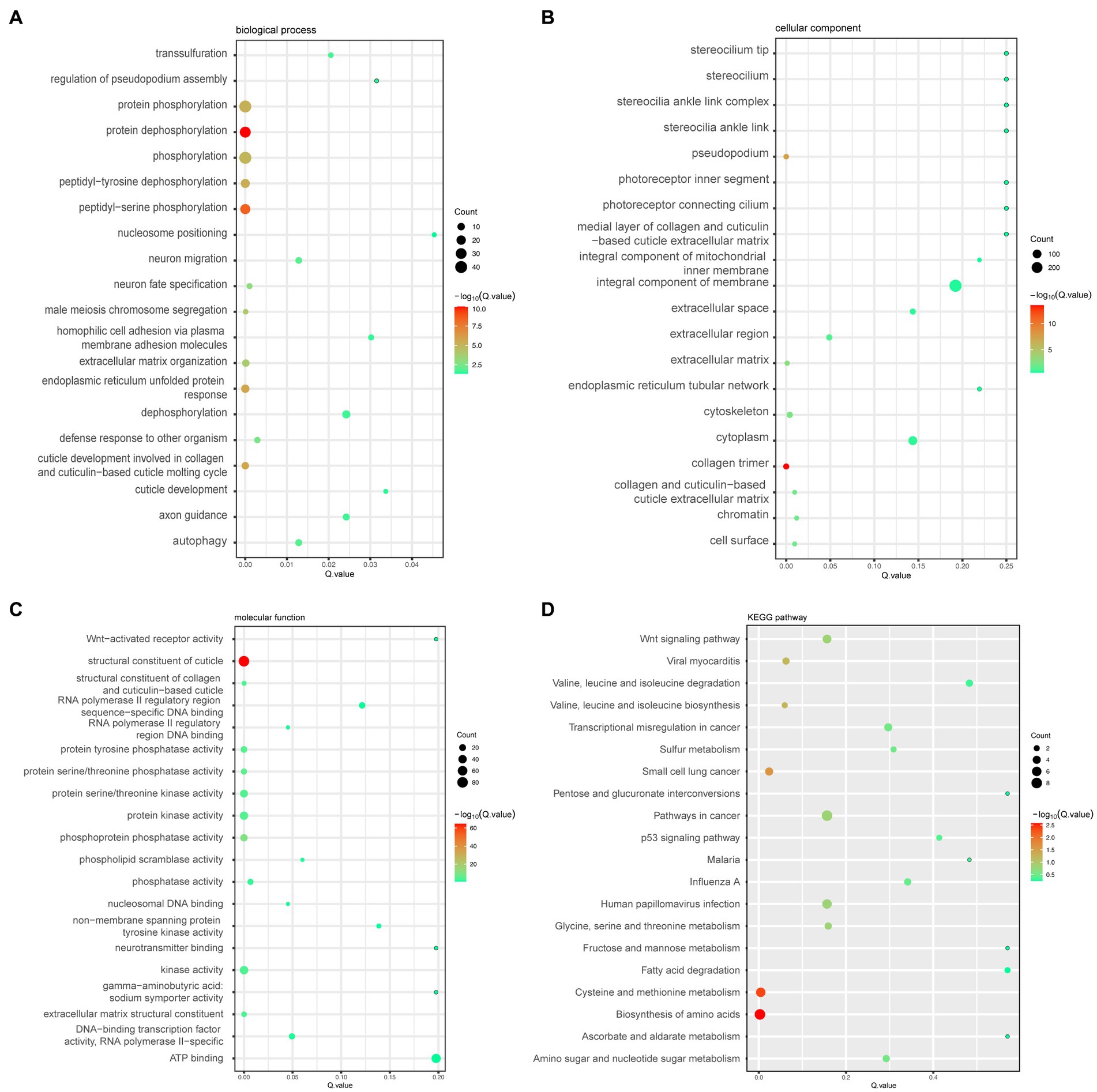
Figure 3. (A) Bubble plots of the top 20 biological processes enriched by the differential genes. (B) Bubble plots of the top 20 cellular components enriched by the differential genes. (C) Bubble plots of the top 20 molecular functions enriched by the differential genes. (D) Bubble plots of the top 20 KEGG pathways enriched by the differential genes.
