- 1College of Chemistry Biology and Environment, Yuxi Normal University, Yuxi, China
- 2Institute of Biology and Environmental Engineering, Yuxi Normal University, Yuxi, China
- 3College of Plant Health and Medicine, Qingdao Agricultural University, Qingdao, China
- 4Yunnan Academy of Tobacco Agricultural Sciences, Kunming, China
A Gram-negative, aerobic, rod-shaped, motile bacterium with multi-flagella, strain RST, was isolated from bacterial wilt of tobacco in Yuxi city of Yunnan province, China. The strain contains the major fatty acids of C16:0, summed feature 3 (C16:1ω7c and/or C16:1ω6c), and summed feature 8 (C18:1ω7c and/or C18:1ω6c). The polar lipid profile of strain RST consists of diphosphatidylglycerol, phosphatidylglycerol, phosphatidylethanolamine, and unidentified aminophospholipid. Strain RST contains ubiquinones Q-7 and Q-8. 16S rRNA gene sequence (1,407 bp) analysis showed that strain RST is closely related to members of the genus Ralstonia and shares the highest sequence identities with R. pseudosolanacearum LMG 9673T (99.50%), R. syzygii subsp. indonesiensis LMG 27703T (99.50%), R. solanacearum LMG 2299T (99.28%), and R. syzygii subsp. celebesensis LMG 27706T (99.21%). The 16S rRNA gene sequence identities between strain RST and other members of the genus Ralstonia were below 98.00%. Genome sequencing yielded a genome size of 5.61 Mbp and a G + C content of 67.1 mol%. The genomic comparison showed average nucleotide identity (ANIb) values between strain RST and R. pseudosolanacearum LMG 9673T, R. solanacearum LMG 2299T, and R. syzygii subsp. indonesiensis UQRS 627T of 95.23, 89.43, and 91.41%, respectively, and the corresponding digital DNA–DNA hybridization (dDDH) values (yielded by formula 2) were 66.20, 44.80, and 47.50%, respectively. In addition, strains belonging to R. solanacearum phylotype I shared both ANIb and dDDH with strain RST above the species cut-off values of 96 and 70%, respectively. The ANIb and dDDH values between the genome sequences from 12 strains of R. solanacearum phylotype III (Current R. pseudosolanacearum) and those of strain RST were below the species cut-off values. Based on these data, we concluded that strains of phylotype I, including RST, represent a novel species of the genus Ralstonia, for which the name Ralstonia nicotianae sp. nov. is proposed. The type strain of Ralstonia nicotianae sp. nov. is RST (=GDMCC 1.3533T = JCM 35814T).
1. Introduction
The genus Ralstonia, a member of the family Burkholderiaceae, was named to accommodate Alcaligenes eutrophus, Burkholderia pickettii, and B. solanacearum (Ralston et al., 1973; Yabuuchi et al., 1995). Later, several novel species were described, and further studies suggested that the Ralstonia strains could be separated into two distinct groups (De Baere et al., 2001). Therefore, the genus Wautersia was created to accommodate the R. eutropha lineage (Vaneechoutte et al., 2004), which has been transferred to the genus Cupriavidus (Vandamme and Coenye, 2004). According to the List of Prokaryotic names with Standing in Nomenclature1 (Parte et al., 2020), the genus Ralstonia currently comprises seven species with validly published names, including R. mannitolilytica, R. insidiosa, R. pickettii, R. pseudosolanacearum, R. solanacearum, R. syzygii, and R. wenshanensis (Safni et al., 2014; Meier-Kolthoff et al., 2022; Lu et al., 2022a). Recently, we proposed three novel species of the genus Ralstonia, including R. chuxiongensis, R. mojiangensis, and R. soli (Lu et al., 2023).
Bacteria from the genus Ralstonia have a broad range of niches, including soil, water, plant tissues, and human specimens. The type strains of R. chuxiongensis, R. mojiangensis, R. soli, and R. wenshanensis have been isolated from tobacco planting soils (Lu et al., 2022a, 2023). The type strains of R. mannitolilytica, R. insidiosa, and R. pickettii were described as pathogens in various human infections (Ralston et al., 1973; De Baere et al., 2001; Coenye et al., 2003). R. pseudosolanacearum, R. solanacearum, and R. syzygii are critical economic pathogens of various plants (Yabuuchi et al., 1995; De Baere et al., 2001; Coenye et al., 2003; Remenant et al., 2010, 2011; Safni et al., 2014; Zulperi et al., 2014; Lu et al., 2021).
The pathogen responsible for plant bacterial wilt was first named Bacillus solanacearum (Smith, 1896) and then successively placed into the genera Bacterium, Phytomonas, Xanthomonas, Pseudomonas, and Burkholderia, before being transferred into the current genus, Ralstonia (Yabuuchi et al., 1995; Paudel et al., 2020). Although classified as R. solanacearum, distinct strains from diverse plants from various regions showed DNA–DNA hybridization values below the species cut-off value of 70%, thus R. solanacearum is considered a species complex (Genin and Denny, 2012). Based on genetic analysis of the 16S-23S internal transcribed spacer (ITS) region, endoglucanase (egl), and hrpB gene sequences, the R. solanacearum species complex was divided into four phylogenetically distinct groups, named phylotypes, which roughly reflect the strains’ geographical origins: phylotype I strains are often sampled from Asia; phylotype II strains are isolated from the Americas, which are further subdivided into three subgroups (IIA, IIB, and IIC) (Poussier et al., 2000; Sharma et al., 2022); phylotype III are obtained from Africa; and phylotype IV are collected from south-east Asia (Prior and Fegan, 2005; Villa et al., 2005). Furthermore, Safni et al. (2014) classified the phylotype II strains as the species R. solanacearum, phylotypes I and III strains as the species R. pseudosolanacearum, and the phylotype IV strains as R. syzygii. The phylotype IV strains were initially proposed as Pseudomonas syzygii by Yabuuchi et al. (1995), transferred into the genus Ralstonia in 2004, and further sub-classified into three subspecies, R. syzygii subsp. syzygii, R. syzygii subsp. indonesiensis, and R. syzygii subsp. celebesensis (Safni et al., 2014). Recently, based on 16S rRNA gene analysis and genome sequence comparisons, Fluit et al. (2021) found that several well-known strains of phylotype I, including GMI1000, OE1-1, EP1, and CQPS-1, might represent an unnamed species within the genus Ralstonia.
In the present study, a novel pathogenic bacterium, RST, belonging to Ralstonia was isolated from tobacco bacterial wilt. It was identified by comparing its physiological and chemical characteristics, its 16S rRNA gene, and its whole genome with those of closely related type strains. Consequently, the name Ralstonia nicotianae sp. nov. is proposed for phylotype I strains and the type strain is RST (=GDMCC 1.3533T = JCM 35814T).
2. Materials and methods
2.1. Sampling site and bacterial culture
Strain RST was isolated from bacterial wilt of flue-cured tobacco sampled in 2018 in Yuxi, Yunnan, China. The approximate geographical coordinates were 102°30′2.22″E, 24°14′22.5″N. The site is 1,630 m above sea level and has a central semi-humid subtropical cool winter plateau monsoon climate, with an average annual temperature of 15–16°C. We used the symptomatic leaves for pathogen isolation. First, the petiole part was surface-disinfected with 75% (v/v) ethanol for 5 min. Second, we removed tissues, including the epidermis layers, palisade, and spongy tissues, other than the leaf vein vascular bundle. Third, after removing the 2–3 cm part at the base, the remaining leaf vein vascular bundle was cut into several pieces and soaked in a sterile petri dish containing 10 ~ 20 mL of distilled water for 5–10 min. Finally, 100 μL of bacterial suspension with 5–6 times 10-fold dilution were spread onto a Tripheny tetrazolium chloride (TZC) agar plate. The dish was incubated under aerobic conditions at 28°C. After 36–48 h, typical circular colonies had formed, with broad white edges, strong fluidity, and a pink to light thin red liquid in the middle. The isolate was named RST and streaked onto TZC agar at 28°C and purified three times (Figure 1). Strain RST was stored in 20% (v/v) glycerol at −80°C for long-term preservation. It has been deposited at the Guangdong Microbial Culture Collection Center (GDMCC 1.3533T) and the Japan Collection of Microorganisms (JCM 35814T). The species type strains, including R. solanacearum LMG 2299T, R. pseudosolanacearum LMG 9673T, R. syzygii subsp. indonesiensis LMG 27703T, R. syzygii subsp. syzygii LMG 10661T, and R. syzygii subsp. celebesensis LMG 27706T, were collected from the BCCM/LMG Bacteria Collection (Table 1).
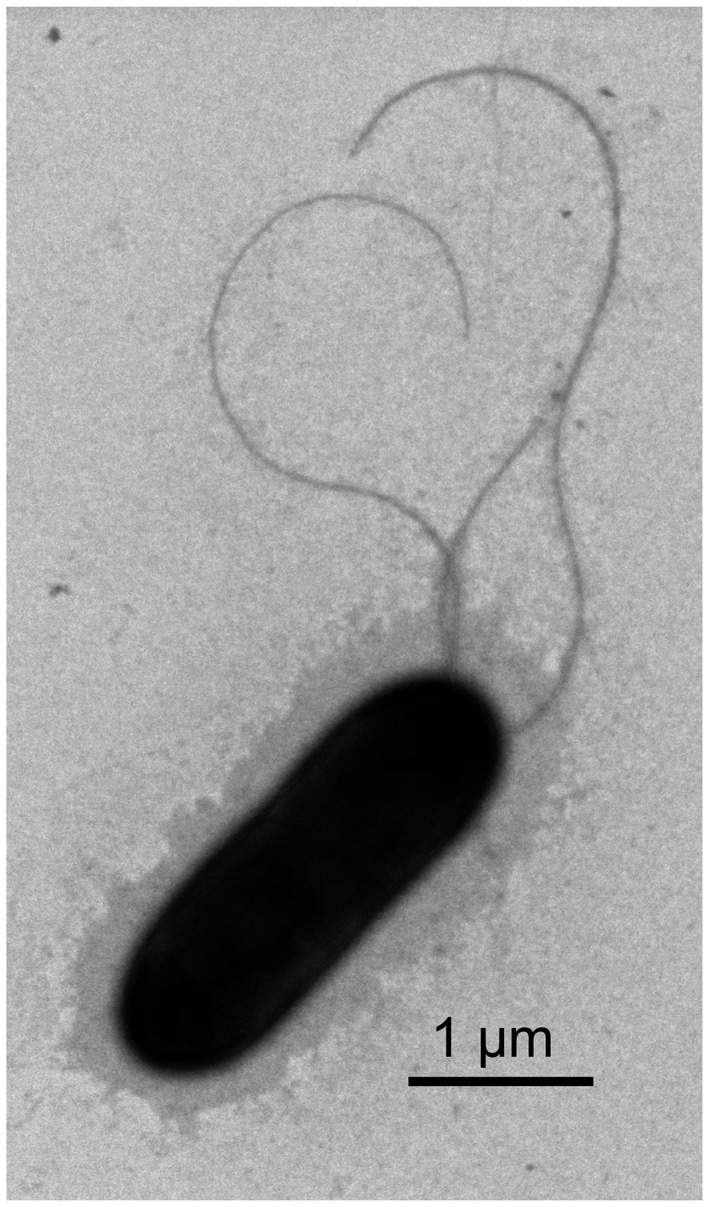
Figure 1. Transmission electron micrograph showing the morphology of strain RST. The bar represents 1 μm.
