
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 30 March 2023
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1128905
This article is part of the Research TopicAntimicrobial Resistance: Agriculture, Environment and Public Health within One Health FrameworkView all 17 articles
 Jiangang Ma1,2†
Jiangang Ma1,2† Juan Wang2†
Juan Wang2† Hua Yang1
Hua Yang1 Mengru Su2
Mengru Su2 Ruichao Li3
Ruichao Li3 Li Bai4
Li Bai4 Jie Feng2
Jie Feng2 Yuting Huang1
Yuting Huang1 Zengqi Yang2*
Zengqi Yang2* Biao Tang1*
Biao Tang1*The tigecycline resistance gene tet(X4) was widespread in various bacteria. However, limited information about the plasmid harboring the tet(X4) gene spread among the different species is available. Here, we investigated the transmission mechanisms of the tet(X4) gene spread among bacteria in a pig farm. The tet(X4) positive Escherichia coli, Klebsiella pneumoniae, Enterobacter cloacae and Enterobacter hormaeche were identified in the same farm. The whole genome sequencing (WGS) analysis showed that the K. pneumoniae belonged to ST727 (n = 11) and ST3830 (n = 1), E. cloacae and E. hormaeche belonged to ST524 (n = 1) and ST1862 (n = 1). All tet(X4) genes were located on the IncHI1 plasmids that could be conjugatively transferred into the recipient E. coli C600 at 30°C. Moreover, a fusion plasmid was identified that the IncHI1 plasmid recombined with the IncN plasmid mediated by ISCR2 during the conjugation from strains B12L to C600 (pB12L-EC-1). The fusion plasmid also has been discovered in a K. pneumoniae (K1L) that could provide more opportunities to spread antimicrobial resistance genes. The tet(X4) plasmids in these bacteria are derived from the same plasmid with a similar structure. Moreover, all the IncHI1 plasmids harboring the tet(X4) gene in GenBank belonged to the pST17, the newly defined pMLST. The antimicrobial susceptibility testing was performed by broth microdilution method showing the transconjugants acquired the most antimicrobial resistance from the donor strains. Taken together, this report provides evidence that IncHI1/pST17 is an important carrier for the tet(X4) spread in Enterobacteriaceae species, and these transmission mechanisms may perform in the environment.
Tigecycline, a member of tetracyclines, is one of the last-resort antibiotics to treat infections caused by Carbapenem-Resistant Enterobacteriaceae (CRE). The tigecycline still exhibits antibacterial activities in the bacteria containing the earlier tetracyclines resistance genes. The plasmid-mediated tet(X3) and tet(X4) genes conferring tigecycline resistance were discovered in various Gram-negative bacteria, including carbapenem-resistant and colistin-resistant bacterial strains (He T. et al., 2019; Sun et al., 2019; Tang et al., 2022a; Ma et al., 2022b). The tet(X) variant-positive isolates from animals, retail meat, and humans have been identified (Zheng et al., 2020; Tang et al., 2021; Umar et al., 2021). Despite the large numbers of tet(X) variant genes were discovered, tet(X3) and tet(X4) were the most popular tigecycline resistance genes, especially tet(X4) (Cheng et al., 2020; Guan et al., 2022). The tet(X4) gene was widelydetected in Escherichia coli, Klebsiella pneumoniae, Aeromonas caviae, Citrobacter freundii, Acinetobacter indicus, Enterobacter cloacae and so on (Chen et al., 2019; Fang et al., 2020; Li et al., 2021; Zeng et al., 2021; Wu et al., 2022; Zhai et al., 2022).
IncHI plasmid is an important vector for the tet(X4) gene, which belongs to the H incompatibility (IncH) group, including IncHI1 to IncHI5 subgroups (Phan and Wain, 2008; Cui et al., 2022). The IncHI1 plasmid is a conjugative plasmid usually larger than 200 kb. IncHI1 plasmid usually contains three replication genes (repHI1A, repHI1B and repFIA-like) (Liang et al., 2018). IncHI1 plasmids are thermosensitive for conjugative transfer, and the efficiency is optimal between 22 and 30°C (Phan and Wain, 2008). It is one of the most common plasmids carrying antimicrobial resistance genes (ARGs) in Salmonella (Kubasova et al., 2016). Moreover, IncHI1 plasmids also have been discovered in other Enterobacteriaceae, such as E. coli, K. pneumoniae and C. freundii (Dolejska et al., 2013; Hüttener et al., 2019). Significantly, the tet(X4) positive IncHI1 plasmids were discovered in several species of Enterobacteriaceae (Feng et al., 2022; Gao et al., 2022; Wu et al., 2022).
There are few reports about the mechanism of the tet(X4) gene spread between bacterial species. Here, we screened the tigecycline resistance bacteria from a large-scale pig farm in Guangxi province, China. The tigecycline-resistant E. coli, K. pneumoniae, E. cloacae and Enterobacter hormaechei were isolated at the same time. The mechanisms of the tet(X4) gene transferred among the spaces were unknown. We analyzed the characterization of these strains and compared the ability of conjugative transfer. To the best of our knowledge, this is the first evidence for the IncHI1 and IncHI1-N plasmid harboring the tet(X4) gene transferred in several bacteria spp. in a farm.
Eighty-nine fecal samples were collected from a pig farm in Guangxi province, China, in 2019. These samples were distributed in several stages of the pig’s life, including the piglets, weanling piglets, fattening pigs, and sows. The samples were sent to the laboratory in a cryogenic incubator and screened by the MacConkey agar containing the tigecycline (4 mg/l). The tet(X) gene was detected by PCR as the primer (F: 5’-TGGACCCGTTGGACTGACTA-3′, R: 5’-CACTTCTTCTTACCAGGTTC-3′) and sequenced by Sanger Sequencing for the tigecycline resistant strains. Then the tet(X)-positive strains were identified by 16S rDNA PCR and sequencing.
Whole genome sequencing (WGS) of all isolates was performed using the Illumina HiSeq platform (Yu et al., 2018). The sequences were assembled with SPAdes and analyzed via the CGE server.1 To further characterize the tet(X4) gene in the isolates, three K. pneumoniae strains, one E. cloacae strain and one E. hormaechei were sequenced by the Nanopore MinION platform and assembled by Unicycler. Then the sequence was annotated with the RAST server.2 The novel plasmid multilocus sequence typing MLST (pMLST) of IncHI1 was assigned by PubMLST.
