- School of Ecology and Nature Conservation, Beijing Forestry University, Beijing, China
Brown-rot fungi account for a small portion of the wood-decaying fungi. There are a few corticioid genera causing brown rot of wood, and their species diversity is still under investigated and studied, especially in subtropical and tropical areas. Two new brown-rot corticioid fungi, Coniophora beijingensis and Veluticeps subfasciculata were found during the investigation of corticioid fungi in China. Phylogenetic analyses of the two genera were carried out separately based on ITS-28S sequence data. Coniophora beijingensis was collected from Beijing, north China, from different kinds of angiosperm and gymnosperm trees, and is characterized by possessing a monomitic hyphal system with colorless hyphae and relatively small pale yellow basidiospores 7–8.6 μm× 4.5–6 μm. Veluticeps subfasciculata was collected from Guizhou and Sichuan Provinces, southwestern China, on Cupressus and is characterized by the resupinate to effused-reflexed basidiomes with a colliculose hymenophore, nodose-septate generative hyphae, fasciculate skeletocystidia and subcylindrical to subfusiform basidiospores 8–11 μm × 2.5–3.5 μm. Descriptions and illustrations are provided for the two new species, and identification keys to Coniophora and Veluticeps species in China are given. Coniophora fusispora is reported in China for the first time.
Introduction
Modern phylogenetic analyses indicated that corticioid brown-rot fungi distributed in several main lineages of Agaricomycetes, such as, Boletales, Polyporales, Gloeophyllales, and Amylocorticiales (Larsson, 2007; Hibbett et al., 2014). The total number of this group of fungi is small with some relatively small but important genera, among which Coniophora DC. and Veluticeps Cooke are two typical ones.
Coniophora is an old genus and includes 170 names (Index of Fungorum)1 but many of them are synonyms (Ginns, 1982). Morphologically, species of Coniophora are characterized by having resupinate basidiomes with a smooth to tuberculate hymenophore, usually simple-septate (occasionally nodose-septate) generative hyphae, thick-walled, pale yellow to brown basidiospores with dextrinoid and cyanophilous reactions, and lacking cystidia (present in one species, Ginns, 1982; Bernicchia and Gorjón, 2010). Leucogyrophana Pouzar and Serpula (Pers.) Gray are two similar genera but differ in having merulioid to irpicoid hymenophores and nodose-septate generative hyphae (Bernicchia and Gorjón, 2010). Phylogenetically, Coniophora formed a well-supported lineage in Boletales and was closely related to Coniophoropsis Hjortstam & Ryvarden, which have distinct ornamented basidiospores (Hjortstam and Ryvarden, 1986; Zhao et al., 2018).
Veluticeps Cooke, typified by V. berkeleyana Cooke, is characterized by the resupinate, effused-reflexed or pileate basidiomes with a smooth, warted or odontioid hymenophore, a monomitic hyphal system with simple- or nodose-septate hyphae, single or fasciculate cystidia and ellipsoid or cylindrical basidiospores negative in Melzer’s reagent (Nakasone, 1990; Bernicchia and Gorjón, 2010). Phylogenetic studies indicated that Veluticeps is a member of the small order Gloeophyllales, which include the famous brown-rot poroid genus, Gloeophyllum P. Karst. (Larsson, 2007; Garcia-Sandoval et al., 2011; He and Li, 2013; Yang et al., 2016). Nakasone (1990, 2004) did monographic studies on the genus and other similar genera. The genus now comprises about 11 species with several new species were described from China based on morphological and molecular evidence recently (He and Li, 2013; Yang et al., 2016; Yang and He, 2016).
Although the poroid brown-rot fungi in China have been intensively studied in recent years (Han et al., 2016; Shen et al., 2019; Liu et al., 2021), the species diversity of the corticioid ones are still largely unknown. Preliminary morphological and molecular studies on the corticioid specimens collected from China recently revealed two undescribed species. In this paper, we carried out two independent phylogenetic analyses based on ITS-28S sequence data, and describe and illustrate the two new species as Coniophora beijingensis and Veluticeps subfasciculata.
Materials and methods
Specimen collection
In situ photos of specimens were taken with a Canon camera EOS 70D (Canon Corporation, Japan). Specimens were dried with a portable dryer, labelled, then stored in a freezer at minus 40°C for 2 weeks to kill the insects and their eggs before proceeding with morphological and molecular studies. Voucher specimens are deposited at the herbarium of Beijing Forestry University, Beijing, China (BJFC). Herbarium code designations follow Index Herbarium.2
Morphological studies
Thin, freehand sections were made from dried basidiomes and mounted in 2% (weight/volume) aqueous potassium hydroxide (KOH) and 1% (w/v) aqueous phloxine. Amyloidity and dextrinoidity of hyphae and basidiospores were checked in Melzer’s reagent (IKI). Cyanophily of hyphal and basidiospore walls was observed in 1% (w/v) cotton blue in 60% (w/v) lactic acid (CB). Microscopic examinations were carried out with a Nikon Eclipse 80i microscope (Nikon Corporation, Japan) at magnifications up to 1,000 ×. Drawings were made with the aid of a drawing tube. The following abbreviations are used: L = mean basidiospore length, W = mean basidiospore width, Q = L/W ratio, n (a/b) = number of basidiospores (a) measured from number of specimens (b). Color codes and names follow Kornerup and Wanscher (1978).