Meanwhile, the enrichment results of KEGG signaling pathways for the differential genes showed that the enriched genes were significantly enriched in the following pathways: the biosynthesis of amino acids, cysteine and methionine metabolism, small cell lung cancer pathway, and cancer pathways (Figure 3D). Additionally, we looked for signaling pathways in all the enriched pathways that are known to be associated with the longevity regulation pathway, where gene cfz-2 was enriched in the mTOR signaling pathway (map04150) and gene catp-4 was enriched in the insulin secretion pathway (map04911) (Table 3).
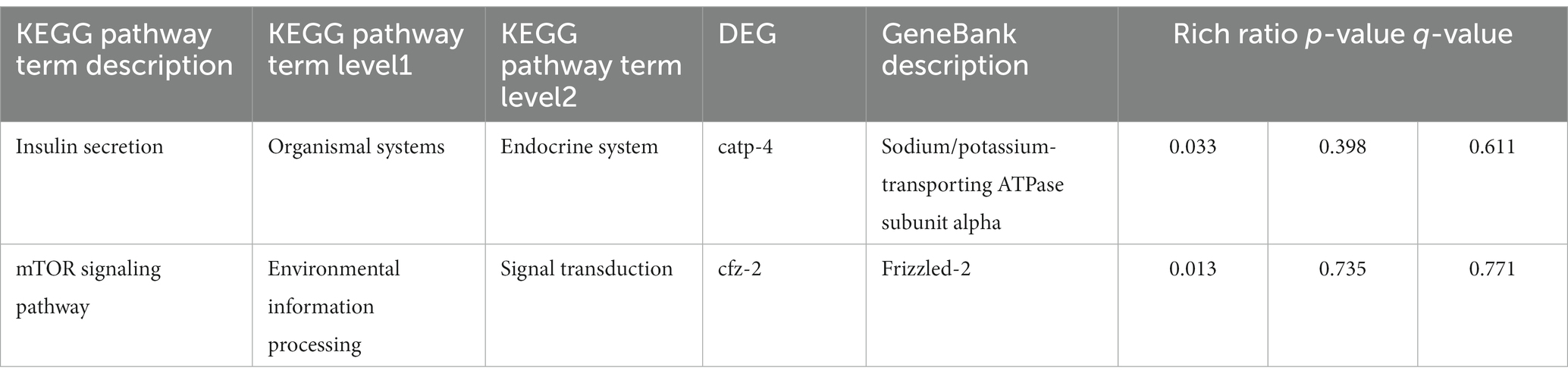
Table 3. KEGG pathway enrichment results in longevity regulation-related pathways and differential genes.
To avoid missing genes that are not significantly differentially expressed but are biologically important, the experiment was performed simultaneously with GSEA of all genes. GSEA resulted in 13 upregulated pathways, 38 downregulated pathways (Q-value <0.05), and 3 upregulated modules (value of p <0.05; Q-value <0.25) for the overall expression level. The set of genes contained in the pathway was highly expressed in the purE or BW25113 group, and the details of the top 10 are shown in Table 4.