The tet(X4) positive K. pneumoniae, E. cloacae and E. hormaechei were used as the donor strains, and the E. coli C600 (rifampin-resistant) was used as the recipient. In addition, E. coli J53 (sodium azide-resistant) was the recipient when the transconjugants used as the donor (Tang et al., 2019, Tang, B. et al., 2020; Lin et al., 2022). The cultures of donor and recipient strains were mixed in fresh LB stationary at 30°C or 37°C overnight. The mixed cultures were collected by centrifugation, then diluted with PBS. The transconjugants were selected by the LB plate containing 4 μg/ml of tigecycline with rifampin (100 μg/ml) at 37°C overnight. Each experiment was repeated three times.
S1-PFGE was used to detect plasmid size in strains performed as previously described (Ma et al., 2022a,b; Tang et al., 2022b). In brief, the plugs were made from the fresh cultures embedded in 2% gold agarose and lysed with cell lysis buffer. Then S1 nuclease was used to cut the plugs, and the Salmonella H9812 was restricted with XbaI as the marker. The plasmids were separated with the CHEF Mapper XA system.
The minimum inhibitory concentrations (MICs) for 13 antimicrobial agents (ampicillin (AMP), amoxicillin-clavulanate (A/C), gentamicin (GEN), florfenicol (FFC), tetracycline (TET), tigecycline (TIG), ceftiofur (CEF), ceftazidime (CAZ), enrofloxacin (ENR), sulfisoxazole (SUL), imipenem (IMP), meropenem (MEM), colistin (COL)) were determined by the broth microdilution method according to the Clinical and Laboratory Standards Institute (CLSI) as previously described (Ma et al., 2020; Tang et al., 2022a). The wild strains and transconjugants were tested.
A total of twelve (13.48%) K. pneumoniae, one (1.12%) E. cloacae and one (1.12%) E. hormaechei were screened by MacConkey agar with tigecycline (4 mg/l) from 89 swine feces samples in Guangxi Province in China. Furthermore, 26 (29.21%) tet(X4)-positive E. coli had been isolated and reported in a previous study (Feng et al., 2022). All of these strains carrying the tet(X4) genes were identified by PCR and sequencing.
The Multi-Locus Sequence Typing (MLST) analysis showed that 12 isolates of K. pneumoniae belong to ST727 (11/12) and ST3830 (1/12), the E. cloacae belong to ST524 and the E. hormaechei belongs to novel type (ST1862), respectively. The K. pneumoniae showed a clone spread with the ST727 type.
The ARGs prediction showed the strains have various resistance genes, including blaTEM-1, blaOXA-10, blaSHV-11, floR, cmlA1, fosA, mef(B), oqxA, oqxB, qnrS1, sul3, tet(A), tet(X4), aadA12, aph(3′)-Ia, dfrA14, arr-2 and so on (Figure 1). The K. pneumoniae strains 16 l, 29 l, 38 l, 39 l, 312 l, 313 l, 421 l, 423 l, K1L, 3Z1L and B12L belonged to ST727 that have a similar ARGs. They were different from the K. pneumoniae 3Z5L (ST3830), E. cloacae GX1Z-1 l (ST524) and E. hormaechei GX4-8 l (ST1862).
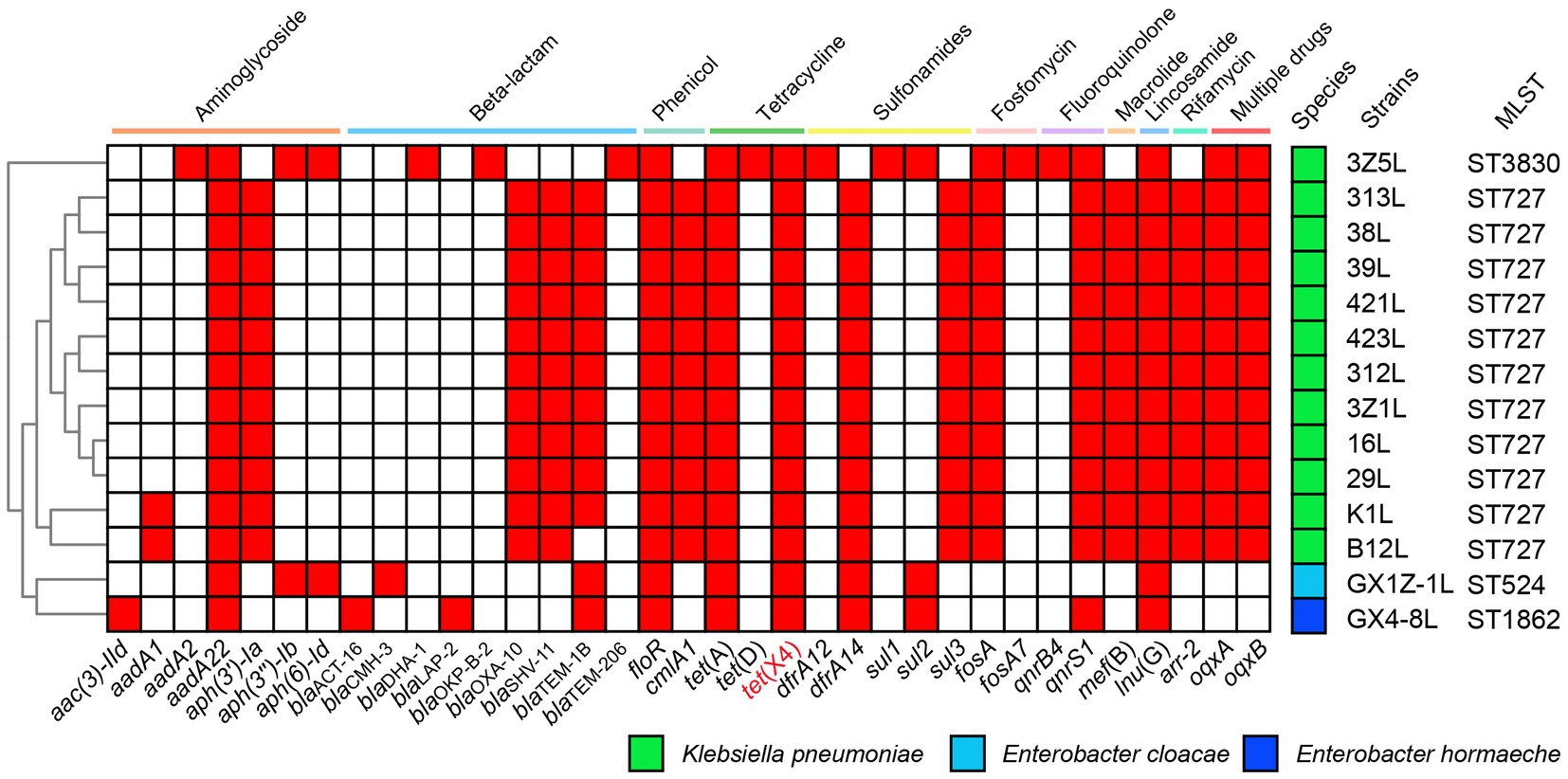
Figure 1. The heat map of antimicrobial resistance genes of 14 strains in this study. The positive genes in strains are marked with the red box. The names of genes are labeled below the heat map.