DNA extraction and sequencing
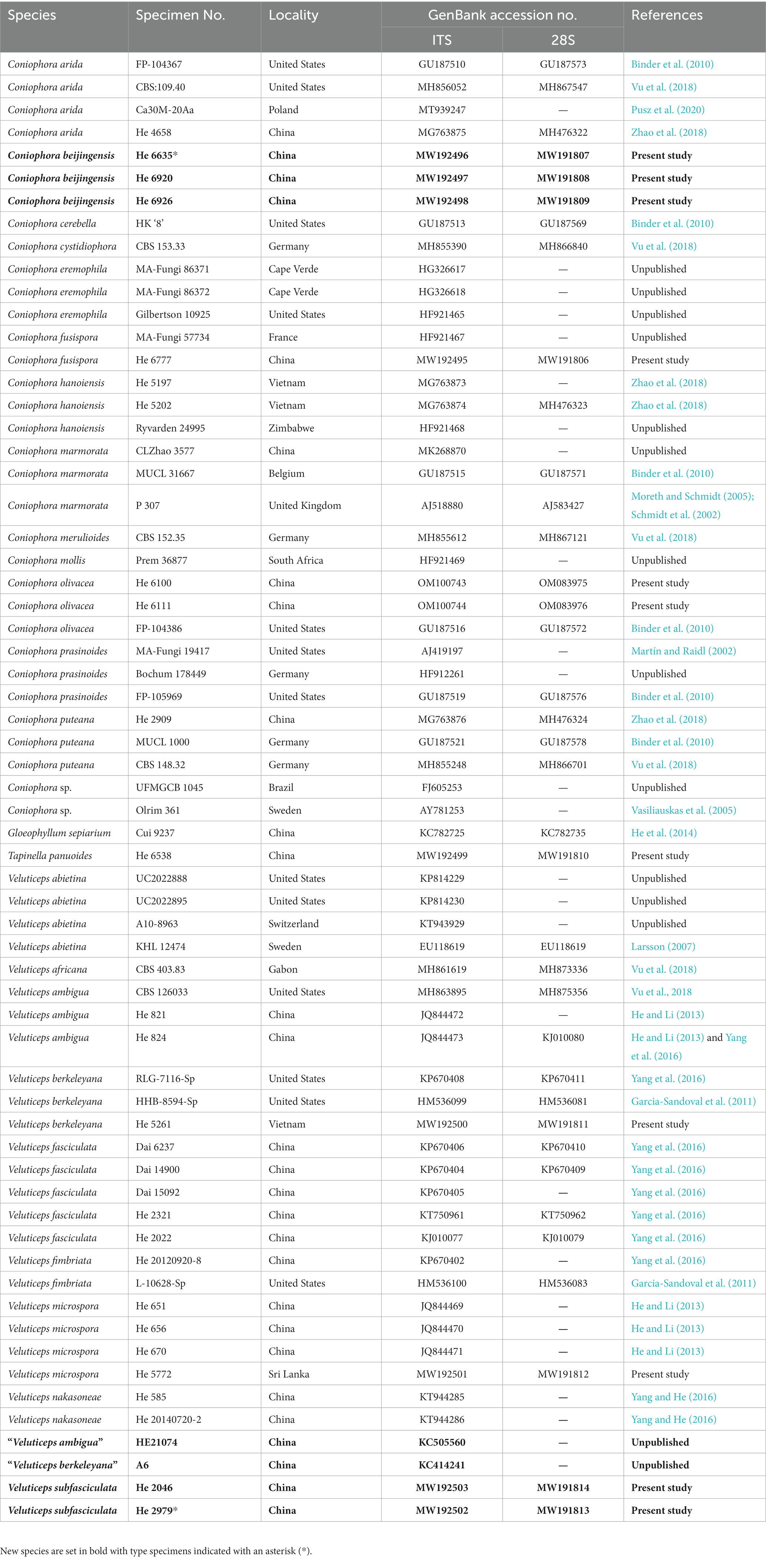
A CTAB plant genomic DNA extraction Kit DN14 (Aidlab Biotechnologies Co., Ltd., Beijing, China) was used to extract total genomic DNA from dried specimens then amplified by the polymerase chain reaction (PCR), according to the manufacturer’s instructions. The ITS1-5.8S-ITS2 region (ITS) was amplified with the primer pair ITS5/ITS4 (White et al., 1990) using the following protocol: initial denaturation at 95°C for 4 min, followed by 34 cycles at 94°C for 40 s, 58°C for 45 s and 72°C for 1 min, and final extension at 72°C for 10 min. The nrLSU D1-D2 region (28S) was amplified with the primer pair LR0R/LR73 employing the following procedure: initial denaturation at 94°C for 1 min, followed by 34 cycles at 94°C for 30 s, 50°C for 1 min and 72°C for 1.5 min, and final extension at 72°C for 10 min. DNA sequencing was performed at Beijing Genomics Institute, and the sequences were deposited in GenBank4 (Table 1). BioEdit v.7.0.5.3 (Hall, 1999) and Geneious Basic v.11.1.15 (Kearse et al., 2012) were used to review the chromatograms and for contig assembly.
Phylogenetic analyses
Two separate datasets of concatenated ITS-28S sequences of the Coniophora and Veluticeps were analyzed. Tapinella panuoides (Fr.) E.-J. Gilbert and Gloeophyllum sepiarium (Wulfen) P. Karst. were selected as the outgroup for the two datasets, respectively. The ITS and 28S sequences were aligned separately using MAFFT v.75 (Katoh et al., 2017) with the G-INS-I iterative refinement algorithm and optimized manually in BioEdit v.7.0.5.3. Then, the separate alignments were concatenated using Mesquite v.3.5.1 (Maddison and Maddison, 2018).
Maximum parsimony (MP), maximum likelihood (ML) analyses and Bayesian inference (BI) were carried out by using PAUP* v.4.0b10 (Swofford, 2002), RAxML v.8.2.10 (Stamatakis, 2014) and MrBayes 3.2.6 (Ronquist et al., 2012), respectively. In MP analysis, trees were generated using 100 replicates of random stepwise addition of sequence and tree-bisection reconnection (TBR) branch-swapping algorithm with all characters given equal weight. Branch supports for all parsimony analyses were estimated by performing 1,000 bootstrap replicates with a heuristic search of 10 random-addition replicates for each bootstrap replicate. In ML analysis, statistical support values were obtained using rapid bootstrapping with 1,000 replicates, with default settings used for other parameters. For BI, the best-fit substitution model was estimated with jModeltest v.2.17 (Darriba et al., 2012). Four Markov chains were run for 2 million generations for the Coniophora dataset and 1 million generations for the Veluticeps dataset until the split deviation frequency value was lower than 0.01. Trees were sampled every 100th generation. The first quarter of the trees, which represented the burn-in phase of the analyses, were discarded, and the remaining trees were used to calculate posterior probabilities (BPP) in the majority rule consensus tree.
Results
Phylogenetic analyses
The Coniophora dataset contained 34 ITS and 21 28S sequences from 34 samples representing 15 ingroup taxa and the outgroup (Table 1). The aligned length was 2040 characters, of which 269 were parsimony informative. MP analysis yielded eight equally parsimonious trees (TL = 784, CI = 0.699, RI = 0.834, RC = 0.583, HI = 0.301). The Veluticeps dataset composed of 29 ITS and 16 28S sequences from 29 samples representing 9 ingroup taxa and the outgroup (Table 1). This dataset had an aligned length of 1,884 characters, of which 277 characters were parsimony informative. MP analysis yielded two equally parsimonious tree (TL = 638, CI = 0.743, RI = 0.893, RC = 0.663, HI = 0.257). jModelTest suggested GTR + I + G and GTR + G as the best-fit models of nucleotide evolution for the Coniophora and Veluticeps datasets, respectively. The average standard deviation of split frequencies of BI was 0.005158 (for the Coniophora dataset) and 0.006744 (for the Veluticeps dataset) at the end of the run. MP and BI analyses resulted in almost identical tree topologies compared to the ML analysis for both datasets. The ML trees of the two genera are shown in Figures 1, 2, respectively, with the parsimony bootstrap values (≥50%, first value), likelihood bootstrap values (≥50%, second value) and Bayesian posterior probabilities (≥0.95, third value) labelled along the branches.