2.2. 16S rRNA sequencing, and phylogenetic and genomic comparisons
The genomic DNA of strain RST was extracted from a 2.0 mL bacterial culture using a bacterial DNA isolation kit (catalog number D3107; GBCBIO Technologies Inc., Guangzhou, China). The universal primer pair 27F and 1492R was used to amplify the 16S rRNA gene sequence (Clayton et al., 1995). The amplicon was ligated into the pMD19-T vector (Takara, Dalian, China) and sequenced following the Sanger method at Sangon Biotech Inc. (Shanghai, China). The sequences were assembled using Contig Express (Invitrogen, Waltham, MA, United States). The sequence similarity of the 16S rRNA gene from RST was calculated using the EzBiocloud database2 (Yoon et al., 2017). Closely related sequences of type species within the genus Ralstonia were collected from the GenBank database3 (Sayers et al., 2021) and aligned using MUSCLE mode in the Unipro UGENE version 33.0 (Unipro, Novosibirsk, Russia) (Table 1). Trimmed sequences were used for sequence identity calculations employing the Clustal Omega multiple alignment package in the EMBL-EBI database (Sievers and Higgins, 2021). Phylogenetic trees were constructed using the maximum-likelihood (ML), neighbor-joining (NJ), and minimum-evolutions (ME) algorithms, with bootstrap values of 1,000 replications in MEGA version 11 software (Kumar et al., 2016).
For genome sequencing, the glycerol suspension was streaked onto an agar plate containing Casamino Acid-peptone-glucose (CPG) medium (Kelman, 1954). A single colony was inoculated into CPG broth and grown for 24 h at 28°C at 180 r/min, followed by centrifugation at 10,000 × g for 10 min and washing with distilled water twice (Lu et al., 2021). The total DNA of strain RST was isolated using an EZNA bacterial DNA kit (Omega Bio-tek, Winooski, VT, United States). The genome of strain RST was sequenced using the PacBio RS II platform (Menlo Park, CA, United States) and the Illumina HiSeq 4,000 platform (San Diego, CA, United States) at the Beijing Genomics Institute (BGI, Shenzhen, China). Four SMRT cell Zero-Mode Waveguide arrays for sequencing were used by the PacBio platform to generate the subreads set. PacBio subreads of length < 1 kb were removed (Utturkar et al., 2014). The program Pbdagcon was used for self-correction (Chin et al., 2013). The draft genomic unitigs, which are uncontested groups of fragments, were assembled using the Celera Assembler against a high quality corrected circular consensus sequence subread set (Berlin et al., 2015). To improve the accuracy of the genome sequences, GATK (Poplin et al., 2018) and SOAP tool packages (SOAP2, SOAPsnp, SOAPindel) (Li et al., 2008, 2009) were used to make single-base corrections. Finally, to trace the presence of any plasmid, the filtered Illumina reads were mapped using SOAP to the bacterial plasmid database.4
Genetic comparisons between strain RST and type strains of the genus Ralstonia were tested based on average nucleotide identity (ANI) and digital DNA–DNA hybridization (dDDH) values using the webserver JSpecies WS version 3.9.6 (Richter et al., 2015) and the Genome-to-Genome Distance Calculator version 3.0 (Meier-Kolthoff et al., 2022), respectively. We conducted the phylogenomic analysis based on genome sequences of strain RST and thirteen closely related strains in the genus Ralstonia using the Type (Strain) Genome Server (Meier-Kolthoff et al., 2022). Strain Cupriavidus necator N-1T served as an outgroup. To confirm the systematic classification of all Ralstonia strains in the GenBank database that were closely related to RST, ANI and dDDH values were tested between genome sequences of strain RST and R. pseudosolanacearum LMG 9673T and those from the strains mentioned above. For a quick calculation, ANI values were primarily tested using the FastANI calculator in the Genome Taxonomy Database (GTDB) (Jain et al., 2018; Parks et al., 2021). Strains with FastANI values between 93 and 97% were selected further to calculate BLAST-based ANI (ANIb), MUMmer-based ANI (ANIm), and dDDH values as described above.
For gene content analysis, the whole genome sequence of strain RST and those of closely related species in the genus Ralstonia were obtained from the GenBank database (Sayers et al., 2021), genes were predicted using GeneMarkS version 4.28 (Besemer et al., 2001) and the genome comparisons were performed using OrthoVenn2, with a threshold e-value of 1e-5 and inflation of 1.5 (Xu et al., 2019).
The type three effectors (T3Es) of strain RST were scanned using the Ralsto T3E database (Peeters et al., 2013; Sabbagh et al., 2019). To analyze the core effector repertoires of plant pathogenic Ralstonia species, we compared the T3Es in Ralstonia strains with the complete genome sequences from four phylotypes, including five strains of phylotype I, four strains of phylotype II, five strains of phylotype III, and three strains of phylotype IV. In addition, the secondary metabolites and antibiotic gene clusters were analyzed in silico using the online web server antiSMASH bacterial version 6.1.1 (Blin et al., 2021).
2.3. Phenotypic and chemotaxonomic analysis
According to the 16S rRNA gene analysis and genomic comparisons, we used eight Ralstonia strains, including RST, GMI1000, CQPS-1, R. pseudosolanacearum LMG 9673T, R. solanacearum LMG 2299T, R. syzygii subsp. celebesensis LMG 27706T, R. syzygii subsp. indonesiensis LMG 27703T, and R. syzygii subsp. syzygii LMG 10661T, to compare the phenotypic properties and chemotaxonomic features.
The protocol used to analyze the strains’ physiological and chemical characteristics was as described previously (Lu et al., 2022a). Culture media, including CPG agar (Kelman, 1954), Kelman’s TZC (García et al., 2019), Luria-Bertani (LB) agar (LA), modified selective medium South Africa (mSMSA) agar (Elphinstone et al., 1996), nutrient agar (NA), potato dextrose agar (PDA), and trypticase soy agar (TSA, BD Difco, Franklin Lakes, NJ, United States), were used to analyze the growth of the strains when incubated at 28°C for 5 days. Cells grown on TSA at 28°C for 48 h were subjected to morphological observation using transmission electron microscopy (HT7700; HITACHI, Tokyo, Japan). The motilities of the tested strains were detected on modified semi-solid motility media (SMM) containing 0.35% agar (Kelman and Hruschka, 1973). A Gram-staining kit (D008-1-1; Nanjing Jiancheng Bioengineering Institute, Nanjing, China) was used stain the strains, which was detected under a light microscope (Axio imager.Z2; Zeiss, Oberkochen, Germany). The oxygen requirement assay was conducted by streaking the test strain on TSA slope covered with mineral oil and incubating at 28°C for 5 days (Lu et al., 2022b).
To analyze the chemical sensitivity, enzymatic activity, carbon source, and substrate utilization of the tested strains, the Biolog GEN III Microplate system, API 20NE, and API ZYM kits (bioMérieux, Marcy-l’Étoile, France), respectively, were used according to the manufacturer’s instructions. Oxidase activities of the strains were tested using 1% (w/v) tetramethyl-p-phenylenediamine, with Escherichia coli K-12 strain MG 1655 as the negative control and Pseudomonas lijiangensis LJ2T as the positive control. The production of bubbles detected catalase activity after adding a drop of 3% (v/v) H2O2 (Lu et al., 2022a). Cells from the tested strains grown on TSA at 28°C for 48 h were harvested to analyze their fatty acid profile (Sasser, 1990). Polar lipids and isoprenoid quinones of three proposed type strains were extracted and analyzed as described previously (Minnikin and Abdolrahimzadeh, 1971; Collins and Jones, 1981).
3. Results and discussion
3.1. 16S rRNA phylogeny
16S rRNA gene sequence analysis showed that strain RST was closely related to the type species in the genus Ralstonia and shared the highest 16S rRNA sequence identity (99.50%) with those of R. pseudosolanacearum LMG 9673T and R. syzygii subsp. indonesiensis LMG 27703T, followed by R. solanacearum LMG 2299T (99.28%) and R. syzygii subsp. celebesensis LMG 27706T (99.21%). The 16S rRNA gene sequence identities between strain RST and the other type strains of the genus Ralstonia were all below 98.00% (Table 2). The ML phylogenetic tree based on 16S rRNA gene sequences revealed that strain RST formed a coherent cluster with type species of R. pseudosolanacearum LMG 9673T, R. solanacearum LMG 2299T, and three subspecies, including R. syzygii subsp. indonesiensis LMG 27703T, R. syzygii subsp. celebesensis LMG 27706T, and R. syzygii subsp. syzygii LMG 10661T (Figure 2). The NJ and MP trees also indicated that strain RST was consistently separated from its closely related type species of R. pseudosolanacearum, R. solanacearum, and R. syzygii (Supplementary Figures 1, 2).
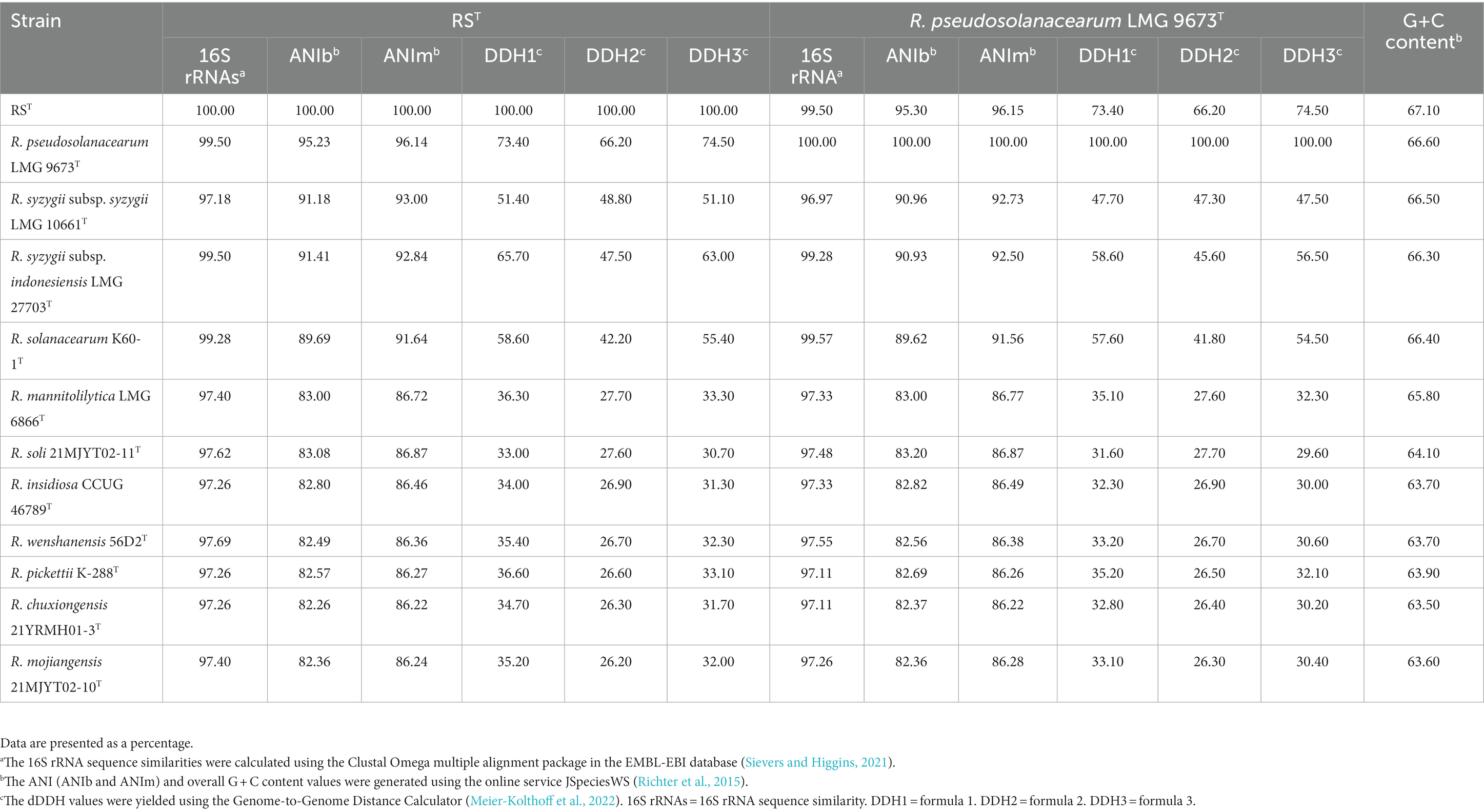
Table 2. Genetic comparisons between strain RST and R. pseudosolanacearum LMG 9673T, and type species in the genus Ralstonia.