All strains carried multiple replicons (IncHI1A, IncHI1B and IncFIA), and most K. pneumoniae strains carried IncN replicon (Table 1). The complete sequence of strains K1L, 3Z5L, B12L, GX1Z-1 l and GX4-8 l showed that the tet(X4) gene are located on the IncHI1 plasmid and belongs to a novel pMLST. The IncHI1 plasmid was assigned to pST17, which contained two novel alleles, HCM1_043 (4) and HCM1_116 (5). Moreover, we analyzed 100 IncHI1 plasmid sequences from GenBank database that showed all tet(X4)-positive plasmids belonged to pST17. Although these plasmids were discovered in different spaces, more commonly E. coli, a small amount of K. pneumoniae (MW940615), Salmonella enterica (CP060586), and Citrobacter sp. (MW940627) that have relatively close consanguinity based on the core genes analysis. Similarly, the mcr and blaNDM positive plasmids have a closer relationship (Figure 2). This suggests that the tet(X4), mcr, and blaNDM-positive IncHI1 plasmid in several species were mainly transmitted by cloning.
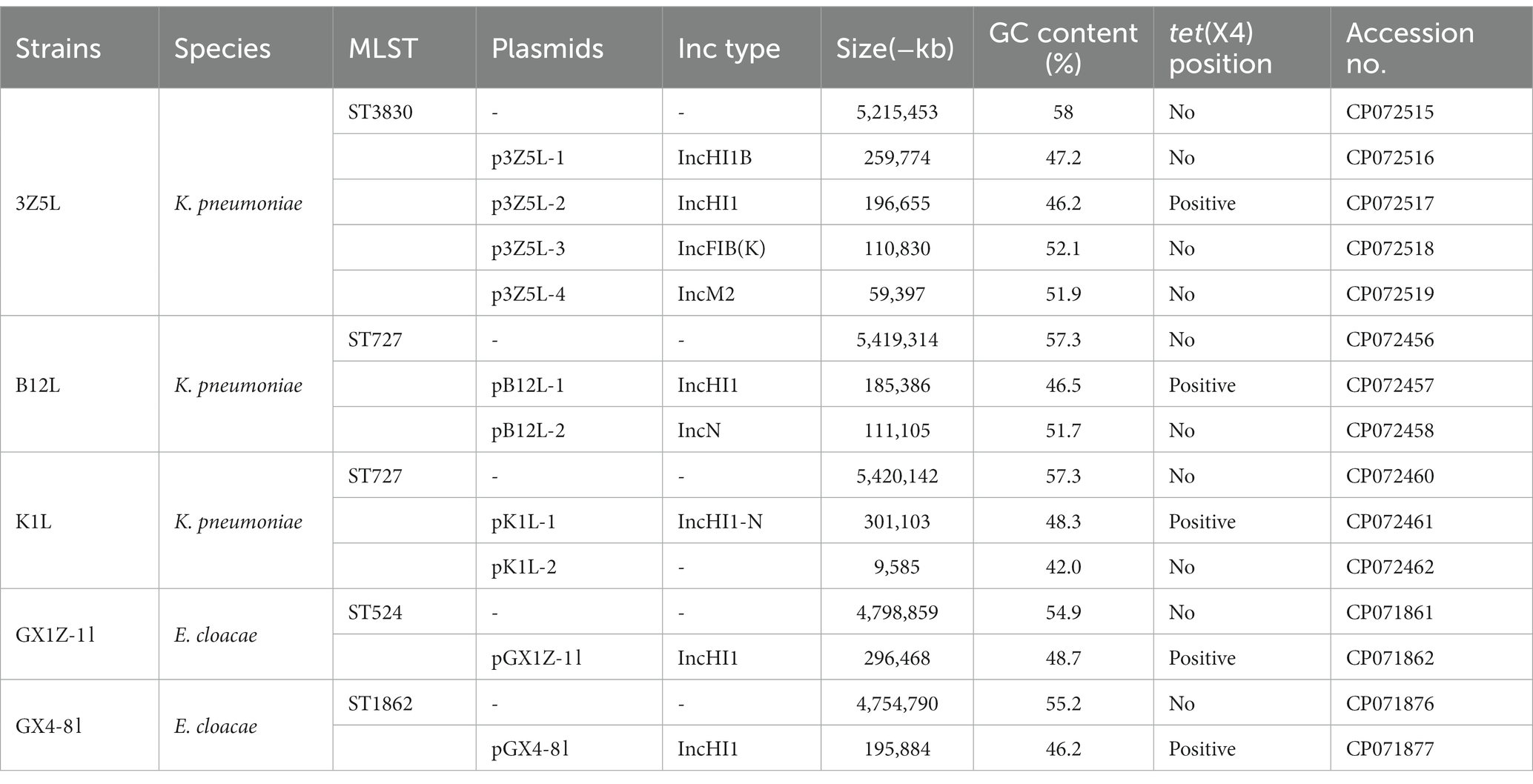
Table 1. The genome characteristics of the K. pneumoniae, E. cloacae and Enterobacter hormaechei strains.