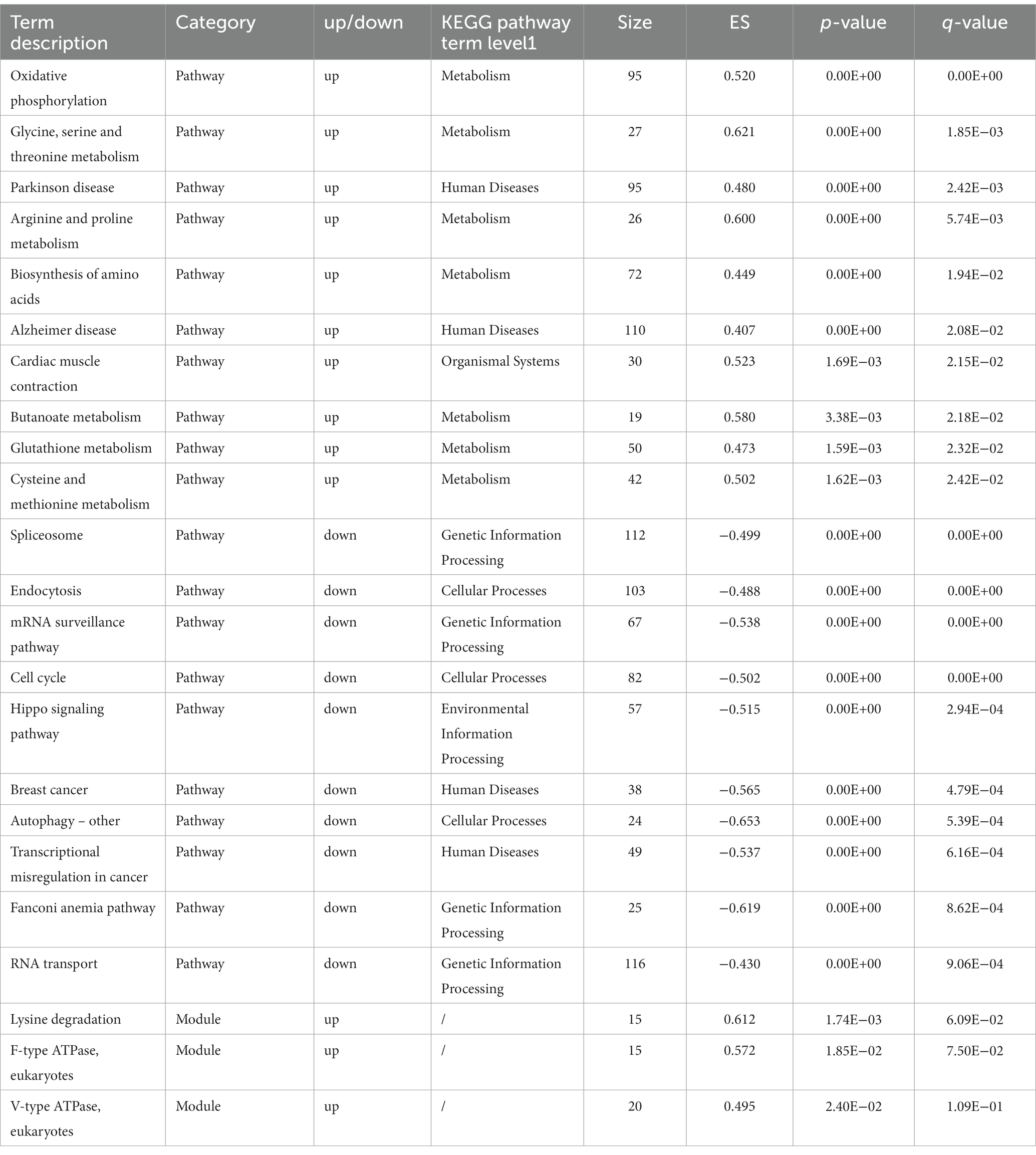
Table 4. Details of pathways and modules with significant changes in overall expression level by GSEA.
Note that the mTOR signaling pathway is also enriched here (Figure 4A), and the results show that the overall expression of the pathway is downregulated.