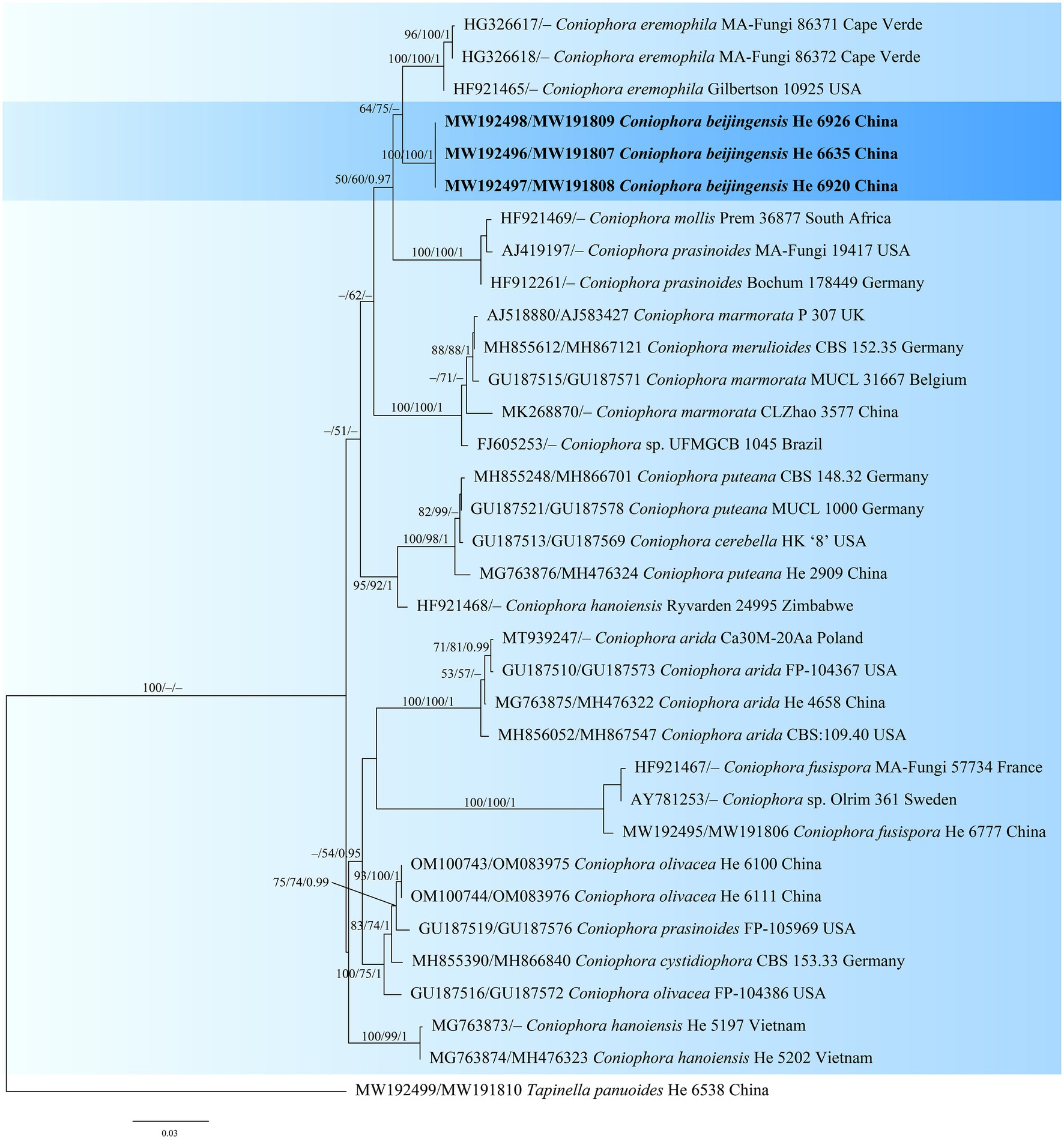
Figure 1. Phylogenetic tree obtained from ML analysis of the ITS-28S sequences of Coniophora. Branches are labelled with parsimony bootstrap values (≥50%, first), likelihood bootstrap values (≥50%, second) and Bayesian posterior probabilities (≥0.95, third). New species are set in bold and highlighted.
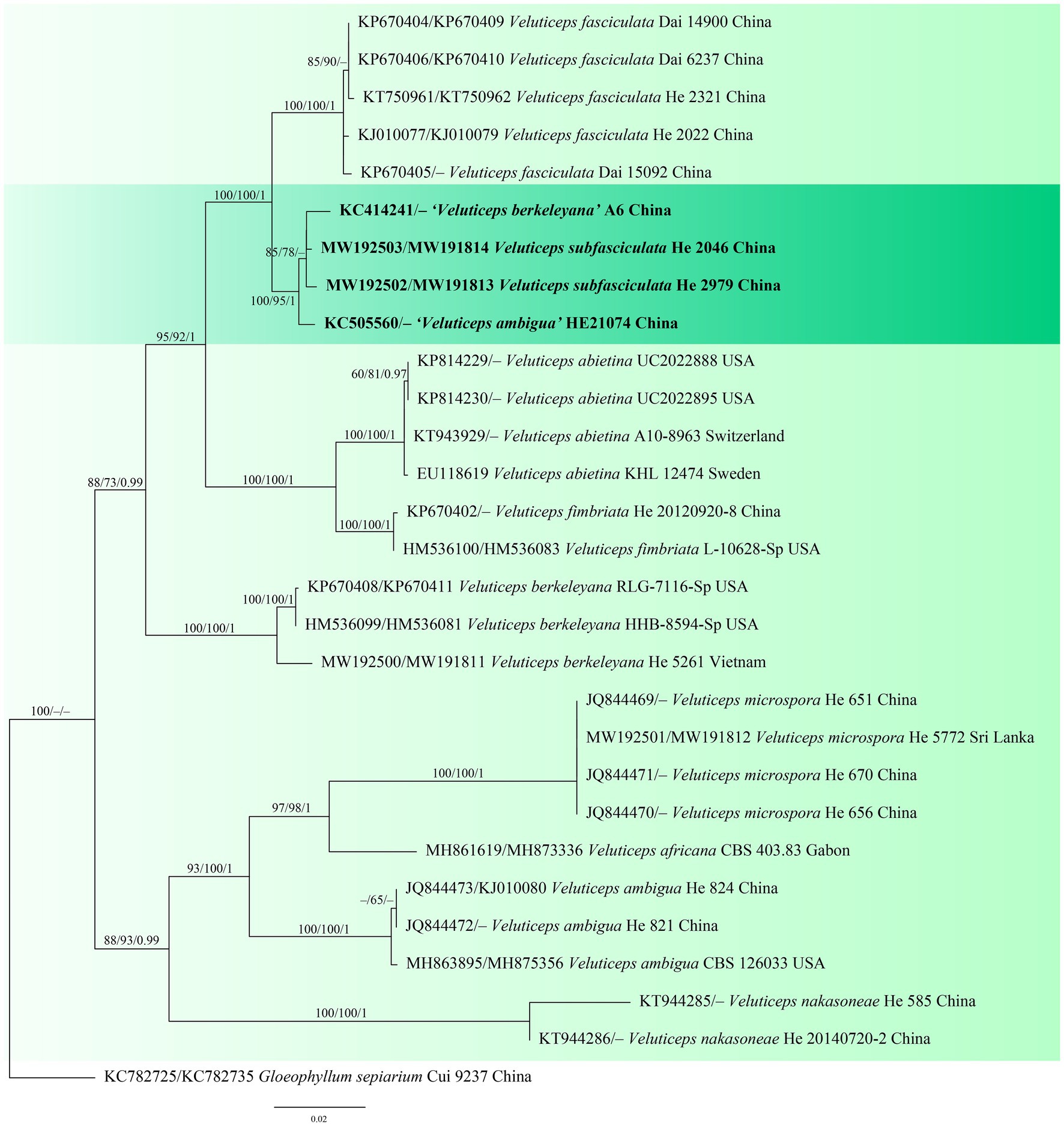
Figure 2. Phylogenetic tree obtained from ML analysis of the ITS-28S sequences of Veluticeps. Branches are labelled with parsimony bootstrap values (≥50%, first), likelihood bootstrap values (≥50%, second) and Bayesian posterior probabilities (≥0.95, third). New species are set in bold and highlighted.