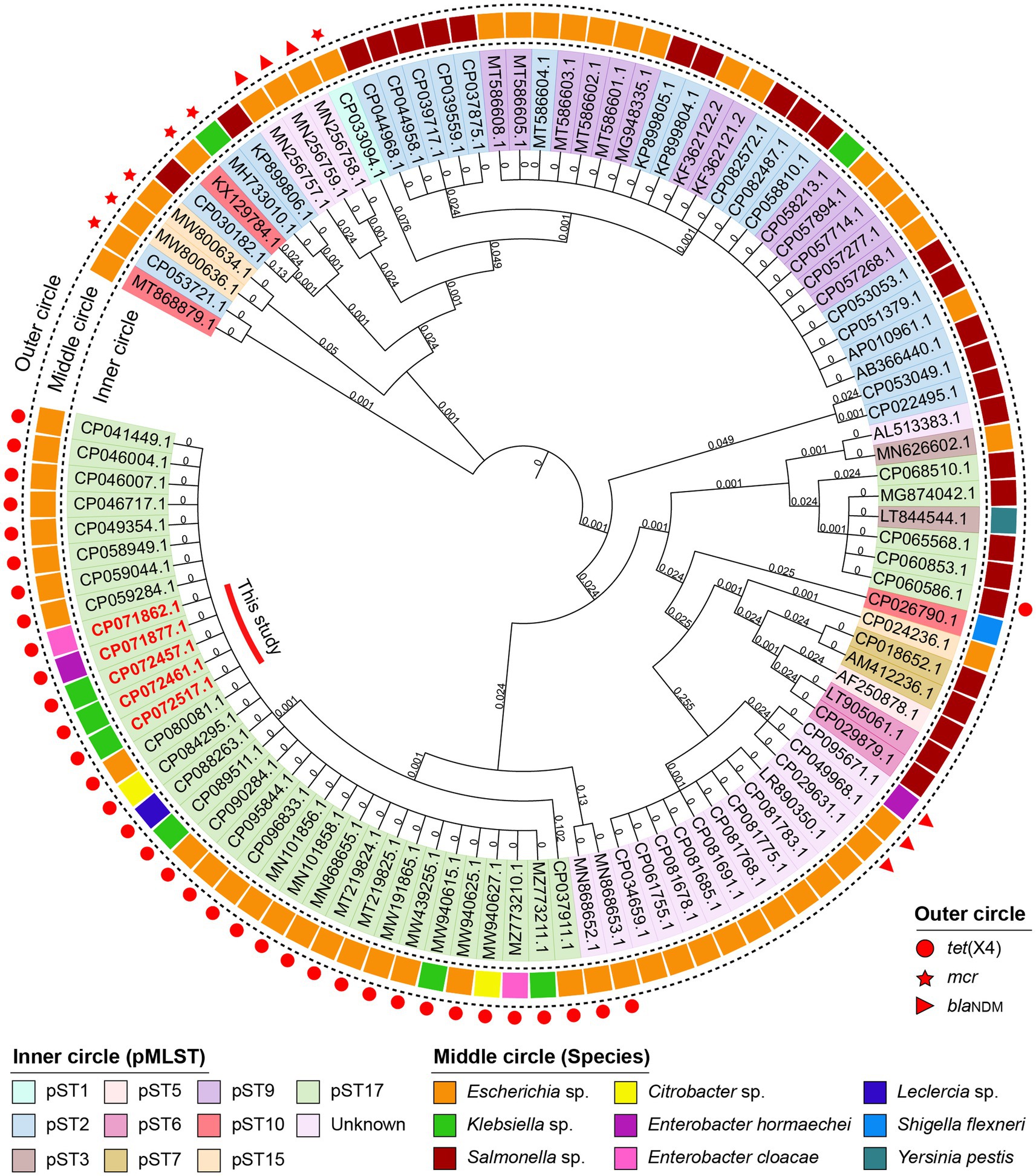
Figure 2. Phylogenetic tree of 100 IncHI1 plasmids with complete sequence from GenBank database. The accession numbers in red are discovered in this study. The innermost to outermost circles indicated the pMLST of plasmids, the species of isolates and the important ARGs on the plasmids.
Klebsiella pneumoniae and E. cloacae tet(X4) positive plasmids have similar backbone structures (Figure 3A). All the IncHI1 plasmids contained the completed conjugation transfer elements including oriT, T4SS and T4CP, but without a conjugation transfer element was identified in IncN plasmid. IncHI1 plasmid-mediated tet(X4) transfer risk in different species is underestimated. The core genetic structures of tet(X4) remained in the conserved sequence as abh-tet(X4)-ISCR2 (Li et al., 2020b). Interestingly, the pK1L (301 kb) is a complex plasmid (IncHI1-N) that is derived by homologous recombination of the IncHI1 (~190 kb) and IncN (~110 kb) from other K. pneumoniae strains. The fusion plasmid IncHI1-N was also detected in the transconjugant (pB12L-EC-1) harboring tet(X4) from the K. pneumoniae B12L (pB12L-1 and pB12L-2) to E. coli C600 mediated by ISCR2 with a ~ 13 kb homologous sequence (ISCR2-virD-floR-lysR-tet(A)-blaTEM-1B-IS26-dfrA14-aadA1-blaOXA-10-cmlA-aadA1-IntI1-IS26) (Figure 3B). This is similar to the previously reported that the mcr-1-bearing plasmids pD72-mcr1 (IncF33: A-: B-) recombined with pD72-F33 (IncN) that was mediated by IS26 (He D. et al., 2019). The fusion plasmid pB12L-EC-1 was highly similar to pK1L.
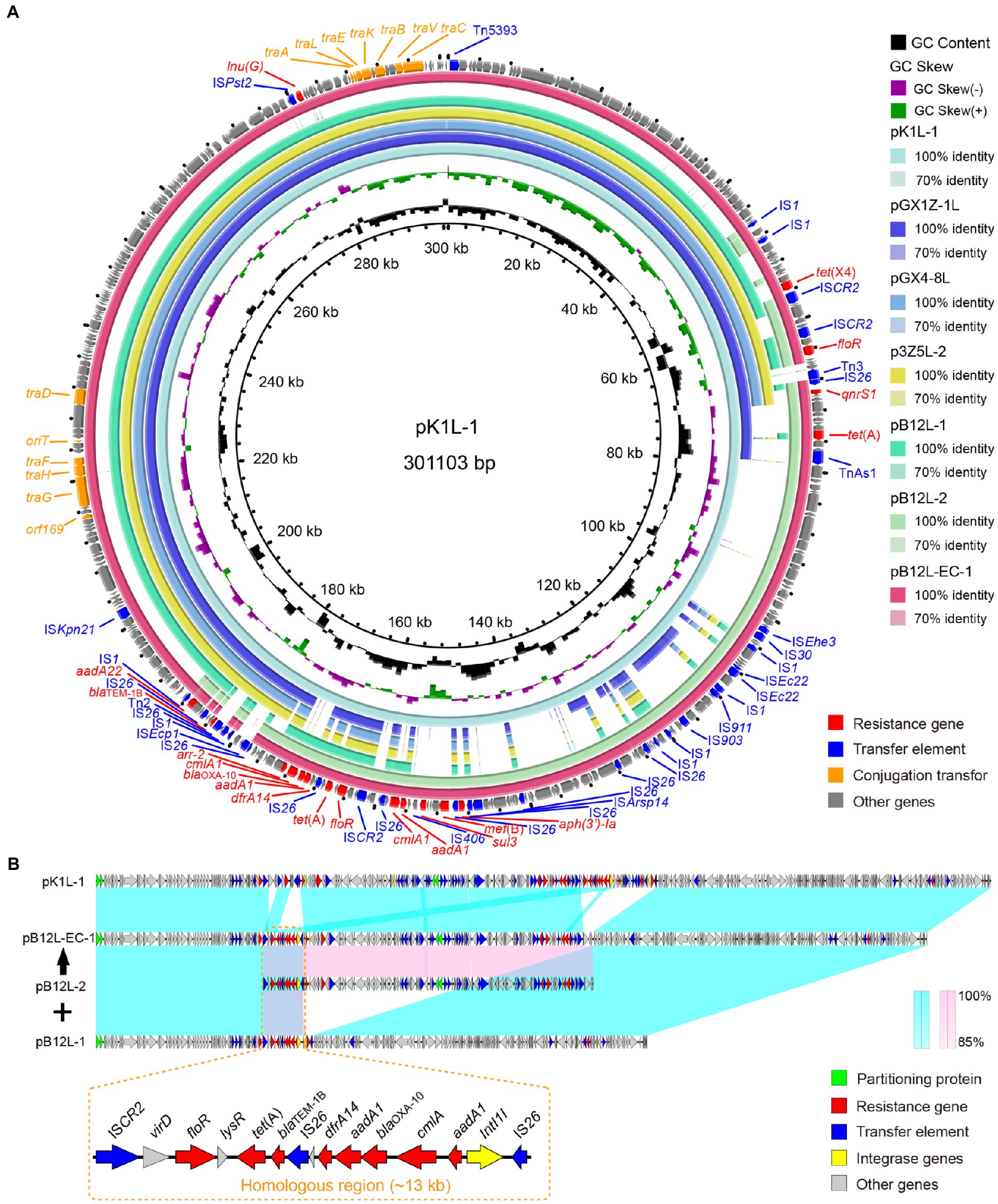