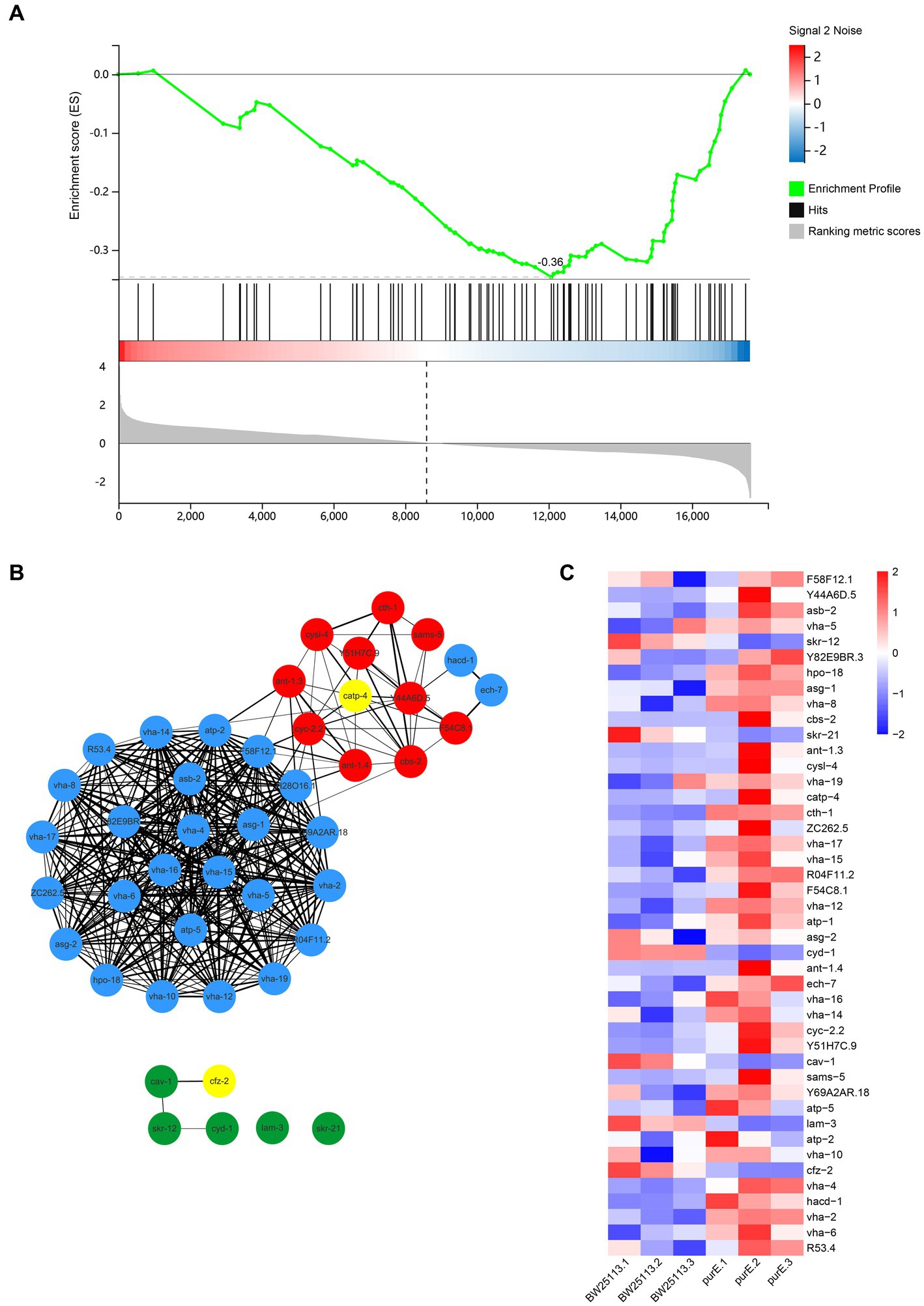
Figure 4. (A) GSEA plot of the mTOR signaling pathway. All genes are arranged according to decreasing differential expression ploidy. Genes are heavily enriched downstream. Enrichment score (ES) = −0.36; size = 78; subsets gene number = 39; normalized enrichment score (NES) = −1.66; nominal value of p = 5.88e−3; FDR Q-value = 0.05. (B) Network interactions of meaningful genes in GSEA. The red circles in the graph are upregulated by a factor of 2 or more DEGs. The green circles are DEGs downregulated by a factor of 2. The yellow circles indicate genes associated with known lifespan-related pathways. The thickness of the line represents the score between genes. The minimum required interaction score is a medium confidence level (0.400). (C) Heat map of meaningful genes in GSEA.