In the trees, the two new species Coniophora beijingensis and Veluticeps subfasciculata formed distinct lineages. Coniophora beijingensis is sister to C. eremophila Lindsey & Gilb. and C. prasinoides (Bourdot & Galzin) Bourdot & Galzin, while Veluticeps subfasciculata is closely related to V. fasciculata Jiao Yang & S.H. He.
Taxonomy
Coniophora beijingensis Yue Li & S.H. He, sp. nov. Figure 3
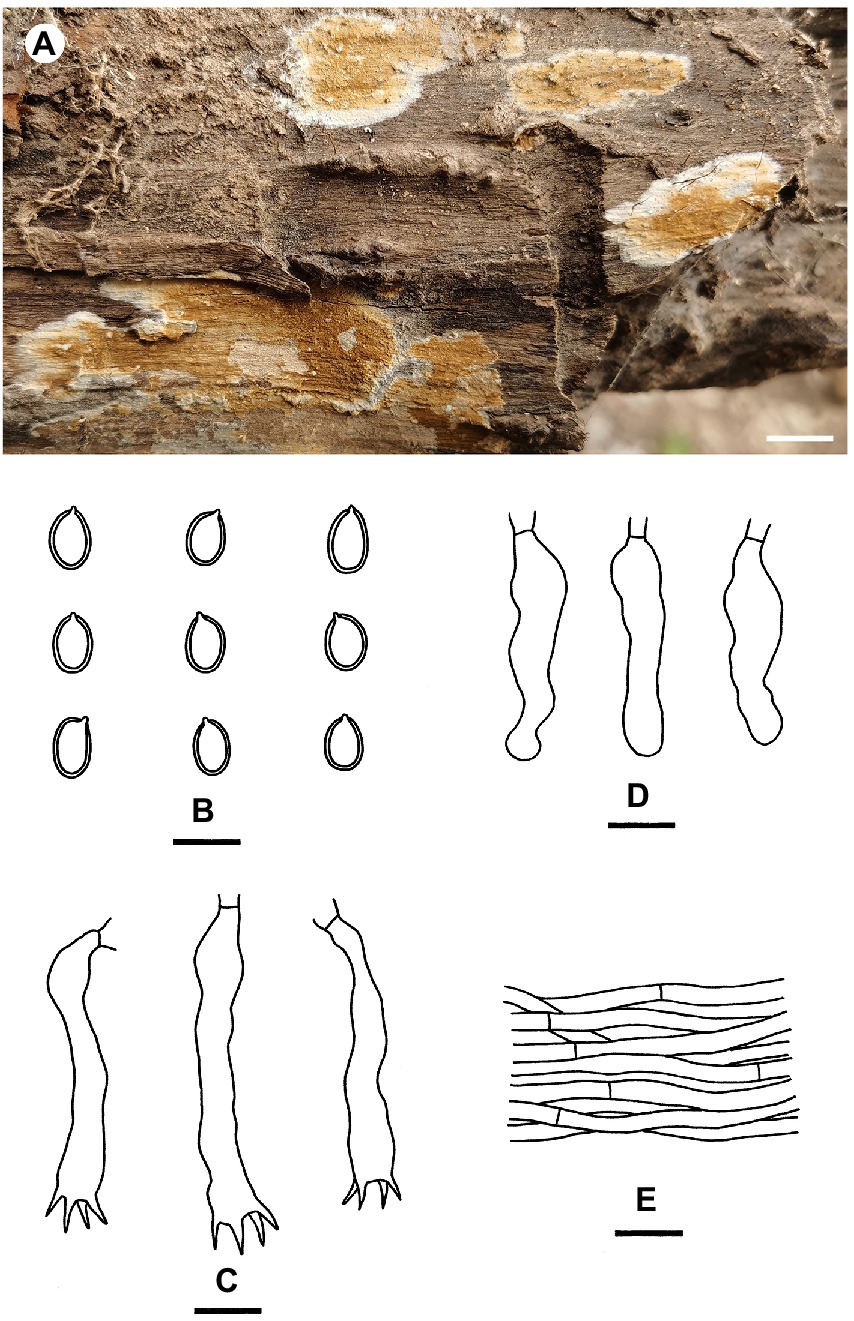
Figure 3. Coniophora beijingensis (from the holotype He 6635; scale bars: a = 1 cm; b–e = 10 μm). (A) Basidiomes; (B) Basidiospores; (C) Basidia; (D) Basidioles; (E) Hyphae from subiculum.
MycoBank: MB847029
Type: China, Beijing, Shijingshan District, Badachu Park, on fallen angiosperm trunk, 30 July 2020, He 6635 (BJFC 033583, holotype).
Etymology: Refers to the type locality in Beijing, China.
Fruiting body: Basidiomes annual, resupinate, effused, closely adnate, inseparable from substrate, membranaceous, first as small patches, later confluent up to 15 cm long, 4 cm wide, 350 μm thick in section. Hymenophore smooth to slightly tuberculate, brownish orange [6C(4–8)] to light brown [6D(4–8)], turning reddish brown in KOH, not cracked; margin thinning out, adnate, without hyphal strands, white to pale yellow, distinct, up to 0.4 cm wide. Context pale yellow.
Microscopic structures: Hyphal system monomitic; all hyphae without clamps. Subiculum distinct, usually with numerous crystals; hyphae colorless, thin- to slightly thick-walled, smooth, rarely branched, moderately septate, tightly interwoven, usually collapsed, 2–5 μm in diam. Subhymenium thickening, composed of collapsed hymenial elements, basidiospores and usually crystals; hyphae colorless, thin-walled, smooth, rarely branched, moderately septate, densely interwoven, slightly agglutinated, 1.5–4 μm in diam. Cystidia absent. Basidia clavate to subcylindrical, colorless, thin-walled, smooth, with four sterigmata and a basal septum, 15–35 × 4–7 μm; basidioles in shape similar to basidia but slightly smaller. Basidiospores ellipsoid to ovoid, with a distinct apiculus, pale yellow, distinctly thick-walled, smooth, strongly dextrinoid in Melzer’s reagent, cyanophilous (6.7–) 7–8.6 (−9.7) × 4.5–6 μm, L = 7.5 μm, W = 5.2 μm, Q = 1.37–1.57 (n = 60/2).