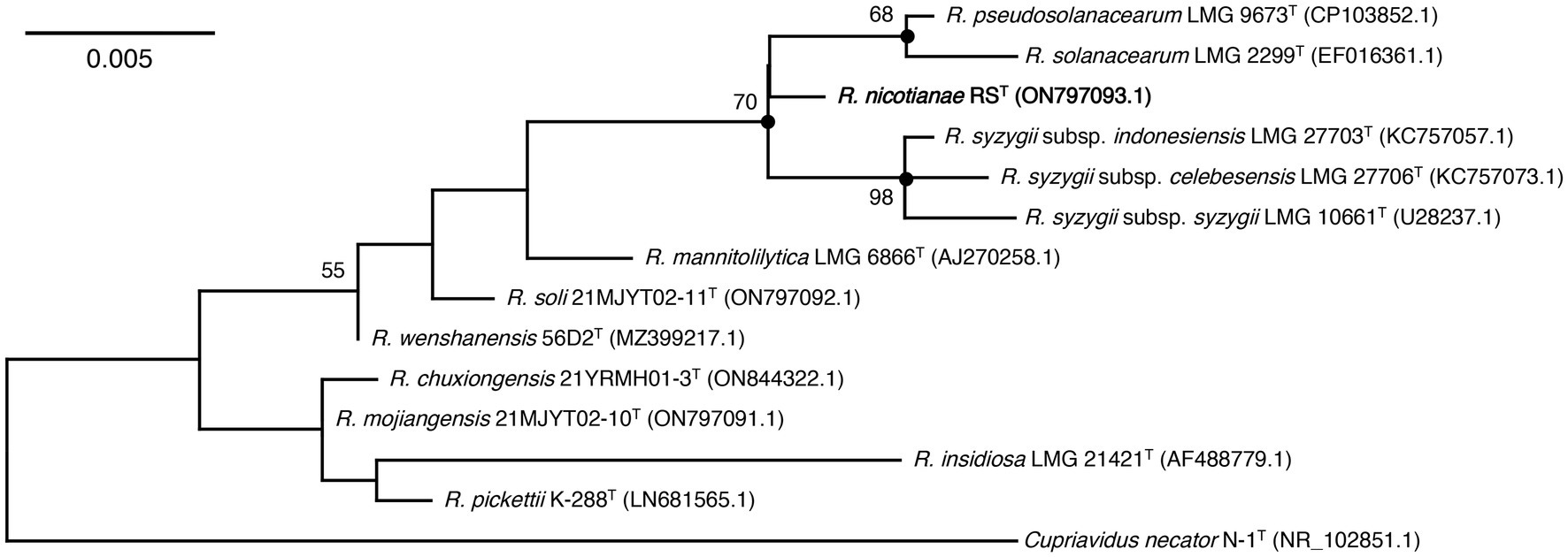
Figure 2. Maximum-likelihood phylogenetic tree based on 16S rRNA sequences of strain RST and type species in the genus Ralstonia. The tree was generated using MEGA 11.0. Bootstrap values (≥50) are shown at branching points as percentages of 1,000 replicates. T indicates the type strain. Closed circles indicate that the corresponding nodes were also recovered in the trees constructed with neighbor-joining and maximum-parsimony. Strain RST is indicated in bold. Cupriavidus necator N-1T is used as an outgroup. The GenBank accession numbers are shown in parentheses. Bar, 0.005 substitutions per nucleotide position.
3.2. Genome features
Genome sequencing yielded 3,971,364 reads and 492 Mb of clean data, with a genome coverage depth of 87×. The overall G + C content of strain RST was 67.10 mol%, which is in the range of 63.0–67.3 mol% (avg. 65.86 mol%) for Ralstonia species (Lu et al., 2022a). The assembled genome of strain RST comprised 5,613,239 bp in two replicons: a circular chromosome of 3,618,724 bp and a megaplasmid of 1,994,515 bp. Strain RST contained 5,105 predicted genes; 58 transfer RNA (tRNA) genes; four sets of 5S rRNA, 16S rRNA, and 23S rRNA genes; and 23 small RNA (sRNA) genes (Figure 3).
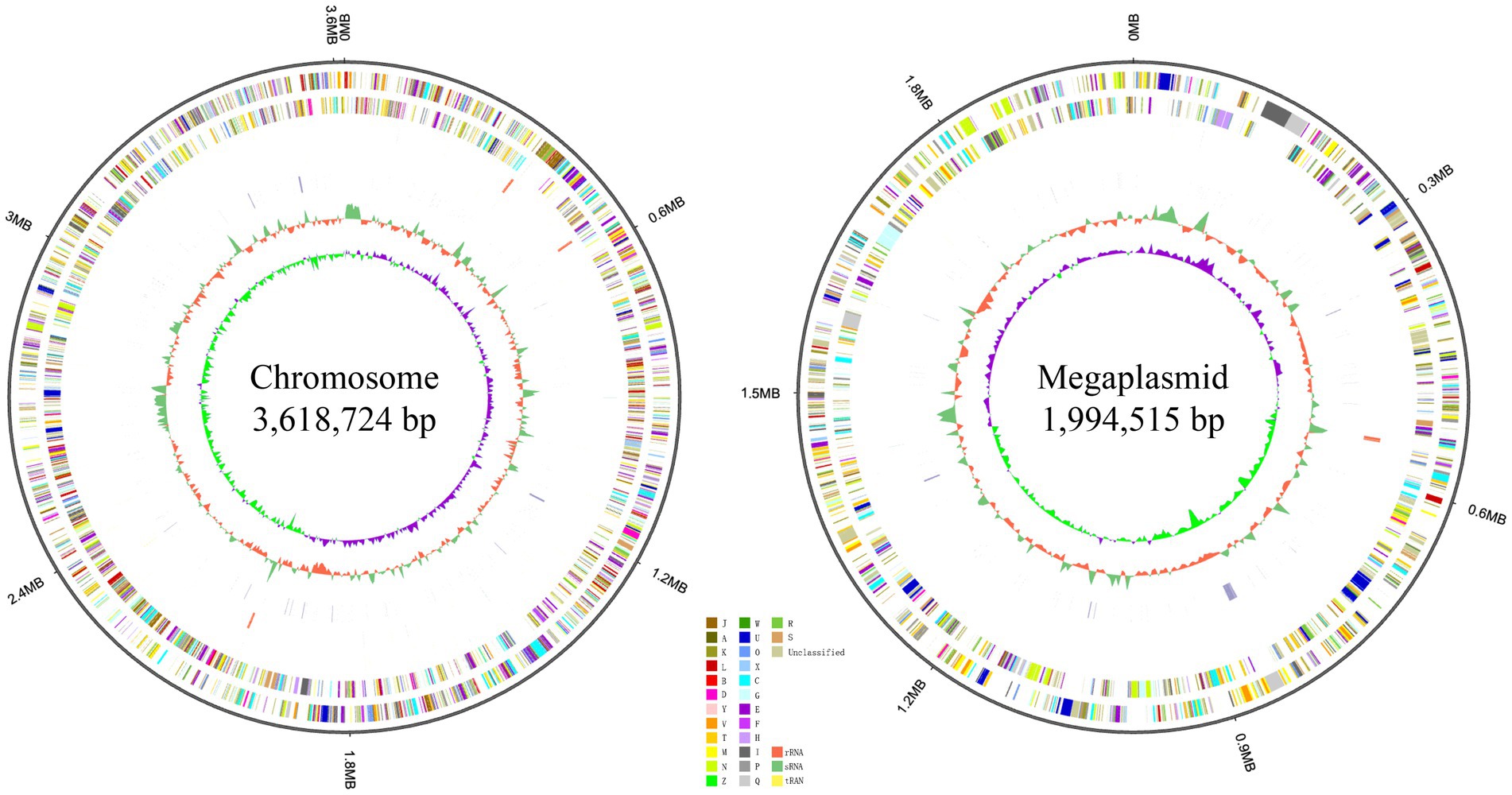
Figure 3. Localization of the principal genomic elements predicted in strain RST. For each figure, from outer to inner: ring 1–genome size; ring 2–forward strand gene, colored according to the cluster of orthologous groups (COG) classification; ring 3–reverse strand gene, colored according to the cluster of orthologous groups (COG) classification; ring 4–forward strand ncRNA; ring 5–reverse strand ncRNA; ring 6–repeat elements; ring 7–G + C content; ring 8–GC-SKEW. Chromosome and megaplasmid of strain RST are presented on the left and right, respectively.
Genome comparisons between strain RST and type strains of the genus Ralstonia showed that R. pseudosolanacearum LMG 9673T had the highest ANIb of 95.23% and ANIm of 96.14% in comparison with strain RST, which was located in the species’ border cut-off value of 95–96%, followed by R. syzygii subsp. syzygii LMG 10661T (ANIb = 91.18% and ANIm = 93.00%), R. syzygii subsp. indonesiensis LMG 27703T (ANIb = 91.41% and ANIm = 92.84%), and R. solanacearum K60-1T (ANIb = 89.69% and ANIm = 91.64%) (Table 2). The ANI values between the seven other type strains of the genus Ralstonia and strain RST were below 90.00%. The dDDH values yielded by formula two were used for analysis (Meier-Kolthoff et al., 2022). Data revealed that R. pseudosolanacearum LMG 9673T shared the highest dDDH value with strains RST at 66.20%, which was below the 70% threshold value recommended for species description. The other 10 type strains had dDDH values with strain RST below 66.00% (Table 2).
The phylogenomic analysis showed that 14 Ralstonia strains were divided into two main phylogenomic branches. Strain RST was positioned in the branch that contained three species, including R. pseudosolanacearum, R. syzygii, and R. solanacearum, which are the main causal agents of bacterial wilts in a wide range of plants. Strain RST formed a coherent cluster with type species of R. pseudosolanacearum LMG 9673T at a supported bootstrap value of 100% (Figure 4). The results allowed us to conclude that strain RST represents a novel species in the genus Ralstonia. Interestingly, compared with strain R. solanacearum K60-1T, R. syzygii strains were genetically closer to strain RST and R. pseudosolanacearum LMG 9673T.