In the GSEA, the core genes that made significant contributions to the enrichment score were selected. Sixteen differentially expressed genes, including cfz-2 and catp-4, were obtained in 3.2.1, and there are 28 core genes in 3 modules. We constructed the reciprocal network of these genes using STRING and Cytoscape 3.9.0 (Figure 4B) and mapped the expression heat map (Figure 4C). The results showed a stronger association between the upregulated genes and the other genes than the downregulated genes. By looking at the heat map, one can see some differential genes between the two groups.
It has been shown that purE knockout E. coli can extend the lifespan of C. elegans by 21% compared with BW25113 wild type (Han et al., 2018). We studied the effects of 25 E. coli mutant strains, including purE, on lifespan and locomotion in Drosophila based on this provided list, and determined the anti-aging properties of PurE E. coli mutant on its host. The gene purE is N (5)-carboxyaminoimidazole ribonucleotide mutase, belonging to E. coli str. K-12 substr. PurE was previously thought to be the catalytic subunit of phosphoribosylaminoimidazole carboxylase, with ATPase subunit PurK (Tiedeman et al., 1989).
Studies have shown that diet is an influential factor in the lifespan of Drosophila. For example, dietary restrictions can reduce mortality in Drosophila at specific ages (Mair et al., 2003; Rizza et al., 2014), and changing the proportion of nutrients consumed affects their lifespan (Mair et al., 2005; Lee et al., 2008; Skorupa et al., 2008; Grandison et al., 2009; Solon-Biet et al., 2020). Moreover, this experiment further confirmed that feeding Drosophila with E. coli purE extended their lifespan. The mean lifespan could be extended by more than 10%, and the climbing ability was correspondingly improved, showing a better anti-aging ability. Notably, compared to live bacteria, UV-inactivated bacteria induce increase survival rate and average lifespan of the host. We reasoned that it may be because the mutant bacteria under UV stimulation activated the stress mechanism and produced some active substances that are beneficial for longevity, just like the discovery of active yeast derivatives (Levin, 1998). Then, to further explore the mechanism of PurE-dependent lifespan extension in flies and nematodes, we performed sequencing analysis of nematodes fed with strain purE and wild type BW25113, respectively.
After performing bioinformatics analysis, 1,001 DEGs with more than twofold significant changes were obtained (FDR Q < 0.05), including 810 upregulated DEGs and 191 downregulated DEGs. After performing an enrichment analysis of DEGs, the following nutritional signaling pathways that have been shown to extend lifespan were identified: insulin/IGF-1 signaling, which maintains glucose homeostasis; mTOR signaling, which senses and transmits amino acid signals; GCN2/ATF4. The following genes and pathways were found to be valuable for the follow-up studies, which may help us identify the anti-aging active substances produced by E. coli and the molecular mechanisms behind their action. Among them, two differential genes are of interest, although the scores were not significant when KEGG was enriched, that may be because the number of differential genes was large so the significance was reduced, but these two genes have strong research significance due to their close relationship with longevity-related pathways, so they are taken out for discussion. In addition, we also used GSEA to screen for additional missing genes and modules. Overall, these bioinformatic analyses focused two longevity related genes for us: cfz-2 and catp-4, which may be potential genes to extend lifespan.