Additional specimens examined: China, Beijing, Changping District, Mangshan Forest Park, on dead angiosperm branch, 1 September 2020, He 6920 (BJFC 033869); on fallen decorticated trunk of Pinus, 1 September 2020, He 6926 (BJFC 033875); Haidian District, Baiwangshan Forest Park, on dead Platycladus orientalis tree, 20 August 2022, He 7732 (BJFC 038868); on dead angiosperm branch, 25 August 2022, He 7754 (BJFC 038890); Xishan Forest Park, on fallen angiosperm trunk, 13 August 2022, He 7685 (BJFC 038821); Yangtaishan Forest Park, on fallen trunk of Syringa, 19 August 2022, He 7722 (BJFC 038858).
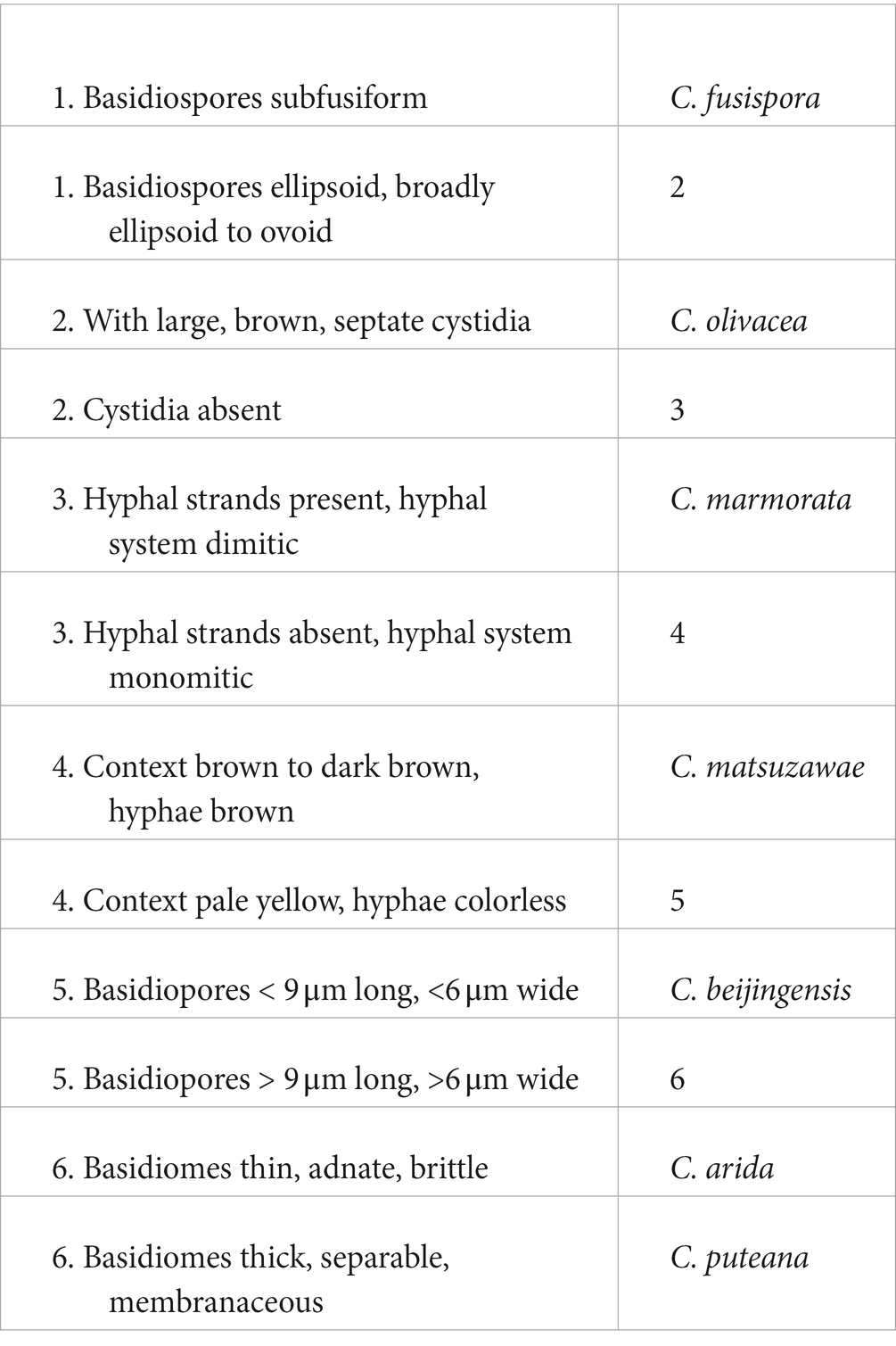
Notes: Coniophora beijingensis is characterized by possessing a monomitic hyphal system with colorless hyphae and relatively small pale yellow basidiospores. Coniophora arida (Fr.) P. Karst. is similar to C. beijingensis in the basidiomes configurations but differs in having larger basidiospores (10.4–13 μm × 6–8 μm, Ginns, 1982). Coniophora prasinoides (Bourdot & Galzin) Bourdot & Galzin is similar to C. beijingensis by sharing the colorless hyphae and small basidiospores (7.4–10 μm × 4.3–6.9 μm) but differs in having hyphal strands, scattered verticillate clamps on hyphae and weakly dextrinoid basidiospores (Ginns, 1982). In the phylogenetic tree (Figure 1), C. beijingensis formed a distinct lineage sister to C. eremophila, which differs in having hyphal strands, loosely interwoven hyphae and larger basidiospores (9.6–11 μm × 6.8–7.6 μm, Ginns, 1982).