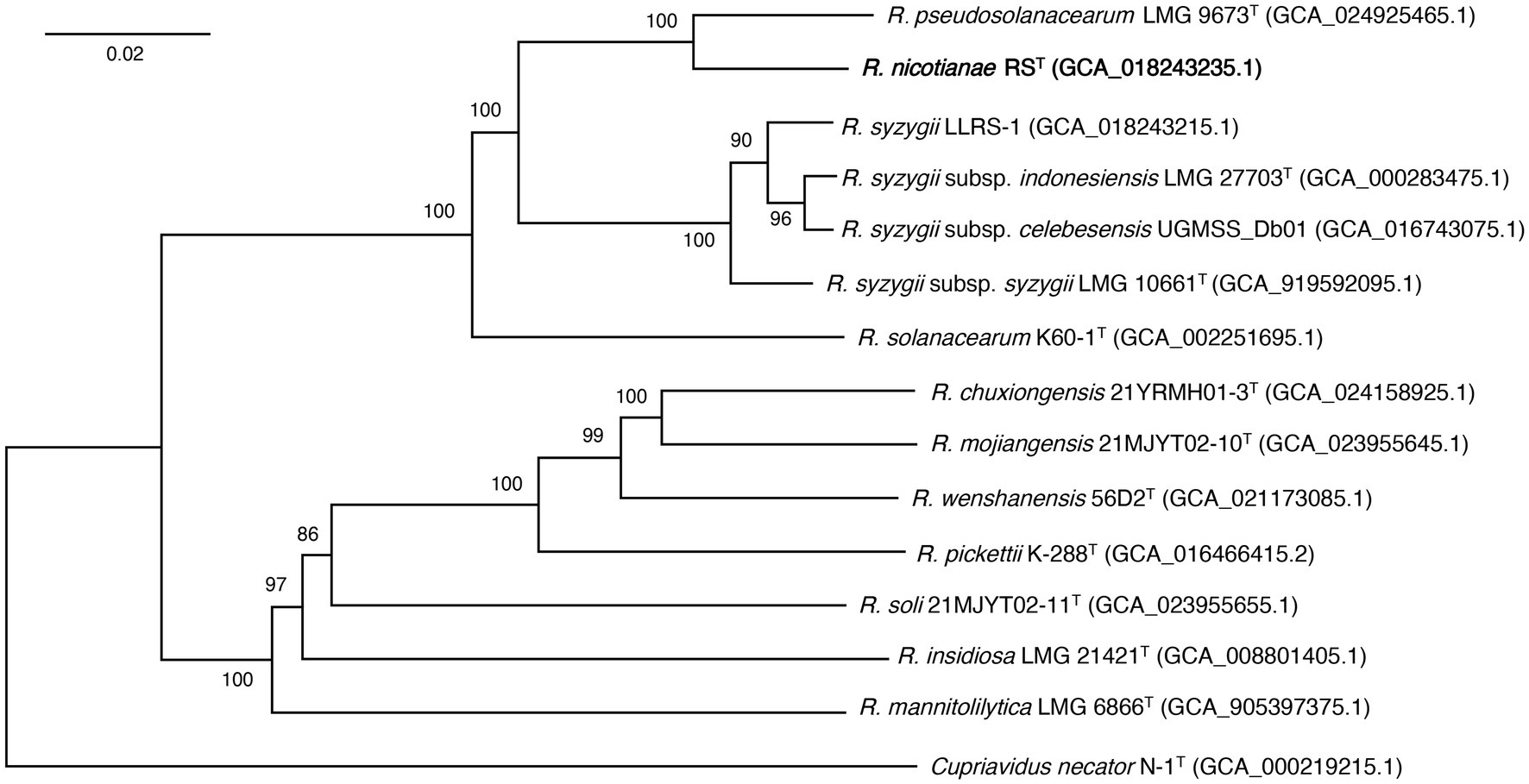
Figure 4. The phylogenomic tree showing the relationship between strain RST and its closely related strains in the genus Ralstonia. The phylogenomic tree was generated using the Type (Strain) Genome Server (Meier-Kolthoff and Göker, 2019). The numbers above branches are Genome BLAST Distance Phylogeny approach (GBDP) pseudo-bootstrap support values >50% from 100 replicates. T indicates the type strain. Strain RST is shown in bold. Cupriavidus necator N-1T is used as an outgroup. Genomic assembly numbers are shown in parentheses. Bar, 0.02 changes per nucleotide position.
To confirm their systematic classifications, all Ralstonia strains closely related to RST whose genome sequences were available in the GenBank database were subjected to genome comparisons using the FastANI calculator in the Genome Taxonomy Database (GTDB) (Jain et al., 2018; Parks et al., 2021). Data showed that strain RST had FastANI values >95.00% with 211 Ralstonia strains. Among them, two hundred and one genomes shared FastANI values >97.00% and dDDH values ≥88.70% with RST. In addition, they had FastANI values of 94.44–96.11% and dDDH values of 63.90–66.90% with R. pseudosolanacearum LMG 9673T (Supplementary Table 1). These data suggested that these strains (phylotype I) are closely related to strain RST. The systematic classifications of 10 strains of phylotype III, including CFBP3059, CMR15, NCPPB 216, UW386, RUN2279, RUN2587_UW776, RUN2474, NCPPB332_UW654, DGBBC1138_UW685, and CIP296_UW472, showed that they shared FastANI values below 97% with strain RST, which was further confirmed using ANI (ANIb and ANIm) and dDDH tests. The results revealed that all strains had ANIb values ≤95.33% and dDDH values ≤67.10% with strain RST and shared ANIb values ≥95.65% and dDDH values ≥70.50% with R. pseudosolanacearum LMG 9673T (Supplementary Tables 1, 2), supporting the view that these strains belong to the species of R. pseudosolanacearum. Based on the genome sequences of strains RST, R. pseudosolanacearum LMG 9673T, and their closely related strains using the Type (Strain) Genome Server (Meier-Kolthoff et al., 2022), a phylogenomic tree also supported the view that strain RST and its closely associated strains of phylotype I formed a coherent cluster with five complete genome-sequenced R. pseudosolanacearum strains (Supplementary Figure 3). These results suggested that strain RST isolated from bacterial wilt of tobacco and its closely related strains of phylotype I represent a novel species in the genus Ralstonia.
3.3. Pan-genome profile and accessory genes
Data revealed that 11 whole genome sequences from strains RST and type species in the genus Ralstonia had a total of 55,346 protein-coding genes, with an average of 5031.45 genes for each species. The protein coding sequences were grouped into 7,159 clusters, containing 4,564 orthologous clusters and 2,595 single-copy gene clusters. All strains shared 2,657 core genome orthologs, which are mainly involved in translation, cell shape regulation, phenylacetate catabolic process, and DNA binding transcription factor activity. Strains from seven non-plant pathogenic type species within the genus Ralstonia shared 252 specific core orthologous clusters, and strains from four plant pathogenic bacteria, including RST, R. pseudosolanacearum LMG 9673T, R. solanacearum K60-1T, and R. syzygii PSI 7T, had 301 unique core orthologous gene clusters, which were absent in non-pathogenic Ralstonia. When we compared the four plant pathogenic bacteria, type strains formed 4,468 clusters, which included 2,304 core orthologs. In addition, strain RST had 16 specific protein family clusters consisting of 47 proteins, which were not present in type strains of the other three plant pathogenic Ralstonia. Eight unique clusters in strain RST were assigned to biological processes such as nucleobase-containing compounds, cellular aromatic and nitrogen compounds, macromolecules, cellular, primary, and heterocycle metabolic processes, and DNA transposition. Strain RST shared 158, 160, and 100 orthologous clusters with its closely related type strains R. pseudosolanacearum LMG 9673T, R. syzygii subsp. indonesiensis PSI 7T, and R. solanacearum K60-1T, respectively, suggesting that strain RST is more closely related to R. pseudosolanacearum LMG 9673T and R. syzygii subsp. indonesiensis PSI 7T than to R. solanacearum K60-1T (Figure 5).
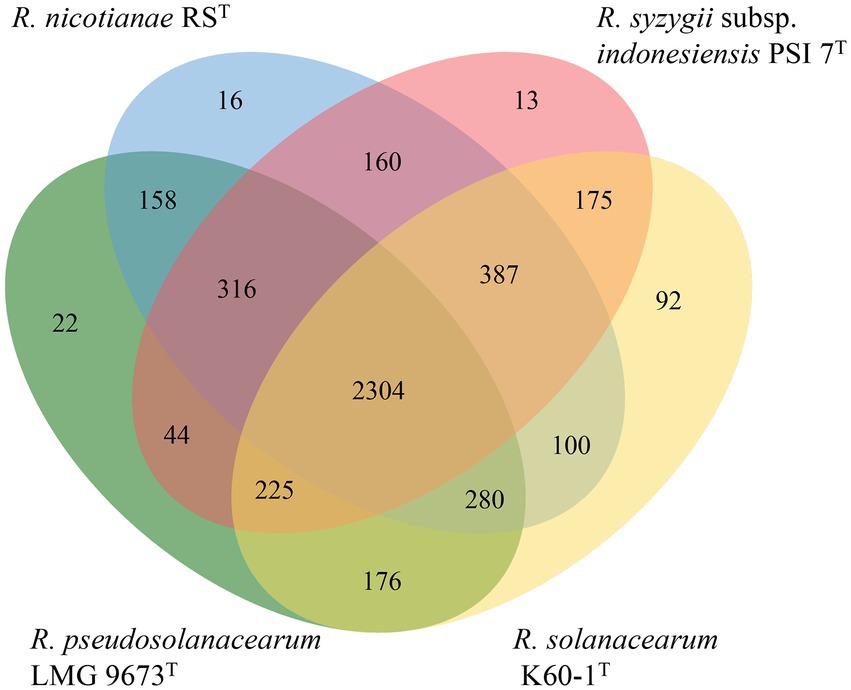
Figure 5. Venn diagram of the distribution of shared gene families (orthologous clusters) among four plant pathogenic strains of the Ralstonia (Xu et al., 2019).
3.4. Core repertoire of T3Es
A total of 57 candidate T3Es, three pseudogenes (RipB, RipS5, and RipAT), and seven hypothetical effectors without assignment were found in strain RST, among which 37 core T3Es were also found in five other closely related strains (Supplementary Table 3). A unique effector, RipAK, was present in strains of phylotype I, but absent in those belonging to phylotypes II, III, and IV. RipAK acts as a plant hypersensitive response suppressor by interacting with host pyruvate decarboxylases (PDCs), inhibiting their oligomerization and enzymatic activities (Sun et al., 2017; Wang et al., 2021). Two T3E candidates, RipBI and Hyp9, were conserved and unique among strains of phylotype II. The functional role of RipBI in Ralstonia is undefined; however, its homologous core effector XopX in Xanthomonas is a virulence factor and can both activate and suppress host immune responses in rice (Metz et al., 2005; Stork et al., 2015). Further study found that XopX induces immune responses by interacting with XopQ (Deb et al., 2020). RipBF is a phylotype IV-specific effector, which was only found in the tested R. syzygii strains; however, its functional roles remain to be determined. RipAP was absent from all R. syzygii strains, but was present in all tested strains of phylotypes I, II, and III. Although 35 candidate T3Es were present in all tested R. pseudosolanacearum strains, no specific effector was found among phylotype III strains, including LMG 9673T, CMR15, RUN2279, RUN2474, and UW386.
In a Venn diagram analysis, we also found that 15 core T3Es, including RipAE, RipAJ, RipC1, RipAI, RipAC, RipB, RipAB, RipX, RipZ, RipU, RipAO, RipR, RipAQ, RipAM, and RipAN, were conserved among 18 tested Ralstonia strains (Figure 6). This suggested that these effectors play essential roles in pathogenicity. Indeed, Cong et al. (2022) have heterologously expressed the five core effectors of phylotype I strain P380 in the polymutant Pseudomonas syringae DC3000D36E. Their data showed that RipAE, RipC1, RipU, and RipW inhibited the reactive oxygen species (ROS) burst, while RipAQ did not. The core effectors, including RipAE, RipU, and RipW, induced cell death as well as upregulating mitogen-activated protein kinase (MAPK) cascades in Nicotiana benthamiana (Cong et al., 2022). Furthermore, RipAC suppresses the hypersensitive response triggered by avirulence effector RipAA of phylotype I strain GMI1000 (Nakano et al., 2020). Moreover, effector RipAC suppresses Nod-like receptor (NLR)-triggered immune responses by inhibiting the interaction between MAPKs and SGT1 (suppressor of G2 allele of SKP1 homolog) targets, and the phosphorylation of a MAPK target motif in the C-terminal domain of SGT1 (Nakano et al., 2020). Recently, Yu et al. (2022) found that RipAC inhibits pattern-triggered immunity by targeting E3 ubiquitin ligase plant U-box 4 (PUB4). Furthermore, core effector RipB acts as a major avirulence factor in Nicotiana species and suppresses the basal defense in susceptible hosts to promote pathogen infection (Nakano and Mukaihara, 2019; Cao et al., 2022). In addition, the nuclear-localized effectors RipAB and RipX are involved in plant immune response (Zheng et al., 2019; Sun et al., 2020). However, the functions of the other core effectors, including RipAJ, RipC1, RipAI, RipZ, RipAO, RipR, RipAM, and RipAN, still need to be determined.