DEG cfz-2 was found to be enriched in the pathway mTOR signaling pathway and its associated longevity regulatory pathway. Cfz-2 expression was downregulated by 2.08 fold in the PurE-treated worms, while the cfz-2 gene is conserved in humans. The C. elegans Frizzled cfz-2 has been reported to be involved in cell migration, neuronal migration, and positive regulation of motor neuron migration and is required for cell migration and interacts with multiple Wnt signaling pathways (Zinovyeva and Forrester, 2005). cfz-2 may non-autonomously direct cell migrations, whereas the Wnt gene CWN-2 may act through cfz-2 for specific cell migrations (Bhanot et al., 1996; Lin et al., 1997; Hsieh et al., 1999; Dann et al., 2001). In addition, we found that cfz-2 is enriched in the mTOR signaling pathway through KEGG analysis, which may be because the Wnt signaling pathway is one of the upstream signaling pathways of mTOR. Thus, a substantial reduction in cfz-2 expression may further affect the host’s lifespan by affecting Wnt and mTOR signaling pathway-related cell migrations.
Another DEG is catp-4, which is enriched in the insulin secretion pathway. Catp-4 expression is upregulated by 2.78 fold in the PurE-treated worms. Catp-4 was predicted to enable P-type sodium: potassium-exchanging transporter activity and be an integral component of the membrane. It is in direct lineage with human ATP12A and ATP4A. The Alliance of Genome Resources mentions that catp-4 regulates potassium-sodium ion homeostasis. This may provide us with an idea for subsequent studies focused on the fact that catp-4 may further regulate ATP levels in vivo by regulating ion homeostasis and ion transport in the cell membrane, thereby affecting the host’s lifespan.
Furthermore, to avoid missing the regulation of non-differential genes in the overall expression level of the pathway, GSEA further obtained critical pathways and modules for the overall expression level changes including the following: oxidative phosphorylation; glycine, serine, and threonine metabolism; spliceosome; endocytosis; autophagy; upregulation of the overall expression of lysine degradation module and F-type/V-type ATPase module. The changes in these pathways and modules may significantly contribute to lifespan regulation. For example, the V-type ATPase module contains all VHA genes. The putative V-type proton ATPase is involved in the positive regulation of programmed cell death and developmental apoptosis. The vacuolar (H+) ATPase is involved in the positive regulation of oocyte development and protein-targeted membranes. These are important candidate genes for further studies on anti-aging mechanisms.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/geo/, GSE214285.
Ethical review and approval was not required for the study on animals in accordance with the local legislation and institutional requirements.
FZ, LW, and JJ have contributed equally to this work and responsible for the conceptualization, methodology, visualization, writing, reviewing, editing, and funding acquisition. HS, ZF, YP, and HW were responsible for the methodology, software, validation, and investigation. YD, YH, and YZ were responsible for the formal analysis. XD and ZZ were responsible for the ideas, writing, reviewing, editing, and project administration. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (81701390), the Natural Science Foundation of Jiangsu Province (BK20170250), the Xuzhou Science and Technology Innovation Project (KC19057), the Jiangsu Postgraduate Innovation Program (730221059), and the Jiangsu University Students Innovation and Entrepreneurship Training Program (202010313026Z).