Veluticeps subfasciculata Yue Li & S.H. He, sp. nov. Figure 4
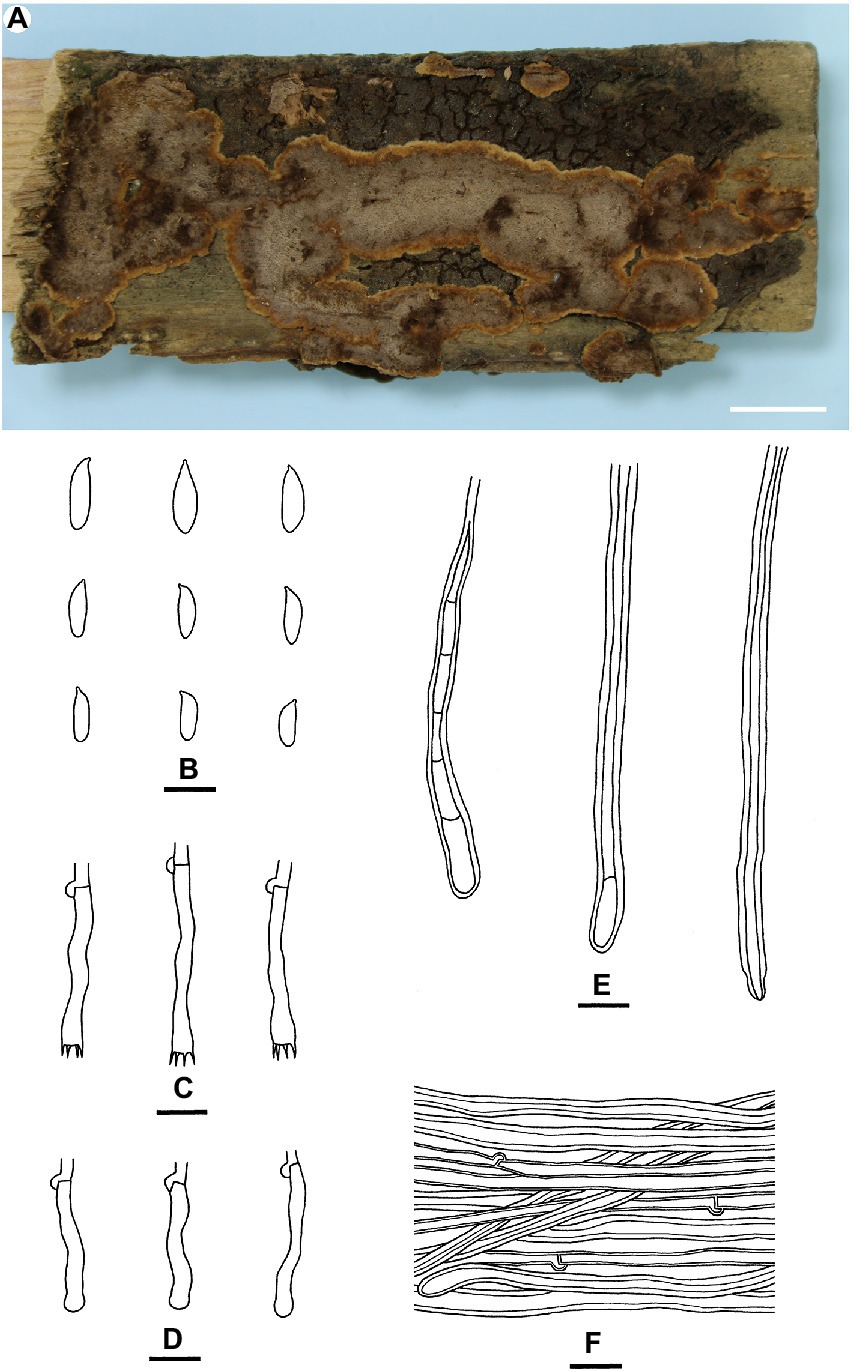
Figure 4. Veluticeps subfasciculata (from the holotype He 2979; scale bars: a = 1 cm; b–f = 10 μm). (A) Basidiomes; (B) Basidiospores; (C) Basidia; (D) Basidioles; (E) Skeletocystidia; (F) Hyphae from subiculum.
MycoBank: MB847030
Type: China, Guizhou Province, Guiyang, Huaxi Park, on fallen trunk of Cupressus, 25 August 2010, He 2979 (BJFC 022046, holotype).
Etymology: Refers to the morphological similarity and close phylogenetic relationship to V. fasciculata Jiao Yang & S.H. He.
Fruiting body: Basidiomes perennial, resupinate to effused-reflexed effused, adnate, separable from substrate, coriaceous to soft corky, first as small patches, later confluent. Reflexed parts protruding up to 2 cm long, 4 cm wide, up to 0.4 cm thick at base; abhymenial surface glabrous, brown [6E(4–8)] to dark brown [6F(4–8)], sulcate and zonate; margin blunt, paler than abhymenial surface. Hymenophore uneven, colliculose to slightly grandinioid with protruding hyphal pegs, greyish brown [6(D–E)3], light brown [6D(4–6)] to brown [6E(4–6)], turning black in KOH, not cracked or deeply cracked with age; margin thinning out, distinct, velvety, adnate or slightly elevated with age, paler than hymenophore, brownish orange [6C(3–5)] to light brown [6D(4–6)], up to 0.1 cm wide. Context light brown.
Microscopic structures: Hyphal system monomitic; generative hyphae with clamps. Abhymenial tomentum usually present, composed of loosely interwoven brown hyphae. Cutis present in aged parts, composed of densely interwoven brown hyphae. Subiculum distinct, thick, composed of more or less horizontally arranged hyphae; subicular hyphae of two types (1) generative hyphae colorless, slightly thick-walled, smooth, moderately septate and branched, 2–4 μm in diam; (2) sclerified hyphae pale yellow to golden yellow, distinctly thick-walled, smooth, with rare clamps, usually with secondary septa, unbranched; transitions of the two kinds of hyphae present. Subhymenium thickening, composed of more or less vertically arranged hyphae, collapsed hymenial elements and skeletocystidia; subhymenial hyphae colorless, slightly thick-walled, smooth, 2–3.5 μm in diam. Skeletocystidia hyphoid, tubular to clavate, originated from subhymenium or subiculum, single or usually with several to many fasciculate together forming the hyphal pegs, colorless, yellow to yellowish brown, distinctly thick-walled, smooth or encrusted with crystals with age, some with one to several secondary septa, with a basal clamp, up to 150 μm long, 3–8 μm wide, protruding up to 80 μm above the hymenium. Basidia subclavate to subcylindrical, colorless, thin-walled, smooth, with four sterigmata and a basal clamp, 20–35 μm × 4–6 μm; basidioles numerous, in shape similar to basidia, but slightly smaller. Basidiospores subcylindrical to subfusiform, colorless, thin-walled, smooth, negative in Melzer’s reagent, acyanophilous, 8–11 × 2.5–3.5 (−4) μm, L = 9.7 μm, W = 3.1 μm, Q = 3–3.2 (n = 90/3).
Additional specimens examined: China, Sichuan Province, Jiange County, Cuiyunlang scenic spot, on fallen trunk of Cupressus, 18 June 2011, He 2046 (BJFC 016644); Jianmenguan scenic spot, on trunk of dead Cupressus, 9 November 2018, Dai 19384 (BJFC 027852).