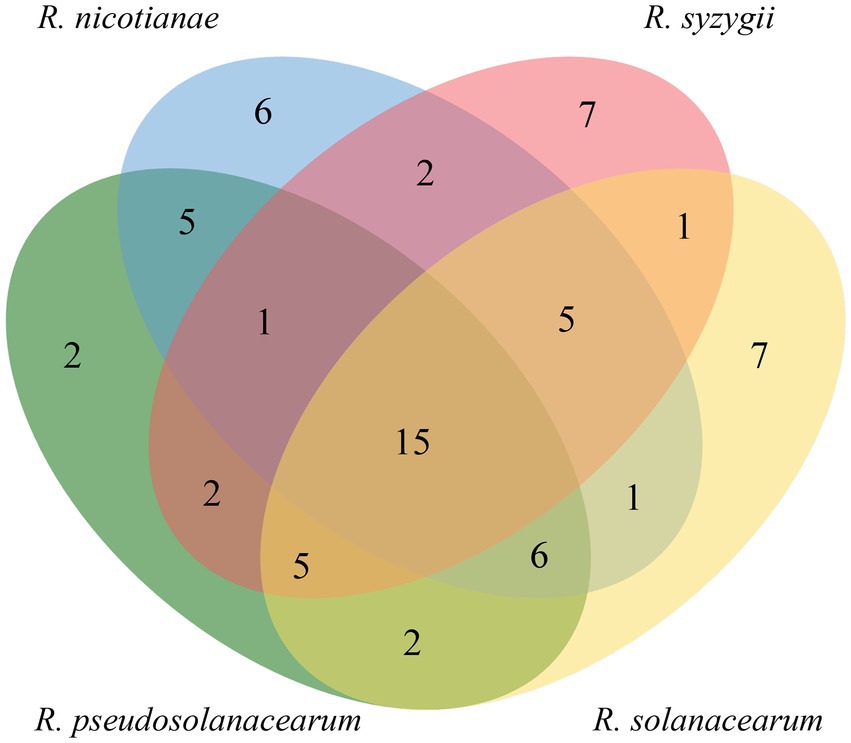
Figure 6. Venn diagram of the distribution of shared candidate T3Es (core effectors) among strains of four phylotypes (https://bioinfogp.cnb.csic.es/tools/venny/index.html).
3.5. Gene prediction involved in secondary metabolites
In silico secondary metabolite analysis predicted 12 gene clusters in the genome of strain RST, referring to seven cluster types, including aryl polyene, furan, homoserine lactone (hserlactone), non-ribosomal peptide synthetase (NRPS), NRPS-like, ribosomally synthesized and post-translationally modified peptide product-like (RiPP-like), terpene, and type I polyketide synthase (T1PKS) (Supplementary Table 4). Among them, the taxonomically widespread aryl polyene, RiPP-like, and terpene biosynthetic gene clusters were also present in all tested Ralstonia strains (Nishida et al., 2005; Arnison et al., 2013; Cimermancic et al., 2014). Interestingly, more known secondary metabolite gene clusters were detected in the plant pathogenic Ralstonia species (10.87 clusters/strain) than in the non-pathogenic group (6.28 clusters/strain), which might suggest that more secondary metabolites remain to be uncovered in the latter group. Data revealed that the hserlactone and furan types were found in pathogenic Ralstonia strains. Both metabolite clusters shared less than 20% similarity to known gene clusters, suggesting that these gene clusters might encode novel metabolites and play significant roles in pathogenicity. The homoserine lactone gene cluster, which is usually involved in signal molecule production, participates in cell-to-cell communication processes. A recent study showed that the novel RasI/R quorum sensing system regulates phylotype I strain EP1 virulence. When the signal molecule N-(3-hydroxydodecanoyl)-homoserine lactone synthetic gene rasI was knocked out, the mutant lost virulence toward eggplants (Yan et al., 2022). The metabolites of NRPS, NRPS-like, and T1PKS types were also only found in plant pathogenic Ralstonia, except for R. pickettii K-288T, which only contained a T1PKS gene cluster. In silico metabolite analysis found that the beta-lactone-containing protease inhibitor (betalactone) and redox-cofactor types appeared to be conserved in non-pathogenic Ralstonia strains.
Siderophores are essential factors for both plant pathogens and beneficial microorganisms (Gu et al., 2020). Based on their chemical nature, siderophores are mainly classified into hydroxamate, catecholate, and carboxylate (Khan et al., 2018). Data revealed that strain RST carried three putative siderophore biosynthetic clusters. Firstly, the chromosome of strain RST encodes a micacocidin biosynthetic cluster (Kreutzer et al., 2011), which is conserved among phylotypes I, III, and IV strains, but absent in strains of phylotype II. However, a gene cluster accounting for siderophore yersiniabactin biosynthesis was only present in strains of phylotype II. Secondly, we detected a gene cluster that shared 18% similarity to the biosynthesis gene cluster of carboxylate siderophore staphyloferrin B on the megaplasmid of strain RST (Bhatt and Denny, 2004). Thirdly, a specific NRPS and/or NRPS-like gene cluster was conserved among tested five strains of phylotype I, with a size range of 48.51–51.11 kb. It consisted of two core biosynthetic genes, seven additional biosynthetic genes, two transport-related genes, six regulatory genes, and 16 other genes on the megaplasmid of strain RST. It shared no sequence similarity with known gene clusters; however, the MIBiG comparison showed that it had the highest similarity score of 0.65 with the catecholate siderophore Amphi-enterobactin 1 biosynthetic gene cluster in the marine organism Vibrio harveyi BAA-1116 (McRose et al., 2018). However, the functional roles of this secondary metabolite biosynthetic cluster are uncharacterized.
In addition, nine out of 10 tested strains from phylotypes I and III shared a specific ralsolamycin biosynthetic cluster (100% similarity), consistent with a previous report that R. solanacearum K60-1T does not encode this specific gene cluster (Spraker et al., 2016). The diffusible lipopeptide ralsolamycin was found in a survey of secondary metabolites that induced chlamydospore formation of fungi when co-cultured with strain GMI1000 (Spraker et al., 2016). The biosynthesis locus of ralsolamycin comprises a non-ribosomal peptide synthetase and polyketide synthase hybrid gene (RmyA) and an NRPS gene (RmyB) (Spraker et al., 2016). Data showed that the PhcBSR quorum sensing (QS) system regulates ralsolamycin production (Li et al., 2017, 2022). A recent study found that ralsolamycin induces the production of the fungal secondary metabolite bikaverin when Fusarium fujikuroi was exposed to ralsolamycin (Spraker et al., 2018), suggesting that this secondary metabolite can act as a communication chemical between bacteria and fungi.
3.6. Physiology and chemotaxonomy
In the API assays, strains of phylotype I, including RST, GMI1000, and CQPS-1, shared more similarity to R. pseudosolanacearum LMG 9673T and R. solanacearum LMG 2299T for many phenotypic characteristics, e.g., all strains were positive for the reduction of potassium nitrate; hydrolysis of eculin; assimilation of D-glucose, potassium gluconate, malic acid, and trisodium citrate; and catalysis activity; and negative for indole production; the assimilation of arabinose, mannose, maltose, and adipic acid; and the activities of arginine dihydrolase, urease, protease, α-galactosidase, β-galactosidase, esterase lipase, valine arylamidase, cystine arylamidase, trypsin, α-chymotrypsin, β-glucuronidase, α-glucosidase, β-glucosidase, N-acetyl-β-glucosaminidase, α-mannosidase, and α-fucosidase. However, strains of phylotype I had unique phenotypic features. For example, R. pseudosolanacearum LMG 9673T and R. solanacearum LMG 2299T were negative for leucine arylamidase and acid phosphatase activity individually, whereas phylotype I strains were positive. In addition, R. pseudosolanacearum LMG 9673T and R. solanacearum LMG 2299T were positive for lipase and Naphthol-AS-BI-phosphohydrolase activity, respectively, and both of them were positive for glucose fermentation; however, all tested phylotype I strains were negative.
In the Biolog GEN III assays, tested phylotype I strains were more similar to R. pseudosolanacearum LMG 9673T and R. solanacearum LMG 2299T for several phenotypic characteristics. All strains were positive for the utilization of D-fructose-6-PO4, D-galacturonic acid, L-galactonic acid lactone, D-glucuronic acid and glucuronamide; were able to grow under conditions supplemented with rifamycin SV, lincomycin, vancomycin, tetrazolium violet, tetrazolium blue and potassium tellurite; were negative for the utilization of carbon sources such as D-maltose, D-trehalose, D-cellobiose, gentiobiose, sucrose, D-turanose, stachyose, D-raffinose, α-D-lactose, D-melibiose, β-methyl-D-glucoside, D-salicin, N-acetyl-D-glucosamine, N-acetyl-β-D-mannosamine, N-acetyl-D-galactosamine, N-acetyl neuraminic acid, L-rhamnose, inosine, D-sorbitol, D-mannitol, D-arabitol, myo-inositol, glycerol, D-serine, Gelatin, glycyl-L-proline, L-alanine, L-arginine, L-pyroglutamic acid, L-serine, D-gluconic acid, mucic acid, quinic acid, D-saccharic acid, p-hydroxy-phenylacetic acid, methyl pyruvate, D-lactic acid methyl ester, L-lactic acid, D-malic acid, bromo-succinic acid, α-hydroxy-butyric acid, α-keto-butyric acid, propionic acid, formic acid; and were unable to grow under conditions supplemented with inhibitory chemicals, including pH 5, 1% NaCl, 4% NaCl, 8% NaCl, 1% sodium lactate, fusidic acid, D-serine, troleandomycin, minocycline, guanidine HCl, niaproof 4, lithium chloride, sodium butyrate, sodium bromate. Furthermore, the data also showed that the tested phylotype I strains, including RST, GMI1000, and CQPS-1, were more similar to R. pseudosolanacearum LMG 9673T. For example, these four strains were unable to use D-aspartic acid, L-histidine, α-keto-glutaric acid, γ-amino-butyric acid, β-hydroxy-D,L-butyric acid, and acetoacetic acid, whereas strain R. solanacearum LMG 2299T was able to use these carbon sources. However, R. pseudosolanacearum LMG 9673T tested positive for the use of two carbon sources, D-Mannose and L-Fucose and one inhibitory chemical (nalidixic acid), whereas RST, GMI1000, CQPS-1, and R. solanacearum LMG 2299T tested negative. Although the tested strains shared several phenotypic features, RST, GMI1000, and CQPS-1 had unique characteristics. Strains of phylotype I tested negative for the use of dextrin, α-D-glucose, 3-methyl glucose, L-aspartic acid, L-glutamic acid, pectin, citric acid, L-malic acid, tween 40, and acetic acid. However, R. pseudosolanacearum LMG 9673T and R. solanacearum LMG 2299T could use these carbon sources (Supplementary Table 5).
The conserved fatty acids present in all tested strains were C14:0, C16:0, C18:0, cyclo-C17:0, C18:1 2-OH, summed feature 2 (C14:0 3-OH/C16:1 iso I), summed feature 3 (C16:1 ω7c and/or C16:1 ω6c), and summed feature 8 (C18:1 ω7c and/or C18:1 ω6c). The major fatty acids of strains RST, GMI1000, and CQPS-1 were C16:0, summed feature 3 (C16:1 ω7c and/or C16:1 ω6c), and summed feature 8 (C18:1 ω7c and/or C18:1 ω6c), which were closely related to those of R. pseudosolanacearum LMG 9673T and R. solanacearum LMG 2299T (Table 3). The major fatty acids of these strains were similar to three recently proposed strains R. wenshanensis 56D2T (Lu et al., 2022a), R. chuxiongensis 21YRMH01-3T, and R. mojiangensis 21MJYT02-10T from our laboratory (Lu et al., 2023). The main fatty acids in tested phylotype I strains were slightly different from those of R. soli 21MJYT02-11T, which included the major fatty acids of C16:0, cyclo-C17:0, and summed features 3 (Lu et al., 2023).