The experiments in this article were completed in the Public Experimental Research Center of Xuzhou Medical University. The authors thank the teachers for their support and help during the experiments.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1138979/full#supplementary-material
Barcena, C., Valdes-Mas, R., Mayoral, P., Garabaya, C., Durand, S., Rodriguez, F., et al. (2019). Healthspan and lifespan extension by fecal microbiota transplantation into progeroid mice. Nat. Med. 25, 1234–1242. doi: 10.1038/s41591-019-0504-5
Bhanot, P., Brink, M., Samos, C. H., Hsieh, J. C., Wang, Y., Macke, J. P., et al. (1996). A new member of the frizzled family from Drosophila functions as a wingless receptor. Nature 382, 225–230. doi: 10.1038/382225a0
Charalambous, A., Grivogiannis, E., Dieronitou, I., Michael, C., Rahme, L., and Apidianakis, Y. (2022). Proteobacteria and Firmicutes secreted factors exert distinct effects on Pseudomonas aeruginosa infection under Normoxia or mild hypoxia. Meta 12:449. doi: 10.3390/metabo12050449
Dann, C. E., Hsieh, J. C., Rattner, A., Sharma, D., Nathans, J., and Leahy, D. J. (2001). Insights into Wnt binding and signalling from the structures of two frizzled cysteine-rich domains. Nature 412, 86–90. doi: 10.1038/35083601
Gao, Y. D., Zhao, Y., and Huang, J. (2014). Metabolic modeling of common Escherichia coli strains in human gut microbiome. Biomed. Res. Int. 2014:694967. doi: 10.1155/2014/694967
Gargano, J. W., Martin, I., Bhandari, P., and Grotewiel, M. S. (2005). Rapid iterative negative geotaxis (RING): a new method for assessing age-related locomotor decline in Drosophila. Exp. Gerontol. 40, 386–395. doi: 10.1016/j.exger.2005.02.005
Grandison, R. C., Piper, M. D., and Partridge, L. (2009). Amino-acid imbalance explains extension of lifespan by dietary restriction in Drosophila. Nature 462, 1061–1064. doi: 10.1038/nature08619
Grotewiel, M. S., Martin, I., Bhandari, P., and Cook-Wiens, E. (2005). Functional senescence in Drosophila melanogaster. Ageing Res. Rev. 4, 372–397. doi: 10.1016/j.arr.2005.04.001
Guarente, L., and Kenyon, C. (2000). Genetic pathways that regulate ageing in model organisms. Nature 408, 255–262. doi: 10.1038/35041700
Guo, C. J., Allen, B. M., Hiam, K. J., Dodd, D., Van Treuren, W., Higginbottom, S., et al. (2019). Depletion of microbiome-derived molecules in the host using Clostridium genetics. Science 366:1282. doi: 10.1126/science.aav1282
Hahn, D. A., and Denlinger, D. L. (2011). Energetics of insect diapause. Annu. Rev. Entomol. 56, 103–121. doi: 10.1146/annurev-ento-112408-085436
Han, B., Sivaramakrishnan, P., Lin, C.-C. J., Neve, I. A. A., He, J., Tay, L. W. R., et al. (2018). Microbial genetic composition Tunes host longevity. Cells 173:1058. doi: 10.1016/j.cell.2018.04.026
Hsieh, J. C., Rattner, A., Smallwood, P. M., and Nathans, J. (1999). Biochemical characterization of Wnt-frizzled interactions using a soluble, biologically active vertebrate Wnt protein. Proc. Natl. Acad. Sci. U. S. A. 96, 3546–3551. doi: 10.1073/pnas.96.7.3546
Iliadi, K. G., Knight, D., and Boulianne, G. L. (2012). Healthy aging – insights from Drosophila. Front. Physiol. 3:106. doi: 10.3389/fphys.2012.00106
Kaletta, T., and Hengartner, M. O. (2006). Finding function in novel targets: C. elegans as a model organism. Nat. Rev. Drug Discov. 5, 387–399. doi: 10.1038/nrd2031
Lee, K. P., Simpson, S. J., Clissold, F. J., Brooks, R., Ballard, J. W. O., Taylor, P. W., et al. (2008). Lifespan and reproduction in Drosophila: new insights from nutritional geometry. Proc. Natl. Acad. Sci. U. S. A. 105, 2498–2503. doi: 10.1073/pnas.0710787105