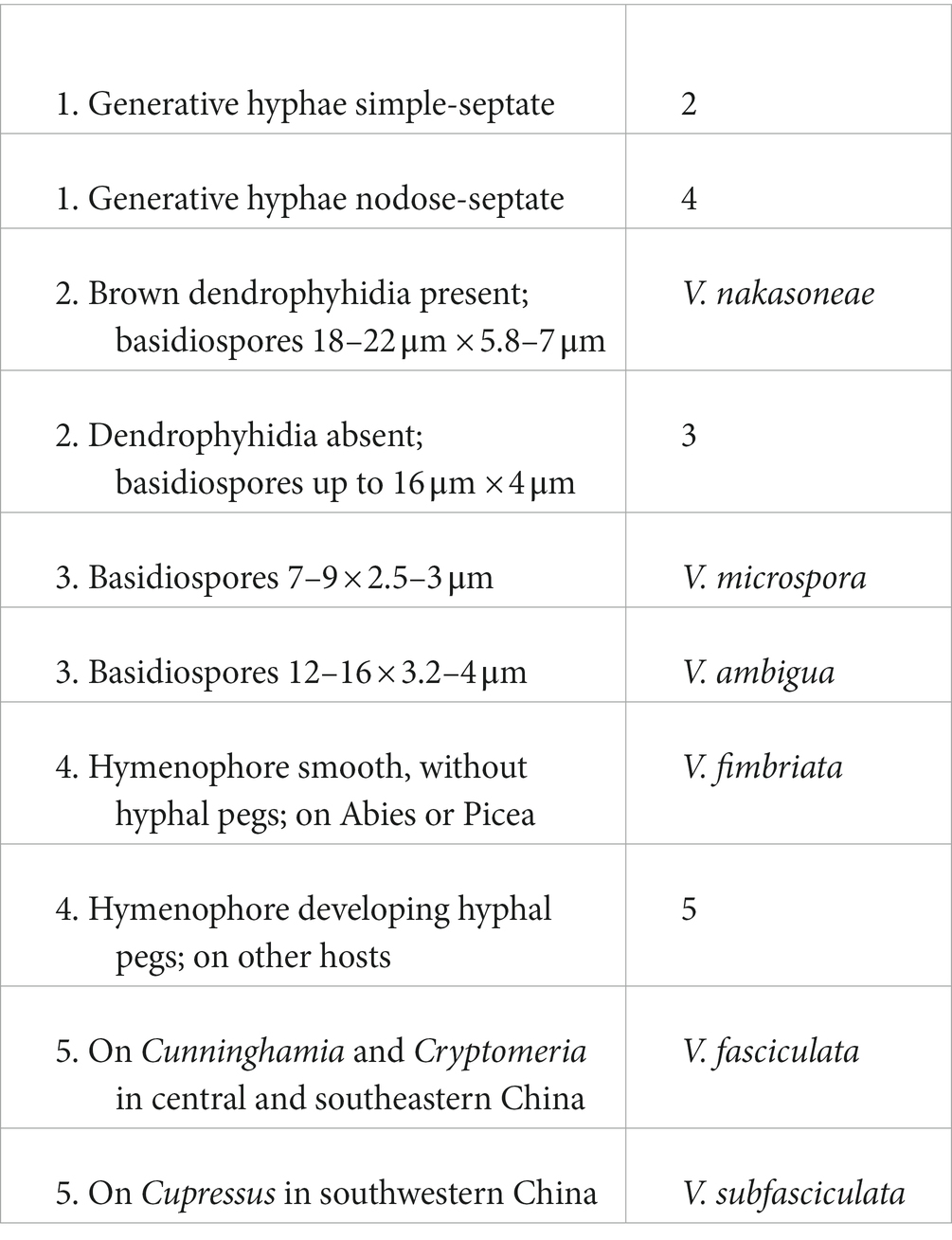
Notes: Veluticeps subfasciculata is characterized by the resupinate to effused-reflexed basidiomes with a colliculose hymenophore, nodose-septate generative hyphae, fasciculate skeletocystidia and growing on trunks of Cupressus in southwestern China. Veluticeps fasciculata is similar to V. subfasciculata by sharing the colliculose hymenophore, nodose-septate generative hyphae and fasciculate skeletocystidia but differs in having longer basidia (30–65 μm) and growing on Cunninghamia and Cryptomeria in central and southeastern China (Yang et al., 2016). Veluticeps berkeleyana is also similar to V. subfasciculata by sharing fasciculate cystidia and nodose-septate generative hyphae but differs in having larger basidiospores (12–14.5 μm × 4–5 μm) and growing on Pinus (Nakasone, 1990). In the phylogenetic tree (Figure 2), two samples from GenBank, “V. berkeleyana isolate A6 (KC414241)” and “V. ambigua isolate HE21074 (KC505560)” both from Sichuan Province, southwestern China clustered with the type specimens with strong support values and formed the V. subfasciculata lineage, which is closely related to V. fasciculata.
Discussion
Although the brown-rot corticioid fungi only account for a small portion of the wood-decaying fungi in nature, they also play important roles in the cycling of substances and renewal of ecosystem. Our present and previous studies (He and Li, 2013; Yang et al., 2016; Yang and He, 2016) indicated that the species diversity of this group of fungi in China is rich and needs more investigations and studies. Future works should be emphasized on the special habitats and hosts, for example, the gymnosperm trees in areas of high altitudes. With more specimens collected and sequenced, the number of this group fungi in China will be largely increased in future.
Coniophora and Veluticeps are two common brown-rot corticoid genera in China. Morphologically, both genera have brown basidiomes, but Coniophora in Boletales can be easily distinguished from Veluticeps in Gloeophyllales by having yellowish-brown, thick-walled basidiospores. Hymenochaete Lév. in Hymenochaetales is similar to Coniophora and Veluticeps by sharing resupinate brown basidiomes, but differs in having characteristic brown setae.
Coniophora fusispora, a characteristic and widely distributed species, is reported from China for the first time based on two specimens collected on the bases of living Pinus tree in Beijing. The species numbers of Coniophora and Veluticeps in China have now become to seven and six with two new species and a new Chinese record reported above. Herein, we present two identification keys to all species of the two genera as follows.
A key to Coniophora species in China
A key to Veluticeps species in China
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/genbank/, MW192496–MW192498; MW191807–MW191809; MW192503; MW192502; MW191814; and MW191813.
Author contributions
YL performed the phylogenetic analyses, did most of the measurements, descriptions and illustrations, and wrote the draft of the manuscript. S-HH designed the research, collected most of the specimens, and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
Financial support was provided by the National Natural Science Foundation of China (nos. 32270014 and 31750001).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
ITS, Internal transcribed spacer; 28S, Nuclear ribosomal large subunit; BJFC, Herbarium of Beijing Forestry University, Beijing, China; KOH, 2% (w/v) potassium hydroxide; IKI, Melzer’s reagent; CB, Cotton blue; IKI–, Neither amyloid nor dextrinoid; CB–, Acyanophilous; L, Mean basidiospore length; W, Mean basidiospore width; Q, L/W ratio, n (a/b), number of basidiospores (a) measured from number of specimens (b); CTAB, Cetyltrimethylammonium bromide; DNA, Deoxyribonucleic acid; PCR, Polymerase chain reaction; MP, Maximum parsimony; ML, Maximum likelihood; BI, Bayesian inference; BPP, Bayesian posterior probability.
Footnotes
1. ^https://www.indexfungorum.org
2. ^http://sweetgum.nybg.org/science/ih/
3. ^https://sites.duke.edu/vilgalyslab/rdna_primers_for_fungi/
References
Bernicchia, A., and Gorjón, S. P. (2010). Corticiaceae s.l. Fungi Europaei 12. Alassio, Italia: Edizioni Candusso.