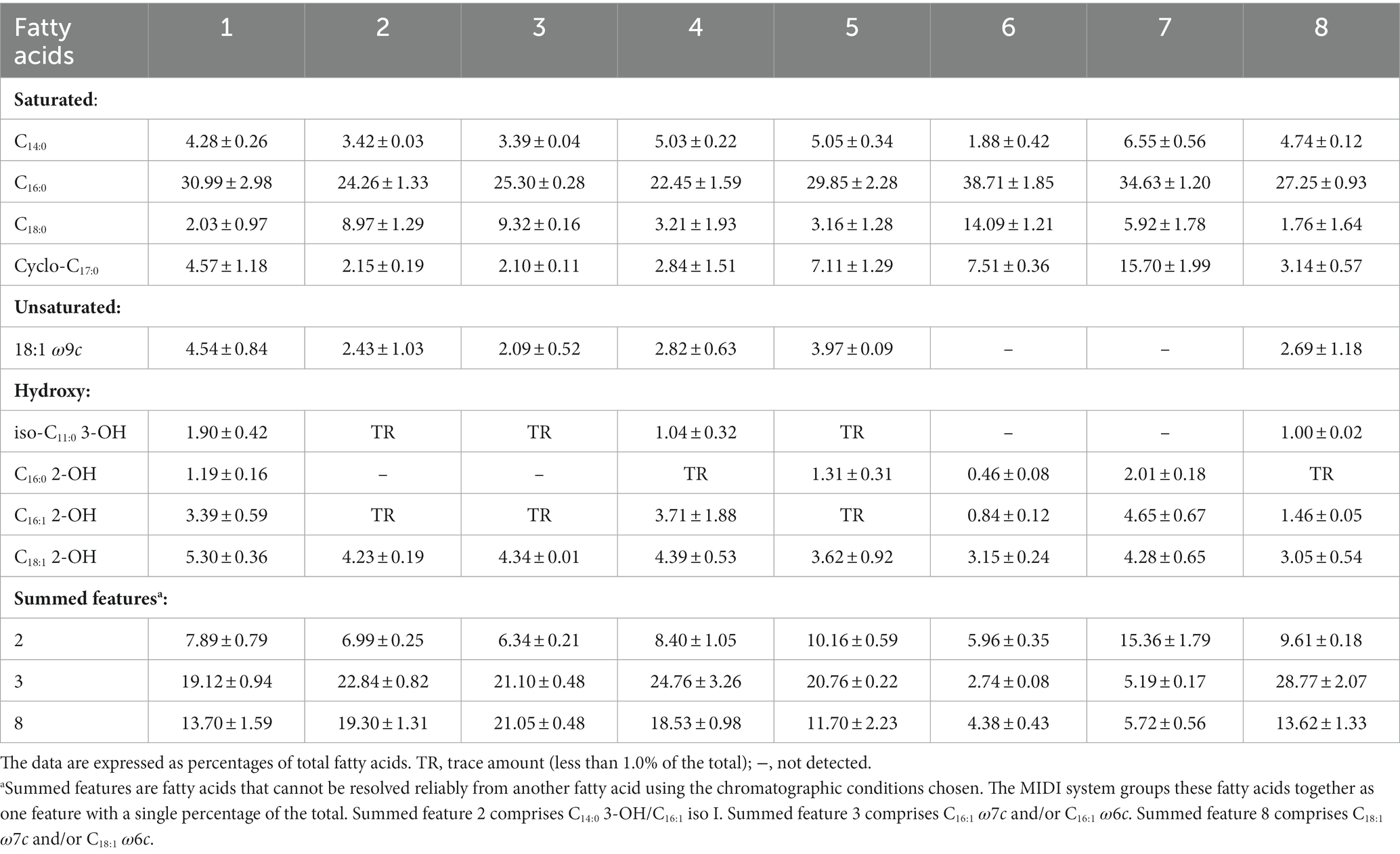
Table 3. Major fatty acids of strain RST and its closely related strains in the genus Ralstonia. Strain: 1. RST; 2. GMI1000; 3. CQPS-1; 4. R. pseudosolanacearum LMG 9673T; 5. R. solanacearum LMG 2299T; 6. R. syzygii subsp. syzygii LMG 10661T; 7. R. syzygii subsp. celebesensis LMG 27706T; 8. R. syzygii subsp. indonesiensis LMG 27703T.
The polar lipids analysis showed that strain RST contained phosphatidylethanolamine, diphosphatidylglycerol, phosphatidylglycerol, and an unidentified aminophospholipid (Supplementary Figure 4). Data from this study and our recent reports suggest that the major polar lipids of Ralstonia species are phosphatidylethanolamine and diphosphatidylglycerol (Lu et al., 2022a, 2023). In addition, isoprenoid quinone analysis showed that strain RST contained ubiquinone Q-7 (17.87%) and Q-8 (82.13%). Data from this study and our recent reports also indicated that Q-8 is the major respiratory quinone in Ralstonia species (Lu et al., 2022a, 2023).
4. Conclusion
Based on the above polyphasic analysis, we concluded that the Ralstonia strains could be phylogenetically separated into two main clusters: non-pathogenic and plant-pathogenic bacteria. The current R. solanacearum phylotype I strain RST and its close relatives represent a novel species in the genus Ralstonia, for which the name Ralstonia nicotianae sp. nov. is proposed.
5. Description of Ralstonia nicotianae sp. nov.
Ralstonia nicotianae (ni.co.ti.a’nae. N.L. gen. fem. n. nicotianae, of Nicotiana, the tobacco plant, from which the type strain (RST) was isolated).
Cells are Gram-negative, aerobic, no-spore, motile with polar flagella, rod-shape, 1.60–3.60 μm long, and 0.68–1.44 μm wide (avg. 2.40 ± 0.52 μm × 0.93 ± 0.18 μm; n = 46). They can grow on LA, NA, PDA, TSA, CPG, TZC, and MacConkey agars, with better growth on CPG and TZC media. Colonies on TSA plates after incubation for 2 days at 28°C are circular, light yellow and convex, with a smooth surface, and 1.5 mm in diameter. Cells grow at 15–40°C (optimum, 25–35°C), 0–5% NaCl (w/v) (optimum, 0–2%), and at pH 5.0–10.0 (optimum, pH 7.0–8.0). Cells are positive for catalase and oxidase activities. In API 20NE assays, cells are positive for potassium nitrate reduction; esculin hydrolysis; and glucose, potassium gluconate, malate, and trisodium citrate assimilation; and negative for indole production; glucose fermentation; arginine dihydrolase; urease; β-galactosidase activities; gelatin hydrolysis; and arabinose, mannose, maltose and adipic acid assimilation. In the API ZYM analysis, strains are positive for enzymatic activities of leucine arylamidase and acid phosphatase; and negative for esterase lipase, lipase, valine arylamidase, cystine arylamidase, trypsin, α-chymotrypsin, naphthol-AS-BI-phosphohydrolase, α-galactosidase, β-glucuronidase, α-glucosidase, β-glucosidase, N-acetyl-β-glucosaminidase, α-mannosidase, and α-fucosidase activities. In the Biolog GEN III analysis, strains were positive for the utilization of D-galacturonic acid, L-galactonic acid lactone, D-glucuronic acid, glucuronamide, and D-fructose-6-PO4; and were resistant to rifamycin SV, lincomycin, vancomycin, tetrazolium violet, tetrazolium blue, and potassium tellurite. The primary fatty acids of RST are saturated fatty acid C16:0, summed feature 3 (C16:1 ω7c and/or C16:1 ω6c), and summed feature 8 (C18:1 ω7c and/or C18:1 ω6c). The polar lipid profile of strain RST consists of phosphatidylethanolamine, diphosphatidylglycerol, phosphatidylglycerol, and an unidentified aminophospholipid. Phosphatidylethanolamine is the major polar lipid. The strain contains ubiquinones Q-7 (17.87%) and Q-8 (82.13%).
The type strain, RST (= GDMCC 1.3533T = JCM 35814T), was isolated from a bacterial wilt of flue-cured tobacco sampled in 2018 from Yuxi, Yunnan, China. The genomic DNA G + C content of type strain RST is 67.1 mol%. The GenBank accession number for the 16S rRNA gene sequence of the type strain is ON797093, and the genome sequence accession numbers are CP046674 and CP046675.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: The GenBank accession numbers for the chromosome, megaplasmid and 16S rRNA gene sequences of strain RST are CP046674.1, CP046675.1, and ON797093.1, respectively.
Author contributions
J-YL, H-ML, Y-QZ, and C-HL designed the research and project outline. J-YL collected the tobacco samples. J-YL, ZC, and H-LW performed the deposition and polyphasic taxonomy. J-YL, J-FZ, C-PZ, S-YL, and C-HL conducted the genome analysis. J-YL, C-HL, and Y-QZ drafted the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the Yunnan Fundamental Research Projects (202001BA0000-091 and 202201AT071023), the Yuxi Normal College Student Innovation and Entrepreneurship Training Program (2021A056), the Yunnan Provincial Tobacco Monopoly Bureau Projects (2020530000241013 and 2018530000241006), and the National Natural Science Foundation of China (32260702).
Acknowledgments
We thank Prof. Wei Ding at Southwest University, and Dr. Peng Li at Hainan Normal University for their generous gifts of the Rasltonia strains CQPS-1 and GMI1000, respectively.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1135872/full#supplementary-material
Abbreviations
ANI, average nucleotide identity; betalactone, beta-lactone-containing protease inhibitor; BGI, Beijing Genomics Institute; COG, cluster of orthologous groups; CPG, casamino acid peptone glucose; egl, endoglucanase; GDMCC, Guangdong Microbial Culture Collection Center; GTDB, Genome Taxonomy Database; hserlactone, homoserine lactone; dDDH, digital DNA–DNA hybridization; ITS, internal transcribed spacer region; JCM, Japan Collection of Microorganisms; LA, Luria-Bertani agar; ME, minimum-evolutions; ML, maximum-likelihood; NA, nutrient agar; NJ, neighbor-joining; NRPS, non-ribosomal peptide synthetase; PDA, potato dextrose agar; PDCs, pyruvate decarboxylases; QS, quorum sensing; RiPP-like, ribosomally synthesized and post-translationally modified peptide product-like; rRNA, ribosomal RNA; sRNA, small RNA; T1PKS, type I polyketide synthase; tRNA, transfer RNA; TSA, trypticase soy agar; TYGS, the type (strain) genome server; TZC, Kelman’s tetrazolium chloride.