Figure 3. Sequence alignment of tet(X) harboring plasmids in this study. (A) The circular genetic map of plasmids pK1L, pGX1Z-1 l, pGX4-8 l, p3Z5L-2, pB12L-1, pB12L-2 and pB12L-EC-1. The pK1L is used as the reference. (B) Linear comparison of plasmids pK1L, pB12L-1 and pB12L-2 with the transconjugant plasmid pB12L-EC-1. The corresponding alignments are represented in wathet blue and pink. The orange dashed boxes indicated the genes in the homologous region. The arrows and triangles represent genes of different function categories (blue: transfer element; red: AMR gene; green: partitioning protein; golden: integrase and resolvase; gray: other functions).
The conjugation assay was performed to investigate the transferability of the tet(X4) gene in Enterobacteriaceae. As with the characteristics of IncHI1 plasmids, conjugation experiments showed that all strains successfully transfer the tet(X4) gene to recipient strain E. coli C600 with a higher conjugation frequency at 30°C, and a significantly reduced conjugation frequency observed at 37°C (Figure 4A). This is consistent with the characteristics of the thermosensitive plasmids. As mentioned above, the fusion plasmid pB12L-C600-1 containing two replicons, IncHI1 and IncN generated by the conjugation transfer of the K. pneumoniae B12L to E. coli C600. The plasmid profiles of the donor and transconjugants detected by S1-PFGE showed the two sizes plasmids (about 200 kb and 300 kb) in the transconjugants (Figure 4B).
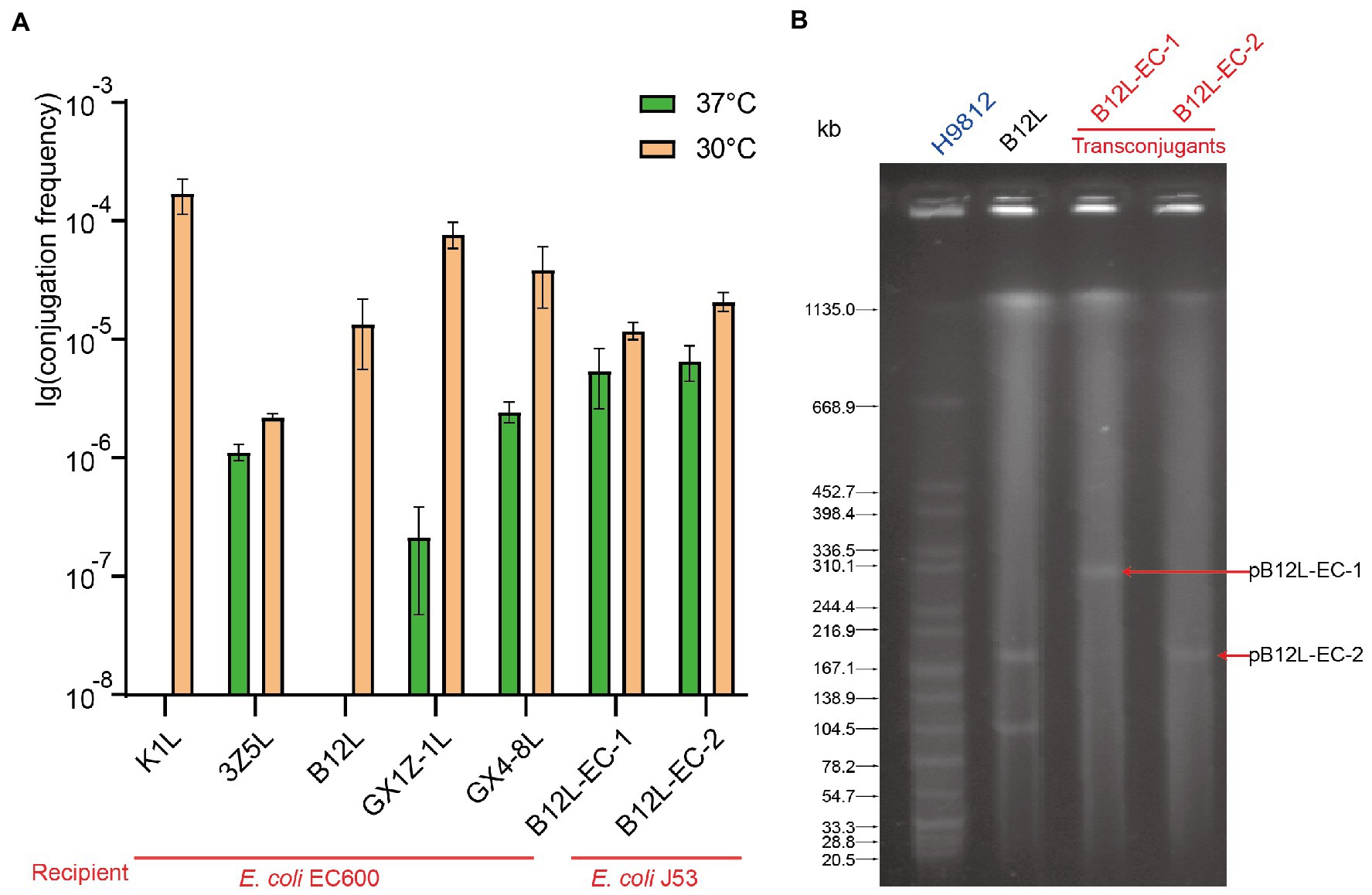
Figure 4. The conjugation transfer frequency of different bacteria and the plasmid profiles of fusion plasmid. (A) The conjugation transfer frequency of five tet(X4) positive strains and the transconjugants of B12L. (B) The S1-PFGE of the donor strain B12L and two types transconjugants (B12L-EC-1 and B12L-EC-2).