Lin, K., Wang, S., Julius, M. A., Kitajewski, J., Moos, M., and Luyten, F. P. (1997). The cysteine-rich frizzled domain of Frzb-1 is required and sufficient for modulation of Wnt signaling. Proc. Natl. Acad. Sci. U. S. A. 94, 11196–11200. doi: 10.1073/pnas.94.21.11196
Mair, W., Goymer, P., Pletcher, S. D., and Partridge, L. (2003). Demography of dietary restriction and death in Drosophila. Science 301, 1731–1733. doi: 10.1126/science.1086016
Mair, W., Piper, M. D. W., and Partridge, L. (2005). Calories do not explain extension of life span by dietary restriction in Drosophila. PLoS Biol. 3:e223. doi: 10.1371/journal.pbio.0030223
Partridge, L., Alic, N., Bjedov, I., and Piper, M. D. W. (2011). Ageing in Drosophila: the role of the insulin/Igf and TOR signalling network. Exp. Gerontol. 46, 376–381. doi: 10.1016/j.exger.2010.09.003
Piper, M. D. W., and Partridge, L. (2018). Drosophila as a model for ageing. Biochim. Biophys. Acta Mol. basis Dis. 1864, 2707–2717. doi: 10.1016/j.bbadis.2017.09.016
Pletcher, S. D., Macdonald, S. J., Marguerie, R., Certa, U., Stearns, C. S., Goldstein, D. B., et al. (2002). Genome-wide transcript profiles in aging and calorically restricted Drosophila melanogaster. Curr. Biol. 12, 712–723. doi: 10.1016/S0960-9822(02)00808-4
Rizza, W., Veronese, N., and Fontana, L. (2014). What are the roles of calorie restriction and diet quality in promoting healthy longevity? Ageing Res. Rev. 13, 38–45. doi: 10.1016/j.arr.2013.11.002
Skorupa, D. A., Dervisefendic, A., Zwiene, J., and Pletcher, S. D. (2008). Dietary composition specifies consumption, obesity, and lifespan in Drosophila melanogaster. Aging Cell 7, 478–490. doi: 10.1111/j.1474-9726.2008.00400.x
Solon-Biet, S. M., McMahon, A. C., Ballard, J. W. O., Ruohonen, K., Wu, L. E., Cogger, V. C., et al. (2020). The ratio of macronutrients, not caloric intake, dictates cardiometabolic health, aging, and longevity in ad libitum-fed mice. Cell Metab. 31:654. doi: 10.1016/j.cmet.2020.01.010
Tamura, T., Chiang, A.-S., Ito, N., Liu, H.-P., Horiuchi, J., Tully, T., et al. (2003). Aging specifically impairs amnesiac – dependent memory in Drosophila. Neuron 40, 1003–1011. doi: 10.1016/S0896-6273(03)00732-3
Tiedeman, A. A., Keyhani, J., Kamholz, J., Daum, H. A., Gots, J. S., and Smith, J. M. (1989). Nucleotide sequence analysis of the purEK operon encoding 5′-phosphoribosyl-5-aminoimidazole carboxylase of Escherichia coli K-12. J. Bacteriol. 171, 205–212. doi: 10.1128/jb.171.1.205-212.1989
Zhang, J., Li, X., Olmedo, M., Holdorf, A. D., Shang, Y., Artal-Sanz, M., et al. (2019). A delicate balance between bacterial Iron and reactive oxygen species supports Optimal C. elegans development. Cell Host Microbe 26, 400–411.e3. doi: 10.1016/j.chom.2019.07.010
Keywords: aging, Escherichia coli , Drosophila melanogaster , Caenorhabditis elegans , lifespan extension
Citation: Zhang F, Wang L, Jin J, Pang Y, Shi H, Fang Z, Wang H, Du Y, Hu Y, Zhang Y, Ding X and Zhu Z (2023) Insights into the genetic influences of the microbiota on the life span of a host. Front. Microbiol. 14:1138979. doi: 10.3389/fmicb.2023.1138979
Received: 06 January 2023; Accepted: 20 July 2023;
Published: 03 August 2023.
Edited by:
Elisa Michelini, University of Bologna, ItalyReviewed by:
Yiorgos Apidianakis, University of Cyprus, CyprusCopyright © 2023 Zhang, Wang, Jin, Pang, Shi, Fang, Wang, Du, Hu, Zhang, Ding and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zuobin Zhu, emh1enVvYmluQHh6aG11LmVkdS5jbg==; Xiaoyue Ding, ZGluZ3hpYW95dWVAeHpobXUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.