Footnotes
2. ^https://www.ezbiocloud.net/
3. ^www.ncbi.nlm.nih.gov/genbank
4. ^http://www.ebi.ac.uk/genomes/plasmid.html, last accessed July 8, 2016
References
Arnison, P. G., Bibb, M. J., Bierbaum, G., Bowers, A. A., Bugni, T. S., Bulaj, G., et al. (2013). Ribosomally synthesized and post-translationally modified peptide natural products: Overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 30, 108–160. doi: 10.1039/C2NP20085F
Berlin, K., Koren, S., Chin, C.-S., Drake, J. P., Landolin, J. M., and Phillippy, A. M. (2015). Assembling large genomes with single-molecule sequencing and locality-sensitive hashing. Nat. Biotechnol. 33, 623–630. doi: 10.1038/nbt.3238
Besemer, J., Lomsadze, A., and Borodovsky, M. (2001). GeneMarkS: A self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 29, 2607–2618. doi: 10.1093/nar/29.12.2607
Bhatt, G., and Denny, T. P. (2004). Ralstonia solanacearum iron scavenging by the siderophore staphyloferrin B is controlled by PhcA, the global virulence regulator. J. Bacteriol. 186, 7896–7904. doi: 10.1128/JB.186.23.7896-7904.2004
Blin, K., Shaw, S., Kloosterman, A. M., Charlop-Powers, Z., van Wezel, G. P., Medema, M. H., et al. (2021). antiSMASH 6.0: Improving cluster detection and comparison capabilities. Nucleic Acids Res. 49, W29–W35. doi: 10.1093/nar/gkab335
Cao, P., Chen, J., Wang, R., Zhao, M., Zhang, S., An, Y., et al. (2022). A conserved type III effector RipB is recognized in tobacco and contributes to Ralstonia solanacearum virulence in susceptible host plants. Biochem. Biophys. Res. Commun. 631, 18–24. doi: 10.1016/j.bbrc.2022.09.062
Cao, Y., Tian, B., Liu, Y., Cai, L., Wang, H., Lu, N., et al. (2013). Genome sequencing of Ralstonia solanacearum FQY_4, isolated from a bacterial wilt nursery used for breeding crop resistance. Genome Announc. 1:e00125-13. doi: 10.1128/genomeA.00125-13
Chin, C.-S., Alexander, D. H., Marks, P., Klammer, A. A., Drake, J., Heiner, C., et al. (2013). Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 10, 563–569. doi: 10.1038/nmeth.2474
Cimermancic, P., Medema, M. H., Claesen, J., Kurita, K., Brown, W., Laura, C., et al. (2014). Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cells 158, 412–421. doi: 10.1016/j.cell.2014.06.034
Clayton, R. A., Sutton, G., Hinkle, P. S., Bult, C., and Fields, C. (1995). Intraspecific variation in small-subunit rRNA sequences in GenBank: Why single sequences may not adequately represent prokaryotic taxa. Int. J. Syst. Evol. Microbiol. 45, 595–599. doi: 10.1099/00207713-45-3-595
Coenye, T., Goris, J., De Vos, P., Vandamme, P., and LiPuma, J. J. (2003). Classification of Ralstonia pickettii-like isolates from the environment and clinical samples as Ralstonia insidiosa sp. nov. Int. J. Syst. Evol. Microbiol. 53, 1075–1080. doi: 10.1099/ijs.0.02555-0
Collins, M. D., and Jones, D. (1981). A note on the separation of natural mixtures of bacterial ubiquinones using reverse-phase partition thin-layer chromatography and high performance liquid chromatography. J. Appl. Bacteriol. 51, 129–134. doi: 10.1111/j.1365-2672.1981.tb00916.x
Cong, S., Li, J.-Z., Xiong, Z.-Z., and Wei, H.-L. (2022). Diverse interactions of five core type III effectors from Ralstonia solanacearum with plants. J. Genet. Genomics. doi: 10.1016/j.jgg.2022.04.018
Daligault, H. E., Davenport, K. W., Minogue, T. D., Broomall, S. M., Bruce, D. C., Chain, P. S., et al. (2014). Draft genome assembly of Ralstonia pickettii type strain K-288 (ATCC 27853). Genome Announc. 2:e00973-14. doi: 10.1128/genomeA.00973-14
De Baere, T., Steyaert, S., Wauters, G., Des Vos, P., Goris, J., Coenye, T., et al. (2001). Classification of Ralstonia pickettii biovar 3/‘thomasii’ strains (Pickett 1994) and of new isolates related to nosocomial recurrent meningitis as Ralstonia mannitolytica sp. nov. Int. J. Syst. Evol. Microbiol. 51, 547–558. doi: 10.1099/00207713-51-2-547
Deb, S., Ghosh, P., Patel, H. K., and Sonti, R. V. (2020). Interaction of the Xanthomonas effectors XopQ and XopX results in induction of rice immune responses. Plant J. 104, 332–350. doi: 10.1111/tpj.14924
Elphinstone, J. G., Hennessy, J., Wilson, J. K., and Stead, D. E. (1996). Sensitivity of different methods for the detection of Ralstonia solanacearum in potato tuber extracts. EPPO Bull. 26, 663–678. doi: 10.1111/j.1365-2338.1996.tb01511.x
Fluit, A. C., Bayjanov, J. R., Aguilar, M. D., Cantón, R., Tunney, M. M., Elborn, J. S., et al. (2021). Characterization of clinical Ralstonia strains and their taxonomic position. Antonie Van Leeuwenhoek 114, 1721–1733. doi: 10.1007/s10482-021-01637-0
García, R. O., Kerns, J. P., and Thiessen, L. (2019). Ralstonia solanacearum species complex: A quick diagnostic guide. Plant Health Progress 20, 7–13. doi: 10.1094/php-04-18-0015-dg
Genin, S., and Denny, T. P. (2012). Pathogenomics of the Ralstonia solanacearum species complex. Annu. Rev. Phytopathol. 50, 67–89. doi: 10.1146/annurev-phyto-081211-173000
Gu, S., Wei, Z., Shao, Z., Friman, V.-P., Cao, K., Yang, T., et al. (2020). Competition for iron drives phytopathogen control by natural rhizosphere microbiomes. Nat. Microbiol. 5, 1002–1010. doi: 10.1038/s41564-020-0719-8
Jain, C., Rodriguez, R. L. M., Phillippy, A. M., Konstantinidis, K. T., and Aluru, S. (2018). High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 9:5114. doi: 10.1038/s41467-018-07641-9
Kelman, A. (1954). The relationship of pathogenicity of Pseudomonas solanacearum to colony appearance in a tetrazolium medium. Phytopathology 44, 693–695.
Kelman, A., and Hruschka, J. (1973). The role of motility and aerotaxis in the selective increase of avirulent bacteria in still broth cultures of Pseudomonas solanacearum. J. Gen. Microbiol. 76, 177–188. doi: 10.1099/00221287-76-1-177
Khan, A., Singh, P., and Srivastava, A. (2018). Synthesis, nature and utility of universal iron chelator – Siderophore: A review. Microbiol. Res. 212-213, 103–111. doi: 10.1016/j.micres.2017.10.012
Kreutzer, M. F., Kage, H., Gebhardt, P., Wackler, B., Saluz, H. P., Hoffmeister, D., et al. (2011). Biosynthesis of a complex Yersiniabactin-like natural product via the mic locus in phytopathogen Ralstonia solanacearum. Appl. Environ. Microbiol. 77, 6117–6124. doi: 10.1128/AEM.05198-11
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Li, P., Cao, X., Zhang, L., Lv, M., and Zhang, L.-H. (2022). PhcA and PhcR regulate ralsolamycin biosynthesis oppositely in Ralstonia solanacearum. Front. Plant Sci. 13:903310. doi: 10.3389/fpls.2022.903310
Li, H., Ruan, J., and Durbin, R. (2008). Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18, 1851–1858. doi: 10.1101/gr.078212.108
Li, P., Wang, D., Yan, J., Zhou, J., Deng, Y., Jiang, Z., et al. (2016). Genomic analysis of phylotype I strain EP1 reveals substantial divergence from other strains in the Ralstonia solanacearum species complex. Front. Microbiol. 7:1719. doi: 10.3389/fmicb.2016.01719
Li, P., Yin, W., Yan, J., Chen, Y., Fu, S., Song, S., et al. (2017). Modulation of inter-kingdom communication by PhcBSR quorum sensing system in Ralstonia solanacearum phylotype I strain GMI1000. Front. Microbiol. 8:1172. doi: 10.3389/fmicb.2017.01172
Li, R., Yu, C., Li, Y., Lam, T.-W., Yiu, S.-M., Kristiansen, K., et al. (2009). SOAP2: An improved ultrafast tool for short read alignment. Bioinformatics 25, 1966–1967. doi: 10.1093/bioinformatics/btp336
Liu, Y., Tang, Y., Qin, X., Yang, L., Jiang, G., Li, S., et al. (2017). Genome sequencing of Ralstonia solanacearum CQPS-1, a phylotype I strain collected from a highland area with continuous cropping of tobacco. Front. Microbiol. 8:974. doi: 10.3389/fmicb.2017.00974
Lu, C.-H., Chen, W., Yin, H.-H., Lin, Z.-L., Li, J.-Y., Ma, J.-H., et al. (2022a). Ralstonia wenshanensis sp. nov., a novel bacterium isolated from a tobacco field in Yunnan, China. Int. J. Syst. Evol. Microbiol. 72:5622. doi: 10.1099/ijsem.0.005622
Lu, C.-H., Han, T.-H., Jiang, N., Gai, X.-T., Cao, Z.-H., Zou, S.-Y., et al. (2022b). Pseudomonas lijiangensis sp. nov., a novel phytopathogenic bacterium isolated from black spots of tobacco. Int. J. Syst. Evol. Microbiol. 72:5591. doi: 10.1099/ijsem.0.005591
Lu, C.-H., Li, J.-Y., Mi, M.-G., Lin, Z.-L., Jiang, N., Gai, X.-T., et al. (2021). Complete genome sequence of Ralstonia syzygii subsp. indonesiensis strain LLRS-1, isolated from wilted tobacco in China. Phytopathology 111, 2392–2395. doi: 10.1094/phyto-04-21-0138-a
Lu, C.-H., Zhang, Y.-Y., Jiang, N., Chen, W., Shao, X., Zhao, Z.-M., et al. (2023). Ralstonia chuxiongensis sp. nov., Ralstonia mojiangensis sp. nov. and Ralstonia soli sp. nov., three novel species in the family Burkholderiaceae isolated from tobacco fields. Front. Microbiol. Submitted
McRose, D. L., Baars, O., Seyedsayamdost, M. R., and Morel, F. M. M. (2018). Quorum sensing and iron regulate a two-for-one siderophore gene cluster in Vibrio harveyi. Proc. Natl. Acad. Sci. 115, 7581–7586. doi: 10.1073/pnas.1805791115
Meier-Kolthoff, J. P., Carbasse, J. S., Peinado-Olarte, R. L., and Goker, M. (2022). TYGS and LPSN: A database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acids Res. 50, D801–D807. doi: 10.1093/nar/gkab902
Meier-Kolthoff, J. P., and Göker, M. (2019). TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 10:2182. doi: 10.1038/s41467-019-10210-3
Metz, M., Dahlbeck, D., Morales, C. Q., Sady, B. A., Clark, E. T., and Staskawicz, B. J. (2005). The conserved Xanthomonas campestris pv. vesicatoria effector protein XopX is a virulence factor and suppresses host defense in Nicotiana benthamiana. Plant J. 41, 801–814. doi: 10.1111/j.1365-313X.2005.02338.x
Minnikin, D. E., and Abdolrahimzadeh, H. (1971). Thin-layer chromatography of bacterial lipids on sodium acetate-impregnated silica gel. J. Chromatogr. A 63, 452–454. doi: 10.1016/S0021-9673(01)85672-7
Nakano, M., Ichinose, Y., and Mukaihara, T. (2020). Ralstonia solanacearum type III effector RipAC targets SGT1 to suppress effector-triggered immunity. Plant Cell Physiol. 61, 2067–2076. doi: 10.1093/pcp/pcaa122
Nakano, M., and Mukaihara, T. (2019). The type III effector RipB from Ralstonia solanacearum RS1000 acts as a major avirulence factor in Nicotiana benthamiana and other Nicotiana species. Mol. Plant Pathol. 20, 1237–1251. doi: 10.1111/mpp.12824
Nishida, Y., Adachi, K., Kasai, H., Shizuri, Y., Shindo, K., Sawabe, A., et al. (2005). Elucidation of a carotenoid biosynthesis gene cluster encoding a novel enzyme, 2,2′-β-hydroxylase, from Brevundimonas sp. strain SD212 and combinatorial biosynthesis of new or rare xanthophylls. Appl. Environ. Microbiol. 71, 4286–4296. doi: 10.1128/AEM.71.8.4286-4296.2005
Parks, D. H., Chuvochina, M., Rinke, C., Mussig, A. J., Chaumeil, P.-A., and Hugenholtz, P. (2021). GTDB: An ongoing census of bacterial and archaeal diversity through a phylogenetically consistent, rank normalized and complete genome-based taxonomy. Nucleic Acids Res. 50, D785–D794. doi: 10.1093/nar/gkab776
Parte, A. C., Sardà Carbasse, J., Meier-Kolthoff, J. P., Reimer, L. C., and Göker, M. (2020). List of prokaryotic names with standing in nomenclature (LPSN) moves to the DSMZ. Int. J. Syst. Evol. Microbiol. 70, 5607–5612. doi: 10.1099/ijsem.0.004332
Paudel, S., Dobhal, S., Alvarez, A. M., and Arif, M. (2020). Taxonomy and phylogenetic research on Ralstonia solanacearum species complex: A complex pathogen with extraordinary economic consequences. Pathogens 9:886. doi: 10.3390/pathogens9110886
Peeters, N., Carrère, S., Anisimova, M., Plener, L., Cazalé, A.-C., and Genin, S. (2013). Repertoire, unified nomenclature and evolution of the type III effector gene set in the Ralstonia solanacearum species complex. BMC Genomics 14:859. doi: 10.1186/1471-2164-14-859
Poehlein, A., Kusian, B., Friedrich, B., Daniel, R., and Bowien, B. (2011). Complete genome sequence of the type strain Cupriavidus necator N-1. J. Bacteriol. 193:5017. doi: 10.1128/JB.05660-11
Poplin, R., Ruano-Rubio, V., DePristo, M. A., Fennell, T. J., Carneiro, M. O., Van der Auwera, G. A., et al. (2018). Scaling accurate genetic variant discovery to tens of thousands of samples. bioRxiv 201178. doi: 10.1101/201178
Poussier, S., Prior, P., Luisetti, J., Hayward, C., and Fegan, M. (2000). Partial sequencing of the hrpB and endoglucanase genes confirms and expands the known diversity within the Ralstonia solanacearum species complex. Syst. Appl. Microbiol. 23, 479–486. doi: 10.1016/S0723-2020(00)80021-1
Prakoso, A. B., Joko, T., Soffan, A., Sari, J. P., Ray, J. D., Drenth, A., et al. (2022). Draft genome sequence of Ralstonia syzygii subsp. celebesensis from Indonesia, the causal agent of blood disease of banana. Phytopathology 112, 1584–1586. doi: 10.1094/PHYTO-10-21-0443-A
Prior, P., and Fegan, M. (2005). Recent developments in the phylogeny and classification of Ralstonia solanacearum. Leuven, Belgium: International Society for Horticultural Science (ISHS), 127–136.