In order to analyze the ability of conjugation transfer of the IncHI1 and IncHI1-N plasmids. The transconjugants containing the pB12L-EC-1(IncHI1-N) and pB12L-EC-2(IncHI1) were further conjugated transfer to recipient E. coli J53 at 30°C or 37°C. The result showed that the pB12L-EC-1 and pB12L-EC-2 had no significant difference in conjugation frequency from EC600 to J53, both at 30°C and 37°C (Figure 4A).
The K. pneumoniae, E. cloacae and E. hormaechei, as well as their transconjugants were detected the antimicrobial susceptibility. All of the wild-type strains exhibited resistance to AMP, A/C, FFC, TET, TIG, ENR and SUL, and sensitive to CAZ, IMP, MEM and COL. Only one strain GX4-8 l was resistant to GEN. The transconjugants acquired the most of ARGs that have similar resistant profiles (Supplementary Table S1).
The tet(X4) is the critical resistant gene for the tetracycline family, especially tigecycline. An increasing number of tet(X4)-positive bacteria species have been discovered since it was first discovered in E. coli (He T. et al., 2019; Sun et al., 2019). The E. coli remains the most common carrier for the tet(X4) gene, and the Citrobacter sp., Acinetobacter sp., K. pneumoniae, E. cloacae and E. hormaechei carrying tet(X4) gene were reported occasionally. The different bacteria spp. carrying tet(X4) gene were always isolated from different farms or regions. Here, we identified the tet(X4)-positive K. pneumoniae, E. cloacae and E. hormaechei in a pig farm at the same time. These strains carried the same plasmid that belonged to pST17 IncHI1 plasmid. The tet(X4)-positive IncHI1 plasmid was also identified in E. coli from this farm in our previous study (Feng et al., 2022). It is strong evidence that the IncHI1plasmid mediated the tet(X4) gene transfer among different Enterobacteriaceae.
The tet(X4) gene was always located on the plasmid in Enterobacteriaceae and on chromosomes in other bacteria. The IncX1 type plasmid was considered the most common vector for the tet(X4) gene, followed closely by the IncHI1 plasmid (Cui et al., 2022). The IncHI1 harbored tet(X4) gene is becoming increasingly common in Enterobacteriaceae (Fang et al., 2020; Gao et al., 2022). Notice that it has become the most important type for the spread of the tet(X4) gene among Enterobacteriaceae, except E. coli, which has been discovered in K. pneumoniae and E. cloacae.
pMLST is an important method for tracing the spread of plasmids based on molecular typing. Phan et al. established the typing method for IncHI1 plasmids using variation in six conserved loci (Phan et al., 2009). Seventeen types that have been identified in IncHI1 plasmids and recorded in PubMLST.3 The tet(X4) gene is only located in the pST17 IncHI1 plasmid, and similar results were observed in mcr, blaNDM and blaCTX-M located in a specific plasmid (Valcek et al., 2021). This suggests that there is some association between resistance genes and plasmid typing. The probability of insertion of resistance genes into plasmids is much lower than plasmid transfer.
The plasmid is the most important vector for transferring the ARGs. The ARGs and the virulence genes are always located on the specific Inc-type plasmids, such as the mcr-1 mainly in IncX4, IncI2 and IncHI2 plasmid (Rodríguez-Santiago et al., 2021). Hybrid plasmids are becoming increasingly common, which could contribute to the ARGs and virulence genes co-translocation and assist the non-conjugative plasmids transferred (Li et al., 2020a; Tang, M. et al., 2020). The most of hybrid plasmids were derived during the conjugation transfer, and the insertion sequence IS26 was the most common element for guided the plasmids recombination by the homologous sequence (Liu et al., 2020; Peng et al., 2022). Conversely, the doner plasmids containing the highly similar homologous sequences suggested that the small plasmids may be produced by decomposition of fusion plasmid from other hosts. The mechanism of initial fusion remains to be investigated. Note that the IncHI1 plasmid could remove the temperature restrictions for conjugative transfer by fusion with the IncN plasmid and obtain the resistance and virulence genes at the same time. This study is the first report of the IncHI1 fusion with IncN plasmid that may increase the ability to spread tet(X4).
In summary, we reported the IncHI1 plasmid harboring the tet(X4) gene discovered in K. pneumoniae, E. cloacae, and E. hormaechei from the same farm. It provided further evidence that the tet(X4) gene transfer among bacteria by IncHI1 plasmid in livestock farm. The tet(X4)-positive IncHI1 plasmids belonged to pST17, a novel subtype. Furthermore, the IncHI1 and IncN plasmid tended to fuse by the ISCR2, which could increase the risk of co-transfer the ARGs among bacteria. The study highlights that the IncHI1 plasmid is a risk factor for transfer tet(X4) among Enterobacteriaceae.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
JM and JW designed the study. BT, HY, MS, and JF collected the samples and conducted the experiments. RL, LB, and YH analyzed and interpreted the data. JM, BT, and ZY drafted the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the China Postdoctoral Science Foundation [grant number 2021 M702904], China Wool Sheep Industry and Technology System project [grant number CARS-39], the Natural Science Basic Research Plan in Shaanxi Province of China [grant number 2021KW-41], State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-products [grant number 2010DS700124-ZZ2008 and 2021DG700024-KF202213], “Leading Goose” R&D Program of Zhejiang Province [2023C03045] and Key Research and Development Program of Zhejiang Province [LY23C180001].