Binder, M., Larsson, K. H., Matheny, P. B., and Hibbett, D. S. (2010). Amylocorticiales Ord. Nov. and Jaapiales Ord. Nov.: early diverging clades of Agaricomycetidae dominated by corticioid forms. Mycologia 102, 865–880. doi: 10.3852/09-288
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9:772. doi: 10.1038/nmeth.2109
Garcia-Sandoval, R., Wang, Z., Binder, M., and Hibbett, D. S. (2011). Molecular phylogenetics of the Gloeophyllales and relative ages of clades of Agaricomycotina producing a brown rot. Mycologia 103, 510–524. doi: 10.3852/10-209
Ginns, J. (1982). A monograph of the genus Coniophora (Aphyllophorales, basidiomycetes). Opera Botanica 61, 1–61.
Hall, T. A. (1999). Bioedit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98.
Han, M. L., Chen, Y. Y., Shen, L. L., Song, J., Vlasák, J., Dai, Y. C., et al. (2016). Taxonomy and phylogeny of the brown-rot fungi: Fomitopsis and its related genera. Fungal Divers. 80, 343–373. doi: 10.1007/s13225-016-0364-y
He, S. H., and Li, H. J. (2013). Veluticeps microspora sp. nov. and V. ambigua new to Asia with a preliminary phylogenetic study on the genus. Mycol. Prog. 12, 367–374. doi: 10.1007/s11557-012-0842-x
He, S. H., Vlasák, J., and Dai, Y. C. (2014). Hispidaedalea gen. Nov. and Griseoporia taiwanense sp. nov. (Gloeophyllales, Basidiomycota) based on morphological and molecular characters. Mycol. Prog. 13, 833–839. doi: 10.1007/s11557-014-0966-2
Hibbett, D. S., Bauer, R., Binder, M., Giachini, A. J., Hosaka, K., Justo, A., et al. (2014). “14 Agaricomycetes” in Systematics and evolution. The Mycota VII part a. eds. D. J. McLaughlin and J. W. Spatafora (Berlin, Heidelberg: Springer), 373–429.
Hjortstam, K., and Ryvarden, L. (1986). Some new and noteworthy fungi (Aphyllophorales, basidiomycetes) from Iguazu, Argentina. Mycotaxon 25, 539–567.
Katoh, K., Rozewicki, J., and Yamada, K. D. (2017). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 20, 1160–1166. doi: 10.1093/bib/bbx108
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Kornerup, A., and Wanscher, J. H. (1978). Methuen handbook of colour. 3rd edn. E. Methuen and Co., Ltd., London.
Larsson, K. H. (2007). Re-thinking the classification of corticioid fungi. Mycol. Res. 111, 1040–1063. doi: 10.1016/j.mycres.2007.08.001
Liu, S., Han, M. L., Xu, T. M., Wang, Y., Wu, D. M., and Cui, B. K. (2021). Taxonomy and phylogeny of the Fomitopsis pinicola complex with descriptions of six new species from East Asia. Front. Microbiol. 12:644979. doi: 10.3389/fmicb.2021.644979
Maddison, W. P., and Maddison, D. R. (2018). Mesquite: a modular system for evolutionary analysis. Version 3.5.1. Available at: http://www.mesquiteproject.org
Martín, M. P., and Raidl, S. (2002). The taxonomic position of Rhizopogon melanogastroides (Boletales). Mycotaxon 84, 221–228.
Moreth, U., and Schmidt, O. (2005). Investigations on ribosomal DNA of indoor wood decay fungi for their characterization and identification. Holzforschung 59, 90–93. doi: 10.1515/HF.2005.014
Nakasone, K. K. (1990). Taxonomic study of Veluticeps (Aphyllophorales). Mycologia 82, 622–641. doi: 10.1080/00275514.1990.12025937
Nakasone, K. K. (2004). Morphological studies in Veluticeps, Pileodon, and related taxa. Sydowia 56, 38–60.
Pusz, W., Baturo-Cieniewska, A., Kaczmarek, A., and Zwijacz-Kozica, T. (2020). The health status of silver fir (Abies alba mill.) in selected locations of the Tatra National Park and the Karkonosze National Park. Acta Scientiarum Polonorum 19, 145–152. doi: 10.17306/J.AFW.2020.3.15
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Hőhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Schmidt, O., Grimm, K., and Moreth, U. (2002). Molecular identity of species and isolates of the Coniphora cellar fungi. Holzforschung 56, 563–571. doi: 10.1515/HF.2002.086
Shen, L. L., Wang, M., Zhou, J. L., Xing, J. H., Cui, B. K., and Dai, Y. C. (2019). Taxonomy and phylogeny of Postia. Multi-gene phylogeny and taxonomy of the brown-rot fungi: Postia and its related genera. Persoonia 42, 101–126. doi: 10.3767/persoonia.2019.42.05
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Swofford, D. L. (2002). PAUP*: Phylogenetic analysis using parsimony (*and other methods). Version 4.0b10. Sinauer Associates, Sunderland, Massachusetts.
Vasiliauskas, R., Larsson, E., Larsson, K. H., and Stenlid, J. (2005). Persistence and long-term impact of rotstop biological control agent on mycodiversity in Picea abies stumps. Biol. Control 32, 295–304. doi: 10.1016/j.biocontrol.2004.10.008
Vu, D., Groenewald, M., De, V. M., Gehrmann, T., Stielow, B., Eberhardt, U., et al. (2018). Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Stud. Mycol. 92, 135–154. doi: 10.1016/j.simyco.2018.05.001
White, T. J., Bruns, T., Lee, S., and Taylor, J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics” in PCR protocols: A guide to methods and applications. eds. M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (San Diego: Academic Press), 315–322.
Yang, J., and He, S. H. (2016). Veluticeps nakasoneae sp.nov. (Gloeophyllales, Basidiomycota) from tropical China. Nova Hedwigia 103, 71–78. doi: 10.1127/nova_hedwigia/2016/0331
Yang, J., Nakasone, K. K., and He, S. H. (2016). Veluticeps fasciculata sp. nov. (Gloeophyllaceae, Basidiomycota), a close relative to V. berkeleyi. Phytotaxa 243, 163–169. doi: 10.11646/phytotaxa.243.2.6
Keywords: Coniophoraceae, Gloeophyllaceae, species diversity, taxonomy, wood-decaying
Citation: Li Y and He S-H (2023) Taxonomy and phylogeny of brown-rot corticioid fungi in China: Coniophora beijingensis and Veluticeps subfasciculata spp. nov. Front. Microbiol. 14:1133236. doi: 10.3389/fmicb.2023.1133236
Edited by:
Jian-Kui Liu, University of Electronic Science and Technology of China, ChinaReviewed by:
Rui-Heng Yang, Shanghai Academy of Agricultural Sciences, ChinaGang He, Jiujiang University, China
Copyright © 2023 Li and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuang-Hui He, heshuanghui@bjfu.edu.cn
 Yue Li
Yue Li Shuang-Hui He
Shuang-Hui He