Ralston, E., Palleroni, N., and Doudoroff, M. (1973). Pseudomonas pickettii, a new species of clinical origin related to Pseudomonas solanacearum. Int. J. Syst. Bacteriol. 23, 15–19. doi: 10.1099/00207713-23-1-15
Remenant, B., Babujee, L., Lajus, A., Médigue, C., Prior, P., and Allen, C. (2012). Sequencing of K60, type strain of the major plant pathogen Ralstonia solanacearum. J. Bacteriol. 194, 2742–2743. doi: 10.1128/JB.00249-12
Remenant, B., Coupat-Goutaland, B., Guidot, A., Cellier, G., Wicker, E., Allen, C., et al. (2010). Genomes of three tomato pathogens within the Ralstonia solanacearum species complex reveal significant evolutionary divergence. BMC Genomics 11:379. doi: 10.1186/1471-2164-11-379
Remenant, B., de Cambiaire, J. C., Cellier, G., Jacobs, J. M., Mangenot, S., Barbe, V., et al. (2011). Ralstonia syzygii, the blood disease bacterium and some Asian R. solanacearum strains form a single genomic species despite divergent lifestyles. PLoS One 6:e24356. doi: 10.1371/journal.pone.0024356
Richter, M., Rosselló-Móra, R., Oliver Glöckner, F., and Peplies, J. (2015). JSpeciesWS: A web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32, 929–931. doi: 10.1093/bioinformatics/btv681
Sabbagh, C. R. R., Carrere, S., Lonjon, F., Vailleau, F., Macho, A. P., Genin, S., et al. (2019). Pangenomic type III effector database of the plant pathogenic Ralstonia spp. PeerJ 7:e7346. doi: 10.7717/peerj.7346
Safni, I., Cleenwerck, I., De Vos, P., Fegan, M., Sly, L., and Kappler, U. (2014). Polyphasic taxonomic revision of the Ralstonia solanacearum species complex: Proposal to emend the descriptions of Ralstonia solanacearum and Ralstonia syzygii and reclassify current R. syzygii strains as Ralstonia syzygii subsp. syzygii subsp. nov., R. solanacearum phylotype IV strains as Ralstonia syzygii subsp. indonesiensis subsp. nov., banana blood disease bacterium strains as Ralstonia syzygii subsp. celebesensis subsp. nov. and R. solanacearum phylotype I and III strains as Ralstonia pseudosolanacearum sp. nov. Int. J. Syst. Evol. Microbiol. 64, 3087–3103. doi: 10.1099/ijs.0.066712-0
Salanoubat, M., Genin, S., Artiguenave, F., Gouzy, J., Mangenot, S., Arlat, M., et al. (2002). Genome sequence of the plant pathogen Ralstonia solanacearum. Nature 415, 497–502. doi: 10.1038/415497a
Sasser, M. (1990). MIDI technical note 101. Identification of bacteria by gas chromatography of cellular fatty acids. Newark, DE: MIDI, 1–7.
Sayers, E. W., Cavanaugh, M., Clark, K., Pruitt, K. D., Schoch, C. L., Sherry, S. T., et al. (2021). GenBank. Nucleic Acids Res. 49, D92–D96. doi: 10.1093/nar/gkaa1023
Sharma, P., Johnson, M. A., Mazloom, R., Allen, C., Heath, L. S., Lowe-Power, T. M., et al. (2022). Meta-analysis of the Ralstonia solanacearum species complex (RSSC) based on comparative evolutionary genomics and reverse ecology. Microb. Genomics 8:000791. doi: 10.1099/mgen.0.000791
Sievers, F., and Higgins, D. G. (2021). “The clustal omega multiple alignment package” in Multiple sequence alignment: Methods and protocols. ed. K. Katoh (New York, NY: Springer US), 3–16.
Smith, E. F. (1896). A bacterial disease of the tomato, eggplant and Irish potato (Bacillus solanacearum nov. sp.). Bull. Divis. Veg. Physiol. Pathol. 12, 1–28.
Spraker, J. E., Sanchez, L. M., Lowe, T. M., Dorrestein, P. C., and Keller, N. P. (2016). Ralstonia solanacearum lipopeptide induces chlamydospore development in fungi and facilitates bacterial entry into fungal tissues. ISME J. 10, 2317–2330. doi: 10.1038/ismej.2016.32
Spraker, J. E., Wiemann, P., Baccile, J. A., Venkatesh, N., Schumacher, J., Schroeder, F. C., et al. (2018). Conserved responses in a war of small molecules between a plant-pathogenic bacterium and fungi. mBio 9:e00820-18. doi: 10.1128/mBio.00820-18
Steidl, O. R., Truchon, A. N., Hayes, M. M., and Allen, C. (2021). Complete genome resources for Ralstonia bacterial wilt strains UW763 (Phylotype I); Rs5 and UW700 (Phylotype II); and UW386, RUN2474, and RUN2279 (Phylotype III). Mol. Plant-Microbe Interact. 34, 1212–1215. doi: 10.1094/mpmi-04-21-0086-a
Stork, W., Kim, J.-G., and Mudgett, M. B. (2015). Functional analysis of plant defense suppression and activation by the Xanthomonas core type III effector XopX. Mol. Plant Microbe Interact. 28, 180–194. doi: 10.1094/mpmi-09-14-0263-r
Sun, Y., Li, P., Deng, M., Shen, D., Dai, G., Yao, N., et al. (2017). The Ralstonia solanacearum effector RipAK suppresses plant hypersensitive response by inhibiting the activity of host catalases. Cell. Microbiol. 19:e12736. doi: 10.1111/cmi.12736
Sun, T., Wu, W., Wu, H., Rou, W., Zhou, Y., Zhuo, T., et al. (2020). Ralstonia solanacearum elicitor RipX induces defense reaction by suppressing the mitochondrial atpA gene in host plant. Int. J. Mol. Sci. 21:2000. doi: 10.3390/ijms21062000
Utturkar, S. M., Klingeman, D. M., Land, M. L., Schadt, C. W., Doktycz, M. J., Pelletier, D. A., et al. (2014). Evaluation and validation of de novo and hybrid assembly techniques to derive high-quality genome sequences. Bioinformatics 30, 2709–2716. doi: 10.1093/bioinformatics/btu391
Vandamme, P., and Coenye, T. (2004). Taxonomy of the genus Cupriavidus: A tale of lost and found. Int. J. Syst. Evol. Microbiol. 54, 2285–2289. doi: 10.1099/ijs.0.63247-0
Vaneechoutte, M., Kampfer, P., De Baere, T., Falsen, E., and Verschraegen, G. (2004). Wautersia gen. nov., a novel genus accommodating the phylogenetic lineage including Ralstonia eutropha and related species, and proposal of Ralstonia [Pseudomonas] syzygii (Roberts et al. 1990) comb. nov. Int. J. Syst. Evol. Microbiol. 54, 317–327. doi: 10.1099/ijs.0.02754-0
Villa, J. E., Tsuchiya, K., Horita, M., Natural, M., Opina, N., and Hyakumachi, M. (2005). Phylogenetic relationships of Ralstonia solanacearum species complex strains from Asia and other continents based on 16S rDNA, endoglucanase, and hrpB gene sequences. J. Gen. Plant Pathol. 71, 39–46. doi: 10.1007/s10327-004-0156-1
Wang, Y., Zhao, A., Morcillo, R. J. L., Yu, G., Xue, H., Rufian, J. S., et al. (2021). A bacterial effector protein uncovers a plant metabolic pathway involved in tolerance to bacterial wilt disease. Mol. Plant 14, 1281–1296. doi: 10.1016/j.molp.2021.04.014
Xu, L., Dong, Z., Fang, L., Luo, Y., Wei, Z., Guo, H., et al. (2019). OrthoVenn2: A web server for whole-genome comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res. 47, W52–W58. doi: 10.1093/nar/gkz333
Xu, J., Zheng, H.-J., Liu, L., Pan, Z.-C., Prior, P., Tang, B., et al. (2011). Complete genome sequence of the plant pathogen Ralstonia solanacearum strain Po82. J. Bacteriol. 193, 4261–4262. doi: 10.1128/JB.05384-11
Yabuuchi, E., Kosako, Y., Yano, I., Hotta, H., and Nishiuchi, Y. (1995). Transfer of two Burkholderia and an Alcaligenes species to Ralstonia gen. nov. Microbiol. Immunol. 39, 897–904. doi: 10.1111/j.1348-0421.1995.tb03275.x
Yan, J., Li, P., Wang, X., Zhu, M., Shi, H., Yu, G., et al. (2022). RasI/R quorum sensing system controls the virulence of Ralstonia solanacearum strain EP1. Appl. Environ. Microbiol. 88:e0032522. doi: 10.1128/aem.00325-22
Yoon, S.-H., Ha, S.-M., Kwon, S., Lim, J., Kim, Y., Seo, H., et al. (2017). Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 67, 1613–1617. doi: 10.1099/ijsem.0.001755
Yu, G., Derkacheva, M., Rufian, J. S., Brillada, C., Kowarschik, K., Jiang, S., et al. (2022). The Arabidopsis E3 ubiquitin ligase PUB4 regulates BIK1 and is targeted by a bacterial type-III effector. EMBO J. 41:e107257. doi: 10.15252/embj.2020107257
Zheng, X., Li, X., Wang, B., Cheng, D., Li, Y., Li, W., et al. (2019). A systematic screen of conserved Ralstonia solanacearum effectors reveals the role of RipAB, a nuclear-localized effector that suppresses immune responses in potato. Mol. Plant Pathol. 20, 547–561. doi: 10.1111/mpp.12774
Keywords: Burkholderiaceae, Ralstonia, sp. nov., phylotype I, phylogeny, Ralstonia solanacearum species complex, average nucleotide identity (ANI), digital DNA–DNA hybridization (dDDH)
Citation: Liu J-Y, Zhang J-F, Wu H-L, Chen Z, Li S-Y, Li H-M, Zhang C-P, Zhou Y-Q and Lu C-H (2023) Proposal to classify Ralstonia solanacearum phylotype I strains as Ralstonia nicotianae sp. nov., and a genomic comparison between members of the genus Ralstonia. Front. Microbiol. 14:1135872. doi: 10.3389/fmicb.2023.1135872
Edited by:
Kanika Bansal, Institute of Microbial Technology (CSIR), IndiaReviewed by:
Sanjeet Kumar, Max Planck Institute for the Science of Human History (MPI-SHH), GermanyTeresa Ann Coutinho, University of Pretoria, South Africa
Copyright © 2023 Liu, Zhang, Wu, Chen, Li, Li, Zhang, Zhou and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Can-Hua Lu, bHVjYW5odWExOTg1QDE2My5jb20=; Yuan-Qing Zhou, eXF6aG91QHl4bnUuZWR1LmNu
†ORCID: Jun-Ying Liu https://orcid.org/0000-0002-8388-1557
Can-Hua Lu https://orcid.org/0000-0002-9165-5138
 Jun-Ying Liu
Jun-Ying Liu Jian-Feng Zhang3
Jian-Feng Zhang3 Can-Hua Lu
Can-Hua Lu