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1128905/full#supplementary-material
Chen, C., Chen, L., Zhang, Y., Cui, C. Y., Wu, X. T., He, Q., et al. (2019). Detection of chromosome-mediated tet(X4)-carrying Aeromonas caviae in a sewage sample from a chicken farm. J. Antimicrob. Chemother. 74, 3628–3630. doi: 10.1093/jac/dkz387
Cheng, Y., Chen, Y., Liu, Y., Guo, Y., Zhou, Y., Xiao, T., et al. (2020). Identification of novel tetracycline resistance gene tet(X14) and its co-occurrence with tet(X2) in a tigecycline-resistant and colistin-resistant Empedobacter stercoris. Emerg. Microbes Infect. 9, 1843–1852. doi: 10.1080/22221751.2020.1803769
Cui, C. Y., Li, X. J., Chen, C., Wu, X. T., He, Q., Jia, Q. L., et al. (2022). Comprehensive analysis of plasmid-mediated tet(X4)-positive Escherichia coli isolates from clinical settings revealed a high correlation with animals and environments-derived strains. Sci. Total Environ. 806:150687. doi: 10.1016/j.scitotenv.2021.150687
Dolejska, M., Villa, L., Poirel, L., Nordmann, P., and Carattoli, A. (2013). Complete sequencing of an IncHI1 plasmid encoding the carbapenemase NDM-1, the ArmA 16S RNA methylase and a resistance-nodulation-cell division/multidrug efflux pump. J. Antimicrob. Chemother. 68, 34–39. doi: 10.1093/jac/dks357
Fang, L. X., Chen, C., Cui, C. Y., Li, X. P., Zhang, Y., Liao, X. P., et al. (2020). Emerging high-level tigecycline resistance: novel tetracycline destructases spread via the mobile tet(X). BioEssays 42:e2000014. doi: 10.1002/bies.202000014
Feng, J., Su, M., Li, K., Ma, J., Li, R., Bai, L., et al. (2022). Extensive spread of tet(X4) in multidrug-resistant Escherichia coli of animal origin in Western China. Vet. Microbiol. 269:109420. doi: 10.1016/j.vetmic.2022.109420
Gao, X., He, X., Lv, L., Cai, Z., Liu, Y. Y., and Liu, J. H. (2022). Detection of Tet(X4)-producing Klebsiella pneumoniae from the environment and wide spread of IncFIA-IncHI1A-IncHI1B plasmid carrying tet(X4) in China. J. Glob. Antimicrob. Resist. 30, 130–132. doi: 10.1016/j.jgar.2022.05.028
Guan, C., Tang, B., Yang, H., Ma, J., Huang, Y., and Liu, C. (2022). Emergence of plasmid-mediated tigecycline resistance gene, tet(X4), in Escherichia fergusonii from pigs. J. Glob. Antimicrob. Resist. 30, 249–251. doi: 10.1016/j.jgar.2022.06.029
He, T., Wang, R., Liu, D., Walsh, T. R., Zhang, R., Lv, Y., et al. (2019). Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat. Microbiol. 4, 1450–1456. doi: 10.1038/s41564-019-0445-2
He, D., Zhu, Y., Li, R., Pan, Y., Liu, J., Yuan, L., et al. (2019). Emergence of a hybrid plasmid derived from IncN1-F33:A-:B- and mcr-1-bearing plasmids mediated by IS26. J. Antimicrob. Chemother. 74, 3184–3189. doi: 10.1093/jac/dkz327
Hüttener, M., Prieto, A., Aznar, S., Bernabeu, M., Glaría, E., Valledor, A. F., et al. (2019). Expression of a novel class of bacterial Ig-like proteins is required for IncHI plasmid conjugation. PLoS Genet. 15:e1008399. doi: 10.1371/journal.pgen.1008399
Kubasova, T., Cejkova, D., Matiasovicova, J., Sekelova, Z., Polansky, O., Medvecky, M., et al. (2016). Antibiotic resistance, core-genome and protein expression in IncHI1 plasmids in Salmonella Typhimurium. Genome Biol. Evol. 8, 1661–1671. doi: 10.1093/gbe/evw105
Li, R., Cheng, J., Dong, H., Li, L., Liu, W., Zhang, C., et al. (2020a). Emergence of a novel conjugative hybrid virulence multidrug-resistant plasmid in extensively drug-resistant Klebsiella pneumoniae ST15. Int. J. Antimicrob. Agents 55:105952. doi: 10.1016/j.ijantimicag.2020.105952
Li, R., Li, Y., Peng, K., Yin, Y., Liu, Y., He, T., et al. (2021). Comprehensive genomic investigation of tigecycline resistance gene tet(X4)-bearing strains expanding among different settings. Microbiol. Spectr. 9:e0163321. doi: 10.1128/spectrum.01633-21
Li, R., Lu, X., Peng, K., Liu, Z., Li, Y., Liu, Y., et al. (2020b). Deciphering the structural diversity and classification of the mobile tigecycline resistance gene tet(X)-bearing plasmidome among bacteria. mSystems 5, e00134–e00120. doi: 10.1128/mSystems.00134-20
Liang, Q., Jiang, X., Hu, L., Yin, Z., Gao, B., Zhao, Y., et al. (2018). Sequencing and genomic diversity analysis of IncHI5 plasmids. Front. Microbiol. 9:3318. doi: 10.3389/fmicb.2018.03318
Lin, J., Tang, B., Zheng, X., Chang, J., Ma, J., He, Y., et al. (2022). Emergence of Incl2 plasmid-mediated colistin resistance in avian Escherichia fergusonii. FEMS Microbiol. Lett. 369:fnac016. doi: 10.1093/femsle/fnac016
Liu, Z., Xiao, X., Liu, Y., Li, R., and Wang, Z. (2020). Recombination of NDM-5-producing plasmids mediated by IS26 among Escherichia coli. Int. J. Antimicrob. Agents 55:105815. doi: 10.1016/j.ijantimicag.2019.09.019
Ma, J., Tang, B., Lin, J., Ed-Dra, A., Lin, H., Wu, J., et al. (2022a). Genome assessment of carbapenem- and colistin-resistant Escherichia coli from patients in a sentinel hospital in China. Cells 11:3480. doi: 10.3390/cells11213480
Ma, J., Wang, J., Feng, J., Liu, Y., Yang, B., Li, R., et al. (2020). Characterization of three porcine Acinetobacter towneri strains co-harboring tet(X3) and Bla(OXA-58). Front. Cell. Infect. Microbiol. 10:586507. doi: 10.3389/fcimb.2020.586507
Ma, J., Zhou, W., Wu, J., Liu, X., Lin, J., Ji, X., et al. (2022b). Large-scale studies on antimicrobial resistance and molecular characterization of Escherichia coli from food animals in developed areas of eastern China. Microbiol. Spectr. 10:e0201522. doi: 10.1128/spectrum.02015-22
Peng, K., Yin, Y., Li, Y., Qin, S., Liu, Y., Yang, X., et al. (2022). QitanTech nanopore long-read sequencing enables rapid resolution of complete genomes of multi-drug resistant pathogens. Front. Microbiol. 13:778659. doi: 10.3389/fmicb.2022.778659
Phan, M. D., Kidgell, C., Nair, S., Holt, K. E., Turner, A. K., Hinds, J., et al. (2009). Variation in Salmonella enterica serovar typhi IncHI1 plasmids during the global spread of resistant typhoid fever. Antimicrob. Agents Chemother. 53, 716–727. doi: 10.1128/AAC.00645-08
Phan, M. D., and Wain, J. (2008). IncHI plasmids, a dynamic link between resistance and pathogenicity. J. Infect. Dev. Ctries. 2, 272–278. doi: 10.3855/jidc.221
Rodríguez-Santiago, J., Cornejo-Juárez, P., Silva-Sánchez, J., and Garza-Ramos, U. (2021). Polymyxin resistance in Enterobacterales: overview and epidemiology in the Americas. Int. J. Antimicrob. Agents 58:106426. doi: 10.1016/j.ijantimicag.2021.106426
Sun, J., Chen, C., Cui, C. Y., Zhang, Y., Liu, X., Cui, Z. H., et al. (2019). Plasmid-encoded tet(X) genes that confer high-level tigecycline resistance in Escherichia coli. Nat. Microbiol. 4, 1457–1464. doi: 10.1038/s41564-019-0496-4
Tang, B., Chang, J., Cao, L., Luo, Q., Xu, H., Lyu, W., et al. (2019). Characterization of an NDM-5 carbapenemase-producing Escherichia coli ST156 isolate from a poultry farm in Zhejiang, China. BMC Microbiol. 19:82. doi: 10.1186/s12866-019-1454-2
Tang, B., Chang, J., Luo, Y., Jiang, H., Liu, C., Xiao, X., et al. (2022a). Prevalence and characteristics of the mcr-1 gene in retail meat samples in Zhejiang Province, China. J. Microbiol. 60, 610–619. doi: 10.1007/s12275-022-1597-y
Tang, B., Chang, J., Zhang, L., Liu, L., Xia, X., Hassan, B. H., et al. (2020). Carriage of distinct mcr-1-harboring plasmids by unusual serotypes of Salmonella. Adv Biosyst. 4:e1900219. doi: 10.1002/adbi.201900219
Tang, M., Kong, X., Hao, J., and Liu, J. (2020). Epidemiological characteristics and formation mechanisms of multidrug-resistant hypervirulent Klebsiella pneumoniae. Front. Microbiol. 11:581543. doi: 10.3389/fmicb.2020.581543
Tang, B., Wang, J., Zheng, X., Chang, J., Ma, J., Wang, J., et al. (2022b). Antimicrobial resistance surveillance of Escherichia coli from chickens in the Qinghai plateau of China. Front. Microbiol. 13:885132. doi: 10.3389/fmicb.2022.885132
Tang, B., Yang, H., Jia, X., and Feng, Y. (2021). Coexistence and characterization of Tet(X5) and NDM-3 in the MDR-Acinetobacter indicus of duck origin. Microb. Pathog. 150:104697. doi: 10.1016/j.micpath.2020.104697
Umar, Z., Chen, Q., Tang, B., Xu, Y., Wang, J., Zhang, H., et al. (2021). The poultry pathogen Riemerella anatipestifer appears as a reservoir for Tet(X) tigecycline resistance. Environ. Microbiol. 23, 7465–7482. doi: 10.1111/1462-2920.15632
Valcek, A., Sismova, P., Nesporova, K., Overballe-Petersen, S., Bitar, I., Jamborova, I., et al. (2021). Horsing around: Escherichia coli ST1250 of equine origin harbouring epidemic IncHI1/ST9 plasmid with Bla(CTX-M-1) and an operon for short-chain fructooligosaccharides metabolism. Antimicrob. Agents Chemother. 65, e02556–e02520. doi: 10.1128/AAC.02556-20
Wu, Y., He, R., Qin, M., Yang, Y., Chen, J., Feng, Y., et al. (2022). Identification of plasmid-mediated tigecycline-resistant gene tet(X4) in Enterobacter cloacae from pigs in China. Microbiol. Spectr. 10:e0206421. doi: 10.1128/spectrum.02064-21
Yu, Y., Tang, B., Dai, R., Zhang, B., Chen, L., Yang, H., et al. (2018). Identification of the streptothricin and tunicamycin biosynthetic gene clusters by genome mining in Streptomyces sp. strain fd1-xmd. Appl. Microbiol. Biotechnol. 102, 2621–2633. doi: 10.1007/s00253-018-8748-4
Zeng, Y., Dong, N., Liu, C., Lu, J., and Zhang, R. (2021). Presence of tet(X4)-positive Citrobacter freundii in a cancer patient with chemotherapy-induced persistent diarrhoea. J. Glob. Antimicrob. Resist. 24, 88–89. doi: 10.1016/j.jgar.2020.11.007
Zhai, W., Tian, Y., Lu, M., Zhang, M., Song, H., Fu, Y., et al. (2022). Presence of mobile tigecycline resistance gene tet(X4) in Clinical Klebsiella pneumoniae. Microbiol. Spectr. 10:e0108121. doi: 10.1128/spectrum.01081-21
Keywords: tigecycline resistance, tet(X4), IncHI1, pMLST, Enterobacteriaceae
Citation: Ma J, Wang J, Yang H, Su M, Li R, Bai L, Feng J, Huang Y, Yang Z and Tang B (2023) IncHI1 plasmids mediated the tet(X4) gene spread in Enterobacteriaceae in porcine. Front. Microbiol. 14:1128905. doi: 10.3389/fmicb.2023.1128905
Received: 21 December 2022; Accepted: 14 March 2023;
Published: 30 March 2023.
Edited by:
Tao Li, Shanghai Veterinary Research Institute (CAAS), ChinaReviewed by:
Xiang-Dang Du, Henan Agricultural University, ChinaCopyright © 2023 Ma, Wang, Yang, Su, Li, Bai, Feng, Huang, Yang and Tang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zengqi Yang, eXpxODE2MkAxNjMuY29t; Biao Tang, dGFuZ2JpYW9AemFhcy5hYy5jbg==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.